
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Cell Dev. Biol. , 18 February 2021
Sec. Stem Cell Research
Volume 9 - 2021 | https://doi.org/10.3389/fcell.2021.645335
This article is part of the Research Topic Chromatin Regulation in Cell Fate Decisions View all 15 articles
 Ian C. MacArthur1,2,3
Ian C. MacArthur1,2,3 Meelad M. Dawlaty1,2,3*
Meelad M. Dawlaty1,2,3*Studies of tissue-specific epigenomes have revealed 5-hydroxymethylcytosine (5hmC) to be a highly enriched and dynamic DNA modification in the metazoan nervous system, inspiring interest in the function of this epigenetic mark in neurodevelopment and brain function. 5hmC is generated by oxidation of 5-methylcytosine (5mC), a process catalyzed by the ten–eleven translocation (TET) enzymes. 5hmC serves not only as an intermediate in DNA demethylation but also as a stable epigenetic mark. Here, we review the known functions of 5hmC and TET enzymes in neural progenitor cell biology and embryonic and postnatal neurogenesis. We also discuss how TET enzymes and 5hmC regulate neuronal activity and brain function and highlight their implications in human neurodevelopmental and neurodegenerative disorders. Finally, we present outstanding questions in the field and envision new research directions into the roles of 5hmC and TET enzymes in neurodevelopment.
Precise temporal and spatial control of gene expression is essential for metazoan neurogenesis. This is achieved, in part, by reversible covalent modifications of DNA and histones which influence the accessibility and recruitment of transcription factors to target genes. Methylation of the 5-position carbon of cytosine (5mC) is one DNA modification influencing the transcriptional state of chromatin. DNA methylation is largely believed to be a suppressive mark achieved by de novo methyltransferases DNMT3A/B and maintained by maintenance methyltransferase DNMT1 (Wu and Zhang, 2014). In 2009, the discovery that ten-eleven translocation (TET) proteins (TET1, TET2, and TET3) are dioxygenases capable of oxidizing 5mC to 5-hydroxymethylcytosine (5hmC) (Tahiliani et al., 2009) ushered in interest to study this modified base not only as an intermediate in DNA demethylation but also as a novel epigenetic mark. Oxidation of 5mC to 5hmC by TETs facilitates passive and active DNA demethylation (Tahiliani et al., 2009; Ito et al., 2011; Wu and Zhang, 2014), the latter via iterative conversion of 5hmC to 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) and subsequent removal by DNA glycosylases and the base excision repair pathway (He et al., 2011; Ito et al., 2011). In addition to being an intermediate in DNA demethylation, 5hmC has been recognized as a stable epigenetic mark. This is supported by initial findings that 5hmC is enriched in Purkinje neurons (Kriaucionis and Heintz, 2009), and subsequent studies confirming the presence of 5hmC and the expression of TET enzymes across many neural cell types and tissues (Globisch et al., 2010; Szwagierczak et al., 2010; Ruzov et al., 2011; Mellen et al., 2012). It has been demonstrated that 5hmC may persist for months without turnover in the brain (Bachman et al., 2014), further supporting a potential role for 5hmC as a bona fide epigenetic mark with regulatory roles in the nervous system.
TET enzymes are required for mammalian development, as loss of all three enzymes in embryonic stem cells compromises differentiation (Dawlaty et al., 2014) and in mice leads to early embryonic lethality due to gastrulation arrest (Dai et al., 2016). Loss of TET3 leads to perinatal lethality (Gu et al., 2011), though individual loss of TET1 and TET2 is compatible with development of viable mice (Dawlaty et al., 2011; Li et al., 2011; Moran-Crusio et al., 2011). Combined loss of TET1 and TET2 leads to partial perinatal lethality, with a subset of neonates exhibiting exencephaly and other developmental abnormalities (Dawlaty et al., 2013). Similarly, combined loss of TET1 and TET3 leads to early developmental arrest and holoprosencephaly (Kang et al., 2015). This phenotypic variability suggests potential compensatory roles among TET paralogs. However, owing to the early embryonic lethality of triple TET deficiency, the absolute molecular and physiological requirements of TETs and 5hmC in neurogenesis has not yet been well-defined.
To understand the roles of 5hmC and TETs in regulation of neural gene expression, several studies have mapped the genomic distribution of 5hmC in various neural cell types and tissues over the course of embryonic and postnatal development (Jin et al., 2011; Szulwach et al., 2011; Khare et al., 2012; Hahn et al., 2013; Lister et al., 2013). In the embryonic mouse cortex, 5hmC levels increase as neural progenitor cells develop into mature neurons (Hahn et al., 2013). Interestingly, this increase is not necessarily accompanied by an increase in unmodified cytosine levels, suggesting that 5hmC can be a stable epigenetic mark in neurons and not merely a DNA demethylation intermediate (Hahn et al., 2013). Consistent with the notion that 5hmC is derived from 5mC, genomic regions in the fetal mouse brain that are enriched for 5hmC also tend to be enriched for 5mC. Notably, many of these regions become depleted of both marks in the adult mouse (Lister et al., 2013), demonstrating that 5hmC facilitates DNA demethylation in the developing brain.
5hmC levels increase in various mouse and human brain tissues over the course of life (Szulwach et al., 2011), and may have implications for neurodegenerative diseases. 5hmC is enriched in gene bodies and promoters, depleted from intergenic regions and transcription start sites, and is deposited at brain-specific enhancers (Jin et al., 2011; Szulwach et al., 2011; Hahn et al., 2013; Lister et al., 2013; Cui et al., 2020). The presence of 5hmC in gene bodies is associated with increased gene expression, suggesting that TET enzymes and 5hmC contribute to a transcriptionally permissive state of chromatin in the brain (Jin et al., 2011; Szulwach et al., 2011; Hahn et al., 2013; Lister et al., 2013). 5hmC also demarcates intron-exon boundaries in human brain cells and marks constitutively expressed exons, suggesting a potential role in control of splicing (Khare et al., 2012). Genes with high levels of 5hmC, for example Syt1 and Nav2, belong to functional categories critical for nervous system function, such as synaptic transmission and neurogenesis (Khare et al., 2012; Hahn et al., 2013). 5hmC is also associated with repetitive elements as it is enriched at SINE and LTR elements in the cerebellum and hippocampus, and depleted from LINE elements in the cerebellum (Szulwach et al., 2011). Enrichment at SINE and LTRs increases over postnatal life in the cerebellum (Szulwach et al., 2011), indicating a possible role in regulation of repetitive element activity in the brain. Indeed, Tet2/3 knockdown reverses loss-of-Uhrf1-mediated increased DNA hydroxymethylation and activation of IAP elements in NPCs (Ramesh et al., 2016). Together, these observations support important roles for 5hmC and TETs in mammalian neurogenesis and brain function.
