- 1Department of Genetics, Faculty of Medical Sciences, University of Kragujevac, Kragujevac, Serbia
- 2Department of Dentistry, Faculty of Medical Sciences, University of Kragujevac, Kragujevac, Serbia
- 3Laboratory for Bioengineering, Department of Science, Institute for Information Technologies, University of Kragujevac, Kragujevac, Serbia
- 4SPEBO Medical Fertility Hospital, Leskovac, Serbia
For a long time, animal models were used to mimic human biology and diseases. However, animal models are not an ideal solution due to numerous interspecies differences between humans and animals. New technologies, such as human-induced pluripotent stem cells and three-dimensional (3D) cultures such as organoids, represent promising solutions for replacing, refining, and reducing animal models. The capacity of organoids to differentiate, self-organize, and form specific, complex, biologically suitable structures makes them excellent in vitro models of development and disease pathogenesis, as well as drug-screening platforms. Despite significant potential health advantages, further studies and considerable nuances are necessary before their clinical use. This article summarizes the definition of embryoids, gastruloids, and organoids and clarifies their appliance as models for early development, diseases, environmental pollution, drug screening, and bioinformatics.
Bioinformatics
Various bioinformatics and computational biology analyses are used to evaluate disease model accuracy (Bock et al., 2020; Gendoo, 2020). Different omic profiling technologies are used to discover molecular and functional alterations in organoids (Figure 1). Transcriptomics copy number and structural variation changes (whole-genome sequencing—WGS, whole-exome sequencing—WES), proteomics (protein expression changes), epigenomics, (toxico) genomics, and metabolomics (enrichment of biological pathways) are applied to comprehend or to predict toxicity (Stojkovic et al., 2021b). However, bioinformatics tools are used to analyze omics data (Wu et al., 2018; Mincarelli et al., 2018; Brazovskaja et al., 2019; Bock et al., 2020; Gendoo, 2020; Zink et al., 2020; Figure 1). To elucidate the exact mechanism and understand how various types of pollution particles influence gene changes and signaling pathways in organoids, a reliable method to estimate the activity within pathways is necessary. By estimating the level of activity of stimulus-response sub-pathways (signaling circuits) within signaling pathways, which ultimately trigger cell responses, we can investigate interactions between various environmental and intracellular pollutions to explore the mechanism and origination of human disease, but also prediction of clinical outcomes (Hidalgo et al., 2017; Peña-Chilet et al., 2019; Bojic et al., 2020; Zeng et al., 2021; Wu et al., 2018). A platform like HiPathia (Bojic et al., 2020), with vast computational data, enables insight into modeling of various diseases (Figure 1). Results obtained in such a manner may serve as a competent tool for further clinical trials (Hidalgo et al., 2017; Bojic et al., 2020; Falco et al., 2020). HiPathia enabled an examination of the effect of carboxyl-modified fluorescent nanosized plastic (polystyrene) items on gene alterations and signaling pathways as reported by Bojic et al. (2020). Moreover, HiPathia pointed to several altered circuits, such as the peroxisome proliferator-activated receptor pathway that has a crucial role in lipid metabolism, but also prediction of the clinical outcome which included the APOC3 circuit, which induces hyperalphalipoproteinemia, thus raising the risk of ischemic cardiovascular disease (Bojic et al., 2020).
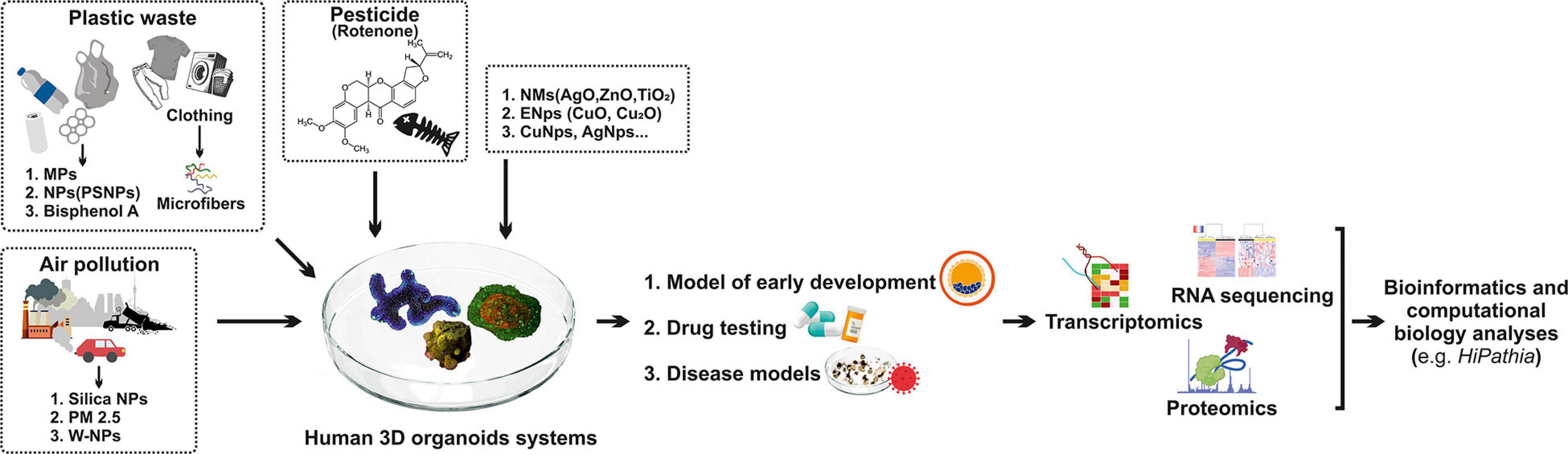
Figure 1. Human 3D organoid systems: a promising platform to study the effect of environmental pollution. Plastic waste, air pollution, pesticides, and various nanomaterials have a negative impact on human health, which is mimicked by organoid systems that serve as a proper model for early development, drug testing, and disease models. As a result of organoids’ exposure to these pollutants, molecular and functional alterations in organoids are noted, so different omic profiling technologies are used to discover these alterations, such as transcriptomic, RNA sequencing, and proteomics. Hence, different bioinformatics tools are used for analyzing omics such as HiPathia. MPFs—microplastic fibers; MPs—microplastics; PSNPs—polystyrene nanoplastics; PM 2.5—particulate matter 2.5; PSCs—pluripotent stem cells; W-Nps—tungsten nanoparticles; NMs—nanomaterials; ENps—engineered nanoparticles; Nps—nanoparticles; AgO—silver oxide; TiO2—titanium dioxide; ZnO—zinc oxide; CuO—copper oxide.
RNA massive sequencing (RNA-seq) is the most used technique for gene expression profiling in a single assay (Garrido-Rodriguez et al., 2021). Although it is possible to compare relative gene expression, RNA-seq cannot warrant function at the protein level (Li et al., 2021). Even though throughput is restricted, single-cell RNA-seq identifies infrequent populations of cells with functional significance (Li et al., 2021; Yan et al., 2013). On the other hand, the transcriptome is more significant because it provides information about the specific biological function or gene expression, compared to separate analyses of the genome, the epigenome, and the proteome (Bar and Benvenisty, 2019; Song et al., 2019; Zeng et al., 2021). In the study by Zeng et al. (2021), transcriptome analysis was used to estimate the effect of particulate matter 2.5 (PM2.5) on human embryonic stem cell derived retinal organoids (hEROs) and showed that mitogen-activated protein kinase (MAPK) and phosphoinositide 3-kinase (PI3K)/AKT pathways were involved significantly, while fibroblast growth factors (FGFs), especially FGF8 and FGF10, were decreased, thereby inducing abnormal human retinal development.
In the previously described study (van Dijk et al., 2021; the contribution is a preprint), the RNA-seq analysis has been used to confirm that nylon fibers were less inhibitory for the growth of alveolar organoids (AO) than treatment with component leaching of the polymer or lower numbers of nylon fibers. The Notch1 and Notch2 signaling pathways were downregulated, as well as their ligands Jag1 and Jag2, which are responsible for the development of airway epithelial cells, and club cells (van Dijk et al., 2021; the contribution is a preprint). Winkler and coworkers used human lung organoids to examine the effect of microplastic fibers (MPFs) on organoid growth and their inflammatory effects on the established lung organoids (Winkler et al., 2021; the contribution is a preprint). Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was used for gene expression analysis of oxidative stress-related genes, lung-specific genes, and inflammatory cytokines (Winkler et al., 2021; the contribution is a preprint). No significant differences in the gene expression of cytokines or oxidative stress-related genes such as superoxide dismutase family genes (SOD1 and SOD2), glutathione detox-related genes [glutathione detox-related genes (GSTA1 and GPX1)], catalase (CAT), and ROS-controlling genes [(NADPH oxidase-2 (NOX2), cyclo-oxygenase 1 (COX1), NADH dehydrogenase 1 (ND1)] were noticed in human lung organoids after exposure to MPFs (Winkler et al., 2021; the contribution is a preprint). The authors also confirmed no significant difference in gene expression responsible for epithelial lung markers such as NK2 homeobox 1 (NKX2.1) and Claudin 1 (CLDN1) as well as the specific airway lung markers surfactant protein A1 (SFTPA1) and surfactant protein C (SFTPC) in alveolar-type 2 progenitor cells (AT2 cells), secretoglobin family 1A member 1 (SCGB1A1) (club cells), nephrocystin 1 (NPHP1), dynein axonemal heavy chain 5 (DNAH5) (ciliated cells), and keratin 5 (KRT5) (basal cells) in human lung organoids exposed to MPFs and the control group (Winkler et al., 2021; the contribution is a preprint). Schwartz et al. (2015) studied developmental neurotoxicity by using reproducible 3D neural constructs containing vascular and microglial components on synthetic hydrogels after exposure to toxic or nontoxic chemicals. In this study, 3D neural constructs were exposed to a set of 31 control compounds and 39 toxins through day 16 or day 21 and then collected for RNA-seq (Schwartz et al., 2015). RNA-Seq identified differentially expressed genes that included neurogenesis such as GABAergic neurons [e.g., gamma-aminobutyric acid receptor (GABA receptors)], glutamatergic neurons [(e.g., vesicular GABA transporter (VGAT) and vesicular glutamate transporters (VGLUT2)], cortical neurons [(POU class 3 homeobox 2 (BRN2/POU3F2), reelin (RELN), BAF chromatin remodeling complex subunit BCL11B (CTIP2/BCL11B)], synaptic markers (e.g., synapsins and synaptic vesicle components), and glial cells [solute carrier family 1 member 3 (GLAST/SLC1A3), glial fibrillary acidic protein (GFAP), platelet-derived growth factor receptor alpha (PDGFRA)] in neural constructs in relation to undifferentiated hESCs (Schwartz et al., 2015).
Proteomics is determined as wide-ranging protein analysis enabling recognition, quantification, and posttranslational modification between other related facts in terms of proteins in a cell, tissue, or biofluid (Lindoso et al., 2019). Among other “omics” analysis, proteomics is one of the widely employed methods in bioinformatics and liquid chromatography linked with mass spectrometry and mainly applied in the examination of induced pluripotent stem cells (iPSCs; Walther and Mann, 2010; Lindoso et al., 2019), hiPSC-derived organoids (Hale et al., 2018), or hESC-derived organoids (Nascimento et al., 2019). Using proteomics analysis, Hale et al. (2018) compared organoid-derived glomeruli (OrgGloms) with conditionally immortalized human podocyte cell lines. They elucidated that OrgGloms displayed higher-level matrix extracellular components and an α5(IV) chain of type IV collagen network, while α3 and α4(IV) chains were less expressed. These outcomes emphasize the importance of OrgGloms as a proper 3D model of the human glomerulus in physiological and pathological conditions (Hale et al., 2018). By using shotgun proteomics to examine human cerebral organoids, Nascimento et al. (2019) identified 3,073 proteins associated with various brain developmental stages, especially with neurogenesis, axon guidance, synaptogenesis, and cortical brain development. If there is a need for analyzing alterations in metabolites at the system level, this type of omics is termed metabolomics (Wishart, 2019; Neef et al., 2020; Garrido-Rodriguez et al., 2021).
