- 1McArdle Laboratory for Cancer Research, University of Wisconsin-Madison, Madison, WI, United States
- 2Department of Cell and Regenerative Biology, University of Wisconsin-Madison, Madison, WI, United States
Ras proteins control a complex intracellular signaling network. Gain-of-function mutations in RAS genes lead to RASopathy disorders in humans, including Noonan syndrome (NS). NS is the second most common syndromic cause of congenital heart disease. Although conditional expression of the NrasG12D/+ mutation in adult hematopoietic system is leukemogenic, its effects on embryonic development remain unclear. Here, we report that pan-embryonic expression of endogenous NrasG12D/+ by Mox2-Cre in mice caused embryonic lethality from embryonic day (E) 15.5 and developmental defects predominantly in the heart. At E13.5, NrasG12D/+; Mox2Cre/+ embryos displayed a moderate expansion of hematopoietic stem and progenitor cells without a significant impact on erythroid differentiation in the fetal liver. Importantly, the mutant embryos exhibited cardiac malformations resembling human congenital cardiac defects seen in NS patients, including ventricular septal defects, double outlet right ventricle, the hypertrabeculation/thin myocardium, and pulmonary valve stenosis. The mutant heart showed dysregulation of ERK, BMP, and Wnt pathways, crucial signaling pathways for cardiac development. Endothelial/endocardial-specific expression of NrasG12D/+ caused the cardiac morphological defects and embryonic lethality as observed in NrasG12D/+; Mox2Cre/+ mutants, but myocardial-specific expression of NrasG12D/+ did not. Thus, oncogenic NrasG12D mutation may not be compatible with embryonic survival.
Introduction
Normal cardiac development is critical for proper cardiac function and embryo viability, which is regulated by complex biological processes including signaling transduction pathways. Congenital cardiac defects are one of the most common forms of human birth defects (Benjamin et al., 2017). However, the genetic basis responsible for congenital cardiac defects is not fully understood. The identification of molecular pathways critical for normal heart development could potentially lead to mechanistic discoveries underlying human congenital heart disease as well as novel therapies. Noonan syndrome (NS) is an autosomal dominant disorder with an incident of 1:1,000-2,500 live births (Baban et al., 2019). It is characterized by multiple defects with variable penetrance, including cranial facial abnormalities and congenital heart defects. The most frequent cardiac manifestation is pulmonic stenosis resulting from dysplastic valve leaflets, but atrioventricular or ventricular septal defects, mitral valve abnormalities and hypertrophic cardiomyopathy are also observed (Lauriol et al., 2015). NS patients have significantly higher risk to develop juvenile myelomonocytic leukemia (JMML). NS, along with a few other developmental diseases, belongs to RASopathy disorders, which are caused by genetic mutations hyperactivating the RAS/RAF/MEK/ERK pathway (Rauen, 2013). The mutated genes include all three RAS isoforms (HRAS, NRAS, and KRAS), RAS function modulators (PTPN11, SOS1, SHOC2, NF1, and SPRED1), and downstream signaling transducers (RAF1, BRAF, MEK1, and MEK2) (Cirstea et al., 2010).
Ras proteins are highly conserved small GTPases and act as master signaling switches that couple extracellular stimuli to the intracellular response machinery. They cycle between active GTP-bound state and inactive GDP-bound state (Bourne et al., 1990). Activated Ras proteins subsequently activate multiple downstream signaling pathways, including the RAF/MEK/ERK pathway. Canonical oncogenic mutations at RAS G12, G13, and Q61 severely compromise the hydrolysis of GTP to GDP, leading to the accumulation of Ras-GTP and the hyperactivation of Ras signaling. These strong mutations are identified essentially in all human cancers (Bos, 1989). Notably, more than 80% of patients with Costello syndrome (CS), a RASopathy disorder, harbor a heterozygous germline G12 mutation in HRAS (Aoki et al., 2005; Gripp et al., 2006; Kerr et al., 2006). A mouse CS model with endogenous expression of HrasG12V/+ induces developmental defects and neoplasms as seen in CS patients (Chen et al., 2009). In addition to CS-like phenotypes, mice with the HrasG12S/+ mutation are resistant to high-fat diet-induced obesity and exhibit impaired hepatic energy homeostasis (Oba et al., 2018).
In contrast to HRAS, relatively weak mutations in the KRAS gene are identified in RASopathy disorders (e.g., NS) (Schubbert et al., 2006; Cirstea et al., 2010). Five weak KRAS mutations (V14I, P34R, T58I, D153V, and F156L) associated with NS were previously characterized (Schubbert et al., 2007). P34R and D153V have normal intrinsic GTPase activity, while V14I and T58I have less impaired intrinsic GTPase activity compared to oncogenic G12D. Although F156L has similarly impaired intrinsic GTPase activity as G12D, this defect is partially rescued by its increased nucleotide exchange activity. These observations suggest that strong oncogenic KRAS mutations are not tolerated during human development. Indeed, expression of KrasG12D/+ in the mouse germline is embryonic lethal, with mutants dying at midgestation stages (Tuveson et al., 2004). To date, numerous unrelated individuals and NS families have been identified with NRAS mutations, including T50I, G60E, and a few G12 mutations (Cirstea et al., 2010; Denayer et al., 2012; Kraoua et al., 2012; Altmuller et al., 2017; Garren et al., 2020). Similar to KRAS mutations, NS-associated NRAS T50I and G60E mutations are weaker mutations compared to G12V as they only marginally activate MEK/ERK signaling in serum-deprived cells (Cirstea et al., 2010; Runtuwene et al., 2011; Denayer et al., 2012; Kraoua et al., 2012; Ekvall et al., 2015). By contrast, the NS patients with NRAS G12 mutations (G12N, G12R, G12S, and G12D) display more severe phenotypes than typical NS, including JMML, brain tumors, or neonatal lethality (Altmuller et al., 2017; Garren et al., 2020).
We and others previously showed that mice with the knock-in NrasG12D mutation are leukemogenic (Wang et al., 2010a, 2011b; Li et al., 2011; Xu et al., 2013). The conditional expression of endogenous NrasG12D in adult hematopoietic system by Mx1-Cre (Wang et al., 2010a, 2011b; Li et al., 2011; Xu et al., 2013) induces a fully penetrant hematologic malignancy after a prolonged latency. However, when early pan-embryonic expression of NrasG12D/+ was induced using Mox2-Cre that expresses Cre in epiblasts beginning at E5, we could not obtain any live pups, suggesting that early ubiquitous embryonic expression of NrasG12D leads to embryonic lethality (Wang et al., 2011b). However, the roles of NrasG12D remain unknown during embryonic development. Here, we demonstrate that pan-embryonic expression of NrasG12D/+ driven by Mox2-Cre results in embryonic lethality between E15.5 and E17.5. At E13.5, mutant embryos appear grossly normal. However, further analyses indicate a moderate expansion of hematopoietic stem and progenitor cells in the fetal liver. Importantly, prominent heart morphological defects are detected from E13.5, which resemble human congenital heart defects such as NS. E13.5 mutant hearts showed downregulated non-canonical Wnt and BMP pathways, and hyper-activated MEK/ERK signaling in the heart. Using different Cre lines to drive NrasG12D expression in different cardiac cell types, we discovered that the cardiac defects and embryonic lethality observed in NrasG12D/+; Mox2Cre/+ mutants originated from endothelial/endocardial cells, but not from myocardial cells.
