- 1Division of Developmental Biology, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, United States
- 2Division of Plastic Surgery, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, United States
- 3Department of Surgery, University of Cincinnati College of Medicine, Cincinnati, OH, United States
- 4Department of Pediatrics, University of Cincinnati College of Medicine, Cincinnati, OH, United States
- 5Division of Biomedical Informatics, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, United States
Proper development of tendons is crucial for the integration and function of the musculoskeletal system. Currently little is known about the molecular mechanisms controlling tendon development and tendon cell differentiation. The transcription factor Scleraxis (Scx) is expressed throughout tendon development and plays essential roles in both embryonic tendon development and adult tendon healing, but few direct target genes of Scx in tendon development have been reported and genome-wide identification of Scx direct target genes in vivo has been lacking. In this study, we have generated a ScxFlag knockin mouse strain, which produces fully functional endogenous Scx proteins containing a 2xFLAG epitope tag at the carboxy terminus. We mapped the genome-wide Scx binding sites in the developing limb tendon tissues, identifying 12,097 high quality Scx regulatory cis-elements in-around 7,520 genes. Comparative analysis with previously reported embryonic tendon cell RNA-seq data identified 490 candidate Scx direct target genes in early tendon development. Furthermore, we characterized a new Scx gene-knockout mouse line and performed whole transcriptome RNA sequencing analysis of E15.5 forelimb tendon cells from Scx–/– embryos and control littermates, identifying 68 genes whose expression in the developing tendon tissues significantly depended on Scx function. Combined analysis of the ChIP-seq and RNA-seq data yielded 32 direct target genes that required Scx for activation and an additional 17 target genes whose expression was suppressed by Scx during early tendon development. We further analyzed and validated Scx-dependent tendon-specific expression patterns of a subset of the target genes, including Fmod, Kera, Htra3, Ssc5d, Tnmd, and Zfp185, by in situ hybridization and real-time quantitative polymerase chain reaction assays. These results provide novel insights into the molecular mechanisms mediating Scx function in tendon development and homeostasis. The ChIP-seq and RNA-seq data provide a rich resource for aiding design of further studies of the mechanisms regulating tendon cell differentiation and tendon tissue regeneration. The ScxFlag mice provide a valuable new tool for unraveling the molecular mechanisms involving Scx in the protein interaction and gene-regulatory networks underlying many developmental and disease processes.
Introduction
Tendons and ligaments are specialized connective tissues densely packed with collagen fibers, composed primarily of type I collagen fascicles with several other collagens, elastin, and various proteoglycans making up the remainder of the extracellular matrix (ECM) surrounding the resident tenocytes (Birch et al., 2013; Davis et al., 2013). Tendons connect skeletal muscles to bones and transmit mechanical forces generated from muscle contraction whereas the ligaments align bones within joints and reinforce their stability and flexibility. Many musculoskeletal diseases involve injuries and tissue degeneration in tendons and ligaments (Yang et al., 2013; Nourissat et al., 2015). Although it is expected that many genes and molecular pathways are shared during the processes of tendon development and tendon healing/regeneration, the molecular mechanisms controlling tendon development, including specification of tendon progenitor cells from mesenchymal stem cells, migration to local domains and differentiation into mature tenocytes, secretion of ECM molecules and organization of fibrils and fibers into tendon bundles, are not well understood. With the development and wide application of gene knockout technologies, thousands of mutant mouse lines carrying loss-of-function mutations in individual genes have been generated and analyzed in the last 30 years. However, only loss of function of the Scleraxis (Scx) transcription factor and TGFβ signaling led to severe disruption of tendon development whereas mice deficient in several other genes individually or in combination, including genes encoding the Mohawk (Mkx) transcription factor, the early growth response (EGR) 1 and 2 transcription factors, various proteoglycans, and tenomodulin, exhibited mild postnatal tendon defects (Docheva et al., 2005; Murchison et al., 2007; Kilts et al., 2009; Pryce et al., 2009; Ito et al., 2010; Liu et al., 2010; Lejard et al., 2011; Dourte et al., 2013; Guerquin et al., 2013; Dunkman et al., 2014; Delgado Caceres et al., 2018; Shukunami et al., 2018). While these data suggest that the tendon developmental processes are well orchestrated with built-in compensatory regulatory mechanisms (Delgado Caceres et al., 2018), better understanding of the molecular mechanisms controlling tendon development and tendon cell differentiation will be instrumental for the development of effective methods for tendon repair and regeneration.
Scleraxis is a basic helix-loop-helix (bHLH) transcription factor that is expressed in early embryonic tendon progenitor cells and stays strongly expressed throughout tendon cell differentiation into mature tenocytes (Cserjesi et al., 1995; Schweitzer et al., 2001). Mice homozygous for Scx null mutation exhibit severe hypoplasia or complete absence of force transmitting tendons, while the muscle anchoring tendons and ligaments are less affected (Murchison et al., 2007; Shukunami et al., 2018). Further studies showed that Scx function is not required for tendon progenitor cell initiation but is crucial for tendon cell differentiation (Murchison et al., 2007). Studies in mouse and chick embryos showed that TGFβ and FGF signaling pathways are essential for induction of tendon progenitor cells by activating expression of Scx and other tendon genes (Kuo et al., 2008; Pryce et al., 2009; Hasson, 2011; Havis et al., 2016). Moreover, a recent study demonstrated that Scx function is also required for adult tendons in response to mechanical loading (Gumucio et al., 2020). How Scx regulates tendon cell differentiation is still unclear. Currently very few genes, including Col1a1 and Tnmd, have been identified as potential direct target genes of Scx in tenocytes primarily through in vitro functional analysis of putative Scx-binding promoter elements (Shukunami et al., 2006, 2018; Lejard et al., 2007), whereas large scale identification of Scx direct transcriptional target genes during tendon cell differentiation in vivo is still lacking.
In this study, we first generated a novel ScxFlag knockin mouse line, which expresses the endogenous Scx protein with a 2xFLAG epitope tag at the carboxy terminus, through CRISPR/cas9-mediated genome editing. Using the ScxFlag mice, we mapped the genome-wide Scx binding sites in embryonic tendon progenitors and differentiating tendon cells using chromatin immunoprecipitation followed by high throughput DNA sequencing, which mapped high quality Scx-binding sites in the promoter and/or enhancer regions in 7,520 genes. Furthermore, we characterized a new Scx gene-knockout mouse resource that is accessible to the wide biomedical research community and performed whole transcriptome RNA sequencing analysis of early differentiating limb tendons in Scx–/– and wildtype littermates. These ChIP-seq and RNA-seq datasets provide a rich resource for further studies of the molecular mechanisms regulating tendon formation and tendon cell differentiation. Combined analyses of the ChIP-seq and RNA-seq data provide novel insights into the molecular mechanisms mediating Scx regulation of tendon cell differentiation.
Materials and Methods
Mice
Scx-GFP transgenic mice (Pryce et al., 2007) were generously provided by Dr. Ronen Schweitzer (Shriners Hospitals for Children, Portland, Oregon). To generate ScxFlag knock-in mice, pretested guide RNAs targeting the genomic sequence containing the endogenous Scx translational STOP codon were co-injected with a single-stranded oligonucleotide donor template (Figure 1A) and humanized Cas9 mRNAs into zygotes of B6D2F1 (C57BL/6 X DBA2) mice. Genome-modified founder mice were identified by using polymerase chain reaction (PCR) assay and crossed to CD-1 mice to generate ScxFlag/+ hemizygous mice. Genotypically verified G1 ScxFlag/+ hemizygous mice were intercrossed to generate ScxFlag/Flag homozygous mice. Scx+/– mutant mice were purchased from the Mutant Mouse Research and Resource Center at the University of California Davis (KOMP catalog#13478). All mouse strains were maintained by crossing to CD-1 wildtype mice (Charles River, Stock# 022), or by intercrossing between siblings. All animal procedures were approved by the Institutional Animal Care and Use Committee (IACUC) at Cincinnati Children’s Hospital Medical Center (CCHMC). The mice were housed in AAALAC accredited barrier housing conditions at CCHMC.
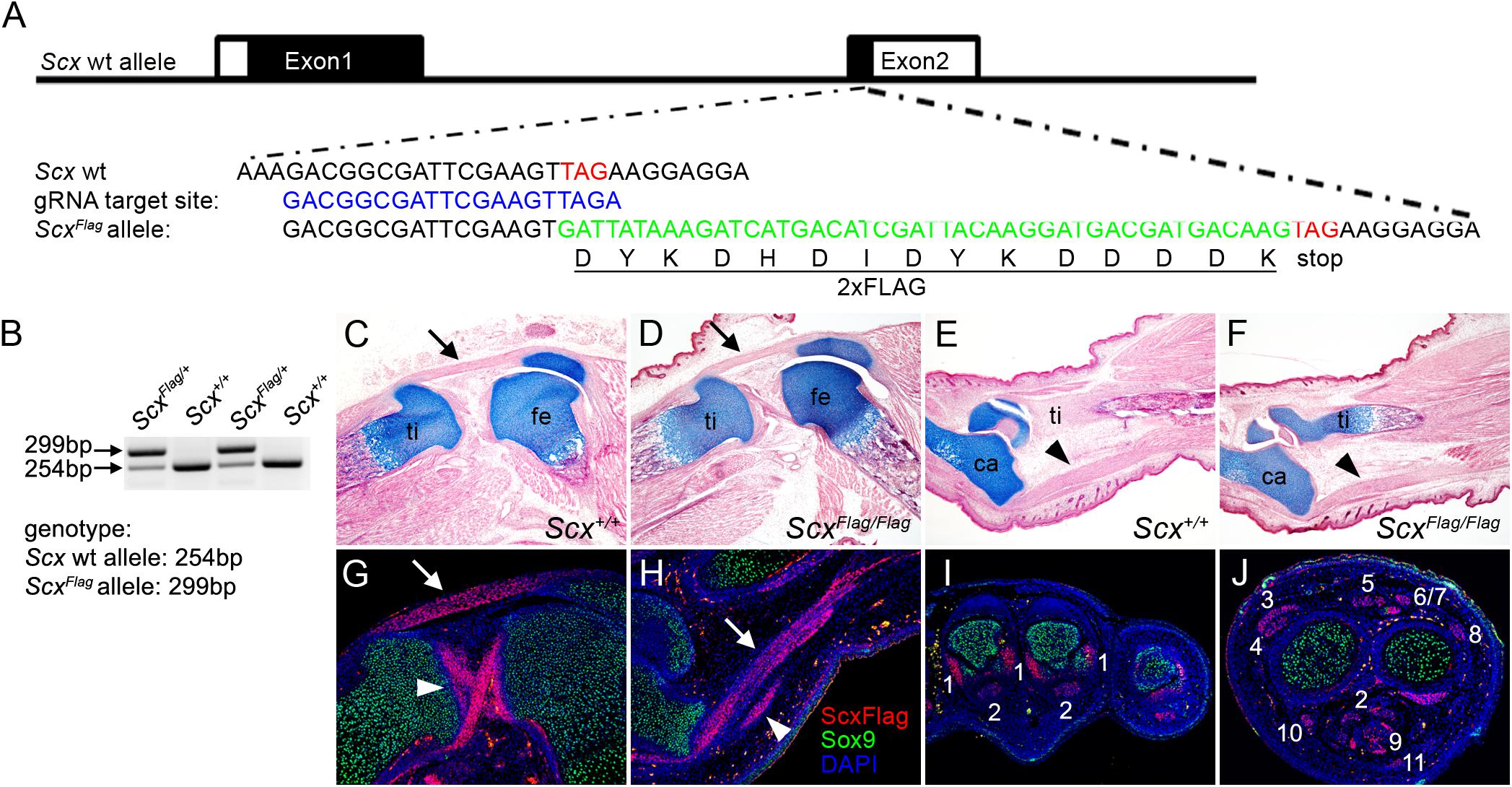
Figure 1. Generation of ScxFlag mice. (A) Schematics of the strategy for generating ScxFlag mice using the CRISPR/CAS9 approach. The top row shows the genomic organization of the mouse Scx locus. The two exons are boxed with coding regions filled in black and untranslated regions in white. The second row shows the sequence around the translation STOP codon (TAG, labeled in red font). The third row shows the selected guide RNA target site sequence. The fourth row shows part of the correctly edited ScxFlag allele sequence containing the insertion of the 2xFLAG coding sequence (in green font) immediately 5′ to the STOP codon. The fifth row shows the amino acid sequence of the 2xFLAG tag. (B) PCR genotyping result from a litter of ScxFlag/+ mouse intercross. (C–F) Images of HE and alcian blue-stained longitudinal sections through the middle of the knee (C,D) and ankle (E,F) of E18.5 wildtype (C,E) and ScxFlag/Flag (D,F) embryos. Arrow in panels (C,D) point to patellar tendon. Arrowheads in panels (E,F) point to Achilles tendon. ca, calcaneus; fe, femur; ti, tibia. (G–J) immunofluorescent staining of sections through the knee (G), ankle (H), forelimb autopod (I), and forelimb zeugopod (J) regions of E16.5 ScxFlag/+ embryos. Immunofluorescence of Sox9 is shown in green color, Immunofluorescence of the FLAG epitope is shown in red color, and DAPI counterstaining of cellular nuclei is shown in blue color. Arrow in panels G,H points to patellar and Achilles tendons, respectively. Arrowhead in panel (G) point to the cruciate ligaments, whereas arrowhead in panel (H) point to a smaller tendon next to the Achilles. The numbers in panels (I,J) mark distinct forelimb tendons as: 1 - Collateral Ligament Metacarpophalangeal joint; 2 - Flexor Digitorium Profundus; 3 - Extensor Pollicis; 4 - Extensor Carpi Radialis Longus; 5 - Extensor Digitorium Communis; 6 - Extensor Digiti Quarti; 7 - Extensor Digiti Quinti; 8 - Extensor Carpi Ulnaris; 9 - Flexor Digitorium Sublimis; 10 - Flexor Carpi Radialis; 11 - Palmaris Longus.
