- 1Key Laboratory of Yak Breeding Engineering Gansu Province, Lanzhou Institute of Husbandry and Pharmaceutical Sciences, Chinese Academy of Agricultural Sciences, Lanzhou, China
- 2State Key Laboratory of Grassland Agro-Ecosystems, College of Pastoral Agriculture Science and Technology, Lanzhou University, Lanzhou, China
Yak (Bos grunniens) is considered an iconic symbol of Tibet and high altitude, but they suffer from malnutrition during the cold season that challenges the metabolism of energy. Adipocytes perform a crucial role in maintaining the energy balance, and adipocyte differentiation is a complex process involving multiple changes in the expression of genes. N6-methyladenosine (m6A) plays a dynamic role in post-transcription gene expression regulation as the most widespread mRNA modification of the higher eukaryotes. However, currently there is no research existing on the m6A transcriptome-wide map of bovine animals and their potential biological functions in adipocyte differentiation. Therefore, we performed methylated RNA immunoprecipitation sequencing (MeRIP-seq) and RNA sequencing (RNA-seq) to determine the distinctions in m6A methylation and gene expression during yak adipocyte differentiation. In yak adipocyte and preadipocyte the content of m6A and m6A-associated enzymes was substantially different. In the two groups, a total of 14,710 m6A peaks and 13,388 m6A peaks were identified. For the most part, m6A peaks were enriched in stop codons, 3′-untranslated regions, and coding regions with consensus motifs of GGACU. The functional enrichment exploration displayed that differentially methylated genes participated in some of the pathways associated with adipogenic metabolism, and several candidate genes (KLF9, FOXO1, ZNF395, and UHRF1) were involved in these pathways. In addition to that, there was a positive association between m6A abundance and levels of gene expression, which displayed that m6A may play a vital role in modulating gene expression during yak adipocyte differentiation. Further, in the adipocyte group, several methylation gene protein expression levels were significantly higher than in preadipocytes. In short, it can be concluded that the current study provides a comprehensive explanation of the m6A features in the yak transcriptome, offering in-depth insights into m6A topology and associated molecular mechanisms underlying bovine adipocyte differentiation, which might be helpful for further understanding its mechanisms.
Introduction
N6-methyladenosine (m6A) was first discovered in the 1970s as the most prevalent internal modification of polyadenylated mRNAs and long noncoding RNAs (lncRNAs) in higher eukaryotes (Desrosiers et al., 1974; Perry and Kelley, 1974; Adams and Cory, 1975; Furuichi et al., 1975; Lavi and Shatkin, 1975; Wei and Moss, 1975). The modification of m6A methylation is mounted by a series of m6A methyltransferases labeled as writers: methyltransferases such as 3 and 14 (METTL3 and METTL14), Wilms Tumor 1-associated protein (WTAP), VIRMA, vir-Like m6A methyltransferase associated (KIAA1429), RNA binding motif protein 15 (RBM15), and zinc finger CCCH domain 13 (ZC3H13) (Bokar et al., 1997; Agarwala et al., 2012; Liu et al., 2014; Ping et al., 2014; Schwartz et al., 2014; Patil et al., 2016; Knuckles et al., 2018; Wen et al., 2018). Besides this, m6A demethylases eliminate methylation from RNAs to enable a delicately dynamic equilibrium modification and are named erasers: fat mass and obesity-associated protein (FTO) and α-ketoglutarate-dependent dioxygenase alkB homolog 5 (ALKBH5) (Jia et al., 2011; Zheng et al., 2013). Further, specific proteins, including the YTH domain family (YTHDF1-3) and IGF2BPs (IGF2BP1-3) (Dominissini et al., 2012; Luo and Tong, 2014; Wang et al., 2014; Wang et al., 2015), were identified as a category of proteins called readers that recognize the information of RNA methylation modifications and engage in downstream mRNA translation, degradation, microRNA binding, and RNA-protein interactions (Liu and Pan, 2016; Roundtree I. A. et al., 2017; Nachtergaele and He, 2017; Zhao et al., 2017). Notably, two independent studies established an m6A RNA immunoprecipitation accompanied with high-throughput sequencing (MeRIP-seq) and subsequently identified the first N6-methyladenosine modification map to methylomes with a resolution of 100-nucleotides (Dominissini et al., 2012; Meyer et al., 2012). Meanwhile, MeRIP-seq has been used to identify the m6A profile in humans and mice. These results reveal that m6A is predominantly located close to stop codons, 3′-untranslated regions (3′-UTRs), and also in long internal exons and transcription start sites, suggesting that m6A plays a crucial role in the post-transcriptional regulation of gene expression. These innovative studies reflect that the construction of transcriptome-wide m6A methylome profiles is of great importance to further investigate the characteristics and functions of such modification.
Currently, m6A modifications are reported in several areas of RNA metabolism, such as RNA localization, transport, splicing, stability, and translation (Liu and Pan, 2016; Roundtree I. A. et al., 2017; Nachtergaele and He, 2017; Zhao et al., 2017). Previous studies describe that m6A modification of mRNA plays an important biological function in controlling cellular metabolic processes, and it is reportedly involved in determining mammalian embryonic stem cell fate (Batista et al., 2014), regulating the initiation and differentiation of meiosis in murine spermatogonial stem cells (Xu et al., 2017), and maintaining the myogenic potential of proliferating skeletal muscle progenitors (Kudou et al., 2017). In particular, FTO facilitates the differentiation of mouse preadipocytes by regulating alternative splicing of pre-mRNAs for genes associated with adipogenesis (Zhao et al., 2014). Zhong et al. report that knockdown of METTL3 or YTHDF2 in vitro enhanced the stability and expression of peroxisome proliferator-activator receptor alpha (PPARα) mRNA, leading to decreased lipid accumulation in a hepatocellular carcinoma cell line (HepG2) (Zhong et al., 2018). Besides this, a recent study reveals that RNA m6A modification has a potential function in the deposition of porcine adipose tissue (Tao et al., 2017), and the modification of m6A on the mRNA of mitochondrial carrier homology 2 (MTCH2) promotes the differentiation of pig intermuscular preadipocytes (Jiang et al., 2019). Thus, we assume that m6A modification may also refer to bovine adipocyte differentiation according to the notable functions of m6A modification described above. However, our knowledge about the relationship between m6A modification and bovine adipocyte differentiation is still scarce.
The yak is the major bovine livestock breed on the Qinghai-Tibet Plateau and is the only large ruminant domestic species that enables daily necessities, such as meat, milk, wool, skins, fuel, and economic benefits, for local herders (Long et al., 1999; Dong et al., 2006). On the Qinghai-Tibet Plateau, domestic yaks mainly grow on natural pastures under typical grazing conditions (Long et al., 2008). Owing to seasonal variations in forage, yaks must constantly undergo insufficient feeding during the harsh winter season (October–May), which leads to the large seasonal weight changes and a circular rhythm of “live in summer, weighty in autumn, thin in winter, and dead in spring” (Shikui et al., 2003). Consequently, the subcutaneous adipose layer of yak accumulates rapidly in summer and early autumn to provide essential energy requirements and withstand severe cold through selective fat catabolism during the cold season (Ding et al., 2012). The distinctive metabolic pattern makes the yak a fascinating model for studying adipose metabolism in plateau domestic animals. Adipocytes are a major component of adipose tissue and are considered to be the cornerstone of metabolic homeostasis regulation throughout the body (Ali et al., 2013). Therefore, it is necessary to assay m6A sites at the transcriptome-wide level to identify the potential biological functions of RNA m6A modification during yak adipocyte differentiation.
In the present study, we initially isolated preadipocytes from yak adipose tissue and differentiated them into mature adipocytes successfully. We obtained the first transcriptome-wide m6A methylome profile in yak by MeRIP-seq and elucidated the features of m6A modification during yak adipocyte differentiation. We found that the different m6A RNA modifications between yak preadipocytes and mature adipocytes have potential regulatory roles in gene expression and pathways related to adipose energy metabolism. This study explores the role of m6A modification in bovine adipose metabolism and complements m6A studies in plateau domestic livestock, which may be a breakthrough point for exploring energy metabolism in yaks.
Materials and Methods
Ethics Statement
Animal treatment during research was carried out in complete accordance with the protocols and guidelines for animal ethics of the People’s Republic of China, and all operations were approved by the Animal Administration and Ethics Committee of Lanzhou Institute of Husbandry and Pharmaceutical Sciences, Chinese Academy of Agricultural Sciences (Permit No. SYXK-2014–0002).
Preadipocyte Isolation
The Datong Yak Breeding Center (Datong County, Qinghai, China) provided three healthy 3-day-old Datong yaks. The night before slaughter, yaks were not fed. On the next morning, the yaks were humanely sacrificed by the way of electrical stunned (90 V, 10 s, and 50 Hz) at a commercial slaughter facility and exsanguinated as necessary to ameliorate the suffering, according to standard approved industry protocols. The subcutaneous adipose tissue was harvested according to the protocols and guidelines for animal ethics of the People’s Republic of China. Primary yak preadipocytes were cultured from subcutaneous adipose tissue according to our previous study (Zhang Y. et al., 2018; Zhang et al., 2020). Briefly, the subcutaneous fat tissue was flushed with penicillin (200 U/mL) and streptomycin (200 U/mL) added to the phosphate saline buffer (HyClone, Thermo Fisher Scientific, Carlsbad, CA, United States). After that, they were finely minced into about 1 mm3 piece in an aseptic setting. The segments were digested by Type I collagenase in a continuously agitated water bath at 37°C for 60–90 min. With a 40-μm nylon mesh film, indigestible material was screened, and the filtrate was resuspended for 5 min at 1400 g. The sediment was subsequently incubated at room temperature for 10 min with the erythrocyte lysis buffer (0.154 M NH4Cl, 10 mM KHCO3, 0.1 mM EDTA). The cells were then filtered with 200-μm nylon mesh film and rinsed twice with a serum-free medium. After 5 min of centrifugation at 1400 g, preadipocytes were harvested and solubilized in the growth media, including DMEM-F12 (Hyclone, UT, United States) supplemented with 10% fetal bovine serum (FBS, Gibco, MA, United States).
Adipogenic Differentiation and Staining of Oil Red O
The adipogenic differentiation was performed according to our previous study (Zhang Y. et al., 2018; Zhang et al., 2020). Preadipocyte was induced for 2 days by adipogenic compounds composed of 3-isobutyl-methylxanthine (MIX) (Sigma, MO, United States), dexamethasone (Sigma, MO, United States), rosiglitazone (Sigma, MO, United States), and insulin (Sigma, MO, United States) after cell confluence approached 70% in growth media. The medium was replaced after 2 days with DMEM-F12 containing 10% FBS, penicillin (200 U/mL), streptomycin (200 U/mL), and 5 ng/ml of insulin and updated with cycles of 2 days until day 12. The cells were usually flushed twice with PBS and set for 1 h in 4% formalin. Cells were then reacted at room temperature for 30 min with a saturated solution of Oil Red O. Then, cells were rinsed three times with sterile water, and photographs were acquired from light microscopy.
