
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol., 02 July 2021
Sec. Stem Cell Research
Volume 9 - 2021 | https://doi.org/10.3389/fcell.2021.687117
This article is part of the Research TopicTendon Stem Cells from Basic Biology to Clinical ApplicationView all 12 articles
Tendons connect the muscle abdomen of skeletal muscles to the bone, which transmits the force generated by the muscle abdomen contraction and pulls the bone into motion. Tendon injury is a common clinical condition occurring in certain populations, such as repeated tendon strains in athletes. And it can lead to substantial pain and loss of motor function, in severe cases, significant disability. Tendon healing and regeneration have attracted growing interests. Some treatments including growth factors, stem cell therapies and rehabilitation programs have been tried to improve tendon healing. However, the basic cellular biology and pathology of tendons are still not fully understood, and the management of tendon injury remains a considerable challenge. Regulating gene expression at post-transcriptional level, microRNA (miRNA) has been increasingly recognized as essential regulators in the biological processes of tendon healing and regeneration. A wide range of miRNAs in tendon injury have been shown to play vital roles in maintaining and regulating its physiological function, as well as regulating the tenogenic differentiation potential of stem cells. In this review, we show the summary of the latest information on the role of miRNAs in tendon healing and regeneration, and also discuss potentials for miRNA-directed diagnosis and therapy in tendon injuries and tendinopathy, which may provide new theoretical foundation for tenogenesis and tendon healing.
Tendon is a connective tissue composed of closely arranged bundles of parallel collagen fibers (Wang et al., 2012). It plays an important role in the skeletal muscle system. Tendons are located between muscles and bones and can cushion the pressure caused by the direct interaction between muscles and bones, thereby stabilizing the joints (Laurencin and Freeman, 2005; Thorpe and Screen, 2016). As a long-term pressure-bearing tissue, tendons are easy to form a variety of acute and chronic injuries (Khan et al., 1999). Since the beginning of this century, the global prevalence of tendon disease has been on the rise. Tendon disease is one of the most common diagnoses of people engaged in sports professions, accounting for 30% of the total number of injuries diagnosed (Millar et al., 2021). At the late stage of tendon healing, it is easy to form scar tissue with decreased strength. And it is easy to adhere to the surrounding tissue, which increases the risk of tendon injury again (Bruns et al., 2000; Sharma and Maffulli, 2005). On the other hand, there are fewer blood vessels and poor blood flow inside the tendon, which makes the tendon heal more slowly (Whalen, 1951; Miyashita et al., 1997; Tempfer and Traweger, 2015; Nichols et al., 2019). The high frequency and slow healing of tendon injury not only seriously affects the normal life of patients, but also increases the social and economic burden. Therefore, how to promote tendon repair and regeneration is a great challenge in tissue engineering. miRNA is a small non-coding ribonucleic acid that participates in the regulation of cellular processes by inhibiting the translation and stability of messenger ribonucleic acid. Since the discovery of miRNA in the 1990s, people’s understanding of miRNAs has been deepened (Lee et al., 1993; Di Leva et al., 2014), and its application has become more and more extensive. miRNAs play an important role in inflammation, cell cycle regulation, stress response, cell growth, differentiation, proliferation, apoptosis and migration etc. (Lee et al., 1993; Hata, 2013; Saliminejad et al., 2019), recent studies have shown that they also participate in tendon regeneration and repair.
Here, we briefly described the structure and function of tendon, and the biology of miRNA. Then the roles of miRNA in tendon repair and regeneration was summarized; and the potentials and challenges of miRNA-directed diagnosis and therapy in tendon injuries and tendinopathy were also discussed.
Tendon is a connective tissue rich in matrix, which is mainly composed of closely arranged bundles of parallel collagen fibers (Wang et al., 2012; Tsai et al., 2021). They are attached to muscles and bones, and stabilize joints by cushioning the pressure caused by direct interaction between muscles and bones (Laurencin and Freeman, 2005; Thorpe and Screen, 2016). The place where the tendon is inserted into the muscle is called the tendon joint, while the joint between the tendon and the bone is called the bone-tendon junction (O’Brien, 1997; Thorpe and Screen, 2016). The tendon, as a tissue that transmits and loads, transfers strength from the muscle to the bone and drives the joint movement by contracting the muscle (Wang, 2006; Voleti et al., 2012). There are usually no blood vessels inside the tendon, while there are blood vessels and nerves passing through the sheath around it (Kannus, 2000; Tempfer and Traweger, 2015).
Tendon is mainly composed of type I collagen (also known as collagen I, COL I), which contains a small amount of proteoglycans, glycoproteins and minor collagens (Voleti et al., 2012). The dry mass of human tendons accounts for about 30% of the total tendon mass, and the remaining 70% is water (Sharma and Maffulli, 2005). Type I collagen accounts for 65–80%, and elastin accounts for about 2% of the dry weight of tendons (Sharma and Maffulli, 2006). Collagen is mainly a helical structure formed by the intertwining of three chains (Voleti et al., 2012). It is constantly cross-linked to form the specific spatial structure of the tendon (Gaut and Duprez, 2016). The collagen fibers are considered to be the basic force transmission units of tendons, which are densely arranged in the extracellular matrix (ECM) and parallel to the bone-muscle axis (Nourissat et al., 2015). Among the tendon cells, tenoblasts and tenocytes accounts for about 90%, while the remaining 10% are mainly composed of chondrocytes, synovial cells, vascular endothelial cells and smooth muscle cells (Sharma and Maffulli, 2006; Wang, 2006).