Studies of embryonic stem cell (ESC) differentiation have suggested a critical role for TET enzymes and 5hmC in neural lineage commitment. Deficiency of all three TETs in ESCs compromises pluripotency and Tet1/2/3 triple knockout (TKO) ESCs fail to form neural pigmented epithelium in teratoma assays, though they are able to form other neural tissue types (Dawlaty et al., 2014). These cells fail to contribute to nervous system structures when injected into wild type blastocysts to form chimeras (Dawlaty et al., 2014). Consistently, Tet TKO mouse embryos and ESCs differentiated toward the neural lineage have reduced neuroectodermal and increased mesodermal gene expression, in part due to failure to inhibit Wnt signaling (Li et al., 2016). Likewise, TET TKO human ESCs exhibit aberrant neuroectodermal gene expression when differentiated toward the neural lineage and fail to demethylate the PAX6 promoter, a transcription factor critical for neurodevelopment (Verma et al., 2018). These studies support a requirement for TET enzymes in the commitment of ESCs to a neural fate, an idea further supported by studies of TET genes in ESC specification to neural progenitor cells (NPCs). TET enzymes, in particular TET2, regulate enhancer methylation during differentiation of ESCs to NPCs (Hon et al., 2014). Though Tet2 knockout ESCs can successfully differentiate into NPCs, these cells exhibit delayed induction of neural gene expression programs accompanied by enhancer hypermethylation and reduced histone H3 lysine 27 acetylation (Hon et al., 2014). This is in line with DNA hypermethylation in the embryonic cerebral cortex of Tet2 knockout mice (Lister et al., 2013). TET3 plays a role in the epigenetic regulation of NPC specification and maintenance of cellular identity (Montibus et al., 2020; Santiago et al., 2020). During differentiation of mouse ESCs to NPCs, the catalytic activity of TET3 promotes expression of histone demethylase Kdm6b, an epigenetic regulator critical for gene regulation during neurogenesis (Montibus et al., 2020), and loss of TET3 promotes NPC apoptosis (Li T. et al., 2015). Knockdown of Tet3 in NPCs promotes de-repression of pluripotency genes Oct4 and Nanog, implicating TET3 in the maintenance of NPC identity (Santiago et al., 2020). These studies implicate TETs in the epigenetic regulation of NPC biology.
In adult NPCs, different TET paralogs have unique functions, highlighting some non-redundant and context-specific roles. Loss of TET2 increases the proliferation of adult NPCs and reduces their differentiation into neurons and astrocytes in vivo, indicating that TET2 promotes NPC differentiation (Li et al., 2017). Deletion of Tet3 decreases NPC proliferation in the subventricular zone of the mouse cortex and promotes astrocytic differentiation, consistent with a role for TET3 in maintaining NPC identity (Montalban-Loro et al., 2019). Tet1 knockout mice have fewer NPCs in the dentate gyrus of the hippocampus and conditional deletion of Tet1 or Tet2 in NPCs compromises hippocampal neurogenesis (Zhang et al., 2013; Gontier et al., 2018). While most functions of TETs in NPCs are attributed to their enzymatic activity (Zhang et al., 2013; Li et al., 2017; Montibus et al., 2020), some functions are independent of enzymatic activity, such as transcriptional repression of the imprinted gene Snrnp by TET3 (Montalban-Loro et al., 2019). Investigating these dual roles of TETs and dissecting their requirements in NPC biology and neurogenesis will be essential.
Although global or neural-specific loss of each Tet gene in mice influences NPC biology, it does not block neurogenesis or cause gross neuroanatomical defects (Rudenko et al., 2013; Zhang et al., 2013). However, combined loss of TET1/2 and TET1/3 causes exencephaly and holoprosencephaly in some embryos, respectively (Dawlaty et al., 2013; Kang et al., 2015) suggesting redundancy between TETs in neurogenesis that warrants further investigation. Findings from other organisms have also supported a role for TET enzymes in embryonic neurogenesis. Xenopus laevis embryos depleted of TET3 are microcephalic and eyeless with deregulation of neurodevelopmental programs leading to aberrant expression of neuronal, eye, neural crest, and sonic hedgehog signaling genes (Xu et al., 2012). Moreover, tet2/3 mutant zebrafish exhibit abnormal brain and eye morphology (Li C. et al., 2015) and impaied retinal neurogenesis, partly due to overactive Notch and Wnt signaling (Seritrakul and Gross, 2017). Aberrant expression of mesodermal genes was also observed in tet2/3 mutant retinas (Seritrakul and Gross, 2017), a finding similar to those in Tet1/2/3 knockout embryos (Li et al., 2016). Importantly, TET enzymes mediate demethylation of conserved developmental enhancers in brain during the phylotypic stage of vertebrates, as demonstrated in zebrafish, Xenopus tropicalis, and mouse (Bogdanovic et al., 2016). Together, these findings support highly conserved and overlapping functions for TETs in neurodevelopment.
In addition to roles in regulation of NPC biology, TETs and 5hmC are important in postnatal neurodevelopment and mature neurons. As previously mentioned, 5hmC accumulates over the course of life (Szulwach et al., 2011). During development of mouse olfactory bulb neurons, which occurs throughout life, 5hmC is enriched in neurons relative to immature cells and is associated with increased neurodevelopmental gene expression (Colquitt et al., 2013). Likewise, 5hmC increases over the course of postnatal retinal maturation, and is enriched at neurogenesis genes (Perera et al., 2015). In the cerebellum, 5hmC increases during an important period of neuronal circuit formation, and TET1 and TET3 are required for proper branching of granule cell dendrites (Zhu et al., 2016). Chimeric Tet3 knockout mice generated by injection of Tet3 sgRNAs in one cell of a two-cell-stage embryo develop histologically normal cerebral cortices composed of Tet3 wild type and knockout cells but exhibit abnormal electrophysiology in recordings of excitatory and inhibitory neurotransmission, suggesting that TET3 is required for developmental synapse and circuit formation (Wang et al., 2017). These studies implicate TETs in shaping the epigenetic landscape during specification of mature neural cell types after birth and in the development of higher order structures, including neuronal circuits.
