- 1State Key Laboratory of Subtropical Silviculture, School of Forestry and Biotechnology, Zhejiang A&F University, Hangzhou, China
- 2Department of Chemistry and Molecular Biology, University of Gothenburg, Gothenburg, Sweden
- 3Faculty of Science, Center for Large-Scale Cell-Based Screening, University of Gothenburg, Gothenburg, Sweden
Stress granules (SGs) are highly dynamic cytoplasmic foci formed in response to stress. The formation of SGs is reported to be regulated by diverse post-translational protein modifications (PTMs). Among them, ADP-ribosylation is of emerging interest due to its recently identified roles in SG organization. In this review, we summarized the latest advances on the roles of poly(ADP-ribose) (PAR) in the regulation of SG formation and dynamics, including its function in modulating nucleocytoplasmic trafficking and SG recruitment of SG components, as well as its effects on protein phase separation behavior. Moreover, the functional role of PAR chain diversity on dynamic of SG composition is also introduced. Potential future developments on investigating global ADP-ribosylation networks, individual roles of different PARPs, and interactions between ADP-ribosylation and other PTMs in SGs are also discussed.
Introduction
Stress granules (SGs) are cytoplasmic membraneless structures that rapidly assemble in cells in response to a variety of stresses, including heat, oxidative stress, and virus infection (Buchan and Parker, 2009). These liquid-like, higher-ordered condensates consist of stalled untranslated mRNA, ribosome proteins, initiation factors, diverse RNA-binding proteins (RBPs) and non-RBPs (Kedersha et al., 2005). Formation of SGs enhances cell survival by protecting certain RNAs and proteins from decay upon exposure to adverse environmental conditions, as well as by regulating intracellular signal transduction to overcome detrimental environmental conditions. As soon as the stress is relieved, SGs are disassembled to allow the mRNAs back into the translation machinery so that protein synthesis can be rapidly re-initiated (Kedersha et al., 2013; Cao et al., 2020). Continuous formation of SGs or disrupted disassembly of SGs results in aberrant conversion of SG into a pathogenic state. This type of pathogenic state is associated with a barrage of diseases, including amyotrophic lateral sclerosis (ALS), Alzheimer’s disease, and Parkinson’s disease (Alberti and Dormann, 2019; Falahati and Haji-Akbari, 2019). Thus, precise modulation of SG dynamics is critical to the maintenance of normal physiological functioning of cells. Previous studies have reported that protein post-translational modification (PTM) is one of the mechanisms by which cells control the assembly and disassembly of SGs (Buchan and Parker, 2009; Cao et al., 2020). In this mini-review, we focus on how ADP-ribosylation, an important PTM, participates in the regulation of the phase behavior of SG components and influences the formation and dynamics of SGs.
ADP-Ribosylation
ADP-Ribosylation is a conserved, reversible PTM of proteins best known for its function in a multitude of cellular processes, such as stress response, DNA repair, signal transduction, and apoptosis (Corda and Di Girolamo, 2003; Perina et al., 2014; Bai, 2015; Qi et al., 2019). This PTM regulates protein functions via two manners: covalent modification of substrates and ADP-ribose-mediated non-covalent association with substrates (Grimaldi et al., 2019). The first relies on the covalent transfer of ADP-ribose (one or more units) to targets; namely, mono(ADP-ribosylation) (MARylation) or PARylation (Leung, 2020). Alternatively, some proteins can non-covalently bind to MARylated or PARylated targets through ADP-ribose-binding domains. This ADP-ribose-binding property is also important for their functional regulation (Krietsch et al., 2013; Cheruiyot et al., 2015; Zhang et al., 2019). PAR formation is catalyzed by a family of PAR polymerases (PARPs) (Otto et al., 2005). Some PARPs function to transfer a single ADP-ribose onto the target, while others have the capacity to add subsequent ADP-ribose units to extend MARylation (Vyas et al., 2014). The reverse process is fulfilled by PAR glycohydrolase (PARG), which cleaves the ribose-ribose bond and removes the ADPr unit from the PAR chain (Slade et al., 2011). However, this enzyme cannot remove the terminal ADP-ribose; the final ADP-ribose on the target needs to be hydrolyzed by several degraders. There are three macrodomain-containing proteins, MacroD1, MacroD2, and TARG1, that can specifically remove the terminal ADPr on Glu and Asp. In addition, ARH1 (Arg specific) and ARH3 (both PAR chains and Ser specific for the terminal ADPr) are also relevant (Jankevicius et al., 2013; Rosenthal et al., 2013; Sharifi et al., 2013). PARylated proteins can be covalently modified on different amino acids, including Glu, Asp, Arg, Ser, Cys, and Tyr (Gupte et al., 2017; Palazzo et al., 2017; Crawford et al., 2018; Leslie Pedrioli et al., 2018; Luscher et al., 2018; O’Sullivan et al., 2019). PAR-binding proteins can bind to free PAR or PARylated proteins via multiple domains, including macrodomain, PAR-binding motif (PBM), poly(ADP-ribose)-binding zinc-finger (PBZ) domain, tryptophan-tryptophan-glutamate (WWE) domain, oligosaccharide-binding fold domain (OB fold), RNA recognition motif (RRM), arginine-glycine-glycine motif (RGG), PilT N-terminus (PIN) domain, and WD40 domain (Grimaldi et al., 2019). For example, macrodomains are evolutionarily conserved domains which have high-affinity to ADP-ribose, which usually bind to mono-ADP-ribose and the terminal ADP-ribose (Karras et al., 2005; Ahel et al., 2009). PBM is enriched of basic and hydrophobic amino acids, and the positively charged residues in this domain could provide favorable electrostatic interactions with PAR (Pleschke et al., 2000; Fahrer et al., 2007). PBM-containing protein, Werner syndrome protein, can bind to PAR to regulate its enzymatic activities (Popp et al., 2013). PBZ domains usually recognize adjacent ADP-ribose groups through binding to adenines in two adjacent ADP-ribose units while WWE domain binds to iso-ADP-ribose (Li et al., 2010; Wang et al., 2012; Leung, 2017). Several PARPs possess WWE domains, such as PARP12. It requires its first WWE domain to bind ADP-ribose to regulate its translocation (Catara et al., 2017). It has also been found that the DNA-binding motif OB fold binds to PAR. OB fold-containing human ssDNA-binding protein 1 (hSSB1) has a high affinity with iso-ADP-ribose and this binding promote its recruitment to DNA damage sites (Zhang et al., 2014). RRM and RGG are abundant protein domains in eukaryotes, and many proteins containing these domains are involved in SG assembly, such as G3BP and FUS. They bind to PAR to regulate their functions (Isabelle et al., 2012; Mastrocola et al., 2013). Detailed structural features and more examples of proteins with these specialized binding domains have been extensively reviewed by Grimaldi et al. (2019).
