- 1Division of Developmental Biology, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, United States
- 2Division of Plastic Surgery, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, United States
- 3Departments of Pediatrics and Surgery, University of Cincinnati College of Medicine, Cincinnati, OH, United States
- 4Shriners Hospitals for Children, Cincinnati, OH, United States
Disruption of FOXF2, encoding a member of the Forkhead family transcription factors, has been associated with cleft palate in humans and mice. FOXF2 is located in a conserved gene cluster containing FOXQ1, FOXF2, and FOXC1. We found that expression of Foxq1 is dramatically upregulated in the embryonic palatal mesenchyme in Foxf2–/– mouse embryos. We show here that the Foxf2 promoter-deletion mutation caused dramatically increased expression of the cis-linked Foxq1 allele but had little effect on the Foxq1 allele in trans. We analyzed effects of the Foxf2 mutation on the expression of other neighboring genes and compared those effects with the chromatin domain structure and recently identified enhancer-promoter associations as well as H3K27ac ChIP-seq data. We show that the Foxf2 mutation resulted in significantly increased expression of the Foxq1 and Exoc2 genes located in the same topologically associated domain with Foxf2 but not the expression of the Foxc1 and Gmds genes located in the adjacent chromatin domain. We inactivated the Foxq1 gene in mice homozygous for a Foxf2 conditional allele using CRISPR genome editing and generated (Foxf2/Foxq1)+/– mice with loss-of-function mutations in Foxf2 and Foxq1 in cis. Whereas the (Foxf2/Foxq1)–/– mice exhibited cleft palate at birth similar as in the Foxf2–/– mice, systematic expression analyses of a large number of Foxf2-dependent genes revealed that the (Foxf2/Foxq1)–/– embryos exhibited distinct effects on the domain-specific expression of several important genes, including Foxf1, Shox2, and Spon1, in the developing palatal shelves compared with Foxf2–/– embryos. These results identify a novel cis-regulatory effect of the Foxf2 mutation and demonstrate that cis-regulation of Foxq1 contributed to alterations in palatal gene expression in Foxf2–/– embryos. These results have important implications for interpretation of results and mechanisms from studies of promoter- or gene-deletion alleles. In addition, the unique mouse lines generated in this study provide a valuable resource for understanding the cross-regulation and combinatorial functions of the Foxf2 and Foxq1 genes in development and disease.
Introduction
The secondary palate separates the nasal cavity from the oral cavity and consists of the bony hard palate anteriorly and muscular soft palate posteriorly (Bush and Jiang, 2012). In mammals, the development of the secondary palate initiates as a pair of outgrowths from the oral side of embryonic maxillary processes, which grow vertically to form the palatal shelves flanking the developing tongue. As development proceeds, the palatal shelves reorient to the horizontal position above the tongue, grow toward and fuse with each other at the midline to form the roof of the oral cavity. Genetic or environmental perturbations of any of these developmental processes, including palatal shelf growth, elevation, and fusion, could cause cleft palate, one of the most common structural defects in humans (Chai and Maxson, 2006; Dixon et al., 2011; Bush and Jiang, 2012; Lan et al., 2015).
The development of secondary palate is regulated by a complex molecular network containing multiple signaling pathways and transcription factors. The Shh and Fgf signaling pathways have been shown to play a key role in regulating the palatal epithelial-mesenchymal interactions governing palatal shelf growth and patterning (Rice et al., 2004; Han et al., 2009; Lan and Jiang, 2009). Shh produced by the palatal epithelial cells signals to the palatal mesenchyme and forms a positive feedback loop with Fgf10 to coordinate cell proliferation in both the epithelium and mesenchyme during palate development (Rice et al., 2004; Lan and Jiang, 2009). In addition, Fgf7 produced by the palatal mesenchyme restricts the expression of Shh mRNAs to the oral side palatal epithelium to control the oral-nasal patterning (Han et al., 2009). Shh signaling pathway is also required to maintain the expression of the Forkhead genes Foxf1 and Foxf2 in the palatal mesenchyme (Lan and Jiang, 2009). We recently showed that a Shh-Foxf1/Foxf2-Fgf18-Shh molecular circuit regulates the proliferation of palatal mesenchymal cells during palatal shelf growth (Xu et al., 2016).
Foxf1 and Foxf2 are paralogous transcription factors of the Forkhead family with highly conserved amino acid sequences in the Forkhead DNA binding domain (Hellqvist et al., 1996). During palate development, Foxf2 is expressed throughout the anterior-posterior axis of the palatal shelves while the expression of Foxf1 is more restrict in the middle portion of palatal shelves (Nik et al., 2016; Xu et al., 2016). Foxf2–/– mouse embryos display complete cleft secondary palate with multiple cellular defects. The proliferation of palatal mesenchymal cell is reduced, with the palatal shelf growth most significantly affected in the posterior portion in the Foxf2–/– embryos (Nik et al., 2016; Xu et al., 2016). In addition, changes of extracellular matrix organization have also been shown to contribute to the cleft palate phenotype in Foxf2–/– embryos (Nik et al., 2016; Xu et al., 2020). Mutations in FOXF2 have been associated with cleft palate in humans (Jochumsen et al., 2008; Bu et al., 2015). A recent study reported a familial palate defect with absent uvula, short posterior border of the soft palate, and abnormal tonsillar pillar (Seselgyte et al., 2019). Further genetic studies identified a missense variant in FOXF2 as the likely cause of this condition (Seselgyte et al., 2019). Thus, better understanding of the molecular mechanisms mediating Foxf2 function in palate development in mice will improve our understanding of cleft palate pathogenesis in humans.
To investigate the molecular mechanisms mediating Foxf2 function in palate development, we have used a combination of whole transcriptome RNA sequencing (RNA-seq) and chromatin immunoprecipitation-sequencing (ChIP-seq) mediated genome wide mapping of Foxf2 binding sites to identify direct Foxf2 target genes in the developing palatal mesenchyme (Xu et al., 2016, 2020). In addition to identifying Fgf18 as a direct Foxf2 target gene that acts in the Shh-Foxf2-Fgf18-Shh molecular circuit to control palatal shelf growth (Xu et al., 2016), we showed that a number of genes encoding components of the extracellular matrix and a group of genes encoding transcription factors are direct Foxf2 target genes in the developing palatal mesenchyme cells (Xu et al., 2020). Among these, Foxq1, which encodes a Forkhead transcription factor with high amino acid sequence similarity with the Foxf2 protein, is one of the most significantly up-regulated genes in the developing palatal mesenchyme in Foxf2–/– embryos (Xu et al., 2020). Foxq1 is expressed at very low levels in the palatal mesenchyme in wildtype mouse embryos (Xu et al., 2020). Foxq1–/– mutant mice exhibited defects in hair follicle development and gastric acid secretion but no palatal defect has been reported (Hong et al., 2001; Goering et al., 2008). Remarkably, Foxq1 and Foxf2 are closely linked genes in an evolutionarily conserved gene cluster in all vertebrate genomes (Wotton and Shimeld, 2006). In both human and mouse genomes, the Foxq1 gene is located directly upstream of Foxf2. Previous studies using breast cancer cell lines have suggested that FOXF2 and FOXQ1 have opposite functions in regulating epithelial-mesenchymal transition and that they repress the expression of each other (Zhang et al., 2011; Wang et al., 2015; Kang et al., 2019). Thus, the significant upregulation of Foxq1 expression in the developing palatal mesenchyme in Foxf2–/– embryos suggest that Foxf2-mediated regulation of Foxq1 expression may play an important role in palatogenesis. To address this possibility and gain better understanding of Foxf2-mediated regulation of Foxq1 expression during palate development, we have generated mice carrying mutations in both Foxf2 and Foxq1 in cis using CRISPR/Cas9 mediated genome editing (Cong et al., 2013; Wang et al., 2013) and our results identify an unexpected cis-regulation of Foxq1 by Foxf2 in the developing palate.
Materials and Methods
Mice
Foxf2tm1Rhc (Foxf2c/c) and Foxf2+/– mice have been described previously (Hoggatt et al., 2013; Bolte et al., 2015) and were maintained by intercrossing or by crossing with C57BL/6J inbred mice. To generate Foxf2c/c; Foxq1+/– mice, two synthetic guide RNAs (sgRNAs) (50 ng/μl each) targeting the genomic sequence flanking the Forkhead domain-coding sequence in the Foxq1 gene were co-injected with humanized Cas9 mRNAs (50 ng/μl) into zygotes of Foxf2c/c mice (Figure 2A). The target sequences of the two sgRNAs are: 5′-GCAGCAAGCCGTACACGCGG-3′ and 5′-GCGAATACACCTTCGCCGAC-3′. Injected eggs were transferred on the same day into the oviductal ampulla of pseudopregnant CD-1 female mice at approximately 25 eggs per recipient. Foxq1 gene-modified founder mice were identified by PCR assay and then the exact nucleotide changes at the edited Foxq1 locus were determined by Sanger sequencing. Mice carrying two independent Foxq1-deletion alleles, lacking 509 bp and 494 bp, respectively, of the Foxq1 coding region (Foxq1D509 and Foxq1D494) were used in this study. Founder mice were crossed to Foxf2c/c mice to generate the Foxf2c/c; Foxq1+/– mice. Genotypically verified G1 Foxf2c/c; Foxq1+/– mice were intercrossed to generate Foxf2c/c; Foxq1–/– homozygotes. In addition, Foxf2c/c; Foxq1+/– mice were crossed to EIIa-Cre transgenic mice (Lakso et al., 1996) to inactivate the Foxf2c allele and generate the (Foxf2/Foxq1)+/– mice. (Foxf2/Foxq1)+/– mice were intercrossed to generate (Foxf2/Foxq1)–/– homozygous embryos for analyses. For timed mating, noon of the day on which a vaginal plug was identified was designated as embryonic day (E) 0.5. All animal work procedures were performed following recommendations in the Guide for Care and Use of Laboratory Animals by the National Institutes of Health and approved by the Institutional Animal Care and Use Committee (IACUC) at Cincinnati Children’s Hospital Medical Center. This study conformed with the ARRIVE (Animal Research: Reporting of in vivo Experiments) guidelines for preclinical animal studies.
Histology, in situ Hybridization, and Immunofluorescent Staining
Embryos were collected and processed for histology, immunostaining, or in situ hybridization as described previously (Xu et al., 2016). For histology and immunofluorescent staining, the embryos were fixed in 4% paraformaldehyde (PFA), dehydrated through an ethanol series, embedded in paraffin, and sectioned at 7 μm thickness. The goat anti-Foxf1 (AF4798; R&D) antibody was used to detect the Foxf1 protein. Images were taken using a Nikon DS-Qi2 microscope (Nikon Instruments Inc., Melville, NY, United States).
RNA Extraction, Real-Time Polymerase Chain Reaction (RT-qPCR), and Enzyme Digestion Assay
Whole palatal shelves of E13.5 embryos were manually dissected in ice-cold phosphate buffered saline. Total RNAs were extracted using the RNeasy micro kit (74004; Qiagen Inc., Germantown, Maryland). First-strand cDNAs were prepared using the SuperScript III First-Strand Synthesis System (18080-051; Invitrogen, Carlsbad, California), and real-time qPCR was performed using a CFX96 Real-Time System (Bio-Rad, Hercules, California) with conditions recommended by the manufacturer. Relative levels of mRNAs in each sample were normalized to that of Hprt mRNAs. For restriction enzyme digestion assay to measure allele specific expression of Foxq1 mRNAs, a pair of primers was designed to flank a single nucleotide polymorphism in the 3′ untranslated region (UTR) of the Foxq1 gene (rs29587452) between the C57BL/6J and 129X1/SvJ mouse strains. Purified PCR products amplified from wildtype, Foxf2+/–, and Foxf2–/– cDNA samples were digested overnight using AciI (NEB, R0551L) at 37oC. Quantification of the digested and undigested DNA fragments was performed by QIAxcel Advanced using QIAxcel® ScreenGel software (QIAGEN, Cat# 9021163).
