
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol., 28 June 2021
Sec. Epigenomics and Epigenetics
Volume 9 - 2021 | https://doi.org/10.3389/fcell.2021.699374
This article is part of the Research TopicEpigenetic Regulation in Cardiovascular DiseasesView all 16 articles
Vascular aging is a pivotal risk factor promoting vascular dysfunction, the development and progression of vascular aging-related diseases. The structure and function of endothelial cells (ECs), vascular smooth muscle cells (VSMCs), fibroblasts, and macrophages are disrupted during the aging process, causing vascular cell senescence as well as vascular dysfunction. DNA methylation, an epigenetic mechanism, involves the alteration of gene transcription without changing the DNA sequence. It is a dynamically reversible process modulated by methyltransferases and demethyltransferases. Emerging evidence reveals that DNA methylation is implicated in the vascular aging process and plays a central role in regulating vascular aging-related diseases. In this review, we seek to clarify the mechanisms of DNA methylation in modulating ECs, VSMCs, fibroblasts, and macrophages functions and primarily focus on the connection between DNA methylation and vascular aging-related diseases. Therefore, we represent many vascular aging-related genes which are modulated by DNA methylation. Besides, we concentrate on the potential clinical application of DNA methylation to serve as a reliable diagnostic tool and DNA methylation-based therapeutic drugs for vascular aging-related diseases.
Vascular aging is characterized by gradual changes in the vasculature structure and function (Laina et al., 2018; Ding et al., 2020). With aging, the structure and mechanical properties of vascular wall alter, i.e., lumen dilation, wall thickening, decreased arterial compliance, and increased arterial stiffness (North and Sinclair, 2012). The anatomical structure of vascular includes intima, media, and adventitia. Significant changes occur in the intima and media in the vascular aging progression (Lakatta and Levy, 2003a). Vascular intima primarily comprises endothelial cells (ECs), media is composed of vascular smooth muscle cells (VSMCs), and vascular adventitia is primarily composed of fibroblasts. Vascular cell senescence triggers cell morphological and functional changes, hence, ECs dysfunction, phenotypic transition of VSMCs, macrophage polarization, and fibroblast differentiation to myofibroblast (Chi et al., 2019). Age is an independent risk factor for vascular disorders (Morgan et al., 2018). To date, the aging population is significantly increasing, and research estimated that by 2040, 22% of people will be over the age of 65 (Heidenreich et al., 2011). Several lines of studies indicated that vascular aging enhanced the incidence and mortality of atherosclerosis (Mahmood et al., 2014), Alzheimer’s disease (AD) (Lakatta and Levy, 2003b), stroke (Lakatta and Levy, 2003a), etc. Vascular aging-related diseases are the leading causes of death among the elderly. Thus, there is an urgent need to identify reliable and efficient diagnosis and treatment for vascular aging-related diseases.
DNA methylation is an epigenetic mechanism involving multiple biological processes such as aging, metabolism, and autoimmune (Jones, 2012). Scholars believe that epigenetics is based on alterations in gene expression levels and does not involve DNA sequence changes (Brunet and Berger, 2014). DNA methylation is a dynamically reversible process regulated by methyltransferases and demethyltransferases and it regulates gene expression by recruiting proteins implicated in gene repression or inhibiting the binding of transcription factors to DNA (Moore et al., 2013). DNA methylation is tightly associated with vascular aging and related disorders. Although studies on the link between DNA methylation and vascular disease have got much attention, the underlying mechanisms and roles of DNA methylation in vascular aging are still not well elucidated.
Since the prevalence and mortality of vascular disorders are closely related to vascular aging, the diagnosis and treatment of vascular aging and related diseases have received significant research attention. Therefore, this review summarizes the current research and recent advances on DNA methylation in vascular aging, revealing the involvement of DNA methylation in ECs, VSMCs, fibroblasts, and macrophages functions. We review the physiological and pathological processes involving DNA methylation in vascular aging-related diseases and represent many vascular disease-related genes that are regulated by DNA methylation. Additionally, we concentrate primarily on the clinical prospect of DNA methylation as an early diagnostic tool and potential DNA methylation-based therapies for vascular aging-related diseases.
DNA methylation is a modification of DNA. In 1942, Waddington first proposed the concept of “epigenotype,” which was used to explain the complex progression process between genotype and phenotype (Waddington, 2012). To date, it is generally accepted that epigenetics primarily focuses on regulating gene expression, including DNA methylation, histone modification, non-coding RNA modification, chromatin remodeling, gene imprinting, etc. Here, we majorly focus on DNA methylation and its vital role in vascular aging-related diseases, including cardiovascular disease (CVD), cerebrovascular disease, and kidney diseases. DNA methylation regulates gene expression by recruiting proteins involved in gene repression or inhibiting the binding of transcription factors to DNA (Moore et al., 2013). DNA methylation is a dynamically reversible process modulated by methyltransferases and demethyltransferases (Chen and Riggs, 2011). There exist three forms of DNA methylation, including N4-methylcytosine (4mC), N5-methylcytosine (5mC), and N6-methyladenine (6mA) (Ratel et al., 2006) (Figure 1).
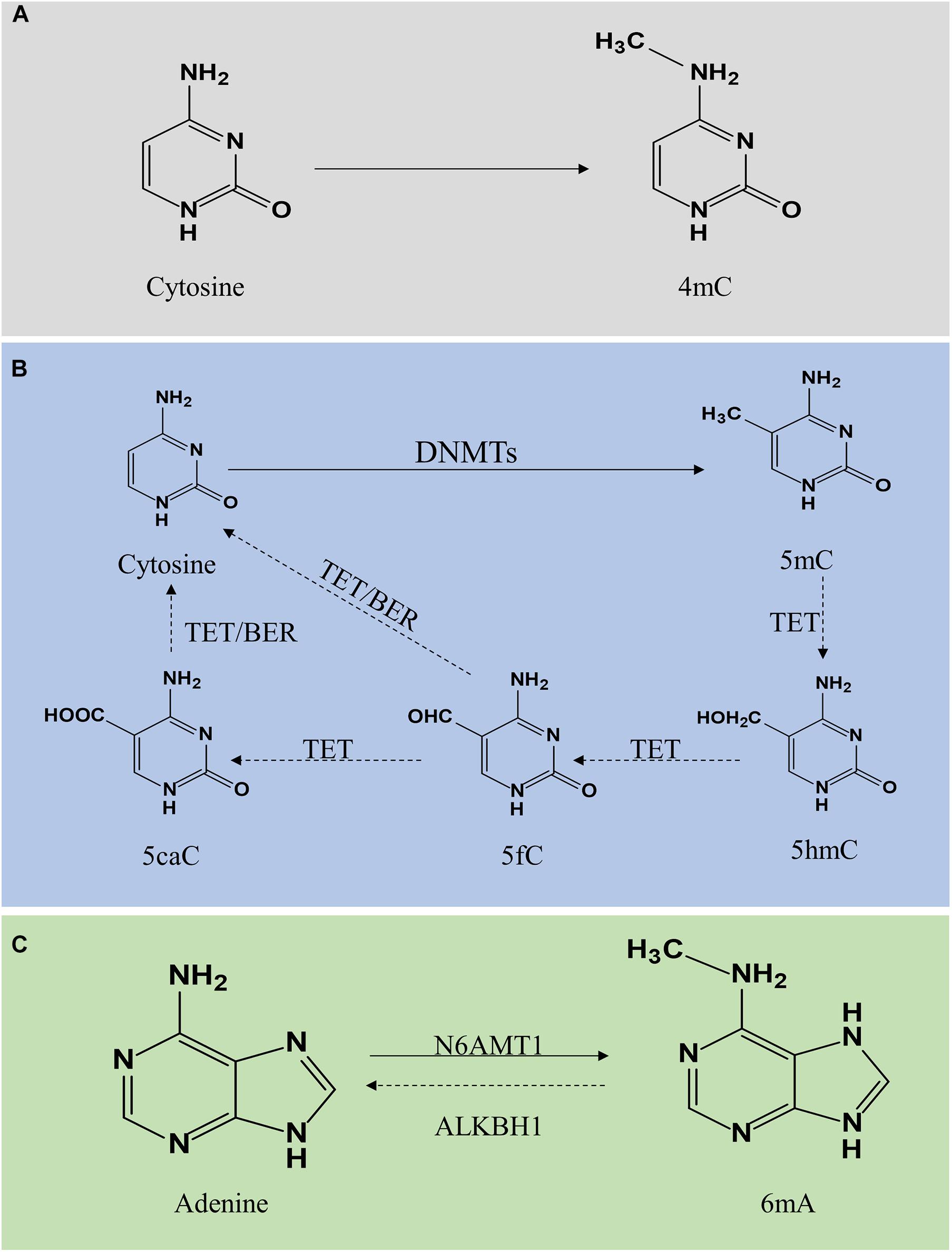
Figure 1. Molecular mechanism of DNA methylation and demethylation. DNA methylation is divided into 4mC, 5mC, and 6mA. Panel (A) shows 4mC, a type of DNA methylation in prokaryotes. Panel (B) shows 5mC. A cytosine base can be methylated by DNMT1, DNMT3a, and DNMT3b to form 5mC, while TETs catalyze the oxidation of 5mC to 5hmC, 5fC, and 5caC. 5fC and 5caC are modulated by TDG/BER pathway. Panel (C) shows 6mA. Adenine is catalyzed by N6AMT1 to form 6 mA. In contrast, ALKBH1 mediates N6-demethyladenine. 4mC, N4-methylcytosine; 5mC, N5-methylcytosine; 6mA, N6-methyladenine; DNMTs, DNA methyltransferases; TETs, 10–11 translocations; 5hmC, 5-hydroxymethylcytosine; 5fC, 5-formylcytosine; 5caC, 5-carboxylcytosine; BER, base excision repair.
N4-methylcytosine (4mC) modification principally exists in the DNA of bacteria, hence, a subject ignored by this review (Ehrlich et al., 1985, 1987). DNA methylation mainly occurs on the CpG dinucleotide in vertebrates. In mammals, global DNA methylation is a dynamic process, thus there exist DNA methylation and demethylation. 5mC is generated by DNA methyltransferases (DNMTs) through transfer a methyl group from S-adenyl methionine to the fifth carbon atom of the cytosine (Moore et al., 2013; Lyko, 2018). The DNMTs family includes DNMT1, DNMT2, DNMT3a, DNMT3b, and DNMT3L (Lyko, 2018). The patterns of 5mC are categorized into two major groups, including maintenance methylation and de novo methylation. DNMT1 is implicated in the DNA methylation through copy 5mC from parental DNA strand onto the newly synthesized daughter strand during DNA replication, thus, DNMT1 is known as maintenance DNMT (Mortusewicz et al., 2005; Nelissen et al., 2011). The de novo DNMTs included DNMT3a and DNMT3b can generate methylation in unmethylated DNA (Law and Jacobsen, 2010; Moore et al., 2013). Generally, DNA methylation inhibits gene expression through two different mechanisms. One is on the basis of intervene the binding of transcription factors included E2F or CREB by DNA methylation to represses gene transcription. On the other hand, DNMTs interact with methyl-CpG binding domain proteins to suppress transcription through establish a repressive chromatin environment (Bogdanović and Veenstra, 2009). DNA hypermethylation is strongly associated with the development of vascular aging-related disorders. Besides, DNA demethylation refers to two different pathways, including active demethylation and passive demethylation. Active DNA demethylation involves the ten-eleven translocations (TETs) induced methylated base oxidation and the activation induced deaminase induced methylated or a nearby base deamination, respectively (Bochtler et al., 2017). Activation induced deaminase deaminates cytosine to uracil. Additionally, several sources of evidence confirmed that 5mC can be converted into 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC) under the activation of TETs (Rasmussen and Helin, 2016; Wu and Zhang, 2017). The level of TETs is tightly associated with DNA methylation. A high generation of TETs significantly alleviated the level of 5mC while a lack of TETs induced DNA hypermethylation. TET-thymine DNA glycosylase (TDG)-base excision repair (BER) mechanism is involved in regulating active DNA demethylation (Weber et al., 2016). Additionally, passive DNA demethylation occurs in DNA replication via the dilution of methylation marks (Ooi and Bestor, 2008). DNMT1 is implicated in the process of maintenance methylation, inhibiting the expression or activation of DNMT1 reduces the level of DNA methylation.
DNA N6-methyladenine (6mA) was previously considered the most prevalent form of DNA methylation in prokaryotes (Xiao et al., 2018). Surprisingly, recent reports suggested that 6mA also exists in eukaryotes, including Caenorhabditis elegans (Greer et al., 2015), Drosophila (Zhang G. et al., 2015), and Chlamydomonas (Fu et al., 2015). In C. elegans, 6mA is mediated by DAMT-1 and NMAD-1 (Greer et al., 2015). In Drosophila, DMAD is implicated in the regulation of 6mA demethylation (Zhang G. et al., 2015). In addition, a study revealed that 6mA was extensively present in the human genome and enriched in the coding region, modulating gene transcription activation (Xiao et al., 2018). Furthermore, human genome 6mA modification is modulated by the methyltransferase N6AMT1 while N6-demethyladenine is mediated by ALKBH1. Several studies indicated that 6mA was involved in human diseases with controversy, thus, further exploration and in-depth studies on the roles of 6mA in the field of mammalian biomedicine are necessary to address the controversy.
Aging is a decline of the biological system, accompanied by a decrease in function. Cell senescence is a permanent state of cell cycle arrest (Khor and Wong, 2020), causing tissue dysfunction and closely associated with aging-related diseases (Bitar, 2019). Accumulating evidence suggested that DNA methylation plays a significant role in regulating ECs, VSMCs, fibroblasts, and macrophages functions, and is implicated in the process of vascular aging and related disorders. Recent studies have identified a series of genes regulated through DNA methylation in the initiation and development of vascular aging (Table 1). This section primarily focuses on the mechanisms and roles of DNA methylation in the functions of ECs and VSMCs (Figure 2).
