- 1The Affiliated People’s Hospital of Jiangsu University, Zhenjiang, China
- 2Jiangsu Key Laboratory of Medical Science and Laboratory Medicine, School of Medicine, Jiangsu University, Zhenjiang, China
RNA N6-methyladenosine (m6A) modification has important regulatory roles in determining cell fate. The reversible methylation process of adding and removing m6A marks is dynamically regulated by a fine-tuned coordination of many enzymes and binding proteins. Stem cells have self-renewal and pluripotent potential and show broad prospects in regenerative medicine and other fields. Stem cells have also been identified in cancer, which is linked to cancer metastasis, therapy resistance, and recurrence. Herein, we aimed to review the molecular mechanism that controls the reversible balance of m6A level in stem cells and the effect of m6A modification on the balance between pluripotency and differentiation. Additionally, we also elaborated the association between aberrant m6A modification and the maintenance of cancer stem cells in many cancers. Moreover, we discussed about the clinical implications of m6A modification in cancer stem cells for cancer diagnosis and therapy.
Introduction
Although the definition of stem cells has been constantly revised, the key properties of stem cells are self-renewal and multidifferentiation potency. Stem cells have differentiation plasticity in a specific microenvironment to make up tissues and organs, thus, playing important roles in regenerative medicine and tissue engineering.
The coordinated gene expression for stem cell maintenance and differentiation are established by epigenetic changes (Berdasco and Esteller, 2011). Increasing studies have shown that m6A modification, the most abundant form of methylation modification in eukaryotic mRNA, can dynamically regulate the expression balance of pluripotency or lineage development factors, which determines the fate of stem cells (self-renewal or differentiation) (Zhao and He, 2015).
Here, we concentrated on the current advances of m6A modification in stem cell self-renewal and differentiation, especially the specific mechanism of regulation. We highlighted the potential of m6A machinery as novel strategies for stem cell expansion in tissue regeneration and stem cell clearance in cancer.
Regulatory Factors for m6A Modification
The m6A modification is deposited by methyltransferase (writers), removed by demethylase (erasers), and recognized by binding proteins (readers). The “writers” include METTL3/5/14/16, KIAA1429, WTAP, VIRMA, HAKAI, RBM15/15B, ZC3H13, and PCIF1/CAPAM, while FTO and ALKBH5 perform as “erasers.” The known “readers” are YTHDF1/2/3, YTHDC1/2, IGF2BP1/2/3, HNRNPA2B1/C/G, eIF3, and FMR1/FMRP. These factors either act independently, or form complexes, or cooperate with other cofactors to participate in almost all steps of RNA metabolism, including structural switch, splicing, export, stability, decay, translation, and miRNA mature (Figure 1A).
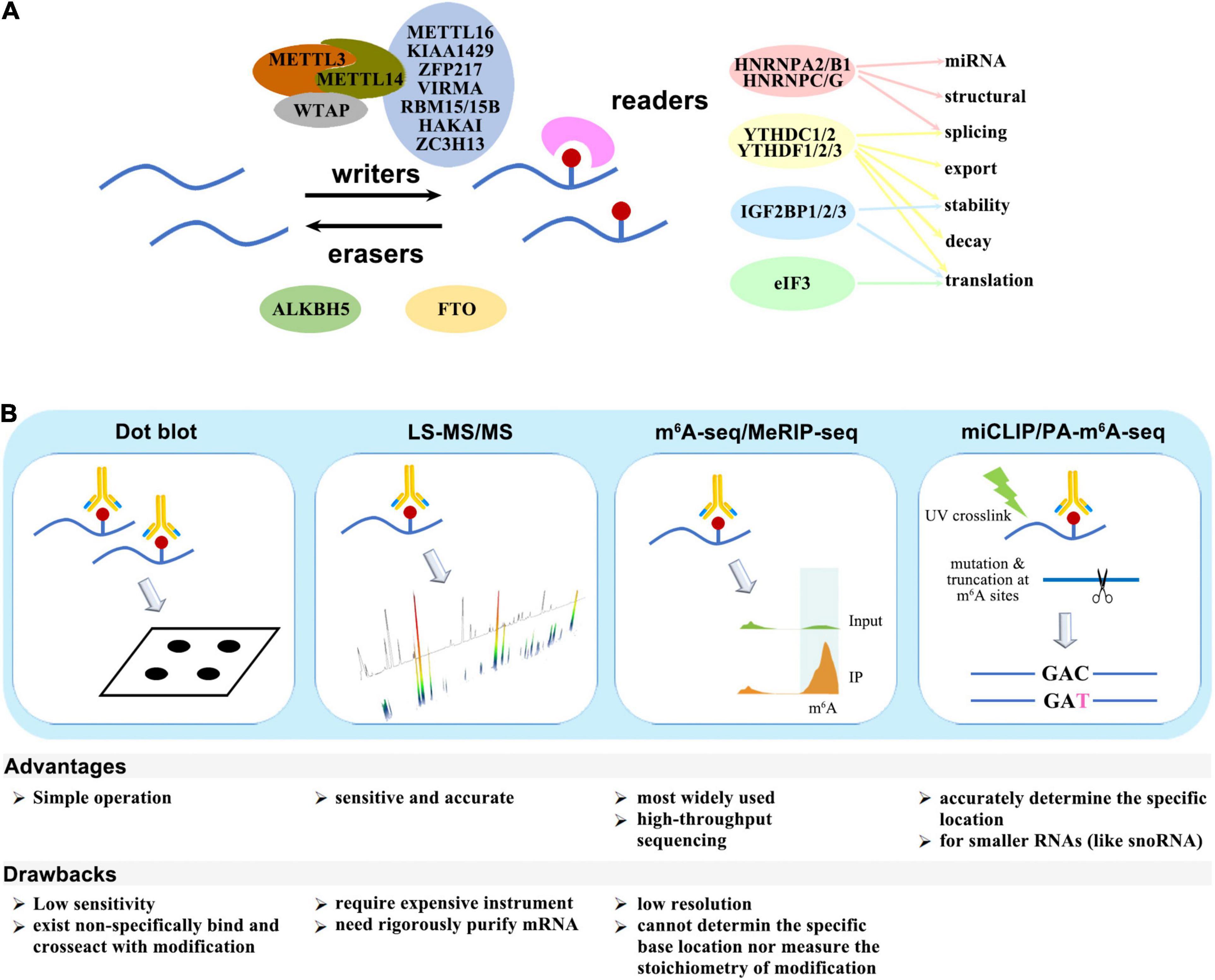
Figure 1. (A) Writers, erasers, and readers of the m6A modification. (B) Schematic diagram of m6A detection methods.
Technology in m6A Research
Several quantitative or site-specific methods have been developed to detect the amount or location of m6A modification (Figure 1B). The methods for global quantitative measurement of m6A include dot blot by using m6A-specific antibodies or LC-MS/MS. The procedure for dot blot is relatively simple, but the sensitivity is low, and there is non-specific binding between existing antibodies and other modifications. In comparison, LC-MS/MS is more sensitive and accurate, though it requires expensive instrument.
Methods for detection of the m6A modification site include m6A−seq, MeRIP-seq, PA-m6A−CLIP, miCLIP, m6A-LAIC-SEQ, SCARLET, and SELECT. m6A−seq and MeRIP-seq is the earliest and most widely used technologies. Purified mRNA fragments are immunoprecipitated by m6A-specific antibodies and subjected to high-throughput sequencing, but these methods have a low resolution and cannot measure the stoichiometry. PA-m6A−CLIP and miCLIP can accurately determine the specific location of m6A modification site. The purified RNA are incorporated with photoactivity ribonucleoside analogs, and the data are analyzed after immunoprecipitation and UV-induced cross-linking.
m6A Modification in Fate Decision of Stem Cells
Embryonic Stem Cells
Embryonic stem cells (ESCs) are derived from the embryo cell mass or primordial germ cells. They can be induced to differentiate into all kinds of cells in the body and expanded, screened, frozen, and resuscitated in vitro without losing their original characteristics. Considerable efforts have been devoted to characterize the epigenetic networks that control the reprogramming of ESCs, which requires precise coordination of transcription factors, chromatin regulators, and RNA modifiers.
