
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol., 05 October 2021
Sec. Morphogenesis and Patterning
Volume 9 - 2021 | https://doi.org/10.3389/fcell.2021.736813
This article is part of the Research TopicRegeneration from Cells to Limbs: Past, Present, and FutureView all 28 articles
 Katia Suarez-Berumen1,2†
Katia Suarez-Berumen1,2† Henry Collins-Hooper3†
Henry Collins-Hooper3† Anastasia Gromova1,4
Anastasia Gromova1,4 Robyn Meech5
Robyn Meech5 Alessandra Sacco4
Alessandra Sacco4 Phil R. Dash3
Phil R. Dash3 Robert Mitchell3
Robert Mitchell3 Valery I. Shestopalov6,7
Valery I. Shestopalov6,7 Thomas E. Woolley8
Thomas E. Woolley8 Sakthivel Vaiyapuri9
Sakthivel Vaiyapuri9 Ketan Patel3*‡
Ketan Patel3*‡ Helen P. Makarenkova1*‡
Helen P. Makarenkova1*‡Adult skeletal muscle has robust regenerative capabilities due to the presence of a resident stem cell population called satellite cells. Muscle injury leads to these normally quiescent cells becoming molecularly and metabolically activated and embarking on a program of proliferation, migration, differentiation, and fusion culminating in the repair of damaged tissue. These processes are highly coordinated by paracrine signaling events that drive cytoskeletal rearrangement and cell-cell communication. Pannexins are a family of transmembrane channel proteins that mediate paracrine signaling by ATP release. It is known that Pannexin1 (Panx1) is expressed in skeletal muscle, however, the role of Panx1 during skeletal muscle development and regeneration remains poorly understood. Here we show that Panx1 is expressed on the surface of myoblasts and its expression is rapidly increased upon induction of differentiation and that Panx1–/– mice exhibit impaired muscle regeneration after injury. Panx1–/– myoblasts activate the myogenic differentiation program normally, but display marked deficits in migration and fusion. Mechanistically, we show that Panx1 activates P2 class purinergic receptors, which in turn mediate a lipid signaling cascade in myoblasts. This signaling induces bleb-driven amoeboid movement that in turn supports myoblast migration and fusion. Finally, we show that Panx1 is involved in the regulation of cell-matrix interaction through the induction of ADAMTS (Disintegrin-like and Metalloprotease domain with Thrombospondin-type 5) proteins that help remodel the extracellular matrix. These studies reveal a novel role for lipid-based signaling pathways activated by Panx1 in the coordination of myoblast activities essential for skeletal muscle regeneration.
Adult mammalian skeletal muscle is composed of multinucleated fibers that cannot divide. Regeneration after an injury is mediated by a resident stem cell population called satellite cells (Lepper et al., 2011; Sambasivan et al., 2011). In undamaged adult muscles, satellite cells are quiescent; however, upon injury, they become activated and divide both asymmetrically and symmetrically. This allows self-renewal of the satellite cell pool, as well as the generation of a transit-amplifying population called myoblasts that become progressively differentiation-competent and eventually withdraw from the cell cycle and differentiate. Muscle regeneration is dependent on the ability of satellite cells to migrate to the point of a lesion, which relies on dynamic cytoskeletal rearrangements and cell-cell communication and is controlled in part by nitric oxide and purinergic signaling pathways (Irintchev et al., 1994; Irizarry et al., 2003; Hutcheson and Kardon, 2009; Makarenkova et al., 2009; Collins-Hooper et al., 2012; Webster et al., 2016). Myoblast migration is characterized by the formation of surface protrusions, called “blebs,” especially on its native substrate, the muscle fiber. Blebs have been implicated in the amoeboid movement of cells that lack mature focal adhesions and stress fibers (Friedl et al., 2001; Friedl and Wolf, 2009; Otto et al., 2011; Khajah et al., 2015). We have previously shown that blebbing is essential to support the rapid migration of myoblasts (Otto et al., 2011). Adhesion of myoblasts to each other and to the extracellular matrix (ECM) and their synthesis of ECM components are also critical for myoblast differentiation (Hantai et al., 1985; Melo et al., 1996; Schwander et al., 2003; Lukjanenko et al., 2016). Myoblasts express a pericellular matrix rich in versican (Ermakova et al., 2011). During differentiation, the pericellular matrix is remodeled by enzymatic cleavage and new matrix synthesis. In particular, the reduction of hyaluronidase-sensitive pericellular coats by the ADAMTS family of versicanases is necessary for myoblast fusion (Hattori et al., 2011; Stupka et al., 2013; Taye et al., 2020).
Muscle fibers and myoblasts express the pannexin family of gap junction-like membrane channel proteins (Panchin et al., 2000; Bruzzone et al., 2003; Baranova et al., 2004; Langlois et al., 2014; Jorquera et al., 2021). The pannexin family consists of three members (Panx1–3; Baranova et al., 2004; Panchin, 2005; D’hondt et al., 2010). Pannexin hemichannels are activated through interaction with purinergic receptors (Iglesias et al., 2008) and by various stimuli such as high intracellular calcium (Locovei et al., 2006), high extracellular potassium (Bunse et al., 2009), membrane stretch via the activation of Piezo-1 channel and submembrane increase in Ca2+ signal (Lopez et al., 2021), and membrane depolarization (Bruzzone et al., 2003; Pelegrin and Surprenant, 2006). Panx1 is ubiquitously expressed in mammalian tissues and is elevated in the central nervous system in the CNS and skeletal muscle (Cea et al., 2012, 2014). In humans and mice, high levels of Panx1 gene transcripts have been found in the nervous system, heart, gonads, kidney, and skeletal muscles (Baranova et al., 2004; Jorquera et al., 2013; Riquelme et al., 2013; Cea et al., 2014; Langlois et al., 2014; Pham et al., 2018). Panx1 forms membrane hemichannels that, upon activation, mediate small molecule communication with the extracellular environment. As a proven conduit for paracrine signaling, Panx1 has been associated with calcium wave propagation, and nucleotide release, including pre-apoptotic “Find Me” signaling, which is transduced via activation of P2 purinergic receptors (Iglesias et al., 2008; Chekeni et al., 2010; Makarenkova et al., 2018). In this context, direct interaction between the P2X7 receptors and Panx1 has been shown to be necessary for ATP release from the cell (Locovei et al., 2006; Schenk et al., 2008; Qiu and Dahl, 2009). A growing number of studies demonstrate that Panx1-mediated ATP release is involved in essential physiological functions such as vasoregulation (Lohman et al., 2012; Gaynullina et al., 2015), and long-range Ca2+ wave propagation in endothelial cells (Iglesias et al., 2008) and astrocytes (Iglesias et al., 2009). Panx1-mediated ATP release is essential for paracrine or autocrine activation of purinergic receptors in many tissues, including brain and skeletal muscles (Bruzzone et al., 2003; Baranova et al., 2004; Prochnow et al., 2012). In skeletal muscle, Panx1 has been implicated in potentiation of myofiber contraction by altering ATP and calcium flux (Riquelme et al., 2013; Cea et al., 2014); however, the role of pannexins in muscle regeneration is not well defined. A recent report suggested that Panx1 is important for primary myoblast differentiation in vitro (Langlois et al., 2014); however, the role of Panx1 in muscle regeneration in vivo has not been studied, and the mechanism(s) by which it controls myoblast differentiation are unknown.
In this study, we show that Panx1 is expressed both by undifferentiated satellite cells/myoblasts and differentiated skeletal muscle fibers. Genetic ablation of Panx1 or pharmacological perturbation of Panx1 or its downstream signaling partners disrupted myoblast differentiation leading to the formation of smaller myofibers. Loss of Panx1 inhibited cell migration by attenuating cell blebbing, and also inhibited myoblast fusion. These findings are consistent with our previous findings that bleb formation is required for myoblast migration and fusion (Makarenkova et al., 2009). Mechanistic studies show that Panx1 activates the P2 (P2X7) purinergic receptor which initiates a lipid signaling cascade that culminates in the activation of myosin-based contraction of the cortical cytoskeleton which supports migration and fusion through the formation of plasma membrane blebs. In addition, Panx1 is required for the expression of several adhesion molecules and a Disintegrin-like and Metalloprotease domain with Thrombospondin-type 5 motifs (ADAMTS5), which have been implicated in ECM remodeling essential for the myoblast fusion process (Stupka et al., 2013).
This study reveals a novel pathway for regulation of myoblast migration and fusion involving Panx1 activated purinergic signaling, lipid metabolism and ECM remodeling, providing important new insights into the molecular mechanisms that underpin muscle regeneration.
In this study we used two Panx1 null mouse strains: the Panx1KO/B6 (Genetech Inc.) and the CMV-Cre/Panx1 strain. The Panx1KO/B6 (Qu et al., 2011) was backcrossed to the C57BL/6 background (see Supplementary Methods) for 11 generations. The Panx1 null strain CMV-Cre/Panx1fl/fl carries a floxed Panx1 allele that was activated with CMV-Cre to create a global knockout. This strain also carries a “passenger” Casp11 mutation (Dvoriantchikova et al., 2012). Caspase11 is induced in macrophages in response to injury or exposure to bacterial metabolites but is not detectable in homeostatic conditions in tissues such as muscle (Kang et al., 2000; Py et al., 2014). The Casp11–/– [B6.129S4(D2)-Casp4tm1Yuan/J] mouse strain in the C57BL/6 background (Jackson Labs) was used as a control for any effects that might be mediated by loss of Casp11 in the CMV-Cre/Panx1 strain. Wildtype C57BL/6 mice were used as an additional control.
Mice were housed under standard conditions of temperature and humidity, with a 12-h light/dark cycle and free access to food and water. All experiments were performed in compliance with the Guidelines for the Care and Use of Laboratory Animals published by the United States National Institutes of Health (NIH Publication No. 85-23, revised 1996) and were pre-approved by the Scripps Research Institute Animal Care and Use Committee.
Primary myoblasts were isolated from limb muscles of 3-week-old mice. Satellite cells were isolated using gentleMACSTM Octo Dissociator and either the MACS Microbid satellite cell isolation kit (Miltenyl Biotech, CA, United States) or Fluorescence Activated cell sorting (FACS) as described previously (Sacco et al., 2010; Meech et al., 2011). Cells were grown on collagen-coated plates or chamber slides in growth medium (GM: 1:1 Ham’s F10/DMEM, supplemented with 20% FBS, and 2.5 ng/ml of basic FGF; Rando and Blau, 1994; Makarenkova et al., 2009). To induce myoblast differentiation, the GM was replaced with a differentiation medium (DM: DMEM supplemented with 2% horse serum).
Tibialis anterior (TA) muscles of anesthetized 3-month-old Panx1-null and wild-type mice were injected with cardiotoxin (CTX, 100 ng) to induce muscle injury, or with saline (control) as described previously (Sacco et al., 2010; Meech et al., 2011). Muscles were dissected at 2–10 days post-injury, fixed with buffered 4% paraformaldehyde (PFA). Paraffin sections prepared from the muscle midsection were stained with Hematoxylin and Eosin (H&E), and Masson’s Trichrome to detect collagen deposition, and imaged using a Leica scanner.
For gene expression analysis, RNA was extracted using Trizol or Zymo Direct-Zol RNA Kit (Zymo Research Corporation, Irvine, CA, United States) either from dissected TA tissues or from myoblast cultures. The amount of total RNA was estimated using a Nanodrop ND-1000 spectrophotometer and RNA purity and integrity (RIN) was assessed using a Bioanalyzer-2100 device (Agilent Technologies, Inc., Santa Clara, CA, United States). RNAs with RIN between 9 and 10 were used in all experiments. RNA was reverse transcribed to cDNA using the RT2 First Strand Kit (#330404, Qiagen, Valencia, CA, United States). The Mouse ECM & Adhesion Molecules RT2 Profiler PCR Array (PAMM-013Z; Qiagen) and RT2 SYBR Green ROX qPCR Mastermix (#330522; Qiagen, Carlsbad, CA, United States) were used to measure expression levels of 84 individual genes involved in cell–cell and cell–matrix interactions. All other gene-specific primers were obtained from Qiagen. Quantitative real-time PCR was performed using an ABI 7300 Real-Time PCR System (Applied Biosystems, Life Technologies, Carlsbad, CA, United States). Statistically significant differences in threshold cycle (Ct) for each reaction were determined using the RT2 Profiler PCR Array Data Analysis software v3.5 (Qiagen). Gene expression differences were filtered for >1.5-fold change and significance P < 0.05. Genes with highly variable Ct values were excluded from the final analysis. Reference genes for normalization of real-time PCR data were b-actin (ACTB), β-2 microglobulin (B2M), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
Myoblast cultures were fixed with 2% PFA in PBS pH 7.5 for 20 min, permeabilized in Tris–buffered Saline with 0.1% Tween 20 (TBST), and blocked with 5% goat serum. Cells were immunostained with primary antibodies overnight at 4°C, followed by staining with appropriate secondary antibodies (antibodies are detailed in Supplementary Methods) and nuclear staining with DAPI. Images were taken using Zeiss LSM 710 laser scanning confocal microscope (LSCM).
Primary antibodies used: affinity purified rabbit Panx1 antibody CT-395 (Px-34; Penuela et al., 2007) was kindly provided by Dr. D. W. Laird (University of Western Ontario, Canada), rabbit polyclonal anti-human Panx1 antibodies (Chemicon, Inc.); mouse monoclonal anti-Myogenin (clone F5D, BD Bioscience Pharmingen, San Diego, CA, United States), mouse monoclonal anti-MyoD (clone MoAb5.8A, BD PharMingen, San Diego, CA, United States), pan myosin heavy chain (MyHC; A4.1025 Developmental Studies Hybridoma Bank, Iowa City, IA, United States), and mouse monoclonal anti-sarcomere myosin (clone MF20, Developmental Studies Hybridoma Bank, Iowa City, IA, United States), antibodies.
