
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Cell Dev. Biol. , 18 February 2021
Sec. Cell Adhesion and Migration
Volume 9 - 2021 | https://doi.org/10.3389/fcell.2021.634759
This article is part of the Research Topic Evolution, Emerging Functions and Structure of Actin-Binding Proteins View all 32 articles
Caldesmon, an actin-binding protein, can inhibit myosin binding to actin and regulate smooth muscle contraction and relaxation. However, caldesmon has recently attracted attention due to its importance in cancer. The upregulation of caldesmon in several solid cancer tissues has been reported. Caldesmon, as well as its two isoforms, is considered as a biomarker for cancer and a potent suppressor of cancer cell invasion by regulating podosome/invadopodium formation. Therefore, caldesmon may be a promising therapeutic target for diseases such as cancer. Here, we review new studies on the gene transcription, isoform structure, expression, and phosphorylation regulation of caldesmon and discuss its clinical implications in cancer.
Caldesmon, an actin-binding protein of 150 kDa, was first isolated and purified from chicken gizzard muscle in 1981 (Sobue et al., 1981). Caldesmon was named from a combination (desmos is a Greek word that means binding) of calmodulin due to its ability to bind with calmodulin at different Ca2+ concentrations (Sobue et al., 1981). Caldesmon has two different molecular weight isoforms: high-molecular-weight caldesmon (H-caldesmon, 120–150 kDa) found in smooth muscle and low-molecular-weight caldesmon (L-caldesmon, 70–80 kDa) found in non-muscle cells (Hayashi et al., 1991; Mayanagi and Sobue, 2011). By cloning and sequencing the cDNA, H-caldesmon and L-caldesmon were determined to be derived by alternative splicing from a single gene (Hayashi et al., 1991). H-caldesmon and L-caldesmon conserve completely identical sequences in the N- and C-terminal domains, and the central repeating sequence of H-caldesmon is deleted in L-caldesmon (Hayashi et al., 1991). Although H-caldesmon and L-caldesmon have similar functional domains, their tissue and cell distributions are distinct (Ball and Kovala, 1988; Sobue et al., 1988).
Caldesmon has recently attracted attention due to its roles in cancer (Mayanagi and Sobue, 2011). Caldesmon can be a biomarker for the pathological diagnosis of tumors and prediction of the chemoradiotherapy response. H-caldesmon is considered a specific marker for tumor with smooth muscle differentiation (Watanabe et al., 1999; Nucci et al., 2001). L-caldesmon-positive human colon cancer cell lines are more resistant to chemoradiotherapy than L-caldesmon-negative cell lines (Kim et al., 2012). Second, caldesmon can also suppress cancer metastasis by regulating the podosome/invadopodium formation in transformed cancer cells, and the suppressive effect has been verified in a variety of cancers (Yoshio et al., 2007). In prostate cancer cells, a twofold increase in migratory capability and a threefold increase in invasion capability were found by scratch and invasion assays after the knockdown of L-caldesmon expression (Dierks et al., 2015). In addition, caldesmon can reversibly and cooperatively inhibit myosin binding actin to regulate smooth muscle contraction (Sobue et al., 1982; Ngai and Walsh, 1984). The phosphorylation of caldesmon plays an important role in the regulation of smooth muscle contraction (Huang et al., 2003). Therefore, this review analyzes the gene transcription, isoform structure, expression, and phosphorylation regulation of caldesmon and its clinical implications in cancer and gastrointestinal motility disorders.
The caldesmon gene is located on human chromosome 7q33 (Ensembl ID of the human caldesmon gene is ENSG00000122786) (Yates et al., 2020; Figure 1A). The caldesmon gene has 17 exons, and its isoforms (H-caldesmon and L-caldesmon) are mainly generated by the selective splicing of exons 7 and 8 (Lin et al., 2009). Exon 7 of selective translation encodes the central repeating sequence, and this central repeating sequence is specific to H-caldesmon (Transcripts 201,793 aa) (Mayanagi and Sobue, 2011). The caldesmon gene has 24 transcripts (201–224). Transcript 201 can generate H-caldesmon, while transcripts 202–206 and 222 can generate L-caldesmon. According to the different promoters, L-caldesmon can be further classified as a Fibro-type (WI-38) or HeLa-type (Hayashi et al., 1991). Two different distinct promoters are used in different cell types or tissues to generate L-caldesmon isoforms with distinct N-terminal domains (Yano et al., 1994). Alternative splicing of the caldesmon gene determines the different structures and expression of isoforms (Hayashi et al., 1991).
