- 1Shenzhen University-Friedrich Schiller Universität Jena Joint Ph.D. Program in Biomedical Sciences, Shenzhen University School of Medicine, Shenzhen, China
- 2Guangdong Key Laboratory for Genome Stability and Disease Prevention and Marshall Laboratory of Biomedical Engineering, Shenzhen University School of Medicine, Shenzhen, China
- 3Leibniz Institute on Aging—Fritz Lipmann Institute (FLI), Jena, Germany
- 4Faculty of Biological Sciences, Friedrich-Schiller-University Jena, Jena, Germany
Iron–sulfur (Fe/S) clusters (ISCs) are redox-active protein cofactors that their synthesis, transfer, and insertion into target proteins require many components. Mitochondrial ISC assembly is the foundation of all cellular ISCs in eukaryotic cells. The mitochondrial ISC cooperates with the cytosolic Fe/S protein assembly (CIA) systems to accomplish the cytosolic and nuclear Fe/S clusters maturation. ISCs are needed for diverse cellular functions, including nitrogen fixation, oxidative phosphorylation, mitochondrial respiratory pathways, and ribosome assembly. Recent research advances have confirmed the existence of different ISCs in enzymes that regulate DNA metabolism, including helicases, nucleases, primases, DNA polymerases, and glycosylases. Here we outline the synthesis of mitochondrial, cytosolic and nuclear ISCs and highlight their functions in DNA metabolism.
Introduction
Iron–sulfur (Fe/S) clusters (ISCs) are extremely ancient, small inorganic protein cofactors found in almost all organisms. Ferredoxin was discovered in the early 1960s, since then, the number of known Fe/S clusters-containing proteins has steadily increased. Until now, over 120 unique types of enzymes and proteins have been identified as ISC-containing proteins (Johnson et al., 2005). Until now, there are more than 200 known Fe/S proteins in human cells according to the UniProt database1. And bacteria contain a great variety of such proteins (Andreini et al., 2017). ISC proteins are found in the nucleus, cytosol, and mitochondria. The essentials of ISC proteins are reflected in the fact that they are required for many fundamental biochemical processes. For example, within mitochondria, the respiratory complexes I, II and III use many ISCs to transfer electrons which reduces ubiquinone by NADH or FADH, respectively. Within the nucleus, ISCs are functionally related to the maintenance of genome stability, RNA modification, and gene regulation. Specifically, ISCs are inserted into DNA repair enzymes, which fix DNA lesions according to the diffusing ability of an electron from an ISC along DNA (Arnold et al., 2016). Defects in mitochondrial ISC biogenesis can result in nuclear genomic instability (Veatch et al., 2009). Various nuclear DNA metabolic enzymes require ISCs to carry out DNA metabolism, including DNA primase, DNA polymerases (Klinge et al., 2007), DNA glycosylases (Alseth et al., 1999), and ATP-dependent DNA helicases (Rudolf et al., 2006; Gari et al., 2012; Stehling et al., 2012).
Mitochondrial Iron–Sulfur (Fe/S) Cluster Biogenesis
There are three independent mechanisms that can synthesize ISCs in bacteria: the ISC assembly, methanoarchaeal sulfur mobilization (SUF) (Takahashi and Tokumoto, 2002), and nitrogen fixation (NIF) pathways (Mettert and Kiley, 2015). Each of these mechanisms shares the same steps: iron and sulfur ions are assembled at scaffold complexes. And then, the transfer system delivers the clusters to target proteins (Roche et al., 2013).
Eukaryotic mitochondria have one dedicated assembly pathway that inherits the ISC pathway from bacteria and integrate the NIF system components (Gisselberg et al., 2013; Roche et al., 2013). The ISCs in the cytoplasm and nucleus are assembled by the CIA pathway. In mammalian cells, there are two major forms of ISCs: 2Fe–2S and 4Fe–4S clusters (Figure 1). These two types of cofactors are generated by two related biochemical machineries in the cytosol (CIA pathway) and mitochondria (ISC pathway), respectively (Maio et al., 2020). The mitochondrial machinery assembly a necessary sulfur-containing intermediate that is exported to the cytoplasm and utilized for extramitochondrial ISCs assembly.
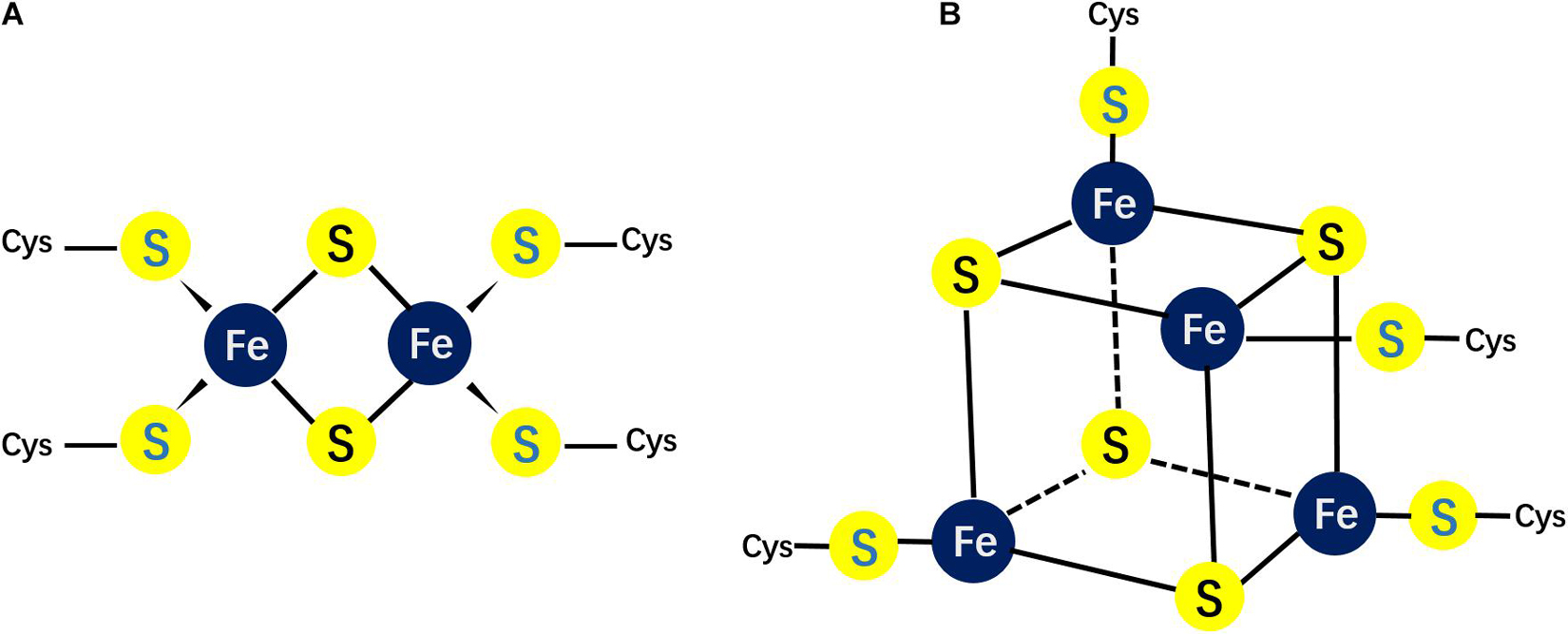
Figure 1. Different possible structures of Fe/S clusters. (A) The structure of the rhombic 2Fe–2S cluster; (B) the structure of the cubane 4Fe–4S cluster.
Mitochondrial ISC biogenesis has two functions: (1) to synthesize functional clusters in the mitochondria, and (2) to provide an essential precursor to the CIA pathway via the inner membrane exporter ABCB7 (Lill et al., 2015). Briefly, persulfide ions are generated by cysteine desulfurase (NFS1). Iron and sulfide ions are then delivered to a scaffold protein ISCU2 to form an initial 2Fe–2S cluster. Then chaperones transfer this 2Fe–2S cluster to a glutaredoxin, which subsequently delivers the 2Fe–2S cluster to the target protein or to the next Iron-sulfur assembly protein (ISA) complex. The ISA complex can condense two 2Fe–2S clusters into one 4Fe–4S center (Beilschmidt and Puccio, 2014). Major components involved in the ISC pathway are shown in Table 1. In sum, 2Fe-2S and 4Fe-4S proteins are made differently, with de novo 2Fe-2S clusters forming on the ISCU scaffold and 4Fe-4S clusters forming subsequently in a downstream step utilizing the ISA complex of proteins.
2Fe–2S Cluster Biogenesis
The 2Fe–2S clusters in rhombic form possess 2 sulfide ions and 2 irons, which coordinate four cysteinyl sulfhydryl side chains (Figure 1A). This rhombic-form cluster exhibits two oxidation states: the oxidized status with two Fe3++, and the reduced status with one Fe3+ and one Fe2+. As mentioned, the mitochondrial ISC pathway is essential for ISC biogenesis. This pathway starts with delivering iron and sulfur ions to scaffold protein ISCU2. ISCU has two isoforms: the mitochondrial isoform ISCU2 and cytosolic and nuclear isoform ISCU1. Cysteine desulfurase NFS1, which interacts with ISD11 and ACP to form a stable complex provides sulfur. However, the iron source of ISC is unknown yet. Frataxin (FXN) and ferredoxin2 (FDX2) are also important for ISCs de novo assembly. The former is thought to regulate NFS1 activity (Fox et al., 2019), while the latter is proposed to donate electrons for reduction (Cai et al., 2017; Gervason et al., 2019).
The eukaryotic cysteine desulfurase NFS1 is a pyridoxal phosphate-dependent enzyme. Sulfur is transferred from cysteine and activated into a persulfide form, which can be used for ISC assembly. In Cory et al. (2017), the first investigation into the eukaryotic NFS1 crystal structure revealed some key features: First, NFS1 binding to its substrate cysteine relies on a pyridoxal phosphate (PLP) cofactor. Second, there is a metal-binding cysteine site located in the C-terminus of NFS1. The activated sulfur abstracted is transferred to this cysteine site. Third, similar to the prokaryotic version of the enzyme, NFS1 forms a dimer. In the field of the enzymatic cycle, the PLP cofactor mediated the interaction of NFS1 and substrate cysteine. Then, NFS1 conformational change results in closing the activity site of cysteine with substrate and proceeding a nucleophilic attach (Johnson et al., 2005). During this second step, additional eukaryotic-specific subunits (ISD11 and ACP) interact with NFS1 and form a stable complex. ISD11 is a small protein of the LYR (Leu-Tyr-Arg motif) family, and ACP, which is an acyl carrier protein, regulates fatty acid synthesis. The long chain fatty acid of ACP is inserted into the helical center of the ISD11 subunit (Pandey et al., 2012; Parent et al., 2015; Cory et al., 2017). Finally, the persulfide sulfur is moved from NFS1 to the scaffold protein ISCU2 for ISC assembly.
