- 1Cancer and Anticancer Drug Research Center, School of Pharmaceutical Sciences, Wenzhou Medical University, Wenzhou, China
- 2Comprehensive Cancer Center, Department of Cancer Biology and Genetics, The Ohio State University, Columbus, OH, United States
Lung cancer is the leading cause of cancer-related deaths worldwide and non-small cell lung cancer (NSCLC) accounts for more than 80% of all lung cancer cases. Recent advancements in diagnostic tools, surgical treatments, chemotherapies, and molecular targeted therapies that improved the therapeutic efficacy in NSCLC. However, the 5-years relative survival rate of NSCLC is only about 20% due to the inadequate screening methods and late onset of clinical symptoms. Dysregulation of microRNAs (miRNAs) was frequently observed in NSCLC and closely associated with NSCLC development, progression, and metastasis through regulating their target genes. In this review, we provide an updated overview of aberrant miRNA signature in NSCLC, and discuss the possibility of miRNAs becoming a diagnostic and therapeutic tool. We also discuss the possible causes of dysregulated miRNAs in NSCLC.
Introduction
MiRNAs are a class of small single-stranded, endogenous non-coding RNAs with approximately 20~22 nucleotide length. miRNAs act as post-transcriptional gene regulators by binding to the complementary 3′- untranslated regions (3′-UTRs) of target mRNAs, resulting in translational inhibition or degradation of mRNAs (Bartel, 2009). The first miRNA, lin-4, was discovered in 1993 by Ambros and colleagues from the Caenorhabditis elegans (C. elegans) (Lee et al., 1993). In 2000, another miRNA in C. elegans, let-7, was reported by Reinhart et al. let-7 capable of being inhibited the expression of heterochronic gene lin-41 by sequencing specific RNA - RNA interaction with the 3′-UTRs of its mRNA (Pasquinelli et al., 2000; Reinhart et al., 2000). In 2002, Dr. Croce group provided the first evidence that miRNA was involved in human cancer pathogenesis. They revealed that miR-15a and miR-16-1, located at 13q14 chromosome region, were frequently deleted in B-cell chronic lymphocytic leukemia (CLL) (Calin et al., 2002). Over the past decade, growing evidences have indicated that the dysregulation of miRNAs is implicated in the development, progression, and metastasis of various cancers (Iorio and Croce, 2012a).
Lung cancer is the most commonly diagnosed cancer and the leading cause of cancer-related deaths worldwide (Bray et al., 2018). Lung cancer is classified into two main histological groups, including NSCLC (~85%) and small cell lung cancer (SCLC, ~15%) (Osmani et al., 2018). miRNAs dysregulation is frequently found in NSCLC, and aberrant expressions of miRNAs play a key role in NSCLC proliferation, invasion, and metastasis through regulating their target genes (Peng et al., 2013; Cortez et al., 2015; Liang et al., 2020). In this review, we briefly introduce aberrant miRNA signatures in NSCLC and summarize how miRNAs act as oncogene or tumor suppressor to regulate NSCLC progression and metastasis by modulating their target genes. We also discuss the possibility of miRNAs becoming therapeutic targets or biomarkers in NSCLC.
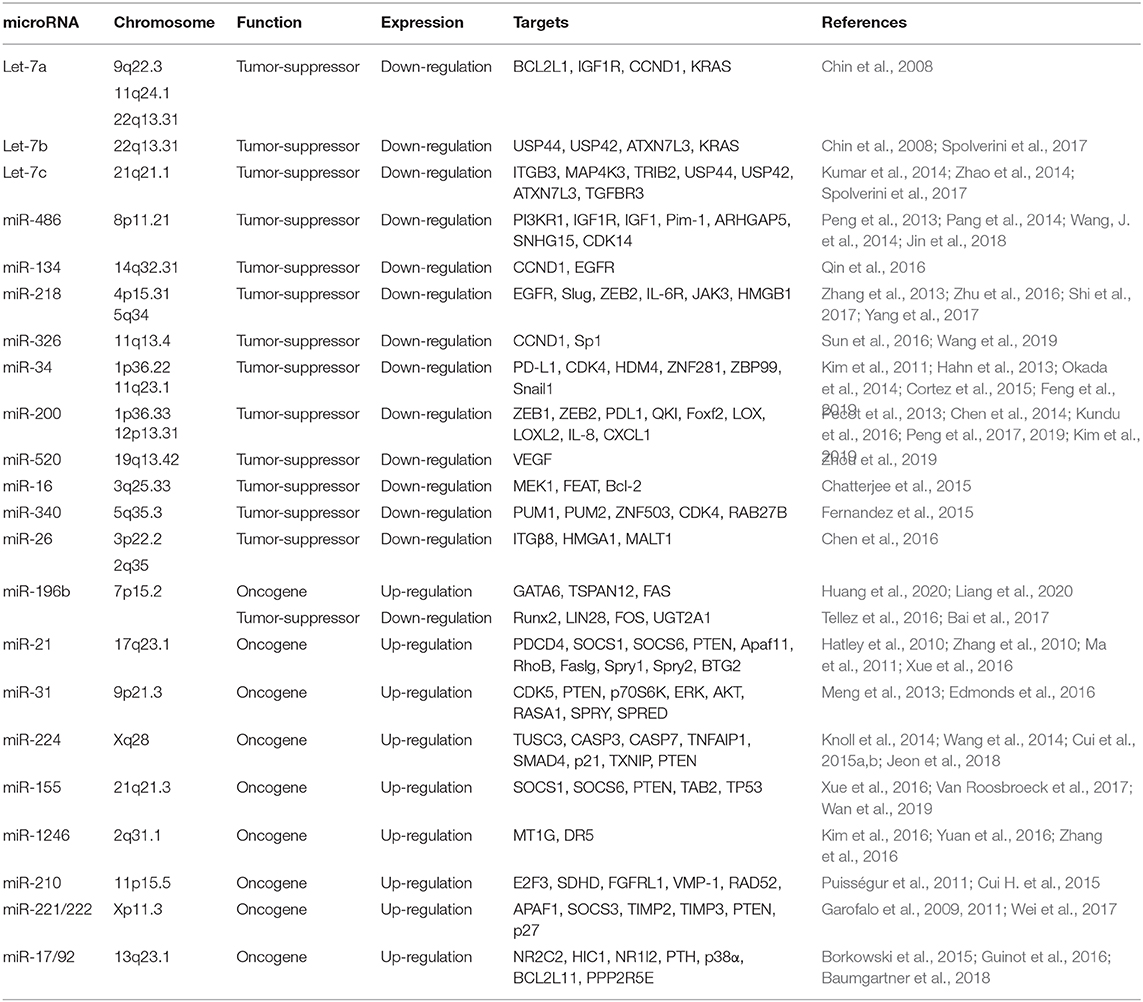
Tumor Suppressor and Oncogenic miRNAs in NSCLC
miRNA dysregulation causes aberrant expression of their target genes, thus involves various aspects of cancer cells including cell growth, apoptosis, metabolism, and invasion (Bartel, 2004; Bracken et al., 2016). A number of studies have shown that miRNAs function as tumor suppressors or oncogenes in NSCLC by inhibiting their target genes (Table 1). Here, we introduce several prominent miRNAs that play critical roles in NSCLC.
