- Institute of Neurology (Edinger Institute), University Hospital Frankfurt, Goethe University, Frankfurt, Germany
Transcription factors (TFs) guide effector proteins like chromatin-modifying or -remodeling enzymes to distinct sites in the genome and thereby fulfill important early steps in translating the genome’s sequence information into the production of proteins or functional RNAs. TFs of the same family are often highly conserved in evolution, raising the question of how proteins with seemingly similar structure and DNA-binding properties can exert physiologically distinct functions or respond to context-specific extracellular cues. A good example is the TALE superclass of homeodomain-containing proteins. All TALE-homeodomain proteins share a characteristic, 63-amino acid long homeodomain and bind to similar sequence motifs. Yet, they frequently fulfill non-redundant functions even in domains of co-expression and are subject to regulation by different signaling pathways. Here we provide an overview of posttranslational modifications that are associated with murine and human TALE-homeodomain proteins and discuss their possible importance for the biology of these TFs.
Introduction
TFs recognize specific DNA sequences, often depending on DNA shape or methylation status, to control the local assembly of larger protein complexes that induce the transcriptional activation or repression of nearby genes. Transcription factors (TFs) are thus vital to determining which gene product is produced when, where, in which quantities, and in response to what external signal(s). In human, these multifaceted tasks are performed by an estimated ∼1,600 different TFs (Lambert et al., 2018). Although this seems like an impressive repertoire, TFs use a limited number of DNA binding domain (DBD) types, with most metazoan TFs belonging to the C2H2 zinc- finger-, homeodomain (HD)-, basic helix-loop-helix (bHLH)-, basic leucine zipper-, forkhead-, nuclear hormone receptor-, or high-mobility group (HMG)/SRY-related HMG-box (SOX)-superclasses. DBD-types are highly variable across classes but very similar in TFs belonging to the same class. Evolutionary related TFs often also share extensive sequence similarity outside of the DBD. This raises the conundrum how physiologically distinct functions may be carried out by proteins that possess the same overall structure and, at least in vitro, nearly identical DNA-binding properties.
TFs almost always function as ensembles, consistent with the concept that the composition of the multiprotein complex dictates the affinity and specificity of DNA binding (Slattery et al., 2011; Bridoux et al., 2020). The ability of a TF to interact with DNA or with other proteins depends on the biochemical properties of the amino acids involved in binding, which in turn can be profoundly altered by the attachment of additional chemical moieties in a process known as posttranslational modification (PTM). Consequently, the type of binding partners a TF assembles with, the sequence motif recognized by the complex, and the strength of interaction with this motif are sensitive to PTMs (Filtz et al., 2014; Draime et al., 2018). These are important features for any TF, because the composition of transcriptional multiprotein complexes determines the cellular and physiological context in which the TF acts, while recognition of motif variations can lead to high- or low affinity DNA binding, which in turn may result in dynamic gene expression levels (Crocker et al., 2016). In this minireview, we manually surveyed high-throughput proteomics studies, published in peer-reviewed journals or deposited to open-source platforms, to compile PTMs that were recorded in TALE-HD TFs isolated from various murine and human sources. Comparing these PTMs between paralog and ortholog proteins revealed general principles by which PTMs may shape the activity of individual members of conserved TF protein families.
TALE-HD Proteins
Three amino acid loop extension-homeodomain (TALE-HD) TFs are evolutionary highly conserved and found in single-cell eukaryotes (e.g., Mata1/Mata2 in yeast), plants (e.g., KNOX and BELL), and animals (see below; Mukherjee and Bürglin, 2007). The TALE-HD differs from the canonical, 60 amino acid-long HD by the insertion of three extra residues between helix 1 and helix 2 of the HD. This motif, known as the TALE-motif, forms a hydrophobic pocket to mediate protein-protein interactions (Figure 1A; Bürglin, 1997; Piper et al., 1999; LaRonde-LeBlanc and Wolberger, 2003; Mukherjee and Bürglin, 2007). For this feature, TALE-HD proteins have been classified as “atypical” HD proteins. In animals, they have been grouped into five classes, PBC, MEINOX, TGIF, IRO and MKX, based on the sequence of the HD itself and conserved, class-specific motifs flanking the HD (Figure 1B). The developmental functions of individual TALE-HD genes and the defects associated with their mutation in animal models or in human diseases have been covered by a series of excellent recent reviews and will therefore not be discussed in detail (Kim et al., 2012; Blasi et al., 2017; Schulte and Geerts, 2019; Selleri et al., 2019). Instead, we here provide an overview of the different PTMs detected in mouse and human TALE-HD TFs and explore how such PTMs may help to convey functional specificity among these structurally similar proteins.