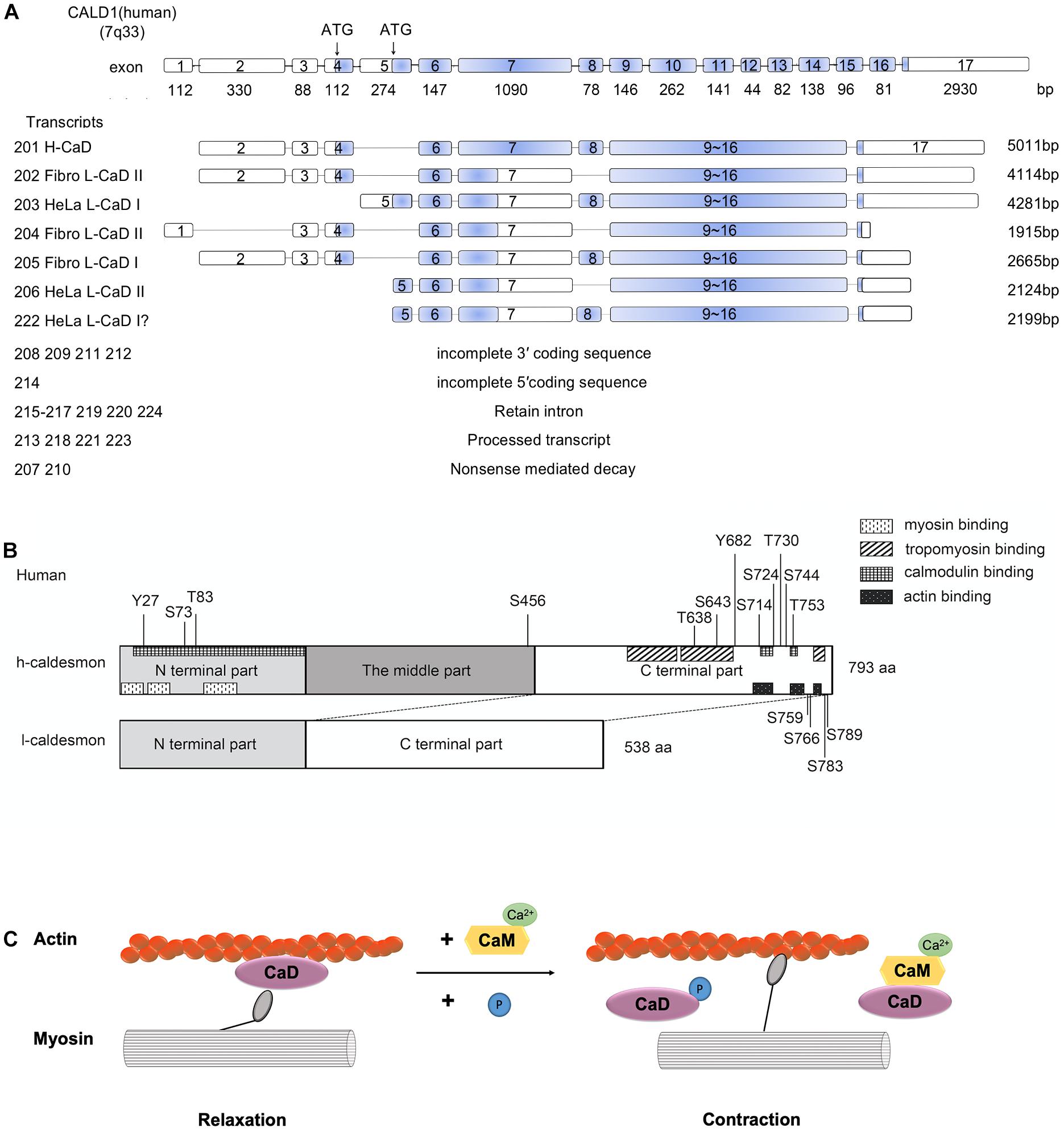
Figure 1. (A) Human Caldesmon Gene and Transcripts. Caldesmon (CALD1) structure showing exon (numbered boxes) and intron (line) regions and sizes. The caldesmon gene has 24 transcripts (201–224). Transcripts 201–206 and 222 are shown with the translated regions, and the blue regions are the coding sequences of the transcripts. The other transcripts cannot generate caldesmon due to the incomplete 3′/5′ coding sequence, retained introns, processed transcripts, or nonsense-mediated decay. Caldesmon isoforms are mainly generated by the selective spliced exons (exons 7 and 8) and distinct promoters (starting with exon 4 or 5). Exon 4 encodes the N-terminal domains of Fibro L-caldesmon and exon 5 encodes HeLa L-caldesmon. Exon 8 encodes 26 amino acids, including an extension of the α-helical motif. Transcripts 202–206 and 222 can generate Fibro-type and HeLa-type L-caldesmon, transcript 205 (Fibro L-caldesmonI, 563aa), transcript 202 and transcript 204 (Fibro L-caldesmonII, 538aa), transcript 222 and transcript 203 (HeLa L-caldesmonI (557–558aa), and transcript 206 (HeLa L-caldesmonII 532aa). Of these, transcript 222 has not been reported previously and was identified by a database search of Ensembl. (B) The domain structures of H-caldesmon and L-caldesmon. Human caldesmon contains an N-terminal domain, a C-terminal domain, and a middle part (repeating domain). The difference between L-caldesmon and H-caldesmon is the deletion of the repeating domain due to alternative splicing. The N-terminal domain contains a myosin-binding site and interacts weakly with actin and calmodulin. The C-terminal part contains an actin-binding site, calmodulin-binding site, tropomyosin-binding site, and phosphorylation sites. Human H-caldesmon (793 aa) is regulated by phosphorylation at Tyr-27, Ser-73, Thr-83, Ser-456, Thr-638, Ser-643, Tyr-682, Ser-714, Ser-724, Thr-730, Ser-744, Thr-753, Ser-759, Ser-766, Ser-783, and Ser-789 through multiple kinases (Cdc2,PAK,PKC, CamKII, CKII, and v-erbB tyrosine kinase). (C) The role of caldesmon in smooth-muscle contraction. The mechanism of reversing the putative inhibition by caldesmon of smooth muscle contraction by caldesmon depends on Ca2+/calmodulin and phosphorylation. Caldesmon can bind to actin filaments at less than 1 μM free Ca2+, whereas at a higher concentration of Ca2+ (>1 μM), calmodulin activated by Ca2+ forms a complex with caldesmon, and this complex is freed from actin filaments. Phosphorylation of caldesmon can attenuate its inhibitory activity, allowing actomyosin interaction and thereby resulting in muscle contraction.
