
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol., 21 May 2021
Sec. Cell Growth and Division
Volume 9 - 2021 | https://doi.org/10.3389/fcell.2021.673258
This article is part of the Research TopicMeiosis: from Molecular Basis to MedicineView all 29 articles
 Fangfang Dong1,2,3†
Fangfang Dong1,2,3† Min Chen1,3†
Min Chen1,3† Min Chen4
Min Chen4 Lin Jiang5
Lin Jiang5 Zhiming Shen1,2,3
Zhiming Shen1,2,3 Longfei Ma1,2,3
Longfei Ma1,2,3 Chunsheng Han1,2,3*
Chunsheng Han1,2,3* Xudong Guo6*
Xudong Guo6* Fei Gao1,2,3*
Fei Gao1,2,3*Protein arginine methyltransferase 5 (PRMT5) catalyzes the formation of mono- or symmetric dimethylarginine residues on histones and non-histone substrates and has been demonstrated to play important roles in many biological processes. In the present study, we observed that PRMT5 is abundantly expressed in spermatogonial stem cells (SSCs) and that Prmt5 deletion results in a progressive loss of SSCs and male infertility. The proliferation of Prmt5-deficient SSCs cultured in vitro exhibited abnormal proliferation, cell cycle arrest in G0/G1 phase and a significant increase in apoptosis. Furthermore, PLZF expression was dramatically reduced in Prmt5-deficient SSCs, and the levels of H3K9me2 and H3K27me2 were increased in the proximal promoter region of the Plzf gene in Prmt5-deficient SSCs. Further study revealed that the expression of lysine demethylases (JMJD1A, JMJD1B, JMJD1C, and KDM6B) was significantly reduced in Prmt5-deficient SSCs and that the level of permissive arginine methylation H3R2me2s was significantly decreased at the upstream promoter region of these genes in Prmt5-deficient SSCs. Our results demonstrate that PRMT5 regulates spermatogonial stem cell development by modulating histone H3 lysine modifications.
Spermatogenesis is a highly precise cellular process, which consists of the self-renewal and differentiation of spermatogonial stem cells, spermatocyte meiosis and post-meiotic development of spermatids (He et al., 2008; Wu et al., 2016). Spermatogonia contain a small population of germline-specific stem cells with the ability to self-renew and differentiation. The differentiation of spermatogonia is stimulated by both intrinsic and extrinsic factors, which subsequently generate differentiating spermatogonia, spermatocytes, spermatids, and mature sperm (Chen and Liu, 2015; Mäkelä and Hobbs, 2019). In mammals, both genetic and epigenetic modifications are involved in the development of male germ cells. Plzf, Gfra1, Pou5f1, Lin28A, and Nanos3 are expressed in undifferentiated spermatogonia, and c-Kit is considered a marker gene of differentiating spermatogonia. Several Sertoli cell-produced growth factors, such as glial cell line-derived neurotrophic factor (GDNF), fibroblast growth factor (FGF2), and insulin-like growth factor (IGF), are also crucial for the maintenance and proliferation of spermatogonial stem cells (SSCs) (Oatley and Brinster, 2008; Zhang et al., 2012; Wang et al., 2016; Chen et al., 2017). Homozygous deletion of Gdnf leads to major defects in neonatal death. The function of Gdnf in SSC maintenance was first discovered in Gdnf heterozygous mutant (Meng et al., 2000).
Plzf (also known as Zfp145, Zbtb16, and promyelocytic leukemia zinc-finger) belongs to the Kruppel family and is expressed in hematopoietic stem/progenitor cells (Reid et al., 1995; Liu et al., 2016; Hai et al., 2019; Poplineau et al., 2019), bone marrow progenitor cells (Shaknovich et al., 1998), mesenchymal stem cells (Agrawal Singh et al., 2019) and other somatic cells (Cook et al., 1995; Barna et al., 2000). In mammalian testes, PLZF was first detected in prospermatogonia at E17.5 and is continually expressed in SSCs and spermatogonial progenitor cells (SPCs) at the adult stage (Avantaggiato et al., 1995; Buaas et al., 2004; Costoya et al., 2004). PLZF has been reported to play an essential role in SSCs pool maintenance and in regulating the self-renewal of SSCs. Plzf knockout leads to progressive germ cell loss after birth, and the spermatogonia are visible in only a few seminiferous tubules of adult mice (Buaas et al., 2004; Costoya et al., 2004).
PRMT5, a type II protein arginine methyltransferase, catalyzes the transfer of a methyl group from S-adenosylmethionine (SAM or AdoMet) to histones and non-histone substrates to form mono- or symmetric dimethylarginine (MMA or sDMA, respectively) (Rho et al., 2001; Bedford, 2007; Stopa et al., 2015). PRMT5 plays important roles in diverse cellular processes, such as cell differentiation, cell cycle, apoptosis, tumorigenesis and spliceosome assembly (Pahlich et al., 2006; Stopa et al., 2015; Hamard et al., 2018; Raposo and Piller, 2018). Previous studies have demonstrated that PRMT5 is required for germ cell development, as the loss of Prmt5 in primordial germ cells (PGCs) causes male and female sterility. PRMT5 represses the activation of LINEs and IAP transposons via symmetric dimethylation of arginine 3 on histone H2A and H4 (H2A/H4R3me2s) (Kim et al., 2014). PRMT5 is also required for PGC survival by promoting methylation of Sm spliceosomal proteins (Li et al., 2015). The results of our previous study demonstrated that inactivation of Prmt5 in male germ cells using Stra8-Cre causes aberrant spermatogenesis and male infertility (Wang et al., 2015c), suggesting that PRMT5 is essential for the development of male germ cells. PRMT5 is also expressed in mouse spermatogonial stem cells (SSCs). However, whether PRMT5 is involved in the development of SSCs is unknown. In the present study, we demonstrated that the deletion of Prmt5 in germ cells resulted in loss of spermatogonial stem cells (SSCs) and male infertility. Prmt5-deficient SSCs cultured in vitro exhibited abnormal proliferation, and the cell cycle was arrested in G0/G1 phase. Further study revealed that the expression of PLZF was dramatically reduced in Prmt5-deficient SSCs. We also observed that inactivation of Prmt5 resulted in enrichment of H3K9me2 and H3K27me2 in the promoter region of the Plzf gene, which in turn caused downregulation of Plzf expression and defects in SSC development.
PRMT5 was previously reported to be expressed in PGCs during the embryonic stage (Ancelin et al., 2006; Kim et al., 2014; Li et al., 2015) and in spermatocytes postnatally (Wang et al., 2015c; Dong et al., 2019). In the present study, PRMT5 expression in spermatogonia was examined by immunofluorescence and western blot assays. As shown in Supplementary Figure 1, PRMT5 was primarily detected in the cytoplasm of PLZF-positive germ cells (Supplementary Figure 1C, white arrowheads). PRMT5 was also detected in both the nucleus and cytoplasm of SSCs cultured in vitro by both immunofluorescence (Supplementary Figure 1D, white arrows) and western blot analysis. Lamin a/c and GAPDH served as loading controls for the nuclear and cytoplasmic extracts, respectively (Supplementary Figure 1G).
To assess the functions of PRMT5 in the development of SSCs, Prmt5 was deleted in prospermatogonia from E15 by crossing with Mvh-Cre transgenic mice (Gallardo et al., 2007). The immunofluorescence showed that PRMT5 was completely deleted in the PLZF-positive germ cells of Prmt5Δ/flox;Mvh-Cre testes (Supplementary Figures 2D–F, white arrows). No obvious developmental abnormalities were observed in adult Prmt5Δ/flox;Mvh-Cre mice (Figure 1A), and their body weights were comparable to those of control littermates (Figure 1B). However, the size of testes from Prmt5Δ/flox;Mvh-Cre mice was significantly reduced (Figures 1C,D). The histological results showed that germ cells at different developmental stages were observed in control testes (Figure 1E), whereas most of the seminiferous tubules in adult Prmt5Δ/flox;Mvh-Cre testes lacked germ cells (Figure 1F, asterisks). The cauda epididymis of control males was filled with mature sperm (Figure 1G), but no mature sperm were observed in Prmt5-deficient males (Figure 1H, asterisks). PLZF-positive spermatogonia were located at the peripheral region of the seminiferous tubules in control mice (Figure 1I, black arrows), whereas no PLZF-positive spermatogonia were observed in Prmt5Δ/flox;Mvh-Cre mice (Figure 1J, asterisks). SOX9-positive Sertoli cells were observed in the seminiferous tubules of both control (Figure 1K, black arrows) and Prmt5Δ/flox;Mvh-Cre mice (Figure 1L, black arrows). These results indicate that PRMT5 is important for the survival and maintenance of spermatogonial stem cells.