Tumor Suppressor miRNAs in NSCLC
let-7 Family
The let-7 family, a first known human miRNA (Pasquinelli et al., 2000; Reinhart et al., 2000), contains 10 isoforms that functions as tumor suppressors by targeting a wide variety of mRNAs encoding oncogenes (Roush and Slack, 2008), including RAS (Johnson et al., 2005). Takamizawa et al. reported that significantly shorter survival after potentially curative resection was observed in cases with reduced let-7 expression (Takamizawa et al., 2004). let-7c, a member of the let-7 family, prevents migration and invasion of NSCLC cells by degrading ITGB3 and MAP4K3. Low expression of let-7 was associated with metastasis, venous invasion, advanced TNM stages, and poor survival of NSCLC patients (Zhao et al., 2014). Loss of let-7 function enhances the lung tumor formation in mouse models, whereas exogenous delivery of let-7 to established tumors in mouse models of NSCLC significantly reduces the tumor burden (Trang et al., 2010). Let-7g effectively induces cell cycle arrest and cell death in K-RasG12D expressing murine lung cancer cells by targeting KRAS oncogene (Kumar et al., 2008). These evidences indicate that let-7 family may serve as prognostic marker and therapeutic target in certain type of NSCLC.
miR-34
The miR-34 family consists of miR-34a, miR-34b, and miR-34c, and is directly regulated by p53, a tumor suppressor gene (He et al., 2007). miR-34a is a direct proapoptotic transcriptional target of p53 that mediates part of the biological functions of p53. Down-regulated miR-34a expression is frequently seen in NSCLC, which may thus contribute to tumorigenesis by attenuating p53-dependent apoptosis (Raver-Shapira et al., 2007). Cortez et al. found that p53 regulates PDL1 expression by transcriptionally up-regulating miR-34 in NSCLC. miR-34 mimics alone or in combination with radiotherapy reduced PDL1 expression in the tumor and antagonized T-cell exhaustion (Cortez et al., 2015). MRX34, a liposomal formulation of miR-34a, is a potential first-in-class miRNA mimic for cancer therapy (Daige et al., 2014; Beg et al., 2017). Phase 1 study of MRX34 in patients with advanced solid tumors, including lung cancer, was conducted and demonstrated manageable toxicity in most patients and some clinical activity. Dose-dependent inhibition of related target genes provides proof-of-concept for miRNA-based cancer therapy (Hong et al., 2020).
miR-486
The low level of miR-486 was observed in various types of human cancer including NSCLC, and considered as an ideal biomarker in cancer diagnosis (Jiang et al., 2018). miR-486 directly targets components related to insulin growth factor (IGF) signaling, including IGF1, IGF1R, and PI3KR1, and functions as a tumor suppressor in NSCLC (Peng et al., 2013). Pim-1, a proto-oncogene, is up-regulated in NSCLC and its expression was associated with advanced stage and lymph node metastasis. miR-486 directly targets Pim-1 and downregulated miR-486 in NSCLC leads to the overexpression of Pim-1 to promote tumor progression, suggesting that critical tumor suppressive functions of miR-486 in NSCLC (Pang et al., 2014).
miR-218
Down-regulated miR-218 expression in NSCLC is implicated in histological grades and lymph node metastasis by targeting the EMT regulator, Slug and ZEB2 (Shi et al., 2017). Overexpression of miR-218 in NSCLC cells inhibits cell proliferation, invasion and colony formation by targeting the IL-6 receptor and JAK3. Down-regulated miR-218 expression was associated with poor prognosis of patients with NSCLC (Zhang et al., 2013; Yang et al., 2017). In addition, miR-218 functions as a tumor suppressor by targeting the expression of high mobility group box-1 (HMGB1) in NSCLC, suggesting that miR-218 might be a potential therapeutic biomarker for metastatic NSCLC patients (Zhang et al., 2013).
miR-200
The miR-200 family (miR-200a, miR-200b, miR-200c, miR-429, and miR-141) is a well-known tumor suppressor that inhibits EMT by targeting ZEB1 and ZEB2 in a wide range of cancers (Korpal et al., 2008; Park et al., 2008). Chen et al. found that ZEB1, an EMT activator and transcriptional repressor of miR-200, relieves miR-200 mediated repression of PD-L1 on tumor cells, leading to CD8+ T cell immunosuppression and metastasis (Chen et al., 2014). miR-200 mediated inhibition of ZEB1 sensitizes KRAS mutant NSCLC cells to MEK inhibition and reduces in vivo tumor growth of NSCLC (Peng et al., 2019). In addition, miR-200 family inhibits lung cancer cell invasion and metastasis by targeting Foxf2, a transcription factor that elevated in mesenchymal-like lung cancer cells (Kundu et al., 2016). miR-200 has also been reported to inhibit tumor angiogenesis in NSCLC by directly targeting interleukin-8 and CXCL1 secreted from the endothelial and cancer cells (Pecot et al., 2013), suggesting multifunction of miR-200 family in NSCLC progression and metastasis.
Onco-miRNAs in NSCLC
miR-196b
The function of miR-196b in NSCLC is controversial. Our recent study demonstrated that the expression of miR-196b is up-regulated NSCLC, and is negatively regulated by RNA binding protein QKI-5, a tumor suppressor in NSCLC. miR-196b promotes NSCLC progression and metastasis by targeting the tumor suppressors, TSPAN12, GATA6, and FAS (Liang et al., 2020) (Huang et al., 2020). However, Tellez et al. reported that miR-196b-5p was epigenetically silenced in the premalignant stage of lung cancer, suggesting that tumor-suppressive functions of miR-196b in NSCLC (Tellez et al., 2016). These dual functions of miR-196b might be due to differences in stage and treatment received and differences in the ethnic origin of the analyzed patients. Further study needs to validate these results.
miR-221/222
The miR-221/222 Cluster is one of the most commonly upregulated miRNA clusters in various cancers (Di Leva et al., 2014). miR-221/222 plays a key role in tyrosine kinase inhibitors (TKIs) resistance in NSCLC. EGFR and MET regulate miR-221/222 expression to control gefitinib-induced apoptosis and NSCLC tumorigenesis by targeting apoptotic peptidase activating factor 1 (APAF1) (Garofalo et al., 2011). In addition, miR-221/222 is involved in TNF-related apoptosis-inducing ligand (TRAIL)-resistance and tumorigenesis of NSCLC by targeting PTEN and TIMP3 tumor suppressors (Garofalo et al., 2009). Another study also showed that exosomic miR-222-3p promoted cell proliferation, gemcitabine resistance, migration, and invasion of NSCLC by targeting SOCS3 (Wei et al., 2017).
miR-17/92 Cluster
The miR-17/92 cluster encodes six miRNAs, including miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1, and miR-92a-1, and resides in intron 3 of the C13orf 25 gene at human chromosome 13q31.3, a region frequently amplified in various solid tumors (Ota et al., 2004). Hayashita et al. reported for the first time that miR-17/92 cluster is overexpressed in lung cancer cells and promotes cell growth (Hayashita et al., 2005). Inhibition of miR-17/92 cluster exerts cytotoxicity in p53-negative NSCLC cells by suppression of CYP24A1 (Borkowski et al., 2015). miR-19 family enhances Wnt signaling by targeting p38α in NSCLC, and increases NSCLC malignant potential (Guinot et al., 2016). Downregulated miR-19b expression is closely associated with reduced phosphorylation of ERK, AKT and their effector proteins in EGFR mutant NSCLC cells (Baumgartner et al., 2018), suggesting that targeting miR-19b could potentially be alternative therapeutic methods in EGFR mutant NSCLC.
miR-21
miR-21 plays oncogenic functions by suppressing many important tumor suppressor genes (Krichevsky and Gabriely, 2009). Large-scale profiling of miRNA expression in multiple human cancer tissue samples revealed that miR-21 is the only miRNA up-regulated in all types of analyzed tumor samples (Volinia et al., 2006). A high level of miR-21 is associated with advanced clinical stage and metastasis in NSCLC, and stimulates cell growth and invasion by inhibiting tumor suppressor PTEN in NSCLC (Zhang et al., 2010; Xue et al., 2016). The expression of miR-21 is increased and is associated with poor prognosis in NSCLC. miR-21 promotes tumorigenesis through inhibition of negative regulators of the Ras/MEK/ERK signaling pathway (Hatley et al., 2010), suggesting that promising oncogenic functions of miR-21 in NSCLC.
miR-224
Our previous studies demonstrated that miR-224 is up-regulated and promotes tumor progression and metastasis in NSCLC. miRNA-224-mediated tumor suppressor candidate 3 (TUSC3) deficiency enhances the metastatic potential of NSCLC through the regulation of three unfolded protein response pathways and HRD1-dependent endoplasmic reticulum associated degradation (ERAD) (Jeon et al., 2018). In addition, miR-224 directly targets tumor suppressor, TNFAIP1 and SMAD4, to promote tumor growth both in vitro and in vivo in NSCLC. Considering that SMAD4 plays a central role in the TGF-β family signaling pathways, and has low frequency of mutation and/or deletion, miR-224 might be an ideal therapeutic target for patients with certain NSCLC (Cui et al., 2015b).
NSCLC metastasis is a complex, multistep process involving a number of molecular and genetic changes, and is a sign of poor prognosis (Zhu et al., 2020). As described avobe, some miRNAs are closely implicated in fundamental processes of NSCLC metastasis. Therefore, deeply understanding miRNA signaling network will help to identify therapeutic targets for NSCLC metastasis.
