- 1National Research Institute for Family Planning, Beijing, China
- 2National Human Genetic Resources Center, Beijing, China
- 3Graduate School of Peking Union Medical College, Beijing, China
- 4Beijing Chao-Yang Hospital, Capital Medical University, Beijing, China
SAPHO syndrome is a rare chronic inflammatory disease which is characterized by the comprehensive manifestations of bone, joint, and skin. However, little is known about the pathogenesis of SAPHO syndrome. A genome-wide association study (GWAS) of 49 patients and 121 control subjects have primarily focused on identification of common genetic variants associated with SAPHO, the data were analyzed by classical multiple logistic regression. Later, GWAS findings were further validated using whole exome sequencing (WES) in 16 patients and 15 controls to identify potentially functional pathways involved in SAPHO pathogenesis. In general, 40588 SNPs in genomic regions were associated with P < 0.05 after filter process, only 9 SNPs meet the expected cut-off P-value, however, none of them had association with SAPHO syndrome based on published literatures. And then, 15 pathways were found involved in SAPHO pathogenesis, of them, 6 pathways including osteoclast differentiation, bacterial invasion of epithelial cells, et al., had strong association with skin, osteoarticular manifestations of SAPHO or inflammatory reaction based published research. This study identified aberrant osteoclast differentiation and other pathways were involved in SAPHO syndrome. This finding may give insight into the understanding of pathogenic genes of SAPHO and provide the basis for SAPHO research and treatment.
Introduction
SAPHO (synovitis, acne, pustulosis, hyperostosis, and osteitis) syndrome, with the clinical manifestations including auto-inflammatory osteoarticular disorders and dermatological conditions, is a rare disease with an estimated prevalence of less than 1 in 10,000 (Magrey and Khan, 2009). It was first reported by the rheumatologist Chamot in 1987 (Kerrison et al., 2004); however, its etiology is still unknown. Previous research reported that the dysregulation of interleukin-1 (IL-1) signaling promoted sterile osteomyelitis in Pstpip2-deficient mice (Ferguson et al., 2006; Sharma and Ferguson, 2013). However, no specific variants were found using genetic screening in the PSTPIP1, PSTPIP2, NOD2 or LPIN2 genes in SAPHO samples (Hurtado-Nedelec et al., 2010; Colina et al., 2012; Guo et al., 2019). There are several factors considered to have the role in the development of SAPHO syndrome, including Propionibacterium acnes infection (Kotilainen et al., 1996; Colina et al., 2007), impaired immune responses, over-activated TH17 axis (Firinu et al., 2014). It was reported the proportion and absolute counts of Th17 cells in untreated SAPHO patients were significantly higher than in healthy controls, and the proportion and absolute counts of NK cells were significantly reduced in SAPHO patients compared with controls (Xu et al., 2019). Apart from these, the inflammatory factors including IL-18, IL-6, IL-8, IL-17A, TNF-α, and IL-1β were higher in SAPHO patients compared with healthy controls (Przepiera-Bedzak et al., 2016; Zhang et al., 2019). To date, no evidence-based therapeutic option has been proposed because of the elusive pathogenesis of this disease. Actual major therapeutic drugs are glucocorticoids, bisphosphonates, non-steroidal anti-inflammatory drugs (NSAIDs), disease-modifying antirheumatic drugs (DMARDs) to biologics, and antibiotics (Yang et al., 2018).
Genome-wide association studies (GWASs) has showed remarkable success in detecting the genetic factors of complex diseases by identifying multiple variants associated with complex clinical phenotypes (International Multiple Sclerosis Genetics Consortium, 2013). A commonly mentioned strategy in GWASs involves the evaluation of individual markers by setting a genome-wide significance cutoff p-value assuming-independence between markers. In this study, we set up a GWAS study in SAPHO patients, followed by pathway-based analyses of GWAS data that focused on the integrated effects of numerous loci, each making a small direct contribution to estimate of disease susceptibility, which might provide understanding the genetic basis of chronic diseases (International Multiple Sclerosis Genetics Consortium, 2013). The GWAS findings were further validated using whole exome sequencing (WES) to discover genetic variants and abnormal pathways involved in SAPHO patients.
Materials and Methods
Patients and Study Design
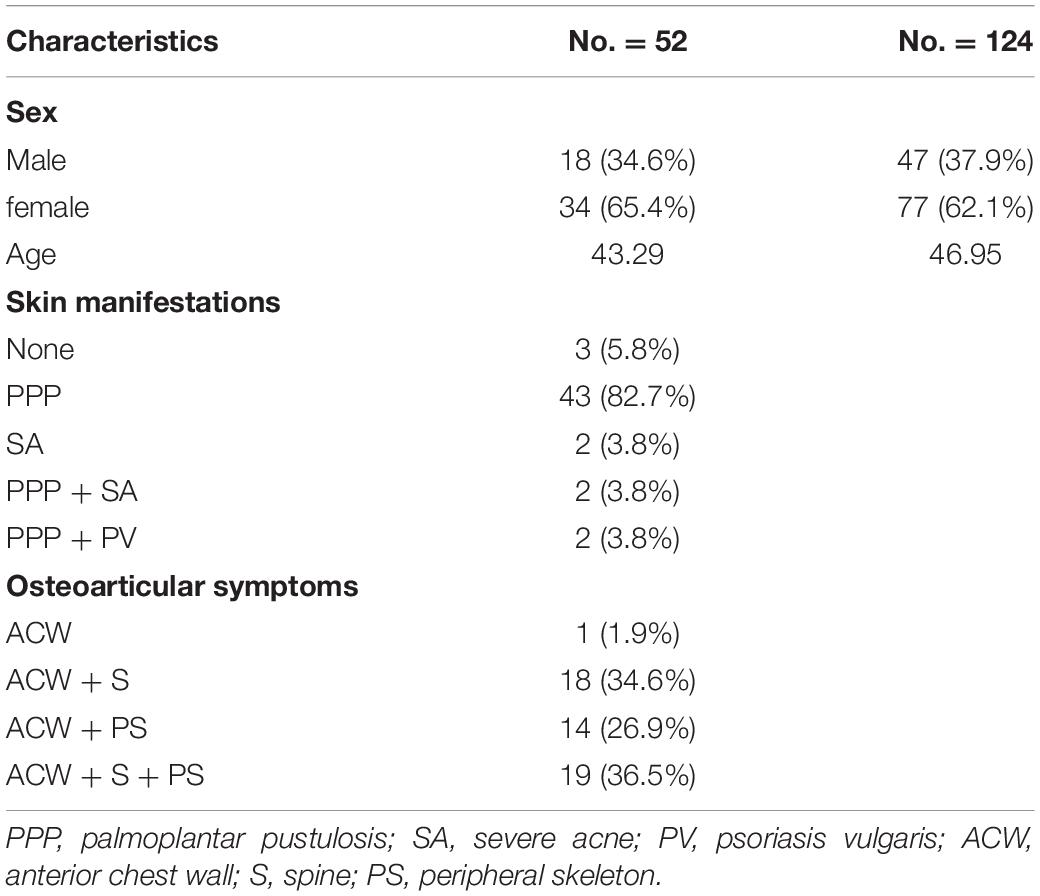
The GWAS study contained 52 SAPHO patients and 124 healthy controls (detailed characteristics are shown in Table 1). All individuals were enrolled from the Beijing Chaoyang Hospital, and were ethnically and geographically matched. SAPHO syndrome was diagnosed according to the Kahn criteria (Kahn et al., 1994). The study was approved by the Ethics Committee of the National Research Institute for Family Planning, and all participants provided written informed consent for participation in this study. The WES study was composed of 16 SAPHO patients (6 males, 10 females; mean age, 41.4 ± 0.08 years, range 33 to 72 years; 12 patients diagnosed with ACW + S + PS (ACW, anterior chest wall; S, spine; PS, peripheral skeleton), 3 patients diagnosed with ACW + S, 1 patient diagnosed with ACW + PS) and 15 healthy controls (sex and age matched).
DNA Isolation
Blood samples were collected from the peripheral blood of individuals into tubes containing EDTA. DNA extraction was carried out using the RelaxGene Blood DNA System Kit (Tiangen Biotech, Beijing, China) according to the manufacturer’s instructions. For the GWAS, all samples were genotyped individually using Illumina Infinium OmniZhongHua-8v1-3_A1 by the BioMiao Biological Technology Company (Beijing, China). For the WES, 3 μg of purified gDNA was fragmented to 180–280 bp and subjected to DNA library creation using established Illumina paired-end protocols. The Agilent SureSelect Human All ExonV6 Kit (Agilent Technologies, Santa Clara, CA, United States) was used for exome capture according to the manufacturer’s instructions. The Illumina Novaseq 6000 platform (Illumina Inc., San Diego, CA, United States) was utilized for genomic DNA sequencing by Novogene Bioinformatics Technology Co., Ltd (Beijing, China) to generate 150-bp paired-end reads with a minimum coverage of 10× for ∼99% of the genome (mean coverage of 100×).
