- 1Children’s Hospital of Eastern Ontario Research Institute, Ottawa, ON, Canada
- 2Department of Cellular and Molecular Medicine, University of Ottawa, Ottawa, ON, Canada
- 3Department of Pediatrics, University of Ottawa, Ottawa, ON, Canada
Cancer predisposition syndromes are rare, typically monogenic disorders that result from germline mutations that increase the likelihood of developing cancer. Although these disorders are individually rare, resulting cancers collectively represent 5–10% of all malignancies. In addition to a greater incidence of cancer, affected individuals have an earlier tumor onset and are frequently subjected to long-term multi-modal cancer screening protocols for earlier detection and initiation of treatment. In vivo models are needed to better understand tumor-driving mechanisms, tailor patient screening approaches and develop targeted therapies to improve patient care and disease prognosis. The zebrafish (Danio rerio) has emerged as a robust model for cancer research due to its high fecundity, time- and cost-efficient genetic manipulation and real-time high-resolution imaging. Tumors developing in zebrafish cancer models are histologically and molecularly similar to their human counterparts, confirming the validity of these models. The zebrafish platform supports both large-scale random mutagenesis screens to identify potential candidate/modifier genes and recently optimized genome editing strategies. These techniques have greatly increased our ability to investigate the impact of certain mutations and how these lesions impact tumorigenesis and disease phenotype. These unique characteristics position the zebrafish as a powerful in vivo tool to model cancer predisposition syndromes and as such, several have already been created, including those recapitulating Li-Fraumeni syndrome, familial adenomatous polyposis, RASopathies, inherited bone marrow failure syndromes, and several other pathogenic mutations in cancer predisposition genes. In addition, the zebrafish platform supports medium- to high-throughput preclinical drug screening to identify compounds that may represent novel treatment paradigms or even prevent cancer evolution. This review will highlight and synthesize the findings from zebrafish cancer predisposition models created to date. We will discuss emerging trends in how these zebrafish cancer models can improve our understanding of the genetic mechanisms driving cancer predisposition and their potential to discover therapeutic and/or preventative compounds that change the natural history of disease for these vulnerable children, youth and adults.
Introduction
Cancer Predisposition Genes
Tumor-specific mutations that cause cancer are usually somatic and are cumulatively acquired over a lifetime, which explains the greater cancer prevalence in the older demographic. In contrast, germline mutations in certain genes, termed cancer predisposition genes (CPGs), are associated with an increased cancer risk. Germline mutations in CPGs often lead to hereditary cancer syndromes, which represent 5–10% of all malignancies and typically have a much earlier tumor onset than their sporadic counterparts (Garber and Offit, 2005). Despite the lack of a generally accepted CPG definition, one study identified 114 CPGs as the genes whose rare mutations increase the relative risk of cancer > 2 fold in at least 5% of individuals (Rahman, 2014). Another extensive study analyzed tumors from 10,389 individuals across 33 different cancer types and identified 152 CPGs using CharGer (Characterization of Germline Variants), an automatic variant classification pipeline they developed that ranks the pathogenicity of a certain mutation (Huang et al., 2018).
Unsurprisingly, many frequently mutated cancer driver genes in sporadic cancers have also been classified as CPGs, such as TP53, APC, BRCA1/2, ATM, and NF1, and as such, the knowledge gained by examining the role of germline CPG mutations will help shed light on the mechanisms of tumor formation for both sporadic and inherited cancers (Lu et al., 2015; Zhang et al., 2015). Additionally, 115 novel ultra-rare cancer-exclusive variants have been associated with known and candidate CPGs (Rasnic et al., 2020). Somatic mutations in 21 of these candidate CPGs were identified in 35% of cancers from 10,953 patients and were associated with a significantly decreased median survival time. This study shows the relevance of CPGs to the broader cancer research field and demonstrates the need to expand the list of known CPGs to assist with personalized diagnostic approaches and prognosis of cancers as well as to better understand the biological mechanisms of tumorigenesis to inform novel targeted therapeutic strategies.
Zebrafish as a Cancer Disease Model System
There are several traditional model systems commonly used to study cancer, including cell culture based, zebrafish and mouse models. These models all have their own unique advantages and limitations (Figure 1) which can be leveraged in different ways to contribute to cancer and developmental biology as well as to human disease research.
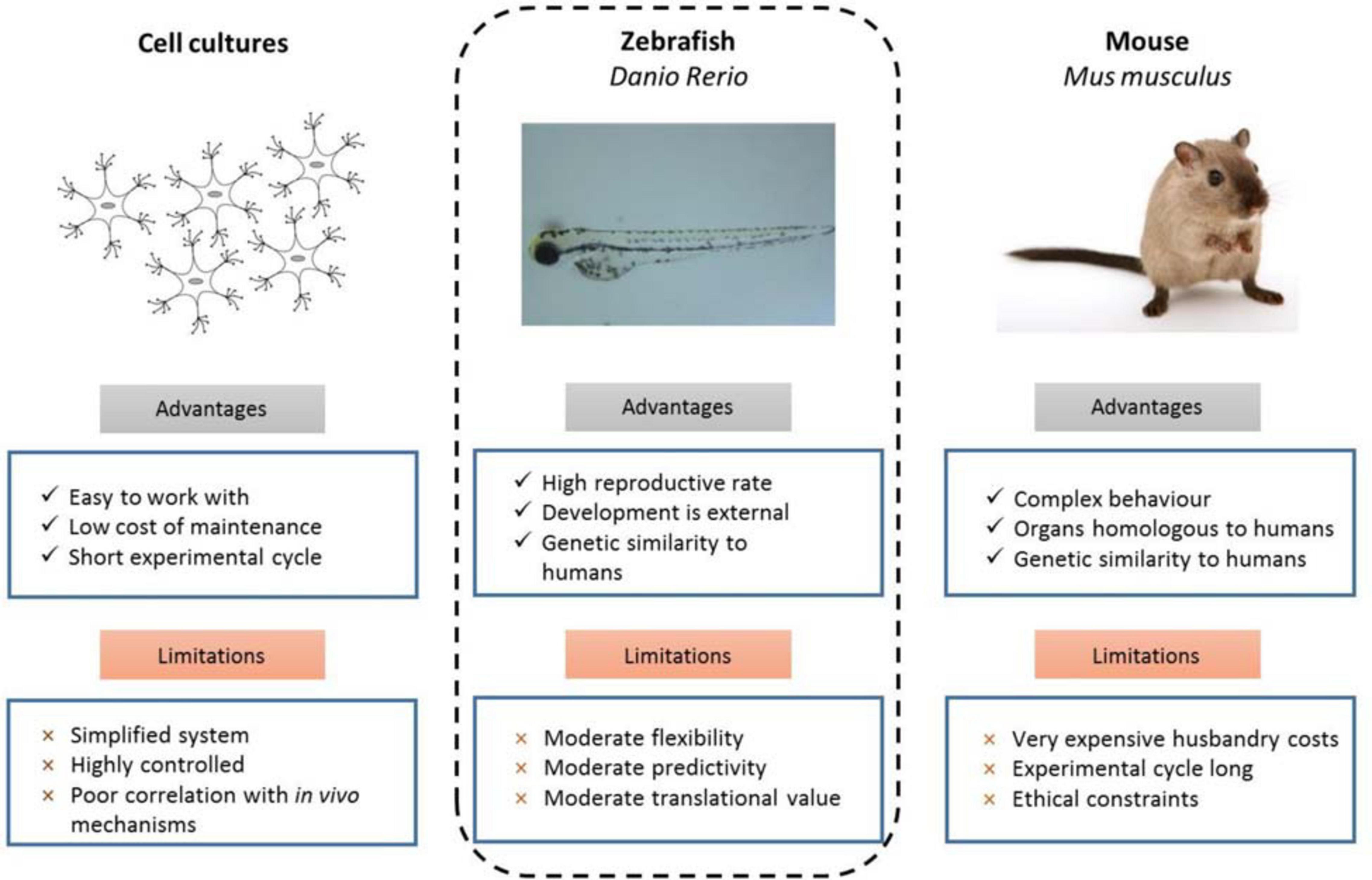
Figure 1. Experimental advantages and limitations of commonly used model organisms (d’Amora and Giordani, 2018).
The many advantages of the zebrafish position it as a powerful in vivo model, especially for preclinical genetic studies to investigate cancer predisposition (Figure 1). Zebrafish share a high degree of genetic conservation with humans, and tumors that develop in zebrafish are histologically similar to their human counterparts. Zebrafish larvae (and some adult strains such as casper White et al., 2008) are transparent, which enables live high-resolution imaging to track single cells and visualize tissues, as well as for large-scale and non-invasive tumor screening studies which is much more feasible than in a mouse model. The high fecundity of zebrafish combined with external fertilization and rapid development to sexual maturity makes the creation of new disease models by genome editing rapid and inexpensive in comparison to the much lower number of offspring, longer time to reach sexual maturity, and higher housing costs of rearing mice. The specificity and adaptability of genome editing technologies available to zebrafish researchers has grown tremendously in the recent years and facilitates both the generation of representative models for specific disease-causing mutations and forward genetic screens to identify genes and pathways involved in cancer predisposition.
Genome Engineering Strategies for Cancer Predisposition Genetic Models in Zebrafish
Genome engineering technologies have been heavily utilized in zebrafish disease modelling. A recent review catalogued and thoroughly discussed available technologies, and we encourage readers to consult this review for information on forward genetic screens using chemical mutagens, TILLING (Targeting Induced Local Lesions IN Genomes), transgenic techniques, gene editing using Zinc Finger Nucleases (ZFN), TAL-effector Endonucleases (TALEN) and Clustered Regularly Interspaced Palindromic Repeats (CRISPR)/Cas9 strategies (Raby et al., 2020).
Recent advances have expanded the range of available CRISPR nucleases, guide RNA design tools and other emerging technologies available to edit the zebrafish genome (Prykhozhij et al., 2018a; Liu et al., 2019). Off-target gene editing is a frequent problem when using CRISPR nucleases and this issue has been recently addressed both by the development of more sophisticated guide RNA design tools (Prykhozhij et al., 2018a; Liu et al., 2019) as well as by generating zebrafish-optimized vectors encoding HiFi Cas9 (Vakulskas et al., 2018) and highly specific but less efficient hypaCas9 (Chen et al., 2017) and testing them as mRNAs as well as commercial wild-type and HiFi Cas9 proteins using the same strategies (Prykhozhij et al., 2020).
Generating point mutants efficiently and precisely is one of the most important areas for optimizing genome editing technologies. The use of CRISPR/Cas9 and chemically synthesized oligonucleotides has been the earliest and primary approach to introduce point mutations into the zebrafish genome. We have optimized this technology in zebrafish to generate cancer-relevant point mutants in the tp53 gene (Prykhozhij et al., 2018b) and have reviewed other studies which applied similar techniques to introduce specific knock-in mutations (Prykhozhij and Berman, 2018). The limitations of this technology are that the double strand breaks and DNA repair machinery are still required for mutant generation, thus limiting efficiency to a maximum of < 10%. CRISPR/Cas9 point mutation knock-ins with enzymatically-synthesized long single-stranded DNA of 300-500 nucleotides as templates have recently been shown to have higher efficiencies in zebrafish compared to shorter oligonucleotides or double-stranded RNA molecules (Bai et al., 2020), but the efficiencies were measured based on single embryo phenotypes, which is prone to over-estimates. The zLOST (zebrafish long single-stranded DNA template) method is directly analogous to a similar Easi-CRISPR method in mice (Miura et al., 2018).
Among newer technologies for introducing precise mutations are base editors and prime editing, which both remove the need for double-strand breaks (DSB) and template oligonucleotides. Base editors use enzymatically inactive dCas9 fused to deaminase enzymes to mediate mutagenesis in defined parts of guide RNA recognition sites (reviewed in Liu et al., 2019). However, base editors are limited by both the inability of one molecule to introduce different types of mutations and a narrow but not completely defined region of activity. The most anticipated technology to be adopted for use in zebrafish is prime editing as developed by the lab of David Liu (Anzalone et al., 2019), who engineered a fusion of a Cas9 nickase with a reverse transcriptase enzyme to perform edits based on the information encoded in a prime editor guide RNA (pegRNA). The basic mechanism of prime editing consists of the following steps: Cas9 nickase cuts the strand opposite from its pegRNA recognition site, the primer-binding site of pegRNA binds to the released single-strand region of target DNA, and the reverse transcriptase enzyme copies the edited sequence from the pegRNA into the single strand of DNA which is then repaired with the edit being stably introduced into the DNA. In another strategy, denoted PE3b, the nickase with the regular guide RNA cuts the unedited strand thus promoting repair using the edited DNA strand (Anzalone et al., 2019).
Additionally, the generation of true null mutants in zebrafish is important for robust gene function analysis. While analyzing the transcriptional adaptation to indel mutants, El-Brolosy identified a strategy of deleting the proximal promoter and the first exon to generate “RNA-less” mutants failing to transcribe the gene (El-Brolosy et al., 2019). This strategy is likely highly robust, based on the published data and our own observations, but its utility has been questioned for some long non-coding RNAs (lncRNA), whose transcription was not fully blocked or altered by RNA-less deletions (Lavalou et al., 2019). Instead, insertion of a poly-adenylation signal was more effective for the malat lncRNA gene. GeneWeld is a CRISPR/Cas9 and microhomology-based gene insertion technology typically used for protein fusion gene generation (Wierson et al., 2020) that can be easily used to insert a fluorescent protein tag or another insert coupled with a stop-codon and a poly-adenylation signal at the 5′ end of a protein-coding gene thus disrupting it coupled with a fluorescent marker. Moreover, GeneWeld allows for a transgenic marker such as fluorescent protein expression in the lens or heart thus marking the mutant allele.
Studies of CPGs in Zebrafish: Developmental and Cancer Predisposition Models
For this review, we focused on zebrafish models of cancer predisposition based on CPGs from published lists (Rahman, 2014; Huang et al., 2018), many of which are implicated in cancer predisposition syndromes such as Li-Fraumeni syndrome (LFS), familial adenomatous polyposis (FAP), Cowden syndrome, Peutz-Jeghers syndrome (PJS), tuberous sclerosis complex (TSC), RASopathies, and certain hematological disorders. We will also discuss CPG mutations that, although not directly linked to a syndrome, are associated with DNA repair and genome stability and thus increase the risk of developing tumors. We have summarized the observations from current zebrafish cancer predisposition models (Supplementary Table 1) and classified all CPGs according to their primary functional category, highlighting those where significant zebrafish studies exist (Table 1).
Li-Fraumeni Syndrome
Li-Fraumeni syndrome is a hereditary cancer predisposition disorder that albeit rare, is one of the most recognized due to the high penetrance and range of associated tumors. Affected individuals develop a spectrum of malignancies (most commonly breast cancers, bone and soft tissue sarcomas, adrenocortical carcinomas, brain tumors, and leukemia) that typically arise much earlier than their sporadic counterparts (Li et al., 1988; Malkin, 2011; Figure 2). More than 30% of LFS patients develop cancer during childhood and adolescence, and those that survive face an 83-fold relative risk of developing additional malignancies (Hisada et al., 1998). Germline mutations in TP53 have been identified as the causative event in 70–80% of individuals with LFS (Malkin, 1993). TP53 is the most commonly mutated gene in all cancers and its corresponding protein, p53, is a potent tumor suppressor that orchestrates the response to cellular stress, such as DNA damage, by activating numerous target genes that induce cell cycle arrest, inhibit proliferation, and promote apoptosis among several other cellular processes (Cadwell and Zambetti, 2001). The most frequent TP53 mutations found in both LFS and sporadic tumors are missense mutations clustered in several hotspot regions in the DNA binding domain at codons 175, 245, 248, 273, and 282, although germline and somatic mutations present at different frequencies (Figure 2). However, the wide spectrum of tumors that develop and the varying time to onset cannot be simply attributed to TP53 mutations but may be partially explained by a number of cooperating factors including copy number variants, shortened telomeres, and somatic mutations in modifier genes (Bond et al., 2004; Tabori et al., 2007; Shlien et al., 2008).
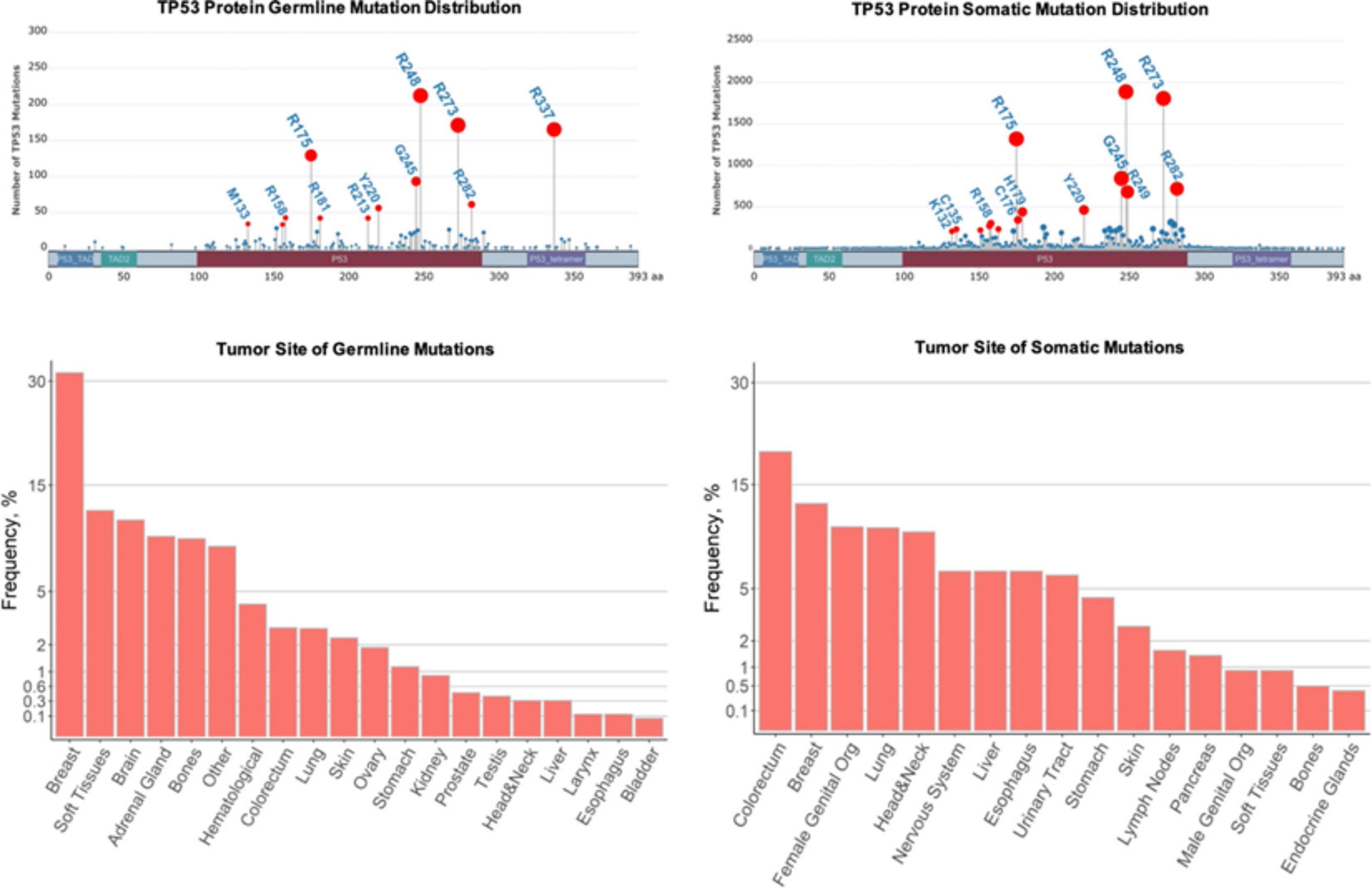
Figure 2. Comparison of germline and sporadic TP53 mutations found in human cancers. Top row: Germline and somatic missense mutations have been mapped to specific TP53 residues and domains. The frequencies of the mutations in selected hot-spot residues are indicated by the height of lollipop graph elements and the size of their circles. The circles of selected mutations are highlighted in red and the residues are labeled. The total numbers of germline and somatic mutations recorded are different in IARC TP53 datasets. Bottom row: Tissues associated with tumors due to germline and somatic TP53 mutations. The frequencies are represented as percentages of the total recorded tumors. In the plot, these percentages are scaled by the square root to visualize less frequent tissues. Plots were constructed using G3viz and ggplot2 R packages based on the IARC TP53 database (https://p53.iarc.fr/) after limiting the datasets to the missense mutations.
TP53 is highly conserved in zebrafish, especially in the protein DNA-binding domain where ∼90% of the mutations identified in human tumors are found (Hainaut and Pfeifer, 2016). Additionally, zebrafish and mammals share induction of the same p53 target genes (e.g., cdkn1a, cycg1, gadd45a/b, mdm2, bax1, puma, and noxa) following ionizing radiation (IR), which is used to induce DNA damage and trigger p53 pathway activation (Parant et al., 2010). Unsurprisingly, several zebrafish tp53 mutants have been developed to investigate both sporadic cancers and LFS. One of the initial approaches used to create tp53 point mutants was through N-ethyl-N-nitrosourea (ENU) targeted mutagenesis. This strategy was employed to generate two zebrafish tp53 mutants with missense mutations in the DNA-binding domain, tp53N168K and tp53M214K, equivalent to human TP53N200K and TP53M246K, the latter being proximal to several hotspot mutations (Berghmans et al., 2005). These mutants were resistant to IR-induced apoptosis, indicating disrupted p53 signaling; however, the tp53N168K mutants were only capable of this phenomenon when raised at 37°C instead of the normal zebrafish homeostatic temperature of 28.5°C. The tp53M214K mutants developed malignant peripheral nerve sheath tumors (MPNSTs) with approximately 28% tumor incidence by 16.5 months. However, a low tumor penetrance combined with a lack of tumor development in heterozygous mutants indicate that this mutant does not completely recapitulate the LFS phenotype (Berghmans et al., 2005).
Another zebrafish mutant, tp53I166T, was identified by screening the progeny of ENU-mutagenized zebrafish for the IR-induced apoptosis resistance phenotype (Parant et al., 2010). The location of this mutation is analogous to human codon 195, which is also implicated in several sporadic cancers (Kanda et al., 2013; Mullany et al., 2015). tp53I166T mutants have characteristics of the LFS phenotype including highly penetrant tumorigenesis, especially sarcomas, and dominant negative activity of mutant p53 (Parant et al., 2010). Both the tp53I166T homozygous and heterozygous mutants developed tumors and the tp53+/I166T mutant tumors displayed a high rate of loss of heterozygosity (LOH) of the wild-type tp53 allele, a characteristic frequently observed in LFS tumors. Mutants experienced aggressive tumorigenesis, with the first tumor appearing by 9 months and overall a dismal 50% survival rate by 15 months. However, the tp53I166T mutation was characterized as a loss of function (LOF) mutation in contrast to gain-of-function (GOF) mutations that are more commonly found in both LFS and sporadic tumors (Parant et al., 2010).