Introduction to Blastoids, Embryoids, Gastruloids, and Organoids—The Definitions
Valuable events of early mammalian development and self-organization are demonstrated in various studies on early mouse and human embryos that could be cultured ex vivo in the absence of maternal tissues (Deglincerti et al., 2016; Shahbazi et al., 2016). However, early human embryos, including the blastocyst stage, are difficult to obtain, have a small number of cells (<100), and are not easy to physically and genetically manipulate. Therefore, many genetically similar structures should be generated, thus opening possibilities for different analyses, including high-throughput screens and biochemistry-based assays. One of the well-known structures is the blastoid—the first version of a preimplantation blastocyst model made by promotion of the self-organization of mouse embryonic stem cells (ESCs) and trophoblast stem cells (TSCs) (Rivron et al., 2018). The trophoblast and embryonic compartments of blastoids can be physically and genetically modified independently from another blastoid, offering enormous technical advantages compared to blastocysts. Recent studies focused on human blastoids to study and comprehend early human development and prevent pregnancy defects and birth loss (Liu et al., 2021; Yu et al., 2021; Zheng and Fu, 2021). Human blastoids can be generated in a two-step culture process—isolation from human blastocysts or by reprogramming adult human cells (Yu et al., 2021), or in a one-step culture process—by reprogramming skin fibroblasts (Liu et al., 2021). Regardless of the in vitro method used for generating human blastoids (one-step or two-step culture), in both cases, it was shown that human blastoids had almost identical size, number of cells, and shape similar to natural blastocysts (Liu et al., 2021; Yu et al., 2021; Zheng and Fu, 2021). In the study of Zheng and Fu (2021), genome-wide expression analysis was used to clearly define molecular similarities of the blastoids with preimplantation human blastocyst. They showed that there are molecular similarities between generated human blastoids and preimplantation human blastocyst. Also, they proved that blastoid cells have crucial characteristics of blastocyst lineages in terms of the ability to generate various stem cell types which are isolated from the blastoids, offering new insights to study early preimplantation and early postimplantation blastocyst development (Zheng and Fu, 2021). However, there are some limitations that need to be reconsidered. For instance, the development of the blastoids is not effective and differs among cell lines from different donors, and between experimental clusters. Also, it was noted that the three lineages in single blastoids developed at different velocities, and their growth in the same dish was not synchronized together with unspecified cell populations with no equivalent in natural human blastocysts (Zheng and Fu, 2021). The other obstacle includes ethical controversies such as the fact that the development of human blastoid in vitro is limited in postimplantation stages until 14 days in vivo (Zheng and Fu, 2021). The findings of Xiang et al. (2020) might help to improve the ability to culture blastoids up to this limit, by 3D systems for culturing human blastocysts, which effectively promote postimplantation development, although bioethical issues need to be addressed.
Embryoid bodies (EBs) are defined as 3D aggregates of pluripotent stem cells (PSCs) or differentiated cells used as a layout of early development and comprising the three embryonic germ layers (Evans, 2011; Simunovic and Brivanlou, 2017). Additionally, EBs go through the initial development phase similarly to pregastrulating embryos and resemble early teratoma (de Jaime-Soguero et al., 2018). In vivo and in vitro, cell differentiation depends on morphogen gradients and signals that supply instructive and positional signs (ten Berge et al., 2008; Simunovic and Brivanlou, 2017). For instance, EBs can be used as a model for teratogen-testing platforms, which includes evaluation of chemically induced effects on EB morphology, effects on the differentiation of particular cell types of interest, estimation of the transcriptome or proteome, effects on specific signaling and developmental pathways, and at last, effects upon cellular physiology (Lee S. et al., 2020).
Unlike embryos, which go through defined stages with typical morphologies, such as blastula, gastrula, and neurula stages, embryoid is an artificial construct made from cultured cells that try to imitate all or part of an embryo, or its specific stage (Simunovic and Brivanlou, 2017). The embryoid can be defined as a more organized EB that develops as a result of cell polarization caused by (i) the extracellular matrix (ECM) in the adjacent medium or (ii) the accurate topology of multiple types of cells that represent an embryo at a specified time of development (Simunovic and Brivanlou, 2017).
Unlike embryoids, gastruloids can be defined as an in vitro multicellular model of a gastrulating embryo, either in 2D (Etoc et al., 2016) or in a 3D culture system (Turner et al., 2016). Gastruloids are also defined as complex 3D structures with the ability to self-organize in vitro and look like developing tissue in vivo (Munsie et al., 2017). They are distinct from organoids because they do not essentially recapitulate an organ but rather a developmental process, offering the possibility to create post-implantation models (Munsie et al., 2017) and models to study (Simunovic and Brivanlou, 2017).
For the purposes of this review, we focus primarily on the opportunities and challenges concerning human organoids, and their ability to serve as a model to study the effects of environmental pollution on human health.
Introduction to Human Organoids
The term “organoids” appeared in the 1950s (Vendrely, 1950) and finally was delineated and systemically elaborated by Eiraku and Sasai (2012). A typical depiction of organoid is explained as a structure in which pluripotent or progenitor stem cells are differentiated into multiple cell populations that self-organize into tissue similar to an organ (Eiraku and Sasai, 2012; Kicheva and Briscoe, 2015; Clevers, 2016) and have the capacity of stable long-term culture and passage (Eiraku and Sasai, 2012). The stem cell sources of the existing cultured organoids are for the most part PSCs—iPSCs, ESCs, and adult stem cells (ASCs). These sources are used to induce various types of organ tissues such as gut (Spence et al., 2011), kidney (Takasato et al., 2015), pancreas (Greggio et al., 2013), brain (Lancaster et al., 2013), retina (Nakano et al., 2012), inner ear (Koehler et al., 2017), lung (Wong et al., 2012), and liver (Takebe et al., 2013). However, the establishment of human AdSC-derived organoids is limited by accessibility to the tissue and prior knowledge of the culture conditions, while an iPSC line, once established from a patient (Brouwer et al., 2016), can be used to repeatedly generate different tissue models without any time limit [that is, beyond the patient’s lifespan (Kim J. et al., 2020; Narsinh et al., 2011)]. Although ASCs can be stimulated to form organoids (Yin et al., 2016), the focus of our review will discuss organoids derived from human iPSCs and ESCs.
Differences Between 2D and 3D Models
The usage of 2D models is limited compared to superior 3D organoid technology (Ho et al., 2018). The major limitation of 2D culture is cells arranged as a monolayer, providing atypical growth kinetics and cell attachments, therefore not completely presenting the natural microenvironment of the cells (Nicolas et al., 2020). To emulate tissue homeostasis and complex interactions, 3D structures, better known as organoids, are preferably used compared to 2D cultures that are unable to sustain intercell communication (Clevers, 2016). Additionally, 3D systems are rather utilized due to improved cellular membrane integrity, and niche manipulation (Ferraz et al., 2016; McCauley and Wells, 2017; Ho et al., 2018; Chen et al., 2019; Stojkovic et al., 2021b). Also, 3D organoids go through multi-lineage differentiation, creating heterogeneous groups of cells that self-assemble into complex tissue-like structures mimicking physiologically more pertinent microenvironment (Cugola et al., 2016). Many studies show that hESCs and hiPSCs are used for generating 2D and 3D organoids allowing the study of differentiation mechanisms (Pamies et al., 2017; Rosca et al., 2020), the processes involved in embryonic development (Lancaster and Knoblich, 2014; Pamies et al., 2017; Rosca et al., 2020), and the mechanisms involved in various diseases (Lancaster and Knoblich, 2014; Pamies et al., 2017; Rosca et al., 2020; Stojkovic et al., 2021a), drug testing (Lancaster and Knoblich, 2014), and toxicity connected with environmental pollutants (Truskey, 2018; Rosca et al., 2020; Stojkovic et al., 2021b). However, it is well known that organoids have tremendous potential in drug screening and personalized medicine (Kondo and Inoue, 2019; Kim J. et al., 2020). By discovering organoids, their use made the drug development faster, more effective, and ethically more justified than using animal models for the same purpose (Aboulkheyr Es et al., 2018). Different new medical treatments that were developed for the human disease often manifest many limitations (e.g., problems with predictions of outcomes, time-consuming drug testing, or differences relating to the patient as an individual) (Eglen and Randle, 2015; Dedhia et al., 2016). Therefore, organoid culture based on a particular individual or disease will advance into the potential instrument for adequate therapy (Lehmann et al., 2019).
Regarding the use of organoids as a suitable 3D model, certain obstacles need to be overcome. (1) There is lack of adequate cellular microenvironment, such as endothelial or immune cells (Koike et al., 2019); this problem can be solved by coculturing additional missing cells with organoids. (2) Prices for organoid establishment are relatively high compared to traditional cell lines (although organoids cost less than mouse or fish models). (3) 3D models mirror only specific organ tissue, not the whole organism, and therefore lack interorgan communication. However, progressive endeavors to solve this problem emerge. For instance, a few organoids have been joined with the aim to examine communication between the liver, pancreas, and gastrointestinal tract (Adhya et al., 2018), or to study the interplay between the brain and hormone-producing organs (Xiang et al., 2020). (4) There are no widely accepted protocols standardized for organoid establishment. (5) Heterogeneity in terms of variation between individuals and protocols results in different outcomes (Truskey, 2018; Kim J. et al., 2020). To overcome this obstacle, single-cell profiling technologies for transcriptome and epigenome analysis might be crucial regarding highly correct assays appropriate for this purpose (Kim J. et al., 2020). It should be emphasized that the advantages of using organoids are much greater compared to their disadvantages—of course, both should be taken into account.
Organoids as Models of Early Development
Besides the improved research of signaling pathways in cell specification and organogenesis, organoids illustrate the physical basis of tissue and organ forming (Lancaster and Knoblich, 2014). Primary sources of organoids are ESCs, iPSCs, and fetal tissues (Spence et al., 2011; Nakano et al., 2012; Wong et al., 2012; Greggio et al., 2013; Lancaster et al., 2013; Takebe et al., 2013; Takasato et al., 2015). The development of tissues such as the stomach, brain, and pancreas has been studied through gradual differentiation of iPSCs and ESCs to organoids by adjusting signaling pathways such as Wingless-related integration site (WNT), bone morphogenetic protein (BMP), and FGF (Greggio et al., 2013; Lancaster et al., 2013; McCracken et al., 2011). Fetal pulmonary organoids are being utilized to demonstrate the signaling interaction between exogenous FGFs (essential for endothelial network assembly) and the vascular endothelial growth factor A (VEGF-A) pathway known to suppress the forming of the endothelial network and the cross talk with the Sonic hedgehog (SHH) pathway that promotes epithelial and endothelial morphogenesis (Mondrinos et al., 2014). When it comes to pancreas development, the loss of F-box and WD repeat domain-containing 7 (Fbw7) in CK19 cytokeratin 19 (Krt19)+ adult pancreatic ductal cells in vivo led to stabilization of the transcription factor Neurog3 (Ngn3), resulting in reprogramming of ductal cells to insulin-secreting beta cells (Sancho et al., 2014). Intestinal organoids–enteroids resulted in structures having leucine-rich repeat-containing G-protein-coupled receptor 5 (Lgr5+) intestinal SCs and other differentiated cells localized equally to the in vivo organization (Cao et al., 2015). The forebrain cerebral cortex was also developed using novel protocols for 3D brain-like tissue development (Lancaster and Knoblich, 2014; Shahbazi et al., 2016), therefore confirming the vast potential of brain organoid research. In 2017, Koehler et al. (2017) reported the derivation of inner ear organoids using human PSCs and modulating FGF, BMP, TGF-β, and WNT signaling, generating organoids with sensory epithelia that are innervated by sensory neurons. This method significantly promoted further studies of human inner ear development and research on regenerative or drug therapies for hearing loss.
