- 1Department of Biochemistry, University of Alberta, Edmonton, AB, Canada
- 2Molecular and Cellular Biology, MacEwan University, Edmonton, AB, Canada
- 3SMALP Network, Edmonton, AB, Canada
Membrane readers take part in trafficking and signaling processes by localizing proteins to organelle surfaces and transducing molecular information. They accomplish this by engaging phosphoinositides (PIs), a class of lipid molecules which are found in different proportions in various cellular membranes. The prototypes are the PX domains, which exhibit a range of specificities for PIs. Our meta-analysis indicates that recognition of membranes by PX domains is specifically controlled by modification of lysine and arginine residues including acetylation, hydroxyisobutyrylation, glycation, malonylation, methylation and succinylation of sidechains that normally bind headgroups of phospholipids including organelle-specific PI signals. Such metabolite-modulated residues in lipid binding elements are named MET-stops here to highlight their roles as erasers of membrane reader functions. These modifications are concentrated in the membrane binding sites of half of all 49 PX domains in the human proteome and correlate with phosphoregulatory sites, as mapped using the Membrane Optimal Docking Area (MODA) algorithm. As these motifs are mutated and modified in various cancers and the responsible enzymes serve as potential drug targets, the discovery of MET-stops as a widespread inhibitory mechanism may aid in the development of diagnostics and therapeutics aimed at the readers, writers and erasers of the PI code.
Introduction
Membrane readers are protein domains that recognize unique PI lipids that mark various organelle membranes found in eukaryotic cells. These conserved modules serve to reversibly recruit cytosolic proteins to membrane surfaces, thus controlling downstream assembly, signaling and trafficking events. The best understood are the FYVE, PH, and PX domain superfamilies, which represent the core of the PI code that underlies eukaryotic membrane recognition (Overduin et al., 2001; Sato et al., 2001). They comprise hundreds of domains and may only represent a small fraction of the total number of membrane readers (Overduin and Kervin, submitted), with the weak, dynamic or temperamental lipid binding activities of many proteins remaining technically difficult to detect. How they are regulated remains obscure, prompting this investigation into PI code control.
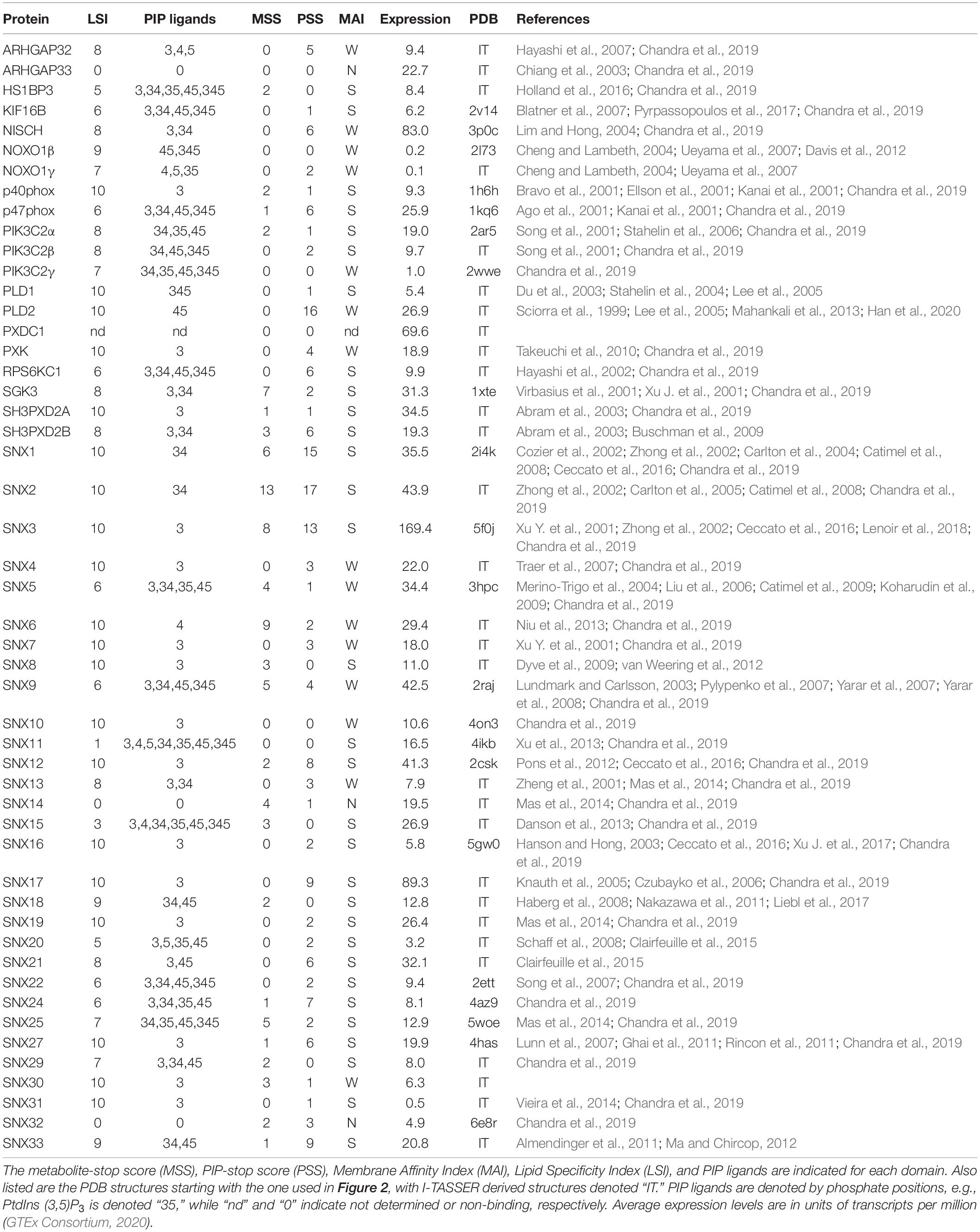
Of the diverse families of membrane readers, PX domains are uniquely able to detect the full spectrum of seven phosphoinositide phosphate (PIP) signals (Table 1). This superfamily includes up to ∼120 members per genome across fungi, protists, viridiplantae, and metazoa (Banerjee et al., 2010), with 49 distinct members in Homo sapiens (Figure 1). The family of human PX domains is the focus here as it comprises the best-defined group of membrane readers in terms of 3D structures, ligand specificities and membrane interactions. Moreover, since the discovery of residues that, when phosphorylated, phosphorylation, block PIP recognition (PIP-stops) in three sorting nexins (Lenoir et al., 2018), this family is ideal for investigating how other protein modifications could control membrane recognition.

Figure 1. Human proteins containing PX domains. The PX domains that contain arginine and lysine-based MET-stops are bounded by dashed purple and green lines, respectively. Modules are color-coded and abbreviated as Agc (AGC-kinase C-terminal), BAR (Bin–Amphiphysin–Rvs), C2 (protein kinase C conserved region 2), CC (coiled coil), GAP (GTPase-activating protein), FHA (forkhead-associated), LRR (leucine-rich repeat), MIT (microtubule interacting and transport), PB1 (Phox and Bem1), PDZ (Postsynaptic density 95, Disk large, Zonula occludens), PH (pleckstrin homology), PIK (phosphoinositide 3-kinase), PXA (PX-associated), PXC (PX C-terminal), RA (Ras-associating), RBD (Ras binding domain), RGS (regulator of G protein signaling), RUN (RPIP8, unc-14 and NESCA), SH3 (src homology 3) and TM (transmembrane) domains.
The regulation of membrane readers could conceivably occur by several different mechanisms. A variety of PIP kinases and phosphatases add and remove phosphates from the lipid’s inositol ring, and are located in different parts of the cell (Balla, 2013). However, altering PIP levels would influence many effector proteins rather than controlling individual players. Coincidence detection allows membrane readers to be recruited only when multiple partners can be simultaneously engaged (Carlton and Cullen, 2005). However, this does not address how the original protein which anchors a larger complex gathers on membranes in the first place. Proteins including those on membranes are downregulated by ubiquitination, but this can be a slow process (Vietri et al., 2020). Protein kinases and phosphatases regulate the lipid-binding activities of membrane readers by activating and deactivating PIP-stops (Lenoir et al., 2018; Kervin and Overduin, 2021). However, the possibility that other modifications control PIP-binding remains unexplored. The addition of metabolites to Arg and Lys residues is of particular interest as these residues are conserved and critical determinants of PIP recognition (Cheever et al., 2001).
Arginine and lysine sidechains are also frequent sites of covalent attachment of metabolites. These post-translational modifications (PTMs) include the addition of acetyl, butyryl, glucosyl, glutaryl, malonyl, methyl, and succinyl groups, which can have profound effects on diverse signaling processes (Figlia et al., 2020). These events are known to regulate protein–protein interactions and cellular metabolism as well as being correlated with progression of cancer and diabetes. However, their role in regulating protein-membrane interactions remains surprisingly unexplored.
Here we report a proteome-wide meta-study of the control of PX domains by metabolite-based modifications of Lys and Arg residues, revealing that such MET-stops are surprisingly frequent in membrane binding sites and are structurally geared to interfere with PIP interactions. Patterns in the distribution of MET-stops are similar to those observed with PIP-stops and are found across eukaryotes, suggesting a conserved developmental function. This supports a widespread role for metabolite modification in toggling PIP binding by membrane readers to regulate local biological events throughout the cell.
Materials and Methods
Comparing PX Domain Sequences
The sequences of human PX domain-containing proteins and their homologs were obtained from UniProt (UniProt, 2019). The sequences were aligned using Clustal Omega (Sievers and Higgins, 2018) and adjusted manually to line up structurally and functionally critical residues, and visualized using Jalview 2 (Waterhouse et al., 2009). Point mutations were identified from the cBioPortal (Cerami et al., 2012) and COSMIC (Tate et al., 2019) databases and references therein.
Membrane Site Definition
The 3D structures of PX domains were calculated with the I-TASSER program to perform structural analysis of the entire human superfamily (Yang et al., 2015). The model with the highest cluster size was chosen as the representative structure. The available PX domain structures in the RCSB Protein Data Bank (PDB) (Berman et al., 2000) were compared and the highest resolution entry with defined membrane binding residues was selected where there were several entries. The structural models were visualized using PyMOL and ICM (Neves et al., 2012) software, and residues that are membrane-interactive and modified were compared. Each PX domain’s membrane binding surface was predicted with the Membrane Optimal Docking Area (MODA) program (Kufareva et al., 2014). This generated a score for each residue in each structure to indicate its likelihood of interacting with membranes, with scores exceeding 30 predicting a probable role in membrane docking. The data from all human PX domains was compared to map the consensus membrane docking sites, which are located in the loop connecting the first and second β strands, in the third β strand and at the beginning of the first α helix, and in the loop encompassing the proline-rich (PR) element and second α helix. This consensus binding area was compared to the experimentally determined PIP-binding modes from NMR and crystal structures of PX domains bound to bicelles, micelles and phospholipids (Table 1). The individual MODA scores of all residues in all three consensus binding sites were added to generate each domain’s total MODA score. For NMR structures, a representative model of the ensemble was selected with MODA scores for such residues nearest to the mean. All PTMs were shown on the alignment and those occurring within the three sites were considered candidate MET-stops as they are positioned to compromise cognate membrane interactions.