Total protein was prepared from primary myoblast cultures using Nonidet P-40 lysis buffer and sonicated. Equal aliquots of protein were resolved by SDS-PAGE using the 4% to 12% Bis-Tris gels (NuPAGE; Invitrogen), transferred to polyvinylidene difluoride membrane, and probed with polyclonal antibodies to Panx1 (Chemicon) and to β-actin, β-tubulin, or GAPDH. Bands were visualized by chemiluminescence using X-ray film exposure and quantified by densitometry. Each experiment was performed in duplicate.
Single muscle fibers were isolated from the extensor digitiorum longus (EDL) muscle of 12-week old male C57Bl6 as described previously (Otto et al., 2008, 2011). Briefly, the EDL was dissected with both tendons intact and myofibers liberated by digestion with (0.2%) type I collagenase (Sigma) in Dulbecco modified eagle medium (DMEM) at 37°C under 5% CO2. Using tapered glass pipettes, single fibers were plated in single fiber culture medium (SFCM, DMEM supplemented with 10% horse serum, 0.5% chick embryo extract, and 1% penicillin/streptomycin).
Following 24 h of culture, myofibers were monitored using a phase contrast time-lapse microscope system. Myofibers grown in chamber sides were imaged in an environmental chamber maintained at 37°C and supplemented with 5% CO2. Time-lapse video was taken at a rate of 1 frame every 15 min over 24 h for satellite cells using a 10X objective, a previously well-studied time course for satellite cell migration following single fiber isolation (Otto et al., 2008, 2011). All video analysis was carried out using freeware package ImageJ (version 1.49 m). Satellite cells were individually manually tracked using the MTrackJ plugin on ImageJ.
For electron microscopy, single myofibers were fixed following 48 h in standard culture with 4% PFA for 15 min, and then processed.
Fixed myofibers were dehydrated through 30, 50, 70, 80, 90, and 100% ethanol solution series (15 min for each step) and transferred to a critical point drier (Balzers CPD 030, using liquid carbon dioxide) thereafter. Dried myofibers were then carefully transferred to scanning electron microscopy (SEM) chucks using micro-forceps under a light microscope. Myofibers were then gold-coated using an Edwards S150B sputter-coater. Coated myofibers were then visualized under an FEI 600F SEM using the accompanying analysis software for image collection.
Mathematical models of particle motion are able to provide us with characterizations, which can be compared with data in order to predict how cells are moving (Woolley et al., 2013, 2014). Although we have shown that the shape of the cell and its blebs are critical to its migration properties (Woolley et al., 2015a,b), here, we simply characterize the motion characteristics in terms of the migration data. Specifically, we model the cells as random walkers on a straight, cylindrical fiber. Previously (Collins-Hooper et al., 2012), we have shown that, under this assumption, we can separate the cellular motion into two components; (i) a purely angular motion of the cell, around the muscle fiber and (ii) a purely longitudinal motion, along the axis of the fiber. We primarily focus of this longitudinal motion as it is the most crucial mode of movement.
Due to the probabilistic nature of the problem, we cannot specify exactly where the cell will be at all times in the future, however, we can predict the probability distribution of the cell locations. We define this probability distribution to be p(x,t), which represents the probability of finding a cell at a position x (measured along the fiber) at a given time t. The distribution satisfies the standard diffusion equation in one dimension (Belmonte-Beitia et al., 2013),
This equation simply encapsulates the idea that the cells are moving randomly, all of the cell’s positions are normalized to start at zero and the fiber has no boundaries. The D parameter is a positive constant that measures that rate at which the cells spread out from the normalized origin. From this equation we derive that the mean squared displacement of the cells is proportional to time,
Thus, taking the trajectory data we normalize the start position of all trajectories to zero and rotate all trajectories such that the fiber lies along the x-axis. The mean square distance moved by the cell along the fiber is calculated, and a straight line is fitted to this derived statistic. A goodness of fit R2 value is also computed to express how well the straight line fits the data. The closer the R2 value is to one, the closer the movement of the cells is to a memoryless, random diffusion.
Primary myoblasts were treated with carbenoxolone (CBX, Sigma-Aldrich # C4790) or vehicle, for 24 h prior and then induced to differentiate in low serum medium also containing CBX or vehicle (control). CBX blocks pannexin and connexin channels at 100 μM, but has high selectivity for pannexin channels at 10–50 μM (Bruzzone et al., 2005). Varying CBX concentrations from 10 to 100 μM were tested and 25 μM was found to be optimal for pannexin inhibition. Myoblast cultures treated with 25 μM CBX in DM were fixed at different time points 6, 12, 24, and 72 h and processed for immunostaining.
A740003 is a potent, selective and competitive P2X7 receptor antagonist that displays selectivity to P2X7 receptor up to a concentration of 100 μM. A740003 (Tocris, # 3701) at 50–100 nM or vehicle (dimethylsulfoxide) was applied to myoblasts 24 h prior to induction of differentiation. GM was replaced with DM containing A740003 or vehicle and cells were fixed at different time points for immunostaining.
To disrupt ATP signaling, cultures were treated with the ATP hydrolyzing enzyme apyrase (2.5 U/ml, Sigma, added to GM) or vehicle 24 h prior to the induction of differentiation. GM was then replaced with DM also containing apyrase or vehicle (control).
To study the role of lipid signaling, single muscle fibers were cultured with the following drugs 24 h post isolation: Butanol (1% v/v Fisher # A383-1), that inhibits the action of PLD in the formation of phosphatidic acid; VPC 32183 (1 μM Avanti Polar Lipids # 857340), a selective inhibitor of the LPA1 and LPA3 receptors; ML-7 (50 μM Sigma # I2764), a selective myosin light chain kinase inhibitor; Y-27632 (10 μM Sigma # Y0503), a potent inhibitor of Rho-associated kinase; and LPA (10 μM Sigma # L7260).
To determine the fusion index, cultures were stained with the MF20 antibody and the numbers of nuclei were counted in MyHC+ cells. Fusion index was calculated as the number of nuclei in multinucleated MyHC+ cells (≥2 nuclei) as a proportion of total nuclei. Three independent experiments were performed for each condition; approximately 500 nuclei were counted per condition.
The erythrocyte exclusion assay was performed as previously described (Hattori et al., 2011; Stupka et al., 2013). Briefly, Panx1 and WT primary myoblasts were plated into two-well chamber slides and cultured for 24–48 h. The culture medium was replaced by freshly prepared erythrocyte suspension (1 × 107 erythrocytes/ml) in serum-free DMEM. After erythrocytes had settled to the bottom, images (20–30 per experimental group) were taken with an inverted microscope. In some assays, Streptomyces hyalurolyticus hyaluronidase (H1136, Sigma) was added prior to erythrocyte addition.
Statistical significance was assessed using unpaired two-tailed Student’s t-test and results were considered significant at P < 0.05, or P < 0.01. Results were expressed as mean ± s.d.m (standard deviation of the mean).
Panx-1 was previously shown to be expressed in skeletal muscle (Langlois et al., 2014); however, its role during muscle differentiation and regeneration remains largely unknown. We examined the temporal pattern of PANX1 protein expression during myoblast differentiation using immunocytochemical, quantitative Western blot and gene expression analyses of primary cultures. Panx1 was expressed in isolated satellite cells (Figure 1A) and was increased rapidly at both mRNA and protein levels after the induction of myoblast differentiation (Figures 1A–C).
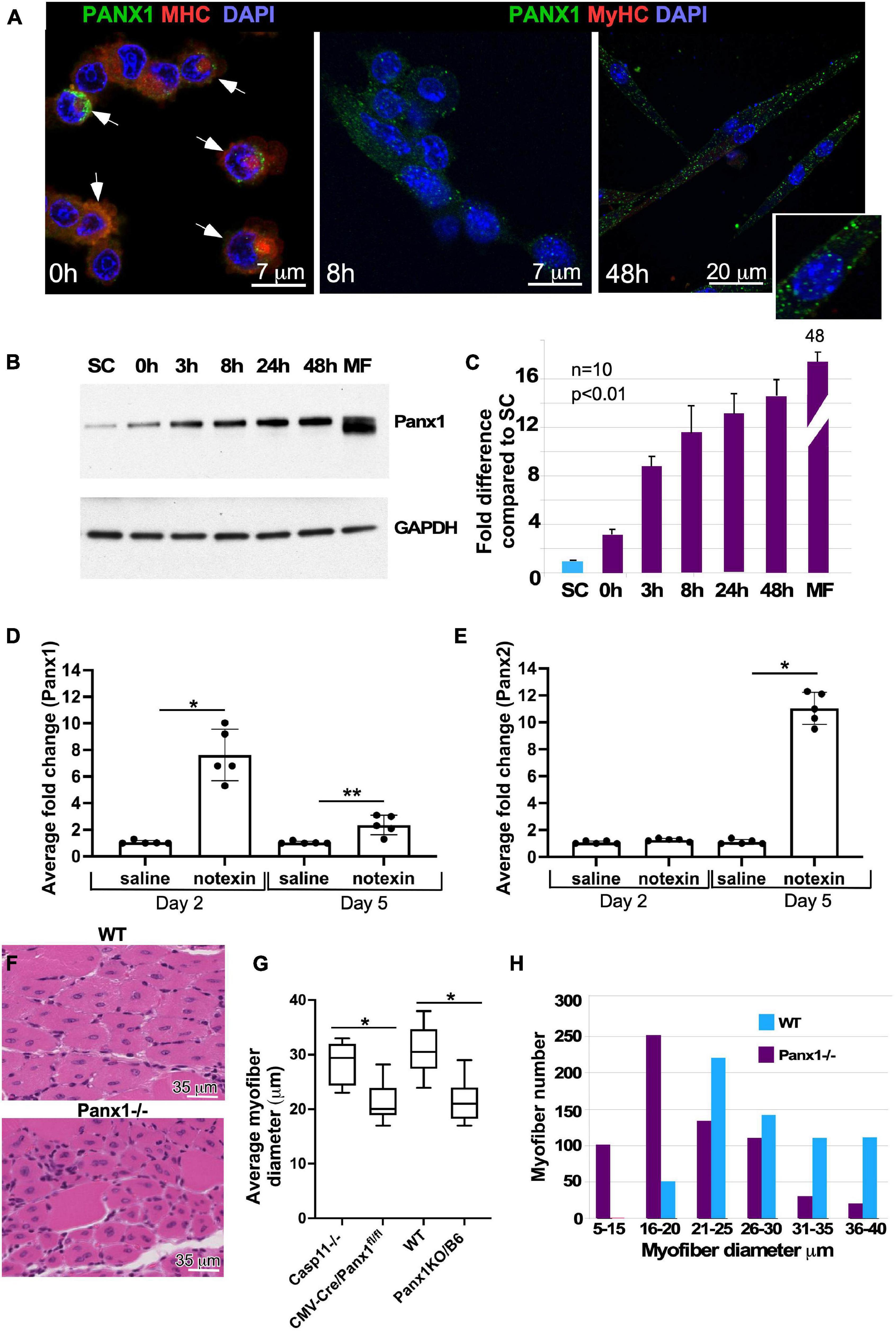
Figure 1. Panx1 expression in primary myoblasts and role of Panx1 in muscle regeneration. (A) Migrating myoblasts express Panx1 (green) in the plasma membrane. Panx1 expression levels are increased in myoblasts forming membrane protrusions, i.e., blebs (indicated by white arrows), stained with SMA (red) to visualize blebbing. Panx1 expression (green) is up-regulated in differentiating myoblasts (8 h in DM) and early myotubes (48 h in DM). An inset shows higher magnification of cells maintained 48 h in DM. Nuclei are stained with DAPI (blue). (B,C) Panx1 expression increases during myoblast differentiation. (B) Western blot showing that undifferentiated freshly isolated SCs show the lowest level of Panx1; Panx1 expression increases during myoblast differentiation (0, 3, 8, 24, and 48 h in DM), peaking in adult myofibers (MF). (C) Quantification of western blotting data (n = 3 independent experiments); P < 0.05. (D,E) Quantitative RT-PCR analysis of Panx1 (D) and Panx2. *p < 0.0001; **p < 0.001. (E) Expression at 2 or 5 days after CTX injection into the TA muscle. Gene expression data from the CTX-treated limb was normalized to that from the vehicle (saline)-treated contralateral limb; n = 3 mice per time point, P < 0.01, *p < 0.0001. (F) Hematoxylin and eosin-stained transverse sections of TA muscle from WT and Panx1–/– (CMV-CrePanx1fl/fl) mice 10 days after muscle injury. Newly formed (centronucleated) Panx1–/– myofibers are smaller in diameter than newly formed WT myofibers. (G) Quantification of myofiber sizes shows that myofibers from both Panx1–/– mouse strains (CMV-CrePanx1fl/fl and Panx1KO/B6) are on average ∼30% smaller than myofibers from Casp11–/– and WT control mice. *p < 0.0001. (H) Histogram demonstrating the distribution of regenerated myofiber sizes in Panx1–/– (CMV-CrePanx1fl/fl) and control mice. Minimal myofiber diameters were measured in transverse sections of TA muscle from three mice for each genotype. n = 647 fibers for Panx1–/– and 636 for WT.
Muscle development and regeneration in the absence of Panx1 have not been previously studied. Basic morphometric analysis of muscle groups (TA, gastrocnemius, and soleus) in WT vs. Panx1–/– mice showed that neither postnatal nor adult Panx1–/– mice have any significant change in the size of limb muscles (data not shown). These results suggest that loss of Panx1 does not significantly affect muscle development and/or postnatal muscle growth.
To determine whether Panx1 may play a role in regeneration, we first examined the dynamics of pannexin expression during muscle regeneration in vivo. The TA muscle of WT mice was injured with notexin and changes in the levels of Panx 1–3 transcripts post-injury were assessed by qRT PCR. The Panx1 transcript levels were higher than those of the Panx2 and Panx3 genes at all stages (data not shown). On day 2 after notexin-injury (muscle inflammation/early regeneration phase), there was a 7.6 ± 1.9-fold increase in the amount of Panx1 transcript in injured vs. saline-injected control TA muscle (Figure 1D), while the amount of Panx2 transcript remained unchanged (Figure 1E). On day 5 after injury (late differentiation phase), Panx1 levels decreased relative to the day 2 peak level and was 2.4 ± 0.7 (Figure 1D). In contrast, Panx2 levels at this time point increased 11 ± 1.2-fold (Figure 1E). These findings suggest that these two pannexins may play different roles in the succession of injury-induced muscle repair phases. As expected (Langlois et al., 2014), the Panx3 mRNA level during muscle regeneration was very low and in some samples undetectable (data not shown).