Since the discovery of the first miRNA in 1993 and another miRNA let-7 7 years later (Lee et al., 1993; Reinhart et al., 2000; Di Leva et al., 2014), people’s understanding of miRNA has been deepened for more than two decades, and the potential therapeutic effects of miRNA have been explored. According to the latest version of the microribonucleic acid database (miRBase) released in 2018, the human genome contains 1,917 annotated hairpin precursors and 2,654 mature sequences (Kozomara et al., 2019). miRNA is a class of endogenous non-coding RNA molecules with a length of about 22 nucleotides (Ambros et al., 2003; Ambros, 2004; Kabekkodu et al., 2018; Lu and Rothenberg, 2018). miRNA regulates gene expression mainly by binding to mRNA, which plays an important role in cell growth, differentiation, proliferation and apoptosis (Vasudevan et al., 2007; Sayed and Abdellatif, 2011). They are widely distributed in many tissues in the body which can be extracted from cells, tissues and body fluids (tears, urine, plasma and serum) (Lu and Rothenberg, 2018). Most of the miRNAs are transcribed by the ribonucleic acid polymerase II (Cai et al., 2004; Lee et al., 2004), but there is a small part of miRNAs that are associated with Alu repeats have been reported to be transcribed by Pol III (Borchert et al., 2006). The production of miRNA is initiated by the transcription of pri-miRNA in the nucleus. Before being transported to the cytoplasm, pri-miRNA is further processed by RNase III Drosha enzyme to form miRNA precursor (pre-miRNA). Then it is digested by another RNAse III enzyme Dicer, and binds with Argonaute protein to form RNA-induced silencing complex (RISC). After that, one of the double strands is degraded and the other forms a mature miRNA (Esteller, 2011; Di Leva et al., 2014; Koturbash et al., 2015; Vidigal and Ventura, 2015). Although the miRNAs do not encode proteins themselves, they could influence the transcription of mRNAs to regulate proteins expression (Selbach et al., 2008). The functions of miRNA are mainly divided into four aspects: (1) participate in epigenetic regulation of gene expression; (2) mRNA degradation; (3) post-transcriptional inhibition of target mRNA translation; (4) act as a ligand to bind to immune receptors and participate in inflammatory response (Carthew and Sontheimer, 2009; Fabbri et al., 2012; Lehmann et al., 2012; Fritz et al., 2016; Kabekkodu et al., 2018). Therefore, exploring new means of treating diseases are made possible by targeting miRNAs to intervene in the activity of specific genes. This strategy is also beneficial for promoting tendon healing and regeneration which is linked to the roles of miRNAs in tendon.
Tendon healing is characterized by scar formation, tissue disorder, and decreased mechanical properties (Andarawis-Puri et al., 2015). Tendon healing can be generally summarized as three stages: inflammatory response stage, proliferative repair stage and remodeling stage, and these three stages overlap each other (Strickland, 2000). Tendinopathy is a failed healing reaction, accompanied by haphazard proliferation of tendinocytes, intracellular abnormalities, destruction of collagen fibers, and subsequent increase in the non-collagen matrix (Longo et al., 2018). Many miRNAs have been shown to be involved in regulating the biological processes of tendon healing (Summarized in Table 1).
Apoptosis of tendinocytes is accelerated within a few days after injury, followed by increased cell proliferation within 2–4 weeks, which activates molecular events to inhibit apoptosis (Wu et al., 2010). One study filtrated differentially expressed genes between diseased and normal tendons, and miR-499 was found to regulate CUGBP2 and MYB which are important regulators of cellular proliferation, apoptosis and differentiation (Cai et al., 2015). Inhibiting miRNA-205-5p could promote tendon-bone healing of rotator cuff tendon (RCT) through increasing cellular viability and proliferation (Xu et al., 2019).
It has been reported that inflammation is involved in the whole process of tissue repair, with both advantages and disadvantages (Eming et al., 2009). The expression levels of miRNAs that are inflammatory targets mediated by the JAK2/STAT3 pathway, such as miR-146a-5p, miR-193b-3p, etc., were significantly reduced in glenohumeral arthritis tendon (Thankam et al., 2018). The network analysis of genes associated with AMPK and TREM-1 signaling revealed miR-31-5p, miR-195-5p and other thirteen miRNAs might be interrelated with the pathogenesis of RCT injury patients (Thankam et al., 2019). Such knowledge has important implications for inflammatory response and proliferative repair stage of tendon healing.
Despite advances in tendon repair and post-operative rehabilitation, tendon adhesion is still considered to be a challenging part of the repair process. The formation of adhesion during tendon healing is regulated by TGF-β, Smad3, p65, etc. (Katzel et al., 2011; Wu et al., 2016; Chen et al., 2017). Using miRNA therapy to modulate TGF-β expression holds great promise in preventing tendon adhesion formation (Chen et al., 2009). Overexpression of miR-29b down-regulated TGF-β1/Smad3 levels, and inhibited fibroblast growth, thereby enhanced tendon healing after rats Achilles tendon injury (Chen et al., 2014). Exosomes are rich in miRNAs, which act as important regulators of intercellular communication and play an irreplaceable role in inflammation, fibrogenesis, and tissue repair (Simeoli et al., 2017). Exosomal miR-21-5p secreted by bone marrow macrophages directly targets Smad7 and leads to increased proliferation and migration ability of tenocytes as well as fibrosis activity, providing a potential target for the prevention and treatment of tendon adhesion (Cui et al., 2019). In addition, a study has shown that human Umbilical Cord Stem Cell-Derived Exosomes may regulate p65 activity through the delivery of miR-21a-3p, and ultimately inhibit tendon adhesion (Yao et al., 2020).
These studies are promising for further research into tendon adhesion, and are critical to determine how to improve repair outcomes and identify new therapeutic strategies to promote tendon healing and prevent adhesion formation.
Tenocytes produce ECM which participates in tendon injury repair to maintain tendons homeostasis. The matrix remodeling rate of pathological tendons is increased, which leads to the decrease of mechanical stability of tendons and more vulnerable to injury (Longo et al., 2018). ECM disrupted by matrix metalloproteinases is another mark for ECM remodeling occurring at the site of the lesion (Xue and Jackson, 2015). Seven miRNAs, including miR-145-5p, miR-151a-3p, miR-382-5p, miR-199a-5p, miR-21-5p, miR-125a-5p, and miR-498 were found to be highly active and are thought to mediate major biological processes related to tendon matrix body integrity, which may be associated with tendon-related pathology (Thankam et al., 2016). COL5A1 encodes the α1 chain of type V collagen which is a minor amount fibrillar collagen (Posthumus et al., 2011). In its 3′-untranslated region (3′-UTR), COL5A1 gene has a BstUI restriction fragment length polymorphisms, which is associated with achilles tendon pathology and more specifically, chronic achilles tendinopathy (Mokone et al., 2006). miRNA can inhibit protein synthesis by binding to COL5A1 3′-UTR to regulate target mRNA stability and/or translation efficiency (Garzon et al., 2009). A functional miRNA site for miR-608 within the COL5A1 3′-UTR was identified, which has important implications for the understanding of the molecular basis of tendinopathy and other exercise-related phenotypes (Abrahams et al., 2013). miR-29a demonstrated a functional role in post-transcriptional regulation of collagen in murine and human (Millar et al., 2015).
