
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol. , 23 March 2021
Sec. Signaling
Volume 9 - 2021 | https://doi.org/10.3389/fcell.2021.636595
The signal transducer and activator of transcription 1 (STAT1) is a transducer protein and acts as a transcription factor but its role in ovarian cancer (OC) is not completely understood. Practically, there are two-faced effects of STAT1 on tumorigenesis in different kinds of cancers. Existing evidence reveals that STAT1 has both tumor-suppressing and tumor-promoting functions involved in angiogenesis, cell proliferation, migration, invasion, apoptosis, drug resistance, stemness, and immune responses mainly through interacting and regulating target genes at multiple levels. The canonical STAT1 signaling pathway shows that STAT1 is phosphorylated and activated by the receptor-activated kinases such as Janus kinase in response to interferon stimulation. The STAT1 signaling can also be crosstalk with other signaling such as transforming growth factor-β signaling involved in cancer cell behavior. OC is often diagnosed at an advanced stage due to symptomless or atypical symptoms and the lack of effective detection at an early stage. Furthermore, patients with OC often develop chemoresistance and recurrence. This review focuses on the multi-faced role of STAT1 and highlights the molecular mechanisms and biological functions of STAT1 in OC.
The signal transducer and activator of transcription (STAT) protein is an essential component of the interferon (IFN)/Janus kinase (JAK) signaling. There are at least seven members in the STAT family, including STAT1, 2, 3, 4, 5A, 5B, and 6, involved in immune surveillance, defense, and homeostasis (Ihle, 2001; O’Shea et al., 2002). STAT1, also known as STAT91, is the first discovered member of the STAT family but it is less understood in tumorigenesis compared to STAT3, a most well-studied member in this family. The gene of STAT1 is located in 2q32.2 and contains 45,215 bp (Yamamoto et al., 1997). STAT1 protein generally exists as an inactive form in the cytoplasm (Zhang and Liu, 2017). In the canonical signaling pathway, STAT1 is initially phosphorylated and activated by the receptor-activated kinases such as JAK in response to IFN stimulation (Ihle and Kerr, 1995). The activated STAT1 forms homodimer or heterodimers with other STATs and translocates from the cytosol to the nucleus where it acts as a transcription factor to regulate its target genes.
Ovarian cancer (OC) is the most lethal gynecological disease in women worldwide. In the United States, 21,750 new-diagnosed cases and 13,940 death cases are estimated to occur in 2020 (Siegel et al., 2020). Because of symptomless or atypical symptoms at the early stage, patients with OC are often diagnosed at an advanced stage due to the lack of effective detection at an early stage, chemoresistance, and even recurrence (Torre et al., 2018). Although the mortality of OC patients keeps a stable or trend down for decades (Malvezzi et al., 2016), the prognosis of OC is still not significantly improved. Therefore, increasing the efficiency of prevention, early diagnosis, and more effective treatment for OC patients is most important (Natanzon et al., 2018; Walker and Sobel, 2018).
The mechanisms of OC development can generally be interpreted by the hallmarks of cancer (Hanahan and Weinberg, 2011), regarding the incidence, progression, recurrence, and drug-resistance (Matulonis, 2018). Hence, most therapeutic drugs are designed to target those molecules involved in tumorigenesis and tumor development (Hainaut and Plymoth, 2013). For example, many inhibitors of poly (ADP-ribose) polymerase (PARP), angiogenesis, RAS/RAF/MEK pathway, PI3K/AKT pathway, and immune checkpoints are currently available to treat patients with OC (Guan and Lu, 2018). In addition to these well-studied pathways and cellular processes, some molecules also contribute to OC progression, including ATP-binding cassette (ABC) transporters, astrocyte-elevated gene-1 (AEG1), BRCA1, BRCA2, CCNG1, C-X-C motif receptor 2 (CXCR2), IGF1R, p53, STAT1, and STAT3 (Blanco et al., 2010; Devapatla et al., 2015; Ween et al., 2015; Zhang and Zou, 2015; Zhou et al., 2015; Liu et al., 2018; Xu et al., 2019; Liang et al., 2020). Most of them are potential therapeutic targets but have not been developed as drugs tested in the clinical trial. Among different molecules indicated, STAT1 has emerged recently as an important STAT. Existing evidence reveals that STAT1 has both tumor-suppressing and tumor-promoting functions involved in angiogenesis, cell proliferation, migration, invasion, apoptosis, drug resistance, stemness, and immune responses mainly through interacting and regulating target genes at multiple levels. More and more studies of STAT1 in cancer unveil the mechanisms of tumor development and focus on the target of the immune response. However, STAT1 has two-faced effects on tumorigenesis and this dual role of STAT1 in OC is not completely understood (Zhang and Liu, 2017).
The gene of STAT1 contains 25 exons (Figure 1A). Because of alternative splicing (Schindler et al., 1992), STAT1 has two transcripts: a full-length STAT1α and a truncated STAT1β (Figure 1B). STAT1α is the main form that has 750 amino acids, whereas STAT1β is a short form that has 712 amino acids (Figure 1C). STAT1α has two important phosphorylation sites at residues tyrosine 701 (Y701) and serine 727 (S727) located at the C-terminal transactivation domain (TAD) (Figure 1D), whereas STAT1β lacks part of the TAD but can be efficiently phosphorylated on Y701 (Oda et al., 2015).

Figure 1. Structure of STAT1. (A) Genomic illustration of STAT1. The STAT1 gene is located in 2q32.2 containing 45,215 bp. There are 25 exons in STAT1α and 23 exons in STAT1β. (B) Transcripts of STAT1α and STAT1β. The mRNA of STAT1α and STAT1β contains 4116 and 2718 bp, respectively. (C) Alignment of protein sequences of STAT1α (750 amino acids) and STAT1β (712 amino acids). (D) Illustration of the functional domains of STAT1. Phosphorylation sites of residues tyrosine 701 (Y701) and serine 727 (S272) are shown. ND, N-terminal domain; CCD, coiled-coil domain; DBD, DNA binding domain; LD, linker domain; SH2D, Src homology-2 domain; TAD, transactivation domain.
The structure of STAT1 protein generally consists of the N-terminal domain (ND), coiled-coil domain (CCD), DNA binding domain (DBD), linker domain (LD), Src homology-2 domain (SH2D), and TAD. From N-terminus to C-terminus, each domain has its own function. The ND is implicated in the protein-protein interaction, nuclear translocation, and deactivation of STAT1 (Morris et al., 2018). The CCD functions as a scaffold for protein-protein interaction which is cooperated with other transcription factors and co-factors (Chen et al., 1998). The DBD is the direct binding site of DNA which helps STAT1 translocation from the cytosol to the nucleus. The LD is involved in regulating transcriptional activity (Mertens et al., 2015). The SH2D is the most conserved region that specifically binds to the phosphorylated-tyrosine site of receptors and is responsible for the dimerization and maintenance of the DNA-binding stability, whereas the TAD is the transactivation site activated after Y701 and/or S727 phosphorylation (Chen et al., 1998).
Signal transducer and activator of transcription 1 can not only be activated by type I and type II IFNs in the canonical JAK/STAT1 signaling pathway but also be activated by other cytokines such as interleukins (e.g., IL-6) and growth factors [e.g., transforming growth factor-β1 (TGF-β1)] through the phosphorylation of Y701 residue in different patterns (Figure 2). As a transducer, STAT1 protein can transduce extracellular signals through transmembrane receptors. Interferon-α receptor 1 (IFNAR1) and IFNAR2 are receptors of type I IFN in response to IFN-α/-β, whereas interferon-γ receptor 1 (IFNGR1) and IFNGR2 are receptors of type II IFN in response to IFN-γ. Despite the confluence of the signals upon ligands binding to their receptors, STAT1 has distinct effects on targeted genes in different cells. The binding of IFNs to their receptors leads to the activation of receptor-associated JAK, which phosphorylates the tyrosine residue of receptors (Stark and Darnell, 2012). The phosphorylated receptors and activated JAKs recruit STAT1 by binding to the SH2D of STAT1 and phosphorylate STAT1 at Y701 residue, leading to the release of STAT1 from receptors and triggering the formation of STAT1/STAT1 homodimer, STAT1/STAT2 or STAT1/STAT3 heterodimers, and in turn, these dimers translocate from the cytosol to the nucleus where they bind to DNA binding sites of IFN-stimulated genes (ISG) (Shuai, 1994). STAT1/STAT2 heterodimer and IFN regulatory factor (IRF) 9 form a complex with ISG factor-3 (ISGF-3) (Ramana et al., 2000), which binds to the IFN-stimulated response element (ISRE) in the promoter of a target gene in response to IFN-α and IFN-β stimulation (Horvath, 2004). However, STAT1 homodimer and STAT1/STAT3 heterodimer bind to the gamma-IFN activation site (GAS), a chromatin open site in the promoter of a target gene in response to IFN-γ stimulation (Figure 2; Ramana et al., 2002).
