- 1Department of Orthopedic Surgery, State Key Laboratory of Complex Severe and Rare Diseases, Peking Union Medical College Hospital, Peking Union Medical College and Chinese Academy of Medical Sciences, Beijing, China
- 2Beijing Key Laboratory for Genetic Research of Skeletal Deformity, Beijing, China
- 3Graduate School of Peking Union Medical College, Beijing, China
- 4Medical Research Center, Peking Union Medical College Hospital, Peking Union Medical College and Chinese Academy of Medical Sciences, Beijing, China
CSF1R encodes the colony-stimulating factor 1 receptor which regulates the proliferation, differentiation, and biological activity of monocyte/macrophage lineages. Pathogenic variants in CSF1R could lead to autosomal dominant adult-onset leukoencephalopathy with axonal spheroids and pigmented glia or autosomal recessive skeletal dysplasia. In this study, we identified three heterozygous deleterious rare variants in CSF1R from a congenital vertebral malformation (CVM) cohort. All of the three variants are located within the carboxy-terminal region of CSF1R protein and could lead to an increased stability of the protein. Therefore, we established a zebrafish model overexpressing CSF1R. The zebrafish model exhibits CVM phenotypes such as hemivertebral and vertebral fusion. Furthermore, overexpression of the mutated CSF1R mRNA depleted of the carboxy-terminus led to a higher proportion of zebrafish with vertebral malformations than wild-type CSF1R mRNA did (p = 0.03452), implicating a gain-of-function effect of the C-terminal variant. In conclusion, variants affecting the C-terminal of CSF1R could cause CVM though a potential gain-of-function mechanism.
Introduction
Colony-stimulating factor 1 receptor (CSF1R) regulates the proliferation, differentiation and biological activity of monocyte/macrophage (Mϕ) lineages (Stanley et al., 1997). CSF1R is also expressed in Paneth cells, renal proximal tubule epithelial cells, and placental trophoblasts, indicating a pleiotropic role of CSF1R protein in embryonic development (Arceci et al., 1989, 1992; Pixley and Stanley, 2004; Huynh et al., 2009; Menke et al., 2009). The CSF1R protein includes an extracellular ligand-binding domain and an intracellular tyrosine kinase domain (PTK domain) (Pixley and Stanley, 2004). The extracellular domain of CSF1R binds to ligands such as CSF1 and IL34, which further induces autophosphorylation and protein dimerization (Wang et al., 2012; Stanley and Chitu, 2014). Downstream molecules interact with the intracellular part of CSF1R and are phosphorylated by the PTK domain. The phosphorylated signal molecules could activate the downstream signaling pathway, promoting the proliferation and differentiation of hematopoietic precursor cells, especially mononuclear phagocytes. Heterozygous deleterious variants in CSF1R have been reported to cause adult-onset leukoencephalopathy with axonal spheroids and pigmented glia (ALSP, MIM: 221820) (Oosterhof et al., 2019), a neurological disease characterized by executive dysfunction, memory decline, personality changes, motor impairments, and seizures (Svensson et al., 2011; Rodríguez-Tornos et al., 2016; Makrythanasis et al., 2018). Notably, most of the reported ALSP-related heterozygous pathogenic variants were located at the intracellular PTK domain (Oosterhof et al., 2019).
More recently, homozygous variants in CSF1R have been linked to brain abnormalities, neurodegeneration, and dysosteosclerosis (MIM: 618476), which is characterized by progressive neurologic deterioration and sclerotic bone dysplasia (Guo et al., 2019; Oosterhof et al., 2019). Csf1r−/− mouse model resembled the skeletal phenotypes of the patients carrying bi-allelic CSF1R variants, implicating the important role of CSF1R in bone development (Erblich et al., 2011). Furthermore, an abnormal vertebral arch compared to normal individuals was observed in the zebrafish model with biallelic loss-of-function mutations in csf1ra and csf1rb, indicating the potential effect of CSF1R variants on vertebral morphology (Oosterhof et al., 2019).
In this study, we analyzed variants in CSF1R in a cohort of congenital vertebral malformation (CVM), which has been partially attributed to genetic defects previously (Wu et al., 2015, 2019; Chen et al., 2016, 2020; Liu et al., 2019; Yang et al., 2019, 2020; Lin et al., 2020; Ren et al., 2020). In vitro and in vivo functional experiments were then performed to explore the effect of these variants on protein expression and vertebral morphology.
Methods
Human Subjects
Five hundred and eighty-three patients diagnosed with CVM were consecutively enrolled and collected in the cohort between 2009 and 2018 at Peking Union Medical College Hospital, as a part of the Deciphering Disorders Involving Scoliosis and COmorbidities (DISCO) study (http://www.discostudy.org/). Detailed phenotypic data was recorded. X-ray, computed tomography (CT), and magnetic resonance imaging (MRI) were also taken. Exome sequencing and bioinformatic analysis were conducted as described previously (Zhao et al., 2021). Variant interpretation was then performed based on the genome Aggregation Database (gnomAD, http://gnomad.broadinstitute.org/). Rare variants in CSF1R were extracted and filtered with the following criteria: (1) truncating (non-sense, frameshift, splice acceptor/donor) and minor allele frequency (MAF) ≤ 0.001 or (2) missense variants absent from public databases in the general population.
Sanger Sequencing
Candidate variants of CSF1R identified in our cohort were validated by Sanger sequencing. The variant-encoding amplicon was amplified by PCR from genomic DNA obtained from patients and purified using an Axygen AP-GX-50 kit (lot no. 05915KE1). Sanger sequencing was then performed on an ABI3730XL instrument.
Plasmid Construction
The pEGFP-C1-based vector was used for the construction of wild-type CSF1R and mutated CSF1R (NM_005211.3: c.2906_2909dupATCA, c.2797G>T, c.2749_2758delGACAGGAGAG) plasmids. All plasmids were verified by DNA sequencing.