Thousands of transcripts, especially those controlling the core network during differentiation, are m6A modified in ESCs (Batista et al., 2014). m6A deposit may target pluripotency or lineage-commitment genes to regulate differentiation in different ESC state (Geula et al., 2015; Zhao and He, 2015). On one side, the absence of methyltransferase led to a dramatic differentiation defect in the naive ESCs. For instance, METTL3-depleted ESCs had enhanced self-renewal but hindered differentiation abilities (Batista et al., 2014; Geula et al., 2015; Figure 2D). In addition, METTL5 depletion led to the reduction of FBXW7 levels, thereby delaying the onset of ESC differentiation (Xing et al., 2020; Figure 2D). Moreover, Linc1281 targeted let-7/Lin28 to ensure ESC differentiation by internal m6A modification (Yang et al., 2018; Figure 2D). Arginine methylation of METTL14 greatly enhanced global m6A levels and promoted ESC endoderm differentiation (Liu et al., 2021b; Wang Z. et al., 2021; Figure 2E). On the other side, removing m6A triggers differentiation when ESCs are at the primed state. Loss of m6A methylation increased the stability of developmental genes by binding to HuR RNA to facilitate ESC differentiation (Wang et al., 2014; Figure 2A). The methyltransferase complex of ZC3H13–WTAP–Virilizer–Hakai could facilitate ESC self-renewal (Wen et al., 2018; Figure 2A). Additionally, ZFP217 modulated m6A deposition on Nanog, Sox2, Klf4, and c-Myc mRNAs by sequestering METTL3 (Aguilo et al., 2015; Figure 2B). Melatonin was shown to prevent m6A-dependent core pluripotency mRNA decay through the MT1-JAK2/STAT3-ZFP217 signal axis (Yang et al., 2020; Figure 2B). YTHDC1 was required for the maintenance of mouse ESCs (Liu et al., 2021; Figure 2C).
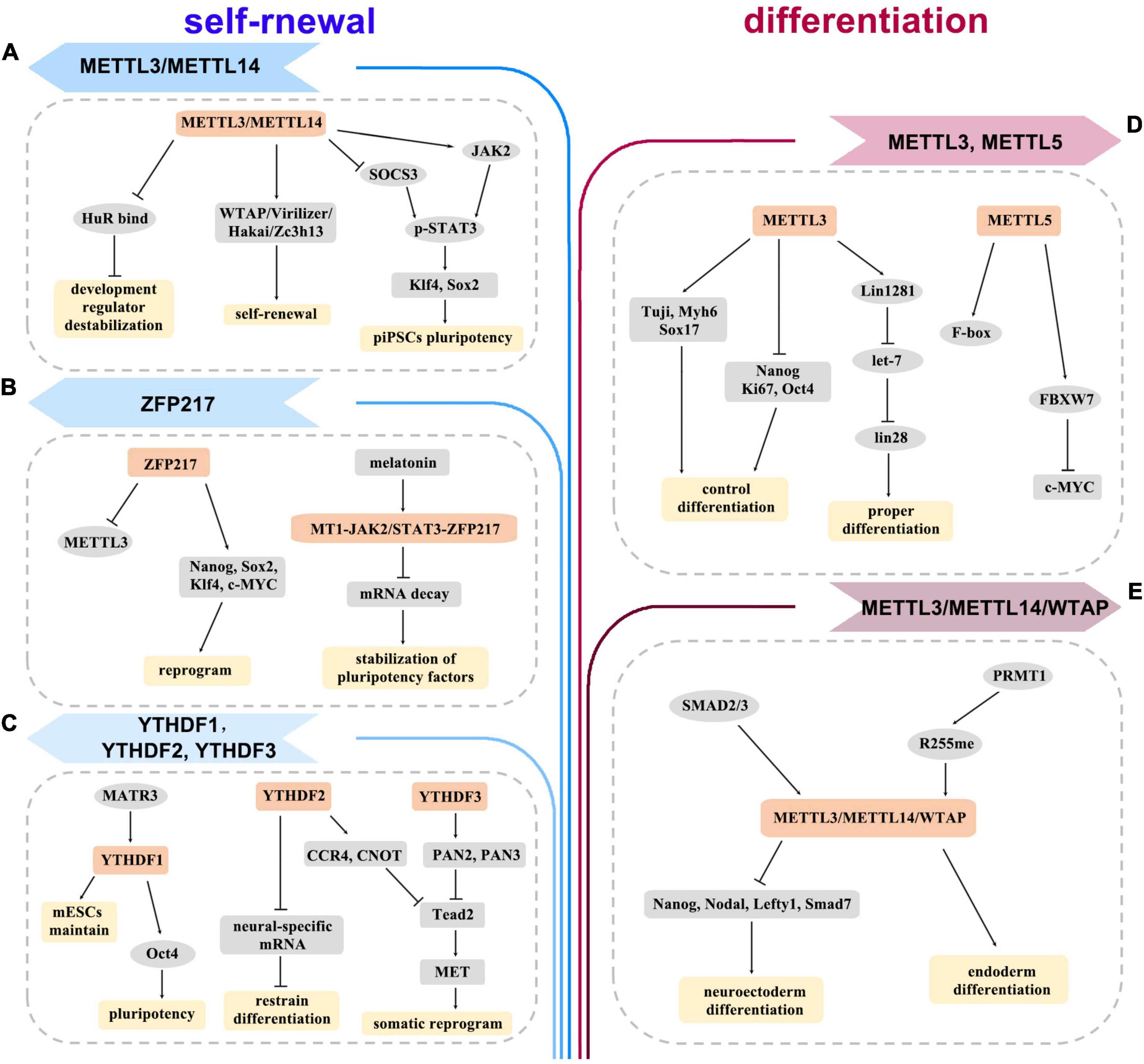
Figure 2. The diagram depicts the roles of m6A methylation in self-renewal (A–C) and differentiation (D,E) of embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs). It proposes how writers, erasers, readers, and other regulators are involved in the regulation of various genes and pathways.
In the field of regeneration, whether transplanting ESCs directly or implanting organoids differentiated from ESCs, the key lies in the precise regulation of ESC proliferation and differentiation. The targets of m6A involve numerous genes in the network of ESC reprogramming. Understanding the mechanism of m6A undoubtedly helps the development of regenerative medicine.
Induced Pluripotent Stem Cells
Induced pluripotent stem cells (iPSCs) are ESC-like cells generated by somatic cell nuclear transfer. They have similar morphology, gene expression, differentiation ability, epigenetic modification status, and function to ESCs. In-depth understanding of the mechanisms controlling self-renewal and transitions to a differentiated state is essential for iPSC in the field of regenerative medicine (Wu et al., 2019b).
However, the effects of m6A modification on the generation of iPSCs are inconsistent. METTL3 maintained pluripotency of porcine iPSCs by sustaining JAK2 and inhibiting SOCS3 expression, and activating STAT3/Klf4/Sox2 signal axis in an m6A-YTHDF1/2-dependent manner (Wu et al., 2019b; Figure 2A). MATR3 favored the pluripotency of iPSCs by regulating the transcriptional and translational efficiency of Nanog, Lin28a, and Oct4 (Pollini et al., 2021; Figure 2C). YTHDF2 was highly expressed in iPSCs and downregulated during neural differentiation. Depletion of YTHDF2 led to loss of pluripotency of iPSCs and stabilization of neural-specific gene expression (Heck et al., 2020; Liu et al., 2020; Figure 2C). SMAD2/3 facilitated cotranscriptional recruitment of METTL3/METTL14/WTAP complex onto nascent transcripts, which allowed iPSCs to timely exit from pluripotency and induced neuroectoderm differentiation (Bertero et al., 2018; Figure 2E). Thus, m6A signaling can induce cellular responses, which provide a basis for the clinical application of iPSCs. iPSCs avoid the ethical barriers of ESCs, which are an important advantage for clinical application. With the in-depth study of the mechanism for m6A regulation, iPSCs will be more widely used in regenerative medicine.