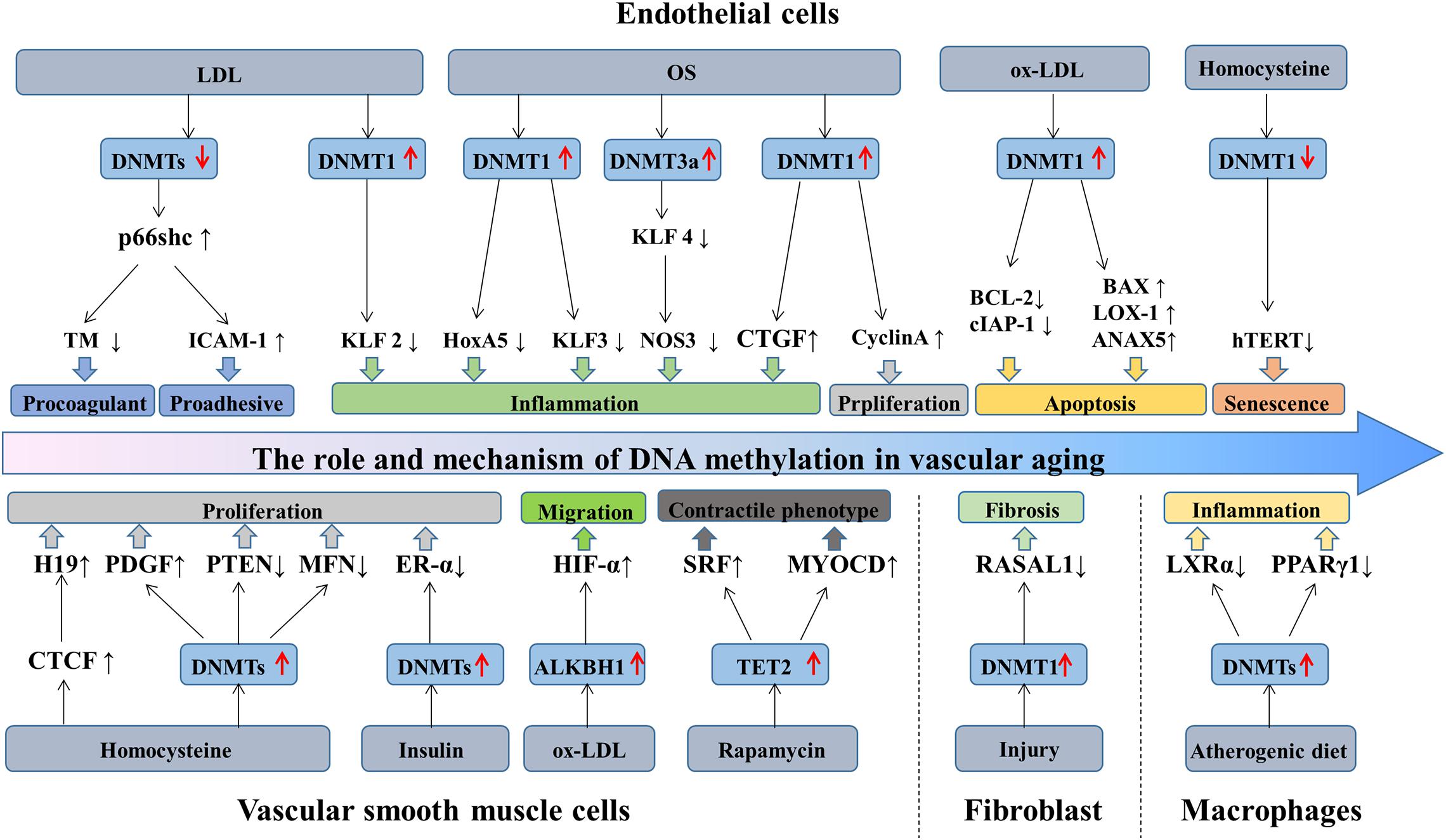
Figure 2. The role and mechanism of DNA methylation in vascular aging. During aging, the structure and function of vascular cells change, causing ECs dysfunction, VSMCs proliferation and migration, fibroblasts differentiation, and macrophages inflammation. LDL stimulates p66shc expression by downregulating the activity of DNMTs and mediating the expression of ICAM-1 and TM, eventually exacerbating ECs dysfunction. Besides, LDL inhibits KLF2 expression via increasing DNMT1 activity, causing ECs inflammation. OS-treated ECs promotes DNMT1 activity and inhibits the generation of HoxA5 and KLF3. OS regulates KLF4, CTGF, and cyclinA methylation patterns by promoting DNMTs activity, causing ECs inflammation and proliferation. Ox-LDL triggers ECs apoptosis through the hypermethylation of cIAP-1 and BCL2 and the hypomethylation of BAX, LOX-1, and ANXA5. Homocysteine promotes ECs senescence by reducing hTERT expression via DNA methylation. Additionally, homocysteine increases CTGF and PDGF expression and decreases PTEN and MFN expression, mediating VSMCs proliferation. Insulin promotes VSMCs proliferation via ER-α hypermethylation. Ox-LDL promotes VSMCs migration by increasing HIF-1α generation through ALKBH1. TET2 overexpression promotes VSMCs differentiation by the upregulation of SRF and MYOCD expression. Besides, in response to injury, fibroblast activation through DNMT1 upregulation and RASAL1 hypermethylation. Furthermore, the hypermethylation of LXRα and PPARγ1 induces macrophages inflammation. DNMTs, DNA methyltransferases; TET, 10—11 translocations; LDL, low-density lipoprotein; OS, oscillatory shear stress; ECs, endothelial cells; VSMCs, vascular smooth muscle cells.
Vascular endothelium is mainly composed of ECs, a barrier between blood and tissues. Endothelial dysfunction is caused by a variety of stimuli such as oxidized low-density lipoprotein (ox-LDL), hypoxia, shear stress, or inflammatory factors (Ni et al., 2020). DNA methylation modulates gene expression and mediates ECs biology in the development of its aging.
Endothelial cells play a crucial role in maintaining vascular homeostasis (Gori, 2018). ECs regulate vasoconstriction and relaxation (Krüger-Genge et al., 2019), involved in physiological processes, including blood coagulation, angiogenesis, and metabolism. Besides, ECs cooperate with VSMCs in modulating blood flow to tissues (Michiels, 2003). ECs aging are characterized by endothelial dysfunction (Jia et al., 2019). Aging causes changes in vascular EC-mediated vasoconstriction and dilation, integrity destruction, increased permeability, destroying the blood–brain barrier, and impaired angiogenesis (Papapetropoulos et al., 1997). Furthermore, ECs senescence stimulates mitochondrial dysfunction and reactive oxygen species (ROS) accumulation, which further aggravates vascular senescence (Ungvari et al., 2008).
Generally, DNA methylation is altered in ECs during aging and upon exposure to stimuli such as shear stress, hypoxia, or ox-LDL. DNA hypermethylation and hypomethylation can result in inhibition and stimulation of gene transcription in ECs in response to injury (Levy et al., 2017). Quite a number of evidence highlighted that DNA methylation is involved in mediating the biological processes of ECs, including inflammation, proliferation, senescence, and apoptosis.
Endothelial cells inflammation is tightly linked with vascular disorders. DNMTs promote ECs inflammation by regulating the methylation level of pertinent gene promoter regions. Under the stimulation of low-density lipoprotein (LDL), KLF2 promoter region was hypermethylated, arousing a down-expression of KLF2 in ECs and causing ECs inflammation and thrombogenesis (Kumar et al., 2013). In addition, the methylation level of relevant genes includes KLF4 (Jiang et al., 2014), SMAD7 (Wei et al., 2018), HoxA5 (Dunn et al., 2014), and CTGF (Zhang et al., 2017) are participating in ECs inflammation and aging-related vascular diseases.
Endothelial cells proliferation and differentiation are modulated by DNA methylation. In vivo, DNMTs promote the hypermethylation of cell cycle regulator cyclin A, stimulating the proliferation and inflammation of ECs. Besides, endothelial nitric oxide synthase (eNOS) expression in ECs senescence declines, leading to a down-expression of NO and impairing vasodilation (Cau et al., 2012). The generation of eNOS can be regulated by DNA methylation (Chan et al., 2004). Moreover, inhibiting DNA methylation in the eNOS promoter region induces the differentiation of human embryonic liver cells into ECs (Lagarkova et al., 2008). In addition, ROS accumulated in aging ECs causes alternation in DNA methylation by changing DNMTs activity and DNA damage (O’Hagan et al., 2011; Tabaei and Tabaee, 2019).
Endothelial cells senescence and apoptosis arouse endothelial dysfunction. Under the condition of ox-LDL, the proapoptosis-related genes such as LOX-1, ANXA5, BAX, and CASP3 are activated due to the hypomethylation of its promoter region. In contrast, the anti-apoptotic BCL2 and cIAP-1 genes are down-regulated by the DNA hypermethylation, eventually resulting in ECs apoptosis (Mitra et al., 2011). Besides, homocysteine promotes ECs senescence through DNA hypomethylation of hTERT (Zhang D. et al., 2015).
Vascular smooth muscle cells, the primary cells of vascular media, mediating blood flow and pressure via vascular contraction and relaxation. Environmental stresses such as oxidative stress, aging, or inflammation stimulate VSMCs contractile state switching into a proliferative and migratory synthetic phenotype, causing cell inflammation, proliferation, and migration. To date, multiple publications supported that DNA methylation plays a role in regulating VSMCs gene expression and VSMCs proliferation, migration, senescence, and apoptosis in response to vascular disorders.
Vascular smooth muscle cells are implicated in maintaining the structural integrity and physiological function of blood vessels, regulating blood pressure and controlling vascular contraction and relaxation (Frismantiene et al., 2018; Chi et al., 2019). Besides, VSMCs and extracellular matrix are the primary regulators of arterial contraction tension and vascular tension (Wang G. et al., 2015; Chi et al., 2019). With aging, VSMCs suffer from mechanical stimulation, chronic inflammation, calcification, epigenetic events, etc. (Lacolley et al., 2018). VSMCs switch into synthetic phenotype, leading to hypertrophy of the vascular wall and collagen deposition, which is pertinent to vascular aging-related diseases.
DNA methylation is implicated in the modulation of VSMCs proliferation, migration, differentiation, and calcification. The proliferation of VSMCs is one of the primary features of vascular aging-related diseases, and DNA methylation is associated with the regulation of gene transcription controlling cell proliferation. A large amount of evidence revealed that DNMTs inhibited gene expression by promoting DNA methylation and regulating the proliferation of VSMCs. DNA hypermethylation of MFN2 (Xu L. et al., 2019), PTEN (Ma et al., 2018), and ER-α (Min et al., 2016) causes the corresponding low expression of MFN2, PTEN, and ER-α and promotes the proliferation of VSMCs. On the contrary, DNA demethylation of PDGF increases PDGF mRNA and protein expression and promotes the proliferation and migration of VSMCs (Zhang et al., 2012). Additionally, the hypomethylation of HIF-1α causes VSMCs proliferation and migration as well (Wu et al., 2019).
Vascular smooth muscle cells have apparent plasticity. TET2, a key enzyme in the DNA demethylation pathway, participates in regulating the differentiation of SMCs. A study demonstrated that in the case of vascular injury and vascular disorders, the expression of TET2 was downregulated, whereas the MYOCD, SRF, and MYH11 were hypermethylated, resulting in SMCs differentiation (Liu et al., 2013).
The phenotypic modulation of SMCs triggers vascular disorders, including vascular calcification and atherosclerosis. Accumulating evidence reported that the contraction and synthetic phenotypes of VSMCs were mediated by DNA methylation. Furthermore, extracellular matrix mediates the phenotypic transition of SMCs via DNA methylation (Liu R. et al., 2015). Conversely, the DNMTs inhibitor 5-aza-2′-deoxycytidine (5Aza) reduces the DNA methylation level of the ALP in VSMCs and promotes the expression and activity of alkaline phosphatase, inducing vascular calcification (Azechi et al., 2014).
The vascular adventitia is primarily composed of fibroblasts. Fibroblasts are thought to exhibit a broad range of physiological functions. Fibroblasts increase vasa vasorum-associated neointima formation and macrophage recruitment by enhancing the expression of vascular endothelial growth factors (Li X. D. et al., 2020). Besides, fibroblasts are involved in vascular remodeling, inflammation, and neointimal formation in vascular disorders (An et al., 2015). Moreover, fibroblasts play a role in providing a supporting framework of the vessel wall through generating and secreting fibrillar collagens, the main components of the adventitial extracellular matrix (Stenmark et al., 2013). Additionally, in response to injury, fibroblasts generate accumulative extracellular matrix, ultimately lead to organ dysfunction, such as liver cirrhosis and chronic renal failure (Thannickal et al., 2014).
It has been identified that aging is a risk factor for fibrosis. DNA methylation is involved in modulating fibroblasts activity. RASSF1A, a regulatory tumor inhibitor, is downregulated by DNMT3a in cardiac fibrosis and fibroblasts activation. Mechanistically, RASSF1A hypermethylation promoted cardiac fibrosis through the activation of the Ras/ERK signal pathway (Tao et al., 2014a). Besides, the hypermethylation of RASAL1 induced by DNMT1 is consistently linked with fibroblast activation and kidney fibrosis (Bechtel et al., 2010). Aberrant hypermethylation of several genes includes RASSF1A, RASAL1, BMP7, and COL1A1 are tightly correlated with fibroblast activation.
Monocytes/macrophages, essential components of the immune system, are heterogeneous and exhibit a vital role in modulating inflammatory responses and vascular functions (Shapouri-Moghaddam et al., 2018). Accumulating studies revealed that monocytes and macrophages exert indispensable roles in the onset and development of chronic inflammatory disorders such as atherosclerosis, diabetes, and cancer. Commonly, macrophages exist in two distinct subpopulations includes classically activated or M1 macrophages and alternatively activated or M2 macrophages. M1 macrophages secrete numerous pro-inflammatory cytokines, such as interleukin-1α (IL-1α), IL-1β, IL-6, IL-12, and cyclooxygenase-2 whereas M2 macrophages generate anti-inflammatory cytokines such as IL-10 and transforming growth factor-β (Shapouri-Moghaddam et al., 2018). Macrophages interact with major histocompatibility complex molecules to present antigen. Moreover, macrophages are implicated in possessing the phagocytosis of pathogens, debris, and dead cells.
Increasing evidence indicated that DNA methylation functions as a significant regulator of monocyte-macrophage phenotypes and functions (Shapouri-Moghaddam et al., 2018). A study compared genome-wide DNA methylation among monocytes and macrophages and found that differential DNA methylation was presented in monocyte to macrophage differentiation, majorly restricted to very short regions (Dekkers et al., 2019). In addition, the loss of DNA methylation was pronounced during monocytes to dendritic cells differentiation and modulated by TET2 (Klug et al., 2013). Furthermore, macrophages polarization and inflammation are regulated by DNMT3b. The expression of DNMT3b was significantly lower in M2 macrophages compared with M1 macrophages. Notably, DNMT3b knockdown causes macrophage polarization to M2 phenotype and inhibits inflammation, whereas DNMT3b overexpression promotes to M1 phenotype and aggravates inflammation. Mechanistically, DNMT3b is implicated in silencing the promoter of PPAR-γ1 (Yang et al., 2014). Besides, TET2 suppresses the expression of IL-6 in mouse macrophages (Hoeksema and de Winther, 2016).
