- School of Life Sciences, Beijing University of Chinese Medicine, Beijing, China
Background: Calcium ions (Ca2+) play an essential role in excitation–contraction coupling in the heart. The association between cardiovascular diseases (CVDs) and genetic polymorphisms in key regulators of Ca2+ homeostasis is well established but still inadequately understood.
Methods: The associations of 11,274 genetic variants located in nine calcium signaling-related genes with 118 diseases of the circulatory system were explored using a large sample from the United Kingdom Biobank (N = 308,366). The clinical outcomes in electronic health records were mapped to the phecode system. Survival analyses were employed to study the role of variants in CVDs incidence and mortality. Phenome-wide association studies (PheWAS) were performed to investigate the effect of variants on cardiovascular risk factors.
Results: The reported association between rs1801253 in β1-adrenergic receptor (ADRB1) and hypertension was successfully replicated, and we additionally found the blood pressure-lowering G allele of this variant was associated with a delayed onset of hypertension and a decreased level of apolipoprotein A. The association of rs4484922 in calsequestrin 2 (CASQ2) with atrial fibrillation/flutter was identified, and this variant also displayed nominal evidence of association with QRS duration and carotid intima-medial thickness. Moreover, our results indicated suggestive associations of rs79613429 in ryanodine receptor 2 (RYR2) with precordial pain.
Conclusion: Multiple novel associations established in our study highlight genetic testing as a useful method for CVDs diagnosis and prevention.
Introduction
Calcium ions (Ca2+) play an essential role in excitation–contraction coupling (EC coupling) in the heart. Dysregulation of intracellular Ca2+ level contributes to the abnormal cardiac contractile function during the pathogenesis of cardiovascular diseases (CVDs), such as coronary artery disease and arrhythmias. For instance, Ca2+ are involved in the initiation of delayed after-depolarization (DAD) and early after-depolarization (EAD), which mediate the initiation of arrhythmias (Clusin, 2003). The regulation of calcium-handling proteins in a well-coordinated manner is required for Ca2+ homeostasis in cardiomyocytes (Eisner et al., 2017). During cardiac action potential, the voltage-gated calcium channels (VGCCs) mediate an influx of external Ca2+ which in turn activate ryanodine receptors (RYRs) to release more Ca2+ from the sarcoplasmic reticulum (SR). The elevated cytosolic Ca2+ cause structural changes in myosin filaments that induce the contraction of muscle fibers. After contraction, Ca2+ can either be pumped back to the SR or pumped out of cardiomyocytes through sarco/endoplasmic reticulum Ca2+-ATPase (SERCA) or sodium–calcium exchanger (NCX), respectively (Eisner et al., 2017). The activities of these crucial Ca2+-handling proteins can be modulated by β-adrenergic signaling and endogenous regulators, such as calmodulin (CALM) and phospholamban (PLN) (Frank and Kranias, 2000; Soltis and Saucerman, 2010). The information of the genes encoding these proteins are summarized in Supplementary Table 1.
Due to the importance of the abovementioned Ca2+-handling proteins in maintaining a proper heart function, genetic polymorphisms of calcium signaling-related genes have been proposed to be associated with the pathogenesis of a certain type of CVDs. For example, a genome-wide association studies (GWAS) study has revealed the critical role of Ca2+-handling proteins in the myocardial repolarization reflected by the QT interval, linking polymorphisms in Ca2+-handling genes to arrhythmias (Arking et al., 2014). However, CVDs represent a group of disorders involving the heart and blood vessels, and the potential associations of genetic polymorphisms in Ca2+ homeostasis regulators with different types of CVDs are still inadequately understood. Thus, we performed this hypothesis-driven study to systematically explore the potential associations between 11,274 genetic variants located in 9 calcium signaling-related genes and 118 diseases of the circulatory system using a large sample from the United Kingdom Biobank (UKBB) (N = 308,366). For the identified genetic variants in genotype–clinical outcome associations, we also conducted a phenome-wide association study (PheWAS) to examine their potential role in modulating cardiovascular risk factors, including blood pressure and biomarkers.
Materials and Methods
The United Kingdom biobank recruited participants aged 40–69 years between 2006 and 2010, and the enrolled sampling persons completed a series of questionnaires and measurements at one of the 22 assessment centers in the United Kingdom. Blood samples were also collected at the time of entry. Around 50,000 samples were genotyped on the BiLEVE Axiom chip, while the rest of the genotyping was performed by the United Kingdom Biobank Axiom array, and these two chips had over 95% markers overlapped (Weng et al., 2017). Phasing and imputation were then conducted, and detailed genotyping and imputation procedures could be found elsewhere (Bycroft et al., 2017). We used the imputed variants for analysis and only included those passing Hardy–Weinberg equilibrium (HWE) test, and with minor allele frequency (MAF) ≥ 0.01, information scores (INFO) ≥ 0.7, and genotyping rate ≥ 90%. For 488,263 participants who had genetic data, we removed individuals who were of non-European ancestry, with sex discrepancies, high heterozygosity/missing rate, sex chromosome aneuploidy, or within a third-degree relatedness reflected by kinship coefficient. Participants who withdrew consent or had insufficient information of covariables and inpatient records were also excluded, leading to a final population of 308,366.
PheWAS R package was employed to examine the associations between each of 11,274 genetic variants and 118 outcomes in the disease category of the circulatory system (Denny et al., 2010). Participants’ clinical outcomes were obtained through the national electronic health records with the International Classification of Diseases (ICD) codes. ICD codes are designed primarily for billing and management purposes, and some of the ICD codes represent a single sub-phenotype of a disease, which are not independent phenotypes (Denny et al., 2010; Rahimi et al., 2019). Thus, ICD-9/10 codes from individual-level hospital episode data were mapped to phecodes according to PheWAS Catalog (Denny et al., 2013). Cases were individuals having at least one record for specific phecodes, and controls were defined as participants who were negative for the investigating phecode and related phecodes. Phecodes with cases exceeding 200 were included in the current study to obtain a reasonable power for detecting the genotype–phenotype associations (Verma et al., 2018). The logistic regression was performed to study the potential genotype–clinical outcome association. Age and sex were treated as covariables, and the first five principal components (PCs) were also adjusted for population structure. For multiple testing correction, the Bonferroni method assumed the tests of association performed were independent. However, the variants within a gene might be highly correlated through linkage disequilibrium, and diseases from the same category might also be related to each other, making Bonferroni correction too conservative. Thus, we estimated the numbers of independent variants and phenotypes by PLINK pairwise pruning methods (Tcheandjieu et al., 2020) and PhenoSpD package (Zheng et al., 2018), respectively, which suggested 2,790 independent variants and 96 independent phenotypes. The p-value threshold was then calculated as 0.05/(2,790 × 96) = 1.87 × 10–7 to define significant association. For the associations surviving a less restrictive threshold (defined as suggestive associations) of 0.05/(the number of independent variants in a gene × 96) (Supplementary Table 1), the GWAS catalog was searched to replicate our results (MacArthur et al., 2017; Tcheandjieu et al., 2020).