For statistical analysis, all results were presented as mean ± SEM. Student’s t test was used for pairwise comparison. One-way ANOVA followed by Newman–Keuls post hoc test was used to compare all pairs when more than two genotypes were included. P < 0.05 was considered significantly different.
Genomic Data Retrieval and Analysis
The whole genome chromosome conformation capture (Hi-C) data and topologically associated domain (TAD) map of mouse embryonic stem cells were retrieved from the 3D Genome Browser1 (Wang et al., 2018). The original Hi-C data was from Bonev_2017- raw (Bonev et al., 2017; Wang et al., 2018), and assembled into mm10 reference genome with the resolution set at 10 kb. The histone H3K27ac ChIP-seq data and the bigwig file of the E12.5 mouse embryonic posterior palatal shelves was obtained from the NCBI GEO database2 (accession number GSE138721) (Xu et al., 2019). The replicated associations between enhancers and gene promoter data were retrieved from the UCSC Genome Browser3 (Gorkin et al., 2020).
Results
Disruption of Foxf2 Causes Significantly Increased Expression of the Linked Foxq1 Allele in Cis in the Developing Palate
Both Foxf2 and Foxq1 are located on mouse Chromosome 13, with Foxq1 at about 65 kb proximal and upstream of Foxf2, and with the two genes in the same transcriptional orientation (Figure 1A). In between Foxq1 and Foxf2, there is an uncharacterized long non-coding RNA (lncRNA) gene, named 1700018A04Rik, which is transcribed from the opposite DNA strand with the most 5′ transcription start site (TSS) located only about 300 bp from the TSS of Foxf2 (Figures 1A,B). To date, three Foxf2 gene-targeted mouse lines have been reported (Wang et al., 2003; Hoggatt et al., 2013; Bolte et al., 2015; Reyahi et al., 2015). In Foxf2tm1Rhc, the Foxf2 conditional allele that we are using, the two loxP sites flank a genomic region containing both Exon-1 of the Foxf2 gene and Exon-1 of the 1700018A04Rik gene (Figure 1B; Hoggatt et al., 2013; Bolte et al., 2015). Cre mediated deletion of the floxed region in this allele deletes the Exon-1 and the promoter of both genes. In another Foxf2 conditional allele, Foxf2tm1Pca, the two loxP sites also flank a genomic region containing the promoter and Exon-1 of the Foxf2 gene (Reyahi et al., 2015), whereas a Foxf2 conventional knockout allele, Foxf2tm1Miu, was generated by replacing the XhoI-EcoRI genomic region containing the promoter and Exon-1 region of the Foxf2 gene with a PGK-Neo expression cassette (Wang et al., 2003). Thus, all three Foxf2 gene-knockout alleles reported to date likely inactivated both Foxf2 and 1700018A04Rik. In situ hybridization analysis showed that 1700018A04Rik mRNAs were expressed in the posterior region of the developing palatal shelves in wildtype embryos (Figure 1C), which overlaps with the posterior palate domain of strong Foxf2 expression as reported previously (Xu et al., 2016). No 1700018A04Rik mRNA expression was detected in the Foxf2–/– mutant embryos whereas Foxf2+/– embryos showed significantly reduced 1700018A04Rik mRNA expression in comparison with the wildtype littermates (Figures 1C–E). Analysis of Foxq1 mRNA expression revealed an almost mirror image pattern, with wildtype embryos exhibiting very low level of Foxq1 mRNA expression in the posterior palatal shelves and Foxf2+/– embryos exhibiting dramatically increased Foxq1 mRNA expression while Foxf2–/– mutant embryos exhibiting even stronger Foxq1 mRNA expression that expanded from the posterior domain to the anterior regions of the palatal shelves (Figures 1F–H).
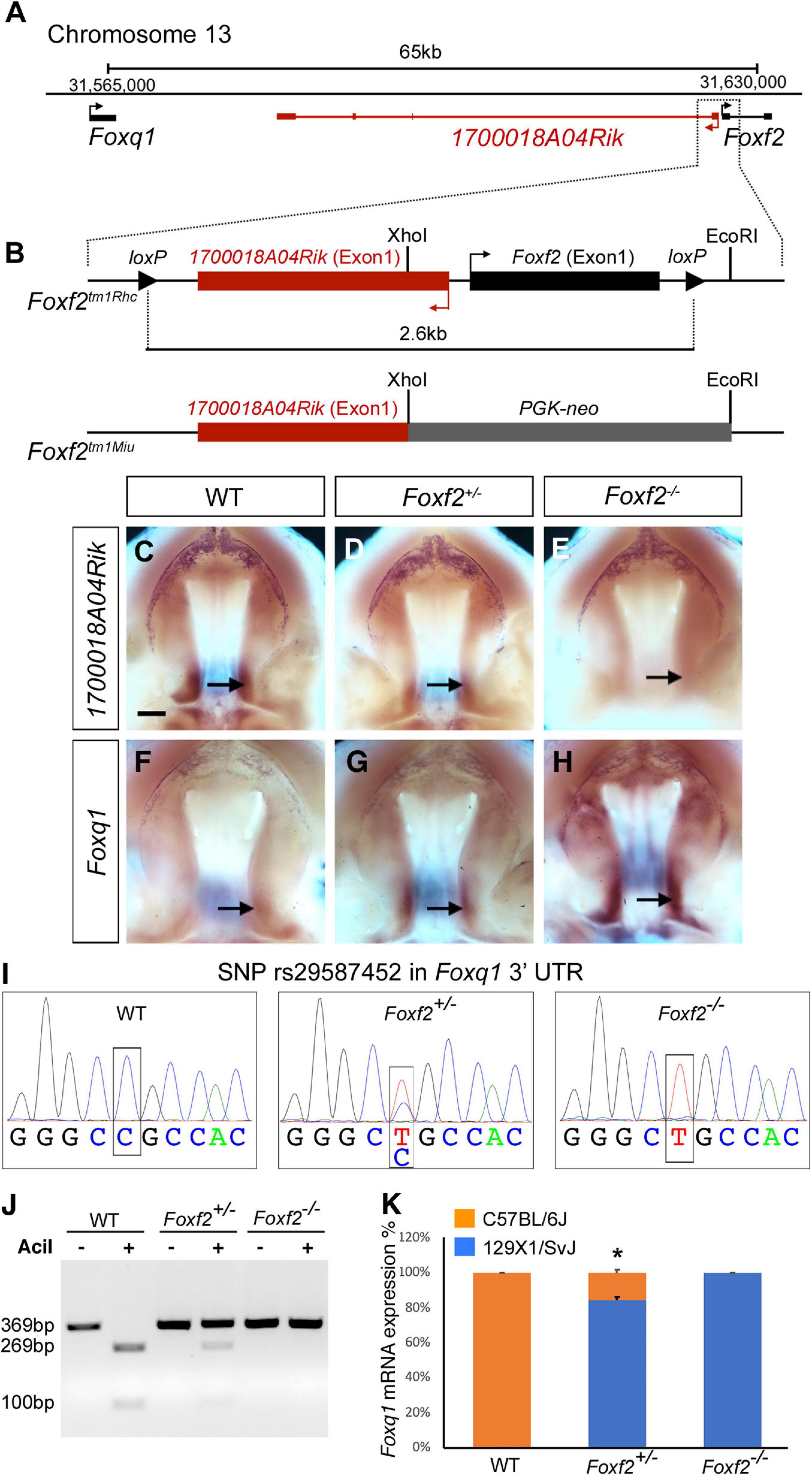
Figure 1. Disruption of Foxf2 causes increased expression of the linked Foxq1 allele in cis in the developing palate. (A) Schematics of the Foxq1-Foxf2 gene locus. Foxq1 is located 65 kb upstream of Foxf2, and the two genes are transcribed from the same strand. The lncRNA gene 1700018A04Rik is located between Foxq1 and Foxf2, and is transcribed from the opposite DNA strand. (B) Schematics of the gene targeting strategies of two Foxf2 mutant mouse lines, Foxf2tm1Rhc and Foxf2tm1Miu. Note that both Foxf2 gene-knockout alleles inactivated both Foxf2 and 1700018A04Rik. (C–H) Comparison of patterns of expression of 1700018A04Rik (C–E) and Foxq1 (F–H) mRNAs in the E13.5 wild-type (WT) (C,F), Foxf2+/– (D,G), and Foxf2–/– (E,H) embryos. Note that 1700018A04Rik mRNAs were downregulated, while Foxq1 mRNAs were upregulated along the anterior-posterior axis, and more significantly increased in the posterior (arrows) subdomains of Foxf2+/– and Foxf2–/– mutant palatal shelves. Scale bar, 400 μm. (I) Sanger sequencing verification of a SNP (rs29587452) in the Foxq1 3′ UTR that could distinguish the mRNAs transcribed from the Foxf2 mutation-linked cis Foxq1 allele (129SvEv background) versus the mRNAs transcribed from the Foxq1 allele linked to the wildtype Foxf2 locus (C57BL/6J background) in heterozygous embryos. (J) DNA electrophoresis gel image showing restriction length polymorphism of RT-PCR products from the 3′ UTR of the Foxq1 mRNAs from E13.5 WT and Foxf2+/– embryonic palatal tissues. (K) Quantification of the ratio of AciI-digested and undigested DNA fragments by QIAxcel advanced system. Note that over 80% of the Foxq1 mRNAs in the Foxf2+/– palatal shelves was expressed from the Foxf2 mutation-linked cis Foxq1 allele (middle column) (n = 5). *p < 0.05.
The expression pattern of Foxq1 in the Foxf2–/– mutant palatal shelves resembles the expression pattern of Foxf2 in wildtype embryos (Xu et al., 2016). The physical linkage of Foxq1 with Foxf2 in the genome and the dose-dependent alteration of Foxq1 expression in the Foxf2+/– and Foxf2–/– embryos raise the question whether the Foxf2 gene disruption directly affects the expression of the linked Foxq1 allele in cis. Since the Foxf2tm1Rhc allele was generated by homologous recombination based gene targeting in mouse embryonic stem cells of the C57BL/6 × 129SvEv hybrid genetic background (Hoggatt et al., 2013; Bolte et al., 2015), we first examined whether a single nucleotide polymorphism (SNP) (rs29587452) in the Foxq1 3′ UTR between C57BL/6J and 129X1/SvJ mouse strains4 could distinguish the mRNAs transcribed from the Foxf2 mutation-linked cis Foxq1 allele versus the mRNAs transcribed from the Foxq1 allele linked to the wildtype Foxf2 locus in heterozygous embryos. Sequencing analysis of RT-PCR products from wildtype, Foxf2+/–, and Foxf2–/– embryos showed that the Foxq1 allele cis-linked with the Foxf2 mutation was of the 129X1/SvJ genotype and distinct from the C57BL/6J allele for rs29587452 (Figure 1I), indicating that the targeted Foxf2tm1Rhc allele was of 129SvEv origin and that the Foxq1 allele in the 129SvEv background shares the same variant in the 3′ UTR as in the 129X1/SvJ background. The C57BL/6J sequence at the SNP site contains a recognition sequence for the AciI endonuclease (CCGC) that is disrupted in the 129SvEv allele. We designed a pairs of PCR primers flanking SNP rs29587452 and used AciI restriction fragment polymorphism to analyze possible differential expression of the two Foxq1 alleles in the Foxf2+/– mutant palate shelves (Figure 1J). We found that over 80% of the Foxq1 mRNAs in the Foxf2+/– palatal shelves was expressed from the Foxf2 mutation-linked cis Foxq1 allele (Figure 1K), suggesting that the increased Foxq1 mRNA expression in the Foxf2+/– and Foxf2–/– embryos resulted primarily from a cis-regulatory effect of the Foxf2 mutation.
