
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol. , 21 September 2021
Sec. Molecular and Cellular Pathology
Volume 9 - 2021 | https://doi.org/10.3389/fcell.2021.728352
This article is part of the Research Topic Hearing Loss: Mechanisms and Prevention View all 35 articles
In the mammalian cochlea, spiral ganglion neurons (SGNs) relay the acoustic information to the central auditory circuits. Degeneration of SGNs is a major cause of sensorineural hearing loss and severely affects the effectiveness of cochlear implant therapy. Cochlear glial cells are able to form spheres and differentiate into neurons in vitro. However, the identity of these progenitor cells is elusive, and it is unclear how to differentiate these cells toward functional SGNs. In this study, we found that Sox2+ subpopulation of cochlear glial cells preserves high potency of neuronal differentiation. Interestingly, Sox2 expression was downregulated during neuronal differentiation and Sox2 overexpression paradoxically inhibited neuronal differentiation. Our data suggest that Sox2+ glial cells are potent SGN progenitor cells, a phenotype independent of Sox2 expression. Furthermore, we identified a combination of small molecules that not only promoted neuronal differentiation of Sox2– glial cells, but also removed glial cell identity and promoted the maturation of the induced neurons (iNs) toward SGN fate. In summary, we identified Sox2+ glial subpopulation with high neuronal potency and small molecules inducing neuronal differentiation toward SGNs.
In the mammalian cochlea, the spiral ganglion neurons (SGNs) relay the acoustic information from inner hair cells (IHCs) to the central auditory circuits (Fettiplace, 2017). SGNs are essential for normal hearing and communication, and degeneration causes sensorineural hearing loss (Sun et al., 2016; Liu et al., 2019; Zhao et al., 2019). Degeneration of SGN nerve terminals or cell bodies can be caused by ototoxicity, noise, or aging (Lang et al., 2005; Bao and Ohlemiller, 2010; Fryatt et al., 2011; Liberman, 2017; Vlajkovic et al., 2017). Because the SGNs lack the ability to regenerate in mammals, damages to the SGNs lead to permanent hearing impairment (Guo et al., 2016, 2020, 2021; Yan et al., 2018; Liu et al., 2021). In addition, the effectiveness of hearing aids and cochlear implants relies on the health and numbers of intact SGNs (Muller and Barr-Gillespie, 2015). If SGNs could be replaced or regenerated, it might be possible to restore the hearing of patients with severely damaged SGNs (Meas et al., 2018b) and benefit individuals treated with hearing aids and cochlear implants.
At the early stage of SGN damage which precedes neuronal cell body degeneration, neurotrophic factors such as neurotrophin 3 (NT3), brain-derived neurotrophic factor (BDNF), and glial-derived neurotrophic factor (GDNF) are used to support the survival of SGNs and their neurite outgrowth to the sensory HCs (Wise et al., 2011; Suzuki et al., 2016; Akil et al., 2019). However, therapeutic strategies are limited to generate induced neurons (iNs) to replace the SGNs once they are lost and SGN regeneration remains a major challenge.
Multiple attempts have been made to replace and regenerate SGNs, including transplantations of iNs differentiated from embryonic stem cells (ESC) or iPSC-derived progenitors (Chen et al., 2012; Koehler et al., 2013; Perny et al., 2017), or neuronal differentiation of cochlear-resident multipotent stem cells/progenitor cells to SGNs (Oshima et al., 2007; Zhang et al., 2011; Diensthuber et al., 2014a,b; Li et al., 2016; McLean et al., 2016; Noda et al., 2018). For induced differentiation of ESC or iPSC-derived progenitors, three-dimensional culture systems have been used to convert mouse ESC into hair cells, supporting cells, and neuronal cells (Koehler et al., 2013; Perny et al., 2017). hESC differentiated into neuronal cells expressed specific neuronal markers with electrophysiological properties characteristic of auditory neurons (Chen et al., 2012). However, transplantation of ESC-derived iNs is hampered by immuno-rejection, tumorigenesis, SGN maturation and functional integration (Lee et al., 2013; Lukovic et al., 2014).
Alternatively, inner ear-resident cells, such as progenitor cells within the utricle (Li et al., 2003) or in the spiral ganglion region (Oshima et al., 2007; Zhang et al., 2011; Li et al., 2016; McLean et al., 2016), could also be induced to neuron-like cells, forming neurites, developing synapses and expressing neuronal markers in vitro. It has been reported that Plp1+ glial cells were cochlear-resident multipotent stem cells/progenitor cells (McLean et al., 2016). Induced neuronal reprogramming of these cochlear-resident progenitors have several advantages over cell transplantations such as enhanced cell survival, physiological relevance of cellular localization and ease of maturation due to lineage similarities. However, Plp1+ glial cells may include heterogenous cell subpopulation. The identity of these progenitor cells is elusive, and it is unclear how to induce these cells to functional SGNs post-injury both in vitro and in vivo.
In this study, we found that Sox2+ subpopulation of cochlear glial cells preserves high potency of neuronal differentiation and identified a combination of small molecules promoted the maturation of the iNs toward SGNs fate. Together, we found that cochlear Sox2+ glial cells subpopulation is highly potent in neuronal differentiation and identified small molecules promoting both neuronal differentiation efficiency and maturity toward the SGN fate.
Plp1CreERT (stock number 005975), Sox2CreERT, (stock number 017593) lines were obtained from Jackson laboratory and crossed with Rosa26-LSL-Cas9-tdTomato (NBRI T002249) mice obtained from Gempharmatech Inc., China. Scrt2-P2A-tdTomato mice were generated as previously reported (Li C. et al., 2020). Mice were injected i.p. with Tamoxifen (Sigma, T5648) at 33 mg/kg for postnatal mice (P1–P3) and 50 mg/kg for juvenile mice (P17–P20). Tamoxifen was dissolved in corn oil.
All mice used in this work were on a mixed background containing C57BL6 and FVB/N strains. Both male and female mice were used. All animal procedures were approved by the Institutional Animal Care and Use Committee of Model Animal Research Center of Nanjing University.
Both sexes of the 6–8 postnatal 4–21 days (P4–P21) mice spiral ganglia were dissected in HBSS (Invitrogen 14065056) at pH 7.4 on ice for tissue harvesting. The stria vascularis, vestibule and the organ of Corti were removed carefully with forceps (Dumont) to dissect the modiolus. The modiolus were digested with Trypsin/EDTA (Sigma 59418C) and DNase I (20 U/ml) at 37°C in a total volume of 50 μl for ∼15 min with shaking at 300 rpm/min (Thermo) in 1.5 ml EP tube. Dissociation was terminated by adding 0.4 ml SCM media containing DMEM/F12 (HyClone 36254) supplemented with B27 (Thermo 17504044) and N2 (Thermo 17502048) supplement, 20 ng/ml EGF (Peprotech 315-09), 10 ng/ml bFGF (Peprotech 450-33), 50 ng/ml IGF (Peprotech 250-19), and 50 ng/ml heparan sulfate (MCE HY-101916). The samples were then carefully triturated with 1 ml pipette tips and next with 200 μl tips, followed by suspension with 1 ml SCM medium.
The cell suspension was then passed through a 70-μm cell strainer, and spun at 300 g for 5–10 min. A small white cell pellet should be observed at the bottom and then carefully aspirate the supernatant. SCM medium were added to resuspend and count cells, and then 200,000–240,000 cells were plated in each well of the six-well dish (Corning 3471). For propagation, the cochlear glial spheres were harvested after 5–7 days and passaged for 3–4 generations.
For P21 mice, the modiolus were calcified and the cochlear glial cells decrease the potential to form spheres. P21 modiolus were digested with Trypsin/EDTA (Sigma 59418C) and DNase I (20 U/ml) at 37°C in a total volume of 50–100 μl for ∼20 min with shaking 300 rpm/min (Thermo) in 1.5 ml EP tube. We used culture media contains DMEM/F12, 10% FBS, B27 in 2D dish (Lang et al., 2011). Firstly, culture media was added to stop trypsin reaction. Tissues were triturated with pipet tips and centrifuged at 300 × g for 5–10 min. The pellet was resuspended in culture media and filtered through a 70 μm cell strainer. Cells were counted, plated and grown to full confluency (5–7 days). Media was then removed and replaced with SCM media for suspension culture.
To induce neuronal differentiation, cochlear glial spheres were plated on 96-well plate (Thermo 310109008) or glass slides (Thermo Fisher 12-545-80) coated with Poly-L-ornithine (Sigma P4957) and 10 ng/ml Laminin (Corning 354232) in SCM for 12–24 h, and then replaced with SCDM containing DMEM/F12 (HyClone 36254) supplemented with B27 (Thermo 17504044) and N2 (Thermo 17502048) supplement, 50 ng/ml BDNF (Stemcell 78005), 50 ng/ml NT3 (Stemcell 78074). Half of the medium was replaced every 2–3 days. Differentiated cells were analyzed after 9 days or more for immunocytochemistry and qPCR. Additional control SCDM/FGF referred to SCDM supplemented with bFGF (100 ng/ml).
The induction media (IM) for small molecule reprogramming contains Neurobasal Medium (Thermo 21103049), supplemented with B27 and N2, GlutaMax (Thermo 35050061), penicillin-streptomycin and bFGF (100 ng/ml), with or without small molecules Forskolin (20 μM), ISX9 (20 μM), I-BET (1–2 μM), Chir99021 (10 μM) (all from Selleck), and LIF (1000 U/ml) (Novus Biologicals).
RNA was isolated using Trizol (Takara 9108) and reverse transcription of total RNA was performed with the Primescript RT reagent kit (Takara RR047A) according to the manufacturer’s protocol. The Quantitative PCR reactions were performed with the Hieff UNICON® qPCR SYBR Green Master Mix (YEASEN 11198ES03) on LightCycler 96 (Roche LightCycler® 96 Instrument). Details of the primers were in Table 1. Data are normalized to GAPDH, and fold changes are calculated by using 2–ΔΔCT method.
Cells were fixed in 4% paraformaldehyde in PBS for 15 min with shaking at room temperature. Inner ear tissues were dissected and fixed in 4% paraformaldehyde in PBS for 2 h with shaking at room temperature, followed by decalcification in 5% EDTA for 4–5 days. Then cells were blocked with 5% heat inactivated horse serum with 0.3% Triton X-100 in PBS for 1 h.
Cells were incubated with primary antibody overnight at 4°C. The primary antibodies used in this study were as follows: anti-Sox2 (goat anti-Sox2;sc-17320, Santa Cruz Biotechnologies; 1:200); anti-Sox10 (rabit anti-Sox10; 69661, Cell Signaling Technology; 1:200); anti-TUJ1 (mouse anti-TUJ1; MMS 435P, Biolegend; 1:2,000); anti-Prox1 (goat anti-Prox1; AF2727, R&D; 1:250); anti-Gata3 (Rabbit anti-Gata3; 5852T, Cell Signaling Technology; 1:1500); anti-Map2 (Mouse anti-Map2; M4403, Sigma-Aldrich; 1:250); anti-Syp (mouse anti-Syp; MA5-14532, Thermofisher; 1:300), and anti-GFP (Rabbit anti-GFP; 31002, Yeasen; 1:400).
Then cells or tissues were incubated with Alexa 488-, Alexa 568-, and/or Alexa 647-labeled secondary antibodies for 1–2 h with shaking at room temperature. Nuclei were visualized with DAPI.
Confocal z-stacks (0.5 μm step size) of cochlear tissues were taken using Leica SP5 microscope equipped with 40× and 63× oil-immersion lens. ImageJ software (version 1.52i, NIH, Bethesda, MD, United States) was used for image processing and three-dimensional reconstruction of z-stacks. All immunofluorescence images shown are representative of at least three individual results. Efficiency of conversion was measured by the number of TUJ1+ cells divided by the total number of plated cells from random 6–10 fields. Axon length was measured with NeuronJ, a plugin of ImageJ to facilitate the tracing and quantification of elongated image structures (Ho et al., 2011).
cDNAs for Sox2 was cloned into lentiviral constructs of pLKO.1 vector (Addgene 10879). We modified the plasmid to replace the PuroR with Sox2-P2A-EGFP. Lentiviruses were produced by transfection of lentiviral backbones containing the indicated transgenes together with packaging plasmids pSPAX2 (Addgene 12260) and pMD2G (Addgene 12259) into HEK293T cells (ATCC® CRL-3216TM). Viruses were concentrated from culture supernatant by ultra-centrifugation (25,000 rpm, 2 h, 4°C). After 24–48 h infection of spheres with 10 μg/ml polybrene in suspension culture, virus-containing medium was replaced with fresh SCM media.
Total RNA was extracted from the tissue using TRIzol® Reagent according the manufacturer’s instructions (TaKara) and genomic DNA was removed using DNase I (TaKara). Then RNA quality was determined by 2100 Bioanalyzer (Agilent) and quantified using the ND-2000 (NanoDrop Technologies). RNA-seq transcriptome library was prepared following TruSeqTM RNA sample preparation Kit from Illumina (San Diego, CA, United States) using 1 μg of total RNA. Libraries were size selected for cDNA target fragments of 200–300 bp on 2% Low Range Ultra Agarose followed by PCR amplified using Phusion DNA polymerase (NEB) for 15 PCR cycles. After quantified by TBS380, paired-end RNA-seq sequencing library was sequenced with the Illumina HiSeq x ten/NovaSeq 6000 sequencer (2 × 150 bp read length). The raw paired end reads were trimmed and quality controlled by SeqPrep and Sickle with default parameters. Then clean reads were separately aligned to reference genome with orientation mode using TopHat (Langmead and Salzberg, 2012) software.