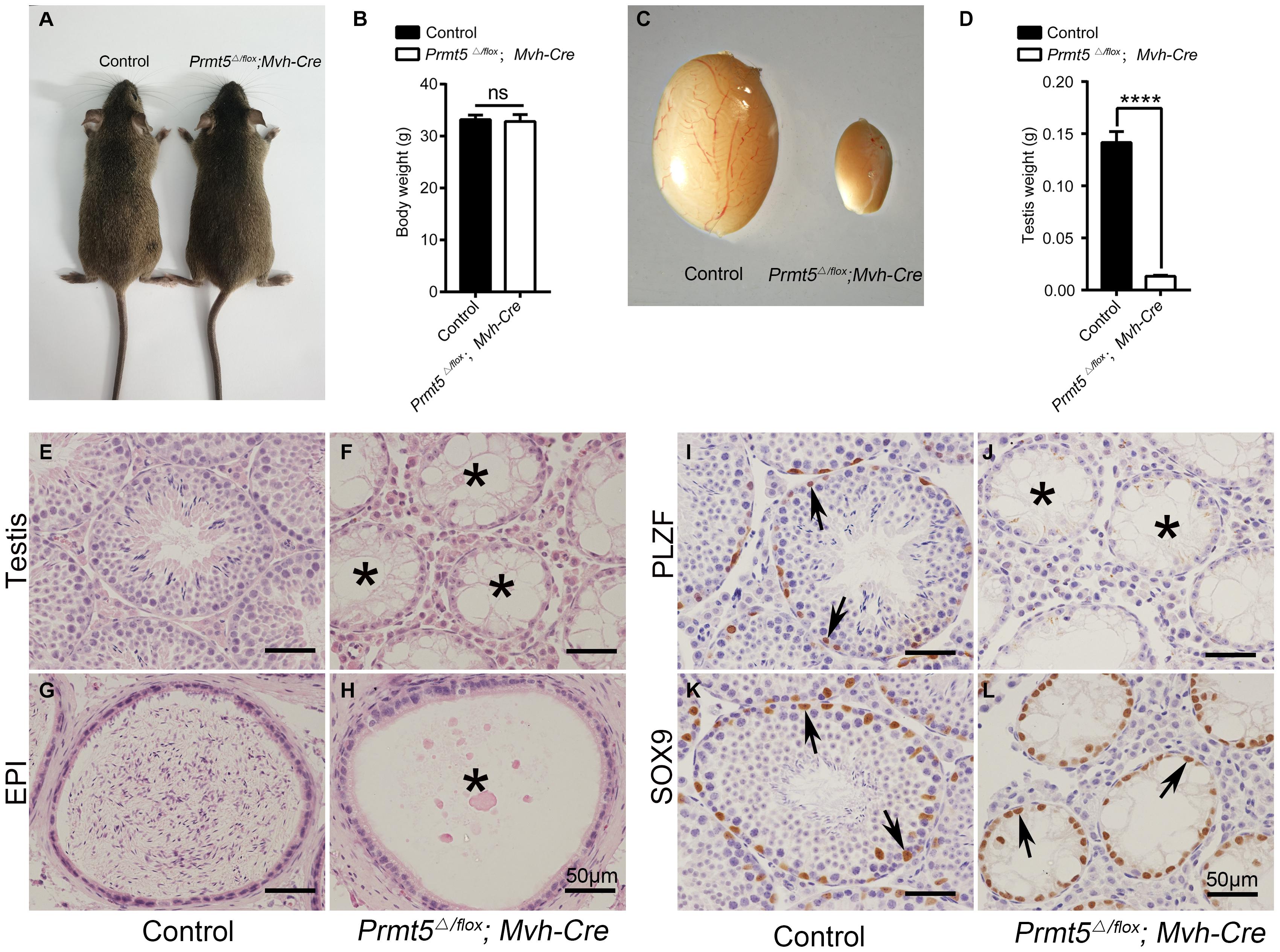
Figure 1. Deletion of Prmt5 causes germ cell loss and male infertility. No obvious developmental abnormalities were observed in adult Prmt5Δ/flox;Mvh-Cre mice (A), and the body weights were comparable to those of control littermates (B). The size of testes from Prmt5Δ/flox;Mvh-Cre mice was significantly smaller than that of control littermates (C,D). Germ cells at different developmental stages were observed in control testes (E), whereas most seminiferous tubules were empty in adult Prmt5Δ/flox;Mvh-Cre testes (F, asterisks). The cauda epididymis of control males was filled with mature sperm (G), but no mature sperm were observed in the epididymis of Prmt5-deficient males (H). PLZF-positive spermatogonia were located at the peripheral region of the seminiferous tubules in control mice [(I), black arrows], whereas no PLZF-positive spermatogonia were observed in Prmt5Δ/flox;Mvh-Cre mice (J). SOX9-positive Sertoli cells were observed in the seminiferous tubules of both control [(K), black arrows] and Prmt5Δ/flox;Mvh-Cre mice [(L), black arrows]. Error bars represent the SEM of triplicate results. ****P < 0.00001 indicates a significant difference (t-test).
Testes from Prmt5Δ/flox;Mvh-Cre mice (Supplementary Figure 3B) were grossly normal at P7 compared to those of control testes (Supplementary Figure 3A). Aberrant seminiferous tubules were first noted in Prmt5Δ/flox;Mvh-Cre testes at P10 (Supplementary Figure 3D, asterisks). Empty tubules were observed in Prmt5-deficient testes at 2 weeks (Supplementary Figure 3F, asterisks), 3 weeks (Supplementary Figure 3H, asterisks), 4 weeks (Supplementary Figure 3J, asterisks) and 6 weeks (Supplementary Figure 3L, asterisks). The development of germ cells at different developmental stages was also examined by immunohistochemistry. MVH-positive germ cells were observed in both control (Figure 2A, black arrows) and Prmt5Δ/flox;Mvh-Cre mice (Figure 2B, black arrows) at P7, and no difference was noted. The number of germ cells in Prmt5Δ/flox;Mvh-Cre mice (Figure 2D, black arrows) was significantly reduced at P10 than that in control mice (Figure 2C, black arrows), while the number of germ cells was significantly increased from 2 to 6 weeks in these mice (Figures 2E,G,I,K, black arrows). In contrast, the number of germ cells in Prmt5Δ/flox;Mvh-Cre mice was gradually reduced from 2 to 4 weeks (Figures 2F,H,J, black arrows), and they were completely absent at 6 weeks of age (Figure 2L, asterisks). PLZF-positive spermatogonia (green) were observed in the seminiferous tubules of control mice at 3 weeks (Supplementary Figure 4A, white arrows), 4 weeks (Supplementary Figure 4B, white arrows), and 6 weeks (Supplementary Figure 4C, white arrows). PLZF-positive spermatogonia were also observed in the seminiferous tubules of Prmt5Δ/flox;Mvh-Cre mice at 3 weeks (Supplementary Figure 4D, white arrows) and 4 weeks (Supplementary Figure 4E, white arrows) but not at 6 weeks (Supplementary Figure 4F, asterisks). The quantitative results showed that the number of PLZF-positive germ cells was dramatically reduced in Prmt5Δ/flox;Mvh-Cre mice at 3 weeks (Supplementary Figure 4G) and 4 weeks (Supplementary Figure 4H), and no germ cells were counted at 6 weeks (Supplementary Figure 4I). These results indicate that PRMT5 is required for the maintenance of the SSC pool.
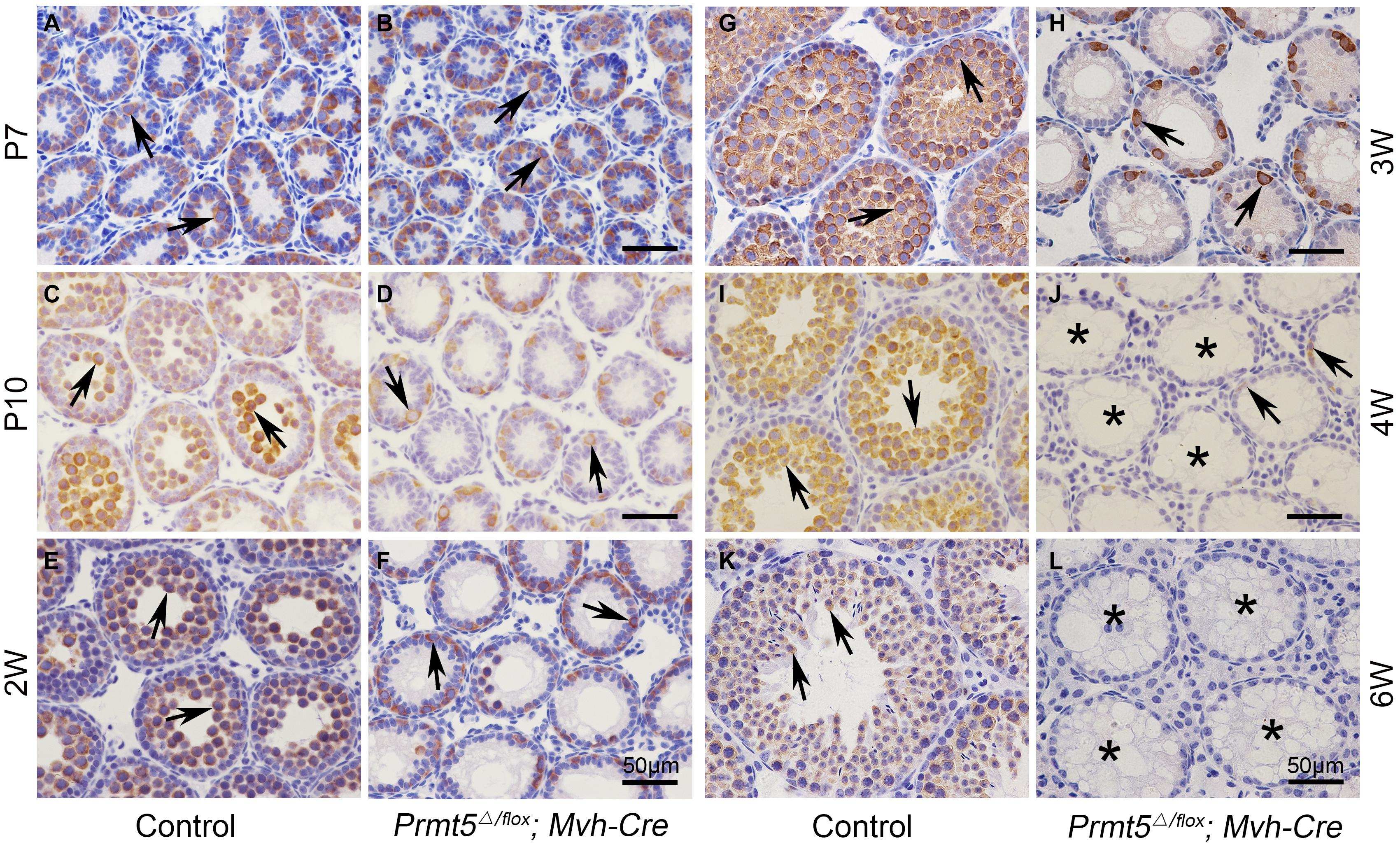
Figure 2. Germ cells from Prmt5Δ/flox;Mvh-Cre mice are gradually lost from P10. The germ cells in control and Prmt5Δ/flox;Mvh-Cre mice were labeled with an anti-MVH antibody. MVH-positive germ cells were observed in both control [(A), black arrows] and Prmt5Δ/flox;Mvh-Cre mice [(B), black arrows] at P7, and no difference was noted. The number of germ cells in Prmt5Δ/flox;Mvh-Cre mice [(D), black arrows] was significantly reduced at P10 compared to that observed in control mice [(C), black arrows]. The number of germ cells was significantly increased from 2 to 6 weeks in control mice [(E,G,I,K), black arrows]. In contrast, the number of germ cells in Prmt5Δ/flox;Mvh-Cre mice was gradually reduced from 2 to 4 weeks [(F,H,J), black arrows] and was completely absent at 6 weeks of age [(L), asterisks].
To test whether the process of meiosis is affected after Prmt5 depletion, the expression of STRA8, SYCP3 and γH2AX at P10 was analyzed by immunofluorescence and western blot assays. A strong STRA8 signal was detected in the germ cells of control testes at P10 (Figure 3A, white arrows), whereas only very weak STRA8 expression was observed in the germ cells of Prmt5Δ/flox;Mvh-Cre testes (Figure 3B, white arrows). SYCP3-positive germ cells were observed in control testes (Figure 3C, white arrows), but no SYCP3 signal was detected in the germ cells of Prmt5Δ/flox;Mvh-Cre testes at P10 (Figure 3D, white asterisks). The western blot results showed that the expression of the meiosis-associated proteins STRA8, SYCP3 and SYCP1 was significantly reduced in Prmt5Δ/flox; Mvh-Cre testes at P10 (Figures 3E,F). These results indicate that meiosis is blocked in Prmt5-deficient germ cells.