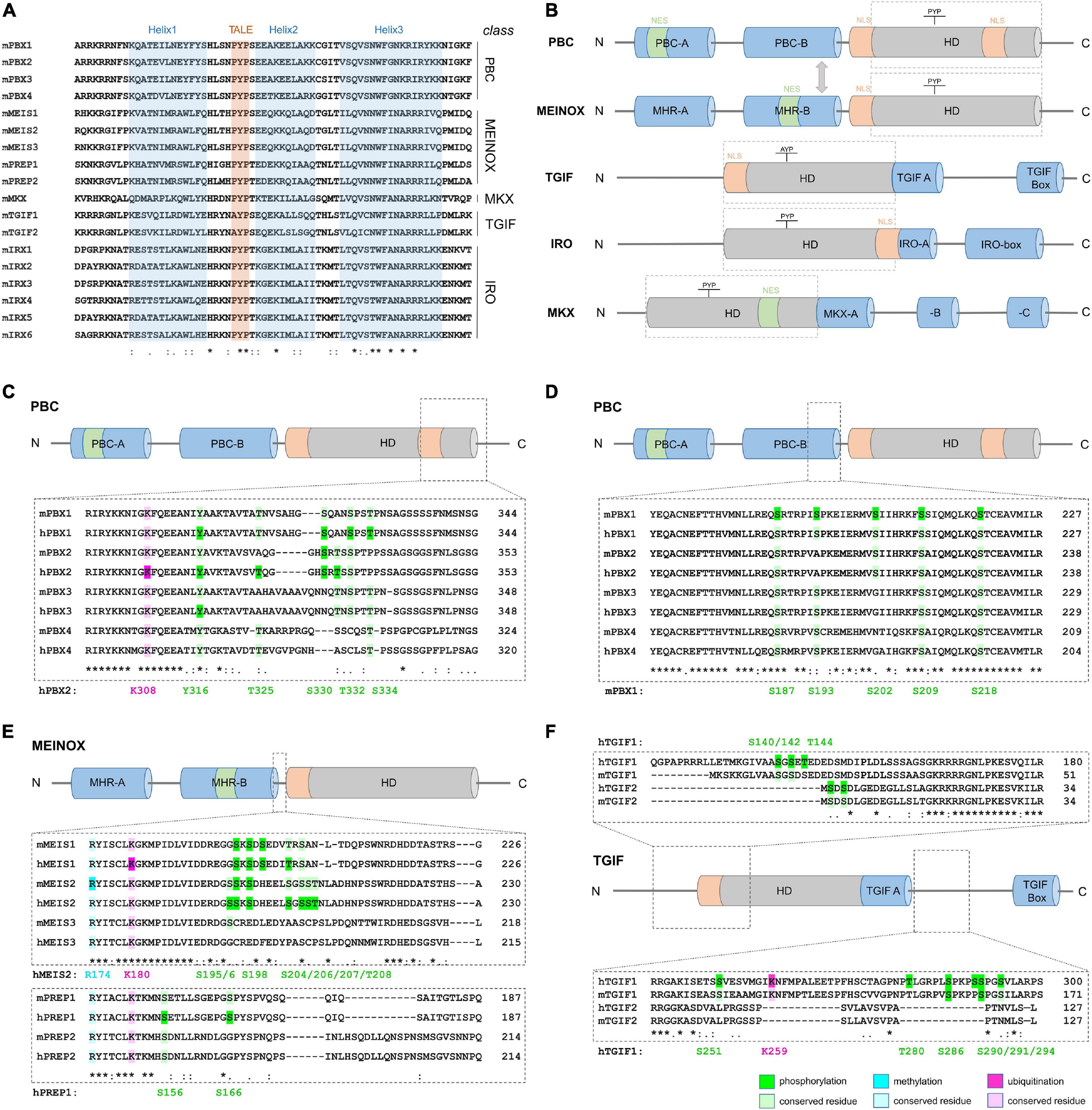
Figure 1. Structure of TALE-HD proteins and examples of abundant, class-specific PTMs. (A) Comparison of the amino acid sequence of mouse TALE-HD domains. Helices 1–3 are highlighted in blue, the name-giving TALE-motif in red. (B) Domain structure of the five TALE-HD protein classes. The HD is shown as gray cylinder, conserved protein domains outside the HD in blue; nuclear export signal (NES): green; nuclear localization signal (NLS): orange. See text for details. Domain sizes are not drawn to scale. (C–E) PTM comparison among paralogs and between human and mouse orthologs. (C) Lysine-ubiquitination and serine-, threonine- and tyrosine-phosphorylation C-terminal to the HD in PBC class proteins. (D) Serine-phosphorylation in PBC class proteins N-terminal to the HD. (E) Arginine-methylation, lysine-ubiquitination and serine/threonine-phosphorylation in MEINOX-proteins. (F) Ubiquitination and phosphorylation in TGIF-class proteins. Numeration of modified amino acids in reference proteins is shown below each amino acid alignment. Color code: green: phosphorylation, blue: methylation, pink: ubiquitination; dark shades indicate PTMs, bright shades indicate residue conservation. A list of PTMs assessed in PBC, MEIS/PREP and TGIF as well as PTMs detected in IRO and MKX can be found in Table 1.
PBC-Class
Mammals have four Pbx (pre-B-cell leukemia homeobox) genes: Pbx1, which was first identified in acute pre-B-cell leukemias, and Pbx2 to Pbx4, which were identified later by sequence homology to Pbx1 (Figure 1A; Kamps et al., 1990; Wagner et al., 2001; Rhee et al., 2004; Selleri et al., 2004). PBC-class TFs dimerize with MEINOX-class proteins through a conserved 180-amino acid-long domain termed PBC-homology domain (Figure 1B; Bürglin and Ruvkun, 1992; Bruckmann et al., 2020). Monomeric PBX proteins or PBX-MEINOX dimers form cell type-specific transcriptional regulatory units with other TFs, including HD transcription factor like HOX-proteins, basic helix-loop-helix (bHLH), nuclear receptors, Smad2,3,4 intracellular signaling proteins of the TGF-β pathway, as well as chromatin modifying enzymes (Peltenburg and Murre, 1996; Wang et al., 2001; Subramaniam et al., 2003; Bailey et al., 2004; Choe et al., 2009, 2014; Merabet and Galliot, 2015). In fact, PBC-class proteins are essential co-factors of HOX-proteins, which themselves are subject to multiple forms of PTM (Draime et al., 2018). Pbx1, Pbx2, and Pbx3 are extensively co-expressed and can partly compensate for each other in domains of co-expression (Selleri et al., 2001, 2004; Rhee et al., 2004; Capellini et al., 2006). Genetic mutant models in rodents are diverse, but defective skeletal patterning and hematopoiesis, as well as hypoplasia and defective development of multiple organs, including pancreas, spleen, face, heart, vascular system, and brain are frequent phenotypes (Selleri et al., 2019).
MEINOX-Class
The vertebrate MEINOX-class is divided into two subclasses, MEIS (Myeloid ecotropic viral integration site) with three genes in mammals, Meis1-3, and PREP/PKNOX (Pbx-regulating protein/PBX-Knotted Homeobox) with Prep1 and Prep2. Meis and Prep are widely co-expressed, compete for heterodimerization with PBC-class proteins and play opposing roles in tumorigenesis (Dardaei et al., 2014). MEIS1 is upregulated in many human cancers, including the majority of acute myeloid leukemias (AML), whereas PREP1 has tumor-suppressive properties (Lawrence et al., 1999; Longobardi et al., 2010; Blasi et al., 2017; Schulte and Geerts, 2019). In addition to the C-terminal HD, MEINOX proteins possess a N-terminal bipartite domain, termed MEIS homology region (MHR) -A and -B, which mediates the binding to their PBX partners (Figure 1B; Bürglin, 1997; Knoepfler et al., 1997; Shanmugam et al., 1999; Bruckmann et al., 2020). Mutant mouse models exist mostly for Meis1 and Prep1. Consistent with MEIS’ or PREP’s heterodimerization with PBX1, many defects associated with Meis1 or Prep1 loss-of-function overlap with those seen in Pbx1 mutants.
