- 1International Institutes of Medicine, The Fourth Affiliated Hospital of Zhejiang University School of Medicine, Yiwu, China
- 2Key Laboratory of Respiratory Disease of Zhejiang Province, Department of Pharmacology and Department of Respiratory and Critical Care Medicine of the Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, China
MUS81 complex, exhibiting endonuclease activity on specific DNA structures, plays an influential part in DNA repair. Research has proved that MUS81 is dispensable for embryonic development and cell viability in mammals. However, an intricate picture has emerged from studies in which discrepant gene mutations completely alter the role of MUS81 in human cancers. Here, we review the recent understanding of how MUS81 functions in tumors with distinct genetic backgrounds and discuss the potential therapeutic strategies targeting MUS81 in cancer.
Introduction
Mus81 was first identified by its cooperation with the homologous recombination (HR) protein Rad54 in yeast (Boddy et al., 2000; Interthal and Heyer, 2000; Haber and Heyer, 2001), indicating its possible role in DNA repair. Owing to its high evolutionary conservation, human MUS81 was subsequently discovered (Chen et al., 2001). The MUS81 protein possesses a characteristic ERCC4 nuclease domain containing the VERKX3D motif, which is indispensable for the endonuclease activity (Chen et al., 2001). The abundance of human MUS81 augments unequivocally when cells are exposed to replication stress (Chen et al., 2001). Human MUS81 localizes to damaged DNA sites under replication stress (Gao et al., 2003). MUS81-depleted U2OS cells exhibit elevated level of chromosomal bridges and micronuclei, characteristic features of DNA damage (Ying et al., 2013). MUS81-deficient cells and mice are intolerant to mitomycin C (MMC), an interstrand crosslinking agent, while MUS81 haploinsufficiency results in genomic instability (McPherson et al., 2004). Altogether, mammalian MUS81 is unambiguously a DNA damage repair protein.
DNA damage response (DDR) defects are common occurrences in multiple cancers, manifesting as mutation and inactivation of DDR-relative proteins. In recent findings, it is becoming increasingly evident that MUS81 has close relationships with cancers. Interestingly, MUS81 is helpful for tumor survival in some cases but lethal to them in other cases. For instance, endonuclease activity of MUS81 is crucial for survival of BRCA2-insufficient cancer cells (Lai et al., 2017; Lemaçon et al., 2017). Nevertheless, in WRN-depleted microsatellite instability (MSI) cancer cells, MUS81 complex shatters chromosomes, which causes apoptosis of cancer cells (van Wietmarschen et al., 2020).
Herein, this review focuses on recent understanding of how MUS81 works in distinct cancer cells and discuss why MUS81 causes such different consequences as black and white in tumors.
Mechanisms of MUS81 in DNA Repair
Molecular Mechanism of Substrate Recognition and Cleavage by MUS81
Human MUS81 is a substrate selective endonuclease and exhibits a wide range of specificity for replication forks, 3′-flap structures, Holliday junctions (HJs), and D-loops (Chen et al., 2001; Constantinou et al., 2002; Zeng et al., 2009).
The crystal structure of human MUS81–essential meiotic structure-specific endonuclease 1 (MUS81–EME1) combined with 3′ flaps demonstrates the recognition and cleavage mechanism of MUS81 complex (Chang et al., 2008; Gwon et al., 2014). Binding of 3′-flap DNA induces rotation of the helix–hairpin–helix (HhH2) heterodimer of the MUS81–EME1 complex. The disordered loop of the EME1 linker becomes ordered, which subsequently unmasks the hydrophobic wedge and forms the 5′ end binding pocket of MUS81 (Gwon et al., 2014). The binding pocket accommodates the 5′ nicked end of the 3′ flap DNA, which is pivotal for the substrate specificity (Tsodikov et al., 2005; Chang et al., 2008). HhH2 of EME1 and HhH2 of MUS81 are able to interact with the pre- and the postnick DNA strands, respectively, as a result of the conformational changes (Gwon et al., 2014). The wedge bends the substrate, which put the 3′ end into the ERCC4 site of MUS81 and assists the substrate cleavage (Enzlin and Schärer, 2002; Gwon et al., 2014).