From a structural perspective, caldesmon contains amino (N)- and carboxy (C)-terminal domains and a middle region (Wang, 2001). The N-terminal part can bind myosin and calmodulin (Lin et al., 2009); the C-terminal part contains actin-binding sites, calmodulin sites, and tropomyosin-binding sites (Wang, 2001; Mayanagi and Sobue, 2011), and the middle region in H-caldesmon (208–462 aa in humans) contains a long α-helix region and separates the N-terminal domain from the C-terminal domain (Wang, 2008; Lin et al., 2009; Figure 1B). The middle region is only present in H-caldesmon and is missing in L-caldesmon due to alternative splicing (Hayashi et al., 1991; Mayanagi and Sobue, 2011). However, the function of the middle region remains unknown. The middle region in H-caldesmon is presumed to fit the specific spatial arrangement of myosin molecules in the smooth muscle thick filament by evolutionary optimization (Wang, 2008).
The tissue and cell distributions of H-caldesmon and L-caldesmon are different. H-caldesmon is expressed in vascular and visceral smooth muscle and not in myofibroblasts, rhabdomyosarcoma, or tumors derived from myofibroblasts (Rush et al., 2001; Fisher et al., 2003). Therefore, H-caldesmon, as a smooth muscle-specific biomarker, can distinguish tumors originating from smooth muscle. In contrast, L-caldesmon is widely distributed in non-muscle tissues, such as the brain, spleen, and lymph nodes (Köhler, 2010, 2011). However, the expression changes of the two isoforms are closely correlated with the phenotypic modulation of smooth muscle cells (Ueki et al., 1987; Yokouchi et al., 2006). The expression of caldesmon can switch from L-caldesmon to H-caldesmon during smooth muscle cell differentiation and the expression turns from H-caldesmon to L-caldesmon during the dedifferentiation of smooth muscle cells (Ueki et al., 1987). Therefore, the different expressional distributions determine the different functions of H-caldesmon and L-caldesmon.
Caldesmon is an actin, myosin, tropomyosin, and Ca2+/calmodulin binding protein capable of regulating actomyosin contraction, actin filament dynamics, and cytoskeleton remodeling in smooth muscle and non-muscle cells (Lin et al., 2009). Posttranslational modification of caldesmon can modify its function and has been studied extensively in vitro (Foster et al., 2004; Ng et al., 2018). The association of caldesmon with tropomyosin-containing actin filaments effectively inhibits actomyosin ATPase activity and in vitro actin filament motility (Lin et al., 2009).
The mechanism of reversing the putative inhibition by caldesmon of smooth muscle contraction by caldesmon depends on Ca2+/calmodulin and phosphorylation (Foster et al., 2004; Mayanagi and Sobue, 2011; Figure 1C). Depending on the concentration of Ca2+, caldesmon shows an alternative binding ability to either calmodulin or actin filaments in vitro (Sobue et al., 1981). Caldesmon can bind to actin filaments at less than 1 μM free Ca2+, whereas at a higher concentration of Ca2+ (>1 μM), calmodulin activated by Ca2+ forms a complex with caldesmon, and this complex is freed from actin filaments (Sobue et al., 1981).
An alternative mechanism calls for phosphorylation of caldesmon in view of the fact that smooth muscles can contract at low Ca2+ concentrations (Foster et al., 2004). The phosphorylation of caldesmon is closely related to smooth muscle contraction (Hai and Gu, 2006). In an in vitro motility assay, unphosphorylated myosin exerted a mechanical load to shorten filaments, suggesting that tethering thick and thin filaments by caldesmon might help maintain some basal force (Horiuchi and Chacko, 1995). Phosphorylation (such as Thr-627, Ser-631, Ser-635, and Ser-642) can attenuate the inhibitory activity of caldesmon and indirectly increase inhibitory activity by weakening binding to Ca2+-calmodulin (Hamden et al., 2010). The interplay between phosphorylation-dependent and Ca2+/calmodulin-dependent mechanisms may be complex. The effect of Ca2+/calmodulin on the activity of caldesmon is dependent on the combination of phosphorylated residues (Hamden et al., 2010).
As a downstream effector of multiple signaling pathways, the inhibition of caldesmon can be reversed by phosphorylation during smooth muscle contraction through multiple kinases, such as ERK and PAK (Hai and Gu, 2006; Lin et al., 2009). Extracellular regulated kinase (ERK)-mediated phosphorylation of caldesmon has been shown to reverse the ability of the actin-binding fragment of caldesmon to stabilize actin filaments (Hai and Gu, 2006). Phosphorylation of caldesmon at ERK sites (Ser-759 and S789) is accompanied by a conformational change that partially dissociates caldesmon from actin (Kordowska et al., 2006). Such a structural change in H-caldesmon exposes the myosin-binding sites on the actin surface and allows actomyosin interactions in smooth muscles (Kordowska et al., 2006). In the case of non-muscle cells, the change in L-caldesmon weakens the stability of the actin filament and facilitates its disassembly (Kordowska et al., 2006). ERK-mediated phosphorylation of caldesmon has been shown to reverse the inhibitory effect of caldesmon on Arp2/3-mediated actin polymerization (Hai and Gu, 2006). The Arp2/3 complex is essential for podosome assembly, which are cytoskeletal adhesion structures that are important for cell invasion and extracellular matrix remodeling (Eves et al., 2006; Morita et al., 2007). Caldesmon is thought to be phosphorylated by ERK during the formation of podosomes (Hai and Gu, 2006). P21-activated kinase (PAK) is emerging as a major regulator of caldesmon-mediated actin dynamics in vivo (Foster et al., 2000; Lin et al., 2009). Reversible caldesmon phosphorylation at PAK-responsive sites is required for normal cell migration and cytokinesis (Lin et al., 2009). PAK phosphorylation sites (Ser-657 and Ser-687) are located close to calmodulin-binding sites (Mayanagi and Sobue, 2011). When caldesmon is phosphorylated by PAK, the ability to bind calmodulin is reduced by approximately 10-fold, and the affinity for actin-tropomyosin and the inhibition of actin-activated myosin ATPase activity are significantly reduced (Mayanagi and Sobue, 2011).
