- 1Department of Colorectal Surgery, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, China
- 2School of Life Sciences, Zhengzhou University, Zhengzhou, China
- 3Henan Academy of Medical and Pharmaceutical Sciences, Zhengzhou University, Zhengzhou, China
- 4School of Basic Medical Sciences, Zhengzhou University, Zhengzhou, China
The tumor microenvironment (TME), which includes immune cells, fibroblasts, and other components, is the site of tumor cell growth and metastasis and significantly impacts tumor development. Among them, N6-methyladenosine RNA modifications (m6A RNA modifications) are the most abundant internal modifications in coding and non-coding RNAs, which can significantly influence the cancer process and have potential as biomarkers and potential therapeutic targets for tumor therapy. This manuscript reviews the role of m6A RNA modifications in TME and their application in tumor therapy. To some extent, an in-depth understanding of the relationship between TME and m6A RNA modifications will provide new approaches and ideas for future cancer therapy.
Introduction
In recent years, cancer is still one of the most important diseases affecting human life and health. As cancer research has progressed, we found that the tumor microenvironment (TME) also profoundly influences the course of tumor development in addition to the tumor cell (Hinshaw and Shevde, 2019). TME is a dynamic, complex system of multiple components, including immune cells, fibroblasts, and lymphocytes, that play a crucial role in cancer development and progression, and its interaction with tumor cells is a critical factor in the success of tumor cell-tolerant immune escape (Ocana et al., 2019). Meanwhile, we have known that hundreds of chemical modifications occur in TME, which play different roles in TME at different stages of cancer development. In addition, N6-methyladenosine RNA modifications (m6A RNA modifications) are the most abundant modification in TME, and the existing studies have shown that m6A RNA modifications occurring in TME are a vital factor mediating tumor progression and influencing tumor treatment outcome (Chen X. Y. et al., 2019; Cheng H. S. et al., 2019; Wang J. et al., 2020). Therefore, enhanced knowledge and understanding of TME may lead to new ideas and approaches for treating cancer patients.
Starting with Crick’s central principle 60 years ago, plenty of previous studies have focused on coding RNA, which make up about 2% of the human genome sequence but are vital to humans (Crick, 1970; Poller et al., 2018). However, non-coding RNA, which accounts for 98% of the human genome, has been ignored as “junk sequences” (Fabbri et al., 2019). Furthermore, with technology development, such as high-throughput sequencing technology, non-coding RNA has entered people’s field of vision with a new role. As the research goes further, we classified non-coding into microRNA, lncRNA, circRNA, and so on (Matsui and Corey, 2017; Momen-Heravi and Bala, 2018). Non-coding RNA lacks the potential to encode proteins or peptides but has a high degree of transcriptional activity, which plays an essential role in gene regulation, structure and performs biological functions at the RNA level (Wong et al., 2018). Recent studies have shown that aberrant expression of non-coding RNA is ubiquitous in different types of cancers, revealing that it may play an essential role in human cancers.
As the most abundant modification in TME, m6A RNA modification is one of the critical factors affecting the biological function of RNA (Ma et al., 2019; Yang G. et al., 2020). In general, m6A RNA modification requires the involvement of three factors: m6A methylase (writer), m6A demethylase (eraser), and m6A binding protein (reader) (Shi et al., 2019). It usually influences cancer progression by regulating biological functions associated with cancer, including proliferation, metastasis, stem cell differentiation, and stabilization (Roskoski, 2019). Currently, m6A RNA modifications are also increasingly used to detect and diagnose cancer (Niu et al., 2019).
This review mainly outlines the links between coding/non-coding RNA and m6A RNA modification in the TME. Meanwhile, we describe the implications of the interactions between the three for the biological functions that influence the cancer process and discuss their possible future role in clinical applications.
Tumor Microenvironment
Tumor microenvironment consists mainly of an immune microenvironment dominated by immune cells and a non-immune microenvironment dominated by fibroblasts, formed by the combined action of malignant tumor cells and non-transformed cells (Balkwill et al., 2012; Chen and Hambardzumyan, 2018; Costa et al., 2018; Baghban et al., 2020). To date, most of our research has focused on the immune microenvironment, and the results have shown that alterations in TME profoundly influence tumorigenesis and progression, not only by causing tumor heterogeneity but also by influencing patient resistance (Pottier et al., 2015; Wang J. J. et al., 2018). For example, in breast cancer, one of the mechanisms of action of TME is the removal or alteration of tumor components, thereby impeding anti-tumor immunity (Jung et al., 2016). In liver cancer, we predict tumor progression based on the dynamics of TME. At the same time, the stromal cells in TME can influence the invasion and migration of tumor cells (Yang et al., 2011). Thus, TME can be used as a therapeutic target for cancer and as a signal to detect cancer progression (Jarosz-Biej et al., 2019).
At the same time, TME, as a metabolic site for the growth of numerous cells, is rich in metabolites such as succinic acid, D-2HG, and fumaric acid, which are essential for the normal development of the cells and the organism. These metabolites can act as cofactors or antagonists of epigenetic modification enzymes, affecting numerous epigenetic processes such as m6A RNA modification (Zhao Y. et al., 2020). METTL3 and METTL14 are important components of the m6A modifying enzyme complex, and they can form a stable dimer involved in the methylation process, which is dynamically regulated by substrates and metabolites. S-adenosylmethionine (SAM/AdoMet) acts as a universal intracellular methyl donor. However, because METTL14 does not have a SAM binding site, only METTL3 is active, but METTL14 makes an important contribution in binding substrates, where SAM is produced by a carbon metabolic pathway consisting of the folate and methionine cycles (Sledz and Jinek, 2016; Wang et al., 2016). Similarly, metabolites within TME can influence the demethylation of m6A RNA represented by FTO and ALKBH5, such as 2-oxoglutarate, a key metabolite of the citric acid cycle. If this product is mutated, the demethylation of FTO and ALKBH5 is drastically reduced or even completely lost. In addition, 2-oxoglutarate can also undergo a series of biological reactions to convert to fumaric acid, and due to their structural similarity, these metabolites can often bind to competing products of 2-oxoglutarate become m6A demethylase inhibitors (Feng et al., 2014; Zhang et al., 2019; Figure 1).
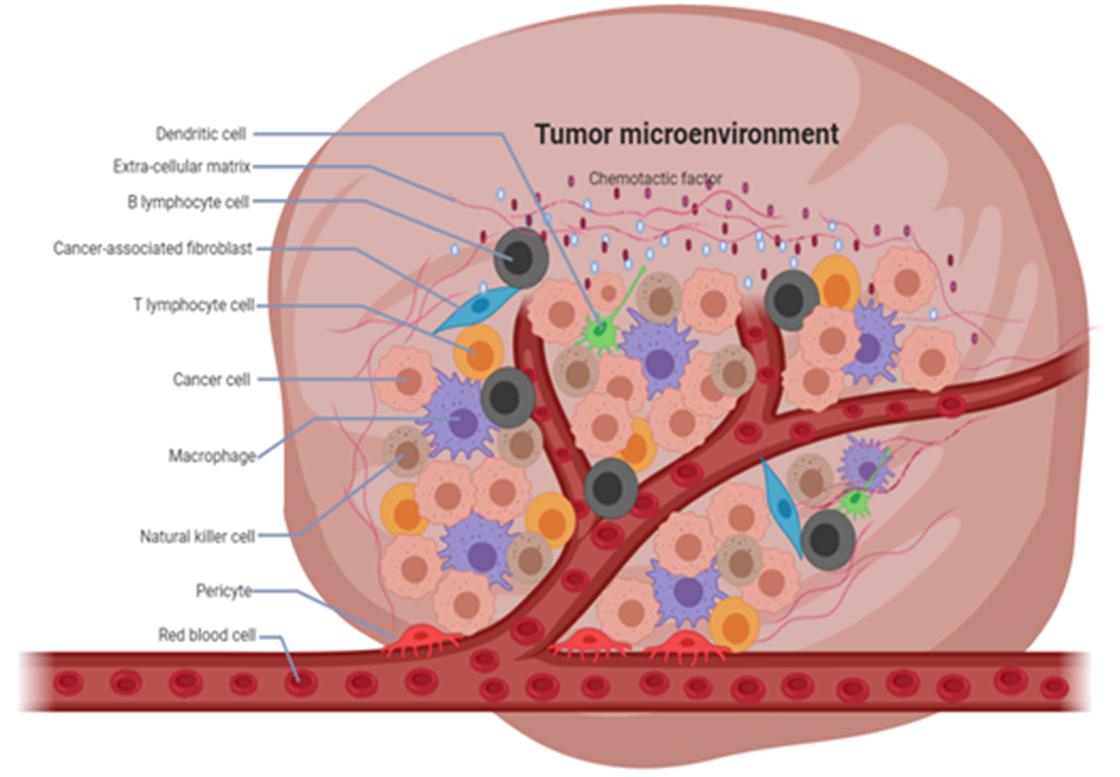
Figure 1. Composition of tumor microenvironment (TME). TME has a complex composition, consisting of immune cells, fibroblasts, and other components which play an important role in the development of cancer.
M6A RNA Modification
To date, hundreds of chemical modifications have been reported, among which m6A RNA modification is the most prevalent in coding and non-coding RNA (Ding et al., 2018; Wang et al., 2019a). m6A RNA modification is a dynamic and reversible process (Wang Q. et al., 2020), it requires the involvement of an m6A modifies enzyme complexes (Lin et al., 2016). In recent years, m6A RNA modification has made great progress in regulating RNA transcription (Wang J. et al., 2020), splicing (Ma et al., 2019), translation (Liu et al., 2018c), and stability (Panneerdoss et al., 2018). Below we summarize the relationship between m6A RNA modification and the above biological processes (Figure 2).
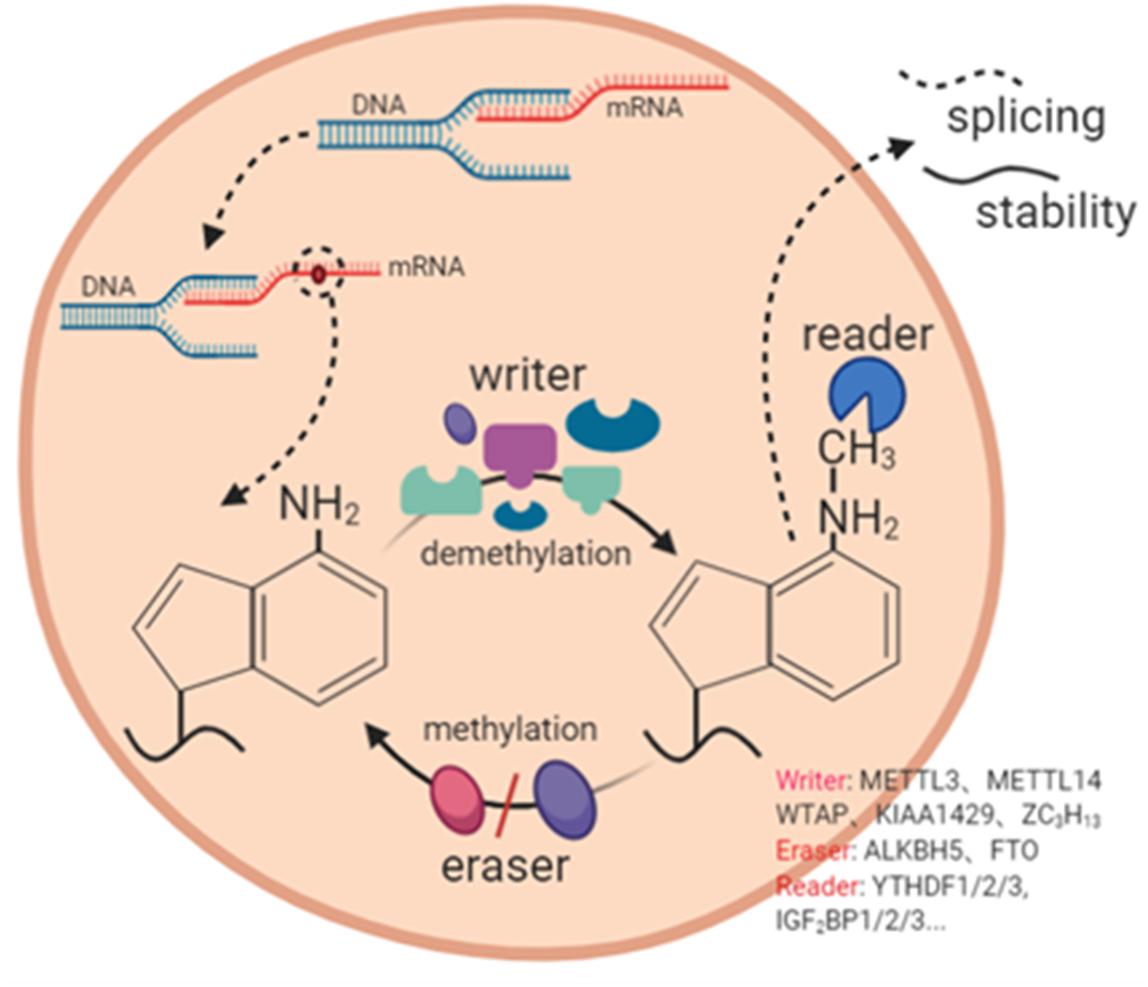
Figure 2. The mechanism of m6A RNA modification. m6A RNA modification is a dynamic and complex process with the participation of m6A methylase (writer), m6A demethylase (eraser), and m6A binding protein (reader). It participates in RNA splicing, RNA stability and other processes.
