- Institute of Biochemistry II, Faculty of Medicine, Goethe University, Frankfurt, Germany
Promyelocytic leukemia nuclear bodies (PML NBs) are multi-protein assemblies representing distinct sub-nuclear structures. As phase-separated molecular condensates, PML NBs exhibit liquid droplet-like consistency. A key organizer of the assembly and dynamics of PML NBs is the ubiquitin-like SUMO modification system. SUMO is covalently attached to PML and other core components of PML NBs thereby exhibiting a glue-like function by providing multivalent interactions with proteins containing SUMO interacting motifs (SIMs). PML NBs serve as the catalytic center for nuclear SUMOylation and SUMO-SIM interactions are essential for protein assembly within these structures. Importantly, however, formation of SUMO chains on PML and other PML NB-associated proteins triggers ubiquitylation and proteasomal degradation which coincide with disruption of these nuclear condensates. To date, a plethora of nuclear activities such as transcriptional and post-transcriptional regulation of gene expression, apoptosis, senescence, cell cycle control, DNA damage response, and DNA replication have been associated with PML NBs. Not surprisingly, therefore, SUMO-dependent PML NB integrity has been implicated in regulating many physiological processes including tumor suppression, metabolism, drug-resistance, development, cellular stemness, and anti-pathogen immune response. The interplay between PML NBs and viral infection is multifaceted. As a part of the cellular antiviral defense strategy, PML NB components are crucial restriction factors for many viruses and a mutual positive correlation has been found to exist between PML NBs and the interferon response. Viruses, in turn, have developed counterstrategies for disarming PML NB associated immune defense measures. On the other end of the spectrum, certain viruses are known to usurp specific PML NB components for successful replication and disruption of these sub-nuclear foci has recently been linked to the stimulation rather than curtailment of antiviral gene repertoire. Importantly, the ability of invading virions to manipulate the host SUMO modification machinery is essential for this interplay between PML NB integrity and viruses. Moreover, compelling evidence is emerging in favor of bacterial pathogens to negotiate with the SUMO system thereby modulating PML NB-directed intrinsic and innate immunity. In the current context, we will present an updated account of the dynamic intricacies between cellular PML NBs as the nuclear SUMO modification hotspots and immune regulatory mechanisms in response to viral and bacterial pathogens.
Introduction
Promyelocytic leukemia nuclear bodies (PML NBs) are ubiquitous, multi-molecular protein condensates associated with the nuclear matrix (Lallemand-Breitenbach and de Thé, 2018; Corpet et al., 2020). By immunofluorescence microscopy, PML NBs are detected as punctate structures in inter-chromatin spaces. Number, size, and morphology of PML NBs vary depending on cell type and cell cycle status as well as on physiological or pathological stimuli. The composition of PML NBs is heterogeneous and most of the constituents remain in dynamic equilibrium between the soluble nucleoplasmic and the insoluble matrix-associated fraction (Lallemand-Breitenbach and de Thé, 2018; Corpet et al., 2020). Core to the compositional heterogeneity and functional maturity of PML NBs is a specific post translational modification (PTM) with small ubiquitin-related modifier (SUMO) proteins (Cheng and Kao, 2013; Sahin et al., 2014a). PML NBs have widely been regarded as the nuclear niche for protein SUMOylation and most of the constitutive and transient PML NB components are covalently or non-covalently associated with SUMO. PML NBs exert crucial modulatory roles in various activities including nuclear protein quality control, control of gene expression, apoptotic cell death and senescence, genome replication and genomic integrity as well as interferon (IFN) response and antiviral defense (Everett and Chelbi-Alix, 2007; Hsu and Kao, 2018; Lallemand-Breitenbach and de Thé, 2018). Accordingly, the impact of SUMO-dependent regulation of PML NBs on cellular physiology is multifaceted. One of these facets is the regulation of intrinsic and innate immune defense in response to invading pathogens. Many reports have enunciated crucial interplays between PML NBs as the hubs of cellular intrinsic immune defenses and viral countermeasures to establish infection and to ensure perpetuation (Geoffroy and Chelbi-Alix, 2011; Scherer and Stamminger, 2016). Emerging evidence, however, is accumulating in favor of viruses to selectively counteract certain components of PML NBs while usurping others (Guion et al., 2019; Guion and Sapp, 2020). In addition to viruses, certain bacterial pathogens also target PML NB components (Ribet and Cossart, 2018). Notably, the interplay between PML NBs and pathogens might involve the host cellular SUMO machinery as an intermediate or might be independent of SUMO-dependent regulations (Everett et al., 2013b; Scherer and Stamminger, 2016; Lowrey et al., 2017; Wilson, 2017; El Motiam et al., 2020). Here, we present an updated overview of how PML NBs can serve as the niches for mounting intrinsic and innate immune defenses against different pathogens and, in turn, how they are antagonized by the pathogens. We have also highlighted how SUMO-dependent regulations can dictate the outcome of such interplays.
Structural Integrity of PML NBs and Sumoylation
Promyelocytic leukemia protein constitutes the primary structural scaffold for PML NBs (Ishov et al., 1999; Lang et al., 2010). The biomedical interest in PML NBs stems from the initial observation that their structural integrity is lost in acute promyelocytic leukemia (APL). Disruption of NBs in APL is caused by expression of the oncogenic fusion protein PML-RARα (PML-retinoic acid receptor alpha) resulting from the t(15; 17) chromosomal translocation (de Thé et al., 1991; Kakizuka et al., 1991; Dyck et al., 1994; Weis et al., 1994). Similarly, experimental ablation of PML results in complete disassembly of PML NBs (Ishov et al., 1999; Zhong et al., 2000). Due to alternative splicing, PML has 7 isoforms (I-VII). All the isoforms possess a conserved N-terminal region which is comprised of a RING finger domain, two B- boxes (B1, B2), and an α-helical coiled-coil domain, defining PML (also known as TRIM19) as a member of the tripartite RBCC/TRIM motif family. With the exception of isoform VII, which lacks a nuclear localization signal (NLS), all other isoforms (PML I-VI) exhibit a predominantly nuclear localization and can take part in forming PML NBs (de Thé et al., 2012; Nisole et al., 2013). The biogenesis of PML NBs is a dynamic biphasic process. The initial nucleation phase includes the polymerization of PML proteins into a peripheral shell. Covalent disulfide linkages between oxidized PML monomers as well as intermolecular non-covalent interactions between RBCC domains of PML proteins have been proposed to contribute to this event (Figure 1; Lallemand-Breitenbach et al., 2001; Jeanne et al., 2010; Sahin et al., 2014c). The subsequent maturation phase of PML NB biogenesis involves the recruitment of a heterogeneous assembly of proteins (client proteins) within the inner core of the scaffold in either a constitutive or a stimulus-dependent fashion (Figure 1; Lallemand-Breitenbach and de Thé, 2018). Importantly, the recruitment and release of these inner core clients are primarily dictated by SUMO-dependent protein-protein interactions.
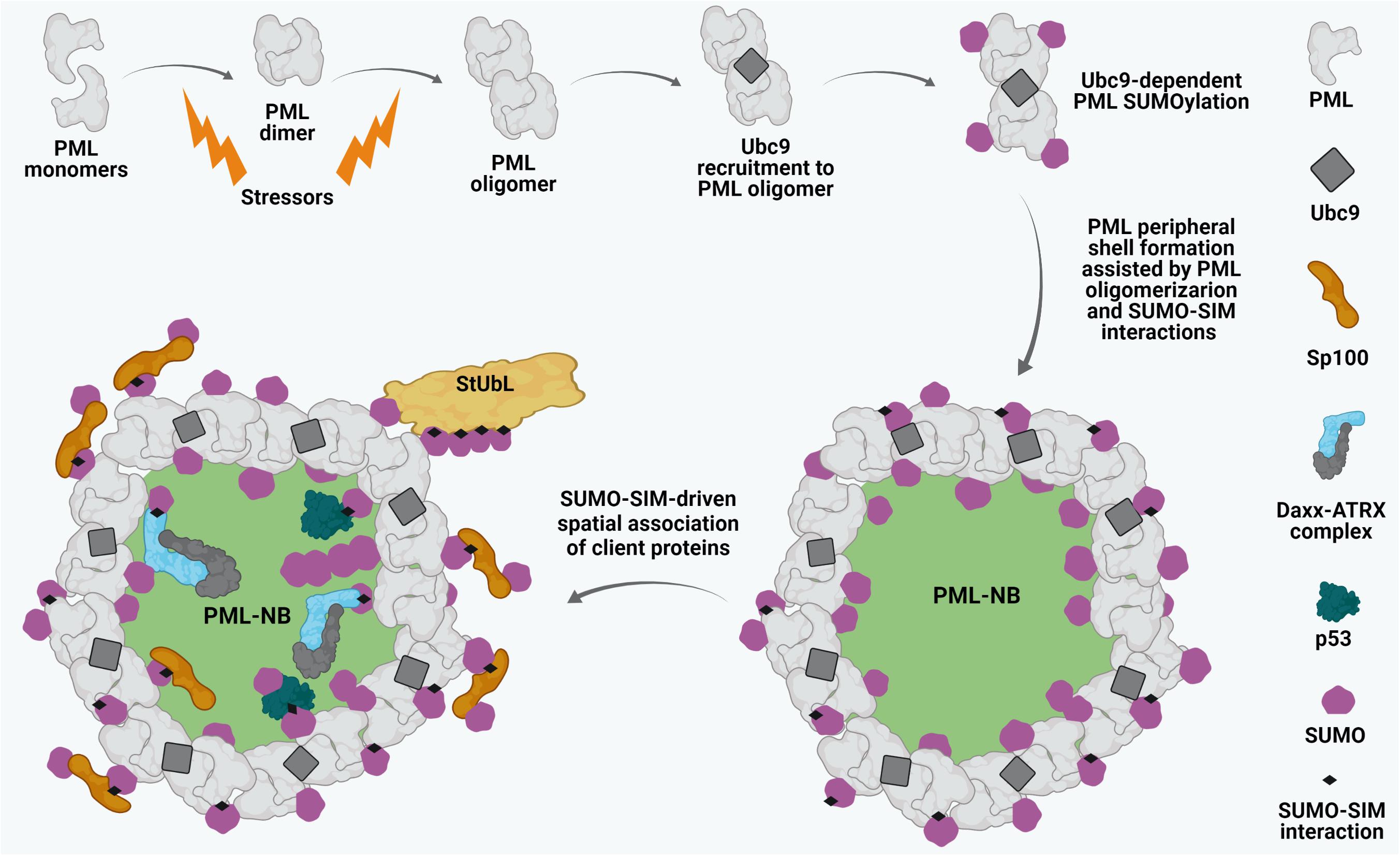
Figure 1. Biogenesis of PML NBs. Oxidative stressors induce PML dimerization and oligomerization through covalent disulphide linkage and intermolecular RBCC motif-driven non-covalent interactions. Subsequently, the SUMO E2 conjugating enzyme Ubc9 is recruited to oligomerized PML and drives PML SUMOylation which further leads to stabilized PML NB shell formation. Sp100 resides along with SUMO1 at the peripheral shell. SUMO-SUMO interacting motif (SIM) affinity also enables recruitment of different client proteins such as Daxx, ATRX, p53 onto mature PML NBs. Ubc9-dependent SUMOylation of the clients may in turn regulate their dynamic interactions with the PML NBs. Poly-SUMO-primed PML might also interact with SUMO targeted ubiquitin ligases (StUbLs), suggesting crucial dynamicity of these matrix-associated sub-nuclear foci.
Small ubiquitin-related modifier proteins are covalently conjugated to their target proteins at lysine (K) residues (Figure 2). In human, three main conjugation-competent SUMO paralogs (SUMO1, SUMO2, SUMO3) are expressed. SUMO2 and SUMO3 differ by 3 amino acids whereas SUMO1 has 50% sequence identity with SUMO2/3 (Saitoh and Hinchey, 2000; Flotho and Melchior, 2013). There is also a SUMO4 paralog which has restricted tissue distribution and cannot be conjugated to target proteins (Bohren et al., 2004; Owerbach et al., 2005). The recently identified SUMO5 is annotated as a pseudogene by the HUGO Gene Nomenclature Committee (HGNC) and therefore it remains under debate whether SUMO5 is expressed at the protein level (Liang et al., 2016). The conjugation of SUMO is a three-step process with sequential involvement of a SUMO E1 activating enzyme (SAE1/SAE2 dimer in human), an E2 conjugating enzyme (Ubc9 in human), and a limited subset of E3 ligases (such as RanBP2, PIAS family members, or ZNF451) (Figure 2). Depending on the type of conjugation, a target can be either mono-SUMOylated at a single K residue, multi-mono SUMOylated at several K residues, or may undergo polySUMOylation by forming SUMO chains on internal K residues within SUMO (Figure 2; Ulrich, 2008; Jansen and Vertegaal, 2021). SUMO-conjugated proteins can make non-covalent interactions with proteins harboring distinct SUMO interacting motifs (SIMs) (Figure 2). The best characterized SIM is composed of a hydrophobic core flanked by acidic amino acids or serine/threonine (S/T) residues. SUMO-SIM affinity is widely used to regulate the dynamics of protein-protein interactions in large multi-molecular complexes, including PML NBs (Ullmann et al., 2012; Raman et al., 2013; Husnjak et al., 2016). Of importance, polymeric SUMO2/3 chains can trigger a specific signaling process known as the SUMO-targeted ubiquitin ligase (StUbL) pathway, where distinct E3 ubiquitin ligases, termed StUbLs, interact with SUMO2/3 multimers/polymers through their tandemly repeated SIM regions (Figure 2). The two characterized mammalian StUbLs, RNF4 and RNF111, trigger proteolytic or non-proteolytic ubiquitylation of poly-SUMO primed substrates (Lallemand-Breitenbach et al., 2008; Tatham et al., 2008; Geoffroy and Hay, 2009; Sriramachandran and Dohmen, 2014; Keiten-Schmitz et al., 2020). SUMO-modifications are reversed by SUMO specific isopeptidases which catalyze cleavage of covalently attached SUMO moieties or SUMO chains from substrates (Figure 2). The best-studied deconjugases belong to the SENP (Sentrin protease) family (Figure 2). Each SENP family member (SENP1-3, SENP5-7 in human) is responsible for deconjugating SUMO residues from a subset of SUMO substrates (Nayak and Müller, 2014; Kunz et al., 2018), thereby providing plasticity within the SUMO system. Dysregulation of the finely tuned balance between SUMO-conjugation and deconjugation can lead to many pathological conditions.
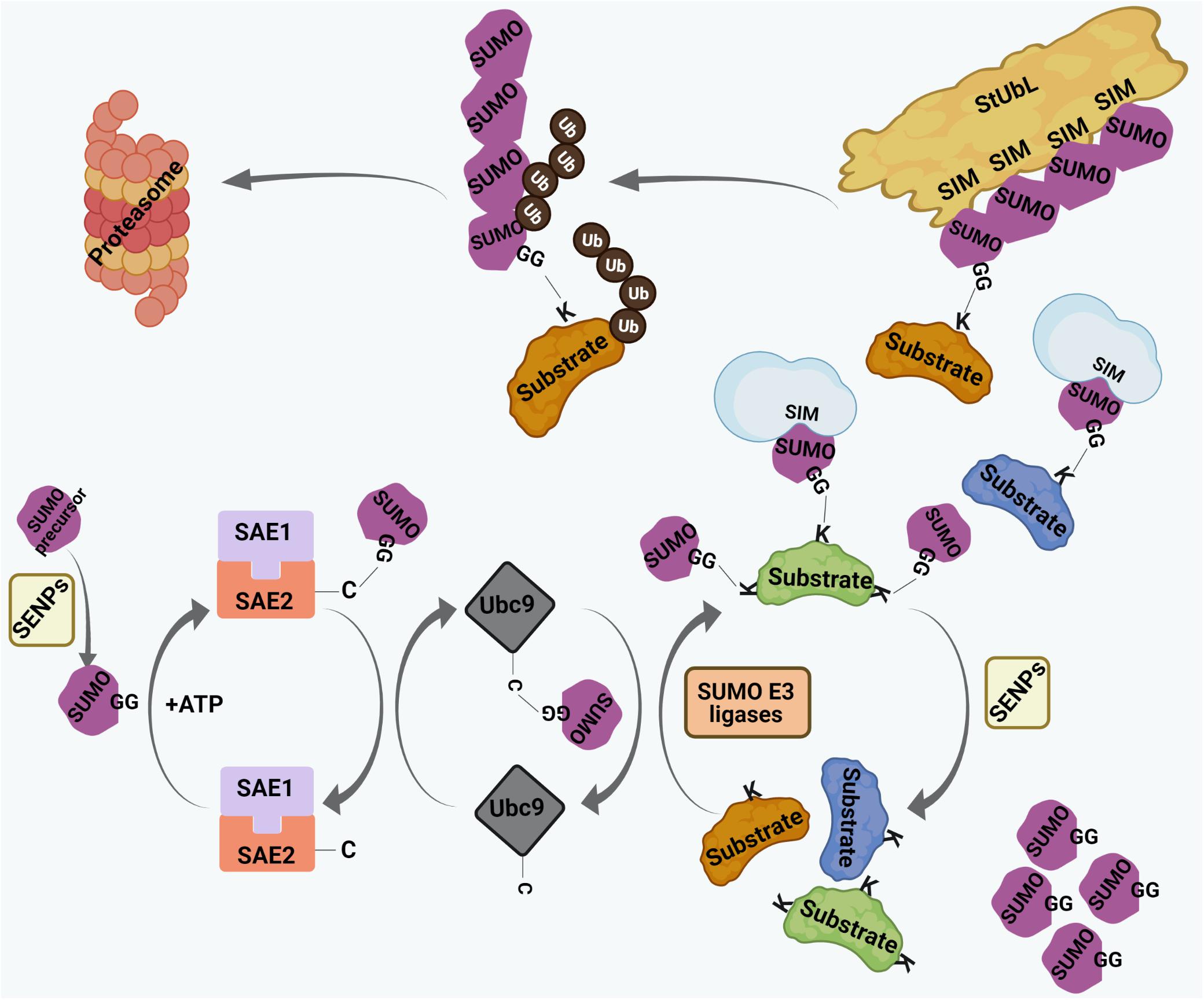
Figure 2. The process of SUMO conjugation and deconjugation. SUMO precursors are proteolytically cleaved by Sentrin-specific proteases (SENPs) to yield conjugation-competent SUMO molecules with terminal di-glycine (GG) motif. SUMO conjugation involves sequential involvement of 3 enzymes – the SUMO E1 activating enzyme (SAE1/SAE2 dimer in human), the SUMO E2 conjugating enzyme (Ubc9 in human), and a limited subset of SUMO E3 ligases. SUMO makes transient, intermediate thioester linkage to cysteine (C) residues of SAE2 and Ubc9 before being finally transferred on to the lysine (K) residues of substrates. The terminally exposed GG of each SUMO forms a covalent isopeptide bond with the ε-amino group on target K residue. Targets can either be monoSUMOylated at single K residue, multi-mono SUMOylated at many K residues or poly-SUMOylated forming SUMO chains. SUMO residues on SUMO-conjugated proteins interact non-covalently with proteins having SUMO interacting motif (SIM). SUMO conjugation can be reversed by SENPs where SUMO residues are cleaved off the substrates. SUMO chains are also targeted by a specific set of E3 ubiquitin ligases, called SUMO targeted ubiquitin ligases (StUbLs), having multiple tandemly arranged SIMs. StUbLs can induce proteolytic/non-proteolytic ubiquitylation of poly-SUMO-primed substrates; the former leads to proteasomal degradation of the substrates.
Interestingly, SUMO-SIM interactions are critical for the assembly of mature PML NBs (Figure 1). A large number of cell-biological studies have demonstrated that SUMO-SIM dependent protein-protein interactions within PML NBs form the scaffold of these structures. More recent in vitro biophysical and biochemical data now underscore this concept by demonstrating that the accumulation of polySUMO-polySIM polymers results in formation of droplet-like structures and also allows interactions with SUMO/SIM-containing clients depending upon their valency (Banani et al., 2016). Moreover, within cells, these SUMO-SIM polymers were shown to form condensates especially at telomeric regions and to regulate partitioning of client proteins (Banani et al., 2016; Min et al., 2019; Zhang et al., 2020). These results advocate for a SUMO-SIM-dependent liquid-liquid phase separation (LLPS) model for PML NB maturation after PML-driven peripheral shell formation.
Promyelocytic leukemia contains three major SUMO conjugation sites at K65, K160 and K490, which are targeted by SUMO1 and SUMO2/3 (Kamitani et al., 1998). PML is polySUMOylated by SUMO2/3 primarily at K160 residue (Tatham et al., 2008). Further, PML isoforms I-V also possess a phospho-regulated SIM (amino acid 556–562) (Stehmeier and Muller, 2009). PML recruits Ubc9 to PML NBs through its RING finger domain, thereby mediating SUMOylation of PML and enabling SUMO-SIM-dependent PML oligomerization (Figure 1; Shen et al., 2006; Sahin et al., 2014c). PML SUMOylation is also important for recruiting additional client proteins to the inner core of mature PML NBs (Figure 1; Shen et al., 2006). Close to 200 client proteins have now been reported to be associated with PML NBs either constitutively or transiently (Van Damme et al., 2010). A subset of proteins associated with PML NBs functions as chromatin and/or transcriptional regulators. One example is Sp100, which was one of the first proteins identified to be localized to PML NBs (Szostecki et al., 1990; Sternsdorf et al., 1995). In human, at least four alternatively spliced isoforms of Sp100-Sp100A, Sp100B, Sp100C, and Sp100HMG-varying at their C-terminal, are present. Sp100B, Sp100C, and Sp100HMG harbor SAND DNA binding domain and are involved with chromatin-dependent transcriptional regulation (Seeler et al., 2001; Negorev et al., 2006, 2009). All Sp100 forms reside in the peripheral shell of PML NBs (Lang et al., 2010) and are extensively SUMOylated (Figure 1). Notably, however, recruitment of Sp100 to PML NBs does not require covalent SUMO conjugation to Sp100, but involves binding of a SIM in Sp100 to SUMO-conjugated PML (Figure 1; Sternsdorf et al., 1997, 1999). Interestingly, lack of Sp100 expression also affects PML expression, underlining the importance of Sp100 in maintaining PML NB integrity (Negorev et al., 2009). Another component of the PML NB core is the chromatin remodeling factor death domain-associated protein (Daxx) which forms a histone chaperone complex with a partner protein alpha-thalassemia retardation syndrome x-linked (ATRX) (Figure 1) and regulates histone H3.3 depositions on DNA in replication-independent manner, thereby modulating transcriptional repression (Lewis et al., 2010). Similar to what is observed for Sp100, recruitment of Daxx to PML NBs depends on the interaction of Daxx SIM with SUMO-conjugated PML (Figure 1; Lin et al., 2006). Accordingly, the SUMOylation status of PML regulates Daxx recruitment to and release from PML NBs (Lehembre et al., 2001; Kawai et al., 2003; Nefkens et al., 2003). SUMO paralogs and SUMO E3 ligases, such as members of the protein inhibitor of activated STAT (PIAS) family, are also compartmentalized in PML NBs (Lang et al., 2010; Rabellino et al., 2012; Brown et al., 2016). Moreover, p53 and a number of p53 regulatory proteins associate with PML NBs, suggesting the significance of these subnuclear foci to transduce anti-proliferative and pro-apoptotic signals (Figure 1; Bernardi et al., 2008).
Certain stimuli such as arsenic trioxide (As2O3) shift the equilibrium of intra-nuclear PML distribution toward the insoluble matrix-associated fraction leading to increased size and number of PML NBs. Importantly, phosphorylation and SUMOylation of PML have been shown to be involved in this dynamic shift in the equilibrium as As2O3 treatment resulted in an enrichment of the phosphorylated pool of PML leading to its recruitment to PML NBs and an increase of PML SUMOylation (Lallemand-Breitenbach et al., 2001; Hayakawa and Privalsky, 2004). As an alternative model, direct binding of As2O3 to PML’s RING domain has been proposed to induce its dimerization and SUMOylation (Zhang et al., 2010). Interestingly, As2O3 also induces SUMO chain formation on PML and the oncogenic fusion protein PML-RARα, thereby activating RNF4-dependent ubiquitylation and proteasomal degradation of PML (Figure 1) and PML-RARα (Lallemand-Breitenbach et al., 2008; Tatham et al., 2008).
Interplay Between PML NBs and Interferon Signaling
Promyelocytic leukemia NBs regulate the IFN-dependent innate immune signaling which constitutes the primary defense arm of the host cells upon encountering a pathogenic entity. The cellular IFN response is initiated upon sensing of pathogen associated molecular patterns (PAMPs) by host cellular pattern recognition receptors (PRRs). The subsequent signal amplification step culminates in the transcriptional upregulation of many IFN stimulated genes (ISGs) through Janus kinase (JAK)-Signal transducer and activator of transcription (STAT) signaling pathway (Schneider et al., 2014). Importantly, PML expression is strongly induced in response to IFNs [type I (IFNα, IFNβ) as well as type II (IFNγ)] (Chelbi-Alix et al., 1995). Consistently, the size and number of PML NBs increase upon IFN stimulation (Chelbi-Alix et al., 1995). The promoter region of the PML gene contains both an IFN-stimulated response element (ISRE) and a gamma activated site (GAS) consensus sequence, explaining induction of PML transcription following IFN stimulation (Stadler et al., 1995). Notably, deletion of ISRE from PML promoter is more derogatory for both type I and II IFN-dependent PML induction than GAS deletion which only modestly decreased PML sensitivity to type II IFN (Stadler et al., 1995). Moreover, downstream of type II IFN, IFN regulatory factor 8 (IRF8) binds to the ISRE within PML promoter leading to transcriptional upregulation of PML (Dror et al., 2007). IFN regulatory factor 3 (IRF3), on the other hand, has been shown to promote PML induction in an ISRE- and GAS-dependent way without the requirement of IFN itself (Kim et al., 2007). The SP100 gene promoter also harbors ISRE and GAS consensus elements and is therefore IFN (both type I and type II)-inducible (Guldner et al., 1992; Grötzinger et al., 1996). Interestingly, IFN treatment also triggers enhanced SUMOylation of PML which coincides with an increase in PML NB size and number (Chelbi-Alix et al., 1995; Maroui et al., 2018). IFN-dependent PML SUMOylation also promotes Daxx recruitment to NBs (Kawai et al., 2003). IFN stimulation also causes a global increase in cellular SUMOylation. Mechanistically, PML, especially the PML III and PML IV isoforms, are important for this process by promoting Ubc9 SUMOylation and Ubc9 recruitment to PML NBs, further amplifying SUMO conjugation (Maroui et al., 2018). Another mechanism behind this global effect involves the induction of an ISG product Lin28B, which represses the expression of microRNAs (miRNAs) belonging to the let-7 family leading to the induction of let-7 targets such as the sumo transcripts (Sahin et al., 2014b).