Cell Culture, Transfection, and Western Blotting Analysis
Cos-7 cells were maintained in Dulbecco's Modified Eagle's medium (DMEM, Invitrogen) supplemented with fetal bovine serum (Gibco), penicillin (50 U/ml), and streptomycin (50 μg/ml) in six-well plates. The full-length wild-type or mutant CSF1R constructs (plasmid pEGFP-C1 1 μg) was transfected into the cells, respectively. After 48 h, western blotting was used to assess the protein expression level in the cell lysates with the following antibodies: rabbit anti-human CSF1R monoclonal antibody (1:1,000, Abcam, ab229188) and mouse anti-GAPDH monoclonal antibody (1:1,000, ZSJQB Co., Ltd.).
Zebrafish Husbandry and Fertilization
Tg (Ola.Sp7:nlsGFP) transgenic zebrafish, where GFP expression was driven by an Sp7 promoter in osteoblasts, was utilized for animal model establishment. The zebrafish were kept at 28°C and fed with brine shrimp twice a day. From the 2–4-cell stage to 3 days post fertilization (dpf), zebrafish were exposed to methylene blue which inhibited fungal contamination.
In vitro mRNA Transcription
A human wild-type CSF1R DNA sequence was cloned into the PCS2+ plasmid, constructing a human wild-type CSF1R plasmid. Then, an indel variant (c.2749_2758delGACAGGAGAG) was constructed to generate a human-mutated CSF1R plasmid containing a mutated gene sequence. After the linearization of plasmids, transcription was performed with mMESSAGE mMACHINE™ SP6 Ultra Transcription Kit (Ambion) to obtain corresponding mRNA.
In vivo mRNA Overexpression Experiment
Human CSF1R mRNA (50 pg for each), i.e., the wild-type mRNA or the mutated mRNA, was dissolved into distilled water (2 nl for each) and injected into embryos at the 1–2-cell stage, respectively. An equal amount of water was injected into the embryos, which were fertilized as the control group. All the individuals in the experimental groups and control group were maintained for phenotype evaluation.
Fluorescence Imaging and Phenotype Evaluation
Fluorescence images were collected at 14 dpf by fluorescent microscopy. The vertebral morphology was observed and recorded for phenotypic evaluation and statistical analysis.
Genetic Analysis of CSF1R-Associated Genes
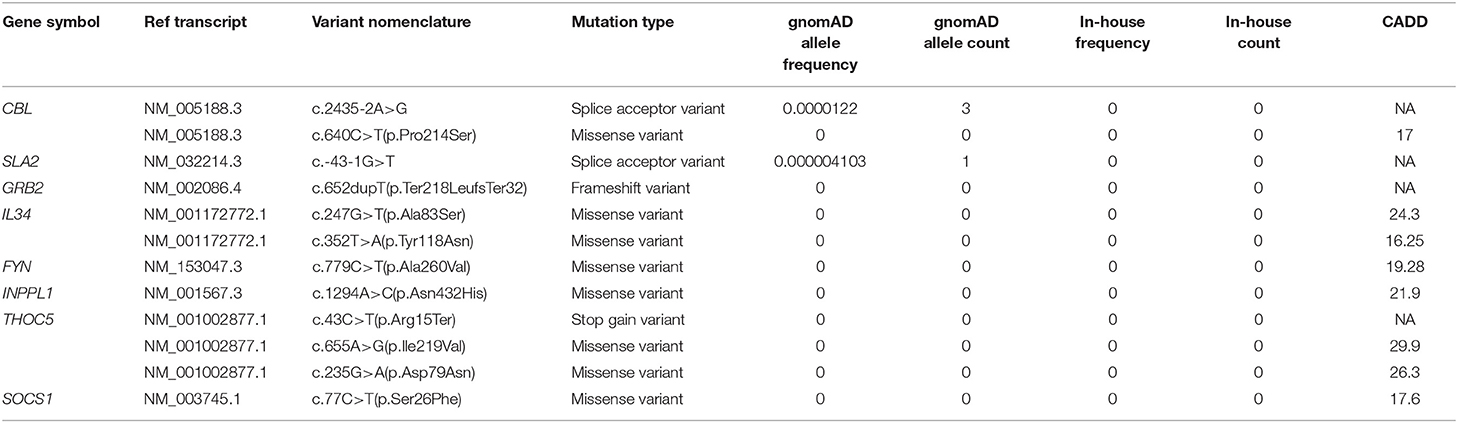
Twenty genes which are biologically linked to CSF1R (IL34, CSF1, SOS, GRB2, INPP5D, INPPL1, PIK3R1, PIK3R2, SOCS1, SOCS3, CBL, FYN, GRAP2, LYN, RASA1, SHC11, THOC5, TES1, CBL, PLCG2, SLA2) were selected for analysis according to public databases including Pubmed (https://pubmed.ncbi.nlm.nih.gov/) and Online Mendelian inheritance in Man (OMIM, https://www.omim.org/). Further, variant filtration of those genes was performed according to the same interpretation method as we did for CSF1R.
Statistics
Statistical differences between different experiment groups and the control group were evaluated with the Chi-square test. All statistical procedures were carried out using GraphPad Prism 8. *, **, ***, and **** denote p < 0.05, < 0.01, < 0.001, and < 0.0001, respectively.
Results
Pathogenic Variants Identified in CSF1R
From three patients with CVM, three deleterious heterozygous variants in CSF1R were identified, including two predicted truncating variants and one novel missense variant (Table 1). DISCO-CSS170368 was a 14-year-old girl admitted for spinal surgery (Figure 1A). The spinal plain radiograph showed that the coronal Cobb angle of the main curve (T2–L1) was 114°. The CT scan and three-dimensional reconstruction showed segmentation defects of T5–T9, dysplasia of T3–9, L4 hemivertebra, and left 6th and 7th rib abnormality (Figure 1B). MRI indicated mild diastematomyelia and syringomyelia. The girl showed no dyspnea, numbness or weakness of the limbs, backache or extremity pain, or marfanoid symptoms during the course of the disease. The clinical diagnoses of this patient were severe congenital scoliosis, pulmonary dysfunction, and diastematomyelia along with syringomyelia. WES identified a heterozygous truncating CSF1R variant (NM_005211.3: c.2749_2758delGACAGGAGAG), which was then confirmed by Sanger sequencing (Figure 1A). This variant is absent from public databases and our in-housed database encompassing 849 exome data (Table 1). Ten nucleotides were deleted between positions 2749 and 2758 in exon 21, resulting in a frameshift mutation in CSF1R. The mRNA harboring this mutation is predicted to escape from non-sense-mediated mRNA decay (NMD) by the NMD Esc Predictor (Figure 2A) (Coban-Akdemir et al., 2018). Hence, this frameshift mutation could consequently generate a truncated protein product (p.Asp917SerfsTer32).