Quantitative Real-Time PCR
Total RNAs were extracted using TRIzol reagent (Invitrogen, CA, United States) from in vitro cultured yak preadipocytes and differentiated adipocytes (three biological replicates for each condition). Concentration and quality were further evaluated using denaturing gel electrophoresis and spectroscopy (Thermo, Waltham, MA, United States). Reverse transcription of mRNA was conducted using commercial kits (Takara, Japan) according to the manufacturer’s protocols. Real-time RT-PCR was accomplished in a CFX Link Real-Time PCR Detection System, and 10 μl volume of reaction consisting of 5 μl 2xSYBR Premix Ex Taq II, 0.4 μl primers (10 μM), and 0.8 μl cDNA. The reaction condition was as follows: denaturation for 30 s at 95°C followed by 35 additional cycles for 15 s at 94°C, annealing for the 30 s at 72°C. A melting procedure with a heating rate of 0.5°C/10 s was performed to create melting curves ranging from 95°C. The gene expression levels were estimated using the 2−ΔΔCt. Supplementary Table S1 lists the sequences used for the primers.
Measuring the m6A Content
The overall content of mRNA m6A was measured by a methylation quantification kit of EpiQuik RNA (Epigentek, P-9005, NY, United States). In short, a standard curve was constructed at concentrations of 0.01–0.5 ng/μl by positive control. The equivalent RNA solution (1–8 μl) and negative control were applied to the strip wells. The plate was wrapped with parafilm, incubating for 1.5 h at 37°C. Then, the wells were washed three times and added to the 1:1000 diluted capture antibody at room temperature for 1 h. After washing thrice, the detection antibody (1:2000 dilution) and enhancer solution were applied to every well incubated at room temperature for 30 min. After five washes, detection solutions were placed on each well and incubated for 10 min at room temperature to protect from light. Finally, a stop solution was applied to each well and absorbance read with a microplate reader at 450 nm.
MeRIP-Seq and mRNA Sequencing
According to the manufacturer’s protocol, the total RNA was extracted using Trizol reagent (Invitrogen, CA, United States). A Bioanalyzer 2100 and RNA 6000 Nano LabChip Kit (Agilent, CA, United States) with RIN number >7.0 were used to evaluate the total RNA quality and quantity. Nearly over 200 μg total RNA was performed to isolate Poly (A) mRNA through magnetic beads (Invitrogen) attached to poly-T oligo. After purifying, poly (A) mRNA fractions are broken into 100-nt-long oligonucleotides using a Magnesium RNA Fragmentation Module (NEB, cat.E6150, United States) under 86°C for 7 min. Then, the fragmentation of broken RNA was incubated in immunoprecipitation (IP) buffer (50 mM Tris-HCl, 750 mM NaCl, and 0.5% Igepal CA-630) supplied with BSA (0.5–1 μg/μl) for 2 h at 4°C with m6A-specific antibody (No. 202003, Synaptic Systems, Germany). Subsequently, the above mixture was incubated with protein-A beads (Thermo Fisher Scientific, MA, United States) and eluted with elution buffer (1 × IP buffer and 6.7 mM m6A). The eluted RNA was extracted by TRIzol reagent (Thermo Fisher Scientific) according to the manufacturer. Then, IP RNA and untreated input control fragment RNA were reverse-transcribed to create the cDNA by SuperScript™ II Reverse Transcriptase (Invitrogen, cat. 1896649, CA, United States), which was then used to synthesize U-labeled second-stranded DNAs with E. coli DNA polymerase I (NEB, cat.M0209, MA, United States), RNase H (NEB, cat.M0297, MA, United States), and dUTP Solution (Thermo Fisher, cat. R0133, MA, United States). Besides this, the A-base was added to the blunt ends of each strand and prepared for linkage to the indexed adapters. Each adapter holds a T-base overhang to link the adapter to the A-tailed fragmented DNA. Single- or dual-index adapters were linked to the fragments, and size selection was performed with AMPureXP beads. After the heat-labile UDG enzyme (NEB, cat. M0280, MA, United States) treatment of the U-labeled second-stranded DNA, the ligated products are amplified with PCR by the following conditions: initial denaturation at 95°C for 3 min, eight denaturation cycles at 98°C for 15 s, annealing at 60°C for 15 s, extension at 72°C for 30 s, and then final extension at 72°C for 5 min. The average insert size of the paired-end libraries was ∼100 ± 50 bp. Finally, the m6A-seq libraries were performed with Tru Standard mRNA Sample Prep Kit (Illumina) along with the published protocol (Huse et al., 2003). The 2 × 150 bp paired-end sequenced (PE150) on Illumina Novaseq™ 6000 (LC-Bio Technology CO., Ltd., Hangzhou, China) in accordance with the vendor’s recommended protocol.
Sequencing Data Analysis
First of all, in-house perl scripts and Cutadapt (Martin, 2011) were performed to eliminate the reads containing contaminants of the adapter, bases of low quality, and indeterminate. Meanwhile, the quality of the sequence was validated using fastp. The reads were mapped to the Bos_mutus genome (Version: BosGru_v2.0) by HISAT2 (Kim et al., 2015) with default parameters. Using R package exomePeak (Meng et al., 2014) identify the m6A peaks from mapped reads of IP and input libraries with bed or bam format to configure for viewing on IGV software (http://www.igv.org/) or the UCSC genome browser. The parameters of the exomePeak R package are as follows: “PEAK_CUTOFF_PVALUE = 0.05, PEAK_CUTOFF_FDR = NA, FRAGMENT_LENGTH = 100.” The examination was performed using the Poisson distribution model, and a p-value < 0.05 was considered as a peak. De novo and defined motifs were identified by MEME (Bailey et al., 2009) and HOMER (Heinz et al., 2010), accompanied by perl scripts in the house seeking the motif concerning peak. Called peaks were annotated using ChIPseeker (Yu et al., 2015) by intersection with gene architecture. The difference peaks were identified using the exomePeak R package with parameters p-value < 0.05 and |log2 (fold change)| ≥ 1. StringTie (Pertea et al., 2015) calculated the expression level of all mRNAs from input libraries, which normalized with FPKM {FPKM = [total exon fragments/mapped reads (millions)]}. The differentially expressed mRNAs were collected by R package edgeR (Robinson et al., 2010) with the |log2 (fold change)| > 1 and p-value < 0.05. GO seq R package was performed on the Gene Ontology (GO, http://www.geneontology.org/) enrichment analysis for the differentially expressed genes (Young et al., 2010). The Kyoto Encyclopedia of Genes and Genomes (KEGG, http://www.genome.jp/kegg/) database is a major resource for learning high-level functions and utilities of biological systems. The statistical enrichment tests for genes of differential expression in the KEGG pathways were used in the KOBAS software (Xie et al., 2011).
Western Blotting
Proteins were extracted from preadipocytes and adipocytes. After detecting the total protein concentration, the protein was denatured at 95°C for 5 min with a protein loading of 50 μg. Subsequently, SDS-PAGE electrophoresis was performed with 10% of the isolate gel and 4% of the concentrate gel, and electrophoresis at 40 V for 25 min in the concentrate gel and 100 V for 80 min in the isolate gel. Then, the protein was transferred to the PVDF membrane and immersed in the closure solution at 37°C for 1.5 h. Then, it was incubated in monoclonal rabbit anti-ENTPD1, anti-USP2, and anti-PGAM2 (1:1000; Abcam, Cambridge, United Kingdom) and monoclonal mouse anti-β-actin (1:5,000; Beyotime, Shanghai, China). Finally, the membranes were incubated for 1.5 h at 37°C by adding an HRP-labeled goat secondary antibody and images captured using a Chemi Doc System (Bio-Rad, Hercules, CA). Grayscale values of proteins were evaluated by ImageJ (https://imagej.nih.gov/ij/).
Statistical Analysis
The SPSS 22 software package was used to evaluate statistics. A one-way test of variance assessed the significance of the differences between all of the groups. Statistically significant was the degree of probability * p < 0.05;** p < 0.01. Values are shown as mean ± SEM.
Results
The Yak Preadipocyte Induced Differentiation and Global m6A Quantification
The results of Oil Red O show that the visibility of lipid droplets in adipocytes increased significantly at day 12 compared to day 0 after induction with adipogenic agents (Figures 1A,B, Supplementary Figure S1). Meanwhile, the expression of adipocyte differentiation-specific marker genes (PPARγ, C/EBPα, and FABP4) was significantly elevated on day 12 (adipocyte) compared with day 0 (preadipocyte) (Figure 1C), suggesting preadipocyte full differentiation into adipocyte. Subsequently, to overview the m6A methylation during yak adipocyte differentiation, the expression of RNA methylation-related genes was contrasted by quantitative real-time PCR (qRT-PCR) detected, including METTL3, WTAP, METTL14, FTO, ALKBH5, and YTHDC1/2. Comparing the group of preadipocytes (Pread0) and adipocytes (Ad), the findings show that the expression level of methyltransferases (METTL14, WTAP, and METTL3) and ALKBH5 were dramatically upregulated, whereas FTO was substantially downregulated, and m6A-binding proteins (YTHDC1 and YTHDC2) were drastically upregulated (Figure 1D). Furthermore, the content of m6A in the group of adipocytes was significantly higher compared with the preadipocyte group (Figure 1E). Thereby, we hypothesized that, during yak adipocyte differentiation, the difference of m6A methylation may exist, which was furtherly discovered using MeRIP-seq.
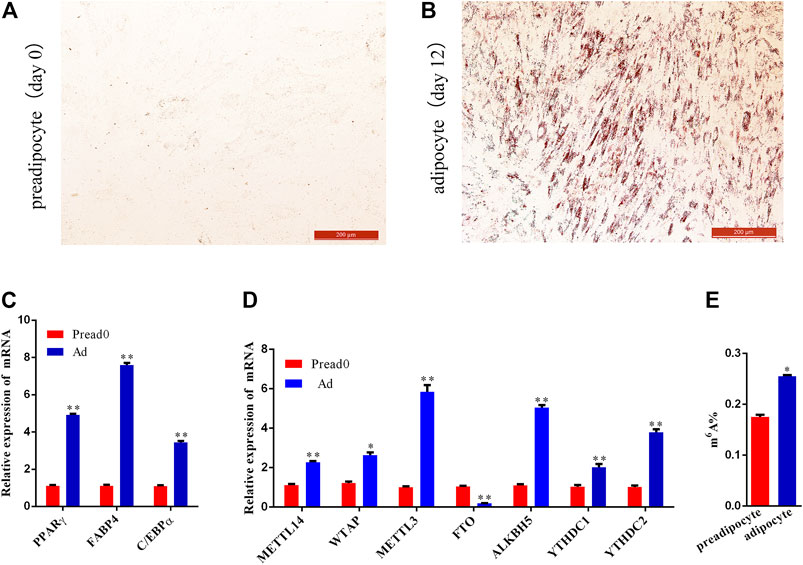
FIGURE 1. Preadipocyte-induced differentiation and detection of m6A level between preadipocyte and adipocyte. (A,B) Preadipocytes and adipocytes dyed with Oil red O. (C) Relative expression of PPARγ, C/EBPα, and FABP4 between preadipocyte (day 0) and adipocyte (day 12). (D) The mRNA expression level of methyltransferases (METTL14, WTAP, and METTL3), demethylases (FTO and ALKBH5) and m6A-binding proteins (YTHDC1and YTHDC2). (E) m6A abundance between the preadipocytes and adipocytes. (SEM, * p < 0.05;** p < 0.01).
