- 1Department of Medical Genetics, School of Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 2Critical Care Quality Improvement Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 3Department of Anatomical Sciences, Faculty of Medicine, Birjand University of Medical Sciences, Birjand, Iran
- 4Department of Anatomical Sciences, Faculty of Medicine, Tabriz University of Medical Sciences, Tabriz, Iran
- 5Urology and Nephrology Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 6Skull Base Research Center, Loghman Hakim Hospital, Shahid Beheshti University of Medical Sciences, Tehran, Iran
Stem cells have two important features, namely the ability for self-renewal and the capacity to differentiate into some cell kinds with specialized functions. These two features are also present in cancer stem cells (CSCs). These cells have been detected in almost all kinds of cancers facilitating their tumorigenicity. Molecular cascades that control self-renewal of stem cells, namely the Wnt, Notch, and Hedgehog pathways have been suggested to influence CSCs functions as well. Moreover, non-coding RNAs can regulate function of CSCs. Function of miRNAs in the regulation of CSCs has been mostly assessed in breast cancer and hepatocellular carcinoma. miR-130a-3p, miR-600, miR-590-5p, miR-142-3p, miR-221, miR-222, miR-638, miR-375, miR-31, and miR-210 are among those regulating this feature in breast cancer. Moreover, miR-206, miR-192-5p, miR-500a-3p, miR-125, miR-125b, miR-613, miR-217, miR-194, and miR-494 regulate function of CSCs in hepatocellular carcinoma. DILC, lncTCF7, MUF, HAND2-AS1, MALAT1, DLX6-AS1, HOTAIR, and XIST are among lncRNAs that regulate function of CSCs. In the present paper, we explain the effects of these two classes of non-coding RNAs in the regulation of activity of CSCs.
Introduction
Stem cells have two important features, namely the ability for self-renewal and the capacity to differentiate into some cell kinds with specialized functions. The latter capacity enables them to produce more stem cells that are kept in an undifferentiated status. However, the latter feature permits production of mature cell types. Several lines of evidence has showed the existence of a group of cells with stem-like features inside tumors (Yu et al., 2012). Being designated as cancer stem cells (CSCs), these cells show features of both stem cells and cancer cells. They not only have self-renewal and differentiation abilities, but also they can give rise to tumors when transferred into an animal (Yu et al., 2012). The primary indication pointing to the presence of CSCs came from Lapidot et al. (1994) study showing organization of human acute myeloid leukemia as a hierarchy that is derived from primitive hematopoietic cells (Lapidot et al., 1994). After nearly a decade, this model was adapted to breast cancer when Al-Hajj et al. (2003) showed the presence of a tumorigenic subpopulation capable of induction of cancer in NOD-SCID mice. This population was characterized by CD44+/CD24−/low feature (Al-Hajj et al., 2003). Subsequent studies have verified the presence of CSCs in other types of cancers as well (Singh et al., 2004). Functionally, signaling cascades that control self-renewal of stem cells, namely the Wnt, Notch, and Hedgehog pathways have been reported to influence CSCs functions as well (Rosen and Jordan, 2009). CSCs features are thought to be affected by several factors among them are certain genetic abnormalities and the stage of cancer progression (Rosen and Jordan, 2009). Meanwhile, function of CSCs is controlled by a number of non-coding RNAs, particularly long non-coding RNAs (lncRNAs) and microRNAs (miRNAs). Both lncRNAs and miRNAs can affect development of cancer through modulation of important cancer-related pathways such as Notch (Ghafouri-Fard et al., 2021c), Rho-GTPase (Ghafouri-Fard et al., 2021d) and NF-κB (Ghafouri-Fard et al., 2021a) pathways, activity of immune-related cascades (Ghafouri-Fard et al., 2021b) and epithelial-mesenchymal transition (EMT) (Hussen et al., 2021). In the present paper, we explain the effects of these two classes of non-coding RNAs in controlling function of CSCs and their impact on the tumorigenesis.
miRNAs and CSCs
Breast Cancer
miR-130a-3p is a putative tumor suppressor miRNA whose expression has been found to be diminished in human breast cancer samples and blood-derived exosomes. Forced up-regulation of miR-130a-3p in breast CSCs has suppressed proliferation, migratory potential, and invasiveness, while its silencing has led to opposite effects. Functionally, miR-130a-3p decreases expression of RAB5B. Besides, down-regulation of exosome-originated miR-130a-3p has been correlated with involvement of lymph nodes and advanced clinical stage. Based on these results, miR-130a-3p has been suggested as a biomarker for monitoring breast cancer progression and a target for treatment of breast cancer (Kong et al., 2018). miR-600 is another miRNA whose silencing enhances expansion of breast CSCs, whereas its up-regulation decreases self-renewal of these cells, resulting in attenuation of tumorigenicity in animal models. miR-600 has been shown to target SCD1 which codes a protein with essential role in the production of active, lipid-altered WNT proteins. Up-regulation of miR-600 suppresses generation of active WNT and increases differentiation of breast CSCs. Down-regulation of miR-600 in clinical samples has been associated with activation of WNT signaling and poor clinical outcome (El Helou et al., 2017). miR-590-5p is another miRNA which decreases breast CSC population. This miRNA inhibits expression of SOX2. Experiments in NOD/SCID mice have shown that miR-590-5p inhibits tumorigenicity of breast cancer cells (Zhou et al., 2017). Table 1 shows the role of miRNAs in regulation of breast CSCs.
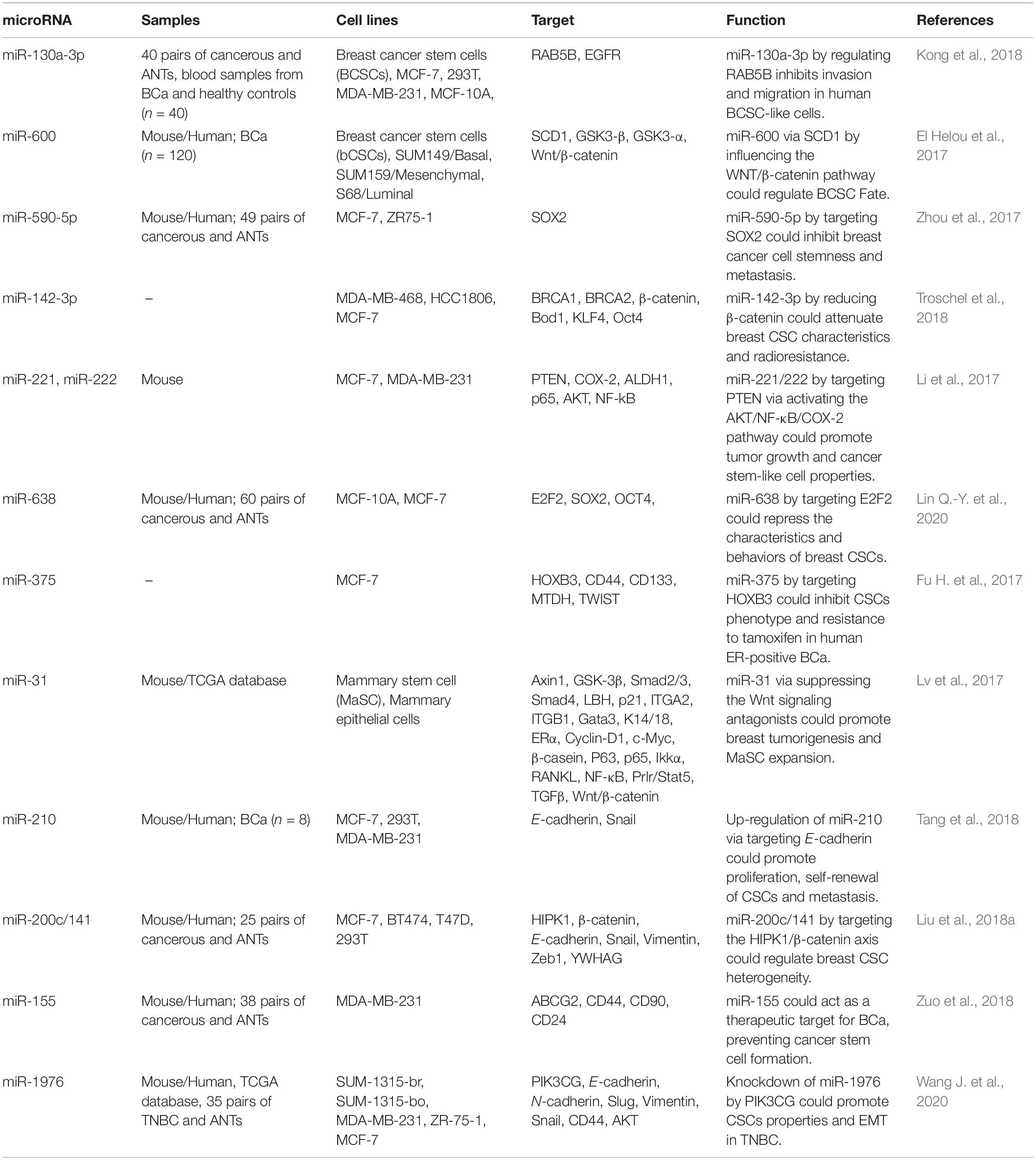
Table 1. Role of miRNAs in regulation of breast CSCs (ANTs, adjacent non-cancerous tissues; BCa, breast cancer).
