- Department of Public Health, China Medical University, Shenyang, China
Eukaryotic messenger mRNAs contain many RNA methyl chemical modifications, in which N6-methyladenosine (m6A) plays a very important role. The modification process of RNA methylation is a dynamic reversible regulatory process that is mainly catalyzed by “Writer” m6A methyltransferase, removed by “Eraser” m6A demethylase, and recognized by the m6A binding protein, thereby, linking m6A modification with other mRNA pathways. At various stages of the life cycle, m6A modification plays an extremely important role in regulating mRNA splicing, processing, translation, as well as degradation, and is associated with gametogenesis and fertility for both sexes. Normal gametogenesis is a basic guarantee of fertility. Infertility leads to trauma, affects harmony in the family and seriously affects the quality of life. We review the roles and mechanisms of RNA m6A methylation modification in infertility and provide a potential target for infertility treatment, which can be used for drug development.
Overview of Infertility
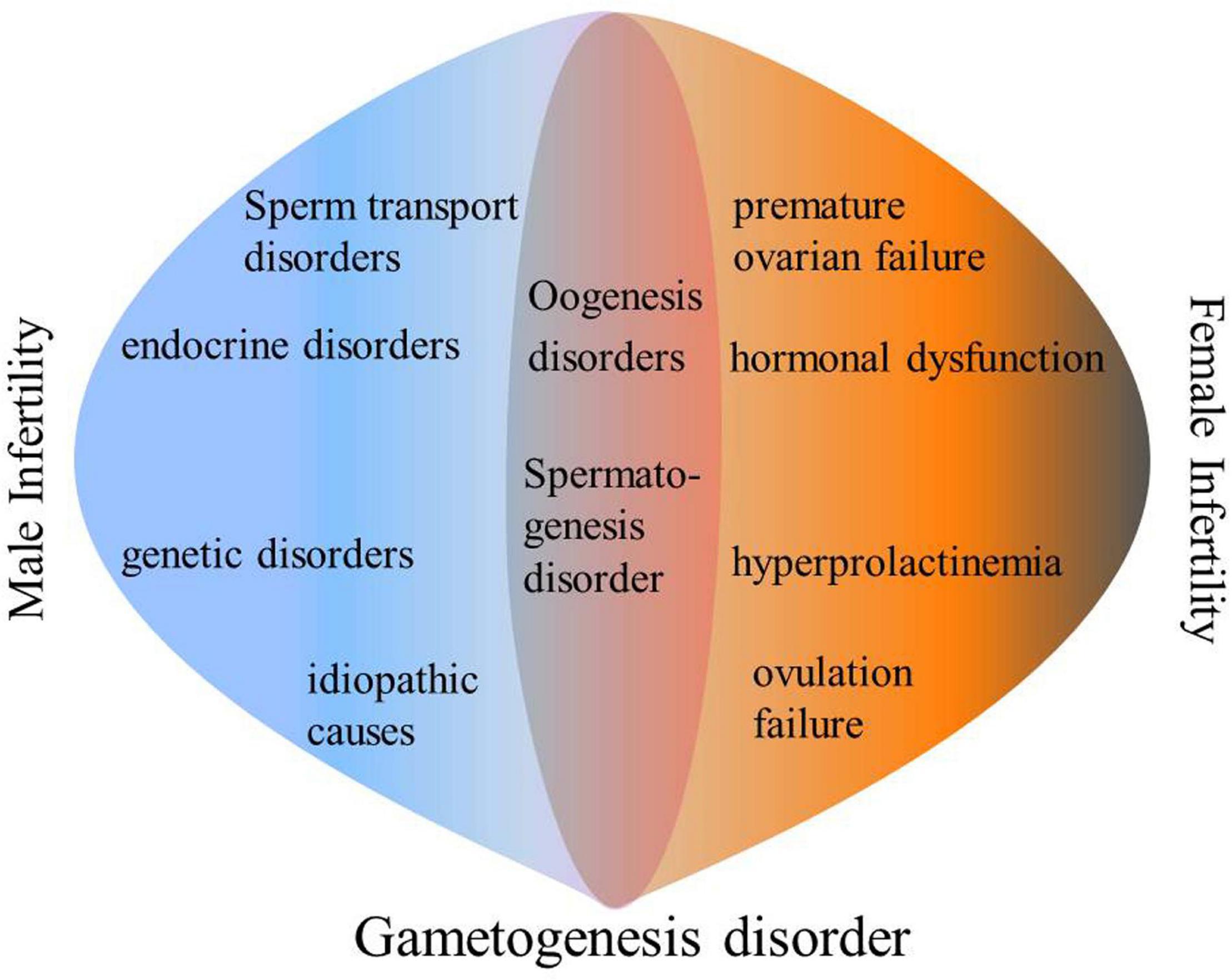
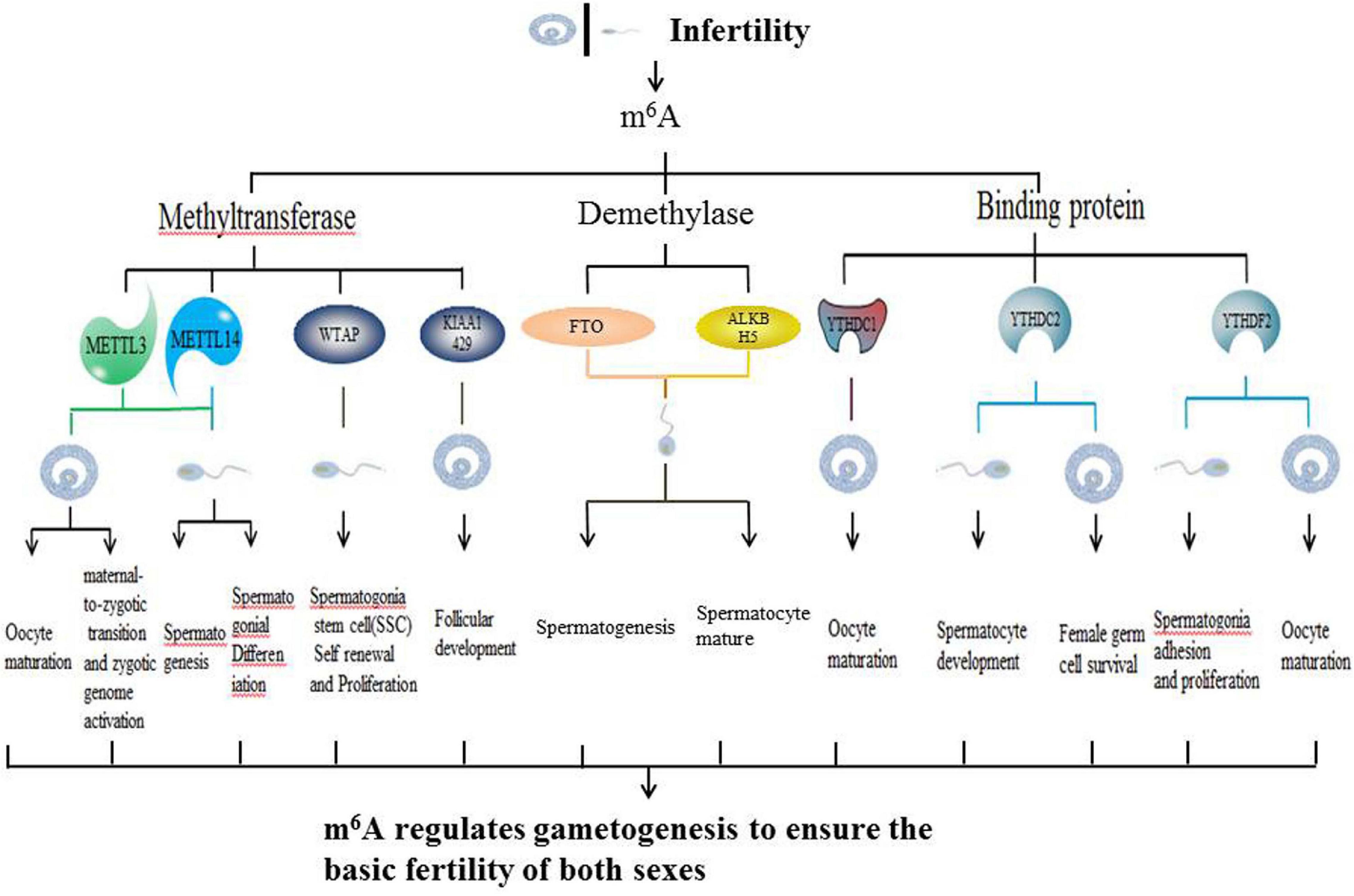
Infertility is a reproductive dysfunction that is characterized by a succession of sexual events and interruption of new life. Accordingly, an inability to get pregnant after 12 months of unprotected intercourse is broadly defined as infertility (Petraglia et al., 2013; Kayode et al., 2020). Infertility can be divided into female infertility and male sterility. Female infertility is defined as: for women under 35 years of age, they cannot get pregnant after 12 months of unprotected sex, or for women over 35, unprotected intercourse for 6 months cannot lead to pregnancy. The most common infertility factor is ovulation disorder, which accounts for about 25% of all infertility factors (“Practice Committee of the American Society for Reproductive Medicine,Practice Committee of the American Society for Reproductive Medicine., 2013” Recent advances in medically assisted conception. Oogenesis is a complex process that is regulated by many internal and external ovarian factors (Sánchez and Smitz, 2012). Oogenesis disorders can lead to ovulation disorders, which in turn lead to female infertility. According to the World Health Organization (WHO), male infertility refers to a person who cannot cause a pregnancy after at least 12 months of unprotected sexual intercourse (La Vignera et al., 2011). The four main causes of male infertility include: sperm transport disorders, endocrine disorders, genetic disorders and idiopathic causes (Kayode et al., 2020). Among them, idiopathic causes account for 40% of male infertility cases (Louis et al., 2013). Spermatogenesis is a sophisticated developmental process in which haploid sperms are continuously produced by diploid spermatogonia. Frequent cell division and differentiation cause epigenetic modification changes and genetic distortion during chromosomal remodeling that eventually inhibit spermatogenesis, leading to low sperm concentrations, low motor abilities and poor morphologies (Clermont, 1972; de Rooij, 2001; Rajender et al., 2011; Battle, 2013; Griswold, 2016). Therefore, a large number of idiopathic male infertility cases are due to genetic aberrations (Figure 1).
Gametogenesis
The process through which mature germ cells are formed is called gametogenesis. Gametogenesis is indispensable to the general health and well-being of various species. Overtime, gametes link the various stages of species’ development. Compared to somatic cells, after a series of mitosis and meiosis during the differentiation process, germ cells produce sperms and oocytes. Sexually mature gametes combine during fertilization to produce the all-powerful oosperm (Carreau and Hess, 2010). An important aspect of gametogenesis is meiosis, during which diploid progenitor cells in mammalian gonads produce heritable haploid gametes, which are controlled by a series of strictly controlled gene expression events that determine key and highly coordinated cellular fates, including ovogenesis and spermatogenesis (Ying et al., 2000; Czerwoniec et al., 2009; Carreau and Hess, 2010; Lesch and Page, 2012; Miyamoto et al., 2012; Sahin et al., 2014).
The transforming growth factor beta-1 proprotein-like (TGF) -β family member proteins (e.g., BMP signal) are essential for primordial germ cells (PGC). They act as precursors of male and female gametes to produce oocytes and sperms. Directional interruption of Bmp2, Bmp4, Bmp8b, or BMP signal transducers, SMAD family member s1, 4 and 5, or protein kinase (ALK2) indicates a loss or decrease of PGCs (Lawson and Hage, 1994; Tremblay et al., 2001; McLaren, 2003; Saitou and Yamaji, 2012). In mammals, the PGC precursor is produced by equipotent epithelial cells, approximately on embryonic day 6 (E6), in response to Bmp4, Bmp8b, and Bmp2 signals emitted by the embryonic ectoderm and intraluminal endoderm (Chiquoine, 1954; Lawson et al., 1999; Loebel et al., 2003; de Sousa Lopes et al., 2004; Ohinata et al., 2009; van Werven and Amon, 2011). Fragilis, a member of interferon-(IFN) inducible transmembrane protein family, is a marker for early appearance of PGC in mice (Ying and Zhao, 2001). Starting from E7, designated PGCs in mice express various markers, including TNAP, SSEA1 and DPPA3 (Ginsburg et al., 1990; Sato et al., 2002; Avilion et al., 2003; Lence et al., 2016). Maintenance of the expression of multiple pluripotency genes, such as SRY- Sox2, Nanog and Oct4, is regulated by mouse PGCs (Yamaguchi et al., 2005; Chambers et al., 2007). Human PGC is first formed around the third week of pregnancy. It originates from precursor cells of the mesoderm, and depends on WNT and BMP pathways (Irie et al., 2015; Tang et al., 2016; Kojima et al., 2017). After formation, PGCs migrate and multiply through the back waves, and then enter the future genital ridge at approximately E7.5 to E10.5 (Anderson et al., 2000; Molyneaux et al., 2001; Richardson and Lehmann, 2010). The two germ cell-somatic signaling pathways of cKIT-STEEL and SDF-CXCR4 promote PGC multiplication and targeted migration (Larose et al., 2019). From the original band to future hindgut and genitals, cKIT-STEEL interactions are essential for PGC proliferation, survival and migration (Ohta et al., 2003; Ewen et al., 2009). In addition, SDF-1, which is expressed on the surrounding stromal genitalia, promotes PGC migration in a specific direction and can be detected by the PGC surface receptor, CXCR4 (Runyan et al., 2006; Gu et al., 2009). During the active migration process, venture capital continues to surge. At E10.5, there are about 500 PGCs in the genital ridge of each embryo. Sex determination begins during PGC migration to the genital ridge, and tends to differentiate into ovaries. Differentiation into testis is due to the effects of the Y-linked gene, SRY, on the XY genital ridge. The absence of such a gene for the XX genital ridge leads to the development of the ovary (Pepling and Spradling, 1998; Bowles and Koopman, 2010). Once prostaglandin cells enter the genital ridge, they are referred to as oogonocytes in females, or gonad cells in males. After colonization of the genital ridge, from E10.5 to E14.5, PGCs enter the mitotic proliferative phase. These embryonic germ cells undergo about 5 phases of mitosis and form “germline cysts” or “germ cell nests” due to incomplete cytokinesis (Pepling, 2006; Lei and Spradling, 2013). These “cysts” or “nests” eventually decompose and produce primary oocytes and spermatogonia in the, respectively, differentiated gonads (Larose et al., 2019).