More Distantly Related TALE-HD Proteins: TGIF-, IRO- and MKX-Classes
Tgif1 and Tgif2 (Transforming growth factor beta (TGF-β)-induced factor/TG-interacting factor) are phylogenetically most closely related to the MEINOX class (Mukherjee and Bürglin, 2007). They carry a distinct variation of the TALE-motif, AYP, instead of the PYP found in all other TALE-HD proteins (Figure 1A) as well as two short sequence motifs C-terminal to the HD (Figure 1B). TGIF proteins are transcriptional repressors that have been implicated in the regulation of various signaling pathways, most prominently TGF-β- and retinoic acid signaling (Bertolino et al., 1995; Wotton et al., 1999; Shen and Walsh, 2005; Guca et al., 2018). Loss-of-function phenotypes for Tgif1 in mice are strain-dependent and range from no overt defect to holoprosencephaly, a brain malformation that has also been linked to TGIF1 mutations in humans (Kuang et al., 2006; Taniguchi et al., 2012). Constituting another TALE-HD class, the six mammalian Irx genes, taking their names from the Iroquois complex in D. melanogaster, are located in two paralogous clusters in the genome and characterized by a bipartite IRO-box C-terminal of the HD (Figure 1B; Peters et al., 2000; Mukherjee and Bürglin, 2007). Loss-of-function models in mice were generated for all six Irx genes and established that Irx3, -4 and -5 are important transcriptional regulators in the developing and adult heart, that Irx1 controls lung- and tooth development, and that Irx5- and -6 participate in retina development (Bruneau et al., 2001; Costantini et al., 2005; Zhang et al., 2011; Gaborit et al., 2012; Star et al., 2012; Yu et al., 2017). Finally, the single gene Mohawk (Mkx, also known as iroquois homeobox protein-like 1) most closely related to IRX but recognized as separate class, plays a prominent role in tendon development (Mukherjee and Bürglin, 2007; Ito et al., 2010).
In short, members of the same class of TALE-HD proteins share a high degree of sequence similarity, are frequently co-expressed, and functionally cooperate in some physiological contexts but fulfill unique developmental functions in others.
PTMs in TALE-HD Proteins
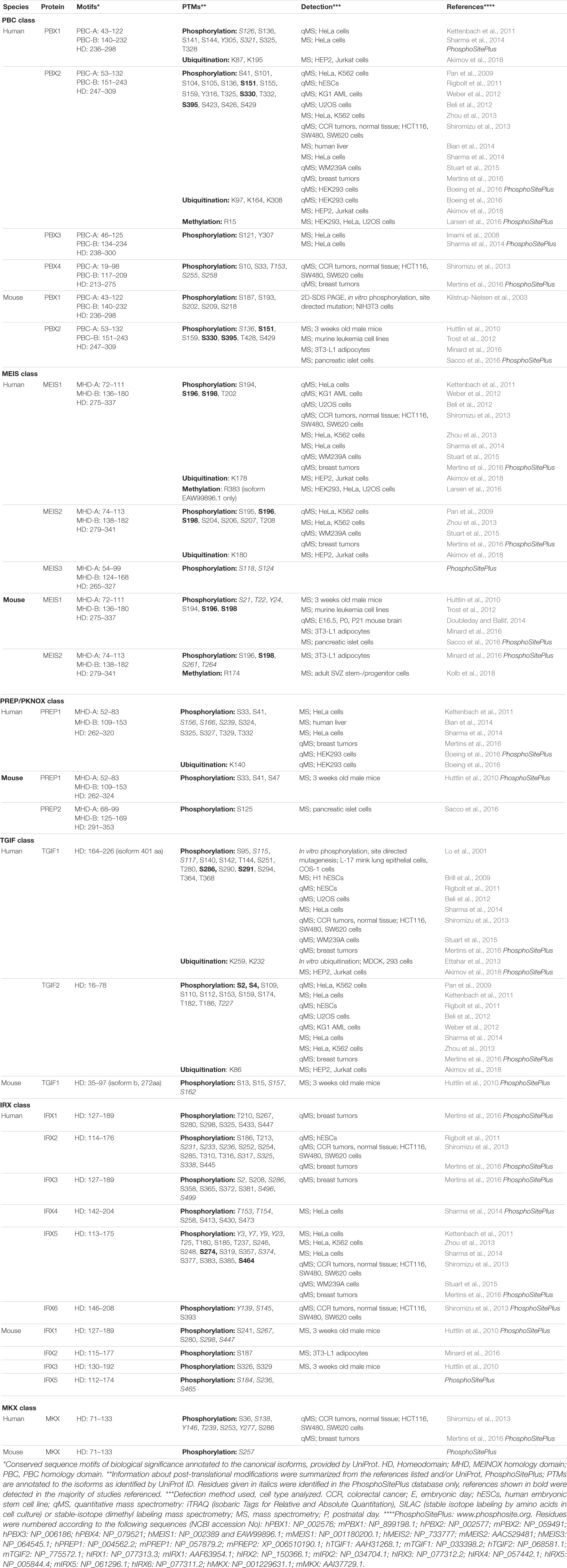
We manually surveyed 26 high-resolution and/or quantitative mass-spectrometry analyses, as well as data deposited in the open-source platform PhosphoSitePlus® to compile PTMs that had been detected in mouse or human TALE-HD proteins (Table 1). Although this information is freely available in the supporting information of the respective publications, it had not been systematically assessed nor had the data been compared among studies or between protein groups. We limited our search to the three PTMs that were most frequently detected in these studies: phosphorylation, lysine-ubiquitination and arginine-methylation. This search identified a total of 187 distinct phosphorylation sites, 11 ubiquitinated and 3 methylated residues. Many of these PTMs were detected in various physiological contexts and across species, suggesting that common regulatory mechanisms apply. Particularly arginine-methylation and lysine-ubiquitination occurred almost exclusively at amino acids that were highly conserved among paralogs, indicating that significant evolutionary pressure may act on these residues (Figures 1C,E,F). The amino acid arginine forms more hydrogen bonds with protein or DNA than any other amino acid, with particularly strong bonds formed with guanine bases and the DNA phosphate backbone (Luscombe et al., 2001). Arginine residues are therefore important to stabilize the intra- and intermolecular interaction of amino acids in proteins and multiprotein complexes as well as the contact of proteins to DNA (Luscombe et al., 2001; Bedford and Clarke, 2009; Lorton and Shechter, 2019). Consequently, methylation of arginine residues in TFs can profoundly alter their function. In fact, although the significance of arginine-methylation in hPBX2 and hMEIS1 is still unknown, methylation of R174 in mMEIS2 controls nucleo-cytoplasmic translocation (Kolb et al., 2018).