To identify differential expression genes (DEGs) between two different samples, the expression level of each transcript was calculated according to the fragments per kilobase of exon per million mapped reads (FRKM) method. RSEM (Li and Dewey, 2011) was used to quantify gene abundances. R statistical package software EdgeR (Empirical analysis of Digital Gene Expression in R (Robinson et al., 2010) was utilized for differential expression analysis. In addition, functional-enrichment analysis including GO and KEGG were performed to identify which DEGs were significantly enriched in GO terms and metabolic pathways at Bonferroni-corrected P-value ≤ 0.05 compared with the whole-transcriptome background.
The transcription data of primary SGN were obtained from previous study (Noda et al., 2018). Raw RNA-seq data have been deposited in the NCBI Gene Expression Omnibus (GEO) under accession number GSE169042.
Statistical tests were performed using Graphpad Prism 8 (Graphpad Software Inc., La Jolla, CA, United States). Results were reported as mean ± SD. Specific statistical tests used in each experiment were described in figure legends. Results were analyzed using Student’s t-test or one-way ANOVA, followed by Bonferroni’s multiple comparisons test.
To expand the population of progenitor cells, we dissociated cochlear modiolus from the postnatal 3–4 days (P3–P4) mice and digested into single cells for 3D suspension culture. The cells grew spheres in vitro and were able to proliferate for more than five generations (Figures 1A,B). We then optimized the growth media by evaluating the sphere numbers with diameters larger than 50 μm. The result showed that bFGF is the primary factor for spheres growth, consistent with the previous report (Diensthuber et al., 2014a). Heparan sulfate has been reported to promote the binding and activation of FGF (Loo and Salmivirta, 2002). Therefore, we cultured the spheres with serum-free media containing IGF, EGF, FGF, and heparan sulfate (Figure 1C). Real-time quantitative PCR (RT-qPCR) results showed increased expression of neuronal stem cell markers such as Sox2 and Nestin in spheres compared to cochlear modiolus (Figure 1D). These results indicated that postnatal cochlear spheres were able to proliferate and preserve the stemness following passages in vitro.
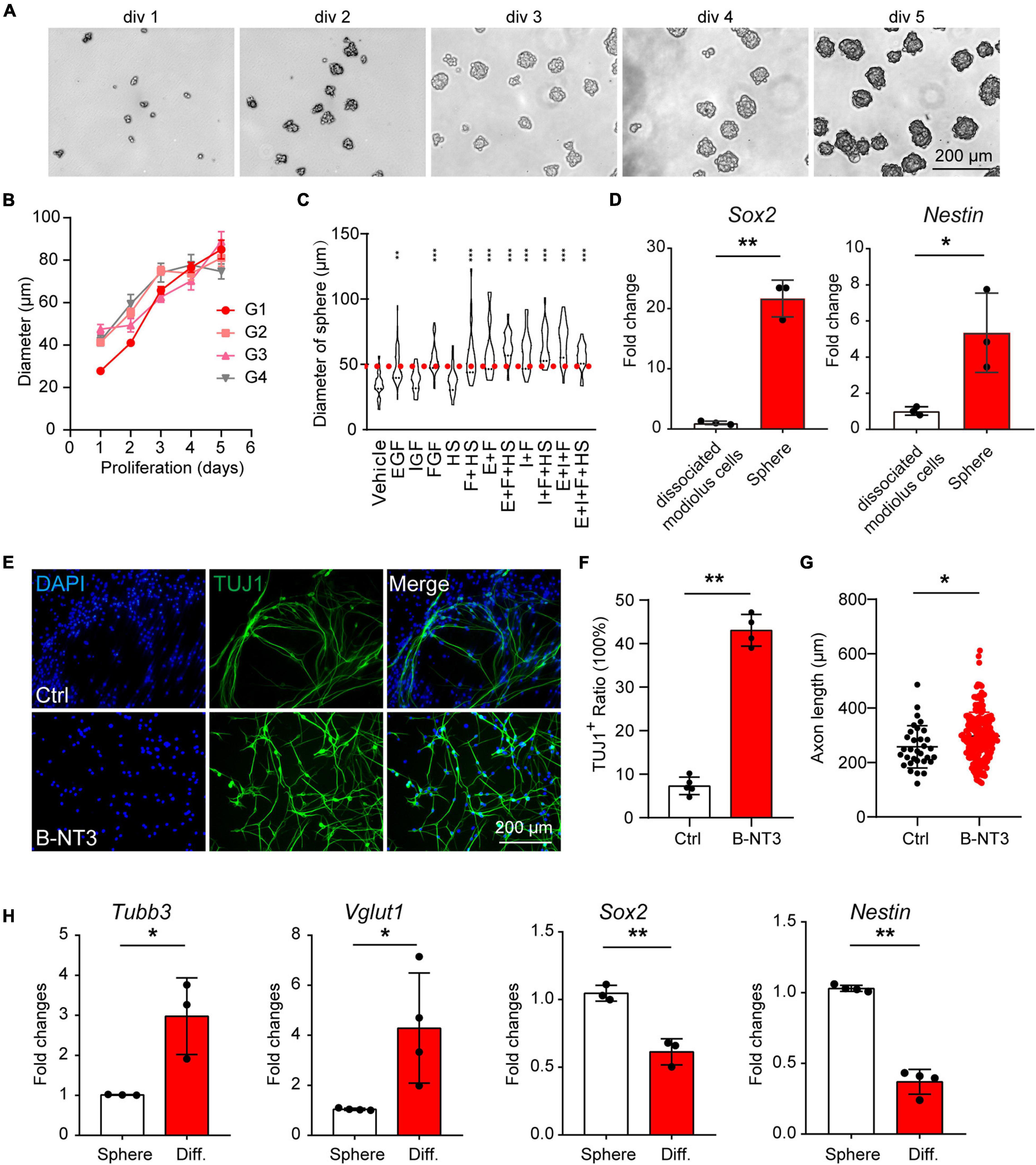
Figure 1. Postnatal cochlear glial cells proliferate as neurospheres and differentiate into neuron-like cells. (A) Bright-field images of the proliferating generation 1 (G1) spheres at 1–5 days in vitro (div). (B) Diameters of G1–G4 spheres during propagation. (C) Quantification of sphere diameters at 5 div with supplementations of E, I, F, H. E, EGF; I, IGF; F, FGF, and H, heparan sulfate. N = 21–48. P-values were calculated against vehicle control. (D) mRNA expression of Sox2 and Nestin in cochlear spheres compared to controls (dissociate modiolus cells). N = 3, error bars represent mean ± SD. (E) Representative TUJ1-positive cells after differentiation at 18 div with BDNF and NT3 (B-NT3). (F) The ratio of TUJ1 positive cells. N = 5 and 4, error bars represent mean ± SD. (G) Axon lengths of the induced neuron-like cells. N = 35 and 220, error bars represent mean ± SD. (H) mRNA expression of Tubb3, Vglut1, Sox2, and Nestin of either spheres or differentiated neuron-like cells (Diff). N = 3 or 4, error bars represent mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001 by one-way ANOVA (C) or unpaired student’s t-test (D,F–H).
To investigate the neuronal potential of the spheres, we induced the spheres for neuronal differentiation. After induction for 8–9 days, the differentiated cells showed typical bipolar neuronal morphology and neuronal marker TUJ1 expression (Figure 1E, top panels). Brain-derived neurotrophic factor and NT3 are important neurotrophic factors for SGN development and function, and are able to promote neuronal differentiation in vitro (Wise et al., 2011; Li et al., 2016; Suzuki et al., 2016; Akil et al., 2019). We found that BDNF and NT3 treatment not only increased the differentiation efficiency (Figures 1E,F) but also promoted neurite extension (Figures 1E,G) during induced differentiation of the spheres. Therefore, BDNF and NT3 were added in the subsequent differentiation experiments. RT-qPCR results also showed that neuronal markers Tubb3, Vglut1 expression were increased, while neuronal stemness markers Nestin and Sox2 were decreased after induced neuronal differentiation (Figure 1H). These results indicate that postnatal cochlear spheres are able to differentiate into neuron-like cells.
Previous studies show the glial cells in association with the SGNs may serve as the source of neuronal progenitors (McLean et al., 2016). Based on the function and localization, cochlear glial cells consist of two major populations including Schwann cells (SCs) and satellite glial cells (SGCs) (Jessen and Mirsky, 2005; Wan and Corfas, 2017). The SCs wrap the axons of SGNs with myelin sheaths and are primarily localized to the osseous spiral lamina (OSL) of the cochlea; while the SGCs are in close association with the SGN cell bodies exclusively localized to the Rosenthal’s canal (RC) of the cochlea (Wan and Corfas, 2017). Despite these differences, the two glial populations share same developmental origin and both serve important roles in survival and function of the SGNs.
It has been reported proteolipid protein 1 (Plp1) was widely expressed in cochlear glial cells, including SCs and SGCs (Jessen and Mirsky, 2005; McLean et al., 2016; Meas et al., 2018b), while Sox2 was expressed in a cochlear glial subpopulation (Zuchero and Barres, 2015; Meas et al., 2018b). To label the cochlear glial cells and the Sox2+ subpopulation of glial cells in mice, we crossed the Plp1CreERT and Sox2CreERT with Rosa26-tdTomato line for lineage tracing. After induction with tamoxifen from P1 to P3, majority of the Sox2+ cells were labeled with tdTomato (70.6 ± 4.3% of the total Sox2+ cells) (Figure 2A). Importantly, Sox2CreERT/tdTomato (Sox2-tdT) positive cells all expressed Sox2 in the cochlea, suggesting the inducible Sox2-tdT specifically labels the Sox2+ subpopulation of glial cells in mice (Figure 2A).
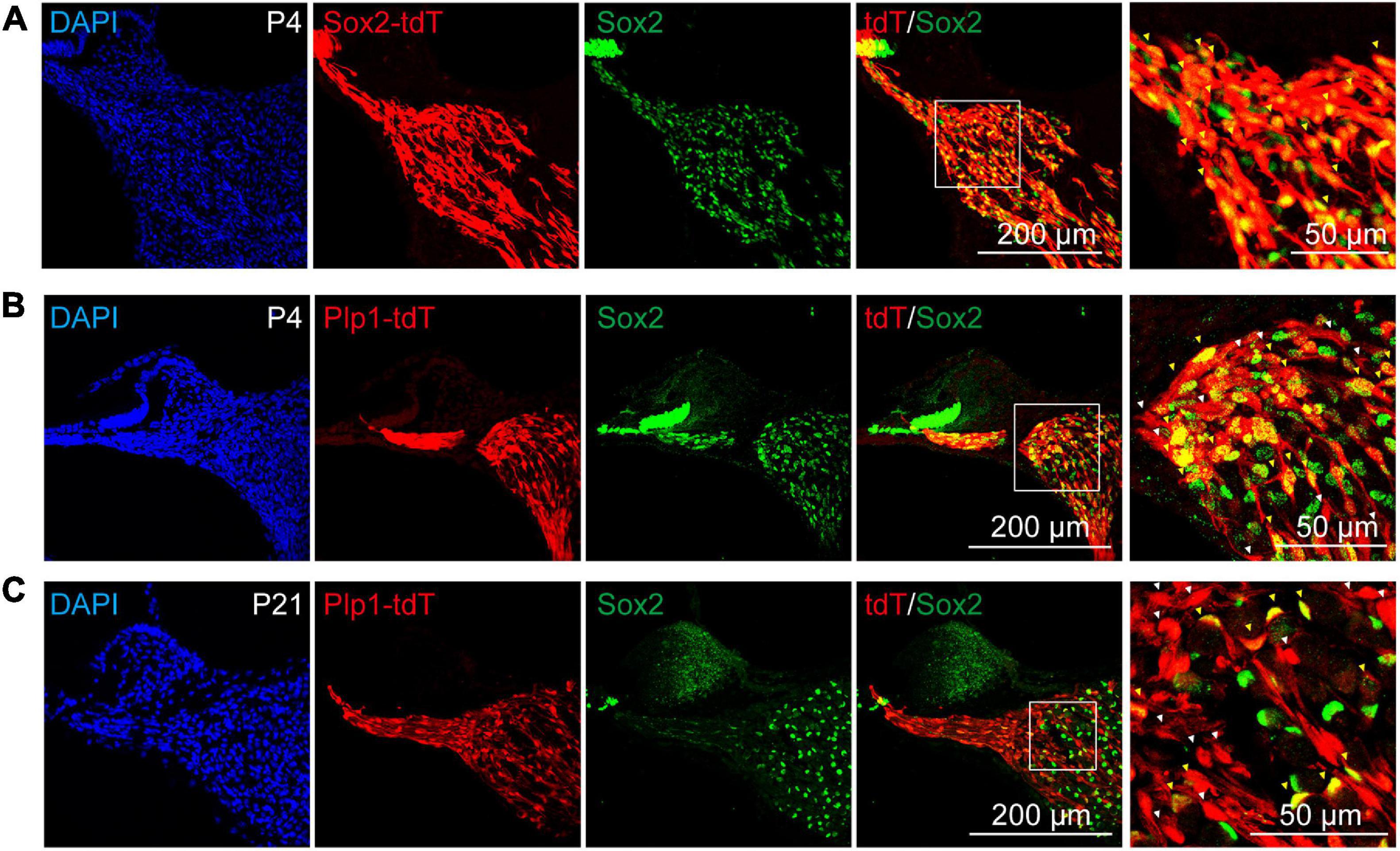
Figure 2. Sox2 labels a subpopulation of cochlear glial cells. (A) Immunocytochemical staining of Sox2-tdT cochlea for Sox2, showed Sox2-Cre:tdTomato labeled the Sox2 positive cochlear glial cells in P4 mice. The yellow arrowheads represent Sox2-tdT+/Sox2+ cells. (B,C) Sox2 were expressed in a subpopulation of Plp1-tdT positive cells in both P4 (B) and P21 (C) cochlea. The white arrowheads represent Plp1-tdT+/Sox2– cells, and the yellow arrows represent Plp1-tdT+/Sox2+ cells. tdTomato expression was induced by tamoxifen from P1 to P3.
