- 1Division of Systems Medicine & Gene Therapy, Saitama Medical University, Saitama, Japan
- 2Department of Systems Aging Science and Medicine, Tokyo Metropolitan Institute of Gerontology, Tokyo, Japan
Ovarian cancer is a health-threatening malignancy of ovary in female reproductive systems and one of the most common gynecological malignancies worldwide. Due to rare early symptoms, ovarian cancers are often diagnosed at advanced stages and exhibit poor prognosis. Thus, efforts have been paid to develop alternative diagnostic and therapeutic strategies for the disease. Recent studies have presented that some long non-coding RNAs (lncRNAs) play roles in apoptosis of ovarian cancer cells through various mechanisms involved in the regulation of transcription factors, histone modification complexes, miRNAs, and protein stability. Because evasion of apoptosis in cancer cells facilitates to promote tumor progression and therapy resistance, apoptosis regulatory mechanisms of lncRNAs may be promising new targets in ovarian cancer. In this review, we introduce the recent findings in regard to the molecular mechanisms of apoptosis-related lncRNAs in ovarian cancer cells.
Ovary and Ovarian Cancer
The ovary is a female reproductive organ where oocyte development occurs (Motta et al., 1997; Virant-Klun, 2015; Yadav et al., 2018) and functions as an endocrine organ involved in the synthesis of the female sex steroid hormones and the regulation of reproduction such as the menstrual cycle, pregnancy, and lactation (Hiller-Sturmhöfel and Bartke, 1998). Thus, the health of ovaries is essential for reproduction and women’s lives, rendering finding cures to ovarian diseases crucial. Ovarian cancer is one of the most common gynecological cancers (Momenimovahed et al., 2019). The GLOBOCAN 2018 data estimates ∼300,000 new cases of ovarian cancer and over 180,000 ovarian cancer–related deaths per year worldwide (Bray et al., 2018). Ovarian cancer is a heterogeneous disease and classified by type of originated cell. Epithelial ovarian cancer (EOC) is the most common ovarian cancer (∼90%) (Rojas et al., 2016; Momenimovahed et al., 2019). The disease is often advanced at diagnosis due to lack of early symptoms and the 5-year cause-specific survival rate is <50% (Siegel et al., 2018; Torre et al., 2018; Trinidad et al., 2020). Based on the current limitations, alternative diagnostic and therapeutic approaches for ovarian cancer remain to be explored.
Apoptosis in Ovary and Ovarian Cancer
Apoptosis is a process of programmed cell death triggered by intrinsic or extrinsic signals (Wong, 2011). Intrinsic signals are initiated by cellular stresses. These signals increase the mitochondrial permeability and release of the pro-apoptotic factors such as cytochrome-c, resulting in activation of cysteine-aspartic acid proteases (caspases), which are essential enzymes for apoptosis execution. Meanwhile, extrinsic signals are mediated by death receptor signaling pathways. Death receptors, their ligands, and adaptor proteins form the death-inducing signaling complex (DISC), which triggers caspase activation (Wong, 2011).
Apoptosis plays physiological roles in normal ovary functions such as follicular atresia and corpus luteum regression (Vaskivuo and Tapanainen, 2003; Yadav et al., 2018). In malignant tumors, evasion of apoptosis facilitates cancer cell survival and tumor progression (Wong, 2011; Binju et al., 2019), thus efforts have been paid for cancer strategies to discover the molecules to exert apoptosis in cancer cells whereas not in normal cells. For ovarian cancer treatment, small chemicals that modulate apoptosis-related proteins such as inhibitors of apoptotic proteins (IAPs) have entered clinical trials (Binju et al., 2019).
In terms of apoptosis pathways in cancers, several mechanisms of apoptosis-related genes have been well characterized. Transcription factors such as E2F family proteins, nuclear factor kappa B (NF-κB) proteins, and signal transducer and activator of transcription (STAT) family proteins modulate apoptosis via regulating transcription of apoptosis-related genes (Bours et al., 2000; Crosby and Almasan, 2004; Karin, 2006; Kim and Lee, 2007; Kent and Leone, 2019; Verhoeven et al., 2020). Histone modification complexes such as polycomb repressive complex 1/2 (PRC1/2) affect transcription of apoptosis-related genes through histone methylation (Cao et al., 2011; Wang W. et al., 2015; Christofides et al., 2016). Apoptosis-related genes are also modulated by post-transcriptional gene regulation mechanism, such as through miRNAs that regulate apoptosis-related gene mRNAs (Di Leva et al., 2014; Pistritto et al., 2016; Si et al., 2019). Ubiquitin-mediated protein degradation systems are also involved in apoptosis (Zhang et al., 2004; Hoeller and Dikic, 2009; Yang et al., 2009), as some E3 ubiquitin ligases are involved in ubiquitination of apoptosis-related proteins (Hoeller and Dikic, 2009; Yang et al., 2009, 2018; Woo and Kwon, 2019).
Considering the importance of apoptosis in cancer pathophysiology, strategies targeting these apoptosis regulatory mechanisms may contribute to the development of novel ovarian cancer therapies.
Long Non-Coding RNA (lncRNA)
Long non-coding RNAs (lncRNAs) are defined as >200-nt transcripts that do not encode proteins and tens of thousands of lncRNA transcripts are identified throughout the human genome, the majority with unknown function. However, functional studies of some lncRNAs have revealed that they have a wide range of functions. For example, lncRNAs regulate transcription and chromatin remodeling by modulating the recruitment of transcription factors and PRC to specific genomic loci. Furthermore, lncRNAs are involved in gene regulation at post-transcriptional levels through interacting with mRNAs, miRNAs, and proteins (Marchese et al., 2017). Intriguingly, lncRNAs play important roles in pathophysiology of various cancers (Takayama and Inoue, 2016; Misawa et al., 2017; Arun et al., 2018; Mitobe et al., 2018; Kamada et al., 2020; Takeiwa et al., 2020). Particularly, several lncRNAs have been suggested to regulate the apoptosis of ovarian cancer cells (Figure 1 and Table 1). In the following sections, we will describe some apoptosis-related lncRNAs in ovarian cancer cells according to their mechanisms.