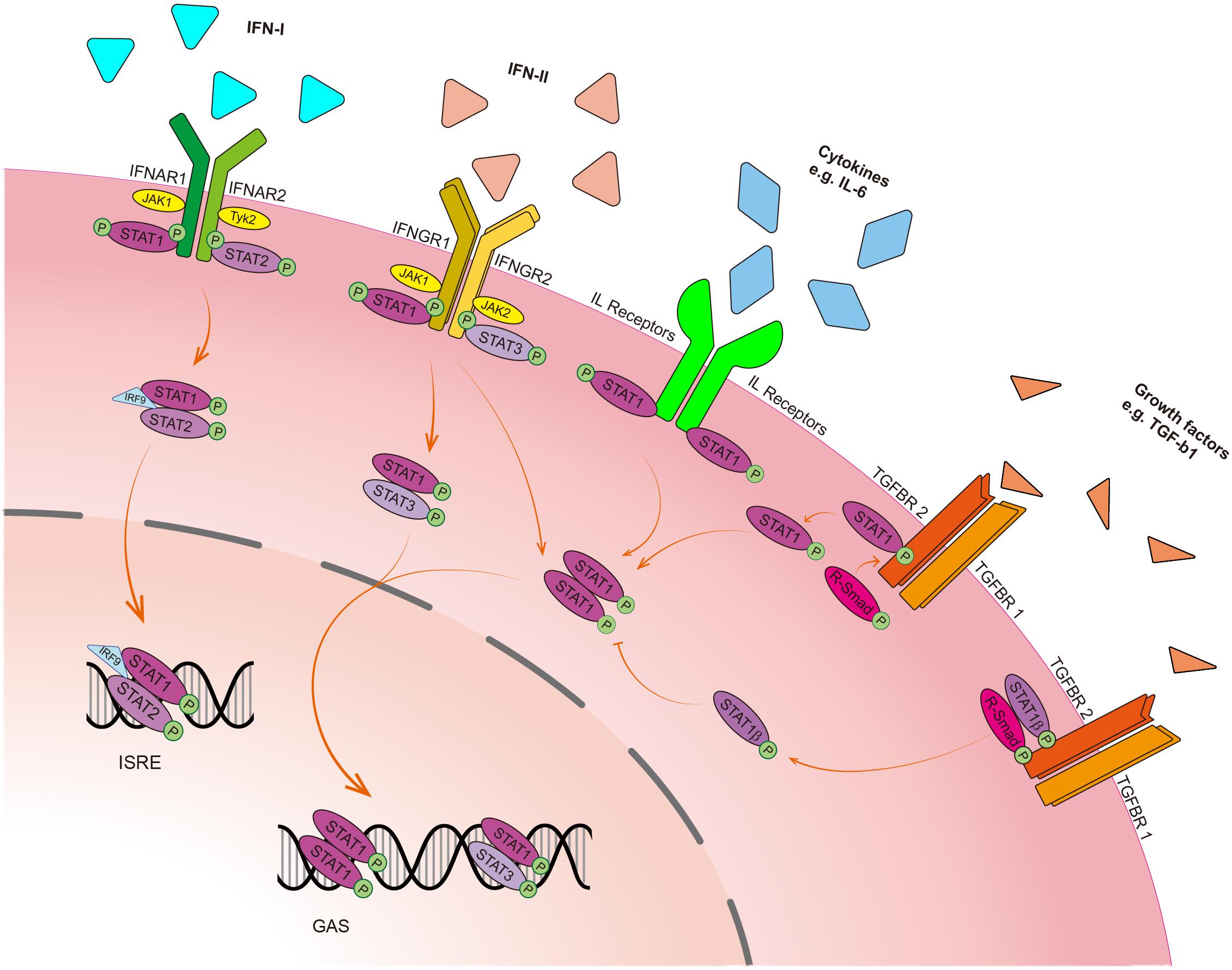
Figure 2. Schematic presentation of the STAT1 signaling pathway. In response to type I IFN (IFN-α and IFN-β), phosphorylated STAT1 binds to STAT2 and forms a heterodimer, which further forms a complex with IFN regulatory factor 9 (IRF9), along with interferon-stimulated gene factor-3 (ISGF-3), and then translocate into the nucleus to bind to the IFN-stimulated response element (ISRE) of target genes to regulate their transcription. In response to type II IFN (IFN-γ), phosphorylated STAT1 forms homodimer with STAT1 or heterodimer with STAT3. These dimers then translocate into the nucleus to bind to the gamma-IFN activation site (GAS) to perform the transcriptional activity. Cytokines such as interleukins (ILs) that bind to their receptors without the help of JAK can activate STAT1 and form STAT1 homodimer, followed by the nucleus translocation. In the crosstalk with the TGF-β signaling pathway, STAT1 constitutively interacts with TGF-β1 receptor. Upon TGF-β1 binding to its receptors (TGFBR1 and TGFBR2), the receptor-complex increases phosphorylation STAT1α on S727. Then, the activated STAT1α dissociates itself from the receptor complex and translocates into the nucleus. STAT1β blocks STAT1α homodimer formation.
The C-terminal TAD has a transactivation function and can recruit transcriptional co-factors to the promoter of IFN-γ-activated genes. For instance, the TAD of STAT1α recruits RNA polymerase II (RNA pol II) to IFN-induced guanylate-binding protein 2 (GBP2) and core Mediator complex to the GAS of the IRF1 and IRF8 promoters (Parrini et al., 2018). The TAD also functions to interact with the histone acetyltransferase CBP/p300 that promotes the DNA binding activity of STAT1 (Wojciak et al., 2009). The phosphorylation of STAT1α at S727 acquires a transactivation function (Chen et al., 1998). Cyclin-dependent kinase 8 (CDK8) has been reported to activate STAT1 by phosphorylation at S727, whereas a CDK8-specific-inhibitor can suppress STAT1 phosphorylation at S727 (Bancerek et al., 2013; Nitulescu et al., 2017). It seems that STAT1β has no transactivational function because of the absence of S727. However, despite lacking the TAD, STAT1β should not be considered as transactivation null. Studies using gene-modified mice suggest that the depletion of the TAD does not eliminate transcriptional activation in responses to IFN-γ signal. The conversion of S727 to alanine (S727A) results in a similar outcome (Semper et al., 2014), indicating that the function of the TAD does not rely on the phosphorylation of S727 alone. Furthermore, Y701 and S727 are two phosphorylated sites of STAT1 which may confer different functions of STAT1 (Nitulescu et al., 2017). Therefore, it is speculated that the ratio of STAT1α/STAT1β may govern the effects of STAT1 in tumorigenesis.
Signal transducer and activator of transcription 1 is overexpressed in OC compared with normal ovarian tissue (Tian et al., 2018; Liu et al., 2020) and is associated with the overall survival of patients (Josahkian et al., 2018). High levels of STAT1 evaluated by immunohistochemistry is associated with longer overall survival and progression-free survival of patients with high-grade serous ovarian cancer (HGSOC) (Table 1; Koti et al., 2015; Au et al., 2016; Josahkian et al., 2018). As a transcription factor and transducer protein, STAT1 can regulate its target genes and can be a target to be regulated by other molecules (Tables 2, 3). These STAT1-related regulators and mediators as well as STAT1 itself are highly important and have played roles in OC development and treatment outcome. However, the molecular mechanisms of the cellular processes underlying such regulations are complex and still not completely understood yet.

Table 1. High-level STAT1 associated with a better outcome of survival in patients with high-grade serous ovarian cancer.
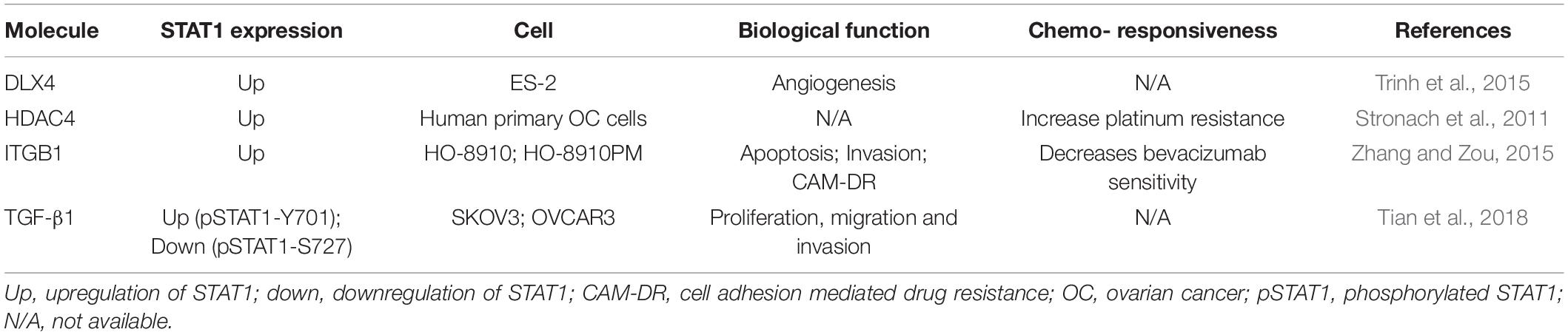
Table 3. Summary of molecules that regulate STAT1 with biological function and chemo-responsiveness in ovarian cancer cells.
It has been reported that STAT1 promotes OC cell proliferation and suppresses apoptosis by up-regulating the expression of inducible nitric oxide synthase (iNOS) in OC tissues compared with normal ovarian tissues (Burke et al., 2017). Distal-less homeobox 4 (DLX4), which is overexpressed in OC and is associated with poor prognosis of patients with OC, can directly interact with STAT1 and activate STAT1, thus triggering the expression of iNOS (Trinh et al., 2015; Kielbik et al., 2019). Furthermore, DLX4-induced iNOS expression is eliminated in STAT1 loss-of-function mutation cells (Wen et al., 1995), indicating that STAT1 is a potential transactivator of iNOS. There is evidence that STAT1α can bind to the GAS of iNOS promoter, which is essential for the transcription of iNOS (Gao et al., 1997), leading to the generation of cytotoxic nitric oxide (NO) (Kielbik et al., 2019), and in turn, promoting OC progression (Li et al., 2017). On the other hand, iNOS-induced NO can promote apoptosis by increasing the expression of p53, a well-known tumor suppression gene (Leung et al., 2008). It has been demonstrated that integrin β 1 (ITGB1), which is upregulated in OC, promotes ovarian tumor growth and progression (Yang et al., 2014) and inhibits apoptosis by upregulating STAT1 expression. Silencing ITGB1 enhances the effect of bevacizumab treatment through inhibiting STAT1 (Zhang and Zou, 2015).
Signal transducer and activator of transcription 1 has been identified to promote cell adhesion, invasion, and migration in a variety of cancers (Greenwood et al., 2012) supported by most studies using adhesion assay, wound healing assay, transwell assay, and Western blot analyses to detect invasion markers such as matrix metallopeptidase 2 (MMP-2) and MMP-9 (Zhang and Zou, 2015). However, the detailed molecular mechanisms underlying STAT1-induced invasion and migration in OC remain unclear. As previously mentioned that ITGB1 is upregulated in OC and promotes ovarian tumor growth and progression (Yang et al., 2014), ITGB1 can upregulate STAT1 through activating focal adhesion kinase (FAK), the downstream kinase of ITGB1, leading to the upregulation of MMP-2 and MMP-9. Besides, STAT1 induces OC cell invasion and migration by suppressing the TGF-β signaling pathway (Tian et al., 2018).
Paradoxically, STAT1 has also been reported to inhibit metastasis of OC. Enhanced transcriptional levels of chemokine C-X-C motif ligand (CXCL) 9 (CXCL9), CXCL10, and CXCL11 are found in STAT1-overexpressed OC cells and may be related to longer overall survival and progression-free survival (Groom and Luster, 2011; Bronger et al., 2016; Au et al., 2017). Apart from that, a study using the OC ID8 cell inoculation mouse model further indicates that CXCL10 reduces ascites formation and tumor burden (Au et al., 2017). Another study using a murine cancer model demonstrates that the release of CXCL9/10 hampers tumor growth and metastasis (Kundu et al., 2005).
Signal transducer and activator of transcription 1 has been reported to be a key regulatory factor of angiogenesis (Hsu et al., 2017) that is one of the hallmarks of cancer (Hanahan and Weinberg, 2011). A study demonstrates that tumor necrosis factor superfamily 15 (TNFSF15) inhibits angiogenesis through IFN-γ-stimulated STAT1 signal (Lu et al., 2014). TNFSF15 functions as a negative modulator of neovascularization by balancing the VEGF/VEGFR ratio (Yang and Li, 2018). Besides, silencing TNFSF15 in the OC ID8 cell inoculation mouse model indeed promotes neovascularization and tumor growth (Deng et al., 2012). Antibody targeting IFN-γ receptor has proved to decrease TNFSF15 expression through IFN-γ/STAT1 signaling. It has been shown that DLX4 promotes proliferation, suppresses apoptosis, and increases microvessel density in OC xenograft models (Trinh et al., 2015). Moreover, DLX4 induces the expression of iNOS by stimulating STAT1 activity, leading to an increase in NO. The major mechanism underlying NO-induced-angiogenesis is the stimulation of VEGF-A (Frank et al., 1999) which can promote angiogenesis and form ascites in OC (Byrne et al., 2003). Furthermore, STAT1 has been proved to suppress neovascularization. For instance, STAT1 upregulates the expression of CXCL9/10/11 that have anti-angiogenesis activities in OC (Weinlander et al., 2008). Another study demonstrates that STAT1 suppresses angiogenesis via inhibiting ITGB1-induced endothelial cell migration (Zhang and Zou, 2015).