Poly(ADP-Ribose) Regulates SG Formation and Dynamics Through Diverse Mechanisms
Recent studies have revealed that PAR functions in the control of SG assembly and disassembly. One interesting example is that the non-structural protein 3 (nsP3) of alphaviruses can remove the PARylation modification from the SG component G3BP1 and thus inhibit SG formation. This ability depends on the conserved macrodomain of nsP3 which function as a mono-ADP-ribosylhydrolase. Therefore, it is more likely that the nsP3 macrodomain prevents MARylation and subsequent indirectly PARylation, instead of PAR chain degradation (Eckei et al., 2017; Jayabalan et al., 2021). The role of PAR in SG regulation is also supported by the following evidence: Several PARPs, including PAR-adding PARP5a, MAR-adding PARP12, PARP14, and PARP15, inactive PARP13.1 and PARP13.2, and two PARGs, PARG99 and PARG102, are localized to cytoplasmic SGs (Leung et al., 2011; Isabelle et al., 2012; Catara et al., 2017). Moreover, overexpression of PARPs, including both MAP-adding and PAR-adding PARPs, can induce the formation of SGs (Leung et al., 2011). Along the same line of evidence, overexpression of PARGs can prevent SG formation. Furthermore, accumulation of PAR in the cell, caused by the absence of PARGs, delays disassembly of SGs (Leung et al., 2011; Leung, 2014; Catara et al., 2017). These evidences indicate that PAR, the product synergistically synthesized by these enzymes, plays a critical role in normal SG assembly, dynamics, and disassembly.
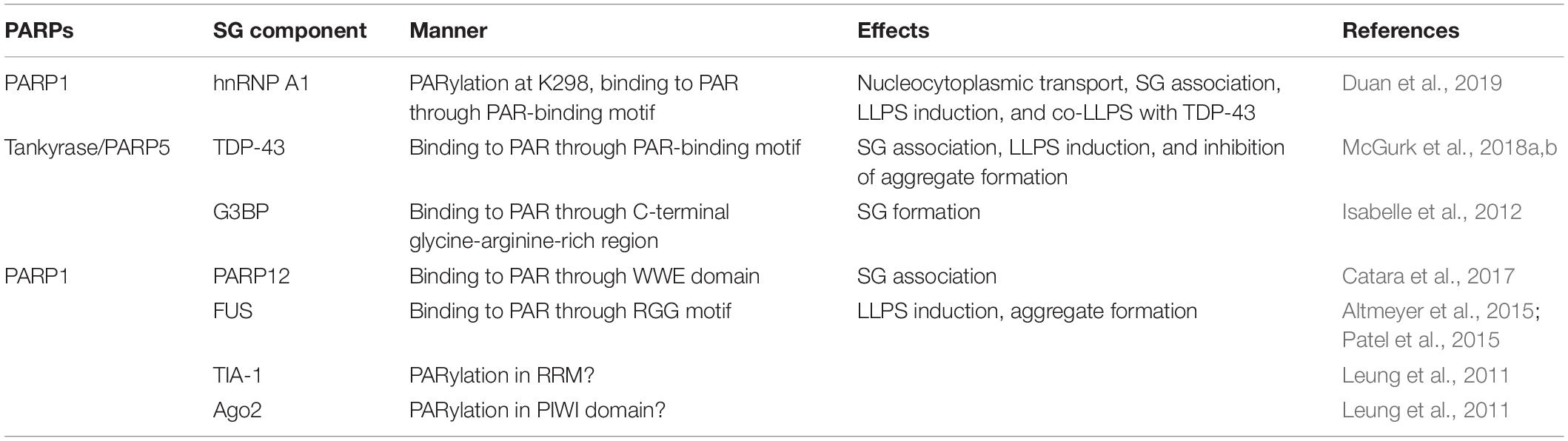
How does PAR regulate these SG related-processes? Recent in-depth studies have shown that PAR controls SG formation and dynamics via different mechanisms (Figure 1). The effects of PAR on SG components are summarized in Table 1.
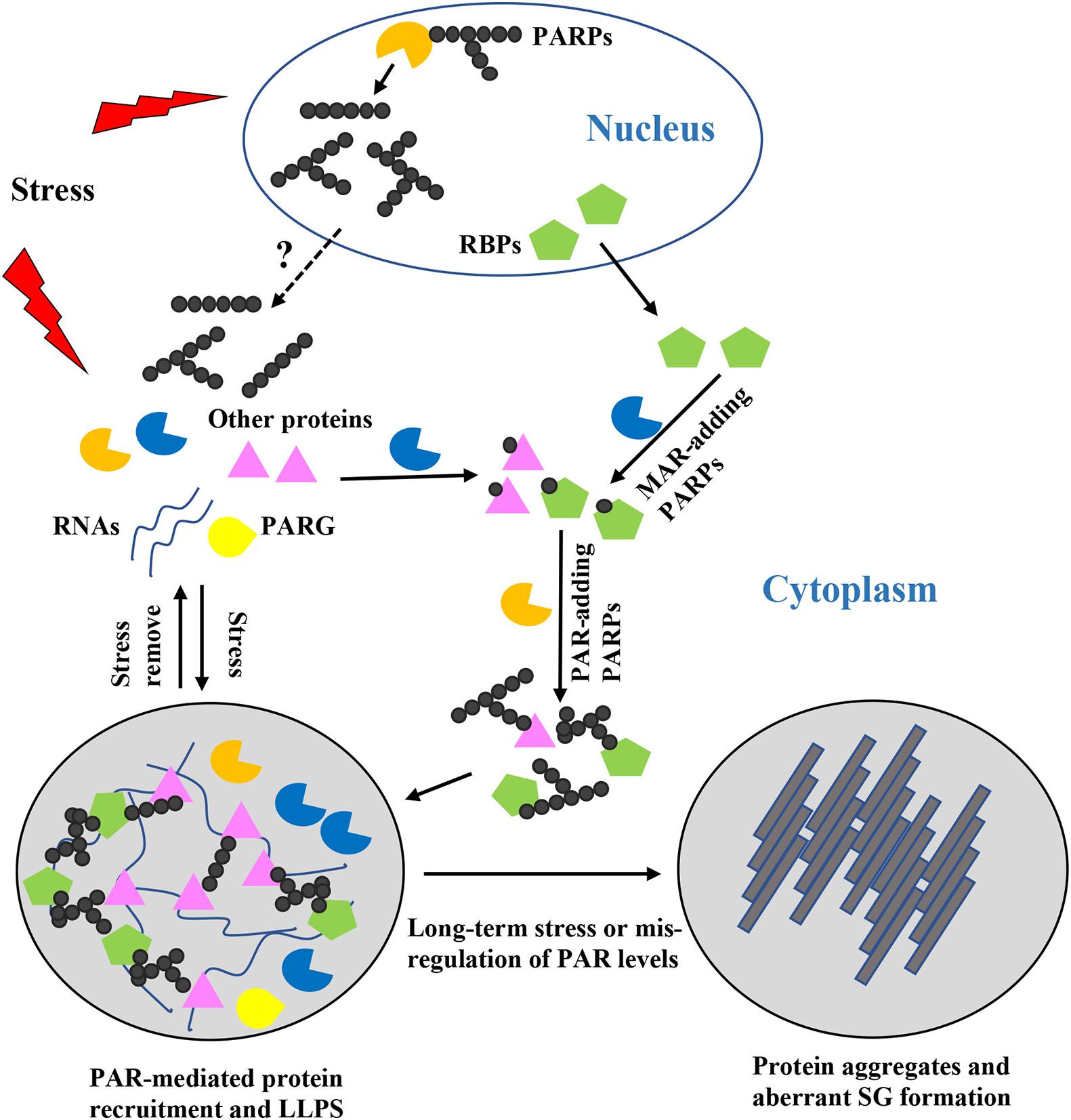
Figure 1. Roles of poly(ADP-ribose) (PAR) in stress granules (SG) formation, composition, and dynamics. When cells are exposed to stress, nuclear and cytoplasmic PARPs are activated, resulting in increased PAR chains with different lengths and structures. But how the PAR chains shuttle between the nucleus and the cytoplasm remains unclear. Formation of PAR chains is mediated through MAR, which might be a rate-limiting step. MAR-adding PARPs may act synergistically with PAR-adding PARPs to regulate stress granule formation. PARylated proteins or proteins bound to PARylated substrates are recruited to the specific sites where SG formation occurs and where they induce liquid-liquid phase separation. Diverse PAR chains contribute to diverse protein-protein and protein-RNA interactions. Once the stress is removed, PAR is degraded by PARG and the SGs are disassembled. However, if cells are exposed to long-term stress or PAR levels are mis-regulated, phase separated proteins are converted into protein aggregates and aberrant pathogenic SGs are formed.
Poly(ADP-Ribose) Regulates Proteins Targeting to SGs
Several studies have found that PAR is present in SGs, and that many proteins in SGs are PARylated or possess PAR-binding domains, such as heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1), TAR DNA binding protein 43 kDa (TDP-43), Ras-GTPase activating protein SH3 domain-binding protein (G3BP), argonaute family member Ago2, and T-cell intracellular antigen-1 (TIA-1) (Leung et al., 2011; Isabelle et al., 2012). For example, RNA-binding protein G3BP is a tunable switch that triggers RNA-dependent liquid-liquid phase separation and SG assembly (Guillén-Boixet et al., 2020; Yang et al., 2020). A recent study found that PAR binds to the G3BP C-terminal glycine-arginine-rich domain via non-covalent PAR binding. Such binding allows G3BP to maintain cytoplasmic localization and