- Institute of Stem Cell and Regenerative Medicine, School of Medicine, Xiamen University, Xiamen, China
Self-renewal of embryonic stem cells (ESCs) is orchestrated by a vast number of genes at the transcriptional and translational levels. However, the molecular mechanisms of post-translational regulatory factors in ESC self-renewal remain unclear. Histidine phosphorylation, also known as hidden phosphorylation, cannot be detected by conventional experimental methods. A recent study defined phospholysine phosphohistidine inorganic pyrophosphate phosphatase (LHPP) as a histidine phosphatase, which regulates various biological behaviors in cells via histidine dephosphorylation. In this study, the doxycycline (DOX)-induced hLHPP-overexpressing mouse ESCs and mouse LHPP silenced mESCs were constructed. Quantitative polymerase chain reaction (qPCR), western blotting analysis, immunofluorescence, Flow cytometry, colony formation assays, alkaline phosphatase (AP) and bromodeoxyuridine (Brdu) staining were performed. We found that the histidine phosphorylation level was strikingly reduced following LHPP overexpression. Besides, the expression of Oct4 and Lefty1, indispensable genes in the process of ESCs self-renewal, was significantly down-regulated, while markers related to the differentiation were markedly elevated. Moreover, LHPP-mediated histidine dephosphorylation induced G0/G1 phase arrest in mESCs, suggesting LHPP was implicated in cell proliferation and cell cycle. Conversely, silencing of Lhpp promoted the self-renewal of mESCs and reversed the RA induced increased expression of genes associated with differentiation. Mechanistically, our findings suggested that the enzymatic active site of LHPP was the cysteine residue at position 226, not 53. LHPP-mediated histidine dephosphorylation lowered the expression levels of β-catenin and the cell cycle-related genes CDK4 and CyclinD1, while it up-regulated the cell cycle suppressor genes P21 and P27. Taken together, our findings reveal that LHPP-mediated histidine dephosphorylation plays a role in the self-renewal of ESCs. LHPP-mediated histidine dephosphorylation inhibited the self-renewal of ESCs by negatively regulating the Wnt/β-catenin pathway and downstream cell cycle-related genes, providing a new perspective and regulatory target for ESCs self-renewal.
Introduction
Embryonic stem cells (ESCs), with the attributes of pluripotency and self-renewal, have the ability to differentiate into multicell lineages (Dogan, 2018). Mounting evidence has shown that ESCs self-renewal is orchestrated by a variety of transcription factors, signaling molecules and RNA regulatory factors (Huang et al., 2015; Abu-Dawud et al., 2018). Previous work has indicated that transcriptional and post-transcriptional modifications are important for the regulation of ESCs self-renewal (Li et al., 2014). It has been reported that Oct4 and NANOG recruit LSD1 and bind to promoters of development-related genes, thereby controlling ESCs self-renewal by modulating H3K4 and H3K27 methylation (Adamo et al., 2011; Whyte et al., 2012). Zinc finger protein ZC3H13 maintains ESCs self-renewal via m6A modifications of mRNA (Wen et al., 2018). In addition, Foxp1-mediated ESCs-specific splicing switches play a crucial role in maintaining self-renewal (Gabut et al., 2011).
Translational regulation and post-translational modifications have a significant impact on ESCs self-renewal (Cassar and Stanford, 2012), where parsimonious translation in the pluripotent state and hierarchical translational regulation during differentiation control self-renewal and determine the fate of ESCs (Sampath et al., 2008). The ubiquitin proteasome pathway is responsible for most of the protein degradation that regulates ESCs self-renewal (Liu et al., 2016; Zhao et al., 2017). Additionally, phosphorylation is the basis for the activation and functional accuracy of a great number of proteins, and is also an important mechanism for the regulation of post-translational temporal and spatial distribution (Watanabe and Osada, 2016; Taracha et al., 2017). Previous studies have primarily focused on the phosphorylation of serine, threonine and tyrosine residues. Protein phosphorylation is related to ESCs self-renewal (Wang et al., 2016). LIF-induced activation of the JAK/STAT3, PI3K/AKT, and SHP2/MAPK pathways facilitates the nuclear translocation of signal factors, leading to gene expression that promotes ESCs self-renewal. Since its discovery, protein phosphorylation has been revealed to play a vital role in drug development, disease pathogenesis and treatment. However, the molecular mechanisms of the maintenance of ESCs self-renewal at the post-translational level remain unclear.
Histidine residues can also be phosphorylated (Boyer et al., 1962). However, histidine phosphorylation, also known as “hidden phosphorylation,” is characterized by an unstable P-N bond, and increased sensitivity to heat and acids. Phosphorylated histidine exists as two isomers, 1-phosphate histidine (1-pHis) and 3-phosphate histidine (3-pHis), which are produced by the phosphorylation of N1 and N3 on histidine residues, respectively (Fuhs et al., 2015). Previous studies have found that LHPP has been defined as a histidine phosphatase that regulates biological behavior in cells via histidine dephosphorylation (Hindupur et al., 2018). In our previous work (unpublished data), during the myocardial differentiation of ESCs and RA-induced differentiation, the expression of Lhpp was significantly upregulated. Meanwhile, compared with ESCs, the expression levels of Lhpp in various adult mouse tissues were upregulated to a much greater degree. However, whether LHPP-mediated histidine dephosphorylation affects ESCs self-renewal remains largely unknown. We hypothesized that LHPP-mediated histidine dephosphorylation may regulate the self-renewal of stem cells. Via construction of an inducible mouse ESCs (mESCs) overexpressing LHPP, and a lentivirus-mediated Lhpp silenced mESCs, we found that LHPP-mediated histidine dephosphorylation suppresses the self-renewal of mESCs, possibly by modulating the Wnt/β-catenin pathway, which provides a new perspective and regulatory target for understanding ESCs self-renewal.
Materials and Methods
Cell Culture
A2Lox-Cre mouse embryonic stem cells (mESCs) which is kindly donated by Kyba et al. (2002; Iacovino et al., 2014) were seeded onto an irradiated MEFs at the density of 5 × 105 cells. R1 mESCs were obtained from the American Type Culture Collection (ATCC). A2Lox-Cre mESCs and R1 mESCs were cultured in DMEM (Gibco, Grand Island, NY, United States) containing 15% FBS (Hyclone, Utah, United States), 0.1 mM non-essential amino acids (Gibco, Grand Island, NY, United States), 1 mM sodium pyruvate (Gibco, Grand Island, NY, United States), 0.1 mM β-mercaptoethanol (Sigma-Aldrich, Saint Louis, MO, United States), 100 U/ml penicillin (Gibco, Grand Island, NY, United States), 100 μg/ml streptomycin (Gibco, Grand Island, NY, United States) and 1000 U/ml leukemia inhibitory factor (LIF) (ESGRO, Millipore, Chemicon, United States). The cells were incubated at 37°C (5% CO2) in a humidified incubator, and the culture medium was changed every 24 h.
Construction of a Doxycycline (DOX)-Induced Human LHPP-Overexpressing Mouse ESC Line
In this study, the construction of a Doxycycline (DOX)-induced human LHPP-overexpressing mouse ESC line was performed as described previously (Zhang et al., 2016). Briefly, human Lhpp (hLhpp) was sequenced and cloned into the p2Lox-Rbm24-Flag-IRES-GFP plasmid to replace Rbm24, which resulted in the p2Lox-hLHPP-IRES-GFP recombinant plasmid. To induce the expression of Cre recombinant enzyme prior to electroporation, the cells were cultured in mouse ESC medium containing 1 μg/ml DOX. The next day, the cells were adjusted to a concentration of 1 × 106 cells per sample and resuspended in 90 μl Opti-MEM (Gibco, Grand Island, NY, United States). The Opti-MEM was equilibrated at room temperature before use. 10 μl p2Lox-hLHPP-IRES-GFP plasmid (10 μg) was used for each sample, which was transferred into a sterile cuvette and electroporated using the NEPA21 Super Electroporator (NEPA GENE, Japan). The procedure was conducted using the following parameters: 130 V, 5 ms; three pulses. Afterward, the cuvettes were immediately placed on ice for 5 min. The mixture was then transferred to a plate containing MEFs. Two days after inoculation, the cells were screened with 300 μg/ml of G418 for a further 7 days, and single clones were selected for amplification and identification.
Construction of a Mouse Lhpp Silenced mESC Line
Short hairpin RNA (shRNA) targeting Lhpp was used to construct LHPP-silenced ESC line. Briefly, sh1 Lhpp sequence (5′-TGGGAAAAGGACGCTATTACAAG-3′); sh2 Lhpp sequence (5′-GCTCAGAATTTGATCAGAT-3′); sh3 Lhpp sequence (5′-GGGAAAAGGACGCTATTACAAGG-3′) was cloned into lentivirus plasmid (plvx), and then transfected into A2Lox-Cre mESCs, followed by incubation with 10 μg/mL puromycin for 3 days. Finally, shLHPP-A2Lox mESCs was validated by qPCR and western blotting. Besides, R1 mESCs infected the above virus were subjected to the same treatment to obtain LHPP-silenced R1 mESCs.
Construction of a Human LHPP-Overexpressing R1 mESCs
To construct R1 mESCs overexpressing human LHPP, the CDS region of human Lhpp was cloned into the polyclonal site of the plvcs plasmid as described above and transfected into 293T cells together with a lentiviral packaging plasmid to produce lentiviral particles. Afterward, R1 mESCs were infected using these lentiviruses. Forty-eight hours later, the cells were selected with 10 μg/mL of puromycin for 3 days to generate stable cell lines. The lentivirus vector carrying a non-targeting shRNA sequence was used as a control.
Construction of LHPP Overexpressing Plasmid and the Mutant Plasmids
To construct the plasmid for transient overexpression of Lhpp, the CDS region of Lhpp was cloned into the pXJ-40 plasmid with the following primers. Lhpp upstream primer (XhoI restriction enzyme site): 5′-CCGCTCGAGATGGCACCGTGGGGCAAG-3′; Lhpp downstream primer (Pst1 restriction enzyme site): 5′-GCTGCAGCTTGTCGGCGTGCTGCAGC-3′. Subsequently, to construct plasmid contained a point mutation in the Lhpp sequence, the mutant Lhpp (C53S) and Lhpp (C226S) were generated using forward primer and reverse primer carrying the mutation, respectively. The primers were as follows: hLhpp (c.158G > C) forward: TGAAGGTGAGGTTCTcCACCAACGAG, hLhpp (c.158G > C) reverse: gAGAACCTCACCTTCAGCCGGGAAC; hLhpp (c.677G > C) forward: GCGGTGCCCAGCGGTcTGGAAT GAGA, hLhpp (c.677G > C) reverse: gACCGCTGGGCACCG CCGACGTCGCC. The products were digested with DMT enzyme for 1 h at 37°C, and then transformed into Escherichia coli for amplification. Mutagenesis was detected by Sanger sequencing. After successful construction of the plasmids, the plasmid vectors were transfected into 293T cells. Forty-eight hours later, western blotting assay was carried out to detect the intracellular pHis content as described below.