M6A RNA Modification and RNA Splicing
The DNA template strand contains introns, and RNA splicing removes introns and joins exons to form a continuous RNA molecule. mRNA splicing produces mRNAs that encode information required for growth and development (Zhao et al., 2014). FTO, a component of the m6A modifying enzyme complex, is responsible for the regulation of exon splicing in the 3′ and 5′ exon regions. Zhao et al. showed that the expression level of FTO was negatively correlated with the level of m6A RNA modification during adipogenesis and that the two could interact to influence the development and treatment process of cancer patients.
M6A RNA Modification and RNA Transcription
RNA transcription is the process of RNA polymerase catalyzing RNA synthesis from a template strand of DNA and four nucleotides, i.e., the transfer of biological information from DNA to RNA, guided by the principle of complementary base pairing (Zhao W. et al., 2020). The m6A RNA modification takes part in and affects the transcriptional process of RNA. KIAA1429 is one of the important components of the m6A modifying enzyme complex and participated in the composition of the m6A writer. Lan et al. showed that in the absence of interference by KIAA1429, the interaction between HuR and GATA3 pre-mRNA put them in dynamic equilibrium in hepatocellular carcinoma again. In contrast, the addition of KIAA1429 affected the progression of tumor development by preferentially inducing m6A methylation on the 3′ UTR of GATA3 pre-mRNA in hepatocellular carcinoma cells, followed by the isolation of HuR and degradation of GATA3 pre-mRNA, and finally, the down-regulation of GATA3 expression (Lan T. et al., 2019).
M6A RNA Modification and RNA Translation
Proteins are the leading performers of biological functions, and RNA translation belongs to the second part of protein biosynthesis, the first part being RNA transcription. According to the central law, RNA translation is deciphering the base sequences of mature messenger RNA molecules to produce specific amino acids. METTL3, which selectively enhances or inhibits mRNA translation, is one of the significant components of m6A methyltransferase (Choe et al., 2018). Meanwhile, insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs) are important components of the m6A-modifying enzyme complex and act as “readers.” Huang et al. (2018) showed that IGF2BP protein recognizes m6A RNA modifications and enhances mRNA stability and translation.
M6A RNA Modification and RNA Stability
The m6A RNA modification has the function of regulating the stability of RNA, which refers to the ability of RNA to maintain its original structure or resist degradation when external conditions or other factors change (Yang L. et al., 2020). The YTH domain family, which includes YTH domain family proteins 1-3 (YTHDF1-3) and YTH domain-containing proteins 1-2 (YTHDC1-2), i.e., the DF family and DC family, is one of the ‘reader” components of the m6A modifying enzyme complex and plays an important function in RNA stability. For example, YTHDF2, as one of the readers of m6A modification, can promote mRNA degradation by recruiting the CCR4-NOT deadenylase complex, thereby reducing the stability of the targeted transcript (Wang et al., 2014; Du et al., 2016). Moreover, Huang et al. (2018) showed that binding of m6A-modified RNA to corresponding binding proteins also could affect RNA stability.
M6A RNA Modifications Can Regulate Biological Functions
m6A RNA modification’s ability to affect cancer is proven in various cancers, and it is likely to be a marker for the molecular diagnosis of tumors. Meanwhile, deepening the understanding of the effects of m6A RNA modification on cancer may provide new targets and ideas for researching and developing clinical molecular targeted therapies (Dai et al., 2018; He et al., 2019; Lan Q. et al., 2019; Zhou et al., 2020). In the following, taking gastric cancer as an example, METTL3 can promote the development and progression of gastric cancer by mediating m6A RNA modification of HDGF mRNA (Wang Q. et al., 2020). In contrast, in hepatocellular carcinoma, Chen and Wong (2020) showed that m6A RNA modification regulates the expression of different downstream targets by regulating mRNA stability and translation efficiency (Figure 3).
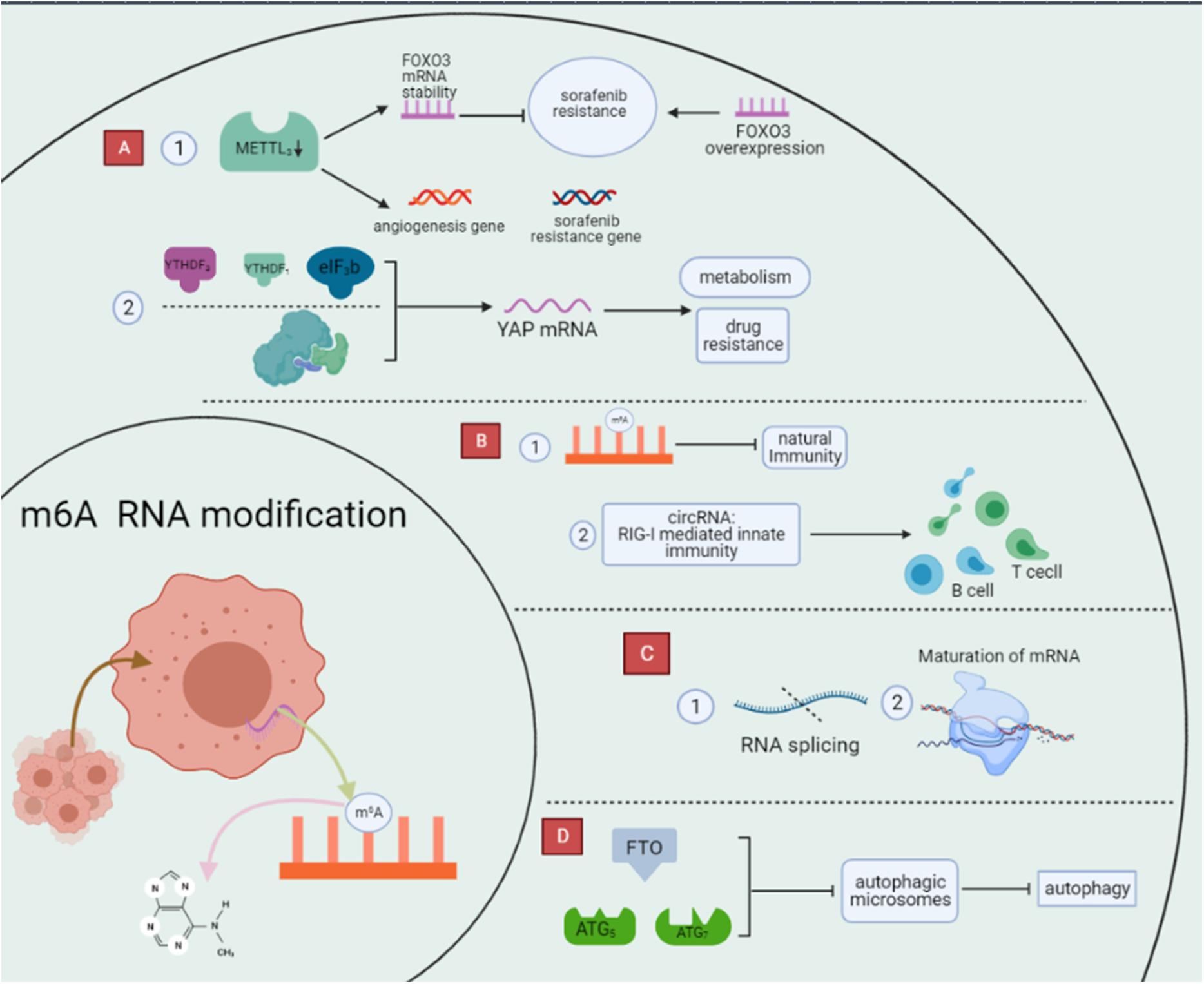
Figure 3. Regulation of biological function of m6A RNA modification. m6A modifications are the most abundant modifications within RNA and can profoundly influence the process of cancer development by regulating biological functions such as drug resistance, immune metabolism, and autophagy through a variety of pathways. (A) m6A RNA modification affect tumor drug resistance. (B) m6A RNA modification affect immunity. (C) m6A RNA modification affect metabolism. (D) m6A RNA modification affect autophagy.
M6A RNA Modification Affect Tumor Drug Resistance
We usually use several approaches in cancer treatment, among which targeted therapies have received more and more attention because of their more significant effectiveness (Li B. et al., 2020). Although targeted therapies are advancing rapidly, they still face tumor resistance that severely affects the efficacy of cancer treatment (Lin et al., 2020). m6A RNA modifications are the most common RNA modifications that can affect RNA splicing, degradation, and translation by regulating cell proliferation, metabolism, metastasis, and ultimately tumor drug resistance (Huang et al., 2018). For example, Liu et al. showed that RNA m6A modification specifically regulates sorafenib resistance in hepatocellular carcinoma through FOXO3-mediated autophagy. The mechanism is that METTL3 depletion under hypoxic conditions promotes sorafenib resistance and angiogenic genes in HCC cells cultured in vitro and subsequently activates the autophagic pathway (Liu et al., 2020b). METTL3 is significantly downregulated in human sorafenib-resistant hepatocellular carcinoma, whereas METTL3 depletion markedly enhances FOXO3 mRNA stability and eliminates METTL3 mediated sensitivity to sorafenib m6A dependence, which could restore resistance to HCC if FOXO3 overexpression was present.
Furthermore, Jin et al. showed that m6A RNA methylation promotes YAP translation and increases YAP activity, thereby inducing drug resistance and metastasis in NSCLC. The mechanism is that m6A methylation causes YTHDF1/3 and eIF3b to join the translation initiation complex, thereby promoting the translation of YAP mRNA. Also, m6A RNA methylation improves YAP mRNA stability through the MALAT1-miR-1914-3p-YAP axis (Jin et al., 2019).
M6A RNA Modification Affect Immunity
The immune system is vital to the body and helps us to defend ourselves against many harmful microorganisms. Recent studies have shown that m6A RNA modification affects the immune system and the function of immune cells to a certain extent, the more obvious of which is that deletion of METTL3 will lead to impaired maturation of these cells in response to lipopolysaccharides and decreased expression of CD40 and CD80, thus inducing a decrease in T cell responsiveness. Also, m6A RNA modification can affect the development of the immune system. In a mouse model, high expression of METTL3 is required for the differentiation of blood-derived endothelial cells into hematopoietic stem cells (Shulman and Stern-Ginossar, 2020). What is more, immune dysregulation is an important cause of several immune diseases, including systemic lupus erythematosus (SLE) and rheumatoid arthritis. It has been reported that m6A RNA modifications can regulate RNA and other gene expressions in immune cells, affecting SLE immune cell pathogenesis (Ma et al., 2019; Sekar and Lakshmanan, 2020). Meanwhile, m6A RNA modifications in circRNA play an essential role in tumor development and anti-tumor immunity (Chen Y. G. et al., 2019). Exogenous circRNA can effectively stimulate immune signaling, and m6A RNA modifications are the main contributors to the immune effects of circRNA. For example, exogenous circRNA activates RIG-I-mediated innate immunity, induces activation of antigen-specific B and T cells, and anti-tumor activity in vivo (Poller et al., 2018). In addition, m6A-modified RNAs are degraded by YTHDF2, suppressing natural immunity. These results indicate that m6A RNA modification plays an essential regulatory role in the tumor immune process (Paramasivam and Vijayashree Priyadharsini, 2020). Therefore, a deeper understanding of the relationship between m6A RNA modifications and immunity may open new doors to treat immune diseases.
M6A RNA Modification Affect Metabolism
Epigenetic regulation of organisms is a complex process. m6A RNA modification is the most prevalent modification in eukaryotic cells and represents a new trajectory of epigenetic modifications. It plays a vital role in regulating RNA metabolisms, such as regulating splicing and translation. One way of RNA editing is converting A to I with the involvement of RNA adenosine deaminase (ADAR). Reports suggest that A-to-I is a crucial factor affecting RNA metabolism and that m6A RNA modification is inversely correlated with A-to-I. A possible reason for this is that m6A RNA modification affects RNA structure (Torsin et al., 2021).