Materials and Methods
Generation of Mutant Mice
All mouse lines were maintained on a pure C57BL/6 genetic background, which was achieved by more than six backcrosses to a C57BL/6 background. Mice bearing the conditional oncogenic Nras mutation (Nrasloxp stop cassett loxp (LSL) G12D/+) (Haigis et al., 2008) were crossed to Mox2Cre/+ mice (Tallquist and Soriano, 2000) (Jackson Laboratory #003755) to generate NrasLSL G12D/+; Mox2Cre/+ (NrasG12D/+; Mox2Cre/+) embryos. Tie2-Cre or cTnt (Tnnt2)-Cre mice (Jackson Laboratory Stock #008863 or 024240) were also employed to generate endothelial/endocardial- or myocardial-specific NrasG12D/+ mutants, respectively. Embryos were genotyped for the Nras mutation and Cre alleles as described (Wang et al., 2011b). All animal experiments were conducted in accordance with the Guide for the Care and Use of Laboratory Animals and approved by an Animal Care and Use Committee at the University of Wisconsin (protocol #M005328). The program is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care. Additional methods are described in Supplementary Materials and Methods.
Results
Pan-Embryonic Expression of NrasG12D/+ Leads to Embryonic Lethality Between E15.5 and E17.5
We used a conditional oncogenic Nras mouse line (NrasLoxp–STOP–Loxp (LSL) G12D/+ line) to induce oncogenic Nras expression from its endogenous locus in somatic cells (Haigis et al., 2008). In this mouse line, the expression of oncogenic Nras is silenced by a floxed stop cassette upstream of the coding sequence in the absence of Cre and induced upon temporally and spatially controlled Cre expression (Figure 1A). We previously reported that pan-embryonic expression of NrasG12D/+ driven by Mox2Cre beginning at E5 resulted in no live neonatal mice (Wang et al., 2011b), suggesting embryonic lethality of the mutants. To determine the exact stage of embryonic lethality, we harvested embryos between E12.5 and E17.5 from crosses of NrasLSL G12D/+ and Mox2Cre/+ mice (Table 1A and see section “Materials and Methods”). Expected numbers of NrasG12D/+; Mox2Cre/+ were recovered between E12.5 and E14.5. However, the total number of mutant embryos as well as the number of live mutants were significantly reduced from E15.5. All NrasG12D/+; Mox2Cre/+ embryos were dead by E17.5. Therefore, our data indicate that pan-embryonic expression of NrasG12D/+ results in embryonic lethality between E15.5 and E17.5.
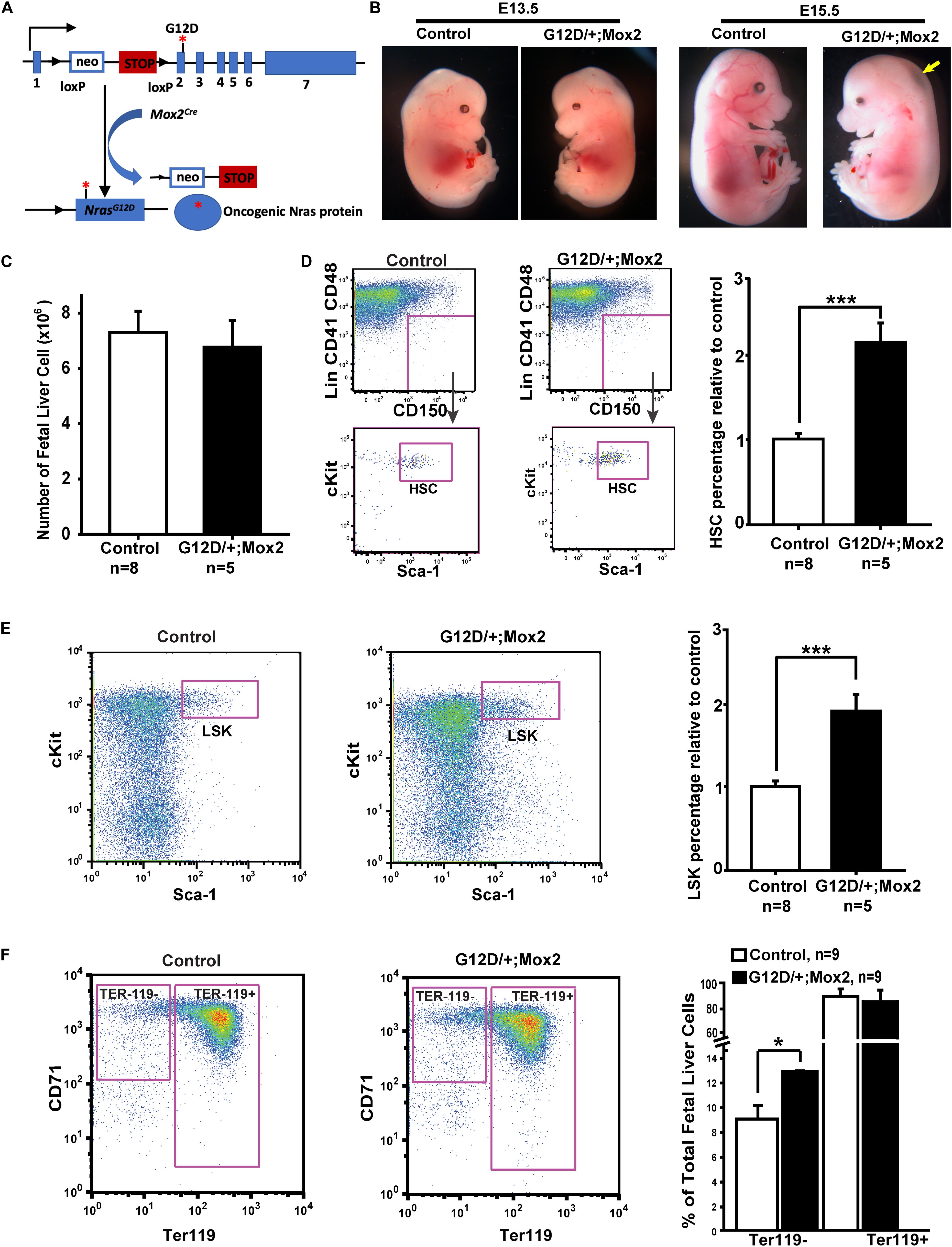
Figure 1. NrasG12D/+; Mox2Cre/+ (G12D/ +; Mox2) embryos exhibit whole body edema and the small liver at E15.5. (A) Schematic illustration of floxed and activated oncogenic Nras alleles. (B) Representative images of E13.5 and E15.5 WT control and G12D/ +; Mox2 mutant. The G12D/ +; Mox2 embryo looks grossly normal at E13.5 but at E15.5 looks pale and shows whole body edema as indicated by an arrow. (C) Quantitative analysis of cell numbers in E13.5 fetal livers from G12D/ +; Mox2 vs. control embryos. (D) Flow cytometric analysis and quantification of HSCs (defined as Lin– CD41– CD48– Mac1+ CD150+ Sca1+ cKit+ cells) in total live fetal liver cells. HSC percentages in control fetal liver cells were arbitrarily set as 1. (E) Flow cytometric analysis and quantification of LSKs (defined as Lin– Sca1+ cKit+ cells) in total fetal liver live cells. LSK percentage in control fetal liver cells were arbitrarily set as 1. (F) Flow cytometric analysis and quantification of erythroid differentiation based on CD71 and Ter119 expression. Unpaired 2-tailed Student’s t tests were used to determine the significance. Results are presented as mean + SD. *p < 0.05; ***p < 0.001.
To determine the mechanisms contributing to embryonic lethality in NrasG12D/+; Mox2Cre/+embryos, we carefully examined the histology of the embryos. The mutant embryos were grossly normal at E13.5 as compared to the wild-type (WT) littermate control (Figure 1B). However, the mutant embryos at E15.5 were smaller and pale with whole-body edema (arrow in Figure 1B) as compared to the control. Evaluation of multiple tissues, including the lung, kidney, and brain did not reveal any gross morphological defects (our unpublished observations). However, the fetal livers from mutant embryos were smaller at E15.5 and hepatic necrosis was evident as indicated by arrow, especially in the regions distal from blood flow (Supplementary Figure 1). Therefore, we investigated whether the embryonic lethality is caused by defective fetal liver hematopoiesis.