Some genes in tumorigenesis are associated with glucose metabolism processes including glycolysis, tricarboxylic acid (TCA) cycle, and oxidative phosphorylation. STAT1 is one of such genes to be essential for the promotion of glycolysis (Pitroda et al., 2009) that takes place under oxygen or hypoxia conditions (Akram, 2013). In normal cells, glycolysis is the first step to generate pyruvate in the presence of oxygen, which is oxidized to CO2 and H2O thereafter (Lunt and Vander Heiden, 2011). On the other hand, cancer cells undergo glycolysis with the increased production of lactate in the presence of oxygen. This interesting phenomenon is termed as Warburg effect or aerobic glycolysis which has been demonstrated in various types of tumors (Lebelo et al., 2019). Cancer cells consume a large amount of intake-glucose through glycolysis in the presence of oxygen. Therefore, glycolysis, also called energy reprogramming, is considered as one of the emerging hallmarks of cancer metabolism (Pavlova and Thompson, 2016) as well as one of the hallmarks of cancer (Hanahan and Weinberg, 2011). A study using the combined transcriptomic-proteomic expression analysis proves that STAT1 promotes the transcription of energy metabolism-associated genes (Pitroda et al., 2009). Among the altered energy metabolism processes, glycolysis shows a significant change of targets both at protein and mRNA levels in response to STAT1 activation. Another study demonstrates that lncRNA ceruloplasmin (NRCP) is associated with cellular metabolism, among which glycolysis is one of the most related metabolic processes (Rupaimoole et al., 2015). NRCP promotes the binding of STAT1 and RNA pol II, leading to an increase in the expression of targeted genes such as glucose-6-phosphate isomerase (GPI). Silencing NRCP results in a significantly decreased expression of GPI, ALDOA, and ALDOC, which further supports the role of the NRCP-STAT1 axis in glycolysis-induced tumorigenesis.
Signal transducer and activator of transcription 1 enhances anti-tumorigenic immune responses and pro-inflammation function mainly through the induction of cytokines secretion, which in turn leads to an increased amount of anti-tumor immune lymphocytes (Zhang and Liu, 2017). Interestingly, STAT1 increases the expression of interleukin-18 binding protein (IL-18BP) and indoleamine 2,3-dioxygenase (IDO), an immunosuppressive enzyme that suppresses anti-tumor immunity. Evading immune destruction is one of the hallmarks of cancer (Hanahan and Weinberg, 2011). Tumor cells escape immune destruction in different ways, including the alteration of antigens, the decrease of immunogenicity, the changes of the tumor microenvironment (TME), and the decrease of immune responses (Beatty and Gladney, 2015). Generally, STAT1 is identified to be associated with enhanced immune responses through upregulating the amount of cytotoxicity immune cells such as cytotoxic T lymphocytes (CTLs) and natural killer (NK) cells (Meissl et al., 2017; Jin et al., 2019). Contrarily, a microarray analysis proves that STAT1 induces the expression of an immunosuppressive gene IDO (Rolvering et al., 2018). The inhibition of phosphorylation and nuclear translocation of STAT1 by bortezomib downregulates IFN-γ-induced IDO expression (Jiang et al., 2017). The IDO gene contains one GAS sequence and two IFN-α-stimulated response element (ISRE) domains (Dai and Gupta, 1990). The activation of STAT1 by IFN-γ forms STAT1-dimers and translocated into the nucleus to bind the GAS sequence of IDO and IRF1. The latter further binds to ISRE-1 and ISRE-2, causing an upregulatory effect on IDO expression (Chon et al., 1995). It has been shown that IDO is constitutively expressed in epithelial OC (EOC) cells isolated from ascites and is induced in vivo by IL-27 through STAT1 phosphorylation (Carbotti et al., 2015; Petretto et al., 2016). IDO has tryptophan metabolic activity which can transform tryptophan to kynurenine (Munn and Mellor, 2016). The decrease of tryptophan stimulates stress-response kinase GCN2 (Munn et al., 2005), which promotes the differentiation of naïve T cells to T regulatory (Treg) cells (Fallarino et al., 2006). Furthermore, IDO is proved to stimulate the aryl hydrocarbon receptor (AhR), which also promotes the differentiation of T cells to Treg cells (Fallarino et al., 2006; Mezrich et al., 2010) that induce immune tolerance by suppressing cytotoxicity of CTLs (Stein et al., 2011). Hence, STAT1-induced IDO plays a central role in the immune escape of OC cells (de Jong et al., 2011). Besides, the inhibition of IDO suppresses tumor growth by removing the inhibition of NK cells (Mougiakakos et al., 2010), suggesting that IDO is a potential therapeutic target of OC (Wang et al., 2012). Moreover, IDO is related to poor prognosis and paclitaxel resistance in patients with serous-type OC (Okamoto et al., 2005).
Similarly, STAT1 promotes IL-18BP expression, which is an endogenous inhibitor of IL-18, in EOC and primary OC cells (Carbotti et al., 2013). As an immune enhancer, IL-18 promotes anti-malignant tumor immune responses and is generally upregulated in the sera of OC patients (Medina et al., 2014). IL-18BP blocks the function of IL-18 by binding with mat-IL-18. On the other hand, IL-18 stimulates the expression of IL-18BP through IFN-γ and IL-27, leading to negative loop feedback (Veenstra et al., 2002). Thus, the ultimate effects depend on the local concentration ratio of IL-18 and IL-18BP (Carbotti et al., 2013; Medina et al., 2014). Concerning the immune therapy based on IL-18 level in OC patients, IL-18BP is a promising target because it can influence the effects of IL-18 therapy.
Elevated STAT1 increases intra-tumor CD8+ T cells, which plays a significant role in tumor inhibition by direct cytotoxicity (Wang et al., 2016; Jifu et al., 2018). Tumor-infiltrating CD8+ T cells suppress OC progression and are associated with a favorable prognosis in OC (Preston et al., 2013; Yeung et al., 2018). The Africa American Cancer Epidemiology Study shows that programmed death ligand-1 (PD-L1) positive and IDO positive tumor cells from HGSOC accompany by increased amounts of CD8+ T cells (Mills et al., 2019). The upregulated expression of PD-L1 and IDO is induced by phosphorylation of STAT3 and STAT1, respectively (Carbotti et al., 2015). PD-L1 and IDO have been identified to promote the immune escape of cancer cells. The blockage of IDO by iNOS-induced-NO may lead to a favorable prognosis (Munn and Mellor, 2016). Our previous study suggested that the elevated PD-L1 and STAT1 may favor the checkpoint immunotherapy in patients with EOC, but may have a limit in paclitaxel-resistant patients because of the low expression of PD-L1 and STAT1 (Liu et al., 2020).
The TME is essential for the secretion of cytokines, growth factors, and immune regulators (Figure 3A). It has been reported that CXCL9/10/11 can promote tissue chemotaxis of CD8+ T cells and NK cells by binding to CXCR3, leading to Th1-dominant anti-tumorigenic responses (Rainczuk et al., 2012; Peng et al., 2015). CD8+ T cells reduce platinum resistance by secreting IFN-γ, which decreases fibroblast glutathione (GSH) and cysteine metabolism by upregulating glutamyl transferases and suppressing transcription of system xc-cystine/glutamate antiporter (xCT) via the JAK/STAT1 pathway in OC cells (Wang et al., 2016). GSH induces cisplatin resistance via facilitating cisplatin efflux, preventing cells from oxidative damage and copper (Cu) coupling (Traverso et al., 2013). xCT is responsible for the increase of intracellular cystine. The accumulation of intracellular cystine is converted into cysteine which is secreted out of fibroblasts and then intake by cancer cells to synthesize GSH that binds cisplatin for its efflux (Wang et al., 2018). IFN-γ-stimulated STAT1 binds to the GAS site near the promoter of xCT, blocking the transcription of xCT in fibroblasts within the OC microenvironment (Wang et al., 2016), which binds cisplatin for its efflux (Figure 3B). Immunohistochemical analysis shows that the main sources of IFN-γ are from tumor-infiltrating NK and CD4+ T cells (Silva et al., 1994; Bach et al., 1997). IFN-γ then activates STAT1 in endothelial cells and suppresses the expression of TNFSF15 that is known to inhibit angiogenesis (Figure 3C). It is predictable that IFN-γ-stimulated unphosphorylated STAT1 contributes to the downregulation of TNFSF15 in endothelial cells (Lu et al., 2014). IFNs, growth factors, and ILs stimulate STAT1 which upregulates and downregulates the targeted genes in OC (Figure 3D).
