- 1Department of Medical Genetics, School of Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 2Men’s Health and Reproductive Health Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 3Skull Base Research Center, Loghman Hakim Hospital, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 4Department of Critical Care Medicine, Imam Hossein Medical and Educational Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran
Being located on 17q25.1, small nucleolar RNA host gene 6 (SNHG16) is a member of SNHG family of long non-coding RNAs (lncRNA) with 4 exons and 13 splice variants. This lncRNA serves as a sponge for a variety of miRNAs, namely miR-520a-3p, miR-4500, miR-146a miR-16–5p, miR-98, let-7a-5p, hsa-miR-93, miR-17-5p, miR-186, miR-302a-3p, miR-605-3p, miR-140-5p, miR-195, let-7b-5p, miR-16, miR-340, miR-1301, miR-205, miR-488, miR-1285-3p, miR-146a-5p, and miR-124-3p. This lncRNA can affect activity of TGF-β1/SMAD5, mTOR, NF-κB, Wnt, RAS/RAF/MEK/ERK and PI3K/AKT pathways. Almost all studies have reported oncogenic effect of SNHG16 in diverse cell types. Here, we explain the results of studies about the oncogenic role of SNHG16 according to three distinct sets of evidence, i.e., in vitro, animal, and clinical evidence.
Introduction
Small nucleolar RNA host gene 6 (SNHG16) is a member of SNHG family of non-coding RNAs. Long non-coding RNAs (lncRNAs) are a class of transcripts that have sizes longer than 200 nt. These transcripts serve as scaffolds for establishment of different complexes of biomolecules. Moreover, the can serve as enhancers, modulators of chromatin structure and decoys for several molecules, particularly miRNAs {Zhang, 2019 #481}. Bioinformatics tools have facilitated identification of several classes of lncRNAs among them is SNHG group of lncRNAs {Li, 2020 #482}.
Being annotated as NC_000017.11, SNHG16 gene is located on 17q25.1 and has 4 exons. Based on the Ensembl database1, 13 splice variants have been identified for this SNHG16 with one of them having a retained intron (ENST00000587743.1) and the rest being categorized as long non-coding RNAs (lncRNAs). These transcripts have sizes ranging from 556 nt (SNHG16-208) to 3607 nt (SNHG16-201). No protein has been recognized for any of these variants. It has been shown to be ubiquitously expressed in ovary, skin and several other tissues. This lncRNA has fundamental roles in the carcinogenesis in numerous types of tissues. Here, we summarize the results of these studies based on three distinct categories of evidence, i.e., in vitro, animal and clinical evidence.
Cell Line Studies
Small nucleolar RNA host gene 6 has been demonstrated to be up-regulated in lung cancer cell lines, where it acts as a sponge for miR-520a-3p. Through decreasing the availability of this miRNA, SNHG16 increases expression of EphA2. SNHG16 silencing has suppressed proliferation, migratory potential and invasiveness of these cells, while stimulating cell apoptosis. Further experiments have shown the prominence of SNHG16/miR-520a-3p/EphA2 axis in the regulation of oncogenicity in lung cancer (Yu et al., 2020). Being transcriptionally regulated by YY1, SNHG16 also sequesters miR-4500 to modulate expression of the deubiquitinase USP21. USP21 can further increase expression of SNHG16 (Xu P. et al., 2020). Another experiment in lung cancer cells has identified miR-146a as the target of SNHG16, through its sequestering SNHG16 enhances proliferation, migration and invasiveness of lung cancer cells. The sponging effect of SNHG16 on this miRNA leads to over-expression of MUC5AC, a protein which accelerates metastasis and recurrence of lung cancer cells (Han et al., 2019). Figure 1 depicts the roles of SNHG16 in lung cancer which are exerted via sponging miR-520a-3p, miR-4500 and miR-146a.
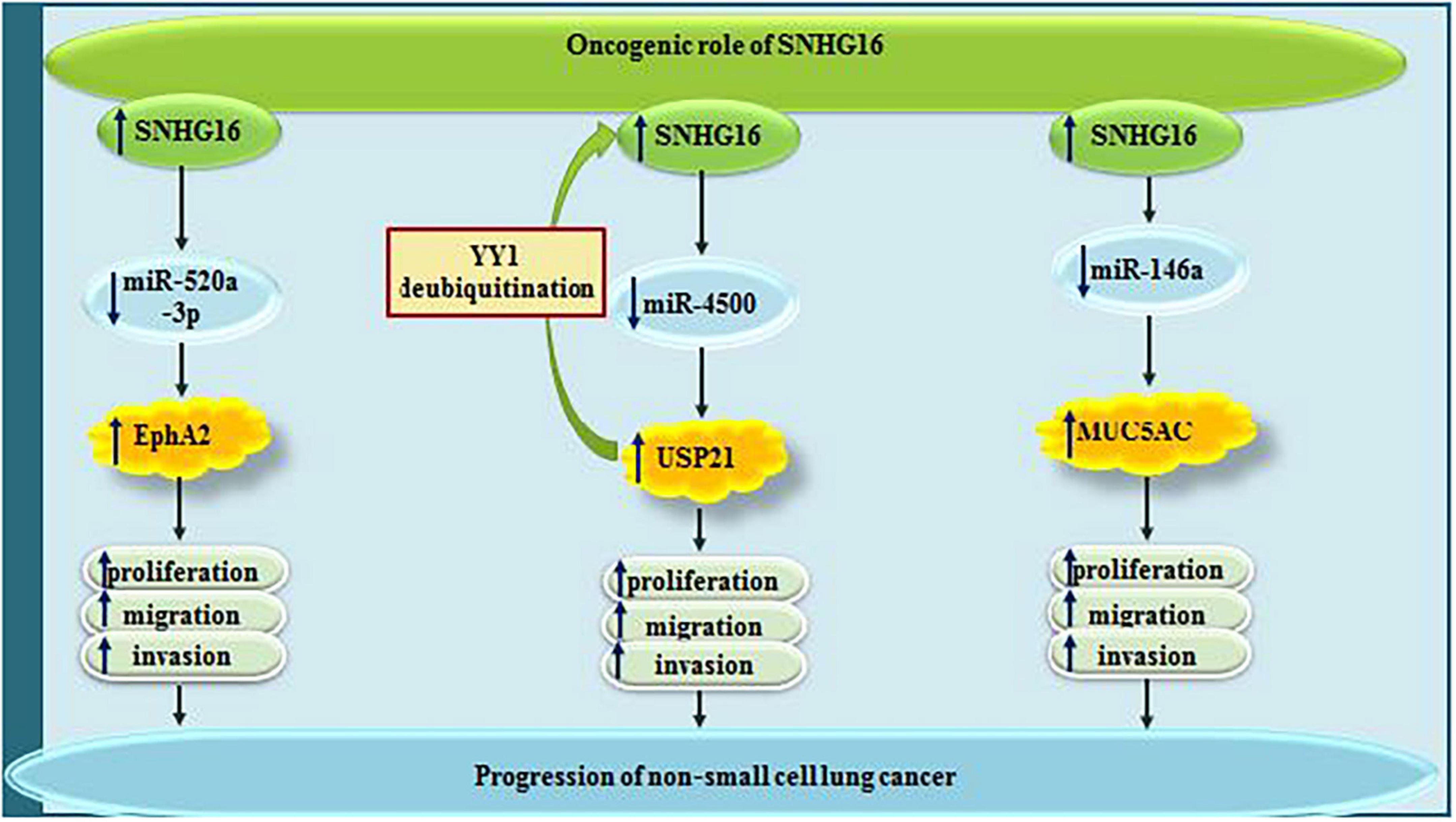
Figure 1. The oncogenic roles of SNHG16 in lung cancer are mainly mediated through sponging miR-520a-3p, miR-4500, and miR-146a.