Some studies have highlighted miRNAs involved in tendon homeostasis during developmental and healing processes. Skeletal muscle has intimate functional adaptations with tendon, and they are called as “tendon units” (Magnusson et al., 2008). miRNAs including miRNA-1, miRNA-133a, miRNA-133a∗ and miRNA-133b, which regulate striated muscle to mechanical loading, unloading and various disease processes (McCarthy et al., 2009; McCarthy, 2011), are all increased in mechanically loaded tendons and TGF-β-treated fibroblasts (Mendias et al., 2012). Mechanosensitive miRNAs, including miR-338, and miR-381, may bind to the 3′-UTR of scleraxis (Scx), which is a key regulator of extremity tendon development (Mendias et al., 2012). miR-100, miR-378, miR-205, miR-21, miR-221, and miR-222 were shown to involve with the accommodation of ECM synthesis and cell proliferation. And the let-7 family (such as let-7a, let-7e, and let-7b) also plays significant role in adjusting postnatal tendon adaptation to mechanical loading and TGF-β treatment (Mendias et al., 2012). Overexpression of mechanical sensitive miR-337-3p mitigates ectopic ossification in rat tendinopathy model via targeting insulin receptor substrate 1 (IRS1) and NADPH oxidase 4 (Nox4) of tendon derived stem cells, which not only provides a new understanding of the molecular mechanism of heterotopic ossification in tendinopathy, but also emphasizes the significance of miR-337-3p as a recognized therapeutic target for the clinical treatment of tendinopathy (Geng et al., 2020). Tenocytes routinely encounter oxidative stress. High glucose combined with oxidative stress lead to the up-regulation of miR28-5p which directly down-regulates the expression of p53 deacetylase sirtuin 3, leading to the increase of p53 acetylation level (Poulsen et al., 2014).
Thrombospondin-4 (Tsp-4) is indispensable for muscle attachment and ECM assembly at muscle-tendon junctions (Subramanian and Schilling, 2014). Targeting Krüppel-like factor 6 (KLF6), miR-148a-3p could affect the expression of TSP-4 in tendonocytes, and is closely related with Tsp-4 levels in tendinopathy tissues, which also promoted endothelial cell (EC) angiogenesis (Ge et al., 2018). Single local injection of double stranded miR-210 accelerated neovascularization through upregulating the expression of VEGF, FGF2 and type I collagen (COL1), and induced a microenvironment conducive to rat Achilles tendon healing during the early phase (Usman et al., 2015). The formation of new capillaries through angiogenesis is the key to tendon healing. However, excessive or dysfunctional angiogenic responses may adversely affect the healing outcome.
Understanding the relationship between miRNAs and the biological elements of tendon development, tendon homeostasis, and healing is the first requirement to determine an effective treatment for tendinopathy.
The aim of tendon regeneration is to restore its inherent structural and functional characteristics, which remains a great challenge (Andarawis-Puri et al., 2015). Stem cell-based therapy has become an important research direction in tissue engineering and regenerative medicine, especially for tendon and bone. Mesenchymal stem cells (MSCs) and tendon stem/progenitor cells (TSPCs) have received increasing attention toward tenogenic differentiation and tendon regeneration. MSCs are self-renewing, cultured and extended adult stem cells isolated from a variety of tissues with the ability of multipotent differentiation (Ferreira et al., 2018). TSPCs referred to as tendon-derived stem cells (TDSCs) or tendon stem cells (TSCs). These cells, residing in the fascicular matrix of tendon, also have self-renewal and multi-lineage differentiation ability, and might open a new field of tenogenesis and replacement of damaged tendons (Zhou et al., 2010). Current studies with respect to the effect of miRNAs known to be involved in tenogenesis were summarized in Table 2.
It is well known that TGF-β1 is produced by fibroblasts and can be upregulated in autosynthesis, and overproduction of TGF-β1 leads to excessive cell proliferation and matrix synthesis (Tomasek et al., 2002; Frangogiannis, 2020). Type I collagen (COL1), forming macromolecular networks, is the most abundant protein in the human body and provides strength and resiliency to tissues such as tendons, and ligaments (Rittié, 2017). miR-29b-3p has been reported to directly inhibit the expression of TGF-β1 and COL1, thereby forming a new regulatory feedback loop between H19 and TGF-β1 and inhibiting tendon differentiation (Lu et al., 2017). Transforming growth factor β2 (TGF-β2) is required for fetal tendon development in mice and had been shown to induce Scx expression in mouse and chick embryos (Pryce et al., 2009; Havis et al., 2016). Embedding in the sequence of this transcriptional regulator of oxidative energy metabolism is miR-378a, which is involved in metabolic pathways, mitochondrial energy homeostasis, and related biological processes such as muscle development, differentiation, and regeneration (Krist et al., 2015). In miR-378a knock-in transgenic mice and their TSPCs, miR-378a impaired tenogenic differentiation and tendon injury healing by inhibition collagen and ECM production via reducing TGF-β2 (Liu et al., 2019). Early growth response-1 (EGR-1) regulated specific differentiation of TSPCs into tenocytes and also induced the expression of tendon marker genes SCX, TNMD, TNC, and COL1 both in vitro and in vivo (Tao et al., 2015). Directly binding with EGR-1, miR-124 prevented collagen formation and tendon differentiation via suppressing EGR-1 expression (Wang et al., 2016). The high-mobility AT-hook 2 (HMGA2) mRNA transcript, involved in many cellular processes, contains seven complementary sites for let-7 binding in its 3′ -UTR and is known for its regulatory role in stem cell self-renewal and differentiation along with oncogenesis (Hammond and Sharpless, 2008). Recent studies have established that HMGA2 overexpression protected hTSPCs against H2O2-induced loss of stemness through autophagy activation, while increased HMGA2 silencing by the miRNA let-7 could induce hTSPC impairments, and thus plays a critical role in tendon homeostasis and regeneration (Sun et al., 2020). Rho-associated coiled-coil protein kinase 1 (ROCK1) plays vital roles in many aspects including cell morphology, mitosis, motility, and even senescence (Julian and Olson, 2014; Guan et al., 2020). miR-135a which directly binded to the 3′-UTR of ROCK1, suppresses proliferation, migration and tenogenic differentiation of TSPCs (Chen et al., 2015b). miR-140-5p has been shown to delay the progression of human TSPCs senescence by targeting Pin1 which is a highly conserved peptidyl-prolyl isomerase (PPI) (Chen et al., 2015a). Therefore, Pin1 overexpression may increases tendon differentiation and inhibits senescence of TSPCs.