Histology, in situ Hybridization, and Immunofluorescent Staining
Mice were euthanized at predetermined stage and embryos were collected in cold PBS, fixed in 4% PFA overnight, washed in PBS for 3 times and processed to 100% methanol (for whole mount in situ hybridization), or paraffin for section (for histology staining and section in situ hybridization). Histology staining and in situ hybridization procedures were performed as previously described (Liu et al., 2013, 2015). In situ probe templates were amplified by PCR from total cDNA sample of E13.5 wildtype forelimbs. Antisense RNA probes were synthesized from templates using T7 RNA polymerase (Promega catalog# P2075). Immunofluorescent staining was performed as previously described (Xu et al., 2014). The primary antibodies used include anti-Sox9 (Santa Cruz, catalog# sc-20095, 1:25 dilution) and anti-FLAG antibody (Sigma, catalog# F1804, 1:50 dilution). The secondary antibodies were Alexa Fluor 568-conjugated goat anti-mouse IgG (H + L) and Alexa Fluor 488-conjugated goat anti-rabbit IgG (H + L) [(Thermo Fisher Scientific, Waltham, MA, United States), catalog# A11004 and A27034, 1:500 dilution].
Skeletal Preparations
Mice were euthanized at postnatal day 0, heated at 65°C in distilled water for 3 min, and de-skinned manually. Whole de-skinned embryos were stained in Alcian Blue solution (15 mg Alcian Blue dissolved in 20 ml glacial acetic acid plus 80 ml 95% ethanol) for 48–72 h, re-fixed in 100% ethanol for 24 h, and stained in Alizarin Red solution (50 mg Alizarin Red dissolved in 2% KOH) for 24 h. The samples were transferred into 50% glycerol solution for imaging and long term storage.
Body Weight Measurement and Statistical Analysis
Mice were weighed at post-natal day 21. Body weights of 10 Scx+/+, 19 Scx+/–, and 12 Scx–/– mice were collected and processed for statistical analysis. To compare the body weight differences among the three genotypes, we used one-way ANOVA followed by Turkey’s multiple comparison posttest. P value less than 0.05 was considered statistically significant and marked as ∗. P value less than 0.01 was marked as ∗∗, whereas P value less than 0.001 was marked as ∗∗∗ when applicable.
ChIP-seq and Data Analysis
Three biological repeats, each containing 10 pairs of forelimbs from E13.5 ScxFlag/Flag embryos, were collected for chromatin immunoprecipitation (ChIP) analysis as previously described (Xu et al., 2020). Briefly, DNA/protein complexes were extracted and incubated with the anti-FLAG antibody (Sigma, catalog# F1804). Sequencing libraries were generated using the ThruPLEX DNA sequencing kit (Rubicon Genomics). Sequencing was performed on Illumina NextSeq500. ChIP-seq reads were aligned to UCSC mouse genome 10 mm using STAR aligner (ver. 2.7.4) (Dobin et al., 2013). To match the sequencing depth across samples, aligned reads were down-sampled to 30 million. After deduplication, peaks were called for each Scx ChIP-seq dataset using HOMER (Hypergeometric Optimization of Motif EnRichment) (v4.11) (Heinz et al., 2010) against a matching input sample. To identify reproducible and unified Scx peaks, we performed second round differential analysis comparing Scx samples versus input samples anchoring on all the detected Scx peaks. First, Scx peaks from the three biological replicates were pooled and merged into a preliminary peak set. Second, read counts were measured in all the Scx ChIP samples and input samples within the peak set. Finally, exact test was performed using EdgeR (Robinson et al., 2010) comparing Scx ChIP and input samples. Final Scx peaks were defined by log2 fold-change (log2FC) over input > 2 and FDR < 0.01. De novo motif analysis of the Scx peaks was performed within a 200 bp window using Homer (v4.11) (Heinz et al., 2010) with default options. Peak-gene association analysis was performed utilizing the online GREAT (Genomic Regions Enrichment of Annotations Tool) program (version 4.0) (McLean et al., 2010). Gene regulatory domains utilized for region annotation were defined as minimum 5 kb upstream and 1 kb downstream of the gene transcription start site (TSS), and extended in both directions to the nearest gene’s basal domain but no more than the maximum extension of 1000 kb in one direction (‘‘Basal plus extension’’ option)1. The ChIP-seq raw data have been deposited into the National Center for Biotechnology Information Gene Expression Omnibus database (NCBI GEO Accession Number GSE173428).
Fluorescence-Activating Cell Sorting (FACS)
E15.5 embryonic forelimbs were dissected from embryos of Scx+/–; Scx-GFP female crossed with Scx+/–; Scx-GFP male mice. The freshly dissected embryonic forelimbs were digested with the trypsin−EDTA solution (Invitrogen) at 37°C for 4 min. After inactivation of trypsin with DMEM containing 10% FBS, cells were dissociated by pipetting. The dissociated limb cells were suspended in PBS with 2% FBS and 10 mM EDTA, and filtered through a 40 μm nylon cell strainer (BD Falcon, 352340). GFP+ cells from each sample were isolated using BD FACSAria II.
RNA−Seq and Data Analysis
RNA-seq were carried out using FACS isolated GFP+ cells from the forelimb tissues of three Scx–/– embryos and three littermate controls at E15.5. Total RNAs were extracted from FACS isolated E15.5 forelimb GFP+ cells using the Qiagen RNeasy Micro Kit (Qiagen catalog# 74004). cDNA amplification was carried out using the Ovation RNA-Seq System V2 kit (Tecan Genomics Inc.). The sequencing libraries were made using the Nextera XT DNA Library Prep kit (Illumina), and sequenced using Illumina NovaSeq 6000 for 75 bp paired-end reads. RNA-seq reads were aligned to UCSC mouse genome 10 mm using STAR aligner (v2.7.4) (Dobin et al., 2013). Read counts for each gene were measured using FeatureCounts in the subread package (v1.6.2) (Liao et al., 2014). To avoid sex-specific bias in the analysis, genes in chromosome Y were discarded. In addition, RUVseq (RUVs, k = 1) (Risso et al., 2014) was applied to account for sex-specific contribution to the gene expression profiles and to identify sex-independent gene expression changes upon Scx deletion. Principal component analysis was applied to normalized FPKM (fragment per kilobase of transcript per million mapped reads) and log-transformed count data. Differential gene expression analysis was performed using DESeq2 (Love et al., 2014). Differentially expressed genes were identified by FDR < 0.05. Gene ontology (GO) analysis was performed using the online ToppGene tools2 (Chen et al., 2009). Hierarchical clustering of the differential gene expression profiles was performed using Pearson correlation coefficient of log-transformed gene expression level (FPKM) as similarity measure under Ward’s criterion. Clustering heatmap was visualized in z-score. The RNA-seq data have been deposited into the National Center for Biotechnology Information Gene Expression Omnibus database (NCBI GEO Accession Number GSE173428).
Quantitative RT-PCR
E13.5 and E15.5 Scx–/– mutant and wildtype control forelimb samples were collected. Total RNAs were extracted using the Qiagen RNeasy Micro Kit (Qiagen catalog# 74004), and reverse transcribed using SuperScriptTM III First-Strand Synthesis System (Thermo Fisher Scientific, Waltham, MA, United States catalog# 18080051). At each stage, at least 5 samples per genotype were used for quantitative RT-PCR. Relative mRNA levels were normalized to that of Hprt mRNAs. Student’s t test was used for pairwise comparison. P < 0.05 was considered significantly different. P value less than 0.05 was considered statistically significant and marked as ∗. P value less than 0.01 was marked as ∗∗, whereas P value less than 0.001 was marked as ∗∗∗ when applicable.
Results
Generation of a Novel ScxFlag Mouse Line and Genome-Wide Mapping of Endogenous Scx Binding Sites in the Tendon Progenitor Cells in vivo
No specific antibody for the Scx protein that allows direct analysis of endogenous Scx-binding at target genes has been reported. To facilitate direct and specific analysis of endogenous Scx protein activity, we used the CRISPR/Cas9-mediated genome editing strategy (Cong et al., 2013; Scott and Hu, 2019) to insert a well-characterized 2xFLAG epitope tag at the carboxy terminus of the endogenous Scx protein in mice (Figure 1A). Detailed description of the procedures for generating the ScxFlag founder mice is provided in the Materials and Methods section. The ScxFlag/+ founder mice were crossed to wildtype CD1 mice and the G1 ScxFlag/+ hemizygous progenies (Figure 1B) were sequence-verified for correct integration and germ-line transmission of the ScxFlag allele as designed. The ScxFlag/+ mice were then intercrossed to generate ScxFlag/Flag homozygous mice, which were born at expected Mendelian ratio and did not display any phenotypic difference from their hemizygous or wildtype littermates (Figures 1C–F). Immunofluorescent staining of sections of mouse embryos using an anti-FLAG antibody (Sigma, catalog# F1804) showed that the Scx-FLAG fusion protein was strongly and specifically expressed in both tendons and ligaments (Figures 1G–J), consistent with previously reported patterns of expression of endogenous Scx mRNAs and of the transgenic Scx-GFP reporter activity (Murchison et al., 2007; Pryce et al., 2007; Watson et al., 2009). Since the ScxFlag/Flag homozygous mice do not have any phenotypic abnormality, the insertion of the FLAG epitope tag at the carboxy-terminus of the endogenous Scx protein did not affect Scx function, providing a valuable new tool for direct analysis of endogenous Scx function during development, tissue homeostasis, and injury repair.