In addition, as one type of novel discovered posttranslational modification, lysine succinylation has been proven to be essential for regulating molecular functions, such as cellular metabolism, in physiological and pathophysiological states (Hirschey and Zhao, 2015). Caldesmon (lysine succinylation position 569) was downregulated in gastric cancer by LC-MS/MS analysis and validated by Western blotting (Song et al., 2017). Lysine succinylation position 569 of caldesmon may function as a potential biomarker in gastric cancer (Song et al., 2017).
Alterations of caldesmon expression level in different types of cancers in the clinic have been investigated (summarized in the Table 1). The upregulated expression of caldesmon is generally observed in different cancers. However, downregulated expression of caldesmon is found in the blood vessels of malignant melanomas compared with both benign melanocytic tumors and normal tissues.
Caldesmon is important for the diagnosis of myoma (Rizzello et al., 2017). H-caldesmon is a highly sensitive and specific marker that shows smooth muscle differentiation and helps identify uterine mesenchymal tumors (Nucci et al., 2001; Saraydaroglu et al., 2008). It is reported that H-caldesmon is negative in normal endometrial stroma (0%, 0 case/25 cases) and endometrial stromal neoplasms (0%, 0 case/24 cases) (Nucci et al., 2001). In contrast, desmin is expressed in endometria (32%, 8 cases/25 cases) and endometrial stromal neoplasms (50%, 12 cases/24 cases) (Nucci et al., 2001). SMA (smooth muscle actin), the other markers of smooth muscle cells, is positive in endometrial stromal sarcoma (44%, 7 cases/16 cases) (Chu et al., 2001). Therefore, H-caldesmon can effectively distinguish endometrial stromal tumors from uterine smooth tumors. However, H-caldesmon is found expressed in some non-myogenic tumors, such as gastrointestinal stromal tumors, malignant pleural mesothelioma, and ovarian adult granulosa cell tumors (Comin et al., 2006; Yu and Qu, 2018; Yu et al., 2019). Therefore, H-caldesmon expression may not be conclusive evidence of myogenic differentiation, and the diagnosis should be referred together with other markers (Yu et al., 2019). In addition, L-caldesmon is also considered a potential serum marker for glioma (Zheng et al., 2005). Taken together, the different isoforms of caldesmon can be promising biomarkers for diagnosis and prognosis prediction.
L-caldesmon is an integral part of the actin-rich core of the podosome (Eves et al., 2006). Caldesmon can suppress cell invasion by regulating the podosome/invadopodium formation of transformed and cancer cells (Yoshio et al., 2007). The overexpression of L-caldesmon suppresses podosome formation, whereas siRNA knockdown of L-caldesmon facilitates its formation (Eves et al., 2006; Gu et al., 2007). By analyzing the relationship between the expression levels of caldesmon and podosome/invadopodium formation in rat fibroblast (3Y1), RSV-transformed 3Y1 (BY1), human colon carcinoma (HCA7), murine melanoma (B16F10), human breast cancer (MB435s), and rat breast cancer (MTC) cell lines, podosome/invadopodium formation increases in transformed and cancer cells when caldesmon is expressed at low levels, and higher levels of caldesmon inhibit their formation (Yoshio et al., 2007). Caldesmon’s decreased expression has been identified in gastric cancer lymph node metastatic cells using a proteomics approach and loss of caldesmon expression could be associated with gastric cancer metastasis progression (Hou et al., 2013). In prostate cancer cells, a twofold increase in migratory capability and a threefold increase in invasion capability were found by scratch and Matrigel invasion assays after the knockdown of L-caldesmon expression (Dierks et al., 2015).
The presence of vessel invasion is considered indicative of a poor prognosis in many malignant tumors (Ekinci et al., 2018). Vascular smooth muscles contain both H-caldesmon (>75%) and L-caldesmon (<25%) (Glukhova et al., 1988). H-caldesmon appears to be the most specific and sensitive marker for vessel wall detection (Ekinci et al., 2018). The structural integrity and functional maturity of blood vessels are determined by the presence of normally functioning endothelial cells as well as the involvement of interendothelial junctions and mural cells (smooth muscle cells or pericytes) (Zheng et al., 2009). The knockdown of caldesmon caused serious defects in vasculogenesis and angiogenesis in zebrafish morphants, and the vascular integrity and blood circulation were concomitantly impaired (Zheng et al., 2009). The level of H-caldesmon expression in the melanoma blood vessels was inversely correlated with the frequency of metastasis (Koganehira et al., 2003). The endothelial cells of blood vessels in melanoma lesions appeared to be fragile compared to the normal tissues under electron microscopy (Koganehira et al., 2003). The fragility of blood vessels may increase metastasis.