Transcriptome-wide m6A-Seq Reveals Global m6A Modification Patterns During Yak Adipocyte Differentiation
The yak adipocyte and preadipocyte of three biological replicates were used for transcriptome-wide m6A-sequencing (m6A-seq) and RNA-sequencing (RNA-seq) assays. In total, 12 libraries were sequenced, comprising three replicates of preadipocyte and adipocyte for input and MeRIP samples (Supplementary Table S2). With each MeRIP library, an average of 9.22 Giga base-pair (Gb) of high-quality data was produced, and 9.49 Gb per input library (RNA-seq data set). Then, we eliminated reads containing adapter pollutants, low quality, and indeterminate bases, an average of 7.17 and 7.11 Gb obtained from per MeRIP and input libraries, respectively. The valid data were mapped to the Bos_mutus genome (Version: BosGru_v2.0) using HISAT2. The proportions of mapped reads ranged from 87.96 to 96.57%, correspondingly (Supplementary Table S2). The RNA species of transcripts included mRNA (19,916), misc_RNA (386), ncRNA (262), pseudogene (916), and tRNA(179) (Supplementary Figure S2). In the yak Ad group, R package exomePeak found a total of 14,710 m6A peaks, containing transcripts of 9633 genes. Likewise, 13,388 m6A peaks were found in the Pread0 group corresponding to transcripts of 9142 genes (Figures 2A,B). In addition, 5848 peaks were consistently observed in the two groups, and 3964 genes within the groups were modified by m6A. Compared with the Pread0 group, the Ad group had 9226 new peaks occurring with the absence of 7904 peaks, reflecting the significant difference between Pread0 and Ad groups in global m6A modification trends (Figures 2A–C). m6A methylomes were ulteriorly mapped by HOMER software to define whether RRACH motifs (R represents purine; A is m6A; and H is U, A, or C) were ubiquitous in our detected m6A. The results of the enrichment analysis in both groups show that the consensus motifs of m6A RRACH were GGACU (Figure 2D) accorded with previous studies, which strengthens the credibility of the m6A peaks and confirms the presence of a prevailing methylated modification mechanism.
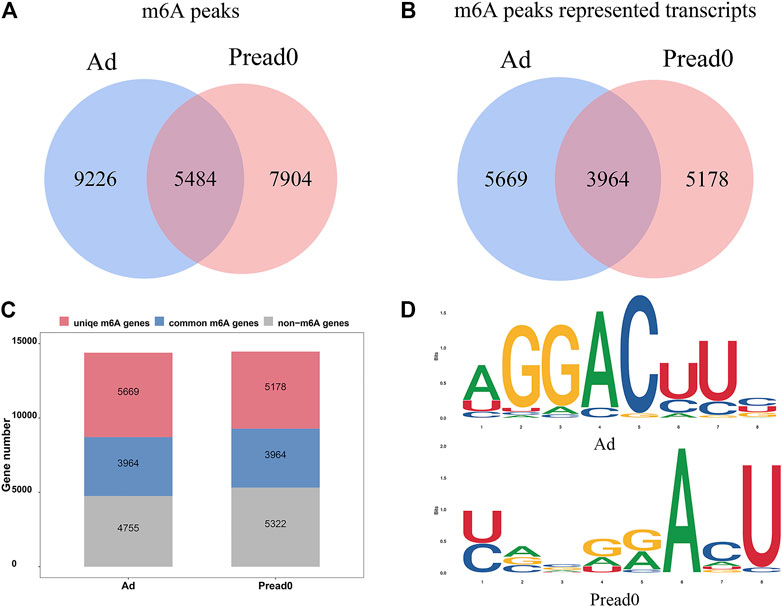
FIGURE 2. m6A-seq transcriptome-wide and m6A peak analysis. (A) Number of common and specific m6A peaks in groups of preadipocyte and adipocyte. (B) Venn diagram showing the m6A peaks for transcripts of two group. (C) Profile of m6A-modified genes discovered in m6A-seq. (D) The motifs enriched from m6A peaks identified from the groups of adipocyte and preadipocyte.
Analysis of m6A Modification Distribution in Yak Transcriptome
We analyzed metagene models of m6A peaks in the global transcriptome to identify the differential distribution of m6A in transcripts. Our findings indicated that m6A peaks were predominantly enriched in the coding sequence (CDS) near the start and stop codons and approach the beginning of the 3′ untranslated region (3′UTRs) in Ad and Pread0 (Figure 3A), which contrast to the pattern found in mice and chickens (Luo et al., 2019; Cheng et al., 2021). Subsequently, to systematically calculate the enrichment, we investigated nonoverlapping transcript segments per m6A peak with 5′UTR, CDS, and 3′UTR (Supplementary Figure S3A), in which most of them were abundant in CDS. Interestingly, m6A peak relative increased at 5′UTR and CDS region in Ad compared with Pread 0 and decreased in 3′UTR region. Afterward, we explored the distribution of m6A modified peaks with each gene, finding that almost 60% of methylated genes hold only one m6A peak, and most genes contain one to three m6A peaks (Figure 2B). Furthermore, we investigated the relationship between m6A peak number and gene length. The results show a global trend that the longer gene length has more m6A peaks (Supplementary Figure S3B).
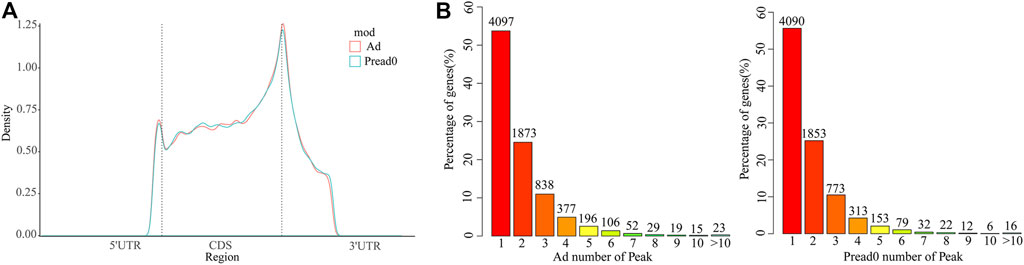
FIGURE 3. m6A methylome distributed on yak transcripts. (A) m6A peaks enriched with transcripts. Transcript is separated into three parts comprising 5′ untranslated region, coding sequence and 3′ untranslated region. (B) Proportion of genes possessing m6A peaks with different numbers.
Analysis of the GO and KEGG Pathways of Differentially Methylated Genes
The comparison was performed for the abundance of m6A peaks between preadipocytes and adipocytes. These findings exposed that 118 markedly hypermethylated m6A peaks and 51 substantially hypomethylated peaks were obtained (|log2 (fold change)| > 1, p< 0.05) (Figure 4A). The residual peaks of the m6A were viewed as unaltered peaks. Moreover, differentially methylated m6A peaks represented genes investigated by GO and KEGG pathway analysis, revealing the biological significance of m6A methylation during yak adipocyte differentiation. GO analysis revealed that differentially methylated genes were mainly implicated with DNA-templated and regulation of transcription by RNA polymerase II (ontology: biological process), cytoplasm, nucleus and integral component of membrane (ontology: cellular component), and transcription factor and microtubule binding (ontology: molecular function) (Figure 4B, Supplementary File S1). Meanwhile, the top 20 biological enrichment of KEGG pathways indicated that the genes differently methylated were substantially related to the adipogenic metabolism regulation pathways, NOD-like receptor signaling pathway, FoxO signaling pathway, Ether lipid metabolism, cAMP signaling pathway, and Hippo signaling pathway (Figure 4C; Supplementary File S2). These results reveal that several genes related to lipid metabolism were modified by m6A methylation during yak adipocyte differentiation. Furthermore, the genes (KLF9, FOXO1, and UHRF1) differentially methylated sites were analyzed by Integrative Genomics Viewer (IGV) software (Figure 4D), located in 5′UTRs, exons, and 3′UTRs. In the 5′UTR region of KLF9, the m6A site was hypermethylated in the adipocyte group compared with the control group, and its mRNA expression was upregulated. In the 3′UTR region of FOXO1, the m6A site was hypomethylated in the adipocyte group compared with the control group, and its mRNA expression was upregulated. In the exon region of UHRF1, the m6A site was hypermethylated in the adipocyte group compared with the control group, and its mRNA expression was downregulated (Supplementary File S3). The different m6A methylation levels of these genes may affect their expression.
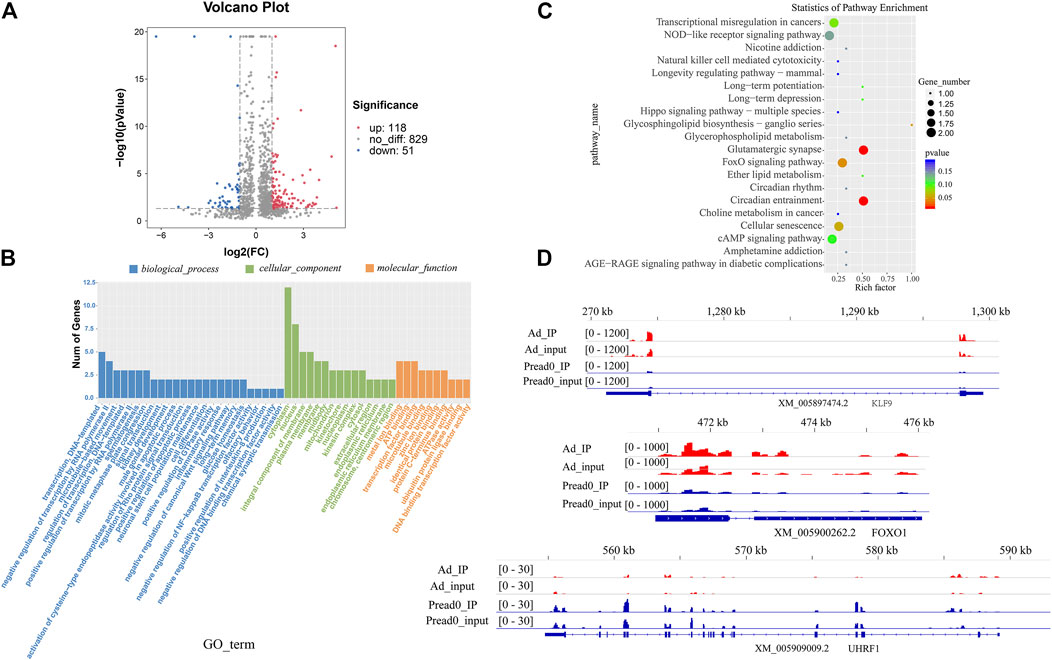
FIGURE 4. The alteration of global m6A modification in adipocyte compared with preadipocyte. (A) Volcano plots showing different m6A peaks (fold change ≥2 and p < 0.05) between preadipocyte and adipocyte. (B) GO terms of genes with differential m6A peaks between preadipocyte and adipocyte. (C) The top 20 markedly enriched pathways for the genes of differential peaks between preadipocyte and adipocyte. (D) The abundance of m6A on KLF9, FOXO1, and UHRF1mRNA transcripts observed by m6A-seq in preadipocyte and adipocyte. Blue boxes represent exons; blue lines represent introns.