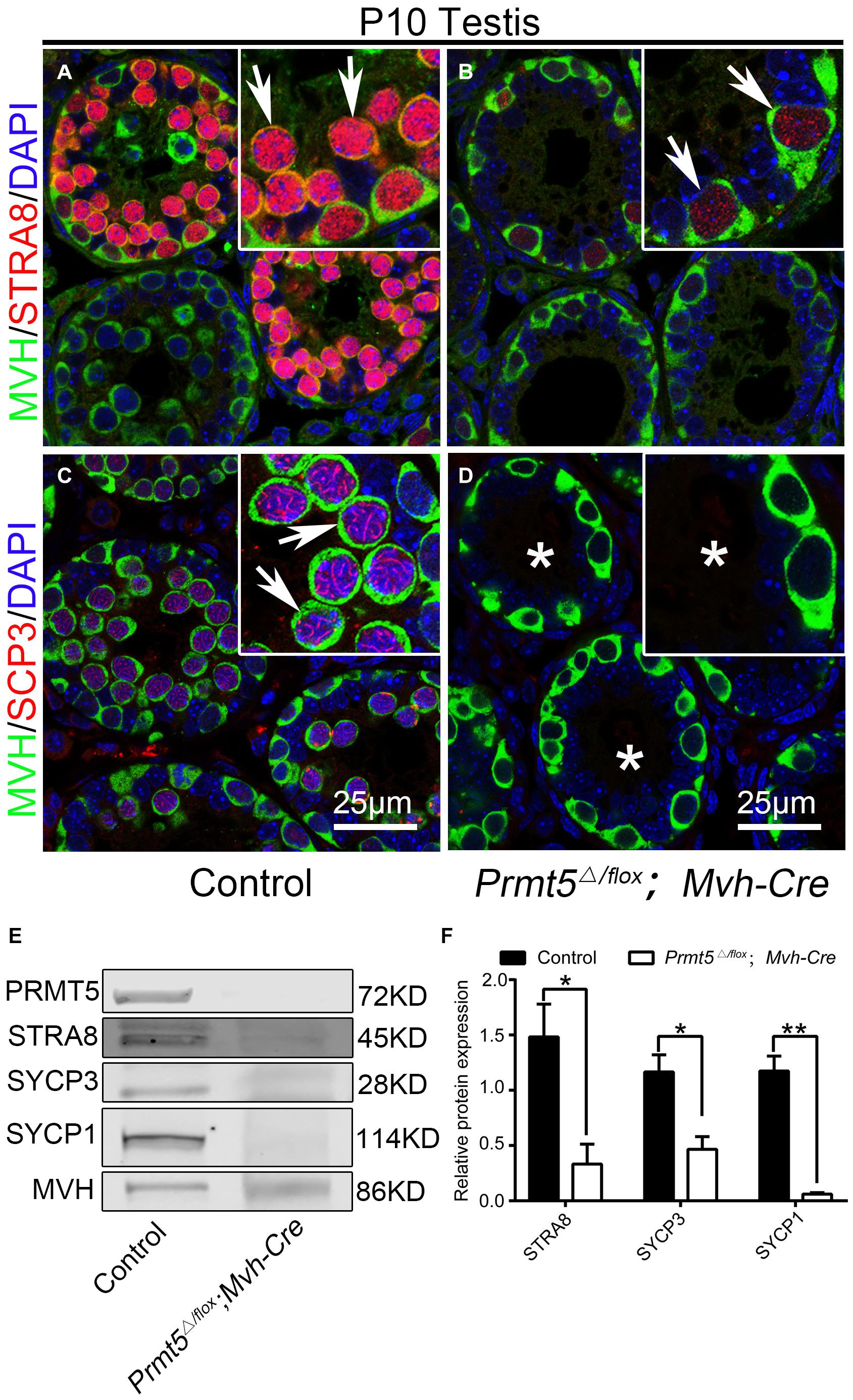
Figure 3. Prmt5-deficient germ cells exhibit a defect in meiosis initiation. A strong STRA8 signal was detected in the germ cells of control testes [(A), white arrows], whereas only a weak STRA8 signal was detected in the germ cells of Prmt5Δ/flox;Mvh-Cre testes [(B), white arrows]. SYCP3-positive germ cells were observed in control testes [(C), white arrows], but no SYCP3 signal was detected in the germ cells of Prmt5Δ/flox;Mvh-Cre testes [(D), asterisks]. The expression of meiosis-associated genes (STRA8, SYCP3 and SYCP1) was significantly reduced in Prmt5Δ/flox;Mvh-Cre testes at P10 (E,F), The total protein detected from control testes was 10 μg, and the total protein detected from Prmt5Δ/flox;Mvh-Cre testes was 40 μg. MVH was used as a loading control in (E), protein values were normalized to MVH and expressed as the mean ± SEM (n = 3), *P < 0.01, **P < 0.001 indicates a significant difference (t-test).
In the present study, we observed that PLZF-positive spermatogonial stem cells were gradually lost in Prmt5 knockout mice. To test whether PRMT5 is involved in the proliferation or self-renewal of SSCs, germ cells were labeled with Ki67 and PH3. Ki67 and PH3 were detected in MVH-positive germ cells in both control (Figures 4A,D, white arrows) and Prmt5Δ/flox;Mvh-Cre testes (Figures 4B,E, white arrows) at P10. The quantitative results showed that the percentage of Ki67- and PH3-positive germ cells was dramatically reduced in Prmt5Δ/flox;Mvh-Cre mice (Figures 4C,F). These results indicate that the deletion of Prmt5 leads to defects in germ cell proliferation. To further confirm these results, SSCs from Prmt5flox/flox;Cre-ERTM mice were cultured in vitro, and Prmt5 was deleted by treatment with 1 μM tamoxifen. Bright-field images showed that the clone size of Prmt5-deficient SSCs was significantly smaller (Figures 4Gc,d) than that of SSCs treated with ethanol (Figures 4Ga,b). In addition, the MTT assay results also showed that knockout of Prmt5 resulted in a significant decrease in cell proliferation (Figure 4H). The flow cytometry results showed that the loss of Prmt5 resulted in G0/G1 phase arrest with a concomitant decrease in S phase (Figures 4I,J). These results indicate that deletion of Prmt5 results in cell cycle arrest of SSCs cultured in vitro.
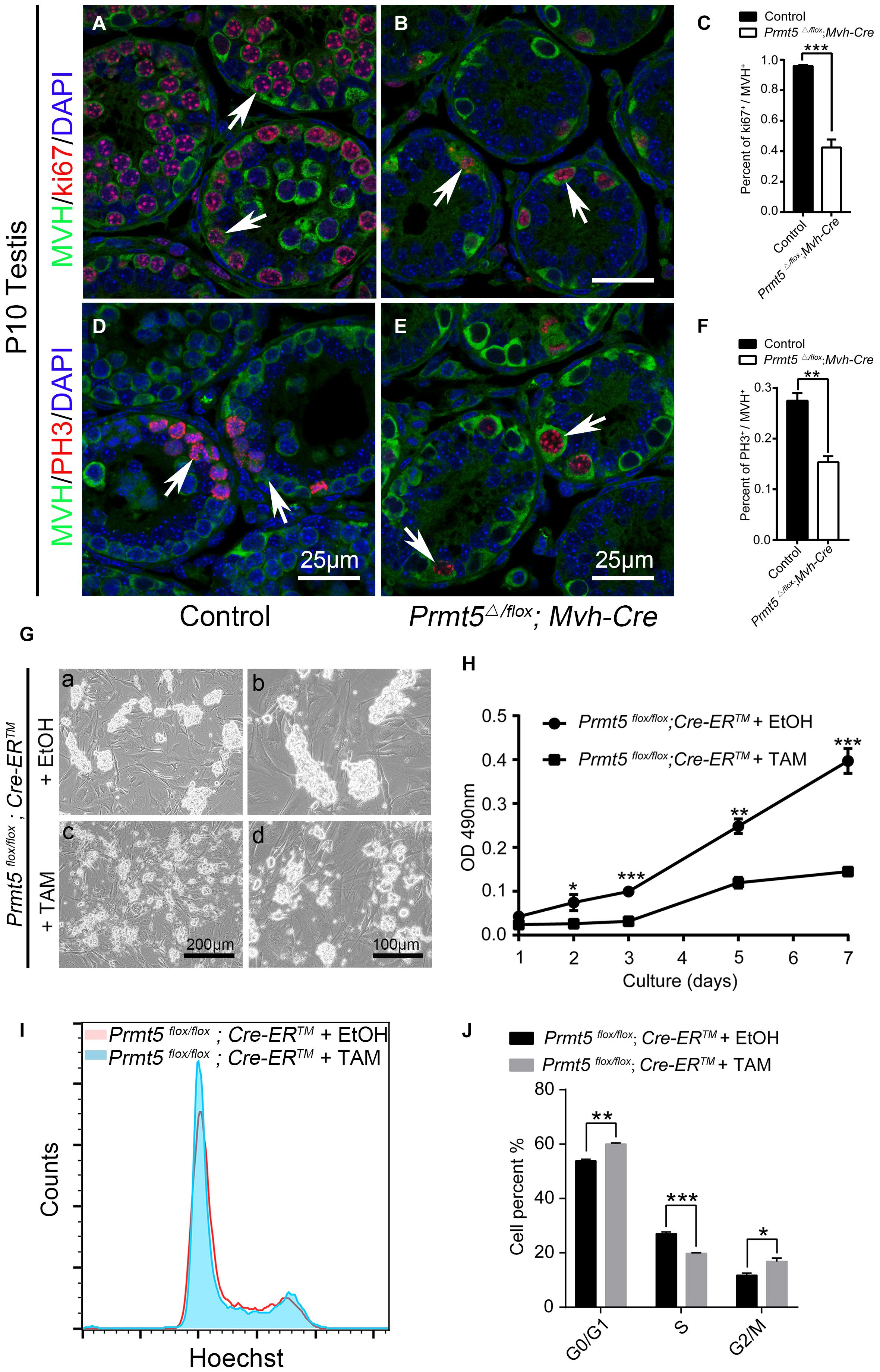
Figure 4. Prmt5-deficient spermatogonial stem cells exhibit abnormal proliferation. Ki67-positive (red) germ cells were detected in both control [(A), white arrows] and Prmt5Δ/flox;Mvh-Cre testes [(B), white arrows] at P10. The percentage of Ki67-positive germ cells was dramatically reduced in Prmt5Δ/flox;Mvh-Cre mice (C). PH3 (red) was detected in MVH-positive germ cells of both control [(D), white arrows] and Prmt5Δ/flox;Mvh-Cre testes [(E), white arrows] at P10. The percentage of PH3-positive germ cells was dramatically reduced in Prmt5Δ/flox;Mvh-Cre mice (F). (G) Bright-field images of control and Prmt5-deficient SSCs cultured in vitro. Prmt5flox/flox;Cre-ERTM SSCs were treated with ethanol or 1 μM tamoxifen to induce Cre activation. (H) MTT assay of cultured control and Prmt5-deficient SSCs. (I) FACS analysis of cultured control and Prmt5-deficient SSCs. (J) Quantitative analysis of cells in G0/G1, S, and G2 phase. Error bars represent the SEM of triplicate results. *P < 0.05, **P < 0.005, ***P < 0.0005 indicates a significant difference (t-test).