Differentiation of mESCs Into Cardiomyocytes
mESCs were digested with trypsin and resuspended in mESC differentiation medium containing the following: DMEM supplemented with 10% FBS, 0.1 mM non-essential amino acids, 1 mM sodium pyruvate, 0.1 mM β-mercaptoethanol, 100 U/ml penicillin, 100 μg/ml streptomycin and 50 μg/ml ascorbic acid (Sigma-Aldrich, Merck KGaA, Darmstadt, Germany). The cell suspension was seeded into gelatin-coated (0.1%) culture dishes at the density of 5 × 106 cells and left to rest for 20 min to remove the irradiated MEFs; the non-adherent cells were then collected and adjusted to 40 cells/μl in mESC differentiation medium. In order to prepare suspended droplets, the cells were transferred into a 50 ml sterile container, and 25 μl homogenous cell suspension was dropped onto the lid of a bacterial culture dish (150 mm) using a multi-channel pipette. A2Lox-Cre mESCs were then incubated at 37°C (5% CO2) to differentiate into embryonic bodies (EBs) via the hanging drop method (Han et al., 2013); each drop contained an average of 1,000 aggregated cells. After 4 days, the EBs were harvested from the droplets and seeded into a gelatin-coated petri dish (0.1%) for further differentiation.
Quantitative Polymerase Chain Reaction (qPCR)
Total RNA was extracted from the cells or tissues using TRIzol® reagent (Life Technologies, CA, United States) according to the manufacturer’s instructions. The purity and concentration of the RNA were determined using an ultraviolet spectrophotometer, and an A260/A280 ratio in the range of 1.8–2.0 was considered to indicate acceptable purity. 1 μg total RNA was reverse transcribed into cDNA at 37°C for 15 min, 85°C for 5 s, and then stored at 4°C for 30 min. The cDNA was subsequently used as the template for qPCR, which was conducted using the ABI7500 Real-Time PCR System (Applied Biosystems, Thermo Fisher Scientific, Inc.) with GAPDH as the internal reference. The thermocycling conditions were as follows: pre-denaturation at 95°C for 2 min; denaturation at 95°C for 15 s; annealing at 60°C for 15 s; all for 40 cycles. Each sample was run in triplicate and relative expression of the target genes was calculated using the 2–ΔΔCt method (Livak and Schmittgen, 2001). The sequences of all primers are represented in Table 1.
Western Blotting Analysis
Standard western blot assay: Briefly, the total protein was extracted from cells using RIPA buffer (Solarbio, Beijing, China) on ice, and protein concentration was determined via the BCA method. Following denaturation, the proteins (30 μg of protein loaded per lane) were separated by 10% SDS-PAGE, and then transferred onto PVDF membranes. The membranes were blocked with 5% non-fat dried milk for 2 h, washed 3 times with TBST, and incubated overnight at 4°C with the following primary antibodies: anti-β-actin (dilution, 1:4000; Catalog No. BM0627; Boster Biological Technology), anti-LHPP (dilution, 1:1000; Catalog No. sc-376648, Santa Cruz Biotechnology, Inc.), anti-β-catenin (6B3) Rabbit Monoclonal Antibody (dilution, 1:1000; #9582, Cell Signaling Technology), anti-CDK4 Mouse Monoclonal Antibody (dilution, 1:1000; Catalog No. 66950-1-Ig; Eenchmark Proteintech Group, Wuhan, China), Anti-p21 antibody (dilution, 1:1000; Catalog No. ab188224; Abcam), Anti-p27 Antibody (dilution, 1:1000; Catalog No. ab92741; Abcam), anti-CyclinD1 Mouse Monoclonal Antibody (dilution, 1:1000; Catalog No. 60186-1-Ig; Eenchmark Proteintech, Wuhan, China). The membranes were washed with TBST (5 min × 10 min) and subsequently incubated with Goat Anti-Mouse IgG (H + L) horseradish peroxidase-conjugated Secondary Antibody (dilutions 1:5000; Catalog No. BM3895; Boster Biological Technology) or Goat Anti-Rabbit IgG (H + L) horseradish peroxidase-conjugated Secondary Antibody (dilutions 1:5000; Catalog No. BA1054; Boster Biological Technology) for 3 h at room temperature. Following further washing with TBST (5 min × 10 min), the bands were visualized using an enhanced chemiluminescence detection kit (Advansta, Menlo Park, CA, United States). Quantity One Version 4.6.7 (Bio-Rad Laboratories, Inc., Hercules, CA, United States) was used to quantify the gray values of bands.
Detection of histidine phosphorylation: All procedures were conducted at 4°C, and all reagents were pre-cooled before use. The same number of cells was directly lysed in 2× pHis buffer (250 mM Tris-HCl at pH 8.8, 0.02% bromophenol blue, 50% glycerol, 50 mM EDTA, 500 mM DTT, and 10% SDS) on ice, followed by ultrasonication (3 times for 15 s each). The sediment was removed by centrifugation (10,000 × g for 15 min, 4°C). The supernatant was immediately separated at 4°C by modified SDS-PAGE (using a modified stacking gel at pH 7.2, and 10% separation gel at pH 8.8), and then transferred onto PVDF membranes. The membranes were blocked in 2.5% BSA buffer (pH 8.8) (Solarbio, Beijing, China) and then incubated overnight at 4°C with the following primary antibodies: anti-N3-Phosphohistidine (3-pHis) Monoclonal Antibody (Catalog No. MABS1352; Merk KGaA, Germany) and anti-N1-Phosphohistidine (1-pHis) Monoclonal Antibody (Catalog No. MABS1330; Merk KGaA, Germany), diluted in BSA buffer (0.1% Tween-20) to 0.5 μg/ml. The membranes were washed three times (at 4°C for 10 min) with TBST (pH8.8) and incubated with an anti-rabbit HRP-conjugated secondary antibody (dilutions 1:5000) at 4°C. As the control, cell lysates were heated at 100°C for 20 min in loading buffer (pH 8.8) to remove phosphohistidine. Lanes loaded with the heated lysates indicated either heat-resistant phosphohistidine or non-specific signals. Absolute gray value of intracellular phosphohistidine = gray value of samples - gray value of the control sample. Relative gray value = absolute gray value/gray value of reference gene.
Immunofluorescence Assay
Firstly, sterile slides were placed into each well of a 6-well plate. Cells in the logarithmic growth phase were digested with 0.25% trypsin, and then seeded into the plates at a density of 1 × 105 cells per well. The following day, the slides were washed 3 times with PBS, and then fixed with 4% paraformaldehyde for 30 min at room temperature prior to additional washing as aforementioned. The slides were then treated with 0.25% Triton for 10 min, and washed again prior to blocking with 5% BSA solution for 1 h. Following overnight incubation with the primary antibodies: anti-Oct3/4 (C-10) Mouse monoclonal Antibody (dilution, 1:300; Catalog No. sc-5279; Santa Cruz Biotechnology, Inc.) and anti-Lefty (F-11) Mouse monoclonal Antibody (dilution, 1:300; Catalog No. sc-166708; Santa Cruz Biotechnology, Inc.) at 4°C, the slides were washed again with PBS (3 times), and incubated with Goat anti-Mouse IgG2b Cross-Adsorbed Secondary Antibody, Alexa Fluor 555 (dilution, 1:300; Catalog No. A-21147; Life Technology) and Goat anti-Rabbit IgG (H + L) Cross-Adsorbed Secondary Antibody (dilution, 1:300; Catalog No. A-21428; Life Technology) at room temperature for 3 h. The slides were then washed with PBS 3 more times, counterstained with DAPI solution (dilution, 1:10,000; Catalog No. C0060; Solarbio, Beijing, China) for 5 min and washed with PBS a further 3 times. Finally, the slides were sealed with 50% glycerol and observed under a fluorescence microscope (Olympus Corporation, Tokyo, Japan). Magnification, 100×.
Cell Cycle Assay
mESCs were collected by centrifugation at 200 × g for 3 min at room temperature, and fixed with 70% pre-cooled ethanol at 4°C overnight. The following day, the cells were centrifuged at 700 × g for 3 min at room temperature, washed with pre-cooled PBS and resuspended (also in pre-cooled PBS) to a final concentration of 1.0 × 106 cells/ml. RNase A (10 μg/ml) was added to 100 μl cell suspension and the samples were harvested by centrifugation (as aforementioned). The cells were washed with pre-cooled PBS, resuspended and filtered using a 75 μm nylon mesh; Cell cycle analysis was detected using a flow cytometer (Guava EasyCyte 8, Merk), and the data were analyzed using FlowJo 7.6 Software (FlowJo LLC, Ashland, OR, United States).
Cell Proliferation Assay
mESCs in the logarithmic growth phase were digested with trypsin, resuspended in culture medium, and then seeded into 96-well plates at a density of 1 × 104 cells per well. 10% of Cell Counting Kit-8 (CCK-8) reagent (Cat. C0038, Beyotime Biotechnology, China) was added at 12, 24, 36, 48, 60, and 72 h time points, followed by incubation at 37°C for 2 h. Absorbance values were detected with a microplate reader (Bio-Rad) at 450 nm.
Alkaline Phosphatase (AP) Staining
Stem cell properties were validated by AP staining. The cells treated with RA were used as a positive control, while untreated cells (receiving neither RA nor DOX) were used as a negative control. Briefly, mESCs were fixed with 4% formaldehyde for 2 min at room temperature. The cells were subsequently stained using the Alkaline Phosphatase Kit (SiDanSai Biotechnology, Shanghai, China) according to the manufacturer’s instructions. After washing 3 times with PBS, the cells were observed under an optical microscope (Leica DM750). Magnification, 100×.
Colony Formation Assay
mRSCs were inoculated into 6-well plates at a density of 500 cells per well and incubated for 10 consecutive days. Culture medium was changed every 2 days. Clones were observed, and the culture was terminated when the clones grew to the naked eye. After fixation with 75% ethanol for 10 min at room temperature, the cells were stained with 0.1% crystal violet solution for 15 min. Images were photographed under a microscope (Leica DM750).
Bromodeoxyuridine (Brdu) Staining
For BrdU staining, when mESCs grown to 60% confluence, Brdu solution (10 μg/ml, Sigma) was added into culture medium. After 6 h of incubation, the cells were collected and fixed with 75% ethanol for 30 min at room temperature. Next, the cells were treated with 1 nM HCl for 30 min. After discarding the HCl, the cells were rinsed with 10 μM boric acid, treated with 0.1% Triton for 30 min, blocked with 5% BSA for 1 h, incubated with anti-BrdU primary antibody (ab6326, 1:200 dilution, Abcam) diluted with 5% BSA for 12 h at 4°C, and then labeled with corresponding fluorophore-conjugated secondary antibody (#4416, 1:400 dilution, Cell Signaling Technology) for 2 h at room temperature. Flow cytometric data were analyzed using FlowJo 7.6 Software (FlowJo LLC, Ashland, OR, United States).