To investigate Scx-mediated transcriptional regulation during early tendon development, we dissected forelimbs from E13.5 ScxFlag/Flag embryos and performed ChIP-seq experiments in triplicates using the anti-FLAG antibody (Sigma, catalog# F1804). Analysis of the ChIP-seq data identified 12,097 highly enriched Scx-associated chromatin regions (ChIP-seq peaks) (Figures 2A,B). About 77% (9,272 of 12,097) of the Scx ChIP-seq peaks were located in intronic or intergenic regions associated with one or two genes in the immediate vicinity within 500 kb from the gene’s transcription start site (TSS), whereas only about 14% (1,670 of 12,097) of the Scx-binding sites were located within gene promoter regions (Figures 2A–C). De novo motif analysis revealed that the most enriched endogenous Scx-binding domain contains a core sequence C-A-G/T-A/C-T-G (Figure 2D), which was present in >45% of the ChIP-seq peak regions and matches the Scx-binding core sequence previously determined by electrophoresis mobility shift assays (Shukunami et al., 2018). The second most enriched motif, which is present in 15.8% of the ChIP-seq peaks, contains a core sequence that matches the consensus binding motif of the Nfat family transcription factors, TGGAAA (Yu et al., 2015; Klein-Hessling et al., 2017; Figure 2D). It has been reported that Scx and Nfatc4, a member of the Nfat family, act cooperatively to regulate the transcription of Col1a1 in tendon fibroblast cells (Lejard et al., 2007). The significant enrichment of the Nfat binding motif in the Scx ChIP-seq peaks indicate that Scx and Nfat family proteins act together to regulate expression of many genes during tendon development.
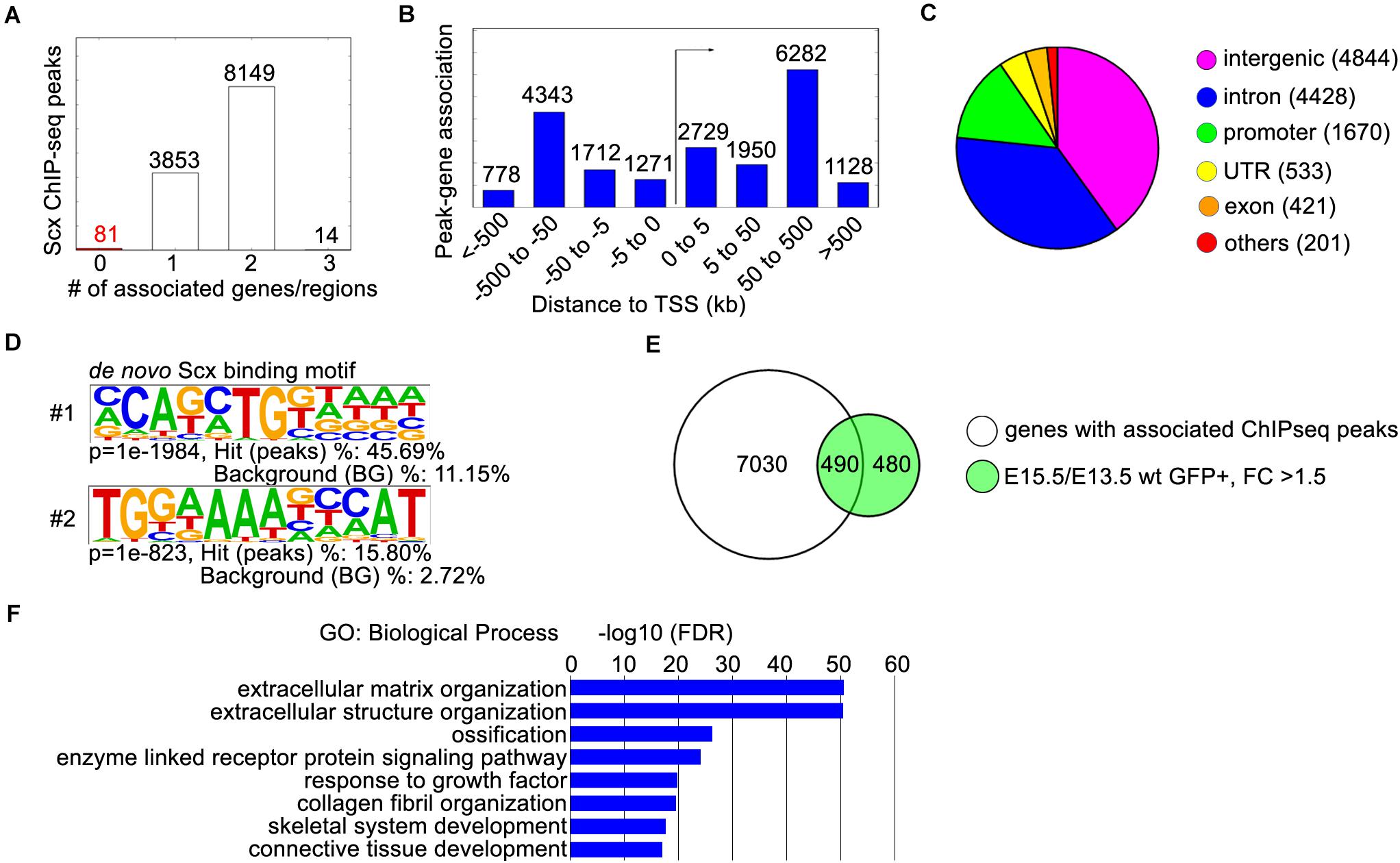
Figure 2. ChIP-seq analysis of E13.5 ScxFlag/Flag forelimbs. (A) Peak-gene association analysis with the GREAT online tool (http://great.stanford.edu). The “basal plus extension” parameter was used to associate the ChIP-seq peaks to genes within 1,000 kb. The majority of peaks were associated with 1 or 2 genes. (B) Distribution of peak-gene associations by their distances to the transcription start site (TSS). (C) Distribution of ChIP-seq peaks by their locations. The majority of peaks are located in intronic (4428) or intergenic (4844) regions, while 1670 peaks were located in promoter regions. (D) The two most enriched Scx binding motifs from ChIP-seq peaks. (E) Comparison of 7520 genes with associated ChIP-seq peaks, with 970 genes that were up-regulated in GFP+ hindlimb cells from E13.5 to E15.5 by at least 1.5-fold. (F) Gene ontology analysis of the 490 differentially expressed genes with associated peaks from panel (E). Representative GO terms are listed in the order of their p-values, with the most enriched GO term on top.
Genomic Regions Enrichment of Annotations Tool analysis of the ChIP-seq data recovered 7,520 genes associated with the Scx-binding genomic regions. To gain insight into Scx-mediated transcriptional regulation during early tendon development, we compared the ChIP-seq data with the previously reported RNA-seq data analyzing transcriptome profiles of developing mouse hindlimb tendons (Liu et al., 2015). We found that high quality Scx ChIP-seq peaks were detected in 490 of the 970 genes whose expression was upregulated by more than 1.5-fold in Scx-GFP+ cells during early tendon cell differentiation in the mouse hindlimb from E13.5 to E15.5 (Figure 2E). GO analysis showed that this group is highly enriched with genes playing roles in “extracellular matrix organization,” “collagen fibril organization,” and “connective tissue development” (Figure 2F), consistent with a major role of Scx in regulating tendon formation.
Characterization of a New Scx Mutant Mouse Resource
Whereas several distinct Scx mutant mouse lines have been reported (Murchison et al., 2007; Yoshimoto et al., 2017; Shukunami et al., 2018), none of these are available to the wide biomedical research community through the Mutant Mouse Resource Centers. On the other hand, the United States National Institutes of Health-funded Knock-out Mouse Project (KOMP) Consortium has generated a Scx-knockout mouse line [C57BL/6N-Scxtm1.1(KOMP)Vlcg/MbpMmucd] and made available through the Mouse Biology Program at the University of California Davis, but the tendon developmental defects in mice homozygous for this particular Scx knockout allele has not been described. We obtained the C57BL/6N-Scxtm1.1(KOMP)Vlcg/MbpMmucd mouse line and established a breeding colony. The Scxtm1.1(KOMP) allele (abbreviated as Scx– in the rest of the report) contains an insertion of the VelociGene ZEN-Ub1 LacZ reporter cassette replacing the entire protein-coding sequences from the translation start site in Exon-1 to 56 bp 3′ to the translation STOP codon in Exon-2 of the Scx gene (Figure 3A). We confirmed the structure and correct integration of the lacZ reporter cassette at the Scx locus through Sanger sequencing of genomic PCR products. However, no specific beta-galactosidase reporter activity was detected in developing tendon tissues in the heterozygous and homozygous mutant mouse embryos, likely due to reporter gene silencing described previously in multiple other KOMP mouse lines (Kirov et al., 2015). To confirm that Scx function is lost in the Scx–/– embryos, we carried out in situ hybridization assay using a probe specific for the Scx coding sequence and found that Scx mRNA expression was completely absent in the Scx–/– embryos (Figures 3B,C).
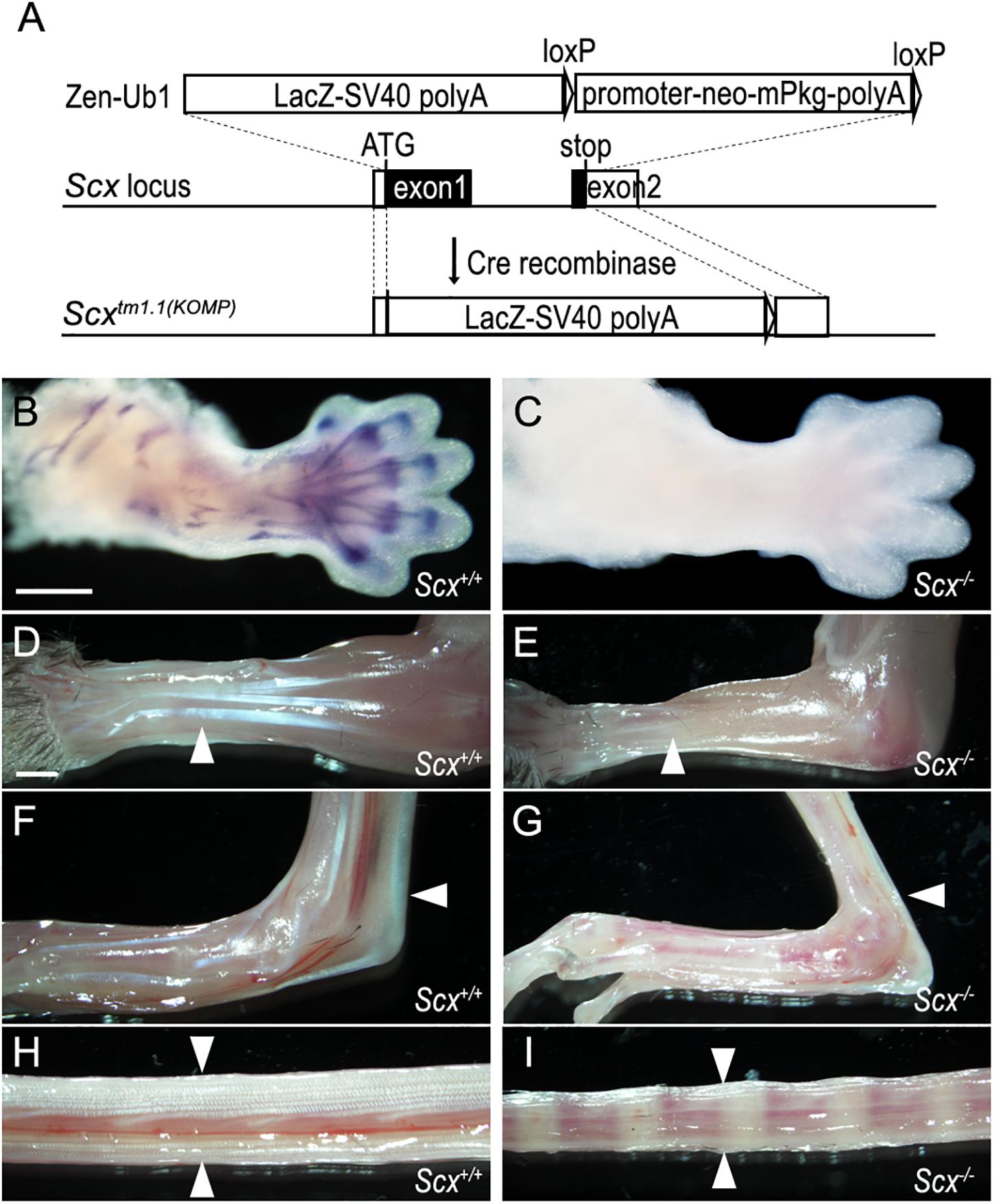
Figure 3. Characterization of a Scx null mutant mouse strain. (A) Schematic diagram and strategy of generating Scx null mutant allele. A VelociGene ZEN-Ub1 LacZ reporter cassette was inserted after the endogenous Scx ATG codon, by homologous recombination, and replaced the entire Scx coding sequence plus a 56 bp 3′ UTR sequence. (B,C) In situ hybridization on E13.5 Scx–/– mutant and control forelimbs, with an anti-sense RNA probe against the protein coding region of the Scx mRNAs. (D–I) Whole mount view of tendons in forelimb (D,E), hindlimb (F,G), and tail (H,I) from Scx–/– mutant (E,G,I) and control littermates (D,F,H) at P21. White arrowheads point to extensor digitorium communis in panels (D,E), Achilles tendons in panels (F,G), and tail tendons in panels (H,I). Scale bar is 500 μm in panel (B) and 1000 μm in panel (D).