Hepatocellular Carcinoma (HCC)
Expression of miR-206 has been found to be diminished in chemoresistant and recurrent HCC tumors. Notably, expression of this miRNA has also been reduced in CD133 or EpCAM-positive hepatic CSCs and in CSC-enriched spheres in hepatoma. Forced up-regulation of miR-206 has been shown to inhibit expansion of hepatic CSCs through blocking dedifferentiation of hepatoma cells and decreasing self-these cells. Mechanistically, EGFR has been described to be targeted by miR-206 (Liu et al., 2020). Expression profile of 14 miRNAs has been found to be altered among five groups of CSC-positive HCC tissues, namely EpCAM+, CD90+, CD133+, CD44+, and CD24+ HCC samples. Among these miRNA, miR-192-5p has been identified as the most important miRNA being under-expressed in all five groups of CSC-positive HCC samples, while being abundant in liver tissues. miR-192-5p silencing has facilitated expansion of CSC populations and CSC-associated characteristics via influencing expression of PABPC4. TP53 mutations and excessive methylation of the promoter area of this miRNA have been identified as underlying mechanisms of inactivation of miR-192-5p transcription in HCC cells and primary CSC-positive HCC (Gu et al., 2019). On the other hand, expression of miR-500a-3p has been significantly increased in HCC tissues and related cell lines. Over-expression of miR-500a-3p has been correlated with poor outcome of these patients. Over-expression of miR-500a-3p has enhanced the spheroid formation capacity, proportion of side population and levels of CSC markers. Besides, this miRNA has increased in vivo tumorigenicity of HCC cells. Functionally, miR-500a-3p enhances CSC features through influencing expression of numerous negative modulators of JAK/STAT3 signaling, such as SOCS2, SOCS4, and PTPN11, resulting in constituent activation of STAT3 cascade (Jiang et al., 2017a). Table 2 shows the role of miRNAs in regulation of HCC CSCs.
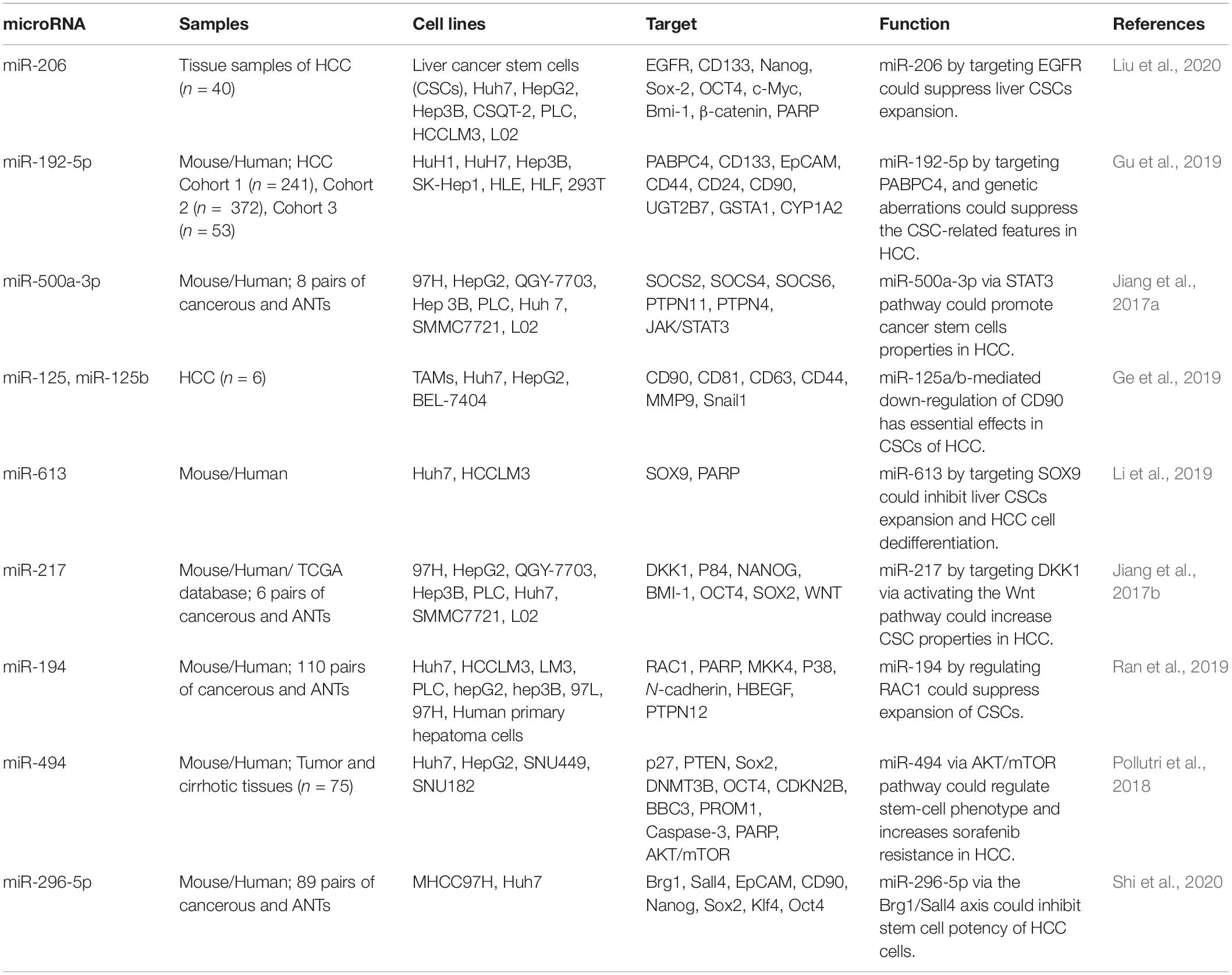
Table 2. Role of miRNAs in regulation of CSCs in hepatocellular carcinoma (ANTs, adjacent non-cancerous tissues; HCC, hepatocellular carcinoma).
Colorectal Cancer (CRC)
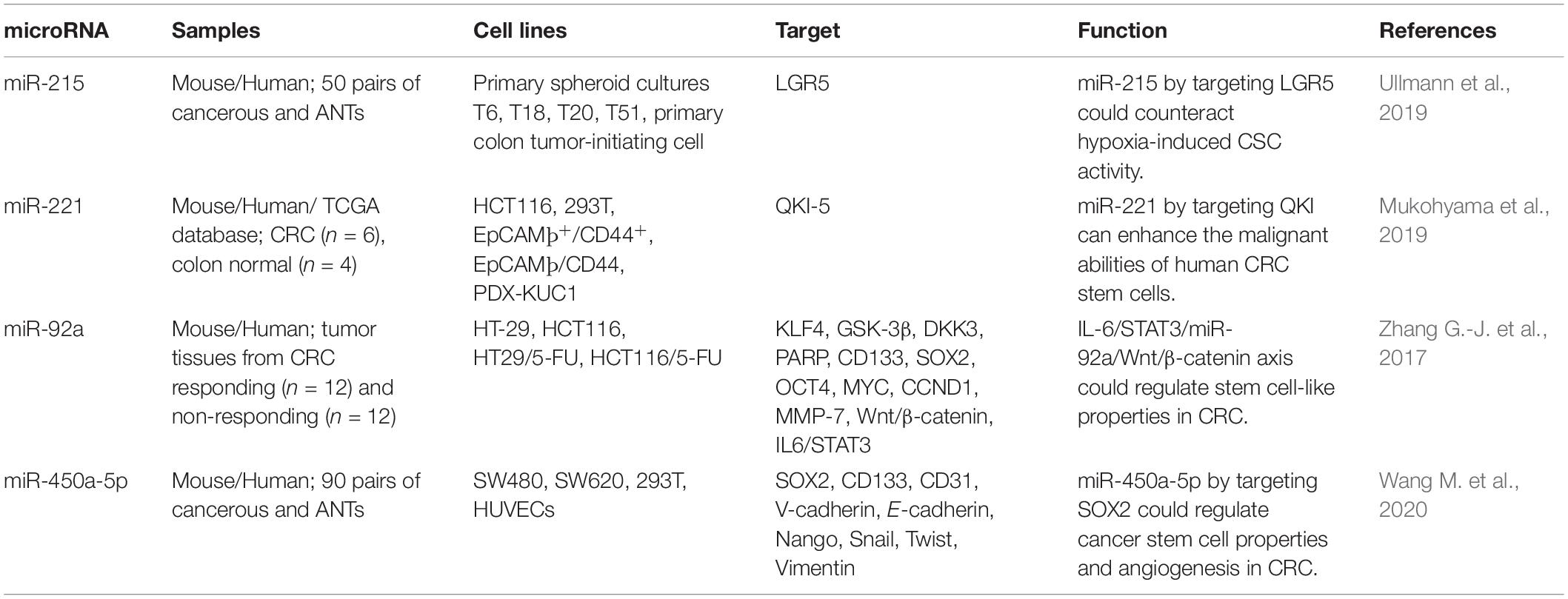
Unrestrained proliferation of cancer cells has been shown to induce hypoxia within the tumor mass, supporting activity of CSCs through induction of certain hypoxia-responsive routes. Expansion of CSCs in hypoxic conditions is associated with high sphere and colony construction. miR-215 has been identified as one of the principal hypoxia-associated miRNAs in primary colon CSCs. miR-215 is a negative modulator of CSC-enhancing influences of hypoxia. LGR5 has been acknowledged as a downstream molecule in hypoxia/miR-215 cascade. This miRNA has a prominent tumor suppressive effect in CRC and a target for anti-CSCs modalities (Ullmann et al., 2019).