NrasG12D/+; Mox2Cre/+ Embryos Display an Expansion of Hematopoietic Stem and Progenitor Cells in the E13.5 Fetal Liver
We examined fetal liver hematopoiesis in E13.5 NrasG12D/+; Mox2Cre/+ and control embryos. At this stage, the number of fetal liver cells in mutant embryos was indistinguishable from WT control littermates (Figure 1C). However, detailed quantification of fetal liver hematopoietic stem cells (HSCs, defined as Lin– Mac1+ CD41– CD48– CD150+ Sca1+ cKit+) and LSK cells (hematopoietic stem and progenitor cells, defined as Lin– Sca1+ cKit+) indicated that both populations were expanded in mutant embryos (Figures 1D,E). This result is consistent with our prior analysis of hematopoietic stem and progenitor cells in adult NrasG12D/+; Mx1-Cre mice (Wang et al., 2010b, 2013a). We also examined fetal liver erythroid differentiation based on CD71 and Ter119 expression patterns as previously described (Zhang et al., 2003). Our results showed a moderate expansion of Ter119– CD71mid/hi cells, which were enriched with colony forming unit-erythroid (CFU-E) progenitors and early erythroblasts, while the percentage of terminally differentiating Ter119+ erythroid cells appeared comparable to that of control embryos (Figure 1F). This result is in sharp contrast to the blocked erythroid differentiation previously reported in KrasG12D/+ embryos (Zhang et al., 2007; Zhang and Lodish, 2007). Our data indicate that endogenous NrasG12D/+ signaling caused moderate effects on fetal liver hematopoiesis at E13.5. We previously reported that NrasQ61R/+; Vav-Cre mice, in which NrasQ61R/+ signaling is activated by Vav-Cre in the fetal liver from E11.5 (Ogilvy et al., 1999; Damnernsawad et al., 2016), were born at the expected Mendelian ratio (You et al., 2018). Since NrasQ61R is a much stronger mutant than NrasG12D (Kong et al., 2016), we believe that the mild perturbation of fetal liver hematopoiesis in NrasG12D/+; Mox2Cre/+ embryos is unlikely to cause a lethal phenotype.
NrasG12D/+; Mox2Cre/+ Embryos Exhibit Cardiac Developmental Defects
We systematically examined serial sections of the lung, kidney, and brain, and did not find any significant morphological abnormalities (data not shown). Given that normal cardiac morphogenesis is critical for proper cardiac function and embryonic survival, and that hyperactivation of Ras signaling is associated with cardiac abnormalities in humans such as NS, we set out to determine whether cardiac development was defective in mutant embryos.
Cardiac development of NrasG12D/+; Mox2Cre/+embryos was analyzed by examining serial H&E stained transverse sections (Figure 2). Since the mutants exhibited embryonic lethality between E15.5 and E17.5, we examined the mutant hearts at E13.5 for cardiac morphological defects. At E13.5, the interventricular septum in the control heart fused to the endocardial cushion and thus the right ventricle was separated from the left ventricle (right panel in Figure 2A, indicated by arrow). Conversely, the interventricular septum in the mutant heart failed to fuse with the endocardial cushion, leading to ventricular septal defects (VSDs) (right panels in Figures 2B,C, indicated by arrows), which is a common human congenital cardiac defect. In the normal heart, the aorta exits the left ventricle and the pulmonary trunk exits the right ventricle as shown in the left and middle panels of Figure 2A. However, the mutant heart showed a double outlet right ventricle (DORV) where both the aorta and pulmonary trunk exited the right ventricle (arrowheads, middle panels in Figures 2B,C), which is one of the human congenital cardiac defects. The mutant hearts showed an aorta that was connected to the right ventricle (middle panels in Figures 2B,C) in contrast to the control, which showed no connection between aorta and right ventricle (middle panel in Figure 2A). In addition, the mutant heart exhibited thinning of the ventricular wall, where the thin compact layer and hypertrabeculation were observed compared to controls as indicated by asterisk (right panels in Figures 2B,C vs. right panel in Figure 2A). The compact layer thickness in the right and left ventricle in the mutant hearts was significantly reduced by 62.5 and 45.5%, respectively (Supplementary Figure 2D). Furthermore, pulmonary valve stenosis was observed in the mutant heart as compared to the control (as indicated by pound sign in left panels in Figures 2B,C vs. left panel in Figure 2A), indicating defects in pulmonary valve remodeling. VSDs were fully penetrant (6/6 mutant hearts), while DORV, thin myocardium, or pulmonary stenosis was partially penetrant (4/6 mutant hearts).
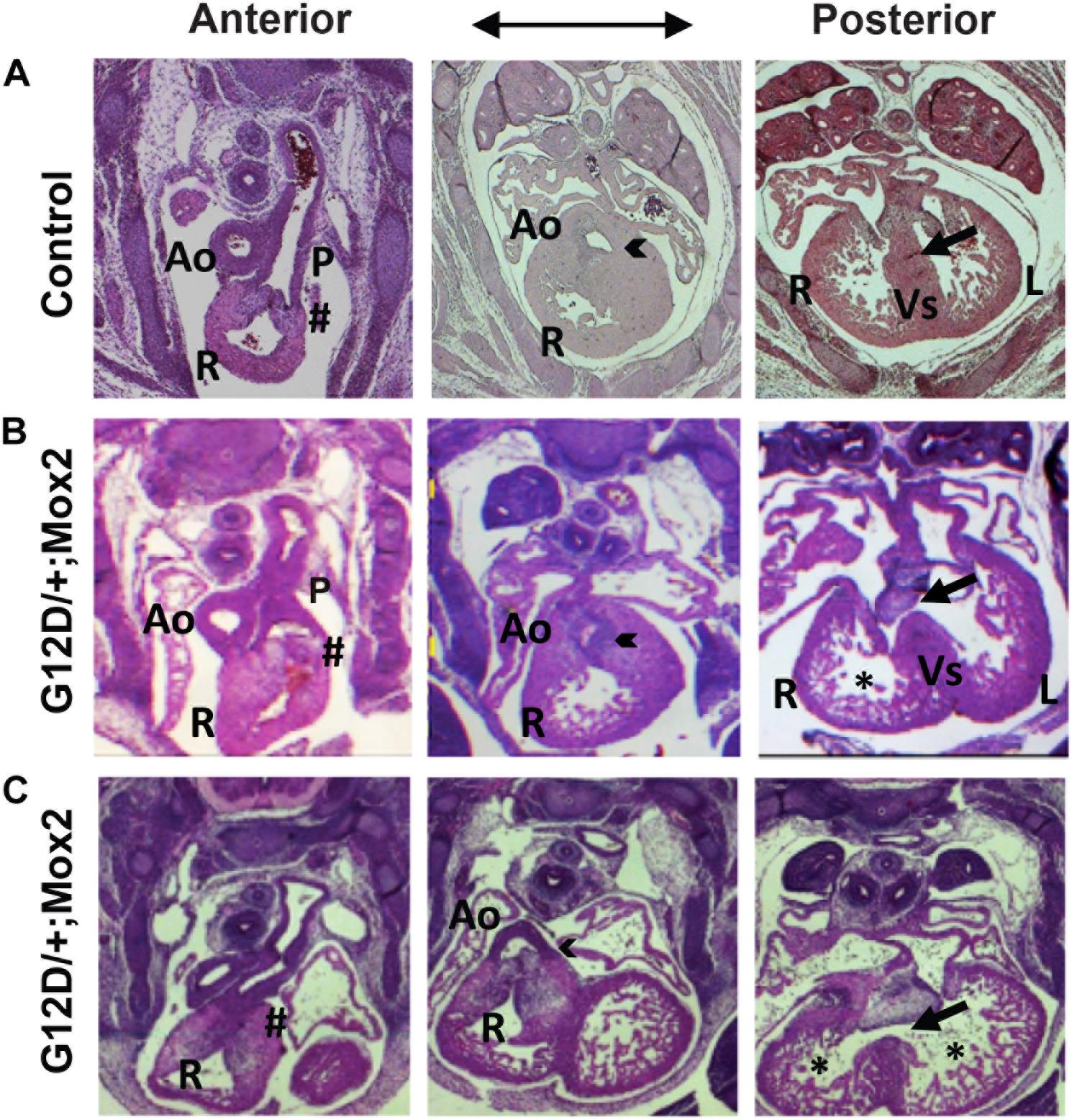
Figure 2. NrasG12D/+; Mox2Cre/+ (G12D/ +; Mox2) embryos exhibit cardiac developmental defects. Representative images of H&E stained transverse sections of control (A) vs. two G12D/ +; Mox2 mutant (B,C) embryos at E13.5. The mutant hearts show VSD (indicated by arrow in right panels B,C), DORV (indicated by arrowhead in middle panels B,C) and the thin ventricular wall (indicated by * in right panels B,C). The pulmonary valves were not remodeled properly (indicated by # in left panels B,C). Ao, Aorta; P, Pulmonary valve; R, Right ventricle; L, Left ventricle; Vs, Ventricular septum.
