- 1Department of Biomedical Science and Bateson Centre, The University of Sheffield, Sheffield, England
- 2SmartImmune Ltd., Cambridge, United Kingdom
Epithelial tissues rely on the adhesion between participating cells to retain their integrity. The transmembrane protein E-cadherin is the major protein that mediates homophilic adhesion between neighbouring cells and is, therefore, one of the critical components for epithelial integrity. E-cadherin downregulation has been described extensively as a prerequisite for epithelial-to-mesenchymal transition and is a hallmark in many types of cancer. Due to this clinical importance, research has been mostly focused on understanding the mechanisms leading to transcriptional repression of this adhesion molecule. However, in recent years it has become apparent that re-expression of E-cadherin is a major step in the progression of many cancers during metastasis. Here, we review the currently known molecular mechanisms of E-cadherin transcriptional activation and inhibition and highlight complex interactions between individual mechanisms. We then propose an additional mechanism, whereby the competition between adhesion complexes and heterochromatin protein-1 for binding to STAT92E fine-tunes the levels of E-cadherin expression in Drosophila but also regulates other genes promoting epithelial robustness. We base our hypothesis on both existing literature and our experimental evidence and suggest that such feedback between the cell surface and the nucleus presents a powerful paradigm for epithelial resilience.
Introduction
Cells in epithelia adhere to both a common substrate and neighbouring cells (Hagios et al., 1998). The molecules mediating this adhesion (cell adhesion molecules, CAMs) are the driving force behind the tissue architecture (Gumbiner, 1996; Buckley et al., 1998; Hagios et al., 1998; Schock and Perrimon, 2002), whereas adhesion defects often occur during tumour formation and metastasis (Zetter, 1993; Janiszewska et al., 2020). Among CAMs, the calcium-dependent adhesion (cadherin) proteins mediate the direct cell–cell adhesion through homophilic binding of extracellular domains (Perez and Nelson, 2004; Nishiguchi et al., 2016). In this perspective, we focus on Epithelial cadherin (E-cadherin, E-cad), as in epithelial cells, it plays a major role in tissue formation and maintenance (Takeichi, 1977; Halbleib and Nelson, 2006; Nelson, 2008; Kaszak et al., 2020). In contrast, other cadherins such as P-cadherin do also contribute to cell–cell adhesion in epithelia but their actions are restricted to specific areas and developmental stages (Paredes et al., 2012).
The strength of adhesion correlates with the number of E-cad molecules on cell surfaces (Chu et al., 2004). Consequently, the regulation of E-cad surface levels in epithelia is instrumental in many processes, and even mild changes in E-cad levels profoundly affect many processes such as cell rearrangements, proliferation, and tissue architecture (Ciesiolka et al., 2004; Lecuit and Yap, 2015; Mohan et al., 2018; Greig and Bulgakova, 2020). Multiple mechanisms regulate E-cad surface levels including intracellular trafficking, transcriptional regulation, post-translational modifications, and protein degradation (Palacios et al., 2005; Bertocchi et al., 2012; Serrano-Gomez et al., 2016; Brüser and Bogdan, 2017; Cai et al., 2018). In this perspective, we discuss transcriptional regulation, as the decrease in E-cad protein levels often correlates with reduced mRNA abundance (Bringuier et al., 1999).
Transcriptional Repression
The best-studied regulation of E-cad transcription in mammalian cells is silencing by transcription factors (TFs) including SNAIL, SLUG (also known as SNAI1 and SNAI2 in mammals), ZEB1/2, and Twist1/2 (Figure 1A), all of which directly bind conserved E-boxes (CANNTG sequences) in the E-cad promoter (Giroldi et al., 1997; Comijn et al., 2001; Bolós et al., 2003; Wong et al., 2014; Vergara et al., 2016; Russell and Pranjol, 2018). All events of epithelial-to-mesenchymal transition (EMT) during development involve at least one of these TFs, and they were all linked to the tumour progression (Gasparotto et al., 2011; Muenst et al., 2013; da Silva et al., 2014; Vergara et al., 2016; Yang et al., 2020). Despite the overall similarity in the action of these TFs, their different affinities and variable expression allow for a dynamic but tightly regulated expression of E-cad (Bolós et al., 2003; Eger et al., 2005; Schmidt et al., 2005; Vesuna et al., 2008; Mazda et al., 2011). Regulation of E-cad transcription by these TFs is extremely conserved across evolution. While Drosophila and mammalian E-cad differ in their extracellular domains and are products of independent evolution from a common N-cadherin-like ancestor, transcription of the Drosophila E-cad (encoded by the shotgun gene) is also inhibited by the Snail and Twist TFs (Figure 1B; Oda et al., 1998, 2005; Oda and Takeichi, 2011; Nishiguchi et al., 2016). This could be either due to this regulation already being present in the ancestor or a result of the parallel evolution due to the same functional requirements. In Drosophila, while Snail represses E-cad during gastrulation in the embryo, it does not inhibit it in the adult midguts (Oda et al., 1998; Campbell et al., 2019), potentially due to the combinatorial action of other regulators (Figure 1B).
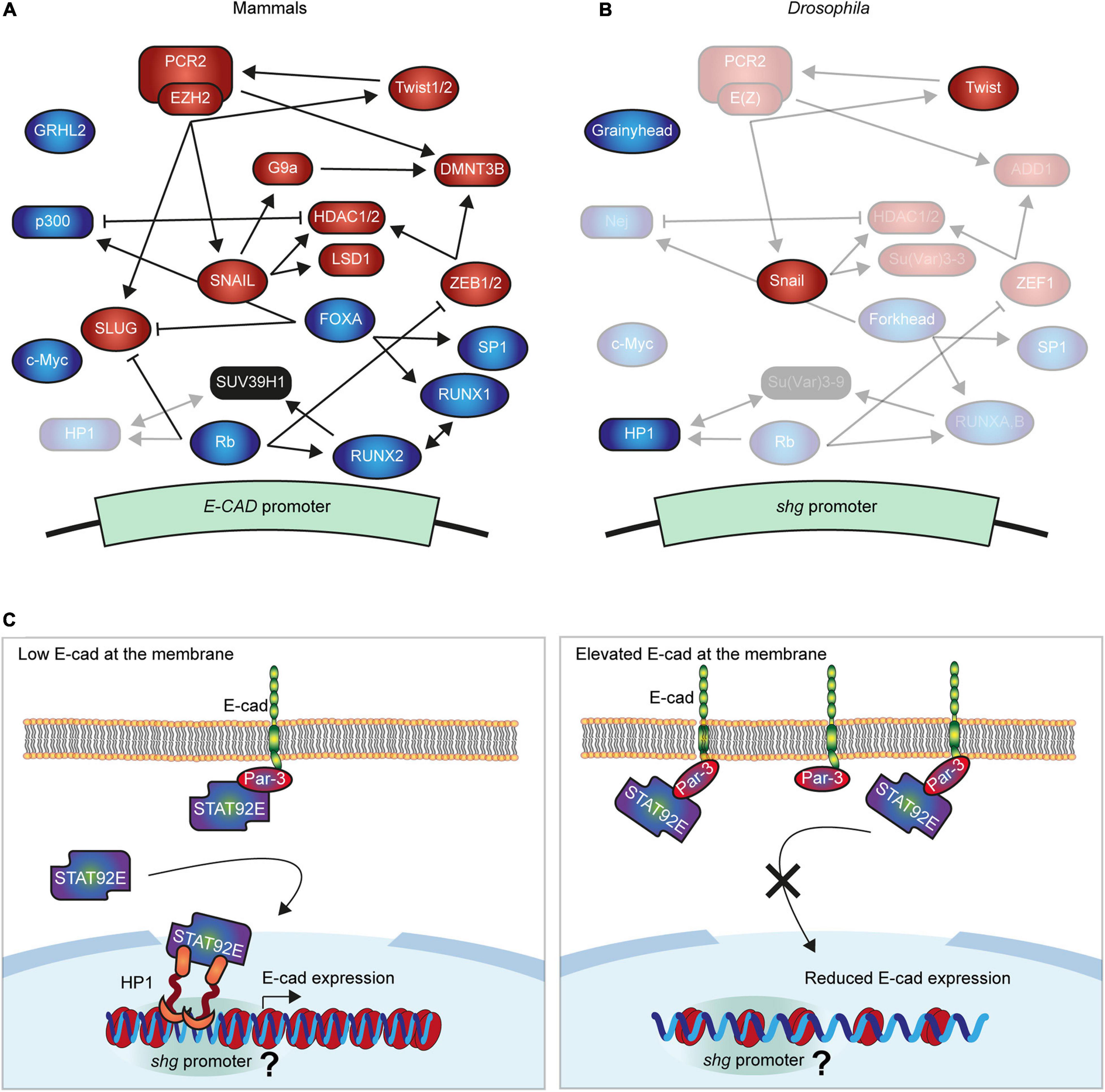
Figure 1. Molecular mechanisms regulating E-cadherin gene expression. (A,B) Summary of the best known negative and positive regulators of E-cad in mammals (A) and Drosophila (B). The expression of E-cad gene (shotgun, shg, in Drosophila) is controlled by transcription factors (ellipses), which inhibit (red) or promote it (green) and interact with the E-cad promoter with varying affinities. Several further proteins modify chromatin at the promoter (squircles) silencing the E-cad expression (red) or increasing it (green). These proteins cooperate, compete, and regulate each other, establishing the intricated network that fine-tune the expression of E-cad. In panel (B) the non-conserved proteins were removed while conserved maintained their relative positions but their names were replaced with those of the Drosophila orthologs. The regulatory roles of transparentized proteins in panels (A,B) were not demonstrated in the respective systems. (C) The proposed model of E-cad modulation of its expression. A subpool of STAT92E translocates into the nucleus independently of the canonical JAK/STAT signalling, where it interacts with heterochromatin protein 1 (HP1). Within euchromatin, HP1 localises at specific loci corresponding to gene promoters, including that of the shg gene. Concurrently, STAT92E is recruited to the cell surface by E-cad and its binding partner Par-3. Therefore, elevated levels of E-cad outcompete STAT92E from the nucleus inhibiting its function in heterochromatin formation and promoter regulation. As the result, elevated E-cad at the cell surface reduces shg expression, ultimately restoring its levels. It is unclear whether this is accompanied by a change in chromatin organisation around shg promoter, indicated by ‘?.’