Adult Stem Cells
Hematopoietic Stem/Progenitor Cells
m6A signal could regulate the generation of hematopoietic stem/progenitor cells (HSCs/HSPCs). In METTL3-deficient zebrafish embryos, the emergence of HSPCs was blocked (Zhang C. et al., 2017). Similar phenomena could also be observed in mouse embryonic development model. METTL3 facilitated m6A methylation on Notch1 to inhibit Notch activity, thereby promoting HSPC generation (Lv et al., 2018).
m6A signal could also regulate the development of HSCs/HSPCs. METTL3 knockdown in HSPCs promoted differentiation, while reducing proliferation (Vu et al., 2017). Conversely, another study showed that deletion of METTL3 led to an accumulation of HSCs in the bone marrow and a blockade of HSC differentiation (Lee et al., 2019). Furthermore, METTL14 knockdown substantially promoted terminal myeloid differentiation of HSPCs (Weng et al., 2018).
YTHDF2 had an important role in long-term HSC maintenance. Repression of YTHDF2 resulted in loss of lymphoid potential and HSCs expansion, as shown by the increased expression of multiple key transcription factors critical for self-renewal (Li et al., 2018; Mapperley et al., 2021). In addition, METTL3 maintained the symmetric commitment and identity of HSPCs by regulating MYC mRNA stability (Cheng et al., 2019).
Spermatogonial Stem Cells
Spermatogonial stem cells (SSCs) are primordial spermatogonial cells locating on the basement membrane of convoluted tubules, which can directionally differentiate into spermatocytes. METTL3 or METTL14 maintained SSC homeostasis through methylating transcripts of key regulators governing proliferation and differentiation (Lin et al., 2017). METTL3 deletion in germ cells prevented spermatogonial differentiation and meiosis (Xu et al., 2017). WTAP-mediated m6A modification sustained the SSC niche and governed normal spermatogenesis (Jia et al., 2020).
Neural Stem Cells
Neural stem cells (NSCs) exist in the nervous system and have the potential to differentiate into neurons, astrocytes, and oligodendrocytes, thus, producing a large number of brain tissue cells. m6A depletion by lacking METTL14 or METTL3 displayed markedly decreased proliferation in NSCs (Yoon et al., 2017; Wang et al., 2018; Chen et al., 2019). However, another study revealed that loss of METTL14 resulted in the nuclear accumulation of neural differentiation-related mRNAs, resulting in delayed neural progenitor differentiation (Edens et al., 2019).
Mesenchymal Stem Cells
Mesenchymal stem cells (MSCs) exist in a variety of tissues, such as the bone marrow, umbilical cord blood, placenta tissue, adipose tissue, etc. They have the potential to differentiate into mesenchymal or non-mesenchymal cells and have a unique cytokine profile. METTL3 was upregulated in bone marrow MSCs (BM-MSCs) undergoing osteogenic induction (Tian et al., 2019). It induced BM-MSC osteogenic differentiation by facilitating M1 macrophage differentiation and through the PTH/Pth1r–PI3K–AKT signaling axis (Wu Y. et al., 2018; Lei et al., 2021). ALKBH5 accelerated the degradation of PRMT6 mRNA and inhibited the osteogenic differentiation of MSCs (Li Z. et al., 2021). Another study showed that deletion of METTL3 in porcine BM-MSCs could promote the activation of the JAK1/STAT5/C/EBPβ pathway and adipogenesis in an m6A-YTHDF2-dependent manner (Yao et al., 2019). Furthermore, METTL3 regulated the cell cycle and mitosis by targeting PLK1, and its knockdown induced cell apoptosis and senescence in immature dental pulp MSCs (Luo et al., 2021).
Adipose-Derived Stem Cell
Increasing evidence show that m6A regulates adipose-derived stem cell (ADSC) fate decision. FTO controlled RUNX1T1 expression by regulating m6A levels around splice sites and thereby promoted adipogenesis (Zhao et al., 2014). Similarly, FTO depletion decreased the expression of CCNA2 and CDK2, impaired cell cycle progression, and activated JAK2-STAT3-C/EBPβ signaling, which inhibited the adipogenesis of preadipocytes (Wu R. et al., 2018; Wu et al., 2019a). In addition, ZFP217 knockdown increased METTL3 expression and the m6A level of CCND1, leading to the mitotic clonal expansion and adipogenesis inhibition (Liu et al., 2019).
Other Adult Stem Cells
Knockdown of METTL3 promoted muscle stem cell proliferation and muscle regeneration by activating Notch signaling pathway to regulate mRNA translation (Liang et al., 2021). METTL14-mediated cytoplasmic export of circGFRa1 promoted female germline stem cell self-renewal through the miR-449/GDNF signal (Li X. et al., 2021. METTL3 or YTHDF1 enhanced the translation of TEAD1 and sustained the stemness of intestinal stem cells (Jiang et al., 2021).
Cancer Stem Cells
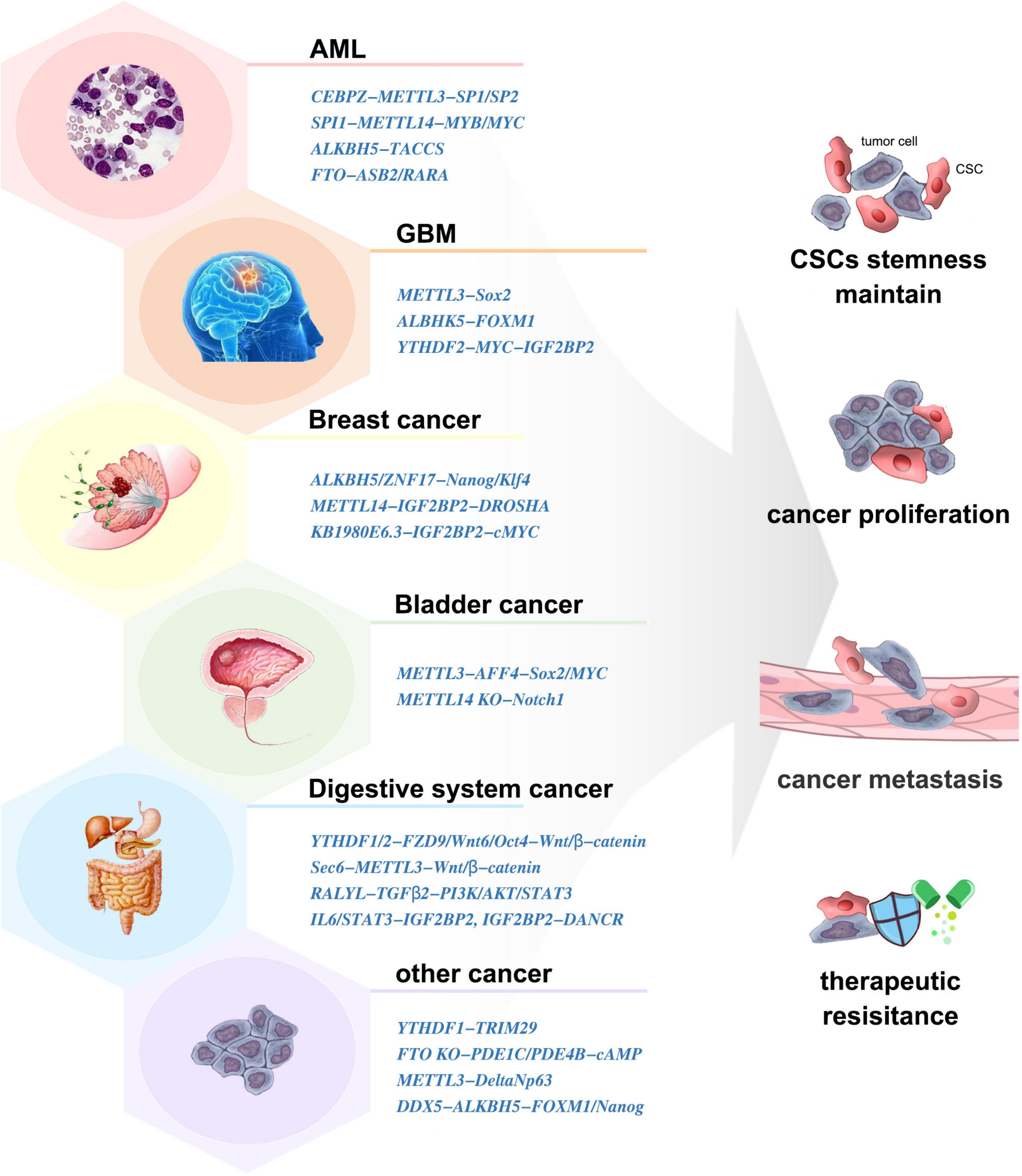
Cancer stem cells (CSCs) are a small group of cells with the unique property of infinite proliferation, transient amplifying, and remaining in dormancy for a long time. They express a variety of drug-resistant molecules and are not sensitive to external physical and chemical factors, which is the root of tumor proliferation, migration, and drug resistance. Dynamic m6A modification has been demonstrated to be involved in CSC generation and maintenance (Figure 3).