The differentially expressed genes associated with stemness maintenance, proliferation and the cell cycle in both control and Prmt5-deficient SSCs were examined by real-time PCR and western blot analysis. Plzf, Oct4, Nanos3, and Gfra1 are well-characterized pluripotent factors that are important for the maintenance of SSC stemness (Oatley and Brinster, 2008; Chen et al., 2017; Mäkelä and Hobbs, 2019). As shown in Figure 5, the expression of Prmt5 was dramatically reduced in Prmt5flox/flox;Cre-ERTM SSCs treated with 1 μM tamoxifen compared to those treated with ethanol. Both the mRNA and protein levels of stemness-related genes, such as Oct4 and Plzf, were significantly decreased in Prmt5 knockout SSCs. We also observed that the expression of the germ cell marker gene Mvh was significantly decreased in Prmt5-deficient SSCs. The mRNA level of c-Myc was decreased in Prmt5-deficient SSCs (Figure 5A), whereas the protein level was not changed (Figures 5B,C). The expression of cell cycle-associated genes was also examined. As shown in Figures 5D–F, the mRNA and protein levels of A-type cyclins (Ccna1 and Ccna2) and B-type cyclins (Ccnb1, Ccnb2, and Ccnb3) were all significantly reduced in Prmt5-deficient SSCs. In contrast, the expression of cyclin-D, cyclin-E1, and cyclin-G was not altered after Prmt5 inactivation. It is worth noting that p21 and p53 expression dramatically increased after inactivation Prmt5 at both the mRNA and protein levels. These results indicate that PRMT5 is important for stemness maintenance and the proliferation of spermatogonial stem cells.
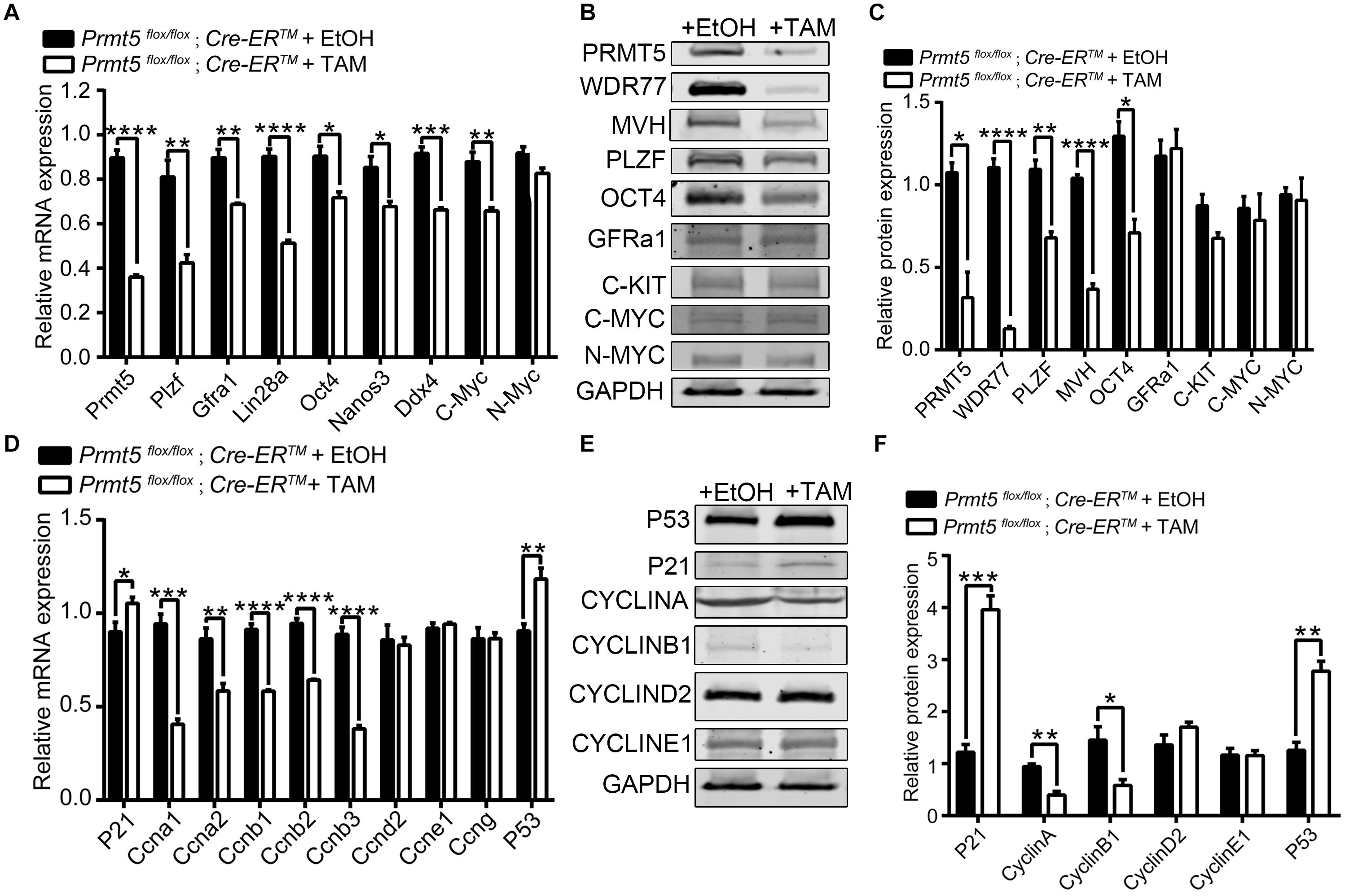
Figure 5. Differentially expressed genes in Prmt5-deficient spermatogonial stem cells. The expression of genes related to self-renewal and proliferation was analyzed by real-time PCR (A) and western blotting (B) in Prmt5flox/flox;Cre-ERTM SSCs treated with ethanol or 1 μM tamoxifen. The quantitative results of western blot analysis (C). The expression of cell cycle-associated genes was analyzed by real-time PCR (D) and western blot (E). The quantitative results of western blot analysis (F). GAPDH was used as a loading control, and the protein values were normalized. Error bars represent the SEM of triplicate results. *P < 0.05, **P < 0.005, ***P < 0.0005, ****P < 0.0005 indicates a significant difference (t-test).
PRMT5 has been shown to regulate the expression of target genes suchas Blimp1 (Ancelin et al., 2006), c-Myc (Liu M. et al., 2020), p21 (Zhang et al., 2015), and androgen receptor (Deng et al., 2017) via symmetric dimethylation of arginine residues of histones H4 (H4R3), H3 (H3R2 and H3R8), and H2A (H2AR3). The western blot results showed that loss of Prmt5 led to a dramatic decrease in H4R3me2s, H3R2me2s, and H2AR3me2s levels. Interestingly, the levels of the repressive histone lysine modifications H3K9me2 and H3K27me2 were significantly increased (Figures 6A,B). Previous studies have reported that histone H3 lysine modifications, especially the methylation of H3K9 and H3K27, play important roles in the prospermatogonia to spermatogonia transition and in the development of SPCs/SSCs (Mu et al., 2014; Tseng et al., 2015; Kuroki et al., 2020). To test whether the downregulation of PLZF in Prmt5-deficient SSCs is due to a decrease in H4R3me2s, H3R2me2s, and H2AR3me2s levels or an increase in H3K9me2 and H3K27me2, ChIP assays were performed to analyze the enrichment of H4R3me2s, H3R2me2s, H2AR3me2s, H3K9me2, and H3K27me2 at the promoter region (TSS-1 kb upstream of TSS) of the Plzf gene. The ChIP-qPCR results showed that H4R3me2s, H3R2me2s, and H2AR3me2s levels remained unchanged (data not shown), whereas those of H3K27me2 and H3K9me2 were significantly increased at the proximal promoter region (Site 2: −175∼−347 bp) of Plzf in Prmt5-deficient SSCs (Figures 6C,D). These results indicate that the downregulation of Plzf in Prmt5-deficient SSCs is not directly regulated by Prmt5 via histone arginine methylation, which is most likely caused by increased H3K27me2 and H3K9me2 levels.
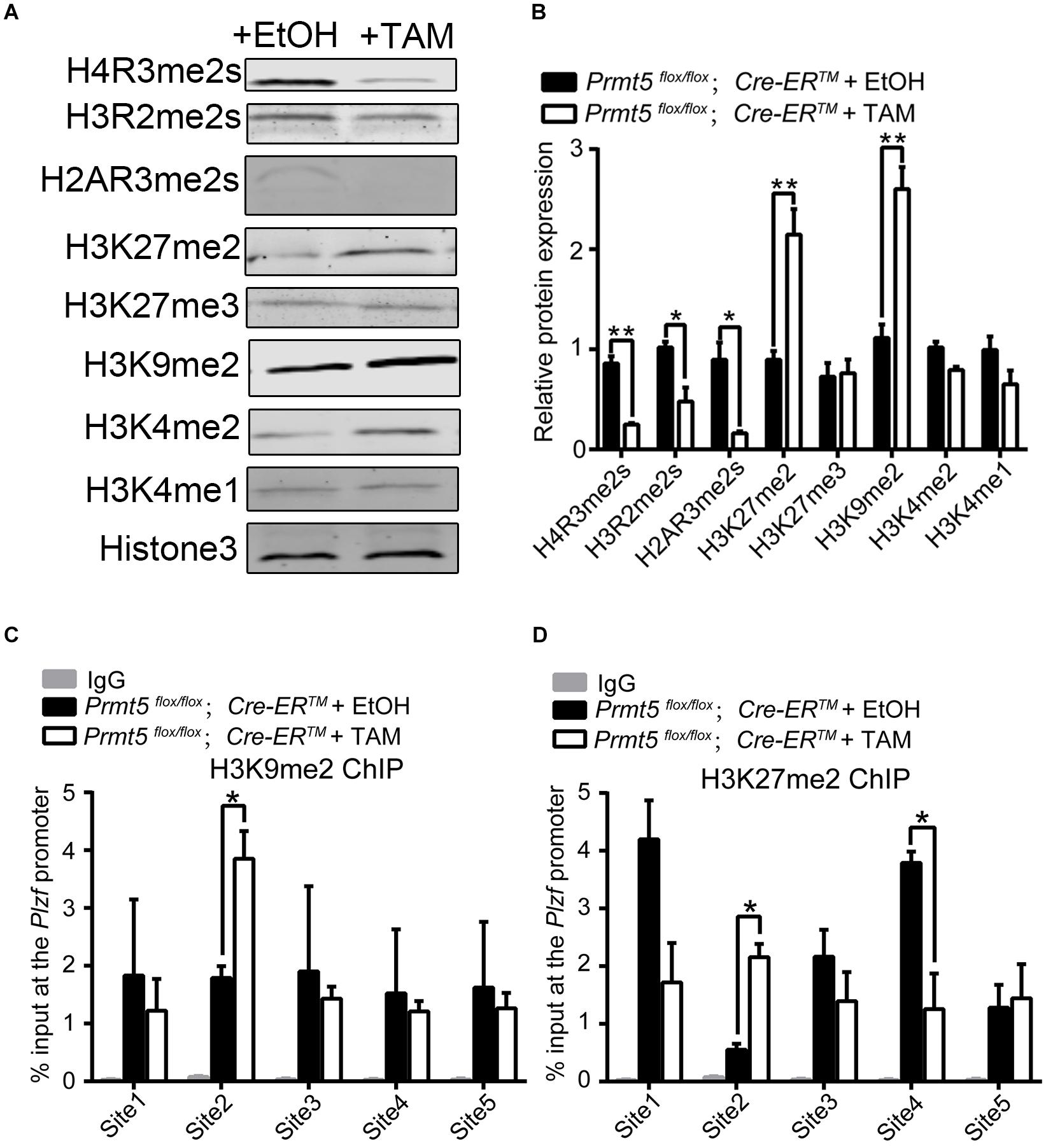
Figure 6. H3K9me2 and H3K27me2 levels are significantly increased in Prmt5-deficient spermatogonial stem cells. (A) The level of histone modification in control and Prmt5-deficient spermatogonial stem cells was examined by western blot. (B) Quantitative results of western blot analysis. ChIP assays were performed to analyze the enrichment of H3K27me2 (C) and H3K9me2 (D) modifications in the promoter regions (TSS-1 kb upstream of TSS) of the Plzf (site 1∼ site 5) gene in control and Prmt5-deficient SSCs. The relative enrichment of the Plzf promoter region was examined by real-time PCR using sequence-specific primer sets. IgG was used as a negative control. Site 1: −114∼−284 bp, Site 2: −175∼−347 bp, Site 3: −381∼−552 bp, Site 4: −573∼−743 bp, Site 5: −755∼−948 bp. Quantitative data are presented as the enrichment of the ChIP to the input DNA. TSS, transcription start site. Error bars present SEM of three ChIP experiments. *P < 0.05, **P < 0.005 indicates a significant difference (t-test).