Interestingly, PML is not only a downstream target of the IFN pathway, but has also emerged as a positive regulator of IFN signaling. PML promotes both IFN synthesis and IFN-induced expression of ISGs. PML II has specifically been reported to buttress type I IFN signaling through association with the transcription factor complexes containing IRF3, NFκB, and STAT1, thereby facilitating their DNA binding and subsequent expression of IFNβ and ISGs (Chen et al., 2015). PML also promotes STAT1/STAT2 activation in IFNβ treated cells and is possibly involved in recruiting STAT1/STAT2 containing interferon stimulated gene factor 3 (ISGF3) complex (containing phospho-STAT1, phospho-STAT2, and IRF9) to ISG promoters along with histone deacetylases (HDAC1, HDAC2) to favor transactivation (Kim and Ahn, 2015). Similarly, PML isoforms I–VI promote IFNγ-induced STAT1 phosphorylation leading to higher DNA binding of STAT1 and increased production of ISGs. In line with these findings, PML–/– mouse embryonic fibroblasts (MEFs) were less efficient in IFNγ-mediated STAT1 DNA-binding activity compared to wild-type MEFs (El Bougrini et al., 2011). PML II also stabilizes the transcription factor CIITA (class II transactivator) which regulates IFNγ–induced major histocompatibility complex II (MHC II) gene transcription by protecting it from proteasomal degradation (Ulbricht et al., 2012). Interestingly, the MHC II gene locus is also spatially reoriented adjacent to PML NBs upon IFNγ stimulation, thereby facilitating a transcriptionally favorable environment for the MHC II expression (Gialitakis et al., 2010). Notably, PML-mediated IFN signaling involves its SUMOylation and the RING finger domain (El Bougrini et al., 2011). Additionally, many regulators of IFN synthesis and IFN-dependent signal amplification have now been reported to be modified by SUMO conjugation (El Motiam et al., 2020). Of importance, SUMO1 and SUMO3 exert distinct effects on the positive feedback loop between IFN signaling and PML, possibly by differentially promoting STAT1 SUMOylation and PML polySUMOylation (Maarifi et al., 2015). In addition to IFNs, production of other pro-inflammatory cytokines such as interleukin 1β (IL1-β) and interleukin 6 (IL-6) are positively regulated by PML as PML deficient cells are markedly inefficient in mounting this acute phase cytokine response (Lunardi et al., 2011; Lo et al., 2013). Consistently, PML knockout mice are insensitive to lipopolysaccharide (LPS)-induced acute shock response and also exhibit a dysregulated immune reaction to bacterial infection (Lunardi et al., 2011).
PML NBs and Pathogens: A Dynamic Intricacy
Many DNA and RNA viruses exhibit cross-regulations with PML NBs. The spectrum of such interplays is wide with certain PML NB constituents curtailing viral gene expression while others are usurped for efficient viral propagation. Viruses have also evolved multifaceted measures to evade PML NB dependent intrinsic immune responses, thereby triggering viral gene expression required for their perpetuation. Moreover, PML NB integrity can also dictate the disease severity upon infection with pathogenic bacteria. In the following sections, we will describe the dynamic intricacies of viral and bacterial pathogens in modulating PML NBs. A profile of this intricate pathogen-host interaction is summarized briefly in Table 1.
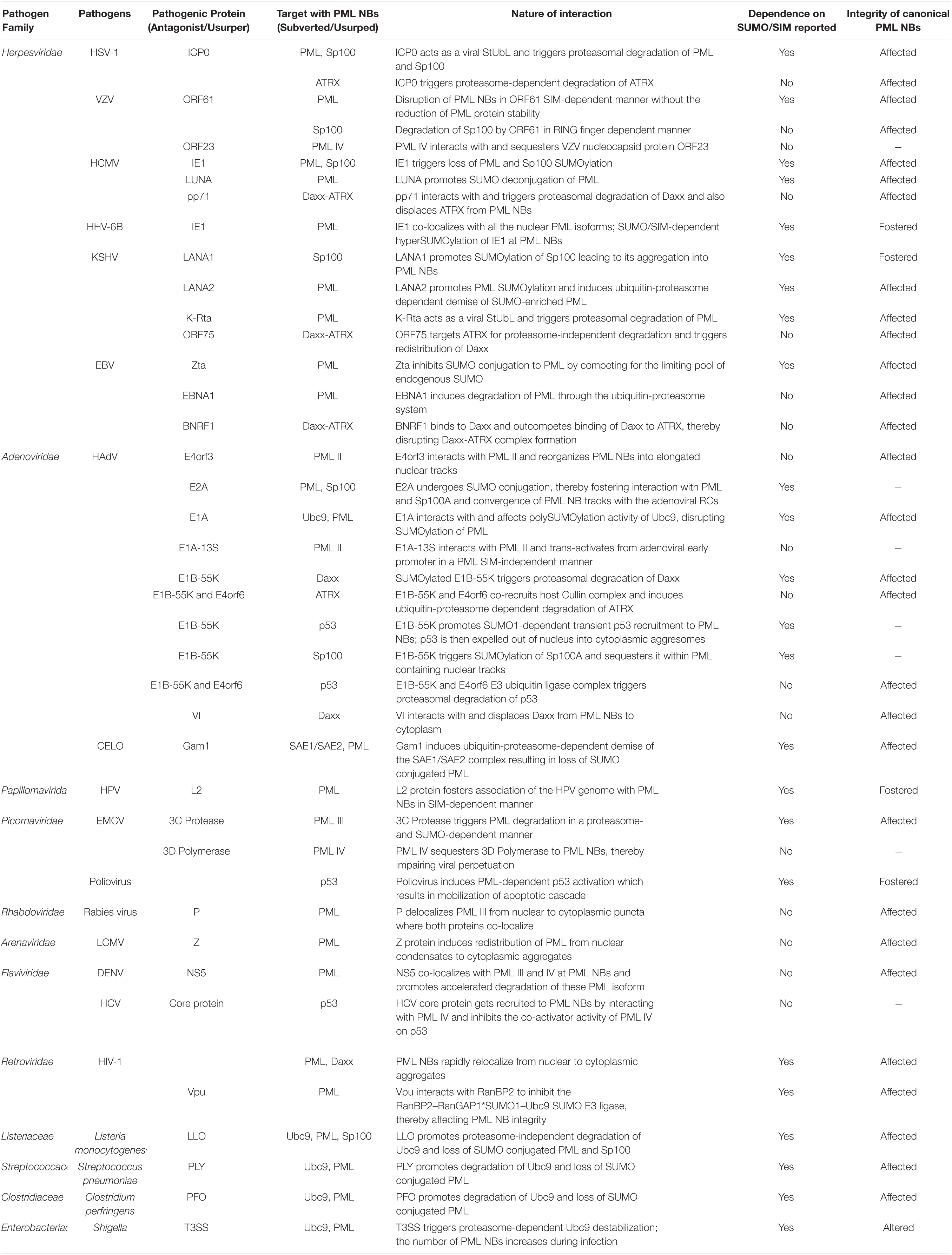
Table 1. SUMO/SIM-dependent/independent interaction profiles of different pathogens with PML NB components.
DNA Viruses
Herpesviruses
Herpes Simplex Virus-1 (HSV-1)
Small ubiquitin-related modifier-dependent dynamics of PML NBs shapes the outcome of infection with HSV-1, an alphaherpesvirus of the Herpesviridae family which generally causes oral sores. Nuclear entry of the viral genomic DNA triggers host cells to mount an intrinsic antiviral response. An essential facet of this response is the silencing of the viral gene expression and onset of latency. This latent reservoir of virus signifies an evolutionary achievement of the virus as it bypasses immune clearance and has the potential to get reactivated periodically leading to disease manifestation and viral transmission. Interestingly, SUMO-dependent regulations are important for initiating this latency. The incoming viral genome, which is a linear molecule of DNA, adsorbs a plethora of both pro- and antiviral host cellular factors (Dembowski and DeLuca, 2018). These include the H3.3 histone chaperone complex Daxx/ATRX which further enables subsequent entrapment of the viral DNA (vDNA) within PML NBs. Other PML NB resident proteins, such as PML and Sp100, as well as the nuclear DNA sensing protein IFI16 also deposit onto the viral genome, where they co-localize with ICP4, an immediate early gene that serves as a marker for incoming HSV-1 genomes (Everett, 2015; Diner et al., 2016; Alandijany et al., 2018; Cabral et al., 2018). Interestingly, SUMO modification of PML and SIMs of PML, Sp100 and Daxx are pivotal for the association of vDNA with PML NBs (Cuchet-Lourenço et al., 2011). Engulfment of the vDNA within PML NBs coincides with the repressive epigenetic modification of the histones (H3K9me3, H3K27me3) on vDNA leading to a transcriptionally inactive chromatin environment (Lee et al., 2016; Cabral et al., 2018; Cohen et al., 2018). The importance of SUMO conjugation to PML and PML-associated NB components is further supported by the finding that host proteins, which promote SUMO conjugation to PML and PML-NB associated proteins curtail HSV-1 infection (Boutell et al., 2011; Brown et al., 2016; Conn et al., 2016). For example, PIAS1, which localizes to PML NBs and acts as a SUMO E3 ligase on PML, contributes to HSV-1 restriction (Brown et al., 2016; Li et al., 2020).
During infection with the wild type virus, the PML NB association of vDNA is transient as the HSV-1 protein ICP0 counteracts this quiescence by dispersal of PML NBs. ICP0 is dispensable for the establishment and maintenance of HSV-1 latency but very critical for the productive rejuvenation of the viral genome from latency and further onset of lytic replication (Leib et al., 1989; Cai et al., 1993; Halford and Schaffer, 2001). Infection with an ICP0 null mutant results in a more persistent association between HSV-1 genome and PML NBs, whereas expression of ICP0 triggers dismantling of PML NB associated proteins from vDNA (Everett et al., 2013a; Alandijany et al., 2018; Cabral et al., 2018). This results in a reduction of histone H3 deposition and enhanced acetylation of remaining histone H3 on vDNA to promote viral transcription (Cliffe and Knipe, 2008; Ferenczy et al., 2011; Lee et al., 2016; Cabral et al., 2018). These observations support a model, in which sequestration of the HSV-1 genome in PML NBs induces repression of viral gene expression, while ICP0-mediated destruction of NBs counters this process.
Core to the functions of ICP0 is its ability to act as a viral StUbL, thereby triggering RING finger-dependent proteolytic ubiquitylation of SUMO-conjugated proteins (Figures 3, 4), including PML (Boutell et al., 2003, 2011; Lanfranca et al., 2014). Accordingly, mutants of HSV-1 which lack ICP0 or express a ligase deficient ICP0, have a dramatically lower probability of initiating a productive infection in restrictive cell types (Everett et al., 2004). Reminiscent to the StUbLs RNF4 or RNF111, ICP0 harbors a RING-domain and 7 putative SIM-Like Sequences (SLS). Accordingly, IPC0 possesses biochemical properties which are similar to host cellular StUbLs (Everett et al., 1998; Boutell et al., 2003, 2011; Sloan et al., 2015). The structure of ICP0 SLS4 (residue 362–367) with SUMO has been resolved by NMR, revealing a cooperation between ICP0 phosphorylation domains (FHA [67-pTELF-70] and Phos2) for recognition of the SUMOylated proteins (Boutell et al., 2008; Mostafa et al., 2013; Hembram et al., 2020). Therefore, both the ICP0 RING-finger and phosphorylation are important for the degradation of SUMOylated proteins and successful reactivation of HSV-1 from latency (Mostafa et al., 2011, 2013; Vanni et al., 2012). One specific SIM in the central region of ICP0 is absolutely essential for degradation of an array of SUMOylated host proteins, including PML and Sp100 (Figure 3; Everett et al., 1998, 2009, 2014; Müller and Dejean, 1999).
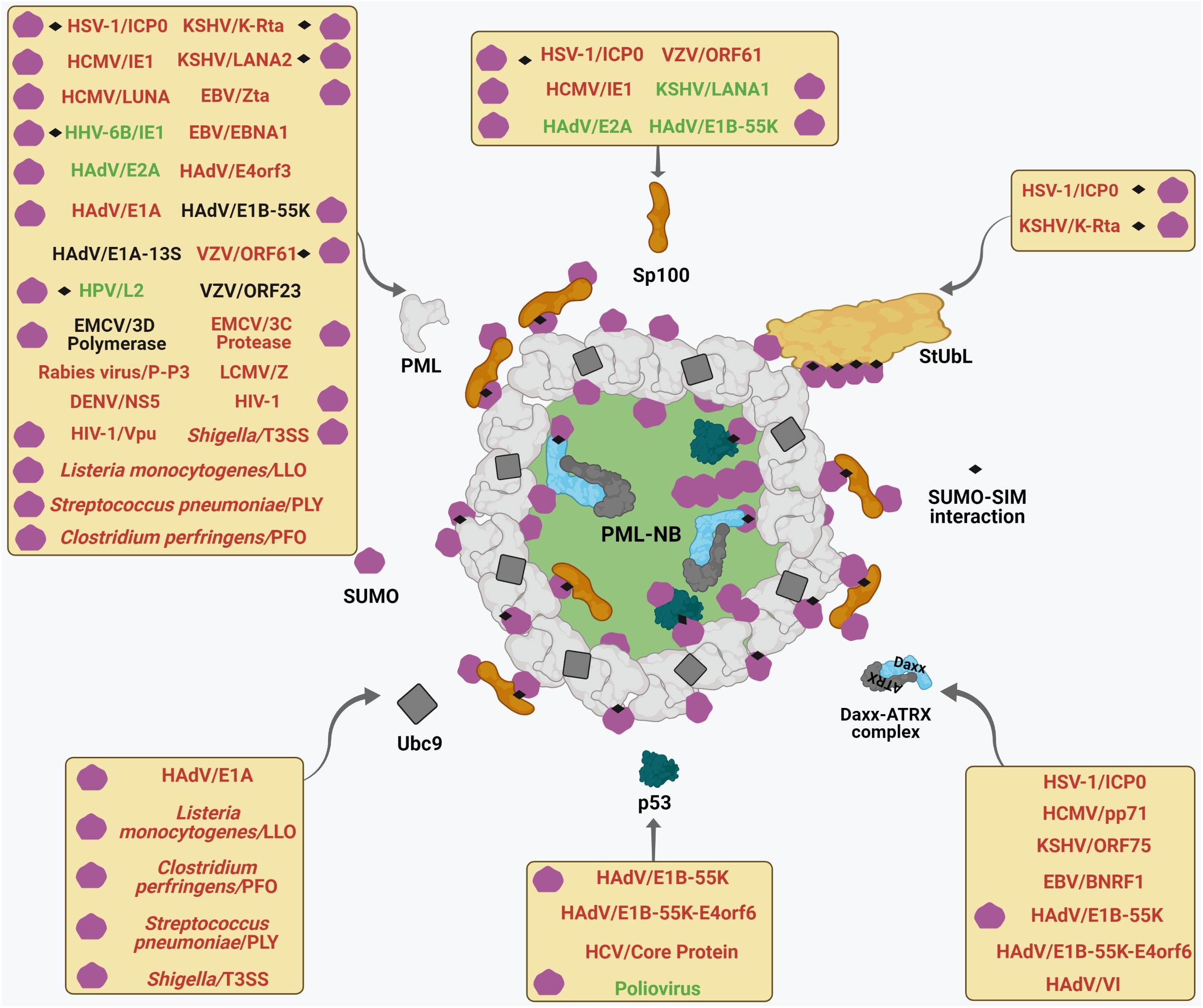
Figure 3. Multifaceted interactions of viruses and bacteria with PML NBs. Different viruses and bacteria target mature PML NBs. Each box represents viral and bacterial components which target specific PML NB components (shown by curved arrows) to either foster (text-colored Green) or perturb (text-colored Red) PML NB integrity or are neutral (text colored Black) on PML NB. The pathogen-PML NB interactions which are SUMO-regulated and/or SUMO-SIM affinity driven are designated by using corresponding symbols within each box. For further details, please refer to the main body of the article.
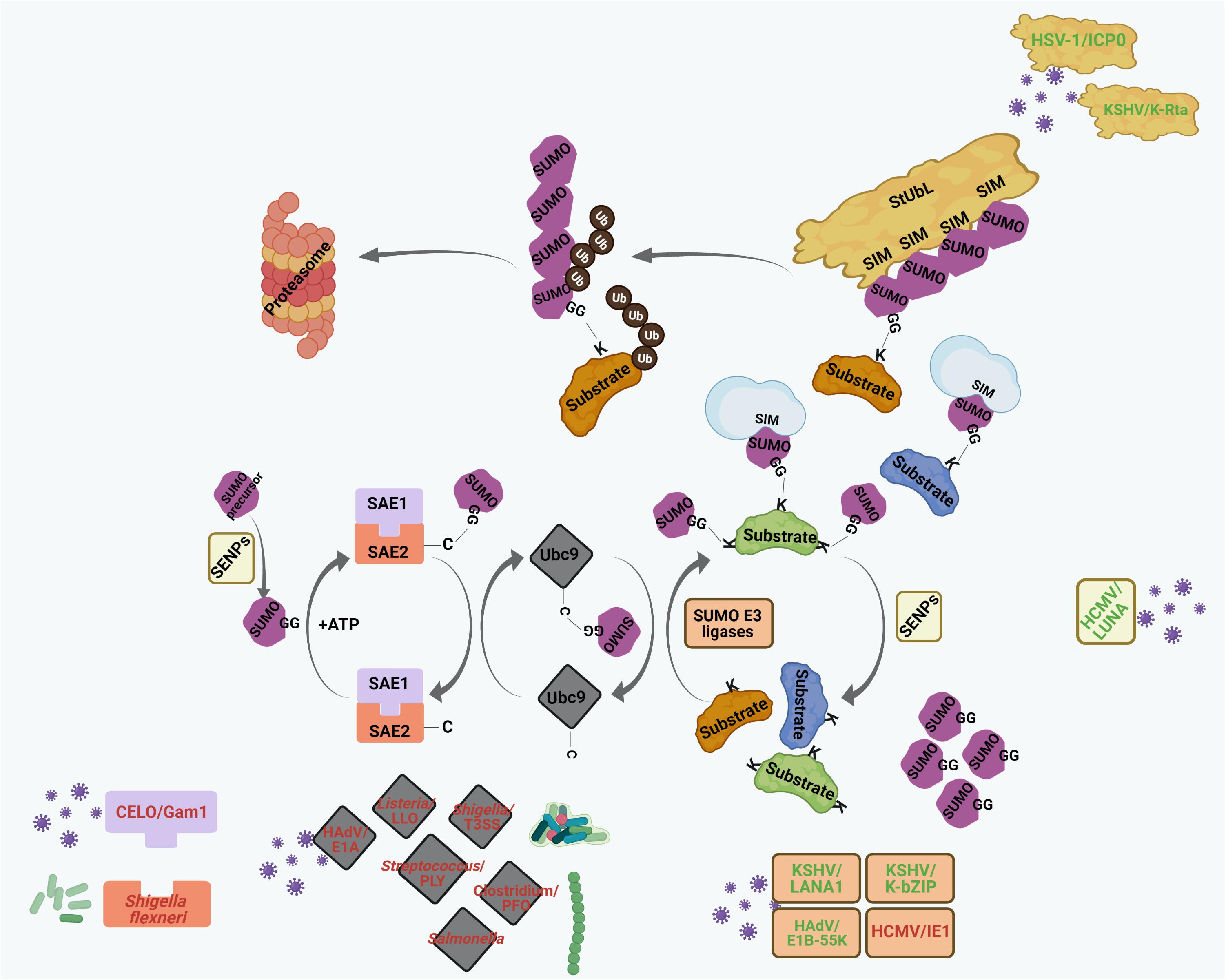
Figure 4. Interplays between different pathogenic components and host cellular SUMO machinery regulating PML NB integrity. Many viral and bacterial factors manipulate with the host cellular SUMO conjugation/deconjugation system to modulate PML NBs. The process of SUMO conjugation and deconjugation as in Figure 2 is shown here. Components of different pathogens are represented within specific colored shapes which are look-alike with their targets within the SUMO system (such as E1, E2, E3, SENPs, and StUbLs). The components which mimic targets within the SUMO machinery are text-colored green whereas those which destabilize/antagonize targets within the SUMO system are text-colored red. For further details, please refer to the main body of the article.
ICP0 also induces the degradation of ATRX and IFI16 (Figure 3; Jurak et al., 2012; Orzalli et al., 2012, 2016; Diner et al., 2015b), but turnover of ATRX and IFI16 occurs with delayed kinetics relative to that of PML (Jurak et al., 2012; Cuchet-Lourenço et al., 2013), possibly indicating that degradation of host components occurs sequentially as infection progresses (Merkl and Knipe, 2019). This is consistent with the step-wise accumulation of cellular factors on vDNA throughout the initiating cycle of HSV- 1 infection (Dembowski and DeLuca, 2015, Dembowski and Deluca, 2017). Recent studies have identified the histone H3.3 chaperone protein HIRA to restrict HSV-1 infection following the onset of vDNA replication, a host response antagonized by ICP0 through the nuclear dispersal of HIRA (McFarlane et al., 2019). Thus, multiple histone H3.3 chaperone proteins (Daxx/ATRX and HIRA) can restrict the replication of ICP0 null HSV-1 at independent phases of infection (Rodríguez et al., 2020). Notably, ICP0 null mutants exhibit reduced plaque forming efficiency which can be reversed by co-depletion of either PML, Sp100, Daxx or ATRX (Everett et al., 2006, 2008; Lukashchuk and Everett, 2010). Absence of a single component does not affect the recruitment of other factors onto HSV-1 genomes, indicating their independent deposition onto the viral genome (Everett et al., 2006, 2008; Lukashchuk and Everett, 2010). Consistent with this, simultaneous depletion of PML and Sp100 enhances ICP0-null HSV-1 propagation greater than single depletion of each factor (Everett et al., 2008). However, another study reported knockdown of PML to enhance wild type HSV-1 propagation (Diner et al., 2015a). Interestingly, SIM-deficient mutants of PML and Daxx are not recruited to vDNA and are incapable of reproducing the repression of ICP0 null mutant HSV-1 viruses (Cuchet-Lourenço et al., 2011), indicating that SIM-dependent PML NB recruitment to HSV-1 DNA during the initial phase of infection is needed to create a transcriptionally repressive environment. Of note, though Sp100 also contributes to the repression of gene expression from null ICP0 HSV-1 virus (Everett et al., 2008, 2009; Glass and Everett, 2013), direct implications of Sp100 on the epigenetic regulation of viral chromatin or modulating innate immune signaling are yet to be determined.
Apart from PML NB resident proteins, a number of host cellular SUMO-modified proteins have now been identified to be targeted by ICP0 (Everett et al., 1998; Chelbi-Alix and de Thé, 1999; Boutell et al., 2011; Sloan et al., 2015). For many of these targets, however, specificity and relevance to HSV-1 replication have yet to be determined, implicating a scenario where at least some of these proteins may be the subjects of “bystander” degradation. Importantly, upon degradation of MORC3 which is a SUMO-conjugated PML NB client targeted by ICP0, recruitment of other PML NB components PML, Sp100, Daxx on to vDNA is also inhibited, substantiating the importance of these foci in defense against invading pathogens (Sloan et al., 2016).
In addition to disarming PML NB-oriented intrinsic immunity, ICP0 also inactivates innate immune defenses early in the infectious cycle through the degradation of cellular PRRs. Detection of viral nucleic acid by host PRRs can elicit a signal transduction cascade leading to generation of IFNs and mobilization of IFN-dependent downstream signaling to produce hundreds of ISGs with antiviral potency. Interestingly, null ICP0 HSV-1 mutants are hypersensitive to IFN (Leib et al., 1999; Mossman et al., 2000; Härle et al., 2002; Everett et al., 2004), indicating that ICP0 limits the IFN-dependent innate immune response. PML II and IV directly influence the induction of cytokines and ISGs by facilitating the loading of transcription factors (including IRF3, NF-κB, and STAT1) onto cellular gene promoters, possibly explaining the IFN hypersensitivity phenotype of null ICP0 HSV-1 mutants (Chee et al., 2003; El Asmi et al., 2014; Chen et al., 2015; McFarlane et al., 2019). Subsequent studies revealed that ICP0 disarms nuclear PRR by degrading IFI16 which recognizes viral dsDNA and signals via STING-dependent pathway and also by triggering decay of DNA-PKcs (Orzalli et al., 2012, 2015, 2016; Cuchet-Lourenço et al., 2013; Diner et al., 2016; Burleigh et al., 2020). Interestingly, a recent study showed a distinct temporal segregation between the entrapment of HSV-1 DNA by PML NBs upon its nuclear entry and the subsequent deposition of IFI16 leading to ISG expression (Alandijany et al., 2018). While the former contributes to the host cellular intrinsic defense measure by triggering viral latency, the latter is related to the host innate immune response. Moreover, PML NB entrapment of vDNA upon its nuclear entry was shown to be independent of IFI16 deposition onto the viral genome which was shown to require vDNA replication. PRR sensing by IFI16 during null ICP0 HSV-1 infection correlates with IFI16 forming nuclear filaments on vDNA in association with PML following the saturation of PML NBs under high genome loads (Cuchet-Lourenço et al., 2013; Alandijany et al., 2018; Merkl and Knipe, 2019). A close functional relationship between innate and intrinsic immunity is further evidenced when resident PML NB proteins (PML, Daxx and ATRX) have been shown to act cooperatively with IFI16 to restrict null ICP0 HSV-1 gene expression that correlates with repressive histone signatures (H3K9me3 and H3K27me3) on viral chromatin (Everett et al., 2008; Lee et al., 2016; Cabral et al., 2018; Merkl et al., 2018).