5hmC and TET enzymes have also been shown to be highly dynamic within post-mitotic neurons. Cortical 5hmC has cell-type specific distributions associated with differential gene expression (Kozlenkov et al., 2018), and the ability of TETs to promote active DNA demethylation and alter gene expression in response to neuronal activity and to influence behavior has been the subject of extensive study (Guo et al., 2011; Kaas et al., 2013; Rudenko et al., 2013; Zhang et al., 2013; Li et al., 2014; Yu et al., 2015). Tet1 expression is downregulated in response to neuronal activity in hippocampus where it regulates spatial memory and fear memory extinction (Guo et al., 2011; Kaas et al., 2013; Rudenko et al., 2013; Zhang et al., 2013). Hippocampal neurons upregulate Tet3 to initiate active DNA demethylation in response to neuronal stimulation (Yu et al., 2015). In cortical neurons, Tet3 is upregulated during fear extinction learning and, like Tet1 in the hippocampus, is required for fear memory extinction (Li et al., 2014). Fear extinction learning is accompanied by Tet3-mediated upregulation of the Gephyrin gene and a transcriptionally-permissive reshaping of chromatin around this locus (Li et al., 2014). Loss of Tet3 is sufficient to produce anxiety-like behaviors in mice, partly due to increased expression of immediate early genes like Npas4 (Antunes et al., 2020), a role that is opposite to the anxiolytic and anti-depressant effects of Tet1 (Feng et al., 2017). In general, the mechanistic basis by which TET enzymes influence behavior is, in part, due to reshaping of neuronal 5mC and 5hmC landscapes in response to activity. This remodeling of the epigenome is required for proper expression of genes involved in memory consolidation and synaptic function, such as Bdnf and Arc, and is sufficient to alter the electrophysiological properties of neurons (Guo et al., 2011; Kaas et al., 2013; Rudenko et al., 2013; Zhang et al., 2013; Li et al., 2014; Yu et al., 2015). In post-mitotic cerebellar neurons, 5hmC dynamics can influence recruitment of key gene regulatory factors. For example, 5hmC in gene bodies is associated with reduced MeCP2 occupancy and increased gene expression, possibly due to loss of MeCP2 repression (Mellen et al., 2017). Of note, reduced MeCP2 occupancy is specifically associated with 5hmC at gene body CpG dinucleotides, whereas 5hmC at CpA sites flanking enhancers retains MeCP2 binding. This highlights the ability of 5hmC to influence recruitment of gene regulatory factors in a sequence-dependent manner (Mellen et al., 2017). Moreover, findings that common MECP2 mutations in Rett syndrome disrupt MeCP2 binding to 5hmC has implications for this mark in Rett syndrome pathogenesis (Mellen et al., 2012; Brown et al., 2016). Together, these studies propose crucial roles for 5hmC and TET enzymes in mature neuronal function.
Compelling evidence for the importance of TETs in neurodevelopment and brain function is the identification of TET gene mutations and alterations in 5hmC levels in human neurodevelopmental and neurodegenerative disorders. Mutations in TET3 were recently identified to underlie an inherited syndrome of intellectual disability and craniofacial abnormalities (Beck et al., 2020). While most mutations are in the catalytic domain and are sufficient to impair enzymatic activity, some are outside of this domain, and one mutation does not affect catalytic activity (Beck et al., 2020), underscoring the importance of TET3 catalytic and non-catalytic functions in human neurodevelopment. Interestingly, the clinical characteristics of patients with TET3 deficiency resemble those of patients with Tatton-Brown-Rahman syndrome and Sotos syndrome, caused by mutations in DNMT3A and NSD1, respectively (Kurotaki et al., 2002; Tatton-Brown et al., 2014). This highlights the general importance of DNA and histone methylation dynamics in human craniofacial and neural development. Other TET mutations have been observed in individuals with intellectual disability. Mutations in TET1 were identified in consanguineous Pakistani and Iranian families with familial intellectual disability syndromes (Harripaul et al., 2018), and a germline TET2 variant in an individual diagnosed with intellectual disability and delayed verbal comprehension in the absence of any other known genetic causes (Kaasinen et al., 2019). Together, these findings support an important role for TETs in the etiology of neurodevelopmental disorders and intellectual disability.
In addition TET enzymes and 5hmC are recurrently dysregulated in neurodegenerative conditions and in aging brain. Induced pluripotent stem cell-derived NPCs and neurons from Alzheimer’s disease (AD) patients exhibit differential hydroxymethylation at genes associated with neurodevelopment and synaptic function, including at known AD susceptibility loci, compared to cells derived from healthy controls (Fetahu et al., 2019). Consistently, presumptive loss-of-function mutations in TET2 have been identified in patients with early onset AD and frontotemporal dementia (Cochran et al., 2020). Interestingly, TET2 promotes proinflammatory gene expression in microglia and TET2 expression is increased in microglia associated with amyloid beta plaques in the brains of AD patients and mouse models (Carrillo-Jimenez et al., 2019). Thus, the positive and negative roles of TET2 in AD are likely specific to distinct stages in clinical course and cell types. TET variants or dysregulation have also been implicated in Parkinson’s disease (PD). TET1 mutations were reported in a Chinese cohort of PD patients (Shu et al., 2019). Intriguingly, increased expression of TET2 and increased 5hmC at neural enhancers is observed in prefrontal cortex of patients with PD and Tet2 knockout mice are protected from inflammatory damage to the substantia nigra (Marshall et al., 2020). Conversely, Tet2 expression declines in the hippocampus of aging mice and is associated with age-related cognitive decline (Gontier et al., 2018). Remarkably, restoration of hippocampal Tet2 expression by stereotactic lentivirus injection is sufficient to improve cognitive function in aged mice (Gontier et al., 2018). Moreover, recent findings that TET1 and TET2 are required for axonal regeneration by reprogramming factor expression highlights their potential as therapeutic targets (Lu et al., 2020). These observations in human disease and mouse models warrant further studies to clarify the discordant roles of TET enzymes in the etiology of neurodegenerative diseases and aging.