To assess the underlying mechanisms that cause the increase in H3K9me2 and H3K27me2 in Prmt5-deficient SSCs, we analyzed the expression of lysine methylases and demethylases for H3K9me2 and H3K27me2 by real-time PCR and western blot analysis. As shown in Figure 7, the mRNA level of lysine demethylase for H3K9me2 (JMJD1A, JMJD1B, and JMJD1C) was significantly decreased after Prmt5 inactivation, whereas the expression of lysine methylases for H3K9me2 (KMT1A, KMT1B, KMT1E, and KMT1F) remained unchanged. The mRNA levels of lysine methylases for H3K27me2 (NSD3, EZH2, and NSD2) were not increased in Prmt5-deficient SSCs, while the expression of demethylase for H3K27me2 (KDM6B) was significantly deceased (Figures 7A,B). The western blot results further demonstrated that the expression of the demethylases for H3K9me2 or H3K27me2 was dramatically reduced in Prmt5-deficient SSCs (Figures 7C,D). These results suggest that the increase in H3K9me2 and H3K27me2 in Prmt5-deficient SSCs is most likely due to the downregulation of demethylases.
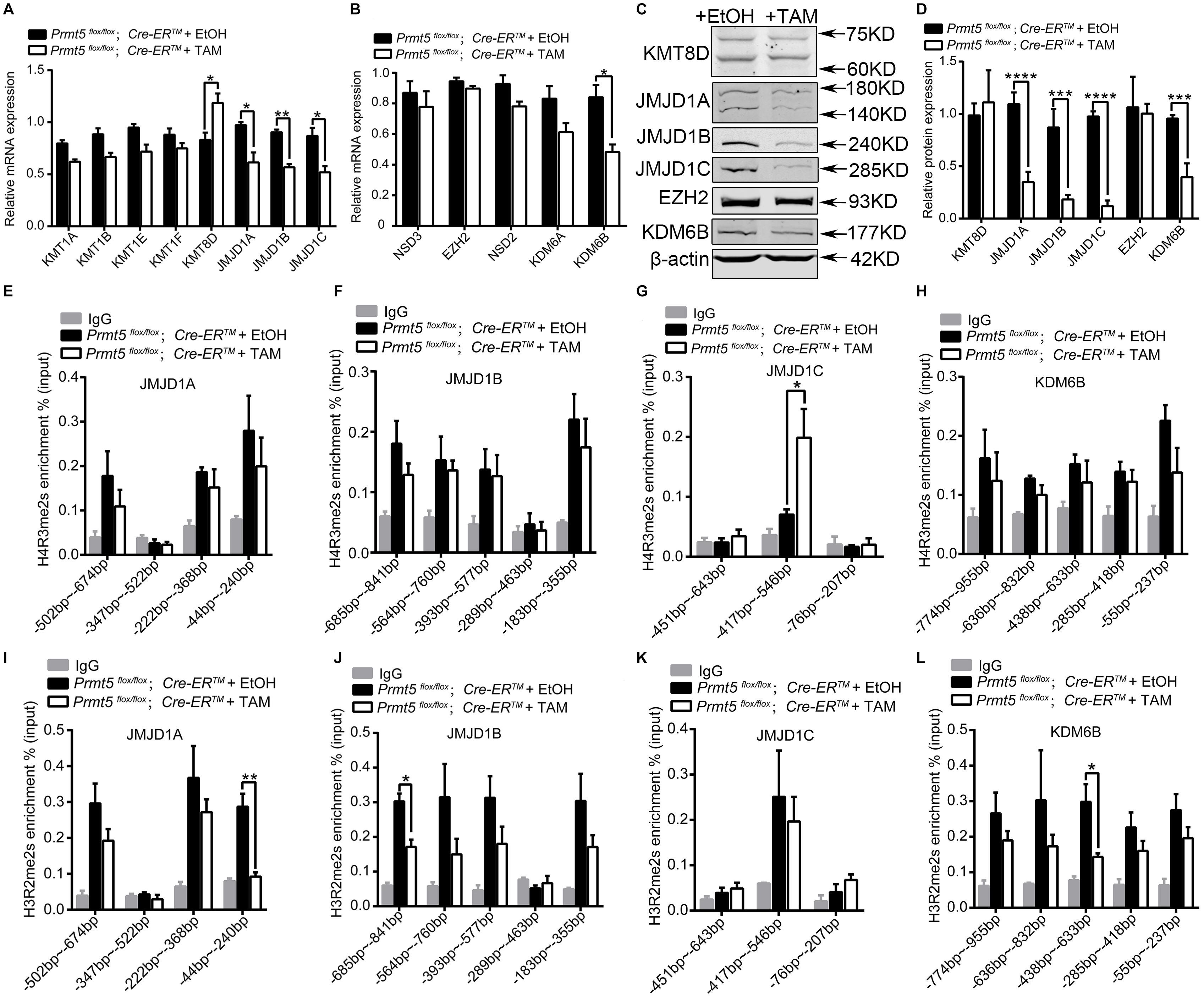
Figure 7. The expression of lysine demethylases of H3K9me2 and H3K27me2 is regulated by PRMT5 via histone arginine modifications. The differential expression of lysine methylases and demethylases of H3K9me2 and H3K27me2 in Prmt5-deficient SSCs was analyzed by real-time PCR (A,B) and western blot (C,D). β-Actin was used as a loading control, and the protein values were normalized. ChIP-qPCR assays were performed to examine the enrichment of H4R3me2s or H3R2me2s modifications at the upstream regions of JMJD1A (E,I), JMJD1B (F,J), JMJD1C (G,K), and KDM6B (H,L) in Prmt5-deficient SSCs. The immunoprecipitated DNA fragments were analyzed by real-time PCR with sequence-specific primer sets. IgG was used as a negative control. Quantitative data are presented as the enrichment of the ChIP to the input DNA. Error bars represent the SEM of triplicate results. *P < 0.05, **P < 0.005, ***P < 0.0005, ****P < 0.00005 indicates a significant difference (t-test).
To further examine whether the downregulation of lysine demethylase for H3K9me2 (JMJD1A, JMJD1B, and JMJD1C) and H3K27me2 (KDM6B) is caused by changes in histone arginine methylation in Prmt5-deficient SSCs, we analyzed the level of H4R3me2s or H3R2me2s at the promoter regions (TSS-1 kb upstream of TSS) of JMJD1A, JMJD1B, JMJD1C, and KDM6B. The ChIP-qPCR results showed that inactivation of Prmt5 led to a significant increase in the repressive histone modification H4R3me2s at the promoter region (−417 ∼−546 bp) of JMJD1C (Figure 7G), whereas the occupancy of H4R3me2s at the upstream promoters of JMJD1A, JMJD1B, KDM6B was not changed (Figures 7E,F,H). Moreover, the occupancy of permissive histone modification H3R2me2s at the upstream regions (TSS-1 kb upstream of TSS) of the JMJD1A, JMJD1B, and KDM6B genes was significantly reduced in Prmt5-deficient SSCs (Figures 7I,J,L). The level of H3R2me2s at the proximal promoter region of JMJD1C was not decreased (Figure 7K). These results suggest that the expression of histone lysine demethylases for H3K9me2 and H3K27me2 is regulated by PRMT5 via histone arginine modifications.
As an epigenetic modifier, PRMT5 has been demonstrated to play important roles in PGC development, and inactivation of this gene caused loss of germ cells during the embryonic stage (Kim et al., 2014; Li et al., 2015; Wang et al., 2015b). The results of our previous study revealed that the postnatal knockout of Prmt5 in male germ cells using Stra8-Cre led to defects in meiosis and male infertility (Wang et al., 2015c). However, the functions of PRMT5 in SSC development have not been previously investigated. In the present study, we demonstrated that PRMT5 is essential for the survival and maintenance of SSCs. The inactivation of Prmt5 resulted in cell cycle arrest and progressive loss of SSCs at 3 weeks of age.
PLZF has been reported to play important roles in the regulation of diverse cellular processes, including stemness maintenance, differentiation, cell cycle, proliferation and apoptosis (Suliman et al., 2012; Liu et al., 2016). PLZF is also highly expressed in spermatogonial stem cells and is considered a marker gene for undifferentiated spermatogonial stem cells. The deletion of Plzf was shown to lead to a progressive loss of spermatogonial stem cells in a mouse model (Buaas et al., 2004; Costoya et al., 2004). Moreover, Plzf is considered a cell cycle regulator, and PLZF overexpression in hematopoietic stem cells or the hematopoietic cell line 32Dcl3 was observed to block cells in G1/S phase, resulting in defects in cell growth and differentiation with an increase in apoptosis (Shaknovich et al., 1998; Yeyati et al., 1999; Vincent-Fabert et al., 2016). In the present study, we showed that the expression was dramatically reduced after the loss of Prmt5. Based on these results, we speculated that the defect in SSC development in Prmt5-deficient mice is most likely due to the downregulation of PLZF.