Comparison of miRNA profile between CSC-enriched CRC cells (EpCAM+/CD44+) and CSC-depleted cells has led to identification of miR-221 as the most abundantly expressed miRNA in EpCAM+/CD44+ CRC cells. Over-expression of miR-221 has been associated with expression of Lgr5 in mouse colon crypts and poor clinical outcome of CRC in human subjects. Constitutive up-regulation of miR-221 has increased organoid-forming ability of both CRC cell lines and patient-originated xenograft in vitro. Notably, suppression of miR-221 has inhibited these features. QKI-5 has been identified as miR-221 target (Mukohyama et al., 2019).
miR-92a is another CSC-related miRNA which has been found to be over-expressed in chemoresistant CRC cell lines and tissues. Forced up-regulation of miR-92a has enhanced resistance to cytotoxic effects of 5-fluorouracil on CRC cells, while its silencing has remarkably promoted chemosensitivity in vivo. Most notably, up-regulation of miR-92a has increased tumor sphere construction and enhanced expression of stem cell biomarkers. miR-92a has been reported to activate Wnt/β-catenin signaling through inhibiting KLF4, GSK3β, and DKK3, which negatively regulate Wnt/β-catenin signaling at multiple levels. Besides, IL-6/STAT3 cascade has been shown to increase miR-92a levels through targeting its promoter, leading to activation of Wnt/β-catenin cascade and subsequent enhancement of stem-like features in CRC (Zhang G.-J. et al., 2017). Table 3 shows the role of miRNAs in regulation of CSCs in CRC.
Gastric Cancer
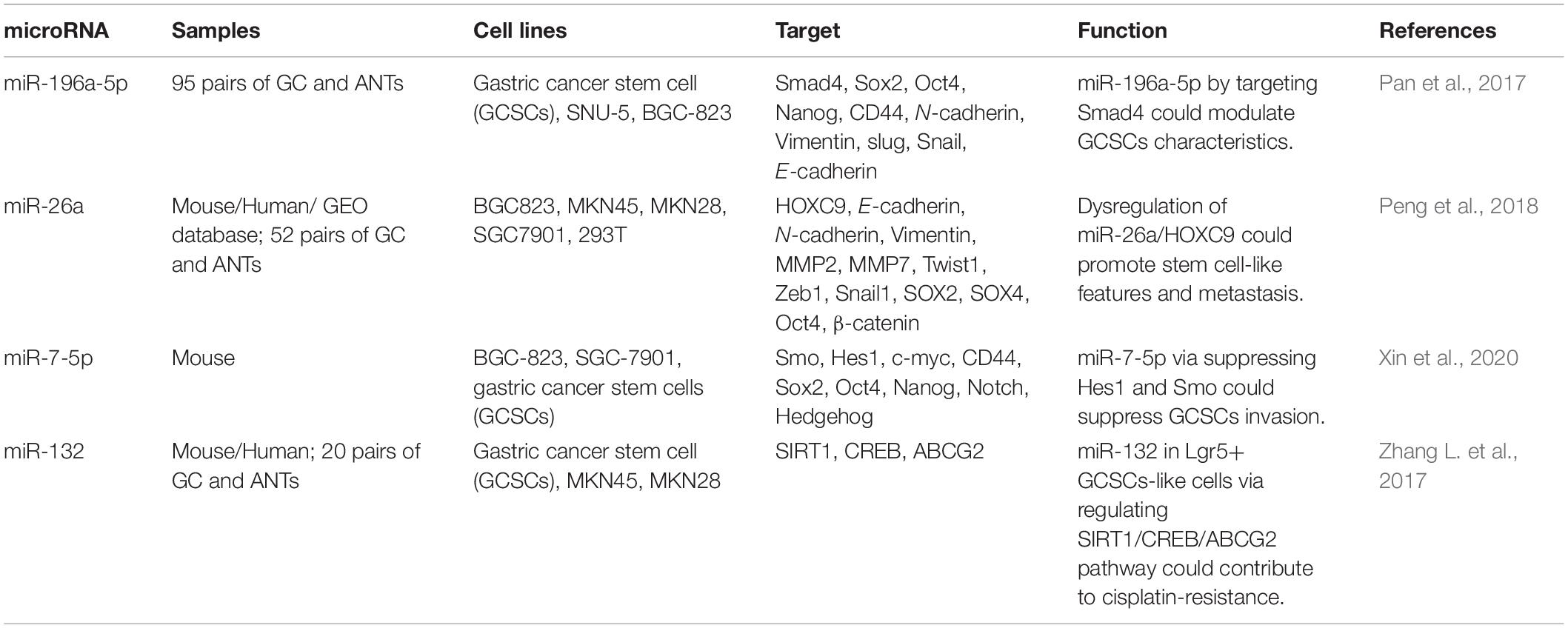
Gastric CSCs are described by expression of the stem cell marker CD44. CD44(+) cells have been shown to form more sphere colonies and pose higher level of invasiveness compared with gastric cancer cells lacking this marker. One of the supreme up-regulated miRNAs in gastric CSCs is miR-196a-5p. Inhibition of expression of miR-196a-5p has reduced colony formation and invasiveness of gastric CSCs. Functionally, miR-196a-reduces Smad4 levels through targeting its 3′-UTR. Smad4 levels in gastric cancer tissues have been associated with levels of differentiation, TNM stage and deepness of invasion. Notably, up-regulation of Smad4 has abolished miR-196a-5p-associated EMT in CSCs (Pan et al., 2017). miR-26a has been shown to be down-regulated in gastric cancer. This miRNA targets HOXC9, an up-regulated gene and a prognosticator of poor clinical outcome in gastric cancer. Over-expression of miR-26a in gastric cancer cells has suppressed HOXC9 levels and inverted its effects on self-renewal of CSCs (Peng et al., 2018). miR-7-5p is another down-regulated miRNA in gastric CSCs. Notably, expression of this miRNA has been enhanced in the methionine-deprived medium. Excessive DNA methylation in the promoter region has been found to be the main cause of down-regulation of miR-7-5p in gastric cancer. Up-regulation of miR-7-5p has decreased colony formation and invasiveness of gastric CSCs via targeting Smo and Hes1 and consequent suppression of Notch and Hedgehog cascades. Up-regulation of miR-7-5p has suppressed growth of gastric cancer in the animal models (Xin et al., 2020). Table 4 shows the role of miRNAs in gastric CSCs.
Glioma
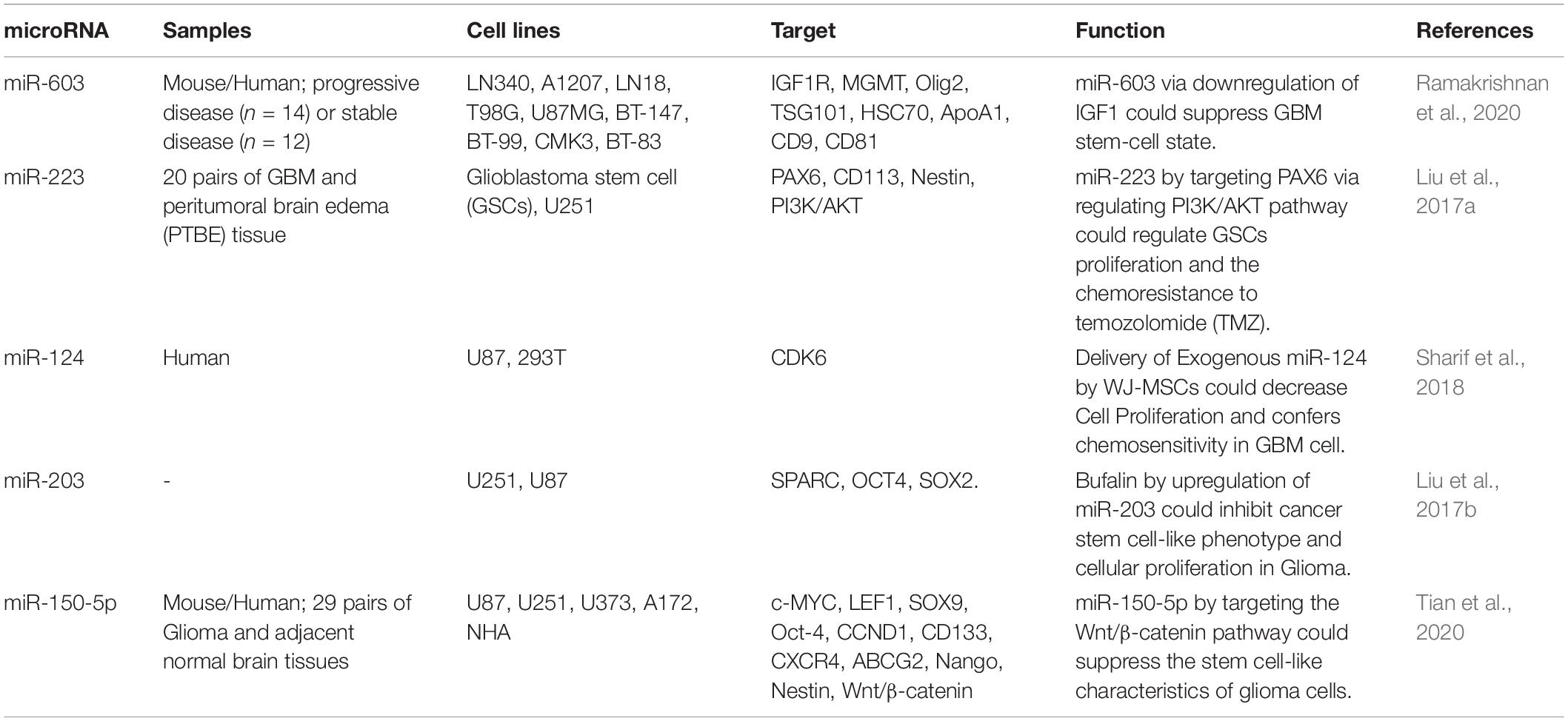
Ramakrishnan et al. (2020) have profiles miRNA signature of glioblastoma samples before and after radiotherapy. All assessed samples had unmethylated MGMT promoter and wild-type IDH (Ramakrishnan et al., 2020). MGMT acts as a DNA “suicide” repair enzyme. The encoded protein amends impaired guanine nucleotides through transporting the methyl at O6 site of guanine to its cysteine residues, therefore preventing gene mutation, apoptosis and carcinogenic processes induced by alkylating agents (Yu et al., 2020). Although expression of most of miRNAs did not change after treatment, expression of a number of miRNAs reduced. miR-603 has been identified as the most altered miRNA. This miRNA has been found to target IGF1 and IGF1R. Cellular transfer of miR-603 via release of extracellular vesicles has been increased following exposure with ionizing radiation, leading to de-repression of IGF1 and IGF1R and enhancement of expansion of CSCs and acquired resistance to radiotherapy. Moreover, miR-603 export has de-repressed MGMT (Ramakrishnan et al., 2020).