Animal Experiments
C57BL/6 mice (n = 10), aged 6–8 weeks and weighing 18.5 ± 1.9 g each, were purchased from the animal center of Xiamen University. The mice were euthanized by cervical dislocation. Adult mouse tissues, including muscle tissue, brain tissue, kidney, gut, spleen, bladder, lung, and liver were carefully excised and kept in liquid nitrogen immediately. All animal experiments were conducted with the approval of the animal ethics and use committee (IACUC) of Xiamen university (Xiamen, Fujian, China; Approval ID: scxk2013-0006).
Statistical Analysis
SPSS 21.0 (IBM Corporation, Armonk, NY, United States) and GraphPad Prism 6.0 (GraphPad Software, Inc., La Jolla, CA, United States) were used for data analysis and graphical representation, respectively. The difference between two groups was determined using the Student’s t-test, and comparisons among multiple groups were measured using one-way ANOVA with Dunnett’s test. Data were expressed as mean ± Standard Deviation (SD). Each experiment was independently repeated three times, and P < 0.05 was considered as statistically significant.
Results
Lhpp Expression Is Up-Regulated During mESCs Differentiation
To understand the role of LHPP in mouse ESCs differentiation, differentiation of A2Lox-Cre mESCs to cardiomyocytes was induced via embryoid bodies (EBs). We observed a weakened self-renewal ability and loss of stemness in mESCs after RA treatment for 72 h, in which Lhpp expression was significant upregulated (Figures 1A–C). Besides, the expression of Lhpp was increased during myocardial differentiation of mESCs at transcriptional and translational levels (Figures 1D–F). Furthermore, data indicated that Lhpp was more highly expressed in adult mouse tissues than in mESCs (Figure 1G).
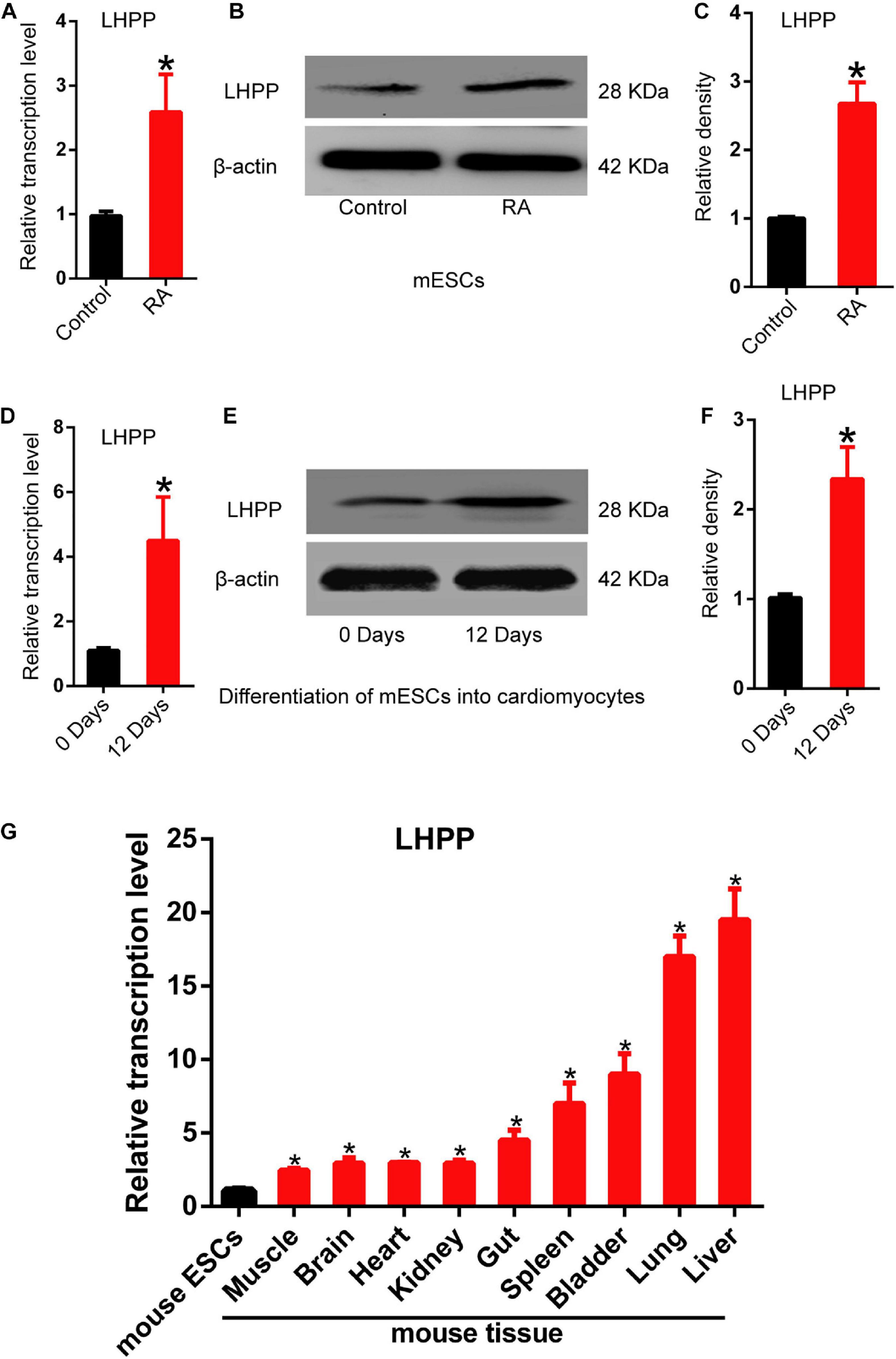
Figure 1. Up-regulation of Lhpp expression during mES cell differentiation. (A–C) Up-regulated expression of mLhpp in mESCs treated with 10 mM of RA for 3 days. (A) Expression of mLhpp mRNA; (B) representative blots of mLHPP expression; (C) statistical analysis of the protein expression of mLHPP. (D–F) Increased expression of mLhpp in mESCs during myocardial differentiation, as validated by qPCR and western blotting analysis. (D) Statistical results of qPCR; (E) representative blots; (F) quantification of protein expression. (G) Up-regulated transcription of mLhpp in adult mouse tissues, compared with mESCs. GAPDH was used as the internal control. All experiments were independently repeated at least three times, and the data are presented as the mean ± SD. *P < 0.05. LHPP, phospholysine phosphohistidine inorganic pyrophosphate phosphatase; mLhpp, mouse Lhpp; mESCs, mouse embryonic stem cells; RA, retinoic acid.
Successful Construction of DOX-Induced Lhpp-Overexpressing and Lhpp-Silenced mESC Lines
To explore the role of LHPP in mESCs, DOX-induced hLHPP-overexpressing mESC line was constructed via homologous recombination (Figure 2A). Following DOX treatment, immunofluorescence, qPCR, and western blotting revealed that the successful construction of human Lhpp (hLhpp)-overexpressing mESC line, in which the expression of DOX-induced hLhpp was up-regulated, compared with that in the untreated cells at different time points (Figures 2B,C); meanwhile, flow cytometry analysis suggested that DOX induction influenced the fluorescence intensity of GFP in a time dependent manner (Figure 2D). To determine the biological function of endogenous LHPP in mESCs, we constructed three shRNAs targeting mouse Lhpp and the silencing efficiency of these 3 shRNAs in A2Lox-Cre ESCs cells was verified. sh2 RNA with the highest silencing efficiency was selected for subsequent experiments (Figure 2E). These results confirmed the successful construction of DOX-induced Lhpp-overexpressing mESCs and Lhpp knockdown mESCs.
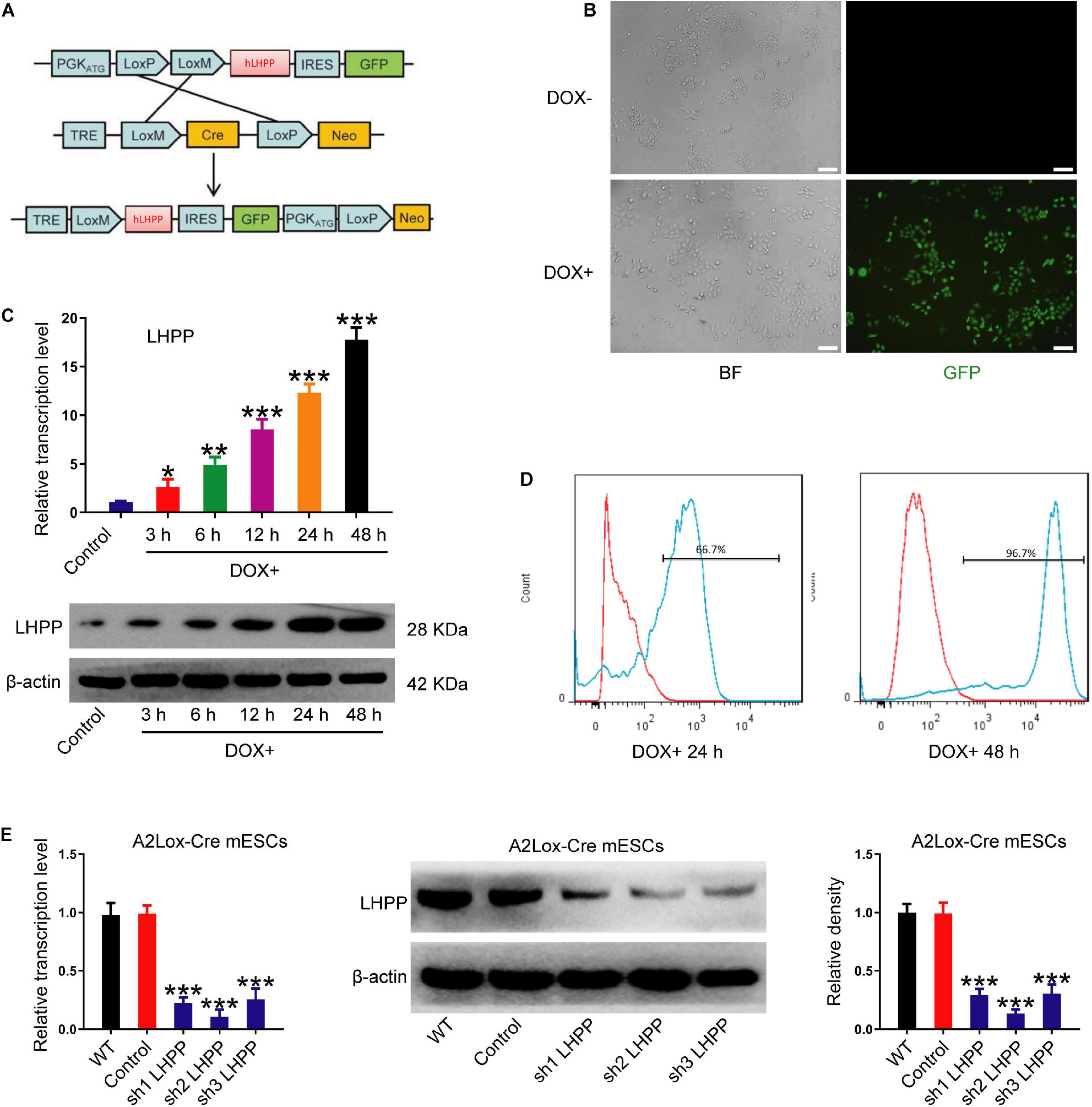
Figure 2. Construction and identification of DOX-induced hLhpp-overexpressing and -silenced mESCs. (A) Schematic diagram depicting the production of DOX-induced hLHPP-overexpressing mESCs. (B) Fluorescence detection of the mESCs following DOX treatment (1 μg/ml) for 48 h. Scale bar = 100 μm. (C) The mRNA and protein expression levels of Lhpp following DOX treatment (1 μg/ml) at different time points. (D) GFP expression in mESCs following DOX treatment (1 μg/ml) for 24 or 48 h was detected by flow cytometry. (E) The mRNA and protein expression levels of Lhpp were determined by qPCR and western blotting analysis in Lhpp-silenced mESCs. All experiments were independently repeated at least three times, and the data are presented as the mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001. DOX, doxycycline; LHPP, human phospholysine phosphohistidine inorganic pyrophosphate phosphatase; hLhpp, human Lhpp; mESCs, mouse embryonic stem cells; RA, retinoic acid.