Varicella-Zoster Virus (VZV)
Another alphaherpesvirus VZV is the etiological agent of chickenpox and the reactivation disease known as shingles (Gilden et al., 2015). Disruption of PML NBs without the reduction of PML protein stability itself has been evidenced in VZV infected cells both in cell culture based studies and in vivo (Kyratsous and Silverstein, 2009; Reichelt et al., 2011). The viral trigger has been found to be the protein ORF61 which is homologous to HSV-1 ICP0 in having a RING finger and SIMs (Figure 3; Moriuchi et al., 1992; Everett et al., 2010; Wang et al., 2011). Unlike ICP0, however, StUbL activity of ORF61 has not yet been formally determined. Nonetheless, Sp100 is targeted for degradation by ORF61 especially during early hours of infection in a ORF61 RING finger-dependent manner (Figure 3; Kyratsous et al., 2009; Kyratsous and Silverstein, 2009; Walters et al., 2010). The significance of this process on VZV life cycle has not been addressed yet. Interestingly, three SIMs within ORF61 were found to be indispensable for targeting of PML NBs. In response to the ORF61 SIM mutant virus, viral replication in skin was severely hampered, the typical VZV lesions were reduced, and viral spread in the epidermis was limited (Wang et al., 2011), suggesting that the SUMO binding function of ORF61 is critical for overcoming the anti-viral effects of the PML bodies. Of interest, as a part of the dynamic host-virus interactions, the persisting PML NBs in VZV infected cells can also mount a potent anitiviral response by sequestering viral nucleocapsids. Mechanistic studies revealed that PML IV interacts with the VZV capsid protein ORF23 to enable this sequestration and to prevent nuclear egress of viral nucleocapsids, thereby inhibiting formation of infectious virions (Figure 3; Reichelt et al., 2011). However, a direct involvement of host cellular SUMO system in this antiviral defense mechanism has remained elusive.
Human Cytomegalovirus (HCMV)
Human cytomegalovirus is a betaherpesvirus which also has the potency to go into latency or to resume lytic reactivation and proliferation. Importantly, transcription of the viral immediate early (IE) genes is crucial for switching the viral infection cycle toward lytic proliferation. Major HCMV IE genes are transcribed under the control of the major immediate early promoter (MIEP), which is only transactivated by a viral protein residing within the tegument layer of infectious virions.
Dynamic SUMO modifications have been reported to contribute to the regulations of intrinsic and innate immune defenses against infection with HCMV. The initial observation that herpesviral DNA associates with PML NBs upon its nuclear entry led to the hypothesis that these condensates are usurped by cytomegalovirus and other herpesviruses for productive initiation of the viral IE gene expression (Maul et al., 1996; Ishov and Maul, 1996; Ishov et al., 1997). Subsequent investigations revealed that these spatial proximity and entrapment of vDNA within PML NBs during initial hours of infection constitute a major arm of the cellular intrinsic antiviral defense (Saffert and Kalejta, 2006; Tavalai et al., 2006, 2008; Woodhall et al., 2006; Adler et al., 2011). This is enabled by creating a transcriptionally–incompetent chromatin environment on vDNA leading to silencing of viral IE gene expression. Agreeably, knocking down expression of individual PML NB components such as PML, Daxx, and Sp100 in primary human fibroblasts showed a de-repressing effect on HCMV IE gene expression at low multiplicity of infection (Saffert and Kalejta, 2006; Tavalai et al., 2006, 2008; Woodhall et al., 2006; Adler et al., 2011). Moreover, simultaneous depletion of more than one PML NB component has additive effects on the number of IE-positive cells than when single knock down was achieved, suggesting that multiple NB components independently contribute to restriction of HCMV lytic replication (Tavalai et al., 2008; Adler et al., 2011). Interestingly, PML NB components play a crucial role in epigenetic modulations of the MIEP, leading to repression of the viral IE gene expression (Groves et al., 2009; Sinclair, 2010). The contribution of Daxx to trigger epigenetic quiescence of viral gene expression is evident from analyzing the chromatin signatures around the MIEP in Daxx-deficient cells (Woodhall et al., 2006). Daxx has also been shown to recruit HDACs and ATRX to the viral promoter to execute transcriptional repression (Saffert and Kalejta, 2006; Lukashchuk et al., 2008). Not surprisingly, therefore, inhibition of HDACs by trichostatin A or depletion of ATRX had a de-repressing effect on HCMV IE gene expression (Saffert and Kalejta, 2006; Lukashchuk et al., 2008). In addition to Daxx, PML has also been reported to interact with HDACs and histone methyl transferases (HMTs) and Sp100 with heterochromatin protein 1 (HP1) (Seeler et al., 1998; Wu et al., 2001; Di Croce et al., 2002; Wilcox et al., 2005; Carbone et al., 2006). However, implications of such interactions in mediating epigenetic silencing of viral genome are yet to be addressed experimentally. Although some mechanistic details are still unclear, the ensemble of these data indicate that - in analogy to HSV1- the sequestration of the HCMV genome in PML NBs mediates the repression of viral gene expression through epigenetic silencing of the genome.
Importantly, however, HCMV has also developed countermeasures to rejuvenate its life cycle and to maintain perpetuation. There are three effector proteins, the tegument protein pp71, the Latency Unique Natural Antigen (LUNA) protein and the major immediate-early protein IE1, which can affect the integrity of PML NBs (Figure 3). The mechanisms employed, however, are different. After being transported to the nucleus during infection of terminally differentiated cells such as fibroblasts and epithelial cells, pp71 interacts with and triggers proteasomal degradation of Daxx and also displaces ATRX from the PML NB condensates (Figure 3; Hofmann et al., 2002; Saffert and Kalejta, 2006; Lukashchuk et al., 2008). This further relieves the Daxx-mediated transcriptional repression on the viral genome leading to expression of viral IE1 from MIEP and productive viral infection. A pp71-null HCMV virus has a major defect in IE gene expression and productive replication (Bresnahan and Shenk, 2000; Cantrell and Bresnahan, 2006). Following infection of incompletely differentiated myeloid cells such as CD34+ hematopoietic progenitor cells (HPCs) and monocytes, pp71 remains trapped in cytoplasmic endosomes (Penkert and Kalejta, 2010, 2013; Lee et al., 2019; Lee and Kalejta, 2019; Kalejta and Albright, 2020). As a result, Daxx is not degraded, thereby facilitating histone deposition and assembly of repressive heterochromatin on viral genomes in conjunction with ATRX and the KAP1/SetDB1/HDAC co-repressor complex. Heterochromatin assembly results in repression of the MIEP allowing for the establishment of latency (Murphy et al., 2002; Groves et al., 2009; Sourvinos et al., 2014; Lee et al., 2015; Rauwel et al., 2015; Albright and Kalejta, 2016; Kalejta and Albright, 2020). Importantly, pp71 mutants which cannot interact with Daxx are also unable to activate an MIEP reporter (Hofmann et al., 2002), substantiating the importance of pp71-Daxx interaction for Daxx repression and transactivation from MIEP. Apart from Daxx degradation, pp71 also triggers Daxx SUMOylation (Hwang and Kalejta, 2009). SUMOylated Daxx is unable to interact with NFκB and therefore fails to prevent NFκB acetylation and activation (Croxton et al., 2006; Park et al., 2007; Kim et al., 2017). This leads to NFκB-dependent enhanced MIEP activity (DeMeritt et al., 2004; Liu et al., 2010; Yuan et al., 2015) and efficient IE gene expression at least in certain contexts.
Although the pro-viral relevance of pp71 in initiating viral IE gene expression to induce a de novo lytic infection is well studied, importance of pp71 in mediating viral rejuvenation from latency and its role of NB remodeling in this process remains questionable (Kalejta and Albright, 2020). Interestingly, a recent report has highlighted disruption of PML NBs in a pp71-independent manner by the viral LUNA protein (Poole et al., 2018). LUNA is encoded from a transcript antisense to the viral UL81 and UL82 genes (which also codes for pp71) (Bego et al., 2005) and is expressed not exclusively during latency but also during lytic infection (Keyes et al., 2012). LUNA has been shown to promote HCMV reactivation (Keyes et al., 2012) by triggering dismantling of PML NBs through a proposed deSUMOylase activity (Figures 3, 4; Poole et al., 2018). Mechanistically, LUNA was proposed to act as a cysteine protease, thereby catalyzing SUMO deconjugation from PML and possibly explaining the loss of PML NB integrity in presence of this HCMV protein (Figure 3). Substituting a potential cysteine residue in the predicted catalytic center completely abrogated isopeptidase activity as well as PML NB disruption by LUNA and this phenomenon coincides with impaired viral reactivation from latency. Of interest, reactivation deficiency of LUNA-null or LUNA catalytically dead virus mutants is rescued in PML-deficient cells, further substantiating LUNA-dependent deSUMOylase activity on PML to be important for HCMV rejuvenation from latency (Poole et al., 2018).
During a productive lytic infection, tegument-delivered pp71 initiates and sustains MIEP transcription long enough for the de novo synthesized IE1 protein to promote prolonged lytic phase gene expression (Vardi et al., 2018) and the completion of productive replication. IE1 protein employs a SUMO-dependent mechanism to disrupt PML NBs (Figure 3). IE1 accumulates at PML NB foci and depletes specifically the SUMO-modified isoforms of PML and Sp100 in a proteasome-independent manner without perturbing global SUMOylation (Figure 3; Müller and Dejean, 1999; Xu et al., 2001; Lee et al., 2004; Tavalai et al., 2011; Everett et al., 2013a; Scherer et al., 2013). IE1 itself is SUMOylated on K450 (Xu et al., 2001; Lee et al., 2004; Nevels et al., 2004; Sadanari et al., 2005), but the functional significance of this modification is poorly understood. Expression of an IE1 K450R mutant shows decreased transcripts and protein expression of IE2, the primary viral transactivating protein for HCMV early and late gene expression, suggesting SUMO modification of IE1 to possibly impact IE2 expression (Nevels et al., 2004). Interestingly, SUMOylation of IE1 is dispensable for its PML NB disrupting function (Xu et al., 2001; Spengler et al., 2002; Lee et al., 2004). However, this disruption requires a direct physical interaction between an extended interaction interface of IE1 globular core domain and the coiled-coil domain of PML (Ahn et al., 1998; Lee et al., 2004; Scherer et al., 2014). How IE1 triggers loss SUMO-conjugated isoforms of PML and Sp100 has recently been elucidated in both cell-based assays and in vitro SUMOylation assay. IE1 was found to sensitize only the de novo SUMOylation at K160 of PML after binding to PML but not the deSUMOylation of pre-SUMO-modified PML (Schilling et al., 2017). Moreover, IE1 also prevented As2O3-mediated hyperSUMOylation of PML at K160, thereby blocking PML degradation. Since IE1 did not interfere with the coiled-coil-mediated PML dimerization, it has been speculated that IE1 impairs PML autoSUMOylation either by directly abrogating the E3 ligase function of PML (Figure 4) or by preventing access to SUMO sites (Schilling et al., 2017). Besides PML and Sp100, IE1 also targets Daxx to antagonize Daxx-mediated transcriptional repression of the viral genome (Reeves et al., 2010). However, the mechanistic details of IE1-Daxx interaction has not been addressed so far. Moreover, IE1 also induces degradation of SUMO unmodified Sp100 in a proteasome-sensitive way in late stages of infection. This event was revealed to be independent of PML NB disruption (Kim et al., 2011).
Interestingly, IE1-mediated PML NB disruption not only has antiviral significance from the perspective of disarming host cellular intrinsic defense measures but also has implications in abrogating innate immune response (Kim and Ahn, 2015; Scherer et al., 2015). SUMOylation status of IE1 regulates IE1-dependent inhibition of innate immune signaling. Unmodified IE1 interacts with STAT1 and STAT2 in the nucleus and decreases binding of these transcription factors to target promoters, leading to repression of ISG transcription (Paulus et al., 2006; Huh et al., 2008; Krauss et al., 2009). Therefore, in comparison to wild-type HCMV, IE1-deleted HCMV shows hypersensitivity to IFN. Interestingly, SUMOylated IE1 has reduced affinity for STAT2 and is less efficient in inhibiting STAT2-dependent ISG transcription (Huh et al., 2008). In addition to STATs, IE1 also antagonizes type I IFN signaling by interacting with PML, thereby sequestering ISGF3 into a functionally inactive complex incapable of DNA binding and ISG transactivation (Kim and Ahn, 2015).
Structural characterization of the IE1 core (Scherer et al., 2014) has revealed it to be a compact, contiguous domain that is highly sensitive to small deletions and point mutations. A single leucine-to-proline exchange (L174P) within the core domain of IE1 at residue 174 hampers the structural integrity and stability of the protein. Consequently, the L174P mutant is incapable of triggering PML NB disruption and has a growth defect that phenocopies IE1-deleted virus (Scherer et al., 2015, 2017). Only very recently, a novel IE1 mutant (IE1cc172–176) has been characterized, which is selectively deficient in interacting with PML, inhibiting PML SUMOylation, and therefore incapable of PML NB localization and disruption without losing overall structural stability (Paulus et al., 2020). Very interestingly, this mutant has been shown to support viral replication almost as efficiently as its wild-type counterpart. Subsequent functional analysis of IE1cc172–176 revealed that this protein is hypermodified by mixed SUMO chains and that IE1 SUMOylation depends on nucleosome rather than PML binding. IE1 mutant which cannot bind to nucleosome (IE1dl476–491) and show reduced chromatin association was revealed to be poorly SUMO-modified, possibly suggesting that IE1 SUMOylation takes place on chromatin. The chromatin-associated E3 SUMO ligase PIAS1 was proposed to SUMOylate IE1 (Paulus et al., 2020). Indeed, PIAS1 has been reported to interact with IE1, to enhance IE1 SUMOylation (Kim et al., 2014), and to act as a SUMO E3 ligase for many transcription factors and chromatin associated proteins (Wu et al., 2014). It was speculated that PIAS1-directed SUMOylation might promote dissociation of the viral protein IE1 from chromatin surface (Paulus et al., 2020). This also explains why IE1 is SUMOylated at nucleosomes but localizes throughout the nucleus. Of interest, with the use of this selective IE1 mutant, not only HCMV IE1-induced deSUMOylation of Sp100 was uncoupled from that of PML, but also upregulation of IFN signaling and ISG expression were observed (Paulus et al., 2020). However, speculation such as disruption of PML NBs upon viral infection is linked to activation rather than inhibition of innate immunity is subject to further studies in other virus infection scenario.
Human Herpesvirus-6 (HHV-6)
Promyelocytic leukemia and PML NBs have also proven to be critical regulators of the betaherpesvirus human herpesvirus 6A and B (HHV-6A/B). HHV-6 infects peripheral blood mononuclear cells (PBMCs) and various T-cell lines where it can either establish latent infection or can replicate as a part of its lytic programme. Notably, HHV6 integrates its genome into the telomeric regions of host chromosomes for prolonged maintenance. Importantly, PML NBs and the SUMO machinery were shown to be involved in controlling viral latency as well as viral integration. In case of HHV6A, it was demonstrated that knocking down components of PML NB such as PML, Daxx, and Sp100 significantly increased viral lytic replication and late gene expression. Consistently, progressive lytic infection was found to reduce the number of PML NBs within infected cells with concomitant increase in the puncta size without triggering complete disassembly of these speckled structures (Sanyal et al., 2018). These findings are in line with a role of PML NBs in restricting lytic replication of HHV-6A.
Work on HHV-6B has recently revealed a surprising facet of PML and the SUMO machinery in integrating the HHV-6B genome into the telomeric regions of host chromosomes for prolonged maintenance (Collin et al., 2020). Further, the HHV-6B trans-activator protein IE1 which undergoes SUMOylation at K802 and localizes to PML NBs has emerged as the viral regulator in this process (Figure 3; Gravel et al., 2002, 2004; Stanton et al., 2002; Collin et al., 2020). Mechanistically, IE1 co-localizes with all the nuclear PML isoforms to subsequently get hyperSUMOylated by SUMO1 (involving K802 and possibly other SUMO conjugation sites), but not SUMO2/3, in presence of PML and intact PML NBs. A putative SIM as well as the primary SUMO accepting site K802 proved to be essential for IE1 hyperSUMOylation and PML NB recruitment. Moreover, PML was also found to mediate IE1 localization to telomeres and PML depletion sensitized telomeric recruitment of IE1 as well as HHV-6B genomic integration into the host chromosomes (Collin et al., 2020).
Kaposi’s Sarcoma-associated Herpesvirus (KSHV)
Kaposi’s sarcoma-associated herpesvirus is a gammaherpesvirus which is capable of establishing latent infection and can get periodically activated to undergo lytic replication and production of progeny viruses. Much alike to the other herpesviruses, PML NB-mediated restriction of the infecting viral genome has been shown to be important for triggering quiescence of KSHV genome. Latency-associated nuclear antigen LANA1 is the primary viral product during latency and has been reported to bind to a series of cellular gene promoters to modulate gene transcription. LANA1 promotes SUMOylation of Sp100 leading to its aggregation into PML NBs (Figure 3; Günther et al., 2014), thereby fostering structural integrity of these structures. LANA1 has a SUMO2-specific SIM which aids in SUMOylation of LANA1 at K1140 residue and also enables recruitment of SUMO2-modified KAP1 together with the Sin3A-containing transcriptional repressor complex. LANA1-KAP1 association maintains viral latency by silencing the expression of viral lytic trans-activator K-Rta. Deletion of SIM from LANA1 abrogates this association, thereby triggering loss of viral episomal genome maintenance and loss of repression of K-Rta expression (Cai et al., 2013). A recent study also demonstrated that inhibition of the LANA SIM motif by a small molecule Cambogin exerted significant antagonistic effects on KSHV persistent infection and proliferation of KSHV-latently infected cells both in vitro and in vivo (Ding et al., 2019). Notably, LANA1 also exerts E3 SUMO ligase-like activity (Figure 4) by enhancing SUMOylation of histones H2A-H2B in a SIM-dependent manner through recruiting the SUMO-Ubc9 intermediate. This likely contributes to chromatin condensation, repression of viral genes and maintenance of viral latency (Campbell and Izumiya, 2012). LANA1 SUMOylation is controlled by the SUMO isopeptidase SENP6 and LANA1 and SENP6 expression are mutually interdependent (Lin et al., 2017). SUMOylated LANA1 further increases LANA1 expression whereas SENP6-mediated deSUMOylation reduces LANA1 expression. Interestingly, LANA1 itself binds to SENP6 promoter to reduce SENP6 expression. This leads to an increased pool of SUMO-modified LANA1 which further maintains its high expression and promotes viral latency. During de novo infection, overexpressing SENP6 has been shown to impair viral latency owing to diminished LANA protein levels which further led to enhanced viral gene expression (Lin et al., 2017).
Another KSHV protein K-bZIP that belongs to the basic region-leucine zipper family of transcription factors, usurps host cellular SUMOylation machinery to exert its function (Izumiya et al., 2005). K-bZIP acts as a transcriptional repressor of the viral genome and aids in maintaining viral latency. Interestingly, K-bZIP has been shown to interact with Ubc9 and to potentially serve as a viral SUMO E3 ligase (Figure 4). K-bZIP’s proposed SUMO E3 ligase activity was revealed to be specific for SUMO2/3 conjugation, possibly implicating a role in promoting polySUMOylation of targets. K-bZIP is itself modified at K158 both in vitro and in vivo (Chang et al., 2010); but a SUMO-deficient mutant of K-bZIP does not show altered protein stability or localization to PML NBs. Further, SUMOylation of K-bZIP is mostly dispensable for the trans-repression activity. However, trans-repression requires interaction of K-bZIP with the catalytically active Ubc9 (Izumiya et al., 2005). Though detailed molecular explanations are lacking, K-bZIP has been shown to induce SUMO2/3 conjugation of transcription factors at the viral promoters, resulting in repression of viral transcription and establishment of viral latency (Yang et al., 2015a). During KSHV reactivation, enrichment of SUMO2/3 association with the promoter regions of both the viral and cellular genomes has been reported (Chang et al., 2013; Yang et al., 2015a). Though awaiting experimental affirmation, K-bZIP may act as a SUMO conjugating agent at cellular promoters thereby repressing cellular innate immune response (such as expression of ISGs) (Chang et al., 2013). At viral genomes, K-bZIP fosters enhanced association of SUMO2/3 with the viral DNA, thereby providing another check for the virus to skew its life cycle back toward latency before lytic commitment. Infection with a virus harboring a SUMO E3 ligase mutant of K-bZIP does not show this effect and has increased proliferation capabilities. Interestingly, knocking down SUMO2/3 expression phenocopied this effect (Yang et al., 2015a). Of note, K-bZIP also harbors a SIM that binds exclusively to SUMO2/3 (Chang et al., 2010). Direct implications of K-bZIP in usurping PML NB components for SUMOylation to induce KSHV latency, however, are lacking so far.
Kaposi’s sarcoma-associated herpesvirus employs several strategies to target PML NB integrity for inducing viral reactivation and lytic rejuvenation. The first KSHV protein which can antagonize PML NBs during de novo infection is one of the viral tegument proteins ORF75 which specifically targets ATRX for degradation in a proteasome-independent manner and triggers redistribution of Daxx, without hampering other PML NB constituents (Figure 3; Full et al., 2014). PML NBs become lesser in number and appear as irregular-shaped larger structures in cells where KSHV-ORF75 is expressed alone. A mutant KSHV lacking the entire ORF75 gene is replication-deficient in cell culture and cannot promote viral gene expression, though much of it can be attributable to the importance of this KSHV protein in structural integrity of the virus (Full et al., 2014).
A robust SUMO-dependent way that KSHV adopts to disrupt PML NB aggregates during the onset of lytic replication requires the viral protein K-Rta. This KSHV trans-activating protein exerts StUbL activity by targetting SUMOylated proteins for ubiquitylation and proteasomal degradation (Figures 3, 4; Izumiya et al., 2013). K-Rta possesses a RING finger domain and multiple SIMs with affinity toward SUMO2/3 multi-/polymers (Figure 4). SUMOylated PML is one of the primary targets of K-Rta, explaining the disruption of PML NBs by K-Rta (Figure 3). Accordingly, SIM-mutated K-Rta was shown to be incapable of triggering PML NB disruption (Izumiya et al., 2013). Another important target of K-Rta is K-bZIP. K-bZIP acts as a transcriptional repressor of K-Rta, thereby impairing viral transactivation and lytic rejuvenation. Therefore, degradation of K-bZIP by the StUbL activity of K-Rta counteracts the repressive action of K-bZIP. Indeed, SIM-mutated K-Rta showed reduced replication potential and attenuated potency of viral transactivation by K-Rta (Izumiya et al., 2013).
Another KSHV protein which disrupts PML NBs is LANA2 (vIRF-3). LANA2 promotes PML SUMOylation and induces proteolytic ubiquitylation followed by proteasomal demise of SUMO-enriched PML, again leading to dissolution of PML NBs (Figure 3; Marcos-Villar et al., 2009, 2011). Mutation of either the SUMO conjugation site of PML or deletion of SIM within LANA2 significantly impaired PML NB disruption, but the mechanism of this process has remained unclear (Marcos-Villar et al., 2009, 2011). Since LANA2 lacks typical features of a ubiquitin ligase, intrinsic StUbL activity of LANA-2 is unlikely. In addition to the significance of SIM, SUMOylation of LANA2 at multiple K residues are also found to be required for this PML NB disruptive function (Marcos-Villar et al., 2009, 2011). Notably, as LANA2 is a viral latency associated protein, the functional relevance of PML NB disruption by a viral latent protein has not been ascertained experimentally. Altogether, KSHV adopts multiple SUMO-dependent strategies to usurp PML NBs for maintaining latency and further disrupts their structural integrity for rejuvenation and lytic proliferation.
Epstein-Barr Virus (EBV)
Epstein-Barr virus, a gammaherpesvirus with a double stranded DNA genome, can cause B cell lymphoma, nasopharyngeal cancer, Hodgkin’s and non-Hodgkin’s lymphomas, and a subset of gastric cancers. It can either be maintained in a latent state and lead to unrestricted cell proliferation and tumorigenesis or can switch to lytic proliferation, especially in the infected epithelial cells. Interestingly, the SUMO system may be involved in maintenance of EBV latent infection. The primary EBV oncoprotein latent membrane protein 1 (LMP1), which is pivotal in modulating viral quiescence, interacts with Ubc9 leading to SUMOylation of many host proteins (Li and Chang, 2003; Bentz et al., 2011). Two important targets of LMP1-mediated SUMOylation are IRF7 and KAP1 (Bentz et al., 2012, 2015). SUMO-conjugated IRF7 at K452 has increased nuclear localization and stability but decreased transactivation potency and therefore is incapable of mounting a robust innate immune response in cells latently infected with EBV (Bentz et al., 2012). SUMOylation of KAP1 increases its transcriptional repression on EBV early promoters and the lytic origin of replication of the virus (Bentz et al., 2015), thereby decreasing transcription of Zta and Rta (EBV lytic trans-activators) and contributing to viral latency (Bentz et al., 2015). None of the LMP1-directed SUMOylation targets resides within PML NB, but LMP1 increases PML expression and immunofluorescence intensity of PML NBs. Disruption of PML NBs results in EBV lytic replication, suggesting that NB disruption is involved in the switch from latent to lytic infection (Sides et al., 2011a, b).
The EBV tegument protein BNRF1 can target PML NBs by interacting with Daxx and disrupting the formation of the Daxx-ATRX Histone H3.3 chaperone complex (Figure 3). Mechanistically, BNRF1 binds to Daxx and outcompetes binding of Daxx to ATRX. Subsequent failure of Histone H3.3 loading onto the viral chromatin leads to enhancement of viral gene expression during primary infection (Tsai et al., 2011, 2014). In addition to preventing Daxx/ATRX/Histone H3.3-mediated heterochromatinization of the viral genome, BNRF1 also facilitates formation of the stabilized ternary complex with Daxx, and histone variants H3.3/H4 and alters the histone dynamics in favor of expression of viral genes at pre-latency stage (Tsai et al., 2014). This further allows expression of latency associated genes and establishment of latent infection.
Disruption of the integrity of PML NBs is crucial for lytic rejuvenation of EBV (Bell et al., 2000). The EBV protein EBNA1 induces degradation of PML through the ubiquitin-proteasome system (Figure 3; Sivachandran et al., 2008). Degradation involves binding to the deubiquitylase USP7 and the kinase casein kinase 2 (CK2), which promotes phosphorylation and subsequent degradation of PML. Disruption of PML NBs also impairs p53 activation, thereby likely contributing to tumorigenesis (Sivachandran et al., 2008, 2010). Interestingly, PML disruption by EBNA1 occurs in epithelial cells, which promote lytic infection, but has not been observed in B lymphocytes, which are predominantly responsible for latent infection, further supporting the idea that dismantling of NBs predisposes latently infected cells to lytic rejuvenation (Sivachandran et al., 2012). Consistently, depleting PML was shown to trigger EBV lytic cycle and supplementing PML depleted cells with single PML isoform (PML I-VI) suppressed EBV lytic reactivation except in case of PML IV which was revealed to be the most vulnerable PML isoform to EBNA1-mediated degradation (Sivachandran et al., 2012).