We next investigated the role of Panx1 in muscle regeneration using two different Panx1 null models: CMV-Cre/Panx1fl/fl mice for which Casp11–/– mice served as the relevant control, and Panx1KO/B6 mice with WT mice as the control. TA muscles were injured with notexin and regeneration studied 10 days later. All mice showed regeneration as evidenced by appearance of new myofibers with centrally located nuclei (Figure 1F). However, the newly regenerated myofibers in both Panx1–/– models were significantly smaller in diameter when compared to those of the control strains (Figure 1G). In addition, we found that regenerated TA muscles of Panx1–/– mice were depleted for large myofibers and enriched in smaller myofibers, as compared to wild-type TA muscles (Figure 1H).
The reduction in myofibre size in regenerating Panx1–/– muscle suggested a possible myoblast fusion defect. We thus compared differentiation of Panx1–/– (both Panx1-null strains) and control (Casp11–/– and/or WT) myoblasts in culture. Control myoblasts cultured in DM formed numerous multinucleated myotubes (Figures 2A,B and Supplementary Figure 1A) and by 72 h the cultures were almost free of unfused myoblasts (Figures 2C,D and Supplementary Figure 1A). In contrast, Panx1–/– myoblasts developed very few multinucleated cells over 72 h in DM (Figures 2E–H and Supplementary Figure 1B). Analysis of Casp11–/– myoblast differentiation showed that they differentiated normally and did not differ morphologically from WT myoblasts. Both differentiating control and Panx1–/– cultures expressed MyHC protein (Figures 2A–H).
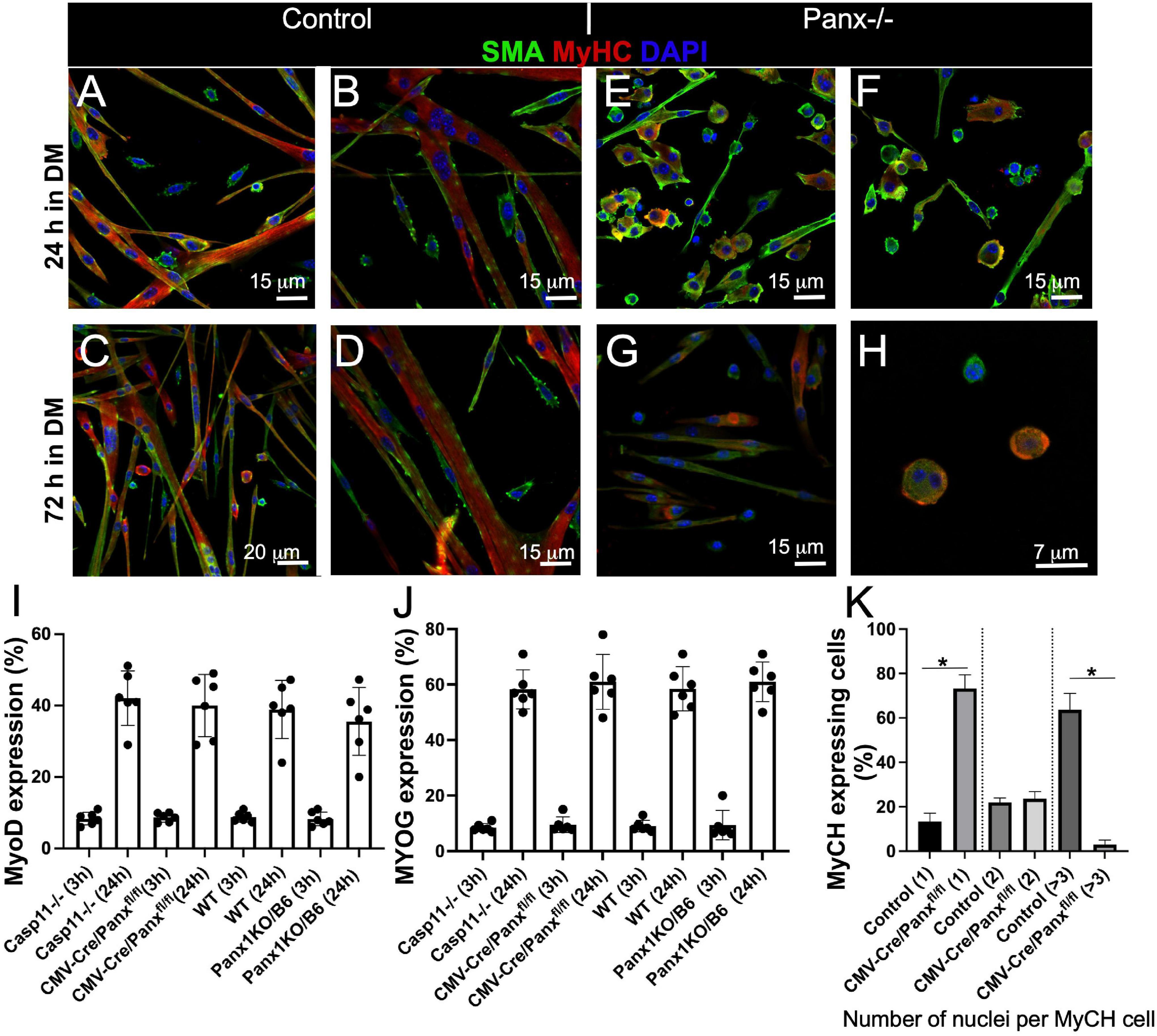
Figure 2. Loss of Panx1 function impairs myoblast fusion but not activation of the molecular differentiation program. WT myoblasts form myotubes after 24 h in DM (A,B), while the majority of Panx1–/– myoblasts remain unfused (C,D). After 72 h in DM, WT cells form long myotubes and only a few single cells remain (E,F). In contrast Panx1–/– cultures show fewer myotubes and many more single myoblasts (G). (H) Higher magnification of cultures shown in (G) shows that even single myoblasts in these Panx1–/– cultures express MyHC (red). In all panesl SMA (green), MyHC (red), DAPI (blue). Quantification of MyoD (I) and MYOG (J) expressing cells in WT and Casp11–/– controls and Panx1–/– (CMV-CrePanx1fl/fl and Panx1KO/b6) myoblasts 3 and 24 h after induction of differentiation. MyoD and MYOG expression is calculated as the percentage of MyoD and MYOG positive nuclei. (K) Fusion index is calculated as the percentage of MyHC-positive cells containing one, two, or ≥3 nuclei at 48 h after induction of differentiation. Data are presented as mean ± standard deviation. n = 3 independent experiments. *P < 0.05 compared with controls.
The process of myoblast differentiation is tightly controlled by myogenic regulatory factors (MRFs). Cultured myoblasts normally express MyoD in GM and initiate expression of myogenin as early as 1–3 h after induction of differentiation (Makarenkova et al., 2009). There was no difference in the number of MyoD expressing cells in Panx1–/– relative to control myoblasts at 3 and 24 h-hours post induction of differentiation (Figure 2I). Moreover, the proportion of myogenin (MYOG)-positive nuclei in Panx1–/– and control myoblast cultures 3 and 24 h after induction of differentiation was not significantly different (Figure 2J). The time to induction of MyHC and percentage of MyHC expressing cells were also similar in both cultures (Figures 2A–H). However, relative to control (WT and Casp11–/–) myoblasts, Panx1–/– myoblasts formed significantly fewer multi-nucleated (3 or more nuclei per cell) MyHC positive myotubes (Figure 2K and Supplementary Figure 1C). Interestingly, unfused myoblasts in Panx1–/– cultures also expressed MyHC despite the impaired myotube formation (Figures 2G,H). In summary, our results suggest that activation of the myogenic differentiation program is not affected in Panx1–/– myoblasts. This finding corroborates previous reports that myoblast fusion is not required for the expression of MyHC (Devlin and Emerson, 1978, 1979).
To form myofibers, differentiating myoblasts must migrate and establish stable cell-cell contacts (Asakura et al., 2001). Real-time in vivo imaging has revealed rapid migration of myoblasts to the site of myofiber damage in vivo (Ishido and Kasuga, 2011, 2012; Webster et al., 2016). Several research groups, including ours, reported that rapid movement of myoblasts requires formation of dynamic membrane blebs (Charras and Paluch, 2008; Makarenkova et al., 2009; Otto et al., 2011), as they utilize an amoeboid-based rather than lamellipodia-mediated propulsion mechanism (Otto et al., 2011). Morphometric analysis showed that cultured primary Panx1–/– myoblasts had fewer blebs than control myoblasts (Figure 3A and Supplementary Figure 1, compare A and B). Analysis of Panx1–/– myoblasts showed that they were significantly less migratory and stayed clustered, in contrast to Control (WT and Casp11–/–) myoblasts that readily spread throughout the dish (Figures 3C,E). Moreover, while control myoblasts had wide, rounded surface blebs, the blebs on Panx1–/– myoblasts were thinner. The average length-to-width ratio of Panx1–/– myoblast blebs was 2-fold less than that of controls (1.23 ± 0.42 for CMV-CrePanx1fl/fl vs. 0.79 ± 0.18 for Casp11–/– and 1.45 ± 0.54 for Panx1KO/B6 vs. for WT myoblasts, p > 0.001), (Figures 3B,D,F).
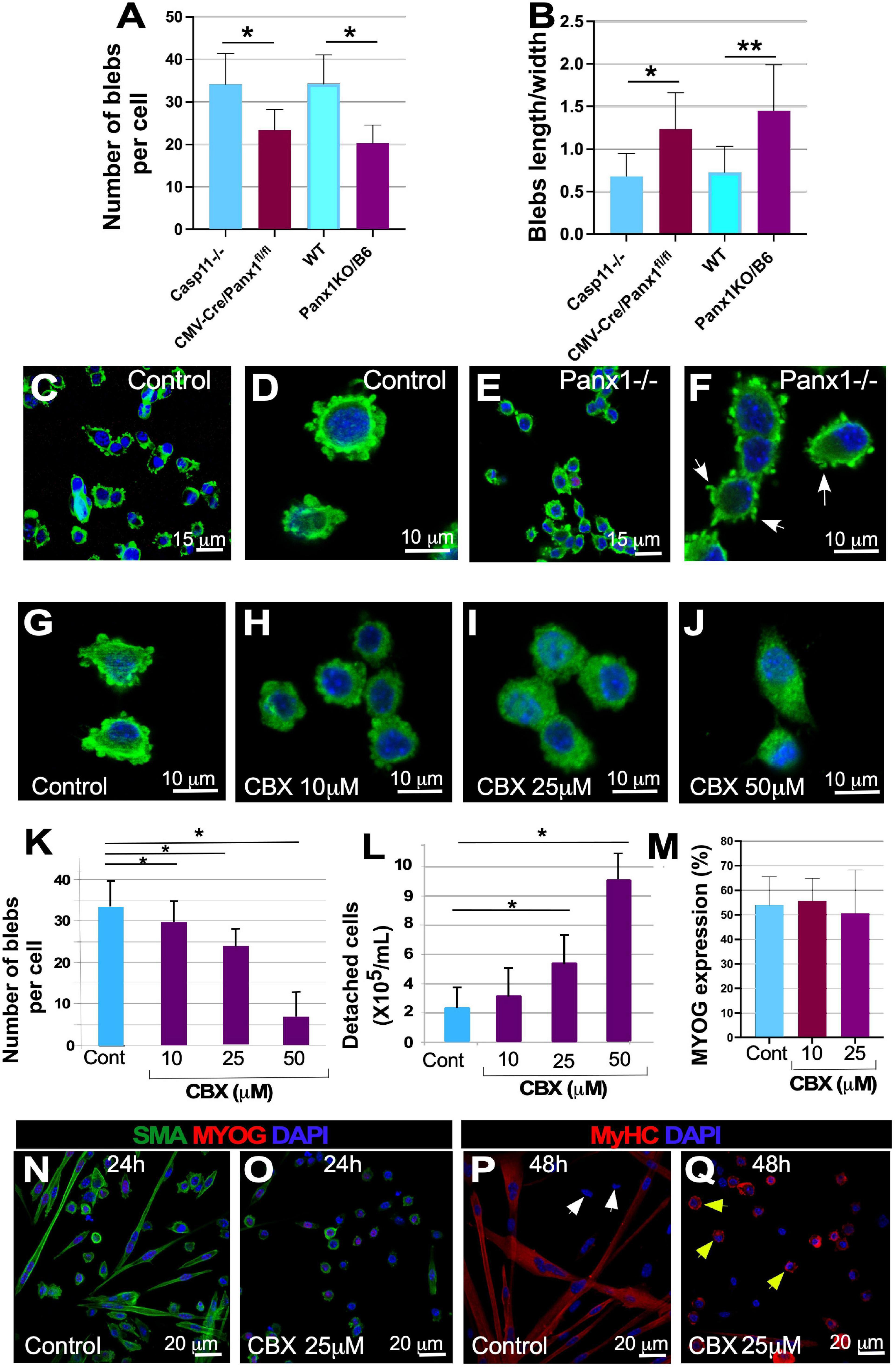
Figure 3. Panx1 signaling regulates myoblast blebbing, migration, myoblast fusion and myofiber formation Panx1–/– myoblasts have fewer surface blebs (A) and altered bleb shape (B), as shown by comparison of bleb length/width ratio in control and Panx1–/– myoblasts. Data are presented as mean ± standard deviation. n = 3 independent experiments. *P < 0.05. The appearance of blebs in control (C,D) and Panx1–/– myoblasts stained with αSMA (E,F). (G–J) Blocking Panx1 signaling in myoblast cultures with carbenoxolone (CBX) decreases blebbing: myoblasts treated with vehicle (G), 10 μm (H), 25 μm (I), 50 μm (J) of CBX for 24 h. Quantification of bleb numbers in control and CBX treated myoblasts (K). (L) High-dose CBX treatment decreases cell adhesion and induces cell detachment in plate washing assays. Analysis of the differentiation program in CBX treated myoblasts: (M–Q): MYOG expression was calculated as the percentage of MYOG-positive nuclei (M). (N–Q) Cultures were treated with vehicle (N,P). and 25 μm of CBX in DM (O,Q) and MYOG (N,O) and MyHC (P,Q) expression was assessed by immunostaining. MyHC expression was found in the majority of cells in the vehicle (P) and CBX-treated (R) cultures. Single migrating cells in control cultures (P, white arrows) did not express MyHC, while almost all single cells in CBX treated cultures expressed MyHC (Q, yellow arrows). Myotube formation was inhibited in CBX-treated (O,Q), compared to control (N,P) cultures. Data in (K–M) is presented as mean ± standard deviation. n = 3 independent experiments. *P < 0.05. **P < 0.01.