Aged TSPCs showed substantial upregulation of senescence marker p16INK4A (Kohler et al., 2013). Osteogenic and adipogenic differentiation capacity is significantly restored by p16 INK4A knockdown in aging MSCs (Feng et al., 2014). Aged TSPCs showed decreased tenogenic differentiation capacity and upregulation of p16 which is a direct target of miR-217 (Han et al., 2017). Furthermore, TSPCs senescence may lead to defective self-renewal and altered fate (Zhou et al., 2010). TOB1 (Transducer of ERBB2, 1) is one of the TOB/B-cell translocation gene family. A study has suggested that the expression of TOB1 increased with aging during skeletal muscle development and the proliferative potential of myoblasts decreased with muscle development and aging (Yuan et al., 2011). What’s more, TOB1 is a negative regulator of BMP/Smad signaling, which negatively regulates proliferation and differentiation of osteoblasts by inhibiting the activity of receptor-regulated Smad protein (Xiong et al., 2006). A study investigated that miR-218 was found to promote the role of BMSCs in tendon-bone healing by inhibiting TOB1 in a rat supraspinatus repair model (Gao et al., 2016).
These studies showed that miRNAs may affect the differentiation of tendon stem cells and mesenchymal stem cells through different pathways and mechanisms (Figure 1). Understanding the functional of miRNAs and their roles in physiological and pathological processes of tendons is crucial for the development of new therapies for tenogenic differentiation and repair.
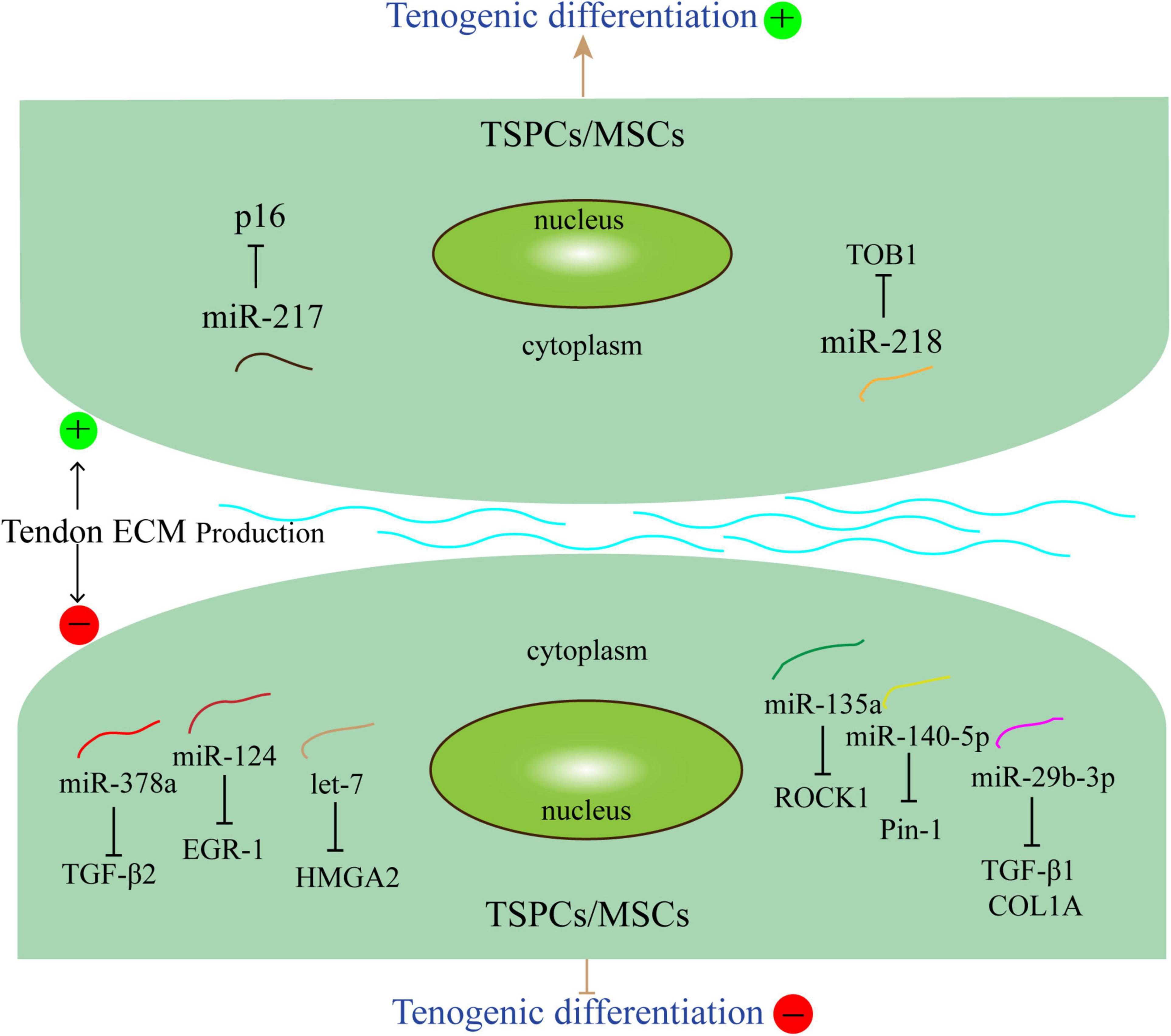
Figure 1. miRNAs related to tenogenic differentiation. Several miRNAs, such as miR-140-5p, miR-29b-3p, miR-378a, miR124, let-7, and miR-135a could inhibit tenogenic differentiation. Other miRNAs, including miR-217 and miR-218 could promote tenogenic differentiation. The arrows and T-shaped lines point toward mechanisms that represent promotion and inhibition, respectively.
miRNAs have been shown to regulate many potential genes related with tendon healing and tenogensis. There is great hope that tendon stem cell research could be applied to improve tendon injuries. TSPCs and MSCs are expected to be the mediums for tendon repair and regeneration, which is regulated by some director genes and a set of miRNAs coupled with some niche factors such as VEGF, TGF-β, ECM, oxidative stress, and mechanical stimuli, etc. Therefore, the study of these miRNAs may provide some potential targets for the diagnosis of tendon diseases and regeneration therapy in the future.
However, miRNAs reaching the target tendon is limited because of the poor pharmacokinetic properties of miRNAs, which means a need to produce adjuvant carrier systems that increase the stability of miRNAs. Future studies should identify how these miRNAs act on other stem cells and their extracellular microenvironments, and discover miRNAs which are responsible for tendon healing and tendon regeneration. And more importantly, future research should focus on finding the methods and approaches applicable to clinical practice.