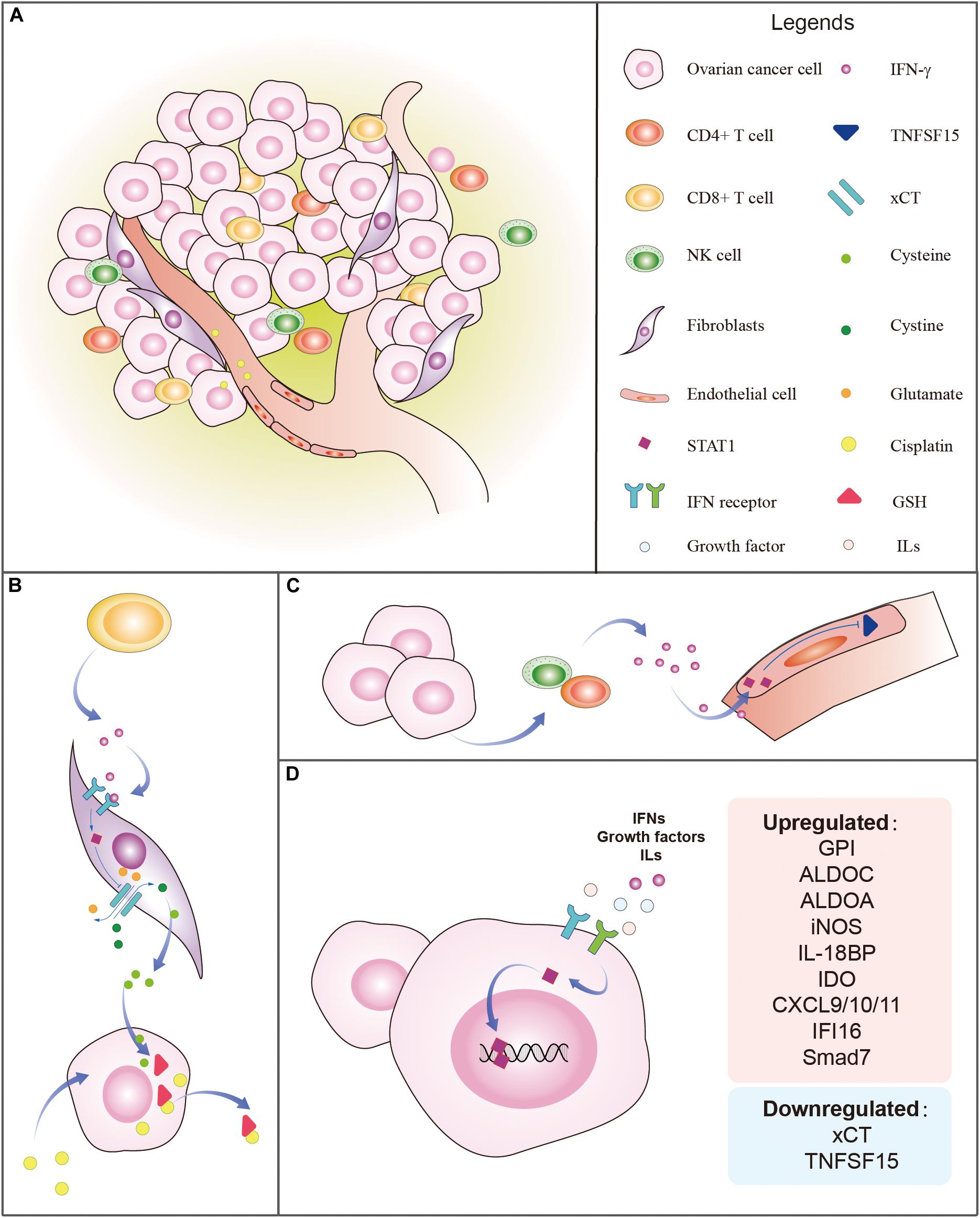
Figure 3. Tumor microenvironment (TME) and regulation of STAT1 in ovarian cancer. (A) The typical TME includes ovarian cancer cells, fibroblasts, endothelial cells, vessels, tumor-infiltrating CD4+ T cells, CD8+ T cells, NK cells, immune regulators, growth factors, and cytokines, etc. (B) Example of the influence of the TME on the STAT1 regulatory mechanism. CD8+ T cells from the ovarian TME release IFN-γ which binds to IFN-γ receptors to stimulate STAT1. The activated STAT1, in turn, suppresses the expression of xCT which is responsible for the transportation of outer cellular cystine into cells and intracellular glutamate out of the cells. The accumulation of intracellular cystine is then converted into cysteine which is secreted out of fibroblasts. Ovarian cancer intakes cysteine to synthesize GSH which binds to cisplatin to accelerate the efflux of cisplatin. (C) Example of the effect of the TME on STAT1 regulatory relationship. Ovarian cancer cells attract tumor-infiltrating CD4+ T cells and NK cells that release IFN-γ which activates STAT1 in endothelial cells and suppresses the expression of TNFSF15 that is known to inhibit angiogenesis. (D) IFNs, growth factors, and ILs stimulate STAT1 which upregulates and downregulates the targeted genes in ovarian cancer. The genes regulated by STAT1 are listed.
It has been shown that high levels of STAT1 and STAT1-induced chemokines such as CXCL9/10/11 and CD8A are associated with chemosensitivity in HGSOC (Au et al., 2016). Silenced NRCP can reduce tumor burden and enhance cisplatin sensitivity in vivo (Rupaimoole et al., 2015). With a deeper understanding of molecule mechanisms concerning tumorigenesis, anti-tumor activity, tumor microenvironment, and cancer stem cells (CSCs), some mechanisms of drug resistance in OC are emerging (Norouzi-Barough et al., 2018). Typically, drug resistance includes innate and acquired. As to acquired resistance, for example, we can use ABC transporter antibody or artificially pack drugs to avoid being recognized by those ABC transporters. Besides, there are many other measures to cope with drug resistance. It has been proved that STAT1 plays different roles in drug resistance including platinum-, doxorubicin-, and paclitaxel-resistance through transcriptional stimulation of target genes. However, it has a conflicting concern about STAT1-induced drug resistance. For example, interferon-inducible protein 16 (IFI16), which is induced by STAT1, exert anti-tumor effects but protect cancer cells from being killed by chemo-drugs at the same time (Bani et al., 2004). IFI16 is an anti-tumor factor in OC and is also proved to be associated with drug resistance by microarray analysis, which is further verified by immunohistochemistry at the protein level (Ju et al., 2009; Januchowski et al., 2017).
Experiments using OC cell lines in vitro come out with different results that iNOS is upregulated in cisplatin-sensitive OV2800 cells. Contrarily, it has been reported that NO induces cisplatin-resistance by increasing levels of glutathione (Turchi, 2006). Concerning the contradictory facts of iNOS and the outcomes of inhibiting iNOS are various (Malone et al., 2006; Kielbik et al., 2019), it is hard to determine whether iNOS is doing good or bad things to OC. Despite the above conflicting results, STAT1 indeed regulates the transcription of iNOS. Although most reports support the idea that OC with high iNOS expression is more sensitive rather than resistant, in general, it is still not clear about iNOS to be considered as a prognosis marker of OC yet (Yu et al., 2017).
Our group is first to demonstrate the involvement of STAT1 in stemness and paclitaxel-resistance in OC cells (Wang et al., 2020). We have found that a clone of paclitaxel-resistant cells shares the characteristics of CSCs and has stemness properties. The stem cell-like paclitaxel-resistant cells have a low expression level of STAT1 and high expression levels of CSC-related markers such as OCT4, CD44, CD133, SOX 2, and NANOG. Moreover, the overexpression of STAT1 in monoclonal paclitaxel-resistant OC cells suppresses the stemness. It is well known that CSCs generally have self-renew and differentiation abilities (Pattabiraman and Weinberg, 2014) and are proved to be associated with drug resistance in OC (Mihanfar et al., 2019). It has also been shown that STAT1 is positively associated with breast CSCs which are characterized by CD24 expression, while CD24 can suppress STAT1 expression (Suyama et al., 2016). It has been reported that CD95 (APO-1/Fas) can maintain stemness in breast cancer cells (Ceppi et al., 2014) and increase stemness by activating STAT1-dependent type I IFN signaling (Qadir et al., 2017). Additionally, the TME plays a significant role in regulating CSCs (Ahmed et al., 2018). Tumor-associated macrophages produce and secret IL-10 which promotes cancer stemness via the JAK/STAT1 and NF-κB/Notch1 pathways in non-small cell lung cancer (Yang et al., 2019). However, whether STAT1 can be a therapeutic target of CSCs is not clear. STAT1-regulating mechanisms in ovarian CSCs remain to explore in the future.
The TGF-β signaling pathway is important in regulating cell proliferation, apoptosis, and differentiation and is involved in cellular processes by cross-talking with many other signaling pathways such as epidermal growth factor receptor (EGFR), mitogen-activated protein kinase (MAPK), PI3K/AKT, Notch, Wnt, Hedgehog, and IFN-γ/STAT signaling pathways (Tzavlaki and Moustakas, 2020). For instance, the IFN-γ/STAT1 signaling pathway inhibits TGF-β signaling by upregulating the expression of an inhibitory Smad, Smad7, and then by suppressing TGF-β-induced phosphorylation and nuclear translocation of Smad3 (Ulloa et al., 1999). A previous study also reveals that the TGF-β/Smad3 signaling pathway is enhanced and the production of Smad7 is decreased in STAT1-null mice (Jeong et al., 2006). STAT1 suppresses TGF-β signaling by attenuating the cytoplasmic expression of runt-related transcription factor 2 (Runx2) and blocking the nuclear translocation of Runx2 (Miyazono et al., 2004). A transcriptional activation co-factor p300 is involved in the competition process between IFN-γ/STAT1 and TGF-β/Smad3 signalings in human glomerular endothelial cells (Yang et al., 2005). STAT1 and Smad3 both bind to p300 to enhance the DNA binding ability but IFN-γ attenuates the binding between Smad3 and p300 while TGF-β do not inhibit the binding of STAT1 to p300. This regulatory relationship between STAT1 and TGF-β signaling pathways have been demonstrated in melanoma, ovarian, lung, liver, and prostate cancer cells (Alvarez Mde et al., 2009; Penafuerte et al., 2011; Ayub and Kaul, 2017; Kaowinn et al., 2018; Tian et al., 2018). It has been known that the TGF-β1 signaling pathway has dual effects on tumor growth. TGF-β1 functions as a tumor suppressor inhibiting cell proliferation and stimulate normal cell differentiation at the early stage of cancer (Bierie and Moses, 2006). However, advanced OC cells hijack the TGF-β pathway to avoid an inhibitory effect on tumor cell proliferation. Hence, it is advised that the TGF-β signaling pathway suppresses tumor growth and induces apoptosis at the early stages but promotes metastasis at the advanced stage (Hao et al., 2019). A recent study suggests that TGF-β signaling and STAT1 regulate each other and come into being a negative feedback loop in OC (Tian et al., 2018). Our previous study showed that the overexpression of STAT1 in OC induces cell proliferation through downregulating the TGF-β signaling pathway, whereas TGF-β1 activates the STAT1 pathway by phosphorylating S727 residue of STAT1α which in turn couples with STAT1β and blocks the activation of the TGF-β signaling pathway (Tian et al., 2018). Furthermore, we are first to define the direct interaction of STAT1 with TGF-β receptors physically, which brings the crosstalk at the cell membrane level.
Chemotherapeutic agents such as inhibitors of glycolysis, glucose transporters, hexokinase, pyruvate kinase M2, glutaminase, and isocitrate dehydrogenase have been demonstrated to be effective in cellular experiments and some have already been tested in a clinical trial (Akins et al., 2018). For example, anti-glycolysis agent 3-bromopyruvate selectively targets tumor cells (Gill et al., 2016). As described previously, STAT1 is essential for the promotion of glycolysis; hence, the inhibition of STAT1 might be developed as a strategy for the treatment of OC. Fludarabine has been allowed to treat patients with OC for decades and has been demonstrated to be able to inhibit the phosphorylation of STAT1 (Zhang and Zou, 2015). Bortezomib is a potent and selective proteasome inhibitor and has been used in clinical trials for several cancers. Bortezomib can induce apoptosis and phosphorylate JAK, leading to the phosphorylation and activation of STAT1 (Kao et al., 2013). However, a phase II clinical trial using bortezomib to treat recurrent platinum-sensitive OC shows minimal anti-tumor activity (Aghajanian et al., 2009). Other studies show that the suppression of STAT1 by AG490 significantly inhibits the expression of IDO that avoids immune tolerance (Kobayashi et al., 2015) and induces B-cell chronic lymphocytic leukemia cell apoptosis (Martinez-Lostao et al., 2005). Concerning the role of STAT1 in OC, targeting STAT1 and its-related genes may have clinical potentials.
Signal transducer and activator of transcription 1 is an important transducer and transcript factor which exerts contradictory effects by directly regulating target genes or cooperating with other transcription factors to mediate tumor progression and drug resistance. STAT1 has a dual role and is involved in the anti-tumor process, tumor progression, drug sensitivity, chemoresistance, and stemness. The crosstalk between the STAT1 signaling pathway and other signaling pathways may enable STAT1 to perform contrary functions during tumorigenesis.
