- 1Department of Biochemistry, School of Dentistry, Kyungpook National University, Daegu, South Korea
- 2Department of Oral and Maxillofacial Radiology, School of Dentistry, Kyungpook National University, Daegu, South Korea
- 3Department of Dental Hygiene, College of Health Science, Gachon University, Incheon, South Korea
- 4Department of Histology and Developmental Biology, Tokyo Dental College, Tokyo, Japan
- 5School of Life Sciences, BK21 Plus KNU Creative BioResearch Group, Kyungpook National University, Daegu, South Korea
- 6Pre-Major of Cosmetics and Pharmaceutics, Daegu Haany University, Gyeongsan-si, South Korea
- 7Department of Biochemistry and Cell Biology, Stony Brook University, Stony Brook, NY, United States
miRNAs are conserved short non-coding RNAs that play a role in the modulation of various biological pathways during tissue and organ morphogenesis. In this study, the function of miRNA-221-3p in tooth development, through its loss or gain in function was evaluated. A variety of techniques were utilized to evaluate detailed functional roles of miRNA-221-3p during odontogenesis, including in vitro tooth cultivation, renal capsule transplantation, in situ hybridization, real-time PCR, and immunohistochemistry. Two-day in vitro tooth cultivation at E13 identified altered cellular events, including cellular proliferation, apoptosis, adhesion, and cytoskeletal arrangement, with the loss and gain of miRNA-221-3p. qPCR analysis revealed alterations in gene expression of tooth-related signaling molecules, including β-catenin, Bmp2, Bmp4, Fgf4, Ptch1, and Shh, when inhibited with miRNA-221-3p and mimic. Also, the inhibition of miRNA-221-3p demonstrated increased mesenchymal localizations of pSMAD1/5/8, alongside decreased expression patterns of Shh and Fgf4 within inner enamel epithelium (IEE) in E13 + 2 days in vitro cultivated teeth. Moreover, 1-week renal transplantation of in vitro cultivated teeth had smaller tooth size with reduced enamel and dentin matrices, along with increased cellular proliferation and Shh expression along the Hertwig epithelial root sheath (HERS), within the inhibitor group. Similarly, in 3-week renal calcified teeth, the overexpression of miRNA-221-3p did not affect tooth phenotype, while the loss of function resulted in long and slender teeth with short mesiodistal length. This study provides evidence that a suitable level of miRNA-221-3p is required for the modulation of major signaling pathways, including Wnt, Bmp, and Shh, during tooth morphogenesis.
Introduction
The well-defined stages in which tooth development progresses require an intricate and reciprocal signaling regulation between both the dental epithelium and neural-crest-derived mesenchyme (Balic and Thesleff, 2015). This reciprocal signaling frequently involves signaling via Bmp, Shh, Wnt, and Fgfs throughout the various stages of tooth development including initiation, bud, bell, cap, and root morphogenesis (Jussila and Thesleff, 2012). In particular during bud to cap transition Bmp, Shh, Wnt, and Fgf signaling play a key role in maintaining the tooth crown structure through enamel knot (EK) morphogenesis (Balic and Thesleff, 2015). In addition, the very same signaling governs hard tissue formation via regulation of differentiation and secretion stages, and tooth root morphogenesis by maintenance and proliferation of HERS, which occurs at the cervical loop of the enamel organ (Lungová et al., 2011; Li et al., 2017). In particular, Wnt signaling is associated with the determination of tooth number, shape, and hard tissue formation; whereas cellular behavior, such as proliferation, apoptosis, differentiation, and migration of cells, is regulated by Fgf, Shh, and Bmp signaling (Liu et al., 2008; Balic and Thesleff, 2015). Disruption of the precise signaling pathways during tooth development will lead to abnormal tooth crown and root morphogenesis (Liu and Millar, 2010; Wang et al., 2012; Wang and Feng, 2017). Therefore, the use of activators and inhibitors on the signaling regulations, such as overexpression vectors or siRNAs, are being investigated in a range of model organs, including the tooth which may provide crucial knowledge related to tooth crown and root morphogenesis (Aryal et al., 2020; Neupane et al., 2020). To date, the signaling involved that modulates the tooth crown and root has yet to be broadened to specific molecules, such as miRNA. Consequently, functional studies including miRNA, siRNA, lncRNA, exosome, and so on have been utilized to define the developmental processes in tooth organogenesis (Jiang et al., 2017; Aryal et al., 2020; Huang et al., 2020) and therefore, require further research.
miRNAs are conserved short non-coding RNAs, an average of 22 nucleotides in length. Mature miRNA are up to several hundred nucleotides in length, called pri-miRNA, before being processed by the enzymes Drosha and Dicer (Ha and Kim, 2014; Wang et al., 2016). The biogenesis of miRNA is a complex and dynamic process, and any disruption to these processes could result in impaired production of mature miRNA/this, in turn, can lead to an imbalance in tissue homeostasis and disease progression (Trionfini et al., 2015; Barwari et al., 2016; Wang et al., 2016). The role of miRNAs in the modulation of various biological pathways including tissue and organ morphogenesis, have been extensively studied (Bartel, 2004; Andl et al., 2006; Yi et al., 2006; Cao et al., 2010; Wang et al., 2016; Rahmanian et al., 2019; Barbu et al., 2020). The involvement of miRNAs during tooth morphogenesis, which involves complex and reciprocal interactions between epithelial and mesenchymal tissue (Jussila and Thesleff, 2012; Huang et al., 2020), has also been identified. During mammalian tooth development, miRNAs regulate dental epithelial and mesenchymal cell differentiation during the early and secretory stages of odontogenesis (Cao et al., 2010; Li et al., 2015; Neupane et al., 2020). Functional studies conducted on different miRNAs highlight their roles during tooth morphogenesis. For example, miR-135a regulates Bmp signaling (Kim et al., 2014), miR-31, and miR-138 regulate differentiation of adult stem cell niche in mice incisors (Jheon et al., 2011), and miR-31, miR-140, MiR-141, miR-143, miR-145, miR-455, miR-689, miR-711, miR-720, and miR-875 regulate ameloblast and odontoblast differentiation (Michon et al., 2010; Liu et al., 2013, 2014; Funada et al., 2020). More than 700 miRNAs have been identified to date (Rahmanian et al., 2019), and it is pertinent to identify the function of each of these miRNAs to understand their precise roles within the signaling pathways during tooth development. In this study, we selected miRNA-221-3p as one of the candidate miRNA due to its high expression identified by miRNA-Microarray experiment utilizing the embryonic mice molar at E14 (unpublished data). This prompted the investigation into the role of miRNA-221-3p in signaling modulation during tooth development, which is important for understanding the regulatory pathways involved in tooth development.
In this study, miRNA-221-3p, one of the conserved miRNA, was investigated via an in vitro organ cultivation system, and a loss and gain of function assay using miRNA-221-3p specific inhibitors and its mimics. miRNA-221-3p is has been well studied in cancer (Liu et al., 2014; Tao et al., 2014; Zheng et al., 2014; Ihle et al., 2015; Wang et al., 2019), wound healing (Xu et al., 2020), and bone metabolism (Maeda et al., 2017), through modulation of signaling pathways. More importantly, miRNA-221-3p is predicted to regulate Shh and Wnt signaling through Ptch1 and Dkk2 inhibition, respectively (Agarwal et al., 2015). In cancer cells, miRNA also regulates proliferation and apoptosis (Ihle et al., 2015; Yang et al., 2019), and these cellular events are also important for tooth morphogenesis. Given the important role of Wnt and Shh signaling in tooth development (Jussila and Thesleff, 2012; Balic and Thesleff, 2015), it is pertinent to investigate signaling modulation of miRNA-221-3p in tooth development. Therefore, in this study, we have elucidated the functional roles of miRNA-221-3p during tooth crown and root morphogenesis.
Materials and Methods
Animals
All experiments involving animals were performed according to the guidelines of the Kyungpook National University, School of Dentistry, Intramural Animal Use and Care Committee (KNU-2020-0107). Mice were kept in an optimal environment, maintaining relative humidity (55%) and temperature (25°C) with access to food and water ad libitum. For time-mated pregnant mice, embryonic day 0 (E0) was assigned to the day in which a vaginal plug was confirmed. All experiments described in this study were independently performed in triplicate as a minimum.
In situ Hybridization
mRNA and miRNA in situ hybridizations were performed as described previously (Neupane et al., 2020). For miRNA in situ hybridizations, a 5′- and 3′- DIG-labeled miRCURY LNATM miRNA custom detection probe was hybridized using the miRNA ISH buffer set according to the manufacturer’s instructions (Exiqon, Skelstedet, Denmark, cat no. 90000). The miRCURY LNATM miRNA custom detection probe for 221-3P (/5DigN/AAACCCAGCAGACAATGTAGCT/3Dig_N/) was purchased from Qiagen (Qiagen, Hilden, Germany, cat. no. 339115 YCD002344-BCG). For section in situ hybridizations, digoxigenin (DIG)-labeled antisense RNA probes were hybridized overnight to examine the detailed expression patterns of genes as described previously (Aryal et al., 2020; Neupane et al., 2020).
In vitro Organ Cultivation and Renal Capsule Transplantation
E13 embryos that are at the bud stage of tooth development, underwent microdissection isolation of the embryonic tooth germs. These were subsequently cultured in DMEM (HyClone, Logan, UT, United States; cat. no.-SH30243.01) with 10% fetal bovine serum (Hyclone, Logan, UT, United States) and antibiotics, and cultivated using a modified Trowell’s culture method for 2 days, as previously described (Aryal et al., 2020; Neupane et al., 2020). To check the loss and gain of function of miRNA-221-3p, tooth germs were cocultivated with transfecting 400 nM of the inhibitor (Qiagen, Hilden, Germany, miCURY LNATM miRNA Power Inhibitor, cat no. 339131 YI04102471-DDB) or (mimic miCURY LNATM miRNA Mimic, cat no. 339173 YM00471077-ADB) of the miRNA-221-3p as previously described (Neupane et al., 2020). After transfection of miRNA at E13 for 2 days, the cultivated tooth germs were transplanted into the kidney capsule of adult ICR male mice as previously described (Aryal et al., 2020; Neupane et al., 2020). Differing groups of mice were sacrificed at either 1-week or 3-week post-transfection, and the calcified teeth were harvested.