LX and SQ conceived, designed, supervised, and commented on all the drafts of this manuscript. LD and MW contributed to the manuscript completion. All authors read and approved the final manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
AMPK, adenosine 5′-monophosphate-activated protein kinase; BMSCs, bone marrow mesenchymal stem cells; COL 1, Type I Collagen; COL5A1, collagen type V α-1 chain; CUGBP2, CUG triplet repeat-binding protein 2; EC, endothelial cell; ECM, Extracellular Matrix; EGR-1, early growth response-1; FGF2, Fibroblast Growth Factor 2; HMGA2, high-mobility AT-hook 2; hTSPCs, human tendon-derived stem/progenitor cells; IRS1, insulin receptor substrate 1; KLF6, krüppel-like factor 6; miRBase, Microribonucleic Acid Database; miRNA/miR, microRNA; mRNA, messenger RNA; MSCs, mesenchymal stem cells; Nox4, NADPH oxidase 4; PPI, peptidyl-prolyl isomerase; pre-miRNA, miRNA precursor; RCT, rotator cuff tendon; RISC, RNA-induced silencing complex; ROCK1, Rho-associated coiled-coil protein kinase 1; Scx, scleraxis; Smad3, SMAD family member 3; TDSCs, tendon-derived stem cells; TGF-β, transforming growth factor-β; TNC, tenascin-C; TNMD, tenomodulin; TOB1, transducer of ERBB2 1; TREM-1, triggering receptor expressed on myeloid cells-1; TSCs, tendon stem cells; Tsp-4, thrombospondin-4; TSPCs, tendon stem/progenitor cells; VEGFA, vascular endothelial growth factor-A; UTR, untranslated region.
Abrahams, Y., Laguette, M. J., Prince, S., and Collins, M. (2013). Polymorphisms within the COL5A1 3’-UTR that alters mRNA structure and the MIR608 gene are associated with Achilles tendinopathy. Ann. Hum. Genet. 77, 204–214. doi: 10.1111/ahg.12013
Ambros, V., Bartel, B., Bartel, D. P., Burge, C. B., Carrington, J. C., Chen, X., et al. (2003). A uniform system for microRNA annotation. RNA 9, 277–279. doi: 10.1261/rna.2183803
Andarawis-Puri, N., Flatow, E. L., and Soslowsky, L. J. (2015). Tendon basic science: development, repair, regeneration, and healing. J. Orthop. Res. 33, 780–784. doi: 10.1002/jor.22869
Borchert, G. M., Lanier, W., and Davidson, B. L. (2006). RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 13, 1097–1101. doi: 10.1038/nsmb1167
Bruns, J., Kampen, J., Kahrs, J., and Plitz, W. (2000). Achilles tendon rupture: experimental results on spontaneous repair in a sheep-model. Knee Surg. Sports Traumatol. Arthrosc. 8, 364–369. doi: 10.1007/s001670000149
Cai, X., Cai, M., and Lou, L. (2015). Identification of differentially expressed genes and small molecule drugs for the treatment of tendinopathy using microarray analysis. Mol. Med. Rep. 11, 3047–3054. doi: 10.3892/mmr.2014.3081
Cai, X., Hagedorn, C. H., and Cullen, B. R. (2004). Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 10, 1957–1966. doi: 10.1261/rna.7135204
Carthew, R. W., and Sontheimer, E. J. (2009). Origins and mechanisms of miRNAs and siRNAs. Cell 136, 642–655. doi: 10.1016/j.cell.2009.01.035
Chen, C. H., Zhou, Y. L., Wu, Y. F., Cao, Y., Gao, J. S., and Tang, J. B. (2009). Effectiveness of microRNA in Down-regulation of TGF-beta gene expression in digital flexor tendons of chickens: in vitro and in vivo study. J. Hand Surg. Am. 34, 1777–1784.e1.
Chen, L., Liu, J., Tao, X., Wang, G., Wang, Q., and Liu, X. (2015a). The role of Pin1 protein in aging of human tendon stem/progenitor cells. Biochem. Biophys. Res. Commun. 464, 487–492. doi: 10.1016/j.bbrc.2015.06.163
Chen, L., Wang, G. D., Liu, J. P., Wang, H. S., Liu, X. M., Wang, Q., et al. (2015b). miR-135a modulates tendon stem/progenitor cell senescence via suppressing ROCK1. Bone 71, 210–216. doi: 10.1016/j.bone.2014.11.001
Chen, Q., Lu, H., and Yang, H. (2014). Chitosan inhibits fibroblasts growth in Achilles tendon via TGF-β1/Smad3 pathway by miR-29b. Int. J. Clin. Exp. Pathol. 7, 8462–8470.
Chen, S., Jiang, S., Zheng, W., Tu, B., Liu, S., Ruan, H., et al. (2017). RelA/p65 inhibition prevents tendon adhesion by modulating inflammation, cell proliferation, and apoptosis. Cell Death Dis. 8:e2710. doi: 10.1038/cddis.2017.135
Cui, H., He, Y., Chen, S., Zhang, D., Yu, Y., and Fan, C. (2019). Macrophage-derived miRNA-containing exosomes induce peritendinous fibrosis after tendon injury through the miR-21-5p/Smad7 pathway. Mol. Ther. Nucleic Acids 14, 114–130. doi: 10.1016/j.omtn.2018.11.006
Di Leva, G., Garofalo, M., and Croce, C. M. (2014). MicroRNAs in cancer. Ann. Rev. Pathol. 9, 287–314.
Eming, S. A., Hammerschmidt, M., Krieg, T., and Roers, A. (2009). Interrelation of immunity and tissue repair or regeneration. Semin. Cell Dev. Biol. 20, 517–527. doi: 10.1016/j.semcdb.2009.04.009
Fabbri, M., Paone, A., Calore, F., Galli, R., Gaudio, E., Santhanam, R., et al. (2012). MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc. Natl. Acad. Sci. U.S.A. 109, E2110–E2116.
Feng, X., Xing, J., Feng, G., Huang, D., Lu, X., Liu, S., et al. (2014). p16(INK4A) mediates age-related changes in mesenchymal stem cells derived from human dental pulp through the DNA damage and stress response. Mech. Ageing Dev. 14, 46–55. doi: 10.1016/j.mad.2014.09.004
Ferreira, J. R., Teixeira, G. Q., Santos, S. G., Barbosa, M. A., Almeida-Porada, G., and Gonçalves, R. M. (2018). Mesenchymal stromal cell secretome: influencing therapeutic potential by cellular pre-conditioning. Front. Immunol. 9:2837. doi: 10.3389/fimmu.2018.02837
Frangogiannis, N. (2020). Transforming growth factor-β in tissue fibrosis. J. Exp. Med. 217:e20190103.
