
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol. , 08 September 2021
Sec. Molecular and Cellular Pathology
Volume 9 - 2021 | https://doi.org/10.3389/fcell.2021.626045
This article is part of the Research Topic Bacteria and Bacterial Metabolites: Molecular Interplay with Gut Immunity View all 11 articles
The role of gut microbiota in the development of various tumors has been a rising topic of public interest, and in recent years, many studies have reported a close relationship between microbial groups and tumor development. Gut microbiota play a role in host metabolism, and the positive and negative alterations of these microbiota have an effect on tumor treatment. The microbiota directly promote, eliminate, and coordinate the efficacy of chemotherapy drugs and the toxicity of adjuvant drugs, and enhance the ability of patients to respond to tumors in adjuvant immunotherapy. In this review, we outline the significance of gut microbiota in tumor development, reveal its impacts on chemotherapy and immunotherapy, and discover various potential mechanisms whereby they influence tumor treatment. This review demonstrates the importance of intestinal microbiota-related research for clinical tumor treatment and provides additional strategy for clinical assistance in cancer treatment.
The homeostasis of the intestinal microbiota plays an important role in the normal physiological activities of the hosts, and microbiome imbalance greatly promotes tumorigenesis. In the past decade, substantial progress has been made in the investigation of the relationship between tumorigenesis and the role of microbiota. Investigations have shown that causes of cancer are associated with obesity, cardiovascular disease, type 2 diabetes, and carcinogenic chemicals, which are proven affected by the microbial community (Viennois et al., 2017; Cremonesi et al., 2018; Qin et al., 2018; Wu et al., 2019). Moreover, intestinal microbiota play a role in the pathogenesis of colorectal cancer. Studies have shown that microbiota indirectly affect the occurrence and development of colorectal cancer through inflammation and immune response (Honda and Littman, 2016; Chiaro et al., 2017). At present, the pathogenesis of colon cancer is mainly affected by epithelial gene mutation, mucosal integrity, intestinal microbiota, and inflammation. Gallimore and Godkin (2013) describe, in detail, the intestinal microorganism-mediated carcinogenic model and propose that the damage to the intestinal mucosal barrier integrity is key in the occurrence and development of colon cancer (Grivennikov et al., 2012; Gallimore and Godkin, 2013). Due to the destruction of the intestinal mucosal barrier, bacteria and their metabolites in the intestinal cavity are translocated to the lamina propria through the intercellular space of the epithelial cells, thus triggering an adaptive inflammatory response and the release of cytokines, such as IL-1, IL-6, and IL-23. This will activate downstream Th17 cells, promoting the release of IL-17 and further activating the inflammatory and proliferative pathways of epithelial cells, such as the STAT3 and NF-κB pathways, in turn, promoting cancer cell proliferation and invasion. This results in further destruction of mucosal integrity, aggravation of inflammatory reaction, and repeated inflammation of the colon, which directly factors in the occurrence and development of colon cancer (Figure 1) (Gallimore and Godkin, 2013; Chen et al., 2017; Dai et al., 2019).
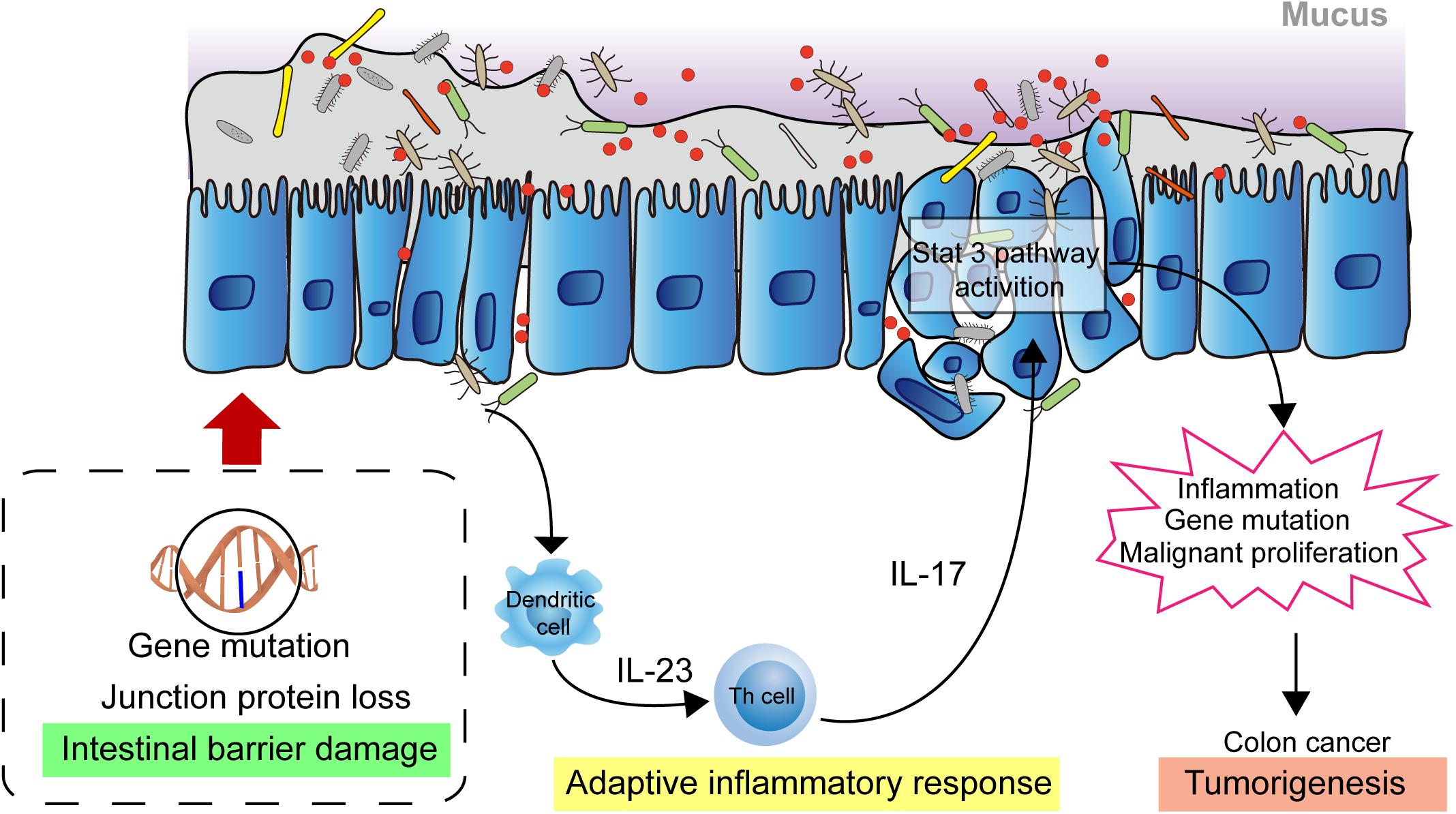
Figure 1. Intestinal microbial facilitating colon cancer progression. The damaged intestinal mucosal barrier permits the transposition of intestinal bacteria and metabolites from the lumen to the lamina propria which activates adaptive immune response. The releasing cytokines drives development of Th17 cells response. The response activates the STAT3, NF-κB signaling pathway in epithelial cells, promoting cell proliferation and invasion, leading to further destruction of mucosal integrity and increased inflammatory response. These biological events contribute to the development of colon cancer.
The mucous layer is the first line of defense of the intestinal mucous barrier. Most of the microbial symbiosis in the human body occurs in the intestinal epithelial barrier, which greatly influences intestinal health. Researchers have found that microbes promote the normal structure and function of the intestine in aseptic mice, that is, the intestinal mucosa of aseptic mice is considerably thin, and intestinal epithelial cell proliferation is significantly reduced. Moreover, the production of mucin and other intestinal epithelial cell derivatives is also impaired. The thinner mucin layer negatively affects the protective function of the epithelial barrier, which allows contact between the host’s intestinal epithelium and exogenous substances, making it more vulnerable to foreign chemicals and pathogenic microbiota, thereby increasing the risk of colon cancer (Allaire et al., 2019).
The outer mucosal later is the habitat of symbiotic microbiota and the nutrient source of some microbiota. The microbial species inhabiting, and physiological functions of, the outer mucosal layer can affect the composition and structure of microbiota in the intestinal cavity. Nineteen strains from the phyla Bacteroidetes, Firmicutes, Actinomycetes, and Verrucobacteria have mucin-degrading ability (Tramontano et al., 2018); these strains mainly include Akkermansia muciniphila (Geerlings et al., 2018), Barnesiella intestinihominis (Desai et al., 2016), Bacteroides thetaiotaomicron (Xu et al., 2003; Comstock, 2009), Bifidobacterium bifidum (Turroni et al., 2011), Bacteroides fragilis (Hecht et al., 2017), Bacteroides vulgatus (Png et al., 2010), Ruminococcus gnavus (Owen et al., 2017), and Ruminococcus torques (Halmos et al., 2015). In addition to the physical mechanical barrier, the intestinal mucus layer also has an immuno-barrier function. Pathogenic microbiota are sensed by various types of pattern recognition receptors and antigen-presenting cells, and the colonic mucus is a repository of Immunoglobulin A (IgA). There are also some bactericidal proteins in the mucus that directly kill displaced microbiota (Xu et al., 2015; Honda and Littman, 2016; Chiaro et al., 2017; Allaire et al., 2019).