Cardiovascular disease is the leading cause of death among the elderly. By 2050, the global population aging above 60 years will approximate 2.1 billion (Wyss-Coray, 2016). Therefore, the incidence and mortality of CVD will have a gradual increase. Vascular aging is a major trigger of many vascular disorders, while vascular cell senescence shows significant effect on the progression of CVD. Increasing evidence revealed the role of epigenetic mechanisms in vascular aging-related diseases. It’s well-known that abnormal DNA methylation and DNMTs expression are tightly associated with vascular diseases (Hiltunen et al., 2002). In this part, we mainly focus on the roles and mechanisms of DNA methylation in aging-related atherosclerosis, hypertension, and other vascular disorders. Besides, we discuss the connection between clonal hematopoiesis and CVD.
Atherosclerosis is a vascular disorder with complicated processes comprising ECs dysfunction, VSMCs proliferation and migration, macrophages inflammation, and collagen matrix accumulation. Age is a fundamental risk factor for atherosclerosis (Uryga and Bennett, 2016). DNA methylation plays a vital role in the initiation and progression of atherosclerosis. Besides, DNMTs act as regulators of vascular structure and functions (Table 2).
Endothelial cells senescence are accompanied by ECs dysfunction, decreased angiogenesis, and damaged eNOS activity, responsible for atherosclerosis progression (Colpani and Spinetti, 2019). Additionally, VSMCs are an important part of the fibrous cap and stromal cells of the artery (Chi et al., 2019). Accumulating evidence demonstrated that collagen secretion decreased in the aging of VSMCs, forming unstable fibrous caps (Gardner et al., 2015). VSMCs senescence enhances plaque vulnerability by secreting matrix metalloproteinases degrading the matrix and releases numerous pro-inflammatory cytokines promoting plaque inflammation and impairing the stability of plaques (Wang J. et al., 2015; Grootaert et al., 2018). Besides, Macrophages are defined as key modulators in atherosclerosis with aging and chronic inflammation (Moore and Tabas, 2011). Activated macrophages and foam cells trigger a cascade of inflammatory responses and induce atherosclerotic plaque formation. Notably, macrophages proliferation is the predominant mechanism in atherosclerotic plaques (Xu H. et al., 2019).
Numerous lines of evidence supported that atherosclerosis is manifested by global DNA hypomethylation and regional DNA hypermethylation. The level of DNA methylation in atherosclerotic arteries in rabbit models is lower compared to normal arteries (Laukkanen et al., 1999). Scholars confirmed that aberrant DNA methylation in atherosclerosis influence the transcription of key regulatory genes, inducing the pro-atherosclerotic cell phenotype (Castillo-Díaz et al., 2010). Gene hypermethylation including ER-α (Post et al., 1999; Huang et al., 2007), DDAH2 (Niu et al., 2014), and Foxp3 (Zhu et al., 2019) are tightly associated with the occurrence and development of atherosclerosis. Additionally, ROS accumulates in the aging of VSMCs and ECs (Chi et al., 2019; Tabaei and Tabaee, 2019), inducing DNA methylation changes via the altered activity of DNMTs and DNA damage, which modulates the formation and development of atherosclerotic plaques.
Hypertension, defined as average systolic blood pressure ≥140 mm Hg and/or average diastolic blood pressure ≥90 mm Hg, is the leading cause of CVD worldwide (Mills et al., 2020). Aging triggers a functional decline of various organ systems in the body. Vascular aging is significantly linked to hypertension prevalence and mortality among the elderly (Cheng et al., 2017; Donato et al., 2018). Besides, hypertension is an important risk factor for other vascular aging-related diseases, such as dementia, and cognitive decline (Cortes-Canteli and Iadecola, 2020; Fuchs and Whelton, 2020). Emerging documents demonstrated that DNA methylation exhibits a significant impact on hypertension development through modulating gene expression and vascular cell functions (Table 3).
It has been recognized that endothelial dysfunction contributed to the initiation and progression of hypertension. ECs release endothelin-1, prostacyclin, NO, and other vasoactive substances regulating vasoconstriction and relaxation (Sandoo et al., 2010). The dysfunction of aging ECs creates an imbalance between vasoconstriction and relaxation, leading to an increase in blood pressure (Jia et al., 2019). In addition, VSMCs mediate arterial compliance and total peripheral resistance (Chi et al., 2019). VSMCs senescence stimulates hypertension by the accumulation of oxidative stress and inflammation. Besides, vascular macrophages trigger endothelial dysfunction by increasing the expression of ROS and inflammatory cytokines, leading to vascular oxidative stress and blood pressure elevation (Justin Rucker and Crowley, 2017). The senescence of vascular cells promotes arterial stiffness, causing hypertension (Qiu et al., 2010; Chi et al., 2019).
DNA methylation is involved in hypertension (Gao et al., 2018; Guo et al., 2020; Amenyah et al., 2021). Kazmi et al. investigated the relationship between hypertension and DNA methylation in European men and discovered that 7 CpG sites were related to diastolic blood pressure (Kazmi et al., 2020). In addition, a study surveyed the association between blood pressure and DNA methylation among Europeans, Hispanics, and African Americans and identified 14 relevant CpG sites (Richard et al., 2017). Moreover, DNA methylation of the natriuretic peptide-A gene was decreased in patients diagnosed with hypertension among the Chinese community (Li J. et al., 2020). As we all know, the renin–angiotensin–aldosterone system (RAAS) is vital to the occurrence and progression of hypertension (Drummond et al., 2019). For instance, AT1aR promoter region hypomethylation in hypertensive rats upregulates the expression of AT1aR, critical to the development of hypertension (Pei et al., 2015). Administration of angiotensin receptor antagonists hinders the progression of hypertension in the early stages (Bogdarina et al., 2007). Besides, the level of 6mA is decreased in hypertensive mice and rat models, causing phenotypic transformation and migration of VSMCs (Guo et al., 2020).
Heart failure (HF) is a clinical syndrome caused by impairment of the systolic and diastolic functions. With the aging population, the incidence and mortality of HF are gradually increasing. In developed countries, the prevalence of HF among the elderly aged above 65 is approximately 11.8% (Groenewegen et al., 2020). Aging induces cardiovascular senescence and myocardial fibrosis (Horn and Trafford, 2016), leading to cardiac dysfunction and promoting HF progression (Triposkiadis et al., 2020). Notably, cardiovascular senescence, atherosclerosis (Triposkiadis et al., 2019), hypertension (Fuchs and Whelton, 2020), and ischemic cardiomyopathy (Napoli et al., 2020) are critical risk factors for HF (Li H. et al., 2020). DNA hypermethylation regulates cardiometabolism by destroying nuclear respiratory factor 1 dependent oxidative metabolism (Pepin et al., 2019). Additionally, in HF patients, the expression of DNMT3a and DNMT3b were upregulated, inhibiting the mRNA levels of several oxidative metabolism genes (Pepin et al., 2019). DNMT3b knockout can induce cardiac contractile insufficiency, ventricular wall thinning, and accelerating the deterioration of contractile function during HF (Vujic et al., 2015). Moreover, aberrant DNA methylation in dilated cardiomyopathies patients is associated with significant ADORA2A and LY75 mRNA expression changes, but not in HOXB13 and ERBB3 (Haas et al., 2013).
Acute myocardial infarction (AMI) primarily occurs based on atherosclerotic stenosis of the coronary arteries. It is attributed to certain triggers of plaque rupture, including platelets gathering on the surface of the ruptured plaque, forming a thrombus that suddenly blocks the lumen of the coronary artery, leading to myocardial ischemic necrosis. The hypermethylation of the ABO gene is seemingly linked with an increased risk of AMI in Pakistani (Yousuf et al., 2020). Besides, the hypomethylation of ZBTB12 gene and LINE-1 gene are early biomarkers of MI in peripheral blood white cells (Guarrera et al., 2015). Besides, the methylation of ZFHX3 and SMARCA4 are independently and significantly related to MI (Nakatochi et al., 2017).
Coronary heart disease (CHD) is a heart condition caused by atherosclerotic lesions in the coronary arteries, causing ischemia, hypoxia or necrosis of the myocardium. DNA methylation is connected with the risk of future CHD. ATP2B2, GUCA1B, CASR, and HPCAL1 are genes regulating calcium modulation (Agha et al., 2019). DNA methylation and hydroxymethylation among the elderly CHD patients were significantly upgraded. Report supported that a lower methylation level in the SOAT1 gene might enhance the risk of CHD (Abuzhalihan et al., 2019).
Cardiac fibrosis is defined as the accumulation of extracellular matrix proteins in the cardiac interstitium (Tao et al., 2014b). Several studies indicated that DNA methylation is related to the onset and development of tissue fibrosis. Increased RASAL1 and RASSF1A promoter methylation is involved in cardiac fibrosis (Tao et al., 2014a; Xu et al., 2015b). Besides, transforming growth factor-beta 1 can induce COL1A1 demethylation and collagen type I expression by suppressing the generation and activity of DNMT1 and DNMT3a (Pan et al., 2013).
Human aging is linked with an increased frequency of somatic mutations in hematopoietic system. This clonal hematopoiesis is associated with CVD (Evans et al., 2020). Age-related clonal hematopoiesis is major occurred in DNMT3a and TET2 and is associated with CVD (Jaiswal et al., 2014). TET2, highly expressed in murine macrophage differentiation, inhibits inflammatory gene expression in macrophages, reducing macrophages inflammation. In contrast, TET2-deficient macrophages affect phenotype of macrophages to promote chronic inflammation in vasculature, resulting in the progression of CVD (Cull et al., 2017). Studies in LDL receptor-deficient mice with TET2-deficient cells induce an increase in atherosclerotic plaque size. Besides, TET2-deficient macrophages enhance the secretion of interleukin-1β (IL-1β), regulated by inflammasome NLRP3 (Fuster et al., 2017). Additionally, DNMT3a is implicated in regulating inflammatory pathways and macrophage functions. Accumulating evidence revealed that hematopoietic DNMT3a mutation can promote HF through exacerbating inflammatory responses. HF Patients with monocytes carrying DNMT3a mutations show an increasing expression of inflammation genes, including IL-1B, IL-6, IL-8, NLRP3, CCL3, and CCL4, which may be contributing to exacerbating HF (Abplanalp et al., 2021). In addition, mice with mutations in TET2 or DNMT3a following an infusion of angiotensin II diminished cardiac function, increased fibrosis and inflammation (Sano et al., 2018). IL-1β was upregulated in TET2-deficient cells, while CXCL1 and CXCL2 were upregulated in DNMT3a-deficient cells. Research between clonal hematopoiesis and CVD is very much in its infancy. To date, only atherosclerosis and HF have been evaluated with clonal hematopoiesis in TET2 or DNMT3a mutation. Further studies should examine the role of clonal hematopoiesis in CVD beyond atherosclerosis and HF (Evans et al., 2020).
Cerebrovascular diseases are a group of diseases causing damage to brain tissue due to blood circulation disorder in the brain. Arterial stiffness and vascular aging trigger cerebrovascular dysfunction and blood–brain barrier contraction. Subsequently, a series of cerebrovascular diseases occur (Toth et al., 2017; Thorin-Trescases et al., 2018; Kalaria and Hase, 2019). Besides, small blood vessel diseases are common in the aging process, manifested as brain and parenchymal microcirculation changes (De Silva and Faraci, 2020), decreasing cerebral blood flow and damaging the blood–brain barrier, eventually lead to an aging-related functional decline of the brain (Iadecola et al., 2019). DNA methylation regulates various cerebrovascular diseases, such as stroke, dementia, and AD (Table 4).
Stroke is a leading cause of death and disability globally. Endothelial dysfunction and macrophages polarization contribute to stroke (Blum et al., 2012). Besides, the vascular stiffness of the elderly population increases with aging. Studies indicated that carotid artery stiffness is a crucial factor in the onset and development of stroke (Mattace-Raso et al., 2006; van Sloten et al., 2015). Pulse wave velocity is used to analyze age-related changes in vascular structure and function as well as to evaluate the endothelial function and vascular stiffness (Stoner et al., 2012). In a cohort study based on a Chinese community population, assessing vascular aging might help stroke risk assessment (Yang et al., 2019). DNA methylation changes with age and is linked to stroke during aging (Soriano-Tárraga et al., 2014). In contrast with healthy subjects, the methylation level of the TP53 promoter region increased among stroke patients (Wei et al., 2019). Besides, the methylation of MTRNR2L8 is a potential therapeutic target for stroke (Shen et al., 2019).
Dementia is a decline in intelligence that severely disrupts daily life (Iadecola et al., 2019). The aging population is crucial to age-related cognitive abilities and important public health challenges (Deak et al., 2016). Research estimated that by 2050, 115 million people will be globally diagnosed with dementia (Prince et al., 2013). Endothelial dysfunction and damage might cause neurovascular dysfunction, resulting in microvascular thrombosis and destruction of the blood–brain barrier (Yamazaki et al., 2016). AD is the most prevalent cause of dementia, while age is an independent risk factor for AD (Delaye et al., 1975), and it has been proved that the progression of AD is tightly related to the alteration of DNA methylation (Qazi et al., 2018; Huo et al., 2019). Although AD is a neurodegenerative disease, it is also attributed to cerebrovascular aging (Iturria-Medina et al., 2016), and most AD patients suffer from Aβ amyloid angiopathy (Love and Miners, 2016). Studies suggest that genes including ANK1 (Lunnon et al., 2014), RHBDF2 (De Jager et al., 2014), ABCA7 (Yamazaki et al., 2017), RPL13, and CDH23 were hypermethylated in AD (Prasad and Jho, 2019). Among them, ANK1, ABCA7, and RHBDF2 hypermethylation were associated with the formation of Aβ plaques. Higher DNA methylation levels in the promoter region of APOE promote the odds of dementia and AD (Karlsson et al., 2018). Besides, ANKRD30B is hypermethylated among AD patients, further implying that DNA methylation regulates the progression of AD (Semick et al., 2019).
Chronic kidney disease (CKD) refers to abnormalities in chronic kidney structure or function caused by various reasons for more than 3 months (Ingrosso and Perna, 2020). CKD is characterized by the development of renal fibrosis subsequent renal failure. Macrophage polarization and fibroblasts differentiate into myofibroblasts are central processes of renal fibrosis (Tang et al., 2019). CKD and renal fibrosis affect approximately 10% of the world’s population and half of the adults aged over 70 (Humphreys, 2018). DNA methylation changes in the renal cortex of patients with CKD (Chu et al., 2017). Besides, a study found that a low DNA methylation in patients with CKD (Zinellu et al., 2017). DNA methylation of genes is linked with CKD and renal fibrosis, including PTPN6, CEBPB, EBF1, Klotho (Chu et al., 2017; Yin et al., 2017), SMAD7 (Yang et al., 2020), sFRP5 (Yu et al., 2017), and RASAL1. Notably, DNA methylation inhibits erythropoietin expression, causing anemia, a prevalent complication of CKD (Yin and Blanchard, 2000).
