
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol. , 30 August 2021
Sec. Membrane Traffic and Organelle Dynamics
Volume 9 - 2021 | https://doi.org/10.3389/fcell.2021.620498
This article is part of the Research Topic Coordination of mRNA Transport and Translation with Vesicle and Organelle Trafficking and Dynamics View all 5 articles
Extracellular vesicles (EVs), including exosomes and microvesicles, are extracellular nanovesicles released by most cells. EVs play essential roles in intercellular communication via the transport of a large variety of lipids, proteins, and nucleic acids to recipient cells. Nucleic acids are the most commonly found molecules inside EVs, and due to their small size, microRNAs and other small RNAs are the most abundant nucleic acids. However, longer molecules, such as messenger RNAs (mRNAs), have also been found. mRNAs encapsulated within EVs have been shown to be transferred to recipient cells and translated into proteins, altering the behavior of the cells. Secretion of EVs is maintained not only through multiple normal physiological conditions but also during aberrant pathological conditions, including cancer. Recently, the mRNAs carried by EVs in cancer have attracted great interest due to their broad roles in tumor progression and microenvironmental remodeling. This review focuses on the biological functions driven by mRNAs carried in EVs in cancer, which include supporting tumor progression by activating cancer cell growth, migration, and invasion; inducing microenvironmental remodeling via hypoxia, angiogenesis, and immunosuppression; and promoting modulation of the microenvironment at distant sites for the generation of a premetastatic niche, collectively inducing metastasis. Furthermore, we describe the potential use of mRNAs carried by EVs as a noninvasive diagnostic tool and novel therapeutic approach.
Intercellular communication is essential for the appropriate performance of multicellular organisms. Recently, extracellular vesicles (EVs) have emerged as a novel mechanism of horizontal gene transfer. EVs are lipid bilayer membrane-enclosed vesicles released by most, if not all, cell types. They carry a wide range of fragile RNA molecules, DNA molecules and proteins within their core (Valadi et al., 2007; Skog et al., 2008; Balaj et al., 2011; Li et al., 2013; Raposo and Stoorvogel, 2013). These molecules are protected by EV membranes from nucleases, proteases, fluctuations in pH and osmolality, and other environmental factors that would otherwise rapidly degrade them (El-Hefnawy et al., 2004; Skog et al., 2008; Cheng et al., 2014; Kim et al., 2017). Thanks to this protection, the cargo is successfully transported, enters into recipient cells, and affects cell behavior, having a significant impact on natural physiological processes. Thus, EVs are considered important mediators of intercellular communication, both in physiological and pathological states, including cancer (Kosaka et al., 2013b; Berardocco et al., 2017; Kim et al., 2017).
Exosomes were first observed in the early 1980s by the Stahl and Johnstone groups—who published within a week of each other—in a study regarding the maturation of reticulocytes into erythrocytes by the removal of transferrin receptors (Pan and Johnstone, 1983; Harding et al., 1984). In that study, it was observed that transferrin receptors were internalized within multivesicular bodies (MVB) and that later, instead of being degraded, they were externalized. At that time, exosomes were described as a mechanism to discard unwanted cellular debris (Johnstone et al., 1987; Harding et al., 2013). One decade later, Raposo et al. (1996) found that exosomes derived from B lymphocytes activated T lymphocytes. Soon thereafter, it was shown that dendritic cells also produced EVs (Raposo et al., 1996; Théry et al., 2001), suggesting a role for EVs in antigen presentation. Later research identified messenger RNA (mRNA) and microRNA (miRNA) as exosome cargo and recognized their transfer into recipient cells, suggesting the biological activity of EVs (Ratajczak et al., 2006; Valadi et al., 2007; Wysocynski and Ratajczak, 2009; Raposo and Stoorvogel, 2013) and revealing their role as essential cell-to-cell communication vehicles.
The International Society for Extracellular Vesicles (ISEV) was founded in 2012. This entity provided the criteria to classify EVs into three groups according to their biogenesis: exosomes, microvesicles (MVs), and apoptotic bodies (AB) (Gould and Raposo, 2013; Yáñez-Mó et al., 2015; Théry et al., 2018).
Among the three subtypes, exosomes are the most thoroughly studied. Exosomes are the smallest of the subtypes, with a diameter of between 30 and 150 nm, and are primarily formed via an endocytic pathway. Endocytic invagination forms early endosomes (Bissig and Gruenberg, 2013), and during their maturation into MVB, inward budding results in the generation of intraluminal vesicles (ILVs). In this process, cytosolic RNAs are taken up into ILVs. Because ILVs become exosomes, this inward budding process is designated “RNA loading into exosomes” (Janas et al., 2015). Then, most MVBs are directed to lysosomes for degradation. However, a subset of MVBs moves along microtubules toward the cell membrane by several Rab-GTPase molecules (Fevrier et al., 2004). When it reaches the cell membrane, the limiting membrane of the MVB fuses with the plasma membrane, and ILVs are released into the extracellular space (Pant et al., 2012; Yáñez-Mó et al., 2015). After secretion, ILVs are termed exosomes. As a consequence of their origin, exosomes contain a set of specific proteins and lipids. For instance, exosomes contain endosome-associated proteins involved in MVB biogenesis and RNA sorting, such as the Endosomal Sorting Complex Required for Transport (ESCRT) complex, Alix and Tsg101 (Nabhan et al., 2012). Other exosome markers are the tetraspanins CD9, CD63, and CD81 (Kowal et al., 2014). Although exosomes are derived from the cell membrane, they are formed during the spontaneous inward budding of raft-like regions, and therefore, their lipid content is slightly different from that of the cell membrane. Exosomes are enriched in cholesterol, sphingomyelin, glycosphingolipids, and phosphatidylcholine with saturated fatty acids (Laulagnier et al., 2004; Raposo and Stoorvogel, 2013; Janas et al., 2015).
Microvesicles have a diameter between 100 and 1000 nm (Bobrie et al., 2011). In contrast to exosomes, MVs are produced directly from the cell surface by outward budding and fission of the plasma membrane. Hence, their surface markers and lipids are primarily dependent on the composition of the cell membrane (Ratajczak et al., 2006; Lee et al., 2012). MVs also contain transmembrane proteins commonly found in the cell membrane, such as integrins and selectins (Bobrie et al., 2011; Raposo and Stoorvogel, 2013).
Apoptotic bodies are the largest EVs, 1–5 μm in diameter on average. These vesicles are products of cell disassembly during apoptosis and are released via blebbing of the plasma membrane (Ivan et al., 2014). Their cargo is quite distinct from that of exosomes and MVs, since in addition to proteins, lipids, DNA, and ribosomal RNA (rRNA), they harbor cytoplasm, organelles and histones with or without a nuclear fragment (Ivan et al., 2014; Van Niel et al., 2018).
A summary of the comparison among the three subtypes of EVs is shown in Figure 1.
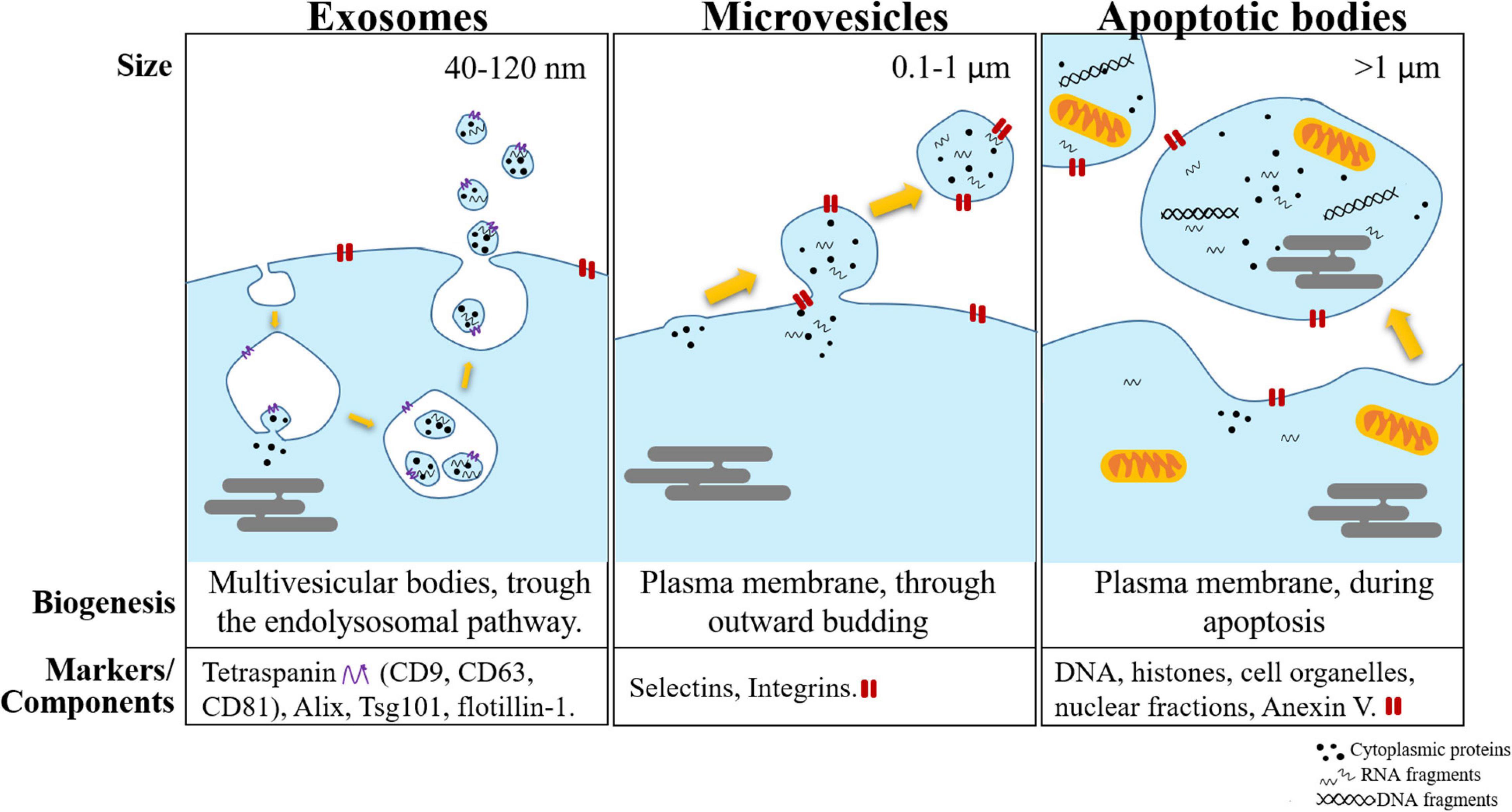
Figure 1. Schematic representation of the different subtypes of extracellular vesicles (exosomes, microvesicles, and apoptotic bodies). These vesicles are separated mainly by their size, biogenesis, and the most common markers used to identify them.