RNA-Seq Identification of Genes Differentially Expressed in Both Groups
An analysis of the RNA-seq data set (m6A-seq input library) displayed that the trends of global mRNA expression between preadipocyte and adipocyte were considerably different. There were 648 significantly different mRNAs, including 300 upregulated and 348 downregulated (|log2 (fold change)| > 1, p < 0.05) as shown in Figure 5A. Then, we conducted a clustered heat map to further explore the potential roles of the genes (Figure 5B; Supplementary File S4). Furthermore, GO ontology and KEGG pathway were performed to analyze the differentially expressed genes. As Figure 5C; Supplementary File S5 display, the top 20 most notable functional annotations include regulation of glucose metabolic process, canonical Wnt signaling pathway, positive regulation of cell proliferation, and insulin-like growth factor ternary complex, which influence adipocyte differentiation. Meanwhile, the pathway exploration revealed that signaling pathways regulating pluripotency of stem cells, ECM-receptor interaction, PI3K-Akt signaling pathway, and FoxO signaling pathway were significantly enriched (Figure 5D; Supplementary File S6), revealing that differentially expressed genes potentially participated in adipogenic metabolism.
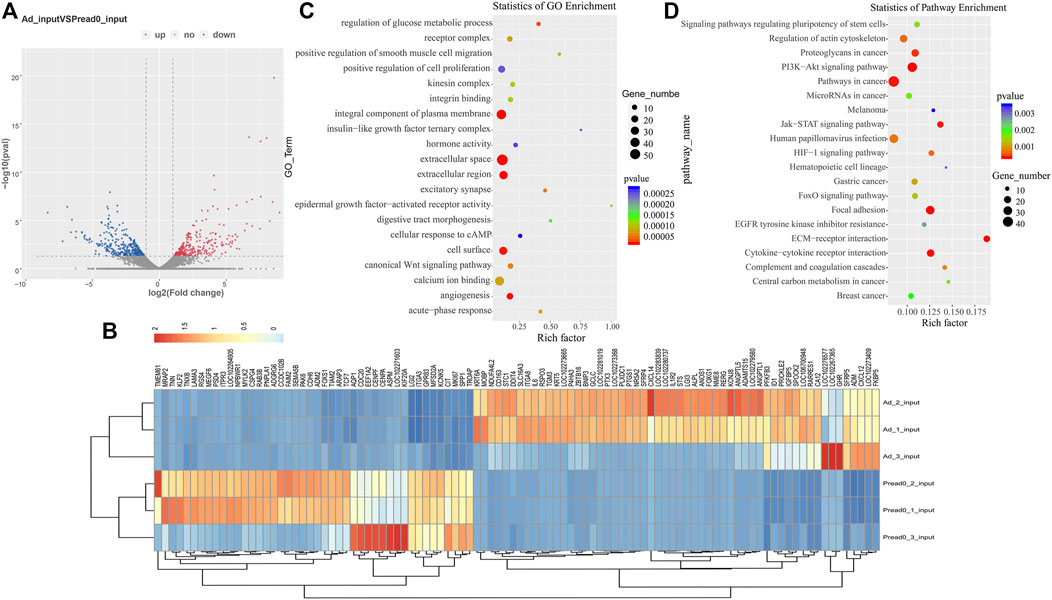
FIGURE 5. RNA-seq analysis of genes differentially expressed between preadipocyte and adipocyte. (A) Volcano plots illustrating the differentially expressed genes. (B) Clustered heat map for comparing preadipocyte and adipocyte with the top 100 most differentially expressed genes. (C) The top 20 significantly GO enrichment terms for significantly differentially expressed genes. (D) The top 20 enriched pathways of dramatically differentially expressed genes.
Conjoint Analysis of RIP-Seq and RNA-Seq Data With Both Groups
We found an interesting relationship of differentially methylated m6A peaks and gene expression patterns in preadipocytes and adipocytes through cross-analysis of the m6A-seq and RNA-seq results, in which a positive correlation existed in differentially methylated m6A peaks and gene expression levels (Figure 6A). Otherwise, all genes were segregated into mainly four types: eight hypermethylated and upregulated genes termed “hyper-up”; seven hypomethylated and downregulated genes termed “hypo-down”; 12 hypermethylated while downregulated genes termed “hyper-down”; and two hypomethylated while upregulated genes termed “hypo-up” (Figure 6B). There were slightly more hyper-up and hypo-down than hyper-down and hypo-up. Table 1 lists the expression of genes that were significantly differently (|log2 (fold change)| > 1, p<.05), comprising significantly differently methylated peaks. Then, both groups were evaluated for the overall expression levels of the m6A-methylated and non-m6A-methylated transcripts (Figure 6C); the expression of methylated transcripts was higher than that of nonmethylated transcripts. These suggest that, in yak adipocyte differentiation, m6A modifications appear to have a positive association with mRNA expression. Furthermore, we were wondering if the position of m6A peaks on RNA transcripts or the number of m6A peaks per transcript is correlated with the levels of gene expression. Based on m6A modification sites, RNA transcripts were classified into subgroups. As shown in (Figure 6D), m6A modifications of RNA transcripts in CDS, 5′UTR or 3′UTR do not differ with gene expression. Through studying m6A-modified sites and relative expression levels of genes, revealing that the genes have three or four modified sites appears to be more abundant in contrast with other m6A-modified sites (Figure 6E). Furthermore, we implemented qRT-PCR to confirm the expression of differentially methylated genes between adipocyte and preadipocyte. The mRNA expression pattern was consistent with the RNA-seq data (Supplementary Figure S4A–B), which confirms the validity of our transcriptome results.
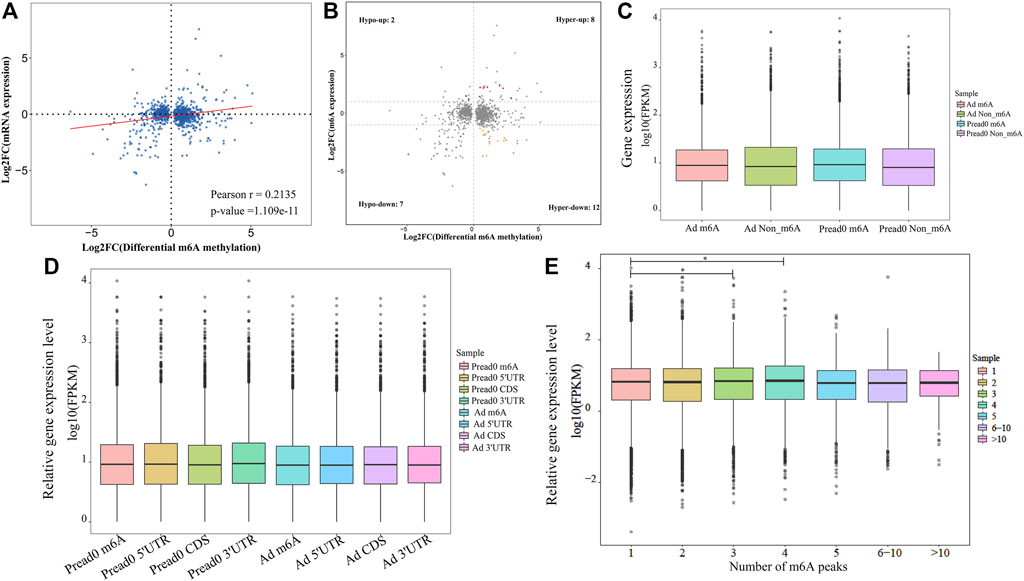
FIGURE 6. Conjoint analysis of differentially methylated genes and differentially expressed genes. (A) Dot plot of Log2 fold change (FC) (mRNA expression) versus Log2 FC (differential m6A methylation) revealing a positive association between total m6A methylation and level of mRNA expression (Pearson r = 0.2135; p-value = 1.109e-11). (B) Four-quadrant plot showing differentially expressed genes with differentially methylated m6A peaks. (C) The holistic expression levels of the transcripts with m6A-methylated and non-m6A-methylated in different groups. (D) For all m6A peaks, the position of m6A peaks for RNA transcripts and gene relative expression level. (E) Relative mRNA transcript expression level with different number of m6A peaks. *p < 0.05 compared with the m6A peak = 1.