Mapping Modifications
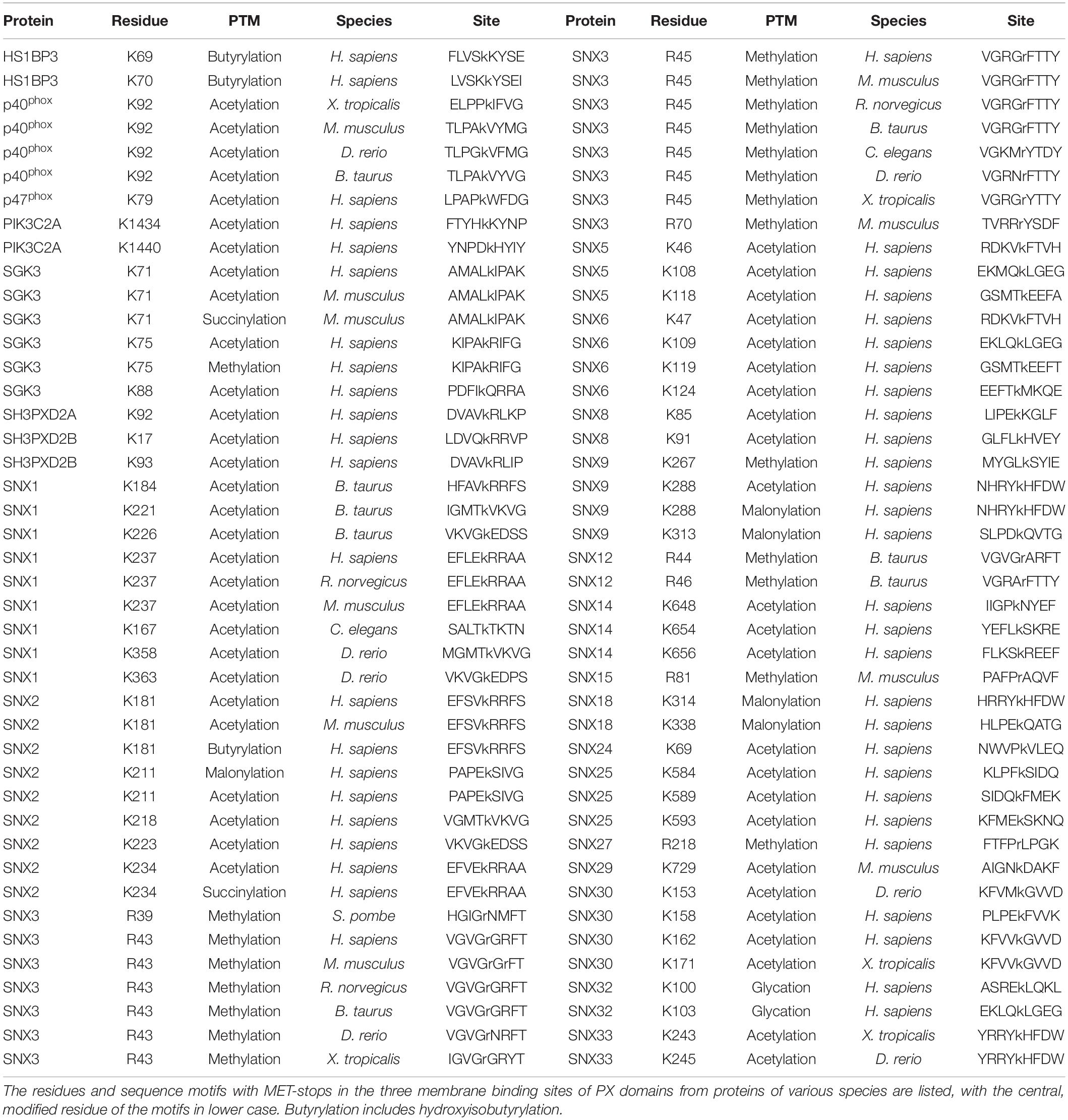
A standardized measure of confidence that membrane binding by a domain is modulated by metabolite-based modifications was needed. We propose a MET-stop score (MSS), which quantifies this likelihood based on existing data, and complements the PIP-stop score (PSS), which relates to Ser/Thr/Tyr phosphorylation occurrences in membrane binding sites (Lenoir et al., 2018; Kervin and Overduin, 2021). Relevant protein modifications based on experimental data were obtained from cBioportal (Cerami et al., 2012), dbPTM (Lee et al., 2006), iProteinDB (Hu et al., 2019), iPTMnet (Huang et al., 2018), PhosPhAT (Heazlewood et al., 2008), PhosphoGrid (Stark et al., 2010), PhosphoSite (Hornbeck et al., 2015), PLMD (Xu H. et al., 2017), PTMcode2 (Minguez et al., 2015), qPTM (Yu et al., 2019), SuperPhos databases (Lanz et al., 2021), and related studies (Hansen et al., 2019), providing broad proteome coverage. Each instance of a candidate MET-stop in the consensus membrane binding sites contributed to a domain’s total MSS value. These modified residues were also compared to the membrane binding residues identified in PIP-complexed PX domains of Grd19, p40phox, p47phox, SNX3, SNX9, and SNX11 PX domains (Bravo et al., 2001; Karathanassis et al., 2002; Zhou et al., 2003; Pylypenko et al., 2007; Stampoulis et al., 2012; Lenoir et al., 2018; Xu et al., 2020) to investigate whether they could compromise ligand recognition. To optimize the MSS we tested various sequence and spatial distance bounds around the MODA maxima and spans in which modified Lys or Arg residues can be considered candidate MET-stops. We opted for sequence alignment-based boundaries that encompass the binding elements consistently identified by MODA as this was determined to be the most accessible approach. All candidate MET-stops added to the MSS of a domain. Each unique study reporting a metabolite-based modification of a Lys or Arg residue in any of the three sites added 1 to the MSS, while modifications with 2–4 references added 2, and modifications with 5 or more references added 3. This scoring function is intended to reduce bias due to varying tissue-specific protein expression levels by integrating and standardizing PTM data from many studies. Manual curation of the collated data ensured that each original dataset is only cited once per modification. Hence the MSS indicates the relative frequency of metabolite-based modifications of a domain that could modulate membrane binding and allows for the comparison of Lys and Arg modifications with PIP-stops as well as PIP specificities and membrane affinities.
Lipid Specificity and Affinity
The ligand specificities of PX domains were quantified using a broadly applicable Lipid Specificity Index (LSI). A domain with relatively high affinity for only a single PIP was assigned a LSI value of 10. In contrast, PX domains that displayed no discernible binding to any PIP were given LSI values of 0. All other PX domains were given an intermediate LSI value equal to 10 minus 1 for each additional in-class PIP ligand and minus 2 for each additional PIP ligand not in their class. Each of the seven PIPs were divided into two classes depending on whether they had 1 or over 1 terminal phosphates, respectively, and PX domains were assigned to the same class as their predominate ligands. Hence a LSI of 1 indicates a perfectly non-specific domain that binds all seven PIPs. The data from all published reports of lipid binding by each PX domain were analyzed to generate a database of PX domain lipid specificities and membrane affinities. In cases where there was conflicting evidence, we gave precedence to quantitative data utilizing liposomes and lipid-binding assays. The subcellular localization of PX domains to organelle membranes was also considered as support for binding to relevant PIPs. The membrane affinity index (MAI) was used to classify the membranes affinities of PX domains as strong, weak and none (S, W, or N) based on evaluation of the relevant studies showing approximately nanomolar affinity, micromolar affinity or no binding to lipid bilayers containing cognate PIP ligands, respectively.
Statistical Analysis
Statistical tests were applied following the collection of MSS, PSS, LSI and expression levels for all human PX domains, as described above. Linear regression, correlation and paired t-tests were performed using JMP 15 (JMP Version, 2019) in order to investigate potential relationships between scores (P < 0.05). Candidate MET-stops and PIP-stops were assigned values from 1 to 3, LSI was scaled from 1 to 10 and average expression levels were obtained (GTEx Consortium, 2020). A linear regression was performed to calculate whether MSS or PSS could be predicted based on LSI, or whether MSS or PSS could be predicted based on expression level. An additional regression test was performed to see whether MSS could predict PSS in order to enhance our understanding of how metabolite-based modifications might be involved in membrane binding. Pairwise correlation tests were applied to further test for positive or negative associations between each set of variables, thus providing information about which variables might be influencing the ability of membrane protein readers to bind PIP ligands. Finally, a paired t-test was performed to determine whether the proportion of modified arginine and lysine residues is higher inside the membrane recognition sites of PX domains as opposed to outside these sites (P < 0.05).
Results
Structural Basis of Membrane Binding
The membrane binding and post-translational modification sites in the structures of all human PX domains were compared. This involved using I-TASSER to calculate 29 structures, all of which had acceptable qualities with average TM and confidence scores of 0.81 ± 0.09 and 0.38, respectively (Table 1). The remaining PX domains had complete 3D structures from previous NMR spectroscopy and X-ray crystallography studies. As expected, the PX structures converged well and display a common fold and membrane binding area (Cheever and Overduin, 2004; Figure 2). This common structural framework provides a straightforward basis for comparing the various lipid binding and regulatory properties.