Quality Control and Data Mining
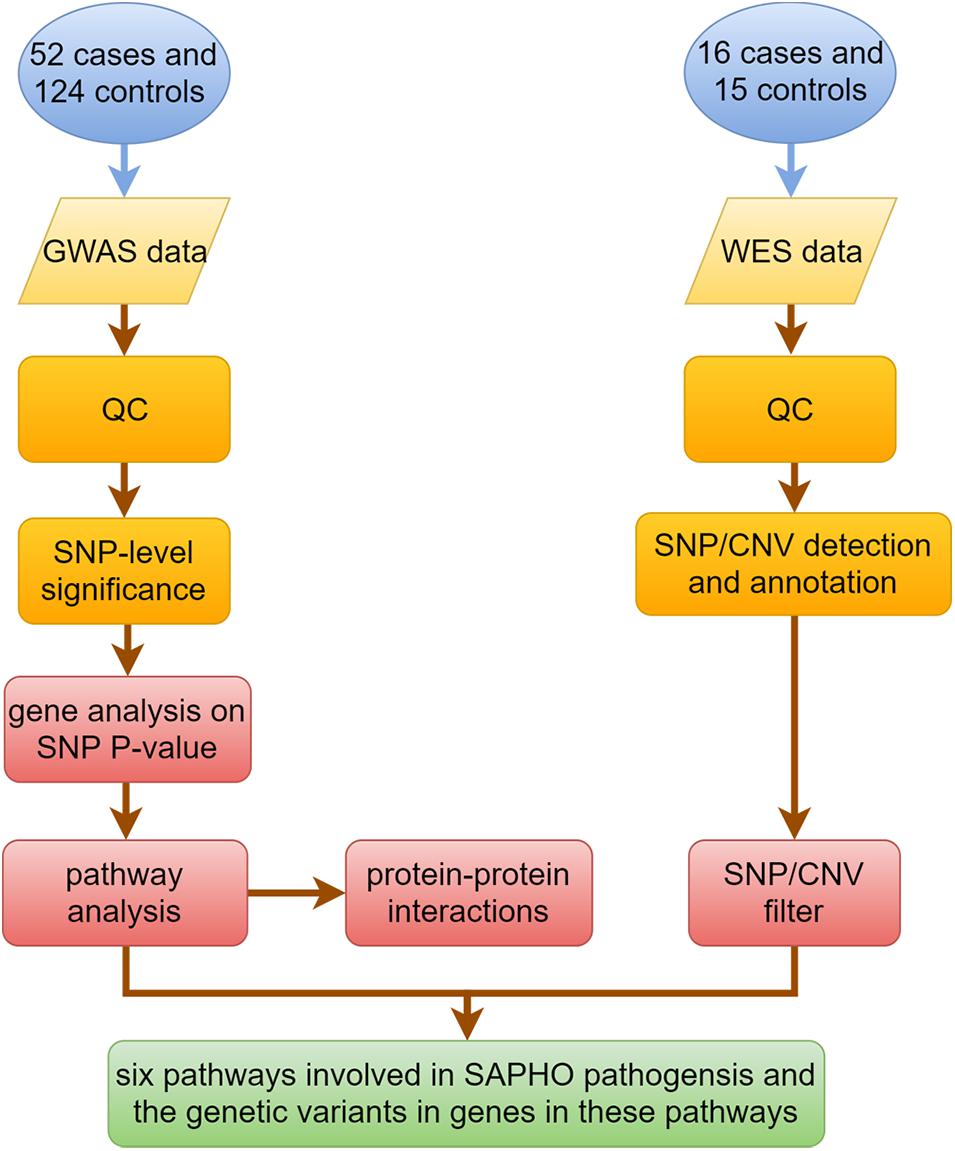
Figure 1 shows the key steps in our analysis method. For the GWAS study, quality control (QC) and data analysis were performed using the software packages R version 3.6.01 and PLINK version 1.90 beta2. Genotype data were cleaned before analysis by excluding SNPs or individuals that did not fulfill the QC criteria, which included: SNP call proportion ≥ 95%, subject completeness proportion ≥ 95%, SNP minor allele frequency ≥ 0.01, and SNP conformity with Hardy-Weinberg equilibrium expectations (P ≥ 0.01 in controls). A comparison of cases and controls was made using Pearson’s chi-square tests or Fisher’s exact test. Because this study examined the functional relationships of genes and proteins, we considered gene-level significance rather than that of single SNP in the traditional GWAS studies. To that end, SNPs in the GWAS were mapped to functional genes according to SNP locations and gene locations by MAGMA software (v1.07beta) (de Leeuw et al., 2015). In order to capture gene regulatory regions, gene boundaries were defined as 5 kb beyond the 5′- UTRs and 1.5 kb beyond 3′-UTRs of each gene. Gene analysis on SNP P-value data was performed by MAGMA and candidate genes were listed according to the gene P-value. P < 0.05 was considered statistically significant. Genes with P < 0.05 were selected for pathway analysis by DAVID software (v6.8) (Huang da et al.,2009a,b), and protein-protein interactions (PPI) by String software (v11) (Szklarczyk et al., 2019).
For WES, after quality control (QC) and preprocessing of sequencing data, the clean data in fastq format was aligned to the human reference genome hg19 (GRCh37) using the Burrows-Wheeler Aligner (bwa) (Li and Durbin, 2009) along with Samtools (Li et al., 2009). Single nucleotide variants (SNVs) and indels were detected with the best-practices GATK/Picard Pipeline (McKenna et al., 2010; Van der Auwera et al., 2013). The VCF data of all samples were merged by bcftools software for further analysis. Annotation was performed using Ensembl Variant Effect Predictor (v91.3) (McLaren et al., 2016) and ANNOVAR (Wang et al., 2010). The annotation information used for further filtering included minor allele frequencies from public databases, deleteriousness and conservation scores, assessment of the likely pathogenicity of variants and consequence of every single variant identified. After preliminary filtering, we extracted the SNPs in genes involved in disease-related pathways selected by pathway analysis. The copy number variants (CNVs) were detected with ExomeDepth software (Plagnol et al., 2012) after being processed by Samtools and annotated with AnnotSV (Geoffroy et al., 2018).
Results
Data Analysis of GWAS
This GWAS study contained 52 SAPHO patients (18 males, 34 females; mean age 43.29 years) and 124 controls (47 males, 77 females; mean age 46.95 years). The number of SNPs were reduced from 878,000 to 802,276 after filtering for low call rate (<90%), minor allele frequency (<0.01) and deviation from Hardy-Weinberg equilibrium (P < 0.00001). Six samples were deleted due to quality control, finally 49 cases and 121 controls were left for subsequent analysis. The mean genotyping rate in the remaining individuals was 99.73%.
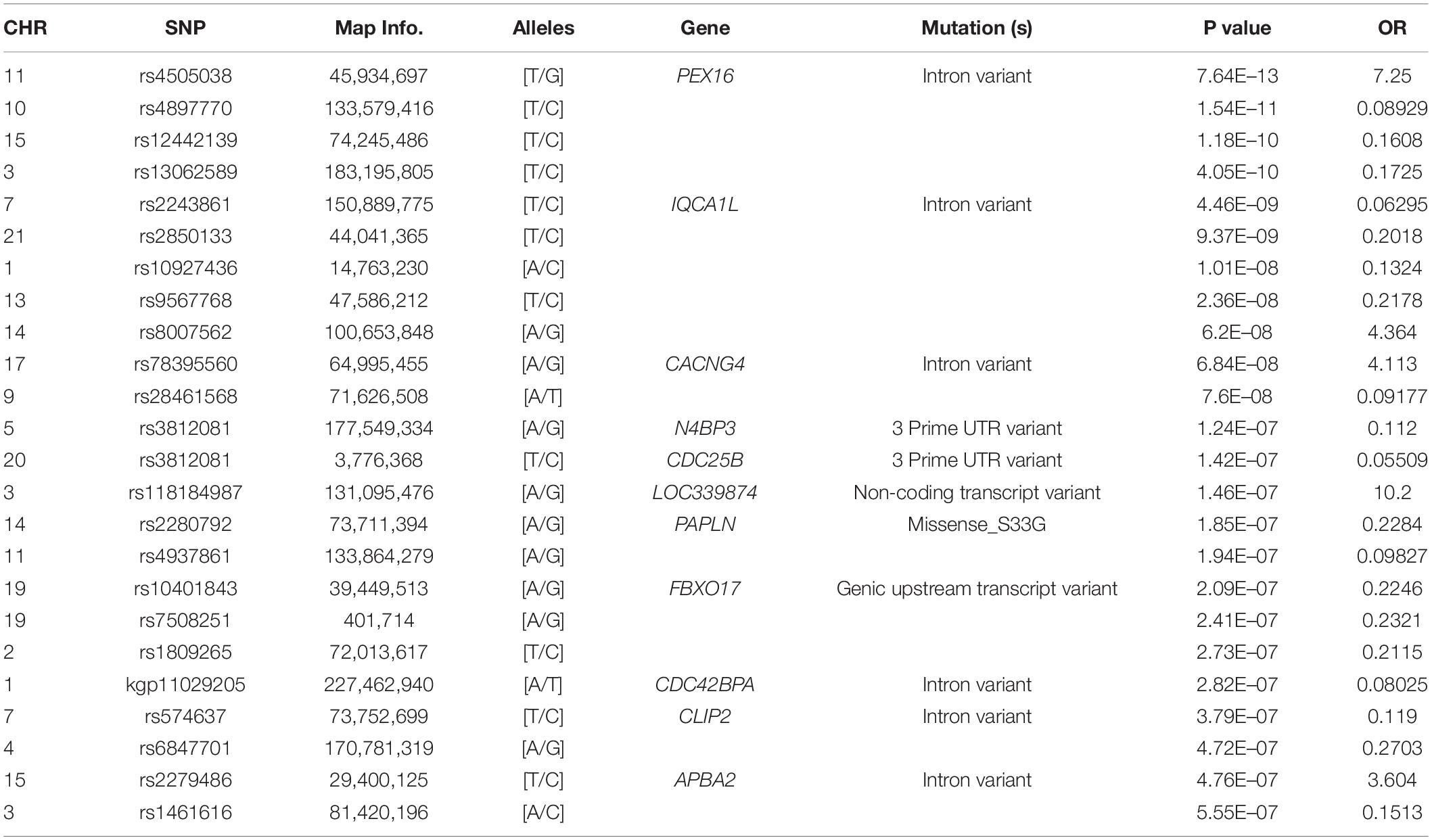
To detect associations, we performed a preliminary analysis by Pearson’s chi-square test. Results were adjusted by multiple test correction and then rank ordered on the basis of their P values. Overall, 40,588 SNPs in genomic regions were associated with P < 0.05 without correction, 84 SNPs were associated with P < 5.6 × 10–7, and only 9 SNPs met the expected cut-off P-value (P < 6.24 × 10–8, Figure 2). Among 9 SNPs, rs4505038 was located in the intron region of the peroxisomal biogenesis factor 16 gene (PEX16), rs2243861 was located in the intron region of the IQ motif containing with AAA domain 1 like gene (IQCA1L), and the other 7 SNPs (rs4897770, rs12442139, rs13062589, rs2850133, rs10927436, rs9567768, and rs8007562) were mapped to genomic regions with no known functional genes. Based on previously published literature, none of the 9 SNPs had an association with SAPHO syndrome or other inflammatory disease. Next, we lowered the significance threshold and 84 SNPs with a P-value below 5.6 × 10–7, a level roughly 10-times the expected threshold, were selected for further analysis. However, no further SNPs or genes were identified (Table 2). Given the complex symptoms and etiology of SAPHO syndrome, we inferred no single genetic variant accounted for this entire complicated syndrome.