Acute Myeloid Leukemia
Acute myeloid leukemia (AML) is a clonal hematopoietic disorder where the differentiation capacity of HSCs/HPCs is blocked (Coombs et al., 2015). m6A mediates myeloid differentiation, which will provide new ideas for the pathogenesis, clinical diagnosis, and treatment of AML.
METTL3/14 was expressed more abundantly in AML cells than in healthy HSCs/HPCs, which contributed to the leukemia progression in vivo (Vu et al., 2017; Weng et al., 2018). Similarly, downregulation of METTL3 resulted in differentiation of leukemic cells and failure to establish leukemia in mice (Barbieri et al., 2017). Moreover, the SPI1-METTL14-MYB/MYC signaling axis was critical for the maintenance of AML and self-renewal of leukemia stem/initiation cells (LSCs/LICs) (Weng et al., 2018). ALKBH5 and FTO were also aberrantly overexpressed in AML and correlated with poor prognosis (Shen et al., 2020; Huff et al., 2021). They exerted tumor-promoting effects by posttranscriptional regulation of its critical targets such as TACC3 (Li et al., 2017; Shen et al., 2020). Inhibition of YTHDF2 specifically impaired LSCs propagation (Paris et al., 2019).
Targeting m6A-related molecules selectively compromised LSC proliferation and AML initiation without derailing normal hematopoiesis damage, which implied the wide application of m6A in the treatment of this hematological malignancy. Furthermore, targeting m6A-meidated control of AML cell differentiation is a promising strategy for AML therapy given the success of ATRA/arsenic trioxide (ATO)-based differentiation therapy.
Glioblastoma
The rapid growth and recurrence of glioblastoma (GBM) is closely related to the existence of glioblastoma stem cells (GSCs). m6A modification peaks were enriched at metabolic pathway-related transcripts in GSCs compared with neural progenitor cells (Li et al., 2019), which implied a crucial role of m6A methylation for GSCs.
The elevated expression of METTL3 was indispensable in the execution of oncogenic pathways and clinical aggressiveness (Li et al., 2019; Visvanathan et al., 2019). METTL3-silenced GSCs showed significant rescue in neurosphere formation and inhibited tumor growth in vivo (Visvanathan et al., 2018). Mechanically, METTL3 sustained its oncogenic role by enhancing the stability and expression of Sox2 (Visvanathan et al., 2018), altering A-to-I and C-to-U RNA-editing events (Visvanathan et al., 2019), and modulating mRNA decay of splicing factors and alternative splicing isoform switches (Li et al., 2019). However, some studies showed that knockdown of METTL3/14 dramatically increased GSC growth and self-renewal (Cui et al., 2017). The abnormally high expression of ALKBH5 has been detected in GSCs. ALKBH5 demethylates FOXM1 nascent transcripts, leading to GSC tumorigenesis (Zhang S. et al., 2017). In contrast to NSCs, GSCs displayed preferential expression of YTHDF2, laying the foundation for the YTHDF2–MYC–IGF2BP3 axis as a novel therapeutic target in GBM (Dixit et al., 2021). The FTO inhibitors could prevent neurosphere formation in GSCs without inhibiting the growth of healthy neural stem cells (Huff et al., 2021). m6A induced neuroshere formation and γ-irradiation resistance and promoted GBM growth, which makes it an important target for GBM diagnosis and treatment. Clarifying the expression and mechanism of m6A-related molecules in GCS formation is helpful in the finding and design of small-molecule inhibitors. Verifying the roles of these inhibitors in clinical application is an important research direction in the future.
Breast Cancer
Breast cancer stem cells (BCSCs) is specified by the expression of core pluripotency factors, including Oct4, Sox2, Klf4, and Nanog. ALKBH5 was stimulated in breast cancer cells when exposed to hypoxia, which induced the BCSC phenotype and increased the number of BCSCs by demethylating Nanog (Zhang et al., 2016a). Exposure of BCSCs to hypoxia induced ZNF217-dependent inhibition of m6A methylation on Nanog and Klf4 (Zhang et al., 2016b). LncRNA KB-1980E6.3 recruited IGF2BP2 to enhance c-MYC stability to maintain the stemness of BCSCs (Zhu et al., 2021). The AURKA–METTL14–IGF2BP2 complex promoted and stabilized DROSHA methylation to maintain BCSC stemness (Peng et al., 2021). Thus, m6A played an irreplaceable role in maintaining the phenotype and quantity of BCSCs. Targeting m6A-related molecules or mechanisms is an effective way to clear BCSCs and inhibit breast cancer.
Bladder Cancer
Global RNA m6A abundance and the expression of METTL3 was higher in CSCs than those in non-CSCs of bladder cancer cells. METTL3 regulated the m6A modification and expression of AFF4, which bound to the promoter regions and sustained the transcription of Sox2 and MYC, to promote self-renewal of bladder cancer CSCs (Gao et al., 2020). In contrast, METTL14 was lowly expressed in bladder tumor-initiating cells (TICs). METTL14 inhibited the proliferation, self-renewal, metastasis, and tumor-initiating capacity of TICs by participating in the RNA stability of Notch1 mRNA (Gu et al., 2019).
Digestive System Tumor
Recent studies have shown that m6A plays important roles in the regulation of digestive system CSCs. In colorectal cancer, YTHDF1 promoted the translation of m6A-modified FZD9 and Wnt6, leading to aberrant activation of Wnt/β-catenin signaling and ultimately increasing the number of CSCs (Bai et al., 2019). Low FTO expression elevated m6A levels, resulting in enhanced CSC tumorigenicity and chemoresistance (Relier et al., 2021). METTL3 upregulated Sec62 to promote the stemness and chemoresistance of CSCs by enhancing Wnt/β-catenin signaling (Liu et al., 2021a). In hepatocellular carcinoma, YTHDF2 promoted cancer metastasis and the emergence of CSCs by modulating the m6A methylation of Oct4 (Zhang et al., 2020). RALYL enhanced CSC stemness by sustaining TGF-β2 mRNA stability (Wang X. et al., 2021). In pancreatic cancer, IGF2BP2 and DANCR worked together to promote CSC properties (Hu et al., 2020). In cholangiocarcinoma, IL-6/STAT3 triggered inflammatory response to facilitate CSC stemness through regulating m6A writers (Ye et al., 2021). These results showed that m6A-related molecules might provide potential therapeutic targets in these tumors. However, further research is needed to explore the roles and mechanisms of m6A in CSCs of more digestive system tumors.
Other Tumors
Recruitment of YTHDF1 to m6A-modified TRIM29 was involved in promoting TRIM29 translation and enhancing the CSC characteristics in cisplatin-resistant ovarian cancer cells (Hao et al., 2021). Conversely, FTO could augment cAMP signaling and suppressed CSC features of ovarian cancer (Huang et al., 2020). m6A methylation in multidrug-resistant osteosarcoma cells was found to correlate with the emergence and maintenance of osteosarcoma CSCs (Wang et al., 2019). The deletion of METTL3 impaired cutaneous squamous cell carcinoma CSC properties, including colony-forming ability in vitro and tumorigenicity in vivo (Zhou et al., 2019). In oral squamous cell carcinoma, ALKBH5 regulated by DDX3 led to decreased m6A methylation in FOXM1 and Nanog, which contributes to increased CSC population (Shriwas et al., 2020).