Organoids in Drug Screening
Many studies showed how healthy organoids can be used in the assessment of drug toxicity such as cardiotoxicity (Eder et al., 2016), nephrotoxicity (Takasato et al., 2015), and hepatotoxicity (Kostadinova et al., 2013; Katsuda et al., 2017). On the other hand, organoids are used to study the effect of some drugs on preexisting diseases, such as primary tumors (Jabs et al., 2017; Abbasi, 2018; Vlachogiannis et al., 2018), rare genetic diseases including cystic fibrosis (Fleischer et al., 2020), neurological diseases (Lee S. E. et al., 2020; Costamagna et al., 2021), and infectious diseases (Zhou et al., 2017; Heo et al., 2018). Other studies reported the use of cancer organoid lines, for example colorectal cancer (CRC) organoid lines to screen 83 drugs (van de Wetering et al., 2015), breast cancer organoid lines for testing inhibitors human epidermal growth factor receptor (HER) signaling pathway (Reid et al., 2018), or bladder cancer organoid lines for testing 26 drugs (Lee et al., 2018). Also, organoid technology aims to supply functional biological structures that can be transplanted into patients in the near future, although precise characterization and validation of organoids as accurate models of human biology are required (Bock et al., 2020). AOs and proximal airway air–liquid interface cell culture systems are advantageous for the examination of antiviral compounds against severe acute respiratory syndrome coronavirus clade 2 (SARS-CoV-2). The next drug for the treatment of COVID-19 has been examined heretofore: IFN type I, IFN type III, remdesivir, camostat mesylate [a cofactor transmembrane protease serine 2 (TMPRSS2) inhibitor], E-64d (an inhibitor of the endosomal cysteine proteases cathepsin B and L), and a library of FDA-approved drugs (the Prestwick collection) (Han et al., 2021; Huang et al., 2020; Lamers et al., 2021). Both remdesivir and camostat mesylate showed antiviral capacity and decreased SARS-CoV-2 N levels (Huang et al., 2020). Imatinib, mycophenolic acid, and quinacrine dihydrochloride diminished SARS-CoV-2 infection of hPSC-derived lung organoids (Han et al., 2021). Pretreatment with IFN-λ1 abolished viral replication in bronchioalveolar organoids. Angiotensin-converting enzyme 2 (ACE2) expression is regulated by androgen signaling and represents an important risk factor of adverse COVID-19 outcome in male adults. This is confirmed by antiandrogenic drugs, which diminished ACE2 expression and prevented SARS-CoV-2 infection of human hESC-derived lung organoids (Samuel et al., 2020).
Embryos, Their “Surrogates” and Organoids as a Model for Environmental Pollution
The systematic assessment of the global effects of environmental pollution on human health has become increasingly quantitative in the last decades. Humans and animals are constantly exposed to many environmental pollutants and stressors—at least those associated with air pollutants, modern chemicals in the home, food, and beverages/water (Briggs, 2003; Manisalidis et al., 2020). Despite the properly planned high-throughput screening that tried to illuminate the model of action of pollutants (Kavlock et al., 2012), further examinations are necessary to completely understand the exact mechanism by which pollutants cause pathology (Kavlock et al., 2012). There is a wide range of environmental pollutants (Table 1). Due to their omnipotent presence and extensive usage in every aspect of the industry, plastics have become one of the most severe environmental pollutants (Bouwmeester et al., 2015). Plastics in the environment have two forms depending on the size, microplastics (MPs, diameter <1 mm), and nanoplastics (NPs, diameter <100 nm), and can be found in water, ground, food, and various objects and materials (Bouwmeester et al., 2015; Prata, 2018; Toussaint et al., 2019). Hence, polystyrene, as one of the most utilized sorts of plastic, especially in packing food and drinks, construction, computer printers, and other industries, requires further research (Gu et al., 2019; Li-Juan et al., 2019). A detailed study on the possible effects of polystyrene NPs (PSNPs) on the transcription profile of preimplantation human embryos and hiPSCs has recently been, for the first time, conducted by our group (Bojic et al., 2020). Applying the gene set enrichment analysis and HiPathia, this study showed that PSNPs led to downregulation of LEFTY1 and LEFTY2, pluripotency genes, and upregulation of CA4 and OCLM, genes related to eye development. Also, there was a significant impact of PSNPs on genes responsible for the development of atrioventricular valve and cellular components. The RNA-seq analysis showed that PSNP intracellular pollution might cause different clinical outcomes, including abnormal early development and several detrimental diseases (Bojic et al., 2020). MPs are omnipresent in the environment (Prata et al., 2020) and are continuously released into the atmosphere. Most MPs consist of MPFs coming from synthetic clothing, fabric, and upholstery (Henry et al., 2019), but mostly from polyester (Dris et al., 2017). One study examined the effect of MPFs on lung organoids derived from tissue-resident ASCs of healthy donors. The organoids were exposed to various MPF concentrations (1, 10, and 50 mg L– 1) and analyzed by optical microscopy, scanning electron microscopy (SEM), and confocal microscopy. Gene expression assessment of lung-specific genes, inflammatory cytokines, and oxidative stress-related genes was performed by qRT-PCR and showed no significant differences when compared to the control group (Winkler et al., 2021; the contribution is a preprint). Even though MPFs did not have an adverse effect on lung organoids, there was the polarization of the cell growth along the fibers, similarly to organoid-covered plastic fibers with a cellular layer in the study of van Dijk et al. (2021; the contribution is a preprint). Such outcomes implied possible negative effects of MPFs. Hence, a recent study by van Dijk et al. (2021) showed that the growth of murine and human lung organoids was inhibited 14 days after exposure to nylon microfibers. This was confirmed by light and fluorescence microscopy. However, the effect of polyester on human organoid growth was less profound. In the same study, it has been proved that nylon microplastics did not affect fully develop 14-day organoids, suggesting that nylon microplastics have a huge impact on developing organoids (van Dijk et al., 2021; the contribution is a preprint). This is explained by the negative impact of nylon microplastics on the top five enriched pathways for downregulated and upregulated genes crucial for epithelial development and function. Even though van Dijk et al. (2021) supposed that bisphenol A is the main reason for lung organoid growth inhibition, incubation of lung organoids with bisphenol-A did not affect organoid growth. Eventually, this study suggested that nylon microplastics can negatively affect children and people with chronic or seasonal respiratory diseases. However, very few studies show the detrimental effect of plastic waste on an individual’s development and metabolism. Bisphenol A, also known as the most examined endocrine disruptor (Corrales et al., 2015), is a widely used chemical that can be found almost everywhere—soft plastic bottles, the lining of aluminum food cans—and can harm metabolic and reproductive function (Choi et al., 2016). Choi et al. (2016) examined the effect of acute exposures to bisphenol A on bovine embryo development in vitro at environmentally proper concentrations (1 and 10 ng mL– 1) at 3.5–7.5 days post-fertilization. They showed that blastocyst development was impaired, embryo quality was decreased, and glucose utilization was increased, although cell number was not altered after exposure to 10 ng mL– 1 bisphenol A (Choi et al., 2016). Some studies displayed the effect of low-dose bisphenol A on the early differentiation of hESCs into mammary epithelial cells in 3D conditions (Yang et al., 2013). Another study showed that a low dose of bisphenol A negatively affected hESCs’ differentiation into prostate organoids (Calderon-Gierszal and Prins, 2015). These two studies addressed the toxic effects of bisphenol A on the reproductive systems using hESCs differentiated into mammary epithelial cells and human prostate organoids in 3D conditions (Yang et al., 2013; Calderon-Gierszal and Prins, 2015).
Besides the plastics, there are numerous environmental pollutants whose effect was examined on organoids (Table 2). The respiratory tract is the first target of numerous professional noxas, such as silica, especially present in industries of glass, foundries, construction, ceramics, chemical, plastics, rubber, water filtration, and agriculture. Di Cristo et al. (2020) confirmed the suitability of the 3D airway model regarding the simulation of working conditions of people exposed to silica nanoparticles (SiO2 Nps). A 3D mucociliary tissue model of the primary human bronchial epithelium was exposed to SiO2 Nps for 12 weeks; the viability of the 3D airway model was assessed by AlamarBlue (resazurin) assay, whereas the integrity of the tissue was measured by transepithelial electrical resistance (TEER) and assessment of the membrane proteins’ expression was performed by Western blot analysis. Interestingly, no adverse effect of SiO2 Np exposition in vitro was confirmed, suggesting the effectiveness of the 3D airway model regarding mucociliary system clearance and respiratory defense mechanisms after SiO2 Np exposition (Di Cristo et al., 2020). When it comes to inhalation pollutants, besides silica, particulate matter 2.5 (PM2.5), an air pollutant of very small size (≤2.5 μm), is present mostly in car gas emission and is related to numerous lung pathologies, such as asthma, COPD, or lung cancer (Li et al., 2018). For that reason, one study examined whether organic PM2.5 extract caused the same cytotoxic effect in in vitro and in vivo conditions (Ferraz et al., 2016). Results displayed that PM2.5 had a cytotoxic effect on A549 cells cultured in a monolayer or 3D, by reducing mitochondrial dehydrogenase activity and cell membrane integrity, respectively (Ferraz et al., 2016). However, the exact mechanism by which PM2.5 influences lung development and leads to various lung pathologies remains unclear. Thereupon, a study was conducted in order to decipher the developmental toxicity of fine diesel PM (dPM2.5) exposure during hPSC-derived alveolar epithelial cell (AEC) differentiation and 3D multicellular AO development (Kim J. H. et al., 2020). Results showed that dPM2.5 harmed the AEC differentiation and led to upregulation of NADP oxidases and inflammation. Also, exposition to PM2.5 caused epithelial-to-mesenchymal transition during AEC and AO development. Remarkably, for the first time, there was an upregulation of two important molecules—ACE-2 and TMRPSS2—in both hPSC-AECs and AOs treated with dPM2.5 (Kim J. H. et al., 2020). Importantly, ACE-2 is a protein that enables the entry point for the coronavirus to invade and infect a wide range of human cells and causes the SARS-CoV-2 (Hoffmann et al., 2020). This study displayed the alveolar development toxicity and the rise of SARS-CoV-2 permissiveness of PM2.5 exposed cells, making this hPSC-based 2D and 3D alveolar induction model beneficial in terms of environmental toxicity and SARS-CoV-2 virus examination (Hoffmann et al., 2020). Not just lung organoids, but also retina, turned out as suitable for PM2.5 toxicity research, which was confirmed by Zeng et al. (2021). They examined the effect of PM2.5 on the development of the human retina by using hEROs. In this study, it was shown that the development of hEROs was influenced by PM2.5 exposure in a dose-dependent manner (25, 50, and 100 μg/mL), by reducing cell proliferation and supporting cell apoptosis, which resulted in abnormal human retinal development (Zeng et al., 2021). Finally, to encircle a wide range of lung organoid employment, it should be noted that the effect of tungsten nanoparticles (W-NPs), utilized in nanotechnology, metallurgy, and fusion technology, can be also successfully examined on organoids. The International Thermonuclear Experimental Reactor (ITER) is a project that examined potential effects of W-Nps that could be emitted in air and environment and subsequently affect the respiratory tract by inhalation (George et al., 2019). The latest study examined the acute toxicity of W-Nps on MucilAirTM, a 3D in vitro model of the human airway epithelium. W-Nps had a restricted influence in terms of toxic effects, cellular absorption, and W transfer over the lung epithelium leading to a decrease in barrier integrity, whereas there was no effect on metabolic activity or cell viability, except a transient increase in IL-8 secretion (George et al., 2019). This research might offer initial data about biokinetic lung models for ITER-like tritiated W-Nps. All of these outcomes can be utilized to settle novel safety policies and radiation protection modes. Besides lungs, certain toxicants may affect the neurological system. For instance, rotenone, a well-deciphered, widely utilized pesticide to control fish populations, is proved to be neurotoxic and leads to Parkinson’s disease (Richardson et al., 2019). To examine that in more detail, the immortalized cell und human mesencephalic (LUHMES) cells were used to study cellular toxicity, resurgence, and adaptability subsequently to rotenone exposition (Harris et al., 2018). 