Circulating miRNAs in NSCLC
Circulating miRNA was initially discovered in the cell-free blood plasma and serum, and was considered as a novel class of biomarkers for diagnosis and prognosis of various diseases including cancer (Chen et al., 2008; Lawrie et al., 2008). The plasma miR-590-5p was significantly down-regulated in NSCLC patients and serves as a potential prognostic marker in NSCLC (Khandelwal et al., 2020). Hypoxic lung cancer-cell-derived extracellular vesicle miR-103a induces oncogenic M2 macrophages polarization by targeting PTEN, it results in activation of AKT and STAT3 signaling as well as expression of several immunosuppressive and pro-angiogenic factors to facilitate cancer progression (Hsu et al., 2018). Circulating miR-320a secreted from neutrophils of high-risk heavy-smokers induces immunosuppressive macrophages M2 phenotype through downregulation of STAT4 to increase lung cancer risk (Fortunato et al., 2019). Although a growing number of studies have reported that circulating miRNAs may serve as diagnostic and prognostic biomarkers in NSCLC, concordance between the experimental results is extremely low due to the lack of standardized measurement methods. Accordingly, establishing a standard guideline is extremely important to screening circulating miRNA biomarkers in NSCLC.
Causes of miRNA Dysregulation in NSCLC
Aberrant miRNA signature was considered as an important biomarker for diagnosis and prognosis of NSCLC (Iorio and Croce, 2012a). Accumulating evidence suggests that genetic and epigenetic alterations, and transcriptional regulations are involved in miRNA dysregulation in cancers (Figure 1) (Iorio and Croce, 2012b).
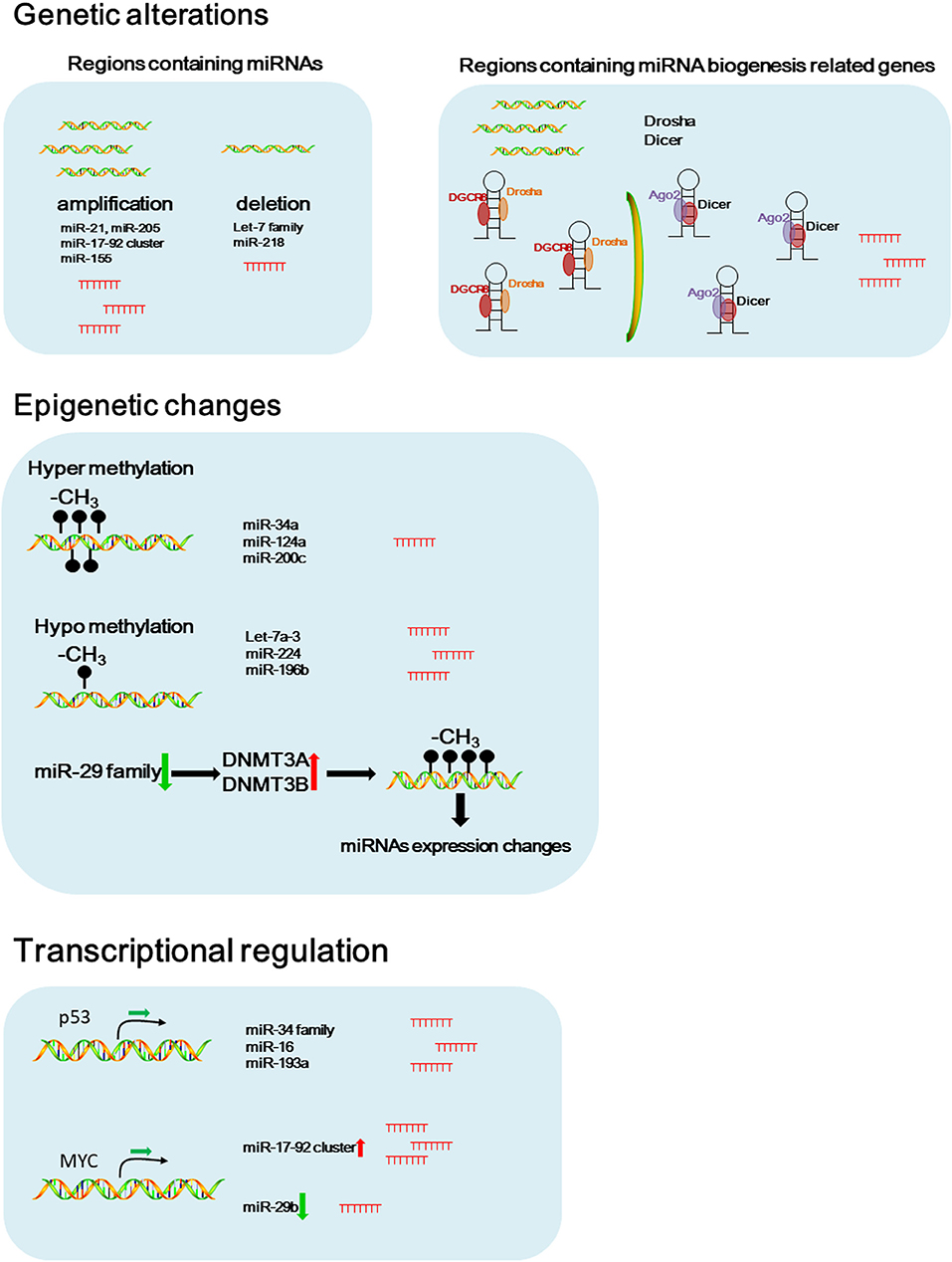
Figure 1. Potential causes of dysregulated miRNAs in NSCLC. Schematic figure of the possible pathways that involved in dysregulated miRNA expressions in NSCLC.
Genetic Alteration
Genetic alterations including chromosomal rearrangements, genomic amplifications, deletions, and gene mutations contribute to the miRNA dysregulation (Di Leva et al., 2014). In 2002, the earliest evidence of miRNAs deletion in human cancer was provided by Dr. Croce's group. They showed that the region containing miR-15a and miR-16-1 in chromosome 13q14 was frequently deleted in patients with B cell chronic lymphocytic leukemia (CLL) (Calin et al., 2002).
Amplification of miRNAs as a result of being located in amplified genomic regions is associated with carcinogenesis and tumor progression. miR-21 and miR-205, known as the oncogenic miRNAs, are located at the regions commonly amplified in lung cancer (Yanaihara et al., 2006). Another oncogenic miRNA, miR-17-92 cluster, is located within the third intron of the open reading frame 13 (C13orf25) at 13q31.3 genomic region. The region containing miR-17-92 cluster is frequently amplified in NSCLC, that results in upregulated miR-17-92 family expression contributing to NSCLC progression (Hayashita et al., 2005; Calin and Croce, 2006). A large scale copy number variation analysis using NSCLC tissue samples also showed that miR-21, miR-17, and miR-155 are the most frequently amplified miRNAs in NSCLC (Czubak et al., 2015). Inversesly, the let-7 family are located in regions commonly deleted in lung cancer, including let-7g at 3p21.1-21.2, let-7a-2 at 11q23-q24, and let-7c at 21q11.1 (Diederichs and Haber, 2006).
Aberrant expression or mutations in the genes that encoding key components of the miRNA biogenesis pathway contribute to the global downregulation of miRNAs in cancer (van Kouwenhove et al., 2011). Czubak et al. reported that the frequent amplification of DICER and DROSHA was observed in NSCLC (Czubak et al., 2015). In addition, the phosphorylation of argonaute 2 (AGO2) by c-Src inhibits the binding of DICER to AGO2 and promotes tumorigenesis of lung cancer, suggesting that change of these miRNA biogenesis related genes might cause alternation of miRNA signature in NSCLC (Liu et al., 2020). Under hypoxic stress, epidermal growth factor receptor (EGFR) suppresses the maturation of specific tumor-suppressor miRNAs by phosphorylation of argonaute 2 (AGO2). Phosphorylated AGO2 protein reduces the binding between Dicer and AGO2, results in inhibition of miRNA processing from precursor miRNAs to mature miRNAs in NSCLC cells (Shen et al., 2013).
Epigenetic Changes
In addition to genetic alterations, epigenetic changes such as aberrant DNA methylation and histone modifications may contribute to the dysregulated miRNA expression in cancers (Egger et al., 2004).