As a protein arginine methyltransferase, PRMT5 catalyzes MMA or sDMA in histones and non-histone substrates (Rho et al., 2001; Bedford, 2007). Interestingly, we observed that the level of global histone lysine modifications (H3K9me2 and H3K27me2) was significantly increased in Prmt5-deficient SSCs. PRMT5-induced H4R3 methylation has been reported to regulate lysine methylation modification (H3K27) via recruitment of Polycomb protein in a Pax2/Grg4-dependent manner (Patel et al., 2012). In hematopoietic cells, Prmt5 depletion resulted in the upregulation of global H3K27 dimethylation and trimethylation (Liu F. et al., 2020). In the present study, the global level of H3K27me2 was significantly increased, whereas the level of H3K27me3 was not increased in Prmt5-deficient SSCs (Figures 6A,B). Moreover, unlike in a previous study (Tae et al., 2011; Patel et al., 2012; Li et al., 2018), the expression of methylases for H3K9me2 and H3K27me2 was not increased in Prmt5-deficient SSCs. However, the expression of demethylases (JMJD1A, JMJD1B, JMJD1C, and KDM6B) was significantly reduced at both the protein and mRNA levels, indicating that the increase in H3K9me2 and H3K27me2 was most likely due to the downregulation of histone H3 lysine demethylase expression. These results suggested that the expression of target genes regulated by PRMT5 is cell context dependent.
The functions of histone lysine methylation in SSCs development have been previously reported. The histone lysine demethylase JMJD1 isozymes targeting H3K9me2 play essential roles in the development of SSCs and spermatogenesis. The depletion of both JMJD1A and JMJD1B results in defects in the prospermatogonia to spermatogonia transition and causes abnormal spermatogenesis (Kuroki et al., 2020). The loss of JMJD1C also results in a progressive reduction of SSCs/SPCs and male infertility (Kuroki et al., 2013). The histone H3K27 demethylase KDM6B is involved in regulating the fragmentation of spermatogonial cysts, but the differentiation of SSCs is not affected (Iwamori et al., 2013). EED is a core subunit of Polycomb-repressive complex (PRC2), which is responsible for catalyzing H3K27me2/H3K27me3, and deletion of EED by Mvh-Cre leads to defects in SSC maintenance (Mu et al., 2014). Therefore, appropriate epigenetic modification is crucial for the maintenance of the SSC pool and the support of long-term sperm production. In the present study, we concluded that aberrant histone H3 lysine methylation leads to the downregulation of PLZF, which in turn causes defects in SSC maintenance. However, other unknown mechanisms that are regulated by histone modification are probably also involved in this process.
H4R3me2s is a repressive histone arginine modification that is catalyzed by PRMT5 (Zhao et al., 2009; Xu et al., 2010; Deng et al., 2017; Li et al., 2018; Liu M. et al., 2020). The global level of H4R3me2s was dramatically reduced in Prmt5-deficient SSCs, consistent with the results of previous studies (Zhao et al., 2009; Wang et al., 2015c; Zhu et al., 2019). Surprisingly, H4R3me2s was enriched at the promoter region of the JMJD1C gene in Prmt5-deficient SSCs. These results suggest that H4R3me2s is probably also catalyzed by other arginine methyltransferases and that the increase in H4R3me2s at the promoter region of the JMJD1C gene is probably not directly regulated by PRMT5. H3R2me2s is a permissive histone arginine modification that is also catalyzed by PRMT5 (Kirmizis et al., 2007; Migliori et al., 2012; Yuan et al., 2012; Chiang et al., 2017; Yang et al., 2020). The global level of H3R2me2s was dramatically reduced in Prmt5-deficient SSCs, and the levels of H3R2me2s at the promoter regions of JMJD1A, JMJD1B, and KDM6B were all significantly decreased. Based on these results, we concluded that the change in histone arginine methylation in Prmt5-deficient SSCs causes the downregulation of histone lysine demethylases. The downregulation of histone lysine demethylase expression causes an increase in H3K9me2 and H3K27me2.
In the present study, we also observed that meiosis was completely blocked in Prmt5-deficient germ cells, and no SYCP3 signal was detected in Prmt5Δ/flox;Mvh-Cre mice at P10. The results of our previous study revealed that deletion of Prmt5 in male germ cells using Stra8-Cre (∼P3) results in aberrant meiotic progression. However, the expression of meiosis-associated genes, such as Stra8, Sycp3, Dmc1, and γH2AX, was not affected (Wang et al., 2015c). Although meiosis was blocked in Prmt5-deficient germ cells, we could not conclude that PRMT5 is required for meiosis initiation. The defect of meiosis is probably a consequence of cell cycle arrest of Prmt5-deficient SSCs, and the underlying mechanism needs further investigation.
Taken together, the results of the present study reveal that PRMT5 is involved in regulating the development of spermatogonia and that deletion of Prmt5 results in depletion of the spermatogonial stem cells pool. We also demonstrated that loss of Prmt5 caused downregulation of demethylases for H3K9me2 and H3K27me2, which in turn led to an increase in H3K9me2 and H3K27me2 and downregulation of the Plzf gene (Figure 8). Our results demonstrated that the crosstalk between histone arginine methylations and histone lysine methylation plays an important role in regulating SSCs development.
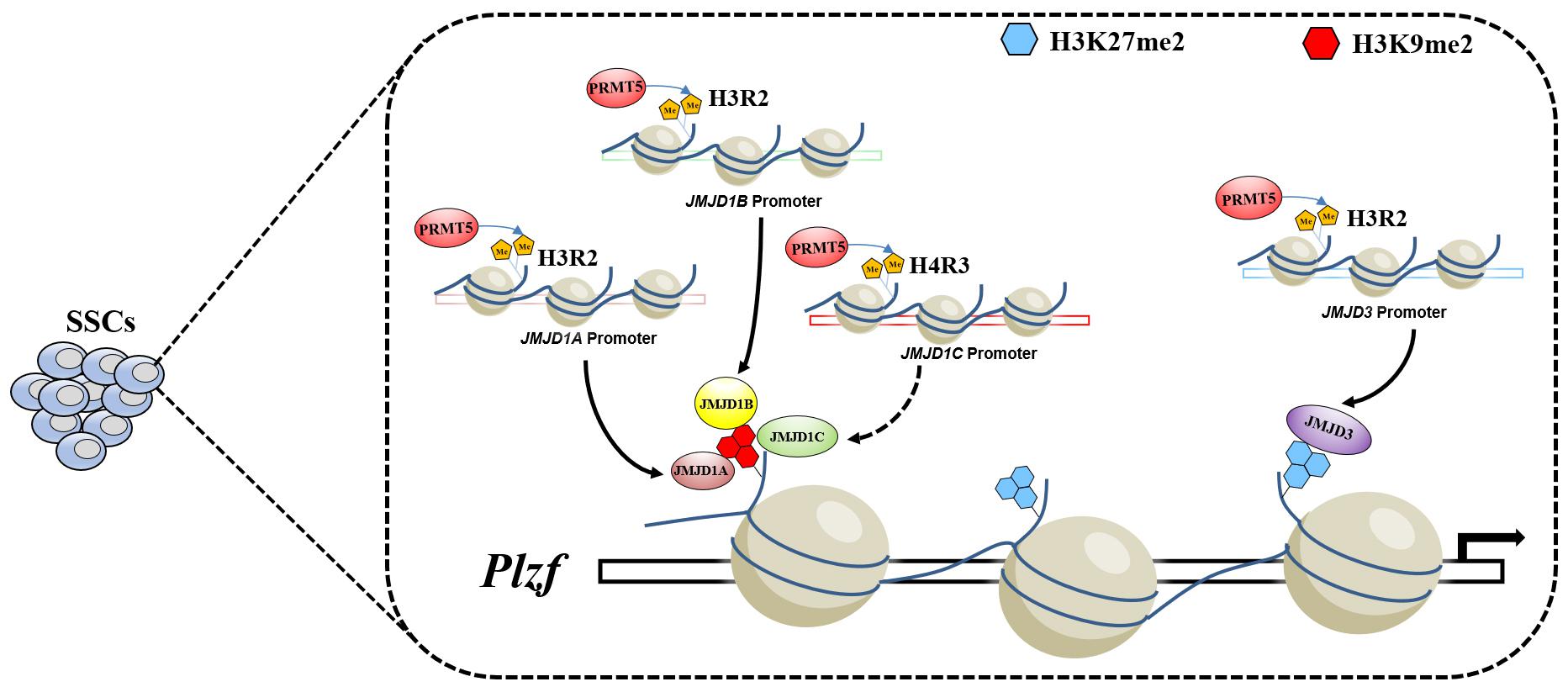
Figure 8. Model of PRMT5 regulating Plzf expression via histone modifications in SSCs. In SSCs, PRMT5 promotes the expression of demethylases for H3K9me2 and H3K27me2 by catalyzing permissive histone H3R2me2s. The high expression of the corresponding lysine demethylases leads to low levels of H3K9me2 and H3K27me2 at the upstream promoter of Plzf, which in turn leads to the upregulation of the Plzf gene.
All mice experiments were carried out in accordance with institutional animal care and the use committee regulations of Institute of Zoology, CAS. All mice were maintained in a C57BL/6;129/SvEv mixed background. Prmt5flox mice were obtained from the European Conditional Mouse Mutagenesis Program (EUCOMM; Prmt5tm2a(EUCOMM)Wtsi) (Bezzi et al., 2013), Prmt5+/Δ mice were obtained by crossing with ZP3-Cre mice. Prmt5Δ/flox;Mvh-Cre mice were obtained by crossing Prmt5+/Δ;Mvh-Cre males with Prmt5flox/flox females. Genotyping was performed by PCR as described previously using DNA isolated from tail tips (Gao et al., 2006; Bezzi et al., 2013; Gao et al., 2014).
Testes were dissected from Prmt5+/Δ;Mvh-Cre and control mice immediately after euthanasia, fixed in 4% paraformaldehyde for up to 24 h, stored in 70% ethanol, and embedded in paraffin. Five-micrometer-thick sections were cut and mounted on glass slides. Then, the tissue sections were processed for immunohistochemistry and immunofluorescence.
IHC and IF procedures were performed as described previously (Chen et al., 2019; Qin et al., 2019). Antibodies were diluted as follows: MVH (1:500, Abcam, ab13840), PLZF (1:100, R&D, AF2944), PRMT5 (1:200, Millipore, 07-405), SOX9 (1:500, Millipore, AB5535), STRA8 (1:200, Abcam, ab49405), SYCP3 (1:200, Abcam, ab15093), Ki67 (1:400, Abcam, ab15580), and PH3 (1:400, Millipore, 2605439). After staining, the sections were examined with a Nikon microscopy, and images were captured with a Nikon DS-Ri1 CCD camera. The IF sections were examined using a confocal laser scanning microscope (Carl Zeiss Inc., Thornwood, NY, United States). TUNEL assay was performed using the Dead-End Fluorometric TUNEL System (Promega, G3250).