The transfer of persulfur to the ISCU2 scaffold seems to be mediated by frataxin (Brancaccio et al., 2014; Fox et al., 2015). Frataxin is the earliest identified as a positive modulatory factor for NFS1 and ISC assembly. Frataxin interacts with NFS1 and enhance NFS1 cysteine desulfurase activity. Human neurodegenerative disease Friedreich’s ataxia results from the deficient of frataxin. Deletion of the yeast orthologous gene, Yfh1, leads to excess iron accumulation in mitochondria (Campuzano et al., 1996; Babcock et al., 1997; Karthikeyan et al., 2002). Because of frataxin’s weak Fe(II)-binding ability, it might be a potential iron donor to scaffold protein ISCU2 (Adamec et al., 2000; Yoon and Cowan, 2003). Structural studies have revealed that neither iron nor ISU oligomerization is essential for the interaction between bacterial frataxin, CyaY, and the ISU complex. Prokaryotic frataxin directly interacts with the bacterial desulfurase, IscS, but not the scaffold, IscU (Prischi et al., 2010). There are two apparently different reports for frataxin functions. One report suggested that frataxin promotes the interaction between NFS1 and substrate (Pandey et al., 2013), while frataxin was reported to promote sulfide transfer from NFS1 to ISCU1 (Parent et al., 2015). Furthermore, frataxin enhances sulfide transfer to ISCU1, forming a 2Fe–2S product on the ISCU scaffold (Bridwell-Rabb et al., 2014).
Transfer of 2Fe–2S Clusters
Once the initial 2Fe–2S cluster has been generated by the ISC system in mitochondria, it is transferred to the glutaredoxin 5 (GLRX5) dimer with the help of HSC20 and HSPA9. This process requires energy, which is supplied by ATP hydrolysis carried out by HSPA9 (Dutkiewicz et al., 2003). The binding of the chaperones HSC20 and HSPA9 leads to the dissociation of the assembly complex consisting NFS1/ISD11/ISCU/ACP/FXN. Competition of FXN with HSPA9 for the LPPVK binding site on ISCU acts as a molecular switch between assembly and transfer complexes (Majewska et al., 2013; Manicki et al., 2014). Cluster–bound GLRX5 includes a 2Fe–2S cluster connecting a GLRX5 dimer. Each GLRX5 contributes one cysteine ligand to the 2Fe–2S cluster, while a second thiolate ligand coming from a GLRX5–bound glutathione stably binds GLRX5 (Banci et al., 2014). glrx5 deletion in yeast cells has dysfunctional phenotypes in both mitochondrial ISC de novo biogenesis and cytosolic ISC assembly (Muhlenhoff et al., 2003; Uzarska et al., 2013).
Mitochondrial 4Fe–4S Cluster Formation
The cubane-type cluster comprises 4 iron and 4 sulfide ions coordinated to four sulfhydryl side chains (Figure 1), which can be subdivided into low- or high-potential clusters39. The oxidation states of the low-potential clusters are the oxidized [2Fe3+, 2Fe2+] and the reduced [Fe3+, 3Fe2+] forms, while the oxidation states for the high potential clusters switch between the reduced [2Fe3+, 2Fe2+] and the oxidized [3Fe3+, Fe2+] forms. Hence, the two ferric-two ferrous state is shared by the two families. Within a cell, the most common clusters are 4Fe–4S clusters. The ISA system mediates the transformation of 2Fe–2S clusters into 4Fe–4S clusters in the mitochondrial matrix. Unlikely to mitochondrial ISC biogenesis system, the cytoplasm contains different machineries to assembly 4Fe–4S clusters. The biogenesis of cytoplasmic 4Fe–4S clusters rely on substrate exported from the matrix by ABCB7 (Lill et al., 2015).
In brief, ISCA1, ISCA2, and IBA57 are responsible for the generation of mitochondrial 4Fe–4S clusters. ISCA1 and ISCA2 interact with each other (Muhlenhoff et al., 2011; Beilschmidt et al., 2017), and structure study reveals that ISCA2, but not ISCA1, is able to bind IBA57 (Muhlenhoff et al., 2011; Beilschmidt et al., 2017; Gourdoupis et al., 2018). Two GLRX5-derived 2Fe–2S clusters are converted to a 4Fe–4S cluster on the ISCA1-ISCA2 complex. This process required the presence of IBA57 and the electron transfer chain NADPH-FDXR-FDX2 (Weiler et al., 2020). Finally, NFU1 promotes the 4Fe–4S cluster transfer from the ISCA1-ISCA2-IBA57 complex to apoproteins. However, one biochemical study indicated that only ISCA1, but neither ISCA2 nor IBA57, is needed for the maturation of the 4Fe–4S cluster in mouse skeletal muscle and in primary neurons (Beilschmidt et al., 2017).
ISA system is only responsible for mitochondrial 4Fe–4S clusters biogenesis. In the absence of the ISA complex, cells showed mitochondrial function defects with uncompromised cellular viability, since the sulfur-containing component required for cytoplasmic Fe/S biogenesis still can be exported from mitochondria.
Biogenesis of Cytosolic and Nuclear Iron–Sulfur (Fe/S) Clusters
Except mitochondrial, there are also abundant ISC proteins located in cytosolic and nuclear in eukaryotic cells. And these proteins are involved in multiple biological processes. For example, DNA metabolism, iron regulation and metabolic catalysis. These processes are catalyzed by the CIA machinery. As discussed, the mitochondrial ISC assembly system generates an initial sulfur-containing intermediate and export the compound from mitochondria by the inner membrane ABC transporter, ABCB7 (Kispal et al., 1999; Gerber et al., 2004; Fosset et al., 2006; Lill et al., 2014). The exported intermediate is necessary for cytoplasmic ISC synthesis by the CIA system. However, chemical characterization and isolation of the intermediate is a subject of ongoing research. Glutathione (GSH) and the intermembrane space protein, ALR are important for this process (Kispal et al., 1999; Sipos et al., 2002; Pondarre et al., 2006; Cavadini et al., 2007). ALR is a FAD-dependent sulfhydryl oxidase. ALR inserts disulfide bridges into mitochondrial preproteins during their import into the intermembrane space (Mesecke et al., 2005). But another group reported that Cytosolic ISC protein maturation and iron regulation are independent of the mitochondrial Erv1/Mia40 import system. After the sulfur-containing compound was transferred to cytosolic, nine proteins of the CIA system are responsible for generating the cytosolic ISCs and inserting them into target proteins (Sharma et al., 2010; Basu et al., 2014; Netz et al., 2014). Major components involved in the CIA system are shown in Table 2. In the next section, the two essential steps of cytosolic and nuclear ISC assembly will be described.
Step 1 of Cytosolic and Nuclear Iron–Sulfur (Fe/S) Cluster Assembly
Similar to ISC biogenesis in mitochondria, the initial step of cytosolic and nuclear ISC synthesis is transient transfer a 4Fe–4S cluster to the cytosolic scaffold protein complex. This complex comprises two P-loop NTPases CFD1 and NBP35 (Roy et al., 2003; Hausmann et al., 2005; Netz et al., 2007; Stehling et al., 2018). CFD1 interact with NBP35 and form a heterotetrameric complex. This complex is able to coordinate two different types of 4Fe–4S clusters (Netz et al., 2012). One type of 4Fe–4S cluster can loosely bind to a conserved CX2C motif that is located at the C-termini of CFD1 and NBP35. The second type of 4Fe–4S clusters bind at a ferredoxin-like CX13CX2CX5C motif, which is located at the N terminus of NBP35. This motif is essential for NBP35 function. Interestingly, a pulse-chase experiment with 55Fe labeled yeast cells revealed the different labilities of the two ISCs associated with the CFD1–NBP35 complex (Pallesen et al., 2013). The 4Fe–4S cluster that binds to the N terminus of NBP35 is more stable than the 4Fe–4S cluster that binds the C-terminus, which transfers the loose-binding 4Fe–4S cluster to target proteins.
Another feature of cytosolic and nuclear ISC biogenesis that is similar to mitochondrial ISCs biogenesis is the dependency on a supply of electrons (Webert et al., 2014). The electron transfer chain of the CIA system comprises NADPH, NDOR1, and the Fe/S protein CIAPIN1 (Netz et al., 2010; Banci et al., 2013). NDOR1 is a key member of the electron transfer chain. It contains NADPH-, FAD- and FMN- binding domains. Protein-protein interaction and high-throughput studies have demonstrated that CIAPIN1 physically interacts with NDOR1 (Vernis et al., 2009). Dre2 is the CIAPIN1 yeast homolog: it is a crucial component of the cytosolic and nuclear ISC biogenesis system. The synthetically lethal effect was observed when deletion Dre2 and mitochondrial iron importers (Mrs3 and Mrs4) (Zhang et al., 2008). Dre2 contains a conserved C-terminal Fe/S domain which is responsible for coordinating one 2Fe–2S or one 4Fe–4S cluster by cysteine residues. The N-terminal of Dre2 is a SAM methyl-transferase-like domain, the middle linker domain of Dre2 mediate the connection of the N-terminus and C-terminus (Zhang et al., 2008; Netz et al., 2010, 2014).
Step 2 of Cytosolic and Nuclear Iron–Sulfur (Fe/S) Cluster Assembly
The second step of cytosolic ISC biogenesis is initialed by releasing the newly assembled 4Fe–4S cluster from the CFD1-NBP35 scaffold complex. Then, the 4Fe–4S cluster is inserted into targeted proteins (Balk et al., 2004, 2005; Song and Lee, 2008, 2011). Both the CIA targeting complex and the iron-only hydrogenase-like protein, IOP1, are essential for this step reaction. Nar1 is the yeast ortholog of IOP1. Structure study of Nar1 revealed that the four conserved Cys residues, which are in the C-terminal of Nar1 are responsible for binding ISCs. The CIA targeting complex comprises CIA1, CIA2B, and human ortholog for the yeast methyl methanesulfonate-sensitivity protein 19 (MMS19). And these components physically interact with a large number of target proteins in the cytoplasm and nucleus (Srinivasan et al., 2007; Weerapana et al., 2010; van Wietmarschen et al., 2012; Stehling et al., 2013; Kassube and Thoma, 2020). The cytosolic ISC biogenesis contains two stages. In yeast, inactivation of early stage CIA assembles complex leads to the immaturity of Fe/S protein Nar1, while defect of late-stage proteins CIA1, CIA2B, and MMS19 do not affect ISC insert to target proteins (Balk et al., 2004). Base on this study, the early and late stage of CIA system is connected by Nar1via an unknown mode of action (Stehling et al., 2013).
