
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol. , 07 July 2021
Sec. Cell Growth and Division
Volume 9 - 2021 | https://doi.org/10.3389/fcell.2021.684352
This article is part of the Research Topic Zebrafish Germ Cell Development View all 9 articles
Fish gonads develop in very diverse ways different from mammalian gonads. This diversity is contributed by species-specific factors. Gonadal somatic cell-derived factor (Gsdf) is one such factor. The gsdf gene exists mostly in teleosts and is absent in many tetrapods, probably as a result of two gene losses during evolution. The gsdf transcript is expressed mainly in gonadal somatic cells, including Sertoli cell in testis and granulosa cells in ovary; however, these gonadal somatic cells can surround many types of germ cells at different developmental stages depending on the fish species. The function of gsdf is also variable. It is involved in germ cell proliferation, testicular formation, ovarian development and even male sex determination. Here, we summarize the common and diverse expression, regulation and functions of gsdf among different fish species with aspect of evolution.
The development of gonads is characterized by its diversity among species, while the development of other organs in different species follows similar rules. The best studied mechanism of gonad development is dictated by the XX/XY sex-determination system in many mammals. In humans and mice, gonadal sex is determined by SRY located on the Y chromosome. In males, SRY triggers the expression of downstream male factors including DMRT1 and SOX9 that control testicular development. In females, the absence of SRY facilitates the expression of female factors including FOXL2 and WNT4, which further trigger ovarian development (Bashamboo and McElreavey, 2016; She and Yang, 2017).
Fishes exhibit very diverse mechanisms of gonadal development, which can be common or distinct from other vertebrates. Fishes such as Nile tilapia (Li et al., 2015) and medaka (Matsuda et al., 2002) can exist as a single sex at a time (gonochoristic), while other fishes like black porgy (Wu et al., 2021) and clownfish (Fricke and Fricke, 1977) exist as hermaphrodites. The factors controlling fish sex can be genetic factors (Matsuda et al., 2002) or environmental factors, such as temperature for European sea bass (Koumoundouros et al., 2002) and social activity for anemone fish (Fricke and Fricke, 1977). To achieve this diversity, fishes preserve unique genes related to gonadal development during evolution.
A unique factor that controls gonad development in fish is gonadal somatic cell-derived factor (Gsdf). Gsdf is a secretory protein in the transforming growth factor β (TGFβ) family expressed mainly in teleost gonads. It is composed of a mature TGFβ domain and a precursor domain, which contains a signal peptide (Figure 1). The precursor domain is cleaved upon secretion, giving rise to mature Gsdf with the TGFβ domain. The TGFβ domain of Gsdf contains a characteristic cystine knot, but it lacks a glycine residue between 2nd and 3rd cystine. This signature is different from many other TGFβ proteins including Amh, Gdf9, and Inha (Vitt et al., 2001; Shibata et al., 2010). Phylogenetic tree further shows that Gsdf forms a unique clade different from other TGFβ proteins also known to be important for gonadal development (Figure 2).
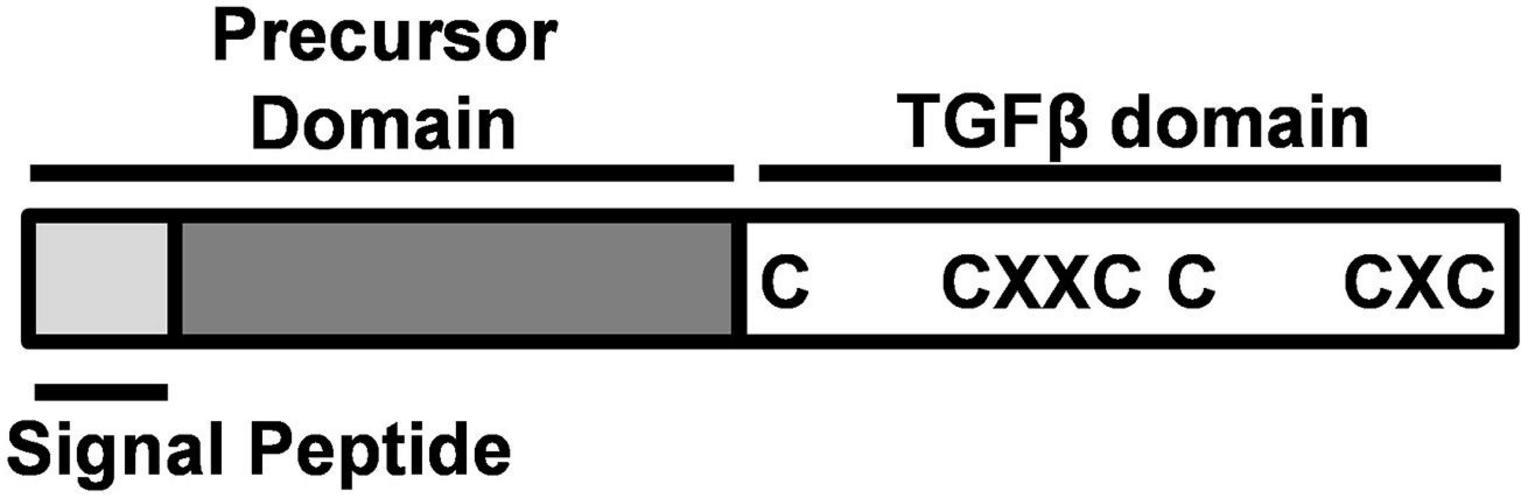
Figure 1. Structure of Gonadal somatic cell-derived factor (Gsdf). Gsdf is composed of a signal peptide and a precursor domain, which are cleaved after protein processing. The mature TGFβ domain is characterized by a cystine knot with three disulfide bonds. C, cystine; X, any amino acid.