Establishment and maintenance of SSCs were performed as previously described (Wang et al., 2015a; Chen et al., 2017). In brief, seminiferous tubules from Prmt5flox/flox;Cre-ERTM mice at 5–7 days postpartum (dpp) were digested with collagenase IV and DNase I for 5 min into small fragments and then centrifuged at 400 rpm for 2 min. The seminiferous fragments were suspended in mouse embryo fibroblast (MEF) medium containing 10% FBS (fetal bovine serum). Twenty-four hours later, the SSCs were collected and then transferred to mitomycin C-treated MEF feeder cells. The specific medium for SSCs contained GDNF and FGF2 for later culture. Prmt5flox/flox;Cre-ERTM SSCs treated with ethanol or 1 μM tamoxifen were harvested for western blot, real-time PCR or MTT assay.
Western blotting procedures were performed as described previously (Zhou et al., 2018). Tissues and cells were lysed in cold RIPA buffer, supplemented with 1 mM phenylmethylsulfonyl fluoride and protease inhibitor cocktail (Roche, Indianapolis, IN, United States). The protein lysates were subjected to SDS-PAGE, transferred onto a nitrocellulose membrane and probed with the primary antibodies. The images were captured with the ODYSSEY Sa Infrared Imaging System (LI-COR Biosciences, Lincoln, NE, United States). Primary antibodies used were shown in Supplementary Table 1.
For Real-Time PCR, RNA was isolated from cultured SSCs using EASYspin Plus Tissue/Cells RNA Rapid Extraction kit following manufacturer’s instructions. The relative expression level was calculated using the formula 2–ΔΔCT. Gapdh was used as an internal control for quantification. The primers used were listed in Supplementary Table 2.
The relative number and viability of SSCs were evaluated by MTT assays. In brief, SSCs seeded in 24-well plates were washed with PBS twice and incubated with MTT solution (0.5 mg/ml) for at least 4 h at 37°C in a CO2 incubator. Then, the medium containing MTT was removed, and 750 μl DMSO was added. After an incubation for 10 min on a shaking table at 75 rpm/min, the OD value at 490 nm was measured.
ChIP assays were conducted using a SimpleChIP® Plus Sonication Chromatin IP Kit (# 56383) following the manufacturer’s instructions. For ChIP-qPCR, the input genomic DNA or immunoprecipitated DNA was used as a template. Quantitative real-time PCR was performed according to the manufacturer’s instructions. The input DNA was used as a normalization control. The primers used are listed in Supplementary Table 2.
Experiments were repeated at least three times. GraphPad Prism7 software was used for analysis of P-values based on three to six independent experiments in PCR reactions or western blotting assay. Multiple t-test-one per row was used to compare the differences between two groups. P < 0.05 or P < 0.01 was considered statistically significant.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of the Institute of Zoology, CAS (AEI-09-02-2014).
FD and MC (co-first author): formal analysis and investigation. FG: funding acquisition. FD, MC (co-first author), MC (third author), LJ, ZS, LM, CH, and XG: methodology. FG: project administration. MC (co-first author) and LM: resources. FD: writing—original draft. FD and FG: writing—review and editing.
This work was supported by the National Key R&D Program of China (2016YFA0500901, 2018YFA0107702, and 2018YFC1004200), Strategic Priority Research Program of the Chinese Academy of Sciences (XDB19000000), the National Science Fund for Distinguished Young Scholars (81525011), and the National Natural Science Foundation of China (31970785 and 31671496).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.673258/full#supplementary-material
Agrawal Singh, S., Lerdrup, M., Gomes, A. R., van de Werken, H. J., Vilstrup Johansen, J., Andersson, R., et al. (2019). PLZF targets developmental enhancers for activation during osteogenic differentiation of human mesenchymal stem cells. Elife 8:e40364. doi: 10.7554/eLife.40364
Ancelin, K., Lange, U. C., Hajkova, P., Schneider, R., Bannister, A. J., Kouzarides, T., et al. (2006). Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat. Cell Biol. 8, 623–630. doi: 10.1038/ncb1413
Avantaggiato, V., Pandolfi, P. P., Ruthardt, M., Hawe, N., Acampora, D., Pelicci, P. G., et al. (1995). Developmental Analysis of Murine Promyelocyte Leukemia Zinc Finger (PLZF) gene expression_ implications for the neuromeric model of the forebrain organization. J. Neurosci. 15(7 Pt. 1), 4927–4942. doi: 10.1523/JNEUROSCI.15-07-04927.1995
Barna, M., Hawe, N., Niswander, L., and Pandolfi, P. P. (2000). Plzf regulates limb and axial skeletal patterning. Nat. Genet. 25, 166–172. doi: 10.1038/76014
Bedford, M. T. (2007). Arginine methylation at a glance. J. Cell Sci. 120, 4243–4246. doi: 10.1242/jcs.019885
Bezzi, M., Teo, S. X., Muller, J., Mok, W. C., Sahu, S. K., Vardy, L. A., et al. (2013). Regulation of constitutive and alternative splicing by PRMT5 reveals a role for Mdm4 pre-mRNA in sensing defects in the spliceosomal machinery. Genes Dev. 27, 1903–1916. doi: 10.1101/gad.219899.113
Buaas, F. W., Kirsh, A. L., Sharma, M., McLean, D. J., Morris, J. L., Griswold, M. D., et al. (2004). Plzf is required in adult male germ cells for stem cell self-renewal. Nat. Genet. 36, 647–652. doi: 10.1038/ng1366
Chen, J., Cai, T., Zheng, C., Lin, X., Wang, G., Liao, S., et al. (2017). MicroRNA-202 maintains spermatogonial stem cells by inhibiting cell cycle regulators and RNA binding proteins. Nucleic Acids Res. 45, 4142–4157. doi: 10.1093/nar/gkw1287
Chen, M., Chen, M., Chen, S., Zhou, J., Dong, F., Shen, Z., et al. (2019). Abnormal meiosis initiation in germ cell caused by aberrant differentiation of gonad somatic cell. Oxid. Med. Cell. Longev. 2019:8030697. doi: 10.1155/2019/8030697
Chen, S. R., and Liu, Y. X. (2015). Regulation of spermatogonial stem cell self-renewal and spermatocyte meiosis by Sertoli cell signaling. Reproduction 149, R159–R167. doi: 10.1530/REP-14-0481
Chiang, K., Zielinska, A. E., Shaaban, A. M., Sanchez-Bailon, M. P., Jarrold, J., Clarke, T. L., et al. (2017). PRMT5 is a critical regulator of breast cancer stem cell function via histone methylation and FOXP1 expression. Cell Rep. 21, 3498–3513. doi: 10.1016/j.celrep.2017.11.096
Cook, M., Gould, A., Brand, N., Davies, J., Strutt, P., Shaknovich, R., et al. (1995). Expression of the zinc-finger gene PLZF at rhombomere boundaries in the vertebrate hindbrain. Proc. Natl. Acad. Sci. U.S.A. 92, 2249–2253. doi: 10.1073/pnas.92.6.2249
Costoya, J. A., Hobbs, R. M., Barna, M., Cattoretti, G., Manova, K., Sukhwani, M., et al. (2004). Essential role of Plzf in maintenance of spermatogonial stem cells. Nat. Genet. 36, 653–659. doi: 10.1038/ng1367
Deng, X., Shao, G., Zhang, H. T., Li, C., Zhang, D., Cheng, L., et al. (2017). Protein arginine methyltransferase 5 functions as an epigenetic activator of the androgen receptor to promote prostate cancer cell growth. Oncogene 36, 1223–1231. doi: 10.1038/onc.2016.287
Dong, J., Wang, X., Cao, C., Wen, Y., Sakashita, A., Chen, S., et al. (2019). UHRF1 suppresses retrotransposons and cooperates with PRMT5 and PIWI proteins in male germ cells. Nat. Commun. 10:4705. doi: 10.1038/s41467-019-12455-4
Gallardo, T., Shirley, L., John, G. B., and Castrillon, D. H. (2007). Generation of a germ cell-specific mouse transgenic Cre line, Vasa-Cre. Genesis 45, 413–417. doi: 10.1002/dvg.20310
Gao, F., Maiti, S., Alam, N., Zhang, Z., Deng, J. M., Behringer, R. R., et al. (2006). The Wilms tumor gene, Wt1, is required for Sox9 expression and maintenace of tubular architecture in the developing testis. Proc. Natl. Acad. Sci. U.S.A. 103, 11987–11992. doi: 10.1073/pnas.0600994103
Gao, F., Zhang, J., Wang, X., Yang, J., Chen, D., Huff, V., et al. (2014). Wt1 functions in ovarian follicle development by regulating granulosa cell differentiation. Hum. Mol. Genet. 23, 333–341. doi: 10.1093/hmg/ddt423
Hai, L., Szwarc, M. M., Lanza, D. G., Heaney, J. D., and Lydon, J. P. (2019). Using CRISPR/Cas9 engineering to generate a mouse with a conditional knockout allele for the promyelocytic leukemia zinc finger transcription factor. Genesis 57:e23281. doi: 10.1002/dvg.23281
Hamard, P. J., Santiago, G. E., Liu, F., Karl, D. L., Martinez, C., Man, N., et al. (2018). PRMT5 regulates DNA repair by controlling the alternative splicing of histone-modifying enzymes. Cell Rep. 24, 2643–2657. doi: 10.1016/j.celrep.2018.08.002
He, Z., Jiang, J., Kokkinaki, M., Golestaneh, N., Hofmann, M. C., and Dym, M. (2008). Gdnf upregulates c-Fos transcription via the Ras/Erk1/2 pathway to promote mouse spermatogonial stem cell proliferation. Stem Cells 26, 266–278. doi: 10.1634/stemcells.2007-0436
Iwamori, N., Iwamori, T., and Matzuk, M. M. (2013). H3K27 demethylase, JMJD3, regulates fragmentation of spermatogonial cysts. PLoS One 8:e72689. doi: 10.1371/journal.pone.0072689
Kim, S., Gunesdogan, U., Zylicz, J. J., Hackett, J. A., Cougot, D., Bao, S., et al. (2014). PRMT5 protects genomic integrity during global DNA demethylation in primordial germ cells and preimplantation embryos. Mol. Cell 56, 564–579. doi: 10.1016/j.molcel.2014.10.003
Kirmizis, A., Santos-Rosa, H., Penkett, C. J., Singer, M. A., Vermeulen, M., Mann, M., et al. (2007). Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature 449, 928–932. doi: 10.1038/nature06160