Histology and Immunohistochemistry
Histology and immunostaining were carried out as described previously (Aryal et al., 2020; Neupane et al., 2020). Hematoxylin and eosin (H&E) staining were employed to examine the detailed morphology of the in vivo and in vitro tooth. Primary antibodies were directed against Ki67 (Cat# RM-9106-s, Thermo Scientific, United States); ROCK1 (Cat# ab45171, Abcam); E-cadherin (Cat# AF748, R&D Systems, United States); β-catenin (Cat# 8814S, Cell Signaling Technology, MA, United States); pSMAD (Cat# 9511S, Cell Signaling Technology, MA, United States); AMELX (Cat# ab153915, Abcam, United States) and NESTIN (Cat# ab11306, Abcam, United States). The secondary antibodies used in this study were biotinylated goat anti-rabbit or anti-mouse IgG. Immunocomplexes were visualized using a diaminobenzidine tetrahydrochloride (DAB) reagent kit (Cat# 00-2014; Zymed Laboratories, United States) and goat Anti-Rabbit Alexa Fluor@488 (Cat# ab150077, Abcam, United States); goat Anti-Mouse Alexa Fluor@647 (Cat# ab150115, Abcam, United States). The DAB stained slides were mounted with xylene based mounting medium Malinol and immunofluorescent slides were mounted with VECTASHIELD® Antifade Mounting Medium containing DAPI (Cat# H-1200-10, Vector laboratories) and cover slipped.
Histological sections were photographed using a Leica DM2500 with and without fluorescent setting. ImageJ1 was used to measure the fluorescence of the images as described previously (Neupane et al., 2021). Briefly, fluorescence images were split for different channels and integrated density and background fluorescent were measured in defined region of interests. The total corrected cell fluorescence was measured as: [Integrated density-(area X background fluorescence)]. Fluorescence was measured in sections obtained from minimum of three independent experiments (n = 3) for each group. Data were evaluated for significance using unpaired, two-tailed t-test. A p-value of <0.05 indicates significance.
For colocalization analysis the correlation index (Pearson’s correlation coefficient) of colocalized pixels between the green (β-Catenin) and blue (DAPI for nucleus) colors were measured in ImageJ and plotted as graph in GraphPad Prism as described previously (Neupane et al., 2021). Briefly, the color of image was split into component channels and the green and blue channel were used to evaluate the colocalization using ImageJ plugin “Colocalization.” If the green (β-Catenin) was localized in the nucleus, it will colocalize with the blue (DAPI for nucleus) that will produce the correlation index of colocalization. Higher the correlation index, higher will be the colocalization and vice versa.
The number of Ki67 positive cells in DAB stained sections were counted in defined area of 200 × 200 μm2 in mesenchyme adjacent to the enamel knot. Whereas number of Ki67 positive cells from fluorescent labeled sections were presented as % of Ki67 positive cells (green) in total numbers of nuclei (blue) in the given field. GraphPad Prism (version 8) was used to calculate the test statistics and plot the graph. Briefly, after splitting the channels to blue (DAPI for nuclei) and green (Ki67) in ImageJ, the individual channels were processed through thresholding, binary processing (close, open, and watershed) and finally particle measurement (Supplementary Figure 1). This will give an approximate number of DAPI positive cells from blue channel and Ki67 + cells from the green channel which was used to calculate the % Ki67 positive cells in total number of cell.
Quantitative RT-PCR (qPCR)
Quantitative RT-PCR (qPCR) was performed as previously described (Aryal et al., 2020; Neupane et al., 2020). RNA was extracted from E13 + 36 and 48 h incubation with miRNA-221-3p inhibited and mimic transfected tooth germs, and qPCR was subsequently performed to examine the loss and gain of function effect of miRNA-221-3p. The mRNA and the miRNA qPCR results for each sample were normalized to those of Hprt and snoRNA135, respectively. Data were represented as means ± SD. Mean expression levels were determined by comparing experimental and control groups using the Student’s t-test. A p-value of <0.05 indicates significance. The experiments are repeated at least three times to check the reproducibility of the data. Primer sequences used in the qPCR assays are detailed in Supplementary Table 1.
Results
miRNA-221-3p Modulates Cellular Events During Tooth Development
The developing tooth germ is at the bud and cap stage at the embryonic stage E13 and E14.5, respectively (Figures 1A,B). During bud stage, the expression of miRNA-221-3p was restricted along the invaginated epithelium (Figure 1C) with intense expression noted along the enamel knot (EK), IEE, outer enamel epithelium (OEE), and dental papilla (DP) at cap stage (Figure 1D). These specific expression patterns suggest a role for miRNA- 221-3p in the bud to cap stage transition of tooth development that ultimately regulates the signaling of the tooth crown and root morphogenesis. To evaluate the functional role of miRNA-221-3p in tooth development, the in vitro cultivation of tooth germ with miRNA-221-3p inhibitor and its mimic was performed. This significantly altered the expression level of miRNA at E13 for 36 h (Figures 1E,F). According to the database TargetScan, miRNA-221-3p is broadly conserved among vertebrates and predicted to target Ptch1 (Figure 1G and Supplementary Figure 2; Agarwal et al., 2015). Inhibition and mimic of miRNA-221-3p increased and decreased the level of Ptch1, which is in agreement with the seed sequence prediction of Agarwal et al. (2015) (Figures 1G–I). A 2-day cultivation of tooth with an inhibitor and mimic resulted in a decrease and increase in tooth size, respectively, along the buccolingual and mesiodistal axis, in comparison to the control (Figures 1J–O). This altered tooth size during in vitro organ cultivation, suggests a possible change in cellular behaviors, including cell proliferation, apoptosis, and cell migration, which are all crucial for determining tooth size (Setkova et al., 2006). Therefore, to understand the precise role of miRNA-221-3p in the cellular events including proliferation, apoptosis, and cell adhesion, we utilized Ki67, E-cadherin, and ROCK1 immunohistochemistry and TUNEL assay. Compared to the control (Figure 2A), the cellular proliferation, in particular at the IEE and dental mesenchyme adjacent to the IEE, was decreased in the inhibitor-treated specimens (Figures 2B,D and Supplementary Figure 3). However, an increase in the mimic-treated specimens was observed (Figures 2C,D). Similarly, the TUNEL positive cells are significantly decreased in the inhibitor-treated specimens compared to control and mimic (Figures 2E–G and Supplementary Figure 3). Meanwhile, the number of apoptotic cells in the EK was similar to that in the control and mimic-treated specimens (Figures 2E,G,H). To understand these cellular events further, we evaluated the localization patterns of E-cadherin and ROCK1 (Figures 2I–L). ROCK1, which is a downstream target of RhoA small GTP-binding proteins, regulates cell adhesion, cytokinesis, cell proliferation, and apoptosis and regulates actin cytoskeleton dynamics forming a complex with E-cadherin (Zohrabian et al., 2009; Smith et al., 2012). In addition, ROCK1 and E-cadherin primarily localize in the enamel organ and regulate the cellular proliferation in the IEE (Aryal et al., 2020). In our study, the colocalization of ROCK1 and E-cadherin was evenly distributed along the IEE in the control specimen (Figures 2I,M,Q). In contrast, inhibiting and mimicking of miRNA-221-3p decreased the intensity of fluorescence of E-cadherin and ROCK1 in the IEE (Figures 2J–L,N–P,R,S). These observations suggest that miRNA-221-3p modulates proliferation and determines the size of the developing tooth through ROCK1 and E-cadherin mediated cell adhesion. This will eventually regulate the tooth crown and root morphogenesis in later stages of tooth development.