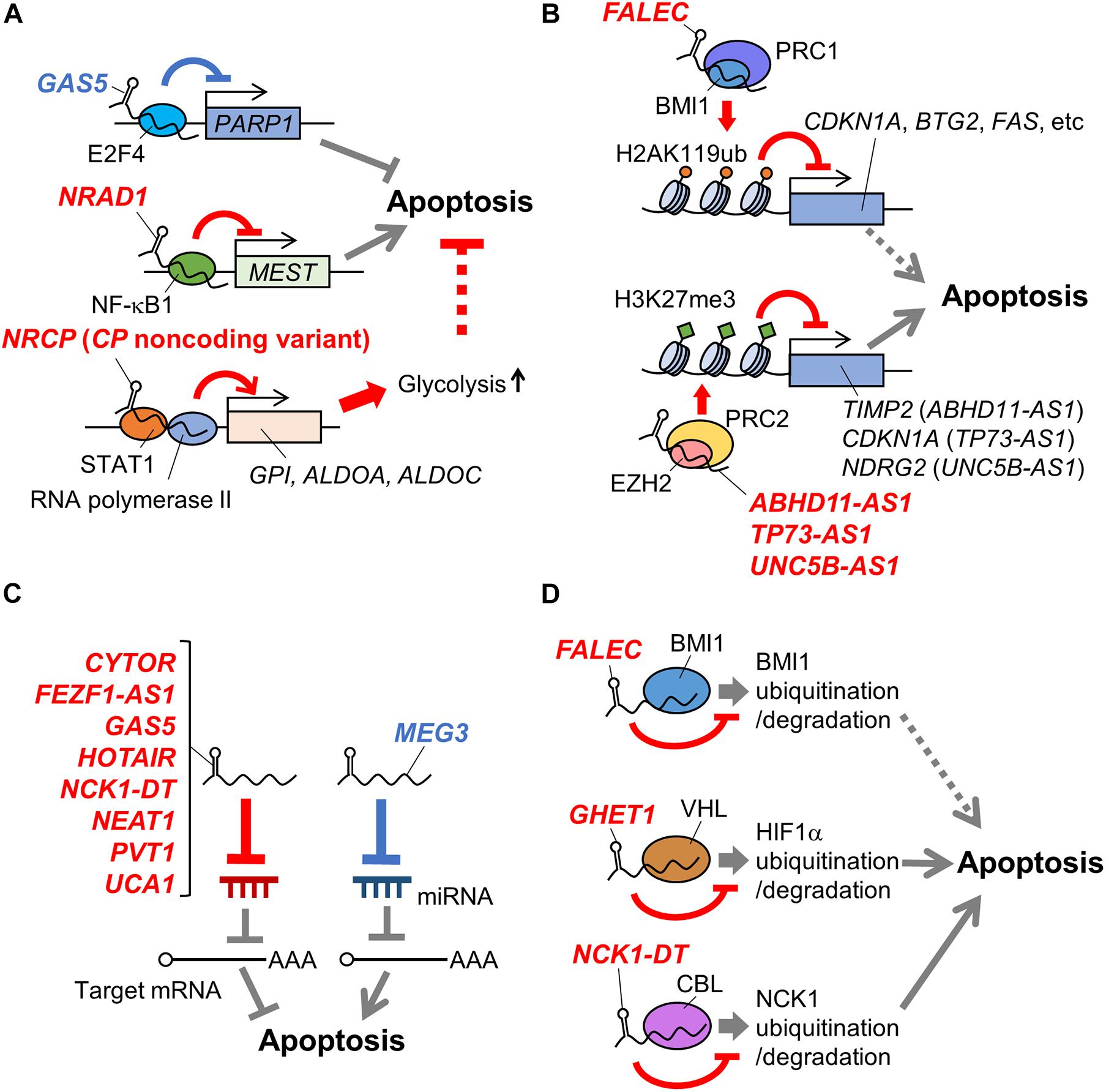
Figure 1. Schematic representation of mechanisms of apoptosis-related lncRNAs in ovarian cancer. LncRNAs involved in apoptosis of ovarian cancer cells via regulating transcription factors (A), histone modification complexes (B), miRNAs (C), and protein stability (D) are shown. Names of lncRNAs promoting and suppressing apoptosis are shown in blue and red, respectively. In addition, apoptosis-promotive or suppressive functions of lncRNAs are shown in blue or red lines, respectively. Potential apoptosis-associated biological pathways are shown in dotted lines. ABHD11-AS1, abhydrolase domain containing 11 antisense RNA 1; ALDOA, aldolase, fructose-bisphosphate A; ALDOC, aldolase, fructose-bisphosphate C; BMI1, B lymphoma Mo-MLV insertion region 1 homolog; BTG2, B-cell translocation gene 2; CBL, casitas B-lineage lymphoma: CDKN1A, cyclin dependent kinase inhibitor 1A; CP, ceruloplasmin; CYTOR, cytoskeleton regulator RNA; EZH2, enhancer of zeste homolog 2; FALEC, focally amplified long non-coding RNA in epithelial cancer; FEZF1-AS1, fasciculation and elongation protein zeta family zinc finger 1 antisense RNA 1; GAS5, growth arrest-specific 5; GHET1, gastric carcinoma high expressed transcript 1; GPI, glucose-6-phosphate isomerase; H2AK119ub, the ubiquitination at the 119th lysine residue of the histone H2A; H3K27me3, the tri-methylation at the 27th lysine residue of the histone H3; HIF1α, hypoxia-inducible factor 1α; HOTAIR, HOX transcript antisense RNA; MEG3, maternally expressed gene 3; MEST, mesoderm specific transcript; NCK1, non-catalytic region of tyrosine kinase adaptor protein 1; NCK1-DT, NCK1 divergent transcript; NDRG2, n-myc downstream-regulated gene 2; NEAT1, nuclear enriched abundant transcript 1; NF-κB1, nuclear factor kappa B subunit 1; NRAD1, non-coding RNA in the aldehyde dehydrogenase 1A pathway; NRCP, lncRNA ceruloplasmin; PARP1, poly(ADP-ribose) polymerase 1; PVT1, plasmacytoma variant translocation 1; STAT1, signal transducer and activator of transcription 1; TIMP2, tissue inhibitor of metalloproteinase 2; TP73-AS1, tumor protein p73 antisense RNA 1; UCA1, urothelial carcinoma associated 1; UNC5B-AS1, uncoordinated 5 netrin receptor B antisense RNA 1.
Apoptosis-Related LncRNAs Regulating Transcription Factors
In this section, we will introduce an apoptosis-promotive lncRNA growth arrest-specific 5 (GAS5) and apoptosis-suppressive lncRNAs non-coding RNA in the aldehyde dehydrogenase 1A pathway (NRAD1)/long intergenic non-coding RNA 00284 (LINC00284) and a non-coding variant of ceruloplasmin (CP) (lncRNA ceruloplasmin; NRCP).