In addition to the direct effect on the intestinal tract, intestinal microbiota can indirectly affect tumor occurrence by influencing inflammation, immune function, and systemic metabolic function, in addition to their direct effect on the intestinal tract. Zhang Guodong’s team found that triclosan (dichlorophenoxy chlorophenol) could change the intestinal microorganism composition to reduce intestinal micromicrobiota diversity and the abundance of beneficial bacteria (Bifidobacterium, a known bacteria with anti-colitis), thereby promoting tumorigenesis through the development of colitis. Experimental results have shown that triclosan can disrupt the intestinal barrier function in the body, increase immune cell infiltration, cause the transfer of TLR4 ligands (such as lipopolysaccharides and other bacterial products) from the intestinal tract to the systemic circulation, and promote the activation of TLR4 signal, thus promoting the occurrence and development of colitis and colon cancer (Thaiss et al., 2016; Yang et al., 2018).
The intestinal microbiota composition grows and develops with the host and is affected by the complex interaction of the host genome, nutrition, and lifestyle. Intestinal microbiota is involved in the regulation of various host metabolic pathways, which affects the co-metabolism of host-micromicrobiota interaction (Nicholson et al., 2012). The host will rely on gut microbiota to increase the production of digestive and metabolic enzymes. Intestinal microbiota produce various metabolic components, which include fermentation products of undigested food and endogenous compounds produced by the host, to construct a natural metabolite microenvironment-intestinal epithelial mucus layer, so that microbial metabolites can enter and interact with the host cell to affect the host immune response and disease occurrence and development (Rooks and Garrett, 2016). The microbial community in the human body can prevent pathogen growth by producing beneficial microbial metabolites; however, the imbalance of metabolites in the body will result in side effects that induce the occurrence and development of cancer (Del Carmen Martínez-Jiménez et al., 2018). Further research on the co-metabolism of host and microbiota is required to optimize treatments that control the intestinal microbiota and is a potential treatment strategy in the prevention and cure of various diseases.
The intestinal microbiota can use substances that cannot be digested by the small intestine, such as dietary fiber, and certain undigested sugars, proteins, and peptides. Consequently, the intestinal microbiota can also cooperate with the human body to form intestinal microbiota-host co-metabolites, such as short-chain fatty acids (SCFAs), vitamin H, and vitamin K (Lokody, 2014). These metabolites can be used as energy materials for intestinal microbes and can be transported to other parts of the body to stimulate cell growth, inhibit harmful microorganism growth, and participate in disease defense (Wu et al., 2016). When the composition, ratio, and quantity of the intestinal microbiota undergo pathological changes, it can lead to corresponding changes in the content and types of metabolites related to the microbiota, affecting the physiological functions of the host, and promoting the occurrence and development of tumors (Han et al., 2018). Therefore, changes in intestinal microbiota metabolites have important potential in tumor disease diagnosis.
SCFAs are mainly produced by the microbial fermentation of undigested food, and its products are mainly acetate, propionate, and butyrate. Among the three major SCFAs, butyrate is the most important component that maintains colon health; it is primarily used as a direct energy source for colon cells. SCFAs plays a beneficial role in human intestinal health and affect colon health through various mechanisms. In vivo and in vitro studies have shown that SCFAs have anti-inflammatory and anticancer effects and plays an important role in maintaining the homeostasis of colon cell metabolism, protecting colon cells from external damage (McNabney and Henagan, 2017).
SCFAs can have a direct effect against cancer by inhibiting histone deacetylase (HDAC) and activating G protein-coupled receptors (GPCRs). HDAC inhibition by SCFAs is related to cell cycle arrest. Moreover, the microarray analysis of human colonic epithelial cells reveals that most butyrate-related genes are indeed involved. Butyrate can reduce the expression levels of the anti-apoptotic gene Bcl-2 and the pro-apoptotic protein Bax. Further, butyric and propionic acids can promote adenoma and cancer cell apoptosis by stimulating the expression of the cell cycle regulatory genes p53 and p21. Finally, butyric and propionic acids can promote cancer cell differentiation, inhibit colon cancer cell migration in vitro, and reduce the invasiveness of colon cancer cells. In addition, both activated GPR43 and GPR109a have anti-tumor effects by inhibiting proliferation and promoting the apoptosis of colon cancer cells, not related to HDAC inhibition (van der Beek et al., 2017). Furthermore, the high expression of SCFAs receptors in immune cells has indicated that SCFAs affect the expansion and production of Treg cells through the inhibition of SCFA-GPCR or HDAC, participating in intestinal immune regulation, and regulating and colon cancer development (El Kaoutari et al., 2013; Johansson et al., 2015).
Based on the above analysis, the role of SCFAs seems contradictory. It could provide energy for the growth and proliferation of normal cells, and inhibit cancer cell proliferation. However, current studies have shown that butyric acid generally could not reach the crypt cells, mainly providing energy for the colon cells in the anterior segment of the crypt. While the crypt structure is destroyed in the state of colon cancer, butyric acid could mediate the inhibitory effect of colon stem cells on colon cancer cells. SCFAs produced by intestinal microbiota play an active role in maintaining the normal state of the body, based on the cooperation between micromicrobiota and the body. Propionic and butyric acids may be involved in the above-mentioned tumor inhibition pathway (Alexander et al., 2017). Exploring the addition of probiotics and prebiotics in preventing or treating cancer can provide new ideas for its clinical treatment.
In the distal human gut, proteins and peptides have three possible prospects: assimilated by the microbiota; as the substrate of microbial alienation metabolism, wherein its products enter the host portal circulation; or as intermediates of extensive microbial crosstalk and fecal excretion (Krautkramer et al., 2021). To a large extent, the degree of amino acid metabolism of intestinal microbiota depends on the utilization of substrate and cavity environments. It has been reported that the bacterial fermentation rate of protein (relative to carbohydrates) is greater in higher colonic pH and lower carbohydrate utilization (Krautkramer et al., 2021). SCFA production caused by the microbial protein degradation being significantly lower than that of carbohydrates. In addition, the decrease of organic acids leads to a higher pH value of the lumen, which in turn alters microbiota structure and function (Ratzke and Gore, 2018). In contrast, the low lumen pH, which is a result of the presence of SCFAs, is considered to inhibit bacterial proteases; eventually, fermentable carbohydrates drive bacterial growth, and bacterial protein assimilation then increases at the expense of fermentation (Stephen and Cummings, 1980; Birkett et al., 1996). To date, due to the complexity of intestinal microbial content, the complex interdependence between many hosts and these substrate metabolic pathways, and the technical limitations of metabolite source classification (host and microbiota), has limited research on intestinal microbial histone degradation to a certain extent (Sridharan et al., 2014). However, in recent decades, intestinal microbiota has recovered large amounts of energy from proteins and peptides that escape host digestion; this results in the synthesis of various bioactive compounds, some of which are potentially toxic, including SCFAs, bifunctional chelating agents, ammonia, phenol, indole, amines, sulfides, and N-nitroso compounds (Smith and Macfarlane, 1996).
Tryptophan is an essential amino acid that ismetabolized into indole derivatives, 5- hydroxytryptophan (5-HT) and kynurenines (kynurenine and its derivatives) by different pathways. Indolepropionic acid (IPA) inhibit the early development of breast cancer by acting on AHR and PXR (Sari et al., 2020), which is converted by Bacteroides spp., Clostridium spp., Lactobacillus spp., Parabacteroides distasonis, Peptostreptococcus spp. etc. (Dodd et al., 2017; Agus et al., 2018). Recent studies have shown that 5-HT enhances the activation of NLRP3 inflammasomes by acting on its ion channel receptor HTR3A and promotes tumor progression in colitis associated colorectal cancer mouse models (Li et al., 2021). The microbiota is also involved in regulating host 5-HT, and Some species grown in culture can produce 5-HT (Tsavkelova et al., 2006). Kynurenines are ligands for arylhydrocarbon receptor (AhR) ligands to promote cell migration and immune tolerance, thereby driving cancer progression (Cervenka et al., 2017). Indoleamine 2, 3-dioxygenase 1 (IDO1) is the rate-limiting step of tryptophan conversion to kynurenines. The gut microbiota, such as Lactobacillus spp., Pseudomonas aeruginosa, Pseudomonas, Fluorescens (Vujkovic-Cvijin et al., 2013; Agus et al., 2018), promotes the expression of IDO1, and IDO1 activity can also regulate the composition of the microbiome.