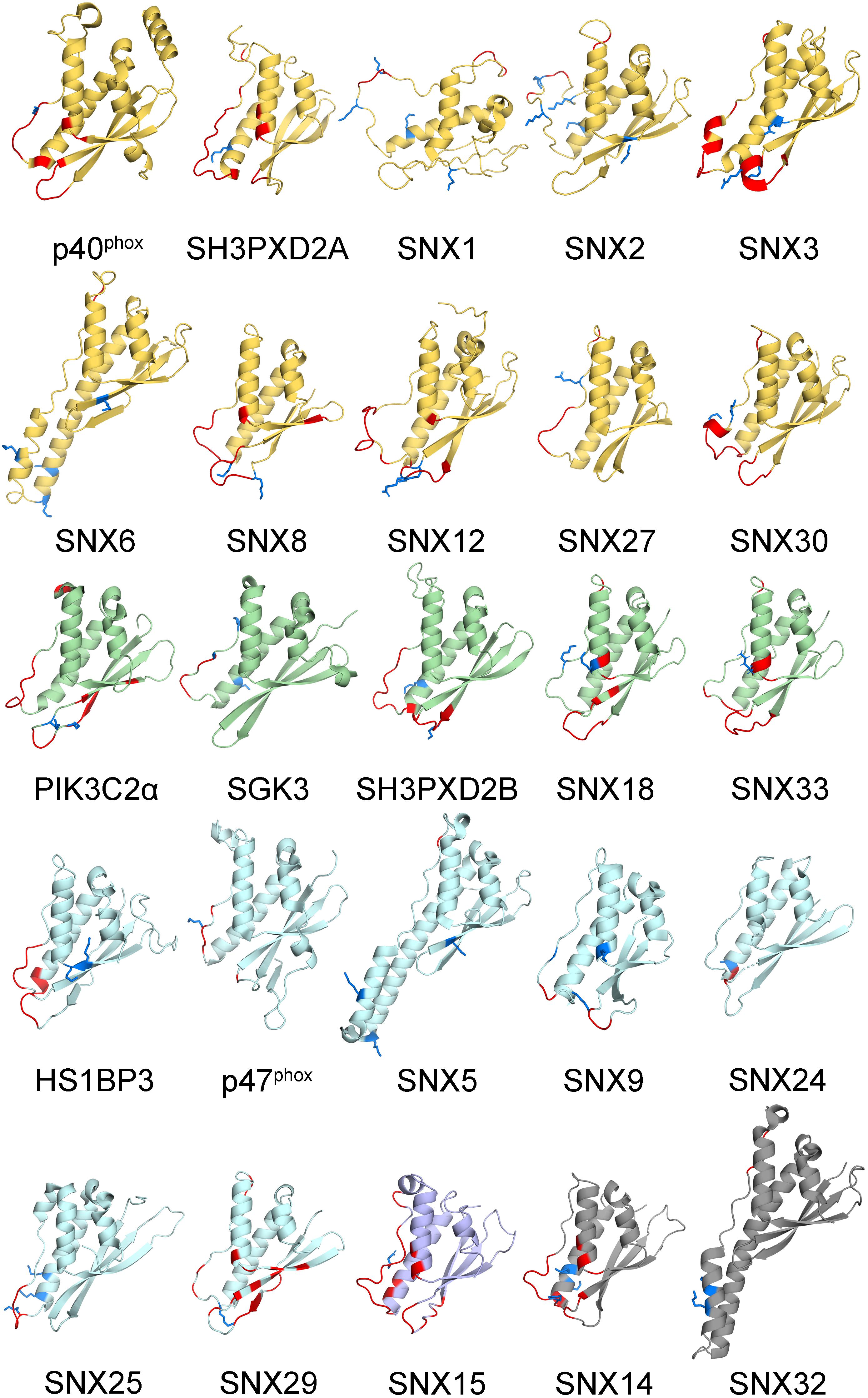
Figure 2. Human PX domain structures with MET-stops. Metabolite-modified Arg and Lys residues are shown with sidechains in blue. Membrane docking residues with MODA scores greater than 30 are shown in red. Backbone ribbon colors correspond to the specificity indices of 10 (yellow), 8–9 (green), 5–7 (light blue), 1–4 (purple), and non-binding (gray).
All human PX domains were categorized based on their PIP ligands using a lipid specificity index (LSI) in order to investigate possible relationships with metabolite-based PTMs (Table 1). The twenty domains which specifically recognize only a single type of membrane-bound PIP were assigned the maximum LSI value of 10. Three domains exhibit no detectable membrane binding activity and were given a LSI of 0. Those domains with intermediate lipid specificities were assigned LSI values equal to 10–n1–2n2, where n1 is the number of PIP ligands of the same class and n2 is the number of PIPs ligands of a different class (see section “Materials and Methods”). The LSI values of the PX domains of SNX30 and SNX33, which have not been experimentally determined, were inferred to be similar to that of SNX7 and SNX18, respectively, as they have the most similar sequences, binding motifs and multidomain structures. Altogether, this represents the best-defined and most diverse family of membrane readers in terms of PIP-binding, allowing us to explore which types of PIP interactions are likely to be affected by the various protein modifications.
In order to investigate how PIP recognition might be regulated, we structurally defined the relevant binding sites using the MODA algorithm, which pinpoints all residues in protein structures that are likely to interact with membranes (Kufareva et al., 2014). These sites were consistent with experimentally mapped contacts from complexed structures of PX domains of Grd19, p40phox, p47phox, SNX3 and SNX9 proteins (Bravo et al., 2001; Karathanassis et al., 2002; Zhou et al., 2003; Pylypenko et al., 2007; Stampoulis et al., 2012; Lenoir et al., 2018). Three elements that are commonly involved in binding PIP-containing membranes were identified. Site 1 is a membrane-insertion loop that connects the β1 and β2 strands, with the exception of SNX5, SNX6, and SNX32 where it instead forms a protein binding element (Paul et al., 2017). Site 2 contains the canonical regulatory PIP-stop that was discovered in SNX1, SNX3, and SNX12 (Lenoir et al., 2018). In the case of the PX domain of p47phox this site is particularly extensive, being capable of binding additional acidic phospholipids in addition to a PIP molecule (Karathanassis et al., 2002; Stampoulis et al., 2012; Takeuchi et al., 2012). Site 3 spans a long irregular loop that can form a secondary PIP-binding site (Chandra et al., 2019) and includes a proline rich element (PRE), which can also serve as a SH3 domain docking site (Hiroaki et al., 2001; Ago et al., 2003). These three sites constitute a common PIP-binding surface that is found across the superfamily and includes dynamic loops and an accessible set of basic and hydrophobic sidechains. These features are known to attract proteins to membranes where they stereospecifically recognize PIP headgroups and insert stably into phospholipid bilayers, thus endowing PX domains with the ability to read PI codes.
Analysis of Membrane Binding Properties
Membrane reader function is associated with a domain’s affinity and specificity for lipid ligands including PIP molecules and is subject to regulatory influences. The ligand affinities of PX domains varies widely, which is to be expected given their diverse biological roles and multidomain architectures (Figure 1). Each PX domain was assigned to the membrane affinity index (MAI) based on the available literature. There are 31, 15 and 3 PX domains with strong, weak and no apparent affinity for membranes, respectively. PIP specificity diverges across the family, with 20, 11, 13, and 2 PX domains that exhibit absolute, high, medium and low membrane selectivity, respectively, based on LSI values of 10, 9–8, 7–5, and 4–1. The lipid specificities and membrane affinities of human PX domains are not significantly correlated based on the currently available experimental data. This indicates that these properties are essentially independent dimensions of function which are tailored to various protein, pathway and organelle needs. We next explored whether these functional properties could be subject to control by various types of PTMs.
Modifications of Membrane Binding Sites
The discovery that phosphorylating a conserved serine in the PIP binding sites of SNX1, SNX3, and SNX12 PX domains blocks membrane recruitment led to the concept of a PIP-stop (Lenoir et al., 2018; Kervin and Overduin, 2021). These are residues that, when phosphorylated, compromise binding to PIP-containing membranes. While this concept has since been independently validated with SNX1 (Feng et al., 2020), it has not been extended to other types of modifications, prompting a deeper analysis here. Mapping of all the known acetylation, butyrylation, glycation, malonylation, methylation and succinylation sites to the PX structures revealed that there are 87 residues in membrane-binding sites with such modifications, of which 51 are found in human proteins (Table 2).
Residues that directly contact lipid headgroups and insert into membrane mimics are heavily modified, including in the membrane insertion loop (MIL), PIP-binding RRY motif in the β3 strand, KxLF motif after the PRE, and RR element in the α2 helix (Figure 3). Modifications in these places would directly interfere with hydrogen bonding and electrostatic interactions of Arg and Lys residues that normally mediate phospholipid recognition and membrane attraction while introducing steric impediments to lipid headgroup docking.
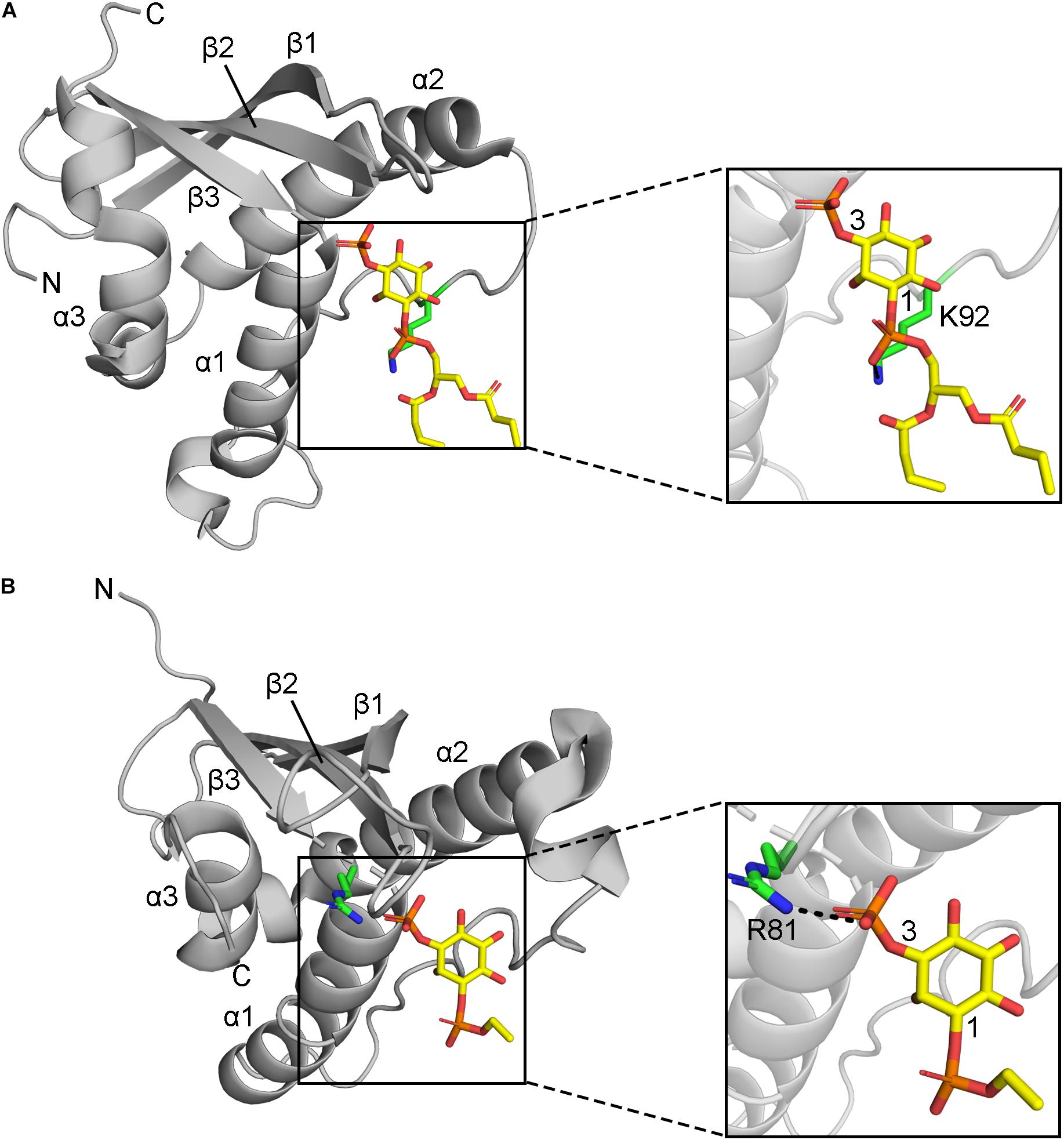
Figure 3. Positions of MET-stop residues in PIP-complexed PX domain structures. (A) The p40phox K92 sidechain in Site 3 hydrogen bonds with the 1-phosphate and is acetylated. (B) Grd19 R81 sidechain in Site 2 hydrogen bonds with the 3-phosphate in PI3P and the equivalent residue in SNX3 is methylated.
