
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol. , 23 September 2021
Sec. Developmental Epigenetics
Volume 9 - 2021 | https://doi.org/10.3389/fcell.2021.740937
This article is part of the Research Topic Monoallelic Expression in Development and Diseases View all 10 articles
X-chromosome inactivation (XCI) and random monoallelic expression of autosomal genes (RMAE) are two paradigms of gene expression regulation where, at the single cell level, genes can be expressed from either the maternal or paternal alleles. X-chromosome inactivation takes place in female marsupial and placental mammals, while RMAE has been described in mammals and also other species. Although the outcome of both processes results in random monoallelic expression and mosaicism at the cellular level, there are many important differences. We provide here a brief sketch of the history behind the discovery of XCI and RMAE. Moreover, we review some of the distinctive features of these two phenomena, with respect to when in development they are established, their roles in dosage compensation and cellular phenotypic diversity, and the molecular mechanisms underlying their initiation and stability.
In diploid organisms, the two alleles of a gene are usually expressed. However, the expression levels of each allele are not necessarily equal, and allelic imbalance (AI) in transcript levels can occur due to genetic differences in the regulatory sequences or the stability of the transcripts. There are, however, special cases not explained by in cis differences in the sequence of the alleles. These have been lumped under the umbrella term “monoallelic expression.” In a broad sense, all genetic expression is epigenetic, but if we use a conservative definition of epigenetics to include all heritable (during mitosis or meiosis) changes in gene expression that occur without any changes in the underlying DNA sequence, then monoallelic expression becomes the poster child of epigenetics. Known cases of monoallelic expression include genomic imprinting, X-chromosome inactivation (XCI), and random monoallelic autosomal expression (RMAE). Genomic imprinting affects all cells of an organism the same way, i.e., it is always the same allele that is expressed, depending on the parent of origin (Barton et al., 1984; McGrath and Solter, 1984; Surani et al., 1984). The fate (expression or silencing) is defined during the formation of the gametes in the progenitor. Thus, despite the associated fascinating molecular mechanisms, evolutionary theory (Haig, 1993), and relevance for development and diseases (Ferguson-Smith and Bourc’his, 2018), genomic imprinting is merely a case of transgenerational gene expression that is reset each generation during the formation of the oocyte and sperm cells. XCI and RMAE differ from genomic imprinting because they give rise to mosaicism: in the same organism, some cells express the maternal allele and other cells express the paternal allele. Over the last decades, this common feature has recurrently tempted many to draw parallels between XCI and RMAE, both in reviews or opinion pieces [e.g., (Efstratiadis, 1995; Goldmit and Bergman, 2004; Chess, 2016; Gendrel et al., 2016)] and original articles [e.g., (Mostoslavsky et al., 2001; Pereira et al., 2003)]. But much like the confusion created by false cognates or “faux amis” between two languages, the parallels between two phenomena often prevent us from seeing the obvious and meaningful differences; parallels can illuminate but also deceive. Thus, here we propose to critically evaluate the relevance of the parallels drawn between XCI and RMAE, and expose their key differences at the cellular and molecular levels.
XCI and RMAE were described in the same decade. X-chromosome inactivation, also named “Lyonisation,” was first proposed in 1961 by mouse geneticist Mary Lyon in a short report with no figures, where she laid the fundamental principles of XCI based solely on mouse genetics and earlier cytological evidence (Lyon, 1961). A few years later, individual B cells were shown to express only one immunoglobulin allele, both for the heavy and the kappa chains (Cebra and Goldstein, 1965; Pernis et al., 1965), two autosomal genes. Retrospectively, these three papers were seminal, but Lyon’s work immediately created a new field, whereas the RMAE of antigen receptors remained essentially a pet subject for a niche of scientists.
Mary Lyon was examining the inheritance and the phenotype of different mutations in X-linked genes affecting coat color in mice. She observed that heterozygous females had mosaic or variegated phenotypes, with patches of normal and mutant color, unlike males. This, coupled with the knowledge that female mice with only one X chromosome were viable and fertile (Welshons and Russell, 1959) and that female cells exhibit one condensed X chromosome in their nuclei (Ohno et al., 1959; Ohno and Hauschka, 1960), led her to the XCI hypothesis. The key principles underlying this hypothesis were the genetic inactivation of the X chromosome of either paternal or maternal origin, the early inactivation during embryogenesis, and the clonal inheritance of the inactive state through cell division (Lyon, 1961). Soon after, other scientists correlated the genetic observations made by Mary Lyon with experimental studies, such as the presence of two red blood cell populations or protein variants associated with mutations in the G6pd X-linked gene in female cells (Beutler et al., 1962; Davidson et al., 1963). In 1962, Mary Lyon published a much longer report focusing on human X-linked syndromes, providing evidence that XCI is present in other mammals, such as humans, and is the basis for dosage compensation between the sex chromosomes (Lyon, 1962). XCI is still often referred nowadays to as the “Lyon hypothesis,” although it should be considered as a fully established law (Gendrel and Heard, 2011).
The finding that the immunoglobulin chains are expressed monoallelically at the cellular level predates the discovery of the mechanism of V(D)J recombination that sets apart the antigen receptor genes (Hozumi and Tonegawa, 1976), including the immunoglobulin and T-cell receptor genes. Over the years, it was found that monoallelic expression – more commonly described in this literature as “allelic exclusion” – is a feature of most antigen receptor genes only partly explained by the relatively high frequency of non-productive sequences (with frameshifts leading to premature stop codons) generated by V(D)J recombination and that the percentage of cells with monoallelic expression varies considerably depending on the antigen receptor gene [reviewed in Vettermann and Schlissel (2010)].
The collection of genes under allelic expression expanded beyond the antigen receptor genes only in the 1990s. The olfactory receptor (OR) genes form the largest gene family in mammals; in the mouse, there are 1,296 OR genes (Zhang and Firestein, 2002). Remarkably, each neuron expresses only one gene and, taking advantage of the OR gene polymorphisms found in Mus musculus x M. spretus F1 mice, it was found that, in a given neuron, only one of the two alleles of the chosen expressed gene is transcribed (Chess et al., 1994). Soon after this finding, using allele-specific antibodies, the Ly49 genes of natural killer cells were also shown to display a monoallelic expression pattern in mice (Held et al., 1995) and a few years later, a pheromone receptor, similar to the OR genes, was shown to display RMAE in the neurons of the accessory olfactory system (Rodriguez et al., 1999).
The end of the 1990s would mark the beginning of a short controversy on the expression patterns of the interleukins (ILs). In T cells, IL-2 was reported to be monoallelically expressed in T cells (Hollander et al., 1998), whereas for IL-4 the expression was described as biallelic or monoallelic, depending on the clone (Bix and Locksley, 1998) or the strength of the signal delivered through the TCR (Rivière et al., 1998). Four subsequent studies on IL-2 reached different conclusions: whereas IL-2 was found to be biallelically expressed in human T cell clones (Chiodetti et al., 2000; Bayley et al., 2003), in mice heterozygous for an IL-2-GFP transgene (Naramura et al., 1998) and in single-cell RT-PCR experiments (Rhoades et al., 2000), most cells expressed both IL-2 alleles but there were also single expressors. Overall, the data for IL-2 seem consistent with the data for IL-4: under optimal and continuous stimulation, cells will tend to express both alleles, but at suboptimal levels of expression, there may be cells expressing only one of the alleles.
The list of genes under monoallelic expression grew slowly until the mid-2000s based on additional reports focused on single genes [e.g., (Endo et al., 1995); Table 1], but in 2007, the application of genome-wide transcriptomics to a collection of human lymphoblast clonal cell lines revealed that over 5% of expressed genes display patterns of random monoallelic expression across the collection of clones (Gimelbrant et al., 2007). Over the subsequent years, several independent reports on clonal cell lines confirmed that the number of genes under random monoallelic expression was higher than previously thought (Table 2).
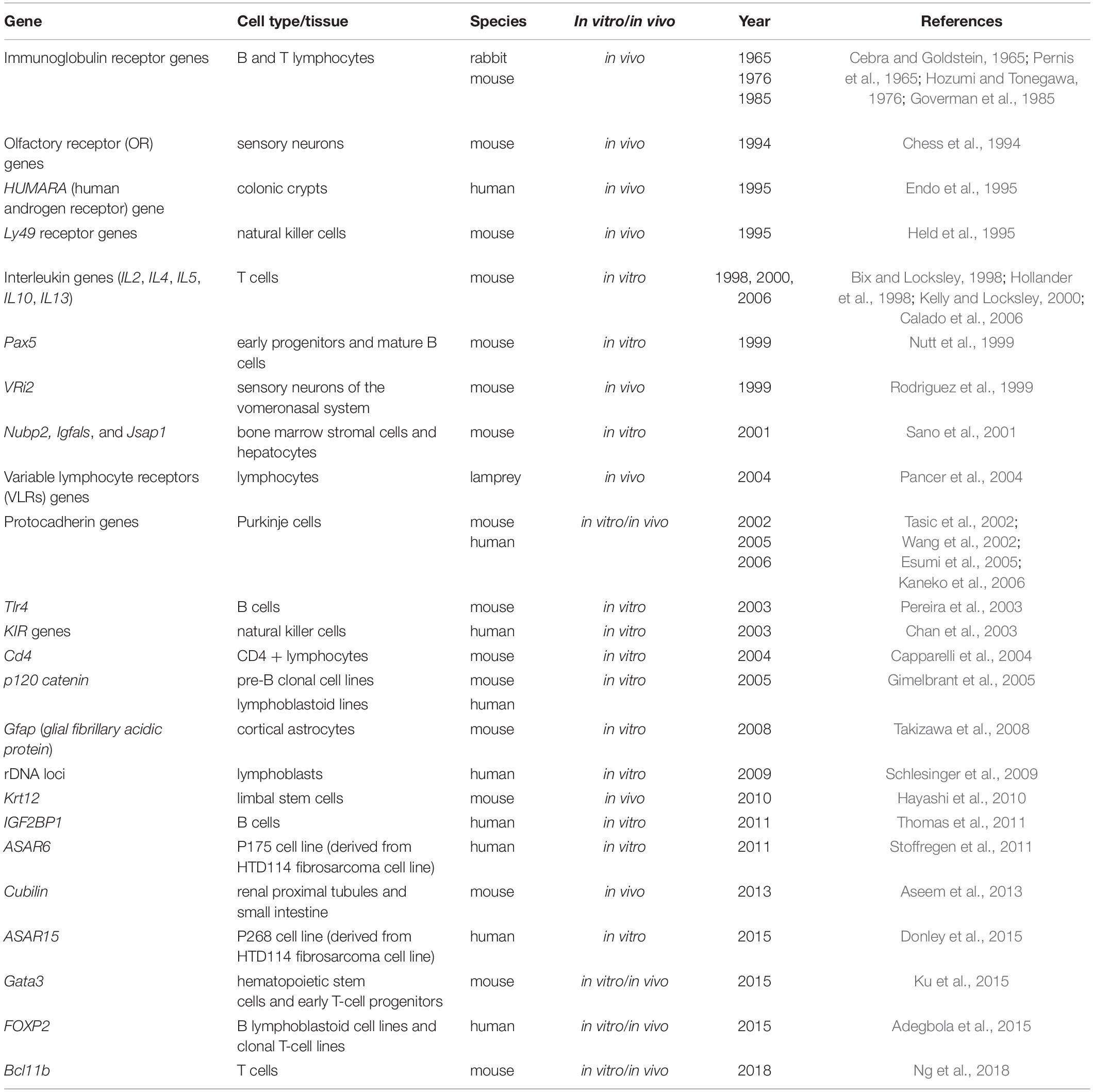
Table 1. List of autosomal genes under random monoallelic expression reported in studies focused on single genes.