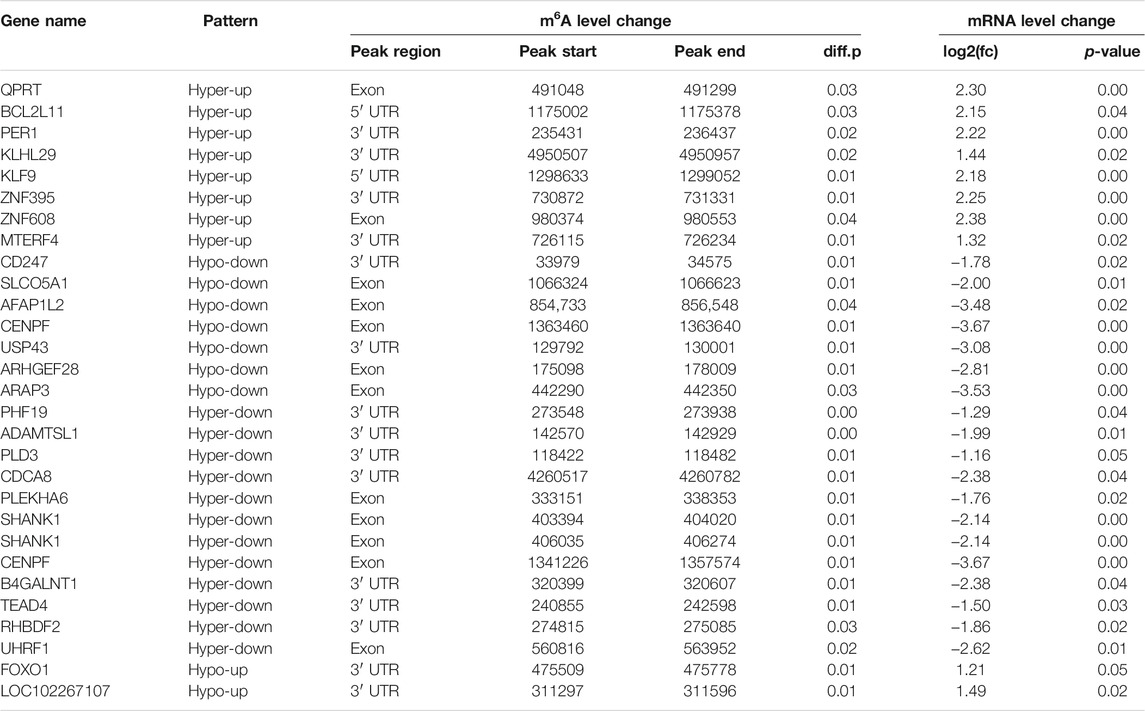
TABLE 1. List of 28 genes with significant changes in m6A and mRNA transcript abundance in yak adipocyte as compared with preadipocytes.
Differentially Methylation Modification is Linked to the Translation of Genes
Previous research indicates that RNA methylation plays an essential role in the translation of mRNA. Therefore, to reveal the influence of RNA methylation on mRNA translation, we explored the metagene with significant differences for methylation and nonsignificant differences in gene expression during yak preadipocyte differentiation. There were 155 genes with significant differences in methylation, and nonsignificant differences in expression existed in preadipocytes and adipocytes (Figure 7A; Supplementary File S7). To predict the function of these genes, GO and KEGG analyses were performed. These genes are mainly allocated to organism development, DNA binding, canonical Wnt signaling pathway, citrate cycle (TCA cycle), and calcium signaling pathway (Supplementary Figure S5A–B; Supplementary Files S8, S9). Therefore, the candidate genes were selected from the top 10 genes (Table 2) with the peck fold change for Western blot. Interestingly, the protein expression levels (ENTPD1, USP2, and PGAM2) were substantially higher in the adipocyte than the preadipocyte group (Figure 7B,C). The findings indicate that RNA methylation not only may regulate mRNA expression, but also effect mRNA translation during yak preadipocyte differentiation.
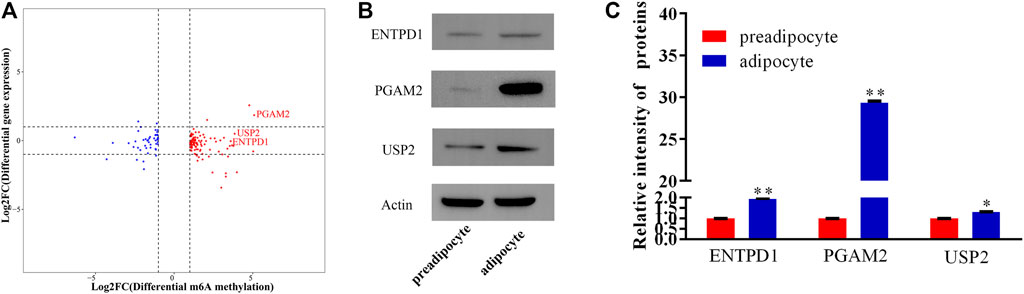
FIGURE 7. The analysis of genes with significant differences in methylation and non-significant differences expression during yak adipocytes differentiation. (A) Four quadrant plot showing the genes with significant differences in methylation and non-significant differences expression. (B,C) The proteins of USP2, ENTPD1, and PGAM2 were expressed in preadipocytes and adipocytes (SEM, * p < 0.05;** p < 0.01).
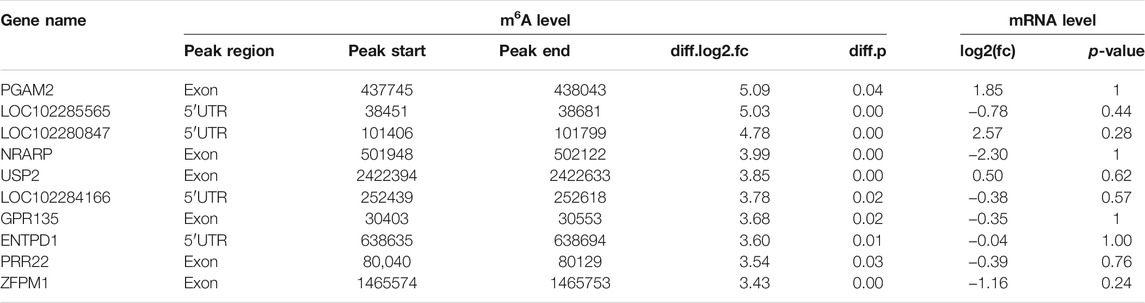
TABLE 2. List of 10 genes with significant difference in m6A and nonsignificant difference expression in yak adipocyte as compared with preadipocyte.
Discussion
The harsh environment of the Qinghai-Tibet Plateau encourages the yak to develop a special mechanism for energy metabolism. As an organ for energy metabolism, adipose tissue plays a crucial role in this process. To date, it is found that epigenetic regulation is engaged in various biological processes, including embryo development, stem cell self-renewal, DNA damage response, primary miRNA processing, and energy metabolism (Wu and Sun, 2006; Shi and Wu, 2009; Donohoe and Bultman, 2012; Li et al., 2013; Wang et al., 2013). In recent years, as the most extensive and plentiful internal modification on mRNAs, m6A modification is a major focus in the area of epigenetic regulation (Niu et al., 2013). Furthermore, the potential roles of m6A modification in most domestic animals, and especially for adipogenic differentiation, remained largely unknown. For the first time, our study establishes a comprehensive transcriptome-wide pattern of m6A modification in yak preadipocyte and adipocyte using MeRIP-Seq technology to explore the function of m6A modification in bovine adipogenic differentiation. Our findings show that yak mRNA m6A sites were primarily located in CDS, 5′UTRs and 3′UTRs, and the distribution semblable with humans and mice (Dominissini et al., 2012; Meyer et al., 2012), suggesting that, in mammalian transcriptomes, the overall distribution of m6A sites is similar. Besides this, Luo et al. reveal that m6A modifications were also enriched near the start codons of Arabidopsis (Luo et al., 2014). Thus, the distribution of m6A modification has various forms in different species. The m6A located at mRNA 5′UTR and 3′UTR of yak differ from mice and chickens (Luo et al., 2019; Cheng et al., 2021). We found m6A more enrichment in 3′UTR compared with 5′UTR, which contrasts with other mammals (Luo et al., 2019; Wang et al., 2019). The high-level of m6A methylation located in 3′UTR may be associated with mRNA stability, selective polyadenylation, signaling transport, and translocation (Shen et al., 2016; Yue et al., 2018). In addition, the m6A modification on the 3′UTR plays a regulatory element role for protein translation by recruiting specific factors to these m6A sites for RNA transport or protein synthesis (Niu et al., 2013; Wang et al., 2014). This may be one of the reasons causing a potential positive correlation between the degree of m6A methylation and transcript levels. Otherwise, the current study finds an m6A peak relatively increased at mRNA 5′UTR in Ad compared with Pread 0. The m6A located at mRNA 5′UTR can improve its cap-independent translation under heat shock (Meyer et al., 2015; Zhou et al., 2015). This indicates that the higher m6A signal at 5′UTR may promote mRNA translation during yak preadipocyte differentiation. Further, in our study, approximately 80% of the methylated transcripts included one or two m6A peaks, and about 20% of the methylated transcripts included three or more than three m6A peaks. The ratio is higher than in humans (5.5%) (Dominissini et al., 2012), pigs (10%) (Wang et al., 2018), chickens (5%) (Cheng et al., 2021), and mice (10%) (Luo et al., 2019). This phenomenon may be due to the more rapid rate of lipid metabolism in yaks, which is consistent with a previous study that cells and tissues with greater proliferation and differentiation capacity may require higher levels of m6A methylation to adapt to faster growth and development (Tao et al., 2017). According to previous studies, the consistent motif pattern of “RRACH” was over-represented in the m6A motif sequence area (Harper et al., 1990; Dominissini et al., 2012; Meyer et al., 2012). Accordingly, in comparison with previous studies (Dominissini et al., 2012; Meyer et al., 2012), the consensus motif GGACU sequence in the yak transcriptome was appropriately identified, revealing that RNA adenosine methylation was conserved in mammals.
Earlier studies indicate that m6A modification is closely related to gene expression (Meyer et al., 2012; Fu et al., 2014; Yue et al., 2015; Chen et al., 2020). Jean-Michel Fustin et al. report that METTL3 depletion inhibited the export mRNA (Jean-Michel et al., 2013), and Guanqun Zheng et al. report that depletion of ALKBH5 increased the export of mRNA to the cytoplasm (Zheng et al., 2013), suggesting m6A promotes the export of mRNA and modulates gene expression (Zhao et al., 2017). In HeLa cells, YTHDC1 was discovered to interact with SRSF3 and nuclear RNA export factor 1 (NXF1) to promote the export of m6A-modified mRNA out of the nucleus (Roundtree IA. et al., 2017). These results indicate a potential positive association between the degree of m6A methylation and the transcript level. In the present study, the genes METTL3, WTAP, METTL14, FTO, ALKBH5, and YTHDC1/2 were dramatically upregulated in adipocytes than the preadipocytes, and the majority of modified m6A genes were expressed at a medium level with a positive relationship in gene expression and m6A methylated modification. Our findings are in agreement with Chen et al., who reveal that m6A modifications tend to have a positive correlation with mRNA expression in clear cell renal cell carcinoma (Chen et al., 2020). These findings show that m6A methylation affects gene expression by controlling post-transcription regulation. The m6A-reader protein-containing YTH structural domain 2 (YTHDC2) can preferentially bind m6A within the consensus motif and improve the translation efficiency of mRNA (Yang et al., 2018). Interestingly, YTHDC2 was significantly upregulated during yak preadipocyte differentiation. Therefore, we speculate that m6A methylation modification not only influences mRNA expression but also may regulate mRNA translation during yak preadipocyte differentiation. Consequently, the genes with significant differences in methylation and nonsignificant differences in expression were detected in this study. Intriguingly, the results of Western blot revealed that the expression of ectonucleotidases CD39 (ENTPD1), ubiquitin-specific protease-2 (USP2), and phosphoglycerate mutase 2 (PGAM2) were significantly elevated in adipocytes compared with preadipocytes. Previous studies report that USP2 can influence the stabilization of fatty acid synthase (FAS), and 3,3′-diindolylmethane inhibits adipogenesis in preadipocytes by targeting USP2 activity (Graner et al., 2004; Yang et al., 2017). Enjyoji et al. reveal that entpd1-deficient mice have impaired glucose tolerance, reduced insulin sensitivity, and significantly elevated plasma insulin levels (Enjyoji et al., 2008). PGAM2 plays an important role in glycolysis, muscle growth and development, and organism physiological balance (Qiu et al., 2008; Mikawa et al., 2021). Accordingly, it is logical to conclude that m6A methylation modification exerts an essential role through affecting the translation of mRNA during yak preadipocyte differentiation. Nevertheless, further study is needed to verify the conjecture.