Methylation in the miR-34a promoter region was frequently seen in various cancers including NSCLC, that results in down-regulated miR-34a expression (Lodygin et al., 2008). Hypermethylation in the promoter region of miR-124a significantly reduces miR-124a expression in lung cancer cells, results in increased expression of CDK6 and Rb phosphorylation (Lujambio et al., 2007). miR-200c plays a critical role in regulating EMT and inhibits cell invasion and metastasis. Hypermethylation in the promoter region of miR-200c significantly reduces miR-200a expression, and that results in induction of an aggressive, invasive, and chemoresistant phenotype in NSCLC (Ceppi et al., 2010).
Conversely, the promoter region of let-7a-3 gene was hypomethylated in lung adenocarcinomas, that results in elevated let-7a-3 expression and subsequent enhanced tumor phenotypic changes in lung cancer (Brueckner et al., 2007). Recently, our study reported that the hypomethylation in the CpG islands of the miR-224 promoter region increased the expression of miR-224, and promoted tumor progression and metastasis of NSCLC by targeting SMAD4 and TNFα-induced protein 1 (TNFAIP1) (Cui et al., 2015b). In another study, we found that the expression of miR-196b-5p was partially controlled by hypomethylation in its promoter region and up-regulated miR-196b-5p promoted NSCLC cell migration, proliferation, and cell cycle through directly targeting the tumor suppressors, GATA6 and TSPAN12 (Liang et al., 2020).
The miR-29 family, including miR-29a, miR-29b, and miR-29c, is a key regulator of DNA methyltransferases, DNMT3A and DNMT3B. In NSCLC, reduced miR-29 expression causes hypermethylation in the promoter region of some tumor suppressor genes by regulating DNMT3A and DNMT3B, leading to elevated expression of tumor suppressor genes (Fabbri et al., 2007).
In addition to DNA methylation, histone modifications play an important role in chromatin remodeling, and cooperate with DNA methylation to regulate miRNA expression in cancers (Chuang and Jones, 2007). KDM5B, a histone H3 lysine 4 (H3K4) demethylase, promotes epithelial-mesenchymal transition (EMT) of lung cancer cells by repressing the expression of the miR-200 family (Enkhbaatar et al., 2013).
Transcriptional Control of miRNAs
Myc, an oncogenic transcription factor, has been reported to positively or negatively regulate the expression of many protein-coding genes and miRNAs (Croce, 2009). The overexpression of miR-17-92 cluster in NSCLC is associated with gene amplification of the miRNA cluster itself and enhanced expression of the myc gene (O'Donnell et al., 2005; Osada and Takahashi, 2011). In NSCLC, c-Myc suppresses miR-29b to promote tumor aggressiveness and poor outcomes by targeting tumor suppressor, FHIT (Wu et al., 2015).
p53, a well-known tumor suppressor, binds to the specific DNA sequence, termed the p53-responsive element (RE) to regulate p53 target genes. p53 promotes the transcription of the miR-34 family by binding to p53 REs in its promoter region (Bommer et al., 2007). p53 mediated increased expression of miR-34a promotes p53 induced apoptosis in NSCLC cells (Hollstein et al., 1991; He et al., 2007; Raver-Shapira et al., 2007). The upregulation of p53 simultaneously activates miR-34a and miR-16, which in turn targets Bcl2 to induce apoptosis in NSCLC cells (Upadhyay et al., 2019). Wang et al. found that miR-193a was directly activated by p53 at the transcriptional level and miR-193a targets EGFR through directly binding to 3′-UTR of the EGFR mRNA in NSCLC (Wang, W. et al., 2019). These facts indicate that the aberrant expression of transcription factors consistently contributes to the dysregulated miRNAs expression in NSCLC.
miRNAs Contribute to Drug Resistance of NSCLC
Platinum-based chemotherapy is a standard first-line treatment for NSCLC patients. However, most patients with NSCLC eventually develop resistance to chemotherapy (Rizvi et al., 2016). The changes in the type and amount of miRNAs in exosomes are associated with the resistance of NSCLC cells to chemotherapeutic drugs (Zhao et al., 2015). Qin et al. demonstrated that the miRNAs were differentially expressed in the exosomes of cisplatin (CDDP)-resistant and CDDP-sensitive NSCLC cells. In CDDP-resistant NSCLC cell lines, the amount of miR-100-5p was significantly decreased in exosomes and was functionally involved in CDDP resistance of NSCLC (Qin et al., 2017). In addition, cancer-associated fibroblasts (CAFs) derived exosomes confer cisplatin resistance of NSCLC cells through transferring miRNA-130a (Zhang et al., 2021).
Aberrant EGFR signaling is the main cause of metastatic NSCLC. Molecular targeted therapy for EGFR in NSCLC has achieved certain effects. However, mutations in EGFR and feedback activation of other signaling pathways limited the therapeutic efficacy of targeted therapy (Lai-Kwon et al., 2021). Abnormal expression of miRNAs has been considered as one of the causes of tyrosine kinase inhibitors (TKIs) resistance (Calin and Croce, 2006). Inhibition of miR-483-3p promotes gefitinib resistance in EGFR-mutant NSCLC, suggesting that miR-483-3p is a promising target to overcome acquired EGFR TKI resistance in EGFR-mutant NSCLC (Yue et al., 2018). Garofalo et al. reported that miR-30b-c and 221-222 are involved in gefitinib, an EGFR inhibitor, resistance of NSCLC cells by modulating APAF-1 and BIM (Garofalo et al., 2011). Zhou et al. found that miR-34a restored the gefitinib sensitivity in HGF/MET activation-mediated gefitinib-resistant NSCLC cells by targeting MET (Zhou et al., 2014). These facts suggest that manipulating the expression of miRNAs could sensitize drug resistance in NSCLC.
Conclusions and Future Directions
Recent advancements in whole-genome profiles of miRNAs for blood samples and biopsies of NSCLC patients facilitate discovery of new biomarkers for diagnosis, prognosis, aggressiveness, metastasis, and drug resistance of NSCLC. However, inconsistent results were also reported between different groups. These might be due to the differences in stage and treatment received for the analyzed samples. Thus, uniformed sample collection and detection methods, and adequate number of samples are necessary to obtain best results.
Preclinical studies of miRNA replacement therapy using miRNA mimics or inhibition of miRNA by antimiRs have shown promising results with minimal toxicity, indicating that targeting miRNAs might be potential novel therapeutics for NSCLC. Nevertheless, only few of miRNA therapeutics advanced into clinical trials. To achieve success in miRNA-based therapeutics, big challenge is identification of best target miRNAs in specific disease type. Other challenges include minimizing the off-target effects of miRNAs, preventing potential toxicities and developing more efficient and specific miRNA delivery methods.