LHPP-Mediated Histidine Dephosphorylation Regulates the Expression of Pluripotent- and Differentiation-Related Genes in mESCs
To unveil the mechanism by which LHPP regulates the biological behavior of mESCs, the level of intracellular histidine phosphorylation was measured using modified SDS-PAGE and immunofluorescence assay. As illustrated in Figures 3A,B, the levels of 1-pHis and 3-pHis were found to be reduced in mESCs overexpressing LHPP, compared with those not induced with DOX. Protein samples that were pretreated at 100°C for 20 min exhibited non-specific bands, which may have been due to the low specificity of the antibody, or an excessive incubation time. For loading control, Ponceau’s staining was conducted (Figures 3C,D). Besides, similar results of 1-pHis and 3-pHis levels were obtained using immunofluorescence (Figures 3E,F).
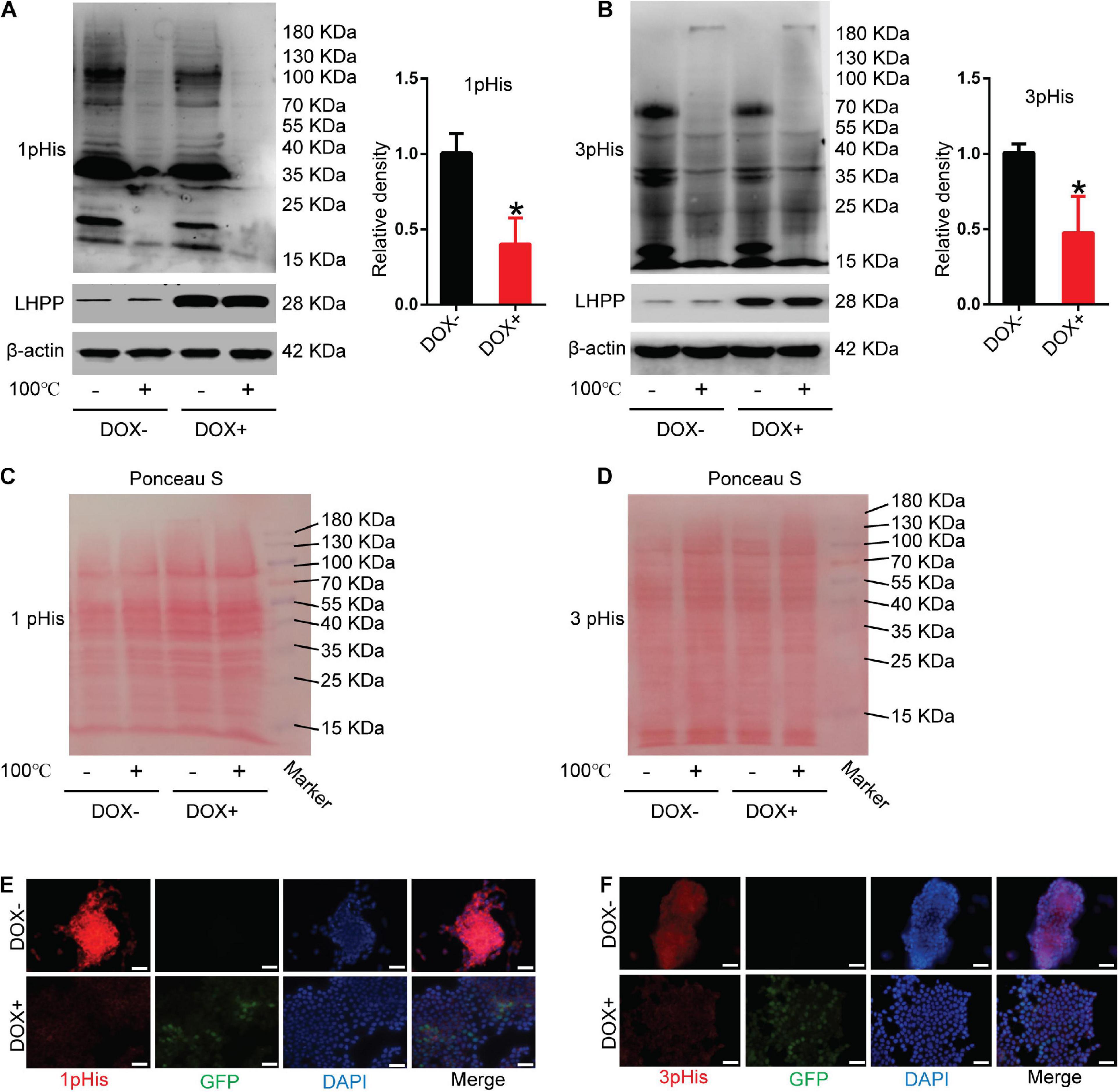
Figure 3. Effect of human LHPP-overexpression on histidine phosphorylation in mESCs. (A) Intracellular 1-pHis levels were detected by modified western blotting in mESCs treated with 1 μg/ml DOX for 72 h. (B) Intracellular 3-pHis levels in mESCs were detected by modified western blotting following treatment with 1 μg/ml DOX for 72 h. (C,D) Ponceau’s staining was used to validate the amount of protein loaded per well. (E,F) Immunofluorescence was carried out to observe the (E) 1-pHis and (F) 3-pHis levels in mESCs treated with 1 μg/ml DOX for 72 h. Red, 1-pHis or 3-pHis; green, GFP; blue, DAPI. Scale bar = 100 μm. All experiments were independently repeated at least three times, and the data are presented as the mean ± SD. *P < 0.05. LHPP, human phospholysine phosphohistidine inorganic pyrophosphate phosphatase; mESCs, mouse embryonic stem cells; DOX, doxycycline.
To further understand the role of LHPP in mESC self-renewal, stem cell proprieties were validated by AP staining. Seventy-two hours after DOX induction (Figure 4A) or RA treatment (Figure 4B), the cell colonies were decentralized with irregular edges, and AP staining was reduced compared with the control (Figures 4C,D). Colony formation experiments further confirmed that the clonogenic capacity of both DOX-induced A2Lox-Cre mESCs and R1 mESCs overexpressing LHPP by lentiviral infection was significantly reduced (Figures 4E,F). Immunofluorescence, qPCR and western blotting analysis revealed that the expression levels of stem cell marker genes Oct4 and Lefty1 were significantly down-regulated (Figures 4G–I and Supplementary Figures 1A–C, 2A) in mESCs with Lhpp overexpression compared with the control cells, where there was no significant change in endogenous mouse Lhpp expression following the exogenous overexpression of human Lhpp. Meanwhile, the mRNA levels of T-Bra, Mesp1, Nestin, SOX1, SOX17, Afp were significantly elevated in LHPP-overexpressed mESCs (Supplementary Figure 1D). Conversely, silencing of Lhpp in A2Lox-Cre mESCs and R1 mESCs enhanced the clonogenic ability of mESCs (Figures 4J,K). Besides, Lhpp knockdown upregulated Lefty1 expression at the mRNA and protein levels, but reduced the mRNA levels of T-Bra, Mesp1, Nestin, SOX1, SOX17, Afp in mESCs as compared to the control group (Figures 4L–N and Supplementary Figures 1E,F, 2B). No significant difference in Oct4 expression was observed in DOX-induced LHPP overexpressing mESCs, while Lhpp knockdown by shRNA upregulated Oct 4 mRNA expression in R1 mESCs compared to the control. Overall, these findings suggested that LHPP plays role in differentiation of mESCs.