We further characterized the phenotypes of the Scx–/– mutant mice. Scx–/– mice were born at Mendelian ratio, but with reduced body size and severely impaired limbs, with the autopod of both fore- and hind-limbs locked in a dorsal flexure (Supplementary Figure 1). All neonatal Scx–/– mutant mice exhibited severe hypoplasia of major limb and tail tendons (Figures 3D–I). Analysis of skeletal preparations showed that Scx–/– mutants lacked the deltoid tuberosity of the humerus in the forelimb and exhibited reduced size of the patella and the entheseal cartilage of the calcaneus in the hindlimb (Supplementary Figures 2A–D). In addition, the transverse processes of the lumbar vertebrae were reduced in the Scx–/– mutants (Supplementary Figures 2E, F). Using a previously reported Scx-GFP transgenic reporter (Pryce et al., 2007), we found that tendon differentiation and condensation in the developing limbs were disrupted by E15.5 in the homozygous mutant embryos (Supplementary Figure 3). The tendon and skeletal defects in these Scx–/– mice are similar to those reported previously in two other independent Scx mutant mouse lines (Murchison et al., 2007; Shukunami et al., 2018). Thus, this KOMP-generated Scx knockout mouse line is a valuable resource for the biomedical research community for further studies of Scx function in vivo.
Analysis of Scx-Dependent Transcriptome Expression Profiles During Early Tendon Cell Differentiation in vivo
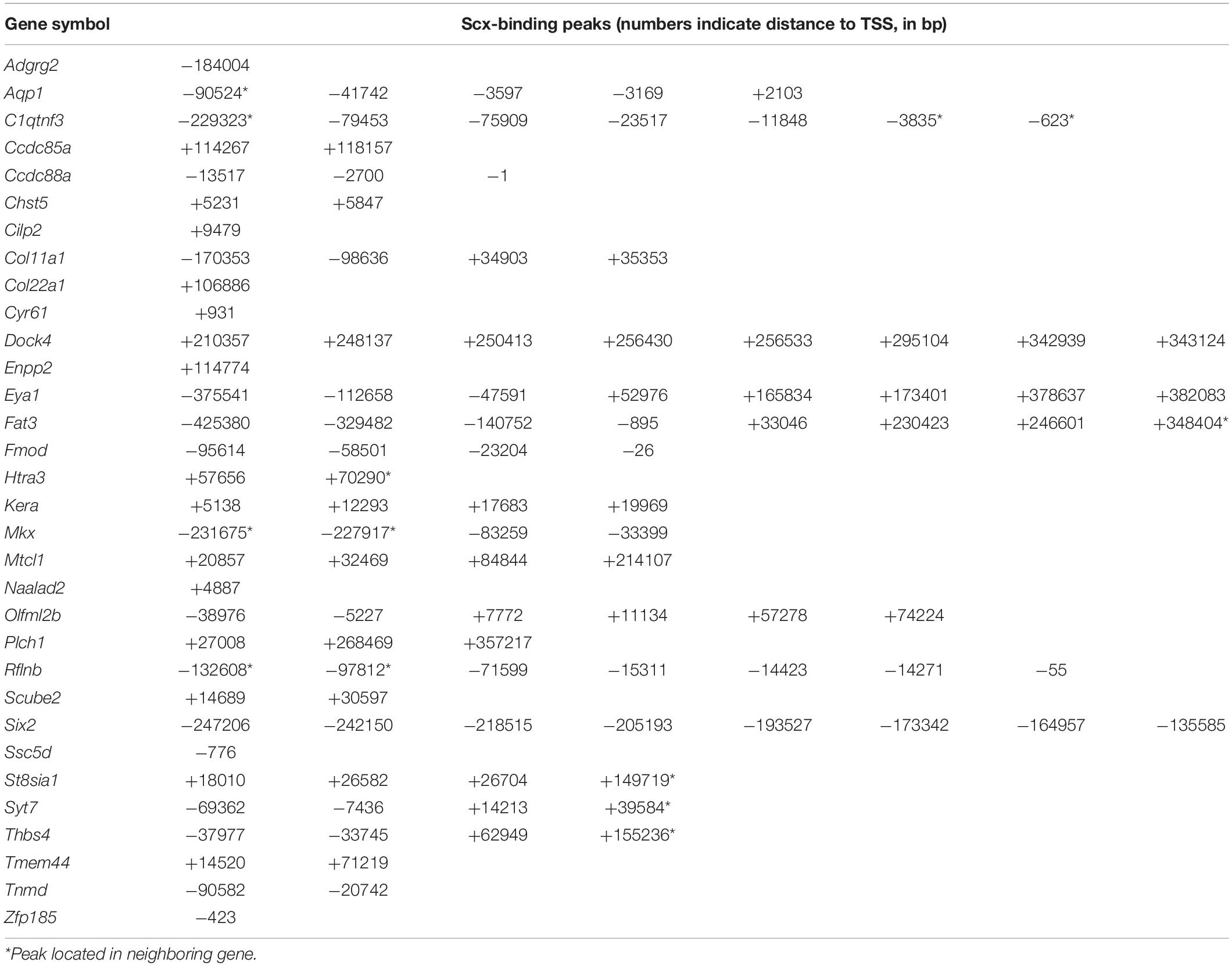
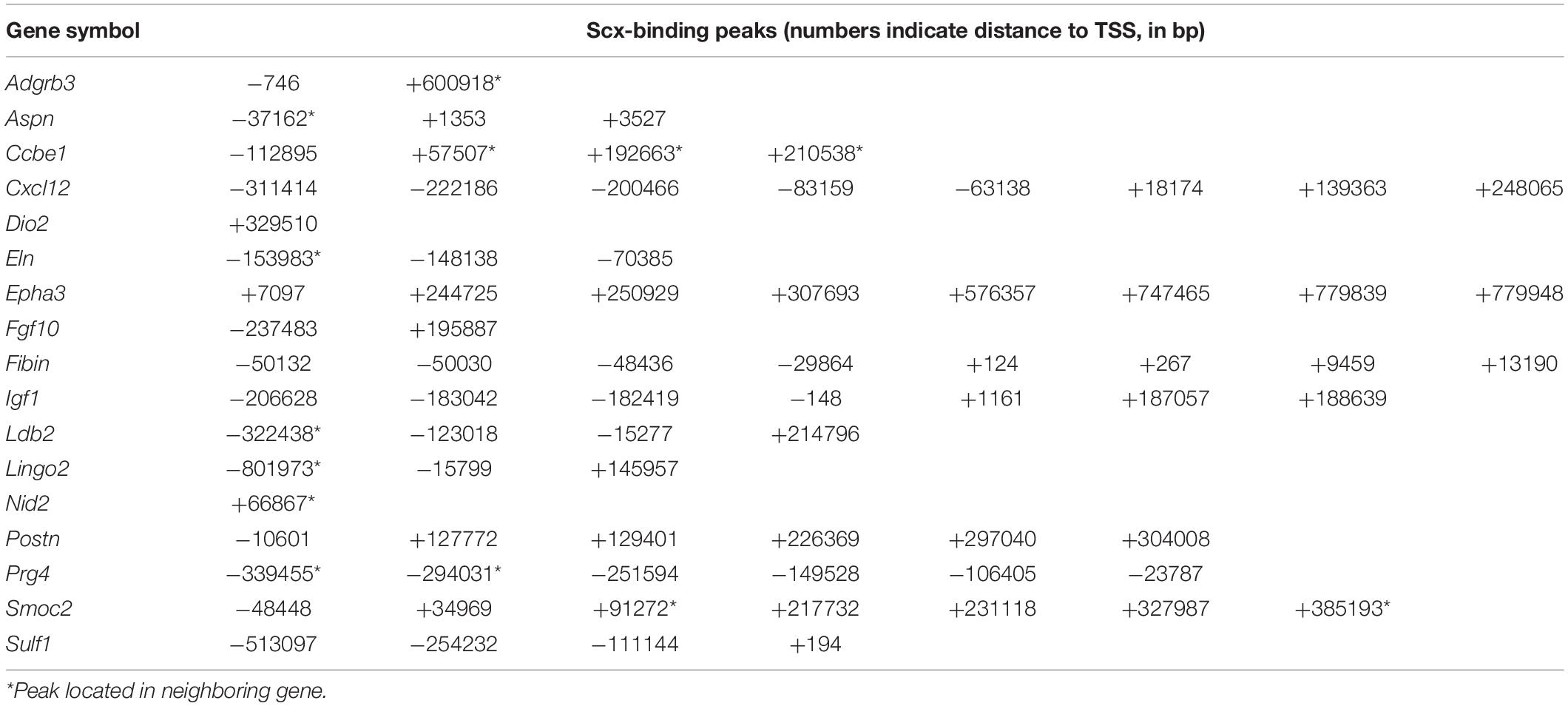
We crossed the Scx-GFP transgenic reporter (Pryce et al., 2007) into the new Scx+/– mouse line and then intercrossed Scx+/–; Scx-GFP mice for analysis of transcriptomic effects of Scx during early tendon cell differentiation. Scx-GFP+ cells were isolated by FACS from freshly dissected forelimb tissues from E15.5 wildtype, Scx+/–, and Scx–/– mutant embryos for RNA-seq analysis. Analysis of the RNA-seq results from three Scx–/– embryos and three control littermates identified 68 genes that exhibited significant changes in expression levels (FDR < 0.05) in the forelimb Scx-GFP+ cells in E15.5 Scx–/– embryos compared with their control littermates (Figures 4A–D). Comparison of the Scx-dependent differentially expressed genes with the Scx ChIP-seq data revealed that 32 of the significantly down-regulated genes were associated with Scx occupancy in the tendon progenitor cells (Figure 4E and Table 1). These genes are likely critical direct target genes mediating Scx function in tendon formation and early tendon cell differentiation. Indeed, GO analysis of this gene group showed that “tendon development,” “tendon cell differentiation,” “keratan sulfate biosynthesis,” and “ECM organization” are among the most significantly enriched biological processes (Figure 4F). The most significantly down-regulated Scx target genes in the E15.5 Scx–/– forelimb tendon cells include Fmod, Tnmd, Kera, and Col11a1 (Figure 4C and Table 1), of which each plays important roles in tendon cell differentiation and/or collagen fibrillogenesis (Svensson et al., 1999; Pellegata et al., 2000; Docheva et al., 2005; Kilts et al., 2009; Wenstrup et al., 2011; Sun et al., 2020) but only Tnmd has previously been identified as a Scx transcriptional target gene (Li et al., 2015; Shukunami et al., 2018). The Scx-dependent target genes also include three genes encoding transcription factors, Mkx, Six2, and Eya1 (Table 1). Mkx function is required for tendon fibril growth and tendon homeostasis (Ito et al., 2010; Liu et al., 2010, 2019; Suzuki et al., 2016).