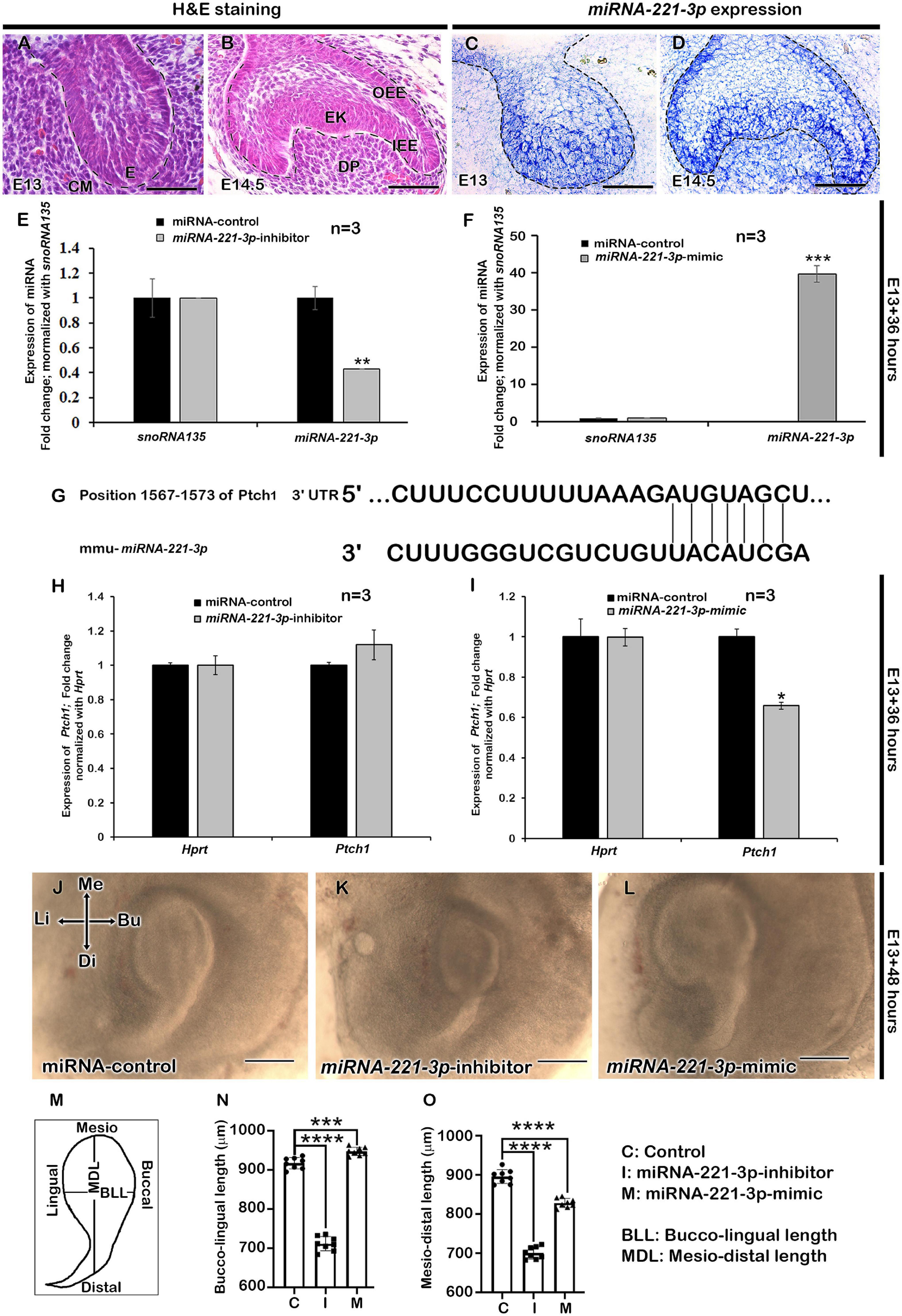
Figure 1. Expression of miRNA-221-3p and loss and gain of function in developing tooth germ. H&E staining showing the developing tooth germ at the bud and cap stage (A,B). Section in situ hybridization showing expression of miRNA-221-3p along the invaginated epithelium at the bud stage (C) and along the IEE, OEE, EK, DP, and cervical loop at the cap stage (D). Mimic and inhibitor significantly altered the miRNA expression level of miRNA-221-3p at E13 for 36 h (E,F, n = 3) (corresponding to data in Supplementary Table 2A). Conserved seed sequence of miRNA-221-3p with Ptch1 gene (G). The expression level of Ptch1 increased with miRNA-221-3p inhibition (n = 3), while decreased with mimic (H,I, n = 3) (corresponding to data in Supplementary Table 2B). The culture of tooth germs with mimic and inhibitor at E13 for 2 days show alteration in the length of developing tooth mesio-distally and bucco-lingually (J–O, n = 8). The dotted lines define the boundary of epithelium and mesenchyme (A–D). H&E, hematoxylin and eosin staining; E, enamel organ; IEE, inner enamel epithelium; OEE, outer enamel epithelium; EK, enamel knot; DP, dental papilla; Me, mesial; Di, distal; Bu, buccal; Li, lingual. Data were evaluated for statistical significance using unpaired, two-tailed t-test. *, **, ***, and **** indicate p < 0.05, 0.01, 0.001, and 0.0001, respectively. Scale bars: 500 μm (J–L), 50 μm (A–D).
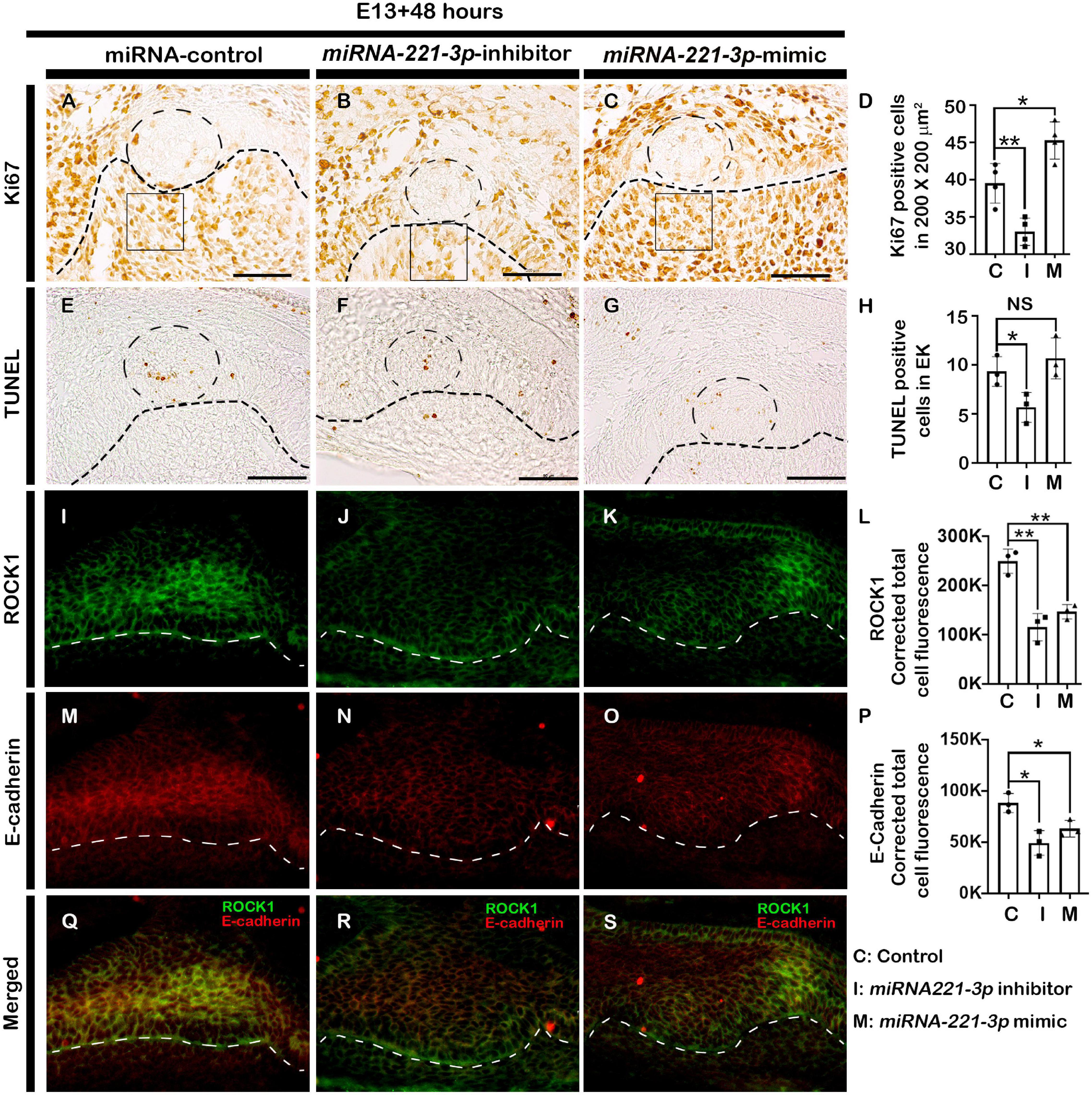
Figure 2. Altered cellular events after loss and gain of function of miRNA-221-3p. The cellular proliferations especially at the EK and dental mesenchyme adjacent to the IEE increased in the mimic and decreased in the inhibitor-treated tooth germs (A–D, n = 4). Loss and gain of miRNA-221-3p at E13 for 2 days show decreased apoptotic cells along the EK of inhibitor-group compared to control and mimic (E–H, n = 3). Localizations of ROCK1 (green) (I–L) (n = 3) and E-cadherin (red) (M–O, n = 3) along the enamel organ, showing decreased intensities in the inhibitor and mimic specimens, compared to control (Q–S, n = 3). The dotted circles in the IEE (A–C,E–G) define the EK, the dotted lines define the boundary of epithelium and mesenchyme (A–O) and the squares (A–C) defines the area where Ki67 positive cells are counted. EK, enamel knot. Data were evaluated for statistical significance using unpaired, two-tailed t-test. * and ** indicate p < 0.05 and 0.01, respectively, NS, not significant. Scale bars: 50 μm (A–O).