Aging is an inevitable process and significantly associated with many vascular aging-related disorders. Vascular aging is the structural and functional changes of the vasculature, including vascular cells senescence, inflammation, oxidative stress, and calcification (Ding et al., 2020). With aging, there is a gradual increase in the prevalence and mortality of vascular aging-related diseases. A high percentage of vascular aging-related diseases progress to functional failure because no available drugs can reverse vascular aging progression. Therefore, there is an urgent need to discover tools and methods for the early diagnosis and treatment of vascular aging-related diseases. Improving vascular cell senescence might ameliorate vascular aging and related diseases, providing novel ideas for clinical research as well as new prevention and treatment for vascular aging-related diseases.
The evidence mentioned above reveals that aberrant DNA methylation modification exists in aging-related vascular diseases and might be a potential biomarker for the diagnosis and prognosis of vascular aging-related diseases (Liu and Tang, 2019). Gene at DNA methylation status based on monocyte/macrophage might work as a diagnostic biomarker for clinical application (Bakshi et al., 2019). Notably, hyperhomocysteine is associated with CVD, potentially influencing DNA methylation modification, suggesting that DNA methylation is a biomarker for the increased risk of CVD (Kim et al., 2010). For example, BRCA1 and CRISP2 specific site methylation changes are associated with atherosclerosis, indicating that differentially methylated regions of BRCA1 and CRISP2 emerge as biomarkers for CVD (Istas et al., 2017). Moreover, the DNA methylation levels of LINE-1 in the blood of 742 elderly men and discovered that LINE-1 hypomethylation is linked to elevated serum vascular cell adhesion molecule-1, related to atherosclerosis progression and high cardiovascular risk (Baccarelli et al., 2010). Additionally, Additionally, DNMT3a expression can be used as novel diagnostic biomarkers for cerebrovascular aging-related diseases (Martínez-Iglesias et al., 2020). Besides, the methylation status of LINE-1 and MTRNR2L8 act as epigenetic biomarkers in stroke patients (Baccarelli et al., 2010; Shen et al., 2019). Specific gene methylation of diseases, combined with changes in DNMTs and TETs levels, are diagnostic and prognostic biomarkers providing broad clinical prospects. Further studies should concentrate on the clinical application of DNA methylation as a potential biomarker of vascular aging-related diseases.
DNA methylation is a dynamically reversible process, providing potential therapeutic targets for delaying or enhancing vascular aging-related diseases. Several agents targeting epigenetic modulators are currently undergoing preclinical or clinical evaluation to treat cancers and might have potential applications in treating cardiovascular events (Gorabi et al., 2020). Drugs that inhibit DNMTs have identified to be promising treatment strategies for many vascular disorders. There are many natural and synthetic compounds able to suppress DNMT activity (Table 5).
Nutrition and dietary compounds are identified to regulate DNA methylation. Nutrients implicated in one-carbon metabolism, including folates (vitamin B9), vitamins B6 and B12, choline, and methionine, are involved in DNA methylation for their essential role in the generation of S-adenyl methionine, the methyl group donor for DNA methylation (Park et al., 2012). Folic acid and folate drugs are used as therapeutics. For instance, folic acid or vitamins B6 and B12 deficiencies can increase homocysteine levels, induce endothelial dysfunction, and aggravate atherosclerosis. On the contrary, dietary supplementation with folic acid can improve DNA methylation status, decrease inflammatory molecule levels, benefit atherosclerosis and reduce the risk of stroke (Wang et al., 2007; Hou and Zhao, 2021). Additionally, diet nutrition with folate and vitamin B6 has been recognized to improve memory and daily activities in AD patients (Chan et al., 2008). In addition, supplement methionine can increase DNA methylation (Waterland, 2006). Furthermore, vitamin C, as an antioxidant, can regulate the activity of TET and is involved in TET-mediated DNA methylation (DiTroia et al., 2019). Previous studies demonstrated that polyphenols [catechin, epicatechin, epigallocatechin-3-O-gallate (EGCG), and resveratrol] and bioflavonoids (quercetin, fisetin, and myricetin) inhibit DNA methylation by repressing DNMTs (Chistiakov et al., 2017). Catechin and epicatechin serve as DNMTs inhibitors through inhibiting human liver catechol-O-methyltransferase-mediated O-methylation of catechol estrogens (Nagai et al., 2004). EGCG is a major green tea polyphenol and can inhibit the activity of DNMTs such as DNMT1, DNMT3a, and DNMT3b (Wong et al., 2011). Besides, resveratrol has been recognized as DNMTs inhibitors to inhibit DNMTs expression (Aldawsari et al., 2016). Quercetin can protect against nickel-induced liver injury by suppressing DNMTs activity and decreasing the DNA methylation level of the NF-E2 related factor 2 (Liu C. M. et al., 2015). The ability of dietary nutrition offers promising therapies for vascular aging-related diseases by modulating DNMTs activities and DNA methylation.
DNA methyltransferases inhibitors are divided into two classes included nucleoside analogs inhibitors and non-nucleoside analogs inhibitors (Nicorescu et al., 2019). Nucleoside analogs inhibitors can incorporate into DNA during cell cycle and sequester DNMTs by regulating their proteasomal degradation. 5-azacytosine (azacytidine) and 5-Aza- 2′-deoxycytidine (5Aza, decitabine), DNA hypomethylation agents, may beneficial in CVD, cerebrovascular disease, and kidney diseases. Azacytidine serves as a DNMT1 inhibitor, upregulated the expression of PTEN, reduced inflammatory factors secretion, and inhibited platelet-derived growth factor stimulated SMCs de-differentiation (Strand et al., 2020). Besides, silencing DNMTs in ECs by 5Aza or siRNA, decreased the level of DNA methylation and attenuated ECs inflammation. Administration of 5Aza in mouse atherosclerosis models decreases the formation of atherosclerotic lesions and promotes the prognosis of atherosclerosis (Dunn et al., 2014). Administering 5Aza combined with specific task training helps recover chronic stroke [144]. Decitabine can ameliorate atherosclerotic lesion, inhibit DNMT1 activity, and downregulate global DNA methylation level (Zhuang et al., 2017). Non-nucleoside analogs such as RG108 and hydralazine are developed to overcome the non-specificity and cytotoxicity of the nucleoside inhibitors (Nicorescu et al., 2019). RG108 can inhibit DNMT1 activity by binding to its active site. Stenzig et al. (2018) indicated that RG108 significantly attenuated global DNA methylation in cardiomyocytes and is associated with decreased cardiac hypertrophy. Hydralazine, an anti-hypertensive drug, can repress DNMT1 and DNMT3a mRNA expression and activity by suppressing Erk signaling pathway (Deng et al., 2003). Hydralazine-mediated the promoter region of SERCA2a demethylation can improve cardiac function (Kao et al., 2011).
Diabetes is characterized by hyperglycemia and act as a significantly risk factor of cardiovascular and kidney disorders. Numerous lines of evidence supported that diabetes is linked with DNA methylation (Salameh et al., 2020). Therefore, DNA methylation-based therapies can benefit diabetes and reduce diabetes-related vascular complications. Cells treated with high glucose exhibited lower DNA methylation levels of NF-κB and SOD2, while co-treatment with GLP-1 agonists reversed these effects by decreasing DNMT1 and DNMT3a mRNA and protein levels and increasing TET2 mRNA and protein levels (Scisciola et al., 2020). Additionally, it has been reported that SGLT2 gene is hypomethylated in kidney proximal tubules, which suggest that SGLT2 inhibitors may be a DNA methylation modulators (Marumo et al., 2015).
Therefore, a better understanding of the mechanisms and roles of DNA methylation in the physiological and pathological process of vascular aging-related diseases might lead to the identification of promising biomarkers and therapeutic drugs. Much work is needed to validate the potential application of DNA methylation in vascular disorders.
With the increase in the aging population, there is an urgent need to identify reliable and effective diagnostic tools and therapies for early diagnosis and treatment of vascular aging. Endothelial dysfunction, VSMCs proliferation and migration, macrophages polarization, and aberrant DNA methylation are implicated in the development and progression of vascular inflammation and vascular aging-related diseases. Notably, DNA methylation is involved in regulating the expression of genes in these mechanisms. In this review, we summarize the cellular and functional alterations in the vascular system during the aging process and concentrated on the roles and mechanisms of DNA methylation in vascular aging-related CVD, cerebrovascular diseases, and kidney diseases. DNA methylation plays a vital role in vascular aging progression and might be a potential biomarker for diagnosis and therapeutic target in treating vascular aging-related diseases. DNA methylation promotes a better understanding of the underlying mechanisms of vascular aging and related diseases and ultimately facilitates the development of effective interventions for vascular aging-related disorders. Regarding the future clinical prospect of DNA methylation, numerous biological explorations should be conducted to clarify the feasibility of DNA methylation in the diagnosis, treatment, and prognosis of vascular aging-related diseases.
HX collected the literature and wrote the manuscript. SL drew the figures and supervised the manuscript. Y-SL conceived the idea and had been involved in manuscript conception and drafting. All authors read and approved the final manuscript.
This work was supported by the National Natural Science Foundation of China (Nos. 82071593, 81770833, and 81974223) and National Key R&D (or Research and Development) Program of China (Grant 2020YFC 2009000).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
ECs, endothelial cells; VSMCs, vascular smooth muscle cells; CVD, cardiovascular disease; AD, Alzheimer’s disease; 4mC, N4-methylcytosine; 5mC, N5-methylcytosine; 6mA, N6-methyladenine; DNMTs, DNA methyltransferases; TETs, 10–11 translocations; 5hmC, 5-hydroxymethylcytosine; 5fC, 5-formylcytosine; 5caC, 5-carboxylcytosine; TDG, thymine DNA glycosylase; BER, base excision repair; ox-LDL, oxidized low-density lipoprotein; ROS, reactive oxygen species; LDL, low-density lipoprotein; eNOS, endothelial nitric oxide synthase; IL-1α, interleukin-1 α; 5Aza, 5-aza-2 ′ -deoxycytidine; RAAS, rennin–angiotensin–aldosterone system; HF, heart failure; AMI, acute myocardial infarction; CHD, coronary heart disease; CKD, chronic kidney disease; EGCG, epigallocatechin-3-O-gallate.
Abplanalp, W. T., Cremer, S., John, D., Hoffmann, J., Schuhmacher, B., Merten, M., et al. (2021). Clonal hematopoiesis-driver DNMT3A mutations alter immune cells in heart failure. Circ. Res. 128, 216–228. doi: 10.1161/circresaha.120.317104
Abuzhalihan, J., Wang, Y. T., Ma, Y. T., Fu, Z. Y., Yang, Y. N., Ma, X., et al. (2019). SOAT1 methylation is associated with coronary heart disease. Lipids Health Dis. 18:192.
Agha, G., Mendelson, M. M., Ward-Caviness, C. K., Joehanes, R., Huan, T., Gondalia, R., et al. (2019). Blood Leukocyte DNA methylation predicts risk of future myocardial infarction and coronary heart disease. Circulation 140, 645–657. doi: 10.1161/CIRCULATIONAHA.118.039357
Aldawsari, F. S., Aguayo-Ortiz, R., Kapilashrami, K., Yoo, J., Luo, M., Medina-Franco, J. L., et al. (2016). Resveratrol-salicylate derivatives as selective DNMT3 inhibitors and anticancer agents. J. Enzyme Inhib. Med. Chem. 31, 695–703. doi: 10.3109/14756366.2015.1058256
Alikhani-Koopaei, R., Fouladkou, F., Frey, F. J., and Frey, B. M. (2004). Epigenetic regulation of 11 beta-hydroxysteroid dehydrogenase type 2 expression. J. Clin. Invest. 114, 1146–1157. doi: 10.1172/jci21647
Amenyah, S. D., Ward, M., McMahon, A., Deane, J., McNulty, H., Hughes, C., et al. (2021). DNA methylation of hypertension-related genes and effect of riboflavin supplementation in adults stratified by genotype for the MTHFR C677T polymorphism. Int. J. Cardiol. 322, 233–239. doi: 10.1016/j.ijcard.2020.09.011
An, S. J., Liu, P., Shao, T. M., Wang, Z. J., Lu, H. G., Jiao, Z., et al. (2015). Characterization and functions of vascular adventitial fibroblast subpopulations. Cell Physiol. Biochem. 35, 1137–1150. doi: 10.1159/000373939
Azechi, T., Sato, F., Sudo, R., and Wachi, H. (2014). 5-aza-2’-Deoxycytidine, a DNA methyltransferase inhibitor, facilitates the inorganic phosphorus-induced mineralization of vascular smooth muscle cells. J. Atheroscler. Thromb. 21, 463–476. doi: 10.5551/jat.20818
Baccarelli, A., Tarantini, L., Wright, R. O., Bollati, V., Litonjua, A. A., Zanobetti, A., et al. (2010). Repetitive element DNA methylation and circulating endothelial and inflammation markers in the VA normative aging study. Epigenetics 5, 222–228. doi: 10.4161/epi.5.3.11377
Bakshi, C., Vijayvergiya, R., and Dhawan, V. (2019). Aberrant DNA methylation of M1-macrophage genes in coronary artery disease. Sci. Rep. 9:1429.
Bao, X. J., Mao, S. Q., Gu, T. L., Zheng, S. Y., Zhao, J. S., and Zhang, L. N. (2018). Hypomethylation of the interferon γ gene as a potential Risk factor for essential hypertension: a case-control study. Tohoku J. Exp. Med. 244, 283–290. doi: 10.1620/tjem.244.283
Bechtel, W., McGoohan, S., Zeisberg, E. M., Müller, G. A., Kalbacher, H., Salant, D. J., et al. (2010). Methylation determines fibroblast activation and fibrogenesis in the kidney. Nat. Med. 16, 544–550. doi: 10.1038/nm.2135
Bitar, M. S. (2019). Diabetes impairs angiogenesis and induces endothelial cell senescence by Up-regulating thrombospondin-CD47-dependent signaling. Int. J. Mol. Sci. 20:673. doi: 10.3390/ijms20030673
Blum, A., Vaispapir, V., Keinan-Boker, L., Soboh, S., Yehuda, H., and Tamir, S. (2012). Endothelial dysfunction and procoagulant activity in acute ischemic stroke. J. Vasc. Interv. Neurol. 5, 33–39.