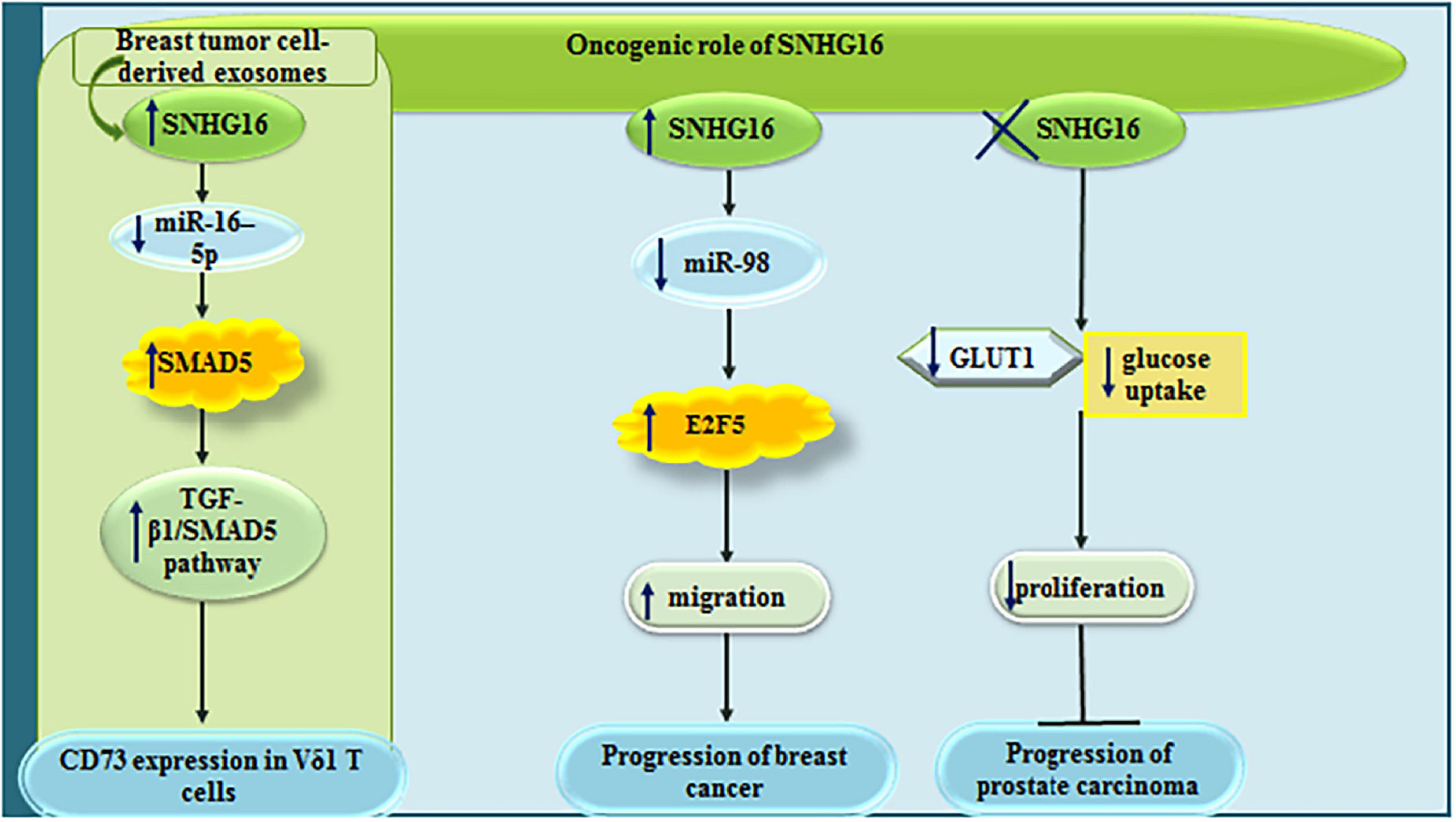
Small nucleolar RNA host gene 6 has also important impacts on the modulation of tumor microenvironment through influencing function of γδ immunosuppressive T cells. Mechanistically, SNHG16 works as a sponge for miR-16-5p, thus augmenting expression of SMAD5 and potentiating the TGF-β1/SMAD5 pathway to increase expression of CD73 in Vδ1 T cells (Ni et al., 2020). In addition, SNHG16 can enhance migratory potential of breast cancer cells via sequestering miR-98 and releasing E2F5 from its inhibitory effects (Cai et al., 2017). In prostate cancer cells, siRNA-mediated silencing of SNHG16 results in down-regulation of GLUT-1, reduction of glucose uptake and inhibition of proliferation of cancerous cells without affecting normal prostate cells (Shao et al., 2020). Figure 2 shows the oncogenic roles of SNHG6 in breast and prostate cancers.
In hepatocellular carcinoma (HCC), SNHG16 has diverse oncogenic as well as tumor suppressor roles (Figures 3, 4). SNHG16 has been shown to accelerate proliferation, migratory aptitude and invasiveness of HCC cells through sequestering miR-186 and enhancing expression of ROCK1 (Chen et al., 2019). Moreover, miR-4500 is another sponged miRNA by SNHG16 through which this lncRNA promotes development of HCC (Lin et al., 2019). In this type of cancer, SNHG16 also interacts with miR-302a-3p to increase expression of FGF19 and enhance cell proliferation (Li W. et al., 2019). Metastatic ability of HCC cells can be regulated by SNHG16 through sequestering miR-605-3p. This miRNA can suppress epithelial-mesenchymal transition (EMT) and metastatic ability of HCC via directly suppressing TRAF6 expression and further modulating NF-κB signaling. Being up-regulated by SNHG16, TRAF6 can in turn increase activity of SNHG16 promoter through activation of NF-κB, thus constructing an positive feedback loop in favor of HCC progression (Hu et al., 2020).
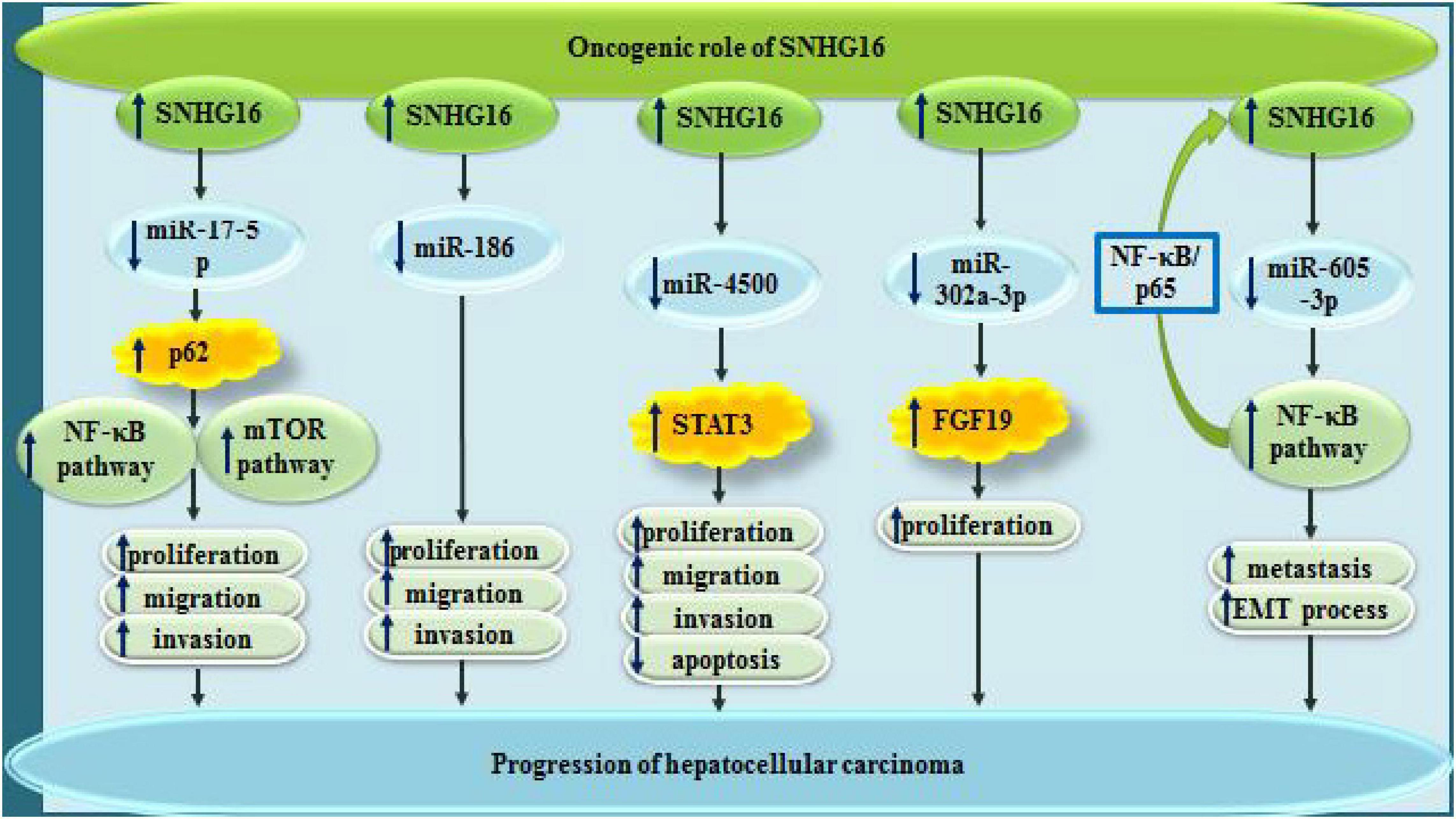
Figure 3. Oncogenic roles of SNHG16 in hepatocellular carcinoma via sponging miR-17-5p, miR-186, miR-4500, miR-302a-3p, and miR-605-3p.