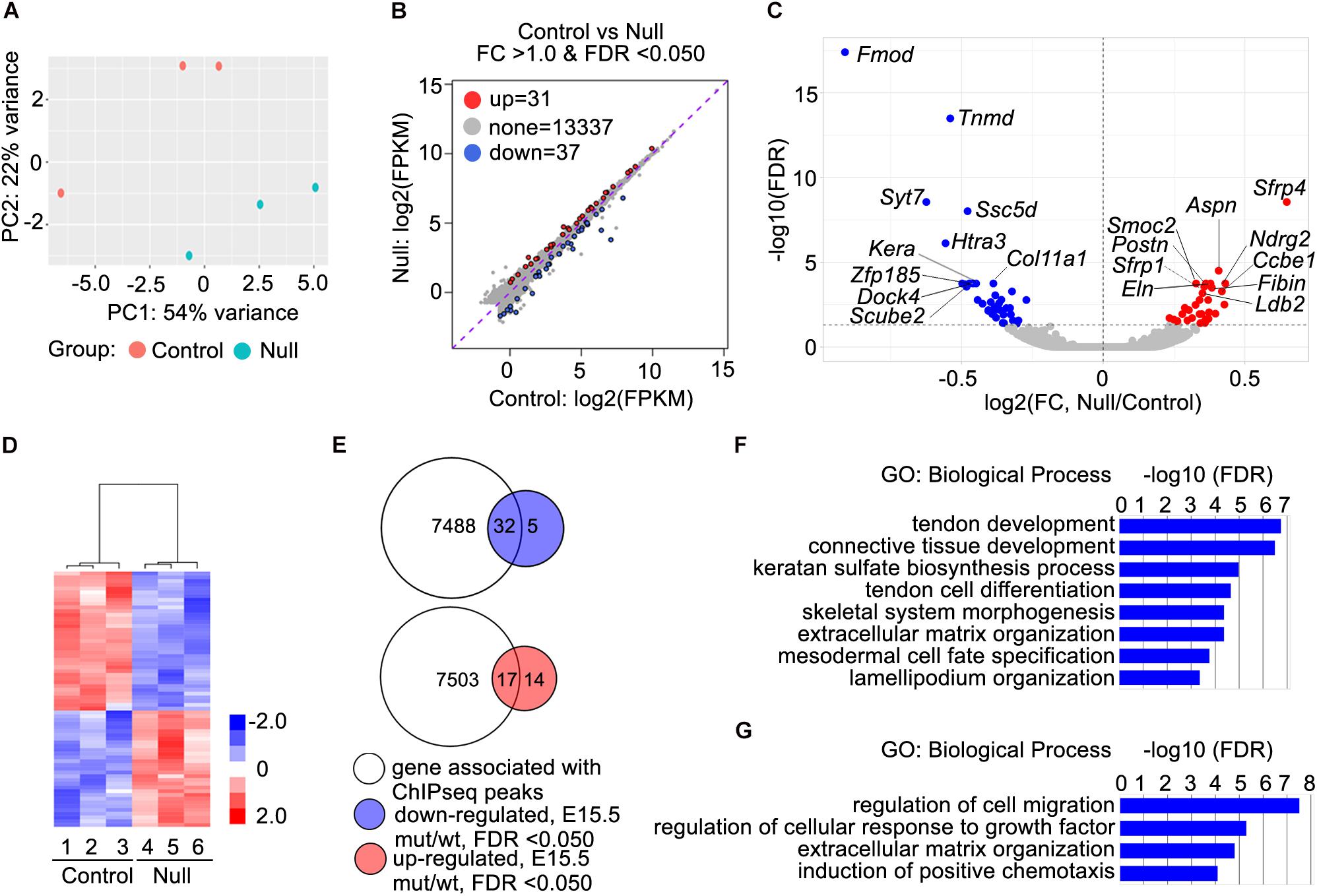
Figure 4. RNA-seq data analysis and identification of candidate Scx direct target genes. (A) Principal component plot displaying the three Scx–/– and three control samples along PC1 and PC2, which account for 54 and 22%, respectively, of the variability within the RNA-seq dataset. (B) MA plot displaying differential expression profiles of the Scx–/– and control samples. Up-regulated genes are marked in red whereas down-regulated genes are marked in blue. (C) Volcano plot displaying the log2 fold change against the log10 value of the FDR adjusted P value. Up-regulated genes are shown in red while down-regulated genes shown in blue. Top 10 down-regulated and top 10 upregulated genes are marked by with symbol. (D) Heatmap showing hierarchical clustering on the 68 differentially expressed genes. The z-score scale bar represents relative expression+/- 2SD from the mean. (E) Comparisons of 7520 genes with associated Scx binding peaks with 37 down-regulated genes in Scx–/– samples at E15.5 and 31 up-regulated genes in Scx–/– samples at E15.5 (B). (F,G) Gene ontology analyses of 32 down-regulated genes associated with Scx-binding peaks and 17 up-regulated genes associated with Scx-binding peaks, respectively. Representative GO terms are listed in the order of their P values, with the most enriched GO term on top.
Analysis of RNA-seq data also revealed 31 genes that were significantly up-regulated in the Scx-GFP+ forelimb cells of E15.5 Scx–/– embryos compared with control littermates (Figures 4B–D). Among these, 17 genes were associated with Scx ChIP-seq peaks in the tendon progenitor cells (Figure 4E and Table 2). GO analysis of this group of genes showed that many of these, including Ccbe1, Cxcl12, Epha3, Fgf10, Igf1, Ldb2, Postn, Smoc2, and Surf1, are involved in “regulation of cell migration” (Figure 4G). In addition, Surf1, Postn, Smoc2, Eln, and Nid2 are involved in regulation of “extracellular matrix organization,” whereas Cxcl12 and Fgf10 have been implicated in “induction of chemotaxis” (Figure 4G and Table 2). Taken together, Scx regulates early tendon cell differentiation by controlling the expression of multiple ECM components as well as signaling pathways controlling cell behavior.
Multiple Scx Target Genes Exhibited Specific Scx-Dependent Expression During Tendon Differentiation
We further analyzed the patterns of expression of the top 10 down-regulated genes in the E15.5 Scx–/– embryos from the RNA-seq data. Whole mount in situ hybridization analyses showed that the Fmod, Htra3, Kera, Ssc5d, Tnmd, and Zfp185 genes each exhibited highly specific patterns of expression in the developing tendon tissues in E14.5 wildtype embryos and their expression in the developing limb tendons was dramatically reduced in the Scx–/– littermates (Figures 5A–L). Furthermore, quantitative real-time RT-PCR analysis revealed that expression of each of these genes was already significantly reduced in the Scx-GFP+ forelimb mesenchyme cells in Scx–/– embryos by E13.5, in comparison with their control littermates (Figure 5M). Among those, Fmod and Tnmd have been reported to play important roles in tendon development whereas Kera plays crucial roles in other connective tissues including the cornea (Svensson et al., 1999; Pellegata et al., 2000; Docheva et al., 2005; Kilts et al., 2009). Htra3 and Zfp185 exhibited specific expression in the developing tendons, similar to Tnmd, whereas Ssc5d exhibited a similar pattern of expression as Fmod in both tendon cells and in joint cells (Figure 5). Further in situ hybridization analysis of serial transverse sections through the digit, metacarpal, and zeugopod regions of the E15.5 forelimb samples confirmed the specificity of expression of Htra3, Zfp185, and Tnmd throughout the differentiating tendon cells (Supplementary Figure 4). Examination of the Scx ChIP-seq profiles associated with these genes showed high quality enrichment of Scx binding at the basal promoter regions of Fmod, Ssc5d, and Zfp185 whereas Scx binding was also specifically enriched at distal regulatory elements associated with Fmod, Kera, Htra3, and Tnmd (Figure 6). Whereas the roles of Htra3, Ssc5d, and Zfp185 in tendon development remain unknown, our finding that they are direct Scx targets that exhibit Scx-dependent activation during early tendon differentiation suggests that they play crucial roles downstream of Scx in tendon development.
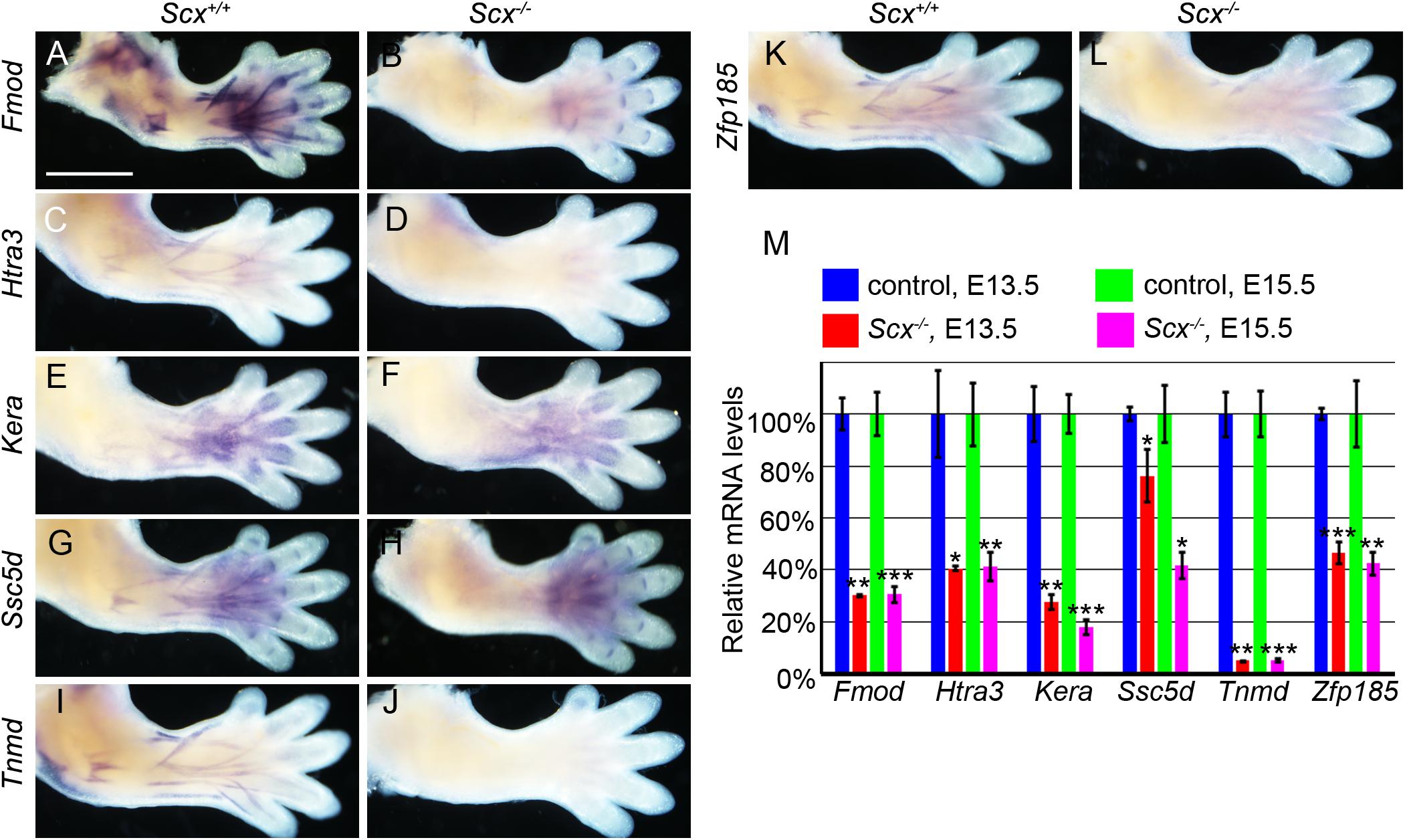
Figure 5. Characterization of candidate genes from ChIP-seq and RNA-seq. (A–L) Whole mount in situ hybridization of six candidate genes in E14.5 Scx–/– mutant (B,D,F,H,J,L) and control (A,C,E,G,I,K) forelimbs. Expression of Fmod, Kera, Ssc5d, and Zfp185 was dramatically reduced in Scx–/– mutant limbs (B,F,H,L), while expression of Htra3 and Tnmd were nearly completely absent in Scx–/– mutant (D,J). Expression of Fmod and Ssc5d persisted in digit ligaments in the mutant limbs (B,H). (M) Quantitative RT-PCR analysis of expression of the six candidate genes in Scx–/– mutant and control forelimb samples at E13.5 and E15.5. All genes were significantly reduced in Scx–/– mutants at both stages, compared with control littermates. *p < 0.05, **p < 0.01, ***p < 0.001.