The specific role of STAT1 in ovarian tumorigenesis is complex. Typically, receptors-associated JAK phosphorylates Y701 of STAT1α/β in response to IFNs and ILs stimulation, whereas the phosphorylation of S727 of STAT1α initiates and executes specific functions of STAT1. Whether STAT1β has an opposite effect of STAT1α on tumorigenesis in OC is still unclear. A previous study reported that STAT1β can block the phosphorylation of STAT1α in B lymphocytes (Baran-Marszak et al., 2004). Therefore, it is predictable that STAT1α performs tumorigenesis function in OC cells while STAT1β balances these malignant-transforming effects of STAT1α in normal cells. Interestingly, TGF-β1 increases the phosphorylation of both Y701 and S727 in normal ovarian surface epithelial cells; while in cancer cells, TGF-β1 only increases S727 phosphorylation but decreases Y701 phosphorylation (Tian et al., 2018). The mechanism for the differential regulation by TGF-β signaling between normal and cancer cells and between Y701 and S727 in OC cells remains unknown and still needs further investigation.
Signal transducer and activator of transcription 1 has been reported to enhance anti-tumor immune response and is considered to inhibit tumor growth (Avalle et al., 2012). However, during tumorigenesis, malignant cells avoid the above impairment through downregulating STAT1 by the methylation of STAT1 in its promoter; the low expression level of STAT1 is observed in a chemoresistant cell line (Wang et al., 2020). Thus, more attention should be paid to the effects of STAT1 on the significance of the TME, the integration of various signals, and the nature of tumor cells. More evidence in the study of stem cell biology is merging but how a drug-resistance is triggered by these cells remains unclear. The role and detailed mechanisms of STAT1 in OC stem cells or the functional process, as well as therapeutic approaches, are needed to explore further.
XL and GX were involved in the conception and design of the work and designed the figures. XL performed the literature search, critically analyzed the existing knowledge, and wrote the draft. XL and FW prepared the figures and provided the critical revisions. XX and JZ performed the bibliographic research and provided the critical revisions, also contributed to editing the manuscript. GX contributed to the conception of the work, edited the manuscript, and provided the critical revisions. All authors were involved in manuscript writing, read and approved the final manuscript.
This work was supported by grants from the National Natural Science Foundation of China (grant no. 81872121), the Natural Science Foundation of Shanghai (grant no. 17ZR1404100) to GX.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
ABC, ATP-binding cassette; AEG1, astrocyte-elevated gene-1; AhR, aryl hydrocarbon receptor; AKT, Akt serine/threonine kinase 1; ALDO, aldolase, fructose-bisphosphate; CA, cortistatin A; CCD, coiled-coil domain; CD, cluster of differentiation; CDK, cyclin-dependent kinase; CSC, cancer stem cell; CTLs, cytotoxic T lymphocytes; CXCL, C-X -C motif ligand; CXCR, C-X -C motif receptor; DBD, DNA binding domain; DLX4, distal-less homeobox 4; EGFR, epidermal growth factor receptor; EMT, epithelial to mesenchymal transition; EOC, epithelial ovarian cancer; FAK, focal adhesion kinase; GAS, gamma-interferon activation site; GPI, glucose-6-phosphate isomerase; GSH, glutathione; HGSOC, high-grade serous ovarian cancer; IDO, indoleamine 2,3-dioxygenase; IFI16, interferon-inducible protein 16; IFN, interferon; IFNAR, interferon- α receptor; IFNGR, interferon- γ receptor; IL, interleukin; IL-18BP, interleukin-18 binding protein; iNOS, inducible nitric oxide synthase; IRF, interferon regulatory factor; ISG, interferon-stimulated genes; ISGF, interferon-stimulated gene factor; ISRE, IFN(alpha)-stimulated response element; ITGB1, integrin beta-1; JAK, Janus kinase; LD, linker domain; MAPK, mitogen-activated protein kinase; miR, microRNA; MMP, matrix metallopeptidase; NADH, nicotinamide adenine dinucleotide; ND, N-terminal; NO, nitric oxide; ND, N-terminal; NRCP, lncRNA ceruloplasmin; OCT4, octamer-binding transcription factor 4; OC, ovarian cancer; PARP, poly (ADP-ribose) polymerase; PD-L1, programmed death ligand-1; PI3K, phosphatidylinositol 3-kinase; RNA pol II, RNA polymerase II; Runx2, runt-related transcription factor 2; S727, serine 727 residue; SH2D, Src homology-2 domain; STAT, signal transducer and activator of transcription; TAD, transactivation domain; TCA, tricarboxylic acid; TGF-β, transforming growth factor-β; TME, tumor microenvironment; TNFSF15, tumor necrosis factor superfamily 15; Treg, T regulatory cells; VEGE, vascular endothelial growth factor; xCT, xc-cystine/glutamate antiporter; Y701, tyrosine 701 residue.
Aghajanian, C., Blessing, J. A., Darcy, K. M., Reid, G., DeGeest, K., Rubin, S. C., et al. (2009). A phase II evaluation of bortezomib in the treatment of recurrent platinum-sensitive ovarian or primary peritoneal cancer: a Gynecologic Oncology Group study. Gynecol. Oncol. 115, 215–220. doi: 10.1016/j.ygyno.2009.07.023
Ahmed, N., Escalona, R., Leung, D., Chan, E., and Kannourakis, G. (2018). Tumour microenvironment and metabolic plasticity in cancer and cancer stem cells: perspectives on metabolic and immune regulatory signatures in chemoresistant ovarian cancer stem cells. Semin. Cancer Biol. 53, 265–281. doi: 10.1016/j.semcancer.2018.10.002
Akins, N. S., Nielson, T. C., and Le, H. V. (2018). Inhibition of glycolysis and glutaminolysis: an emerging drug discovery approach to combat Cancer. Curr. Top. Med. Chem. 18, 494–504. doi: 10.2174/1568026618666180523111351
Akram, M. (2013). Mini-review on glycolysis and cancer. J. Cancer Educ. 28, 454–457. doi: 10.1007/s13187-013-0486-9
Alvarez Mde, L., Quiroga, A. D., Parody, J. P., Ronco, M. T., Frances, D. E., Carnovale, C. E., et al. (2009). Cross-talk between IFN-alpha and TGF-beta1 signaling pathways in preneoplastic rat liver. Growth Fact. 27, 1–11. doi: 10.1080/08977190802547357
Au, K. K., Le Page, C., Ren, R., Meunier, L., Clement, I., Tyrishkin, K., et al. (2016). STAT1-associated intratumoural TH1 immunity predicts chemotherapy resistance in high-grade serous ovarian cancer. J. Pathol. Clin. Res. 2, 259–270. doi: 10.1002/cjp2.55
Au, K. K., Peterson, N., Truesdell, P., Reid-Schachter, G., Khalaj, K., Ren, R., et al. (2017). CXCL10 alters the tumour immune microenvironment and disease progression in a syngeneic murine model of high-grade serous ovarian cancer. Gynecol. Oncol. 145, 436–445. doi: 10.1016/j.ygyno.2017.03.007
Avalle, L., Pensa, S., Regis, G., Novelli, F., and Poli, V. (2012). STAT1 and STAT3 in tumorigenesis: a matter of balance. JAKSTAT 1, 65–72. doi: 10.4161/jkst.20045
Ayub, S. G., and Kaul, D. (2017). miR-2909 regulates ISGylation system via STAT1 signalling through negative regulation of SOCS3 in prostate cancer. Andrology 5, 790–797. doi: 10.1111/andr.12374
Bach, E. A., Aguet, M., and Schreiber, R. D. (1997). The IFN gamma receptor: a paradigm for cytokine receptor signaling. Annu. Rev. Immunol. 15, 563–591. doi: 10.1146/annurev.immunol.15.1.563
Bancerek, J., Poss, Z. C., Steinparzer, I., Sedlyarov, V., Pfaffenwimmer, T., Mikulic, I., et al. (2013). CDK8 kinase phosphorylates transcription factor STAT1 to selectively regulate the interferon response. Immunity 38, 250–262. doi: 10.1016/j.immuni.2012.10.017
Bani, M. R., Nicoletti, M. I., Alkharouf, N. W., Ghilardi, C., Petersen, D., Erba, E., et al. (2004). Gene expression correlating with response to paclitaxel in ovarian carcinoma xenografts. Mol. Cancer Ther. 3, 111–121.
Baran-Marszak, F., Feuillard, J., Najjar, I., Le Clorennec, C., Bechet, J. M., Dusanter-Fourt, I., et al. (2004). Differential roles of STAT1alpha and STAT1beta in fludarabine-induced cell cycle arrest and apoptosis in human B cells. Blood 104, 2475–2483. doi: 10.1182/blood-2003-10-3508
Beatty, G. L., and Gladney, W. L. (2015). Immune escape mechanisms as a guide for cancer immunotherapy. Clin. Cancer Res. 21, 687–692. doi: 10.1158/1078-0432.CCR-14-1860
Bierie, B., and Moses, H. L. (2006). Tumour microenvironment: TGFbeta: the molecular Jekyll and Hyde of cancer. Nat. Rev. Cancer 6, 506–520. doi: 10.1038/nrc1926
Blanco, A., Grana, B., Fachal, L., Santamarina, M., Cameselle-Teijeiro, J., Ruiz-Ponte, C., et al. (2010). Beyond BRCA1 and BRCA2 wild-type breast and/or ovarian cancer families: germline mutations in TP53 and PTEN. Clin. Genet. 77, 193–196. doi: 10.1111/j.1399-0004.2009.01309.x
Bronger, H., Singer, J., Windmuller, C., Reuning, U., Zech, D., Delbridge, C., et al. (2016). CXCL9 and CXCL10 predict survival and are regulated by cyclooxygenase inhibition in advanced serous ovarian cancer. Br. J. Cancer 115, 553–563. doi: 10.1038/bjc.2016.172
Burke, A. J., Garrido, P., Johnson, C., Sullivan, F. J., and Glynn, S. A. (2017). Inflammation and Nitrosative Stress Effects in Ovarian and Prostate Pathology and Carcinogenesis. Antioxid. Redox Signal. 26, 1078–1090. doi: 10.1089/ars.2017.7004
Byrne, A. T., Ross, L., Holash, J., Nakanishi, M., Hu, L., Hofmann, J. I., et al. (2003). Vascular endothelial growth factor-trap decreases tumor burden, inhibits ascites, and causes dramatic vascular remodeling in an ovarian cancer model. Clin. Cancer Res. 9, 5721–5728.
Carbotti, G., Barisione, G., Airoldi, I., Mezzanzanica, D., Bagnoli, M., Ferrero, S., et al. (2015). IL-27 induces the expression of IDO and PD-L1 in human cancer cells. Oncotarget 6, 43267–43280. doi: 10.18632/oncotarget.6530
Carbotti, G., Barisione, G., Orengo, A. M., Brizzolara, A., Airoldi, I., Bagnoli, M., et al. (2013). The IL-18 antagonist IL-18-binding protein is produced in the human ovarian cancer microenvironment. Clin. Cancer Res. 19, 4611–4620. doi: 10.1158/1078-0432.CCR-13-0568
Cardenas, H., Jiang, G., Thomes Pepin, J., Parker, J. B., Condello, S., Nephew, K. P., et al. (2019). Interferon-gamma signaling is associated with BRCA1 loss-of-function mutations in high grade serous ovarian cancer. NPJ Precis Oncol. 3:32. doi: 10.1038/s41698-019-0103-4
Ceppi, P., Hadji, A., Kohlhapp, F. J., Pattanayak, A., Hau, A., Liu, X., et al. (2014). CD95 and CD95L promote and protect cancer stem cells. Nat. Commun. 5:5238. doi: 10.1038/ncomms6238
Chen, X., Vinkemeier, U., Zhao, Y., Jeruzalmi, D., Darnell, J. E. Jr., and Kuriyan, J. (1998). Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell 93, 827–839.