For mortality analyses, death information, including the date and cause of death, was obtained from the death registries, and follow-up time was calculated from the date of participants’ first visit to the assessment center to the date of death or the last follow-up date (July 31, 2020). Disease-related mortality was defined when the investigating disease was a primary or contributory cause of death (Batty et al., 2019). For disease incidence analyses, participants with disease onset before the start date were excluded from the cohort, and the person-time for each individual was censored at the date of first disease occurrence, date of death, or the last follow-up date (June 30, 2020 in England, October 31, 2016 in Scotland and February 29, 2016 in Wales), whichever came first. R was used to generate survival curves.
Phenome scan analysis tool (PHESANT) is an opensource tool that can scan the relationships between the variable of interest and a large range of phenotypes and traits in UKBB (Millard et al., 2018; Ripatti et al., 2019). We used PHESANT to scan the causal effect of identified genetic variation on various continuous variables, including blood pressure, birth weight, and blood biochemical indexes. Inverse normal rank transformation was performed for continuous variables before regression to ensure their normality. Age, sex, genotype chip, and the first 10 PCs were used as covariables for adjustments. The conservative Bonferroni corrected p-value of 0.05/1,646 = 3.04 × 10–5 was employed as the threshold for evaluating the results.
Results
Demographics of the unrelated UKBB participants involved in the current study are presented in Supplementary Table 2. The number of genetic variants in each of nine calcium signaling-related genes and the description and number of cases for each of 118 diseases of the circulatory system (in terms of phecodes) are provided in Supplementary Tables 1, 3, respectively. To systematically investigate the potential associations between all 11,274 genetic variants and all 118 clinical outcomes, association tests were performed (Figures 1, 2), and the results indicated significant associations between rs1801253 in β1-adrenergic receptor (ADRB1) and hypertension/essential hypertension. Moreover, 11 additional associations reached the suggestive level of significance. Due to the high levels of pairwise linkage disequilibrium (LD) of single nucleotide polymorphisms (SNPs) in one gene (Supplementary Table 4), only the leading SNPs with the smallest p-value in genotype–phenotype associations are shown in Table 1. It is worthy of note that the association between rs62420492 in TRDN and hypertension/essential hypertension did not reach the suggestive level after further adjusting the status of overweight (BMI ≥ 25 kg/m2), smoking, and drinking as covariables in the model (Supplementary Table 5). We next searched the GWAS catalog to replicate our finding, and the results indicated the association between rs1801253 (or its proxy in high LD with R2 > 0.8) and blood pressure had been reported, and the direction of effect was consistent with our observations (Supplementary Table 6). rs4484922 and rs4074536 in calsequestrin 2 (CASQ2) gene were in high LD (Supplementary Table 4) and were reported to be associated with QRS duration and atrial fibrillation, which was also replicated by our finding (Table 1 and Supplementary Table 7). However, the suggestive association of rs79613429 in ryanodine receptor 2 (RYR2) with precordial pain was not found in the GWAS catalog, representing novel genotype–phenotype association.
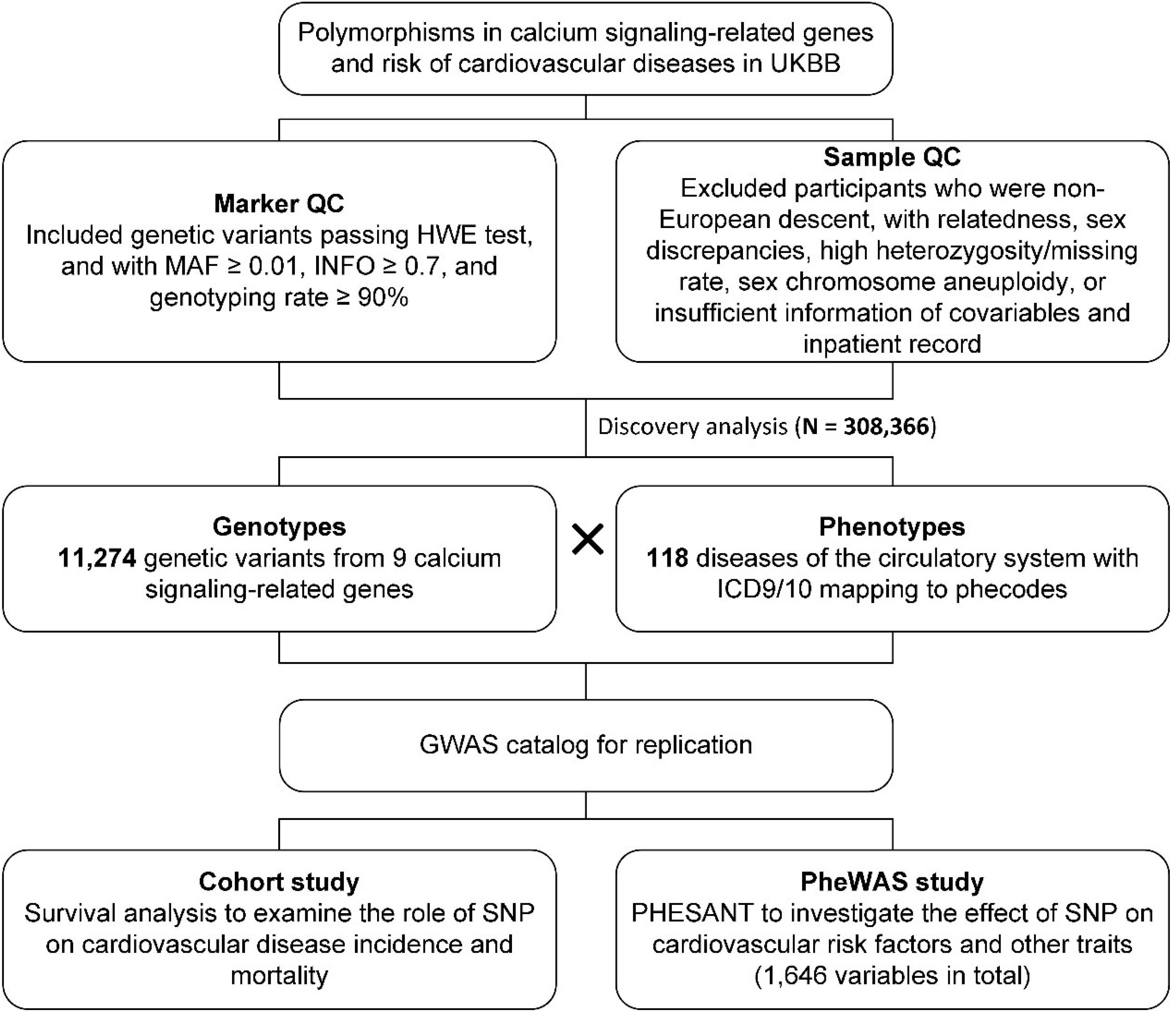
Figure 1. Workflow diagram for discovery and validation of associations between genetic variants in calcium signaling-related genes and CVDs.