miRNA-221-3p Modulates Tooth Development-Related Signaling Molecules
There is high conservation of signaling molecules belonging to Bmp, Fgf, Shh, and Wnt pathways to achieve proper tooth development from initiation to eruption (Jussila and Thesleff, 2012). To evaluate whether the inhibition and mimicking of miRNA-221-3p during odontogenesis altered tooth development-related signaling molecules, we employed qPCR at E13 + 2 days to evaluate the expressions of these signaling molecules as described previously (Aryal et al., 2020; Neupane et al., 2020). Tooth germ culture at E13 + 2 days with inhibitor upregulated the Ptch1 and with the mimic downregulated the Ptch1 expression consistent with our hypothesis that miRNA-221 targets Ptch1. Meanwhile Axin2, β-catenin, Bmp2, Fgf4, Shh, Smo, Gli1, Gli2, and Gli3 were downregulated, whereas Bmp4 was upregulated in the inhibitor-treated specimen (Figure 3A). In contrast, β-catenin, Lef1, Fgf4, Shh, Smo, Gli1, Gli2, and Gli3 were upregulated, with Bmp4 downregulated in the mimic-treated specimen (Figure 3B). Except Shh target genes, the expressions of Wnt, Bmp, and Fgf-related signaling molecules were almost similar in both 36 and 48 h in in vitro cultivated tooth germs (Figures 3A,B and Supplementary Figure 4). Wnt/β-catenin signaling negatively regulates the odontogenic epithelial cell proliferation during the bell stage of tooth development (Lu et al., 2019) and has a role in crown and tooth root morphogenesis. To determine the role of miRNA-221-3p in Wnt signaling modulation, immunohistochemistry with β-catenin was performed in E13 + 2 days in vitro cultivated tooth germs. The active form of β-catenin was localized in the cytoplasm and nucleus of IEE in the control tooth at E13 + 2 days (Figures 3C,I,J). However, an increase in the localization of β-catenin was observed comparatively within the nucleus in the inhibitor and mimic-treated specimen (Figures 3D,E,I,J and Supplementary Figure 5) suggesting that Wnt signaling is negatively regulated by miRNA-221-3p during tooth cap and bell stages. Previous study suggested that Wnts and Bmp4 are the key mediators of odontogenic epithelial–mesenchymal interactions, and that Wnt signaling is upstream of Fgf, Shh, and Bmp signaling during tooth development (O’Connell et al., 2012). This suggests that Bmp signaling would be indirectly affected when the levels of miRNA-221-3p are altered. Therefore, immunohistochemistry of pSMAD1/5/8 was performed on inhibitor and mimic to miRNA-221-3p-treated teeth. The epithelial localization of pSMAD1/5/8 was almost similar in all specimens, however, the mesenchymal localizations of pSMAD1/5/8 were stronger in the inhibitor-treated specimen compared to control and mimic-treated specimens (Figures 3F–H,K,L). This suggests a role for miRNA-221-3p during the cap and bell stages of tooth development that also affects the tooth crown and root morphogenesis. Similarly, section in situ hybridization was performed to examine the Shh and Fgf signaling after inhibition and mimicking of miRNA-221-3p. Shh activity in the developing tooth is very dynamic throughout tooth development, including tooth crown and root morphogenesis (Seppala et al., 2017). The expression of Shh was expanded along the entire IEE of the control specimen (Figure 3M), however, the inhibitor-treated tooth germ had confined expression patterns along the EK only (Figure 3N). However, the expression of Shh in the mimic-treated specimen was expanded along the EK, although its expression pattern was not continuous along the entire IEE and ectopically expressed in the dental papilla (Figure 3O), suggesting miRNA-221-3p modulates Shh in the developing tooth potentially through Ptch1 inhibition. Meanwhile, Fgf4, which determines invagination of dental epithelium, tooth shape, and cusps formation, and also ameloblast and odontoblast differentiation (Li et al., 2014) was expressed along the EK of the in vitro cultivated tooth germs. However, the expression pattern of Fgf4 is extended in the EK of mimic-treated tooth germ compared to the control and inhibitor (Figures 3P–R), suggesting the potential role of miRNA-221-3p in crown morphogenesis that ultimately affecting the root morphogenesis.
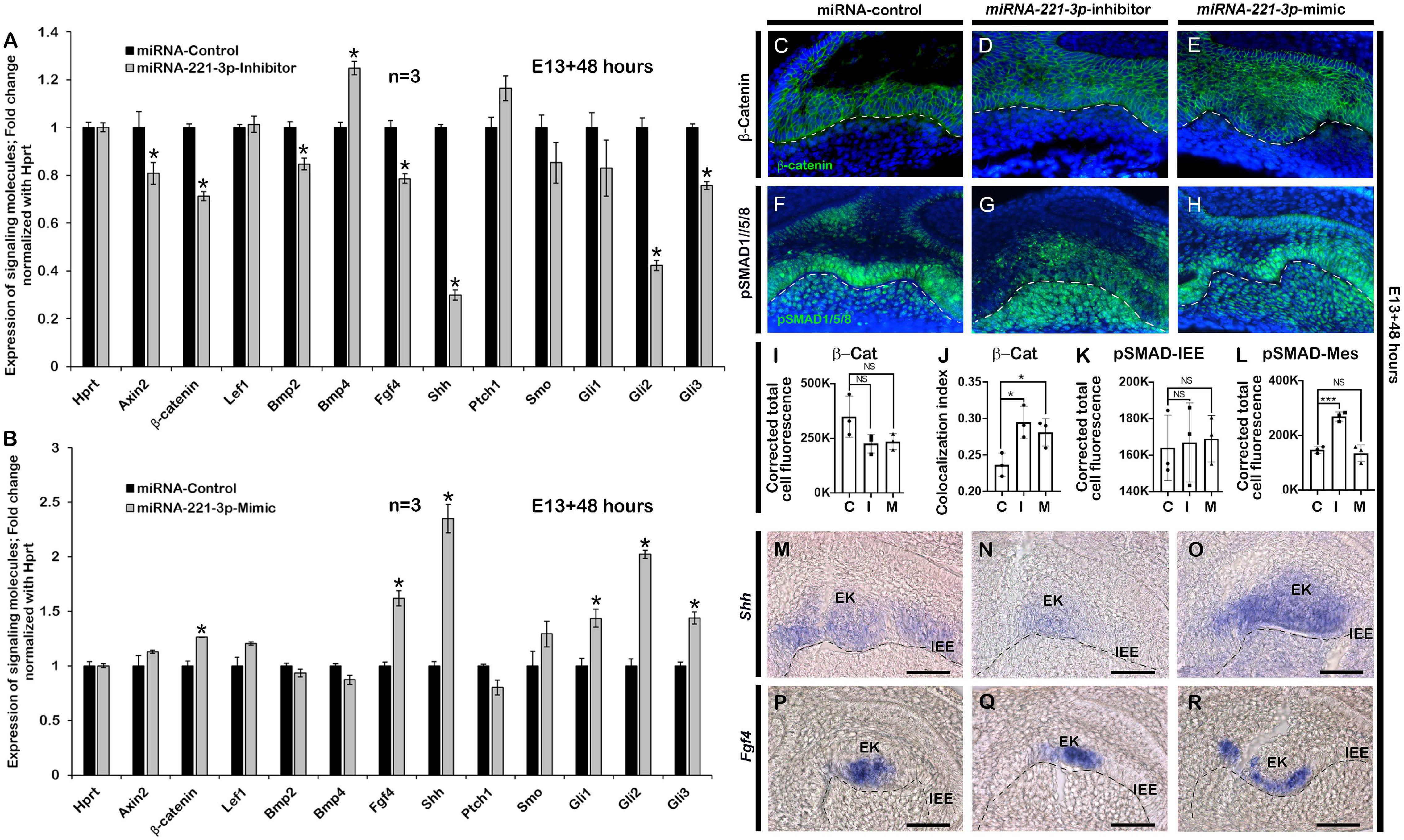
Figure 3. Altered expressions of signaling molecules after loss and gain of function of miRNA-221-3p. Signaling molecules including Axin2, β-catenin, Bmp2, Bmp4, Fgf4, Shh, Gli1, Gli2, and Gli3 are significantly altered in the inhibitor and mimic-treated specimens (A,B, n = 3) (corresponding to data in Supplementary Table 2B). Epithelial localization of β-catenin (green) along IEE, showing comparatively increased nuclear localization of β-catenin in the inhibitor and mimic-treated specimens compared to control (C–E,I,J, n = 3). DAPI (blue) stains the nuclei. Epithelial and mesenchymal localization pattern of pSMAD1/5/8 (green) after inhibition and mimic of miRNA-221-3p, showing intense mesenchymal localization in the inhibitor-treated specimens compared to control and mimic (F–H,K,L, n = 3). Section in situ hybridization of in vitro cultivated tooth germs showing expressions of Shh (M–O, n = 3) and Fgf4 (P–R, n = 3) along the EK and IEE. β-Cat, β-Catenin; EK, enamel knot; IEE, inner enamel epithelium; Mes, mesenchyme. Dotted line demarcate the boundary of epithelium and mesenchyme (C–H,M–R). Data were evaluated for statistical significance using unpaired, two-tailed t-test. * indicate p < 0.05; NS, not significant. Scale bars: 50 μ (C–N).
Loss of miRNA-221-3p Alters Tooth Crown and Root Morphogenesis
Renal capsule transplantation was utilized to determine whether inhibition and mimicking of miRNA-221-3p occurred during tooth crown and root development, a technique previously used (Aryal et al., 2020; Neupane et al., 2020). The developmental timeline of in vitro (E13 + 2 days) and 1-week renal calcified teeth was similar to the in vivo post-natal 3 (PN3) mice. The double-layered HERS grew apically and meets OEE and IEE leaving no interposing layers of stellate reticulum (SR) and stratum intermedium (SI) (Figures 4A–C). The ameloblasts (Am) and odontoblasts (Od) are in their secretory phase and secrete extracellular matrix (Em) (Figures 4A–C). Consistently, the inhibition of miRNA-221-3p demonstrated a smaller tooth with a reduction in Em secretion, while mimicking the miRNA did not show an alteration in tooth size (Figures 4A–C). Therefore, we evaluated the localization patterns of major extracellular matrix protein of enamel: amelogenin (AMELX) and odontoblast differentiation marker: NESTIN. The localization patterns of AMELX and NESTIN were almost similar in both control and mimic-treated specimens, whereas the localization intensity decreased in the inhibitor-treated specimen (Figures 4D–F). Meanwhile, the localization of NESTIN had an increase in intensity in the dental pulp cells along with secretory odontoblasts in the inhibitor-treated specimen, in comparison to the control and mimic specimen (Figures 4D–F). In addition, the epithelial cells of HERS in 1-week renal calcified teeth were columnar shaped in the inhibitor-treated specimen in comparison to control and mimic, which were cuboidal in shape (Figures 4G–I). This led us to examine the cellular proliferations along the HERS, and it was identified that cellular proliferation of HERS was comparatively higher in the inhibitor-treated specimen compared to mimic and control (Figures 4J–M and Supplementary Figure 1). As HERS play an important role in tooth root formation, we further examined the expression of the signaling molecule Shh, which plays an important role in HERS during tooth root formation (Nakatomi et al., 2006; Ota, 2008; Li et al., 2017). The section in situ hybridization demonstrated intense expression of Shh along the HERS within the inhibitor group compared to control and mimic (Supplementary Figure 6). To confirm whether altered proliferation and signaling in HERS in the inhibitor-treated specimen would have impacted tooth morphology, we employed 3-week renal capsule calcified teeth, and their morphometric measurements were taken (Figure 5). The particular measurements taken were crown length, mesiodistal length, root length, and absolute tooth length. Inhibition of miRNA-221-3p results in a longer and slenderer tooth compared to control and mimic (Figures 5A–H). However, in the inhibitor-treated specimen the mesiodistal length is significantly shorter, while the root and absolute tooth length is significantly longer (Figures 5B,E,G).