subsequent formation of SGs (Table 1; Isabelle et al., 2012). Moreover, PAR is required for PARP12 re-localization to SGs. Upon exposure to stress, nuclear PARP1 is activated and synthetic PAR polymers bind to the first WWE domain of PARP12, contributing to the translocation of PARP12 from the Golgi complex to SGs. This leads to the disassembly of Golgi membranes and blockage of anterograde-membrane traffic (Table 1; Catara et al., 2017). But so far, there is no clear evidence showing how the PAR chain is generated in the cytoplasm or whether the PAR chain is released from the nucleus. Translocation of TDP-43 is also PAR-dependent. Downregulation of a PARP in Drosophila reduces TDP-43 cytoplasmic location, and in mammalian cells and neurons, PAR- or PAR-scaffold-binding through PBM is required for TDP-43 accumulation in cytoplasmic SGs (Table 1; McGurk et al., 2018a). hnRNP A1 is another predominantly nuclear protein that can translocated to the cytoplasm upon exposure to stress (Geuens et al., 2016; Mohagheghi et al., 2016). It can be PARylated, and can also bind to PAR or PARylated proteins. Inhibition of PARP or PARG affects hnRNP A1 recruitment to or retrieval from SGs, indicating that PAR is critical for hnRNP A1’s SG location. Further analysis shows that PARylation and PAR-binding have different functions, as PARylation on K298 of hnRNP A1 is required for its cytoplasmic trafficking, while association with PAR or PARylated targets controls its sorting to SGs (Table 1; Duan et al., 2019). These studies provide strong support for the importance of PAR in regulating proteins targeting to SGs (Figure 1).
Poly(ADP-Ribose) Affects Protein Phase Separation Behavior of SG Components
It has been reported that SG formation is induced by liquid-liquid phase separation (LLPS) of intrinsically disordered proteins (IDPs) and RNAs (Hyman et al., 2014; Gomes and Shorter, 2019). IDPs lack defined 3D structures and are highly flexible, which allows diverse, promiscuous interactions that drive LLPS formation and protein condensation (Deiana et al., 2019). And a previous study demonstrated that certain RNAs can act as seeds that initiate phase separation and FUS-containing assembly formation (Schwartz et al., 2013). Recently, a new mechanism has arisen for this seeding event. Several groups have shown that PAR, a nucleic acid-mimicking biopolymer, can stimulate liquid demixing of IDPs or proteins with low-complexity regions in vitro or in vivo. For instance, a recent publication on apoptosis signal-regulating kinase 3 (ASK3) indicated that PAR could keep ASK3 condensates in the liquid phase and enable cells to sense osmotic stress (Watanabe et al., 2021). LLPS of SG components including fused in sarcoma (FUS), TDP-43, and hnRNP A1 are also regulated by PAR (Altmeyer et al., 2015; Patel et al., 2015; Kam et al., 2018; McGurk et al., 2018a; Duan et al., 2019; Singatulina et al., 2019). For instance, in vitro addition of PAR polymers promotes hnRNP A1 LLPS (Table 1). And the positive effect of PAR on phase separation of hnRNP A1 was not observed in a PAR-binding-deficient mutant, demonstrating that PAR-binding ability is required for this process. In addition, PAR-binding also regulates interactions between hnRNP A1 and TDP-43, and in vitro assays have shown that PAR addition enhances co-LLPS of these two proteins (Table 1; Duan et al., 2019). Moreover, PAR not only elevates LLPS of TDP-43, but also mitigates TDP-43 granulo-filamentous aggregation, which is predominantly found during disease (Table 1; McGurk et al., 2018b). Thus, association with PARylated scaffolds or PAR chains not only ensures the proper targeting of proteins to SGs, but also facilitates their LLPS (Figure 1; Duan et al., 2019).