GO analysis explored the differentially methylated genes, which participated in the transcript regulation with a variety of transcription factors by RNA polymerase II. For example, FOXO1 identified as a Forkhead transcription factor controlling the differentiation of adipocytes (Nakae et al., 2003) and many members of the ZNF family considered as the crucial eukaryotic transcription factors involved in adipogenic metabolism (Wei et al., 2013), indicating m6A methylation participates in lipid metabolism. The KEGG pathway analysis revealed that the signaling pathway of differentially methylated genes is closely related to adipose metabolisms, such as the FoxO signaling pathway, Ether lipid metabolism, Glycerophospholipid metabolism, and Hippo signaling pathway-multiple species. In particular, FOXO1 was further found to be involved in the FoxO signaling pathway, which demonstrated the importance of adipocyte differentiation (Nakae et al., 2003). As a TEA domain family transcription factor, TEAD4 was selected from Hippo signaling pathway-multiple species, which recruits the cofactors VGLL4 and CtBP2 to inhibit murine adipogenesis (Zhang W. et al., 2018). To summarize the above findings, we concluded that activating the FoxO and Hippo signaling pathways through m6A methylated gene may perform a key function during the differentiation of yak adipocytes.
Integrated analysis of m6A-seq and mRNA-seq data exposed that 28 significant change genes exist in the adipocyte group with differently methylated m6A sites compared with preadipocyte. Several of the genes are confirmed to regulate adipose metabolism and adipogenic differentiation, such as ZNF395, KLF9, TEAD4, FOXO1, and UHRF1. ZNF395, the mRNA of which is hypermethylated and the expression upregulated in the adipocyte group compared with the preadipocyte group. As a member of the C2H-type Zinc finger proteins, ZNF395 is classified as Papillomavirus-binding factor and Huntington disease gene regulatory region binding protein 2 (Tanaka et al., 2004). Experiments of loss and gain function demonstrate that ZNF395 interacts with PPARG2 to modulate the transcriptional regulatory pathway that may be necessary for preadipocyte differentiation (Hasegawa et al., 2013). Besides this, previous literature reports that mesenchymal stem cells were cotransduced with ZNF395 and PPARG2 enhanced the endogenous expression of PPARG2 and C/EBPα, which are necessary for adipocyte differentiation (Sichtig et al., 2007; Hasegawa et al., 2013). In addition to that, it is reported that Krüppel-like factor 9 (KLF9), deemed to be the basic transcription element-binding protein-1 (BTEB1), could transactivate PPARγ2 to regulate adipogenesis in the 3T3-L1 cell line (Pei et al., 2011). Besides this, Kimura Hiroko et al. find that KLF9 triggered the early stage of adipogenesis by promoting the C/EBPβ gene expression in 3T3-L1 cells (Kimura and Fujimori, 2014). Ubiquitin-like with PHD and RING finger domains 1 (UHRF1) is widely documented to promote cell proliferation. Additionally, a study revealed that UHRF1 facilitates the proliferation of human adipose-derived stem cells and represses adipogenesis via inhibiting peroxisome proliferator-activated receptor γ (Chen et al., 2019). These findings suggest that m6A modifications may perform an essential role during yak adipocyte differentiation.
Conclusion
Current findings display that the m6A profiles and distribution patterns in the yak transcriptome. Besides this, functional enrichment analysis of differentially methylated genes reveal that several candidate genes participated in lipid metabolic pathways, suggesting that m6A methylation modifications are involved in the modulation of yak preadipocyte differentiation. Furthermore, we also explore the correlation between m6A methylation and the level of gene expression or mRNA translation, indicating a potential regulatory mechanism for m6A in adipocyte differentiation. These results provide additional knowledge of m6A methylation in adipose tissues, and it set the foundation for further understanding its possible roles and regulatory mechanisms, which could be helpful for exploration the yak adaptive mechanism in the harsh environment.
Data Availability Statement
The data was submitted to the data base of the Sequence Read Archive (SRA). The appropriate number for accession is PRJNA649748.
Ethics Statement
The animal study was reviewed and approved by the Animal Administration and Ethics Committee of Lanzhou Institute of Husbandry and Pharmaceutical Sciences, Chinese Academy of Agricultural Sciences. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author Contributions
The experiments were conceived and designed by YZ and PY. The experiments were performed by YZ. The experiments were assisted with JP, XW, XG, MC, PB, XD, CL. The paper was written by YZ and revised by QK. All authors have read and agreed to the published version of the article.
Funding
The National Natural Science Foundation of China (31972561), The Agricultural Science and Technology Innovation Program (CAASASTIP-2014-LIHPS-01) and The National Beef Cattle Industry Technology & System (CARS-37). The National Natural Science Foundation of China (31972561) supplied sequencing funding and laboratory verification cost, the Agricultural Science and Technology Innovation Program (CAASASTIP-2014-LIHPS-01) and the National Beef Cattle Industry Technology & System (CARS-37) supplied the source of experimental animals. The funding bodies played no role in the design of the study and collection, analysis, and interpretation of data and in writing the article.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the Key laboratory of yak Breeding Engineering Gansu Province for experimental condition.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.689067/full#supplementary-material
References
Adams, J. M., and Cory, S. (1975). Modified Nucleosides and Bizarre 5′-termini in Mouse Myeloma mRNA. Nature 255 (5503), 28–33. doi:10.1038/255028a0
Agarwala, S. D., Blitzblau, H. G., Hochwagen, A., and Fink, G. R. (2012). RNA Methylation by the MIS Complex Regulates a Cell Fate Decision in Yeast. Plos Genet. 8 (6), e1002732. doi:10.1371/journal.pgen.1002732
Ali, A. T., Hochfeld, W. E., Myburgh, R., and Pepper, M. S. (2013). Adipocyte and Adipogenesis. Eur. J. Cel Biol. 92 (6-7), 229–236. doi:10.1016/j.ejcb.2013.06.001
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: Tools for Motif Discovery and Searching. Nucleic Acids Res. 37 (Suppl. l_2), W202–W208. doi:10.1093/nar/gkp335
Batista, P. J., Molinie, B., Wang, J., Qu, K., Zhang, J., Li, L., et al. (2014). m6A RNA Modification Controls Cell Fate Transition in Mammalian Embryonic Stem Cells. Cell stem cell 15 (6), 707–719. doi:10.1016/j.stem.2014.09.019
Bokar, J. A., Shambaugh, M. E., Polayes, D., Matera, A. G., and Rottman, F. M. (1997). Purification and cDNA Cloning of the AdoMet-Binding Subunit of the Human mRNA (N6-Adenosine)-Methyltransferase. Rna 3 (11), 1233–1247.
Chen, K., Guo, Z., Luo, Y., Yuan, J., and Mo, Z. (2019). UHRF1 Promotes Proliferation of Human Adipose-Derived Stem Cells and Suppresses Adipogenesis via Inhibiting Peroxisome Proliferator-Activated Receptor γ. Biomed. Research International. 2019, 9456847 doi:10.1155/2019/9456847
Chen, Y., Zhou, C., Sun, Y., He, X., and Xue, D. (2020). m6A RNA Modification Modulates Gene Expression and Cancer-Related Pathways in clear Cell Renal Cell Carcinoma. Epigenomics 12 (2), 87–99. doi:10.2217/epi-2019-0182
Cheng, B., Leng, L., Li, Z., Wang, W., Jing, Y., Li, Y., et al. (2021). Profiling of RNA N6-Methyladenosine Methylation Reveals the Critical Role of m6A in Chicken Adipose Deposition. Front. Cel Dev. Biol. 9, 590468. doi:10.3389/fcell.2021.590468
Desrosiers, R., Friderici, K., and Rottman, F. (1974). Identification of Methylated Nucleosides in Messenger RNA from Novikoff Hepatoma Cells. Proc. Natl. Acad. Sci. 71 (10), 3971–3975. doi:10.1073/pnas.71.10.3971
Ding, X. Z., Guo, X., Yan, P., Liang, C. N., Bao, P. J., and Chu, M. (2012). Seasonal and Nutrients Intake Regulation of Lipoprotein Lipase (LPL) Activity in Grazing Yak (Bos Grunniens) in the Alpine Regions Around Qinghai Lake. Livestock Sci. 143 (1), 29–34. doi:10.1016/j.livsci.2011.08.004
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., et al. (2012). Topology of the Human and Mouse m6A RNA Methylomes Revealed by m6A-Seq. Nature 485 (7397), 201–206. doi:10.1038/nature11112