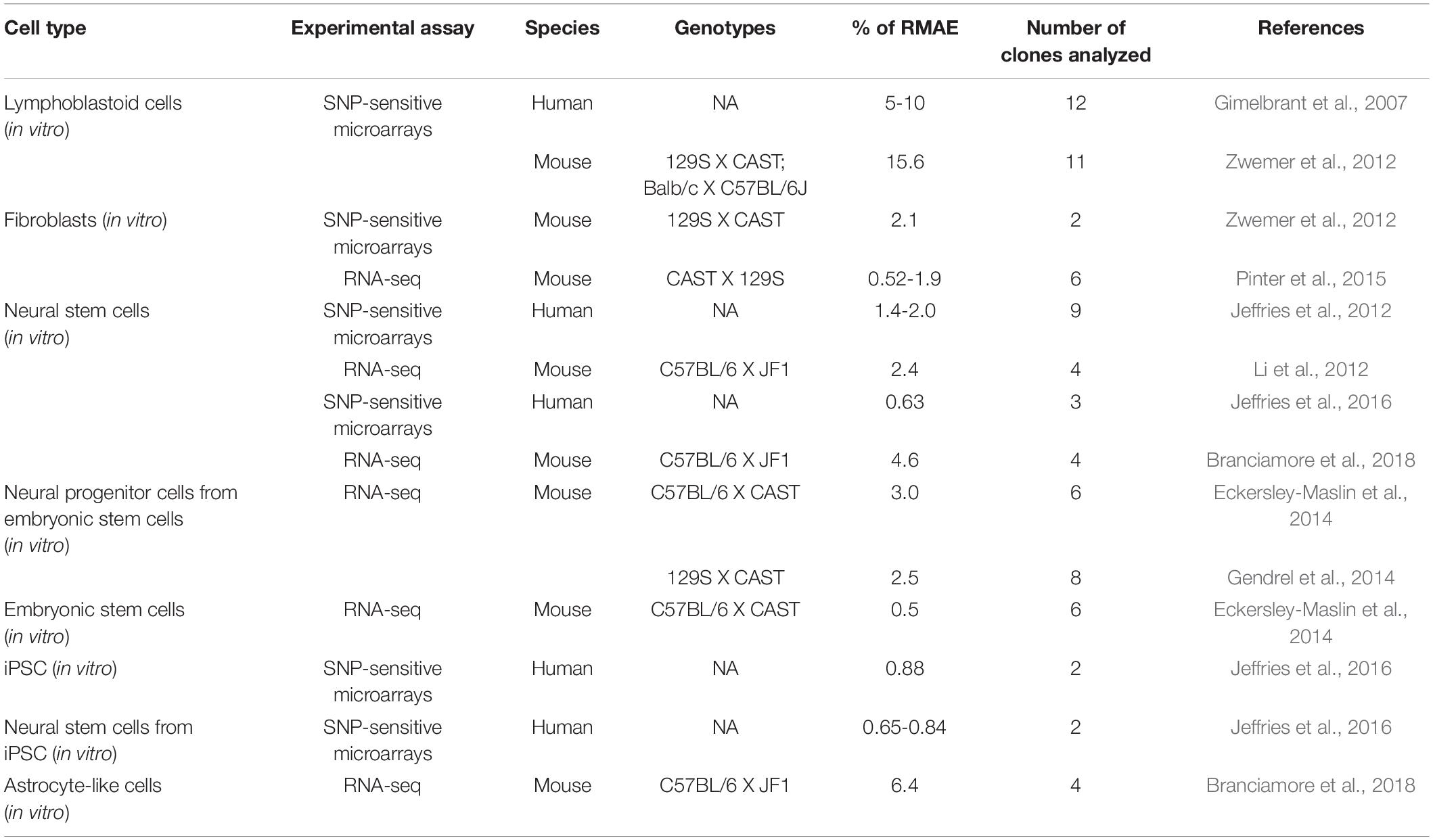
Table 2. A summary of reports based on genome-wide transcriptomics analysis in different cell types.
Technological progress allowing transcriptomics at the single-cell level revealed that stochastic bursts of transcription occurring independently at the allelic level may lead to the presence of RNA from only one of the alleles at a given time (Deng et al., 2014). We will not cover these cases because RMAE due to transcriptional bursting is not expected to be stable over time and is not observed at the clonal level.
XCI is usually perceived as a homogeneous process. It affects an entire chromosome leading to silencing of nearly all genes, with a few notable exceptions, and therefore sets the basis for a robust monoallelic expression of these genes. The inactivation is established in all cells in a random manner early during embryogenesis and is then stably inherited during mitotic cell divisions throughout life; all cells therefore carry an inactive X (Xi) and active X (Xa) chromosome, and females are mosaic individuals with cell populations expressing genes from either the maternal or the paternal X chromosome (Figure 1). Most genes that are subject to XCI stay stably repressed during development and adulthood, and rarely become biallelically expressed, except under specific circumstances discussed below (Galupa and Heard, 2018).
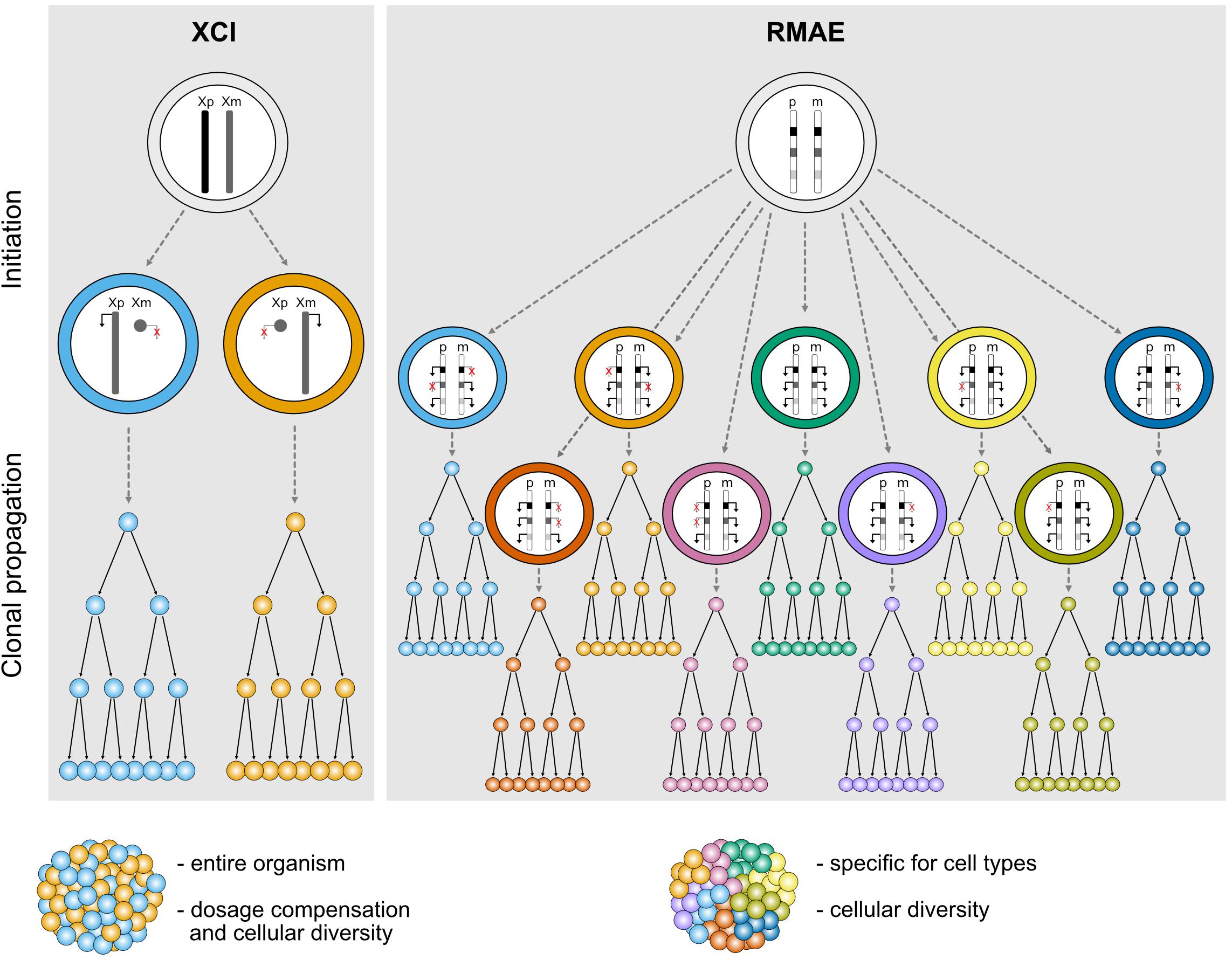
Figure 1. Schematic representation illustrating the features of different types of random monoallelic expression: X-chromosome inactivation (XCI) and random monoallelic autosomal expression (RMAE). Xp, X chromosome of paternal origin, Xm, X chromosome of maternal origin; p, paternal autosome, m, maternal autosome.
In clear contrast with XCI, which has defined physical boundaries (the X chromosome), a precise timing during development, a phylogenetic association with female marsupial and placental mammals, and a master player (the Xist long non-coding RNA), genes under RMAE are scattered throughout autosomal chromosomes, are expressed at different times and in different tissues, can be biallelically expressed and are found in animals other than mammals, including jawless vertebrates (Pancer et al., 2004), trypanosomes (Borst, 2002), and perhaps even in diatom algae (Hoguin et al., 2021) (Figure 1). Additionally, there is no evidence that they are regulated by a single factor and no clear molecular signature that could suggest the existence of a common mechanism regulating RMAE has emerged. Although the first examples of genes under RMAE were of cell surface receptors, which remain an over-represented class, this group is diverse in terms of function.
XCI is initiated during early embryogenesis in mammals. In mice, random XCI starts around the time of implantation (E5.5) and is complete by E6.5 in all cells in embryonic tissues (Mak et al., 2004). In humans, the timing of XCI has been complicated to address owing to obvious difficulties to access relevant material. Nevertheless, studies showed that random XCI is initiated around the implantation stage and, compared to mice, appears to be a much more gradual process during the first four weeks of embryonic development (Tang et al., 2015; Zhou et al., 2019). This process is absolutely essential, as failure to induce XCI results in early embryonic lethality at around day 10 of development for mouse embryos (Takagi and Abe, 1990).
Whereas XCI is established early in development, RMAE can occur early or late, depending on when the gene is first expressed. For instance, in the case of OR genes, the critical events leading to RMAE, namely the stochastic expression of a single OR allele from a pool of OR genes silenced with heterochromatic marks, occurs in the maturing olfactory sensory neurons of the mouse olfactory epithelium (Magklara et al., 2011). However, for most genes under RMAE, it is unknown if the choice of which allele to express is pre-determined in progenitor cells long before the gene becomes expressed. In 2001, that possibility was suggested for the antigen receptor genes based on the finding that these genes replicate asynchronously (Mostoslavsky et al., 2001). Asynchronous replication is a feature of monoallelically expressed genes because, typically, transcribed genes (or alleles) undergo replication before silenced genes (or alleles). The authors drew parallels between XCI and the pattern of asynchronous replication in autosomal genes in their study. In both processes, the epigenetic marks are erased in the morula, re-established around the time of implantation randomly and then clonally maintained. However, in subsequent publications from the same group and others, these parallels fell apart (Farago et al., 2012; Alves-Pereira et al., 2014). Notably, in mice reconstituted with a single hematopoietic stem cell, it was shown that the immunoglobulin heavy chain alleles rearrange independently, i.e., without any evidence for an epigenetic mark (Alves-Pereira et al., 2014). Whether such mark is eventually established later in development and before V(D)J rearrangement is an open question. In the kappa light chain, this pre-determination has been proposed (Farago et al., 2012), but for the heavy chain no evidence was found (Alves-Pereira et al., 2014). In any case, the advantage of such clonal pre-determination long before the genes are expressed is not obvious.
The main purpose of XCI is to enable dosage compensation of X-linked genes products to correct for the imbalance between XX females and XY males in mammals (Graves, 2016). The lethality resulting from failure of XCI is a consequence of the absence of dosage compensation. However, it remains unclear whether dosage compensation is critical for all X-linked genes or only a fraction of them. It is also not known whether compensation of dosage-sensitive genes is necessary in all tissues and all developmental stages. Transcriptome analysis of pre-implantation embryos and differentiating embryonic stem cells indicate that absence of XCI leads to failure to exit the pluripotent state, aberrant expression of extra-embryonic factors, and inappropriate expression of developmental genes, which leads to compromised development and differentiation, hence early lethality (Schulz et al., 2014; Chen et al., 2016; Borensztein et al., 2017).
