- 1Fujian Key Laboratory of Developmental and Neural Biology & Southern Center for Biomedical Research, College of Life Sciences, Fujian Normal University, Fuzhou, China
- 2Key Laboratory of Stem Cell Engineering Societ and Regenerative Medicine, School of Basic Medical Science, Fujian Medical University, Fuzhou, China
Palate-derived growth factor receptor α (Pdgfrα) signaling has been reported to play important roles in the cardiac development. A previous study utilizing Pdgfrα conventional knockout mice reported hypoplasia of the sinus venous myocardium including the sinoatrial node (SAN) accompanied by increased expression of Nkx2.5. This mouse line embryos die by E11.5 due to embryonic lethality, rendering them difficult to investigate the details. To elucidate the underlying mechanism, in this study, we revisited this observation by generation of specific ablation of Pdgfrα in the SAN by Shox2-Cre at E9.5, using a Shox2-Cre;Pdgfrαflox/flox conditional mouse line. Surprisingly, we found that resultant homozygous mutant mice did not exhibit any malformation in SAN morphology as compared to their wild-type littermates. Further analysis revealed the normal cardiac function in adult mutant mice assessed by the record of heart rate and electrocardiogram and unaltered expression of Nkx2.5 in the E13.5 SAN of Pdgfrα conditional knockout mice. Our results unambiguously demonstrate that Pdgfrα is dispensable for SAN development after its fate commitment in mice.
Introduction
The mature sinoatrial node (SAN), composed of an Nkx2.5– head and an Nkx2.5+ sinoatrial junction or tail, is a group of specialized cells with obvious heterogeneity located on the dorsal wall of the right atrium close to the entrance of superior vena cava, functioning as the primary cardiac pacemaker that regulate the rhythm of the heartbeat by spontaneously producing electrical impulses (Christoffels et al., 2006; Yamamoto et al., 2006; Liu et al., 2007; Liang et al., 2013). The first heartbeat is recorded in the left inflow tract region as early as embryonic day (E) 8.0-E8.5 in mice (Yi et al., 2012), then the pace-making activity is localized to the right inflow (Dyer and Kirby, 2009; Csepe et al., 2015). These pacemaker cells will contribute to the definitive SAN and become morphologically discernible at E10.5 and form the comma-like structure with the SAN-like action potential configurations at E12.5 (Virágh and Challice, 1982). Deficiencies in its pacemaking function leads to cardiac arrhythmias, cardiac arrest, sinoatrial exit block, and even sudden cardiac death (Durham and Worthley, 2002).
A number of genes, including Hcn4 (hyperpolarization-activated cyclic nucleotide-gated channel subtype 4), Tbx3, Shox2, and Nkx2.5, work as cardiac markers and are critical for the development of SAN. For instance, mutation in Shox2 results in a severely hypoplastic SAN due to ectopic Nkx2.5 expression in the head of SAN (Blaschke et al., 2007; Espinoza-Lewis et al., 2009). Nkx2.5 is expressed in the precardiac tissues as one of early cardiac markers and is a key transcription factor for the development of working myocardium (Lints et al., 1993; Moses et al., 2001). Nkx2.5 mutant display early embryonic lethality due to heart defects. However, Nkx2.5 is only expressed in the junction of a developing SAN but absent in the head because of the transcription repression by Shox2 and Tbx3 (Christoffels et al., 2006; Espinoza-Lewis et al., 2009; Ye et al., 2015a,b). Mice lacking Nkx2.5 in the SAN junction adopt normal morphology but physiological dysfunction, indicating that Nkx2.5 is essential for SAN function but is dispensable for SAN morphogenesis (Li et al., 2019).
Platelet-derived growth factor receptor (PDGFR), located on the surface of a wide range of cell types, is bound by certain isoforms of PDGF ligands and thereby becomes active in stimulating cell signaling pathways that elicit responses including growth, development, and differentiation. There are two types of PDGFR, α and β, functioning in the form of homodimer, α-α (termed as Pdgfrα) and β-β (termed as Pdgfrβ), or heterodimer, α-β (termed as Pdgfrα-β) (Heldin and Westermark, 1999). Each type of the dimmer plays its distinct physiological function and does not display functional redundancy among them (Rosenkranz and Kazlauskas, 1999). Pdgfrα signaling plays a critical role in cardiac development, including the development of primary cardiac muscle, venous, and mesenchymal myocardium (Bax et al., 2010). Deletion of Pdgfrα causes neurological stenosis-related malformations at the heart outflow tract, such as persistent truncus arteriosus, double outlet right ventricle, ventricular myocardial insufficiency, and abnormal pulmonary veins (Morrison-Graham et al., 1992; Schatteman et al., 1995).
A previous study using green fluorescent protein (GFP) as a reporter in a knock-in mouse line (PdgfrαGFP) reported that Pdgfrα is markedly expressed at the venous pole in the mesocardium, the myocardium of the sinus venosus, the proepicardial organ, and the coelomic mesothelium in the E9.5 heart, as well as in the developing SAN at E10.5. Several cardiac malformations including the significantly smaller SAN accompanied by increased expression of Nkx2.5 were further observed in Pdgfrα conventional knockout mouse line (Bax et al., 2010). Unfortunately, most Pdgfrα–/– mice, even with intraperitoneal injection of isoprenaline in maternal mice, suffer severe early embryonic lethality by E11.5 when crossed in C57/B6 background in our animal facility, preventing detailed assessment of cellular and molecular events contributing to the abnormal morphology and function of sinus node, atrium, and ventricle. Here we attempted to further address this question using a conditional Pdgfrα knockout mouse line Shox2-Cre;Pdgfrαflox/flox, in which Pdgfrα expression was specifically ablated in the myocardium of the sinus venosus at E9.5 when the Cre activity is initiated (Sun et al., 2013). Unexpectedly, our results showed that homozygous mutant mice exhibit not only normal SAN morphology with normal heartbeat rate and electrocardiogram, but also unaltered Nkx2.5 expression pattern in the SAN, indicating dispensability of Pdgfrα signaling after E9.5 for the development and differentiation of the SAN.
Materials and Methods
Mouse Line
Shox2-Cre, Pdgfrαflox/flox and Rosa26mTmG mice were reported previously and obtained from Jackson Laboratory (Soriano, 1997; Muzumdar et al., 2007; Sun et al., 2013). To specifically inactivate Pdgfrα in the SAN, we crossed the mice carrying Shox2-Cre with Pdgfrαflox/flox mice to generate Pdgfrα conditional knockout mouse line Shox2-Cre;Pdgfrαflox/flox. We also compound Shox2-Cre allele with Rosa26mTmG reporter mice for the lineage tracing of Shox2-Cre positive cells. All animal experiments in this study were approved by The Fujian Normal University Institutional Animal Care and Use Committee.