Author Contributions
RC conceived the structure of manuscript. XZ, MK, and XH collected the articles and made a table. HS and HT guide XZ and XH to collect articles and made a figure. MK and CC pre-revised the manuscript. RC and CC wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (81672305 to RC), National Cancer Institute (5R35CA197706-06 to CC), and Zhejiang Province Natural Science Fundation of China (LY19H160029 to HT).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Bai, X., Meng, L., Sun, H., Li, Z., Zhang, X., and Hua, S. (2017). MicroRNA-196b inhibits cell growth and metastasis of lung cancer cells by targeting runx2. Cell. Physiol. Biochem. 43, 757–767. doi: 10.1159/000481559
Bartel, D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297. doi: 10.1016/S0092-8674(04)00045-5
Bartel, D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. doi: 10.1016/j.cell.2009.01.002
Baumgartner, U., Berger, F., Hashemi Gheinani, A., Burgener, S. S., Monastyrskaya, K., and Vassella, E. (2018). miR-19b enhances proliferation and apoptosis resistance via the EGFR signaling pathway by targeting PP2A and BIM in non-small cell lung cancer. Mol. Cancer 17:44. doi: 10.1186/s12943-018-0781-5
Beg, M. S., Brenner, A. J., Sachdev, J., Borad, M., Kang, Y. K., Stoudemire, J., et al. (2017). Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Invest. New Drugs 35, 180–188. doi: 10.1007/s10637-016-0407-y
Bommer, G. T., Gerin, I., Feng, Y., Kaczorowski, A. J., Kuick, R., Love, R. E., et al. (2007). p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr. Biol. 17, 1298–1307. doi: 10.1016/j.cub.2007.06.068
Borkowski, R., Du, L., Zhao, Z., McMillan, E., Kosti, A., Yang, C. R., et al. (2015). Genetic mutation of p53 and suppression of the miR-17~92 cluster are synthetic lethal in non-small cell lung cancer due to upregulation of vitamin D Signaling. Cancer Res. 75, 666–675. doi: 10.1158/0008-5472.CAN-14-1329
Bracken, C. P., Scott, H. S., and Goodall, G. J. (2016). A network-biology perspective of microRNA function and dysfunction in cancer. Nat. Rev. Genet. 17, 719–732. doi: 10.1038/nrg.2016.134
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A., and Jemal, A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424. doi: 10.3322/caac.21492
Brueckner, B., Stresemann, C., Kuner, R., Mund, C., Musch, T., Meister, M., et al. (2007). The human let-7a-3 locus contains an epigenetically regulated microRNA gene with oncogenic function. Cancer Res. 67, 1419–1423. doi: 10.1158/0008-5472.CAN-06-4074
Calin, G. A., and Croce, C. M. (2006). MicroRNA signatures in human cancers. Nat. Rev. Cancer 6, 857–866. doi: 10.1038/nrc1997
Calin, G. A., Dumitru, C. D., Shimizu, M., Bichi, R., Zupo, S., Noch, E., et al. (2002). Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. U. S. A. 99, 15524–15529. doi: 10.1073/pnas.242606799
Ceppi, P., Mudduluru, G., Kumarswamy, R., Rapa, I., Scagliotti, G. V., Papotti, M., et al. (2010). Loss of miR-200c expression induces an aggressive, invasive, and chemoresistant phenotype in non-small cell lung cancer. Mol. Cancer Res. 8, 1207–1216. doi: 10.1158/1541-7786.MCR-10-0052
Chatterjee, A., Chattopadhyay, D., and Chakrabarti, G. (2015). MiR-16 targets Bcl-2 in paclitaxel-resistant lung cancer cells and overexpression of miR-16 along with miR-17 causes unprecedented sensitivity by simultaneously modulating autophagy and apoptosis. Cell. Signal 27, 189–203. doi: 10.1016/j.cellsig.2014.11.023
Chen, C. Y., Chang, J. T., Ho, Y. F., and Shyu, A. B. (2016). MiR-26 down-regulates TNF-α/NF-κB signalling and IL-6 expression by silencing HMGA1 and MALT1. Nucl. Acids Res. 44, 3772–3787. doi: 10.1093/nar/gkw205
Chen, L., Gibbons, D. L., Goswami, S., Cortez, M. A., Ahn, Y. H., Byers, L. A., et al. (2014). Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat. Commun. 5:5241. doi: 10.1038/ncomms6241
Chen, X., Ba, Y., Ma, L., Cai, X., Yin, Y., Wang, K., et al. (2008). Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 18, 997–1006. doi: 10.1038/cr.2008.282
Chin, L. J., Ratner, E., Leng, S., Zhai, R., Nallur, S., Babar, I., et al. (2008). A SNP in a let-7 microRNA complementary site in the KRAS 3' untranslated region increases non-small cell lung cancer risk. Cancer Res. 68, 8535–8540. doi: 10.1158/0008-5472.CAN-08-2129
Chuang, J. C., and Jones, P. A. (2007). Epigenetics and microRNAs. Pediatr. Res. 61, 24–29. doi: 10.1203/pdr.0b013e3180457684
Cortez, M. A., Ivan, C., Valdecanas, D., Wang, X., Peltier, H. J., Ye, Y., et al. (2015). PDL1 regulation by p53 via miR-34. J. Natl. Cancer Inst. 108:djv303. doi: 10.1093/jnci/djv303
Croce, C. M. (2009). Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 10, 704–714. doi: 10.1038/nrg2634
Cui, H., Seubert, B., Stahl, E., Dietz, H., Reuning, U., Moreno-Leon, L., et al. (2015). Tissue inhibitor of metalloproteinases-1 induces a pro-tumourigenic increase of miR-210 in lung adenocarcinoma cells and their exosomes. Oncogene 34, 3640–3650. doi: 10.1038/onc.2014.300
Cui, R., Kim, T., Fassan, M., Meng, W., Sun, H. L., Jeon, Y. J., et al. (2015a). MicroRNA-224 is implicated in lung cancer pathogenesis through targeting caspase-3 and caspase-7. Oncotarget 6, 21802–21815. doi: 10.18632/oncotarget.5224
Cui, R., Meng, W., Sun, H. L., Kim, T., Ye, Z., Fassan, M., et al. (2015b). MicroRNA-224 promotes tumor progression in nonsmall cell lung cancer. Proc. Natl. Acad. Sci. U. S. A. 112, E4288–4297. doi: 10.1073/pnas.1502068112
Czubak, K., Lewandowska, M. A., Klonowska, K., Roszkowski, K., Kowalewski, J., Figlerowicz, M., et al. (2015). High copy number variation of cancer-related microRNA genes and frequent amplification of DICER1 and DROSHA in lung cancer. Oncotarget 6, 23399–23416. doi: 10.18632/oncotarget.4351
Daige, C. L., Wiggins, J. F., Priddy, L., Nelligan-Davis, T., Zhao, J., and Brown, D. (2014). Systemic delivery of a miR34a mimic as a potential therapeutic for liver cancer. Mol. Cancer Ther. 13, 2352–2360. doi: 10.1158/1535-7163.MCT-14-0209
Di Leva, G., Garofalo, M., and Croce, C. M. (2014). MicroRNAs in cancer. Annu. Rev. Pathol. 9, 287–314. doi: 10.1146/annurev-pathol-012513-104715
Diederichs, S., and Haber, D. A. (2006). Sequence variations of microRNAs in human cancer: alterations in predicted secondary structure do not affect processing. Cancer Res. 66, 6097–6104. doi: 10.1158/0008-5472.CAN-06-0537
Edmonds, M. D., Boyd, K. L., Moyo, T., Mitra, R., Duszynski, R., Arrate, M. P., et al. (2016). MicroRNA-31 initiates lung tumorigenesis and promotes mutant KRAS-driven lung cancer. J. Clin. Invest. 126, 349–364. doi: 10.1172/JCI82720
Egger, G., Liang, G., Aparicio, A., and Jones, P. A. (2004). Epigenetics in human disease and prospects for epigenetic therapy. Nature 429, 457–463. doi: 10.1038/nature02625
Enkhbaatar, Z., Terashima, M., Oktyabri, D., Tange, S., Ishimura, A., Yano, S., et al. (2013). KDM5B histone demethylase controls epithelial-mesenchymal transition of cancer cells by regulating the expression of the microRNA-200 family. Cell Cycle 12, 2100–2112. doi: 10.4161/cc.25142
Fabbri, M., Garzon, R., Cimmino, A., Liu, Z., Zanesi, N., Callegari, E., et al. (2007). MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc. Natl. Acad. Sci. U. S. A. 104, 15805–15810. doi: 10.1073/pnas.0707628104