L-caldesmon can decrease the chemoradiotherapy susceptibility of cancer cells. L-caldesmon-positive human colon cancer cell lines were more resistant to 5-fluorouracil (5-FU) and radiation treatment than L-caldesmon-negative cell lines (Kim et al., 2012). The expression level of L-caldesmon is therefore helpful in predicting the response of upper gastrointestinal carcinomas to neoadjuvant chemotherapy (Kim et al., 2012).
In addition, caldesmon can reversibly and cooperatively inhibit myosin binding actin to regulate smooth muscle contraction (Sobue et al., 1982; Ngai and Walsh, 1984). Smooth muscle dysmotility is the main pathogenic driver of gastrointestinal motility disorders. H-caldesmon can affect the contraction and relaxation of intestinal smooth muscle by binding to Ca2+/calmodulin and via phosphorylation (Wang, 2001). Structurally, H-caldesmon tethers myosin filaments to actin filaments to maintain the orderly arrangement of the thick and thin filaments. Functionally, H-caldesmon, as a “molecular brake,” sterically blocks actomyosin interactions in the resting state to modulate the development of contractile force (Guo et al., 2013). The expression of caldesmon was to be downregulated in rat models of chronic gastrointestinal motility hypofunction (Wang et al., 2001). Disruption of the normal inhibitory function of H-caldesmon could enhance intestinal peristalsis in both wild-type zebrafish larvae and mutant larvae that lack enteric nerves (Abrams et al., 2012). The detection of H-caldesmon phosphorylation sites by phosphorylation site-specific antibodies in colonic smooth muscle showed that H-caldesmon phosphorylation occurred on Ser-789 (Hedges et al., 2000). Ser-789 is phosphorylated by activated ERK, resulting in the C-terminal portion of H-caldesmon dissociating from actin and releasing the inhibition of ATPase activity, resulting in muscle contraction (Somara and Bitar, 2008). Expression levels of caldesmon in the gastric antrum were negatively correlated to gastric motility in rats treated by electroacupuncture (Yang et al., 2014). Expression of caldesmon was upregulated when gastrointestinal motility was inhibited. On the contrary, expression of caldesmon was downregulated when gastrointestinal motility was promoted (Yang et al., 2014). At present, the evidence correlating caldesmon and gastrointestinal motility disorders is not sufficient. However, whether caldesmon can regulate the contraction and relaxation of intestinal smooth muscle to treat gastrointestinal motility disorders needs further study.
The biochemical features of caldesmon and its clinical implications in cancer have been reviewed in this article. The following main points are noted: (1) Alternative splicing of the caldesmon gene determines its different structures and the expression of its isoforms. (2) H-caldesmon and L-caldesmon conserve the completely identical sequences in the N- and C-terminal domains, and the central repeating sequence of H-caldesmon is deleted in L-caldesmon. (3) Although H-caldesmon and L-caldesmon have similar functional domains, their tissue and cell distributions are different. (4) Caldesmon can be a biomarker for the pathological diagnosis of tumors and the prediction of chemoradiotherapy response. (5) Caldesmon can suppress tumor metastasis by regulating podosome/invadopodium formation and vasculogenesis. Future research aspects may include (1) clinical data about the relationship between expression of the two isoforms in cancers (primary and metastasis) and patient survival; (2) the effects of the expression of upregulated or downregulated isoform in cancers (primary and metastasis) on cell motility and invasive characteristics; and (3) evidence-based clinical studies or animal models on the role of caldesmon in gastrointestinal motility disorders are critically required.
Y-BY and C-FX prepared literature research and wrote the manuscript. J-GL conceived the ideas and wrote the manuscript. CW and Y-BY designed project, wrote the manuscript, and prepared figures. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (Nos. 81603625 and 81603618), the Program for Xinglin Scholar at Shanghai University of Traditional Chinese Medicine (No. RC-2017-02-08), Cao Yongqing’s Shanghai Famous Traditional Chinese Medicine Academic Experience Research Studio (SHGZS-2017017), the Three-year action program for further accelerating the development of traditional Chinese medicine in Shanghai [ZY(2018-2020)-CCCX-1007], and the Second round construction project of national traditional Chinese medicine academic school inheritance studio (No. 2019-62).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abrams, J., Davuluri, G., Seiler, C., and Pack, M. (2012). Smooth muscle caldesmon modulates peristalsis in the wild type and non-innervated zebrafish intestine. Neurogastroenterol. Motil. 24, 288–299. doi: 10.1111/j.1365-2982.2011.01844.x