An immediate-early protein of EBV Zta (BZLF1), which is involved in lytic replication of the virus and transactivation of EBV early genes, can also disrupt PML NBs even under exogenously expressed condition (Figure 3; Adamson and Kenney, 2001). Zta itself can be SUMOylated at K12 (Adamson and Kenney, 2001; Hagemeier et al., 2010; Murata et al., 2010) and it has been proposed that Zta inhibits SUMO conjugation to PML by competing for the limiting pool of endogenous SUMO. Loss of SUMO-modified PML induces the dissolution of PML NBs and a diffuse nuclear distribution of PML (Figure 3; Adamson and Kenney, 2001). Notably, however, partial disruption of PML NBs was still observed upon expression of a SUMOylation-deficient mutant of Zta, implying other viral regulators to have PML NB disruptive functions (Hagemeier et al., 2010). Consistently, another study revealed overexpression of many EBV-encoded proteins such as BDLF1, EBNA3B, BRLF1, BFLF2, BLLF2, and BZLF1 to have the potency to reduce the average number of PML NBs in infected cells (Salsman et al., 2008). Therefore, EBV has developed multiple mechanisms to disintegrate PML NB integrity which emphasizes the significance of these structures during the lytic cycle of this gammaherpesvirus.
There are also reports of viral protein such as the EBV-encoded protein kinase (EBV-PK), and host regulatory protein such as the cellular scaffolding protein RanBPM to dictate Zta SUMOylation status and therefore to influence Zta-mediated lytic reactivation of EBV (Hagemeier et al., 2010; Yang et al., 2015b). Whether these regulatory aspects of Zta SUMOylation have any direct impact on Zta’s ability to disrupt PML NBs has not been addressed yet.
Therefore, in general, herpesviruses exploit the molecular assembly within PML NBs to heterochromatinize their genome and to go into the latent phase of infection. During rejuvenation, they actively disrupt PML NB integrity by specific lytic proteins to trigger lytic proliferation.
Adenoviruses
Human Adenovirus (HAdV)
In contrast to the early recruitment of herpesviral DNA to PML NBs, adenoviral DNA is associated with PML NBs at a relatively late stage of infection (≥4 h) (Komatsu et al., 2015). Interestingly, this targeting does not require the incoming viral DNA itself but is mediated by the viral single-strand DNA binding protein DBP (also called E2A) which is involved in viral DNA replication (Komatsu et al., 2015). During infection with adenovirus type 5, constituents of PML NBs, including PML and Daxx, are re-organized from their typical punctate appearance to elongated nuclear tracks (Figure 3; Carvalho et al., 1995; Puvion-Dutilleul et al., 1995). Mechanistically, this reorganization involves an interaction of the adenoviral protein E4orf3 with the PML (Figure 3; Doucas et al., 1996; Hoppe et al., 2006). Elegant structural work revealed that E4orf3 forms a multivalent polymer thereby creating avidity-driven interactions with PML and other components of these tracks (Ou et al., 2012). Reorganization of PML NBs fosters viral replication as an E4orf3 mutant that is defective in reorganization is severely growth-compromised. Further, this E4orf3 mutant virus is less potent in antagonizing the IFN-induced antiviral state. Interestingly, however, this phenotype can be rescued upon depletion of PML, underlining the importance of PML in anti-viral defense (Ullman et al., 2007; Ullman and Hearing, 2008). E4orf3-induced nuclear tracks also sequester other antiviral host proteins, such as Mre11 and Nbs1, which are part of the MRN (Mre11-Rad50-Nbs1) complex that mediates the DNA damage response (DDR) and DNA repair (Sohn and Hearing, 2012). The DDR hampers viral DNA replication and HAdV has evolved complementary mechanisms to inhibit the MRN complex. In addition to the sequestration of MRN components within the tracks of infected cell nuclei, the virus mediates ubiquitin-proteasome-dependent degradation of Mre11 through an E3 cullin-RING ligase (CRL) complex assembled by adenoviral proteins E1B-55K in conjunction with E4orf6. Importantly, E4orf3-mediated track sequestration of Mre11 and Nbs1 triggers their SUMOylation potentially facilitating their relocation to cytosolic aggresomes and subsequent degradation (Stracker et al., 2002; Araujo et al., 2005; Liu et al., 2005; Karen et al., 2009; Sohn and Hearing, 2012). Along this line, the transcriptional regulators TIF1γ and TFII-I are also sequestered in PML containing nuclear tracks, where they undergo E4orf3-mediated polySUMOylation and subsequent ubiquitin-proteasom-dependent degradation, most likely through cellular StUbL activity (Bridges et al., 2016; Sohn and Hearing, 2016). Removal of TIF1γ and TFII-I might cause de-repression of specific adenoviral promoters, thereby favoring viral gene transcription. The inactivation of antiviral host factors by virus-induced sequestration or proteasomal degradation is also exemplified by inhibition of Daxx function (Figure 3; Schreiner et al., 2010). In this case it has been proposed that, the adenoviral capsid protein VI interacts with and displaces Daxx from PML NBs to the cytoplasm, thereby de-repressing Daxx-mediated silencing of the viral E1A promoter (Figure 3; Schreiner et al., 2012). Further, SUMO conjugated E1B-55K in conjunction with the cellular StUbL RNF4 was shown to induce proteasomal degradation of Daxx (Endter et al., 2001; Schreiner et al., 2011; Wimmer et al., 2013; Müncheberg et al., 2018). Altogether, these data indicate that reorganization of PML NBs and E4Orf3-mediated formation of tracks in the course of adenovirus infection is a key process in repurposing or inactivating cellular regulators of transcription and DNA repair. Notably, these nuclear tracks also harbor PIAS3 SUMO E3 ligase which might further explain the stimulatory effect of E4orf3 on SUMO conjugation and SUMO chain formation on many host proteins (Higginbotham and O’Shea, 2015; Sohn et al., 2015). Alternatively, E4ORF3 functions as a SUMO E3 ligase and/or E4 chain elongase on selected substrates (Sohn and Hearing, 2016).
Interestingly, the E4orf3-positive tracks reside in close spatial proximity of the adenoviral replication centers (RCs) and a recent report has unveiled the pivotal role of SUMOylation for this positioning (Stubbe et al., 2020). In particular, E2A was found to undergo SUMO conjugation, thereby fostering interaction with both PML and Sp100A and convergence of PML NB tracks with the adenoviral RCs (Figure 3). A SUMOylation-deficient E2A mutant exhibits reduced binding to PML and Sp100A, and accordingly, viral RCs and PML tracks remain separated. Compared to their wild type counterpart these mutant viruses are less efficient in yielding progeny virions (Stubbe et al., 2020). Juxtaposition of viral RCs to PML NBs may promote adenoviral early gene transcription through PML-mediated recruitment of the viral E1A-13S protein (Figure 3; Berscheminski et al., 2013). The compartmentalization of PML NB components within the adenoviral RCs or E4orf3-containing tracks is a very selective process that is, at least partially, regulated by SUMO-dependent mechanisms (Berscheminski et al., 2014). Sp100B, Sp100C, and Sp100HMG are delocalized to the adenoviral RCs possibly as part of a viral strategy to counteract the restrictive functions of these specific Sp100 isoforms on chromatin architecture. Sp100A, on the other hand, is retained around the periphery of the RCs as a part of the non-canonical track-like PML NB structures, where it favors transcriptional activation of the adenoviral genome (Berscheminski et al., 2014). Mechanistically, adenoviral infection triggers loss of SUMO2 conjugated Sp100A leading to reduced association between Sp100A and HP1, explaining chromatin decondensation by Sp100A on adenoviral promoters (Berscheminski et al., 2014).
Apart from E4orf3, E1B-55K also exploits the SUMO system to target host cell proteins. E1B-55K itself is modified by SUMO and the modification facilitates its nuclear retention by blocking a CRM-dependent nuclear export signal (Kindsmüller et al., 2007; Wimmer et al., 2013). Further, SUMOylation of E1B-55K is important for its interaction with PML IV (Figure 3; Wimmer et al., 2010). Interestingly, E1B-55K can also induce SUMOylation of several PML NB-associated host proteins, including p53 and Sp100. Inhibition of p53 activity by E1B-55K is a hallmark of HAdV infection and E1B-55K appears to apply different strategies to inhibit the antiproliferative and pro-apoptotic activity of p53. One strategy is the ubiquitin-proteasome-mediated degradation of p53 by the above-mentioned CRL/E1B-55K/E4orf6 ubiquitin ligase complex (Figure 3; Querido et al., 2001). Intriguingly, E1B-55K also induces SUMOylation of p53 thereby rendering it transcriptionally inactive by facilitating its nuclear export through transient interactions with PML NBs (Figures 3, 4; Muller and Dobner, 2008; Pennella et al., 2010). Whether E1B-55K exerts E3 SUMO ligase activity by its own or stimulates SUMOylation by co-operating with host E3 ligases is a matter of debate (Muller and Dobner, 2008; Pennella et al., 2010). In addition to p53, E1B-55K also triggers SUMOylation of Sp100A and sequesters it within PML containing nuclear tracks (Figures 3, 4), thereby limiting stimulatory effects of Sp100A on transactivation potential of p53 (Berscheminski et al., 2016). Moreover, ATRX is targeted by the CRL/E1B-55K/E4orf6 complex for proteasomal degradation (Figure 3; Schreiner et al., 2013).
The adenoviral protein E1A also has PML NB destabilizing properties (Figure 3). The mechanism involves a direct interaction between the conserved region 2 of E1A and the N-terminal region of Ubc9 which binds SUMO and regulates polySUMOylation (Figures 3, 4; Yousef et al., 2010). Therefore, interaction of Ubc9 with E1A might affect polySUMOylation, which might further destabilize PML NB integrity (Figure 3; Yousef et al., 2010). At least in a polySUMOylation assay in yeast, wild-type E1A, but not a mutant defective in Ubc9 binding, inhibited polySUMOylation. Moreover, the authors also speculated that E1A binding might affect the ability of Ubc9 to be SUMOylated at K14 residue. As SUMO modification at this residue affects Ubc9 substrate specificity, this interaction might also alter SUMOylation of host proteins in general (Yousef et al., 2010).
Therefore, HAdV selectively usurps certain components of PML NB components at the expense of antagonizing others to favor its own perpetuation.
Chicken Embryo Lethal Orphan (CELO) Adenovirus
Chicken embryo lethal orphan adenovirus adopts a robust SUMO-dependent mechanism of PML NB disruption. The viral early protein Gam1 induces ubiquitin-proteasome-dependent degradation of the dimeric SUMO activating enzyme SAE1/SAE2 (Figure 4) by recruiting a cullin-RING E3 ubiquitin ligase (Cul2/5- Rbx1-Elongin B/C) complex (Boggio et al., 2004, 2007). The resulting loss of SUMO conjugation is transduced to the disruption of PML NBs possibly as a result of absence of PML SUMOylation (Colombo et al., 2002; Boggio et al., 2007). As SUMOylation usually leads to transcriptional repression, this global loss of SUMOylation might have a pro-viral significance (Boggio et al., 2004).
Human Papillomavirus (HPV)
Human papillomavirus, belonging to the Papillomaviridae family, possess limited arsenal to counteract host immune responses because of their small genome size and therefore employ a fundamentally different strategy to manipulate with the PML NBs. Unlike other viruses where PML NBs are associated with restrictive chromatin environment and curtailment of viral lytic gene expression and lytic proliferation, HPVs exploit PML NBs for replication. Accordingly, HPV pseudoviruses are more potent in infecting PML expressing cells than PML depleted cells (Day et al., 2004). Consistently, in absence of PML, transcript and genome levels of bovine papillomavirus 1 (BPV1) were greatly reduced. Moreover, depletion of PML in HaCaT cells triggered significant reduction of both viral transcripts and nuclear load of viral genomes. Interestingly, co-inhibition of JAK/STAT signaling restored the loss of viral genomes, but not viral transcription, suggesting possible antiviral implications of IFN-mediated immune response in absence of PML. In consistence with this, PML depleted HeLa cells, which are intrinsically transformed with HPV 18 oncogenes E6 and E7 and therefore might sustain a perturbed JAK/STAT response, were found to be as supportive as their wild type counterpart in sustaining HPV infection (Hong et al., 2011; Reiser et al., 2011; Bienkowska-Haba et al., 2017). Altogether, PML may provide a protective environment for the HPV genome against innate immune recognition and subsequent degradation and also favors HPV transcription. Agreeably, HPV 16 pseudogenomes colocalized with PML protein in both HaCaT and HeLa cells (Bienkowska-Haba et al., 2017).
Unlike other viruses that disrupt PML NBs, HPVs usurp molecular condensates of PML upon their reformation after mitosis. Mitotic entry coincides with PML deSUMOylation and PML NB disintegration. Subsequently, cytoplasmic PML aggregates, called mitotic accumulations of PML proteins (MAPPs), are formed by homo-multimerization of PML proteins through the RBCC/TRIM motif. These cytosolic PML condensates which even persist in the cytosol after completion of mitosis and nuclear envelope reformation, lack typical PML NB components. When PML translocates back to the daughter cell nuclei, canonical nuclear PML NBs comprising of SUMOylated PML and NB clients are reassembled (Everett et al., 1999; Dellaire et al., 2006). Notably, the HPV genome which remains within an uncharacterized vesicular compartment along with the major viral capsid protein L1 requires disassembly of the nuclear membrane during mitosis to get delivered within the nucleus (Pyeon et al., 2009; Aydin et al., 2014; DiGiuseppe et al., 2016; DiGiuseppe et al., 2017). Another capsid protein L2 has a transmembrane localization and the extra-membrane domain of L2 mediates transport of the genome-containing vesicle along mitotic microtubules toward the condensed chromosomes (DiGiuseppe et al., 2016; Aydin et al., 2017; Day et al., 2019). The viral genome remains in the transport vesicle shortly after completion of mitosis and nuclear envelope reformation (DiGiuseppe et al., 2016; Day et al., 2019). HPV has been shown to interact with PML NBs through L2 after nuclear delivery as part of the viral life cycle (Figure 3). L2 of BPV 1 was initially identified to localize to PML NBs (Day et al., 1998). Subsequent studies also detected HPV 33 L2 protein at PML NBs and reported displacement of Sp100, but not of PML, from these structures (Florin et al., 2002; Becker et al., 2003). Moreover, HPV 16 L2 protein and BPV 1 pseudogenomes were also found to localize to PML NBs (Day et al., 2004). In PML depleted cells, L2 protein shows diffused pattern in the nucleus.
The mechanism of how HPV L2 protein fosters association of the HPV genome with PML NBs is still poorly understood, but SUMO-dependent regulation appears to be involved in this process. L2 protein itself contains one SUMO conjugation motif at K35 which was evidenced to modulate viral capsid assembly (Marusic et al., 2010), but its contribution to interact with PML has not been investigated yet. L2 also has one highly conserved SIM and two additional putative SIMs (Bund et al., 2014). The conserved SIM has been experimentally shown to be important for interaction with PML as mutation of the SIM prevented localization to PML NBs and reduced infectivity independent of the L2 SUMOylation status. A recent report shed light on the nature of this recruitment where PML protein was found to get recruited to the viral genomes and assemble around them, rather than the viral genomes targeting preformed PML NBs (Guion et al., 2019). This engulfment temporally coincides with post-mitotic early interphase while PML protein is translocating back to the nucleus. Spatially, too, this entrapment is unique as PML protein co-localizes with viral genomes that are still residing within transport vesicles. This observation suggests that viral genomes are continuously protected from cellular sensors, first by the transport vesicle, then by PML protein which may provide a protective environment for the genome in the nucleus of host cells to prevent innate immune recognition (Guion et al., 2019). It seemed also plausible that HPV usurps the dissociation of PML NB during mitosis to modify the composition of PML NB rather than reorganizing preassembled structures (Guion et al., 2019; Guion and Sapp, 2020).
Interestingly, unlike PML, Sp100 has been observed to act as a restriction factor for HPV transcription and replication but to be unimportant for the maintenance phase of the HPV life cycle (Stepp et al., 2013, 2017). Consistently, viral transcription and replication from transfected HPV 18 genome in primary human keratinocytes were greatly enhanced under the condition of Sp100 depletion (Stepp et al., 2013). Also in line with this is the observation that recruitment of Sp100 to the HPV genome occurs at late interphase and in a temporally delayed manner compared to PML recruitment (Guion et al., 2019). Furthermore, Sp100 was only observed to co-localize with viral genomes after the loss of the transport vesicles and only when PML protein had already been recruited. A justification has been put forward that the delayed recruitment of Sp100 perhaps allows a window where a burst of initial viral transcription ensues in absence of Sp100 before Sp100-mediated repression of viral transcription sets in to promote the skewing of viral life cycle toward the maintenance phase (Guion et al., 2019). Of note, knocking down Daxx expression did not have an impact on the HPV18 transcription and replication implying dispensability of Daxx in this process (Stepp et al., 2013).
RNA Viruses
Picornaviruses
Encephalomyocarditis Virus (EMCV)
Though not as elaborately studied as DNA viruses, there are examples of RNA viruses interacting with the host cellular SUMOylation machinery and PML NBs. One classical example includes anti-PML activities of encephalomyocarditis virus (EMCV) protease 3C. EMCV which belongs to the family Picornaviridae can cause neurological and reproductive complicacies in many mammals, likely including humans through zoonosis. Interestingly, primary fibroblasts derived from PML knockout mice are more vulnerable to infection with EMCV, suggesting that the PML protein might act as a restriction factor for EMCV replication (El McHichi et al., 2010). Moreover, progressive infection with EMCV was found to cause a reduction in PML protein levels both in IFN-treated cells and in PML III-expressing cells. The degradation event was preceded by translocation of PML from nucleoplasm to the nuclear matrix and SUMO1, SUMO2/3 conjugation to PML, leading to an increase in PML NB size. Subsequently, the viral protease 3C was found to localize to the PML NBs along with proteasome components to trigger PML degradation in a proteasome- and SUMO-dependent manner (Figure 3); PML SIM, however, proved to be dispensable for the degradation (El McHichi et al., 2010). Unlike PML III which is antagonized by EMCV protease 3C, SUMO-conjugated PML isoform IV has been reported to interact with the EMCV 3D polymerase via the isoform’s unique C-terminal domain and to sequester the viral protein to PML NBs (Figure 3), thereby impairing viral perpetuation (Maroui et al., 2011). Consistently, selective depletion of PML IV also undermines the anti-EMCV potency of IFN (Maroui et al., 2011).
Poliovirus
SUMOylation of PML has been reported to have indirect effects on the life cycle of poliovirus, another Picornaviridae family member etiologically responsible for paralytic poliomyelitis. The mechanism includes virus-mediated activation of the extracellular-signal-regulated kinase (ERK), which phosphorylates PML, thereby facilitating PML SUMOylation and translocation to the nuclear matrix. Subsequently, p53 gets recruited to PML NBs and becomes phosphorylated on Ser15 in a PML-dependent way (Figure 3; Pampin et al., 2006). Therefore, poliovirus infection results in PML-dependent p53 activation and mobilization of apoptotic cascade downstream of activated p53, resulting in inhibition of viral replication. PML III overexpression conferred resistance to poliovirus in p53 wild-type cells, but not in p53-inactive cells, while depletion of PML III abolishes p53-dependent apoptosis and antiviral effects. Interestingly, as a countermeasure against this transient cyto-protective effects, poliovirus induces degradation of p53 through proteasome via the E3 ubiquitin ligase Mdm2 (Pampin et al., 2006).
Rhabdoviruses
Vesicular Stomatitis Virus (VSV)
Antiviral effects of PML have also been extended to VSV, a member of the Rhabdoviridae family which infects human zoonotically, as exogenous PML III conferred resistance to VSV by inhibiting viral mRNA and protein synthesis (Chelbi-Alix et al., 1998), while PML knockout mice were found to be more susceptible to VSV infection than wild type mice (Bonilla et al., 2002). Disruption of PML NBs, however, has not been observed in VSV infected cells. The mechanism of how PML exerts antagonistic activity against a virus, whose replication takes place entirely in the cytoplasm and which does not disrupt PML NBs, is yet to be addressed. The observation that only the coiled-coil domain deletion mutant of PML, but not the other RING finger PML mutant and cytoplasmic PML III mutant, fails to show the antiviral activity, emphasizes the importance of the coiled-coil region of PML for anti-VSV effects (Chelbi-Alix et al., 1998). Another report has also elucidated PML IV to have anti-VSV activity (El Asmi et al., 2014). Mechanistic profiling elucidated PML IV to potentiate both the intrinsic and innate immune arms of the antiviral defense pathway. The innate immune response involves SUMOylated PML IV to interact with and recruit peptidyl-prolyl cis/trans isomerase (Pin1) to PML NBs. This results in activated IRF3 to escape from Pin1-mediated degradation and to potentiate its canonical functions of IFN-β synthesis. Interestingly, though depletion of IFR3 abrogated PML IV-induced IFN synthesis, it could not entirely inhibit PML IV-induced inhibition of viral proteins, suggesting parallel IFN-dependent and independent pathways to operate downstream of PML IV. Moreover, the agonistic effects of SUMOylated PML IV to stimulate IFN-β synthesis was found to be a broader effect on innate immune regulation rather than a VSV infection-specific scenario (El Asmi et al., 2014).
Rabies Virus
Unlike VSV, another Rhabdoviridae family member rabies virus which causes central nervous system dysfunctions does reorganize PML NBs during infection to form larger and more electron-dense aggregates. The P3 protein, an amino-terminally truncated product of the viral P mRNA which also codes for phosphoprotein P, causes this effect. Interestingly, sole expression of P delocalizes PML III from nuclear to cytoplasmic puncta where both proteins colocalize (Figure 3). A direct interaction was also evidenced between the C-terminal domain of P protein and the PML RING finger. However, the observations that overexpression of PML III isoform does not perturb rabies infection but that PML–/– MEFs are more susceptible to rabies infection than wild-type MEFs, do suggest involvement of other PML isoforms to take part in the anti-rabies defense (Blondel et al., 2002). Indeed, subsequent studies revealed the antiviral importance of PML IV, but not of other nuclear as well as cytoplasmic PML isoforms, to act as a restriction factor against rabies infection independent of IFN signaling axis. Notably, PML IV SUMO conjugation was found to be essential for its antiviral effect as the protective effect of PML IV was lost when the SUMOylation sites were mutated (Blondel et al., 2010).
Arenavirus
Lymphocytic Choriomeningitis Virus (LCMV)
The RING finger protein Z of LCMV, an Arenaviridae family member which transmits zoonotically to human leading to aseptic meningitis, induces redistribution of PML from nuclear condensates to cytoplasmic aggregates during infection (Figure 3). In these aggregates, both proteins interact with the translation initiation factor eukaryotic initiation factor 4E (eIF-4E) and reduce eIF-4E’s affinity for the 5′ mRNA cap structure, thereby inhibiting cellular translation (Borden et al., 1998; Kentsis et al., 2001). Subsequently, PML has been shown to act as a restriction factor during infection with LCMV as PML knockout mice showed more susceptibility to LCMV infection than their wild type counterparts (Bonilla et al., 2002). Consistently, anti-LCMV potency of IFN was also found to be partially sensitive to PML knocking out in MEFs, suggesting PML to be one of the IFN-induced antiviral mediators for defense against LCMV (Djavani et al., 2001). However, neither overexpression of PML III nor Sp100 had any repressive effect on LCMV growth (Asper et al., 2004), implicating PML isoforms other than PML III to have anti-LCMV relevance.
Orthomyxovirus
Influenza Virus (IFV)
Like other RNA viruses, PML exerts antagonistic effects during infection with the Orthomyxoviridae family member influenza A virus in a viral strain-specific manner (Li et al., 2009). Overexpression of different PML isoforms such as PML III, PML IV, and PML VI attenuates influenza A viral propagation in cell culture (Chelbi-Alix et al., 1998; Iki et al., 2005; Li et al., 2009), while PML silencing has an agonistic effect (Iki et al., 2005; Li et al., 2009). However, neither the mechanism by which PML exerts anti-influenza activity nor the significance of PML NB recruitment of viral proteins (the matrix protein M1 and the nonstructural polypeptides NS1 and NS2) (Iki et al., 2005) has been addressed yet. A more recent study suggested induction of a specific host cellular SUMOylation response in influenza virus infected cells. However, PML SUMOylation was insensitive to this infection. Interestingly, confocal imaging showed PML-stained puncta to get smaller and more in number whereas Sp100 and Daxx appeared dispersed in the nuclear compartment of infected cells, suggestive of PML NB dispersion (Domingues et al., 2015).
Flaviviruses
Hepatitis C Virus (HCV)
Interplay of HCV, a member of the family Flaviviridae, with the host cellular PML NBs involves the HCV core protein to get recruited to PML NBs by interacting with PML IV and to inhibit the co-activator activity of PML IV on p53 (Figure 3). This leads to prevention of p53-dependent apoptotic induction and development of HCV-associated hepatocellular carcinoma (Herzer et al., 2005). However, SUMO-dependent regulation of PML NB structures has not yet been reported for HCV.
Dengue Virus (DENV)
Promyelocytic leukemia has recently been identified as an antiviral host cellular determinant capable of restricting all serotypes of DENV, another Flaviviridae family member, as siRNA-mediated depletion of PML resulted in increased virus production (Giovannoni et al., 2015, 2019). Moreover, as a part of viral countermeasures, PML NBs were found to get significantly reduced in number during infection with DENV serotypes. The causative agent was identified to be the non-structural viral protein NS5 which co-localized with PML III and IV at PML NBs and promoted accelerated degradation of PML III and IV (Figure 3; Giovannoni et al., 2019). Consistently, over expression of these specific PML isoforms curtailed DENV replication and progeny yield. Of note, Sp100 and Daxx were proved to be dispensable for DENV progeny production, suggesting specific anti-DENV implications of PML III and IV (Giovannoni et al., 2019). Interestingly, DENV NS5 interacts with Ubc9 and undergoes SUMOylation. This SUMOylation requires a SIM domain within NS5 (Su et al., 2016). Any correlation of SUMO conjugation of NS5 on its PML NB disruptive property, however, is yet to be addressed experimentally.