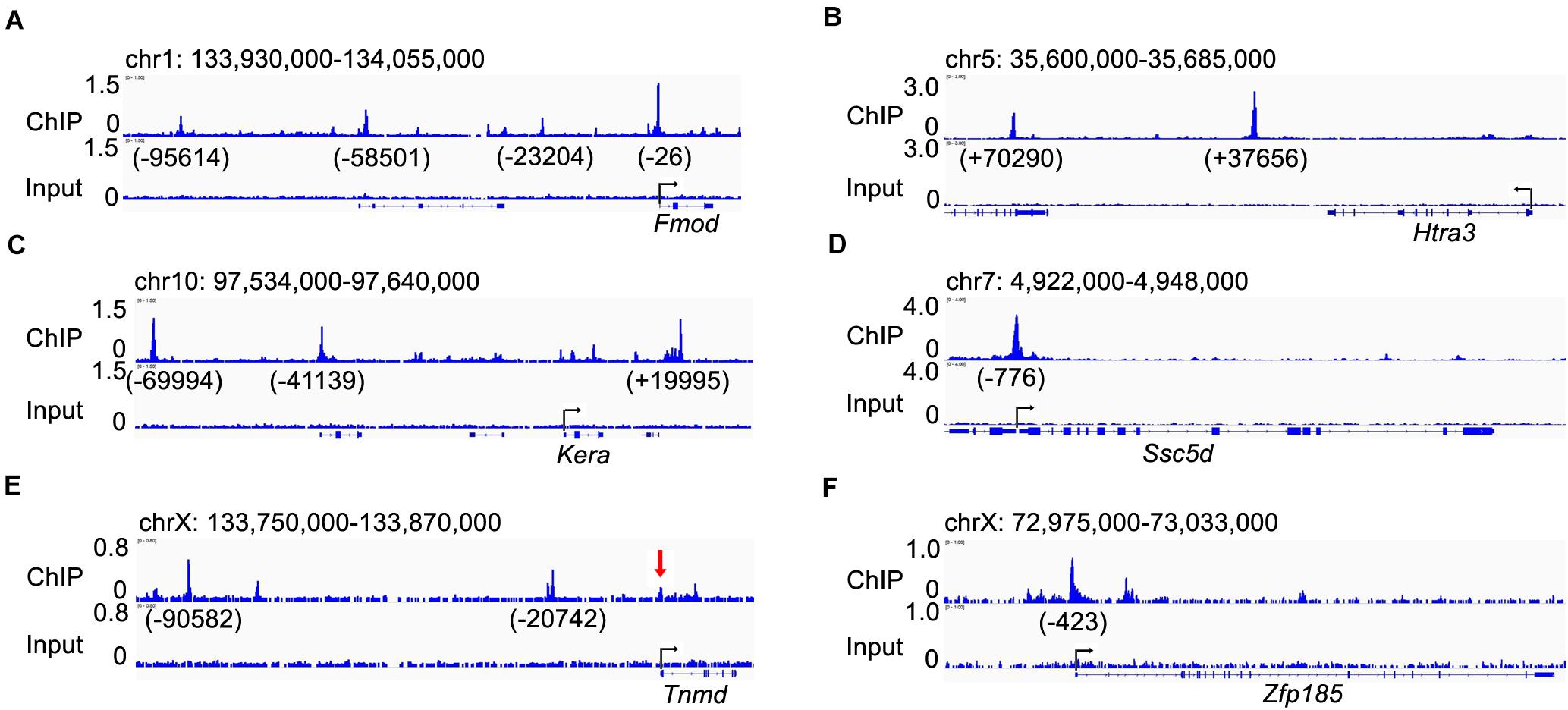
Figure 6. Visualization of the Scx-binding peaks associated with the candidate Scx direct target genes. (A–F) genome browser views of genomic regions containing, Fmod (A), Htra3 (B), Kera (C), Ssc5d (D), Tnmd (E), and Zfp185 (F) genes. Numbers under each peak indicate the distance to the transcription start site (TSS) the marked gene. The red arrow in panel (E) point to a Scx-binding peak at the Tnmd gene promoter region that is recognizable upon careful examination but was below the FDR < 0.01 threshold for identifying genome-wide Scx binding sites in our ChIP-seq data analysis.
Discussion
Previous genetic studies have identified Scx as the most crucial transcriptional regulator of tendon development as well as tendon homeostasis and repair (Murchison et al., 2007; Yoshimoto et al., 2017; Shukunami et al., 2018; Gumucio et al., 2020). Mice lacking Scx exhibited loss or severe disruption of force-transmitting tendons throughout the body as well as defects in entheseal development (Murchison et al., 2007; Killian and Thomopoulos, 2016; Yoshimoto et al., 2017). However, the molecular mechanisms mediating Scx function in tendon development is not well understood and very few Scx target genes in tendon cells have been identified. The lack of large scale identification of direct target genes of Scx in tendon development is largely due to the lack of a reliable specific antibody for mapping genome-wide Scx binding in tendon cells in vivo. Scx belongs to the class II bHLH family of transcription factors, of which many have been shown to bind to an E-box (CANNTG) motif (Cserjesi et al., 1995; Massari and Murre, 2000). However, this short redundant sequence can be found throughout the genome. Thus, previous studies of Scx-mediated transcriptional regulation of putative target genes primarily relied on in vitro biochemical assays including electrophoresis mobility shift assay of recombinant Scx protein binding to E-box containing promoter sequences and promoter reporter assays in cell transfection studies (Lejard et al., 2007; Shukunami et al., 2018; Paterson et al., 2020), and more recently ChIP-qPCR assays of candidate promoter regions (Paterson et al., 2020). In this study, we have generated ScxFlag mice and demonstrate that this new mouse line enables direct analysis of endogenous Scx protein function in the normal developmental and physiological context. Whereas we have used this mouse line for genome-wide mapping of Scx binding sites in early developing limb tendon tissues, this mouse line will provide a valuable tool for direct analysis of Scx function in many developmental and disease processes where Scx plays a role.
Our ChIP-seq analysis identified 12,097 high quality Scx binding sites in the E13.5 mouse forelimb, with the most highly enriched de novo motif identified as 5′ CAG/TA/CTG 3′. Whereas previous in vitro studies have shown Scx binding to various E-boxes including CACGTG in the Col1a1 promoter (Lejard et al., 2007), CAGGTG in the Col2a1 promoter (Furumatsu et al., 2010), and CAAATG and CAGATG in the Tnmd promoter (Li et al., 2015), direct comparative EMSA analysis of Scx binding to five distinct E-box containing elements from the mouse Tnmd promoter region showed that Scx preferentially bound to CAGATG and CATCTG, but not the others (Shukunami et al., 2018). Thus, the consensus Scx binding motif from our ChIP-seq data matches perfectly with the EMSA-determined preferential Scx binding motif, which affirms our genome-wide mapped Scx-binding sites. Whereas several in vitro studies have repeatedly demonstrated Scx binding to the Tnmd promoter regions and Scx activated expression of Tnmd promoter-reporter constructs in co-transfected cells (Li et al., 2015; Shukunami et al., 2018), our ChIP-seq results showed that endogenous Scx binding was primarily enriched at two upstream locations at about 20 and 90 kb, respectively, from the Tnmd TSS (Figure 6E). A minor Scx binding peak was detected at the Tnmd basal promoter region (Figure 6E), which is enriched over 4-fold over the input but the FDR value, at 0.025, was below the stringent threshold (4-fold enrichment over input and FDR < 0.01) used for identifying the genome-wide Scx binding peaks in our analysis. It is possible that Scx binding to both the distal enhancers and the promoter region to synergistically activate Tnmd gene expression in tenocytes.
Whereas previous studies have shown that activation of Tnmd expression during tendon development depends on Scx function and that Tnmd plays crucial roles in tenocyte proliferation and maturation, Tnmd null mice exhibited a much milder tendon phenotype than Scx–/– mice (Docheva et al., 2005; Murchison et al., 2007; Yoshimoto et al., 2017; Shukunami et al., 2018). Our ChIP-seq and RNA-seq results demonstrate that Scx directly regulates expression of a large number of genes during early tendon development, including activation of expression of many tendon cell-specific genes. In particular, we validated Fmod, Htra3, Ssc5d, and Zfp185 as new direct target genes that dependent on Scx for their expression in the differentiating tendon cells. Fmod-deficient mice exhibited defects in tendon collagen fibrillogenesis (Chakravarti, 2002), but the roles of Htra3, Ssc5d, and Zfp185 in tendon development are unknown. Htra3 encodes a serine peptidase that cleaves proteoglycans, thus may function in ECM remodeling (Nie et al., 2003; Glaza et al., 2015). Ssc5d (scavenger receptor cysteine rich family member with 5 domains) has been implicated in playing a role at the interface between adaptive and innate immunity and in placental function (Goncalves et al., 2009). Zfp185 encodes LIM domain type zinc finger protein (Heiss et al., 1997; Zhang et al., 2016). Whereas the Zfp185 protein was originally hypothesized to reside in the cell nucleus, subsequent in vitro cell biological studies suggested that Zfp185 binds to F-actin and may be involved in modulating dynamics of actin filaments (Wang et al., 2008; Zhang et al., 2016). Thus, these gene products have diverse cellular functions and loss of their expression during early tendon cell differentiation in the Scx–/– embryos likely contributed to the severe disruption of tendon condensation phenotype.
Whereas our ChIP-seq analysis identified more than 12,000 high quality Scx-binding regions associated with over 7,500 genes in the embryonic forelimb tissues, our RNA-seq analysis uncovered a relatively small number of genes that exhibited significant differential expression in the Scx-GFP+ forelimb tendon cells between E15.5 Scx–/– and control littermates. These results suggest that Scx participates in the regulation of a large number of genes during early tendon development, but other factors likely partly compensate for Scx function in the Scx–/– mice. Nevertheless, the differential gene expression profiles uncovered by our RNA-seq data provide new insights into the molecular mechanisms underlying Scx-mediated regulation of tendon formation. It has been shown that the tendon microfibrils are highly disorganized in the Scx–/– mouse embryos (Murchison et al., 2007). We found that expression of Col11a1 was among the most down-regulated genes in the E15.5 Scx–/– forelimb tendon cells. Recent studies have shown that ColXI, the gene product of Col11a1, plays an essential role in tendon fibril assembly and organization (Wenstrup et al., 2011; Sun et al., 2020). Mice with tendon-specific disruption of Col11a1 in the Scx-expressing lineages exhibited abnormal tendon fibril structure, smaller fibril diameter and disrupted fibril alignment (Sun et al., 2020). Thus, activation of Col11a1 expression in the differentiating tenocytes is likely an important part of Scx function in tendon formation. In addition to regulating tendon ECM organization and fibrillogenesis, Scx plays a crucial role in regulating tendon cell morphology and organization (Murchison et al., 2007). Whereas wildtype tenocytes develop a complex network of cytoplasmic extensions that engulf the collagen fibril bundles during tendon fibrillogenesis, the tenocytes in Scx–/– mouse embryos exhibited much reduced and less complex cytoplasmic extensions (Murchison et al., 2007). We found that Dock4, encoding a key regulator of filapodia and lamellipodia protrusions (Hiramoto et al., 2006; Abraham et al., 2015), was among the top downregulated genes in the E15.5 tendon cells. As tenocyte cytoplasmic extensions likely also involve spatially regulated changes of the actin cytoskeleton, our finding of significant down-regulation of Zfp185, Ccdc85a and Ccdc88a, which all encode actin binding proteins, in the tendon cells in E15.5 Scx–/– embryos suggests that Scx regulates expression of multiple cytoplasmic proteins to control tenocyte morphology and organization. Moreover, to maintain the stretched tenocyte morphology with a complex network of cytoplasmic extensions during tendon growth and elongation likely requires cell membrane repair mechanisms to maintain plasma membrane integrity. In Scx–/– mice tendon rudiments were detected in the embryonic tail but tail tendon was completely absent in 2-week-old mutant mice due to increased apoptosis (Murchison et al., 2007). Among the most significantly downregulated genes in the Scx–/– tendon cells in our RNA-seq data is Syt7 (Figure 4C), which encodes a transmembrane protein with a crucial role in maintenance of plasma membrane integrity and repair via regulating lysosomal exocytosis (Chakrabarti et al., 2003). Further studies will be needed to investigate whether Syt7 plays an important role in Scx-mediated tenocyte differentiation and maintenance. Furthermore, it has been shown that Scx function is required for proper organization of tendon sheath such as the endotenon and that Epha4-expressing endotenon cells were detected intermixed with tenocytes in the limb tendons in Scx–/– embryos (Murchison et al., 2007). Our RNA-seq data showed that expression of Epha3 was significantly increased in the limb tendon cell in E15.5 Scx–/– embryos. It is possible that the increased Epha3 expression in the tenocytes resulted in disruption of the Eph-Efn signaling involved in regulation of tendon sheath formation.