GAS5
Growth arrest-specific 5 is downregulated in ovarian cancer, with this low expression associated with shorter disease-free period and lower overall survival rate of ovarian cancer patients (Gao et al., 2015; Li et al., 2016; Zhao et al., 2018; Long et al., 2019). GAS5 overexpression promotes apoptosis of ovarian cancer cells such as A2780, HEY, OVCAR3, and SKOV3, and increases the sensitivity of HEY and SKOV3 cells to the anticancer agent cisplatin (Gao et al., 2015; Li et al., 2016; Zhao et al., 2018; Long et al., 2019). A functional study has shown that GAS5 recruits the E2F4 transcription factor to the poly(ADP-ribose) polymerase 1 (PARP1) promoter, repressing PARP1 transcription in HEY and SKOV3 cells (Long et al., 2019; Figure 1A). GAS5-mediated PARP1 repression might contribute to apoptosis by downregulating the mitogen-activated protein kinase (MAPK) pathway, but direct evidence will be required in the future study.
NRAD1/LINC00284
Non-coding RNA in the aldehyde dehydrogenase 1A pathway/long intergenic non-coding RNA 00284 is highly expressed in ovarian cancer. NRAD1 overexpression and knockdown experiments have shown that it suppresses the apoptosis of HO8910 and OVCAR3 cells. Functional analyses using HO8910 cells have suggested that NRAD1 binds to NF-κB subunit 1 (NF-κB1) transcription factor and induces NF-κB1–mediated transcriptional repression of mesoderm specific transcript (MEST), contributing to the suppression of apoptosis (Ruan and Zhao, 2019; Figure 1A). However, since a previous study has reported that HO8910 is a cross-contaminated cell line, this mechanism is needed to be verified using other ovarian cancer models (Ye et al., 2015).
CP Non-coding Variant (NRCP)
NRCP is a non-coding splice variant of the ceruloplasmin-coding gene that is upregulated in ovarian cancer (Rupaimoole et al., 2015). High NRCP expression levels correlate with shorter overall survival in patients with ovarian cancer, while NRCP knockdown induces apoptosis in A2780 and SKOV3 cells (Rupaimoole et al., 2015). NRCP binds to RNA polymerase II and STAT1 transcription factor, and promotes glycolysis in A2780 and SKOV3 cells by upregulating glycolysis pathway genes such as glucose-6-phosphate isomerase (GPI), aldolase, fructose-bisphosphate A (ALDOA), and aldolase, fructose-bisphosphate C (ALDOC) via STAT1 (Rupaimoole et al., 2015; Figure 1A). These results suggest a possibility that NRCP may modulate apoptosis by regulating cancer metabolism. NRCP is not annotated in National Center for Biotechnology Information (NCBI) Reference Sequence (RefSeq) database (on Feb 3rd, 2021) and requires further characterization of sequences and expression profiles.
Apoptosis-Related LncRNAs Regulating Histone Modification Complexes
In this section, we will describe the following apoptosis-suppressive lncRNAs: abhydrolase domain containing 11 antisense RNA 1 (ABHD11-AS1), focally amplified long non-coding RNA in epithelial cancer (FALEC)/focally amplified lncRNA on chromosome 1 (FAL1), tumor protein p73 antisense RNA 1 (TP73-AS1), and uncoordinated 5 netrin receptor B antisense RNA 1 (UNC5B-AS1).
ABHD11-AS1
Abhydrolase domain containing 11 antisense RNA 1 is upregulated in ovarian cancer (Wu et al., 2017; Zeng et al., 2019). A functional study has shown that ABHD11-AS1 modulates the expression of ras homolog family member C (RhoC) by an unknown mechanism, suppressing apoptosis in A2780 and OVCAR3 cells (Wu et al., 2017). Another functional study has shown that ABHD11-AS1 binds to enhancer of zeste homolog 2 (EZH2), a component of PRC2. ABHD11-AS1 facilitates tri-methylation at the 27th lysine residue of the histone H3 protein (H3K27me3) on the tissue inhibitor of metalloproteinase 2 (TIMP2) promoter, as mediated by PRC2, and likewise suppresses TIMP2 expression in HO8910 cells and OVCA429 ovarian cancer cells (Figure 1B). TIMP2 suppression mediated by ABHD11-AS1 promotes the proliferation of OVCA429 cells, suggesting that ABHD11-AS1 may also modulate apoptosis by this mechanism (Zeng et al., 2019).
FALEC/FAL1
Focally amplified lncRNA in epithelial cancer/focally amplified lncRNA on chromosome 1 was initially identified as an lncRNA whose gene copy number increased in multiple types of cancers, including ovarian cancer (Hu et al., 2014). Its high expression level and gain in genomic copy number correlate with a shorter overall survival rate of late-stage ovarian cancer patients (Hu et al., 2014). A functional study using A2780 cells has suggested that FALEC binds to a component of PRC1, B lymphoma Mo-MLV insertion region 1 homolog (BMI1) protein, and recruits PRC1 to the promoters of genes such as cyclin dependent kinase inhibitor 1A (CDKN1A), B-cell translocation gene 2 (BTG2), and FAS. Subsequently, PRC1 mediates the ubiquitination at the 119th lysine residue of the histone H2A (H2AK119ub) on these promoter regions and the suppression of these genes (Figure 1B). The FALEC/PRC1 complex target genes such as CDKN1A, BTG2, and FAS are suggested to be involved in apoptosis regulation (El-Deiry, 2001; Mao et al., 2015). Thus, FALEC can be a regulator of ovarian cancer apoptosis.
TP73-AS1
Tumor protein p73 antisense RNA 1 is upregulated in EOC and associated with poor prognosis in EOC patients (Li Y. et al., 2019). A recent study has shown that TP73-AS1 knockdown induces apoptosis of SKOV3 cells, suppressing the proliferation in in vitro culture and the xenograft tumor formation in athymic mice. In contrast, TP73-AS1 overexpression suppresses apoptosis in CAOV3 ovarian cancer cells. Functional analyses have suggested that TP73-AS1 epigenetically suppresses CDKN1A expression by recruiting PRC2 to its promoter (Figure 1B) and modulates apoptosis of SKOV3 cells through this mechanism (Li Y. et al., 2019).
UNC5B-AS1
Uncoordinated 5 netrin receptor B antisense RNA 1 is highly expressed in ovarian cancer, and a recent study has shown that its knockdown activates caspase 3 in ES2 and SKOV3 cells, suggesting the apoptosis-suppressive role of UNC5B-AS1 (Wang et al., 2020). Moreover, the same study has suggested that UNC5B-AS1 promotes PRC2 to repress the n-myc downstream-regulated gene 2 (NDRG2) expression epigenetically (Figure 1B), which may suppress ovarian cancer cell apoptosis (Wang et al., 2020). This study is limited in the elucidation of the mechanism by which UNC5B-AS1 regulates PRC2 and its in vivo function, and further functional analyses are required.