Retroviruses
Human Immunodeficiency Virus-1 (HIV-1)
During infection with HIV-1, a lentivirus belonging to the family Retroviridae, PML NBs rapidly relocalize from nuclear to cytoplasmic aggregates (Figure 3). Further, PML depletion increased reverse transcription capacity of HIV-1. However, involvement of PML is likely indirect through stabilization of Daxx which sensitized the reverse transcription of HIV-1 RNA by spatially relocating to incoming viral capsids in the cytoplasm. Similar relocation of PML NB components and Daxx-dependent antagonistic impact of PML were also observed in other retroviruses such as simian immunodeficiency virus (SIVmac), murine leukemia virus Moloney and equine infectious anemia virus. Interestingly, SUMOylation inhibitor ginkgolic acid (that impairs the formation of the E1-SUMO intermediates) strongly sensitized formation of cytoplasmic PML-containing aggregates in HIV-1 infected cells, suggesting importance of SUMO conjugation for structural integrity of these cytoplasmic aggregates. However, data based on ginkgolic acid treatment need to be interpreted with a note of caution since it is a rather unspecific SUMO inhibitor. Further, no other specific SUMO target specifically directing this delocalization has been assigned (Dutrieux et al., 2015). An interesting recent report also elucidated another SUMO-dependent PML-NB disruptive strategy by the HIV-1 accessory protein Vpu. In brief, Vpu interacts with RanBP2 at nuclear pore to inhibit the SUMO E3 ligase activity of the RanBP2–RanGAP1∗SUMO1–Ubc9 complex, thereby affecting the PML NB integrity (Figure 3) and SUMOylation of DNA repair proteins BLM, Rad52. As a consequence, Rad52-dependent homologous repair and circularization of non-integrated linear DNA from a superinfecting virus were prevented. Moreover, BLM-dependent nucleolytic attack on those susceptible DNA ends were facilitated (Volcic et al., 2020). On a different note, PML aggregation was observed in syncytia present in the brain or lymph nodes of infected patients or in syncytia elicited by the envelope glycoprotein complex of HIV-1 in vitro (Perfettini et al., 2009). PML was shown to foster phosphorylation of ATM which was co-recruited with PML in nuclear aggregates and mobilized p53-dependent syncytial apoptosis in HIV-infected CD4 cells (Perfettini et al., 2009). However, any mechanistic role of SUMO-mediated regulation has not been addressed in this respect.
Human Foamy Virus (HFV)
Human foamy virus are retroviruses infecting non-human primates but can be transmitted to human through zoonotic infection. Though endogenous PML has been shown not to interfere with HFV replication and latency and also not to get delocalized during infection (Regad et al., 2001; Meiering and Linial, 2003), PMLIII overexpression reportedly attenuates HFV transcription by interacting with the HFV transactivator protein Tas and hindering its binding to DNA (long-terminal repeat and internal promoter) (Regad et al., 2001). However, this interaction was revealed to be independent of SUMO regulation. Anti-viral potency of IFN is also diminished in PML deficient MEFs but other IFN-dependent antiviral effectors such as MxA, Mx1, and PKR could not sensitize HFV replication (Regad et al., 2001), again bolstering the specificity of PML in creating an IFN-induced antiviral state against HFV.
Bacterial Pathogens
Alteration in the SUMOylation landscape is also reported in response to bacterial infection, indicating a broader implication of SUMO proteins in controlling infection and immunity. PML knockout mice have been found to be more sensitive to infection by Listeria monocytogenes, a Gram-positive bacterium that is responsible for the foodborne disease listeriosis (Lunardi et al., 2011). A subsequent study revealed that SUMOylation status of PML largely serves as a sensor of Listeria infection (Ribet et al., 2017). The pore-forming toxin listeriolysin O (LLO) was found to be the determining factor for such response (Ribet et al., 2017). One of the multi-faceted signaling events that ensues upon LLO-mediated perforation of the infected host cell membrane is the proteasome-independent degradation of Ubc9 (Figures 3, 4; Ribet et al., 2010). Impairment of de novo SUMOylation coupled to unrestricted activity of SUMO proteases results in massively deSUMOylated population of PML (Figure 3) which gets multimerized and associated with the nuclear matrix in response to infection-induced oxidative stress. This PML deSUMOylation event has been shown to dampen bacterial multiplication. Functionally potent HCMV IE1 protein which is capable of triggering PML deSUMOylation was shown to substitute LLO action and reduced bacterial replication efficiency. Notably, in addition to PML, Sp100 deSUMOylation was also distinct in presence of LLO (Ribet et al., 2017). Antibacterial activity of PML can also be attributable to a number of immunity and cytokine genes that are transcriptionally regulated by PML independent of LLO (Ribet et al., 2017). In contrast to PML, deSUMOylation of many cellular factors, especially transcription factors, has been shown to be beneficial for the bacteria by favoring bacterial replication or survival in the host cells (Impens et al., 2014). One of such candidates is the SUMO-stabilized SMAD4 which acts as a transactivator during TGFβ signaling. LLO was found to destabilize SMAD4 through deSUMOylation leading to impairment of the cyto-protective TGFβ signaling (Ribet et al., 2010). These results suggest a finely tuned SUMO response that occurs during Listeria infection and controls the bacterial pathogenesis. Of note, similar to LLO, other bacterial pore-forming toxins [perfringolysin O (PFO; from Clostridium perfringens) and pneumolysin (PLY; from Streptococcus pneumoniae)] were also evidenced to induce PML deSUMOylation and multimerization (Figure 3; Ribet et al., 2017). Moreover, in line with the observation that membrane association and the pore-forming activity of LLO are essential for Ubc9 degradation, PFO and PLY show a comparable reduction in cellular levels of Ubc9 (Figures 3, 4; Ribet et al., 2010). Strikingly, however, PML was found to have no role in controlling infection by Salmonella typhimurium, a bacterium that is responsible for gastroenteritis in humans, even though this pathogen was also shown to induce a decrease in Ubc9 level leading to inhibition of host cellular SUMOylation (Figure 4; Decque et al., 2016; Ribet et al., 2017). The reduction in Ubc9 level is due to induction of two microRNAs, miR30c and miR30e, which specifically target Ubc9 within infected host cells (Verma et al., 2015). In case of Shigella flexneri, which is another gastroenteritis-causing bacterium, infection is associated with an influx of calcium into the host cell. This ion flux activates the host calpain proteases which cleave SAE2, one of the two components of the E1 SUMO enzyme (Figure 4; Lapaquette et al., 2017). The resulting inhibition of SUMOylation is associated with increased Shigella entry (Fritah et al., 2014; Lapaquette et al., 2017). Moreover, another mechanism of global impairment of SUMOylated proteins (both SUMO1 and SUMO2/3 conjugated) in Shigella infected cells was revealed to be mediated by the bacterial type 3 Secretion System (T3SS) which triggered a proteasome-dependent Ubc9 destabilization (Figures 3, 4). Whether proteasomal degradation is preceded by proteolytic ubiquitination is, however, not known (Sidik et al., 2015). Not surprisingly, therefore, overexpression of components of the SUMO pathway significantly decreased the ability of Shigella to enter cultured epithelial cells. Corroborative in vivo data in an infection model of haploinsufficient Ubc9+/– mice also showed profound destruction of colonic tissue, increased intestinal permeability, and increased signatures of pro-inflammatory cytokines in comparison to wild type controls, suggesting SUMO response to curtail Shigella infection at multiple steps (Fritah et al., 2014). Indeed, quantitative proteomic studies revealed Shigella infection to be associated with the SUMO-2 modification of multiple transcriptional regulators which control inflammatory response (Fritah et al., 2014).
Notably, the number of PML NBs increases during infection with Shigella (Sidik et al., 2015). However, this increase is not accompanied by the enhanced expression of endogenous PML, suggesting the occurrence of PML NB fission or de novo PML body nucleation from soluble PML. Shigella infection also triggered alteration in the SUMO1 localization and abundance. In cells infected with wild-type Shigella, SUMO1 was visualized as more condensed puncta with sharper edges and brighter staining in comparison to cells that were uninfected or infected with non-invasive Shigella. However, no correlation was found between PML body number and the condensed SUMO1 phenotype (Sidik et al., 2015). Thus, many extracellular pathogens presumably target the SUMO modification system to manipulate with the PML NBs, thereby arguing for an evolutionarily conserved strategy in some pathogenic bacteria to increase infectivity by restricting SUMO-dependent host cell responses.
Conclusion and Perspectives
Promyelocytic leukemia NBs have been emerging to be sub-nuclear niches where SUMO-SIM interactions guide the assembly of a heterogeneous macromolecular complex. The formation involves liquid-liquid phase separation in matrix-associated spherical condensates but allows a dynamic equilibrium with the surrounding nucleoplasmic contents. With the growing list of client proteins which are shown to be transiently associated with PML NBs (Van Damme et al., 2010), functional implications of these sub-nuclear foci are expanding. PML NBs have been traditionally regarded as hubs of intrinsic immune defenses against invading pathogens. In light of some current discoveries, however, a new dimension has emerged indicating components of PML NBs to be differentially regulated during infection with some pathogens such as HAdV and HPV. Consistently, certain PML NB components have been revealed to have anti-pathogenic functions whereas others have shown to foster progression of infection for the same pathogen. This reiterates the classical pathogen-mediated host cellular reprogramming where host determinants are more often the specific targets of manipulation rather than stochastic off targets. From a different perspective, PML NB-mediated repression of gene expression has been proved to be pivotal for triggering and/or maintaining latency in many viral infections, suggesting pro-viral implications of these host determinants in establishing prolonged viral persistence. Of note, many viral proteins which play critical roles in dictating PML NB physiology are now emerging to be the functional (and sometimes, even structural) counterparts of the cellular SUMO machinery such as SUMO E3 ligases, SENPs or StUbLs, suggesting implications of the host cellular SUMO modulations on viral evolutionary dynamics. With the recent advances and refinements at the technological frontier, future studies will likely unveil many uncharacterized SUMO-dependent PML NB regulations in the context of pathogen infected cells. Apart from their obvious scholarly significance, these endeavors will also propel research focused on therapeutic importance. Indeed, Cambogin, a targeted small molecule inhibiting the SIM within KSHV LANA protein has been proved to have anti-KSHV function at sub-cytotoxic dose (Ding et al., 2019). Moreover, in coherence with the agonistic effects of PML in governing innate immune response, novel SUMO-dependent intersections between PML NBs and pathogenic organisms will be of significant relevance in future.
Author Contributions
UP and SM wrote the manuscript. UP prepared the figures. Both authors approved the final version of the manuscript.
Funding
This work was funded by the Cluster project ENABLE funded by the State of Hesse.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank all members of our group for constructive feedback and discussions. The contribution of BioRender (through paid subscription) is also acknowledged for preparing the figures used in the manuscript. We also acknowledge the contribution of researchers whose works are relevant yet exempted from this review because of space constraint.
References
Adamson, A. L., and Kenney, S. (2001). Epstein-barr virus immediate-early protein BZLF1 is SUMO-1 modified and disrupts promyelocytic leukemia bodies. J. Virol. 75, 2388–2399. doi: 10.1128/JVI.75.5.2388-2399.2001
Adler, M., Tavalai, N., Müller, R., and Stamminger, T. (2011). Human cytomegalovirus immediate-early gene expression is restricted by the nuclear domain 10 component Sp100. J. Gen. Virol. 92(Pt 7), 1532–1538. doi: 10.1099/vir.0.030981-0
Ahn, J. H., Brignole, E. J. III, and Hayward, G. S. (1998). Disruption of PML subnuclear domains by the acidic IE1 protein of human cytomegalovirus is mediated through interaction with PML and may modulate a RING finger-dependent cryptic transactivator function of PML. Mol. Cell. Biol. 18, 4899–4913. doi: 10.1128/mcb.18.8.4899
Alandijany, T., Roberts, A., Conn, K. L., Loney, C., McFarlane, S., Orr, A., et al. (2018). Distinct temporal roles for the promyelocytic leukaemia (PML) protein in the sequential regulation of intracellular host immunity to HSV-1 infection. PLoS Pathog. 14:e1006769. doi: 10.1371/journal.ppat.1006769
Albright, E. R., and Kalejta, R. F. (2016). Canonical and variant forms of histone H3 are deposited onto the human cytomegalovirus genome during lytic and latent infections. J. Virol. 90, 10309–10320. doi: 10.1128/JVI.01220-16
Araujo, F. D., Stracker, T. H., Carson, C. T., Lee, D. V., and Weitzman, M. D. (2005). Adenovirus type 5 E4orf3 protein targets the Mre11 complex to cytoplasmic aggresomes. J. Virol. 79, 11382–11391. doi: 10.1128/JVI.79.17.11382-11391.2005
Asper, M., Sternsdorf, T., Hass, M., Drosten, C., Rhode, A., Schmitz, H., et al. (2004). Inhibition of different Lassa virus strains by alpha and gamma interferons and comparison with a less pathogenic arenavirus. J. Virol. 78, 3162–3169. doi: 10.1128/jvi.78.6.3162-3169.2004
Aydin, I., Villalonga-Planells, R., Greune, L., Bronnimann, M. P., Calton, C. M., Becker, M., et al. (2017). A central region in the minor capsid protein of papillomaviruses facilitates viral genome tethering and membrane penetration for mitotic nuclear entry. PLoS Pathog. 13:e1006308. doi: 10.1371/journal.ppat.1006308
Aydin, I., Weber, S., Snijder, B., Samperio Ventayol, P., Kühbacher, A., Becker, M., et al. (2014). Large scale RNAi reveals the requirement of nuclear envelope breakdown for nuclear import of human papillomaviruses. PLoS Pathog. 10:e1004162. doi: 10.1371/journal.ppat.1004162
Banani, S. F., Rice, A. M., Peeples, W. B., Lin, Y., Jain, S., Parker, R., et al. (2016). Compositional control of phase-separated cellular bodies. Cell 166, 651–663. doi: 10.1016/j.cell.2016.06.010
Becker, K. A., Florin, L., Sapp, C., and Sapp, M. (2003). Dissection of human papillomavirus type 33 L2 domains involved in nuclear domains (ND) 10 homing and reorganization. Virology 314, 161–167. doi: 10.1016/s0042-6822(03)00447-1
Bego, M., Maciejewski, J., Khaiboullina, S., Pari, G., and St Jeor, S. (2005). Characterization of an antisense transcript spanning the UL81-82 locus of human cytomegalovirus. J. Virol. 79, 11022–11034. doi: 10.1128/JVI.79.17.11022-11034.2005
Bell, P., Lieberman, P. M., and Maul, G. G. (2000). Lytic but not latent replication of epstein-barr virus is associated with PML and induces sequential release of nuclear domain 10 proteins. J. Virol. 74, 11800–11810. doi: 10.1128/jvi.74.24.11800-11810.2000
Bentz, G. L., Moss, C. R. II, Whitehurst, C. B., Moody, C. A., and Pagano, J. S. (2015). LMP1-induced sumoylation influences the maintenance of epstein-barr virus latency through KAP1. J. Virol. 89, 7465–7477. doi: 10.1128/JVI.00711-15
Bentz, G. L., Shackelford, J., and Pagano, J. S. (2012). Epstein-Barr virus latent membrane protein 1 regulates the function of interferon regulatory factor 7 by inducing its sumoylation. J. Virol. 86, 12251–12261. doi: 10.1128/JVI.01407-12
Bentz, G. L., Whitehurst, C. B., and Pagano, J. S. (2011). Epstein-Barr virus latent membrane protein 1 (LMP1) C-terminal-activating region 3 contributes to LMP1-mediated cellular migration via its interaction with Ubc9. J. Virol. 85, 10144–10153. doi: 10.1128/JVI.05035-11
Bernardi, R., Papa, A., and Pandolfi, P. P. (2008). Regulation of apoptosis by PML and the PML NBs. Oncogene 27, 6299–6312. doi: 10.1038/onc.2008.305
Berscheminski, J., Brun, J., Speiseder, T., Wimmer, P., Ip, W. H., Terzic, M., et al. (2016). Sp100A is a tumor suppressor that activates p53-dependent transcription and counteracts E1A/E1B-55K-mediated transformation. Oncogene 35, 3178–3189. doi: 10.1038/onc.2015.378
Berscheminski, J., Groitl, P., Dobner, T., Wimmer, P., and Schreiner, S. (2013). The adenoviral oncogene E1A-13S interacts with a specific isoform of the tumor suppressor PML to enhance viral transcription. J. Virol. 87, 965–977. doi: 10.1128/JVI.02023-12
Berscheminski, J., Wimmer, P., Brun, J., Ip, W. H., Groitl, P., Horlacher, T., et al. (2014). Sp100 isoform-specific regulation of human adenovirus 5 gene expression. J. Virol. 88, 6076–6092. doi: 10.1128/JVI.00469-14
Bienkowska-Haba, M., Luszczek, W., Keiffer, T. R., Guion, L., DiGiuseppe, S., Scott, R. S., et al. (2017). Incoming human papillomavirus 16 genome is lost in PML protein-deficient HaCaT keratinocytes. Cell. Microbiol. 19:e12708. doi: 10.1111/cmi.12708
Blondel, D., Kheddache, S., Lahaye, X., Dianoux, L., and Chelbi-Alix, M. K. (2010). Resistance to rabies virus infection conferred by the PMLIV isoform. J. Virol. 84, 10719–10726. doi: 10.1128/JVI.01286-10
Blondel, D., Regad, T., Poisson, N., Pavie, B., Harper, F., Pandolfi, P. P., et al. (2002). Rabies virus P and small P products interact directly with PML and reorganize PML nuclear bodies. Oncogene 21, 7957–7970. doi: 10.1038/sj.onc.1205931
Boggio, R., Colombo, R., Hay, R. T., Draetta, G. F., and Chiocca, S. (2004). A mechanism for inhibiting the SUMO pathway. Mol. Cell 16, 549–561. doi: 10.1016/j.molcel.2004.11.007
Boggio, R., Passafaro, A., and Chiocca, S. (2007). Targeting SUMO E1 to ubiquitin ligases: a viral strategy to counteract sumoylation. J. Biol. Chem. 282, 15376–15382. doi: 10.1074/jbc.M700889200
Bohren, K. M., Nadkarni, V., Song, J. H., Gabbay, K. H., and Owerbach, D. (2004). A M55V polymorphism in a novel SUMO gene (SUMO-4) differentially activates heat shock transcription factors and is associated with susceptibility to type I diabetes mellitus. J. Biol. Chem. 279, 27233–27238. doi: 10.1074/jbc.M402273200
Bonilla, W. V., Pinschewer, D. D., Klenerman, P., Rousson, V., Gaboli, M., Pandolfi, P. P., et al. (2002). Effects of promyelocytic leukemia protein on virus-host balance. J. Virol. 76, 3810–3818. doi: 10.1128/jvi.76.8.3810-3818.2002
Borden, K. L., Campbell Dwyer, E. J., and Salvato, M. S. (1998). An arenavirus RING (zinc-binding) protein binds the oncoprotein promyelocyte leukemia protein (PML) and relocates PML nuclear bodies to the cytoplasm. J. Virol. 72, 758–766. doi: 10.1128/JVI.72.1.758-766.1998
Boutell, C., Cuchet-Lourenço, D., Vanni, E., Orr, A., Glass, M., McFarlane, S., et al. (2011). A viral ubiquitin ligase has substrate preferential SUMO targeted ubiquitin ligase activity that counteracts intrinsic antiviral defence. PLoS Pathog. 7:e1002245. doi: 10.1371/journal.ppat.1002245
Boutell, C., Everett, R., Hilliard, J., Schaffer, P., Orr, A., and Davido, D. (2008). Herpes simplex virus type 1 ICP0 phosphorylation mutants impair the E3 ubiquitin ligase activity of ICP0 in a cell type-dependent manner. J. Virol. 82, 10647–10656. doi: 10.1128/JVI.01063-08
Boutell, C., Orr, A., and Everett, R. D. (2003). PML residue lysine 160 is required for the degradation of PML induced by herpes simplex virus type 1 regulatory protein ICP0. J. Virol. 77, 8686–8694. doi: 10.1128/jvi.77.16.8686-8694.2003
Bresnahan, W. A., and Shenk, T. E. (2000). UL82 virion protein activates expression of immediate early viral genes in human cytomegalovirus-infected cells. Proc. Natl. Acad. Sci. U.S.A. 97, 14506–14511. doi: 10.1073/pnas.97.26.14506
Bridges, R. G., Sohn, S. Y., Wright, J., Leppard, K. N., and Hearing, P. (2016). The adenovirus E4-ORF3 protein stimulates SUMOylation of general transcription factor TFII-I to direct proteasomal degradation. mBio 7:e02184-15. doi: 10.1128/mBio.02184-15
Brown, J. R., Conn, K. L., Wasson, P., Charman, M., Tong, L., Grant, K., et al. (2016). SUMO ligase protein inhibitor of activated STAT1 (PIAS1) is a constituent promyelocytic leukemia nuclear body protein that contributes to the intrinsic antiviral immune response to herpes simplex virus 1. J. Virol. 90, 5939–5952. doi: 10.1128/JVI.00426-16
Bund, T., Spoden, G. A., Koynov, K., Hellmann, N., Boukhallouk, F., Arnold, P., et al. (2014). An L2 SUMO interacting motif is important for PML localization and infection of human papillomavirus type 16. Cell. Microbiol. 16, 1179–1200. doi: 10.1111/cmi.12271
Burleigh, K., Maltbaek, J. H., Cambier, S., Green, R., Gale, M. Jr., James, R. C., et al. (2020). Human DNA-PK activates a STING-independent DNA sensing pathway. Sci. Immunol. 5:eaba4219. doi: 10.1126/sciimmunol.aba4219
Cabral, J. M., Oh, H. S., and Knipe, D. M. (2018). ATRX promotes maintenance of herpes simplex virus heterochromatin during chromatin stress. eLife 7:e40228. doi: 10.7554/eLife.40228
Cai, Q., Cai, S., Zhu, C., Verma, S. C., Choi, J. Y., and Robertson, E. S. (2013). A unique SUMO-2-interacting motif within LANA is essential for KSHV latency. PLoS Pathog. 9:e1003750. doi: 10.1371/journal.ppat.1003750
Cai, W., Astor, T. L., Liptak, L. M., Cho, C., Coen, D. M., and Schaffer, P. A. (1993). The herpes simplex virus type 1 regulatory protein ICP0 enhances virus replication during acute infection and reactivation from latency. J. Virol. 67, 7501–7512. doi: 10.1128/JVI.67.12.7501-7512.1993
Campbell, M., and Izumiya, Y. (2012). Post-translational modifications of Kaposi’s sarcoma-associated herpesvirus regulatory proteins - SUMO and KSHV. Front. Microbiol. 3:31. doi: 10.3389/fmicb.2012.00031
Cantrell, S. R., and Bresnahan, W. A. (2006). Human cytomegalovirus (HCMV) UL82 gene product (pp71) relieves hDaxx-mediated repression of HCMV replication. J. Virol. 80, 6188–6191. doi: 10.1128/JVI.02676-05
Carbone, R., Botrugno, O. A., Ronzoni, S., Insinga, A., Di Croce, L., Pelicci, P. G., et al. (2006). Recruitment of the histone methyltransferase SUV39H1 and its role in the oncogenic properties of the leukemia-associated PML-retinoic acid receptor fusion protein. Mol. Cell. Biol. 26, 1288–1296. doi: 10.1128/MCB.26.4.1288-1296.2006
Carvalho, T., Seeler, J. S., Ohman, K., Jordan, P., Pettersson, U., Akusjärvi, G., et al. (1995). Targeting of adenovirus E1A and E4-ORF3 proteins to nuclear matrix-associated PML bodies. J. Cell Biol. 131, 45–56. doi: 10.1083/jcb.131.1.45
Chang, P. C., Cheng, C. Y., Campbell, M., Yang, Y. C., Hsu, H. W., Chang, T. Y., et al. (2013). The chromatin modification by SUMO-2/3 but not SUMO-1 prevents the epigenetic activation of key immune-related genes during Kaposi’s sarcoma associated herpesvirus reactivation. BMC Genomics 14:824. doi: 10.1186/1471-2164-14-824
Chang, P. C., Izumiya, Y., Wu, C. Y., Fitzgerald, L. D., Campbell, M., Ellison, T. J., et al. (2010). Kaposi’s sarcoma-associated herpesvirus (KSHV) encodes a SUMO E3 ligase that is SIM-dependent and SUMO-2/3-specific. J. Biol. Chem. 285, 5266–5273. doi: 10.1074/jbc.M109.088088
Chee, A. V., Lopez, P., Pandolfi, P. P., and Roizman, B. (2003). Promyelocytic leukemia protein mediates interferon-based anti-herpes simplex virus 1 effects. J. Virol. 77, 7101–7105. doi: 10.1128/jvi.77.12.7101-7105.2003
Chelbi-Alix, M. K., and de Thé, H. (1999). Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene 18, 935–941. doi: 10.1038/sj.onc.1202366
Chelbi-Alix, M. K., Pelicano, L., Quignon, F., Koken, M. H., Venturini, L., Stadler, M., et al. (1995). Induction of the PML protein by interferons in normal and APL cells. Leukemia 9, 2027–2033.