Prospect of Clinical Application and Future Research
Stem cells perform complex biological processes through the dynamic changes of pluripotency or pedigree factors. Their gene expression programs need not only sufficient stability to maintain self-renewal and pluripotency over multiple cell generations but also flexibility to change rapidly in response to differentiation cues (Batista et al., 2014). The reversible change in m6A marks turns over transcript expression in a timely fashion, which becomes an effective means to regulate stem cell fate (Batista et al., 2014). m6A maintain the balance between pluripotency and lineage priming factors, thus, ensuring orderly differentiation of stem cells (Batista et al., 2014; Geula et al., 2015; Zhao and He, 2015). m6A may intersect with other preexisting pathways by altering downstream gene expression to regulate the stem cell differentiation network (Geula et al., 2015; Zhao and He, 2015). m6A could also target non-coding RNAs to regulate stem cell fate through existing mechanisms. The detailed classification of stem cells is based on different species or tissue sources, states, external environment, differentiation stages, and so on. These factors may lead to different outcomes of m6A modification. Therefore, further studies are needed to explore the specific molecular mechanism in different types of stem cells (Liang et al., 2020).
As a switch regulator of stem cell self-renewal and differentiation, m6A has great potential in the clinical application of stem cells. (1) ESCs can be used as seed cells for cell therapy and organ replacement therapy. It is a difficult problem for stem cells to overcome aging and maintain long-term self-renewal. METTL3 ablation and m6A loss increase the stability of methylated pluripotent mRNA transcripts and contribute to the pluripotent phenotype of ESCs (Geula et al., 2015). It is possible for damaged tissues or organs to recover completely by regulating the artificial expansion in vitro and directional differentiation in vivo of stem cells through m6A modification. (2) The establishment of human embryonic stem cell line provides an important tool to explore the influencing factors and regulatory mechanisms in the process of body development. It is expected to reveal the molecular mechanism of embryonic development by comparing the differences of m6A status and gene expression between ESCs and differentiated cells in different time and space. (3) Stem cells are ideal carriers for the gene therapy of diseases and have broad application prospects in the fields of severe immune deficiency, genetic diseases, malignant tumors, AIDS, and so on. The progress of stem cell isolation and purification technology, the improvement of gene transfection efficiency, the continuous improvement of gene transfer vector, the expansion and directional differentiation of transgenic cells, the discovery of new target genes, the stable expression and regulation of target genes after transfection, and other fields have attracted the attention of scientists. m6A cannot only regulate the phenotype of transgenic stem cells but also regulates the expression of target genes through posttranscriptional modification. (4) The involvement of m6A in CSCs may be used for predicting cancer risk, achieving early diagnosis, tracking the prognosis of tumor fate, and ultimately, providing novel therapeutic approaches. Evidence has highlighted the potential of both the altered m6A levels and the score model with different methylase as promising biomarkers in various cancers, such as lung cancer, prostatic cancer, bladder cancer, AML, and so on (Ma and Ji, 2020). Currently, inhibitors of FTO and ALKBH5 are being used as candidates for anticancer drug development, especially to inhibit the growth of CSCs by manipulating their m6A modification levels. Although these inhibitors have not been tested in clinical trials, they will provide more possibilities for the treatment of cancer (Xu et al., 2020).
So far, the information on m6A in stem cell fate regulation is still lacking. A wider scope of research should be considered: (1) New m6A writers, erasers, readers, and associated effectors need to be identified. Discovery of more components could help understand the regulatory network of m6A. (2) Once these new members are identified, their cell location, biological structure, physical and chemical properties, physiological functions, and other characteristics need to be further studied. (3) Determining the additional factors that coordinate m6A deposition or removal, and understanding how these factors are regulated and how the specificity of the methylated sites is achieved. (4) Verification of existing m6A components or functions in more biological events. (5) More precise techniques are needed to assist in the study of the distribution and target selection of m6A. (6) Further exploring diagnostic index and therapeutic targets involved in m6A machinery complexes might be very promising for some stubborn diseases, such as cancers or neurological diseases. These extensive studies may unveil more exact mechanisms of m6A in multiple biological processes.
Conclusion
Current studies on posttranscriptional modifications have revealed another layer of complexity in the regulation of cellular processes. In this review, we have summarized current knowledge on m6A and described their roles in stem cell self-renewal and differentiation. We put forward the potential of m6A mechanism in the clinical application of stem cells in future studies. Despite the progress made in recent years, more studies are still needed to provide clear information for the functional roles of m6A modification and the underlying mechanisms in stem cell fate decision.
Author Contributions
RJ wrote the manuscript and designed the figures. XZ revised the manuscript. Both authors read and approved the final manuscript.
Funding
The present study was supported by the National Natural Science Foundation of China (Grant Nos. 81702429, 81672416, and 81972310), the Natural Science Foundation of the Jiangsu Province (Grant No. BK20170561), and the Zhenjiang Science and Technology Program (Grant No. SH2019051).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
METTL3/5/14/16, methyltransferase-like 3/5/14/16; WTAP, WT1-associated protein; RBM15/15B, RNA-binding motif protein 15/15B; ZC3H13, zinc finger CCCH-type-containing 13; FTO, alpha-ketoglutarate-dependent dioxygenase; ALKBH5, AlkB homolog 5, RNA demethylase; YTHDF1/2/3, YTH N6-methyladenosine RNA-binding protein 1/2/3; YTHDC1/2, YTH domain-containing 1/2; IGF2BP1/2/3, insulin-like growth factor 2 mRNA-binding protein 1/2/3; HNRNPA2B1/C/G, heterogeneous nuclear ribonucleoprotein A2B1/C/G; LC-MS/MS, liquid chromatography coupled to tandem mass spectrometry; MeRIP-seq, methylated RNA immunoprecipitation sequencing; PA-m6A-CLIP, photo-crosslinking-assisted m6A sequencing strategy; miCLIP, m6A individual-nucleotide-resolution crosslinking and immunoprecipitation; m6A-LAIC-SEQ, m6A-level and isoform-characterization sequencing; SCARLET, site-specific cleavage and radioactive-labeling followed by ligation-assisted extraction and thin-layer chromatography; SELECT, single-base elongation and ligation-based qPCR amplification method; ESC, embryonic stem cell; Nanog, Nanog homeobox; Klf4, Kruppel-like factor 4; MYC, MYC proto-oncogene, BHLH transcription factor; Lin28, Lin-28 homolog; Med1, mediator complex subunit 1; Jarid2, jumonji and AT-rich interaction domain-containing 2; Eed, embryonic ectoderm development; FBXW7, F-box and WD repeat domain-containing 7; ZFP217/ZNF217, zinc finger protein 217; Sox2, SRY-box transcription factor 2; JAK2, Janus kinase 2; STAT3, signal transducer and activator of transcription 3; iPSC, induced pluripotent stem cell; PAN2/3, poly(A) specific ribonuclease subunit PAN2/3; Tead2, TEA domain transcription factor 2; TGFβ1, transforming growth factor beta 1; SOCS3, suppressor of cytokine signaling 3; SMAD2/3, SMAD family member 2/3; HSC/HSPC, Hematopoietic stem/progenitor cell; HE, hemogenic endothelial; DA, dorsal aorta; EHT, endothelial-to-hematopoietic transition; SSC, spermatogonial stem cell; NSC, neural stem cell; FMRP, fragile X mental retardation protein; MSC, mesenchymal stem cell; PTH/Pth1r, parathyroid hormone/parathyroid hormone receptor-1; PI3K, phosphatidylinositol-4,5-bisphosphate 3-kinase; AKT, AKT serine/threonine kinase 1; PLK1, polo-like kinase 1; EBPβ, CCAAT enhancer-binding protein beta; ADSC, adipose-derived stem cell; RUNX1T1, RUNX1 partner transcriptional corepressor 1; CCNA2, cyclin A2; CDK2, cyclin-dependent kinase 2; CCND1, cyclin D1; CSC, cancer stem cell; AML, acute myeloid leukemia; CEBPZ, CCAAT enhancer-binding protein zeta; ATRA/ATO, all-trans-retinoic acid/arsenic trioxide; LSC/LIC, leukemia stem/initiation cell; TACC3, transforming acidic coiled-coil-containing protein 3; ASB2, ankyrin repeat and SOCS box containing 2; RARA, retinoic acid receptor alpha; GBM, glioblastoma; GSC, glioblastoma stem cell; HuR, human antigen R; ADAR, adenosine deaminase RNA specific; APOBEC3A, apolipoprotein B MRNA-editing enzyme catalytic subunit 3A; FOXM1, forkhead box M1; FOXM1-AS, a long non-coding RNA antisense to FOXM1; Oct4, octamer-binding transcription factor 4; BCSC, breast cancer stem cell; HIF, hypoxia inducible factor; DROSHA, Drosha ribonuclease III; STC1, stanniocalcin 1; AURKA, Aurora kinase A; AFF4, AF4/FMR2 family member 4; FZD9, frizzled class receptor 9; PCIF1/CAPAM, phosphorylated CTD-interacting factor 1; TRIM29, tripartite motif containing 29; PDE1C/4B, phosphodiesterase 1C/4B; cAMP, cyclic adenosine monophosphate; DDX3, DEAD-box helicase 3.