Histology and Immunofluorescence
Mouse embryos were harvested from timed pregnant females. Bodies of collected embryos were fixed in 4% Paraformaldehyde at 4°C for 24 h and further processed for paraffin embedding and sectioning at 7 μm. Slides were subjected to Hematoxylin/eosin staining for histology analysis or immunofluorescent staining as described previously (Hu et al., 2014). The mean fluorescence intensity of Pdgfrα expression in SAN was quantified by using ImageJ software (National Institutes of Health). Antibodies used for this study include anti-Pdgfrα (R&D, AF1062), anti-Hcn4 (Abcam, ab32675), anti-Nkx2.5 (Santa Cruz, sc8697), and anti-GFP (Abcam, ab13970).
3D Reconstruction
Images of consecutive H&E stained sections (7 μm) of SAN and surrounding tissues were captured, stacked, and loaded into Amira 6.0.1, in which subsequent alignment, segmentation, and 3D model generation were performed.
Surface Electrocardiography
Adult Shox2-Cre;Pdgfrαflox/flox mice and their wild-type littermates were anesthetized with 1.5% isoflurane supplemented with O2 and placed in prone position on a Mouse Monitor S (Indus Instruments) to be recorded as reported previously (Ye et al., 2015a,b).
scRNA-seq Data Analysis
The protocol of the E13.5 SAN scRNA-seq was described in a previous study (Li et al., 2019) and this dataset is accessible under the GEO series accession number GSE130461. The E16.5 SAN scRNA-seq raw data was downloaded from NCBI/GEO datasets under accession number GSE32658. The visualization of Pdgfrα and Nkx2.5 distributions was obtained with Uniform Manifold Approximation and Projection (UMAP) embedding.
Statistical Analysis
Statistical analysis on the volume measurement were performed with independent unpaired Student’s t test. All data of the volume measurements are presented as mean ± SEM. Graphics of statistical analysis were performed with GraphPad Prism 5.0 software. P < 0.05 was considered statistically significant.
Results
Expression of Pdgfrα in the Developing SAN
It has been reported that Pdgfrα is expressed as early as E7.5 in several cardiac progenitor tissues including SAN during early heart development (Prall et al., 2007; Bax et al., 2010; Bloomekatz et al., 2017). To comprehensively document the Pdgfrα expression pattern during SAN morphogenesis, we further performed immunostaining of anti-Pdgfrα antibody in the developing SAN of wild-type mice from E11.5 to E16.5. Since the SAN is a structurally heterogeneous tissue composed of a head and junction with distinct gene expression profile (Li et al., 2019), we examined the expression of Pdgfrα in both domains, respectively. We found that, at E11.5, a weak expression of Pdgfrα was seen in both the head and junction of the SAN (Figures 1A,B). Whereas at E12.5 and E13.5, the expression of Pdgfrα reached to a relatively higher expression level in both of the domains (Figures 1C–F). At subsequent E14.5 to E16.5, Pdgfrα expression became dramatically decreased and manifested dim and sporadic pattern in the SAN (Figures 1G–L). By virtue of semi-quantitative analysis of fluorescence intensity at the different stage (N = 4/each stage), this pattern of Pdgfrα expression was confirmed (Figure 1M). Together, immunostaining data revealed that the expression of Pdgfrα in the developing SAN was weak at E11.5, then gradually upregulated to maximal levels at E13.5, and tapered to a faint level at E14.5 thereafter when the morphology and function of SAN is well established. In addition, Pdgfrα is evenly expressed in the whole SAN without obviously distinct pattern in both regions of the SAN. These observations implied that Pdgfrα may play a regulatory role during SAN development.
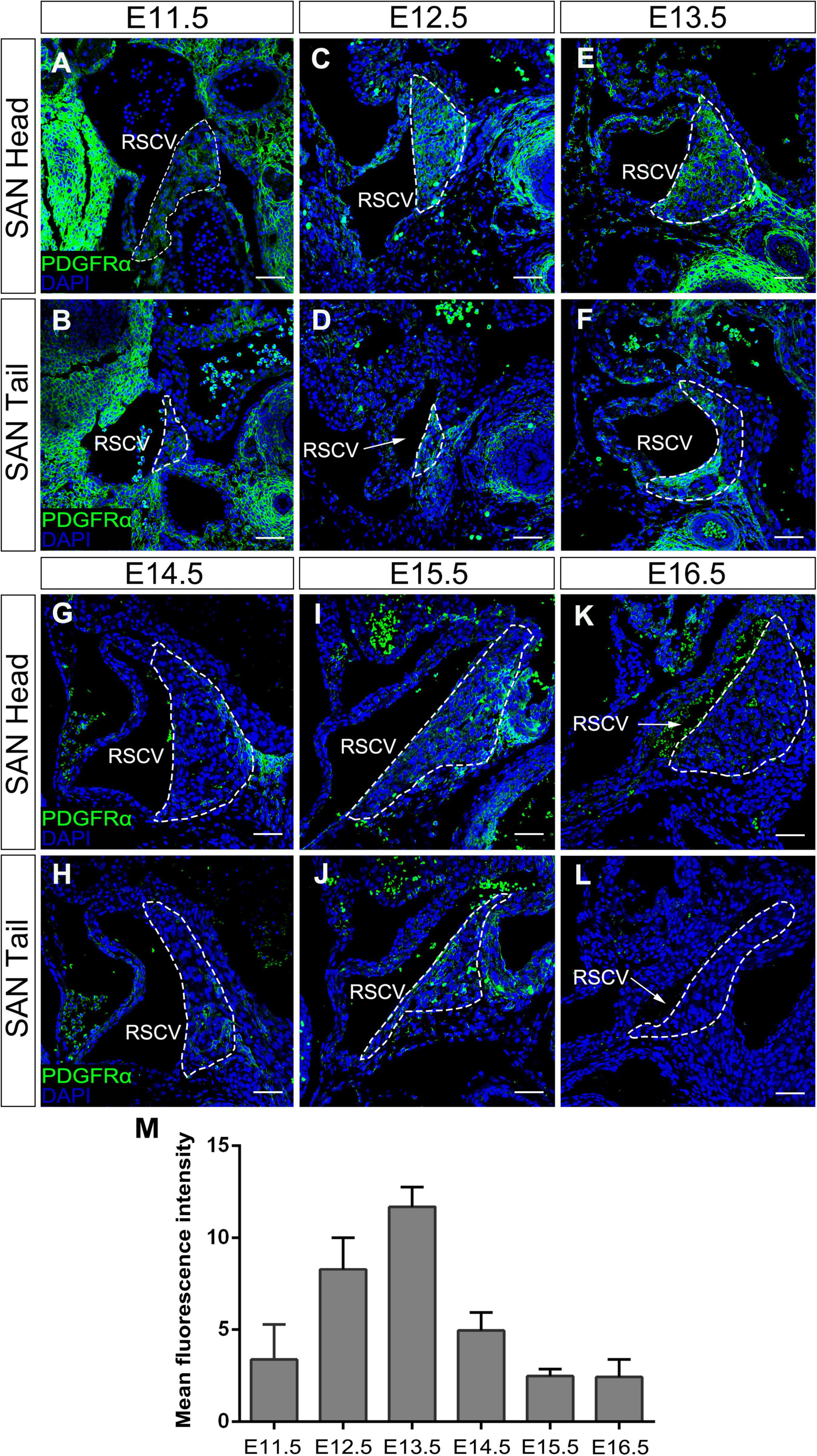
Figure 1. The expression pattern of Pdgfrα in the developing SAN. Immunostaining showing the expression of Pdgfrα in the head of the developing SAN (dotted lines) at E11.5 (A), E12.5 (C), E13.5 (E), E14.5 (G), E15.5 (I), and E16.5 (K) and in the junction of the developing SAN at E11.5 (B), E12.5 (D), E13.5 (F), E14.5 (H), E15.5 (J), and E16.5 (L). (M) Semi-quantitative analysis of Pdgfrα fluorescent intensity in the developing SAN. RSVC, right superior vena cava. Scale bar = 100 μm.