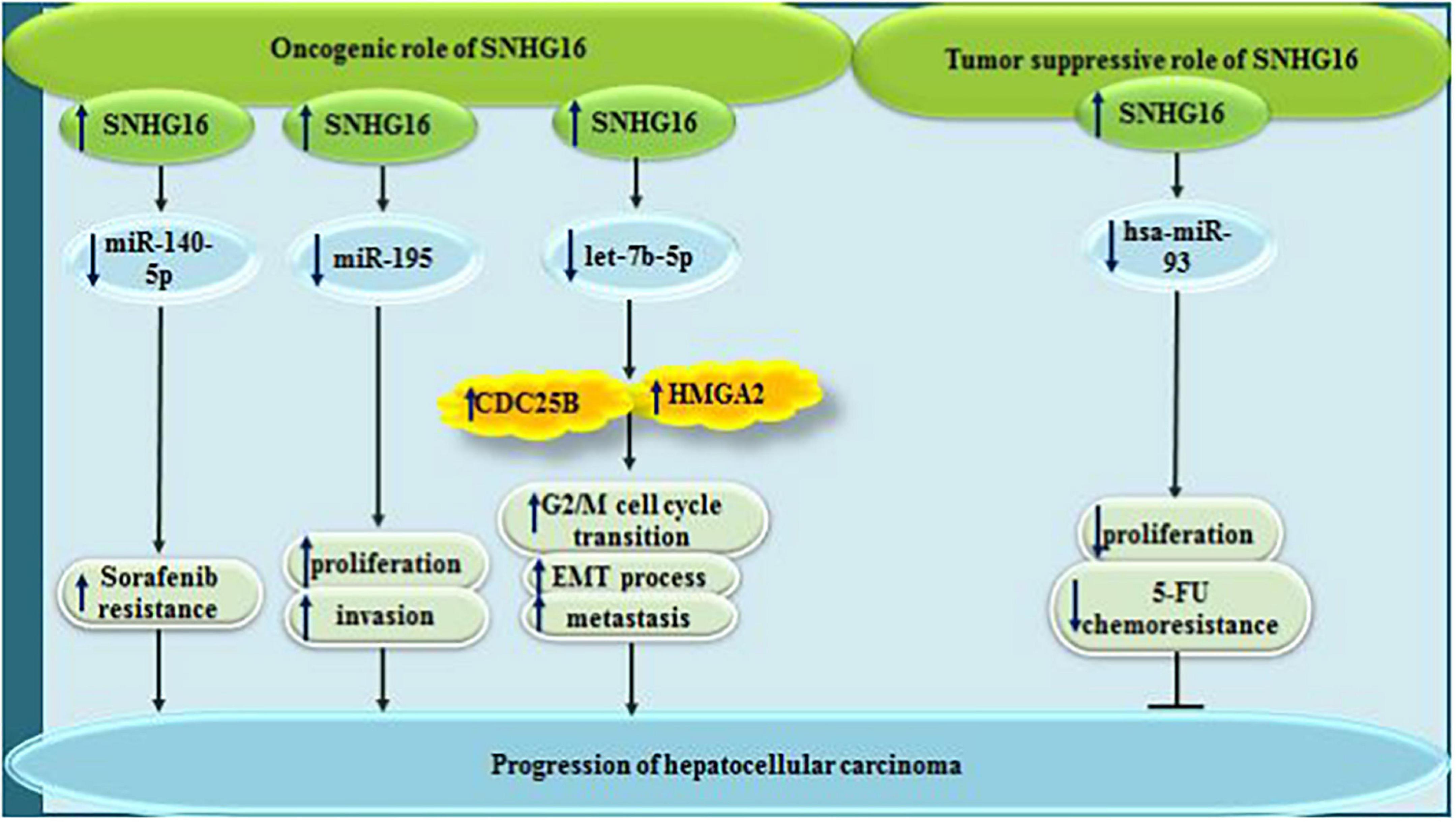
Figure 4. In hepatocellular carcinoma, while SNHG16 exerts oncogenic effect via sponging miR-140-5p, miR-195, and let7b-5p, it can have tumor suppressor effect via sponging has-miR-93.
Contrary to the mentioned studies which reported the oncogenic effects of SNHG16 in the development of HCC, a single study has revealed down-regulation of SNHG16 in HCC cell lines. Ectopic virus-mediated over-expression of SNHG16 has repressed proliferation of HCC cells and attenuated their resistance to 5-FU through sponging hsa-miR-93 (Xu et al., 2018).
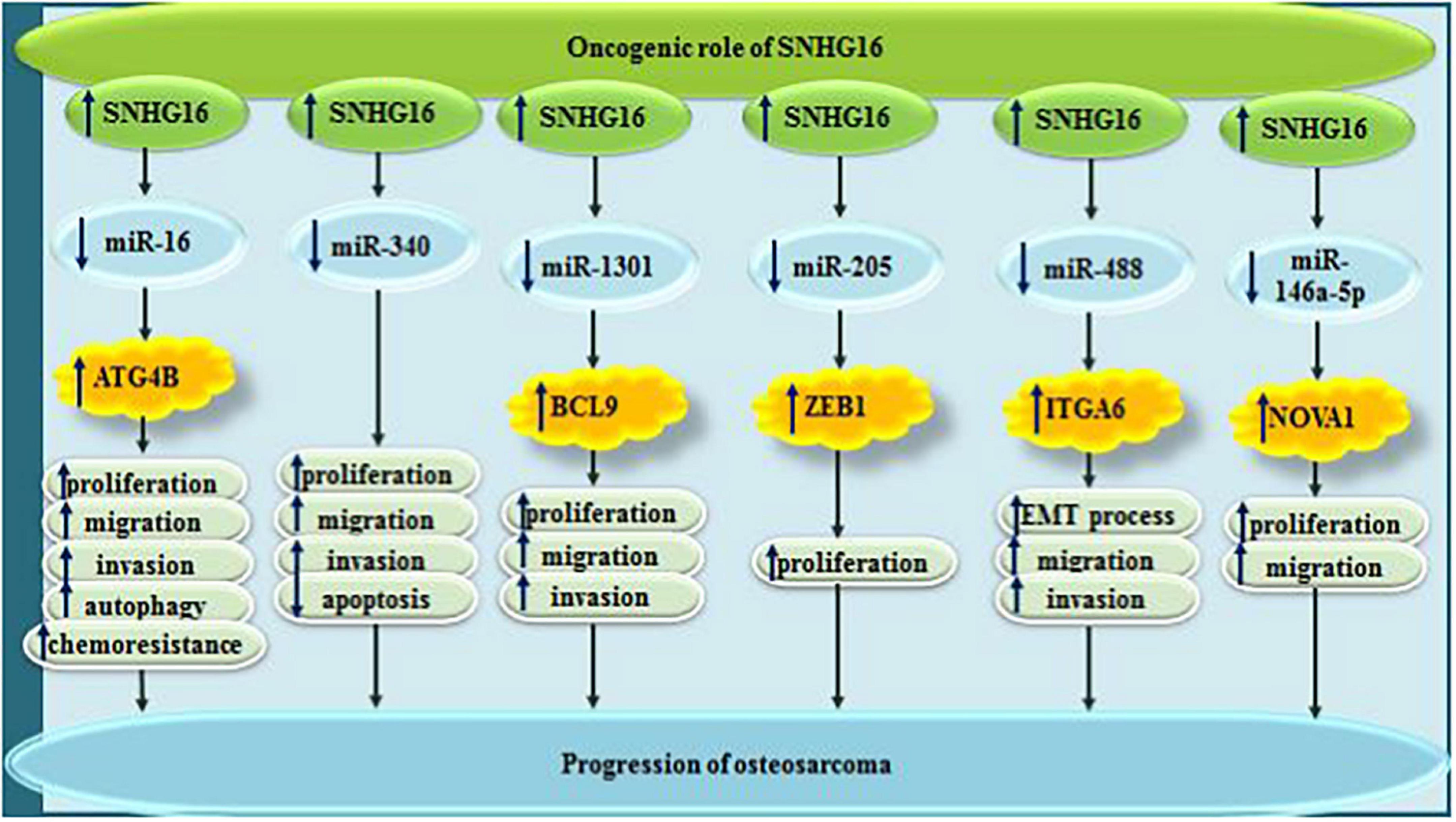
In osteosarcoma, sponging impact of SNHG16 on miR-98-5p has an essential impact on proliferation, migration and invasive aptitude of cancer cell. Simultaneously, it can enhance cell cycle progression and decease cell apoptosis (Liao et al., 2019). Meanwhile, through sponging miR-16 and up-regulating ATG4B levels, SNHG16 can induce resistance to cisplatin in these cells (Liu Y. et al., 2019). SNHG16 can also promote proliferation of osteosarcoma cells through sponging miR-205 and enhancing expression of ZEB1 (Zhu C. et al., 2018). Finally, SNHG16 can facilitate EMT of osteosarcoma cells through miR-488/ITGA6 axis (Bu et al., 2021). Figure 5 depicts the oncogenic roles of SNHG16 in osteosarcoma.
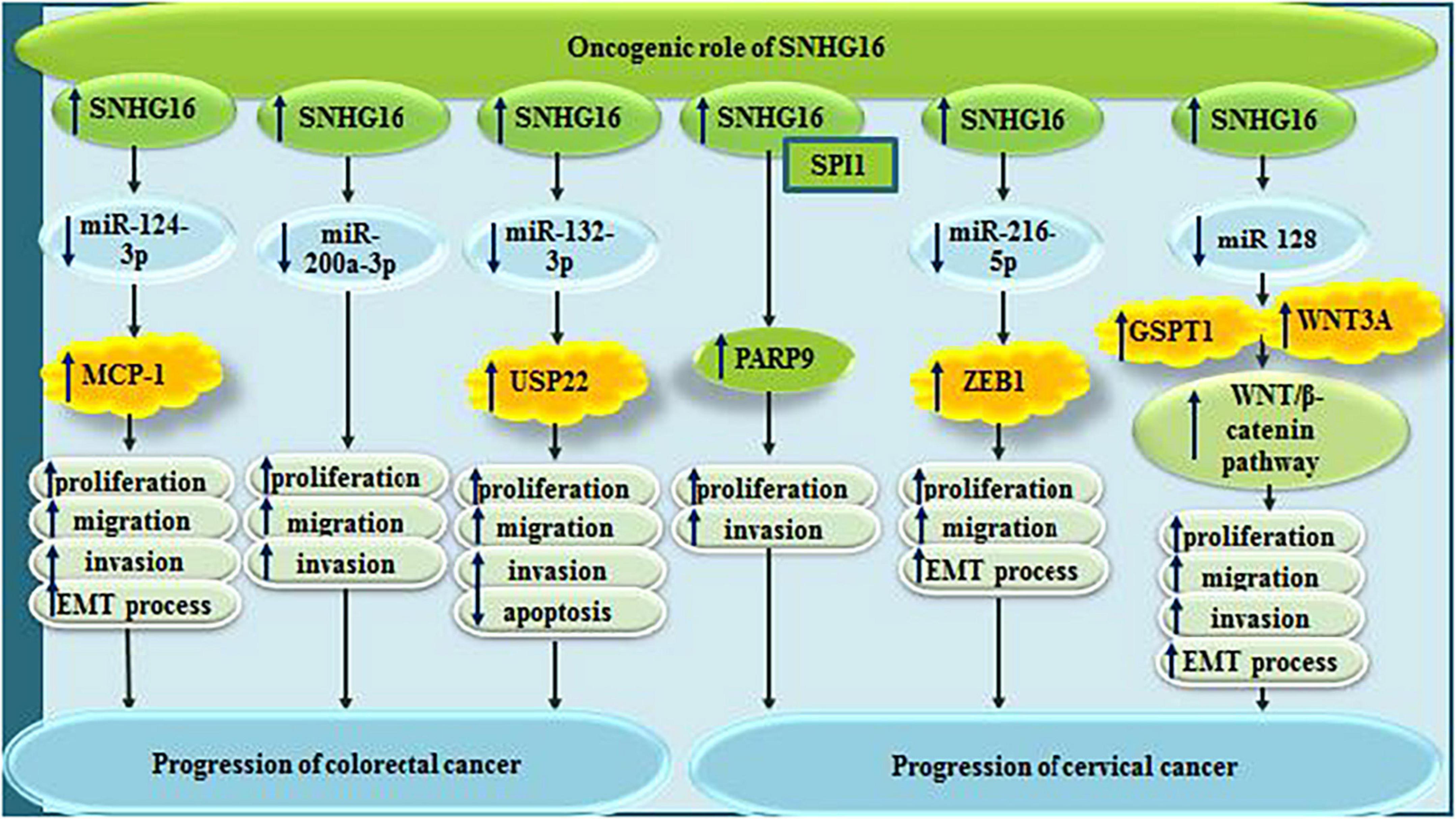
Small nucleolar RNA host gene 6/miR-124-3p/MCP-1 has an important role in induction of cell proliferation and EMT in colorectal cancer (Chen et al., 2020). The sponging effect of SNHG16 on miR-200a-3p (Li Y. et al., 2019), miR-132-3p (He et al., 2020), and miR-302a-3p (Ke et al., 2019), also promotes tumorigenicity of colorectal cancer.