References
Aguilo, F., Zhang, F., Sancho, A., Fidalgo, M., Di Cecilia, S., Vashisht, A., et al. (2015). Coordination of m(6)A mRNA methylation and gene transcription by ZFP217 regulates pluripotency and reprogramming. Cell Stem Cell 17, 689–704. doi: 10.1016/j.stem.2015.09.005
Bai, Y., Yang, C., Wu, R., Huang, L., Song, S., Li, W., et al. (2019). YTHDF1 regulates tumorigenicity and cancer stem cell-like activity in human colorectal carcinoma. Front. Oncol. 9:332. doi: 10.3389/fonc.2019.00332
Barbieri, I., Tzelepis, K., Pandolfini, L., Shi, J., Millán-Zambrano, G., Robson, S. C., et al. (2017). Promoter-bound METTL3 maintains myeloid leukaemia by m6A-dependent translation control. Nature 552, 126–131. doi: 10.1038/nature24678
Batista, P. J., Molinie, B., Wang, J., Qu, K., Zhang, J., Li, L., et al. (2014). m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 15, 707–719. doi: 10.1016/j.stem.2014.09.019
Berdasco, M., and Esteller, M. (2011). DNA methylation in stem cell renewal and multipotency. Stem Cell Res. Ther. 2:42. doi: 10.1186/scrt83
Bertero, A., Brown, S., Madrigal, P., Osnato, A., Ortmann, D., Yiangou, L., et al. (2018). The SMAD2/3 interactome reveals that TGFbeta controls m(6)A mRNA methylation in pluripotency. Nature 555, 256–259. doi: 10.1038/nature25784
Chen, J., Zhang, Y. C., Huang, C., Shen, H., Sun, B., Cheng, X., et al. (2019). m6A regulates neurogenesis and neuronal development by modulating histone methyltransferase Ezh2. Genomics Proteomics Bioinformatics 17, 154–168. doi: 10.1016/j.gpb.2018.12.007
Cheng, Y., Luo, H., Izzo, F., Pickering, B. F., Nguyen, D., Myers, R., et al. (2019). m(6)A RNA methylation maintains hematopoietic stem cell identity and symmetric commitment. Cell Rep. 28, 1703–1716. doi: 10.1016/j.celrep.2019.07.032
Coombs, C. C., Tavakkoli, M., and Tallman, M. S. (2015). Acute promyelocytic leukemia: where did we start, where are we now, and the future. Blood Cancer J. 5:e304. doi: 10.1038/bcj.2015.25
Cui, Q., Shi, H., Ye, P., Li, L., Qu, Q., Sun, G., et al. (2017). m6A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 18, 2622–2634. doi: 10.1016/j.celrep.2017.02.059
Dixit, D., Prager, B. C., Gimple, R. C., Poh, H. X., Wang, Y., Wu, Q., et al. (2021). The RNA m6A reader YTHDF2 maintains oncogene expression and is a targetable dependency in glioblastoma stem cells. Cancer Discov. 11, 480–499. doi: 10.1158/2159-8290.CD-20-0331
Edens, B. M., Vissers, C., Su, J., Arumugam, S., Xu, Z., Shi, H., et al. (2019). FMRP modulates neural differentiation through m(6)A-dependent mRNA nuclear export. Cell Rep. 28, 845–854. doi: 10.1016/j.celrep.2019.06.072
Gao, Q., Zheng, J., Ni, Z., Sun, P., Yang, C., Cheng, M., et al. (2020). The m(6)A methylation-regulated AFF4 promotes self-renewal of bladder cancer stem cells. Stem Cells Int. 2020:8849218. doi: 10.1155/2020/8849218
Geula, S., Moshitch-Moshkovitz, S., Dominissini, D., Mansour, A. A., Kol, N., Salmon-Divon, M., et al. (2015). Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science 347, 1002–1006. doi: 10.1126/science.1261417
Gu, C., Wang, Z., Zhou, N., Li, G., Kou, Y., Luo, Y., et al. (2019). Mettl14 inhibits bladder TIC self-renewal and bladder tumorigenesis through N6-methyladenosine of Notch1. Mol. Cancer 18:168. doi: 10.1186/s12943-019-1084-1
Hao, L., Wang, J. M., Liu, B. Q., Yan, J., Li, C., Jiang, J. Y., et al. (2021). m6A-YTHDF1-mediated TRIM29 upregulation facilitates the stem cell-like phenotype of cisplatin-resistant ovarian cancer cells. Biochim. Biophys. Acta Mol. Cell Res. 1868:118878. doi: 10.1016/j.bbamcr.2020.118878
Heck, A. M., Russo, J., Wilusz, J., Nishimura, E. O., and Wilusz, C. J. (2020). YTHDF2 destabilizes m(6)A-modified neural-specific RNAs to restrain differentiation in induced pluripotent stem cells. RNA 26, 739–755. doi: 10.1261/rna.073502.119
Hu, X., Peng, W. X., Zhou, H., Jiang, J., Zhou, X., Huang, D., et al. (2020). IGF2BP2 regulates DANCR by serving as an N6-methyladenosine reader. Cell Death Differ. 27, 1782–1794. doi: 10.1038/s41418-019-0461-z
Huang, H., Wang, Y., Kandpal, M., Zhao, G., Cardenas, H., Ji, Y., et al. (2020). FTO-dependent N(6)-methyladenosine modifications inhibit ovarian cancer stem cell self-renewal by blocking cAMP signaling. Cancer Res. 80, 3200–3214. doi: 10.1158/0008-5472.CAN-19-4044
Huff, S., Tiwari, S. K., Gonzalez, G. M., Wang, Y., and Rana, T. M. (2021). m(6)A-RNA demethylase FTO inhibitors impair self-renewal in glioblastoma stem cells. ACS Chem. Biol. 16, 324–333. doi: 10.1021/acschembio.0c00841
Jia, G. X., Lin, Z., Yan, R. G., Wang, G. W., Zhang, X. N., Li, C., et al. (2020). WTAP function in Sertoli cells is essential for sustaining the spermatogonial stem cell niche. Stem Cell Rep. 15, 968–982. doi: 10.1016/j.stemcr.2020.09.001
Jiang, D., Hou, J., Qian, Y., Gao, Y., Gao, X., and Wei, S. (2021). YTHDF1-regulated expression of TEAD1 contributes to the maintenance of intestinal stem cells. Biochem. Biophys. Res. Commun. 557, 85–89. doi: 10.1016/j.bbrc.2021.03.175
Lee, H., Bao, S., Qian, Y., Geula, S., Leslie, J., Zhang, C., et al. (2019). Stage-specific requirement for Mettl3-dependent m(6)A mRNA methylation during haematopoietic stem cell differentiation. Nat. Cell Biol. 21, 700–709. doi: 10.1038/s41556-019-0318-1
Lei, H., He, M., He, X., Li, G., Wang, Y., Gao, Y., et al. (2021). METTL3 induces bone marrow mesenchymal stem cells osteogenic differentiation and migration through facilitating M1 macrophage differentiation. Am. J. Transl. Res. 13, 4376–4388.