To further investigate the effect of p53 loss on tumorigenesis, a tp53 knockout zebrafish mutant was engineered using TALEN endonucleases (Ignatius et al., 2018). Although most LFS mutations are missense mutations, a small subset of LFS patients possess deletions at the TP53 locus. The tp53–/– fish are resistant to IR-induced apoptosis and developed a wider spectrum of malignancies than the tp53M214K and tp53I166T mutants, including MPNSTs, angiosarcomas, germ cell tumors, and a natural killer cell-like leukemia. Homozygous tp53–/– mutants developed tumors as early as 5 months with a 37% tumor incidence by 12 months. Heterozygous tp53+/– mutants also developed tumors, although infrequently, suggesting LOH also contributes to tumorigenesis in these mutants.
Recently, our group optimized the oligonucleotide-based CRISPR/Cas9 point mutation insertion strategy to generate specific knock-in mutations and employed this strategy to engineer a zebrafish tp53R217H mutant, equivalent to the human GOF TP53R248H mutation, representing one of the most frequent hotspot mutations found in both LFS and sporadic tumors (Prykhozhij et al., 2018b). Further characterization of these mutants will help shed light on the specific mechanisms and pathways involved in LFS.
CPGs in the Wnt/β-Catenin and PI3K/Akt/mTOR Pathways
Both the Wnt/β-catenin pathway and the phosphatidylinositol-3-kinase/Akt/mechanistic target of rapamycin complex 1 (PI3K/Akt/mTOR) pathway are upregulated in many cancers. The Wnt/β-catenin signaling pathway is essential for development and plays an important regulatory role in cell proliferation, polarity, and fate determination as well as for adult tissue maintenance (Logan and Nusse, 2004; MacDonald et al., 2009; Martin-Orozco et al., 2019). The PI3K/Akt/mTOR signaling pathway regulates many key cellular processes including proliferation, growth, metabolism, and survival (Jiang et al., 2020). Numerous studies have implicated crosstalk between these pathways as contributing to both tumorigenesis and therapeutic resistance in several cancer types including colorectal cancer, T-cell acute lymphoblastic leukemia, gastric cancer, and cervical cancer (Choi et al., 2019; Evangelisti et al., 2020; Guerra et al., 2020; Prossomariti et al., 2020). Oncogenic aberrations in these pathways are implicated in a wide spectrum of malignancies and are present in many syndromes associated with germline mutations in CPGs including FAP(APC), PJS (LKB1); Cowden syndrome (PTEN), and TSC (TSC1/2) (Figure 3).
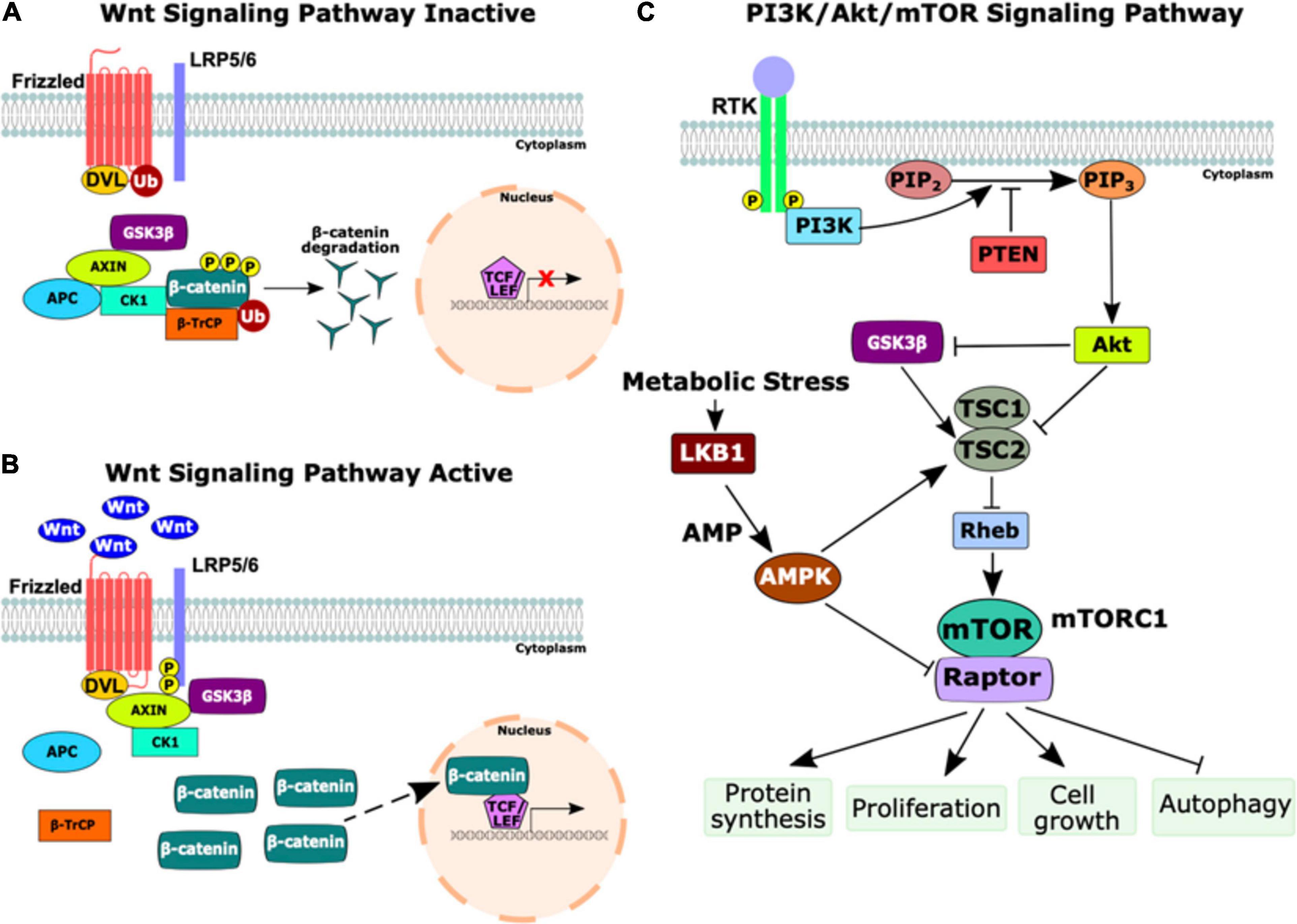
Figure 3. Schematic of the Wnt and PI3K/Akt/mTOR signaling pathways. (A) The inactive state of the Wnt signaling pathway. In the absence of Wnt ligands, the destruction complex (Axin, adenomatous polyposis coli (APC), casein kinase 1 (CK1), and GSK3β) phosphorylates β-catenin, marking it for ubiquitination (Ub) by beta-transducin repeat-containing protein (β-TrCP). β-catenin is degraded by proteasomes, thus preventing transcription of Wnt target genes. (B) The activation of the Wnt signaling pathway occurs upon the binding of Wnt ligands to Frizzled receptors. Next, CK1 and GSK3β phosphorylate the low-density lipoprotein receptor-related protein 5/6 (LRP5/6) co-receptor, activating DVL, which inhibits the destruction complex and prevents the degradation of β-catenin. Stabilized β-catenin accumulates in the cytoplasm and translocates to the nucleus as a transcriptional co-activator with T-cell factor/lymphoid enhancer factor (TCF/LEF) to induce transcription of Wnt target genes. (C) The PI3K/Akt/mTOR signaling pathway. Growth factors bind to a receptor tyrosine kinase (RTK), causing dimerization and autophosphorylation of the receptors. PI3K is recruited to the membrane and catalyzes the production of phosphatidylinositol-3, 4, 5-triphosphate (PIP3) from phosphatidylinositol-4, 5-bisphosphate (PIP2) which is negatively regulated by phosphatase and tensin homolog (PTEN). PIP3 activates Akt, a serine/threonine kinase that regulates numerous cell survival and cell cycle processes, including the inhibition of GSK3β and TSC1/2. The inhibition of TSC1/2 releases Rheb to activate mTOR complex 1 (mTORC1), comprised of mTOR and the regulatory associated protein of mTOR (Raptor). The upregulation of mTORC1 activates several important cellular processes including protein synthesis, proliferation, cell growth, and autophagy. During times of cellular metabolic stress, LKB1, a serine-threonine kinase, activates AMP-activated protein kinase (AMPK) which activates TSC2 and inhibits Raptor to downregulate mTORC1 signaling. This figure was constructed using information from several reviews (Shaw, 2009; Kaidanovich-Beilin and Woodgett, 2011; Porta et al., 2014; Martin-Orozco et al., 2019; Prossomariti et al., 2020).
Familial Adenomatous Polyposis
Germline truncating mutations in the APC tumor suppressor gene are implicated in FAP, a cancer predisposition syndrome, as loss of APC transforms the Wnt/β-catenin pathway into a constitutively active state. APC normally regulates the stability of β-catenin and the loss of APC causes the accumulation of β-catenin, greatly increasing transcription of Wnt target genes and ultimately leading to cancer in many cases (Fodde et al., 2001; Figure 3). Mouse models with different truncating Apc mutations develop varying numbers of polyps in the small intestine, from 3 to 300 (Moser et al., 1990; Fodde et al., 1994; Oshima et al., 1995), while an Apc null mutant engineered with concurrent Cdx2 mutations, a modifier gene that encodes a transcription factor important for intestinal differentiation, instead developed polyps in the colon which is more representative of human FAP (Aoki et al., 2003). These models show that mice with different truncating APC mutations develop tumors, although at different frequencies, and that modifier mutations influence polyp formation.
The first zebrafish apc mutant was created using ENU mutagenesis and contains a truncating mutation in the mutation cluster region (MCR; defined by mutations identified in human FAP tumors) (Hurlstone et al., 2003). Homozygous mutant embryos (apcMCR) exhibit a curved body type, impaired cardiac valve development, and do not survive past 96 hours post-fertilization (hpf). Cardiac valve defects were rescued by overexpression of APC or Dickkopf-1, a Wnt inhibitor. The heterozygous apcMCR mutants developed intestinal adenomas that histologically resembled mammalian polyps with high levels of β-catenin in proliferating cells, as well as liver and intestinal tumors by 15 months following treatment with the carcinogen 7,12-dimethylbenz[a]anthracene (Haramis et al., 2006). LOH of apc was not detected in these lesions, which is normally found in human FAP tumors, but the accumulation of β-catenin was suggestive that APC functions were disrupted. A later study demonstrated that homozygous apcMCR zebrafish embryos had a 34% reduction in intestinal epithelial cells and revealed that β-catenin was upregulated in these cells but was restricted to the cytoplasm (Phelps et al., 2009). Further, they showed that through a β-catenin-independent process, the apcMCR embryos had suppressed intestinal cell differentiation associated with increased expression of ctbp1, a transcriptional co-repressor. Injection of oncogenic KRASG12D mRNA into apcMCR embryos caused the nuclear accumulation of β-catenin, which greatly increased proliferation and resulted in a four-fold increase of the total intestinal cell numbers compared to non-injected apcMCR embryos. This was not observed in KRASG12D injected wild-type embryos, suggesting that additional mutations may be required following the loss of APC to trigger the nuclear accumulation of β-catenin and robust proliferation. These characteristics are often observed in the malignant transformation of FAP adenomas as well as sporadic carcinomas (Phelps et al., 2009).
For years, the consensus was that APC mutations alone were responsible for the initiation of adenomas, but whole-genome sequencing of adenomas from FAP patients has implicated other non-Wnt genes (Delacruz et al., 2019). CCR4-NOT Transcription Complex Subunit 3 (CNOT3), a gene important for regulating mRNA transcription, was identified as the most commonly mutated gene (excluding APC) in 20% of 37 FAP adenomas compared to only 1% in the sporadic/inherited colon cancer subset from the TGCA dataset (Gao J. et al., 2013; Delacruz et al., 2019). Two of the most frequent mutations identified were CNOT3E20K and CNOT3E70K. Knockdown of cnot3a in apcMCR zebrafish embryos decreased intestinal development and differentiation (Delacruz et al., 2019). Additionally, introducing mutated human CNOT3E20K mRNA into apcMCR embryos rescued intestinal differentiation while CNOT3E70K mRNA did not, suggesting that E70K is an inactivating mutation. CNOT3E70K is commonly found in adenoma tissues and a variety of cancers, but the discovery of its inhibitory effect on intestinal differentiation is novel and suggests that CNOT3E70K may contribute to the progression of adenomas to carcinomas (Delacruz et al., 2019).
One main tumor-suppressive function of APC is to promote intestinal differentiation through regulation of the retinoic acid (RA) biosynthesis pathway (Jette et al., 2004; Nadauld et al., 2004). Zebrafish apcMCR embryos demonstrate a lack of colonocyte differentiation which is rescued by treatment with RA (Nadauld et al., 2005). Further, CYP26A1, a gene encoding one of the major RA catabolic enzymes, is upregulated in human FAP adenomas and sporadic colon carcinomas that have truncating APC mutations as well as in the intestine of zebrafish apcMCR embryos (Shelton et al., 2006). Inhibition of cyp26a1 in apcMCR zebrafish restored intestinal differentiation, further demonstrating the role of APC in RA biosynthesis regulation and implicates CYP26A1 as a potential therapeutic target. A follow-up study demonstrated that deficiencies in the RA biosynthesis pathway in apcMCR embryos led to the upregulation of cebpb, a transcription factor gene, which was associated with increased cyclooxygenase-2 (cox-2) expression, also known as ptgs2a (prostaglandin-endoperoxide synthase 2a) (Eisinger et al., 2006). Elevated COX-2 expression is observed in colorectal carcinomas, is associated with the loss of APC, and ultimately drives the activation of Wnt target genes. Additionally, numerous genes important for intestinal cell fate determination are hypomethylated in homozygous apcMCR embryos and this methylation loss was reversed upon treatment with RA (Rai et al., 2010). The hypomethylation of these key genes was associated with impaired differentiation, as intestinal cells were maintained in a progenitor-like state.
Given the role of the Wnt/β-catenin pathway in tumorigenesis, it has been a target for therapy development. Zebrafish embryos were exposed to 6-bromoindirubin-3′-oxime, a GSK3 inhibitor that activates Wnt signaling, and underwent a drug screen where axitinib, a clinically approved VEGF inhibitor, was identified as the strongest inhibitor of the Wnt pathway (Qu et al., 2016). Importantly, treatment of adult zebrafish with axitinib for 6 days revealed no observable defects or morphological changes at the concentration required to inhibit Wnt signaling. This provides preclinical evidence that axitinib could be repurposed as a cancer therapy. Further, cross regulation between the Wnt/β-catenin and the mTOR (previously known as zTOR in zebrafish) pathways has been demonstrated in the apcMCR zebrafish mutants. In vitro studies have revealed that APC activates GSK3β which inhibits mTORC1 activity (Inoki et al., 2006; Valvezan et al., 2012). Consistent with this, mTORC1 activity is strongly upregulated in apcMCR zebrafish mutants (Valvezan et al., 2014) and inhibition of mTORC1 partially rescues embryonic apcMCR defects, with further rescue of curved body types by inhibiting both mTORC1 and Wnt signaling (Valvezan et al., 2014). This suggests that simultaneous targeting of these pathways is a potentially robust therapeutic strategy.
Peutz-Jeghers Syndrome
Peutz-Jeghers syndrome is a cancer predisposition disorder caused by germline inactivating mutations in LKB1 (Hemminki et al., 1998). Affected individuals with PJS develop both benign hamartomas and early-onset cancers, especially in the gastrointestinal tract, and females with PJS have an increased risk of developing breast cancer (Giardiello et al., 1987; Hearle et al., 2006). Somatic LKB1 mutations have been implicated in a spectrum of malignancies, including lung cancer, cervical cancer, breast cancer, malignant melanoma, non-small cell lung carcinomas, and acute myeloid leukemia (AML) (Guldberg et al., 1999; Fenton et al., 2006; Ji et al., 2007; Matsumoto et al., 2007; Wingo et al., 2009; Marinaccio et al., 2020). During periods of cellular metabolic stress, such as low levels of ATP, glucose, or oxygen, LKB1 plays a major role in energy metabolism control to maintain homeostasis (Kullmann and Krahn, 2018). Normally following a metabolic stress event, LKB1 activates AMP-activated protein kinase (AMPK), leading to a signaling cascade that inhibits the mTOR pathway (Figure 3C), thus preventing cell replication and growth in unfavorable conditions (Hardie, 2007; Kullmann and Krahn, 2018).
Zebrafish lkb1–/– mutants exhibit a starvation phenotype that is lethal by 7–8 dpf (Van Der Velden et al., 2011). Following depletion of the yolk which occurs around 5–7 dpf, homozygous mutants develop an accelerated metabolism that results in rapid depletion of their energy reserves. Treatment with rapamycin, an mTOR inhibitor, slowed the metabolic rate and prolonged survival, but did not fully rescue the mutant phenotype. These mutants demonstrate that lkb1 is crucial for metabolic control during energetic stress, like yolk depletion, and represent a potential screening tool for compounds that decrease the accelerated metabolic rate, a common characteristic of cancer cells. The lkb1 null mutants have also demonstrated that loss of lkb1 prevents the activation of autophagy during the metabolic stress that ensues from the switch of nutrient absorption from the yolk to external nutrients (Mans et al., 2017). Autophagy is a cellular mechanism of self-digestion important for maintaining cellular homeostasis both under basal conditions and during metabolic stress, and defects in this process have been implicated in cancer (Rabinowitz and White, 2010). Impaired autophagy in the lkb1 mutants was associated with an accumulation of p62, a multifunctional signaling protein important for mTORC1 activation and autophagy regulation (Duran et al., 2011), as well as decreased survival (Mans et al., 2017).
Additionally, zebrafish lkb1–/– mutant embryos have glucose homeostasis defects, enhanced glycolysis, and increased lactate production (Kuang et al., 2016). This is similar to the metabolic state observed in cancer cells which often rely on aerobic glycolysis and fermentation of pyruvate to lactate to produce ATP instead of oxidative phosphorylation. This is termed the Warburg effect which, although inefficient for generating ATP, preserves carbon to support anabolic processes for increased proliferation (Vander Heiden et al., 2009). As a proof-of-principle, treatment with dichloroacetate, an aerobic glycolysis inhibitor, decreased lactate production in lkb1–/– embryos, showing the potential of these mutants to be used as a drug screening platform to identify compounds that inhibit aerobic glycolysis (Kuang et al., 2016).
Cowden Syndrome
Cowden syndrome is a rare hamartoma tumor syndrome that is characterized by mutations in PTEN and is associated with an increased susceptibility to benign and malignant tumors including breast, skin, and thyroid cancers (Eng, 2003). PTEN is a tumor suppressor gene that is frequently inactivated in many cancers and normally functions as a negative regulator of the PI3K/Akt/mTOR signaling pathway (Sansal and Sellers, 2004; Figure 3C). PTEN has two homologs in zebrafish, ptena and ptenb, and while single homozygous mutants develop normally, double homozygous mutants do not survive past 5 dpf (Faucherre et al., 2008). The ptenb–/– single mutants developed spontaneous eye tumors, identified as neuroepitheliomas, around seven months of age. The presence of just one pten wild-type allele, as either ptena–/–; ptenb+/– or ptena+/–; ptenb–/–, is sufficient for survival to adulthood and the three mutant alleles are associated with spontaneous tumor growth (Faucherre et al., 2008; Choorapoikayil et al., 2012). Double homozygous pten embryos have increased proliferation and decreased apoptosis, and treatment with LY294002, a PI3K inhibitor, rescued all developmental phenotypes (Faucherre et al., 2008). Compared to other single and double mutants, the ptena+/–; ptenb–/– mutants were more susceptible to tumor development as they had a 10% incidence rate with 26/30 tumors found near the eye, while only 1/42 ptena–/–; ptenb+/– mutants developed a tumor (Choorapoikayil et al., 2012). The tumors were diagnosed as hemangiosarcomas, malignant hamartomas made up of vascular tissue, consistent with the increased frequency of hamartomas found in individuals with Cowden syndrome, and were characterized by increased proliferation, upregulated Akt signaling, and haploinsufficiency of either single pten allele (Choorapoikayil et al., 2012; Stumpf et al., 2015). A follow-up study from this group revealed that ptena–/–; ptenb–/– embryos have enhanced angiogenesis which was rescued by treatment with either LY294002 or sunitinib, a receptor tyrosine kinase (RTK) inhibitor used to reduce angiogenesis in cancer patients (Choorapoikayil et al., 2013). However, treatment with sunitinib greatly increased vegfaa expression in both the ptena–/–; ptenb–/– embryos and the hemangiosarcomas that developed in the adult haploinsufficient mutants (Choorapoikayil et al., 2013). VEGFA, the human ortholog of zebrafish vegfaa, encodes a ligand crucial for angiogenesis and thus its elevated expression may explain the relapse that is clinically observed following sunitinib treatment in some cases (Tonini et al., 2010; Kikuchi et al., 2012). Importantly, combination treatment with both LY294002 and sunitinib was successful in reducing neovascularization while maintaining normal levels of vegfaa in the ptena–/–; ptenb–/– embryos and thus has potential as a novel treatment strategy for the prevention of tumor angiogenesis (Choorapoikayil et al., 2013).
Tuberous Sclerosis Complex
Tuberous sclerosis complex is an autosomal dominant disorder associated with an increased susceptibility to renal cell carcinomas, as well as benign hamartomas that appear in multiple organs (Crino et al., 2006). This disorder is characterized by mutations in either the TSC1 or TSC2 CPGs which normally function to inhibit Rheb, a GTPase that activates the mTOR pathway (Figure 3C). As such, upregulation of the mTOR pathway is commonly observed in individuals with TSC (Zhang et al., 2003). Interestingly, TSC mutations are also associated with decreased cell proliferation and increased apoptosis which may counteract the effects of increased mTOR signaling enough to prevent the malignant transformation (Kim et al., 2013). Indeed, individuals with TSC mostly develop benign tumors compared to individuals with Cowden’s syndrome who also experience mTOR upregulation but develop multiple malignancies.
Morpholino knockdown of tsc1a, one zebrafish paralog of TSC1, led to kidney cyst formation, left-right asymmetry defects, increased ciliary length, and upregulation of the mTOR pathway (DiBella et al., 2009). Homozygous tsc2 mutant embryos (tsc2vu242) had greatly increased mTORC1 signaling, consistent with TSC patients, but do not survive past 11 dpf (Kim et al., 2011). Heterozygous mutants had only a moderate increase of mTORC1 signaling and showed no development defects, suggesting there may be a minimum threshold of pathogenic mTORC1 signaling. As both tsc1a and tsc2 homozygous mutations were embryonically lethal and heterozygous mutants did not develop tumors, Kim et al. (2013) introduced the heterozygous tsc2vu242 mutation into a mutant tp53 background to investigate the tumorigenic potential of tsc2 mutations. Compared to the tp53M214K/M214K mutants, the tsc2vu242/+; tp53M214K/M214K compound mutants had an increased rate of multiple malignancies, mTORC1 signaling, and angiogenesis while treatment with rapamycin caused regression of the tumor and tumor-associated blood vessels.