Spermatogenesis
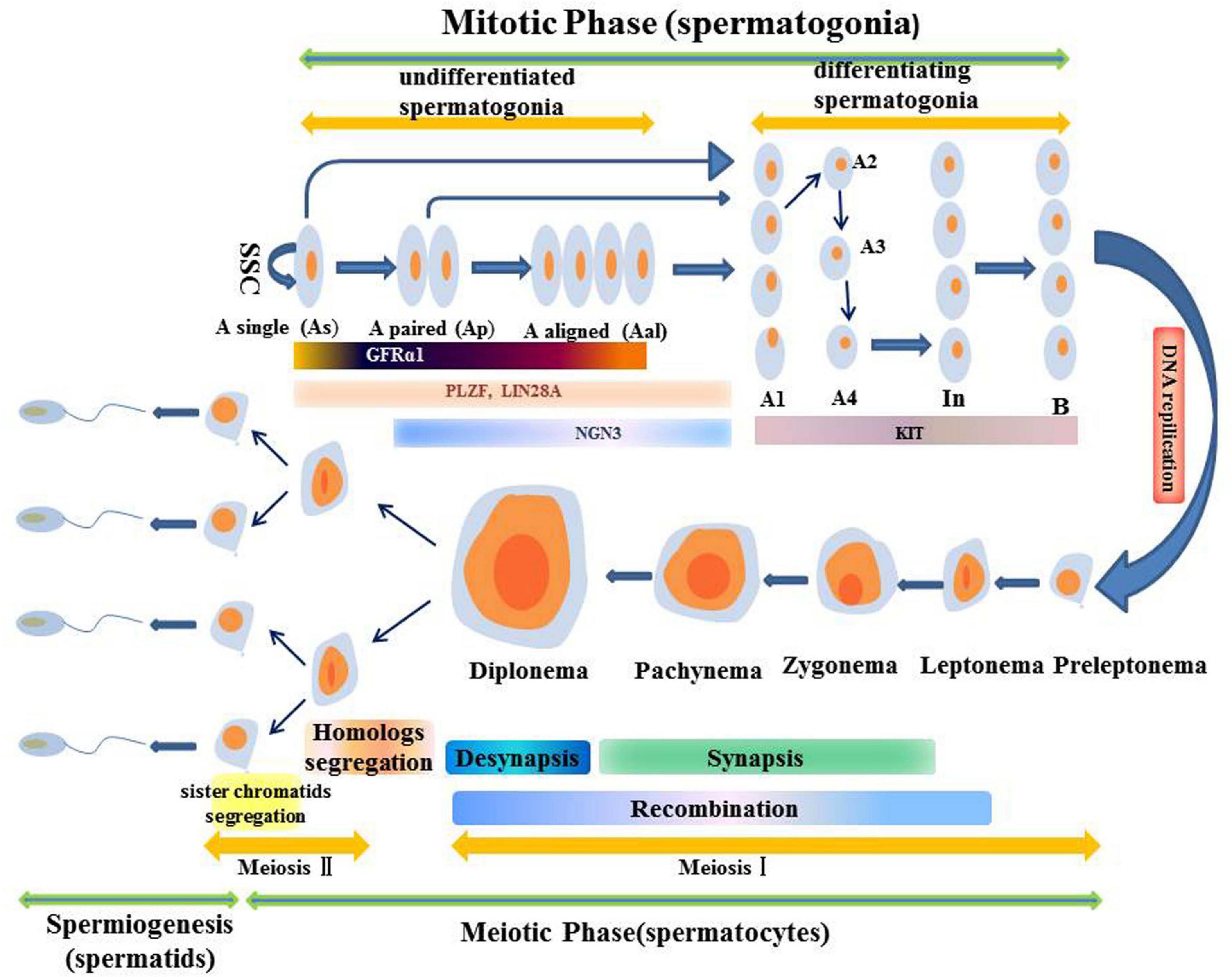
Sperm production is a complex asynchronous differentiation process that is divided into three stages based on the cell types found: Spermatogonia are formed through mitosis, spermatocytes are formed through meiosis, and the haploid stage of sperm cells or sperm formation (Shinohara et al., 2000; Wolgemuth et al., 2013; Griswold, 2016). Continuous sperm production depends on normal SSC functions. Spermatogonia with actual SSC functions can either self-renew to maintain the stem cell pool or can differentiate into Ap spermatogonia for spermatogenesis. The Ap spermatogonia stay connected through the intercellular bridge caused by incomplete cytokinesis. After repeated mitosis, AP spermatogonia divide to produce Aal spermatogonia (Lin and Tong, 2019). The Ap and Aal sperms are indiscriminate sperms, and many genes associated with self-renewal and proliferation, including Gfrα1, Nanos2, and Plzf among others are expressed in them to maintain the balance between self-renewal and differentiation. In addition, the GDNF/GFRα1/RET pathway is a key signaling pathway that regulates self-renewal and differentiation of undifferentiated spermatogonia. It can activate multiple signaling pathways such as the PI3K/AKT signaling pathway (Lin and Tong, 2019). Aal spermatogonia differentiates into A1 spermatogonia without mitosis. Then, they undergo a series of mitosis to produce A2, A3, A4, intermediate and B spermatogonia (Ohbo et al., 2003; Barroca et al., 2009; Nakagawa et al., 2010; Oatley and Brinster, 2012). Type A 1 to B Spermatogonia are referred to as “differentiated spermatogonia” (Battle, 2013). During the subsequent spermatogonia differentiation, genes such as Dnmt3b, Stra8, Kit and Ccnd2 among others are expressed. Among them, the genes associated with self-renewal are downregulated in undifferentiated sperms while those associated with differentiation are upregulated (Lin and Tong, 2019). The RA signal is essential for the differentiation of spermatogonia because it controls the expression of direct target genes (Hist1 cluster, Stra8 and Kit) (Lin and Tong, 2019). Type B spermatogonia pass through a long-lasting S phase and differentiate into pre-leptotene stage spermatocytes, followed by pre-meiotic stage I spermatocytes. This stage is highly regulated, and can be subdivided into four phases, that is, leptotene, zygotene, pachytene, and diplotene. The most complex and critical events of spermatogenesis, such as recombination and synapse, occur in the first stage before meiosis (Huckins, 1971; Handel and Schimenti, 2010). After prostatitis I, sperm cells are separated by two chromosomes, leading to doubling of round sperm cells. Spermiogenesis is the last stage of spermatogenesis, during which sperm cell nucleus undergo unique chromatin remodeling, including extreme compaction of the genome. Along with drastic reduction in nuclear volumes, its shape changes from round to rod-shaped, and finally pear-shaped, eventually forming mature elongated sperms (Ahmed and de Rooij, 2009) (Figure 2).
Oogenesis
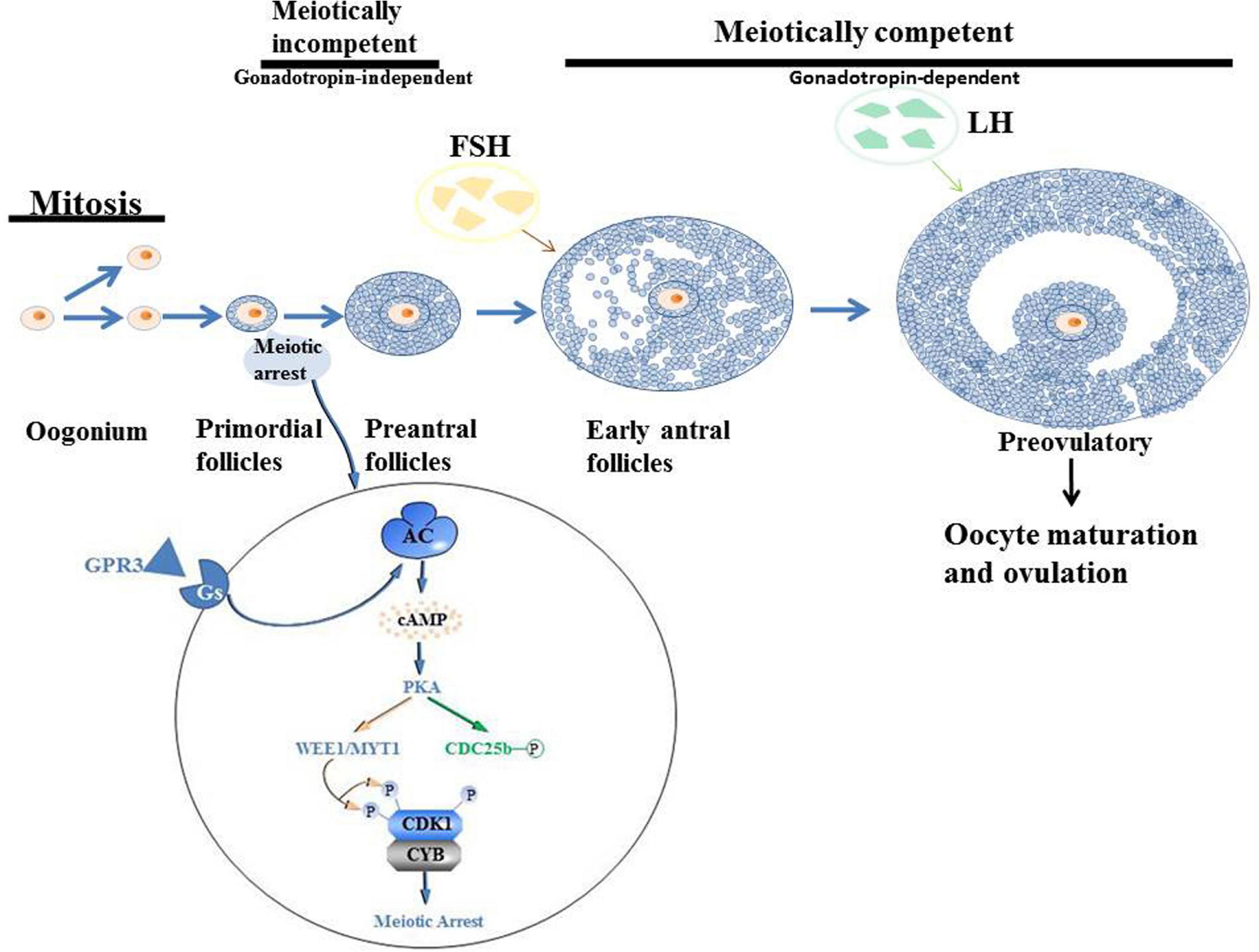
Oogenesis is a long-term process in mammals that begins in the embryonic period and ends in menopause. In oogenesis, two basic processes are necessary; the formation and maturation of oocytes and folliculogenesis. Follicles are reproductive units composed of oocytes, somatic cells and follicles. Oteocytes are made up of cumulus granulosa, mural granulosa and theca cells. In the embryonic stage, oogonia are the precursors of female gametes, expanding in number through mitosis. They differentiate into primary oocytes, grow in the follicle and undergo meiosis. Meiosis has a complex early stage that can be divided into five phases: leptotene, zygotene, pachytene, diplotene and diakinesis. Various key events, including homologous chromosome pairing, synapsis and recombination or crossover initially occur in the first period of the prophase (Andronico et al., 2019). In female mammals, meiosis has been shown to occur for a very long time. After the initiation of meiosis, oocytes are surrounded by a layer of granular cells that form the original follicles. When oocytes grow to more than 20 μm in size, granulosa cells become columnar and undergo mitosis to form multi-layered stratum granulosum (Sánchez and Smitz, 2012; Paci et al., 2018). Primordial follicles limit oocyte growth as well as the development of granulosa cells to seven layers. When oocytes progress from the diplotene phase, they enter a prolonged resting phase, referred to as the dictyate phase (Porras-Gómez and Moreno-Mendoza, 2017). Elevated cAMP levels in the oocyte determine meiosis arrest (Mehlmann, 2005). Oocytes activate Gs proteins through the G protein-coupled receptor 3 (GPR3) to stimulate cAMP production by AC (Mehlmann, 2005). cAMP-activated protein kinase A, promotes the phosphorylation (P) of cell cycle regulatory complex CDK1/cyclin B (CYB), leading to its inactivation. Through direct or indirect mechanisms, PKA leads to the phosphorylation and inactivation of CDC25b phosphatase (CDC25b-P) (Mehlmann, 2005). In addition, there is a possibility that PKA affects the activity of WEE1/MYT1 kinase. Phosphorylation of CDK1 by WEE1/MYT1 kinase leads to its inactivation, resulting in the failure to resume meiosis (Mehlmann, 2005). Elevated levels of LH hormone, produced by the pituitary, stimulate immature oocytes to resume meiosis (Liang et al., 1997; Rajkovic et al., 2004; Mehlmann, 2005; Porras-Gómez and Moreno-Mendoza, 2017). When oocytes complete the first meiosis phase and undergo cytoplasmic changes, they proceed to the metaphase II stage, at which time the oocyte mature (Figure 3).