Fritz, J. V., Heintz-Buschart, A., Ghosal, A., Wampach, L., Etheridge, A., Galas, D., et al. (2016). Sources and functions of extracellular small RNAs in human circulation. Annu. Rev. Nutr. 36, 301–336. doi: 10.1146/annurev-nutr-071715-050711
Gao, Y., Zhang, Y., Lu, Y., Wang, Y., Kou, X., Lou, Y., et al. (2016). TOB1 deficiency enhances the effect of bone marrow-derived mesenchymal stem cells on tendon-bone healing in a rat rotator cuff repair model. Cell Physiol. Biochem. 38, 319–329. doi: 10.1159/000438632
Garzon, R., Calin, G. A., and Croce, C. M. (2009). MicroRNAs in cancer. Annu. Rev. Med. 60, 167–179.
Gaut, L., and Duprez, D. (2016). Tendon development and diseases. Wiley Interdiscip. Rev. Dev. Biol. 5, 5–23. doi: 10.1002/wdev.201
Ge, H., Shrestha, A., Liu, C., Wu, P., and Cheng, B. (2018). MicroRNA 148a-3p promotes Thrombospondin-4 expression and enhances angiogenesis during tendinopathy development by inhibiting Krüppel-like factor 6. Biochem. Biophys. Res. Commun. 502, 276–282. doi: 10.1016/j.bbrc.2018.05.167
Geng, Y., Zhao, X., Xu, J., Zhang, X., Hu, G., Fu, S. C., et al. (2020). Overexpression of mechanical sensitive miR-337-3p alleviates ectopic ossification in rat tendinopathy model via targeting IRS1 and Nox4 of tendon-derived stem cells. J. Mol. Cell Biol. 12, 305–317. doi: 10.1093/jmcb/mjz030
Guan, X., Guan, X., Dong, C., and Jiao, Z. (2020). Rho GTPases and related signaling complexes in cell migration and invasion. Exp. Cell. Res. 388:111824. doi: 10.1016/j.yexcr.2020.111824
Hammond, S. M., and Sharpless, N. E. (2008). HMGA2, microRNAs, and stem cell aging. Cell 135, 1013–1016. doi: 10.1016/j.cell.2008.11.026
Han, W., Wang, B., Liu, J., and Chen, L. (2017). The p16/miR-217/EGR1 pathway modulates age-related tenogenic differentiation in tendon stem/progenitor cells. Acta Biochim. Biophys. Sin. (Shanghai) 49, 1015–1021. doi: 10.1093/abbs/gmx104
Hata, A. (2013). Functions of microRNAs in cardiovascular biology and disease. Annu. Rev. Physiol. 75, 69–93. doi: 10.1146/annurev-physiol-030212-183737
Havis, E., Bonnin, M. A., Esteves de Lima, J., Charvet, B., Milet, C., and Duprez, D. (2016). TGFβ and FGF promote tendon progenitor fate and act downstream of muscle contraction to regulate tendon differentiation during chick limb development. Development 143, 3839–3851.
Julian, L., and Olson, M. F. (2014). Rho-associated coiled-coil containing kinases (ROCK): structure, regulation, and functions. Small GTPases 5:e29846. doi: 10.4161/sgtp.29846
Kabekkodu, S. P., Shukla, V., Varghese, V. K., D’ Souza, J., Chakrabarty, S., and Satyamoorthy, K. (2018). Clustered miRNAs and their role in biological functions and diseases. Biol. Rev. Camb. Philos. Soc. 93, 1955–1986. doi: 10.1111/brv.12428
Kannus, P. (2000). Structure of the tendon connective tissue. Scand. J. Med. Sci. Sports 10, 312–320. doi: 10.1034/j.1600-0838.2000.010006312.x
Katzel, E. B., Wolenski, M., Loiselle, A. E., Basile, P., Flick, L. M., Langstein, H. N., et al. (2011). Impact of Smad3 loss of function on scarring and adhesion formation during tendon healing. J. Orthop. Res. 29, 684–693. doi: 10.1002/jor.21235
Khan, K. M., Cook, J. L., Bonar, F., Harcourt, P., and Astrom, M. (1999). Histopathology of common tendinopathies. Update and implications for clinical management. Sports Med. 27, 393–408. doi: 10.2165/00007256-199927060-00004
Kohler, J., Popov, C., Klotz, B., Alberton, P., Prall, W. C., Haasters, F., et al. (2013). Uncovering the cellular and molecular changes in tendon stem/progenitor cells attributed to tendon aging and degeneration. Aging Cell 12, 988–999. doi: 10.1111/acel.12124
Koturbash, I., Tolleson, W. H., Guo, L., Yu, D., Chen, S., Hong, H., et al. (2015). microRNAs as pharmacogenomic biomarkers for drug efficacy and drug safety assessment. Biomark. Med. 9, 1153–1176. doi: 10.2217/bmm.15.89
Kozomara, A., Birgaoanu, M., and Griffiths-Jones, S. (2019). miRBase: from microRNA sequences to function. Nucleic Acids Res. 47, D155–D162.
Krist, B., Florczyk, U., Pietraszek-Gremplewicz, K., Józkowicz, A., and Dulak, J. (2015). The role of miR-378a in metabolism, angiogenesis, and muscle biology. Int. J. Endocrinol. 2015:281756.
Laurencin, C. T., and Freeman, J. W. (2005). Ligament tissue engineering: an evolutionary materials science approach. Biomaterials 26, 7530–7536. doi: 10.1016/j.biomaterials.2005.05.073
Lee, R. C., Feinbaum, R. L., and Ambros, V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854. doi: 10.1016/0092-8674(93)90529-y
Lee, Y., Kim, M., Han, J., Yeom, K. H., Lee, S., Baek, S. H., et al. (2004). MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 23, 4051–4060. doi: 10.1038/sj.emboj.7600385
Lehmann, S. M., Krüger, C., Park, B., Derkow, K., Rosenberger, K., Baumgart, J., et al. (2012). An unconventional role for miRNA: let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat. Neurosci. 15, 827–835. doi: 10.1038/nn.3113
Liu, Y., Feng, L., Xu, J., Yang, Z., Wu, T., Zhang, J., et al. (2019). MiR-378a suppresses tenogenic differentiation and tendon repair by targeting at TGF-β2. Stem Cell Res Ther. 10:108.
Longo, U. G., Ronga, M., and Maffulli, N. (2018). Achilles tendinopathy. Sports Med. Arthrosc. Rev. 26, 16–30.