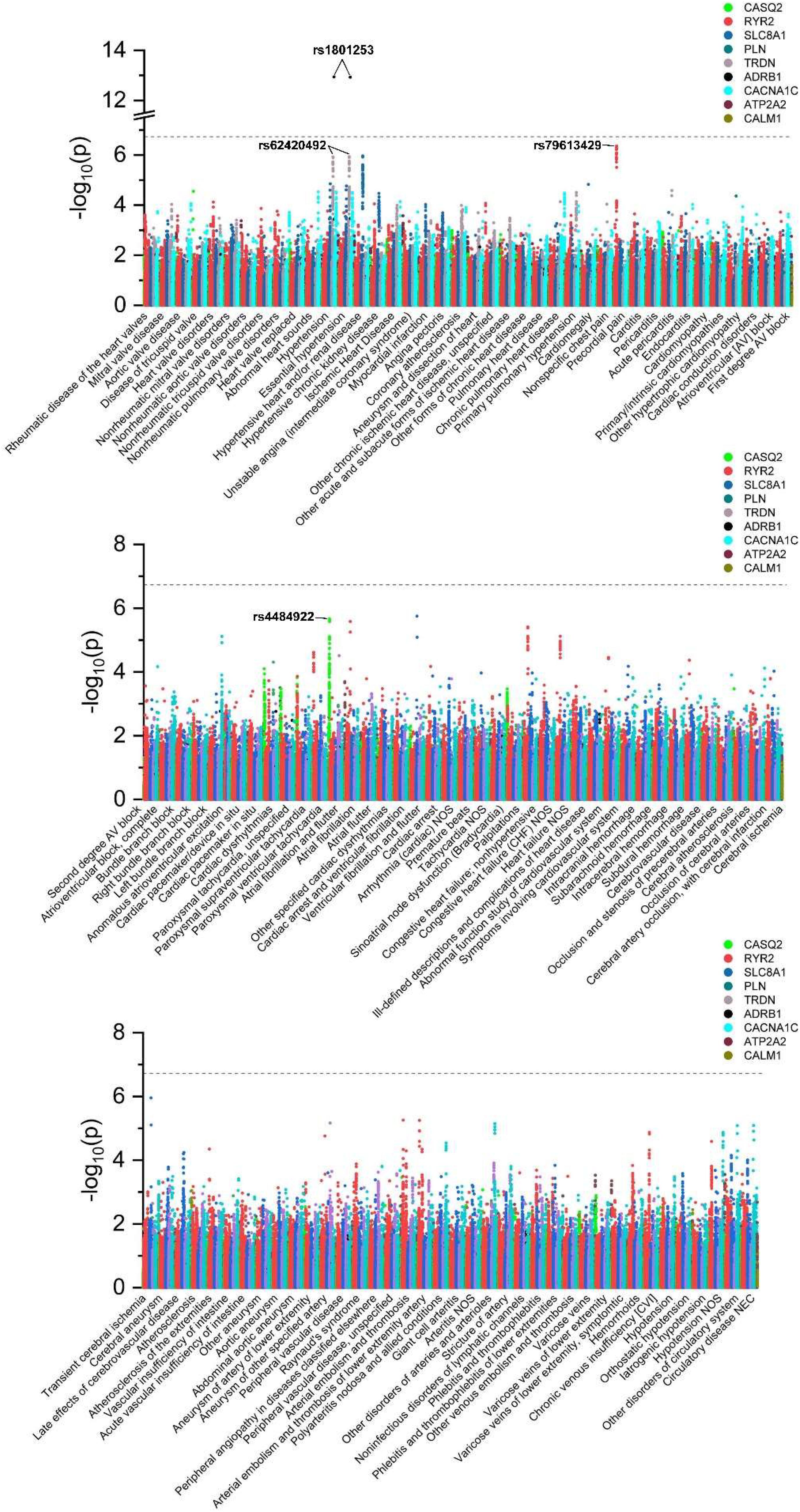
Figure 2. Manhattan plots showing p-values of all tested associations between 11,274 genetic variants and 118 diseases of the circulatory system. The threshold indicating a significant level of association is marked by a dotted line. The leading SNPs of significant or suggestive genotype–phenotype associations are annotated.
To further investigate the effects of variants in replicated associations on CVDs incidence and mortality, survival analyses were performed (Figure 3). The results consistently revealed that rs1801253 and rs4484922 were associated with the incidences of essential hypertension and atrial fibrillation/flutter, respectively, but not all-cause mortality or mortality-related to these diseases (Figure 3). We next studied whether these two variants were correlated to other risk factors that could potentially alter the incidence of their associated clinical outcomes. One thousand six hundred forty-six variables, including 30 biochemistry markers, were screened by PHESANT. Besides the known association between rs1801253 and blood pressure, we also reported a novel association of the blood pressure-lowering G allele of rs1801253 with a delayed onset of hypertension (Figure 4 and Supplementary Table 8). For rs4484922, no association survived Bonferroni or false discovery rate (FDR) correction, but this variant displayed nominal evidence of association with multiple electrocardiogram (ECG) parameters, such as QRS duration (Supplementary Table 7).