Apoptosis-Related LncRNAs Regulating miRNAs
In the section, we will introduce an apoptosis-promotive lncRNA GAS5 and the following apoptosis-suppressive lncRNAs: fasciculation and elongation protein zeta family zinc finger 1 antisense RNA 1 (FEZF1-AS1), HOX transcript antisense RNA (HOTAIR), non-catalytic region of tyrosine kinase adaptor protein 1 (NCK1) divergent transcript (NCK1-DT)/NCK1 antisense RNA 1 (NCK1-AS1), nuclear enriched abundant transcript 1 (NEAT1), and urothelial carcinoma associated 1 (UCA1).
FEZF1-AS1
High levels of FEZF1-AS1 are detected in tumor tissues and the serum of EOC patients, with its high expression associated with shorter overall survival of EOC patients (Sun et al., 2020). Moreover, its knockdown promotes apoptosis in COC1 and SKOV3 ovarian cancer cells, suggesting the apoptosis-suppressive role of FEZF1-AS1. In vitro analyses of FEZF1-AS1 have shown that it functions as a competing endogenous RNA (ceRNA) for miR-130a-5p, or sponges miR-130a-5p (Figure 1C). Consequently, FEZF1-AS1 upregulates the expression of a miR-130a-5p target gene, sex-determining region Y (SRY)-box transcription factor 4 (SOX4), that promotes proliferation of COC1 and SKOV3 cells and may contribute to apoptosis suppression (Sun et al., 2020). Further analysis of FEZF1-AS1 function, especially in vivo, will clarify its role and significance in apoptosis of ovarian cancer cells.
GAS5
A recent functional study has suggested that GAS5 functions as a ceRNA for miR-196a-5p to upregulate homeobox A5 (HOXA5), promoting apoptosis of primary tumor cells from high-grade serous ovarian cancer tissues as well as A2780 and OVCAR3 cells (Zhao et al., 2018; Figure 1C).
HOTAIR
HOX transcript antisense RNA is upregulated in ovarian cancer, and the elevated expression level correlates with the shorter overall survival of ovarian cancer patients (Qiu et al., 2015; Wang Y. et al., 2015; Zhang et al., 2016; Luo et al., 2017; Yu et al., 2018). HOTAIR knockdown induces apoptosis in ovarian cancer cells such as A2780, HeyC2, and OVCA429, and decreases the cisplatin sensitivity of A2780 and SKOV3 cells (Qiu et al., 2015; Wang Y. et al., 2015; Zhang et al., 2016, 2020; Yu et al., 2018). A recent functional study using A2780 and SKOV3 cells has suggested that HOTAIR acts as a ceRNA for miR-138-5p, leading to cisplatin resistance of these cells (Zhang et al., 2020; Figure 1C). This study has shown that HOTAIR/miR-138-5p axis modulates EZH2 and sirtuin 1 (SIRT1) expression, but its biological significance has not been elucidated.
NCK1-DT/NCK1-AS1
Non-catalytic region of tyrosine kinase adaptor protein 1 divergent transcript is highly expressed in ovarian cancer. Mechanistically, it acts as a ceRNA for miR-137 to upregulate NCK1, which suppresses apoptosis of CAOV3 and SKOV3 cells and enhances their cisplatin resistance (Chang et al., 2020; Figure 1C).
NEAT1
Nuclear enriched abundant transcript 1 is upregulated in ovarian cancer and is associated with shorter overall survival of ovarian cancer patients (Chen et al., 2016). NEAT1 acts as a ceRNA for miR-34a-5p to upregulate BCL2 and suppresses apoptosis of OVCAR3 and SKOV3 cells (Ding et al., 2017). In addition, NEAT1 acts as a ceRNA for miR-4500, to upregulate basic leucine zipper and W2 domain-containing protein 1 (BZW1) that suppresses apoptosis of CAOV3 and ES2 cells (Xu et al., 2020), and miR-194 to upregulate the transcription factor zinc finger E-box binding homeobox 1 (ZEB1), promoting resistance to the anticancer agent paclitaxel (PTX) in HeyA8 and SKOV3 cells (An et al., 2017). Furthermore, NEAT1 sponges miR-770-5p, to upregulate PARP1 and increase cisplatin resistance in A2780 cells in vivo (Zhu et al., 2020; Figure 1C).
UCA1
The lncRNA UCA1 is upregulated in ovarian cancer and is detected in exosomes derived from the serum of ovarian cancer patients (Li Z. et al., 2019; Li et al., 2020). Functional studies have shown that UCA1 acts as a ceRNA for miR-129 and miR-654-5p to upregulate ATP binding cassette subfamily B member 1 (ABCB1) and SALT INDUCIBLE KINASE 2 (SIK2), respectively, which contribute to the suppression of apoptosis and the enhancement of PTX resistance in HeyA8 and SKOV3 cells (Wang et al., 2018; Li et al., 2020). In addition, UCA1 functions as a ceRNA for miR-143 to increase Fos-related antigen 2 (FOSL2), and enhances cisplatin resistance in A2780 and SKOV3 cells (Li Z. et al., 2019; Figure 1C). However, the importance of the function of UCA1 as a ceRNA in vivo has not been fully analyzed.
Recent studies have found that many other lncRNAs modulate ovarian cancer apoptosis through regulating miRNAs. For example, CYTOR/LINC00152 acts as a ceRNA of miR-125b to upregulate an antiapoptotic protein MCL1 in A2780 and SKOV3 cells (Chen et al., 2018). PVT1 suppresses apoptosis in OVACAR3 and TOV112D cells by inhibiting miR-543 and increasing a miR-543 target SERPIN1 (Qu et al., 2020). In contrast, MEG3 promotes apoptosis in OVCAR8 and SKOV3 cells by sponging miR-205-5p (Tao et al., 2020). The detail of lncRNAs regulating miRNAs is also reviewed in other articles (Braga et al., 2020; Salamini-Montemurri et al., 2020).
Apoptosis-Related LncRNAs Regulating Protein Stability
In the section, we will introduce the following apoptosis-suppressive lncRNAs: FALEC/FAL1, gastric carcinoma high expressed transcript 1 (GHET1), and NCK1-DT/NCK1-AS1.
FALEC/FAL1
As described above, FALEC binds to BMI1 and modulates PRC1 function in A2780 cells. In addition, FALEC stabilizes BMI1 by suppressing ubiquitin-mediated BMI1 protein degradation (Hu et al., 2014; Figure 1D).