The results of the immunofluorescence showed that Plp1CreERT/tdTomato (Plp1-td) not only labels the Sox2+ glial cells, but also Sox2– glial cells at both P4 and P21 (Figures 2B,C), indicating that Sox2 positive cells are a subpopulation of Plp1 positive cochlear glial cells. Thus, Plp1-tdT labels both SCs and SGCs while Sox2-tdT positive glial cells represents a lineage population of cochlear glial cells.
To directly compare the potency of Plp1+ and Sox2+ glial cells as neuronal progenitors, the lineage-traced cells (Plp1-tdT and Sox2-tdT) were subjected to sphere formation and neuronal differentiation assays (Figure 3A). Although both Plp1-tdT and Sox2-tdT cells were able to form spheres, >95% of the spheres were Plp1-positive while only about 60% of the spheres were Sox2-positive (Figures 3B,C), suggesting that both Plp1+/Sox2+ and Plp1+/Sox2– glial cells were able to proliferate as spheres. Interestingly, Plp1-tdT+ or Sox2-tdT+ cells were clustered at the periphery of the spheres with the non-glial cells proliferating at the center of spheres (Figure 3B). These non-glial cells were also highly proliferative and Plp1-tdT or Sox2-tdT cells contribute to only about 20 or 12% of the total cells after sphere culture, respectively (Supplementary Figure 1A). To investigate the identity of these non-glial cells, Plp1-tdT– cells were separated from Plp1-tdT+ cells by FACS followed by continuous sphere culture (Supplementary Figure 1B). While the non-glial markers failed to express Sox10 or Sox2 as expected, they expressed high levels of Sox9 and Pou3f4 (Supplementary Figure 1C), both of which are enriched in otic mesenchyme cells (Coate et al., 2012; Brooks et al., 2020).
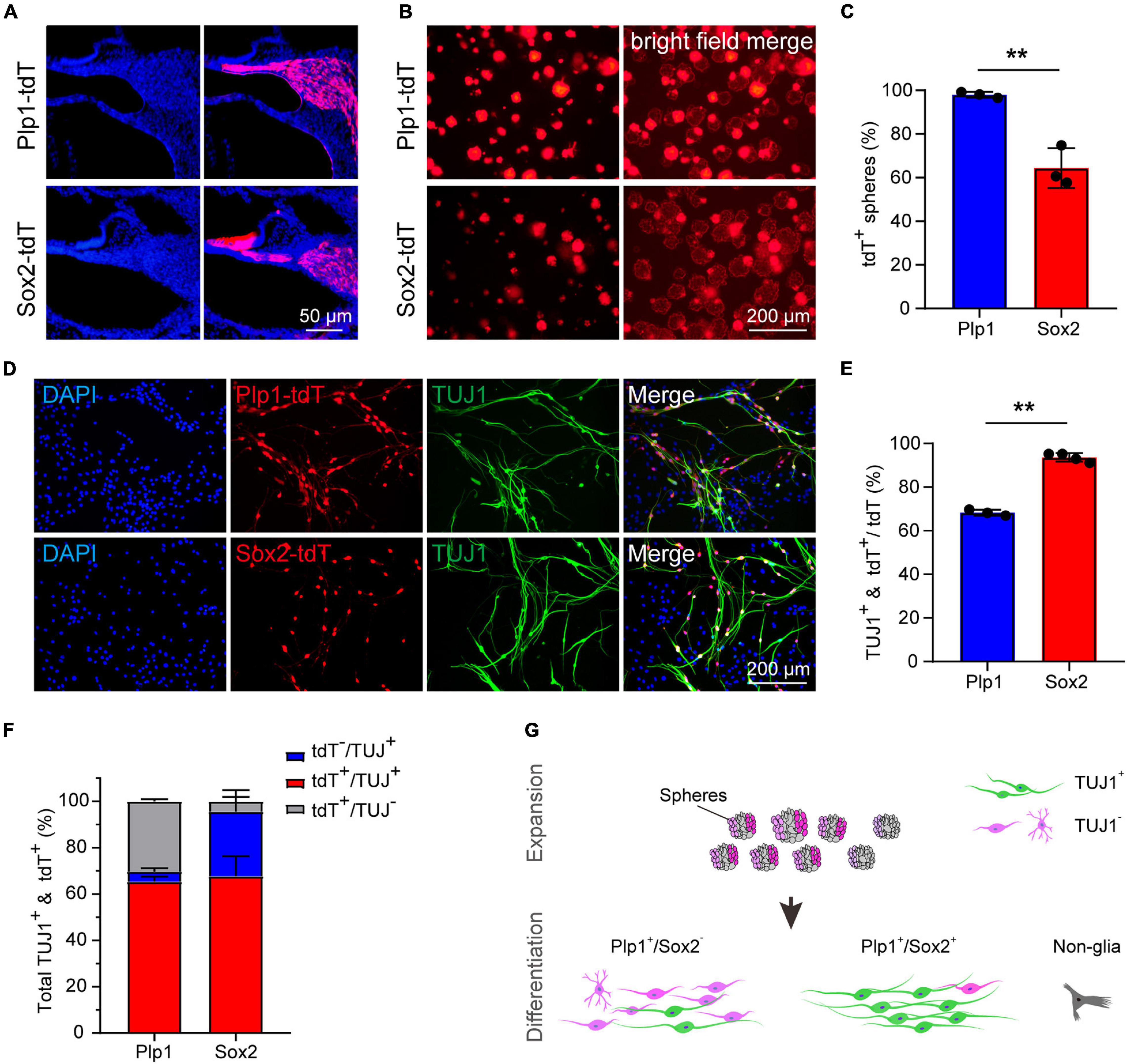
Figure 3. The Sox2+ cochlear glial cells were highly efficient in neuronal differentiation. (A) Distributions of Sox2-tdT and Plp1-tdT positive cochlear glial cells in P4 cochlea. tdTomato expression was induced by tamoxifen from P1 to P3. (B,C) Representative images and quantification of the Sox2-tdT and Plp1-tdT positive spheres at 5 div. N = 3, error bars represent mean ± SD. (D,E) Representative images and ratios of TUJ1+ neurons differentiated from Sox2-tdT and Plp1-tdT positive cells at 8–9 div. N = 3 or 4, error bars represent mean ± SD. Data from 3 to 4 independent experiments. (F) Percentages of TUJ1+ neurons derived from Sox2-tdT or Plp1-tdT positive cells relative to the total amount of TUJ1+ plus tdT+ cells. (G) A model showing Sox2+ glial cells exhibit high potential for neuronal differentiation. **p < 0.01 by unpaired student’s t-test.
To address if Plp1+/Sox2+ and Plp1+/Sox2– glial cells display different potential for neuronal differentiation, spheres from Plp1-tdT and Sox2-tdT mice were induced to neuronal differentiation for 8–9 days. Firstly, both Plp1-tdT and Sox2-tdT positive cells developed typical bipolar neuronal morphology and expressed neuronal marker TUJ1 (Figure 3D). Intriguingly, while only ∼68% Plp1-tdT cells expressed TUJ1, more than 90% Sox2-tdT cells were co-labeled with TUJ1 (Figure 3E). Further analyses indicated that almost all TUJ1+ neurons were derived from Plp1+ glial cells but a large proportion of Plp1+ cells were unable to differentiate (Figure 3F, left). In contrast, although Sox2+ glial cells only contributed to ∼70% of total iNs, almost all Sox2+ cells were successfully differentiated (Figure 3F, right). These results indicate that Plp1+ cochlear glial cells are the major source of neuronal differentiation in vitro, and that Plp1+/Sox2+ glial subpopulation displays much higher potency/efficiency in neuronal differentiation compared to the Plp1+/Sox2– glial cells (Figure 3G).
As Sox2 expression marks a subpopulation of cochlear glial cells with high neuronal differentiation potency, we next examined the expression of Sox2 at different postnatal ages. The ratio of Sox2+ glial cells were calculated based on co-labeling with Sox10, a generic marker of cochlear glial cells similar to Plp1 (Jessen and Mirsky, 2005). We found that the percentage of Sox2+ glial cells gradually declined after birth (Figures 4A,B). Specifically, at P4, ∼70% glial cells were Sox2+, and the Sox2+ ratio decreased to ∼47 and ∼38% at P7 and P10. At hearing onset (P14), the percentage of Sox2+ glial cells reduced further to ∼30%, which was maintained at P21 (Figure 4C).
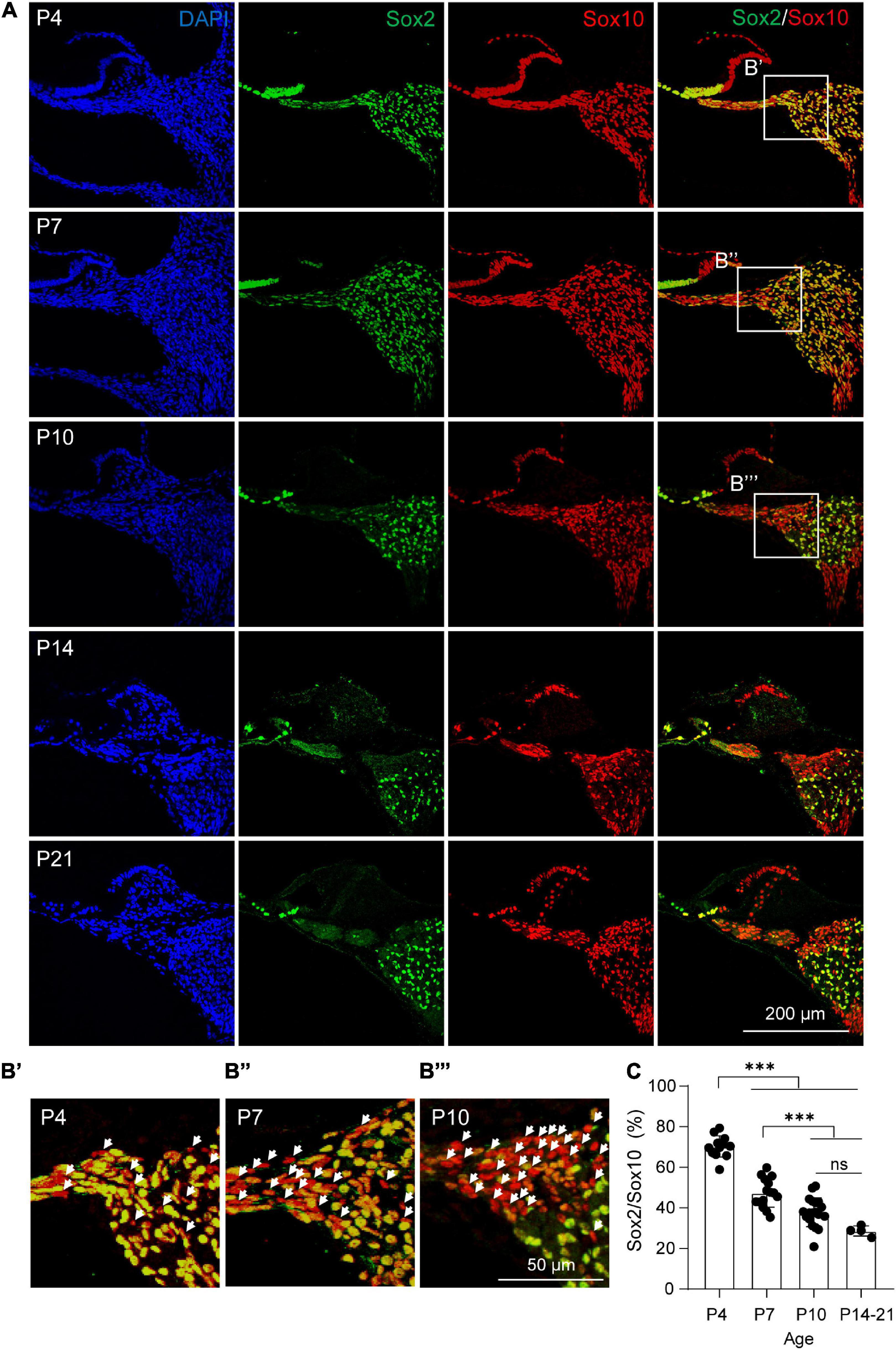
Figure 4. Dynamic changes of Sox2+ cochlear glial cells during postnatal development. (A,B) Immunocytochemical staining of Sox2 and Sox10 from P4, P7, P10, P14, and P21 cochleae. The white arrow represents the Sox2–/Sox10+ cell. (C) Percentages of Sox2+/Sox10+ cells from P4, P7, P10, and P14 to P21 cochleae. N = 4–13 sections from 3 cochleae, error bars represent mean ± SD. ***p < 0.001 by one-way ANOVA.