Lu, Y. F., Liu, Y., Fu, W. M., Xu, J., Wang, B., Sun, Y. X., et al. (2017). Long noncoding RNA H19 accelerates tenogenic differentiation and promotes tendon healing through targeting miR-29b-3p and activating TGF-β1 signaling. FASEB J. 31, 954–964. doi: 10.1096/fj.201600722r
Magnusson, S. P., Narici, M. V., Maganaris, C. N., and Kjaer, M. (2008). Human tendon behaviour and adaptation, in vivo. J. Physiol. 586, 71–81. doi: 10.1113/jphysiol.2007.139105
McCarthy, J. J. (2011). The MyomiR network in skeletal muscle plasticity. Exerc. Sport Sci. Rev. 39, 150–154. doi: 10.1097/jes.0b013e31821c01e1
McCarthy, J. J., Esser, K. A., Peterson, C. A., and Dupont-Versteegden, E. E. (2009). Evidence of MyomiR network regulation of beta-myosin heavy chain gene expression during skeletal muscle atrophy. Physiol. Genomics 39, 219–226. doi: 10.1152/physiolgenomics.00042.2009
Mendias, C. L., Gumucio, J. P., and Lynch, E. B. (2012). Mechanical loading and TGF-β change the expression of multiple miRNAs in tendon fibroblasts. J. Appl. Physiol. (1985) 113, 56–62. doi: 10.1152/japplphysiol.00301.2012
Millar, N. L., Gilchrist, D. S., Akbar, M., Reilly, J. H., Kerr, S. C., Campbell, A. L., et al. (2015). MicroRNA29a regulates IL-33-mediated tissue remodelling in tendon disease. Nat. Commun. 6:6774.
Millar, N. L., Silbernagel, K. G., Thorborg, K., Kirwan, P. D., Galatz, L. M., Abrams, G. D., et al. (2021). Tendinopathy. Nat. Rev. Dis. Primers 7:1. doi: 10.1002/9780470757987.ch1
Miyashita, H., Ochi, M., and Ikuta, Y. (1997). Histological and biomechanical observations of the rabbit patellar tendon after removal of its central one-third. Arch. Orthop. Trauma Surg. 116, 454–462. doi: 10.1007/s004020050162
Mokone, G. G., Schwellnus, M. P., Noakes, T. D., and Collins, M. (2006). The COL5A1 gene and Achilles tendon pathology. Scand. J. Med. Sci. Sports 16, 19–26. doi: 10.1111/j.1600-0838.2005.00439.x
Nichols, A. E. C., Best, K. T., and Loiselle, A. E. (2019). The cellular basis of fibrotic tendon healing: challenges and opportunities. Transl. Res. 209, 156–168. doi: 10.1016/j.trsl.2019.02.002
Nourissat, G., Berenbaum, F., and Duprez, D. (2015). Tendon injury: from biology to tendon repair. Nat. Rev. Rheumatol. 11, 223–233. doi: 10.1038/nrrheum.2015.26
O’Brien, M. (1997). Structure and metabolism of tendons. Scand. J. Med. Sci. Sports 7, 55–61. doi: 10.1111/j.1600-0838.1997.tb00119.x
Posthumus, M., Schwellnus, M. P., and Collins, M. (2011). The COL5A1 gene: a novel marker of endurance running performance. Med. Sci. Sports Exerc. 43, 584–589. doi: 10.1249/mss.0b013e3181f34f4d
Poulsen, R. C., Knowles, H. J., Carr, A. J., and Hulley, P. A. (2014). Cell differentiation versus cell death: extracellular glucose is a key determinant of cell fate following oxidative stress exposure. Cell Death Dis. 5:e1074. doi: 10.1038/cddis.2014.52
Pryce, B. A., Watson, S. S., Murchison, N. D., Staverosky, J. A., Dünker, N., and Schweitzer, R. (2009). Recruitment and maintenance of tendon progenitors by TGFbeta signaling are essential for tendon formation. Development 136, 1351–1361. doi: 10.1242/dev.027342
Reinhart, B. J., Slack, F. J., Basson, M., Pasquinelli, A. E., Bettinger, J. C., Rougvie, A. E., et al. (2000). The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403, 901–906. doi: 10.1038/35002607
Rittié, L. (2017). Type I collagen purification from rat tail tendons. Methods Mol. Biol. 1627, 287–308. doi: 10.1007/978-1-4939-7113-8_19
Saliminejad, K., Khorram Khorshid, H. R., Soleymani Fard, S., and Ghaffari, S. H. (2019). An overview of microRNAs: biology, functions, therapeutics, and analysis methods. J. Cell. Physiol. 234, 5451–5465. doi: 10.1002/jcp.27486
Sayed, D., and Abdellatif, M. (2011). MicroRNAs in development and disease. Physiol. Rev. 91, 827–887.
Selbach, M., Schwanhäusser, B., Thierfelder, N., Fang, Z., Khanin, R., and Rajewsky, N. (2008). Widespread changes in protein synthesis induced by microRNAs. Nature 455, 58–63. doi: 10.1038/nature07228
Sharma, P., and Maffulli, N. (2005). Tendon injury and tendinopathy: healing and repair. J. Bone Joint Surg. Am. 87, 187–202. doi: 10.2106/00004623-200501000-00030
Sharma, P., and Maffulli, N. (2006). Biology of tendon injury: healing, modeling and remodeling. J. Musculoskelet. Neuronal Interact. 6, 181–190.
Simeoli, R., Montague, K., Jones, H. R., Castaldi, L., Chambers, D., Kelleher, J. H., et al. (2017). Exosomal cargo including microRNA regulates sensory neuron to macrophage communication after nerve trauma. Nat. Commun. 8:1778.
Strickland, J. W. (2000). Development of flexor tendon surgery: twenty-five years of progress. J. Hand Surg. Am. 25, 214–235. doi: 10.1053/jhsu.2000.jhsu25a0214
Subramanian, A., and Schilling, T. F. (2014). Thrombospondin-4 controls matrix assembly during development and repair of myotendinous junctions. Elife 3:e02372.