CIA targeting complex component CIA1 contains seven WD40-repeat domains. The structural analysis demonstrated that these seven WD40-repeats distribute around a central axis, which functions as binding region docking site of the CIA targeting complex (Srinivasan et al., 2007). Point mutation of CIA1 has revealed that the conserved, surface-exposed residue R127 is responsible for assembling other subunits of cytosolic Fe/S protein (Paul and Lill, 2015). The conserved Cys residue is important for CIA2 function, which is also conserved in eukaryotes (Weerapana et al., 2010; Luo et al., 2012; Stehling et al., 2013). Knockout human CIA2B or its ortholog Cia2 suppresses the Fe/S proteins maturation (Chen et al., 2012). MMS19 contains 4 HEAT repeats at N-terminal. As the largest component of CIA, MMS19 is associated with the multitude of biological processes. For example, impaired chromosome segregation, defective double-strand break repair via homologous recombination, and immature cytosolic and nuclear Fe/S proteins (Prakash and Prakash, 1977; Lauder et al., 1996; Kou et al., 2008; Ito et al., 2010). For a long time, it was difficult to associate these phenotypes of MMS19-deficient cells with one molecular function. Until known that MMS19 is involved in cytosolic ISC biogenesis, this problem was resolved (Gari et al., 2012; Stehling et al., 2012). As a major determinant of the CIA targeting complex, MMS19 interacts with numerous target proteins and promotes the insertion of ISCs into them, including key enzymes in DNA synthesis (POLD1, PRIM2), DNA repair [XPD, DNA2 (DNA replication helicase/nuclease 2)], and telomere length regulation (RTEL1). Deletion of these enzymes, respectively, phenocopied variant MMS19 depletion defects.
The Close Link Between Iron–Sulfur (Fe/S) Clusters and Genome Integrity
Mitochondria are organelles with a double-layer membrane found in most eukaryotic organisms. They generate most of the cellular chemical energy via oxidative phosphorylation. In addition to supplying energy, mitochondria are also involved in multitude of cellular biochemical processes such as programmed cell death, reactive oxygen species (ROS) production, and ISCs biogenesis. Biochemical studies revealed that mitochondrial DNA (mtDNA) defection result in nuclear genome instability and reduction of cells’ viability in yeast. This effect is due to the important role of mitochondria ISC biogenesis (Veatch et al., 2009). As mentioned, down-regulation of Nar1 is sufficient to alter nuclear genome stability (Gari et al., 2012). Consistent with this study, deletion of Zim17, an important component of ISC biogenesis, leading to genomic instability (Diaz de la Loza Mdel et al., 2011; Stehling et al., 2013). Given that many enzymes that are required for DNA synthesis and repair harbor ISC cofactors, these observations suggest that defects in Fe/S biogenesis and distribution are likely to be the origin of genomic instability (Ben-Aroya et al., 2008; Veatch et al., 2009; Gari et al., 2012; Stehling et al., 2012; Stehling et al., 2013). For example, MMS19 was identified as a gene involved in transcription conducted by RNA polymerase II, nucleotide excision repair (NER), and methionine biosynthesis (Prakash and Prakash, 1977; Thomas et al., 1992). In yeast, the essential transcription factor IIH (TFIIH) complex is required for transcription-coupled NER (Lauder et al., 1996). MMS19 is not a component of the TFIIH complex, while numerous studies demonstrate that MMS19 is crucial to maintain the cellular Rad3 (XPD in human) protein level, a component of the TFIIH (Kou et al., 2008). Consistent with these findings, human MMS19 homolog also is reported to be involved in the NER pathway by regulating TFIIH function. In addition to regulating TFIIH function, MMS19 also directly interacts with CIA components CIA1 and CIA2B. And this interaction is important for regulating chromosome segregation and telomere length (Askree et al., 2004; Ito et al., 2010). All these MMS19 functional studies reveal the different phenotypes observed in MMS19 deficient cells (Gari et al., 2012; Stehling et al., 2012).
The function of MMS19 in DNA metabolism has been reported in multitude of ways. Many biochemical studies demonstrate that MMS19 and other CIA complex components directly interact with diverse DNA metabolism enzymes, such as DNA helicases [XPD, FANCJ (Fanconi anemia complementation group J)], and RTEL1, DNA polymerase subunits (POLD1, POLA1, and POLE1), the nuclease DNA2, the DNA glycosylase NTHL1, and the DNA primase PRI2. With the help of MMS19, these enzymes coordinate an ISC. Biochemical studies have revealed that MMS19 mediates the interaction of XPD and TFIIH, which is important for DNA metabolism. In yeast, deletion of Met18/Mms19, CIA complex components, increases phosphorylation of Rad3 and promotes Rad3-dependent gene expression (Gari et al., 2012; Stehling et al., 2012). Consist with this finding, cells lacking CIA complex proteins are very sensitive to DNA damage events, e.g., UV and chemical agents. Based on these studies, MMS19 not only is involved in the CIA complex for the maturity of target ISC proteins, but also plays a crucial role in DNA metabolism.
Recently, one biochemical study also indicated that inhibition of ISCs synthesis via NFS1 depletion in elevated O2 environment led to decreased POLE protein level. This perturbation reduces Pol ε activity and causes replication stress (Sviderskiy et al., 2020).
Iron–Sulfur (Fe/S) Clusters and DNA Replication
High-fidelity DNA replication ensures the accurate transmission of parental genetic information to daughter cells. This process is coordinated by numerous enzymes (Bell and Dutta, 2002). Firstly, the DNA helicases open the double-stranded DNA. Then, the DNA primases initiate DNA synthesis via assembling short RNA primers, which are extended by DNA polymerases. DNA polymerases then utilize the two parental DNA strands as templates to synthesize complementary strands, but not to start de novo DNA replication (Bell and Dutta, 2002). During the DNA replication process, DNA2, a helicase/nuclease, is critical for lagging strand DNA replication via processing Okazaki fragment (Kang et al., 2010). The ISCs are critical for the proper functions of all three types of enzymes (Table 3). In the next section, we will describe the three types of replication factors that coordinate these crucial ISCs.
Iron–Sulfur (Fe/S) Clusters and Helicases
Helicases are a class of motor proteins that can unwind structured nucleic acids in an ATP-dependent manner. In this way, helicases can regulate many different processes that depend on strand separation during DNA metabolism (Lohman and Bjornson, 1996), including transcription, DNA replication, DNA repair, and telomere length regulation. Thus, helicases are important for genomic stability (Patel and Donmez, 2006; Brosh and Bohr, 2007; Lohman et al., 2008; Pyle, 2008). Helicases are classified into six super-families according to their primary amino acid sequences, and ISCs exist in numerous helicases.
In the helicase super-family 1, DNA2 is a multifunction enzyme not only involved in DNA replication, but also in double-strand DNA break (DSB) repair and telomere maintenance (Budd et al., 2005; Kang et al., 2010; Balakrishnan and Bambara, 2011). AddAB which contains a 4Fe–4S cluster, is a helicase-nuclease complex in bacteria. Eukaryotic helicase-nuclease DNA2 putative metal-binding motif was identified by sequence alignment with AddAB. Interestingly, structure studies revealed that four conserved Cys residues which are coordinated ISCs exist in the nuclease domain. This suggests that ISCs might function in stabilizing the nuclease domain conformation (Yeeles et al., 2009). Biochemical studies confirmed that yeast DNA2 coordinates ISCs by its conserved Cys residues (Pokharel and Campbell, 2012). ISC binding cysteine residues mutation results in nuclease activity and ATPase function defects in DNA2. However, the DNA binding ability of DNA2 is normal. Another biochemical study revealed that pro residue at position 504 of DNA2 is crucial to stabilize the ISC. These studies confirmed that the ISC regulates DNA2 nuclease and helicase activities by mediating conformational changes.
In the helicase super-family 2, an XPD homolog from Archaea was the first DNA repair helicase to be identified. A sequence alignment revealed that all the XPD helicase family members contain four highly conserved Cys residues. These conserved Cys residues which coordinate an ISC are crucial for 5′–3′ DNA helicases activity. The XPD helicase family comprises XPD and several related super-family 2 DNA helicases including DDX11/ChlR1 (DEAD/DEAH box helicase 11), RTEL1 (regulator of telomere elongation 1), and FANCJ (Fanconi anemia complementation group J). Many human diseases are linked to mutations in these three proteins (Table 3; White, 2009; Wu and Brosh, 2012).
XPD is a crucial subunit of the transcription initiation factor TFIIH, which is involved in NER and transcription (Compe and Egly, 2012). TFIIH comprises two major functional subcomplexes, a core complex (XPB, p8, p34, p44, p52, and p62), and a CAK (CDK–activating kinase) complex (cyclin H, CDK7, and MAT1). Helicase XPD is an important bridge between these two subcomplexes.
The mutations of XPD gene are related to three genetic diseases: xeroderma pigmentosum (XP), Cockayne syndrome (CS), and trichothiodystrophy (TTD) (White, 2009; Wu and Brosh, 2012). All three disorders have similar characteristics, with patients’ skin being hypersensitive to sun exposure. This is due to the defect of the NER pathway (Lehmann, 2003). In 2006, biochemical and spectroscopic analyses indicated that XPD coordinates a 4Fe–4S cluster, which is a key determinant of XPD helicase activity. This finding significantly contributes to revealing the molecular differences of how mutations in a single gene result in different diseases (Stehling et al., 2012). Subsequently, structural analysis showed that the 4Fe–4S domain forms a channel with an arch domain, that can accommodate single-stranded DNA (ssDNA) (Fan et al., 2008; Liu et al., 2008; Wolski et al., 2008). Mutational analysis of conserved cysteine residues in the Fe/S domain of XPD indicated that an intact Fe/S domain is essential for helicase activity and/or stabilizing the protein structure (Liu et al., 2008; Pugh et al., 2008). For patients with XP, mutations in XPD primarily inhibit helicase activity without affecting the protein structure. Interestingly, all XP-causing mutations are conserved in archaeal XPD (Fan et al., 2008; Liu et al., 2008). However, most of the mutated residues in TTD are not conserved in the archaeal protein (Liu et al., 2008). In TTD patients, R112H exchange is the most common mutation. This amino acid substitution leads to loss of XPD helicase activity and deficiency of NER (Dubaele et al., 2003). Biochemical and structural studies demonstrated that this Arg residue is essential for the Fe/S domain. These findings underscore the structural role played by the ISC in helicase activity and highlight the close relationship between ISCs and DNA replication. In addition to these disease-associated mutations, other mutations of XPD destabilize the helicase structure and compromise interactions between the two TFIIH sub-complexes (Dubaele et al., 2003; Liu et al., 2008).
FANCJ, which was able to interact with the breast cancer C-terminal (BRCT) repeats of BRCA1, is another important member of the XPD helicase family (Cantor et al., 2001). FANCJ has been identified as the gene that is mutated in the J complementation group of Fanconi anemia (FA), a genome instability disorder with an elevated risk of developing cancer. FANCJ is known as an anti-oncogene because of its functions in DNA repair (Wu and Brosh, 2009). The substitution A349P in FANCJ is a common mutation seen in patients with FA (Wu et al., 2010). Although this alanine is not a conserved site in the XPD helicase family, the residue is near the fourth highly conserved cysteine residue in the ISC. Consistent with this, recombinant FANCJ-A349P protein was shown to decrease iron content and inhibit the separation of double-stranded DNA (dsDNA) (Wu et al., 2010). This finding indicates that, like XPD helicase, the catalytic activities of FANCJ critically rely on an intact Fe/S domain.