Kuroki, S., Akiyoshi, M., Tokura, M., Miyachi, H., Nakai, Y., Kimura, H., et al. (2013). JMJD1C, a JmjC domain-containing protein, is required for long-term maintenance of male germ cells in mice. Biol. Reprod. 89:93. doi: 10.1095/biolreprod.113.108597
Kuroki, S., Maeda, R., Yano, M., Kitano, S., Miyachi, H., Fukuda, M., et al. (2020). H3K9 demethylases JMJD1A and JMJD1B control prospermatogonia to spermatogonia transition in mouse germline. Stem Cell Rep. 15, 424–438. doi: 10.1016/j.stemcr.2020.06.013
Li, S., Ali, S., Duan, X., Liu, S., Du, J., Liu, C., et al. (2018). JMJD1B demethylates H4R3me2s and H3K9me2 to facilitate gene expression for development of hematopoietic stem and progenitor cells. Cell Rep. 23, 389–403. doi: 10.1016/j.celrep.2018.03.051
Li, Z., Yu, J., Hosohama, L., Nee, K., Gkountela, S., Chaudhari, S., et al. (2015). The Sm protein methyltransferase PRMT5 is not required for primordial germ cell specification in mice. EMBO J. 34, 748–758. doi: 10.15252/embj.201489319
Liu, F., Xu, Y., Lu, X., Hamard, P. J., Karl, D. L., Man, N., et al. (2020). PRMT5-mediated histone arginine methylation antagonizes transcriptional repression by polycomb complex PRC2. Nucleic Acids Res. 48, 2956–2968. doi: 10.1093/nar/gkaa065
Liu, M., Yao, B., Gui, T., Guo, C., Wu, X., Li, J., et al. (2020). PRMT5-dependent transcriptional repression of c-Myc target genes promotes gastric cancer progression. Theranostics 10, 4437–4452. doi: 10.7150/thno.42047
Liu, T. M., Lee, E. H., Lim, B., and Shyh-Chang, N. (2016). Concise Review: balancing stem cell self-renewal and differentiation with PLZF. Stem Cells 34, 277–287. doi: 10.1002/stem.2270
Mäkelä, J. A., and Hobbs, R. M. (2019). Molecular regulation of spermatogonial stem cell renewal and differentiation. Reproduction 158, R169–R187. doi: 10.1530/REP-18-0476
Meng, X., Lindahl, M., Hyvönen, M. E., Parvinen, M., de Rooij, D. G., Hess, M. W., et al. (2000). Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science 287, 1489–1493. doi: 10.1126/science.287.5457.1489
Migliori, V., Muller, J., Phalke, S., Low, D., Bezzi, M., Mok, W. C., et al. (2012). Symmetric dimethylation of H3R2 is a newly identified histone mark that supports euchromatin maintenance. Nat. Struct. Mol. Biol. 19, 136–144. doi: 10.1038/nsmb.2209
Mu, W., Starmer, J., Fedoriw, A. M., Yee, D., and Magnuson, T. (2014). Repression of the soma-specific transcriptome by Polycomb-repressive complex 2 promotes male germ cell development. Genes Dev. 28, 2056–2069. doi: 10.1101/gad.246124.114
Oatley, J. M., and Brinster, R. L. (2008). Regulation of spermatogonial stem cell self-renewal in mammals. Annu. Rev. Cell Dev. Biol. 24, 263–286. doi: 10.1146/annurev.cellbio.24.110707.175355
Pahlich, S., Zakaryan, R. P., and Gehring, H. (2006). Protein arginine methylation: cellular functions and methods of analysis. Biochim. Biophys. Acta 1764, 1890–1903. doi: 10.1016/j.bbapap.2006.08.008
Patel, S. R., Bhumbra, S. S., Paknikar, R. S., and Dressler, G. R. (2012). Epigenetic mechanisms of Groucho/Grg/TLE mediated transcriptional repression. Mol. Cell 45, 185–195. doi: 10.1016/j.molcel.2011.11.007
Poplineau, M., Vernerey, J., Platet, N., N’Guyen, L., Herault, L., Esposito, M., et al. (2019). PLZF limits enhancer activity during hematopoietic progenitor aging. Nucleic Acids Res. 47, 4509–4520. doi: 10.1093/nar/gkz174
Qin, Y., Zhou, Y., Shen, Z., Xu, B., Chen, M., Li, Y., et al. (2019). WDR62 is involved in spindle assembly by interacting with CEP170 in spermatogenesis. Development 146:dev174128. doi: 10.1242/dev.174128
Raposo, A. E., and Piller, S. C. (2018). Protein arginine methylation: an emerging regulator of the cell cycle. Cell Div. 13:3. doi: 10.1186/s13008-018-0036-2
Reid, A., Gould, A., Brand, N., Cook, M., Strutt, P., Li, J., et al. (1995). Leukemia translocation gene, PLZF, is expressed with a speckled nuclear pattern in early hematopoietic progenitors. Blood 86, 4544–4552. doi: 10.1182/blood.V86.12.4544.bloodjournal86124544
Rho, J., Choi, S., Seong, Y. R., Cho, W. K., Kim, S. H., and Im, D. S. (2001). Prmt5, which forms distinct homo-oligomers, is a member of the protein-arginine methyltransferase family. J. Biol. Chem. 276, 11393–11401. doi: 10.1074/jbc.M008660200
Shaknovich, R., Yeyati, P. L., Ivins, S., Melnick, A., Lempert, C., Waxman, S., et al. (1998). The promyelocytic leukemia zinc finger protein affects myeloid cell growth, differentiation, and apoptosis. Mol. Cell. Biol. 18, 5533–5545. doi: 10.1128/MCB.18.9.5533
Stopa, N., Krebs, J. E., and Shechter, D. (2015). The PRMT5 arginine methyltransferase: many roles in development, cancer and beyond. Cell Mol. Life Sci. 72, 2041–2059. doi: 10.1007/s00018-015-1847-9
Suliman, B. A., Xu, D., and Williams, B. R. (2012). The promyelocytic leukemia zinc finger protein: two decades of molecular oncology. Front. Oncol. 2:74. doi: 10.3389/fonc.2012.00074
Tae, S., Karkhanis, V., Velasco, K., Yaneva, M., Erdjument-Bromage, H., Tempst, P., et al. (2011). Bromodomain protein 7 interacts with PRMT5 and PRC2, and is involved in transcriptional repression of their target genes. Nucleic Acids Res. 39, 5424–5438. doi: 10.1093/nar/gkr170
Tseng, Y. T., Liao, H. F., Yu, C. Y., Mo, C. F., and Lin, S. P. (2015). Epigenetic factors in the regulation of prospermatogonia and spermatogonial stem cells. Reproduction 150, R77–R91. doi: 10.1530/REP-14-0679
Vincent-Fabert, C., Platet, N., Vandevelde, A., Poplineau, M., Koubi, M., Finetti, P., et al. (2016). PLZF mutation alters mouse hematopoietic stem cell function and cell cycle progression. Blood 127, 1881–1885. doi: 10.1182/blood-2015-09-666974
Wang, S., Wang, X., Ma, L., Lin, X., Zhang, D., Li, Z., et al. (2016). Retinoic acid is sufficient for the in vitro induction of mouse spermatocytes. Stem Cell Rep. 7, 80–94. doi: 10.1016/j.stemcr.2016.05.013
Wang, S., Wang, X., Wu, Y., and Han, C. (2015a). IGF-1R signaling is essential for the proliferation of cultured mouse spermatogonial stem cells by promoting the G2/M progression of the cell cycle. Stem Cells Dev. 24, 471–483. doi: 10.1089/scd.2014.0376
Wang, Y., Li, Q., Liu, C., Han, F., Chen, M., Zhang, L., et al. (2015b). Protein arginine methyltransferase 5 (Prmt5) is required for germ cell survival during mouse embryonic development. Biol. Reprod. 92:104. doi: 10.1095/biolreprod.114.127308
Wang, Y., Zhu, T., Li, Q., Liu, C., Han, F., Chen, M., et al. (2015c). Prmt5 is required for germ cell survival during spermatogenesis in mice. Sci. Rep. 5:11031. doi: 10.1038/srep11031
Wu, Y., Hu, X., Li, Z., Wang, M., Li, S., Wang, X., et al. (2016). Transcription factor RFX2 is a key regulator of mouse spermiogenesis. Sci. Rep. 6:20435. doi: 10.1038/srep20435
Xu, X., Hoang, S., Mayo, M. W., and Bekiranov, S. (2010). Application of machine learning methods to histone methylation ChIP-Seq data reveals H4R3me2 globally represses gene expression. BMC Bioinformatics 11:396. doi: 10.1186/1471-2105-11-396
Yang, M., Lin, X., Segers, F., Suganthan, R., Hildrestrand, G. A., Rinholm, J. E., et al. (2020). OXR1A, a coactivator of PRMT5 regulating histone arginine methylation. Cell Rep. 30, 4165–4178.e7. doi: 10.1016/j.celrep.2020.02.063
Yeyati, P. L., Shaknovich, R., Boterashvili, S., Li, J., Ball, H. J., Waxman, S., et al. (1999). Leukemia translocation protein PLZF inhibits cell growth and expression of cyclin A. Oncogene 18, 925–934. doi: 10.1038/sj.onc.1202375
Yuan, C. C., Matthews, A. G., Jin, Y., Chen, C. F., Chapman, B. A., Ohsumi, T. K., et al. (2012). Histone H3R2 symmetric dimethylation and histone H3K4 trimethylation are tightly correlated in eukaryotic genomes. Cell Rep. 1, 83–90. doi: 10.1016/j.celrep.2011.12.008
Zhang, T., Gunther, S., Looso, M., Kunne, C., Kruger, M., Kim, J., et al. (2015). Prmt5 is a regulator of muscle stem cell expansion in adult mice. Nat. Commun. 6:7140. doi: 10.1038/ncomms8140
Zhang, Y., Wang, S., Wang, X., Liao, S., Wu, Y., and Han, C. (2012). Endogenously produced FGF2 is essential for the survival and proliferation of cultured mouse spermatogonial stem cells. Cell Res. 22, 773–776. doi: 10.1038/cr.2012.17
Zhao, Q., Rank, G., Tan, Y. T., Li, H., Moritz, R. L., Simpson, R. J., et al. (2009). PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nat. Struct. Mol. Biol. 16, 304–311. doi: 10.1038/nsmb.1568
Zhou, Y., Qin, Y., Qin, Y., Xu, B., Guo, T., Ke, H., et al. (2018). Wdr62 is involved in female meiotic initiation via activating JNK signaling and associated with POI in humans. PLoS Genet. 14:e1007463. doi: 10.1371/journal.pgen.1007463
Keywords: PRMT5, spermatogonial stem cells, histone lysine modification, lysine demethylase, PLZF
Citation: Dong F, Chen M, Chen M, Jiang L, Shen Z, Ma L, Han C, Guo X and Gao F (2021) PRMT5 Is Involved in Spermatogonial Stem Cells Maintenance by Regulating Plzf Expression via Modulation of Lysine Histone Modifications. Front. Cell Dev. Biol. 9:673258. doi: 10.3389/fcell.2021.673258
Received: 27 February 2021; Accepted: 26 April 2021;
Published: 21 May 2021.
Edited by:
Liangran Zhang, Shandong University, ChinaReviewed by:
Wei Shen, Qingdao Agricultural University, ChinaCopyright © 2021 Dong, Chen, Chen, Jiang, Shen, Ma, Han, Guo and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chunsheng Han, aGFuY3NAaW96LmFjLmNu; Xudong Guo, Yml4ZGd1b0BpbXUuZWR1LmNu; Fei Gao, Z2FvZkBpb3ouYWMuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.