Meanwhile, in vivo immunofluorescence analysis revealed that METTL3, a component of the m6A methyltransferase complex, is located on the mRNA splicing factor, revealing that m6A RNA modifications may play a regulatory role in RNA metabolism and have an impact on cellular reprogramming (Yang et al., 2018). As the study progressed, Liu Y. et al. (2019) found that m6A methylation affected multiple aspects of mRNA metabolism, from expression in the nucleus to processing of pre-mRNA to attenuation of the translational machinery of mRNA in the cytoplasm. The transition from pre-mRNA to mature mRNA requires a splicing process, and there is evidence that m6A RNA modification is a vital splicing regulator (Liu et al., 2014; Ping et al., 2014). First, based on PAR-CLIP analysis, it was observed that mRNAs with selective splicing have more METTL3 binding and methylation sites. A recent study also showed that METTL3 could play a role in spermatogenesis by initiating selective splicing of related mRNAs to regulate sperm differentiation and meiosis (Dominissini et al., 2012). Secondly, m6A erasers and readers also differentially affect RNA splicing, with FTO binding to pre-mRNAs in the nucleus to initiate selective spliced exons and YTHDF1 binding directly to m6A-modified variable splice exons to facilitate their incorporation into mRNAs (Meyer et al., 2012; Bartosovic et al., 2017).
M6A RNA Modification Affect Autophagy
Autophagy is a complex biological process that involves the engulfment of organelles and proteins by autophagosomes (Doria et al., 2013; Galluzzi et al., 2017; Russo and Russo, 2018), followed by digestion by lysosomes, and finally, entry and recycling of cellular processes (Kimura et al., 2007; Yuan et al., 2020). It can influence multiple aspects of cancer, such as regulating resistance to sorafenib in hepatocellular carcinoma. The mechanism is the downregulation of the m6A demethylase component FTO, which affects the expression of ATG5 and ATG7, ultimately reducing the ability of autophagic microsomes and inhibiting autophagy. That is, after FTO Silencing, YTHDF2, modified with high levels of m6A RNA, binds to ATG5 and ATG7 transcripts, and mRNA degradation increases, leading to reduced protein expression and affecting the autophagic process (Jin et al., 2018; Wang C. Y. et al., 2020, Wang X. et al., 2020).
M6A RNA Modification Affects the Infiltration Characteristics of Cancer Cells in Tumor Microenvironment
m6A RNA Modification Can Activate Oncogenes Within Tumor Microenvironment
m6A RNA modifications can influence cancer development by activating tumor-associated genes (He L. et al., 2018; Zheng W. et al., 2019). Depending on the outcome, we can classify m6A RNA modification into promoter and suppressor effects. Next, we will specifically summarize the dual role of m6A RNA modification in cancer.
On the one hand, m6A RNA modification can promote tumorigenesis and development. Take bladder cancer, lung cancer, acute myeloid leukemia, ovarian cancer, breast cancer, pancreatic cancer, colorectal cancer, and nasopharyngeal cancer. Gu et al. showed that in bladder cancer, the expression of METTL3, METTL14, and the oncogene CDCP1 influenced the progression of tumor development. In bladder cancer, m6A expression showed upregulation, and inhibition of METTL3 inhibited proliferation, invasion, and migration of bladder cancer tumor cells. In addition, METTL14 expression is downregulated in bladder cancer, and some studies have shown that eliminating METTL14 promotes capsule proliferation (Gu et al., 2019). In lung cancer, METTL3 expression increases. In a mouse model, deletion of METTL3 promoted apoptosis in transplanted tumor cells. Related experiments showed that METTL3 expression levels in A549 cells responded to the induction of simvastatin. Chen et al. (2020) showed that METTL3 positively regulates the level of EZH2 and modifies its mRNA via m6A, thereby affecting tumor progression (Choe et al., 2018). In acute myeloid leukemia, increased expression of METTL3 promotes the translation of SP1, which regulates the expression of the oncogene C-MYC, and METTL14 enhances the translation of MYB and MYC by maintaining its high expression (Barbieri et al., 2017; You et al., 2017). In ovarian cancer, increased expression levels of IGF2BP1 enhanced the expression of SRF and inhibited its degradation, thereby increasing the expression levels of SRF, FOXK1, and PDLIM7 (Ro, 2016; Muller et al., 2018, 2019). Meanwhile, METTL3 promotes growth, invasion, and migration in ovarian cancer by stimulating peripheral mRNA transfer and epithelial-mesenchymal transition. In breast cancer, increased expression levels of METTL3 would promote HBXIP expression and thus affect breast cancer phenotype. In addition, upregulation of ALKBH5 expression promotes mRNA stability and expression of the pluripotent silver NANOG gene, affecting breast cancer development (Deng et al., 2018). In pancreatic cancer, METTL3 regulates tumor progression and drug resistance through MAPK cascade, RNA splicing, and cellular regulation, and therefore further studies METTL3’s interaction with these processes is essential to understand the functional mechanisms of METTL3 (Taketo et al., 2018). The results of Song et al. (2020) showed increased expression of the YTHDF1 gene in colorectal cancer, where activation of the WNT/β-catenin signaling pathway may initiate transcription-dependent oncogenic effects, promote gross cell cycle progression, and regulate resistance. Knockdown of the YTHDF1 gene would affect the activity of the WNT/β-catenin signaling pathway and inhibit the tumorigenicity of colorectal cancer cells (Nishizawa et al., 2018; Bai et al., 2019). Multiple research teams have found that Nasopharyngeal carcinoma m6A RNA modification affects the overall tumorigenicity of FAM225A and ultimately affects the proliferation of nasopharyngeal carcinoma tumor cells. In addition, METTL3 promotes the expression of EZH2 protein by mediating m6A modification of EZH2 mRNA, which increases the malignancy of nasopharyngeal carcinoma cells by silencing CDKN1C, thereby affecting the development and progression of nasopharyngeal carcinoma (Lai et al., 2018; Wang et al., 2019b; Zheng Z. Q. et al., 2019; Meng et al., 2020).
On the other hand, m6A RNA modification inhibits tumorigenesis and progression. We take endometrial cancer, hepatocellular carcinoma, breast cancer, lung squamous cell carcinoma, and glioblastoma. In endometrial tumors, METTL3 expression decreased, and METTL14 mutated to some extent. m6A methylation reduction promoted cell proliferation, clone formation, migration, and invasion, affecting endometrial tumorigenesis and progression. In addition, FTO catalyzes the demethylation of HOXB13 mRNA in the 3′UTR region, thereby eliminating the recognition of YTHDF2 protein-modified m6A that affects tumor metastasis (Zhang L. et al., 2020). In hepatocellular carcinoma, Hou et al. (2019) showed a unique m6A-mRNA editing process in the hypoxic state. m6A RNA modification through YTHDF2-mediated EGFR degradation plays a role in tumor suppression. In addition, YTHDF2 inhibits ERK/MAPK signaling by destabilizing EGFR mRNA and affecting hepatocellular carcinoma progression (Ma et al., 2017). In breast cancer, Zhang et al. (2016a,b) showed that ALKBH5 expression increases, that NANOG can maintain and regulate tumor stem cells, and that increased ALKBH5 expression correlates with decreased NANOG mRNA m6A levels increased mRNA stability. In lung squamous cell carcinoma, the expression level of FTO is lower. By inhibiting m6A methylation, FTO enhances mRNA stability, promotes the expression of the oncogene MZF1, and affects the progression of lung squamous carcinoma. In addition to FTO, METTL3 expression was also abnormal in lung squamous cell carcinoma (Liu et al., 2018b; Cayir et al., 2019). Wang M. et al. (2020)) showed that ALKBH5 expression increases in glioblastoma and that reduced m6A methylation promoted the expression of the oncogene FOX1, enhancing the self-renewal ability and tumorigenicity of glioblastoma stem cells (Zhang et al., 2017; Figure 4).
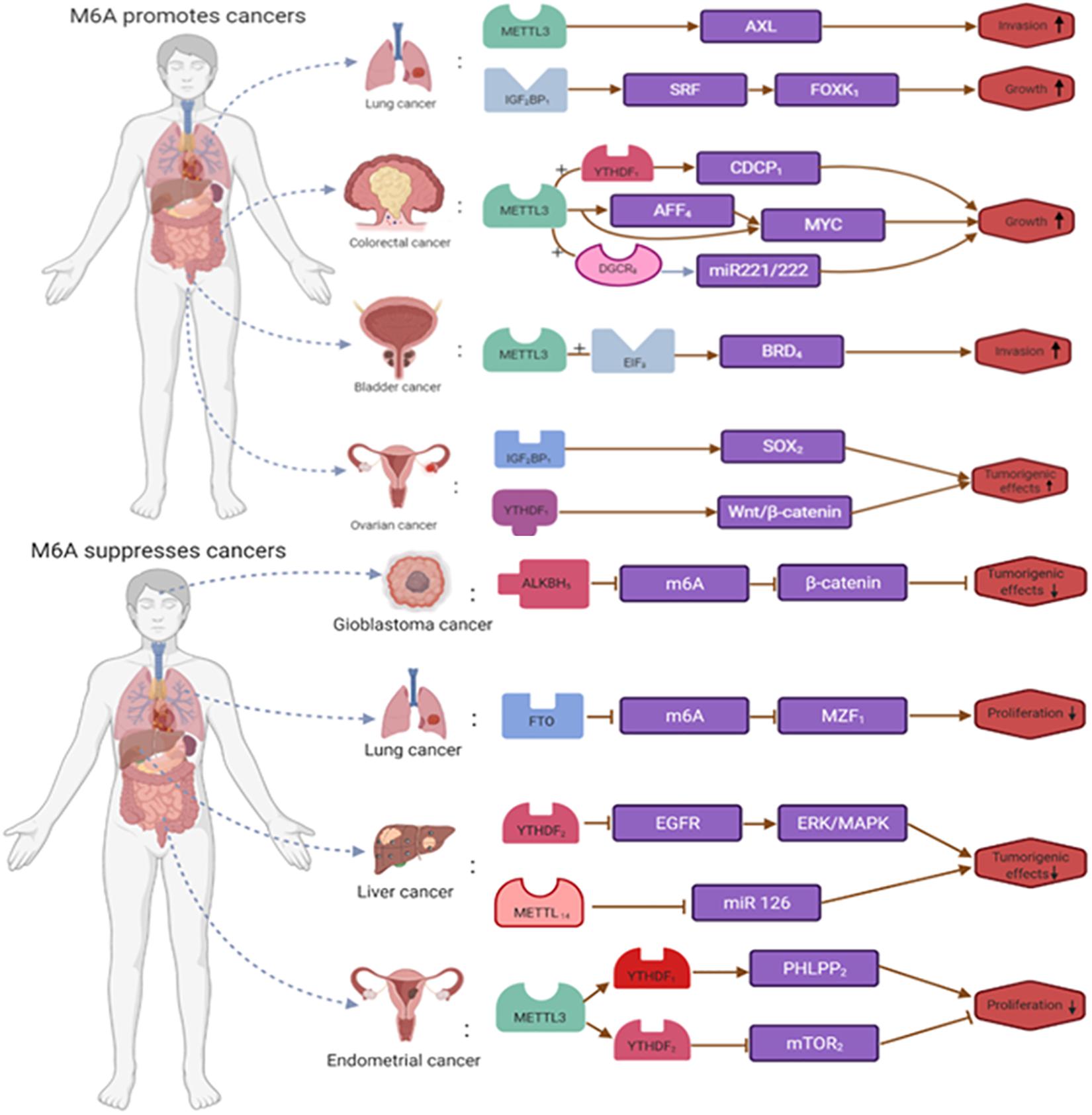
Figure 4. The dual roles of m6A RNA modification in cancer. m6A is modified to regulate the expression of tumor-related genes. m6A RNA modification promote the occurrence and development of tumors by enhancing the expression of tumor-related genes or inhibit the occurrence and development of tumors by inhibiting the expression of tumor-related genes.
m6A RNA Modification Can Activate Immune Cells
m6A RNA modifications can influence cancer development by activating immune cells. For example, immune checkpoint blockade therapy is a revolutionary change in cancer treatment, but many patients do not respond to or are resistant to immune checkpoint blockade therapy (Zhang et al., 2017). ALKBH5 is an important component of the m6A modifying enzyme complex, and it has been reported that deletion of the m6A demethylase ALKBH5 can make patients more sensitive to cancer immunotherapy (Fang et al., 2020). The mechanism is that ALKBH5 affects the efficacy of immunotherapy during immune checkpoint blockade by regulating the expression and lactate content of MCT4/Slc16a3 in TME and the composition of tumor-infiltrating T cells and myeloid suppressor cells. Among these, growth factor-α is a target gene for ALKBH5, MCT4/Slc16a3 and is involved in regulating extracellular lactate concentration, regulatory T cells, and MDSC aggregation in TME (Li N. et al., 2020).
m6A RNA Modification Can Suppress Immune Cells
In addition to activating immune cells, m6A RNA modifications can also affect the cancer process by suppressing the expression of immune cells. CD4 regulatory T cells are involved in and resolve immune suppression in TME. The transcription factor Foxp3, a marker molecule of Treg cells, is regulated by the Suppressor of cytokine signaling (SOCS) family and activates STAT5 (Tong et al., 2018). It has been reported that in a mouse model that has been constructed with METTL3 deletion, elevated expression of the SOCS gene compared to controls inhibited IL2-STAT5 signaling pathway and maintained the inhibitory function of Treg (Lou et al., 2021). Secondly, in hepatocellular carcinoma, METTL3 overexpression then inhibited the expression of suppressor of cytokine signaling factor 2 (SOCS2) through an m6A-YTHDF2-dependent mechanism, which ultimately promoted tumor growth (Tuncel and Kalkan, 2019).