A paired t-test was performed to determine if there is a significant difference in the relative proportion of modified arginine and lysine residues within membrane binding sites compared to those outside such sites (Figure 4B). The data revealed that the mean proportion of modified residues is significantly higher inside sites than outside sites (t = –3.2360, df = 49, P = 0.0022). Arginine modifications are rarely reported, yet are found in sorting nexins 3, 12, and 15 (which do not exhibit any lysine modifications) and are exclusively located in guanidinium groups which are positioned to contact phospholipid headgroups directly. In contrast lysine acetylations are found in other PX domains where they are more widely distributed, while protein phosphorylations offer more frequent modifications that are typically adjacent to points of stereospecific PIP contacts. Thus, we propose that each residue-specific modification offers a differentiated handle for complementary erasure of membrane reader functions.
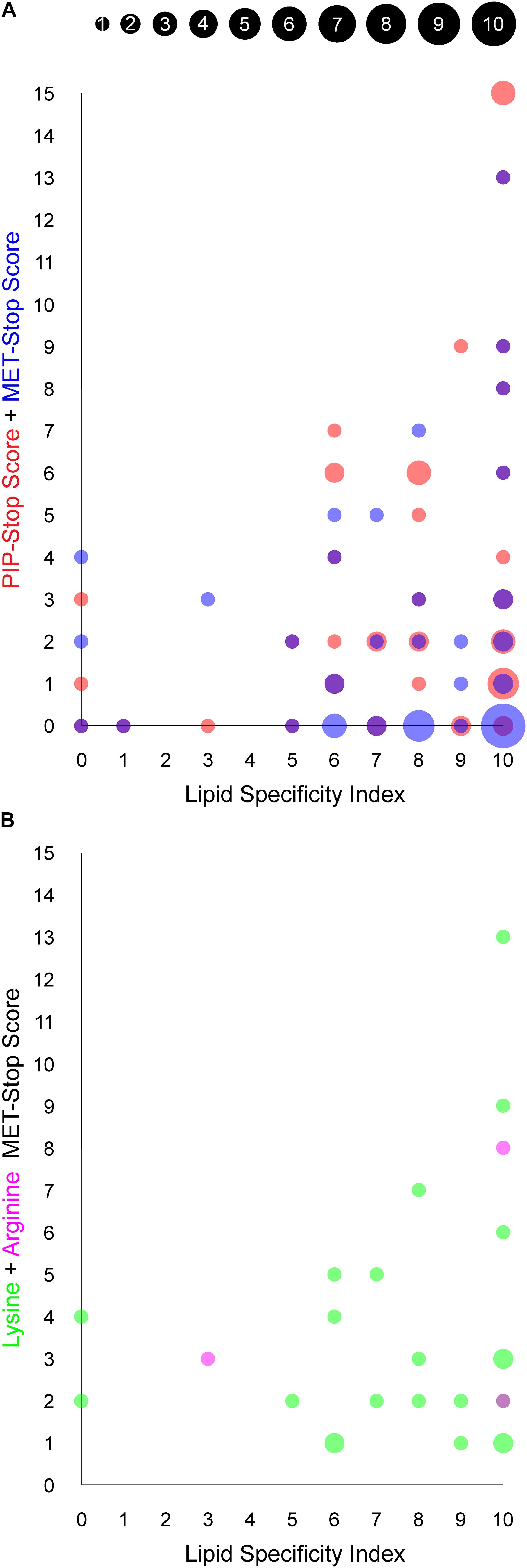
Figure 4. Relationship between PIP binding specificity and post-translational modifications of PX domains. The area of each circle is proportional to the number of PX domains that occupy the position of the circle, as indicated in the inset and are based on values in Table 1. (A) The PIP-stop score (PSS) and MET-stop score (MSS) of each human PX domain is plotted against its Lipid Specificity Index (LSI), shown in red and blue, respectively, while overlapping circles appear purple. (B) The contributions to the MSS from lysine and arginine modifications for each PX domain, discounting scores of zero, are shown in green and magenta, respectively, while overlapping circles appear purple.
The membrane binding sites of PX domains are modified in many species, suggesting conserved regulatory mechanisms. For example, the RGR motif in the β1-β2 loop of SNX3 is heavily methylated in human, mouse and rat cells and also modified in frog, bull, nematode, yeast and zebrafish homologs. The conserved PIP-coordinating Lys residue after the PRE is modified in p40phox, SGK3, SNX2, SNX9, SNX18, and SNX30, while Lys residues N-terminal to the PIP-binding RR motif in α2 are modified in SGK3, SH3PXD2A, SH3PXD2B, SNX1, SNX2, SNX6, SNX14, and SNX25. Hence membrane binding elements and proximal residues appear to be preferentially modified throughout evolution. This suggests a longstanding mechanism to regulate to PX domains by abolishing or weakening canonical PIP-membrane recognition through metabolite-based inhibition.
Influences on Modification Frequency
We explored whether Arg and Lys modifications in PX domains are related to PIP-stops and ligand specificities using the MSS, PSS and LSI parameters, respectively. Higher MSS values appear visually to be concentrated in PX domains with high LSI values, reminiscent of the pattern observed with PSS and LSI (Kervin and Overduin, 2021, Figure 4A). A significant linear regression equation was found between MSS and PSS [F(1, 48) = 8.3586, P = 0.0058, R2 = 0.148311], where the predicted PSS is equal to 2.5472 + 0.5939 (MSS). A correlation test also indicated that MSS and PSS are related [r(48) = 0.3851, P = 0.0058]. Together this suggests that cells may generally be inclined to regulate specific PX domains with both MET-stop and PIP-stop mechanisms.
Although PIP-stops are more likely to be found in PX domains with high specificity, a pronounced relationship between MET-stops and lipid specificity has not yet emerged. A significant linear regression equation was found between PSS and LSI [F(1, 47) = 5.1535, P = 0.0278, R2 = 0.0988], where the predicted PSS is equal to 0.0904 + 0.4735 (LSI), indicating that lipid specificity is a predictor of PIP-stops. In contrast, a linear regression equation relating MSS and LSI indicates that MET-stops are not related to lipid specificity (P > 0.05). Likewise, correlation tests indicated that PSS and LSI are related [r(48) = 0.3143, P = 0.0278] while MSS and LSI are not (r = 0.0809, P = 0.5803). A caveat is that MET-stops are 2.5-fold less abundant than PIP-stops. Hence their analysis would benefit by inclusion of richer data on such membrane reader modifications. Arg-based MET-stops are particularly rare yet are highly abundant in SNX3 and are conserved in SNX12. This suggests that arginine methylation may have evolved to control whether SNX3 and its close relative SNX12 localize to endosomal membranes, while lysine modifications could serve to regulate PX domains more broadly inside the cell with less regard for lipid specificity. Thus both Arg- and Lys-based of MET-stops appear to differentially complement PIP-stops, which serve to more frequently and selectively down-regulate the most PIP-specific domains.
Several factors could influence susceptibility of membrane readers to metabolite modification. The highest MSS values were found in SGK3 and sorting nexins 1, 2, 3, and 6, which selectively recognize only PI3P, PI4P or PI(3,4)P2 lipids, suggesting that endosomal and Golgi compartments are likely locations for metabolite attachment. The PX proteins that are uniquely plasma membrane localized, that is, NOXO1β, PLD1 and PLD2, lack any reported MET-stops in their PX domains, suggesting that this environment may be less fertile for such modifications. Two PX proteins, SNX14 and SNX32, do not bind membranes yet contain candidate MET-stops and PIP-stops, suggesting that some of these PTMs may influence events other than PIP-mediated membrane interactions. A further 19 PX domains exhibit intermediate MET-stop scores and display a wide range of PIP specificities, although all visit endocytic routes. In contrast, 20 membrane-binding PX domains do not exhibit known metabolite-based modifications, with 17 of these containing PIP-stops instead. The remainder including sorting nexins 10 and 11 may not be regulated by such modifications in vivo or could mediate membrane interactions that are constitutive or dependent on coincidence detection.
The broad dynamic range of the expression of PX domain-containing proteins is also a factor. We found that their average mRNA expression levels, as shown in Table 1, could be predicted by both MSS [F(1, 48) = 7.3181, P = 0.0094, R2 = 0.1323] and PSS [F(1, 48) = 14.7109, P = 0.0004, R2 = 0.2346]. The predicted expression is equal to either 16.9500 + 3.9156 (MSS) or 12.3204 + 3.2080 (PSS). Correlation tests further revealed that expression is related to both MSS [r(48) = 0.3637, P = 0.0094] and PSS [r(48) = 0.4843, P = 0.0004]. This suggests that cells tend to regulate their most highly expressed PX domains through such PTMs, although this relationship is not absolute. For example, the PX domains of highly expressed NISCH and SNX17 do not exhibit any discernable MET-stops despite contributing to endocytic pathways, although they do carry PIP-stops. In contrast, the highly expressed PXDC1 lacks a known function or any such PTMs yet is an established tumor marker in endothelial cells (St Croix et al., 2000). Thus, the presence of a PTM that perturbs a lipid binding site is influenced by not only the activity of the responsible enzymes but also membrane reader expression level and co-localization in the context of a network of regulatory pathways that are only beginning to be mapped and understood.
Discussion
The discovery of MET-stops exposes a new dimension of ways by which cells could regulate their diverse membrane interactions. These events are carried out by enzymes including methyl and acyl transferases, while a complementary set of demethylases and deacetylases such as HDACs and sirtuins can remove such modifications. While historically these PTMs have been thought to regulate histone interactions with DNA (Figlia et al., 2020), here we show that they are also frequently positioned to regulate recognition of PI codes by a large number of membrane readers. Undoubtably many more pathways are governed by such mechanisms, although some modifications will represent metabolic noise with no functional consequences, necessitating careful structure-based assessment. The preferential modification of lipid binding sites revealed here suggests broad selective pressure to control PIP recognition, with membrane reader sites representing significant biological targets. This is consistent with earlier studies showing that such modifications control the subcellular localization and activity of proteins (Sundaresan et al., 2011; Thandapani et al., 2013; Su et al., 2017). In addition to metabolite addition, ubiquitination of lysine residues also occurs in PX domains and is found predominantly in and C-terminal to membrane binding Site 3. While generally thought to maintain quality control of proteins including sorting nexins (Hanley and Cooper, 2020), such ubiquitin-based modifications would also alter lipid interactions via proximal membrane docking surfaces. The presence of a terminal acetyl group prevents ubiquination from occurring on a lysine and promotes protein stability (Caron et al., 2005). Thus MET-stops could promote the formation of relatively long-lived reservoirs of negatively regulated PI readers which are dislodged from membrane surfaces, although functional studies would be needed to validate this hypothesis.
The putative regulatory mechanism mediated by MET-stops appears to be highly conserved across many membrane readers. For example, the presence of a conserved and methylated glycine-arginine-rich (GAR) motif (Thandapani et al., 2013) in SNX3 homologs (Table 2) could infer the presence of an ancient and critical switch. The most recurrent MET-stop in human PX domains occurs at a Lys residue immediately C-terminal to the PRE (Figure 5), indicating that this is a dominant regulatory feature of the superfamily. Interestingly this feature has been supplanted by an Arg-based MET-stop in SNX15, suggesting regulatory convergence. The diversity of modification types across the three Sites of this superfamily suggests that several enzymatic pathways evolved in parallel to directly toggle residues responsible for lipid recognition. While this could be taken to suggest redundant control of crucial PI reader functions, the complexity of the actual patterns indicate complementary roles of each distinct enzyme-mediated regulatory pathway.