In ubiquitination, the 76-amino acid protein ubiquitin is covalently attached to lysine residues of protein substrates. Ubiquitination generates conjugates that widely differ in structure, size, composition, and function (Pickart, 2001). The many ways by which lysine-ubiquitination impacts on gene expression include modification of histone tails and the subsequent change in chromatin structure and the ubiquitin-guided partial processing or full degradation of TFs (Rape, 2018). The presence of several, highly conserved ubiquitination sites in TALE-HD proteins argues for important regulatory roles, although it is presently unexplored what type(s) of ubiquitin modification TALE-HD proteins carry (e.g., monomeric, polymeric, linear, branched, carrying additional PTMs or not), whether ubiquitin-conjugation targets TALE-HD proteins for degradation, and what the cellular consequences of TALE-HD protein ubiquitination are.
Compared to arginine-methylation and lysine-ubiquitination, protein phosphorylation emerges as more wide-spread and diverse type of PTM in TALE-HD proteins. Protein phosphorylation, the covalent attachment of phosphate groups on serine, threonine, or tyrosine residues, acts within milliseconds to seconds to control protein function by primarily two mechanisms: it locally changes the electrochemical properties of a protein and by this its conformation, and it creates docking sites for intermolecular protein interactions, which in turn can propagate cellular signals or create recognition sites for other post-translationally modifying enzymes that catalyze the deposition of further PTMs nearby (Filtz et al., 2014). Phosphorylation of TFs can thereby increase or decrease protein stability, control nuclear import or export, alter the secondary structure of the TF to expose or hide its DBD, and modify the DBD’s affinity to distinct sequences in the DNA resulting in high-affinity or low-affinity binding (Filtz et al., 2014). In TALE-HD proteins, phosphosites often cluster together, frequently in regions anterior or posterior of the HD (Figures 1C–F). For instance, several studies identified phosphorylated serine, threonine, and tyrosine residues in PBX family proteins just C-terminal to the TALE HD (Figure 1C). In particular phosphorylation at T325 and S330 (numeration according to hPBX2, NCBI# NP_002577) had been detected in different mouse tissues (Huttlin et al., 2010), murine pancreatic cells following glucose exposure (Sacco et al., 2016), EGF-stimulated HeLa cells (Pan et al., 2009; Sharma et al., 2014), FGF-stimulated adipocytes (Minard et al., 2016), mouse AML models and human AML cell lines (Trost et al., 2012; Weber et al., 2012), breast cancer samples (Mertins et al., 2016), human embryonic stem cells during differentiation (Rigbolt et al., 2011), and etoposide-treated human osteosarcoma (U2OS) cells during DNA damage response (Beli et al., 2012). Interestingly, only some of these residues are conserved among paralogs. For instance, while phosphorylation is frequent at S330 in PBX2 and at the corresponding S321 in PBX1, PBX3 carries an asparagine residue and PBX4 bears a microdeletion at this position, suggesting that PBX3 and PBX4 may be insensitive to the kinase networks that impact on S321/S330 in PBX1 and PBX2, respectively (Figure 1C). In addition, these differentially phosphorylated sites are close to a NLS (KRIRYKKNI; Saleh et al., 2000). Given that controlled nuclear import is an important mechanism by which the transcriptional activity of TALE-HD proteins is regulated, these observations raise the intriguing possibility that differential phosphorylation at these residues may influence nuclear localization (Mann and Abu-Shaar, 1996; Abu-Shaar et al., 1999; Berthelsen et al., 1999; Huang et al., 2003; Kolb et al., 2018). Supporting this view, protein kinase A (PKA)-mediated phosphorylation of mammalian PBX1 at S187, S193, S202, S209, and S218, all located near a second NLS (RRKRR, N-terminal to helix 1 of the HD), affect nuclear export of PBX1 (Figure 1D; Saleh et al., 2000; Kilstrup-Nielsen et al., 2003).
MEIS1 and MEIS2 proteins exhibit a striking accumulation of phosphosites clustered between the MHR-B domain and the TALE-HD, with frequent phosphorylation at serines 195, 196, 198, 204, 206, 207, and threonine 208 (numeration according to hMEIS2, NCBI# NP_733777; Figure 1E). In fact, phosphorylation at S195/S196/S198 was detected in virtually all phosphoproteomic studies that were examined for this minireview (Table 1). Nestled between these phosphosites are several aspartate and glutamate residues, amino acids with electrically changed, acidic side chains. Phosphorylation at these serine or threonine residues is therefore expected to create a strong, focal negative charge in this region of the MEIS1 and MEIS2 polypeptide.
PTMs, a Way to Generate Functional Diversity?
Although the physiological relevance of these phosphorylation events and the signaling pathways that induce them remain to be elucidated, it is worth pointing out that none of these phosphosites are conserved in MEIS3, PREP1, or PREP2 (Figure 1E). Similarly, most of the phosphorylated amino acids that were detected in TGIF1 are not conserved in TGIF2, and vice versa (Figure 1F). Whether or not TALE-HD paralogous proteins are subject to regulation by shared kinase pathways thus appears to be dictated by the substitution of few key residues. It should be pointed out, however, that phosphorylation is a dynamic process in which phosphorylation and dephosphorylation may alternate in rather rapid cycles (Gelens and Saurin, 2018). Phosphoproteomic data hence only reflect a snapshot of a transient phosphorylation state. Lack of evidence in literature for a specific phosphorylation event can thus very well just reflect the inability of detection at a specific moment and in that specific cellular context.