Secondary bile acid is a primary bile acid catalyzed by intestinal microbiota, which is a substance that is converted by dehydroxyl group by the de-binding reaction. Secondary bile acids have a potential DNA-damaging ability, that is, a carcinogenic effect (Cao et al., 2017). In people with obesity, the dominant Clostridium members in the intestines can convert primary bile acids (such as chenodeoxycholic acid and cholic acid) into secondary bile acids (such as lithocholic acid and deoxycholic acid), which have a high affinity for bile acid receptors that affect multiple metabolism-associated processes. Hepatic sinusoidal endothelial cells expressed chemokine CXCL16 (CXCR6 ligand) to regulate the accumulation of NKT cells. Intestinal microbiota then enzymatically modify primary bile acidsto secondary bile acids, which could affect the process: primary bile acids increased the expression of CXCL16, while secondary bile acids had the opposite effect (Ma et al., 2018). Intestinal bacteria with this enzymatic reaction capacity have BSH enzymes, including Lactobacillus (Wang et al., 2012), Bifidobacterium (Kim et al., 2004), Clostridium spp. (Kim et al., 2004), Listeria (Begley et al., 2005), and Enterococcus (Wijaya et al., 2004). In addition, secondary bile acids can directly or indirectly affect the composition of intestinal microbiota. Current studies have shown that reduced abundance of Clostridium labile has an inhibitory effect on secondary bile acids, which can prevent liver tumors in mice (Winston and Theriot, 2016).
Spontaneous tumor remission in patients with severe bacterial infection has been reported for the past two centuries. Coley, a surgical oncologist from the United States, used Streptococcus pyogenes extract named Coley’s toxin to treat tumor patients at the late nineteenth century. Approximately 30% of lymphoma and sarcoma patients were cured, thus opening the door to tumor immunotherapy. In the history of tumor therapy development, chemotherapy agents emerged as the mainstay of present tumor therapy (Hoffman, 2012). Exploring the combination of intestinal microbiota with chemotherapeutic drugs and the interaction mechanism between them can provide better treatment innovations in clinical settings.
Since the discovery of nitrogen mustard cytotoxicity in World War II, researchers have gradually developed cytotoxic chemotherapeutic agents, such as chemotherapeutic drugs. Nowadays, chemotherapeutic drugs remain as the main treatment of tumors in most clinics (Einhorn, 1985). Lehouritis et al. (2015) observed the potential effect of bacteria on the effectiveness of chemotherapeutic drugs against cancer cells in vitro, whereby the activities of 10 out of 30 tested drugs were found to be specifically inhibited by one or two bacteria, and the correlation analysis of HPLC and mass spectrometry revealed that bacterial contact leads to the biotransformation of drugs. Therefore, experimental results show a complex and dynamic interaction between chemotherapeutic drugs and microbiota.
Intestinal microbiota directly affect drug absorption and metabolism, and indirectly affect oral drug metabolism by regulating host gene expression. Compared with ordinary mice, it was found that the expression of certain members of the cytochrome P450 (Cyp450) gene family increased in aseptic mice livers. The expression of proteins from the Cyp2a, Cyp2b, and Cyp3a families, which are involved in heterogeneous steroid metabolism was increased, but that of other cytochromes was decreased. The fatty acid and arachidonic acid metabolism associated with members from the Cyp4a family involves heterogeneous biosensing receptors and transcription factors, such as androgen receptor, aryl hydrogen carbon receptor, and P450 oxidoreductase, which regulate target gene overexpression. Interestingly, the colonization of microbiota taken from routinely cultured ordinary mice in aseptic mice can restore the normal expression of related genes, and probiotic use in aseptic mice can also improve certain gene expression. These gene changes accelerate the metabolism of multiple drugs in aseptic mice (Jourová et al., 2017; Li et al., 2017). The role of microbiota in regulating drug metabolism and detoxification was also indirectly proven. Therefore, the heterogeneity of the therapeutic effect and toxicity of drugs on tumor patients can be exhibited from the differences in the composition and activity of intestinal microbiota among individual patients (Björkholm et al., 2009; Selwyn et al., 2015). In addition to oral drugs, several injected drugs are metabolized in the liver, and then excreted into the intestinal tract through bile, in which they are further metabolized and reabsorbed in the intestinal microbiota’ environment.
Additionally, intestinal microbiota are involved in the biochemical conversion of various drugs, including reduction, hydrolysis and functional group removal, such as N-oxide cleavage, proteolysis, denitrification, amine formation, hydrolysis, thiazole ring opening, and acetylation (Wilson and Nicholson, 2017). Microbiota also reduce drug absorption through physical binding and separation. At present, it has been shown that more than 40 types of exogenous chemicals (non-natural foreign chemicals) are metabolized by intestinal microbiota. However, among anticancer drugs, only misonidazole, a radiosensitizer, and irinotecan (also known as CPT-11), a topoisomerase I inhibitor for hydrolysis and depolymerization of methotrexate, are affected by intestinal microbiota (Haiser and Turnbaugh, 2013).
Most chemotherapeutic drugs have no specificity and generally produce significant toxicity for all cells and tissues exhibiting accelerated renewal (Sancho-Martínez et al., 2012). Platinum anti-tumor drugs, such as oxaliplatin and cisplatin, kill tumor cells by inhibiting DNA replication and targeting the cell membranes and mitochondria. In addition, they can cause severe enterotoxicity, nephrotoxicity, ototoxicity, and peripheral neuropathy. Chemotherapeutic drugs have a strong toxic effect, particularly on intestinal mucosal cells which show acute regeneration, and are damaging to intestinal barrier function, moreover, this damage causes microbiota and pathogens to enter the mesenteric lymph nodes and blood circulation, leading to septicemia and systemic inflammation (Hooper and Macpherson, 2010). Therefore, generating better methods to combine and operate biospecific molecules on the surface of the chemotherapeutic drugs in order to safely and effectively deliver the drugs to the tumor site is a limitation of the current research. The latest research uses genetic engineering to modify bacterial protoplasts and develop nano-vesicles without toxic outer membrane components, which can aid in the specific targeting of the tumor tissue by chemotherapeutic agents to improve drug safety and efficacy (Kim et al., 2017).
Briefly, intestinal microbiota play a critical role in drug metabolism, and the interaction between chemotherapeutic drugs and intestinal microbiota has a major effect. A significant reference for the development of chemotherapeutic drugs can be provided through a deeper understanding of the role and function of intestinal microbiota in the pathology and treatment of cancer.
Tumor immunotherapy kills cancer cells and inhibits their proliferation through artificial intervention and mobilization of the body’s own immune system. It is the fourth tumor therapy after surgery, radiotherapy, and chemotherapy, with great potential for further development. Particularly, immune checkpoint inhibitors (ICIs), particularly CTLA-4 and PD-1 protein inhibitors, and adoptive cell therapy show good prospects in various tumor treatment.
Recently, ICIs have played an important role in tumor therapy. CTLA-4 inhibitor, the first immune checkpoint inhibitor, was discovered in 2011, with the approval of ipilimumab by the United States FDA. In 2014, nivolumab was approved as the first PD-1 inhibitor on the market worldwide. Within the next few years, a number of ICIs, including the antibody against PD-1 and PD-L1, were approved. The main indications include metastatic non-small cell lung cancer, melanoma, and urothelial carcinoma. At present, five PD-1 and PD-L1 antibodies have been approved by the US FDA, namely, nivolumab, pembrolizumab, atezolizumab, avelumab, and durvalumab (Clarke et al., 2018; Gong et al., 2018) (Table 1).
With immunotherapy development, it has been found that small molecule drugs have greater advantages that might be complementary and potentially synergistic to large biological molecules in the immune system (Weinmann, 2016) (Table 1).
In addition, the role of microbiota is essential in immunotherapy. In 2015, the Gustave Roussy Cancer Center and Laurence found that gut microbiota composition determines the effectiveness of cancer immunotherapy represented by ICIs. In 2017, the two teams proved that microbiota in vivo play a decisive role in immunotherapy based on a large-scale analysis of patients with different types of cancer treated with PD-1 inhibitors (Sivan et al., 2015; Vétizou et al., 2015; Gopalakrishnan et al., 2018; Routy et al., 2018).
However, certain problems remain in tumor immunotherapy, such as the uncertainty of the curative effect, the narrow application scope, and immune system-related complications. Therefore, finding an optimal approach to immunotherapy is an important direction of clinical research. In 2015, Vétizou et al. discovered anticancer immunotherapy by CTLA-4 blockade that relies on the gut microbiota. Since then, the relationship between tumor immunotherapy and intestinal microbiota has become a popular research topic (Vesely and Schreiber, 2013; Bachireddy et al., 2015; Garrett, 2015; Segre, 2015; Topalian et al., 2015; Drewes et al., 2016; Tran et al., 2017).