Chelbi-Alix, M. K., Quignon, F., Pelicano, L., Koken, M. H., and de Thé, H. (1998). Resistance to virus infection conferred by the interferon-induced promyelocytic leukemia protein. J. Virol. 72, 1043–1051. doi: 10.1128/JVI.72.2.1043-1051.1998
Chen, Y., Wright, J., Meng, X., and Leppard, K. N. (2015). Promyelocytic leukemia protein isoform II promotes transcription factor recruitment to activate interferon beta and interferon-responsive gene expression. Mol. Cell. Biol. 35, 1660–1672. doi: 10.1128/MCB.01478-14
Cheng, X., and Kao, H. Y. (2013). Post-translational modifications of PML: consequences and implications. Front. Oncol. 2:210. doi: 10.3389/fonc.2012.00210
Cliffe, A. R., and Knipe, D. M. (2008). Herpes simplex virus ICP0 promotes both histone removal and acetylation on viral DNA during lytic infection. J. Virol. 82, 12030–12038. doi: 10.1128/JVI.01575-08
Cohen, C., Corpet, A., Roubille, S., Maroui, M. A., Poccardi, N., Rousseau, A., et al. (2018). Promyelocytic leukemia (PML) nuclear bodies (NBs) induce latent/quiescent HSV-1 genomes chromatinization through a PML NB/Histone H3.3/H3.3 Chaperone Axis. PLoS Pathog. 14:e1007313. doi: 10.1371/journal.ppat.1007313
Collin, V., Gravel, A., Kaufer, B. B., and Flamand, L. (2020). The Promyelocytic Leukemia Protein facilitates human herpesvirus 6B chromosomal integration, immediate-early 1 protein multiSUMOylation and its localization at telomeres. PLoS Pathog. 16:e1008683. doi: 10.1371/journal.ppat.1008683
Colombo, R., Boggio, R., Seiser, C., Draetta, G. F., and Chiocca, S. (2002). The adenovirus protein Gam1 interferes with sumoylation of histone deacetylase 1. EMBO Rep. 3, 1062–1068. doi: 10.1093/embo-reports/kvf213
Conn, K. L., Wasson, P., McFarlane, S., Tong, L., Brown, J. R., Grant, K. G., et al. (2016). Novel role for protein inhibitor of activated STAT 4 (PIAS4) in the restriction of herpes simplex virus 1 by the cellular intrinsic antiviral immune response. J. Virol. 90, 4807–4826. doi: 10.1128/JVI.03055-15
Corpet, A., Kleijwegt, C., Roubille, S., Juillard, F., Jacquet, K., Texier, P., et al. (2020). PML nuclear bodies and chromatin dynamics: catch me if you can! Nucleic Acids Res. 48, 11890–11912. doi: 10.1093/nar/gkaa828
Croxton, R., Puto, L. A., de Belle, I., Thomas, M., Torii, S., Hanaii, F., et al. (2006). Daxx represses expression of a subset of antiapoptotic genes regulated by nuclear factor-kappaB. Cancer Res. 66, 9026–9035. doi: 10.1158/0008-5472.CAN-06-1047
Cuchet-Lourenço, D., Anderson, G., Sloan, E., Orr, A., and Everett, R. D. (2013). The viral ubiquitin ligase ICP0 is neither sufficient nor necessary for degradation of the cellular DNA sensor IFI16 during herpes simplex virus 1 infection. J. Virol. 87, 13422–13432. doi: 10.1128/JVI.02474-13
Cuchet-Lourenço, D., Boutell, C., Lukashchuk, V., Grant, K., Sykes, A., Murray, J., et al. (2011). SUMO pathway dependent recruitment of cellular repressors to herpes simplex virus type 1 genomes. PLoS Pathog. 7:e1002123. doi: 10.1371/journal.ppat.1002123
Day, P. M., Baker, C. C., Lowy, D. R., and Schiller, J. T. (2004). Establishment of papillomavirus infection is enhanced by promyelocytic leukemia protein (PML) expression. Proc. Natl. Acad. Sci. U.S.A. 101, 14252–14257. doi: 10.1073/pnas.0404229101
Day, P. M., Roden, R. B., Lowy, D. R., and Schiller, J. T. (1998). The papillomavirus minor capsid protein, L2, induces localization of the major capsid protein, L1, and the viral transcription/replication protein, E2, to PML oncogenic domains. J. Virol. 72, 142–150. doi: 10.1128/JVI.72.1.142-150.1998
Day, P. M., Weisberg, A. S., Thompson, C. D., Hughes, M. M., Pang, Y. Y., Lowy, D. R., et al. (2019). Human papillomavirus 16 capsids mediate nuclear entry during infection. J. Virol. 93:e00454-19. doi: 10.1128/JVI.00454-19
de Thé, H., Lavau, C., Marchio, A., Chomienne, C., Degos, L., and Dejean, A. (1991). The PML-RAR alpha fusion mRNA generated by the t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell 66, 675–684. doi: 10.1016/0092-8674(91)90113-d
de Thé, H., Le Bras, M., and Lallemand-Breitenbach, V. (2012). The cell biology of disease: acute promyelocytic leukemia, arsenic, and PML bodies. J. Cell Biol. 198, 11–21. doi: 10.1083/jcb.201112044
Decque, A., Joffre, O., Magalhaes, J. G., Cossec, J. C., Blecher-Gonen, R., Lapaquette, P., et al. (2016). Sumoylation coordinates the repression of inflammatory and anti-viral gene-expression programs during innate sensing. Nat. Immunol. 17, 140–149. doi: 10.1038/ni.3342
Dellaire, G., Eskiw, C. H., Dehghani, H., Ching, R. W., and Bazett-Jones, D. P. (2006). Mitotic accumulations of PML protein contribute to the re-establishment of PML nuclear bodies in G1. J. Cell Sci. 119(Pt 6), 1034–1042. doi: 10.1242/jcs.02817
Dembowski, J. A., and DeLuca, N. A. (2015). Selective recruitment of nuclear factors to productively replicating herpes simplex virus genomes. PLoS Pathog. 11:e1004939. doi: 10.1371/journal.ppat.1004939
Dembowski, J. A., and Deluca, N. A. (2017). Purification of viral DNA for the identification of associated viral and cellular proteins. J. Vis. Exp. 126:56374. doi: 10.3791/56374
Dembowski, J. A., and DeLuca, N. A. (2018). Temporal viral genome-protein interactions define distinct stages of productive herpesviral infection. mBio 9:e01182-18. doi: 10.1128/mBio.01182-18
DeMeritt, I. B., Milford, L. E., and Yurochko, A. D. (2004). Activation of the NF-kappaB pathway in human cytomegalovirus-infected cells is necessary for efficient transactivation of the major immediate-early promoter. J. Virol. 78, 4498–4507. doi: 10.1128/jvi.78.9.4498-4507.2004
Di Croce, L., Raker, V. A., Corsaro, M., Fazi, F., Fanelli, M., Faretta, M., et al. (2002). Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science 295, 1079–1082. doi: 10.1126/science.1065173
DiGiuseppe, S., Bienkowska-Haba, M., Guion, L., Keiffer, T. R., and Sapp, M. (2017). Human papillomavirus major capsid protein L1 remains associated with the incoming viral genome throughout the entry process. J. Virol. 91:e00537-17. doi: 10.1128/JVI.00537-17
DiGiuseppe, S., Luszczek, W., Keiffer, T. R., Bienkowska-Haba, M., Guion, L. G., and Sapp, M. J. (2016). Incoming human papillomavirus type 16 genome resides in a vesicular compartment throughout mitosis. Proc. Natl. Acad. Sci. U.S.A. 113, 6289–6294. doi: 10.1073/pnas.1600638113
Diner, B. A., Li, T., Greco, T. M., Crow, M. S., Fuesler, J. A., Wang, J., et al. (2015a). The functional interactome of PYHIN immune regulators reveals IFIX is a sensor of viral DNA. Mol. Syst. Biol. 11:787. doi: 10.15252/msb.20145808
Diner, B. A., Lum, K. K., Javitt, A., and Cristea, I. M. (2015b). Interactions of the antiviral factor interferon gamma-inducible protein 16 (IFI16) mediate immune signaling and herpes simplex virus-1 immunosuppression. Mol. Cell. Proteomics 14, 2341–2356. doi: 10.1074/mcp.M114.047068
Diner, B. A., Lum, K. K., Toettcher, J. E., and Cristea, I. M. (2016). Viral DNA sensors IFI16 and cyclic GMP-AMP synthase possess distinct functions in regulating viral gene expression, immune defenses, and apoptotic responses during herpesvirus infection. mBio 7:e01553-16. doi: 10.1128/mBio.01553-16
Ding, L., Zhu, Q., Zhou, F., Tan, H., Xu, W., Pan, C., et al. (2019). Identification of viral SIM-SUMO2-interaction inhibitors for treating primary effusion lymphoma. PLoS Pathog. 15:e1008174. doi: 10.1371/journal.ppat.1008174
Djavani, M., Rodas, J., Lukashevich, I. S., Horejsh, D., Pandolfi, P. P., Borden, K. L., et al. (2001). Role of the promyelocytic leukemia protein PML in the interferon sensitivity of lymphocytic choriomeningitis virus. J. Virol. 75, 6204–6208. doi: 10.1128/JVI.75.13.6204-6208.2001
Domingues, P., Golebiowski, F., Tatham, M. H., Lopes, A. M., Taggart, A., Hay, R. T., et al. (2015). Global reprogramming of host SUMOylation during influenza virus infection. Cell Rep. 13, 1467–1480. doi: 10.1016/j.celrep.2015.10.001
Doucas, V., Ishov, A. M., Romo, A., Juguilon, H., Weitzman, M. D., Evans, R. M., et al. (1996). Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes dev. 10, 196–207. doi: 10.1101/gad.10.2.196
Dror, N., Rave-Harel, N., Burchert, A., Azriel, A., Tamura, T., Tailor, P., et al. (2007). Interferon regulatory factor-8 is indispensable for the expression of promyelocytic leukemia and the formation of nuclear bodies in myeloid cells. J. Biol. Chem. 282, 5633–5640. doi: 10.1074/jbc.M607825200
Dutrieux, J., Maarifi, G., Portilho, D. M., Arhel, N. J., Chelbi-Alix, M. K., and Nisole, S. (2015). PML/TRIM19-dependent inhibition of retroviral reverse-transcription by Daxx. PLoS Pathog. 11:e1005280. doi: 10.1371/journal.ppat.1005280
Dyck, J. A., Maul, G. G., Miller, W. H. Jr., Chen, J. D., Kakizuka, A., and Evans, R. M. (1994). A novel macromolecular structure is a target of the promyelocyte-retinoic acid receptor oncoprotein. Cell 76, 333–343. doi: 10.1016/0092-8674(94)90340-9
El Asmi, F., Maroui, M. A., Dutrieux, J., Blondel, D., Nisole, S., and Chelbi-Alix, M. K. (2014). Implication of PMLIV in both intrinsic and innate immunity. PLoS Pathog. 10:e1003975. doi: 10.1371/journal.ppat.1003975
El Bougrini, J., Dianoux, L., and Chelbi-Alix, M. K. (2011). PML positively regulates interferon gamma signaling. Biochimie 93, 389–398. doi: 10.1016/j.biochi.2010.11.005
El McHichi, B., Regad, T., Maroui, M. A., Rodriguez, M. S., Aminev, A., Gerbaud, S., et al. (2010). SUMOylation promotes PML degradation during encephalomyocarditis virus infection. J. Virol. 84, 11634–11645. doi: 10.1128/JVI.01321-10
El Motiam, A., Vidal, S., Seoane, R., Bouzaher, Y. H., González-Santamaría, J., and Rivas, C. (2020). SUMO and cytoplasmic RNA viruses: from enemies to best friends. Adv. Exp. Med. Biol. 1233, 263–277. doi: 10.1007/978-3-030-38266-7_11
Endter, C., Kzhyshkowska, J., Stauber, R., and Dobner, T. (2001). SUMO-1 modification required for transformation by adenovirus type 5 early region 1B 55-kDa oncoprotein. Proc. Natl. Acad. Sci. U.S.A. 98, 11312–11317. doi: 10.1073/pnas.191361798
Everett, R. D. (2015). Dynamic response of IFI16 and promyelocytic leukemia nuclear body components to herpes simplex virus 1 infection. J. Virol. 90, 167–179. doi: 10.1128/JVI.02249-15
Everett, R. D., Bell, A. J., Lu, Y., and Orr, A. (2013a). The replication defect of ICP0-null mutant herpes simplex virus 1 can be largely complemented by the combined activities of human cytomegalovirus proteins IE1 and pp71. J. Virol. 87, 978–990. doi: 10.1128/JVI.01103-12
Everett, R. D., Boutell, C., and Hale, B. G. (2013b). Interplay between viruses and host sumoylation pathways. Nat. Rev. Microbiol. 11, 400–411. doi: 10.1038/nrmicro3015
Everett, R. D., Boutell, C., McNair, C., Grant, L., and Orr, A. (2010). Comparison of the biological and biochemical activities of several members of the alphaherpesvirus ICP0 family of proteins. J. Virol. 84, 3476–3487. doi: 10.1128/JVI.02544-09
Everett, R. D., Boutell, C., and Orr, A. (2004). Phenotype of a herpes simplex virus type 1 mutant that fails to express immediate-early regulatory protein ICP0. J. Virol. 78, 1763–1774. doi: 10.1128/jvi.78.4.1763-1774.2004
Everett, R. D., Boutell, C., Pheasant, K., Cuchet-Lourenço, D., and Orr, A. (2014). Sequences related to SUMO interaction motifs in herpes simplex virus 1 protein ICP0 act cooperatively to stimulate virus infection. J. Virol. 88, 2763–2774. doi: 10.1128/JVI.03417-13
Everett, R. D., and Chelbi-Alix, M. K. (2007). PML and PML nuclear bodies: implications in antiviral defence. Biochimie 89, 819–830. doi: 10.1016/j.biochi.2007.01.004
Everett, R. D., Freemont, P., Saitoh, H., Dasso, M., Orr, A., Kathoria, M., et al. (1998). The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J. Virol. 72, 6581–6591. doi: 10.1128/JVI.72.8.6581-6591.1998
Everett, R. D., Lomonte, P., Sternsdorf, T., van Driel, R., and Orr, A. (1999). Cell cycle regulation of PML modification and ND10 composition. J. Cell Sci. 112(Pt 24), 4581–4588. doi: 10.1242/jcs.112.24.4581
Everett, R. D., Parada, C., Gripon, P., Sirma, H., and Orr, A. (2008). Replication of ICP0-null mutant herpes simplex virus type 1 is restricted by both PML and Sp100. J. Virol. 82, 2661–2672. doi: 10.1128/JVI.02308-07
Everett, R. D., Parsy, M. L., and Orr, A. (2009). Analysis of the functions of herpes simplex virus type 1 regulatory protein ICP0 that are critical for lytic infection and derepression of quiescent viral genomes. J. Virol. 83, 4963–4977. doi: 10.1128/JVI.02593-08
Everett, R. D., Rechter, S., Papior, P., Tavalai, N., Stamminger, T., and Orr, A. (2006). PML contributes to a cellular mechanism of repression of herpes simplex virus type 1 infection that is inactivated by ICP0. J. Virol. 80, 7995–8005. doi: 10.1128/JVI.00734-06
Ferenczy, M. W., Ranayhossaini, D. J., and Deluca, N. A. (2011). Activities of ICP0 involved in the reversal of silencing of quiescent herpes simplex virus 1. J. Virol. 85, 4993–5002. doi: 10.1128/JVI.02265-10
Florin, L., Sapp, C., Streeck, R. E., and Sapp, M. (2002). Assembly and translocation of papillomavirus capsid proteins. J. Virol. 76, 10009–10014. doi: 10.1128/jvi.76.19.10009-10014.2002
Flotho, A., and Melchior, F. (2013). Sumoylation: a regulatory protein modification in health and disease. Annu. Rev. biochem. 82, 357–385. doi: 10.1146/annurev-biochem-061909-093311
Fritah, S., Lhocine, N., Golebiowski, F., Mounier, J., Andrieux, A., Jouvion, G., et al. (2014). Sumoylation controls host anti-bacterial response to the gut invasive pathogen Shigella flexneri. EMBO Rep. 15, 965–972. doi: 10.15252/embr.201338386
Full, F., Jungnickl, D., Reuter, N., Bogner, E., Brulois, K., Scholz, B., et al. (2014). Kaposi’s sarcoma associated herpesvirus tegument protein ORF75 is essential for viral lytic replication and plays a critical role in the antagonization of ND10-instituted intrinsic immunity. PLoS Pathog. 10:e1003863. doi: 10.1371/journal.ppat.1003863
Geoffroy, M. C., and Chelbi-Alix, M. K. (2011). Role of promyelocytic leukemia protein in host antiviral defense. J. Interferon Cytokine Res. 31, 145–158. doi: 10.1089/jir.2010.0111
Geoffroy, M. C., and Hay, R. T. (2009). An additional role for SUMO in ubiquitin-mediated proteolysis. Nat. Rev. Mol. Cell biol. 10, 564–568. doi: 10.1038/nrm2707
Gialitakis, M., Arampatzi, P., Makatounakis, T., and Papamatheakis, J. (2010). Gamma interferon-dependent transcriptional memory via relocalization of a gene locus to PML nuclear bodies. Mol. Cell. Biol. 30, 2046–2056. doi: 10.1128/MCB.00906-09
Gilden, D., Nagel, M., Cohrs, R., Mahalingam, R., and Baird, N. (2015). Varicella zoster virus in the nervous system. F1000Res. 4:F1000 Faculty Rev-1356. doi: 10.12688/f1000research.7153.1
Giovannoni, F., Damonte, E. B., and García, C. C. (2015). Cellular promyelocytic leukemia protein is an important dengue virus restriction factor. PLoS One 10:e0125690. doi: 10.1371/journal.pone.0125690
Giovannoni, F., Ladelfa, M. F., Monte, M., Jans, D. A., Hemmerich, P., and García, C. (2019). Dengue non-structural protein 5 polymerase complexes with promyelocytic leukemia protein (PML) isoforms III and IV to disrupt PML-nuclear bodies in infected cells. Front. Cell. Infect. Microbiol. 9:284. doi: 10.3389/fcimb.2019.00284
Glass, M., and Everett, R. D. (2013). Components of promyelocytic leukemia nuclear bodies (ND10) act cooperatively to repress herpesvirus infection. J. Virol. 87, 2174–2185. doi: 10.1128/JVI.02950-12
Gravel, A., Dion, V., Cloutier, N., Gosselin, J., and Flamand, L. (2004). Characterization of human herpesvirus 6 variant B immediate-early 1 protein modifications by small ubiquitin-related modifiers. J. Gen. Virol. 85(Pt 5), 1319–1328. doi: 10.1099/vir.0.19610-0
Gravel, A., Gosselin, J., and Flamand, L. (2002). Human herpesvirus 6 immediate-early 1 protein is a sumoylated nuclear phosphoprotein colocalizing with promyelocytic leukemia protein-associated nuclear bodies. J. Biol. Chem. 277, 19679–19687. doi: 10.1074/jbc.M200836200
Grötzinger, T., Jensen, K., and Will, H. (1996). The interferon (IFN)-stimulated gene Sp100 promoter contains an IFN-gamma activation site and an imperfect IFN-stimulated response element which mediate type I IFN inducibility. J. Biol. Chem. 271, 25253–25260. doi: 10.1074/jbc.271.41.25253
Groves, I. J., Reeves, M. B., and Sinclair, J. H. (2009). Lytic infection of permissive cells with human cytomegalovirus is regulated by an intrinsic ‘pre-immediate-early’ repression of viral gene expression mediated by histone post-translational modification. J. Gen. Virol. 90(Pt 10), 2364–2374. doi: 10.1099/vir.0.012526-0
Guion, L., Bienkowska-Haba, M., DiGiuseppe, S., Florin, L., and Sapp, M. (2019). PML nuclear body-residing proteins sequentially associate with HPV genome after infectious nuclear delivery. PLoS Pathog. 15:e1007590. doi: 10.1371/journal.ppat.1007590
Guion, L. G., and Sapp, M. (2020). The role of promyelocytic leukemia nuclear bodies during HPV infection. Front. Cell. Infect. Microbiol. 10:35. doi: 10.3389/fcimb.2020.00035
Guldner, H. H., Szostecki, C., Grötzinger, T., and Will, H. (1992). IFN enhance expression of Sp100, an autoantigen in primary biliary cirrhosis. J. Immunol. 149, 4067–4073.