Differential ultracentrifugation is the most commonly used approach for EV purification (Théry et al., 2006). Ultracentrifugation can remove soluble components from EV preparations; however, similarly sized particles (membrane vesicles and protein aggregates) copurify (Vergauwen et al., 2017). Therefore, due to the overlap in size between MVs and exosomes, common methods of vesicle isolation do not allow the proper separation of these two populations. The use of markers to separate these two populations is also unreliable since there is still a lack of specific markers to distinguish these populations (Tauro et al., 2012). Furthermore, exosomes and MVs, to a large extent, have a similar molecular composition (Lee et al., 2012; Crescitelli et al., 2013). For these reasons, in this article, we review all articles mentioning both exosomes and MVs. To unify the nomenclature throughout this review, we refer to them collectively as EVs, a term that has been recommended by the ISEV for use as a generic term (Théry et al., 2018; Witwer and Théry, 2019).
A large body of evidence has shown that all organisms, despite their complexity and diversity, secrete EVs. This secretion appears to be a conserved mechanism across all three domains of living organisms: Archaea, Bacteria, and Eukarya (Gill et al., 2019). EVs have been shown to mediate cell–cell communication, playing a key role in the regulation of various physiological processes and the pathogenesis of many diseases, being one of them, cancer (Jasmina et al., 2008; Kosaka et al., 2013b, 2016). Cancer cells secrete EVs to both cells in proximity and distal sites, transferring a variety of molecules that modify the microenvironment, which in turn favors cancer cell growth, invasion, and metastasis (Skog et al., 2008; Hood et al., 2011; Yang and Robbins, 2011; Peinado, 2012; He et al., 2015). The amount of shed EVs rises sharply in disease states compared with nondisease states (Knijff-Dutmer et al., 2002; Noerholm et al., 2012). The interest of scientists and physicians in EVs has increased logarithmically in response to the discoveries that EVs can be isolated from body fluids such as serum, plasma, urine, and cerebrospinal fluid and can be used as biomarkers (Lee et al., 2012; Raposo and Stoorvogel, 2013; Tewari, 2015). Here, we review the biological effects of EVs and, more specifically, their mRNA cargo in the context of human cancers. We will further discuss their involvement in tumor progression, invasion, microenvironmental remodeling, and metastasis.
Extracellular vesicles contain an abundant cargo of different RNA species that can modulate physiological responses in recipient cells. Since EVs have a small size in comparison to cells, it is easy to understand that most previous studies on EV content focused on miRNAs and other smaller RNA species, such as Y-RNA, rRNA, transfer RNA (tRNA), piwi-interacting RNA (piRNA), and small nucleolar RNA (snoRNA) (Kosaka et al., 2010; Nolte’T Hoen et al., 2012; Lefebvre et al., 2016; Wei et al., 2017). However, larger RNA species, including mRNA, long non-coding RNA (lncRNA), and circular RNA (circRNA), have also been described in EVs (Valadi et al., 2007; Nolte’T Hoen et al., 2012; Dong et al., 2016; Lefebvre et al., 2016). Despite efforts to describe the different RNA species and their abundance in EVs, this topic remains controversial. For instance, while Bellingham et al. (2012) described that close to 90% of the RNA content was tRNA, Wei et al. (2017) only found between 11.6 and 17.4%, depending on the EV subtype (exosome and MV, respectively). Nonetheless, this value was even lower in the study done by Huang et al. (2013), in which they found only 1.24% of tRNA. Similar discrepancies in amount have been described mainly with microRNA and rRNA (Nolte’T Hoen et al., 2012; Huang et al., 2013; Wei et al., 2017). These dissimilarities are mainly due to the differences in the sequencing methodologies, the species of EVs analyzed, and the cell type of origin. However, it seems to be the consensus that smaller RNAs are more abundant than larger species, especially in exosomes compared with MVs (Dong et al., 2016; Lefebvre et al., 2016; Wei et al., 2017). This pattern makes sense since those types of EVs are also smaller than the other types and their biogenesis is different.
Due to the fragile nature of RNA and the large amount of ribonucleases (RNases) in the extracellular milieu, naked RNA fragments are rapidly degraded. However, the ability to detect RNAs in serum suggests that they are found in structures and protected from degradation. Those structures include Argonaute2 complexes and EVs (Arroyo et al., 2011). miRNAs encapsulated in EVs can resist digestion by RNases, with a decrease in the RNA content of less than 7%, showing that most miRNAs are inside EVs (Skog et al., 2008). This protection by the EV membrane is especially important for mRNAs, since compared to miRNAs, they are longer and are therefore more fragile.
It has been shown that mRNA and other small RNAs loading is not a passive event but rather a selective process (Guduric-fuchs et al., 2012). Although the mRNAs carried by EVs are the same as those that are expressed in the cell of origin and the cargo of EVs is thus representative of their cell of origin (Jenjaroenpun et al., 2013), it has been found that the relative amount of each mRNA is not completely correlated (Nolte’T Hoen et al., 2012). The similitude with the cellular content is higher in MVs rather than exosomes (Wei et al., 2017). Many RNAs isolated from EVs were found to be enriched in EVs in relation to the RNA profiles of the cells of origin, while others were specifically excluded or not found at all. This indicates that RNA molecules are selectively incorporated into EVs (Skog et al., 2008; Nolte’T Hoen et al., 2012; Kim et al., 2017). For instance, Hong et al. (2009) reported that despite the similarities at the global level between EVs and cellular mRNAs, they identified 241 mRNAs enriched and 1461 diminished in EVs. Moreover, of the enriched mRNAs, 27 were involved in the cell cycle, suggesting that EV shedding might be cell cycle-dependent (Hong et al., 2009). Specifically, packaging during the S phase of cell division has been proposed due to the specific transcript content in EVs (Conley et al., 2017).
The enrichment of certain types of mRNA has also been explained by zip codes. Zip-codes are sequences typically found in the 3′-untranslated regions (3′UTRs) of mRNA transcripts that mediate the binding of a ribonuclear protein complex and eventually mediate mRNA localization within EVs (Martin and Ephrussi, 2009). A 25 nt sequence with a “CTGCC” core sequence was found in all mRNAs highly enriched in EVs derived from glioblastoma (GBM) and melanoma cells. Interestingly, this zip code could be used to increase the levels of mRNAs in EVs, as shown by the increased amount of GFP mRNA, while a mutation in the core sequence inhibited the loading (Bolukbasi et al., 2012). A similar zip code was found in miRNAs and called EXOmotif; consisting of the sequence “GGAG” (Villarroya-Beltri et al., 2013). Furthermore, the sequence “UGCA” was found in miRNAs specifically excluded from EVs and was called the CLmotif (Villarroya-Beltri et al., 2013). These motifs, in both mRNAs and miRNAs, control their loading into EVs, modulating the cargo in these vesicles.
Certain proteins, such as ESCRT, are well-known machinery for the loading of several molecules into EVs (Kowal et al., 2014). More specifically, the so-called Arc proteins assist in mRNA transfer into EVs through ionic interactions with this nucleic acid molecule (Pastuzyn et al., 2018). Other proteins compressed within the autophagy pathway, such as the LC-3/ATG8 complex, were found to regulate the loading of several RNAs into EVs (Leidal et al., 2020). LC-3 protein interacts with RNA-binding proteins (RBP), which at the same time, bind to snoRNA increasing their loading. Interestingly, this complex also required neutral sphingomyelinase 2 (nSMase2), another protein known to regulated EV secretion by the synthesis of ceramides (Trajkovic et al., 2008). However, many important mechanisms for mRNA packaging remain unknown.
Whether the various RNA molecules in EVs are intact or partially degraded is an essential consideration, because intact mRNAs could act as templates for functional proteins in recipient cells. Although EVs contain long RNA molecules, their average length is shorter than that of RNA molecules found within cells. It was found that the cargo of EVs derived from hepatocellular carcinoma cells spanned a range of between 25–50 and 4000 nt (He et al., 2015; Berardocco et al., 2017). These lengths are considerably different from those of cellular mRNAs, which have lengths of between 400 and 12,000 nt (Batagov and Kurochkin, 2013; Enderle et al., 2015). One possible explanation for these observations could be that exosomes are enriched in mRNAs encoding very short proteins. In addition, fragmented mRNA has been found in EVs, along with full-length mRNA (Batagov and Kurochkin, 2013). In contrast to the initial view, both full-length and truncated mRNAs are transferred into recipient cells and can be translated into proteins, but the resulting proteins have different functions (Batagov and Kurochkin, 2013). Fragmented mRNAs are translated mainly into enzyme modulators and proteins participating in extracellular transport, while full-length mRNAs are translated into cell surface receptor, cell communication and system development proteins (Batagov and Kurochkin, 2013). The difference in function suggests a specialization mechanism rather than a random phenomenon. Indeed, mRNA fragments and their loading into EVs was correlated with the proximity of the probes to the 3′UTRs of their transcripts. The 3′UTRs of mRNAs are rich in regulatory sequences and seem to also be involved in the translational ability of mRNA fragments.
Extracellular vesicle mRNAs are not only stable mRNA molecules that are accessible for use as biomarkers, they can also be delivered to recipient cells and be translated into functional proteins (Valadi et al., 2007). EVs were first described as a cellular mechanism to discard unwanted cellular debris (Johnstone et al., 1987; Harding et al., 2013), and therefore, they were thought to not have other functions. A few years later, the antigen-presenting function of B lymphocyte-derived EVs to activate T cells was described, followed by the equivalent activation by dendritic cell-derived EVs. Therefore, it was thought that EVs played a role in the immune system (Bobrie et al., 2011). It was not until the reports by Ratajczak et al. (2006) and Valadi et al. (2007) the function of mRNAs contained in EVs and their ability to be translated into proteins was demonstrated.