Expression of miR-223 has been increased in glioma tissues. Up-regulation of miR-223 has increased survival of these cells treated with Temozolomide (TMZ), while its suppression reversed this feature. PAX6 has been recognized to be targeted by miR-223. miR-223/PAX6 axis has been shown to regulate growth, invasiveness, and resistance TMZ via modulating PI3K/Akt signaling (Liu et al., 2017a).
The Chinese traditional medicine, Bufalin has been shown to inhibit proliferation, colony construction, and CSC features and enhance cell apoptosis and expression of miR-203 by glioblastoma cells. Notably, up-regulation of miR-203 results in similar effects as treatment with bufalin. SPARC has been acknowledged as a target of miR-203. Taken together, miR-203 mediates to effects of bufalin in suppression of growth of glioma cells and CSCs development (Liu et al., 2017b). SPARC gene codes for a cysteine-rich acidic matrix-associated protein which has essential role in the biogenesis of extracellular matrix, induction of alterations to cell shape and invasiveness of tumor cells (Brekken et al., 2003). Table 5 summarizes the role of miRNAs in regulation of glioma CSCs.
Leukemia
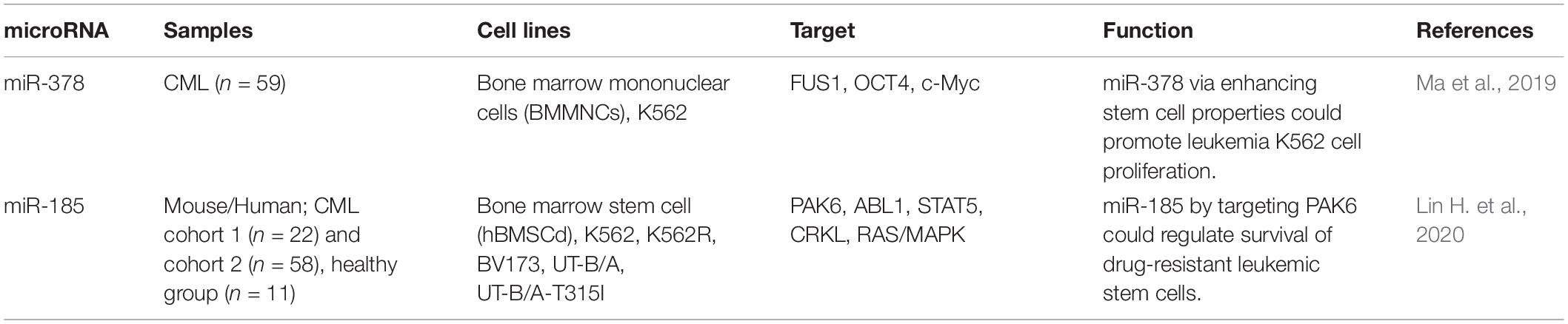
miR-378 is an up-regulated miRNA in chronic myeloid leukemia (CML) patients compared with normal persons. Up-regulation of miR-378 has enhanced proliferation and drug-resistance of CML cells, while inhibiting their apoptosis. Notably, transfection of cell with miR-378 has led to expansion of stem cell spheres and up-regulation of OCT4 and c-Myc. Moreover, miR-378 suppresses levels of FUS1 (Ma et al., 2019), a tumor suppressor that activates intrinsic apoptotic pathway via induction of Apaf-1-related mechanisms and hinders activities of protein tyrosine kinases (Ji and Roth, 2008). A high throughput transcriptome analysis of treatment-naive CML stem/progenitor cells has led to identification of miR-185 as a predictor of response to ABL tyrosine kinase inhibitors (TKIs). miR-185 has a tumor suppressive effect. Forced over-expression of miR-185 has impaired survival of drug-resistant cells, enhanced their response to TKIs, and noticeably eradicated long-term repopulating leukemic stem cells and infiltrating blasts. These effects have been accompanied by higher survival of xenotransplantation models. miR-185 has been shown to target PAK6. Suppression of PAK6 expression has interfered with activity of the RAS/MAPK pathway and mitochondria, enhancing sensitivity of resistant cells to TKIs (Lin H. et al., 2020). Table 6 shows the role of miRNAs in regulation of leukemic CSCs.
Other Cancers
In renal cell carcinoma, expression of miR-106b-5p has been found to elevated compared with normal controls. Up-regulation of miR-106b-5p in these cells has enhanced spheres formation capability and the quantity of side population cells. Further experiments in an orthotopic renal cancer model and a tail vein injection have shown that up-regulation of miR-106b-5p enhances tumor growth and metastasis to lungs, respectively. miR-106b-5p has been recognized as an activator of Wnt/β-catenin signaling which instantaneously inhibits numerous negative regulators of this pathway, including LZTFL1, SFRP1, and DKK2 (Lu et al., 2017b).
miR-328-3p is an up-regulated miRNA in ovarian CSCs. Over-expression of miR-328 results in better maintenance of CSC features through affecting expression of DNA damage binding protein 2, a protein that suppresses activity of ovarian CSCs. Attenuation of activity of ERK pathway in ovarian CSCs contributes in up-regulation of miR-328 and expansion of CSCs. miR-328 silencing in mouse orthotopic ovarian xenograft has blocked tumor growth and suppressed metastatic ability (Srivastava et al., 2019).
miR-335 has been revealed to be down-regulated in osteosarcoma CSCs compared with differentiated cells. Up-regulation of miR-335 has been associated with reduced stem cell-like features. On the other hand, miR-335 silencing has inhibited stem cell-like features and invasiveness. POU5F1 has been shown to be targeted by miR-335. The inhibitory effects of miR-335 on CSCs has exerted a synergic effect with traditional chemotherapeutic substances in the treatment of osteosarcoma (Guo et al., 2017). Since POU5F1 is an essential regulator of pluripotency (Nichols et al., 1998), miR-335 can affect pluripotency of stem cells through regulating its expression.
Expression of miR-1275 has been increased in lung cancer cell lines and tissues. Up-regulation of miR-1275 in clinical samples has been associated poor clinical outcome. Expression of miR-1275 is induced by the proto-oncogene HIF-1α. miR-1275 enhances the activity of Wnt/β-catenin and Notch pathways and consequently increases the stemness of lung adenocarcinoma cells. miR-1275 can suppress expression of multiple negative regulators of Wnt/β-catenin and Notch pathways, namely DKK3, SFRP1, GSK3β, RUNX3, and NUMB (Jiang et al., 2020). Table 7 shows the impact of miRNAs on CSCs in different cancers.
lncRNAs and CSCs
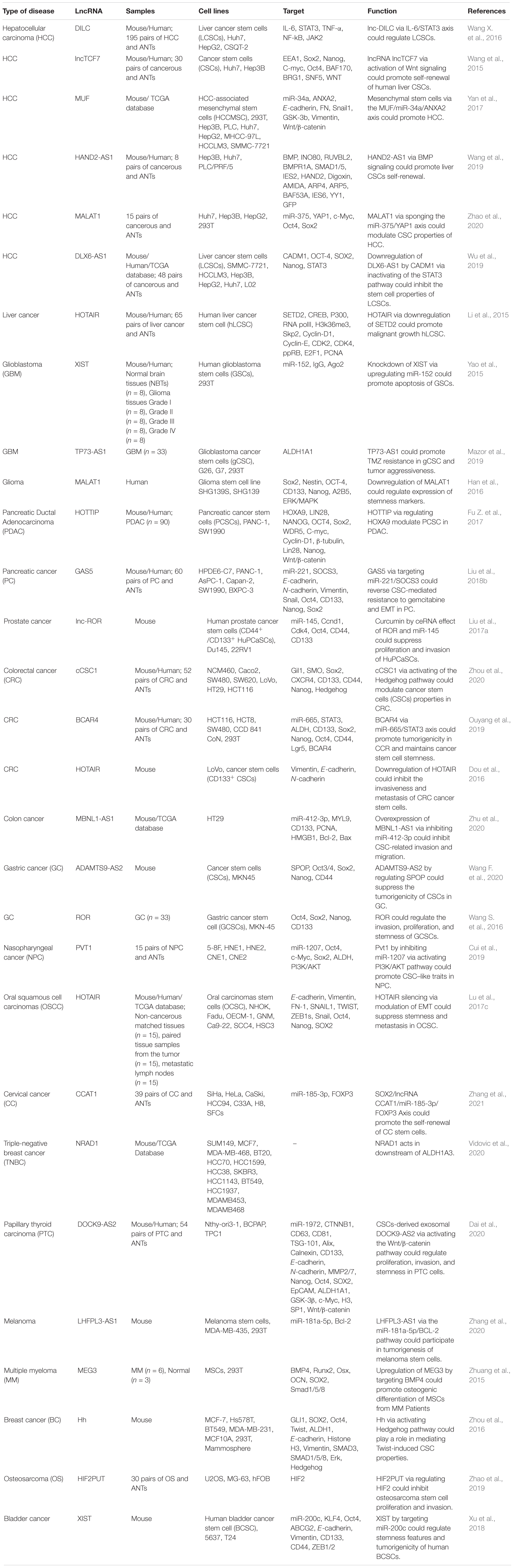
Contribution of lncRNAs in the expansion of CSCs has been verified in several investigations. A number of lncRNAs have been reported to regulate this phenotype in different types of cancers. MALAT1, HOTAIR, and XIST are among the mostly assessed lncRNAs in this regard.