Li, F., Yi, Y., Miao, Y., Long, W., Long, T., Chen, S., et al. (2019). N(6)-methyladenosine modulates nonsense-mediated mRNA decay in human glioblastoma. Cancer Res. 79, 5785–5798. doi: 10.1158/0008-5472.CAN-18-2868
Li, X., Tian, G., and Wu, J. (2021). Novel circGFRα1 promotes self-renewal of female germline stem cells mediated by m(6)A writer METTL14. Front. Cell Dev. Biol. 9:640402. doi: 10.3389/fcell.2021.640402
Li, Z., Qian, P., Shao, W., Shi, H., He, X. C., Gogol, M., et al. (2018). Suppression of m(6)A reader Ythdf2 promotes hematopoietic stem cell expansion. Cell Res. 28, 904–917. doi: 10.1038/s41422-018-0072-0
Li, Z., Wang, P., Li, J., Xie, Z., Cen, S., Li, M., et al. (2021). The N(6)-methyladenosine demethylase ALKBH5 negatively regulates the osteogenic differentiation of mesenchymal stem cells through PRMT6. Cell Death Dis. 12:578. doi: 10.1038/s41419-021-03869-4
Li, Z., Weng, H., Su, R., Weng, X., Zuo, Z., Li, C., et al. (2017). FTO plays an oncogenic role in acute myeloid leukemia as a N6-methyladenosine RNA demethylase. Cancer Cell 31, 127–141. doi: 10.1016/j.ccell.2016.11.017
Liang, W., Lin, Z., Du, C., Qiu, D., and Zhang, Q. (2020). mRNA modification orchestrates cancer stem cell fate decisions. Mol. Cancer 19:38. doi: 10.1186/s12943-020-01166-w
Liang, Y., Han, H., Xiong, Q., Yang, C., Wang, L., Ma, J., et al. (2021). METTL3-mediated m(6)A methylation regulates muscle stem cells and muscle regeneration by notch signaling pathway. Stem Cells Int. 2021:9955691. doi: 10.1155/2021/9955691
Lin, Z., Hsu, P. J., Xing, X., Fang, J., Lu, Z., Zou, Q., et al. (2017). Mettl3-/Mettl14-mediated mRNA N(6)-methyladenosine modulates murine spermatogenesis. Cell Res. 27, 1216–1230. doi: 10.1038/cr.2017.117
Liu, J., Gao, M., He, J., Wu, K., Lin, S., Jin, L., et al. (2021). The RNA m(6)A reader YTHDC1 silences retrotransposons and guards ES cell identity. Nature 591, 322–326. doi: 10.1038/s41586-021-03313-9
Liu, J., Gao, M., Xu, S., Chen, Y., Wu, K., Liu, H., et al. (2020). YTHDF2/3 are required for somatic reprogramming through different RNA deadenylation pathways. Cell Rep. 32:108120. doi: 10.1016/j.celrep.2020.108120
Liu, Q., Zhao, Y., Wu, R., Jiang, Q., Cai, M., Bi, Z., et al. (2019). ZFP217 regulates adipogenesis by controlling mitotic clonal expansion in a METTL3-m(6)A dependent manner. RNA Biol. 16, 1785–1793. doi: 10.1080/15476286.2019.1658508
Liu, X., Su, K., Sun, X., Jiang, Y., Wang, L., Hu, C., et al. (2021a). Sec62 promotes stemness and chemoresistance of human colorectal cancer through activating Wnt/beta-catenin pathway. J. Exp. Clin. Cancer Res. 40:132. doi: 10.1186/s13046-021-01934-6
Liu, X., Wang, H., Zhao, X., Luo, Q., Wang, Q., Tan, K., et al. (2021b). Arginine methylation of METTL14 promotes RNA N(6)-methyladenosine modification and endoderm differentiation of mouse embryonic stem cells. Nat. Commun. 12:3780. doi: 10.1038/s41467-021-24035-6
Luo, H., Liu, W., Zhang, Y., Yang, Y., Jiang, X., Wu, S., et al. (2021). METTL3-mediated m(6)A modification regulates cell cycle progression of dental pulp stem cells. Stem Cell Res. Ther. 12:159. doi: 10.1186/s13287-021-02223-x
Lv, J., Zhang, Y., Gao, S., Zhang, C., Chen, Y., Li, W., et al. (2018). Endothelial-specific m6A modulates mouse hematopoietic stem and progenitor cell development via Notch signaling. Cell Res. 28, 249–252. doi: 10.1038/cr.2017.143
Ma, Z., and Ji, J. (2020). N6-methyladenosine (m6A) RNA modification in cancer stem cells. Stem Cells. doi: 10.1002/stem.3279
Mapperley, C., van de Lagemaat, L. N., Lawson, H., Tavosanis, A., Paris, J., Campos, J., et al. (2021). The mRNA m6A reader YTHDF2 suppresses proinflammatory pathways and sustains hematopoietic stem cell function. J. Exp. Med. 218:e20200829. doi: 10.1084/jem.20200829
Paris, J., Morgan, M., Campos, J., Spencer, G. J., Shmakova, A., Ivanova, I., et al. (2019). Targeting the RNA m(6)A reader YTHDF2 selectively compromises cancer stem cells in acute myeloid leukemia. Cell Stem Cell 25, 137–148. doi: 10.1016/j.stem.2019.03.021
Peng, F., Xu, J., Cui, B., Liang, Q., Zeng, S., He, B., et al. (2021). Oncogenic AURKA-enhanced N(6)-methyladenosine modification increases DROSHA mRNA stability to transactivate STC1 in breast cancer stem-like cells. Cell Res. 31, 345–361. doi: 10.1038/s41422-020-00397-2
Pollini, D., Loffredo, R., Maniscalco, F., Cardano, M., Micaelli, M., Bonomo, I., et al. (2021). Multilayer and MATR3-dependent regulation of mRNAs maintains pluripotency in human induced pluripotent stem cells. iScience 24:102197. doi: 10.1016/j.isci.2021.102197
Relier, S., Ripoll, J., Guillorit, H., Amalric, A., Achour, C., Boissière, F., et al. (2021). FTO-mediated cytoplasmic m(6)A(m) demethylation adjusts stem-like properties in colorectal cancer cell. Nat. Commun. 12:1716. doi: 10.1038/s41467-021-21758-4
Shen, C., Sheng, Y., Zhu, A. C., Robinson, S., Jiang, X., Dong, L., et al. (2020). RNA demethylase ALKBH5 selectively promotes tumorigenesis and cancer stem cell self-renewal in acute myeloid leukemia. Cell Stem Cell 27, 64–80. doi: 10.1016/j.stem.2020.04.009
Shriwas, O., Priyadarshini, M., Samal, S. K., Rath, R., Panda, S., Das Majumdar, S. K., et al. (2020). DDX3 modulates cisplatin resistance in OSCC through ALKBH5-mediated m(6)A-demethylation of FOXM1 and NANOG. Apoptosis 25, 233–246. doi: 10.1007/s10495-020-01591-8
Tian, C., Huang, Y., Li, Q., Feng, Z., and Xu, Q. (2019). Mettl3 regulates osteogenic differentiation and alternative splicing of vegfa in bone marrow mesenchymal stem cells. Int. J. Mol. Sci. 20:551. doi: 10.3390/ijms20030551
Visvanathan, A., Patil, V., Abdulla, S., Hoheisel, J., and Somasundaram, K. N. (2019). (6)-methyladenosine landscape of glioma stem-like cells: METTL3 is essential for the expression of actively transcribed genes and sustenance of the oncogenic signaling. Genes 10:141. doi: 10.3390/genes10020141
Visvanathan, A., Patil, V., Arora, A., Hegde, A. S., Arivazhagan, A., Santosh, V., et al. (2018). Essential role of METTL3-mediated m6A modification in glioma stem-like cells maintenance and radioresistance. Oncogene 37, 522–533. doi: 10.1038/onc.2017.351
Vu, L. P., Pickering, B. F., Cheng, Y., Zaccara, S., Nguyen, D., Minuesa, G., et al. (2017). The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat. Med. 23, 1369–1376. doi: 10.1038/nm.4416
Wang, X., Wang, J., Tsui, Y. M., Shi, C., Wang, Y., Zhang, X., et al. (2021). RALYL increases hepatocellular carcinoma stemness by sustaining the mRNA stability of TGF-beta2. Nat. Commun. 12:1518. doi: 10.1038/s41467-021-21828-7