DDX11/CHLR is the third member of the XPD helicase family. The genetic disease Warsaw breakage syndrome (WABS) arises from a mutation in the human CHLR1 gene (van der Lelij et al., 2010). In S. cerevisiae, a mutation in chl1 causes chromosome loss and unusual mating phenotypes (Liras et al., 1978). Consistent with this finding, mutations in chl1 or CHLR1, the human homolog, show similar results (Skibbens, 2004; Parish et al., 2006). Unsurprisingly, patient-derived mutations also abolish helicase activity due to their perturbance of DNA binding and DNA-dependent ATPase activity (Wu et al., 2012).
Iron–Sulfur (Fe/S) Clusters and DNA Primase
A common feature of all DNA polymerases is that they are unable to initiate de novo synthesis of a DNA strand; they can only elongate an existing strand. Synthesis of a new strand can only begin from a primer with the 3′-OH end. Hence, a primase is required to catalyze the priming, form a primer, and initiate DNA replication. Primase in eukaryotic cells comprises two subunits, the catalytic PRIM1 subunit, and a large subunit PRIM2, both interacted with DNA polymerase-α (Frick and Richardson, 2001; Kang et al., 2010). Although only the PRIM1 subunit possesses catalytic activity, PRIM2 is also crucial for primase function (Zerbe and Kuchta, 2002). Spectroscopic analysis indicates that PRIM2 is able to bind a 4Fe–4S cluster, which is conserved from Archaea to eukaryotic cells (Weiner et al., 2007). Without this ISC, its enzymatic activity is compromised. High-resolution structural studies show that the conserved Lys314 in the C-terminal domain of human PRIM2 is supported by the 4Fe–4S cluster. This Lys314 mutant abolishes primer synthesis and DNA binding (Vaithiyalingam et al., 2010). This finding suggests that ISCs facilitate DNA binding via organizing the protein surface (Vaithiyalingam et al., 2010).
In addition, ISCs serve as a major determinant for regulation through their physical interactions with other proteins involved in DNA replication, the DNA damage response, stalled replication fork, and telomere maintenance (Weiner et al., 2007). The N- and C-terminal domains of PRIM2 folded together and are connected by a flexible 18-residue linker (Baranovskiy et al., 2018). Crystal structure studies reviewed that there are three metal-binding sites in the DNA primase, a Zn2+-binding site, a PRIM1 catalytic site which coordinates two Mg2+ (or Mn2+) ions, and a 4Fe–4S binding site in PRIM2. Furthermore, PRIM2 has four conserved Cys residues: Cys287, Cys367, Cys384, and Cys424, which are important for coordinating ISC. Point mutation of these Cys residues cause instability of both PRIM1 and PRIM2. The unstable structure of PRIM1 and PRIM2 lead to dysfunction of DNA polymerase-α primase complex and stalled replication fork (Liu and Huang, 2015). In fact, even a single point mutation of the conserved Cys residues is sufficient to reduce the activities of DNA primase and DNA polymerase. This result indicates that ISCs have an important role in enzyme functions(12).
Iron–Sulfur (Fe/S) Clusters and Polymerases
In eukaryotes, four types of class B family DNA polymerase complexes mediate replication and replication-associated genome maintenance. During normal replication, DNA polymerases (Pol) α, δ, and ε are responsible for replication fork extension. While the fourth polymerase, Polζ, is required for DNA synthesis at damaged sites (Johansson and Macneill, 2010). These polymerases are comprised of catalytic, regulatory, and accessory subunits (Burgers et al., 2001). Biochemical and structural studies demonstrate that there are two metal-binding motifs with conserved cysteine (CysA and CysB) located at Pol α, Pol δ, and Pol ε C-terminal catalytic subunits. At first, it was reported that these two metal-binding motifs coordinate Zn2+ ions (Evanics et al., 2003; Klinge et al., 2009). And they are essential for the stability of replisome. However, synthetically lethal effects are observed in yeast containing a single point mutant in the Pol3 CysB motif with essential components (DRE2, NBP35, and TAH18) of CIA complex (Chanet and Heude, 2003). Furthermore, pulse-chase 55Fe experiment, UV–Vis, and electron paramagnetic resonance (EPR) spectroscopic studies proved that the CysB motifs of all B-family DNA polymerases coordinate ISCs rather than Zn2+ (Netz et al., 2011; Suwa et al., 2015; Ter Beek et al., 2019). Overexpression S. cerevisiae Polδ subunit Pol31 enhances the ability of binding ISCs (Sanchez Garcia et al., 2004). Consist with this study, Polζ catalytic subunit Rev3 also coordinates the 4Fe–4S cluster in CysB. And the 4Fe–4S cluster is crucial for stabilizing the polymerase complex (Baranovskiy et al., 2018). Together, these findings suggest that the proper activities of DNA polymerases require 4Fe–4S cluster coordination. In addition to CysB, CysA is also important for the interaction between PCNA with Polδ on DNA. PCNA is a major determinant for regulating DNA replication and cell cycle. Notably, the DNA polymerase and exonuclease of Polδ were regulated by coordinated ISC (Jozwiakowski et al., 2019). In addition to class B-family polymerase complex, biochemical studies revealed that D-family polymerases also coordinate ISCs in their CysB motif. Furthermore, ISCs are important for polymerase complex formation. Point mutation of conserved Cys residue in Pol3 results in the reduction of coordinated ISC and disassociation with Polδ subunits Pol31 and Pol32. Moreover, Pol3 and the Fe/S biosynthetic genes are synthetic lethal, indicate that ISC is an essential cofactor for DNA polymerase to regulate its structure and functions (Chanet and Heude, 2003).
Iron–Sulfur (Fe/S) Clusters Protein and DNA Repair
Oxidation, deamination, and alkylation are likely to induce single base damage in DNA. Base excision repair (BER) is a highly conserved cellular biochemical process that repairs damaged bases throughout the cell cycle (Krokan and Bjoras, 2013). BER is started from DNA glycosylases, which recognize and remove damaged or inappropriate bases by forming AP sites. Then, these AP sites are cleaved by an AP endonuclease. Finally, according to the length of the resulting single-strand break, the damaged DNA can be repaired by short-patch or long-patch BER (Wallace, 2013). During this process, many DNA glycosylases contain Fe/S cofactor (Guan et al., 1998; Alseth et al., 1999; Hinks et al., 2002). The E. coli endonuclease III (Endo III) is the first known DNA glycosylase that coordinates a 4Fe–4S cluster. The interaction of Endo III with the DNA phosphate backbone is dependent on its ISC (Kuo et al., 1992). The E. coli MutY is an adenine DNA glycosylase involved in BER. Structurally like Endo III, MutY coordinates a 4Fe–4S cluster (Guan et al., 1998), which is important for MutY structure stability and recognition of substrates (Porello et al., 1998; Lu and Wright, 2003). Electrochemical studies showed that DNA binding of Endo III and MutY shifts the redox potentials of the 4Fe–4S clusters, which sense DNA lesions via electron transfer (Boal et al., 2005). Consist with MutY, the mammalian homolog MUTYH also functions in fixing oxidation caused DNA lesions (McGoldrick et al., 1995).
To date, there are no reports to suggest that DNA topoisomerase or ligase is coordinated with the ISC. However, both have been linked to cellular ISCs metabolism. Eukaryotic DNA topoisomerase II (Topo II) is able to modulate negative supercoiling DNA in an ATP-dependent manner. The inhibition of Topo II leads to a loss of chromosomal supercoiling and furthermore results in the upregulation of oxidative phosphorylation (Dahan-Grobgeld et al., 1998), which increases ROS levels (Nosal et al., 2014). The inhibition of Topo II induces the DNA damage response, upregulation of iron uptake, and ISC biosynthesis (Dwyer et al., 2007).
Concluding Remarks and Future Perspectives
Much research conducted over the past decade has greatly advanced our understanding of how the ISCs assemble and insert into target proteins in mitochondria, cytoplasm, and nucleus. However, there is still much to learn. For example, most of the proteins functioning in these pathways have been identified, but a complete picture of how ISC formation is regulated remains unclear. Recently, one group reported that acylated ACP1 may regulate ISCs de novo assembly via its dynamic interaction with ISD11. Upon high acetyl-CoA, mtFAS promotes long fatty acyl chain synthesis and acylated ACP1 binds to NFS1-ISD11. The long fatty acyl chain is able to stabilize the NFS1-ISD11-ACP1 complex and promote ISCs de novo assembly. On the contrary, cells that lack acylated ACP1 exhibit lower efficiency of ISCs assembly (Van Vranken et al., 2018). It is undeniable, however, that the regulation of ISC formation is crucial for cell survival.
ISC formation is controlled by several comprehensive mechanisms, including that (1) the Fe/S machinery requires delicate allosteric control, (2) the ISC delivery variations are regulated by carrier proteins, and (3) the expression levels of the Fe/S assembly protein are transcriptionally regulated. Despite great progress has been made, more research is needed to gain further insights into these processes.
Recent studies using mass spectrometry have identified many phosphorylation sites of NFS1; it has also been shown that mitochondria contribute to NFS1’s phosphorylation, which is required for its activity. Since lacking sulfur from cysteine stops the ISC synthesis, this Nfs1 phosphorylation in mitochondria has the great potential to regulate the entire ISC assembly process (Rocha et al., 2018). However, although the crystal structure of the human NFS1/ISD11/ACP complex has been observed, the phosphorylated residues were not detected and remain unclear (Cory et al., 2017).
Similarly, in the cytoplasm of mammalian cells, ISCU is phosphorylated by mTORC1. This phosphorylation event enhances the stability of the protein and promotes ISC assembly (La et al., 2013).
The degradation mechanism of the ISC is also unclear. One unique feature of 4Fe–4S is that it can be cleaved to either one 3Fe–4S cluster or two 2Fe–2S clusters. For instance, the 4Fe–4S cluster in a nitrogenase Fe-protein can be converted into two 2Fe–2S clusters (Sen et al., 2004). ISCs can also serve as sulfur donors for other sulfur-containing protein cofactors, such as biotin and lipoic acid, in a self-sacrificing fashion. Exploring the mechanism of this cleavage is essential to understanding the function of these proteins. We believe that a combination of the developing approaches in the structural, biochemical, and cell biological fields will deepen our knowledge of the molecular mechanisms of assembly, insertion, and regulation of the ISCs in target proteins.