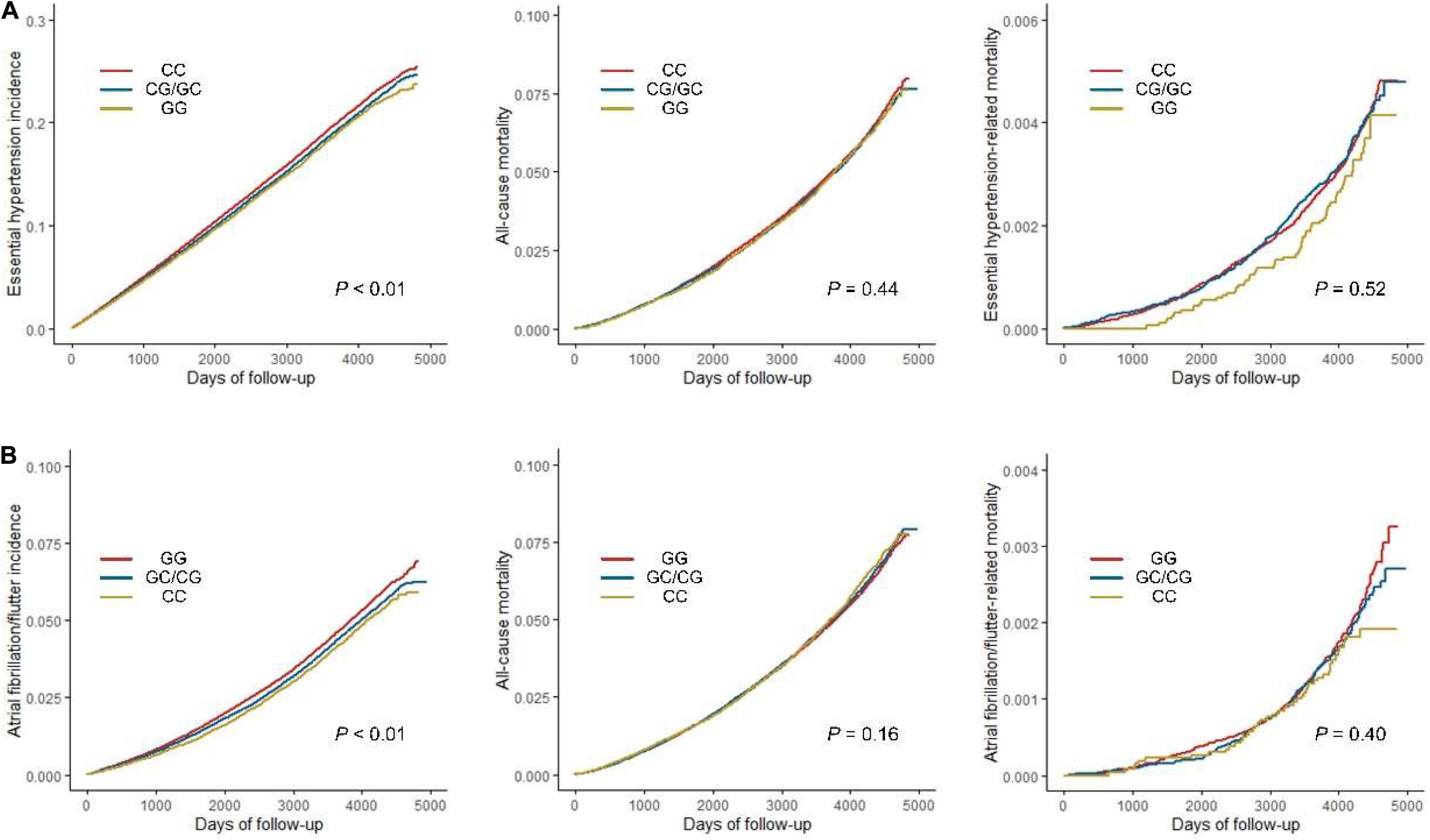
Figure 3. Survival analyses of essential hypertension (A) or atrial fibrillation/flutter (B) incidence, all-cause and disease-related mortality by genotypes of rs1801253 (A) or rs4484922 (B).
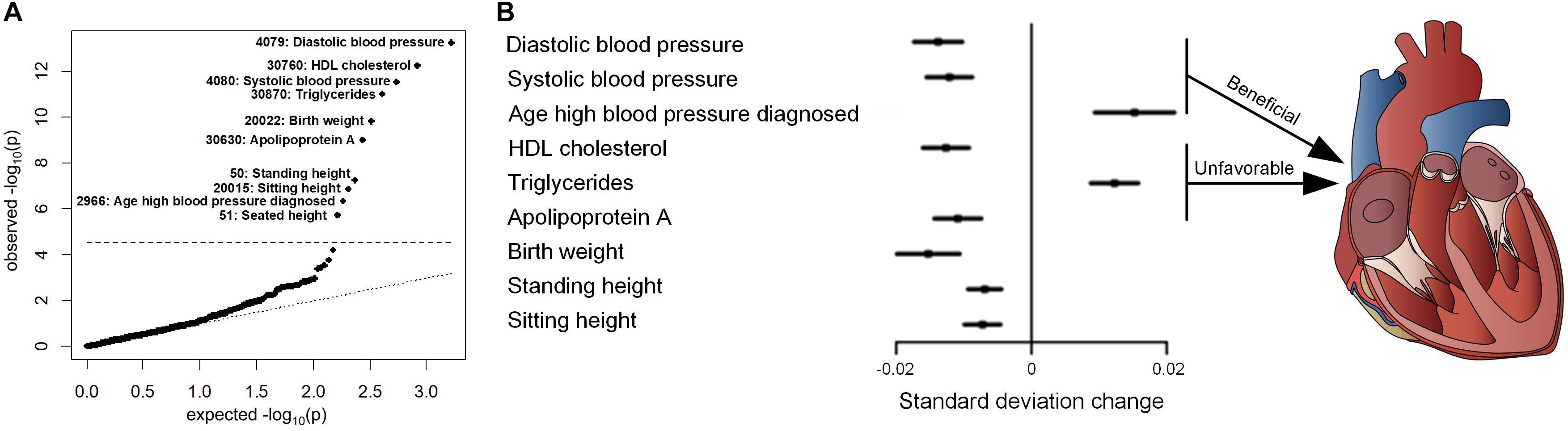
Figure 4. PheWAS of rs1801253 in UKBB. (A) QQ plot. The black dashed line represents the Bonferroni-corrected threshold (p = 0.05/1,646 = 3.04 × 10– 5). The number indicates the field ID of variables in UKBB. (B) Forest plot of variables that were significantly associated with rs1801253. The estimates reflect the standard deviation change in the inverse ranked transformed variables per increase of rs1801253 G allele.
Discussion
The excitability and contraction of cardiomyocytes are largely regulated by Ca2+. Ca2+ not only participate in the normal myocardial contraction but also enroll in the occurrence of CVDs. Indeed, accumulating evidence suggests a significant impact of genetic polymorphisms and mutations of Ca2+ signaling-related genes on CVDs pathogenesis. For example, a literature review evaluated and summarized the effect of 53 SNPs on sudden cardiac death (SCD) and found that rs6684209, rs3814843, and rs35594137 in CASQ2, CALM1, and GJA5, respectively, had the most significant association with SCD (Tamariz et al., 2019). Furthermore, mutations of RYR2, which is responsible for SR Ca2+ release, are associated with various CVDs, including dilated cardiomyopathy and hypertrophic cardiomyopathy (Bhuiyan et al., 2007; Tang et al., 2012). Mutation of PLN also induced dilated cardiomyopathy (van der Zwaag et al., 2012). Experiments with human induced pluripotent stem cell (hiPSC) with inserted PLN R9C mutations indicated these cells had a weak response to isoproterenol compared to wild type cells and displayed abnormal Ca2+ handling properties, reflecting a pathological state (Ceholski et al., 2018). Moreover, abnormal Ca2+ handling in failing cardiomyocytes could promote arrhythmia, and a genetic variant in SERCA2 that is responsible for SR Ca2+ uptake has been linked to a decreased arrhythmia risk in patients with heart failure (Francia et al., 2013). GWAS or PheWAS has been widely applied to identify genotype–phenotype associations. However, strict control of false positives by multiple testing corrections in these unbiased estimations may result in the missing of true positive correlation (Johnson et al., 2011). To reduce the multiple testing burden, we included a predefined subset of genotypes (11,274 genetic variants of calcium signaling-related genes) and phenotypes (118 diseases of the circulatory system) based on a priori hypotheses that genetic variations of genes related to cardiac Ca2+ handling play an essential role in the pathogenesis of a wide range of CVDs. For the identified genotype–clinical outcome association, we employed PHESANT to investigate the potential effect of SNPs on cardiovascular risk factors, including blood pressure and biochemical indexes, which helps to illustrate the mechanisms underlying the development and progression of heart diseases.