RASopathies Associated With CPG Mutations
RASopathies are a group of diseases caused by germline mutations in genes that regulate the Ras/MAPK pathway. Several RASopathies are associated with CPG mutations, including Noonan, LEOPARD, CBL-mutated, and Costello syndromes as well as neurofibromatosis type 1 (NF1). The Ras/Raf/Mek/Erk (MAPK) pathway is a key signaling cascade that promotes cell growth, division and differentiation (Molina and Adjei, 2006; Figure 4). Ras mutations and aberrant Ras signaling have been extensively studied in the context of malignancy as mutated Ras is found in about 20% of cancers (Prior et al., 2012). Patients with inherited or de novo congenital RASopathies are prone to variety of malignancies during childhood as well as later in life through second hit events that eliminate their remaining wild-type allele. Affected individuals also suffer from a variety of developmental phenotypes affecting multiple organs and tissue types which often leads to the initial RASopathy diagnosis.
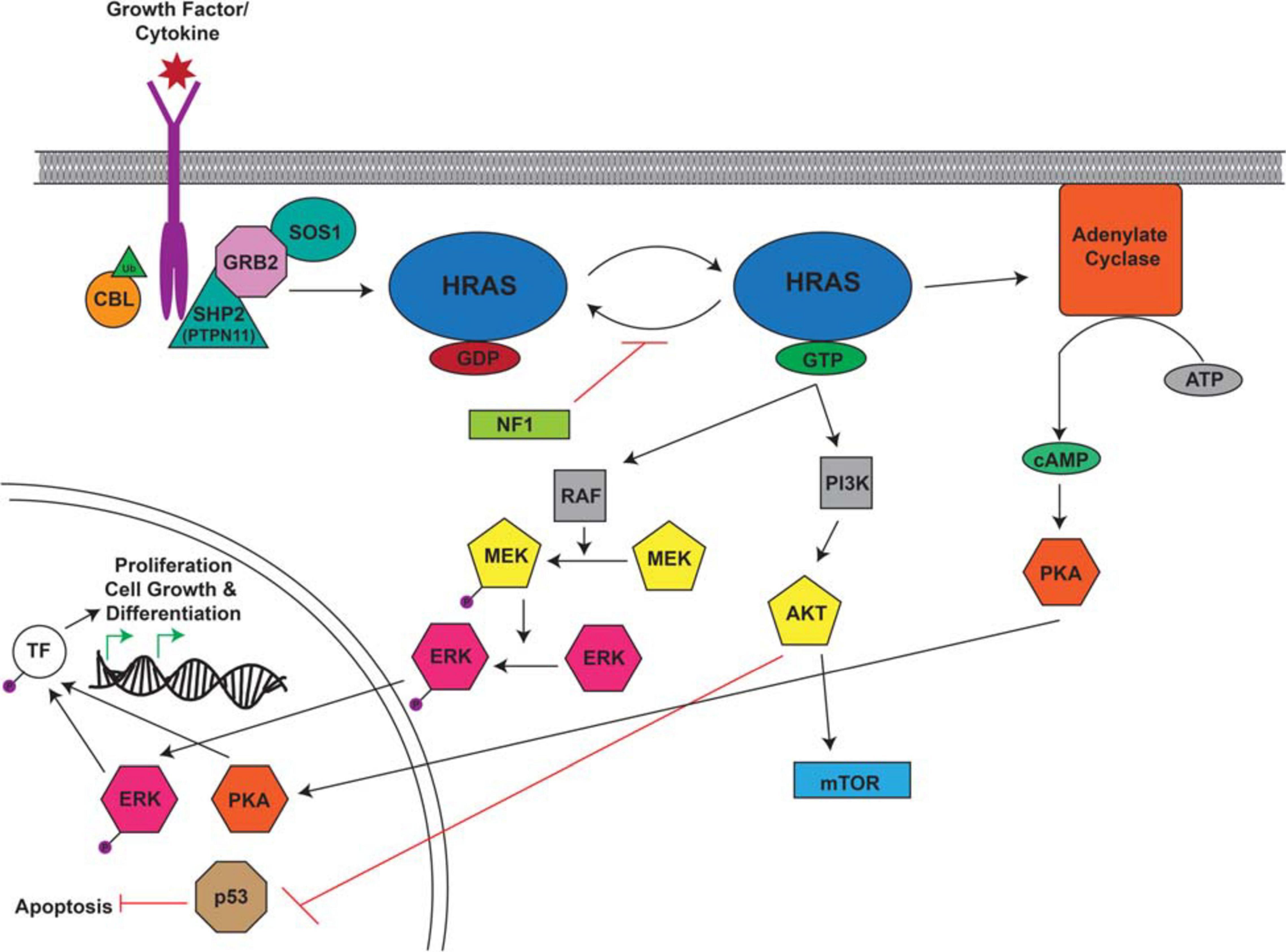
Figure 4. Overview of the RAS Signaling Pathway. The Ras signaling cascade is stimulated by the binding of a growth factor/cytokine to the appropriate RTK. C-CBL (Casitas B-lineage lymphoma) is an E3 ubiquitin ligase responsible for the ubiquitination of RTKs leading to their internalization and degradation. The activated receptor binds the adaptor proteins GRB2 (growth factor receptor bound protein 2) and SHP2/PTPN11 (protein tyrosine phosphatase non-receptor type 11) and recruits SOS1 (son of sevenless homolog 1). SOS1 facilitates the conversion of RAS from its inactive to active confirmation through the binding of GTP. RAS-GTP activates RAF, PI3K, and adenylate cyclase to cause a signaling cascade. RAF activates MEK which in turn activates ERK. Adenylate cyclase facilitates the production of cAMP which ultimately activates PKA. Activated ERK and PKA are protein kinases that translocate to the nucleus where they phosphorylate and activate transcription factors (TF) which drive the transcription of target genes involved in proliferation, cell growth, and differentiation. PI3K activates AKT which can activate mTOR and inhibit p53, leading to the inhibition of apoptosis. This figure was generated using information from several research articles and reviews (Ries et al., 2000; Kratz et al., 2006; Malaquias and Jorge, 2014; Wang et al., 2018).
Noonan/LEOPARD Syndrome
Noonan syndrome (NS) and LEOPARD syndrome (LS) are RASopathies that are typically grouped together as they are genotypically and phenotypically similar (Tartaglia et al., 2011; Martínez-Quintana and Rodríguez-González, 2013) and both syndromes predispose to malignancies such as AML (Jongmans et al., 2011). NS is characterized by growth retardation, skeletal deformities, facial dysmorphia and congenital heart defects, among other abnormalities (Tartaglia et al., 2001) and shares many overlapping features with LS which is defined by multiple lentigines, electrocardiographic abnormalities, ocular hypertelorism, pulmonary stenosis, abnormal genitalia, retardation of growth, and sensorineural deafness (Tartaglia et al., 2011).
Noonan syndrome and LS are heterogenous autosomal dominant diseases caused by mutations in several genes, including the CPG, PTPN11. PTPN11 encodes the ubiquitously expressed Src homology region 2 domain containing phosphatase-2 (SHP2), a protein tyrosine phosphatase involved in relaying growth signals and activating RAS and the MAPK/ERK pathway (Neel et al., 2003; Figure 4). PTPN11 is essential for many developmental pathways and is frequently deregulated in several types of cancer (Jongmans et al., 2011). Although phenotypically similar, PTPN11 mutations in LS and NS have biologically distinct functions. GOF PTPN11 mutations are found in roughly half of NS patients typically as missense mutations that activate PTPN11, while PTPN11 mutations found in LS mainly affect catalytic residues in the PTP domain that suppress phosphatase activity (Tartaglia et al., 2011). Studies of NS and LS in zebrafish have focused on examining the underlying disease mechanisms of ptpn11 deregulation and exploring the ways in which overlapping syndromic features can be caused by opposing effects on ptpn11 catalytic activity. One such explanation implicates the shared activation of RAS/MAPK signaling in overlapping symptoms whereas distinct NS and LS features are the result of differences in PI3K/Akt pathway expression which is uniquely upregulated in LS patients and mouse models (Kontaridis et al., 2006; Serra-Nedelec et al., 2012).
The first of these studies overexpressed either NS or LS mutant mRNA, which most notably resulted in shorter zebrafish larvae that were affected by heart abnormalities and a ‘hammer-head’ like phenotype reminiscent of both patients and mouse models (Jopling et al., 2007). Furthermore, the simultaneous overexpression of NS and LS mutant mRNA did not increase the severity of the phenotypes; a finding consistent with the established opposing effects of the two mutations. Similar phenotypes have been observed in subsequent studies using morpholinos to knockdown endogenous expression of zebrafish PTPN11 orthologues, ptpn11a and ptpn11b, injecting NS or LS mutant mRNA, and in ptpn11a–/– and ptpn11a–/–; ptpn11b–/– mutant fish (Stewart et al., 2010; Bonetti et al., 2014a, b). In contrast to homozygous knockout mice that die pre-implantation, zebrafish ptpn11a–/– and ptpn11a–/–; ptpn11b–/– mutants live to 5dpf, while ptpn11b–/– mutants survive to adulthood and are fertile (Yang et al., 2006; Bonetti et al., 2014b). Together, mortality and expression data, which demonstrated lack of significant ptpn11b expression during early embryogenesis, implicate ptpn11a as the more influential ortholog (Bonetti et al., 2014b). Decreased expression of erk was observed in morphant and double homozygous knockout models while PI3K signaling remained unaffected (Stewart et al., 2010; Bonetti et al., 2014b). Congruent with previous observations, phospho-Akt levels increased only in embryos injected with LS mutant mRNA (Bonetti et al., 2014a). Additionally, NS/LS zebrafish models have been used to investigate the role of PTPN11 mutations in both congenital sensorineural hearing loss and heart defects which are found in individuals with NS and LS (Sarkozy et al., 2008; Bonetti et al., 2014a; Gao et al., 2020).
c-CBL Mutation Associated Syndrome
Clinical features of patients with germline c-CBL (Casitas B-lineage Lymphoma) mutations partially overlap those of NS, including developmental delays, growth defects, facial dysmorphia, and a predisposition to hematological malignancies, specifically juvenile myelomonocytic leukemia (JMML) (Niemeyer et al., 2010; Perez et al., 2010). CBL is an E3 ubiquitin ligase that targets both active β-catenin and FLT3 (fms related receptor tyrosine kinase 3): two factors important for tumor-induced angiogenesis which have also been implicated in AML (Lyle et al., 2019).
The c-cblH382Y zebrafish ENU-induced line (Peng et al., 2015) harbors a missense mutation within a highly conserved domain critical for transfer of ubiquitin to target molecules (Thien and Langdon, 2001; Deshaies and Joazeiro, 2009). c-cblH382Y zebrafish displayed a myeloproliferative phenotype with increased numbers of hematopoietic stem cells (HSCs), erythrocytes and neutrophils at 5 dpf and a median survival time of only 15 dpf (Peng et al., 2015). phospho-S10 histone H3 (pH3) antibody/staining revealed that these HSCs were highly proliferative and remained more abundant in older fish, despite no obvious impairments in HSC differentiation. In contrast to the c-cblH382Y line, zebrafish injected with human phospho-tyrosine-inactive mutation c-CBLY731F mRNA, an amino acid residue that when phosphorylated suppresses Wnt signalling, had enhanced angiogenesis and increased expression vegfa while those injected with the phosphomimetic mutation c-CBLY731E mRNA displayed impaired angiogenesis and decreased expression vegfa (Shivanna et al., 2015). Thus, the different phenotypes were likely observed as the different missense mutations affect the binding of c-cbl to different proteins.
Costello Syndrome
Costello syndrome (CS), another NS-like syndrome, is caused by inherited activating mutations in HRAS, typically in the form of missense mutations (Aoki et al., 2005). CS has phenotypic features similar to NS, as well as the distinctive and common formation of benign cutaneous papillomata within the perinasal and perianal regions (Aoki et al., 2005). Santoriello and colleagues developed a zebrafish model of CS that ubiquitously expresses the constituently active HRASG12V gene (Santoriello et al., 2009). HRASG12V transgenic fish were shorter, had a smaller heart with thicker walls, a flattened head and suffered from scoliosis; features indicative of Costello syndrome. Tumors frequently formed in fish between 5–12 months and included melanoma, gastrointestinal carcinoma, hepatocarcinoma and rhabdomyosarcoma. Congruent with mouse models (Schuhmacher et al., 2008), ERK1/2 and Akt phosphorylation were not upregulated in HRASG12V transgenic zebrafish and remain unlikely drivers of CS pathology; rather, the ability of HRAS to induce hyperproliferation, DNA damage, and cellular senescence through the DNA damage response (DDR) is a well-documented cause (Di Micco et al., 2006). Indeed, HRASG12V transgenic zebrafish demonstrated oncogene-induced senescence in regions of the heart and brain as well as increased expression of DDR markers within various tissues (Santoriello et al., 2009).
Neurofibromatosis Type 1
Neurofibromatosis type 1 is an autosomal dominant disorder caused by mutations to the RAS GTPase activating domain of the NF1 gene. This mutation renders NF1 unable to hydrolyze RAS-bound GTP to GDP, ultimately activating the RAS signaling pathway (Cichowski and Jacks, 2001; Figure 4). Affected individuals develop cardiovascular abnormalities, nervous system defects including cognitive defects, and an increased incidence of malignancies, including JMML, gliomas, and MPNSTs. Homozygous deletion of murine Nf1 results in embryonic lethality mid-gestation while heterozygous mice remain viable and fertile, but this mutant does not fully recapitulate the human phenotype (Brannan et al., 1994; Silva et al., 1997). More complex mouse models that use Cre/lox technology to express mutant NF1 within specific cell populations better reproduce specific features of NF1 but lack the ability to study the disease as a whole (Zhu, 2002; Gitler et al., 2003).
Zebrafish have been successfully used to widen the scope of in vivo NF1 research using ZFN-mediated knockout of NF1 paralogues, nf1a and nf1b, and allogenic transplantation techniques. Double homozygous nf1 mutant zebrafish display macrocephaly as well as an increase in oligodendrocyte precursor cell (OPC) numbers and migration within the spinal cord (Shin et al., 2012). Further, defects in myelin structure formation and axon wrapping lead to decreased myelination in double homozygous nf1 mutant fish, despite no change in myb (myelin basic protein) expression within the central nervous system. Several studies have postulated NF1 as a positive regulator of adenylate cyclase (Guo et al., 2000; Tong et al., 2002; Dasgupta et al., 2003) and linked cAMP signaling to the expression of myelin components. NF1 mutations associated with cognitive dysfunction have been linked to regions outside of the GTPase-activating protein-related domain (GRD) (Fahsold et al., 2000), suggesting that some features of NF1 may be caused by aberrant cAMP signaling rather than activated Ras. The memory formation of nf1 mutant larvae was tested by evaluating the O-bend response to light/dark stimulus (Wolman et al., 2014). Trained nf1 mutant larvae had a delayed O-bend response which was improved upon treatment with chemical stimulants for the cAMP pathway, but not MAPK or PI3K inhibitors. Similarly, when testing memory recall, nf1 mutants again showed deficits, however these fish showed improvements over the course of the experiment, suggesting that cognitive defects are reversible and that patients may benefit from specialized learning environments and cAMP pathway effectors. Additionally, when crossed into a p53 null background, nf1 mutants develop high-grade gliomas and MPNSTs, similar to what is commonly observed in NF1 patients (Shin et al., 2012).
Neurofibromatosis type 1 modulates neuronal differentiation (Hegedus et al., 2007), and NF1 patients frequently develop benign neurofibromas that can undergo malignant transformation to MPNSTs, commonly through the loss of CDKN2A and p53 as well as amplification of PDGFRA (platelet-derived growth factor receptor A) (Mantripragada et al., 2008; Zietsch et al., 2010; Lee et al., 2014). The over-expression of PDGFRA has been linked to the malignant transformation of MPNSTs (Holtkamp et al., 2004). When modeled in transgenic zebrafish, overexpressing PDGFRA in a nf1a+/–; nf1b–/–; tp53M214K/M214K background accelerated the onset of MPNSTs (Ki et al., 2017). Interestingly, over-expression of wild-type PDGFRA, was better at accelerating tumor development than the constitutively active PDGFRA which activated Ras signaling to levels higher than optimal for tumor growth. MPNST tumors from wild-type PDGFRA transgenic fish were harvested and transplanted into nf1a+/–; nf1b–/–; tp53M214K/M214K larvae, establishing an allogenic transplant model that can rapidly test effectiveness of therapies. Indeed, sunitinib treatment effectively arrested progression of the transplanted MPNSTs in these mutants, and treatment with both sunitinib and trametinib, a MEK inhibitor, enhanced this therapeutic effect. Additionally, both DNA topoisomerase I (Topo I) targeting drugs and mTOR kinase inhibitors were identified as the most effective single agent therapies that avoided excessive toxicity (Ki et al., 2019). Treating with Topo I inhibitor Irinotecan and mTOR inhibitor AZD2014, acted synergistically to induce apoptosis and block protein synthesis resulting in tumor cell death.
Hematopoietic Disorders Associated With CPG Mutations
Several familial hematologic syndromes are associated with mutations in CPGs. Inherited bone marrow failure syndromes (IBMFS), including dyskeratosis congenita (DC), Schwachman-Diamond syndrome (SDS) and Fanconi anemia (FA) (discussed in a later section) are rare heterogeneous disorders caused by genetic defects that impair normal hematopoiesis, leading to anemia, cytopenia, and a variety of solid tissues/organ abnormalities. Additionally, these disorders predispose patients to develop malignancies. Zebrafish have long been used to study blood development and model hematologic diseases due to their high level of genetic conservation, especially within gene pathways critical to hematopoiesis (Jing and Zon, 2011). Similar to humans, zebrafish blood development occurs in two waves and within analogous structures recapitulating all blood cell types (Figure 5) (Davidson and Zon, 2004).
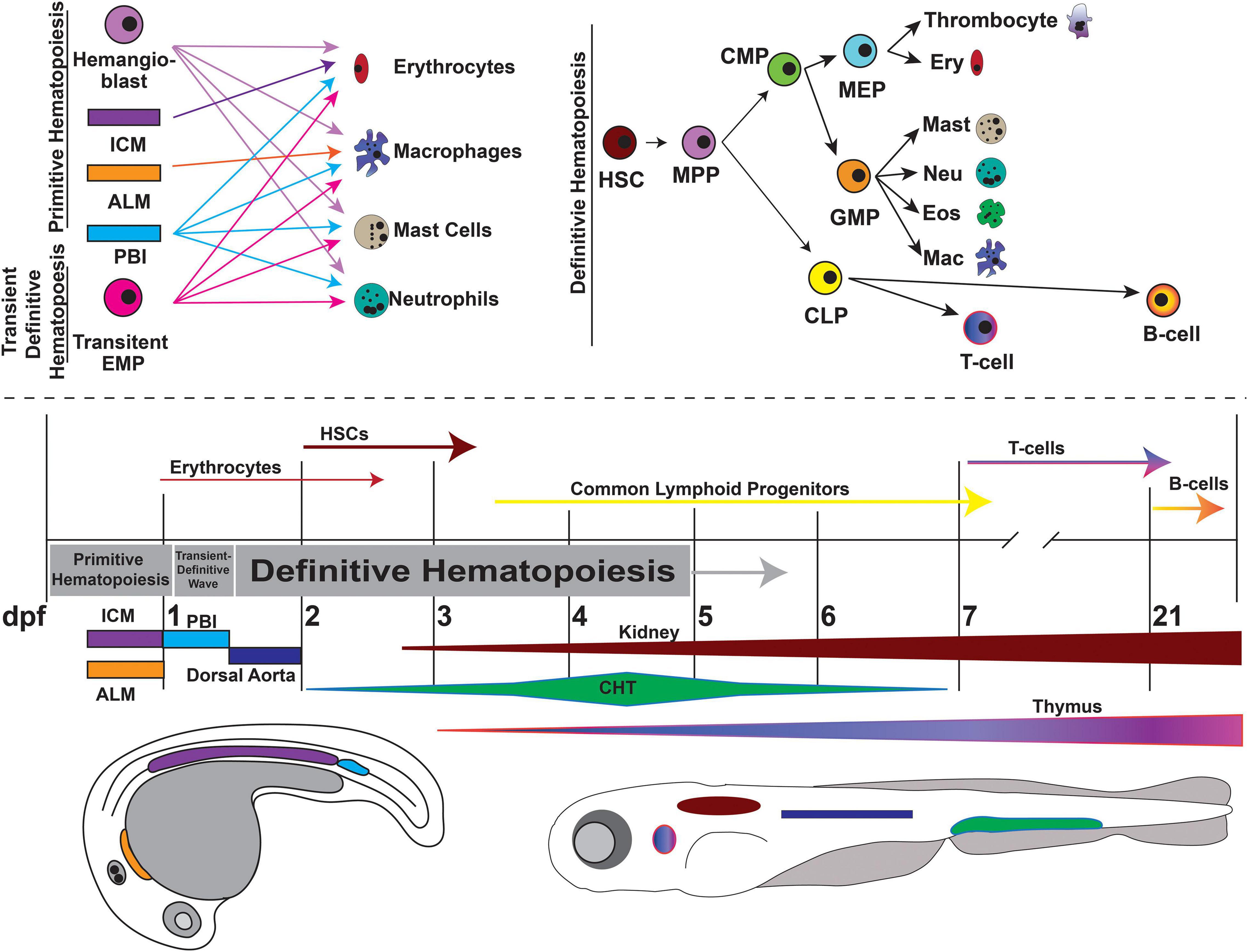
Figure 5. Overview of zebrafish hematopoiesis. Zebrafish hematopoiesis occurs in two waves, primitive and definitive, the latter of which recapitulates all the same blood cell types as humans. During the primitive wave, hemangioblasts give rise to a portion of primitive erythrocytes and granulocytes. The main source of primitive granulocyte production occurs within the anterior lateral mesoderm (ALM) (orange), while primitive erythrocyte productions occurs in the intermediate cell mass (ICM) (purple). Once circulation begins at approximately 24 hpf, primitive blood production transitions to the posterior blood island (PBI) (blue). Between 24 and 36 hpf a transient wave occurs in which transient bipotential erythromyeloid progenitor cells (EMPs). By 36 hpf, the first hematopoietic stem cells (HSCs) arise from the ventral wall of the dorsal aorta (navy), a site analogous to the aorta-gonad-mesonephros (AGM) in mammals. Once HSCs arise, they migrate to the caudal hematopoietic tissue (CHT) (previously the PBI), a structure analogous to the mammalian fetal liver. It is in the CHT that definitive erythroid and myeloid cells are produced and replace their primitive counterparts. By 4dpf HSCs migrate to and seed the kidney marrow (dark red), the equivalent of bone marrow in mammals. The first T-cells originate at approximately 7 dpf and mature in the thymus (purple/pink), while B-cells are not present until 21 dpf. This figure was constructed using information from published reviews (Vogeli et al., 2006; Rasighaemi et al., 2015; Gore et al., 2018). (MPP, Multipotent Progenitor; CLP, Common Lymphoid Progenitor; CMP, Common Myeloid Progenitor; MEP, Megakaryocyte-Erythroid Progenitor; GMP, Granulocyte-Monocyte Progenitor).