Clinical studies have confirmed that the progression-free survival and overall survival of patients with malignant tumors using antibiotics are significantly shorter than those of patients undergoing immunotherapy that did not use antibiotics, indicating the important role of the intestine in the tumor immunotherapy process (Derosa et al., 2018; Routy et al., 2018). Intestinal microbiota analysis revealed that the abundance of various types of bifidobacteria spp. increased significantly in mice with slow tumor growth, and anti-PD-1 therapy could have a significant effect. Useful microbiota in mice could be transferred to mice that lack them by fecal microbiota transplantation (FMT), or through feeding in the same nest. In addition, the anti-tumor effect of PD-L1 blockade was improved in mice that had an unfavorable intestinal microbiota through the oral administration of Bifidobacterium-containing probiotics. This effect predominantly benefits from the enhancement of DC maturation, which increases tumor-specific CD8+ T cell activity (Sivan et al., 2015). After anti-CTLA-4 treatment, the richness of intestinal micromicrobiota in mice changed significantly, mainly manifesting as a relative increase in Bacteroidales and Burkholderiales abundance, and a decrease in that of Clostridiales. Oral administration of Bacteroides fragilis with cyclophosphamide (Bacteroides thetaiotaomicron) or cephalosporin (Burkholderia cepacia) could trigger Th1 reaction and promote DC maturation, thus improving the efficacy of anti-CTLA-4 therapy. In germ-free and specific-pathogen-free mice treated with broad-spectrum antibiotics, the effect of anti-CTLA-4 therapy was significantly reduced, which could reverse the trend of the dominant microbiota of patients by FMT (Vétizou et al., 2015).
Researchers from the University of Texas MD Anderson Cancer Center have found that intestinal microbiota regulate the response of melanoma patients to PD-1 immunotherapy (Beaver et al., 2018). Through fecal microbial sample analysis and the study of different intestinal microorganism functions in the metagenome of patients, they found that there was a significant difference in intestinal micromicrobiota composition between melanoma patients who responded, and did not respond, to PD-1 immunotherapy and the immune response to tumors was significantly enhanced. To verify the conjecture, they transplanted the relevant beneficial bacteria and the feces of the respondent into the sterile mice with melanoma, and the same result was obtained.
Several species of intestinal bacteria have been associated with enhanced efficacy of immune-checkpoint blockade (ICB). Akkermansia muciniphila was correlated with increased immune cell infiltration in lung and kidney cancers, as CCR9+CXCR3+CD4+ T cells were recruited to the tumor bed in an interleukin-12-dependent manner and the ratio of CD4+ T cells to CD4+FoxP3+ T cells (Tregs) was increased (Routy et al., 2018). Oral administration of pasteurized A. muciniphila or its purified membrane protein Amuc_1100 reduced colon infiltration of macrophages and cytotoxic T lymphocytes (CTL) and improve colitis in mice. Moreover, they increased the number of CTL in colon and mesenteric lymph nodes, up-regulate the expression of TNF-α, inhibit the expression of PD-1, and increase the activation of CTL, which has an inhibitory effect on colitis associated colorectal cancer in mice (Wang et al., 2020). Furthermore, Mager et al. (2020) isolated three bacterial species, including Bifidobacterium pseudolongum, Lactobacillus johnsonii, and Olsenella species, that significantly enhanced the efficacy of immune checkpoint inhibitors (anti-CTLA-4 and anti-PD-1). Notably, intestinal B. pseudolongum modulated enhanced immunotherapy response through the production of the metabolite inosine (Mager et al., 2020) (Figure 2). However, the mechanisms whereby the microbiome enhances anti-tumor immunity are unclear.
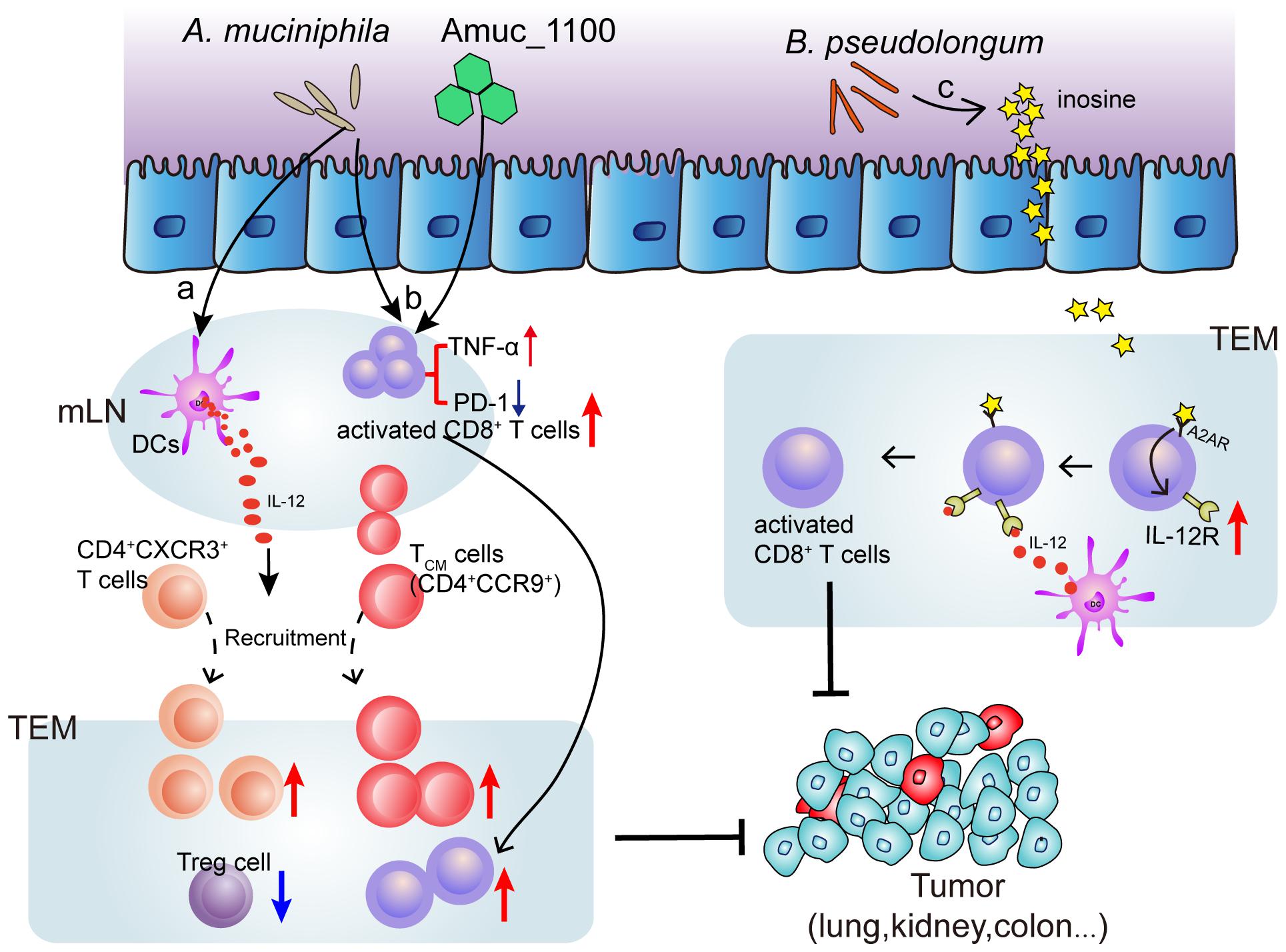
Figure 2. Mechanism of bacteria (Akkermansia muciniphila and Bifidobacterium pseudolongum) induced ICB efficacy enhancement. (a) Oral supplementation with A. muciniphila accelerates IL-12 secretion by dendritic cells (DCs) leading to restoring the recruitment of CXCR3+CD4+ and CCR9+ CD4+ T lymphocytes into tumor microenvironment (TEM), so that restoring the efficacy of PD-1 blockade. (b) A. muciniphila or a A purified membrane protein (Amuc_1100) from A. muciniphila treatment activates CD8+ T cells in mesenteric lymph nodes (mLN) and promote the recruitment of CD8+ T cells into TEM, representing by TNF-α upregulation and PD-1 downregulation. (c) Inosine, the metabolite from B. pseudolongum, co-stimulates A2A receptor (A2AR) leading to increased expression of IL-12 receptor on CD8+T cells, which further increases the T cell activation.