No Morphological and Physiological Anomaly in the SAN of Shox2-Cre; Pdgfrαflox/flox Mice
To further investigate the role of Pdgfrα during SAN development and avoid embryonic lethality, we next generated a conditional knockout mouse line Shox2-Cre;Pdgfrαflox/flox, which exhibit Cre activity in the sinus venosus as early as E9.5 (Sun et al., 2013). To validate the efficiency of Cre recombinase in inactivating floxed gene, we crossed Shox2-Cre mice to Cre reporter Rosa26mTmG allele. Immunostaining for GFP and Pdgfrα showed that Pdgfrα was clearly present in the sinus venosus of E9.5 Shox2Cre/+;Rosa26mTmG control embryos (Figures 2A,C) but absent from that of the mutant ones (Figures 2B,D), confirming a successful knockout of Pdgfrα in the sinus venosus of Shox2-Cre;Pdgfrαflox/flox mice at this early stage. But to our surprise, analysis of electrocardiogram (ECG) in adult Shox2-Cre;Pdgfrαflox/flox mice showed that the electrocardiograms are normal with the P waves corresponding to atrial depolarization clearly identical to their adult wild-type littermates and each P wave followed by a normal QRS complex corresponding to ventricular depolarization (Figures 2E,F; N = 6/each group). This normal electrocardiogram from Shox2-Cre;Pdgfrαflox/flox mice indicated that cardiac function, particularly the pacemaking function of the SAN, was normal in the Pdgfrα conditional knockout mice. Given the fact that a smaller SAN was acquired with 37% reduction in volume at E13.5 in Pdgfrα conventional knockout mice in the previous study (Bax et al., 2010), we further performed morphological analysis and 3D reconstruction of the SAN from E13.5 mutant embryos. As shown in Figure 3, we did not find any morphological malformation and volume difference in the mutants as compared to their littermate controls (Figure 3, N = 3/each group). Therefore, our results provided compelling evidence that Pdgfrα is dispensable for SAN morphogenesis and physiological function in mice when it is ablated with Shox2-Cre at E9.5.
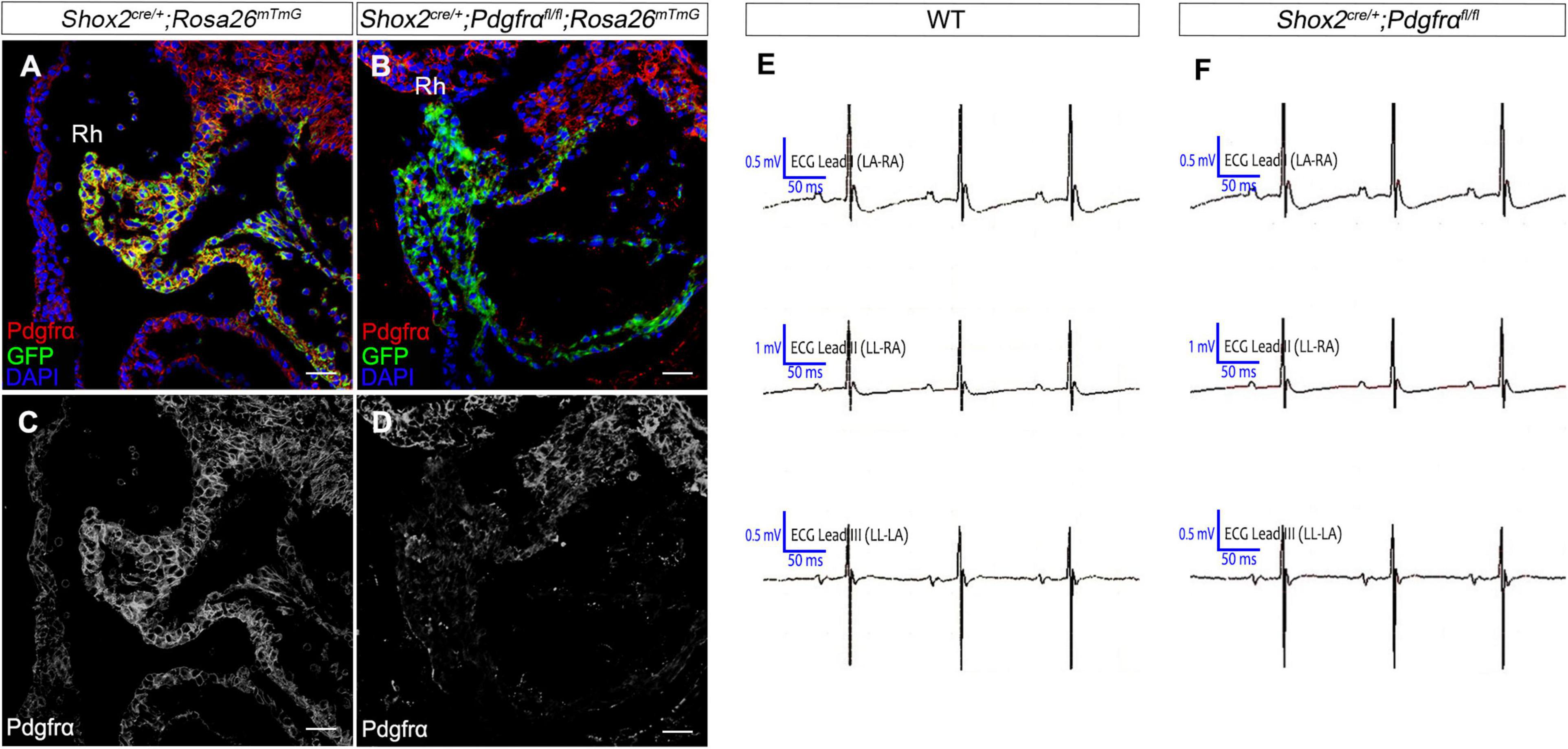
Figure 2. Normal cardiac function in Pdgfrα conditional knockout mice. (A–D) Co-immunostaining of Cre reporter mGFP and Pdgfrα verifies specific deletion of Pdgfrα in the E9.5 Shox2-Cre;Pdgfrαflox/flox;Rosa26mTmG sinus venosus as compared to the positive staining in the Shox2cre/+;Rosa26mTmG controls. (E,F) Representative recording of electrocardiogram (ECG) reveals normal cardiac function from Pdgfrα conditional knockout mice compared to that from the wild-type. Rh, right horn of sinus venosus. Scale bar = 50 μm.