Chon, S. Y., Hassanain, H. H., Pine, R., and Gupta, S. L. (1995). Involvement of two regulatory elements in interferon-gamma-regulated expression of human indoleamine 2,3-dioxygenase gene. J. Interf. Cytokine Res. 15, 517–526. doi: 10.1089/jir.1995.15.517
Dai, W., and Gupta, S. L. (1990). Regulation of indoleamine 2,3-dioxygenase gene expression in human fibroblasts by interferon-gamma. Upstream control region discriminates between interferon-gamma and interferon-alpha. J. Biol. Chem. 265, 19871–19877.
de Jong, R. A., Nijman, H. W., Boezen, H. M., Volmer, M., Ten Hoor, K. A., Krijnen, J., et al. (2011). Serum tryptophan and kynurenine concentrations as parameters for indoleamine 2,3-dioxygenase activity in patients with endometrial, ovarian, and vulvar cancer. Int. J. Gynecol. Cancer 21, 1320–1327. doi: 10.1097/IGC.0b013e31822017fb
Deng, W., Gu, X., Lu, Y., Gu, C., Zheng, Y., Zhang, Z., et al. (2012). Down-modulation of TNFSF15 in ovarian cancer by VEGF and MCP-1 is a pre-requisite for tumor neovascularization. Angiogenesis 15, 71–85. doi: 10.1007/s10456-011-9244-y
Devapatla, B., Sharma, A., and Woo, S. (2015). CXCR2 inhibition combined with sorafenib improved antitumor and antiangiogenic response in preclinical models of ovarian cancer. PLoS One 10:e0139237. doi: 10.1371/journal.pone.0139237
Fallarino, F., Grohmann, U., You, S., McGrath, B. C., Cavener, D. R., Vacca, C., et al. (2006). The combined effects of tryptophan starvation and tryptophan catabolites down-regulate T cell receptor zeta-chain and induce a regulatory phenotype in naive T cells. J. Immunol. 176, 6752–6761. doi: 10.4049/jimmunol.176.11.6752
Frank, S., Stallmeyer, B., Kampfer, H., Kolb, N., and Pfeilschifter, J. (1999). Nitric oxide triggers enhanced induction of vascular endothelial growth factor expression in cultured keratinocytes (HaCaT) and during cutaneous wound repair. FASEB J. 13, 2002–2014.
Gao, J., Morrison, D. C., Parmely, T. J., Russell, S. W., and Murphy, W. J. (1997). An interferon-gamma-activated site (GAS) is necessary for full expression of the mouse iNOS gene in response to interferon-gamma and lipopolysaccharide. J. Biol. Chem. 272, 1226–1230. doi: 10.1074/jbc.272.2.1226
Gill, K. S., Fernandes, P., O’Donovan, T. R., McKenna, S. L., Doddakula, K. K., Power, D. G., et al. (2016). Glycolysis inhibition as a cancer treatment and its role in an anti-tumour immune response. Biochim. Biophys. Acta 1866, 87–105. doi: 10.1016/j.bbcan.2016.06.005
Greenwood, C., Metodieva, G., Al-Janabi, K., Lausen, B., Alldridge, L., Leng, L., et al. (2012). Stat1 and CD74 overexpression is co-dependent and linked to increased invasion and lymph node metastasis in triple-negative breast cancer. J. Proteom. 75, 3031–3040. doi: 10.1016/j.jprot.2011.11.033
Groom, J. R., and Luster, A. D. (2011). CXCR3 in T cell function. Exp. Cell Res. 317, 620–631. doi: 10.1016/j.yexcr.2010.12.017
Guan, L. Y., and Lu, Y. (2018). New developments in molecular targeted therapy of ovarian cancer. Discov. Med. 26, 219–229.
Hainaut, P., and Plymoth, A. (2013). Targeting the hallmarks of cancer: towards a rational approach to next-generation cancer therapy. Curr. Opin. Oncol. 25, 50–51. doi: 10.1097/CCO.0b013e32835b651e
Hanahan, D., and Weinberg, R. A. (2011). Hallmarks of cancer: the next generation. Cell 144, 646–674. doi: 10.1016/j.cell.2011.02.013
Hao, Y., Baker, D., and Ten Dijke, P. (2019). TGF-beta-mediated epithelial-mesenchymal transition and cancer metastasis. Int. J. Mol. Sci. 20:2767. doi: 10.3390/ijms20112767
Horvath, C. M. (2004). The Jak-STAT pathway stimulated by interferon alpha or interferon beta. Sci. STKE 2004:tr10. doi: 10.1126/stke.2602004tr10
Hsu, K. S., Zhao, X., Cheng, X., Guan, D., Mahabeleshwar, G. H., Liu, Y., et al. (2017). Dual regulation of Stat1 and Stat3 by the tumor suppressor protein PML contributes to interferon alpha-mediated inhibition of angiogenesis. J. Biol. Chem. 292, 10048–10060. doi: 10.1074/jbc.M116.771071
Ihle, J. N. (2001). The Stat family in cytokine signaling. Curr. Opin. Cell Biol. 13, 211–217. doi: 10.1016/s0955-0674(00)00199-x
Ihle, J. N., and Kerr, I. M. (1995). Jaks and Stats in signaling by the cytokine receptor superfamily. Trends Genet. 11, 69–74. doi: 10.1016/s0168-9525(00)89000-9
Januchowski, R., Sterzynska, K., Zawierucha, P., Rucinski, M., Swierczewska, M., Partyka, M., et al. (2017). Microarray-based detection and expression analysis of new genes associated with drug resistance in ovarian cancer cell lines. Oncotarget 8, 49944–49958. doi: 10.18632/oncotarget.18278
Jeong, W. I., Park, O., Radaeva, S., and Gao, B. (2006). STAT1 inhibits liver fibrosis in mice by inhibiting stellate cell proliferation and stimulating NK cell cytotoxicity. Hepatology 44, 1441–1451. doi: 10.1002/hep.21419
Jiang, G. M., Wang, H. S., Du, J., Ma, W. F., Wang, H., Qiu, Y., et al. (2017). Bortezomib relieves immune tolerance in nasopharyngeal carcinoma via STAT1 suppression and indoleamine 2,3-dioxygenase downregulation. Cancer Immunol. Res. 5, 42–51. doi: 10.1158/2326-6066.CIR-16-0102
Jifu, E., Yan, F., Kang, Z., Zhu, L., Xing, J., and Yu, E. (2018). CD8(+)CXCR5(+) T cells in tumor-draining lymph nodes are highly activated and predict better prognosis in colorectal cancer. Hum. Immunol. 79, 446–452. doi: 10.1016/j.humimm.2018.03.003
Jin, Y., Sun, Z., Geng, J., Yang, L., Song, Z., Song, H., et al. (2019). IL-21 reinvigorates exhausted natural killer cells in patients with HBV-associated hepatocellular carcinoma in STAT1-depedent pathway. Int. Immunopharmacol. 70, 1–8. doi: 10.1016/j.intimp.2019.02.007
Josahkian, J. A., Saggioro, F. P., Vidotto, T., Ventura, H. T., Candido Dos Reis, F. J., de Sousa, C. B., et al. (2018). Increased STAT1 expression in high grade serous ovarian cancer is associated with a better outcome. Int. J. Gynecol. Cancer 28, 459–465. doi: 10.1097/IGC.0000000000001193
Ju, W., Yoo, B. C., Kim, I. J., Kim, J. W., Kim, S. C., and Lee, H. P. (2009). Identification of genes with differential expression in chemoresistant epithelial ovarian cancer using high-density oligonucleotide microarrays. Oncol. Res. 18, 47–56. doi: 10.3727/096504009789954672
Kao, C., Chao, A., Tsai, C. L., Lin, C. Y., Chuang, W. C., Chen, H. W., et al. (2013). Phosphorylation of signal transducer and activator of transcription 1 reduces bortezomib-mediated apoptosis in cancer cells. Cell Death Dis. 4:e512. doi: 10.1038/cddis.2013.38
Kaowinn, S., Kaewpiboon, C., Koh, S. S., Kramer, O. H., and Chung, Y. H. (2018). STAT1HDAC4 signaling induces epithelialmesenchymal transition and sphere formation of cancer cells overexpressing the oncogene, CUG2. Oncol. Rep. 40, 2619–2627. doi: 10.3892/or.2018.6701
Kielbik, M., Szulc-Kielbik, I., and Klink, M. (2019). The potential role of iNOS in ovarian cancer progression and chemoresistance. Int. J. Mol. Sci. 20:1751. doi: 10.3390/ijms20071751
Kobayashi, A., Tanizaki, Y., Kimura, A., Ishida, Y., Nosaka, M., Toujima, S., et al. (2015). AG490, a Jak2 inhibitor, suppressed the progression of murine ovarian cancer. Eur. J. Pharmacol. 766, 63–75. doi: 10.1016/j.ejphar.2015.09.039
Koti, M., Siu, A., Clement, I., Bidarimath, M., Turashvili, G., Edwards, A., et al. (2015). A distinct pre-existing inflammatory tumour microenvironment is associated with chemotherapy resistance in high-grade serous epithelial ovarian cancer. Br. J. Cancer 112, 1215–1222. doi: 10.1038/bjc.2015.81
Kundu, N., Walser, T. C., Ma, X., and Fulton, A. M. (2005). Cyclooxygenase inhibitors modulate NK activities that control metastatic disease. Cancer Immunol. Immunother. CII 54, 981–987. doi: 10.1007/s00262-005-0669-2
Lebelo, M. T., Joubert, A. M., and Visagie, M. H. (2019). Warburg effect and its role in tumourigenesis. Arch. Pharm. Res. 42, 833–847. doi: 10.1007/s12272-019-01185-2
Leung, E. L., Fraser, M., Fiscus, R. R., and Tsang, B. K. (2008). Cisplatin alters nitric oxide synthase levels in human ovarian cancer cells: involvement in p53 regulation and cisplatin resistance. Br. J. Cancer 98, 1803–1809. doi: 10.1038/sj.bjc.6604375
Li, L., Zhu, L., Hao, B., Gao, W., Wang, Q., Li, K., et al. (2017). iNOS-derived nitric oxide promotes glycolysis by inducing pyruvate kinase M2 nuclear translocation in ovarian cancer. Oncotarget 8, 33047–33063. doi: 10.18632/oncotarget.16523
Liang, R., Chen, X., Chen, L., Wan, F., Chen, K., Sun, Y., et al. (2020). STAT3 signaling in ovarian cancer: a potential therapeutic target. J. Cancer 11, 837–848. doi: 10.7150/jca.35011
Liu, F., Liu, J., Zhang, J., Shi, J., Gui, L., and Xu, G. (2020). Expression of STAT1 is positively correlated with PD-L1 in human ovarian cancer. Cancer Biol. Ther. 21, 963–971. doi: 10.1080/15384047.2020.1824479
Liu, L., Wang, X., Li, X., Wu, X., Tang, M., and Wang, X. (2018). Upregulation of IGF1 by tumor-associated macrophages promotes the proliferation and migration of epithelial ovarian cancer cells. Oncol. Rep. 39, 818–826. doi: 10.3892/or.2017.6148
Lu, Y., Gu, X., Chen, L., Yao, Z., Song, J., Niu, X., et al. (2014). Interferon-gamma produced by tumor-infiltrating NK cells and CD4+ T cells downregulates TNFSF15 expression in vascular endothelial cells. Angiogenesis 17, 529–540. doi: 10.1007/s10456-013-9397-y
Lunt, S. Y., and Vander Heiden, M. G. (2011). Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu. Rev. Cell Dev. Biol. 27, 441–464. doi: 10.1146/annurev-cellbio-092910-154237