Ratajczak et al.’s (2006) provided the first evidence regarding the function of mRNAs transferred to recipient cells, that is to be translated into proteins. They found that embryonic stem (ES) cells and their derived EVs contained high levels of Wnt-3 mRNA. When parental cells were cultured with ES cell-derived EVs, several stemness-related genes were upregulated (Ratajczak et al., 2006). In another study using mouse mast cell-derived EVs and the machinery in rabbit reticulocyte lysates as an in vitro translation system, mouse-derived proteins (COX5B, HSPA8, SHMT1, LDH1, ZFP125, GPI1, and RAD23B) were detected by mass spectrometry analysis, confirming the possibility of the translation of EV-derived mRNAs (Valadi et al., 2007). However, Skog et al. (2008) showed the complete process. GBM cells were transduced with a lentiviral vector encoding a secreted luciferase from Gaussia (Gluc) (Tannous et al., 2005), and EVs from these cells were collected and confirmed to contain the mRNA but not the protein. When those EVs were added to endothelial cells (ECs), Gluc activity produced by recipient cells was detected, confirming that the mRNA delivered into recipient cells generated a functional protein (Skog et al., 2008). Similar studies have used GFP instead of Gluc, allowing detection of transfected cells by confocal microscopy after a 6-h incubation (Deregibus et al., 2007). Moreover, to further confirm that mRNA derived from EVs and not other elements within the EVs were the main effector of function, mRNA was isolated from EVs and transfected into recipient EC with lipofectamine (Deregibus et al., 2007). Overall, all these in vitro assays showed that EV mRNA is functional in recipient cells, leading to behavioral changes in recipient cells in vitro.
The following step should then be to assess the translational capacity in vivo. Zomer et al. (2015) elegantly demonstrated the transfer of functional cargo in EVs in vivo using the Cre-LoxP system from an a priori-defined tumor cell population. The Cre-LoxP system induced a color switch (from red to green) in cells that internalized EVs released from specific cells expressing Cre recombinase (Ridder et al., 2014). The switch could be observed by in vivo imaging. They demonstrated that highly malignant cancer cells communicated with their microenvironment and other less malignant cancer cells and induced, through transfer of EVs, increases in migratory behavior and metastatic capacity (Zomer et al., 2015). Via this system, the authors also demonstrated that although B16 melanoma cells had the ability to take up EVs from healthy cells, this transfer was not frequent. The opposite phenomenon—transfer from cancer cells to healthy cells—was more common.
A summarized information of all the events can be found in Table 1.
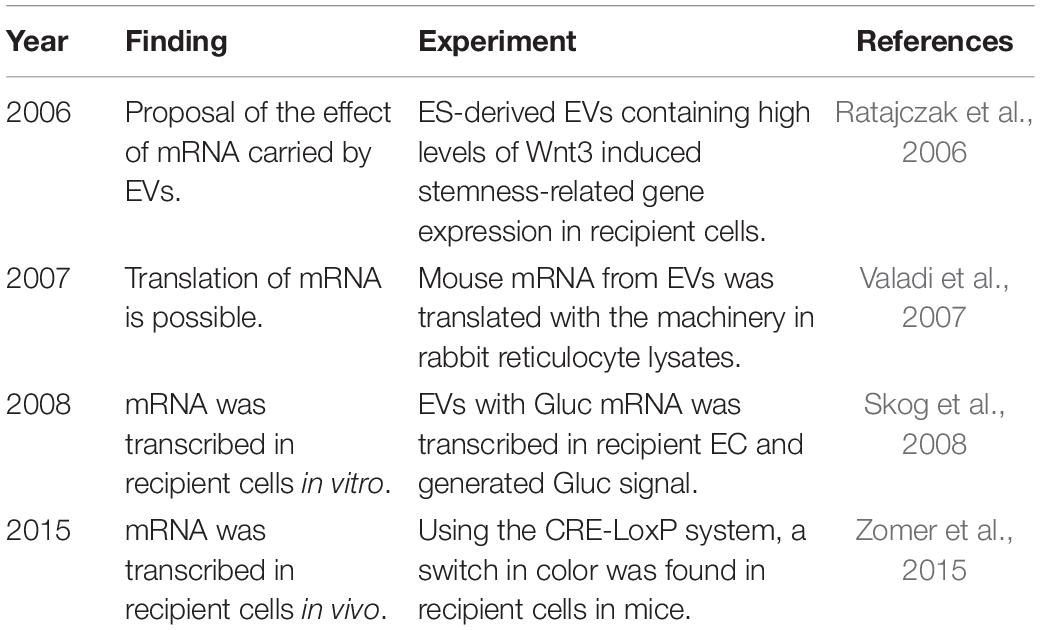
Table 1. Historiological order of essential research to prove functional loading of mRNA into recipient cells via EVs.
Extracellular vesicles play an essential role in cellular communication under both physiological and pathological conditions. However, cancer cells constitutively secrete EVs in greater numbers than their healthy counterparts (Knijff-Dutmer et al., 2002; Noerholm et al., 2012). This pattern can be inferred from biological fluids obtained from patients with cancer. For instance, a study reported that while the blood of healthy patients contained approximately 2 × 1012 EVs/mL, the blood of cancer patients contained 4 × 1012 EVs/mL (Kalluri, 2016), although further studies are needed to confirm these values since other reports showed the contrary effect (Fang et al., 2016). Nonetheless, it has been shown that under stress, cells increase the secretion of EVs (Harmati et al., 2019; O’Neill et al., 2019). Such stress could be hypoxia (King et al., 2012), laser irradiation inducing autophagy (Bagheri et al., 2018), or oxidative stress (Harmati et al., 2019). The tumoral microenvironment contains most of the cellular stresses mentioned before and some more, including low levels of oxygen, nutrients, low pH, oxidative stress, and others (Chen and Xie, 2018). These factors could explain the general tendency of an increased EV secretion. Importantly, since EVs can mirror their cell of origin to a certain extent, their cargo is oncogenic. Tumor-derived EVs ensure crosstalk between tumor cells and their microenvironment for the benefit of cancer cells. Tumor-derived EVs have been reported to play major roles in the modulation of tumorigenesis, angiogenesis, the immune response, drug resistance, and invasion. Moreover, since EVs are secreted in large amounts into the bloodstream, they can travel to distant sites, where they promote premetastatic niche formation, extravasation, and the survival of metastatic cells at these distant sites (Hood et al., 2011; Peinado, 2012; Hoshino et al., 2015; Tominaga et al., 2015; Yokoi et al., 2017; Costa-silva et al., 2018).
Tumors are not composed only of cancer cells; they are located within an environment containing numerous types of stromal cells, including fibroblasts, EC, mesenchymal cells, and immune cells. This environment is called the tumor microenvironment and plays multiple roles in tumor progression (Whiteside, 2008). The tumor microenvironment also includes the extracellular matrix and factors secreted by these cells, such as growth factors, cytokines, and EVs (Whiteside, 2008; Hu et al., 2018). Cancer cells release EVs into the tumor microenvironment, and these EVs are taken up by other cells, altering their signaling pathways and gene regulation programs. Thus, EVs reshape the local tumor environment into a more favorable niche for tumor growth, invasion, and metastatic spread (Taylor and Gerçel-Taylor, 2005; Skog et al., 2008; Wysocynski and Ratajczak, 2009; Peinado, 2012; Salomon et al., 2013; He et al., 2015; Costa-silva et al., 2018). Via these mechanisms, EVs can promote EC growth and thus induce tumor angiogenesis (Deregibus et al., 2007), cause immune tolerance by activating suppressive pathways in immune cells (Valenti et al., 2007), transform fibroblasts into cancer-associated fibroblasts (Webber et al., 2010) and directly modify and disrupt the extracellular matrix (Dolo et al., 1999). Importantly, since EVs are secreted into body fluids, they can also reach distal organs and affect the environment, facilitating metastasis at those sites (Hood et al., 2011; Peinado, 2012; Tominaga et al., 2015; Yokoi et al., 2017; Costa-silva et al., 2018).
Following, several biological functions activated by the different tumor microenvironment components as a response to cancer cell-derived EVs are described. A summary of the same processes is provided in Figure 2.
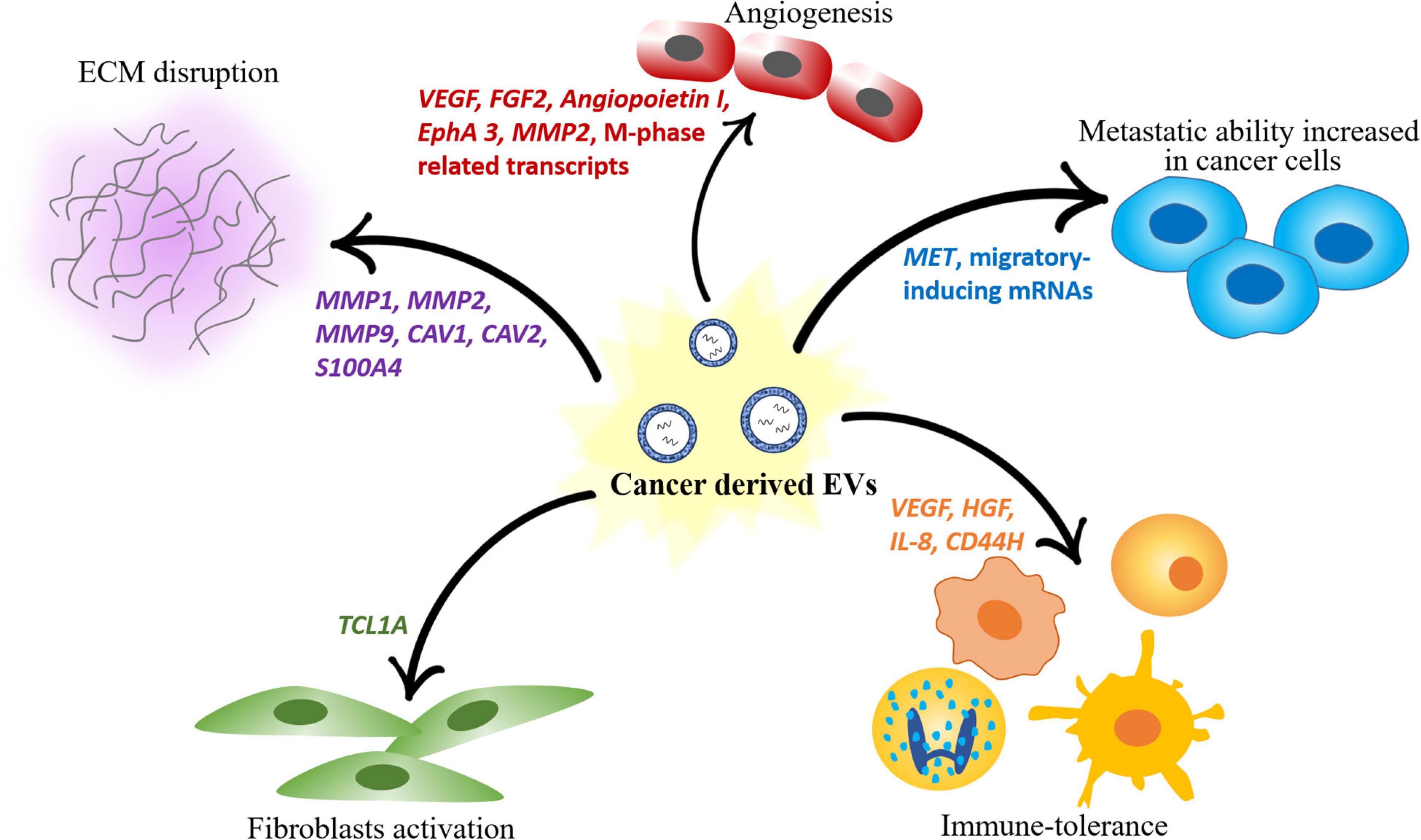
Figure 2. Scheme of the different roles of mRNAs contained in EVs in modulating the components of the tumor microenvironment. Cancer-derived EVs induce angiogenesis, disrupt the ECM, promote fibroblast activation and immune tolerance and increase the metastatic ability of other cancer cells.