Generation of Foxf2/Foxq1 Double Mutant Mice and Validation of Cis-Regulation of Foxq1 Expression by Foxf2 Disruption
To further investigate the regulation and function of Foxq1 in the Foxf2 mutant mice, we used the CRISPR/Cas9 genome editing technology to inactivate the Foxq1 gene in the Foxf2c/c mice. Two independent mouse lines, carrying a deletion of 509 (Foxf2c; Foxq1D509) and 494 (Foxf2c; Foxq1D494) bp, respectively, spanning the entire Forkhead domain-coding region of the Foxq1 gene were established and used in this study (Figure 2A). Both Foxq1D509 and Foxq1D494 alleles resulted in identical silky coat phenotypes in the homozygous mutants (Supplementary Figure 1), which are similar to the previously reported phenotype of Foxq1 null mutant mice (Hong et al., 2001; Goering et al., 2008). Thus, we refer to these alleles as Foxq1–. We crossed the Foxf2c/c; Foxq1+/– mice with the EIIa-Cre transgenic mice (Lakso et al., 1996) to delete the floxed Foxf2 region in the early embryo and subsequently crossed the (Foxf2/Foxq1)+/– progeny to C57BL/6J mice to establish the (Foxf2/Foxq1)+/– mouse colony. These (Foxf2/Foxq1)+/– mice allowed us to further investigate allele-specific effects of the Foxf2 mutation on the linked Foxq1 allele by direct quantitative comparison of allele-specific Foxq1 expression between different embryos of distinct genotypes (Figure 2B). As shown in Figures 2B,C, we found that the total amount of mRNAs from the two Foxq1 alleles expressed in the E13.5 palatal shelves were increased by more than twofold in both Foxf2+/– and (Foxf2/Foxq1)+/– embryos in comparison with their wildtype littermates. However, the amount of mRNAs produced from the wildtype Foxq1 allele in the (Foxf2/Foxq1)+/– embryos was only about 50% of the amount of Foxq1 mRNAs in the wildtype sample (Figures 2B,C). Together with the result that the Foxq1 mRNAs in the palatal mesenchyme in the Foxf2+/– embryos was predominantly produced by the Foxf2 mutation-linked cis Foxq1 allele (Figures 1J,K), these results indicate that the Foxf2 mutation caused the significantly increased expression of the cis-linked Foxq1 allele but had little effect on the Foxq1 allele in trans.
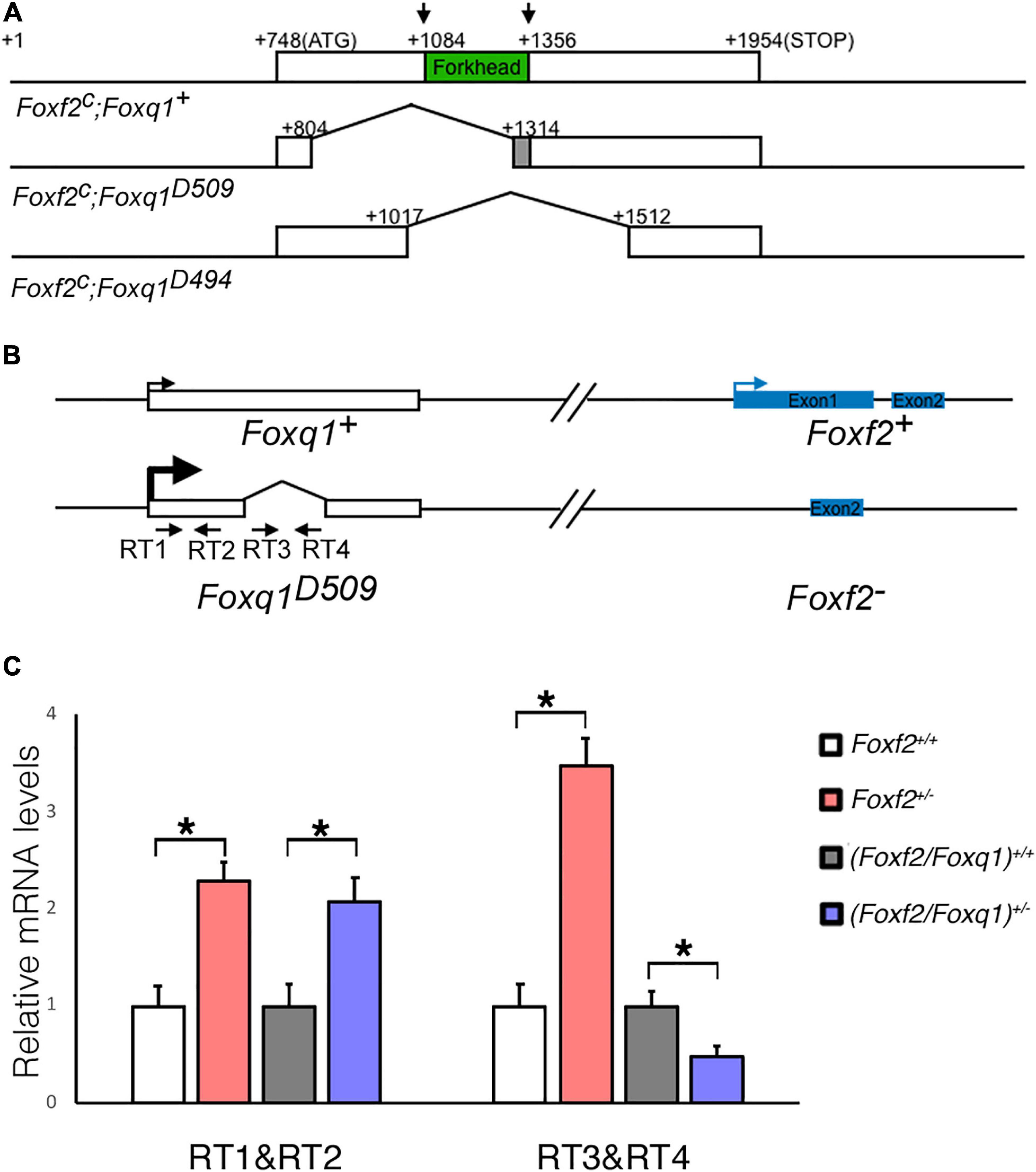
Figure 2. Generation of Foxf2/Foxq1 double mutant mouse. (A) Schematics of the strategy for generating Foxf2c; Foxq1– mice using the CRISPR/cas9 approach. The top row shows the genomic organization of the mouse Foxq1 locus. The coding sequence is boxed with the Forkhead-domain coding region filled in green. The two vertical arrows indicate the positions of recognition sites of the two sgRNAs used for CRISPR genome editing. The second and third rows show the two edited Foxf2c; Foxq1– mouse alleles, Foxf2c; Foxq1D509, and Foxf2c; Foxq1D494, respectively. (B) Schematics of the strategy for investigating the allele-specific effects of the Foxf2 mutation on the linked Foxq1 gene. A pair of primers (RT1&RT2) was designed to detect the Foxq1 mRNAs expressed from both alleles. Another pair of primers (RT3&RT4) was designed to detect the Foxq1 mRNAs expressed from only the wildtype Foxq1 allele. (C) Real time RT-qPCR analysis of the levels of expression of Foxq1 mRNAs in E13.5 palatal shelves in Foxf2+/+, Foxf2+/–,(Foxf2/Foxq1)+/+, and (Foxf2/Foxq1)+/– embryos (n ≥ 4). *p < 0.05.
The Cis-Regulatory Effect of the Foxf2 Mutation on Foxq1 Expression Is Highly Specific and Correlates With Local Genome Organization
Genome-wide chromosome conformation capture and sequencing (Hi-C) studies have demonstrated that mammalian genomes are organized into a series of topologically associated domains (TADs), megabase-scale genomic intervals where interactions between enhancers and gene promoters take place more frequently within than across adjacent TADs (Dixon et al., 2012; Nora et al., 2012; Bonev et al., 2017). Although the Foxc1 gene is located immediately downstream of the Foxf2 gene in the evolutionarily conserved Foxq1-Foxf2-Foxc1 gene cluster, analysis of previously generated Hi-C data from mouse and human embryonic stem cells showed that the Foxq1 and Foxf2 genes are located in the same TAD while the Foxc1 gene is located in a separate adjacent TAD (Figure 3A; Dixon et al., 2012; Haliburton et al., 2016). RT-qPCR analysis showed that, in contrast to significantly increased expression of Foxq1 in E13.5 Foxf2+/– and Foxf2–/– palatal tissues, the levels of Foxc1 mRNA expression was not significantly altered in Foxf2+/– and Foxf2–/– palatal tissues in comparison with the wildtype littermates (Figure 3B). Expression of Gmds, which is located downstream of but in the same TAD with Foxc1, was not significantly altered either (Figure 3B). On the other hand, expression of Exoc2, a gene located about 600 kb upstream of Foxf2, was significantly increased in the palatal tissues in Foxf2+/– embryos and further increased in Foxf2–/– embryos in comparison with wildtype littermates (Figure 3B). In situ hybridization analysis showed that Exoc2 mRNA expression was increased throughout the anterior-posterior axis of the palatal shelves, with particularly strong upregulation in the posterior region of the palatal shelves in the Foxf2+/– and Foxf2–/– embryos (Figures 3C–E). Analysis of the Hi-C data (Bonev et al., 2017) indicated that Exoc2 is located in the same TAD with Foxf2 and Foxq1 (Figure 3A). Furthermore, recent analysis of data from systematic epigenomic and transcriptome profiling of mouse embryonic tissues at multiple developmental stages in the Encyclopedia of DNA Elements (ENCODE) project identified many thousands of enhancer-promoter interactions, among which three long distance enhancers located between Exoc2 and Foxq1 exhibited strong replicated associations with the Foxf2 gene promoter but not with the Foxq1 gene promoter (Gorkin et al., 2020; Figure 3A). Remarkably, analysis of recently generated ChIP-seq data for histone H3K27 acetylation chromatin marks in the posterior palatal shelves of E12.5 mouse embryos (Xu et al., 2019) showed that one prominent H3K27ac peak colocalized with one of the distal enhancers, e8937 located in intron-1 of the Exoc2 gene, that showed strong replicated association with the Foxf2 gene promoter in multiple embryonic tissues (Gorkin et al., 2020; Figure 3A). In addition, analysis of the H3K27ac ChIP-seq data (Xu et al., 2019) revealed another strong peak in the intergenic region between Exoc2 and Foxq1, which likely marks an active enhancer in the E12.5 mouse palatal tissues (Figure 3A). Together, these data suggest that expression of Foxf2 in the palatal mesenchyme in wildtype embryos is controlled by distant enhancers located close to the Exoc2 gene and deletion of the Foxf2 gene promoter likely resulted in increased activation of the nearby Foxq1 and Exoc2 genes within the same TAD in the Foxf2+/– and Foxf2–/– palatal mesenchyme by those enhancers.