Dyskeratosis Congenita
Dyskeratosis congenita is a heterogenous disease caused by mutations in multiple genes involved with telomere maintenance including telomerase reverse transcriptase (TERT) and members of the H/ACA ribonucleoprotein (RNP) complex. H/ACA RNPs, NOP10 and DKC1 (dyskerin pseudouridine synthase 1) are involved in ribosome biogenesis, splicing and processing of various RNAs, and are also part of the telomerase complex together with TERT and the telomerase RNA component (Kiss et al., 2010). Telomerase is important for counteracting the progressive shortening of replicated DNA, especially in highly proliferative cell types and stem cells, and prevents activation of DDR pathways that could result in cellular senescence or apoptosis. Affected individuals develop nail dystrophy, mucosal leukoplakia, skin hyperpigmentation, increased risk of both hematological and solid cancers as well as bone marrow failure (BMF) (Cole et al., 1930; Alter et al., 2009). DC mortality is predominantly due to BMF, although the cumulative incidence of cancer is high with roughly 40% of patients developing a malignancy by the age of 50 (Alter et al., 2009). Mouse models of DC are limited because mouse telomeres are roughly 5–10 times longer than their human counterparts (10–15 kb) (Kipling and Cooke, 1990) while zebrafish telomeres are more similar to humans at 15–20 kb (Carneiro et al., 2016).
Zebrafish represent an advantageous platform for modeling DC and telomere deficiencies, and tert mutant zebrafish have reduced telomere length, premature aging, decreased fertility, and shorter lifespans (Anchelin et al., 2013). Dramatic telomere shortening triggered p53 activation, whereas p53 knockdown rescued apoptosis in larvae although telomere length remained short. Adult tert mutants had increased levels of apoptosis within the kidney marrow (the equivalent of human bone marrow), which also showed signs of depletion upon histopathological analysis (Henriques et al., 2013). Notably, reintroducing tert into second-generation mutants which typically died by 7dpf prevented both mortality and further telomere shortening (Anchelin et al., 2013). This suggests that drugs that activate wild-type TERT expression could be a potential therapy for DC.
The most severe form of DC, Hoyeraal-Hreidarsson syndrome, is caused by mutations to the catalytic domain of dyskerin (DKC1), suggesting that telomere-independent mechanisms contribute to DC (Knight et al., 1999; Zhang et al., 2012). Morpholino-mediated knockdown of zebrafish dkc1 significantly reduced the development of all blood cell lineages during definitive hematopoiesis (Zhang et al., 2012). Similar to the knockdown of tert, dkc1 morphants had a significant increase in the expression of p53 and pro-apoptotic genes such as bax1. Hematopoietic defects could be reversed by inhibiting p53 expression in dkc1 morphants, again suggesting that DC-associated cytopenia is partially p53 dependent. Interestingly, investigators observed significant defects in 18S rRNA production prior to the onset of abnormal blood development in dkc1I morphants and nop10 mutant fish (Pereboom et al., 2011; Zhang et al., 2012). Extensive p53-mediated apoptosis in nop10 mutants was linked to increased binding of Mdm2 to Rps7 resulting in degradation of Mdm2 protein which is normally responsible for p53 degradation. Furthermore, nop10 mutants failed to form HSCs despite no evidence of telomere shortening, and cytopenia was rescued through the inactivation of p53, providing additional evidence that p53-mediated apoptosis of HSCs is driven by ribosomal biogenesis defects and not solely by telomere shortening (Pereboom et al., 2011).
Shwachman-Diamond Syndrome
Schwachman-Diamond syndrome is an autosomal recessive disorder caused by mutations in SBDS that either interrupt transcript splice sites or result in a truncated protein (Boocock et al., 2003). Affected individuals develop exocrine pancreatic insufficiency, impaired hematopoiesis, skeletal abnormalities, gastrointestinal dysfunction and an increased risk of leukemia with ∼36% of patients developing myelodysplastic syndrome (MDS) (a syndrome which represents a pre-leukemia state) or AML by age 30 (Burroughs et al., 2009; McReynolds and Savage, 2017). SBDS is involved in ribosome biogenesis and function with a role in 40S and 60S ribosomal subunit maturation and assembly (Stepensky et al., 2017; Warren, 2018; Tan et al., 2019). Zebrafish sbds shares 86% amino acid identity with humans and retains the same intron/exon boundaries, making it an advantageous model for SDS modelling (Oyarbide et al., 2019). Morpholino-mediated knockdown of sbds significantly decreases neutrophil proliferation and migration as well as results in pancreatic hypoplasia and abnormal morphology (Venkatasubramani and Mayer, 2008; Provost et al., 2012). Additionally, sbds morphant embryos displayed broad changes in ribosomal gene expression. Simultaneous knockdown of p53 did not restore normal neutrophil numbers or normal pancreatic morphology, suggesting SDS pathology is not p53 dependent. Two CRISPR/Cas9 generated sbds mutant zebrafish, sbdsnu167 producing a truncated protein and sbdsnu132 with a 7 amino acid in-frame deletion, had reduced length and weight and died by 21 dpf, which was not rescued by crossing sbds mutants into a tp53 mutant (p53M214K/M214K) background (Oyarbide et al., 2020). Similar to the morphant studies, sbdsnu132 displayed neutropenia, while the number of macrophages and hemoglobin production remained normal. In addition, kidney marrow morphology remained normal at 21 dpf. In contrast to morphant studies, neither pancreatic hypoplasia nor changes to ptf1a expression were observed in sbds mutant larvae up to 15 dpf but rather pancreatic atrophy began in 15–21 dpf mutant fish after seemingly normal development. Therefore, zebrafish sbds models recapitulate several patient phenotypes including neutropenia and pancreatic defects, although the mechanisms that lead to pancreatic atrophy have not yet been explained.
Wiskott-Aldrich Syndrome and X-Linked Congenital Neutropenia
Wiskott-Aldrich syndrome (WAS) and X-linked congenital neutropenia (XLN) are severe immunodeficiency diseases caused by both LOF and GOF mutations in the Wiskott-Aldrich syndrome protein (WASp), respectively. WASp is an important regulator of the actin cytoskeleton in hematopoietic cells and is therefore a key player in leukocyte motility, phagocytosis, and a productive immune response (Tsuboi and Meerloo, 2007; Thrasher and Burns, 2010). Individuals with WAS or XLN develop frequent and recurrent infections while WAS patients also suffer from autoimmunity, bleeding disorders, and various types of leukemia and lymphoma at an incidence of roughly 13–22%, typically before 18 years of age (Sullivan et al., 1994; Imai et al., 2004; Keszei et al., 2018). One explanation for such predisposition is a failure to identify and eradicate malignant cells prior to tumor formation due to compromised immune function. Zebrafish offer a unique advantage over mouse models as the optical clarity of zebrafish larvae allows for the live imaging and analysis of immune cell response and migration.
Both zebrafish homologs of WASp, wasp1 (waspb) and wasp2 (waspa), are well conserved with 52% and 41% amino acid similarity to humans, respectively, and are hematopoietically restricted (Cvejic et al., 2008). Live imaging of lyz:eGFP transgenic zebrafish (fluorescently labelled neutrophils and macrophages) with knockdown of wasp1 revealed a reduction in neutrophils and macrophage migration to wounds in morphant larvae, while the number of immune cells remained normal. Furthermore, wasp morphants had decreased survival when inoculated with pneumococci (Rounioja et al., 2012), resembling life-threatening pneumococcal infections seen in individuals with WAS (Sullivan et al., 1994). Homozygous wasp1 mutant zebrafish recapitulated the ineffective immune responses to both inflammatory wounds and bacterial challenges seen in morphant studies and further revealed that that the ineffective formation, maintenance and retraction of neutrophil pseudopods and uropods contributed to the defective immune response (Jones et al., 2013). Expression of wasp was replaced in the wasp1 null background using a variety of patient-specific WASp transgenes and the expression of wild-type human WASp rescued neutrophil migration and wound recruitment and restored phagocytosis when challenged with S. aureus. In comparison, phospho-dead WASpY291F provided minimal improvement to neutrophil migration and did not improve microbe uptake by macrophages. Moreover, expression of constitutively active WASpI294T, observed in patients with XLN, resulted in an overall reduction of neutrophils but upon wound-response, WASpI294T expressing neutrophils had significantly increased cell velocity (Jones et al., 2013).
GATA2 Deficiency Syndromes
GATA2 is a transcription factor critical to both primitive and definitive blood development and lymphatic vessel formation. GATA2 haploinsufficiency, from inherited or spontaneous heterozygous mutations, produces a range of hematopoietic disorders in humans including monocytopenia, mycobacterium avium complex (MonoMAC) syndrome; dendritic cell, monocyte, B and NK lymphoid (DCML) deficiency; congenital neutropenia; and familial MDS/AML (Bigley and Collin, 2011; Dickinson et al., 2011; Hahn et al., 2011; Hsu et al., 2011). Affected individuals often present with infection, cytopenias, aplastic anemia and hypocellular bone marrow, of whom 75% will develop MDS/AML with a median age of onset of 20 years (Wlodarski et al., 2016, 2017). Control of Gata2 expression has been linked to a conserved +9.5 kb enhancer region (Hsu et al., 2013), where deletions are found in ∼10% of patients with MonoMAC syndrome (Wlodarski et al., 2016). Zebrafish have two highly conserved homologs of GATA2, gata2a and gata2b, with 57% and 67% conserved amino acid identity (Jin et al., 2004; Lutskiy et al., 2005; Gillis et al., 2009; Butko et al., 2015). Comprehensive expression and lineage tracing analysis indicates gata2a and gata2b zebrafish have distinct patterns of expression; gata2a is expressed throughout the endothelium and is more broadly required for vascular morphogenesis while gata2b is only present within the hemogenic endothelium of the dorsal aorta and has a more specialized role to activate runx1 expression required for HSC formation (Butko et al., 2015). Thus, this study suggests segregated endothelial and hematopoietic functions of the two zebrafish gata2 paralogs.
Using CRISPR/Cas9, Dobrzycki and colleagues introduced a deletion to the conserved +9.5 enhancer within intron 4 of zebrafish gata2a (gata2aΔi4/Δi4) (Dobrzycki et al., 2020). Unlike mouse Gata2Δ+9.5 models (Gao X. et al., 2013), gata2aΔi4/Δi4 fish survive past embryogenesis, and had decreased expression of HSC markers, runx1 and cmyb, during primitive hematopoiesis. Despite the recovery of runx1 and cmyb expression to wild-type levels by 48hpf, adult gata2aΔi4/Δi4 fish had half the number of blood cells within their kidney marrow and a surplus of immature blasts was found in 10% mutants. Additionally, these fish had a high incidence of cardiac edema and infection within less than 6 months of age, suggestive of the immune deficiency and lymphatic defects seen in MonoMAC syndrome (Dobrzycki et al., 2020).
Preliminary results from two gata2b mutant zebrafish models highlight how zebrafish can elucidate the link between GATA2 deficiencies and MDS/AML predisposition as gata2b+/– mutants have reduced myeloid differentiation and dysplastic myeloid cells in older animals (Avagyan et al., 2017; Gioacchino et al., 2019). Cre/Lox fluorescent lineage tracing technology is being used to assess the expansion of single-color clones within the kidney marrow of 3dpf gata2b+/– larvae with/without concurrent mutations (Avagyan et al., 2017). Juvenile gata2b+/– zebrafish had a decreased myeloid compartment in the kidney marrow of which 30% also had an expansion of single-color clones by 3 months post-fertilization, an event not observed in wild-type fish. This myelocytopenic phenotype is more profound in gata2b+/– fish when challenged with similar concurrent mutations, as compared to wild-type, and is indicative of a predisposition to MDS/AML.
Familial AML Associated With CEBPA Mutations
CEBPA, a transcription factor important for normal myelopoiesis and granulocyte differentiation, is mutated in ∼10% of individuals with AML (Pabst et al., 2001; Gombart et al., 2002). Of these, about half have biallelic mutations consisting of mutations in both hotspot N- and C- terminal regions that are typically acquired somatically. Mutations in the N-terminal region frequently produce frameshifts that lead to premature stop codons and translation of a dominant negative proteins, while C-terminal mutations are usually in-frame indels (Pabst et al., 2001; Nerlov, 2004). Germline mutations typically occur at the N-terminal hotspot and these individuals acquire a second C-terminal mutation somatically (Fuchs et al., 2008; Tawana et al., 2015). Although a rare occurrence, individuals with germline CEBPA mutations develop AML much earlier than sporadic AML at a median age of 25 versus 67 years (McReynolds and Savage, 2017).
Zebrafish studies reveal a conserved role for cebpa and its involvement in primitive erythropoiesis and HSC formation (Liu et al., 2007; Yuan et al., 2011, 2015). Myeloid defects, including a lack of embryonic neutrophils and macrophages were observed in three different cebpa mutant lines (molihkz7, cebpasum2, cebpasum3) (Dai et al., 2016). Myeloid defects in cebpa deficient molihkz7 mutants could be explained, in part, by the aberrant cell cycle arrest of myeloid progenitors once the definitive wave of hematopoiesis began, suggesting that cebpa is more important for myeloid progenitor migration and maintenance than initiation. To more accurately model CEBPA-associated MDS/AML, two new cebpa mutant zebrafish lines were engineered with either N- or C- terminal mutations (Hockings et al., 2018). Preliminary evidence demonstrates striking defects in mature myelocytes and monocytes in all double mutant larvae, while leukemic transformation was only observed in mutants between 4-6 weeks of age that carried at least one N-terminal mutation.
Familial Platelet Disorder With Predisposition to AML
Patients with familial platelet disorder with predisposition to AML (FPD/AML) suffer from thrombocytopenia, platelet dysfunction, increased risk of bleeding and of developing hematological malignancies, as 35–40% will develop MDS/AML by 33 years of age (Sood et al., 2017). FPD/AML is associated with germline mutations in RUNX1, a master transcription factor regulator of hematopoiesis and blood vessel development, most commonly small indels or point mutations that can lead to haploinsufficiency or dominant negative effects (Latger-Cannard et al., 2016). Monoallelic RUNX1 mutations alone are insufficient to initiation leukemogenesis, rather secondary somatic mutations are required within RUNX1 or other AML-associated genes (Song et al., 1999; Preudhomme et al., 2009; Ripperger et al., 2009; Schmit et al., 2015; Antony-Debré et al., 2016). Neither heterozygous RUNX1 murine or zebrafish models develop FPD phenotypes, however a germline homozygous Runx1 deletion is embryonically lethal in mice (Okuda et al., 1996) while zebrafish runx1W84X/W84X mutants are viable to adulthood (Jin et al., 2012). Like human patients, homozygous runx1W84X larvae have a severe reduction in both immature and mature neutrophils as well as in mature thrombocytes (Sood et al., 2010; Jin et al., 2012), mimicking findings in conditional knockout mice (Ichikawa et al., 2004; Growney et al., 2005). Furthermore, adult runx1W84X/W84X fish display reduced number of B-cells (Chi et al., 2018), a phenotype reminiscent of common variable immunodeficiency disorder (Li et al., 2015). Critically, neither myelocyte dysplasia nor an MDS-like phenotype have been reported in runx1W84X/W84X fish and it is likely that additional models involving second-hit mutations are needed to more accurately model FPD/AML in zebrafish (Chi et al., 2018).
Congenital Amegakaryocytic Thrombocytopenia
Congenital amegakaryocytic thrombocytopenia (CAMT) is a rare IBMFS that often presents in infancy with mutations in the gene MPL which encodes the receptor for thrombopoietin. Affected individuals develop severe thrombocytopenia, aplastic anemia, hemorrhage and a predisposition to leukemia (Germeshausen et al., 2006; Al-Qahtani, 2010). To date, only one CAMT zebrafish model exists as mplsmu3, a LOF mutant that experiences severe thrombocytopenia during embryonic development (Lin et al., 2017). Although the development of HSC and progenitor populations are unaffected, there was a significant reduction in thrombocyte precursor proliferation and thrombocyte markers remained significantly low in adult fish. Similar to the high risk of prolonged bleeding observed in CAMT, mplsmu3 larvae displayed reduced hemostasis and abnormal bleeding. To further adapt their model as a platform for high-throughput drug screening, Lin et al. generated the thrombocyte-specific fluorescent transgenic reporter line Tg(mpl:eGFP)smu4; mplsmu3, and as proof-of-principle, showed that treatment with recombinant human IL-11, a cytokine important for megakaryopoiesis (Peeters et al., 2008), modestly increased thrombocyte populations in mplsmu3 larvae.
CPG Mutations Involved in DNA Repair and Genome Stability Maintenance
Genes linked to DNA repair and genome stability comprise the single most numerous group of CPGs and represent 50 out of 152 genes currently identified, further emphasizing the fundamental importance of genome stability maintenance in cancer development (Table 1). We undertook an analysis of the interactions within this group of 50 genes using the STRING database (Szklarczyk et al., 2015). The inclusion of all known associations results in a highly dense interaction network with only the DICER1-DROSHA-DIS3L2 group, VHL, and BAP1 exhibiting minimal links (Figure 6A) while focusing on well-established physical interactions results in a sparser network highlighting the best-known interactions (Figure 6B). Further, these 50 proteins are part of many dynamic protein complexes with several hundred members, but their discussion is beyond the scope of this review. Instead, we will focus on the contributions that zebrafish model research has made towards understanding the fundamental biology and cancer models associated with these CPGs.
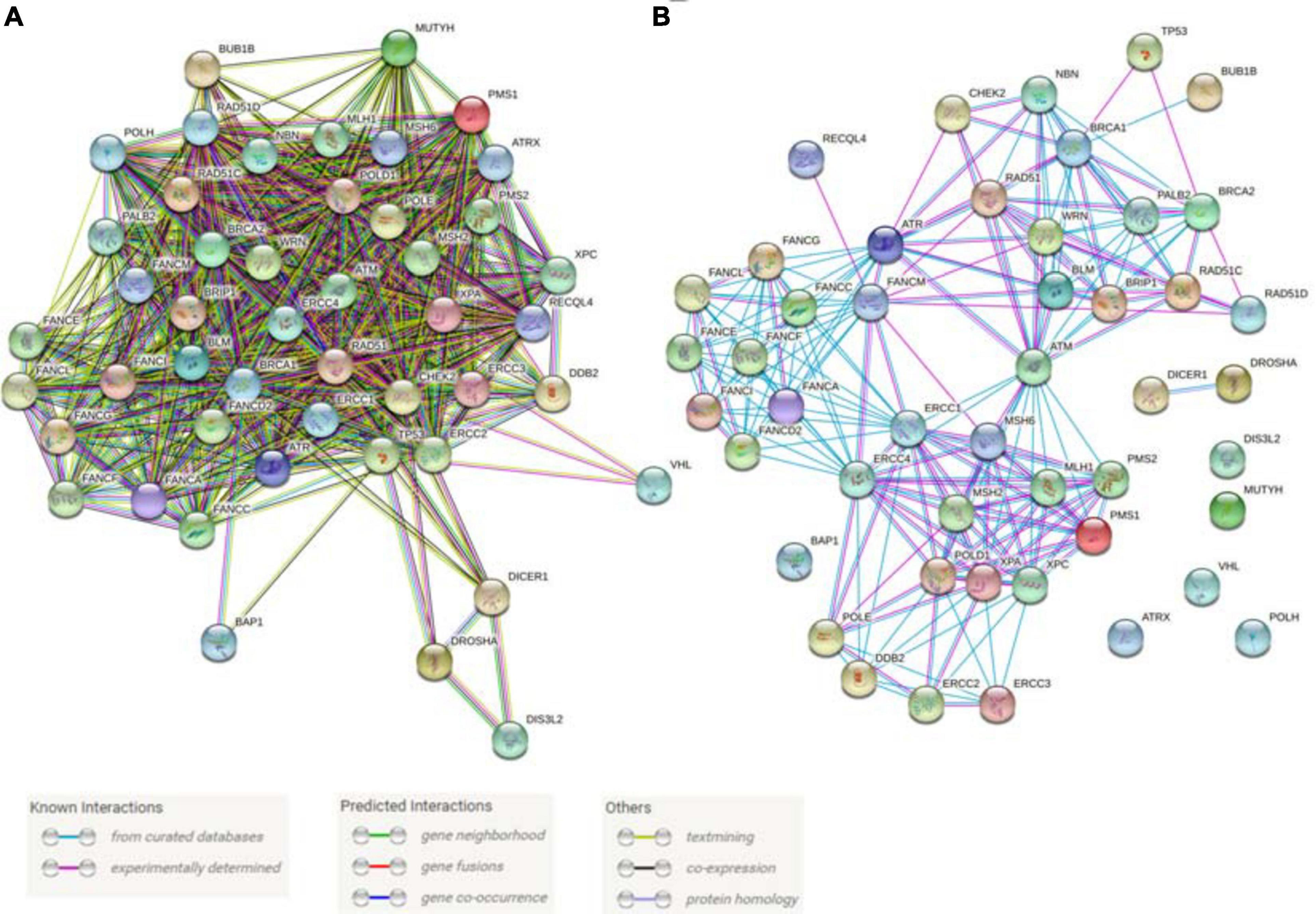
Figure 6. Predicted and known interactions among the cancer predisposition genes involved in DNA repair and genome stability maintenance. (A) All known interactions produce a very dense network. (B) A physical network with only experimentally verified results and curated database interactions were included. Both panels of the figure were produced using STRING database online tool (https://string-db.org/).
Ataxia-Telangiectasia (A-T) Genes (ATM, ATR and CHEK2)
ATM (A-T mutated) and ATR (A-T and Rad3 mutated) are key regulators of the initiation of the DDR (Awasthi et al., 2016). ATM mutations are responsible for Ataxia-Telangectasia (A-T) which is characterized by progressive ataxia, dilatation of small vessels (telangiectasia), immune defects, genome instability and malignancy while ATR mutations are implicated in Seckel syndrome which is associated with dwarfism, microcephaly, growth defects and intellectual disabilities (Weber and Ryan, 2015). ATM is activated by DNA DSBs while ATR is activated primarily by single-stranded DNA filaments resulting from replication stress (Figure 7A). Both ATM and ATR belong to the PI3K-related kinase (PIKK) family and upon activation they phosphorylate many target proteins including p53, Chk2 and Chk1 kinases, which mediate many of the downstream effects of ATM and ATR (Awasthi et al., 2016; Figure 7). CHEK2 is also an established CPG confirming the importance of this pathway in cancer.