In cervical cancer cells, SNHG16 has been found to recruit transcriptional factor SPI1 to increase expression of PARP9, thus promoting malignant behaviors of cells (Tao et al., 2020). Moreover, through sponging miR-216-5p, SNHG16 can increase expression of ZEB1, therefore increasing both cell proliferation and EMT process (Zhu H. et al., 2018). Finally, through sponging miR-128, it affects activity Wnt/β-catenin pathway (Wu et al., 2020). Figure 6 summarizes the role of SNHG16 in colorectal and cervical cancers.
In neuroblastoma cells, SNHG16 has been revealed to sequester miR-542-3p (Deng et al., 2020), miR-128-3p (Bao et al., 2020) and miR-338-3p (Xu Z. et al., 2020), thus increasing expressions of HNF4α, HOXA7, and PLK4, respectively (Figure 7).
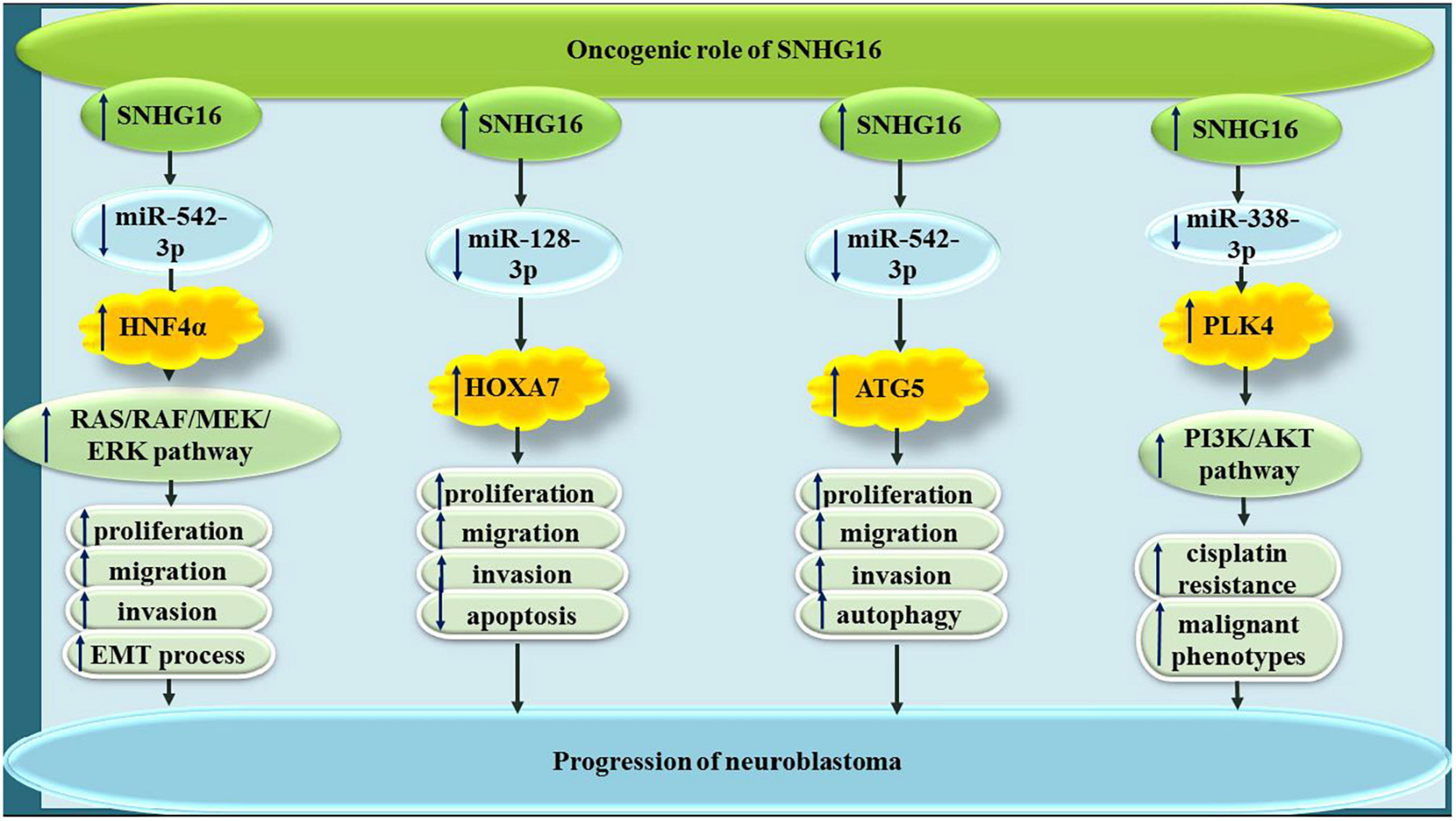
Figure 7. In neuroblastoma, SNHG16 has been revealed to sponge miR-542-3p, miR-128-3p, and miR-338-3p.
In other types of cancers, including retinoblastoma, oral squamous cell carcinoma, nasopharyngeal carcinoma, SNHG16 sequesters a number of miRNAs, namely miR-140-5p, miR-182-5p, miR-128-3p, miR-183-5p, miR-17-5p, and miR-520a-3p (Figure 8).
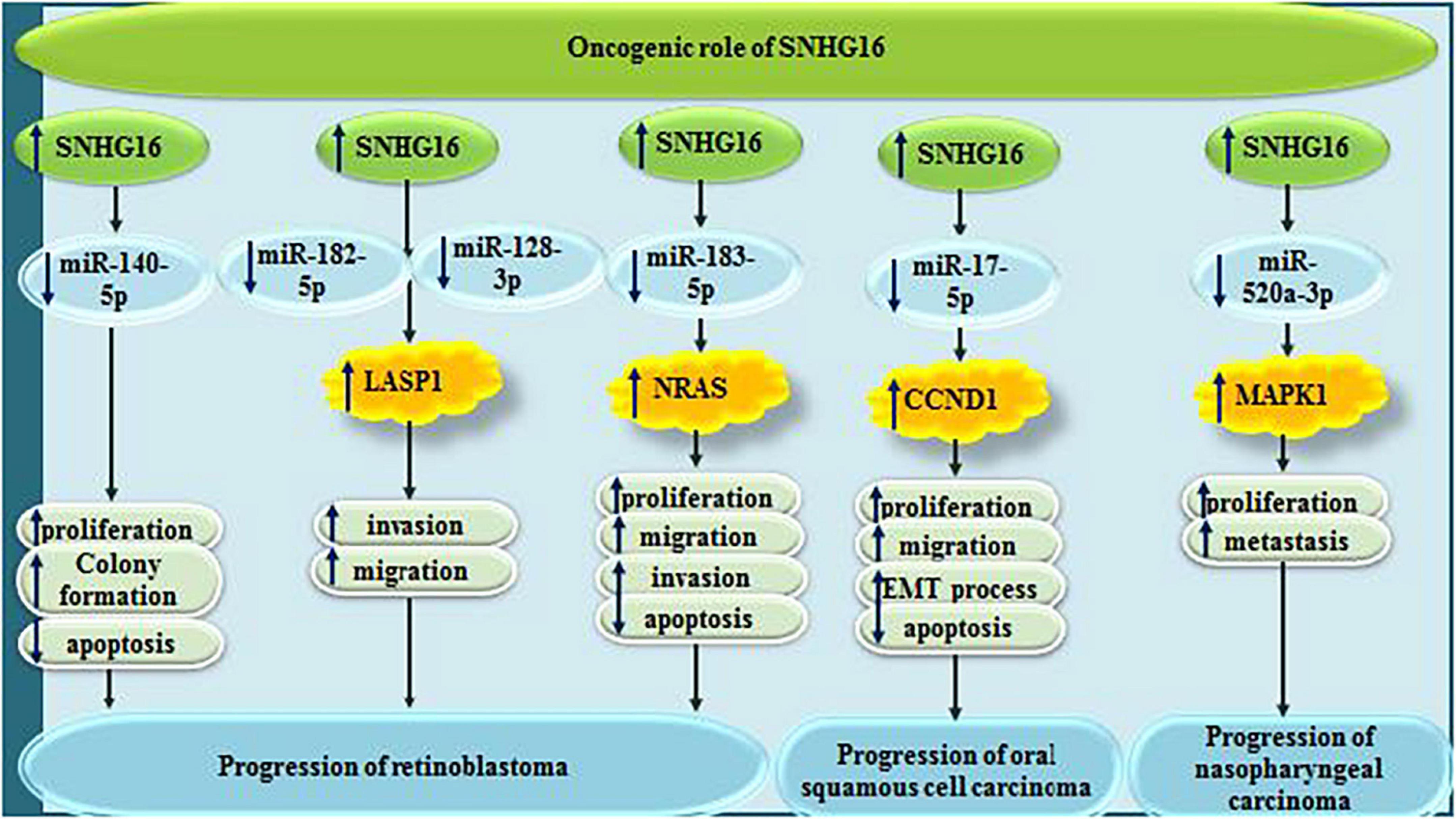
Figure 8. Oncogenic roles of SNHG16 in retinoblastoma, oral squamous cell carcinoma and nasopharyngeal carcinoma.