GHET1
The lncRNA GHET1 is upregulated in ovarian cancer and higher expression correlates with increased tumor size and distant metastasis (Liu and Li, 2019). Conversely, its knockdown induces apoptosis and downregulates glycolysis in A2780 and SKOV3 cells, where GHET1 binds to an E3 ubiquitin ligase, von Hippel–Lindau tumor suppressor (VHL), and prevents VHL-mediated degradation of hypoxia-inducible factor 1α (HIF1α) (Figure 1D). Since the GHET1 function in ovarian cancer cells has been only examined by in vitro assays, in vivo analyses of GHET1 are needed. Although the role of the GHET1/VHL/HIF1α axis in apoptosis has not yet been elucidated, HIF1α and cancer metabolism have been shown to play important roles in apoptosis regulation, suggesting the possibility that this axis may also be involved in the phenomenon (Zhou et al., 2006; Matsuura et al., 2016).
NCK1-DT/NCK1-AS1
In addition to the function as a ceRNA, NCK1-AS1 increases the stability of NCK1: NCK1-AS1 binds to an E3 ubiquitin ligase, casitas B-lineage lymphoma (CBL), and prevents CBL-mediated degradation of NCK1 (Chang et al., 2020; Figure 1D). The functions of NCK-AS1 in ovarian cancer have been suggested based on in vitro experiments, and thus needs to be evaluated using ovarian tumor specimens or in vivo ovarian cancer models.
Conclusion
In this review, we introduced the mechanisms of apoptosis-related lncRNAs in ovarian cancer cells. Considering that dysregulation of apoptosis is involved in the resistance to ovarian cancer therapies, small molecule inhibitors/siRNAs targeting apoptosis-suppressing lncRNAs, or apoptosis-promoting lncRNAs themselves may be applicable to ovarian cancer therapies. For nucleic acid-based therapeutics, it is important to develop the drug delivery systems (DDSs) with high target specificity and less non-specific toxicity in vivo. Particularly, for ovarian cancer, DDSs will be useful to treat metastatic cancer cells in peritoneal cavity (Amreddy et al., 2018; van den Brand et al., 2018). Moreover, apoptosis-related lncRNAs may be potential diagnostic and prognostic biomarkers. Especially, FEZF1-AS1 and UCA1 are detected in serum and exosomes recovered from serum of ovarian cancer patients, respectively, which suggested their potential as liquid biopsy markers for ovarian cancer.
Apoptosis-related lncRNAs have basically been studied using conventional ovarian cancer cell lines, and the functions of some lncRNAs have been examined by in vitro assays alone. For clinical application, it is required to elucidate the lncRNA functions in vivo. Moreover, previous studies have indicated some discrepancies between ovarian cancer cell lines and the original tumor clinical tissues in terms of genomic and histological features and gene expression profiles (Domcke et al., 2013; Beaufort et al., 2014). Thus, lncRNA studies using ovarian tumor specimens or other ovarian cancer models are strongly demanded. Three-dimensional cultures of patient-derived cancer cells (PDCs) and cancer models established by transplanting tumor specimens into host mice (patient-derived xenograft [PDX] models) retain the properties of original tumors and have attracted attention as promising models for cancer research and drug screening (Ishiguro et al., 2016; Maru and Hippo, 2019; Namekawa et al., 2019; Shiba et al., 2019). Further studies using PDC and PDX models would advance the application of apoptosis-related lncRNAs to ovarian cancer diagnosis, prognosis, and therapies.
Author Contributions
All authors contributed to the conception and provided the data and design. TT contributed to manuscript writing. KI, KH-I, and SI contributed to the conception and final approval of the manuscript.
Funding
This work was supported by grants from the Japan Society for the Promotion of Science (20K21667 to SI, 17H04205 and 20H03734 to KH-I, and 20K21636 to KI) and Takeda Science Foundation to SI and KI.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Amreddy, N., Babu, A., Muralidharan, R., Panneerselvam, J., Srivastava, A., Ahmed, R., et al. (2018). Recent advances in nanoparticle-based cancer drug and gene delivery. Adv. Cancer Res. 137, 115–170. doi: 10.1016/bs.acr.2017.11.003
An, J., Lv, W., and Zhang, Y. (2017). LncRNA NEAT1 contributes to paclitaxel resistance of ovarian cancer cells by regulating ZEB1 expression via miR-194. Onco Targets Ther. 10, 5377–5390. doi: 10.2147/OTT.S147586
Arun, G., Diermeier, S. D., and Spector, D. L. (2018). Therapeutic targeting of long non-coding RNAs in cancer. Trends Mol. Med. 24, 257–277. doi: 10.1016/j.molmed.2018.01.001
Beaufort, C. M., Helmijr, J. C., Piskorz, A. M., Hoogstraat, M., Ruigrok-Ritstier, K., Besselink, N., et al. (2014). Ovarian cancer cell line panel (OCCP): clinical importance of in vitro morphological subtypes. PLoS One 9:e103988. doi: 10.1371/journal.pone.0103988
Binju, M., Amaya-Padilla, M. A., Wan, G., Gunosewoyo, H., Suryo Rahmanto, Y., and Yu, Y. (2019). Therapeutic inducers of apoptosis in ovarian cancer. Cancers (Basel) 11:1786. doi: 10.3390/cancers11111786
Bours, V., Bentires-Alj, M., Hellin, A. C., Viatour, P., Robe, P., Delhalle, S., et al. (2000). Nuclear factor-kappa B, cancer, and apoptosis. Biochem. Pharmacol. 60, 1085–1089. doi: 10.1016/s0006-2952(00)00391-9
Braga, E. A., Fridman, M. V., Moscovtsev, A. A., Filippova, E. A., Dmitriev, A. A., and Kushlinskii, N. E. (2020). LncRNAs in ovarian cancer progression, metastasis, and main pathways: ceRNA and alternative mechanisms. Int. J. Mol. Sci. 21:8855. doi: 10.3390/ijms21228855
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A., and Jemal, A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424. doi: 10.3322/caac.21492
Cao, L., Bombard, J., Cintron, K., Sheedy, J., Weetall, M. L., and Davis, T. W. (2011). BMI1 as a novel target for drug discovery in cancer. J. Cell. Biochem. 112, 2729–2741. doi: 10.1002/jcb.23234
Chang, H., Li, B., Zhang, X., and Meng, X. (2020). NCK1-AS1 promotes NCK1 expression to facilitate tumorigenesis and chemo-resistance in ovarian cancer. Biochem. Biophys. Res. Commun. 522, 292–299. doi: 10.1016/j.bbrc.2019.11.014
Chen, P., Fang, X., Xia, B., Zhao, Y., Li, Q., and Wu, X. (2018). Long noncoding RNA LINC00152 promotes cell proliferation through competitively binding endogenous miR-125b with MCL-1 by regulating mitochondrial apoptosis pathways in ovarian cancer. Cancer Med. 7, 4530–4541. doi: 10.1002/cam4.1547
Chen, Z. J., Zhang, Z., Xie, B. B., and Zhang, H. Y. (2016). Clinical significance of up-regulated lncRNA NEAT1 in prognosis of ovarian cancer. Eur. Rev. Med. Pharmacol. Sci. 20, 3373–3377.