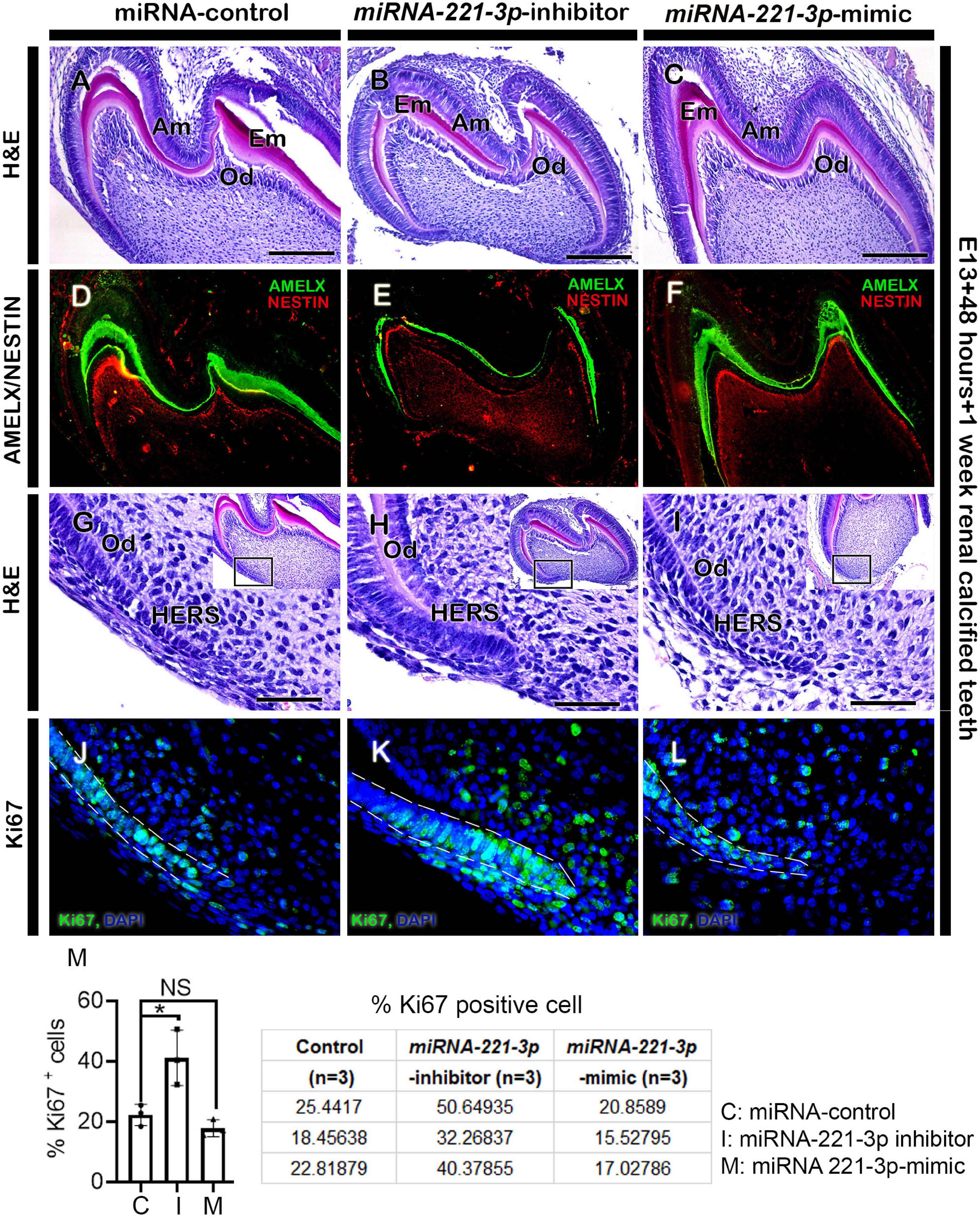
Figure 4. Altered histogenesis and cellular events in 1-week renal calcified teeth. H&E staining showing 1 week renal calcified teeth after loss and gain of function of miRNA-221-3p (A–C, n = 3). The inhibitor-treated tooth (B) is comparatively smaller with reduced extracellular matrix secretion compared to control and mimic (A,C). The localizations of AMELX and NESTIN are weaker in inhibitor-treated specimen compared to mimic and control (D–F, n = 3). H&E staining showing HERS in 1-week calcified teeth (G–I). The cellular proliferation (green) along the HERS is higher in the inhibitor-treated specimen compared to mimic and control (J–M, n = 3). DAPI (blue) stains the nuclei. Boxes in G–I are magnified views. Dotted lines (J–L) indicate the HERS. H&E, hematoxylin and eosin; Am, ameloblasts; Od, odontoblasts; AMELX, amelogenin; HERS, hertwig’s epithelial root sheath. Data were evaluated for statistical significance using unpaired, two-tailed t-test. ∗Indicates p < 0.05; NS, not significant. Scale bars: 200 μm (A–F), 50 μm (G–L).
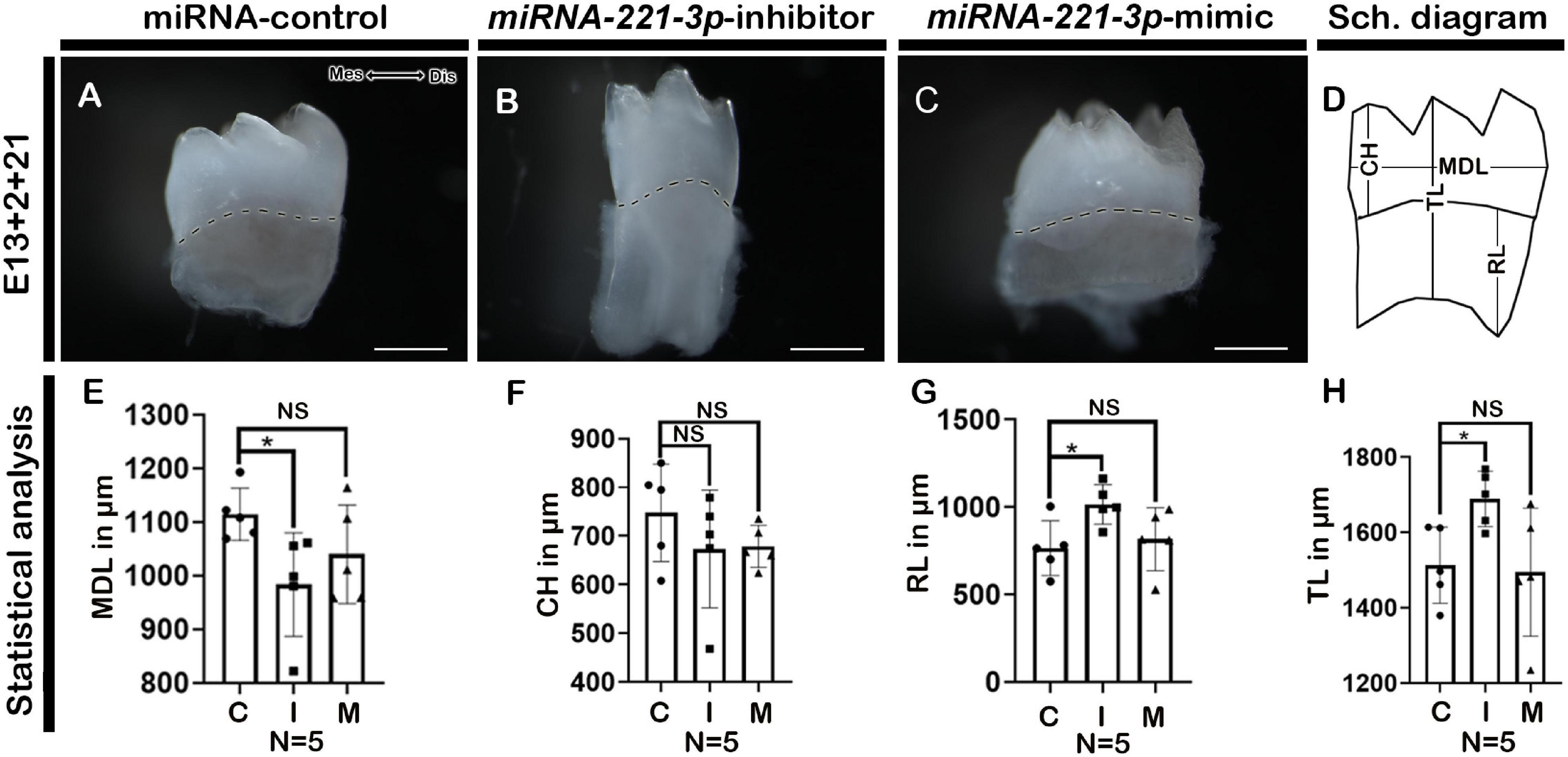
Figure 5. Altered tooth morphology in 3-week renal calcified teeth. Three weeks renal calcified teeth showing buccal view (A–C). Schematic diagram showing MDL, CH, RL, and TL of renal calcified teeth (D). Statistical analysis showing MDL, CH, RL, and TL of 3-week renal calcified tooth (E–H, n = 5). Inhibition of miRNA-221-3p makes tooth longer and slenderer compared to control and mimic (A–C). The mesio-distal length is significantly shorter, while root length is significantly longer in the inhibitor-treated specimen (B,E,G). The dotted lines demarcate the boundary of crown and root (A–C). MDL, mesio-distal length; CH, crown height; RL, root length; TL, total length; Mes, mesial; Dis, distal. Data were evaluated for statistical significance using unpaired, two-tailed t-test. ∗Indicates p < 0.05; NS, not significant. Scale bars: 500 μm (A–C).