We next tested whether pharmacological blockade of Panx1 channels in culture could phenocopy Panx1 genetic ablation. Carbexonolone (CBX) inhibits ATP release through pannexin channels (Hayoz et al., 2012; Xia et al., 2012) and has high specificity for Panx1 channels at <100 μM (Bao et al., 2012). Undifferentiated wildtype myoblasts were exposed to CBX (10, 25, and 50 μM) for 24 h prior to induction of differentiation and then transferred into DM also containing CBX. Myoblast treatment with CBX for 24 h produced significant changes in the morphology and number of surface blebs. With increasing CBX concentration, the number of surface blebs per cell decreased significantly after 10 μM CBX applicationammount of protein; at 25–50 μM CBX myoblasts retained only a few small protrusions (Figures 3G–K).
The morphological changes induced by CBX negatively correlated with adhesion of treated myoblasts to the collagen substrate, causing cells to detach in plate-washing assays more readily (Figure 3L and Supplementary Methods). Thus, 50 μM CBX treatment caused a 5-fold increase in myoblast detachment relative to control (Figure 3L). In contrast, treatment with 10–25 μM of CBX did not induce significant myoblast detachment and we used these drug concentrations in all further experiments. After 24 h exposure to 10 and 25 μM CBX, myoblasts were induced to differentiate in DM also containing CBX, and expression of MYOG and MyHC was assessed by immunocytochemistry (Figures 3M–Q). Similar to Panx1–/– myoblasts, the number of MYOG expressing cells remained unchanged in CBX treated myoblasts (Figures 3M–O), implying that the core transcriptional differentiation program remained unaltered. The induction of MyHC protein expression also appeared unaffected in control and CBX treated cultures (Figures 3P,Q and Supplementary Figures 2A,B). However, unlike control myoblasts (Figures 3N,P), myoblasts treated with CBX failed to elongate and showed almost no fusion (Figures 3O,Q). The similarities in differentiation deficiencies of CBX-treated and Panx1–/– cells, suggest that Panx1 and ATP mediated signaling controls myoblast migration and fusion.
Next, we examined whether signaling downstream of Panx1, particularly release of intracellular ATP, is involved in myoblast migration and fusion. We treated myoblasts with apyrase, an enzyme that catalyzes the breakdown of ATP into AMP and inorganic phosphate. Apyrase treatment did not alter the timing of MYOG expression or the number of MYOG-expressing cells (Figures 4A,B, white arrows, C), but dramatically inhibited myotube formation (Figures 4D–L). After 24 h in DM, apyrase treated myoblasts elongated but did not form myotubes (Figure 4, compare E, F with G, H) and after 48 h the percentage of myotubes with 3 or more nuclei was much lower than in control cultures (Figure 4D, compare I, J with K, L). This effect phenocopied Panx1–/– and CBX-treated myoblasts (see Figures 2, 3). Thus, both ablation of Panx1 and pharmacological blockade of ATP lead to altered cell shape, migration, and fusion properties, suggesting that Panx1 mediates these events through ATP signaling.
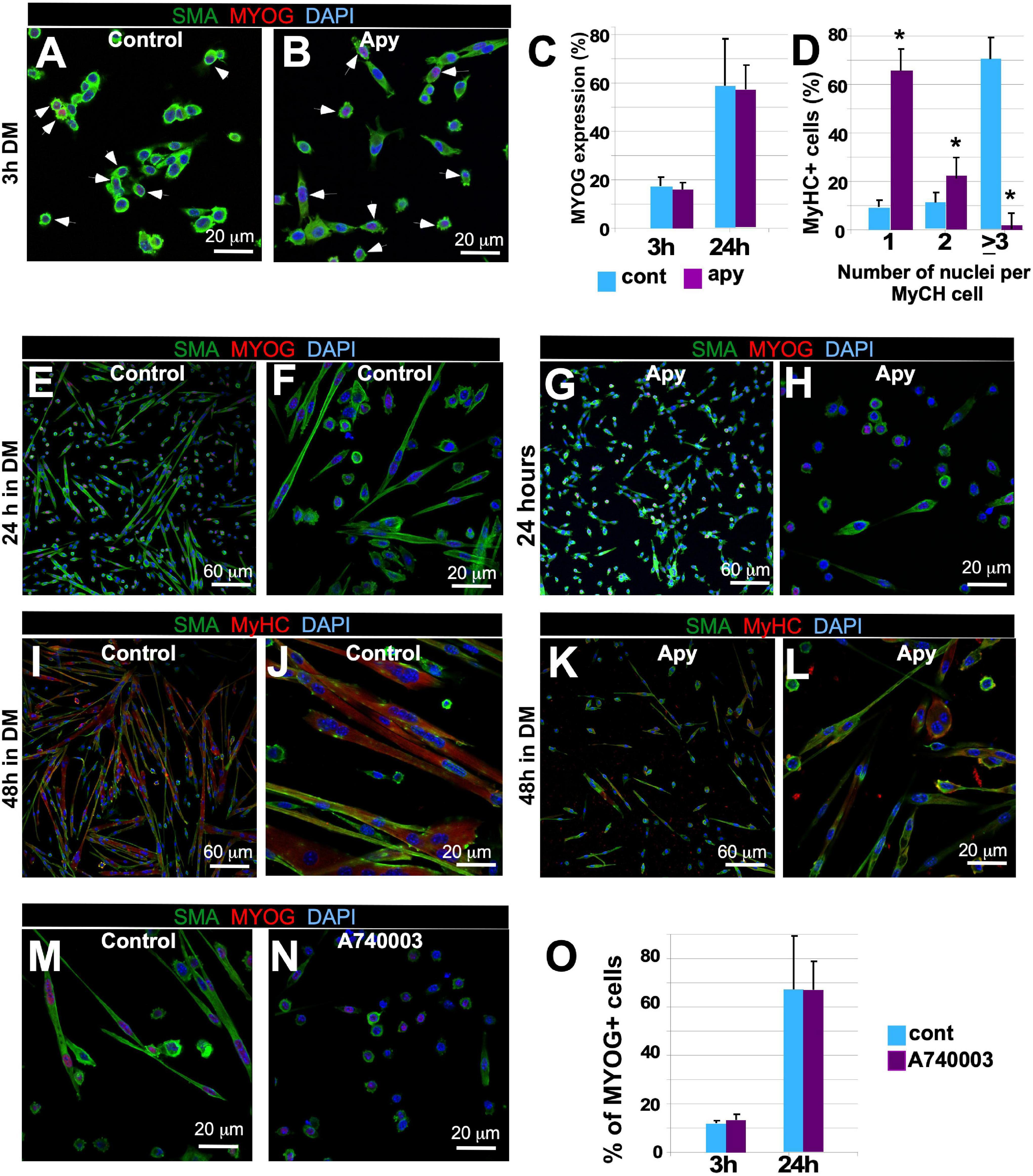
Figure 4. Disruption of ATP or P2X7 receptor signaling inhibits myoblast fusion but not the molecular differentiation program. (A–L) Myoblast cultures were treated with ATP hydrolyzing enzyme apyrase (2.5 U/ml) or vehicle 1 day prior to the induction of differentiation. GM was then replaced with the DM also containing apyrase or vehicle. Myogenin (MYOG) expression (A-control, B-apyrase, white arrows) was assessed by immunostaining; MYOG – red, SMA – green, DAPI – blue, scale bar is 20 μm. The percentage of MYOG expressing cells was quantified (C) 3 h after induction of differentiation with and without apyrase treatment (compare I, J with K, L). (D) The percentage of MyHC-positive cells containing one, two, or ≥3 nuclei was quantified after 48 h of differentiation. Data are presented as mean ± standard deviation. N = 3 independent experiments *p < 0.0001. Control (vehicle-treated; E,F – 24 h in DM; I,J – 48 h in DM) and apyrase treated (G,H – 24 h in DM; K,L – 48 h in DM) primary myoblast cultures. Myoblast fusion is impaired in apyrase treated cultures (E–H; SMA – green; MYOG – red, and DAPI – blue; I–L; SMA – green, MyHC – red, and DAPI – blue). Scale bars: in (E,G,I,K) – 50 μm; in (F,H,J,L) – 20 μm. The A740003 treatment inhibited myotube formation (N) relative to vehicle (M) treated cultures; activation of the differentiation program as assessed by quantification of the percentage of MYOG expressing cells (O) was unaffected at 3 and 24 h after induction of differentiation. In (M,N), SMA – green, MYOG – red, and DAPI – blue. Scale bars are 20 μm.
Extracellular ATP is known to transmit signals by activating purinergic ionotropic P2X or metabotropic P2Y receptors (Wilkinson et al., 1993; Meyer et al., 1999; Ryten et al., 2004; Martinello et al., 2011); involvement of P2X receptors in C2C12 myoblast differentiation was documented previously (Araya et al., 2004). To test whether purinergic signaling via P2X7 is involved in myoblast migration and differentiation, we blocked this receptor in WT primary myoblasts with P2X7 receptor antagonist A740003. A740003 treatment inhibited myotube formation relative to control (Figures 4M,N), while again MYOG expression was unaffected (Figure 4O). These data suggest an important role for P2X7-mediated signaling in myotube formation.
We next examined the effect of inhibiting the P2X7 channel on cell blebbing and migration using single myofiber cultures where individual satellite cells migrate along the myofibers. Treatment of fiber associated-satellite cells with A740003 significantly reduced bleb formation and migration speed (Figures 5A–E). Lipid signaling mediated by LPA was shown to be a major determinant in bleb formation in a variety of cell types (Hagmann et al., 1999; Jia et al., 2006). Moreover, P2X7 receptors activate a signaling pathway that induces Phospholipase D (PLD) to convert phosphatidylcholine to phosphatidic acid (PA) which in turn is transformed into lysophosphatidic acid (LPA) by Phospholipase A2 (Panupinthu et al., 2007). The hydrolytic reaction mediated by PLD requires water and is inhibited by primary but not tertiary alcohols. To assess whether P2X7 may control blebbing through (L)PA production, we repressed PLD activity by addition of 1-butanol to satellite cells on their native substrate and found that it resulted in a significant decrease in the number of blebs displayed on the cell surface, as well as a reduction in migration speed (Figures 5F–J). In a control experiment, an identical concentration of tertiary butanol had no effect on either parameter (Figures 5G–J). These data indicate that P2X7-facilitated blebbing and migration is reliant on PLD activity to form PA. The action of lipids to induce signaling cascades is heavily dependent on the conversion of PA into LPA by phospholipase A2, which in turn activates the LPA receptor, a G-coupled protein that controls a diverse activity including cell migration. We examined whether LPA activity was important in blebbing and satellite cell migration using VPC-32182, a selective LPA1 and LPA3 receptor antagonist. Treatment with VPC-32182 dramatically reduced bleb formation and rate of satellite cell migration (Figures 5K–O), showing a role for LPA activity in these processes. LPA receptors can activate Rho, which ultimately leads to myosin light chain phosphorylation hence controlling actin/myosin-based contraction which may drive bleb dynamics. We found that inhibition of myosin light chain kinase with a ML7 inhibitor reduced satellite cell blebbing and migration speed (Figures 5P–T).