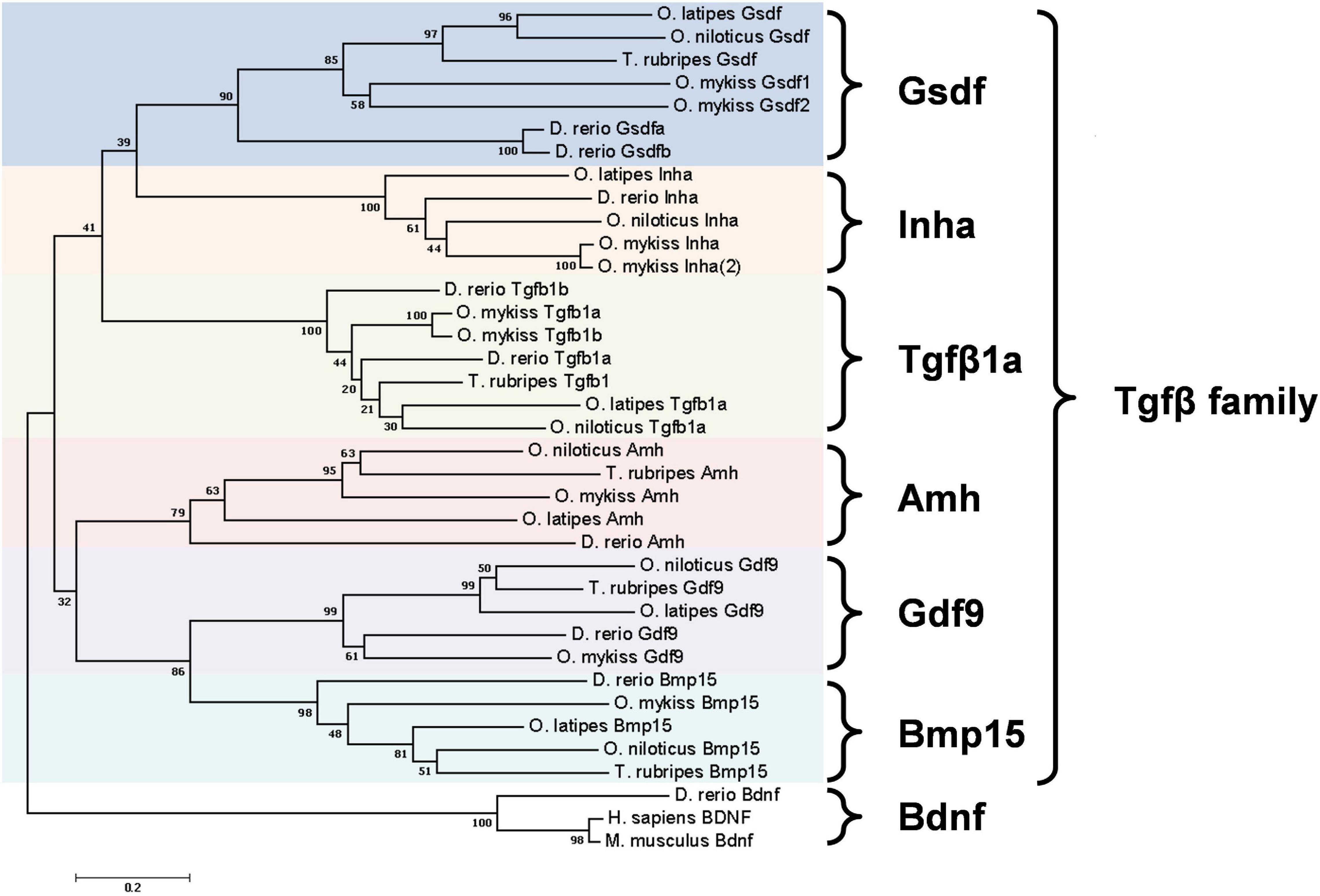
Figure 2. Phylogenetic tree of six Tgfβ family members in five teleost species. Danio rerio (zebrafish), Oryzias latipes (medaka), Orechromis niloticus (Nile tilapia), Oncorhynchus mykiss (rainbow trout), and Takifugu rubripes (Fugu). Phylogenetic analysis was performed by MEGA7 using nearest neighbor-joining method with 1000 bootstrap replicates. The unit of scale bar refers to the number of amino acid substitutions per site. Bdnf proteins were used as the outgroup. Amino acid sequence extracted from GenBank or Ensembl database are: D. rerio Gsdfa, ENSDARP00000149345; Gsdfb, ENSDARP00000134025; O. latipes Gsdf, ENSORLP00000042059; O. niloticus Gsdf, ENSONIP00000009618; O. mykiss Gsdf1, ENSOMYP00000032402; Gsdf2, ENSOMYP00000004997; T. rubripes Gsdf, ENSTRUP00000036138; D. rerio Inha, ENSDARP00000057347; O. latipes Inha, ENSORLP00000002713; O. niloticus Inha, ENSONIP00000016791; O. mykiss Inha, ENSOMYP00000007000; Inha(2), ENSOMYP00000010980; D. rerio Tgfb1a, ENSDARP00000060838; Tgfb1b, ENSDARP00000122056; O. latipes Tgfb1a, ENSORLP00000001563; O. niloticus Tgfb1a, ENSONIP00000007415; O. mykiss Tgfb1a, ENSOMYP00000099035; Tgfb1b, ENSOMYP00000101842; T. rubripes Tgfb1, ENSTRUP00000040849; D. rerio Amh, ENSDARP00000015395; O. niloticus Amh, ENSONIP00000006018; O. mykiss Amh, ENSOMYP00000058709; O. latipes Amh, ENSORLP00000006358; T. rubripes Amh ENSTRUP00000051375; D. rerio Gdf9, ENSDARP00000008475; O. latipes Gdf9, ENSORLP00000042760; O. niloticus Gdf9, ENSONIP00000016237; O. mykiss Gdf9, ENSOMYP00000052293; T. rubripes Gdf9, ENSTRUP00000050821; D. rerio Bmp15, ENSDARP00000054590; O. latipes Bmp15, ENSORLP00000010817; O. niloticus Bmp15, ENSONIP00000042526; O. mykiss Bmp15, XP_021470286; T. rubripes Bmp15, ENSTRUP00000048048; D. rerio Bdnf, ENSDARP00000069872; O. latipes Bdnf, ENSORLP00000031660; O. niloticus Bdnf, ENSONIP00000025919; O. mykiss Bdnf, ENSOMYP00000079299; T. rubripes Bdnf, ENSTRUP00000044236; M. musculus Bdnf, NP_031566.4; H. sapiens BDNF, NP-001137277.1.
Gonadal somatic cell-derived factor has attracted attention because it exists only in limited species to promote gonad development. In this review, we have compared the expression, regulation and functions of Gsdf in different species, and have delineated that Gsdf controls gonadal development via very diverse pathways.
First found in rainbow trout (Oncorhynchus mykiss) (Sawatari et al., 2007), gsdf exists mainly in teleosts (a branch of Osteichthyes) (Figure 3). BLAST and transcriptome analysis have further identified gsdf in non-teleost jawed fishes such as Latimeria menadoensis (Coelacanthiformes), Protopterus annectens (Dipnoi), and Callorhinchus mili (Chondrichthyes) (Forconi et al., 2013; Biscotti et al., 2018). However, whether gsdf exists in jawless fishes (Agnatha), the remaining class of extant fish, is still unclear. In addition, it is present in such tetrapods as Cynops orientalis (Urodela) and Microcaecilia unicolor (Gymnophiona), but absent in other tetrapods such as Xenopus (Anura), mammals and reptiles (Amniota). Phylogenetic analysis of various species that do or do not contain gsdf leads to the hypothesis that there are at least two losses of gsdf during the evolution of Tetrapoda (Figure 3) (Biscotti et al., 2020). One event of gsdf loss is in Anura, the other is in Amniota. The evolution of gsdf in ancient genome is an interesting question worth further investigation.