Discussion
The complex and dynamic molecular interactions involved in tooth development are well understood and have highlighted the importance of spatiotemporal functions of signaling molecules during odontogenesis (Jussila and Thesleff, 2012; Huang et al., 2020). Experimental approaches, such as in vitro cultivation and loss and gain of function of signaling molecules, demonstrate their precise roles during tissue and organ morphogenesis (Jiang et al., 2017; Aryal et al., 2020; Huang et al., 2020). In this study, miRNA-221-3p, a conserved miRNA within the animal kingdom (Agarwal et al., 2015; Mari et al., 2016; Abak et al., 2018), was evaluated through knockdown and mimicking of function, during mice molar development. The precise expression of miRNA-221-3p has been identified to overlap with the expression of known transcription and paracrine factors, such as Bmp2, Bmp4, Bmp7, Wnt3, Wnt10a, Wnt10b, Fgf4, and Shh (Jussila and Thesleff, 2012; Balic and Thesleff, 2015), during important stages of tooth development (Figure 1). This demonstrates its modulating role during odontogenesis that prompted us to examine its role during molar development in detail.
The epithelial–mesenchymal interactions up to, and beyond the bud stage plays a significant role in normal tooth progression and development (Jussila and Thesleff, 2012; Huang et al., 2020). The altered signaling molecules that arise as a result of inhibition and mimicking of miRNA-221-3p during the bud stage of tooth development, highlighted its role in signal modulation during odontogenesis. Normally, the activation of Wnt/β-catenin signaling occurs in the dental epithelial and adjacent mesenchymal cells. However, upon inhibition, it resulted in the developmental arrest at the bud stage and short root phenotype (Liu et al., 2008; Liu and Millar, 2010; Han et al., 2011). In our study, the inhibition of miRNA-221-3p during bud stage showed downregulation of β-catenin along the dental epithelium. Therefore, it can be suggested that miRNA-221-3p plays a modulating role in Wnt/β-catenin signaling during crosstalk between epithelium and mesenchyme, due to the optimal level of Wnt/β-catenin signaling being essential for the developmental progression of the tooth (Wang and Feng, 2017). Similarly, the developmental role of Bmp, a member of the TGF- β- superfamily, also plays a vital role during early and late tooth differentiation (Thesleff, 2003; Jussila and Thesleff, 2012; Wang et al., 2012). In our study, the inhibition of miRNA-221-3p resulted in increased Bmp signaling and intense mesenchymal localization of pSMAD1/5/8 in the inhibitor-treated specimen. This altered Bmp signaling resulted in decreased cellular proliferation and apoptosis in the dental epithelium and adjacent mesenchyme, as in previous reports (Wang et al., 2012; Kim et al., 2014). This would result in a smaller tooth size at E13 + 2 days (Figure 1). A miRNA target prediction tool miRDB (Chen and Wang, 2020) predicts more than 530 targets of miRNA-221-3p. Among some of the notable targets are cyclin-dependent kinase inhibitors, Bcl2 like 11, Bmpr1a, and Lrp10 suggesting that change in tooth size resulted from change in combination of different signaling pathways related to cell cycle arrest, apoptosis, BMP and WNT signaling. Future works are needed for the validation of putative role of miRNA 221-3p on modulation of these molecules.
Inhibition of miRNA-221-3p decreased the expression of Fgf4, which in general, promotes proliferation, determines tooth cusps (Jernvall et al., 1994; Du et al., 2017), and prevents cell apoptosis in the dental epithelium and mesenchyme (Vaahtokari et al., 1996; Jernvall et al., 1998). Kratochwil et al., 2002 showed that Fgf4 is completely lost in Lef1 knockout teeth which is arrested at bud stage and recombinant Fgf4 rescued this phenotype. Consistent with this, we expected smaller tooth size with decreased level of Fgf4 in inhibitor treated specimen. Pharmacological inhibition of Fgf signaling by SU5402 blocked the epithelial morphogenesis and tooth mineralization in zebrafish (Jackman et al., 2004) which also suggest that Fgf4 is important signaling for proper tooth crown morphogenesis. It also contributed to decreased tooth size through decreased cell proliferation and increased apoptosis. At the same time, Shh induced proliferation in a differential manner and determines tooth shape and affects ameloblast polarization through odontoblast differentiation and loss of Shh did not change the expression of other signaling molecules like Fgf4, Bmp2, and Wnt10b (Dassule et al., 2000). However, loss of Shh altered the expression patterns in developing dental tissues suggesting that the loss of Shh results in a change of cell fate, from proliferative to a less proliferative enamel knot-like population (Dassule et al., 2000) which reinforce our hypothesis that miRNA modulation of Shh signaling is crucial for crown morphogenesis and evident in inhibitor-treated specimen. Consistent with our results, inhibition of Shh by 5E1 and Forskolin during tooth organ culture resulted decreased proliferation, increased apoptosis leading to altered tooth phenotype (Cobourne et al., 2001). In quiescent state, Ptch1 suppress the Smoothened which in turn controls the Gli transcriptional effectors in the repressive forms. In the presence of SHH ligand, the Ptch1 suppression of Smoothened is relieved and the signal is transduced through activation of Gli transcription factors (Cohen et al., 2015) which is tightly regulated process. When tooth are cultivated in presence of miRNA-221-3p inhibitor, Ptch1 is elevated which may interfere the regulation of Shh signaling through extended Smoothened suppression. On the other hand, mimicking the miRNA-221-3p lowers the Ptch1 in the system and Smoothened may no longer be suppressed which may bypass the early step of Ptch1-Shh binding which ultimately affects the Shh signaling cascade resulting altered Shh expression in the cultured tissues. Also, these altered expression area would be explained by spatiotemporal specific expression of genes which are required for harmonious organogenesis rather than just changing in expression levels during development. These results demonstrate the tight regulation of signaling molecules during the early stages of tooth development, which lay the foundation for the complete morphogenesis of crown and root. In addition, mesenchymal expressed Bmp signaling is also important for HERS development and tooth root formation (Hosoya et al., 2008), further suggesting that the signaling modulation by miRNA-221-3p will guide the tooth root formation possibly through modulation of Wnt/β-catenin and Bmp signaling. Previous studies have demonstrated the role of ROCK1 in ameloblast differentiation (Otsu et al., 2011) and the association of ROCK1 and cadherin complex for maintaining cellular adhesion (Smith et al., 2012). However, E-cadherin-mediated cellular adhesion was decreased with inhibition and mimicking of miRNA-221-3p in our study (Figure 2), suggesting that miRNA-221-3p modulates ROCK1 and E-cadherin-mediated maintenance of actin cytoskeleton and cellular adhesion that ultimately regulates the differentiation stage of tooth and crown morphogenesis.
During mice molar development, at around PN2, crown formation is almost completed and the root formation initiates with the emergence of HERS (Lungová et al., 2011). The corono-apical extension of the OEE and IEE along the cervical loop forms HERS and guides for the development of roots (Lungová et al., 2011; Li et al., 2017). HERS, which is regarded as the signaling center for tooth root formation and contains various signaling molecules and transcription factors (Huang et al., 2009; Lungová et al., 2011; Li et al., 2017). For tooth root progression, the cellular proliferation of HERS together with adjacent mesenchymal cells is evident (Lungová et al., 2011; Sohn et al., 2014; Li et al., 2017). In this study, the inhibition of miRNA-221-3p showed increased proliferative cells along the HERS and its adjacent mesenchymal cells (Figure 4). The epithelial cells of HERS had Shh expression, and the reciprocal interactions of Shh signaling along the HERS epithelium and adjacent mesenchymal cells, would play a significant role in tooth root formation (Nakatomi et al., 2006; Ota, 2008; Li et al., 2017). However, this study demonstrated an increased expression of Shh along the HERS in the inhibitor group compared to control and mimic (Supplementary Figure 6). This may have contributed to increased proliferation of HERS and ultimately affected tooth height, which is consistent with previous studies that showed the partial rescue of root development by Shh in the rootless K14-Cre;Smad4fl/fl (Huang et al., 2010; Hosoya et al., 2020). In addition, cell proliferation is reduced around the HERS in homozygous Ptcmes mutants affecting the tooth root formation (Nakatomi et al., 2006) further reiterating that the long and slender teeth in 3-week renal calcified miRNA-221-3p inhibitor-treated teeth are a result of increased proliferation and intense Shh expression. The inhibition of miRNA-221-3p at the bud and cap stage resulted in a decreased expression of Shh after 2 days (Figure 2), however, the knockdown effect at this stage would result in a return of its expression patterns after the inhibitor was removed. Therefore, it can be speculated that 1-week renal calcified teeth had comparatively increased expression patterns of Shh along the HERS in the inhibitor group compared to control and mimic (Supplementary Figure 6). As the transient knockdown of genes can result in a regain of its expression at a certain time point (Eadon et al., 2017), it can also be deduced that the inhibition of miRNA-221-3p may also be regained and upregulate the Shh expression in the HERS after 1-week.
In conclusion, miRNA-221-3p modulates the Shh and Wnt signaling during cap and bell stages of tooth development, and the reciprocal interaction between the mesenchyme and epithelium, in turn, regulates other signaling molecules, such as Fgf and Bmp. These changes in signaling eventually determine the fate of tooth crown and root morphogenesis through hard tissue formation. Therefore, we conclude that the modulation of miRNA-221-3p signaling may explain some of the processes involved in the integrative and fine-tuning networks of signaling molecules involved in tooth development. In addition, it is required to examine the detailed signaling regulations of SHH signaling through miRNA-221-3p using pharmacological modulators of miRNA inhibited tooth culture and, rescue of β-catenin and Fgf signaling. Furthermore it also would be necessary to reveal the consequence signaling including p-ERK, Etv4, Etv5, and so on using genome wide screening methods to define the detailed signaling modulations in tooth morphogenesis.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The animal study was reviewed and approved by KNU-2020-0107.