Ras signaling pathways are known to regulate cell proliferation and survival, which are crucial for normal cardiac development. Thus, cell proliferation and apoptosis were investigated in the embryonic heart. To examine cell proliferation, immunohistochemistry was performed on heart sections at E13.5 using Ki-67 antibodies (Supplementary Figure 2A). Our results showed no significant alterations in cell proliferation in the mutant heart compared to the control heart. When the trabecular and compact layers of the right and left ventricles were analyzed separately, we did not detect any significant changes in cell proliferation between the mutant heart and the control heart (Supplementary Figures 2B,C). Furthermore, the mutant heart did not show any significant changes in apoptosis compared to the control at E13.5 by immunofluorescence staining using Caspase3 antibodies (Supplementary Figure 3).
NrasG12D/+; Mox2Cre/+ Hearts Exhibit Defective Developmental Signaling Pathways and the Hyperactivated MEK/ERK Pathway
Our analyses of embryonic organ development indicate that the most significant defects were observed in the developing heart. To understand the molecular mechanisms underlying cardiac defects in NrasG12D/+; Mox2Cre/+ embryos, we performed gene profiling analysis on E13.5 control and mutant hearts. We identified 147 genes that were differentially expressed in mutant hearts (p < 0.01 and fold change >2), 98 genes downregulated and 49 genes upregulated (Figure 3A). One of the downregulated genes in mutant hearts is Islet1 (Isl1) (24-fold reduction), which is known to play critical roles in cardiac development (Moretti et al., 2006) and is also involved in conduction system development (Liang et al., 2015). Gene set enrichment analysis (GSEA) showed that non-canonical Wnt and BMP signaling pathways were significantly downregulated in mutant hearts (Figure 3B). Both Wnt (canonical and non-canonical) and BMP signaling pathways play critical roles in early embryonic as well as cardiac development (Tian et al., 2010; Wang et al., 2011a). By contrast, the MEK/ERK pathway downstream of oncogenic Ras was hyperactivated as expected due to the expression of NrasG12D/+. The hyperactivation of MEK/ERK signaling in mutant hearts was further validated using Western blot analysis (Figure 3C). Our finding is supported by previous reports that hyperactivated RAS/RAF/MEK/ERK pathway affects cardiac development in mice and humans [reviewed in Wang (2007)]. Phosphorylation of AKT (p-AKT), another signaling molecule downstream of Ras, was also moderately increased in the mutant heart, indicating that both ERK and AKT axes are activated.
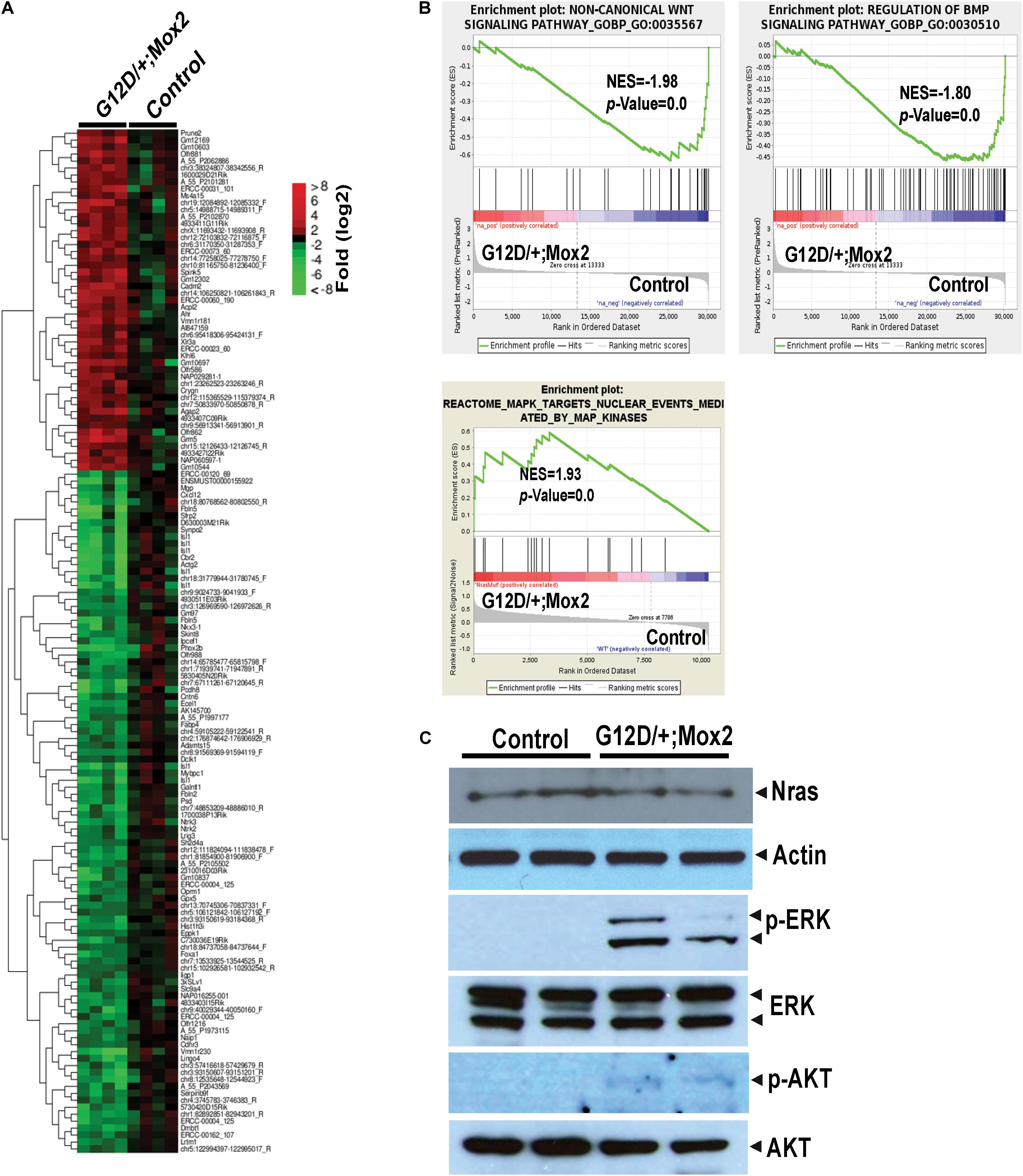
Figure 3. Global gene expression analyses of E13.5 NrasG12D/+; Mox2Cre/+ (G12D/+; Mox2) heart. Agilent microarray analysis was performed on E13.5 hearts from control and G12D/ +; Mox2 embryos. (A) Heat map depicting differentially expressed genes (p < 0.01 and fold change >2) between control and mutant hearts. (B) Gene Set Enrichment Analysis identified altered gene expression profiles in mutant hearts. Representative examples in non-canonical Wnt signaling pathway, BMP signaling pathway, and MAPK cascade are shown here. NES, normalized enrichment score. (C) Analysis of MEK/ERK and AKT signaling in E13.5 mutant hearts by Western blotting. Actin antibody was used for a loading control.