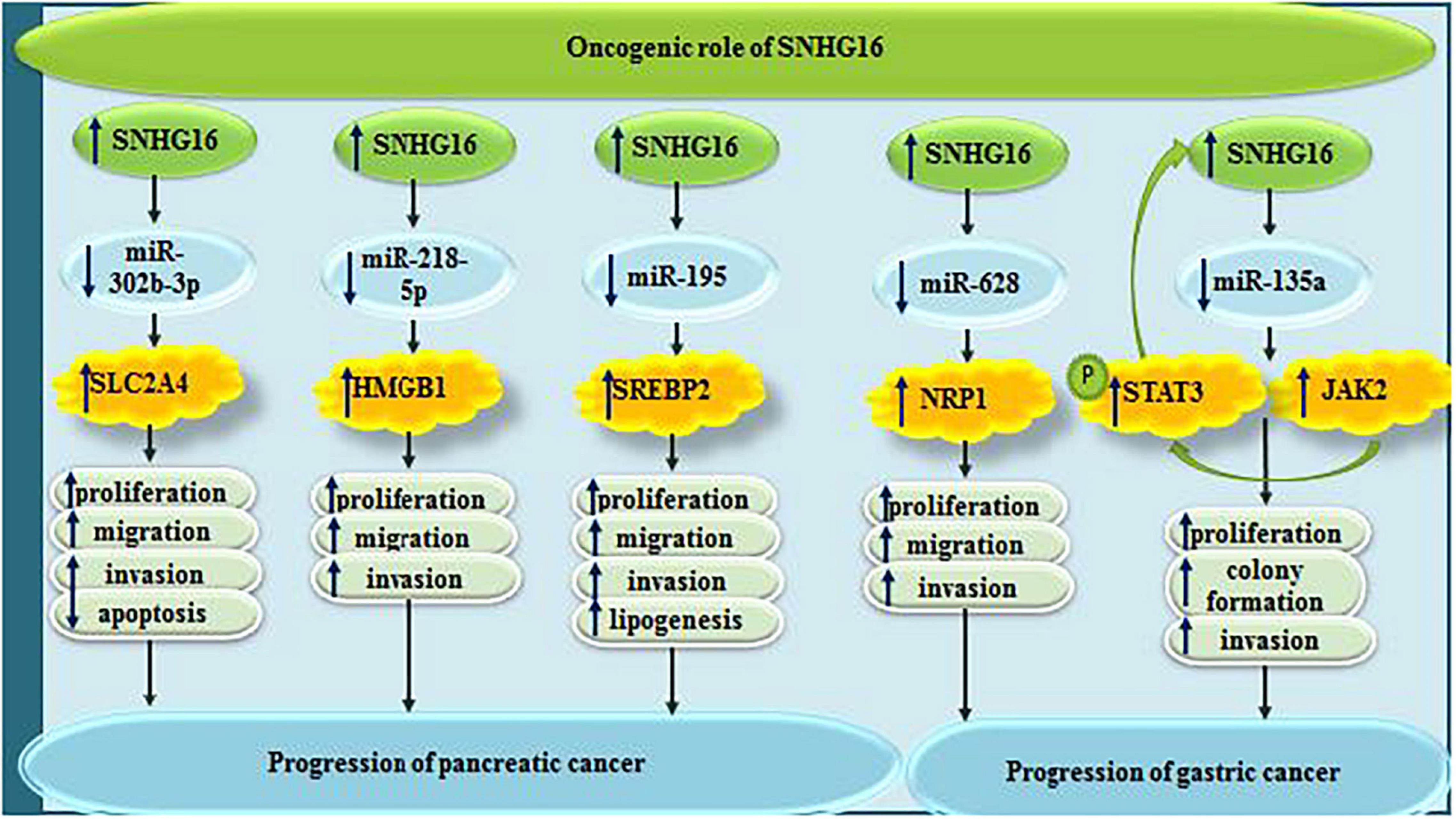
In pancreatic cancer, SNHG16 acts in favor of tumor progression through sponging miR-302b-3p and subsequently increasing expression of SLC2A4 (Xu et al., 2021). Moreover, it can contribute in this process through sponging miR-218-5p (Liu S. et al., 2019). Finally, SNHG16-mediated enhancement of lipogenesis through affecting expression of SREBP2 facilitates progression of pancreatic cancer (Yu et al., 2019b).
Small nucleolar RNA host gene 6 participates in the progression of gastric cancer via sequestering miR-628-3p and consequently decreasing expression of NRP1 (Pang et al., 2019). In this type of cancer, SNHG16 also sponges miR-135a and activates JAK2/STAT3 signaling (Wang et al., 2019b; Figure 9).
Table 1 summarizes the results of in vitro studies regarding the role of SNHG16 in carcinogenesis.
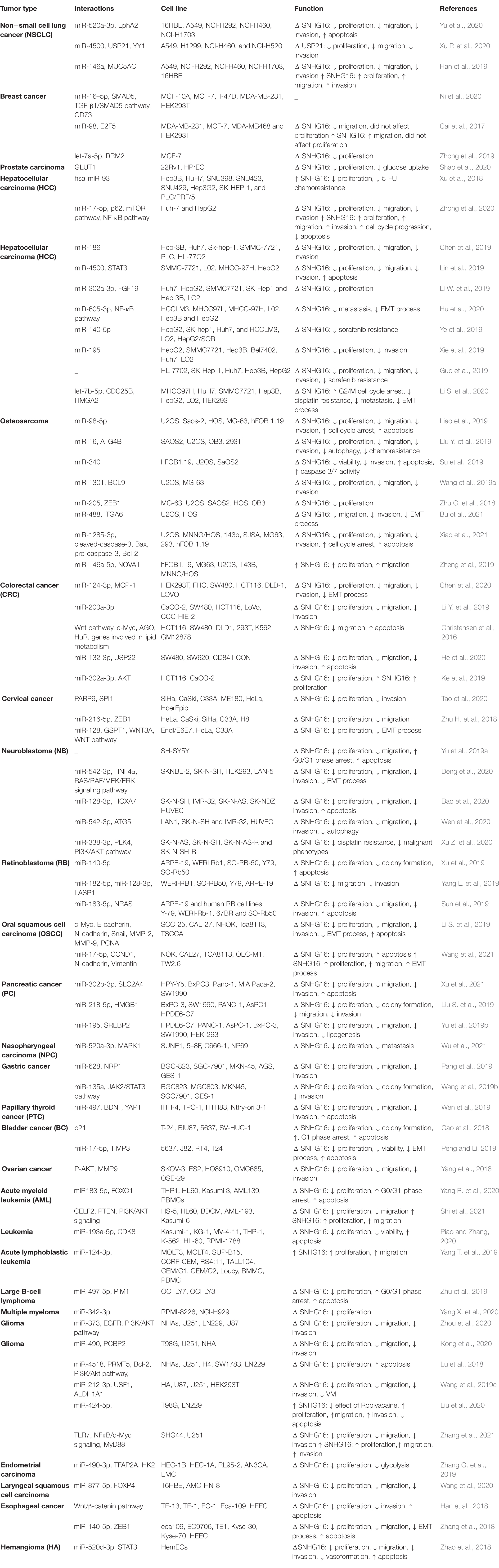
Table 1. Outline of researches which measured expression of SNHG16 in cell lines (Δ, knock-down or deletion; 5-FU, 5-fluorouracil; VM, vasculogenic mimicry).
Animal Studies
Animal studies have consistently shown that SNHG16 silencing decreases malignant feature of the grafted cancer cells (Table 2). The only exception has been reported in HCC where SNHG16 over-expression has significantly suppressed the in vivo expansion of grafted HuH7 cells (Xu et al., 2018). Another study in HCC xenograft model has shown that SNHG16 silencing enhances response of HepG2/SOR cells to cytotoxic effect of sorafenib and attenuates tumor growth (Ye et al., 2019). In xenograft models of retinoblastoma, up-regulation SNHG16 (Xu et al., 2019) or its downstream target NRAS (Sun et al., 2019) can increase tumor growth. Finally, in gastric cancer where SNHG16 sponges miR-628, in vivo studies have shown that up-regulation of miR-628 can decrease tumor expansion (Pang et al., 2019).