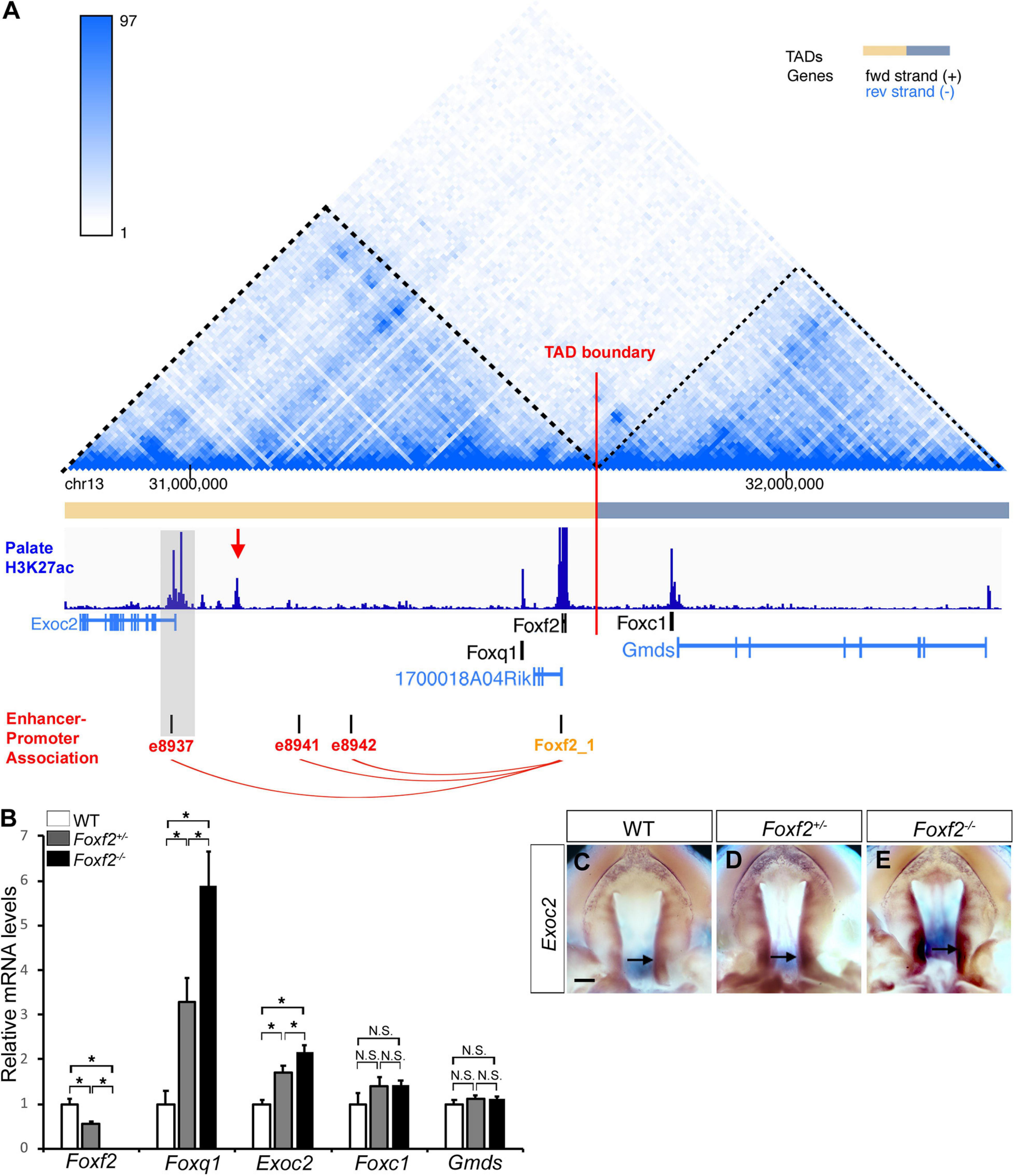
Figure 3. Analysis of genome organization around the Foxf2-Foxq1 gene cluster and the effect of the Foxf2 mutation on expression of neighboring genes in palate development. (A) Hi-C map and histone H3K27ac ChIP-seq peaks in the genomic region containing the Exoc2, Foxq1, 1700018A04Rik, Foxf2, Foxc1, and Gmds genes (Chr13: 30,790,000-32,370,000). The top panel shows the chromatin interaction frequency heatmap from Hi-C analysis of mouse embryonic stem cells (3dgenome.fsm.northwestern.edu) (Bonev et al., 2017; Wang et al., 2018). The color of the heatmap indicates the level of normalized interaction. The identified topological association domains (TADs) are indicated by orange and gray color, respectively. The middle panel shows histone H3K27ac ChIP-seq peak signal plot in deep blue color [data from Xu et al. (2019)]. A red arrow points to a highly enriched H3K27ac peak in the intergenic region between Exoc2 and Foxq1. Transcription orientation of the genes is indicated by black (“ + ” strand) or blue color (“-” strand). The bottom panel shows the replicated associations between enhancers and the Foxf2 gene promoter, retrieved from ENCODE3 and EPDnew (https://genome.ucsc.edu). Three enhancers (e8937, e8941 and e8942) were annotated to show replicated associations with the Foxf2 promoter in mouse fetal tissues. Note that enhancer e8937 is located in intron-1 of the Exoc2 gene and colocalized with a H3K27ac peak from the E12.5 mouse posterior palatal tissues (highlighted in gray). (B) Real time RT-qPCR analysis of the levels of expression of Foxf2, Foxq1, Exoc2, Foxc1, and Gmds mRNAs in the E13.5 palatal shelves in wildtype (WT), Foxf2+/– and Foxf2–/– embryos (n ≥ 4). *p < 0.05. N.S., not significantly different. (C–E) Palatal view of whole mount embryonic upper jaws showing Exoc2 mRNA expression in the palatal tissues in E13.5 wildtype (WT) (C), Foxf2+/– (D), and Foxf2–/– (E) embryos. Arrow points to posterior region of the palatal shelves. Scale bar, 400 μm.
Analysis of the Function of Foxq1 in Foxf2-Mediated Regulation of Palate Development
Intercrossing of (Foxf2/Foxq1)+/– mice generated (Foxf2/Foxq1)–/– mutant mice with over 80% of the homozygous mutants exhibited complete cleft palate (Figures 4A,B). Analysis of the (Foxf2/Foxq1)–/– embryos at multiple developmental stages by histology or skeletal preparations revealed that craniofacial anomalies, including cleft palate, were similar as the phenotypes of Foxf2–/– embryos described previously (Xu et al., 2016). Similar with Foxf2–/– mutant embryos, (Foxf2/Foxq1)–/– embryos displayed defects in palatal shelf elevation (Figures 4C–H) and malformed pterygoid processes (Figures 4A,B).
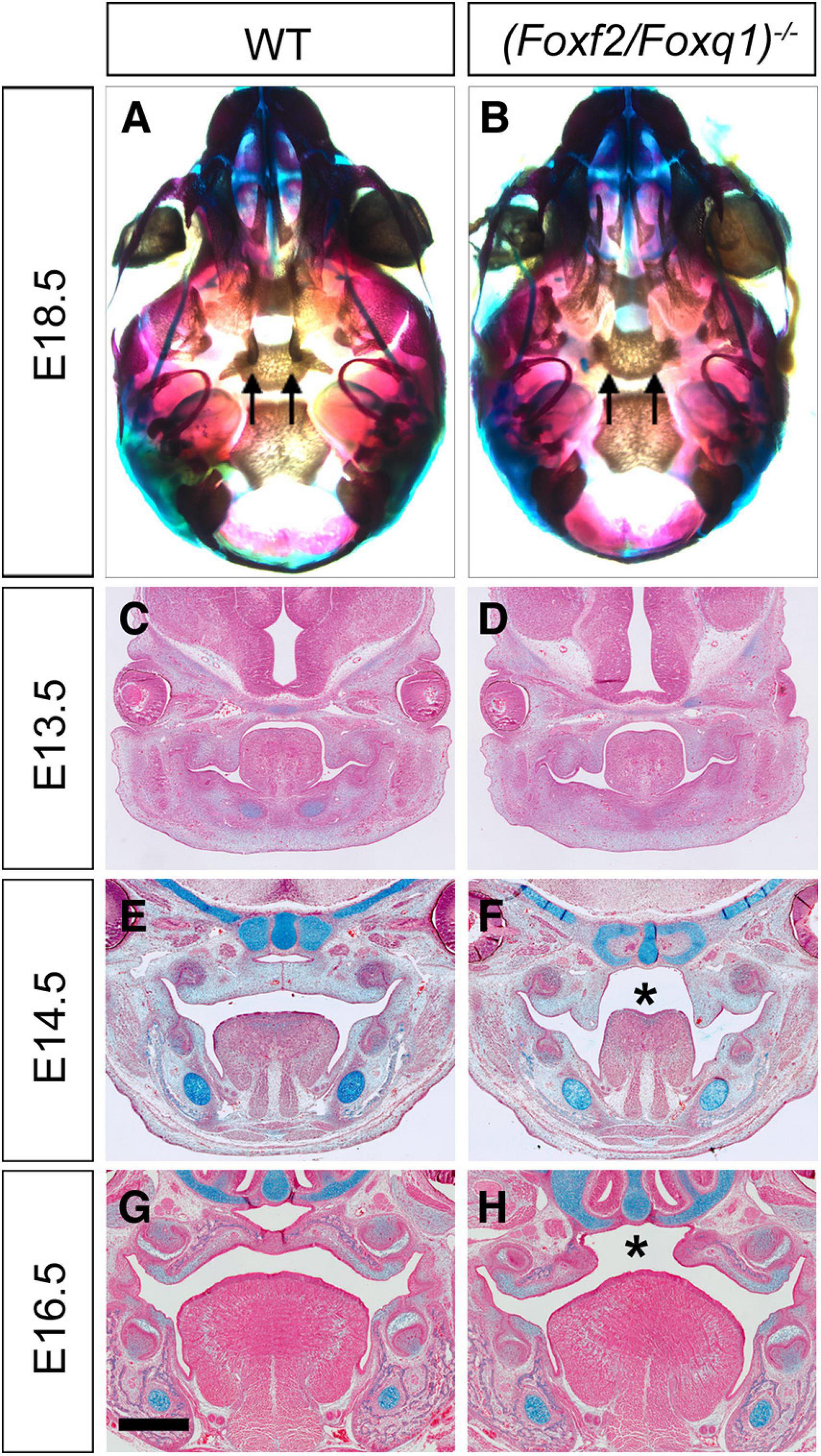
Figure 4. Phenotypical analysis of palate developmental defects in (Foxf2/Foxq1)–/– mutant mouse embryos. (A) Skeletal preparations of heads of E18.5 wildtype (WT) (A) and (Foxf2/Foxq1)–/– (B) embryos. Note that the pterygoid processes (arrows) were extremely hypoplastic in (Foxf2/Foxq1)–/– (B) compare with WT (A) embryos. (C–H) Representative frontal sections of wildtype (C,E,G) and (Foxf2/Foxq1)–/– (D,F,H) embryos at E13.5 (C,D), E14.5 (E,F), and E16.5 (G,H). Note that (Foxf2/Foxq1)–/– embryos displayed cleft palate (“*” in F and H marks the gap between the bilateral palatal shelves). Scale bar, 500 μm.
We then investigated whether the increased expression of Foxq1 contributed to changes in gene expression as previously reported in Foxf2–/– embryos. We examined both levels and patterns of expression of multiple previously identified differentially expressed genes between the Foxf2–/– and control embryos. For most of these genes, the differential expression changes were similarly observed in (Foxf2/Foxq1)–/– embryos as previously reported for Foxf2–/– mutant embryos (Xu et al., 2020; Figure 5). The expression of Foxd1, Exoc2, Fgf18, Chst2, Corin, Adamts9, Pcdh19, Dusp6, Tbx15, Jazf1, Creb5, Smoc2, Lrrc32, and Lmcd1 were increased in the posterior palatal shelves in (Foxf2/Foxq1)–/– embryos in comparison with wildtype littermates at E13.5 (Figure 5). The expression of Shh mRNAs was down-regulated in the posterior palate as well as in the anterior domain corresponding to the most anterior palatal rugae (Figures 5Q,Q′), which is also similar to the pattern of Shh expression in Foxf2–/– embryos (Xu et al., 2016). Nevertheless, we found that expression of Spon1, which was significantly increased in the posterior palatal shelves in Foxf2–/– mutant embryos at E13.5 (Figures 6A,B,I) but was not significantly increased in the E13.5 palatal shelves in (Foxf2/Foxq1)–/– embryos compared with the wildtype littermates (Figures 6C,D,I). In addition, while Shox2 was strongly expressed throughout the anterior half of the palatal shelves in wildtype embryos (Figures 6E,G) and was significantly decreased in the most anterior region of the Foxf2–/– mutant palatal shelves at E13.5 (Figures 6F,I), Shox2 expression in the anterior region of the palatal shelves appeared partly restored in the (Foxf2/Foxq1)–/– mutant embryos (Figures 6H,I). These results indicate that the increased expression of Foxq1 affected expression of some previously identified Foxf2-dependent gene expression patterns in both the anterior and posterior regions of the palatal shelves in the Foxf2–/– embryos.