Taken together, we here compiled a broad collection of PTMs in TALE-HD proteins that had been identified in unbiased, high-resolution mass-spectrometry analyses (Table 1). Few of these PTMs have been assigned a physiological function. Yet, by taking the evolutionary conservation of modification sites into account we identified both class-specific and paralog-specific PTMs. From comparing these, concepts emerge about how the combinatorial use of such PTMs may generate functional diversity from evolutionarily conserved protein structures. Specifically, we propose that the vast repertoire of PTMs, shared or not, in paralogous and orthologous TALE-HD proteins, forms the structural backbone by which individual proteins can acquire the ability to respond to context-specific extracellular signals and exert physiologically diverse functions. Although explored here only by the example of the TALE-HD superclass, similar principles may very well also apply to other evolutionarily conserved TFs. Assays based on mutational approaches now need to be developed to test these PTMs alone and in combination for their functionality and physiological relevance. Ultimately, such information can pave the way for future studies, help unravel disease processes and facilitate rational drug design.
Author Contributions
MR, LE, and DS jointly developed and wrote the review. All authors contributed to the article and approved the submitted version.
Funding
Research leading to this review was supported by the Deutsche Forschungsgemeinschaft (Grants SCHU1218/3-3 and SCHU1218/4-1), the Wilhelm-Sander-Foundation and the Paul and Ursula Klein-Foundation.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the members of the Schulte lab for discussions and critical reading of the manuscript.
References
Abu-Shaar, M., Ryoo, H. D., and Mann, R. S. (1999). Control of the nuclear localization of Extradenticle by competing nuclear import and export signals. Genes Dev. 13, 935–945. doi: 10.1101/gad.13.8.935
Akimov, V., Barrio-Hernandez, I., Hansen, S. V. F., Hallenborg, P., Pedersen, A.-K., Bekker-Jensen, D. B., et al. (2018). UbiSite approach for comprehensive mapping of lysine and N-terminal ubiquitination sites. Nat. Struct. Mol. Biol. 25, 631–640. doi: 10.1038/s41594-018-0084-y
Bailey, J. S., Rave-Harel, N., McGillivray, S. M., Coss, D., and Mellon, P. L. (2004). Activin regulation of the follicle-stimulating hormone beta-subunit gene involves Smads and the TALE homeodomain proteins Pbx1 and Prep1. Mol. Endocrinol. 18, 1158–1170. doi: 10.1210/me.2003-0442
Bedford, M. T., and Clarke, S. G. (2009). Protein arginine methylation in mammals: who, what, and why. Mol. Cell 33, 1–13. doi: 10.1016/j.molcel.2008.12.013
Beli, P., Lukashchuk, N., Wagner, S. A., Weinert, B. T., Olsen, J. V., Baskcomb, L., et al. (2012). Proteomic investigations reveal a role for RNA processing factor THRAP3 in the DNA damage response. Mol. Cell 46, 212–225. doi: 10.1016/j.molcel.2012.01.026
Berthelsen, J., Kilstrup-Nielsen, C., Blasi, F., Mavilio, F., and Zappavigna, V. (1999). The subcellular localization of PBX1 and EXD proteins depends on nuclear import and export signals and is modulated by association with PREP1 and HTH. Genes Dev. 13, 946–953. doi: 10.1101/gad.13.8.946
Bertolino, E., Reimund, B., Wildt-Perinic, D., and Clerc, R. G. (1995). A novel homeobox protein which recognizes a TGT core and functionally interferes with a retinoid-responsive motif. J. Biol. Chem. 270, 31178–31188. doi: 10.1074/jbc.270.52.31178
Bian, Y., Song, C., Cheng, K., Dong, M., Wang, F., Huang, J., et al. (2014). An enzyme assisted RP-RPLC approach for in-depth analysis of human liver phosphoproteome. J. Proteomics 96, 253–262. doi: 10.1016/j.jprot.2013.11.014
Blasi, F., Bruckmann, C., Penkov, D., and Dardaei, L. (2017). A tale of TALE, PREP1, PBX1, and MEIS1: interconnections and competition in cancer. Bioessays 39:1600245. doi: 10.1002/bies.201600245
Boeing, S., Williamson, L., Encheva, V., Gori, I., Saunders, R. E., Instrell, R., et al. (2016). Multiomic analysis of the UV-Induced DNA damage response. Cell Rep. 15, 1597–1610. doi: 10.1016/j.celrep.2016.04.047
Bridoux, L., Zarrineh, P., Mallen, J., Phuycharoen, M., Latorre, V., Ladam, F., et al. (2020). HOX paralogs selectively convert binding of ubiquitous transcription factors into tissue-specific patterns of enhancer activation. PLoS Genet. 16:e1009162. doi: 10.1371/journal.pgen.1009162
Brill, L. M., Xiong, W., Lee, K.-B., Ficarro, S. B., Crain, A., Xu, Y., et al. (2009). Phosphoproteomic analysis of human embryonic stem cells. Cell Stem Cell 5, 204–213. doi: 10.1016/j.stem.2009.06.002
Bruckmann, C., Tamburri, S., Lorenzi, V., Doti, N., Monti, A., Mathiasen, L., et al. (2020). Mapping the native interaction surfaces of PREP1 with PBX1 by cross-linking mass-spectrometry and mutagenesis. Sci. Rep. 10:16809. doi: 10.1038/s41598-020-74032-w
Bruneau, B. G., Bao, Z. Z., Fatkin, D., Xavier-Neto, J., Georgakopoulos, D., Maguire, C. T., et al. (2001). Cardiomyopathy in Irx4-deficient mice is preceded by abnormal ventricular gene expression. Mol. Cell Biol. 21, 1730–1736.