Besides dosage compensation, XCI is also able to generate a significant level of biological diversity both within and between female individuals (Figure 1). This was exemplified by a study that built topographic maps of XCI mosaicism at single cell resolution, using female mice carrying X-linked fluorescent reporters on each X chromosome (Wu et al., 2014). The authors observed that some organs or tissues are particularly prone to deviations from the expected 50:50 inactivation ratio. In particular, the brain stands out as one organ where the diversity conferred by XCI could have an important functional impact for the stimulus-response amplitude of neuronal networks, especially when considering heterozygosity for an X-linked gene expressed in the brain (Wu et al., 2014). Given the level of genetic variation on the human genome including the X chromosome, XCI could generate a remarkable level of intra- and inter-individual differences in the human central nervous system.
RMAE is thought to have evolved exclusively to increase the biological (or phenotypic) diversity at the cellular level. Assuming polymorphisms within a gene, heterozygous cells with a biallelic pattern of expression will be phenotypically identical, whereas partial RMAE will produce three types of cells within the organism: single paternal allele-expressing cells, single maternal-expressing cells, and biallelic expressing cells (Figure 1). The most exuberant cases of phenotypic diversity are found in the OR and antigen receptor genes. In the former, RMAE is coupled with the selection of a single gene from the largest gene family for expression at the single-cell level, leading to the generation of hundreds or thousands of different sensory olfactory neurons, depending on the species (Niimura et al., 2014). In the latter, thousands of allelic forms are generated during the organism’s life by V(D)J recombination that will be monoallelically expressed to ensure the single cell-single receptor rule, thus facilitating the processes of negative and positive selection that shape the immune repertoires. The potential phenotypic diversity for the average gene served by only two alleles is much lower than that of OR and antigen receptor genes, but for phenotypes determined by multiple genes under RMAE there is a considerable combinatory potential (3n phenotypes, where n is the number of relevant polymorphic genes under RMAE). However, outside of OR and antigen receptor genes, the importance of RMAE-driven phenotypic diversity remains to be demonstrated, and it is a complicated problem to tackle experimentally. As explained below, particular cis-regulatory sequences play a role in RMAE. Thus, a feasible approach would be to replace these sequences with regular promoters, but even in this case the interpretation of the data would not be clear-cut because the expression of multiple receptors at the surface of the cell would decrease the density of any particular receptor compared to its density in a cell with RMAE. Since the manipulation of master epigenetic regulators is unlikely to be sufficiently specific, a proof of principle will probably be obtained using CRISPR/Cas tools that allow epigenetic manipulations at the allele-specific level. An alternative approach would be to generate aggregation chimeras of cells expressing different receptors or other relevant proteins for the quantitative response to be tested. In the absence of such data, other hypotheses can be raised, such as a role for RMAE in dosage compensation (Gendrel et al., 2016). However, the finding that genes under RMAE have increased genetic diversity (polymorphisms) in humans compared to biallelically expressed genes remains a powerful indication that RMAE evolved to increase phenotypic diversity at the cellular level (Savova et al., 2016).
One has to recognize that stochasticity is the key feature common to XCI and RMAE: how come identical or quasi-identical sequences (the X chromosomes or autosomal alleles) sharing the same nuclear environment undergo completely opposite fates (expression or silencing)? This stochastic component is at the core of the appeal these phenomena have to biologists, but there is a critical difference between XCI and RMAE worth mentioning. It has been proposed that each individual X chromosome has an independent probability to be inactivated that is directly proportional to the X: ploidy ratio. Selection in favor of cells keeping one active X chromosome per diploid genome eliminates cells with two inactive X or two active X chromosomes (Monkhorst et al., 2008; Sousa et al., 2018). Thus, XCI involves the inactivation of one X chromosome and counterselection at the cellular level. In contrast, the stochastic component in OR genes, antigen receptor genes, and possibly other autosomal genes under RMAE involves the activation of alleles in a default state of silencing, and cell counterselection is not thought to play a major role in shaping the pattern of monoallelic expression; in fact, B lymphocytes genetically engineered to express two different immunoglobulin heavy chains at the surface were shown to be fit and able to generate a normal B cell compartment (Sonoda et al., 1997).
A key aspect of RMAE in antigen receptor genes is the feedback mechanism that prevents the recombination of the second allele once the protein encoded by the first allele to rearrange productively is expressed at the surface. When the exon encoding the transmembrane domain of the immunoglobulin chain is disrupted, the cell is no longer able to trigger this feedback mechanism and the second allele is given the chance to recombine (Kitamura and Rajewsky, 1992). A similar mechanism has been described for the beta chain of the TCR gene (Aifantis et al., 1997) and the OR genes (Serizawa et al., 2003). The overall picture, then, is the coupling of a stochastic process of gene activation that is sufficiently slow for negative feedback mechanisms to act, preventing further rearrangements (antigen receptors) or gene activation (OR genes). Because the feedback mechanism implies a time-window during which the two alleles can be activated, it also explains the generation of biallelic cells; a slow feedback mechanism will produce many biallelic cells, whereas biallelic cells are rare when the time-window is narrow. However, it is not clear whether such feedbacks are involved for other genes under RMAE and additional mechanisms have been described, which we discuss below.
The process of XCI can be divided in two distinct stages: initiation and maintenance. During the initiation phase, XCI is dependent on the expression of the long non-coding RNA Xist, which induces transcriptional silencing in cis and ultimately coats the entire inactive X chromosome (Loda and Heard, 2019). However, Xist is no longer essential for the maintenance of XCI, as deletion of Xist in somatic cells in culture does not lead to Xi reactivation (Brown and Willard, 1994). Following Xist accumulation on the Xi, one of the first observable events is the formation of a 3D silent nuclear compartment excluding RNA polymerase II and transcription factors, likely to be important for Xist spreading and the initiation of gene silencing (Chaumeil et al., 2006; Clemson et al., 2006; Pandya-Jones et al., 2020). Xist interacts with several RNA-binding proteins, in particular SPEN, which acts as a bridge between Xist RNA and repressor complexes that mediates the removal of histone modifications associated with active genes (H3K27ac, H3K9ac, H4ac), a crucial early step for the initiation of gene silencing (Żylicz et al., 2019; Dossin et al., 2020). Following this, a number of chromosome-wide chromatin changes occur on the Xi to lock in the silenced state, such as deposition of repressive histone modifications (H2AK119Ub and H3K27me3) mediated by the Polycomb repressive complexes 1 and 2 [reviewed in Boeren and Gribnau (2021)]. The late or maintenance phase is characterized by a switch to late replication timing, incorporation of the histone variant macroH2A and DNA methylation of X-linked gene promoter regions by the DNA methyltransferase Dnmt3b [reviewed in Strehle and Guttman (2020)]. These changes ensure the stable and heritable silencing of the majority of genes on the Xi, over hundreds of cell divisions.
By definition, epigenetic modifications, namely histone modifications and DNA methylation, and non-coding RNAs, have been shown to be associated with genes under RMAE (Gendrel et al., 2016). However, unlike XCI, there is no master regulator, and several scenarios have been reported, such the initial repression of both alleles followed by activation (e.g., OR genes and murine Ly49 genes) or the initial activation of both alleles followed by the inactivation of one allele (e.g., human KIR genes).
One puzzling question in XCI has been the nature of the X-linked cis-acting elements important for the binding and spreading of Xist along the X chromosome, prior to gene silencing. Because of the higher density of long interspersed nuclear element-1 (LINE-1) retrotransposons in the X chromosome compared to autosomes (Boyle et al., 1990; Ross et al., 2005), with her so-called “repeat hypothesis,” Mary Lyon postulated that these sequences could act as booster elements for the spreading of the inactive signal along the chromosome and efficient silencing (Lyon, 1998). However, we now know that Xist does not bind directly LINE-1 sequences nor associate with LINE-1-enriched regions. Xist rather exploits the 3D conformation of the X chromosome to spread first to sites that are spatially proximal to the Xist gene at the onset of XCI and is then found enriched over gene-dense regions that are depleted of LINE-1 sequences (Engreitz et al., 2013; Simon et al., 2013). Yet, studies of Xist spreading on autosomal chromatin in X:autosome translocations (Sharp et al., 2002; Popova et al., 2006) or using Xist transgenes on autosomes (Chow et al., 2010; Tang et al., 2010; Loda et al., 2017) all show a good correlation between LINE-1 density, efficiency of spreading, and gene silencing. These observations suggest that LINE-1 elements may contribute to the process of XCI, either by facilitating gene silencing locally in some regions, reinforcing the long-term maintenance of XCI and/or influencing heterochromatin reorganization. This, however, remains to be formally tested using, for example, functional approaches to perturb LINE-1 expression or enrichment on the X chromosome.
Whether genes under RMAE have a DNA sequence signature remains unclear. It has been proposed that these genes are surrounded by an increased density of LINE-1 elements, which are evolutionarily more recent and less truncated than the LINE-1 elements around biallelically expressed genes (Allen et al., 2003). How exactly LINE-1 elements could contribute to RMAE is not known, but their association to RMAE would be one of the few potential parallels with XCI. However, the overlap between predicted RMAE genes based on the presence of LINE-1 elements and the collection of genes under RMAE generated by a genome-wide approach is not statistically significant (Allen et al., 2003; Gimelbrant et al., 2007).
It has been known for decades that the X chromosome and autosomal genes under RMAE replicate asynchronously (Taylor, 1960; Chess et al., 1994) and that this mitotically stable pattern is established early in development. Asynchronous replication was even found to be a property of autosomal chromosomes (Singh et al., 2003), reinforcing the parallel with the X chromosome. Whether the asynchronous replication of autosomes is absolutely stable is not clear, and it has been found that RMAE is not coordinated at the chromosome level, i.e., the alleles from different genes under RMAE on the same chromosome can be active or silent (Gimelbrant et al., 2007). However, an autosomal gene named asynchronous replication and autosomal RNA on chromosome 6 (ASAR6) was shown to encode a non-coding RNA under RMAE, which when expressed leads to the silencing of nearby alleles and remains associated with the chromosome from which it is expressed. Moreover, the disruption of this locus results in delayed replication timing and reactivation of previously silent alleles of nearby genes (Stoffregen et al., 2011; Donley et al., 2013). There is an obvious parallel with XIST, which is also monoallelically expressed, silences most of the genes on the X chromosome in cis and, when deleted, also alters replication timing (Diaz-Perez et al., 2005). Interestingly, a LINE-1 retrotransposon located within ASAR6, in antisense orientation, was then found to control the replication timing (Platt et al., 2018). This constitutes one of the most solid evidence that LINE-1 elements could be involved in the spreading of inactivation also on autosomal chromosomes. Another locus, ASAR15, displays features similar with ASAR6 (Donley et al., 2015). However, it is not known how frequent this type of regional silencing occurs on autosomes, and, unlike XIST, the ASAR6 RNA does not seem to coat the entire chromosome 6 and is not expressed in adult tissues (Stoffregen et al., 2011). A thorough and granular reappreciation of the impact of LINE-1 elements in RMAE would be welcomed.
Several histone modifications influence gene expression, including H3K4me3 and H3K27me3, which are associated with gene activation and repression, respectively. Although active and repressive histone marks are typically imagined as being mutually exclusive, in 2006 two groups reported the existence of regulatory regions – named bivalent domains – that have both (Azuara et al., 2006; Bernstein et al., 2006). Genes with bivalent promoters in embryonic stem cells are expressed at low levels but thought to be poised for rapid activation upon differentiation cues. Interestingly, about 80% of the genes under RMAE in differentiated cells, identified by transcriptomics or the presence of activation and repression histone marks on different alleles, were found to have bivalent promoters in precursor cells (Nag et al., 2013) (Figure 2). Thus, the rapid and timely activation ensured by bivalent promoters seems to increase the probability of RMAE, as if the alleles resolve their status stochastically, leading to cells that activate only the paternal or maternal allele and cells that activate both (Nag et al., 2013).