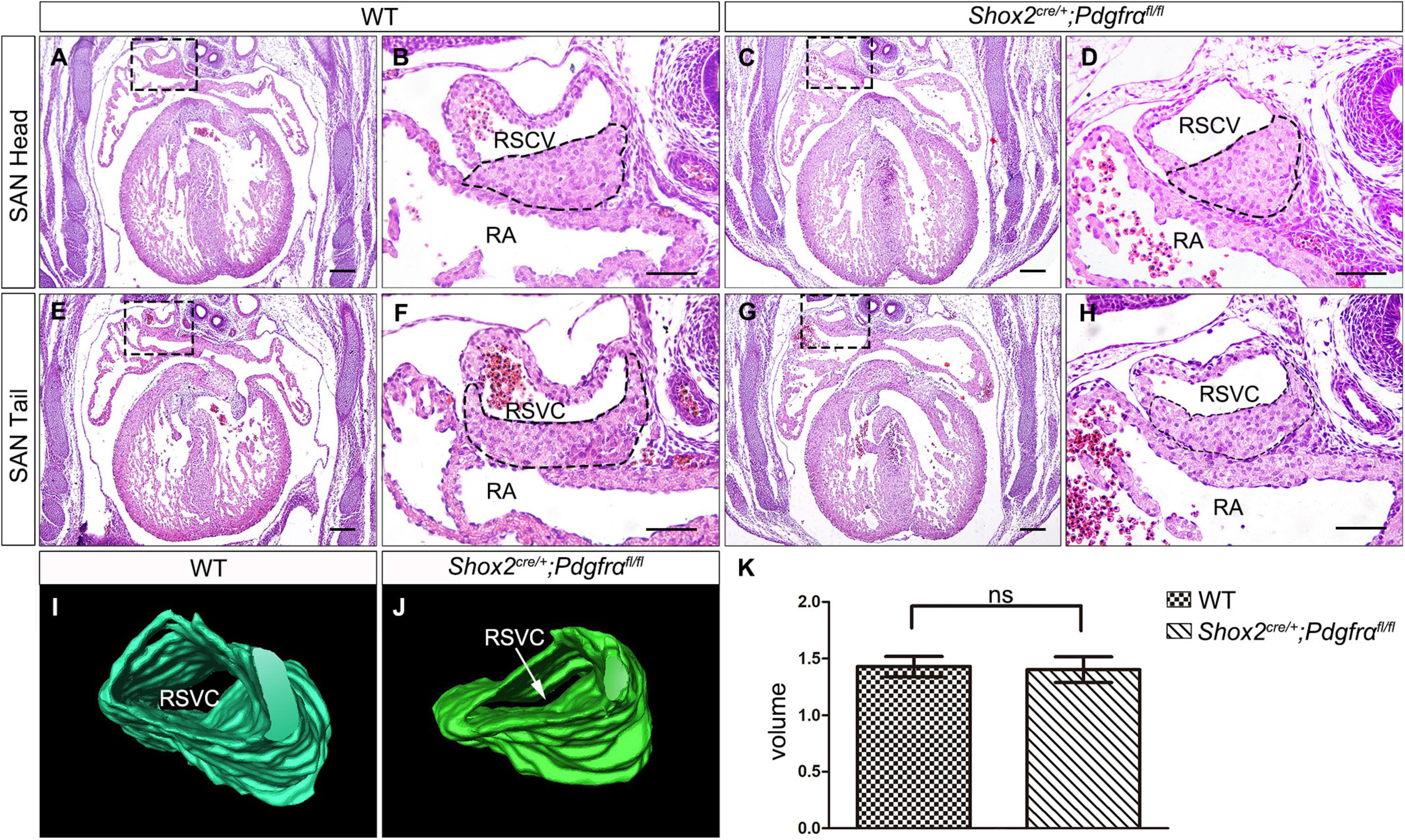
Figure 3. Formation of normal SAN in Pdgfrα conditional knockout mice. (A–H) H&E staining reveals the SAN with normal morphological characteristics in the E13.5 Shox2-Cre;Pdgfrαflox/flox developing heart as compared to the wild-type. (I,J) 3D reconstruction of the SAN from E13.5 wild-type and mutant heart. (K) Statistical analysis based on 3D reconstruction shows no significant difference in cardiac volume between Pdgfrα conditional knockout mice and controls. RA, right atrium; RSVC, right superior vena cava. Scale bar = 50 μm.
Unaltered Expression of Nkx2.5 in the SAN of Shox2-Cre; Pdgfrαflox/flox Mice
Although the SAN is a heterogenetic tissue with an Nkx2.5– head and an Nkx2.5+ junction (Christoffels et al., 2006; Ye et al., 2015a), the fact that overdosed Nkx2.5 in the head led to severe hypoplasia of the SAN and that the mice with Nkx2.5 deletion in SAN show normal SAN morphogenesis but SAN dysfunction indicates that Nkx2.5 activity is detrimental to formation of the SAN (Espinoza-Lewis et al., 2011; Li et al., 2019). Prior study showed the formation of a significant smaller SAN in conventional Pdgfrα knockout mice was accompanied with increased Nkx2.5 expression (Bax et al., 2010), while our results showed formation of the SAN with normal morphology and physiological function in Shox2-Cre;Pdgfrαflox/flox mice (Figure 3), we reasoned that the expression of Nkx2.5 would be unaltered in the SAN of the mutant mice. We, therefore, subsequently examined the expression of Nkx2.5 in the SAN of Shox2-Cre;Pdgfrαflox/flox mice. Our immunostaining showed that, while there is no Nkx2.5 present in the SAN head marked by Hcn4 (Figures 4A–D), Nkx2.5 is indeed expressed in the junction of the mutants in the pattern similar to that of the wild-type one (Figures 4E–H; Wiese et al., 2009; Liang et al., 2013), indicating unaltered expression of Nkx2.5 in the SAN of Shox2-Cre;Pdgfrαflox/flox mice compared to the controls. Thus, our results demonstrated that the lack of Pdgfrα posed no evidence of ectopic or enhanced expression of Nkx2.5 in both the head and junction of the SAN. In consistent with this observation, analysis of scRNA-seq data from the E13.5 SAN revealed that, of the eight visualized clusters (Li et al., 2019), Pdgfrα was present in smooth muscle cells (SMC), mesenchymal cells (MC), endothelial cells (EC) and cardiomyocytes (CM), but Nkx2.5 was only restricted in CM and appeared not overlapped with Pdgfrα (Figure 5A-left). Further analysis of the cells in the CM cluster identified three clusters, in which the expression of Pdgfrα and Nkx2.5 was indeed not co-expressed in the same individual cells (Figure 5A-right). This non-co-expression pattern of Pdgfrα and Nkx2.5 in the same individual cells of the SAN was further confirmed with the analysis of scRNA-seq data from the E16.5 SAN (Figure 5B). Obviously, Pdgfrα, as a membrane-residing signaling molecule, would not exert its regulatory function on the cells where it is not expressed.