Malone, J. M., Saed, G. M., Diamond, M. P., Sokol, R. J., and Munkarah, A. R. (2006). The effects of the inhibition of inducible nitric oxide synthase on angiogenesis of epithelial ovarian cancer. Am. J. Obstetr. Gynecol. 194, 1110–1116. doi: 10.1016/j.ajog.2005.12.019
Malvezzi, M., Carioli, G., Rodriguez, T., Negri, E., and La Vecchia, C. (2016). Global trends and predictions in ovarian cancer mortality. Ann. Oncol. 27, 2017–2025. doi: 10.1093/annonc/mdw306
Martinez-Lostao, L., Briones, J., Forne, I., Martinez-Gallo, M., Ferrer, B., Sierra, J., et al. (2005). Role of the STAT1 pathway in apoptosis induced by fludarabine and JAK kinase inhibitors in B-cell chronic lymphocytic leukemia. Leukemia Lymphoma 46, 435–442. doi: 10.1080/10428190400018398
Matulonis, U. A. (2018). Ovarian cancer. Hematol. Oncol. Clin. N. Am. 32, 13–14. doi: 10.1016/j.hoc.2018.09.006
Medina, L., Rabinovich, A., Piura, B., Dyomin, V., Levy, R. S., and Huleihel, M. (2014). Expression of IL-18, IL-18 binding protein, and IL-18 receptor by normal and cancerous human ovarian tissues: possible implication of IL-18 in the pathogenesis of ovarian carcinoma. Mediators Inflamm. 2014:914954. doi: 10.1155/2014/914954
Meissl, K., Macho-Maschler, S., Muller, M., and Strobl, B. (2017). The good and the bad faces of STAT1 in solid tumours. Cytokine 89, 12–20. doi: 10.1016/j.cyto.2015.11.011
Mertens, C., Haripal, B., Klinge, S., and Darnell, J. E. (2015). Mutations in the linker domain affect phospho-STAT3 function and suggest targets for interrupting STAT3 activity. Proc. Natl. Acad. Sci. U.S.A. 112, 14811–14816. doi: 10.1073/pnas.1515876112
Mezrich, J. D., Fechner, J. H., Zhang, X., Johnson, B. P., Burlingham, W. J., and Bradfield, C. A. (2010). An interaction between kynurenine and the aryl hydrocarbon receptor can generate regulatory T cells. J. Immunol. 185, 3190–3198. doi: 10.4049/jimmunol.0903670
Mihanfar, A., Aghazadeh Attari, J., Mohebbi, I., Majidinia, M., Kaviani, M., Yousefi, M., et al. (2019). Ovarian cancer stem cell: a potential therapeutic target for overcoming multidrug resistance. J. Cell. Physiol. 234, 3238–3253. doi: 10.1002/jcp.26768
Mills, A. M., Peres, L. C., Meiss, A., Ring, K. L., Modesitt, S. C., Abbott, S. E., et al. (2019). Targetable immune regulatory molecule expression in high-grade serous ovarian carcinomas in african american women: a study of PD-L1 and IDO in 112 Cases From the African American Cancer Epidemiology Study (AACES). Int. J. Gynecol. Pathol. 38, 157–170. doi: 10.1097/PGP.0000000000000494
Miyazono, K., Maeda, S., and Imamura, T. (2004). Coordinate regulation of cell growth and differentiation by TGF-beta superfamily and Runx proteins. Oncogene 23, 4232–4237. doi: 10.1038/sj.onc.1207131
Morris, R., Kershaw, N. J., and Babon, J. J. (2018). The molecular details of cytokine signaling via the JAK/STAT pathway. Protein Sci. 27, 1984–2009. doi: 10.1002/pro.3519
Mougiakakos, D., Choudhury, A., Lladser, A., Kiessling, R., and Johansson, C. C. (2010). Regulatory T cells in cancer. Adv. Cancer Res. 107, 57–117. doi: 10.1016/S0065-230X(10)07003-X
Munn, D. H., and Mellor, A. L. (2016). IDO in the tumor microenvironment: inflammation, counter-regulation, and tolerance. Trends Immunol. 37, 193–207. doi: 10.1016/j.it.2016.01.002
Munn, D. H., Sharma, M. D., Baban, B., Harding, H. P., Zhang, Y., Ron, D., et al. (2005). GCN2 kinase in T cells mediates proliferative arrest and anergy induction in response to indoleamine 2,3-dioxygenase. Immunity 22, 633–642. doi: 10.1016/j.immuni.2005.03.013
Natanzon, Y., Goode, E. L., and Cunningham, J. M. (2018). Epigenetics in ovarian cancer. Semin. Cancer Biol. 51, 160–169. doi: 10.1016/j.semcancer.2017.08.003
Nitulescu, I. I., Meyer, S. C., Wen, Q. J., Crispino, J. D., Lemieux, M. E., Levine, R. L., et al. (2017). Mediator kinase phosphorylation of STAT1 S727 promotes growth of neoplasms with JAK-STAT activation. EBioMedicine 26, 112–125. doi: 10.1016/j.ebiom.2017.11.013
Norouzi-Barough, L., Sarookhani, M. R., Sharifi, M., Moghbelinejad, S., Jangjoo, S., and Salehi, R. (2018). Molecular mechanisms of drug resistance in ovarian cancer. J. Cell. Physiol. 233, 4546–4562. doi: 10.1002/jcp.26289
Oda, K., Matoba, Y., Irie, T., Kawabata, R., Fukushi, M., Sugiyama, M., et al. (2015). Structural basis of the inhibition of STAT1 activity by sendai virus C protein. J. Virol. 89, 11487–11499. doi: 10.1128/JVI.01887-15
Okamoto, A., Nikaido, T., Ochiai, K., Takakura, S., Saito, M., Aoki, Y., et al. (2005). Indoleamine 2,3-dioxygenase serves as a marker of poor prognosis in gene expression profiles of serous ovarian cancer cells. Clin. Cancer Res. 11, 6030–6039. doi: 10.1158/1078-0432.CCR-04-2671
O’Shea, J. J., Gadina, M., and Schreiber, R. D. (2002). Cytokine signaling in 2002: new surprises in the Jak/Stat pathway. Cell 109(Suppl.), S121–S131. doi: 10.1016/s0092-8674(02)00701-8
Parrini, M., Meissl, K., Ola, M. J., Lederer, T., Puga, A., Wienerroither, S., et al. (2018). The C-terminal transactivation domain of STAT1 has a gene-specific role in transactivation and cofactor recruitment. Front. Immunol. 9:2879. doi: 10.3389/fimmu.2018.02879
Pattabiraman, D. R., and Weinberg, R. A. (2014). Tackling the cancer stem cells - what challenges do they pose? Nat. Rev. Drug Discov. 13, 497–512. doi: 10.1038/nrd4253
Pavlova, N. N., and Thompson, C. B. (2016). The emerging hallmarks of cancer metabolism. Cell Metab. 23, 27–47. doi: 10.1016/j.cmet.2015.12.006
Penafuerte, C., Bautista-Lopez, N., Bouchentouf, M., Birman, E., Forner, K., and Galipeau, J. (2011). Novel TGF-beta antagonist inhibits tumor growth and angiogenesis by inducing IL-2 receptor-driven STAT1 activation. J. Immunol. 186, 6933–6944. doi: 10.4049/jimmunol.1003816
Peng, D., Kryczek, I., Nagarsheth, N., Zhao, L., Wei, S., Wang, W., et al. (2015). Epigenetic silencing of TH1-type chemokines shapes tumour immunity and immunotherapy. Nature 527, 249–253. doi: 10.1038/nature15520
Petretto, A., Carbotti, G., Inglese, E., Lavarello, C., Pistillo, M. P., Rigo, V., et al. (2016). Proteomic analysis uncovers common effects of IFN-gamma and IL-27 on the HLA class I antigen presentation machinery in human cancer cells. Oncotarget 7, 72518–72536. doi: 10.18632/oncotarget.12235
Pitroda, S. P., Wakim, B. T., Sood, R. F., Beveridge, M. G., Beckett, M. A., MacDermed, D. M., et al. (2009). STAT1-dependent expression of energy metabolic pathways links tumour growth and radioresistance to the Warburg effect. BMC Med. 7:68. doi: 10.1186/1741-7015-7-68
Preston, C. C., Maurer, M. J., Oberg, A. L., Visscher, D. W., Kalli, K. R., Hartmann, L. C., et al. (2013). The ratios of CD8+ T cells to CD4+CD25+ FOXP3+ and FOXP3- T cells correlate with poor clinical outcome in human serous ovarian cancer. PLoS One 8:e80063. doi: 10.1371/journal.pone.0080063
Qadir, A. S., Ceppi, P., Brockway, S., Law, C., Mu, L., Khodarev, N. N., et al. (2017). CD95/Fas increases stemness in cancer cells by inducing a STAT1-dependent type I interferon response. Cell reports 18, 2373–2386. doi: 10.1016/j.celrep.2017.02.037
Rainczuk, A., Rao, J., Gathercole, J., and Stephens, A. N. (2012). The emerging role of CXC chemokines in epithelial ovarian cancer. Reproduction 144, 303–317. doi: 10.1530/REP-12-0153
Ramana, C. V., Chatterjee-Kishore, M., Nguyen, H., and Stark, G. R. (2000). Complex roles of Stat1 in regulating gene expression. Oncogene 19, 2619–2627. doi: 10.1038/sj.onc.1203525
Ramana, C. V., Gil, M. P., Schreiber, R. D., and Stark, G. R. (2002). Stat1-dependent and -independent pathways in IFN-gamma-dependent signaling. Trends Immunol. 23, 96–101. doi: 10.1016/s1471-4906(01)02118-4
Rolvering, C., Zimmer, A. D., Ginolhac, A., Margue, C., Kirchmeyer, M., Servais, F., et al. (2018). The PD-L1- and IL6-mediated dampening of the IL27/STAT1 anticancer responses are prevented by alpha-PD-L1 or alpha-IL6 antibodies. J. Leukocyte Biol. 104, 969–985. doi: 10.1002/JLB.MA1217-495R
Rupaimoole, R., Lee, J., Haemmerle, M., Ling, H., Previs, R. A., Pradeep, S., et al. (2015). Long noncoding RNA ceruloplasmin promotes cancer growth by altering glycolysis. Cell Rep. 13, 2395–2402. doi: 10.1016/j.celrep.2015.11.047
Schindler, C., Fu, X. Y., Improta, T., Aebersold, R., and Darnell, J. E. Jr. (1992). Proteins of transcription factor ISGF-3: one gene encodes the 91-and 84-kDa ISGF-3 proteins that are activated by interferon alpha. Proc. Natl. Acad. Sci. U.S.A. 89, 7836–7839. doi: 10.1073/pnas.89.16.7836
Semper, C., Leitner, N. R., Lassnig, C., Parrini, M., Mahlakoiv, T., Rammerstorfer, M., et al. (2014). STAT1beta is not dominant negative and is capable of contributing to gamma interferon-dependent innate immunity. Mol. Cell. Biol. 34, 2235–2248. doi: 10.1128/MCB.00295-14
Shuai, K. (1994). Interferon-activated signal transduction to the nucleus. Curr. Opin. Cell Biol. 6, 253–259. doi: 10.1016/0955-0674(94)90144-9
Siegel, R. L., Miller, K. D., and Jemal, A. (2020). Cancer statistics, 2020. CA Cancer J. Clin. 70, 7–30. doi: 10.3322/caac.21590
Silva, C. M., Lu, H., Weber, M. J., and Thorner, M. O. (1994). Differential tyrosine phosphorylation of JAK1, JAK2, and STAT1 by growth hormone and interferon-gamma in IM-9 cells. J. Biol. Chem. 269, 27532–27539.