MUS81–EME2 Promotes DNA Replication Completion in S Phase
Replication forks are prone to stall when encountering obstacles during DNA replication (Liao et al., 2018). The recovery of the replication forks is necessary for faithful DNA replication and cell division (Bryant et al., 2009). MUS81–EME2 can cleave reversed replication forks (Amangyeld et al., 2014) and D-loops (Pepe and West, 2014b) and is responsible for the restart of stalled forks in S phase (Gao et al., 2003; Pepe and West, 2014a). When replication forks are stalled in S phase, MUS81–EME2 complex is recruited to cleave the stalled replication forks and induces transient DNA double-strand break (DSB) formation (Gao et al., 2003; Pepe and West, 2014a). The cleavage of MUS81–EME2 restarts the stalled replication forks and promotes replication recovery (Hanada et al., 2007) via break-induced replication (BIR) pathway (Kramara et al., 2018).
MUS81–EME1 Promotes Faithful Chromosome Disjunction in M Phase
MUS81–EME1 Complex Triggers Common Fragile Site Expression
Common fragile sites (CFSs) are difficult-to-replicate foci that preferentially form breaks in chromosomes under replication stress and frequently rearrange in tumor cells (Durkin and Glover, 2007). We revealed that MUS81–EME1 complex promotes expression of CFSs and faithful disjunction of sister chromosomes in human cells (Naim et al., 2013; Ying et al., 2013). Under replication inhibitors, CFSs remain underreplicated at the end of S phase, are prone to form replication intermediates, and then give rise to sister-chromatid bridging in M phase, which contributes to chromosomal instability and oncogenesis (Chan et al., 2009). Nonetheless, MUS81–EME1 complex is phosphorylated and collected to underreplicated CFS loci in early mitosis. RECQ5 dismantles RAD51 from the single-strand DNA (ssDNA), which promotes the cleavage of MUS81–EME1 on stalled replication forks (Di Marco et al., 2017). MUS81–EME1 then cleaves the intertwined DNA strands, manifesting as breaks observed at CFSs (Naim et al., 2013; Ying et al., 2013). Ultimately, we identified that POLD3-dependent DNA synthesis, promoted by MUS81 cleavage, repairs the expressed CFSs and boosts faithful sister chromatid disjunction (Minocherhomji et al., 2015).
SLX–MUS Complex Contributes to Holliday Junction Resolution
HJs are cruciform-shaped chromosome junctions that arise temporarily during HR, and their resolution is critical for chromosomal disjunction and genome maintenance (West, 2009). There are three major HJ-resolving pathways in human cells, one of which involves synthetic lethal of unknown function protein 1 (SLX1)–SLX4–MUS81–EME1 (SLX–MUS) complex, a backup for Bloom syndrome protein (BLM)–TopoisomeraseIIIa–RecQ-mediated genome instability protein 1 (RMI1)–RMI2 (BTRR) pathway (Fekairi et al., 2009; Wyatt et al., 2013; Sarbajna et al., 2014). Cells, lacking SLX–MUS complex-associated proteins, show defective chromosome morphology and reduced survival (Wyatt et al., 2013; Sarbajna et al., 2014). After SLX4 and EME1 are phosphorylated by cyclin-dependent kinase, MUS81–MEM1 and SLX1–SLX4 associate and combine into a stable SLX–MUS complex at the G2/M phase (Wyatt et al., 2013). At that point, SLX–MUS complex triggers bilateral cleavage of intact HJs by a coordinated nicking and counternicking mechanism (Wyatt et al., 2013). Interestingly, MUS81–EME1 cleaves intact HJs inefficiently (Ciccia et al., 2003), and SLX1–SLX4 introduces nicks into intact HJs optionally. However, SLX–MUS complex mobilizes cleavage activity of both SLX1–SLX4 and MUS81–EME1 and exhibits a more orchestrated reaction for efficient HJs resolution (Wyatt et al., 2013).