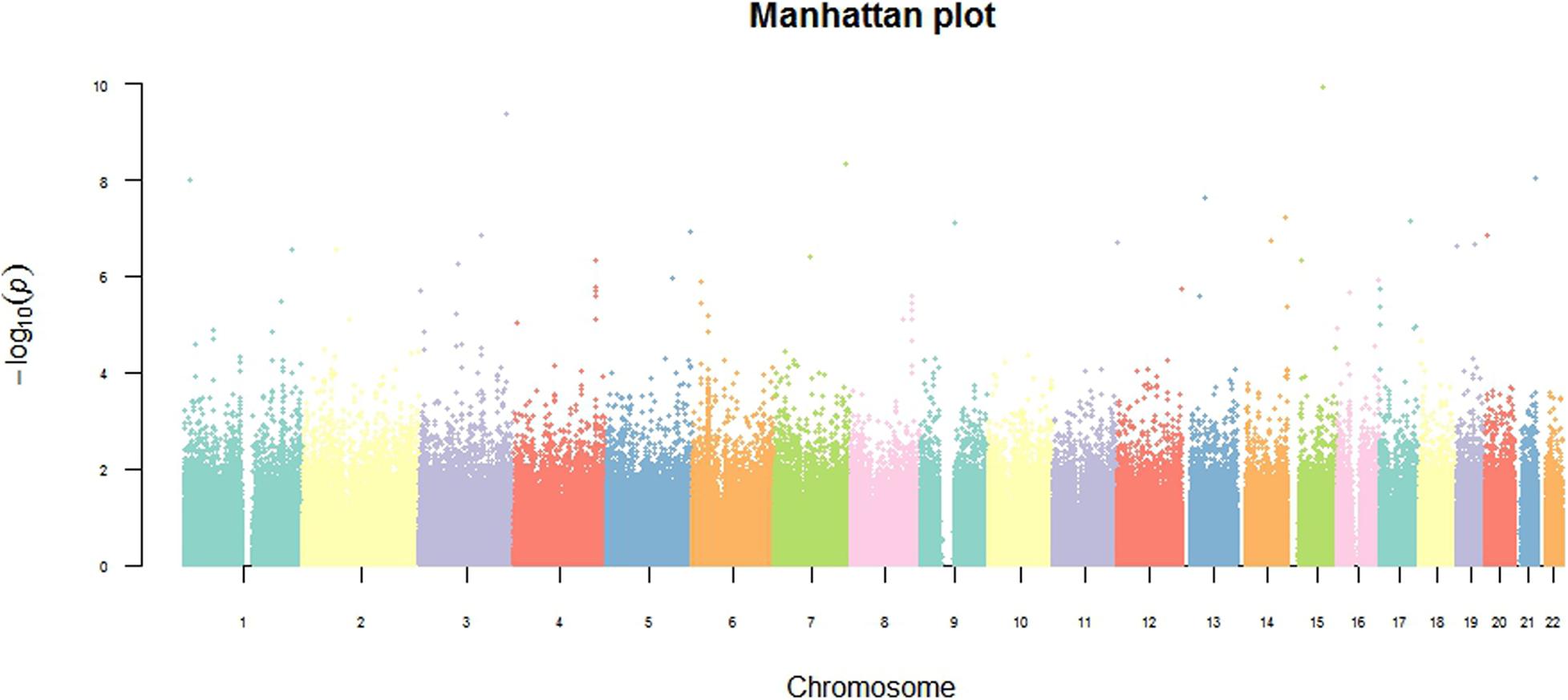
Figure 2. Genome-wide overview of GWAS findings. The Manhattan plot shows genome-wide association analysis of 802,276 single-nucleotide polymorphisms (SNPs) in 49 SAPHO patients and 121 control subjects. The 2 log10 (P-value) for each SNP is plotted against their chromosomal position. All statistical tests were two-sided.
As in other GWAS studies of this kind, many SNPs that did not reach the statistical significance level were abandon in further analysis. There is a certain proportion of rejected associations that are actually false negative; meanwhile, many studies have showed some significant combinations of gene markers with only limited association if they were involved in the same biological pathway or molecular mechanism. Compared to single-locus associations identified by classical genome-wide analysis, this type of analysis is useful for identifying pathways and networks involved in disease susceptibility in accordance with current models of pathogenesis, as well as identifying statistically over-represented but unexpected pathways responsible for novel disease mechanisms (Baranzini et al., 2009; Ritchie, 2009; International Multiple Sclerosis Genetics Consortium, 2013).
Pathway Analysis of GWAS Data
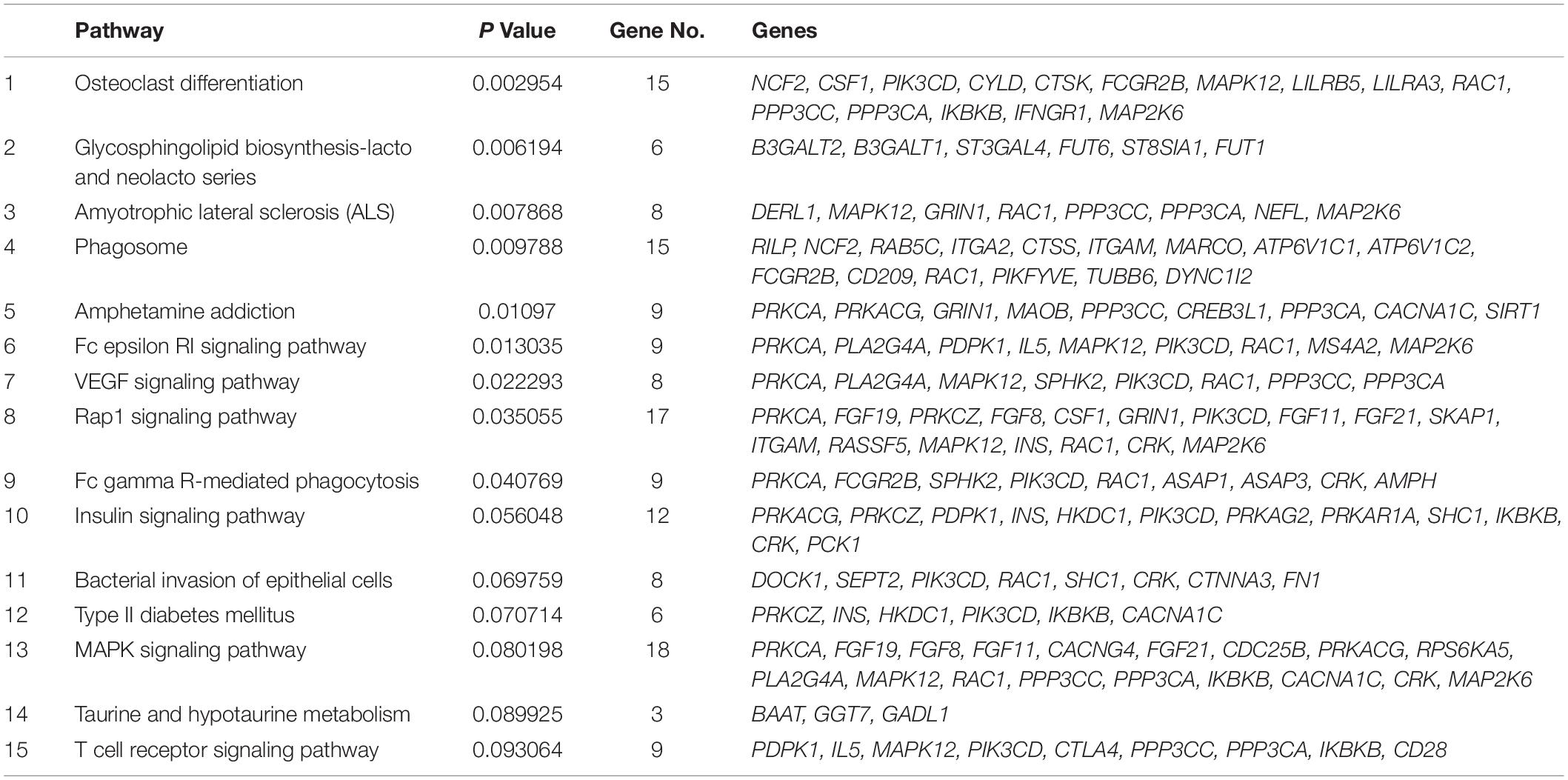
To dissect the pathways involved in SAPHO disease, we proposed a pathway-oriented analysis of the GWAS result. We analyzed a list of differentially expressed genes and a P-value for each gene was performed on SNP that indicated the strength of the gene-disease associations. Many SNPs that were not annotated within gene regulatory regions were excluded from the present analysis. In this step, we computed gene-wise P-values for 18,151 genes for the GWAS, of which, 891 genes reached the significance threshold of P < 0.05 (Figure 3). For the pathway enrichment analysis, we mapped these screened genes to 15 KEGG pathways (Table 3) including osteoclast differentiation, glycosphingolipid biosynthesis, amyotrophic lateral sclerosis, cell-matrix interactions, and inflammatory associations, with a default threshold of the EASE Score (a modified Fisher Exact P-Value). Because of the multiple roles of some genes and complex interactions in cellular activities between protein pathway networks, candidate genes were always involved in different pathways. For example, mitogen-activated protein kinase 12 (MAPK12) gene, an important transduction factor of extracellular signals, was involved in osteoclast differentiation, amyotrophic lateral sclerosis (ALS), Fc epsilon RI signaling pathway, VEGF signaling pathway, Rap1 signaling pathway, MAPK signaling pathway, and T cell receptor signaling pathway (Figure 4). Previous studies report contradictory information regarding which pathways might be related to inflammatory reactions or symptoms of SAPHO syndrome, therefore, it is important to preclude potential misleading pathways. Based on published studies of identified pathways, we inferred osteoclast differentiation pathway (P = 0.002954, 15 genes involved), phagosome (P = 0.009788, 15 genes involved), Fc epsilon RI signaling pathway (P = 0.013035, 9 gene involved), Rap1 signaling pathway (P = 0.035055, 17 genes involved), Fc gamma R-mediated phagocytosis pathway (P = 0.040769, 9 genes involved), and bacterial invasion of epithelial cells pathway (P = 0.069759, 8 genes involved) were highly correlated with SAPHO syndrome based on bone and skin manifestations of SAPHO. Other pathways may affect the pathological process of SAPHO syndrome as well, but their direct association requires further research.