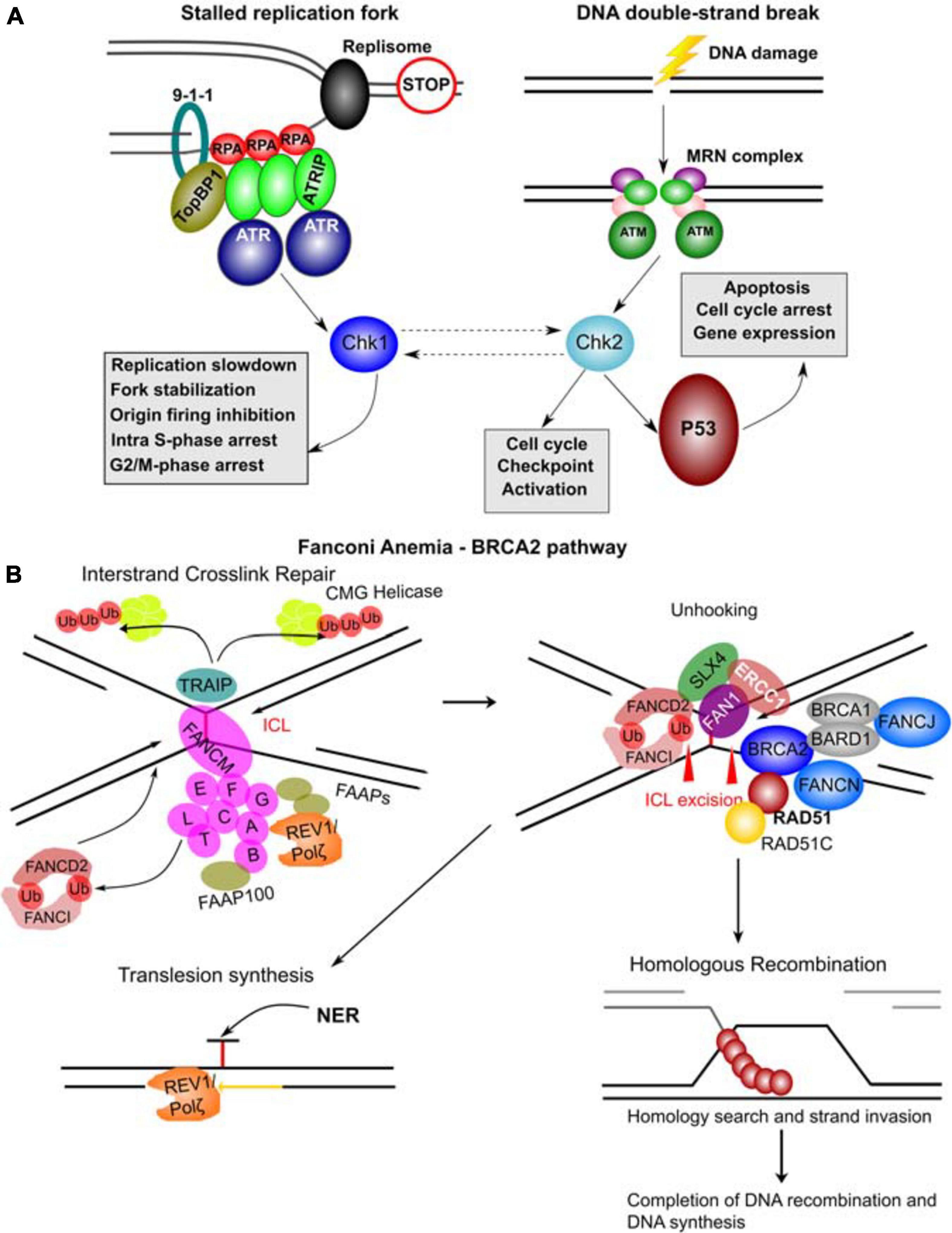
Figure 7. Molecular complexes formed and DNA repair pathways mediated by CPG proteins. (A) Mechanisms of ATR and ATM activation by different types of DNA damage. Stalled replication forks resulting from a blockage of replisome progression (indicated by the “STOP” diagram). The single-stranded DNA (ssDNA) region is recognized by Replication Protein A (RPA) forming the filaments which can then recruit ATR-ATRIP (ATR Interacting Protein) complexes. The RAD17-RFC complex (not shown) loads the 9-1-1 complex (RAD9-RAD1-HUS1) onto the ssDNA-dsDNA junction. RAD9 S387 phosphorylation provides a binding site for DNA topoisomerase 2-binding protein 1 (TopBP1), which in turn binds and activates ATR kinase. ATR phosphorylates and activates Chk1 kinase mediating many of its functional effects and directly phosphorylates many other target proteins. DNA double-strand breaks results in a complex signaling cascade resulting in binding of the MRN complex (MRE11-RAD50-NBN) to the DNA ends. MRN provides a DNA damage sensor platform for recruitment and activation of ATM, which then phosphorylates multiple targets including Chk2 responsible for cell cycle checkpoint activation and p53 activation. Chk2 and Chk1 can also functionally interact (hashed lines with arrows). p53 activation leads to apoptosis, cell cycle arrest and gene expression changes. (B) FA – BRCA2 pathway. A major role of this pathway is to respond to inter-strand crosslinks (ICL), which can be especially problematic during DNA replication since ICLs prevent replication fork convergence. At such blocked replication forks, TRAIP ubiquitin ligase ubiquitinates the CMG (CDC45-MCM-GINS) Helicase leading to its unloading. The ICL itself is recognized by FANCM / FAAP24 complex. FANCM then recruits the core FA complex components and FAAPs (FA associated proteins). E2 ubiquitin-conjugating enzyme FANCT and E3 ubiquitin ligase FANCL then cooperate to perform mono-ubiquitination of FANCD2 and FANCI forming the ID2 complex. Monoubiquitinated ID2 complex gets recruited the ICL lesion. In the process of unhooking, DNA near the ICL gets processed by nucleases to enable specific DNA repair pathways. The strand with the ICL structure gets repaired by Nucleotide Excision Repair (NER) and the translesion synthesis by REV1/Polζ, which gets recruited by the FA core complex. ID2 recruits FANCP (SLX4), FANCQ (ERCC1) and the nuclease FAN1 to mediate the ICL incision. The ID2 also interacts with BRCA2 (FANCD1), which gets recruited to the parts of blocked replication forks that will have to go through DNA recombination. BRCA2 in complex with BRCA1, BARD1, FANCN, FANCJ, RAD51 and RAD51C as well as other factors leads to loading of RAD51 into resected ssDNA regions, which can then participate in homologous recombination (HR). The figure was constructed based on the information in several recent reviews (Awasthi et al., 2016; Fradet-Turcotte et al., 2016; Bonilla et al., 2020; Liu et al., 2020).
Zebrafish ATM is highly conserved in specific domains, exhibits a broad pattern of expression at early stages and morpholino knockdown results in increased sensitivity to γ-irradiation (Imamura and Kishi, 2005). Further, in prim1 (DNA primase subunit 1) zebrafish mutants, knockdown of atm or atr is too toxic for retinal development, but retinal apoptosis due to DNA damage from faulty DNA replication is mostly suppressed with chemical inhibition of ATM (via KU55933) or ATM/ATR (via CGK733), or knockdown of chk2 (Yamaguchi et al., 2008). Similar observations were made in the ugly duckling (udu) zebrafish mutant now mapped to the gon4l gene which has been recently implicated in regulating several pleotropic mechanisms including DNA repair and genome stability (Lim et al., 2009; Tsai et al., 2020). p53-mediated apoptosis in this mutant could be successfully inhibited by either knockdown of tp53, chk2 or chemical inhibition of ATM.
Even though zebrafish ATR does not appear to control the DDR directly, it has been linked to cilia formation in Kuppfer’s vesicle cells involved in establishment of left-right asymmetry (Stiff et al., 2016). ATR knockdown in zebrafish reduces the size of the Kuppfer’s vesicle, shortens cilia in this area, reduces gli1 levels, a Hedgehog pathway target, and induces p53 targets p21 and puma. The authors linked p53 target induction to Hedgehog signaling inhibition, but it is more likely to be a direct ATR knockdown effect due to limited evidence of Hedgehog pathway inhibition. Morphologically, ATR knockdown produced left-right asymmetry defects, curved bodies, small eyes and narrow heads in zebrafish embryos, phenotypes supportive of significant Hedgehog signaling inactivation. As of this review, there are no characterized zebrafish mutants of either atm or atr. Based on the above studies, null mutants may have strong phenotypes and generation of both null and precise point mutants would help identify the spectrum of potential developmental and cancer predisposition phenotypes of ATM and ATR mutations in zebrafish to discern the relative roles of these kinases in genome stability maintenance and tumor suppression.
Fanconi Anemia Genes
Fanconi anemia is a genetic disorder caused by mutations in a group of genes involved in several aspects of DNA repair. Collectively known as the Fanconi anemia pathway, this group (22 known members) of FA proteins forms multiple complexes and mediate DNA interstrand crosslink (ICL) repair resulting from endogenous aldehydes or exogenous crosslinking agents (Liu et al., 2020). Briefly, this involves fork convergence, ICL recognition by the FA core complex, and removal of DNA replication components, involving FA genes such as BRCA2, RAD51 and others (Figure 7B). FA is an IBMFS with disease characteristics that include cancer predisposition, fertility issues, growth retardation, and congenital abnormalities affecting many tissue types (Ramanagoudr-Bhojappa et al., 2018). Individuals with FA have defective HSCs that result in aplastic anemia due to BMF, MDS, and AML (Kutler et al., 2003). The frequency of BMF is estimated at 80% in affected individuals, of whom 30% develop hematologic and solid tumors including leukemias, head and neck carcinomas, liver tumors and gynecologic malignancies by 40 years of age with rates several hundred-fold higher than the general population and the age of onset frequently in childhood or early adulthood (Kutler et al., 2003; Liu et al., 2020).
Most FA genes are highly conserved in zebrafish (Titus et al., 2009) and the most comprehensive study of 17 FA genes (fanca, fancb, fancc, fancd1/brca2, fancd2, fance, fancf, fancg, fanci, fancj/brip1, fancl, fancm, fancn/palb2, fanco/rad51c, fancp/slx4, fancq/ercc4, fanct/ube2t and 2 FA-associated genes: faap100 and faap24) revealed that FA mutants do not exhibit overt morphological phenotypes during embryonic development (Ramanagoudr-Bhojappa et al., 2018). However, adult FA zebrafish experience a female-to-male sex reversal as mutants in 12 genes had a complete reversal while 5 others had a partial reversal. This reversal has complicated certain phenotypic analyses due to the need for crosses with heterozygous females, which prevents generation of maternal-zygotic mutants. Additionally, 11 out of 17 mutants have some degree of hypersensitivity to treatment with diepoxybutane (DEB), which serves as a diagnostic test for FA gene mutations. fancp mutants exhibit a decreased body size but no FA mutants demonstrate blood-related phenotypes or increased cancer susceptibility (Ramanagoudr-Bhojappa et al., 2018). Previous work in fancl zebrafish mutants similarly demonstrated female-to-male sex reversal, which mechanistically was connected to germ cell apoptosis mediated by p53 activation (Rodríguez-Marí et al., 2010). This reversal was blocked in the fancl–/–; tp53–/– mutants, confirming the role of p53 in this process and suggested a model where lack of oocyte survival generates signals for testis development and overall male sex determination in fancl–/– mutants.
Fanconi anemia genes such as BRCA2/FANCD1, RAD51C and RAD51, homologous to the bacterial RecA, are involved in repair of DNA DSBs by homologous recombination (Bonilla et al., 2020). BRCA2 is also more broadly important for genome stability due to its involvement in DNA replication, cell cycle progression and telomere homeostasis (Fradet-Turcotte et al., 2016). Zebrafish brca2/fancd1 has received a significant amount of research attention which has revealed its important functions in genome stability, germ cell development, kidney development and tumor suppression. Female-to-male reversal and male infertility is observed in homozygous brca2 zebrafish mutants (Q658X and ZM_00057434 referred to as brca2–/– and both have a disruption in exon 11 of brca2 which is commonly observed in hereditary breast and ovarian tumors) due to the disruptions in germ cell meiosis after being correctly specified in development (Shive et al., 2010; Rodríguez-Marí et al., 2011). Loss of tp53 rescues ovary development but not male fertility. Apoptosis is dramatically increased after DEB treatment in brca2–/– zebrafish embryos and brca2–/– mutant cell lines have higher rates of chromosomal aberrations, apoptosis and slower growth compared to wild-type (Rodríguez-Marí et al., 2011). These brca2 mutant studies represent the first FA zebrafish cancer susceptibility models as both the infertile brca2–/– males developed testicular cancer, and tp53 and brca2 double heterozygotes (tp53+/M214K; brca2+/–) or homozygotes (tp53M214K/M214K; brca2–/–) had accelerated cancer development by about 5 months (Rodríguez-Marí et al., 2011). To further analyze tumor development, brca2Q658X mutants were crossed to a tp53 mutant background to drive tumor formation (Shive et al., 2014). After a > 2-year follow-up, all lines, brca2+/+; tp53+/M214K, brca2+/Q658X; tp53+/M214K and brca2Q658X/Q658X; tp53+/M214K, had a tumor incidence of 80–100% and tp53 LOH was observed nearly universally in the brca2+/+; tp53+/M214K tumors while it was only observed in 30% of the brca2Q658X/Q658X; tp53+/M214K tumors, suggesting that brca2 homozygous mutants undergo other genetic changes that drive tumorigenesis (Shive et al., 2014). Additionally, the creation of the zebrafish zeppelin (zep) mutant, mapped to brca2 by whole-genome sequencing, has revealed that brca2 also plays a role in kidney development (Kroeger et al., 2017).
RAD51/FANCR has recently been associated with FA and provides genetic support to well-known interactions between RAD51 and BRCA2 (Bonilla et al., 2020). This functional interaction is well-conserved in zebrafish as antibody staining in embryos show that endogenous rad51 forms foci upon induction of DNA DSBs, but these foci do not form in brca2–/– mutants and are reduced in brca2+/– embryos (Vierstraete et al., 2017). A recent zebrafish study showed that rad51 null embryos exhibit both increased chromosomal abnormalities after DEB treatment and increased sensitivity to gamma irradiation, consistent with the designation of RAD51 as an FA pathway gene (Botthof et al., 2017). Zebrafish rad51l1 mutants, a rad51 paralog, are synthetically lethal when combined with rad51 mutant, demonstrating the redundancy of their functions. In contrast to all other FA gene mutants, rad51–/– mutants exhibit decreased staining for HSC markers during larval stages and lower total cell numbers in kidneys in adults, both of which are rescued by tp53 loss (Botthof et al., 2017).
DNA Mismatch Repair
The DNA mismatch repair (MMR) pathway maintains chromosome stability and prevents increases in the rate of mutations, therefore it is of great significance for the maintenance of genome stability (Ijsselsteijn et al., 2020). Indeed, genes involved in the MMR pathway, including MLH1, MSH2, MSH6, PMS1, PMS2, are known CPGs and their mutations are responsible for Lynch syndrome (hereditary non-polyposis colorectal cancer) (Rahman, 2014). The zebrafish mlh1 mutant shows the importance of mlh1 for chromosome segregation in meiosis during gametogenesis as mutant males are infertile and exhibit abnormal testis histology, accumulation of spermatocytes and apoptosis (Feitsma et al., 2007) with a < 1% fertilization rate, as well as higher mutational burden in the resulting embryos (Leal et al., 2008). mlh1 mutant females are fertile when bred with wild-type males, but still frequently produced aneuploid and triploid embryos. These phenotypes are fully consistent with mouse models of MMR defects and possible links of MMR gene defects to human male infertility (Mukherjee et al., 2010). Further characterization of mlh1, msh2 and msh6 mutants shows microsatellite instability in the progeny from mutant males and revealed increased cancer incidence in mutant adults with tumors mainly characterized as neurofibromas and MPNSTs (Feitsma et al., 2008).
CPGs of Diverse Functions Contribute to Genome Stability
The genes discussed so far in this section are directly involved in DNA repair pathways. However, given the complexity of this process and its regulatory mechanisms, it can be expected that additional genes will be implicated in tumor suppression that have indirect or unexpected roles in genome stability. We will discuss several case studies highlighting how the zebrafish model has been employed to examine such genes.
ATRX is a SWI/SNF-like chromatin remodeler specialized in depositing H3.3 histone onto genomic DNA in a complex with the histone chaperone DAXX. Loss of ATRX is associated with both alternative lengthening of telomeres, thus promoting cell immortalization, and a wide range of cancer types, especially those of the nervous system, such as gliomas and neuroendocrine tumors (Nye et al., 2018). However, an recently-identified interaction between ATRX and FANCD2 implicates ATRX and DAXX in repair mechanisms of DNA DSBs, suggesting a direct and active role of ATRX in the DDR (Raghunandan et al., 2020). Although atrx null mutant (atrx–/–) zebrafish do not survive past larval stages, blood development in mutant embryos is relatively normal with the exception of a single globin gene and more spherical erythrocytes (Oppel et al., 2019). Lethality of atrx loss required the authors to investigate its cancer-promoting properties in highly sensitized tumor-prone mutants, tp53–/–; nf1b–/–; nf1a+/–; atrx+/– and tp53–/–; nf1b–/–; nf1a+/+; atrx+/–. Heterozygous atrx loss did not increase the rate of tumor formation but led to a wider spectrum of tumor types, tert down-regulation, lengthening of telomeres and up-regulation of polycomb repressive complex 2 genes compared to atrx+/+ tumors. This model represents an important step for atrx investigation in zebrafish and may be further improved by tissue-specific or conditional approaches to atrx inactivation to circumvent larval lethality of this mutant.
The designation of DICER1 and DROSHA as CPGs is not surprising given their fundamental roles in microRNA (miRNA) biogenesis as miRNA genes are well-known oncogenes and tumor suppressors (Hammond, 2015) and there is a diverse spectrum of cancers observed in DICER1 syndrome (Robertson et al., 2018). However, over the last decade, small RNAs and lncRNAs have been implicated in the DDR (Vågbø and Slupphaug, 2020). Genomic DDR foci produce DICER1- and DROSHA-dependent small RNAs (20–35 nucleotides), which are required for full DDR activation in mammalian cells and zebrafish (Francia et al., 2012). Further, dicer1 knockdown in zebrafish strongly disrupts the DDR based on reductions in ATM and γ-H2AX phosphorylation in a cell-autonomous manner based on the aberrant DDR in transplanted dicer1 morphant cells (Francia et al., 2012). This suggests that DICER1 and DROSHA are part of the DNA repair pathway and further evaluation is needed from the perspectives of both miRNA biogenesis and genome stability maintenance.
The von Hippel-Lindau protein (pVHL) mainly functions in the cellular response to hypoxia where it is responsible for poly-ubiquitination of Hypoxia Inducible Factor (HIF) proteins 1 and 2. Under normoxic conditions, HIF1/2 are hydroxylated and degraded; however, they become less hydroxylated under hypoxic conditions and accumulate, causing transactivation of their target genes. Loss of pVHL leads to a constitutively active hypoxic response and is the cause of von Hippel Lindau disease. Affected individuals develop retinal and central nervous system vascular tumors known as hemangioblastomas, pancreatic and kidneys cysts, pheochromocytoma, and clear cell renal cell carcinomas (ccRCC) (Gossage et al., 2015). Previous work in ccRCC cell lines demonstrates the importance of pVHL for DNA DSB repair (Metcalf et al., 2014). VHL has two paralogs in zebrafish, vhl and vll, and zebrafish pVHL mutants demonstrate that vhl gene loss is mainly responsible for HIF signaling activation, which is significantly enhanced upon vll loss, while vll is important for genome stability maintenance (Kim et al., 2020). The vhl–/–; vll–/– double mutants have reduced p53 activation and are resistance to apoptosis following the induction of genotoxic stimuli which was phenocopied by Hif activation. Additionally, genotoxic-induced apoptosis in brca2–/– mutants is rescued by vhl loss. Thus, these results firmly establish vhl and vll as VHL homologs with divided but complementary functions that can enable the creation of cancer models involving these genes (Kim et al., 2020).
Conclusion
Zebrafish models are amenable to a variety of genetic technologies and represent a powerful in vivo tool to investigate the roles and mechanisms of CPGs in development and tumorigenesis. We have reviewed how the zebrafish is well-positioned for preclinical studies focused on identifying novel mutations and genes, as well as elucidating how their molecular mechanisms contribute to biological functioning and cancer development. One major advantage of the zebrafish model is the more rapid and less expensive generation of genetic disease models than is possible in the mouse. Zebrafish also provide excellent opportunities for in vivo imaging throughout development and to some extent in adults. Additionally, mutations of certain genes are often more tolerable in zebrafish than in the mouse which is partially explained by the partial genome duplication that occurred in zebrafish and as such, zebrafish models often survive longer and allow for more in-depth studies. This phenomenon also provides the ability to investigate the effect of multiple modifier genes through the creation of compound mutants, especially for CPGs. One common CPG/modifier often investigated is tp53 due to the prominent role of tp53 in tumor suppression. Several of the cancer predisposition models we have highlighted investigated the role of p53 in disease progression and have employed tp53 mutants to facilitate tumor development in their models.
In this way, zebrafish models have tremendous potential to provide preclinical insights to change the natural history of life-threatening cancer predisposition disorders. Mutations in CPGs predispose individuals to an increased risk of cancer via their key roles in both development and homeostatic control/maintenance, especially during times of cellular stress like DNA damage or energy depletion. Thus, the generation and characterization of zebrafish models has tremendous utility for identifying novel therapies for these rare but devastating syndromes. Further, knowledge gained from zebrafish models also holds great significance to the broader cancer research community, as biological insights about CPG mechanisms will also reveal the biological processes underlying sporadic tumorigenesis. However, one current limitation is that several of the aforementioned zebrafish disease models are knockouts while their corresponding syndrome is more typically caused by a missense mutation that does not result in protein truncation; thus, these models may not fully represent the genetic underpinnings of the disease nor the resulting protein interactions. This was likely due to the availability/usability of genome editing technologies at the time of model creation, but the recent optimization of several genome editing techniques, as well as soon anticipated-in-zebrafish technologies including prime editing, will aid future researchers to more easily and efficiently develop more representative disease-specific genetic models. Additionally, once established, genetic zebrafish models can serve as a drug screening platform to rapidly and efficiently identify novel therapies. However, to date, zebrafish cancer predisposition models have been employed predominantly in proof-of-principle drug studies with known molecular targets to highlight genotype-phenotype correlations. Thus, this represents an exciting and powerful next step for many of these models to provide clinical insights and reveal new avenues of therapeutic intervention.
Author Contributions
KK, KC, and SP wrote and edited the manuscript and generated the figures and tables. JB edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Canadian Institutes of Health Research (PJT-166081).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.660069/full#supplementary-material
References
Al-Qahtani, F. S. (2010). Congenital amegakaryocytic thrombocytopenia: a brief review of the literature. Clin. Med. Insights Pathol. 3, 25–30. doi: 10.4137/CPath.S4972
Alter, B. P., Giri, N., Savage, S. A., and Rosenberg, P. S. (2009). Cancer in dyskeratosis congenita. Blood 113, 6549–6557. doi: 10.1182/blood-2008-12-192880
Anchelin, M., Alcaraz-Pérez, F., Martínez, C. M., Bernabé-García, M., Mulero, V., and Cayuela, M. L. (2013). Premature aging in telomerase-deficient zebrafish. Dis. Model. Mech. 6, 1101–1112. doi: 10.1242/dmm.011635
Antony-Debré, I., Duployez, N., Bucci, M., Geffroy, S., Micol, J. B., Renneville, A., et al. (2016). Somatic mutations associated with leukemic progression of familial platelet disorder with predisposition to acute myeloid leukemia. Leukemia 30, 999–1002. doi: 10.1038/leu.2015.236
Anzalone, A. V., Randolph, P. B., Davis, J. R., Sousa, A. A., Koblan, L. W., Levy, J. M., et al. (2019). Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157. doi: 10.1038/s41586-019-1711-4
Aoki, K., Tamai, Y., Horiike, S., Oshima, M., and Taketo, M. M. (2003). Colonic polyposis caused by mTOR-mediated chromosomal instability in Apc+/Δ716 Cdx2+/- compound mutant mice. Nat. Genet. 35, 323–330. doi: 10.1038/ng1265
Aoki, Y., Niihori, T., Kawame, H., Kurosawa, K., Ohashi, H., Tanaka, Y., et al. (2005). Germline mutations in HRAS proto-oncogene cause Costello syndrome. Nat. Genet. 37, 1038–1040. doi: 10.1038/ng1641
Avagyan, S., Mannherz, W., and Zon, L. I. (2017). Visualizing clonal hematopoiesis associated with gata2 deficiency in zebrafish using color-barcoding. Blood 130:4237. doi: 10.1182/blood.V130.Suppl_1.4237.4237
Awasthi, P., Foiani, M., and Kumar, A. (2016). ATM and ATR signaling at a glance. J. Cell Sci. 129:1285. doi: 10.1242/jcs.188631
Bai, H., Liu, L., An, K., Lu, X., Harrison, M., Zhao, Y., et al. (2020). CRISPR/Cas9-mediated precise genome modification by a long ssDNA template in zebrafish. BMC Genomics 21:67. doi: 10.1186/s12864-020-6493-4
Berghmans, S., Murphey, R. D., Wienholds, E., Neuberg, D., Kutok, J. L., Fletcher, C. D. M., et al. (2005). Tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. Proc. Natl. Acad. Sci. U.S.A. 102, 407–412.