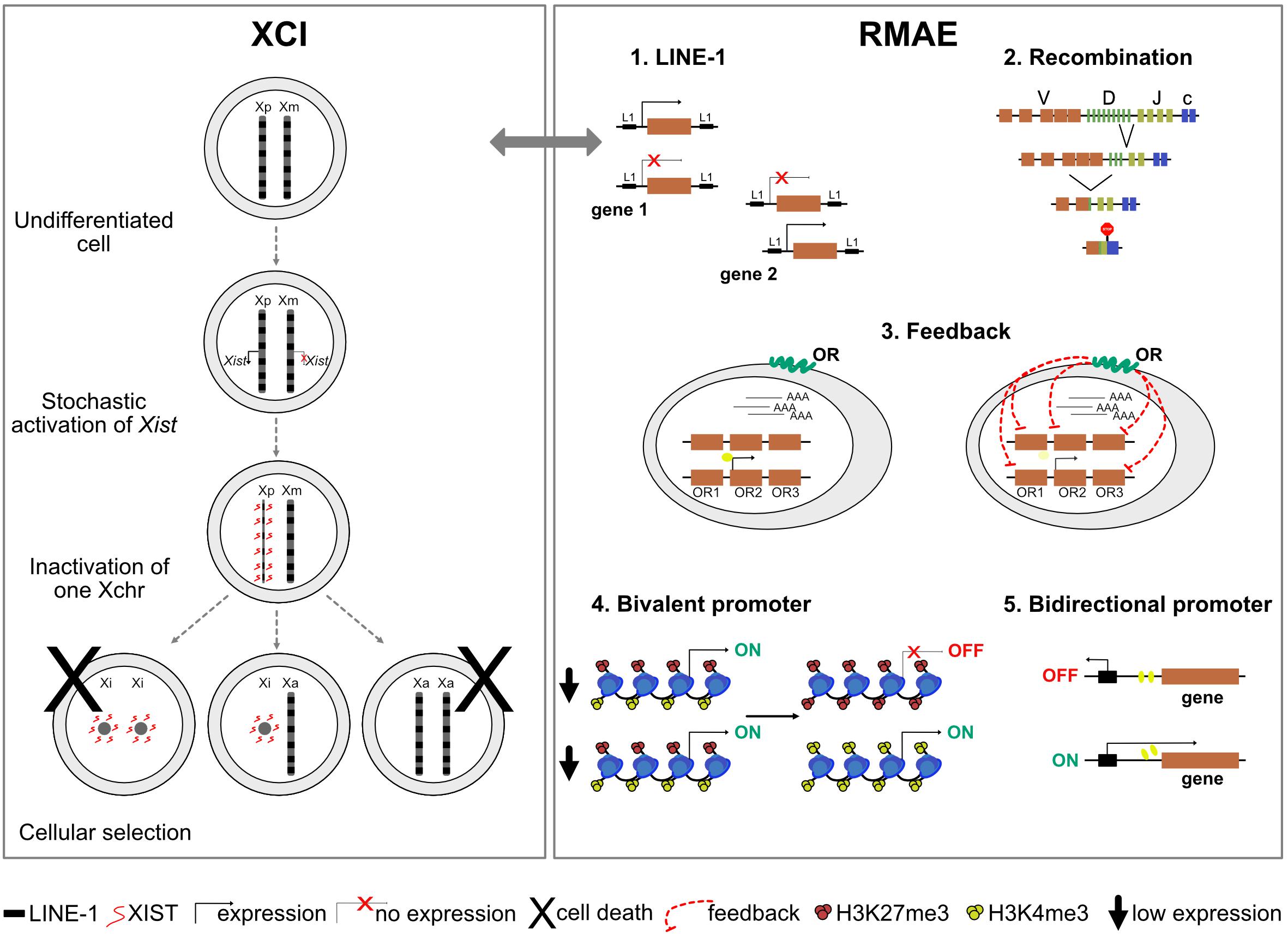
Figure 2. A schematic representation of mechanisms responsible for X-chromosome inactivation (XCI) and mechanisms possibly responsible for random monoallelic autosomal expression (RMAE). The gray arrow represents a potential parallel between XCI and RMAE associated with LINE-1 (L1) elements.
Two cases of RMAE dissected in considerable detail are the murine Ly49 receptor genes of natural killer cells and the human KIR genes [reviewed in Anderson (2014)]. Both rely on cis probabilistic bidirectional promoter switches that produce sense and antisense transcripts associated with expression and silencing, respectively (Figure 2). In the case of the murine Ly49 receptor genes, the default condition is silencing. Transcription starts if the sense non-coding transcripts of a distal bidirectional promoter (Pro1) activate a downstream promoter (Pro2); the antisense transcripts of the bidirectional promoter do not lead to gene activation (Saleh et al., 2002, 2004). In the case of the KIR genes, the default condition is activation and the role of the stochastic switch, located close to the ATG start codon, is to produce a sense transcript that correlates with the maintenance of the activation state or an antisense piRNA that silences the allele. The murine Ly49 and the human KIR genes illustrate how a probabilistic bidirectional promoter can create a mitotically stable asymmetry between two alleles. Whether these are two exceptional cases or examples of a frequent solution to generate RMAE has not been addressed. Notably, divergently transcribed gene pairs represent more than 10% of the human genes (Trinklein et al., 2004; Yang et al., 2007), and thorough analyses of tissue-specific sense and antisense transcripts from the same locus [e.g., (Hu et al., 2014)] could reveal additional candidate genes to be under RMAE due to bidirectional promoter or other complex arrangements of regulatory sequences leading to genetic switches.
Once established, XCI is believed to be extremely stable and irreversible. Genes that are subject to XCI rarely show reactivation and biallelic expression, as silencing is maintained through multiple layers of epigenetic control. However, there are some exceptions and some genes can be expressed from both the Xa and the Xi. This is the case for genes that have a Y-linked homolog, including genes from the pseudoautosomal regions (PAR1 and PAR2, short regions of homology between the X and Y chromosomes, which undergo recombination during meiosis), for which there is no requirement for dosage compensation. Several genes not located in the PAR regions have also retained a functional Y paralog and would thus appear not to require dosage compensation. However, other genes do not have a Y-linked copy yet still have the ability to escape XCI (Berletch et al., 2010). In some cases, this may be due to a highly controlled process permitting escape where the gene product is required in increased dose, while in other cases, it may be due to leaky or inefficient XCI.
Escape from X inactivation is rather limited in the mouse, with around 3% of genes displaying such behavior in somatic cells (Yang et al., 2010; Pinter et al., 2012). In humans, the situation is different, as 15% of genes (excluding the PAR) have been reported to escape XCI (Carrel and Willard, 2005). Intriguingly, an additional 10% of X-linked genes appear to show heterogeneous inactivation and escape, varying between lineages and from one individual to another (Carrel and Willard, 2005). Such candidates seem to display accessible promoter regions on the Xi (Kucera et al., 2011), suggesting that they may be poised for expression in some cell lineages and that the Xi allele becomes active under specific circumstances. In the mouse, lineage-specific escape has also been found, for example in the case of the Atrx gene, which is fully inactivated in embryonic tissues but escapes inactivation in specific subsets of extraembryonic cells (Garrick et al., 2006). The Atrx protein is actually enriched on the Xi in extraembryonic tissues (Baumann and De La Fuente, 2009; Sarma et al., 2014), suggesting that its escape from XCI might occur in a regulated manner in tissues where a higher dose of the protein is necessary (Corbel et al., 2013).
Interestingly, some of the phenotypes observed in Turner (X0) syndrome patients are believed to be due, in part, to the reduced expression levels of escapees given the lack of the second X chromosome (Berletch et al., 2010). This indicates that expression of a double dose is essential for some X-linked genes and that escape for these genes is a highly controlled process. Escape or reactivation of genes from the Xi can also occur more sporadically, but it is currently unknown whether this is caused by inefficient XCI or associated with a controlled mechanism.
Sporadic reactivation of genes from the Xi has been observed in non-pathological contexts, in specific tissues (Gendrel et al., 2014) or lineages (Wang et al., 2016), during aging (Migeon et al., 1988; Sharp et al., 2000) and also in disease contexts (Youness et al., 2021). In normal contexts, both the brain and the lymphoid lineage appear to stand as exceptions. In the brain, the Mecp2 gene, which is associated with Rett syndrome, was shown to display biallelic expression in a significant proportion of neural stem cells in the subventricular zone in the neonatal brain of inbred female mice (Gendrel et al., 2014). This could be indicative of a certain relaxation of epigenetic control of the Xi in these cells at least for this gene or a need for an increased dose of the protein, given that MeCP2 is a highly abundant protein in the brain (Skene et al., 2010). Moreover, Xist conditional deletion in adult mice leads to a global erasure of repressive histone modifications from the Xi, and more importantly, a mild loss of dosage compensation in 2-5% of neurons (Adrianse et al., 2018), highlighting again the peculiarity of the brain and neuronal lineages. Other studies have also reported partial Xi reactivation following Xist conditional deletion in adult tissues (Yildirim et al., 2013; Yang et al., 2016, 2020).
In the female lymphoid lineage, the maintenance of XCI is atypical and it has been hypothesized that this could predispose females to autoimmunity (Wang et al., 2016; Syrett et al., 2019). It was shown that both human and mouse naive B and T lymphocytes miss the typical XIST/Xist RNA domain within the nuclei. Instead, Xist shows an unusual and dispersed pattern, associated with a structure lacking some of the canonical hallmarks of heterochromatin of the Xi, such as H3K27me3, H2AK119ub1 and macroH2A. However, this state appears transient as both Xist and repressive histone marks are relocalized to the Xi upon B/T cell activation (Savarese et al., 2006; Wang et al., 2016; Syrett et al., 2019). This state was shown to be correlated with modest biallelic expression and increased expression of X-linked immunity-related genes (Wang et al., 2016; Souyris et al., 2018). The role of reactivation/increased expression of these genes and whether this is a cause of the atypical maintenance of XCI during lymphocyte differentiation remain unclear. However, enhanced expression of these genes could contribute to higher susceptibility of females to autoimmune disorders, such as systemic lupus erythematosus if not properly regulated (Youness et al., 2021).
Concerning RMAE, by definition all the examples of monoallelic expression discovered in clonal cell lines are mitotically stable. However, most reports have focused on cells that are also phenotypically stable, i.e., cells that during the period of the study do not go through major steps of differentiation. Thus, it remains to be addressed if RMAE is as stable as XCI, which is known to keep the status of the X chromosomes established early in development even after hundreds of cell divisions and extensive differentiation. Unfortunately, the antigen receptor and OR genes, which have been thoroughly investigated over decades, do not shed much light on this issue. The monoallelic expression pattern of antigen receptor genes is in part established by the process of V(D)J recombination and in developing lymphocytes, when recombination is active, the second allele is given the chance to recombine if the rearrangement of the first allele did not lead to the production of a receptor. In other words, the stability is not achieved before the expression of the receptor on the surface. Furthermore, the kappa immunoglobulin undergoes a process of receptor editing during which it can replace at its surface one protein by the protein encoded by the other allele (Casellas et al., 2001, 2007). It is only in mature lymphocytes that the pattern of monoallelic expression is stable, because the process of V(D)J recombination is permanently shut down and the silenced allele is epigenetically repressed and repositioned in the nucleus. With respect to OR genes, the patterns of monoallelic expression are stable, but it should be kept in mind that the cells are post-mitotic and terminally differentiated (Monahan and Lomvardas, 2015).
In mammalian females, under normal physiological conditions cases of two active X chromosomes are only found in undifferentiated cells and primordial germ cells before meiosis entry. All other cells have only one active X chromosome, because the double X dosage interferes with differentiation (Schulz et al., 2014). In contrast, biallelic expression of genes under RMAE is common and ranges from rare cells, such as in the case of the immunoglobulin heavy chain (Barreto and Cumano, 2000), to biallelic populations as frequent as the monoallelic ones (Gimelbrant et al., 2007). In the case of genes under RMAE with a low frequency of biallelic expression, these exceptions could correspond to the rare cases in which the two alleles become activated within the time-window allowed, before a negative feedback is triggered. Cases with a sizable population of biallelic expression could result from a relatively high individual probability of allele activation if the fitness of the cell is not compromised by the dual expression.