Multiple epigenetic mechanisms also inhibit E-cad expression in mammalian cells. Hypermethylation of the large CpG island around the promoter is frequently linked with loss of E-cad expression (Yoshiura et al., 1995; van Roy and Berx, 2008). This DNA hypermethylation is often accompanied by histone modifications associated with inactive chromatin (Serrano-Gomez et al., 2016). Most notably, polycomb repressive complex 2 (PRC2) is recruited to the E-cad promoter where its key component, enhancer of zeste homolog 2 (EZH2), places the repressive H3K27me3 mark, deactivating E-cad transcription (Viré et al., 2006; Cao et al., 2008; Fujii and Ochiai, 2008). The histone methyltransferase G9a places the repressive H3K9me2/3 marks on the E-cad promoter (Dong et al., 2012). Histones are subjected to other post-translational modifications that contribute to E-cad silencing, such as the removal of the H3K4me2 mark by the lysine-specific demethylase 1 (LSD1) (Lin et al., 2010), or H3 and H4 deacetylation by the histone deacetylases 1 and 2 (HDAC1/2) (Peinado et al., 2004a). Histone phosphorylation and ubiquitination were also suggested to alter transcription of the mammalian E-cad (Surapaneni et al., 2020; Wang et al., 2020). While there is only a low level of DNA methylation in Drosophila (Lyko et al., 2000) other epigenetic mechanisms are conserved (Figure 1B) and might, similarly to Snail and Twist, regulate E-cad transcription in invertebrates.
These mechanisms of E-cad silencing closely interact with each other (Figure 1A). For example, SNAIL recruits the repressive complex containing LSD1 and HDAC1/2 (Peinado et al., 2004a; Lin et al., 2014), which may facilitate the subsequent recruitment of PRC2 (Ai et al., 2017; Jin et al., 2017, p. 1). SNAIL also recruits G9a to the E-cad promoter, whereas both G9a and PRC2 interact with the DNA methyltransferases DNMT1/3A/3B, and thus can promote the CpG methylation (Viré et al., 2006; Dong et al., 2012; Lin et al., 2014). Twist1 can also promote the recruitment of PRC2 (Cakouros et al., 2012; Malouf et al., 2013), whereas ZEB1 recruits histone deacetylases, HDAC1/2, and possibly DNMT3B (Aghdassi et al., 2012; Zhang et al., 2019). Concurrently, EZH2 facilitates the expression of SNAIL and SLUG through an unknown mechanism (Zhang et al., 2017), and increases Twist expression by suppressing the miR-361 (Ihira et al., 2017). Through these interactions, we suggest that individual mechanisms act together to ensure the robust silencing of the E-cad.
Transcriptional Activation
A fluctuation in the activity of a repressive mechanism, for example, due to random changes in fundamentally stochastic expression of regulatory genes, could lead to the E-cad loss (Raj and van Oudenaarden, 2008; Wendt et al., 2011). Protein turnover may counteract the resulting short-term E-cad silencing, whereby the balance between recycling and degradation of endocytosed molecules quickly modulates E-cad surface levels (Bryant and Stow, 2004; Huang et al., 2011; Bulgakova et al., 2013; Erami et al., 2015; Bulgakova and Brown, 2016). However, this might be insufficient to protect tissue integrity from a transient but excessive drop in the E-cad levels. Therefore, the robust E-cad expression requires mechanisms for transcriptional activation.
The traditional view is that E-cad expression is activated by constitutive factors in mammalian cells, which are overcome by repressors (Peinado et al., 2004b). This view was contested upon discovery of the binding of the TF FOXA/HNF3 (Forkhead in Drosophila) to the E-cad promoter, increasing its expression and driving re-epithelization of breast cancer cells (Liu et al., 2005). This activation system interacts with other E-cad regulatory factors and epigenetic machinery. FOXA suppresses the expression of SLUG, combining E-cad activation with the release of repression (Anzai et al., 2017). FOXA also interacts with p300, which promotes E-cad expression with the two p300-binding sites in the human E-cad promoter (Liu et al., 2005). This interaction suggests the potential role of histone modifications in E-cad transcriptional activation due to the intrinsic histone acetyltransferase (HAT) activity of p300 (Ogryzko et al., 1996; Chan and Thangue, 2001). While p300 can acetylate all histones, it preferentially modifies H3 and H4 and competes for binding with HDAC1 (Ogryzko et al., 1996; Li et al., 2014), making p300 an excellent candidate to counteract the histone deacetylation at the E-cad promoter by HDAC1/2. Finally, FOXA drives E-cad downregulation by interacting with Sp1 and RUNX1/AML1 (Liu et al., 2005; D’Costa et al., 2012). Of particular interest for the data presented below is that in mammalian cells RUNX1/AML1 forms a complex with SUV39H1 [Su(var)3–9 in Drosophila], whereas methylation of the H3K9 by SUV39H1 creates a binding site for the heterochromatin protein 1 (HP1) (Lachner et al., 2001; Chakraborty et al., 2003).
Retinoblastoma (Rb) and c-Myc also specifically activate the expression of E-cad promoter through an interaction with the TF AP-2, thus maintaining the epithelial phenotype in mammalian cells (Batsché et al., 1998). Rb interacts with the TF RUNX2, which acts redundantly with closely related RUNX1 in chondrocyte differentiation (Kimura et al., 2010; Gündüz et al., 2012; Komori, 2015). This, alongside RUNX2 and RUNX1 cooperation in tumour cells and RUNX1 recruitment to the E-cad promoter, raises a possibility of similar recruitment of RUNX2, and therefore Rb, to the E-cad promoter (Fowler et al., 2006). Concurrently, Rb depletion induces expression of SLUG and ZEB1, suggesting an indirect effect on E-cad expression (Arima et al., 2008). Additionally to these mechanisms, Rb is likely to modulate epigenetic mechanisms controlling E-cad expression, as it targets the HP1 to gene promoters (Nielsen et al., 2001). This complexity of E-cad transcriptional regulation is further amplified by the involvement of post-transcriptional mechanisms. For example, c-Myc activates E-cad expression, but its substantial overexpression in breast cancer cells leads to post-transcriptional repression of E-cad (Batsché et al., 1998; Cowling and Cole, 2007). While roles of Rb, c-Myc, and Forkhead in the expression of Drosophila E-cad are not known, another TF – Grhl2 (Grainyhead in Drosophila) – activates E-cad expression in both mammalian and invertebrate cells. In the former, intron-bound Grhl2 promotes E-cad transcription through chromatin looping, playing an essential role in determining an epithelial phenotype (Werth et al., 2010; Xiang et al., 2012). Similarly in flies, Grainyhead has four binding sites in the shotgun locus and its overexpression promotes E-cad transcription, as well as that of multiple other genes involved in epithelial maturation (Yao et al., 2017).