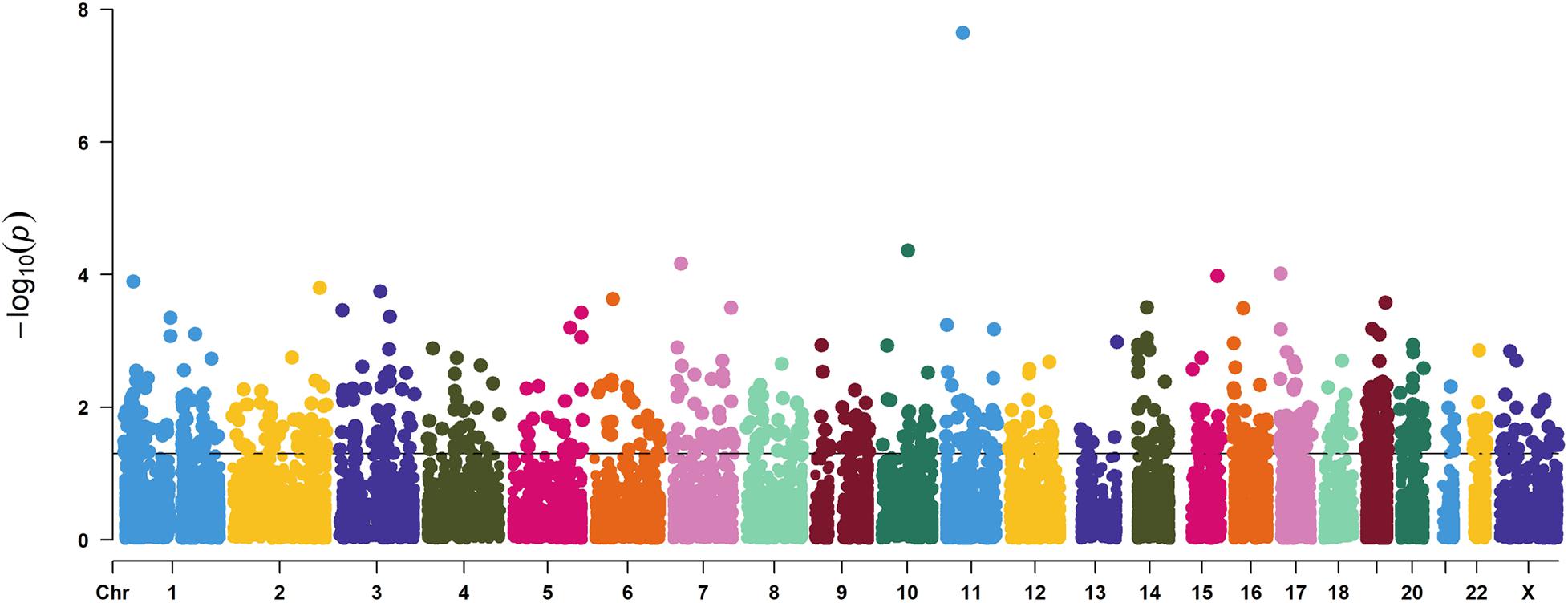
Figure 3. A Manhattan plot showing the gene level P values of GWAS used in this study. Genes in each chromosome are represented by different colors.
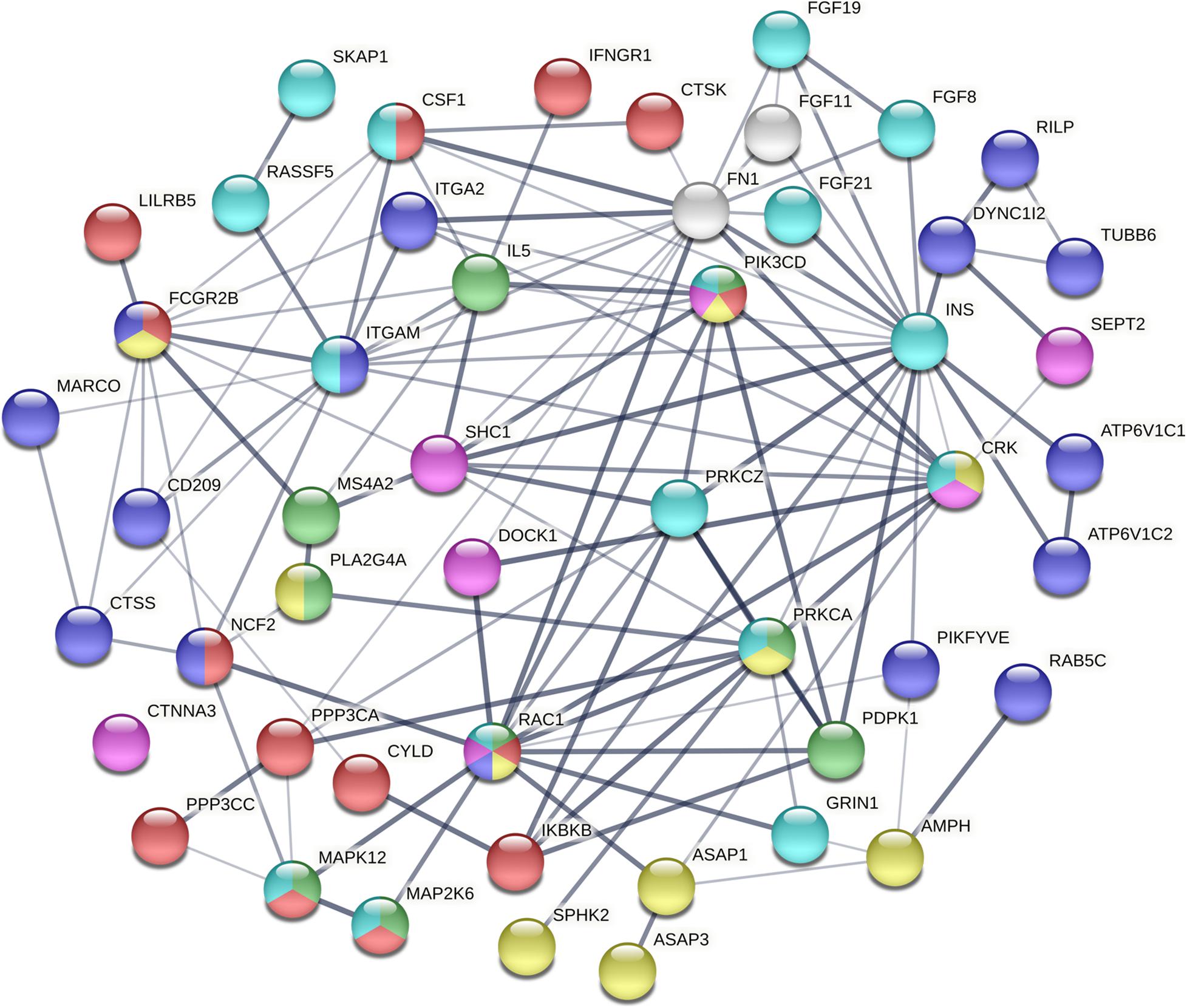
Figure 4. The interaction of mutated genes in SAPHO associated pathways identified in this study. The mutated genes of different pathways are shown by different colors. Each color represents a unique pathway and line thickness indicates the strength of data support (red, osteoclast differentiation; purple, phagosome; green, Fc epsilon RI signaling pathway; yellow, Fc gamma R-mediated phagocytosis; pink, Bacterial invasion of epithelial cells; blue, Rap1 signaling pathway).