Dong, Q., Zhao, X., Ma, Y., Xu, S., and Li, Q. (2006). Live-weight Gain, Apparent Digestibility, and Economic Benefits of Yaks Fed Different Diets during winter on the Tibetan Plateau. Livestock Sci. 101 (1-3), 199–207. doi:10.1016/j.livprodsci.2005.11.009
Donohoe, D. R., and Bultman, S. J. (2012). Metaboloepigenetics: Interrelationships between Energy Metabolism and Epigenetic Control of Gene Expression. J. Cel. Physiol. 227 (9), 3169–3177. doi:10.1002/jcp.24054
Enjyoji, K., Kotani, K., Thukral, C., Blumel, B., Sun, X., Wu, Y., et al. (2008). Deletion of Cd39/entpd1 Results in Hepatic Insulin Resistance. Diabetes 57 (9), 2311–2320. doi:10.2337/db07-1265
Fu, Y., Dominissini, D., Rechavi, G., and He, C. (2014). Gene Expression Regulation Mediated through Reversible m6A RNA Methylation. Nat. Rev. Genet. 15 (5), 293–306. doi:10.1038/nrg3724
Furuichi, Y., Morgan, M., Shatkin, A. J., Jelinek, W., Salditt-Georgieff, M., and Darnell, J. E. (1975). Methylated, Blocked 5 Termini in HeLa Cell mRNA. Proc. Natl. Acad. Sci. 72 (5), 1904–1908. doi:10.1073/pnas.72.5.1904
Graner, E., Tang, D., Rossi, S., Baron, A., Migita, T., Weinstein, L. J., et al. (2004). The Isopeptidase USP2a Regulates the Stability of Fatty Acid Synthase in Prostate Cancer. Cancer cell 5 (3), 253–261. doi:10.1016/s1535-6108(04)00055-8
Harper, J. E., Miceli, S. M., Roberts, R. J., and Manley, J. L. (1990). Sequence Specificity of the Human mRNA N6-Adenosine Methylasein Vitro. Nucl. Acids Res. 18 (19), 5735–5741. doi:10.1093/nar/18.19.5735
Hasegawa, R., Tomaru, Y., de Hoon, M., Suzuki, H., Hayashizaki, Y., and Shin, J. W. (2013). Identification of ZNF395 as a Novel Modulator of Adipogenesis. Exp. Cel. Res. 319 (3), 68–76. doi:10.1016/j.yexcr.2012.11.003
Heinz, S., Benner, C., Spann, N., Bertolino, E., Lin, Y. C., Laslo, P., et al. (2010). Simple Combinations of Lineage-Determining Transcription Factors Prime Cis-Regulatory Elements Required for Macrophage and B Cell Identities. Mol. Cel. 38 (4), 576–589. doi:10.1016/j.molcel.2010.05.004
Huse, J. T., Byant, D., Yang, Y., Pijak, D. S., D'Souza, I., Lah, J. J., et al. (2003). Endoproteolysis of औ-Secretase (औ-Site Amyloid Precursor Protein-Cleaving Enzyme) within its Catalytic Domain. J. Biol. Chem. 278 (19), 17141–17149. doi:10.1074/jbc.m213303200
Jean-Michel, F., Masao, D., Yoshiaki, Y., Hayashi, H., Shinichi, N., Minoru, Y., et al. (2013). RNA-Methylation-Dependent RNA Processing Controls the Speed of the Circadian Clock. Cell 155 (4), 793–806. doi:10.1016/j.cell.2013.10.026
Jia, G., Fu, Y., Zhao, X., Dai, Q., Zheng, G., Yang, Y., et al. (2011). N6-methyladenosine in Nuclear RNA Is a Major Substrate of the Obesity-Associated FTO. Nat. Chem. Biol. 7 (12), 885–887. doi:10.1038/nchembio.687
Jiang, Q., Sun, B., Liu, Q., Cai, M., Wu, R., Wang, F., et al. (2019). MTCH2 Promotes Adipogenesis in Intramuscular Preadipocytes via an M 6 A‐YTHDF1‐dependent Mechanism. FASEB j. 33 (2), 2971–2981. doi:10.1096/fj.201801393RRR
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 12 (4), 357–360. doi:10.1038/nmeth.3317
Kimura, H., and Fujimori, K. (2014). Activation of Early Phase of Adipogenesis through Krüppel-like Factor KLF9-Mediated, Enhanced Expression of CCAAT/enhancer-binding Protein β in 3T3-L1 Cells. Gene 534 (2), 169–176. doi:10.1016/j.gene.2013.10.065
Knuckles, P., Lence, T., Haussmann, I. U., Jacob, D., Kreim, N., Carl, S. H., et al. (2018). Zc3h13/Flacc Is Required for Adenosine Methylation by Bridging the mRNA-Binding Factor Rbm15/Spenito to the m6A Machinery Component Wtap/Fl(2)d. Genes Dev. 32 (5-6), 415–429. doi:10.1101/gad.309146.117
Kudou, K., Komatsu, T., Nogami, J., Maehara, K., Harada, A., Saeki, H., et al. (2017). The Requirement of Mettl3-Promoted MyoD mRNA Maintenance in Proliferative Myoblasts for Skeletal Muscle Differentiation. Open Biol. 7 (9), 170119. doi:10.1098/rsob.170119
Lavi, S., and Shatkin, A. J. (1975). Methylated Simian Virus 40-specific RNA from Nuclei and Cytoplasm of Infected BSC-1 Cells. Proc. Natl. Acad. Sci. 72 (6), 2012–2016. doi:10.1073/pnas.72.6.2012
Li, D.-Q., Nair, S. S., and Kumar, R. (2013). The MORC Family. Epigenetics 8 (7), 685–693. doi:10.4161/epi.24976
Liu, J., Yue, Y., Han, D., Wang, X., Fu, Y., Zhang, L., et al. (2014). A METTL3-METTL14 Complex Mediates Mammalian Nuclear RNA N6-Adenosine Methylation. Nat. Chem. Biol. 10 (2), 93–95. doi:10.1038/nchembio.1432
Liu, N., and Pan, T. (2016). N6-methyladenosine-encoded Epitranscriptomics. Nat. Struct. Mol. Biol. 23 (2), 98–102. doi:10.1038/nsmb.3162
Long, R. J., Apori, S. O., Castro, F. B., and Ørskov, E. R. (1999). Feed Value of Native Forages of the Tibetan Plateau of China. Anim. Feed Sci. Techn. 80(2), 101–113. doi:10.1016/S0377-8401(99)00057-7
Long, R. J., Ding, L. M., Shang, Z. H., and Guo, X. H. (2008). The Yak Grazing System on the Qinghai-Tibetan Plateau and its Status. Rangel. J. 30 (2), 241–246. doi:10.1071/RJ08012
Luo, G.-Z., MacQueen, A., Zheng, G., Duan, H., Dore, L. C., Lu, Z., et al. (2014). Unique Features of the m6A Methylome in Arabidopsis thaliana. Nat. Commun. 5, 5630. doi:10.1038/ncomms6630
Luo, S., and Tong, L. (2014). Molecular Basis for the Recognition of Methylated Adenines in RNA by the Eukaryotic YTH Domain. Proc. Natl. Acad. Sci. 111 (38), 13834–13839. doi:10.1073/pnas.1412742111
Luo, Z., Zhang, Z., Tai, L., Zhang, L., Sun, Z., and Zhou, L. (2019). Comprehensive Analysis of Differences of N6-Methyladenosine RNA Methylomes between High-Fat-Fed and normal Mouse Livers. Epigenomics 11 (11), 1267–1282. doi:10.2217/epi-2019-0009
Martin, M. (2011). Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet j. 17 (1), 10–12. doi:10.14806/ej.17.1.200
Meng, J., Lu, Z., Liu, H., Zhang, L., Zhang, S., Chen, Y., et al. (2014). A Protocol for RNA Methylation Differential Analysis with MeRIP-Seq Data and exomePeak R/Bioconductor Package. Methods 69 (3), 274–281. doi:10.1016/j.ymeth.2014.06.008
Meyer, K. D., Patil, D. P., Zhou, J., Zinoviev, A., Skabkin, M. A., Elemento, O., et al. (2015). 5′ UTR m6A Promotes Cap-independent Translation. Cell 163 (4), 999–1010. doi:10.1016/j.cell.2015.10.012
Meyer, K. D., Saletore, Y., Zumbo, P., Elemento, O., Mason, C. E., and Jaffrey, S. R. (2012). Comprehensive Analysis of mRNA Methylation Reveals Enrichment in 3′ UTRs and Near Stop Codons. Cell 149 (7), 1635–1646. doi:10.1016/j.cell.2012.05.003
Mikawa, T., Shibata, E., Shimada, M., Ito, K., Ito, T., Kanda, H., et al. (2021). Characterization of Genetically Modified Mice for Phosphoglycerate Mutase, a Vitally-Essential Enzyme in Glycolysis. Plos one 16 (4), e0250856. doi:10.1371/journal.pone.0250856
Nachtergaele, S., and He, C. (2017). The Emerging Biology of RNA post-transcriptional Modifications. RNA Biol. 14 (2), 156–163. doi:10.1080/15476286.2016.1267096
Nakae, J., Kitamura, T., Kitamura, Y., Biggs, W. H., Arden, K. C., and Accili, D. (2003). The Forkhead Transcription Factor Foxo1 Regulates Adipocyte Differentiation. Dev. Cel. 4 (1), 119–129. doi:10.1016/s1534-5807(02)00401-x
Niu, Y., Zhao, X., Wu, Y.-S., Li, M.-M., Wang, X.-J., and Yang, Y.-G. (2013). N6-methyl-adenosine (m6A) in RNA: an Old Modification with a Novel Epigenetic Function. Genomics, Proteomics & Bioinformatics 11 (1), 8–17. doi:10.1016/j.gpb.2012.12.002
Patil, D. P., Chen, C.-K., Pickering, B. F., Chow, A., Jackson, C., Guttman, M., et al. (2016). m6A RNA Methylation Promotes XIST-Mediated Transcriptional Repression. Nature 537 (7620), 369–373. doi:10.1038/nature19342
Pei, H., Yao, Y., Yang, Y., Liao, K., and Wu, J.-R. (2011). Krüppel-like Factor KLF9 Regulates PPARγ Transactivation at the Middle Stage of Adipogenesis. Cell Death Differ 18 (2), 315–327. doi:10.1038/cdd.2010.100
Perry, R. P., and Kelley, D. E. (1974). Existence of Methylated Messenger RNA in Mouse L Cells. Cell 1 (1), 37–42. doi:10.1016/0092-8674(74)90153-6
Pertea, M., Pertea, G. M., Antonescu, C. M., Chang, T.-C., Mendell, J. T., and Salzberg, S. L. (2015). StringTie Enables Improved Reconstruction of a Transcriptome from RNA-Seq Reads. Nat. Biotechnol. 33 (3), 290–295. doi:10.1038/nbt.3122
Ping, X.-L., Sun, B.-F., Wang, L., Xiao, W., Yang, X., Wang, W.-J., et al. (2014). Mammalian WTAP Is a Regulatory Subunit of the RNA N6-Methyladenosine Methyltransferase. Cell Res 24 (2), 177–189. doi:10.1038/cr.2014.3
Qiu, H., Zhao, S., Xu, X., Yerle, M., and Liu, B. (2008). Assignment and Expression Patterns of Porcine Muscle-specific Isoform of Phosphoglycerate Mutase Gene. J. Genet. genomics 35 (5), 257–260. doi:10.1016/s1673-8527(08)60036-3
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: a Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 26 (1), 139–140. doi:10.1093/bioinformatics/btp616