3D LUHMES was exposed to rotenone (100 nM, 24 h) which led to a decrease in complex I activity, ATP, mitochondrial diameter, neurite outgrowth, and transcriptomic changes. Subsequently to compound removal, all of these adverse effects were overcome, due to cells’ adaptation to short-term rotenone exposure. In order to test resilience, cells were reexposed to rotenone after the washout and recovery period. There were transcriptomic alterations in genes, such as nuclear factor erythroid-derived 2-like 2 (NEF2L2) which regulates the response to oxidative stress, activating transcription factor 4 (ATF4) branch of the unfolded protein response activated in response to endoplasmic reticulum (ER) disturbances or proteotoxicity, and excitatory amino-acid carrier 1 (EAAC1), a high-affinity Na+-dependent L-glutamate/D,L-aspartate cell-membrane transport protein that were downregulated on day 14 but unaltered in pre-exposed aggregates. Dopamine active transporter (DAT) and Caspase 3 (CASP3) were only changed after reexposure to rotenone, while thymidylate synthetase (TYMS) and centromere protein U (MLF1IP) were downregulated in both single-exposed and pre-exposed aggregates. This study enables insight into the effect of rotenone in neurodegenerative diseases and displayed 3D systems as an excellent tool for neurotoxicity research. Another detailed study examined the neurotoxic profile of 87 compounds (Table 1) widely utilized in various industrial sectors (Sirenko et al., 2019). All of the compounds were tested using hiPSC-based 3D neural cultures. Calcium oscillations—which are related to necrosis or disease progression—are detected in 57% of the analyzed compounds (Sirenko et al., 2019). Characterization of oscillation profiles in 3D neural cultures was performed through multi-parametric analysis while cellular and mitochondrial toxicity was estimated by high-content imaging. This model turned out as beneficial and illuminating toward the exact neurological effect of such a huge complex of compounds. Besides brain and lung organoids, various compounds (Table 1) have been examined on other types of 3D systems—liver, heart, intestine, skin, placenta, and retina (Table 2). The liver, as the metabolic center, represents a perfect target for toxicology research. A study by Forsythe et al. (2018) examined the dose-response toxicity of lead, mercury, glyphosate, and thallium on the liver and cardiac organoids within 48 h. The effects of all compounds were estimated by cytotoxicity and viability staining, ATP activity evaluation, IC50 value, and cardiac beat activity. Likewise, it turned out that all tested compounds have a toxic effect on both liver and cardiac organoids, especially thallium (Forsythe et al., 2018). Not just heavy metals, but also nanomaterials (NMs), due to ubiquitous employment and constant public exposure rose concerns regarding the exact impact of NMs on human health. Therefore, one study examined effects on the liver organoids of single and numerous exposures of NMs [silver oxide (AgO), zinc oxide (ZnO), titanium dioxide (TiO2), and multi-walled carbon nanotubes (MWCNT)] (Kermanizadeh et al., 2014). Results showed that numerous exposures were significantly more harmful, specifically AgO and ZnO. The cytotoxic effect was analyzed by the level of cytotoxicity, cytokine secretion, lipid peroxidation, and genotoxicity. Later on, the same group (Kermanizadeh et al., 2019) examined a 3D human liver microtissue (MT) repeatedly exposed to minimal NM concentrations, including TiO2, ZnO, CeO2, and crystalline silica DQ12. NM cell toxicity effect was observed by analysis of cell membrane integrity and aspartate aminotransferase (AST) activity, pro/anti-inflammatory response, and hepatic function. NM-treated MT displayed a concentration-dependent penetration of NMs profoundly within the tissue, while AST assessment turned out to be unsuitable in this experiment, whereas the cytokine analysis (IL6, IL8, IL10, and TNF-α) proved useful in highlighting recovery periods. Overall, this study emphasized the great advantage of liver MT in nanotoxicology research and highlighted the nanotoxicological assessment on liver MT beyond 2 weeks as unsuitable, due to the aging effect on cells. Since ingestion is a possible route of environmental toxin intake, the gastrointestinal (GI) tract is inevitable for environmental pollution research. Even though toxicological studies on engineered nanoparticles’ (ENps) influence on the GI tract are minimal (McCauley and Wells, 2017), still, there are promising outcomes that could reveal the impact of nanoparticles on the gut. Henson et al. (2019) examined the cytotoxicity of cupric (II) oxide (CuO) and Cu2O-polyvinylpyrrolidone (PVP)-coated Nps and copper ions on rat intestine epithelial cells (IEC-6) and human intestinal cells, 2D and 3D models. The mechanism by which copper nanoparticles cause toxicological properties includes reactive oxygen species (ROS) forming, reduction of cellular glutathione, mitochondrial membrane depolarization (Thit et al., 2015; Wang et al., 2021), mitochondrial membrane damage (Wang Y et al., 2012; Wang Z et al., 2012), and the release of toxic Cu ions (Fröhlich, 2013). In line with these outcomes, Henson et al. (2019) estimated cell viability by MTT assay, H2O2, and glutathione (GSH) detection, and mitochondrial membrane potential. The CuO Nps were more cytotoxic to the rat 2D intestinal model than the human 3D model, probably due to differences between 2D and 3D cultures themselves, and/or differences between species origins (rats vs. humans). Finally, CuO Nps were cytotoxic to rat and human intestinal cells in a dose- and time-dependent manner, proposing therefore that Cu2O-PVP Nps are toxic due to their dissolution to Cu ions, while CuO Nps have innate cytotoxicity, without dissolving to form Cu ions. Another detailed research observed the impact of AgO, CuO, ZnO, and TiO2 Nps on the EpiIntestinal tissues (Markus et al., 2021). Outcomes again displayed a decline in viability and tissue barrier debilitation after exposition to CuO, ZnO, and SWCNT Nps. Additionally, in culture supernatants 24 h after incubation, there was the dosage-dependent release of IL-8 (inflammatory response) for CuO and ZnO, together with 8-isoprostane release (oxidative stress) for CuO. However, Ag Nps had no side effects on the intestinal microtissues in vitro, as it was displayed earlier (Burduşel et al., 2018). Also, AgNp toxicity effect was examined on a 3D epidermal model and a 2D keratinocyte model (Chen et al., 2019; Crosera et al., 2009). In vitro examination displayed that a similar dosage of AgNps emerged in considerable oxidative damage and inflammation-related cytotoxicity. However, only 2D keratinocyte cultures were affected by Ag Np toxicity (Chen et al., 2019), which can be explained by the abovementioned drawbacks of monolayer culture when compared to 3D systems. This distinction regarding the different toxicological effects on 2D and 3D cultures was also confirmed in placental toxicity research. Accordingly, a recent study utilized scaffold-free hanging drop technology and a 3D coculture MT model similar to in vivo placental tissue (Muoth et al., 2016). Outcomes from this study displayed that secretion levels of human chorionic gonadotropin (hCG) were notably more elevated in 3D when compared to 2D cell cultures (Muoth et al., 2016). Also, it was displayed that cadmium telluride (CdTe) and CuO Nps negatively affected MT viability and hCG secretion (Muoth et al., 2016).
Organoids as Disease Models
There have been numerous studies utilizing organoid cultures to research congenital and acquired human diseases (McGuigan and Sefton, 2006; Spence et al., 2011; Dekkers et al., 2013; Lancaster et al., 2013; Schwank et al., 2013; Zhou et al., 2017; Praharaj et al., 2018). Here, the focus will be only on infectious disease, coronavirus precisely, since the current pandemic situation in regard to SARS-CoV-2 infection requires rapid and thorough observations. In such a manner, the accent of this section will be focused on lung organoids in the model of SARS-CoV-2 infection highlighting, again, a wide range of employment of organoids in research of almost every known pathology. Respectively, the susceptibility to SARS-CoV-2 and its impact on the human lung AECs were examined in different in vitro models. In these studies AOs were derived from various cell types: primary small airway basal cells (Lamers et al., 2020), single adult human alveolar epithelial type II or KRT5+ basal cells (Katsura et al., 2020; Salahudeen et al., 2020), multipotent SOX2+SOX9+ lung bud tip progenitor cells, and either HESCs or iPSCs (Huang et al., 2020). In all of these studies, alveolar epithelial type II-like cells were cultured in 3D as monolayered epithelial spheres or as 2D air–liquid interface cultures, distinguished by apical–basal polarization and barrier integrity (Huang et al., 2020; Katsura et al., 2020; Lamers et al., 2021; Han et al., 2021), or as organoids in long-term feeder-free, chemically defined culture systems (Salahudeen et al., 2020). The expression of ACE2 and TMPRSS2 was equal to the adult stage, with more extensive expression of TMPRSS2 (Huang et al., 2020; Katsura et al., 2020; Salahudeen et al., 2020; Lamers et al., 2021; Han et al., 2021). Experiments showed that SARS-CoV-2 could infect and replicate in alveolar epithelial type II cells grown as either 3D organoids, 3D spheres, or 2D air–liquid interface cultures (Huang et al., 2020; Katsura et al., 2020; Salahudeen et al., 2020; Lamers et al., 2021; Han et al., 2021). All of these cell cultures managed to mirror viral infection and release of infectious virus predominantly from the apical side, following the expression of ACE2 protein. Furthermore, SARS-CoV-2 infection was examined in distal-lung basal cell-derived organoids (Salahudeen et al., 2020; Lamers et al., 2021). The SARS-CoV-2 infection led to pathological and apoptotic effects and a vigorous induction of host antiviral response genes, such as IFN type I and type III, IFN receptors and other interferon-stimulated genes (ISGs) referred to as type I and type III IFN responses, and NF-kB-mediated inflammatory signaling and chemokine signaling pathway (Han et al., 2021; Huang et al., 2020; Katsura et al., 2020; Lamers et al., 2021). There was an upregulation of apoptosis-related genes, while in infected cells certain functions of alveolar epithelial type II cells, such as surfactant gene expression, including DNA replication and cell cycle genes, were downregulated (Katsura et al., 2020). Even though primary cell cultures displayed potent IFN response (Lamers et al., 2020, 2021), alveolar epithelial type II cell organoids and PSC-derived 2D air–liquid interface cultures had a mild response (Han et al., 2021; Huang et al., 2020). Since cigarette smoke is displayed to enhance chances of a severe form of SARS-CoV-2 infection (Adams et al., 2020), the lungs were therefore examined in terms of androgens and cigarette consumption models (Purkayastha et al., 2020; Samuel et al., 2020). In a study of the primary human nonsmoker airway basal stem cell-derived air–liquid interface cultures, exposure to cigarette smoke prior to SARS-CoV-2 infection induced a 2- to 3-fold rise in viral load, increased the number of infected and apoptotic cells, hindered the normal airway basal stem cell repair response, and attenuated IFN response (Purkayastha et al., 2020). Besides lung organoids, intestinal organoids can serve as models for various infections such as coronavirus (which can be propagated in vitro so that the small intestine is an alternate infection route) (Zhou et al., 2017). To understand the tissue tropism of SARS-CoV-2, multiple research groups (Ardestani and Maedler, 2020; Yang et al., 2020; Han et al., 2021) resorted to organoid approaches. Previously, Monteil et al. (2020) demonstrated that SARS-CoV-2 could directly infect capillary organoids and kidney organoids, both derived from hiPSCs (Monteil et al., 2020). These observations may explain the spread of the virus through body and kidney function loss in severely ill individuals (Clevers, 2020).