Detect Genetic Variants Based on WES Data
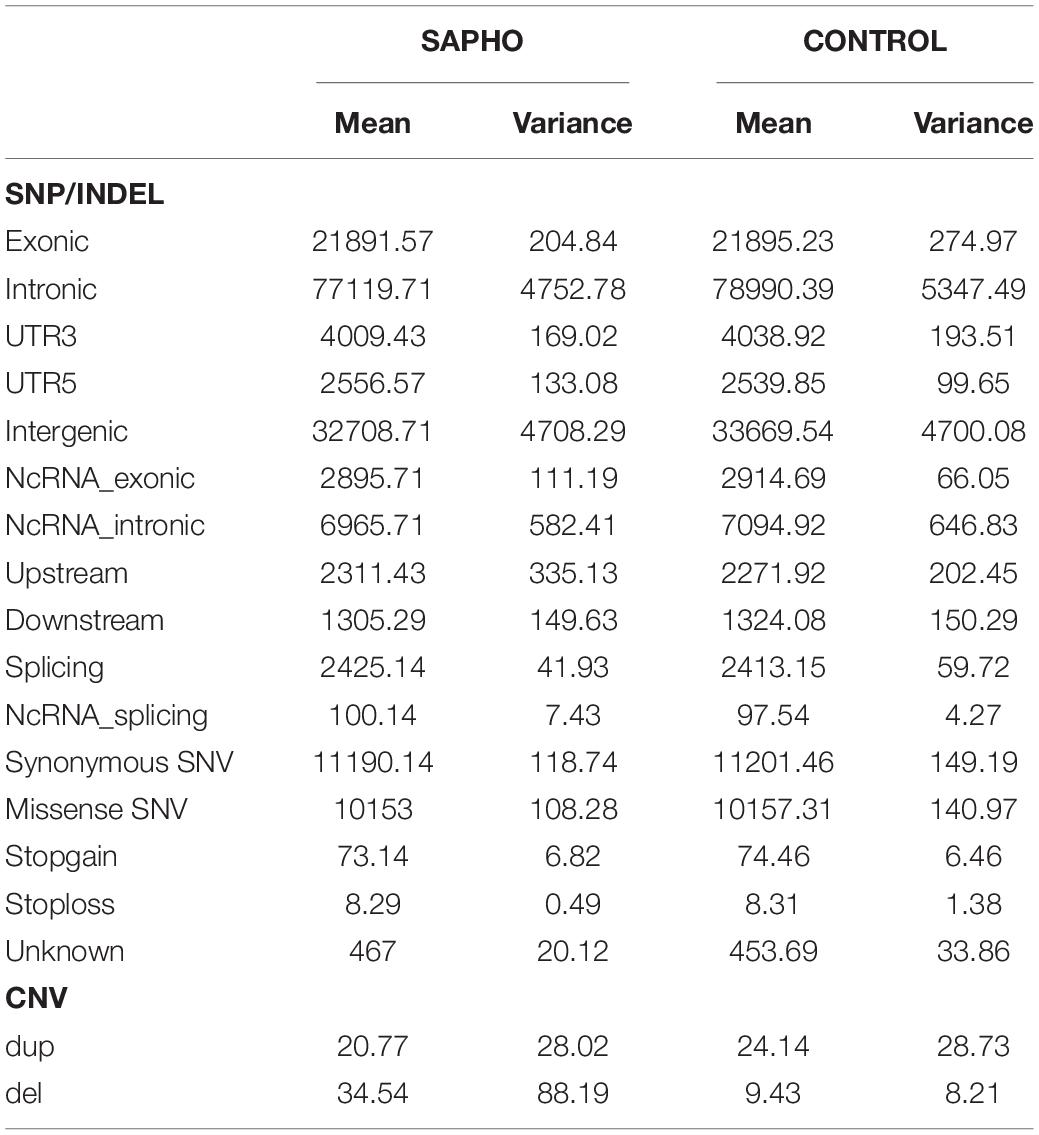
Genome-wide association study has limited power in identifying low frequency and rare causal genetic variants with greater penetrance involved in complex diseases. To verify the association between aberrant signaling pathways and SAPHO syndrome, we performed WES analysis for 16 patients diagnosed with SAPHO and 15 healthy individuals. Results showed the coverage of target region was 99.91%, mean depth on target region was 181.52 ± 21.33× and the target coverage with at least 20× was 94 ± 2% (Table 4). After filtering, sequencing resulted in 176,181 SNP/INDEL and 55.31 CNV variants in each SAPHO cases and 179,145.5 SNP/INDEL and 33.57 CNV variants in healthy control samples (Table 5). First of all, we tried to identify the shared genetic variants or genes in SAPHO and control groups. Again, the result of the WES was inconsistent between individual samples, no single genetic variant or gene was highly conserved in more than three cases when compared with healthy controls. Next, we aligned these variants to KEGG pathways identified in the GWAS analysis. The result showed each SAPHO sample had at least one aberrant pathway involved: 15 samples had gene aberrations in osteoclast differentiation pathway (genetic variants were found in CSF1, FCGR2B, PIK3CD, MAP2K6, LILRB5, PPP3CA and MAPK12, CNV aberrations were found in LILRA3, FCGR2B, and CTSK genes); 7 samples had gene aberrations in the phagosome pathway (genetic variants were found in ITGA2, MARCO, FCGR2B and CD209, CNV aberrations were found in the CTSS gene); 6 samples had gene aberrations in the Fc epsilon RI signaling pathway (genetic variants were found in PIK3CD, MAP2K6 and MAPK12, CNV aberrations were found in the PDPK1 gene); 7 samples had gene aberrations in Rap1 signaling pathway (genetic variants were found in MAP2K6, MAPK12, CRK, CSF1, and GRIN1); 5 samples had gene aberrations in the Fc gamma R-mediated phagocytosis pathway (genetic variants were found in FCGR2B, PIK3CD, AMPH, SPHK2, and CRK); 11 samples had gene aberrations in the Bacterial invasion of epithelial cells pathway (genetic variants were found in PIK3CD, DOCK1, CRK, CTNNA3, and FN1) (Tables 6, 7).
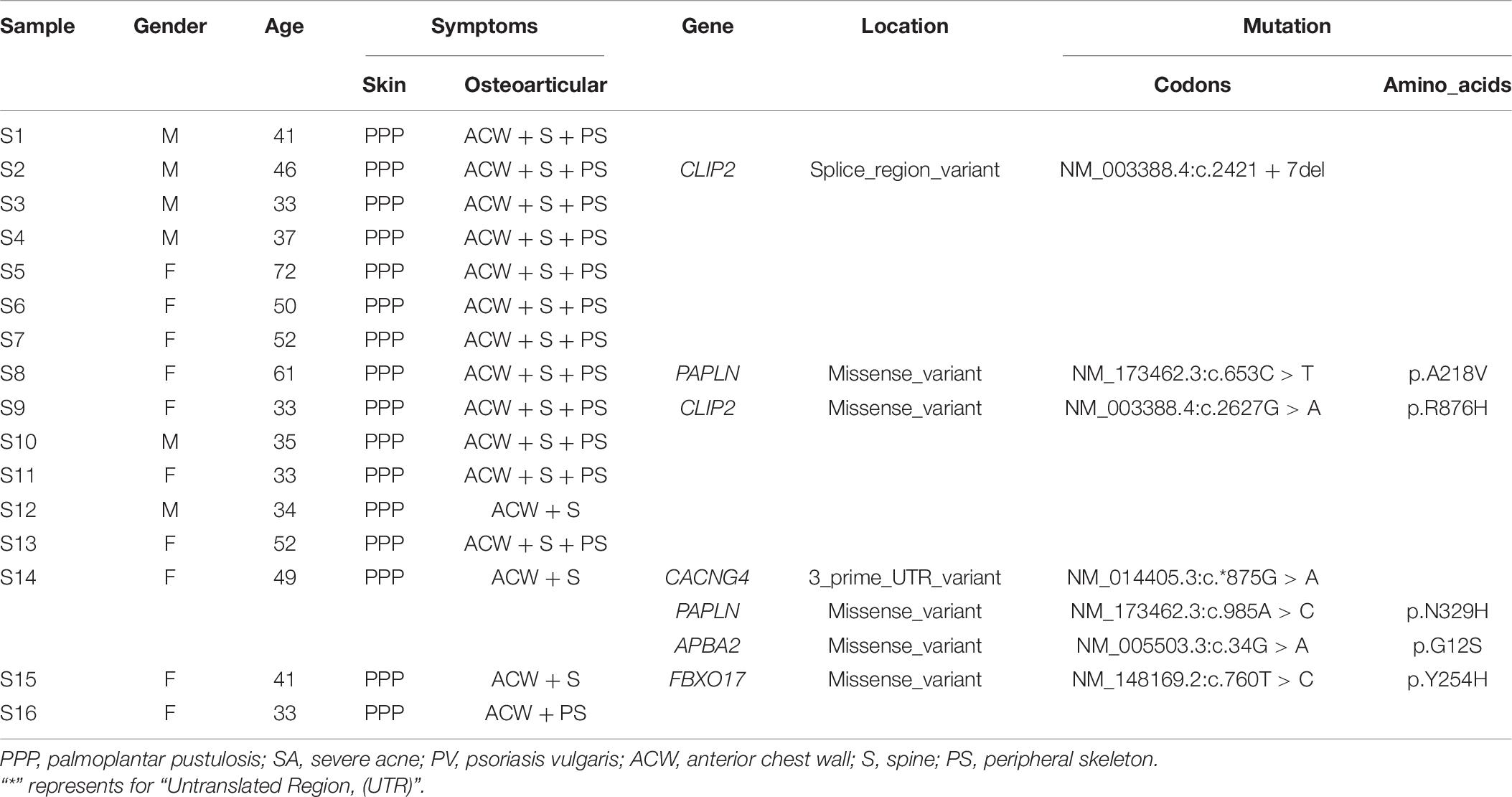
Table 6. The clinical features of 16 SAPHO patients and genetic variants corresponding to GWAS results (P < 6.24 × 10–8).