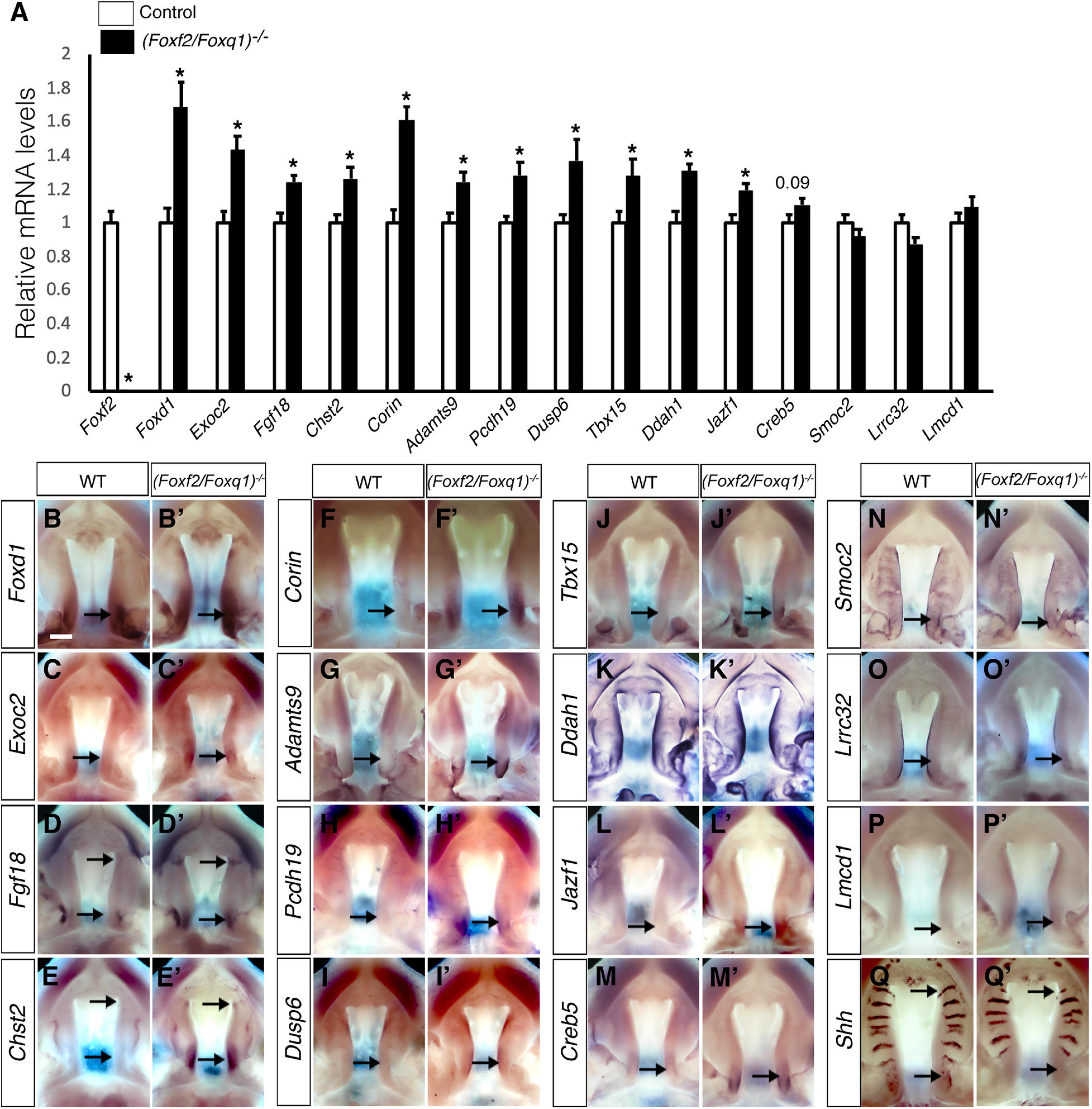
Figure 5. Analysis of expression of Foxf2 target genes in the developing palatal shelves. (A) Real time RT-qPCR analysis of the levels of expression of Foxf2, Foxd1, Exoc2, Fgf18, Chst2, Corin, Adamts9, Pcdh19, Dusp6, Tbx15, Ddah1, Jazf1, Creb5, Smoc2, Lrrc32, and Lmcd1 mRNAs in the E13.5 palatal shelves in control and (Foxf2/Foxq1)–/– embryos (n = 7). *p < 0.05. (B–Q, B′–Q′) Comparison of patterns of expression of Foxd1, Exoc2, Fgf18, Chst2, Corin, Adamts9, Pcdh19, Dusp6, Tbx15, Ddah1, Jazf1, Creb5, Smoc2, Lrrc32, Lmcd1, and Shh mRNAs in E13.5 wild-type (WT) (B–Q) and (Foxf2/Foxq1)–/– (B′–Q′) mutant embryos. Scale bar, 400 μm.
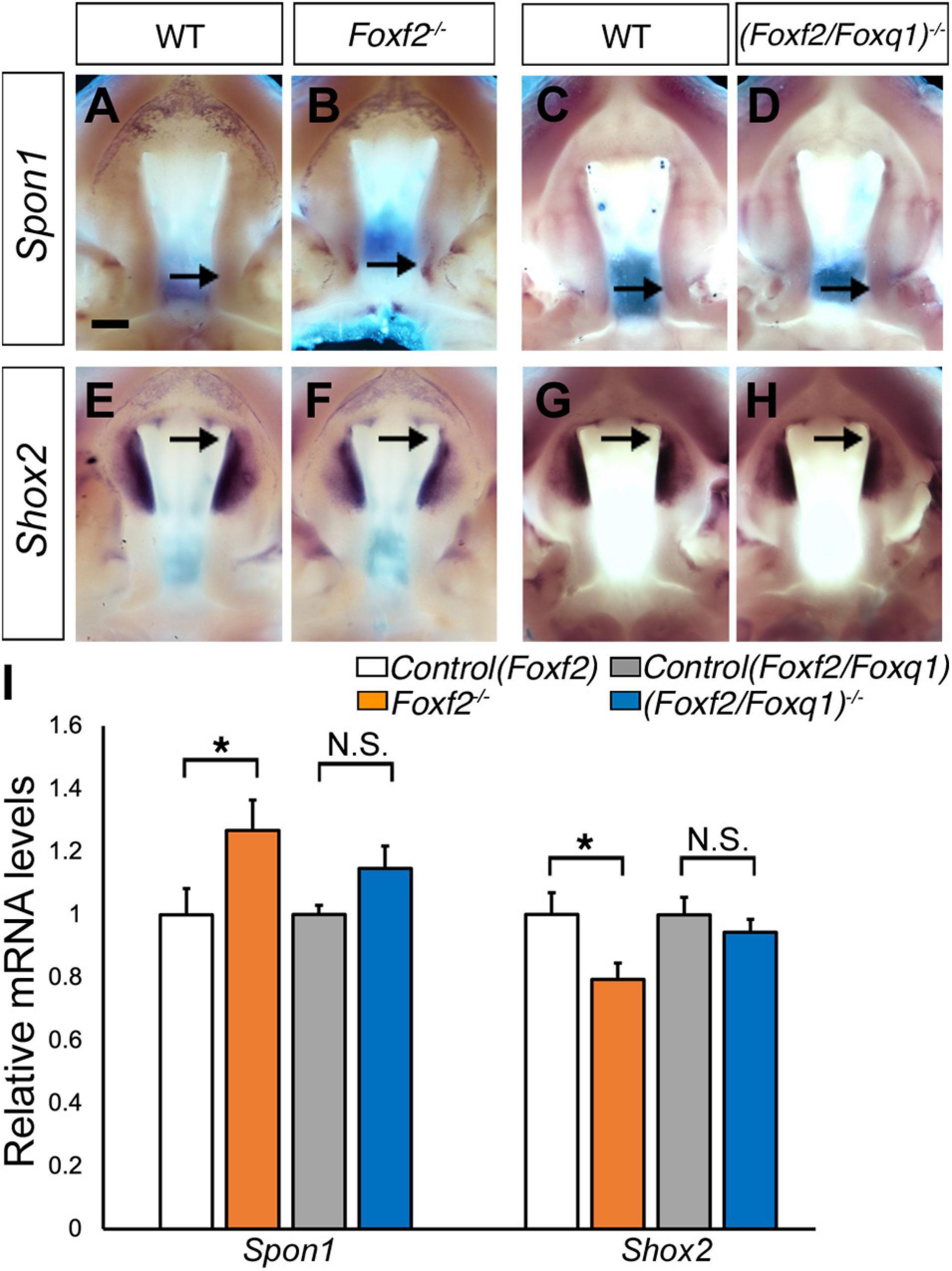
Figure 6. Analysis of expression of Spon1 and Shox2 in the developing palatal shelves. (A–H) Comparison of patterns of expression of Spon1 (A–D) and Shox2 (E–H) mRNAs in E13.5 wild-type (WT) (A,C,E,G), Foxf2–/– (B,F) and (Foxf2/Foxq1)–/– (D,H) mutant embryos. The expression of Spon1 was increased in a specific domain in posterior palatal shelves in Foxf2–/– embryos [arrow in (B)]. The expression of Shox2 was decreased in the anterior palatal shelves in Foxf2–/– embryos (arrow in F). Scale bar, 400 μm. (I) Real time RT-qPCR analysis of the levels of expression of Spon1 and Shox2 mRNAs in E13.5 palatal shelves in the control, Foxf2–/– and (Foxf2/Foxq1)–/– embryos (n = 7) *p < 0.05. N.S., not significantly different.
Foxq1 Partly Complemented Foxf2-Mdiated Regulation of Foxf1 Expression in the Developing Palatal Shelves in Foxf2–/– Embryos
We previously generated and analyzed Foxf1c/c; Foxf2c/c; Wnt1-Cre compound mutant mice and demonstrated that Foxf1 partly compensated Foxf2 function in palate development (Xu et al., 2016). Another study showed that Foxf2–/– mice had increased Foxf1 expression in the colonic smooth muscle cells and that Foxf2 bound and repressed the Foxf1 gene promoter in co-transfection assays (Bolte et al., 2015). Our previous RNA-seq analysis did not detect significant changes in Foxf1 expression in the palatal mesenchyme in Foxf2–/– embryos compared with control littermates. However, we found that Foxf1 expression was significantly increased in the developing palatal shelves in E13.5 (Foxf2/Foxq1)–/– embryos in comparison with the wildtype littermates by both whole mount in situ hybridization and real-time RT-qPCR assays (Figures 7A–C). In the wildtype embryos, expression of Foxf1 was restricted to the middle portion of palatal shelves at E13.5 (Figure 7B). The expression of Foxf1 was increased and extended to the posterior region of the palatal shelves in the (Foxf2/Foxq1)–/– mutant embryos (Figure 7C). We further confirmed these results by immunofluorescent staining using an anti-Foxf1 antibody on frontal sections of wildtype, Foxf2–/–, and (Foxf2/Foxq1)–/– embryos (Figures 7D–I). The Foxf1 protein displayed an oral-to-nasal gradient in the developing palatal mesenchyme, and its levels were increased and extended to the nasal side of the middle portion as well as throughout the posterior region of the palatal shelves in E13.5 (Foxf2/Foxq1)–/– embryos in comparison with the wildtype embryos (Figure 7, compare F and I with D and G, respectively). Interestingly, we also consistently detected moderately increased expression of Foxf1 protein in the posterior region of the palatal shelves in Foxf2–/– embryos compared with the wildtype embryos (Figure 7, compare H with G). These results indicate that Foxf2 negatively regulates Foxf1 gene expression in the developing palatal mesenchyme and that the increased expression of Foxq1 in the Foxf2–/– embryonic palatal mesenchyme partly complemented for Foxf2-mediated regulation of Foxf1 expression. Altogether, although deletion of Foxq1 was ultimately insufficient to rescue the cleft palate defects in the Foxf2–/– mice, the increased expression of Foxq1 resulting from the cis-regulatory effect of the Foxf2 mutation affected the regulation of multiple important genes in palate development.