Author Contributions
YA, SN, and J-YK contributed to conception, design, and data interpretation, critically revised the manuscript. T-YK, E-SL, C-HA, J-YK, HY, SL, YL, and W-JS contributed to data analysis, interpretation, and critically revised the manuscript. All authors gave final approval and agreed to be accountable for all aspects of the work.
Funding
This study was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MEST) (NRF-2017R1D1A1B03034378, NRF-2017R1A5A2015391, NRF-2018R1A2A3075600, and NRF-2018R1D1A1A02086098).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.697243/full#supplementary-material
Footnotes
References
Abak, A., Amini, S., Sakhinia, E., and Abhari, A. (2018). MicroRNA-221: biogenesis, function and signatures in human cancers. Eur. Rev. Med. Pharmacol. Sci. 22, 3094–3117. doi: 10.26355/eurrev_201805_15069
Agarwal, V., Bell, G. W., Nam, J.-W., and Bartel, D. P. (2015). Predicting effective microRNA target sites in mammalian mRNAs. eLife 4:e05005. doi: 10.7554/eLife.05005
Andl, T., Murchison, E. P., Liu, F., Zhang, Y., Yunta-Gonzalez, M., Tobias, J. W., et al. (2006). The miRNA-processing enzyme dicer is essential for the morphogenesis and maintenance of hair follicles. Curr. Biol. 16, 1041–1049. doi: 10.1016/j.cub.2006.04.005
Aryal, Y. P., Neupane, S., Kim, T. Y., Lee, E. S., Pokhrel, N. K., Yeon, C. Y., et al. (2020). Developmental roles of FUSE binding Protein 1 (Fubp1) in tooth morphogenesis. Int. J. Mol. Sci. 21:8079. doi: 10.3390/ijms21218079
Balic, A., and Thesleff, I. (2015). Tissue interactions regulating tooth development and renewal. Curr. Top. Dev. Biol. 157–186. doi: 10.1016/bs.ctdb.2015.07.006
Barbu, M. G., Condrat, C. E., Thompson, D. C., Bugnar, O. L., Cretoiu, D., Toader, O. D., et al. (2020). MicroRNA involvement in signaling pathways during viral infection. Front. Cell Dev. Biol. 8:143. doi: 10.3389/fcell.2020.00143
Bartel, D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297. doi: 10.1016/S0092-8674(04)00045-5
Barwari, T., Joshi, A., and Mayr, M. (2016). MicroRNAs in cardiovascular disease. J. Am. Coll. Cardiol. 68, 2577–2584. doi: 10.1016/j.jacc.2016.09.945
Cao, H., Wang, J., Li, X., Florez, S., Huang, Z., Venugopalan, S. R., et al. (2010). MicroRNAs play a critical role in tooth development. J. Dent. Res. 89, 779–784. doi: 10.1177/0022034510369304
Chen, Y., and Wang, X. (2020). miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 48, D127–D131.
Cobourne, M. T., Hardcastle, Z., and Sharpe, P. T. (2001). Sonic hedgehog regulates epithelial proliferation and cell survival in the developing tooth germ. J. Dent. Res. 80, 1974–1979. doi: 10.1177/00220345010800110501
Cohen, M., Kicheva, A., Ribeiro, A., Blassberg, R., Page, K. M., Barnes, C. P., et al. (2015). Ptch1 and Gli regulate Shh signalling dynamics via multipe mechanisms. Nat. Comm. 6:6709. doi: 10.1038/ncomms7709
Dassule, H. R., Lewis, P., Bei, M., Maas, R., and McMahon, A. P. (2000). Sonic hedgehog regulates growth and morphogenesis of the tooth. Development 127, 4775–4785. doi: 10.1242/dev.127.22.4775
Du, W., Hu, J. K. H., Du, W., and Klein, O. D. (2017). Lineage tracing of epithelial cells in developing teeth reveals two strategies for building signaling centers. J. Biol. Chem. 292, 15062–15069. doi: 10.1074/jbc.M117.785923
Eadon, M. T., Cheng, Y. H., Hato, T., Benson, E. A., Ipe, J., Collins, K. S., et al. (2017). In vivo siRNA delivery and rebound of renal LRP2 in mice. J. Drug Deliv. 2017:4070793. doi: 10.1155/2017/4070793
Funada, K., Yoshizaki, K., MIyazaki, K., Han, X., Yuta, T., Tian, T., et al. (2020). microRNA-875-5p plays critical role for mesenchymal condensation in epithelial-mesenchymal interaction during tooth development. Sci. Rep. 10:4918. doi: 10.1038/s41598-020-61693-w
Ha, M., and Kim, V. N. (2014). Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 15, 509–524. doi: 10.1038/nrm3838
Han, X. L., Liu, M., Voisey, A., Ren, Y. S., Kurimoto, P., Gao, T., et al. (2011). Post-natal effect of overexpressed DKK1 on mandibular molar formation. J. Dent. Res. 90, 1312–1317. doi: 10.1177/0022034511421926
Hosoya, A., Kim, J. Y., Cho, S. W., and Jung, H. S. (2008). BMP4 signaling regulates formation of Hertwig’s epithelial root sheath during tooth root development. Cell Tissue Res. 333, 503–509. doi: 10.1007/s00441-008-0655-z
Hosoya, A., Shalehin, N., Takebe, H., Shimo, T., and Irie, K. (2020). Sonic hedgehog signaling and tooth development. Int. J. Mol. Sci. 21:1587. doi: 10.3390/ijms21051587
Huang, D., Ren, J., Li, R., Guan, C., Feng, Z., Bao, B., et al. (2020). Tooth regeneration: insights from tooth development and spatial-temporal control of bioactive drug release. Stem Cell Rev. Rep. 16, 41–55. doi: 10.1007/s12015-019-09940-0
Huang, X., Bringas, P. Jr., Slavkin, H. C., and Chai, Y. (2009). Fate of HERS during tooth root development. Dev. Biol. 334, 22–30. doi: 10.1016/j.ydbio.2009.06.034
Huang, X., Xu, X., Bringas, P. Jr., Hung, Y. P., and Chai, Y. (2010). Smad4-Shh-Nfic signaling cascade–mediated epithelial-mesenchymal interaction is crucial in regulating tooth root development. J. Bone Miner. Res. 25, 1167–1178. doi: 10.1359/jbmr.091103
Ihle, M. A., Trautmann, M., Kuenstlinger, H., Huss, S., Heydt, C., Fassunke, J., et al. (2015). miRNA-221 and miRNA-222 induce apoptosis via the KIT/AKT signalling pathway in gastrointestinal stromal tumours. Mol. Oncol. 9, 1421–1433. doi: 10.1016/j.molonc.2015.03.013
Jackman, W. R., Draper, B. W., and Stock, D. W. (2004). Fgf signaling is required for zebrafish tooth development. Dev. Biol. 274, 139–157.
Jernvall, J., Aberg, T., Kettunen, P., Keränen, S., and Thesleff, I. (1998). The life history of an embryonic signaling center: BMP-4 induces p21 and is associated with apoptosis in the mouse tooth enamel knot. Development 125, 161–169.
Jernvall, J., Kettunen, P., Karavanova, I., Martin, L. B., and Thesleff, I. (1994). Evidence for the role of the enamel knot as a control center in mammalian tooth cusp formation: non-dividing cells express growth stimulating Fgf-4 gene. Int. J. Dev. Biol. 38, 463–469.