Christofides, A., Karantanos, T., Bardhan, K., and Boussiotis, V. A. (2016). Epigenetic regulation of cancer biology and anti-tumor immunity by EZH2. Oncotarget 7, 85624–85640. doi: 10.18632/oncotarget.12928
Crosby, M. E., and Almasan, A. (2004). Opposing roles of E2Fs in cell proliferation and death. Cancer Biol. Ther. 3, 1208–1211. doi: 10.4161/cbt.3.12.1494
Di Leva, G., Garofalo, M., and Croce, C. M. (2014). MicroRNAs in cancer. Annu. Rev. Pathol. 9, 287–314. doi: 10.1146/annurev-pathol-012513-104715
Ding, N., Wu, H., Tao, T., and Peng, E. (2017). NEAT1 regulates cell proliferation and apoptosis of ovarian cancer by miR-34a-5p/BCL2. Onco Targets Ther. 10, 4905–4915. doi: 10.2147/OTT.S142446
Domcke, S., Sinha, R., Levine, D. A., Sander, C., and Schultz, N. (2013). Evaluating cell lines as tumour models by comparison of genomic profiles. Nat. Commun. 4:2126. doi: 10.1038/ncomms3126
El-Deiry, W. S. (2001). Insights into cancer therapeutic design based on p53 and TRAIL receptor signaling. Cell Death Differ. 8, 1066–1075. doi: 10.1038/sj.cdd.4400943
Gao, J., Liu, M., Zou, Y., Mao, M., Shen, T., Zhang, C., et al. (2015). Long non-coding RNA growth arrest-specific transcript 5 is involved in ovarian cancer cell apoptosis through the mitochondria-mediated apoptosis pathway. Oncol. Rep. 34, 3212–3221. doi: 10.3892/or.2015.4318
Hiller-Sturmhöfel, S., and Bartke, A. (1998). The endocrine system: an overview. Alcohol Health Res. World 22, 153–164.
Hoeller, D., and Dikic, I. (2009). Targeting the ubiquitin system in cancer therapy. Nature 458, 438–444. doi: 10.1038/nature07960
Hu, X., Feng, Y., Zhang, D., Zhao, S. D., Hu, Z., Greshock, J., et al. (2014). A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer Cell 26, 344–357. doi: 10.1016/j.ccr.2014.07.009
Ishiguro, T., Sato, A., Ohata, H., Ikarashi, Y., Takahashi, R. U., Ochiya, T., et al. (2016). Establishment and characterization of an in vitro model of ovarian cancer stem-like cells with an enhanced proliferative capacity. Cancer Res. 76, 150–160. doi: 10.1158/0008-5472.CAN-15-0361
Kamada, S., Takeiwa, T., Ikeda, K., Horie-Inoue, K., and Inoue, S. (2020). Long non-coding RNAs involved in metabolic alterations in breast and prostate cancers. Front. Oncol. 10:593200. doi: 10.3389/fonc.2020.593200
Karin, M. (2006). Nuclear factor-κB in cancer development and progression. Nature 441, 431–436. doi: 10.1038/nature04870
Kent, L. N., and Leone, G. (2019). The broken cycle: E2F dysfunction in cancer. Nat. Rev. Cancer 19, 326–338. doi: 10.1038/s41568-019-0143-7
Kim, H. S., and Lee, M. S. (2007). STAT1 as a key modulator of cell death. Cell. Signal. 19, 454–465. doi: 10.1016/j.cellsig.2006.09.003
Li, J., Huang, H., Li, Y., Li, L., Hou, W., and You, Z. (2016). Decreased expression of long non-coding RNA GAS5 promotes cell proliferation, migration and invasion, and indicates a poor prognosis in ovarian cancer. Oncol. Rep. 36, 3241–3250. doi: 10.3892/or.2016.5200
Li, Y., Jiao, Y., Hao, J., Xing, H., and Li, C. (2019). Long noncoding RNA TP73-AS1 accelerates the epithelial ovarian cancer via epigenetically repressing p21. Am. J. Transl. Res. 11, 2447–2454.
Li, Z., Niu, H., Qin, Q., Yang, S., Wang, Q., Yu, C., et al. (2019). lncRNA UCA1 mediates resistance to cisplatin by regulating the mir-143/FOSL2-signaling pathway in ovarian cancer. Mol. Ther. Nucleic Acids 17, 92–101. doi: 10.1016/j.omtn.2019.05.007
Li, Z. Y., Wang, X. L., Dang, Y., Zhu, X. Z., Zhang, Y. H., Cai, B. X., et al. (2020). Long non-coding RNA UCA1 promotes the progression of paclitaxel resistance in ovarian cancer by regulating the miR-654-5p/SIK2 axis. Eur. Rev. Med. Pharmacol. Sci. 24, 591–603. doi: 10.26355/eurrev_202001_20035
Liu, D., and Li, H. (2019). Long non-coding RNA GEHT1 promoted the proliferation of ovarian cancer cells via modulating the protein stability of HIF1α. Biosci. Rep. 39:BSR20181650. doi: 10.1042/BSR20181650
Long, X., Song, K., Hu, H., Tian, Q., Wang, W., Dong, Q., et al. (2019). Long non-coding RNA GAS5 inhibits DDP-resistance and tumor progression of epithelial ovarian cancer via GAS5-E2F4-PARP1-MAPK axis. J. Exp. Clin. Cancer Res. 38:345. doi: 10.1186/s13046-019-1329-2
Luo, P., Liu, X. F., Wang, Y. C., Li, N. D., Liao, S. J., Yu, M. X., et al. (2017). Prognostic value of abnormally expressed lncRNAs in ovarian carcinoma: a systematic review and meta-analysis. Oncotarget 8, 23927–23936. doi: 10.18632/oncotarget.14760
Mao, B., Zhang, Z., and Wang, G. (2015). BTG2: a rising star of tumor suppressors (review). Int. J. Oncol. 46, 459–464. doi: 10.3892/ijo.2014.2765