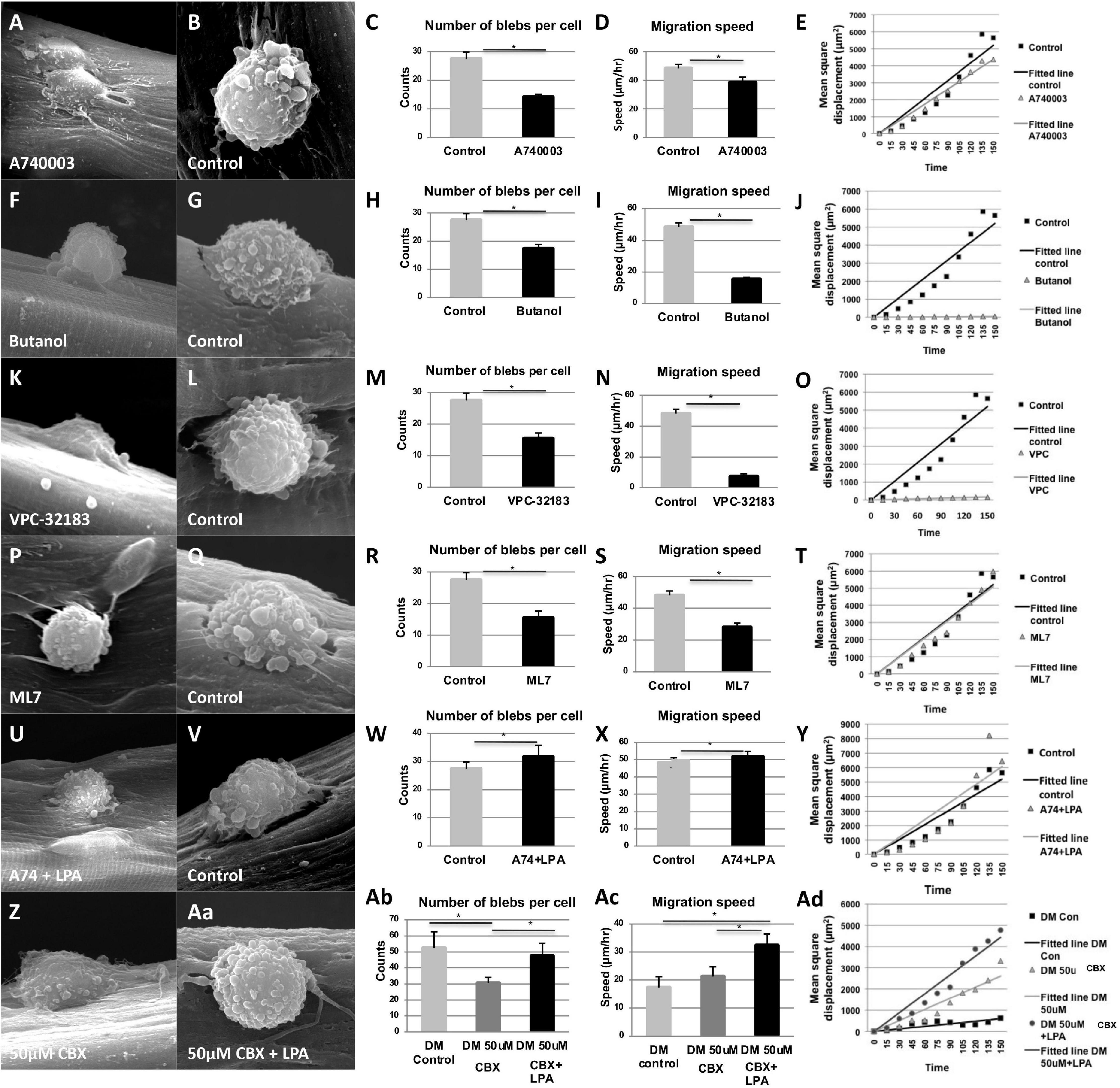
Figure 5. Panx1 regulates a lipid-based signaling pathway that controls bleb formation and satellite cell migration on single myofibers. Quantification of SEM images shows a reduction in the number of blebs per cell, reduced migration speed, but no change in the directionality of movement (plotted as mean square displacement over time) after treatment with A740003 (A–E), Butanol (F–J), VPC-32183 (K–O), and ML7 (O–T). The inhibitory effect of A740003 (U–Y) and CBX on blebbing and migration were (Z–Ad) rescued by LPA. Treatment with LPA resulted in a significantly higher migration speed than the control cells (X,Ac). For movement directionality plots, control data are represented by black squares and treatment data by gray triangles. Black and gray lines represent best fit curves; straight fit curves indicate random migration. *p < 0.05.
To show a more direct link between Panx1/P2X7-mediated ATP signaling and the activity of the LPA receptor in controlling blebbing and migration, we inhibited Panx1 channels with CBX or P2X7 receptors with A740003, and simultaneously introduced LPA. The inhibitory effect of both CBX and A740003 on bleb formation and migration was rescued by LPA (Figures 5U–Y for A740003 and Z-Ac for CBX).
Finally, we used mathematical modeling to examine whether interfering with the Panx1-mediated lipid based signaling pathway in myoblasts influenced not only migration speed but also directionality. We have previously demonstrated that satellite cells from young mice move in a random, directionless manner on the myofiber that contrasts the behavior of cells from old mice, which show directionality (Otto et al., 2011). Plotting the mean square displacement over time showed that although A740003, Butanol, VPC-32183 and ML7 treatments all decreased migration speed of young myoblasts on single myofiber, they still moved in a random manner indicated by a straight-line plot (Figures 5E,J,O,T). Furthermore, rescue of the migration deficit induced by A740003 or CBX with LPA did not alter the migration mode (Figures 5Z, Aa–Ad).
The studies above define a signaling pathway activated by Panx1 that regulates bleb formation and migration of satellite cells. Here we determined whether the same signaling components also regulate myoblast fusion. Freshly isolated satellite cells were cultured at high density permitting cell-cell contact and then maintained in GM or DM for 72 h under a range of drug treatment conditions. In the control conditions, cells in GM remained largely mono-nucleated with few cells expressing MyHC (Figure 6A), while cells in DM formed myotubes with an average of 4.5 nuclei and robust MyHC expression (Figure 6B). Addition of Panx1 channel inhibitor CBX or P2X7 channel inhibitor A740003, to high-density cultures in DM inhibited fusion despite the expression of MyHC (Figures 6C–F). Likewise, fusion was attenuated by addition of butanol, LPA receptor inhibitor VPC32183, or ROCK inhibitor Y-27632 (Figures 6G–L). Finally, inhibition of fusion by blockade of either Panx1 or P2X7 receptor could be reversed by LPA addition (Figures 6M–P). Together these results show Panx1 signaling facilitates myoblast fusion through the P2X7 receptor, downstream lipid intermediaries and the ROCK pathway that activates blebbing.
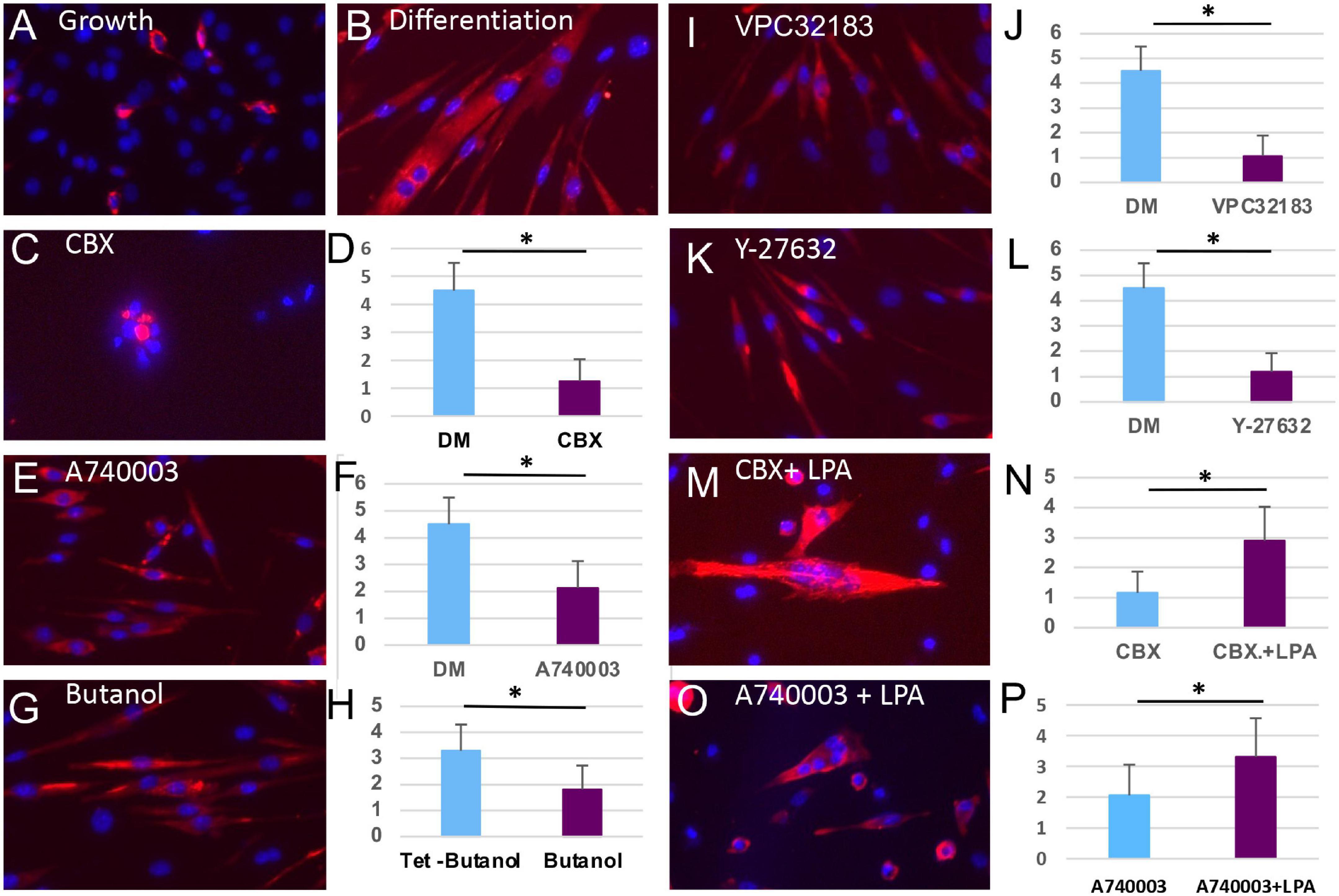
Figure 6. Inhibition of multiple steps of the Panx1 signaling cascade blocs fusion of myoblasts but not the molecular differentiation program. (A) Primary myoblasts cultured in GM do not form myotubes and express little MyHC (red). (B) Culture of primary myoblasts for 72 h in DM induces the formation of MyHC-expressing myotubes. (C–L) Treatment with CBX (C,D), A740003 (E,F), Butanol (G,H), VPC32183 (I,J), and 27632-Y (K,L) all inhibit myotube formation (presented as an average number of nuclei per cells) but not the expression of MyHC. The inhibitory actions of CBX (M,N) or A740003 (O,P) are rescued by the action of LPA (p < 0.05). *p < 0.05.
Pannexins mediate dynamic adhesive contacts and thus regulate diverse functions including cell alignment, adhesion, migration and fusion (Gorbe et al., 2007; Bao et al., 2012). To test whether loss of Panx1 affects expression of key genes involved in these processes, we isolated Panx1–/– and WT myoblasts from 4-month-old mice by FACS and profiled the expression of 84 genes important for cell-cell and cell-matrix interactions. We found nine genes that were significantly altered (>1.5-fold) in Panx1–/– vs. WT myoblasts (Table 1). Among the genes significantly up-regulated in Panx1–/– vs. WT myoblasts were Cadherin-2 (Cdh2), Integrin-β4 (Itgb4), Collagen VI (Col6a1), and sparc/osteonectin (SPOCK1). Among the genes down-regulated in Panx1–/– cultures, we identified transforming growth factor, beta-induced gene (TGFBI/BIGH3; 3.04-fold change, p = 0.008). TGFBI activation has been previously shown to promote myofibril bundling and ECM deposition (Kim and Ingham, 2009). We also found significant downregulation of Elastin Microfibril Interface-Located Protein 1 (Emilin1) that was recently shown to be responsible for the formation of the elastic fibers and anchoring muscle cells to them (Gundry et al., 2009). Multiple Matrix Metalloproteinases (MMPs) were also down-regulated, particularly MMP12 (5.6-fold; p = 0.02), and the disintegrin metallopeptidase with thrombospondin type 5 motif gene (Adamts5, 1.79-fold; p = 0.01; Table 1).
ADAMTS versicanases such as ADAMTS5 have been shown to mediate proteolysis of a hyaluronan and versican-rich matrix leading to pericellular matrix clearance enabling migration, cell-cell interaction, and fusion (D’hondt et al., 2010; Stupka et al., 2013). Moreover, a recent report shows that Adamts5 gene knockdown impairs myoblast fusion in vitro (Stupka et al., 2013). Hypothesizing that this gene may play a role in the migration- and fusion-defective phenotypes of Panx1–/– myoblasts, we tested whether impaired Panx1 signaling disrupts pericellular matrix accumulation using an erythrocyte exclusion assay. Consistent with our hypothesis, we observed significantly more pericellular matrix in Panx1–/– myoblast cultures than in WT (Supplementary Figure 3, compare A and B, D). Treatment of Panx1–/– myoblasts with hyaluronidase blocked erythrocyte exclusion (Supplementary Figures 3C,D) indicating that the pericellular matrix is hyaluronan-based (and thus versican containing). These data suggest that reduced levels of ADAMTS5 and/or other extracellular proteinases in Panx1–/– myoblasts may interfere with fusion by attenuating extracellular protein remodeling.
In this study, we used genetic perturbation to show for the first time that Panx1 is important for efficient skeletal muscle regeneration in vivo. Moreover, in vitro studies provided evidence that Panx1 regulates three major satellite cell/myoblast functions: migration, fusion, and remodeling of the ECM, all of which can contribute to formation of myotubes/myofibers. The signaling events downstream of Panx1 that could control these cellular processes were probed using chemical tools. These data supported a model in which Panx1 functions to induce membrane blebbing and migration via activation of the purinergic receptor P2X7, which in turn leads to production of extracellular ATP. Moreover, several lines of evidence support a role for lipid intermediates in the signaling pathway. In particular, myoblast blebbing and migration could be blocked by inhibition of LPA production, and conversely, P2X7 blockade could be rescued by addition of LPA. Based on this data, we propose that Panx1-regulated P2X7 receptors may signal through PLD and A2 (PLA2); the resulting LPA then causes dynamic membrane blebbing. This pathway could involve Rho-associated kinase (ROCK; Jia et al., 2006; Figure 7). This would be consistent with previous studies showing that ROCK promotes rearrangement of the actin cytoskeleton to facilitate migration and fusion (Signorello and Leoncini, 2014; Hyder et al., 2015).