The Roles of M6A RNA Modification in Tme of Various Cancers
m6A RNA modifications found in TMEs of different cancers influence the onset and progression of cancer. In this review, we have selected several cancers and summarized the role of m6A RNA modifications in TME (Figure 5 and Table 1).
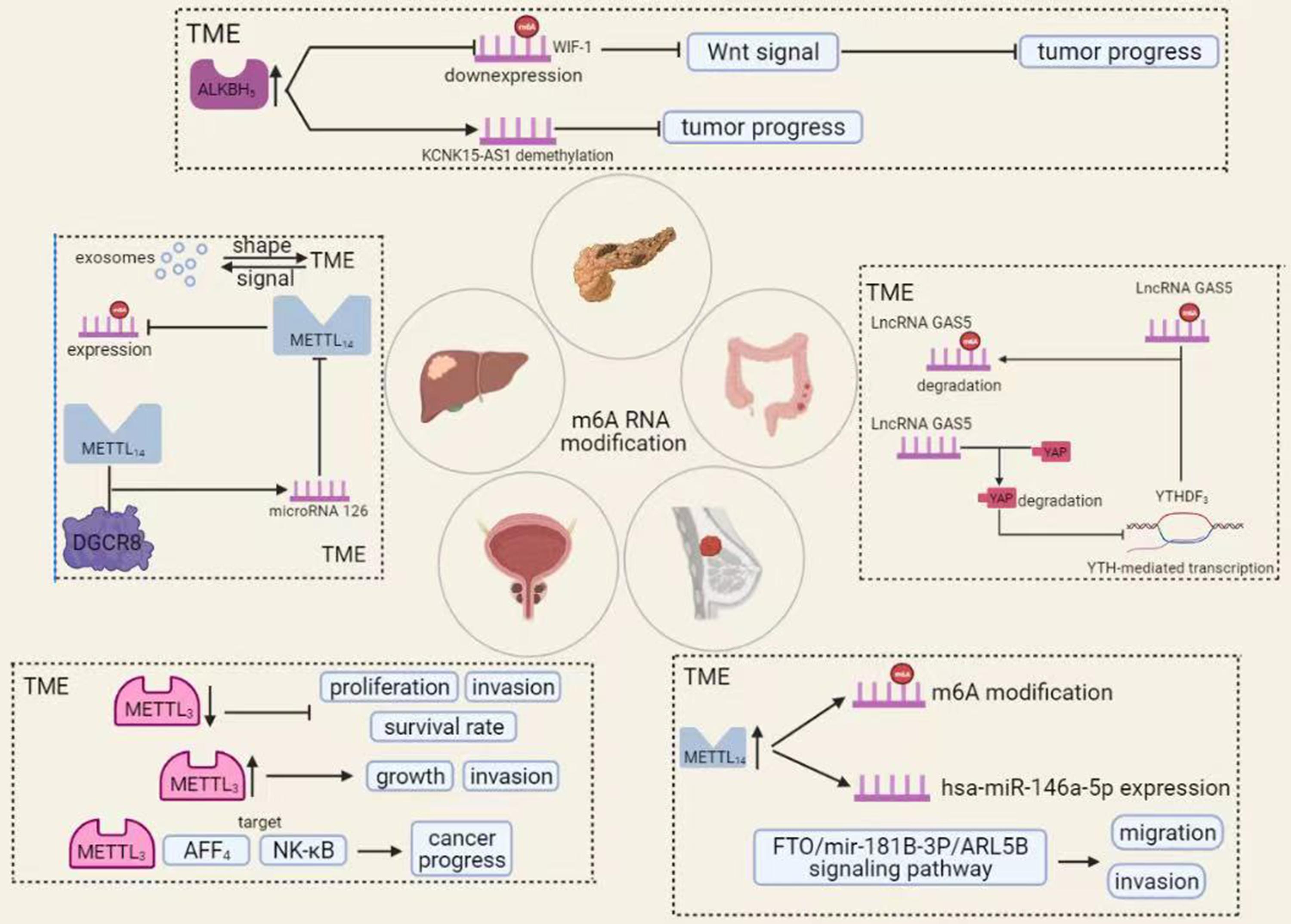
Figure 5. The roles of m6A RNA modification in TME of various cancers. A variety of chemical modifications occur within the tumor microenvironment that is important for tumorigenesis and progression. Among these, m6A RNA modification is the most abundant. In different tumors, m6A RNA modifications can affect cancer progression in various ways.
Colorectal Cancer
Colorectal cancer is the third most deadly cancer in the world (Bray et al., 2018). Currently, we have made some progress in the diagnosis and treatment of this disease, but the prognosis of patients with advanced cancer remains poor (Nishihara et al., 2013). As research into cancer has intensified, TME has gradually attracted attention and attempted to use it in cancer treatment, in addition to malignant tumor cells. TME is the site of growth and transformation of malignant tumor cells, in which it produces and secretes various chemokines and growth factors that promote tumor development and progression (Bahrami et al., 2018). TME contains numerous immune components that can influence tumor development. It is part of the TME and is a very promising therapeutic target. The resulting tumor immune microenvironment has a dynamic character during tumor progression, in which many kinds of cells are involved (Zhang Y. et al., 2020). Ni et al. showed that disruption of the YAP signaling pathway significantly promotes the development and progression of colorectal cancer. The mechanism is that long-stranded non-coding RNA Gas5 is negatively regulated by the m6A reader YTHDF3, forming an adverse regulatory pathway through phosphorylation and degradation of YAP, thereby inhibiting the colorectal carcinogenesis process (Kino et al., 2010; Ni et al., 2019).
Meanwhile, a study by Xu et al. showed that the expression level of long-stranded non-coding RNA SATB2-AS1 was significantly increased in colorectal cancer. This study found that SATB2-AS1 interacted with chromatin regulatory proteins WDR5 and GADD45A in colorectal cancer cell species, with WDR5 catalyzing Lys4 trimethylation and affecting transcription when bound to methylated H3K4. SATB2-AS1 suppressed tumor metastasis in colorectal cancer by regulating STAB2, i.e., regulating cancer progression by affecting the tumor immune microenvironment (Xu et al., 2019).
Hepatocellular Carcinoma
Hepatocellular carcinoma (HCC) is a common malignancy with high morbidity and mortality worldwide (Zhang et al., 2018). Patients with advanced hepatocellular carcinoma prognosis are poor under existing conditions, and new approaches are urgently needed. In recent years, immunotherapy has gained prominence in cancer treatment. Because TME is more variable at different stages of tumor development, it shows more significant variation in other individuals or distant locations of the same individual (Quail and Joyce, 2013). Immune cells can inhibit tumor development, but when they interact with specific cells of TME, the tendency for tumor development to spread will be more pronounced (Zamarron and Chen, 2011). Secondly, HCC can classify into multiple types that share a similar TME, both of which promote immune tolerance and immune escape through various mechanisms (Fu et al., 2019). TME is an essential partner of tumor cells, providing the necessary conditions for the development and progression of tumor cells, such as angiogenesis and tumor formation. Exosomes are information carriers for TME and are the molecular entities involved in building TME (Wu et al., 2019). In hepatocellular carcinoma, exosomes shape TME, provide energy, promote tumor growth, and induce angiogenesis. TME contains exosomes regulating cell line survival and development by giving stimulatory or inhibitory signals. The role of m6A RNA modifications, which occur mainly in TME in HCC, has attracted increasing attention (Bissell and Hines, 2011). For example, microRNA 126 is an essential component of the methyltransferase complex and decreased expression in vitro and in vivo (Ma et al., 2017). MicroRNA 126 is a significant component of METTL14 in HCC. METTL14 can influence the progression of tumor development by m6A-dependent regulation of PRI-microRNA 126 progression, leading to reduced expression of microRNA 126 (Hu et al., 2019).
Bladder Cancer
Bladder cancer is one of the three significant urinary system tumors and is the most common malignancy of the urinary system, with the highest incidence among malignancies (Han et al., 2019). Bladder cancer develops as a result of aberrant genetic alterations and epigenetic abnormalities (Cheng M. et al., 2019). With increased research into bladder cancer and improvements in immunotherapy, methods including intravenous BCG and immune checkpoint inhibitors can influence the development of bladder cancer. At the same time, bladder cancer cells can promote the formation of a tumor immune-suppressive microenvironment. If we can modify or modify TME, it is likely to improve the treatment of bladder cancer. Currently, m6A RNA modification, a very abundant modification in TME, largely influences and alters TME and bladder cancer (Crispen and Kusmartsev, 2020). The level of m6A modification increased significantly in bladder cancer tumor tissues.
Moreover, METTL3 expression levels were similarly upregulated compared to paracancerous tissues. METTL3 can affect the AFF4/NF-κB/myc signaling network in an m6A-dependent manner, promoting bladder cancer progression (Alarcon et al., 2015). Briefly, low METTL3 expression inhibits the in vivo proliferation of bladder cancer and, conversely, METTL3 overexpression promotes its proliferation in vivo. Briefly, common METTL3 expression inhibits the in vivo proliferation of bladder cancer and, conversely, METTL3 overexpression promotes its proliferation in vivo (Cheng M. et al., 2019). The mechanism is that METTL3 interacts with the microprocessor protein Dgcr8 to complete miR221/222 processing, which rescues METTL3-induced proliferation of bladder cancer cells, and its processing exerts an oncogenic effect by positively regulating the pri-miR221/222 process in an m6A-dependent manner (Gu et al., 2016).
Breast Cancer
Breast cancer is the most common malignancy in women and has the second-highest mortality rate in the world (Ahn et al., 2016). Although we have made significant progress in the diagnosis, surgery, and drug development of breast cancer, drug resistance in metastatic cancers remains an insurmountable problem. As research has progressed, we have focused not only on the anti-cancer treatment of tumor cells but also on the TME where the tumor cells reside (Houthuijzen and Jonkers, 2018). Some studies have shown that breast cancer is associated with molecular heterogeneity (Schnitt, 2010). ROS levels in breast cancer cells are associated with the expression and activity of the transcription factor AHR, which controls tumor growth and chemokine production in the TME, and regulating the TME can influence tumor progression (Kubli et al., 2019). The m6A RNA modification in breast cancer TME has a profound impact on the development of breast cancer. In breast cancer, increased expression of METTL14 enhances m6A RNA modifications and has-miR-146a-5p expression, ultimately enhancing breast cancer cell invasion and migration (Yi et al., 2020).
Meanwhile, as an eraser, FTO also plays a vital role in m6A RNA modification. For example, FTO can regulate the migration and invasion of breast cancer cells by regulating the FTO/Mir-181B-3P/ARL5B signaling pathway, which ultimately affects the development of breast cancer. In conclusion, a good grasp of the relationship between m6A RNA modification, TME, and breast cancer will open a new door for breast cancer treatment (Xu et al., 2020).