Hypertension is a prevalent disease that contributes to a significant portion of premature deaths globally (Surendran et al., 2016). Abnormal increase of blood pressure is a known risk factor for multiple CVDs, such as coronary heart disease and stroke (Surendran et al., 2016), and interventions in blood pressure benefit cardiovascular health (Patel et al., 2016). The heritability of blood pressure can reach 30–50%, highlighting the role of genetic variation in blood pressure determination (Hoffmann et al., 2017). Adrenergic receptors, encoded by ADRB gene, belong to the G-protein-coupled receptors family and play a vital role in regulating myocardial contraction and heart rate (Johnson et al., 2011). The rs1801253 genetic variant in ADRB1 gene leads to arginine to glycine replacement at position 389 of β1 adrenergic receptor (Arg389Gly). The location of Arg389Gly polymorphism is proposed to be a binding domain of Gs-protein (Peng et al., 2009). In vitro experiments suggested a higher level of adenylyl cyclase activation in cells with Arg389 β1 adrenergic receptor upon isoprenaline treatment (Rathz et al., 2003). Thus, the enhanced activity caused by Arg389 variant may contribute to a higher cardiac output, which, together with peripheral resistance, determines blood pressure (Peng et al., 2009). Furthermore, polymorphisms in ADRB1 also alter plasma renin activity, which is essential for blood pressure regulation (Petersen et al., 2012). Indeed, the association between Arg389Gly polymorphism and blood pressure/hypertension has been reported across a wide range of ethnicities. For example, the rs1801253 variant was associated with the risk of essential hypertension in a study based on the Chinese Han population (Peng et al., 2009). In a population from Mexico City, participants with homozygous C allele in rs1801253 were more likely to have increased diastolic pressure (Burguete-Garcia et al., 2014). Our results in UKBB with a large sample size consistently revealed that the G allele of rs1801253 was associated with lower diastolic/systolic blood pressure and incidence of hypertension (Figure 4 and Table 1). Moreover, our PheWAS approach identified a delayed onset of hypertension in patients with GG genotype (diagnosed age: 50.99 ± 9.88 years) compared to those with CC genotype (diagnosed age: 50.58 ± 9.94 years) (Figure 4 and Supplementary Table 8). For lipid metabolism traits, the published GWAS meta-analyses from the global lipids genetics consortium (GLGC) indicated that the G allele of rs1801253 was associated with an increased level of triglycerides (TG) and decreased level of high-density lipoprotein (HDL) cholesterol (Willer et al., 2013), which was consistent with our observations (Supplementary Tables 8, 9). We additionally reported that the level of apolipoprotein A, as protein carried in HDL cholesterol, was decreased in participants with G allele (Figure 4 and Supplementary Table 8). Thus, the heart-beneficial blood pressure-lowering rs1801253 G allele is also associated with unfavorable lipid metabolism that is linked to an increased risk of developing CVDs. These double-edged sword effects of Arg389Gly polymorphism may be attributed to its association with fetal growth reflected by birth weight (Horikoshi et al., 2013), which was also replicated in our analyses that G allele of rs1801253 was correlated with a lower birth weight (Figure 4 and Supplementary Table 8). It is worthy of note that when applying the less conservative FDR correction for multiple comparisons, rs1801253 was associated with additional variables, such as apolipoprotein B, pulse rate, and mean sphered cell volume, reflecting the pleiotropy of this variant (Supplementary Table 10).
Arrhythmia is characterized by abnormal heartbeat caused by the dysregulation of electrical impulse generation or conduction (Kennedy et al., 2016). Calcium-related proteins affect cardiac electrophysiology by changing either the electrophysiological properties of cells or heart tissue structure (Kolder et al., 2012). The prolongation of the QT interval measured non-invasively by ECG can potentially promote ventricular arrhythmias, and the effects of genetic polymorphisms in Ca2+-handling proteins in myocardial repolarization reflected by the QT interval has been reported (Arking et al., 2014). Arrhythmias is a well-known risk factor for SCD. The correlation between Ca2+ and SCD has been proved in an animal model, in which a high level of intracellular Ca2+ promoted malignant arrhythmias (Billman et al., 1991). The genetic polymorphisms having the most significant associations with SCD are also from the genes of Ca2+-handling proteins responsible for EC coupling (Tamariz et al., 2019). In chronic heart failure patients from the Chinese Han population, both rs3814843 and rs361508, located on CALM1 and triadin (TRDN), respectively, were associated with increased risk of SCD as examined by survival analysis, suggesting common variants in Ca2+-handling proteins played an important role in the development of SCD during chronic heart failure (Laver et al., 2015). Cardiac calsequestrin 2, encoded by CASQ2 gene, has a high capacity for Ca2+ binding and is responsible for Ca2+ storage in SR (Yano and Zarain-Herzberg, 1994). Calsequestrin can also form a complex with RYR2 to modulate SR Ca2+ release (Flores et al., 2018). It has been known that mutations in CASQ2 lead to catecholaminergic polymorphic ventricular tachycardia (CPVT), which may trigger cardiac arrest (Faggioni et al., 2012). For the common variants in CASQ2, the C allele of non-synonymous variant rs4074536 was associated with decreased QRS duration in participants of European ancestry (Prins et al., 2018). In the Chinese Han population, a cohort study with 379 participants free of CVDs indicated rs4074536 was associated with the PR interval in ECG. More specifically, PR interval prolongation in individuals with CC genotype compared to those with non-CC genotypes was statistically significant (Li et al., 2020). Using a large sample from UKBB, we consistently found the suggestive association of rs4074536, or its proxy SNPs rs4484922 and rs3810998, with atrial fibrillation/flutter (Table 1 and Supplementary Table 4). Moreover, participants with GG genotype had a prolonged QRS duration compared to those with CC genotype (89.00 ± 14.01 vs. 88.48 ± 14.44 ms) (Supplementary Table 7), which was in accordance with the notion that each copy of C allele in rs4074536 was associated with less than 1 ms shortening of QRS duration (Sotoodehnia et al., 2010). Interestingly, our PheWAS approach revealed that rs4484922 showed nominal evidence of association with multiple parameters of carotid intima-medial thickness (cIMT) ultrasound measurement (Supplementary Table 7), and the elevated cIMT observed in individuals with CC genotype may increase the risk of atherosclerosis, which is an underlying cause of multiple CVDs (Strawbridge et al., 2020).