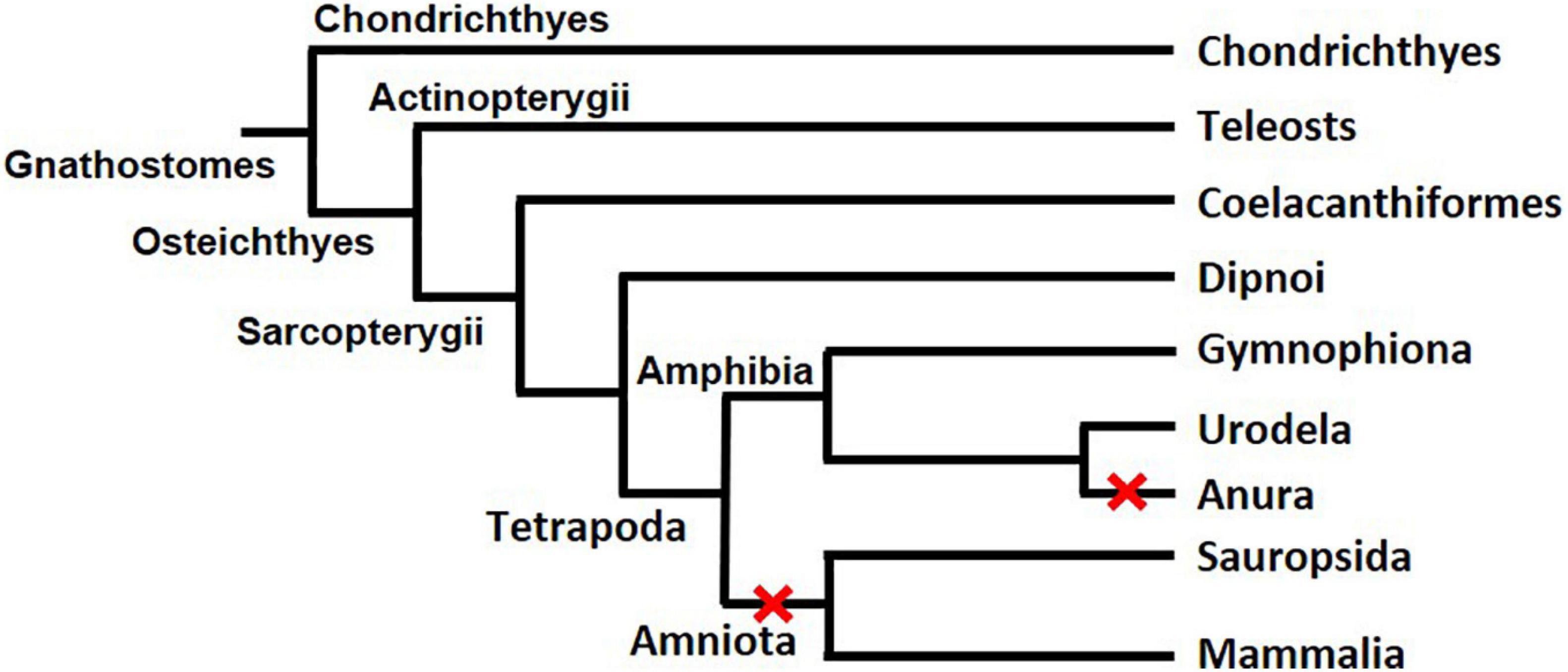
Figure 3. The evolution of gsdf in Gnathostomes. The two red X signs indicate two events of gsdf losses during the evolution of Tetrapoda. The figure is taken from a published article with permission (Biscotti et al., 2020).
The synteny analysis comparing zebrafish (Danio rerio), human, spotted gar (Lepisosteus oculatus), and chicken (Gallus gallus) reveals that zebrafish gsdf locus contains conserved syntenies similar to that of human chromosome 4 (Gautier et al., 2011a; Yan et al., 2017). However, gsdf is within a breakpoint during chromosome rearrangement while other genes within the same syntenic locus are preserved. Synteny analysis comparing zebrafish, human, and spotted gar further indicates that bmp15, gdf9, and gsdf might originally be paralogs. The importance of gsdf, bmp15, and gdf9 in ovarian development suggests that they might retain certain subfunctions of the ancestor gene (Force et al., 1999; Yan et al., 2017; Dalbies-Tran et al., 2020). Moreover, most genes within the syntenies are predominantly expressed in previtellogenic oocytes of Oryzias latipes and ovary of zebrafish (Gautier et al., 2011a). Thus, despite its male-dominant expression, gsdf is located within a conserved synteny inside a cluster of ovarian genes. Further analysis comparing synteny from two Tetrapoda clades that retain or lose gsdf may reveal the process of gsdf gene loss during evolution.
The gsdf gene is expressed in gonads, but the exact location of the gonad and timing of expression diverge among different species (Table 1). In most teleosts, gsdf is mainly expressed in gonadal somatic cells, and there are often more gsdf transcripts in testis than in ovary, suggesting the main role of Gsdf in testicular development. In the testis, gsdf is expressed in the Sertoli cells surrounding spermatogonia in all species studied to date (Sawatari et al., 2007; Shibata et al., 2010; Gautier et al., 2011b; Myosho et al., 2012; Kaneko et al., 2015; Jiang et al., 2019). Thus, Gsdf may support spermatogonial functions including their self-renewal, proliferation, and differentiation. In Monopterus albus, Salmo salar, and Cynoglossus semilaevis, gsdf expression is enriched in immature testis and decreased in mature testis (Zhu et al., 2016; Zhu et al., 2018; Kleppe et al., 2020), suggesting that gsdf functions in testis maturation during development.
In Halichoeres trimaculatus and such Ovalentaria as O. latipes, Oryzias luzonensis, and Oreochromis niloticus, in addition to expression in Sertoli cells, gsdf is also expressed in epithelial cells of the intratesticular efferent duct, where no germ cells reside (Figure 4) (Shibata et al., 2010; Myosho et al., 2012; Horiguchi et al., 2013; Kaneko et al., 2015). For the female counterpart, no studies indicate expression of gsdf in ovarian cavity. The expression in ductal cells suggests that gsdf in these species may acquire additional function in the differentiation of male structure aside from gamete development.