A steadily increasing number of ISC proteins that function in genome integrity maintenance have been identified. Thus, the next major research challenge is elucidating their molecular mechanisms. To date, most of the studies support that the ISC stabilizes the structure of DNA metabolism proteins (White and Dillingham, 2012). Besides that, Barton’s group found that electron transport happens over a long distance of DNA. DNA lesions disrupt this charge transfer, which changes the redox-active status of the ISC in DNA. This proposes that, ISC is the key of how DNA glycosylases distinguish the intact and damaged bases. Similarly, primer synthesis by primase also requires the 4Fe–4S cluster although the underlying mechanisms remain unclear. One DNA-mediated electrochemistry experiment demonstrated that a reversible on/off switch in DNA primase for DNA binding is the oxidation state of the 4Fe–4S cluster. Moreover, primer synthesis is regulated by both the conserved charge transfer pathway through primase and DNA charge transport chemistry. This finding suggests that the primase uses DNA charge transport for redox signaling of 4Fe–4S clusters thus provides a chemical basis for understanding the precise regulation of primase activity and supports the notion of a fundamentally new redox switch model for substrate handoff (O’Brien et al., 2017).
Overall, the ISCs coordinate with key proteins in DNA metabolism. This coordination with the ISC regulates the target proteins via (1) stabilizing their structures, (2) mediating their local conformational changes, and (3) facilitating DNA charge transport. Improving our understanding of the critical roles played by ISCs in DNA replication and repair enzymes will ultimately help us solve the great mysteries around the DNA metabolism enzymes critical to life.
Author Contributions
RS and XX conceived the scope and schemes of this review manuscript. RS wrote the first draft. WH, Z-QW, and XX revised and finalized the manuscript. All authors read and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (Grants 31530016, 32090031, and 31761133012), the National Basic Research Program of China (Grant 2017YFA0503900), and the Shenzhen Science and Technology Innovation Commission (Grants JCYJ20180507182213033 and JCYJ20170412113009742).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank the members of the Xu lab for their insightful discussion and Dr. Jessica Tamanini for language editing prior to submission.
Footnotes
References
Adamec, J., Rusnak, F., Owen, W. G., Naylor, S., Benson, L. M., Gacy, A. M., et al. (2000). Iron-dependent self-assembly of recombinant yeast frataxin: implications for Friedreich ataxia. Am. J. Hum. Genet. 67, 549–562. doi: 10.1086/303056
Alseth, I., Eide, L., Pirovano, M., Rognes, T., Seeberg, E., and Bjoras, M. (1999). The Saccharomyces cerevisiae homologues of endonuclease III from Escherichia coli, Ntg1 and Ntg2, are both required for efficient repair of spontaneous and induced oxidative DNA damage in yeast. Mol. Cell Biol. 19, 3779–3787. doi: 10.1128/mcb.19.5.3779
Andreini, C., Rosato, A., and Banci, L. (2017). The relationship between environmental dioxygen and iron-sulfur proteins explored at the genome level. PLoS One 12:e0171279. doi: 10.1371/journal.pone.0171279
Arnold, A. R., Grodick, M. A., and Barton, J. K. (2016). DNA charge transport: from chemical principles to the cell. Cell Chem. Biol. 23, 183–197. doi: 10.1016/j.chembiol.2015.11.010
Askree, S. H., Yehuda, T., Smolikov, S., Gurevich, R., Hawk, J., Coker, C., et al. (2004). A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc. Natl. Acad. Sci. U.S.A. 101, 8658–8663. doi: 10.1073/pnas.0401263101
Babcock, M., de Silva, D., Oaks, R., Davis-Kaplan, S., Jiralerspong, S., Montermini, L., et al. (1997). Regulation of mitochondrial iron accumulation by Yfh1p, a putative homolog of frataxin. Science 276, 1709–1712. doi: 10.1126/science.276.5319.1709
Balakrishnan, L., and Bambara, R. A. (2011). The changing view of Dna2. Cell Cycle 10, 2620–2621. doi: 10.4161/cc.10.16.16545
Balk, J., Aguilar Netz, D. J., Tepper, K., Pierik, A. J., and Lill, R. (2005). The essential WD40 protein Cia1 is involved in a late step of cytosolic and nuclear iron-sulfur protein assembly. Mol. Cell Biol. 25, 10833–10841. doi: 10.1128/mcb.25.24.10833-10841.2005
Balk, J., Pierik, A. J., Netz, D. J., Muhlenhoff, U., and Lill, R. (2004). The hydrogenase-like Nar1p is essential for maturation of cytosolic and nuclear iron-sulphur proteins. EMBO J. 23, 2105–2115. doi: 10.1038/sj.emboj.7600216
Banci, L., Bertini, I., Calderone, V., Ciofi-Baffoni, S., Giachetti, A., Jaiswal, D., et al. (2013). Molecular view of an electron transfer process essential for iron-sulfur protein biogenesis. Proc. Natl. Acad. Sci. U.S.A. 110, 7136–7141. doi: 10.1073/pnas.1302378110
Banci, L., Brancaccio, D., Ciofi-Baffoni, S., Del Conte, R., Gadepalli, R., Mikolajczyk, M., et al. (2014). [2Fe-2S] cluster transfer in iron-sulfur protein biogenesis. Proc. Natl. Acad. Sci. U.S.A. 111, 6203–6208. doi: 10.1073/pnas.1400102111
Baranovskiy, A. G., Siebler, H. M., Pavlov, Y. I., and Tahirov, T. H. (2018). Iron-sulfur clusters in DNA polymerases and primases of eukaryotes. Methods Enzymol. 599, 1–20. doi: 10.1016/bs.mie.2017.09.003
Basu, S., Netz, D. J., Haindrich, A. C., Herlerth, N., Lagny, T. J., Pierik, A. J., et al. (2014). Cytosolic iron-sulphur protein assembly is functionally conserved and essential in procyclic and bloodstream Trypanosoma brucei. Mol. Microbiol. 93, 897–910. doi: 10.1111/mmi.12706
Beilschmidt, L. K., Ollagnier de Choudens, S., Fournier, M., Sanakis, I., Hograindleur, M. A., Clemancey, M., et al. (2017). ISCA1 is essential for mitochondrial Fe4S4 biogenesis in vivo. Nat. Commun. 8:15124.
Beilschmidt, L. K., and Puccio, H. M. (2014). Mammalian Fe-S cluster biogenesis and its implication in disease. Biochimie 100, 48–60. doi: 10.1016/j.biochi.2014.01.009
Bell, S. P., and Dutta, A. (2002). DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71, 333–374.
Ben-Aroya, S., Coombes, C., Kwok, T., O’Donnell, K. A., Boeke, J. D., and Hieter, P. (2008). Toward a comprehensive temperature-sensitive mutant repository of the essential genes of Saccharomyces cerevisiae. Mol. Cell 30, 248–258. doi: 10.1016/j.molcel.2008.02.021
Boal, A. K., Yavin, E., Lukianova, O. A., O’Shea, V. L., David, S. S., and Barton, J. K. (2005). DNA-bound redox activity of DNA repair glycosylases containing [4Fe-4S] clusters. Biochemistry 44, 8397–8407. doi: 10.1021/bi047494n
Brancaccio, D., Gallo, A., Mikolajczyk, M., Zovo, K., Palumaa, P., Novellino, E., et al. (2014). Formation of [4Fe-4S] clusters in the mitochondrial iron-sulfur cluster assembly machinery. J. Am. Chem. Soc. 136, 16240–16250. doi: 10.1021/ja507822j
Bridwell-Rabb, J., Fox, N. G., Tsai, C. L., Winn, A. M., and Barondeau, D. P. (2014). Human frataxin activates Fe-S cluster biosynthesis by facilitating sulfur transfer chemistry. Biochemistry 53, 4904–4913. doi: 10.1021/bi500532e
Brosh, R. M. Jr., and Bohr, V. A. (2007). Human premature aging, DNA repair and RecQ helicases. Nucleic Acids Res. 35, 7527–7544. doi: 10.1093/nar/gkm1008
Budd, M. E., Tong, A. H., Polaczek, P., Peng, X., Boone, C., and Campbell, J. L. (2005). A network of multi-tasking proteins at the DNA replication fork preserves genome stability. PLoS Genet. 1:e61. doi: 10.1371/journal.pgen.0010061
Burgers, P. M., Koonin, E. V., Bruford, E., Blanco, L., Burtis, K. C., Christman, M. F., et al. (2001). Eukaryotic DNA polymerases: proposal for a revised nomenclature. J. Biol. Chem. 276, 43487–43490.
Cai, K., Tonelli, M., Frederick, R. O., and Markley, J. L. (2017). Human mitochondrial ferredoxin 1 (FDX1) and ferredoxin 2 (FDX2) both bind cysteine desulfurase and donate electrons for iron-sulfur cluster biosynthesis. Biochemistry 56, 487–499. doi: 10.1021/acs.biochem.6b00447
Campuzano, V., Montermini, L., Molto, M. D., Pianese, L., Cossee, M., Cavalcanti, F., et al. (1996). Friedreich’s ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science 271, 1423–1427.
Cantor, S. B., Bell, D. W., Ganesan, S., Kass, E. M., Drapkin, R., Grossman, S., et al. (2001). BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell 105, 149–160. doi: 10.1016/s0092-8674(01)00304-x
Cavadini, P., Biasiotto, G., Poli, M., Levi, S., Verardi, R., Zanella, I., et al. (2007). RNA silencing of the mitochondrial ABCB7 transporter in HeLa cells causes an iron-deficient phenotype with mitochondrial iron overload. Blood 109, 3552–3559. doi: 10.1182/blood-2006-08-041632
Chanet, R., and Heude, M. (2003). Characterization of mutations that are synthetic lethal with pol3-13, a mutated allele of DNA polymerase delta in Saccharomyces cerevisiae. Curr. Genet. 43, 337–350. doi: 10.1007/s00294-003-0407-2
Chen, K. E., Richards, A. A., Ariffin, J. K., Ross, I. L., Sweet, M. J., Kellie, S., et al. (2012). The mammalian DUF59 protein Fam96a forms two distinct types of domain-swapped dimer. Acta Crystallogr. D Biol. Crystallogr. 68, 637–648. doi: 10.1107/s0907444912006592
Compe, E., and Egly, J. M. (2012). TFIIH: when transcription met DNA repair. Nat. Rev. Mol. Cell Biol. 13, 343–354. doi: 10.1038/nrm3350
Cory, S. A., Van Vranken, J. G., Brignole, E. J., Patra, S., Winge, D. R., Drennan, C. L., et al. (2017). Structure of human Fe-S assembly subcomplex reveals unexpected cysteine desulfurase architecture and acyl-ACP-ISD11 interactions. Proc. Natl. Acad. Sci. U.S.A. 114, E5325–E5334.