Pancreatic Cancer
Pancreatic cancer is cancer with a high mortality rate in the world today. Its late onset makes early diagnosis difficult, and it is often diagnosed late in the patient’s life (Cheng et al., 2017). Although we have made significant advances in the genetics and biology of pancreatic cancer, the results in prolonging the survival time of pancreatic cancer are still not substantial (Ren et al., 2018). In pancreatic cancer, the stability of TME refers to a complex balance between pro-and anti-tumor components (Melstrom et al., 2017). We are combined with clinical trials showing that TME in pancreatic cancer results from the interaction between pancreatic epithelial and mesenchymal cells, which influences the efficacy of radiotherapy, chemotherapy, and immunotherapy. Two distinguishing features of TME in pancreatic cancer are extensive immunosuppression and intensive adhesion formation, accelerating the proliferation of tumor cells (Neesse et al., 2015). m6A RNA modifications also play an essential role in pancreatic cancer, with METTL3 promoting chemo- and radiation resistance in pancreatic cancer cells (Taketo et al., 2018). In TME, ALKBH5 overexpression can inhibit pancreatic cancer by decreasing the level of m6A RNA modification of WIF-1, blocking the activation of Wnt signaling, and increasing the sensitivity of pancreatic cancer cells to drugs (Tang et al., 2020). Meanwhile, ALKBH5 could demethylate the long non-coding RNA KCNK15-AS1 and inhibit pancreatic carcinogenesis. In addition, aberrant activation of the Wnt signaling pathway is also a critical factor in the development and progression of pancreatic cancer, directly or indirectly affecting tumor cells’ proliferation and drug resistance (Morris et al., 2010; Bailey et al., 2016).
The Application of M6A RNA Modification in Cancer
m6A RNA modifications are among the most abundant chemical modifications within RNA, and we can now exploit their range of properties in RNA, such as abundance, to explore their potential function in cancer therapy (Liu X. et al., 2019). Below we summarize the present and future applications of m6A RNA modifications and the difficulties we have encountered.
Current Clinical Applications of M6A RNA Modifications
m6A RNA Modification Might a Biomarker in Cancer
A growing number of studies have shown that m6A RNA modification has excellent cancer diagnosis and prognosis potential (Wu et al., 2020). It relies mainly on the m6A modifying enzyme complex and plays a vital role in the biological processes of cancer cell proliferation, invasion, and migration (Wang Q. et al., 2018). Many experimental and clinical results show that m6A RNA modifications are closely related to the clinical characteristics of patients and show great potential in the diagnosis of tumors as diagnostic biomarkers for human cancers (Meng et al., 2017). For example, the expression level of m6A is significantly higher in circulating tumor cells than in whole blood cells. Suppose the expression level of m6A changed in circulating tumor cells. In that case, the significance of tumorigenesis and progression can be known and used to predict the diagnosis and prognosis of patients (Liu et al., 2020a).
In patients with hepatocellular carcinoma, upregulation of METTL3 and YTHDF expression decreases patient survival (Ma et al., 2017). Patients in the low METTL3/YTHDF1 group have a better prognosis, so the combination of METTL3 and YTHDF1 serves as a biomarker reflecting the malignancy and prognostic outcome of hepatocellular carcinoma. In gastric cancer, abnormal expression of FTO correlates with the progression and metastasis of gastric cancer. The presentation of FTO plays a vital role in promoting the development of gastric cancer. Hence, FTO is an important molecular marker for the diagnosis and prognosis of gastric cancer. In pancreatic cancer, ALKBH also has an important prognostic molecular marker (Yue et al., 2019).
m6A RNA Modification Might Be a Therapeutic Target in Cancer
In addition to being a biomarker for cancer, m6A RNA modifications have great potential in cancer therapy. Radiotherapy and chemotherapy remain the main treatments for cancer patients, but in recent studies, m6A RNA modifications have shown great potential in cancer therapy (Poller et al., 2018). Analyzing the relevant TCGA dataset, we found that aberrant expression of m6A associated with TP53 gene mutations in AML patients and that m6A mutations reduced survival in AML patients. YTHDF1 is an essential component of the m6A modifying enzyme complex. Its deletion significantly improves the therapeutic effect of PD-L1 checkpoint blockade, revealing that it may be a molecular target for anti-cancer immunotherapy. In addition, m6A RNA modification may also serve as an essential indicator of patients’ sensitivity to radiotherapy. For example, METTL3 promoted resistance to radiotherapy and reduced survival in pancreatic cancer patients, while FTO was expressed at significantly higher levels in CSCC tissues than in pre-cancerous tissues (Liu et al., 2020b). Also, FTO promoted resistance to radiotherapy in vivo and in vitro by reducing the levels of its target m6A RNA modifications. In summary, m6A RNA modification has the potential to be an effective potential therapeutic target for cancer therapy (Wouters and Delwel, 2016; Yang et al., 2017).
Future Clinical Applications of m6A RNA Modifications
In recent years, a growing number of studies have demonstrated the ability of m6A RNA modifications to act as diagnostic biomarkers and therapeutic targets. Based on these studies, we should aim to understand and apply them to clinical treatment more fully. For example, the main clinical treatments for cancer are still chemotherapy and radiotherapy, so drug resistance is an inevitable problem. Since m6A RNA modifications are associated with drug resistance, we can link radiotherapy and m6A RNA modifications to explore a promising therapeutic approach. Secondly, RNA and protein are inextricably linked, and the importance of protein as the main performer of biological functions cannot be overstated. If we affect the relevant biological functions of protein through m6A RNA modification, it may bring us unexpected gains in clinical treatment. At the same time, there are still few readers, writers and erasers identified, and we need more research to deepen our understanding of the biological properties of m6A RNA modifications for human health and disease.
Challenges in Clinical Application
Although m6A RNA modifications are abundantly present in the TME of RNA, the vast majority have no detectable specific biological function, which makes it difficult to further explore their application in clinical aspects. Secondly, the RNA status varies considerably between individuals or different tissues of the same individual, making it difficult to target specific RNA modifications to cells alone and without guaranteeing therapeutic efficacy. Thirdly, little is known about m6A RNA modification, and further understanding of its regulatory mechanisms or possible biological functions in different cancers is necessary. Similarly, the limited number of readers, writers, and erasers involved in m6A RNA modification has also posed a certain obstacle to our research. Finally, most studies on m6A RNA modification have remained in vitro analyses, and translational application of the results remains a great challenge due to the dynamic and complex nature of m6A RNA modification and the organism.
Conclusion
Tumor microenvironment is a complex dynamic system consisting of immune cells, fibroblasts, and lymphocytes, whose interaction with malignant cells influences the development and progression of the cancer process. At the same time, there are hundreds of chemical modifications in TME, and different chemical modifications affect the cancer development process to different degrees. As the most abundant modifications in TME, m6A RNA modifications, in addition to regulating various biological functions such as metabolism, are also clinically meaningful and have the potential to become biomarkers or targets for intervention in tumor therapy. In conclusion, an in-depth understanding of the relationship between TME and m6A RNA modifications is essential for exploring the expression and regulation of oncogenes.
Author Contributions
All authors conceptualized, wrote, edited, read, and approved the final manuscript.
Funding
This study was supported by The National Natural Science Foundation of China (81972663 and U2004112); Key Scientific Research Projects of Institutions of Higher Education in Henan Province (19A310024); The National Natural Science Foundation of Henan Province (182300410342); and The Health Commission Technology Talents Overseas Training Project of Henan Province (2018140).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
TME, tumor microenvironment; m6A RNA modification, the N6-methyladenosine RNA modification; FTO, obesity-associated protein; METTL3/14, methyltransferase-like 3/14; IGF2BP protein, insulin-like growth factor 2 mRNA-binding protein; HDGF, hepatoma-derived growth factor; FOXO3, forkhead box O3; HCC, hepatocellular carcinoma; NSCLC, non-small cell lung cancer; eIF3b, eukaryotic translation initiation factor 3B; SLE, systemic lupus erythematosus; YTHDF, YTH domain-containing family; FTO, obesity-associated protein; YTHDC1, YTH domain-containing protein 1; ALKBH5, alkB homolog 5; CDCP1, CUB-domain containing protein 1; AML, acute myeloid leukemia; HBXIP, hepatitis B X-interacting protein; WTAP, WT1-associated protein; HNRNPG, heterogeneous nuclear ribonucleoprotein G.
References
Ahn, S. G., Kim, S. J., Kim, C., and Jeong, J. (2016). Molecular classification of triple-negative breast cancer. J. Breast Cancer 19, 223–230. doi: 10.4048/jbc.2016.19.3.223
Alarcon, C. R., Lee, H., Goodarzi, H., Halberg, N., and Tavazoie, S. F. (2015). N6-methyladenosine marks primary microRNAs for processing. Nature 519, 482–485. doi: 10.1038/nature14281
Baghban, R., Roshangar, L., Jahanban-Esfahlan, R., Seidi, K., Ebrahimi-Kalan, A., Jaymand, M., et al. (2020). Tumor microenvironment complexity and therapeutic implications at a glance. Cell Commun. Signal. 18:59. doi: 10.1186/s12964-020-0530-4
Bahrami, A., Khazaei, M., Hassanian, S. M., ShahidSales, S., Joudi-Mashhad, M., Maftouh, M., et al. (2018). Targeting the tumor microenvironment as a potential therapeutic approach in colorectal cancer: rational and progress. J. Cell Physiol. 233, 2928–2936. doi: 10.1002/jcp.26041
Bai, Y., Yang, C., Wu, R., Huang, L., Song, S., Li, W., et al. (2019). YTHDF1 regulates tumorigenicity and cancer stem cell-like activity in human colorectal carcinoma. Front. Oncol. 9:332. doi: 10.3389/fonc.2019.00332
Bailey, P., Chang, D. K., Nones, K., Johns, A. L., Patch, A. M., Gingras, M. C., et al. (2016). Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 531, 47–52. doi: 10.1038/nature16965
Balkwill, F. R., Capasso, M., and Hagemann, T. (2012). The tumor microenvironment at a glance. J. Cell Sci. 125(Pt 23), 5591–5596. doi: 10.1242/jcs.116392
Bansal, H., Yihua, Q., Iyer, S. P., Ganapathy, S., Proia, D. A., Penalva, L. O., et al. (2014). WTAP is a novel oncogenic protein in acute myeloid leukemia. Leukemia 28, 1171–1174. doi: 10.1038/leu.2014.16
Barbieri, I., Tzelepis, K., Pandolfini, L., Shi, J., Millan-Zambrano, G., Robson, S. C., et al. (2017). Promoter-bound METTL3 maintains myeloid leukaemia by m(6)A-dependent translation control. Nature 552, 126–131. doi: 10.1038/nature24678
Bartosovic, M., Molares, H. C., Gregorova, P., Hrossova, D., Kudla, G., and Vanacova, S. (2017). N6-methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3’-end processing. Nucleic Acids Res. 45, 11356–11370. doi: 10.1093/nar/gkx778
Bissell, M. J., and Hines, W. C. (2011). Why don’t we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat. Med. 17, 320–329. doi: 10.1038/nm.2328
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A., and Jemal, A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424. doi: 10.3322/caac.21492
Cayir, A., Barrow, T. M., Guo, L., and Byun, H. M. (2019). Exposure to environmental toxicants reduces global N6-methyladenosine RNA methylation and alters expression of RNA methylation modulator genes. Environ. Res. 175, 228–234. doi: 10.1016/j.envres.2019.05.011
Chen, M., Wei, L., Law, C. T., Tsang, F. H., Shen, J., Cheng, C. L., et al. (2018). RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology 67, 2254–2270. doi: 10.1002/hep.29683
Chen, M., and Wong, C. M. (2020). The emerging roles of N6-methyladenosine (m6A) deregulation in liver carcinogenesis. Mol. Cancer 19:44. doi: 10.1186/s12943-020-01172-y
Chen, W. W., Qi, J. W., Hang, Y., Wu, J. X., Zhou, X. X., Chen, J. Z., et al. (2020). Simvastatin is beneficial to lung cancer progression by inducing METTL3-induced m6A modification on EZH2 mRNA. Eur. Rev. Med. Pharmacol. Sci. 24, 4263–4270.