We next examined if the Sox2+ glial population remains highly efficient in neuronal differentiation at P21. Due to difficulties in obtaining sufficient cochlear glial cells in the calcified modiolus of P21 cochlea, the P21 cochlear glial cells were initially cultured 2D in SCM-FBS medium (Lang et al., 2011) and then transferred to 3D sphere suspension culture. We used the same inducible Plp1CreERT and Sox2CreERT to cross with Rosa26-tdTomato line to specific label the total glial cell population and Sox2+ glial cell subpopulation, respectively (Figure 3A). Lineage tracing was induced by tamoxifen injection from P17 to P20, and the cochlear glial cells isolated at P21. Both P21 Plp1-tdT and Sox2-tdT positive cells distributed at the periphery of spheres (Figure 5A), similar to the observation from the P4 spheres (Figure 3B). Followed by induction of neuronal differentiation, ∼55% Plp1+ glial cells were TUJ1 positive and ∼90% Sox2+ glial cells were TUJ1 positive (Figures 5B,C). In summary, the cochlear Sox2+ glial cell subpopulation preserves high potency of neuronal differentiation in both neonatal (P4) and P21 mice.
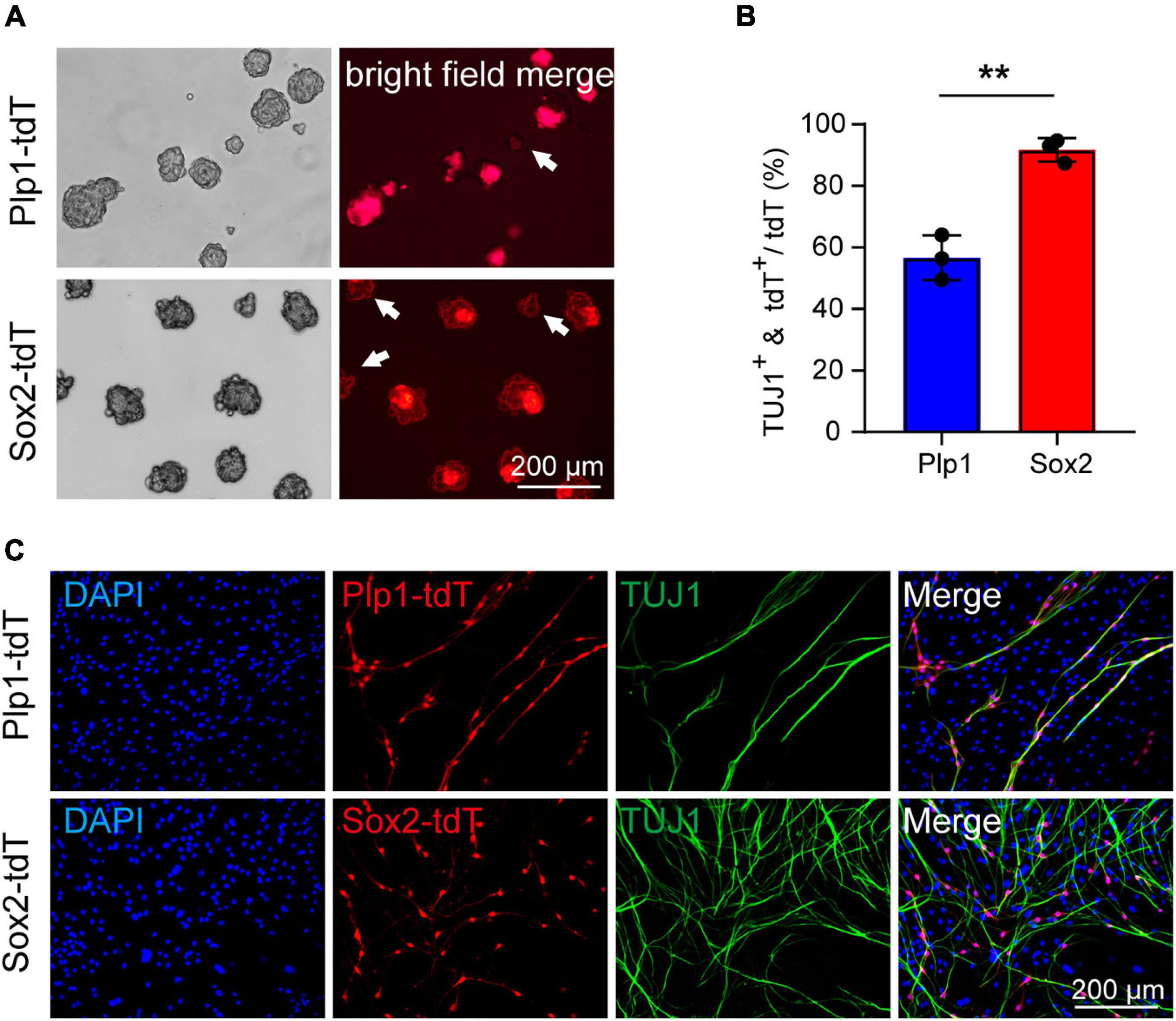
Figure 5. Cochlear Sox2+ glial subpopulation preserves high potency of neuronal differentiation in P21 mice. (A) Representative images of the Sox2-tdT and Plp1-tdT positive spheres culture 5 div. The white arrow represents the Plp1-tdT or Sox2-tdT negative spheres. (B,C) Representative images (C) and ratios (B) of TUJ1+ neurons differentiated from Sox2-tdT or Plp1-tdT positive cells. N = 3, error bars represent mean ± SD. **p < 0.01 by unpaired student’s t-test.
To further investigate if the Sox2 expression contributes to the high neuronal differentiation potential of the Sox2+ glial cells, we overexpressed Sox2 in both Sox2+ and Sox2– glial cells during 3D suspension culture of the spheres. Spheres were infected with lentivirus encoding Sox2-GFP or GFP control and the Sox2 overexpression was validated by both RT-qPCR and immunofluorescence (Figures 6A,B).
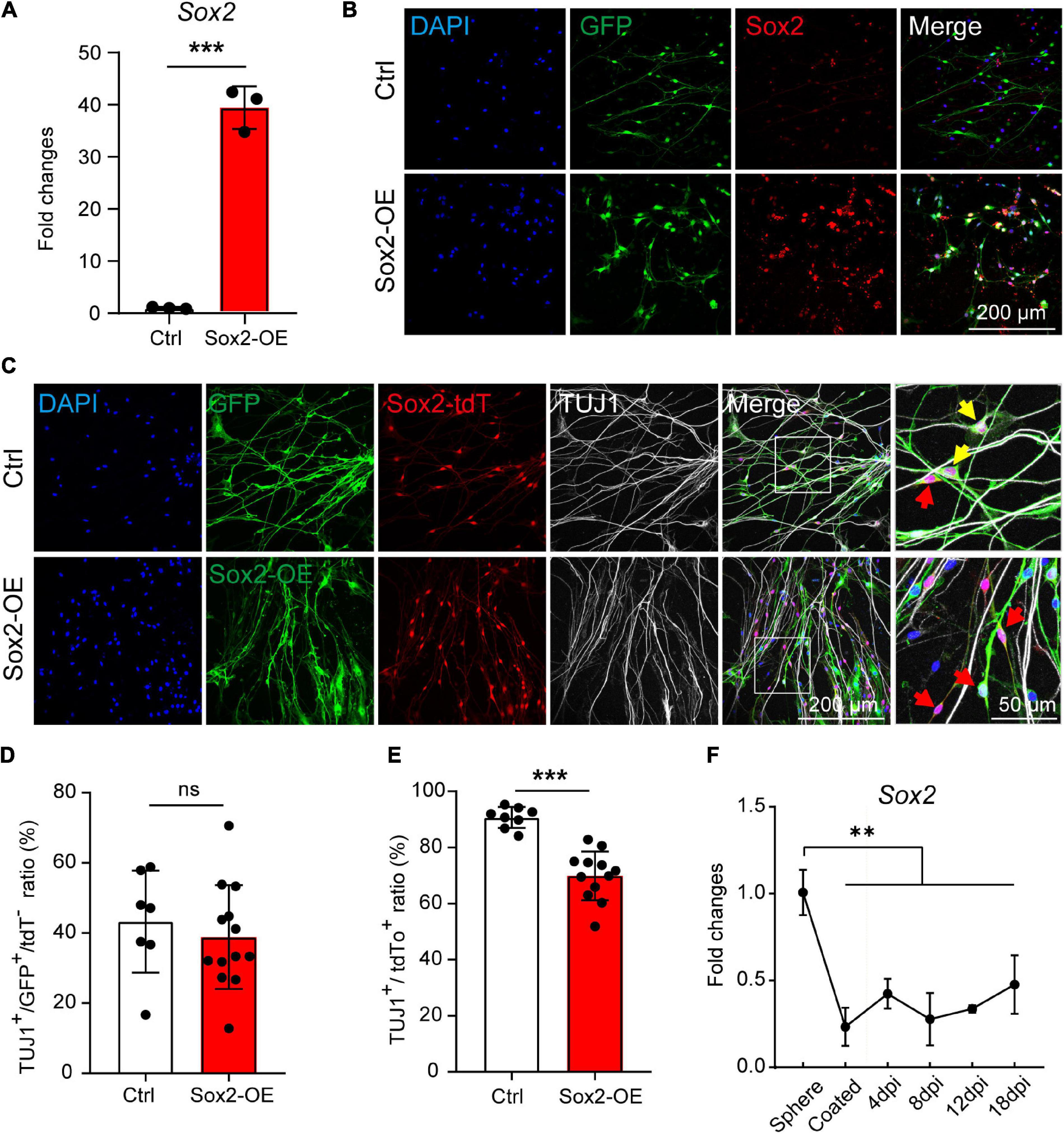
Figure 6. Sox2 overexpression inhibits the neuronal differentiation of Sox2+ glial cells. (A,B) mRNA expression (A) and immunocytochemical staining (B) of Sox2 overexpressed in cochlear glial cells. N = 3, error bars represent mean ± SD. (C) Immunocytochemical staining of TUJ1+ neurons differentiated from cochlear glial cells of Sox2-tdT mice. The yellow arrows represent TUJ1+/GFP+/tdT+ cells, and the red arrows represent TUJ1–/GFP+/tdT+ glial cells. (D) Quantification of TUJ1+/GFP+ cells from Sox2-tdT negative glial cells. N = 8–13 images from 3 coverslips, error bars represent mean ± SD. (E) Quantification of TUJ1+/Sox2-tdT+ cells from glial cells overexpressing either GFP or Sox2. N = 7–11 images from 3 coverslips, error bars represent mean ± SD. (F) mRNA expression of Sox2 during induced neuronal differentiation of the cochlear glial cells at different conditions and time points. N = 3, error bars represent mean ± SD. **p < 0.01, ***p < 0.001 by one-way ANOVA (F) or unpaired student’s t-test (A,E).
Next, the spheres were induced to neuronal differentiation for 8–9 days and were immuno-stained with TUJ1 antibody. We first evaluated the effect of Sox2 overexpression on neuronal differentiation of Sox2– glial cells. Neuronal differentiation of Sox2– glial cells, as marked by TUJ1+/Sox2-tdT– cells, was not affect by Sox2 overexpression compared to GFP controls (Figures 6C,D). This result suggest that the incompetence of Sox2– glial cells in neuronal differentiation is not due to lack of Sox2 expression. To our surprise, neuronal differentiation of the Sox2-tdT (Sox2+) glial cells was significantly reduced after Sox2 overexpression (Figures 6C,E). Consistently, Sox2 expression was decreased dramatically from the proliferative stage in 3D culture to the differentiation stage in 2D culture, and then maintained at a stably low level during the entire process of neuronal differentiation (Figure 6F). These results suggest that Sox2+ glial cells exhibit high potential for neuronal differentiation independent of Sox2 expression, and that downregulation of Sox2 is required for efficient neuronal differentiation.
Although cochlear glial cells can be differentiated into iNs, the specific markers for SGNs were rarely detected, indicative a major huddle in SGN fate conversion and maturation. Small molecules have been reported to promote neuronal reprogramming from somatic and glial cells (Hu et al., 2015; He et al., 2017; Belin-Rauscent et al., 2018). To promote the neuronal differentiation and maturation to SGNs from cochlear glial cells, we next screened 10 neurogenic small molecules that were shown to activate neuronal signaling pathways, inhibit glial signaling pathways, or modulate epigenetics to promote neuronal reprogramming. Small molecules selected for our initial screening were as follows: SB431542, CHIR99021, Y27632, retinoic acid (RA), valproic acid (VPA), Forskolin, ISX9, I-BET151, Vitamin C (Vc), and LIF.