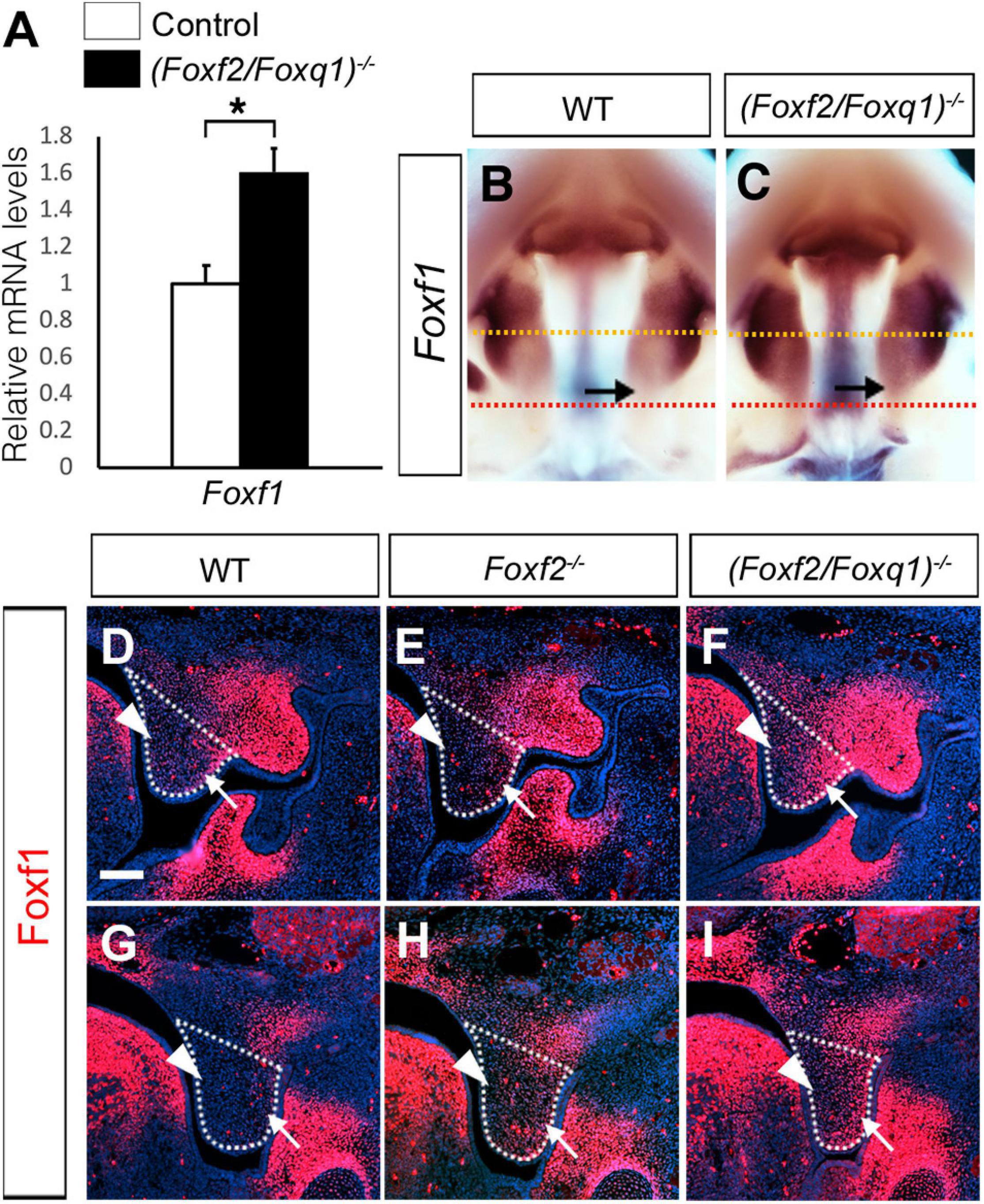
Figure 7. Analysis of expression of Foxf1 in the developing palatal shelves. (A) Real time RT-qPCR analysis of the levels of expression of Foxf1 mRNAs in E13.5 palatal shelves in control and (Foxf2/Foxq1)–/– embryos (n = 7). *p < 0.05. (B,C) Comparison of patterns of expression of Foxf1 mRNAs in E13.5 wild-type (WT) (B) and (Foxf2/Foxq1)–/– (C) mutant embryos. Note that the expression of Foxf1 mRNAs (detected in purple color) was extended to the posterior region of the palatal shelves in the (Foxf2/Foxq1)–/– embryos. Arrow points to the posterior region of the palatal shelves. Yellow dashed line marks the approximate position of the plain of frontal sections of different embryos shown in panels (D–F), whereas red dashed line indicates the approximate position of the plain of frontal sections of different embryos shown in panels (G–I). (D–I) Immunofluorescent staining showing Foxf1 protein (red) on comparable frontal sections from the middle (D–F) and posterior (G–I) regions of the palatal shelves in E13.5 wildtype (WT) (D,G), Foxf2–/– (E,H), and (Foxf2/Foxq1)–/– (F,I) embryos. The palatal shelf was outlined with white dashed lines. Arrow points to the lateral side whereas arrowhead points to the medial side of the palatal shelves. Scale bar, 100 μm.
Discussion
FOXF2 is located in an evolutionarily conserved gene cluster containing FOXQ1, FOXF2, and FOXC1 (Wotton and Shimeld, 2006). Disruption of FOXF2 is associated with cleft palate and posterior palate defects in humans (Jochumsen et al., 2008; Bu et al., 2015; Seselgyte et al., 2019), making Foxf2–/– mutant mouse a good model to study mechanisms of human palate development. Our RNA-seq and in vivo expression assays showed that expression of Foxq1 was the most dramatically upregulated gene in the embryonic palatal mesenchyme in Foxf2–/– mouse embryos. Using CRISPR/Cas9 genome editing, we generated (Foxf2/Foxq1)+/– mice carrying null alleles of Foxf2 and Foxq1 in cis, and identified a novel cis-regulation of Foxq1 expression by the Foxf2 mutation during palate development. Our results indicate that Foxq1 affected expression of several previously identified Foxf2-dependent genes in palate development, which needs to be taken into consideration for elucidating the molecular mechanisms involving Foxf2 in regulating palate development.
While recent in vitro study suggested that FOXF2 binds to the promoter and represses the expression of FOXQ1 in a breast cancer cell line (Kang et al., 2019), our in vivo data demonstrated a cis-regulation of Foxq1 expression by the Foxf2 gene disruption. Cis-regulation of gene expression is primarily mediated by interactions of enhancer and repressor sequences with their target gene promoters. Enhancers can control gene expression in a distance- and orientation-independent manner (Furlong and Levine, 2018). On the other hand, the interactions of enhancers and promoters are controlled by the three-dimensional organization of the genome, which in mammals is organized into a series of TADs that exhibit frequent intra-domain chromatin interactions but relatively rare inter-domain interactions (Dixon et al., 2012; Sexton and Cavalli, 2015). TADs are conserved between different cell types and across species, suggesting that TADs are important for directing enhancer-promoter interactions for controlling spatiotemporal gene expression during animal development (Dixon et al., 2012; Nora et al., 2012). Analysis of the Hi-C data of mouse embryonic stem cells (Bonev et al., 2017; Wang et al., 2018) showed that Foxf2, 1700018A04Rik, Foxq1, and Exoc2 genes are located within the same TAD, whereas Foxc1 is located in a separate TAD (Figure 3A). We found that, in contrast to the significant increases in expression of Foxq1 and Exoc2 mRNAs in the palatal mesenchyme in Foxf2+/– and Foxf2–/– embryos, expression of Foxc1 was not significantly altered in the palatal mesenchyme of those mutant embryos compared with wildtype littermates, indicating that the Foxf2 mutation did not affect the TAD boundary-mediated restriction of enhancer-promoter interactions. The exact molecular mechanism underlying the cis-regulatory effect of the Foxf2 mutation requires further investigation. Previous studies indicated that closely linked gene promoters may compete for the activity of a shared enhancer (Choi and Engel, 1988; Dillon et al., 1997; Lower et al., 2009) and that the sequence composition of core promoters plays a critical role in the specificity of enhancer responsiveness (Merli et al., 1996; Haberle et al., 2019). The ENCODE and Eukaryotic Promoter Database (EPD) projects annotated replicated promoter-enhancer associations between the Foxf2 gene promoter and three distant enhancers in the genomic region between the Exoc2 and Foxq1 genes (Gorkin et al., 2020; Figure 3A), indicating that those enhancers preferentially activated the Foxf2 gene promoter even though the Exoc2 and Foxq1 gene promoters are closer. One of those distant enhancers colocalized with a strong histone H3K27ac peak identified in ChIP-seq analysis of E12.5 mouse palatal tissues (Xu et al., 2019). Another highly enriched H3K27ac peak in the intergenic region between Exoc2 and Foxq1 likely marked another active enhancer in the developing palatal mesenchyme (Figure 3A). One possible mechanism is that the distant enhancers that activated Foxf2 gene expression in the palatal mesenchyme in wildtype embryos interacted with and activated the Foxq1 gene promoter in cis with the Foxf2 mutant allele (Figure 8A) in Foxf2+/– and Foxf2–/– embryos. A similar cis-regulatory mechanism has been shown to drive increased expression of the NME4 gene located ∼300 kb away from the α-globin gene cluster in humans when one or more copies of the α-globin gene was deleted (Lower et al., 2009). In addition, the co-transcription of the 1700018A04Rik lncRNA gene with Foxf2 may play a role in repressing Foxq1 gene expression in the developing palatal mesenchyme in wildtype embryos (Figure 8A). A few lncRNAs have been shown to recruit regulatory complexes through RNA-protein interactions to repress nearby genes in cis (Maamar et al., 2013; McHugh et al., 2015; Cipriano et al., 2021). Transcription of some lncRNA genes has also been shown to repress expression of a nearby gene in cis through transcriptional interference independently of the lncRNA transcript (Houseley et al., 2008; Latos et al., 2012). Whether 1700018A04Rik plays a detectable role in the transcriptional regulation of Foxf2 and/or Foxq1 remains to be determined.