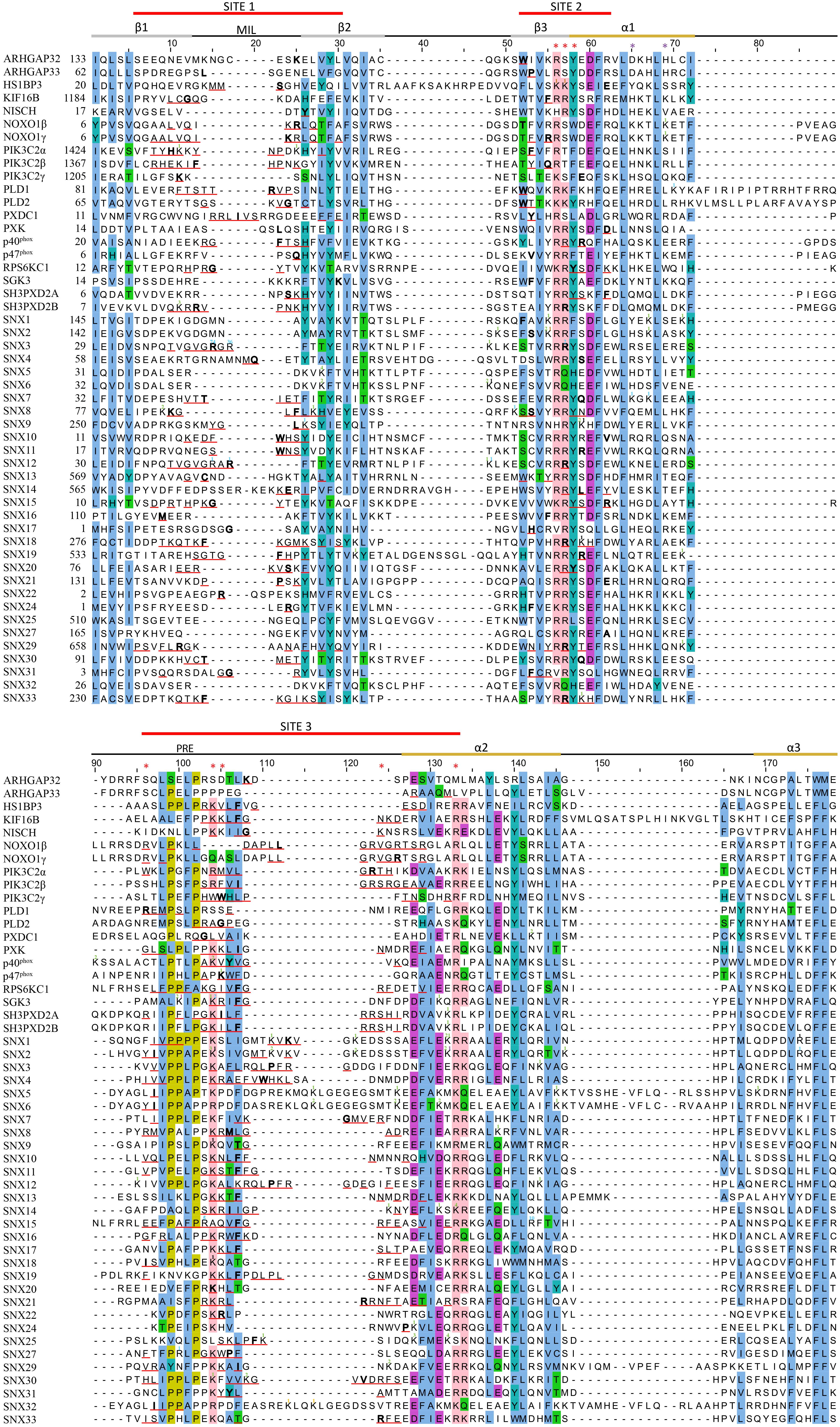
Figure 5. Sequence alignment of all human PX domains. Citations of acetylated, butyrylated, glycated, malonylated, methylated and succinylated residues are indicated with a green, brown, orange, black, blue and purple superscript, respectively. The secondary structure, membrane insertion loop (MIL) and proline rich element (PRE) are indicated below the three red-lined membrane docking sequences. PIP headgroup and phosphatidic acid binding positions are indicated with red and purple asterisks, respectively. The residue with the highest MODA score in each site is bolded. Aligned conserved hydrophobic, proline, polar, acidic, and basic residues are highlighted in blue, olive, green, purple, and pink.
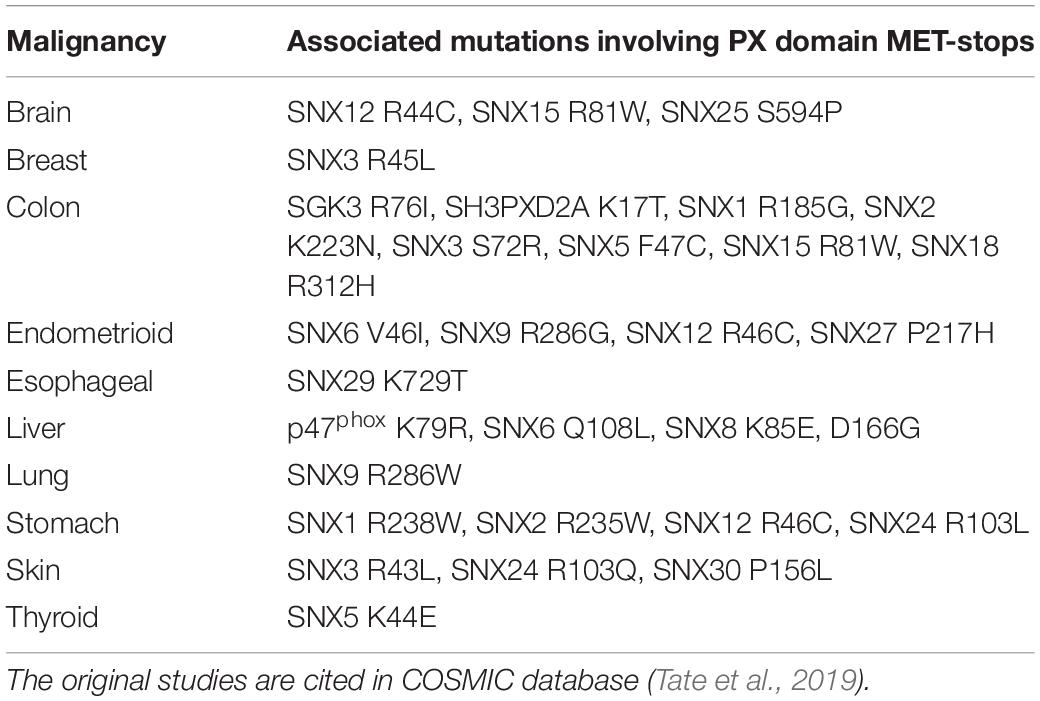
There are likely candidates responsible for MET-stop activation. These include protein arginine methyltransferases such as PRMT8, which localizes to the plasma membrane through myristoylation (Toma-Fukai et al., 2016), as well as lysine acetylases that are known to block membrane interactions of the Akt kinase by modifying its PH domain (Sundaresan et al., 2011). Proteins that promote lysine acetylation and organelle biogenesis engage sorting nexins at endosomes in plants and mammals, suggesting longstanding interactions (Zhang et al., 2014). The enzymes controlling these PTMs are emerging as cancer targets, and with clinical trials of inhibitors underway, there is a need to clarify their mechanisms of action (Dang and Wei, 2021; Samuel et al., 2021). The data presented here indicate that negative regulation of endosome recruitment of SNX3 and its associated retromer assembly (Lucas et al., 2016; Leneva et al., 2021) would be blocked by methyl transferase inhibitors, while a broad range of membrane reader interactions could be affected by acetyltransferase and deacetylase inhibitors. Disease-linked mutations found at MET-stop residues now have more predictable effects. For example, various mutations in human PX domains have been associated with malignancies (Tate et al., 2019; Table 3). As these point mutations alter MET-stop motifs they are positioned to block or deregulate PIP binding activity, inferring potential signaling or trafficking defects that could contribute to pathogenic effects.
Other domains and proteins may be impacted by the discovery of MET-stops. Larger assemblies including those with pathogen proteins (Paul et al., 2017) and retromers (Lucas et al., 2016; Leneva et al., 2021) are anchored by PX domain binding to PIPs and could be dislodged. Multi-subunit complexes are also influenced by phosphorylation events that alter protein-protein interactions (Ago et al., 2003), and some of the metabolite-based PTMs identified here could also affect protein-protein interactions rather than solely influencing membrane binding. The presence of so many MET-stops in the PX superfamily suggests that thousands of other membrane readers (Overduin and Kervin, submitted) could also utilize such regulatory mechanisms, including the hundreds of FYVE and PH domains that also recognize PIPs (Lenoir et al., 2015; Eitzen et al., 2019). While their unique folds, binding sites and lipid specificities will require further analysis, it appears that direct control of PI code readers by a growing variety of specialized PTMs could provide a unifying principle for ensuring high-fidelity membrane recognition in any eukaryotic cell.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the methods section of this article.
Author Contributions
BCW, MO, and TAK collected, analyzed the data, and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by NSERC Discovery Grant (RGPIN-2018-04994) and Campus Alberta Innovates Program (#RCP-12-002C) grants to MO, a BioTalent Canada student work placement program grant to BCW and MO and an Alberta Innovates Summer Research Studentship to TAK.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Molsoft LLC for making the ICM program (MODA’s engine) and MODA available, and Catharine Trieber, Gwen Craddock, Irina Kufareva, Jean Gruenberg, Jeannette Kunz, Marc Lenoir and Ruben Abagyan for discussions.
References
Abram, C. L., Seals, D. F., Pass, I., Salinsky, D., Maurer, L., Roth, T. M., et al. (2003). The adaptor protein fish associates with members of the ADAMs family and localizes to podosomes of Src-transformed cells. J. Biol. Chem. 278, 16844–16851. doi: 10.1074/jbc.m300267200
Ago, T., Kuribayashi, F., Hiroaki, H., Takeya, R., Ito, T., Kohda, D., et al. (2003). Phosphorylation of p47phox directs phox homology domain from SH3 domain toward phosphoinositides, leading to phagocyte NADPH oxidase activation. Proc. Natl. Acad. Sci. U. S. A. 100, 4474–4479. doi: 10.1073/pnas.0735712100
Ago, T., Takeya, R., Hiroaki, H., Kuribayashi, F., Ito, T., Kohda, D., et al. (2001). The PX domain as a novel phosphoinositide- binding module. Biochem. Biophys. Res. Commun. 287, 733–738. doi: 10.1006/bbrc.2001.5629
Almendinger, J., Doukoumetzidis, K., Kinchen, J. M., Kaech, A., Ravichandran, K. S., and Hengartner, M. O. (2011). A conserved role for SNX9-family members in the regulation of phagosome maturation during engulfment of apoptotic cells. PLoS One. 6:e18325. doi: 10.1371/journal.pone.0018325
Balla, T. (2013). Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol. Rev. 93, 1019–1137. doi: 10.1152/physrev.00028.2012
Banerjee, S., Basu, S., and Sarkar, S. (2010). Comparative genomics reveals selective distribution and domain organization of FYVE and PX domain proteins across eukaryotic lineages. BMC Genomics. 11:83. doi: 10.1186/1471-2164-11-83
Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., et al. (2000). The Protein Data Bank. Nucleic Acids Res. 28, 235–242.