Bigley, V., and Collin, M. (2011). Dendritic cell, monocyte, B and NK lymphoid deficiency defines the lost lineages of a new GATA-2 dependent myelodysplastic syndrome. Haematologica 96, 1081–1083. doi: 10.3324/haematol.2011.048355
Bond, G. L., Hu, W., Bond, E. E., Robins, H., Lutzker, S. G., Arva, N. C., et al. (2004). A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell 119, 591–602. doi: 10.1016/j.cell.2004.11.022
Bonetti, M., Overman, J. P., Tessadori, F., Noël, E., Bakkers, J., and den Hertog, J. (2014a). Noonan and LEOPARD syndrome Shp2 variants induce heart displacement defects in zebrafish. Development 141, 1961–1970. doi: 10.1242/dev.106310
Bonetti, M., Rodriguez-Martinez, V., Overman, J. P., Overvoorde, J., Van Eekelen, M., Jopling, C., et al. (2014b). Distinct and overlapping functions of ptpn11 genes in zebrafish development. PLoS One 9:e94884. doi: 10.1371/journal.pone.0094884
Bonilla, B., Hengel, S., Grundy, M., and Bernstein, K. (2020). RAD51 gene family structure and function. Annu. Rev. Genet. 54, 1–22. doi: 10.1146/annurev-genet-021920-092410
Boocock, G. R. B., Morrison, J. A., Popovic, M., Richards, N., Ellis, L., Durie, P. R., et al. (2003). Mutations in SBDS are associated with shwachman–diamond syndrome. Nat. Genet. 33, 97–101. doi: 10.1038/ng1062
Botthof, J. G., Bielczyk-Maczynska, E., Ferreira, L., and Cvejic, A. (2017). Loss of the homologous recombination gene rad51 leads to Fanconi anemia-like symptoms in zebrafish. Proc. Natl. Acad. Sci. U. S. A. 114, E4452–E4461. doi: 10.1073/pnas.1620631114
Brannan, C. I., Perkins, A. S., Vogel, K. S., Ratner, N., Nordlund, M. L., Reid, S. W., et al. (1994). Targeted disruption of the neurofibromatosis type-1 gene leads to developmental abnormalities in heart and various neural crest-derived tissues. Genes Dev. 8, 1019–1029. doi: 10.1101/gad.8.9.1019
Burroughs, L., Woolfrey, A., and Shimamura, A. (2009). Shwachman-diamond syndrome: a review of the clinical presentation, molecular pathogenesis, diagnosis, and treatment. Hematol. Oncol. Clin. North Am. 23, 233–248. doi: 10.1016/j.hoc.2009.01.007
Butko, E., Distel, M., Pouget, C., Weijts, B., Kobayashi, I., Ng, K., et al. (2015). Gata2b is a restricted early regulator of hemogenic endothelium in the zebrafish embryo. Development 142, 1050–1061. doi: 10.1242/dev.119180
Cadwell, C., and Zambetti, G. P. (2001). The effects of wild-type p53 tumor suppressor activity and mutant p53 gain-of-function on cell growth. Gene 277, 15–30. doi: 10.1016/S0378-1119(01)00696-5
Carneiro, M. C., De Castro, I. P., and Ferreira, M. G. (2016). Telomeres in aging and disease: lessons from zebrafish. Dis. Model. Mech. 9, 737–748. doi: 10.1242/dmm.025130
Chen, J. S., Dagdas, Y. S., Kleinstiver, B. P., Welch, M. M., Sousa, A. A., Harrington, L. B., et al. (2017). Enhanced proofreading governs CRISPR–Cas9 targeting accuracy. Nature 550, 407–410. doi: 10.1038/nature24268
Chi, Y., Huang, Z., Chen, Q., Xiong, X., Chen, K., Xu, J., et al. (2018). Loss of runx1 function results in B cell immunodeficiency but not T cell in adult zebrafish. Open Biol. 8:180043. doi: 10.1098/rsob.180043
Choi, M. Y., Cho, M.-H., Chang, H. J., Lee, S.-N., and Lee, K. E. (2019). Investigation of enhanced antitumor effects via co-inhibition of Wnt/β-catenin and PI3K/Akt/mTOR signaling pathways in human gastric cancer. J. Clin. Oncol. 37, (15_suppl):e15553. doi: 10.1200/JCO.2019.37.15_suppl.e15553
Choorapoikayil, S., Kuiper, R. V., de Bruin, A., and den Hertog, J. (2012). Haploinsufficiency of the genes encoding the tumor suppressor Pten predisposes zebrafish to hemangiosarcoma. Dis. Model. Mech. 5, 241–247. doi: 10.1242/dmm.008326
Choorapoikayil, S., Weijts, B., Kers, R., de Bruin, A., and den Hertog, J. (2013). Loss of Pten promotes angiogenesis and enhanced vegfaa expression in zebrafish. Dis. Model. Mech. 6, 1159–1166. doi: 10.1242/dmm.012377
Cichowski, K., and Jacks, T. (2001). NF1 tumor suppressor gene function. Cell 104, 593–604. doi: 10.1016/S0092-8674(01)00245-8
Cole, H. N., Rauschkolb, J. E., and Toomey, J. (1930). Dyskeratosis congenita with pigmentation, dystrophia unguis and leukokeratosis oris. Arch. Derm. Syphilol. 21, 71–95. doi: 10.1001/archderm.1930.01440070079008
Crino, P. B., Nathanson, K. L., and Henske, E. P. (2006). The tuberous sclerosis complex. N. Engl. J. Med. 355, 1345–1356. doi: 10.1056/NEJMra055323
Cvejic, A., Hall, C., Bak-Maier, M., Flores, M. V., Crosier, P., Redd, M. J., et al. (2008). Analysis of WASp function during the wound inflammatory response - live-imaging studies in zebrafish larvae. J. Cell Sci. 121, 3196–3206. doi: 10.1242/jcs.032235
Dai, Y., Zhu, L., Huang, Z., Zhou, M., Jin, W., Liu, W., et al. (2016). Cebpα is essential for the embryonic myeloid progenitor and neutrophil maintenance in zebrafish. J. Genet. Genomics 43, 593–600. doi: 10.1016/j.jgg.2016.09.001
d’Amora, M., and Giordani, S. (2018). The utility of zebrafish as a model for screening developmental neurotoxicity. Front. Neurosci. 12:976. doi: 10.3389/fnins.2018.00976
Dasgupta, B., Dugan, L. L., and Gutmann, D. H. (2003). The neurofibromatosis 1 gene product neurofibromin regulates pituitary adenylate cyclase-activating polypeptide-mediated signaling in astrocytes. J. Neurosci. 23, 8949–8954. doi: 10.1523/JNEUROSCI.23-26-08949.2003
Davidson, A. J., and Zon, L. I. (2004). The ‘definitive’ (and ‘primitive’) guide to zebrafish hematopoiesis. Oncogene 23, 7233–7246. doi: 10.1038/sj.onc.1207943
Delacruz, R. G. C., Sandoval, I. T., Chang, K., Miller, B. N., Reyes-Uribe, L., Borras, E., et al. (2019). Functional characterization of CNOT3 variants identified in familial adenomatous polyposis adenomas. Oncotarget 10, 3939–3951. doi: 10.18632/oncotarget.27003
Deshaies, R. J., and Joazeiro, C. A. P. (2009). RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434. doi: 10.1146/annurev.biochem.78.101807.093809
Di Micco, R., Fumagalli, M., Cicalese, A., Piccinin, S., Gasparini, P., Luise, C., et al. (2006). Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 444, 638–642. doi: 10.1038/nature05327
DiBella, L. M., Park, A., and Sun, Z. (2009). Zebrafish Tsc1 reveals functional interactions between the cilium and the TOR pathway. Hum. Mol. Genet. 18, 595–606. doi: 10.1093/hmg/ddn384
Dickinson, R. E., Griffin, H., Bigley, V., Reynard, L. N., Hussain, R., Haniffa, M., et al. (2011). Exome sequencing identifies GATA-2 mutation as the cause of dendritic cell, monocyte, B and NK lymphoid deficiency. Blood 118, 2656–2658. doi: 10.1182/blood-2011-06-360313
Dobrzycki, T., Mahony, C. B., Krecsmarik, M., Koyunlar, C., Rispoli, R., Peulen-Zink, J., et al. (2020). Deletion of a conserved Gata2 enhancer impairs haemogenic endothelium programming and adult zebrafish haematopoiesis. Commun. Biol. 3:71. doi: 10.1038/s42003-020-0798-3
Duran, A., Amanchy, R., Linares, J. F., Joshi, J., Abu-Baker, S., Porollo, A., et al. (2011). p62 is a key regulator of nutrient sensing in the mTORC1 pathway. Mol. Cell 44, 134–146. doi: 10.1016/j.molcel.2011.06.038
Eisinger, A. L., Nadauld, L. D., Shelton, D. N., Peterson, P. W., Phelps, R. A., Chidester, S., et al. (2006). The adenomatous polyposis coli tumor suppressor gene regulates expression of cyclooxygenase-2 by a mechanism that involves retinoic acid. J. Biol. Chem. 281, 20474–20482. doi: 10.1074/jbc.M602859200
El-Brolosy, M. A., Kontarakis, Z., Rossi, A., Kuenne, C., Günther, S., Fukuda, N., et al. (2019). Genetic compensation triggered by mutant mRNA degradation. Nature 568, 193–197. doi: 10.1038/s41586-019-1064-z
Evangelisti, C., Chiarini, F., Cappellini, A., Paganelli, F., Fini, M., Santi, S., et al. (2020). Targeting Wnt/β-catenin and PI3K/Akt/mTOR pathways in T-cell acute lymphoblastic leukemia. J. Cell. Physiol. 235, 5413–5428. doi: 10.1002/jcp.29429
Fahsold, R., Hoffmeyer, S., Mischung, C., Gille, C., Ehlers, C., Kücükceylan, N., et al. (2000). Minor lesion mutational spectrum of the entire NF1 gene does not explain its high mutability but points to a functional domain upstream of the GAP-related domain. Am. J. Hum. Genet. 66, 790–818. doi: 10.1086/302809
Faucherre, A., Taylor, G. S., Overvoorde, J., Dixon, J. E., and den Hertog, J. (2008). Zebrafish pten genes have overlapping and non-redundant functions in tumorigenesis and embryonic development. Oncogene 27, 1079–1086. doi: 10.1038/sj.onc.1210730
Feitsma, H., Kuiper, R. V., Korving, J., Nijman, I. J., and Cuppen, E. (2008). Zebrafish with mutations in mismatch repair genes develop neurofibromas and other tumors. Cancer Res. 68, 5059–5066. doi: 10.1158/0008-5472.CAN-08-0019
Feitsma, H., Leal, M. C., Moens, P. B., Cuppen, E., and Schulz, R. W. (2007). Mlh1 deficiency in zebrafish results in male sterility and aneuploid as well as triploid progeny in females. Genetics 175, 1561–1569. doi: 10.1534/genetics.106.068171
Fenton, H., Carlile, B., Montgomery, E. A., Carraway, H., Herman, J., Sahin, F., et al. (2006). LKB1 protein expression in human breast cancer. Appl. Immunohistochem. Mol. Morphol. 14, 146–153. doi: 10.1097/01.pai.0000176157.07908.20
Fodde, R., Edelmann, W., Yang, K., van Leeuwen, C., Carlson, C., Renault, B., et al. (1994). A targeted chain-termination mutation in the mouse Apc gene results in multiple intestinal tumors. Proc. Natl. Acad. Sci. U.S.A. 91, 8969–8973. doi: 10.1073/pnas.91.19.8969
Fodde, R., Smits, R., and Clevers, H. (2001). APC, Signal transduction and genetic instability in colorectal cancer. Nat. Rev. Cancer 1, 55–67. doi: 10.1038/35094067
Fradet-Turcotte, A., Sitz, J., Grapton, D., and Orthwein, A. (2016). BRCA2 functions: from DNA repair to replication fork stabilization. Endocr. Relat. Cancer 23, T1–T17. doi: 10.1530/ERC-16-0297
Francia, S., Michelini, F., Saxena, A., Tang, D., De Hoon, M., Anelli, V., et al. (2012). Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature 488, 231–235. doi: 10.1038/nature11179
Fuchs, O., Provaznikova, D., Kocova, M., Kostecka, A., Cvekova, P., Neuwirtova, R., et al. (2008). CEBPA polymorphisms and mutations in patients with acute myeloid leukemia, myelodysplastic syndrome, multiple myeloma and non-Hodgkin’s lymphoma. Blood Cells Mol. Dis. 40, 401–405. doi: 10.1016/j.bcmd.2007.11.005
Gao, J., Aksoy, B. A., Dogrusoz, U., Dresdner, G., Gross, B., Sumer, S. O., et al. (2013). Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6:l1. doi: 10.1126/scisignal.2004088
Gao, X., Johnson, K. D., Chang, Y. I., Boyer, M. E., Dewey, C. N., Zhang, J., et al. (2013). Gata2 cis-element is required for hematopoietic stem cell generation in the mammalian embryo. J. Exp. Med. 210, 2833–2842. doi: 10.1084/jem.20130733
Gao, X., Huang, S.-S., Qiu, S.-W., Su, Y., Wang, W.-Q., Xu, H.-Y., et al. (2020). Congenital sensorineural hearing loss as the initial presentation of PTPN11 -associated Noonan syndrome with multiple lentigines or Noonan syndrome: clinical features and underlying mechanisms. J. Med. Genet. doi: 10.1136/jmedgenet-2020-106892 Online ahead of print
Garber, J. E., and Offit, K. (2005). Hereditary cancer predisposition syndromes. J. Clin. Oncol. 23, 276–292. doi: 10.1200/JCO.2005.10.042
Germeshausen, M., Ballmaier, M., and Welte, K. (2006). MPL mutations in 23 patients suffering from congenital amegakaryocytic thrombocytopenia: the type of mutation predicts the course of the disease. Hum. Mutat. 27:296. doi: 10.1002/humu.9415
Giardiello, F. M., Welsh, S. B., Hamilton, S. R., Offerhaus, G. J. A., Gittelsohn, A. M., Booker, S. V., et al. (1987). Increased risk of cancer in the peutz–jeghers syndrome. N. Engl. J. Med. 316, 1511–1514. doi: 10.1056/NEJM198706113162404
Gillis, W. Q., St John, J., Bowerman, B., and Schneider, S. Q. (2009). Whole genome duplications and expansion of the vertebrate GATA transcription factor gene family. BMC Evol. Biol. 9:207. doi: 10.1186/1471-2148-9-207
Gioacchino, E., Koyunlar, C., de looper, H., Peulen, J., Bosch, D., Hoogenboezem, R., et al. (2019). Pf346 zebrafish as a novel model to study Gata2 haploinsufficiency syndromes. HemaSphere 3, 124–125.
Gitler, A. D., Zhu, Y., Ismat, F. A., Lu, M. M., Yamauchi, Y., Parada, L. F., et al. (2003). Nf1 has an essential role in endothelial cells. Nat. Genet. 33, 75–79. doi: 10.1038/ng1059
Gombart, A. F., Hofmann, W. K., Kawano, S., Takeuchi, S., Krug, U., Kwok, S. H., et al. (2002). Mutations in the gene encoding the transcription factor CCAAT/enhancer binding protein α in myelodysplastic syndromes and acute myeloid leukemias. Blood 99, 1332–1340. doi: 10.1182/blood.V99.4.1332
Gore, A. V., Pillay, L. M., Venero Galanternik, M., and Weinstein, B. M. (2018). The zebrafish: a fintastic model for hematopoietic development and disease. Wiley Interdiscip. Rev. Dev. Biol. 7:e312. doi: 10.1002/wdev.312
Gossage, L., Eisen, T., and Maher, E. R. (2015). VHL, the story of a tumour suppressor gene. Nat. Rev. Cancer 15, 55–64. doi: 10.1038/nrc3844
Growney, J. D., Shigematsu, H., Li, Z., Lee, B. H., Adelsperger, J., Rowan, R., et al. (2005). Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood 106, 494–504. doi: 10.1182/blood-2004-08-3280
Guerra, F., Rocher, A., Díaz, L., and Palaoro, L. (2020). Wnt/Beta-catenin and EGFR/PI3K/pAKT/mTOR signaling pathways and their relation with cervical cancer. J. Gynecol. Oncol. 3:1035.
Guldberg, P., Straten, P., Ahrenkiel, V., Seremet, T., Kirkin, A. F., and Zeuthen, J. (1999). Somatic mutation of the Peutz-Jeghers syndrome gene, LKB1/STK11, in malignant melanoma. Oncogene 18, 1777–1780. doi: 10.1038/sj.onc.1202486
Guo, H.-F., Tong, J., Hannan, F., Luo, L., and Zhong, Y. (2000). A neurofibromatosis-1-regulated pathway is required for learning in Drosophila. Nature 403, 895–898. doi: 10.1038/35002593
Hahn, C. N., Chong, C. E., Carmichael, C. L., Wilkins, E. J., Brautigan, P. J., Li, X. C., et al. (2011). Heritable GATA2 mutations associated with familial myelodysplastic syndrome and acute myeloid leukemia. Nat. Genet. 43, 1012–1019. doi: 10.1038/ng.913
Hainaut, P., and Pfeifer, G. P. (2016). Somatic TP53 mutations in the era of genome sequencing. Cold Spring Harb. Perspect. Med. 6:a026179. doi: 10.1101/cshperspect.a026179
Hammond, S. M. (2015). An overview of microRNAs. Adv. Drug Deliv. Rev. 87, 3–14. doi: 10.1016/j.addr.2015.05.001
Haramis, A.-P. G., Hurlstone, A., van der Velden, Y., Begthel, H., van den Born, M., Offerhaus, G. J. A., et al. (2006). Adenomatous polyposis coli-deficient zebrafish are susceptible to digestive tract neoplasia. EMBO Rep. 7, 444–449. doi: 10.1038/sj.embor.7400638
Hardie, D. G. (2007). AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat. Rev. Mol. Cell Biol. 8, 774–785. doi: 10.1038/nrm2249
Hearle, N., Schumacher, V., Menko, F., Olschwang, S., Boardman, L., and Gille, J. (2006). Frequency and Spectrum of Cancers in the Peutz-Jeghers Syndrome. Clin. Cancer Res. 12, 3209–3215. doi: 10.1158/1078-0432.CCR-06-0083
Hegedus, B., Dasgupta, B., Shin, J. E., Emnett, R. J., Hart-mahon, E. K., Elghazi, L., et al. (2007). Article neurofibromatosis-1 regulates neuronal and glial cell differentiation from neuroglial progenitors in vivo by Both cAMP- and Ras-dependent mechanisms. Cell Stem Cell 1, 443–457. doi: 10.1016/j.stem.2007.07.008
Hemminki, A., Markie, D., Tomlinson, I., Avizienyte, E., Roth, S., Loukola, A., et al. (1998). A serine/threonine kinase gene defective in Peutz–Jeghers syndrome. Nature 391, 184–187. doi: 10.1038/34432
Henriques, C. M., Carneiro, M. C., Tenente, I. M., Jacinto, A., and Ferreira, M. G. (2013). Telomerase is required for zebrafish lifespan. PLoS Genet. 9:e1003214. doi: 10.1371/journal.pgen.1003214
Hisada, M., Garber, J. E., Fung, C. Y., Joseph, F., and Li, F. P. (1998). Multiple primary cancers in families with Li-Fraumeni syndrome. J. Natl. Cancer Inst. 90, 606–611.