In addition to their roles in neurodevelopmental and neurodegenerative disorders, TET enzymes are associated with addictive behaviors in humans. TET1 expression is decreased in the nucleus accumbens (NAc) of humans suffering from cocaine addiction and cocaine administration to mice is sufficient to alter 5hmC at enhancers in NAc (Feng et al., 2015). Tet expression in NAc is also responsive to methamphetamine administration in rats (Jayanthi et al., 2018). Further work is necessary to clarify the role of TETs in mediating addictive behaviors.
A dozen years since the discovery that TET enzymes promote DNA hydroxylation and demethylation and the first studies reporting the abundance of 5hmC in the mammalian nervous system (Kriaucionis and Heintz, 2009; Tahiliani et al., 2009), work in the field has shed some light on their functions in neural physiology. TETs are dynamically expressed during development and in particular during embryonic and adult neurogenesis. 5hmC is a highly enriched mark in the brain and its levels increase over the course of embryonic neurogenesis and postnatal life where it is associated with neural gene expression. TET enzymes are required for various aspects of neurodevelopment, NPC biology, and neuronal activity. Findings that 5hmC and TETs are dysregulated in human neurodevelopmental and neurodegenerative disorders and addiction open new frontiers for utilizing them in clinical diagnostics and therapeutics. Despite this progress, several fundamental questions remain unanswered. These pertain to: (1) mechanisms of TET recruitment to target sites, (2) functional redundancy between TET paralogs, (3) gene activation and silencing by the dual enzymatic and non-enzymatic functions of TET enzymes, (4) relevance of 5hmC readers and interactomes of TETs in gene regulation, (5) re-establishment of 5hmC upon active DNA demethylation at activity-dependent genes in post-mitotic neurons, and (6) involvement of 5fC and 5caC, the other oxidized derivatives of 5mC, in DNA demethylation and beyond.
It has been substantiated that increases in gene body 5hmC is associated with activation of neural genes over the course of development, but the mechanism by which 5hmC influences gene expression remains incompletely understood. One possibility is that 5hmC recruits specific factors to promote transcription, and several groups have sought to identify such readers of 5hmC (Yildirim et al., 2011; Mellen et al., 2012; Spruijt et al., 2013). For example, MeCP2 binds 5hmC at neuronal genes to facilitate transcription, a finding with implications in the pathogenesis of Rett syndrome, where MECP2 is mutated and 5hmC levels are altered (Mellen et al., 2012). Alternatively, evidence also supports a role for MeCP2 and MBD2 in protecting 5mC from conversion to 5hmC (Szulwach et al., 2011; Ludwig et al., 2017). In addition, UHRF2 is a specific reader of 5hmC in NPCs while THAP11 interacts with 5hmC in brain tissue (Spruijt et al., 2013). The functions of these and other 5hmC readers, including WDR76 and THY96, in the nervous system remain to be further explored (Spruijt et al., 2013). In contrast, the observation that elevated 5hmC levels may persist at genes after silencing suggests that any factor recruitment by this mark is not sufficient to maintain gene expression in the presence of antagonistic or in the absence of agonistic transcriptional cues (Colquitt et al., 2013). This would be consistent with a necessary but not sufficient role for TET-mediated 5hmC deposition and DNA demethylation for potent gene transcription (Baumann et al., 2019). This could also indicate a requirement for higher-order formation of 5fC and 5caC for transcription factor recruitment, given the fact that a growing number of proteins bind 5fC more specifically than 5hmC (Iurlaro et al., 2013). Additional work is needed to substantiate a causal role for 5hmC in neural gene regulation.
Although functional studies have defined important roles for individual TETs in neural development and physiology, the absolute requirements of these enzymes and 5hmC is not fully established. This is in part due to the possible compensatory effects of TETs in studies involving deletion of individual TET genes. Leveraging conditional genetic systems for spatial and temporal deletion of all three TET genes in specific cell types may allow for identification of their novel functions in the brain. While TET enzymes certainly influence gene expression through their enzymatic functions, non-enzymatic activities of these proteins involving formation and recruitment of chromatin regulatory complexes have also been described (Chen et al., 2013; Ito et al., 2019; Montalban-Loro et al., 2019). Further work is necessary to fully elucidate protein-protein interactions by which TETs influence transcription and chromatin state, such as those between TET2 and FOXO3 in NPCs (Li et al., 2017) and TET3 and NSD3 in mature neurons (Perera et al., 2015). Comparison of the neural phenotypes associated with loss of TET enzymes vs. loss of their enzymatic activity alone will help dissect key enzymatic-dependent and independent functions of TET proteins in the brain. Use of existing and development of new Tet catalytic mutant mouse models will facilitate the in vivo study of TET catalytic-independent functions. Furthermore, the observation that loss of Tet in Drosophila results in aberrant brain development and reduced RNA hydroxymethylation warrants investigation into the role of TET-mediated RNA modifications in mammalian brains (Delatte et al., 2016). In summary, as illustrated in Figure 1, TET enzymes and 5hmC play crucial roles in various aspects of neurobiology, from regulation of NPCs and neurogenesis to adult brain function and human diseases. Gaining further insights into their roles will enhance our understanding of metazoan nervous system development and the etiology of human neurological disorders.