Wang, Y., Li, Y., Toth, J. I., Petroski, M. D., Zhang, Z., and Zhao, J. C. (2014). N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 16, 191–198. doi: 10.1038/ncb2902
Wang, Y., Li, Y., Yue, M., Wang, J., Kumar, S., Wechsler-Reya, R. J., et al. (2018). N(6)-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications. Nat. Neurosci. 21, 195–206. doi: 10.1038/s41593-017-0057-1
Wang, Y., Zeng, L., Liang, C., Zan, R., Ji, W., Zhang, Z., et al. (2019). Integrated analysis of transcriptome-wide m(6)A methylome of osteosarcoma stem cells enriched by chemotherapy. Epigenomics 11, 1693–1715. doi: 10.2217/epi-2019-0262
Wang, Z., Pan, Z., Adhikari, S., Harada, B. T., Shen, L., Yuan, W., et al. (2021). m(6)A deposition is regulated by PRMT1-mediated arginine methylation of METTL14 in its disordered C-terminal region. EMBO J. 40:e106309. doi: 10.15252/embj.2020106309
Wen, J., Lv, R., Ma, H., Shen, H., He, C., Wang, J., et al. (2018). Zc3h13 regulates nuclear RNA m(6)A methylation and mouse embryonic stem cell self-renewal. Mol. Cell 69, 1028–1038. doi: 10.1016/j.molcel.2018.02.015
Weng, H., Huang, H., Wu, H., Qin, X., Zhao, B. S., Dong, L., et al. (2018). METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m(6)A modification. Cell Stem Cell 22, 191–205. doi: 10.1016/j.stem.2017.11.016
Wu, R., Guo, G., Bi, Z., Liu, Y., Zhao, Y., Chen, N., et al. (2019a). m(6)A methylation modulates adipogenesis through JAK2-STAT3-C/EBPbeta signaling. Biochim. Biophys. Acta Gene. Regul. Mech. 1862, 796–806. doi: 10.1016/j.bbagrm.2019.06.008
Wu, R., Liu, Y., Yao, Y., Zhao, Y., Bi, Z., Jiang, Q., et al. (2018). FTO regulates adipogenesis by controlling cell cycle progression via m(6)A-YTHDF2 dependent mechanism. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1863, 1323–1330. doi: 10.1016/j.bbalip.2018.08.008
Wu, R., Liu, Y., Zhao, Y., Bi, Z., Yao, Y., Liu, Q., et al. (2019b). m(6)A methylation controls pluripotency of porcine induced pluripotent stem cells by targeting SOCS3/JAK2/STAT3 pathway in a YTHDF1/YTHDF2-orchestrated manner. Cell Death Dis. 10:171. doi: 10.1038/s41419-019-1417-4
Wu, Y., Xie, L., Wang, M., Xiong, Q., Guo, Y., Liang, Y., et al. (2018). Mettl3-mediated m(6)A RNA methylation regulates the fate of bone marrow mesenchymal stem cells and osteoporosis. Nat. Commun. 9:4772. doi: 10.1038/s41467-018-06898-4
Xing, M., Liu, Q., Mao, C., Zeng, H., Zhang, X., Zhao, S., et al. (2020). The 18S rRNA m6A methyltransferase METTL5 promotes mouse embryonic stem cell differentiation. EMBO Rep. 21:e49863. doi: 10.15252/embr.201949863
Xu, K., Yang, Y., Feng, G. H., Sun, B. F., Chen, J. Q., Li, Y. F., et al. (2017). Mettl3-mediated m(6)A regulates spermatogonial differentiation and meiosis initiation. Cell Res. 27, 1100–1114. doi: 10.1038/cr.2017.100
Xu, Y., Liu, J., Chen, W. J., Ye, Q. Q., Chen, W. T., Li, C. L., et al. (2020). Regulation of N6-methyladenosine in the differentiation of cancer stem cells and their fate. Front. Cell Dev. Biol. 8:561703. doi: 10.3389/fcell.2020.561703
Yang, D., Qiao, J., Wang, G., Lan, Y., Li, G., Guo, X., et al. (2018). N6-methyladenosine modification of lincRNA 1281 is critically required for mESC differentiation potential. Nucleic Acids Res. 46, 3906–3920. doi: 10.1093/nar/gky130
Yang, L., Liu, X., Song, L., Su, G., Di, A., Bai, C., et al. (2020). Melatonin restores the pluripotency of long-term-cultured embryonic stem cells through melatonin receptor-dependent m6A RNA regulation. J. Pineal Res. 69:e12669. doi: 10.1111/jpi.12669
Yao, Y., Bi, Z., Wu, R., Zhao, Y., Liu, Y., Liu, Q., et al. (2019). METTL3 inhibits BMSC adipogenic differentiation by targeting the JAK1/STAT5/C/EBPbeta pathway via an m(6)A-YTHDF2-dependent manner. FASEB J. 33, 7529–7544. doi: 10.1096/fj.201802644R
Ye, H., Chen, T., Zeng, Z., He, B., Yang, Q., Pan, Q., et al. (2021). The m6A writers regulated by the IL-6/STAT3 inflammatory pathway facilitate cancer cell stemness in cholangiocarcinoma. Cancer Biol. Med. doi: 10.20892/j.issn.2095-3941.2020.0661
Yoon, K. J., Ringeling, F. R., Vissers, C., Jacob, F., Pokrass, M., Jimenez-Cyrus, D., et al. (2017). Temporal control of mammalian cortical neurogenesis by m(6)A methylation. Cell 171, 877–889. doi: 10.1016/j.cell.2017.09.003
Zhang, C., Chen, Y., Sun, B., Wang, L., Yang, Y., Ma, D., et al. (2017). m6A modulates haematopoietic stem and progenitor cell specification. Nature 549, 273–276. doi: 10.1038/nature23883
Zhang, C., Huang, S., Zhuang, H., Ruan, S., Zhou, Z., Huang, K., et al. (2020). YTHDF2 promotes the liver cancer stem cell phenotype and cancer metastasis by regulating OCT4 expression via m6A RNA methylation. Oncogene 39, 4507–4518. doi: 10.1038/s41388-020-1303-7
Zhang, C., Samanta, D., Lu, H., Bullen, J. W., Zhang, H., Chen, I., et al. (2016a). Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m6A-demethylation of NANOG mRNA. Proc. Natl. Acad. Sci. U.S.A. 113, E2047–E2056. doi: 10.1073/pnas.1602883113
Zhang, C., Zhi, W. I., Lu, H., Samanta, D., Chen, I., Gabrielson, E., et al. (2016b). Hypoxia-inducible factors regulate pluripotency factor expression by ZNF217-and ALKBH5-mediated modulation of RNA methylation in breast cancer cells. Oncotarget 7, 64527–64542. doi: 10.18632/oncotarget.11743
Zhang, S., Zhao, B. S., Zhou, A., Lin, K., Zheng, S., Lu, Z., et al. (2017). m6A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell 31, 591–606. doi: 10.1016/j.ccell.2017.02.013
Zhao, B. S., and He, C. (2015). Fate by RNA methylation: m6A steers stem cell pluripotency. Genome Biol. 16:43. doi: 10.1186/s13059-015-0609-1
Zhao, X., Yang, Y., Sun, B. F., Shi, Y., Yang, X., Xiao, W., et al. (2014). FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 24, 1403–1419. doi: 10.1038/cr.2014.151
Zhou, R., Gao, Y., Lv, D., Wang, C., Wang, D., and Li, Q. (2019). METTL3 mediated m(6) A modification plays an oncogenic role in cutaneous squamous cell carcinoma by regulating DeltaNp63. Biochem. Biophys. Res. Commun. 515, 310–317. doi: 10.1016/j.bbrc.2019.05.155
Keywords: N6-methyladenosine, post-transcriptional modifications, cancer, stem cell, cell differentiation
Citation: Ji R and Zhang X (2021) The Roles of RNA N6-Methyladenosine in Regulating Stem Cell Fate. Front. Cell Dev. Biol. 9:765635. doi: 10.3389/fcell.2021.765635
Received: 27 August 2021; Accepted: 20 September 2021;
Published: 05 November 2021.
Edited by:
Dong-Hua Yang, St. John’s University, United StatesReviewed by:
Feng Wang, Affiliated Hospital of Nantong University, ChinaChuanli Ren, Yangzhou University, China
Copyright © 2021 Ji and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xu Zhang, eHV6aGFuZ0B1anMuZWR1LmNu
 Runbi Ji
Runbi Ji Xu Zhang
Xu Zhang