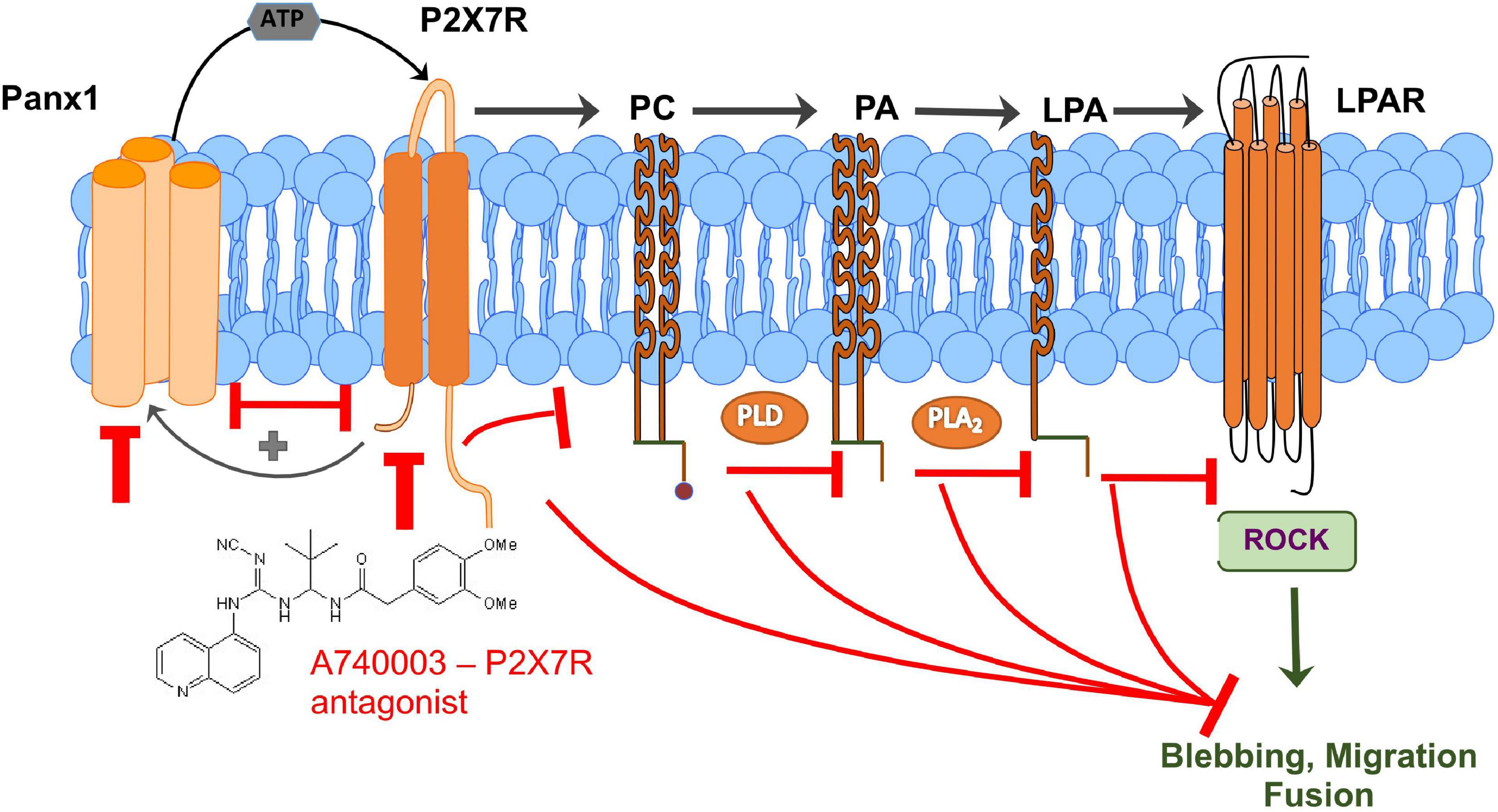
Figure 7. Schematic representation of a new signaling mechanism that connects Panx1/P2X7R signaling with lipid metabolism and lipid-mediated ROCK signaling and regulation of myoblast blebbing/migration/fusion which is important for efficient muscle regeneration. Panx1-regulated P2X7 receptors can signal through PLD and A2 (PLA2); the resulting lysophosphatidic acid (LPA) then causes dynamic membrane blebbing via a pathway dependent on Rho-associated kinase (ROCK). Interaction between Panx1 and P2X7 receptors leads to further activation of the pannexin channel through a positive feedback loop that has been reported previously (Pelegrin and Surprenant, 2006; Velasquez and Eugenin, 2014). Perturbation of any component of these signaling pathways results in decreased blebbing and reduced myoblast migration and fusion.
Blebbing involves changes in the membrane and the underlying actin cytoskeleton. As reported previously, blebs on the surface of myoblasts and other cells are spherical membrane protrusions that appear due to hydrostatic pressure in the cytoplasm but their retraction involves actomyosin complexes of the cell cortex (Charras and Paluch, 2008; Tinevez et al., 2009; Otto et al., 2011). The profound effect of Panx1 gene ablation or P2X channel blockade on the shape and length of blebs, suggests that Panx1 influences the actomyosin bleb cortex. In addition to the potential for ROCK mediated effects on actin, Panx1/P2X7R may function via direct physical links to the cytoskeleton (Wicki-Stordeur and Swayne, 2013; Boyce et al., 2014). In particular, this complex was shown to directly interact with actin filaments (Kuehnel et al., 2009a,b; Wicki-Stordeur and Swayne, 2013; Boyce et al., 2014), and in BICR-M1Rk cells, Panx1 has been reported to co-localize and interact with actin at the leading edge of the lamellipodia and filopodia (Jaenisch and Bird, 2003). It was also shown that in the brain PANX1 controls actin cytoskeleton rearrangement during cell migration via interaction with the ARP2/3 complex (Wicki-Stordeur and Swayne, 2013; Boyce et al., 2014). The question of whether Panx1 interacts with directly or indirectly with actin in myoblasts and whether it helps control bleb retraction would be interesting to study in the future.
In addition to cytoskeletal changes that could modulate migration and fusion, the blebbing process might play a direct role in promoting interaction between the plasma membranes of adjacent cells. This idea is supported by recent studies, which demonstrate that blebbing promotes fusion of cancer cells exposed to chemical stress. In this context, bleb-mediated fusion results in the formation of blebbishields that ensure cell survival and growth (Kuehnel et al., 2009b; Jinesh et al., 2013, 2016; Jinesh and Kamat, 2016). We suggest that bleb-mediated myoblast fusion could involve a process that is normally found in cells undergoing apoptosis: plasma membrane scrambling (Marino and Kroemer, 2013). This scrambling involves the formation of dynamic membrane blebs with phosphatidylserine translocated from the inner to the outer membrane. In contrast to the apoptotic pathway, we suggest that in myoblasts these molecules could become available to bind the phosphatidylserine receptor BAI1 on adjacent myoblasts to activate the ELMO/DOCK180/Rac1 pathway leading to fusion (Hochreiter-Hufford et al., 2013).
The precise role of ATP production in Panx1/P2X channel mediated signaling in myoblasts is yet to be defined. ATP is known to mediate paracrine cell-cell signaling (Iglesias and Spray, 2012) and is involved in multiple important biological functions, such as sympathetic nerve activation in the ischemic heart (Dong et al., 2016), leukocyte emigration through blood endothelium (Lohman et al., 2015), and response to mechanical pressure (Lopez et al., 2021). Our new data suggests that ATP release can also promote cell fusion, however, further studies are required to confirm this hypothesis.
While this discussion has focused in the role of Panx1 signaling in blebbing and actin/membrane changes that may promote migration and fusion, it is important to note that myoblast differentiation involves multiple steps including cell cycle exit and commitment to differentiation, myoblast migration and fusion, and sequential expression of differentiation factors (Zammit et al., 2006). A previous study (Langlois et al., 2014), reported that Panx1 is involved in muscle differentiation, in part because they found that the formation of MyHC-expressing myotubes was perturbed by CBX application. However, the study did not define the specific stage of myoblast differentiation controlled by Panx1. Our data indicate that Panx1 inhibition impairs myoblast migration and fusion, but does not alter the expression of differentiation markers. Further work will be required to clarify whether Panx1 has a role in regulation of the muscle regulatory factor (MRF)-driven gene expression program during differentiation.
While levels of core differentiation factors did not seem to be altered in our genetic and chemical Panx1 perturbation studies, we did find that Panx1–/– myoblasts had reduced expression of matrix remodeling genes including Adamts5 and MMPs. There was also dysregulation of several adhesion related molecules. A publication from Stupka and co-authors showed the importance of Adamts versicanases and especially Adamts5 in myoblast fusion. Moreover, our data suggested that Panx1 inhibition leads to the failure to remodel pericellular matrix during fusion. Adhesive interactions also play critical roles in migration, proliferation, and fusion of myoblasts, and aberrations in such interactions can lead to compromised function and pathology (Siegel et al., 2009; Lukjanenko et al., 2016). Integrin β (Itgb1) expression was reduced in Panx1–/– myoblasts; Igtb1-deficient myoblasts (see Table 1) were previously shown to have defective plasma membrane breakdown and fusion (Schwander et al., 2003). It is possible that reduced expression of several adhesion and matrix remodeling factors (such as Itgb1 and Adamts5) jointly contribute to delayed fusion of Panx1–/– myoblasts.
In conclusion, our study provides multiple lines of evidence that Panx1 mediates purinergic signaling, which can be mediated via lipid intermediates, to control key processes underlying myoblast migration and fusion including membrane blebbing, ECM remodeling, migration, and adhesion. These pathways are essential for robust skeletal muscle regeneration.
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author/s.
The animal study was reviewed and approved by Scripps Research Institute Animal Care and Use Committee.
KS-B, HC-H, AG, and TEW: collection and/or assembly of data, data analysis and interpretation. RM: collection and/or assembly of data, data analysis and interpretation, manuscript writing, and final approval of manuscript. AS: collection and/or assembly of data, data analysis and interpretation, and final approval of manuscript. PRD and SV: data analysis and interpretation. VIS: data analysis and interpretation, conception and design. KP: conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing, and final approval of manuscript. HPM: conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript, financial support, administrative support, and final approval of manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by a grant from the French Muscular Dystrophy Association, grants 13899 and 15654, National Eye Institute (NIH NEI) grants 1R01EY026202 and R01EY028983, and NIH NIDCR grant 1R01DE031044 to HPM, BBSRC BB/J016454/1 to HC-H, NIH NEI grant EY032261, Department of Defense grant VR200079 and Russian Science Foundation grant N17-15-01433 to VIS, Center Grant P30-EY014801 and unrestricted Department of Defense grant DAMB W81XWH-13-1-0048 and RPB grants to Bascom Palmer Eye Institute, and National Institute of Arthritis and Musculoskeletal and Skin Disease (NIH NIAMS) grant R21 AR075205 to AS and KP wishes to acknowledge the generous support of the Freiburg Institute for Advanced Studies (FRIAS) through the senior fellowship program. TEW would like to thank St John’s College Oxford for its financial support. RM was supported by an Australian Research Council Future Fellowship and by Australian Research Council (ARC) Discovery Project DP210103065.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We are also grateful to the Scripps Research Institute Core Microscopy Facility and in particular to Drs. William B. Kiosses and Scott Henderson for help and support.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.736813/full#supplementary-material
Supplementary Figure 1 | Panx1–/– myoblasts obtained from Panx1KO/B6 mice show significant delays in myotube formation, confirming the phenotype of Panx1–/– myoblasts from CMV-Cre/Panx1flf/fl mice; Casp11–/– myoblasts show normal differentiation. (A) Casp11–/– myoblasts: Casp11–/– myoblasts have prominent surface blebs and differentiate similarly to WT myoblasts at 24 and 48 h in DM. (B) Panx1–/– myoblasts: Panx1–/– myoblasts have fewer surface blebs and do not spread well through the surface of the dish. Panx1–/– myoblasts show a delay in myotube formation at 24 h (B,C) and 48 h after replacement of GM with the DM. (E–H,I) Fusion index quantification of Panx1–/– (Panx1KO/B6), WT, and Casp11–/– myoblasts. Myoblast fusion was calculated as the percentage of MyHC expressing cells containing one, two, or ≥3 nuclei at 48 h after induction of differentiation. Data are presented as mean ± standard deviation. n = 3 independent experiments. ∗P < 0.05 compared with WT.
Supplementary Figure 2 | Western blot analysis (A) and quantification (B) of the MyCH in control (WT untreated) myoblasts and myoblasts treated with the 10 mm and 25 mm of CBX. Analysis of MyCH expression was perfomed 1 and 48 h after induction of differentiation. Control is vehicle treated myoblasts. Myoblast Lysate was probed with anti-MyCH antibody at 1:1000 dilution (#05-716, clone A4.1025, Sigma-Aldrich).
Supplementary Figure 3 | Loss of Panx1 function in primary myoblasts results in accumulation of pericellular matrix (A,B), Panx1–/– myoblasts (A-red dashed line) accumulate more pericellular matrix as shown by the exclusion of erythrocytes (yellow dashed line) than WT myoblasts (B). Treatment of Panx1–/– myoblasts with hyaluronidase (hyalur) reduces the area of erythrocyte exclusion to be similar to WT myoblasts (C). Scale bar = 15 μm. (D) Quantitation of the area of pericellular matrix exclusion around WT myoblasts and Panx1–/– myoblasts with and without hyaluronidase. Data is the ratio of exclusion area to cellular area; images are taken randomly from three independent experiments (n ∼20 cells per condition). ∗p < 0.001.