Endothelial/Endocardial Origin of Cardiac Defects in NrasG12D/+; Mox2Cre/+ Embryos
It is believed that crosstalk between the endocardium and myocardium is essential for normal heart development (MacGrogan et al., 2018) and the endocardium/endothelium plays an important role in underlying myocardial development (Mysliwiec et al., 2011). Thus, we investigated whether the cardiac defects of the mutant heart were due to endothelial/endocardial- or myocardial-specific defects by employing Tie2-Cre (Kisanuki et al., 2001) or Tnnt2-Cre (Chen et al., 2004) mice, respectively (Tables 1B,C and Supplementary Figure 4). NrasLSL G12D/+; Tie2-Cre (NrasG12D/+; Tie2-Cre) mutants showed the same embryonic lethality as NrasG12D/+; Mox2Cre/+ embryos (Table 1B). All NrasG12D/+; Tie2-Cre mutant embryos died by E17.5. Histological analyses at E13.5 revealed that the NrasG12D/+; Tie2-Cre mutant heart showed VSDs (2/5 mutants; indicated by arrow) and the thin compact layer with the deeply invaginated trabecular layer (3/5 mutants; indicated by asterisk) compared to the control heart (Supplementary Figure 4). DORV or pulmonary stenosis was also detected (2/5 mutants). Interestingly, NrasLSL G12D/+; Tnnt2-Cre (NrasG12D/+; Tnnt2-Cre) mutants showed neither embryonic lethality (Table 1C) nor any gross cardiac malformations (data not shown), indicating that the myocardial-specific expression of NrasG12D does not contribute to cardiac malformations or lethality during embryonic development. These results indicate that cardiac defects observed in NrasG12D/+; Mox2Cre/+ embryos have originated from endothelial/endocardial defects, but not from myocardial defects.
Discussion
Here we report that pan-embryonic expression of NrasG12D/+ in mice caused embryonic lethality at E15.5-E17.5 and developmental defects predominantly in the heart. The E13.5 NrasG12D/+; Mox2Cre/+ mutants displayed a moderate expansion of hematopoietic stem and progenitor cells without significant impact on erythroid differentiation in the fetal liver. The development of other tissues, including the lung, kidney, and brain, appeared morphologically normal. Importantly, the mutant hearts showed cardiac malformations resembling human congenital cardiac defects seen in NS, such as VSDs, DORV, pulmonary valve stenosis, and hypertrabeculation/thin myocardium. Consistent with cardiac functional insufficiency, mutant embryos started to die at E15.5 with whole body edema and pale body. Thus, our data indicate that ubiquitous expression of NrasG12D/+ in the early mouse embryo results in a spectrum of cardiac malformations in the mid to late gestation period. These cardiac defects have been well accepted as lethal congenital heart defects in various mouse models (Shou et al., 1998; Lee et al., 2000; He et al., 2012).
Somatic mutations in NRAS are involved in the development of hematological malignancies and a variety of solid tumors (COSMIC database)1. However, these oncogenic mutations have rarely been identified as germline mutations in human live births, and the majority of identified NRAS mutations in NS patients are biochemically weaker mutations instead (Runtuwene et al., 2011). These data suggest that like KRAS mutations, strong oncogenic NRAS mutations would cause embryonic lethality in humans. In support of this notion, we showed that pan-embryonic expression of NrasG12D/+ at its endogenous level caused embryonic lethality in mice (Table 1), while the expression of a hypomorphic allele of NrasG12D at up to 80% of NrasG12D endogenous level did not cause any detectable phenotypes (Wang et al., 2011c).
NrasG12D/+; Mox2Cre/+ embryos at E13.5 were grossly normal (Figure 1B) and showed mild phenotypes in fetal liver hematopoiesis (Figure 1). We detected that hematopoietic stem and progenitor cells underwent moderate expansion without significantly affecting terminal erythroid differentiation. We previously reported that NrasQ61R/+; Vav-Cre mice, in which the hematopoietic-specific expression of NrasQ61R [a much stronger mutant than NrasG12D (Kong et al., 2016)] was induced in the fetal liver from E11.5 (Ogilvy et al., 1999; Damnernsawad et al., 2016), were born at normal Mendelian ratios with hematopoietic phenotypes (You et al., 2018) similar to NrasG12D/+; Mox2Cre/+ embryos or NrasG12D/+; Mx1-Cre adult mice (Wang et al., 2011b). Therefore, we believe that the hematopoietic phenotypes would not cause or contribute to the embryonic lethality observed in NrasG12D/+; Mox2Cre/+ mutants. It is likely that the smaller liver and liver necrosis observed in E15.5 NrasG12D/+; Mox2Cre/+ embryos are a secondary effect caused by cardiac defects, due to limited blood flow to support the survival of fetal liver cells.
Our data indicate that the endothelial/endocardial expression of NrasG12D/+ in NrasG12D/+; Tie2-Cre embryos caused embryonic lethality (Table 1B) and cardiac malformations (Supplementary Figure 4), recapitulating the defects observed in NrasG12D/+; Mox2Cre/+ mutant embryos. By contrast, the myocardial-specific expression of NrasG12D/+ in NrasG12D/+; Tnnt2-Cre mutants showed neither embryonic lethality (Table 1C) nor gross cardiac malformations, indicating a non-cardiomyocyte origin of the cardiac defects observed in the NrasG12D/+; Mox2Cre/+ mutant mice. The ventricular myocardial wall consists of three layers: the endocardium (the inner layer), the myocardium (the middle layer) and the epicardium (the outer layer). During cardiac development, the one-layered myocardium must expand to the multilayered ventricular myocardial wall, consisting of the inner trabecular and outer compact layer (Wagner and Siddiqui, 2007). Myocardial trabeculation refers to the process by which the endocardium induces the underlying cardiomyocytes to proliferate, migrate to form finger-like projections in the heart. We and others demonstrated that endocardial signaling to the underlying myocardium plays crucial roles in the myocardial growth and development (Grego-Bessa et al., 2007; Mysliwiec et al., 2011). Our study indicates that Ras signaling in endothelial/endocardial cells is critical for normal cardiac morphogenesis including ventricular septation and the ventricular myocardial formation. These findings reiterate the importance of crosstalk between endocardial/endothelial cells and myocardial cells for normal ventricle development. Our conclusion is supported by other reports that the cell types responsible for the cardiac defects in a NS mouse model with the Ptpn11 D61 mutation are of endothelial/endocardial, but not cardiomyocyte origin (Araki et al., 2004; Araki et al., 2009). Although we have not observed significant changes in cell proliferation and apoptosis in the mutant ventricular wall at E13.5, it is plausible that cell proliferation or apoptosis may have been altered transiently and regionally within the mutant heart. Since Tie2-Cre drives oncogenic Nras expression in all endothelial cells, including both endocardium and endothelium of the heart, it cannot be used to determine direct changes in the endocardium vs. indirect changes in the myocardium upon activating Ras signaling. Future experiments employing an endocardial-specific Cre driver (Wu et al., 2012) may help address this question.