X-chromosome inactivation (XCI) is a well-established specific silencing mechanism that ensures dosage compensation between the sexes in marsupial and placental mammals. At the heart of this process lies the long non-coding RNA Xist, which is capable of orchestrating structural changes and recruiting chromatin and repressor complexes to ensure transcriptional gene silencing at the level of an entire chromosome, early in development. XCI has clear implications in disease, as illustrated by the Turner (X0) and Klinefelter (XXY) syndromes, as well as the severe phenotypes or lethality in males and variable phenotypes in females associated with X-linked disorders (e.g., Duchenne muscular dystrophy, hemophilia, and Rett syndrome). In contrast, RMAE evolved independently in a wide range of organisms beyond mammals, mostly to increase phenotypic diversity at the cellular level. RMAE lacks a master regulator and various mechanisms can establish it at different times during cellular differentiation. The collection of target genes encompass many cell types, showing some degree of overlap (Figure 3 and Supplementary File), but have been reported to be largely tissue-specific (Gendrel et al., 2016). Finally, the extent of the bias in monoallelic expression varies widely amongst RMAE genes. In addition, there is so far no obvious link between the RMAE of autosomal genes and disease, although a number of RMAE genes are associated with autosomal dominant diseases. X-chromosome inactivation and RMAE are essentially different phenomena that share the stochastic component and perhaps the asymmetric silencing of chromosomal regions dependent on the presence of LINE-1 elements. But it is not known whether and how exactly LINE-1 elements boost XCI and if a similar process explains a considerable fraction of genes under RMAE.
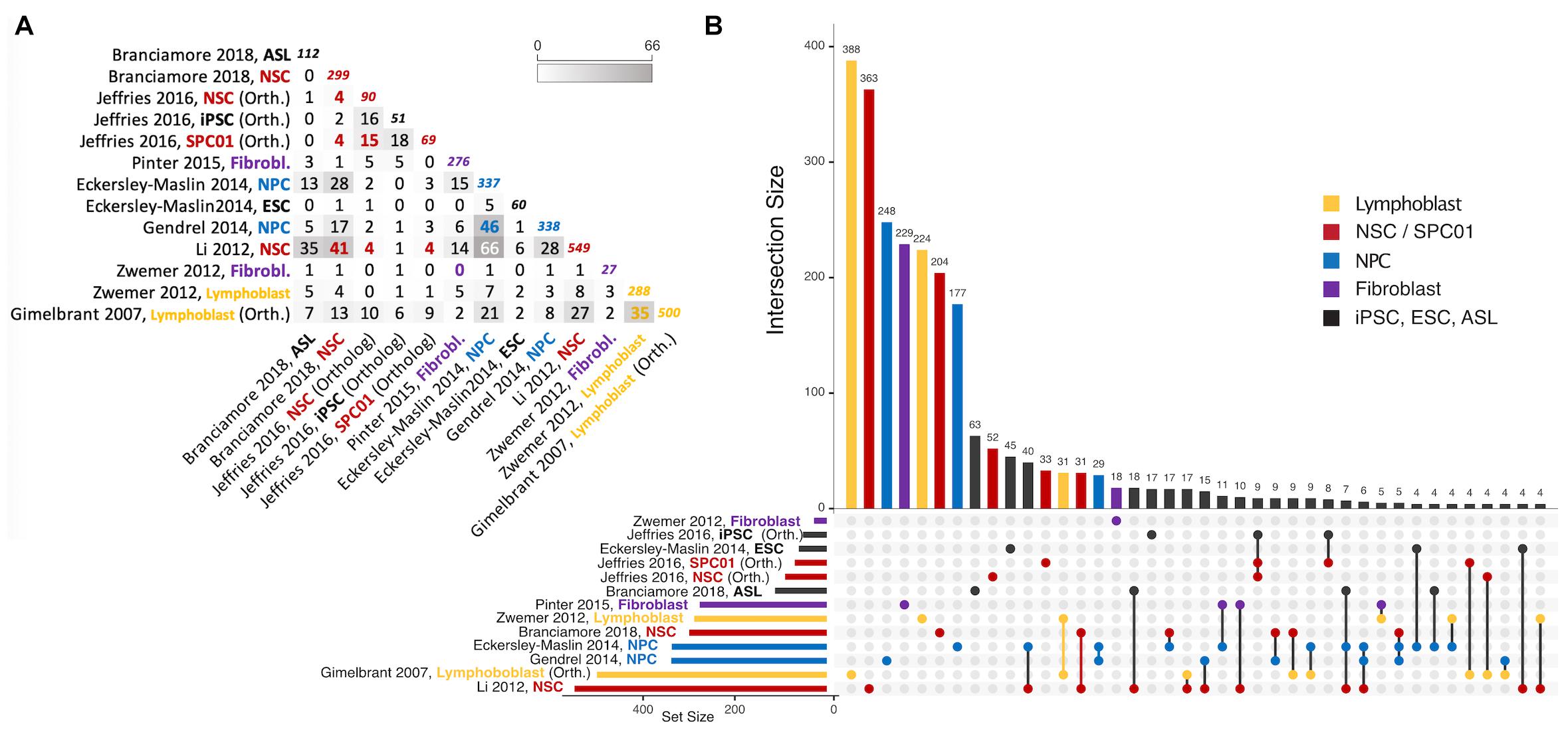
Figure 3. Intersections of autosomal gene collections identified as random monoallelically expressed in the genome-wide studies described in Table 2 (except Jeffries et al., 2012, which is not publicly available). (A) Half-matrix showing all pairwise intersections. (B) Upset plot (Conway et al., 2017) showing gene collection intersections of the same studies. The lower part of the panel has a horizontal bar plot showing the number of elements on each study collection, and a right section with a dot matrix. Each dot represents unique gene intersections, i.e., each gene is represented only once in the dot matrix. The upper vertical bar plot is related to the dot matrix, showing the number of unique genes in each intersection (for instance, there are 500 MAE genes in the Gimelbrant et al. (2007) dataset, but only 388 of those are uniquely present in that dataset; similarly, the Li et al. (2012) dataset shares more than 40 MAE genes with Eckerley-Maslin (2014) NPC dataset, but those 40 are uniquely shared between those two sets). Intersections of size smaller than 4 are not represented. For a complete description of the intersections and gene listing, see the Supplementary File provided with this review. ASL, Astrocyte-like cells; NSC, Neural stem cells; NPC, Neural progenitor cells; ESC, Embryonic stem cells; SPC01, Clonal Neural stem cells (before epigenetic reprogramming); iPSC, induced Pluripotent stem cells after epigenetic reprogramming of SPC01. Note that “NPC” on Jeffries et al. (2016) are derived from iPSC. Colors represent instances where a different cell/tissue type was studied more than once. To obtain intersections, gene ids were manually curated for immediate inconsistencies (e.g., gene name-to-date conversions when data was originally provided in microsoft excel format). All gene sets were then parsed with the gprofiler2 R package (Raudvere et al., 2019) for gene id consistency, using transcript ids as query whenever possible, and ENSEMBL gene ids as target (performed July 12th, 2021). Orthology conversion (from human to mouse) was performed with the same package for datasets involving human data. For Gimelbrant et al. (2007) and Zwemer et al. (2012) gene collections, MAE classes I, II and III were used to retrieve RMAE genes, and for Gendrel et al. (2014), the “NPC_random_catalog” classification was retrieved as RMAE.
VMB and A-VG wrote the manuscript. NK and CFA-P prepared the tables and figures. All authors contributed to the review content.
The work of NK and VMB was funded by iNOVA4Health – UIDB/Multi/04462/2020 and UIDP/Multi/04462/2020, a program financially supported by Fundação para a Ciência e Tecnologia (FCT)/Ministério da Educação e Ciência through national funds, and the FCT grant PTDC/BEX-BCM/5900/2014. CFA-P has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No. 752806. A-VG was supported by Funda o para a Ci ncia e Tecnologia (FCT), Portugal, through an assistant research contract (CEECIND/02085/2018) and the project grant PTDC/MED-OUT/4301/2020 IC&DT.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank the reviewers for providing constructive comments to this review.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.740937/full#supplementary-material
AI, allelic imbalance; IL, interleukin; iPSC, induced pluripotent stem cell; LINE-1, long interspersed nuclear element-1; OR, olfactory receptor; PAR, pseudoautosomal region; RMAE, random monoallelic autosomal expression; SNP, single nucleotide polymorphism; TCR, T-cell receptor; Xa, active X; XCI, X-chromosome inactivation; Xi, inactive X.