It has been reported that Forskolin, ISX9, I-BET151, and CHIR99021 (FIBC) combination induced neuronal differentiation from mouse embryonic fibroblasts (MEFs) and astrocytes efficiently (Hu et al., 2015; He et al., 2017). Firstly, we used the FIBC combination to test the concentration for neuronal differentiation from cochlear glial cells (Supplementary Figure 2A). Based on the bipolar neuronal morphology of TUJ1+ cells and minimal cytotoxicity, we identified the optimal concentrations of Forskolin (20 μM), ISX9 (20 μM), I-BET151 (2 μM), and CHIR99021 (10 μM). Next, we induced the neuronal differentiation with FIBC in the presence of SB431542, Y27632 (Y), RA, VPA, Vc, or LIF (L). The results showed FIBC and Y27632 (FIBCY) and FIBC and LIF (FIBCL) increased the expression of Prox1 (Supplementary Figure 3E), an SGN marker, compared with other small molecules (Supplementary Figure 2B). Thus, we used FIBCY and FIBCL as the small molecules cocktail for neuronal induction and maturation.
RT-qPCR results also showed increased expression of generic neuronal markers such as Tubb3 (Supplementary Figures 3A,B), and SGN-specific markers including Prox1, Islet1, and Scrt2 (Supplementary Figure 3G) of the iNs by small molecules FIBCL compared with SCDM or IM control medium (Figure 7A). Furthermore, the expression of Tubb3, Scrt2, and Islet1 were further increased in a temporal manner during neuronal differentiation (Figure 7A, 7 dpi vs. 18 dpi). FIBCY and FIBCL increased the number of TUJ1+ cells and induced both co-labeling of TUJ1 and Prox1 (Figure 7B). Quantitative analyses indicate that FIBCY and FIBCL significantly increased the percentage of overall TUJ1+ cells (Figure 7C) and percentage of TUJ1+ cells co-expressing Prox1 (Figure 7D). FIBCL appeared to perform better in inducing TUJ1+ iNs than FIBCY (Figure 7C). These results suggested that FIBCY/FIBCL promoted neuronal differentiation and maturation toward SGNs from neonatal cochlear glial cells.
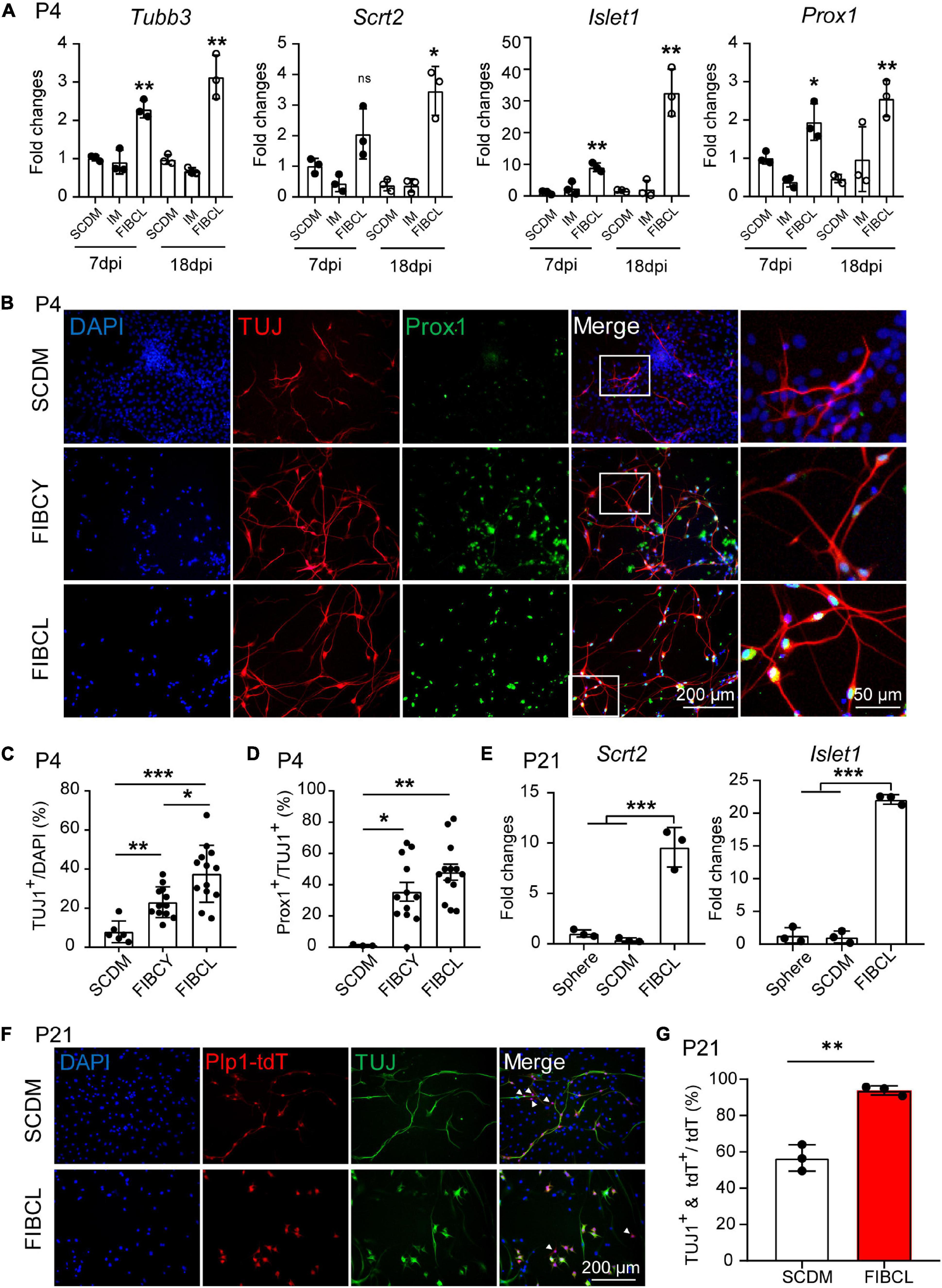
Figure 7. Small molecules promote neuronal differentiation and maturation of cochlear glial cells. (A) RT-qPCR gene expression analyses of P4 cochlear glial cells differentiated in SCDM, IM or FIBCL at 7 and 18 div. FIBCL, Forskolin, ISX9, I-BET151, CHIR99021, and LIF. N = 3, error bars represent mean ± SD. (B) Immunocytochemical stainings of TUJ1 and Prox1 in neurons induced from P4 cochlear glial cells at different conditions. (C,D) Percentages of TUJ1 positive cells (C) and TUJ1/Prox1 double positive cells (D) differentiated in SCDM, FIBCY, or FIBCL. N = 3–13 images from 2 wells of each condition, error bars represent mean ± SD. (E) mRNA expression of SGN markers Scrt2 and Islet1. N = 3, error bars represent mean ± SD. (F) Immunocytochemical staining of TUJ1 of induced neurons from P21 Plp1-tdT cochlear glial cells. The white arrowheads represent the Plp1-tdT+/TUJ1– cells. (G) Percentages of TUJ1 positive cells differentiated from P21 Plp1-tdT+ cochlear glial cells. N = 3, error bars represent mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001 by one-way ANOVA (A,C–E) or unpaired student’s t-test (G).
We then induced neuronal differentiation of P21 Plp1-tdT glial cells in the presence or absence of FIBCL. For P21 glial cells, FIBCL also promoted the expression of SGN markers Scrt2, Islet1 (Figure 7E). Remarkably, FIBCL treatment resulted in a significant increase in the percentage of TUJ1+/Plp1-tdT+ cells (∼90%) compared to the control group (∼55%) (Figures 7F,G). As Plp1+/Sox2+ but not Plp1+/Sox2– cells showed potent neuronal differentiation in control condition, FIBCL treatment may have also promoted neuronal differentiation of Plp1+/Sox2– glial cells. Together, these results highlight the effectiveness of small molecule combinations FIBCY and FIBCL in promoting neuronal differentiation and SGN maturation of neonatal and P21 cochlear glial cells.
To evaluate the maturity under different induction conditions of iNs, we performed transcriptome sequencing analyses. Pearson correlation analyses showed that the expression profiles of neurons induced by SCDM and SCDM/FGF were relatively closer to that of the spheres (Figure 8A). However, the overall expression profile of FIBCL-iNs was distinct from those of the spheres, SCDM or SCDM/FGF, and was more similar to that of the primary SGNs (Figure 8A). In addition, the results of principal component analyses (PCA) showed that FIBCL-iNs were similar to the SGNs (Figure 8B). Comparing the differentially expressed genes between SCDM and FIBCL-iNs, we found about 3200 genes in the FIBCL group were significantly upregulated and 3000 genes were significantly downregulated (Figure 8C). The GO and KEGG enrichment analyses of these genes, respectively showed that the upregulated genes were mainly enriched in neuron development, neurotransmitter, synapse and calcium ion channels; while the downregulated genes contained glial cell migration and myelination, as well as GPCR and cytokine signaling pathways (Figures 8D–G).
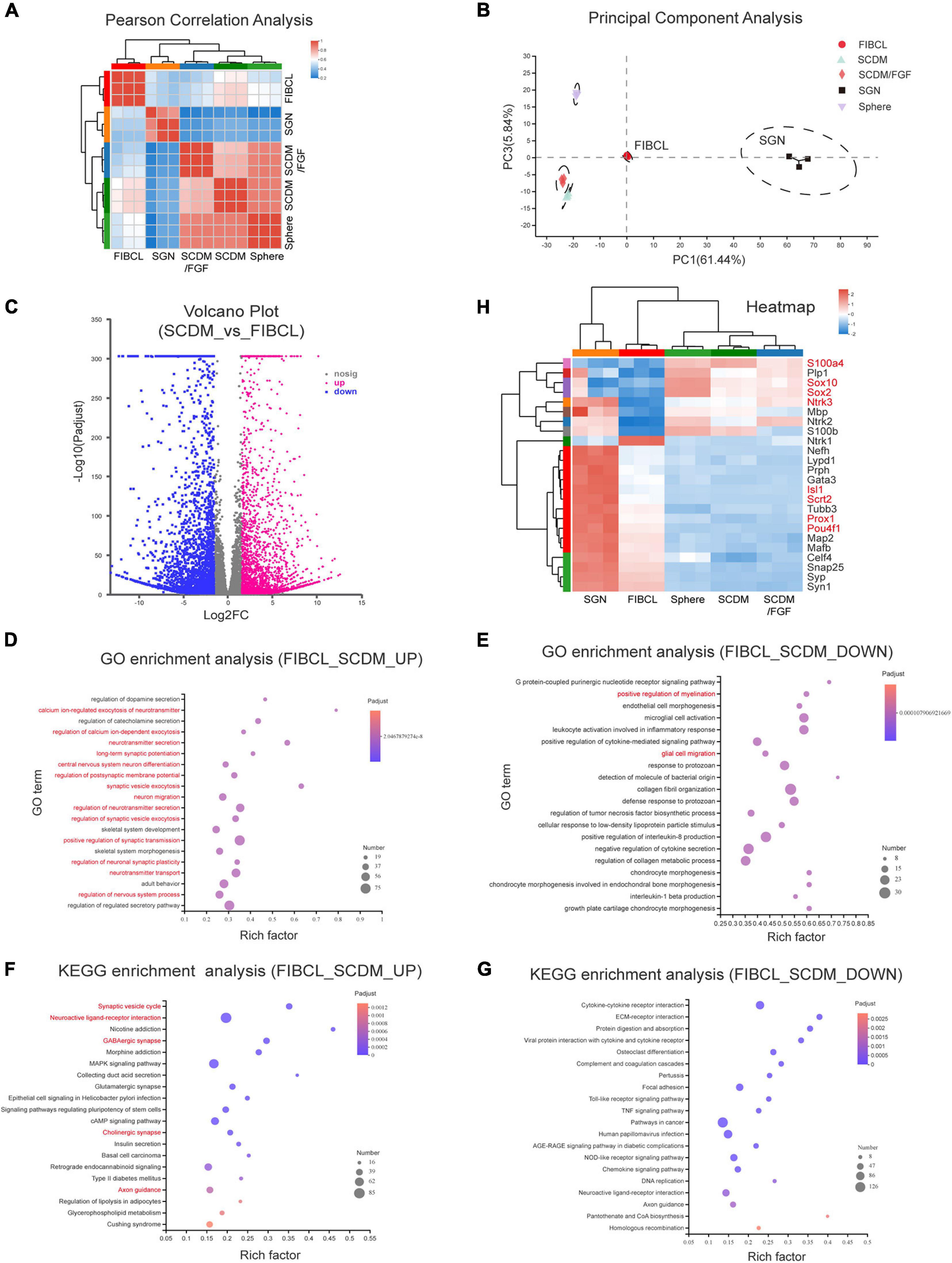
Figure 8. Transcriptomic analyses of the induced neurons. (A,B) Pearson correlation analysis (A) and principal component analysis (B) of neurospheres, differentiated culture in SCDM, SCDM/FGF, FIBCL media, and primary SGNs. (C) Volcano graph of the genes upregulated and downregulated in the FIBCL-induced neurons compared to the SCDM media. (D,E) GO enrichment analysis of upregulated (D) and downregulated (E) functional pathways in the FIBCL-induced neurons compared to the SCDM media. (F,G) KEGG enrichment analysis of upregulated (D) and downregulated (E) cellular processes in the FIBCL-induced neurons compared to the SCDM media. (H) Heatmap graph of specific glial and neuronal genes expressed in spheres, differentiated culture in SCDM, SCDM/FGF, FIBCL media, and primary SGNs. Changes in expression of selected genes (gene symbol highlighted in red) were validated by RT-qPCR in Supplementary Figure 4.