Similar to NrasG12D, germline KrasG12D mutation in mice also caused embryonic lethality (Tuveson et al., 2004). The mutants died much earlier, between E9.5 and E11.5, with profound developmental arrest and widespread apoptosis, including cardiomegaly and abnormal brain development. Interestingly, although mice with endogenous cardiomyocyte-specific KrasG12D expression showed hyperactivation of myocardial ERK and AKT signaling, they were born in expected Mendelian ratios and appeared healthy with normal function, size, and histology of the heart (Dalin et al., 2014). These phenotypes are highly consistent with our mutants containing the myocardial-specific expression of NrasG12D. In addition to NRAS and KRAS, embryonic expression of the most common and potent Ptpn11 mutation (Ptpn11E76K/+) also results in embryonic lethality at E11.5 (Xu et al., 2011). In contrast to NRAS and KRAS, strong oncogenic HRAS mutations are identified in CS (Aoki et al., 2005). Corroborating with human results, mice with germline expression of HrasG12V/+ were born at the expected Mendelian frequency with CS-like phenotypes (Chen et al., 2009). HrasG12V/+ embryos at E19 do not display any cardiopulmonary anomalies. However, most of them (>80%) died within 14 days after birth. The cause of this neonatal lethality is unknown since there are no abnormalities in the heart and lungs and no evidence of bleeding.
Future studies will be aimed at determining how dysregulated gene expression and signaling pathways cause cardiac defects and embryonic lethality, which will advance mechanistic insights into congenital heart defects including NS associated heart defects. Isl1 was significantly downregulated in mutant hearts (24-fold). Isl-1 plays critical roles in early cardiac development and is expressed in the second heart field that gives rise to the right ventricle, outflow tract and atria and thus considered as a cardiac progenitor marker (Moretti et al., 2006). Furthermore, a new ISL1 mutation predisposes to congenital DORV (Wang et al., 2019). Thus, it would be interesting to investigate the effect of Isl1 downregulation on cardiac abnormalities in the mutant heart. Our GSEA identified that non-canonical Wnt and BMP signaling pathways were significantly downregulated in mutant hearts (Figure 3B). Since both Wnt and BMP signaling play critical roles in cardiac development (Tian et al., 2010; Wang et al., 2011a), future direction would be to determine their effects on gene regulation and thus cardiac malformations. Interestingly, our gene expression profiling showed the significant reduction of a Wnt regulator Sfrp2 (Lin et al., 2016) and Foxa1(Wang et al., 2013b), a transcription factor targeted by BMP or Wnt (11.3- and 14.9-fold reduction, respectively). However, the mechanisms by which these alterations cause cardiac malformations remain to be elucidated. Wnt signaling plays key roles in early cardiac development and disease, but their functions are complex (Wagner and Siddiqui, 2007). Non-canonical Wnt such as Wnt11 induces cardiogenesis while other canonical Wnt3a and 8c inhibit this process during early development. BMPs are also involved in early embryonic patterning and cardiogenesis (Wagner and Siddiqui, 2007; Wang et al., 2011a). In particular, BMPs 2 and 4 are necessary for early cardiac mesoderm development, and BMP10 is crucial for ventricular trabeculation (Chen et al., 2004). Trabeculation is a critical step for the formation of normal thickened ventricular wall. It is plausible that reduced BMP signaling contributes to the thin ventricular wall in NrasG12D/+; Mox2Cre/+ heart. Furthermore, it would be interesting to investigate whether dysregulated Isl1, BMP, or Wnt signaling also plays a role in cardiac defects manifested in NS patients with RAS activation, which will help develop treatment options.
Taken together, our and others’ studies suggest that hyperactivation of Ras signaling beyond a certain threshold would cause cardiac defects and more severely, embryonic lethality. Furthermore, endothelial/endocardial-specific expression of the NrasG12D mutant gene results in embryonic lethality and contributes to the cardiac morphological defects observed in the NrasG12D/+; Mox2Cre/+ mutant hearts. Our findings would provide the important basis to advance our understanding of developmental defects associated with hyperactivated Ras signaling.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Ethics Statement
All animal experiments were conducted in accordance with the Guide for the Care and Use of Laboratory Animals and approved by an Animal Care and Use Committee at the University of Wisconsin (protocol #M005328). The program is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care.
Author Contributions
XY, M-JR, JZ, and YLe: conception, design, writing, review, and/or revision of the manuscript. XY, M-JR, EC, YS, and AD: acquisition of data, analysis, and interpretation of data. YLi and YZ: technical or material support. JZ and YLe: study supervision. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by R01 grants CA152108 and HL113066 to JZ. This work was also supported in part by NIH/NCI P30 CA014520, UWCCC Support. YLe was supported in part by American Heart Association 12GRNT 12070021 and UW VCRGE with the Wisconsin Alumni Research Foundation.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Dr. Christopher Bradfield for the microarray experiments and analyses, to Dr. Ruth Sullivan for pathological evaluation of various embryonic tissues, and to Dr. Xin Sun and Dr. Zhen Zhang for their help to dissect and take pictures of embryos. We also thank the University of Wisconsin Carbone Cancer Center (UWCCC) for use of its Shared Services (Flow Cytometry Laboratory, Genome Editing and Animal Models Shared Resource, and Experimental Pathology Laboratory) to conduct various experiments.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.633661/full#supplementary-material
Footnotes
References
Altmuller, F., Lissewski, C., Bertola, D., Flex, E., Stark, Z., Spranger, S., et al. (2017). Genotype and phenotype spectrum of NRAS germline variants. Eur. J. Hum. Genet. 25, 823–831. doi: 10.1038/ejhg.2017.65
Aoki, Y., Niihori, T., Kawame, H., Kurosawa, K., Ohashi, H., Tanaka, Y., et al. (2005). Germline mutations in HRAS proto-oncogene cause costello syndrome. Nat. Genet. 37, 1038–1040. doi: 10.1038/ng1641
Araki, T., Chan, G., Newbigging, S., Morikawa, L., Bronson, R. T., and Neel, B. G. (2009). Noonan syndrome cardiac defects are caused by PTPN11 acting in endocardium to enhance endocardial-mesenchymal transformation. Proc. Natl. Acad. Sci. U.S.A. 106, 4736–4741. doi: 10.1073/pnas.0810053106
Araki, T., Mohi, M. G., Ismat, F. A., Bronson, R. T., Williams, I. R., Kutok, J. L., et al. (2004). Mouse model of Noonan syndrome reveals cell type- and gene dosage-dependent effects of Ptpn11 mutation. Nat. Med. 10, 849–857. doi: 10.1038/nm1084
Baban, A., Olivini, N., Lepri, F. R., Cali, F., Mucciolo, M., Digilio, M. C., et al. (2019). SOS1 mutations in Noonan syndrome: cardiomyopathies and not only congenital heart defects! Report of six patients including two novel variants and literature review. Am. J. Med. Genet. A 179, 2083–2090. doi: 10.1002/ajmg.a.61312
Benjamin, E. J., Blaha, M. J., Chiuve, S. E., Cushman, M., Das, S. R., Deo, R., et al. (2017). Heart disease and stroke statistics-2017 update: a report from the american heart association. Circulation 135, e146–e603. doi: 10.1161/CIR.0000000000000485
Bourne, H. R., Sanders, D. A., and McCormick, F. (1990). The GTPase superfamily: a conserved switch for diverse cell functions. Nature 348, 125–132. doi: 10.1038/348125a0
Chen, H., Shi, S., Acosta, L., Li, W., Lu, J., Bao, S., et al. (2004). BMP10 is essential for maintaining cardiac growth during murine cardiogenesis. Development 131, 2219–2231. doi: 10.1242/dev.01094
Chen, X., Mitsutake, N., LaPerle, K., Akeno, N., Zanzonico, P., Longo, V. A., et al. (2009). Endogenous expression of Hras(G12V) induces developmental defects and neoplasms with copy number imbalances of the oncogene. Proc. Natl. Acad. Sci. U.S.A. 106, 7979–7984. doi: 10.1073/pnas.0900343106
Cirstea, I. C., Kutsche, K., Dvorsky, R., Gremer, L., Carta, C., Horn, D., et al. (2010). A restricted spectrum of NRAS mutations causes Noonan syndrome. Nat. Genet. 42, 27–29. doi: 10.1038/ng.497
Dalin, M. G., Zou, Z., Scharin-Tang, M., Safari, R., Karlsson, C., and Bergo, M. O. (2014). Myocardial KRAS(G12D) expression does not cause cardiomyopathy in mice. Cardiovasc. Res. 101, 229–235. doi: 10.1093/cvr/cvt260
Damnernsawad, A., Kong, G., Wen, Z., Liu, Y., Rajagopalan, A., You, X., et al. (2016). Kras is required for adult hematopoiesis. Stem Cells 34, 1859–1871. doi: 10.1002/stem.2355
Denayer, E., Peeters, H., Sevenants, L., Derbent, M., Fryns, J. P., and Legius, E. (2012). NRAS mutations in noonan syndrome. Mol Syndromol 3, 34–38.