Immune checkpoint inhibitors enhance the activity of anti-tumor T cells and are widely used in cancer therapy, but also cause immune-related adverse reactions, especially in the gastrointestinal tract (Soularue et al., 2018). The effect of intestinal microbiota on toxicity was studied in animal models and clinical cohorts. The melanoma patients treated with anti-CTLA-4 were rich in Bacteroidetes and various genetic pathways, involving polyamine transport and B vitamin synthesis, and no colitis occurred. This lack of toxicity could be related to the known effects of these bacteria. Interestingly, the bacteria related to reaction and toxicity have different taxa, some of which have overlapping characteristics. According to related studies, there was a higher risk of colitis in patients with a higher abundance of Faecalibacterium prausnitzii and a lower abundance of Bacteroides after anti-CTLA-4 treatment (Chaput et al., 2017). However, other studies have shown that a greater abundance of Ruminococcaceae family (including Faecalibacterium spp.) have been found in patients who responded to ICIs therapy (Gopalakrishnan et al., 2018). In contrast, bacterial taxa related to a poor response to ICIs are included in the order Bacteroidales of Bacteroidetes; however, a higher abundance of these classifications usually reduces the incidence of toxicity. In addition, a series of studies have shown that the intestinal microbiota can affect the therapeutic effects of immune checkpoints through a series of metabolites. Members of Firmicutes and F. prausnitzii produce SCFAs. These SCFAs enter the bloodstream through the damaged intestinal mucosa (Coutzac et al., 2020). It can reduce the effect of immune checkpoint suppression via the upregulation of CD80/CD86 on DCs and promoting the accumulation of tumor-specific T cells and memory T cells.
In summary, intestinal microbiota undeniably affect immunotherapy. Intestinal microbiota can regulate the anti-tumor immune response and the response to immune checkpoint inhibitors. These findings explain the therapeutic potential of intestinal microbiota in checkpoint blocking immunotherapy, which can greatly contribute to the treatment of tumors through further clinical and experimental studies (Gopalakrishnan et al., 2018).
Since the FDA first approved CAR-T therapy in 2017, adoptive cell therapy (ACT) has become a hotspot in current tumor immunotherapy research due to its remarkable efficacy in treating various tumors. Due to the recent approval of ACT, there are relatively few clinical studies on its effects on the intestinal microbiota. In 2007, Paulos et al. (2007) first demonstrated that in mouse tumor models, intestinal microbiota can be induced to translocate to the mesenteric lymph nodes under the action of systemic radiotherapy; then, the efficacy of ACT was enhanced through TLR4 signal transduction. After antibiotic treatment in mice, the efficacy of ACT was greatly reduced. After supplementation with bacterial lipopolysaccharide, the anti-tumor response of ACT was enhanced (Paulos et al., 2007). This phenomenon was found in clinical studies of metastatic melanoma patients, and the efficacy of ACT was better in patients who received radiotherapy pretreatment. Further, Uribe-Herranz et al. (2018) reported that in the process of ACT for treating mouse tumors, the treatment efficacy in vancomycin-treated mice was significantly better than that of untreated mice. However, the effect of ACT after treatment with neomycin and metronidazole was lower than that of untreated mice. Compared with the treated group, there was no significant change; 16S rRNA sequencing of mouse feces found that several genes from the members of Bacteroidetes were significantly different following vancomycin treatment, including bacteria from Bacteroides and Parabacteroides. Further research found that the mechanism underlying the action of ACT may involve the increased abundance of systemic CD8α + DCs, which further promotes IL-12 expression and enhances the efficacy of ACT. In summary, these results revealed a certain correlation between the intestinal microbiota and the therapeutic effects of ACT; however, the specific microbiota that exert these effects remain unknown. With the continuous expansion of ACT-related research and the wide range of clinical applications, more in-depth research will reveal potential gut microbiota-related targets for improving the therapeutic effects of ACT.
With the in-depth study of intestinal microbiota, the correlation between intestinal microbiota and tumorigenesis will be revealed. The intestinal microbiota plays an important role in tumorigenesis and tumor progression, and simultaneously affects the effectiveness of chemotherapy. In recent years, increasing attention has been paid to the relationship between intestinal microbiota and tumor-related health problems, which mainly benefits from the rapid development of microtechnology, including the second and third generations of high-throughput sequencing, such as 16S rRNA sequencing and PCR-DGGE, that can determine the DNA of microbial specimens. Real-time fluorescence quantitative PCR was used to quantitatively study the intestinal microbiota and explore the microbial diversity. The bacteria in intestinal microbiota were obtained by anaerobic culture, and intestinal microorganism function was studied at the strain level. Fecal transplantation technology verifies the mechanism of intestinal microbiota through the forward and reverse experiment.
In current studies, the effects of intestinal microbiota on tumors and immunity have been observed; however, the mechanism underlying these effects remain unclear. In addition, there are still challenges associated with studying how intestinal microbiota regulation improves the effects of tumor immunotherapy. At present, the components of intestinal microbiota that are most conducive to promoting anti-tumor immune response remain unclear. Furthermore, there are various treatments to modify the intestinal microbiota, which need to be carefully tested in clinical trials. After fully understanding these interactions, optimization of intestinal microbiota regulation for enhancing the host’s anti-tumor immunity and for potentially improving immune surveillance can be achieved.
At present, only a few types of bacteria have been reported to be related to cancer, and great potential exists to explore the relationship between microbiota and tumorigenesis and tumor progression. The technologies to study these aspects are improving with the emergence of new treatments, such as the above-mentioned fecal transplantation, i.e., transplanting feces from healthy people to patients, which can have a good therapeutic effect and broaden the ideas of clinical treatment.
In future research, with improvements in microtechnology and analytical technology, we believe that more cancer-related target microbiota will be discovered, providing new methods and ideas for the clinical treatment of malignancies.
ZW and YL: conceptualization and design the review, review final version approval, and funding acquisition. PC, RD, YS, and WC: bibliographic research. PC, PS, and YY: writing—original draft preparation. PS, PC, YS, and ZW: the table and figure design. WC and YL: supervision. All authors contributed to the article and approved the submitted version.
This work was financially supported by National Natural Science Foundation of China (82004124, 81961128020, and 81973734), China Postdoctoral Science Foundation (2020M671551), Science and Technology Development Foundation of Nanjing Medical University (NMUB2019186), Jiangsu Province Traditional Chinese Medicine Leading Talents Program (SLJ0229), The Open Project of Chinese Materia Medica First-Class Discipline of Nanjing University of Chinese Medicine (2020YLXK20).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Agus, A., Planchais, J., and Sokol, H. (2018). Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe 23, 716–724. doi: 10.1016/j.chom.2018.05.003
Alexander, J. L., Wilson, I. D., Teare, J., Marchesi, J. R., Nicholson, J. K., and Kinross, J. M. (2017). Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat. Rev. Gastroenterol. Hepatol. 14, 356–365. doi: 10.1038/nrgastro.2017.20
Alhalabi, O., Rafei, H., Shah, A., Siefker-Radtke, A., Campbell, M., and Gao, J. (2019). Targeting advanced urothelial carcinoma-developing strategies. Curr. Opin. Oncol. 31, 207–215. doi: 10.1097/cco.0000000000000532
Allaire, J. M., Crowley, S. M., Law, H. T., Chang, S.-Y., Ko, H.-J., and Vallance, B. A. (2019). The intestinal epithelium: central coordinator of mucosal immunity: (trends in immunology 39, 677-696, 2018). Trends Immunol. 40:174. doi: 10.1016/j.it.2018.12.008
Antonioli, L., Haskó, G., Fornai, M., Colucci, R., and Blandizzi, C. (2014). Adenosine pathway and cancer: where do we go from here? Expert Opin. Ther. Targets 18, 973–977.
Bachireddy, P., Burkhardt, U. E., Rajasagi, M., and Wu, C. J. (2015). Haematological malignancies: at the forefront of immunotherapeutic innovation. Nat. Rev. Cancer 15, 201–215. doi: 10.1038/nrc3907
Bastid, J., Cottalorda-Regairaz, A., Alberici, G., Bonnefoy, N., Eliaou, J. F., and Bensussan, A. (2013). ENTPD1/CD39 is a promising therapeutic target in oncology. Oncogene 32, 1743–1751. doi: 10.1038/onc.2012.269
Beaver, J. A., Hazarika, M., Mulkey, F., Mushti, S., Chen, H., He, K., et al. (2018). Patients with melanoma treated with an anti-PD-1 antibody beyond RECIST progression: a US food and drug administration pooled analysis. Lancet Oncol. 19, 229–239. doi: 10.1016/s1470-2045(17)30846-x
Begley, M., Sleator, R. D., Gahan, C. G., and Hill, C. (2005). Contribution of three bile-associated loci, bsh, pva, and btlB, to gastrointestinal persistence and bile tolerance of Listeria monocytogenes. Infect. Immun. 73, 894–904. doi: 10.1128/iai.73.2.894-904.2005
Bhattarai, S., Freundlieb, M., Pippel, J., Meyer, A., Abdelrahman, A., Fiene, A., et al. (2015). α,β-Methylene-ADP (AOPCP) derivatives and analogues: development of potent and selective ecto-5′-nucleotidase (CD73) inhibitors. J. Med. Chem. 58, 6248–6263. doi: 10.1021/acs.jmedchem.5b00802
Billan, S., Kaidar-Person, O., and Gil, Z. (2020). Treatment after progression in the era of immunotherapy. Lancet Oncol. 21, e463–e476.