The above mechanisms overlook the potential of the cell adhesion molecules (CAMs) in triggering changes in gene expression (Robinson and Moberg, 2011; Dasgupta and McCollum, 2019; Hannezo and Heisenberg, 2019). For example, β-catenin is a TF activated by canonical Wnt signalling (Shang et al., 2017) and induces expression of SLUG in human carcinoma cells (Conacci-Sorrell et al., 2003), meaning that the release of β-catenin from junctions due to E-cad downregulation can reinforce this downregulation. Via α-catenin, E-cad regulates the Hippo pathway by sequestering the YAP protein outside the nucleus, thus explaining the antiproliferative properties of E-cad (Kim et al., 2011; Hannezo and Heisenberg, 2019; Sarpal et al., 2019). α-catenin counteracts β-catenin by inhibiting Wnt-activated transcription (Daugherty et al., 2014). Similarly, p120-catenin antagonises β-catenin-mediated transcriptional activation by interacting with the repressive TF Kaiso (Daniel and Reynolds, 1999; Kourtidis et al., 2013; Schackmann et al., 2013). Curiously, β-catenin expression is inhibited by the Kaiso–p120-catenin complex (Liu et al., 2014), highlighting the reciprocal interactions between transcriptional mechanisms and adhesion molecules. In Drosophila, although Kaiso is not present, p120-catenin is nevertheless involved in transcriptional regulation (Stefanatos et al., 2013). As catenins are recruited by E-cad, their availability for either performing nuclear functions or sequestering other TFs outside the nucleus is dependent on E-cad levels. Indeed, E-cad levels impact on transcription of multiple genes involved in a wide range of processes (Soncin et al., 2011). Furthermore, β-catenin recruits the p300 protein, which promotes E-cad expression (Mosimann et al., 2009). This raises the questions of whether E-cad contributes to own gene expression.
A Feedback Loop Regulating E-Cad Expression Through Par-3, HP1, and STAT92E
The polarity determinant Par-3 (Bazooka in Drosophila) is an evolutionarily conserved protein recruited to E-cad adhesion in both mammals and Drosophila (Harris and Peifer, 2005; Wei et al., 2005; Iden et al., 2006; Wang et al., 2009; Bulgakova et al., 2013; Xue et al., 2013). Par-3 has six protein–protein interaction domains, making it a versatile scaffold for protein recruitment (McKinley et al., 2012). In Drosophila, one of the proteins recruited by Par-3 is the only fly member of the signal transducer and activator of transcription (STAT) protein family, STAT92E (Hou et al., 1996; Yan et al., 1996). This recruitment of STAT92E is necessary for the efficient Janus kinase (JAK)/STAT signalling in Drosophila epithelial cells (Sotillos et al., 2008). In human and mouse cells, the loss of Par-3 activates STAT3 signalling, although it is not known if the loss of STAT3 recruitment to E-cad adhesion sites by Par-3 contributes to this activation (McCaffrey et al., 2012; Guyer and Macara, 2015). However, this is very likely, as the loss of E-cad adhesion also leads to STAT3 activation (del Valle et al., 2013).
The canonical signalling pathway comprises STAT translocation into the nucleus after its phosphorylation by the ligand-activated JAK. However, there is growing evidence of non-canonical signalling in both Drosophila and mammals, whereby non-phosphorylated STAT stabilises heterochromatin by binding HP1 (Li, 2008; Hixson et al., 2019). HP1 is an evolutionarily conserved non-histone chromatin protein whose best-characterised role is the formation and propagation of heterochromatin (Lomberk et al., 2006; Dialynas et al., 2008; Xu et al., 2014). HP1 is recruited to H3K9me2/3 through SUV39H1, and then itself recruits more SUV39H1 molecules, propagating H3K9 methylation (Lomberk et al., 2006). This is accompanied by DNA methylation in mammalian cells, as both HP1 and SUV39H1 bind DNMTs (Fuks et al., 2003). HP1 was also linked to other functions including transcriptional regulation, where HP1 usually represses transcription by facilitating methylation of H3K9. However, HP1 also localises at specific loci within euchromatin (Dialynas et al., 2008), and there is growing evidence of its association with transcriptionally active regions from C. elegans to Drosophila and humans (Lu et al., 2000; Piacentini et al., 2003; Mateescu et al., 2008; McMurchy et al., 2017). In Drosophila, the active euchromatic regions with HP1 recruitment include developmentally-regulated and heat-shock genes, whose activity correlates with HP1 dosage (Piacentini et al., 2003). Non-phosphorylated STAT92E and HP1 co-localise at euchromatic regions of Drosophila polytene chromosomes (Shi et al., 2008). HP1 disperses from chromosomes following hyperactivation of the canonical signalling, suggesting an intricate cross-talk between the two modes of signalling, likely through the availability of non-phosphorylated STAT92E (Xu et al., 2014).
As STAT92E promotes heterochromatin gene silencing in a dose-dependent manner (Shi et al., 2008), we hypothesised that an increase of E-cad levels would lead to STAT92E sequestration at adhesion sites, making it unavailable to bind HP1 and promote heterochromatin formation alongside other functions (Figure 1C). We examined the effects of E-cad overexpression on heterochromatic gene silencing monitored using position-effect variegation (PEV). In this assay, a gene is translocated into a peri-heterochromatic region where its expression is silenced in a stochastic pattern (Figure 2A). This silencing depends on the spreading of heterochromatin: if a protein normally promotes heterochromatin formation, its loss would reduce the amount of heterochromatin and suppress the variegation (=loss of silencing of the translocated gene, Figure 2A; Elgin and Reuter, 2013). We overexpressed E-cad in the Drosophila eye using the UAS–Gal4 system with GMR::Gal4, which expresses Gal4 in all retinal cells (Hay et al., 1997). We used the variegating allele of the brown gene (bwPEV), necessary to produce the red pigment pteridine. Overexpression of E-cad increased the normalised eye pigmentation with pteridine to 104% from 69% in the control (Figures 2B,C and Supplementary Figure 1). Therefore, E-cad suppresses PEV in the eye, supporting that elevated E-cad inhibits heterochromatin formation.
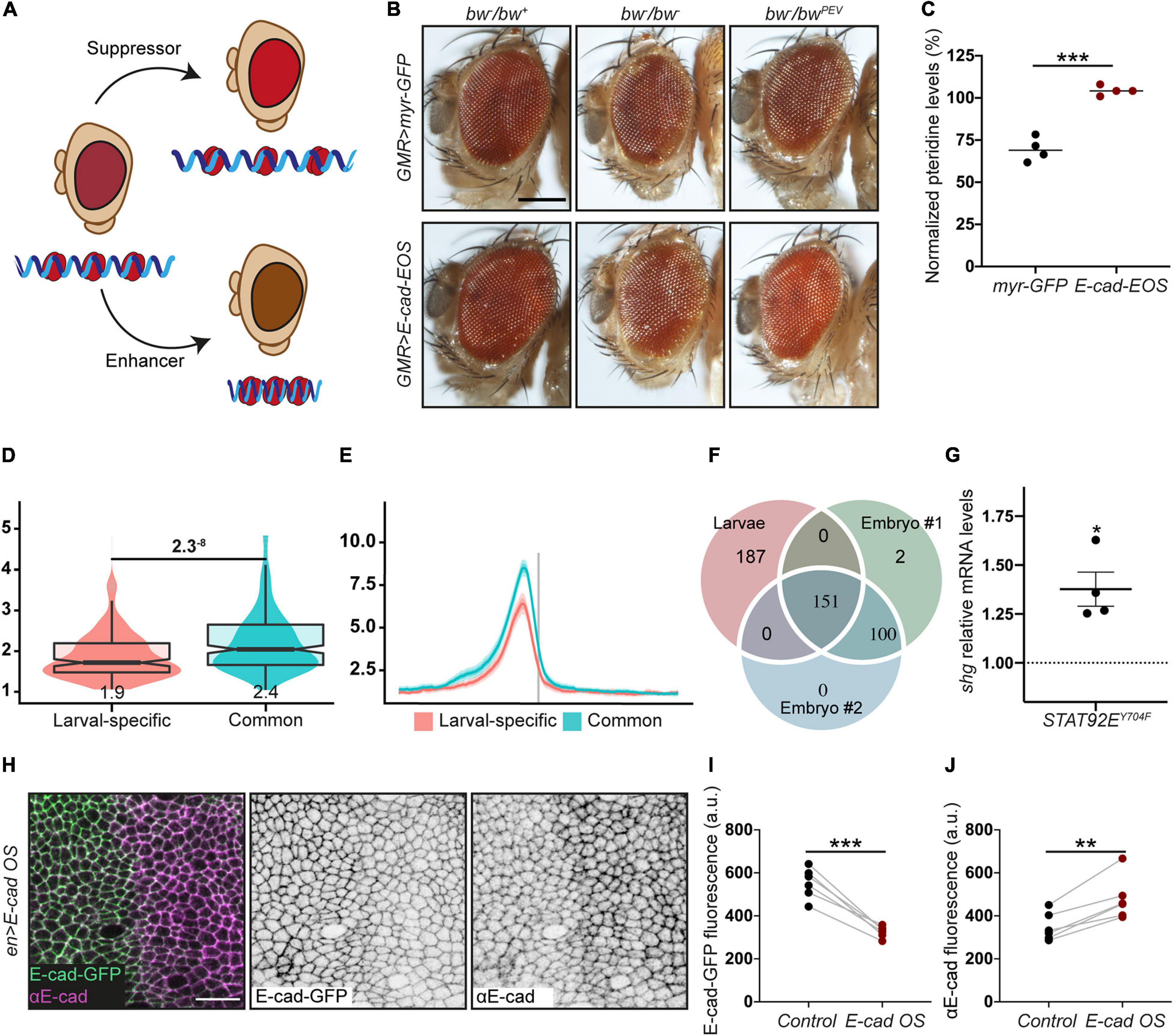
Figure 2. The crosstalk between E-cadherin, STAT92E and HP1. (A) A diagram of position effect variegation (PEV) in relation to chromatin compaction. (B,C) E-cad overexpression suppresses PEV: examples (B) and quantification (C). Scale bar – 0.2 mm, ***p = 0.0001 (unpaired t-test). (D) Overlapping box-and-violin plots showing the quantification of rBEADS normalised HP1 signal on larval-specific (red) and common (blue) promoters. The p-value above – the significance calculated using U-test. The value below the boxplots – the mean of the signal. (E) The distribution of the rBEADS normalised HP1 signal over transcription start sites (TSS) loci of larval-specific (red) and common (blue) genes. The vertical grey line represents the location of TSS, the plots span 1 kb upstream and downstream from annotated TSS. (F) A Venn diagram showing the overlaps between peak loci in the larval dataset and two independent embryonic (16–24 h) replicates. All three peak sets were filtered from peaks overlapping centromeric heterochromatin clusters and non-mappable, repetitive regions, defined based on GEM mappability analyses. modENCODE peak calls have been lifted from dm3 to dm6 genome assembly to match the analyses of larval HP1 peaks. The IDR method with 0.05 p-vale threshold was used to collapse the replicates to a common, high confidence peak set. (G) shotgun expression following overexpression of non-phosphorylatable STAT92EY704F measured using RT-qPCR and normalised to the expression of house-keeping gene RpL32. Four biological replicates are shown as distinct dots with three technical replicates each. *p = 0.02 (one-sample t-test comparing to 1). (H–J) E-cad-GFP expressed from the endogenous promoter is downregulated following E-cad overexpression (E-cad OS) from a heterologous UAS promoter. A representative image (H) shows endogenously expressed E-cad-GFP visualised with native GFP fluorescence (green, left; grey, middle) and total E-cad visualised with antibody staining (magenta, left; grey, right), scale bar – 10 μm. Levels of endogenously expressed E-cad-GFP are quantified in panel (I) and total E-cad levels in panel (J). **p = 0.0018 and ***p = 0.0003 (paired t-test).