We found suggestive association of rs79613429 located in the intron of RYR2 gene with precordial pain (p = 4.39 × 10–7). Genetic variants located in introns are well recognized to regulate mRNA splicing and gene expression (Wang and Cooper, 2007; Faraday et al., 2011). However, how these variants lead to an altered risk of clinical outcomes needs to be further elucidated. The major strength of our study is that multiple genotype–clinical outcome associations are systematically tested to investigate the role of calcium signaling-related genetic variants in CVDs. Furthermore, we performed the analyses in a large sample from the UKBB to avoid the decreased power in detecting association between CVDs and common variants with MAF below 5% (Altshuler et al., 2008). For the classification of clinical outcomes in electronic health records, we employed the phecode system that is more representative of independent phenotypes compared to ICD codes (Denny et al., 2010; Rahimi et al., 2019; King et al., 2020). Moreover, excluding participants with phecodes/diseases related to the investigating phecode/disease in the analysis prevented the contamination of the control population by cases, leading to an increased statistical power (Wei et al., 2017). Our genetic variant-based PheWAS is also advantageous because the genotype is fixed at birth and the results are less susceptible to reverse causality (Shen et al., 2020). The present study also has several limitations. First, using a single SNP in the correlation analyses can predict only a small effect size of the association (Cai et al., 2018). Second, the novel associations observed in our study are needed to be replicated in independent datasets of similar ancestry to enhance their credibility. Moreover, these associations were solely based on the European population, and replication in non-European populations will further validate our results. Also, cell and animal-based experiments could be applied to examine the functions of the reported variants to discover novel biological mechanisms.
Conclusion
Multiple associations of genetic variants in calcium signaling-related genes with CVDs and cardiovascular risk factors have been identified, highlighting genetic testing as a useful method for disease diagnosis and prevention.
Data Availability Statement
The data used in this study is publicly available and can be obtained from United Kingdom Biobank (http://www.ukbiobank.ac.uk).
Ethics Statement
All participants provided written informed consent, and the United Kingdom Biobank is ethically approved by the National Research Ethics Service (ref 11/NW/0382).
Author Contributions
SL designed the study. ZJ and ZZ performed the statistical analyses. SL, ZZ, and YL drafted the manuscript. MY and TY critically reviewed the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (Grant Nos. 81703942 and 81973698), the Young Elite Scientists Sponsorship Program by CACM (Grant No. 2019-QNRC2-B08), the Science Fund for Distinguished Young Scholars in BUCM (Grant No. BUCM-2019-JCRC004), and the BUCM Research Start-Up Fund (to SL).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
This research has been conducted using data from United Kingdom Biobank, a major biomedical database.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.642141/full#supplementary-material
Abbreviations
EC, coupling, excitation–contraction coupling; RYRs, ryanodine receptors; SR, sarcoplasmic reticulum; SERCA, sarco/endoplasmic reticulum Ca2+-ATPase; NCX, sodium–calcium exchanger; CALM, calmodulin; PLN, phospholamban; CVDs, cardiovascular diseases; DAD, delayed after-depolarization; EAD, early after-depolarization; CPVT, catecholaminergic polymorphic ventricular tachycardia; UKBB, United Kingdom Biobank; PheWAS, phenome-wide association study; HWE, Hardy–Weinberg equilibrium; MAF, minor allele frequency; ICD, the International Classification of Diseases; PCs, principal components; PHESANT, phenome scan analysis tool; LD, linkage disequilibrium; FDR, false discovery rate; ECG, electrocardiogram; SCD, sudden cardiac death; hiPSC, human induced pluripotent stem cell; GWAS, genome-wide association studies; TRDN, triadin; GLGC, global lipids genetics consortium; cIMT, carotid intima-medial thickness; BMI, body mass index; HDL, high-density lipoprotein; TG, triglycerides; OR, odds ratio; N, number; SD, standard deviation; 95% CI, 95% confidence interval.
References
Altshuler, D., Daly, M. J., and Lander, E. S. (2008). Genetic mapping in human disease. Science 322, 881–888. doi: 10.1126/science.1156409
Arking, D. E., Pulit, S. L., Crotti, L., van der Harst, P., Munroe, P. B., Koopmann, T. T., et al. (2014). Genetic association study of QT interval highlights role for calcium signaling pathways in myocardial repolarization. Nat. Genet. 46, 826–836. doi: 10.1038/ng.3014
Batty, G. D., Gale, C. R., Kivimäki, M., and Bell, S. (2019). Assessment of relative utility of underlying vs contributory causes of death. JAMA Netw. Open 2:e198024. doi: 10.1001/jamanetworkopen.2019.8024
Bhuiyan, Z. A., van den Berg, M. P., van Tintelen, J. P., Bink-Boelkens, M. T., Wiesfeld, A. C., Alders, M., et al. (2007). Expanding spectrum of human RYR2-related disease: new electrocardiographic, structural, and genetic features. Circulation 116, 1569–1576. doi: 10.1161/CIRCULATIONAHA.107.711606
Billman, G. E., McIlroy, B., and Johnson, J. D. (1991). Elevated myocardial calcium and its role in sudden cardiac death. FASEB J. 5, 2586–2592. doi: 10.1096/fasebj.5.11.1714409
Burguete-Garcia, A. I., Martinez-Nava, G. A., Valladares-Salgado, A., Bermudez Morales, V. H., Estrada-Velasco, B., Wacher, N., et al. (2014). Association of β1 and β3 adrenergic receptors gene polymorphisms with insulin resistance and high lipid profiles related to type 2 diabetes and metabolic syndrome. Nutr. Hosp. 29, 1327–1334. doi: 10.3305/nh.2014.29.6.7367
Bycroft, C., Freeman, C., Petkova, D., Band, G., Elliott, L. T., Sharp, K., et al. (2017). Genome-wide genetic data on ~500,000 UK Biobank participants. Biorxiv [Preprint] doi: 10.1101/166298
Cai, T., Zhang, Y., Ho, Y.-L., Link, N., Sun, J., Huang, J., et al. (2018). Association of interleukin 6 receptor variant with cardiovascular disease effects of interleukin 6 receptor blocking therapy. JAMA Cardiol. 3, 849–857. doi: 10.1001/jamacardio.2018.2287
Ceholski, D. K., Turnbull, I. C., Kong, C.-W., Koplev, S., Mayourian, J., Gorski, P. A., et al. (2018). Functional and transcriptomic insights into pathogenesis of R9C phospholamban mutation using human induced pluripotent stem cell-derived cardiomyocytes. J. Mol. Cell. Cardiol. 119, 147–154. doi: 10.1016/j.yjmcc.2018.05.007
Clusin, W. T. (2003). Calcium and cardiac arrhythmias: dads, eads, and alternans. Crit. Rev. Clin. Lab. Sci. 40, 337–375.