Analyses of specific gene expression revealed that glial cell-specific genes such as Plp1, Sox10, S100β, and MBP still maintained high expression in sphere, SCDM and SCDM/FGF groups, but the expression levels of these genes were significantly downregulated in FIBCL-iNs group (Figure 8H). Interestingly, Sox2 expression was also further reduced after FIBCL treatment (Figure 8H), consistent with an inhibitory role of Sox2 in neuronal differentiation (Figure 6). In addition, expression levels of neuron-specific genes such as Tubb3, Nefh, Snap25, Syp (Supplementary Figure 3D), and Map2 (Supplementary Figure 3F) were induced in SCDM and SCDM/FGF groups compared to the spheres, which were further upregulated in the FIBCL group (Figure 8H). Finally, the expression levels of SGN-specific genes such as Isl1, Pou4f1, Prox1, Gata3 (Supplementary Figure 3C), and Mafb were also significantly upregulated, while cochlear glial genes such as Sox10, Sox2, and S100a4 were downregulated in the FIBCL-iNs (Figure 8H and Supplementary Figure 4). Overall, the RNA-seq results indicate that iNs in control medium were at the immature state, retaining some of the glial cell characteristics; while small molecules FIBCL removed glial cell barriers and further promoted neuronal maturation.
Spiral ganglion neurons lack the ability to regenerate after damage in the mammalian cochlea, which is a major cause of auditory neuropathy and may compromise the therapeutic effects of cochlear implants (Guo et al., 2019). Extensive studies have been performed using neural stem cells for regeneration of neurons and the SGNs (Fang et al., 2019; Li et al., 2019; Tang et al., 2019; Han et al., 2020; Xia et al., 2020; Yang et al., 2020, 2021; Yuan et al., 2021). Yet, functional regeneration of SGNs from the resident glial cells may represent a novel strategy for hearing restoration caused by SGN damages. In this study, we found that the cochlear Sox2+ glial cell subpopulation exhibits high potency of neuronal differentiation, and the efficiency of neuronal differentiation requires Sox2 downregulation. Furthermore, we identified a small molecule combination that promotes neuronal differentiation and maturation toward SGN fate (Figure 9).
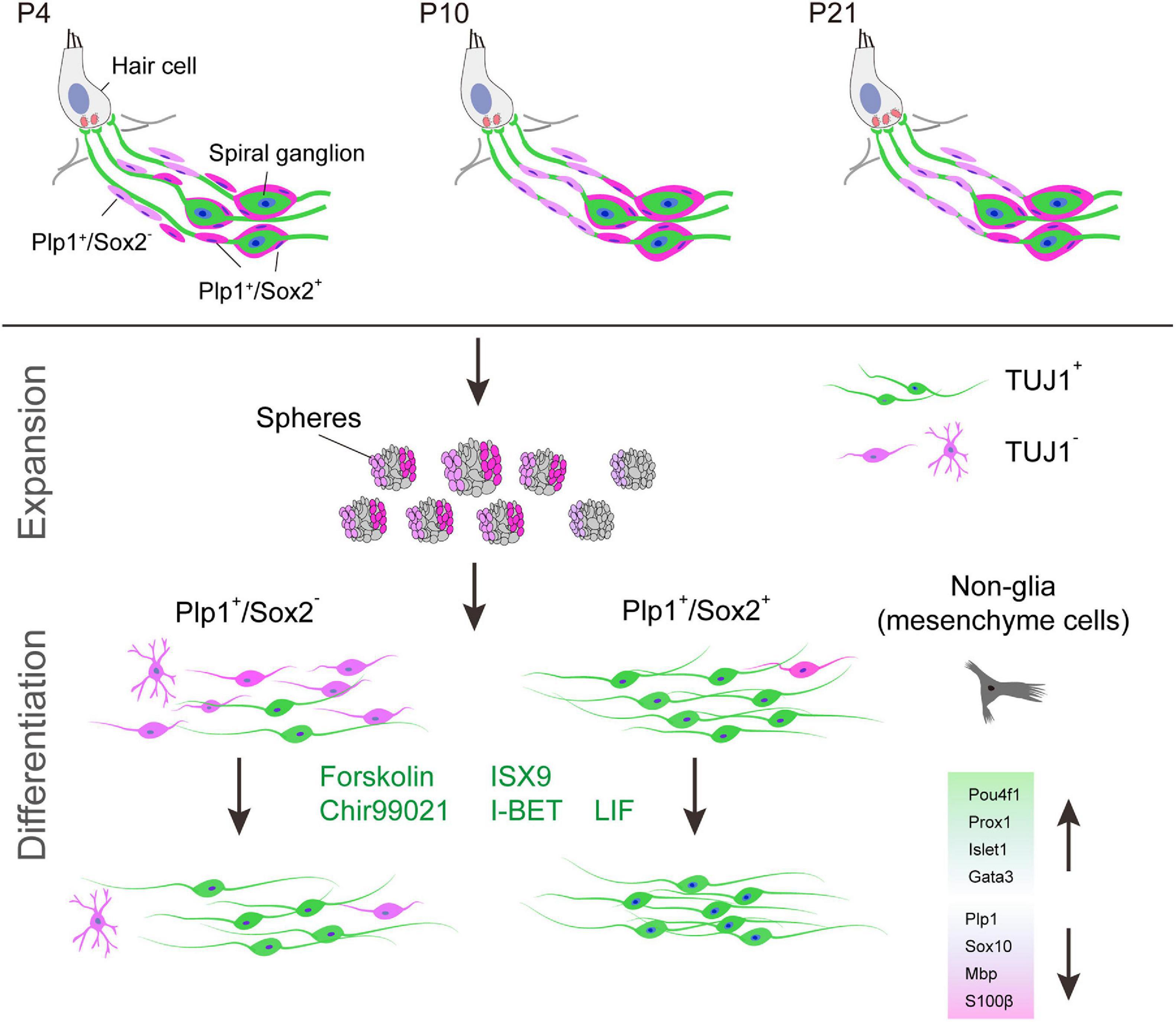
Figure 9. Graphic summary of cochlear Sox2+ glial cells as potent progenitors for spiral ganglion neuron reprogramming induced by small molecules. Sox2+ cells are a subpopulation of Plp1+ cochlear glial cells. Although both Sox2+ and Sox2– glial cells can form neurospheres, Plp1+/Sox2+ spheres are more neurogenic compared to Plp1+/Sox2– spheres. Efficiency and maturity of the induced neurons can be further improved by a small molecule cocktail.
The starting progenitor population is one of the key considerations for successful cell fate reprogramming (Gascon et al., 2017). In the brain and retina, astrocytes (Guo et al., 2014; Qian et al., 2020) and Müller glial cells (Qian et al., 2020) have been shown to convert to new neurons and retinal ganglion cells under specific conditions. Cochlear glial cells were regarded as specialized cells and exhibited characteristics as well as corresponding functions of glial cells. Similar to the central glial cells, cochlear glial cells can also proliferate and regenerate themselves upon injury in vivo (Lang et al., 2015; Wan and Corfas, 2017), suggesting that these glial cells may also function as progenitors for SGNs.
Cochlear glial cells are heterogeneous and include SCs and Satellite cells (Wan and Corfas, 2017). While Plp1 and Sox10 are generic markers for cochlear glial cells, Sox2 is only expressed in a subset of glial cells. Sox2+ glial cells are mainly located around the SGN cell body, which overlapped with SGCs. A few Sox2+ glial cells are also located along SGN axons at OSL after P14 in mice. In this study, we identified Sox2+ glial cells sub-population as potent neuronal progenitors compared to Sox2– glial cells, suggesting that Sox2+ glial cells may be the target progenitor population for SGN regeneration in future studies.
In recent years, mechanisms of proliferation and neuronal differentiation of the cochlear glial progenitor has been under close investigation. The Plp1+ cochlear glial cells serve as potent progenitors for neurons, astrocytes and oligodendrocytes, but not hair cells in vitro (McLean et al., 2016). Cochlear glial cells isolated from Sox2-eGFP reporter mice also displayed potent neurogenic potential in vitro (Lang et al., 2015; Meas et al., 2018a). However, a direct comparison of glial subpopulations is lacking and the iNs did not appear to mature as SGNs. Importantly, two groups recently reported that iNs can be generated from Plp1+ glial cell by Ng1/Nd1 or Lin28 overexpression in vivo post-SGN injury, and that iNs expressed both pan-neuronal and SGN markers (Kempfle et al., 2020; Li X. et al., 2020). However, the efficiency of neuronal conversion and neuronal maturity is still limited. Based on our findings, we believe that direct reprogramming of the Sox2+ glial cells may be key to enhance functional glia-to-neuron conversion in vivo.
Developmentally, Sox2 is generally expressed in a variety of cells types including neuronal stem cells (Graham et al., 2003; Zhang and Cui, 2014; Cui et al., 2018) and tissue specific cells, such as DRG satellite cells and cochlear supporting cells (Kiernan et al., 2005; Koike et al., 2015). Our finding on higher potential of Sox2+ glial cells poses a question on whether Sox2 expression is required for efficient neuronal induction or merely serves as a marker for progenitors with high neurogenic potential. Our results point to the later scenario and may also suggest that Sox2 serves as an inhibitory signal for efficient neuronal induction. This notion is consistent with the observations in CNS, whereby Sox2 was highly expressed in neuronal progenitor cells (NPCs) and inhibited the neuronal differentiation of these progenitor cells (Graham et al., 2003; Cavallaro et al., 2008). Sox2 expression was gradually downregulated during the process of neuronal differentiation (Cavallaro et al., 2008; Cui et al., 2018; Mercurio et al., 2019). Knockdown of Sox2 partially rescued the impairment of neuronal differentiation induced by miR-145 downregulation (Morgado et al., 2016).
Intriguingly, although Sox2 was downregulated during neuronal differentiation of the cochlear glial cells, we observed that the iNs still maintained the expression of Sox2 at a specific level. Sox2 expression of iNs was higher than that of SGNs, which may present a barrier for further neuronal maturation. This is in congruence with our finding that the small molecule cocktail promoted further neuronal maturation while also significantly reduced the expression of Sox2.
Neuronal reprogramming may not be initiated due to the stable barriers of glial cells identity and lack of neuronal factors to regulate specific transcriptional program (Mertens et al., 2016; Black and Gersbach, 2018; Li et al., 2018). Small molecules that modulate specific signaling pathways have been shown to promote neuronal reprogramming. For example, small molecules CHIR99021, Forskolin, and ISX9 were shown to improve the neuronal conversion and differentiation (Schneider et al., 2008; Dworkin and Mantamadiotis, 2010; Liu et al., 2013; Li et al., 2015; Gao et al., 2017; Gascon et al., 2017; Yang et al., 2019). IBET151 serves to erase the initial cell-fate specific gene expression pattern (Di Micco et al., 2014; Li et al., 2015; Wu et al., 2015; Marazzi et al., 2018). VPA has been reported to enhanced neuronal induction of cochlear glial cells (Moon et al., 2018). However, more small molecules were hardly studied in cochlear glia-to-neuron induction.
In this study, the small molecule cocktail we identified can promote neuronal maturation and cell fate conversion toward SGNs. The results showed upregulation of generic neuronal genes and SGN specific genes such as Prox1, Islet1, Pou4f1, and Scrt2. Consistent with previous reports that Ng1/Nd1 or Lin28 induced glial cells-to-auditory neuron conversion in the cochlea (Kempfle et al., 2020; Li X. et al., 2020), the iNs also exhibited increased expression of both Nd1 and Lin28. Furthermore, we observed the downregulation of glial cell markers such as Plp1, Sox10, S100β, and MBP. Thus, the small molecule cocktail induces neuron differentiation and maturation by removal of glial identity and establishment of neuronal identity.
Although the small molecule cocktail greatly promoted neuronal induction toward SGN, expression levels of SGN-specific genes were still lower than those in primary SGNs and some key SGN markers, such as Ntrk3, were not upregulated. It is possible that means to upregulate these developmental and functional relevant genes may further promote the maturation of the iNs to functional SGNs in vivo.
Other challenges in functional maturation of the iNs remain to be addressed. For example, synaptic connections between the iNs and hair cells or cochlear nucleus are the fundamental elements of the cochlear neuronal circuitry in vivo. Previous studies showed synapse formation between ESC-derived iNs and sensory epithelium or cochlear nucleus in vitro with limited efficiency (Liu et al., 2018; Meas et al., 2018b) and the de novo synapses were not observed in vivo (Kempfle et al., 2020; Li X. et al., 2020). Secondly, SGNs are heterogenous in nature and display distinct molecular signature and spontaneous firing rates (Petitpre et al., 2018). The determination of SGN identity is induced by interaction of different spontaneous activity from hair cells (Petitpre et al., 2018; Shrestha et al., 2018; Sun et al., 2018). How the iNs may adopt specific SGN subtype and integrate into the cochlear neuronal circuitry needs to be explored in future studies.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The animal study was reviewed and approved by Institutional Animal Care and Use Committee of Model Animal Research Center of Nanjing University.