MALAT1
MALAT1 expression has been revealed to be increased in cancer spheroids compared with parental liver cancer cells. MALAT1 silencing has decreased sphere construction and reduced expression of stem cell markers in liver cancer cells. The effects of MALAT1 on CSCs are mediated through sponging miR-375, and subsequently up-regulation of YAP1 (Zhao et al., 2020), a protein that regulates expression of genes contributing in cell proliferation and blocks expression of apoptotic genes (Sudol, 1994). Thus, MALAT1/miR-375/YAP1 represents a functional axis in liver carcinogenesis and a target for elimination of CSCs (Zhao et al., 2020). Similarly, in glioma cells, MALAT1 silencing has inhibited expression of two stemness-related factors, namely Sox2 and Nestin (Han et al., 2016). Since Nestin regulates assembly and disassembly of intermediate filaments (Guérette et al., 2007), MALAT1-mediated regulation of Nestin can affect cell remodeling. In these cells, MALAT1 mainly regulates activity of ERK/MAPK pathway (Han et al., 2016).
HOTAIR
HOTAIR is another lncRNA which enhances expansion of CSCs. In liver cancer cells, HOTAIR increases stemness features via suppressing expression of SETD2. From a mechanistical point of view, HOTAIR decreases the recruitment of the CREB, P300, RNA polII on promoter of SETD2 gene, reducing its expression and phosphorylation. Thus, the ability of SETD2 for binding with substrate histone H3 is attenuated and trimethylation of lysine 36th of histone H3 is decreased, leading to reduction of H3K36me3-hMSH2-hMSH6-SKP2 complex. Notably, the complex tenancy on chromosome is decreased and mismatch DNA repair function is reduced (Li et al., 2015). Consistent with this study, HOTAIR silencing in CD133(+) CRC CSCs has significantly reduced the tumor growth and metastatic ability in xenograft model of CRC (Dou et al., 2016). In oral squamous cell carcinomas, HOTAIR has been found to be up-regulated in tumor samples, particularly in the metastatic ones. HOTAIR silencing has remarkably suppressed stemness, invasiveness and tumorigenicity in xenografts. On the other hand, up-regulation of HOTAIR has led to promotion of metastatic ability and EMT. Notably, HOTAIR levels have been positively correlated with levels of mesenchymal markers and negatively related with epithelial markers (Lu et al., 2017c).
XIST
Expression of XIST has been increased in glioma tissues and CSCs. XIST silencing has reduced cell proliferation, migratory potential and invasiveness, while increasing apoptosis. XIST knock down has also inhibited in vivo growth of tumor and enhanced survival of affected animals. Mechanistically, XIST interacts with miR-152. Through modulation of this miRNA, XIST regulates functions of glioma stem cells (Yao et al., 2015). In bladder cancer, XIST sponges miR-200c and enhances clone formation, self-renewal aptitude and EMT in bladder CSCs (Xu et al., 2018).
GAS5
GAS5 is a tumor suppressor lncRNA with acknowledged roles in several tissues (Ji et al., 2019). In pancreatic cancer cells, GAS5 regulates chemoresistance to gemcitabine and metastatic ability of transformed cells. Up-regulation of GAS5 could suppress proliferation, migratory potential, resistance to gemcitabine, stem cell-like features, and EMT process through sequestering miR-221 and releasing SOCS3 from its inhibitory effects. Moreover, GAS5 could enhance effects of gemcitabine on suppression of tumor growth and metastasis. The GAS5/miR-221/SOCS3 cascade has been identified as an important modulator of EMT and CSC function in pancreatic cancer (Liu et al., 2018b).
DILC
lnc-DILC has an acknowledged role in suppression of expansion of CSCs, since its silencing has led to enhancement of expansion of liver CSCs and induction of initiation and progression of liver cancer. This lncRNA can inhibit autocrine activity of IL-6/STAT3 cascade through directly binding with IL-6 promoter. Besides, lnc-DILC can mediate the interaction between TNF-α/NF-κB pathway and IL-6/STAT3 axis. Consistent with these findings, expression of lnc-DILC has been found to be reduced in patients with liver cancer. Moreover, expression of lnc-DILC has been correlated with IL-6, EpCAM or CD24 levels. Down-regulation of lnc-DILC in these patients has been correlated with risk of early recurrence and poor clinical outcome. Thus, lnc-DILC can connect liver inflammation with expansion of CSCs through mediating the interplay between mentioned pathways (Wang X. et al., 2016). Table 8 demonstrates the role of lncRNAs in CSCs.
Discussion
Cancer stem cells have especial properties that potentiate them for anti-cancer treatment, since it is expected that treatments targeting these cells preclude cancer metatstasis and recurrence. Therefore, identification of molecules that regulate their function has practical significance. miRNAs and lncRNAs have been shown to influence activity and expansion of CSCs. The impact of these regulatory ranscripts on CSCs has been mostly assessed in breast cancer and HCC.
A number of studies have demonstrated the role of CSC-related miRNAs/lncRNAs such as miR-1976, miR-196a-5p, miR-221, HOTAIR, and GAS5 in EMT, emphasizing on the multifaceted functions of these transcripts in the carcinogenesis and connection between these cancer-related processes. The impact of CSC-related miRNAs/lncRNAs on radio/chemoresistance has also been assessed. miR-142-3p, miR-375, miR-494, miR-223, miR-132, miR-185, miR-181b, TP73-AS1 and GAS5 are among non-coding RNAs with appreciated roles in this aspect. Notably, miRNAs/lncRNAs that attenuate expansion of CSCs cab have synergic effects with conventional anti-cancer drugs, therefore these targeted therapies migh have promisisng effects in the clinical settings.
Among lncRNAs, MALAT1, HOTAIR, and XIST have been the mostly assessed ones in CSCs. Independent studies have verified the effects of these lncRNAs in expansion of CSCs. The effects of lncRNAs in modulation of function of CSCs are mediated through different routes, among them is their miRNA spoging function. MUF/miR-34a, MALAT1/miR-375, XIST/miR-152, XIST/miR-200c,
GAS5/miR-221, lnc-ROR/miR-145, BCAR4/miR-665, MBNL1-AS1/miR-412-3p, PVT1/miR-1207, CCAT1/miR-185-3p, DOCK9-AS2/miR-1972 and LHFPL3-AS1/miR-181a-5p are examples of lncRNAs/miRNAs axes which contribute in the regulation of CSCs.
miRNAs can simultaneously regulate CSC-related pathways such as Wnt/β-catenin signaling through targeting multiple regulators of these pathways. miR-92a (Zhang G.-J. et al., 2017), miR-106b-5p (Lu et al., 2017b) and miR-1275 (Jiang et al., 2020) are examples of miRNAs with this kind of aptitude. Identification of additional miRNAs with the ability to target CSC-related pathways at multiple levels would facilitate enhancing efficiency of therapeutic interventions.
Wnt/β-catenin pathway as one of the most important pathways in determination of stem cells fate is regulated by several miRNAs such as miR-600, miR-142-3p, miR-31, miR-217, miR-92a, miR-150-5p, miR-106b-5p, miR-1275, miR-708-5p, miR-1301-3p as well as a number of lncRNAs such as lncTCF7, MUF, HOTTIP and DOCK9-AS2. Activity of Notch pathway is modulated by miR-7-5p, miR-26a, miR-1275, and miR-181b.
The underlying cause of abnormal expression of these non-coding RNAs has not been clarified completely. However, methylation marks in the promoter region can explain down-regulation of a number of these non-coding RNAs. miR-7-5p is an exmaple of these non-coding RNAs (Xin et al., 2020). Meanwhile, some lncRNAs have pivotal roels in the modulation of methylation of target genes. Therefore, a complicated interaction network exists between epigenetic factors that regulate methylation marks, miRNAs, lncRNAs and transcription factors that modulate expression of lncRNAs/miRNAs or are regulated by these transcripts. Understanding this multifaceted network is the prerequisite of design of targeted therapies against CSCs.
Taken together, several miRNAs and lncRNAs have been identified that regulate expansion of CSCs via different mechanisms, particularly regulation of cancer-related signaling pathways. Based on the importance of CSCs in the metastatic ability of malignnat cells and their resistance to therapeutic regimens, these transcripts represent promising targets in cancer management. Most of accomplished studies have assessed the impact of lncRNA/miRNA silencing or up-regulation in cell lines. However, a number of eminnet studies in this field have validated the results of in silico and in vitro studies in animal models of cancers providing more valuable clues for implementation of these methods in clinical settings. miR-500a-3p, miR-92a and XIST are among non-coding RNAs with sufficient in vivo studies.
Author Contributions
MT and SG-F wrote the draft and revised it. HS, ZB, MH, and GS collected the data and designed the tables and study. All the authors read and approved submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Al-Hajj, M., Wicha, M. S., Benito-Hernandez, A., Morrison, S. J., and Clarke, M. F. (2003). Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci.U.S.A. 100, 3983–3988.