Bochtler, M., Kolano, A., and Xu, G. L. (2017). DNA demethylation pathways: additional players and regulators. Bioessays 39, 1–13. doi: 10.1002/bies.201600178
Bogdanović, O., and Veenstra, G. J. (2009). DNA methylation and methyl-CpG binding proteins: developmental requirements and function. Chromosoma 118, 549–565. doi: 10.1007/s00412-009-0221-9
Bogdarina, I., Welham, S., King, P. J., Burns, S. P., and Clark, A. J. (2007). Epigenetic modification of the renin-angiotensin system in the fetal programming of hypertension. Circ. Res. 100, 520–526. doi: 10.1161/01.res.0000258855.60637.58
Boström, A. E., Mwinyi, J., Voisin, S., Wu, W., Schultes, B., Zhang, K., et al. (2016). Longitudinal genome-wide methylation study of Roux-en-Y gastric bypass patients reveals novel CpG sites associated with essential hypertension. BMC Med. Genom. 9:20. doi: 10.1186/s12920-016-0180-y
Brunet, A., and Berger, S. L. (2014). Epigenetics of aging and aging-related disease. J. Gerontol. A Biol. Sci. Med. Sci. 69, S17–S20. doi: 10.1093/gerona/glu042
Cao, Q., Wang, X., Jia, L., Mondal, A. K., Diallo, A., Hawkins, G. A., et al. (2014). Inhibiting DNA Methylation by 5-Aza-2’-deoxycytidine ameliorates atherosclerosis through suppressing macrophage inflammation. Endocrinology 155, 4925–4938. doi: 10.1210/en.2014-1595
Castillo-Díaz, S. A., Garay-Sevilla, M. E., Hernández-González, M. A., Solís-Martínez, M. O., and Zaina, S. (2010). Extensive demethylation of normally hypermethylated CpG islands occurs in human atherosclerotic arteries. Int. J. Mol. Med. 26, 691–700. doi: 10.3892/ijmm-00000515
Cau, S. B., Carneiro, F. S., and Tostes, R. C. (2012). Differential modulation of nitric oxide synthases in aging: therapeutic opportunities. Front. Physiol. 3:218. doi: 10.3389/fphys.2012.00218
Chan, A., Paskavitz, J., Remington, R., Rasmussen, S., and Shea, T. B. (2008). Efficacy of a vitamin/nutriceutical formulation for early-stage Alzheimer’s disease: a 1-year, open-label pilot study with an 16-month caregiver extension. Am. J. Alzheimers Dis. 23, 571–585. doi: 10.1177/1533317508325093
Chan, Y., Fish, J. E., D’Abreo, C., Lin, S., Robb, G. B., Teichert, A. M., et al. (2004). The cell-specific expression of endothelial nitric-oxide synthase: a role for DNA methylation. J. Biol. Chem. 279, 35087–35100. doi: 10.1074/jbc.M405063200
Chen, Z. X., and Riggs, A. D. (2011). DNA methylation and demethylation in mammals. J. Biol. Chem. 286, 18347–18353. doi: 10.1074/jbc.r110.205286
Cheng, H. M., Park, S., Huang, Q., Hoshide, S., Wang, J. G., Kario, K., et al. (2017). Vascular aging and hypertension: implications for the clinical application of central blood pressure. Int. J. Cardiol. 230, 209–213. doi: 10.1016/j.ijcard.2016.12.170
Chi, C., Li, D. J., Jiang, Y. J., Tong, J., Fu, H., Wu, Y. H., et al. (2019). Vascular smooth muscle cell senescence and age-related diseases: state of the art. Biochim. Biophys. Acta Mol. Basis Dis. 1865, 1810–1821. doi: 10.1016/j.bbadis.2018.08.015
Chistiakov, D. A., Orekhov, A. N., and Bobryshev, Y. V. (2017). Treatment of cardiovascular pathology with epigenetically active agents: focus on natural and synthetic inhibitors of DNA methylation and histone deacetylation. Int. J. Cardiol. 227, 66–82. doi: 10.1016/j.ijcard.2016.11.204
Cho, H. M., Lee, H. A., Kim, H. Y., Han, H. S., and Kim, I. K. (2011). Expression of Na+-K+ -2Cl- cotransporter 1 is epigenetically regulated during postnatal development of hypertension. Am. J. Hypertens. 24, 1286–1293. doi: 10.1038/ajh.2011.136
Chu, A. Y., Tin, A., Schlosser, P., Ko, Y. A., Qiu, C., Yao, C., et al. (2017). Epigenome-wide association studies identify DNA methylation associated with kidney function. Nat. Commun. 8:1286. doi: 10.1038/s41467-017-01297-7
Colpani, O., and Spinetti, G. (2019). MicroRNAs orchestrating senescence of endothelial and vascular smooth muscle cells. Vasc. Biol. 1, H75–H81. doi: 10.1530/VB-19-0017
Cortes-Canteli, M., and Iadecola, C. (2020). Alzheimer’s disease and vascular aging: JACC focus seminar. J. Am. Coll. Cardiol. 75, 942–951. doi: 10.1016/j.jacc.2019.10.062
Cull, A. H., Snetsinger, B., Buckstein, R., Wells, R. A., and Rauh, M. J. (2017). Tet2 restrains inflammatory gene expression in macrophages. Exp. Hematol. 55, 56–70.e13. doi: 10.1016/j.exphem.2017.08.001
Dasgupta, C., Chen, M., Zhang, H., Yang, S., and Zhang, L. (2012). Chronic hypoxia during gestation causes epigenetic repression of the estrogen receptor-α gene in ovine uterine arteries via heightened promoter methylation. Hypertension 60, 697–704. doi: 10.1161/hypertensionaha.112.198242
De Jager, P. L., Srivastava, G., Lunnon, K., Burgess, J., Schalkwyk, L. C., Yu, L., et al. (2014). Alzheimer’s disease: early alterations in brain DNA methylation at ANK1, BIN1, RHBDF2 and other loci. Nat. Neurosci. 17, 1156–1163. doi: 10.1038/nn.3786
De Silva, T. M., and Faraci, F. M. (2020). Contributions of aging to cerebral small vessel disease. Annu. Rev. Physiol. 82, 275–295. doi: 10.1146/annurev-physiol-021119-034338
Deak, F., Freeman, W. M., Ungvari, Z., Csiszar, A., and Sonntag, W. E. (2016). Recent developments in understanding brain aging: implications for Alzheimer’s disease and vascular cognitive impairment. J. Gerontol. A Biol. Sci. Med. Sci. 71, 13–20. doi: 10.1093/gerona/glv206
Dekkers, K. F., Neele, A. E., Jukema, J. W., Heijmans, B. T., and de Winther, M. P. J. (2019). Human monocyte-to-macrophage differentiation involves highly localized gain and loss of DNA methylation at transcription factor binding sites. Epigenetics Chromatin 12:34.
Delaye, J., Pourchaire, J., and Gonin, A. (1975). [Hemodynamic effects of the intravenous form of acebutolol]. Nouv. Presse Med. 46(Suppl.), 3239–3243.
Deng, C., Lu, Q., Zhang, Z., Rao, T., Attwood, J., Yung, R., et al. (2003). Hydralazine may induce autoimmunity by inhibiting extracellular signal-regulated kinase pathway signaling. Arthritis Rheum. 48, 746–756. doi: 10.1002/art.10833
Ding, Q., Shao, C., Rose, P., and Zhu, Y. Z. (2020). Epigenetics and vascular senescence-potential new therapeutic targets? Front. Pharmacol. 11:535395. doi: 10.3389/fphar.2020.535395
DiTroia, S. P., Percharde, M., Guerquin, M. J., Wall, E., Collignon, E., Ebata, K. T., et al. (2019). Maternal vitamin C regulates reprogramming of DNA methylation and germline development. Nature 573, 271–275. doi: 10.1038/s41586-019-1536-1
Donato, A. J., Machin, D. R., and Lesniewski, L. A. (2018). Mechanisms of dysfunction in the aging vasculature and role in age-related disease. Circ. Res. 123, 825–848 doi: 10.1161/circresaha.118.312563
Drummond, G. R., Vinh, A., Guzik, T. J., and Sobey, C. G. (2019). Immune mechanisms of hypertension. Nat. Rev. Immunol. 19, 517–532. doi: 10.1038/s41577-019-0160-5
Dunn, J., Qiu, H., Kim, S., Jjingo, D., Hoffman, R., Kim, C. W., et al. (2014). Flow-dependent epigenetic DNA methylation regulates endothelial gene expression and atherosclerosis. J. Clin. Invest. 124, 3187–3199. doi: 10.1172/jci74792
Dunn, J., Thabet, S., and Jo, H. (2015). Flow-dependent epigenetic DNA methylation in endothelial gene expression and atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 35, 1562–1569. doi: 10.1161/atvbaha.115.305042
Ehrlich, M., Gama-Sosa, M. A., Carreira, L. H., Ljungdahl, L. G., Kuo, K. C., and Gehrke, C. W. (1985). DNA methylation in thermophilic bacteria: N4-methylcytosine, 5-methylcytosine, and N6-methyladenine. Nucleic Acids Res. 13, 1399–1412. doi: 10.1093/nar/13.4.1399
Ehrlich, M., Wilson, G. G., Kuo, K. C., and Gehrke, C. W. (1987). N4-methylcytosine as a minor base in bacterial DNA. J. Bacteriol. 169, 939–943. doi: 10.1128/jb.169.3.939-943.1987
Evans, M. A., Sano, S., and Walsh, K. (2020). Cardiovascular disease, aging, and clonal hematopoiesis. Annu. Rev. Pathol. 15, 419–438. doi: 10.1146/annurev-pathmechdis-012419-032544
Fan, R., Mao, S. Q., Gu, T. L., Zhong, F. D., Gong, M. L., Hao, L. M., et al. (2017). Preliminary analysis of the association between methylation of the ACE2 promoter and essential hypertension. Mol. Med. Rep. 15, 3905–3911. doi: 10.3892/mmr.2017.6460
Fan, R., Mao, S., Zhong, F., Gong, M., Yin, F., Hao, L., et al. (2015). Association of AGTR1 promoter methylation levels with essential hypertension risk: a matched case-control study. Cytogenet. Genome Res. 147, 95–102. doi: 10.1159/000442366
Fisslthaler, B., Zippel, N., Abdel Malik, R., Delgado Lagos, F., Zukunft, S., Thoele, J., et al. (2019). Myeloid-specific deletion of the AMPKα2 subunit alters monocyte protein expression and atherogenesis. Int. J. Mol. Sci. 20:3005. doi: 10.3390/ijms20123005
Frismantiene, A., Philippova, M., Erne, P., and Resink, T. J. (2018). Smooth muscle cell-driven vascular diseases and molecular mechanisms of VSMC plasticity. Cell. Signal. 52, 48–64. doi: 10.1016/j.cellsig.2018.08.019
Fu, Y., Luo, G. Z., Chen, K., Deng, X., Yu, M., Han, D., et al. (2015). N6-methyldeoxyadenosine marks active transcription start sites in Chlamydomonas. Cell 161, 879–892. doi: 10.1016/j.cell.2015.04.010
Fuchs, F. D., and Whelton, P. K. (2020). High blood pressure and cardiovascular disease. Hypertension 75, 285–292.
Fuster, J. J., MacLauchlan, S., Zuriaga, M. A., Polackal, M. N., Ostriker, A. C., Chakraborty, R., et al. (2017). Clonal hematopoiesis associated with TET2 deficiency accelerates atherosclerosis development in mice. Science 355, 842–847. doi: 10.1126/science.aag1381
Gallego-Fabrega, C., Carrera, C., Reny, J. L., Fontana, P., Slowik, A., Pera, J., et al. (2016a). PPM1A methylation is associated with vascular recurrence in aspirin-treated patients. Stroke 47, 1926–1929. doi: 10.1161/strokeaha.116.013340
Gallego-Fabrega, C., Carrera, C., Reny, J. L., Fontana, P., Slowik, A., Pera, J., et al. (2016b). TRAF3 epigenetic regulation is associated with vascular recurrence in patients with ischemic stroke. Stroke 47, 1180–1186. doi: 10.1161/strokeaha.115.012237
Gao, X., Colicino, E., Shen, J., Just, A. C., Nwanaji-Enwerem, J. C., Wang, C., et al. (2018). Accelerated DNA methylation age and the use of antihypertensive medication among older adults. Aging 10, 3210–3228. doi: 10.18632/aging.101626
Gardner, S. E., Humphry, M., Bennett, M. R., and Clarke, M. C. (2015). Senescent vascular smooth muscle cells drive inflammation through an interleukin-1α-dependent senescence-associated secretory phenotype. Arterioscler. Thromb. Vasc. Biol. 35, 1963–1974. doi: 10.1161/atvbaha.115.305896
Gorabi, A. M., Penson, P. E., Banach, M., Motallebnezhad, M., Jamialahmadi, T., and Sahebkar, A. (2020). Epigenetic control of atherosclerosis via DNA methylation: a new therapeutic target? Life Sci. 253:117682. doi: 10.1016/j.lfs.2020.117682
Gori, T. (2018). Endothelial function: a short guide for the interventional cardiologist. Int. J. Mol. Sci. 19:3838. doi: 10.3390/ijms19123838
Greer, E. L., Blanco, M. A., Gu, L., Sendinc, E., Liu, J., Aristizábal-Corrales, D., et al. (2015). DNA methylation on N6-Adenine in C. elegans. Cell 161, 868–878. doi: 10.1016/j.cell.2015.04.005
Groenewegen, A., Rutten, F. H., Mosterd, A., and Hoes, A. W. (2020). Epidemiology of heart failure. Eur. J. Heart Fail. 22, 1342–1356.
Grootaert, M. O. J., Moulis, M., Roth, L., Martinet, W., Vindis, C., Bennett, M. R., et al. (2018). Vascular smooth muscle cell death, autophagy and senescence in atherosclerosis. Cardiovasc. Res. 114, 622–634. doi: 10.1093/cvr/cvy007
Guarrera, S., Fiorito, G., Onland-Moret, N. C., Russo, A., Agnoli, C., Allione, A., et al. (2015). Gene-specific DNA methylation profiles and LINE-1 hypomethylation are associated with myocardial infarction risk. Clin. Epigenet. 7:133.
Guay, S. P., Brisson, D., Lamarche, B., Marceau, P., Vohl, M. C., Gaudet, D., et al. (2013). DNA methylation variations at CETP and LPL gene promoter loci: new molecular biomarkers associated with blood lipid profile variability. Atherosclerosis 228, 413–420. doi: 10.1016/j.atherosclerosis.2013.03.033
Guo, Y., Pei, Y., Li, K., Cui, W., and Zhang, D. (2020). DNA N(6)-methyladenine modification in hypertension. Aging 12, 6276–6291. doi: 10.18632/aging.103023
Haas, J., Frese, K. S., Park, Y. J., Keller, A., Vogel, B., Lindroth, A. M., et al. (2013). Alterations in cardiac DNA methylation in human dilated cardiomyopathy. EMBO Mol. Med. 5, 413–429.