Feng, H., Ge, F., Du, L., Zhang, Z., and Liu, D. (2019). MiR-34b-3p represses cell proliferation, cell cycle progression and cell apoptosis in non-small-cell lung cancer (NSCLC) by targeting CDK4. J. Cell. Mol. Med. 23, 5282–5291. doi: 10.1111/jcmm.14404
Fernandez, S., Risolino, M., Mandia, N., Talotta, F., Soini, Y., Incoronato, M., et al. (2015). miR-340 inhibits tumor cell proliferation and induces apoptosis by targeting multiple negative regulators of p27 in non-small cell lung cancer. Oncogene 34, 3240–3250. doi: 10.1038/onc.2014.267
Fortunato, O., Borzi, C., Milione, M., Centonze, G., Conte, D., Boeri, M., et al. (2019). Circulating mir-320a promotes immunosuppressive macrophages M2 phenotype associated with lung cancer risk. Int. J. Cancer 144, 2746–2761. doi: 10.1002/ijc.31988
Garofalo, M., Di Leva, G., Romano, G., Nuovo, G., Suh, S. S., Ngankeu, A., et al. (2009). miR-221and222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell 16, 498–509. doi: 10.1016/j.ccr.2009.10.014
Garofalo, M., Romano, G., Di Leva, G., Nuovo, G., Jeon, Y. J., Ngankeu, A., et al. (2011). EGFR and MET receptor tyrosine kinase-altered microRNA expression induces tumorigenesis and gefitinib resistance in lung cancers. Nat. Med. 18, 74–82. doi: 10.1038/nm.2577
Guinot, A., Oeztuerk-Winder, F., and Ventura, J. J. (2016). miR-17-92/p38α dysregulation enhances wnt signaling and selects lgr6+ cancer stem-like cells during lung adenocarcinoma progression. Cancer Res. 76, 4012–4022. doi: 10.1158/0008-5472.CAN-15-3302
Hahn, S., Jackstadt, R., Siemens, H., Hünten, S., and Hermeking, H. (2013). SNAIL and miR-34a feed-forward regulation of ZNF281/ZBP99 promotes epithelial-mesenchymal transition. EMBO J. 32, 3079–3095. doi: 10.1038/emboj.2013.236
Hatley, M. E., Patrick, D. M., Garcia, M. R., Richardson, J. A., Bassel-Duby, R., van Rooij, E., et al. (2010). Modulation of k-ras-dependent lung tumorigenesis by microRNA-21. Cancer Cell 18, 282–293. doi: 10.1016/j.ccr.2010.08.013
Hayashita, Y., Osada, H., Tatematsu, Y., Yamada, H., Yanagisawa, K., Tomida, S., et al. (2005). A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 65, 9628–9632. doi: 10.1158/0008-5472.CAN-05-2352
He, L., He, X., Lim, L. P., de Stanchina, E., Xuan, Z., Liang, Y., et al. (2007). A microRNA component of the p53 tumour suppressor network. Nature 447, 1130–1134. doi: 10.1038/nature05939
Hollstein, M., Sidransky, D., Vogelstein, B., and Harris, C. C. (1991). p53 mutations in human cancers. Science 253, 49–53. doi: 10.1126/science.1905840
Hong, D. S., Kang, Y. K., Borad, M., Sachdev, J., Ejadi, S., Lim, H. Y., et al. (2020). Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. Br. J. Cancer 122, 1630–1637. doi: 10.1038/s41416-020-0802-1
Hsu, Y. L., Hung, J. Y., Chang, W. A., Jian, S. F., Lin, Y. S., Pan, Y. C., et al. (2018). Hypoxic lung-cancer-derived extracellular vesicle microRNA-103a increases the oncogenic effects of macrophages by targeting PTEN. Mol. Ther. 26, 568–581. doi: 10.1016/j.ymthe.2017.11.016
Huang, X., Xiao, S., Zhu, X., Yu, Y., Cao, M., Zhang, X., et al. (2020). miR-196b-5p-mediated downregulation of FAS promotes NSCLC progression by activating IL6-STAT3 signaling. Cell Death Dis. 11:785. doi: 10.1038/s41419-020-02997-7
Iorio, M. V., and Croce, C. M. (2012a). MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol. Med. 4, 143–159. doi: 10.1002/emmm.201100209
Iorio, M. V., and Croce, C. M. (2012b). Causes and consequences of microRNA dysregulation. Cancer J. 18, 215–222. doi: 10.1097/PPO.0b013e318250c001
Jeon, Y. J., Kim, T., Park, D., Nuovo, G. J., Rhee, S., Joshi, P., et al. (2018). miRNA-mediated TUSC3 deficiency enhances UPR and ERAD to promote metastatic potential of NSCLC. Nat. Commun. 9:5110. doi: 10.1038/s41467-018-07561-8
Jiang, M., Li, X., Quan, X., Yang, X., Zheng, C., Hao, X., et al. (2018). MiR-486 as an effective biomarker in cancer diagnosis and prognosis: a systematic review and meta-analysis. Oncotarget 9, 13948–13958. doi: 10.18632/oncotarget.24189
Jin, B., Jin, H., Wu, H.-B., Xu, J.-J., and Li, B. (2018). Long non-coding RNA SNHG15 promotes CDK14 expression via miR-486 to accelerate non-small cell lung cancer cells progression and metastasis. J. Cell. Physiol. 233, 7164–7172. doi: 10.1002/jcp.26543
Johnson, S. M., Grosshans, H., Shingara, J., Byrom, M., Jarvis, R., Cheng, A., et al. (2005). RAS is regulated by the let-7 microRNA family. Cell 120, 635–647. doi: 10.1016/j.cell.2005.01.014
Khandelwal, A., Seam, R. K., Gupta, M., Rana, M. K., Prakash, H., Vasquez, K. M., et al. (2020). Circulating microRNA-590-5p functions as a liquid biopsy marker in non-small cell lung cancer. Cancer Sci. 111, 826–839. doi: 10.1111/cas.14199
Kim, E. J., Kim, J. S., Lee, S., Lee, H., Yoon, J. S., Hong, J. H., et al. (2019). QKI, a miR-200 target gene, suppresses epithelial-to-mesenchymal transition and tumor growth. Int. J. Cancer 145, 1585–1595. doi: 10.1002/ijc.32372
Kim, G., An, H. J., Lee, M. J., Song, J. Y., Jeong, J. Y., Lee, J. H., et al. (2016). Hsa-miR-1246 and hsa-miR-1290 are associated with stemness and invasiveness of non-small cell lung cancer. Lung Cancer 91, 15–22. doi: 10.1016/j.lungcan.2015.11.013
Kim, N. H., Kim, H. S., Li, X.-Y., Lee, I., Choi, H.-S., Kang, S. E., et al. (2011). A p53/miRNA-34 axis regulates Snail1-dependent cancer cell epithelial-mesenchymal transition. J. Cell Biol. 195, 417–433. doi: 10.1083/jcb.201103097
Knoll, S., Fürst, K., Kowtharapu, B., Schmitz, U., Marquardt, S., Wolkenhauer, O., et al. (2014). E2F1 induces miR-224/452 expression to drive EMT through TXNIP downregulation. EMBO Rep. 15, 1315–1329. doi: 10.15252/embr.201439392
Korpal, M., Lee, E. S., Hu, G., and Kang, Y. (2008). The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J. Biol. Chem. 283, 14910–14914. doi: 10.1074/jbc.C800074200
Krichevsky, A. M., and Gabriely, G. (2009). miR-21: a small multi-faceted RNA. J. Cell. Mol. Med. 13, 39–53. doi: 10.1111/j.1582-4934.2008.00556.x
Kumar, M. S., Armenteros-Monterroso, E., East, P., Chakravorty, P., Matthews, N., Winslow, M. M., et al. (2014). HMGA2 functions as a competing endogenous RNA to promote lung cancer progression. Nature 505, 212–217. doi: 10.1038/nature12785
Kumar, M. S., Erkeland, S. J., Pester, R. E., Chen, C. Y., Ebert, M. S., Sharp, P. A., et al. (2008). Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc. Natl. Acad. Sci. U. S. A. 105, 3903–3908. doi: 10.1073/pnas.0712321105
Kundu, S. T., Byers, L. A., Peng, D. H., Roybal, J. D., Diao, L., Wang, J., et al. (2016). The miR-200 family and the miR-183~96~182 cluster target Foxf2 to inhibit invasion and metastasis in lung cancers. Oncogene 35, 173–186. doi: 10.1038/onc.2015.71
Lai-Kwon, J., Tiu, C., Pal, A., Khurana, S., and Minchom, A. (2021). Moving beyond epidermal growth factor receptor resistance in metastatic non-small cell lung cancer - a drug development perspective. Crit. Rev. Oncol. Hematol. 159:103225. doi: 10.1016/j.critrevonc.2021.103225
Lawrie, C. H., Gal, S., Dunlop, H. M., Pushkaran, B., Liggins, A. P., Pulford, K., et al. (2008). Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br. J. Haematol. 141, 672–675. doi: 10.1111/j.1365-2141.2008.07077.x