It has been observed that PAR levels can affect protein phase separation behavior McGurk et al., 2018a; Duan et al., 2019). PAR levels are strictly regulated in cells by PARPs and PARGs, and their chain length varies from 2 to 200 ADP-ribose units. Therefore, PAR chains may serve as multivalent platforms for non-covalent binding of proteins (Leung, 2020). Upon exposure to stress, PARPs are activated and local PAR levels are rapidly increased; thus, PBM-containing proteins can sense the concentrated PAR and are recruited to specific sites. This is possibly why PARPs are included in SGs and PAR-binding ability is important for components anchoring to SGs (McGurk et al., 2018a; Duan et al., 2019). The longer the PAR chain and the more binding sites it provides, the more abundant the proteins are recruited and bind to the chain. Similarly, if a PAR is covalently conjugated on different amino acids of a single protein, it is possible that this covalently modified protein can create a scaffold for other PAR-binding proteins to increase the local concentration of macromolecules. Once the concentration exceeds threshold, LLPS starts and a membraneless condensate will be formed (Molliex et al., 2015; Cao et al., 2020). This is consistent with the fact that the addition of PAR promotes TDP-43 and hnRNP A1 LLPS in a dose-dependent manner (McGurk et al., 2018a; Duan et al., 2019). If two or more factors that initiate SG formation can interact with each other and be covalently modified at the same stage, they would provide a larger platform for subsequent interaction network formation. Since the PARylation of certain proteins depends on their PAR-binding ability (Fischbach et al., 2018; Kim et al., 2019), covalent modification events may initially occur and serve as triggers for the recruitment of PAR-binding proteins. Subsequently, these PAR-binding proteins are further covalently modified to finally form a complex protein-PAR interaction network. Once the stress is removed, PAR is degraded by PARG and higher-ordered condensates are disassembled (Figure 1). PAR can be produced in a short time in the nucleus, usually from seconds to minutes. For example, upon DNA damage, PARPs are recruited within seconds and PAR are rapidly synthesized at the DNA damage sites to repair DNA (Gupte et al., 2017; Palazzo et al., 2017; Luscher et al., 2018; O’Sullivan et al., 2019). It might give the cell an ability to respond quickly to environmental changes. However, little information is available concerning the PAR chain formation in SG. It might take longer time than the synthesis in the nucleus but this needs to be further investigated.
Mutations of RBPs such as TDP-43 and hnRNP A1 have been identified as associated with many neurodegenerative diseases, including ALS, frontotemporal degeneration (FTD), and myopathy. This has been correlated with the aggregation-prone propensities of these mutants in the cytoplasm, resulting in SGs nucleation and pathological SGs formation (Sreedharan et al., 2008; Kim et al., 2013; Prasad et al., 2019). Although PAR can promote LLPS, it is worth noting that when PAR levels increase in an in vitro phase separation system, the dynamics of the hnRNP A1 droplet are reduced (Duan et al., 2019). Moreover, PAR can accelerate the formation of pathogenic solid phase aggregates of an ALS-linked mutation of TDP-43 that is defective in LLPS ability (McGurk et al., 2018a). Other groups found that long-term incubation of RBPs with PAR accelerates the formation of disease-related solid phase aggregates (Altmeyer et al., 2015; Kam et al., 2018). The above-mentioned proteins are known to contain disordered domains, and these disordered domains can also cause them to be more aggregation-prone (Alberti et al., 2009). Therefore, in the early stage, PAR may have the ability to initiate and accelerate LLPS. But with time, if PAR continues to exist in high concentrations or under disease conditions, it may enhance the intrinsic aggregation propensity of proteins especially disease-associated mutants (Altmeyer et al., 2015; McGurk et al., 2018a; Duan et al., 2019; Liu and Fang, 2019). Hyperactivation of PARP and/or increased levels of PARylation have been found in patients or animal models of some neurodegenerative diseases (Liu and Fang, 2019; McGurk et al., 2019). Under long-term stress or disease conditions, when PARylation and PAR levels are dysregulated, PAR has the tendency to alter protein phase separation behavior and further develop into solid protein aggregates, which may result in pathological SG formation (Figure 1). This is supported by the observation of decreased PARylation levels following PARP knockdown or treatment with PARP inhibitor can antagonize cytoplasmic aggregation of the disease-related proteins hnRNP A1 and TDP-43 and their mediated neurotoxicity (Duan et al., 2019).
Taken together, PAR can serve as a scaffold for inducing LLPS of macromolecules and SG assembly. However, when PARP activity and PAR levels are mis-regulated, the proteins and granules may condense into irreversible solid phase aggregations that contribute to neuropathies (Figure 1).
Poly(ADP-Ribose) Regulates the Dynamic of SG Composition
The PAR chain can vary from a few to hundreds of ADP-ribose units (Leung, 2020). Studies suggested that different proteins have different PAR chain length preference. Some proteins prefer long PAR chains while other proteins bind to short chains more efficiently (Fahrer et al., 2007, 2010; Popp et al., 2013). Meanwhile, proteins containing different PAR-binding domains can associate with different types of ADP-ribose groups. For example, terminal ADP-ribose units are recognized by macrodomains while adjacent ADP-ribose units are recognized by the PBZ domain (Li et al., 2010; Leung, 2017). Therefore, the PAR chain provides the possibility to recruit diverse proteins in one place, and this ability is enzymatically controlled through regulating polymer size and structures (linear or branched) in a precise temporal order (Figure 1). SGs are highly dynamic and include different components that are utilized to respond to different types of stress. During the SG formation process, the composition of an SG may vary dramatically (Buchan and Parker, 2009; Protter and Parker, 2016). Distinct PARPs and PARGs resident in SGs function in the recognition of their individual targets–either to pull them together into the SG or to exclude unwanted proteins from the SG, according to the cell’s needs. Also, as many PAR-binding proteins are RBPs, which have high affinities for RNA, PAR may compete with RNA for protein binding (Leung, 2020). Therefore, covalent PAR formation on targets and PAR-binding contribute to diverse protein-protein interactions and protein-RNA interactions, providing another means of regulation of SG formation and controlling of SG components’ dynamic.