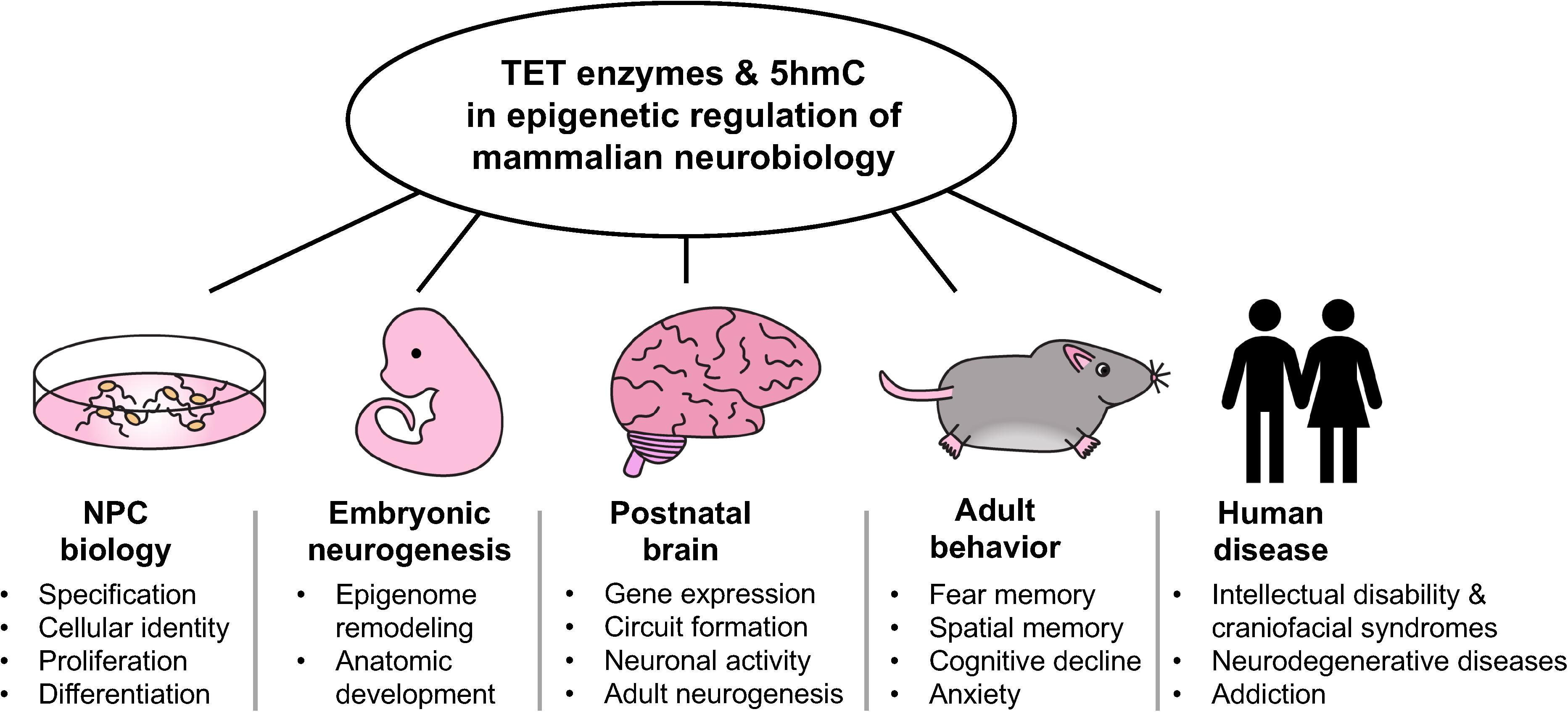
Figure 1. Overview of the multifaceted roles of TET enzymes and 5hmC in the epigenetic regulation of mamalian neurobiology.
IM wrote the first draft and prepared the figure. MD edited and revised the draft and figure, and finalized the final version with input from IM. Both authors contributed to the article and approved the submitted version.
The Dawlaty laboratory was supported by the National Institutes of Health (R01GM122839, R01HL148852), the New State Department of Health (NYSTEM IIRP Award), the Feldstein Medical Foundation, and the Hirschl and Weill-Caulier Charitable Trust Funds to study the biology of Tet enzymes in stem cells and development. MD’s salary was in part supported by these funds. IM was supported by the Medical Scientist Training Program training grant T32GM007288-48 from the National Institutes of Health to Albert Einstein College of Medicine.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Drs. Frank Soldner and Jean Hébert of the Albert Einstein College of Medicine’s Neuroscience and Genetics Departments for critical reading of the manuscript, and members of the Dawlaty laboratory for helpful discussions. We apologize to colleagues whose work we could not discuss here due to space limitations.
Antunes, C., Da Silva, J. D., Guerra-Gomes, S., Alves, N. D. F., Ferreira, F., Loureiro-Campos, E., et al. (2020). Tet3 ablation in adult brain neurons increases anxiety-like behavior and regulates cognitive function in mice. Mol. Psychiatry. doi: 10.1038/s41380-020-0695-7 [Epub ahead of print].
Bachman, M., Uribe-Lewis, S., Yang, X., Williams, M., Murrell, A., and Balasubramanian, S. (2014). 5-Hydroxymethylcytosine is a predominantly stable DNA modification. Nat. Chem. 6, 1049–1055. doi: 10.1038/nchem.2064
Baumann, V., Wiesbeck, M., Breunig, C. T., Braun, J. M., Koferle, A., Ninkovic, J., et al. (2019). Targeted removal of epigenetic barriers during transcriptional reprogramming. Nat. Commun. 10:2119. doi: 10.1038/s41467-019-10146-8
Beck, D. B., Petracovici, A., He, C., Moore, H. W., Louie, R. J., Ansar, M., et al. (2020). Delineation of a human mendelian disorder of the DNA demethylation machinery: TET3 deficiency. Am. J. Hum. Genet. 106, 234–245. doi: 10.1016/j.ajhg.2019.12.007
Bogdanovic, O., Smits, A. H., de la Calle Mustienes, E., Tena, J. J., Ford, E., and Williams, R. (2016). Active DNA demethylation at enhancers during the vertebrate phylotypic period. Nat. Genet. 48, 417–426. doi: 10.1038/ng.3522
Brown, K., Selfridge, J., Lagger, S., Connelly, J., De Sousa, D., Kerr, A., et al. (2016). The molecular basis of variable phenotypic severity among common missense mutations causing Rett syndrome. Hum. Mol. Genet. 25, 558–570. doi: 10.1093/hmg/ddv496
Carrillo-Jimenez, A., Deniz, O., Niklison-Chirou, M. V., Ruiz, R., Bezerra-Salomao, K., Stratoulias, V., et al. (2019). TET2 regulates the neuroinflammatory response in microglia. Cell Rep. 29, 697–713.e8. doi: 10.1016/j.celrep.2019.09.013
Chen, Q., Chen, Y., Bian, C., Fujiki, R., and Yu, X. (2013). TET2 promotes histone O-GlcNAcylation during gene transcription. Nature 493, 561–564. doi: 10.1038/nature11742
Cochran, J. N., Geier, E. G., Bonham, L. W., Newberry, J. S., Amaral, M. D., Thompson, M. L., et al. (2020). Non-coding and loss-of-function coding variants in TET2 are associated with multiple neurodegenerative diseases. Am. J. Hum. Genet. 106, 632–645. doi: 10.1016/j.ajhg.2020.03.010
Colquitt, B. M., Allen, E. W., Barnea, G., and Lomvardas, S. (2013). Alteration of genic 5-hydroxymethylcytosine patterning in olfactory neurons correlates with changes in gene expression and cell identity. Proc. Natl. Acad. Sci. U.S.A. 110, 14682–14687. doi: 10.1073/pnas.1302759110
Cui, X. L., Nie, J., Ku, J., Dougherty, U., West-Szymanski, D. C., Collin, F., et al. (2020). A human tissue map of 5-hydroxymethylcytosines exhibits tissue specificity through gene and enhancer modulation. Nat. Commun. 11:6161. doi: 10.1038/s41467-020-20001-w
Dai, H. Q., Wang, B. A., Yang, L., Chen, J. J., Zhu, G. C., Sun, M. L., et al. (2016). TET-mediated DNA demethylation controls gastrulation by regulating Lefty-Nodal signalling. Nature 538, 528–532. doi: 10.1038/nature20095
Dawlaty, M. M., Breiling, A., Le, T., Barrasa, M. I., Raddatz, G., Gao, Q., et al. (2014). Loss of Tet enzymes compromises proper differentiation of embryonic stem cells. Dev. Cell 29, 102–111. doi: 10.1016/j.devcel.2014.03.003