Relevance Between MUS81 and Cancer
Involvement of MUS81 in Tumor Suppression
As defects in DNA damage repair are frequently interrelated with high predisposition to cancer, research has been carried out to demonstrate whether MUS81 suppresses tumors in vivo. Mus81-deficient mice are born at expected Mendelian frequencies (McPherson et al., 2004), indicating a non-essential role of MUS81 in murine meiotic recombination and oncogenesis. However, MUS81 deficiency, even MUS81 insufficiency, leads to a dramatic susceptibility to cancers, especially lymphomas in mice through the first year of life (McPherson et al., 2004). These tumors show a high frequency of DNA aneuploid via cytogenetic analysis (McPherson et al., 2004). Strikingly, concomitant deficiency of MUS81 and P53 leads to an extremely high frequency in sarcoma development in mice, indicating the collaboration of MUS81 and P53 in tumor suppression (Pamidi et al., 2007). There is, however, a different voice suggesting that murine MUS81 is unnecessary for tumor suppression, based on evidence that no increased sign of tumors was detected in MUS81-deficient mice during a 15-month monitoring (Dendouga et al., 2005). The reason for the discrepant performance of murine MUS81 in tumor predisposition is still unclear. Thus, determining the role of MUS81 in tumor suppression requires further studies. Nonetheless, a growing number of evidence prove that MUS81 plays a dominant role in some specific gene-mutated cancer cells.
MUS81 Is Essential in BRCA2-Deficient Cancer Cells
Inheritance of BRCA2 mutations is responsible for predisposing humans to breast cancer (Ford et al., 1998). Primary human cells with BRCA2 deletion accumulate spontaneous DNA damage and go toward senescence and apoptosis (Carlos et al., 2013). Mice with homozygous BRCA2 mutation show embryonic lethality (Jonkers et al., 2001). However, BRCA2-deficient cancer cells can survive in virtue of high tolerance of endogenous DNA damage (Pardo et al., 2020). Dual loss of BRCA2 and MUS81 results in obvious cancer cell death (Lai et al., 2017), indicating a pivotal role of MUS81 in BRCA2-mutated cancer cells (Figure 1A). Replication forks stall when they encounter DNA lesions upon drug treatment, and RAD51 promotes reversed fork formation (Zellweger et al., 2015; Lemaçon et al., 2017). Subsequently, BRCA2 is recruited to protect the regressed arms of nascent DNA strands at stalled forks by stabilizing RAD51 filaments from nucleolytic degradation (Schlacher et al., 2011; Lemaçon et al., 2017; Mijic et al., 2017). In the lack of BRCA2, MRE11, initiated by CtIP, targets unprotected reversed forks and starts fork resection by its endonuclease activity (Lemaçon et al., 2017). MRE11 resection leads to the ssDNA flap formation in reversed forks (Lemaçon et al., 2017). MUS81 cleaves these resected regressed forks and leads to transient DSB accumulation (Lemaçon et al., 2017). Finally, POLD3-dependent DNA synthesis repairs DSBs and restarts the MUS81-cleaved forks (Lemaçon et al., 2017). Conversely, in cancer cells with dual loss of BRCA2 and MUS81, resected forks cannot be restarted, which is supported by the observation that frequency of reversed forks with ssDNA is increased dramatically (Lemaçon et al., 2017).
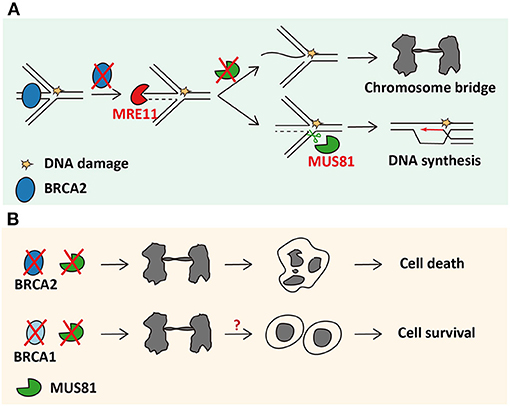
Figure 1. MUS81 sustains survival of BRCA2-deficient cancer cells through rescuing reversed forks. (A) The mechanism of MUS81 resolving chromosomal interlinks in cells lacking BRCA2. (B) MUS81 is dispensable in BRCA1-deficient cells.