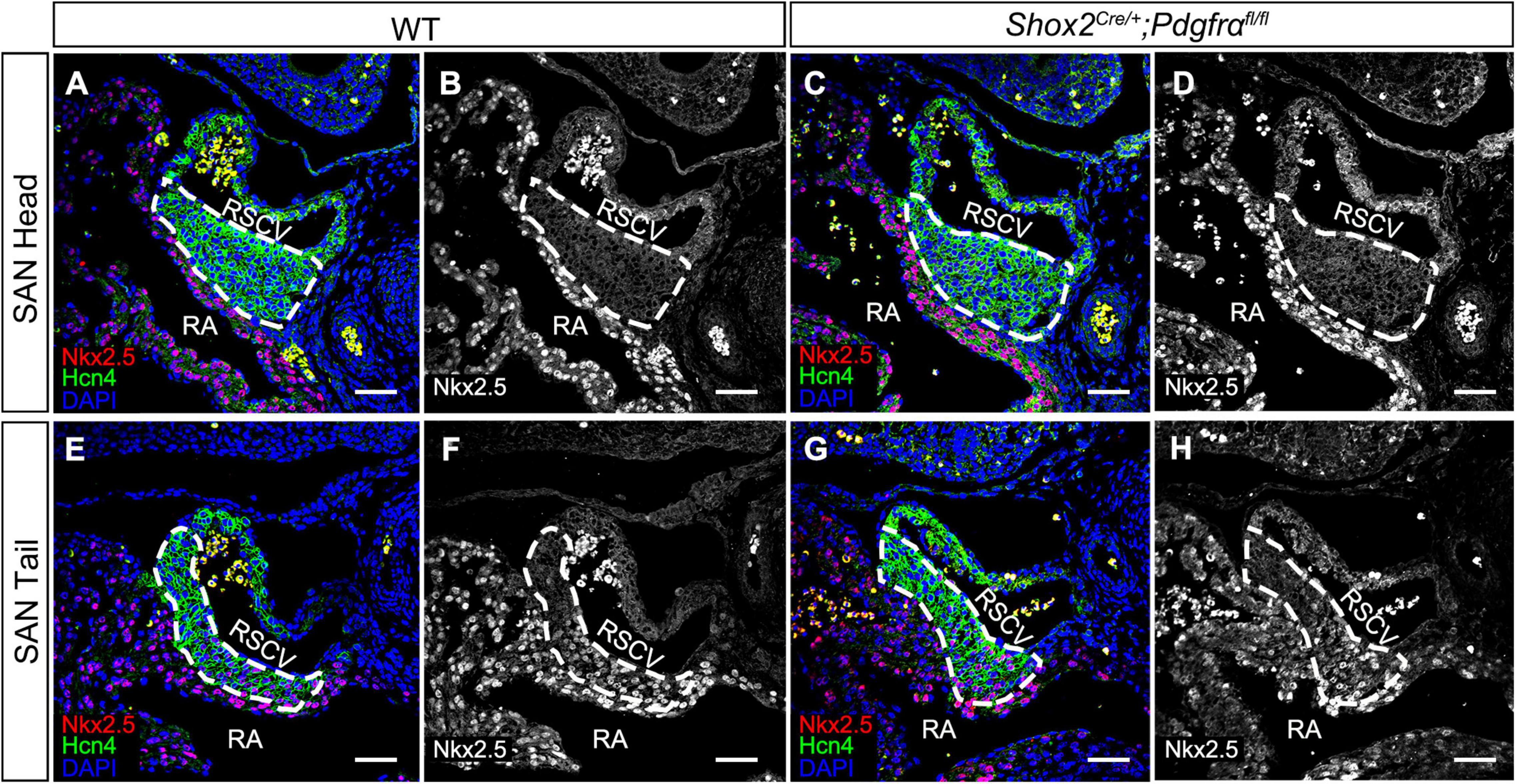
Figure 4. Unaltered Nkx2.5 expression in the SAN of Pdgfrα conditional knockout mice. (A–D) Negative staining of Nkx2.5 in the SAN head, marked by Hcn4, of the E13.5 wild-type and mutant heart. (E–H) A similar expression pattern of Nkx2.5 is detected in the SAN junction, marked by Hcn4, of the E13.5 mutant heart as compared to the wild-type. RA, right atrium; RSVC, right superior vena cava. Scale bars = 50 μm.