Dawlaty, M. M., Breiling, A., Le, T., Raddatz, G., Barrasa, M. I., Cheng, A. W., et al. (2013). Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Dev. Cell 24, 310–323. doi: 10.1016/j.devcel.2012.12.015
Dawlaty, M. M., Ganz, K., Powell, B. E., Hu, Y. C., Markoulaki, S., Cheng, A. W., et al. (2011). Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell 9, 166–175. doi: 10.1016/j.stem.2011.07.010
Delatte, B., Wang, F., Ngoc, L. V., Collignon, E., Bonvin, E., Deplus, R., et al. (2016). RNA biochemistry. transcriptome-wide distribution and function of RNA hydroxymethylcytosine. Science 351, 282–285. doi: 10.1126/science.aac5253
Feng, J., Pena, C. J., Purushothaman, I., Engmann, O., Walker, D., Brown, A. N., et al. (2017). Tet1 in nucleus accumbens opposes depression- and anxiety-like behaviors. Neuropsychopharmacology 42, 1657–1669. doi: 10.1038/npp.2017.6
Feng, J., Shao, N., Szulwach, K. E., Vialou, V., Huynh, J., Zhong, C., et al. (2015). Role of Tet1 and 5-hydroxymethylcytosine in cocaine action. Nat. Neurosci. 18, 536–544. doi: 10.1038/nn.3976
Fetahu, I. S., Ma, D., Rabidou, K., Argueta, C., Smith, M., Liu, H., et al. (2019). Epigenetic signatures of methylated DNA cytosine in Alzheimer’s disease. Sci. Adv. 5:eaaw2880. doi: 10.1126/sciadv.aaw2880
Globisch, D., Munzel, M., Muller, M., Michalakis, S., Wagner, M., Koch, S., et al. (2010). Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS One 5:e15367. doi: 10.1371/journal.pone.0015367
Gontier, G., Iyer, M., Shea, J. M., Bieri, G., Wheatley, E. G., Ramalho-Santos, M., et al. (2018). Tet2 rescues age-related regenerative decline and enhances cognitive function in the adult mouse brain. Cell Rep. 22, 1974–1981. doi: 10.1016/j.celrep.2018.02.001
Gu, T. P., Guo, F., Yang, H., Wu, H. P., Xu, G. F., Liu, W., et al. (2011). The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature 477, 606–610. doi: 10.1038/nature10443
Guo, J. U., Su, Y., Zhong, C., Ming, G. L., and Song, H. (2011). Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell 145, 423–434. doi: 10.1016/j.cell.2011.03.022
Hahn, M. A., Qiu, R., Wu, X., Li, A. X., Zhang, H., Wang, J., et al. (2013). Dynamics of 5-hydroxymethylcytosine and chromatin marks in mammalian neurogenesis. Cell Rep. 3, 291–300. doi: 10.1016/j.celrep.2013.01.011
Harripaul, R., Vasli, N., Mikhailov, A., Rafiq, M. A., Mittal, K., Windpassinger, C., et al. (2018). Mapping autosomal recessive intellectual disability: combined microarray and exome sequencing identifies 26 novel candidate genes in 192 consanguineous families. Mol. Psychiatry 23, 973–984. doi: 10.1038/mp.2017.60
He, Y. F., Li, B. Z., Li, Z., Liu, P., Wang, Y., Tang, Q., et al. (2011). Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333, 1303–1307. doi: 10.1126/science.1210944
Hon, G. C., Song, C. X., Du, T., Jin, F., Selvaraj, S., Lee, A. Y., et al. (2014). 5mC oxidation by Tet2 modulates enhancer activity and timing of transcriptome reprogramming during differentiation. Mol. Cell 56, 286–297. doi: 10.1016/j.molcel.2014.08.026
Ito, K., Lee, J., Chrysanthou, S., Zhao, Y., Josephs, K., Sato, H., et al. (2019). Non-catalytic roles of Tet2 are essential to regulate hematopoietic stem and progenitor cell homeostasis. Cell Rep. 28, 2480–2490.e4. doi: 10.1016/j.celrep.2019.07.094
Ito, S., Shen, L., Dai, Q., Wu, S. C., Collins, L. B., Swenberg, J. A., et al. (2011). Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333, 1300–1303. doi: 10.1126/science.1210597
Iurlaro, M., Ficz, G., Oxley, D., Raiber, E. A., Bachman, M., Booth, M. J., et al. (2013). A screen for hydroxymethylcytosine and formylcytosine binding proteins suggests functions in transcription and chromatin regulation. Genome Biol. 14:R119. doi: 10.1186/gb-2013-14-10-r119
Jayanthi, S., Gonzalez, B., McCoy, M. T., Ladenheim, B., Bisagno, V., and Cadet, J. L. (2018). Methamphetamine induces TET1- and TET3-dependent DNA hydroxymethylation of Crh and Avp genes in the rat nucleus accumbens. Mol. Neurobiol. 55, 5154–5166. doi: 10.1007/s12035-017-0750-9
Jin, S. G., Wu, X., Li, A. X., and Pfeifer, G. P. (2011). Genomic mapping of 5-hydroxymethylcytosine in the human brain. Nuclei. Acid. Res. 39, 5015–5024. doi: 10.1093/nar/gkr120
Kaas, G. A., Zhong, C., Eason, D. E., Ross, D. L., Vachhani, R. V., Ming, G. L., et al. (2013). TET1 controls CNS 5-methylcytosine hydroxylation, active DNA demethylation, gene transcription, and memory formation. Neuron 79, 1086–1093. doi: 10.1016/j.neuron.2013.08.032