Heidenreich, P. A., Trogdon, J. G., Khavjou, O. A., Butler, J., Dracup, K., Ezekowitz, M. D., et al. (2011). Forecasting the future of cardiovascular disease in the United States: a policy statement from the American Heart Association. Circulation 123, 933–944. doi: 10.1161/cir.0b013e31820a55f5
Hiltunen, M. O., Turunen, M. P., Häkkinen, T. P., Rutanen, J., Hedman, M., Mäkinen, K., et al. (2002). DNA hypomethylation and methyltransferase expression in atherosclerotic lesions. Vasc. Med. 7, 5–11. doi: 10.1191/1358863x02vm418oa
Hoeksema, M. A., and de Winther, M. P. (2016). Epigenetic regulation of monocyte and macrophage function. Antioxid. Redox. Signal. 25, 758–774. doi: 10.1089/ars.2016.6695
Horn, M. A., and Trafford, A. W. (2016). Aging and the cardiac collagen matrix: novel mediators of fibrotic remodelling. J. Mol. Cell Cardiol. 93, 175–185. doi: 10.1016/j.yjmcc.2015.11.005
Hou, H., and Zhao, H. (2021). Epigenetic factors in atherosclerosis: DNA methylation, folic acid metabolism, and intestinal microbiota. Clin. Chim. Acta 512, 7–11. doi: 10.1016/j.cca.2020.11.013
Hua, X., Chen, L. M., Zhu, Q., Hu, W., Lin, C., Long, Z. Q., et al. (2019). Efficacy of controlled-release oxycodone for reducing pain due to oral mucositis in nasopharyngeal carcinoma patients treated with concurrent chemoradiotherapy: a prospective clinical trial. Support Care Cancer 27, 3759–3767. doi: 10.1007/s00520-019-4643-5
Huang, Y., Peng, K., Su, J., Huang, Y., Xu, Y., and Wang, S. (2007). Different effects of homocysteine and oxidized low density lipoprotein on methylation status in the promoter region of the estrogen receptor alpha gene. Acta Biochim. Biophys. Sin. 39, 19–26.
Huo, Z., Zhu, Y., Yu, L., Yang, J., De Jager, P., Bennett, D. A., et al. (2019). DNA methylation variability in Alzheimer’s disease. Neurobiol. Aging 76, 35–44.
Iadecola, C., Duering, M., Hachinski, V., Joutel, A., Pendlebury, S. T., Schneider, J. A., et al. (2019). Vascular cognitive impairment and dementia: JACC scientific expert panel. J. Am. Coll. Cardiol. 73, 3326–3344.
Ingrosso, D., and Perna, A. F. (2020). DNA methylation dysfunction in chronic kidney disease. Genes 11:811. doi: 10.3390/genes11070811
Istas, G., Declerck, K., Pudenz, M., Szic, K. S. V., Lendinez-Tortajada, V., Leon-Latre, M., et al. (2017). Identification of differentially methylated BRCA1 and CRISP2 DNA regions as blood surrogate markers for cardiovascular disease. Sci. Rep. 7:5120.
Iturria-Medina, Y., Sotero, R. C., Toussaint, P. J., Mateos-Pérez, J. M., and Evans, A. C. (2016). Early role of vascular dysregulation on late-onset Alzheimer’s disease based on multifactorial data-driven analysis. Nat. Commun. 7:11934.
Jaiswal, S., Fontanillas, P., Flannick, J., Manning, A., Grauman, P. V., Mar, B. G., et al. (2014). Age-related Clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 371, 2488–2498.
Jia, G., Aroor, A. R., Jia, C., and Sowers, J. R. (2019). Endothelial cell senescence in aging-related vascular dysfunction. Biochim. Biophys. Acta Mol. Basis Dis. 1865, 1802–1809. doi: 10.1016/j.bbadis.2018.08.008
Jiang, Y. Z., Jiménez, J. M., Ou, K., McCormick, M. E., Zhang, L. D., and Davies, P. F. (2014). Hemodynamic disturbed flow induces differential DNA methylation of endothelial Kruppel-Like Factor 4 promoter in vitro and in vivo. Circ. Res. 115, 32–43. doi: 10.1161/circresaha.115.303883
Jones, P. A. (2012). Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 13, 484–492. doi: 10.1038/nrg3230
Justin Rucker, A., and Crowley, S. D. (2017). The role of macrophages in hypertension and its complications. Pflugers Arch. 469, 419–430. doi: 10.1007/s00424-017-1950-x
Kalaria, R. N., and Hase, Y. (2019). Neurovascular ageing and age-related diseases. Subcell. Biochem. 91, 477–499. doi: 10.1007/978-981-13-3681-2_17
Kao, Y. H., Cheng, C. C., Chen, Y. C., Chung, C. C., Lee, T. I., Chen, S. A., et al. (2011). Hydralazine-induced promoter demethylation enhances sarcoplasmic reticulum Ca2+ -ATPase and calcium homeostasis in cardiac myocytes. Lab. Invest. 91, 1291–1297. doi: 10.1038/labinvest.2011.92
Karlsson, I. K., Ploner, A., Wang, Y., Gatz, M., Pedersen, N. L., and Hägg, S. (2018). Apolipoprotein E DNA methylation and late-life disease. Int. J. Epidemiol. 47, 899–907. doi: 10.1093/ije/dyy025
Kazmi, N., Elliott, H. R., Burrows, K., Tillin, T., Hughes, A. D., Chaturvedi, N., et al. (2020). Associations between high blood pressure and DNA methylation. PLoS One 15:e0227728. doi: 10.1371/journal.pone.0227728
Khor, E. S., and Wong, P. F. (2020). The roles of MTOR and miRNAs in endothelial cell senescence. Biogerontology 21, 517–530. doi: 10.1007/s10522-020-09876-w
Kim, J., Kim, J. Y., Song, K. S., Lee, Y. H., Seo, J. S., Jelinek, J., et al. (2007). Epigenetic changes in estrogen receptor beta gene in atherosclerotic cardiovascular tissues and in-vitro vascular senescence. Biochim. Biophys. Acta 1772, 72–80. doi: 10.1016/j.bbadis.2006.10.004
Kim, M., Long, T. I., Arakawa, K., Wang, R., Yu, M. C., and Laird, P. W. (2010). DNA methylation as a biomarker for cardiovascular disease risk. PLoS One 5:e9692. doi: 10.1371/journal.pone.0009692
Kim, Y. R., Kim, C. S., Naqvi, A., Kumar, A., Kumar, S., Hoffman, T. A., et al. (2012). Epigenetic upregulation of p66shc mediates low-density lipoprotein cholesterol-induced endothelial cell dysfunction. Am. J. Physiol. Heart Circ. Physiol. 303, H189–H196.
Klug, M., Schmidhofer, S., Gebhard, C., Andreesen, R., and Rehli, M. (2013). 5-Hydroxymethylcytosine is an essential intermediate of active DNA demethylation processes in primary human monocytes. Genome Biol. 14:R46.
Krüger-Genge, A., Blocki, A., Franke, R. P., and Jung, F. (2019). Vascular endothelial cell biology: an update. Int. J. Mol. Sci. 20:4411. doi: 10.3390/ijms20184411
Kumar, A., Kumar, S., Vikram, A., Hoffman, T. A., Naqvi, A., Lewarchik, C. M., et al. (2013). Histone and DNA methylation-mediated epigenetic downregulation of endothelial Kruppel-like factor 2 by low-density lipoprotein cholesterol. Arterioscler. Thromb. Vasc. Biol. 33, 1936–1942. doi: 10.1161/atvbaha.113.301765
Lacolley, P., Regnault, V., and Avolio, A. P. (2018). Smooth muscle cell and arterial aging: basic and clinical aspects. Cardiovasc. Res. 114, 513–528. doi: 10.1093/cvr/cvy009
Lagarkova, M. A., Volchkov, P. Y., Philonenko, E. S., and Kiselev, S. L. (2008). Efficient differentiation of hESCs into endothelial cells in vitro is secured by epigenetic changes. Cell Cycle 7, 2929–2935. doi: 10.4161/cc.7.18.6700
Laina, A., Stellos, K., and Stamatelopoulos, K. (2018). Vascular ageing: underlying mechanisms and clinical implications. Exp. Gerontol. 109, 16–30. doi: 10.1016/j.exger.2017.06.007
Lakatta, E. G., and Levy, D. (2003a). Arterial and cardiac aging: major shareholders in cardiovascular disease enterprises: Part I: aging arteries: a “set up” for vascular disease. Circulation 107, 139–146. doi: 10.1161/01.cir.0000048892.83521.58
Lakatta, E. G., and Levy, D. (2003b). Arterial and cardiac aging: major shareholders in cardiovascular disease enterprises: Part II: the aging heart in health: links to heart disease. Circulation 107, 346–354. doi: 10.1161/01.cir.0000048893.62841.f7
Laukkanen, M. O., Mannermaa, S., Hiltunen, M. O., Aittomäki, S., Airenne, K., Jänne, J., et al. (1999). Local hypomethylation in atherosclerosis found in rabbit ec-sod gene. Arterioscler. Thromb. Vasc. Biol. 19, 2171–2178.
Law, J. A., and Jacobsen, S. E. (2010). Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11, 204–220. doi: 10.1038/nrg2719
Levy, E., Spahis, S., Bigras, J. L., Delvin, E., and Borys, J. M. (2017). The epigenetic machinery in vascular dysfunction and hypertension. Curr. Hypertens Rep. 19:52.
Li, H., Hastings, M. H., Rhee, J., Trager, L. E., Roh, J. D., and Rosenzweig, A. (2020). Targeting age-related pathways in heart failure. Circ. Res. 126, 533–551. doi: 10.1161/circresaha.119.315889
Li, J., Zhu, J., Ren, L., Ma, S., Shen, B., Yu, J., et al. (2020). Association between NPPA promoter methylation and hypertension: results from Gusu cohort and replication in an independent sample. Clin. Epigenet. 12:133.
Li, L., Xie, J., Zhang, M., and Wang, S. (2009). Homocysteine harasses the imprinting expression of IGF2 and H19 by demethylation of differentially methylated region between IGF2/H19 genes. Acta Biochim. Biophys. Sin. 41, 464–471. doi: 10.1093/abbs/gmp033
Li, X. D., Hong, M. N., Chen, J., Lu, Y. Y., Ye, M. Q., Ma, Y., et al. (2020). Adventitial fibroblast-derived vascular endothelial growth factor promotes vasa vasorum-associated neointima formation and macrophage recruitment. Cardiovasc. Res. 116, 708–720. doi: 10.1093/cvr/cvz159
Liu, C. F., and Tang, W. H. W. (2019). Epigenetics in cardiac hypertrophy and heart failure. JACC Basic Transl. Sci. 4, 976–993.
Liu, C. M., Ma, J. Q., Xie, W. R., Liu, S. S., Feng, Z. J., Zheng, G. H., et al. (2015). Quercetin protects mouse liver against nickel-induced DNA methylation and inflammation associated with the Nrf2/HO-1 and p38/STAT1/NF-κB pathway. Food Chem. Toxicol. 82, 19–26. doi: 10.1016/j.fct.2015.05.001
Liu, C., Xu, D., Sjöberg, J., Forsell, P., Björkholm, M., and Claesson, H. E. (2004). Transcriptional regulation of 15-lipoxygenase expression by promoter methylation. Exp. Cell Res. 297, 61–67. doi: 10.1016/j.yexcr.2004.02.014
Liu, R., Jin, Y., Tang, W. H., Qin, L., Zhang, X., Tellides, G., et al. (2013). Ten-eleven translocation-2 (TET2) is a master regulator of smooth muscle cell plasticity. Circulation 128, 2047–2057. doi: 10.1161/circulationaha.113.002887
Liu, R., Leslie, K. L., and Martin, K. A. (2015). Epigenetic regulation of smooth muscle cell plasticity. Biochim. Biophys. Acta 1849, 448–453. doi: 10.1016/j.bbagrm.2014.06.004
Love, S., and Miners, J. S. (2016). Cerebrovascular disease in ageing and Alzheimer’s disease. Acta Neuropathol. 131, 645–658.
Lunnon, K., Smith, R., Hannon, E., De Jager, P. L., Srivastava, G., Volta, M., et al. (2014). Methylomic profiling implicates cortical deregulation of ANK1 in Alzheimer’s disease. Nat. Neurosci. 17, 1164–1170. doi: 10.1038/nn.3782
Luz, I., Soukri, M., and Lail, M. (2018). Transformation of single MOF nanocrystals into single nanostructured catalysts within mesoporous supports: a platform for pioneer fluidized-nanoreactor hydrogen carriers. Chem. Commun. 54, 8462–8465. doi: 10.1039/c8cc04562c
Lyko, F. (2018). The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 19, 81–92. doi: 10.1038/nrg.2017.80
Ma, S. C., Cao, J. C., Zhang, H. P., Jiao, Y., Zhang, H., He, Y. Y., et al. (2017). Aberrant promoter methylation of multiple genes in VSMC proliferation induced by Hcy. Mol. Med. Rep. 16, 7775–7783. doi: 10.3892/mmr.2017.7521
Ma, S. C., Zhang, H. P., Jiao, Y., Wang, Y. H., Zhang, H., Yang, X. L., et al. (2018). Homocysteine-induced proliferation of vascular smooth muscle cells occurs via PTEN hypermethylation and is mitigated by resveratrol. Mol. Med. Rep. 17, 5312–5319.
Ma, S. C., Zhang, H. P., Kong, F. Q., Zhang, H., Yang, C., He, Y. Y., et al. (2016). Integration of gene expression and DNA methylation profiles provides a molecular subtype for risk assessment in atherosclerosis. Mol. Med. Rep. 13, 4791–4799. doi: 10.3892/mmr.2016.5120
Mahmood, S. S., Levy, D., Vasan, R. S., and Wang, T. J. (2014). The framingham heart study and the epidemiology of cardiovascular disease: a historical perspective. Lancet 383, 999–1008. doi: 10.1016/s0140-6736(13)61752-3
Martínez-Iglesias, O., Carrera, I., Carril, J. C., Fernández-Novoa, L., Cacabelos, N., and Cacabelos, R. (2020). DNA methylation in neurodegenerative and cerebrovascular disorders. Int. J. Mol. Sci. 21:2220. doi: 10.3390/ijms21062220
Mao, S. Q., Sun, J. H., Gu, T. L., Zhu, F. B., Yin, F. Y., and Zhang, L. N. (2017). Hypomethylation of interleukin-6 (IL-6) gene increases the risk of essential hypertension: a matched case-control study. J. Hum. Hypertens. 31, 530–536. doi: 10.1038/jhh.2017.7
Mao, S., Gu, T., Zhong, F., Fan, R., Zhu, F., Ren, P., et al. (2017). Hypomethylation of the Toll-like receptor-2 gene increases the risk of essential hypertension. Mol. Med. Rep. 16, 964–970. doi: 10.3892/mmr.2017.6653
Marumo, T., Yagi, S., Kawarazaki, W., Nishimoto, M., Ayuzawa, N., Watanabe, A., et al. (2015). Diabetes induces aberrant DNA Methylation in the proximal tubules of the kidney. J. Am. Soc. Nephrol. 26, 2388–2397. doi: 10.1681/asn.2014070665
Mattace-Raso, F. U., van der Cammen, T. J., Hofman, A., van Popele, N. M., Bos, M. L., Schalekamp, M. A., et al. (2006). Arterial stiffness and risk of coronary heart disease and stroke: the rotterdam study. Circulation 113, 657–663. doi: 10.1161/circulationaha.105.555235
Mills, K. T., Stefanescu, A., and He, J. (2020). The global epidemiology of hypertension. Nat. Rev. Nephrol. 16, 223–237.