Blatner, N. R., Wilson, M. I., Lei, C., Hong, W., Murray, D., Williams, R. L., et al. (2007). The structural basis of novel endosome anchoring activity of KIF16B kinesin. EMBO J. 26, 3709–3719. doi: 10.1038/sj.emboj.7601800
Bravo, J., Karathanassis, D., Pacold, C. M., Pacold, M. E., Ellson, C. D., Anderson, K. E., et al. (2001). The crystal structure of the PX domain from p40(phox) bound to phosphatidylinositol 3-phosphate. Mol. Cell. 8, 829–839. doi: 10.1016/s1097-2765(01)00372-0
Buschman, M. D., Bromann, P. A., Cejudo-Martin, P., Wen, F., Pass, I., and Courtneidge, S. A. (2009). The novel adaptor protein Tks4 (SH3PXD2B) is required for functional podosome formation. Mol. Biol. Cell. 20, 1302–1311. doi: 10.1091/mbc.e08-09-0949
Carlton, J. G., and Cullen, P. J. (2005). Coincidence detection in phosphoinositide signaling. Trends Cell Biol. 15, 540–547. doi: 10.1016/j.tcb.2005.08.005
Carlton, J. G., Bujny, M. V., Peter, B. J., Oorschot, V. M., Rutherford, A., Arkell, R. S., et al. (2005). Sorting nexin-2 is associated with tubular elements of the early endosome, but is not essential for retromer-mediated endosome-to-TGN transport. J. Cell Sci. 118, 4527–4539. doi: 10.1242/jcs.02568
Carlton, J., Bujny, M., Peter, B. J., Oorschot, V. M., Rutherford, A., Mellor, H., et al. (2004). Sorting nexin-1 mediates tubular endosome-to-TGN transport through coincidence sensing of high- curvature membranes and 3-phosphoinositides. Curr. Biol. 14, 1791–1800. doi: 10.1016/j.cub.2004.09.077
Caron, C., Boyault, C., and Khochbin, S. (2005). Regulatory cross-talk between lysine acetylation and ubiquitination: role in the control of protein stability. Bioessays 27, 408–415. doi: 10.1002/bies.20210
Catimel, B., Schieber, C., Condron, M., Patsiouras, H., Connolly, L., Catimel, J., et al. (2008). The PI(3,5)P2 and PI(4,5)P2 interactomes. J. Proteome Res. 7, 5295–5313. doi: 10.1021/pr800540h
Catimel, B., Yin, M. X., Schieber, C., Condron, M., Patsiouras, H., Catimel, J., et al. (2009). PI(3,4,5)P3 Interactome. J. Proteome Res. 8, 3712–3726. doi: 10.1021/pr900320a
Ceccato, L., Chicanne, G., Nahoum, V., Pons, V., Payrastre, B., Gaits-Iacovoni, F., et al. (2016). PLIF: A rapid, accurate method to detect and quantitatively assess protein-lipid interactions. Sci. Signal. 9:rs2. doi: 10.1126/scisignal.aad4337
Cerami, E., Gao, J., Dogrusoz, U., Gross, B. E., Sumer, S. O., Aksoy, B. A., et al. (2012). The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404. doi: 10.1158/2159-8290.cd-12-0095
Chandra, M., Chin, Y. K., Mas, C., Feathers, J. R., Paul, B., Datta, S., et al. (2019). Classification of the human phox homology (PX) domains based on their phosphoinositide binding specificities. Nat. Commun. 10:1528.
Cheever, M. L., and Overduin, M. (2004). “PX domains,” in Modular Protein Domains, eds G. Cesareni, M. Gimona, M. Sudol, and M. Yaffe (Wiley), 389–408. doi: 10.1002/3527603611.ch19
Cheever, M. L., Sato, T. K., de Beer, T., Kutateladze, T. G., Emr, S. D., and Overduin, M. (2001). Phox domain interaction with PtdIns(3)P targets the Vam7 t-SNARE to vacuole membranes. Nat. Cell Biol. 3, 613–618. doi: 10.1038/35083000
Cheng, G., and Lambeth, J. D. (2004). NOXO1, regulation of lipid binding, localization, and activation of Nox1 by the Phox homology (PX) domain. J. Biol. Chem. 279, 4737–4742. doi: 10.1074/jbc.m305968200
Chiang, S. H., Hwang, J., Legendre, M., Zhang, M., Kimura, A., and Saltiel, A. R. (2003). TCGAP, a multidomain Rho GTPase-activating protein involved in insulin-stimulated glucose transport. EMBO J. 22, 2679–2691. doi: 10.1093/emboj/cdg262
Clairfeuille, T., Norwood, S. J., Qi, X., Teasdale, R. D., and Collins, B. M. (2015). Structure and Membrane Binding Properties of the Endosomal Tetratricopeptide Repeat (TPR) Domain-containing Sorting Nexins SNX20 and SNX21. J. Biol. Chem. 290, 14504–14517. doi: 10.1074/jbc.m115.650598
Cozier, G. E., Carlton, J., McGregor, A. H., Gleeson, P. A., Teasdale, R. D., Mellor, H., et al. (2002). The phox homology (PX) domain-dependent, 3-phosphoinositide-mediated association of sorting nexin-1 with an early sorting endosomal compartment is required for its ability to regulate epidermal growth factor receptor degradation. J. Biol. Chem. 277, 48730–48736. doi: 10.1074/jbc.m206986200
Czubayko, M., Knauth, P., Schluter, T., Florian, V., and Bohnensack, R. (2006). Sorting nexin 17, a non-self-assembling and a PtdIns(3)P high class affinity protein, interacts with the cerebral cavernous malformation related protein KRIT1. Biochem. Biophys. Res. Commun. 345, 1264–1272. doi: 10.1016/j.bbrc.2006.04.129
Dang, F., and Wei, W. (2021). Targeting the acetylation signaling pathway in cancer therapy. Semin. Cancer Biol. doi: 10.1016/j.semcancer.2021.03.001 [Epub Online ahead of print].
Danson, C., Brown, E., Hemmings, O. J., McGough, I. J., Yarwood, S., Heesom, K. J., et al. (2013). SNX15 links clathrin endocytosis to the PtdIns3P early endosome independently of the APPL1 endosome. J. Cell Sci. 126, 4885–4899.
Davis, N. Y., McPhail, L. C., and Horita, D. A. (2012). The NOXO1beta PX domain preferentially targets PtdIns(4,5)P2 and PtdIns(3,4,5)P3. J. Mol. Biol. 417, 440–453. doi: 10.1016/j.jmb.2012.01.058
Du, G., Altshuller, Y. M., Vitale, N., Huang, P., Chasserot-Golaz, S., Morris, A. J., et al. (2003). Regulation of phospholipase D1 subcellular cycling through coordination of multiple membrane association motifs. J. Cell Biol. 162, 305–315. doi: 10.1083/jcb.200302033
Dyve, A. B., Bergan, J., Utskarpen, A., and Sandvig, K. (2009). Sorting nexin 8 regulates endosome-to-Golgi transport. Biochem. Biophys. Res. Commun. 390, 109–114. doi: 10.1016/j.bbrc.2009.09.076
Eitzen, G., Smithers, C. C., Murray, A. G., and Overduin, M. (2019). Structure and function of the Fgd family of divergent FYVE domain proteins (1). Biochem. Cell Biol. 97, 257–264. doi: 10.1139/bcb-2018-0185
Ellson, C. D., Gobert-Gosse, S., Anderson, K. E., Davidson, K., Erdjument-Bromage, H., Tempst, P., et al. (2001). PtdIns(3)P regulates the neutrophil oxidase complex by binding to the PX domain of p40(phox). Nat. Cell Biol. 3, 679–682. doi: 10.1038/35083076
Feng, Z., Kovalev, N., and Nagy, P. D. (2020). Key interplay between the co-opted sorting nexin-BAR proteins and PI3P phosphoinositide in the formation of the tombusvirus replicase. PLoS Pathog. 16:e1009120. doi: 10.1371/journal.ppat.1009120
Figlia, G., Willnow, P., and Teleman, A. A. (2020). Metabolites Regulate Cell Signaling and Growth via Covalent Modification of Proteins. Dev. Cell. 54, 156–170. doi: 10.1016/j.devcel.2020.06.036
Ghai, R., Mobli, M., Norwood, S. J., Bugarcic, A., Teasdale, R. D., King, G. F., et al. (2011). Phox homology band 4.1/ezrin/radixin/moesin-like proteins function as molecular scaffolds that interact with cargo receptors and Ras GTPases. Proc. Natl. Acad. Sci. U. S. A. 108, 7763–7768. doi: 10.1073/pnas.1017110108
GTEx Consortium (2020). The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369, 1318–1330. doi: 10.1126/science.aaz1776
Haberg, K., Lundmark, R., and Carlsson, S. R. (2008). SNX18 is an SNX9 paralog that acts as a membrane tubulator in AP-1-positive endosomal trafficking. J. Cell Sci. 121, 1495–1505. doi: 10.1242/jcs.028530
Han, K., Pastor, R. W., and Fenollar-Ferrer, C. (2020). PLD2-PI(4,5)P2 interactions in fluid phase membranes: Structural modeling and molecular dynamics simulations. PLoS One. 15:e0236201. doi: 10.1371/journal.pone.0236201
Hanley, S. E., and Cooper, K. F. (2020). Sorting Nexins in Protein Homeostasis. Cells 10:17. doi: 10.3390/cells10010017
Hansen, B. K., Gupta, R., Baldus, L., Lyon, D., Narita, T., Lammers, M., et al. (2019). Analysis of human acetylation stoichiometry defines mechanistic constraints on protein regulation. Nat. Commun. 10:1055.
Hanson, B. J., and Hong, W. (2003). Evidence for a role of SNX16 in regulating traffic between the early and later endosomal compartments. J. Biol. Chem. 278, 34617–34630. doi: 10.1074/jbc.m300143200
Hayashi, S., Okada, T., Igarashi, N., Fujita, T., Jahangeer, S., and Nakamura, S. (2002). Identification and characterization of RPK118, a novel sphingosine kinase-1-binding protein. J. Biol. Chem. 277, 33319–33324. doi: 10.1074/jbc.m201442200
Hayashi, T., Okabe, T., Nasu-Nishimura, Y., Sakaue, F., Ohwada, S., Matsuura, K., et al. (2007). PX-RICS, a novel splicing variant of RICS, is a main isoform expressed during neural development. Genes Cells 12, 929–939. doi: 10.1111/j.1365-2443.2007.01101.x
Heazlewood, J. L., Durek, P., Hummel, J., Selbig, J., Weckwerth, W., Walther, D., et al. (2008). PhosPhAt: a database of phosphorylation sites in Arabidopsis thaliana and a plant-specific phosphorylation site predictor. Nucleic Acids Res. 36, D1015–D1021.