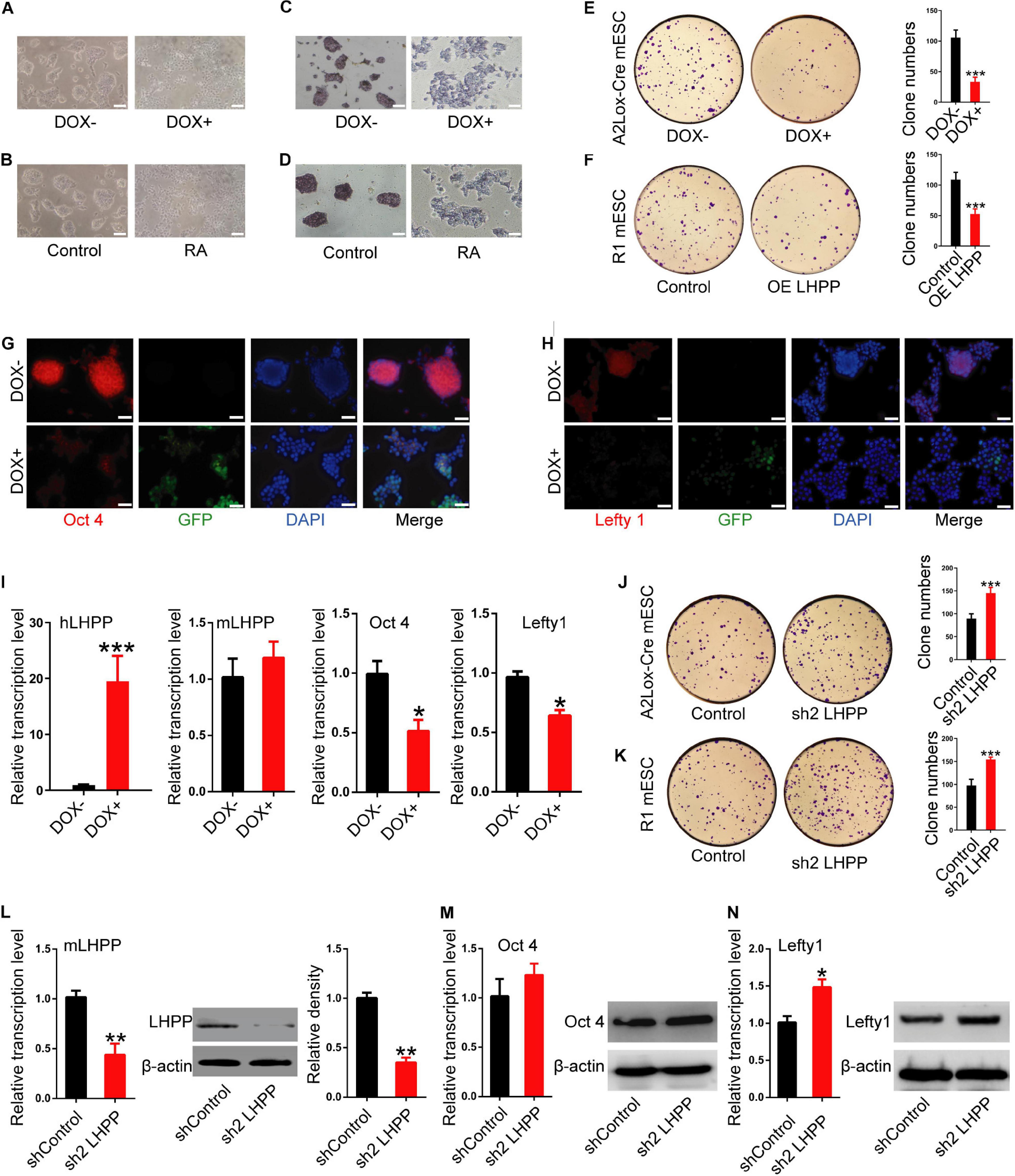
Figure 4. Effects of LHPP on the expression of cellular pluripotency and differentiation-related genes in mESCs. (A) mES cell colonies were observed with or without DOX treatment (1 μg/ml) for 72 h. (B) mES cell colonies after incubation with or without 10 mM RA for 72 h. (C,D) AP staining was performed in mESCs with or without (C) DOX (1 μg/ml) or (D) 10 mM RA treatment for 72 h. (E,F) Colony-formation assay was performed to detected the effects of Lhpp overexpression on (E) A2Lox-Cre mESCs and (F) R1 mESCs. (G,H) Immunofluorescent labeling of (G) Oct4 and (H) Lefty1 in DOX-induced hLhpp-overexpressing mESCs. Red, Oct4 or Lefty1; green, GFP; blue, DAPI. Scale bar = 100 μm. (I) qPCR quantification of the mRNA levels of human Lhpp, mouse Lhpp, Oct4 and Lefty1 in mESCs treated with or without DOX (1 μg/ml). (J,K) Colony-formation assay was performed to evaluate the effects of Lhpp knockdown on (J) A2Lox-Cre mESCs and (K) R1 mESCs. (L) qPCR detection of mLhpp expression in mESCs. (M,N) The mRNA and protein levels of (L) Oct4 and (M) Lefty1 in Lhpp-silenced mESCs. All experiments were independently repeated at least three times, and the data are represented as the mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001. DOX, doxycycline; LHPP, human phospholysine phosphohistidine inorganic pyrophosphate phosphatase; hLhpp, human Lhpp; mESCs, mouse embryonic stem cells; RA, retinoic acid; AP, alkaline phosphatase; sh, short hairpin RNA.
The Enzymatic Active Site of LHPP Is the Cysteine Residue at Position 226, Not 53, Which Is Critical for mESCs Self-Renewal
Although previous literature has speculated that the specific enzymatic active site of LHPP is the cysteine residues at position 53 and 226 (Yokoi et al., 2003), so far, no study has confirmed this. In present study, we constructed two mutant plasmids of Lhpp as described in the methods, resulting in mutant proteins LHPP (C53S) and LHPP (C226S) (Supplementary Figures 3A,B). After overexpression of these two mutants in human 293T cells, respectively, we detected that the LHPP (C53S) mutant was able to significantly reduce intracellular histidine phosphorylation, but the LHPP (C226S) mutant did not produce this effect (Figures 5A–D).
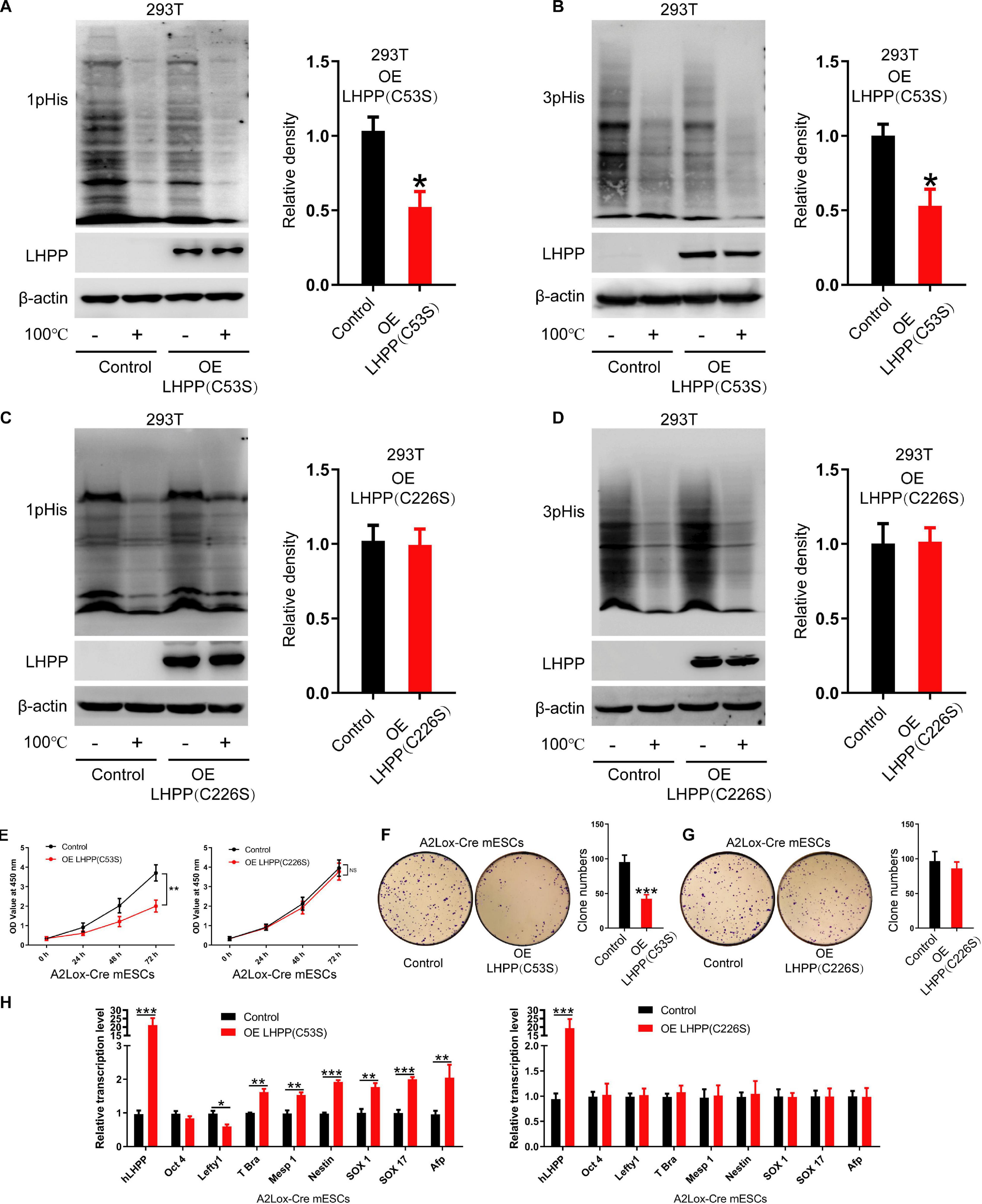
Figure 5. The cysteine at position 226 of LHPP is essential for its histidine phosphatase activity. (A,B) Western blotting analysis for intracellular (A) 1-pHis and (B) 3-pHis levels after overexpression of LHPP (C53S) in 293T cells. (C,D) Western blotting analysis for intracellular (C) 1-pHis and (D) 3-pHis levels after overexpression of LHPP (C226S) in 293T cells. (E) Cell proliferative ability was detected by CCK-8 assay after overexpression of LHPP (C53S) mutant or LHPP (C226S) mutant in A2Lox-Cre mESCs. (F,G) Cell colony formation capability was assayed by colony formation assay after overexpression of (F) LHPP (C53S) mutant or (G) LHPP (C226S) mutant in A2Lox-Cre mESCs. (H) After overexpression of LHPP (C53S) mutant or LHPP (C226S) mutant in A2Lox-Cre mESCs, the expression levels of pluripotency and differentiation-associated genes were evaluated by qPCR analysis. All experiments were independently repeated at least three times, and the data are represented as the mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001. NS, not significant difference. LHPP, phospholysine phosphohistidine inorganic pyrophosphate phosphatase.
To determine the effects of overexpression of these two catalytic mutants induced by DOX on the self-renewal of mESCs, CCK-8 assay, qPCR analysis for pluripotency and differentiation markers and colony formation assay was performed in A2Lox-Cre mESCs. We found that overexpression of LHPP (C53S) mutant significantly attenuated cell proliferative ability, while there was no significant difference between LHPP (C226S) mutant-overexpressed mESCs and the control group (Figure 5E). Besides, colony formation assay confirmed that overexpression of LHPP (C53S) mutant resulted in significantly reduced colony formation capability in mESCs; however, mutating LHPP (C226S) had no significant effect (Figures 5F,G). Meanwhile, qPCR analysis indicated that both mutants were successfully overexpressed in the mESCs. Similar to the aforementioned results, LHPP (C53S) mutant markedly decreased the mRNA expression of pluripotency gene Lefty1, but elevated the expression of differentiation-associated genes T-box transcription factor Brachyury (T-Bra), Mesp1, Nestin, SOX1, SOX17, and Afp (Figure 5H). In contrast, although LHPP (C226S) mutant was also significantly upregulated in mESCs, it did not produce similar results. Cumulatively, these data suggest that the enzymatic active site of LHPP is the cysteine residue at position 226, which is essential for maintaining its function to regulate mESCs self-renewal.
LHPP-Mediated Histidine Dephosphorylation Modulates the Proliferation of mESCs by Cell Cycle Arrest
To investigate how LHPP exerts its biological effects, flow cytometry was performed to detect changes of cell-cycle progression. It was revealed that when cultured with medium containing LIF, the proliferative capacity of DOX-induced LHPP-overexpressing mESCs was dramatically attenuated (Figures 6A,C), and that the proliferation of mESCs slowed down, compared with cells of the control group (Figures 6D,E), which was consistent with what was observed in RA treated (RA+) group (Figures 6F,G). Conversely, proliferation of Lhpp knockdown mESCs was significantly enhanced compared with that in the control cells (Figure 6B). Moreover, compared with wild-type cells, Lhpp knockdown mESCs cultured in LIF medium showed significant changes in cell cycle distribution, with an increase in the proportion of G2/M cells, but a decrease in the proportion of G0/G1 cells (Figures 6H,I). In addition, DOX-induced overexpression of LHPP exhibits a lower proportion of pHH3-positive cells than the control cells. These results were consistent with the results from RA-induced ESCs differentiation. Conversely, Lhpp knockdown caused an increased proportion of pHH3-positive cells (Figure 6J). Further, Brdu staining showed that overexpression of LHPP in A2Lox-Cre mESCs and R1 mESCs significantly reduced the percentage of Brdu-positive cells, whereas silencing of Lhpp in both cells yielded opposing effects (Figures 6K–N).