GW conceived the study. ZC and GW designed the experiments, analyzed the results, and wrote the manuscript. ZC, YH, CY, QL, and CQ performed the experiments. All authors have read and agreed to the published version of the manuscript.
This work was supported by grants from the National Natural Science Foundation of China (31771153 and 81970888 to GW) and the Fundamental Research Funds for the Central Universities (0903-1480601101 to GW).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.728352/full#supplementary-material
Supplementary Figure 1 | The non-glial cells in the spheres are likely derived from cochlear mesenchyme cells. (A) Percentage of Plp1-tdT+ or Sox2-tdT+ cells to the total number of cells after sphere culture. N = 8–10, error bars represent mean ± SD. (B) Representative images of Plp1-tdT+ glial and Plp1-tdT– non-glial cells after FACS followed by sphere culture. (C) RT-qPCR analyses of the glial and non-glial spheres with glial markers (Sox10, Sox2) and mesenchymal markers (Sox9, Pou3f4). N = 3, error bars represent mean ± SD. ***p < 0.001 by unpaired student’s t-test.
Supplementary Figure 2 | Small molecules screening and optimization. (A) Representative images of TUJ1 immunostaining of iNs treated with various concentrations of small molecules, I-BET, Forskolin, ISX9, Chir99021 at 15 div. (B) Representative images of Prox1 immunostaining of iNs treated with small molecules Y27632, SB431542, RA, LIF, Vitamin C, and VPA at 15 div. The white arrowheads represent Prox1+ cells.
Supplementary Figure 3 | Specific expression of SGN markers. (A) Representative cartoon showing cochlear localization of SGNs and surrounding glial cells. (B–G) Specific expressions of TUJ1 (B), Gata3 (C), Syp (D), Prox1 (E), Map2 (F), and Scrt2 (G) in P3–P42 cochlear SGNs.
Supplementary Figure 4 | RT-qPCR validations of glial and neuronal genes regulated during FIBCL-induced differentiation. mRNA expressions of Islet1, Scrt2, Pou4f1, Prox1, Sox10, Sox2, Ntrk3, and S100a4 (as highlighted in Figure 8H) in non-differentiated spheres or differentiated cultures treated with SCDM, SCDM/FGF, or FIBCL were analyzed by RT-qPCR. N = 3, error bars represent mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001 by one-way ANOVA.
Akil, O., Blits, B., Lustig, L. R., and Leake, P. A. (2019). Virally mediated overexpression of glial-derived neurotrophic factor elicits age- and dose-dependent neuronal toxicity and hearing loss. Hum. Gene Ther. 30, 88–105. doi: 10.1089/hum.2018.028
Bao, J., and Ohlemiller, K. K. (2010). Age-related loss of spiral ganglion neurons. Hear Res. 264, 93–97. doi: 10.1016/j.heares.2009.10.009
Belin-Rauscent, A., Lacoste, J., Hermine, O., Moussy, A., Everitt, B. J., and Belin, D. (2018). Decrease of cocaine, but not heroin, self-administration and relapse by the tyrosine kinase inhibitor masitinib in male Sprague Dawley rats. Psychopharmacology 235, 1545–1556. doi: 10.1007/s00213-018-4865-0
Black, J. B., and Gersbach, C. A. (2018). Synthetic transcription factors for cell fate reprogramming. Curr. Opin. Genet. Dev. 52, 13–21. doi: 10.1016/j.gde.2018.05.001
Brooks, P. M., Rose, K. P., MacRae, M. L., Rangoussis, K. M., Gurjar, M., Hertzano, R., et al. (2020). Pou3f4-expressing otic mesenchyme cells promote spiral ganglion neuron survival in the postnatal mouse cochlea. J. Comp. Neurol. 528, 1967–1985. doi: 10.1002/cne.24867
Cavallaro, M., Mariani, J., Lancini, C., Latorre, E., Caccia, R., Gullo, F., et al. (2008). Impaired generation of mature neurons by neural stem cells from hypomorphic Sox2 mutants. Development 135, 541–557. doi: 10.1242/dev.010801
Chen, W., Jongkamonwiwat, N., Abbas, L., Eshtan, S. J., Johnson, S. L., Kuhn, S., et al. (2012). Restoration of auditory evoked responses by human ES-cell-derived otic progenitors. Nature 490, 278–282. doi: 10.1038/nature11415
Coate, T. M., Raft, S., Zhao, X., Ryan, A. K., Crenshaw, E. B. III, and Kelley, M. W. (2012). Otic mesenchyme cells regulate spiral ganglion axon fasciculation through a Pou3f4/EphA4 signaling pathway. Neuron 73, 49–63. doi: 10.1016/j.neuron.2011.10.029
Cui, C. P., Zhang, Y., Wang, C., Yuan, F., Li, H., Yao, Y., et al. (2018). Dynamic ubiquitylation of Sox2 regulates proteostasis and governs neural progenitor cell differentiation. Nat. Commun. 9:4648. doi: 10.1038/s41467-018-07025-z
Di Micco, R., Fontanals-Cirera, B., Low, V., Ntziachristos, P., Yuen, S. K., Lovell, C. D., et al. (2014). Control of embryonic stem cell identity by BRD4-dependent transcriptional elongation of super-enhancer-associated pluripotency genes. Cell Rep. 9, 234–247. doi: 10.1016/j.celrep.2014.08.055
Diensthuber, M., Zecha, V., Wagenblast, J., Arnhold, S., Edge, A. S., and Stover, T. (2014a). Spiral ganglion stem cells can be propagated and differentiated into neurons and glia. Biores. Open. Access 3, 88–97. doi: 10.1089/biores.2014.0016
Diensthuber, M., Zecha, V., Wagenblast, J., Arnhold, S., and Stover, T. (2014b). Clonal colony formation from spiral ganglion stem cells. Neuroreport 25, 1129–1135. doi: 10.1097/WNR.0000000000000240
Dworkin, S., and Mantamadiotis, T. (2010). Targeting CREB signalling in neurogenesis. Expert Opin. Ther. Targets 14, 869–879. doi: 10.1517/14728222.2010.501332
Fang, Q., Zhang, Y., Chen, X., Li, H., Cheng, L., Zhu, W., et al. (2019). Three-dimensional graphene enhances neural stem cell proliferation through metabolic regulation. Front. Bioeng. Biotechnol. 7:436. doi: 10.3389/fbioe.2019.00436
Fettiplace, R. (2017). Hair cell transduction, tuning, and synaptic transmission in the mammalian cochlea. Compr. Physiol. 7, 1197–1227. doi: 10.1002/cphy.c160049
Fryatt, A. G., Mulheran, M., Egerton, J., Gunthorpe, M. J., and Grubb, B. D. (2011). Ototrauma induces sodium channel plasticity in auditory afferent neurons. Mol. Cell Neurosci. 48, 51–61. doi: 10.1016/j.mcn.2011.06.005
Gao, L., Guan, W., Wang, M., Wang, H., Yu, J., Liu, Q., et al. (2017). Direct generation of human neuronal cells from adult astrocytes by small molecules. Stem Cell Rep. 8, 538–547. doi: 10.1016/j.stemcr.2017.01.014
Gascon, S., Masserdotti, G., Russo, G. L., and Gotz, M. (2017). Direct neuronal reprogramming: achievements, hurdles, and new roads to success. Cell Stem Cell 21, 18–34. doi: 10.1016/j.stem.2017.06.011
Graham, V., Khudyakov, J., Ellis, P., and Pevny, L. (2003). SOX2 functions to maintain neural progenitor identity. Neuron 39, 749–765. doi: 10.1016/S0896-6273(03)00497-5
Guo, R., Li, J., Chen, C., Xiao, M., Liao, M., Hu, Y., et al. (2021). Biomimetic 3D bacterial cellulose-graphene foam hybrid scaffold regulates neural stem cell proliferation and differentiation. Colloids Surf. B Biointerfaces 200:111590. doi: 10.1016/j.colsurfb.2021.111590
Guo, R., Ma, X., Liao, M., Liu, Y., Hu, Y., Qian, X., et al. (2019). Development and application of cochlear implant-based electric-acoustic stimulation of spiral ganglion neurons. ACS Biomater. Sci. Eng. 5, 6735–6741. doi: 10.1021/acsbiomaterials.9b01265
Guo, R., Xiao, M., Zhao, W., Zhou, S., Hu, Y., Liao, M., et al. (2020). 2D Ti3C2TxMXene couples electrical stimulation to promote proliferation and neural differentiation of neural stem cells. Acta Biomater. S1742–7061, 30749–30752. doi: 10.1016/j.actbio.2020.12.035
Guo, R., Zhang, S., Xiao, M., Qian, F., He, Z., Li, D., et al. (2016). Accelerating bioelectric functional development of neural stem cells by graphene coupling: implications for neural interfacing with conductive materials. Biomaterials 106, 193–204. doi: 10.1016/j.biomaterials.2016.08.019
Guo, Z., Zhang, L., Wu, Z., Chen, Y., Wang, F., and Chen, G. (2014). In vivo direct reprogramming of reactive glial cells into functional neurons after brain injury and in an Alzheimer’s disease model. Cell Stem Cell 14, 188–202. doi: 10.1016/j.stem.2013.12.001
Han, S., Xu, Y., Sun, J., Liu, Y., Zhao, Y., Tao, W., et al. (2020). Isolation and analysis of extracellular vesicles in a Morpho butterfly wing-integrated microvortex biochip. Biosens. Bioelectron. 154:112073. doi: 10.1016/j.bios.2020.112073
He, Z., Guo, L., Shu, Y., Fang, Q., Zhou, H., Liu, Y., et al. (2017). Autophagy protects auditory hair cells against neomycin-induced damage. Autophagy 13, 1884–1904. doi: 10.1080/15548627.2017.1359449
Ho, S.-Y., Chao, C.-Y., Huang, H.-L., Chiu, T.-W., Charoenkwan, P., and Hwang, E. (2011). NeurphologyJ: an automatic neuronal morphology quantification method and its application in pharmacological discovery. BMC Bioinformatics 12:230. doi: 10.1186/1471-2105-12-230
Hu, W., Qiu, B., Guan, W., Wang, Q., Wang, M., Li, W., et al. (2015). Direct conversion of normal and Alzheimer’s disease human fibroblasts into neuronal cells by small molecules. Cell Stem Cell 17, 204–212. doi: 10.1016/j.stem.2015.07.006
Jessen, K. R., and Mirsky, R. (2005). The origin and development of glial cells in peripheral nerves. Nat. Rev. Neurosci. 6, 671–682. doi: 10.1038/nrn1746
Kempfle, J. S., Luu, N. C., Petrillo, M., Al-Asad, R., Zhang, A., and Edge, A. S. B. (2020). Lin28 reprograms inner ear glia to a neuronal fate. Stem Cells 38, 890–903. doi: 10.1002/stem.3181
Kiernan, A. E., Pelling, A. L., Leung, K. K. H., Tang, A. S. P., Bell, D. M., Tease, C., et al. (2005). Sox2 is required for sensory organ development in the mammalian inner ear. Nature 434, 1031–1035. doi: 10.1038/nature03487
Koehler, K. R., Mikosz, A. M., Molosh, A. I., Patel, D., and Hashino, E. (2013). Generation of inner ear sensory epithelia from pluripotent stem cells in 3D culture. Nature 500, 217–221. doi: 10.1038/nature12298
Koike, T., Wakabayashi, T., Mori, T., Hirahara, Y., and Yamada, H. (2015). Sox2 promotes survival of satellite glial cells in vitro. Biochem. Biophys. Res. Commun. 464, 269–274. doi: 10.1016/j.bbrc.2015.06.141
Lang, H., Li, M., Kilpatrick, L. A., Zhu, J., Samuvel, D. J., Krug, E. L., et al. (2011). Sox2 up-regulation and glial cell proliferation following degeneration of spiral ganglion neurons in the adult mouse inner ear. J. Assoc. Res. Otolaryngol. 12, 151–171. doi: 10.1007/s10162-010-0244-1
Lang, H., Schulte, B. A., and Schmiedt, R. A. (2005). Ouabain induces apoptotic cell death in type I spiral ganglion neurons, but not type II neurons. J. Assoc. Res. Otolaryngol. 6, 63–74. doi: 10.1007/s10162-004-5021-6
Lang, H., Xing, Y., Brown, L. N., Samuvel, D. J., Panganiban, C. H., Havens, L. T., et al. (2015). Neural stem/progenitor cell properties of glial cells in the adult mouse auditory nerve. Sci. Rep. 5:13383. doi: 10.1038/srep13383
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Lee, A. S., Tang, C., Rao, M. S., Weissman, I. L., and Wu, J. C. (2013). Tumorigenicity as a clinical hurdle for pluripotent stem cell therapies. Nat. Med. 19, 998–1004. doi: 10.1038/nm.3267
Li, B., and Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323
Li, C., Li, X., Bi, Z., Sugino, K., Wang, G., Zhu, T., et al. (2020). Comprehensive transcriptome analysis of cochlear spiral ganglion neurons at multiple ages. Elife 9:e50491. doi: 10.7554/eLife.50491