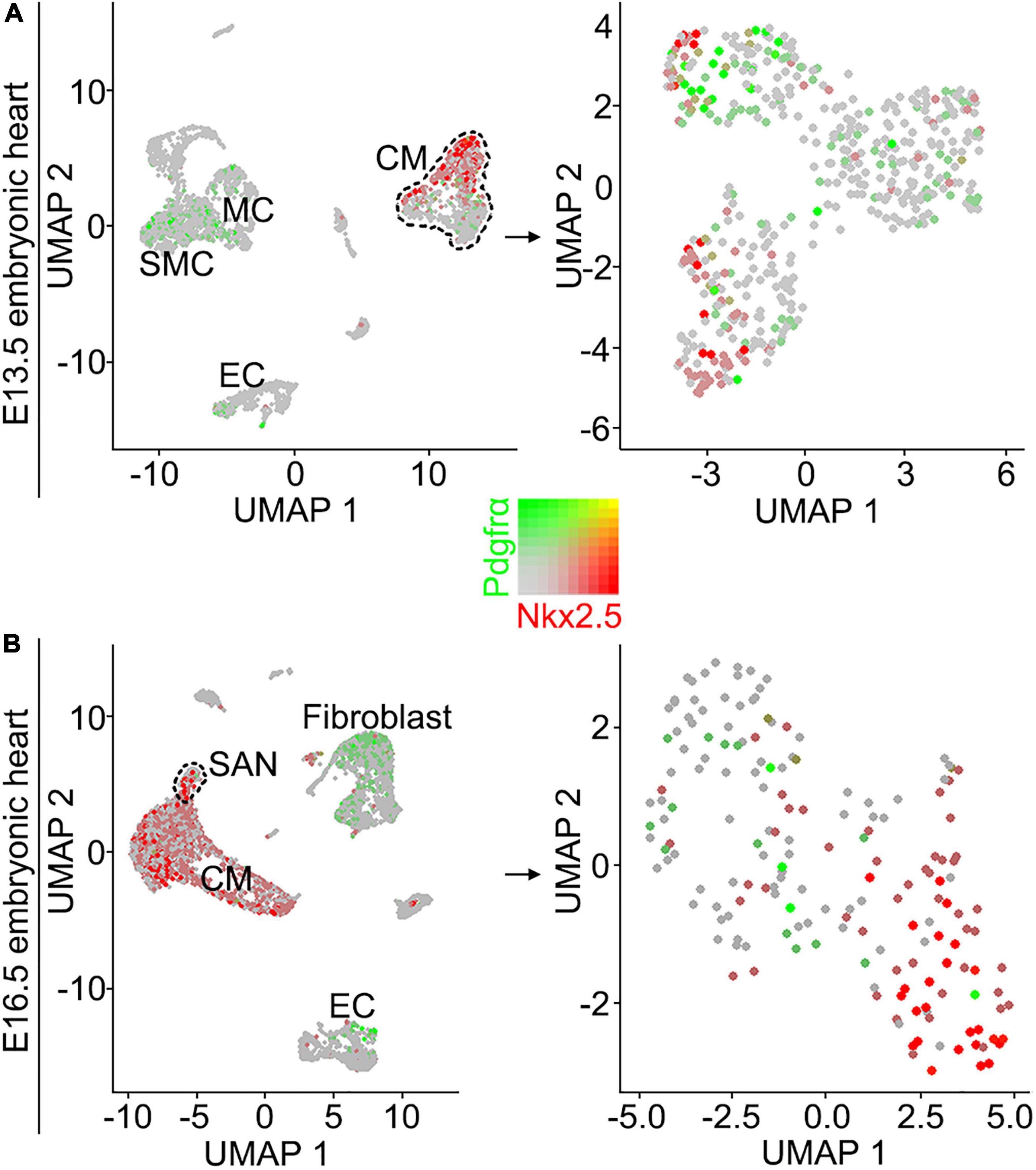
Figure 5. Analysis of scRNA-seq data reveals that Pdgfrα and Nkx2.5 are not co-expressed in the same individual cells of the E13.5 and E16.5 SAN. (A) UMAP plots showing the distribution of Pdgfrα (green) and Nkx2.5 (red) in the eight defined clusters (left) and three subset of the CM cluster (right) on the SAN and atrial cells from E13.5 Shox2cre/+;R26RmTmG mice. (B) UMAP plots showing the distribution of Pdgfrα and Nkx2.5 in the eight defined clusters (left) and the subset of SAN cluster (right) on the SAN and atrial cells from E16.5 WT mice. SMC, smooth muscle cells; MC, mesenchymal cells; CM, cardiomyocyte; EC, endothelial cells.
Discussion
As a heterogeneous structure, the SAN is composed of a head and junction and several specialized cell types including myocardial pacemaker cells and a substantial number of non-pace maker cells. At E8.0-8.5, the dominant pacemaking activity originates from the left inflow tract region (Yi et al., 2012). By heart looping stage, pacemaking activity is localized to the right inflow tract (Dyer and Kirby, 2009; Csepe et al., 2015). These cells are eventually restricted to the dorsal wall of the right atrium and become a morphologically discernible SAN at E10.5, which further proliferates and matures at E13.5 (Virágh and Challice, 1982). Lineage studies indicate that the Nkx2.5– head and Nkx2.5+ junction are derived from distinct progenitor population and represent separate regulatory domain expressing distinctive gene programs at E7.5 to E8.5 (Liang et al., 2013; Später et al., 2013). Formation of the SAN is tightly regulated by a complex network comprized of various signaling molecules, their receptors, and the intracellular signal transduction and transcription system (Wiese et al., 2009; Munshi, 2012). Reciprocal interactions among cardiac progenitor cells, mediated via secreted growth factor activated signaling including Pdgfrα signaling, are critical players in establishment of such a network (Bax et al., 2010).
The expression of Pdgfrα can be observed in the anterior lateral plate mesoderm as early as E7.5 and in caudal the second heart field (SHF) progenitors and dorsal mesocardium at E8.0 (Prall et al., 2007; Bloomekatz et al., 2017). Its expression then identifies cardiac progenitor cells in the posterior part of SHF at E8.5 and can be found at the venous pole in the mesocardium, the sinus venosus myocardium at E9.5 in mouse (Van Den Akker et al., 2005; Bax et al., 2010). At E10.5 when SAN is morphologically discernible, the developing SAN is also positive for the receptor (Van Den Akker et al., 2005). Our immunostaining reveals that Pdgfrα remains apparently expressed until E13.5 when the SAN is becoming mature. Conventional deficiency of Pdgfrα signaling by targeted disruption of this gene in mice led to atrial and sinus venosus myocardium hypoplasia, including formation of hypoplastic SAN with 37% reduced volume and increased myocardial expression of Nkx2.5 (Bax et al., 2010). However, in this study, we found that a SAN was formed with normal morphology and ECG in Shox2-Cre;Pdgfrαflox/flox mice where Pdgfrα was specifically ablated using Shox2-Cre allele.
Several Cre lines have been designed previously for cardiac-specific excision of loxP-flanked target genes (Chien, 2001; Hoesl et al., 2008; Arnolds and Moskowitz, 2011; Sun et al., 2013). Among them, Hcn4-CreERT2 allele represents the first tool for deletion of floxed genes selectively in the cardiac conduction system since Hcn4 is expressed specifically in both the sinoatrial and atrioventricular node (Hoesl et al., 2008). In comparison with Hcn4, Shox2 expression is restricted to the inflow tract and sinus venosus in the posterior heart field that give rise to the SAN and the sinus valves. Expression of Shox2 initiates in a restricted pattern at the junction formed by the common cardinal vein, the common atrial chamber and the sinus venosus at E8.5, restricted to the forming sinus horns region at the junction between the common cardinal vein and the common atrium (the sinus venosus) at E9.5, extends to the sinus valves originating from the SAN at E10.5, and becomes restricted to the entire SAN including the head and tail regions at E11.5 and later (Espinoza-Lewis et al., 2009; Ye et al., 2015b). Moreover, Sun et al. (2013) constructed the Shox2-Cre knock-in mice, and they found the Cre activity was first detected in the developing sinus venosus at E9.5 and subsequently at E10.5 and E11.5 in all tissues with the expression of Shox2, including the SAN, the proximal domain of the limb, the anterior region of the palatal shelves, the temporomandibular junction, as well as the dorsal root ganglia and brain. These results indicate that the Shox2-Cre allele is another suitable Cre line to manipulate gene function in SAN development and the ablation of Pdgfrα with Shox2-Cre will specifically inactivate the Pdgfrα signaling in the tissue that give rise to the SAN and the sinus valve at not earlier than E9.5 when Shox2 is expressed. Whereas, ablation of Pdgfrα in conventional knockout mouse line will inactivate Pdgfrα signaling in all tissue including cardiac progenitor cells and their derived tissues as early as E7.5. Given the importance of the interaction among cardiac progenitor cells and the fact that the production of electrical impulse by early pacemaker cells as early as E8.0-8.5, we believe that there is a cell population designated for the fate of SAN and their developmental fate has been determined at E8.5 when Shox2 is expressed. Pdgfrα signaling is crucial for commitment of these SAN progenitor cells, and once the fate of these cells are committed, Pdgfrα signaling is no longer required.