Stark, G. R., and Darnell, J. E. Jr. (2012). The JAK-STAT pathway at twenty. Immunity 36, 503–514. doi: 10.1016/j.immuni.2012.03.013
Stein, P., Weber, M., Prufer, S., Schmid, B., Schmitt, E., Probst, H. C., et al. (2011). Regulatory T cells and IL-10 independently counterregulate cytotoxic T lymphocyte responses induced by transcutaneous immunization. PLoS One 6:e27911. doi: 10.1371/journal.pone.0027911
Stronach, E. A., Alfraidi, A., Rama, N., Datler, C., Studd, J. B., Agarwal, R., et al. (2011). HDAC4-regulated STAT1 activation mediates platinum resistance in ovarian cancer. Cancer Res. 71, 4412–4422. doi: 10.1158/0008-5472.CAN-10-4111
Suyama, K., Onishi, H., Imaizumi, A., Shinkai, K., Umebayashi, M., Kubo, M., et al. (2016). CD24 suppresses malignant phenotype by downregulation of SHH transcription through STAT1 inhibition in breast cancer cells. Cancer Lett. 374, 44–53. doi: 10.1016/j.canlet.2015.12.013
Tian, X., Guan, W., Zhang, L., Sun, W., Zhou, D., Lin, Q., et al. (2018). Physical interaction of STAT1 isoforms with TGF-beta receptors leads to functional crosstalk between two signaling pathways in epithelial ovarian cancer. J. Exp. Clin. Cancer Res. CR 37:103. doi: 10.1186/s13046-018-0773-8
Torre, L. A., Trabert, B., DeSantis, C. E., Miller, K. D., Samimi, G., Runowicz, C. D., et al. (2018). Ovarian cancer statistics, 2018. CA Cancer J. Clin. 68, 284–296. doi: 10.3322/caac.21456
Traverso, N., Ricciarelli, R., Nitti, M., Marengo, B., Furfaro, A. L., Pronzato, M. A., et al. (2013). Role of glutathione in cancer progression and chemoresistance. Oxid. Med. Cell. Longev. 2013:972913. doi: 10.1155/2013/972913
Trinh, B., Ko, S. Y., Haria, D., Barengo, N., and Naora, H. (2015). The homeoprotein DLX4 controls inducible nitric oxide synthase-mediated angiogenesis in ovarian cancer. Mol. Cancer 14:97. doi: 10.1186/s12943-015-0368-3
Turchi, J. J. (2006). Nitric oxide and cisplatin resistance: no easy answers. Proc. Natl. Acad. Sci. U.S.A. 103, 4337–4338. doi: 10.1073/pnas.0601001103
Tzavlaki, K., and Moustakas, A. (2020). TGF-beta signaling. Biomolecules 10:487. doi: 10.3390/biom10030487
Ulloa, L., Doody, J., and Massague, J. (1999). Inhibition of transforming growth factor-beta/SMAD signalling by the interferon-gamma/STAT pathway. Nature 397, 710–713. doi: 10.1038/17826
Veenstra, K. G., Jonak, Z. L., Trulli, S., and Gollob, J. A. (2002). IL-12 induces monocyte IL-18 binding protein expression via IFN-gamma. J. Immunol. 168, 2282–2287. doi: 10.4049/jimmunol.168.5.2282
Walker, M., and Sobel, M. (2018). Diagnosing ovarian cancer. CMAJ 190:E1259. doi: 10.1503/cmaj.180499
Wang, D., Saga, Y., Mizukami, H., Sato, N., Nonaka, H., Fujiwara, H., et al. (2012). Indoleamine-2,3-dioxygenase, an immunosuppressive enzyme that inhibits natural killer cell function, as a useful target for ovarian cancer therapy. Int. J. Oncol. 40, 929–934. doi: 10.3892/ijo.2011.1295
Wang, F., Zhang, L., Liu, J., Zhang, J., and Xu, G. (2020). Highly expressed STAT1 contributes to the suppression of stemness properties in human paclitaxel-resistant ovarian cancer cells. Aging 12, 11042–11060. doi: 10.18632/aging.103317
Wang, S. F., Wung, C. H., Chen, M. S., Chen, C. F., Yin, P. H., Yeh, T. S., et al. (2018). Activated integrated stress response induced by salubrinal promotes cisplatin resistance in human gastric cancer cells via enhanced xCT expression and glutathione biosynthesis. Int. J. Mol. Sci. 19:3389. doi: 10.3390/ijms19113389
Wang, W., Kryczek, I., Dostal, L., Lin, H., Tan, L., Zhao, L., et al. (2016). Effector T cells abrogate stroma-mediated chemoresistance in ovarian cancer. Cell 165, 1092–1105. doi: 10.1016/j.cell.2016.04.009
Ween, M. P., Armstrong, M. A., Oehler, M. K., and Ricciardelli, C. (2015). The role of ABC transporters in ovarian cancer progression and chemoresistance. Crit. Rev. Oncol. Hematol. 96, 220–256. doi: 10.1016/j.critrevonc.2015.05.012
Weinlander, K., Naschberger, E., Lehmann, M. H., Tripal, P., Paster, W., Stockinger, H., et al. (2008). Guanylate binding protein-1 inhibits spreading and migration of endothelial cells through induction of integrin alpha4 expression. FASEB J. 22, 4168–4178. doi: 10.1096/fj.08-107524
Wen, Z., Zhong, Z., and Darnell, J. E. Jr. (1995). Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell 82, 241–250.
Wojciak, J. M., Martinez-Yamout, M. A., Dyson, H. J., and Wright, P. E. (2009). Structural basis for recruitment of CBP/p300 coactivators by STAT1 and STAT2 transactivation domains. EMBO J. 28, 948–958. doi: 10.1038/emboj.2009.30
Xu, Y., Zhang, Q., Miao, C., Dongol, S., Li, Y., Jin, C., et al. (2019). CCNG1 (Cyclin G1) regulation by mutant-P53 via induction of Notch3 expression promotes high-grade serous ovarian cancer (HGSOC) tumorigenesis and progression. Cancer Med. 8, 351–362. doi: 10.1002/cam4.1812
Yamamoto, K., Kobayashi, H., Arai, A., Miura, O., Hirosawa, S., and Miyasaka, N. (1997). cDNA cloning, expression and chromosome mapping of the human STAT4 gene: both STAT4 and STAT1 genes are mapped to 2q32.2–>q32.3. Cytogenet. Cell Genet. 77, 207–210. doi: 10.1159/000134578
Yang, G. L., and Li, L. Y. (2018). Counterbalance: modulation of VEGF/VEGFR activities by TNFSF15. Signal. Transduct. Target Ther. 3:21. doi: 10.1038/s41392-018-0023-8
Yang, L., Dong, Y., Li, Y., Wang, D., Liu, S., Wang, D., et al. (2019). IL-10 derived from M2 macrophage promotes cancer stemness via JAK1/STAT1/NF-kappaB/Notch1 pathway in non-small cell lung cancer. Int. J. Cancer 145, 1099–1110. doi: 10.1002/ijc.32151
Yang, W. S., Han, N. J., Kim, C. S., Ahn, H., Lee, S. K., Lee, K. U., et al. (2005). STAT1-independent down-regulation of interferon-gamma-induced class II transactivator and HLA-DR expression by transforming growth factor beta-1 in human glomerular endothelial cells. Nephron Exp. Nephrol. 100, e124–e131. doi: 10.1159/000085058
Yang, Z., Zhou, X., Liu, Y., Gong, C., Wei, X., Zhang, T., et al. (2014). Activation of integrin beta1 mediates the increased malignant potential of ovarian cancer cells exerted by inflammatory cytokines. Anticancer Agents Med. Chem. 14, 955–962. doi: 10.2174/1871520614666140613123108
Yeung, T. L., Tsai, C. C., Leung, C. S., Au Yeung, C. L., Thompson, M. S., Lu, K. H., et al. (2018). ISG15 Promotes ERK1 ISGylation, CD8+ T cell activation and suppresses ovarian cancer progression. Cancers 10:464. doi: 10.3390/cancers10120464
Yu, Y., Xie, Q., Liu, W., Guo, Y., Xu, N., Xu, L., et al. (2017). Increased intracellular Ca(2+) decreases cisplatin resistance by regulating iNOS expression in human ovarian cancer cells. Biomed. Pharmacother. 86, 8–15. doi: 10.1016/j.biopha.2016.11.135
Zhang, L., and Zou, W. (2015). Inhibition of integrin beta1 decreases the malignancy of ovarian cancer cells and potentiates anticancer therapy via the FAK/STAT1 signaling pathway. Mol. Med. Rep. 12, 7869–7876. doi: 10.3892/mmr.2015.4443
Keywords: chemoresistance, immune response, microenvironment, tumorigenesis, signaling pathway, stemness
Citation: Li X, Wang F, Xu X, Zhang J and Xu G (2021) The Dual Role of STAT1 in Ovarian Cancer: Insight Into Molecular Mechanisms and Application Potentials. Front. Cell Dev. Biol. 9:636595. doi: 10.3389/fcell.2021.636595
Received: 10 December 2020; Accepted: 01 March 2021;
Published: 23 March 2021.
Edited by:
Albena Todorova Dinkova-Kostova, University of Dundee, United KingdomReviewed by:
Gillian Smith, University of Dundee, United KingdomCopyright © 2021 Li, Wang, Xu, Zhang and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guoxiong Xu, Z3VveGlvbmcueHVAZnVkYW4uZWR1LmNu; orcid.org/0000-0002-9074-8754
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.