Lee, R. C., Feinbaum, R. L., and Ambros, V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854. doi: 10.1016/0092-8674(93)90529-Y
Liang, G., Meng, W., Huang, X., Zhu, W., Yin, C., Wang, C., et al. (2020). miR-196b-5p-mediated downregulation of TSPAN12 and GATA6 promotes tumor progression in non-small cell lung cancer. Proc. Natl. Acad. Sci. U. S. A. 117, 4347–4357. doi: 10.1073/pnas.1917531117
Liu, T., Zhang, H., Fang, J., Yang, Z., Chen, R., Wang, Y., et al. (2020). AGO2 phosphorylation by c-Src kinase promotes tumorigenesis. Neoplasia 22, 129–141. doi: 10.1016/j.neo.2019.12.004
Lodygin, D., Tarasov, V., Epanchintsev, A., Berking, C., Knyazeva, T., Körner, H., et al. (2008). Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle 7, 2591–2600. doi: 10.4161/cc.7.16.6533
Lujambio, A., Ropero, S., Ballestar, E., Fraga, M. F., Cerrato, C., Setién, F., et al. (2007). Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 67, 1424–1429. doi: 10.1158/0008-5472.CAN-06-4218
Ma, X., Kumar, M., Choudhury, S. N., Becker Buscaglia, L. E., Barker, J. R., Kanakamedala, K., et al. (2011). Loss of the miR-21 allele elevates the expression of its target genes and reduces tumorigenesis. Proc. Natl. Acad. Sci. U. S. A. 108, 10144–10149. doi: 10.1073/pnas.1103735108
Meng, W., Ye, Z., Cui, R., Perry, J., Dedousi-Huebner, V., Huebner, A., et al. (2013). MicroRNA-31 predicts the presence of lymph node metastases and survival in patients with lung adenocarcinoma. Clin. Cancer Res. 19, 5423–5433. doi: 10.1158/1078-0432.CCR-13-0320
O'Donnell, K. A., Wentzel, E. A., Zeller, K. I., Dang, C. V., and Mendell, J. T. (2005). c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435, 839–843. doi: 10.1038/nature03677
Okada, N., Lin, C.-P., Ribeiro, M. C., Biton, A., Lai, G., He, X., et al. (2014). A positive feedback between p53 and miR-34 miRNAs mediates tumor suppression. Genes Dev. 28, 438–450. doi: 10.1101/gad.233585.113
Osada, H., and Takahashi, T. (2011). let-7 and miR-17-92: small-sized major players in lung cancer development. Cancer Sci. 102, 9–17. doi: 10.1111/j.1349-7006.2010.01707.x
Osmani, L., Askin, F., Gabrielson, E., and Li, Q. K. (2018). Current WHO guidelines and the critical role of immunohistochemical markers in the subclassification of non-small cell lung carcinoma (NSCLC): moving from targeted therapy to immunotherapy. Semin. Cancer Biol. 52, 103–109. doi: 10.1016/j.semcancer.2017.11.019
Ota, A., Tagawa, H., Karnan, S., Tsuzuki, S., Karpas, A., Kira, S., et al. (2004). Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res. 64:3087. doi: 10.1158/0008-5472.CAN-03-3773
Pang, W., Tian, X., Bai, F., Han, R., Wang, J., Shen, H., et al. (2014). Pim-1 kinase is a target of miR-486-5p and eukaryotic translation initiation factor 4E, and plays a critical role in lung cancer. Mol. Cancer 13:240. doi: 10.1186/1476-4598-13-240
Park, S. M., Gaur, A. B., Lengyel, E., and Peter, M. E. (2008). The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 22, 894–907. doi: 10.1101/gad.1640608
Pasquinelli, A. E., Reinhart, B. J., Slack, F., Martindale, M. Q., Kuroda, M. I., Maller, B., et al. (2000). Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 408, 86–89. doi: 10.1038/35040556
Pecot, C. V., Rupaimoole, R., Yang, D., Akbani, R., Ivan, C., Lu, C., et al. (2013). Tumour angiogenesis regulation by the miR-200 family. Nat. Commun. 4:2427. doi: 10.1038/ncomms3427
Peng, D. H., Kundu, S. T., Fradette, J. J., Diao, L., Tong, P., Byers, L. A., et al. (2019). ZEB1 suppression sensitizes KRAS mutant cancers to MEK inhibition by an IL17RD-dependent mechanism. Sci. Transl. Med. 11:483. doi: 10.1126/scitranslmed.aaq1238
Peng, D. H., Ungewiss, C., Tong, P., Byers, L. A., Wang, J., Canales, J. R., et al. (2017). ZEB1 induces LOXL2-mediated collagen stabilization and deposition in the extracellular matrix to drive lung cancer invasion and metastasis. Oncogene 36, 1925–1938. doi: 10.1038/onc.2016.358
Peng, Y., Dai, Y., Hitchcock, C., Yang, X., Kassis, E. S., Liu, L., et al. (2013). Insulin growth factor signaling is regulated by microRNA-486, an underexpressed microRNA in lung cancer. Proc. Natl. Acad. Sci. U. S. A. 110, 15043–15048. doi: 10.1073/pnas.1307107110
Puisségur, M. P., Mazure, N. M., Bertero, T., Pradelli, L., Grosso, S., Robbe-Sermesant, K., et al. (2011). miR-210 is overexpressed in late stages of lung cancer and mediates mitochondrial alterations associated with modulation of HIF-1 activity. Cell Death Differ. 18, 465–478. doi: 10.1038/cdd.2010.119
Qin, Q., Wei, F., Zhang, J., Wang, X., and Li, B. (2016). miR-134 inhibits non-small cell lung cancer growth by targeting the epidermal growth factor receptor. J. Cell. Mol. Med. 20, 1974–1983. doi: 10.1111/jcmm.12889
Qin, X., Yu, S., Zhou, L., Shi, M., Hu, Y., Xu, X., et al. (2017). Cisplatin-resistant lung cancer cell-derived exosomes increase cisplatin resistance of recipient cells in exosomal miR-100-5p-dependent manner. Int. J. Nanomed. 12, 3721–3733. doi: 10.2147/IJN.S131516
Raver-Shapira, N., Marciano, E., Meiri, E., Spector, Y., Rosenfeld, N., Moskovits, N., et al. (2007). Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol. Cell 26, 731–743. doi: 10.1016/j.molcel.2007.05.017
Reinhart, B. J., Slack, F. J., Basson, M., Pasquinelli, A. E., Bettinger, J. C., Rougvie, A. E., et al. (2000). The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403, 901–906. doi: 10.1038/35002607
Rizvi, N. A., Hellmann, M. D., Brahmer, J. R., Juergens, R. A., Borghaei, H., Gettinger, S., et al. (2016). Nivolumab in combination with platinum-based doublet chemotherapy for first-line treatment of advanced non-small-cell lung cancer. J. Clin. Oncol. 34, 2969–2979. doi: 10.1200/JCO.2016.66.9861
Roush, S., and Slack, F. J. (2008). The let-7 family of microRNAs. Trends Cell Biol. 18, 505–516. doi: 10.1016/j.tcb.2008.07.007
Shen, J., Xia, W., Khotskaya, Y. B., Huo, L., Nakanishi, K., Lim, S. O., et al. (2013). EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature 497, 383–387. doi: 10.1038/nature12080
Shi, Z. M., Wang, L., Shen, H., Jiang, C. F., Ge, X., Li, D. M., et al. (2017). Downregulation of miR-218 contributes to epithelial-mesenchymal transition and tumor metastasis in lung cancer by targeting Slug/ZEB2 signaling. Oncogene 36, 2577–2588. doi: 10.1038/onc.2016.414
Spolverini, A., Fuchs, G., Bublik, D. R., and Oren, M. (2017). let-7b and let-7c microRNAs promote histone H2B ubiquitylation and inhibit cell migration by targeting multiple components of the H2B deubiquitylation machinery. Oncogene 36, 5819–5828. doi: 10.1038/onc.2017.187
Sun, C., Huang, C., Li, S., Yang, C., Xi, Y., Wang, L., et al. (2016). Hsa-miR-326 targets CCND1 and inhibits non-small cell lung cancer development. Oncotarget 7, 8341–8359. doi: 10.18632/oncotarget.7071
Takamizawa, J., Konishi, H., Yanagisawa, K., Tomida, S., Osada, H., Endoh, H., et al. (2004). Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 64, 3753–3756. doi: 10.1158/0008-5472.CAN-04-0637
Tellez, C. S., Juri, D. E., Do, K., Picchi, M. A., Wang, T., Liu, G., et al. (2016). miR-196b is epigenetically silenced during the premalignant stage of lung carcinogenesis. Cancer Res. 76, 4741–4751. doi: 10.1158/0008-5472.CAN-15-3367