Next, we sought to determine what genes might be regulated through our hypothetical mechanism and examined the HP1 localisation using publicly available ChIP-seq datasets generated using third instar larvae (Riddle et al., 2011), which include epithelial monolayers of imaginal discs. There were distinct peaks of HP1 overlapping with promoters of 456 genes outside of heterochromatin clusters and non-mappable (non-unique, repeatable) regions (Figures 2D,E and Supplementary Figure 2). We compared the larval HP1 peaks with two independent replicates from an earlier developmental stage – embryos at 16–24 h of development (modENCODE Consortium, Roy et al., 2010). Roughly half of the HP1 peaks in the larval dataset (155 peaks mapping to 151 loci) were common to all three datasets, while another half (187 peaks/loci) were larval-specific (Figure 2F). The common peaks (potentially, developmentally conserved) mostly mapped to bi-directional promoters, so that 155 peaks mapped to a total of 280 genes. These genes are enriched for housekeeping terms: regulation of primary and macromolecule metabolic processes (Supplementary Figure 3, terms 7–11); and mitotic cell cycle and cell cycle regulation (Supplementary Figure 3, terms 2–6). In contrast, the larval-specific peaks, which map to promoters of 178 genes, are enriched for terms related to epithelial tissue: morphogenesis of epithelium, actin filament organisation, epithelium development and imaginal disc development (Supplementary Figure 3, terms 19–24). The shotgun promoter overlapped a larval-specific HP1 peak with a signal value of 9.83 (207th promoter rank). This peak is consistent with two potential mechanisms for HP1 recruitment – via Su(var)3–9 or one of the two fly Rb-family proteins. Therefore, the modulation of HP1 function downstream of E-cad and non-canonical STAT signalling might alter the E-cad expression.
Indeed, overexpression of the non-phosphorylatable STAT92E (STAT92EY704F), which acts only in the non-canonical signalling (Karsten et al., 2006; Xu et al., 2014), increased expression of the shotgun gene (Figure 2G). Such an increase is consistent with the model whereby an unprogrammed drop in E-cad levels releases STAT92E, which leads to a consequent increase of E-cad expression and thus restores E-cad levels (Figure 1C). We observed that overexpression of E-cad from a heterologous UAS promoter leads to downregulation of the protein expressed from the endogenous locus (Figures 2H–J). This might be accompanied by simultaneous upregulation of the canonical JAK/STAT signalling as observed for STAT3 activation in some human cancers (Loh et al., 2019). We postulate that other genes with larval-specific HP1 peaks behave similarly to E-cad. Then, the activation of non-canonical STAT signalling following a stochastic drop in E-cad levels would reinforce the epithelial nature of the cells as many of these genes are involved in epithelial morphogenesis.
Outlook/Perspective
E-cad levels at the cell surface play an important role in determining cellular properties: cell shape, response to signalling, cell division, and neighbour exchange to name just a few (Lecuit and Lenne, 2007; Rübsam et al., 2017; Mendonsa et al., 2018). Not surprisingly, E-cad expression is subjected to complex regulation by multiple interconnected mechanisms. However, to respond to the cell’s needs and modulate E-cad levels accordingly, information about the E-cad levels at the surface must be transferred to the nucleus. Here, we propose one such mechanism, whereby surface E-cad feeds back to the nucleus through competition for STAT92E binding. We hypothesise that this feedback stabilises E-cad transcription enabling robust and resilient cell–cell adhesion. Importantly, this provides the cell with a tumour-suppressive mechanism, as heterochromatin formation, which is likely to follow a drop in E-cad levels, is linked with cellular senescence (Narita et al., 2003). This contrasts another feedback mechanism whereby the release of β-catenin from adhesion sites leads to SLUG expression and reinforces E-cad silencing in human cells. Therefore, depending on the context E-cad expression may be stabilised or silenced through feedback interactions. It is tempting to speculate that further mechanisms of information transfer from the cell surface to the nucleus exist – not unlike the multiple and intertwined mechanisms for regulation of E-cad transcription. A comprehensive understanding of E-cad transcriptional regulation requires the discovery and in-depth analysis of such mechanisms.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: http://data.modencode.org/, 3187; http://data.modencode.org/, 3188; http://data.modencode.org/, 3391; http://data.modencode.org/, 3392.
Author Contributions
NAB and MRM performed the experiments and wrote the manuscript. NAB and PAS did the bioinformatics analyses. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by a grant from the UKRI BBSRC (BB/P007503/1) to NAB.
Conflict of Interest
PAS was a shareholder and the managing director of the company SmartImmune Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Martin Zeidler for STAT92EY704F flies and the University of Sheffield’s Wolfson Light Microscopy and Drosophila Facillities for their technical support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.701175/full#supplementary-material
References
Aghdassi, A., Sendler, M., Guenther, A., Mayerle, J., Behn, C.-O., Heidecke, C.-D., et al. (2012). Recruitment of histone deacetylases HDAC1 and HDAC2 by the transcriptional repressor ZEB1 downregulates E-cadherin expression in pancreatic cancer. Gut 61, 439–448. doi: 10.1136/gutjnl-2011-300060
Ai, S., Peng, Y., Li, C., Gu, F., Yu, X., Yue, Y., et al. (2017). EED orchestration of heart maturation through interaction with HDACs is H3K27me3-independent. eLife 6:e24570.
Anzai, E., Hirata, K., Shibazaki, M., Yamada, C., Morii, M., Honda, T., et al. (2017). FOXA1 induces E-Cadherin expression at the protein level via suppression of slug in epithelial breast cancer cells. Biol. Pharm. Bull. 40, 1483–1489. doi: 10.1248/bpb.b17-00307
Arima, Y., Inoue, Y., Shibata, T., Hayashi, H., Nagano, O., Saya, H., et al. (2008). Rb depletion results in deregulation of E-cadherin and induction of cellular phenotypic changes that are characteristic of the epithelial-to-mesenchymal transition. Cancer Res. 68, 5104–5112. doi: 10.1158/0008-5472.can-07-5680
Batsché, E., Muchardt, C., Behrens, J., Hurst, H. C., and Crémisi, C. (1998). RB and c-Myc activate expression of the E-cadherin gene in epithelial cells through interaction with transcription factor AP-2. Mol. Cell. Biol. 18, 3647–3658. doi: 10.1128/mcb.18.7.3647
Bertocchi, C., Vaman Rao, M., and Zaidel-Bar, R. (2012). Regulation of adherens junction dynamics by phosphorylation switches. J. Signal. Transduct. 2012:125295.