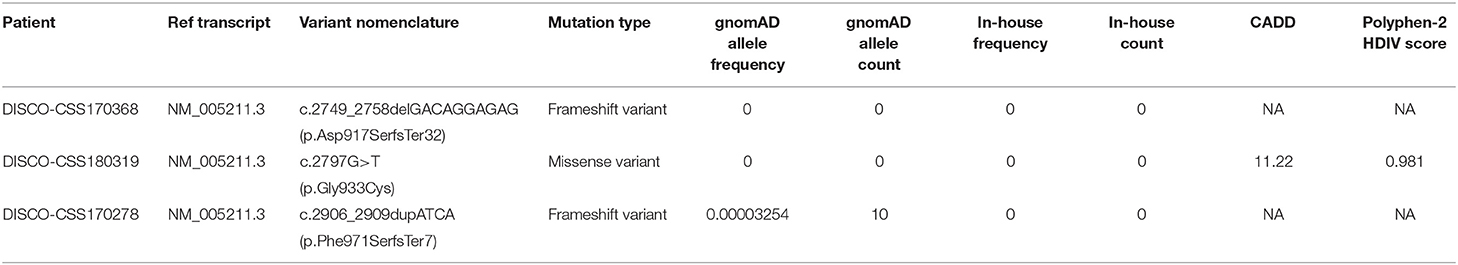
Table 1. Deleterious and rare CSF1R variants identified in patients with congenital vertebral malformation.
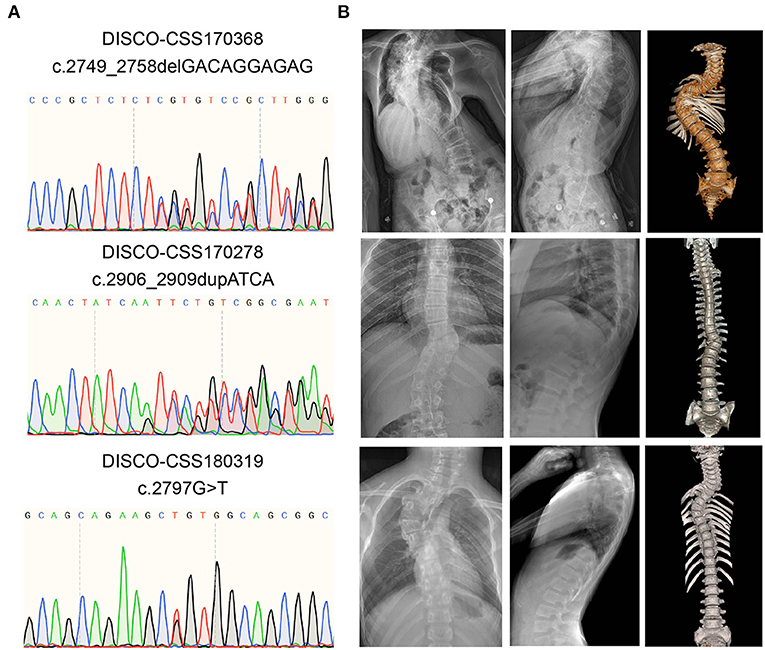
Figure 1. Variant information and clinical phenotype of CSF1R carboxy-terminal variants in three patients with CVM. (A) Results of Sanger sequencing of the rare CSF1R carboxy-terminal variants. (B) Spinal plain radiograph and computed tomography (CT) scan three-dimensional reconstruction of those CVM patients. CVM, congenital vertebral malformation.
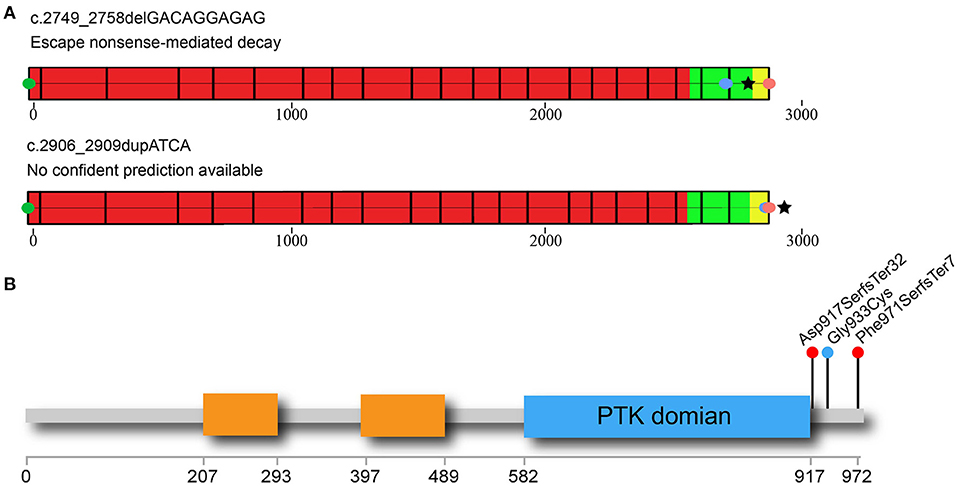
Figure 2. Non-sense-mediated decay (NMD) prediction and distribution of rare CSF1R C-terminal variants in CVM patients. (A) NMD prediction of two CSF1R-truncating variants identified in the CVM cohort. Green circle indicates the CDS start site; red circle indicates the canonical site of CSF1R protein; blue circle indicates the location of the truncating variant. Black star symbol indicates the predicted termination site of the mutated protein. The red part of protein-coding exons indicates the region in which truncating variants lead to NMD. On the contrary, the alleles with truncating variants located in the green part of protein could escape from NMD. The variants located in the yellow region of the protein may lead to non-stop RNA decay. One of the C-terminal CSF1R variants in the CVM cohort (c.2749_2758delGACAGGAGAG) is predicted to escape NMD. The other variant (c.2906_2909dupATCA) located within the yellow region could generate a new stop codon in the 3′ UTR region of mRNA, adding several amino acids on the C-terminus of the CSF1R protein (p.Phe971SerfsTer7). Hence, this variant allele is also predicted to escape from NMD. (B) Simplified diagram of the CSF1R protein is shown in the figure, with important domains and deleterious variants indicated. The immunoglobin domains are indicated by orange rectangles. The intracellular protein tyrosine kinase (PTK) domain is indicated by the blue rectangle. The truncating variants identified in DISCO-CSS170368 and DISCO-CSS170278 are indicated by the red circles. The missense variant identified in DISCO-CSS180319 is indicated by the blue circle. CVM, congenital vertebral malformation.