Sun, Y., Chen, H., Ye, H., Liang, W., Lam, K. K., Cheng, B., et al. (2020). Nudt21-mediated alternative polyadenylation of HMGA2 3’-UTR impairs stemness of human tendon stem cell. Aging (Albany N. Y.) 12, 18436–18452. doi: 10.18632/aging.103771
Tao, X., Liu, J., Chen, L., Zhou, Y., and Tang, K. (2015). EGR1 induces tenogenic differentiation of tendon stem cells and promotes rabbit rotator cuff repair. Cell Physiol. Biochem. 35, 699–709. doi: 10.1159/000369730
Tempfer, H., and Traweger, A. (2015). Tendon vasculature in health and disease. Front. Physiol. 6:330. doi: 10.3389/fphys.2015.00330
Thankam, F. G., Boosani, C. S., Dilisio, M. F., and Agrawal, D. K. (2018). MicroRNAs associated with inflammation in shoulder tendinopathy and glenohumeral arthritis. Mol. Cell. Biochem. 437, 81–97. doi: 10.1007/s11010-017-3097-7
Thankam, F. G., Boosani, C. S., Dilisio, M. F., Dietz, N. E., and Agrawal, D. K. (2016). MicroRNAs associated with shoulder tendon matrisome disorganization in glenohumeral arthritis. PLoS One 11:e0168077. doi: 10.1371/journal.pone.0168077
Thankam, F. G., Boosani, C. S., Dilisio, M. F., Gross, R. M., and Agrawal, D. K. (2019). Genes interconnecting AMPK and TREM-1 and associated microRNAs in rotator cuff tendon injury. Mol Cell. Biochem 454, 97–109. doi: 10.1007/s11010-018-3456-z
Thorpe, C. T., and Screen, H. R. (2016). Tendon structure and composition. Adv. Exp. Med. Biol. 920, 3–10. doi: 10.1007/978-3-319-33943-6_1
Tomasek, J. J., Gabbiani, G., Hinz, B., Chaponnier, C., and Brown, R. A. (2002). Myofibroblasts and mechano-regulation of connective tissue remodelling. Nat. Rev. Mol. Cell Biol. 3, 349–363. doi: 10.1038/nrm809
Tsai, S. L., Nödl, M. T., and Galloway, J. L. (2021). Bringing tendon biology to heel: leveraging mechanisms of tendon development, healing, and regeneration to advance therapeutic strategies. Dev. Dyn. 250, 393–413. doi: 10.1002/dvdy.269
Usman, M. A., Nakasa, T., Shoji, T., Kato, T., Kawanishi, Y., Hamanishi, M., et al. (2015). The effect of administration of double stranded MicroRNA-210 on acceleration of Achilles tendon healing in a rat model. J. Orthop. Sci. 20, 538–546. doi: 10.1007/s00776-015-0709-5
Vasudevan, S., Tong, Y., and Steitz, J. A. (2007). Switching from repression to activation: microRNAs can up-regulate translation. Science 318, 1931–1934. doi: 10.1126/science.1149460
Vidigal, J. A., and Ventura, A. (2015). The biological functions of miRNAs: lessons from in vivo studies. Trends Cell Biol. 25, 137–147. doi: 10.1016/j.tcb.2014.11.004
Voleti, P. B., Buckley, M. R., and Soslowsky, L. J. (2012). Tendon healing: repair and regeneration. Annu. Rev. Biomed. Eng. 14, 47–71.
Wang, B., Guo, J., Feng, L., Suen, C. W., Fu, W. M., Zhang, J. F., et al. (2016). MiR124 suppresses collagen formation of human tendon derived stem cells through targeting egr1. Exp. Cell Res. 347, 360–366. doi: 10.1016/j.yexcr.2016.08.018
Wang, J. H. (2006). Mechanobiology of tendon. J. Biomech. 39, 1563–1582. doi: 10.1016/j.jbiomech.2005.05.011
Wang, J. H., Guo, Q., and Li, B. (2012). Tendon biomechanics and mechanobiology–a minireview of basic concepts and recent advancements. J. Hand Ther. 25, 133–140; quiz141.
Watts, A. E., Millar, N. L., Platt, J., Kitson, S. M., Akbar, M., Rech, R., et al. (2017). MicroRNA29a treatment improves early tendon injury. Mol. Ther. 25, 2415–2426. doi: 10.1016/j.ymthe.2017.07.015
Whalen, W. P. (1951). Utilization of scar tissue in bridging tendon defects. Ann. Surg. 133, 567–571. doi: 10.1097/00000658-195113340-00019
Wu, Y. F., Chen, C. H., Cao, Y., Avanessian, B., Wang, X. T., and Tang, J. B. (2010). Molecular events of cellular apoptosis and proliferation in the early tendon healing period. J. Hand Surg. Am. 35, 2–10. doi: 10.1016/j.jhsa.2009.10.021
Wu, Y. F., Mao, W. F., Zhou, Y. L., Wang, X. T., Liu, P. Y., and Tang, J. B. (2016). Adeno-associated virus-2-mediated TGF-β1 microRNA transfection inhibits adhesion formation after digital flexor tendon injury. Gene Ther. 23, 167–175. doi: 10.1038/gt.2015.97
Xiong, B., Rui, Y., Zhang, M., Shi, K., Jia, S., Tian, T., et al. (2006). Tob1 controls dorsal development of zebrafish embryos by antagonizing maternal beta-catenin transcriptional activity. Dev. Cell 11, 225–238. doi: 10.1016/j.devcel.2006.06.012
Xu, Q., Sun, W. X., and Zhang, Z. F. (2019). High expression of VEGFA in MSCs promotes tendon-bone healing of rotator cuff tear via microRNA-205-5p. Eur. Rev. Med. Pharmacol. Sci. 23, 4081–4088.
Xue, M., and Jackson, C. J. (2015). Extracellular matrix reorganization during wound healing and its impact on abnormal scarring. Adv. Wound Care (New Rochelle) 4, 119–136. doi: 10.1089/wound.2013.0485
Yao, Z., Li, J., Wang, X., Peng, S., Ning, J., Qian, Y., et al. (2020). MicroRNA-21-3p engineered umbilical cord stem cell-derived exosomes inhibit tendon adhesion. J. Inflamm. Res. 13, 303–316. doi: 10.2147/jir.s254879
Yuan, J., Cao, J. Y., Tang, Z. L., Wang, N., and Li, K. (2011). Molecular characterization of Tob1 in muscle development in pigs. Int. J. Mol. Sci. 12, 4315–4326. doi: 10.3390/ijms12074315
Keywords: microRNA, tendon healing, tendinopathy, tenogenesis, stem cells
Citation: Ding L, Wang M, Qin S and Xu L (2021) The Roles of MicroRNAs in Tendon Healing and Regeneration. Front. Cell Dev. Biol. 9:687117. doi: 10.3389/fcell.2021.687117
Received: 29 March 2021; Accepted: 11 June 2021;
Published: 02 July 2021.
Edited by:
Gang Li, The Chinese University of Hong Kong, ChinaReviewed by:
Erick Omar Hernandez-Ochoa, University of Maryland, Baltimore, United StatesCopyright © 2021 Ding, Wang, Qin and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shengnan Qin, cWlucWluc25AMTYzLmNvbQ==; Liangliang Xu, eHVsbC0yMDE2QGd6dWNtLmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.