Ball, E. H., and Kovala, T. (1988). Mapping of caldesmon: relationship between the high and low molecular weight forms. Biochemistry 27, 6093–6098. doi: 10.1021/bi00416a039
Chang, K. P., Wang, C. L., Kao, H. K., Liang, Y., Liu, S. C., Huang, L. L., et al. (2013). Overexpression of caldesmon is associated with lymph node metastasis and poorer prognosis in patients with oral cavity squamous cell carcinoma. Cancer 119, 4003–4011. doi: 10.1002/cncr.28300
Chu, P. G., Arber, D. A., Weiss, L. M., and Chang, K. L. (2001). Utility of CD10 in distinguishing between endometrial stromal sarcoma and uterine smooth muscle tumors: an immunohistochemical comparison of 34 cases. Mod. Pathol. 14, 465–471. doi: 10.1038/modpathol.3880335
Comin, C. E., Dini, S., Novelli, L., Santi, R., Asirelli, G., and Messerini, L. (2006). h-Caldesmon, a useful positive marker in the diagnosis of pleural malignant mesothelioma, epithelioid type. Am. J. Surg. Pathol. 30, 463–469. doi: 10.1097/00000478-200604000-200604006
Dierks, S., von Hardenberg, S., Schmidt, T., Bremmer, F., Burfeind, P., and Kaulfuß, S. (2015). Leupaxin stimulates adhesion and migration of prostate cancer cells through modulation of the phosphorylation status of the actin-binding protein caldesmon. Oncotarget 6, 13591–13606. doi: 10.18632/oncotarget.3792
Ekinci, Ö, Öğüt, B., Çelik, B., and Dursun, A. (2018). Compared with elastin stains, h-caldesmon and desmin offer superior detection of vessel invasion in gastric, pancreatic, and colorectal adenocarcinomas. Int. J. Surg. Pathol. 26, 318–326. doi: 10.1177/1066896917752442
Eves, R., Webb, B. A., Zhou, S., and Mak, A. S. (2006). Caldesmon is an integral component of podosomes in smooth muscle cells. J. Cell Sci. 119, 1691–1702. doi: 10.1242/jcs.02881
Fisher, C., Montgomery, E., and Healy, V. (2003). Calponin and h-caldesmon expression in synovial sarcoma; the use of calponin in diagnosis. Histopathology 42, 588–593. doi: 10.1046/j.1365-2559.2003.01652.x
Foster, D. B., Huang, R., Hatch, V., Craig, R., Graceffa, P., Lehman, W., et al. (2004). Modes of caldesmon binding to actin: sites of caldesmon contact and modulation of interactions by phosphorylation. J. Biol. Chem. 279, 53387–53394. doi: 10.1074/jbc.M410109200
Foster, D. B., Shen, L. H., Kelly, J., Thibault, P., Van Eyk, J. E., and Mak, A. S. (2000). Phosphorylation of caldesmon by p21-activated kinase. implications for the Ca(2+) sensitivity of smooth muscle contraction. J. Biol. Chem. 275, 1959–1965. doi: 10.1074/jbc.275.3.1959
Glukhova, M. A., Kabakov, A. E., Frid, M. G., Ornatsky, O. I., Belkin, A. M., Mukhin, D. N., et al. (1988). Modulation of human aorta smooth muscle cell phenotype: a study of muscle-specific variants of vinculin, caldesmon, and actin expression. Proc. Natl. Acad. Sci. U S A. 85, 9542–9546. doi: 10.1073/pnas.85.24.9542
Gu, Z., Kordowska, J., Williams, G. L., Wang, C. L., and Hai, C. M. (2007). Erk1/2 MAPK and caldesmon differentially regulate podosome dynamics in A7r5 vascular smooth muscle cells. Exp. Cell Res. 313, 849–866. doi: 10.1016/j.yexcr.2006.12.005
Guo, H., Huang, R., Semba, S., Kordowska, J., Huh, Y. H., Khalina-Stackpole, Y., et al. (2013). Ablation of smooth muscle caldesmon affects the relaxation kinetics of arterial muscle. Pflugers Arch. 465, 283–294. doi: 10.1007/s00424-012-1178-1178
Hai, C. M., and Gu, Z. (2006). Caldesmon phosphorylation in actin cytoskeletal remodeling. Eur. J. Cell Biol. 85, 305–309. doi: 10.1016/j.ejcb.2005.08.008
Hamden, S. S., Schroeter, M. M., and Chalovich, J. M. (2010). Phosphorylation of caldesmon at sites between residues 627 and 642 attenuates inhibitory activity and contributes to a reduction in Ca2+-calmodulin affinity. Biophys. J. 99, 1861–1868. doi: 10.1016/j.bpj.2010.07.018
Hayashi, K., Fujio, Y., Kato, I., and Sobue, K. (1991). Structural and functional relationships between h- and l-caldesmons. J. Biol. Chem. 266, 355–361. doi: 10.1016/s0021-9258(18)52442-x
Hedges, J. C., Oxhorn, B. C., Carty, M., Adam, L. P., Yamboliev, I. A., and Gerthoffer, W. T. (2000). Phosphorylation of caldesmon by ERK MAP kinases in smooth muscle. Am. J. Physiol. Cell Physiol. 278, C718–C726. doi: 10.1152/ajpcell.2000.278.4.C718