Birkett, A., Muir, J., Phillips, J., Jones, G., and O’dea, K. (1996). Resistant starch lowers fecal concentrations of ammonia and phenols in humans. Am. J. Clin. Nutr. 63, 766–772. doi: 10.1093/ajcn/63.5.766
Björkholm, B., Bok, C. M., Lundin, A., Rafter, J., Hibberd, M. L., and Pettersson, S. (2009). Intestinal microbiota regulate xenobiotic metabolism in the liver. PLoS One 4:e6958. doi: 10.1371/journal.pone.0006958
Cao, H., Xu, M., Dong, W., Deng, B., Wang, S., Zhang, Y., et al. (2017). Secondary bile acid-induced dysbiosis promotes intestinal carcinogenesis. Int. J. Cancer 140, 2545–2556. doi: 10.1002/ijc.30643
Cervenka, I., Agudelo, L. Z., and Ruas, J. L. (2017). Kynurenines: tryptophan’s metabolites in exercise, inflammation, and mental health. Science 357:eaaf9794. doi: 10.1126/science.aaf9794
Chaput, N., Lepage, P., Coutzac, C., Soularue, E., Le Roux, K., Monot, C., et al. (2017). Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann. Oncol. 28, 1368–1379. doi: 10.1093/annonc/mdx108
Chen, J., Pitmon, E., and Wang, K. (2017). Microbiome, inflammation and colorectal cancer. Semin. Immunol. 32, 43–53. doi: 10.1016/j.smim.2017.09.006
Chiaro, T. R., Soto, R., Zac Stephens, W., Kubinak, J. L., Petersen, C., Gogokhia, L., et al. (2017). A member of the gut mycobiota modulates host purine metabolism exacerbating colitis in mice. Sci. Transl. Med. 9:eaaf9044. doi: 10.1126/scitranslmed.aaf9044
Clarke, J. M., George, D. J., Lisi, S., and Salama, A. K. S. (2018). Immune checkpoint blockade: the new frontier in cancer treatment. Target. Oncol. 13, 1–20. doi: 10.1007/s11523-017-0549-7
Comstock, L. E. (2009). Importance of glycans to the host-bacteroides mutualism in the mammalian intestine. Cell Host Microbe 5, 522–526. doi: 10.1016/j.chom.2009.05.010
Coutzac, C., Jouniaux, J. M., Paci, A., Schmidt, J., Mallardo, D., Seck, A., et al. (2020). Systemic short chain fatty acids limit antitumor effect of CTLA-4 blockade in hosts with cancer. Nat. Commun. 11:2168.
Cremonesi, E., Governa, V., Garzon, J. F. G., Mele, V., Amicarella, F., Muraro, M. G., et al. (2018). Gut microbiota modulate T cell trafficking into human colorectal cancer. Gut 67, 1984–1994.
Dai, Z., Zhang, J., Wu, Q., Chen, J., Liu, J., Wang, L., et al. (2019). The role of microbiota in the development of colorectal cancer. Int. J. Cancer 145, 2032–2041.
Del Carmen Martínez-Jiménez, V., Méndez-Mancilla, A., and Patricia Portales-Pérez, D. (2018). miRNAs in nutrition, obesity, and cancer: the biology of miRNAs in metabolic disorders and its relationship with cancer development. Mol. Nutr. Food Res. 62:1600994. doi: 10.1002/mnfr.201600994
Derosa, L., Hellmann, M. D., Spaziano, M., Halpenny, D., Fidelle, M., Rizvi, H., et al. (2018). Negative association of antibiotics on clinical activity of immune checkpoint inhibitors in patients with advanced renal cell and non-small-cell lung cancer. Ann. Oncol. 29, 1437–1444. doi: 10.1093/annonc/mdy103
Desai, M. S., Seekatz, A. M., Koropatkin, N. M., Kamada, N., Hickey, C. A., Wolter, M., et al. (2016). A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell 167, 1339–1353.e21.
Dodd, D., Spitzer, M. H., Van Treuren, W., Merrill, B. D., Hryckowian, A. J., Higginbottom, S. K., et al. (2017). A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 551, 648–652. doi: 10.1038/nature24661
Dounay, A. B., Tuttle, J. B., and Verhoest, P. R. (2015). Challenges and opportunities in the discovery of new therapeutics targeting the kynurenine pathway. J. Med. Chem. 58, 8762–8782. doi: 10.1021/acs.jmedchem.5b00461
Drewes, J. L., Housseau, F., and Sears, C. L. (2016). Sporadic colorectal cancer: microbial contributors to disease prevention, development and therapy. Br. J. Cancer 115, 273–280. doi: 10.1038/bjc.2016.189
Dubensky, T. W., Kanne, D. B., and Leong, M. L. (2013). Rationale, progress and development of vaccines utilizing STING-activating cyclic dinucleotide adjuvants. Ther. Adv. Vaccines 1, 131–143. doi: 10.1177/2051013613501988
Einhorn, J. (1985). Nitrogen mustard: the origin of chemotherapy for cancer. Int. J. Radiat. Oncol. Biol. Phys. 11, 1375–1378. doi: 10.1016/0360-3016(85)90254-8
El Kaoutari, A., Armougom, F., Gordon, J. I., Raoult, D., and Henrissat, B. (2013). The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat. Rev. Microbiol. 11, 497–504. doi: 10.1038/nrmicro3050
Fridlender, Z. G., Jassar, A., Mishalian, I., Wang, L. C., Kapoor, V., Cheng, G., et al. (2013). Using macrophage activation to augment immunotherapy of established tumours. Br. J. Cancer 108, 1288–1297.
Gallimore, A. M., and Godkin, A. (2013). Epithelial barriers, microbiota, and colorectal cancer. N. Engl. J. Med. 368, 282–284. doi: 10.1056/nejmcibr1212341
Geerlings, S. Y., Kostopoulos, I., De Vos, W. M., and Belzer, C. (2018). Akkermansia muciniphila in the human gastrointestinal tract: When, Where, and How? Microorganisms 6:75. doi: 10.3390/microorganisms6030075
Gong, J., Chehrazi-Raffle, A., Reddi, S., and Salgia, R. (2018). Development of PD-1 and PD-L1 inhibitors as a form of cancer immunotherapy: a comprehensive review of registration trials and future considerations. J. Immunother. Cancer 6:8.
Gopalakrishnan, V., Spencer, C. N., Nezi, L., Reuben, A., Andrews, M. C., Karpinets, T. V., et al. (2018). Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 359, 97–103.
Gostner, J. M., Becker, K., Berall, F., and Fuchs, D. (2015). The potential of targeting indoleamine 2,3-dioxygenase for cancer treatment. Expert Opin. Ther. Targets 19, 605–615.
Gravekamp, C., and Chandra, D. (2015). Targeting STING pathways for the treatment of cancer. Oncoimmunology 4:e988463. doi: 10.4161/2162402x.2014.988463
Grivennikov, S. I., Wang, K., Mucida, D., Stewart, C. A., Schnabl, B., Jauch, D., et al. (2012). Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature 491, 254–258. doi: 10.1038/nature11465
Haiser, H. J., and Turnbaugh, P. J. (2013). Developing a metagenomic view of xenobiotic metabolism. Pharmacol. Res. 69, 21–31. doi: 10.1016/j.phrs.2012.07.009
Halmos, E. P., Christophersen, C. T., Bird, A. R., Shepherd, S. J., Gibson, P. R., and Muir, J. G. (2015). Diets that differ in their FODMAP content alter the colonic luminal microenvironment. Gut 64, 93–100. doi: 10.1136/gutjnl-2014-307264
Hamm, S., Rath, S., Michel, S., and Baumgartner, R. (2009). Cancer immunotherapeutic potential of novel small molecule TLR7 and TLR8 agonists. J. Immunotoxicol. 6, 257–265. doi: 10.3109/15476910903286733
Han, A., Bennett, N., Ahmed, B., Whelan, J., and Donohoe, D. R. (2018). Butyrate decreases its own oxidation in colorectal cancer cells through inhibition of histone deacetylases. Oncotarget 9, 27280–27292. doi: 10.18632/oncotarget.25546
Hecht, A. L., Casterline, B. W., Choi, V. M., and Bubeck Wardenburg, J. (2017). A two-component system regulates Bacteroides fragilis toxin to maintain intestinal homeostasis and prevent lethal disease. Cell Host Microbe 22, 443–448.e5.