Discussion
SAPHO syndrome is a systemic and recurrent disease with unknown etiology, characterized by chronic inflammatory osteoarticular lesions and dermatological disorders. The diagnosis and treatment of this rare disease have been limited by its complex etiology and phenotypic heterogeneity. In the last decade, many studies have demonstrated that comprehensive analyses of GWAS data and protein-protein interaction (PPI) networks can provide valuable biological clues (Cong et al., 2017). In this study, to identify genetic variants of SAPHO syndrome, we performed a novel genome-wide network-based integrative analysis of SAPHO syndrome based on GWAS and WES data. Using a basic GWAS analysis after filtering process, we found 9 SNPs (rs4505038, rs4897770, rs12442139, rs13062589, rs2850133, rs10927436, rs9567768, rs2243861, and rs8007562) that met the expected significance threshold (P < 6.24 × 10–8). Among them, 7 SNPs located in the regions without known functional genes, rs4505038 (PEX16) and rs2243861 (IQCA1L) located in the introns of known genes. Pex16 plays an critical role in adipose tissue peroxisomal biogenesis, and mice deficient for the Pex16 gene showed increased diet-induced obesity and impaired thermogenesis ability without skin or osteoarticular manifestations (Suzuki et al., 2001; Park et al., 2019). The IQCA1L gene is specifically expressed in the testis and has not been reported with immunity (Gaudet et al., 2011). Based on previous studies, none of the 9 SNPs had an association with SAPHO syndrome or other inflammatory diseases. Then we lowered the significance threshold to approximately 10-times the expected threshold, 84 SNPs with a P-value below 5.6 × 10–7 were selected for further analysis. However, no valuable SNP or candidate genes were identified.
Given the complex symptom and etiology of SAPHO syndrome, we speculate that no single genetic variant accounts for all the complicated manifestations of this disease. Thus, in the following analysis, we reanalyzed the GWAS data by adopting pathway and network-based analysis. We found several pathways were altered in SAPHO samples, and six of these had evidence with skin, osteoarticular manifestations of SAPHO syndrome or inflammatory reactions, including osteoclast differentiation pathway, phagosome pathway, Fc epsilon RI signaling pathway, Rap1 signaling pathway, Fc gamma R-mediated phagocytosis pathway, and bacterial invasion of epithelial cells pathway.
The osteoclast differentiation pathway, a key regulator of resorption and formation of bone tissue, was the most significant aberrant pathway in SAPHO patients. Previous studies reported disruption of the osteoclast differentiation or function leads to inhibited bone resorption, which further can result in bone marrow deficiency and no teething (Grigoriadis et al., 1994; Kong et al., 1999). On the contrary,enhancement of osteoclast differentiation or function in patients with osteoporosis and metastatic bone cancer resulted in the decrease of bone mass and destruction of bone, respectively (Miyamoto, 2011). Some important signaling molecules are essential for the correct fulfillment of osteoclastogenesis, for example, monocyte colony-stimulating factor (M-CSF) exert a proliferative and survival effect on early pre-monocyte phase and the entire process, respectively (Boyle et al., 2003; Edwards and Mundy, 2011; Anesi et al., 2019). The function of the second signaling molecule receptor activator NF-κB ligand (RANKL) is differentiation in the late post-monocyte phase of the process that is necessary to transform monocytes into osteoclasts (Takayanagi,2007b,a,c; Kim and Kim, 2016; Kim et al.,2016a,b; Anesi et al., 2019). Osteoarticular involvement, a characteristic sign of disease, was observed in nearly all SAPHO patients and mainly involved the anterior chest wall and lumbosacral and peripheral skeletal regions (Cao et al., 2019). Zhang et al. (2019) reported RANKL levels were significantly higher in active SAPHO patients than in non-active or healthy samples, suggesting the aberrant osteoclast differentiation pathway plays pivotal role in the pathology of SAPHO. Our findings reconfirmed the foregoing conclusion. In addition to the molecules mentioned above, mary other signaling molecules play important role in regulating osteoclastic differentiation process as well. Osteoclastogenic cytokines are represented by inflammatory cytokines including tumor necrosis factor α (TNF-α), interleukin-1 (IL-1), IL-6, IL-7, IL-8, IL-11, IL-15, IL-17, IL-23, and IL-34. Anti-osteoclastogenic cytokines are represented by IFN-α, IFN-β, IFN-γ as well as IL-3, IL-4, IL-10, IL-12, IL-27, and IL-33 (Amarasekara et al., 2018). Published research reported some inflammatory factors including IL-1β, IL-17A, IL-6, IL-8, IL-18, and TNF-α were higher in SAPHO patients than in healthy controls (Przepiera-Bedzak et al., 2016; Zhang et al., 2019), based on these findings, it is plausible that the combined actions of elevated cytokines and a disrupted osteoclast differentiation pathway might aggravate bone devastation and reconstruction, resulting in the osteoarticular symptoms.
Phagocytosis is an evolutionarily ancient process whereby cells engulf large particles. It is an important core mechanism in some immune processes, including defense against infectious agents, inflammation, tissue remodeling, and antigen degradation and presentation (Dean et al., 2019). Phagocytic cells such as monocytes and macrophages participate in host defense by forming phagosomes. During phagocytosis, the membrane on the surface of a phagocyte forms a phagosome when the receptors on it bind to the ligands on the surface of the particle surface. After its formation, the new phagosome gradually acquire digestive properties. In the process of phagosome maturation, there are other membrane organelles involved, including circulating endosomes, late endosomes, and lysosomes. By fusing lysosomes, phagosomes can activate enzymes and lower the pH value in the lumen that eventually degrades phagocytized micro-organisms into fragments (Kanehisa et al., 2017). Accordingly, disruptions to this process cause some bacteria such as Mycobacterium tuberculosis to escape bacterial killing and survive within host phagocytes (Ehrt and Schnappinger, 2009; Kanehisa et al., 2017). In this study, two phagocytosis-related pathways (phagosome, P = 0.0098, 15 genes; Fc gamma R-mediated phagocytosis, P = 0.041, 9 genes) were highly associated with SAPHO syndrome, suggesting phagocytosis has an important role in SAPHO syndrome. James et al. (2010) found the phagocytosis of disease-relevant particles (PMMA, titanium, and silica) inhibited the RANKL-mediated osteoclastogenesis of human monocytes. They demonstrated phagocytosis mediates this effect by down-regulation of RANK and c-Fms, receptors for the essential osteoclastogenic cytokines RANKL and M-CSF (James et al., 2010). However, the mechanisms involved in phagocytosis and SAPHO required further research.
Fc epsilon RI is the specific receptor for IgE, which has an important role in IgE-associated allergic reactions. A cascade of signaling events can be induced by the cross-linking of Fc epsilon RI on mast cells, leading to degranulation, proinflammatory cytokine production, and leukotriene release, which contribute to the emergence of allergic symptomology (Klemm and Ruland, 2006; Kambayashi and Koretzky, 2007). IFN-γ activates mast cells through FceRI to induce PGD2 and LTC4 release, and the subsequent up-regulation of mRNAs for IL-1a, IL-3, IL-8, G-CSF, LIF, CSF1, oncostatin M (OSM), SCF, TGF-β1, IP-10, I-309, MIP-1α, and MIP-1β (Okayama et al., 2001). Our results showed that the Fc epsilon RI signaling pathway was involved in the pathogenesis of SAPHO syndrome (P = 0.013035, 9 genes). In accordance, Li et al. reported a SAPHO patient with elevated serum immunoglobulin E levels, and demonstrated methylprednisolone treatment achieve long-term remarkable remission on clinical manifestations (Wang et al., 2019), which is consistent with our finding.
In this study, we found two pathways associated with aberrant cell barrier function in SAPHO patients, Rap1 signaling pathway (P = 0.035055, 17 genes) and bacterial invasion of epithelial cells (P = 0.069759, 8 genes), suggesting damage to the cell barrier contributes to the complicated manifestation of SAPHO syndrome. The function of the small G-protein Rap1 is to regulate endothelial barrier function controlled by cell–cell adhesion and the actin cytoskeleton. When this process is activated, numerous signaling cascades are induced by Rap1 to enhance the endothelial barrier function. Of note, Rap1 activation results inhibit of Rho to decrease radial stress fibers and activate Cdc42 to increase junctional actin (Pannekoek et al., 2014). These are some studies has proven the above results in human umbilical endothelial cells (Cullere et al., 2005; Citalan-Madrid et al., 2013) and retinal vascular endothelial cells (Ramos et al., 2018). Moreover, Rap1 deletion in mature osteoclasts caused osteopetrosis by reducing talin/β integrin recognition (Zou et al., 2013).