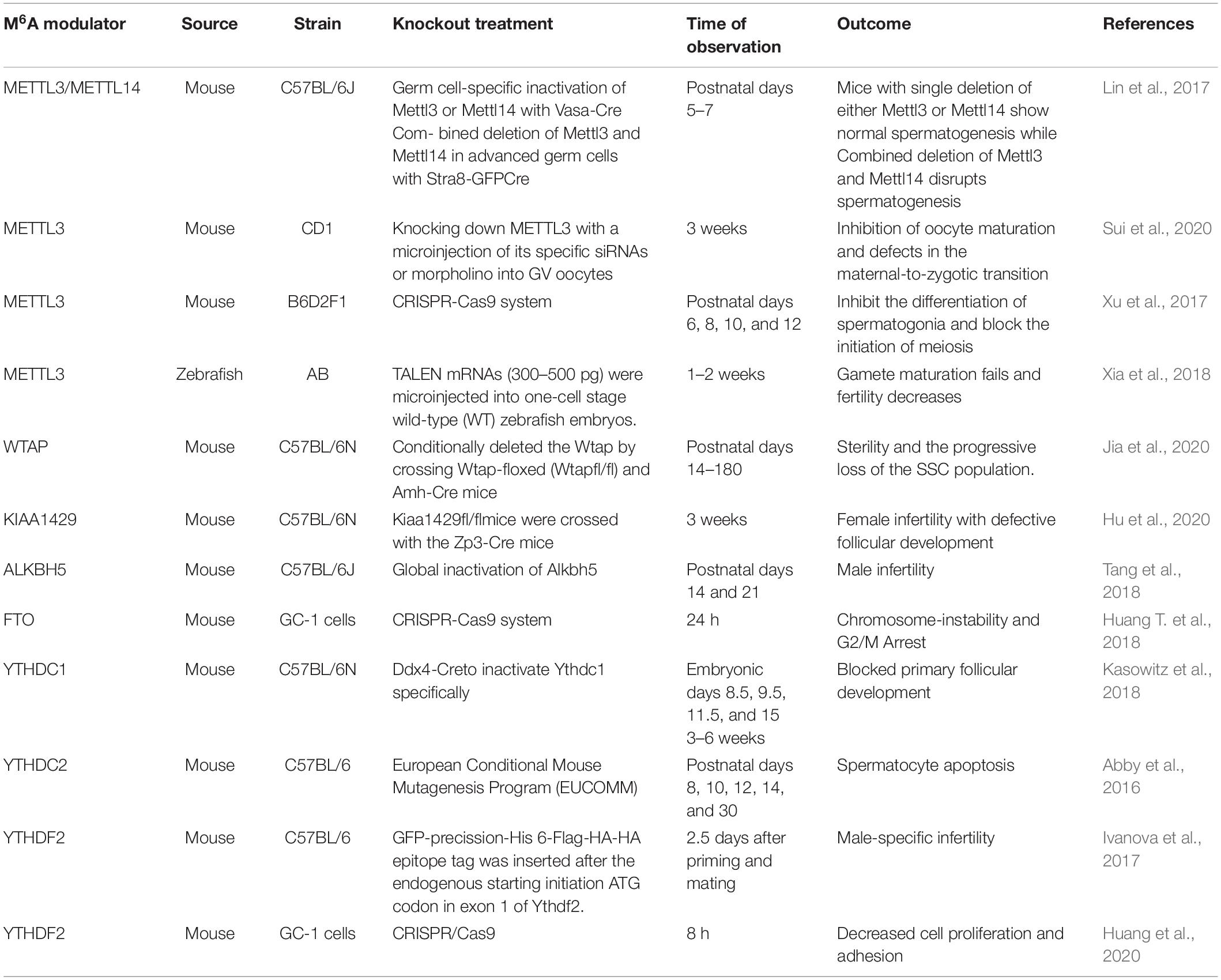
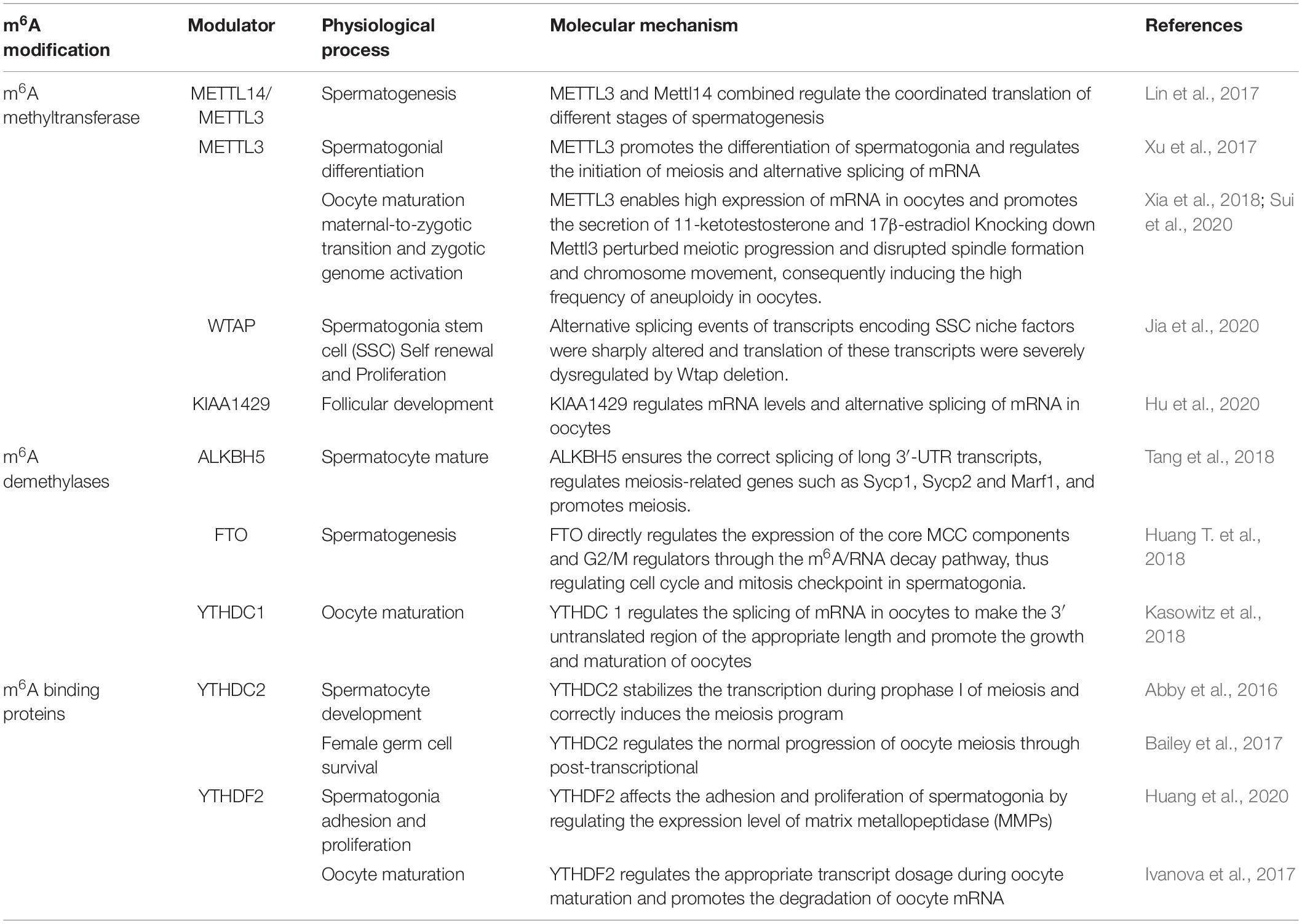
In this review, we summarized up to date knowledge on studies of infertility in mammals. Normal gametogenesis is a basic guarantee of fertility, therefore, we discussed the impact of gametogenesis disorders on infertility. Then, we elucidated on RNA methylation m6A modification and its functions as a whole, briefly introduced m6A methyl transferase (“author”), dimethyl enzyme (“eraser”) and binding protein (“reader”) (Figure 4). Then, we discussed the roles of methyltransferase, demethylase and binding proteins in regulating gametogenesis (Figure 5 and Table 1). Finally, we evaluated the mechanisms through which m6A is involved in regulation of post-transcriptional gene expression as well as its role in infertility.
RNA Methylation m6A Modification
Both DNA and histone proteins can control gene expression through dynamic reversible chemical modification. Similar to DNA and proteins, RNA, which form the center of the central dogma transmit genetic information. Chemical modification of RNA molecules is more diverse and abundant when compared to DNA. Apart from the four basic bases, there are more than 170 chemically modified nucleotides, which play various roles in many different types of cellular RNAs, including rRNA, tRNA and mRNA, and snRNAamong others (Cantara et al., 2011; Boccaletto et al., 2018; Xuan et al., 2018). Approximately two-thirds of the discovered chemical RNA modifications involve the addition of methyl groups, and these modifications are typically introduced in a post-transcriptional modification reaction. Co-transcriptional modifications are very few (Cantara et al., 2011).
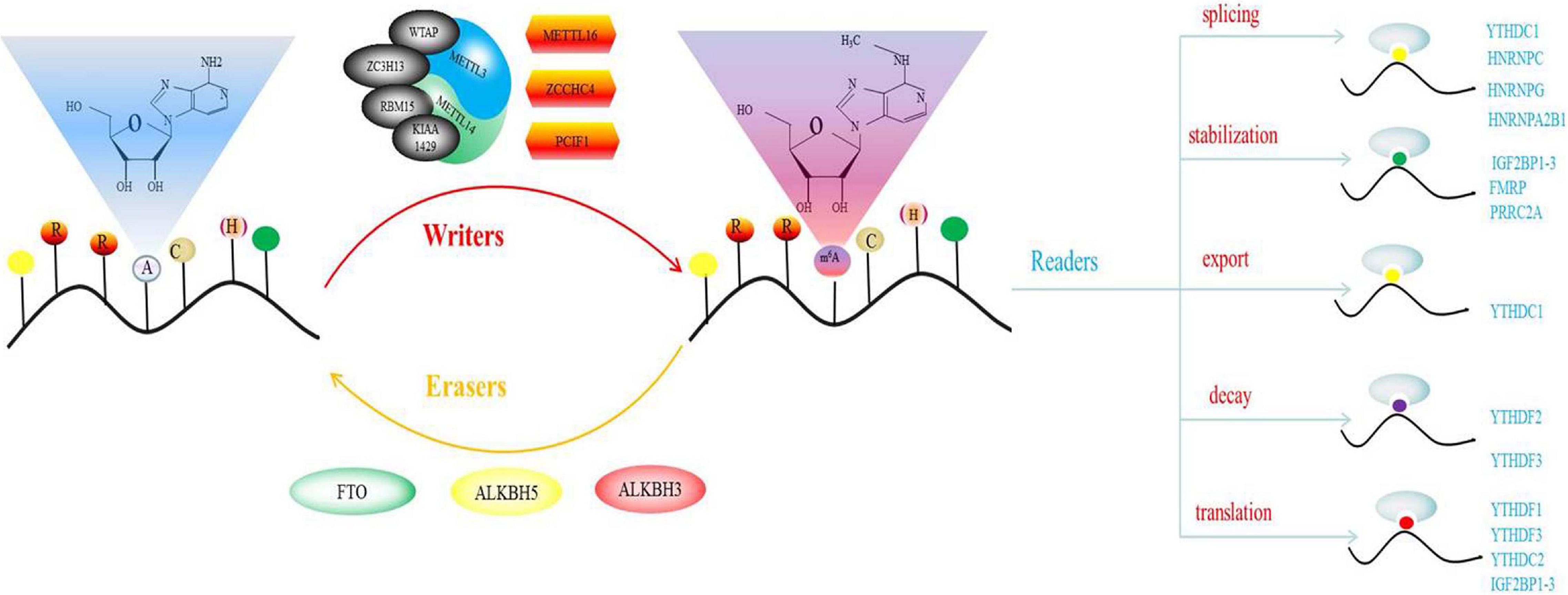
Among these modifications, N6-methyladenosine (m6A) exerts the most significant effect on gene expression, and is the first modification that has been shown to regulate mRNA abundance. N6-methyladenosine (M6A) modification was first described 40 years ago. M6A was first discovered in mouse L cell polyA-RNA, and was subsequently confirmed to be the most widely distributed and highest internal modification in mRNA (Schäfer, 1982). It was first detected in mRNA isolated from eukaryotes and in viral RNA replicating in the nucleus (Zhao and He, 2015). Methylation has also been shown to occur in the intron region of pre-mRNA, confirming that methylation occurs shortly after co-transcription or transcription (Roundtree et al., 2017a). m6A modification can modulate mRNA fate at splicing, stability, subcellular localization, output, decay, and transformation levels (Dominissini et al., 2012; Wang et al., 2015). m6A, which is ubiquitous in many transcripts, is uniquely and conservatively distributed. The consensus sequence, RRACH, has the highest frequency of occurrence in m6A, where R represents purine, A represents m6A site, while H represents a non-guanine base. However, the M6A on RRACH is not randomly distributed on the entire report card. It mainly appears in the coding sequence (CDS) of 3′-UTR, especially in the area near the stop codon (Perry et al., 1975; Wan et al., 2015). About 0.1–0.4% of adenosine residues in cellular mRNA are m6A. The average content of m6A is estimated to account for 3–5 residues in mammalian mRNA, 1–15 residues in RSV RNA, and 1.4–2.0 residues in Arabidopsis (Heck and Wilusz, 2019). M6A regulates RNA gene expression and metabolism through the action of methyltransferase (writer), demethylase (eraser) and m6A binding protein (reader). Demethylase is a proof that m6A modification is dynamically reversible.