DISCO-CSS180319 was a 10-year-old female patient with CVM. Imaging examination revealed a 56° Cobb angle of the main curve (T3–T7). Besides, a structure disorder of T3–T7 was also observed in CT scan and three-dimensional reconstruction (Figure 1B). A missense variant (NM_005211.3: c.2797G>T) was identified in this girl (Figure 1A) and was also absent from the public and in-house databases. This variant is predicted to be deleterious by Polyphen-2 (Polyphen2 HDIV score: 0.981).
DISCO-CSS170278 was a 16-year-old boy with thoracic and lumbar scoliosis. The spinal plain radiograph revealed the presence of T10 hemivertebra (Figure 1B). WES analysis revealed a deleterious variant (NM_005211.3: c.2906_2909dupATCA) in CSF1R, which has been reported in the East Asian population with low frequency (0.0005) (Figure 1A). This deleterious variant could lead to a reading-frame shift. As a result, the last two amino acids of CSF1R were deleted and six extra amino acids were added to the C-terminus of the protein.
Interestingly, all of the three rare CSF1R variants identified in our cohort were located within or near the carboxy-terminal region downstream of the PTK domain (Figure 2B), which has been reported to be responsible for protein degradation mediated through the ubiquitination–proteasome pathway (Pawson, 1995; Schlessinger, 2000). No pathogenic variant in this region has ever been reported to be related to human diseases. Hence, those newly identified deleterious CSF1R variants might contribute to CVM through a distinct mechanism, which remains to be further investigated.
Variants Affecting the C-Terminal of CSF1R Increased the Stability of Protein and Could Induce Vertebral Malformation in Zebrafish
To determine the functional consequence of the C-terminal CSF1R variants in vitro, we transfected wild-type and mutant CSF1R constructs into cos-7 cells and examined the cellular levels of protein products with Western blotting. As a result, all of the three mutants accumulated at greater levels than the wild type, suggesting increased stability of mutant CSF1R proteins (Figures 3A,B).
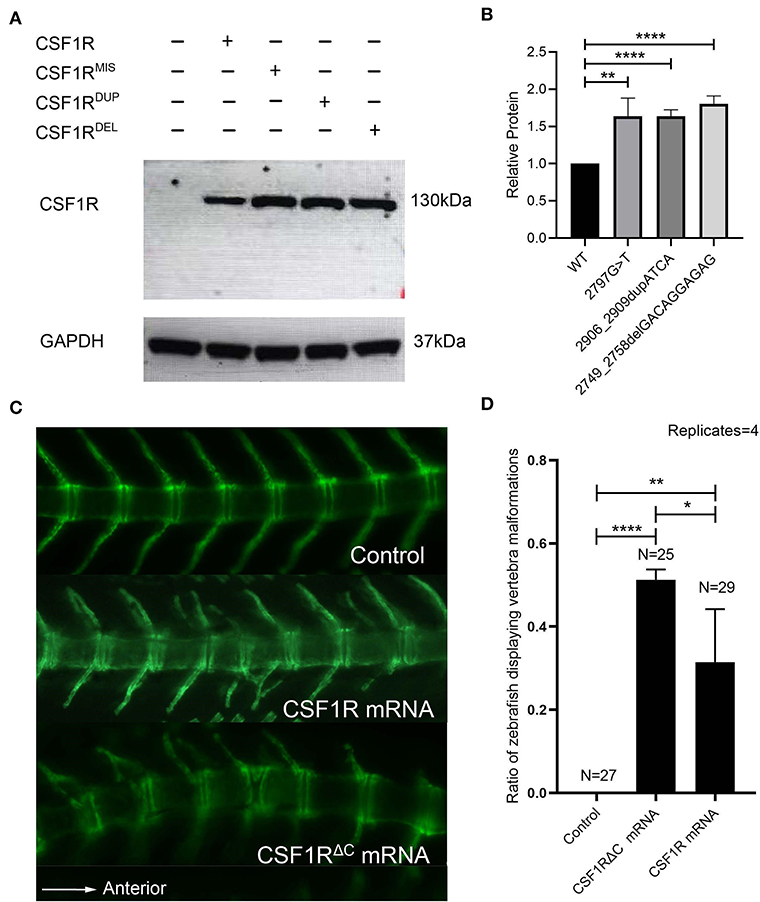
Figure 3. In vitro and in vivo functional study of CSF1R C-terminal variants. (A) Human CSF1R protein in total cell lysates made from cos-7 cells transfected with wild-type or mutant CSF1R constructs. GAPDH is used as a loading control. CSF1RMIS: c.2797G>T; CSF1RDUP: c.2906_2909dupATCA; and CSF1RDEL: c.2749_2758delGACAGGAGAG. (B) Quantification of relative wild-type and mutant CSF1R protein levels (n = 3). Protein levels of mutants were significantly increased compared to wild type (c.2797G>T, p = 0.00259; c.2906_2909dupATCA, p = 0.00005; c.2749_2758delGACAGGAGAG, p = 0.00004). Protein levels were normalized to GAPDH, and CSF1R was set to 1. (C) Fluorescence imaging (side view, anterior to the right) of Tg(Ola.Sp7:nlsGFP) transgenic zebrafish injected with 50 pg of wild-type CSF1R or CSF1RΔC human mRNA at 21 days post fertilization (dpf). The transgenic zebrafish injected with distilled water are fertilized and imagined as the control group. Note the vertebra labeled with green fluorescence. (D) A significant difference of the percentage of zebrafish displaying CVM phenotypes exists between experimental groups (CSF1R mRNA: 24%, 7/29; CSF1RΔC mRNA: 52%, 13/25) and control group (0%) (Chi-square test, CSF1R mRNA: p = 0.00635; CSF1RΔC mRNA: p = 0.00002). Compared to the zebrafish injected with CSF1R mRNA in the period of embryo, the zebrafish injected with CSF1RΔC mRNA have a significantly higher percentage of vertebral malformation phenotypes (Chi-square test p = 0.03452). Error bars represent one SD, n = 4 replicates. A different clutch of zebrafish was observed for each replicate. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; CVM, congenital vertebral malformation.