Jheon, A. H., Li, C. Y., Wen, T., Michon, F., and Klein, O. D. (2011). Expression of microRNAs in the stem cell niche of the adult mouse incisor. PLoS One 6:e24536. doi: 10.1371/journal.pone.0024536
Jiang, N., Xiang, L., He, L., Yang, G., Zheng, J., Wang, C., et al. (2017). Exosomes mediate epithelium–mesenchyme crosstalk in organ development. A.C.S. Nano. 11, 7736–7746. doi: 10.1021/acsnano.7b01087
Jussila, M., and Thesleff, I. (2012). Signaling networks regulating tooth organogenesis and regeneration, and the specification of dental mesenchymal and epithelial cell lineages. Cold Spring Harb. Perspect. Biol. 4:a008425. doi: 10.1101/cshperspect.a008425
Kim, E. J., Lee, M. J., Li, L. I., Yoon, K. S., Kim, K. S., and Jung, H. S. (2014). Failure of tooth formation mediated by miR-135a overexpression via BMP signaling. J. Dent. Res. 93, 571–575. doi: 10.1177/0022034514529303
Kratochwil, K., Galcern, J., Tontsch, S., Roth, W., and Grosschedl, R. (2002). FGF4, a direct target of LEF1 and Wnt signaling, can rescue the arrest of tooth organogenesis in Lef1−/− mice. Genes Dev. 16, 3173–3185. doi: 10.1101/gad.1035602
Li, A., Li, Y., Song, T., Wang, F., Liu, D., Fan, Z., et al. (2015). Identification of differential microRNA expression during tooth morphogenesis in the heterodont dentition of miniature pigs, SusScrofa. B.M.C. Dev. Biol. 15:51. doi: 10.1186/s12861-015-0099-0
Li, C. Y., Prochazka, J., Goodwin, A. F., and Klein, O. D. (2014). Fibroblast growth factor signaling in mammalian tooth development. Odontology 102, 1–13. doi: 10.1007/s10266-013-0142-1
Li, J., Parada, C., and Chai, Y. (2017). Cellular and molecular mechanisms of tooth root development. Development 144, 374–384. doi: 10.1242/dev.137216
Liu, F., Chu, E. Y., Watt, B., Zhang, Y., Gallant, N. M., Andl, T., et al. (2008). Wnt/β-catenin signaling directs multiple stages of tooth morphogenesis. Dev. Biol. 313, 210–224. doi: 10.1016/j.ydbio.2007.10.016
Liu, F., and Millar, S. E. (2010). Wnt/β-catenin signaling in oral tissue development and disease. J. Dent. Res. 89, 318–330. doi: 10.1177/0022034510363373
Liu, H., Lin, H., Zhang, L., Sun, Q., Yuan, G., Zhang, L., et al. (2013). miR-145 and miR-143 regulate odontoblast differentiation through targeting Klf4 and Osx genes in a feedback loop. J. Biol. Chem. 288, 9261–9271. doi: 10.1074/jbc.M112.433730
Liu, M., Liu, J., Wang, L., Wu, H., Zhou, C., Zhu, H., et al. (2014). Association of serum microRNA expression in hepatocellular carcinomas treated with transarterial chemoembolization and patient survival. PLoS One 9:e109347. doi: 10.1371/journal.pone.0109347
Lu, X., Yang, J., Zhao, S., and Liu, S. (2019). Advances of Wnt signalling pathway in dental development and potential clinical application. Organogenesis 15, 101–110. doi: 10.1080/15476278.2019.1656996
Lungová, V., Radlanski, R. J., Tucker, A. S., Renz, H., Míšek, I., and Matalová, E. (2011). Tooth-bone morphogenesis during postnatal stages of mouse first molar development. J. Anat. 218, 699–716. doi: 10.1111/j.1469-7580.2011.01367.x
Maeda, Y., Farina, N. H., Matzelle, M. M., Fanning, P. J., Lian, J. B., and Gravallese, E. M. (2017). Synovium-derived microRNAs regulate bone pathways in rheumatoid arthritis. J. Bone Miner. Res. 32, 461–472. doi: 10.1002/jbmr.3005
Mari, E., Zicari, A., Fico, F., Massimi, I., Martina, L., and Mardente, S. (2016). Action of HMGB1 on miR-221/222 cluster in neuroblastoma cell lines. Oncol. Lett. 12, 2133–2138. doi: 10.3892/ol.2016.4876
Michon, F., Tummers, M., Kyyrönen, M., Frilander, M. J., and Thesleff, I. (2010). Tooth morphogenesis and ameloblast differentiation are regulated by micro-RNAs. Dev. Biol. 340, 355–368. doi: 10.1016/j.ydbio.2010.01.019
Nakatomi, M., Morita, I., Eto, K., and Ota, M. S. (2006). Sonic hedgehog signaling is important in tooth root development. J. Dent. Res. 85, 427–431. doi: 10.1177/154405910608500506
Neupane, S., Aryal, Y. P., Kim, T. Y., Yeon, C. Y., An, C. H., Kim, J. Y., et al. (2020). Signaling Modulations of miR-206-3p in Tooth Morphogenesis. Int. J. Mol. Sci. 21:5251. doi: 10.3390/ijms21155251
Neupane, S., Goto, J., Berardinelli, S. J., Ito, A., Haltiwanger, R. S., and Holdener, B. C. (2021). Hydrocephalus in mouse B3glct mutants is likely caused by defects in multiple B3glct substrates in ependymal cells and subcommissural organ. Glycobiology cwab033. doi: 10.1093/glycob/cwab033 [Epub ahead of print].
O’Connell, D. J., Ho, J. W., Mammoto, T., Turbe-Doan, A., O’Connell, J. T., Haseley, P. S., et al. (2012). A Wnt-Bmp feedback circuit controls intertissue signaling dynamics in tooth organogenesis. Sci. Signal. 5:ra4. doi: 10.1126/scisignal.2002414
Ota, M. S. (2008). The role of Sonic hedgehog signaling and fibroblast growth factors in tooth development in mice. J. Oral Biosci. 50, 167–174. doi: 10.1016/S1349-0079(08)80004-7
Otsu, K., Kishigami, R., Fujiwara, N., Ishizeki, K., and Harada, H. (2011). Functional role of rho-kinase in ameloblast differentiation. J. Cell. Physiol. 226, 2527–2534. doi: 10.1002/jcp.22597
Rahmanian, S., Murad, R., Breschi, A., Zeng, W., Mackiewicz, M., Williams, B., et al. (2019). Dynamics of microRNA expression during mouse prenatal development. Genome Res. 29, 1900–1909. doi: 10.1101/gr.248997.119
Seppala, M., Fraser, G. J., Birjandi, A. A., Xavier, G. M., and Cobourne, M. T. (2017). Sonic hedgehog signaling and development of the dentition. J. Dev. Biol. 5:6. doi: 10.3390/jdb5020006
Setkova, J., Lesot, H., Matalova, E., Witter, K., Matulova, P., and Misek, I. (2006). Proliferation and apoptosis in early molar morphogenesis-voles as models in odontogenesis. Int. J. Dev. Biol. 50, 481–489. doi: 10.1387/ijdb.052067js
Sohn, W.-J., Choi, M.-A., Yamamoto, H., Lee, S., Lee, Y., Jung, J.-K., et al. (2014). Contribution of mesenchymal proliferation in tooth root morphogenesis. J. Dent. Res. 93, 78–83. doi: 10.1177/0022034513511247
Smith, A. L., Dohn, M. R., Brown, M. V., and Reynolds, A. B. (2012). Association of Rho-associated protein kinase 1 with E-cadherin complexes is mediated by p120-catenin. Mol. Biol. Cell. 23, 99–110. doi: 10.1091/mbc.E11-06-0497
Tao, K., Yang, J., Guo, Z., Hu, Y., Sheng, H., Gao, H., et al. (2014). Prognostic value of miRNA-221-3p, miR-342-3p and miR-491-5p expression in colon cancer. Am. J. Transl. Res. 6, 391–401.
Thesleff, I. (2003). Epithelial-mesenchymal signalling regulating tooth morphogenesis. J. Cell Sci. 116, 1647–1648. doi: 10.1242/jcs.00410
Trionfini, P., Benigni, A., and Remuzzi, G. (2015). MicroRNAs in kidney physiology and disease. Nat. Rev. Nephrol. 11, 23–33. doi: 10.1038/nrneph.2014.202
Vaahtokari, A., Aberg, T., and Thesleff, I. (1996). Apoptosis in the developing tooth: association with an embryonic signaling center and suppression by EGF and FGF-4. Development 122, 121–129.
Wang, J., Chen, J., and Sen, S. (2016). MicroRNA as biomarkers and diagnostics. J. Cell. Physiol. 231, 25–30. doi: 10.1002/jcp.25056
Wang, J., and Feng, J. Q. (2017). Signaling pathways critical for tooth root formation. J. Dent. Res. 96, 1221–1228. doi: 10.1177/0022034517717478
Wang, X., Liao, X., Huang, K., Zeng, X., Liu, Z., Zhou, X., et al. (2019). Clustered microRNAs hsa-miRNA-221-3p/hsa-miR-222-3p and their targeted genes might be prognostic predictors for hepatocellular carcinoma. J. Cancer 10, 2520–2533. doi: 10.7150/jca.29207
Wang, Y., Li, L., Zheng, Y., Yuan, G., Yang, G., He, F., et al. (2012). BMP activity is required for tooth development from the lamina to bud stage. J. Dent. Res. 91, 690–695. doi: 10.1177/0022034512448660
Xu, J., Bai, S., Cao, Y., Liu, L., Fang, Y., Du, J., et al. (2020). miRNA-221-3p in endothelial progenitor cell-derived exosomes accelerates skin wound healing in diabetic mice. Diabetes Metab. Syndr. Obes. 13, 1259–1270. doi: 10.2147/DMSO.S243549
Yang, Y., Cui, H., and Wang, X. (2019). Downregulation of EIF5A2 by miRNA-221-3p inhibits cell proliferation, promotes cell cycle arrest and apoptosis in medulloblastoma cells. Biosci. Biotechnol. Biochem. 83, 400–408. doi: 10.1080/09168451.2018.1553604
Yi, R., O’Carroll, D., Pasolli, H. A., Zhang, Z., Dietrich, F. S., Tarakhovsky, A., et al. (2006). Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat. Genet. 38, 356–362. doi: 10.1038/ng1744
Zheng, Q., Peskoe, S. B., Ribas, J., Rafiqi, F., Kudrolli, T., Meeker, A. K., et al. (2014). Investigation of miR-21, miR-141, and miR-221 expression levels in prostate adenocarcinoma for associated risk of recurrence after radical prostatectomy. Prostate 74, 1655–1662. doi: 10.1002/pros.22883
Keywords: miRNA, tooth development stage, signaling modulation, cellular events, tooth morphogenesis
Citation: Aryal YP, Kim T-Y, Lee E-S, An C-H, Kim J-Y, Yamamoto H, Lee S, Lee Y, Sohn W-J, Neupane S and Kim J-Y (2021) Signaling Modulation by miRNA-221-3p During Tooth Morphogenesis in Mice. Front. Cell Dev. Biol. 9:697243. doi: 10.3389/fcell.2021.697243
Received: 19 April 2021; Accepted: 03 August 2021;
Published: 25 August 2021.
Edited by:
Dorothea Schulte, University Hospital Frankfurt, GermanyReviewed by:
Timo Lüdtke, Medizinische Hochschule Hannover, GermanyTsuyoshi Hirashima, Kyoto University, Japan
Copyright © 2021 Aryal, Kim, Lee, An, Kim, Yamamoto, Lee, Lee, Sohn, Neupane and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sanjiv Neupane, c2Fuaml2LmtudUBnbWFpbC5jb20=; c2Fuaml2Lm5ldXBhbmVAc3Rvbnlicm9vay5lZHU=; Jae-Young Kim, anlraW05MUBrbnUuYWMua3I=
†These authors have contributed equally to this work and share last authorship
 Yam Prasad Aryal1
Yam Prasad Aryal1 Hitoshi Yamamoto
Hitoshi Yamamoto Youngkyun Lee
Youngkyun Lee Sanjiv Neupane
Sanjiv Neupane Jae-Young Kim
Jae-Young Kim