Angiogenesis is the formation of new blood vessels from a preexisting vascular network by the proliferation of ECs, which occurs naturally during organismal growth and development and during tissue restoration. Angiogenesis is a vital process for the development of tumors, supplying cancer cells with the nutrients, and oxygen necessary for their growth (Carmeliet, 2005). The newly formed tubes also provide a route for metastatic cells to escape (Nishida et al., 2006; Kikuchi et al., 2019). Therefore, cancer cells, especially those in rapidly growing solid tumors, promote angiogenesis (Carmeliet, 2005; Todorova et al., 2017).
Paradoxically, hypoxia is a common feature in many cancers (Finger and Giaccia, 2010). Angiogenesis and hypoxia frequently co-occur. Under hypoxic conditions, cells secrete multiple proangiogenic molecules that facilitate the proliferation of ECs toward the tumor. Among the large variety of proangiogenic molecules, which commonly include cytokines and growth factors directly secreted into the extracellular space, EVs, and their cargo have also been shown to favor tumor vascularization (Salomon et al., 2013; Kikuchi et al., 2019). However, it is especially during hypoxia that this process is enhanced (Wysocynski and Ratajczak, 2009). There is evidence in several systems for boosted EV release under hypoxic (King et al., 2012) and anoxic conditions (Gupta and Knowlton, 2007). Developmental studies have shown that the release and bioactivity of placental mesenchymal stem cell (pMSC)-derived EVs is oxygen concentration-dependent. Under hypoxic conditions (1 or 3% O2), EV release from pMSCs increased by up to sevenfold compared to that from cells incubated under normoxic conditions (Salomon et al., 2013). Similarly, exposure of breast cancer cells to moderate (1%) and severe (0.1%) hypoxia resulted in increases of 30 and 90%, respectively, in the number of particles in the conditioned medium. EV secretion is regulated by the transcriptional factor HIF-1α (King et al., 2012); however, the specific mechanisms by which hypoxia induces EV release remain to be clarified.
As mentioned, angiogenesis is a common process under physiological conditions and occurs during the whole lifespan of organisms. Developmental studies have shown that pMSC-derived EVs promote EC migration and the formation of capillary-like structures in vitro (Salomon et al., 2013). In adult organisms, cells with some extent of stemness have also been proven to induce angiogenesis. EVs derived from endothelial progenitor cells (EPCs) induced proliferation and in vitro tube formation by horizontal transfer of mRNA associated with the PI3K/AKT signaling pathway (Deregibus et al., 2007). The PI3K/AKT pathway promotes EC proliferation, a requirement for angiogenesis (Karar and Maity, 2011). Indeed, this important pathway was also activated by endothelial colony-forming cell-derived EVs. In this situation, the authors found angiogenesis stimulation both in vitro and in vivo through the delivery of mRNAs associated with eNOS and the PI3K/AKT signaling pathway (Deregibus et al., 2007).
Tumor cells can also hijack the normal vasculature and stimulate the rapid formation of new blood vessels to supply the tumor with nutrients (Carmeliet, 2005; Al-Nedawi et al., 2009; Grange et al., 2011). Pioneer studies in tumor angiogenesis focused on the whole cargo of EVs, including mRNA and proteins such as VEGF, FGF, IL-6, and IL-8 (Skog et al., 2008) that could stimulate the angiogenic phenotype in glioblastoma. Other proteins induced the same phenotype in an indirect manner in lung cancer (Al-Nedawi et al., 2009). The expression of the oncogenic epidermal growth factor receptor (EGFR) molecule on EVs is transferred to ECs, triggering the expression VEGF and contributing to an overall increased angiogenesis.
In general terms, angiogenesis is promoted by the transfer of mRNAs related to cell cycle progression and EC proliferation. For instance, EVs derived from the colorectal cancer cell line SW480 were enriched with M phase-related transcripts such as CENPE, KIF15, CEP55, CCNA2, NEK2, PBK, and CDK8, which initiated angiogenesis via stimulation of EC proliferation (Hong et al., 2009).
Overall, the hypoxic tumor microenvironment induces tumor cells to increase the release of EVs containing molecules that induce EC proliferation and facilitate angiogenesis. The new vasculature supplies nutrients and oxygen to cancer cells and provides an escape route for tumor cell migration and metastasis. An example of this process is a population of renal carcinoma cells that were found to secrete EVs containing the proangiogenic VEGF, FGF2, Angiopoietin1, Ephrin A3, MMP2, and MMP9 mRNAs, which promoted in vivo angiogenesis and lung metastasis (Grange et al., 2011).
Extracellular vesicles mediate a wide range of immunological processes. Indeed, the first function of EVs described, contrary to the previous assumption that EVs were merely cell waste, was their function as antigen-presenting vesicles to recipient T cells (Raposo et al., 1996; Théry et al., 1999). Subsequent studies revealed that EVs participate in multiple activities, including T cell activation (Skokos et al., 2001; Taylor and Gerçel-Taylor, 2005), dendritic cell maturation (Skokos et al., 2003), and antigen presentation and macrophage activation (Schorey and Bhatnagar, 2008). The specific biological function depends on the immune cell type. For instance, EVs from B lymphocytes have potential antitumor effects, since they are potent immunostimulators. However, patients with cancer, especially those with advanced disease, often have depressed antitumor immunity (Penn, 1994). To grow, cancer cells are capable of modulating immune responses, which typically cause suppression of the immune system (Skokos et al., 2001; Taylor et al., 2003; Taylor and Gerçel-Taylor, 2005; Wen et al., 2016). Several mechanisms, including inactivation of the cytotoxic activity of T lymphocytes or natural killer (NK) cells, contribute to the ability of tumor cells to escape the immune response (Huang-Ge and William, 2012). NK cells are cytotoxic lymphocytes of the innate immune system that kill cancer cells directly (Liu et al., 2006; Vivier et al., 2017). However, under contact with EVs derived from murine mammary tumors, their cytolytic activity was reduced by a decrease in perforin gene expression, and IL-2 stimulated cell growth, allowing the implanted tumors to grow rapidly (Liu et al., 2006). Induction of T cell apoptosis by cancer cell-derived EVs has also been found in cancer (Taylor and Gerçel-Taylor, 2005; Muller et al., 2016). Ovarian cancer EVs carry the death ligands FasL and tumor necrosis factor–related apoptosis-inducing ligand (TRAIL), which cause significant DNA fragmentation and trigger the apoptotic pathway in T cells (Taylor et al., 2003; Jeong et al., 2005; Taylor and Gerçel-Taylor, 2005). Other mechanisms of immunosuppression include inhibition of cell differentiation, such as the differentiation of monocytes into dendritic cells (Valenti et al., 2007). This differentiation, in turn, is necessary for T cell activation. Since cancer cell-derived EVs block cell maturation, those cells instead become immunosuppressive cells by secreting factors that inhibit T lymphocyte proliferation, thus facilitating tumor progression (Wieckowski and Whiteside, 2006; Liu et al., 2010).
The crosstalk between cancer cells and immune cells via EVs has been gaining importance and is currently widely studied. However, most studies have focused on EV membrane proteins (Jeong et al., 2005; Taylor and Gerçel-Taylor, 2005; Muller et al., 2016), partially neglecting the role of packaged mRNAs in inhibiting the immune system. While it has been reported that T cells specifically show very low uptake of EVs, suggesting that the main effect of EVs is mediated by receptor binding on the surface membrane, other cells such as B lymphocytes and monocytes highly internalize EVs (Muller et al., 2016). Accordingly, the role of EV mRNA in monocytes has been described.
Tumor-derived EVs contain mRNAs encoding growth factors, including VEGF, HGF, IL-8, and CD44H. When EVs are phagocytosed by monocytes, these molecules are transferred and activate the Akt signaling pathway, resulting in altered biological activity (Baj-Krzyworzeka et al., 2006).
Although most reports have focused on membrane proteins of EVs, a report suggest the biological effects of mRNAs contained in EVs on T cells. Under non-pathological conditions, T cell responses are critical for antitumor immunity. However, EVs diminish the proliferation and activation and increase the apoptosis of T cells, ultimately resulting in progressive loss of T cell function, a process called T cell exhaustion (Yi et al., 2010; Liu et al., 2015). Muller et al. (2016) reported alterations in the recipient T cell transcriptome upon treatment with cancer cell-derived EVs. An increase in the levels of several immunosuppressive genes and the levels of the corresponding proteins was found. Moreover, decreased levels of CD69, a T cell activation marker, were observed, suggesting that mRNA transcripts delivered by EVs to recipient T cells caused immunosuppression (Muller et al., 2016).
Cancer cell-derived EVs are taken up by noncancerous cells and facilitate tumor progression by remodeling the microenvironment, promoting a more favorable environment for growth, as explained previously. However, it has also been shown that malignant cancer cells secrete EVs that can be taken up by less malignant cancer cells and affect their behavior. These behaviors often involve the acquisition of a more migratory and invasive phenotype.