Given that BRCA2 is best known for its function in HR repair, whether the synthetic lethality between MUS81 and BRCA2 can be stretched to other proteins involved in HR pathway, like BRCA1, was further explored. Surprisingly, BRCA1, RAD51, or RAD51C depletion, unlike BRCA2 depletion, cannot lead to synthetic lethality upon concomitant loss of MUS81 in cancer cells (Lai et al., 2017). BRCA1 functions upstream of BRCA2 in a common HR pathway (Roy et al., 2011) and is capable of protecting the reversed forks as BRCA2 does (Lemaçon et al., 2017). Nevertheless, MUS81 and POLD3 foci do not accumulate in BRCA1-insufficient cancer cells (Lemaçon et al., 2017). It would be interesting to uncover the mechanism by which MUS81 insufficiency leads to such different phenotypes between BRCA1- and BRCA2-deficient cancer cells (Figure 1).
MUS81 Induces Chromosome Shattering in WRN-Deficient MSI Cancer Cells
MSI is characterized by a hypermutable state of nucleotide repeat regions, which is promoted by defects in DNA mismatch repair (MMR) (Kim et al., 2013). MSI facilitates occurrence of multiple cancers (Pal et al., 2008; Kim et al., 2013). Considering the vulnerabilities of MSI, WRN is identified as an essential gene in MSI cells (Chan et al., 2019). WRN depletion causes DSBs and chromosome shattering (van Wietmarschen et al., 2020) and reduces the cell viability (Chan et al., 2019) in MSI cells but neither in microsatellite stable (MSS) cancer cells nor in primary human cells. However, MUS81 exhaustion before WRN depletion notably cuts down the chromosome shattering and DSBs formation at TA repeats (van Wietmarschen et al., 2020), indicating that MUS81 cleavage contributes to apoptosis of MSI cells with WRN deficiency (Figure 2).
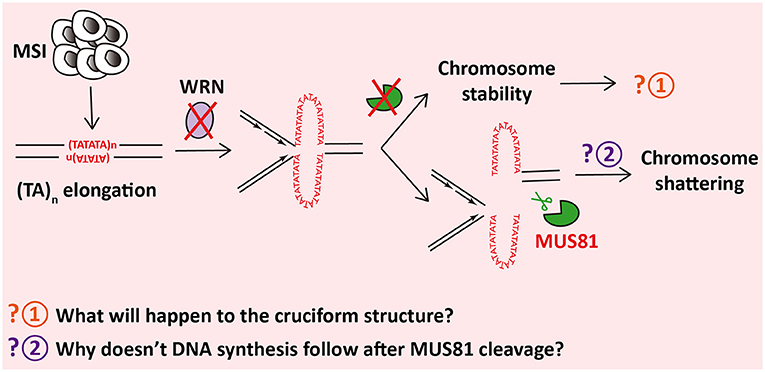
Figure 2. MUS81 shatters chromosomes in WRN-insufficient microsatellite instability (MSI) cells by cleaving (TA)n-formed cruciform structures.
In MSI cells, TA repeats, which are highly unstable, encounter and accumulate large-scale expansions for some reason (van Wietmarschen et al., 2020). Cruciform structures form at the expanded TA repeats (Inagaki et al., 2009), which are tended to stall replication forks. WRN is then recruited and unwinds the secondary structures in virtue of its helicase activity rather than its exonuclease activity, which allows restart of the stalled replication forks (Chan et al., 2019), whereas, in the absence of WRN, cruciform structures of TA repeats cannot be unfolded. Instead, MUS81–EME1 cleaves the cruciform structures and causes DSBs. Interestingly, the cleavage sites are exactly adjacent to the border of expanded TA repeats (van Wietmarschen et al., 2020). Furthermore, cleavage of MUS81 and accumulation of DSBs lead to massive chromosome shattering. Finally, WRN-deficient MSI cells ends in cell cycle arrest and apoptosis (van Wietmarschen et al., 2020).
A question, which remains to be answered, will be of considerable interest. Why does MUS81 cleavage in TA repeats lead to chromosome shattering rather than triggering POLD3-dependent DNA synthesis for DNA repair? It has been found that these TA repeats cannot be amplified in vitro and have extremely low sequencing depth (van Wietmarschen et al., 2020), indicating that secondary structures formed at TA repeats might restrain polymerase extension and inhibit DNA synthesis.