Min, J., Weitian, Z., Peng, C., Yan, P., Bo, Z., Yan, W., et al. (2016). Correlation between insulin-induced estrogen receptor methylation and atherosclerosis. Cardiovasc. Diabetol. 15:156.
Mitra, S., Khaidakov, M., Lu, J., Ayyadevara, S., Szwedo, J., Wang, X. W., et al. (2011). Prior exposure to oxidized low-density lipoprotein limits apoptosis in subsequent generations of endothelial cells by altering promoter methylation. Am. J. Physiol. Heart Circ. Physiol. 301, H506–H513.
Moore, K. J., and Tabas, I. (2011). Macrophages in the pathogenesis of atherosclerosis. Cell 145, 341–355. doi: 10.1016/j.cell.2011.04.005
Moore, L. D., Le, T., and Fan, G. (2013). DNA methylation and its basic function. Neuropsychopharmacology 38, 23–38. doi: 10.1038/npp.2012.112
Morgan, R. G., Donato, A. J., and Walker, A. E. (2018). Telomere uncapping and vascular aging. Am. J. Physiol. Heart Circ. Physiol. 315, H1–H5.
Mortusewicz, O., Schermelleh, L., Walter, J., Cardoso, M. C., and Leonhardt, H. (2005). Recruitment of DNA methyltransferase I to DNA repair sites. Proc. Natl. Acad. Sci. U. S. A. 102, 8905–8909.
Nagai, M., Conney, A. H., and Zhu, B. T. (2004). Strong inhibitory effects of common tea catechins and bioflavonoids on the O-methylation of catechol estrogens catalyzed by human liver cytosolic catechol-O-methyltransferase. Drug Metab. Dispos. 32, 497–504. doi: 10.1124/dmd.32.5.497
Nakatochi, M., Ichihara, S., Yamamoto, K., Naruse, K., Yokota, S., Asano, H., et al. (2017). Epigenome-wide association of myocardial infarction with DNA methylation sites at loci related to cardiovascular disease. Clin. Epigenet. 9:54.
Napoli, C., Benincasa, G., Donatelli, F., and Ambrosio, G. (2020). Precision medicine in distinct heart failure phenotypes: focus on clinical epigenetics. Am. Heart J. 224, 113–128. doi: 10.1016/j.ahj.2020.03.007
Nelissen, E. C., van Montfoort, A. P., Dumoulin, J. C., and Evers, J. L. (2011). Epigenetics and the placenta. Hum. Reprod. Update 17, 397–417.
Ni, Y. Q., Zhan, J. K., and Liu, Y. S. (2020). Roles and mechanisms of MFG-E8 in vascular aging-related diseases. Ageing Res. Rev. 64:101176. doi: 10.1016/j.arr.2020.101176
Nicorescu, I., Dallinga, G. M., de Winther, M. P. J., Stroes, E. S. G., and Bahjat, M. (2019). Potential epigenetic therapeutics for atherosclerosis treatment. Atherosclerosis 281, 189–197. doi: 10.1016/j.atherosclerosis.2018.10.006
Niu, P. P., Cao, Y., Gong, T., Guo, J. H., Zhang, B. K., and Jia, S. J. (2014). Hypermethylation of DDAH2 promoter contributes to the dysfunction of endothelial progenitor cells in coronary artery disease patients. J. Transl. Med. 12:170. doi: 10.1186/1479-5876-12-170
North, B. J., and Sinclair, D. A. (2012). The intersection between aging and cardiovascular disease. Circ. Res. 110, 1097–1108. doi: 10.1161/circresaha.111.246876
O’Hagan, H. M., Wang, W., Sen, S., Destefano Shields, C., Lee, S. S., Zhang, Y. W., et al. (2011). Oxidative damage targets complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter CpG Islands. Cancer Cell 20, 606–619. doi: 10.1016/j.ccr.2011.09.012
Ooi, S. K., and Bestor, T. H. (2008). The colorful history of active DNA demethylation. Cell 133, 1145–1148. doi: 10.1016/j.cell.2008.06.009
Pan, X., Chen, Z., Huang, R., Yao, Y., and Ma, G. (2013). Transforming growth factor β1 induces the expression of collagen type I by DNA methylation in cardiac fibroblasts. PLoS One 8:e60335. doi: 10.1371/journal.pone.0060335
Papapetropoulos, A., García-Cardeña, G., Madri, J. A., and Sessa, W. C. (1997). Nitric oxide production contributes to the angiogenic properties of vascular endothelial growth factor in human endothelial cells. J. Clin. Invest. 100, 3131–3139. doi: 10.1172/jci119868
Park, L. K., Friso, S., and Choi, S. W. (2012). Nutritional influences on epigenetics and age-related disease. Proc. Nutr. Soc. 71, 75–83. doi: 10.1017/s0029665111003302
Pei, F., Wang, X., Yue, R., Chen, C., Huang, J., Huang, J., et al. (2015). Differential expression and DNA methylation of angiotensin type 1A receptors in vascular tissues during genetic hypertension development. Mol. Cell. Biochem. 402, 1–8. doi: 10.1007/s11010-014-2295-9
Pepin, M. E., Drakos, S., Ha, C. M., Tristani-Firouzi, M., Selzman, C. H., Fang, J. C., et al. (2019). DNA methylation reprograms cardiac metabolic gene expression in end-stage human heart failure. Am. J. Physiol. Heart Circ. Physiol. 317, H674–H684.
Post, W. S., Goldschmidt-Clermont, P. J., Wilhide, C. C., Heldman, A. W., Sussman, M. S., Ouyang, P., et al. (1999). Methylation of the estrogen receptor gene is associated with aging and atherosclerosis in the cardiovascular system. Cardiovasc. Res. 43, 985–991. doi: 10.1016/s0008-6363(99)00153-4
Prasad, R., and Jho, E. H. (2019). A concise review of human brain methylome during aging and neurodegenerative diseases. BMB Rep. 52, 577–588. doi: 10.5483/bmbrep.2019.52.10.215
Prince, M., Bryce, R., Albanese, E., Wimo, A., Ribeiro, W., and Ferri, C. P. (2013). The global prevalence of dementia: a systematic review and metaanalysis. Alzheimers Dement. 9, 63–75.e62.
Qazi, T. J., Quan, Z., Mir, A., and Qing, H. (2018). Epigenetics in Alzheimer’s disease: perspective of DNA Methylation. Mol. Neurobiol. 55, 1026–1044. doi: 10.1007/s12035-016-0357-6
Qin, X., Li, J., Wu, T., Wu, Y., Tang, X., Gao, P., et al. (2019). Overall and sex-specific associations between methylation of the ABCG1 and APOE genes and ischemic stroke or other atherosclerosis-related traits in a sibling study of Chinese population. Clin. Epigenet. 11:189.
Qiu, H., Zhu, Y., Sun, Z., Trzeciakowski, J. P., Gansner, M., Depre, C., et al. (2010). Short communication: vascular smooth muscle cell stiffness as a mechanism for increased aortic stiffness with aging. Circ. Res. 107, 615–619. doi: 10.1161/circresaha.110.221846
Rajan, R., Raj, N. A., Madeswaran, S., and Babu, D. R. (2015). Dielectric studies on struvite urinary crystals, a gateway to the new treatment modality for urolithiasis. Spectrochim. Acta A Mol. Biomol. Spectrosc. 148, 266–270. doi: 10.1016/j.saa.2015.03.136
Rasmussen, K. D., and Helin, K. (2016). Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 30, 733–750. doi: 10.1101/gad.276568.115
Ratel, D., Ravanat, J. L., Berger, F., and Wion, D. (2006). N6-methyladenine: the other methylated base of DNA. Bioessays 28, 309–315. doi: 10.1002/bies.20342
Richard, M. A., Huan, T., Ligthart, S., Gondalia, R., Jhun, M. A., Brody, J. A., et al. (2017). DNA Methylation analysis identifies loci for blood pressure regulation. Am. J. Hum. Genet. 101, 888–902. doi: 10.1016/j.ajhg.2017.09.028
Rivière, G., Lienhard, D., Andrieu, T., Vieau, D., Frey, B. M., and Frey, F. J. (2011). Epigenetic regulation of somatic angiotensin-converting enzyme by DNA methylation and histone acetylation. Epigenetics 6, 478–489. doi: 10.4161/epi.6.4.14961
Salameh, Y., Bejaoui, Y., and El Hajj, N. (2020). DNA methylation biomarkers in aging and age-related diseases. Front. Genet. 11:171.
Sandoo, A., van Zanten, J. J., Metsios, G. S., Carroll, D., and Kitas, G. D. (2010). The endothelium and its role in regulating vascular tone. Open Cardiovasc. Med. J. 4, 302–312. doi: 10.2174/1874192401004010302
Sano, S., Oshima, K., Wang, Y., Katanasaka, Y., Sano, M., and Walsh, K. (2018). CRISPR-mediated gene editing to assess the roles of Tet2 and Dnmt3a in clonal hematopoiesis and Cardiovascular disease. Circ. Res. 123, 335–341. doi: 10.1161/circresaha.118.313225
Scisciola, L., Rizzo, M. R., Cataldo, V., Fontanella, R. A., Balestrieri, M. L., D’Onofrio, N., et al. (2020). Incretin drugs effect on epigenetic machinery: new potential therapeutic implications in preventing vascular diabetic complications. FASEB J. 34, 16489–16503. doi: 10.1096/fj.202000860rr
Semick, S. A., Bharadwaj, R. A., Collado-Torres, L., Tao, R., Shin, J. H., Deep-Soboslay, A., et al. (2019). Integrated DNA methylation and gene expression profiling across multiple brain regions implicate novel genes in Alzheimer’s disease. Acta Neuropathol. 137, 557–569. doi: 10.1007/s00401-019-01966-5
Shapouri-Moghaddam, A., Mohammadian, S., Vazini, H., Taghadosi, M., Esmaeili, S. A., Mardani, F., et al. (2018). Macrophage plasticity, polarization, and function in health and disease. J. Cell. Physiol. 233, 6425–6440. doi: 10.1002/jcp.26429
Shen, Y., Peng, C., Bai, Q., Ding, Y., Yi, X., Du, H., et al. (2019). Epigenome-wide association study indicates Hypomethylation of MTRNR2L8 in large-artery atherosclerosis stroke. Stroke 50, 1330–1338. doi: 10.1161/strokeaha.118.023436
Soriano-Tárraga, C., Jiménez-Conde, J., Giralt-Steinhauer, E., Mola, M., Ois, A., Rodríguez-Campello, A., et al. (2014). Global DNA methylation of ischemic stroke subtypes. PLoS One 9:e96543. doi: 10.1371/journal.pone.0096543
Stenmark, K. R., Yeager, M. E., El Kasmi, K. C., Nozik-Grayck, E., Gerasimovskaya, E. V., Li, M., et al. (2013). The adventitia: essential regulator of vascular wall structure and function. Annu. Rev. Physiol. 75, 23–47. doi: 10.1146/annurev-physiol-030212-183802
Stenzig, J., Schneeberger, Y., Löser, A., Peters, B. S., Schaefer, A., Zhao, R. R., et al. (2018). Pharmacological inhibition of DNA methylation attenuates pressure overload-induced cardiac hypertrophy in rats. J. Mol. Cell Cardiol. 120, 53–63. doi: 10.1016/j.yjmcc.2018.05.012
Stoner, L., Young, J. M., and Fryer, S. (2012). Assessments of arterial stiffness and endothelial function using pulse wave analysis. Int. J. Vasc. Med. 2012:903107.
Strand, K. A., Lu, S., Mutryn, M. F., Li, L., Zhou, Q., Enyart, B. T., et al. (2020). High throughput screen identifies the DNMT1 (DNA Methyltransferase-1) Inhibitor, 5-Azacytidine, as a potent inducer of PTEN (Phosphatase and Tensin Homolog): central role for PTEN in 5-Azacytidine protection against pathological vascular remodeling. Arterioscler. Thromb. Vasc. Biol. 40, 1854–1869. doi: 10.1161/atvbaha.120.314458
Tabaei, S., and Tabaee, S. S. (2019). DNA methylation abnormalities in atherosclerosis. Artif. Cells Nanomed. Biotechnol. 47, 2031–2041. doi: 10.1080/21691401.2019.1617724
Tang, P. M., Nikolic-Paterson, D. J., and Lan, H. Y. (2019). Macrophages: versatile players in renal inflammation and fibrosis. Nat. Rev. Nephrol. 15, 144–158. doi: 10.1038/s41581-019-0110-2
Tao, H., Yang, J. J., Chen, Z. W., Xu, S. S., Zhou, X., Zhan, H. Y., et al. (2014a). DNMT3A silencing RASSF1A promotes cardiac fibrosis through upregulation of ERK1/2. Toxicology 323, 42–50. doi: 10.1016/j.tox.2014.06.006
Tao, H., Yang, J. J., Shi, K. H., Deng, Z. Y., and Li, J. (2014b). DNA methylation in cardiac fibrosis: new advances and perspectives. Toxicology 323, 125–129. doi: 10.1016/j.tox.2014.07.002
Thannickal, V. J., Zhou, Y., Gaggar, A., and Duncan, S. R. (2014). Fibrosis: ultimate and proximate causes. J. Clin. Invest. 124, 4673–4677. doi: 10.1172/jci74368
Thorin-Trescases, N., de Montgolfier, O., Pinçon, A., Raignault, A., Caland, L., Labbé, P., et al. (2018). Impact of pulse pressure on cerebrovascular events leading to age-related cognitive decline. Am. J. Physiol. Heart Circ. Physiol. 314, H1214–H1224. doi: 10.1152/ajpheart.00637.2017
Toth, P., Tarantini, S., Csiszar, A., and Ungvari, Z. (2017). Functional vascular contributions to cognitive impairment and dementia: mechanisms and consequences of cerebral autoregulatory dysfunction, endothelial impairment, and neurovascular uncoupling in aging. Am. J. Physiol. Heart Circ. Physiol. 312, H1–H20. doi: 10.1152/ajpheart.00581.2016
Triposkiadis, F., Xanthopoulos, A., and Butler, J. (2019). Cardiovascular aging and heart failure: JACC review topic of the week. J. Am. Coll. Cardiol. 74, 804–813. doi: 10.1016/j.jacc.2019.06.053
Triposkiadis, F., Xanthopoulos, A., Parissis, J., Butler, J., and Farmakis, D. (2020). Pathogenesis of chronic heart failure: cardiovascular aging, risk factors, comorbidities, and disease modifiers. Heart Fail Rev. doi: 10.1007/s10741-020-09987-z Online ahead of print.