Hiroaki, H., Ago, T., Ito, T., Sumimoto, H., and Kohda, D. (2001). Solution structure of the PX domain, a target of the SH3 domain. Nat. Struct. Biol. 8, 526–530.
Holland, P., Knaevelsrud, H., Soreng, K., Mathai, B. J., Lystad, A. H., Pankiv, S., et al. (2016). HS1BP3 negatively regulates autophagy by modulation of phosphatidic acid levels. Nat. Commun. 7:13889.
Hornbeck, P. V., Zhang, B., Murray, B., Kornhauser, J. M., Latham, V., and Skrzypek, E. (2015). PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 43, D512–D520.
Hu, Y., Sopko, R., Chung, V., Foos, M., Studer, R. A., Landry, S. D., et al. (2019). iProteinDB: An Integrative Database of Drosophila Post-translational Modifications. G3 9, 1–11. doi: 10.1534/g3.118.200637
Huang, H., Arighi, C. N., Ross, K. E., Ren, J., Li, G., Chen, S. C., et al. (2018). iPTMnet: an integrated resource for protein post-translational modification network discovery. Nucleic Acids Res. 46, D542–D550.
Kanai, F., Liu, H., Field, S. J., Akbary, H., Matsuo, T., Brown, G. E., et al. (2001). The PX domains of p47phox and p40phox bind to lipid products of PI(3)K. Nat. Cell Biol. 3, 675–678. doi: 10.1038/35083070
Karathanassis, D., Stahelin, R. V., Bravo, J., Perisic, O., Pacold, C. M., Cho, W., et al. (2002). Binding of the PX domain of p47phoxto phosphatidylinositol 3, 4-bisphosphate and phosphatidic acid is masked by an intramolecular interaction. EMBO J. 21, 5057–5068. doi: 10.1093/emboj/cdf519
Kervin, T. A., and Overduin, M. (2021). Regulation of the Phosphoinositide Code by Phosphorylation of Membrane Readers. Cells 10:1205. doi: 10.3390/cells10051205
Knauth, P., Schluter, T., Czubayko, M., Kirsch, C., Florian, V., Schreckenberger, S., et al. (2005). Functions of sorting nexin 17 domains and recognition motif for P-selectin trafficking. J. Mol. Biol. 347, 813–825. doi: 10.1016/j.jmb.2005.02.004
Koharudin, L. M., Furey, W., Liu, H., Liu, Y. J., and Gronenborn, A. M. (2009). The phox domain of sorting nexin 5 lacks phosphatidylinositol 3-phosphate (PtdIns(3)P) specificity and preferentially binds to phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2). J. Biol. Chem. 284, 23697–23707. doi: 10.1074/jbc.m109.008995
Kufareva, I., Lenoir, M., Dancea, F., Sridhar, P., Raush, E., Bissig, C., et al. (2014). Discovery of novel membrane binding structures and functions. Biochem. Cell Biol. 92, 555–563. doi: 10.1139/bcb-2014-0074
Lanz, M. C., Yugandhar, K., Gupta, S., Sanford, E. J., Faca, V. M., Vega, S., et al. (2021). In-depth and 3-dimensional exploration of the budding yeast phosphoproteome. EMBO Rep. 22:e51121.
Lee, J. S., Kim, J. H., Jang, I. H., Kim, H. S., Han, J. M., Kazlauskas, A., et al. (2005). Phosphatidylinositol (3,4,5)-trisphosphate specifically interacts with the phox homology domain of phospholipase D1 and stimulates its activity. J. Cell Sci. 118, 4405–4413. doi: 10.1242/jcs.02564
Lee, T. Y., Huang, H. D., Hung, J. H., Huang, H. Y., Yang, Y. S., and Wang, T. H. (2006). dbPTM: an information repository of protein post-translational modification. Nucleic Acids Res. 34, D622–D627.
Leneva, N., Kovtun, O., Morado, D. R., Briggs, J. A. G., and Owen, D. J. (2021). Architecture and mechanism of metazoan retromer:SNX3 tubular coat assembly. Sci Adv. 7:13.
Lenoir, M., Kufareva, I., Abagyan, R., and Overduin, M. (2015). Membrane and Protein Interactions of the Pleckstrin Homology Domain Superfamily. Membranes 5, 646–663. doi: 10.3390/membranes5040646
Lenoir, M., Ustunel, C., Rajesh, S., Kaur, J., Moreau, D., Gruenberg, J., et al. (2018). Phosphorylation of conserved phosphoinositide binding pocket regulates sorting nexin membrane targeting. Nat. Commun. 9:993.
Liebl, D., Qi, X., Zhe, Y., Barnett, T. C., and Teasdale, R. D. (2017). SopB-Mediated Recruitment of SNX18 Facilitates Salmonella Typhimurium Internalization by the Host Cell. Front. Cell Infect. Microbiol. 7:257.
Lim, K. P., and Hong, W. (2004). Human Nischarin/imidazoline receptor antisera-selected protein is targeted to the endosomes by a combined action of a PX domain and a coiled-coil region. J. Biol. Chem. 279, 54770–54782. doi: 10.1074/jbc.m411315200
Liu, H., Liu, Z. Q., Chen, C. X., Magill, S., Jiang, Y., and Liu, Y. J. (2006). Inhibitory regulation of EGF receptor degradation by sorting nexin 5. Biochem. Biophys. Res. Commun. 342, 537–546. doi: 10.1016/j.bbrc.2006.01.179
Lucas, M., Gershlick, D. C., Vidaurrazaga, A., Rojas, A. L., Bonifacino, J. S., and Hierro, A. (2016). Structural Mechanism for Cargo Recognition by the Retromer Complex. Cell 167, 1623.e–1635.e.
Lundmark, R., and Carlsson, S. R. (2003). Sorting nexin 9 participates in clathrin-mediated endocytosis through interactions with the core components. J. Biol. Chem. 278, 46772–46781. doi: 10.1074/jbc.m307334200
Lunn, M. L., Nassirpour, R., Arrabit, C., Tan, J., McLeod, I., Arias, C. M., et al. (2007). A unique sorting nexin regulates trafficking of potassium channels via a PDZ domain interaction. Nat. Neurosci. 10, 1249–1259. doi: 10.1038/nn1953
Ma, M. P., and Chircop, M. (2012). SNX9, SNX18 and SNX33 are required for progression through and completion of mitosis. J. Cell Sci. 125, 4372–4382.
Mahankali, M., Henkels, K. M., and Gomez-Cambronero, J. (2013). A GEF-to-phospholipase molecular switch caused by phosphatidic acid, Rac and JAK tyrosine kinase that explains leukocyte cell migration. J. Cell Sci. 126, 1416–1428.
Mas, C., Norwood, S. J., Bugarcic, A., Kinna, G., Leneva, N., Kovtun, O., et al. (2014). Structural basis for different phosphoinositide specificities of the PX domains of sorting nexins regulating G-protein signaling. J. Biol. Chem. 289, 28554–28568. doi: 10.1074/jbc.m114.595959
Merino-Trigo, A., Kerr, M. C., Houghton, F., Lindberg, A., Mitchell, C., Teasdale, R. D., et al. (2004). Sorting nexin 5 is localized to a subdomain of the early endosomes and is recruited to the plasma membrane following EGF stimulation. J. Cell Sci. 117, 6413–6424. doi: 10.1242/jcs.01561
Minguez, P., Letunic, I., Parca, L., Garcia-Alonso, L., Dopazo, J., Huerta-Cepas, J., et al. (2015). PTMcode v2: a resource for functional associations of post-translational modifications within and between proteins. Nucleic Acids Res. 43, D494–D502.
Nakazawa, S., Gotoh, N., Matsumoto, H., Murayama, C., Suzuki, T., and Yamamoto, T. (2011). Expression of sorting nexin 18 (SNX18) is dynamically regulated in developing spinal motor neurons. J. Histochem. Cytochem. 59, 202–213. doi: 10.1369/0022155410392231
Neves, M. A., Totrov, M., and Abagyan, R. (2012). Docking and scoring with ICM: the benchmarking results and strategies for improvement. J. Comput. Aided Mol. Des. 26, 675–686. doi: 10.1007/s10822-012-9547-0
Niu, Y., Zhang, C., Sun, Z., Hong, Z., Li, K., Sun, D., et al. (2013). PtdIns(4)P regulates retromer-motor interaction to facilitate dynein-cargo dissociation at the trans-Golgi network. Nat. Cell Biol. 15, 417–429. doi: 10.1038/ncb2710
Overduin, M., Cheever, M. L., and Kutateladze, T. G. (2001). Signaling with phosphoinositides: better than binary. Mol. Interv. 1, 150–159.
Paul, B., Kim, H. S., Kerr, M. C., Huston, W. M., Teasdale, R. D., and Collins, B. M. (2017). Structural basis for the hijacking of endosomal sorting nexin proteins by Chlamydia trachomatis. Elife 6:e22311.