Trang, P., Medina, P. P., Wiggins, J. F., Ruffino, L., Kelnar, K., Omotola, M., et al. (2010). Regression of murine lung tumors by the let-7 microRNA. Oncogene 29, 1580–1587. doi: 10.1038/onc.2009.445
Upadhyay, P., Sarker, S., Ghosh, A., Gupta, P., Das, S., Ahir, M., et al. (2019). Transferrin-decorated thymoquinone-loaded PEG-PLGA nanoparticles exhibit anticarcinogenic effect in non-small cell lung carcinoma via the modulation of miR-34a and miR-16. Biomater. Sci. 7, 4325–4344. doi: 10.1039/C9BM00912D
van Kouwenhove, M., Kedde, M., and Agami, R. (2011). MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat. Rev. Cancer 11, 644–656. doi: 10.1038/nrc3107
Van Roosbroeck, K., Fanini, F., Setoyama, T., Ivan, C., Rodriguez-Aguayo, C., Fuentes-Mattei, E., et al. (2017). Combining anti-mir-155 with chemotherapy for the treatment of lung cancers. Clin. Cancer Res. 23, 2891–2904. doi: 10.1158/1078-0432.CCR-16-1025
Volinia, S., Calin, G. A., Liu, C. G., Ambs, S., Cimmino, A., Petrocca, F., et al. (2006). A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. U. S. A. 103, 2257–2261. doi: 10.1073/pnas.0510565103
Wan, J., Yang, X., Ren, Y., Li, X., Zhu, Y., Haddock, A. N., et al. (2019). Inhibition of miR-155 reduces impaired autophagy and improves prognosis in an experimental pancreatitis mouse model. Cell Death Dis. 10:303. doi: 10.1038/s41419-019-1545-x
Wang, H., Zhu, L. J., Yang, Y. C., Wang, Z. X., and Wang, R. (2014). MiR-224 promotes the chemoresistance of human lung adenocarcinoma cells to cisplatin via regulating G1/S transition and apoptosis by targeting p21(WAF1/CIP1). Br. J. Cancer 111, 339–354. doi: 10.1038/bjc.2014.157
Wang, J., Tian, X., Han, R., Zhang, X., Wang, X., Shen, H., et al. (2014). Downregulation of miR-486-5p contributes to tumor progression and metastasis by targeting protumorigenic ARHGAP5 in lung cancer. Oncogene 33, 1181–1189. doi: 10.1038/onc.2013.42
Wang, R., Xu, J., Xu, J., Zhu, W., Qiu, T., Li, J., et al. (2019). MiR-326/Sp1/KLF3: a novel regulatory axis in lung cancer progression. Cell Prolif. 52:e12551. doi: 10.1111/cpr.12551
Wang, W., Shen, X. B., Jia, W., Huang, D. B., Wang, Y., and Pan, Y. Y. (2019). The p53/miR-193a/EGFR feedback loop function as a driving force for non-small cell lung carcinoma tumorigenesis. Ther. Adv. Med. Oncol. 11:1758835919850665. doi: 10.1177/1758835919850665
Wei, F., Ma, C., Zhou, T., Dong, X., Luo, Q., Geng, L., et al. (2017). Exosomes derived from gemcitabine-resistant cells transfer malignant phenotypic traits via delivery of miRNA-222-3p. Mol. Cancer 16:132. doi: 10.1186/s12943-017-0694-8
Wu, D. W., Hsu, N. Y., Wang, Y. C., Lee, M. C., Cheng, Y. W., Chen, C. Y., et al. (2015). c-Myc suppresses microRNA-29b to promote tumor aggressiveness and poor outcomes in non-small cell lung cancer by targeting FHIT. Oncogene 34, 2072–2082. doi: 10.1038/onc.2014.152
Xue, X., Liu, Y., Wang, Y., Meng, M., Wang, K., Zang, X., et al. (2016). MiR-21 and MiR-155 promote non-small cell lung cancer progression by downregulating SOCS1, SOCS6, and PTEN. Oncotarget 7, 84508–84519. doi: 10.18632/oncotarget.13022
Yanaihara, N., Caplen, N., Bowman, E., Seike, M., Kumamoto, K., Yi, M., et al. (2006). Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 9, 189–198. doi: 10.1016/j.ccr.2006.01.025
Yang, Y., Ding, L., Hu, Q., Xia, J., Sun, J., Wang, X., et al. (2017). MicroRNA-218 functions as a tumor suppressor in lung cancer by targeting IL-6/STAT3 and negatively correlates with poor prognosis. Mol. Cancer 16:141. doi: 10.1186/s12943-017-0710-z
Yuan, D., Xu, J., Wang, J., Pan, Y., Fu, J., Bai, Y., et al. (2016). Extracellular miR-1246 promotes lung cancer cell proliferation and enhances radioresistance by directly targeting DR5. Oncotarget 7, 32707–32722. doi: 10.18632/oncotarget.9017
Yue, J., Lv, D., Wang, C., Li, L., Zhao, Q., Chen, H., et al. (2018). Epigenetic silencing of miR-483-3p promotes acquired gefitinib resistance and EMT in EGFR-mutant NSCLC by targeting integrin β3. Oncogene 37, 4300–4312. doi: 10.1038/s41388-018-0276-2
Zhang, C., Ge, S., Hu, C., Yang, N., and Zhang, J. (2013). MiRNA-218, a new regulator of HMGB1, suppresses cell migration and invasion in non-small cell lung cancer. Acta Biochim. Biophys. Sin. (Shanghai) 45, 1055–1061. doi: 10.1093/abbs/gmt109
Zhang, J. G., Wang, J. J., Zhao, F., Liu, Q., Jiang, K., and Yang, G. H. (2010). MicroRNA-21 (miR-21) represses tumor suppressor PTEN and promotes growth and invasion in non-small cell lung cancer (NSCLC). Clin. Chim. Acta 411, 846–852. doi: 10.1016/j.cca.2010.02.074
Zhang, T., Zhang, P., and Li, H. X. (2021). CAFs-derived exosomal mirna-130a confers cisplatin resistance of NSCLC cells through pum2-dependent packaging. Int. J. Nanomed. 16, 561–577. doi: 10.2147/IJN.S271976
Zhang, W. C., Chin, T. M., Yang, H., Nga, M. E., Lunny, D. P., Lim, E. K., et al. (2016). Tumour-initiating cell-specific miR-1246 and miR-1290 expression converge to promote non-small cell lung cancer progression. Nat. Commun. 7:11702. doi: 10.1038/ncomms11702
Zhao, B., Han, H., Chen, J., Zhang, Z., Li, S., Fang, F., et al. (2014). MicroRNA let-7c inhibits migration and invasion of human non-small cell lung cancer by targeting ITGB3 and MAP4K3. Cancer Lett. 342, 43–51. doi: 10.1016/j.canlet.2013.08.030
Zhao, L., Liu, W., Xiao, J., and Cao, B. (2015). The role of exosomes and “exosomal shuttle microRNA” in tumorigenesis and drug resistance. Cancer Lett. 356, 339–346. doi: 10.1016/j.canlet.2014.10.027
Zhou, J. Y., Chen, X., Zhao, J., Bao, Z., Chen, X., Zhang, P., et al. (2014). MicroRNA-34a overcomes HGF-mediated gefitinib resistance in EGFR mutant lung cancer cells partly by targeting MET. Cancer Lett. 351, 265–271. doi: 10.1016/j.canlet.2014.06.010
Zhou, W., Wu, Y., Pan, M., Liu, D., and Liu, B. (2019). Proliferation and migration of lung cancer could be inhibited by oxymatrine through the regulation for miR-520/VEGF. Am. J. Chin. Med. 47, 865–878. doi: 10.1142/S0192415X19500459
Zhu, K., Ding, H., Wang, W., Liao, Z., Fu, Z., Hong, Y., et al. (2016). Tumor-suppressive miR-218-5p inhibits cancer cell proliferation and migration via EGFR in non-small cell lung cancer. Oncotarget 7, 28075–28085. doi: 10.18632/oncotarget.8576
Keywords: NSCLC, miRNA, miRNA therapeutics, circulatory miRNAs, miRNA dysregulation
Citation: Zhu X, Kudo M, Huang X, Sui H, Tian H, Croce CM and Cui R (2021) Frontiers of MicroRNA Signature in Non-small Cell Lung Cancer. Front. Cell Dev. Biol. 9:643942. doi: 10.3389/fcell.2021.643942
Received: 19 December 2020; Accepted: 15 March 2021;
Published: 07 April 2021.
Edited by:
Boris Zhivotovsky, Karolinska Institutet (KI), SwedenReviewed by:
Alexey Zamaraev, Lomonosov Moscow State University, RussiaAmmad Farooqi, Institute of Biomedical and Genetic Engineering (IBGE), Pakistan
Copyright © 2021 Zhu, Kudo, Huang, Sui, Tian, Croce and Cui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ri Cui, d3ptdWN1aXJpQDE2My5jb20=; Carlo M. Croce, Y2FybG8uY3JvY2VAb3N1bWMuZWR1
†These authors have contributed equally to this work
 Xinping Zhu1†
Xinping Zhu1† Masahisa Kudo
Masahisa Kudo Carlo M. Croce
Carlo M. Croce Ri Cui
Ri Cui