In breast cancer, it was found that when the less-malignant T47D cells were located close to the highly malignant MDA-MB-231 cells, T47D cells migrated faster. Moreover, via the Cre-Lox system, it was shown that this increased motility was restricted to cells that had taken up MDA-MB-23-derived EVs, as shown by the switch in fluorescence (from red to green) (Zomer et al., 2015). Further analyses showed that this behavior was consistent with the EV cargo, which was enriched with migration-inducing mRNAs (Zomer et al., 2015), although the exact mRNAs were not specified in the original article. Because activation of migration and invasion often results in acquisition of a more metastatic phenotype (Nguyen et al., 2009), it is not surprising that the transfer of migration-inducing mRNAs also increases the metastatic potential of tumor cells. As a result, T47D cells that had taken up EVs released by the more malignant cell line and showed a more migratory phenotype also showed an increase in metastatic potential. These cells were 52-fold more abundant in the lungs than control cells (Zomer et al., 2015). Other studies also showed the increased invasive capacity of cancer cells upon contact with EVs derived from more metastatic cells. Malignant ovarian cancer cells expressing the stemness marker LIN28A promoted the invasion of HEK293 cells by activating epithelial-to-mesenchymal transition (Enriquez et al., 2015). Interestingly, it has been described that EVs derived from not only highly malignant cancer cells but also drug-resistant cells are able to induce migratory and invasive behaviors. Transfer of EVs derived from icotinib-resistant lung cancer cells into icotinib-sensitive lung cancer cells induced the expression of MET, which plays a role in migration. MET expression was correlated with increased migration and invasion of lung cancer cells (Yu et al., 2019).
The potential of cancer cells for invasion and migration is also dependent on their ability to degrade the ECM in which they are anchored. Accordingly, highly metastatic hepatocellular carcinoma cells induced migratory and invasive behaviors of nonmotile hepatocytes, providing the ability to degrade the ECM. EVs carrying CAV1, CAV2, and S100A4 mRNA molecules, upon internalization, induced the PI3K/AKT and MAPK signaling pathways and eventually increased the secretion of the matrix-degrading proteases MMP-2 and MMP-9 (He et al., 2015). Other studies have shown that some cancer cell-derived EVs carry both the protein and mRNA of metalloproteinases such as MMP1, MMP2, and MMP9 (Dolo et al., 1999; Hakulinen et al., 2008; He et al., 2015; Yokoi et al., 2017).
Metastasis is the ultimate consequence of cancer propagation and remains the leading cause of cancer-related mortality (Weigelt et al., 2005). However, to metastasize, tumor cells must manipulate the microenvironment in distant organs to optimize the conditions for the establishment and further growth of metastatic cells in a so-called premetastatic niche (Hood et al., 2011; Peinado, 2012; Sleeman, 2012; Chin and Wang, 2016; Costa-silva et al., 2018). These processes support cancer cell engraftment and survival (Sceneay et al., 2013). Since EVs are nanoparticles secreted into the extracellular space, they often enter body fluids circulating throughout the body. This precise systemic distribution of EVs and their capacity to alter the behavior of multiple types of cells have been shown to promote cancer metastasis, mainly via their contribution to generating a premetastatic niche (Hood et al., 2011; Peinado, 2012). Cancer cell-derived EVs contribute directly to this event by remodeling and degrading the ECM and organ parenchyma (Mu et al., 2013; Yokoi et al., 2017; Costa-silva et al., 2018) and via the recruitment of cells that promote metastasis, such as bone marrow-derived cells and macrophages (Peinado, 2012; Costa-silva et al., 2018), creating a microenvironment conducive to metastasis.
A clear example was described by Yokoi et al. (2017), who described the mechanisms of peritoneal metastasis promotion by EVs. Highly metastatic ovarian cancer cells secrete EVs with high concentrations of MMP1 mRNA and are transferred to mesothelial cells, the most prevalent cells in the peritoneal cavity. Upon uptake, MMP1 induces apoptosis in mesothelial cells, breaking the mesothelial-peritoneal barrier and promoting metastasis in vivo. Cells with knockdown of nSMase2, a protein required for EV secretion (Kosaka et al., 2013a), had a significant reduction in metastasis in the peritoneal cavity, although no major difference was found in the primary tumor size (Yokoi et al., 2017).
Moreover, Hood et al. (2011) demonstrated that premetastatic niche formation by EVs is a selective phenomenon mediated by the generation of liposomes to mimic empty EVs (these liposomes are nanovesicles lacking proteins, mRNAs, and miRNAs) but with a lipid content similar to that of the membrane. Their experiments showed the selective homing of melanoma EVs to sentinel lymph nodes and nonselective homing by liposomes (Hood et al., 2011). Moreover, EVs recruited melanoma cells to sentinel lymph nodes by altering the ECM, which promoted the trapping of melanoma cells within sentinel nodes. These studies helped to explain metastatic organotropism of melanoma. However, in most cancer types, the specificity of the metastatic organ cannot be explained by the anatomy of the blood and lymphatic circulatory networks (Fidler, 2003). Organotropism of EVs derived from cancer cells is logical if we take into consideration the internalization of EVs, which depends mainly on their structure and composition. Therefore, it is reasonable to think that organotropism is regulated mainly by molecules in the EV membrane. For instance, some integrins were found to be responsible for organotropism. EVs expressing integrin αvβ5 were associated with liver metastasis, while EVs expressing α6β4 and α6β1 were associated with lung metastasis (Hoshino et al., 2015). Since mRNAs have been found mainly inside EVs, we expect that these mRNAs have almost no function in organotropism. However, once EVs are taken up, their contained mRNAs play key roles in metastasis.
The discovery of EVs and their function allowed us to address one more layer of complexity in cancer. Thus, subsequent research should be directed at the use of this knowledge against cancer cells. One strategy is the use of EVs as biomarkers for cancer diagnosis. Another strategy is to use them as a therapeutic treatment. Both options will be further discussed below.
Currently, there is a need for biomarkers that can be utilized to diagnose cancers accurately and to predict drug sensitivities effectively (Collins and Varmus, 2015). A promising new approach for cancer diagnosis is focused on the use of biomarkers available in body fluids and able to be assessed in a noninvasive manner. In this area, EVs have great potential as biomarkers for cancer therapeutic monitoring since they are easily accessible, and their contents have been shown to be enriched with key molecules characteristic of the cells of origin (Atay and Godwin, 2014; Melo et al., 2015).
While it is true that most cells in the body secrete EVs, it has been demonstrated that tumors are especially potent producers of EVs; thus, cancer cell-derived EVs can be detected. Moreover, EVs have been found not only in blood (Caby et al., 2005; Dong et al., 2019) but also in most, if not all, body fluids, including urine (Pisitkun et al., 2004; Murakami et al., 2018), saliva (Yang et al., 2014; Han et al., 2018), cerebrospinal fluid (Chen et al., 2013; Akers et al., 2016), seminal fluid (Aalberts et al., 2012; Höög and Lötvall, 2015), breast milk (Admyre et al., 2007), amniotic fluid (Asea et al., 2008), ascites (Andre et al., 2002; Yokoi et al., 2017), and bile (Masyuk et al., 2010). Thus, EVs from most body fluids are a source for noninvasive or minimally invasive diagnosis of different cancers and, subsequently, as a tool for longitudinal disease monitoring (Collins and Varmus, 2015; Di Meo et al., 2017).
As mentioned previously, mRNA encapsulated within the phospholipid bilayer structure of EVs is protected from the harsh external environment, which would otherwise degrade it (El-Hefnawy et al., 2004; Skog et al., 2008; Cheng et al., 2014; Kim et al., 2017). Furthermore, in comparison with other EV cargo molecules, mRNA is very suitable as a biomarker due to the high sensitivity of PCR detection. Although it has been shown that the mRNA cargo in EVs is not an exact copy of the mRNA content in the cell of origin and that some mRNAs are enriched while others are depleted, the transcriptomic profile of EVs still reflects that of the cell of origin (Jenjaroenpun et al., 2013). Indeed, the transcriptomic signatures of EVs can differentiate healthy controls and patients with certain types of cancer, demonstrating their potential value as biomarkers (Noerholm et al., 2012).
Glioblastoma is a type of cancer that is very difficult to biopsy and requires invasive and dangerous brain surgery; thus, a biomarker for this kind of tumor is greatly required. The most prominent study in GBM found a variant of the gene EGFR, EGFRvIII, to be enriched in EVs isolated from patients, as has been described in 30% of GBM tumors (Johnson et al., 2012). EGFRvIII mRNA was easily detected by RT-PCR in patient serum (Skog et al., 2008). Other studies reported MDK, COX8C, GALR3, and LOC653602 to be enriched in EVs from GBM patients (Skog et al., 2008).
Biomarkers in urine from patients with bladder cancer are another promising approach. Due to the high recurrence rate of bladder cancer, which is higher than 50% (Botterman et al., 2003), a noninvasive lifelong monitoring biomarker is needed. A study conducted on EVs secreted during several stages of urothelial cancer narrowed the field of promising urinary EV biomarkers to three mRNA molecules, SLC2A1, GPRC5A, and KRT17. Importantly, the performance of these biomarkers was equivalent to that of conventional urine cytology and bladder tumor antigen assays (Murakami et al., 2018).
Of special interest in the finding of salivary EVs derived from cancer cells is their potential for identification by a routine sampling method that would be even less invasive than blood analysis. Human saliva is a slightly acidic biofluid (pH 6.0–7.0) that contains many proteins, enzymes, bacteria, and unique mRNAs, hindering the identification of biomarkers (Li et al., 2004; Lee and Wong, 2009). However, several studies have succeeded in discovering saliva-based biomarkers for oral and systemic cancers (Li et al., 2004). Yang et al. (2014) demonstrated the presence of distal cancer EVs in mouse saliva. To this end, they generated a lung cancer model in which cancer cells constitutively expressed the hCD63-GFP protein. The authors identified human CD63-GFP-containing EVs and GAPDH mRNA molecules in the blood and saliva of tumor-bearing mice (Yang et al., 2014). Furthermore, the lung cancer-specific mRNA molecules BRAF, EGFR, LZTS1, and FGF19 were also found in the isolated salivary EVs (Yang et al., 2014). In addition, other researchers have reported the presence of human pancreatic cancer markers in salivary EVs (Lau et al., 2013), suggesting that the presence of cancer EVs in saliva may be a common factor in cancers that should be properly addressed in the future.
Ascites is a condition commonly found in patients with gastric and ovarian cancers and consists of fluid accumulation in the peritoneal cavity. This fluid has been described to contain cancer cells and promote metastasis. Moreover, EVs derived from cancer cells have been found (Yokoi et al., 2017). These EVs contained high levels of MMP1 mRNA that induced apoptosis in mesothelial cells (Yokoi et al., 2017) MMP1 expression levels in EVs from ascites fluid were strongly related to patient prognosis in early stages of ovarian cancer and thus are a promising biomarker for the degree of ovarian cancer malignancy.