Hirschey, M. D., and Zhao, Y. (2015). Metabolic regulation by lysine malonylation, succinylation, and glutarylation. Mol. Cell Proteom. 14, 2308–2315. doi: 10.1074/mcp.R114.046664
Horiuchi, K. Y., and Chacko, S. (1995). Effect of unphosphorylated smooth muscle myosin on caldesmon-mediated regulation of actin filament velocity. J. Muscle Res. Cell Motil. 16, 11–19. doi: 10.1007/bf00125306
Hou, Q., Tan, H. T., Lim, K. H., Lim, T. K., Khoo, A., Tan, I. B., et al. (2013). Identification and functional validation of caldesmon as a potential gastric cancer metastasis-associated protein. J. Proteome Res. 12, 980–990. doi: 10.1021/pr3010259
Huang, R., Li, L., Guo, H., and Wang, C. L. (2003). Caldesmon binding to actin is regulated by calmodulin and phosphorylation via different mechanisms. Biochemistry 42, 2513–2523. doi: 10.1021/bi0268605
Kim, K. H., Yeo, S. G., Kim, W. K., Kim, D. Y., Yeo, H. Y., Hong, J. P., et al. (2012). Up-regulated expression of l-caldesmon associated with malignancy of colorectal cancer. BMC Cancer 12:601. doi: 10.1186/1471-2407-12-601
Koganehira, Y., Takeoka, M., Ehara, T., Sasaki, K., Murata, H., Saida, T., et al. (2003). Reduced expression of actin-binding proteins, h-caldesmon and calponin h1, in the vascular smooth muscle inside melanoma lesions: an adverse prognostic factor for malignant melanoma. Br. J. Dermatol. 148, 971–980. doi: 10.1046/j.1365-2133.2003.05238.x
Köhler, C. N. (2010). The actin-binding protein caldesmon is in spleen and lymph nodes predominately expressed by smooth-muscle cells, reticular cells, and follicular dendritic cells. J. Histochem. Cytochem. 58, 183–193. doi: 10.1369/jhc.2009.954651
Köhler, C. N. (2011). Histochemical localization of caldesmon in the CNS and ganglia of the mouse. J. Histochem. Cytochem. 59, 504–517. doi: 10.1369/0022155411400875
Kordowska, J., Huang, R., and Wang, C. L. (2006). Phosphorylation of caldesmon during smooth muscle contraction and cell migration or proliferation. J. Biomed. Sci. 13, 159–172. doi: 10.1007/s11373-005-9060-9068
Lee, M. S., Lee, J., Kim, J. H., Kim, W. T., Kim, W. J., Ahn, H., et al. (2015). Overexpression of caldesmon is associated with tumor progression in patients with primary non-muscle-invasive bladder cancer. Oncotarget 6, 40370–40384. doi: 10.18632/oncotarget.5458
Lin, J. J., Li, Y., Eppinga, R. D., Wang, Q., and Jin, J. P. (2009). Roles of caldesmon in cell motility and actin cytoskeleton remodeling. Int. Rev. Cell Mol. Biol. 274, 1–68. doi: 10.1016/s1937-6448(08)02001-2007
Martinez-Ciarpaglini, C., Agustí, J., Alvarez, E., Hueso, L., Terrádez, L., and Monteagudo, C. (2018). h-caldesmon immunoreactivity in atypical fibroxanthoma: implications for the differential diagnosis. Pathology 50, 358–361. doi: 10.1016/j.pathol.2017.09.020
Mayanagi, T., and Sobue, K. (2011). Diversification of caldesmon-linked actin cytoskeleton in cell motility. Cell Adh. Migr. 5, 150–159. doi: 10.4161/cam.5.2.14398
Morita, T., Mayanagi, T., Yoshio, T., and Sobue, K. (2007). Changes in the balance between caldesmon regulated by p21-activated kinases and the Arp2/3 complex govern podosome formation. J. Biol. Chem. 282, 8454–8463. doi: 10.1074/jbc.M609983200
Ng, C. T., Fong, L. Y., Yong, Y. K., Hakim, M. N., and Ahmad, Z. (2018). Interferon-γ induces biphasic changes in caldesmon localization as well as adherens junction organization and expression in HUVECs. Cytokine 111, 541–550. doi: 10.1016/j.cyto.2018.06.010
Ngai, P. K., and Walsh, M. P. (1984). Inhibition of smooth muscle actin-activated myosin Mg2+-ATPase activity by caldesmon. J. Biol. Chem. 259, 13656–13659. doi: 10.1016/s0021-9258(18)89793-9
Nucci, M. R., O’Connell, J. T., Huettner, P. C., Cviko, A., Sun, D., and Quade, B. J. (2001). h-Caldesmon expression effectively distinguishes endometrial stromal tumors from uterine smooth muscle tumors. Am. J. Surg. Pathol. 25, 455–463. doi: 10.1097/00000478-200104000-200104004
Rizzello, A., Franck, J., Pellegrino, M., De Nuccio, F., Simeone, P., Fiore, G., et al. (2017). A proteomic analysis of human uterine myoma. Curr. Protein. Pept. Sci. 18, 167–174. doi: 10.2174/1389203717666160322150603
Rush, D. S., Tan, J., Baergen, R. N., and Soslow, R. A. (2001). h-Caldesmon, a novel smooth muscle-specific antibody, distinguishes between cellular leiomyoma and endometrial stromal sarcoma. Am. J. Surg. Pathol. 25, 253–258. doi: 10.1097/00000478-200102000-200102014
Saraydaroglu, O., Ozuysal, S., Kasap, M., and Ozerkan, K. (2008). The importance of CD10 and h-Caldesmon in the distinction of smooth muscle tumors of the uterus and endometrial stromal sarcoma. Saudi. Med. J. 29, 1349–1350.