Brekken, R. A., Puolakkainen, P., Graves, D. C., Workman, G., Lubkin, S. R., and Sage, E. H. (2003). Enhanced growth of tumors in SPARC null mice is associated with changes in the ECM. J. Clin. Invest. 111, 487–495. doi: 10.1172/jci16804
Cui, M., Chang, Y., Fang, Q.-G., Du, W., Wu, J.-F., Wang, J.-H., et al. (2019). Non-coding RNA Pvt1 promotes cancer stem cell–like traits in nasopharyngeal cancer via inhibiting miR-1207. Pathol. Oncol. Res. 25, 1411–1422. doi: 10.1007/s12253-018-0453-1
Dai, W., Jin, X., Han, L., Huang, H., Ji, Z., Xu, X., et al. (2020). Exosomal lncRNA DOCK9-AS2 derived from cancer stem cell-like cells activated Wnt/β-catenin pathway to aggravate stemness, proliferation, migration, and invasion in papillary thyroid carcinoma. Cell Death Dis. 11, 1–17. doi: 10.1016/j.biochi.2018.02.009
Dong, L., Pu, Y., Zhang, L., Qi, Q., Xu, L., Li, W., et al. (2018). Human umbilical cord mesenchymal stem cell-derived extracellular vesicles promote lung adenocarcinoma growth by transferring miR-410. Cell Death Dis. 9, 1–13. doi: 10.1155/2019/8108576
Dou, J., Ni, Y., He, X., Wu, D., Li, M., Wu, S., et al. (2016). Decreasing lncRNA HOTAIR expression inhibits human colorectal cancer stem cells. Am. J. Transl. Res. 8:98.
El Helou, R., Pinna, G., Cabaud, O., Wicinski, J., Bhajun, R., Guyon, L., et al. (2017). miR-600 acts as a bimodal switch that regulates breast cancer stem cell fate through WNT signaling. Cell Rep. 18, 2256–2268. doi: 10.1016/j.celrep.2017.02.016
Fu, H., Fu, L., Xie, C., Zuo, W.-S., Liu, Y.-S., Zheng, M.-Z., et al. (2017). miR-375 inhibits cancer stem cell phenotype and tamoxifen resistance by degrading HOXB3 in human ER-positive breast cancer. Oncol. Rep. 37, 1093–1099. doi: 10.3892/or.2017.5360
Fu, Z., Chen, C., Zhou, Q., Wang, Y., Zhao, Y., Zhao, X., et al. (2017). LncRNA HOTTIP modulates cancer stem cell properties in human pancreatic cancer by regulating HOXA9. Cancer Lett. 410, 68–81. doi: 10.1016/j.canlet.2017.09.019
Ge, B., Liu, H., Liang, Q., Shang, L., Wang, T., and Ge, S. (2019). Oxytocin facilitates the proliferation, migration and osteogenic differentiation of human periodontal stem cells in vitro. Arch. Oral Biol. 99, 126–133. doi: 10.1016/j.archoralbio.2019.01.007
Ghafouri-Fard, S., Abak, A., Fattahi, F., Hussen, B. M., Bahroudi, Z., Shoorei, H., et al. (2021a). The interaction between miRNAs/lncRNAs and nuclear factor-κB (NF-κB) in human disorders. Biomed. Pharmacother. 138:111519. doi: 10.1016/j.biopha.2021.111519
Ghafouri-Fard, S., Abak, A., Shoorei, H., Talebi, S. F., Mohaqiq, M., Sarabi, P., et al. (2021b). Interaction between non-coding RNAs and Toll-like receptors. Biomed. Pharmacother. 140:111784. doi: 10.1016/j.biopha.2021.111784
Ghafouri-Fard, S., Glassy, M. C., Abak, A., Hussen, B. M., Niazi, V., and Taheri, M. (2021c). The interaction between miRNAs/lncRNAs and Notch pathway in human disorders. Biomed. Pharmacother. 138:111496. doi: 10.1016/j.biopha.2021.111496
Ghafouri-Fard, S., Noroozi, R., Abak, A., Taheri, M., and Salimi, A. (2021d). Emerging role of lncRNAs in the regulation of Rho GTPase pathway. Biomed. Pharmacother. 140:111731. doi: 10.1016/j.biopha.2021.111731
Gu, Y., Wei, X., Sun, Y., Gao, H., Zheng, X., Wong, L. L., et al. (2019). miR-192-5p silencing by genetic aberrations is a key event in hepatocellular carcinomas with cancer stem cell features. Cancer Res. 79, 941–953. doi: 10.1158/0008-5472.can-18-1675
Guérette, D., Khan, P. A., Savard, P. E., and Vincent, M. (2007). Molecular evolution of type VI intermediate filament proteins. BMC Evol. Biol. 7:164. doi: 10.1186/1471-2148-7-164
Guo, X., Yu, L., Zhang, Z., Dai, G., Gao, T., and Guo, W. (2017). miR-335 negatively regulates osteosarcoma stem cell-like properties by targeting POU5F1. Cancer Cell Int. 17:29.
Han, Y., Zhou, L., Wu, T., Huang, Y., Cheng, Z., Li, X., et al. (2016). Downregulation of lncRNA-MALAT1 affects proliferation and the expression of stemness markers in glioma stem cell line SHG139S. Cell. Mol. Neurobiol. 36, 1097–1107. doi: 10.1007/s10571-015-0303-6
Hu, T., Chong, Y., Lu, S., Wang, R., Qin, H., Silva, J., et al. (2018). miR-339 promotes development of stem cell leukemia/lymphoma syndrome via downregulation of the BCL2L11 and BAX proapoptotic genes. Cancer Res. 78, 3522–3531.
Hussen, B. M., Shoorei, H., Mohaqiq, M., Dinger, M. E., Hidayat, H. J., Taheri, M., et al. (2021). The impact of non-coding RNAs in the epithelial to mesenchymal transition. Front. Mol. Biosci. 8:665199.
Ji, J., Dai, X., Yeung, S.-C. J., and He, X. (2019). The role of long non-coding RNA GAS5 in cancers. Cancer Manag. Res. 11, 2729–2737. doi: 10.2147/cmar.s189052
Ji, L., and Roth, J. A. (2008). Tumor suppressor FUS1 signaling pathway. J. Thorac. Oncol. 3, 327–330. doi: 10.1097/jto.0b013e31816bce65
Jiang, C., Long, J., Liu, B., Xu, M., Wang, W., Xie, X., et al. (2017a). miR-500a-3p promotes cancer stem cells properties via STAT3 pathway in human hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 36:99. doi: 10.3390/cells7080099
Jiang, C., Yu, M., Xie, X., Huang, G., Peng, Y., Ren, D., et al. (2017b). miR-217 targeting DKK1 promotes cancer stem cell properties via activation of the Wnt signaling pathway in hepatocellular carcinoma. Oncol. Rep. 38, 2351–2359. doi: 10.3892/or.2017.5924
Jiang, N., Zou, C., Zhu, Y., Luo, Y., Chen, L., Lei, Y., et al. (2020). HIF-1α-regulated miR-1275 maintains stem cell-like phenotypes and promotes the progression of LUAD by simultaneously activating Wnt/β-catenin and Notch signaling. Theranostics 10:2553. doi: 10.7150/thno.41120
Jiang, P., Xu, C., Chen, L., Chen, A., Wu, X., Zhou, M., et al. (2018). Epigallocatechin-3-gallate inhibited cancer stem cell–like properties by targeting hsa-mir-485-5p/RXRα in lung cancer. J. Cell. Biochem. 119, 8623–8635. doi: 10.1002/jcb.27117
Kong, X., Zhang, J., Li, J., Shao, J., and Fang, L. (2018). MiR-130a-3p inhibits migration and invasion by regulating RAB5B in human breast cancer stem cell-like cells. Biochem. Biophys. Res. Commun. 501, 486–493. doi: 10.1016/j.bbrc.2018.05.018
Lapidot, T., Sirard, C., Vormoor, J., Murdoch, B., Hoang, T., Caceres-Cortes, J., et al. (1994). A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature 367, 645–648. doi: 10.1038/367645a0
Li, B., Liu, D., Yang, P., Li, H.-Y., and Wang, D. (2019). miR-613 inhibits liver cancer stem cell expansion by regulating SOX9 pathway. Gene 707, 78–85. doi: 10.1016/j.gene.2019.05.015
Li, B., Lu, Y., Yu, L., Han, X., Wang, H., Mao, J., et al. (2017). miR-221/222 promote cancer stem-like cell properties and tumor growth of breast cancer via targeting PTEN and sustained Akt/NF-κB/COX-2 activation. Chem. Biol. Int. 277, 33–42. doi: 10.1016/j.cbi.2017.08.014
Li, H., An, J., Wu, M., Zheng, Q., Gui, X., Li, T., et al. (2015). LncRNA HOTAIR promotes human liver cancer stem cell malignant growth through downregulation of SETD2. Oncotarget 6:27847. doi: 10.18632/oncotarget.4443
Lin, H., Rothe, K., Chen, M., Wu, A., Babaian, A., Yen, R., et al. (2020). The miR-185/PAK6 axis predicts therapy response and regulates survival of drug-resistant leukemic stem cells in CML. Blood J. Am. Soc. Hematol. 136, 596–609. doi: 10.1182/blood.2019003636