Hockings, C., Deaner, V., Hoade, Y., Dace, P., Lubin, A., and Payne, E. (2018). A zebrafish model of cooperating C and N terminal CEBPA mutations reveals defects in early myelopoeisis and HSPCs leading to leukaemogenesis. Blood 132:1343. doi: 10.1182/blood-2018-99-119336
Holtkamp, N., Mautner, V. F., Friedrich, R. E., Harder, A., Hartmann, C., Theallier-Janko, A., et al. (2004). Differentially expressed genes in neurofibromatosis 1-associated neurofibromas and malignant peripheral nerve sheath tumors. Acta Neuropathol. 107, 159–168. doi: 10.1007/s00401-003-0797-8
Hsu, A. P., Johnson, K. D., Falcone, E. L., Sanalkumar, R., Sanchez, L., Hickstein, D. D., et al. (2013). GATA2 haploinsufficiency caused by mutations in a conserved intronic element leads to MonoMAC syndrome. Blood 121, 3830–3837. doi: 10.1182/blood-2012-08-452763
Hsu, A. P., Sampaio, E. P., Khan, J., Calvo, K. R., Lemieux, J. E., Patel, S. Y., et al. (2011). Mutations in GATA2 are associated with the autosomal dominant and sporadic monocytopenia and mycobacterial infection (MonoMAC) syndrome. Blood 118, 2653–2655. doi: 10.1182/blood-2011-05-356352
Huang, K., Mashl, R. J., Wu, Y., Ritter, D. I., Wang, J., Oh, C., et al. (2018). Pathogenic germline variants in 10,389 adult cancers. Cell 173, 355.e14–370.e14. doi: 10.1016/j.cell.2018.03.039
Hurlstone, A. F. L., Haramis, A.-P. G., Wienholds, E., Begthel, H., Korving, J., van Eeden, F., et al. (2003). The Wnt/β-catenin pathway regulates cardiac valve formation. Nature 425, 633–637. doi: 10.1038/nature02028
Ichikawa, M., Asai, T., Saito, T., Yamamoto, G., Seo, S., Yamazaki, I., et al. (2004). AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nat. Med. 10, 299–304. doi: 10.1038/nm997
Ignatius, M. S., Hayes, M. N., Moore, F. E., Tang, Q., Garcia, S. P., Blackburn, P. R., et al. (2018). tp53 Deficiency causes a wide tumor spectrum and increases embryonal rhabdomyosarcoma metastasis in zebrafish. Elife 7:e37202. doi: 10.7554/eLife.37202.001
Ijsselsteijn, R., Jansen, J. G., and de Wind, N. (2020). DNA mismatch repair-dependent DNA damage responses and cancer. DNA Repair 93:102923. doi: 10.1016/j.dnarep.2020.102923
Imai, K., Morio, T., Zhu, Y., Jin, Y., Itoh, S., Kajiwara, M., et al. (2004). Clinical course of patients with WASP gene mutations. Blood 103, 456–464. doi: 10.1182/blood-2003-05-1480
Imamura, S., and Kishi, S. (2005). Molecular cloning and functional characterization of zebrafish ATM. Int. J. Biochem. Cell Biol. 37, 1105–1116. doi: 10.1016/j.biocel.2004.10.015
Inoki, K., Ouyang, H., Zhu, T., Lindvall, C., Wang, Y., Zhang, X., et al. (2006). TSC2 integrates wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell 126, 955–968. doi: 10.1016/j.cell.2006.06.055
Jette, C., Peterson, P. W., Sandoval, I. T., Manos, E. J., Hadley, E., Ireland, C. M., et al. (2004). The tumor suppressor adenomatous polyposis coli and caudal related homeodomain protein regulate expression of retinol dehydrogenase L. J. Biol. Chem. 279, 34397–34405. doi: 10.1074/jbc.M314021200
Jopling, C., van Geemen, D., and den Hertog, J. (2007). Shp2 knockdown and Noonan/LEOPARD mutant Shp2-Induced gastrulation defects. PLoS Genet. 3, e225–e225. doi: 10.1371/journal.pgen.0030225
Ji, H., Ramsey, M. R., Hayes, D. N., Fan, C., McNamara, K., Kozlowski, P., et al. (2007). LKB1 modulates lung cancer differentiation and metastasis. Nature 448, 807–810. doi: 10.1038/nature06030
Jiang, N., Dai, Q., Su, X., Fu, J., Feng, X., and Peng, J. (2020). Role of PI3K/AKT pathway in cancer: the framework of malignant behavior. Mol. Biol. Rep. 47, 4587–4629. doi: 10.1007/s11033-020-05435-1
Jin, H., Li, L., Xu, J., Zhen, F., Zhu, L., Liu, P. P., et al. (2012). Runx1 regulates embryonic myeloid fate choice in zebrafish through a negative feedback loop inhibiting Pu.1 expression. Blood 119, 5239–5249. doi: 10.1182/blood-2011-12-398362
Jin, Y., Mazza, C., Christie, J. R., Giliani, S., Fiorini, M., Mella, P., et al. (2004). Mutations of the Wiskott-Aldrich Syndrome Protein (WASP): hotspots, effect on transcription, and translation and phenotype/genotype correlation. Blood 104, 4010–4019. doi: 10.1182/blood-2003-05-1592
Jing, L., and Zon, L. I. (2011). Zebrafish as a model for normal and malignant hematopoiesis. Dis. Model. Mech. 4, 433–438. doi: 10.1242/dmm.006791
Jones, R. A., Feng, Y., Worth, A. J., Thrasher, A. J., Burns, S. O., and Martin, P. (2013). Modelling of human Wiskott-Aldrich syndrome protein mutants in zebrafish larvae using in vivo live imaging. J. Cell Sci. 126, 4077–4084. doi: 10.1242/jcs.128728
Jongmans, M. C. J., van der Burgt, I., Hoogerbrugge, P. M., Noordam, K., Yntema, H. G., Nillesen, W. M., et al. (2011). Cancer risk in patients with Noonan syndrome carrying a PTPN11 mutation. Eur. J. Hum. Genet. 19, 870–874. doi: 10.1038/ejhg.2011.37
Kaidanovich-Beilin, O., and Woodgett, J. R. (2011). GSK-3: functional insights from cell biology and animal models. Front. Mol. Neurosci. 4:40. doi: 10.3389/fnmol.2011.00040
Kanda, M., Sadakari, Y., Borges, M., Topazian, M., Farrell, J., Syngal, S., et al. (2013). Mutant TP53 in duodenal samples of pancreatic juice from patients with pancreatic cancer or high-grade dysplasia. Clin. Gastroenterol. Hepatol. 11, 719.e5–730.e5. doi: 10.1016/j.cgh.2012.11.016
Keszei, M., Record, J., Kritikou, J. S., Wurzer, H., Geyer, C., Thiemann, M., et al. (2018). Constitutive activation of WASp in X-linked neutropenia renders neutrophils hyperactive. J. Clin. Invest. 128, 4115–4131. doi: 10.1172/JCI64772
Ki, D. H., He, S., Rodig, S., and Look, A. T. (2017). Overexpression of PDGFRA cooperates with loss of NF1 and p53 to accelerate the molecular pathogenesis of malignant peripheral nerve sheath tumors. Oncogene 36, 1058–1068. doi: 10.1038/onc.2016.269
Ki, D. H., Oppel, F., Durbin, A. D., and Look, A. T. (2019). Mechanisms underlying synergy between DNA topoisomerase I-targeted drugs and mTOR kinase inhibitors in NF1-associated malignant peripheral nerve sheath tumors. Oncogene 38, 6585–6598. doi: 10.1038/s41388-019-0965-5
Kikuchi, H., Miyazaki, S., Setohuchi, T., Hiramatsu, Y., Ohta, M., Kamiya, K., et al. (2012). Rapid relapse after resection of a sunitinib-resistant gastrointestinal stromal tumor harboring a secondary mutation in exon 13 of the c-KIT gene. Anticancer Res. 32, 4105–4109.
Kim, H. R., Santhakumar, K., Markham, E., Baldera, D., Greenald, D., Bryant, H. E., et al. (2020). Investigation of the role of VHL-HIF signaling in DNA repair and apoptosis in zebrafish. Oncotarget 11, 1109–1130. doi: 10.18632/oncotarget.27521
Kim, S.-H., Kowalski, M. L., Carson, R. P., Bridges, L. R., and Ess, K. C. (2013). Heterozygous inactivation of tsc2 enhances tumorigenesis in p53 mutant zebrafish. Dis. Model. Mech. 6, 925–933. doi: 10.1242/dmm.011494
Kim, S.-H., Speirs, C. K., Solnica-Krezel, L., and Ess, K. C. (2011). Zebrafish model of tuberous sclerosis complex reveals cell-autonomous and non-cell-autonomous functions of mutant tuberin. Dis. Model. Mech. 4, 255–267. doi: 10.1242/dmm.005587
Kipling, D., and Cooke, H. J. (1990). Hypervariable ultra-long telomeres in mice. Nature 347, 400–402. doi: 10.1038/347400a0
Kiss, T., Fayet-Lebaron, E., and Jády, B. E. (2010). Box H/ACA small ribonucleoproteins. Mol. Cell 37, 597–606. doi: 10.1016/j.molcel.2010.01.032
Knight, S. W., Heiss, N. S., Vulliamy, T. J., Aalfs, C. M., McMahon, C., Richmond, P., et al. (1999). Unexplained aplastic anaemia, immunodeficiency, and cerebellar hypoplasia (Hoyeraal-Hreidarsson syndrome) due to mutations in the dyskeratosis congenita gene. DKC1. Br. J. Haematol. 107, 335–339. doi: 10.1046/j.1365-2141.1999.01690.x
Kontaridis, M. I., Swanson, K. D., David, F. S., Barford, D., and Neel, B. G. (2006). PTPN11 (Shp2) mutations in LEOPARD syndrome have dominant negative, not activating, effects. J. Biol. Chem. 281, 6785–6792. doi: 10.1074/jbc.M513068200
Kratz, C. P., Schubbert, S., Bollag, G., Niemeyer, C. M., Shannon, K. M., and Zenker, M. (2006). Germline mutations in components of the ras signaling pathway in noonan syndrome and related disorders. Cell Cycle 5, 1607–1611. doi: 10.4161/cc.5.15.3128
Kroeger, P. T., Drummond, B. E., Miceli, R., McKernan, M., Gerlach, G. F., Marra, A. N., et al. (2017). The zebrafish kidney mutant zeppelin reveals that brca2/fancd1 is essential for pronephros development. Dev. Biol. 428, 148–163. doi: 10.1016/j.ydbio.2017.05.025
Kuang, X., Liu, C., Fang, J., Ma, W., Zhang, J., and Cui, S. (2016). The tumor suppressor gene lkb1 is essential for glucose homeostasis during zebrafish early development. FEBS Lett. 2076–2085. doi: 10.1002/1873-3468.12237
Kullmann, L., and Krahn, M. P. (2018). Controlling the master—upstream regulation of the tumor suppressor LKB1. Oncogene 37, 3045–3057. doi: 10.1038/s41388-018-0145-z
Kutler, D. I., Singh, B., Satagopan, J., Batish, S. D., Berwick, M., Giampietro, P. F., et al. (2003). A 20-year perspective on the International Fanconi Anemia Registry (IFAR). Blood 101, 1249–1256. doi: 10.1182/blood-2002-07-2170
Latger-Cannard, V., Philippe, C., Bouquet, A., Baccini, V., Alessi, M. C., Ankri, A., et al. (2016). Haematological spectrum and genotype-phenotype correlations in nine unrelated families with RUNX1 mutations from the French network on inherited platelet disorders. Orphanet J. Rare Dis. 11:49. doi: 10.1186/s13023-016-0432-0
Lavalou, P., Eckert, H., Damy, L., Constanty, F., Majello, S., Bitetti, A., et al. (2019). Strategies for genetic inactivation of long noncoding RNAs in zebrafish. RNA 25, 897–904. doi: 10.1261/rna.069484.118
Leal, M. C., Feitsma, H., Cuppen, E., França, L. R., and Schulz, R. W. (2008). Completion of meiosis in male zebrafish (Danio rerio) despite lack of DNA mismatch repair gene mlh1. Cell Tissue Res. 332, 133–139. doi: 10.1007/s00441-007-0550-z
Lee, W., Teckie, S., Wiesner, T., Ran, L., Prieto Granada, C. N., Lin, M., et al. (2014). PRC2 is recurrently inactivated through EED or SUZ12 loss in malignant peripheral nerve sheath tumors. Nat. Genet. 46, 1227–1232. doi: 10.1038/ng.3095
Li, F. P., Fraumeni, J. F., Mulvihill, J. J., Blattner, W. A., Dreyfus, M. G., Tucker, M. A., et al. (1988). A cancer family syndrome in twenty-four kindreds. Cancer Res. 48, 5358–5362.
Li, J., Jørgensen, S. F., Maggadottir, S. M., Bakay, M., Warnatz, K., Glessner, J., et al. (2015). Association of CLEC16A with human common variable immunodeficiency disorder and role in murine B cells. Nat. Commun. 6:6804. doi: 10.1038/ncomms7804
Lim, C.-H., Chong, S.-W., and Jiang, Y.-J. (2009). Udu deficiency activates DNA damage checkpoint. Mol. Biol. Cell 20, 4183–4193. doi: 10.1091/mbc.E09
Lin, Q., Zhang, Y., Zhou, R., Zheng, Y., Zhao, L., Huang, M., et al. (2017). Establishment of a congenital amegakaryocytic thrombocytopenia model and a thrombocyte-specific reporter line in zebrafish. Leukemia 31, 1206–1216. doi: 10.1038/leu.2016.320
Liu, K., Petree, C., Requena, T., Varshney, P., and Varshney, G. K. (2019). Expanding the CRISPR toolbox in zebrafish for studying development and disease. Front. Cell Dev. Biol. 7:13. doi: 10.3389/fcell.2019.00013
Liu, T. X., Rhodes, J., Deng, M., Hsu, K., Radomska, H. S., Kanki, J. P., et al. (2007). Dominant-interfering C/EBPα stimulates primitive erythropoiesis in zebrafish. Exp. Hematol. 35, 230–239. doi: 10.1016/j.exphem.2006.10.008
Liu, W., Palovcak, A., Li, F., Zafar, A., Yuan, F., and Zhang, Y. (2020). Fanconi anemia pathway as a prospective target for cancer intervention. Cell Biosci. 10:39. doi: 10.1186/s13578-020-00401-7
Logan, C. Y., and Nusse, R. (2004). The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 20, 781–810. doi: 10.1146/annurev.cellbio.20.010403.113126
Lu, C., Xie, M., Wendl, M. C., Wang, J., McLellan, M. D., Leiserson, M. D. M., et al. (2015). Patterns and functional implications of rare germline variants across 12 cancer types. Nat. Commun. 6:10086. doi: 10.1038/ncomms10086
Lutskiy, M. I., Rosen, F. S., and Remold-O’Donnell, E. (2005). Genotype-proteotype linkage in the wiskott-aldrich syndrome. J. Immunol. 175, 1329–1336. doi: 10.4049/jimmunol.175.2.1329
Lyle, C. L., Belghasem, M., and Chitalia, V. C. (2019). c-Cbl: an important regulator and a target in angiogenesis and tumorigenesis. Cells 8:498. doi: 10.3390/cells8050498
MacDonald, B. T., Tamai, K., and He, X. (2009). Wnt/β-catenin signaling: components, mechanisms, and diseases. Dev. Cell 17, 9–26. doi: 10.1016/j.devcel.2009.06.016
Malaquias, A. C., and Jorge, A. A. L. (2014). Developmental Syndromes of Ras/MAPK Pathway Dysregulation in eLS. Chichester: John Wiley & Sons, Ltd, doi: 10.1002/9780470015902.a0021426
Malkin, D. (1993). p53 and the Li-Fraumeni syndrome. Cancer Genet. Cytogenet. 66, 83–92. doi: 10.1016/0165-4608(93)90233-c
Mans, L. A., Querol Cano, L., van Pelt, J., Giardoglou, P., Keune, W.-J., and Haramis, A.-P. G. (2017). The tumor suppressor LKB1 regulates starvation-induced autophagy under systemic metabolic stress. Sci. Rep. 7:7327. doi: 10.1038/s41598-017-07116-9
Mantripragada, K. K., Spurlock, G., Kluwe, L., Chuzhanova, N., Ferner, R. E., Frayling, I. M., et al. (2008). High-resolution DNA copy number profiling of malignant peripheral nerve sheath tumors using targeted microarray-based comparative genomic hybridization. Clin. Cancer Res. 14, 1015–1024. doi: 10.1158/1078-0432.CCR-07-1305
Marinaccio, C., Suraneni, P. K., Celik, H., Volk, A., Wen, J. Q., Ling, T., et al. (2020). Loss of LKB1/STK11 facilitates leukemic progression of the myeloproliferative neoplasms. Blood 136, 1–1. doi: 10.1182/blood-2020-140557
Martínez-Quintana, E., and Rodríguez-González, F. (2013). RASopathies: from noonan to LEOPARD syndrome. Rev. Española Cardiol. 66, 756–757. doi: 10.1016/j.rec.2013.05.005
Martin-Orozco, E., Sanchez-Fernandez, A., Ortiz-Parra, I., and Ayala-San Nicolas, M. (2019). WNT signaling in tumors: the way to evade drugs and immunity. Front. Immunol. 10:2854. doi: 10.3389/fimmu.2019.02854
Matsumoto, S., Iwakawa, R., Takahashi, K., Kohno, T., Nakanishi, Y., Matsuno, Y., et al. (2007). Prevalence and specificity of LKB1 genetic alterations in lung cancers. Oncogene 26, 5911–5918. doi: 10.1038/sj.onc.1210418
McReynolds, L. J., and Savage, S. A. (2017). Pediatric leukemia susceptibility disorders: manifestations and management. Hematology 2017, 242–250. doi: 10.1182/asheducation-2017.1.242
Metcalf, J. L., Bradshaw, P. S., Komosa, M., Greer, S. N., Stephen Meyn, M., and Ohh, M. (2014). K63-Ubiquitylation of VHL by SOCS1 mediates DNA double-strand break repair. Oncogene 33, 1055–1065. doi: 10.1038/onc.2013.22
Miura, H., Quadros, R. M., Gurumurthy, C. B., and Ohtsuka, M. (2018). Easi-CRISPR for creating knock-in and conditional knockout mouse models using long ssDNA donors. Nat. Protoc. 13, 195–215. doi: 10.1038/nprot.2017.153
Molina, J. R., and Adjei, A. A. (2006). The Ras/Raf/MAPK Pathway. J. Thorac. Oncol. 1, 7–9. doi: 10.1016/s1556-0864(15)31506-9
Moser, A., Pitot, H., and Dove, W. (1990). A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science 247, 322–324. doi: 10.1126/science.2296722
Mukherjee, S., Ridgeway, A. D., and Lamb, D. J. (2010). DNA mismatch repair and infertility. Curr. Opin. Urol. 20, 525–532. doi: 10.1097/MOU.0b013e32833f1c21
Mullany, L. K., Wong, K.-K., Marciano, D. C., Katsonis, P., King-Crane, E. R., Ren, Y. A., et al. (2015). Specific TP53 mutants overrepresented in ovarian cancer impact CNV, TP53 activity, responses to Nutlin-3a, and cell survival. Neoplasia 17, 789–803. doi: 10.1016/j.neo.2015.10.003
Nadauld, L. D., Sandoval, I. T., Chidester, S., Yost, H. J., and Jones, D. A. (2004). Adenomatous polyposis coli control of retinoic acid biosynthesis is critical for zebrafish intestinal development and differentiation. J. Biol. Chem. 279, 51581–51589. doi: 10.1074/JBC.M408830200
Nadauld, L. D., Shelton, D. N., Chidester, S., Yost, H. J., and Jones, D. A. (2005). The zebrafish retinol dehydrogenase, rdh1l, Is essential for intestinal development and is regulated by the tumor suppressor adenomatous polyposis coli. J. Biol. Chem. 280, 30490–30495. doi: 10.1074/JBC.M504973200
Neel, B. G., Gu, H., and Pao, L. (2003). The ‘Shp’ing news: SH2 domain-containing tyrosine phosphatases in cell signaling. Trends Biochem. Sci. 28, 284–293. doi: 10.1016/S0968-0004(03)00091-4
Nerlov, C. (2004). C/EBPα mutations in acute myeloid leukaemias. Nat. Rev. Cancer 4, 394–400. doi: 10.1038/nrc1363
Niemeyer, C. M., Kang, M. W., Shin, D. H., Furlan, I., Erlacher, M., Bunin, N. J., et al. (2010). Germline CBL mutations cause developmental abnormalities and predispose to juvenile myelomonocytic leukemia. Nat. Genet. 42, 794–800. doi: 10.1038/ng.641
Nye, J., Melters, D. P., and Dalal, Y. (2018). The art of War: harnessing the epigenome against cancer. F1000Res. 7:141. doi: 10.12688/f1000research.12833.1
Okuda, T., Van Deursen, J., Hiebert, S. W., Grosveld, G., and Downing, J. R. (1996). AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 84, 321–330. doi: 10.1016/S0092-8674(00)80986-1
Oppel, F., Tao, T., Shi, H., Ross, K. N., Zimmerman, M. W., He, S., et al. (2019). Loss of atrx cooperates with p53-deficiency to promote the development of sarcomas and other malignancies. PLoS Genet. 15:e1008039. doi: 10.1371/journal.pgen.1008039
Oshima, M., Oshima, H., Kitagawa, K., Kobayashi, M., Itakura, C., and Taketo, M. (1995). Loss of Apc heterozygosity and abnormal tissue building in nascent intestinal polyps in mice carrying a truncated Apc gene. Proc. Natl. Acad. Sci. U.S.A. 92, 4482–4486. doi: 10.1073/pnas.92.10.4482
Oyarbide, U., Shah, A. N., Amaya-Mejia, W., Snyderman, M., Kell, M. J., Allende, D. S., et al. (2020). Loss of Sbds in zebrafish leads to neutropenia and pancreas and liver atrophy. JCI Insight 5:e134309. doi: 10.1172/jci.insight.134309
Oyarbide, U., Topczewski, J., and Corey, S. J. (2019). Peering through zebrafish to understand inherited bone marrow failure syndromes. Haematologica 104, 13–24. doi: 10.3324/haematol.2018.196105
Pabst, T., Mueller, B. U., Zhang, P., Radomska, H. S., Narravula, S., Schnittger, S., et al. (2001). Dominant-negative mutations of CEBPA, encoding CCAAT/enhancer binding protein-α (C/EBPα), in acute myeloid leukemia. Nat. Genet. 27, 263–270. doi: 10.1038/85820
Parant, J. M., George, S. A., Holden, J. A., and Yost, H. J. (2010). Genetic modeling of Li-Fraumeni syndrome in zebrafish. Dis. Model. Mech. 3, 45–56. doi: 10.1242/dmm.003749
Peeters, K., Stassen, J.-M., Collen, D., Van Geet, C., and Freson, K. (2008). Emerging treatments for thrombocytopenia: increasing platelet production. Drug Discov. Today 13, 798–806. doi: 10.1016/j.drudis.2008.06.002
Peng, X., Dong, M., Ma, L., Jia, X. E., Mao, J., Jin, C., et al. (2015). A point mutation of zebrafish c-cbl gene in the ring finger domain produces a phenotype mimicking human myeloproliferative disease. Leukemia 29, 2355–2365. doi: 10.1038/leu.2015.154
Pereboom, T. C., Van Weele, L. J., Bondt, A., and MacInnes, A. W. (2011). A zebrafish model of dyskeratosis congenita reveals hematopoietic stem cell formation failure resulting from ribosomal protein-mediated p53 stabilization. Blood 118, 5458–5465. doi: 10.1182/blood-2011-04-351460
Perez, B., Mechinaud, F., Galambrun, C., Ben Romdhane, N., Isidor, B., Philip, N., et al. (2010). Germline mutations of the CBL gene define a new genetic syndrome with predisposition to juvenile myelomonocytic leukaemia. J. Med. Genet. 47, 686–691. doi: 10.1136/jmg.2010.076836
Phelps, R. A., Chidester, S., Dehghanizadeh, S., Phelps, J., Sandoval, I. T., Rai, K., et al. (2009). A two-step model for colon adenoma initiation and progression caused by APC loss. Cell 137, 623–634. doi: 10.1016/j.cell.2009.02.037