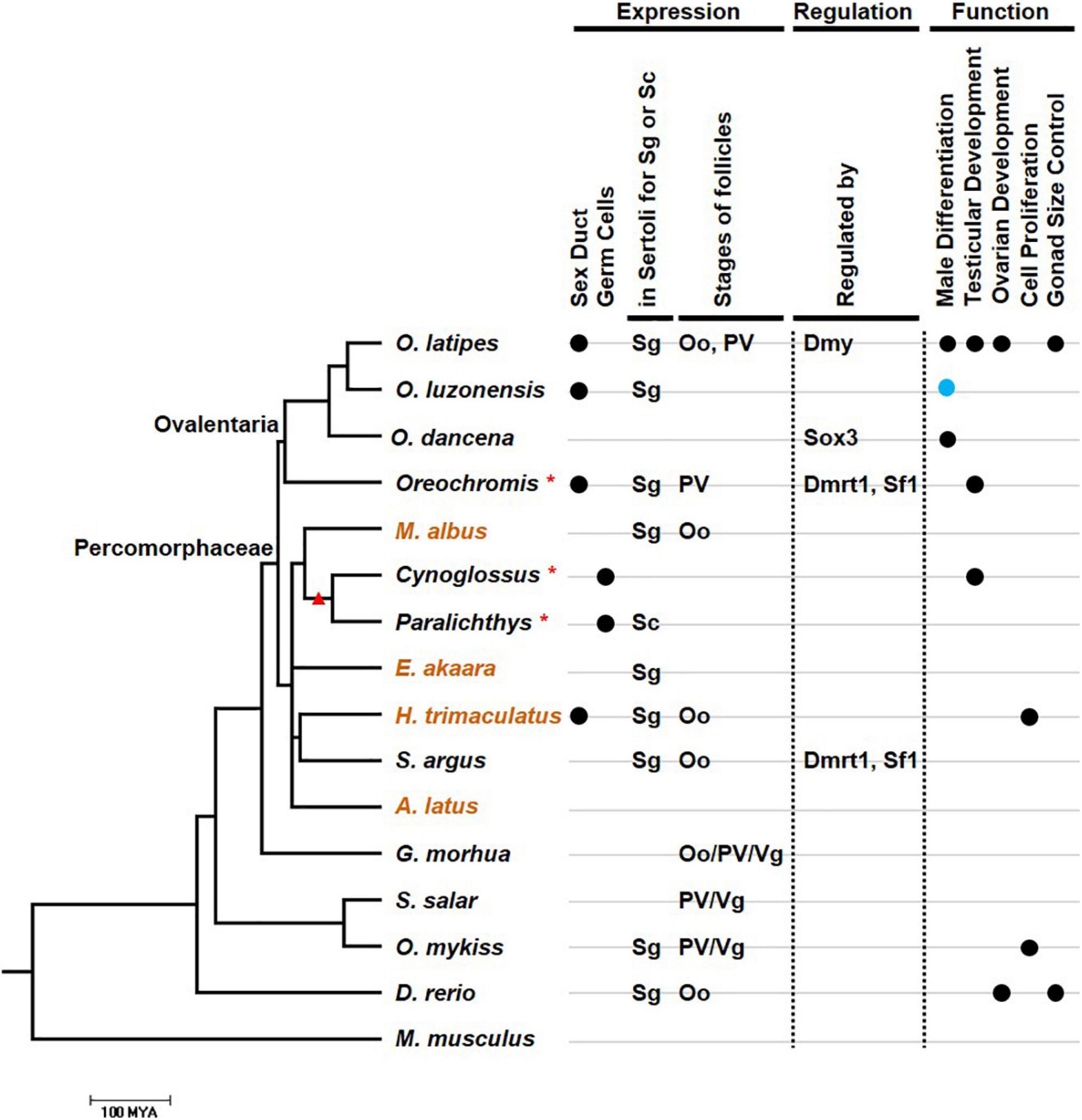
Figure 4. The relationship between the expression, regulation, function, and evolution of gsdf. The phylogenetic tree was drawn using information from the TimeTree database (http://www.timetree.org/resources). Species in the tree including those with gsdf expression in either Sertoli cell in testis or granulose cell in ovary. Mus musculus which does not have gsdf is an outgroup. Scale bar refers to 100 million years ago (MYA). When the evolutionary data of a particular species is missing in the TimeTree database, its genus is used for the compilation of the phylogenic tree and marked with an asterisk. These species include O. niloticus (Oreochromis), Cynoglossus semilaevis (Cynoglossus), and Paralichthys olivaceus (Paralichthys). Orange species names indicate hermaphroditic fish. The branch with a red triangle indicates Pleuronectiformes. The condition of additional expression of gsdf, including expression in efferent duct and germ cells, is labeled with black dots at the right part of first and second lane, respectively. The types of germ cells with predominant surrounding gsdf expression are listed in third and fourth lane. Oo, oogonia; PV, pre-vitellogenic oocyte; Sc, spermatocyte; Sg, spermatogonia; Vg, vitellogenic oocytes. The regulators are list in fifth lane while the functions are listed in lanes 6–9. The function of sex determination is labeled with a blue dot.
In addition to somatic cells, gsdf is also expressed in germ cells of some fish species. In Paralichthys olivaceus, gsdf is additionally expressed in the cytoplasm of oocytes (Liu et al., 2017). This expression pattern is compatible with that of genes clustered in the gsdf locus of O. latipes, which are also expressed in previtellogenic oocytes (Gautier et al., 2011a). Thus, gsdf may participate in oogenesis in P. olivaceus. In the testis of C. semilaevis, gsdf is additionally expressed in spermatogonia and spermatids, suggesting gsdf function during spermatogenesis (Zhu et al., 2018). Both C. semilaevis and P. olivaceus are flatfish (Pleuronectiformes) (Figure 4). The additional expression in germ cells can be a feature either preserved or acquired during evolution. It will be interesting to know whether gsdf is also expressed in germ cells in other Pleuronectiformes.
In females, gsdf transcripts are present in granulosa cells. However, these granulosa cells can surround germs cells at early or late ovarian developmental stages depending on the species (Figure 4). The gsdf transcript is expressed at all follicular stages of Gadus morhua (Nagasawa et al., 2014), in vitellogenic follicles of S. salar (Kleppe et al., 2020), in previtellogenic follicles of Oncorhynchus kisutch (Luckenbach et al., 2008), and surround oogonia of Scatophagus argus (Jiang et al., 2019). The diverse expression in females suggests that Gsdf may have roles in early oogenesis, folliculogenesis and follicle maturation; and the regulatory elements controlling gsdf expression may be differentially acquired in different species.
The gsdf transcripts have been detected in undifferentiated XY gonad during the critical sex-differentiating period in juvenile S. salar (Lubieniecki et al., 2015), O. niloticus (Kaneko et al., 2015), O. latipes (Shibata et al., 2010), Oryzias dancena (Takehana et al., 2014), and Oryzias sakaizumii (Horie et al., 2016). Moreover, gsdf expression is correlated with the expression of the sex-determining genes, dmy in O. latipes and O. sakaizumii, sox3 in O. dancena, and sdY in S. salar (Shibata et al., 2010; Takehana et al., 2014; Lubieniecki et al., 2015; Horie et al., 2016). This implies a role of gsdf in male sex differentiation downstream from initial sex determination.
In hermaphroditic species, expression of gsdf is also correlated with testicular development (Table 2). In protogynous teleosts, H. trimaculatus, Epinephelus akaara, and M. albus, gsdf is highly enriched during the transition from the ovarian phase to the testicular phase (Horiguchi et al., 2013; Chen et al., 2016; Zhu et al., 2016). This implies that gsdf may be involved in the transition into the male fate. In H. trimaculatus, the expression is in supporting cells surrounding gonial cell at early transition stage and in Sertoli cells surrounding spermatogonia at later transition stage, suggesting that gsdf might be involved in self-renewal and differentiation of spermatogonia during sex change (Horiguchi et al., 2013). In a protandrous teleost, Acanthopagrus latus, gsdf is highly expressed in the testicular zone of the ovo-testis, especially at the spermatogonia-dominant stage during spermatogenesis. This suggest that gsdf might be involved in the proliferation and differentiation of spermatogonia (Chen Y. et al., 2015).
In non-teleost species, C. orientalis, P. annectens, and L. menadoensis, gsdf mRNA is enriched in the male gonad similar to that in teleosts (Forconi et al., 2013; Biscotti et al., 2018; Biscotti et al., 2020). This implies that gsdf may retain common functions in testicular development of different species.
The restricted but variable expression of gsdf in gonadal cells reflects its diverse roles during spermatogenesis and oogenesis (Figure 4). It can promote germ cell proliferation in some fish species, while inhibit proliferation in other fish species. It activates testicular development in many teleost species, and is a sex-determining gene for O. luzonensis.
The gsdf transcript is present in Sertoli cells surrounding spermatogonia of several fish species (Table 1). It is an early gonadal marker when germ cells proliferate in European bass, Dicentrarchus labrax. However, gsdf transcript is reduced in germ cells of some males that enter meiosis precociously (Crespo et al., 2013). Knockdown of gsdf in the rainbow trout O. mykiss results in a decrease of primordial germ cells in larvae (Sawatari et al., 2007), indicating that gsdf might be important for the proliferation of germ cells especially spermatogonia. Proliferation assays further demonstrate this hypothesis. Recombinant Gsdf increases the number of BrdU-positive spermatogonia in a dose-dependent manner in vitro (Sawatari et al., 2007). BrdU staining further shows that most of the BrdU-positive cells are surrounded by Gsdf-positive supporting cells during female-to-male sex change of H. trimaculatus (Horiguchi et al., 2013). These reports all show that Gsdf is essential for the proliferation of spermatogonia for those fish during testicular development.