Li, G., Chen, K., You, D., Xia, M., Li, W., Fan, S., et al. (2019). Laminin-coated electrospun regenerated silk fibroin mats promote neural progenitor cell proliferation, differentiation, and survival in vitro. Front. Bioeng. Biotechnol. 7:190. doi: 10.3389/fbioe.2019.00190
Li, H., Liu, H., and Heller, S. (2003). Pluripotent stem cells from the adult mouse inner ear. Nat. Med. 9, 1293–1299. doi: 10.1038/nm925
Li, X., Aleardi, A., Wang, J., Zhou, Y., Andrade, R., and Hu, Z. (2016). Differentiation of spiral ganglion-derived neural stem cells into functional synaptogenetic neurons. Stem Cells Dev 25, 803–813. doi: 10.1089/scd.2015.0345
Li, X., Bi, Z., Sun, Y., Li, C., Li, Y., and Liu, Z. (2020). In vivo ectopic Ngn1 and Neurod1 convert neonatal cochlear glial cells into spiral ganglion neurons. FASEB J. 34, 4764–4782. doi: 10.1096/fj.201902118R
Li, X., Xu, J., and Deng, H. (2018). Small molecule-induced cellular fate reprogramming: promising road leading to Rome. Curr. Opin. Genet. Dev. 52, 29–35. doi: 10.1016/j.gde.2018.05.004
Li, X., Zuo, X., Jing, J., Ma, Y., Wang, J., Liu, D., et al. (2015). Small-molecule-driven direct reprogramming of mouse fibroblasts into functional neurons. Cell Stem Cell 17, 195–203. doi: 10.1016/j.stem.2015.06.003
Liberman, M. C. (2017). Noise-induced and age-related hearing loss: new perspectives and potential therapies. F1000Res. 6:927. doi: 10.12688/f1000research.11310.1
Liu, M. L., Zang, T., Zou, Y., Chang, J. C., Gibson, J. R., Huber, K. M., et al. (2013). Small molecules enable neurogenin 2 to efficiently convert human fibroblasts into cholinergic neurons. Nat. Commun. 4:2183. doi: 10.1038/ncomms3183
Liu, W., Xu, L., Wang, X., Zhang, D., Sun, G., Wang, M., et al. (2021). PRDX1 activates autophagy via the PTEN-AKT signaling pathway to protect against cisplatin-induced spiral ganglion neuron damage. Autophagy 1–23. doi: 10.1080/15548627.2021.1905466
Liu, W., Xu, X., Fan, Z., Sun, G., Han, Y., Zhang, D., et al. (2019). Wnt signaling activates TP53-induced glycolysis and apoptosis regulator and protects against cisplatin-induced spiral ganglion neuron damage in the mouse cochlea. Antioxid. Redox Signal. 30, 1389–1410. doi: 10.1089/ars.2017.7288
Liu, Z., Jiang, Y., Li, X., and Hu, Z. (2018). Embryonic stem cell-derived peripheral auditory neurons form neural connections with mouse central auditory neurons in vitro via the alpha2delta1 receptor. Stem Cell Rep. 11, 157–170. doi: 10.1016/j.stemcr.2018.05.006
Loo, B.-M., and Salmivirta, M. (2002). Heparin/Heparan sulfate domains in binding and signaling of fibroblast growth factor 8b. J. Biol. Chem. 277, 32616–32623. doi: 10.1074/jbc.M204961200
Lukovic, D., Stojkovic, M., Moreno-Manzano, V., Bhattacharya, S. S., and Erceg, S. (2014). Perspectives and future directions of human pluripotent stem cell-based therapies: lessons from Geron’s clinical trial for spinal cord injury. Stem Cells Dev. 23, 1–4. doi: 10.1089/scd.2013.0266
Marazzi, I., Greenbaum, B. D., Low, D. H. P., and Guccione, E. (2018). Chromatin dependencies in cancer and inflammation. Nat. Rev. Mol. Cell Biol. 19, 245–261. doi: 10.1038/nrm.2017.113
McLean, W. J., McLean, D. T., Eatock, R. A., and Edge, A. S. (2016). Distinct capacity for differentiation to inner ear cell types by progenitor cells of the cochlea and vestibular organs. Development 143, 4381–4393. doi: 10.1242/dev.139840
Meas, S. J., Nishimura, K., Scheibinger, M., and Dabdoub, A. (2018a). In vitro methods to cultivate spiral ganglion cells, and purification of cellular subtypes for induced neuronal reprogramming. Front. Neurosci. 12:822. doi: 10.3389/fnins.2018.00822
Meas, S. J., Zhang, C. L., and Dabdoub, A. (2018b). Reprogramming glia into neurons in the peripheral auditory system as a solution for sensorineural hearing loss: lessons from the central nervous system. Front. Mol. Neurosci. 11:77. doi: 10.3389/fnmol.2018.00077
Mercurio, S., Serra, L., and Nicolis, S. K. (2019). More than just stem cells: functional roles of the transcription factor Sox2 in differentiated glia and neurons. Int. J. Mol. Sci. 20:4540. doi: 10.3390/ijms20184540
Mertens, J., Marchetto, M. C., Bardy, C., and Gage, F. H. (2016). Evaluating cell reprogramming, differentiation and conversion technologies in neuroscience. Nat. Rev. Neurosci. 17, 424–437. doi: 10.1038/nrn.2016.46
Moon, B. S., Lu, W., and Park, H. J. (2018). Valproic acid promotes the neuronal differentiation of spiral ganglion neural stem cells with robust axonal growth. Biochem. Biophys. Res. Commun. 503, 2728–2735. doi: 10.1016/j.bbrc.2018.08.032
Morgado, A. L., Rodrigues, C. M., and Sola, S. (2016). MicroRNA-145 regulates neural stem cell differentiation through the Sox2-Lin28/let-7 signaling pathway. Stem Cells 34, 1386–1395. doi: 10.1002/stem.2309
Muller, U., and Barr-Gillespie, P. G. (2015). New treatment options for hearing loss. Nat. Rev. Drug Discov. 14, 346–365. doi: 10.1038/nrd4533
Noda, T., Meas, S. J., Nogami, J., Amemiya, Y., Uchi, R., Ohkawa, Y., et al. (2018). Direct reprogramming of spiral ganglion non-neuronal cells into neurons: toward ameliorating sensorineural hearing loss by gene therapy. Front. Cell Dev. Biol. 6:16. doi: 10.3389/fcell.2018.00016
Oshima, K., Grimm, C. M., Corrales, C. E., Senn, P., Martinez Monedero, R., Geleoc, G. S., et al. (2007). Differential distribution of stem cells in the auditory and vestibular organs of the inner ear. J. Assoc. Res. Otolaryngol. 8, 18–31. doi: 10.1007/s10162-006-0058-3
Perny, M., Ting, C. C., Kleinlogel, S., Senn, P., and Roccio, M. (2017). Generation of otic sensory neurons from mouse embryonic stem cells in 3D culture. Front. Cell Neurosci. 11:409. doi: 10.3389/fncel.2017.00409
Petitpre, C., Wu, H., Sharma, A., Tokarska, A., Fontanet, P., Wang, Y., et al. (2018). Neuronal heterogeneity and stereotyped connectivity in the auditory afferent system. Nat. Commun. 9:3691. doi: 10.1038/s41467-018-06033-3
Qian, H., Kang, X., Hu, J., Zhang, D., Liang, Z., Meng, F., et al. (2020). Reversing a model of Parkinson’s disease with in situ converted nigral neurons. Nature 582, 550–556. doi: 10.1038/s41586-020-2388-4
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Schneider, J. W., Gao, Z., Li, S., Farooqi, M., Tang, T. S., Bezprozvanny, I., et al. (2008). Small-molecule activation of neuronal cell fate. Nat. Chem. Biol. 4, 408–410. doi: 10.1038/nchembio.95
Shrestha, B. R., Chia, C., Wu, L., Kujawa, S. G., Liberman, M. C., and Goodrich, L. V. (2018). Sensory neuron diversity in the inner ear is shaped by activity. Cell 174, 1229–1246.e17. doi: 10.1016/j.cell.2018.07.007
Sun, G., Liu, W., Fan, Z., Zhang, D., Han, Y., Xu, L., et al. (2016). The three-dimensional culture system with matrigel and neurotrophic factors preserves the structure and function of spiral ganglion neuron in vitro. Neural Plast. 2016:4280407. doi: 10.1155/2016/4280407
Sun, S., Babola, T., Pregernig, G., So, K. S., Nguyen, M., Su, S. M., et al. (2018). Hair cell mechanotransduction regulates spontaneous activity and spiral ganglion subtype specification in the auditory system. Cell 174, 1247–1263.e15. doi: 10.1016/j.cell.2018.07.008
Suzuki, J., Corfas, G., and Liberman, M. C. (2016). Round-window delivery of neurotrophin 3 regenerates cochlear synapses after acoustic overexposure. Sci. Rep. 6:24907. doi: 10.1038/srep24907
Tang, M., Li, J., He, L., Guo, R., Yan, X., Li, D., et al. (2019). Transcriptomic profiling of neural stem cell differentiation on graphene substrates. Colloids Surf. B Biointerfaces 182:110324. doi: 10.1016/j.colsurfb.2019.06.054
Vlajkovic, S. M., Ambepitiya, K., Barclay, M., Boison, D., Housley, G. D., and Thorne, P. R. (2017). Adenosine receptors regulate susceptibility to noise-induced neural injury in the mouse cochlea and hearing loss. Hear Res. 345, 43–51. doi: 10.1016/j.heares.2016.12.015
Wan, G., and Corfas, G. (2017). Transient auditory nerve demyelination as a new mechanism for hidden hearing loss. Nat. Commun. 8:14487. doi: 10.1038/ncomms14487
Wise, A. K., Tu, T., Atkinson, P. J., Flynn, B. O., Sgro, B. E., Hume, C., et al. (2011). The effect of deafness duration on neurotrophin gene therapy for spiral ganglion neuron protection. Hear Res. 278, 69–76. doi: 10.1016/j.heares.2011.04.010
Wu, T., Pinto, H. B., Kamikawa, Y. F., and Donohoe, M. E. (2015). The BET family member BRD4 interacts with OCT4 and regulates pluripotency gene expression. Stem Cell Rep. 4, 390–403. doi: 10.1016/j.stemcr.2015.01.012
Xia, L., Shang, Y., Chen, X., Li, H., Xu, X., Liu, W., et al. (2020). Oriented neural spheroid formation and differentiation of neural stem cells guided by anisotropic inverse opals. Front. Bioeng. Biotechnol. 8:848. doi: 10.3389/fbioe.2020.00848
Yan, W., Liu, W., Qi, J., Fang, Q., Fan, Z., Sun, G., et al. (2018). A three-dimensional culture system with matrigel promotes purified spiral ganglion neuron survival and function in vitro. Mol. Neurobiol. 55, 2070–2084. doi: 10.1007/s12035-017-0471-0
Yang, Y., Chen, R., Wu, X., Zhao, Y., Fan, Y., Xiao, Z., et al. (2019). Rapid and efficient conversion of human fibroblasts into functional neurons by small molecules. Stem Cell Rep. 13, 862–876. doi: 10.1016/j.stemcr.2019.09.007
Yang, Y., Gao, B., Hu, Y., Wei, H., Zhang, C., Chai, R., et al. (2021). Ordered inverse-opal scaffold based on bionic transpiration to create a biomimetic spine. Nanoscale 13, 8614–8622. doi: 10.1039/D1NR00731A
Yang, Y., Zhang, Y., Chai, R., and Gu, Z. (2020). A polydopamine-functionalized carbon microfibrous scaffold accelerates the development of neural stem cells. Front. Bioeng. Biotechnol. 8:616. doi: 10.3389/fbioe.2020.00616
Yuan, T. F., Dong, Y., Zhang, L., Qi, J., Yao, C., Wang, Y., et al. (2021). Neuromodulation-based stem cell therapy in brain repair: recent advances and future perspectives. Neurosci. Bull. 37, 735–745. doi: 10.1007/s12264-021-00667-y
Zhang, L., Jiang, H., and Hu, Z. (2011). Concentration-dependent effect of nerve growth factor on cell fate determination of neural progenitors. Stem Cells Dev. 20, 1723–1731. doi: 10.1089/scd.2010.0370
Zhang, S., and Cui, W. (2014). Sox2, a key factor in the regulation of pluripotency and neural differentiation. World J. Stem Cells 6, 305–311. doi: 10.4252/wjsc.v6.i3.305
Zhao, J., Tang, M., Cao, J., Ye, D., Guo, X., Xi, J., et al. (2019). Structurally tunable reduced graphene oxide substrate maintains mouse embryonic stem cell pluripotency. Adv. Sci. (Weinh.) 6:1802136. doi: 10.1002/advs.201802136
Keywords: Sox2+ glial cells, glia-to-neuron conversion, small molecules reprogramming, SGN regeneration, lineage tracing
Citation: Chen Z, Huang Y, Yu C, Liu Q, Qiu C and Wan G (2021) Cochlear Sox2+ Glial Cells Are Potent Progenitors for Spiral Ganglion Neuron Reprogramming Induced by Small Molecules. Front. Cell Dev. Biol. 9:728352. doi: 10.3389/fcell.2021.728352
Received: 21 June 2021; Accepted: 30 August 2021;
Published: 21 September 2021.
Edited by:
Renjie Chai, Southeast University, ChinaReviewed by:
Tao Xue, Fourth Military Medical University, ChinaCopyright © 2021 Chen, Huang, Yu, Liu, Qiu and Wan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guoqiang Wan, Z3VvcWlhbmd3YW5Abmp1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.