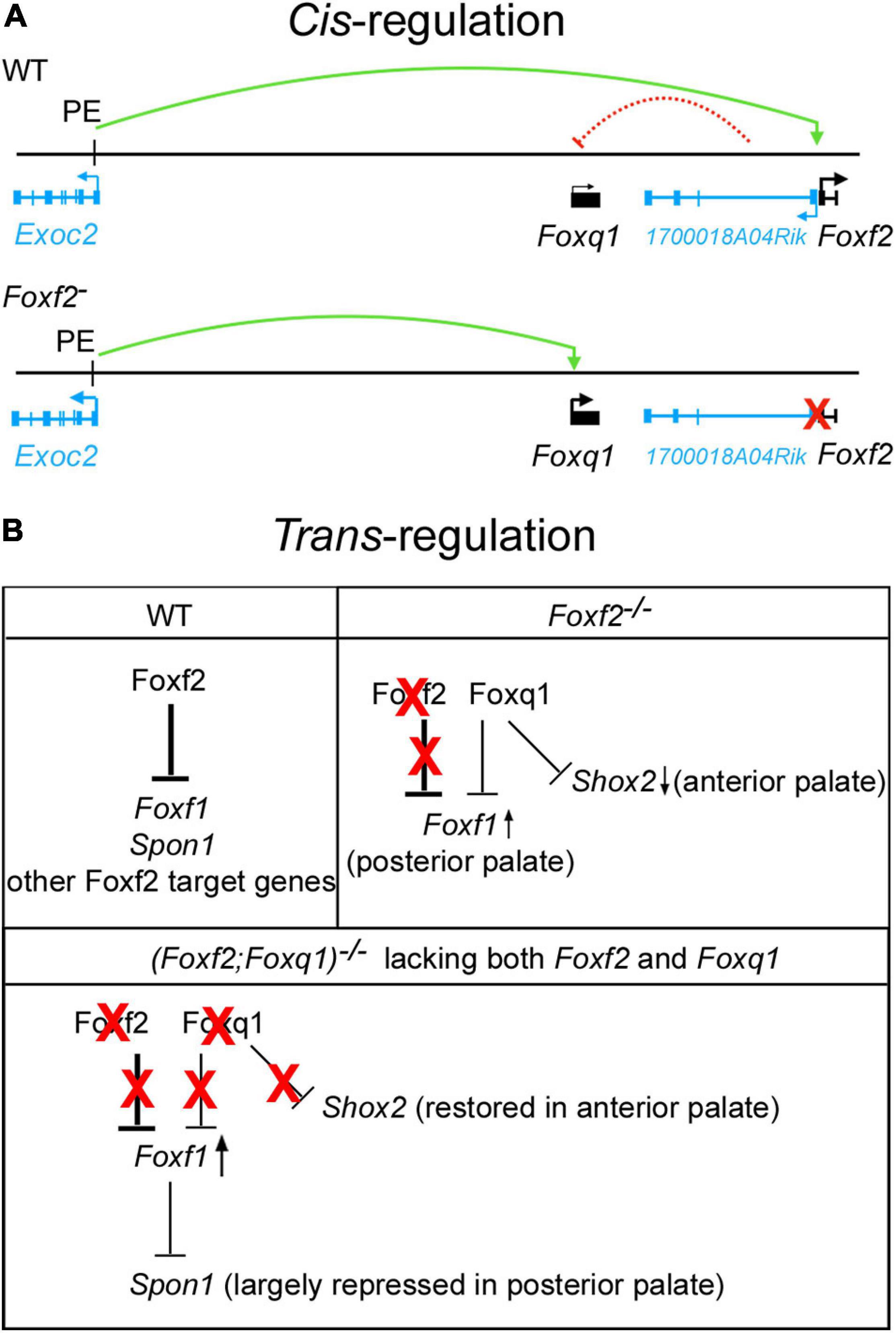
Figure 8. Schematic models of cis-regulation of Foxf2 and Foxq1 expression and of trans-regulation of Foxf1, Shox2, Spon1 by Foxf2, and Foxq1 in palate development. (A) Model of mechanisms of cis-regulation of Foxq1 and Foxf2 in palate development. Top panel, in wildtype (WT) embryonic palate, expression of Foxf2 and the 1700018A04Rik lncRNA gene is activated through the bidirectional promoter by distal enhancers, including the one identified in intron-1 of the Exoc2 gene (labeled as PE, for palatal enhancer). Expression of Foxq1 is not activated and may be repressed by the lncRNA transcript or by transcriptional interference from the 1700018A04Rik gene in wildtype embryonic palate. The green curved line with an arrow indicates transcriptional activation by the palatal enhancer. The red dashed curved line from the 1700018A04Rik gene to the Foxq1 gene indicates a hypothetical cis-repression. Bottom panel depicts activation of Foxq1 gene expression in Foxf2+/– and Foxf2–/– embryos by the palatal enhancer (PE) due to deletion of the Foxf2 gene promoter (marked by a red X), which also abolished any cis-repressive effect of the 1700018A04Rik gene on the Foxq1 allele. (B) Models of trans-regulation involving Foxf1, Foxf2, and Foxq1. In wildtype embryonic palate, Foxf2 is strongly expressed in the posterior region of palatal shelves and represses expression of Foxf1, Spon1, and multiple other target genes as previously identified (Xu et al., 2020) in the posterior palatal mesenchyme. In Foxf2–/– embryonic palate, Foxf2 protein is absent (marked by red X over “Foxf2”) but Foxq1 was activated in both the anterior and posterior regions of the palatal shelves. The increased Foxq1 expression at least partly accounted for the reduction in Shox2 expression in the anterior palate in Foxf2–/– embryos since Shox2 expression was largely restored in the (Foxf2/Foxq1) –/– embryos. Foxf1 expression was moderately increased in the posterior palate in Foxf2–/– embryos and more strongly expressed in the (Foxf2/Foxq1)–/– embryonic palate, indicating that Foxq1 had a repressive effect on Foxf1 expression in the Foxf2–/– palate. Spon1 expression was significantly increased in the posterior palate in Foxf2–/– embryos but not in the (Foxf2/Foxq1)–/– embryos, likely due to the increased Foxf1 complementing Foxf2-mediated repression of Spon1 in the (Foxf2/Foxq1)–/– embryos. Red X indicates absence of the protein and its regulatory action. Arrow pointing up indicates increased expression, whereas arrow pointing down indicates decreased expression. Note that expression of many Foxf2-dependent genes was similarly altered in the Foxf2–/– and (Foxf2/Foxq1)–/– embryos and are not depicted.
A recent study reported a congenital palate defect with absent uvula, shortened posterior border of the soft palate, and abnormal tonsillar pillar, that cosegregated with a heterozygous missense mutation in FOXF2 in a large family (Seselgyte et al., 2019). Moreover, submicroscopic 6p25 deletion and/or duplication, including or near the FOXF2 gene locus, have been associated with syndromic defects including palatal abnormalities in patients (Descipio et al., 2005; Nakane et al., 2013; Seselgyte et al., 2019). Therefore, studying the cis-regulation network of this genomic locus will provide valuable insight into the mechanisms of FOXF2-related human diseases.
The expression of Foxq1 was dramatically up-regulated in the palatal mesenchyme cells in Foxf2–/– embryos compared with wildtype littermates, but deletion of Foxq1 didn’t significantly rescue the craniofacial defects or expression of most of the previously identified Foxf2 target genes in the (Foxf2/Foxq1)–/– embryos. This is possibly due to the relatively low level of expression of Foxq1 in the Foxf2–/– palatal mesenchyme compared with the amount of Foxf2 mRNAs expressed in the wildtype palatal mesenchyme. On the other hand, previous studies have suggested that FOXF2 and FOXQ1 play opposite roles in controlling epithelial-mesenchymal transition and visceral metastasis in basal-like breast cancer cells (Qiao et al., 2011; Zhang et al., 2011; Cai et al., 2015; Wang et al., 2015; Kang et al., 2019). Other studies showed that FOXF1 and FOXQ1 was able to bind to the same region of the telokin promoter, but they exhibited opposing effects on the promoter activity in colonic smooth muscle cells (Hoggatt et al., 2013). We found that inactivation of Foxq1 partly restored Shox2 expression in the anterior palate and reduced the aberrant overexpression of Spon1 in the posterior palate, but also caused obviously enhanced increase in Foxf1 expression in both the middle and posterior regions of the palatal mesenchyme, in (Foxf2/Foxq1)–/– embryos compared with the Foxf2–/– and wildtype control embryos (Figure 7). While these data may suggest that Foxq1 acted to repress Shox2 and Foxf1 expression while also activating Spon1 expression in a domain specific manner, the most logical explanation is that the increased expression of Foxq1 directly or indirectly repressed Shox2 and Foxf1 expression in the palatal mesenchyme in the Foxf2–/– embryos whereas the more significantly increased expression of Foxf1 compensated for Foxf2-mediated repression of Spon1 in the posterior palatal mesenchyme in the (Foxf2/Foxq1)–/– embryos (Figure 8B). In this scenario, Foxq1 acted as a repressor similar as Foxf2, but also had distinct activity with regards to the regulation of Shox2 and Spon1 expression. Furthermore, although Foxf1 expression was extended into throughout the posterior region of the palatal shelves in the (Foxf2/Foxq1)–/– embryos, the increased expression of Foxf1 was insufficient to rescue the cleft palate phenotype or the altered expression of many genes in the posterior palatal mesenchyme that resulted from Foxf2 disruption, suggesting that Foxf2 has distinct molecular functions in palate development that could not be complemented by either Foxq1 or Foxf1.
In summary, this study demonstrates that disruption of the Foxf2 gene promoter had a cis-regulatory effect on the expression of nearby genes. Our results clearly demonstrate that the increased expression of Foxq1 in the Foxf2+/– and Foxf2–/– embryos was due to the cis-regulatory effect. Although (Foxf2/Foxq1)–/– mice exhibited similar cleft palate phenotypes as Foxf2–/– mice, Foxq1 exerted a regulatory effect on the expression of several previously identified Foxf2-dependent genes in palate development. We show that Exoc2, located about 600 kb upstream of Foxf2, was also significantly upregulated in the developing palatal shelves in Foxf2+/– embryos and was further significantly upregulated in the Foxf2–/– embryos. Whether the increase in Exoc2 expression in the palatal tissues in Foxf2+/– and Foxf2–/– embryos was solely due to the cis-regulatory effect and whether the altered expression of Exoc2 contributed significantly to the craniofacial and other developmental defects in the Foxf2–/– mice remain to be investigated. Since many gene knockout studies assigned gene function based on promoter-deletion alleles, our finding of a cis-regulatory effect of the Foxf2 mutation calls for caution in interpretation of results and underlying mechanisms from those alleles. Potential cis-regulatory effects on neighboring genes should also be taken into consideration when analyzing pathogenicity of human gene deletion variants. In addition, since Foxf2 mutant mouse studies have shown that multiple tissues and developmental processes, including pericyte development and maturation of the blood-brain barrier, development of the respiratory and digestive organs, in addition to craniofacial and palate tissues, depend on Foxf2 function (reviewed by He et al., 2020), the (Foxf2/Foxq1)+/– mice provide a valuable resource for understanding the cross-regulation and combinatorial functions of the Foxf2 and Foxq1 genes in multiple developmental and disease processes.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: The accession number for the RNA-seq data is GSE67015. The accession number for the palate Foxf2 ChIP-seq data is GSE137585.
Ethics Statement
The animal study was reviewed and approved by Institutional Animal Care and Use Committee (IACUC) at Cincinnati Children’s Hospital Medical Center.
Author Contributions
YL and RJ designed the research. JX and HL conducted the experiments and collected the data. JX and RJ wrote the manuscript. All authors analyzed the data, revised the manuscript, and agreed and approved the manuscript.
Funding
This work was supported by Shriners Hospitals for Children grant #85900 and by the National Institutes of Health/National Institute of Dental and Craniofacial Research grant R01DE027046 to RJ.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the Transgenic Animal and Genome Editing Core facility at Cincinnati Children’s Hospital Medical Center for assistance with the generation of the Foxq1 gene-edited mouse lines.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.665109/full#supplementary-material
Footnotes
- ^ http://3dgenome.fsm.northwestern.edu
- ^ https://www.ncbi.nlm.nih.gov/geo/
- ^ https://genome.ucsc.edu/
- ^ http://www.informatics.jax.org/snp/rs29587452
References
Bolte, C., Ren, X., Tomley, T., Ustiyan, V., Pradhan, A., Hoggatt, A., et al. (2015). Forkhead box F2 regulation of platelet-derived growth factor and myocardin/serum response factor signaling is essential for intestinal development. J. Biol. Chem. 290, 7563–7575. doi: 10.1074/jbc.m114.609487
Bonev, B., Mendelson Cohen, N., Szabo, Q., Fritsch, L., Papadopoulos, G. L., Lubling, Y., et al. (2017). Multiscale 3D genome rewiring during mouse neural development. Cell 171, 557–572 e524.
Bu, L., Chen, Q., Wang, H., Zhang, T., Hetmanski, J. B., Schwender, H., et al. (2015). Novel evidence of association with nonsyndromic cleft lip with or without cleft palate was shown for single nucleotide polymorphisms in FOXF2 gene in an Asian population. Birth Defects Res. A Clin. Mol. Teratol. 103, 857–862.
Bush, J. O., and Jiang, R. (2012). Palatogenesis: morphogenetic and molecular mechanisms of secondary palate development. Development 139, 231–243. doi: 10.1242/dev.067082
Cai, J., Tian, A. X., Wang, Q. S., Kong, P. Z., Du, X., Li, X. Q., et al. (2015). FOXF2 suppresses the FOXC2-mediated epithelial-mesenchymal transition and multidrug resistance of basal-like breast cancer. Cancer Lett. 367, 129–137. doi: 10.1016/j.canlet.2015.07.001
Chai, Y., and Maxson, R. E. (2006). Recent advances in craniofacial morphogenesis. Dev. Dyn. 235, 2353–2375. doi: 10.1002/dvdy.20833
Choi, O. R., and Engel, J. D. (1988). Developmental regulation of beta-globin gene switching. Cell 55, 17–26. doi: 10.1016/0092-8674(88)90005-0
Cipriano, A., Macino, M., Buonaiuto, G., Santini, T., Biferali, B., Peruzzi, G., et al. (2021). Epigenetic regulation of Wnt7b expression by the cis-acting long noncoding RNA Lnc-Rewind in muscle stem cells. Elife 10:e54782.