Chen, X. Y., Zhang, J., and Zhu, J. S. (2019). The role of m(6)A RNA methylation in human cancer. Mol. Cancer 18:103. doi: 10.1186/s12943-019-1033-z
Chen, Y. G., Chen, R., Ahmad, S., Verma, R., Kasturi, S. P., Amaya, L., et al. (2019). N6-methyladenosine modification controls circular rna immunity. Mol. Cell 76, 96–109. doi: 10.1016/j.molcel.2019.07.016
Chen, Z., and Hambardzumyan, D. (2018). Immune microenvironment in glioblastoma subtypes. Front. Immunol. 9:1004. doi: 10.3389/fimmu.2018.01004
Cheng, H., Liu, C., Jiang, J., Luo, G., Lu, Y., Jin, K., et al. (2017). Analysis of ctDNA to predict prognosis and monitor treatment responses in metastatic pancreatic cancer patients. Int. J. Cancer 140, 2344–2350. doi: 10.1002/ijc.30650
Cheng, H. S., Lee, J. X. T., Wahli, W., and Tan, N. S. (2019). Exploiting vulnerabilities of cancer by targeting nuclear receptors of stromal cells in tumor microenvironment. Mol. Cancer 18:51. doi: 10.1186/s12943-019-0971-9
Cheng, M., Sheng, L., Gao, Q., Xiong, Q., Zhang, H., Wu, M., et al. (2019). The m(6)A methyltransferase METTL3 promotes bladder cancer progression via AFF4/NF-kappaB/MYC signaling network. Oncogene 38, 3667–3680. doi: 10.1038/s41388-019-0683-z
Choe, J., Lin, S., Zhang, W., Liu, Q., Wang, L., Ramirez-Moya, J., et al. (2018). mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature 561, 556–560. doi: 10.1038/s41586-018-0538-8
Costa, A., Kieffer, Y., Scholer-Dahirel, A., Pelon, F., Bourachot, B., Cardon, M., et al. (2018). Fibroblast heterogeneity and immunosuppressive environment in human breast cancer. Cancer Cell 33, 463–479. doi: 10.1016/j.ccell.2018.01.011
Crispen, P. L., and Kusmartsev, S. (2020). Mechanisms of immune evasion in bladder cancer. Cancer Immunol. Immunother. 69, 3–14. doi: 10.1007/s00262-019-02443-4
Cui, Q., Shi, H., Ye, P., Li, L., Qu, Q., Sun, G., et al. (2017). m(6)A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 18, 2622–2634. doi: 10.1016/j.celrep.2017.02.059
Dai, D., Wang, H., Zhu, L., Jin, H., and Wang, X. (2018). N6-methyladenosine links RNA metabolism to cancer progression. Cell Death Dis. 9:124. doi: 10.1038/s41419-017-0129-x
Deng, X., Su, R., Feng, X., Wei, M., and Chen, J. (2018). Role of N(6)-methyladenosine modification in cancer. Curr. Opin. Genet. Dev. 48, 1–7. doi: 10.1016/j.gde.2017.10.005
Ding, C., Zou, Q., Ding, J., Ling, M., Wang, W., Li, H., et al. (2018). Increased N6-methyladenosine causes infertility is associated with FTO expression. J. Cell Physiol. 233, 7055–7066. doi: 10.1002/jcp.26507
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206. doi: 10.1038/nature11112
Doria, A., Gatto, M., and Punzi, L. (2013). Autophagy in human health and disease. N. Engl. J. Med. 368, 1845. doi: 10.1056/NEJMc1303158
Du, H., Zhao, Y., He, J., Zhang, Y., Xi, H., Liu, M., et al. (2016). YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun. 7:12626. doi: 10.1038/ncomms12626
Fabbri, M., Girnita, L., Varani, G., and Calin, G. A. (2019). Decrypting noncoding RNA interactions, structures, and functional networks. Genome Res. 29, 1377–1388. doi: 10.1101/gr.247239.118
Fang, J., Hu, M., Sun, Y., Zhou, S., and Li, H. (2020). Expression profile analysis of m6a rna methylation regulators indicates they are immune signature associated and can predict survival in kidney renal cell carcinoma. DNA Cell Biol. 39, 1–8. doi: 10.1089/dna.2020.5767
Feng, C., Liu, Y., Wang, G., Deng, Z., Zhang, Q., Wu, W., et al. (2014). Crystal structures of the human RNA demethylase Alkbh5 reveal basis for substrate recognition. J. Biol. Chem. 289, 11571–11583. doi: 10.1074/jbc.M113.546168
Fu, Y., Liu, S., Zeng, S., and Shen, H. (2019). From bench to bed: the tumor immune microenvironment and current immunotherapeutic strategies for hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 38:396. doi: 10.1186/s13046-019-1396-4
Galluzzi, L., Baehrecke, E. H., Ballabio, A., Boya, P., Bravo-San Pedro, J. M., Cecconi, F., et al. (2017). Molecular definitions of autophagy and related processes. EMBO J. 36, 1811–1836. doi: 10.15252/embj.201796697
Gu, C., Wang, Z., Zhou, N., Li, G., Kou, Y., Luo, Y., et al. (2019). Mettl14 inhibits bladder TIC self-renewal and bladder tumorigenesis through N(6)-methyladenosine of Notch1. Mol. Cancer 18:168. doi: 10.1186/s12943-019-1084-1
Gu, J., Wang, D., Zhang, J., Zhu, Y., Li, Y., Chen, H., et al. (2016). GFRalpha2 prompts cell growth and chemoresistance through down-regulating tumor suppressor gene PTEN via Mir-17-5p in pancreatic cancer. Cancer Lett. 380, 434–441. doi: 10.1016/j.canlet.2016.06.016
Han, J., Wang, J. Z., Yang, X., Yu, H., Zhou, R., Lu, H. C., et al. (2019). METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol. Cancer 18:110. doi: 10.1186/s12943-019-1036-9
He, L., Li, H., Wu, A., Peng, Y., Shu, G., and Yin, G. (2019). Functions of N6-methyladenosine and its role in cancer. Mol. Cancer 18:176. doi: 10.1186/s12943-019-1109-9
He, L., Li, J., Wang, X., Ying, Y., Xie, H., Yan, H., et al. (2018). The dual role of N6-methyladenosine modification of RNAs is involved in human cancers. J. Cell Mol. Med. 22, 4630–4639. doi: 10.1111/jcmm.13804
He, Y., Hu, H., Wang, Y., Yuan, H., Lu, Z., Wu, P., et al. (2018). ALKBH5 inhibits pancreatic cancer motility by decreasing long non-coding RNA KCNK15-AS1 methylation. Cell Physiol. Biochem. 48, 838–846. doi: 10.1159/000491915
Hinshaw, D. C., and Shevde, L. A. (2019). The tumor microenvironment innately modulates cancer progression. Cancer Res. 79, 4557–4566. doi: 10.1158/0008-5472.CAN-18-3962
Hou, J., Zhang, H., Liu, J., Zhao, Z., Wang, J., Lu, Z., et al. (2019). YTHDF2 reduction fuels inflammation and vascular abnormalization in hepatocellular carcinoma. Mol. Cancer 18:163. doi: 10.1186/s12943-019-1082-3
Houthuijzen, J. M., and Jonkers, J. (2018). Cancer-associated fibroblasts as key regulators of the breast cancer tumor microenvironment. Cancer Metastasis Rev. 37, 577–597. doi: 10.1007/s10555-018-9768-3
Hu, B. B., Wang, X. Y., Gu, X. Y., Zou, C., Gao, Z. J., Zhang, H., et al. (2019). N(6)-methyladenosine (m(6)A) RNA modification in gastrointestinal tract cancers: roles, mechanisms, and applications. Mol. Cancer 18:178. doi: 10.1186/s12943-019-1099-7
Huang, H., Weng, H., Sun, W., Qin, X., Shi, H., Wu, H., et al. (2018). Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 20, 285–295. doi: 10.1038/s41556-018-0045-z
Jarosz-Biej, M., Smolarczyk, R., Cichon, T., and Kulach, N. (2019). Tumor microenvironment as a “game changer” in cancer radiotherapy. Int. J. Mol. Sci. 20:3212. doi: 10.3390/ijms20133212
Jin, D., Guo, J., Wu, Y., Du, J., Yang, L., Wang, X., et al. (2019). m(6)A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1-miR-1914-3p-YAP axis to induce NSCLC drug resistance and metastasis. J. Hematol. Oncol. 12:135. doi: 10.1186/s13045-019-0830-6
Jin, S., Zhang, X., Miao, Y., Liang, P., Zhu, K., She, Y., et al. (2018). m(6)A RNA modification controls autophagy through upregulating ULK1 protein abundance. Cell Res. 28, 955–957. doi: 10.1038/s41422-018-0069-8
Jung, Y. Y., Kim, H. M., and Koo, J. S. (2016). The role of cancer-associated fibroblasts in breast cancer pathobiology. Histol. Histopathol. 31, 371–378.
Kimura, S., Noda, T., and Yoshimori, T. (2007). Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 3, 452–460. doi: 10.4161/auto.4451
Kino, T., Hurt, D. E., Ichijo, T., Nader, N., and Chrousos, G. P. (2010). Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 3:ra8. doi: 10.1126/scisignal.2000568
Kubli, S. P., Bassi, C., Roux, C., Wakeham, A., Gobl, C., Zhou, W., et al. (2019). AhR controls redox homeostasis and shapes the tumor microenvironment in BRCA1-associated breast cancer. Proc. Natl. Acad. Sci. U.S.A. 116, 3604–3613. doi: 10.1073/pnas.1815126116
Kwok, C. T., Marshall, A. D., Rasko, J. E., and Wong, J. J. (2017). Genetic alterations of m(6)A regulators predict poorer survival in acute myeloid leukemia. J. Hematol. Oncol. 10:39. doi: 10.1186/s13045-017-0410-6
Lai, W., Jia, J., Yan, B., Jiang, Y., Shi, Y., Chen, L., et al. (2018). Baicalin hydrate inhibits cancer progression in nasopharyngeal carcinoma by affecting genome instability and splicing. Oncotarget 9, 901–914. doi: 10.18632/oncotarget.22868
Lan, Q., Liu, P. Y., Haase, J., Bell, J. L., Huttelmaier, S., and Liu, T. (2019). The critical role of RNA m(6)A methylation in cancer. Cancer Res. 79, 1285–1292. doi: 10.1158/0008-5472.CAN-18-2965
Lan, T., Li, H., Zhang, D., Xu, L., Liu, H., Hao, X., et al. (2019). KIAA1429 contributes to liver cancer progression through N6-methyladenosine-dependent post-transcriptional modification of GATA3. Mol. Cancer 18:186. doi: 10.1186/s12943-019-1106-z
Li, B., Jiang, J., Assaraf, Y. G., Xiao, H., Chen, Z. S., and Huang, C. (2020). Surmounting cancer drug resistance: new insights from the perspective of N(6)-methyladenosine RNA modification. Drug Resist. Updat. 53:100720. doi: 10.1016/j.drup.2020.100720
Li, N., Kang, Y., Wang, L., Huff, S., Tang, R., Hui, H., et al. (2020). ALKBH5 regulates anti-PD-1 therapy response by modulating lactate and suppressive immune cell accumulation in tumor microenvironment. Proc. Natl. Acad. Sci. U.S.A. 117, 20159–20170. doi: 10.1073/pnas.1918986117
Li, Z., Weng, H., Su, R., Weng, X., Zuo, Z., Li, C., et al. (2017). FTO plays an oncogenic role in acute myeloid leukemia as a N(6)-methyladenosine RNA demethylase. Cancer Cell. 31, 127–141. doi: 10.1016/j.ccell.2016.11.017
Lin, S., Choe, J., Du, P., Triboulet, R., and Gregory, R. I. (2016). The m(6)A methyltransferase METTL3 promotes translation in human cancer cells. Mol. Cell 62, 335–345. doi: 10.1016/j.molcel.2016.03.021
Lin, Z., Niu, Y., Wan, A., Chen, D., Liang, H., Chen, X., et al. (2020). RNA m(6) A methylation regulates sorafenib resistance in liver cancer through FOXO3-mediated autophagy. EMBO J. 39:e103181. doi: 10.15252/embj.2019103181
Liu, Z. X., Li, L. M., Sun, H. L., and Liu, S. M. (2018c). Link between m6A modification and cancers. Front. Bioeng. Biotechnol. 6:89. doi: 10.3389/fbioe.2018.00089
Liu, J., Eckert, M. A., Harada, B. T., Liu, S. M., Lu, Z., Yu, K., et al. (2018a). m(6)A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat. Cell Biol. 20, 1074–1083. doi: 10.1038/s41556-018-0174-4
Liu, J., Ren, D., Du, Z., Wang, H., Zhang, H., and Jin, Y. (2018b). m(6)A demethylase FTO facilitates tumor progression in lung squamous cell carcinoma by regulating MZF1 expression. Biochem. Biophys. Res. Commun. 502, 456–464. doi: 10.1016/j.bbrc.2018.05.175
Liu, J., Yue, Y., Han, D., Wang, X., Fu, Y., Zhang, L., et al. (2014). A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10, 93–95. doi: 10.1038/nchembio.1432
Liu, S., Li, Q., Li, G., Zhang, Q., Zhuo, L., Han, X., et al. (2020b). The mechanism of m(6)A methyltransferase METTL3-mediated autophagy in reversing gefitinib resistance in NSCLC cells by beta-elemene. Cell Death Dis. 11:969. doi: 10.1038/s41419-020-03148-8
Liu, S., Li, Q., Chen, K., Zhang, Q., Li, G., Zhuo, L., et al. (2020a). The emerging molecular mechanism of m(6)A modulators in tumorigenesis and cancer progression. Biomed. Pharmacother. 127:110098. doi: 10.1016/j.biopha.2020.110098
Liu, X., Liu, L., Dong, Z., Li, J., Yu, Y., Chen, X., et al. (2019). Expression patterns and prognostic value of m(6)A-related genes in colorectal cancer. Am. J. Transl. Res. 11, 3972–3991.