In addition, conventional deficiency of Pdgfrα results in increased myocardial expression of Nkx2.5, leading to formation of hypoplastic SAN (Bax et al., 2010). Nkx2.5 is a cardiac differentiation marker, which is expressed in the SAN junction (Espinoza-Lewis et al., 2009; Ye et al., 2015b). Although mice lacking Nkx2.5 in the SAN junction adopt normal morphology, the physiological function is impaired, indicating Nkx2.5 is dispensable for SAN morphogenesis but required for physiological function. Formation of hypoplastic SAN caused by overexpression of Nkx2.5 in the heart and overdosed Nkx2.5 in the SAN head when Shox2 repressive function is deficient further supports the idea that overdosed Nkx2.5 is detrimental to SAN morphogenesis (Espinoza-Lewis et al., 2009, 2011; Liu et al., 2012). Our results showed that Nkx2.5 expression in Pdgfrα conditional knock-out mice was unaltered and that Pdgfrα and Nkx2.5 were not co-expressed in the same individual cells of the mature SAN, suggesting that Pdgfrα signaling exerts only its early function on repression of Nkx2.5 during development of posterior heart field derived cardiac structures (Bax et al., 2010).
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics Statement
The animal study was reviewed and approved by The Animal Ethical and Welfare Committee of Fujian Normal University.
Author Contributions
XuH designed the study and administrated the whole study. XZ and FW conducted experiments and collected data and drafted the manuscript. HL provided the E13.5 SAN scRNA-seq raw data. XiH analyzed the scRNA-seq data. ZG involved in methodology and statistical analysis. YZ and XH performed manuscript reviewing and polishing. All authors gave final approval of the version to be published, and agreed to be accountable for all aspects of this work.
Funding
This study was supported by the National Natural Science Foundation of China (Grant Nos. 81870739, 81271102, and 81771034) and the Natural Science Foundation of Fujian Province (Grant No. 2015I0011).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Arnolds, D. E., and Moskowitz, I. P. (2011). Inducible recombination in the cardiac conduction system of minK: creert2 BAC transgenic mice. Genesis 49, 878–884. doi: 10.1002/dvg.20759
Bax, N. A. M., Bleyl, S. B., Gallini, R., Wisse, L. J., Hunter, J., Van Oorschot, A. A. M., et al. (2010). Cardiac malformations in Pdgfrα mutant embryos are associated with increased expression of WT1 and Nkx2.5 in the second heart field. Dev. Dynam. 239, 2307–2317. doi: 10.1002/dvdy.22363
Blaschke, R. J., Hahurij, N. D., Kuijper, S., Just, S., Wisse, L. J., Deissler, K., et al. (2007). Targeted mutation reveals essential functions of the homeodomain transcription factor Shox2 in sinoatrial and pacemaking development. Circulation 115, 1830–1838. doi: 10.1161/CIRCULATIONAHA.106.637819
Bloomekatz, J., Singh, R., Prall, O. W., Dunn, A. C., Vaughan, M., Loo, C. S., et al. (2017). Platelet-derived growth factor (PDGF) signaling directs cardiomyocyte movement toward the midline during heart tube assembly. Elife 6:e21172. doi: 10.7554/eLife.21172
Chien, K. R. (2001). To Cre or not to Cre: the next generation of mouse models of human cardiac diseases. Circ. Res. 88, 546–549. doi: 10.1161/01.res.88.6.546
Christoffels, V. M., Mommersteeg, M. T. M., Trowe, M.-O., Prall, O. W. J., Vries, C. D. G.-D., Soufan, A. T., et al. (2006). Formation of the venous pole of the heart from an Nkx2-5-negative precursor population requires Tbx18. Circ. Res. 98, 1555–1563. doi: 10.1161/01.RES.0000227571.84189.65
Csepe, T. A., Kalyanasundaram, A., Hansen, B. J., Zhao, J., and Fedorov, V. V. (2015). Fibrosis: a structural modulator of sinoatrial node physiology and dysfunction. Front. Physiol. 6:37. doi: 10.3389/fphys.2015.00037
Durham, D., and Worthley, L. I. G. (2002). Cardiac arrhythmias: diagnosis and management. Bradycardias. Crit. Care Resusc. 4:54.
Dyer, L. A., and Kirby, M. L. (2009). The role of secondary heart field in cardiac development. Dev. Biol. 336, 137–144.
Espinoza-Lewis, R. A., Liu, H., Sun, C., Chen, C., Jiao, K., and Chen, Y. (2011). Ectopic expression of Nkx2.5 suppresses the formation of the sinoatrial node in mice. Dev. Biol. 356, 359–369.
Espinoza-Lewis, R. A., Yu, L., He, F., Liu, H., Tang, R., Shi, J., et al. (2009). Shox2 is essential for the differentiation of cardiac pacemaker cells by repressing Nkx2-5. Dev. Biol. 327, 376–385. doi: 10.1016/j.ydbio.2008.12.028
Heldin, C.-H., and Westermark, B. (1999). Mechanism of action and in vivo role of platelet-derived growth factor. Physiol. Rev. 79, 1283–1316. doi: 10.1152/physrev.1999.79.4.1283
Hoesl, E., Stieber, J., Herrmann, S., Feil, S., Tybl, E., Hofmann, F., et al. (2008). Tamoxifen-inducible gene deletion in the cardiac conduction system. J. Mol. Cell Cardiol. 45, 62–69. doi: 10.1016/j.yjmcc.2008.04.008
Hu, X., Lin, C., Shen, B., Ruan, N., Guan, Z., Chen, Y., et al. (2014). Conserved odontogenic potential in embryonic dental tissues. J. Dent Res. 93, 490–495. doi: 10.1177/0022034514523988
Li, H., Li, D., Wang, Y., Huang, Z., Xu, J., Yang, T., et al. (2019). Nkx2-5 defines a subpopulation of pacemaker cells and is essential for the physiological function of the sinoatrial node in mice. Development 146:dev178145. doi: 10.1242/dev.178145
Liang, X., Wang, G., Lin, L., Lowe, J., Zhang, Q., Bu, L., et al. (2013). HCN4 dynamically marks the first heart field and conduction system precursors. Circ. Res. 113, 399–407. doi: 10.1161/CIRCRESAHA.113.301588
Lints, T. J., Parsons, L. M., Hartley, L., Lyons, I., and Harvey, R. P. (1993). Nkx-2.5-a novel murine homeobox gene expressed in early heart progenitor cells and their myogenic descendants. Development 119, 419–431.