The characteristics of these effectors in various biological systems emphasize the versatility and tunability of their functions, and confirms that the local environment is an important determinant of their biological effects (Yue et al., 2015). For example, reproductive functions of mammals have a very high probability of being affected by acquired dispositions, such as from the external environment, drugs and heavy metals. Therefore, these factors have an impact on epigenetic m6A modification, which in turn affect reproductive functions, leading to infertility (Figure 4).
M6A Methyltransferase
Methyltransferase complexes, which consists of two sub-complexes (MAC and MACOM) catalyzes m6A modification. MAC is the core component of MTC. It is composed of METTL3 and METTL14 and is a heterodimer with a size of 200 kDa (Liu et al., 2014; Wang et al., 2016). Both METTL3 and METTL14 have methyltransferase structures. METTL14 is degenerate compared to METTL3 and has important implications for the stability of METTL3 conformation and binding of the RNA substrate. METTL3 is the only subunit that requires the donor substrate, S-adenosylmethionine, to bind the catalytic site to exert its catalytic activity, which involves mRNA biogenesis, decay and translation (Śledź and Jinek, 2016; Choe et al., 2018). METTL3 and METTL14 are essential in properly maintaining m6A in the body. Abnormal expressions or mutations of METTL 3 and/or METTL 14 are associated with the development of various human diseases, including infertility. MACOM contains WTAP and its cofactor, KIAA1429. KIAA1429 plays an important role in maintaining the metabolic capacity of oocytes (Ping et al., 2014; Schwartz et al., 2014; Hu et al., 2020). HAKAI, RBM15 and ZC3H13 anchor the MTC to nuclear speckles and U-rich regions near the m6A site (Patil et al., 2016; Knuckles et al., 2018; Wen et al., 2018; Akichika et al., 2019). A new methyltransferase, PCIF1, has been shown to affect the methylation of adenosine N6 sites. However, PCIF1 is not involved in internal m6A methylation, it mainly catalyzes m6AM methylation of mRNA (Pendleton et al., 2017; Sun et al., 2019). Two other methyltransferase proteins, METTL16 and ZCCHC4 have also been reported. METTL16 can catalyze U6 splicing of RNA and m6A in some structural RNAs, while ZCCHC4 can mediate rRNA methylation in the AAC motif (Brown et al., 2016; Warda et al., 2017; Mendel et al., 2018; Ma et al., 2019).
M6A Demethylases
Fat-mass and obesity-associated protein was the first m6A demethylase to be discovered in 2011. As a result, the mechanisms and biological functions of m6A have been widely evaluated. The second m6A demethylase, ALKBH5, was discovered in 2013 (Zheng et al., 2013). These demethylases can exert a series of complex intermediate reactions to reverse the m6A methylation process of mRNA. The priority target of FTO is m6Am instead of m6A (Jia et al., 2011). Unlike FTO, knocking out ALKBH5 elevates m6A levels but not m6Am, implying that ALKBH5 selectively demethylates m6A, but has no affinity for m6Am. In addition, ALKBH5-deficient male mice were shown to exhibit abnormal fertilization characteristics. This was associated with abnormal sperm production and apoptosis in mouse testicles. Another m6A demethylase, ALKBH3, has been shown to be common in tRNA than in mRNA and in rRNA (Ueda et al., 2017). We believe that there are more m6A demethylases that are yet to be discovered.
M6A Binding Proteins
Discovery of the m6A binding protein provides a basis for studying the functions of m6A modification. Currently, two different types of m6A binding proteins have been characterized. This characterization is based on the mechanism of recognizing and binding RNA with m6A markers. From a structural aspect, binding proteins include YTH YTHDF1-3 and YTHDC1-2 (Meyer et al., 2015). In addition to the yth domain protein, eIF3 is also a binding protein for m6A, which promotes hat-independent translation when inducing cellular stress (Alarcón et al., 2015). There are two types of heteronuclear ribonucleoproteins; HNRPA2B1 and HNRNPC. In addition, IGF2BP1-3 was has also been reported (Liu et al., 2015; Huang H. et al., 2018). Other m6A binding proteins, such as ABCF1, FMRP, ELAV1, PRRC2A, and G3BPs have also been discovered (Wu et al., 2019). It has been shown that in pre-mRNA, m6A, YTHDC1, HNRNPC, HNRNPG, and HNRPA2B1 are involved in the regulation of mRNA splicing (Mauer et al., 2017; Zhou et al., 2019). Moreover, YTHDC1 can mediate the export of processed RNA from the nucleus to the cytoplasm (Roundtree et al., 2017b). YTHDC2 or YTHDF2, and YTHDF3 bind transcripts containing m6A, thereby causing it to be degraded in P bodies in the cytoplasm (Li et al., 2017). PRRC2A, FMRP and IGF2BP1/2/3 play the opposite role, and have been shown to be important in maintaining the stability of m6A modified transcripts (Wang et al., 2014). The combination of YTHDC2, YTHDF1 and m6A promotes mRNA translation. In addition, YTHDF3 and IGF2BP1/2/3 play the same role (Xiao et al., 2016; Hsu et al., 2017; Li et al., 2017; Shi et al., 2017; Zhao and He, 2017; Zhang et al., 2018).
M6A Modification and Gametogenesis Disorders
Sexual reproduction begins when parent gametes form functional gametes through meiosis, and ends with the formation of oosperms. In this extremely complex and highly precisely regulated process. Sperm production requires a high degree of coordination between transcription and conversion levels. Therefore, there are extremely complex regulatory procedures in this process, which ensure that spermatogenic cells at different developmental stages can correctly express specific gene sets (Lin et al., 2017). Spermatogenesis exhibits various features that are divided into different phases. First, the transcription program is turned on. The time at which the transcriptional program is started may be earlier when compared to the time at which the biological process is finally determined. The second phase involves the detection a higher level of alternative spliceosomes in spermatogenic cells. In the third phase, the transcriptional activity is significantly reduced in the early stages of meiosis I and stopped in the latter stages of spermiogenesis. Oocyte maturation is also a key step in sexual reproduction. It is directly related to the quality of oocytes and the subsequent reproductive processes (Mehlmann, 2005). Since transcriptional activities of oocyte DNA are inhibited during cell maturation, and the whole genome of the oocyte is only reactivated at the metaphase blastocyst stage, it is important to precisely regulate post-transcriptional levels of intracellular mRNA during oocyte maturation (Mehlmann, 2005). m6A plays an important role in different gametogenesis stages (Table 2 and Figure 6), however, it has not been established whether they are closely related. For example, a decrease in METTL3 affects the addition of m6A. In cases of decreased m6A, downstream YTHDC2 may not perform its normal mRNA translation functions, thereby affecting germ cell proliferation and differentiation. More studies should be performed to elucidate on these relations. Manipulation of specific m6A regulators maybe potential therapeutic targets. A limited number of studies have evaluated the role of m6A regulator inhibitors in infertility. These inhibitors may provide a scientific basis for targeted treatment of infertility.
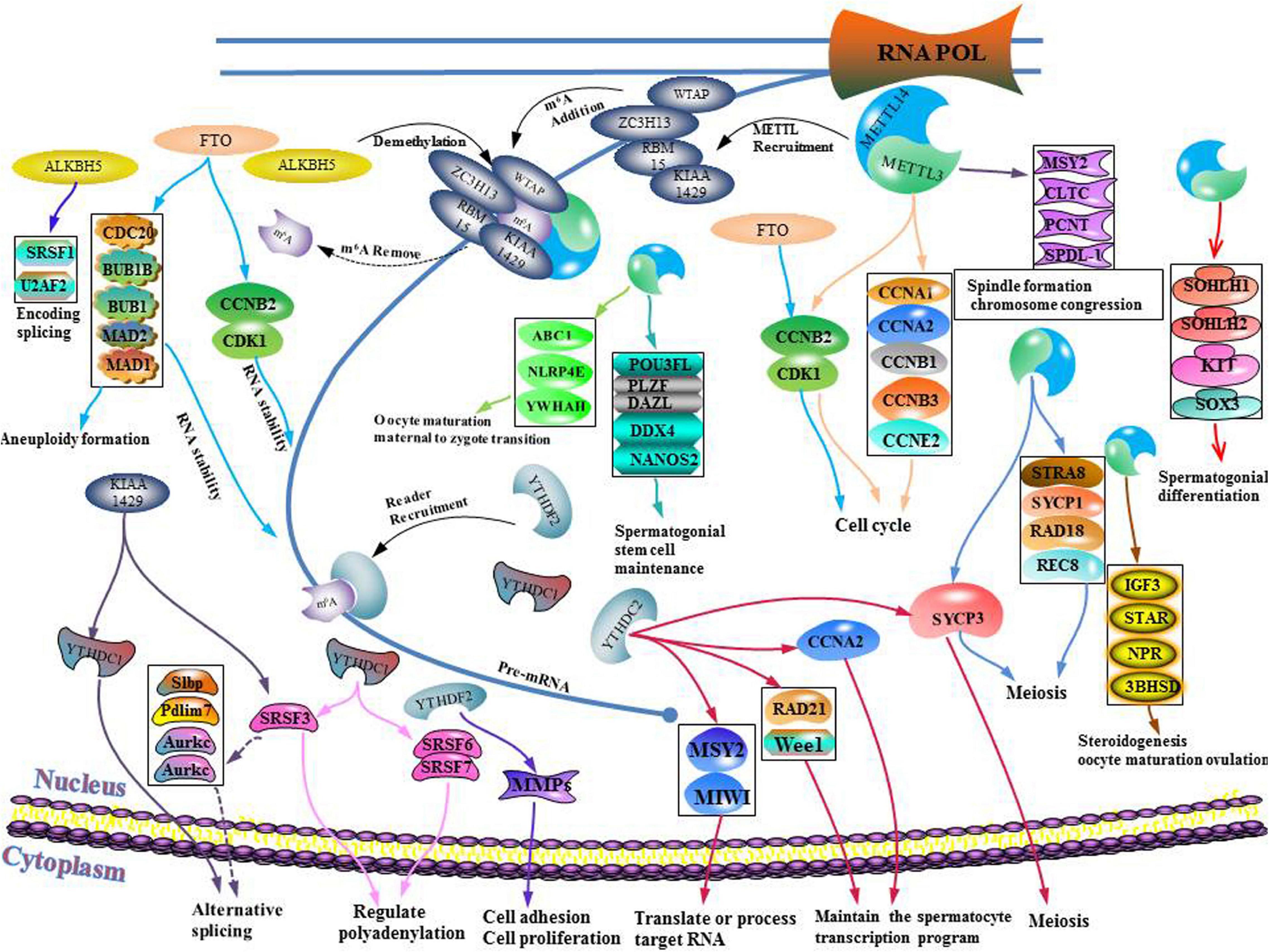
Figure 6. Schematic illustration of the molecular mechanism between m6A writer, eraser, binding protein, and gametogenesis disorder.
M6A Methyltransferase and Gametogenesis Disorders
The methyltransferases (METTL3 and METTL14) are modulators that regulate germ cells. After knocking out METTL3 or METTL14, it was found that m6A levels in undifferentiated spermatogonia were significantly suppressed (Lin et al., 2017). Moreover, undifferentiated spermatogonia in METTL3 or METL14 knockout testis were found to exhibit obvious translational disorders. Before the appearance of obvious morphological defects, mRNA levels of undifferentiated spermatogonia were found to have been altered. These findings indicate that m6A modification mediated by METTL3/METTL14 affects the fate of SSC (Lin et al., 2017). The knockout of either METTL3 or METTL14 genes by Stra8-GFPCre in mice did not exert any effect on spermatogenesis. Combined knockout of METTL3 and METTL14 in mice was shown to lead to normal meiosis, but abnormal spermatogenesis (Lin et al., 2017). These results imply that m6A requires the involvement of METTL3 and METTL14 to timely regulate the translation of methylation transcription and the production of proteins required for spermatogenesis (Lin et al., 2017).