Günther, T., Schreiner, S., Dobner, T., Tessmer, U., and Grundhoff, A. (2014). Influence of ND10 components on epigenetic determinants of early KSHV latency establishment. PLoS Pathog. 10:e1004274. doi: 10.1371/journal.ppat.1004274
Hagemeier, S. R., Dickerson, S. J., Meng, Q., Yu, X., Mertz, J. E., and Kenney, S. C. (2010). Sumoylation of the epstein-barr virus BZLF1 protein inhibits its transcriptional activity and is regulated by the virus-encoded protein kinase. J. Virol. 84, 4383–4394. doi: 10.1128/JVI.02369-09
Halford, W. P., and Schaffer, P. A. (2001). ICP0 is required for efficient reactivation of herpes simplex virus type 1 from neuronal latency. J. Virol. 75, 3240–3249. doi: 10.1128/JVI.75.7.3240-3249.2001
Härle, P., Sainz, B. Jr., Carr, D. J., and Halford, W. P. (2002). The immediate-early protein, ICP0, is essential for the resistance of herpes simplex virus to interferon-alpha/beta. Virology 293, 295–304. doi: 10.1006/viro.2001.1280
Hayakawa, F., and Privalsky, M. L. (2004). Phosphorylation of PML by mitogen-activated protein kinases plays a key role in arsenic trioxide-mediated apoptosis. Cancer cell 5, 389–401. doi: 10.1016/s1535-6108(04)00082-0
Hembram, D., Negi, H., Biswas, P., Tripathi, V., Bhushan, L., Shet, D., et al. (2020). The viral SUMO-targeted ubiquitin ligase ICP0 is phosphorylated and activated by host kinase Chk2. J. Mol. Biol. 432, 1952–1977. doi: 10.1016/j.jmb.2020.01.021
Herzer, K., Weyer, S., Krammer, P. H., Galle, P. R., and Hofmann, T. G. (2005). Hepatitis C virus core protein inhibits tumor suppressor protein promyelocytic leukemia function in human hepatoma cells. Cancer Res. 65, 10830–10837. doi: 10.1158/0008-5472.CAN-05-0880
Higginbotham, J. M., and O’Shea, C. C. (2015). Adenovirus E4-ORF3 targets PIAS3 and together with E1B-55K remodels SUMO interactions in the nucleus and at virus genome replication domains. J. Virol. 89, 10260–10272. doi: 10.1128/JVI.01091-15
Hofmann, H., Sindre, H., and Stamminger, T. (2002). Functional interaction between the pp71 protein of human cytomegalovirus and the PML-interacting protein human Daxx. J. Virol. 76, 5769–5783. doi: 10.1128/jvi.76.11.5769-5783.2002
Hong, S., Mehta, K. P., and Laimins, L. A. (2011). Suppression of STAT-1 expression by human papillomaviruses is necessary for differentiation-dependent genome amplification and plasmid maintenance. J. Virol. 85, 9486–9494. doi: 10.1128/JVI.05007-11
Hoppe, A., Beech, S. J., Dimmock, J., and Leppard, K. N. (2006). Interaction of the adenovirus type 5 E4 Orf3 protein with promyelocytic leukemia protein isoform II is required for ND10 disruption. J. Virol. 80, 3042–3049. doi: 10.1128/JVI.80.6.3042-3049.2006
Hsu, K. S., and Kao, H. Y. (2018). PML: regulation and multifaceted function beyond tumor suppression. Cell Biosci. 8:5. doi: 10.1186/s13578-018-0204-8
Huh, Y. H., Kim, Y. E., Kim, E. T., Park, J. J., Song, M. J., Zhu, H., et al. (2008). Binding STAT2 by the acidic domain of human cytomegalovirus IE1 promotes viral growth and is negatively regulated by SUMO. J. Virol. 82, 10444–10454. doi: 10.1128/JVI.00833-08
Husnjak, K., Keiten-Schmitz, J., and Müller, S. (2016). Identification and characterization of SUMO-SIM interactions. Methods Mol. Biol. 1475, 79–98. doi: 10.1007/978-1-4939-6358-4_6
Hwang, J., and Kalejta, R. F. (2009). Human cytomegalovirus protein pp71 induces Daxx SUMOylation. J. Virol. 83, 6591–6598. doi: 10.1128/JVI.02639-08
Iki, S., Yokota, S., Okabayashi, T., Yokosawa, N., Nagata, K., and Fujii, N. (2005). Serum-dependent expression of promyelocytic leukemia protein suppresses propagation of influenza virus. Virology 343, 106–115. doi: 10.1016/j.virol.2005.08.010
Impens, F., Radoshevich, L., Cossart, P., and Ribet, D. (2014). Mapping of SUMO sites and analysis of SUMOylation changes induced by external stimuli. Proc. Natl. Acad. Sci. U.S.A. 111, 12432–12437. doi: 10.1073/pnas.1413825111
Ishov, A. M., and Maul, G. G. (1996). The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J. Cell Biol. 134, 815–826. doi: 10.1083/jcb.134.4.815
Ishov, A. M., Sotnikov, A. G., Negorev, D., Vladimirova, O. V., Neff, N., Kamitani, T., et al. (1999). PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 147, 221–234. doi: 10.1083/jcb.147.2.221
Ishov, A. M., Stenberg, R. M., and Maul, G. G. (1997). Human cytomegalovirus immediate early interaction with host nuclear structures: definition of an immediate transcript environment. J. Cell Biol. 138, 5–16. doi: 10.1083/jcb.138.1.5
Izumiya, Y., Ellison, T. J., Yeh, E. T., Jung, J. U., Luciw, P. A., and Kung, H. J. (2005). Kaposi’s sarcoma-associated herpesvirus K-bZIP represses gene transcription via SUMO modification. J. Virol. 79, 9912–9925. doi: 10.1128/JVI.79.15.9912-9925.2005
Izumiya, Y., Kobayashi, K., Kim, K. Y., Pochampalli, M., Izumiya, C., Shevchenko, B., et al. (2013). Kaposi’s sarcoma-associated herpesvirus K-Rta exhibits SUMO-targeting ubiquitin ligase (STUbL) like activity and is essential for viral reactivation. PLoS Pathog. 9:e1003506. doi: 10.1371/journal.ppat.1003506
Jansen, N. S., and Vertegaal, A. (2021). A chain of events: regulating target proteins by SUMO polymers. Trends Biochem. Sci. 46, 113–123. doi: 10.1016/j.tibs.2020.09.002
Jeanne, M., Lallemand-Breitenbach, V., Ferhi, O., Koken, M., Le Bras, M., Duffort, S., et al. (2010). PML/RARA oxidation and arsenic binding initiate the antileukemia response of As2O3. Cancer Cell 18, 88–98. doi: 10.1016/j.ccr.2010.06.003
Jurak, I., Silverstein, L. B., Sharma, M., and Coen, D. M. (2012). Herpes simplex virus is equipped with RNA- and protein-based mechanisms to repress expression of ATRX, an effector of intrinsic immunity. J. Virol. 86, 10093–10102. doi: 10.1128/JVI.00930-12
Kakizuka, A., Miller, W. H. Jr., Umesono, K., Warrell, R. P. Jr., Frankel, S. R., Murty, V. V., et al. (1991). Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RAR alpha with a novel putative transcription factor, PML. Cell 66, 663–674. doi: 10.1016/0092-8674(91)90112-c
Kalejta, R. F., and Albright, E. R. (2020). Expanding the known functional repertoire of the human cytomegalovirus pp71 protein. Front. Cell. Infect. Microbiol. 10:95. doi: 10.3389/fcimb.2020.00095
Kamitani, T., Kito, K., Nguyen, H. P., Wada, H., Fukuda-Kamitani, T., and Yeh, E. T. (1998). Identification of three major sentrinization sites in PML. J. Biol. Chem. 273, 26675–26682. doi: 10.1074/jbc.273.41.26675
Karen, K. A., Hoey, P. J., Young, C. S., and Hearing, P. (2009). Temporal regulation of the Mre11-Rad50-Nbs1 complex during adenovirus infection. J. Virol. 83, 4565–4573. doi: 10.1128/JVI.00042-09
Kawai, T., Akira, S., and Reed, J. C. (2003). ZIP kinase triggers apoptosis from nuclear PML oncogenic domains. Mol. Cell. Biol. 23, 6174–6186. doi: 10.1128/mcb.23.17.6174-6186.2003
Keiten-Schmitz, J., Schunck, K., and Müller, S. (2020). SUMO chains rule on chromatin occupancy. Front. Cell Dev. Biol. 7:343. doi: 10.3389/fcell.2019.00343
Kentsis, A., Dwyer, E. C., Perez, J. M., Sharma, M., Chen, A., Pan, Z. Q., et al. (2001). The RING domains of the promyelocytic leukemia protein PML and the arenaviral protein Z repress translation by directly inhibiting translation initiation factor eIF4E. J. Mol. Biol. 312, 609–623. doi: 10.1006/jmbi.2001.5003
Keyes, L. R., Hargett, D., Soland, M., Bego, M. G., Rossetto, C. C., Almeida-Porada, G., et al. (2012). HCMV protein LUNA is required for viral reactivation from latently infected primary CD14+ cells. PLoS One 7:e52827. doi: 10.1371/journal.pone.0052827
Kim, E. T., Kim, Y. E., Kim, Y. J., Lee, M. K., Hayward, G. S., and Ahn, J. H. (2014). Analysis of human cytomegalovirus-encoded SUMO targets and temporal regulation of SUMOylation of the immediate-early proteins IE1 and IE2 during infection. PLoS One 9:e103308. doi: 10.1371/journal.pone.0103308
Kim, J. A., Choi, M. S., Min, J. S., Kang, I., Oh, J., Kim, J. C., et al. (2017). HSV-1 ICP27 represses NF-κB activity by regulating Daxx sumoylation. BMB Rep. 50, 275–280. doi: 10.5483/bmbrep.2017.50.5.010
Kim, T. K., Lee, J. S., Oh, S. Y., Jin, X., Choi, Y. J., Lee, T. H., et al. (2007). Direct transcriptional activation of promyelocytic leukemia protein by IFN regulatory factor 3 induces the p53-dependent growth inhibition of cancer cells. Cancer Res. 67, 11133–11140. doi: 10.1158/0008-5472.CAN-07-1342
Kim, Y. E., and Ahn, J. H. (2015). Positive role of promyelocytic leukemia protein in type I interferon response and its regulation by human cytomegalovirus. PLoS Pathog. 11:e1004785. doi: 10.1371/journal.ppat.1004785
Kim, Y. E., Lee, J. H., Kim, E. T., Shin, H. J., Gu, S. Y., Seol, H. S., et al. (2011). Human cytomegalovirus infection causes degradation of Sp100 proteins that suppress viral gene expression. J. Virol. 85, 11928–11937. doi: 10.1128/JVI.00758-11
Kindsmüller, K., Groitl, P., Härtl, B., Blanchette, P., Hauber, J., and Dobner, T. (2007). Intranuclear targeting and nuclear export of the adenovirus E1B-55K protein are regulated by SUMO1 conjugation. Proc. Natl. Acad. Sci. U.S.A. 104, 6684–6689. doi: 10.1073/pnas.0702158104
Komatsu, T., Nagata, K., and Wodrich, H. (2015). An adenovirus DNA replication factor, but not incoming genome complexes, targets PML nuclear bodies. J. Virol. 90, 1657–1667. doi: 10.1128/JVI.02545-15
Krauss, S., Kaps, J., Czech, N., Paulus, C., and Nevels, M. (2009). Physical requirements and functional consequences of complex formation between the cytomegalovirus IE1 protein and human STAT2. J. Virol. 83, 12854–12870. doi: 10.1128/JVI.01164-09
Kunz, K., Piller, T., and Müller, S. (2018). SUMO-specific proteases and isopeptidases of the SENP family at a glance. J. Cell Sci. 131:jcs211904. doi: 10.1242/jcs.211904
Kyratsous, C. A., and Silverstein, S. J. (2009). Components of nuclear domain 10 bodies regulate varicella-zoster virus replication. J. Virol. 83, 4262–4274. doi: 10.1128/JVI.00021-09
Kyratsous, C. A., Walters, M. S., Panagiotidis, C. A., and Silverstein, S. J. (2009). Complementation of a herpes simplex virus ICP0 null mutant by varicella-zoster virus ORF61p. J. Virol. 83, 10637–10643. doi: 10.1128/JVI.01144-09
Lallemand-Breitenbach, V., and de Thé, H. (2018). PML nuclear bodies: from architecture to function. Curr. Opin. Cell Biol. 52, 154–161. doi: 10.1016/j.ceb.2018.03.011
Lallemand-Breitenbach, V., Jeanne, M., Benhenda, S., Nasr, R., Lei, M., Peres, L., et al. (2008). Arsenic degrades PML or PML-RARalpha through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat. Cell Biol. 10, 547–555. doi: 10.1038/ncb1717
Lallemand-Breitenbach, V., Zhu, J., Puvion, F., Koken, M., Honoré, N., Doubeikovsky, A., et al. (2001). Role of promyelocytic leukemia (PML) sumolation in nuclear body formation, 11S proteasome recruitment, and As2O3-induced PML or PML/retinoic acid receptor alpha degradation. J. Exp. Med. 193, 1361–1371. doi: 10.1084/jem.193.12.1361
Lanfranca, M. P., Mostafa, H. H., and Davido, D. J. (2014). HSV-1 ICP0: an E3 ubiquitin ligase that counteracts host intrinsic and innate immunity. Cells 3, 438–454. doi: 10.3390/cells3020438
Lang, M., Jegou, T., Chung, I., Richter, K., Münch, S., Udvarhelyi, A., et al. (2010). Three-dimensional organization of promyelocytic leukemia nuclear bodies. J. Cell Sci. 123(Pt 3), 392–400. doi: 10.1242/jcs.053496
Lapaquette, P., Fritah, S., Lhocine, N., Andrieux, A., Nigro, G., Mounier, J., et al. (2017). Shigella entry unveils a calcium/calpain-dependent mechanism for inhibiting sumoylation. Elife 6:e27444. doi: 10.7554/eLife.27444
Lee, H. R., Kim, D. J., Lee, J. M., Choi, C. Y., Ahn, B. Y., Hayward, G. S., et al. (2004). Ability of the human cytomegalovirus IE1 protein to modulate sumoylation of PML correlates with its functional activities in transcriptional regulation and infectivity in cultured fibroblast cells. J. Virol. 78, 6527–6542. doi: 10.1128/JVI.78.12.6527-6542.2004
Lee, J. H., and Kalejta, R. F. (2019). Human cytomegalovirus enters the primary CD34+ hematopoietic progenitor cells where it establishes latency by macropinocytosis. J. Virol. 93:e00452-19. doi: 10.1128/JVI.00452-19
Lee, J. H., Pasquarella, J. R., and Kalejta, R. F. (2019). Cell line models for human cytomegalovirus latency faithfully mimic viral entry by macropinocytosis and endocytosis. J. Virol. 93:e01021-19. doi: 10.1128/JVI.01021-19
Lee, J. S., Raja, P., and Knipe, D. M. (2016). Herpesviral ICP0 protein promotes two waves of heterochromatin removal on an early viral promoter during lytic infection. mBio 7:e02007-15. doi: 10.1128/mBio.02007-15
Lee, S. H., Albright, E. R., Lee, J. H., Jacobs, D., and Kalejta, R. F. (2015). Cellular defense against latent colonization foiled by human cytomegalovirus UL138 protein. Sci. Adv. 1:e1501164. doi: 10.1126/sciadv.1501164
Lehembre, F., Müller, S., Pandolfi, P. P., and Dejean, A. (2001). Regulation of Pax3 transcriptional activity by SUMO-1-modified PML. Oncogene 20, 1–9. doi: 10.1038/sj.onc.1204063
Leib, D. A., Coen, D. M., Bogard, C. L., Hicks, K. A., Yager, D. R., Knipe, D. M., et al. (1989). Immediate-early regulatory gene mutants define different stages in the establishment and reactivation of herpes simplex virus latency. J. Virol. 63, 759–768. doi: 10.1128/JVI.63.2.759-768.1989
Leib, D. A., Harrison, T. E., Laslo, K. M., Machalek, M. A., Moorman, N. J., and Virgin, H. W. (1999). Interferons regulate the phenotype of wild-type and mutant herpes simplex viruses in vivo. J. Exp. Med. 189, 663–672. doi: 10.1084/jem.189.4.663
Lewis, P. W., Elsaesser, S. J., Noh, K. M., Stadler, S. C., and Allis, C. D. (2010). Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc. Natl. Acad. Sci. U.S.A. 107, 14075–14080. doi: 10.1073/pnas.1008850107
Li, C., McManus, F. P., Plutoni, C., Pascariu, C. M., Nelson, T., Alberici Delsin, L. E., et al. (2020). Quantitative SUMO proteomics identifies PIAS1 substrates involved in cell migration and motility. Nat. Commun. 11:834. doi: 10.1038/s41467-020-14581-w
Li, H. P., and Chang, Y. S. (2003). Epstein-Barr virus latent membrane protein 1: structure and functions. J. Biomed. Sci. 10, 490–504. doi: 10.1007/BF02256110
Li, W., Wang, G., Zhang, H., Zhang, D., Zeng, J., Chen, X., et al. (2009). Differential suppressive effect of promyelocytic leukemia protein on the replication of different subtypes/strains of influenza A virus. Biochem. Biophys. Res. Commun. 389, 84–89. doi: 10.1016/j.bbrc.2009.08.091
Liang, Y. C., Lee, C. C., Yao, Y. L., Lai, C. C., Schmitz, M. L., and Yang, W. M. (2016). SUMO5, a novel poly-SUMO isoform, regulates PML nuclear bodies. Sci. Rep. 6:26509. doi: 10.1038/srep26509
Lin, D. Y., Huang, Y. S., Jeng, J. C., Kuo, H. Y., Chang, C. C., Chao, T. T., et al. (2006). Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol. Cell 24, 341–354. doi: 10.1016/j.molcel.2006.10.019
Lin, X., Sun, R., Zhang, F., Gao, Y., Bin, L., and Lan, K. (2017). The latency-associated nuclear antigen of Kaposi’s sarcoma-associated herpesvirus inhibits expression of SUMO/sentrin-specific peptidase 6 to facilitate establishment of latency. J. Virol. 91:e00806-17. doi: 10.1128/JVI.00806-17
Liu, X., Yuan, J., Wu, A. W., McGonagill, P. W., Galle, C. S., and Meier, J. L. (2010). Phorbol ester-induced human cytomegalovirus major immediate-early (MIE) enhancer activation through PKC-delta, CREB, and NF-kappaB desilences MIE gene expression in quiescently infected human pluripotent NTera2 cells. J. Virol. 84, 8495–8508. doi: 10.1128/JVI.00416-10
Liu, Y., Shevchenko, A., Shevchenko, A., and Berk, A. J. (2005). Adenovirus exploits the cellular aggresome response to accelerate inactivation of the MRN complex. J. Virol. 79, 14004–14016. doi: 10.1128/JVI.79.22.14004-14016.2005
Lo, Y. H., Huang, Y. W., Wu, Y. H., Tsai, C. S., Lin, Y. C., Mo, S. T., et al. (2013). Selective inhibition of the NLRP3 inflammasome by targeting to promyelocytic leukemia protein in mouse and human. Blood 121, 3185–3194. doi: 10.1182/blood-2012-05-432104
Lowrey, A. J., Cramblet, W., and Bentz, G. L. (2017). Viral manipulation of the cellular sumoylation machinery. Cell Commun. Signal. 15:27. doi: 10.1186/s12964-017-0183-0
Lukashchuk, V., and Everett, R. D. (2010). Regulation of ICP0-null mutant herpes simplex virus type 1 infection by ND10 components ATRX and hDaxx. J. Virol. 84, 4026–4040. doi: 10.1128/JVI.02597-09
Lukashchuk, V., McFarlane, S., Everett, R. D., and Preston, C. M. (2008). Human cytomegalovirus protein pp71 displaces the chromatin-associated factor ATRX from nuclear domain 10 at early stages of infection. J. Virol. 82, 12543–12554. doi: 10.1128/JVI.01215-08
Lunardi, A., Gaboli, M., Giorgio, M., Rivi, R., Bygrave, A., Antoniou, M., et al. (2011). A role for PML in innate immunity. Genes Cancer 2, 10–19. doi: 10.1177/1947601911402682
Maarifi, G., Maroui, M. A., Dutrieux, J., Dianoux, L., Nisole, S., and Chelbi-Alix, M. K. (2015). Small ubiquitin-like modifier alters IFN response. J. Immunol. 195, 2312–2324. doi: 10.4049/jimmunol.1500035
Marcos-Villar, L., Campagna, M., Lopitz-Otsoa, F., Gallego, P., González-Santamaría, J., González, D., et al. (2011). Covalent modification by SUMO is required for efficient disruption of PML oncogenic domains by Kaposi’s sarcoma-associated herpesvirus latent protein LANA2. J. Gen. Virol. 92(Pt 1), 188–194. doi: 10.1099/vir.0.024984-0
Marcos-Villar, L., Lopitz-Otsoa, F., Gallego, P., Muñoz-Fontela, C., González-Santamaría, J., Campagna, M., et al. (2009). Kaposi’s sarcoma-associated herpesvirus protein LANA2 disrupts PML oncogenic domains and inhibits PML-mediated transcriptional repression of the survivin gene. J. Virol. 83, 8849–8858. doi: 10.1128/JVI.00339-09
Maroui, M. A., Maarifi, G., McManus, F. P., Lamoliatte, F., Thibault, P., and Chelbi-Alix, M. K. (2018). Promyelocytic leukemia protein (PML) requirement for interferon-induced global cellular SUMOylation. Mol. Cell. Proteomics 17, 1196–1208. doi: 10.1074/mcp.RA117.000447
Maroui, M. A., Pampin, M., and Chelbi-Alix, M. K. (2011). Promyelocytic leukemia isoform IV confers resistance to encephalomyocarditis virus via the sequestration of 3D polymerase in nuclear bodies. J. Virol. 85, 13164–13173. doi: 10.1128/JVI.05808-11
Marusic, M. B., Mencin, N., Licen, M., Banks, L., and Grm, H. S. (2010). Modification of human papillomavirus minor capsid protein L2 by sumoylation. J. Virol. 84, 11585–11589. doi: 10.1128/JVI.01269-10
Maul, G. G., Ishov, A. M., and Everett, R. D. (1996). Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology 217, 67–75. doi: 10.1006/viro.1996.0094
McFarlane, S., Orr, A., Roberts, A., Conn, K. L., Iliev, V., Loney, C., et al. (2019). The histone chaperone HIRA promotes the induction of host innate immune defences in response to HSV-1 infection. PLoS Pathog. 15:e1007667. doi: 10.1371/journal.ppat.1007667
Meiering, C. D., and Linial, M. L. (2003). The promyelocytic leukemia protein does not mediate foamy virus latency in vitro. J. Virol. 77, 2207–2213. doi: 10.1128/jvi.77.3.2207-2213.2003
Merkl, P. E., and Knipe, D. M. (2019). Role for a filamentous nuclear assembly of IFI16, DNA, and host factors in restriction of herpesviral infection. mBio 10:e02621-18. doi: 10.1128/mBio.02621-18
Merkl, P. E., Orzalli, M. H., and Knipe, D. M. (2018). Mechanisms of host IFI16, PML, and Daxx protein restriction of herpes simplex virus 1 replication. J. Virol. 92:e00057-18. doi: 10.1128/JVI.00057-18
Min, J., Wright, W. E., and Shay, J. W. (2019). Clustered telomeres in phase-separated nuclear condensates engage mitotic DNA synthesis through BLM and RAD52. Genes Dev. 33, 814–827. doi: 10.1101/gad.324905.119
Moriuchi, H., Moriuchi, M., Smith, H. A., Straus, S. E., and Cohen, J. I. (1992). Varicella-zoster virus open reading frame 61 protein is functionally homologous to herpes simplex virus type 1 ICP0. J. Virol. 66, 7303–7308. doi: 10.1128/JVI.66.12.7303-7308.1992
Mossman, K. L., Saffran, H. A., and Smiley, J. R. (2000). Herpes simplex virus ICP0 mutants are hypersensitive to interferon. J. Virol. 74, 2052–2056. doi: 10.1128/jvi.74.4.2052-2056.2000
Mostafa, H. H., Thompson, T. W., and Davido, D. J. (2013). N-terminal phosphorylation sites of herpes simplex virus 1 ICP0 differentially regulate its activities and enhance viral replication. J. Virol. 87, 2109–2119. doi: 10.1128/JVI.02588-12
Mostafa, H. H., Thompson, T. W., Kushnir, A. S., Haenchen, S. D., Bayless, A. M., Hilliard, J. G., et al. (2011). Herpes simplex virus 1 ICP0 phosphorylation site mutants are attenuated for viral replication and impaired for explant-induced reactivation. J. Virol. 85, 12631–12637. doi: 10.1128/JVI.05661-11
Müller, S., and Dejean, A. (1999). Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J. Virol. 73, 5137–5143. doi: 10.1128/JVI.73.6.5137-5143.1999
Muller, S., and Dobner, T. (2008). The adenovirus E1B-55K oncoprotein induces SUMO modification of p53. Cell Cycle 7, 754–758. doi: 10.4161/cc.7.6.5495
Müncheberg, S., Hay, R. T., Ip, W. H., Meyer, T., Weiß, C., Brenke, J., et al. (2018). E1B-55K-mediated regulation of RNF4 SUMO-targeted ubiquitin ligase promotes human adenovirus gene expression. J. Virol. 92:e00164-18. doi: 10.1128/JVI.00164-18
Murata, T., Hotta, N., Toyama, S., Nakayama, S., Chiba, S., Isomura, H., et al. (2010). Transcriptional repression by sumoylation of Epstein-Barr virus BZLF1 protein correlates with association of histone deacetylase. J. Biol. Chem. 285, 23925–23935. doi: 10.1074/jbc.M109.095356
Murphy, J. C., Fischle, W., Verdin, E., and Sinclair, J. H. (2002). Control of cytomegalovirus lytic gene expression by histone acetylation. EMBO J. 21, 1112–1120. doi: 10.1093/emboj/21.5.1112
Nayak, A., and Müller, S. (2014). SUMO-specific proteases/isopeptidases: SENPs and beyond. Genome Biol. 15:422. doi: 10.1186/s13059-014-0422-2
Nefkens, I., Negorev, D. G., Ishov, A. M., Michaelson, J. S., Yeh, E. T., Tanguay, R. M., et al. (2003). Heat shock and Cd2+ exposure regulate PML and Daxx release from ND10 by independent mechanisms that modify the induction of heat-shock proteins 70 and 25 differently. J. Cell Sci. 116(Pt 3), 513–524. doi: 10.1242/jcs.00253
Negorev, D. G., Vladimirova, O. V., Ivanov, A., Rauscher, F. III, and Maul, G. G. (2006). Differential role of Sp100 isoforms in interferon-mediated repression of herpes simplex virus type 1 immediate-early protein expression. J. Virol. 80, 8019–8029. doi: 10.1128/JVI.02164-05
Negorev, D. G., Vladimirova, O. V., and Maul, G. G. (2009). Differential functions of interferon-upregulated Sp100 isoforms: herpes simplex virus type 1 promoter-based immediate-early gene suppression and PML protection from ICP0-mediated degradation. J. Virol. 83, 5168–5180. doi: 10.1128/JVI.02083-08
Nevels, M., Brune, W., and Shenk, T. (2004). SUMOylation of the human cytomegalovirus 72-kilodalton IE1 protein facilitates expression of the 86-kilodalton IE2 protein and promotes viral replication. J. Virol. 78, 7803–7812. doi: 10.1128/JVI.78.14.7803-7812.2004
Nisole, S., Maroui, M. A., Mascle, X. H., Aubry, M., and Chelbi-Alix, M. K. (2013). Differential roles of PML isoforms. Front. Oncol. 3:125. doi: 10.3389/fonc.2013.00125
Orzalli, M. H., Broekema, N. M., Diner, B. A., Hancks, D. C., Elde, N. C., Cristea, I. M., et al. (2015). cGAS-mediated stabilization of IFI16 promotes innate signaling during herpes simplex virus infection. Proc. Natl. Acad. Sci. U.S.A. 112, E1773–E1781. doi: 10.1073/pnas.1424637112
Orzalli, M. H., Broekema, N. M., and Knipe, D. M. (2016). Relative contributions of herpes simplex virus 1 ICP0 and vhs to loss of cellular IFI16 vary in different human cell types. J. Virol. 90, 8351–8359. doi: 10.1128/JVI.00939-16
Orzalli, M. H., DeLuca, N. A., and Knipe, D. M. (2012). Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proc. Natl. Acad. Sci. U.S.A. 109, E3008–E3017. doi: 10.1073/pnas.1211302109
Ou, H. D., Kwiatkowski, W., Deerinck, T. J., Noske, A., Blain, K. Y., Land, H. S., et al. (2012). A structural basis for the assembly and functions of a viral polymer that inactivates multiple tumor suppressors. Cell 151, 304–319. doi: 10.1016/j.cell.2012.08.035
Owerbach, D., McKay, E. M., Yeh, E. T., Gabbay, K. H., and Bohren, K. M. (2005). A proline-90 residue unique to SUMO-4 prevents maturation and sumoylation. Biochem. Biophys. Res. Commun. 337, 517–520. doi: 10.1016/j.bbrc.2005.09.090
Pampin, M., Simonin, Y., Blondel, B., Percherancier, Y., and Chelbi-Alix, M. K. (2006). Cross talk between PML and p53 during poliovirus infection: implications for antiviral defense. J. Virol. 80, 8582–8592. doi: 10.1128/JVI.00031-06
Park, J., Lee, J. H., La, M., Jang, M. J., Chae, G. W., Kim, S. B., et al. (2007). Inhibition of NF-kappaB acetylation and its transcriptional activity by Daxx. J. Mol. Biol. 368, 388–397. doi: 10.1016/j.jmb.2007.02.047
Paulus, C., Harwardt, T., Walter, B., Marxreiter, A., Zenger, M., Reuschel, E., et al. (2020). Revisiting promyelocytic leukemia protein targeting by human cytomegalovirus immediate-early protein 1. PLoS Pathog. 16:e1008537. doi: 10.1371/journal.ppat.1008537
Paulus, C., Krauss, S., and Nevels, M. (2006). A human cytomegalovirus antagonist of type I IFN-dependent signal transducer and activator of transcription signaling. Proc. Natl. Acad. Sci. U.S.A. 103, 3840–3845. doi: 10.1073/pnas.0600007103