Conclusion and Perspective
Poly(ADP-ribose) has become an emerging research topic due to its recently identified roles in the organization of SGs and pathogenesis of SG-related neurodegenerative diseases (Grimaldi et al., 2019; McGurk et al., 2019). We summarized the possible mechanisms by which PAR mediates SG formation and dynamics as follows. PAR can promote the proper localization of SG components to SGs through ADP-ribosylation or PAR-binding, and this PTM assists in protein LLPS and SG formation. Various PAR chain lengths and structures could help in diverse SG composition. More importantly, when PAR levels are mis-regulated, the physical properties of SGs may become altered, and aberrant liquid-solid phase separation and pathogenic SGs may form (Figure 1). Besides these investigations on the roles of PAR in localizing SG components and regulating their LLPS behaviors, mechanisms underlying the interplay between PAR and SGs under different types of stress, individual roles of various PARPs in SGs, and the links between ADP-ribosylation and other PTMs remain unclear.
Further in-depth investigations are needed to clarify the interplay between PAR and SGs under different conditions. Such as what are the PAR-mediated interaction networks in SGs. This includes systematically determination of SG components that are PARylated or PAR-bound under different stresses. Different groups have used diverse approaches to identify PARylated vs. PAR-binding proteins (Daniels et al., 2015; Vivelo and Leung, 2015; Bonfiglio et al., 2017; Ando et al., 2019; Dasovich et al., 2021), which could be used as valuable tools on deciphering which SG components are PARylated and whether they undergo the same ADP-ribosylation modifications under different types of stress. For example, in a recently published paper, hundreds of novel PAR-binding proteins were identified using photoaffinity-based proteomics, which provides us with a valuable resource for exploring proteins involved in LLPS and SG formation (Dasovich et al., 2021). Moreover, innovative approaches are needed to systematically identify the modified sites on SG components and even the PAR chain length and structure on each site. It would be beneficial for identifying SG scaffold proteins and their binding partners, and building the overall PAR-mediated interaction networks in SGs. However, there are studies that failed to identify PARPs in SGs (Markmiller et al., 2018; Youn et al., 2018). Therefore, whether the initiation and dynamic maintenance of diverse SGs absolutely require PAR? Up to now, the mechanism of SG formation is not fully resolved, and the diversity of SG is stress or cell-type dependent. Does SG formation under different stress conditions have different requirements for PAR? Systematic analysis on the involvement of PARPs/PAR in SG under different conditions using different models will provide a global view on the importance of PAR in SG formation, and such systematic studies will increase our understanding on the formation mechanisms of SGs.
The other remaining question is what are the individual roles played by various PARPs in SGs, and whether or not they affect each other. Six reported SG-localized PARPs might have their own specific targets and functions. They might specifically regulate the localization, LLPS behavior, and interaction network of their substrates. For example, previous study indicated that long-term stress result in excessive cytoplasmic accumulation of the disease-related protein TDP-43, and reduction of PARP5a/5b activity could antagonize cytoplasmic aggregation of TDP-43 and its mediated neurotoxicity (McGurk et al., 2018a). Therefore, determination of which PARP responsible for the specific modification of different SG components could help in the development of novel therapeutic avenues, which could specifically regulate the formation and composition of pathogenic SGs.
Moreover, what are the links between ADP-ribosylation and other PTMs. Do they work together synergistically or compete with each other on substrates via conjunction on the same or adjacent motifs in SG regulation? A previous study indicated that PAR-dependent anchoring of TDP-43 to SGs inhibits its disease-associated phosphorylation. If TDP-43 were to accumulate excessively in the cytoplasm and become excluded from SGs, it could become phosphorylated and form irreversible aggregates (McGurk et al., 2018a). Therefore, as a single SG component may be modified by multiple PTMs, there might be interplays between ADP-ribosylation and other PTMs. PAR synthesis and degradation is a highly dynamic process, whose timely regulation ensures proper formation and dynamics of SGs. But how do cells finely regulate these processes? Should other PTMs such as phosphorylation block continued ADP-ribosylation on same substrate to ensure that the high levels of PAR are transient, preventing PARylated or PAR-binding proteins from transformation into pathogenic aggregates under long-term stress. Thus, more details of the mechanism of interplay between ADP-ribosylation and other PTMs need to be revealed.
Poly(ADP-ribose) as an important novel regulator of SG formation and dynamics, in-depth investigations on the remaining questions discussed above will help us to acquire a clear mechanistic view on global PAR-mediated interaction networks, specific functions of PARPs and links between ADP-ribosylation and other PTMs in SGs. This will eventually contribute on the development of novel therapeutic approaches targeting the aberrant pathogenic SG formation and further pave the way for effective SG-related neurodegenerative disease treatments.
Author Contributions