Araya, R., Riquelme, M. A., Brandan, E., and Saez, J. C. (2004). The formation of skeletal muscle myotubes requires functional membrane receptors activated by extracellular ATP. Brain Res. Rev. 47, 174–188. doi: 10.1016/j.brainresrev.2004.06.003
Asakura, A., Komaki, M., and Rudnicki, M. (2001). Muscle satellite cells are multipotential stem cells that exhibit myogenic, osteogenic, and adipogenic differentiation. Differentiation 68, 245–253. doi: 10.1046/j.1432-0436.2001.680412.x
Bao, B. A., Lai, C. P., Naus, C. C., and Morgan, J. R. (2012). Pannexin1 drives multicellular aggregate compaction via a signaling cascade that remodels the actin cytoskeleton. J. Biol. Chem. 287, 8407–8416. doi: 10.1074/jbc.M111.306522
Baranova, A., Ivanov, D., Petrash, N., Pestova, A., Skoblov, M., Kelmanson, I., et al. (2004). The mammalian pannexin family is homologous to the invertebrate innexin gap junction proteins. Genomics 83, 706–716. doi: 10.1016/j.ygeno.2003.09.025
Belmonte-Beitia, J., Woolley, T. E., Scott, J. G., Maini, P. K., and Gaffney, E. A. (2013). Modelling biological invasions: individual to population scales at interfaces. J. Theor. Biol. 334, 1–12. doi: 10.1016/j.jtbi.2013.05.033
Boyce, A. K., Wicki-Stordeur, L. E., and Swayne, L. A. (2014). Powerful partnership: crosstalk between pannexin 1 and the cytoskeleton. Front. Physiol. 5:27. doi: 10.3389/fphys.2014.00027
Bruzzone, R., Barbe, M. T., Jakob, N. J., and Monyer, H. (2005). Pharmacological properties of homomeric and heteromeric pannexin hemichannels expressed in Xenopus oocytes. J. Neurochem. 92, 1033–1043. doi: 10.1111/j.1471-4159.2004.02947.x
Bruzzone, R., Hormuzdi, S. G., Barbe, M. T., Herb, A., and Monyer, H. (2003). Pannexins, a family of gap junction proteins expressed in brain. Proc. Natl. Acad. Sci. U.S.A. 100, 13644–13649. doi: 10.1073/pnas.2233464100
Bunse, S., Locovei, S., Schmidt, M., Qiu, F., Zoidl, G., Dahl, G., et al. (2009). The potassium channel subunit Kvbeta3 interacts with pannexin 1 and attenuates its sensitivity to changes in redox potentials. FEBS J. 276, 6258–6270. doi: 10.1111/j.1742-4658.2009.07334.x
Cea, L. A., Riquelme, M. A., Cisterna, B. A., Puebla, C., Vega, J. L., Rovegno, M., et al. (2012). Connexin- and pannexin-based channels in normal skeletal muscles and their possible role in muscle atrophy. J. Membr. Biol. 245, 423–436. doi: 10.1007/s00232-012-9485-8
Cea, L. A., Riquelme, M. A., Vargas, A. A., Urrutia, C., and Saez, J. C. (2014). Pannexin 1 channels in skeletal muscles. Front. Physiol. 5:139. doi: 10.3389/fphys.2014.00139
Charras, G., and Paluch, E. (2008). Blebs lead the way: how to migrate without lamellipodia. Nat. Rev. Mol. Cell Biol. 9, 730–736. doi: 10.1038/nrm2453
Chekeni, F. B., Elliott, M. R., Sandilos, J. K., Walk, S. F., Kinchen, J. M., Lazarowski, E. R., et al. (2010). Pannexin 1 channels mediate ‘find-me’ signal release and membrane permeability during apoptosis. Nature 467, 863–867. doi: 10.1038/nature09413
Collins-Hooper, H., Woolley, T. E., Dyson, L., Patel, A., Potter, P., Baker, R. E., et al. (2012). Age-related changes in speed and mechanism of adult skeletal muscle stem cell migration. Stem Cells 30, 1182–1195. doi: 10.1002/stem.1088
Devlin, R. B., and Emerson, C. P. Jr. (1978). Coordinate regulation of contractile protein synthesis during myoblast differentiation. Cell 13, 599–611. doi: 10.1016/0092-8674(78)90211-8
Devlin, R. B., and Emerson, C. P. Jr. (1979). Coordinate accumulation of contractile protein mRNAs during myoblast differentiation. Dev. Biol. 69, 202–216. doi: 10.1016/0012-1606(79)90286-0
D’hondt, C., Ponsaerts, R., De Smedt, H., Vinken, M., De Vuyst, E., De Bock, M., et al. (2010). Pannexin channels in ATP release and beyond: an unexpected rendezvous at the endoplasmic reticulum. Cell. Signal. 23, 305–316. doi: 10.1016/j.cellsig.2010.07.018
Dong, F., Yang, X. J., Jiang, T. B., and Chen, Y. (2016). Ischemia triggered ATP release through Pannexin-1 channel by myocardial cells activates sympathetic fibers. Microvasc. Res. 104, 32–37. doi: 10.1016/j.mvr.2015.11.005
Dvoriantchikova, G., Ivanov, D., Barakat, D., Grinberg, A., Wen, R., Slepak, V. Z., et al. (2012). Genetic ablation of Pannexin1 protects retinal neurons from ischemic injury. PLoS One 7:e31991. doi: 10.1371/journal.pone.0031991
Ermakova, Ii, Sakuta, G. A., Potekhina, M. A., Fedorova, M. A., Hoffmann, R., and Morozov, V. I. (2011). Major chondroitin sulfate proteoglycans identified in L6J1 myoblast culture. Biochemistry 76, 359–365. doi: 10.1134/S0006297911030102
Friedl, P., and Wolf, K. (2009). Plasticity of cell migration: a multiscale tuning model. J. Cell Biol. 188, 11–19. doi: 10.1083/jcb.200909003
Friedl, P., Borgmann, S., and Brocker, E. B. (2001). Amoeboid leukocyte crawling through extracellular matrix: lessons from the Dictyostelium paradigm of cell movement. J. Leukoc. Biol. 70, 491–509.
Gaynullina, D., Shestopalov, V. I., Panchin, Y., and Tarasova, O. S. (2015). Pannexin 1 facilitates arterial relaxation via an endothelium-derived hyperpolarization mechanism. FEBS Lett. 589, 1164–1170. doi: 10.1016/j.febslet.2015.03.018
Gorbe, A., Krenacs, T., Cook, J. E., and Becker, D. L. (2007). Myoblast proliferation and syncytial fusion both depend on connexin43 function in transfected skeletal muscle primary cultures. Exp. Cell Res. 313, 1135–1148. doi: 10.1016/j.yexcr.2007.01.012
Gundry, R. L., Raginski, K., Tarasova, Y., Tchernyshyov, I., Bausch-Fluck, D., Elliott, S. T., et al. (2009). The mouse C2C12 myoblast cell surface N-linked glycoproteome: identification, glycosite occupancy, and membrane orientation. Mol. Cell. Proteomics 8, 2555–2569. doi: 10.1074/mcp.M900195-MCP200
Hagmann, J., Burger, M. M., and Dagan, D. (1999). Regulation of plasma membrane blebbing by the cytoskeleton. J. Cell Biochem. 73, 488–499. doi: 10.1002/(SICI)1097-4644(19990615)73:4<488::AID-JCB7>3.0.CO;2-P
Hantai, D., Tassin, A. M., Gautron, J., and Labat-Robert, J. (1985). Biosynthesis of laminin and fibronectin by rat satellite cells during myogenesis in vitro. Cell Biol. Int. Rep. 9, 647–654. doi: 10.1016/0309-1651(85)90057-8
Hattori, N., Carrino, D. A., Lauer, M. E., Vasanji, A., Wylie, J. D., Nelson, C. M., et al. (2011). Pericellular versican regulates the fibroblast-myofibroblast transition: a role for ADAMTS5 protease-mediated proteolysis. J. Biol. Chem. 286, 34298–34310. doi: 10.1074/jbc.M111.254938
Hayoz, S., Jia, C., and Hegg, C. (2012). Mechanisms of constitutive and ATP-evoked ATP release in neonatal mouse olfactory epithelium. BMC Neurosci. 13:53. doi: 10.1186/1471-2202-13-53
Hochreiter-Hufford, A. E., Lee, C. S., Kinchen, J. M., Sokolowski, J. D., Arandjelovic, S., Call, J. A., et al. (2013). Phosphatidylserine receptor BAI1 and apoptotic cells as new promoters of myoblast fusion. Nature 497, 263–267. doi: 10.1038/nature12135
Hutcheson, D. A., and Kardon, G. (2009). Genetic manipulations reveal dynamic cell and gene functions: cre-ating a new view of myogenesis. Cell Cycle 8, 3675–3678. doi: 10.4161/cc.8.22.9992
Hyder, C. L., Kemppainen, K., Isoniemi, K. O., Imanishi, S. Y., Goto, H., Inagaki, M., et al. (2015). Sphingolipids inhibit vimentin-dependent cell migration. J. Cell Sci. 128, 2057–2069. doi: 10.1242/jcs.160341
Iglesias, R. M., and Spray, D. C. (2012). Pannexin1-mediated ATP release provides signal transmission between Neuro2A cells. Neurochem. Res. 37, 1355–1363. doi: 10.1007/s11064-012-0720-6
Iglesias, R., Dahl, G., Qiu, F., Spray, D. C., and Scemes, E. (2009). Pannexin 1: the molecular substrate of astrocyte “hemichannels”. J. Neurosci. 29, 7092–7097. doi: 10.1523/JNEUROSCI.6062-08.2009
Iglesias, R., Locovei, S., Roque, A., Alberto, A. P., Dahl, G., Spray, D. C., et al. (2008). P2X7 receptor-Pannexin1 complex: pharmacology and signaling. Am. J. Physiol. Cell Physiol. 295, C752–C760. doi: 10.1152/ajpcell.00228.2008
Irintchev, A., Zeschnigk, M., Starzinski-Powitz, A., and Wernig, A. (1994). Expression pattern of M-cadherin in normal, denervated, and regenerating mouse muscles. Dev. Dyn. 199, 326–337. doi: 10.1002/aja.1001990407
Irizarry, R. A., Hobbs, B., Collin, F., Beazer-Barclay, Y. D., Antonellis, K. J., Scherf, U., et al. (2003). Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264. doi: 10.1093/biostatistics/4.2.249
Ishido, M., and Kasuga, N. (2011). In situ real-time imaging of the satellite cells in rat intact and injured soleus muscles using quantum dots. Histochem. Cell Biol. 135, 21–26. doi: 10.1007/s00418-010-0767-x
Ishido, M., and Kasuga, N. (2012). in vivo real-time imaging of exogenous HGF-triggered cell migration in rat intact soleus muscles. Acta Histochem Cytochem 45, 193–199. doi: 10.1267/ahc.11058
Jaenisch, R., and Bird, A. (2003). Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 33(Suppl.), 245–254. doi: 10.1038/ng1089
Jia, Z., Vadnais, J., Lu, M. L., Noel, J., and Nabi, I. R. (2006). Rho/ROCK-dependent pseudopodial protrusion and cellular blebbing are regulated by p38 MAPK in tumour cells exhibiting autocrine c-Met activation. Biol. Cell 98, 337–351. doi: 10.1042/BC20050088
Jinesh, G. G., and Kamat, A. M. (2016). Endocytosis and serpentine filopodia drive blebbishield-mediated resurrection of apoptotic cancer stem cells. Cell Death Discov. 2:15069. doi: 10.1038/cddiscovery.2015.69
Jinesh, G. G., Choi, W., Shah, J. B., Lee, E. K., Willis, D. L., and Kamat, A. M. (2013). Blebbishields, the emergency program for cancer stem cells: sphere formation and tumorigenesis after apoptosis. Cell Death Differ. 20, 382–395. doi: 10.1038/cdd.2012.140
Jinesh, G. G., Taoka, R., Zhang, Q., Gorantla, S., and Kamat, A. M. (2016). Novel PKC-zeta to p47 phox interaction is necessary for transformation from blebbishields. Sci. Rep. 6:23965. doi: 10.1038/srep23965
Jorquera, G., Altamirano, F., Contreras-Ferrat, A., Almarza, G., Buvinic, S., Jacquemond, V., et al. (2013). Cav1.1 controls frequency-dependent events regulating adult skeletal muscle plasticity. J. Cell Sci. 126, 1189–1198. doi: 10.1242/jcs.116855
Jorquera, G., Meneses-Valdes, R., Rosales-Soto, G., Valladares-Ide, D., Campos, C., Silva-Monasterio, M., et al. (2021). High extracellular ATP levels released through pannexin-1 channels mediate inflammation and insulin resistance in skeletal muscle fibres of diet-induced obese mice. Diabetologia 64, 1389–1401. doi: 10.1007/s00125-021-05418-2
Kang, S. J., Wang, S., Hara, H., Peterson, E. P., Namura, S., Amin-Hanjani, S., et al. (2000). Dual role of caspase-11 in mediating activation of caspase-1 and caspase-3 under pathological conditions. J. Cell Biol. 149, 613–622. doi: 10.1083/jcb.149.3.613
Khajah, M. A., Mathew, P. M., Alam-Eldin, N. S., and Luqmani, Y. A. (2015). Bleb formation is induced by alkaline but not acidic pH in estrogen receptor silenced breast cancer cells. Int. J. Oncol. 46, 1685–1698. doi: 10.3892/ijo.2015.2884
Kim, H. R., and Ingham, P. W. (2009). The extracellular matrix protein TGFBI promotes myofibril bundling and muscle fibre growth in the zebrafish embryo. Dev. Dyn. 238, 56–65. doi: 10.1002/dvdy.21812
Kuehnel, M. P., Reiss, M., Anand, P. K., Treede, I., Holzer, D., Hoffmann, E., et al. (2009a). Sphingosine-1-phosphate receptors stimulate macrophage plasma-membrane actin assembly via ADP release. ATP synthesis and P2X7R activation. J. Cell Sci. 122, 505–512. doi: 10.1242/jcs.034207
Kuehnel, M. P., Rybin, V., Anand, P. K., Anes, E., and Griffiths, G. (2009b). Lipids regulate P2X7-receptor-dependent actin assembly by phagosomes via ADP translocation and ATP synthesis in the phagosome lumen. J. Cell Sci. 122, 499–504. doi: 10.1242/jcs.034199
Langlois, S., Xiang, X., Young, K., Cowan, B. J., Penuela, S., and Cowan, K. N. (2014). Pannexin 1 and pannexin 3 channels regulate skeletal muscle myoblast proliferation and differentiation. J. Biol. Chem. 289, 30717–30731. doi: 10.1074/jbc.M114.572131
Lepper, C., Partridge, T. A., and Fan, C.-M. (2011). An absolute requirement for Pax7-positive satellite cells in acute injury-induced skeletal muscle regeneration. Development 138, 3639–3646. doi: 10.1242/dev.067595
Locovei, S., Bao, L., and Dahl, G. (2006). Pannexin 1 in erythrocytes: function without a gap. Proc. Natl. Acad. Sci. U.S.A. 103, 7655–7659. doi: 10.1073/pnas.0601037103
Lohman, A. W., Billaud, M., and Isakson, B. E. (2012). Mechanisms of ATP release and signalling in the blood vessel wall. Cardiovasc. Res. 95, 269–280. doi: 10.1093/cvr/cvs187
Lohman, A. W., Leskov, I. L., Butcher, J. T., Johnstone, S. R., Stokes, T. A., Begandt, D., et al. (2015). Pannexin 1 channels regulate leukocyte emigration through the venous endothelium during acute inflammation. Nat. Commun. 6:7965. doi: 10.1038/ncomms8965
Lopez, X., Palacios-Prado, N., Guiza, J., Escamilla, R., Fernandez, P., Vega, J. L., et al. (2021). A physiologic rise in cytoplasmic calcium ion signal increases pannexin1 channel activity via a C-terminus phosphorylation by CaMKII. Proc. Natl. Acad. Sci. U.S.A. 118:e2108967118. doi: 10.1073/pnas.2108967118
Lukjanenko, L., Jung, M. J., Hegde, N., Perruisseau-Carrier, C., Migliavacca, E., Rozo, M., et al. (2016). Loss of fibronectin from the aged stem cell niche affects the regenerative capacity of skeletal muscle in mice. Nat. Med. 22, 897–905. doi: 10.1038/nm.4126
Makarenkova, H. P., Gonzalez, K. N., Kiosses, W. B., and Meech, R. (2009). Barx2 controls myoblast fusion and promotes MyoD-mediated activation of the smooth muscle alpha actin gene. J. Biol. Chem. 284, 14866–14874. doi: 10.1074/jbc.M807208200
Makarenkova, H. P., Shah, S. B., and Shestopalov, V. I. (2018). The two faces of pannexins: new roles in inflammation and repair. J. Inflamm. Res. 11, 273–288. doi: 10.2147/JIR.S128401
Marino, G., and Kroemer, G. (2013). Mechanisms of apoptotic phosphatidylserine exposure. Cell Res. 23, 1247–1248. doi: 10.1038/cr.2013.115
Martinello, T., Baldoin, M. C., Morbiato, L., Paganin, M., Tarricone, E., Schiavo, G., et al. (2011). Extracellular ATP signaling during differentiation of C2C12 skeletal muscle cells: role in proliferation. Mol. Cell. Biochem. 351, 183–196. doi: 10.1007/s11010-011-0726-4
Meech, R., Gonzalez, K. N., Barro, M., Gromova, A., Zhuang, L., Hulin, J. A., et al. (2011). Barx2 is expressed in satellite cells and is required for normal postnatal muscle growth and regeneration. Stem Cells 23, 1247–1248.