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823.
Descipio, C., Schneider, L., Young, T. L., Wasserman, N., Yaeger, D., Lu, F., et al. (2005). Subtelomeric deletions of chromosome 6p: molecular and cytogenetic characterization of three new cases with phenotypic overlap with Ritscher-Schinzel (3C) syndrome. Am. J. Med. Genet. A 134A, 3–11. doi: 10.1002/ajmg.a.30573
Dillon, N., Trimborn, T., Strouboulis, J., Fraser, P., and Grosveld, F. (1997). The effect of distance on long-range chromatin interactions. Mol. Cell. 1, 131–139. doi: 10.1016/s1097-2765(00)80014-3
Dixon, J. R., Selvaraj, S., Yue, F., Kim, A., Li, Y., Shen, Y., et al. (2012). Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380. doi: 10.1038/nature11082
Dixon, M. J., Marazita, M. L., Beaty, T. H., and Murray, J. C. (2011). Cleft lip and palate: understanding genetic and environmental influences. Nat. Rev. Genet. 12, 167–178. doi: 10.1038/nrg2933
Furlong, E. E. M., and Levine, M. (2018). Developmental enhancers and chromosome topology. Science 361, 1341–1345. doi: 10.1126/science.aau0320
Goering, W., Adham, I. M., Pasche, B., Manner, J., Ochs, M., Engel, W., et al. (2008). Impairment of gastric acid secretion and increase of embryonic lethality in Foxq1-deficient mice. Cytogenet. Genome Res. 121, 88–95. doi: 10.1159/000125833
Gorkin, D. U., Barozzi, I., Zhao, Y., Zhang, Y., Huang, H., Lee, A. Y., et al. (2020). An atlas of dynamic chromatin landscapes in mouse fetal development. Nature 583, 744–751.
Haberle, V., Arnold, C. D., Pagani, M., Rath, M., Schernhuber, K., and Stark, A. (2019). Transcriptional cofactors display specificity for distinct types of core promoters. Nature 570, 122–126. doi: 10.1038/s41586-019-1210-7
Haliburton, G. D., Mckinsey, G. L., and Pollard, K. S. (2016). Disruptions in a cluster of computationally identified enhancers near FOXC1 and GMDS may influence brain development. Neurogenetics 17, 1–9. doi: 10.1007/s10048-015-0458-9
Han, J., Mayo, J., Xu, X., Li, J., Bringas, P., and Maas, R. L. (2009). Indirect modulation of Shh signaling by Dlx5 affects the oral-nasal patterning of palate and rescues cleft palate in Msx1-null mice. Development 136, 4225–4233. doi: 10.1242/dev.036723
He, W., Kang, Y., Zhu, W., Zhou, B., Jiang, X., Ren, C., et al. (2020). FOXF2 acts as a crucial molecule in tumours and embryonic development. Cell Death Dis. 11:424.
Hellqvist, M., Mahlapuu, M., Samuelsson, L., Enerback, S., and Carlsson, P. (1996). Differential activation of lung-specific genes by two forkhead proteins, FREAC-1 and FREAC-2. J. Biol. Chem. 271, 4482–4490. doi: 10.1074/jbc.271.8.4482
Hoggatt, A. M., Kim, J. R., Ustiyan, V., Ren, X., Kalin, T. V., Kalinichenko, V. V., et al. (2013). The transcription factor Foxf1 binds to serum response factor and myocardin to regulate gene transcription in visceral smooth muscle cells. J. Biol. Chem. 288, 28477–28487. doi: 10.1074/jbc.m113.478974
Hong, H. K., Noveroske, J. K., Headon, D. J., Liu, T., Sy, M. S., Justice, M. J., et al. (2001). The winged helix/forkhead transcription factor Foxq1 regulates differentiation of hair in satin mice. Genesis 29, 163–171. doi: 10.1002/gene.1020
Houseley, J., Rubbi, L., Grunstein, M., Tollervey, D., and Vogelauer, M. (2008). A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Mol. Cell. 32, 685–695. doi: 10.1016/j.molcel.2008.09.027
Jochumsen, U., Werner, R., Miura, N., Richter-Unruh, A., Hiort, O., and Holterhus, P. M. (2008). Mutation analysis of FOXF2 in patients with disorders of sex development (DSD) in combination with cleft palate. Sex Dev. 2, 302–308. doi: 10.1159/000195679
Kang, L. J., Yu, Z. H., Cai, J., He, R., Lu, J. T., Hou, C., et al. (2019). Reciprocal transrepression between FOXF2 and FOXQ1 controls basal-like breast cancer aggressiveness. FASEB J. 33, 6564–6573. doi: 10.1096/fj.201801916r
Lakso, M., Pichel, J. G., Gorman, J. R., Sauer, B., Okamoto, Y., Lee, E., et al. (1996). Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc. Natl. Acad. Sci. U.S.A. 93, 5860–5865. doi: 10.1073/pnas.93.12.5860
Lan, Y., and Jiang, R. (2009). Sonic hedgehog signaling regulates reciprocal epithelial-mesenchymal interactions controlling palatal outgrowth. Development 136, 1387–1396. doi: 10.1242/dev.028167
Lan, Y., Xu, J., and Jiang, R. (2015). Cellular and molecular mechanisms of palatogenesis. Curr. Top. Dev. Biol. 115, 59–84. doi: 10.1016/bs.ctdb.2015.07.002
Latos, P. A., Pauler, F. M., Koerner, M. V., Senergin, H. B., Hudson, Q. J., Stocsits, R. R., et al. (2012). Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science 338, 1469–1472. doi: 10.1126/science.1228110
Lower, K. M., Hughes, J. R., De Gobbi, M., Henderson, S., Viprakasit, V., Fisher, C., et al. (2009). Adventitious changes in long-range gene expression caused by polymorphic structural variation and promoter competition. Proc. Natl. Acad. Sci. U.S.A. 106, 21771–21776. doi: 10.1073/pnas.0909331106
Maamar, H., Cabili, M. N., Rinn, J., and Raj, A. (2013). linc-HOXA1 is a noncoding RNA that represses Hoxa1 transcription in cis. Genes Dev. 27, 1260–1271. doi: 10.1101/gad.217018.113
McHugh, C. A., Chen, C. K., Chow, A., Surka, C. F., Tran, C., Mcdonel, P., et al. (2015). The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 521, 232–236. doi: 10.1038/nature14443
Merli, C., Bergstrom, D. E., Cygan, J. A., and Blackman, R. K. (1996). Promoter specificity mediates the independent regulation of neighboring genes. Genes Dev. 10, 1260–1270. doi: 10.1101/gad.10.10.1260
Nakane, T., Kousuke, N., Sonoko, H., Yuko, K., Sato, H., Kubota, T., et al. (2013). 6p subtelomere deletion with congenital glaucoma, severe mental retardation, and growth impairment. Pediatr. Int. 55, 376–381. doi: 10.1111/j.1442-200x.2012.03729.x
Nik, A. M., Johansson, J. A., Ghiami, M., Reyahi, A., and Carlsson, P. (2016). Foxf2 is required for secondary palate development and Tgfbeta signaling in palatal shelf mesenchyme. Dev. Biol. 415, 14–23. doi: 10.1016/j.ydbio.2016.05.013
Nora, E. P., Lajoie, B. R., Schulz, E. G., Giorgetti, L., Okamoto, I., Servant, N., et al. (2012). Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385. doi: 10.1038/nature11049
Qiao, Y., Jiang, X., Lee, S. T., Karuturi, R. K., Hooi, S. C., and Yu, Q. (2011). FOXQ1 regulates epithelial-mesenchymal transition in human cancers. Cancer Res. 71, 3076–3086. doi: 10.1158/0008-5472.can-10-2787
Reyahi, A., Nik, A. M., Ghiami, M., Gritli-Linde, A., Ponten, F., Johansson, B. R., et al. (2015). Foxf2 is required for brain pericyte differentiation and development and maintenance of the blood-brain barrier. Dev. Cell 34, 19–32. doi: 10.1016/j.devcel.2015.05.008
Rice, R., Spencer-Dene, B., Connor, E. C., Gritli-Linde, A., Mcmahon, A. P., Dickson, C., et al. (2004). Disruption of Fgf10/Fgfr2b-coordinated epithelial-mesenchymal interactions causes cleft palate. J. Clin. Invest. 113, 1692–1700. doi: 10.1172/jci20384
Seselgyte, R., Bryant, D., Demetriou, C., Ishida, M., Peskett, E., Moreno, N., et al. (2019). Disruption of FOXF2 as a likely cause of absent uvula in an egyptian family. J. Dent. Res. 98, 659–665. doi: 10.1177/0022034519837245
Sexton, T., and Cavalli, G. (2015). The role of chromosome domains in shaping the functional genome. Cell 160, 1049–1059. doi: 10.1016/j.cell.2015.02.040
Wang, H., Yang, H., Shivalila, C. S., Dawlaty, M. M., Cheng, A. W., Zhang, F., et al. (2013). One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918. doi: 10.1016/j.cell.2013.04.025
Wang, Q. S., Kong, P. Z., Li, X. Q., Yang, F., and Feng, Y. M. (2015). FOXF2 deficiency promotes epithelial-mesenchymal transition and metastasis of basal-like breast cancer. Breast Cancer Res. 17:30.
Wang, T., Tamakoshi, T., Uezato, T., Shu, F., Kanzaki-Kato, N., Fu, Y., et al. (2003). Forkhead transcription factor Foxf2 (LUN)-deficient mice exhibit abnormal development of secondary palate. Dev. Biol. 259, 83–94. doi: 10.1016/s0012-1606(03)00176-3
Wang, Y., Song, F., Zhang, B., Zhang, L., Xu, J., Kuang, D., et al. (2018). The 3D genome browser: a web-based browser for visualizing 3D genome organization and long-range chromatin interactions. Genome Biol. 19:151.
Wotton, K. R., and Shimeld, S. M. (2006). Comparative genomics of vertebrate Fox cluster loci. BMC Genomics 7:271.
Xu, J., Liu, H., Lan, Y., Aronow, B. J., Kalinichenko, V. V., and Jiang, R. (2016). A Shh-Foxf-Fgf18-Shh molecular circuit regulating palate development. PLoS Genet. 12:e1005769. doi: 10.1371/journal.pgen.1005769
Xu, J., Liu, H., Lan, Y., Park, J. S., and Jiang, R. (2020). Genome-wide identification of Foxf2 target genes in palate development. J. Dent. Res. 99, 463–471. doi: 10.1177/0022034520904018
Xu, J., Wang, L., Li, H., Yang, T., Zhang, Y., Hu, T., et al. (2019). Shox2 regulates osteogenic differentiation and pattern formation during hard palate development in mice. J. Biol. Chem. 294, 18294–18305. doi: 10.1074/jbc.ra119.008801
Keywords: bidirectional promoter, cleft palate, cis regulation, CRISPR, Forkhead, Foxf2, gene cluster, lincRNA
Citation: Xu J, Liu H, Lan Y and Jiang R (2021) Cis-Repression of Foxq1 Expression Affects Foxf2-Mediated Gene Expression in Palate Development. Front. Cell Dev. Biol. 9:665109. doi: 10.3389/fcell.2021.665109
Received: 07 February 2021; Accepted: 19 March 2021;
Published: 08 April 2021.
Edited by:
Shinji Takada, Graduate University for Advanced Studies (Sokendai), JapanReviewed by:
Li Cai, Rutgers, The State University of New Jersey, United StatesTadashi Okubo, Kitasato University, Japan
Copyright © 2021 Xu, Liu, Lan and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rulang Jiang, UnVsYW5nLkppYW5nQGNjaG1jLm9yZw==
 Jingyue Xu
Jingyue Xu Han Liu
Han Liu Yu Lan
Yu Lan Rulang Jiang
Rulang Jiang