XJ, XC, and BL contributed to the design and wrote the manuscript. BL and SL supervised the overall direction, planning, and editing of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Grants from the National Natural Science Foundation of China (31800163) to XJ and Zhejiang Provincial Natural Science Foundation of China (LQ19C070001) to XC, as well as the Swedish Cancer Fund (Cancerfonden) [CAN 2017/643 and 19 0069] and the Swedish Natural Research Council (Vetenskapsrådet) [VR 2015-04984 and VR 2019-03604] to BL.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Ahel, D., Horejsí, Z., Wiechens, N., Polo, S. E., Garcia-Wilson, E., Ahel, I., et al. (2009). Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science 325, 1240–1243. doi: 10.1126/science.1177321
Alberti, S., and Dormann, D. (2019). Liquid-liquid phase separation in disease. Annu. Rev. Genet. 53, 171–194. doi: 10.1146/annurev-genet-112618-043527
Alberti, S., Halfmann, R., King, O., Kapila, A., and Lindquist, S. (2009). A Systematic Survey Identifies Prions and Illuminates Sequence Features of Prionogenic Proteins. Cell 137, 146–158. doi: 10.1016/j.cell.2009.02.044
Altmeyer, M., Neelsen, K. J., Teloni, F., Pozdnyakova, I., Pellegrino, S., Grofte, M., et al. (2015). Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose). Nat. Commun. 6:8088. doi: 10.1038/ncomms9088
Ando, Y., Elkayam, E., McPherson, R. L., Dasovich, M., Cheng, S. J., Voorneveld, J., et al. (2019). ELTA: enzymatic labeling of terminal ADP-ribose. Mol. Cell 73, 845–856.e5. doi: 10.1016/j.molcel.2018.12.022
Bai, P. (2015). Biology of poly(ADP-ribose) polymerases: the factotums of cell maintenance. Mol. Cell 58, 947–958. doi: 10.1016/j.molcel.2015.01.034
Bonfiglio, J. J., Colby, T., and Matic, I. (2017). Mass spectrometry for serine ADP-ribosylation? Think o-glycosylation!. Nucleic Acids Res. 45, 6259–6264. doi: 10.1093/nar/gkx446
Buchan, J. R., and Parker, R. (2009). Eukaryotic stress granules: the ins and outs of translation. Mol. Cell 36, 932–941. doi: 10.1016/j.molcel.2009.11.020
Cao, X., Jin, X., and Liu, B. (2020). The involvement of stress granules in aging and aging-associated diseases. Aging Cell 19:e13136. doi: 10.1111/acel.13136
Catara, G., Grimaldi, G., Schembri, L., Spano, D., Turacchio, G., Lo Monte, M., et al. (2017). PARP1-produced poly-ADP-ribose causes the PARP12 translocation to stress granules and impairment of Golgi complex functions. Sci. Rep. 7:e14035. doi: 10.1038/s41598-017-14156-8
Cheruiyot, A., Paudyal, S. C., Kim, I. K., Sparks, M., Ellenberger, T., Piwnica-Worms, H., et al. (2015). Poly(ADP-ribose)-binding promotes Exo1 damage recruitment and suppresses its nuclease activities. DNA Repair 35, 106–115. doi: 10.1016/j.dnarep.2015.09.021
Corda, D., and Di Girolamo, M. (2003). Functional aspects of protein mono-ADP-ribosylation. EMBO J. 22, 1953–1958. doi: 10.1093/emboj/cdg209
Crawford, K., Bonfiglio, J. J., Mikoc, A., Matic, I., and Ahel, I. (2018). Specificity of reversible ADP-ribosylation and regulation of cellular processes. Crit. Rev. Biochem. Mol. Biol. 53, 64–82. doi: 10.1080/10409238.2017.1394265
Daniels, C. M., Ong, S. E., and Leung, A. K. (2015). The promise of proteomics for the study of ADP-ribosylation. Mol. Cell 58, 911–924. doi: 10.1016/j.molcel.2015.06.012
Dasovich, M., Beckett, M. Q., Bailey, S., Ong, S. E., Greenberg, M. M., and Leung, A. K. L. (2021). Identifying poly(ADP-ribose)-binding proteins with photoaffinity-based proteomics. J. Am. Chem. Soc. 143, 3037–3042. doi: 10.1021/jacs.0c12246
Deiana, A., Forcelloni, S., Porrello, A., and Giansanti, A. (2019). Intrinsically disordered proteins and structured proteins with intrinsically disordered regions have different functional roles in the cell. PLoS One 14:e0217889. doi: 10.1371/journal.pone.0217889
Duan, Y., Du, A., Gu, J., Duan, G., Wang, C., Gui, X., et al. (2019). PARylation regulates stress granule dynamics, phase separation, and neurotoxicity of disease-related RNA-binding proteins. Cell Res. 29, 233–247. doi: 10.1038/s41422-019-0141-z
Eckei, L., Krieg, S., Bütepage, M., Lehmann, A., Gross, A., Lippok, B., et al. (2017). The conserved macrodomains of the non-structural proteins of Chikungunya virus and other pathogenic positive strand RNA viruses function as mono-ADP-ribosylhydrolases. Sci. Rep. 7:e41746. doi: 10.1038/srep41746
Fahrer, J., Kranaster, R., Altmeyer, M., Marx, A., and Burkle, A. (2007). Quantitative analysis of the binding affinity of poly(ADP-ribose) to specific binding proteins as a function of chain length. Nucleic Acids Res. 35:e143. doi: 10.1093/nar/gkm944
Fahrer, J., Popp, O., Malanga, M., Beneke, S., Markovitz, D. M., Ferrando-May, E., et al. (2010). High-affinity interaction of poly(ADP-ribose) and the human DEK oncoprotein depends upon chain length. Biochemistry 49, 7119–7130. doi: 10.1021/bi1004365
Falahati, H., and Haji-Akbari, A. (2019). Thermodynamically driven assemblies and liquid-liquid phase separations in biology. Soft Matter 15, 1135–1154. doi: 10.1039/c8sm02285b
Fischbach, A., Krüger, A., Hampp, S., Assmann, G., Rank, L., Hufnagel, M., et al. (2018). The C-terminal domain of p53 orchestrates the interplay between non-covalent and covalent poly(ADP-ribosyl)ation of p53 by PARP1. Nucleic Acids Res. 46, 804–822. doi: 10.1093/nar/gkx1205
Geuens, T., Bouhy, D., and Timmerman, V. (2016). The hnRNP family: insights into their role in health and disease. Hum. Genet. 135, 851–867. doi: 10.1007/s00439-016-1683-5
Gomes, E., and Shorter, J. (2019). The molecular language of membraneless organelles. J. Biol. Chem. 294, 7115–7127. doi: 10.1074/jbc.TM118.001192
Grimaldi, G., Catara, G., Palazzo, L., Corteggio, A., Valente, C., and Corda, D. (2019). PARPs and PAR as novel pharmacological targets for the treatment of stress granule-associated disorders. Biochem. Pharmacol. 167, 64–75. doi: 10.1016/j.bcp.2019.05.019
Guillén-Boixet, J., Kopach, A., Holehouse, A. S., Wittmann, S., Jahnel, M., Schlüßler, R., et al. (2020). RNA-induced conformational switching and clustering of G3BP drive stress granule assembly by condensation. Cell 181, 346–361.e17. doi: 10.1016/j.cell.2020.03.049