Therefore, we proposed that the heterozygous deleterious CSF1R variants might exert a gain-of-function effect on CSF1R biological function through disturbing the carboxy-terminal region of the protein. We then overexpressed the mutated CSF1R alleles (NM_005211.3: c.2749_2758delGACAGGAGAG) which is depleted of all the carboxy-terminal regions downstream of the PTK domain in the zebrafish model. This mutated mRNA-deleted carboxy-terminal region is referred to as CSF1RΔC mRNA in the following text.
The human CSF1R and CSF1RΔC mRNAs were injected into Tg (Ola.Sp7:nlsGFP) zebrafish embryos, respectively. Fluorescence images of the spine were collected at 21 dpf by fluorescent microscopy. The zebrafish of the CSF1R mRNA overexpression group exhibited vertebral malformations including vertebral fusion, hemivertebra, and fused vertebral arches (Figure 3C), which recapitulated the human CVM phenotype. These phenotypes indicate that the dose effect of CSF1R expression might be related to vertebral malformation. Intriguingly, injection of human CSF1RΔC mRNA led to a higher proportion of zebrafish with vertebral malformations than the wild-type mRNA did (p = 0.03452) (Figure 3D). These results revealed that the carboxy-terminus-mutated CSF1R alleles might exert gain-of-function effects and thereby cause vertebral malformations.
Mutational Spectrum in Genes Interacting With CSF1R
As an essential signaling transductor, CSF1R participates in several biological processes in individual development and interact with many other genes. To explore whether the variants in CSF1R-related genes may also be related with clinical phenotypes like vertebral abnormalities, we identified 21 genes of which the protein products interacted with CSF1R (IL34, CSF1, SOS, GRB2, INPP5D, INPPL1, PIK3R1, PIK3R2, SOCS1, SOCS3, CBL, FYN, GRAP2, LYN, RASA1, SHC11, THOC5, TES1, CBL, PLCG2, SLA2) and performed a genetic spectrum analysis. Twelve potential deleterious rare variants were identified in different patients, including four truncating variants and eight missense variants (Table 2). Among those CSF1R-related genes, CBL encodes an adaptor protein c-Cbl for CSF1R. It positively regulates CSF1R ubiquitination in a manner dependent on its variant SH2 and RING finger domains, then marking active CSF1R for degradation (Pawson, 1995; Schlessinger, 2000). One of the binding sites of c-Cbl on CSF1R is 969aa, which lies in the carboxy-terminal region of CSF1R where all three pathogenic variants identified in CSF1R were located (Wilhelmsen et al., 2002). We prioritized two heterozygous CBL variants, including a rare truncating variant and a novel missense variant, from two patients in our CVM cohort. Those deleterious variants of CBL may affect the interaction between CSF1R and c-Cbl and lead to CVM by a similar mechanism as carboxy-terminal region variants of CSF1R do.
Discussion
In this study, we used an exome-level genomic approach to identify potentially pathogenic variants of CSF1R genes in a CVM cohort. We identified three rare deleterious heterozygous C-terminal CSF1R variants, which were proved to increase the stability of protein in vitro. The CVM patients carrying those CSF1R variants exhibited vertebral malformations and early onset of scoliosis clinically. The zebrafish overexpression model resembled those vertebral phenotypes of CVM patients, indicating a gain-of-function mechanism.
In previous studies, CSF1R has been related to ALSP, which was inherited in an autosomal dominant pattern (Rademakers et al., 2011). ALSP is a fatal neurological disease characterized by progressive cognitive and motor impairment and seizures. The majority of the reported variants related with ALSP are missense or in-frame indels within the PTK domain (Guo et al., 2019). Recent studies indicated that bi-allelic CSF1R variants contribute to skeletal abnormalities in an autosomal recessive pattern (Guo et al., 2019; Oosterhof et al., 2019). The skeletal phenotypes of affected patients were referred to as the DOS-Pyle disease spectrum, which is characterized by sclerotic skull and flat and diffusely dense vertebral bodies (Guo et al., 2019). The pleiotropic effects of CSF1R could be attributed to the different inheritance modes and the variety of variants. Unlike the reported pathogenic CSF1R mutations, the variants identified in our cohort were located within the carboxy-terminal region of protein. This terminal region contains two autophosphorylation sites (923aa and 969aa), which offer binding sites for c-Cbl and participate in the degradation of CSF1R (Wilhelmsen et al., 2002; Ho et al., 2020). We proposed that the biological function of the protein could be affected when the carboxy-terminal region is mutated, which contributes to different clinical phenotypes like vertebral malformation.