Marchese, F. P., Raimondi, I., and Huarte, M. (2017). The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 18:206. doi: 10.1186/s13059-017-1348-2
Maru, Y., and Hippo, Y. (2019). Current status of patient-derived ovarian cancer models. Cells 8:505. doi: 10.3390/cells8050505
Matsuura, K., Canfield, K., Feng, W., and Kurokawa, M. (2016). Metabolic regulation of apoptosis in cancer. Int. Rev. Cell Mol. Biol. 327, 43–87. doi: 10.1016/bs.ircmb.2016.06.006
Misawa, A., Takayama, K. I., and Inoue, S. (2017). Long non-coding RNAs and prostate cancer. Cancer Sci. 108, 2107–2114. doi: 10.1111/cas.13352
Mitobe, Y., Takayama, K. I., Horie-Inoue, K., and Inoue, S. (2018). Prostate cancer-associated lncRNAs. Cancer Lett. 418, 159–166. doi: 10.1016/j.canlet.2018.01.012
Momenimovahed, Z., Tiznobaik, A., Taheri, S., and Salehiniya, H. (2019). Ovarian cancer in the world: epidemiology and risk factors. Int. J. Womens Health 11, 287–299. doi: 10.2147/IJWH.S197604
Motta, P. M., Makabe, S., and Nottola, S. A. (1997). The ultrastructure of human reproduction. I. The natural history of the female germ cell: origin, migration and differentiation inside the developing ovary. Hum. Reprod. Update 3, 281–295. doi: 10.1093/humupd/3.3.281
Namekawa, T., Ikeda, K., Horie-Inoue, K., and Inoue, S. (2019). Application of prostate cancer models for preclinical study: advantages and limitations of cell lines, patient-derived xenografts, and three-dimensional culture of patient-derived cells. Cells 8:74. doi: 10.3390/cells8010074
Pistritto, G., Trisciuoglio, D., Ceci, C., Garufi, A., and D’Orazi, G. (2016). Apoptosis as anticancer mechanism: function and dysfunction of its modulators and targeted therapeutic strategies. Aging (Albany NY) 8, 603–619. doi: 10.18632/aging.100934
Qiu, J. J., Wang, Y., Ding, J. X., Jin, H. Y., Yang, G., and Hua, K. Q. (2015). The long non-coding RNA HOTAIR promotes the proliferation of serous ovarian cancer cells through the regulation of cell cycle arrest and apoptosis. Exp. Cell Res. 333, 238–248. doi: 10.1016/j.yexcr.2015.03.005
Qu, C., Dai, C., Guo, Y., Qin, R., and Liu, J. (2020). Long non-coding RNA PVT1-mediated miR-543/SERPINI1 axis plays a key role in the regulatory mechanism of ovarian cancer. Biosci. Rep. 40:BSR20200800. doi: 10.1042/BSR20200800
Rojas, V., Hirshfield, K. M., Ganesan, S., and Rodriguez-Rodriguez, L. (2016). Molecular characterization of epithelial ovarian cancer: implications for diagnosis and treatment. Int. J. Mol. Sci. 17:2113. doi: 10.3390/ijms17122113
Ruan, Z., and Zhao, D. (2019). Long intergenic noncoding RNA LINC00284 knockdown reduces angiogenesis in ovarian cancer cells. FASEB J. 33, 12047–12059. doi: 10.1096/fj.201900101RR
Rupaimoole, R., Lee, J., Haemmerle, M., Ling, H., Previs, R. A., Pradeep, S., et al. (2015). Long noncoding RNA ceruloplasmin promotes cancer growth by altering glycolysis. Cell Rep. 13, 2395–2402. doi: 10.1016/j.celrep.2015.11.047
Salamini-Montemurri, M., Lamas-Maceiras, M., Barreiro-Alonso, A., Vizoso-Vázquez, Á, Rodríguez-Belmonte, E., Quindós-Varela, M., et al. (2020). The challenges and opportunities of lncRNAs in ovarian cancer research and clinical use. Cancers (Basel) 12:1020. doi: 10.3390/cancers12041020
Shiba, S., Ikeda, K., Suzuki, T., Shintani, D., Okamoto, K., Horie-Inoue, K., et al. (2019). Hormonal regulation of patient-derived endometrial cancer stem-like cells generated by three-dimensional culture. Endocrinology 160, 1895–1906. doi: 10.1210/en.2019-00362
Si, W., Shen, J., Zheng, H., and Fan, W. (2019). The role and mechanisms of action of microRNAs in cancer drug resistance. Clin. Epigenetics 11:25. doi: 10.1186/s13148-018-0587-8
Siegel, R. L., Miller, K. D., and Jemal, A. (2018). Cancer statistics, 2018. CA Cancer J. Clin. 68, 7–30. doi: 10.3322/caac.21442
Sun, Z., Gao, S., Xuan, L., and Liu, X. (2020). Long non-coding RNA FEZF1-AS1 induced progression of ovarian cancer via regulating miR-130a-5p/SOX4 axis. J. Cell. Mol. Med. 24, 4275–4285. doi: 10.1111/jcmm.15088
Takayama, K., and Inoue, S. (2016). The emerging role of noncoding RNA in prostate cancer progression and its implication on diagnosis and treatment. Brief. Funct. Genomics 15, 257–265. doi: 10.1093/bfgp/elv057
Takeiwa, T., Ikeda, K., Mitobe, Y., Horie-Inoue, K., and Inoue, S. (2020). Long noncoding rnas involved in the endocrine therapy resistance of breast cancer. Cancers (Basel) 12:1424. doi: 10.3390/cancers12061424
Tao, P., Yang, B., Zhang, H., Sun, L., Wang, Y., and Zheng, W. (2020). The overexpression of lncRNA MEG3 inhibits cell viability and invasion and promotes apoptosis in ovarian cancer by sponging miR-205-5p. Int. J. Clin. Exp. Pathol. 13, 869–879.