In contrast to a positive role in cell proliferation, Gsdf modulates cell division and prevents hyperplasia in some other species. Gsdf can prevent hyperproliferation of germ cells in O. latipes testis (Zhang et al., 2021). In gsdf-deficient juvenile ovaries, cystic germ cells undergo abnormal type-II division (Wu et al., 2019). These results suggest that Gsdf may modulate mitotic cell division. In gsdf mutant larvae of O. latipes, germ cell number in XY gonads is increased similar to that in XX gonad (Imai et al., 2015). Excessive number of germ cells is also present in adult testis. The mutant testis of D. rerio is fertile but hyperplastic, indicating the importance of gsdf in the regulation of testis size (Yan et al., 2017). Thus, Gsdf might play contradictory roles by promoting or curtailing germ cell proliferation in different species.
The sex of medaka is determined by a master sex-determining gene on the Y chromosome (Myosho et al., 2015). The most well-known fish sex-determining gene is dmy first found in Japanese medaka, O. latipes (Matsuda et al., 2002). In Philippine medaka, O. luzonensis, in which dmy is absent, gsdfY on the Y chromosome replaces dmy to act as a master gene for sex determination (Myosho et al., 2012). XX transgenic fish with overexpression of gsdfY develop into males (Myosho et al., 2012). This gsdfY is also functional in medaka species with other master sex-determining genes. For O. latipes and O. dancena, in which sex is determined by dmy and sox3, respectively, transgenic overexpression of O. luzonensis gsdfY leads to male development in most of XX fish (Myosho et al., 2012; Takehana et al., 2014). This indicates that downstream from GsdfY, Dmy, and Sox3, the steps involved in sex-specific gonad differentiation are probably similar in different medaka species. The promoter sequences of gsdfY in O. luzonensis differ from that of autosomal O. latipes gsdf in nine places. The gsdfY sequences at these sites are required for gene activation in a promoter reporter assay (Myosho et al., 2012). Thus, the unique proximal promoter sequence of gsdfY contributes to its predominant expression in the XY gonad. In sablefish, Anoplopoma fimbria, the gsdf locus is located downstream from sex-specific region, and is expressed in the XY gonad during the larval stage. Thus, gsdf may be the master sex-determining gene in A. fimbria (Rondeau et al., 2013).
The gsdf transcript is predominantly expressed in testis over ovary in most fish species. EE2 treatment of O. latipes causes male-to-female sex reversal, and expression of gsdf in XY gonad is decreased (Shibata et al., 2010). On the contrary, gsdf is highly expressed in male-like XX gonads in O. latipes with cyp19a1 mutation or in O. niloticus with foxl2 mutation (Zhang et al., 2017; Nakamoto et al., 2018). In addition, gsdf expression increases when ovarian follicles start to degenerate (Nakamoto et al., 2018). All these data imply that Gsdf has a role in testicular development.
All gain-of-function and loss-of-function studies show that Gsdf triggers testicular formation. Knocking down gsdf in a C. semilaevis testicular cell line, CSGC, leads to expression of female-related genes, wnt4a, foxl2, star, and cyp19a1a (Zhu et al., 2018). Knocking down gsdf in XY O. niloticus leads to ovarian differentiation (Kaneko et al., 2015). In knockout studies, all gsdf mutants of O. latipes and O. niloticus develop into females despite their genetic sex. Their secondary sex characteristics are also female. Their ovaries express female marker genes such as foxl2 and cyp19a1a, and downregulates male marker genes, dmrt1 and cyp11b2 (Jiang et al., 2016; Zhang et al., 2016).
Male and female germ cells of O. latipes undergo different types of cell division. In XY larvae, germ cells carry out only intermittent type I division, in which only one or two germ cell is present in one cyst. In XX larvae, germ cells undergo both type I and continuous type II division forming clusters of germ cells (Saito et al., 2007). The gsdf mutant larvae contain more germ cells and their germ cell division is more female-type, indicating the induction of female development (Imai et al., 2015; Zhang et al., 2016). Besides, overexpression of gsdf in O. niloticus and O. latipes leads to testis morphology and male secondary sex characteristics. This indicates that gsdf is sufficient to initiate testicular differentiation (Kaneko et al., 2015; Zhang et al., 2016).
Although gsdf is mainly expressed in the testis, the weak expression in the ovary suggests its other role in ovarian development. When gsdf is mutated, the ovaries of D. rerio and O. latipes become hyperplastic with most follicles arrested in the primary growth and previtellogenic stage (Guan et al., 2017; Yan et al., 2017). In O. latipes, gsdf mutation leads to restrained oocyte growth, and ovarian maturation is compromised (Guan et al., 2017). In gsdf-deficient juvenile females, cystic germ cells undergo abnormal type-II division, causing ovarian hyperplasia (Wu et al., 2019).
In D. rerio, ovarian defects of gsdf mutants include a decrease of estrogen production, downregulation of genes for steroid biosynthesis, and decreased estrogen action. Furthermore, granulosa marker genes including cyp19a1a and gata4 are downregulated in mutants (Yan et al., 2017). Therefore, gsdf is essential for the maturation or maintenance of granulosa cells, which are essential for the secretion of estrogen and the ensuing vitellogenin synthesis (Yan et al., 2017).
Although the mechanism of gsdf regulation is not fully understood, there are some hints. Some of the regulatory pathway is common, while others are distinct among species (Figure 4). In Ovalentaria species, gsdf can act downstream from and be regulated by the master sex-determination genes. The sex-determining gene on the Y chromosome of O. dancena is sox3. The gsdf transcript is upregulated in XX gonad by sox3 overexpression while downregulated in the gonad of XY sox3 mutants (Takehana et al., 2014). In O. latipes, the gsdf promoter contains two putative binding sites for the sex-determining protein, Dmy (Zhang et al., 2016). ChIP and luciferase assay show that Dmy can directly bind to the gsdf promoter and enhance its activity in a dose-dependent manner (Chakraborty et al., 2016). It is interesting to investigate whether in other Ovalentaria species, Gsdf also act as the downstream factor of master sex-determining genes.
The regulation of gsdf among different species shares some common elements. Putative binding elements were found. In mammals, Sf1 and Dmrt1 are required for early testicular development after the initiation of sex determination (She and Yang, 2017). The proximal promoter of gsdf in S. argus and O. niloticus genomes also contain binding sites for Sf1 and Dmrt1 required for the activation of gsdf promoter (Jiang et al., 2016; Jiang et al., 2019). Orechromis niloticus and S. argus are species from two evolutionarily distant clades among Percomorphaceae (Figure 4). This implies that these gsdf regulators are conserved among these fish species during evolution.
In D. rerio, six DNA binding motifs, E-box, SOX, GATA, SF1, CEBP/AP2/IL6RE, and one unknown motif, were found within 2-kb proximal promoter of gsdf, which are conserved among three other teleosts, Gasterosteus aculeatus, Takifugu rubripes, and O. latipes. The transcription factors of most of the motifs are expressed in Sertoli cells or granulosa cells (Gautier et al., 2011b), but their functions in regulating gsdf expression has not been shown. The functions of gsdf are diverse among different species. Thus, each species might acquire its own regulatory motif resulting in distinct regulation of gsdf.