As part of the methyl transferase complex, METTL3 was the first to be discovered. METTL3 was found to modulate neurological functions in mice and sex determination in Drosophila (Chen et al., 2018), and had an important impact on the pluripotency of stem cells and early embryonic development in mice (Aguilo et al., 2015; Geula et al., 2015; Haussmann et al., 2016). Moreover, it has an important effect on gamete maturation and fertility of female zebrafish (Xia et al., 2018). In their study, Sui et al. microinjected siRNA targeting METTL3 into GV oocytes to knock down METTL3. They found that meiotic maturation of mammalian oocytes is modulated by MEtTL3-mediated m6A (Sui et al., 2020). Spindle formation and chromosomal aggregation occur during the maturation of mouse oocytes. These processes require the expression of genes such as cltc, Msy2, Pcnt and Spdl-1. Western blot analysis revealed that their protein abundance decreased after METTL3 knockdown (Sui et al., 2020). After METTL3 knockdown, HPG incorporation experiments revealed that the translation efficiency of maternal mRNA in oocytes was suppressed, implying that oocyte maturation can be affected by mRNA translational efficiency. When the efficiency is low, oocyte maturation is inhibited, which leads to defects in mother-to-enzyme transition (Sui et al., 2020).
In addition, METTL3 has been shown to regulate the maturation process of oocytes by regulating the mRNA levels of oocytes. In METTL3 cKO zebrafish, oocytes were found to stagnate during early development while the follicular maturation rate was significantly low than that of normal zebrafish (Xia et al., 2018). Suppressed m6A modification levels in Zmettl3 m/m zebrafish oocytes lacking METTL3 was found to lead to abnormal expressions of key genes associated with sex hormone synthesis and gonadotropin signal transduction in zebrafish. In turn, levels of 11-ketotestosterone and 17-β-estradiol, secreted by offspring embryos, were found to have significantly decreased, which led to impaired gamete maturation disorders and weakened fertility (Xia et al., 2018).
Testicular spermatogonia of METTL3 cKO mice did not reach the pachytene phase of meiosis (Xu et al., 2017). Xu et al. proved that m6A mediated by METTL3 affects spermiogenesis, which is crucial to male fertility. They found that spermatogonia differentiation and spermatocyte meiosis processes during spermatogenesis are mediated by METTL3 (Xu et al., 2017). Transcriptome analysis and qRT-PCR analysis revealed that after METTL3 knockout, there was an alternative splicing dysregulation that was not conducive for spermatogenesis (Xu et al., 2017). Four types of genes related to spermatogenesis were identified the METTL3 cKO testis. These genes regulate cell cycles, sperm stem cell maintenance, spermatogonia differentiation and spermatocyte meiosis, respectively (Xu et al., 2017). They were significantly down-regulated after METTL3 knockout. Genes that regulate the cell cycle include Ccna1, Cdk1, and RAD51 among others; genes involved in maintenance of spermatogonial stem cells include Dazl, Plzf and Bcl6 among others; genes associated with differentiation of spermatogonia include Sohlh1, Kit and Sox3 among others; genes involved in meiosis include Stra8, Sycp3, and Smc1b. After METTL3 knockout, initial spermatogonia differentiation and spermatocyte meiosis were found to be severely blocked. Therefore, METTL3 significantly affects sperm production from undifferentiated sperms. This outcome is attributed to significant suppression of the abundance of STRA8-positive cells and synaptic complex protein SYCP3 levels (Xu et al., 2017).
A highly effective and selective METTL3 inhibitor, STM2457, has been reported. This small molecule inhibitor is highly specific for METTL3, but has no inhibitory effects on other RNA methyltransferases (Yankova et al., 2021). Various chemical and biological molecules that can affect METTL3 have also been identified. For example, cigarette smoke condensates induce METTL3 overexpression. The intestinal microbial metabolite, butyrate, promotes the downregulation of METTL3. Several micro-RNAs, including miR-186, miR-4429, and miR-600 have also been shown to target METL3 mRNA, inhibiting its expression (Zeng et al., 2020).
Wilms tumor 1-associated protein is a subunit of m6A-METTL related complexes. During spermatogenesis, its gene was found to be significantly expressed in mouse Sertoli cells. In Sertoli cells, WTAP was found to significantly affect SSC proliferation and self-renewal (Jia et al., 2020). By conditionally eliminating WTAP in supporting cells, it was found that SSC was progressively lost and eventually led to infertility (Jia et al., 2020). WTAP knockout was also found to lead to progressive loss of SSC and depletion of germ cells, which reduced sperm concentrations in mice, thereby contributing to the sterile phenotype of mice. Moreover, by making WTAP ineffective, expression levels of GDNF, which is essential for maintaining the SCC pool, were also found to be down-regulated, therefore, sertoli cells cannot maintain the SSC pool, leading to SSC depletion. In addition, transcripts associated with SCC maintenance and spermatogonia differentiation in sertoli cells are severely affected by gene transcription and translation changes (Jia et al., 2020). Knockout of WTAP was found to alter splicing events in sertoli cells, with a large number of abnormal splicing events occurring in m6A-rich genes. If the WTAP function is lost, splicing events in genes that regulate spermatogenesis are altered, which eventually affects spermatogenesis. The WTAP-mediated m6A regulates mRNA transcription and translation, thereby coordinating the expression of essential genes, so that SSC maintenance of sertoli cells and differentiation of spermatogonia occur normally (Jia et al., 2020).
The downregulation and overexpression of METTL3 protein leads to upregulation of WTAP. In particular, METTL3 levels regulate the expression of WTAP at multiple levels through direct and indirect mechanisms, including mRNA translation and stability (Sorci et al., 2018). Therefore, the dynamic balance of WTAP expression is indirectly maintained through METTL3 to ensure normal progression of SSC proliferation and self-renewal, which is essential for preventive treatment of spermatogenesis disorders.
Virus-like m6A methyltransferase associated protein is also an important component of m6A-METTL related complex. It has been shown to mediate the deposition of m6A in the 3′UTR and the region close to the stop codon. It also affects the alternating polyadenylation in HeLa cells. KIAA1429 has been found to be highly expressed in mouse oocytes, implying that it plays a specific function in mouse oocytes (Hu et al., 2020). Hu et al. (2020) found that the expression of KIAA1429 mRNA in GV oocytes is dominant, and that the role of KIAA1429 during oocyte growth is essential for female fertility. HE staining and TUNEL analysis confirmed that the expression of KIAA1429 is essential for folliculogenesis. The loss of KIAA1429 function in oocytes is associated with follicular growth defects, leading to abnormal proliferation and apoptosis of granulosa cells (Hu et al., 2020). KIAA1429cKO mice were found to exhibit multiple oocyte follicles. In addition, compared to the control group mice, there were no significant differences in the number of follicles at different stages. KIAA1429cKO female mice have abnormal follicular morphologies. In preovulatory follicles, abnormal RNA granules were found in the cytoplasm of KIAA1429cKO oocytes, and the GV oocytes were much smaller than those of the control group (Hu et al., 2020). Moreover, KIAA1429 is essential for oocyte competence in mediating chromatin configuration and for GV oocyte RNA metabolism (Hu et al., 2020). Srsf3 mediates Pdlim7 exon inclusion bodies to maintain proper GVBD during meiosis in mouse oocytes (Hu et al., 2020). KIAA1429 may also affect the location of YTHDC1 through Srsf3 to regulate alternative mRNA splicing. Therefore, it is involved in post-transcriptional regulation, promotion of follicular development and in ensuring fertility in women.
M6A Demethylases and Gametogenesis Disorder
Fat-mass and obesity-associated protein is a member of the α-ketoglutaric acid dependent dioxygenase alkb family. It is encoded by fat and obesity-related genes located on human chromosome 16 (Huang T. et al., 2018). The reduction in semen quality among azoospermia patients has been attributed to the occurrence of two missense mutations in FTO. Huang et al. found that by knocking out FTO, the expression levels of MCC in spermatogonia were upregulated, thereby inhibiting chromosomal segregation and inducing aneuploidy. Moreover, the expression of CDK1 and CCNB2 were found to be significantly downregulated in FTO-KO cells. FTO was shown to regulate G2/M transition under the influence of the Cdk1/Ccnb2 complex, and expression levels of CDK1 and CCNB2 were significantly downregulated in FTOcKO cells, implying that FTO knockout can cause chromosomal instability in mouse spermatogonia and arrest the G2/M transition (Huang T. et al., 2018).
Several inhibitors of FTO have been identified targets for Rhein was the first effective FTO inhibitor to be identified. However, its selectivity is not strong and it also exhibits a certain inhibitory activity against ALKBH5. Entacapone has also been identified as a potential FTO inhibitor. As a drug for the treatment of Parkinson’s disease, it may have a certain effect on metabolic disorders (Zhao et al., 2020). Studies on m6A target inhibitors in infertility are limited. Some FTO inhibitors may have potential therapeutic effects on infertility. For example, Meclofenamic acid (MA) is a non-steroidal anti-inflammatory drug that is considered to be a highly selective FTO inhibitor. As an ethyl ester MA derivative, MA-2 can effectively inhibit the growth of glioblastoma stem cells (GSC) and tumor formation induced by GSC. It also has a significant inhibitory effect on the expression of FTO (Zhao et al., 2020).
Alk B homolog 5 has been found to be the second M6A demethylase with the highest expression levels in mouse testis. Its deletion was found to significantly elevate m6A levels in testicular cells. Male mice lacking ALKBH5 exhibit abnormal spermatogenesis, leading to spermatogonia apoptosis and formation of abnormal sperms, which are not fertile. Meiosis and haploid phases of spermatogenesis were found to be significantly affected by ALKBH5 in cKO mouse (Mauer et al., 2017). The 3′-UTR mRNA length affects the enrichment and levels of m6A. A longer 3′-UTR mRNA has elevated m6A levels than the shorter 3′-UTR mRNA. In the longer 3′-UTR mRNA, M6A is enriched in the 3′-UTR region near the stop codon. ALKBH5 knockout leads to abnormal splicing of the genes that encode splicing factors such as Khdrbs3, Sfswap, Snrnp70, Srsf1 and U2af2. Due to abnormal splicing of these genes, more target gene splicing errors occur, then, a vicious cycle of abnormal splicing occurs, amplifying the initial adverse effects. In mature spermatocytes and round spermatocytes, deletion of ALKBH5 can lead to abnormal splicing of long 3′-UTR mRNA while the short mRNA is produced with increasing m6A levels, indicating that the correct splicing of long 3′-UTR mRNA may require ALKBH5-mediated m6A demethylation (Tang et al., 2018). The m6A of Alkbh5 cKO spermatocytes cannot be demethylated while various genes, such as ycp1, Sycp2 and Marf1, which are essential for meiosis are dysregulated, leading to meiotic defects.
M6A Binding Proteins and Gametogenesis Disorder
YTH domain-containing 1 has a significant regulatory effect on the development of spermatogonia and oocyte growth and maturation. In oocytes, the failure of YTHDC1 blocks the primary follicular phase. Kasowitz et al. found that 3′-terminal pro-mRNA processing factors (CPSF6, Srsf3, and Srsf7) are closely associated with YTHDC1, a key nuclear factor in the processing of precursor mRNA transcripts. Knockout of YTHDC1 changes the length of the 3′UTR. This is because, extensive selective polyadenylation occurs in oocytes, leading to alternative splicing defects in oocytes.
GO analysis showed that YTHDC2 in mouse testes is highly expressed. In vitro pulldown assays proved that YTHDC2 preferentially binds m6A (Abby et al., 2016). After YTHDC2 knockout, both male and female mice exhibited infertility, implying that YTHDC2 is essential for fertility. In YTHDC2cKO mice, germ cells could not develop to the zygotene stage of meiotic prophase I (Abby et al., 2016). YTHDC2cKO mice have normal spermatogonia and sertoli cells, indicating that YTHDC2cKO mice only exhibit spermatogenesis defects at the spermatocyte stage (Abby et al., 2016). To stabilize transcription in the prophase I of meiosis, and to ensure that meiosis is correctly induced, YTHDC2 interacts with MEIOC in an RNA-independent manner. Therefore, YTHDC2 is necessary for normal development of spermatocytes (Abby et al., 2016). YTHDC2 interacts with RNA particles and post-transcriptionally regulates the meiotic process of germ cells by binding Ccna2, other mitotic transcripts and specific piRNA precursors (Bailey et al., 2017). Adult females with YTHDC2cKO were also found to have thin uterine walls and small ovaries, without developed follicles in the ovaries, implying that YTHDC2 knockout in female mice leads to infertility (Bailey et al., 2017). Moreover, YTHDC2cKO female mice were found to have much less DDX4 and germ cells in the ovaries at birth (P0.5), no primordial follicles were detected in the ovaries at postnatal 5 days (P5), and no germ-vesicles were detected on P21. These findings suggest that YTHDC2 is essential for female germ cell survival during the embryonic period (Bailey et al., 2017). After YTHDC2 knockout, germ cells were found to exhibit a nuclear morphology that is similar to that of premeiotic cells without chromosomal changes in the prophase stage of meiosis. Without YTHDC2 regulation, female germ cells cannot correctly reach the pre-meiotic phase in the fetal ovary (Bailey et al., 2017).
YTHDF2 is also involved in degrading target mRNA. YTHDF2 is differentially expressed in tissues, with the testis exhibiting the highest expression levels, especially during spermatogenesis. YTHDF2cKO male mice have been shown to be fertile with normal seminiferous tubule histologies. After YTHDF2 knockout, MMPs were found to be downregulated, affecting cell adhesion and proliferation. Edu analysis revealed that the positive rate of Edu in the Wild type (WT) group was significantly higher than that in the YTHDF2cKO group, implying that proliferation of spermatogonia is inhibited after YTHDF2 knockout. Morphologically, wild-type spermatogonia are polygons while YTHDF2cKO spermatogonia are fusiform or round. Cell-adhesion assays revealed that YTHDF2 has a significant effect on the adhesion and spread of spermatogonia (Huang et al., 2020).