Melo, F., Carey, D. J., and Brandan, E. (1996). Extracellular matrix is required for skeletal muscle differentiation but not myogenin expression. J. Cell. Biochem. 62, 227–239. doi: 10.1002/(SICI)1097-4644(199608)62:2<227::AID-JCB11>3.0.CO;2-I
Meyer, M. P., Groschel-Stewart, U., Robson, T., and Burnstock, G. (1999). Expression of two ATP-gated ion channels. P2X5 and P2X6, in developing chick skeletal muscle. Dev. Dyn. 216, 442–449. doi: 10.1002/(SICI)1097-0177(199912)216:4/5<442::AID-DVDY12>3.0.CO;2-Z
Otto, A., Collins-Hooper, H., Patel, A., Dash, P. R., and Patel, K. (2011). Adult skeletal muscle stem cell migration is mediated by a blebbing/amoeboid mechanism. Rejuvenation Res. 14, 249–260. doi: 10.1089/rej.2010.1151
Otto, A., Schmidt, C., Luke, G., Allen, S., Valasek, P., Muntoni, F., et al. (2008). Canonical Wnt signalling induces satellite-cell proliferation during adult skeletal muscle regeneration. J. Cell Sci. 121, 2939–2950. doi: 10.1242/jcs.026534
Panchin, Y. V. (2005). Evolution of gap junction proteins–the pannexin alternative. J. Exp. Biol. 208, 1415–1419. doi: 10.1242/jeb.01547
Panchin, Y., Kelmanson, I., Matz, M., Lukyanov, K., Usman, N., and Lukyanov, S. (2000). A ubiquitous family of putative gap junction molecules. Curr. Biol. 10, R473–R474. doi: 10.1016/S0960-9822(00)00576-5
Panupinthu, N., Zhao, L., Possmayer, F., Ke, H. Z., Sims, S. M., and Dixon, S. J. (2007). P2X7 nucleotide receptors mediate blebbing in osteoblasts through a pathway involving lysophosphatidic acid. J. Biol. Chem. 282, 3403–3412. doi: 10.1074/jbc.M605620200
Pelegrin, P., and Surprenant, A. (2006). Pannexin-1 mediates large pore formation and interleukin-1beta release by the ATP-gated P2X7 receptor. EMBO J. 25, 5071–5082. doi: 10.1038/sj.emboj.7601378
Penuela, S., Bhalla, R., Gong, X. Q., Cowan, K. N., Celetti, S. J., Cowan, B. J., et al. (2007). Pannexin 1 and pannexin 3 are glycoproteins that exhibit many distinct characteristics from the connexin family of gap junction proteins. J. Cell Sci. 120, 3772–3783. doi: 10.1242/jcs.009514
Pham, T. L., St-Pierre, M. E., Ravel-Chapuis, A., Parks, T. E. C., Langlois, S., Penuela, S., et al. (2018). Expression of Pannexin 1 and Pannexin 3 during skeletal muscle development, regeneration, and Duchenne muscular dystrophy. J. Cell. Physiol. 233, 7057–7070. doi: 10.1002/jcp.26629
Prochnow, N., Abdulazim, A., Kurtenbach, S., Wildforster, V., Dvoriantchikova, G., Hanske, J., et al. (2012). Pannexin1 stabilizes synaptic plasticity and is needed for learning. PLoS One 7:e51767. doi: 10.1371/journal.pone.0051767
Py, B. F., Jin, M., Desai, B. N., Penumaka, A., Zhu, H., Kober, M., et al. (2014). Caspase-11 controls interleukin-1beta release through degradation of TRPC1. Cell Rep. 6, 1122–1128. doi: 10.1016/j.celrep.2014.02.015
Qiu, F., and Dahl, G. (2009). A permeant regulating its permeation pore: inhibition of pannexin 1 channels by ATP. Am. J. Physiol. Cell Physiol. 296, C250–C255. doi: 10.1152/ajpcell.00433.2008
Qu, Y., Misaghi, S., Newton, K., Gilmour, L. L., Louie, S., Cupp, J. E., et al. (2011). Pannexin-1 is required for ATP release during apoptosis but not for inflammasome activation. J Immunol 186, 6553–6561. doi: 10.4049/jimmunol.1100478
Rando, T. A., and Blau, H. M. (1994). Primary mouse myoblast purification, characterization, and transplantation for cell-mediated gene therapy. J. Cell Biol. 125, 1275–1287. doi: 10.1083/jcb.125.6.1275
Riquelme, M. A., Cea, L. A., Vega, J. L., Boric, M. P., Monyer, H., Bennett, M. V., et al. (2013). The ATP required for potentiation of skeletal muscle contraction is released via pannexin hemichannels. Neuropharmacology 75, 594–603. doi: 10.1016/j.neuropharm.2013.03.022
Ryten, M., Yang, S. Y., Dunn, P. M., Goldspink, G., and Burnstock, G. (2004). Purinoceptor expression in regenerating skeletal muscle in the mdx mouse model of muscular dystrophy and in satellite cell cultures. FASEB J. 18, 1404–1406. doi: 10.1096/fj.03-1175fje
Sacco, A., Mourkioti, F., Tran, R., Choi, J., Llewellyn, M., Kraft, P., et al. (2010). Short telomeres and stem cell exhaustion model Duchenne muscular dystrophy in mdx/mTR mice. Cell 143, 1059–1071. doi: 10.1016/j.cell.2010.11.039
Sambasivan, R., Yao, R., Kissenpfennig, A., Van Wittenberghe, L., Paldi, A., Gayraud-Morel, B., et al. (2011). Pax7-expressing satellite cells are indispensable for adult skeletal muscle regeneration. Development 138, 3647–3656. doi: 10.1242/dev.067587
Schenk, U., Westendorf, A. M., Radaelli, E., Casati, A., Ferro, M., Fumagalli, M., et al. (2008). Purinergic control of T cell activation by ATP released through pannexin-1 hemichannels. Sci. Signal. 1:ra6. doi: 10.1126/scisignal.1160583
Schwander, M., Leu, M., Stumm, M., Dorchies, O. M., Ruegg, U. T., Schittny, J., et al. (2003). Beta1 integrins regulate myoblast fusion and sarcomere assembly. Dev. Cell 4, 673–685. doi: 10.1016/S1534-5807(03)00118-7
Siegel, A. L., Atchison, K., Fisher, K. E., Davis, G. E., and Cornelison, D. D. (2009). 3D timelapse analysis of muscle satellite cell motility. Stem cells 27, 2527–2538. doi: 10.1002/stem.178
Signorello, M. G., and Leoncini, G. (2014). Effect of 2-arachidonoylglycerol on myosin light chain phosphorylation and platelet activation: the role of phosphatidylinositol 3 kinase/AKT pathway. Biochimie 105, 182–191. doi: 10.1016/j.biochi.2014.07.014
Stupka, N., Kintakas, C., White, J. D., Fraser, F. W., Hanciu, M., Aramaki-Hattori, N., et al. (2013). Versican processing by a disintegrin-like and metalloproteinase domain with thrombospondin-1 repeats proteinases-5 and -15 facilitates myoblast fusion. J. Biol. Chem. 288, 1907–1917. doi: 10.1074/jbc.M112.429647
Taye, N., Stanley, S., and Hubmacher, D. (2020). Stable Knockdown of Genes Encoding Extracellular Matrix Proteins in the C2C12 Myoblast Cell Line Using Small-Hairpin (sh)RNA. J. Vis. Exp. 156:10.3791/60824. doi: 10.3791/60824
Tinevez, J. Y., Schulze, U., Salbreux, G., Roensch, J., Joanny, J. F., and Paluch, E. (2009). Role of cortical tension in bleb growth. Proc. Natl. Acad. Sci. U.S.A. 106, 18581–18586. doi: 10.1073/pnas.0903353106
Velasquez, S., and Eugenin, E. A. (2014). Role of Pannexin-1 hemichannels and purinergic receptors in the pathogenesis of human diseases. Front. Physiol. 5:96. doi: 10.3389/fphys.2014.00096
Webster, M. T., Manor, U., Lippincott-Schwartz, J., and Fan, C. M. (2016). Intravital imaging reveals ghost fibers as architectural units guiding myogenic progenitors during regeneration. Cell Stem Cell 18, 243–252. doi: 10.1016/j.stem.2015.11.005
Wicki-Stordeur, L. E., and Swayne, L. A. (2013). Panx1 regulates neural stem and progenitor cell behaviours associated with cytoskeletal dynamics and interacts with multiple cytoskeletal elements. Cell Commun. Signal. 11:62. doi: 10.1186/1478-811X-11-62
Wilkinson, G. F., Purkiss, J. R., and Boarder, M. R. (1993). The regulation of aortic endothelial cells by purines and pyrimidines involves co-existing P2y-purinoceptors and nucleotide receptors linked to phospholipase C. Br. J. Pharmacol. 108, 689–693. doi: 10.1111/j.1476-5381.1993.tb12862.x
Woolley, T. E., Gaffney, E. A., Oliver, J. M., Baker, R. E., Waters, S. L., and Goriely, A. (2013). Cellular blebs: pressure-driven, axisymmetric, membrane protrusions. Biomech. Model. Mechanobiol. 13, 463–476. doi: 10.1007/s10237-013-0509-9
Woolley, T. E., Gaffney, E. A., Oliver, J. M., Waters, S. L., Baker, R. E., and Goriely, A. (2014). Three mechanical models for blebbing and multi-blebbing. IMA J. App. Math. 79, 636–660. doi: 10.1093/imamat/hxu028
Woolley, T. E., Gaffney, E. A., and Goriely, A. (2015a). Membrane shrinkage and cortex remodelling are predicted to work in harmony to retract blebs. Roy. Soc. Open Sci 2:150184. doi: 10.1098/rsos.150184
Woolley, T. E., Gaffney, E. A., Oliver, J. M., Waters, S. L., Baker, R. E., and Goriely, A. (2015b). Global contraction or local growth, bleb shape depends on more than just cell structure. J. Theor. Biol. 380, 83–97. doi: 10.1016/j.jtbi.2015.04.023
Xia, J., Lim, J. C., Lu, W., Beckel, J. M., Macarak, E. J., Laties, A. M., et al. (2012). Neurons respond directly to mechanical deformation with pannexin-mediated ATP release and autostimulation of P2X7 receptors. J. Physiol. 590, 2285–2304. doi: 10.1113/jphysiol.2012.227983
Keywords: Panx1, lipid signaling, myoblast migration, pannexins, blebbing, myoblast fusion
Citation: Suarez-Berumen K, Collins-Hooper H, Gromova A, Meech R, Sacco A, Dash PR, Mitchell R, Shestopalov VI, Woolley TE, Vaiyapuri S, Patel K and Makarenkova HP (2021) Pannexin 1 Regulates Skeletal Muscle Regeneration by Promoting Bleb-Based Myoblast Migration and Fusion Through a Novel Lipid Based Signaling Mechanism. Front. Cell Dev. Biol. 9:736813. doi: 10.3389/fcell.2021.736813
Received: 05 July 2021; Accepted: 13 September 2021;
Published: 05 October 2021.
Edited by:
Pamela Imperadore, Zoological Station Anton Dohrn, ItalyReviewed by:
Juan C. Saez, Pontificia Universidad Católica de Chile, ChileCopyright © 2021 Suarez-Berumen, Collins-Hooper, Gromova, Meech, Sacco, Dash, Mitchell, Shestopalov, Woolley, Vaiyapuri, Patel and Makarenkova. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ketan Patel, a2V0YW4ucGF0ZWxAcmVhZGluZy5hYy51aw==; Helen P. Makarenkova, aG1ha2FyZW5rQHNjcmlwcHMuZWR1
†These authors have contributed equally to this work
‡These authors have contributed equally to this work and share senior authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.