Kaasinen, E., Kuismin, O., Rajamaki, K., Ristolainen, H., Aavikko, M., Kondelin, J., et al. (2019). Impact of constitutional TET2 haploinsufficiency on molecular and clinical phenotype in humans. Nat. Commun. 10:1252. doi: 10.1038/s41467-019-09198-7
Kang, J., Lienhard, M., Pastor, W. A., Chawla, A., Novotny, M., Tsagaratou, A., et al. (2015). Simultaneous deletion of the methylcytosine oxidases Tet1 and Tet3 increases transcriptome variability in early embryogenesis. Proc. Natl. Acad. Sci. U.S.A. 112, E4236–E4245. doi: 10.1073/pnas.1510510112
Khare, T., Pai, S., Koncevicius, K., Pal, M., Kriukiene, E., Liutkeviciute, Z., et al. (2012). 5-hmC in the brain is abundant in synaptic genes and shows differences at the exon-intron boundary. Nat. Struct. Mol. Biol. 19, 1037–1043. doi: 10.1038/nsmb.2372
Kozlenkov, A., Li, J., Apontes, P., Hurd, Y. L., Byne, W. M., Koonin, E. V., et al. (2018). A unique role for DNA (hydroxy)methylation in epigenetic regulation of human inhibitory neurons. Sci. Adv. 4:eaau6190. doi: 10.1126/sciadv.aau6190
Kriaucionis, S., and Heintz, N. (2009). The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324, 929–930. doi: 10.1126/science.1169786
Kurotaki, N., Imaizumi, K., Harada, N., Masuno, M., Kondoh, T., Nagai, T., et al. (2002). Haploinsufficiency of NSD1 causes Sotos syndrome. Nat. Genet. 30, 365–366. doi: 10.1038/ng863
Li, C., Lan, Y., Schwartz-Orbach, L., Korol, E., Tahiliani, M., Evans, T., et al. (2015). Overlapping requirements for Tet2 and Tet3 in normal development and hematopoietic stem cell emergence. Cell Rep. 12, 1133–1143. doi: 10.1016/j.celrep.2015.07.025
Li, T., Yang, D., Li, J., Tang, Y., Yang, J., and Le, W. (2015). Critical role of Tet3 in neural progenitor cell maintenance and terminal differentiation. Mol. Neurobiol. 51, 142–154. doi: 10.1007/s12035-014-8734-5
Li, X., Wei, W., Zhao, Q. Y., Widagdo, J., Baker-Andresen, D., Flavell, C. R., et al. (2014). Neocortical Tet3-mediated accumulation of 5-hydroxymethylcytosine promotes rapid behavioral adaptation. Proc. Natl. Acad. Sci. U.S.A. 111, 7120–7125. doi: 10.1073/pnas.1318906111
Li, X., Yao, B., Chen, L., Kang, Y., Li, Y., Cheng, Y., et al. (2017). Ten-eleven translocation 2 interacts with forkhead box O3 and regulates adult neurogenesis. Nat. Commun. 8:15903. doi: 10.1038/ncomms15903
Li, X., Yue, X., Pastor, W. A., Lin, L., Georges, R., Chavez, L., et al. (2016). Tet proteins influence the balance between neuroectodermal and mesodermal fate choice by inhibiting Wnt signaling. Proc. Natl. Acad. Sci. U.S.A. 113, E8267–E8276. doi: 10.1073/pnas.1617802113
Li, Z., Cai, X., Cai, C. L., Wang, J., Zhang, W., Petersen, B. E., et al. (2011). Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood 118, 4509–4518. doi: 10.1182/blood-2010-12-325241
Lister, R., Mukamel, E. A., Nery, J. R., Urich, M., Puddifoot, C. A., Johnson, N. D., et al. (2013). Global epigenomic reconfiguration during mammalian brain development. Science 341:1237905. doi: 10.1126/science.1237905
Lu, Y., Brommer, B., Tian, X., Krishnan, A., Meer, M., Wang, C., et al. (2020). Reprogramming to recover youthful epigenetic information and restore vision. Nature 588, 124–129. doi: 10.1038/s41586-020-2975-4
Ludwig, A. K., Zhang, P., Hastert, F. D., Meyer, S., Rausch, C., Herce, H. D., et al. (2017). Binding of MBD proteins to DNA blocks Tet1 function thereby modulating transcriptional noise. Nucleic Acids Res. 45, 2438–2457. doi: 10.1093/nar/gkw1197
Marshall, L. L., Killinger, B. A., Ensink, E., Li, P., Li, K. X., Cui, W., et al. (2020). Epigenomic analysis of Parkinson’s disease neurons identifies Tet2 loss as neuroprotective. Nat. Neurosci. 23, 1203–1214. doi: 10.1038/s41593-020-0690-y
Mellen, M., Ayata, P., Dewell, S., Kriaucionis, S., and Heintz, N. (2012). MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell 151, 1417–1430. doi: 10.1016/j.cell.2012.11.022
Mellen, M., Ayata, P., and Heintz, N. (2017). 5-hydroxymethylcytosine accumulation in postmitotic neurons results in functional demethylation of expressed genes. Proc. Natl. Acad. Sci. U.S.A. 114, E7812–E7821. doi: 10.1073/pnas.1708044114
Montalban-Loro, R., Lozano-Urena, A., Ito, M., Krueger, C., Reik, W., Ferguson-Smith, A. C., et al. (2019). TET3 prevents terminal differentiation of adult NSCs by a non-catalytic action at Snrpn. Nat. Commun. 10:1726. doi: 10.1038/s41467-019-09665-1
Montibus, B., Cercy, J., Bouschet, T., Charras, A., Maupetit-Mehouas, S., Nury, D., et al. (2020). TET3 controls the expression of the H3K27me3 demethylase Kdm6b during neural commitment. Cell Mol. Life Sci. doi: 10.1007/s00018-020-03541-8 [Epub ahead of print].