Whereas Scx is expressed in all tendon progenitor cells and throughout of tendon development (Schweitzer et al., 2001; Brent et al., 2003), only a subset of tendon tissues were significantly disrupted in Scx–/– mice (Murchison et al., 2007). Huang et al. (2019) demonstrated that Scx function is required for recruitment of mesenchymal progenitor cells into the initially formed tendon rudiments during the growth and elongation of the limbs and tail (Huang et al., 2019). Remarkably, lineage-specific genetic analysis and cell transplantation assays demonstrated that Scx function is exclusively required in the recruited mesenchymal cells, but not in the recruiting tendon, for the recruitment and integration of the mesenchymal progenitor cells during tendon elongation (Huang et al., 2019). The molecular mechanism acting downstream of Scx in mediating the tendon cell recruitment and elongation is currently unknown. Although our RNA-seq data show significant changes in expression of several signaling molecules involved in regulation of cell migration and/or chemotaxis in the Scx–/– Scx-GFP+ forelimb cells, it is not clear whether and how some of these molecules may mediate Scx function in tendon cell recruitment. Further analysis of the detailed spatiotemporal patterns of expression of these genes during tendon elongation and/or single-cell transcriptomic profiling combined with lineage-specific functional studies will help resolve the underlying molecular mechanism.
In a recent report, Gumucio et al. (2020) performed RNA-seq analysis to identify genes and signaling pathways that respond differently to mechanical overloads in the plantaris tendons in adult mice due to conditional inactivation of Scx (Gumucio et al., 2020). We compared our RNA-seq results from embryonic tendon cells with their RNA-seq results from the adult tenocytes and found that there were a number of overlapping genes in both the down-regulated and up-regulated groups of Scx-dependent differentially expressed genes. In particular, the Fmod, Kera, Ssc5d, Tnmd, and Zfp185 genes were significantly down-regulated in the adult Scx-deficient mutant tendon tissues. These genes likely play important roles in tendon cell differentiation at both embryonic and adult stages. Thus, further elucidation of the molecular mechanisms of tendon development will facilitate investigation of adult tendon cell behaviors during injuries and regeneration and contribute ultimately to improvement in strategies for tendon therapies. In this regard, our ChIP-seq and RNA-seq datasets provide a rich resource for aiding the design of new studies of the molecular and cellular mechanisms of tendon development and repair.
Data Availability Statement
ChIP-seq and RNA-seq data was deposited in the NCBI GEO database under the accession number(s) GSE173428.
Ethics Statement
The animal study was reviewed and approved by Cincinnati Children’s Research Foundation Institutional Animal Care and Use Committee (IACUC).
Author Contributions
HL, YL, and RJ conceptualized and designed the research. HWL helped revise the research design and data analysis during the manuscript revision. HL and JX performed the research. HL, JX, YL, HWL, and RJ analyzed the data and critically revised the manuscript. HL and RJ wrote the manuscript. All authors approved the final manuscript and agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Funding
This research was supported by Cincinnati Children’s Hospital Medical Center institutional funds to RJ and Cincinnati Children’s Research Foundation Trustee Award to HWL.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The Scxtm1.1(KOMP)Vlcg/+ mouse strain used for this research project was created from ES cell clone 14378A-H1, generated by Regeneron Pharmaceuticals, Inc., and made into live mice by the KOMP Repository (www.KOMP.org) and the Mouse Biology Program (www.mousebiology.org) at the University of California Davis. We thank Ronen Schweitzer at Shriners Hospitals for Children (Portland, Oregon) for providing the Scx-GFP transgenic mouse strain. We thank the Transgenic Animal and Genome Editing Core facility at Cincinnati Children’s Hospital Medical Center for assistance with the generation of the ScxFlag mouse strain.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.654397/full#supplementary-material
Footnotes
References
Abraham, S., Scarcia, M., Bagshaw, R. D., McMahon, K., Grant, G., Harvey, T., et al. (2015). A Rac/Cdc42 exchange factor complex promotes formation of lateral filopodia and blood vessel lumen morphogenesis. Nat. Commun. 6:7286. doi: 10.1038/ncomms8286
Birch, H. L., Thorpe, C. T., and Rumian, A. P. (2013). Specialisation of extracellular matrix for function in tendons and ligaments. Muscles Ligaments Tendons J. 3, 12–22. doi: 10.11138/mltj/2013.3.1.012
Brent, A. E., Schweitzer, R., and Tabin, C. J. (2003). A somitic compartment of tendon progenitors. Cell 113, 235–248.
Chakrabarti, S., Kobayashi, K. S., Flavell, R. A., Marks, C. B., Miyake, K., Liston, D. R., et al. (2003). Impaired membrane resealing and autoimmune myositis in synaptotagmin VII-deficient mice. J. Cell Biol. 162, 543–549. doi: 10.1083/jcb.200305131
Chakravarti, S. (2002). Functions of lumican and fibromodulin: lessons from knockout mice. Glycoconj J. 19, 287–293. doi: 10.1023/A:1025348417078
Chen, J., Bardes, E. E., Aronow, B. J., and Jegga, A. G. (2009). ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 37, 305–311. doi: 10.1093/nar/gkp427
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. doi: 10.1126/science.1231143
Cserjesi, P., Brown, D., Ligon, K. L., Lyons, G. E., Copeland, N. G., Gilbert, D. J., et al. (1995). Scleraxis: a basic helix-loop-helix protein that prefigures skeletal formation during mouse embryogenesis. Development 121, 1099–1110.
Davis, M. E., Gumucio, J. P., Sugg, K. B., Bedi, A., and Mendias, C. L. (2013). MMP inhibition as a potential method to augment the healing of skeletal muscle and tendon extracellular matrix. J. Appl. Physiol. 115, 884–891. doi: 10.1152/japplphysiol.00137.2013
Delgado Caceres, M., Pfeifer, C. G., and Docheva, D. (2018). Understanding tendons: lessons from transgenic mouse models. Stem Cells Dev. 27, 1161–1174. doi: 10.1089/scd.2018.0121
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. doi: 10.1093/bioinformatics/bts635
Docheva, D., Hunziker, E. B., Fassler, R., and Brandau, O. (2005). Tenomodulin is necessary for tenocyte proliferation and tendon maturation. Mol. Cell. Biol. 25, 699–705. doi: 10.1128/MCB.25.2.699-705.2005
Dourte, L. M., Pathmanathan, L., Mienaltowski, M. J., Jawad, A. F., Birk, D. E., and Soslowsky, L. J. (2013). Mechanical, compositional, and structural properties of the mouse patellar tendon with changes in biglycan gene expression. J. Orthop. Res. 31, 1430–1437. doi: 10.1002/jor.22372
Dunkman, A. A., Buckley, M. R., Mienaltowski, M. J., Adams, S. M., Thomas, S. J., Kumar, A., et al. (2014). The injury response of aged tendons in the absence of biglycan and decorin. Matrix Biol. 35, 232–238. doi: 10.1016/j.matbio.2013.10.008
Furumatsu, T., Shukunami, C., Amemiya-Kudo, M., Shimano, H., and Ozaki, T. (2010). Scleraxis and E47 cooperatively regulate the Sox9-dependent transcription. Int. J. Biochem. Cell. Biol. 42, 148–156. doi: 10.1016/j.biocel.2009.10.003
Glaza, P., Osipiuk, J., Wenta, T., Zurawa-Janicka, D., Jarzab, M., Lesner, A., et al. (2015). Structural and functional analysis of human HtrA3 protease and its subdomains. PLoS One 10:e0131142. doi: 10.1371/journal.pone.0131142
Goncalves, C. M., Castro, M. A., Henriques, T., Oliveira, M. I., Pinheiro, H. C., Oliveira, C., et al. (2009). Molecular cloning and analysis of SSc5D, a new member of the scavenger receptor cysteine-rich superfamily. Mol. Immunol. 46, 2585–2596. doi: 10.1016/j.molimm.2009.05.006
Guerquin, M. J., Charvet, B., Nourissat, G., Havis, E., Ronsin, O., Bonnin, M. A., et al. (2013). Transcription factor EGR1 directs tendon differentiation and promotes tendon repair. J. Clin. Invest. 123, 3564–3576. doi: 10.1172/JCI67521
Gumucio, J. P., Schonk, M. M., Kharaz, Y. A., Comerford, E., and Mendias, C. L. (2020). Scleraxis is required for the growth of adult tendons in response to mechanical loading. JCI Insight 5:e138295. doi: 10.1172/jci.insight.138295
Hasson, P. (2011). “Soft” tissue patterning: muscles and tendons of the limb take their form. Dev. Dyn. 240, 1100–1107. doi: 10.1002/dvdy.22608
Havis, E., Bonnin, M. A., Esteves de Lima, J., Charvet, B., Milet, C., and Duprez, D. (2016). TGFbeta and FGF promote tendon progenitor fate and act downstream of muscle contraction to regulate tendon differentiation during chick limb development. Development 143, 3839–3851. doi: 10.1242/dev.136242
Heinz, S., Benner, C., Spann, N., Bertolino, E., Lin, Y. C., Laslo, P., et al. (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 38, 576–589. doi: 10.1016/j.molcel.2010.05.004
Heiss, N. S., Gloeckner, G., Bachner, D., Kioschis, P., Klauck, S. M., Hinzmann, B., et al. (1997). Genomic structure of a novel LIM domain gene (ZNF185) in Xq28 and comparisons with the orthologous murine transcript. Genomics 43, 329–338. doi: 10.1006/geno.1997.4810
Hiramoto, K., Negishi, M., and Katoh, H. (2006). Dock4 is regulated by RhoG and promotes Rac-dependent cell migration. Exp. Cell Res. 312, 4205–4216. doi: 10.1016/j.yexcr.2006.09.006
Huang, A. H., Watson, S. S., Wang, L., Baker, B. M., Akiyama, H., Brigande, J. V., et al. (2019). Requirement for scleraxis in the recruitment of mesenchymal progenitors during embryonic tendon elongation. Development 146:dev182782. doi: 10.1242/dev.182782
Ito, Y., Toriuchi, N., Yoshitaka, T., Ueno-Kudoh, H., Sato, T., Yokoyama, S., et al. (2010). The Mohawk homeobox gene is a critical regulator of tendon differentiation. Proc. Natl. Acad. Sci. U S A 107, 10538–10542. doi: 10.1073/pnas.1000525107