Bolós, V., Peinado, H., Pérez-Moreno, M. A., Fraga, M. F., Esteller, M., and Cano, A. (2003). The transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: a comparison with Snail and E47 repressors. J. Cell Sci. 116, 499–511. doi: 10.1242/jcs.00224
Bringuier, P. P., Giroldi, L. A., Umbas, R., Shimazui, T., and Schalken, J. A. (1999). Mechanisms associated with abnormal E-cadherin immunoreactivity in human bladder tumors. Int. J. Cancer 83, 591–595. doi: 10.1002/(sici)1097-0215(19991126)83:5<591::aid-ijc3>3.0.co;2-6
Brüser, L., and Bogdan, S. (2017). Adherens junctions on the move—membrane trafficking of E-Cadherin. Cold Spring Harb. Perspect. Biol. 9:a029140. doi: 10.1101/cshperspect.a029140
Bryant, D. M., and Stow, J. L. (2004). The ins and outs of E-cadherin trafficking. Trends Cell Biol. 14, 427–434. doi: 10.1016/j.tcb.2004.07.007
Buckley, C. D., Rainger, G. E., Bradfield, P. F., Nash, G. B., and Simmons, D. L. (1998). Cell adhesion: more than just glue (Review). Mol. Membr. Biol. 15, 167–176. doi: 10.3109/09687689709044318
Bulgakova, N. A., and Brown, N. H. (2016). Drosophila p120-catenin is crucial for endocytosis of the dynamic E-cadherin-Bazooka complex. J. Cell Sci. 129, 477–482.
Bulgakova, N. A., Grigoriev, I., Yap, A. S., Akhmanova, A., and Brown, N. H. (2013). Dynamic microtubules produce an asymmetric E-cadherin-Bazooka complex to maintain segment boundaries. J. Cell Biol. 201, 887–901. doi: 10.1083/jcb.201211159
Cai, J., Culley, M. K., Zhao, Y., and Zhao, J. (2018). The role of ubiquitination and deubiquitination in the regulation of cell junctions. Protein Cell 9, 754–769. doi: 10.1007/s13238-017-0486-3
Cakouros, D., Isenmann, S., Cooper, L., Zannettino, A., Anderson, P., Glackin, C., et al. (2012). Twist-1 Induces Ezh2 recruitment regulating histone methylation along the Ink4A/Arf locus in mesenchymal stem cells. Mol. Cell. Biol. 32, 1433–1441. doi: 10.1128/mcb.06315-11
Campbell, K., Rossi, F., Adams, J., Pitsidianaki, I., Barriga, F. M., Garcia-Gerique, L., et al. (2019). Collective cell migration and metastases induced by an epithelial-to-mesenchymal transition in Drosophila intestinal tumors. Nat. Commun. 10:2311.
Cao, Q., Yu, J., Dhanasekaran, S. M., Kim, J. H., Mani, R.-S., Tomlins, S. A., et al. (2008). Repression of E-cadherin by the polycomb group protein EZH2 in cancer. Oncogene 27, 7274–7284. doi: 10.1038/onc.2008.333
Chakraborty, S., Sinha, K. K., Senyuk, V., and Nucifora, G. (2003). SUV39H1 interacts with AML1 and abrogates AML1 transactivity. AML1 is methylated in vivo. Oncogene 22, 5229–5237. doi: 10.1038/sj.onc.1206600
Chan, H. M., and Thangue, N. B. L. (2001). p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J. Cell Sci. 114, 2363–2373. doi: 10.1242/jcs.114.13.2363
Chu, Y.-S., Thomas, W. A., Eder, O., Pincet, F., Perez, E., Thiery, J. P., et al. (2004). Force measurements in E-cadherin–mediated cell doublets reveal rapid adhesion strengthened by actin cytoskeleton remodeling through Rac and Cdc42. J. Cell Biol. 167, 1183–1194. doi: 10.1083/jcb.200403043
Ciesiolka, M., Delvaeye, M., Imschoot, G. V., Verschuere, V., McCrea, P., Roy, F., et al. (2004). p120 catenin is required for morphogenetic movements involved in the formation of the eyes and the craniofacial skeleton in Xenopus. J. Cell Sci. 117, 4325–4339. doi: 10.1242/jcs.01298
Comijn, J., Berx, G., Vermassen, P., Verschueren, K., van Grunsven, L., Bruyneel, E., et al. (2001). The two-handed e box binding zinc finger protein SIP1 downregulates E-Cadherin and induces invasion. Mol. Cell 7, 1267–1278. doi: 10.1016/s1097-2765(01)00260-x
Conacci-Sorrell, M., Simcha, I., Ben-Yedidia, T., Blechman, J., Savagner, P., and Ben-Ze’ev, A. (2003). Autoregulation of E-cadherin expression by cadherin-cadherin interactions: the roles of beta-catenin signaling. Slug, and MAPK. J. Cell Biol. 163, 847–857. doi: 10.1083/jcb.200308162
Cowling, V. H., and Cole, M. D. (2007). E-cadherin repression contributes to c-Myc-induced epithelial cell transformation. Oncogene 26, 3582–3586. doi: 10.1038/sj.onc.1210132
D’Costa, Z. J., Jolly, C., Androphy, E. J., Mercer, A., Matthews, C. M., and Hibma, M. H. (2012). Transcriptional repression of E-Cadherin by human papillomavirus Type 16 E6. PLoS One 7:e48954. doi: 10.1371/journal.pone.0048954
da Silva, S. D., Alaoui-Jamali, M. A., Soares, F. A., Carraro, D. M., Brentani, H. P., Hier, M., et al. (2014). TWIST1 is a molecular marker for a poor prognosis in oral cancer and represents a potential therapeutic target. Cancer 120, 352–362. doi: 10.1002/cncr.28404
Daniel, J. M., and Reynolds, A. B. (1999). The Catenin p120ctn interacts with Kaiso, a Novel BTB/POZ domain zinc finger transcription factor. Mol. Cell Biol. 19, 3614–3623. doi: 10.1128/mcb.19.5.3614
Dasgupta, I., and McCollum, D. (2019). Control of cellular responses to mechanical cues through YAP/TAZ regulation. J. Biol. Chem. 294, 17693–17706. doi: 10.1074/jbc.rev119.007963
Daugherty, R. L., Serebryannyy, L., Yemelyanov, A., Flozak, A. S., Yu, H.-J., Kosak, S. T., et al. (2014). α-Catenin is an inhibitor of transcription. Proc. Natl. Acad. Sci. U. S. A. 111, 5260–5265.
del Valle, I., Rudloff, S., Carles, A., Li, Y., Liszewska, E., Vogt, R., et al. (2013). E-cadherin is required for the proper activation of the Lifr/Gp130 signaling pathway in mouse embryonic stem cells. Development 140, 1684–1692. doi: 10.1242/dev.088690
Dialynas, G. K., Vitalini, M. W., and Wallrath, L. L. (2008). Linking heterochromatin protein 1 (HP1) to cancer progression. Mutat. Res. 647, 13–20. doi: 10.1016/j.mrfmmm.2008.09.007
Dong, C., Wu, Y., Yao, J., Wang, Y., Yu, Y., Rychahou, P. G., et al. (2012). G9a interacts with Snail and is critical for Snail-mediated E-cadherin repression in human breast cancer. J. Clin. Invest. 122, 1469–1486. doi: 10.1172/jci57349
Eger, A., Aigner, K., Sonderegger, S., Dampier, B., Oehler, S., Schreiber, M., et al. (2005). DeltaEF1 is a transcriptional repressor of E-cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene 24, 2375–2385. doi: 10.1038/sj.onc.1208429
Elgin, S. C. R., and Reuter, G. (2013). Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila. Cold Spring Harb. Perspect. Biol. 5:a017780. doi: 10.1101/cshperspect.a017780
Erami, Z., Timpson, P., Yao, W., Zaidel-Bar, R., and Anderson, K. I. (2015). There are four dynamically and functionally distinct populations of E-cadherin in cell junctions. Biol. Open 4, 1481–1489. doi: 10.1242/bio.014159
Fowler, M., Borazanci, E., McGhee, L., Pylant, S. W., Williams, B. J., Glass, J., et al. (2006). RUNX1 (AML-1) and RUNX2 (AML-3) cooperate with prostate-derived Ets factor to activate transcription from the PSA upstream regulatory region. J. Cell. Biochem. 97, 1–17. doi: 10.1002/jcb.20664
Fujii, S., and Ochiai, A. (2008). Enhancer of zeste homolog 2 downregulates E-cadherin by mediating histone H3 methylation in gastric cancer cells. Cancer Sci. 99, 738–746. doi: 10.1111/j.1349-7006.2008.00743.x
Fuks, F., Hurd, P. J., Deplus, R., and Kouzarides, T. (2003). The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 31, 2305–2312. doi: 10.1093/nar/gkg332
Gasparotto, D., Polesel, J., Marzotto, A., Colladel, R., Piccinin, S., Modena, P., et al. (2011). Overexpression of TWIST2 correlates with poor prognosis in head and neck squamous cell carcinomas. Oncotarget 2, 1165–1175. doi: 10.18632/oncotarget.390
Giroldi, L. A., Bringuier, P.-P., de Weijert, M., Jansen, C., van Bokhoven, A., and Schalken, J. A. (1997). Role of E boxes in the repression of E-cadherin expression. Biochem. Biophys. Res. Commun. 241, 453–458. doi: 10.1006/bbrc.1997.7831
Greig, J., and Bulgakova, N. A. (2020). Interplay between actomyosin and E-cadherin dynamics regulates cell shape in the Drosophila embryonic epidermis. J. Cell Sci. 133:jcs242321.