Bürglin, T. R. (1997). Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res. 25, 4173–4180. doi: 10.1093/nar/25.21.4173
Bürglin, T. R., and Ruvkun, G. (1992). New motif in PBX genes. Nat. Genet. 1, 319–320. doi: 10.1038/ng0892-319
Capellini, T. D., Di Giacomo, G., Salsi, V., Brendolan, A., Ferretti, E., Srivastava, D., et al. (2006). Pbx1/Pbx2 requirement for distal limb patterning is mediated by the hierarchical control of Hox gene spatial distribution and Shh expression. Development 133, 2263–2273. doi: 10.1242/dev.02395
Choe, S.-K., Ladam, F., and Sagerström, C. G. (2014). TALE factors poise promoters for activation by Hox proteins. Dev. Cell 28, 203–211. doi: 10.1016/j.devcel.2013.12.011
Choe, S.-K., Lu, P., Nakamura, M., Lee, J., and Sagerström, C. G. (2009). Meis cofactors control HDAC and CBP accessibility at Hox-regulated promoters during zebrafish embryogenesis. Dev. Cell 17, 561–567. doi: 10.1016/j.devcel.2009.08.007
Costantini, D. L., Arruda, E. P., Agarwal, P., Kim, K.-H., Zhu, Y., Zhu, W., et al. (2005). The homeodomain transcription factor Irx5 establishes the mouse cardiac ventricular repolarization gradient. Cell 123, 347–358. doi: 10.1016/j.cell.2005.08.004
Crocker, J., Noon, E. P.-B., and Stern, D. L. (2016). The soft touch: low-affinity transcription factor binding sites in development and evolution. Curr. Top Dev. Biol. 117, 455–469. doi: 10.1016/bs.ctdb.2015.11.018
Dardaei, L., Longobardi, E., and Blasi, F. (2014). Prep1 and Meis1 competition for Pbx1 binding regulates protein stability and tumorigenesis. Proc. Natl. Acad. Sci. U.S.A. 111, E896–E905. doi: 10.1073/pnas.1321200111
Doubleday, P. F., and Ballif, B. A. (2014). Developmentally-Dynamic murine brain proteomes and phosphoproteomes revealed by quantitative proteomics. Proteomes 2, 197–207. doi: 10.3390/proteomes2020191
Draime, A., Bridoux, L., Graba, Y., and Rezsohazy, R. (2018). Post-translational modifications of HOX proteins, an underestimated issue. Int. J. Dev. Biol. 62, 733–744. doi: 10.1387/ijdb.180178rr
Ettahar, A., Ferrigno, O., Zhang, M.-Z., Ohnishi, M., Ferrand, N., Prunier, C., et al. (2013). Identification of PHRF1 as a tumor suppressor that promotes the TGF-β cytostatic program through selective release of TGIF-driven PML inactivation. Cell Rep. 4, 530–541. doi: 10.1016/j.celrep.2013.07.009
Filtz, T. M., Vogel, W. K., and Leid, M. (2014). Regulation of transcription factor activity by interconnected post-translational modifications. Trends Pharmacol. Sci. 35, 76–85. doi: 10.1016/j.tips.2013.11.005
Gaborit, N., Sakuma, R., Wylie, J. N., Kim, K.-H., Zhang, S.-S., Hui, C.-C., et al. (2012). Cooperative and antagonistic roles for Irx3 and Irx5 in cardiac morphogenesis and postnatal physiology. Development 139, 4007–4019. doi: 10.1242/dev.081703
Gelens, L., and Saurin, A. T. (2018). Exploring the function of dynamic phosphorylation-dephosphorylation cycles. Dev. Cell 44, 659–663. doi: 10.1016/j.devcel.2018.03.002
Guca, E., Suñol, D., Ruiz, L., Konkol, A., Cordero, J., Torner, C., et al. (2018). TGIF1 homeodomain interacts with Smad MH1 domain and represses TGF-β signaling. Nucleic Acids Res. 46, 9220–9235. doi: 10.1093/nar/gky680
Huang, H., Paliouras, M., Rambaldi, I., Lasko, P., and Featherstone, M. (2003). Nonmuscle myosin promotes cytoplasmic localization of PBX. Mol. Cell Biol. 23, 3636–3645. doi: 10.1128/MCB.23.10.3636-3645.2003
Huttlin, E. L., Jedrychowski, M. P., Elias, J. E., Goswami, T., Rad, R., Beausoleil, S. A., et al. (2010). A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174–1189. doi: 10.1016/j.cell.2010.12.001
Imami, K., Sugiyama, N., Kyono, Y., Tomita, M., and Ishihama, Y. (2008). Automated phosphoproteome analysis for cultured cancer cells by two-dimensional nanoLC-MS using a calcined titania/C18 biphasic column. Anal Sci. 24, 161–166. doi: 10.2116/analsci.24.161
Ito, Y., Toriuchi, N., Yoshitaka, T., Ueno-Kudoh, H., Sato, T., Yokoyama, S., et al. (2010). The Mohawk homeobox gene is a critical regulator of tendon differentiation. Proc. Natl. Acad. Sci. U.S.A. 107, 10538–10542. doi: 10.1073/pnas.1000525107
Kamps, M. P., Murre, C., Sun, X. H., and Baltimore, D. (1990). A new homeobox gene contributes the DNA binding domain of the t(1;19) translocation protein in pre-B ALL. Cell 60, 547–555. doi: 10.1016/0092-8674(90)90658-2
Kettenbach, A. N., Schweppe, D. K., Faherty, B. K., Pechenick, D., Pletnev, A. A., and Gerber, S. A. (2011). Quantitative phosphoproteomics identifies substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells. Sci. Signal 4:rs5. doi: 10.1126/scisignal.2001497
Kilstrup-Nielsen, C., Alessio, M., and Zappavigna, V. (2003). PBX1 nuclear export is regulated independently of PBX-MEINOX interaction by PKA phosphorylation of the PBC-B domain. EMBO J. 22, 89–99. doi: 10.1093/emboj/cdg010
Kim, K.-H., Rosen, A., Bruneau, B. G., Hui, C.-C., and Backx, P. H. (2012). Iroquois homeodomain transcription factors in heart development and function. Circ. Res. 110, 1513–1524. doi: 10.1161/CIRCRESAHA.112.265041
Knoepfler, P. S., Calvo, K. R., Chen, H., Antonarakis, S. E., and Kamps, M. P. (1997). Meis1 and pKnox1 bind DNA cooperatively with Pbx1 utilizing an interaction surface disrupted in oncoprotein E2a-Pbx1. Proc. Natl. Acad. Sci. U.S.A. 94, 14553–14558. doi: 10.1073/pnas.94.26.14553
Kolb, J., Anders-Maurer, M., Müller, T., Hau, A.-C., Grebbin, B. M., Kallenborn-Gerhardt, W., et al. (2018). Arginine methylation regulates meis2 nuclear localization to promote neuronal differentiation of adult svz progenitors. Stem Cell Rep. 10, 1184–1192. doi: 10.1016/j.stemcr.2018.03.010