Ekvall, S., Wilbe, M., Dahlgren, J., Legius, E., van Haeringen, A., Westphal, O., et al. (2015). Mutation in NRAS in familial Noonan syndrome–case report and review of the literature. BMC Med. Genet. 16:95. doi: 10.1186/s12881-015-0239-1
Garren, B., Stephan, M., and Hogue, J. S. (2020). NRAS associated RASopathy and embryonal rhabdomyosarcoma. Am. J. Med. Genet. A 182, 195–200. doi: 10.1002/ajmg.a.61395
Grego-Bessa, J., Luna-Zurita, L., del Monte, G., Bolos, V., Melgar, P., Arandilla, A., et al. (2007). Notch signaling is essential for ventricular chamber development. Dev. Cell 12, 415–429. doi: 10.1016/j.devcel.2006.12.011
Gripp, K. W., Lin, A. E., Stabley, D. L., Nicholson, L., Scott, C. I. Jr., Doyle, D., et al. (2006). HRAS mutation analysis in Costello syndrome: genotype and phenotype correlation. Am. J. Med. Genet. A 140, 1–7. doi: 10.1002/ajmg.a.31047
Haigis, K. M., Kendall, K. R., Wang, Y., Cheung, A., Haigis, M. C., Glickman, J. N., et al. (2008). Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nat. Genet. 40, 600–608. doi: 10.1038/ng.115
He, A., Ma, Q., Cao, J., von Gise, A., Zhou, P., Xie, H., et al. (2012). Polycomb repressive complex 2 regulates normal development of the mouse heart. Circ. Res. 110, 406–415. doi: 10.1161/CIRCRESAHA.111.252205
Kerr, B., Delrue, M. A., Sigaudy, S., Perveen, R., Marche, M., Burgelin, I., et al. (2006). Genotype-phenotype correlation in Costello syndrome: HRAS mutation analysis in 43 cases. J. Med. Genet. 43, 401–405. doi: 10.1136/jmg.2005.040352
Kisanuki, Y. Y., Hammer, R. E., Miyazaki, J., Williams, S. C., Richardson, J. A., and Yanagisawa, M. (2001). Tie2-Cre transgenic mice: a new model for endothelial cell-lineage analysis in vivo. Dev. Biol. 230, 230–242. doi: 10.1006/dbio.2000.0106
Kong, G., Chang, Y. I., You, X., Ranheim, E. A., Zhou, Y., Burd, C. E., et al. (2016). The ability of endogenous Nras oncogenes to initiate leukemia is codon-dependent. Leukemia 30, 1935–1938. doi: 10.1038/leu.2016.89
Kraoua, L., Journel, H., Bonnet, P., Amiel, J., Pouvreau, N., Baumann, C., et al. (2012). Constitutional NRAS mutations are rare among patients with Noonan syndrome or juvenile myelomonocytic leukemia. Am. J. Med. Genet. A 158A, 2407–2411. doi: 10.1002/ajmg.a.35513
Lauriol, J., Jaffre, F., and Kontaridis, M. I. (2015). The role of the protein tyrosine phosphatase SHP2 in cardiac development and disease. Semin. Cell Dev. Biol. 37, 73–81. doi: 10.1016/j.semcdb.2014.09.013
Lee, Y., Song, A. J., Baker, R., Micales, B., Conway, S. J., and Lyons, G. E. (2000). Jumonji, a nuclear protein that is necessary for normal heart development. Circ. Res. 86, 932–938. doi: 10.1161/01.res.86.9.932
Li, Q., Haigis, K. M., McDaniel, A., Harding-Theobald, E., Kogan, S. C., Akagi, K., et al. (2011). Hematopoiesis and leukemogenesis in mice expressing oncogenic NrasG12D from the endogenous locus. Blood 117, 2022–2032. doi: 10.1182/blood-2010-04-280750
Liang, X., Zhang, Q., Cattaneo, P., Zhuang, S., Gong, X., Spann, N. J., et al. (2015). Transcription factor ISL1 is essential for pacemaker development and function. J. Clin. Invest. 125, 3256–3268. doi: 10.1172/JCI68257
Lin, H., Angeli, M., Chung, K. J., Ejimadu, C., Rosa, A. R., and Lee, T. (2016). sFRP2 activates Wnt/beta-catenin signaling in cardiac fibroblasts: differential roles in cell growth, energy metabolism, and extracellular matrix remodeling. Am. J. Physiol. Cell Physiol. 311, C710–C719. doi: 10.1152/ajpcell.00137.2016
MacGrogan, D., Munch, J., and de la Pompa, J. L. (2018). Notch and interacting signalling pathways in cardiac development, disease, and regeneration. Nat. Rev. Cardiol. 15, 685–704. doi: 10.1038/s41569-018-0100-2
Moretti, A., Caron, L., Nakano, A., Lam, J. T., Bernshausen, A., Chen, Y., et al. (2006). Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell 127, 1151–1165. doi: 10.1016/j.cell.2006.10.029
Mysliwiec, M. R., Bresnick, E. H., and Lee, Y. (2011). Endothelial Jarid2/Jumonji is required for normal cardiac development and proper Notch1 expression. J. Biol. Chem. 286, 17193–17204. doi: 10.1074/jbc.M110.205146
Oba, D., Inoue, S. I., Miyagawa-Tomita, S., Nakashima, Y., Niihori, T., Yamaguchi, S., et al. (2018). Mice with an oncogenic HRAS mutation are resistant to high-fat diet-induced obesity and exhibit impaired hepatic energy homeostasis. EBioMedicine 27, 138–150. doi: 10.1016/j.ebiom.2017.11.029
Ogilvy, S., Metcalf, D., Gibson, L., Bath, M. L., Harris, A. W., and Adams, J. M. (1999). Promoter elements of vav drive transgene expression in vivo throughout the hematopoietic compartment. Blood 94, 1855–1863.