Discussion
DDR pathways play crucial roles in genomic stability, and compromised DDR pathways are common in multiple tumors. Recent studies have unveiled the link between DDR deficiency and innate/adaptive immunity against tumor cells (Barber, 2015; Reisländer et al., 2020). Cyclic GMP–AMP synthase (cGAS) detects the cytosolic DNA caused by DDR deficiency in cancers and activates stimulator of interferon genes (STING), which triggers interferons (IFNs) signaling and antitumor immunity (Chen et al., 2016). DNA mismatch repair (MMR) inactivation-induced neoantigens in cancers boost adaptive immunity as well, which is independent of the cGAS–STING pathway (Germano et al., 2017). Therefore, targeting DDR pathways holds therapeutic potential against cancers.
DDR-directed therapies have been introduced in clinical trials recently (Pilié et al., 2019). However, chromosomal instability (CIN) is the dominant drawback of this strategy, which conversely underpins evolution and growth of tumor cells (Bakhoum and Cantley, 2018; Calzetta et al., 2020). Thus, knowledge of the interactions among multiple DDR pathways and understanding the discrepant functions of DDR-relevant proteins in various cancers contribute to efficient cancer therapy and needs to be further elucidated.
As this review demonstrates, human MUS81 is closely related to cancers but results in totally different fates of cancers bearing distinct mutated genes, indicating that the interactions between MUS81 and other DDR proteins master the cancer cell fate and MUS81 is a potential and intriguing target in specific cancer therapies. A further understanding of the precise mechanisms of MUS81 and the interactions among DDR pathways in discrepant cancers may pave the way toward improved cancer therapeutic strategies.
Author Contributions
SC wrote the manuscript. XG, MS, ZH, CZ, and SY provided guidance and supervised the final version of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the Ministry of Science and Technology of the People's Republic of China (2016YFA0100301), the Natural Science Foundation of China (81870007, 81920108001), the Zhejiang Provincial Natural Science Foundation (LD19H160001), and the Zhejiang Provincial Program for the Cultivation of High-Level Innovative Health Talents (2016-63).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Amangyeld, T., Shin, Y. K., Lee, M., Kwon, B., and Seo, Y. S. (2014). Human MUS81-EME2 can cleave a variety of DNA structures including intact Holliday junction and nicked duplex. Nucleic Acids Res. 42, 5846–5862. doi: 10.1093/nar/gku237
Bakhoum, S. F., and Cantley, L. C. (2018). The multifaceted role of chromosomal instability in cancer and its microenvironment. Cell 174, 1347–1360. doi: 10.1016/j.cell.2018.08.027
Barber, G. N. (2015). STING: infection, inflammation, and cancer. Nat. Rev. Immunol. 15, 760–770. doi: 10.1038/nri3921
Boddy, M. N., Lopez Girona, A., Shanahan, P., Interthal, H., Heyer, W. D., and Russell, P. (2000). Damage tolerance protein Mus81 associates with the FHA1 domain of checkpoint kinase Cds1. Mol. Cell. Biol. 20, 8758–8766. doi: 10.1128/MCB.20.23.8758-8766.2000
Bryant, H. E., Petermann, E., Schultz, N., Jemth, A. S., Loseva, O., Issaeva, N., et al. (2009). PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J. 28, 2601–2615. doi: 10.1038/emboj.2009.206
Calzetta, N. L., Besteiro, M. A. G., and Gottifredi, V. (2020). Mus81-Eme1-dependent aberrant processing of DNA replication intermediates in mitosis impairs genome integrity. Sci. Adv. 6:eabc8257. doi: 10.1126/sciadv.abc8257