Killian, M. L., and Thomopoulos, S. (2016). Scleraxis is required for the development of a functional tendon enthesis. FASEB J. 30, 301–311. doi: 10.1096/fj.14-258236
Kilts, T., Ameye, L., Syed-Picard, F., Ono, M., Berendsen, A. D., Oldberg, A., et al. (2009). Potential roles for the small leucine-rich proteoglycans biglycan and fibromodulin in ectopic ossification of tendon induced by exercise and in modulating rotarod performance. Scand J. Med. Sci. Sports 19, 536–546. doi: 10.1111/j.1600-0838.2009.00909.x
Kirov, J. V., Adkisson, M., Nava, A. J., Cipollone, A., Willis, B., Engelhard, E. K., et al. (2015). Reporter gene silencing in targeted mouse mutants is associated with promoter CpG Island Methylation. PLoS One 10:e0134155. doi: 10.1371/journal.pone.0134155
Klein-Hessling, S., Muhammad, K., Klein, M., Pusch, T., Rudolf, R., Floter, J., et al. (2017). NFATc1 controls the cytotoxicity of CD8(+) T cells. Nat. Commun. 8:511. doi: 10.1038/s41467-017-00612-6
Kuo, C. K., Petersen, B. C., and Tuan, R. S. (2008). Spatiotemporal protein distribution of TGF-betas, their receptors, and extracellular matrix molecules during embryonic tendon development. Dev. Dyn. 237, 1477–1489. doi: 10.1002/dvdy.21547
Lejard, V., Blais, F., Guerquin, M. J., Bonnet, A., Bonnin, M. A., Havis, E., et al. (2011). EGR1 and EGR2 involvement in vertebrate tendon differentiation. J. Biol. Chem. 286, 5855–5867. doi: 10.1074/jbc.M110.153106
Lejard, V., Brideau, G., Blais, F., Salingcarnboriboon, R., Wagner, G., Roehrl, M. H., et al. (2007). Scleraxis and NFATc regulate the expression of the pro-alpha1(I) collagen gene in tendon fibroblasts. J. Biol. Chem. 282, 17665–17675. doi: 10.1074/jbc.M610113200
Li, Y., Ramcharan, M., Zhou, Z., Leong, D. J., Akinbiyi, T., Majeska, R. J., et al. (2015). The role of scleraxis in fate determination of mesenchymal stem cells for tenocyte differentiation. Sci. Rep. 5:13149. doi: 10.1038/srep13149
Liao, Y., Smyth, G. K., and Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656
Liu, H., Lan, Y., Xu, J., Chang, C. F., Brugmann, S. A., and Jiang, R. (2013). Odd-skipped related-1 controls neural crest chondrogenesis during tongue development. Proc. Natl. Acad. Sci. U S A 110, 18555–18560. doi: 10.1073/pnas.1306495110
Liu, H., Xu, J., and Jiang, R. (2019). Mkx-Deficient mice exhibit hedgehog signaling-dependent ectopic ossification in the achilles tendons. J. Bone Miner Res. 34, 557–569. doi: 10.1002/jbmr.3630
Liu, H., Xu, J., Liu, C. F., Lan, Y., Wylie, C., and Jiang, R. (2015). Whole transcriptome expression profiling of mouse limb tendon development by using RNA-seq. J. Orthop. Res. 33, 840–848. doi: 10.1002/jor.22886
Liu, W., Watson, S. S., Lan, Y., Keene, D. R., Ovitt, C. E., Liu, H., et al. (2010). The atypical homeodomain transcription factor Mohawk controls tendon morphogenesis. Mol. Cell. Biol. 30, 4797–4807. doi: 10.1128/MCB.00207-10
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Massari, M. E., and Murre, C. (2000). Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol. Cell. Biol. 20, 429–440. doi: 10.1128/mcb.20.2.429-440.2000
McLean, C. Y., Bristor, D., Hiller, M., Clarke, S. L., Schaar, B. T., Lowe, C. B., et al. (2010). GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501. doi: 10.1038/nbt.1630
Murchison, N. D., Price, B. A., Conner, D. A., Keene, D. R., Olson, E. N., Tabin, C. J., et al. (2007). Regulation of tendon differentiation by scleraxis distinguishes force-transmitting tendons from muscle-anchoring tendons. Development 134, 2697–2708. doi: 10.1242/dev.001933
Nie, G. Y., Hampton, A., Li, Y., Findlay, J. K., and Salamonsen, L. A. (2003). Identification and cloning of two isoforms of human high-temperature requirement factor A3 (HtrA3), characterization of its genomic structure and comparison of its tissue distribution with HtrA1 and HtrA2. Biochem. J. 371, 39–48. doi: 10.1042/BJ20021569
Nourissat, G., Berenbaum, F., and Duprez, D. (2015). Tendon injury: from biology to tendon repair. Nat. Rev. Rheumatol. 11, 223–233. doi: 10.1038/nrrheum.2015.26
Paterson, Y. Z., Evans, N., Kan, S., Cribbs, A., Henson, F. M. D., and Guest, D. J. (2020). The transcription factor scleraxis differentially regulates gene expression in tenocytes isolated at different developmental stages. Mech. Dev. 163:103635. doi: 10.1016/j.mod.2020.103635
Pellegata, N. S., Dieguez-Lucena, J. L., Joensuu, T., Lau, S., Montgomery, K. T., Krahe, R., et al. (2000). Mutations in KERA, encoding keratocan, cause cornea plana. Nat. Genet. 25, 91–95. doi: 10.1038/75664
Pryce, B. A., Brent, A. E., Murchison, N. D., Tabin, C. J., and Schweitzer, R. (2007). Generation of transgenic tendon reporters, ScxGFP and ScxAP, using regulatory elements of the scleraxis gene. Dev. Dyn. 236, 1677–1682. doi: 10.1002/dvdy.21179
Pryce, B. A., Watson, S. S., Murchison, N. D., Staverosky, J. A., Dunker, N., and Schweitzer, R. (2009). Recruitment and maintenance of tendon progenitors by TGFbeta signaling are essential for tendon formation. Development 136, 1351–1361. doi: 10.1242/dev.027342
Risso, D., Ngai, J., Speed, T. P., and Dudoit, S. (2014). Normalization of RNA-seq data using factor analysis of control genes or samples. Nat. Biotechnol. 32, 896–902. doi: 10.1038/nbt.2931
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Schweitzer, R., Chyung, J. H., Murtaugh, L. C., Brent, A. E., Rosen, V., Olson, E. N., et al. (2001). Analysis of the tendon cell fate using Scleraxis, a specific marker for tendons and ligaments. Development 128, 3855–3866.
Scott, M. A., and Hu, Y. C. (2019). Generation of CRISPR-edited rodents using a Piezo-Driven Zygote injection technique. Methods Mol. Biol. 1874, 169–178. doi: 10.1007/978-1-4939-8831-0_9
Shukunami, C., Takimoto, A., Nishizaki, Y., Yoshimoto, Y., Tanaka, S., Miura, S., et al. (2018). Scleraxis is a transcriptional activator that regulates the expression of Tenomodulin, a marker of mature tenocytes and ligamentocytes. Sci. Rep. 8:3155. doi: 10.1038/s41598-018-21194-3
Shukunami, C., Takimoto, A., Oro, M., and Hiraki, Y. (2006). Scleraxis positively regulates the expression of tenomodulin, a differentiation marker of tenocytes. Dev. Biol. 298, 234–247. doi: 10.1016/j.ydbio.2006.06.036
Sun, M., Luo, E. Y., Adams, S. M., Adams, T., Ye, Y., Shetye, S. S., et al. (2020). Collagen XI regulates the acquisition of collagen fibril structure, organization and functional properties in tendon. Matrix Biol. 94, 77–94. doi: 10.1016/j.matbio.2020.09.001
Suzuki, H., Ito, Y., Shinohara, M., Yamashita, S., Ichinose, S., Kishida, A., et al. (2016). Gene targeting of the transcription factor Mohawk in rats causes heterotopic ossification of Achilles tendon via failed tenogenesis. Proc. Natl. Acad. Sci. U S A 113, 7840–7845. doi: 10.1073/pnas.1522054113
Svensson, L., Aszodi, A., Reinholt, F. P., Fassler, R., Heinegard, D., and Oldberg, A. (1999). Fibromodulin-null mice have abnormal collagen fibrils, tissue organization, and altered lumican deposition in tendon. J. Biol. Chem. 274, 9636–9647. doi: 10.1074/jbc.274.14.9636
Wang, N., Zheng, Q., Zhang, J. S., and Zhao, Y. (2008). Molecular cloning and characterization of a novel mouse actin-binding protein Zfp185. J. Mol. Histol. 39, 295–302. doi: 10.1007/s10735-008-9165-2
Watson, S. S., Riordan, T. J., Pryce, B. A., and Schweitzer, R. (2009). Tendons and muscles of the mouse forelimb during embryonic development. Dev. Dyn. 238, 693–700. doi: 10.1002/dvdy.21866
Wenstrup, R. J., Smith, S. M., Florer, J. B., Zhang, G., Beason, D. P., Seegmiller, R. E., et al. (2011). Regulation of collagen fibril nucleation and initial fibril assembly involves coordinate interactions with collagens V and XI in developing tendon. J. Biol. Chem. 286, 20455–20465. doi: 10.1074/jbc.M111.223693
Xu, J., Liu, H., Lan, Y., Park, J. S., and Jiang, R. (2020). Genome-wide identification of Foxf2 target genes in palate development. J. Dent. Res. 99, 463–471. doi: 10.1177/0022034520904018
Xu, J., Liu, H., Park, J. S., Lan, Y., and Jiang, R. (2014). Osr1 acts downstream of and interacts synergistically with Six2 to maintain nephron progenitor cells during kidney organogenesis. Development 141, 1442–1452. doi: 10.1242/dev.103283
Yang, G., Rothrauff, B. B., and Tuan, R. S. (2013). Tendon and ligament regeneration and repair: clinical relevance and developmental paradigm. Birth Defects Res. C Embryo Today 99, 203–222. doi: 10.1002/bdrc.21041
Yoshimoto, Y., Takimoto, A., Watanabe, H., Hiraki, Y., Kondoh, G., and Shukunami, C. (2017). Scleraxis is required for maturation of tissue domains for proper integration of the musculoskeletal system. Sci. Rep. 7:45010. doi: 10.1038/srep45010
Yu, H. B., Yurieva, M., Balachander, A., Foo, I., Leong, X., Zelante, T., et al. (2015). NFATc2 mediates epigenetic modification of dendritic cell cytokine and chemokine responses to dectin-1 stimulation. Nucleic Acids Res. 43, 836–847. doi: 10.1093/nar/gku1369
Keywords: bHLH, cell differentiation, ChIP-seq, CRISPR, RNA-seq, Scx, tendon development
Citation: Liu H, Xu J, Lan Y, Lim H-W and Jiang R (2021) The Scleraxis Transcription Factor Directly Regulates Multiple Distinct Molecular and Cellular Processes During Early Tendon Cell Differentiation. Front. Cell Dev. Biol. 9:654397. doi: 10.3389/fcell.2021.654397
Received: 16 January 2021; Accepted: 05 May 2021;
Published: 03 June 2021.
Edited by:
Hongwei Ouyang, Zhejiang University, ChinaReviewed by:
Lin Junxin, Zhejiang University, ChinaAlice Huang, Icahn School of Medicine at Mount Sinai, United States
Copyright © 2021 Liu, Xu, Lan, Lim and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rulang Jiang, UnVsYW5nLkppYW5nQGNjaG1jLm9yZw==
 Han Liu
Han Liu Jingyue Xu
Jingyue Xu Yu Lan
Yu Lan Hee-Woong Lim4,5
Hee-Woong Lim4,5 Rulang Jiang
Rulang Jiang