Denny, J. C., Bastarache, L., Ritchie, M. D., Carroll, R. J., Zink, R., Mosley, J. D., et al. (2013). Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat. Biotechnol. 31, 1102–1110.
Denny, J. C., Ritchie, M. D., Basford, M. A., Pulley, J. M., Bastarache, L., Brown-Gentry, K., et al. (2010). PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics 26, 1205–1210. doi: 10.1093/bioinformatics/btq126
Eisner, D. A., Caldwell, J. L., Kistamás, K., and Trafford, A. W. (2017). Calcium and excitation-contraction coupling in the heart. Circ. Res. 121, 181–195. doi: 10.1016/b978-012436570-4/50007-8
Faggioni, M., Kryshtal, D. O., and Knollmann, B. C. (2012). Calsequestrin mutations and catecholaminergic polymorphic ventricular tachycardia. Pediatr. Cardiol. 33, 959–967. doi: 10.1007/s00246-012-0256-1
Faraday, N., Yanek, L. R., Yang, X. P., Mathias, R., Herrera-Galeano, J. E., Suktitipat, B., et al. (2011). Identification of a specific intronic PEAR1 gene variant associated with greater platelet aggregability and protein expression. Blood 118, 3367–3375. doi: 10.1182/blood-2010-11-320788
Flores, D. J., Duong, T., Brandenberger, L. O., Mitra, A., Shirali, A., Johnson, J. C., et al. (2018). Conditional ablation and conditional rescue models for Casq2 elucidate the role of development and of cell-type specific expression of Casq2 in the CPVT2 phenotype. Hum. Mol. Genet. 27, 1533–1544. doi: 10.1093/hmg/ddy060
Francia, P., Adduci, C., Ricotta, A., Stanzione, R., Sensini, I., Uccellini, A., et al. (2013). Common genetic variants in selected Ca2+ signaling genes and the risk of appropriate ICD interventions in patients with heart failure. J. Interv. Card. Electrophysiol. 38, 169–177.
Frank, K., and Kranias, E. G. (2000). Phospholamban and cardiac contractility. Ann. Med. 32, 572–578. doi: 10.3109/07853890008998837
Hoffmann, T. J., Ehret, G. B., Nandakumar, P., Ranatunga, D., Schaefer, C., Kwok, P. Y., et al. (2017). Genome-wide association analyses using electronic health records identify new loci influencing blood pressure variation. Nat. Genet. 49, 54–64. doi: 10.1038/ng.3715
Horikoshi, M., Yaghootkar, H., Mook-Kanamori, D. O., Sovio, U., Taal, H. R., Hennig, B. J., et al. (2013). New loci associated with birth weight identify genetic links between intrauterine growth and adult height and metabolism. Nat. Genet. 45, 76–82. doi: 10.1038/ng.2477
Johnson, A. D., Newton-Cheh, C., Chasman, D. I., Ehret, G. B., Johnson, T., Rose, L., et al. (2011). Association of hypertension drug target genes with blood pressure and hypertension in 86 588 individuals. Hypertension 57, 903–910. doi: 10.1161/HYPERTENSIONAHA.110.158667
Kennedy, A., Finlay, D. D., Guldenring, D., Bond, R., Moran, K., and McLaughlin, J. (2016). The cardiac conduction system: generation and conduction of the cardiac impulse. Crit. Care Nurs. Clin. North Am. 28, 269–279.
King, C., Mulugeta, A., Nabi, F., Walton, R., Zhou, A., and Hyppönen, E. (2020). Mendelian randomization case-control PheWAS in UK Biobank shows evidence of causality for smoking intensity in 28 distinct clinical conditions. Eclinicalmedicine 26:100488. doi: 10.1016/j.eclinm.2020.100488
Kolder, I. C., Tanck, M. W., and Bezzina, C. R. (2012). Common genetic variation modulating cardiac ECG parameters and susceptibility to sudden cardiac death. J. Mol. Cell Cardiol. 52, 620–629. doi: 10.1016/j.yjmcc.2011.12.014
Laver, D., Liu, Z., Liu, X., Yu, H., Pei, J., Zhang, Y., et al. (2015). Common variants in TRDN and CALM1 are associated with risk of sudden cardiac death in chronic heart failure patients in chinese han population. PLoS One 10:e0132459. doi: 10.1371/journal.pone.0132459
Li, X., Guo, L.-Z., Liu, N., Du, X., Bai, R., Dong, J.-Z., et al. (2020). Association of T66A polymorphism in CASQ2 with PR interval in a Chinese population. Herz 46(Suppl 1), 123–129. doi: 10.1007/s00059-020-04913-3
MacArthur, J., Bowler, E., Cerezo, M., Gil, L., Hall, P., Hastings, E., et al. (2017). The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 45, D896–D901.
Millard, L. A. C., Davies, N. M., Gaunt, T. R., Davey Smith, G., and Tilling, K. (2018). Software application profile: phesant: a tool for performing automated phenome scans in UK Biobank. Int. J. Epidemiol. 47, 29–35. doi: 10.1093/ije/dyx204
Patel, P., Ordunez, P., DiPette, D., Escobar, M. C., Hassell, T., Wyss, F., et al. (2016). Improved blood pressure control to reduce cardiovascular disease morbidity and mortality: the standardized hypertension treatment and prevention project. J. Clin. Hypertens 18, 1284–1294. doi: 10.1111/jch.12861
Peng, Y., Xue, H., Luo, L., Yao, W., and Li, R. (2009). Polymorphisms of the beta1-adrenergic receptor gene are associated with essential hypertension in Chinese. Clin. Chem. Lab. Med. 47, 1227–1231.