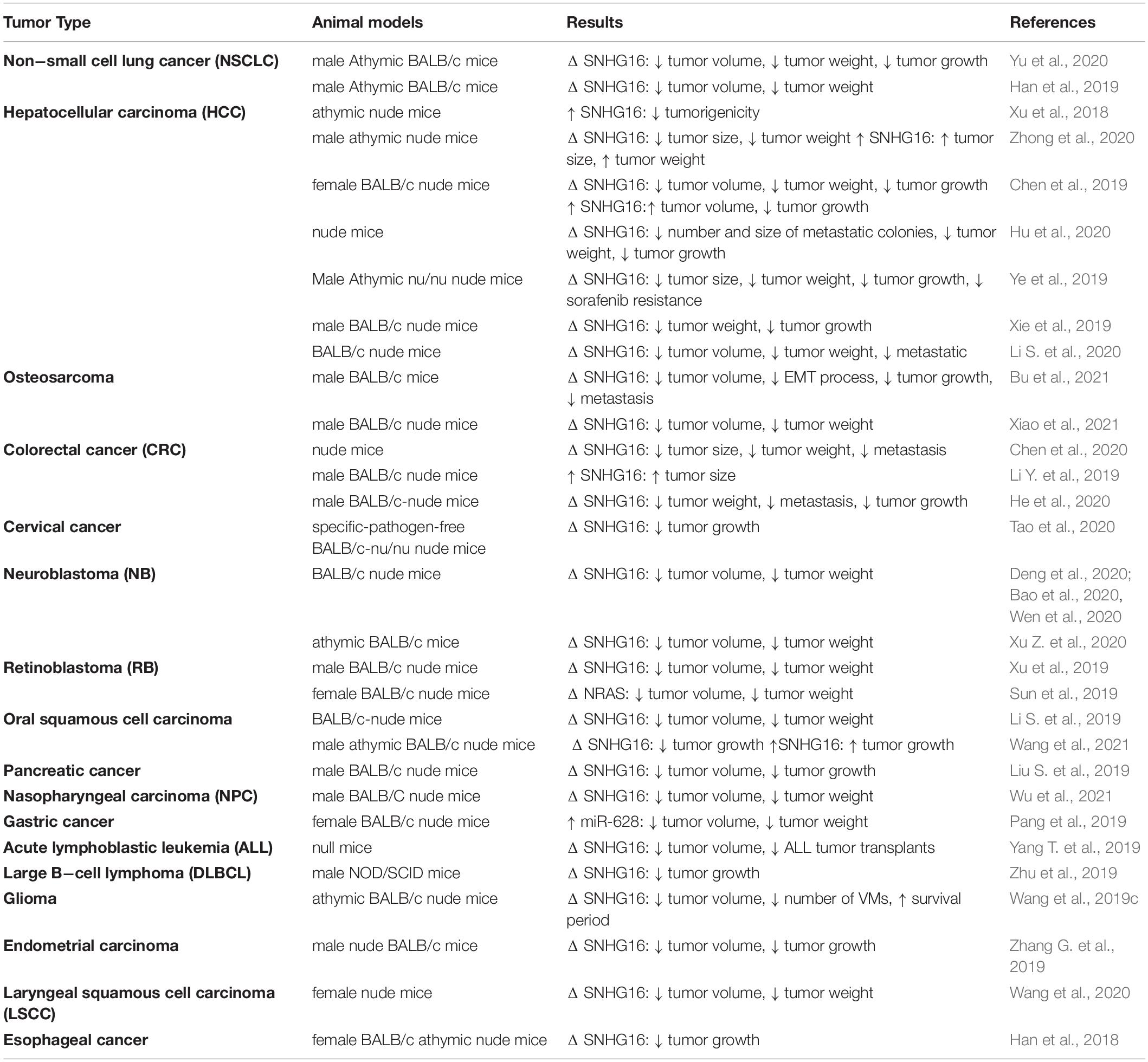
Table 2. Outline of studies which judged function of SNHG16 in animal models (Δ, knock-down or deletion; VM, vasculogenic mimicry).
Clinical Studies
Except for a single study which demonstrated down-regulation of SNHG16 in HCC samples versus nearby non-malignant hepatic tissues (Xu et al., 2018), other studies have indicated up-regulation of SNHG16 in malignant tissues of different origins compared with non-neoplastic samples (Supplementary Table 1). Consistent with these findings, up-regulation of SNHG16 has been revealed to predict poor survival of patients. Moreover, its expression has been related with greater chance of distant metastasis, lymph node involvement and low differentiation of tumor cells.
Discussion
Small nucleolar RNA host gene 6 has been regarded as an oncogenic lncRNA in almost all tissues. This lncRNA affect carcinogenesis through multifaceted mechanisms including mechanisms related to both tumor cells and their niche. In fact, it can both affect cellular functions and processes, particularly those related with proliferation, survival and apoptosis as well as microenvironmental aspects of cancer progression.
More than 20 miRNAs have been found to interact with SNHG16. The sponging effects of SNHG16 on miRNAs have been well studied. miR-520a-3p, miR-4500, miR-146a miR-16–5p, miR-98, let-7a-5p, hsa-miR-93, miR-17-5p, miR-186, miR-302a-3p, miR-605-3p, miR-140-5p, miR-195, let-7b-5p, miR-16, miR-340, miR-1301, miR-205, miR-488, miR-1285-3p, miR-146a-5p, and miR-124-3p are examples of miRNAs sponged by this lncRNA in different types of cancers. Verification of interaction between this lncRNA and a number of miRNAs such as miR-98 in different tissues raises the possibility of independence of such interactions from the tissue type. TGF-β1/SMAD5, mTOR, NF-κB, RAS/RAF/MEK/ERK, PI3K/AKT, and Wnt/β-catenin pathways are among cancer-related pathways being affected by this lncRNA. Moreover, SNHG16 has been shown to affect expression of a number of EMT-associated transcription factors and enhance this process. SNHG16 has also been found to affect response of cancer cells to 5-FU and sorafenib.
Based on the results of functional studies that confirmed the ability of siRNA-mediated SNHG16 silencing in reduction of cancer cell proliferation and invasiveness, this strategy can be proposed as a therapeutic strategy for cancer. In vivo studies have also confirmed applicability of these methods; however no clinical study has applied these methods yet. Antisense oligonucleotides as a promising strategy for suppression of expression of SNHG16 should be appraised in clinical settings considering the bioavailability and safety issues.
Although over-expression of SNHG16 has been verified in tissue samples of different types of tumors, application of this lncRNA as a circulatory marker for early detection of cancer has not been assessed. Since clinical studies have revealed correlation between expression amounts of SNHG16 and malignant features, one can suppose that SNHG16 can be used as both diagnostic and prognostic marker. However, this speculation should be verified in future.
Author Contributions
MT and SG-F wrote the draft and revised it. TK and SS collected the data and designed the tables and figures. All authors read and approved submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.741684/full#supplementary-material
Footnotes
References
Bao, J., Zhang, S., Meng, Q., and Qin, T. (2020). SNHG16 silencing inhibits neuroblastoma progression by downregulating HOXA7 via sponging miR-128-3p. Neurochem. Res. 45, 825–836. doi: 10.1007/s11064-020-02955-x
Bu, J., Guo, R., Xu, X.-Z., Luo, Y., and Liu, J.-F. (2021). LncRNA SNHG16 promotes epithelial-mesenchymal transition by upregulating ITGA6 through miR-488 inhibition in osteosarcoma. J. Bone Oncol. 27:100348. doi: 10.1016/j.jbo.2021.100348
Cai, C., Huo, Q., Wang, X., Chen, B., and Yang, Q. (2017). SNHG16 contributes to breast cancer cell migration by competitively binding miR-98 with E2F5. Biochem. Biophys. Res. Commun. 485, 272–278. doi: 10.1016/j.bbrc.2017.02.094
Cao, X., Xu, J., and Yue, D. (2018). LncRNA-SNHG16 predicts poor prognosis and promotes tumor proliferation through epigenetically silencing p21 in bladder cancer. Cancer Gene Ther. 25, 10–17. doi: 10.1038/s41417-017-0006-x
Chen, H., Li, M., and Huang, P. (2019). LncRNA SNHG16 promotes hepatocellular carcinoma proliferation, migration and invasion by regulating miR-186 expression. J. Cancer 10:3571. doi: 10.7150/jca.28428
Chen, Z.-Y., Wang, X.-Y., Yang, Y.-M., Wu, M.-H., Yang, L., Jiang, D.-T., et al. (2020). LncRNA SNHG16 promotes colorectal cancer cell proliferation, migration, and epithelial–mesenchymal transition through miR-124-3p/MCP-1. Gene ther. 1–13. doi: 10.1038/s41434-020-0176-2 [Epub ahead of print].
Christensen, L. L., True, K., Hamilton, M. P., Nielsen, M. M., Damas, N. D., Damgaard, C. K., et al. (2016). SNHG16 is regulated by the Wnt pathway in colorectal cancer and affects genes involved in lipid metabolism. Mol. Oncol. 10, 1266–1282. doi: 10.1016/j.molonc.2016.06.003
Deng, D., Yang, S., and Wang, X. (2020). Long non-coding RNA SNHG16 regulates cell behaviors through miR-542-3p/HNF4α axis via RAS/RAF/MEK/ERK signaling pathway in pediatric neuroblastoma cells. Biosci. Rep. 40:BSR20200723.
Guo, Z., Zhang, J., Fan, L., Liu, J., Yu, H., Li, X., et al. (2019). Long noncoding RNA (lncRNA) small nucleolar RNA host gene 16 (SNHG16) predicts poor prognosis and sorafenib resistance in hepatocellular carcinoma. Med. Sci. Monit. 25:2079. doi: 10.12659/msm.915541
Han, G., Lu, K., Wang, P., Ye, J., Ye, Y., and Huang, J. (2018). LncRNA SNHG16 predicts poor prognosis in ESCC and promotes cell proliferation and invasion by regulating Wnt/beta-catenin signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 22, 3795–3803.