Penkert, R. R., and Kalejta, R. F. (2010). Nuclear localization of tegument-delivered pp71 in human cytomegalovirus-infected cells is facilitated by one or more factors present in terminally differentiated fibroblasts. J. Virol. 84, 9853–9863. doi: 10.1128/JVI.00500-10
Penkert, R. R., and Kalejta, R. F. (2013). Human embryonic stem cell lines model experimental human cytomegalovirus latency. mBio 4:e00298-13. doi: 10.1128/mBio.00298-13
Pennella, M. A., Liu, Y., Woo, J. L., Kim, C. A., and Berk, A. J. (2010). Adenovirus E1B 55-kilodalton protein is a p53-SUMO1 E3 ligase that represses p53 and stimulates its nuclear export through interactions with promyelocytic leukemia nuclear bodies. J. Virol. 84, 12210–12225. doi: 10.1128/JVI.01442-10
Perfettini, J. L., Nardacci, R., Séror, C., Bourouba, M., Subra, F., Gros, L., et al. (2009). The tumor suppressor protein PML controls apoptosis induced by the HIV-1 envelope. Cell Death Differ. 16, 298–311. doi: 10.1038/cdd.2008.158
Poole, E. L., Kew, V. G., Lau, J., Murray, M. J., Stamminger, T., Sinclair, J. H., et al. (2018). A virally encoded DeSUMOylase activity is required for cytomegalovirus reactivation from latency. Cell Rep. 24, 594–606. doi: 10.1016/j.celrep.2018.06.048
Puvion-Dutilleul, F., Chelbi-Alix, M. K., Koken, M., Quignon, F., Puvion, E., and de Thé, H. (1995). Adenovirus infection induces rearrangements in the intranuclear distribution of the nuclear body-associated PML protein. Exp. Cell Res. 218, 9–16. doi: 10.1006/excr.1995.1125
Pyeon, D., Pearce, S. M., Lank, S. M., Ahlquist, P., and Lambert, P. F. (2009). Establishment of human papillomavirus infection requires cell cycle progression. PLoS Pathog. 5:e1000318. doi: 10.1371/journal.ppat.1000318
Querido, E., Blanchette, P., Yan, Q., Kamura, T., Morrison, M., Boivin, D., et al. (2001). Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 15, 3104–3117. doi: 10.1101/gad.926401
Rabellino, A., Carter, B., Konstantinidou, G., Wu, S. Y., Rimessi, A., Byers, L. A., et al. (2012). The SUMO E3-ligase PIAS1 regulates the tumor suppressor PML and its oncogenic counterpart PML-RARA. Cancer Res. 72, 2275–2284. doi: 10.1158/0008-5472.CAN-11-3159
Raman, N., Nayak, A., and Muller, S. (2013). The SUMO system: a master organizer of nuclear protein assemblies. Chromosoma 122, 475–485. doi: 10.1007/s00412-013-0429-6
Rauwel, B., Jang, S. M., Cassano, M., Kapopoulou, A., Barde, I., and Trono, D. (2015). Release of human cytomegalovirus from latency by a KAP1/TRIM28 phosphorylation switch. Elife 4:e06068. doi: 10.7554/eLife.06068
Reeves, M., Woodhall, D., Compton, T., and Sinclair, J. (2010). Human cytomegalovirus IE72 protein interacts with the transcriptional repressor hDaxx to regulate LUNA gene expression during lytic infection. J. Virol. 84, 7185–7194. doi: 10.1128/JVI.02231-09
Regad, T., Saib, A., Lallemand-Breitenbach, V., Pandolfi, P. P., de Thé, H., and Chelbi-Alix, M. K. (2001). PML mediates the interferon-induced antiviral state against a complex retrovirus via its association with the viral transactivator. EMBO J. 20, 3495–3505. doi: 10.1093/emboj/20.13.3495
Reichelt, M., Wang, L., Sommer, M., Perrino, J., Nour, A. M., Sen, N., et al. (2011). Entrapment of viral capsids in nuclear PML cages is an intrinsic antiviral host defense against varicella-zoster virus. PLoS Pathog. 7:e1001266. doi: 10.1371/journal.ppat.1001266
Reiser, J., Hurst, J., Voges, M., Krauss, P., Münch, P., Iftner, T., et al. (2011). High-risk human papillomaviruses repress constitutive kappa interferon transcription via E6 to prevent pathogen recognition receptor and antiviral-gene expression. J. Virol. 85, 11372–11380. doi: 10.1128/JVI.05279-11
Ribet, D., and Cossart, P. (2018). Ubiquitin, SUMO, and NEDD8: key targets of bacterial pathogens. Trends Cell Biol. 28, 926–940. doi: 10.1016/j.tcb.2018.07.005
Ribet, D., Hamon, M., Gouin, E., Nahori, M. A., Impens, F., Neyret-Kahn, H., et al. (2010). Listeria monocytogenes impairs SUMOylation for efficient infection. Nature 464, 1192–1195. doi: 10.1038/nature08963
Ribet, D., Lallemand-Breitenbach, V., Ferhi, O., Nahori, M. A., Varet, H., de Thé, H., et al. (2017). Promyelocytic leukemia protein (PML) controls listeria monocytogenes infection. mBio 8:e02179-16. doi: 10.1128/mBio.02179-16
Rodríguez, M. C., Dybas, J. M., Hughes, J., Weitzman, M. D., and Boutell, C. (2020). The HSV-1 ubiquitin ligase ICP0: modifying the cellular proteome to promote infection. Virus Res. 285:198015. doi: 10.1016/j.virusres.2020.198015
Sadanari, H., Yamada, R., Ohnishi, K., Matsubara, K., and Tanaka, J. (2005). SUMO-1 modification of the major immediate-early (IE) 1 and 2 proteins of human cytomegalovirus is regulated by different mechanisms and modulates the intracellular localization of the IE1, but not IE2, protein. Arch. Virol. 150, 1763–1782. doi: 10.1007/s00705-005-0559-0
Saffert, R. T., and Kalejta, R. F. (2006). Inactivating a cellular intrinsic immune defense mediated by Daxx is the mechanism through which the human cytomegalovirus pp71 protein stimulates viral immediate-early gene expression. J. Virol. 80, 3863–3871. doi: 10.1128/JVI.80.8.3863-3871.2006
Sahin, U., de Thé, H., and Lallemand-Breitenbach, V. (2014a). PML nuclear bodies: assembly and oxidative stress-sensitive sumoylation. Nucleus 5, 499–507. doi: 10.4161/19491034.2014.970104
Sahin, U., Ferhi, O., Carnec, X., Zamborlini, A., Peres, L., Jollivet, F., et al. (2014b). Interferon controls SUMO availability via the Lin28 and let-7 axis to impede virus replication. Nat. Commun. 5:4187. doi: 10.1038/ncomms5187
Sahin, U., Ferhi, O., Jeanne, M., Benhenda, S., Berthier, C., Jollivet, F., et al. (2014c). Oxidative stress-induced assembly of PML nuclear bodies controls sumoylation of partner proteins. J. Cell Biol. 204, 931–945. doi: 10.1083/jcb.201305148
Saitoh, H., and Hinchey, J. (2000). Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 275, 6252–6258. doi: 10.1074/jbc.275.9.6252
Salsman, J., Zimmerman, N., Chen, T., Domagala, M., and Frappier, L. (2008). Genome-wide screen of three herpesviruses for protein subcellular localization and alteration of PML nuclear bodies. PLoS Pathog. 4:e1000100. doi: 10.1371/journal.ppat.1000100
Sanyal, A., Wallaschek, N., Glass, M., Flamand, L., Wight, D. J., and Kaufer, B. B. (2018). The ND10 complex represses lytic human herpesvirus 6A replication and promotes silencing of the viral genome. Viruses 10:401. doi: 10.3390/v10080401
Scherer, M., Klingl, S., Sevvana, M., Otto, V., Schilling, E. M., Stump, J. D., et al. (2014). Crystal structure of cytomegalovirus IE1 protein reveals targeting of TRIM family member PML via coiled-coil interactions. PLoS Pathog. 10:e1004512. doi: 10.1371/journal.ppat.1004512
Scherer, M., Otto, V., Stump, J. D., Klingl, S., Müller, R., Reuter, N., et al. (2015). Characterization of recombinant human cytomegaloviruses encoding IE1 mutants L174P and 1-382 reveals that viral targeting of PML bodies perturbs both intrinsic and innate immune responses. J. Virol. 90, 1190–1205. doi: 10.1128/JVI.01973-15
Scherer, M., Reuter, N., Wagenknecht, N., Otto, V., Sticht, H., and Stamminger, T. (2013). Small ubiquitin-related modifier (SUMO) pathway-mediated enhancement of human cytomegalovirus replication correlates with a recruitment of SUMO-1/3 proteins to viral replication compartments. J. Gen. Virol. 94(Pt 6), 1373–1384. doi: 10.1099/vir.0.051078-0
Scherer, M., Schilling, E. M., and Stamminger, T. (2017). The human CMV IE1 protein: an offender of PML nuclear bodies. Adv. Anat. Embryol. Cell Biol. 223, 77–94. doi: 10.1007/978-3-319-53168-7_4
Scherer, M., and Stamminger, T. (2016). Emerging role of PML nuclear bodies in innate immune signaling. J. Virol. 90, 5850–5854. doi: 10.1128/JVI.01979-15
Schilling, E. M., Scherer, M., Reuter, N., Schweininger, J., Muller, Y. A., and Stamminger, T. (2017). The human cytomegalovirus IE1 protein antagonizes PML nuclear body-mediated intrinsic immunity via the inhibition of PML de novo SUMOylation. J. Virol. 91:e02049-16. doi: 10.1128/JVI.02049-16
Schneider, W. M., Chevillotte, M. D., and Rice, C. M. (2014). Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 32, 513–545. doi: 10.1146/annurev-immunol-032713-120231
Schreiner, S., Bürck, C., Glass, M., Groitl, P., Wimmer, P., Kinkley, S., et al. (2013). Control of human adenovirus type 5 gene expression by cellular Daxx/ATRX chromatin-associated complexes. Nucleic Acids Res. 41, 3532–3550. doi: 10.1093/nar/gkt064
Schreiner, S., Martinez, R., Groitl, P., Rayne, F., Vaillant, R., Wimmer, P., et al. (2012). Transcriptional activation of the adenoviral genome is mediated by capsid protein VI. PLoS Pathog. 8:e1002549. doi: 10.1371/journal.ppat.1002549
Schreiner, S., Wimmer, P., Groitl, P., Chen, S. Y., Blanchette, P., Branton, P. E., et al. (2011). Adenovirus type 5 early region 1B 55K oncoprotein-dependent degradation of cellular factor Daxx is required for efficient transformation of primary rodent cells. J. Virol. 85, 8752–8765. doi: 10.1128/JVI.00440-11
Schreiner, S., Wimmer, P., Sirma, H., Everett, R. D., Blanchette, P., Groitl, P., et al. (2010). Proteasome-dependent degradation of Daxx by the viral E1B-55K protein in human adenovirus-infected cells. J. Virol. 84, 7029–7038. doi: 10.1128/JVI.00074-10
Seeler, J. S., Marchio, A., Losson, R., Desterro, J. M., Hay, R. T., Chambon, P., et al. (2001). Common properties of nuclear body protein SP100 and TIF1alpha chromatin factor: role of SUMO modification. Mol. Cell. Biol. 21, 3314–3324. doi: 10.1128/MCB.21.10.3314-3324.2001
Seeler, J. S., Marchio, A., Sitterlin, D., Transy, C., and Dejean, A. (1998). Interaction of SP100 with HP1 proteins: a link between the promyelocytic leukemia-associated nuclear bodies and the chromatin compartment. Proc. Natl. Acad. Sci. U.S.A. 95, 7316–7321. doi: 10.1073/pnas.95.13.7316
Shen, T. H., Lin, H. K., Scaglioni, P. P., Yung, T. M., and Pandolfi, P. P. (2006). The mechanisms of PML-nuclear body formation. Mol. Cell 24, 331–339. doi: 10.1016/j.molcel.2006.09.013
Sides, M. D., Block, G. J., Chadwick, R. W., Shan, B., Flemington, E. K., and Lasky, J. A. (2011a). Epstein–barr virus latent membrane protein 1 suppresses reporter activity through modulation of promyelocytic leukemia protein-nuclear bodies. Virol. J. 8:461. doi: 10.1186/1743-422X-8-461
Sides, M. D., Block, G. J., Shan, B., Esteves, K. C., Lin, Z., Flemington, E. K., et al. (2011b). Arsenic mediated disruption of promyelocytic leukemia protein nuclear bodies induces ganciclovir susceptibility in Epstein-Barr positive epithelial cells. Virology 416, 86–97. doi: 10.1016/j.virol.2011.04.005
Sidik, S. M., Salsman, J., Dellaire, G., and Rohde, J. R. (2015). Shigella infection interferes with SUMOylation and increases PML NB number. PLoS One 10:e0122585. doi: 10.1371/journal.pone.0122585
Sinclair, J. (2010). Chromatin structure regulates human cytomegalovirus gene expression during latency, reactivation and lytic infection. Biochim. Biophys. Acta 1799, 286–295. doi: 10.1016/j.bbagrm.2009.08.001
Sivachandran, N., Cao, J. Y., and Frappier, L. (2010). Epstein-Barr virus nuclear antigen 1 Hijacks the host kinase CK2 to disrupt PML nuclear bodies. J. Virol. 84, 11113–11123. doi: 10.1128/JVI.01183-10
Sivachandran, N., Sarkari, F., and Frappier, L. (2008). Epstein-Barr nuclear antigen 1 contributes to nasopharyngeal carcinoma through disruption of PML nuclear bodies. PLoS Pathog. 4:e1000170. doi: 10.1371/journal.ppat.1000170
Sivachandran, N., Wang, X., and Frappier, L. (2012). Functions of the epstein-barr virus EBNA1 protein in viral reactivation and lytic infection. J. Virol. 86, 6146–6158. doi: 10.1128/JVI.00013-12
Sloan, E., Orr, A., and Everett, R. D. (2016). MORC3, a component of PML nuclear bodies, has a role in restricting herpes simplex virus 1 and human cytomegalovirus. J. Virol. 90, 8621–8633. doi: 10.1128/JVI.00621-16
Sloan, E., Tatham, M. H., Groslambert, M., Glass, M., Orr, A., Hay, R. T., et al. (2015). Analysis of the SUMO2 Proteome during HSV-1 Infection. PLoS Pathog. 11:e1005059. doi: 10.1371/journal.ppat.1005059
Sohn, S. Y., Bridges, R. G., and Hearing, P. (2015). Proteomic analysis of ubiquitin-like posttranslational modifications induced by the adenovirus E4-ORF3 protein. J. Virol. 89, 1744–1755. doi: 10.1128/JVI.02892-14
Sohn, S. Y., and Hearing, P. (2012). Adenovirus regulates sumoylation of Mre11-Rad50-Nbs1 components through a paralog-specific mechanism. J. Virol. 86, 9656–9665. doi: 10.1128/JVI.01273-12
Sohn, S. Y., and Hearing, P. (2016). The adenovirus E4-ORF3 protein functions as a SUMO E3 ligase for TIF-1γ sumoylation and poly-SUMO chain elongation. Proc. Natl. Acad. Sci. U.S.A. 113, 6725–6730. doi: 10.1073/pnas.1603872113
Sourvinos, G., Morou, A., Sanidas, I., Codruta, I., Ezell, S. A., Doxaki, C., et al. (2014). The downregulation of GFI1 by the EZH2-NDY1/KDM2B-JARID2 axis and by human cytomegalovirus (HCMV) associated factors allows the activation of the HCMV major IE promoter and the transition to productive infection. PLoS Pathog. 10:e1004136. doi: 10.1371/journal.ppat.1004136
Spengler, M. L., Kurapatwinski, K., Black, A. R., and Azizkhan-Clifford, J. (2002). SUMO-1 modification of human cytomegalovirus IE1/IE72. J. Virol. 76, 2990–2996. doi: 10.1128/jvi.76.6.2990-2996.2002
Sriramachandran, A. M., and Dohmen, R. J. (2014). SUMO-targeted ubiquitin ligases. Biochim. Biophys. Acta 1843, 75–85. doi: 10.1016/j.bbamcr.2013.08.022
Stadler, M., Chelbi-Alix, M. K., Koken, M. H., Venturini, L., Lee, C., Saïb, A., et al. (1995). Transcriptional induction of the PML growth suppressor gene by interferons is mediated through an ISRE and a GAS element. Oncogene 11, 2565–2573.
Stanton, R., Fox, J. D., Caswell, R., Sherratt, E., and Wilkinson, G. (2002). Analysis of the human herpesvirus-6 immediate-early 1 protein. J. Gen. Virol. 83(Pt 11), 2811–2820. doi: 10.1099/0022-1317-83-11-2811
Stehmeier, P., and Muller, S. (2009). Phospho-regulated SUMO interaction modules connect the SUMO system to CK2 signaling. Mol. cell 33, 400–409. doi: 10.1016/j.molcel.2009.01.013
Stepp, W. H., Meyers, J. M., and McBride, A. A. (2013). Sp100 provides intrinsic immunity against human papillomavirus infection. mBio 4:e00845-13. doi: 10.1128/mBio.00845-13
Stepp, W. H., Stamos, J. D., Khurana, S., Warburton, A., and McBride, A. A. (2017). Sp100 colocalizes with HPV replication foci and restricts the productive stage of the infectious cycle. PLoS Pathog. 13:e1006660. doi: 10.1371/journal.ppat.1006660
Sternsdorf, T., Guldner, H. H., Szostecki, C., Grötzinger, T., and Will, H. (1995). Two nuclear dot-associated proteins, PML and Sp100, are often co-autoimmunogenic in patients with primary biliary cirrhosis. Scand. J. Immunol. 42, 257–268. doi: 10.1111/j.1365-3083.1995.tb03652.x
Sternsdorf, T., Jensen, K., Reich, B., and Will, H. (1999). The nuclear dot protein sp100, characterization of domains necessary for dimerization, subcellular localization, and modification by small ubiquitin-like modifiers. J. Biol. Chem. 274, 12555–12566. doi: 10.1074/jbc.274.18.12555
Sternsdorf, T., Jensen, K., and Will, H. (1997). Evidence for covalent modification of the nuclear dot-associated proteins PML and Sp100 by PIC1/SUMO-1. J. Cell Biol. 139, 1621–1634. doi: 10.1083/jcb.139.7.1621
Stracker, T. H., Carson, C. T., and Weitzman, M. D. (2002). Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature 418, 348–352. doi: 10.1038/nature00863
Stubbe, M., Mai, J., Paulus, C., Stubbe, H. C., Berscheminski, J., Karimi, M., et al. (2020). Viral DNA binding protein SUMOylation promotes PML nuclear body localization next to viral replication centers. mBio 11:e00049-20. doi: 10.1128/mBio.00049-20
Su, C. I., Tseng, C. H., Yu, C. Y., and Lai, M. (2016). SUMO modification stabilizes dengue virus nonstructural protein 5 to support virus replication. J. Virol. 90, 4308–4319. doi: 10.1128/JVI.00223-16
Szostecki, C., Guldner, H. H., Netter, H. J., and Will, H. (1990). Isolation and characterization of cDNA encoding a human nuclear antigen predominantly recognized by autoantibodies from patients with primary biliary cirrhosis. J. Immunol. 145, 4338–4347.
Tatham, M. H., Geoffroy, M. C., Shen, L., Plechanovova, A., Hattersley, N., Jaffray, E. G., et al. (2008). RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat. Cell Biol. 10, 538–546. doi: 10.1038/ncb1716
Tavalai, N., Adler, M., Scherer, M., Riedl, Y., and Stamminger, T. (2011). Evidence for a dual antiviral role of the major nuclear domain 10 component Sp100 during the immediate-early and late phases of the human cytomegalovirus replication cycle. J. Virol. 85, 9447–9458. doi: 10.1128/JVI.00870-11
Tavalai, N., Papior, P., Rechter, S., Leis, M., and Stamminger, T. (2006). Evidence for a role of the cellular ND10 protein PML in mediating intrinsic immunity against human cytomegalovirus infections. J. Virol. 80, 8006–8018. doi: 10.1128/JVI.00743-06
Tavalai, N., Papior, P., Rechter, S., and Stamminger, T. (2008). Nuclear domain 10 components promyelocytic leukemia protein and hDaxx independently contribute to an intrinsic antiviral defense against human cytomegalovirus infection. J. Virol. 82, 126–137. doi: 10.1128/JVI.01685-07
Tsai, K., Chan, L., Gibeault, R., Conn, K., Dheekollu, J., Domsic, J., et al. (2014). Viral reprogramming of the Daxx histone H3.3 chaperone during early Epstein-Barr virus infection. J. Virol. 88, 14350–14363. doi: 10.1128/JVI.01895-14
Tsai, K., Thikmyanova, N., Wojcechowskyj, J. A., Delecluse, H. J., and Lieberman, P. M. (2011). EBV tegument protein BNRF1 disrupts DAXX-ATRX to activate viral early gene transcription. PLoS Pathog. 7:e1002376. doi: 10.1371/journal.ppat.1002376
Ulbricht, T., Alzrigat, M., Horch, A., Reuter, N., von Mikecz, A., Steimle, V., et al. (2012). PML promotes MHC class II gene expression by stabilizing the class II transactivator. J. Cell Biol. 199, 49–63. doi: 10.1083/jcb.201112015
Ullman, A. J., and Hearing, P. (2008). Cellular proteins PML and Daxx mediate an innate antiviral defense antagonized by the adenovirus E4 ORF3 protein. J. Virol. 82, 7325–7335. doi: 10.1128/JVI.00723-08
Ullman, A. J., Reich, N. C., and Hearing, P. (2007). Adenovirus E4 ORF3 protein inhibits the interferon-mediated antiviral response. J. Virol. 81, 4744–4752. doi: 10.1128/JVI.02385-06
Ullmann, R., Chien, C. D., Avantaggiati, M. L., and Muller, S. (2012). An acetylation switch regulates SUMO-dependent protein interaction networks. Mol. Cell 46, 759–770. doi: 10.1016/j.molcel.2012.04.006
Ulrich, H. D. (2008). The fast-growing business of SUMO chains. Mol. Cell 32, 301–305. doi: 10.1016/j.molcel.2008.10.010
Van Damme, E., Laukens, K., Dang, T. H., and Van Ostade, X. (2010). A manually curated network of the PML nuclear body interactome reveals an important role for PML NBs in SUMOylation dynamics. Int. J. Biol. Sci. 6, 51–67. doi: 10.7150/ijbs.6.51
Vanni, E., Gatherer, D., Tong, L., Everett, R. D., and Boutell, C. (2012). Functional characterization of residues required for the herpes simplex virus 1 E3 ubiquitin ligase ICP0 to interact with the cellular E2 ubiquitin-conjugating enzyme UBE2D1 (UbcH5a). J. Virol. 86, 6323–6333. doi: 10.1128/JVI.07210-11
Vardi, N., Chaturvedi, S., and Weinberger, L. S. (2018). Feedback-mediated signal conversion promotes viral fitness. Proc. Natl. Acad. Sci. U.S.A. 115, E8803–E8810. doi: 10.1073/pnas.1802905115
Verma, S., Mohapatra, G., Ahmad, S. M., Rana, S., Jain, S., Khalsa, J. K., et al. (2015). Salmonella engages host MicroRNAs to modulate SUMOylation: a new arsenal for intracellular survival. Mol. Cell. Biol. 35, 2932–2946. doi: 10.1128/MCB.00397-15
Volcic, M., Sparrer, K., Koepke, L., Hotter, D., Sauter, D., Stürzel, C. M., et al. (2020). Vpu modulates DNA repair to suppress innate sensing and hyper-integration of HIV-1. Nat. Microbiol. 5, 1247–1261. doi: 10.1038/s41564-020-0753-6
Walters, M. S., Kyratsous, C. A., and Silverstein, S. J. (2010). The RING finger domain of Varicella-Zoster virus ORF61p has E3 ubiquitin ligase activity that is essential for efficient autoubiquitination and dispersion of Sp100-containing nuclear bodies. J. Virol. 84, 6861–6865. doi: 10.1128/JVI.00335-10
Wang, L., Oliver, S. L., Sommer, M., Rajamani, J., Reichelt, M., and Arvin, A. M. (2011). Disruption of PML nuclear bodies is mediated by ORF61 SUMO-interacting motifs and required for varicella-zoster virus pathogenesis in skin. PLoS Pathog. 7:e1002157. doi: 10.1371/journal.ppat.1002157
Weis, K., Rambaud, S., Lavau, C., Jansen, J., Carvalho, T., Carmo-Fonseca, M., et al. (1994). Retinoic acid regulates aberrant nuclear localization of PML-RAR alpha in acute promyelocytic leukemia cells. Cell 76, 345–356. doi: 10.1016/0092-8674(94)90341-7
Wilcox, K. W., Sheriff, S., Isaac, A., and Taylor, J. L. (2005). SP100B is a repressor of gene expression. J. Cell. Biochem. 95, 352–365. doi: 10.1002/jcb.20434
Wilson, V. G. (2017). Viral interplay with the host sumoylation system. Adv. Exp. Med. Biol. 963, 359–388. doi: 10.1007/978-3-319-50044-7_21
Wimmer, P., Blanchette, P., Schreiner, S., Ching, W., Groitl, P., Berscheminski, J., et al. (2013). Cross-talk between phosphorylation and SUMOylation regulates transforming activities of an adenoviral oncoprotein. Oncogene 32, 1626–1637. doi: 10.1038/onc.2012.187
Wimmer, P., Schreiner, S., Everett, R. D., Sirma, H., Groitl, P., and Dobner, T. (2010). SUMO modification of E1B-55K oncoprotein regulates isoform-specific binding to the tumour suppressor protein PML. Oncogene 29, 5511–5522. doi: 10.1038/onc.2010.284
Woodhall, D. L., Groves, I. J., Reeves, M. B., Wilkinson, G., and Sinclair, J. H. (2006). Human Daxx-mediated repression of human cytomegalovirus gene expression correlates with a repressive chromatin structure around the major immediate early promoter. J. biol. Chem. 281, 37652–37660. doi: 10.1074/jbc.M604273200
Wu, J., Kim, S., Kwak, M. S., Jeong, J. B., Min, H. J., Yoon, H. G., et al. (2014). High mobility group nucleosomal binding domain 2 (HMGN2) SUMOylation by the SUMO E3 ligase PIAS1 decreases the binding affinity to nucleosome core particles. J. Biol. Chem. 289, 20000–20011. doi: 10.1074/jbc.M114.555425
Wu, W. S., Vallian, S., Seto, E., Yang, W. M., Edmondson, D., Roth, S., et al. (2001). The growth suppressor PML represses transcription by functionally and physically interacting with histone deacetylases. Mol. Cell. Biol. 21, 2259–2268. doi: 10.1128/MCB.21.7.2259-2268.2001
Xu, Y., Ahn, J. H., Cheng, M., apRhys, C. M., Chiou, C. J., Zong, J., et al. (2001). Proteasome-independent disruption of PML oncogenic domains (PODs), but not covalent modification by SUMO-1, is required for human cytomegalovirus immediate-early protein IE1 to inhibit PML-mediated transcriptional repression. J. Virol. 75, 10683–10695. doi: 10.1128/JVI.75.22.10683-10695.2001
Yang, W. S., Hsu, H. W., Campbell, M., Cheng, C. Y., and Chang, P. C. (2015a). K-bZIP Mediated SUMO-2/3 specific modification on the KSHV genome negatively regulates lytic gene expression and viral reactivation. PLoS Pathog. 11:e1005051. doi: 10.1371/journal.ppat.1005051
Yang, Y. C., Feng, T. H., Chen, T. Y., Huang, H. H., Hung, C. C., Liu, S. T., et al. (2015b). RanBPM regulates Zta-mediated transcriptional activity in Epstein-Barr virus. J. Gen. Virol. 96, 2336–2348. doi: 10.1099/vir.0.000157
Yousef, A. F., Fonseca, G. J., Pelka, P., Ablack, J. N., Walsh, C., Dick, F. A., et al. (2010). Identification of a molecular recognition feature in the E1A oncoprotein that binds the SUMO conjugase UBC9 and likely interferes with polySUMOylation. Oncogene 29, 4693–4704. doi: 10.1038/onc.2010.226
Yuan, J., Li, M., Torres, Y. R., Galle, C. S., and Meier, J. L. (2015). Differentiation-coupled induction of human cytomegalovirus replication by union of the major enhancer retinoic acid, cyclic AMP, and NF-κB response elements. J. Virol. 89, 12284–12298. doi: 10.1128/JVI.00965-15
Zhang, H., Zhao, R., Tones, J., Liu, M., Dilley, R. L., Chenoweth, D. M., et al. (2020). Nuclear body phase separation drives telomere clustering in ALT cancer cells. Mol. Biol. Cell 31, 2048–2056. doi: 10.1091/mbc.E19-10-0589
Zhang, X. W., Yan, X. J., Zhou, Z. R., Yang, F. F., Wu, Z. Y., Sun, H. B., et al. (2010). Arsenic trioxide controls the fate of the PML-RARalpha oncoprotein by directly binding PML. Science 328, 240–243. doi: 10.1126/science.1183424
Keywords: promyelocytic leukemia nuclear bodies, small ubiquitin-related modifier, viruses, bacteria, intrinsic and innate immune response
Citation: Patra U and Müller S (2021) A Tale of Usurpation and Subversion: SUMO-Dependent Integrity of Promyelocytic Leukemia Nuclear Bodies at the Crossroad of Infection and Immunity. Front. Cell Dev. Biol. 9:696234. doi: 10.3389/fcell.2021.696234
Received: 16 April 2021; Accepted: 30 July 2021;
Published: 27 August 2021.
Edited by:
Carmen Rivas, Consejo Superior de Investigaciones Científicas (CSIC), SpainReviewed by:
Mounira K. Chelbi-Alix, Centre National de la Recherche Scientifique (CNRS), FranceYingqiu Li, Sun Yat-sen University, China
Sabrina Schreiner, Hannover Medical School, Germany
Copyright © 2021 Patra and Müller. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Upayan Patra, cGF0cmFAbWVkLnVuaS1mcmFua2Z1cnQuZGU=; Stefan Müller, c3RlLm11ZWxsZXJAZW0udW5pLWZyYW5rZnVydC5kZQ==
 Upayan Patra
Upayan Patra Stefan Müller
Stefan Müller