Gumbiner, B. M. (1996). Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell 84, 345–357. doi: 10.1016/s0092-8674(00)81279-9
Gündüz, V., Kong, E., Bryan, C. D., and Hinds, P. W. (2012). Loss of the retinoblastoma tumor suppressor protein in murine calvaria facilitates immortalization of osteoblast-adipocyte bipotent progenitor cells characterized by low expression of N-Cadherin. Mol. Cell Biol. 32, 2561–2569. doi: 10.1128/mcb.06453-11
Guyer, R. A., and Macara, I. G. (2015). Loss of the polarity protein PAR3 activates STAT3 signaling via an atypical protein Kinase C (aPKC)/NF-κB/Interleukin-6 (IL-6) Axis in mouse mammary cells. J. Biol. Chem. 290, 8457–8468. doi: 10.1074/jbc.m114.621011
Hagios, C., Lochter, A., and Bissell, M. J. (1998). Tissue architecture: the ultimate regulator of epithelial function? Philos. Trans. R Soc. Lond. B Biol. Sci. 353, 857–870. doi: 10.1098/rstb.1998.0250
Halbleib, J. M., and Nelson, W. J. (2006). Cadherins in development: cell adhesion, sorting, and tissue morphogenesis. Genes Dev. 20, 3199–3214. doi: 10.1101/gad.1486806
Hannezo, E., and Heisenberg, C.-P. (2019). Mechanochemical feedback loops in development and disease. Cell 178, 12–25. doi: 10.1016/j.cell.2019.05.052
Harris, T. J. C., and Peifer, M. (2005). The positioning and segregation of apical cues during epithelial polarity establishment in Drosophila. J. Cell Biol. 170, 813–823. doi: 10.1083/jcb.200505127
Hay, B. A., Maile, R., and Rubin, G. M. (1997). P element insertion-dependent gene activation in the Drosophila eye. PNAS 94, 5195–5200. doi: 10.1073/pnas.94.10.5195
Hixson, K. M., Cogswell, M., Brooks-Kayal, A. R., and Russek, S. J. (2019). Evidence for a non-canonical JAK/STAT signaling pathway in the synthesis of the brain’s major ion channels and neurotransmitter receptors. BMC Genom. 20:677.
Hou, X. S., Melnick, M. B., and Perrimon, N. (1996). Marelle acts downstream of the Drosophila HOP/JAK kinase and encodes a protein similar to the mammalian STATs. Cell 84, 411–419. doi: 10.1016/s0092-8674(00)81286-6
Huang, J., Huang, L., Chen, Y.-J., Austin, E., Devor, C. E., Roegiers, F., et al. (2011). Differential regulation of adherens junction dynamics during apical–basal polarization. J. Cell Sci. 124, 4001–4013. doi: 10.1242/jcs.086694
Iden, S., Rehder, D., August, B., Suzuki, A., Wolburg-Buchholz, K., Wolburg, H., et al. (2006). A distinct PAR complex associates physically with VE-cadherin in vertebrate endothelial cells. EMBO Rep. 7, 1239–1246. doi: 10.1038/sj.embor.7400819
Ihira, K., Dong, P., Xiong, Y., Watari, H., Konno, Y., Hanley, S. J., et al. (2017). EZH2 inhibition suppresses endometrial cancer progression via miR-361/Twist axis. Oncotarget 8, 13509–13520. doi: 10.18632/oncotarget.14586
Janiszewska, M., Primi, M. C., and Izard, T. (2020). Cell adhesion in cancer: beyond the migration of single cells. J. Biol. Chem. 295, 2495–2505. doi: 10.1074/jbc.rev119.007759
Jin, Y., Huo, B., Fu, X., Hao, T., Zhang, Y., Guo, Y., et al. (2017). LSD1 collaborates with EZH2 to regulate expression of interferon-stimulated genes. Biomed. Pharmacother. 88, 728–737. doi: 10.1016/j.biopha.2017.01.055
Karsten, P., Plischke, I., Perrimon, N., and Zeidler, M. P. (2006). Mutational analysis reveals separable DNA binding and trans-activation of Drosophila STAT92E. Cell. Signal. 18, 819–829. doi: 10.1016/j.cellsig.2005.07.006
Kaszak, I., Witkowska-Piłaszewicz, O., Niewiadomska, Z., Dworecka-Kaszak, B., Ngosa Toka, F., and Jurka, P. (2020). Role of cadherins in cancer-a review. Int. J. Mol. Sci. 21:7624. doi: 10.3390/ijms21207624
Kim, N.-G., Koh, E., Chen, X., and Gumbiner, B. M. (2011). E-cadherin mediates contact inhibition of proliferation through Hippo signaling-pathway components. PNAS 108, 11930–11935. doi: 10.1073/pnas.1103345108
Kimura, A., Inose, H., Yano, F., Fujita, K., Ikeda, T., Sato, S., et al. (2010). Runx1 and Runx2 cooperate during sternal morphogenesis. Development 137, 1159–1167. doi: 10.1242/dev.045005
Komori, T. (2015). The functions of Runx family transcription factors and Cbfb in skeletal development. Oral Sci. Int. 12, 83–93.
Kourtidis, A., Ngok, S. P., and Anastasiadis, P. Z. (2013). p120 catenin: an essential regulator of cadherin stability, adhesion-induced signaling, and cancer progression. Prog. Mol. Biol. Transl. Sci. 116, 409–432.
Lachner, M., O’Carroll, D., Rea, S., Mechtler, K., and Jenuwein, T. (2001). Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410, 116–120. doi: 10.1038/35065132
Lecuit, T., and Lenne, P.-F. (2007). Cell surface mechanics and the control of cell shape, tissue patterns and morphogenesis. Nat. Rev. Mol. Cell Biol. 8, 633–644. doi: 10.1038/nrm2222
Lecuit, T., and Yap, A. S. (2015). E-cadherin junctions as active mechanical integrators in tissue dynamics. Nat. Cell Biol. 17, 533–539. doi: 10.1038/ncb3136
Li, W. X. (2008). Canonical and non-canonical JAK–STAT signaling. Trends Cell Biol. 18, 545–551. doi: 10.1016/j.tcb.2008.08.008
Li, X., Yang, H., Huang, S., and Qiu, Y. (2014). Histone deacetylase 1 and p300 can directly associate with chromatin and compete for binding in a mutually exclusive manner. PLoS One 9:e94523. doi: 10.1371/journal.pone.0094523
Lin, T., Ponn, A., Hu, X., Law, B. K., and Lu, J. (2010). Requirement of the histone demethylase LSD1 in snai1-mediated transcriptional repression during epithelial-mesenchymal transition. Oncogene 29, 4896–4904. doi: 10.1038/onc.2010.234
Lin, Y., Dong, C., and Zhou, B. P. (2014). Epigenetic regulation of EMT: the snail story. Curr. Pharm. Des. 20, 1698–1705. doi: 10.2174/13816128113199990512
Liu, Y., Dong, Q.-Z., Wang, S., Xu, H.-T., Miao, Y., Wang, L., et al. (2014). Kaiso interacts with p120-catenin to regulate β-catenin expression at the transcriptional level. PLoS One 9:e87537. doi: 10.1371/journal.pone.0087537
Liu, Y.-N., Lee, W.-W., Wang, C.-Y., Chao, T.-H., Chen, Y., and Chen, J. H. (2005). Regulatory mechanisms controlling human E-cadherin gene expression. Oncogene 24, 8277–8290. doi: 10.1038/sj.onc.1208991
Loh, C.-Y., Chai, J. Y., Tang, T. F., Wong, W. F., Sethi, G., Shanmugam, M. K., et al. (2019). The E-Cadherin and N-Cadherin switch in epithelial-to-mesenchymal transition: signaling, therapeutic implications, and challenges. Cells 8:1118. doi: 10.3390/cells8101118
Lomberk, G., Wallrath, L., and Urrutia, R. (2006). The heterochromatin protein 1 family. Genome Biol. 7:228.