Kuang, C., Xiao, Y., Yang, L., Chen, Q., Wang, Z., Conway, S. J., et al. (2006). Intragenic deletion of Tgif causes defectsin brain development. Hum. Mol. Genet. 15, 3508–3519. doi: 10.1093/hmg/ddl427
Lambert, S. A., Jolma, A., Campitelli, L. F., Das, P. K., Yin, Y., Albu, M., et al. (2018). The human transcription factors. Cell 172, 650–665. doi: 10.1016/j.cell.2018.01.029
LaRonde-LeBlanc, N. A., and Wolberger, C. (2003). Structure of HoxA9 and Pbx1 bound to DNA: hox hexapeptide and DNA recognition anterior to posterior. Genes Dev. 17, 2060–2072. doi: 10.1101/gad.1103303
Larsen, S. C., Sylvestersen, K. B., Mund, A., Lyon, D., Mullari, M., Madsen, M. V., et al. (2016). Proteome-wide analysis of arginine monomethylation reveals widespread occurrence in human cells. Sci. Signal 9:rs9. doi: 10.1126/scisignal.aaf7329
Lawrence, H. J., Rozenfeld, S., Cruz, C., Matsukuma, K., Kwong, A., Kömüves, L., et al. (1999). Frequent co-expression of the HOXA9 and MEIS1 homeobox genes in human myeloid leukemias. Leukemia 13, 1993–1999. doi: 10.1038/sj.leu.2401578
Lo, R. S., Wotton, D., and Massagué, J. (2001). Epidermal growth factor signaling via Ras controls the Smad transcriptional co-repressor TGIF. EMBO J. 20, 128–136. doi: 10.1093/emboj/20.1.128
Longobardi, E., Iotti, G., Di Rosa, P., Mejetta, S., Bianchi, F., Fernandez-Diaz, L. C., et al. (2010). Prep1 (pKnox1)-deficiency leads to spontaneous tumor development in mice and accelerates EmuMyc lymphomagenesis: a tumor suppressor role for Prep1. Mol. Oncol. 4, 126–134. doi: 10.1016/j.molonc.2010.01.001
Lorton, B. M., and Shechter, D. (2019). Cellular consequences of arginine methylation. Cell Mol. Life Sci. 76, 2933–2956. doi: 10.1007/s00018-019-03140-2
Luscombe, N. M., Laskowski, R. A., and Thornton, J. M. (2001). Amino acid-base interactions: a three-dimensional analysis of protein-DNA interactions at an atomic level. Nucleic Acids Res. 29, 2860–2874. doi: 10.1093/nar/29.13.2860
Mann, R. S., and Abu-Shaar, M. (1996). Nuclear import of the homeodomain protein extradenticle in response to Wg and Dpp signalling. Nature 383, 630–633. doi: 10.1038/383630a0
Merabet, S., and Galliot, B. (2015). The TALE face of Hox proteins in animal evolution. Front. Genet. 6:267. doi: 10.3389/fgene.2015.00267
Mertins, P., Mani, D. R., Ruggles, K. V., Gillette, M. A., Clauser, K. R., Wang, P., et al. (2016). Proteogenomics connects somatic mutations to signalling in breast cancer. Nature 534, 55–62. doi: 10.1038/nature18003
Minard, A. Y., Tan, S.-X., Yang, P., Fazakerley, D. J., Domanova, W., Parker, B. L., et al. (2016). mTORC1 is a major regulatory node in the FGF21 signaling network in adipocytes. Cell Rep. 17, 29–36. doi: 10.1016/j.celrep.2016.08.086
Mukherjee, K., and Bürglin, T. R. (2007). Comprehensive analysis of animal TALE homeobox genes: new conserved motifs and cases of accelerated evolution. J. Mol. Evol. 65, 137–153. doi: 10.1007/s00239-006-0023-0
Pan, C., Olsen, J. V., Daub, H., and Mann, M. (2009). Global effects of kinase inhibitors on signaling networks revealed by quantitative phosphoproteomics. Mol. Cell Proteomics 8, 2796–2808. doi: 10.1074/mcp.M900285-MCP200
Peltenburg, L. T., and Murre, C. (1996). Engrailed and Hox homeodomain proteins contain a related Pbx interaction motif that recognizes a common structure present in Pbx. EMBO J. 15, 3385–3393.
Peters, T., Dildrop, R., Ausmeier, K., and Rüther, U. (2000). Organization of mouse Iroquois homeobox genes in two clusters suggests a conserved regulation and function in vertebrate development. Genome Res. 10, 1453–1462. doi: 10.1101/gr.144100
Pickart, C. M. (2001). Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70, 503–533. doi: 10.1146/annurev.biochem.70.1.503
Piper, D. E., Batchelor, A. H., Chang, C. P., Cleary, M. L., and Wolberger, C. (1999). Structure of a HoxB1-Pbx1 heterodimer bound to DNA: role of the hexapeptide and a fourth homeodomain helix in complex formation. Cell 96, 587–597. doi: 10.1016/s0092-8674(00)80662-5
Rape, M. (2018). Ubiquitylation at the crossroads of development and disease. Nat. Rev. Mol. Cell Biol. 19, 59–70. doi: 10.1038/nrm.2017.83
Rhee, J. W., Arata, A., Selleri, L., Jacobs, Y., Arata, S., Onimaru, H., et al. (2004). Pbx3 deficiency results in central hypoventilation. Am. J. Pathol. 165, 1343–1350. doi: 10.1016/S0002-9440(10)63392-5
Rigbolt, K. T. G., Prokhorova, T. A., Akimov, V., Henningsen, J., Johansen, P. T., Kratchmarova, I., et al. (2011). System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci. Signal 4:rs3. doi: 10.1126/scisignal.2001570
Sacco, F., Humphrey, S. J., Cox, J., Mischnik, M., Schulte, A., Klabunde, T., et al. (2016). Glucose-regulated and drug-perturbed phosphoproteome reveals molecular mechanisms controlling insulin secretion. Nat. Commun. 7:13250. doi: 10.1038/ncomms13250
Saleh, M., Huang, H., Green, N. C., and Featherstone, M. S. (2000). A conformational change in PBX1A is necessary for its nuclear localization. Exp. Cell Res. 260, 105–115. doi: 10.1006/excr.2000.5010
Schulte, D., and Geerts, D. (2019). MEIS transcription factors in development and disease. Development 146:dev174706. doi: 10.1242/dev.174706
Selleri, L., Depew, M. J., Jacobs, Y., Chanda, S. K., Tsang, K. Y., Cheah, K. S., et al. (2001). Requirement for Pbx1 in skeletal patterning and programming chondrocyte proliferation and differentiation. Development 128, 3543–3557.