Ungvari, Z., Labinskyy, N., Gupte, S., Chander, P. N., Edwards, J. G., and Csiszar, A. (2008). Dysregulation of mitochondrial biogenesis in vascular endothelial and smooth muscle cells of aged rats. Am. J. Physiol. Heart Circ. Physiol. 294, H2121–H2128. doi: 10.1152/ajpheart.00012.2008
Uryga, A. K., and Bennett, M. R. (2016). Ageing induced vascular smooth muscle cell senescence in atherosclerosis. J. Physiol. 594, 2115–2124. doi: 10.1113/jp270923
van Sloten, T. T., Sedaghat, S., Laurent, S., London, G. M., Pannier, B., Ikram, M. A., et al. (2015). Carotid stiffness is associated with incident stroke: a systematic review and individual participant data meta-analysis. J. Am. Coll. Cardiol. 66, 2116–2125. doi: 10.1016/j.jacc.2015.08.888
Vujic, A., Robinson, E. L., Ito, M., Haider, S., Ackers-Johnson, M., See, K., et al. (2015). Experimental heart failure modelled by the cardiomyocyte-specific loss of an epigenome modifier. DNMT3B. J. Mol. Cell Cardiol. 82, 174–183. doi: 10.1016/j.yjmcc.2015.03.007
Waddington, C. H. (2012). The epigenotype. 1942. Int. J. Epidemiol. 41, 10–13. doi: 10.1093/ije/dyr184
Wang, C., Xu, G., Wen, Q., Peng, X., Chen, H., Zhang, J., et al. (2019). CBS promoter hypermethylation increases the risk of hypertension and stroke. Clinics 74:e630.
Wang, F., Demura, M., Cheng, Y., Zhu, A., Karashima, S., Yoneda, T., et al. (2014). Dynamic CCAAT/enhancer binding protein-associated changes of DNA methylation in the angiotensinogen gene. Hypertension 63, 281–288. doi: 10.1161/hypertensionaha.113.02303
Wang, G., Jacquet, L., Karamariti, E., and Xu, Q. (2015). Origin and differentiation of vascular smooth muscle cells. J. Physiol. 593, 3013–3030. doi: 10.1113/jp270033
Wang, J., Uryga, A. K., Reinhold, J., Figg, N., Baker, L., Finigan, A., et al. (2015). Vascular smooth muscle cell senescence promotes atherosclerosis and features of plaque vulnerability. Circulation 132, 1909–1919. doi: 10.1161/circulationaha.115.016457
Wang, X., Falkner, B., Zhu, H., Shi, H., Su, S., Xu, X., et al. (2013). A genome-wide methylation study on essential hypertension in young African American males. PLoS One 8:e53938. doi: 10.1371/journal.pone.0053938
Wang, X., Qin, X., Demirtas, H., Li, J., Mao, G., Huo, Y., et al. (2007). Efficacy of folic acid supplementation in stroke prevention: a meta-analysis. Lancet 369, 1876–1882. doi: 10.1016/s0140-6736(07)60854-x
Waterland, R. A. (2006). Assessing the effects of high methionine intake on DNA methylation. J. Nutr. 136, 1706s–1710s. doi: 10.1093/jn/136.6.1706S
Weber, A. R., Krawczyk, C., Robertson, A. B., Kuśnierczyk, A., Vågbø, C. B., Schuermann, D., et al. (2016). Biochemical reconstitution of TET1-TDG-BER-dependent active DNA demethylation reveals a highly coordinated mechanism. Nat. Commun. 7:10806. doi: 10.1038/ncomms10806
Wei, L., Zhao, S., Wang, G., Zhang, S., Luo, W., Qin, Z., et al. (2018). SMAD7 methylation as a novel marker in atherosclerosis. Biochem. Biophys. Res. Commun. 496, 700–705. doi: 10.1016/j.bbrc.2018.01.121
Wei, Y., Sun, Z., Wang, Y., Xie, Z., Xu, S., Xu, Y., et al. (2019). Methylation in the TP53 promoter is associated with ischemic stroke. Mol. Med. Rep. 20, 1404–1410. doi: 10.3892/mmr.2019.10348
Wong, C. P., Nguyen, L. P., Noh, S. K., Bray, T. M., Bruno, R. S., and Ho, E. (2011). Induction of regulatory T cells by green tea polyphenol EGCG. Immunol. Lett. 139, 7–13. doi: 10.1016/j.imlet.2011.04.009
Wu, L., Pei, Y., Zhu, Y., Jiang, M., Wang, C., Cui, W., et al. (2019). Association of N(6)-methyladenine DNA with plaque progression in atherosclerosis via myocardial infarction-associated transcripts. Cell Death Dis. 10:909. doi: 10.1038/s41419-019-2152-6
Wu, X., and Zhang, Y. (2017). TET-mediated active DNA demethylation: mechanism, function and beyond. Nat. Rev. Genet. 18, 517–534. doi: 10.1038/nrg.2017.33
Wyss-Coray, T. (2016). Ageing, neurodegeneration and brain rejuvenation. Nature 539, 180–186. doi: 10.1038/nature20411
Xiao, C. L., Zhu, S., He, M., Chen, D., Zhang, Q., Chen, Y., et al. (2018). N(6)-Methyladenine DNA modification in the human genome. Mol. Cell. 71, 306–318.e307. doi: 10.1016/j.molcel.2018.06.015
Xu, G., Wang, C., Ying, X., Kong, F., Ji, H., Zhao, J., et al. (2019). Serine hydroxymethyltransferase 1 promoter hypermethylation increases the risk of essential hypertension. J. Clin. Lab. Anal. 33:e22712. doi: 10.1002/jcla.22712
Xu, H., Jiang, J., Chen, W., Li, W., and Chen, Z. (2019). Vascular macrophages in atherosclerosis. J. Immunol. Res. 2019:4354786.
Xu, L., Hao, H., Hao, Y., Wei, G., Li, G., Ma, P., et al. (2019). Aberrant MFN2 transcription facilitates homocysteine-induced VSMCs proliferation via the increased binding of c-Myc to DNMT1 in atherosclerosis. J. Cell Mol. Med. 23, 4611–4626. doi: 10.1111/jcmm.14341
Xu, M., Li, J., Chen, X., Han, L., Li, L., and Liu, Y. (2019). MTHFD1 promoter hypermethylation increases the risk of hypertension. Clin. Exp. Hypertens. 41, 422–427. doi: 10.1080/10641963.2018.1501057
Xu, X., Friehs, I., Zhong, Hu, T., Melnychenko, I., Tampe, B., et al. (2015a). Endocardial fibroelastosis is caused by aberrant endothelial to mesenchymal transition. Circ. Res. 116, 857–866. doi: 10.1161/circresaha.116.305629
Xu, X., Tan, X., Tampe, B., Nyamsuren, G., Liu, X., Maier, L. S., et al. (2015b). Epigenetic balance of aberrant Rasal1 promoter methylation and hydroxymethylation regulates cardiac fibrosis. Cardiovasc. Res. 105, 279–291. doi: 10.1093/cvr/cvv015
Yamada, Y., Nishida, T., Horibe, H., Oguri, M., Kato, K., and Sawabe, M. (2014). Identification of hypo- and hypermethylated genes related to atherosclerosis by a genome-wide analysis of DNA methylation. Int. J. Mol. Med. 33, 1355–1363. doi: 10.3892/ijmm.2014.1692
Yamazaki, K., Yoshino, Y., Mori, T., Yoshida, T., Ozaki, Y., Sao, T., et al. (2017). Gene expression and methylation analysis of ABCA7 in patients with Alzheimer’s disease. J. Alzheimers Dis. 57, 171–181. doi: 10.3233/jad-161195
Yamazaki, Y., Baker, D. J., Tachibana, M., Liu, C. C., van Deursen, J. M., Brott, T. G., et al. (2016). Vascular cell senescence contributes to blood-brain barrier breakdown. Stroke 47, 1068–1077. doi: 10.1161/strokeaha.115.010835
Yang, Q., Chen, H. Y., Wang, J. N., Han, H. Q., Jiang, L., Wu, W. F., et al. (2020). Alcohol promotes renal fibrosis by activating Nox2/4-mediated DNA methylation of Smad7. Clin. Sci. 134, 103–122. doi: 10.1042/cs20191047
Yang, Q., Zhao, Y., Zhang, Z., and Chen, J. (2016). Association of interleukin-6 methylation in leukocyte DNA with serum level and the risk of ischemic heart disease. Scand. J. Clin. Lab. Invest. 76, 291–295. doi: 10.3109/00365513.2016.1149616
Yang, X., Wang, X., Liu, D., Yu, L., Xue, B., and Shi, H. (2014). Epigenetic regulation of macrophage polarization by DNA methyltransferase 3b. Mol. Endocrinol. 28, 565–574. doi: 10.1210/me.2013-1293
Yang, Y., Wang, A., Yuan, X., Zhao, Q., Liu, X., Chen, S., et al. (2019). Association between healthy vascular aging and the risk of the first stroke in a community-based Chinese cohort. Aging 11, 5807–5816. doi: 10.18632/aging.102170
Yang, Z., Wang, L., Zhang, W., Wang, X., and Zhou, S. (2016). Plasma homocysteine involved in methylation and expression of thrombomodulin in cerebral infarction. Biochem. Biophys. Res. Commun. 473, 1218–1222. doi: 10.1016/j.bbrc.2016.04.042
Yin, H., and Blanchard, K. L. (2000). DNA methylation represses the expression of the human erythropoietin gene by two different mechanisms. Blood 95, 111–119. doi: 10.1182/blood.v95.1.111.001k20_111_119
Yin, S., Zhang, Q., Yang, J., Lin, W., Li, Y., Chen, F., et al. (2017). TGFβ-incurred epigenetic aberrations of miRNA and DNA methyltransferase suppress Klotho and potentiate renal fibrosis. Biochim. Biophys. Acta Mol. Cell Res. 1864, 1207–1216. doi: 10.1016/j.bbamcr.2017.03.002
Yousuf, F. A., Kazmi, K., Iqbal, J., Ahmed, N., and Iqbal, M. P. (2020). Higher DNA methylation of ABO gene promoter is associated with acute myocardial infarction in a hospital-based population in Karachi. Pak. J. Med. Sci. 36, 505–510.
Yu, Y., Guan, X., Nie, L., Liu, Y., He, T., Xiong, J., et al. (2017). DNA hypermethylation of sFRP5 contributes to indoxyl sulfate-induced renal fibrosis. J. Mol. Med. 95, 601–613. doi: 10.1007/s00109-017-1538-0
Zhang, D., Chen, Y., Xie, X., Liu, J., Wang, Q., Kong, W., et al. (2012). Homocysteine activates vascular smooth muscle cells by DNA demethylation of platelet-derived growth factor in endothelial cells. J. Mol. Cell Cardiol. 53, 487–496. doi: 10.1016/j.yjmcc.2012.07.010
Zhang, D., Sun, X., Liu, J., Xie, X., Cui, W., and Zhu, Y. (2015). Homocysteine accelerates senescence of endothelial cells via DNA hypomethylation of human telomerase reverse transcriptase. Arterioscler. Thromb. Vasc. Biol. 35, 71–78. doi: 10.1161/atvbaha.114.303899
Zhang, G., Huang, H., Liu, D., Cheng, Y., Liu, X., Zhang, W., et al. (2015). N6-methyladenine DNA modification in Drosophila. Cell 161, 893–906.
Zhang, H., Zhao, X., Wang, C., Du, R., Wang, X., Fu, J., et al. (2019). A preliminary study of the association between apolipoprotein E promoter methylation and atherosclerotic cerebral infarction. J. Stroke Cerebrovasc. Dis. 28, 1056–1061. doi: 10.1016/j.jstrokecerebrovasdis.2018.12.027
Zhang, L. N., Liu, P. P., Wang, L., Yuan, F., Xu, L., Xin, Y., et al. (2013). Lower ADD1 gene promoter DNA methylation increases the risk of essential hypertension. PLoS One 8:e63455. doi: 10.1371/journal.pone.0063455
Zhang, Y. P., Huang, Y. T., Huang, T. S., Pang, W., Zhu, J. J., Liu, Y. F., et al. (2017). The mammalian target of rapamycin and DNA methyltransferase 1 axis mediates vascular endothelial dysfunction in response to disturbed flow. Sci. Rep. 7:14996.
Zhong, Q., Liu, C., Fan, R., Duan, S., Xu, X., Zhao, J., et al. (2016). Association of SCNN1B promoter methylation with essential hypertension. Mol. Med. Rep. 14, 5422–5428. doi: 10.3892/mmr.2016.5905
Zhou, S., Cai, B., Zhang, Z., Zhang, Y., Wang, L., Liu, K., et al. (2017). CDKN2B Methylation and Aortic Arch calcification in patients with ischemic stroke. J. Atheroscler. Thromb. 24, 609–620. doi: 10.5551/jat.36897
Zhu, L., Jia, L., Liu, Z., Zhang, Y., Wang, J., Yuan, Z., et al. (2019). Elevated Methylation of FOXP3 (Forkhead Box P3)-TSDR (Regulatory T-Cell-Specific Demethylated Region) is associated with increased risk for adverse outcomes in patients with acute coronary syndrome. Hypertension 74, 581–589. doi: 10.1161/hypertensionaha.119.12852
Zhuang, J., Luan, P., Li, H., Wang, K., Zhang, P., Xu, Y., et al. (2017). The Yin-Yang dynamics of DNA Methylation is the key regulator for smooth muscle cell phenotype switch and vascular remodeling. Arterioscler. Thromb. Vasc. Biol. 37, 84–97. doi: 10.1161/atvbaha.116.307923
Keywords: DNA methylation, aging, vascular diseases, endothelial cells, vascular smooth muscle cells
Citation: Xu H, Li S and Liu Y-S (2021) Roles and Mechanisms of DNA Methylation in Vascular Aging and Related Diseases. Front. Cell Dev. Biol. 9:699374. doi: 10.3389/fcell.2021.699374
Received: 23 April 2021; Accepted: 07 June 2021;
Published: 28 June 2021.
Edited by:
Suowen Xu, University of Science and Technology of China, ChinaCopyright © 2021 Xu, Li and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: You-Shuo Liu, bGl1eW91c2h1b0Bjc3UuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.