Apart from the variety of variants and different pathogenic mechanisms, the age of onset is another factor which potentially affects the clinical symptoms of CSF1R variant carriers. The average age of symptom onset for ALSP is about 40 years (Sundal and Wszolek, 1993; Adams et al., 2018), while the average age of the patients with CSF1R deleterious variants identified in our cohort is around 10 years. The CVM patients may be too young to exhibit neurological phenotypes, which makes vertebral malformation-related scoliosis the only clinical symptom of those patients.
As an indispensable regulator of the monocyte/macrophage lineage, CSF1R has already been studied in variable animal models with the assistance of the transgenic technique. For instance, bi-allelic Csf1r deficiency in mouse was reported to cause brain development abnormalities and sclerosing skeletal dysplasia which leads to death within 6 weeks (Dai et al., 2004). Besides, the zebrafish model with biallelic loss-of-function mutations in csf1ra and csf1rb was also established in recent studies. This csf1rDM (csf1ra−/−, csf1rb−/−) zebrafish also exhibited neurological abnormalities primarily, along with skeletal phenotypes like osteopetrosis (Oosterhof et al., 2019). Considering the gain-of -function effect of the newly identified heterozygous CSF1R variants, the overexpression experiment could be used to prove the pathogenicity of the CSF1R mono-allelic mutations identified in patients with spine abnormalities like CVM.
Furthermore, we focused on the detailed structural and biological effects of the variants and hypothesized how these variants affected the biological function of CSF1R. One of the CSF1R variants (NM_005211.3: c.2749_2758delGACAGGAGAG) identified in our cohort was related to higher protein expression level and led to more severe clinical phenotypes as compared with that of the other two deleterious variants. This variant (NM_005211.3: c.2749_2758delGACAGGAGAG) deletes the entire carboxy-terminus region sequence (p.Asp917SerfsTer32), which is important for the binding of c-Cbl and thus the ubiquitination-mediated protein degradation. However, the other two CSF1R variants only lead to minor structural disturbance of the protein carboxy terminus and consequently confer milder effects on the protein expression and human orthopedic manifestations. This phenomenon indicates a genotype–phenotype correlation in CSF1R-related CVM.
Correspondingly, considering the potential role c-Cbl plays in the etiology of CSF1R-related CVM, we also focused on this protein and identified two deleterious rare CBL variants in our cohort. It is demonstrated that c-Cbl binds to activated CSF1R and mediates its degradation through the tyrosine-kinase binding (TKB) domain (Wilhelmsen et al., 2002). Interestingly, the novel deleterious CBL missense variant (NM_005188.3: c.640C>T) identified in our CVM patient is located within the TKB domain exactly, which potentially interrupts the interaction between c-Cbl and CSF1R, further affecting CSF1R degradation. Besides, another variant (NM_005188.3: c.2435-2A>G) of CBL is predicted to affect the splicing process of c-Cbl and create a critical structure disorder, which could also inhibit c-Cbl biological function. All those CBL deleterious variants were identified in CVM patients, who exhibited similar clinical phenotypes as CSF1R carboxy-terminus variant carriers did. This phenomenon indicates that the CSF1R ubiquitination and degradation might be one of the important biological processes related to CVM. The inhibition of CSF1R degradation may contribute to the enhancement of CSF1R biological function, further affecting bone metabolism and vertebral development, leading to vertebral malformation.
Besides, the variants in our cohort may also affect the biological function of CSF1R through generating structural disorders potentially. For instance, the missense variant (NM_005211.3: c.2797G>T) leads to amino acid change (p.Gly933Cys), for which glycine was replaced by cystine. Glycine is a relatively stable amino acid with low molecule weight. The amino acid is replaced by cystine, which is much more active and easily oxidizable. The oxidized cystine tends to form a disulfide bond with each other, potentially promoting the formation and stabilization of the CSF1R homodimer. The biological activity of protein could be enhanced by such structure disorder, which contributes to vertebral malformation. All the hypothesized mechanisms mentioned before remains to be explored further.
In conclusion, our study reveals the pathogenicity of heterozygous carboxy-terminal variants of CSF1R in CVM. The variants affect the biological function of the CSF1R product through potential gain-of-function effects.
Data Availability Statement
The datasets generated during the current study are available in the Mendeley repository, https://data.mendeley.com/datasets/wzjxyz99st/1.
Ethics Statement
The studies involving human participants were reviewed and approved by ethics committee at Peking Union Medical College Hospital (JS-098). Written informed consent to participate in this study was provided by the participants' legal guardian/next of kin. The animal study was reviewed and approved by ethics committee at Peking Union Medical College Hospital (JS-098). Written informed consent was obtained from the individual(s), and minor(s)' legal guardian/next of kin, for the publication of any potentially identifiable images or data included in this article.
Author Contributions
NW, ZW, TZ, BL, SZ, and GQ conceived of the project and designed the study. NW, BL, SZ, ZY, SW, and YN recruited the patients and collected and interpreted the data. BL and SZ conducted the statistical analysis and bioinformatic analyses. BL and JL performed the in vivo experiments. BL, LZ, and XL performed the in vitro experiments. BL, SZ, and NW wrote the first draft of the manuscript and critically revised the work for important intellectual content. All authors contributed to the article and approved the submitted version.
Funding
This research was funded in part by the National Key Research & Development Program of China (No. 2017YFC1104902), the National Natural Science Foundation of China (81672123 and 81972037 to TZ, 81930068 and 81772299 to ZW, 81822030 and 82072391 to NW, 81902178 to SW), Beijing Natural Science Foundation (L192015 to JZ, JQ20032 to NW, 7191007 to ZW), Tsinghua University-Peking Union Medical College Hospital Initiative Scientific Research Program, Innovation Fund for Medical Sciences (CIFMS, 2016-I2M-3-003 to GQ and NW, 2016-I2M-2-006 and 2017-I2M-2-001 to ZW), Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences (No. 2019PT320025), Fundamental Research Funds for the Central Universities (No. 3332019021), China Postdoctoral Science Foundation (No. 2020M670008ZX), and the PUMC Youth Fund & the Fundamental Research Funds for the Central Universities (No. 3332019021 to SW).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We appreciate all the patients, their families, and clinical research coordinators who participated in this project. We thank GeneSeeq Inc. for exome sequencing technical support. We thank Beijing Ekitech Co. Ltd. for the technical support and bioinformatic analysis.