Dahan-Grobgeld, E., Livneh, Z., Maretzek, A. F., Polak-Charcon, S., Eichenbaum, Z., and Degani, H. (1998). Reversible induction of ATP synthesis by DNA damage and repair in Escherichia coli. In vivo NMR studies. J. Biol. Chem. 273, 30232–30238. doi: 10.1074/jbc.273.46.30232
Diaz de la Loza Mdel, C., Gallardo, M., Garcia-Rubio, M. L., Izquierdo, A., Herrero, E., Aguilera, A., et al. (2011). Zim17/Tim15 links mitochondrial iron-sulfur cluster biosynthesis to nuclear genome stability. Nucleic Acids Res. 39, 6002–6015. doi: 10.1093/nar/gkr193
Dubaele, S., Proietti De Santis, L., Bienstock, R. J., Keriel, A., Stefanini, M., Van Houten, B., et al. (2003). Basal transcription defect discriminates between xeroderma pigmentosum and trichothiodystrophy in XPD patients. Mol. Cell 11, 1635–1646. doi: 10.1016/s1097-2765(03)00182-5
Dutkiewicz, R., Schilke, B., Knieszner, H., Walter, W., Craig, E. A., and Marszalek, J. (2003). Ssq1, a mitochondrial Hsp70 involved in iron-sulfur (Fe/S) center biogenesis. Similarities to and differences from its bacterial counterpart. J. Biol. Chem. 278, 29719–29727. doi: 10.1074/jbc.m303527200
Dwyer, D. J., Kohanski, M. A., Hayete, B., and Collins, J. J. (2007). Gyrase inhibitors induce an oxidative damage cellular death pathway in Escherichia coli. Mol. Syst. Biol. 3:91. doi: 10.1038/msb4100135
Evanics, F., Maurmann, L., Yang, W. W., and Bose, R. N. (2003). Nuclear magnetic resonance structures of the zinc finger domain of human DNA polymerase-alpha. Biochim. Biophys. Acta 1651, 163–171. doi: 10.1016/s1570-9639(03)00266-8
Fan, L., Fuss, J. O., Cheng, Q. J., Arvai, A. S., Hammel, M., Roberts, V. A., et al. (2008). XPD helicase structures and activities: insights into the cancer and aging phenotypes from XPD mutations. Cell 133, 789–800. doi: 10.1016/j.cell.2008.04.030
Fosset, C., Chauveau, M. J., Guillon, B., Canal, F., Drapier, J. C., and Bouton, C. (2006). RNA silencing of mitochondrial m-Nfs1 reduces Fe-S enzyme activity both in mitochondria and cytosol of mammalian cells. J. Biol. Chem. 281, 25398–25406. doi: 10.1074/jbc.m602979200
Fox, N. G., Das, D., Chakrabarti, M., Lindahl, P. A., and Barondeau, D. P. (2015). Frataxin accelerates [2Fe-2S] cluster formation on the human Fe-S assembly complex. Biochemistry 54, 3880–3889. doi: 10.1021/bi5014497
Fox, N. G., Yu, X., Feng, X., Bailey, H. J., Martelli, A., Nabhan, J. F., et al. (2019). Structure of the human frataxin-bound iron-sulfur cluster assembly complex provides insight into its activation mechanism. Nat. Commun. 10:2210.
Frick, D. N., and Richardson, C. C. (2001). DNA primases. Annu. Rev. Biochem. 70, 39–80. doi: 10.1146/annurev.biochem.70.1.39
Gari, K., Leon Ortiz, A. M., Borel, V., Flynn, H., Skehel, J. M., and Boulton, S. J. (2012). MMS19 links cytoplasmic iron-sulfur cluster assembly to DNA metabolism. Science 337, 243–245. doi: 10.1126/science.1219664
Gerber, J., Neumann, K., Prohl, C., Muhlenhoff, U., and Lill, R. (2004). The yeast scaffold proteins Isu1p and Isu2p are required inside mitochondria for maturation of cytosolic Fe/S proteins. Mol. Cell Biol. 24, 4848–4857. doi: 10.1128/mcb.24.11.4848-4857.2004
Gervason, S., Larkem, D., Mansour, A. B., Botzanowski, T., Muller, C. S., Pecqueur, L., et al. (2019). Physiologically relevant reconstitution of iron-sulfur cluster biosynthesis uncovers persulfide-processing functions of ferredoxin-2 and frataxin. Nat. Commun. 10:3566.
Gisselberg, J. E., Dellibovi-Ragheb, T. A., Matthews, K. A., Bosch, G., and Prigge, S. T. (2013). The suf iron-sulfur cluster synthesis pathway is required for apicoplast maintenance in malaria parasites. PLoS Pathog. 9:e1003655. doi: 10.1371/journal.ppat.1003655.g004
Gourdoupis, S., Nasta, V., Calderone, V., Ciofi-Baffoni, S., and Banci, L. (2018). IBA57 Recruits ISCA2 to form a [2Fe-2S] cluster-mediated complex. J. Am. Chem. Soc. 140, 14401–14412. doi: 10.1021/jacs.8b09061
Guan, Y., Manuel, R. C., Arvai, A. S., Parikh, S. S., Mol, C. D., Miller, J. H., et al. (1998). MutY catalytic core, mutant and bound adenine structures define specificity for DNA repair enzyme superfamily. Nat. Struct. Biol. 5, 1058–1064. doi: 10.1038/4168
Hausmann, A., Aguilar Netz, D. J., Balk, J., Pierik, A. J., Muhlenhoff, U., and Lill, R. (2005). The eukaryotic P loop NTPase Nbp35: an essential component of the cytosolic and nuclear iron-sulfur protein assembly machinery. Proc. Natl. Acad. Sci. U.S.A. 102, 3266–3271. doi: 10.1073/pnas.0406447102
Hinks, J. A., Evans, M. C., De Miguel, Y., Sartori, A. A., Jiricny, J., and Pearl, L. H. (2002). An iron-sulfur cluster in the family 4 uracil-DNA glycosylases. J. Biol. Chem. 277, 16936–16940. doi: 10.1074/jbc.m200668200
Ito, S., Tan, L. J., Andoh, D., Narita, T., Seki, M., Hirano, Y., et al. (2010). MMXD, a TFIIH-independent XPD-MMS19 protein complex involved in chromosome segregation. Mol. Cell 39, 632–640. doi: 10.1016/j.molcel.2010.07.029
Johansson, E., and Macneill, S. A. (2010). The eukaryotic replicative DNA polymerases take shape. Trends Biochem. Sci. 35, 339–347. doi: 10.1016/j.tibs.2010.01.004
Johnson, D. C., Dean, D. R., Smith, A. D., and Johnson, M. K. (2005). Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 74, 247–281. doi: 10.1146/annurev.biochem.74.082803.133518
Jozwiakowski, S. K., Kummer, S., and Gari, K. (2019). Human DNA polymerase delta requires an iron-sulfur cluster for high-fidelity DNA synthesis. Life Sci. Alliance 2:e201900321. doi: 10.26508/lsa.201900321
Kang, Y. H., Lee, C. H., and Seo, Y. S. (2010). Dna2 on the road to Okazaki fragment processing and genome stability in eukaryotes. Crit. Rev. Biochem. Mol. Biol. 45, 71–96. doi: 10.3109/10409230903578593
Karthikeyan, G., Lewis, L. K., and Resnick, M. A. (2002). The mitochondrial protein frataxin prevents nuclear damage. Hum. Mol. Genet. 11, 1351–1362. doi: 10.1093/hmg/11.11.1351
Kassube, S. A., and Thoma, N. H. (2020). Structural insights into Fe-S protein biogenesis by the CIA targeting complex. Nat. Struct. Mol. Biol. 27, 735–742. doi: 10.1038/s41594-020-0454-0
Kispal, G., Csere, P., Prohl, C., and Lill, R. (1999). The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. EMBO J. 18, 3981–3989. doi: 10.1093/emboj/18.14.3981
Klinge, S., Hirst, J., Maman, J. D., Krude, T., and Pellegrini, L. (2007). An iron-sulfur domain of the eukaryotic primase is essential for RNA primer synthesis. Nat. Struct. Mol. Biol. 14, 875–877. doi: 10.1038/nsmb1288
Klinge, S., Nunez-Ramirez, R., Llorca, O., and Pellegrini, L. (2009). 3D architecture of DNA Pol alpha reveals the functional core of multi-subunit replicative polymerases. EMBO J. 28, 1978–1987. doi: 10.1038/emboj.2009.150
Kou, H., Zhou, Y., Gorospe, R. M., and Wang, Z. (2008). Mms19 protein functions in nucleotide excision repair by sustaining an adequate cellular concentration of the TFIIH component Rad3. Proc. Natl. Acad. Sci. U.S.A. 105, 15714–15719. doi: 10.1073/pnas.0710736105
Krokan, H. E., and Bjoras, M. (2013). Base excision repair. Cold Spring Harb. Perspect. Biol. 5:a012583.
Kuo, C. F., McRee, D. E., Cunningham, R. P., and Tainer, J. A. (1992). Crystallization and crystallographic characterization of the iron-sulfur-containing DNA-repair enzyme endonuclease III from Escherichia coli. J. Mol. Biol. 227, 347–351. doi: 10.1016/0022-2836(92)90703-m
La, P., Yang, G., and Dennery, P. A. (2013). Mammalian target of rapamycin complex 1 (mTORC1)-mediated phosphorylation stabilizes ISCU protein: implications for iron metabolism. J. Biol. Chem. 288, 12901–12909. doi: 10.1074/jbc.m112.424499
Lauder, S., Bankmann, M., Guzder, S. N., Sung, P., Prakash, L., and Prakash, S. (1996). Dual requirement for the yeast MMS19 gene in DNA repair and RNA polymerase II transcription. Mol. Cell Biol. 16, 6783–6793. doi: 10.1128/mcb.16.12.6783
Lehmann, A. R. (2003). DNA repair-deficient diseases, xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy. Biochimie 85, 1101–1111. doi: 10.1016/j.biochi.2003.09.010
Lill, R., Dutkiewicz, R., Freibert, S. A., Heidenreich, T., Mascarenhas, J., Netz, D. J., et al. (2015). The role of mitochondria and the CIA machinery in the maturation of cytosolic and nuclear iron-sulfur proteins. Eur. J. Cell Biol. 94, 280–291. doi: 10.1016/j.ejcb.2015.05.002
Lill, R., Srinivasan, V., and Muhlenhoff, U. (2014). The role of mitochondria in cytosolic-nuclear iron-sulfur protein biogenesis and in cellular iron regulation. Curr. Opin. Microbiol. 22, 111–119. doi: 10.1016/j.mib.2014.09.015
Liras, P., McCusker, J., Mascioli, S., and Haber, J. E. (1978). Characterization of a mutation in yeast causing nonrandom chromosome loss during mitosis. Genetics 88, 651–671. doi: 10.1093/genetics/88.4.651
Liu, H., Rudolf, J., Johnson, K. A., McMahon, S. A., Oke, M., Carter, L., et al. (2008). Structure of the DNA repair helicase XPD. Cell 133, 801–812.