Liu, Y., You, Y., Lu, Z., Yang, J., Li, P., Liu, L., et al. (2019). N (6)-methyladenosine RNA modification-mediated cellular metabolism rewiring inhibits viral replication. Science 365, 1171–1176. doi: 10.1126/science.aax4468
Lou, X., Wang, J. J., Wei, Y. Q., and Sun, J. J. (2021). Emerging role of RNA modification N6-methyladenosine in immune evasion. Cell Death Dis. 12:300. doi: 10.1038/s41419-021-03585-z
Ma, J. Z., Yang, F., Zhou, C. C., Liu, F., Yuan, J. H., Wang, F., et al. (2017). METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N(6) -methyladenosine-dependent primary MicroRNA processing. Hepatology 65, 529–543. doi: 10.1002/hep.28885
Ma, S., Chen, C., Ji, X., Liu, J., Zhou, Q., Wang, G., et al. (2019). The interplay between m6A RNA methylation and noncoding RNA in cancer. J. Hematol. Oncol. 12:121. doi: 10.1186/s13045-019-0805-7
Matsui, M., and Corey, D. R. (2017). Non-coding RNAs as drug targets. Nat. Rev. Drug Discov. 16, 167–179. doi: 10.1038/nrd.2016.117
Mauer, J., Luo, X., Blanjoie, A., Jiao, X., Grozhik, A. V., Patil, D. P., et al. (2017). Reversible methylation of m(6)Am in the 5’ cap controls mRNA stability. Nature 541, 371–375. doi: 10.1038/nature21022
Melstrom, L. G., Salazar, M. D., and Diamond, D. J. (2017). The pancreatic cancer microenvironment: a true double agent. J. Surg. Oncol. 116, 7–15. doi: 10.1002/jso.24643
Meng, Q. Z., Cong, C. H., Li, X. J., Zhu, F., Zhao, X., and Chen, F. W. (2020). METTL3 promotes the progression of nasopharyngeal carcinoma through mediating M6A modification of EZH2. Eur. Rev. Med. Pharmacol. Sci. 24, 4328–4336.
Meng, S., Zhou, H., Feng, Z., Xu, Z., Tang, Y., Li, P., et al. (2017). CircRNA: functions and properties of a novel potential biomarker for cancer. Mol. Cancer 16:94. doi: 10.1186/s12943-017-0663-2
Meyer, K. D., Saletore, Y., Zumbo, P., Elemento, O., Mason, C. E., and Jaffrey, S. R. (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell 149, 1635–1646. doi: 10.1016/j.cell.2012.05.003
Momen-Heravi, F., and Bala, S. (2018). Emerging role of non-coding RNA in oral cancer. Cell Signal. 42, 134–143. doi: 10.1016/j.cellsig.2017.10.009
Morris, J. P. T., Wang, S. C., and Hebrok, M. (2010). KRAS, hedgehog, Wnt and the twisted developmental biology of pancreatic ductal adenocarcinoma. Nat. Rev. Cancer 10, 683–695. doi: 10.1038/nrc2899
Muller, S., Bley, N., Glass, M., Busch, B., Rousseau, V., Misiak, D., et al. (2018). IGF2BP1 enhances an aggressive tumor cell phenotype by impairing miRNA-directed downregulation of oncogenic factors. Nucleic Acids Res. 46, 6285–6303. doi: 10.1093/nar/gky229
Muller, S., Glass, M., Singh, A. K., Haase, J., Bley, N., Fuchs, T., et al. (2019). IGF2BP1 promotes SRF-dependent transcription in cancer in a m6A- and miRNA-dependent manner. Nucleic Acids Res. 47, 375–390. doi: 10.1093/nar/gky1012
Neesse, A., Algul, H., Tuveson, D. A., and Gress, T. M. (2015). Stromal biology and therapy in pancreatic cancer: a changing paradigm. Gut 64, 1476–1484. doi: 10.1136/gutjnl-2015-309304
Ni, W., Yao, S., Zhou, Y., Liu, Y., Huang, P., Zhou, A., et al. (2019). Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m(6)A reader YTHDF3. Mol. Cancer 18:143. doi: 10.1186/s12943-019-1079-y
Nishihara, R., Wu, K., Lochhead, P., Morikawa, T., Liao, X., Qian, Z. R., et al. (2013). Long-term colorectal-cancer incidence and mortality after lower endoscopy. N. Engl. J. Med. 369, 1095–1105. doi: 10.1056/NEJMoa1301969
Nishizawa, Y., Konno, M., Asai, A., Koseki, J., Kawamoto, K., Miyoshi, N., et al. (2018). Oncogene c-Myc promotes epitranscriptome m(6)A reader YTHDF1 expression in colorectal cancer. Oncotarget 9, 7476–7486. doi: 10.18632/oncotarget.23554
Niu, Y., Lin, Z., Wan, A., Chen, H., Liang, H., Sun, L., et al. (2019). RNA N6-methyladenosine demethylase FTO promotes breast tumor progression through inhibiting BNIP3. Mol. Cancer 18:46. doi: 10.1186/s12943-019-1004-4
Ocana, M. C., Martinez-Poveda, B., Quesada, A. R., and Medina, M. A. (2019). Metabolism within the tumor microenvironment and its implication on cancer progression: an ongoing therapeutic target. Med. Res. Rev. 39, 70–113. doi: 10.1002/med.21511
Panneerdoss, S., Eedunuri, V. K., Yadav, P., Timilsina, S., Rajamanickam, S., Viswanadhapalli, S., et al. (2018). Cross-talk among writers, readers, and erasers of m(6)A regulates cancer growth and progression. Sci. Adv. 4:eaar8263. doi: 10.1126/sciadv.aar8263
Paramasivam, A., and Vijayashree Priyadharsini, J. (2020). Novel insights into m6A modification in circular RNA and implications for immunity. Cell Mol. Immunol. 17, 668–669. doi: 10.1038/s41423-020-0387-x
Ping, X. L., Sun, B. F., Wang, L., Xiao, W., Yang, X., Wang, W. J., et al. (2014). Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 24, 177–189. doi: 10.1038/cr.2014.3
Poller, W., Dimmeler, S., Heymans, S., Zeller, T., Haas, J., Karakas, M., et al. (2018). Non-coding RNAs in cardiovascular diseases: diagnostic and therapeutic perspectives. Eur. Heart J. 39, 2704–2716. doi: 10.1093/eurheartj/ehx165
Pottier, C., Wheatherspoon, A., Roncarati, P., Longuespee, R., Herfs, M., Duray, A., et al. (2015). The importance of the tumor microenvironment in the therapeutic management of cancer. Expert. Rev. Anticancer Ther. 15, 943–954. doi: 10.1586/14737140.2015.1059279
Quail, D. F., and Joyce, J. A. (2013). Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 19, 1423–1437. doi: 10.1038/nm.3394
Ren, B., Cui, M., Yang, G., Wang, H., Feng, M., You, L., et al. (2018). Tumor microenvironment participates in metastasis of pancreatic cancer. Mol. Cancer 17:108. doi: 10.1186/s12943-018-0858-1
Ro, S. (2016). Multi-phenotypic role of serum response factor in the gastrointestinal system. J. Neurogastroenterol. Motil. 22, 193–200. doi: 10.5056/jnm15183
Roskoski, R. Jr. (2019). Small molecule inhibitors targeting the EGFR/ErbB family of protein-tyrosine kinases in human cancers. Pharmacol. Res. 139, 395–411. doi: 10.1016/j.phrs.2018.11.014
Russo, M., and Russo, G. L. (2018). Autophagy inducers in cancer. Biochem. Pharmacol. 153, 51–61. doi: 10.1016/j.bcp.2018.02.007
Schnitt, S. J. (2010). Classification and prognosis of invasive breast cancer: from morphology to molecular taxonomy. Mod. Pathol. 23(Suppl. 2) S60–S64. doi: 10.1038/modpathol.2010.33
Sekar, D., and Lakshmanan, G. (2020). Methylation of N6-adenosine (m6A) modification in miRNAs and its implications in immunity. Epigenomics 12, 1083–1085. doi: 10.2217/epi-2020-0131
Shi, H., Wei, J., and He, C. (2019). Where, when, and how: context-dependent functions of rna methylation writers, readers, and erasers. Mol. Cell 74, 640–650. doi: 10.1016/j.molcel.2019.04.025
Shulman, Z., and Stern-Ginossar, N. (2020). The RNA modification N(6)-methyladenosine as a novel regulator of the immune system. Nat. Immunol. 21, 501–512. doi: 10.1038/s41590-020-0650-4
Sledz, P., and Jinek, M. (2016). Structural insights into the molecular mechanism of the m(6)A writer complex. Elife 5:e18434. doi: 10.7554/eLife.18434
Song, P., Feng, L., Li, J., Dai, D., Zhu, L., Wang, C., et al. (2020). beta-catenin represses miR455-3p to stimulate m6A modification of HSF1 mRNA and promote its translation in colorectal cancer. Mol. Cancer 19:129. doi: 10.1186/s12943-020-01244-z
Su, R., Dong, L., Li, C., Nachtergaele, S., Wunderlich, M., Qing, Y., et al. (2018). R-2HG exhibits anti-tumor activity by targeting FTO/m(6)A/MYC/CEBPA signaling. Cell 172, 90–105. doi: 10.1016/j.cell.2017.11.031
Taketo, K., Konno, M., Asai, A., Koseki, J., Toratani, M., Satoh, T., et al. (2018). The epitranscriptome m6A writer METTL3 promotes chemo- and radioresistance in pancreatic cancer cells. Int. J. Oncol. 52, 621–629. doi: 10.3892/ijo.2017.4219
Tang, B., Yang, Y., Kang, M., Wang, Y., Wang, Y., Bi, Y., et al. (2020). m(6)A demethylase ALKBH5 inhibits pancreatic cancer tumorigenesis by decreasing WIF-1 RNA methylation and mediating Wnt signaling. Mol. Cancer 19:3. doi: 10.1186/s12943-019-1128-6
Tang, J., Wang, F., Cheng, G., Si, S., Sun, X., Han, J., et al. (2018). Wilms’ tumor 1-associating protein promotes renal cell carcinoma proliferation by regulating CDK2 mRNA stability. J. Exp. Clin. Cancer Res. 37:40. doi: 10.1186/s13046-018-0706-6
Tong, J., Cao, G., Zhang, T., Sefik, E., Amezcua Vesely, M. C., Broughton, J. P., et al. (2018). m(6)A mRNA methylation sustains treg suppressive functions. Cell Res. 28, 253–256. doi: 10.1038/cr.2018.7
Torsin, L. I., Petrescu, G. E. D., Sabo, A. A., Chen, B., Brehar, F. M., Dragomir, M. P., et al. (2021). Editing and chemical modifications on non-coding rnas in cancer: a new tale with clinical significance. Int. J. Mol. Sci. 22:581. doi: 10.3390/ijms22020581
Tuncel, G., and Kalkan, R. (2019). Importance of m N(6)-methyladenosine (m(6)A) RNA modification in cancer. Med. Oncol. 36:36. doi: 10.1007/s12032-019-1260-6
Visvanathan, A., Patil, V., Arora, A., Hegde, A. S., Arivazhagan, A., Santosh, V., et al. (2018). Essential role of METTL3-mediated m(6)A modification in glioma stem-like cells maintenance and radioresistance. Oncogene 37, 522–533. doi: 10.1038/onc.2017.351
Vu, L. P., Pickering, B. F., Cheng, Y., Zaccara, S., Nguyen, D., Minuesa, G., et al. (2017). The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat. Med. 23, 1369–1376. doi: 10.1038/nm.4416
Wang, C. Y., Lin, T. A., Ho, M. Y., Yeh, J. K., Tsai, M. L., Hung, K. C., et al. (2020). Regulation of autophagy in leukocytes through RNA N(6)-adenosine methylation in chronic kidney disease patients. Biochem. Biophys. Res. Commun. 527, 953–959. doi: 10.1016/j.bbrc.2020.04.138
Wang, H., Hu, X., Huang, M., Liu, J., Gu, Y., Ma, L., et al. (2019a). Mettl3-mediated mRNA m(6)A methylation promotes dendritic cell activation. Nat. Commun. 10:1898. doi: 10.1038/s41467-019-09903-6
Wang, H., Zhou, Y., Oyang, L., Han, Y., Xia, L., Lin, J., et al. (2019b). LPLUNC1 stabilises PHB1 by counteracting TRIM21-mediated ubiquitination to inhibit NF-kappaB activity in nasopharyngeal carcinoma. Oncogene 38, 5062–5075. doi: 10.1038/s41388-019-0778-6
Wang, J., Wang, J., Gu, Q., Ma, Y., Yang, Y., Zhu, J., et al. (2020). The biological function of m6A demethylase ALKBH5 and its role in human disease. Cancer Cell. Int. 20:347. doi: 10.1186/s12935-020-01450-1
Wang, J. J., Lei, K. F., and Han, F. (2018). Tumor microenvironment: recent advances in various cancer treatments. Eur. Rev. Med. Pharmacol. Sci. 22, 3855–3864.