Lin, Q.-Y., Wang, J.-Q., Wu, L.-L., Zheng, W.-E., and Chen, P.-R. (2020). miR-638 represses the stem cell characteristics of breast cancer cells by targeting E2F2. Breast Cancer 27, 147–158. doi: 10.1007/s12282-019-01002-0
Liu, B., Du, R., Zhou, L., Xu, J., Chen, S., Chen, J., et al. (2018a). miR-200c/141 regulates breast cancer stem cell heterogeneity via targeting HIPK1/β-catenin axis. Theranostics 8:5801. doi: 10.7150/thno.29380
Liu, B., Wu, S., Ma, J., Yan, S., Xiao, Z., Wan, L., et al. (2018b). lncRNA GAS5 reverses EMT and tumor stem cell-mediated gemcitabine resistance and metastasis by targeting miR-221/SOCS3 in pancreatic cancer. Mol. Ther. Nucleic Acids 13, 472–482. doi: 10.1016/j.omtn.2018.09.026
Liu, C., Li, J., Wang, W., Zhong, X., Xu, F., and Lu, J. (2020). miR-206 inhibits liver cancer stem cell expansion by regulating EGFR expression. Cell Cycle 19, 1077–1088. doi: 10.1080/15384101.2020.1739808
Liu, T., Chi, H., Chen, J., Chen, C., Huang, Y., Xi, H., et al. (2017a). Curcumin suppresses proliferation and in vitro invasion of human prostate cancer stem cells by ceRNA effect of miR-145 and lncRNA-ROR. Gene 631, 29–38. doi: 10.1016/j.gene.2017.08.008
Liu, T., Wu, C., Weng, G., Zhao, Z., He, X., Fu, C., et al. (2017b). Bufalin inhibits cellular proliferation and cancer stem cell-like phenotypes via upregulation of MiR-203 in glioma. Cell. Physiol. Biochem. 44, 671–681. doi: 10.1159/000485279
Liu, T., Wu, X., Chen, T., Luo, Z., and Hu, X. (2018c). Downregulation of DNMT3A by miR-708-5p inhibits lung cancer stem cell–like phenotypes through repressing Wnt/β-catenin signaling. Clin. Cancer Res. 24, 1748–1760. doi: 10.1158/1078-0432.ccr-17-1169
Lu, J., Song, G., Tang, Q., Yin, J., Zou, C., Zhao, Z., et al. (2017a). MiR-26a inhibits stem cell-like phenotype and tumor growth of osteosarcoma by targeting Jagged1. Oncogene 36, 231–241. doi: 10.1038/onc.2016.194
Lu, J., Wei, J.-H., Feng, Z.-H., Chen, Z.-H., Wang, Y.-Q., Huang, Y., et al. (2017b). miR-106b-5p promotes renal cell carcinoma aggressiveness and stem-cell-like phenotype by activating Wnt/β-catenin signalling. Oncotarget 8:21461. doi: 10.18632/oncotarget.15591
Lu, M.-Y., Liao, Y.-W., Chen, P.-Y., Hsieh, P.-L., Fang, C.-Y., Wu, C.-Y., et al. (2017c). Targeting LncRNA HOTAIR suppresses cancer stemness and metastasis in oral carcinomas stem cells through modulation of EMT. Oncotarget 8:98542. doi: 10.18632/oncotarget.21614
Lv, C., Li, F., Li, X., Tian, Y., Zhang, Y., Sheng, X., et al. (2017). MiR-31 promotes mammary stem cell expansion and breast tumorigenesis by suppressing Wnt signaling antagonists. Nat. Commun. 8:5308.
Ma, J., Wu, D., Yi, J., Yi, Y., Zhu, X., Qiu, H., et al. (2019). MiR-378 promoted cell proliferation and inhibited apoptosis by enhanced stem cell properties in chronic myeloid leukemia K562 cells. Biomed. Pharmacother. 112:108623. doi: 10.1016/j.biopha.2019.108623
Mazor, G., Levin, L., Picard, D., Ahmadov, U., Carén, H., Borkhardt, A., et al. (2019). The lncRNA TP73-AS1 is linked to aggressiveness in glioblastoma and promotes temozolomide resistance in glioblastoma cancer stem cells. Cell Death Dis. 10:246.
Mukohyama, J., Isobe, T., Hu, Q., Hayashi, T., Watanabe, T., Maeda, M., et al. (2019). miR-221 targets QKI to enhance the tumorigenic capacity of human colorectal cancer stem cells. Cancer Res. 79, 5151–5158. doi: 10.1158/0008-5472.can-18-3544
Nichols, J., Zevnik, B., Anastassiadis, K., Niwa, H., Klewe-Nebenius, D., Chambers, I., et al. (1998). Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95, 379–391. doi: 10.1016/s0092-8674(00)81769-9
Ouyang, S., Zhou, X., Chen, Z., Wang, M., Zheng, X., and Xie, M. (2019). LncRNA BCAR4, targeting to miR-665/STAT3 signaling, maintains cancer stem cells stemness and promotes tumorigenicity in colorectal cancer. Cancer Cell Int. 19:72.
Pan, Y., Shu, X., Sun, L., Yu, L., Sun, L., Yang, Z., et al. (2017). miR-196a-5p modulates gastric cancer stem cell characteristics by targeting Smad4. Int. J. Oncol. 50, 1965–1976. doi: 10.3892/ijo.2017.3965
Peng, X., Kang, Q., Wan, R., and Wang, Z. (2018). MiR-26a/HOXC9 dysregulation promotes metastasis and stem cell-like phenotype of gastric cancer. Cell. Physiol. Biochem. 49, 1659–1676. doi: 10.1159/000493502
Pollutri, D., Patrizi, C., Marinelli, S., Giovannini, C., Trombetta, E., Giannone, F. A., et al. (2018). The epigenetically regulated miR-494 associates with stem-cell phenotype and induces sorafenib resistance in hepatocellular carcinoma. Cell Death Dis. 9:4.
Ramakrishnan, V., Xu, B., Akers, J., Nguyen, T., Ma, J., Dhawan, S., et al. (2020). Radiation-induced extracellular vesicle (EV) release of miR-603 promotes IGF1-mediated stem cell state in glioblastomas. EBioMedicine 55:102736. doi: 10.1016/j.ebiom.2020.102736
Ran, R.-Z., Chen, J., Cui, L.-J., Lin, X.-L., Fan, M.-M., Cong, Z.-Z., et al. (2019). miR-194 inhibits liver cancer stem cell expansion by regulating RAC1 pathway. Exp. Cell Res. 378, 66–75. doi: 10.1016/j.yexcr.2019.03.007
Rosen, J. M., and Jordan, C. T. (2009). The increasing complexity of the cancer stem cell paradigm. Science 324, 1670–1673. doi: 10.1126/science.1171837
Sharif, S., Ghahremani, M., and Soleimani, M. (2018). Delivery of exogenous miR-124 to glioblastoma multiform cells by Wharton’s jelly mesenchymal stem cells decreases cell proliferation and migration, and confers chemosensitivity. Stem Cell Rev. Rep. 14, 236–246. doi: 10.1007/s12015-017-9788-3
Shi, D.-M., Shi, X.-L., Xing, K.-L., Zhou, H.-X., Lu, L.-L., and Wu, W.-Z. (2020). miR-296-5p suppresses stem cell potency of hepatocellular carcinoma cells via regulating Brg1/Sall4 axis. Cell. Signall. 72:109650. doi: 10.1016/j.cellsig.2020.109650
Singh, S. K., Hawkins, C., Clarke, I. D., Squire, J. A., Bayani, J., Hide, T., et al. (2004). Identification of human brain tumour initiating cells. Nature 432, 396–401.
Song, X.-L., Huang, B., Zhou, B.-W., Wang, C., Liao, Z.-W., Yu, Y., et al. (2018). miR-1301-3p promotes prostate cancer stem cell expansion by targeting SFRP1 and GSK3β. Biomed. Pharmacother. 99, 369–374. doi: 10.1016/j.biopha.2018.01.086
Srivastava, A. K., Banerjee, A., Cui, T., Han, C., Cai, S., Liu, L., et al. (2019). Inhibition of miR-328–3p impairs cancer stem cell function and prevents metastasis in ovarian cancer. Cancer Res. 79, 2314–2326. doi: 10.1158/0008-5472.can-18-3668
Sudol, M. (1994). Yes-associated protein (YAP65) is a proline-rich phosphoprotein that binds to the SH3 domain of the Yes proto-oncogene product. Oncogene 9, 2145–2152.
Tang, T., Yang, Z., Zhu, Q., Wu, Y., Sun, K., Alahdal, M., et al. (2018). Up-regulation of miR-210 induced by a hypoxic microenvironment promotes breast cancer stem cell metastasis, proliferation, and self-renewal by targeting E-cadherin. FASEB J. 32, 6965–6981. doi: 10.1096/fj.201801013r
Tian, W., Zhu, W., and Jiang, J. (2020). miR-150-5p suppresses the stem cell-like characteristics of glioma cells by targeting the Wnt/β-catenin signaling pathway. Cell Biol. Int. 44, 1156–1167. doi: 10.1002/cbin.11314
Troschel, F. M., Böhly, N., Borrmann, K., Braun, T., Schwickert, A., Kiesel, L., et al. (2018). miR-142-3p attenuates breast cancer stem cell characteristics and decreases radioresistance in vitro. Tumor Biol. 40:1010428318791887.