Petersen, M., Andersen, J. T., Jimenez-Solem, E., Broedbaek, K., Hjelvang, B. R., Henriksen, T., et al. (2012). Effect of the Arg389Gly β1-adrenoceptor polymorphism on plasma renin activity and heart rate, and the genotype-dependent response to metoprolol treatment. Clin. Exp. Pharmacol. Physiol. 39, 779–785. doi: 10.1111/j.1440-1681.2012.05736.x
Prins, B. P., Mead, T. J., Brody, J. A., Sveinbjornsson, G., Ntalla, I., Bihlmeyer, N. A., et al. (2018). Exome-chip meta-analysis identifies novel loci associated with cardiac conduction, including ADAMTS6. Genome Biol. 19:87. doi: 10.1186/s13059-018-1457-6
Rahimi, K., Li, X., Meng, X., He, Y., Spiliopoulou, A., Timofeeva, M., et al. (2019). Genetically determined serum urate levels and cardiovascular and other diseases in UK Biobank cohort: a phenome-wide mendelian randomization study. PLoS Med. 16:e1002937. doi: 10.1371/journal.pmed.1002937
Rathz, D. A., Gregory, K. N., Fang, Y., Brown, K. M., and Liggett, S. B. (2003). Hierarchy of polymorphic variation and desensitization permutations relative to beta 1- and beta 2-adrenergic receptor signaling. J. Biol. Chem. 278, 10784–10789. doi: 10.1074/jbc.M206054200
Ripatti, S., Millard, L. A. C., Davies, N. M., Tilling, K., Gaunt, T. R., and Davey Smith, G. (2019). Searching for the causal effects of body mass index in over 300 000 participants in UK Biobank, using Mendelian randomization. PLoS Genet. 15:e1007951. doi: 10.1371/journal.pgen.1007951
Shen, X., Howard, D. M., Adams, M. J., Hill, W. D., Clarke, T.-K., Deary, I. J., et al. (2020). A phenome-wide association and mendelian randomisation study of polygenic risk for depression in UK Biobank. Nat. Commun. 11: 2301.
Soltis, A. R., and Saucerman, J. J. (2010). Synergy between CaMKII substrates and β-adrenergic signaling in regulation of cardiac myocyte Ca(2+) handling. Biophys. J. 99, 2038–2047. doi: 10.1016/j.bpj.2010.08.016
Sotoodehnia, N., Isaacs, A., de Bakker, P. I., Dörr, M., Newton-Cheh, C., Nolte, I. M., et al. (2010). Common variants in 22 loci are associated with QRS duration and cardiac ventricular conduction. Nat. Genet. 42, 1068–1076. doi: 10.1038/ng.716
Strawbridge, R. J., Ward, J., Bailey, M. E. S., Cullen, B., Ferguson, A., Graham, N., et al. (2020). Carotid intima-media thickness: novel loci, sex-specific effects, and genetic correlations with obesity and glucometabolic traits in UK biobank. Arterioscler. Thromb. Vasc. Biol. 40, 446–461. doi: 10.1161/ATVBAHA.119.313226
Surendran, P., Drenos, F., Young, R., Warren, H., Cook, J. P., Manning, A. K., et al. (2016). Trans-ancestry meta-analyses identify rare and common variants associated with blood pressure and hypertension. Nat. Genet. 48, 1151–1161. doi: 10.1038/ng.3654
Tamariz, L., Balda, J., Pareja, D., Palacio, A., Myerburg, R. J., Conway, D., et al. (2019). Usefulness of single nucleotide polymorphisms as predictors of sudden cardiac death. Am. J. Cardiol. 123, 1900–1905. doi: 10.1016/j.amjcard.2019.02.058
Tang, Y., Tian, X., Wang, R., Fill, M., and Chen, S. R. (2012). Abnormal termination of Ca2+ release is a common defect of RyR2 mutations associated with cardiomyopathies. Circ. Res. 110, 968–977. doi: 10.1161/circresaha.111.256560
Tcheandjieu, C., Aguirre, M., Gustafsson, S., Saha, P., Potiny, P., Haendel, M., et al. (2020). A phenome-wide association study of 26 mendelian genes reveals phenotypic expressivity of common and rare variants within the general population. PLoS Genet. 16:e1008802. doi: 10.1371/journal.pgen.1008802
van der Zwaag, P. A., van Rijsingen, I. A., Asimaki, A., Jongbloed, J. D., van Veldhuisen, D. J., Wiesfeld, A. C., et al. (2012). Phospholamban R14del mutation in patients diagnosed with dilated cardiomyopathy or arrhythmogenic right ventricular cardiomyopathy: evidence supporting the concept of arrhythmogenic cardiomyopathy. Eur. J. Heart Fail. 14, 1199–1207. doi: 10.1093/eurjhf/hfs119
Verma, A., Bradford, Y., Dudek, S., Lucas, A. M., Verma, S. S., Pendergrass, S. A., et al. (2018). A simulation study investigating power estimates in phenome-wide association studies. BMC Bioinformatics 19:120. doi: 10.1186/s12859-018-2135-0
Wang, G. S., and Cooper, T. A. (2007). Splicing in disease: disruption of the splicing code and the decoding machinery. Nat. Rev. Genet. 8, 749–761. doi: 10.1038/nrg2164
Wei, W. Q., Bastarache, L. A., Carroll, R. J., Marlo, J. E., Osterman, T. J., Gamazon, E. R., et al. (2017). Evaluating phecodes, clinical classification software, and ICD-9-CM codes for phenome-wide association studies in the electronic health record. PLoS One 12:e0175508. doi: 10.1371/journal.pone.0175508
Weng, L.-C., Choi, S. H., Klarin, D., Smith, J. G., Loh, P.-R., Chaffin, M., et al. (2017). Heritability of atrial fibrillation. Circ. Cardiovasc. Genet. 10:e001838.
Willer, C. J., Schmidt, E. M., Sengupta, S., Peloso, G. M., Gustafsson, S., Kanoni, S., et al. (2013). Discovery and refinement of loci associated with lipid levels. Nat. Genet. 45, 1274–1283. doi: 10.1038/ng.2797
Yano, K., and Zarain-Herzberg, A. (1994). Sarcoplasmic reticulum calsequestrins: structural and functional properties. Mol. Cell Biochem. 135, 61–70. doi: 10.1007/bf00925961
Keywords: genetic polymorphism, calcium, cardiovascular diseases, cardiovascular risk factors, PheWAS
Citation: Li S, Jia Z, Zhang Z, Li Y, Yan M and Yu T (2021) Association Study of Genetic Variants in Calcium Signaling-Related Genes With Cardiovascular Diseases. Front. Cell Dev. Biol. 9:642141. doi: 10.3389/fcell.2021.642141
Received: 15 December 2020; Accepted: 01 October 2021;
Published: 29 November 2021.
Edited by:
Mario Antonio Bianchet, Johns Hopkins University, United StatesReviewed by:
Monica Trif, Centre for Innovative Process Engineering, GermanyDaniela Rossi, University of Siena, Italy
Copyright © 2021 Li, Jia, Zhang, Li, Yan and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sen Li, c2VubGlAYnVjbS5lZHUuY24=
 Sen Li
Sen Li Zhaoqi Jia
Zhaoqi Jia Zhang Zhang
Zhang Zhang Yuxin Li
Yuxin Li Tingting Yu
Tingting Yu