Sobue, K., Kanda, K., Tanaka, T., and Ueki, N. (1988). Caldesmon: a common actin-linked regulatory protein in the smooth muscle and nonmuscle contractile system. J. Cell Biochem. 37, 317–325. doi: 10.1002/jcb.240370306
Sobue, K., Morimoto, K., and Inui, M. (1982). Control of actin-myosin interaction of gizzard smooth muscle by calmodulin- and caldesmon linked flip-flop mechanism. Biomed. Res. 3, 188–196. doi: 10.2220/biomedres.3.188
Sobue, K., Muramoto, Y., Fujita, M., and Kakiuchi, S. (1981). Purification of a calmodulin-binding protein from chicken gizzard that interacts with F-actin. Proc. Natl. Acad. Sci. U S A. 78, 5652–5655. doi: 10.1073/pnas.78.9.5652
Somara, S., and Bitar, K. N. (2008). Direct association of calponin with specific domains of PKC-alpha. Am. J. Physiol. Gastrointest. Liver Physiol. 295, G1246–G1254. doi: 10.1152/ajpgi.90461.2008
Song, Y., Wang, J., Cheng, Z., Gao, P., Sun, J., Chen, X., et al. (2017). Quantitative global proteome and lysine succinylome analyses provide insights into metabolic regulation and lymph node metastasis in gastric cancer. Sci. Rep. 7:42053. doi: 10.1038/srep42053
Ueki, N., Sobue, K., Kanda, K., Hada, T., and Higashino, K. (1987). Expression of high and low molecular weight caldesmons during phenotypic modulation of smooth muscle cells. Proc. Natl. Acad. Sci. U S A. 84, 9049–9053. doi: 10.1073/pnas.84.24.9049
Wang, C. L. (2001). Caldesmon and smooth-muscle regulation. Cell Biochem. Biophys. 35, 275–288. doi: 10.1385/cbb
Wang, C. L. (2008). Caldesmon and the regulation of cytoskeletal functions. Adv. Exp. Med. Biol. 644, 250–272. doi: 10.1007/978-0-387-85766-4_19
Wang, X., Wu, K., Zhang, Z., Lan, M., Jin, J., and Fan, D. (2001). [The effect of calponin and caldesmon in regulation of the gastrointestinal motility during pathophysiological adaptation]. Zhonghua Nei Ke Za Zhi 40, 459–462.
Watanabe, K., Kusakabe, T., Hoshi, N., Saito, A., and Suzuki, T. (1999). h-Caldesmon in leiomyosarcoma and tumors with smooth muscle cell-like differentiation: its specific expression in the smooth muscle cell tumor. Hum. Pathol. 30, 392–396. doi: 10.1016/s0046-8177(99)90113-90112
Watanabe, K., Tajino, T., Sekiguchi, M., and Suzuki, T. (2000). h-Caldesmon as a specific marker for smooth muscle tumors. Comparison with other smooth muscle markers in bone tumors. Am. J. Clin. Pathol. 113, 663–668. doi: 10.1309/jnqx-f4km-q0q0-7xk8
Yang, Q., Xie, Y. D., Zhang, M. X., Huang, B., Zhang, C., Li, H. Y., et al. (2014). Effect of electroacupuncture stimulation at Zusanli acupoint (ST36) on gastric motility: possible through PKC and MAPK signal transduction pathways. BMC Comp. Altern. Med. 14:137. doi: 10.1186/1472-6882-14-137
Yano, H., Hayashi, K., Haruna, M., and Sobue, K. (1994). Identification of two distinct promoters in the chicken caldesmon gene. Biochem. Biophys. Res. Commun. 201, 618–626. doi: 10.1006/bbrc.1994.1746
Yates, A. D., Achuthan, P., Akanni, W., Allen, J., Allen, J., Alvarez-Jarreta, J., et al. (2020). Ensembl 2020. Nucleic Acids Res. 48, D682–D688. doi: 10.1093/nar/gkz966
Yokouchi, K., Numaguchi, Y., Kubota, R., Ishii, M., Imai, H., Murakami, R., et al. (2006). l-Caldesmon regulates proliferation and migration of vascular smooth muscle cells and inhibits neointimal formation after angioplasty. Arterioscler. Thromb. Vasc. Biol. 26, 2231–2237. doi: 10.1161/01.Atv.0000239441.29687.97
Yoshio, T., Morita, T., Kimura, Y., Tsujii, M., Hayashi, N., and Sobue, K. (2007). Caldesmon suppresses cancer cell invasion by regulating podosome/invadopodium formation. FEBS Lett. 581, 3777–3782. doi: 10.1016/j.febslet.2007.06.073
Yu, G., and Qu, G. (2018). High molecular weight caldesmon expression in ovarian adult granulosa cell tumour and fibrothecoma. Histopathology 72, 359–361. doi: 10.1111/his.13365
Yu, G., Xu, J., Jiang, L., Cai, L., Zohar, Y., Wu, S., et al. (2019). Expression and clinical significance of H-caldesmon in gastrointestinal stromal tumor: is it a specific marker for myogenic differentiation? Int. J. Clin. Exp. Pathol. 12, 2566–2571.
Zheng, P. P., Hop, W. C., Sillevis Smitt, P. A., van den Bent, M. J., Avezaat, C. J., Luider, T. M., et al. (2005). Low-molecular weight caldesmon as a potential serum marker for glioma. Clin. Cancer Res. 11, 4388–4392. doi: 10.1158/1078-0432.Ccr-04-2512
Keywords: actin binding protein, caldesmon, isoform, smooth muscle, cancer
Citation: Yao Y-B, Xiao C-F, Lu J-G and Wang C (2021) Caldesmon: Biochemical and Clinical Implications in Cancer. Front. Cell Dev. Biol. 9:634759. doi: 10.3389/fcell.2021.634759
Received: 28 November 2020; Accepted: 18 January 2021;
Published: 18 February 2021.
Edited by:
Chang-Duk Jun, Gwangju Institute of Science and Technology, South KoreaReviewed by:
Daniele Vergara, University of Salento, ItalyCopyright © 2021 Yao, Xiao, Lu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yi-Bo Yao, ZWxldmVuem9lQDE2My5jb20=; Chen Wang, d2FuZ2NoZW5fbG9uZ2h1YUAxNjMuY29t
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.