Torre, L. A., Trabert, B., DeSantis, C. E., Miller, K. D., Samimi, G., Runowicz, C. D., et al. (2018). Ovarian cancer statistics, 2018. CA Cancer J. Clin. 68, 284–296. doi: 10.3322/caac.21456
Trinidad, C. V., Tetlow, A. L., Bantis, L. E., and Godwin, A. K. (2020). Reducing ovarian cancer mortality through early detection: approaches using circulating biomarkers. Cancer Prev. Res. (Phila.) 13, 241–252. doi: 10.1158/1940-6207.CAPR-19-0184
van den Brand, D., Mertens, V., Massuger, L. F. A. G., and Brock, R. (2018). siRNA in ovarian cancer – delivery strategies and targets for therapy. J. Control. Release 283, 45–58. doi: 10.1016/j.jconrel.2018.05.012
Vaskivuo, T. E., and Tapanainen, J. S. (2003). Apoptosis in the human ovary. Reprod. Biomed. Online 6, 24–35. doi: 10.1016/s1472-6483(10)62052-4
Verhoeven, Y., Tilborghs, S., Jacobs, J., De Waele, J., Quatannens, D., Deben, C., et al. (2020). The potential and controversy of targeting STAT family members in cancer. Semin. Cancer Biol. 60, 41–56. doi: 10.1016/j.semcancer.2019.10.002
Virant-Klun, I. (2015). Postnatal oogenesis in humans: a review of recent findings. Stem Cells Cloning 8, 49–60. doi: 10.2147/SCCAA.S32650
Wang, H., Su, H., and Tan, Y. (2020). UNC5B-AS1 promoted ovarian cancer progression by regulating the H3K27me on NDRG2 via EZH2. Cell Biol. Int. 44, 1028–1036. doi: 10.1002/cbin.11300
Wang, J., Ye, C., Liu, J., and Hu, Y. (2018). UCA1 confers paclitaxel resistance to ovarian cancer through miR-129/ABCB1 axis. Biochem. Biophys. Res. Commun. 501, 1034–1040. doi: 10.1016/j.bbrc.2018.05.104
Wang, W., Qin, J. J., Voruganti, S., Nag, S., Zhou, J., and Zhang, R. (2015). Polycomb group (PcG) proteins and human cancers: multifaceted functions and therapeutic implications. Med. Res. Rev. 35, 1220–1267. doi: 10.1002/med.21358
Wang, Y., Wang, H., Song, T., Zou, Y., Jiang, J., Fang, L., et al. (2015). HOTAIR is a potential target for the treatment of cisplatin-resistant ovarian cancer. Mol. Med. Rep. 12, 2211–2216. doi: 10.3892/mmr.2015.3562
Wong, R. S. (2011). Apoptosis in cancer: from pathogenesis to treatment. J. Exp. Clin. Cancer Res. 30:87. doi: 10.1186/1756-9966-30-87
Woo, S. M., and Kwon, T. K. (2019). E3 ubiquitin ligases and deubiquitinases as modulators of TRAIL-mediated extrinsic apoptotic signaling pathway. BMB Rep. 52, 119–126. doi: 10.5483/BMBRep.2019.52.2.011
Wu, D. D., Chen, X., Sun, K. X., Wang, L. L., Chen, S., and Zhao, Y. (2017). Role of the lncRNA ABHD11-AS1 in the tumorigenesis and progression of epithelial ovarian cancer through targeted regulation of RhoC. Mol. Cancer 16:138. doi: 10.1186/s12943-017-0709-5
Xu, H., Sun, X., Huang, Y., Si, Q., and Li, M. (2020). Long non-coding RNA NEAT1 modifies cell proliferation, colony formation, apoptosis, migration and invasion via the miR-4500/BZW1 axis in ovarian cancer. Mol. Med. Rep. 22, 3347–3357. doi: 10.3892/mmr.2020.11408
Yadav, P. K., Tiwari, M., Gupta, A., Sharma, A., Prasad, S., Pandey, A. N., et al. (2018). Germ cell depletion from mammalian ovary: possible involvement of apoptosis and autophagy. J. Biomed. Sci. 25:36. doi: 10.1186/s12929-018-0438-0
Yang, L., Chen, J., Huang, X., Zhang, E., He, J., and Cai, Z. (2018). Novel insights into E3 ubiquitin ligase in cancer chemoresistance. Am. J. Med. Sci. 355, 368–376. doi: 10.1016/j.amjms.2017.12.012
Yang, Y., Kitagaki, J., Wang, H., Hou, D. X., and Perantoni, A. O. (2009). Targeting the ubiquitin-proteasome system for cancer therapy. Cancer Sci. 100, 24–28. doi: 10.1111/j.1349-7006.2008.01013.x
Ye, F., Chen, C., Qin, J., Liu, J., and Zheng, C. (2015). Genetic profiling reveals an alarming rate of cross-contamination among human cell lines used in China. FASEB J. 29, 4268–4272. doi: 10.1096/fj.14-266718
Yu, Y., Zhang, X., Tian, H., Zhang, Z., and Tian, Y. (2018). Knockdown of long non-coding RNA HOTAIR increases cisplatin sensitivity in ovarian cancer by inhibiting cisplatin-induced autophagy. J. BUON 23, 1396–1401.
Zeng, X. Y., Jiang, X. Y., Yong, J. H., Xie, H., Yuan, J., Zeng, D., et al. (2019). lncRNA ABHD11-AS1, regulated by the EGFR pathway, contributes to the ovarian cancer tumorigenesis by epigenetically suppressing TIMP2. Cancer Med. 8, 7074–7085. doi: 10.1002/cam4.2586
Zhang, H. G., Wang, J., Yang, X., Hsu, H. C., and Mountz, J. D. (2004). Regulation of apoptosis proteins in cancer cells by ubiquitin. Oncogene 23, 2009–2015. doi: 10.1038/sj.onc.1207373
Zhang, Y., Ai, H., Fan, X., Chen, S., Wang, Y., and Liu, L. (2020). Knockdown of long non-coding RNA HOTAIR reverses cisplatin resistance of ovarian cancer cells through inhibiting miR-138-5p-regulated EZH2 and SIRT1. Biol. Res. 53:18. doi: 10.1186/s40659-020-00286-3
Zhang, Z., Cheng, J., Wu, Y., Qiu, J., Sun, Y., and Tong, X. (2016). LncRNA HOTAIR controls the expression of Rab22a by sponging miR-373 in ovarian cancer. Mol. Med. Rep. 14, 2465–2472. doi: 10.3892/mmr.2016.5572
Zhao, H., Yu, H., Zheng, J., Ning, N., Tang, F., Yang, Y., et al. (2018). Lowly-expressed lncRNA GAS5 facilitates progression of ovarian cancer through targeting miR-196-5p and thereby regulating HOXA5. Gynecol. Oncol. 151, 345–355. doi: 10.1016/j.ygyno.2018.08.032
Zhou, J., Schmid, T., Schnitzer, S., and Brüne, B. (2006). Tumor hypoxia and cancer progression. Cancer Lett. 237, 10–21. doi: 10.1016/j.canlet.2005.05.028
Keywords: ovarian cancer, ovary, apoptosis, long non-coding RNA, transcriptional regulation, histone modification complex, competing endogenous RNA
Citation: Takeiwa T, Ikeda K, Horie-Inoue K and Inoue S (2021) Mechanisms of Apoptosis-Related Long Non-coding RNAs in Ovarian Cancer. Front. Cell Dev. Biol. 9:641963. doi: 10.3389/fcell.2021.641963
Received: 15 December 2020; Accepted: 06 April 2021;
Published: 29 April 2021.
Edited by:
Tang Zhonglin, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, ChinaCopyright © 2021 Takeiwa, Ikeda, Horie-Inoue and Inoue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Satoshi Inoue, c2lub3VlQHRtaWcub3IuanA=
 Toshihiko Takeiwa
Toshihiko Takeiwa Kazuhiro Ikeda
Kazuhiro Ikeda Kuniko Horie-Inoue1
Kuniko Horie-Inoue1 Satoshi Inoue
Satoshi Inoue