References
Adams, S. J., Kirk, A., and Auer, R. N. (2018). Adult-onset leukoencephalopathy with axonal spheroids and pigmented glia (ALSP): integrating the literature on hereditary diffuse leukoencephalopathy with spheroids (HDLS) and pigmentary orthochromatic leukodystrophy (POLD). J. Clin. Neurosci. 48, 42–49. doi: 10.1016/j.jocn.2017.10.060
Arceci, R. J., Pampfer, S., and Pollard, J. W. (1992). Expression of CSF-1/c-fms and SF/c-kit mRNA during preimplantation mouse development. Dev. Biol. 151, 1–8. doi: 10.1016/0012-1606(92)90207-W
Arceci, R. J., Shanahan, F., Stanley, E. R., and Pollard, J. W. (1989). Temporal expression and location of colony-stimulating factor 1 (CSF-1) and its receptor in the female reproductive tract are consistent with CSF-1-regulated placental development. Proc. Natl. Acad. Sci. U.S.A. 86, 8818–8822. doi: 10.1073/pnas.86.22.8818
Chen, W., Lin, J., Wang, L., Li, X., Zhao, S., Liu, J., et al. (2020). TBX6 missense variants expand the mutational spectrum in a non-Mendelian inheritance disease. Hum. Mutat. 41, 182–195. doi: 10.1002/humu.23907
Chen, Y., Liu, Z., Chen, J., Zuo, Y., Liu, S., Chen, W., et al. (2016). The genetic landscape and clinical implications of vertebral anomalies in VACTERL association. J. Med. Genet. 53, 431–437. doi: 10.1136/jmedgenet-2015-103554
Coban-Akdemir, Z., White, J. J., Song, X., Jhangiani, S. N., Fatih, J. M., Gambin, T., et al. (2018). Identifying genes whose mutant transcripts cause dominant disease traits by potential gain-of-function alleles. Am. J. Hum. Genet. 103, 171–187. doi: 10.1016/j.ajhg.2018.06.009
Dai, X. M., Zong, X. H., Akhter, M. P., and Stanley, E. R. (2004). Osteoclast deficiency results in disorganized matrix, reduced mineralization, and abnormal osteoblast behavior in developing bone. J. Bone Miner. Res. 19, 1441–1451. doi: 10.1359/JBMR.040514
Erblich, B., Zhu, L., Etgen, A. M., Dobrenis, K., and Pollard, J. W. (2011). Absence of colony stimulation factor-1 receptor results in loss of microglia, disrupted brain development and olfactory deficits. PLoS ONE 6:e26317. doi: 10.1371/journal.pone.0026317
Guo, L., Bertola, D. R., Takanohashi, A., Saito, A., Segawa, Y., Yokota, T., et al. (2019). Bi-allelic CSF1R mutations cause skeletal dysplasia of dysosteosclerosis-pyle disease spectrum and degenerative encephalopathy with brain malformation. Am. J. Hum. Genet. 104, 925–935. doi: 10.1016/j.ajhg.2019.03.004
Ho, J., Peters, T., Dickson, B. C., Swanson, D., Fernandez, A., Frova-Seguin, A., et al. (2020). Detection of CSF1 rearrangements deleting the 3' UTR in tenosynovial giant cell tumors. Genes Chromosomes Cancer 59, 96–105. doi: 10.1002/gcc.22807
Huynh, D., Dai, X. M., Nandi, S., Lightowler, S., Trivett, M., Chan, C. K., et al. (2009). Colony stimulating factor-1 dependence of paneth cell development in the mouse small intestine. Gastroenterology 137, 136–144, 144.e1–e3. doi: 10.1053/j.gastro.2009.03.004
Lin, M., Zhao, S., Liu, G., Huang, Y., Yu, C., Zhao, Y., et al. (2020). Identification of novel FBN1 variations implicated in congenital scoliosis. J. Hum. Genet. 65, 221–230. doi: 10.1038/s10038-019-0698-x
Liu, J., Wu, N., Yang, N., Takeda, K., Chen, W., Li, W., et al. (2019). TBX6-associated congenital scoliosis (TACS) as a clinically distinguishable subtype of congenital scoliosis: further evidence supporting the compound inheritance and TBX6 gene dosage model. Genet. Med. 21, 1548–1558. doi: 10.1038/s41436-018-0377-x
Makrythanasis, P., Maroofian, R., Stray-Pedersen, A., Musaev, D., Zaki, M. S., Mahmoud, I. G., et al. (2018). Biallelic variants in KIF14 cause intellectual disability with microcephaly. Eur. J. Hum. Genet. 26, 330–339. doi: 10.1038/s41431-017-0088-9
Menke, J., Iwata, Y., Rabacal, W. A., Basu, R., Yeung, Y. G., Humphreys, B. D., et al. (2009). CSF-1 signals directly to renal tubular epithelial cells to mediate repair in mice. J. Clin. Invest. 119, 2330–2342. doi: 10.1172/JCI39087
Oosterhof, N., Chang, I. J., Karimiani, E. G., Kuil, L. E., Jensen, D. M., Daza, R., et al. (2019). Homozygous mutations in CSF1R cause a pediatric-onset leukoencephalopathy and can result in congenital absence of microglia. Am. J. Hum. Genet. 104, 936–947. doi: 10.1016/j.ajhg.2019.03.010
Pawson, T. (1995). Protein modules and signalling networks. Nature 373, 573–580. doi: 10.1038/373573a0
Pixley, F. J., and Stanley, E. R. (2004). CSF-1 regulation of the wandering macrophage: complexity in action. Trends Cell Biol. 14, 628–638. doi: 10.1016/j.tcb.2004.09.016