Conclusion
Rapid improvement of technology and growing urbanization require constant exposure to a plethora of various compounds, many of which are toxic. In such a manner, revealing the exact impact of pollutants on humans is imperative. Even though some studies elaborated precise mechanisms of pollutants via organoids including cytotoxic effects and decrease of cell viability, membrane integrity disruption, upregulation and downregulation of genes involved in cell growth differentiation and homeostasis, and increased levels of ROS, calcium oscillations, etc. (Yang et al., 2013; Kermanizadeh et al., 2014; Ferraz et al., 2016; Forsythe et al., 2018; Harris et al., 2018; Chen et al., 2019; Henson et al., 2019; Kim J. H. et al., 2020; Zeng et al., 2021; van Dijk et al., 2021, the contribution is a preprint; Winkler et al., 2021, the contribution is a preprint), other studies showed no adverse effects, or minimal cellular alterations (George et al., 2019; Di Cristo et al., 2020). Of great importance is consideration of specific features when it comes to organoids—lack of vasculature and immune system cells, cell-to-cell communication, ECM, and natural cell niche, even few, but are detrimental regarding the narrow portrayal of organoids’ reaction to pollutants. Also, the process of humans’ pollutant intake and exposure, even though mimicked to some extent, on the other hand, is not ideal. Despite of all these limitations, organoids will be a breakthrough in pollution research due to closer mirroring of human physiology, dodging of animal killing, comfortable manipulation with various compounds and dosage, excellent possibility to examine even the slightest changes in signal pathways, gene expression, and toxical effects of pollutants by different bioinformatics tools for analyzing omics such as HiPathia or others. Also, a wide range of applications regarding early development research, disease modeling (especially current SARS-CoV-2 virus infection), and testing of the vast majority of drug and toxicants overcome these limitations. In the aftermath, future directions should be aimed at overcoming limitations of 2D and 3D cell cultures, spreading and deepening the research with a much larger group of pollutants and cell types.
Author Contributions
MS devised the idea. DM and DP wrote the manuscript. MP prepared the figure. MS and BLj provided the expert comments and edited the manuscript. MJ, SN, and NM wrote sections of the manuscript. All the authors contributed to the manuscript revision and read and approved the submitted version, which was completed by MS and BLj.
Funding
This study was supported by the Serbian Ministry of Sciences (project number ON 175103), by the Serbian Ministry of Education, Science, and Technological Development [project number 451-03-9/2021-14/200378 (Institute for Information Technologies, University of Kragujevac)], and by the Faculty of Medical Sciences, University of Kragujevac, Serbia (JP 25/19, JP 06/20, and JP 24/20).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Sanja Bojic from the University of Newcastle for the proofread as a native English speaker in our manuscript, and Fransien van Dijk and Anna Winkler who are mentioned in our reference list, for their articles which are still preprints, which helped us to elucidate the effects of environmental pollutants on human organoids.
Abbreviations
ACE-2, angiotensin-converting enzyme 2; AEC, alveolar epithelial cell; AgNps, silver nanoparticles; AgO, silver oxide; AO, alveolar organoid; APOC3, apolipoprotein C3; ASCs, adult stem cells; AST, aspartate aminotransferase; AT2, alveolar type 2 progenitor cells; ATF4, activating transcription factor 4; BMP, bone morphogenetic protein; BRN2/POU3F2, POU class 3 homeobox 2; CASP3, caspase 3; CAT, catalase; CdTe, cadmium telluride; CeO2, cerium oxide; CF, cystic fibrosis; CLDN1, claudin 1; COPD, chronic obstructive pulmonary disease; COX1, cyclo-oxygenase 1; CRC, colorectal cancer; CTIP2/BCL11B, BAF chromatin remodeling complex subunit BCL11B; CuO, copper oxide; DAT, dopamine active transporter; DNAH5, dynein axonemal heavy chain 5; dPM2.5, diesel particulate matter 2.5; EAAC1, excitatory amino-acid carrier 1; EBs, embryoid bodies; ECM, extracellular matrix; ENps, engineered nanoparticles; ESCs, embryonic stem cells; Fbw7, F-box and WD repeat domain-containing 7; FGF, fibroblast growth factor; GABA receptor, gamma-aminobutyric acid receptor; GFAP, glial fibrillary acidic protein; GI, gastrointestinal; GLAST/SLC1A3, solute carrier family 1 member 3; GSH, glutathione; GSTA1 and GPX1, glutathione detox-related genes; hCG, human chorionic gonadotropin; HER, human epidermal growth factor receptor; hEROs, human embryonic stem cell derived retinal organoids; iHIOs, induced human intestinal organoids; iPSCs, induced pluripotent stem cells; ITER, international thermonuclear experimental reactor; Krt19, CK19 cytokeratin 19 protein; KRT5, keratin 5; LGR5, leucine-rich repeat-containing G-protein coupled receptor 5; LUHMES, Lund human mesencephalic cells; MLF1IP, centromere protein U; MPFs, microplastic fibers; MPs, microplastics; MT, microtissue; MWCNT, multiwalled carbon nanotubes; NADP, nicotinamide adenine dinucleotide phosphate; ND1, NADH dehydrogenase 1; NEF2L2, nuclear factor erythroid-derived 2-like 2); Ngn3, neurogenin 3; NKX2.1, NK2 homeobox 1; NM, nanomaterial; NOX2, NADPH oxidase-2; NPHP1, nephrocystin 1; NPs, nanoplastics; Nps, nanoparticles; OrgGloms, organoid-derived glomeruli; PAHs, polycyclic aromatic hydrocarbons; PDGFRA, platelet-derived growth factor receptor alpha; PM2.5, particulate matter 2.5; PSCs, pluripotent stem cells; PSNPs, polystyrene nanoplastics; qRT-PCR, quantitative reverse transcription–polymerase chain reaction; RELN, reelin; RNA-seq, RNA sequencing; ROS, reactive oxygen species; SARS-CoV-2, severe acute respiratory syndrome coronavirus clade 2; SCGB1A1, secretoglobin family 1A member 1; SFTPA1, surfactant protein A1; SFTPC, surfactant protein C; SHH, Sonic hedgehog pathway; SOD1 and SOD2, superoxide dismutase family genes; TEER, transepithelial electrical resistance; TiO2, titanium dioxide; TMRPSS2, cofactor transmembrane protease serine 2; TSCs, trophoblast stem cells; TYMS, thymidylate synthetase; VEGF, vascular endothelial growth factor; VGAT, vesicular GABA transporter; VGLUT2, vesicular glutamate transporters; WES, whole-exome sequencing; WGS, whole-genome sequencing; W-Nps, tungsten nanoparticles; WNT, wingless-related integration site; ZnO, zinc oxide.
References
Abbasi, J. (2018). Patient-derived organoids predict cancer treatment response. JAMA 319:1427. doi: 10.1001/jama.2018.3760
Aboulkheyr Es, H., Montazeri, L., Aref, A. R., Vosough, M., and Baharvand, H. (2018). Personalized cancer medicine: an organoid approach. Trends Biotechnol. 36, 358–371. doi: 10.1016/j.tibtech.2017.12.005
Adams, S. H., Park, M. J., Schaub, J. P., Brindis, C. D., and Irwin, C. E. Jr. (2020). medical vulnerability of young adults to severe COVID-19 illness-data from the national health interview survey. J. Adolesc. Health 67, 362–368. doi: 10.1016/j.jadohealth.2020.06.025
Adhya, D., Annuario, E., Lancaster, M. A., Price, J., Baron-Cohen, S., and Srivastava, D. P. (2018). Understanding the role of steroids in typical and atypical brain development: advantages of using a “brain in a dish” approach. J. Neuroendocrinol. 30, e12547. doi: 10.1111/jne.12547
Ardestani, A., and Maedler, K. (2020). Commentary: a human pluripotent stem cell-based platform to study SARS-CoV-2 tropism and model virus infection in human cells and organoids. Front. Endocrinol. 11:585922. doi: 10.3389/fendo.2020.585922
Bar, S., and Benvenisty, N. (2019). Epigenetic aberrations in human pluripotent stem cells. EMBO J. 38:e101033. doi: 10.15252/embj.2018101033
Bojic, S., Falco, M. M., Stojkovic, P., Ljujic, B., Gazdic Jankovic, M., Armstrong, L., et al. (2020). Platform to study intracellular polystyrene nanoplastic pollution and clinical outcomes. Stem Cells 38, 1321–1325. doi: 10.1002/stem.3244
Bock, C., Boutros, M., Camp, J. G., Clarke, L., Clevers, H., Knoblich, J. A. K., et al. (2020). The Organoid Cell Atlas: A Rosetta Stone for Biomedical Discovery and Regenerative Therapy. Geneva: Zenodo, doi: 10.5281/zenodo.4001718
Bouwmeester, H., Hollman, P. C., and Peters, R. J. (2015). Potential health impact of environmentally released micro- and nanoplastics in the human food production chain: experiences from nanotoxicology. Environ. Sci. Technol. 49, 8932–8947. doi: 10.1021/acs.est.5b01090
Brazovskaja, A., Treutlein, B., and Camp, J. G. (2019). High-throughput single-cell transcriptomics on organoids. Curr. Opin. Biotechnol. 55, 167–171. doi: 10.1016/j.copbio.2018.11.002
Briggs, D. (2003). Environmental pollution and the global burden of disease. Br. Med. Bull. 68, 1–24. doi: 10.1093/bmb/ldg019
Brouwer, M., Zhou, H., and Kasri, N. N. (2016). Choices for induction of pluripotency: recent developments in human induced pluripotent stem cell reprogramming strategies. Stem Cell Rev. Rep. 12, 54–72. doi: 10.1007/s12015-015-9622-8
Burduşel, A. C., Gherasim, O., Grumezescu, A. M., Mogoantă, L., Ficai, A., and Andronescu, E. (2018). Biomedical applications of silver nanoparticles: an up-to-date overview. Nanomaterials 8:681. doi: 10.3390/nano8090681
Calderon-Gierszal, E. L., and Prins, G. S. (2015). Directed differentiation of human embryonic stem cells into prostate organoids in vitro and its perturbation by low-dose bisphenol a exposure. PLoS One 10:e0133238. doi: 10.1371/journal.pone.0133238
Cao, L., Kuratnik, A., Xu, W., Gibson, J. D., Kolling, F. IV, Falcone, E. R., et al. (2015). Development of intestinal organoids as tissue surrogates: cell composition and the epigenetic control of differentiation. Mol. Carcinog. 54, 189–202. doi: 10.1002/mc.22089
Katsura, H., Sontake, V., Tata, A., Kobayashi, Y., Edwards, C. E., Heaton, B. E., et al. (2020). Human lung stem cell-based alveolospheres provide insights into SARS-CoV-2-mediated interferon responses and pneumocyte dysfunction. Cell Stem Cell 27, 890–904.e8. doi: 10.1016/j.stem.2020.10.005
Choi, B. I., Harvey, A. J., and Green, M. P. (2016). Bisphenol A affects early bovine embryo development and metabolism that is negated by an oestrogen receptor inhibitor. Sci. Rep. 6:29318. doi: 10.1038/srep29318
Chen, L., Wu, M., Jiang, S., Zhang, Y., Li, R., Lu, Y., et al. (2019). Skin toxicity assessment of silver nanoparticles in a 3D Epidermal model compared to 2D keratinocytes. Int. J. Nanomed. 14, 9707–9719. doi: 10.2147/IJN.S225451
Clevers, H. (2020). COVID-19: organoids go viral. Nat. Rev. Mol. Cell Biol. 21, 355–356. doi: 10.1038/s41580-020-0258-4
Clevers, H. (2016). Modeling development and disease with organoids. Cell 165, 1586–1597. doi: 10.1016/j.cell.2016.05.082
Corrales, J., Kristofco, L. A., Steele, W. B., Yates, B. S., Breed, C. S., Williams, E. S., et al. (2015). Global assessment of bisphenol a in the environment: review and analysis of its occurrence and bioaccumulation. Dose Response. 13:1559325815598308. doi: 10.1177/1559325815598308
Costamagna, G., Comi, G. P., and Corti, S. (2021). Advancing drug discovery for neurological disorders using ipsc-derived neural organoids. Int. J. Mol. Sci. 22:2659.