Many studies have been performed on cell lines as preliminary screens. For instance, EVs extracted from conditioned medium of several hepatocellular carcinoma cell lines were found to have a rather large variation in the candidate mRNAs (Berardocco et al., 2017). Among the mRNAs, TGM2 was contained in all EV cell lines and is a candidate for use as a hepatocellular carcinoma marker. Similarly, in prostate cancer cell lines, the expressed mRNAs in cancer cell-derived EVs were characteristic of those in the parental cells, especially with the differential expression of androgen metabolism genes such as ETV1 and FASN (Shen and Abate-Shen, 2010; Huang et al., 2016), suggesting the use of those markers to distinguish the androgen sensitivity of prostate cancer (Lázaro-Ibáñez et al., 2017). Studies in cell lines, as previously shown, have provided insight into possible biomarkers, but these results still have to be confirmed in patients to ascertain the real potential of these molecules as biomarkers. As an example, Cha et al. (2020) selected a total of 12 mRNAs that were differentially expressed in EVs from 4 colorectal cancer cell lines, and through a series of comparisons, identified that the combination of only two of these mRNAs—CD133 and VEGF—was able to distinguish colorectal cancer patients from healthy controls. However, the combination of these two mRNAs achieved 100% sensitivity with extremely high specificity and accuracy.
To date, numerous EV mRNAs have been reported as specific cancer biomarkers in diverse cancer types and body fluids, especially serum. An extensive list of these mRNAs is provided in Table 2.
Cancer EVs can be used not only as a diagnostic tool but also as a prognostic factor for cancer treatment outcomes. For example, measurement of PD-L1 mRNA in plasma-derived EVs from melanoma and non-small cell lung cancer patients allowed monitoring of the therapeutic response to nivolumab and pembrolizumab, agents that are anti-PD-1 antibodies (Del Re et al., 2018). Patients who responded to the treatment presented EVs with decreased PD-L1 mRNA levels compared to initial levels, while no difference was found in nonresponding patients. Monitoring of implanted treatments is very useful to identify the patient population responding to the treatment and to adjust the treatment of no-responding patients to achieve a proper treatment regimen.
Multiple important functions by which cancer cell-derived EVs promote their growth and metastasis through both the local and systemic transfer of genetic material have been described. Thus, suppression of EVs impairs the metastatic ability of cancer cells. Moreover, similar to the utility of EVs as biomarkers, their ability to circulate systemically throughout the human body makes EVs an interesting target for novel therapeutic approaches.
Because EVs play roles in multiple biological functions, including invasion, migration, angiogenesis, immunosuppression, and metastasis promotion, one potential strategy is to reduce the release of EVs from tumor cells or deplete them from the circulation. Although the complete mechanism of exosome release is not fully understood, it is known that the Rab GTPases Rab27a and Rab27b, as well as ceramides (e.g., nSMase2), play important roles in regulating exosome secretion (Ostrowski et al., 2010; Kosaka et al., 2013a). Knockdown of nSMase2 in cancer cells was found to reduce the level of EV secretion. Importantly, the metastatic ability of breast and ovarian cancer cells in vivo was dramatically reduced (Kosaka et al., 2013a; Yokoi et al., 2017).
On the other hand, depletion of circulating EVs has been attempted by dialysis (Marleau et al., 2012) and antibody targeting. Nishida-Aoki et al. (2017) took advantage of the opsonization effect of macrophages. When human-specific anti-CD9 or anti-CD63 antibodies were administered in a mouse model, macrophages phagocytized human cancer cell-derived EVs, and lung metastasis was significantly decreased. Although the results were promising, those experiments were performed in mouse models and used human antibodies. Thus, this methodology is not suitable for direct use in humans, since CD9 and CD63 are ubiquitous in the human body, and their eradication would generate several side effects (Nishida-Aoki et al., 2017). Future approaches should be directed toward cancer-specific molecules (Raposo and Stoorvogel, 2013), which were also found in EVs. This approach has been proposed as well by Zaborowski et al. (2019), who developed a methodology to identify such markers.
Because EVs are taken up by cancer cells, an alternative therapeutic approach is to reengineer naturally derived EVs for targeted gene therapy and drug delivery (Arslan et al., 2013; Katsuda et al., 2013; Ohno et al., 2013; Mao et al., 2018). Furthermore, exosomes have a long circulating half-life, and the lipid bilayer protects the mRNA cargo from degradation, facilitating its delivery to the target cells (Vickers and Remaley, 2012). For instance, ECRG4 is a tumor suppressor gene commonly downregulated in many cancers, including breast, prostate, lung, liver, colon, brain, and head and neck cancers (Götze et al., 2009; Li et al., 2009; Xu et al., 2013). Transfer of ECRG4 mRNA by EVs into human tongue squamous carcinoma cells increased its basal expression level and eventually inhibited malignant phenotypes in vitro and suppressed tumor growth and metastasis in vivo (Mao et al., 2018). To obtain EVs with high levels of ECGR4, the authors previously overexpressed this gene in HEK293 cells, from which they obtained the EVs. Overexpression in a cell line such as HEK293 may not be a recommended method for clinical use due to poor control of the complete EV cargo. Thus, the use of this method for clinical purposes is controversial. Moreover, there are other issues facing EV therapy. The main one is the process of gene loading into EVs, which has not been very successful to date. Electroporation, the main approach, usually results in fragmented mRNA (Hung and Leonard, 2016). To overcome this problem, novel approaches must be studied. A different method was offered by Bolukbasi et al. (2012), who found a 25-nt zip code-like sequence in mRNA that promoted encapsulation within EVs (discussed previously). Addition of this zip code in the 3′UTR of a specific mRNA resulted in enrichment of this mRNA in EVs (Bolukbasi et al., 2012). The use of this zip code or a similar one that promotes the loading of a specific mRNA into EVs for their later use in cancer treatment is very promising.
Messenger RNA in EVs provides an extensive resource for biomarkers that are highly sensitive and easily accessible in body fluids. Due to encapsulation within the lipid bilayer membrane, mRNA molecules remain stable in the circulation and are a promising tool for cancer diagnosis. Moreover, both native and modified EVs can be used as biological therapeutics. However, although many biomarkers have been reported, especially in cell lines, their function is often neglected. mRNA carried by EVs can be translated into proteins within target cells, inducing several phenotypes that are beneficial for cancer cells. Considering the functions of mRNAs in EVs that are already understood, including the modulation of tumorigenesis and the tumor microenvironment by the promotion of angiogenesis, the immune response, drug resistance and invasion, as well as the remodeling at distant sites that induces metastasis, we expect that EV-derived mRNAs perform many additional important functions in cancer pathogenesis. We believe that an understanding of these molecules will provide a novel target for anticancer therapies.
MP-V: conceptualization and writing. YY and TO: review and supervision. All authors contributed to the article and approved the submitted version.
This work was supported by the Grant-in-Aid for JSPS Fellows (20F20112) from the Japan Society for the Promotion of Science to MP-V, CREST grant (JPMJCR19H1) from the Japan Science and Technology Agency to YY, and Project for Cancer Research and Therapeutic Evolution grant (JP20cm0106402) from the Japan Agency for Medical Research, Development (AMED) to TO.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aalberts, M., van Dissel-Emiliani, F. M., van Adrichem, N. P., van Wijnen, M., Wauben, M. H., Stout, T. A., et al. (2012). Identification of distinct populations of prostasomes that differentially express prostate stem cell antigen, Annexin A1, and GLIPR2 in Humans1. Biol. Reprod. 86:82. doi: 10.1095/biolreprod.111.095760
Admyre, C., Johansson, S. M., Qazi, K. R., Filén, J.-J., Lahesmaa, R., Norman, M., et al. (2007). Exosomes with immune modulatory features are present in human breast milk. J. Immunol. 179, 1969–1978. doi: 10.4049/jimmunol.179.3.1969
Akers, J. C., Ramakrishnan, V., Yang, I., Hua, W., Mao, Y., Bob, S., et al. (2016). Optimizing preservation of extracellular vesicular miRNAs. Cancer Biomark. 17, 125–132.