Ullmann, P., Nurmik, M., Schmitz, M., Rodriguez, F., Weiler, J., Qureshi-Baig, K., et al. (2019). Tumor suppressor miR-215 counteracts hypoxia-induced colon cancer stem cell activity. Cancer Lett. 450, 32–41. doi: 10.1016/j.canlet.2019.02.030
Vidovic, D., Huynh, T. T., Konda, P., Dean, C., Cruickshank, B. M., Sultan, M., et al. (2020). ALDH1A3-regulated long non-coding RNA NRAD1 is a potential novel target for triple-negative breast tumors and cancer stem cells. Cell Death Differ. 27, 363–378. doi: 10.1038/s41418-019-0362-1
Wang, F., Tang, C., Xu, D., Tang, Y., Jiang, Y., Gao, X., et al. (2020). LncRNA ADAMTS9-AS2 suppresses the proliferation of gastric cancer cells and the tumorigenicity of cancer stem cells through regulating SPOP. J. Cell. Mol. Med. 24, 4830–4838. doi: 10.1111/jcmm.15161
Wang, J., Li, M., Han, X., Wang, H., Wang, X., Ma, G., et al. (2020). MiR-1976 knockdown promotes epithelial–mesenchymal transition and cancer stem cell properties inducing triple-negative breast cancer metastasis. Cell Death Dis. 11, 1–12.
Wang, M., Li, J., Cai, J., Cheng, L., Wang, X., Xu, P., et al. (2020). Overexpression of MicroRNA-16 alleviates atherosclerosis by inhibition of inflammatory pathways. BioMed. Res. Int. 55:2020.
Wang, S., Liu, F., Deng, J., Cai, X., Han, J., and Liu, Q. (2016). Long noncoding RNA ROR regulates proliferation, invasion, and stemness of gastric cancer stem cell. Cell. Reprogr. 18, 319–326. doi: 10.1089/cell.2016.0001
Wang, X., Meng, Q., Qiao, W., Ma, R., Ju, W., Hu, J., et al. (2018). miR-181b/Notch2 overcome chemoresistance by regulating cancer stem cell-like properties in NSCLC. Stem Cell Res. Ther. 9:327.
Wang, X., Sun, W., Shen, W., Xia, M., Chen, C., Xiang, D., et al. (2016). Long non-coding RNA DILC regulates liver cancer stem cells via IL-6/STAT3 axis. J. Hepatol. 64, 1283–1294. doi: 10.1016/j.jhep.2016.01.019
Wang, Y., He, L., Du, Y., Zhu, P., Huang, G., Luo, J., et al. (2015). The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell. 16, 413–425. doi: 10.1016/j.stem.2015.03.003
Wang, Y., Zhu, P., Luo, J., Wang, J., Liu, Z., Wu, W., et al. (2019). LncRNA HAND2-AS1 promotes liver cancer stem cell self-renewal via BMP signaling. EMBO J. 38:e101110.
Wu, D.-M., Zheng, Z.-H., Zhang, Y.-B., Fan, S.-H., Zhang, Z.-F., Wang, Y.-J., et al. (2019). Down-regulated lncRNA DLX6-AS1 inhibits tumorigenesis through STAT3 signaling pathway by suppressing CADM1 promoter methylation in liver cancer stem cells. J. Exp. Clin. Cancer Res. 38, 1–17.
Xin, L., Liu, L., Liu, C., Zhou, L. Q., Zhou, Q., Yuan, Y. W., et al. (2020). DNA-methylation-mediated silencing of miR-7-5p promotes gastric cancer stem cell invasion via increasing Smo and Hes1. J. Cell. Physiol. 235, 2643–2654. doi: 10.1002/jcp.29168
Xu, R., Zhu, X., Chen, F., Huang, C., Ai, K., Wu, H., et al. (2018). LncRNA XIST/miR-200c regulates the stemness properties and tumourigenicity of human bladder cancer stem cell-like cells. Cancer Cell Int. 18:41.
Yan, X., Zhang, D., Wu, W., Wu, S., Qian, J., Hao, Y., et al. (2017). Mesenchymal stem cells promote hepatocarcinogenesis via lncRNA–MUF interaction with ANXA2 and miR-34a. Cancer Res. 77, 6704–6716. doi: 10.1158/0008-5472.can-17-1915
Yao, Y., Ma, J., Xue, Y., Wang, P., Li, Z., Liu, J., et al. (2015). Knockdown of long non-coding RNA XIST exerts tumor-suppressive functions in human glioblastoma stem cells by up-regulating miR-152. Cancer Lett. 359, 75–86. doi: 10.1016/j.canlet.2014.12.051
Yu, W., Zhang, L., Wei, Q., and Shao, A. (2020). O6-methylguanine-DNA methyltransferase (MGMT): challenges and new opportunities in glioma chemotherapy. Front. Oncol. 9:1547.
Yu, Z., Pestell, T. G., Lisanti, M. P., and Pestell, R. G. (2012). Cancer stem cells. Int. J. Biochem. Cell Biol. 44, 2144–2151.
Zhang, G.-J., Li, L.-F., Yang, G.-D., Xia, S.-S., Wang, R., Leng, Z.-W., et al. (2017). MiR-92a promotes stem cell-like properties by activating Wnt/β-catenin signaling in colorectal cancer. Oncotarget 8:101760. doi: 10.18632/oncotarget.21667
Zhang, L., Guo, C., Ji, T., and Chen, X. (2021). SOX2 Regulates lncRNA CCAT1/MicroRNA-185-3p/FOXP3 axis to affect the proliferation and self-renewal of cervical cancer stem cells. Nanoscale Res. Lett. 16:2.
Zhang, L., Guo, X., Zhang, D., Fan, Y., Qin, L., Dong, S., et al. (2017). Upregulated miR-132 in Lgr5+ gastric cancer stem cell-like cells contributes to cisplatin-resistance via SIRT1/CREB/ABCG2 signaling pathway. Mol. Carcinog. 56, 2022–2034. doi: 10.1002/mc.22656
Zhang, S., Wan, H., and Zhang, X. (2020). LncRNA LHFPL3-AS1 contributes to tumorigenesis of melanoma stem cells via the miR-181a-5p/BCL2 pathway. Cell Death Dis. 11, 1–16. doi: 10.3233/ch-211130
Zhao, D., Wang, S., Chu, X., and Han, D. (2019). LncRNA HIF2PUT inhibited osteosarcoma stem cells proliferation, migration and invasion by regulating HIF2 expression. Artif. Cells Nanomed. Biotechnol. 47, 1342–1348. doi: 10.1080/21691401.2019.1596934
Zhao, L., Lou, G., Li, A., and Liu, Y. (2020). lncRNA MALAT1 modulates cancer stem cell properties of liver cancer cells by regulating YAP1 expression via miR-375 sponging. Mol. Med. Rep. 22, 1449–1457. doi: 10.3892/mmr.2020.11196
Zhou, H., Xiong, Y., Peng, L., Wang, R., Zhang, H., and Fu, Z. (2020). LncRNA-cCSC1 modulates cancer stem cell properties in colorectal cancer via activation of the Hedgehog signaling pathway. J. Cell. Biochem. 121, 2510–2524. doi: 10.1002/jcb.29473
Zhou, L., Zhao, L., Jiang, N., Wang, X., Zhou, X., Luo, X., et al. (2017). MicroRNA miR-590-5p inhibits breast cancer cell stemness and metastasis by targeting SOX2. Eur. Rev. Med. Pharmacol. Sci. 21, 87–94.
Zhou, M., Hou, Y., Yang, G., Zhang, H., Tu, G., and D, Ye, et al. (2016). LncRNA-Hh strengthen cancer stem cells generation in twist-positive breast cancer via activation of hedgehog signaling pathway. Stem Cells 34, 55–66. doi: 10.1002/stem.2219
Zhu, K., Wang, Y., Liu, L., Li, S., and Yu, W. (2020). Long non-coding RNA MBNL1-AS1 regulates proliferation, migration, and invasion of cancer stem cells in colon cancer by interacting with MYL9 via sponging microRNA-412-3p. Clin. Res. Hepatol. Gastroenterol. 44, 101–114. doi: 10.1016/j.clinre.2019.05.001
Zhuang, W., Ge, X., Yang, S., Huang, M., Zhuang, W., Chen, P., et al. (2015). Upregulation of lncRNA MEG3 promotes osteogenic differentiation of mesenchymal stem cells from multiple myeloma patients by targeting BMP4 transcription. Stem Cells 33, 1985–1997. doi: 10.1002/stem.1989
Keywords: lncRNA, miRNA, cancer stem cell, expression, biomarker
Citation: Ghafouri-Fard S, Hajiesmaeili M, Shoorei H, Bahroudi Z, Taheri M and Sharifi G (2021) The Impact of lncRNAs and miRNAs in Regulation of Function of Cancer Stem Cells and Progression of Cancer. Front. Cell Dev. Biol. 9:696820. doi: 10.3389/fcell.2021.696820
Received: 17 April 2021; Accepted: 06 July 2021;
Published: 22 July 2021.
Edited by:
Richard G. Pestell, Baruch S. Blumberg Institute, United StatesReviewed by:
Soichiro Yamamura, University of California, San Francisco, United StatesRezvan Noroozi, Jagiellonian University, Poland
Amin Safa, Complutense University of Madrid, Spain
Copyright © 2021 Ghafouri-Fard, Hajiesmaeili, Shoorei, Bahroudi, Taheri and Sharifi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohammad Taheri, bW9oYW1tYWRfODIzQHlhaG9vLmNvbQ==; Guive Sharifi, Z2libm93QHlhaG9vLmNvbQ==
 Soudeh Ghafouri-Fard
Soudeh Ghafouri-Fard Mohammadreza Hajiesmaeili2
Mohammadreza Hajiesmaeili2 Hamed Shoorei
Hamed Shoorei Mohammad Taheri
Mohammad Taheri Guive Sharifi
Guive Sharifi