Adegbola, A. A., Cox, G. F., Bradshaw, E. M., Hafler, D. A., Gimelbrant, A., and Chess, A. (2015). Monoallelic expression of the human FOXP2 speech gene. Proc. Natl. Acad. Sci. U.S.A. 112, 6848–6854. doi: 10.1073/pnas.1411270111
Adrianse, R. L., Smith, K., Gatbonton-Schwager, T., Sripathy, S. P., Lao, U., Foss, E. J., et al. (2018). Perturbed maintenance of transcriptional repression on the inactive X-chromosome in the mouse brain after Xist deletion. Epigenetics Chromatin 11:50. doi: 10.1186/s13072-018-0219-8
Aifantis, I., Buer, J., von Boehmer, H., and Azogui, O. (1997). Essential role of the pre-T cell receptor in allelic exclusion of the T cell receptor beta locus. Immunity 7, 601–607. doi: 10.1016/s1074-7613(00)80381-7
Allen, E., Horvath, S., Tong, F., Kraft, P., Spiteri, E., Riggs, A. D., et al. (2003). High concentrations of long interspersed nuclear element sequence distinguish monoallelically expressed genes. Proc. Natl. Acad. Sci. U.S.A. 100, 9940–9945. doi: 10.1073/pnas.1737401100
Alves-Pereira, C. F., de Freitas, R., Lopes, T., Gardner, R., Marta, F., Vieira, P., et al. (2014). Independent recruitment of Igh alleles in V(D)J recombination. Nat. Commun. 5:5623. doi: 10.1038/ncomms6623
Anderson, S. K. (2014). Probabilistic bidirectional promoter switches: noncoding RNA takes control. Mol. Ther. Nucleic Acids 3:e191. doi: 10.1038/mtna.2014.42
Aseem, O., Barth, J. L., Klatt, S. C., Smith, B. T., and Argraves, W. S. (2013). Cubilin expression is monoallelic and epigenetically augmented via PPARs. BMC Genomics 14:405. doi: 10.1186/1471-2164-14-405
Azuara, V., Perry, P., Sauer, S., Spivakov, M., Jørgensen, H. F., John, R. M., et al. (2006). Chromatin signatures of pluripotent cell lines. Nat. Cell. Biol. 8, 532–538. doi: 10.1038/ncb1403
Barreto, V., and Cumano, A. (2000). Frequency and characterization of phenotypic Ig heavy chain allelically included IgM-expressing B cells in mice. J. Immunol. 164, 893–899. doi: 10.4049/jimmunol.164.2.893
Barton, S. C., Surani, M. A., and Norris, M. L. (1984). Role of paternal and maternal genomes in mouse development. Nature 311, 374–376. doi: 10.1038/311374a0
Baumann, C., and De La Fuente, R. (2009). ATRX marks the inactive X chromosome (Xi) in somatic cells and during imprinted X chromosome inactivation in trophoblast stem cells. Chromosoma 118, 209–222. doi: 10.1007/s00412-008-0189-x
Bayley, J.-P., Bakker, A. M., Kaijzel, E. L., Wierenga, E. A., van der Pouw-Kraan, T. C. T. M., Huizinga, T. W. J., et al. (2003). Analysis of allelic expression patterns of IL-2, IL-3, IL-4, and IL-13 in human CD4+ T cell clones. Eur. J. Immunol. 33, 2142–2148. doi: 10.1002/eji.200323976
Berletch, J. B., Yang, F., and Disteche, C. M. (2010). Escape from X inactivation in mice and humans. Genome Biol. 11, 213–217. doi: 10.1186/gb-2010-11-6-213
Bernstein, B. E., Mikkelsen, T. S., Xie, X., Kamal, M., Huebert, D. J., Cuff, J., et al. (2006). A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326. doi: 10.1016/j.cell.2006.02.041
Beutler, E., Yeh, M., and Fairbanks, V. F. (1962). The normal human female as a mosaic of X-chromosome activity: studies using the gene for C-6-PD-deficiency as a marker. Proc. Natl. Acad. Sci. U.S.A. 48, 9–16. doi: 10.1073/pnas.48.1.9
Bix, M., and Locksley, R. M. (1998). Independent and epigenetic regulation of the interleukin-4 alleles in CD4+ T cells. Science 281, 1352–1354. doi: 10.1126/science.281.5381.1352
Boeren, J., and Gribnau, J. (2021). Xist-mediated chromatin changes that establish silencing of an entire X chromosome in mammals. Curr. Opin. Cell Biol. 70, 44–50. doi: 10.1016/j.ceb.2020.11.004
Borensztein, M., Syx, L., Ancelin, K., Diabangouaya, P., Picard, C., Liu, T., et al. (2017). Xist-dependent imprinted X inactivation and the early developmental consequences of its failure. Nat. Struct. Mol. Biol. 24, 226–233. doi: 10.1038/nsmb.3365
Borst, P. (2002). Antigenic variation and allelic exclusion. Cell 109, 5–8. doi: 10.1016/s0092-8674(02)00711-0
Boyle, A. L., Ballard, S. G., and Ward, D. C. (1990). Differential distribution of long and short interspersed element sequences in the mouse genome: chromosome karyotyping by fluorescence in situ hybridization. Proc. Natl. Acad. Sci. U.S.A. 87, 7757–7761. doi: 10.1073/pnas.87.19.7757
Branciamore, S., Valo, Z., Li, M., Wang, J., Riggs, A. D., and Singer-Sam, J. (2018). Frequent monoallelic or skewed expression for developmental genes in CNS-derived cells and evidence for balancing selection. Proc. Natl. Acad. Sci. U.S.A. 115, E10379–E10386. doi: 10.1073/pnas.1808652115
Brown, C. J., and Willard, H. F. (1994). The human X-inactivation centre is not required for maintenance of X-chromosome inactivation. Nature 368, 154–156. doi: 10.1038/368154a0
Calado, D. P., Paixão, T., Holmberg, D., and Haury, M. (2006). Stochastic monoallelic expression of IL-10 in T cells. J. Immunol. 177, 5358–5364. doi: 10.4049/jimmunol.177.8.5358
Capparelli, R., Costabile, A., Viscardi, M., and Iannelli, D. (2004). Monoallelic expression of mouse Cd4 gene. Mamm. Genome 15, 579–584. doi: 10.1007/s00335-004-2351-y
Carrel, L., and Willard, H. F. (2005). X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 434, 400–404. doi: 10.1038/nature03479
Casellas, R., Shih, T. A., Kleinewietfeld, M., Rakonjac, J., Nemazee, D., Rajewsky, K., et al. (2001). Contribution of receptor editing to the antibody repertoire. Science 291, 1541–1544. doi: 10.1126/science.1056600
Casellas, R., Zhang, Q., Zheng, N.-Y., Mathias, M. D., Smith, K., and Wilson, P. C. (2007). Igkappa allelic inclusion is a consequence of receptor editing. J. Exp. Med. 204, 153–160. doi: 10.1084/jem.20061918
Cebra, J. J., and Goldstein, G. (1965). Chromatographic purification of tetramethylrhodamine-immune globulin conjugates and their use in the cellular localization of rabbit gamma-globulin polypeptide chains. J. Immunol. 95, 230–245.
Chan, H.-W., Kurago, Z. B., Stewart, C. A., Wilson, M. J., Martin, M. P., Mace, B. E., et al. (2003). DNA methylation maintains allele-specific KIR gene expression in human natural killer cells. J. Exp. Med. 197, 245–255. doi: 10.1084/jem.20021127
Chaumeil, J., Le Baccon, P., Wutz, A., and Heard, E. (2006). A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev. 20, 2223–2237. doi: 10.1101/gad.380906
Chen, G., Schell, J. P., Benitez, J. A., Petropoulos, S., Yilmaz, M., Reinius, B., et al. (2016). Single-cell analyses of X Chromosome inactivation dynamics and pluripotency during differentiation. Genome Res. 26, 1342–1354. doi: 10.1101/gr.201954.115
Chess, A. (2016). Monoallelic gene expression in mammals. Annu. Rev. Genet. 50, 317–327. doi: 10.1146/annurev-genet-120215-035120
Chess, A., Simon, I., Cedar, H., and Axel, R. (1994). Allelic inactivation regulates olfactory receptor gene expression. Cell 78, 823–834. doi: 10.1016/s0092-8674(94)90562-2
Chiodetti, L., Barber, D. L., and Schwartz, R. H. (2000). Biallelic expression of the IL-2 locus under optimal stimulation conditions. Eur. J. Immunol. 30, 2157–2163. doi: 10.1002/1521-4141(2000)30:18<2157::aid-immu2157>3.0.co;2-g
Chow, J. C., Ciaudo, C., Fazzari, M. J., Mise, N., Servant, N., Glass, J. L., et al. (2010). LINE-1 activity in facultative heterochromatin formation during X chromosome inactivation. Cell 141, 956–969. doi: 10.1016/j.cell.2010.04.042
Clemson, C. M., Hall, L. L., Byron, M., McNeil, J., and Lawrence, J. B. (2006). The X chromosome is organized into a gene-rich outer rim and an internal core containing silenced nongenic sequences. Proc. Natl. Acad. Sci. U.S.A. 103, 7688–7693. doi: 10.1073/pnas.0601069103
Conway, J. R., Lex, A., and Gehlenborg, N. (2017). UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics 33, 2938–2940. doi: 10.1093/bioinformatics/btx364
Corbel, C., Diabangouaya, P., Gendrel, A.-V., Chow, J. C., and Heard, E. (2013). Unusual chromatin status and organization of the inactive X chromosome in murine trophoblast giant cells. Development 140, 861–872. doi: 10.1242/dev.087429
Davidson, R. G., Nitowsky, H. M., and Childs, B. (1963). Demonstration of two populations of cells in the human female heterozygous for glucose-6-phosphate dehydrogenase variants. Proc. Natl. Acad. Sci. U.S.A. 50, 481–485. doi: 10.1073/pnas.50.3.481
Deng, Q., Ramsköld, D., Reinius, B., and Sandberg, R. (2014). Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science 343, 193–196. doi: 10.1126/science.1245316
Diaz-Perez, S., Ouyang, Y., Perez, V., Cisneros, R., Regelson, M., and Marahrens, Y. (2005). The element(s) at the nontranscribed Xist locus of the active X chromosome controls chromosomal replication timing in the mouse. Genetics 171, 663–672. doi: 10.1534/genetics.105.043026
Donley, N., Smith, L., and Thayer, M. J. (2015). ASAR15, A cis-acting locus that controls chromosome-wide replication timing and stability of human chromosome 15. PLoS Genet. 11:e1004923. doi: 10.1371/journal.pgen.1004923
Donley, N., Stoffregen, E. P., Smith, L., Montagna, C., and Thayer, M. J. (2013). Asynchronous replication, mono-allelic expression, and long range Cis-effects of ASAR6. PLoS Genet. 9:e1003423. doi: 10.1371/journal.pgen.1003423
Dossin, F., Pinheiro, I., Żylicz, J. J., Roensch, J., Collombet, S., Le Saux, A., et al. (2020). SPEN integrates transcriptional and epigenetic control of X-inactivation. Nature 578, 455–460. doi: 10.1038/s41586-020-1974-9
Eckersley-Maslin, M. A., Thybert, D., Bergmann, J. H., Marioni, J. C., Flicek, P., and Spector, D. L. (2014). Random monoallelic gene expression increases upon embryonic stem cell differentiation. Dev. Cell 28, 351–365. doi: 10.1016/j.devcel.2014.01.017
Efstratiadis, A. (1995). Epigenetics. A new whiff of monoallelic expression. Curr. Biol. 5, 21–24. doi: 10.1016/s0960-9822(95)00007-8
Endo, Y., Sugimura, H., and Kino, I. (1995). Monoclonality of normal human colonic crypts. Pathol. Int. 45, 602–604. doi: 10.1111/j.1440-1827.1995.tb03509.x
Engreitz, J. M., Pandya-Jones, A., McDonel, P., Shishkin, A., Sirokman, K., Surka, C., et al. (2013). The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 341:1237973. doi: 10.1126/science.1237973
Esumi, S., Kakazu, N., Taguchi, Y., Hirayama, T., Sasaki, A., Hirabayashi, T., et al. (2005). Monoallelic yet combinatorial expression of variable exons of the protocadherin-alpha gene cluster in single neurons. Nat. Genet. 37, 171–176. doi: 10.1038/ng1500
Farago, M., Rosenbluh, C., Tevlin, M., Fraenkel, S., Schlesinger, S., Masika, H., et al. (2012). Clonal allelic predetermination of immunoglobulin-κ rearrangement. Nature 490, 561–565. doi: 10.1038/nature11496
Ferguson-Smith, A. C., and Bourc’his, D. (2018). The discovery and importance of genomic imprinting. Elife 7:e42368. doi: 10.7554/eLife.42368
Galupa, R., and Heard, E. (2018). X-chromosome inactivation: a crossroads between chromosome architecture and gene regulation. Annu. Rev. Genet. 52, 535–566. doi: 10.1146/annurev-genet-120116-024611