Unlike other immunologically relevant diseases, except chronic multifocal osteomyelitis, SAPHO patients suffer from recurrent demographic manifestations, including palmoplantar pustulosis, psoriasis vulgaris, and severe acne. On the base of the findings in this study, we inferred a single pathway was not responsible for this complicated syndrome, two or more pathways probably act simultaneously. For example, an impaired cell barrier or inflammatory cytokine release induced by allergic reactions might promote the demographic manifestations and elevated inflammatory factors, moreover aberrant phagocytosis and osteoclast differentiation pathways might cause alterations to bone resorption and formation, ultimately leading to osteoarticular deformation. These pathways are closely linked and might affect each other, for example, an impaired cell barrier and pathogen infection or allergic reaction might lead to the over-expression of inflammatory factors, which increase the permeability of the skin or endothelial cells, thus increasing infection. Moreover, the disruption of phagocytosis might allow bacteria to escape and enhance infection.
Conclusion
In conclusion, this GWAS study combined with pathway-based analysis and WES identified aberrant pathways including the osteoclast differentiation pathway involved in SAPHO syndrome. This finding may provide insights into the pathogenic genes of SAPHO and provide the basis for SAPHO research and treatment. Further studies should be conducted to validate this conclusion in a larger sample size and in other ethnic backgrounds.
Data Availability Statement
The raw sequence data reported in this article have been deposited in the Genome Sequence Archive (Wang et al., 2017) in BIG Data Center (National Genomics Data Center Members and Partners, 2020), Beijing Institute of Genomics, Chinese Academy of Sciences, under accession number(s) HRA000288. All data is available from the corresponding author upon request.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethics Committee of the National Research Institute for Family Planning. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
RC and YD: planning of the project, analysis and interpretation of data, and drafting the manuscript. MF, YF, and JG: analysis and interpretation of data. YZ, ZC, and FH: collecting of the data. CG and XM: putting forward research ideas and planning of the project. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Key Research and Development Program of China (2016YFC1000307, 2016YFC1000300, 2016YFC1000803, and 2017YFC1002000), International Science & Technology Cooperation Program of China (0102012DFB30130), the National Natural Science Foundation of China (81501418), and the National Population and Reproductive Health Science Data Center (grant no. 2005DKA32408), China.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the subjects who agreed to participate in this stud, Genetic Resource Center staff for their support, and Edanz Group China (www.liwenbianji.cn/ac) for editing the English text of a draft of the manuscript.
Footnotes
References
Amarasekara, D. S., Yun, H., Kim, S., Lee, N., Kim, H., and Rho, J. (2018). Regulation of osteoclast differentiation by cytokine networks. Immune Netw. 18:e8. doi: 10.4110/in.2018.18.e8
Anesi, A., Generali, L., Sandoni, L., Pozzi, S., and Grande, A. (2019). From osteoclast differentiation to osteonecrosis of the jaw: molecular and clinical insights. Int. J. Mol. Sci. 20:4925. doi: 10.3390/ijms20194925
Baranzini, S. E., Galwey, N. W., Wang, J., Khankhanian, P., Lindberg, R., Pelletier, D., et al. (2009). Pathway and network-based analysis of genome-wide association studies in multiple sclerosis. Hum. Mol. Genet. 18, 2078–2090. doi: 10.1093/hmg/ddp120
Boyle, W. J., Simonet, W. S., and Lacey, D. L. (2003). Osteoclast differentiation and activation. Nature 423, 337–342. doi: 10.1038/nature01658
Cao, Y., Li, C., Yang, Q., Wu, N., Xu, P., Li, Y., et al. (2019). Three patterns of osteoarticular involvement in SAPHO syndrome: a cluster analysis based on whole body bone scintigraphy of 157 patients. Rheumatology (Oxf.) 58, 1047–1055. doi: 10.1093/rheumatology/key415
Citalan-Madrid, A. F., Garcia-Ponce, A., Vargas-Robles, H., Betanzos, A., and Schnoor, M. (2013). Small GTPases of the Ras superfamily regulate intestinal epithelial homeostasis and barrier function via common and unique mechanisms. Tissue Barriers 1:e26938. doi: 10.4161/tisb.26938
Colina, M., Lo Monaco, A., Khodeir, M., and Trotta, F. (2007). Propionibacterium acnes and SAPHO syndrome: a case report and literature review. Clin. Exp. Rheumatol. 25, 457–460.
Colina, M., Pippucci, T., Moro, M. A., Marconi, C., Magini, P., Ciancio, G., et al. (2012). Synovitis, acne, pustulosis, hyperostosis, osteitis (SAPHO) syndrome: is PTPN22 involved? Clin. Exp. Rheumatol. 30:451.
Cong, W., Meng, X., Li, J., Zhang, Q., Chen, F., Liu, W., et al. (2017). Genome-wide network-based pathway analysis of CSF t-tau/Abeta1-42 ratio in the ADNI cohort. BMC Genomics 18:421. doi: 10.1186/s12864-017-3798-z
International Multiple Sclerosis Genetics Consortium (2013). Network-based multiple sclerosis pathway analysis with GWAS data from 15,000 cases and 30,000 controls. Am. J. Hum. Genet. 92, 854–865. doi: 10.1016/j.ajhg.2013.04.019
Cullere, X., Shaw, S. K., Andersson, L., Hirahashi, J., Luscinskas, F. W., and Mayadas, T. N. (2005). Regulation of vascular endothelial barrier function by Epac, a cAMP-activated exchange factor for Rap GTPase. Blood 105, 1950–1955. doi: 10.1182/blood-2004-05-1987
de Leeuw, C. A., Mooij, J. M., Heskes, T., and Posthuma, D. (2015). MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 11:e1004219. doi: 10.1371/journal.pcbi.1004219
Dean, P., Heunis, T., Hartlova, A., and Trost, M. (2019). Regulation of phagosome functions by post-translational modifications: a new paradigm. Curr. Opin. Chem. Biol. 48, 73–80.
Edwards, J. R., and Mundy, G. R. (2011). Advances in osteoclast biology: old findings and new insights from mouse models. Nat. Rev. Rheumatol. 7, 235–243. doi: 10.1038/nrrheum.2011.23
Ehrt, S., and Schnappinger, D. (2009). Mycobacterial survival strategies in the phagosome: defence against host stresses. Cell. Microbiol. 11, 1170–1178. doi: 10.1111/j.1462-5822.2009.01335.x
Ferguson, P. J., Bing, X., Vasef, M. A., Ochoa, L. A., Mahgoub, A., Waldschmidt, T. J., et al. (2006). A missense mutation in pstpip2 is associated with the murine autoinflammatory disorder chronic multifocal osteomyelitis. Bone 38, 41–47.
Firinu, D., Barca, M. P., Lorrai, M. M., Perra, S., Cabras, S., Muggianu, E., et al. (2014). TH17 cells are increased in the peripheral blood of patients with SAPHO syndrome. Autoimmunity 47, 389–394. doi: 10.3109/08916934.2014.906582
Gaudet, P., Livstone, M. S., Lewis, S. E., and Thomas, P. D. (2011). Phylogenetic-based propagation of functional annotations within the Gene Ontology consortium. Brief. Bioinform. 12, 449–462. doi: 10.1093/bib/bbr042
Geoffroy, V., Herenger, Y., Kress, A., Stoetzel, C., Piton, A., Dollfus, H., et al. (2018). AnnotSV: an integrated tool for structural variations annotation. Bioinformatics 34, 3572–3574. doi: 10.1093/bioinformatics/bty304
Grigoriadis, A. E., Wang, Z. Q., Cecchini, M. G., Hofstetter, W., Felix, R., Fleisch, H. A., et al. (1994). c-Fos: a key regulator of osteoclast-macrophage lineage determination and bone remodeling. Science 266, 443–448. doi: 10.1126/science.7939685
Guo, C., Li, C., Han, F., Gao, J., and Ma, X. (2019). Association analysis of interleukin-23 receptor SNPs and SAPHO syndrome in Chinese people. Int. J. Rheum. Dis. 22, 2178–2184. doi: 10.1111/1756-185X.13741
Huang da, W., Sherman, B. T., and Lempicki, R. A. (2009a). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13. doi: 10.1093/nar/gkn923
Huang da, W., Sherman, B. T., and Lempicki, R. A. (2009b). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. doi: 10.1038/nprot.2008.211
Hurtado-Nedelec, M., Chollet-Martin, S., Chapeton, D., Hugot, J. P., Hayem, G., and Gerard, B. (2010). Genetic susceptibility factors in a cohort of 38 patients with SAPHO syndrome: a study of PSTPIP2, NOD2, and LPIN2 genes. J. Rheumatol. 37, 401–409. doi: 10.3899/jrheum.090456
James, D. E., Nestor, B. J., Sculco, T. P., Ivashkiv, L. B., Ross, F. P., Goldring, S. R., et al. (2010). The relative timing of exposure to phagocytosable particulates and to osteoclastogenic cytokines is critically important in the determination of myeloid cell fate. J. Immunol. 185, 1265–1273. doi: 10.4049/jimmunol.0902808
Kahn, M. F., Hayem, F., Hayem, G., and Grossin, M. (1994). Is diffuse sclerosing osteomyelitis of the mandible part of the synovitis, acne, pustulosis, hyperostosis, osteitis (SAPHO) syndrome? Analysis of seven cases. Oral Surg. Oral Med. Oral Pathol. 78, 594–598.