Although gsdf has been found widely in many species, the identity of Gsdf receptor still remains illusive. Being a Tgfβ family member, Gsdf may bind to known Tgfβ receptors. Thus, studies of other Tgfβ proteins, including Bmp and Amh, and their receptor can provide some clues. In O. latipes, mutation of amhrII results in gonadal hyperplasia and male-to-female sex reversal in some XY fish, similar to gsdf mutant phenotypes (Morinaga et al., 2007; Imai et al., 2015; Zhang et al., 2016). In D. rerio, mutations of genes encoding amh, bmpr1bb, and bmpr2a all lead to gonadal hyperplasia and accumulation of immature oocytes, mimicking the phenotypes of gsdf mutant (Neumann et al., 2011; Yan et al., 2017, 2019; Zhang Z. et al., 2020). Thus, Gsdf might share the same receptors with other Tgfβ proteins. However, these mutants have additional phenotypes including female-biased sex ratio in amh mutant and the accumulation of immature spermatogenic cells in testis of bmpr1bb mutant (Neumann et al., 2011; Yan et al., 2019). This also raises the possibility that Gsdf may bind to its own yet unidentified receptor.
Gonadal somatic cell-derived factor is a factor that exists mostly in teleosts and functions mainly in gonad differentiation. Its expression, regulation and function, however, change substantially in different species. In this review, we summarize the existence, expression, regulation and function of gsdf, comparing their commonality and diversity during species evolution. The presence of gsdf contributes to the difference of gonad development between teleosts and most tetrapods, while its varied upstream and downstream regulation also results in diversity among teleost species.
Gonadal somatic cell-derived factor is present in both male and female gonads. In males, the expression and regulation of gsdf can be common or species-specific. The gsdf gene is usually expressed in Sertoli cells surrounding spermatogonia, suggesting its involvement in self-renewal, proliferation and differentiation of spermatogonia. Moreover, gsdf is additionally expressed in efferent ducts in H. trimaculatus and species belonging to Ovalentaria (Figure 4). In species from two distant clades among Percomorphaceae, gsdf is activated by Sf1 and Dmrt1, implying the conserved common regulation (Figure 4). In Ovalentaria, Gsdf functions in male sex determination, either acting as a sex-determining gene or working downstream from sex-determining factors (Figure 4). Thus, the species-specific regulation of gsdf is preserved in limited clades during evolution.
In females, gsdf is less expressed and mostly in granulose cell surrounding different types of germ cell in different teleost species. Besides participating in testicular development, Gsdf plays a major role in ovarian development in D. rerio (Yan et al., 2017). The distinct ovarian function in D. rerio and the diverse expression suggest that the regulation of gsdf is acquired differently during evolution.
Gonadal somatic cell-derived factor is a gonadal protein of multiple functions widely expressed in many species. The mechanism by which Gsdf functions in different species and developmental stages remains an interesting question. Furthermore, little is known about the upstream and downstream regulation of Gsdf. The identification of Gsdf receptor and the analysis of gsdf promoters in different species will be the key. With this knowledge at hand, one can then better understand the multiple functions of Gsdf as a result of its diverse regulation.
C-WH and B-CC conceived the ideas, organized the literature, analyzed the data, and wrote the manuscript. B-CC conceived the ideas, analyzed the data, and wrote the manuscript.
This work was funded by grants from Academia Sinica, AS-101-TP-B05, NHRI-EX107-10506SI, MOST 107-2321-B-001-034, MOST 108-2311-B-001-038-MY3.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank Taiwan Zebrafish Core Facility and Institute of Molecular Biology, Academia Sinica for providing the infrastructure enabling the execution of our study.
Bashamboo, A., and McElreavey, K. (2016). Mechanism of sex determination in humans: insights from disorders of sex development. Sex Dev. 10, 313–325. doi: 10.1159/000452637
Biscotti, M. A., Adolfi, M. C., Barucca, M., Forconi, M., Pallavicini, A., Gerdol, M., et al. (2018). A comparative view on sex differentiation and gametogenesis genes in lungfish and coelacanths. Genome Biol. Evol. 10, 1430–1444. doi: 10.1093/gbe/evy101
Biscotti, M. A., Carducci, F., Barucca, M., Gerdol, M., Pallavicini, A., Schartl, M., et al. (2020). The transcriptome of the newt Cynops orientalis provides new insights into evolution and function of sexual gene networks in sarcopterygians. Sci. Rep. 10:5445.
Chakraborty, T., Zhou, L. Y., Chaudhari, A., Iguchi, T., and Nagahama, Y. (2016). Dmy initiates masculinity by altering Gsdf/Sox9a2/Rspo1 expression in medaka (Oryzias latipes). Sci. Rep. 6:19480.
Chen, J. J., Xia, X. H., Wang, L. F., Jia, Y. F., Nan, P., Li, L., et al. (2015). Identification and comparison of gonadal transcripts of testis and ovary of adult common carp Cyprinus carpio using suppression subtractive hybridization. Theriogenology 83, 1416–1427. doi: 10.1016/j.theriogenology.2015.01.001
Chen, Y., Hong, W. S., Wang, Q., and Chen, S. X. (2015). Cloning and expression pattern of gsdf during the first maleness reproductive phase in the protandrous Acanthopagrus latus. Gen. Comp. Endocrinol. 21, 71–80. doi: 10.1016/j.ygcen.2015.02.018
Chen, Y., Hong, W. S., Wang, Q., and Chen, S. X. (2016). Cloning and pattern of gsdf mRNA during gonadal development in the protogynous Epinephelus akaara. Anim. Reprod. Sci. 165, 46–55. doi: 10.1016/j.anireprosci.2015.12.004
Crespo, B., Gomez, A., Mazon, M. J., Carrillo, M., and Zanuy, S. (2013). Isolation and characterization of Ff1 and Gsdf family genes in European sea bass and identification of early gonadal markers of precocious puberty in males. Gen. Comp. Endocrinol. 191, 155–167. doi: 10.1016/j.ygcen.2013.06.010
Dalbies-Tran, R., Cadoret, V., Desmarchais, A., Elis, S., Maillard, V., and Monget, P. (2020). A comparative analysis of oocyte development in mammals. Cells 9:1002. doi: 10.3390/cells9041002
Force, A., Lynch, M., Pickett, F. B., Amores, A., Yan, Y. L., and Postlethwait, J. (1999). Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151, 1531–1545. doi: 10.1093/genetics/151.4.1531
Forconi, M., Canapa, A., Barucca, M., Biscotti, M. A., Capriglione, T., and Buonocore, F. (2013). Characterization of sex determination and sex differentiation genes in Latimeria. PLoS One 8:e56006. doi: 10.1371/journal.pone.0056006