Garrick, D., Sharpe, J. A., Arkell, R., Dobbie, L., Smith, A. J. H., Wood, W. G., et al. (2006). Loss of Atrx affects trophoblast development and the pattern of X-inactivation in extraembryonic tissues. PLoS Genet. 2:e58. doi: 10.1371/journal.pgen.0020058
Gendrel, A.-V., Attia, M., Chen, C.-J., Diabangouaya, P., Servant, N., Barillot, E., et al. (2014). Developmental dynamics and disease potential of random monoallelic gene expression. Dev. Cell 28, 366–380. doi: 10.1016/j.devcel.2014.01.016
Gendrel, A.-V., and Heard, E. (2011). Fifty years of X-inactivation research. Development 138, 5049–5055. doi: 10.1242/dev.068320
Gendrel, A.-V., Marion-Poll, L., Katoh, K., and Heard, E. (2016). Random monoallelic expression of genes on autosomes: parallels with X-chromosome inactivation. Semin. Cell Dev. Biol. 56, 100–110. doi: 10.1016/j.semcdb.2016.04.007
Gimelbrant, A., Hutchinson, J. N., Thompson, B. R., and Chess, A. (2007). Widespread monoallelic expression on human autosomes. Science 318, 1136–1140. doi: 10.1126/science.1148910
Gimelbrant, A. A., Ensminger, A. W., Qi, P., Zucker, J., and Chess, A. (2005). Monoallelic expression and asynchronous replication of p120 catenin in mouse and human cells. J. Biol. Chem. 280, 1354–1359. doi: 10.1074/jbc.M411283200
Goldmit, M., and Bergman, Y. (2004). Monoallelic gene expression: a repertoire of recurrent themes. Immunol. Rev. 200, 197–214. doi: 10.1111/j.0105-2896.2004.00158.x
Goverman, J., Minard, K., Shastri, N., Hunkapiller, T., Hansburg, D., Sercarz, E., et al. (1985). Rearranged beta T cell receptor genes in a helper T cell clone specific for lysozyme: no correlation between V beta and MHC restriction. Cell 40, 859–867. doi: 10.1016/0092-8674(85)90345-9
Graves, J. A. M. (2016). Evolution of vertebrate sex chromosomes and dosage compensation. Nat. Rev. Genet. 17, 33–46. doi: 10.1038/nrg.2015.2
Haig, D. (1993). Genetic conflicts in human pregnancy. Q. Rev. Biol. 68, 495–532. doi: 10.1086/418300
Hayashi, Y., Call, M. K., Liu, C.-Y., Hayashi, M., Babcock, G., Ohashi, Y., et al. (2010). Monoallelic expression of Krt12 gene during corneal-type epithelium differentiation of limbal stem cells. Invest. Ophthalmol. Vis. Sci. 51, 4562–4568. doi: 10.1167/iovs.10-5331
Held, W., Roland, J., and Raulet, D. H. (1995). Allelic exclusion of Ly49-family genes encoding class I MHC-specific receptors on NK cells. Nature 376, 355–358. doi: 10.1038/376355a0
Hoguin, A., Rastogi, A., Bowler, C., and Tirichine, L. (2021). Genome-wide analysis of allele-specific expression of genes in the model diatom Phaeodactylum tricornutum. Sci. Rep. 11:2954. doi: 10.1038/s41598-021-82529-1
Hollander, G. A., Zuklys, S., Morel, C., Mizoguchi, E., Mobisson, K., Simpson, S., et al. (1998). Monoallelic expression of the interleukin-2 locus. Science 279, 2118–2121. doi: 10.1126/science.279.5359.2118
Hozumi, N., and Tonegawa, S. (1976). Evidence for somatic rearrangement of immunoglobulin genes coding for variable and constant regions. Proc. Natl. Acad. Sci. U.S.A. 73, 3628–3632. doi: 10.1073/pnas.73.10.3628
Hu, H. Y., He, L., and Khaitovich, P. (2014). Deep sequencing reveals a novel class of bidirectional promoters associated with neuronal genes. BMC Genomics 15:457. doi: 10.1186/1471-2164-15-457
Jeffries, A. R., Perfect, L. W., Ledderose, J., Schalkwyk, L. C., Bray, N. J., Mill, J., et al. (2012). Stochastic choice of allelic expression in human neural stem cells. Stem Cells 30, 1938–1947. doi: 10.1002/stem.1155
Jeffries, A. R., Uwanogho, D. A., Cocks, G., Perfect, L. W., Dempster, E., Mill, J., et al. (2016). Erasure and reestablishment of random allelic expression imbalance after epigenetic reprogramming. RNA 22, 1620–1630. doi: 10.1261/rna.058347.116
Kaneko, R., Kato, H., Kawamura, Y., Esumi, S., Hirayama, T., Hirabayashi, T., et al. (2006). Allelic gene regulation of Pcdh-alpha and Pcdh-gamma clusters involving both monoallelic and biallelic expression in single Purkinje cells. J. Biol. Chem. 281, 30551–30560. doi: 10.1074/jbc.M605677200
Kelly, B. L., and Locksley, R. M. (2000). Coordinate regulation of the IL-4, IL-13, and IL-5 cytokine cluster in Th2 clones revealed by allelic expression patterns. J. Immunol. 165, 2982–2986. doi: 10.4049/jimmunol.165.6.2982
Kitamura, D., and Rajewsky, K. (1992). Targeted disruption of mu chain membrane exon causes loss of heavy-chain allelic exclusion. Nature 356, 154–156. doi: 10.1038/356154a0
Ku, C.-J., Lim, K.-C., Kalantry, S., Maillard, I., Engel, J. D., and Hosoya, T. (2015). A monoallelic-to-biallelic T-cell transcriptional switch regulates GATA3 abundance. Genes Dev. 29, 1930–1941. doi: 10.1101/gad.265025.115
Kucera, K. S., Reddy, T. E., Pauli, F., Gertz, J., Logan, J. E., Myers, R. M., et al. (2011). Allele-specific distribution of RNA polymerase II on female X chromosomes. Hum. Mol. Genet. 20, 3964–3973. doi: 10.1093/hmg/ddr315
Li, S. M., Valo, Z., Wang, J., Gao, H., Bowers, C. W., and Singer-Sam, J. (2012). Transcriptome-wide survey of mouse CNS-derived cells reveals monoallelic expression within novel gene families. PLoS One 7:e31751. doi: 10.1371/journal.pone.0031751
Loda, A., Brandsma, J. H., Vassilev, I., Servant, N., Loos, F., Amirnasr, A., et al. (2017). Genetic and epigenetic features direct differential efficiency of Xist-mediated silencing at X-chromosomal and autosomal locations. Nat. Commun. 8:690. doi: 10.1038/s41467-017-00528-1
Loda, A., and Heard, E. (2019). Xist RNA in action: past, present, and future. PLoS Genet. 15:e1008333. doi: 10.1371/journal.pgen.1008333
Lyon, M. F. (1961). Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 190, 372–373. doi: 10.1038/190372a0
Lyon, M. F. (1962). Sex chromatin and gene action in the mammalian X-chromosome. Am. J. Hum. Genet. 14, 135–148.
Lyon, M. F. (1998). X-chromosome inactivation: a repeat hypothesis. Cytogenet. Cell Genet. 80, 133–137. doi: 10.1159/000014969
Magklara, A., Yen, A., Colquitt, B. M., Clowney, E. J., Allen, W., Markenscoff-Papadimitriou, E., et al. (2011). An epigenetic signature for monoallelic olfactory receptor expression. Cell 145, 555–570. doi: 10.1016/j.cell.2011.03.040
Mak, W., Nesterova, T. B., de Napoles, M., Appanah, R., Yamanaka, S., Otte, A. P., et al. (2004). Reactivation of the paternal X chromosome in early mouse embryos. Science 303, 666–669. doi: 10.1126/science.1092674
McGrath, J., and Solter, D. (1984). Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell 37, 179–183. doi: 10.1016/0092-8674(84)90313-1
Migeon, B. R., Axelman, J., and Beggs, A. H. (1988). Effect of ageing on reactivation of the human X-linked HPRT locus. Nature 335, 93–96. doi: 10.1038/335093a0
Monahan, K., and Lomvardas, S. (2015). Monoallelic expression of olfactory receptors. Annu. Rev. Cell. Dev. Biol. 31, 721–740. doi: 10.1146/annurev-cellbio-100814-125308
Monkhorst, K., Jonkers, I., Rentmeester, E., Grosveld, F., and Gribnau, J. (2008). X inactivation counting and choice is a stochastic process: evidence for involvement of an X-linked activator. Cell 132, 410–421. doi: 10.1016/j.cell.2007.12.036
Mostoslavsky, R., Singh, N., Tenzen, T., Goldmit, M., Gabay, C., Elizur, S., et al. (2001). Asynchronous replication and allelic exclusion in the immune system. Nature 414, 221–225. doi: 10.1038/35102606
Nag, A., Savova, V., Fung, H.-L., Miron, A., Yuan, G.-C., Zhang, K., et al. (2013). Chromatin signature of widespread monoallelic expression. Elife (Cambridge) 2:e01256. doi: 10.7554/eLife.01256
Naramura, M., Hu, R. J., and Gu, H. (1998). Mice with a fluorescent marker for interleukin 2 gene activation. Immunity 9, 209–216. doi: 10.1016/s1074-7613(00)80603-2
Ng, K. K., Yui, M. A., Mehta, A., Siu, S., Irwin, B., Pease, S., et al. (2018). A stochastic epigenetic switch controls the dynamics of T-cell lineage commitment. Elife 7:e37851. doi: 10.7554/eLife.37851
Niimura, Y., Matsui, A., and Touhara, K. (2014). Extreme expansion of the olfactory receptor gene repertoire in African elephants and evolutionary dynamics of orthologous gene groups in 13 placental mammals. Genome Res. 24, 1485–1496. doi: 10.1101/gr.169532.113
Nutt, S. L., Vambrie, S., Steinlein, P., Kozmik, Z., Rolink, A., Weith, A., et al. (1999). Independent regulation of the two Pax5 alleles during B-cell development. Nat. Genet. 21, 390–395. doi: 10.1038/7720
Ohno, S., and Hauschka, T. S. (1960). Allocycly of the X-chromosome in tumors and normal tissues. Cancer Res. 20, 541–545.
Ohno, S., Kaplan, W. D., and Kinosita, R. (1959). Formation of the sex chromatin by a single X-chromosome in liver cells of Rattus norvegicus. Exp. Cell Res. 18, 415–418. doi: 10.1016/0014-4827(59)90031-x
Pancer, Z., Amemiya, C. T., Ehrhardt, G. R. A., Ceitlin, J., Gartland, G. L., and Cooper, M. D. (2004). Somatic diversification of variable lymphocyte receptors in the agnathan sea lamprey. Nature 430, 174–180. doi: 10.1038/nature02740
Pandya-Jones, A., Markaki, Y., Serizay, J., Chitiashvili, T., Mancia Leon, W. R., Damianov, A., et al. (2020). A protein assembly mediates Xist localization and gene silencing. Nature 587, 145–151. doi: 10.1038/s41586-020-2703-0
Pereira, J. P., Girard, R., Chaby, R., Cumano, A., and Vieira, P. (2003). Monoallelic expression of the murine gene encoding Toll-like receptor 4. Nat. Immunol. 4, 464–470. doi: 10.1038/ni917
Pernis, B., Chiappino, G., Kelus, A. S., and Gell, P. G. (1965). Cellular localization of immunoglobulins with different allotypic specificities in rabbit lymphoid tissues. J. Exp. Med. 122, 853–876. doi: 10.1084/jem.122.5.853
Pinter, S. F., Colognori, D., Beliveau, B. J., Sadreyev, R. I., Payer, B., Yildirim, E., et al. (2015). Allelic imbalance is a prevalent and tissue-specific feature of the mouse transcriptome. Genetics 200, 537–549. doi: 10.1534/genetics.115.176263
Pinter, S. F., Sadreyev, R. I., Yildirim, E., Jeon, Y., Ohsumi, T. K., Borowsky, M., et al. (2012). Spreading of X chromosome inactivation via a hierarchy of defined Polycomb stations. Genome Res. 22, 1864–1876. doi: 10.1101/gr.133751.111
Platt, E. J., Smith, L., and Thayer, M. J. (2018). L1 retrotransposon antisense RNA within ASAR lncRNAs controls chromosome-wide replication timing. J. Cell Biol. 217, 541–553. doi: 10.1083/jcb.201707082
Popova, B. C., Tada, T., Takagi, N., Brockdorff, N., and Nesterova, T. B. (2006). Attenuated spread of X-inactivation in an X;autosome translocation. Proc. Natl. Acad. Sci. U.S.A. 103, 7706–7711. doi: 10.1073/pnas.0602021103
Raudvere, U., Kolberg, L., Kuzmin, I., Arak, T., Adler, P., Peterson, H., et al. (2019). g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 47, W191–W198. doi: 10.1093/nar/gkz369
Rhoades, K. L., Singh, N., Simon, I., Glidden, B., Cedar, H., and Chess, A. (2000). Allele-specific expression patterns of interleukin-2 and Pax-5 revealed by a sensitive single-cell RT-PCR analysis. Curr. Biol. 10, 789–792. doi: 10.1016/s0960-9822(00)00565-0