YTHDF2 has the ability to regulate oocytes to reach metaphase stage of meiosis II. During early zygotic development, these oocytes can remain in the metaphase stage of meiosis II. Large amounts of YTHDF2 in GV and MII oocytes imply that it is also expressed during oocyte maturation, spermatogenesis or follicular development. YTHDF2 has been detected in the cytoplasms of germ cells and somatic cells (Ivanova et al., 2017). When the corpus luteum is found in the ovary of YTHDF2cKO female mice, it indicates that ovulation has taken place and that that woman is infertile. Therefore, YTHDF2 knockout leads to female-specific infertility (Ivanova et al., 2017). By producing YTHDF2 maternal conditional deletion (mCKO) oocytes, without affecting the expression of YTHDF2 in somatic granulosa cells, YTHDF2 has been proven to be indispensable for women to have children. MII oocyte analysis showed that after YTHDF2 knockout, oocyte growth or the formation of maternal transcriptome was not seriously affected. In the process of oocyte maturation, YTHDF2 is needed to inform the appropriate transcript dosage (Ivanova et al., 2017).
Discussion
With developments in methylation modifications of RNA, studies on RNA methylation modification have gradually developed from relatively concentrated functional research to studies on disease. To maintain generations, the harm caused by reproductive health and its influencing factors to humans should be urgently resolved. RNA methylation modification is an important epigenetic phenomenon. M6A modification and its methyltransferase and demethylase, binding protein have an extremely important impact on gametogenesis.
We have discussed the role and mechanism of RNA methylation modification in infertility. The mechanism involves the cooperation of METTL3 and METTL14 to control timely translation of methylation transcription, thereby, appropriately regulating the production of proteins that are necessary for spermatogenesis. METTL3 regulates the differentiation of spermatogonia and plays an important regulatory role in oocyte maturation. KIAA1429 plays a vital role in follicle formation and has a regulatory on normal spermatogenesis. The correct regulatory mechanism of YTHDC2 on meiosis in gamete development has not been clearly elucidated. YTHDF2 regulates spermatogonia proliferation and has a role in the maturation of oocytes.
Secondly, by identifying specific m6A regulators, potential preventive and treatment methods for infertility can explored. Proteins for m6A regulation can be used as biomarkers to reflect gametogenesis disorders in both sexes and to achieve preventive treatment of infertility. Precise treatment of gene targets such as METL3, WTAP, KIAA1429, FTO, ALKBH5, YTHDC1, YTHDC2, and YTHDF2, by regulating m6A modification ensures normal gametogenesis and exerts a better therapeutic effect on infertility. Entacapone, MA drugs, STM2457, butyrate, miR-186, miR-4429, miR-600 and let-7G, Rhein, MA-2 inhibitors, as well as folate, betaine and methionine methylation donors can be used in the treatment plans for gametogenesis disorders.
RNA methylation can prevent and treat infertility by regulating gametogenesis. This discovery has important research significance in theory and practice. However, the mechanisms involved have not been clearly established. For example, it has not been determined whether the specific molecular mechanism that causes infertility is associated with other diseases. Moreover, it has not been determined whether other RNA methylation modification proteins affect gametogenesis. To comprehensively understand the regulatory mechanisms of RNA methylation on infertility, studies should determine how RNA methylation affects gametogenesis under the stimulation of the external environment, drugs and poisons.
Author Contributions
XL, HW, BL, ZQ, JL, BX, WL, and ZX: revising the manuscript. YD: conception and design, revising it critically for important intellectual content, and final approval of the version to be published. All authors substantially contributed to the review and read and approved the final manuscript.
Funding
This work was supported by the Natural Science Foundation of Liaoning Province (2020-MS-152), the Basic Research Fund of Young Program of Higher Education of Liaoning Province (grant no. QNK201735), the National Natural Science Foundation of China (grant no. 81302406), and the Funds for Distinguished Young Scientists in School of Public Health, China Medical University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to acknowledge Minghui Li, Xianchao Du, Lei Zhang, and Shuang Wang for their valuable advices.
Abbreviations
WHO, World Health Organization; PGCs, Primordial germ cells; BMP, bone morphogenetic protein; TNAP, tissue non-specific alkaline phosphatase; SSEA1, stage-specific embryonic antigen 1; DPPA3 or STELLA, developmental pluripotency type 3 protein; SRY, sex determining region-Y; Sox2, sex determining region-Y -box2l; SDF-1, stromal cell-derived factor 1; CXCR4, C-X -C motif chemokine receptor 4; SSCs, spermatogonial stem cells; As, single spermatogonia; Ap, A-paired spermatogonia; Aal, A aligned spermatogonia; Gfr α 1, glial cell line-derived neurotrophic receptor α 1; Nanos2, anos homolog 2; Plzf, Zbtb16; Id4, inhibitor of differentiation; Ngn3, Neurog 3; RA, retinoic acid; Hist1 cluster, replication-de pendent core histone genes; FF, follicular flfluid; AC, adenylate cyclase; PKA cAMP activates protein kinase A; CYB, cyclin B; LH, luteinizing hormone; rRNA, ribosomal RNA; tRNA, transfer RNA; mRNA, messenger RNA; snRNA, small nuclear RNA; m6A, N6-methyladenosine; polyA-RNA, polyadenylated RNA; RSV, Rous Sarcoma virus; CDS, coding sequences; 3 ′ -UTRs, 3 ′ -untranslated regions; MTC, methyltransferase complexes; MACOM, m6A-METTL related complex; METTL3, methyltransferase-like protein 3; METTL14, methyltransferase-like protein 14; SAM, S-adenosylmethionine; WTAP, Wilms tumor 1-associated protein; KIAA1429, virus-like m6A methyltransferase associated protein; HAKAI, Cbl proto-oncogene E3 ubiquitin protein ligase-like1; RBM15, RNA binding motif protein 15; ZC3H13, zinc finger CCCH domain-containing protein 13; PCIF1, Phosphorylated CTD Interacting Factor 1; m6Am, N6-2-O-dimethyladenosine; METTL16, methyltransferase-like protein 16; ZCCHC4, CCCH domain-containing protein 4; FTO, Fat-mass and obesity-associated protein; ALKBH5, Alk B homolog 5; ALKBH3, Alk B homolog 3; YTHDF1-3, YTH N6-methyladenosine RNA binding protein 1-3; YTHDC1-2, YTH domain-containing 1-2; eIF3, eukaryotic initiation factor 3; HNRNP, heterogeneous nuclear ribonucleoprotein; HNRNPC, heterogeneous nuclear ribonucleoproteins C; IGF2BP1-3, insulin-like growth factor 2 mRNA-binding proteins 1-3; ABCF1, ATP binding cassette subfamily F member 1; FMRP, fragile X mental retardation protein; ELAV1, ELAV-like protein 1; PRRC2A, proline rich coiled-coil 2A; AS, alternative splicing; Zmettl3 m/m, zygotic deficiency mutant lines; APA, alternative polyadenylation; GV, germinal vesicle; GVBD, germinal vesicle breakdown; MCC, mitotic checkpoint complex; MMPs, matrix metallopeptidase.
References
Abby, E., Tourpin, S., Ribeiro, J., Daniel, K., Messiaen, S., Moison, D., et al. (2016). Implementation of meiosis prophase I programme requires a conserved retinoid-independent stabilizer of meiotic transcripts. Nat. Commun. 7:10324. doi: 10.1038/ncomms10324
Aguilo, F., Zhang, F., Sancho, A., Fidalgo, M., Di Cecilia, S., Vashisht, A., et al. (2015). Coordination of m(6)A mRNA Methylation and Gene Transcription by ZFP217 Regulates Pluripotency and Reprogramming. Cell Stem Cell 17, 689–704. doi: 10.1016/j.stem.2015.09.005
Ahmed, E. A., and de Rooij, D. G. (2009). Staging of mouse seminiferous tubule cross-sections. Methods Mol. Biol. 558, 263–277. doi: 10.1007/978-1-60761-103-5_16
Akichika, S., Hirano, S., Shichino, Y., Suzuki, T., Nishimasu, H., Ishitani, R., et al. (2019). Cap-specific terminal N (6)-methylation of RNA by an RNA polymerase II-associated methyltransferase. Science 363:eaav0080. doi: 10.1126/science.aav0080
Alarcón, C. R., Goodarzi, H., Lee, H., Liu, X., Tavazoie, S., and Tavazoie, S. F. (2015). HNRNPA2B1 is a mediator of m(6)A-dependent nuclear RNA processing events. Cell 162, 1299–1308. doi: 10.1016/j.cell.2015.08.011
Anderson, R., Copeland, T. K., Schöler, H., Heasman, J., and Wylie, C. (2000). The onset of germ cell migration in the mouse embryo. Mech. Dev. 91, 61–68. doi: 10.1016/s0925-4773(99)00271-3
Andronico, F., Battaglia, R., Ragusa, M., Barbagallo, D., Purrello, M., and Di Pietro, C. (2019). Extracellular vesicles in human oogenesis and implantation. Int. J. Mol. Sci. 20:2162. doi: 10.3390/ijms20092162
Avilion, A. A., Nicolis, S. K., Pevny, L. H., Perez, L., Vivian, N., and Lovell-Badge, R. (2003). Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 17, 126–140. doi: 10.1101/gad.224503
Bailey, A. S., Batista, P. J., Gold, R. S., Chen, Y. G., de Rooij, D. G., Chang, H. Y., et al. (2017). The conserved RNA helicase YTHDC2 regulates the transition from proliferation to differentiation in the germline. Elife 6:26116. doi: 10.7554/eLife.26116
Barroca, V., Lassalle, B., Coureuil, M., Louis, J. P., Le Page, F., Testart, J., et al. (2009). Mouse differentiating spermatogonia can generate germinal stem cells in vivo. Nat. Cell. Biol. 11, 190–196. doi: 10.1038/ncb1826
Battle, D. E. (2013). Diagnostic and Statistical Manual of mental disorders (DSM). Codas 25, 191–192. doi: 10.1590/s2317-17822013000200017
Boccaletto, P., Machnicka, M. A., Purta, E., Piatkowski, P., Baginski, B., Wirecki, T. K., et al. (2018). MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 46, D303–D307. doi: 10.1093/nar/gkx1030
Bowles, J., and Koopman, P. (2010). Sex determination in mammalian germ cells: extrinsic versus intrinsic factors. Reproduction 139, 943–958. doi: 10.1530/rep-10-0075
Brown, J. A., Kinzig, C. G., DeGregorio, S. J., and Steitz, J. A. (2016). Methyltransferase-like protein 16 binds the 3’-terminal triple helix of MALAT1 long noncoding RNA. Proc. Natl. Acad. Sci. U S A. 113, 14013–14018. doi: 10.1073/pnas.1614759113
Cantara, W. A., Crain, P. F., Rozenski, J., McCloskey, J. A., Harris, K. A., Zhang, X., et al. (2011). The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Res. 39, 195–201. doi: 10.1093/nar/gkq1028
Carreau, S., and Hess, R. A. (2010). Oestrogens and spermatogenesis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 1517–1535. doi: 10.1098/rstb.2009.0235
Chambers, I., Silva, J., Colby, D., Nichols, J., Nijmeijer, B., Robertson, M., et al. (2007). Nanog safeguards pluripotency and mediates germline development. Nature 450, 1230–1234. doi: 10.1038/nature06403
Chen, Y., Zheng, Y., Gao, Y., Lin, Z., Yang, S., Wang, T., et al. (2018). Single-cell RNA-seq uncovers dynamic processes and critical regulators in mouse spermatogenesis. Cell. Res. 28, 879–896. doi: 10.1038/s41422-018-0074-y
Chiquoine, A. D. (1954). The identification, origin, and migration of the primordial germ cells in the mouse embryo. Anat. Rec. 118, 135–146. doi: 10.1002/ar.1091180202
Choe, J., Lin, S., Zhang, W., Liu, Q., Wang, L., Ramirez-Moya, J., et al. (2018). mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature 561, 556–560. doi: 10.1038/s41586-018-0538-8
Clermont, Y. (1972). Kinetics of spermatogenesis in mammals: seminiferous epithelium cycle and spermatogonial renewal. Physiol. Rev. 52, 198–236. doi: 10.1152/physrev.1972.52.1.198