Han, W., Du, X., Liu, M., Wang, J., Sun, L., and Li, Y. (2019). Increased expression of long non-coding RNA SNHG16 correlates with tumor progression and poor prognosis in non-small cell lung cancer. Int. J. Biol. Macromol. 121, 270–278. doi: 10.1016/j.ijbiomac.2018.10.004
He, X., Ma, J., Zhang, M., Cui, J., and Yang, H. (2020). Long non-coding RNA SNHG16 activates USP22 expression to promote colorectal cancer progression by sponging miR-132-3p. Onco Targets Ther. 13:4283. doi: 10.2147/ott.s244778
Hu, Y. L., Feng, Y., Chen, Y. Y., Liu, J. Z., Su, Y., Li, P., et al. (2020). SNHG16/miR-605-3p/TRAF6/NF-κB feedback loop regulates hepatocellular carcinoma metastasis. J. Cell. Mol. Med. 24, 7637–7651. doi: 10.1111/jcmm.15399
Ke, D., Wang, Q., Ke, S., Zou, L., and Wang, Q. (2019). Long-non coding RNA SNHG16 supports colon cancer cell growth by modulating miR-302a-3p/AKT axis. Pathol. Oncol. Res. 26, 1605–1613. doi: 10.1007/s12253-019-00743-9
Kong, F., Yan, Y., Deng, J., Zhu, Y., Li, Y., Li, H., et al. (2020). LncRNA SNHG16 promotes proliferation, migration, and invasion of glioma cells through regulating the miR-490/PCBP2 Axis. Cancer Biother. Radiopharm. doi: 10.1089/cbr.2019.3535 [Epub ahead of print].
Li, S., Peng, F., Ning, Y., Jiang, P., Peng, J., Ding, X., et al. (2020). SNHG16 as the miRNA let-7b-5p sponge facilitates the G2/M and epithelial-mesenchymal transition by regulating CDC25B and HMGA2 expression in hepatocellular carcinoma. J. Cell. Biochem. 121, 2543–2558. doi: 10.1002/jcb.29477
Li, S., Zhang, S., and Chen, J. (2019). c-Myc induced upregulation of long non-coding RNA SNHG16 enhances progression and carcinogenesis in oral squamous cell carcinoma. Cancer Gene Ther. 26, 400–410. doi: 10.1038/s41417-018-0072-8
Li, W., Xu, W., Song, J. S., Wu, T., and Wang, W. X. (2019). LncRNA SNHG16 promotes cell proliferation through miR-302a-3p/FGF19 axis in hepatocellular carcinoma. Neoplasma 66, 397–404. doi: 10.4149/neo_2018_180720n504
Li, Y., Lu, Y., and Chen, Y. (2019). Long non-coding RNA SNHG16 affects cell proliferation and predicts a poor prognosis in patients with colorectal cancer via sponging miR-200a-3p. Biosci. Rep. 39:BSR20182498.
Liao, S., Xing, S., and Ma, Y. (2019). LncRNA SNHG16 sponges miR-98-5p to regulate cellular processes in osteosarcoma. Cancer Chemother. Pharmacol. 83, 1065–1074. doi: 10.1007/s00280-019-03822-5
Lin, Q., Zheng, H., Xu, J., Zhang, F., and Pan, H. (2019). LncRNA SNHG16 aggravates tumorigenesis and development of hepatocellular carcinoma by sponging miR-4500 and targeting STAT3. J. Cell. Biochem. 120, 11604–11615. doi: 10.1002/jcb.28440
Liu, R., Wu, M., Xu, G., Ju, L., Xiao, J., Zhong, W., et al. (2020). Ropivacaine inhibits proliferation, migration, and invasion while inducing apoptosis of glioma cells by regulating the SNHG16/miR-424-5p axis. Open Life Sci. 15, 988–999. doi: 10.1515/biol-2020-0108
Liu, S., Zhang, W., Liu, K., and Liu, Y. (2019). LncRNA SNHG16 promotes tumor growth of pancreatic cancer by targeting miR-218-5p. Biomed. Pharmacother. 114:108862. doi: 10.1016/j.biopha.2019.108862
Liu, Y., Gu, S., Li, H., Wang, J., Wei, C., and Liu, Q. (2019). SNHG16 promotes osteosarcoma progression and enhances cisplatin resistance by sponging miR-16 to upregulate ATG4B expression. Biochem. Biophys. Res. Commun. 518, 127–133. doi: 10.1016/j.bbrc.2019.08.019
Lu, Y.-F., Cai, X.-L., Li, Z.-Z., Lv, J., Xiang, Y.-A., Chen, J.-J., et al. (2018). LncRNA SNHG16 functions as an oncogene by sponging MiR-4518 and up-regulating PRMT5 expression in glioma. Cell. Physiol. Biochem. 45, 1975–1985. doi: 10.1159/000487974
Ni, C., Fang, Q.-Q., Chen, W.-Z., Jiang, J.-X., Jiang, Z., Ye, J., et al. (2020). Breast cancer-derived exosomes transmit LncRNA SNHG16 to induce CD73+ γδ1 Treg cells. Signal Transduct. Target. Ther. 5:41.
Pang, W., Zhai, M., Wang, Y., and Li, Z. (2019). Long noncoding RNA SNHG16 silencing inhibits the aggressiveness of gastric cancer via upregulation of microRNA-628-3p and consequent decrease of NRP1. Cancer Manag. Res. 11:7263. doi: 10.2147/cmar.s211856
Peng, H., and Li, H. (2019). The encouraging role of long noncoding RNA small nuclear RNA host gene 16 in epithelial-mesenchymal transition of bladder cancer via directly acting on miR-17-5p/metalloproteinases 3 axis. Mol. Carcinog. 58, 1465–1480. doi: 10.1002/mc.23028
Piao, M., and Zhang, L. (2020). Knockdown of SNHG16 suppresses the proliferation and induces the apoptosis of leukemia cells via miR-193a-5p/CDK8. Int. J. Mol. Med. 46, 1175–1185. doi: 10.3892/ijmm.2020.4671
Shao, M., Yu, Z., and Zou, J. (2020). LncRNA-SNHG16 silencing inhibits prostate carcinoma cell growth, downregulate glut1 expression and reduce glucose uptake. Cancer Manag. Res. 12:1751. doi: 10.2147/cmar.s231370
Shi, M., Yang, R., Lin, J., Wei, Q., Chen, L., Gong, W., et al. (2021). LncRNA-SNHG16 promotes proliferation and migration of acute myeloid leukemia cells via PTEN/PI3K/AKT axis through suppressing CELF2 protein. J. Biosci. 46:4.
Su, P., Mu, S., and Wang, Z. (2019). Long noncoding RNA SNHG16 promotes osteosarcoma cells migration and invasion via sponging miRNA-340. DNA Cell Biol. 38, 170–175. doi: 10.1089/dna.2018.4424
Sun, G., Su, G., Liu, F., and Han, W. N. R. A. S. (2019). Contributes to retinoblastoma progression through SNHG16/miR-183-5p/NRAS regulatory network. OncoTargets Ther. 12:10703. doi: 10.2147/ott.s232470
Tao, L., Wang, X., and Zhou, Q. (2020). Long noncoding RNA SNHG16 promotes the tumorigenicity of cervical cancer cells by recruiting transcriptional factor SPI1 to upregulate PARP9. Cell Biol. Int. 44, 773–784. doi: 10.1002/cbin.11272
Wang, Q., Han, J., Xu, P., Jian, X., Huang, X., and Liu, D. (2021). Silencing of LncRNA SNHG16 downregulates cyclin D1 (CCND1) to abrogate malignant phenotypes in oral squamous cell carcinoma (OSCC) through upregulating miR-17-5p. Cancer Manag. Res. 13:1831. doi: 10.2147/cmar.s298236
Wang, X., Kan, J., Han, J., Zhang, W., Bai, L., and Wu, H. (2019b). LncRNA SNHG16 functions as an oncogene by sponging MiR-135a and promotes JAK2/STAT3 signal pathway in gastric cancer. J. Cancer 10:1013. doi: 10.7150/jca.29527
Wang, X., Hu, K., Chao, Y., and Wang, L. (2019a). LncRNA SNHG16 promotes proliferation, migration and invasion of osteosarcoma cells by targeting miR-1301/BCL9 axis. Biomed. Pharmacother. 114:108798. doi: 10.1016/j.biopha.2019.108798
Wang, D., Zheng, J., Liu, X., Xue, Y., Liu, L., Ma, J., et al. (2019c). Knockdown of USF1 inhibits the vasculogenic mimicry of glioma cells via stimulating SNHG16/miR-212-3p and linc00667/miR-429 axis. Mol. Ther. Nucleic Acids 14, 465–482. doi: 10.1016/j.omtn.2018.12.017
Wang, X., Liu, L., Zhao, W., Li, Q., Wang, G., and Li, H. (2020). LncRNA SNHG16 promotes the progression of laryngeal squamous cell carcinoma by mediating miR-877-5p/FOXP4 Axis. Onco Targets Ther. 13:4569. doi: 10.2147/ott.s250752
Wen, Q., Zhao, L., Wang, T., Lv, N., Cheng, X., Zhang, G., et al. (2019). LncRNA SNHG16 drives proliferation and invasion of papillary thyroid cancer through modulation of miR-497. Onco Targets Ther. 12, 699–708. doi: 10.2147/ott.s186923
Wen, Y., Gong, X., Dong, Y., and Tang, C. (2020). Long non coding RNA SNHG16 facilitates proliferation, migration, invasion and autophagy of neuroblastoma cells via sponging miR-542-3p and upregulating ATG5 expression. Onco Targets Ther. 13:263. doi: 10.2147/ott.s226915
Wu, Q., Zhao, Y., Shi, R., and Wang, T. (2021). LncRNA SNHG16 facilitates nasopharyngeal carcinoma progression by acting as ceRNA to sponge miR-520a-3p and upregulate MAPK1 expression. Cancer Manag. Res. 13:4103. doi: 10.2147/cmar.s305544
Wu, W., Guo, L., Liang, Z., Liu, Y., and Yao, Z. (2020). Lnc-SNHG16/miR-128 axis modulates malignant phenotype through WNT/β-catenin pathway in cervical cancer cells. J. Cancer 11:2201. doi: 10.7150/jca.40319
Xiao, X., Jiang, G., Zhang, S., Hu, S., Fan, Y., Li, G., et al. (2021). LncRNA SNHG16 contributes to osteosarcoma progression by acting as a ceRNA of miR-1285-3p. BMC Cancer 21:355.