Rivière, I., Sunshine, M. J., and Littman, D. R. (1998). Regulation of IL-4 expression by activation of individual alleles. Immunity 9, 217–228. doi: 10.1016/s1074-7613(00)80604-4
Rodriguez, I., Feinstein, P., and Mombaerts, P. (1999). Variable patterns of axonal projections of sensory neurons in the mouse vomeronasal system. Cell 97, 199–208. doi: 10.1016/s0092-8674(00)80730-8
Ross, M. T., Grafham, D. V., Coffey, A. J., Scherer, S., McLay, K., Muzny, D., et al. (2005). The DNA sequence of the human X chromosome. Nature 434, 325–337. doi: 10.1038/nature03440
Saleh, A., Davies, G. E., Pascal, V., Wright, P. W., Hodge, D. L., Cho, E. H., et al. (2004). Identification of probabilistic transcriptional switches in the Ly49 gene cluster: a eukaryotic mechanism for selective gene activation. Immunity 21, 55–66. doi: 10.1016/j.immuni.2004.06.005
Saleh, A., Makrigiannis, A. P., Hodge, D. L., and Anderson, S. K. (2002). Identification of a novel Ly49 promoter that is active in bone marrow and fetal thymus. J. Immunol. 168, 5163–5169. doi: 10.4049/jimmunol.168.10.5163
Sano, Y., Shimada, T., Nakashima, H., Nicholson, R. H., Eliason, J. F., Kocarek, T. A., et al. (2001). Random monoallelic expression of three genes clustered within 60 kb of mouse t complex genomic DNA. Genome Res. 11, 1833–1841. doi: 10.1101/gr.194301
Sarma, K., Cifuentes-Rojas, C., Ergun, A., Del Rosario, A., Jeon, Y., White, F., et al. (2014). ATRX directs binding of PRC2 to Xist RNA and Polycomb targets. Cell 159, 869–883. doi: 10.1016/j.cell.2014.10.019
Savarese, F., Flahndorfer, K., Jaenisch, R., Busslinger, M., and Wutz, A. (2006). Hematopoietic precursor cells transiently reestablish permissiveness for X inactivation. Mol. Cell. Biol. 26, 7167–7177. doi: 10.1128/MCB.00810-06
Savova, V., Chun, S., Sohail, M., McCole, R. B., Witwicki, R., Gai, L., et al. (2016). Genes with monoallelic expression contribute disproportionately to genetic diversity in humans. Nat. Genet. 48, 231–237. doi: 10.1038/ng.3493
Schlesinger, S., Selig, S., Bergman, Y., and Cedar, H. (2009). Allelic inactivation of rDNA loci. Genes Dev. 23, 2437–2447. doi: 10.1101/gad.544509
Schulz, E. G., Meisig, J., Nakamura, T., Okamoto, I., Sieber, A., Picard, C., et al. (2014). The two active X chromosomes in female ESCs block exit from the pluripotent state by modulating the ESC signaling network. Cell Stem Cell 14, 203–216. doi: 10.1016/j.stem.2013.11.022
Serizawa, S., Miyamichi, K., Nakatani, H., Suzuki, M., Saito, M., Yoshihara, Y., et al. (2003). Negative feedback regulation ensures the one receptor-one olfactory neuron rule in mouse. Science 302, 2088–2094. doi: 10.1126/science.1089122
Sharp, A., Robinson, D., and Jacobs, P. (2000). Age- and tissue-specific variation of X chromosome inactivation ratios in normal women. Hum. Genet. 107, 343–349. doi: 10.1007/s004390000382
Sharp, A. J., Spotswood, H. T., Robinson, D. O., Turner, B. M., and Jacobs, P. A. (2002). Molecular and cytogenetic analysis of the spreading of X inactivation in X;autosome translocations. Hum. Mol. Genet. 11, 3145–3156. doi: 10.1093/hmg/11.25.3145
Simon, M. D., Pinter, S. F., Fang, R., Sarma, K., Rutenberg-Schoenberg, M., Bowman, S. K., et al. (2013). High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature 504, 465–469. doi: 10.1038/nature12719
Singh, N., Ebrahimi, F. A. W., Gimelbrant, A. A., Ensminger, A. W., Tackett, M. R., Qi, P., et al. (2003). Coordination of the random asynchronous replication of autosomal loci. Nat. Genet. 33, 339–341. doi: 10.1038/ng1102
Skene, P. J., Illingworth, R. S., Webb, S., Kerr, A. R. W., James, K. D., Turner, D. J., et al. (2010). Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol. Cell 37, 457–468. doi: 10.1016/j.molcel.2010.01.030
Sonoda, E., Pewzner-Jung, Y., Schwers, S., Taki, S., Jung, S., Eilat, D., et al. (1997). B cell development under the condition of allelic inclusion. Immunity 6, 225–233. doi: 10.1016/s1074-7613(00)80325-8
Sousa, E. J., Stuart, H. T., Bates, L. E., Ghorbani, M., Nichols, J., Dietmann, S., et al. (2018). Exit from naive pluripotency induces a transient X chromosome inactivation-like state in males. Cell Stem Cell 22, 919–928.e6. doi: 10.1016/j.stem.2018.05.001
Souyris, M., Cenac, C., Azar, P., Daviaud, D., Canivet, A., Grunenwald, S., et al. (2018). TLR7 escapes X chromosome inactivation in immune cells. Sci. Immunol. 3:eaa8855. doi: 10.1126/sciimmunol.aap8855
Stoffregen, E. P., Donley, N., Stauffer, D., Smith, L., and Thayer, M. J. (2011). An autosomal locus that controls chromosome-wide replication timing and mono-allelic expression. Hum. Mol. Genet. 20, 2366–2378. doi: 10.1093/hmg/ddr138
Strehle, M., and Guttman, M. (2020). Xist drives spatial compartmentalization of DNA and protein to orchestrate initiation and maintenance of X inactivation. Curr. Opin. Cell Biol. 64, 139–147. doi: 10.1016/j.ceb.2020.04.009
Surani, M. A., Barton, S. C., and Norris, M. L. (1984). Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature 308, 548–550. doi: 10.1038/308548a0
Syrett, C. M., Paneru, B., Sandoval-Heglund, D., Wang, J., Banerjee, S., Sindhava, V., et al. (2019). Altered X-chromosome inactivation in T cells may promote sex-biased autoimmune diseases. JCI Insight 4:1290. doi: 10.1172/jci.insight.126751
Takagi, N., and Abe, K. (1990). Detrimental effects of two active X chromosomes on early mouse development. Development 109, 189–201. doi: 10.1242/dev.109.1.189
Takizawa, T., Gudla, P. R., Guo, L., Lockett, S., and Misteli, T. (2008). Allele-specific nuclear positioning of the monoallelically expressed astrocyte marker GFAP. Genes Dev. 22, 489–498. doi: 10.1101/gad.1634608
Tang, W. W. C., Dietmann, S., Irie, N., Leitch, H. G., Floros, V. I., Bradshaw, C. R., et al. (2015). A unique gene regulatory network resets the human germline epigenome for development. Cell 161, 1453–1467. doi: 10.1016/j.cell.2015.04.053
Tang, Y. A., Huntley, D., Montana, G., Cerase, A., Nesterova, T. B., and Brockdorff, N. (2010). Efficiency of Xist-mediated silencing on autosomes is linked to chromosomal domain organisation. Epigenetics Chromatin 3, 10–12. doi: 10.1186/1756-8935-3-10
Tasic, B., Nabholz, C. E., Baldwin, K. K., Kim, Y., Rueckert, E. H., Ribich, S. A., et al. (2002). Promoter choice determines splice site selection in protocadherin alpha and gamma pre-mRNA splicing. Mol. Cell 10, 21–33. doi: 10.1016/s1097-2765(02)00578-6
Taylor, J. H. (1960). Asynchronous duplication of chromosomes in cultured cells of Chinese hamster. J. Biophys. Biochem. Cytol. 7, 455–464. doi: 10.1083/jcb.7.3.455
Thomas, B. J., Rubio, E. D., Krumm, N., Broin, P. O., Bomsztyk, K., Welcsh, P., et al. (2011). Allele-specific transcriptional elongation regulates monoallelic expression of the IGF2BP1 gene. Epigenetics Chromatin 4:14. doi: 10.1186/1756-8935-4-14
Trinklein, N. D., Aldred, S. F., Hartman, S. J., Schroeder, D. I., Otillar, R. P., and Myers, R. M. (2004). An abundance of bidirectional promoters in the human genome. Genome Res. 14, 62–66. doi: 10.1101/gr.1982804
Vettermann, C., and Schlissel, M. S. (2010). Allelic exclusion of immunoglobulin genes: models and mechanisms. Immunol. Rev. 237, 22–42. doi: 10.1111/j.1600-065X.2010.00935.x
Wang, J., Syrett, C. M., Kramer, M. C., Basu, A., Atchison, M. L., and Anguera, M. C. (2016). Unusual maintenance of X chromosome inactivation predisposes female lymphocytes for increased expression from the inactive X. Proc. Natl. Acad. Sci. U.S.A. 113, E2029–E2038. doi: 10.1073/pnas.1520113113
Wang, X., Su, H., and Bradley, A. (2002). Molecular mechanisms governing Pcdh-gamma gene expression: evidence for a multiple promoter and cis-alternative splicing model. Genes Dev. 16, 1890–1905. doi: 10.1101/gad.1004802
Welshons, W. J., and Russell, L. B. (1959). The Y-chromosome as the bearer of male determining factors in the mouse. Proc. Natl. Acad. Sci. U.S.A. 45, 560–566. doi: 10.1073/pnas.45.4.560
Wu, H., Luo, J., Yu, H., Rattner, A., Mo, A., Wang, Y., et al. (2014). Cellular resolution maps of X chromosome inactivation: implications for neural development, function, and disease. Neuron 81, 103–119. doi: 10.1016/j.neuron.2013.10.051
Yang, F., Babak, T., Shendure, J., and Disteche, C. M. (2010). Global survey of escape from X inactivation by RNA-sequencing in mouse. Genome Res. 20, 614–622. doi: 10.1101/gr.103200.109
Yang, L., Kirby, J. E., Sunwoo, H., and Lee, J. T. (2016). Female mice lacking Xist RNA show partial dosage compensation and survive to term. Genes Dev. 30, 1747–1760. doi: 10.1101/gad.281162.116
Yang, L., Yildirim, E., Kirby, J. E., Press, W., and Lee, J. T. (2020). Widespread organ tolerance to Xist loss and X reactivation except under chronic stress in the gut. Proc. Natl. Acad. Sci. U.S.A. 117, 4262–4272. doi: 10.1073/pnas.1917203117
Yang, M. Q., Koehly, L. M., and Elnitski, L. L. (2007). Comprehensive annotation of bidirectional promoters identifies co-regulation among breast and ovarian cancer genes. PLoS Comput. Biol. 3:e72. doi: 10.1371/journal.pcbi.0030072
Yildirim, E., Kirby, J. E., Brown, D. E., Mercier, F. E., Sadreyev, R. I., Scadden, D. T., et al. (2013). Xist RNA is a potent suppressor of hematologic cancer in mice. Cell 152, 727–742. doi: 10.1016/j.cell.2013.01.034
Youness, A., Miquel, C.-H., and Guéry, J.-C. (2021). Escape from X chromosome inactivation and the female predominance in autoimmune diseases. Int. J. Mol. Sci. 22:1114. doi: 10.3390/ijms22031114
Zhang, X., and Firestein, S. (2002). The olfactory receptor gene superfamily of the mouse. Nat. Neurosci. 5, 124–133. doi: 10.1038/nn800
Zhou, Q., Wang, T., Leng, L., Zheng, W., Huang, J., Fang, F., et al. (2019). Single-cell RNA-seq reveals distinct dynamic behavior of sex chromosomes during early human embryogenesis. Mol. Reprod. Dev. 86, 871–882. doi: 10.1002/mrd.23162
Zwemer, L. M., Zak, A., Thompson, B. R., Kirby, A., Daly, M. J., Chess, A., et al. (2012). Autosomal monoallelic expression in the mouse. Genome Biol. 13:R10. doi: 10.1186/gb-2012-13-2-r10
Keywords: X-chromosome inactivation, random monoallelic expression, epigenetic silencing, LINE-1 elements, cellular diversity, stochasticity, dosage compensation
Citation: Barreto VM, Kubasova N, Alves-Pereira CF and Gendrel A-V (2021) X-Chromosome Inactivation and Autosomal Random Monoallelic Expression as “Faux Amis”. Front. Cell Dev. Biol. 9:740937. doi: 10.3389/fcell.2021.740937
Received: 13 July 2021; Accepted: 30 August 2021;
Published: 23 September 2021.
Edited by:
Jin Xu, Sun Yat-sen University, ChinaReviewed by:
Bernhard Payer, Centre for Genomic Regulation (CRG), SpainCopyright © 2021 Barreto, Kubasova, Alves-Pereira and Gendrel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vasco M. Barreto, dmFzY28uYmFycmV0b0BubXMudW5sLnB0; Anne-Valerie Gendrel, YW5uZS5nZW5kcmVsQG1lZGljaW5hLnVsaXNib2EucHQ=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.