Fricke, H., and Fricke, S. (1977). Monogamy and sex change by aggressive dominance in coral reef fish. Nature 266, 830–832. doi: 10.1038/266830a0
Gautier, A., Le Gac, F., and Lareyre, J. J. (2011a). The gsdf gene locus harbors evolutionary conserved and clustered genes preferentially expressed in fish previtellogenic oocytes. Gene 472, 7–17. doi: 10.1016/j.gene.2010.10.014
Gautier, A., Sohm, F., Joly, J. S., Le Gac, F., and Lareyre, J. J. (2011b). The proximal promoter region of the zebrafish gsdf gene is sufficient to mimic the spatio-temporal expression pattern of the endogenous gene in Sertoli and granulosa cells. Biol. Reprod. 85, 1240–1251. doi: 10.1095/biolreprod.111.091892
Guan, G., Sun, K., Zhang, X., Zhao, X., Li, M., and Yan, Y. (2017). Developmental tracing of oocyte development in gonadal soma-derived factor deficiency medaka (Oryzias latipes) using a transgenic approach. Mech. Dev. 143, 53–61. doi: 10.1016/j.mod.2016.12.006
Hayman, E. S., Fairgrieve, W. T., and Luckenbach, J. A. (2021). Molecular and morphological sex differentiation in sablefish (Anoplopoma fimbria), a marine teleost with XX/XY sex determination. Gene 764:145093. doi: 10.1016/j.gene.2020.145093
Horie, Y., Myosho, T., Sato, T., Sakaizumi, M., Hamaguchi, S., and Kobayashi, T. (2016). Androgen induces gonadal soma-derived factor, Gsdf, in XX gonads correlated to sex-reversal but not Dmrt1 directly, in the teleost fish, northern medaka (Oryzias sakaizumii). Mol. Cell. Endocrinol. 436, 141–149. doi: 10.1016/j.mce.2016.07.022
Horiguchi, R., Nozu, R., Hirai, T., Kobayashi, Y., Nagahama, Y., and Nakamura, M. (2013). Characterization of gonadal soma-derived factor expression during sex change in the protogynous wrasse. Halichoeres trimaculatus. Dev. Dyn. 242, 388–399. doi: 10.1002/dvdy.23929
Imai, T., Saino, K., and Matsuda, M. (2015). Mutation of Gonadal soma-derived factor induces medaka XY gonads to undergo ovarian development. Biochem. Biophys. Res. Commun. 467, 109–114. doi: 10.1016/j.bbrc.2015.09.112
Jiang, D. N., Mustapha, U. F., Shi, H. J., Huang, Y. Q., Si-Tu, J. X., and Wang, M. (2019). Expression and transcriptional regulation of gsdf in spotted scat (Scatophagus argus). Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 233, 35–45. doi: 10.1016/j.cbpb.2019.04.002
Jiang, D. N., Yang, H. H., Li, M. H., Shi, H. J., Zhang, X. B., and Wang, D. S. (2016). gsdf is a downstream gene of dmrt1 that functions in the male sex determination pathway of the Nile tilapia. Mol. Reprod. Dev. 83, 497–508. doi: 10.1002/mrd.22642
Kaneko, H., Ijiri, S., Kobayashi, T., Izumi, H., Kuramochi, Y., Wang, D. S., et al. (2015). Gonadal soma-derived factor (gsdf), a TGF-beta superfamily gene, induces testis differentiation in the teleost fish Oreochromis niloticus. Mol. Cell. Endocrinol. 415, 87–99. doi: 10.1016/j.mce.2015.08.008
Kleppe, L., Edvardsen, R. B., Furmanek, T., Andersson, E., Skaftnesmo, K. O., Thyri Segafredo, F., et al. (2020). Transcriptomic analysis of dead end knockout testis reveals germ cell and gonadal somatic factors in Atlantic salmon. BMC Genom. 21:99.
Koumoundouros, G., Pavlidis, M., Anezaki, L., Kokkari, C., Sterioti, A., Divanach, P., et al. (2002). Temperature sex determination in the European sea bass, Dicentrarchus labrax (L., 1758) (Teleostei, Perciformes, Moronidae): critical sensitive ontogenetic phase. J. Exp. Zool. 292, 573–579. doi: 10.1002/jez.10095
Li, M., Sun, Y., Zhao, J., Shi, H., Zeng, S., and Ye, K. (2015). A tandem duplicate of anti-mullerian hormone with a missense SNP on the Y chromosome is essential for male sex determination in Nile Tilapia, Oreochromis niloticus. PLoS Genet. 11:e1005678. doi: 10.1371/journal.pgen.1005678
Liu, Y., Zhang, W., Du, X., Zhao, J., Liu, X., Li, X., et al. (2017). Sexually dimorphic expression in developing and adult gonads shows an important role of gonadal soma-derived factor during sex differentiation in olive flounder (Paralichthys olivaceus). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 210, 1–8. doi: 10.1016/j.cbpb.2017.05.003
Lubieniecki, K. P., Botwright, N. A., Taylor, R. S., Evans, B. S., Cook, M. T., and Davidson, W. S. (2015). Expression analysis of sex-determining pathway genes during development in male and female Atlantic salmon (Salmo salar). Physiol. Genom. 47, 581–587. doi: 10.1152/physiolgenomics.00013.2015
Luckenbach, J. A., Iliev, D. B., Goetz, F. W., and Swanson, P. (2008). Identification of differentially expressed ovarian genes during primary and early secondary oocyte growth in coho salmon, Oncorhynchus kisutch. Reprod. Biol. Endocrinol. 6:2.
Matsuda, M., Nagahama, Y., Shinomiya, A., Sato, T., Matsuda, C., and Kobayashi, T. (2002). DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature 417, 559–563. doi: 10.1038/nature751
Morinaga, C., Saito, D., Nakamura, S., Sasaki, T., Asakawa, S., and Shimizu, N. (2007). The hotei mutation of medaka in the anti-Mullerian hormone receptor causes the dysregulation of germ cell and sexual development. Proc. Natl. Acad. Sci. U. S. A. 104, 9691–9696. doi: 10.1073/pnas.0611379104
Myosho, T., Otake, H., Masuyama, H., Matsuda, M., Kuroki, Y., Fujiyama, A., et al. (2012). Tracing the emergence of a novel sex-determining gene in medaka, Oryzias luzonensis. Genetics 191, 163–170. doi: 10.1534/genetics.111.137497
Myosho, T., Takehana, Y., Hamaguchi, S., and Sakaizumi, M. (2015). Turnover of sex chromosomes in celebensis group medaka fishes. G3 5, 2685–2691. doi: 10.1534/g3.115.021543
Nagasawa, K., Presslauer, C., Kirtiklis, L., Babiak, I., and Fernandes, J. M. (2014). Sexually dimorphic transcription of estrogen receptors in cod gonads throughout a reproductive cycle. J. Mol. Endocrinol. 52, 357–371. doi: 10.1530/jme-13-0187
Nakamoto, M., Shibata, Y., Ohno, K., Usami, T., Kamei, Y., and Taniguchi, Y. (2018). Ovarian aromatase loss-of-function mutant medaka undergo ovary degeneration and partial female-to-male sex reversal after puberty. Mol. Cell. Endocrinol. 460, 104–122. doi: 10.1016/j.mce.2017.07.013
Neumann, J. C., Chandler, G. L., Damoulis, V. A., Fustino, N. J., Lillard, K., and Looijenga, L. (2011). Mutation in the type IB bone morphogenetic protein receptor Alk6b impairs germ-cell differentiation and causes germ-cell tumors in zebrafish. Proc. Natl. Acad. Sci. U. S. A. 108, 13153–13158. doi: 10.1073/pnas.1102311108
Robledo, D., Ribas, L., Cal, R., Sanchez, L., Piferrer, F., Martinez, P., et al. (2015). Gene expression analysis at the onset of sex differentiation in turbot (Scophthalmus maximus). BMC Genom. 16:973.