Xie, X., Xu, X., Sun, C., and Yu, Z. (2019). Long intergenic noncoding RNA SNHG16 interacts with miR-195 to promote proliferation, invasion and tumorigenesis in hepatocellular carcinoma. Exp. Cell Res. 383:111501. doi: 10.1016/j.yexcr.2019.111501
Xu, C., Hu, C., Wang, Y., and Liu, S. (2019). Long noncoding RNA SNHG16 promotes human retinoblastoma progression via sponging miR-140-5p. Biomed. Pharmacother. 117:109153. doi: 10.1016/j.biopha.2019.109153
Xu, F., Zha, G., Wu, Y., Cai, W., and Ao, J. (2018). Overexpressing lncRNA SNHG16 inhibited HCC proliferation and chemoresistance by functionally sponging hsa-miR-93. Onco Targets Ther. 11:8855. doi: 10.2147/ott.s182005
Xu, H., Miao, X., Li, X., Chen, H., Zhang, B., and Zhou, W. (2021). LncRNA SNHG16 contributes to tumor progression via the miR-302b-3p/SLC2A4 axis in pancreatic adenocarcinoma. Cancer Cell Int. 21:51.
Xu, P., Xiao, H., Yang, Q., Hu, R., Jiang, L., Bi, R., et al. (2020). The USP21/YY1/SNHG16 axis contributes to tumor proliferation, migration, and invasion of non-small-cell lung cancer. Exp. Mol. Med. 52, 41–55. doi: 10.1038/s12276-019-0356-6
Xu, Z., Sun, Y., Wang, D., Sun, H., and Liu, X. (2020). SNHG16 promotes tumorigenesis and cisplatin resistance by regulating miR-338-3p/PLK4 pathway in neuroblastoma cells. Cancer Cell Int. 20:236.
Yang, L., Zhang, L., Lu, L., and Wang, Y. (2019). Long noncoding RNA SNHG16 sponges miR-182-5p and miR-128-3p to promote retinoblastoma cell migration and invasion by targeting LASP1. Onco Targets Ther. 12:8653. doi: 10.2147/ott.s212352
Yang, R., Ma, D., Wu, Y., Zhang, Y., and Zhang, L. (2020). LncRNA SNHG16 regulates the progress of acute myeloid leukemia through miR183-5p–FOXO1 Axis. Onco Targets Ther. 13:12943. doi: 10.2147/ott.s258684
Yang, T., Jin, X., Lan, J., and Wang, W. (2019). Long non-coding RNA SNHG16 has tumor suppressing effect in acute lymphoblastic leukemia by inverse interaction on hsa-miR-124-3p. IUBMB Life 71, 134–142. doi: 10.1002/iub.1947
Yang, X., Huang, H., Wang, X., Liu, H., Liu, H., and Lin, Z. (2020). Knockdown of lncRNA SNHG16 suppresses multiple myeloma cell proliferation by sponging miR-342-3p. Cancer Cell Int. 20:38.
Yang, X., Wang, G., and Luo, L. (2018). Long non-coding RNA SNHG16 promotes cell growth and metastasis in ovarian cancer. Eur. Rev. Med. Pharmacol. Sci. 22, 616–622.
Ye, J., Zhang, R., Du, X., Chai, W., and Zhou, Q. (2019). Long noncoding RNA SNHG16 induces sorafenib resistance in hepatocellular carcinoma cells through sponging miR-140-5p. Onco Targets Ther. 12:415. doi: 10.2147/ott.s175176
Yu, L., Chen, D., and Song, J. (2020). LncRNA SNHG16 promotes non-small cell lung cancer development through regulating EphA2 expression by sponging miR-520a-3p. Thorac. Cancer 11, 603–611. doi: 10.1111/1759-7714.13304
Yu, Y., Dong, J.-T., He, B., Zou, Y.-F., Li, X.-S., Xi, C.-H., et al. (2019b). LncRNA SNHG16 induces the SREBP2 to promote lipogenesis and enhance the progression of pancreatic cancer. Future Oncol. 15, 3831–3844. doi: 10.2217/fon-2019-0321
Yu, Y., Chen, F., Yang, Y., Jin, Y., Shi, J., Han, S., et al. (2019a). lncRNA SNHG16 is associated with proliferation and poor prognosis of pediatric neuroblastoma. Int. J. Oncol. 55, 93–102.
Zhang, G., Ma, A., Jin, Y., Pan, G., and Wang, C. (2019). LncRNA SNHG16 induced by TFAP2A modulates glycolysis and proliferation of endometrial carcinoma through miR-490-3p/HK2 axis. Am. J. Transl. Res. 11:7137.
Zhang, K., Chen, J., Song, H., and Chen, L. B. (2018). SNHG16/miR-140-5p axis promotes esophagus cancer cell proliferation, migration and EMT formation through regulating ZEB1. Oncotarget 9, 1028–1040. doi: 10.18632/oncotarget.23178
Zhang, R., Li, P., Lv, H., Li, N., Ren, S., and Xu, W. (2021). Exosomal SNHG16 secreted by CSCs promotes glioma development via TLR7. Stem Cell Res. Ther. 12:349.
Zhang, S., Du, L., Wang, L., Jiang, X., Zhan, Y., Li, J., et al. (2019). Evaluation of serum exosomal Lnc RNA-based biomarker panel for diagnosis and recurrence prediction of bladder cancer. J. Cell. Mol. Med. 23, 1396–1405. doi: 10.1111/jcmm.14042
Zhao, W., Fu, H., Zhang, S., Sun, S., and Liu, Y. (2018). LncRNA SNHG16 drives proliferation, migration, and invasion of hemangioma endothelial cell through modulation of miR-520d-3p/STAT3 axis. Cancer Med. 7, 3311–3320. doi: 10.1002/cam4.1562
Zheng, S., Ge, D., Tang, J., Yan, J., Qiu, J., Yin, Z., et al. (2019). LncSNHG16 promotes proliferation and migration of osteosarcoma cells by targeting microRNA-146a-5p. Eur. Rev. Med. Pharmacol. Sci. 23, 96–104.
Zhong, G., Lou, W., Yao, M., Du, C., Wei, H., and Fu, P. (2019). Identification of novel mRNA-miRNA-lncRNA competing endogenous RNA network associated with prognosis of breast cancer. Epigenomics 11, 1501–1518. doi: 10.2217/epi-2019-0209
Zhong, J. H., Xiang, X., Wang, Y. Y., Liu, X., Qi, L. N., Luo, C. P., et al. (2020). The lncRNA SNHG16 affects prognosis in hepatocellular carcinoma by regulating p62 expression. J. Cell. Physiol. 235, 1090–1102. doi: 10.1002/jcp.29023
Zhou, X.-Y., Liu, H., Ding, Z.-B., Xi, H.-P., and Wang, G.-W. (2020). lncRNA SNHG16 promotes glioma tumorigenicity through miR-373/EGFR axis by activating PI3K/AKT pathway. Genomics 112, 1021–1029. doi: 10.1016/j.ygeno.2019.06.017
Zhu, C., Cheng, D., Qiu, X., Zhuang, M., and Liu, Z. (2018). Long noncoding RNA SNHG16 promotes cell proliferation by sponging microRNA-205 and upregulating ZEB1 expression in osteosarcoma. Cell. Physiol. Biochem. 51, 429–440. doi: 10.1159/000495239
Zhu, H., Zeng, Y., Zhou, C.-C., and Ye, W. (2018). SNHG16/miR-216-5p/ZEB1 signal pathway contributes to the tumorigenesis of cervical cancer cells. Arch. Biochem. Biophys. 637, 1–8.
Keywords: SNHG6, lncRNA, cancer, biomarker, expression
Citation: Ghafouri-Fard S, Khoshbakht T, Taheri M and Shojaei S (2021) A Review on the Role of Small Nucleolar RNA Host Gene 6 Long Non-coding RNAs in the Carcinogenic Processes. Front. Cell Dev. Biol. 9:741684. doi: 10.3389/fcell.2021.741684
Received: 15 July 2021; Accepted: 09 September 2021;
Published: 04 October 2021.
Edited by:
Shiv K. Gupta, Mayo Clinic, United StatesReviewed by:
Rezvan Noroozi, Jagiellonian University, PolandAmin Safa, Complutense University of Madrid, Spain
Copyright © 2021 Ghafouri-Fard, Khoshbakht, Taheri and Shojaei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohammad Taheri, bW9oYW1tYWRfODIzQHlhaG9vLmNvbQ==; Seyedpouzhia Shojaei, cHNoMTE4MjAwMkB5YWhvby5jb20=
 Soudeh Ghafouri-Fard
Soudeh Ghafouri-Fard Tayyebeh Khoshbakht
Tayyebeh Khoshbakht Mohammad Taheri
Mohammad Taheri Seyedpouzhia Shojaei4*
Seyedpouzhia Shojaei4*