Porta, C., Paglino, C., and Mosca, A. (2014). Targeting PI3K/Akt/mTOR signaling in cancer. Front. Oncol. 4:64. doi: 10.3389/fonc.2014.00064
Preudhomme, C., Renneville, A., Bourdon, V., Philippe, N., Roche-Lestienne, C., Boissel, N., et al. (2009). High frequency of RUNX1 biallelic alteration in acute myeloid leukemia secondary to familial platelet disorder. Blood 113, 5583–5587. doi: 10.1182/blood-2008-07-168260
Prior, I. A., Lewis, P. D., and Mattos, C. (2012). A comprehensive survey of ras mutations in cancer. Cancer Res. 72, 2457–2467. doi: 10.1158/0008-5472.CAN-11-2612
Prossomariti, A., Piazzi, G., Alquati, C., and Ricciardiello, L. (2020). Are Wnt/β-Catenin and PI3K/AKT/mTORC1 distinct pathways in colorectal cancer? Cell. Mol. Gastroenterol. Hepatol. 10, 491–506. doi: 10.1016/j.jcmgh.2020.04.007
Provost, E., Wehner, K. A., Zhong, X., Ashar, F., Nguyen, E., Green, R., et al. (2012). Ribosomal biogenesis genes play an essential and p53- independent role in zebrafish pancreas development. Development 139, 3232–3241. doi: 10.1242/dev.077107
Prykhozhij, S. V., and Berman, J. N. (2018). Zebrafish knock-ins swim into the mainstream. Dis. Model. Mech. 11:dmm037515. doi: 10.1242/dmm.037515
Prykhozhij, S. V., Caceres, L., and Berman, J. N. (2018a). New developments in CRISPR/Cas-based functional genomics and their implications for research using Zebrafish. Curr. Gene Ther. 17, 286–300. doi: 10.2174/1566523217666171121164132
Prykhozhij, S. V., Fuller, C., Steele, S. L., Veinotte, C. J., Razaghi, B., Robitaille, J. M., et al. (2018b). Optimized knock-in of point mutations in zebrafish using CRISPR/Cas9. Nucleic Acids Res. 46:e102. doi: 10.1093/nar/gky512
Prykhozhij, S. V., Cordeiro-Santanach, A., Caceres, L., and Berman, J. N. (2020). “Genome editing in zebrafish using high-fidelity Cas9 nucleases: choosing the right nuclease for the task. Methods Mol. Biol. 2115, 385–405. doi: 10.1007/978-1-0716-0290-4_21
Qu, Y., Gharbi, N., Yuan, X., Olsen, J. R., Blicher, P., Dalhus, B., et al. (2016). Axitinib blocks Wnt/β-catenin signaling and directs asymmetric cell division in cancer. Proc. Natl. Acad. Sci. U. S. A. 113, 9339–9344. doi: 10.1073/pnas.1604520113
Rabinowitz, J. D., and White, E. (2010). Autophagy and Metabolism. Science 330, 1344–1348. doi: 10.1126/science.1193497
Raby, L., Völkel, P., Le Bourhis, X., and Angrand, P. O. (2020). Genetic engineering of zebrafish in cancer research. Cancers 12:2168. doi: 10.3390/cancers12082168
Raghunandan, M., Yeo, J. E., Walter, R., Saito, K., Harvey, A. J., Ittershagen, S., et al. (2020). Functional cross talk between the Fanconi anemia and ATRX/DAXX histone chaperone pathways promotes replication fork recovery. Hum. Mol. Genet. 29, 1083–1095. doi: 10.1093/hmg/ddz250
Rahman, N. (2014). Realizing the promise of cancer predisposition genes. Nature 505, 302–308. doi: 10.1038/nature12981
Rai, K., Sarkar, S., Broadbent, T. J., Voas, M., Grossmann, K. F., Nadauld, L. D., et al. (2010). DNA demethylase activity maintains intestinal cells in an undifferentiated state following loss of APC. Cell 142, 930–942. doi: 10.1016/j.cell.2010.08.030
Ramanagoudr-Bhojappa, R., Carrington, B., Ramaswami, M., Bishop, K., Robbins, G. M., Jones, M. P., et al. (2018). Multiplexed CRISPR/Cas9-mediated knockout of 19 Fanconi anemia pathway genes in zebrafish revealed their roles in growth, sexual development and fertility. PLoS Genet. 14:e1007821. doi: 10.1371/journal.pgen.1007821
Rasighaemi, P., Basheer, F., Liongue, C., and Ward, A. C. (2015). Zebrafish as a model for leukemia and other hematopoietic disorders. J. Hematol. Oncol. 8:29. doi: 10.1186/s13045-015-0126-4
Rasnic, R., Linial, N., and Linial, M. (2020). Expanding cancer predisposition genes with ultra-rare cancer-exclusive human variations. Sci. Rep. 10:13462. doi: 10.1038/s41598-020-70494-0
Ries, S., Biederer, C., Woods, D., Shifman, O., Shirasawa, S., Sasazuki, T., et al. (2000). Opposing effects of Ras on p53. Cell 103, 321–330. doi: 10.1016/S0092-8674(00)00123-9
Ripperger, T., Steinemann, D., Göhring, G., Finke, J., Niemeyer, C. M., Strahm, B., et al. (2009). A novel pedigree with heterozygous germline RUNX1 mutation causing familial MDS-related AML: can these families serve as a multistep model for leukemic transformation? Leukemia 23, 1364–1366. doi: 10.1038/leu.2009.87
Robertson, J. C., Jorcyk, C. L., and Oxford, J. T. (2018). DICER1 syndrome: DICER1 mutations in rare cancers. Cancers 10:143. doi: 10.3390/cancers10050143
Rodríguez-Marí, A., Cañestro, C., BreMiller, R. A., Nguyen-Johnson, A., Asakawa, K., Kawakami, K., et al. (2010). Sex reversal in zebrafish fancl mutants is caused by Tp53-mediated germ cell apoptosis. PLoS Genet. 6:e1001034. doi: 10.1371/journal.pgen.1001034
Rodríguez-Marí, A., Wilson, C., Titus, T. A., Cañestro, C., BreMiller, R. A., Yan, Y. L., et al. (2011). Roles of brca2 (fancd1) in oocyte nuclear architecture, gametogenesis, gonad tumors, and genome stability in zebrafish. PLoS Genet. 7:e1001357. doi: 10.1371/journal.pgen.1001357
Rounioja, S., Saralahti, A., Rantala, L., Parikka, M., Henriques-Normark, B., Silvennoinen, O., et al. (2012). Defense of zebrafish embryos against Streptococcus pneumoniae infection is dependent on the phagocytic activity of leukocytes. Dev. Comp. Immunol. 36, 342–348. doi: 10.1016/j.dci.2011.05.008
Sansal, I., and Sellers, W. R. (2004). The biology and clinical relevance of the PTEN tumor suppressor pathway. J. Clin. Oncol. 22, 2954–2963. doi: 10.1200/JCO.2004.02.141
Santoriello, C., Deflorian, G., Pezzimenti, F., Kawakami, K., Lanfrancone, L., Di Fagagna, F. D. A., et al. (2009). Expression of H-RASV12 in a zebrafish model of costello syndrome causes cellular senescence in adult proliferating cells. Dis. Model. Mech. 2, 56–67. doi: 10.1242/dmm.001016
Sarkozy, A., Digilio, M. C., and Dallapiccola, B. (2008). Leopard syndrome. Orphanet J. Rare Dis. 3:13. doi: 10.1186/1750-1172-3-13
Schmit, J. M., Turner, D. J., Hromas, R. A., Wingard, J. R., Brown, R. A., Li, Y., et al. (2015). Two novel RUNX1 mutations in a patient with congenital thrombocytopenia that evolved into a high grade myelodysplastic syndrome. Leuk. Res. Rep. 4, 24–27. doi: 10.1016/j.lrr.2015.03.002
Schuhmacher, A. J., Guerra, C., Sauzeau, V., Cañamero, M., Bustelo, X. R., and Barbacid, M. (2008). A mouse model for costello syndrome reveals an Ang II-mediated hypertensive condition. J. Clin. Invest. 118, 2169–2179. doi: 10.1172/JCI34385
Serra-Nedelec, A. D. R., Edouard, T., Treguer, K., Tajan, M., Araki, T., Dance, M., et al. (2012). Noonan syndrome-causing SHP2 mutants inhibit insulin-like growth factor 1 release via growth hormone-induced ERK hyperactivation, which contributes to short stature. Proc. Natl. Acad. Sci. U.S.A. 109, 4257–4262. doi: 10.1073/pnas.1119803109
Shaw, R. J. (2009). LKB1 and AMP-activated protein kinase control of mTOR signalling and growth. Acta Physiol. 196, 65–80. doi: 10.1111/j.1748-1716.2009.01972.x
Shelton, D. N., Sandoval, I. T., Eisinger, A., Chidester, S., Ratnavake, A., Ireland, C. M., et al. (2006). Up-regulation of CYP26A1 in adenomatous polyposis coli-deficient vertebrates via a WNT-dependent mechanism: implications for intestinal cell differentiation and colon tumor development. Cancer Res. 66, 7571–7577. doi: 10.1158/0008-5472.CAN-06-1067.
Shin, J., Padmanabhan, A., De Groh, E. D., Lee, J. S., Haidar, S., Dahlberg, S., et al. (2012). Zebrafish neurofibromatosis type 1 genes have redundant functions in tumorigenesis and embryonic development. Dis. Model. Mech. 5, 881–894. doi: 10.1242/dmm.009779
Shivanna, S., Harrold, I., Shashar, M., Meyer, R., Kiang, C., Francis, J., et al. (2015). The C-Cbl ubiquitin ligase regulates nuclear β-catenin and angiogenesis by its tyrosine phosphorylation mediated through the Wnt signaling pathway. J. Biol. Chem. 290, 12537–12546. doi: 10.1074/jbc.M114.616623
Shive, H. R., West, R. R., Embree, L. J., Azuma, M., Sood, R., Liu, P., et al. (2010). brca2 in zebrafish ovarian development, spermatogenesis, and tumorigenesis. Proc. Natl. Acad. Sci. U.S.A. 107, 19350–19355. doi: 10.1073/pnas.1011630107
Shive, H. R., West, R. R., Embree, L. J., Golden, C. D., and Hickstein, D. D. (2014). Brca2 and Tp53 collaborate in tumorigenesis in Zebrafish. PLoS One 9:e87177. doi: 10.1371/journal.pone.0087177
Shlien, A., Tabori, U., Marshall, C. R., Pienkowska, M., Feuk, L., Novokmet, A., et al. (2008). Excessive genomic DNA copy number variation in the Li-Fraumeni cancer predisposition syndrome. Proc. Natl. Acad. Sci. U.S.A. 105, 11264–11269. doi: 10.1073/pnas.0802970105
Silva, A. J., Frankland, P. W., Marowitz, Z., Friedman, E., Lazlo, G., Cioffi, D., et al. (1997). A mouse model for the learning and memory deficits associated with neurofibromatosis type I. Nat. Genet. 15, 281–284. doi: 10.1038/ng0397-281
Song, W. J., Sullivan, M. G., Legare, R. D., Hutchings, S., Tan, X., Kufrin, D., et al. (1999). Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nat. Genet. 23, 166–175. doi: 10.1038/13793
Sood, R., English, M. A., Belele, C. L., Jin, H., Bishop, K., Haskins, R., et al. (2010). Development of multilineage adult hematopoiesis in the zebrafish with a runx1 truncation mutation. Blood 115, 2806–2809. doi: 10.1182/blood-2009-08-236729
Sood, R., Kamikubo, Y., and Liu, P. (2017). Role of RUNX1 in hematological malignancies. Blood 129, 2070–2082. doi: 10.1182/blood-2016-10-687830
Stepensky, P., Chacón-Flores, M., Kim, K. H., Abuzaitoun, O., Bautista-Santos, A., Simanovsky, N., et al. (2017). Mutations in EFL1, an SBDS partner, are associated with infantile pancytopenia, exocrine pancreatic insufficiency and skeletal anomalies in a Shwachman-Diamond like syndrome. J. Med. Genet. 54, 558–566. doi: 10.1136/jmedgenet-2016-104366
Stewart, R. A., Sanda, T., Widlund, H. R., Zhu, S., Swanson, K. D., Hurley, A. D., et al. (2010). Phosphatase-dependent and -independent functions of Shp2 in neural crest cells underlie LEOPARD syndrome pathogenesis. Dev. Cell 18, 750–762. doi: 10.1016/j.devcel.2010.03.009
Stiff, T., Tena, T. C., O’Driscoll, M., Jeggo, P. A., and Philipp, M. (2016). ATR promotes cilia signalling: links to developmental impacts. Hum. Mol. Genet. 25, 1574–1587. doi: 10.1093/hmg/ddw034
Stumpf, M., Choorapoikayil, S., and den Hertog, J. (2015). Pten function in zebrafish: anything but a fish story. Methods 77–78, 191–196. doi: 10.1016/j.ymeth.2014.11.002
Sullivan, K. E., Mullen, C. A., Blaese, R. M., and Winkelstein, J. A. (1994). A multiinstitutional survey of the Wiskott-Aldrich syndrome. J. Pediatr. 125, 876–885. doi: 10.1016/S0022-3476(05)82002-5
Szklarczyk, D., Franceschini, A., Wyder, S., Forslund, K., Heller, D., Huerta-Cepas, J., et al. (2015). STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452. doi: 10.1093/nar/gku1003
Tabori, U., Nanda, S., Druker, H., Lees, J., and Malkin, D. (2007). Younger age of cancer initiation is associated with shorter telomere length in Li-fraumeni syndrome. Cancer Res. 67, 1415–1418. doi: 10.1158/0008-5472.CAN-06-3682
Tan, S., Kermasson, L., Hoslin, A., Jaako, P., Faille, A., Acevedo-Arozena, A., et al. (2019). EFL1 mutations impair eIF6 release to cause Shwachman-Diamond syndrome. Blood 134, 277–290. doi: 10.1182/blood.2018893404
Tartaglia, M., Gelb, B. D., and Zenker, M. (2011). Noonan syndrome and clinically related disorders. Best Pract. Res. Clin. Endocrinol. Metab. 25, 161–179. doi: 10.1016/j.beem.2010.09.002
Tartaglia, M., Mehler, E. L., Goldberg, R., Zampino, G., Brunner, H. G., Kremer, H., et al. (2001). Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat. Genet. 29, 465–468. doi: 10.1038/ng772
Tawana, K., Wang, J., Renneville, A., Bödör, C., Hills, R., Loveday, C., et al. (2015). Disease evolution and outcomes in familial AML with germline CEBPA mutations. Blood 126, 1214–1223. doi: 10.1182/blood-2015-05-647172
Thien, C. B. F., and Langdon, W. Y. (2001). Cbl: many adaptations to regulate protein tyrosine kinases. Nat. Rev. Mol. Cell Biol. 2, 294–307. doi: 10.1038/35067100
Thrasher, A. J., and Burns, S. O. (2010). WASP: a key immunological multitasker. Nat. Rev. Immunol. 10, 182–192. doi: 10.1038/nri2724
Titus, T. A., Yan, Y. L., Wilson, C., Starks, A. M., Frohnmayer, J. D., Bremiller, R. A., et al. (2009). The Fanconi anemia/BRCA gene network in zebrafish: embryonic expression and comparative genomics. Mutat. Res. Fundam. Mol. Mech. Mutagen. 668, 117–132. doi: 10.1016/j.mrfmmm.2008.11.017
Tong, J., Hannan, F., Zhu, Y., Bernards, A., and Zhong, Y. (2002). Neurofibromin regulates G protein-stimulated adenylyl cyclase activity. Nat. Neurosci. 5, 95–96. doi: 10.1038/nn792
Tonini, G., Intagliata, S., Cagli, B., Segreto, F., Perrone, G., Onetti Muda, A., et al. (2010). Recurrent scrotal hemangiomas during treatment with Sunitinib. J. Clin. Oncol. 28, e737–e738. doi: 10.1200/JCO.2010.30.4865
Tsai, S.-M., Chu, K.-C., and Jiang, Y.-J. (2020). Newly identified Gon4l/Udu-interacting proteins implicate novel functions. Sci. Rep. 10:14213. doi: 10.1038/s41598-020-70855-9
Tsuboi, S., and Meerloo, J. (2007). Wiskott-aldrich syndrome protein is a key regulator of the phagocytic cup formation in macrophages. J. Biol. Chem. 282, 34194–34203. doi: 10.1074/jbc.M705999200
Vågbø, C. B., and Slupphaug, G. (2020). RNA in DNA repair. DNA Repair. 95:102927. doi: 10.1016/j.dnarep.2020.102927
Vakulskas, C. A., Dever, D. P., Rettig, G. R., Turk, R., Jacobi, A. M., Collingwood, M. A., et al. (2018). A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells. Nat. Med. 24, 1216–1224. doi: 10.1038/s41591-018-0137-0
Valvezan, A. J., Huang, J., Lengner, C. J., Pack, M., and Klein, P. S. (2014). Oncogenic mutations in adenomatous polyposis coli (Apc) activate mechanistic target of rapamycin complex 1 (mTORC1) in mice and zebrafish. Dis. Model. Mech. 7, 63–71. doi: 10.1242/dmm.012625
Valvezan, A. J., Zhang, F., Diehl, J. A., and Klein, P. S. (2012). Adenomatous polyposis coli (APC) regulates multiple signaling pathways by enhancing glycogen synthase kinase-3 (GSK-3) activity. J. Biol. Chem. 287, 3823–3832. doi: 10.1074/jbc.M111.323337
Van Der Velden, Y. U., Wang, L., Zevenhoven, J., Van Rooijen, E., Van Lohuizen, M., Giles, R. H., et al. (2011). The serine-threonine kinase LKB1 is essential for survival under energetic stress in zebrafish. Proc. Natl. Acad. Sci. U. S. A. 108, 4358–4363. doi: 10.1073/pnas.1010210108
Vander Heiden, M. G., Cantley, L. C., and Thompson, C. B. (2009). Understanding the warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033. doi: 10.1126/science.1160809
Venkatasubramani, N., and Mayer, A. N. (2008). A zebrafish model for the Shwachman-Diamond Syndrome (SDS). Pediatr. Res. 63, 348–352. doi: 10.1203/PDR.0b013e3181659736
Vierstraete, J., Willaert, A., Vermassen, P., Coucke, P. J., Vral, A., and Claes, K. B. M. (2017). Accurate quantification of homologous recombination in zebrafish: Brca2 deficiency as a paradigm. Sci. Rep. 7:16518. doi: 10.1038/s41598-017-16725-3
Vogeli, K. M., Jin, S.-W., Martin, G. R., and Stainier, D. Y. R. (2006). A common progenitor for haematopoietic and endothelial lineages in the zebrafish gastrula. Nature 443, 337–339. doi: 10.1038/nature05045
Wang, H., Xu, J., Lazarovici, P., Quirion, R., and Zheng, W. (2018). cAMP Response element-binding protein (CREB): a possible signaling molecule link in the pathophysiology of Schizophrenia. Front. Mol. Neurosci. 11:225. doi: 10.3389/fnmol.2018.00255
Warren, A. J. (2018). Molecular basis of the human ribosomopathy Shwachman-Diamond syndrome. Adv. Biol. Regul. 67, 109–127. doi: 10.1016/j.jbior.2017.09.002
Weber, A. M., and Ryan, A. J. (2015). ATM and ATR as therapeutic targets in cancer. Pharmacol. Ther. 149, 124–138. doi: 10.1016/j.pharmthera.2014.12.001
White, R. M., Sessa, A., Burke, C., Bowman, T., LeBlanc, J., Ceol, C., et al. (2008). Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell 2, 183–189. doi: 10.1016/j.stem.2007.11.002
Wierson, W. A., Welker, J. M., Almeida, M. P., Mann, C. M., Webster, D. A., Torrie, M. E., et al. (2020). Efficient targeted integration directed by short homology in zebrafish and mammalian cells. Elife 9:e53968. doi: 10.7554/eLife.53968
Wingo, S. N., Gallardo, T. D., Akbay, E. A., Liang, M.-C., Contreras, C. M., Boren, T., et al. (2009). Somatic LKB1 mutations promote cervical cancer progression. PLoS One 4:e5137. doi: 10.1371/journal.pone.0005137
Wlodarski, M. W., Collin, M., and Horwitz, M. S. (2017). GATA2 deficiency and related myeloid neoplasms. Semin. Hematol. 54, 81–86. doi: 10.1053/j.seminhematol.2017.05.002
Wlodarski, M. W., Hirabayashi, S., Pastor, V., Starý, J., Hasle, H., Masetti, R., et al. (2016). Prevalence, clinical characteristics, and prognosis of GATA2-related myelodysplastic syndromes in children and adolescents. Blood 127, 1387–1397. doi: 10.1182/blood-2015-09-669937
Wolman, M. A., deGroh, E. D., McBride, S. M., Jongens, T. A., Granato, M., and Epstein, J. A. (2014). Modulation of cAMP and ras signaling pathways improves distinct behavioral deficits in a zebrafish model of neurofibromatosis type 1. Cell Rep. 8, 1265–1270. doi: 10.1016/j.celrep.2014.07.054
Yamaguchi, M., Fujimori-Tonou, N., Yoshimura, Y., Kishi, T., Okamoto, H., and Masai, I. (2008). Mutation of DNA primase causes extensive apoptosis of retinal neurons through the activation of DNA damage checkpoint and tumor suppressor p53. Development 135, 1247–1257. doi: 10.1242/dev.011015
Yang, W., Klaman, L. D., Chen, B., Araki, T., Harada, H., Thomas, S. M., et al. (2006). An Shp2/SFK/Ras/Erk signaling pathway controls trophoblast stem cell survival. Dev. Cell 10, 317–327. doi: 10.1016/j.devcel.2006.01.002
Yuan, H., Wen, B., Liu, X., Gao, C., Yang, R., Wang, L., et al. (2015). CCAAT/enhancer-binding protein α is required for hepatic outgrowth via the p53 pathway in zebrafish. Sci. Rep. 5:15838. doi: 10.1038/srep15838
Yuan, H., Zhou, J., Deng, M., Zhang, Y., Chen, Y., Jin, Y., et al. (2011). Sumoylation of CCAAT/enhancer-binding protein α promotes the biased primitive hematopoiesis of zebrafish. Blood 117, 7014–7020. doi: 10.1182/blood-2010-12-325712
Zhang, J., Walsh, M. F., Wu, G., Edmonson, M. N., Gruber, T. A., Easton, J., et al. (2015). Germline mutations in predisposition genes in pediatric cancer. N. Engl. J. Med. 373, 2336–2346. doi: 10.1056/NEJMoa1508054
Zhang, Y., Gao, X., Saucedo, L. J., Ru, B., Edgar, B. A., and Pan, D. (2003). Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat. Cell Biol. 5, 578–581. doi: 10.1038/ncb999
Zhang, Y., Morimoto, K., Danilova, N., Zhang, B., and Lin, S. (2012). Zebrafish models for dyskeratosis congenita reveal critical roles of p53 activation contributing to hematopoietic defects through RNA processing. PLoS One 7:e30188. doi: 10.1371/journal.pone.0030188
Zhu, Y. (2002). Neurofibromas in NF1: schwann cell origin and role of tumor environment. Science 296, 920–922. doi: 10.1126/science.1068452
Keywords: zebrafish, cancer predisposition, genetic models, cancer, model organism, p53
Citation: Kobar K, Collett K, Prykhozhij SV and Berman JN (2021) Zebrafish Cancer Predisposition Models. Front. Cell Dev. Biol. 9:660069. doi: 10.3389/fcell.2021.660069
Received: 28 January 2021; Accepted: 23 March 2021;
Published: 27 April 2021.
Edited by:
Yasuhito Shimada, Mie University, JapanReviewed by:
Caterina Sturtzel, St. Anna Children’s Cancer Research Institute (CCRI), AustriaYasuyuki Hosono, Aichi Cancer Center, Japan
Shuning He, Dana–Farber Cancer Institute, United States
Copyright © 2021 Kobar, Collett, Prykhozhij and Berman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jason Berman, amJlcm1hbkBjaGVvLm9uLmNh
†These authors have contributed equally to this work
 Kim Kobar
Kim Kobar Keon Collett1†
Keon Collett1†