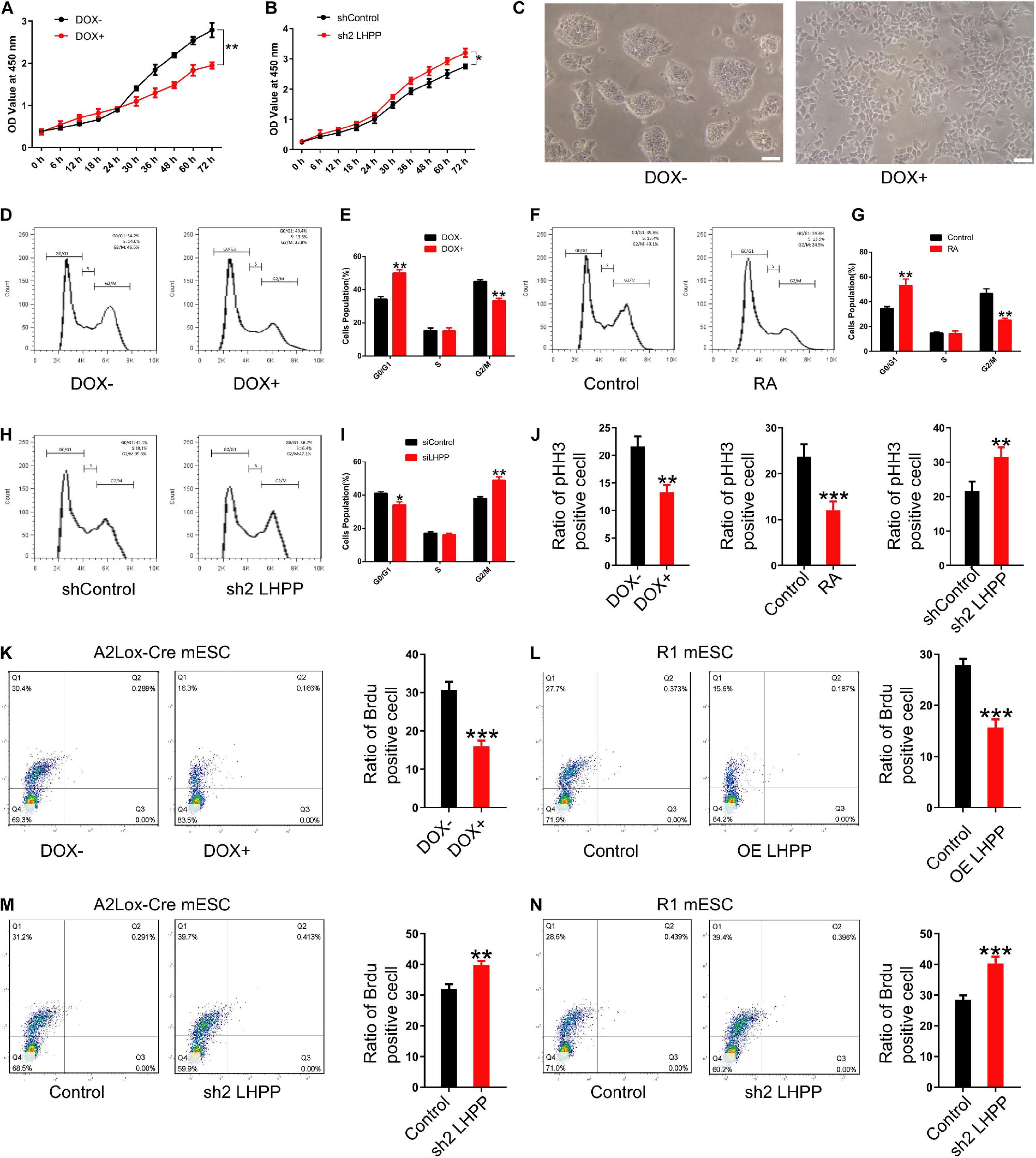
Figure 6. Effect of LHPP on mESC proliferation and cell cycle. (A,B) The Cell Counting Kit-8 assay was used to detect the proliferation of (A) mESCs following treatment with DOX (1 μg/ml) and (B) shRNA targeting Lhpp. (C) mES cell colonies following DOX treatment (1 μg/ml) for 72 h. Scale bar = 100 μm. (D,E) Flow cytometry was conducted to detect changes to the cell cycle in mESCs treated with DOX (1 μg/ml) for 3 days. (E) Quantification. (F,G) RA-treated (10 mM) cells were used as the positive control. Cells without DOX treatment (1 μg/ml) were used as the negative control group. (G) Quantification. (H,I) Flow cytometry was used to determine the changes in the cell cycle in Lhpp knockdown mESCs cultured for 3 days, respectively. (I) Quantification. (J) The percentage of pHH3-positive cells in mESCs. (K,L) The percentage of Brdu-positive cells in (K) DOX-induced (1 μg/ml) LHPP overexpressing mESCs and (L) R1 mESCs with LHPP overexpression. (M,N) The percentage of Brdu-positive cells in Lhpp knockdown (M) A2Lox-Cre mESCs and (N) R1 mESCs. All experiments were independently repeated at least three times, and the data are presented as the mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001. LHPP, phospholysine phosphohistidine inorganic pyrophosphate phosphatase; mESCs, mouse embryonic stem cells; DOX, doxycycline; RA, retinoic acid; shRNA, short hairpin RNA.
We further repeated the above experiments with LIF-free culture medium. Data indicated that the proliferation and cell cycle of mESCs were affected after culturing with LIF-free medium for 3 days, in comparison to cells treated with medium containing LIF; besides, when the cells were cultured in LIF-free medium, the inhibitory effect of DOX-induced LHPP-overexpression on the cell cycle was enhanced, resulting in a slower proliferation of mESCs (Supplementary Figures 4A,C,D). Nevertheless, the proliferation of Lhpp knockdown mESCs cultured in LIF-free medium were promoted (Supplementary Figure 4B) and cell cycle was changed significantly as in LIF medium (Supplementary Figures 4E,F). Collectively, our findings suggest that LHPP suppresses the proliferation of mESCs and induces cell cycle arrest, and that these effects are enhanced in the absence of LIF treatment.
LHPP-Mediated Histidine Dephosphorylation Regulates the Expression of Wnt Signaling Pathway-Related Molecules and Cell Cycle-Related Genes
Mechanistically, we detected the expression of Wnt signaling pathway-related molecules and cell cycle-related genes. Data showed that the mRNA (Supplementary Figure 5A) and protein (Figure 7) expression levels of β-catenin, and the cell cycle-promoting genes CDK4 and CyclinD1 were significantly attenuated, whilst those of the cell cycle-inhibitory genes P21 and P27 were markedly increased in DOX-induced LHPP-overexpressing mESCs, in comparison to the control cells (when both were cultivated with medium containing LIF). By contrast, Lhpp knockdown produced the opposite results (Supplementary Figure 5B).
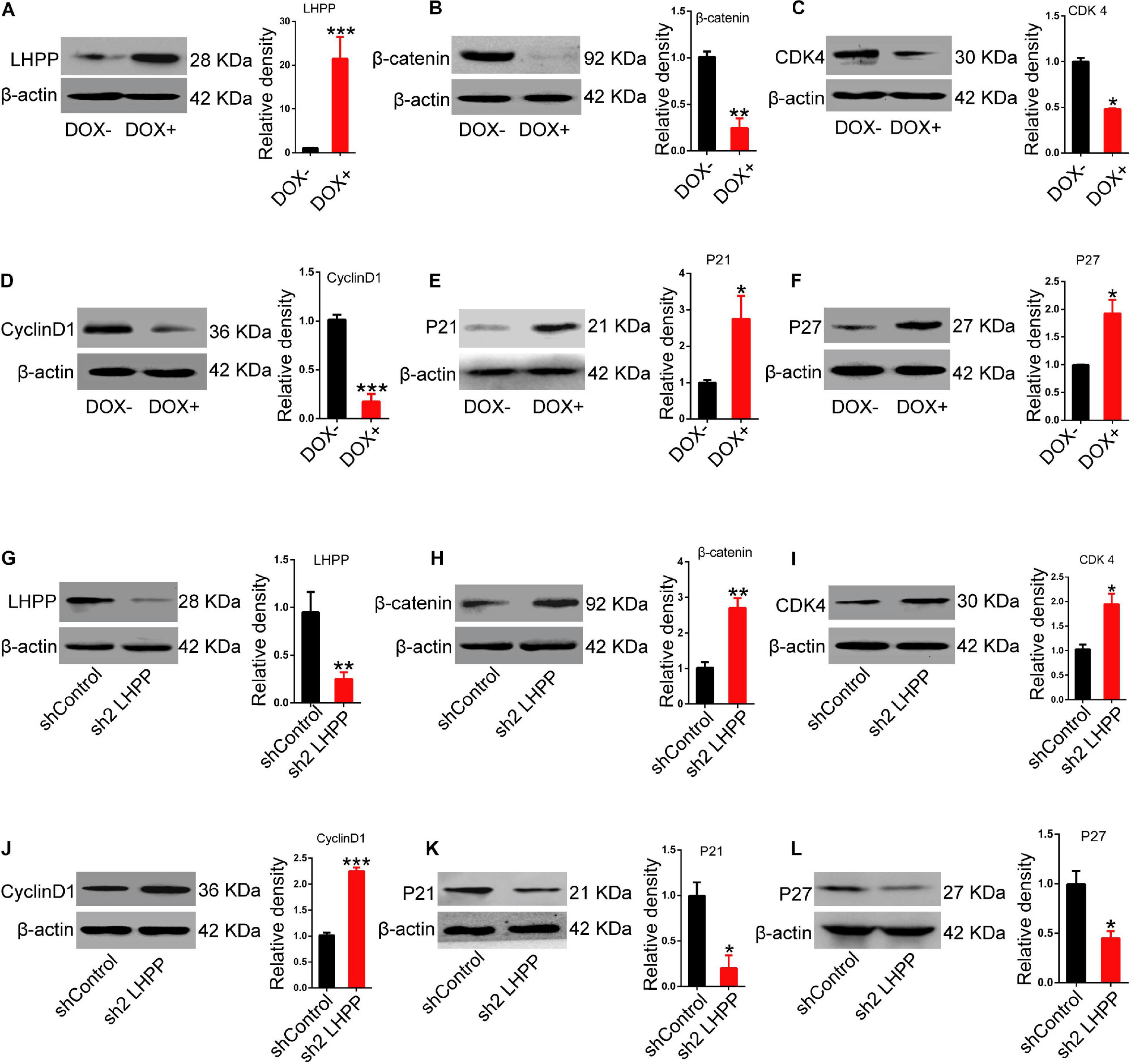
Figure 7. Effect of LHPP-overexpression on the expression levels of Wnt signaling pathway-related molecules and cell cycle-related genes. (A–F) The protein expression levels of (A) hLHPP, (B) β-catenin, (C) CDK4, (D) CyclinD1, (E) P21, and (F) P27 in A2Lox-Cre mESCs treated with DOX (1 μg/ml) for 72 h were detected by western blotting. (G–L) The protein expression levels of (G) mLHPP, (H) β-catenin, (I) CDK4, (J) CyclinD1, (K) P21, and (L) P27 in Lhpp-silenced A2Lox-Cre mESCs cultured for 72 h. All experiments were independently repeated at least three times, and the data are presented as the mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001. LHPP, phospholysine phosphohistidine inorganic pyrophosphate phosphatase; hLHPP, human LHPP; mLHPP, mouse LHPP; mESCs, mouse embryonic stem cells; DOX, doxycycline; shRNA, short hairpin RNA.