Al-Nedawi, K., Meehan, B., Kerbel, R. S., Allison, A. C., and Rak, A. (2009). Endothelial expression of autocrine VEGF upon the uptake of tumor-derived microvesicles containing oncogenic EGFR. Proc. Natl. Acad. Sci. U.S.A. 106, 3794–3799. doi: 10.1073/pnas.0804543106
Andre, F., Schartz, N. E. C., Mosavassagh, M., Flament, C., Pautier, P., Morice, P., et al. (2002). Malignant effusions and immunogenic tumour-derived exosomes. Mech. Dis. 360, 295–305. doi: 10.1016/S0140-6736(02)09552-1
Arroyo, J. D., Chevillet, J. R., Kroh, E. M., Ruf, I. K., Pritchard, C. C., Gibson, D. F., et al. (2011). Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. U.S.A. 108, 5003–5008. doi: 10.1073/pnas.1019055108
Arslan, F., Lai, R. C., Smeets, M. B., Akeroyd, L., Choo, A., Aguor, E. N. E., et al. (2013). Mesenchymal stem cell-derived exosomes increase ATP levels, decrease oxidative stress and activate PI3K/Akt pathway to enhance myocardial viability and prevent adverse remodeling after myocardial ischemia/reperfusion injury. Stem Cell Res. 10, 301–312. doi: 10.1016/j.scr.2013.01.002
Asea, A., Jean-Pierre, C., Kaur, P., Rao, P., Linhares, I. M., Skupski, D., et al. (2008). Heat shock protein-containing exosomes in mid-trimester amniotic fluids. J. Reprod. Immunol. 79, 12–17. doi: 10.1016/j.jri.2008.06.001
Atay, S., and Godwin, A. K. (2014). Tumor-derived exosomes: a message delivery system for tumor progression. Commun. Integr. Biol. 7:e28231. doi: 10.4161/cib.28231
Bagheri, H. S., Mousavi, M., Rezabakhsh, A., Rezaire, J., Rasta, S. H., Nourazarian, A., et al. (2018). Low-level laser irradiation at a high power intensity increased human endothelial cell exosome secretion via Wnt signaling. Lasers Med. Sci. 33, 1131–1145. doi: 10.1007/s10103-018-2495-8
Baj-Krzyworzeka, M., Szatanek, R., Węglarczyk, K., Baran, J., Urbanowicz, B., Brański, P., et al. (2006). Tumour-derived microvesicles carry several surface determinants and mRNA of tumour cells and transfer some of these determinants to monocytes. Cancer Immunol. Immunother. 55, 808–818. doi: 10.1007/s00262-005-0075-9
Balaj, L., Lessard, R., Dai, L., Cho, Y.-J., Pomeroy, S. L., Breakefield, X. O., et al. (2011). Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat. Commun. 2:180. doi: 10.1038/ncomms1180
Batagov, A. O., and Kurochkin, I. V. (2013). Exosomes secreted by human cells transport largely mRNA fragments that are enriched in the 3′-untranslated regions. Biol. Direct 8:12. doi: 10.1186/1745-6150-8-12
Bellingham, S. A., Coleman, B. M., and Hill, A. F. (2012). Small RNA deep sequencing reveals a distinct miRNA signature released in exosomes from prion-infected neuronal cells. Nucleic Acids Res. 40, 10937–10949. doi: 10.1093/nar/gks832
Berardocco, M., Radeghieri, A., Busatto, S., Gallorini, M., Raggi, C., Gissi, C., et al. (2017). RNA-seq reveals distinctive RNA profiles of small extracellular vesicles from different human liver cancer cell lines. Oncotarget 8, 82920–82939. doi: 10.18632/oncotarget.20503
Bissig, C., and Gruenberg, J. (2013). Lipid sorting and multivesicular endosome biogenesis. Cold Spring Harb. Perspect. Biol. 5:a016816. doi: 10.1101/cshperspect.a016816
Bobrie, A., Colombo, M., Raposo, G., and Théry, C. (2011). Exosome secretion: molecular mechanisms and roles in immune responses. Traffic 12, 1659–1668. doi: 10.1111/j.1600-0854.2011.01225.x
Bolukbasi, M. F., Mizrak, A., Ozdener, G. B., Madlener, S., Ströbel, T., Erkan, E. P., et al. (2012). MiR-1289 and “zipcode”-like sequence enrich mRNAs in microvesicles. Mol. Ther. Nucleic Acids 1:e10. doi: 10.1038/mtna.2011.2
Botterman, M. F., Pashos, C. L., Refaelli, A., Laskin, B., and Hauser, R. (2003). The health economics of bladder cancer. Pharmaco Economics. 21, 1315–1330. doi: 10.1007/BF03262330
Caby, M. P., Lankar, D., Vincendeau-Scherrer, C., Raposo, G., and Bonnerot, C. (2005). Exosomal-like vesicles are present in human blood plasma. Int. Immunol. 17, 879–887. doi: 10.1093/intimm/dxh267
Carmeliet, P. (2005). Angiogenesis in life, disease and medicine. Nature 438, 932–936. doi: 10.1038/nature04478
Cha, B. S., Park, K. S., and Park, J. S. (2020). Signature mRNA markers in extracellular vesicles for the accurate diagnosis of colorectal cancer. J. Biol. Eng. 214, 1–9. doi: 10.1186/s13036-020-0225-9
Chen, M., and Xie, S. (2018). Therapeutic targeting of cellular stress responses in cancer. Thoracic Cancer 9, 1575–1582. doi: 10.1111/1759-7714.12890
Chen, W. W., Balaj, L., Liau, L. M., Samuels, M. L., Kotsopoulos, S. K., Maguire, C. A., et al. (2013). BEAMing and droplet digital PCR Analysis of Mutant IDH1 mRNA in gglioma patient serum and cerebrospinal fluid extracellular vesicles. Mol. Ther. Nucleic Acids 2:e109. doi: 10.1038/mtna.2013.28
Cheng, L., Sharples, R. A., Scicluna, B. J., and Hill, A. F. (2014). Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J. Extracell. Vesicles 3, 1–14. doi: 10.3402/jev.v3.23743
Chin, A. R., and Wang, S. E. (2016). Cancer tills the premetastatic field: mechanistic basis and clinical implications. Clin. Cancer Res. 1, 3725–3733. doi: 10.1016/j.physbeh.2017.03.040
Collins, F. S., and Varmus, H. (2015). A new initiative on precision medicine. N. Engl. J. Med. 372, 793–795. doi: 10.1056/NEJMp1500523
Conley, A., Minciacchi, V. R., Lee, D. H., Knudsen, B. S., Karlan, B. Y., Citrigno, L., et al. (2017). High-throughput sequencing of two populations of extracellular vesicles provides an mRNA signature that can be detected in the circulation of breast cancer patients. RNA Biol. 14, 305–316. doi: 10.1080/15476286.2016.1259061
Costa-silva, B., Aiello, N. M., Ocean, A. J., Singh, S., Thakur, B. K., Becker, A., et al. (2018). Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver Bruno. Nat. Cell Biol. 17, 816–826. doi: 10.1038/ncb3169
Crescitelli, R., Lässer, C., Szabó, T. G., Kittel, A., Eldh, M., Dianzani, I., et al. (2013). Distinct RNA profiles in subpopulations of extracellular vesicles: apoptotic bodies, microvesicles and exosomes. J. Extracell. Vesicles 2, 1–10. doi: 10.3402/jev.v2i0.20677
Del Re, M., Marconcini, R., Pasquini, G., Rofi, E., Vivaldi, C., Bloise, F., et al. (2018). PD-L1 mRNA expression in plasma-derived exosomes is associated with response to anti-PD-1 antibodies in melanoma and NSCLC. Br. J. Cancer 118, 820–824. doi: 10.1038/bjc.2018.9
Deregibus, M. C., Cantaluppi, V., Calogero, R., Lo Iacono, M., Tetta, C., Biancone, L., et al. (2007). Endothelial progenitor cell - Derived microvesicles activate an angiogenic program in endothelial cells by a horizontal transfer of mRNA. Blood 110, 2440–2448. doi: 10.1182/blood-2007-03-078709
Di Meo, A., Bartlett, J., Cheng, Y., Pasic, M. D., and Yousef, G. M. (2017). Liquid biopsy: a step forward towards precision medicine in urologic malignancies. Mol. Cancer 16, 1–14. doi: 10.1186/s12943-017-0644-5
Dolo, V., D’Ascenzo, S., Violini, S., Pompucci, L., Festuccia, C., Ginestra, A., et al. (1999). Matrix-degrading proteinases are shed in membrane vesicles by ovarian cancer cells in vivo and in vitro. Clin. Exp. Metastasis 17, 131–140. doi: 10.1023/A:1006500406240
Dong, L., Lin, W., Qi, P., Xu, M. D., Wu, X., Ni, S., et al. (2016). Circulating long RNAs in serum extracellular vesicles: their characterization and potential application as biomarkers for diagnosis of colorectal cancer. Cancer Epidemiol. Biomarkers Prev. 25, 1158–1166. doi: 10.1158/1055-9965.EPI-16-0006
Dong, Z., Sun, X., Xu, J., Han, X., Xing, Z., Wang, D., et al. (2019). Serum membrane type 1-matrix metalloproteinase (MT1-MMP) mRNA protected by exosomes as a potential biomarker for gastric cancer. Med. Sci. Monit. 25, 7770–7783. doi: 10.12659/MSM.918486
El-Hefnawy, T., Raja, S., Kelly, L., Bigbee, W. L., Kirkwood, J. M., Luketich, J. D., et al. (2004). Characterization of amplifiable, circulating RNA in plasma and its potential as a tool for cancer diagnostics. Clin. Chem. 50, 564–573. doi: 10.1373/clinchem.2003.028506
Enderle, D., Spiel, A., Coticchia, C. M., Berghoff, E., Mueller, R., Schlumpberger, M., et al. (2015). Characterization of RNA from exosomes and other extracellular vesicles isolated by a novel spin column-based method. PLoS One 10:e0136133. doi: 10.1371/journal.pone.0136133
Enriquez, V. A., Cleys, E. R., Da Silveira, J. C., Spillman, M. A., Winger, Q. A., and Bouma, G. J. (2015). High LIN28A expressing ovarian cancer cells secrete exosomes that induce invasion and migration in HEK293 cells. Biomed. Res. Int. 2015:701390. doi: 10.1155/2015/701390
Fang, Y., Garnier, D., Lee, T. H., D’Asti, E., Montermini, L., Meehan, B., et al. (2016). PML–RARa modulates the vascular signature of extracellular vesicles released by acute promyelocytic leukemia cells. Angiogenesis 19, 25–38. doi: 10.1007/s10456-015-9486-1
Fevrier, B., Vilette, D., Archer, F., Loew, D., Faigle, W., Vidal, M., et al. (2004). Cells release prions in association with exosomes. PNAS. 101, 9683–9688.
Fidler, I. J. (2003). The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat. Rev. Cancer 3, 453–458. doi: 10.1038/nrc1098
Finger, E. C., and Giaccia, A. J. (2010). Hypoxia, inflammation, and the tumor microenvironment in metastatic disease. Cancer Metastasis Rev. 29, 285–293. doi: 10.1038/jid.2014.371
Gill, S., Catchpole, R., and Forterre, P. (2019). Extracellular membrane vesicles in the three domains of life and beyond. FEMS Microbiol. Rev. 43, 273–303. doi: 10.1093/femsre/fuy042
Götze, S., Feldhaus, V., Traska, T., Wolter, M., Reifenberger, G., Tannapfel, A., et al. (2009). ECRG4 is a candidate tumor suppressor gene frequently hypermethylated in colorectal carcinoma and glioma. BMC Cancer 9:447. doi: 10.1186/1471-2407-9-447
Gould, S. J., and Raposo, G. (2013). As we wait: coping with an imperfect nomenclature for extracellular vesicles. J. Extracell. Vesicles 2:20389. doi: 10.3402/jev.v2i0.20389
Grange, C., Tapparo, M., Collino, F., Vitillo, L., Damasco, C., Deregibus, M. C., et al. (2011). Microvesicles released from human renal cancer stem cells stimulate angiogenesis and formation of lung premetastatic niche. Cancer Res. 71, 5346–5356. doi: 10.1158/0008-5472.CAN-11-0241
Guduric-fuchs, J., Connor, A. O., Camp, B., Neill, C. L. O., Medina, R. J., and Simpson, D. A. (2012). Selective extracellular vesicle-mediated export of an overlapping set of microRNAs from multiple cell types. BMC Genomics 13:357. doi: 10.1186/1471-2164-13-357
Gupta, S., and Knowlton, A. A. (2007). HSP60 trafficking in adult cardiac myocytes: Role of the exosomal pathway. Am. J. Physiol. Hear. Circ. Physiol. 292, 3052–3056. doi: 10.1152/ajpheart.01355.2006
Hakulinen, J., Sankkila, L., Sugiyama, N., Lehti, K., and Keski-Oia, J. (2008). Secretion of active membrane type 1 matrix metalloproteinase (MMP-14) into extracellular space in microvesicular exosomes. JCB. 105, 1211–1218. doi: 10.1002/jcb.21923
Han, Y., Jia, L., Zheng, Y., and Li, W. (2018). Salivary exosomes: Emerging roles in systemic disease. Int. J. Biol. Sci. 14, 633–643. doi: 10.7150/ijbs.25018
Harding, C., Heuser, J., and Stahl, P. (1984). Endocytosis and intracellular processing of transferrin and colloidal gold-transferrin in rat reticulocytes: demonstration of a pathway for receptor shedding. Eur. J. Cell Biol. 35, 256–263.