Gupte, R., Liu, Z., and Kraus, W. L. (2017). PARPs and ADP-ribosylation: recent advances linking molecular functions to biological outcomes. Genes Dev. 31, 101–126. doi: 10.1101/gad.291518.116
Hyman, A. A., Weber, C. A., and Julicher, F. (2014). Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 30, 39–58. doi: 10.1146/annurev-cellbio-100913-013325
Isabelle, M., Gagne, J. P., Gallouzi, I. E., and Poirier, G. G. (2012). Quantitative proteomics and dynamic imaging reveal that G3BP-mediated stress granule assembly is poly(ADP-ribose)-dependent following exposure to MNNG-induced DNA alkylation. J. Cell Sci. 125, 4555–4566. doi: 10.1242/jcs.106963
Jankevicius, G., Hassler, M., Golia, B., Rybin, V., Zacharias, M., Timinszky, G., et al. (2013). A family of macrodomain proteins reverses cellular mono-ADP-ribosylation. Nat. Struct. Mol. Biol. 20, 508–514. doi: 10.1038/nsmb.2523
Jayabalan, A. K., Adivarahan, S., Koppula, A., Abraham, R., Batish, M., Zenklusen, D., et al. (2021). Stress granule formation, disassembly, and composition are regulated by alphavirus ADP-ribosylhydrolase activity. Proc. Natl. Acad. Sci. U. S. A. 118:e2021719118. doi: 10.1073/pnas.2021719118
Kam, T. I., Mao, X., Park, H., Chou, S. C., Karuppagounder, S. S., Umanah, G. E., et al. (2018). Poly(ADP-ribose) drives pathologic alpha-synuclein neurodegeneration in Parkinson’s disease. Science 362:eaat8407. doi: 10.1126/science.aat8407
Karras, G. I., Kustatscher, G., Buhecha, H. R., Allen, M. D., Pugieux, C., Sait, F., et al. (2005). The macro domain is an ADP-ribose binding module. EMBO J. 24, 1911–1920. doi: 10.1038/sj.emboj.7600664
Kedersha, N., Ivanov, P., and Anderson, P. (2013). Stress granules and cell signaling: more than just a passing phase?. Trends Biochem. Sci. 38, 494–506. doi: 10.1016/j.tibs.2013.07.004
Kedersha, N., Stoecklin, G., Ayodele, M., Yacono, P., Lykke-Andersen, J., Fritzler, M. J., et al. (2005). Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169, 871–884. doi: 10.1083/jcb.200502088
Kim, D. S., Camacho, C. V., Nagari, A., Malladi, V. S., Challa, S., and Kraus, W. L. (2019). Activation of PARP-1 by snoRNAs controls ribosome biogenesis and cell growth via the RNA helicase DDX21. Mol. Cell 75, 1270–1285.e14. doi: 10.1016/j.molcel.2019.06.020
Kim, H. J., Kim, N. C., Wang, Y. D., Scarborough, E. A., Moore, J., Diaz, Z., et al. (2013). Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 495, 467–473. doi: 10.1038/nature11922
Krietsch, J., Rouleau, M., Pic, É, Ethier, C., Dawson, T. M., Dawson, V. L., et al. (2013). Reprogramming cellular events by poly(ADP-ribose)-binding proteins. Mol. Aspects Med. 34, 1066–1087. doi: 10.1016/j.mam.2012.12.005
Leslie Pedrioli, D. M., Leutert, M., Bilan, V., Nowak, K., Gunasekera, K., Ferrari, E., et al. (2018). Comprehensive ADP-ribosylome analysis identifies tyrosine as an ADP-ribose acceptor site. EMBO Rep. 19:e45310. doi: 10.15252/embr.201745310
Leung, A. K. L. (2014). Poly(ADP-ribose): an organizer of cellular architecture. J. Cell Biol. 205, 613–619. doi: 10.1083/jcb.201402114
Leung, A. K. L. (2020). Poly(ADP-ribose): a dynamic trigger for biomolecular condensate formation. Trends Cell Biol. 30, 370–383. doi: 10.1016/j.tcb.2020.02.002
Leung, A. K. L., Vyas, S., Rood, J. E., Bhutkar, A., Sharp, P. A., and Chang, P. (2011). Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol. Cell 42, 489–499. doi: 10.1016/j.molcel.2011.04.015
Li, G. Y., McCulloch, R. D., Fenton, A. L., Cheung, M., Meng, L., Ikura, M., et al. (2010). Structure and identification of ADP-ribose recognition motifs of APLF and role in the DNA damage response. Proc. Natl. Acad. Sci. U. S. A. 107, 9129–9134. doi: 10.1073/pnas.1000556107
Liu, C., and Fang, Y. (2019). New insights of poly(ADP-ribosylation) in neurodegenerative diseases: a focus on protein phase separation and pathologic aggregation. Biochem. Pharmacol. 167, 58–63. doi: 10.1016/j.bcp.2019.04.028
Luscher, B., Butepage, M., Eckei, L., Krieg, S., Verheugd, P., and Shilton, B. H. (2018). ADP-ribosylation, a multifaceted posttranslational modification involved in the control of cell physiology in health and disease. Chem. Rev. 118, 1092–1136. doi: 10.1021/acs.chemrev.7b00122
Markmiller, S., Soltanieh, S., Server, K. L., Mak, R., Jin, W., Fang, M. Y., et al. (2018). Context-dependent and disease-specific diversity in protein interactions within stress granules. Cell 172, 590–604.e13. doi: 10.1016/j.cell.2017.12.032
Mastrocola, A. S., Kim, S. H., Trinh, A. T., Rodenkirch, L. A., and Tibbetts, R. S. (2013). The RNA-binding protein fused in sarcoma (FUS) functions downstream of poly(ADP-ribose) polymerase (PARP) in response to DNA damage. J. Biol. Chem. 288, 24731–24741. doi: 10.1074/jbc.M113.497974
McGurk, L., Gomes, E., Guo, L., Mojsilovic-Petrovic, J., Tran, V., Kalb, R. G., et al. (2018a). Poly(ADP-ribose) prevents pathological phase separation of TDP-43 by promoting liquid demixing and stress granule localization. Mol. Cell 71, 703–717.e9. doi: 10.1016/j.molcel.2018.07.002
McGurk, L., Gomes, E., Guo, L., Shorter, J., and Bonini, N. M. (2018b). Poly(ADP-ribose) engages the TDP-43 nuclear-localization sequence to regulate granulo-filamentous aggregation. Biochemistry 57, 6923–6926. doi: 10.1021/acs.biochem.8b00910
McGurk, L., Rifai, O. M., and Bonini, N. M. (2019). Poly(ADP-ribosylation) in age-related neurological disease. Trends Genet. 35, 601–613. doi: 10.1016/j.tig.2019.05.004
Mohagheghi, F., Prudencio, M., Stuani, C., Cook, C., Jansen-West, K., Dickson, D. W., et al. (2016). TDP-43 functions within a network of hnRNP proteins to inhibit the production of a truncated human SORT1 receptor. Hum. Mol. Genet. 25, 534–545. doi: 10.1093/hmg/ddv491
Molliex, A., Temirov, J., Lee, J., Coughlin, M., Kanagaraj, A. P., Kim, H. J., et al. (2015). Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133. doi: 10.1016/j.cell.2015.09.015