Rauen, K. A. (2013). The RASopathies. Annu. Rev. Genomics Hum. Genet. 14, 355–369. doi: 10.1146/annurev-genom-091212-153523
Runtuwene, V., van Eekelen, M., Overvoorde, J., Rehmann, H., Yntema, H. G., Nillesen, W. M., et al. (2011). Noonan syndrome gain-of-function mutations in NRAS cause zebrafish gastrulation defects. Dis. Model Mech. 4, 393–399. doi: 10.1242/dmm.007112
Schubbert, S., Bollag, G., Lyubynska, N., Nguyen, H., Kratz, C. P., Zenker, M., et al. (2007). Biochemical and functional characterization of germ line KRAS mutations. Mol. Cell. Biol. 27, 7765–7770. doi: 10.1128/MCB.00965-07
Schubbert, S., Zenker, M., Rowe, S. L., Boll, S., Klein, C., Bollag, G., et al. (2006). Germline KRAS mutations cause Noonan syndrome. Nat. Genet. 38, 331–336. doi: 10.1038/ng1748
Shou, W., Aghdasi, B., Armstrong, D. L., Guo, Q., Bao, S., Charng, M. J., et al. (1998). Cardiac defects and altered ryanodine receptor function in mice lacking FKBP12. Nature 391, 489–492. doi: 10.1038/35146
Tallquist, M. D., and Soriano, P. (2000). Epiblast-restricted Cre expression in MORE mice: a tool to distinguish embryonic vs. extra-embryonic gene function. Genesis 26, 113–115.
Tian, Y., Cohen, E. D., and Morrisey, E. E. (2010). The importance of Wnt signaling in cardiovascular development. Pediatr. Cardiol. 31, 342–348. doi: 10.1007/s00246-009-9606-z
Tuveson, D. A., Shaw, A. T., Willis, N. A., Silver, D. P., Jackson, E. L., Chang, S., et al. (2004). Endogenous oncogenic K-ras(G12D) stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell 5, 375–387.
Wagner, M., and Siddiqui, M. A. (2007). Signal transduction in early heart development (I): cardiogenic induction and heart tube formation. Exp. Biol. Med. 232, 852–865.
Wang, J., Greene, S. B., and Martin, J. F. (2011a). BMP signaling in congenital heart disease: new developments and future directions. Birth Defects Res. A Clin. Mol. Teratol. 91, 441–448. doi: 10.1002/bdra.20785
Wang, J., Kong, G., Liu, Y., Du, J., Chang, Y.-I., Zhang, X., et al. (2013a). Nras G12D/+ promotes leukemogenesis by aberrantly regulating haematopoietic stem cell functions. Blood 121, 5203–5207.
Wang, J., Liu, Y., Li, Z., Du, J., Ryu, M. J., Taylor, P. R., et al. (2010a). Endogenous oncogenic Nras mutation promotes aberrant GM-CSF signaling in granulocytic/monocytic precursors in a murine model of chronic myelomonocytic leukemia. Blood 116, 5991–6002. doi: 10.1182/blood-2010-04-281527
Wang, J., Liu, Y., Li, Z., Wang, Z., Tan, L. X., Ryu, M. J., et al. (2011b). Endogenous oncogenic Nras mutation initiates hematopoietic malignancies in a dose- and cell type-dependent manner. Blood 118, 368–379. doi: 10.1182/blood-2010-12-326058
Wang, J., Park, J. S., Wei, Y., Rajurkar, M., Cotton, J. L., Fan, Q., et al. (2013b). TRIB2 acts downstream of Wnt/TCF in liver cancer cells to regulate YAP and C/EBPalpha function. Mol. Cell. 51, 211–225. doi: 10.1016/j.molcel.2013.05.013
Wang, J. Y., Liu, Y. G., Li, Z. Y., Du, J., Ryu, M. J., Taylor, P. R., et al. (2010b). Endogenous oncogenic Nras mutation leads to aberrant GM-CSF signaling in granulocytic/monocytic precursors in a murine model of chronic myelomonocytic leukemia. Blood 116, 5991–6002.
Wang, J. Y., Liu, Y. G., Li, Z. Y., Wang, Z. D., Tan, L. X., Ryu, M. J., et al. (2011c). Endogenous oncogenic Nras mutation initiates hematopoietic malignancies in a dose- and cell type-dependent manner. Blood 118, 368–379.
Wang, Y. (2007). Mitogen-activated protein kinases in heart development and diseases. Circulation 116, 1413–1423. doi: 10.1161/CIRCULATIONAHA.106.679589
Wang, Z., Song, H. M., Wang, F., Zhao, C. M., Huang, R. T., Xue, S., et al. (2019). A New ISL1 loss-of-function mutation predisposes to congenital double outlet right ventricle. Int Heart J 60, 1113–1122. doi: 10.1536/ihj.18-685
Wu, B., Zhang, Z., Lui, W., Chen, X., Wang, Y., Chamberlain, A. A., et al. (2012). Endocardial cells form the coronary arteries by angiogenesis through myocardial-endocardial VEGF signaling. Cell 151, 1083–1096. doi: 10.1016/j.cell.2012.10.023
Xu, D., Liu, X., Yu, W. M., Meyerson, H. J., Guo, C., Gerson, S. L., et al. (2011). Non-lineage/stage-restricted effects of a gain-of-function mutation in tyrosine phosphatase Ptpn11 (Shp2) on malignant transformation of hematopoietic cells. J. Exp. Med. 208, 1977–1988. doi: 10.1084/jem.20110450
Xu, J., Haigis, K. M., Firestone, A. J., McNerney, M. E., Li, Q., Davis, E., et al. (2013). Dominant role of oncogene dosage and absence of tumor suppressor activity in Nras-driven hematopoietic transformation. Cancer Discov. 3, 993–1001. doi: 10.1158/2159-8290.CD-13-0096
You, X., Kong, G., Ranheim, E. A., Yang, D., Zhou, Y., and Zhang, J. (2018). Unique dependence on Sos1 in Kras (G12D) -induced leukemogenesis. Blood 132, 2575–2579. doi: 10.1182/blood-2018-09-874107
Zhang, J., Liu, Y., Beard, C., Tuveson, D. A., Jaenisch, R., Jacks, T. E., et al. (2007). Expression of oncogenic K-ras from its endogenous promoter leads to a partial block of erythroid differentiation and hyperactivation of cytokine-dependent signaling pathways. Blood 109, 5238–5241.
Zhang, J., and Lodish, H. F. (2007). Endogenous K-ras signaling in erythroid differentiation. Cell Cycle 6, 1970–1973.
Keywords: Nras mutation, RASopathy, congenital heart defects, fetal liver hematopoiesis, embryonic development, Noonan syndrome, heart development
Citation: You X, Ryu M-J, Cho E, Sang Y, Damnernsawad A, Zhou Y, Liu Y, Zhang J and Lee Y (2021) Embryonic Expression of NrasG 12 D Leads to Embryonic Lethality and Cardiac Defects. Front. Cell Dev. Biol. 9:633661. doi: 10.3389/fcell.2021.633661
Received: 25 November 2020; Accepted: 21 January 2021;
Published: 11 February 2021.
Edited by:
Sigmar Stricker, Freie Universität Berlin, GermanyReviewed by:
Robert Kelly, UMR7288 Institut de Biologie du Développement de Marseille (IBDM), FranceErnesto Diaz-Flores, University of California, San Francisco, United States
Copyright © 2021 You, Ryu, Cho, Sang, Damnernsawad, Zhou, Liu, Zhang and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jing Zhang, emhhbmdAb25jb2xvZ3kud2lzYy5lZHU=; Youngsook Lee, eW91bmdzb29rbGVlQHdpc2MuZWR1
†These authors have contributed equally to this work
 Xiaona You
Xiaona You Myung-Jeom Ryu
Myung-Jeom Ryu Eunjin Cho2
Eunjin Cho2 Jing Zhang
Jing Zhang Youngsook Lee
Youngsook Lee