Discussion
In this study, our findings suggested that Lhpp expression was up-regulated during mouse ESCs differentiation. Via construction of DOX-induced LHPP-overexpressing and LHPP-silenced mouse ESC lines, we further found that Lhpp overexpression contributed to the down-regulation of histidine phosphorylation. Importantly, data indicated that the enzymatic active site of LHPP is the cysteine residue at position 226, not 53. Moreover, LHPP-mediated histidine dephosphorylation modulates the proliferation of mESCs by cell cycle arrest, where the expression of pluripotent- and differentiation-related genes, Wnt signaling pathway-related molecules and cell cycle-related genes were altered as well. Overall, our data provide new insight into how LHPP influences ESCs self-renewal.
Embryonic stem cells are capable of self-renewal and have the potential to differentiate into a variety of different cell types. Their proliferation (self-renewal) and differentiation are precisely regulated at the transcriptional, translational and post-translational levels in a state of dynamic equilibrium (Si et al., 2016; Liou et al., 2017; Ran et al., 2017). Mounting evidences have revealed that post-transcriptional modification plays a vital role in ESCs self-renewal and differentiation. However, the exact molecular mechanisms remain poorly studied. Previous studies have shown that post-translational modifications, such as protein ubiquitination and phosphorylation, are important components of the ESCs self-renewal regulatory network. For protein phosphorylation, most studies have focused on the phosphorylation of serine, threonine and tyrosine residues. However, histidine can also undergo phosphorylation modification. LHPP was identified as a protein histidine phosphatase, which may act as a liver cancer suppressor gene (Hindupur et al., 2018). Mechanistically, LHPP regulates the expression and function of a series of genes via histidine dephosphorylation in cells, thereby inhibiting cellular proliferation (Hindupur et al., 2018; Zheng et al., 2018). Previous studies have reported that LHPP is associated with a variety of neurological diseases (Converge consortium, 2015; Polimanti et al., 2017), and is also implicated in the development of hematological cancers (Gutierrez-Camino et al., 2017; Vijayakrishnan et al., 2017). However, whether LHPP-mediated histidine dephosphorylation can regulate the biological behavior of ESCs remains largely unknown.
In present study, Lhpp expression was markedly upregulated after myocardial differentiation of mESCs or mESCs treated with RA, at both the transcriptional and translational levels. However, the role of Lhpp expression and LHPP-mediated histidine dephosphorylation in ESCs cells deserve further investigation. Herein, we constructed a DOX-induced human LHPP-overexpression mESCs via homologous recombination, as previously reported (Iacovino et al., 2014; Zhang et al., 2016). An shLHPP ESC line was also constructed. Western blotting and immunofluorescence assays demonstrated that following LHPP-overexpression, the level of histidine phosphorylation in ESCs was significantly downregulated. Furthermore, consistent with the observations in the positive control group (RA treatment group), colonies of ESCs overexpressing LHPP were dispersed and the levels of AP were attenuated, suggesting that LHPP inhibits ESCs proliferation. Furthermore, our data revealed that the expression levels of Oct4 and Lefty1 were significantly downregulated, while the expression levels of genes related to the differentiation were markedly upregulated. By contrast, Lhpp silencing was able to reverse the gene expression mentioned above. Our findings indicated that LHPP-mediated histidine dephosphorylation may suppress ESCs self-renewal.
In further experiments, the CCK-8 assay was used to validate the observation that LHPP overexpression significantly inhibited ESCs proliferation, whereas LHPP silencing promoted ESCs proliferation. Cell cycle assays revealed a significant change in the cell cycle distribution of mESCs overexpressing LHPP, and a marked increase in the number of cells in the G0/G1 phase, which was consistent with the results in ESCs cells treated with RA. However, in Lhpp knockdown ESCs, an increased proportion of G2/M phase cells was observed.
In a further mechanistic study, it was found that LHPP-mediated histidine dephosphorylation significantly downregulated the mRNA and protein expression levels of CDK4 and CyclinD1, which promote cell cycle progression, whereas the expression of the cell cycle-related inhibitory genes P21 and P27 was significantly increased. Although previous studies have identified multiple target proteins of LHPP via mass spectrometry, including essential fatty acid synthetase ATP-citrate lyase (ACLY) (Hindupur et al., 2018), how LHPP exerts its biological function through its substrates, which needed future studies. Interestingly, the expression level of β-catenin was considerably downregulated in our study. β-catenin is a key protein of the Wnt signaling pathway (Clevers and Nusse, 2012; Srivastava et al., 2018) that can trigger the expression of a series of genes, including CyclinD1 and CDK4, which subsequently regulate the expression of genes associated with the cell cycle (Yang et al., 2014; Tao et al., 2017). Wnt signaling pathway is one of the mechanisms affecting stem cell proliferation and differentiation (Xu et al., 2016; Selvaraj et al., 2017). Therefore, we hypothesized that LHPP affects the proliferation and differentiation of ESCs (and potentially human pluripotent stem cells) by regulating Wnt signaling, which warrants further investigation.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
All animal experiments were conducted with the approval of the animal ethics and use committee (IACUC) of Xiamen University (Xiamen, Fujian, China; Approval ID: scxk2013-0006).
Author Contributions
RX: conception and design, collection and/or assembly of data, data analysis and interpretation, and manuscript writing. DY and XC: collection and/or assembly of data. XX: conception and design, financial support, administrative support, manuscript writing, and final approval of manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key R&D program of China (Grant No. 2018YFA0107304), National Natural Science Foundation of China (NSFC) (Grant No. 81871744), the Fujian Provincial Health Education Joint Research Project (Grant No. 2019-WJ-16), and the Fundamental Research Funds for the Central Universities (Grant No. 20720200059).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Dr. Michael Kyba for kind donation of the A2Lox ESC (A2Lox-Cre mESC). We would like to thank all members of the Institute of Stem Cell and Regenerative Medicine, School of Medicine of Xiamen University for their assistances in this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.638815/full#supplementary-material
Supplementary Figure 1 | The effects of LHPP overexpression or silencing on gene expressions. (A–C) qPCR and western blotting quantification of (A) mLhpp, (B) Oct4, and (C) Lefty1 levels in A2Lox-Cre mESCs treated with LIF alone or in combination with DOX (1 μg/ml). (D) qPCR quantification of the mRNA levels of SOX17, Afp, T-Bra, Mesp1, Nestin, and SOX1 in mESCs treated with LIF alone or in combination with DOX (1 μg/ml). (E,F) qPCR detection of the mRNA levels of SOX17, Afp, T-Bra, Mesp1, Nestin and SOX1 (E) in mESCs transfected with shRNA targeting Lhpp, or (F) in A2Lox-Cre mESCs treated with RA (10 mM). All experiments were independently repeated at least three times, and the data are represented as the mean ± SD. ∗P < 0.05, **P < 0.01, ***P < 0.001. LHPP, phospholysine phosphohistidine inorganic pyrophosphate phosphatase; mLHPP, mouse LHPP; mESCs, mouse embryonic stem cells; DOX, doxycycline; LIF, leukemia inhibitory factor; RA, retinoic acid.
Supplementary Figure 2 | The expression of Lefty1 and Oct4 in Lhpp-overexpressed and silenced R1 mESCs. (A) Lefty1 and Oct4 mRNA expression were downregulated in Lhpp-overexpressed R1 mESCs. (B) Lefty1 and Oct4 mRNA expression were upregulated in L Lhpp-silenced R1 mESCs. All experiments were independently repeated at least three times, and the data are represented as the mean ± SD. *P < 0.05, **P < 0.01. LHPP, phospholysine phosphohistidine inorganic pyrophosphate phosphatase; mESCs, mouse embryonic stem cells.
Supplementary Figure 3 | Schematic diagram of the mutation sites in Lhpp. (A) Sanger sequencing confirmed the successful mutation of LHPP at position 158, labeled as LHPP (c.158G > C). The corresponding amino acid sequence is LHPP (C53S). (B) Sanger sequencing showed the successful mutation of LHPP at position 677, labeled LHPP (c.677G > C). The corresponding amino acid sequence is LHPP (C226S). The mutation sites are colored in red.
Supplementary Figure 4 | Effect of LHPP on proliferation and cell cycle of mESCs. (A,B) Cell Counting Kit-8 assay was used to detect the proliferation of (A) A2Lox-Cre mESCs following treatment with DOX (1 μg/ml) alone or in combination with LIF and (B) Lhpp knockdown mESCs cultured with LIF-free medium. (C,D) Flow cytometry was used to detect changes to the cell cycle in mESCs cultured with LIF-free medium, with or without DOX (1 μg/ml) for 3 days, respectively. (D) Quantification. (E,F) Flow cytometry was used to determine the changes in the cell cycle in Lhpp knockdown mESCs cultured with LIF-free medium or medium containing LIF for 3 days. (F) Quantification. All experiments were independently repeated at least three times, and the data are presented as the mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001. LHPP, phospholysine phosphohistidine inorganic pyrophosphate phosphatase; mESCs, mouse embryonic stem cells; DOX, doxycycline; LIF, leukemia inhibitory factor.
Supplementary Figure 5 | Effects of Lhpp overexpression or knockdown on gene expressions in mESCs. (A,B) The mRNA levels of hLhpp, mLhpp, β-catenin, CDK4, CyclinD1, P21 and P27 (A) in mESCs treated with DOX (1 μg/ml) for 72 h and (B) in Lhpp-silenced mESCs cultured for 72 h, respectively, were detected by qPCR. *P < 0.05, **P < 0.01. LHPP, phospholysine phosphohistidine inorganic pyrophosphate phosphatase; hLhpp, human Lhpp; mLhpp, mouse Lhpp; mESCs, mouse embryonic stem cells; DOX, doxycyclin.