O’Sullivan, J., Tedim Ferreira, M., Gagne, J. P., Sharma, A. K., Hendzel, M. J., Masson, J. Y., et al. (2019). Emerging roles of eraser enzymes in the dynamic control of protein ADP-ribosylation. Nat. Commun. 10:e1182. doi: 10.1038/s41467-019-08859-x
Otto, H., Reche, P. A., Bazan, F., Dittmar, K., Haag, F., and Koch-Nolte, F. (2005). In silico characterization of the family of PARP-like poly(ADP-ribosyl) transferases (pARTs). BMC Genomics 6:e139. doi: 10.1186/1471-2164-6-139
Palazzo, L., Mikoc, A., and Ahel, I. (2017). ADP-ribosylation: new facets of an ancient modification. FEBS J. 284, 2932–2946. doi: 10.1111/febs.14078
Patel, A., Lee, H. O., Jawerth, L., Maharana, S., Jahnel, M., Hein, M. Y., et al. (2015). A liquid-to-solid phase transition of the als protein fus accelerated by disease mutation. Cell 162, 1066–1077. doi: 10.1016/j.cell.2015.07.047
Perina, D., Mikoc, A., Ahel, J., Cetkovic, H., Zaja, R., and Ahel, I. (2014). Distribution of protein poly(ADP-ribosyl)ation systems across all domains of life. DNA Repair 23, 4–16. doi: 10.1016/j.dnarep.2014.05.003
Pleschke, J. M., Kleczkowska, H. E., Strohm, M., and Althaus, F. R. (2000). Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J. Biol. Chem. 275, 40974–40980. doi: 10.1074/jbc.M006520200
Popp, O., Veith, S., Fahrer, J., Bohr, V. A., Burkle, A., and Mangerich, A. (2013). Site-specific noncovalent interaction of the biopolymer poly(ADP-ribose) with the Werner syndrome protein regulates protein functions. ACS Chem. Biol. 8, 179–188. doi: 10.1021/cb300363g
Prasad, A., Bharathi, V., Sivalingam, V., Girdhar, A., and Patel, B. K. (2019). Molecular mechanisms of TDP-43 misfolding and pathology in amyotrophic lateral sclerosis. Front. Mol. Neurosci. 12:e25. doi: 10.3389/fnmol.2019.00025
Protter, D. S. W., and Parker, R. (2016). Principles and properties of stress granules. Trends Cell Biol. 26, 668–679. doi: 10.1016/j.tcb.2016.05.004
Qi, H., Price, B. D., and Day, T. A. (2019). Multiple roles for mono- and Poly(ADP-ribose) in regulating stress responses. Trends Genet. 35, 159–172. doi: 10.1016/j.tig.2018.12.002
Rosenthal, F., Feijs, K. L., Frugier, E., Bonalli, M., Forst, A. H., Imhof, R., et al. (2013). Macrodomain-containing proteins are new mono-ADP-ribosylhydrolases. Nat. Struct. Mol. Biol. 20, 502–507. doi: 10.1038/nsmb.2521
Schwartz, J. C., Wang, X., Podell, E. R., and Cech, T. R. (2013). RNA seeds higher-order assembly of FUS protein. Cell Rep. 5, 918–925. doi: 10.1016/j.celrep.2013.11.017
Sharifi, R., Morra, R., Appel, C. D., Tallis, M., Chioza, B., Jankevicius, G., et al. (2013). Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease. EMBO J. 32, 1225–1237. doi: 10.1038/emboj.2013.51
Singatulina, A. S., Hamon, L., Sukhanova, M. V., Desforges, B., Joshi, V., Bouhss, A., et al. (2019). PARP-1 activation directs FUS to DNA damage sites to form PARG-reversible compartments enriched in damaged DNA. Cell Rep. 27, 1809–1821.e5. doi: 10.1016/j.celrep.2019.04.031
Slade, D., Dunstan, M. S., Barkauskaite, E., Weston, R., Lafite, P., Dixon, N., et al. (2011). The structure and catalytic mechanism of a poly(ADP-ribose) glycohydrolase. Nature 477, 616–620. doi: 10.1038/nature10404
Sreedharan, J., Blair, I. P., Tripathi, V. B., Hu, X., Vance, C., Rogelj, B., et al. (2008). TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science 319, 1668–1672. doi: 10.1126/science.1154584
Vivelo, C. A., and Leung, A. K. (2015). Proteomics approaches to identify mono-(ADP-ribosyl)ated and poly(ADP-ribosyl)ated proteins. Proteomics 15, 203–217. doi: 10.1002/pmic.201400217
Vyas, S., Matic, I., Uchima, L., Rood, J., Zaja, R., Hay, R. T., et al. (2014). Family-wide analysis of poly(ADP-ribose) polymerase activity. Nat. Commun. 5:e4426. doi: 10.1038/ncomms5426
Wang, Z., Michaud, G. A., Cheng, Z., Zhang, Y., Hinds, T. R., Fan, E., et al. (2012). Recognition of the iso-ADP-ribose moiety in poly(ADP-ribose) by WWE domains suggests a general mechanism for poly(ADP-ribosyl)ation-dependent ubiquitination. Genes Dev. 26, 235–240. doi: 10.1101/gad.182618.111
Watanabe, K., Morishita, K., Zhou, X., Shiizaki, S., Uchiyama, Y., Koike, M., et al. (2021). Cells recognize osmotic stress through liquid-liquid phase separation lubricated with poly(ADP-ribose). Nat. Commun. 12:e1353. doi: 10.1038/s41467-021-21614-5
Yang, P., Mathieu, C., Kolaitis, R. M., Zhang, P., Messing, J., Yurtsever, U., et al. (2020). G3BP1 is a tunable switch that triggers phase separation to assemble stress granules. Cell 181, 325–345.e28. doi: 10.1016/j.cell.2020.03.046
Youn, J. Y., Dunham, W. H., Hong, S. J., Knight, J. D. R., Bashkurov, M., Chen, G. I., et al. (2018). High-density proximity mapping reveals the subcellular organization of mRNA-associated granules and bodies. Mol. Cell 69, 517–532.e11. doi: 10.1016/j.molcel.2017.12.020
Zhang, F., Chen, Y., Li, M., and Yu, X. (2014). The oligonucleotide/oligosaccharide-binding fold motif is a poly(ADP-ribose)-binding domain that mediates DNA damage response. Proc. Natl. Acad. Sci. U. S.A. 111, 7278–7283. doi: 10.1073/pnas.1318367111
Keywords: poly(ADP-ribose), PAR-binding, post-translational modification, stress granules, liquid–liquid phase separation
Citation: Jin X, Cao X, Liu S and Liu B (2021) Functional Roles of Poly(ADP-Ribose) in Stress Granule Formation and Dynamics. Front. Cell Dev. Biol. 9:671780. doi: 10.3389/fcell.2021.671780
Received: 24 February 2021; Accepted: 06 April 2021;
Published: 26 April 2021.
Edited by:
Alexei Degterev, Tufts University, United StatesReviewed by:
Bernhard Lüscher, RWTH Aachen University, GermanyAnthony Fehr, University of Kansas, United States
Guy Poirier, Laval University, Canada
Copyright © 2021 Jin, Cao, Liu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shenkui Liu, c2hlbmt1aWxpdUBuZWZ1LmVkdS5jbg==; Beidong Liu, YmVpZG9uZy5saXVAY21iLmd1LnNl
†These authors have contributed equally to this work
 Xuejiao Jin1†
Xuejiao Jin1† Xiuling Cao
Xiuling Cao Shenkui Liu
Shenkui Liu Beidong Liu
Beidong Liu