Liu, L., and Huang, M. (2015). Essential role of the iron-sulfur cluster binding domain of the primase regulatory subunit Pri2 in DNA replication initiation. Protein Cell 6, 194–210. doi: 10.1007/s13238-015-0134-8
Lohman, T. M., and Bjornson, K. P. (1996). Mechanisms of helicase-catalyzed DNA unwinding. Annu. Rev. Biochem. 65, 169–214. doi: 10.1146/annurev.bi.65.070196.001125
Lohman, T. M., Tomko, E. J., and Wu, C. G. (2008). Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nat. Rev. Mol. Cell Biol. 9, 391–401. doi: 10.1038/nrm2394
Lu, A. L., and Wright, P. M. (2003). Characterization of an Escherichia coli mutant MutY with a cysteine to alanine mutation at the iron-sulfur cluster domain. Biochemistry 42, 3742–3750. doi: 10.1021/bi0269198
Luo, D., Bernard, D. G., Balk, J., Hai, H., and Cui, X. (2012). The DUF59 family gene AE7 acts in the cytosolic iron-sulfur cluster assembly pathway to maintain nuclear genome integrity in Arabidopsis. Plant Cell 24, 4135–4148. doi: 10.1105/tpc.112.102608
Maio, N., Jain, A., and Rouault, T. A. (2020). Mammalian iron-sulfur cluster biogenesis: recent insights into the roles of frataxin, acyl carrier protein and ATPase-mediated transfer to recipient proteins. Curr. Opin. Chem. Biol. 55, 34–44. doi: 10.1016/j.cbpa.2019.11.014
Majewska, J., Ciesielski, S. J., Schilke, B., Kominek, J., Blenska, A., Delewski, W., et al. (2013). Binding of the chaperone Jac1 protein and cysteine desulfurase Nfs1 to the iron-sulfur cluster scaffold Isu protein is mutually exclusive. J. Biol. Chem. 288, 29134–29142. doi: 10.1074/jbc.m113.503524
Manicki, M., Majewska, J., Ciesielski, S., Schilke, B., Blenska, A., Kominek, J., et al. (2014). Overlapping binding sites of the frataxin homologue assembly factor and the heat shock protein 70 transfer factor on the Isu iron-sulfur cluster scaffold protein. J. Biol. Chem. 289, 30268–30278. doi: 10.1074/jbc.m114.596726
McGoldrick, J. P., Yeh, Y. C., Solomon, M., Essigmann, J. M., and Lu, A. L. (1995). Characterization of a mammalian homolog of the Escherichia coli MutY mismatch repair protein. Mol. Cell Biol. 15, 989–996. doi: 10.1128/mcb.15.2.989
Mesecke, N., Terziyska, N., Kozany, C., Baumann, F., Neupert, W., Hell, K., et al. (2005). relay system in the intermembrane space of mitochondria that mediates protein import. Cell 121, 1059–1069. doi: 10.1016/j.cell.2005.04.011
Mettert, E. L., and Kiley, P. J. (2015). How is Fe-S cluster formation regulated? Annu. Rev. Microbiol. 69, 505–526. doi: 10.1146/annurev-micro-091014-104457
Muhlenhoff, U., Gerber, J., Richhardt, N., and Lill, R. (2003). Components involved in assembly and dislocation of iron-sulfur clusters on the scaffold protein Isu1p. EMBO J. 22, 4815–4825. doi: 10.1093/emboj/cdg446
Muhlenhoff, U., Richter, N., Pines, O., Pierik, A. J., and Lill, R. (2011). Specialized function of yeast Isa1 and Isa2 proteins in the maturation of mitochondrial [4Fe-4S] proteins. J. Biol. Chem. 286, 41205–41216. doi: 10.1074/jbc.m111.296152
Netz, D. J., Mascarenhas, J., Stehling, O., Pierik, A. J., and Lill, R. (2014). Maturation of cytosolic and nuclear iron-sulfur proteins. Trends Cell Biol. 24, 303–312. doi: 10.1016/j.tcb.2013.11.005
Netz, D. J., Pierik, A. J., Stumpfig, M., Bill, E., Sharma, A. K., Pallesen, L. J., et al. (2012). A bridging [4Fe-4S] cluster and nucleotide binding are essential for function of the Cfd1-Nbp35 complex as a scaffold in iron-sulfur protein maturation. J. Biol. Chem. 287, 12365–12378. doi: 10.1074/jbc.m111.328914
Netz, D. J., Pierik, A. J., Stumpfig, M., Muhlenhoff, U., and Lill, R. (2007). The Cfd1-Nbp35 complex acts as a scaffold for iron-sulfur protein assembly in the yeast cytosol. Nat. Chem. Biol. 3, 278–286. doi: 10.1038/nchembio872
Netz, D. J., Stith, C. M., Stumpfig, M., Kopf, G., Vogel, D., Genau, H. M., et al. (2011). Eukaryotic DNA polymerases require an iron-sulfur cluster for the formation of active complexes. Nat. Chem. Biol. 8, 125–132. doi: 10.1038/nchembio.721
Netz, D. J., Stumpfig, M., Dore, C., Muhlenhoff, U., Pierik, A. J., and Lill, R. (2010). Tah18 transfers electrons to Dre2 in cytosolic iron-sulfur protein biogenesis. Nat. Chem. Biol. 6, 758–765. doi: 10.1038/nchembio.432
Nosal, R., Drabikova, K., Jancinova, V., Perecko, T., Ambrozova, G., Ciz, M., et al. (2014). On the molecular pharmacology of resveratrol on oxidative burst inhibition in professional phagocytes. Oxid. Med. Cell Longev. 2014:706269.
O’Brien, E., Holt, M. E., Thompson, M. K., Salay, L. E., Ehlinger, A. C., Chazin, W. J., et al. (2017). The [4Fe4S] cluster of human DNA primase functions as a redox switch using DNA charge transport. Science 355:eaag1789. doi: 10.1126/science.aag1789
Pallesen, L. J., Solodovnikova, N., Sharma, A. K., and Walden, W. E. (2013). Interaction with Cfd1 increases the kinetic lability of FeS on the Nbp35 scaffold. J. Biol. Chem. 288, 23358–23367. doi: 10.1074/jbc.m113.486878
Pandey, A., Golla, R., Yoon, H., Dancis, A., and Pain, D. (2012). Persulfide formation on mitochondrial cysteine desulfurase: enzyme activation by a eukaryote-specific interacting protein and Fe-S cluster synthesis. Biochem. J. 448, 171–187. doi: 10.1042/bj20120951
Pandey, A., Gordon, D. M., Pain, J., Stemmler, T. L., Dancis, A., and Pain, D. (2013). Frataxin directly stimulates mitochondrial cysteine desulfurase by exposing substrate-binding sites, and a mutant Fe-S cluster scaffold protein with frataxin-bypassing ability acts similarly. J. Biol. Chem. 288, 36773–36786. doi: 10.1074/jbc.m113.525857
Parent, A., Elduque, X., Cornu, D., Belot, L., Le Caer, J. P., Grandas, A., et al. (2015). Mammalian frataxin directly enhances sulfur transfer of NFS1 persulfide to both ISCU and free thiols. Nat. Commun. 6:5686.
Parish, J. L., Rosa, J., Wang, X., Lahti, J. M., Doxsey, S. J., and Androphy, E. J. (2006). The DNA helicase ChlR1 is required for sister chromatid cohesion in mammalian cells. J. Cell Sci. 119, 4857–4865. doi: 10.1242/jcs.03262
Paul, V. D., and Lill, R. (2015). Biogenesis of cytosolic and nuclear iron-sulfur proteins and their role in genome stability. Biochim. Biophys. Acta 1853, 1528–1539.
Pokharel, S., and Campbell, J. L. (2012). Cross talk between the nuclease and helicase activities of Dna2: role of an essential iron-sulfur cluster domain. Nucleic Acids Res. 40, 7821–7830. doi: 10.1093/nar/gks534
Pondarre, C., Antiochos, B. B., Campagna, D. R., Clarke, S. L., Greer, E. L., Deck, K. M., et al. (2006). The mitochondrial ATP-binding cassette transporter Abcb7 is essential in mice and participates in cytosolic iron-sulfur cluster biogenesis. Hum. Mol. Genet. 15, 953–964. doi: 10.1093/hmg/ddl012
Porello, S. L., Cannon, M. J., and David, S. S. (1998). A substrate recognition role for the [4Fe-4S]2+ cluster of the DNA repair glycosylase MutY. Biochemistry 37, 6465–6475. doi: 10.1021/bi972433t
Prakash, L., and Prakash, S. (1977). Isolation and characterization of MMS-sensitive mutants of Saccharomyces cerevisiae. Genetics 86, 33–55. doi: 10.1093/genetics/86.1.33
Prischi, F., Konarev, P. V., Iannuzzi, C., Pastore, C., Adinolfi, S., Martin, S. R., et al. (2010). Structural bases for the interaction of frataxin with the central components of iron-sulphur cluster assembly. Nat. Commun. 1:95.
Pugh, R. A., Honda, M., Leesley, H., Thomas, A., Lin, Y., Nilges, M. J., et al. (2008). The iron-containing domain is essential in Rad3 helicases for coupling of ATP hydrolysis to DNA translocation and for targeting the helicase to the single-stranded DNA-double-stranded DNA junction. J. Biol. Chem. 283, 1732–1743. doi: 10.1074/jbc.m707064200
Pyle, A. M. (2008). Translocation and unwinding mechanisms of RNA and DNA helicases. Annu. Rev. Biophys. 37, 317–336. doi: 10.1146/annurev.biophys.37.032807.125908
Rocha, A. G., Knight, S. A. B., Pandey, A., Yoon, H., Pain, J., Pain, D., et al. (2018). Cysteine desulfurase is regulated by phosphorylation of Nfs1 in yeast mitochondria. Mitochondrion 40, 29–41. doi: 10.1016/j.mito.2017.09.003
Roche, B., Aussel, L., Ezraty, B., Mandin, P., Py, B., and Barras, F. (2013). Iron/sulfur proteins biogenesis in prokaryotes: formation, regulation and diversity. Biochim. Biophys. Acta 1827, 455–469. doi: 10.1016/j.bbabio.2012.12.010
Roy, A., Solodovnikova, N., Nicholson, T., Antholine, W., and Walden, W. E. (2003). A novel eukaryotic factor for cytosolic Fe-S cluster assembly. EMBO J. 22, 4826–4835. doi: 10.1093/emboj/cdg455
Rudolf, J., Makrantoni, V., Ingledew, W. J., Stark, M. J., and White, M. F. (2006). The DNA repair helicases XPD and FancJ have essential iron-sulfur domains. Mol. Cell 23, 801–808.