Crosera, M., Bovenzi, M., Maina, G., Adami, G., Zanette, C., Florio, C., et al. (2009). Nanoparticle dermal absorption and toxicity: a review of the literature. Int. Arch. Occup. Environ. Health 82, 1043–1055. doi: 10.1007/s00420-009-0458-x
Cugola, F. R., Fernandes, I. R., Russo, F. B., Freitas, B. C., Dias, J. L., Guimarães, K. P., et al. (2016). The Brazilian Zika virus strain causes birth defects in experimental models. Nature 534, 267–271. doi: 10.1038/nature18296
Dedhia, P. H., Bertaux-Skeirik, N., Zavros, Y., and Spence, J. R. (2016). Organoid models of human gastrointestinal development and disease. Gastroenterology 150, 1098–1112. doi: 10.1053/j.gastro.2015.12.042
Deglincerti, A., Croft, G. F., Pietila, L. N., Zernicka-Goetz, M., Siggia, E. D., and Brivanlou, A. H. (2016). Self-organization of the in vitro attached human embryo. Nature 533, 251–254. doi: 10.1038/nature17948
Dekkers, J. F., Wiegerinck, C. L., de Jonge, H. R., Bronsveld, I., Janssens, H. M., de Winter-de Groot, K. M., et al. (2013). A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nat. Med. 19, 939–945. doi: 10.1038/nm.3201
Di Cristo, L., Boccuni, F., Iavicoli, S., and Sabella, S. (2020). A human-relevant 3d in vitro platform for an effective and rapid simulation of workplace exposure to nanoparticles: silica nanoparticles as case study. Nanomaterials 10:1761. doi: 10.3390/nano10091761
Dris, R., Gasperi, J., Mirande, C., Mandin, C., Guerrouache, M., Langlois, V., et al. (2017). A first overview of textile fibers, including microplastics, in indoor and outdoor environments. Environ. Pollut. 221, 453–458. doi: 10.1016/j.envpol.2016.12.013
Eder, A., Vollert, I., Hansen, A., and Eschenhagen, T. (2016). Human engineered heart tissue as a model system for drug testing. Adv. Drug Deliv. Rev. 96, 214–224. doi: 10.1016/j.addr.2015.05.010
Eglen, R. M., and Randle, D. H. (2015). Drug discovery goes three-dimensional: goodbye to flat high-throughput screening? Assay Drug Dev. Technol. 13, 262–265. doi: 10.1089/adt.2015.647
Eiraku, M., and Sasai, Y. (2012). Self-formation of layered neural structures in three-dimensional culture of ES cells. Curr. Opin. Neurobiol. 22, 768–777. doi: 10.1016/j.conb.2012.02.005
Etoc, F., Metzger, J., Ruzo, A., Kirst, C., Yoney, A., Ozair, M. Z., et al. (2016). A balance between secreted inhibitors and edge sensing controls gastruloid self-organization. Dev Cell 39, 302–315. doi: 10.1016/j.devcel.2016.09.016
Evans, M. (2011). Discovering pluripotency: 30 years of mouse embryonic stem cells. Nat. Rev. Mol. Cell Biol. 12, 680–686. doi: 10.1038/nrm3190
Falco, M. M., Chilet, M. P., Loucera, C., Hidalgo, M. R., and Dopazo, J. (2020). Mechanistic models of signaling pathways deconvolute the glioblastoma single-cell functional landscape. NAR Cancer 2:2.
Ferraz, E. R., Rainho, C. R., Fernandes, A. S., and Felzenszwalb, I. (2016). Differential toxicity of an organic PM2.5 extract to human lung cells cultured in three dimensions (3D) and monolayers. J. Toxicol. Environ. Health A 79, 221–231. doi: 10.1080/15287394.2016.1143902
Fleischer, A., Vallejo-Díez, S., Martín-Fernández, J. M., Sánchez-Gilabert, A., Castresana, M., Del Pozo, A., et al. (2020). iPSC-derived intestinal organoids from cystic fibrosis patients acquire CFTR activity upon TALEN-mediated repair of the p.F508del mutation. Mol. Ther. Methods Clin. Dev. 17, 858–870. doi: 10.1016/j.omtm.2020.04.005
Forsythe, S. D., Devarasetty, M., Shupe, T., Bishop, C., Atala, A., Soker, S., et al. (2018). Environmental toxin screening using human-derived 3D Bioengineered liver and cardiac organoids. Front. Public Health 6:103. doi: 10.3389/fpubh.2018.00103
Fröhlich, E. (2013). Cellular targets and mechanisms in the cytotoxic action of non-biodegradable engineered nanoparticles. Curr. Drug Metab. 14, 976–988. doi: 10.2174/1389200211314090004
Garrido-Rodriguez, M., Lopez-Lopez, D., Ortuno, F. M., Peña-Chilet, M., Muñoz, E., Calzado, M. A., et al. (2021). A versatile workflow to integrate RNA-seq genomic and transcriptomic data into mechanistic models of signaling pathways. PLoS Comput. Biol. 17:e1008748. doi: 10.1371/journal.pcbi.1008748
Gendoo, D. M. A. (2020). Bioinformatics and computational approaches for analyzing patient-derived disease models in cancer research. Comput. Struct. Biotechnol. J. 18, 375–380. doi: 10.1016/j.csbj.2020.01.010
George, I., Uboldi, C., Bernard, E., Sobrido, M. S., Dine, S., Hagège, A., et al. (2019). Toxicological assessment of ITER-Like tungsten nanoparticles using an in vitro 3D human airway epithelium model. Nanomaterials 9:1374. doi: 10.3390/nano9101374
Greggio, C., De Franceschi, F., Figueiredo-Larsen, M., Gobaa, S., Ranga, A., Semb, H., et al. (2013). Artificial three-dimensional niches deconstruct pancreas development in vitro. Developmen. 140, 4452–4462. doi: 10.1242/dev.096628
Gu, J., Wensing, M., Uhde, E., and Salthammer, T. (2019). Characterization of particulate and gaseous pollutants emitted during operation of a desktop 3D printer. Environ. Int. 123, 476–485.
Hale, L. J., Howden, S. E., Phipson, B., Lonsdale, A., Er, P. X., Ghobrial, I., et al. (2018). 3D organoid-derived human glomeruli for personalised podocyte disease modelling and drug screening. Nat. Commun. 9:5167. doi: 10.1038/s41467-018-07594-z
Han, Y., Duan, X., Yang, L., Nilsson-Payant, B. E., Wang, P., Duan, F., et al. (2021). Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature 589, 270–275. doi: 10.1038/s41586-020-2901-9
Harris, G., Eschment, M., Orozco, S. P., McCaffery, J. M., Maclennan, R., Severin, D., et al. (2018). Toxicity, recovery, and resilience in a 3D dopaminergic neuronal in vitro model exposed to rotenone. Arch. Toxicol. 92, 2587–2606. doi: 10.1007/s00204-018-2250-8
Henry, B., Laitala, K., and Klepp, I. G. (2019). Microfibres from apparel and home textiles: prospects for including microplastics in environmental sustainability assessment. Sci. Total Environ. 652, 483–494.
Henson, T. E., Navratilova, J., Tennant, A. H., Bradham, K. D., Rogers, K. R., and Hughes, M. F. (2019). In vitro intestinal toxicity of copper oxide nanoparticles in rat and human cell models. Nanotoxicology 13, 795–811. doi: 10.1080/17435390.2019.1578428
Heo, I., Dutta, D., Schaefer, D. A., Iakobachvili, N., Artegiani, B., Sachs, N., et al. (2018). Modelling Cryptosporidium infection in human small intestinal and lung organoids. Nat. Microbiol. 3, 814–823. doi: 10.1038/s41564-018-0177-8
Hidalgo, M. R., Cubuk, C., Amadoz, A., Salavert, F., Carbonell-Caballero, J., and Dopazo, J. (2017). High throughput estimation of functional cell activities reveals disease mechanisms and predicts relevant clinical outcomes. Oncotarget 8, 5160–5178. doi: 10.18632/oncotarget.14107
Ho, B. X., Pek, N. M. Q., and Soh, B. S. (2018). Disease modeling using 3D organoids derived from human induced pluripotent stem cells. Int. J. Mol. Sci. 19:936. doi: 10.3390/ijms19040936
Hoffmann, M., Kleine-Weber, H., Schroeder, S., Krüger, N., Herrler, T., Erichsen, S., et al. (2020). SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181, 271–280.e8. doi: 10.1016/j.cell.2020.02.052
Huang, J., Hume, A. J., Abo, K. M., Werder, R. B., Villacorta-Martin, C., Alysandratos, K. D., et al. (2020). SARS-CoV-2 infection of pluripotent stem cell-derived human lung alveolar type 2 cells elicits a rapid epithelial-intrinsic inflammatory response. Cell Stem Cell 27, 962–973.e7. doi: 10.1016/j.stem.2020.09.013
Jabs, J., Zickgraf, F. M., Park, J., Wagner, S., Jiang, X., Jechow, K., et al. (2017). Screening drug effects in patient-derived cancer cells links organoid responses to genome alterations. Mol. Syst. Biol. 13:955. doi: 10.15252/msb.20177697
de Jaime-Soguero, A., Abreu de Oliveira, W. A., and Lluis, F. (2018). The pleiotropic effects of the canonical wnt pathway in early development and pluripotency. Genes 9:93. doi: 10.3390/genes9020093
Katsuda, T., Kawamata, M., Hagiwara, K., Takahashi, R. U., Yamamoto, Y., Camargo, F. D., et al. (2017). Conversion of terminally committed hepatocytes to culturable bipotent progenitor cells with regenerative capacity. Cell Stem Cell 20, 41–55. doi: 10.1016/j.stem.2016.10.007
Kavlock, R., Chandler, K., Houck, K., Hunter, S., Judson, R., Kleinstreuer, N., et al. (2012). Update on EPA’s ToxCast program: providing high throughput decision support tools for chemical risk management. Chem. Res. Toxicol. 25, 1287–1302. doi: 10.1021/tx3000939
Kermanizadeh, A., Berthing, T., Guzniczak, E., Wheeldon, M., Whyte, G., Vogel, U., et al. (2019). Assessment of nanomaterial-induced hepatotoxicity using a 3D human primary multi-cellular microtissue exposed repeatedly over 21 days - the suitability of the in vitro system as an in vivo surrogate. Part Fibre Toxicol. 16:42. doi: 10.1186/s12989-019-0326-0
Kermanizadeh, A., Løhr, M., Roursgaard, M., Messner, S., Gunness, P., Kelm, J. M., et al. (2014). Hepatic toxicology following single and multiple exposure of engineered nanomaterials utilising a novel primary human 3D liver microtissue model. Part Fibre Toxicol. 11:56. doi: 10.1186/s12989-014-0056-2
Kicheva, A., and Briscoe, J. (2015). Developmental pattern formation in phases. Trends Cell Biol. 25, 579–591. doi: 10.1016/j.tcb.2015.07.006
Kim, J., Koo, B. K., and Knoblich, J. A. (2020). Human organoids: model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 21, 571–584. doi: 10.1038/s41580-020-0259-3
Kim, J. H., Kim, J., Kim, W. J., Choi, Y. H., Yang, S. R., and Hong, S. H. (2020). Diesel particulate matter 2.5 induces epithelial-to-mesenchymal transition and upregulation of SARS-CoV-2 receptor during human pluripotent stem cell-derived alveolar organoid development. Int. J. Environ. Res. Public Health 17:8410. doi: 10.3390/ijerph17228410