Wang, M., Mao, C., Ouyang, L., Liu, Y., Lai, W., Liu, N., et al. (2020). Correction to: long noncoding RNA LINC00336 inhibits ferroptosis in lung cancer by functioning as a competing endogenous RNA. Cell Death Differ. 27:1447. doi: 10.1038/s41418-019-0394-6
Wang, P., Doxtader, K. A., and Nam, Y. (2016). Structural basis for cooperative function of mettl3 and mettl14 methyltransferases. Mol. Cell 63, 306–317. doi: 10.1016/j.molcel.2016.05.041
Wang, Q., Chen, C., Ding, Q., Zhao, Y., Wang, Z., Chen, J., et al. (2020). METTL3-mediated m(6)A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut 69, 1193–1205. doi: 10.1136/gutjnl-2019-319639
Wang, Q., Chen, J., Wang, A., Sun, L., Qian, L., Zhou, X., et al. (2018). Differentially expressed circRNAs in melanocytes and melanoma cells and their effect on cell proliferation and invasion. Oncol. Rep. 39, 1813–1824. doi: 10.3892/or.2018.6263
Wang, X., Feng, J., Xue, Y., Guan, Z., Zhang, D., Liu, Z., et al. (2017). Corrigendum: structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature 542:260. doi: 10.1038/nature21073
Wang, X., Lu, Z., Gomez, A., Hon, G. C., Yue, Y., Han, D., et al. (2014). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120. doi: 10.1038/nature12730
Wang, X., Wu, R., Liu, Y., Zhao, Y., Bi, Z., Yao, Y., et al. (2020). m(6)A mRNA methylation controls autophagy and adipogenesis by targeting Atg5 and Atg7. Autophagy 16, 1221–1235. doi: 10.1080/15548627.2019.1659617
Weng, H., Huang, H., Wu, H., Qin, X., Zhao, B. S., Dong, L., et al. (2018). METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m(6)A modification. Cell Stem Cell 22, 191–205. doi: 10.1016/j.stem.2017.11.016
Wong, C. M., Tsang, F. H., and Ng, I. O. (2018). Non-coding RNAs in hepatocellular carcinoma: molecular functions and pathological implications. Nat. Rev. Gastroenterol. Hepatol. 15, 137–151. doi: 10.1038/nrgastro.2017.169
Wouters, B. J., and Delwel, R. (2016). Epigenetics and approaches to targeted epigenetic therapy in acute myeloid leukemia. Blood 127, 42–52. doi: 10.1182/blood-2015-07-604512
Wu, Q., Zhou, L., Lv, D., Zhu, X., and Tang, H. (2019). Exosome-mediated communication in the tumor microenvironment contributes to hepatocellular carcinoma development and progression. J. Hematol. Oncol. 12:53. doi: 10.1186/s13045-019-0739-0
Wu, X., Xiao, Y., Ma, J., and Wang, A. (2020). Circular RNA: a novel potential biomarker for skin diseases. Pharmacol. Res. 158:104841. doi: 10.1016/j.phrs.2020.104841
Xu, M., Xu, X., Pan, B., Chen, X., Lin, K., Zeng, K., et al. (2019). LncRNA SATB2-AS1 inhibits tumor metastasis and affects the tumor immune cell microenvironment in colorectal cancer by regulating SATB2. Mol. Cancer 18:135. doi: 10.1186/s12943-019-1063-6
Xu, Y., Ye, S., Zhang, N., Zheng, S., Liu, H., Zhou, K., et al. (2020). The FTO/miR-181b-3p/ARL5B signaling pathway regulates cell migration and invasion in breast cancer. Cancer Commun. 40, 484–500. doi: 10.1002/cac2.12075
Yang, G., Sun, Z., and Zhang, N. (2020). Reshaping the role of m6A modification in cancer transcriptome: a review. Cancer Cell Int. 20:353. doi: 10.1186/s12935-020-01445-y
Yang, J. D., Nakamura, I., and Roberts, L. R. (2011). The tumor microenvironment in hepatocellular carcinoma: current status and therapeutic targets. Semin. Cancer Biol. 21, 35–43. doi: 10.1016/j.semcancer.2010.10.007
Yang, L., Liu, X., Song, L., Su, G., Di, A., Bai, C., et al. (2020). Melatonin restores the pluripotency of long-term-cultured embryonic stem cells through melatonin receptor-dependent m6A RNA regulation. J. Pineal. Res. 69:e12669. doi: 10.1111/jpi.12669
Yang, Y., Hsu, P. J., Chen, Y. S., and Yang, Y. G. (2018). Dynamic transcriptomic m(6)A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res. 28, 616–624. doi: 10.1038/s41422-018-0040-8
Yang, Z., Li, J., Feng, G., Gao, S., Wang, Y., Zhang, S., et al. (2017). MicroRNA-145 Modulates N(6)-methyladenosine levels by targeting the 3′-untranslated mRNA region of the N(6)-methyladenosine binding yth domain family 2 protein. J. Biol. Chem. 292, 3614–3623. doi: 10.1074/jbc.M116.749689
Yi, D., Wang, R., Shi, X., Xu, L., Yilihamu, Y., and Sang, J. (2020). METTL14 promotes the migration and invasion of breast cancer cells by modulating N6methyladenosine and hsamiR146a5p expression. Oncol. Rep. 43, 1375–1386. doi: 10.3892/or.2020.7515
You, C., Dai, X., and Wang, Y. (2017). Position-dependent effects of regioisomeric methylated adenine and guanine ribonucleosides on translation. Nucleic Acids Res. 45, 9059–9067. doi: 10.1093/nar/gkx515
Yuan, Y., Du, Y., Wang, L., and Liu, X. (2020). The M6A methyltransferase METTL3 promotes the development and progression of prostate carcinoma via mediating MYC methylation. J. Cancer 11, 3588–3595. doi: 10.7150/jca.42338
Yue, B., Song, C., Yang, L., Cui, R., Cheng, X., Zhang, Z., et al. (2019). METTL3-mediated N6-methyladenosine modification is critical for epithelial-mesenchymal transition and metastasis of gastric cancer. Mol. Cancer 18:142. doi: 10.1186/s12943-019-1065-4
Zamarron, B. F., and Chen, W. (2011). Dual roles of immune cells and their factors in cancer development and progression. Int. J. Biol. Sci. 7, 651–658. doi: 10.7150/ijbs.7.651
Zhang, C., Samanta, D., Lu, H., Bullen, J. W., Zhang, H., Chen, I., et al. (2016a). Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m(6)A-demethylation of NANOG mRNA. Proc. Natl. Acad. Sci. U.S.A. 113, E2047–E2056. doi: 10.1073/pnas.1602883113
Zhang, C., Zhi, W. I., Lu, H., Samanta, D., Chen, I., Gabrielson, E., et al. (2016b). Hypoxia-inducible factors regulate pluripotency factor expression by ZNF217- and ALKBH5-mediated modulation of RNA methylation in breast cancer cells. Oncotarget 7, 64527–64542. doi: 10.18632/oncotarget.11743
Zhang, L., Wan, Y., Zhang, Z., Jiang, Y., Lang, J., Cheng, W., et al. (2020). FTO demethylates m6A modifications in HOXB13 mRNA and promotes endometrial cancer metastasis by activating the WNT signalling pathway. RNA Biol. 18, 1265–1278. doi: 10.1080/15476286.2020.1841458
Zhang, Q., Lou, Y., Bai, X. L., and Liang, T. B. (2018). Immunometabolism: a novel perspective of liver cancer microenvironment and its influence on tumor progression. World J. Gastroenterol. 24, 3500–3512. doi: 10.3748/wjg.v24.i31.3500
Zhang, S., Zhao, B. S., Zhou, A., Lin, K., Zheng, S., Lu, Z., et al. (2017). m(6)A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell 31, 591–606. doi: 10.1016/j.ccell.2017.02.013
Zhang, X., Wei, L. H., Wang, Y., Xiao, Y., Liu, J., Zhang, W., et al. (2019). Structural insights into FTO’s catalytic mechanism for the demethylation of multiple RNA substrates. Proc. Natl. Acad. Sci. U.S.A. 116, 2919–2924. doi: 10.1073/pnas.1820574116
Zhang, Y., Song, J., Zhao, Z., Yang, M., Chen, M., Liu, C., et al. (2020). Single-cell transcriptome analysis reveals tumor immune microenvironment heterogenicity and granulocytes enrichment in colorectal cancer liver metastases. Cancer Lett. 470, 84–94. doi: 10.1016/j.canlet.2019.10.016
Zhao, W., Qi, X., Liu, L., Ma, S., Liu, J., and Wu, J. (2020). Epigenetic regulation of m(6)A modifications in human cancer. Mol. Ther. Nucleic Acids. 19, 405–412. doi: 10.1016/j.omtn.2019.11.022
Zhao, X., Yang, Y., Sun, B. F., Shi, Y., Yang, X., Xiao, W., et al. (2014). FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 24, 1403–1419. doi: 10.1038/cr.2014.151
Zhao, Y., Feng, F., Guo, Q. H., Wang, Y. P., and Zhao, R. (2020). Role of succinate dehydrogenase deficiency and oncometabolites in gastrointestinal stromal tumors. World J. Gastroenterol. 26, 5074–5089. doi: 10.3748/wjg.v26.i34.5074
Zheng, G., Dahl, J. A., Niu, Y., Fedorcsak, P., Huang, C. M., Li, C. J., et al. (2013). ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49, 18–29. doi: 10.1016/j.molcel.2012.10.015
Zheng, W., Dong, X., Zhao, Y., Wang, S., Jiang, H., Zhang, M., et al. (2019). Multiple functions and mechanisms underlying the role of METTL3 in human cancers. Front Oncol 9:1403. doi: 10.3389/fonc.2019.01403
Zheng, Z. Q., Li, Z. X., Zhou, G. Q., Lin, L., Zhang, L. L., Lv, J. W., et al. (2019). Long noncoding RNA FAM225A promotes nasopharyngeal carcinoma tumorigenesis and metastasis by acting as ceRNA to sponge miR-590-3p/miR-1275 and upregulate ITGB3. Cancer Res. 79, 4612–4626. doi: 10.1158/0008-5472.CAN-19-0799
Zhou, S., Bai, Z. L., Xia, D., Zhao, Z. J., Zhao, R., Wang, Y. Y., et al. (2018). FTO regulates the chemo-radiotherapy resistance of cervical squamous cell carcinoma (CSCC) by targeting beta-catenin through mRNA demethylation. Mol. Carcinog. 57, 590–597. doi: 10.1002/mc.22782
Keywords: tumor microenvironment, m6A RNA modification, coding RNA, non-coding RNA, cancer
Citation: Si C, Chen C, Guo Y, Kang Q and Sun Z (2021) Effect, Mechanism, and Applications of Coding/Non-coding RNA m6A Modification in Tumor Microenvironment. Front. Cell Dev. Biol. 9:711815. doi: 10.3389/fcell.2021.711815
Received: 19 May 2021; Accepted: 30 August 2021;
Published: 30 September 2021.
Edited by:
Zong Sheng Guo, Roswell Park Comprehensive Cancer Center, United StatesReviewed by:
Mohammad Imran Khan, King Abdulaziz University, Saudi ArabiaJohn Morton, University of Colorado, Colorado, United States
Copyright © 2021 Si, Chen, Guo, Kang and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhenqiang Sun, ZmNjc3VuenFAenp1LmVkdS5jbg==; Qiaozhen Kang, cXprYW5nQHp6dS5lZHUuY24=
 Chaohua Si1,2
Chaohua Si1,2 Chen Chen
Chen Chen Zhenqiang Sun
Zhenqiang Sun