Carlos, A. R., Escandell, J. M., Kotsantis, P., Suwaki, N., Bouwman, P., Badie, S., et al. (2013). ARF triggers senescence in Brca2-deficient cells by altering the spectrum of p53 transcriptional targets. Nat. Commun. 4:2697. doi: 10.1038/ncomms3697
Chan, E. M., Shibue, T., McFarland, J. M., Gaeta, B., Ghandi, M., Dumont, N., et al. (2019). WRN helicase is a synthetic lethal target in microsatellite unstable cancers. Nature 568, 551–556. doi: 10.1038/s41586-019-1102-x
Chan, K. L., Palmai Pallag, T., Ying, S., and Hickson, I. D. (2009). Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat. Cell Biol. 11, 753–760. doi: 10.1038/ncb1882
Chang, J. H., Kim, J. J., Choi, J. M., Lee, J. H., and Cho, Y. (2008). Crystal structure of the Mus81-Eme1 complex. Genes Dev. 22, 1093–1106. doi: 10.1101/gad.1618708
Chen, Q., Sun, L., and Chen, Z. J. (2016). Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat. Immunol. 17, 1142–1149. doi: 10.1038/ni.3558
Chen, X. B., Melchionna, R., Denis, C. M., Gaillard, P. H. L., Blasina, A., Van de Weyer, I., et al. (2001). Human Mus81-associated endonuclease cleaves Holliday junctions in vitro. Mol. Cell 8, 1117–1127. doi: 10.1016/S1097-2765(01)00375-6
Ciccia, A., Constantinou, A., and West, S. C. (2003). Identification and characterization of the human mus81-eme1 endonuclease. J. Biol. Chem. 278, 25172–25178. doi: 10.1074/jbc.M302882200
Constantinou, A., Chen, X. B., McGowan, C. H., and West, S. C. (2002). Holliday junction resolution in human cells: two junction endonucleases with distinct substrate specificities. EMBO J. 21, 5577–5585. doi: 10.1093/emboj/cdf554
Dendouga, N., Gao, H., Moechars, D., Janicot, M., Vialard, J., and McGowan, C. H. (2005). Disruption of murine Mus81 increases genomic instability and DNA damage sensitivity but does not promote tumorigenesis. Mol. Cell. Biol. 25, 7569–7579. doi: 10.1128/MCB.25.17.7569-7579.2005
Di Marco, S., Hasanova, Z., Kanagaraj, R., Chappidi, N., Altmannova, V., Menon, S., et al. (2017). RECQ5 helicase cooperates with MUS81 endonuclease in processing stalled replication forks at common fragile sites during mitosis. Mol. Cell 66, 658–671. doi: 10.1016/j.molcel.2017.05.006
Durkin, S. G., and Glover, T. W. (2007). Chromosome fragile sites. Annu. Rev. Genet. 41, 169–192. doi: 10.1146/annurev.genet.41.042007.165900
Enzlin, J. H., and Schärer, O. D. (2002). The active site of the DNA repair endonuclease XPF-ERCC1 forms a highly conserved nuclease motif. EMBO J. 21, 2045–2053. doi: 10.1093/emboj/21.8.2045
Fekairi, S., Scaglione, S., Chahwan, C., Taylor, E. R., Tissier, A., Coulon, S., et al. (2009). Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell 138, 78–89. doi: 10.1016/j.cell.2009.06.029
Ford, D., Easton, D. F., Stratton, M., Narod, S., Goldgar, D., Devilee, P., et al. (1998). Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. The breast cancer linkage consortium. Am. J. Hum. Genet. 62, 676–689. doi: 10.1086/301749
Gao, H., Chen, X. B., and McGowan, C. H. (2003). Mus81 endonuclease localizes to nucleoli and to regions of DNA damage in human S-phase cells. Mol. Biol. Cell 14, 4826–4834. doi: 10.1091/mbc.e03-05-0276
Germano, G., Lamba, S., Rospo, G., Barault, L., Magr,ì, A., Maione, F., et al. (2017). Inactivation of DNA repair triggers neoantigen generation and impairs tumour growth. Nature 552, 116–120. doi: 10.1038/nature24673
Gwon, G. H., Jo, A., Baek, K., Jin, K. S., Fu, Y., Lee, J.-B., et al. (2014). Crystal structures of the structure-selective nuclease Mus81-Eme1 bound to flap DNA substrates. EMBO J. 33, 1061–1072. doi: 10.1002/embj.201487820