Czerwoniec, A., Dunin-Horkawicz, S., Purta, E., Kaminska, K. H., Kasprzak, J. M., Bujnicki, J. M., et al. (2009). MODOMICS: a database of RNA modification pathways. 2008 update. Nucleic Acids Res. 37, 118–121. doi: 10.1093/nar/gkn710
de Rooij, D. G. (2001). Proliferation and differentiation of spermatogonial stem cells. Reproduction 121, 347–354. doi: 10.1530/rep.0.1210347
de Sousa Lopes, S. M., Roelen, B. A., Monteiro, R. M., Emmens, R., Lin, H. Y., Li, E., et al. (2004). BMP signaling mediated by ALK2 in the visceral endoderm is necessary for the generation of primordial germ cells in the mouse embryo. Genes Dev. 18, 1838–1849. doi: 10.1101/gad.294004
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206. doi: 10.1038/nature11112
Ewen, K., Baker, M., Wilhelm, D., Aitken, R. J., and Koopman, P. (2009). Global survey of protein expression during gonadal sex determination in mice. Mol. Cell Proteomics 8, 2624–2641. doi: 10.1074/mcp.M900108-MCP200
Geula, S., Moshitch-Moshkovitz, S., Dominissini, D., Mansour, A. A., Kol, N., Salmon-Divon, M., et al. (2015). Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 347, 1002–1006. doi: 10.1126/science.1261417
Ginsburg, M., Snow, M. H., and McLaren, A. (1990). Primordial germ cells in the mouse embryo during gastrulation. Development 110, 521–528. doi: 10.1242/dev.110.2.521
Griswold, M. D. (2016). Spermatogenesis: The commitment to meiosis. Physiol. Rev. 96, 1–17. doi: 10.1152/physrev.00013.2015
Gu, Y., Runyan, C., Shoemaker, A., Surani, A., and Wylie, C. (2009). Steel factor controls primordial germ cell survival and motility from the time of their specification in the allantois, and provides a continuous niche throughout their migration. Development 136, 1295–1303. doi: 10.1242/dev.030619
Handel, M. A., and Schimenti, J. C. (2010). Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat. Rev. Genet. 11, 124–136. doi: 10.1038/nrg2723
Haussmann, I. U., Bodi, Z., Sanchez-Moran, E., Mongan, N. P., Archer, N., Fray, R. G., et al. (2016). m(6)A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature 540, 301–304. doi: 10.1038/nature20577
Heck, A. M., and Wilusz, C. J. (2019). Small changes, big implications: The impact of m(6)A RNA methylation on gene expression in pluripotency and development. Biochim. Biophys. Acta Gene. Regul. Mech. 1862:194402. doi: 10.1016/j.bbagrm.2019.07.003
Hsu, P. J., Zhu, Y., Ma, H., Guo, Y., Shi, X., Liu, Y., et al. (2017). Ythdc2 is an N(6)-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell. Res. 27, 1115–1127. doi: 10.1038/cr.2017.99
Hu, Y., Ouyang, Z., Sui, X., Qi, M., Li, M., He, Y., et al. (2020). Oocyte competence is maintained by m(6)A methyltransferase KIAA1429-mediated RNA metabolism during mouse follicular development. Cell Death Differ. 27, 2468–2483. doi: 10.1038/s41418-020-0516-1
Huang, H., Weng, H., Sun, W., Qin, X., Shi, H., Wu, H., et al. (2018). Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 20, 285–295. doi: 10.1038/s41556-018-0045-z
Huang, T., Gao, Q., Feng, T., Zheng, Y., Guo, J., and Zeng, W. (2018). FTO knockout causes chromosome instability and G2/M arrest in mouse GC-1 cells. Front. Genet. 9:732. doi: 10.3389/fgene.2018.00732
Huang, T., Liu, Z., Zheng, Y., Feng, T., Gao, Q., and Zeng, W. (2020). YTHDF2 promotes spermagonial adhesion through modulating MMPs decay via m(6)A/mRNA pathway. Cell Death Dis. 11:37. doi: 10.1038/s41419-020-2235-4
Huckins, C. (1971). The spermatogonial stem cell population in adult rats. I. Their morphology, proliferation and maturation. Anat. Rec. 169, 533–557. doi: 10.1002/ar.1091690306
Irie, N., Weinberger, L., Tang, W. W., Kobayashi, T., Viukov, S., Manor, Y. S., et al. (2015). SOX17 is a critical specifier of human primordial germ cell fate. Cell 160, 253–268. doi: 10.1016/j.cell.2014.12.013
Ivanova, I., Much, C., Di Giacomo, M., Azzi, C., Morgan, M., Moreira, P. N., et al. (2017). The RNA m(6)A reader YTHDF2 is essential for the post-transcriptional regulation of the maternal transcriptome and Oocyte Competence. Mol. Cell. 67, 1059–1067. doi: 10.1016/j.molcel.2017.08.003
Jia, G., Fu, Y., Zhao, X., Dai, Q., Zheng, G., Yang, Y., et al. (2011). N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887. doi: 10.1038/nchembio.687
Jia, G. X., Lin, Z., Yan, R. G., Wang, G. W., Zhang, X. N., Li, C., et al. (2020). WTAP function in Sertoli Cells is essential for sustaining the spermatogonial stem cell Niche. Stem Cell Rep. 15, 968–982. doi: 10.1016/j.stemcr.2020.09.001
Kasowitz, S. D., Ma, J., Anderson, S. J., Leu, N. A., Xu, Y., Gregory, B. D., et al. (2018). Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development. PLoS Genet. 14:e1007412. doi: 10.1371/journal.pgen.1007412
Kayode, O. T., Rotimi, D. E., Kayode, A. A. A., Olaolu, T. D., and Adeyemi, O. S. (2020). Monosodium Glutamate (MSG)-induced male reproductive dysfunction: A mini review. Toxics 8:7. doi: 10.3390/toxics8010007
Knuckles, P., Lence, T., Haussmann, I. U., Jacob, D., Kreim, N., Carl, S. H., et al. (2018). Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m(6)A machinery component Wtap/Fl(2)d. Genes. Dev. 32, 415–429. doi: 10.1101/gad.309146.117
Kojima, Y., Sasaki, K., Yokobayashi, S., Sakai, Y., Nakamura, T., Yabuta, Y., et al. (2017). Evolutionarily distinctive transcriptional and signaling programs drive human germ cell lineage specification from pluripotent stem cells. Cell Stem Cell 21, 517–532. doi: 10.1016/j.stem.2017.09.005
La Vignera, S., Vicari, E., Condorelli, R. A., D’Agata, R., and Calogero, A. E. (2011). Male accessory gland infection and sperm parameters (review). Int. J. Androl. 34, e330–e347. doi: 10.1111/j.1365-2605.2011.01200.x
Larose, H., Shami, A. N., Abbott, H., Manske, G., Lei, L., and Hammoud, S. S. (2019). Gametogenesis: A journey from inception to conception. Curr. Top. Dev. Biol. 132, 257–310. doi: 10.1016/bs.ctdb.2018.12.006
Lawson, K. A., Dunn, N. R., Roelen, B. A., Zeinstra, L. M., Davis, A. M., Wright, C. V., et al. (1999). Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes. Dev. 13, 424–436. doi: 10.1101/gad.13.4.424
Lawson, K. A., and Hage, W. J. (1994). Clonal analysis of the origin of primordial germ cells in the mouse. Ciba Found Symp. 182, 68–84. doi: 10.1002/9780470514573.ch5
Lei, L., and Spradling, A. C. (2013). Mouse primordial germ cells produce cysts that partially fragment prior to meiosis. Development 140, 2075–2081. doi: 10.1242/dev.093864
Lence, T., Akhtar, J., Bayer, M., Schmid, K., Spindler, L., Ho, C. H., et al. (2016). m(6)A modulates neuronal functions and sex determination in Drosophila. Nature 540, 242–247. doi: 10.1038/nature20568
Lesch, B. J., and Page, D. C. (2012). Genetics of germ cell development. Nat. Rev. Genet. 13, 781–794. doi: 10.1038/nrg3294
Li, A., Chen, Y. S., Ping, X. L., Yang, X., Xiao, W., Yang, Y., et al. (2017). Cytoplasmic m(6)A reader YTHDF3 promotes mRNA translation. Cell. Res. 27, 444–447. doi: 10.1038/cr.2017.10
Liang, L., Soyal, S. M., and Dean, J. (1997). FIGalpha, a germ cell specific transcription factor involved in the coordinate expression of the zona pellucida genes. Development 124, 4939–4947. doi: 10.1242/dev.124.24.4939
Lin, Z., Hsu, P. J., Xing, X., Fang, J., Lu, Z., Zou, Q., et al. (2017). Mettl3-/Mettl14-mediated mRNA N(6)-methyladenosine modulates murine spermatogenesis. Cell. Res. 27, 1216–1230. doi: 10.1038/cr.2017.117
Lin, Z., and Tong, M. H. (2019). m(6)A mRNA modification regulates mammalian spermatogenesis. Biochim. Biophys. Acta Gene. Regul. Mech. 1862, 403–411. doi: 10.1016/j.bbagrm.2018.10.016
Liu, J., Yue, Y., Han, D., Wang, X., Fu, Y., Zhang, L., et al. (2014). A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10, 93–95. doi: 10.1038/nchembio.1432
Liu, N., Dai, Q., Zheng, G., He, C., Parisien, M., and Pan, T. (2015). N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 518, 560–564. doi: 10.1038/nature14234
Loebel, D. A., Watson, C. M., De Young, R. A., and Tam, P. P. (2003). Lineage choice and differentiation in mouse embryos and embryonic stem cells. Dev. Biol. 264, 1–14. doi: 10.1016/s0012-1606(03)00390-7
Louis, J. F., Thoma, M. E., Sørensen, D. N., McLain, A. C., King, R. B., Sundaram, R., et al. (2013). The prevalence of couple infertility in the United States from a male perspective: evidence from a nationally representative sample. Andrology 1, 741–748. doi: 10.1111/j.2047-2927.2013.00110.x
Ma, H., Wang, X., Cai, J., Dai, Q., Natchiar, S. K., Lv, R., et al. (2019). N(6-)Methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat. Chem. Biol. 15, 88–94. doi: 10.1038/s41589-018-0184-3
Mauer, J., Luo, X., Blanjoie, A., Jiao, X., Grozhik, A. V., Patil, D. P., et al. (2017). Reversible methylation of m(6)A(m) in the 5’ cap controls mRNA stability. Nature 541, 371–375. doi: 10.1038/nature21022
McLaren, A. (2003). Primordial germ cells in the mouse. Dev. Biol. 262, 1–15. doi: 10.1016/s0012-1606(03)00214-8
Mehlmann, L. M. (2005). Stops and starts in mammalian oocytes: recent advances in understanding the regulation of meiotic arrest and oocyte maturation. Reproduction 130, 791–799. doi: 10.1530/rep.1.00793
Mendel, M., Chen, K. M., Homolka, D., Gos, P., Pandey, R. R., McCarthy, A. A., et al. (2018). Methylation of structured RNA by the m(6)A writer METTL16 is essential for mouse embryonic development. Mol. Cell. 71, 986–1000. doi: 10.1016/j.molcel.2018.08.004
Meyer, K. D., Patil, D. P., Zhou, J., Zinoviev, A., Skabkin, M. A., Elemento, O., et al. (2015). 5’ UTR m(6)A promotes cap-independent translation. Cell 163, 999–1010. doi: 10.1016/j.cell.2015.10.012
Miyamoto, Y., Boag, P. R., Hime, G. R., and Loveland, K. L. (2012). Regulated nucleocytoplasmic transport during gametogenesis. Biochim. Biophys. Acta 1819, 616–630. doi: 10.1016/j.bbagrm.2012.01.015
Molyneaux, K. A., Stallock, J., Schaible, K., and Wylie, C. (2001). Time-lapse analysis of living mouse germ cell migration. Dev. Biol. 240, 488–498. doi: 10.1006/dbio.2001.0436
Nakagawa, T., Sharma, M., Nabeshima, Y., Braun, R. E., and Yoshida, S. (2010). Functional hierarchy and reversibility within the murine spermatogenic stem cell compartment. Science 328, 62–67. doi: 10.1126/science.1182868
Oatley, J. M., and Brinster, R. L. (2012). The germline stem cell niche unit in mammalian testes. Physiol. Rev. 92, 577–595. doi: 10.1152/physrev.00025.2011
Ohbo, K., Yoshida, S., Ohmura, M., Ohneda, O., Ogawa, T., Tsuchiya, H., et al. (2003). Identification and characterization of stem cells in prepubertal spermatogenesis in mice. Dev. Biol. 258, 209–225. doi: 10.1016/s0012-1606(03)00111-8
Ohinata, Y., Ohta, H., Shigeta, M., Yamanaka, K., Wakayama, T., and Saitou, M. (2009). A signaling principle for the specification of the germ cell lineage in mice. Cell 137, 571–584. doi: 10.1016/j.cell.2009.03.014
Ohta, H., Tohda, A., and Nishimune, Y. (2003). Proliferation and differentiation of spermatogonial stem cells in the w/wv mutant mouse testis. Biol. Reprod. 69, 1815–1821. doi: 10.1095/biolreprod.103.019323
Paci, M., Elkhatib, R., Longepied, G., Bourgeois, P., Ray, P. F., Levy, N., et al. (2018). The involvement of the nuclear lamina in human and rodent spermiogenesis: a systematic review. Basic Clin. Androl. 28:7. doi: 10.1186/s12610-018-0072-4
Patil, D. P., Chen, C. K., Pickering, B. F., Chow, A., Jackson, C., Guttman, M., et al. (2016). m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537, 369–373. doi: 10.1038/nature19342