Roundtree, I. A., Luo, G. Z., Zhang, Z., Wang, X., Zhou, T., Cui, Y., et al. (2017b). YTHDC1 Mediates Nuclear export of N6-Methyladenosine Methylated mRNAs. Elife 6, e31311. doi:10.7554/eLife.31311
Roundtree, I. A., Evans, M. E., Pan, T., and He, C. (2017a). Dynamic RNA Modifications in Gene Expression Regulation. Cell 169 (7), 1187–1200. doi:10.1016/j.cell.2017.05.045
Schwartz, S., Mumbach, M. R., Jovanovic, M., Wang, T., Maciag, K., Bushkin, G. G., et al. (2014). Perturbation of m6A Writers Reveals Two Distinct Classes of mRNA Methylation at Internal and 5′ Sites. Cel Rep. 8 (1), 284–296. doi:10.1016/j.celrep.2014.05.048
Shen, L., Liang, Z., Gu, X., Chen, Y., Teo, Z. W. N., Hou, X., et al. (2016). N 6 -Methyladenosine RNA Modification Regulates Shoot Stem Cell Fate in Arabidopsis. Dev. Cel. 38 (2), 186–200. doi:10.1016/j.devcel.2016.06.008
Shi, L., and Wu, J. (2009). Epigenetic Regulation in Mammalian Preimplantation Embryo Development. Reprod. Biol. Endocrinol. 7 (1), 59. doi:10.1186/1477-7827-7-59
Shikui, D., Ruijun, L., Muyi, K., Xiaogeng, P., and Yanjun, G. (2003). Effect of Urea Multinutritional Molasses Block Supplementation on Liveweight Change of Yak Calves and Productive and Reproductive Performances of Yak Cows. Can. J. Anim. Sci. 83 (1), 141–145. doi:10.4141/a01-097
Sichtig, N., Körfer, N., and Steger, G. (2007). Papillomavirus Binding Factor Binds to SAP30 and Represses Transcription via Recruitment of the HDAC1 Co-repressor Complex. Arch. Biochem. Biophys. 467 (1), 67–75. doi:10.1016/j.abb.2007.08.015
Tanaka, K., Shouguchi-Miyata, J., Miyamoto, N., and Ikeda, J.-E. (2004). Novel Nuclear Shuttle Proteins, HDBP1 and HDBP2, Bind to Neuronal Cell-specific Cis-Regulatory Element in the Promoter for the Human Huntington's Disease Gene. J. Biol. Chem. 279 (8), 7275–7286. doi:10.1074/jbc.m310726200
Tao, X., Chen, J., Jiang, Y., Wei, Y., Chen, Y., Xu, H., et al. (2017). Transcriptome-wide N 6 -methyladenosine Methylome Profiling of Porcine Muscle and Adipose Tissues Reveals a Potential Mechanism for Transcriptional Regulation and Differential Methylation Pattern. BMC Genomics 18 (1), 336. doi:10.1186/s12864-017-3719-1
Wang, X., Lu, Z., Gomez, A., Hon, G. C., Yue, Y., Han, D., et al. (2014). N6-methyladenosine-dependent Regulation of Messenger RNA Stability. Nature 505 (7481), 117–120. doi:10.1038/nature12730
Wang, X., Sun, B., Jiang, Q., Wu, R., Cai, M., Yao, Y., et al. (2018). mRNA m6A Plays Opposite Role in Regulating UCP2 and PNPLA2 Protein Expression in Adipocytes. Int. J. Obes. 42 (11), 1912–1924. doi:10.1038/s41366-018-0027-z
Wang, X., Zhao, B. S., Roundtree, I. A., Lu, Z., Han, D., Ma, H., et al. (2015). N6-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 161 (6), 1388–1399. doi:10.1016/j.cell.2015.05.014
Wang, Y., Zheng, Y., Guo, D., Zhang, X., Guo, S., Hui, T., et al. (2019). m6A Methylation Analysis of Differentially Expressed Genes in Skin Tissues of Coarse and Fine Type Liaoning Cashmere Goats. Front. Genet. 10, 1318. doi:10.3389/fgene.2019.01318
Wang, Z., Yao, H., Lin, S., Zhu, X., Shen, Z., Lu, G., et al. (2013). Transcriptional and Epigenetic Regulation of Human microRNAs. Cancer Lett. 331 (1), 1–10. doi:10.1016/j.canlet.2012.12.006
Wei, C. M., and Moss, B. (1975). Methylated Nucleotides Block 5'-terminus of Vaccinia Virus Messenger RNA. Proc. Natl. Acad. Sci. 72 (1), 318–322. doi:10.1073/pnas.72.1.318
Wei, S., Zhang, L., Zhou, X., Du, M., Jiang, Z., Hausman, G. J., et al. (2013). Emerging Roles of Zinc finger Proteins in Regulating Adipogenesis. Cell. Mol. Life Sci. 70 (23), 4569–4584. doi:10.1007/s00018-013-1395-0
Wen, J., Lv, R., Ma, H., Shen, H., He, C., Wang, J., et al. (2018). Zc3h13 Regulates Nuclear RNA m6A Methylation and Mouse Embryonic Stem Cell Self-Renewal. Mol. Cel. 69 (6), 1028–1038. doi:10.1016/j.molcel.2018.02.015
Wu, H., and Sun, Y. E. (2006). Epigenetic Regulation of Stem Cell Differentiation. Pediatr. Res. 59 (4), 21R–5R. doi:10.1203/01.pdr.0000203565.76028.2a
Xie, C., Mao, X., Huang, J., Ding, Y., Wu, J., Dong, S., et al. (2011). KOBAS 2.0: a Web Server for Annotation and Identification of Enriched Pathways and Diseases. Nucleic Acids Res. 39 (Web Server issue), W316–W322. doi:10.1093/nar/gkr483
Xu, K., Yang, Y., Feng, G.-H., Sun, B.-F., Chen, J.-Q., Li, Y.-F., et al. (2017). Mettl3-mediated m6A Regulates Spermatogonial Differentiation and Meiosis Initiation. Cel Res 27 (9), 1100–1114. doi:10.1038/cr.2017.100
Yang, H., Seo, S. G., Shin, S. H., Min, S., Kang, M. J., Yoo, R., et al. (2017). 3,3'-Diindolylmethane Suppresses High-Fat Diet-Induced Obesity through Inhibiting Adipogenesis of Pre-adipocytes by Targeting USP2 Activity. Mol. Nutr. Food Res. 61 (10), 1700119. doi:10.1002/mnfr.201700119
Yang, Y., Hsu, P. J., Chen, Y.-S., and Yang, Y.-G. (2018). Dynamic Transcriptomic m6A Decoration: Writers, Erasers, Readers and Functions in RNA Metabolism. Cel Res 28 (6), 616–624. doi:10.1038/s41422-018-0040-8
Young, M. D., Wakefield, M. J., Smyth, G. K., and Oshlack, A. (2010). Gene Ontology Analysis for RNA-Seq: Accounting for Selection Bias. Genome Biol. 11 (2), R14. doi:10.1186/gb-2010-11-2-r14
Yu, G., Wang, L.-G., and He, Q.-Y. (2015). ChIPseeker: an R/Bioconductor Package for ChIP Peak Annotation, Comparison and Visualization. Bioinformatics 31 (14), 2382–2383. doi:10.1093/bioinformatics/btv145
Yue, Y., Liu, J., Cui, X., Cao, J., Luo, G., Zhang, Z., et al. (2018). VIRMA Mediates Preferential m6A mRNA Methylation in 3'UTR and Near Stop Codon and Associates with Alternative Polyadenylation. Cell Discov 4 (1), 10–17. doi:10.1038/s41421-018-0019-0
Yue, Y., Liu, J., and He, C. (2015). RNAN6-methyladenosine Methylation in post-transcriptional Gene Expression Regulation. Genes Dev. 29 (13), 1343–1355. doi:10.1101/gad.262766.115
Zhang, W., Xu, J., Li, J., Guo, T., Jiang, D., Feng, X., et al. (2018a). The TEA Domain Family Transcription Factor TEAD4 Represses Murine Adipogenesis by Recruiting the Cofactors VGLL4 and CtBP2 into a Transcriptional Complex. J. Biol. Chem. 293 (44), 17119–17134. doi:10.1074/jbc.ra118.003608
Zhang, Y., Guo, X., Pei, J., Chu, M., Ding, X., Wu, X., et al. (2020). CircRNA Expression Profile during Yak Adipocyte Differentiation and Screen Potential circRNAs for Adipocyte Differentiation. Genes 11 (4), 414. doi:10.3390/genes11040414
Zhang, Y., Wu, X., Liang, C., Bao, P., Ding, X., Chu, M., et al. (2018b). MicroRNA-200a Regulates Adipocyte Differentiation in the Domestic Yak Bos Grunniens. Gene 650, 41–48. doi:10.1016/j.gene.2018.01.054
Zhao, B. S., Roundtree, I. A., and He, C. (2017). Post-transcriptional Gene Regulation by mRNA Modifications. Nat. Rev. Mol. Cel Biol 18 (1), 31–42. doi:10.1038/nrm.2016.132
Zhao, X., Yang, Y., Sun, B.-F., Shi, Y., Yang, X., Xiao, W., et al. (2014). FTO-dependent Demethylation of N6-Methyladenosine Regulates mRNA Splicing and Is Required for Adipogenesis. Cel Res 24 (12), 1403–1419. doi:10.1038/cr.2014.151
Zheng, G., Dahl, J. A., Niu, Y., Fedorcsak, P., Huang, C.-M., Li, C. J., et al. (2013). ALKBH5 Is a Mammalian RNA Demethylase that Impacts RNA Metabolism and Mouse Fertility. Mol. Cel 49 (1), 18–29. doi:10.1016/j.molcel.2012.10.015
Zhong, X., Yu, J., Frazier, K., Weng, X., Li, Y., Cham, C. M., et al. (2018). Circadian Clock Regulation of Hepatic Lipid Metabolism by Modulation of m6A mRNA Methylation. Cel Rep. 25 (7), 1816–1828. doi:10.1016/j.celrep.2018.10.068
Keywords: yak, adipocyte, N6-methyladenosine, MeRIP-seq, regulatory mechanism
Citation: Zhang Y, Liang C, Wu X, Pei J, Guo X, Chu M, Ding X, Bao P, Kalwar Q and Yan P (2021) Integrated Study of Transcriptome-wide m6A Methylome Reveals Novel Insights Into the Character and Function of m6A Methylation During Yak Adipocyte Differentiation. Front. Cell Dev. Biol. 9:689067. doi: 10.3389/fcell.2021.689067
Received: 31 March 2021; Accepted: 01 November 2021;
Published: 03 December 2021.
Edited by:
Stephanie McKay, University of Vermont, United StatesReviewed by:
Narendra Singh, Stowers Institute for Medical Research, United StatesXiaojian Shao, National Research Council Canada (NRC-CNRC), Canada
Copyright © 2021 Zhang, Liang, Wu, Pei, Guo, Chu, Ding, Bao, Kalwar and Yan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ping Yan, cGluZ3lhbmx6QDE2My5jb20=
 Yongfeng Zhang
Yongfeng Zhang Chunnian Liang1
Chunnian Liang1 Xiaoyun Wu
Xiaoyun Wu Xian Guo
Xian Guo Xuezhi Ding
Xuezhi Ding Pengjia Bao
Pengjia Bao Qudratullah Kalwar
Qudratullah Kalwar Ping Yan
Ping Yan