Sanchez Garcia, J., Ciufo, L. F., Yang, X., Kearsey, S. E., and MacNeill, S. A. (2004). The C-terminal zinc finger of the catalytic subunit of DNA polymerase delta is responsible for direct interaction with the B-subunit. Nucleic Acids Res. 32, 3005–3016. doi: 10.1093/nar/gkh623
Sen, S., Igarashi, R., Smith, A., Johnson, M. K., Seefeldt, L. C., and Peters, J. W. (2004). A conformational mimic of the MgATP-bound “on state” of the nitrogenase iron protein. Biochemistry 43, 1787–1797. doi: 10.1021/bi0358465
Sharma, A. K., Pallesen, L. J., Spang, R. J., and Walden, W. E. (2010). Cytosolic iron-sulfur cluster assembly (CIA) system: factors, mechanism, and relevance to cellular iron regulation. J. Biol. Chem. 285, 26745–26751. doi: 10.1074/jbc.r110.122218
Sipos, K., Lange, H., Fekete, Z., Ullmann, P., Lill, R., and Kispal, G. (2002). Maturation of cytosolic iron-sulfur proteins requires glutathione. J. Biol. Chem. 277, 26944–26949. doi: 10.1074/jbc.m200677200
Skibbens, R. V. (2004). Chl1p, a DNA helicase-like protein in budding yeast, functions in sister-chromatid cohesion. Genetics 166, 33–42. doi: 10.1534/genetics.166.1.33
Song, D., and Lee, F. S. (2008). A role for IOP1 in mammalian cytosolic iron-sulfur protein biogenesis. J. Biol. Chem. 283, 9231–9238. doi: 10.1074/jbc.m708077200
Song, D., and Lee, F. S. (2011). Mouse knock-out of IOP1 protein reveals its essential role in mammalian cytosolic iron-sulfur protein biogenesis. J. Biol. Chem. 286, 15797–15805. doi: 10.1074/jbc.m110.201731
Srinivasan, V., Netz, D. J., Webert, H., Mascarenhas, J., Pierik, A. J., Michel, H., et al. (2007). Structure of the yeast WD40 domain protein Cia1, a component acting late in iron-sulfur protein biogenesis. Structure 15, 1246–1257. doi: 10.1016/j.str.2007.08.009
Stehling, O., Jeoung, J. H., Freibert, S. A., Paul, V. D., Banfer, S., Niggemeyer, B., et al. (2018). Function and crystal structure of the dimeric P-loop ATPase CFD1 coordinating an exposed [4Fe-4S] cluster for transfer to apoproteins. Proc. Natl. Acad. Sci. U.S.A. 115, E9085–E9094.
Stehling, O., Mascarenhas, J., Vashisht, A. A., Sheftel, A. D., Niggemeyer, B., Rosser, R., et al. (2013). Human CIA2A-FAM96A and CIA2B-FAM96B integrate iron homeostasis and maturation of different subsets of cytosolic-nuclear iron-sulfur proteins. Cell Metab. 18, 187–198. doi: 10.1016/j.cmet.2013.06.015
Stehling, O., Vashisht, A. A., Mascarenhas, J., Jonsson, Z. O., Sharma, T., Netz, D. J., et al. (2012). MMS19 assembles iron-sulfur proteins required for DNA metabolism and genomic integrity. Science 337, 195–199. doi: 10.1126/science.1219723
Suwa, Y., Gu, J., Baranovskiy, A. G., Babayeva, N. D., Pavlov, Y. I., and Tahirov, T. H. (2015). Crystal structure of the human pol alpha b subunit in complex with the C-terminal domain of the catalytic subunit. J. Biol. Chem. 290, 14328–14337.
Sviderskiy, V. O., Blumenberg, L., Gorodetsky, E., Karakousi, T. R., Hirsh, N., Alvarez, S. W., et al. (2020). Hyperactive CDK2 activity in basal-like breast cancer imposes a genome integrity liability that can be exploited by targeting DNA polymerase epsilon. Mol. Cell 80, 682–698.e7.
Takahashi, Y., and Tokumoto, U. (2002). A third bacterial system for the assembly of iron-sulfur clusters with homologs in archaea and plastids. J. Biol. Chem. 277, 28380–28383. doi: 10.1074/jbc.c200365200
Ter Beek, J., Parkash, V., Bylund, G. O., Osterman, P., Sauer-Eriksson, A. E., and Johansson, E. (2019). Structural evidence for an essential Fe-S cluster in the catalytic core domain of DNA polymerase. Nucleic Acids Res. 47, 5712–5722. doi: 10.1093/nar/gkz248
Thomas, D., Barbey, R., Henry, D., and Surdin-Kerjan, Y. (1992). Physiological analysis of mutants of Saccharomyces cerevisiae impaired in sulphate assimilation. J. Gen. Microbiol. 138, 2021–2028. doi: 10.1099/00221287-138-10-2021
Uzarska, M. A., Dutkiewicz, R., Freibert, S. A., Lill, R., and Muhlenhoff, U. (2013). The mitochondrial Hsp70 chaperone Ssq1 facilitates Fe/S cluster transfer from Isu1 to Grx5 by complex formation. Mol. Biol. Cell 24, 1830–1841. doi: 10.1091/mbc.e12-09-0644
Vaithiyalingam, S., Warren, E. M., Eichman, B. F., and Chazin, W. J. (2010). Insights into eukaryotic DNA priming from the structure and functional interactions of the 4Fe-4S cluster domain of human DNA primase. Proc. Natl. Acad. Sci. U.S.A. 107, 13684–13689. doi: 10.1073/pnas.1002009107
van der Lelij, P., Chrzanowska, K. H., Godthelp, B. C., Rooimans, M. A., Oostra, A. B., Stumm, M., et al. (2010). Warsaw breakage syndrome, a cohesinopathy associated with mutations in the XPD helicase family member DDX11/ChlR1. Am. J. Hum. Genet. 86, 262–266. doi: 10.1016/j.ajhg.2010.01.008
Van Vranken, J. G., Nowinski, S. M., Clowers, K. J., Jeong, M. Y., Ouyang, Y., Berg, J. A., et al. (2018). Is an Acetyl-CoA-dependent modification required for electron transport chain assembly. Mol. Cell 71, 567–580.e4.
van Wietmarschen, N., Moradian, A., Morin, G. B., Lansdorp, P. M., and Uringa, E. J. (2012). The mammalian proteins MMS19, MIP18, and ANT2 are involved in cytoplasmic iron-sulfur cluster protein assembly. J. Biol. Chem. 287, 43351–43358. doi: 10.1074/jbc.m112.431270
Veatch, J. R., McMurray, M. A., Nelson, Z. W., and Gottschling, D. E. (2009). Mitochondrial dysfunction leads to nuclear genome instability via an iron-sulfur cluster defect. Cell 137, 1247–1258. doi: 10.1016/j.cell.2009.04.014
Vernis, L., Facca, C., Delagoutte, E., Soler, N., Chanet, R., Guiard, B., et al. (2009). A newly identified essential complex, Dre2-Tah18, controls mitochondria integrity and cell death after oxidative stress in yeast. PLoS One 4:e4376. doi: 10.1371/journal.pone.0004376
Wallace, S. S. (2013). DNA glycosylases search for and remove oxidized DNA bases. Environ. Mol. Mutagen. 54, 691–704. doi: 10.1002/em.21820
Webert, H., Freibert, S. A., Gallo, A., Heidenreich, T., Linne, U., Amlacher, S., et al. (2014). Functional reconstitution of mitochondrial Fe/S cluster synthesis on Isu1 reveals the involvement of ferredoxin. Nat. Commun. 5:5013.
Weerapana, E., Wang, C., Simon, G. M., Richter, F., Khare, S., Dillon, M. B., et al. (2010). Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature 468, 790–795.
Weiler, B. D., Bruck, M. C., Kothe, I., Bill, E., Lill, R., and Muhlenhoff, U. (2020). Mitochondrial [4Fe-4S] protein assembly involves reductive [2Fe-2S] cluster fusion on ISCA1-ISCA2 by electron flow from ferredoxin FDX2. Proc. Natl. Acad. Sci. U.S.A. 117, 20555–20565.
Weiner, B. E., Huang, H., Dattilo, B. M., Nilges, M. J., Fanning, E., and Chazin, W. J. (2007). An iron-sulfur cluster in the C-terminal domain of the p58 subunit of human DNA primase. J. Biol. Chem. 282, 33444–33451.
White, M. F. (2009). Structure, function and evolution of the XPD family of iron-sulfur-containing 5′–>3′ DNA helicases. Biochem. Soc. Trans. 37, 547–551.
White, M. F., and Dillingham, M. S. (2012). Iron-sulphur clusters in nucleic acid processing enzymes. Curr. Opin. Struct. Biol. 22, 94–100.
Wolski, S. C., Kuper, J., Hanzelmann, P., Truglio, J. J., Croteau, D. L., Van Houten, B., et al. (2008). Crystal structure of the FeS cluster-containing nucleotide excision repair helicase XPD. PLoS Biol. 6:e149. doi: 10.1371/journal.pbio.0060149
Wu, Y., and Brosh, R. M. Jr. (2009). FANCJ helicase operates in the Fanconi Anemia DNA repair pathway and the response to replicational stress. Curr. Mol. Med. 9, 470–482.
Wu, Y., and Brosh, R. M. Jr. (2012). DNA helicase and helicase-nuclease enzymes with a conserved iron-sulfur cluster. Nucleic Acids Res. 40, 4247–4260.
Wu, Y., Sommers, J. A., Khan, I., de Winter, J. P., and Brosh, R. M. Jr. (2012). Biochemical characterization of Warsaw breakage syndrome helicase. J. Biol. Chem. 287, 1007–1021.
Wu, Y., Sommers, J. A., Suhasini, A. N., Leonard, T., Deakyne, J. S., Mazin, A. V., et al. (2010). Fanconi anemia group J mutation abolishes its DNA repair function by uncoupling DNA translocation from helicase activity or disruption of protein-DNA complexes. Blood 116, 3780–3791.
Yeeles, J. T., Cammack, R., and Dillingham, M. S. (2009). An iron-sulfur cluster is essential for the binding of broken DNA by AddAB-type helicase-nucleases. J. Biol. Chem. 284, 7746–7755.
Yoon, T., and Cowan, J. A. (2003). Iron-sulfur cluster biosynthesis. Characterization of frataxin as an iron donor for assembly of [2Fe-2S] clusters in ISU-type proteins. J. Am. Chem. Soc. 125, 6078–6084.
Zerbe, L. K., and Kuchta, R. D. (2002). The p58 subunit of human DNA primase is important for primer initiation, elongation, and counting. Biochemistry 41, 4891–4900.
Keywords: iron-sulfur (Fe-S) clusters, genome stability, DNA replication, DNA repair, DNA metabolism
Citation: Shi R, Hou W, Wang Z-Q and Xu X (2021) Biogenesis of Iron–Sulfur Clusters and Their Role in DNA Metabolism. Front. Cell Dev. Biol. 9:735678. doi: 10.3389/fcell.2021.735678
Received: 03 July 2021; Accepted: 06 September 2021;
Published: 30 September 2021.
Edited by:
Chunlong Chen, Institut Curie, FranceReviewed by:
Jun Huang, Zhejiang University, ChinaAndrew Dancis, University of Pennsylvania, United States
Copyright © 2021 Shi, Hou, Wang and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xingzhi Xu, Xingzhi.Xu@szu.edu.cn
 Ruifeng Shi
Ruifeng Shi