Haber, J. E., and Heyer, W. D. (2001). The fuss about Mus81. Cell 107, 551–554. doi: 10.1016/S0092-8674(01)00593-1
Hanada, K., Budzowska, M., Davies, S. L., van Drunen, E., Onizawa, H., Beverloo, H. B., et al. (2007). The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nat. Struct. Mol. Biol. 14, 1096–1104. doi: 10.1038/nsmb1313
Inagaki, H., Ohye, T., Kogo, H., Kato, T., Bolor, H., Taniguchi, M., et al. (2009). Chromosomal instability mediated by non-B DNA: cruciform conformation and not DNA sequence is responsible for recurrent translocation in humans. Genome Res. 19, 191–198. doi: 10.1101/gr.079244.108
Interthal, H., and Heyer, W. D. (2000). MUS81 encodes a novel helix-hairpin-helix protein involved in the response to UV- and methylation-induced DNA damage in Saccharomyces cerevisiae. Mol. Gen. Genet. 263, 812–827. doi: 10.1007/s004380000241
Jonkers, J., Meuwissen, R., van der Gulden, H., Peterse, H., van der Valk, M., and Berns, A. (2001). Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat. Genet. 29, 418–425. doi: 10.1038/ng747
Kim, T. M., Laird, P. W., and Park, P. J. (2013). The landscape of microsatellite instability in colorectal and endometrial cancer genomes. Cell 155, 858–868. doi: 10.1016/j.cell.2013.10.015
Kramara, J., Osia, B., and Malkova, A. (2018). Break-induced replication: the where, the why, and the how. Trends Genet. 34, 518–531. doi: 10.1016/j.tig.2018.04.002
Lai, X., Broderick, R., Bergoglio, V., Zimmer, J., Badie, S., Niedzwiedz, W., et al. (2017). MUS81 nuclease activity is essential for replication stress tolerance and chromosome segregation in BRCA2-deficient cells. Nat. Commun. 8:15983. doi: 10.1038/ncomms16171
Lemaçon, D., Jackson, J., Quinet, A., Brickner, J. R., Li, S., Yazinski, S., et al. (2017). MRE11 and EXO1 nucleases degrade reversed forks and elicit MUS81-dependent fork rescue in BRCA2-deficient cells. Nat. Commun. 8:860. doi: 10.1038/s41467-017-01180-5
Liao, H., Ji, F., Helleday, T., and Ying, S. (2018). Mechanisms for stalled replication fork stabilization: new targets for synthetic lethality strategies in cancer treatments. EMBO Rep. 19:e46263. doi: 10.15252/embr.201846263
McPherson, J. P., Lemmers, B., Chahwan, R., Pamidi, A., Migon, E., Matysiak Zablocki, E., et al. (2004). Involvement of mammalian Mus81 in genome integrity and tumor suppression. Science (N. Y.) 304, 1822–1826. doi: 10.1126/science.1094557
Mijic, S., Zellweger, R., Chappidi, N., Berti, M., Jacobs, K., Mutreja, K., et al. (2017). Replication fork reversal triggers fork degradation in BRCA2-defective cells. Nat. Commun. 8:859. doi: 10.1038/s41467-017-01164-5
Minocherhomji, S., Ying, S., Bjerregaard, V. A., Bursomanno, S., Aleliunaite, A., Wu, W., et al. (2015). Replication stress activates DNA repair synthesis in mitosis. Nature 528, 286–290. doi: 10.1038/nature16139
Naim, V., Wilhelm, T., Debatisse, M., and Rosselli, F. (2013). ERCC1 and MUS81-EME1 promote sister chromatid separation by processing late replication intermediates at common fragile sites during mitosis. Nat. Cell Biol. 15, 1008–1015. doi: 10.1038/ncb2793
Pal, T., Permuth Wey, J., Kumar, A., and Sellers, T. A. (2008). Systematic review and meta-analysis of ovarian cancers: estimation of microsatellite-high frequency and characterization of mismatch repair deficient tumor histology. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 14, 6847–6854. doi: 10.1158/1078-0432.CCR-08-1387
Pamidi, A., Cardoso, R., Hakem, A., Matysiak Zablocki, E., Poonepalli, A., Tamblyn, L., et al. (2007). Functional interplay of p53 and Mus81 in DNA damage responses and cancer. Cancer Res. 67, 8527–8535. doi: 10.1158/0008-5472.CAN-07-1161