Hoffman, R. M. (2012). The preclinical discovery of bacterial therapy for the treatment of metastatic cancer with unique advantages. Expert Opin. Drug Discov. 7, 73–83. doi: 10.1517/17460441.2012.644534
Holldack, J. (2014). Toll-like receptors as therapeutic targets for cancer. Drug Discov. Today 19, 379–382. doi: 10.1016/j.drudis.2013.08.020
Honda, K., and Littman, D. R. (2016). The microbiota in adaptive immune homeostasis and disease. Nature 535, 75–84. doi: 10.1038/nature18848
Hooper, L. V., and Macpherson, A. J. (2010). Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat. Rev. Immunol. 10, 159–169. doi: 10.1038/nri2710
Johansson, M. E. V., Jakobsson, H. E., Holmén-Larsson, J., Schütte, A., Ermund, A., Rodríguez-Piñeiro, A. M., et al. (2015). Normalization of host intestinal mucus layers requires long-term microbial colonization. Cell Host Microbe 18, 582–592. doi: 10.1016/j.chom.2015.10.007
Jourová, L., Anzenbacher, P., Lišková, B., Matušková, Z., Hermanová, P., Hudcovic, T., et al. (2017). Colonization by non-pathogenic bacteria alters mRNA expression of cytochromes P450 in originally germ-free mice. Folia Microbiol. 62, 463–469.
Kim, G. B., Miyamoto, C. M., Meighen, E. A., and Lee, B. H. (2004). Cloning and characterization of the bile salt hydrolase genes (bsh) from Bifidobacterium bifidum strains. Appl. Environ. Microbiol. 70, 5603–5612. doi: 10.1128/aem.70.9.5603-5612.2004
Kim, O. Y., Dinh, N. T. H., Park, H. T., Choi, S. J., Hong, K., and Gho, Y. S. (2017). Bacterial protoplast-derived nanovesicles for tumor targeted delivery of chemotherapeutics. Biomaterials 113, 68–79. doi: 10.1016/j.biomaterials.2016.10.037
Kim, T. J., Cho, K. S., and Koo, K. C. (2020). Current status and future perspectives of immunotherapy for locally advanced or metastatic urothelial carcinoma: a comprehensive review. Cancers 12:192. doi: 10.3390/cancers12010192
Krautkramer, K. A., Fan, J., and Bäckhed, F. (2021). Gut microbial metabolites as multi-kingdom intermediates. Nat. Rev. Microbiol. 19, 77–94. doi: 10.1038/s41579-020-0438-4
Lehouritis, P., Cummins, J., Stanton, M., Murphy, C. T., Mccarthy, F. O., Reid, G., et al. (2015). Local bacteria affect the efficacy of chemotherapeutic drugs. Sci. Rep. 5:14554.
Li, C. Y., Lee, S., Cade, S., Kuo, L.-J., Schultz, I. R., Bhatt, D. K., et al. (2017). Novel interactions between gut microbiome and host drug-processing genes modify the hepatic metabolism of the environmental chemicals polybrominated diphenyl ethers. Drug Metab. Dispos. 45, 1197–1214. doi: 10.1124/dmd.117.077024
Li, T., Fu, B., Zhang, X., Zhou, Y., Yang, M., Cao, M., et al. (2021). Over-production of gastrointestinal 5-HT promotes colitis-associated colorectal cancer progression via enhancing NLRP3 inflammasome activation. Cancer Immunol. Res. Online ahead of print
Lokody, I. (2014). Microbial genetics: bacterial sensors report on the gut. Nat. Rev. Genet. 15:290. doi: 10.1038/nrg3721
Ma, C., Han, M., Heinrich, B., Fu, Q., Zhang, Q., Sandhu, M., et al. (2018). Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science 360:eaan5931. doi: 10.1126/science.aan5931
Mager, L. F., Burkhard, R., Pett, N., Cooke, N. C. A., Brown, K., Ramay, H., et al. (2020). Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science 369, 1481–1489. doi: 10.1126/science.abc3421
Mancini, R. J., Stutts, L., Ryu, K. A., Tom, J. K., and Esser-Kahn, A. P. (2014). Directing the immune system with chemical compounds. ACS Chem. Biol. 9, 1075–1085.
Mándi, Y., and Vécsei, L. (2012). The kynurenine system and immunoregulation. J. Neural Transm. 119, 197–209. doi: 10.1007/s00702-011-0681-y
McEnaney, P. J., Fitzgerald, K. J., Zhang, A. X., Douglass, E. F., Shan, W., Balog, A., et al. (2014). Chemically synthesized molecules with the targeting and effector functions of antibodies. J. Am. Chem. Soc. 136, 18034–18043. doi: 10.1021/ja509513c
McNabney, S. M., and Henagan, T. M. (2017). Short chain fatty acids in the colon and peripheral tissues: a focus on butyrate, colon cancer, obesity and insulin resistance. Nutrients 9:1348. doi: 10.3390/nu9121348
Muller-Haegele, S., Muller, L., and Whiteside, T. L. (2014). Immunoregulatory activity of adenosine and its role in human cancer progression. Expert Rev. Clin. Immunol. 10, 897–914. doi: 10.1586/1744666x.2014.915739
Nicholson, J. K., Holmes, E., Kinross, J., Burcelin, R., Gibson, G., Jia, W., et al. (2012). Host-gut microbiota metabolic interactions. Science 336, 1262–1267. doi: 10.1126/science.1223813
Owen, C. D., Tailford, L. E., Monaco, S., Šuligoj, T., Vaux, L., Lallement, R., et al. (2017). Unravelling the specificity and mechanism of sialic acid recognition by the gut symbiont Ruminococcus gnavus. Nat. commun. 8:2196.
Paulos, C. M., Wrzesinski, C., Kaiser, A., Hinrichs, C. S., Chieppa, M., Cassard, L., et al. (2007). Microbial translocation augments the function of adoptively transferred self/tumor-specific CD8+ T cells via TLR4 signaling. J. Clin. Invest. 117, 2197–2204. doi: 10.1172/jci32205
Pestana, R. C., Sen, S., Hobbs, B. P., and Hong, D. S. (2020). Histology-agnostic drug development - considering issues beyond the tissue. Nat. Rev. Clin. Oncol. 17, 555–568. doi: 10.1038/s41571-020-0384-0
Png, C. W., Lindén, S. K., Gilshenan, K. S., Zoetendal, E. G., Mcsweeney, C. S., Sly, L. I., et al. (2010). Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am. J. Gastroenterol. 105, 2420–2428. doi: 10.1038/ajg.2010.281
Pradere, J. P., Dapito, D. H., and Schwabe, R. F. (2014). The yin and yang of toll-like receptors in cancer. Oncogene 33, 3485–3495. doi: 10.1038/onc.2013.302
Qin, Y., Roberts, J. D., Grimm, S. A., Lih, F. B., Deterding, L. J., Li, R., et al. (2018). An obesity-associated gut microbiome reprograms the intestinal epigenome and leads to altered colonic gene expression. Genome Biol. 19:7.
Ratzke, C., and Gore, J. (2018). Modifying and reacting to the environmental pH can drive bacterial interactions. PLoS Biol. 16:e2004248. doi: 10.1371/journal.pbio.2004248
Rooks, M. G., and Garrett, W. S. (2016). Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 16, 341–352. doi: 10.1038/nri.2016.42
Routy, B., Le Chatelier, E., Derosa, L., Duong, C. P. M., Alou, M. T., Daillère, R., et al. (2018). Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359, 91–97.
Sancho-Martínez, S. M., Prieto-García, L., Prieto, M., López-Novoa, J. M., and López-Hernández, F. J. (2012). Subcellular targets of cisplatin cytotoxicity: an integrated view. Pharmacol. Ther. 136, 35–55.
Sari, Z., Miko, E., Kovacs, T., Janko, L., Csonka, T., Lente, G., et al. (2020). Indolepropionic acid, a metabolite of the microbiome, has cytostatic properties in breast cancer by activating AHR and PXR receptors and inducing oxidative stress. Cancers 12:2411. doi: 10.3390/cancers12092411
Segre, J. A. (2015). MICROBIOME. microbial growth dynamics and human disease. Science 349, 1058–1059. doi: 10.1126/science.aad0781
Selwyn, F. P., Cui, J. Y., and Klaassen, C. D. (2015). RNA-seq quantification of hepatic drug processing genes in germ-free mice. Drug Metab. Dispos. 43, 1572–1580. doi: 10.1124/dmd.115.063545
Sivan, A., Corrales, L., Hubert, N., Williams, J. B., Aquino-Michaels, K., Earley, Z. M., et al. (2015). Commensal bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 350, 1084–1089. doi: 10.1126/science.aac4255
Smith, E. A., and Macfarlane, G. T. (1996). Enumeration of human colonic bacteria producing phenolic and indolic compounds: effects of pH, carbohydrate availability and retention time on dissimilatory aromatic amino acid metabolism. J. Appl. Bacteriol. 81, 288–302.