Pons, V., Ustunel, C., Rolland, C., Torti, E., Parton, R. G., and Gruenberg, J. (2012). SNX12 role in endosome membrane transport. PLoS One. 7:e38949. doi: 10.1371/journal.pone.0038949
Pylypenko, O., Lundmark, R., Rasmuson, E., Carlsson, S. R., and Rak, A. (2007). The PX-BAR membrane-remodeling unit of sorting nexin 9. EMBO J. 26, 4788–4800. doi: 10.1038/sj.emboj.7601889
Pyrpassopoulos, S., Shuman, H., and Ostap, E. M. (2017). Adhesion force and attachment lifetime of the KIF16B-PX domain interaction with lipid membranes. Mol. Biol. Cell. 28, 3315–3322. doi: 10.1091/mbc.e17-05-0324
Rincon, E., Saez, de Guinoa, J., Gharbi, S. I., Sorzano, C. O., Carrasco, Y. R., et al. (2011). Translocation dynamics of sorting nexin 27 in activated T cells. J. Cell Sci. 124, 776–788. doi: 10.1242/jcs.072447
Samuel, S. F., Barry, A., Greenman, J., and Beltran-Alvarez, P. (2021). Arginine methylation: the promise of a ‘silver bullet’ for brain tumours? Amino Acids. 53, 489–506. doi: 10.1007/s00726-020-02937-x
Sato, T. K., Overduin, M., and Emr, S. D. (2001). Location, location, location: membrane targeting directed by PX domains. Science 294, 1881–1885. doi: 10.1126/science.1065763
Schaff, U. Y., Shih, H. H., Lorenz, M., Sako, D., Kriz, R., Milarski, K., et al. (2008). SLIC-1/sorting nexin 20: a novel sorting nexin that directs subcellular distribution of PSGL-1. Eur. J. Immunol. 38, 550–564. doi: 10.1002/eji.200737777
Sciorra, V. A., Rudge, S. A., Prestwich, G. D., Frohman, M. A., Engebrecht, J., and Morris, A. J. (1999). Identification of a phosphoinositide binding motif that mediates activation of mammalian and yeast phospholipase D isoenzymes. EMBO J. 18, 5911–5921. doi: 10.1093/emboj/18.21.5911
Sievers, F., and Higgins, D. G. (2018). Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 27, 135–145. doi: 10.1002/pro.3290
Song, J., Zhao, K. Q., Newman, C. L., Vinarov, D. A., and Markley, J. L. (2007). Solution structure of human sorting nexin 22. Protein Sci. 16, 807–814. doi: 10.1110/ps.072752407
Song, X., Xu, W., Zhang, A., Huang, G., Liang, X., Virbasius, J. V., et al. (2001). Phox homology domains specifically bind phosphatidylinositol phosphates. Biochemistry 40, 8940–8944. doi: 10.1021/bi0155100
St Croix, B., Rago, C., Velculescu, V., Traverso, G., Romans, K. E., Montgomery, E., et al. (2000). Genes expressed in human tumor endothelium. Science 289, 1197–1202. doi: 10.1126/science.289.5482.1197
Stahelin, R. V., Ananthanarayanan, B., Blatner, N. R., Singh, S., Bruzik, K. S., Murray, D., et al. (2004). Mechanism of membrane binding of the phospholipase D1 PX domain. J. Biol. Chem. 279, 54918–54926. doi: 10.1074/jbc.m407798200
Stahelin, R. V., Karathanassis, D., Bruzik, K. S., Waterfield, M. D., Bravo, J., Williams, R. L., et al. (2006). Structural and membrane binding analysis of the Phox homology domain of phosphoinositide 3-kinase-C2alpha. J. Biol. Chem. 281, 39396–39406. doi: 10.1074/jbc.m607079200
Stampoulis, P., Ueda, T., Matsumoto, M., Terasawa, H., Miyano, K., Sumimoto, H., et al. (2012). Atypical membrane-embedded phosphatidylinositol 3,4-bisphosphate (PI(3,4)P2)-binding site on p47(phox) Phox homology (PX) domain revealed by NMR. J. Biol. Chem. 287, 17848–17859. doi: 10.1074/jbc.m111.332874
Stark, C., Su, T. C., Breitkreutz, A., Lourenco, P., Dahabieh, M., Breitkreutz, B. J., et al. (2010). PhosphoGRID: a database of experimentally verified in vivo protein phosphorylation sites from the budding yeast Saccharomyces cerevisiae. Database 2010:ba026.
Su, H., Yang, F., Wang, Q., Shen, Q., Huang, J., Peng, C., et al. (2017). VPS34 Acetylation Controls Its Lipid Kinase Activity and the Initiation of Canonical and Non-canonical Autophagy. Mol. Cell 90:e7.
Sundaresan, N. R., Pillai, V. B., Wolfgeher, D., Samant, S., Vasudevan, P., Parekh, V., et al. (2011). The deacetylase SIRT1 promotes membrane localization and activation of Akt and PDK1 during tumorigenesis and cardiac hypertrophy. Sci. Signal. 4:ra46. doi: 10.1126/scisignal.2001465
Takeuchi, H., Takeuchi, T., Gao, J., Cantley, L. C., and Hirata, M. (2010). Characterization of PXK as a protein involved in epidermal growth factor receptor trafficking. Mol. Cell Biol. 30, 1689–1702. doi: 10.1128/mcb.01105-09
Takeuchi, H., Zhang, Z., Gao, J., Sugiyama, G., Takeuchi, T., and Hirata, M. (2012). Second basic pockets contribute to the localization of PX domains by binding to phosphatidic acid. Adv. Biol. Regul. 52, 183–194. doi: 10.1016/j.advenzreg.2011.09.006
Tate, J. G., Bamford, S., Jubb, H. C., Sondka, Z., Beare, D. M., Bindal, N., et al. (2019). COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 47, D941–D947.
Thandapani, P., O’Connor, T. R., Bailey, T. L., and Richard, S. (2013). Defining the RGG/RG motif. Mol. Cell. 50, 613–623. doi: 10.1016/j.molcel.2013.05.021
Toma-Fukai, S., Kim, J. D., Park, K. E., Kuwabara, N., Shimizu, N., Krayukhina, E., et al. (2016). Novel helical assembly in arginine methyltransferase 8. J. Mol. Biol. 428, 1197–1208. doi: 10.1016/j.jmb.2016.02.007
Traer, C. J., Rutherford, A. C., Palmer, K. J., Wassmer, T., Oakley, J., Attar, N., et al. (2007). SNX4 coordinates endosomal sorting of TfnR with dynein-mediated transport into the endocytic recycling compartment. Nat. Cell Biol. 9, 1370–1380. doi: 10.1038/ncb1656
Ueyama, T., Lekstrom, K., Tsujibe, S., Saito, N., and Leto, T. L. (2007). Subcellular localization and function of alternatively spliced Noxo1 isoforms. Free Radic Biol. Med. 42, 180–190. doi: 10.1016/j.freeradbiomed.2006.08.024
van Weering, J. R., Verkade, P., and Cullen, P. J. (2012). SNX-BAR-mediated endosome tubulation is co-ordinated with endosome maturation. Traffic 13, 94–107. doi: 10.1111/j.1600-0854.2011.01297.x
Vieira, N., Deng, F. M., Liang, F. X., Liao, Y., Chang, J., Zhou, G., et al. (2014). SNX31: a novel sorting nexin associated with the uroplakin-degrading multivesicular bodies in terminally differentiated urothelial cells. PLoS One. 9:e99644. doi: 10.1371/journal.pone.0099644
Vietri, M., Radulovic, M., and Stenmark, H. (2020). The many functions of ESCRTs. Nat. Rev. Mol. Cell Biol. 21, 25–42. doi: 10.1038/s41580-019-0177-4
Virbasius, J. V., Song, X., Pomerleau, D. P., Zhan, Y., Zhou, G. W., and Czech, M. P. (2001). Activation of the Akt-related cytokine-independent survival kinase requires interaction of its phox domain with endosomal phosphatidylinositol 3-phosphate. Proc. Natl. Acad. Sci. U. S. A. 98, 12908–12913. doi: 10.1073/pnas.221352898
Waterhouse, A. M., Procter, J. B., Martin, D. M. A., Clamp, M., and Barton, G. J. (2009). Jalview version 2: A Multiple Sequence Alignment and Analysis Workbench. Bioinformatics 25, 1189–1191. doi: 10.1093/bioinformatics/btp033
Xu, H., Zhou, J., Lin, S., Deng, W., Zhang, Y., and Xue, Y. (2017). PLMD: An updated data resource of protein lysine modifications. J. Genet Genomics 44, 243–250. doi: 10.1016/j.jgg.2017.03.007
Xu, J., Liu, D., Gill, G., and Songyang, Z. (2001). Regulation of cytokine-independent survival kinase (CISK) by the Phox homology domain and phosphoinositides. J. Cell Biol. 154, 699–705. doi: 10.1083/jcb.200105089
Xu, J., Xu, T., Wu, B., Ye, Y., You, X., Shu, X., et al. (2013). Structure of sorting nexin 11 (SNX11) reveals a novel extended phox homology (PX) domain critical for inhibition of SNX10-induced vacuolation. J. Biol. Chem. 288, 16598–16605. doi: 10.1074/jbc.m112.449306
Xu, J., Zhang, L., Ye, Y., Shan, Y., Wan, C., Wang, J., et al. (2017). SNX16 Regulates the Recycling of E-Cadherin through a Unique Mechanism of Coordinated Membrane and Cargo Binding. Structure 125:e5.
Xu, T., Gan, Q., Wu, B., Yin, M., Xu, J., Shu, X., et al. (2020). Molecular Basis for PI(3,5)P2 Recognition by SNX11, a Protein Involved in Lysosomal Degradation and Endosome Homeostasis Regulation. J. Mol. Biol. 432, 4750–4761. doi: 10.1016/j.jmb.2020.06.010
Xu, Y., Hortsman, H., Seet, L., Wong, S. H., and Hong, W. (2001). SNX3 regulates endosomal function through its PX-domain-mediated interaction with PtdIns(3)P. Nat. Cell Biol. 3, 658–666. doi: 10.1038/35083051
Yang, J., Yan, R., Roy, A., Xu, D., Poisson, J., and Zhang, Y. (2015). The I-TASSER Suite: protein structure and function prediction. Nat. Methods. 12, 7–8. doi: 10.1038/nmeth.3213
Yarar, D., Surka, M. C., Leonard, M. C., and Schmid, S. L. (2008). SNX9 activities are regulated by multiple phosphoinositides through both PX and BAR domains. Traffic 9, 133–146. doi: 10.1111/j.1600-0854.2007.00675.x
Yarar, D., Waterman-Storer, C. M., and Schmid, S. L. (2007). SNX9 couples actin assembly to phosphoinositide signals and is required for membrane remodeling during endocytosis. Dev. Cell 13, 43–56. doi: 10.1016/j.devcel.2007.04.014
Yu, K., Zhang, Q., Liu, Z., Zhao, Q., Zhang, X., Wang, Y., et al. (2019). qPhos: a database of protein phosphorylation dynamics in humans. Nucleic Acids Res. 47, D451–D458.
Zhang, A., He, X., Zhang, L., Yang, L., Woodman, P., and Li, W. (2014). Biogenesis of lysosome-related organelles complex-1 subunit 1 (BLOS1) interacts with sorting nexin 2 and the endosomal sorting complex required for transport-I (ESCRT-I) component TSG101 to mediate the sorting of epidermal growth factor receptor into endosomal compartments. J. Biol. Chem. 289, 29180–29194. doi: 10.1074/jbc.m114.576561
Zheng, B., Ma, Y. C., Ostrom, R. S., Lavoie, C., Gill, G. N., Insel, P. A., et al. (2001). RGS-PX1, a GAP for GalphaS and sorting nexin in vesicular trafficking. Science 294, 1939–1942. doi: 10.1126/science.1064757
Zhong, Q., Lazar, C. S., Tronchere, H., Sato, T., Meerloo, T., Yeo, M., et al. (2002). Endosomal localization and function of sorting nexin 1. Proc. Natl. Acad. Sci. U. S. A. 99, 6767–6772. doi: 10.1073/pnas.092142699
Keywords: lipid specificity, membrane recognition, phosphoinositide binding, lysine acetylation, arginine methylation, PX domain, protein regulation, metabolite signaling
Citation: Kervin TA, Wiseman BC and Overduin M (2021) Phosphoinositide Recognition Sites Are Blocked by Metabolite Attachment. Front. Cell Dev. Biol. 9:690461. doi: 10.3389/fcell.2021.690461
Received: 02 April 2021; Accepted: 18 June 2021;
Published: 22 July 2021.
Edited by:
Cecilia Beatriz Conde, Medical Research Institute Mercedes and Martín Ferreyra (INIMEC), ArgentinaReviewed by:
Gerry Hammond, University of Pittsburgh, United StatesBrett M. Collins, University of Queensland, Australia
Copyright © 2021 Kervin, Wiseman and Overduin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael Overduin, b3ZlcmR1aW5AdWFsYmVydGEuY2E=
 Troy A. Kervin
Troy A. Kervin Brittany C. Wiseman1,2,3
Brittany C. Wiseman1,2,3 Michael Overduin
Michael Overduin