Rademakers, R., Baker, M., Nicholson, A. M., Rutherford, N. J., Finch, N., Soto-Ortolaza, A., et al. (2011). Mutations in the colony stimulating factor 1 receptor (CSF1R) gene cause hereditary diffuse leukoencephalopathy with spheroids. Nat. Genet. 44, 200–205. doi: 10.1038/ng.1027
Ren, X., Yang, N., Wu, N., Xu, X., Chen, W., Zhang, L., et al. (2020). Increased TBX6 gene dosages induce congenital cervical vertebral malformations in humans and mice. J. Med. Genet. 57, 371–379. doi: 10.1136/jmedgenet-2019-106333
Rodríguez-Tornos, F. M., Briz, C. G., Weiss, L. A., Sebastián-Serrano, A., Ares, S., Navarrete, M., et al. (2016). Cux1 enables interhemispheric connections of layer II/III neurons by regulating Kv1-dependent firing. Neuron 89, 494–506. doi: 10.1016/j.neuron.2015.12.020
Schlessinger, J. (2000). Cell signaling by receptor tyrosine kinases. Cell 103, 211–225. doi: 10.1016/S0092-8674(00)00114-8
Stanley, E. R., Berg, K. L., Einstein, D. B., Lee, P. S., Pixley, F. J., Wang, Y., et al. (1997). Biology and action of colony–stimulating factor-1. Mol. Reprod. Dev. 46, 4–10. doi: 10.1002/(SICI)1098-2795(199701)46:1<4::AID-MRD2>3.0.CO;2-V
Stanley, E. R., and Chitu, V. (2014). CSF-1 receptor signaling in myeloid cells. Cold Spring Harb. Perspect. Biol. 6:a021857. doi: 10.1101/cshperspect.a021857
Sundal, C., and Wszolek, Z. K. (1993). “CSF1R-related adult-onset leukoencephalopathy with axonal spheroids and pigmented glia,” in GeneReviews(®), eds M. P. Adam, H. H. Ardinger, R. A. Pagon, S. E. Wallace, L. J. H. Bean, G. Mirzaa, and A. Amemiya (Seattle, WA: University of Washington), 1993–2021.
Svensson, C., Part, K., Künnis-Beres, K., Kaldmäe, M., Fernaeus, S. Z., and Land, T. (2011). Pro-survival effects of JNK and p38 MAPK pathways in LPS-induced activation of BV-2 cells. Biochem. Biophys. Res. Commun. 406, 488–492. doi: 10.1016/j.bbrc.2011.02.083
Wang, Y., Szretter, K. J., Vermi, W., Gilfillan, S., Rossini, C., Cella, M., et al. (2012). IL-34 is a tissue-restricted ligand of CSF1R required for the development of Langerhans cells and microglia. Nat. Immunol. 13, 753–760. doi: 10.1038/ni.2360
Wilhelmsen, K., Burkhalter, S., and van der Geer, P. (2002). C-Cbl binds the CSF-1 receptor at tyrosine 973, a novel phosphorylation site in the receptor's carboxy-terminus. Oncogene 21, 1079–1089. doi: 10.1038/sj.onc.1205166
Wu, N., Ming, X., Xiao, J., Wu, Z., Chen, X., Shinawi, M., et al. (2015). TBX6 null variants and a common hypomorphic allele in congenital scoliosis. N. Engl. J. Med. 372, 341–350. doi: 10.1056/NEJMoa1406829
Wu, N., Wang, L., Hu, J., Zhao, S., Liu, B., Li, Y., et al. (2019). A recurrent rare SOX9 variant (M469V) is associated with congenital vertebral malformations. Curr. Gene Ther. 19, 242–247. doi: 10.2174/1566523219666190924120307
Yang, N., Wu, N., Zhang, L., Zhao, Y., Liu, J., Liang, X., et al. (2019). TBX6 compound inheritance leads to congenital vertebral malformations in humans and mice. Hum. Mol. Genet. 28, 539–547. doi: 10.1093/hmg/ddy358
Yang, Y., Zhao, S., Zhang, Y., Wang, S., Shao, J., Liu, B., et al. (2020). Mutational burden and potential oligogenic model of TBX6-mediated genes in congenital scoliosis. Mol Genet Genomic Med. 8:e1453. doi: 10.1002/mgg3.1453
Keywords: colony stimulating factor 1 receptor, congenital vertebra malformation, C-terminal variant, gain-of-function mechanism, zebrafish model
Citation: Liu B, Zhao S, Yan Z, Zhao L, Lin J, Wang S, Niu Y, Li X, Qiu G, Deciphering Disorders Involving Scoliosis and COmorbidities (DISCO) study, Zhang TJ, Wu Z and Wu N (2021) Variants Affecting the C-Terminal of CSF1R Cause Congenital Vertebral Malformation Through a Gain-of-Function Mechanism. Front. Cell Dev. Biol. 9:641133. doi: 10.3389/fcell.2021.641133
Received: 13 December 2020; Accepted: 10 February 2021;
Published: 19 March 2021.
Edited by:
Liang Cheng, Harbin Medical University, ChinaReviewed by:
Jianqiu Xiao, Washington University in St. Louis, United StatesSean Shishu Huang, Sichuan University, China
Copyright © 2021 Liu, Zhao, Yan, Zhao, Lin, Wang, Niu, Li, Qiu, Deciphering Disorders Involving Scoliosis and COmorbidities (DISCO) study, Zhang, Wu and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Terry Jianguo Zhang, amd6aGFuZ19wdW1jaEB5YWhvby5jb20=; Zhihong Wu, b3J0aG9zY2llbmNlQDEyNi5jb20=; Nan Wu, ZHIud3VuYW5AcHVtY2guY24=
†These authors have contributed equally to this work
 Bowen Liu1,2†
Bowen Liu1,2† Zihui Yan
Zihui Yan Nan Wu
Nan Wu