Koehler, K. R., Nie, J., Longworth-Mills, E., Liu, X. P., Lee, J., Holt, J. R., et al. (2017). Generation of inner ear organoids containing functional hair cells from human pluripotent stem cells. Nat. Biotechnol. 35, 583–589. doi: 10.1038/nbt.3840
Koike, H., Iwasawa, K., Ouchi, R., Maezawa, M., Giesbrecht, K., Saiki, N., et al. (2019). Modelling human hepato-biliary-pancreatic organogenesis from the foregut-midgut boundary. Nature 574, 112–116. doi: 10.1038/s41586-019-1598-0
Kondo, J., and Inoue, M. (2019). Application of cancer organoid model for drug screening and personalized therapy. Cells 8:470. doi: 10.3390/cells8050470
Kostadinova, R., Boess, F., Applegate, D., Suter, L., Weiser, T., Singer, T., et al. (2013). A long-term three dimensional liver co-culture system for improved prediction of clinically relevant drug-induced hepatotoxicity. Toxicol. Appl. Pharmacol. 268, 1–16. doi: 10.1016/j.taap.2013.01.012
Lamers, M. M., Beumer, J., van der Vaart, J., Knoops, K., Puschhof, J., Breugem, T. I., et al. (2020). SARS-CoV-2 Productively Infects Human Gut Enterocytes. bioRxiv [Preprint] doi: 10.1101/2020.04.25.060350
Lamers, M. M., van der Vaart, J., Knoops, K., Riesebosch, S., Breugem, T. I., Mykytyn, A. Z., et al. (2021). An organoid-derived bronchioalveolar model for SARS-CoV-2 infection of human alveolar type II-like cells. EMBO J. 40, e105912. doi: 10.15252/embj.2020105912
Lancaster, M. A., and Knoblich, J. A. (2014). Organogenesis in a dish: modeling development and disease using organoid technologies. Science 345:1247125. doi: 10.1126/science.1247125
Lancaster, M. A., Renner, M., Martin, C. A., Wenzel, D., Bicknell, L. S., Hurles, M. E., et al. (2013). Cerebral organoids model human brain development and microcephaly. Nature. 501, 373–379. doi: 10.1038/nature12517
Lee, S. H., Hu, W., Matulay, J. T., Silva, M. V., Owczarek, T. B., Kim, K., et al. (2018). Tumor evolution and drug response in patient-derived organoid models of bladder cancer. Cell 173, 515–528.e17. doi: 10.1016/j.cell.2018.03.017
Lee, S., Bochanis, A., and Rasmussen, T. P. (2020). “Chapter 7 – pluripotent cells for the assessment of chemically induced teratogenesis and developmental toxicology,” in Engineering Strategies for Regenerative Medicine, eds T. G. Fernandes, M. M. Diogo, and J. M. S. Cabral (Cambridge MA: Academic Press), 243–258. doi: 10.1016/B978-0-12-816221-7.00007-0
Lee, S. E., Shin, N., Kook, M. G., Kong, D., Kim, N. G., Choi, S. W., et al. (2020). Human iNSC-derived brain organoid model of lysosomal storage disorder in Niemann-Pick disease type C. Cell Death Dis. 11:1059. doi: 10.1038/s41419-020-03262-7
Lehmann, R., Lee, C. M., Shugart, E. C., Benedetti, M., Charo, R. A., Gartner, Z., et al. (2019). Human organoids: a new dimension in cell biology. Mol. Biol. Cell. 30, 1129–1137. doi: 10.1091/mbc.E19-03-0135
Li-Juan, F., Li, J.-W., Xu, E. G., Sun, X.-D., Zhu, F.-P., Ding, Z., et al. (2019). Short-term exposure to positively charged polystyrene nanoparticles causes oxidative stress and membrane destruction in cyanobacteria. R. Soc. Chem. 6, 3072–3079. doi: 10.1039/c9en00807a
Li, P. J., Roose, J. P., Jablons, D. M., and Kratz, J. R. (2021). Bioinformatic approaches to validation and functional analysis of 3D lung cancer models. Cancers 13:701. doi: 10.3390/cancers13040701
Li, R., Zhou, R., and Zhang, J. (2018). Function of PM2.5 in the pathogenesis of lung cancer and chronic airway inflammatory diseases. Oncol. Lett. 15, 7506–7514. doi: 10.3892/ol.2018.8355
Lindoso, R. S., Kasai-Brunswick, T. H., Monnerat Cahli, G., Collino, F., Bastos Carvalho, A., Campos de Carvalho, A. C., et al. (2019). Proteomics in the world of induced pluripotent stem cells. Cells 8:703. doi: 10.3390/cells8070703
Liu, X., Tan, J. P., Schröder, J., Aberkane, A., Ouyang, J. F., Mohenska, M., et al. (2021). Modelling human blastocysts by reprogramming fibroblasts into iBlastoids. Nature 591, 627–632. doi: 10.1038/s41586-021-03372-y
Manisalidis, I., Stavropoulou, E., Stavropoulos, A., and Bezirtzoglou, E. (2020). Environmental and health impacts of air pollution: a review. Front. Public Health 8:14. doi: 10.3389/fpubh.2020.00014
Markus, J., Landry, T., Stevens, Z., Scott, H., Llanos, P., Debatis, M., et al. (2021). Human small intestinal organotypic culture model for drug permeation, inflammation, and toxicity assays. Vitro Cell Dev. Biol. Anim. 57, 160–173. doi: 10.1007/s11626-020-00526-6
McCauley, H. A., and Wells, J. M. (2017). Pluripotent stem cell-derived organoids: using principles of developmental biology to grow human tissues in a dish. Development 144, 958–962. doi: 10.1242/dev.140731
McCracken, K. W., Howell, J. C., Wells, J. M., and Spence, J. R. (2011). Generating human intestinal tissue from pluripotent stem cells in vitro. Nat. Protoc. 6, 1920–1928. doi: 10.1038/nprot.2011.410 (for the record this manuscript was available in PMC 2014 Jan 20, but published in 2011).
McGuigan, A. P., and Sefton, M. V. (2006). Vascularized organoid engineered by modular assembly enables blood perfusion. Proc. Natl. Acad. Sci. U.S.A. 103, 11461–11466. doi: 10.1073/pnas.0602740103
Mincarelli, L., Lister, A., Lipscombe, J., and Macaulay, I. C. (2018). Defining cell identity with single-cell omics. Proteomics 18:e1700312. doi: 10.1002/pmic.201700312
Mondrinos, M. J., Jones, P. L., Finck, C. M., and Lelkes, P. I. (2014). Engineering de novo assembly of fetal pulmonary organoids. Tissue Eng. Part A 20, 2892–2907. doi: 10.1089/ten.TEA.2014.0085
Monteil, V., Kwon, H., Prado, P., Hagelkrüys, A., Wimmer, R. A., Stahl, M., et al. (2020). Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell 181, 905–913.e7. doi: 10.1016/j.cell.2020.04.004
Munsie, M., Hyun, I., and Sugarman, J. (2017). Ethical issues in human organoid and gastruloid research. Development 144, 942–945. doi: 10.1242/dev.140111
Muoth, C., Wichser, A., Monopoli, M., Correia, M., Ehrlich, N., Loeschner, K., et al. (2016). A 3D co-culture microtissue model of the human placenta for nanotoxicity assessment. Nanoscale 8, 17322–17332. doi: 10.1039/c6nr06749b
Nakano, T., Ando, S., Takata, N., Kawada, M., Muguruma, K., Sekiguchi, K., et al. (2012). Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell. 10, 771–785. doi: 10.1016/j.stem.2012.05.009
Narsinh, K. H., Plews, J., and Wu, J. C. (2011). Comparison of human induced pluripotent and embryonic stem cells: fraternal or identical twins? Mol. Ther. 19, 635–638. doi: 10.1038/mt.2011.41
Nascimento, J. M., Saia-Cereda, V. M., Sartore, R. C., da Costa, R. M., Schitine, C. S., Freitas, H. R., et al. (2019). Human cerebral organoids and fetal brain tissue share proteomic similarities. Front. Cell Dev. Biol. 7:303. doi: 10.3389/fcell.2019.00303
Neef, S. K., Janssen, N., Winter, S., Wallisch, S. K., Hofmann, U., Dahlke, M. H., et al. (2020). Metabolic drug response phenotyping in colorectal cancer organoids by LC-QTOF-MS. Metabolites 10:494. doi: 10.3390/metabo10120494
Nicolas, J., Magli, S., Rabbachin, L., Sampaolesi, S., Nicotra, F., and Russo, L. (2020). 3D extracellular matrix mimics: fundamental concepts and role of materials chemistry to influence stem cell fate. Biomacromolecules 21, 1968–1994. doi: 10.1021/acs.biomac.0c00045
Pamies, D., Barreras, P., Block, K., Makri, G., Kumar, A., Wiersma, D., et al. (2017). A human brain microphysiological system derived from induced pluripotent stem cells to study neurological diseases and toxicity. ALTEX 34, 362–376.
Peña-Chilet, M., Esteban-Medina, M., Falco, M. M., Rian, K., Hidalgo, M. R., Loucera, C., et al. (2019). Using mechanistic models for the clinical interpretation of complex genomic variation. Sci. Rep. 9:18937. doi: 10.1038/s41598-019-55454-7
Praharaj, P. P., Bhutia, S. K., Nagrath, S., Bitting, R. L., and Deep, G. (2018). Circulating tumor cell-derived organoids: Current challenges and promises in medical research and precision medicine. Biochim. Biophys. Acta Rev. Cancer 1869, 117–127. doi: 10.1016/j.bbcan.2017.12.005
Prata, J. C. (2018). Airborne microplastics: Consequences to human health? Environ Pollut. 234, 115–126. doi: 10.1016/j.envpol.2017.11.043
Prata, J. C., da Costa, J. P., Lopes, I., Duarte, A. C., and Rocha-Santos, T. (2020). Environmental exposure to microplastics: An overview on possible human health effects. Sci. Total Environ. 702:134455. doi: 10.1016/j.scitotenv.2019.134455
Reid, J. A., Mollica, P. A., Bruno, R. D., and Sachs, P. C. (2018). Consistent and reproducible cultures of large-scale 3D mammary epithelial structures using an accessible bioprinting platform. Breast Cancer Res. 20:122. doi: 10.1186/s13058-018-1045-4
Richardson, J. R., Fitsanakis, V., Westerink, R. H. S., and Kanthasamy, A. G. (2019). Neurotoxicity of pesticides. Acta Neuropathol. 138, 343–362. doi: 10.1007/s00401-019-02033-9
Purkayastha, A., Sen, C., Garcia, G. Jr., Langerman, J., Shia, D. W., Meneses, L. K., et al. (2020). Direct exposure to SARS-CoV-2 and cigarette smoke increases infection severity and alters the stem cell-derived airway repair response. Cell Stem Cell 27, 869–875.e4. doi: 10.1016/j.stem.2020.11.010
Rivron, N. C., Frias-Aldeguer, J., Vrij, E. J., Boisset, J. C., Korving, J., Vivié, J., et al. (2018). Blastocyst-like structures generated solely from stem cells. Nature 557, 106–111. doi: 10.1038/s41586-018-0051-0
Rosca, A., Coronel, R., Moreno, M., González, R., Oniga, A., Martín, A., et al. (2020). Impact of environmental neurotoxic: current methods and usefulness of human stem cells. Heliyon 6:e05773. doi: 10.1016/j.heliyon.2020.e05773
Salahudeen, A. A., Choi, S. S., Rustagi, A., Zhu, J., van Unen, V., de la O, S. M., et al. (2020). Progenitor identification and SARS-CoV-2 infection in human distal lung organoids. Nature 588, 670–675.