Liu, H., Espinoza-Lewis, R. A., Chen, C., Hu, X., Zhang, Y., and Chen, Y. P. (2012). The role of Shox2 in SAN development and function. Pediatr. Cardiol. 33, 882–889.
Liu, J., Dobrzynski, H., Yanni, J., Boyett, M. R., and Lei, M. (2007). Organisation of the mouse sinoatrial node: structure and expression of HCN channels. Cardiovasc. Res. 73, 729–738. doi: 10.1016/j.cardiores.2006.11.016
Morrison-Graham, K., Schatteman, G. C., Bork, T., Bowen-Pope, D. F., and Weston, J. A. (1992). A PDGF receptor mutation in the mouse (Patch) perturbs the development of a non-neuronal subset of neural crest-derived cells. Development 115, 133–142.
Moses, K. A., DeMayo, F., Braun, R. M., Reecy, J. L., and Schwartz, R. J. (2001). Embryonic expression of an Nkx2-5/Cre gene using ROSA26 reporter mice. Genesis 31, 176–180. doi: 10.1002/gene.10022
Munshi, N. V. (2012). Gene regulatory networks in cardiac conduction system development. Circ. Res. 110:1525.
Muzumdar, M. D., Tasic, B., Miyamichi, K., Li, L., and Luo, L. (2007). A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605. doi: 10.1002/dvg.20335
Prall, O. W., Menon, M. K., Solloway, M. J., Watanabe, Y., Zaffran, S., Bajolle, F., et al. (2007). An Nkx2-5/Bmp2/Smad1 negative feedback loop controls heart progenitor specification and proliferation. Cell 128, 947–959. doi: 10.1016/j.cell.2007.01.042
Rosenkranz, S., and Kazlauskas, A. (1999). Evidence for distinct signaling properties and biological responses induced by the PDGF receptor α and β subtypes. Growth Factors 16, 201–216.
Schatteman, G. C., Motley, S. T., Effmann, E. L., and Bowen-Pope, D. F. (1995). Platelet-derived growth factor receptor alpha subunit deleted Patch mouse exhibits severe cardiovascular dysmorphogenesis. Teratology 51, 351–366. doi: 10.1002/tera.1420510602
Soriano, P. (1997). The PDGF alpha receptor is required for neural crest cell development and for normal patterning of the somites. Development 124, 2691–2700.
Später, D., Abramczuk, M. K., Buac, K., Zangi, L., Stachel, M. W., Clarke, J., et al. (2013). A HCN4+ cardiomyogenic progenitor derived from the first heart field and human pluripotent stem cells. Nat. Cell Biol. 15:1098.
Sun, C., Zhang, T., Liu, C., Gu, S., and Chen, Y. (2013). Generation of Shox2-Cre allele for tissue specific manipulation of genes in the developing heart, palate, and limb. Genesis 51, 515–522. doi: 10.1002/dvg.22397
Van Den Akker, N. M. S., Lie-Venema, H., Maas, S., Eralp, I., DeRuiter, M. C., Poelmann, R. E., et al. (2005). Platelet-derived growth factors in the developing avian heart and maturating coronary vasculature. Dev. Dynam. 233, 1579–1588. doi: 10.1002/dvdy.20476
Virágh, S., and Challice, C. E. (1982). The development of the conduction system in the mouse embryo heart. I. The first embryonic A-V conduction pathway. Dev. Biol. 89, 25–40. doi: 10.1016/0012-1606(77)90278-0
Wiese, C., Grieskamp, T., Airik, R., Mommersteeg, M. T. M., Gardiwal, A., Vries, C. D. G.-D., et al. (2009). Formation of the sinus node head and differentiation of sinus node myocardium are independently regulated by Tbx18 and Tbx3. Circ. Res. 104, 388–397. doi: 10.1161/CIRCRESAHA.108.187062
Yamamoto, M., Dobrzynski, H., Tellez, J., Niwa, R., Billeter, R., Honjo, H., et al. (2006). Extended atrial conduction system characterised by the expression of the HCN4 channel and connexin45. Cardiovasc. Res. 72, 271–281. doi: 10.1016/j.cardiores.2006.07.026
Ye, W., Song, Y., Huang, Z., Zhang, Y., and Chen, Y. (2015a). Genetic regulation of sinoatrial node development and pacemaker program in the venous pole. J. Cardiovasc. Dev. Dis. 2, 282–298. doi: 10.3390/jcdd2040282
Ye, W., Wang, J., Song, Y., Yu, T., Sun, C., Liu, C., et al. (2015b). A common Shox2-Nkx2-5 antagonistic mechanism primes the pacemaker cell fate in the pulmonary vein myocardium and sinoatrial node. Development 142, 2521–2532. doi: 10.1242/dev.120220
Yi, T., Wong, J., Feller, E., Sink, S., Taghli-Lamallem, O., Wen, J., et al. (2012). Electrophysiological mapping of embryonic mouse hearts: mechanisms for developmental pacemaker switch and internodal conduction pathway. J. Cardiovasc. Electrophysiol. 23, 309–318. doi: 10.1111/j.1540-8167.2011.02191.x
Keywords: Pdgfrα, Nkx2.5, sinoatrial node (SAN), development, conditional knockout
Citation: Zheng X, Wang F, Hu X, Li H, Guan Z, Zhang Y and Hu X (2021) PDGFRα-Signaling Is Dispensable for the Development of the Sinoatrial Node After Its Fate Commitment. Front. Cell Dev. Biol. 9:647165. doi: 10.3389/fcell.2021.647165
Received: 29 December 2020; Accepted: 18 May 2021;
Published: 10 June 2021.
Edited by:
Juan Jose Sanz-Ezquerro, Centro Nacional de Biotecnología, Consejo Superior de Investigaciones Científicas (CSIC), SpainReviewed by:
Stéphane Zaffran, Aix-Marseille Université, FranceGonzalo del Monte-Nieto, Monash University, Australia
Copyright © 2021 Zheng, Wang, Hu, Li, Guan, Zhang and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuefeng Hu, YmlveGZoQGZqbnUuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
 Xi Zheng
Xi Zheng Fengjiao Wang1†
Fengjiao Wang1†