Selleri, L., DiMartino, J., van Deursen, J., Brendolan, A., Sanyal, M., Boon, E., et al. (2004). The TALE homeodomain protein Pbx2 is not essential for development and long-term survival. Mol. Cell Biol. 24, 5324–5331.
Selleri, L., Zappavigna, V., and Ferretti, E. (2019). ‘Building a perfect body’: control of vertebrate organogenesis by PBX-dependent regulatory networks. Genes Dev. 33, 258–275. doi: 10.1101/gad.318774.118
Shanmugam, K., Green, N. C., Rambaldi, I., Saragovi, H. U., and Featherstone, M. S. (1999). PBX and MEIS as non-DNA-binding partners in trimeric complexes with HOX proteins. Mol. Cell Biol. 19, 7577–7588. doi: 10.1128/mcb.19.11.7577
Sharma, K., D’Souza, R. C. J., Tyanova, S., Schaab, C., Wiśniewski, J. R., Cox, J., et al. (2014). Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell Rep. 8, 1583–1594. doi: 10.1016/j.celrep.2014.07.036
Shen, J., and Walsh, C. A. (2005). Targeted disruption of Tgif, the mouse ortholog of a human holoprosencephaly gene, does not result in holoprosencephaly in mice. Mol. Cell Biol. 25, 3639–3647.
Shiromizu, T., Adachi, J., Watanabe, S., Murakami, T., Kuga, T., Muraoka, S., et al. (2013). Identification of missing proteins in the neXtProt database and unregistered phosphopeptides in the PhosphoSitePlus database as part of the chromosome-centric human proteome project. J. Proteome Res. 12, 2414–2421. doi: 10.1021/pr300825v
Slattery, M., Riley, T., Liu, P., Abe, N., Gomez-Alcala, P., Dror, I., et al. (2011). Cofactor binding evokes latent differences in DNA binding specificity between Hox proteins. Cell 147, 1270–1282. doi: 10.1016/j.cell.2011.10.053
Star, E. N., Zhu, M., Shi, Z., Liu, H., Pashmforoush, M., Sauve, Y., et al. (2012). Regulation of retinal interneuron subtype identity by the Iroquois homeobox gene Irx6. Development 139, 4644–4655. doi: 10.1242/dev.081729
Stuart, S. A., Houel, S., Lee, T., Wang, N., Old, W. M., and Ahn, N. G. (2015). A phosphoproteomic comparison of B-RAFV600E and MKK1/2 inhibitors in melanoma cells. Mol. Cell Proteomics 14, 1599–1615. doi: 10.1074/mcp.M114.047233
Subramaniam, N., Campión, J., Rafter, I., and Okret, S. (2003). Cross-talk between glucocorticoid and retinoic acid signals involving glucocorticoid receptor interaction with the homoeodomain protein Pbx1. Biochem. J. 370, 1087–1095. doi: 10.1042/BJ20020471
Taniguchi, K., Anderson, A. E., Sutherland, A. E., and Wotton, D. (2012). Loss of Tgif function causes holoprosencephaly by disrupting the SHH signaling pathway. PLoS Genet. 8:e1002524. doi: 10.1371/journal.pgen.1002524
Trost, M., Sauvageau, M., Hérault, O., Deleris, P., Pomiès, C., Chagraoui, J., et al. (2012). Posttranslational regulation of self-renewal capacity: insights from proteome and phosphoproteome analyses of stem cell leukemia. Blood 120, e17–e27. doi: 10.1182/blood-2011-12-397844
Wagner, K., Mincheva, A., Korn, B., Lichter, P., and Pöpperl, H. (2001). Pbx4, a new Pbx family member on mouse chromosome 8, is expressed during spermatogenesis. Mech. Dev. 103, 127–131. doi: 10.1016/s0925-4773(01)00349-5
Wang, Y., Yin, L., and Hillgartner, F. B. (2001). The homeodomain proteins PBX and MEIS1 are accessory factors that enhance thyroid hormone regulation of the malic enzyme gene in hepatocytes. J. Biol. Chem. 276, 23838–23848. doi: 10.1074/jbc.M102166200
Weber, C., Schreiber, T. B., and Daub, H. (2012). Dual phosphoproteomics and chemical proteomics analysis of erlotinib and gefitinib interference in acute myeloid leukemia cells. J. Proteomics 75, 1343–1356. doi: 10.1016/j.jprot.2011.11.004
Wotton, D., Lo, R. S., Lee, S., and Massagué, J. (1999). A Smad transcriptional corepressor. Cell 97, 29–39. doi: 10.1016/s0092-8674(00)80712-6
Yu, W., Li, X., Eliason, S., Romero-Bustillos, M., Ries, R. J., Cao, H., et al. (2017). Irx1 regulates dental outer enamel epithelial and lung alveolar type II epithelial differentiation. Dev. Biol. 429, 44–55. doi: 10.1016/j.ydbio.2017.07.011
Zhang, S.-S., Kim, K.-H., Rosen, A., Smyth, J. W., Sakuma, R., Delgado-Olguín, P., et al. (2011). Iroquois homeobox gene 3 establishes fast conduction in the cardiac His-Purkinje network. Proc. Natl. Acad. Sci. U.S.A. 108, 13576–13581. doi: 10.1073/pnas.1106911108
Keywords: homeodomain protein, PTM, protein phosphorylation, MEIS, PBX, PREP/PKNOX, TGIF, IRX
Citation: Reichlmeir M, Elias L and Schulte D (2021) Posttranslational Modifications in Conserved Transcription Factors: A Survey of the TALE-Homeodomain Superclass in Human and Mouse. Front. Cell Dev. Biol. 9:648765. doi: 10.3389/fcell.2021.648765
Received: 01 January 2021; Accepted: 09 February 2021;
Published: 09 March 2021.
Edited by:
May Khanna, University of Arizona, United StatesReviewed by:
René Rezsohazy, Catholic University of Louvain, BelgiumFrédéric Marmigere, UMR 5242 Institut de Génomique Fonctionnelle de Lyon (IGFL), France
Copyright © 2021 Reichlmeir, Elias and Schulte. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dorothea Schulte, ZG9yb3RoZWEuc2NodWx0ZUBrZ3UuZGU=
†These authors have contributed equally to this work
 Marina Reichlmeir
Marina Reichlmeir Lena Elias
Lena Elias Dorothea Schulte
Dorothea Schulte