Moran-Crusio, K., Reavie, L., Shih, A., Abdel-Wahab, O., Ndiaye-Lobry, D., Lobry, C., et al. (2011). Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell 20, 11–24. doi: 10.1016/j.ccr.2011.06.001
Perera, A., Eisen, D., Wagner, M., Laube, S. K., Kunzel, A. F., Koch, S., et al. (2015). TET3 is recruited by REST for context-specific hydroxymethylation and induction of gene expression. Cell Rep. 11, 283–294. doi: 10.1016/j.celrep.2015.03.020
Ramesh, V., Bayam, E., Cernilogar, F. M. I, Bonapace, M., Schulze, M., Riemenschneider, M. J., et al. (2016). Loss of Uhrf1 in neural stem cells leads to activation of retroviral elements and delayed neurodegeneration. Genes Dev. 30, 2199–2212. doi: 10.1101/gad.284992.116
Rudenko, A., Dawlaty, M. M., Seo, J., Cheng, A. W., Meng, J., Le, T., et al. (2013). Tet1 is critical for neuronal activity-regulated gene expression and memory extinction. Neuron 79, 1109–1122. doi: 10.1016/j.neuron.2013.08.003
Ruzov, A., Tsenkina, Y., Serio, A., Dudnakova, T., Fletcher, J., Bai, Y., et al. (2011). Lineage-specific distribution of high levels of genomic 5-hydroxymethylcytosine in mammalian development. Cell Res. 21, 1332–1342. doi: 10.1038/cr.2011.113
Santiago, M., Antunes, C., Guedes, M., Iacovino, M., Kyba, M., Reik, W., et al. (2020). Tet3 regulates cellular identity and DNA methylation in neural progenitor cells. Cell Mol. Life Sci. 77, 2871–2883. doi: 10.1007/s00018-019-03335-7
Seritrakul, P., and Gross, J. M. (2017). Tet-mediated DNA hydroxymethylation regulates retinal neurogenesis by modulating cell-extrinsic signaling pathways. PLoS Genet. 13:e1006987. doi: 10.1371/journal.pgen.1006987
Shu, L., Qin, L., Min, S., Pan, H., Zhong, J., Guo, J., et al. (2019). Genetic analysis of DNA methylation and hydroxymethylation genes in Parkinson’s disease. Neurobiol. Aging 84, 242.e13–242.e16. doi: 10.1016/j.neurobiolaging.2019.02.025
Spruijt, C. G., Gnerlich, F., Smits, A. H., Pfaffeneder, T., Jansen, P. W., Bauer, C., et al. (2013). Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell 152, 1146–1159. doi: 10.1016/j.cell.2013.02.004
Szulwach, K. E., Li, X., Li, Y., Song, C. X., Wu, H., Dai, Q., et al. (2011). 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat. Neurosci. 14, 1607–1616. doi: 10.1038/nn.2959
Szwagierczak, A., Bultmann, S., Schmidt, C. S., Spada, F., and Leonhardt, H. (2010). Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res. 38:e181. doi: 10.1093/nar/gkq684
Tahiliani, M., Koh, K. P., Shen, Y., Pastor, W. A., Bandukwala, H., Brudno, Y., et al. (2009). Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935. doi: 10.1126/science.1170116
Tatton-Brown, K., Seal, S., Ruark, E., Harmer, J., Ramsay, E., Vecchio, S. Del, et al. (2014). Mutations in the DNA methyltransferase gene DNMT3A cause an overgrowth syndrome with intellectual disability. Nat. Genet. 46, 385–388. doi: 10.1038/ng.2917
Verma, N., Pan, H., Dore, L. C., Shukla, A., Li, Q. V., Pelham-Webb, B., et al. (2018). TET proteins safeguard bivalent promoters from de novo methylation in human embryonic stem cells. Nat. Genet. 50, 83–95. doi: 10.1038/s41588-017-0002-y
Wang, L., Li, M. Y., Qu, C., Miao, W. Y., Yin, Q., Liao, J., et al. (2017). CRISPR-Cas9-mediated genome editing in one blastomere of two-cell embryos reveals a novel Tet3 function in regulating neocortical development. Cell Res. 27, 815–829. doi: 10.1038/cr.2017.58
Wu, H., and Zhang, Y. (2014). Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell 156, 45–68. doi: 10.1016/j.cell.2013.12.019
Xu, Y., Xu, C., Kato, A., Tempel, W., Abreu, J. G., Bian, C., et al. (2012). Tet3 CXXC domain and dioxygenase activity cooperatively regulate key genes for Xenopus eye and neural development. Cell 151, 1200–1213. doi: 10.1016/j.cell.2012.11.014
Yildirim, O., Li, R., Hung, J. H., Chen, P. B., Dong, X., Ee, L. S., et al. (2011). Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell 147, 1498–1510. doi: 10.1016/j.cell.2011.11.054
Yu, H., Su, Y., Shin, J., Zhong, C., Guo, J. U., Weng, Y. L., et al. (2015). Tet3 regulates synaptic transmission and homeostatic plasticity via DNA oxidation and repair. Nat. Neurosci. 18, 836–843. doi: 10.1038/nn.4008
Zhang, R. R., Cui, Q. Y., Murai, K., Lim, Y. C., Smith, Z. D., Jin, S., et al. (2013). Tet1 regulates adult hippocampal neurogenesis and cognition. Cell Stem Cell 13, 237–245. doi: 10.1016/j.stem.2013.05.006
Keywords: TET enzymes, 5-hydroxymethylcytosine, neural progenitor cells, neurogenesis, neurodevelopmental disorders, epigenetics
Citation: MacArthur IC and Dawlaty MM (2021) TET Enzymes and 5-Hydroxymethylcytosine in Neural Progenitor Cell Biology and Neurodevelopment. Front. Cell Dev. Biol. 9:645335. doi: 10.3389/fcell.2021.645335
Received: 23 December 2020; Accepted: 01 February 2021;
Published: 18 February 2021.
Edited by:
Bruno Di Stefano, Baylor College of Medicine, United StatesReviewed by:
Claudia Doege, Columbia University, United StatesCopyright © 2021 MacArthur and Dawlaty. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Meelad M. Dawlaty, bWVlbGFkLmRhd2xhdHlAZWluc3RlaW5tZWQub3Jn
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.