Lu, B. Y., Emtage, P. C., Duyf, B. J., Hilliker, A. J., and Eissenberg, J. C. (2000). Heterochromatin protein 1 is required for the normal expression of two heterochromatin genes in Drosophila. Genetics 155, 699–708. doi: 10.1093/genetics/155.2.699
Lyko, F., Ramsahoye, B. H., and Jaenisch, R. (2000). DNA methylation in Drosophila melanogaster. Nature 408, 538–540. doi: 10.1038/35046205
Malouf, G. G., Taube, J. H., Lu, Y., Roysarkar, T., Panjarian, S., Estecio, M. R., et al. (2013). Architecture of epigenetic reprogramming following Twist1-mediated epithelial-mesenchymal transition. Genome Biol. 14:R144.
Mateescu, B., Bourachot, B., Rachez, C., Ogryzko, V., and Muchardt, C. (2008). Regulation of an inducible promoter by an HP1beta-HP1gamma switch. EMBO Rep. 9, 267–272. doi: 10.1038/embor.2008.1
Mazda, M., Nishi, K., Naito, Y., and Ui-Tei, K. (2011). E-Cadherin is transcriptionally activated via suppression of ZEB1 transcriptional repressor by small RNA-mediated gene silencing. PLoS One 6:e28688. doi: 10.1371/journal.pone.0028688
McCaffrey, L. M., Montalbano, J., Mihai, C., and Macara, I. G. (2012). Loss of the Par3 polarity protein promotes breast tumorigenesis and metastasis. Cancer Cell 22, 601–614. doi: 10.1016/j.ccr.2012.10.003
McKinley, R. F. A., Yu, C. G., and Harris, T. J. C. (2012). Assembly of Bazooka polarity landmarks through a multifaceted membrane-association mechanism. J. Cell Sci. 125, 1177–1190. doi: 10.1242/jcs.091884
McMurchy, A. N., Stempor, P., Gaarenstroom, T., Wysolmerski, B., Dong, Y., Aussianikava, D., et al. (2017). A team of heterochromatin factors collaborates with small RNA pathways to combat repetitive elements and germline stress. eLife 6:e21666.
Mendonsa, A., Na, T.-Y., and Gumbiner, B. M. (2018). E-cadherin in contact inhibition and cancer. Oncogene 37, 4769–4780. doi: 10.1038/s41388-018-0304-2
modENCODE Consortium, Roy, S., Ernst, J., Kharchenko, P. V., Kheradpour, P., Negre, N., et al. (2010). Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330, 1787–1797.
Mohan, A., Schlue, K. T., Kniffin, A. F., Mayer, C. R., Duke, A. A., Narayanan, V., et al. (2018). Spatial proliferation of epithelial cells is regulated by E-Cadherin force. Biophys. J. 115, 853–864. doi: 10.1016/j.bpj.2018.07.030
Mosimann, C., Hausmann, G., and Basler, K. (2009). β-Catenin hits chromatin: regulation of Wnt target gene activation. Nat. Rev. Mol. Cell Biol. 10, 276–286. doi: 10.1038/nrm2654
Muenst, S., Däster, S., Obermann, E. C., Droeser, R. A., Weber, W. P., von Holzen, U., et al. (2013). Nuclear expression of snail is an independent negative prognostic factor in human breast cancer. Dis. Markers 35, 337–344. doi: 10.1155/2013/902042
Narita, M., Nunez, S., Heard, E., Narita, M., Lin, A. W., Hearn, S. A., et al. (2003). Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113, 703–716. doi: 10.1016/s0092-8674(03)00401-x
Nelson, W. J. (2008). Regulation of cell-cell adhesion by the cadherin-catenin complex. Biochem. Soc. Trans. 36, 149–155. doi: 10.1042/bst0360149
Nielsen, S. J., Schneider, R., Bauer, U. M., Bannister, A. J., Morrison, A., O’Carroll, D., et al. (2001). Rb targets histone H3 methylation and HP1 to promoters. Nature 412, 561–565. doi: 10.1038/35087620
Nishiguchi, S., Yagi, A., Sakai, N., and Oda, H. (2016). Divergence of structural strategies for homophilic E-cadherin binding among bilaterians. J. Cell Sci. 129, 3309–3319.
Oda, H., and Takeichi, M. (2011). Structural and functional diversity of cadherin at the adherens junction. J. Cell Biol. 193, 1137–1146. doi: 10.1083/jcb.201008173
Oda, H., Tagawa, K., and Akiyama-Oda, Y. (2005). Diversification of epithelial adherens junctions with independent reductive changes in cadherin form: identification of potential molecular synapomorphies among bilaterians. Evol. Dev. 7, 376–389. doi: 10.1111/j.1525-142x.2005.05043.x
Oda, H., Tsukita, S., and Takeichi, M. (1998). Dynamic behavior of the cadherin-based cell-cell adhesion system during Drosophila gastrulation. Dev. Biol. 203, 435–450. doi: 10.1006/dbio.1998.9047
Ogryzko, V. V., Schiltz, R. L., Russanova, V., Howard, B. H., and Nakatani, Y. (1996). The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87, 953–959. doi: 10.1016/s0092-8674(00)82001-2
Palacios, F., Tushir, J. S., Fujita, Y., and D’Souza-Schorey, C. (2005). Lysosomal targeting of E-Cadherin: a unique mechanism for the down-regulation of cell-cell adhesion during epithelial to mesenchymal transitions. Mol. Cell Biol. 25, 389–402. doi: 10.1128/mcb.25.1.389-402.2005
Paredes, J., Figueiredo, J., Albergaria, A., Oliveira, P., Carvalho, J., Ribeiro, A. S., et al. (2012). Epithelial E- and P-cadherins: role and clinical significance in cancer. Biochim. Biophys. Acta 1826, 297–311.
Peinado, H., Ballestar, E., Esteller, M., and Cano, A. (2004a). Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol. Cell Biol. 24, 306–319. doi: 10.1128/mcb.24.1.306-319.2004
Peinado, H., Portillo, F., and Cano, A. (2004b). Transcriptional regulation of cadherins during development and carcinogenesis. Int. J. Dev. Biol. 48, 365–375. doi: 10.1387/ijdb.041794hp
Perez, T. D., and Nelson, W. J. (2004). Cadherin adhesion: mechanisms and molecular interactions. Handb. Exp. Pharmacol. 2004, 3–21. doi: 10.1007/978-3-540-68170-0_1
Piacentini, L., Fanti, L., Berloco, M., Perrini, B., and Pimpinelli, S. (2003). Heterochromatin protein 1 (HP1) is associated with induced gene expression in Drosophila euchromatin. J. Cell Biol. 161, 707–714. doi: 10.1083/jcb.200303012
Raj, A., and van Oudenaarden, A. (2008). Stochastic gene expression and its consequences. Cell 135, 216–226. doi: 10.1016/j.cell.2008.09.050
Riddle, N. C., Minoda, A., Kharchenko, P. V., Alekseyenko, A. A., Schwartz, Y. B., Tolstorukov, M. Y., et al. (2011). Plasticity in patterns of histone modifications and chromosomal proteins in Drosophila heterochromatin. Genome Res. 21, 147–163. doi: 10.1101/gr.110098.110
Robinson, B. S., and Moberg, K. H. (2011). Cell–cell junctions: α-Catenin and E-Cadherin help fence in Yap1. Curr. Biol. 21, R890–R892.
Rübsam, M., Mertz, A. F., Kubo, A., Marg, S., Jüngst, C., Goranci-Buzhala, G., et al. (2017). E-cadherin integrates mechanotransduction and EGFR signaling to control junctional tissue polarization and tight junction positioning. Nat. Commun. 8:1250.
Russell, H., and Pranjol, M. Z. I. (2018). Transcription factors controlling E-cadherin down-regulation in ovarian cancer. Biosci. Horizons Int. J. Student Res. 11:hzy010.