Rondeau, E. B., Messmer, A. M., Sanderson, D. S., Jantzen, S. G., von Schalburg, K. R., and Minkley, D. R. (2013). Genomics of sablefish (Anoplopoma fimbria): expressed genes, mitochondrial phylogeny, linkage map and identification of a putative sex gene. BMC Genom. 14:452. doi: 10.1186/1471-2164-14-452
Saito, D., Morinaga, C., Aoki, Y., Nakamura, S., Mitani, H., Furutani-Seiki, M., et al. (2007). Proliferation of germ cells during gonadal sex differentiation in medaka: insights from germ cell-depleted mutant zenzai. Dev. Biol. 310, 280–290. doi: 10.1016/j.ydbio.2007.07.039
Sawatari, E., Shikina, S., Takeuchi, T., and Yoshizaki, G. (2007). A novel transforming growth factor-beta superfamily member expressed in gonadal somatic cells enhances primordial germ cell and spermatogonial proliferation in rainbow trout (Oncorhynchus mykiss). Dev. Biol. 301, 266–275. doi: 10.1016/j.ydbio.2006.10.001
She, Z. Y., and Yang, W. X. (2017). Sry and SoxE genes: how they participate in mammalian sex determination and gonadal development?”. Semin. Cell Dev. Biol. 63, 13–22. doi: 10.1016/j.semcdb.2016.07.032
Shibata, Y., Paul-Prasanth, B., Suzuki, A., Usami, T., Nakamoto, M., Matsuda, M., et al. (2010). Expression of gonadal soma derived factor (GSDF) is spatially and temporally correlated with early testicular differentiation in medaka. Gene Expr. Patterns 10, 283–289. doi: 10.1016/j.gep.2010.06.005
Takehana, Y., Matsuda, M., Myosho, T., Suster, M. L., Kawakami, K., and Shin, I. T. (2014). Co-option of Sox3 as the male-determining factor on the Y chromosome in the fish Oryzias dancena. Nat. Commun. 5:4157.
Vitt, U. A., Hsu, S. Y., and Hsueh, A. J. (2001). Evolution and classification of cystine knot-containing hormones and related extracellular signaling molecules. Mol. Endocrinol. 15, 681–694. doi: 10.1210/mend.15.5.0639
Wang, W., Zhu, H., Dong, Y., Tian, Z., Dong, T., Hu, H., et al. (2017). Dimorphic expression of sex-related genes in different gonadal development stages of sterlet, Acipenser ruthenus, a primitive fish species. Fish Physiol. Biochem. 43, 1557–1569. doi: 10.1007/s10695-017-0392-x
Wu, G. C., Dufour, S., and Chang, C. F. (2021). Molecular and cellular regulation on sex change in hermaphroditic fish, with a special focus on protandrous black porgy, Acanthopagrus schlegelii. Mol. Cell. Endocrinol. 520:111069. doi: 10.1016/j.mce.2020.111069
Wu, X., Zhang, Y., Xu, S., Chang, Y., Ye, Y., and Guo, A. (2019). Loss of Gsdf leads to a dysregulation of Igf2bp3-mediated oocyte development in medaka. Gen. Comp. Endocrinol. 277, 122–129. doi: 10.1016/j.ygcen.2019.04.001
Yan, H., Shen, X., Cui, X., Wu, Y., Wang, L., Zhang, L., et al. (2018). Identification of genes involved in gonadal sex differentiation and the dimorphic expression pattern in Takifugu rubripes gonad at the early stage of sex differentiation. Fish Physiol. Biochem. 44, 1275–1290. doi: 10.1007/s10695-018-0519-8
Yan, Y. L., Batzel, P., Titus, T., Sydes, J., Desvignes, T., BreMiller, R., et al. (2019). A hormone that lost its receptor: anti-mullerian hormone (AMH) in zebrafish gonad development and sex determination. Genetics 213, 529–553. doi: 10.1534/genetics.119.302365
Yan, Y. L., Desvignes, T., Bremiller, R., Wilson, C., Dillon, D., High, S., et al. (2017). Gonadal soma controls ovarian follicle proliferation through Gsdf in zebrafish. Dev. Dyn. 246, 925–945. doi: 10.1002/dvdy.24579
Zeng, Q., Liu, S., Yao, J., Zhang, Y., Yuan, Z., Jiang, C., et al. (2016). Transcriptome display during testicular differentiation of channel catfish (Ictalurus punctatus) as revealed by RNA-Seq analysis. Biol. Reprod. 95:19. doi: 10.1095/biolreprod.116.138818
Zhang, X., Chang, Y., Zhai, W., Qian, F., Zhang, Y., and Xu, S. (2021). A Potential role for the Gsdf-eEF1alpha complex in inhibiting germ cell proliferation: a protein-interaction analysis in Medaka (Oryzias latipes) from a proteomics perspective. Mol. Cell. Proteomics 20:100023. doi: 10.1074/mcp.ra120.002306
Zhang, X., Guan, G., Li, M., Zhu, F., Liu, Q., Naruse, K., et al. (2016). Autosomal gsdf acts as a male sex initiator in the fish medaka. Sci. Rep. 6:19738.
Zhang, X., Li, M., Ma, H., Liu, X., Shi, H., Li, M., et al. (2017). Mutation of foxl2 or cyp19a1a results in female to male sex reversal in XX Nile Tilapia. Endocrinology 158, 2634–2647.
Zhang, X., Zhou, J., Li, L., Huang, W., Ahmad, H. I., Li, H., et al. (2020). Full-length transcriptome sequencing and comparative transcriptomic analysis to uncover genes involved in early gametogenesis in the gonads of Amur sturgeon (Acipenser schrenckii). Front. Zool. 17:11.
Zhang, Y., Wang, J., Lu, L., Li, Y., Wei, Y., and Cheng, Y. (2020). Genotoxic biomarkers and histological changes in marine medaka (Oryzias melastigma) exposed to 17alpha-ethynylestradiol and 17beta-trenbolone. Mar. Pollut. Bull. 150:110601. doi: 10.1016/j.marpolbul.2019.110601
Zhang, Z., Wu, K., Ren, Z., and Ge, W. (2020). Genetic evidence for Amh modulation of gonadotropin actions to control gonadal homeostasis and gametogenesis in zebrafish and its noncanonical signaling through Bmpr2a receptor. Development 147:dev189811.
Zhu, Y., Meng, L., Xu, W., Cui, Z., Zhang, N., Guo, H., et al. (2018). The autosomal Gsdf gene plays a role in male gonad development in Chinese tongue sole (Cynoglossus semilaevis). Sci. Rep. 8:17716.
Keywords: gonad, ovary, oocyte, development, evolution, teleost, testis, gsdf
Citation: Hsu C-w and Chung B-c (2021) Evolution, Expression, and Function of Gonadal Somatic Cell-Derived Factor. Front. Cell Dev. Biol. 9:684352. doi: 10.3389/fcell.2021.684352
Received: 23 March 2021; Accepted: 27 May 2021;
Published: 07 July 2021.
Edited by:
Karuna Sampath, University of Warwick, United KingdomReviewed by:
Andreas Zaucker, University of Warwick, United KingdomCopyright © 2021 Hsu and Chung. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bon-chu Chung, bWJjaHVuZ0BzaW5pY2EuZWR1LnR3
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.