Pardo, B., Moriel-Carretero, M., Vicat, T., Aguilera, A., and Pasero, P. (2020). Homologous recombination and Mus81 promote replication completion in response to replication fork blockage. EMBO Rep. 21:e49367. doi: 10.15252/embr.201949367
Pepe, A., and West, S. C. (2014a). MUS81-EME2 promotes replication fork restart. Cell Rep. 7, 1048–1055. doi: 10.1016/j.celrep.2014.04.007
Pepe, A., and West, S. C. (2014b). Substrate specificity of the MUS81-EME2 structure selective endonuclease. Nucleic Acids Res. 42, 3833–3845. doi: 10.1093/nar/gkt1333
Pilié, P. G., Tang, C., Mills, G. B., and Yap, T. A. (2019). State-of-the-art strategies for targeting the DNA damage response in cancer. Nat. Rev. Clin. Oncol. 16, 81–104. doi: 10.1038/s41571-018-0114-z
Reisländer, T., Groelly, F. J., and Tarsounas, M. (2020). DNA Damage and Cancer Immunotherapy: a STING in the Tale. Mol. Cell 80, 21–28. doi: 10.1016/j.molcel.2020.07.026
Roy, R., Chun, J., and Powell, S. N. (2011). BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat. Rev. Cancer 12, 68–78. doi: 10.1038/nrc3181
Sarbajna, S., Davies, D., and West, S. C. (2014). Roles of SLX1-SLX4, MUS81-EME1, and GEN1 in avoiding genome instability and mitotic catastrophe. Genes Dev. 28, 1124–1136. doi: 10.1101/gad.238303.114
Schlacher, K., Christ, N., Siaud, N., Egashira, A., Wu, H., and Jasin, M. (2011). Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell 145, 529–542. doi: 10.1016/j.cell.2011.03.041
Tsodikov, O. V., Enzlin, J. H., Schärer, O. D., and Ellenberger, T. (2005). Crystal structure and DNA binding functions of ERCC1, a subunit of the DNA structure-specific endonuclease XPF-ERCC1. Proc. Natl. Acad. Sci. U.S.A. 102, 11236–11241. doi: 10.1073/pnas.0504341102
van Wietmarschen, N., Sridharan, S., Nathan, W. J., Tubbs, A., Chan, E. M., Callen, E., et al. (2020). Repeat expansions confer WRN dependence in microsatellite-unstable cancers. Nature 586, 292–298. doi: 10.1038/s41586-020-2769-8
West, S. C. (2009). The search for a human Holliday junction resolvase. Biochem. Soc. Trans. 37, 519–526. doi: 10.1042/BST0370519
Wyatt, H. D. M., Sarbajna, S., Matos, J., and West, S. C. (2013). Coordinated actions of SLX1-SLX4 and MUS81-EME1 for Holliday junction resolution in human cells. Mol. Cell 52, 234–247. doi: 10.1016/j.molcel.2013.08.035
Ying, S., Minocherhomji, S., Chan, K. L., Palmai Pallag, T., Chu, W. K., Wass, T., et al. (2013). MUS81 promotes common fragile site expression. Nat. Cell Biol. 15, 1001–1007. doi: 10.1038/ncb2773
Zellweger, R., Dalcher, D., Mutreja, K., Berti, M., Schmid, J. A., Herrador, R., et al. (2015). Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells. J. Cell Biol. 208, 563–579. doi: 10.1083/jcb.201406099
Keywords: human MUS81, endonuclease, DNA damage response, cancer therapy, chromosomal instability
Citation: Chen S, Geng X, Syeda MZ, Huang Z, Zhang C and Ying S (2021) Human MUS81: A Fence-Sitter in Cancer. Front. Cell Dev. Biol. 9:657305. doi: 10.3389/fcell.2021.657305
Received: 22 January 2021; Accepted: 10 February 2021;
Published: 15 March 2021.
Edited by:
Yunzhou Dong, Boston Children's Hospital and Harvard Medical School, United StatesReviewed by:
Yanbin Zhang, University of Miami, United StatesJianjun Wu, University of Texas Southwestern Medical Center, United States
Copyright © 2021 Chen, Geng, Syeda, Huang, Zhang and Ying. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Songmin Ying, eWluZ3NAemp1LmVkdS5jbg==
 Sisi Chen
Sisi Chen Xinwei Geng
Xinwei Geng Madiha Zahra Syeda
Madiha Zahra Syeda Zhengming Huang2
Zhengming Huang2 Songmin Ying
Songmin Ying