Kambayashi, T., and Koretzky, G. A. (2007). Proximal signaling events in Fc epsilon RI-mediated mast cell activation. J. Allergy Clin. Immunol. 119, 544–552; quiz 553—544.
Kanehisa, M., Furumichi, M., Tanabe, M., Sato, Y., and Morishima, K. (2017). KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45, D353–D361. doi: 10.1093/nar/gkw1092
Kerrison, C., Davidson, J. E., Cleary, A. G., and Beresford, M. W. (2004). Pamidronate in the treatment of childhood SAPHO syndrome. Rheumatology (Oxf.) 43, 1246–1251. doi: 10.1093/rheumatology/keh295
Kim, H. J., Ohk, B., Kang, W. Y., Seong, S. J., Suk, K., Lim, M. S., et al. (2016a). Deficiency of lipocalin-2 promotes proliferation and differentiation of osteoclast precursors via regulation of c-fms expression and nuclear factor-kappa B activation. J. Bone Metab. 23, 8–15. doi: 10.11005/jbm.2016.23.1.8
Kim, J. H., Kim, E. Y., Lee, B., Min, J. H., Song, D. U., Lim, J. M., et al. (2016b). The effects of lycii radicis cortex on RANKL-induced osteoclast differentiation and activation in RAW 264.7 cells. Int. J. Mol. Med. 37, 649–658. doi: 10.3892/ijmm.2016.2477
Kim, J. H., and Kim, N. (2016). Signaling pathways in osteoclast differentiation. Chonnam. Med. J. 52, 12–17. doi: 10.4068/cmj.2016.52.1.12
Klemm, S., and Ruland, J. (2006). Inflammatory signal transduction from the Fc epsilon RI to NF-kappa B. Immunobiology 211, 815–820.
Kong, Y. Y., Yoshida, H., Sarosi, I., Tan, H. L., Timms, E., Capparelli, C., et al. (1999). OPGL is a key regulator of osteoclastogenesis, lymphocyte development and lymph-node organogenesis. Nature 397, 315–323. doi: 10.1038/16852
Kotilainen, P., Merilahti-Palo, R., Lehtonen, O. P., Manner, I., Helander, I., Mottonen, T., et al. (1996). Propionibacterium acnes isolated from sternal osteitis in a patient with SAPHO syndrome. J. Rheumatol. 23, 1302–1304.
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Magrey, M., and Khan, M. A. (2009). New insights into synovitis, acne, pustulosis, hyperostosis, and osteitis (SAPHO) syndrome. Curr. Rheumatol. Rep. 11, 329–333. doi: 10.1007/s11926-009-0047-8
McKenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., Kernytsky, A., et al. (2010). The genome analysis toolkit: a mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. doi: 10.1101/gr.107524.110
McLaren, W., Gil, L., Hunt, S. E., Riat, H. S., Ritchie, G. R., Thormann, A., et al. (2016). The ensembl variant effect predictor. Genome Biol. 17:122. doi: 10.1186/s13059-016-0974-4
Miyamoto, T. (2011). Regulators of osteoclast differentiation and cell-cell fusion. Keio J. Med. 60, 101–105. doi: 10.2302/kjm.60.101
National Genomics Data Center Members and Partners (2020). Database resources of the National genomics data center in 2020. Nucleic Acids Res 48, D24–D33. doi: 10.1093/nar/gkz913
Okayama, Y., Hagaman, D. D., and Metcalfe, D. D. (2001). A comparison of mediators released or generated by IFN-gamma-treated human mast cells following aggregation of Fc gamma RI or Fc epsilon RI. J. Immunol. 166, 4705–4712. doi: 10.4049/jimmunol.166.7.4705
Pannekoek, W. J., Post, A., and Bos, J. L. (2014). Rap1 signaling in endothelial barrier control. Cell Adh. Migr. 8, 100–107.
Park, H., He, A., Tan, M., Johnson, J. M., Dean, J. M., Pietka, T. A., et al. (2019). Peroxisome-derived lipids regulate adipose thermogenesis by mediating cold-induced mitochondrial fission. J. Clin. Invest. 129, 694–711. doi: 10.1172/JCI120606
Plagnol, V., Curtis, J., Epstein, M., Mok, K. Y., Stebbings, E., Grigoriadou, S., et al. (2012). A robust model for read count data in exome sequencing experiments and implications for copy number variant calling. Bioinformatics 28, 2747–2754. doi: 10.1093/bioinformatics/bts526
Przepiera-Bedzak, H., Fischer, K., and Brzosko, M. (2016). Serum interleukin-18, Fetuin-A, soluble intercellular adhesion molecule-1, and endothelin-1 in ankylosing spondylitis, psoriatic arthritis, and SAPHO syndrome. Int. J. Mol. Sci. 17:1255. doi: 10.3390/ijms17081255
Ramos, C. J., Lin, C., Liu, X., and Antonetti, D. A. (2018). The EPAC-Rap1 pathway prevents and reverses cytokine-induced retinal vascular permeability. J. Biol. Chem. 293, 717–730. doi: 10.1074/jbc.M117.815381
Ritchie, M. D. (2009). Using prior knowledge and genome-wide association to identify pathways involved in multiple sclerosis. Genome Med. 1:65. doi: 10.1186/gm65
Sharma, M., and Ferguson, P. J. (2013). Autoinflammatory bone disorders: update on immunologic abnormalities and clues about possible triggers. Curr. Opin. Rheumatol. 25, 658–664. doi: 10.1097/BOR.0b013e328363eb08
Suzuki, Y., Shimozawa, N., Imamura, A., Fukuda, S., Zhang, Z., Orii, T., et al. (2001). Clinical, biochemical and genetic aspects and neuronal migration in peroxisome biogenesis disorders. J. Inherit. Metab. Dis. 24, 151–165. doi: 10.1023/a:1010310816743
Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., et al. (2019). STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613. doi: 10.1093/nar/gky1131
Takayanagi, H. (2007a). Interaction between the immune system and bone metabolism: an emerging field of osteoimmunology. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 83, 136–143. doi: 10.2183/pjab.83.136
Takayanagi, H. (2007c). Osteoimmunology: shared mechanisms and crosstalk between the immune and bone systems. Nat. Rev. Immunol. 7, 292–304.
Van der Auwera, G. A., Carneiro, M. O., Hartl, C., Poplin, R., Del Angel, G., Levy-Moonshine, A., et al. (2013). From FastQ data to high confidence variant calls: the genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinformatics 43, 11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43
Wang, K., Li, M., and Hakonarson, H. (2010). ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38:e164. doi: 10.1093/nar/gkq603
Wang, L., Li, C., Yu, M., Cao, Y., Jing, H., and Li, J. (2019). Long-term remarkable remission of SAPHO syndrome in response to short-term systemic corticosteroids treatment in an immunoglobulin E elevated patient: a case report. Medicine (Baltimore) 98:e16045. doi: 10.1097/MD.0000000000016045
Wang, Y., Song, F., Zhu, J., Zhang, S., Yang, Y., Chen, T., et al. (2017). GSA: genome sequence archive∗. Genom. Proteom. Bioinform. 15, 14–18. doi: 10.1016/j.gpb.2017.01.001
Xu, D., Liu, X., Lu, C., Luo, J., Wang, C., Gao, C., et al. (2019). Reduction of peripheral natural killer cells in patients with SAPHO syndrome. Clin. Exp. Rheumatol. 37, 12–18.
Yang, Q., Zhao, Y., Li, C., Luo, Y., Hao, W., and Zhang, W. (2018). Case report: successful treatment of refractory SAPHO syndrome with the JAK inhibitor tofacitinib. Medicine (Baltimore) 97:e11149. doi: 10.1097/MD.0000000000011149
Zhang, S., Li, C., Li, L., Zhang, W., and Dong, Z. (2019). Serum levels of proinflammatory, anti-inflammatory cytokines, and RANKL/OPG in synovitis, acne, pustulosis, hyperostosis, and osteitis (SAPHO) syndrome. Mod. Rheumatol. 29, 523–530. doi: 10.1080/14397595.2018.1469580
Keywords: SAPHO syndrome, GWAS, WES, pathway analysis, immune-mediated conditions
Citation: Cai R, Dong Y, Fang M, Fan Y, Cheng Z, Zhou Y, Gao J, Han F, Guo C and Ma X (2021) Genome-Wide Association Identifies Risk Pathways for SAPHO Syndrome. Front. Cell Dev. Biol. 9:643644. doi: 10.3389/fcell.2021.643644
Received: 18 December 2020; Accepted: 18 February 2021;
Published: 18 March 2021.
Edited by:
Yu-Hang Zhang, Channing Division of Network Medicine, Brigham and Women’s Hospital, United StatesReviewed by:
Xu Yan, Department of Pathophysiology, Beijing Neurosurgical Institute, ChinaJiangbo Pu, Institute of Biomedical Engineering, Chinese Academy of Medical Sciences, Peking Union Medical College, China
Copyright © 2021 Cai, Dong, Fang, Fan, Cheng, Zhou, Gao, Han, Guo and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Changlong Guo, Z3VvY2hhbmdsb25nQG5yaWZwLm9yZy5jbg==; Xu Ma, bWF4dV9ncm91cEAxNjMuY29t
†These authors have contributed equally to this work and share first authorship
 Ruikun Cai
Ruikun Cai Yichao Dong
Yichao Dong Mingxia Fang1,3
Mingxia Fang1,3 Feifei Han
Feifei Han