Sarpal, R., Yan, V., Kazakova, L., Sheppard, L., Yu, J. C., Fernandez-Gonzalez, R., et al. (2019). Role of α-Catenin and its mechanosensing properties in regulating Hippo/YAP-dependent tissue growth. PLoS Genet. 15:e1008454. doi: 10.1371/journal.pgen.1008454
Schackmann, R. C. J., Tenhagen, M., Ven, R. A. H., van de, and Derksen, P. W. B. (2013). p120-catenin in cancer – mechanisms, models and opportunities for intervention. J. Cell Sci. 126, 3515–3525. doi: 10.1242/jcs.134411
Schmidt, C. R., Gi, Y. J., Patel, T. A., Coffey, R. J., Beauchamp, R. D., and Pearson, A. S. (2005). E-cadherin is regulated by the transcriptional repressor SLUG during Ras-mediated transformation of intestinal epithelial cells. Surgery 138, 306–312. doi: 10.1016/j.surg.2005.06.007
Schock, F., and Perrimon, N. (2002). Molecular mechanisms of epithelial morphogenesis. Annu. Rev. Cell Dev. Biol. 18, 463–493.
Serrano-Gomez, S. J., Maziveyi, M., and Alahari, S. K. (2016). Regulation of epithelial-mesenchymal transition through epigenetic and post-translational modifications. Mol. Cancer 15:18.
Shang, S., Hua, F., and Hu, Z.-W. (2017). The regulation of β-catenin activity and function in cancer: therapeutic opportunities. Oncotarget 8, 33972–33989. doi: 10.18632/oncotarget.15687
Shi, S., Larson, K., Guo, D., Lim, S. J., Dutta, P., Yan, S.-J., et al. (2008). Drosophila STAT is required for directly maintaining HP1 localization and heterochromatin stability. Nat. Cell Biol. 10, 489–496. doi: 10.1038/ncb1713
Soncin, F., Mohamet, L., Ritson, S., Hawkins, K., Bobola, N., Zeef, L., et al. (2011). E-cadherin acts as a regulator of transcripts associated with a wide range of cellular processes in mouse embryonic stem cells. PLoS One 6:e21463. doi: 10.1371/journal.pone.0021463
Sotillos, S., Díaz-Meco, M. T., Moscat, J., and Castelli-Gair Hombría, J. (2008). Polarized subcellular localization of Jak/STAT components is required for efficient signaling. Curr. Biol. 18, 624–629. doi: 10.1016/j.cub.2008.03.055
Stefanatos, R. K., Bauer, C., and Vidal, M. (2013). p120 catenin is required for the stress response in Drosophila. PLoS One 8:e83942. doi: 10.1371/journal.pone.0083942
Surapaneni, S. K., Bhat, Z. R., and Tikoo, K. (2020). MicroRNA-941 regulates the proliferation of breast cancer cells by altering histone H3 Ser 10 phosphorylation. Sci. Rep. 10:17954.
Takeichi, M. (1977). Functional correlation between cell adhesive properties and some cell surface proteins. J. Cell Biol. 75, 464–474. doi: 10.1083/jcb.75.2.464
van Roy, F., and Berx, G. (2008). The cell-cell adhesion molecule E-cadherin. Cell Mol. Life Sci. 65, 3756–3788. doi: 10.1007/s00018-008-8281-1
Vergara, D., Simeone, P., Franck, J., Trerotola, M., Giudetti, A., Capobianco, L., et al. (2016). Translating epithelial mesenchymal transition markers into the clinic: novel insights from proteomics. EuPA Open Proteom 10, 31–41. doi: 10.1016/j.euprot.2016.01.003
Vesuna, F., van Diest, P., Chen, J. H., and Raman, V. (2008). Twist is a transcriptional repressor of E-cadherin gene expression in breast cancer. Biochem. Biophys. Res. Commun. 367, 235–241. doi: 10.1016/j.bbrc.2007.11.151
Viré, E., Brenner, C., Deplus, R., Blanchon, L., Fraga, M., Didelot, C., et al. (2006). The Polycomb group protein EZH2 directly controls DNA methylation. Nature 439, 871–874. doi: 10.1038/nature04431
Wang, D., Wang, Y., Wu, X., Kong, X., Li, J., and Dong, C. (2020). RNF20 is critical for snail-mediated E-Cadherin repression in human breast cancer. Front. Oncol. 10:613470.
Wang, Z., Sandiford, S., Wu, C., and Li, S. S.-C. (2009). Numb regulates cell-cell adhesion and polarity in response to tyrosine kinase signalling. EMBO J. 28, 2360–2373. doi: 10.1038/emboj.2009.190
Wei, S.-Y., Escudero, L. M., Yu, F., Chang, L.-H., Chen, L.-Y., Ho, Y.-H., et al. (2005). Echinoid is a component of adherens junctions that cooperates with DE-Cadherin to mediate cell adhesion. Dev. Cell 8, 493–504. doi: 10.1016/j.devcel.2005.03.015
Wendt, M. K., Taylor, M. A., Schiemann, B. J., and Schiemann, W. P. (2011). Down-regulation of epithelial cadherin is required to initiate metastatic outgrowth of breast cancer. Mol. Biol. Cell 22, 2423–2435. doi: 10.1091/mbc.e11-04-0306
Werth, M., Walentin, K., Aue, A., Schönheit, J., Wuebken, A., Pode-Shakked, N., et al. (2010). The transcription factor grainyhead-like 2 regulates the molecular composition of the epithelial apical junctional complex. Development 137, 3835–3845. doi: 10.1242/dev.055483
Wong, T.-S., Gao, W., and Chan, J. Y.-W. (2014). Transcription regulation of E-Cadherin by Zinc Finger E-Box binding homeobox proteins in solid tumors. Biomed. Res. Int. 2014:921564.
Xiang, X., Deng, Z., Zhuang, X., Ju, S., Mu, J., Jiang, H., et al. (2012). Grhl2 determines the epithelial phenotype of breast cancers and promotes tumor progression. PLoS One 7:e50781. doi: 10.1371/journal.pone.0050781
Xu, N., Emelyanov, A. V., Fyodorov, D. V., and Skoultchi, A. I. (2014). Drosophila linker histone H1 coordinates STAT-dependent organization of heterochromatin and suppresses tumorigenesis caused by hyperactive JAK-STAT signaling. Epigenet. Chromatin 7:16. doi: 10.1186/1756-8935-7-16
Xue, B., Krishnamurthy, K., Allred, D. C., and Muthuswamy, S. K. (2013). Loss of Par3 promotes breast cancer metastasis by compromising cell-cell cohesion. Nat. Cell Biol. 15, 189–200. doi: 10.1038/ncb2663
Yan, R., Small, S., Desplan, C., Dearolf, C. R., and Darnell, J. E. (1996). Identification of a stat gene that functions in Drosophila development. Cell 84, 421–430. doi: 10.1016/s0092-8674(00)81287-8
Yang, J., Antin, P., Berx, G., Blanpain, C., Brabletz, T., Bronner, M., et al. (2020). Guidelines and definitions for research on epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 21, 341–352.
Yao, L., Wang, S., Westholm, J. O., Dai, Q., Matsuda, R., Hosono, C., et al. (2017). Genome-wide identification of Grainy head targets in Drosophila reveals regulatory interactions with the POU domain transcription factor Vvl. Development 144, 3145–3155.
Yoshiura, K., Kanai, Y., Ochiai, A., Shimoyama, Y., Sugimura, T., and Hirohashi, S. (1995). Silencing of the E-cadherin invasion-suppressor gene by CpG methylation in human carcinomas. PNAS 92, 7416–7419. doi: 10.1073/pnas.92.16.7416
Zetter, B. R. (1993). Adhesion molecules in tumor metastasis. Semin. Cancer Biol. 4, 219–229. doi: 10.3109/15419069409004440
Zhang, Q., Dong, P., Liu, X., Sakuragi, N., and Guo, S.-W. (2017). Enhancer of Zeste homolog 2 (EZH2) induces epithelial-mesenchymal transition in endometriosis. Sci. Rep. 7:6804.
Keywords: adhesion, JAK/STAT, heterochromatin, HP1, PAR-3
Citation: Ramirez Moreno M, Stempor PA and Bulgakova NA (2021) Interactions and Feedbacks in E-Cadherin Transcriptional Regulation. Front. Cell Dev. Biol. 9:701175. doi: 10.3389/fcell.2021.701175
Received: 27 April 2021; Accepted: 04 June 2021;
Published: 28 June 2021.
Edited by:
Daiki Umetsu, Tohoku University, JapanReviewed by:
Mohit Kumar Jolly, Indian Institute of Science (IISc), IndiaHiroki Oda, JT Biohistory Research Hall, Japan
Copyright © 2021 Ramirez Moreno, Stempor and Bulgakova. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Natalia A. Bulgakova, bi5idWxnYWtvdmFAc2hlZmZpZWxkLmFjLnVr
 Miguel Ramirez Moreno
Miguel Ramirez Moreno Przemyslaw A. Stempor
Przemyslaw A. Stempor Natalia A. Bulgakova
Natalia A. Bulgakova