Soularue, E., Lepage, P., Colombel, J. F., Coutzac, C., Faleck, D., Marthey, L., et al. (2018). Enterocolitis due to immune checkpoint inhibitors: a systematic review. Gut 67, 2056–2067. doi: 10.1136/gutjnl-2018-316948
Sridharan, G. V., Choi, K., Klemashevich, C., Wu, C., Prabakaran, D., Pan, L. B., et al. (2014). Prediction and quantification of bioactive microbiota metabolites in the mouse gut. Nat. Commun. 5:5492.
Stephen, A. M., and Cummings, J. H. (1980). Mechanism of action of dietary fibre in the human colon. Nature 284, 283–284. doi: 10.1038/284283a0
Thaiss, C. A., Zmora, N., Levy, M., and Elinav, E. (2016). The microbiome and innate immunity. Nature 535, 65–74. doi: 10.1038/nature18847
Topalian, S. L., Drake, C. G., and Pardoll, D. M. (2015). Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell 27, 450–461. doi: 10.1016/j.ccell.2015.03.001
Tramontano, M., Andrejev, S., Pruteanu, M., Klünemann, M., Kuhn, M., Galardini, M., et al. (2018). Nutritional preferences of human gut bacteria reveal their metabolic idiosyncrasies. Nat. Microbiol. 3, 514–522. doi: 10.1038/s41564-018-0123-9
Tran, E., Robbins, P. F., and Rosenberg, S. A. (2017). ‘Final common pathway’ of human cancer immunotherapy: targeting random somatic mutations. Nat. Immunol. 18, 255–262. doi: 10.1038/ni.3682
Tsavkelova, E. A., Klimova, S., Cherdyntseva, T. A., and Netrusov, A. I. (2006). [Hormones and hormone-like substances of microorganisms: a review]. Prikl Biokhim. Mikrobiol. 42, 261–268.
Turroni, F., Milani, C., Van Sinderen, D., and Ventura, M. (2011). Genetic strategies for mucin metabolism in Bifidobacterium bifidum PRL2010: an example of possible human-microbe co-evolution. Gut Microbes 2, 183–189. doi: 10.4161/gmic.2.3.16105
Uribe-Herranz, M., Bittinger, K., Rafail, S., Guedan, S., Pierini, S., Tanes, C., et al. (2018). Gut microbiota modulates adoptive cell therapy via CD8alpha dendritic cells and IL-12. JCI Insight 3:e94952.
van der Beek, C. M., Dejong, C. H. C., Troost, F. J., Masclee, A. A. M., and Lenaerts, K. (2017). Role of short-chain fatty acids in colonic inflammation, carcinogenesis, and mucosal protection and healing. Nutr. Rev. 75, 286–305. doi: 10.1093/nutrit/nuw067
Vesely, M. D., and Schreiber, R. D. (2013). Cancer immunoediting: antigens, mechanisms, and implications to cancer immunotherapy. Ann. N. Y. Acad. Sci. 1284, 1–5. doi: 10.1111/nyas.12105
Vétizou, M., Pitt, J. M., Daillère, R., Lepage, P., Waldschmitt, N., Flament, C., et al. (2015). Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 350, 1079–1084. doi: 10.1126/science.aad1329
Viennois, E., Merlin, D., Gewirtz, A. T., and Chassaing, B. (2017). Dietary emulsifier-induced low-grade inflammation promotes colon carcinogenesis. Cancer Res. 77, 27–40. doi: 10.1158/0008-5472.can-16-1359
Vujkovic-Cvijin, I., Dunham, R. M., Iwai, S., Maher, M. C., Albright, R. G., Broadhurst, M. J., et al. (2013). Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Sci. Transl. Med. 5:193ra191.
Wang, L., Tang, L., Feng, Y., Zhao, S., Han, M., Zhang, C., et al. (2020). A purified membrane protein from Akkermansia muciniphila or the pasteurised bacterium blunts colitis associated tumourigenesis by modulation of CD8(+) T cells in mice. Gut 69, 1988–1997. doi: 10.1136/gutjnl-2019-320105
Wang, Q., Liu, X., Zhou, Q., and Wang, C. (2015). Cytosolic sensing of aberrant DNA: arming STING on the endoplasmic reticulum. Expert Opin. Ther. Targets 19, 1397–1409. doi: 10.1517/14728222.2015.1067303
Wang, Z., Zeng, X., Mo, Y., Smith, K., Guo, Y., and Lin, J. (2012). Identification and characterization of a bile salt hydrolase from Lactobacillus salivarius for development of novel alternatives to antibiotic growth promoters. Appl. Environ. Microbiol. 78, 8795–8802. doi: 10.1128/aem.02519-12
Weinmann, H. (2016). Cancer immunotherapy: selected targets and small-molecule modulators. ChemMedChem 11, 450–466. doi: 10.1002/cmdc.201500566
Wijaya, A., Hermann, A., Abriouel, H., Specht, I., Yousif, N. M., Holzapfel, W. H., et al. (2004). Cloning of the bile salt hydrolase (bsh) gene from Enterococcus faecium FAIR-E 345 and chromosomal location of bsh genes in food enterococci. J. Food Prot. 67, 2772–2778. doi: 10.4315/0362-028x-67.12.2772
Wilson, I. D., and Nicholson, J. K. (2017). Gut microbiome interactions with drug metabolism, efficacy, and toxicity. Transl. Res. 179, 204–222. doi: 10.1016/j.trsl.2016.08.002
Winston, J. A., and Theriot, C. M. (2016). Impact of microbial derived secondary bile acids on colonization resistance against Clostridium difficile in the gastrointestinal tract. Anaerobe 41, 44–50. doi: 10.1016/j.anaerobe.2016.05.003
Wu, G. D., Compher, C., Chen, E. Z., Smith, S. A., Shah, R. D., Bittinger, K., et al. (2016). Comparative metabolomics in vegans and omnivores reveal constraints on diet-dependent gut microbiota metabolite production. Gut 65, 63–72.
Wu, T.-R., Lin, C.-S., Chang, C.-J., Lin, T.-L., Martel, J., Ko, Y.-F., et al. (2019). Gut commensal plays a predominant role in the anti-obesity effects of polysaccharides isolated from. Gut 68, 248–262. doi: 10.1136/gutjnl-2017-315458
Xu, J., Bjursell, M. K., Himrod, J., Deng, S., Carmichael, L. K., Chiang, H. C., et al. (2003). A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299, 2074–2076. doi: 10.1126/science.1080029
Xu, J., Lian, F., Zhao, L., Zhao, Y., Chen, X., Zhang, X., et al. (2015). Structural modulation of gut microbiota during alleviation of type 2 diabetes with a Chinese herbal formula. ISME J. 9, 552–562. doi: 10.1038/ismej.2014.177
Yang, H., Wang, W., Romano, K. A., Gu, M., Sanidad, K. Z., Kim, D., et al. (2018). A common antimicrobial additive increases colonic inflammation and colitis-associated colon tumorigenesis in mice. Sci. Transl. Med. 10:eaan4116. doi: 10.1126/scitranslmed.aan4116
Yau, T., Kang, Y.-K., Kim, T.-Y., El-Khoueiry, A. B., Santoro, A., Sangro, B., et al. (2020). Efficacy and safety of nivolumab plus ipilimumab in patients with advanced hepatocellular carcinoma previously treated with sorafenib: the checkmate 040 randomized clinical trial. JAMA Oncol. 6:e204564. doi: 10.1001/jamaoncol.2020.4564
Young, A., Mittal, D., Stannard, K., Yong, M., Teng, M. W., Allard, B., et al. (2014). Co-blockade of immune checkpoints and adenosine A receptor suppresses metastasis. Oncoimmunology 3:e958952. doi: 10.4161/21624011.2014.958952
Keywords: gut microbiota, cancer, metabolism, gut immunity, immunotherapy, chemotherapy
Citation: Cheng P, Shen P, Shan Y, Yang Y, Deng R, Chen W, Lu Y and Wei Z (2021) Gut Microbiota-Mediated Modulation of Cancer Progression and Therapy Efficacy. Front. Cell Dev. Biol. 9:626045. doi: 10.3389/fcell.2021.626045
Received: 04 November 2020; Accepted: 18 August 2021;
Published: 08 September 2021.
Edited by:
Qiyuan Yang, University of Massachusetts Medifigcal School, United StatesReviewed by:
Salvatrice Mancuso, University of Palermo, ItalyCopyright © 2021 Cheng, Shen, Shan, Yang, Deng, Chen, Lu and Wei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yin Lu, bHV5aW5ncmVlbkBuanVjbS5lZHUuY24=; Zhonghong Wei, d3poMTIyNUBuanVjbS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.