References
Abu-Dawud, R., Graffmann, N., Ferber, S., Wruck, W., and Adjaye, J. (2018). Pluripotent stem cells: induction and self-renewal. Philos. Trans. R. Soc. Lond. B Biol. Sci. 373:20170213. doi: 10.1098/rstb.2017.0213
Adamo, A., Sese, B., Boue, S., Castano, J., Paramonov, I., Barrero, M. J., et al. (2011). LSD1 regulates the balance between self-renewal and differentiation in human embryonic stem cells. Nat. Cell Biol. 13, 652–659. doi: 10.1038/ncb2246
Boyer, P. D., Deluca, M., Ebner, K. E., Hultquist, D. E., and Peter, J. B. (1962). Identification of phosphohistidine in digests from a probable intermediate of oxidative phosphorylation. J. Biol. Chem. 237, PC3306–PC3308.
Cassar, P. A., and Stanford, W. L. (2012). Integrating post-transcriptional regulation into the embryonic stem cell gene regulatory network. J. Cell. Physiol. 227, 439–449. doi: 10.1002/jcp.22787
Clevers, H., and Nusse, R. (2012). Wnt/beta-catenin signaling and disease. Cell 149, 1192–1205. doi: 10.1016/j.cell.2012.05.012
Converge consortium. (2015). Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature 523, 588–591. doi: 10.1038/nature14659
Dogan, A. (2018). Embryonic stem cells in development and regenerative medicine. Adv. Exp. Med. Biol. 1079, 1–15. doi: 10.1007/5584_2018_175
Fuhs, S. R., Meisenhelder, J., Aslanian, A., Ma, L., Zagorska, A., Stankova, M., et al. (2015). Monoclonal 1- and 3-phosphohistidine antibodies: new tools to study histidine phosphorylation. Cell 162, 198–210. doi: 10.1016/j.cell.2015.05.046
Gabut, M., Samavarchi-Tehrani, P., Wang, X., Slobodeniuc, V., O’Hanlon, D., Sung, H. K., et al. (2011). An alternative splicing switch regulates embryonic stem cell pluripotency and reprogramming. Cell 147, 132–146. doi: 10.1016/j.cell.2011.08.023
Gutierrez-Camino, A., Martin-Guerrero, I., and Garcia-Orad, A. (2017). Genetic susceptibility in childhood acute lymphoblastic leukemia. Med. Oncol. 34:179. doi: 10.1007/s12032-017-1038-7
Han, H., Irimia, M., Ross, P. J., Sung, H. K., Alipanahi, B., David, L., et al. (2013). MBNL proteins repress ES-cell-specific alternative splicing and reprogramming. Nature 498, 241–245. doi: 10.1038/nature12270
Hindupur, S. K., Colombi, M., Fuhs, S. R., Matter, M. S., Guri, Y., Adam, K., et al. (2018). The protein histidine phosphatase LHPP is a tumour suppressor. Nature 555, 678–682. doi: 10.1038/nature26140
Huang, G., Ye, S., Zhou, X., Liu, D., and Ying, Q. L. (2015). Molecular basis of embryonic stem cell self-renewal: from signaling pathways to pluripotency network. Cell. Mol. Life Sci. 72, 1741–1757. doi: 10.1007/s00018-015-1833-2
Iacovino, M., Roth, M. E., and Kyba, M. (2014). Rapid genetic modification of mouse embryonic stem cells by inducible cassette exchange recombination. Methods Mol. Biol. 1101, 339–351. doi: 10.1007/978-1-62703-721-1_16
Kyba, M., Perlingeiro, R. C., and Daley, G. Q. (2002). HoxB4 confers definitive lymphoid-myeloid engraftment potential on embryonic stem cell and yolk sac hematopoietic progenitors. Cell 109, 29–37. doi: 10.1016/s0092-8674(02)00680-3
Li, L., Zhang, L., Liu, G., Feng, R., Jiang, Y., Yang, L., et al. (2014). Synergistic transcriptional and post-transcriptional regulation of ESC characteristics by core pluripotency transcription factors in protein-protein interaction networks. PLoS One 9:e105180. doi: 10.1371/journal.pone.0105180
Liou, J. Y., Ko, B. S., and Chang, T. C. (2017). An efficient transfection method for differentiation and cell proliferation of mouse embryonic stem cells. Methods Mol. Biol. 1622, 139–147. doi: 10.1007/978-1-4939-7108-4_11
Liu, X., Yao, Y., Ding, H., Han, C., Chen, Y., Zhang, Y., et al. (2016). USP21 deubiquitylates Nanog to regulate protein stability and stem cell pluripotency. Signal Transduct. Target. Ther. 1:16024. doi: 10.1038/sigtrans.2016.24
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Polimanti, R., Wang, Q., Meda, S. A., Patel, K. T., Pearlson, G. D., Zhao, H., et al. (2017). The interplay between risky sexual behaviors and alcohol dependence: genome-wide association and neuroimaging support for LHPP as a risk gene. Neuropsychopharmacology 42, 598–605. doi: 10.1038/npp.2016.153
Ran, X., Xiao, C. H., Xiang, G. M., and Ran, X. Z. (2017). Regulation of embryonic stem cell self-renewal and differentiation by microRNAs. Cell. Reprogram. 19, 150–158. doi: 10.1089/cell.2016.0048
Sampath, P., Pritchard, D. K., Pabon, L., Reinecke, H., Schwartz, S. M., Morris, D. R., et al. (2008). A hierarchical network controls protein translation during murine embryonic stem cell self-renewal and differentiation. Cell Stem Cell 2, 448–460. doi: 10.1016/j.stem.2008.03.013
Selvaraj, P., Xiao, L., Lee, C., Murthy, S. R., Cawley, N. X., Lane, M., et al. (2017). Neurotrophic factor-alpha1: a key Wnt-beta-catenin dependent anti-proliferation factor and ERK-Sox9 activated inducer of embryonic neural stem cell differentiation to astrocytes in neurodevelopment. Stem Cells 35, 557–571. doi: 10.1002/stem.2511
Si, Y., Zhu, J., Huang, X., Zhu, P., and Xie, C. (2016). Effects of Panax notoginseng saponins on proliferation and differentiation of rat embryonic cortical neural stem cells. J. Chin. Med. Assoc. 79, 256–263. doi: 10.1016/j.jcma.2015.10.011
Srivastava, S., Li, Z., Soomro, I., Sun, Y., Wang, J., Bao, L., et al. (2018). Regulation of KATP channel trafficking in pancreatic beta-cells by protein histidine phosphorylation. Diabetes 67, 849–860. doi: 10.2337/db17-1433
Tao, J., Zhang, R., Singh, S., Poddar, M., Xu, E., Oertel, M., et al. (2017). Targeting beta-catenin in hepatocellular cancers induced by coexpression of mutant beta-catenin and K-Ras in mice. Hepatology 65, 1581–1599. doi: 10.1002/hep.28975
Taracha, A., Kotarba, G., and Wilanowski, T. (2017). [Methods of analysis of protein phosphorylation]. Postepy Biochem. 63, 137–142.
Vijayakrishnan, J., Kumar, R., Henrion, M. Y., Moorman, A. V., Rachakonda, P. S., Hosen, I., et al. (2017). A genome-wide association study identifies risk loci for childhood acute lymphoblastic leukemia at 10q26.13 and 12q23.1. Leukemia 31, 573–579. doi: 10.1038/leu.2016.271
Wang, S., Li, Z., Shen, H., Zhang, Z., Yin, Y., Wang, Q., et al. (2016). Quantitative phosphoproteomic study reveals that protein kinase a regulates neural stem cell differentiation through phosphorylation of catenin Beta-1 and glycogen synthase kinase 3beta. Stem Cells 34, 2090–2101. doi: 10.1002/stem.2387
Watanabe, N., and Osada, H. (2016). Small molecules that target phosphorylation dependent protein-protein interaction. Bioorg. Med. Chem. 24, 3246–3254. doi: 10.1016/j.bmc.2016.03.023
Wen, J., Lv, R., Ma, H., Shen, H., He, C., Wang, J., et al. (2018). Zc3h13 regulates nuclear RNA m(6)A methylation and mouse embryonic stem cell self-renewal. Mol. Cell 69, 1028–1038.e6. doi: 10.1016/j.molcel.2018.02.015
Whyte, W. A., Bilodeau, S., Orlando, D. A., Hoke, H. A., Frampton, G. M., Foster, C. T., et al. (2012). Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature 482, 221–225. doi: 10.1038/nature10805
Xu, Z., Robitaille, A. M., Berndt, J. D., Davidson, K. C., Fischer, K. A., Mathieu, J., et al. (2016). Wnt/beta-catenin signaling promotes self-renewal and inhibits the primed state transition in naive human embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 113, E6382–E6390. doi: 10.1073/pnas.1613849113
Yang, J., Mowry, L. E., Nejak-Bowen, K. N., Okabe, H., Diegel, C. R., Lang, R. A., et al. (2014). beta-catenin signaling in murine liver zonation and regeneration: a Wnt-Wnt situation! Hepatology 60, 964–976. doi: 10.1002/hep.27082
Yokoi, F., Hiraishi, H., and Izuhara, K. (2003). Molecular cloning of a cDNA for the human phospholysine phosphohistidine inorganic pyrophosphate phosphatase. J. Biochem. 133, 607–614. doi: 10.1093/jb/mvg078
Zhang, T., Lin, Y., Liu, J., Zhang, Z. G., Fu, W., Guo, L. Y., et al. (2016). Rbm24 regulates alternative splicing switch in embryonic stem cell cardiac lineage differentiation. Stem Cells 34, 1776–1789. doi: 10.1002/stem.2366
Zhao, H., Han, Z., Liu, X., Gu, J., Tang, F., Wei, G., et al. (2017). The chromatin remodeler Chd4 maintains embryonic stem cell identity by controlling pluripotency- and differentiation-associated genes. J. Biol. Chem. 292, 8507–8519. doi: 10.1074/jbc.M116.770248
Keywords: embryonic stem cell, self-renewal, histidine dephosphorylation, LHPP, post-translational modification
Citation: Xia RM, Yao DB, Cai XM and Xu XQ (2021) LHPP-Mediated Histidine Dephosphorylation Suppresses the Self-Renewal of Mouse Embryonic Stem Cells. Front. Cell Dev. Biol. 9:638815. doi: 10.3389/fcell.2021.638815
Received: 07 December 2020; Accepted: 25 February 2021;
Published: 16 March 2021.
Edited by:
Masatake Osawa, Gifu University, JapanReviewed by:
Yangming Wang, Peking University, ChinaLeonardo Romorini, Fundación Para la Lucha Contra las Enfermedades Neurológicas de la Infancia (FLENI), Argentina
Copyright © 2021 Xia, Yao, Cai and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiu Qin Xu, eHV4cUB4bXUuZWR1LmNu
 Rong Mu Xia
Rong Mu Xia Dong Bo Yao
Dong Bo Yao