Samuel, R. M., Majd, H., Richter, M. N., Ghazizadeh, Z., Zekavat, S. M., Navickas, A., et al. (2020). Androgen signaling regulates SARS-CoV-2 receptor levels and is associated with severe COVID-19 symptoms in men. Cell Stem Cell. 27, 876–889.e12. doi: 10.1016/j.stem.2020.11.009
Sancho, R., Gruber, R., Gu, G., and Behrens, A. (2014). Loss of Fbw7 reprograms adult pancreatic ductal cells into α, δ, and β cells. Cell Stem Cell. 15, 139–153. doi: 10.1016/j.stem.2014.06.019
Schwank, G., Koo, B. K., Sasselli, V., Dekkers, J. F., Heo, I., Demircan, T., et al. (2013). Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 13, 653–658. doi: 10.1016/j.stem.2013.11.002
Schwartz, M. P., Hou, Z., Propson, N. E., Zhang, J., Engstrom, C. J., Santos Costa, V., et al. (2015). Human pluripotent stem cell-derived neural constructs for predicting neural toxicity. Proc. Natl. Acad. Sci. U.S.A. 112, 12516–12521. doi: 10.1073/pnas.1516645112
Shahbazi, M. N., Jedrusik, A., Vuoristo, S., Recher, G., Hupalowska, A., Bolton, V., et al. (2016). Self-organization of the human embryo in the absence of maternal tissues. Nat. Cell Biol. 18, 700–708. doi: 10.1038/ncb3347
Simunovic, M., and Brivanlou, A. H. (2017). Embryoids, organoids and gastruloids: new approaches to understanding embryogenesis. Development 144, 976–985. doi: 10.1242/dev.143529
Sirenko, O., Parham, F., Dea, S., Sodhi, N., Biesmans, S., Mora-Castilla, S., et al. (2019). Functional and mechanistic neurotoxicity profiling using human ipsc-derived neural 3D cultures. Toxicol. Sci. 167, 58–76. doi: 10.1093/toxsci/kfy218
Song, Y., Xu, X., Wang, W., Tian, T., Zhu, Z., and Yang, C. (2019). Single cell transcriptomics: moving towards multi-omics. Analyst 144, 3172–3189. doi: 10.1039/c8an01852a
Spence, J. R., Mayhew, C. N., Rankin, S. A., Kuhar, M. F., Vallance, J. E., Tolle, K., et al. (2011). Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 470, 105–109. doi: 10.1038/nature09691
Stojkovic, M., Han, D., Jeong, M., Stojkovic, P., and Stankovic, K. M. (2021a). Human induced pluripotent stem cells and CRISPR/Cas-mediated targeted genome editing: Platforms to tackle sensorineural hearing loss. Stem Cells 39, 673–696. doi: 10.1002/stem.3353
Stojkovic, M., Stojkovic, P., and Stankovic, K. M. (2021b). Human pluripotent stem cells - Unique tools to decipher the effects of environmental and intracellular plastic pollution on human health. Environ. Pollut. 269:116144. doi: 10.1016/j.envpol.2020.116144
Takasato, M., Er, P. X., Chiu, H. S., Maier, B., Baillie, G. J., Ferguson, C., et al. (2015). Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature. 526, 564–568. doi: 10.1038/nature15695
Thit, A., Selck, H., and Bjerregaard, H. F. (2015). Toxic mechanisms of copper oxide nanoparticles in epithelial kidney cells. Toxicol. Vitro 29, 1053–1059. doi: 10.1016/j.tiv.2015.03.020
Takebe, T., Sekine, K., Enomura, M., Koike, H., Kimura, M., Ogaeri, T., et al. (2013). Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature 499, 481–484. doi: 10.1038/nature12271
ten Berge, D., Koole, W., Fuerer, C., Fish, M., Eroglu, E., and Nusse, R. (2008). Wnt signaling mediates self-organization and axis formation in embryoid bodies. Cell Stem Cell 3, 508–518. doi: 10.1016/j.stem.2008.09.013
Toussaint, B., Raffael, B., Angers-Loustau, A., Gilliland, D., Kestens, V., Petrillo, M., et al. (2019). Review of micro- and nanoplastic contamination in the food chain. Food Addit. Contam Part A Chem. Anal. Control Expo. Risk Assess. 36, 639–673. doi: 10.1080/19440049.2019.1583381
Truskey, G. A. (2018). Human microphysiological systems and organoids as in vitro models for toxicological studies. Front. Public Health 6:185. doi: 10.3389/fpubh.2018.00185
Turner, D. A., Baillie-Johnson, P., and Martinez Arias, A. (2016). Organoids and the genetically encoded self-assembly of embryonic stem cells. Bioessays 38, 181–191. doi: 10.1002/bies.201500111
van de Wetering, M., Francies, H. E., Francis, J. M., Bounova, G., Iorio, F., et al. (2015). Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161, 933–945. doi: 10.1016/j.cell.2015.03.053
van Dijk, F., Song, S., van Eck, G. W. A., Wu, X., Bos, I. S. T., Boom, D. H. A., et al. (2021). Inhalable textile microplastic fibers impair airway epithelial growth. bioRxiv [Preprint] doi: 10.1101/2021.01.25.428144
Vendrely, C. (1950). Contribution à l’etude cytochimique des acides nucléiques de quelques organites cellulaires [Contribution to the cytochemical study of nucleic acids of some cell organoids]. Arch. Anat. Histol. Embryol. 33, 113–155.
Vlachogiannis, G., Hedayat, S., Vatsiou, A., Jamin, Y., Fernández-Mateos, J., Khan, K., et al. (2018). Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 359, 920–926. doi: 10.1126/science.aao2774
Walther, T. C., and Mann, M. (2010). Mass spectrometry-based proteomics in cell biology. J Cell Biol. 190, 491–500. doi: 10.1083/jcb.201004052
Wang, X., Klingan, K., Klingenhof, M., Möller, T., Ferreira de Araújo, J., et al. (2021). Morphology and mechanism of highly selective Cu(II) oxide nanosheet catalysts for carbon dioxide electroreduction. Nat. Commun. 12:794. doi: 10.1038/s41467-021-20961-7
Wang, Y., Zi, X. Y., Su, J., Zhang, H. X., Zhang, X. R., Zhu, H. Y., et al. (2012). Cuprous oxide nanoparticles selectively induce apoptosis of tumor cells. Int. J. Nanomedicine. 7, 2641–2652. doi: 10.2147/IJN.S31133
Wang, Z., Li, N., Zhao, J., White, J. C., Qu, P., and Xing, B. (2012). CuO nanoparticle interaction with human epithelial cells: cellular uptake, location, export, and genotoxicity. Chem. Res. Toxicol. 25, 1512–1521. doi: 10.1021/tx3002093
Winkler, A., Santo, N., Madaschi, L., Cherubini, A., Rusconi, F., Rosso, L., et al. (2021). Lung organoids and microplastic fibers: a new exposure model for emerging contaminants. bioRxiv [Preprint] doi: 10.1101/2021.03.07.434247
Wishart, D. S. (2019). Metabolomics for investigating physiological and pathophysiological processes. Physiol. Rev. 99, 1819–1875. doi: 10.1152/physrev.00035.2018
Wong, A. P., Bear, C. E., Chin, S., Pasceri, P., Thompson, T. O., Huan, L. J., et al. (2012). Directed differentiation of human pluripotent stem cells into mature airway epithelia expressing functional CFTR protein. Nat. Biotechnol. 30, 876–882. doi: 10.1038/nbt.2328
Wu, H., Uchimura, K., Donnelly, E. L., Kirita, Y., Morris, S. A., and Humphreys, B. D. (2018). Comparative analysis and refinement of human PSC-derived kidney organoid differentiation with single-cell transcriptomics. Cell Stem Cell 23, 869–881.e8. doi: 10.1016/j.stem.2018.10.010
Xiang, L., Yin, Y., Zheng, Y., Ma, Y., Li, Y., Zhao, Z., et al. (2020). A developmental landscape of 3D-cultured human pre-gastrulation embryos. Nature 577, 537–542. doi: 10.1038/s41586-019-1875-y
Yang, L., Han, Y., Nilsson-Payant, B. E., Gupta, V., Wang, P., Duan, X., et al. (2020). A human pluripotent stem cell-based platform to study SARS-CoV-2 tropism and model virus infection in human cells and organoids. Cell Stem Cell. 27, 125–136.e7. doi: 10.1016/j.stem.2020.06.015
Yang, L., Luo, L., Ji, W., Gong, C., Wu, D., Huang, H., et al. (2013). Effect of low dose bisphenol A on the early differentiation of human embryonic stem cells into mammary epithelial cells. Toxicol. Lett. 218, 187–193. doi: 10.1016/j.toxlet.2013.01.026
Yan, L., Yang, M., Guo, H., Yang, L., Wu, J., Li, R., et al. (2013). Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 20, 1131–1139. doi: 10.1038/nsmb.2660
Yin, X., Mead, B. E., Safaee, H., Langer, R., Karp, J. M., and Levy, O. (2016). Engineering stem cell organoids. Cell Stem Cell. 18, 25–38. doi: 10.1016/j.stem.2015.12.005
Yu, L., Wei, Y., Duan, J., Schmitz, D. A., Sakurai, M., Wang, L., et al. (2021). Blastocyst-like structures generated from human pluripotent stem cells. Nature. 591, 620–626. doi: 10.1038/s41586-021-03356-y
Zeng, Y., Li, M., Zou, T., Chen, X., Li, Q., Li, Y., et al. (2021). the impact of particulate matter (PM2.5) on human retinal development in hESC-derived retinal organoids. Front. Cell Dev. Biol. 9:607341. doi: 10.3389/fcell.2021.607341
Zheng, Y., and Fu, J. (2021). First complete model of the human embryo. Nature 591, 531–532. doi: 10.1038/d41586-021-00581-3
Zhou, J., Li, C., Zhao, G., Chu, H., Wang, D., Hoi-Ning Yan, H., et al. (2017). Human intestinal tract serves as an alternative infection route for Middle East respiratory syndrome coronavirus. Sci. Adv. 3:eaao4966. doi: 10.1126/sciadv.aao4966
Keywords: organoids, early development, model disease, environmental pollution, drug screening, bioinformatics
Citation: Miloradovic D, Pavlovic D, Jankovic MG, Nikolic S, Papic M, Milivojevic N, Stojkovic M and Ljujic B (2021) Human Embryos, Induced Pluripotent Stem Cells, and Organoids: Models to Assess the Effects of Environmental Plastic Pollution. Front. Cell Dev. Biol. 9:709183. doi: 10.3389/fcell.2021.709183
Received: 13 May 2021; Accepted: 19 July 2021;
Published: 03 September 2021.
Edited by:
Sofia Avnet, University of Bologna, ItalyReviewed by:
Federica Sangiuolo, University of Rome Tor Vergata, ItalyNicole Prior, University of Southampton, United Kingdom
Copyright © 2021 Miloradovic, Pavlovic, Jankovic, Nikolic, Papic, Milivojevic, Stojkovic and Ljujic. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Biljana Ljujic, YmxqdWppYzc0QGdtYWlsLmNvbQ==
†These authors have contributed equally to this work and share first authorship
‡Present address: Miodrag Stojkovic, Eaton Peabody Laboratories and Department of Otolaryngology, Massachusetts Eye and Ear, Boston, MA, United States; Department of Head and Neck Surgery, Harvard Medical School, Boston, MA, United States
 Dragana Miloradovic
Dragana Miloradovic Dragica Pavlovic
Dragica Pavlovic Marina Gazdic Jankovic
Marina Gazdic Jankovic Sandra Nikolic
Sandra Nikolic Milos Papic
Milos Papic Nevena Milivojevic
Nevena Milivojevic Miodrag Stojkovic
Miodrag Stojkovic Biljana Ljujic
Biljana Ljujic