Pendleton, K. E., Chen, B., Liu, K., Hunter, O. V., Xie, Y., Tu, B. P., et al. (2017). The U6 snRNA m(6)A Methyltransferase METTL16 regulates SAM Synthetase Intron Retention. Cell 169, 824–835. doi: 10.1016/j.cell.2017.05.003
Pepling, M. E. (2006). From primordial germ cell to primordial follicle: mammalian female germ cell development. Genesis 44, 622–632. doi: 10.1002/dvg.20258
Pepling, M. E., and Spradling, A. C. (1998). Female mouse germ cells form synchronously dividing cysts. Development 125, 3323–3328. doi: 10.1242/dev.125.17.3323
Perry, R. P., Kelley, D. E., Friderici, K., and Rottman, F. (1975). The methylated constituents of L cell messenger RNA: evidence for an unusual cluster at the 5’ terminus. Cell 4, 387–394. doi: 10.1016/0092-8674(75)90159-2
Petraglia, F., Serour, G. I., and Chapron, C. (2013). The changing prevalence of infertility. Int. J. Gynaecol. Obstet. 123, S4–S8. doi: 10.1016/j.ijgo.2013.09.005
Ping, X. L., Sun, B. F., Wang, L., Xiao, W., Yang, X., Wang, W. J., et al. (2014). Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell. Res. 24, 177–189. doi: 10.1038/cr.2014.3
Porras-Gómez, T. J., and Moreno-Mendoza, N. (2017). Neo-oogenesis in mammals. Zygote 25, 404–422. doi: 10.1017/s0967199417000363
Practice Committee of the American Society for Reproductive Medicine. (2013). Definitions of infertility and recurrent pregnancy loss: a committee opinion. Fertil. Steril. 99:63. doi: 10.1016/j.fertnstert.2012.09.023
Rajender, S., Avery, K., and Agarwal, A. (2011). Epigenetics, spermatogenesis and male infertility. Mutat. Res. 727, 62–71. doi: 10.1016/j.mrrev.2011.04.002
Rajkovic, A., Pangas, S. A., Ballow, D., Suzumori, N., and Matzuk, M. M. (2004). NOBOX deficiency disrupts early folliculogenesis and oocyte-specific gene expression. Science 305, 1157–1159. doi: 10.1126/science.1099755
Richardson, B. E., and Lehmann, R. (2010). Mechanisms guiding primordial germ cell migration: strategies from different organisms. Nat. Rev. Mol. Cell. Biol. 11, 37–49. doi: 10.1038/nrm2815
Roundtree, I. A., Evans, M. E., Pan, T., and He, C. (2017a). Dynamic RNA modifications in gene expression regulation. Cell 169, 1187–1200. doi: 10.1016/j.cell.2017.05.045
Roundtree, I. A., Luo, G. Z., Zhang, Z., Wang, X., Zhou, T., Cui, Y., et al. (2017b). YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. Elife 6:31311. doi: 10.7554/eLife.31311
Runyan, C., Schaible, K., Molyneaux, K., Wang, Z., Levin, L., and Wylie, C. (2006). Steel factor controls midline cell death of primordial germ cells and is essential for their normal proliferation and migration. Development 133, 4861–4869. doi: 10.1242/dev.02688
Sahin, Z., Szczepny, A., McLaughlin, E. A., Meistrich, M. L., Zhou, W., Ustunel, I., et al. (2014). Dynamic hedgehog signalling pathway activity in germline stem cells. Andrology 2, 267–274. doi: 10.1111/j.2047-2927.2014.00187.x
Saitou, M., and Yamaji, M. (2012). Primordial germ cells in mice. Cold Spring Harb. Perspect. Biol. 4:a008375. doi: 10.1101/cshperspect.a008375
Sánchez, F., and Smitz, J. (2012). Molecular control of oogenesis. Biochim. Biophys. Acta 1822, 1896–1912. doi: 10.1016/j.bbadis.2012.05.013
Sato, M., Kimura, T., Kurokawa, K., Fujita, Y., Abe, K., Masuhara, M., et al. (2002). Identification of PGC7, a new gene expressed specifically in preimplantation embryos and germ cells. Mech. Dev. 113, 91–94. doi: 10.1016/s0925-4773(02)00002-3
Schäfer, K. P. (1982). RNA synthesis and processing reactions in a subcellular system from mouse L cells. Hoppe Seylers Z Physiol. Chem. 363, 33–43. doi: 10.1515/bchm2.1982.363.1.33
Schwartz, S., Mumbach, M. R., Jovanovic, M., Wang, T., Maciag, K., Bushkin, G. G., et al. (2014). Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5’ sites. Cell Rep. 8, 284–296. doi: 10.1016/j.celrep.2014.05.048
Shi, H., Wang, X., Lu, Z., Zhao, B. S., Ma, H., Hsu, P. J., et al. (2017). YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 27, 315–328. doi: 10.1038/cr.2017.15
Shinohara, T., Orwig, K. E., Avarbock, M. R., and Brinster, R. L. (2000). Spermatogonial stem cell enrichment by multiparameter selection of mouse testis cells. Proc. Natl. Acad. Sci. U S A. 97, 8346–8351. doi: 10.1073/pnas.97.15.8346
Śledź, P., and Jinek, M. (2016). Structural insights into the molecular mechanism of the m(6)A writer complex. Elife 5:18434. doi: 10.7554/eLife.18434
Sorci, M., Ianniello, Z., Cruciani, S., Larivera, S., Ginistrelli, L. C., Capuano, E., et al. (2018). METTL3 regulates WTAP protein homeostasis. Cell Death Dis. 9:796. doi: 10.1038/s41419-018-0843-z
Sui, X., Hu, Y., Ren, C., Cao, Q., Zhou, S., Cao, Y., et al. (2020). METTL3-mediated m(6)A is required for murine oocyte maturation and maternal-to-zygotic transition. Cell Cycle 19, 391–404. doi: 10.1080/15384101.2019.1711324
Sun, H., Zhang, M., Li, K., Bai, D., and Yi, C. (2019). Cap-specific, terminal N(6)-methylation by a mammalian m(6)Am methyltransferase. Cell. Res. 29, 80–82. doi: 10.1038/s41422-018-0117-4
Tang, C., Klukovich, R., Peng, H., Wang, Z., Yu, T., Zhang, Y., et al. (2018). ALKBH5-dependent m6A demethylation controls splicing and stability of long 3’-UTR mRNAs in male germ cells. Proc. Natl. Acad. Sci. U S A. 115, E325–E333. doi: 10.1073/pnas.1717794115
Tang, W. W., Kobayashi, T., Irie, N., Dietmann, S., and Surani, M. A. (2016). Specification and epigenetic programming of the human germ line. Nat. Rev. Genet. 17, 585–600. doi: 10.1038/nrg.2016.88
Tremblay, K. D., Dunn, N. R., and Robertson, E. J. (2001). Mouse embryos lacking Smad1 signals display defects in extra-embryonic tissues and germ cell formation. Development 128, 3609–3621. doi: 10.1242/dev.128.18.3609
Ueda, Y., Ooshio, I., Fusamae, Y., Kitae, K., Kawaguchi, M., Jingushi, K., et al. (2017). AlkB homolog 3-mediated tRNA demethylation promotes protein synthesis in cancer cells. Sci. Rep. 7:42271. doi: 10.1038/srep42271
van Werven, F. J., and Amon, A. (2011). Regulation of entry into gametogenesis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 366, 3521–3531. doi: 10.1098/rstb.2011.0081
Wan, Y., Tang, K., Zhang, D., Xie, S., Zhu, X., Wang, Z., et al. (2015). Transcriptome-wide high-throughput deep m(6)A-seq reveals unique differential m(6)A methylation patterns between three organs in Arabidopsis thaliana. Genome Biol. 16:272. doi: 10.1186/s13059-015-0839-2
Wang, X., Feng, J., Xue, Y., Guan, Z., Zhang, D., Liu, Z., et al. (2016). Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature 534, 575–578. doi: 10.1038/nature18298
Wang, X., Lu, Z., Gomez, A., Hon, G. C., Yue, Y., Han, D., et al. (2014). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120. doi: 10.1038/nature12730
Wang, X., Zhao, B. S., Roundtree, I. A., Lu, Z., Han, D., Ma, H., et al. (2015). N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell 161, 1388–1399. doi: 10.1016/j.cell.2015.05.014
Warda, A. S., Kretschmer, J., Hackert, P., Lenz, C., Urlaub, H., Höbartner, C., et al. (2017). Human METTL16 is a N(6)-methyladenosine (m(6)A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 18, 2004–2014. doi: 10.15252/embr.201744940
Wen, J., Lv, R., Ma, H., Shen, H., He, C., Wang, J., et al. (2018). Zc3h13 regulates nuclear RNA m(6)A methylation and mouse embryonic stem cell self-renewal. Mol. Cell. 69, 1028–1038. doi: 10.1016/j.molcel.2018.02.015
Wolgemuth, D. J., Manterola, M., and Vasileva, A. (2013). Role of cyclins in controlling progression of mammalian spermatogenesis. Int. J. Dev. Biol. 57, 159–168. doi: 10.1387/ijdb.130047av
Wu, R., Li, A., Sun, B., Sun, J. G., Zhang, J., Zhang, T., et al. (2019). A novel m(6)A reader Prrc2a controls oligodendroglial specification and myelination. Cell. Res. 29, 23–41. doi: 10.1038/s41422-018-0113-8
Xia, H., Zhong, C., Wu, X., Chen, J., Tao, B., Xia, X., et al. (2018). Mettl3 mutation disrupts gamete maturation and reduces fertility in zebrafish. Genetics 208, 729–743. doi: 10.1534/genetics.117.300574
Xiao, W., Adhikari, S., Dahal, U., Chen, Y. S., Hao, Y. J., Sun, B. F., et al. (2016). Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol. Cell. 61, 507–519. doi: 10.1016/j.molcel.2016.01.012
Xu, K., Yang, Y., Feng, G. H., Sun, B. F., Chen, J. Q., Li, Y. F., et al. (2017). Mettl3-mediated m(6)A regulates spermatogonial differentiation and meiosis initiation. Cell. Res. 27, 1100–1114. doi: 10.1038/cr.2017.100
Xuan, J. J., Sun, W. J., Lin, P. H., Zhou, K. R., Liu, S., Zheng, L. L., et al. (2018). RMBase v2.0: deciphering the map of RNA modifications from epitranscriptome sequencing data. Nucleic Acids Res. 46, D327–D334. doi: 10.1093/nar/gkx934
Yamaguchi, S., Kimura, H., Tada, M., Nakatsuji, N., and Tada, T. (2005). Nanog expression in mouse germ cell development. Gene Expr. Patterns 5, 639–646. doi: 10.1016/j.modgep.2005.03.001
Yankova, E., Blackaby, W., Albertella, M., Rak, J., De Braekeleer, E., Tsagkogeorga, G., et al. (2021). Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature 593, 597–601. doi: 10.1038/s41586-021-03536-w
Ying, Y., Liu, X. M., Marble, A., Lawson, K. A., and Zhao, G. Q. (2000). Requirement of Bmp8b for the generation of primordial germ cells in the mouse. Mol. Endocrinol. 14, 1053–1063. doi: 10.1210/mend.14.7.0479
Ying, Y., and Zhao, G. Q. (2001). Cooperation of endoderm-derived BMP2 and extraembryonic ectoderm-derived BMP4 in primordial germ cell generation in the mouse. Dev. Biol. 232, 484–492. doi: 10.1006/dbio.2001.0173
Yue, Y., Liu, J., and He, C. (2015). RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 29, 1343–1355. doi: 10.1101/gad.262766.115
Zeng, C., Huang, W., Li, Y., and Weng, H. (2020). Roles of METTL3 in cancer: mechanisms and therapeutic targeting. J. Hematol. Oncol. 13:117. doi: 10.1186/s13045-020-00951-w
Zhang, F., Kang, Y., Wang, M., Li, Y., Xu, T., Yang, W., et al. (2018). Fragile X mental retardation protein modulates the stability of its m6A-marked messenger RNA targets. Hum. Mol. Genet. 27, 3936–3950. doi: 10.1093/hmg/ddy292
Zhao, B. S., and He, C. (2015). Fate by RNA methylation: m6A steers stem cell pluripotency. Genome Biol. 16:43. doi: 10.1186/s13059-015-0609-1
Zhao, B. S., and He, C. (2017). “Gamete On” for m(6)A: YTHDF2 exerts essential functions in female fertility. Mol. Cell. 67, 903–905. doi: 10.1016/j.molcel.2017.09.004
Zhao, Z., Meng, J., Su, R., Zhang, J., Chen, J., Ma, X., et al. (2020). Epitranscriptomics in liver disease: Basic concepts and therapeutic potential. J. Hepatol. 73, 664–679. doi: 10.1016/j.jhep.2020.04.009
Zheng, G., Dahl, J. A., Niu, Y., Fedorcsak, P., Huang, C. M., Li, C. J., et al. (2013). ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 49, 18–29. doi: 10.1016/j.molcel.2012.10.015
Keywords: m6A modification, methyltransferase, demethylase, binding protein, gametogenesis, infertility
Citation: Liu X, Wang H, Liu B, Qi Z, Li J, Xu B, Liu W, Xu Z and Deng Y (2021) The Latest Research Progress of m6A Modification and Its Writers, Erasers, Readers in Infertility: A Review. Front. Cell Dev. Biol. 9:681238. doi: 10.3389/fcell.2021.681238
Received: 16 March 2021; Accepted: 06 July 2021;
Published: 10 September 2021.
Edited by:
Trygve Tollefsbol, University of Alabama at Birmingham, United StatesCopyright © 2021 Liu, Wang, Liu, Qi, Li, Xu, Liu, Xu and Deng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yu Deng, ydeng@cmu.edu.cn; dengyu.cmu@163.com
 Xuda Liu
Xuda Liu Haiying Wang
Haiying Wang Bingchen Liu
Bingchen Liu Zhipeng Qi
Zhipeng Qi Bin Xu
Bin Xu Yu Deng
Yu Deng