Harding, C. V., Heuser, J. E., and Stahl, P. D. (2013). Exosomes: looking back three decades and into the future. J. Cell Biol. 200, 367–371. doi: 10.1083/jcb.201212113
Harmati, M., Gyukity-Sebestyen, E., Dobra, G., Janovak, L., Dekany, I., Saydam, O., et al. (2019). Small extracellular vesicles convey the stress-induced adaptive responses of melanoma cells. Scien. Rep. 9:15329. doi: 10.1038/s41598-019-51778-6
He, M., Qin, H., Poon, T. C. W., Sze, S. C., Ding, X., Co, N. N., et al. (2015). Hepatocellular carcinoma-derived exosomes promote motility of immortalized hepatocyte through transfer of oncogenic proteins and RNAs. Carcinogenesis 36, 1008–1018. doi: 10.1093/carcin/bgv081
Hong, B. S., Cho, J. H., Kim, H., Choi, E. J., Rho, S., Kim, J., et al. (2009). Colorectal cancer cell-derived microvesicles are enriched in cell cycle-related mRNAs that promote proliferation of endothelial cells. BMC Genomics 10:556. doi: 10.1186/1471-2164-10-556
Hood, J. L., San Roman, S., and Wickline, S. A. (2011). Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Res. 71, 3792–3801. doi: 10.1158/0008-5472.CAN-10-4455
Höög, J. L., and Lötvall, J. (2015). Diversity of extracellular vesicles in human ejaculates revealed by cryo-electron microscopy. J. Extracell. Vesicles 4:28680. doi: 10.3402/jev.v4.28680
Hoshino, A., Costa-Silva, B., Shen, T. L., Rodrigues, G., Hashimoto, A., Tesic Mark, M., et al. (2015). Tumour exosome integrins determine organotropic metastasis. Nature 527, 329–335. doi: 10.1038/nature15756
Hu, C., Chen, M., Jiang, R., Guo, Y., Wu, M., and Zhang, X. (2018). Exosome-related tumor microenvironment. J. Cancer 9, 3084–3092. doi: 10.7150/jca.26422
Huang, M., Koizumi, A., Narita, S., Inoue, T., Tsuchiya, N., Nakanishi, H., et al. (2016). Diet-induced alteration of fatty acid synthase in prostate cancer progression. Oncogenesis 5:e195. doi: 10.1038/oncsis.2015.42
Huang, X., Yuan, T., Tschannen, M., Sun, Z., Jacob, H., Du, M., et al. (2013). Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics 14:319. doi: 10.1186/1471-2164-14-319
Huang-Ge, Z., and William, E. G. (2012). Exosomes and cancer: a newly described pathway of immune suppression. Clin. Cancer Res. 29, 997–1003. doi: 10.1016/j.biotechadv.2011.08.021
Hung, M. E., and Leonard, J. N. (2016). A platform for actively loading cargo RNA to elucidate limiting steps in EV-mediated delivery. J. Extracell. Vesicles 5:31027. doi: 10.3402/jev.v5.31027
Ivan, K. H. P., Lucas, C. D., Rossi, A. G., and Ravichandran, K. S. (2014). Apoptotic cell clearance: basic biology and therapeutic potential. Nat. Rev. Immunol. 14, 166–180.
Janas, T., Janas, M. M., Sapoń, K., and Janas, T. (2015). Mechanisms of RNA loading into exosomes. FEBS Lett. 589, 1391–1398. doi: 10.1016/j.febslet.2015.04.036
Jasmina, S. R., Balaj, L., Vos, K. E., van der, and Breakefield, X. O. (2008). Extracellular RNA mediates and marks cancer progression. Semin. Cancer Biol. 28, 14–23.
Jenjaroenpun, P., Kremenska, Y., Nair, V. M., Kremenskoy, M., Joseph, B., and Kurochkin, I. V. (2013). Characterization of RNA in exosomes secreted by human breast cancer cell lines using next-generation sequencing. PeerJ 1:e201. doi: 10.7717/peerj.201
Jeong, W. K., Wieckowski, E., Taylor, D. D., Reichert, T. E., Watkins, S., and Whiteside, T. L. (2005). Fas ligand-positive membranous vesicles isolated from sera of patients with oral cancer induce apoptosis of activated T lymphocytes. Clin. Cancer Res. 11, 1010–1020.
Johnson, H., Del Rosario, A. M., Bryson, B. D., Schroeder, M. A., Sarkaria, J. N., and White, F. M. (2012). Molecular characterization of EGFR and EGFRvIII signaling networks in human glioblastoma tumor xenografts. Mol. Cell. Proteomics 11, 1724–1740. doi: 10.1074/mcp.M112.019984
Johnstone, R. M., Adam, M., Hammond, J. R., Orr, L., and Turbide, C. (1987). Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J. Biol. Chem. 262, 9412–9420.
Kalluri, R. (2016). The biology and function of urine exosomes in bladder cancer. J. Clin. Invest. 4:2362. doi: 10.1172/JCI81135.The
Karar, J., and Maity, A. (2011). PI3K/AKT/mTOR pathway in angiogenesis. Front. Mol. Neurosci. 4:51. doi: 10.3389/fnmol.2011.00051
Katsuda, T., Tsuchiya, R., Kosaka, N., Yoshioka, Y., Takagaki, K., Oki, K., et al. (2013). Human adipose tissue-derived mesenchymal stem cells secrete functional neprilysin-bound exosomes. Sci. Rep. 3:1197. doi: 10.1038/srep01197
Kikuchi, S., Yoshioka, Y., Prieto-Vila, M., and Ochiya, T. (2019). Involvement of extracellular vesicles in vascular-related functions in cancer progression and metastasis. Int. J. Mol. Sci. 20:2584. doi: 10.3390/ijms20102584
Kim, K. M., Abdelmohsen, K., Mustapic, M., Kapogiannis, D., and Gorospe, M. (2017). RNA in extracellular vesicles. Wiley Interdiscip. Rev. RNA 8:1413.
King, H. W., Michael, M. Z., and Gleadle, J. M. (2012). Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer 12:421. doi: 10.1186/1471-2407-12-421
Knijff-Dutmer, E. A. J., Koerts, J., Nieuwland, R., Kalsbeek-Batenburg, E. M., and Van De Laar, M. A. F. J. (2002). Elevated levels of platelet microparticles are associated with disease activity in rheumatoid arthritis. Arthritis Rheum. 46, 1498–1503. doi: 10.1002/art.10312
Kosaka, N., Fujita, Y., Ochiya, T., Kosaka, N., Yoshioka, Y., Fujita, Y., et al. (2016). Versatile roles of extracellular vesicles in cancer Find the latest version: versatile roles of extracellular vesicles in cancer. J. Clin. Invest. 126, 1163–1172. doi: 10.1172/JCI81130.bolic
Kosaka, N., Iguchi, H., Hagiwara, K., Yoshioka, Y., Takeshita, F., and Ochiya, T. (2013a). Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic micrornas regulate cancer cell metastasis. J. Biol. Chem. 288, 10849–10859. doi: 10.1074/jbc.M112.446831
Kosaka, N., Iguchi, H., and Ochiya, T. (2010). Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 101, 2087–2092. doi: 10.1111/j.1349-7006.2010.01650.x
Kosaka, N., Yoshioka, Y., Hagiwara, K., Tominaga, N., Katsuda, T., and Ochiya, T. (2013b). Trash or Treasure: Extracellular microRNAs and cell-to-cell communication. Front. Genet. 4:173. doi: 10.3389/fgene.2013.00173
Kowal, J., Tkach, M., and Théry, C. (2014). Biogenesis and secretion of exosomes. Curr. Opin. Cell Biol. 6, 116–125. doi: 10.1016/j.ceb.2014.05.004
Lau, C., Kim, Y., Chia, D., Spielmann, N., Eibl, G., Elashoff, D., et al. (2013). Role of pancreatic cancer-derived exosomes in salivary biomarker development. J. Biol. Chem. 288, 2688–2697. doi: 10.1074/jbc.M113.452458
Laulagnier, K., Motta, C., Hamdi, S., Roy, S., Fauvelle, F., Pageaux, J. F., et al. (2004). Mast cell- and dendritic cell-derived display a specific lipid composition and an unusual membrane organization. Biochem. J. 380, 161–171. doi: 10.1042/BJ20031594
Lázaro-Ibáñez, E., Lunavat, T. R., Jang, S. C., Escobedo-Lucea, C., Oliver-De La Cruz, J., Siljander, P., et al. (2017). Distinct prostate cancer-related mRNA cargo in extracellular vesicle subsets from prostate cell lines. BMC Cancer 17:92. doi: 10.1186/s12885-017-3087-x
Lee, Y., El Andaloussi, S., and Wood, M. J. A. (2012). Exosomes and microvesicles: extracellular vesicles for genetic information transfer and gene therapy. Hum. Mol. Genet. 21, 125–134. doi: 10.1093/hmg/dds317
Lee, Y. H., and Wong, D. T. (2009). Saliva: an emerging biofluid for early detection of diseases. Am. J. Dent. 22, 241–248.
Lefebvre, F. A., Benoit Bouvrette, L. P., Perras, L., Blanchet-Cohen, A., Garnier, D., Rak, J., et al. (2016). Comparative transcriptomic analysis of human and Drosophila extracellular vesicles. Sci. Rep. 6:27680. doi: 10.1038/srep27680
Leidal, A. M., Huang, H. H., Marsh, T., Solvik, T., Zhang, D., Ye, J., et al. (2020). The LC3-Conjugation machinery specifies the loading of RNA- binding proteins into extracellular vesicles. Nat. Cell Biol. 22, 187–199. doi: 10.1038/s41556-019-0450-y
Li, C. C. Y., Eaton, S. A., Young, P. E., Lee, M., Shuttleworth, R., Humphreys, D. T., et al. (2013). Glioma microvesicles carry selectively packaged coding and noncoding RNAs which alter gene expression in recipient cells. RNA Biol. 10, 1333–1344. doi: 10.4161/rna.25281
Li, N. W., Yu, X. Y., Yang, Y., Zhang, C. P., Guo, L. P., and Lu, S. H. (2009). Expression of esophageal cancer related gene 4 (ECRG4), a novel tumor suppressor gene, in esophageal cancer and its inhibitory effect on the tumor growth in vitro and in vivo. Int. J. Cancer 125, 1505–1513. doi: 10.1002/ijc.24513
Li, Y., St. John, M. A. R., Zhou, X., Kim, Y., Sinha, U., Jordan, R. C. K., et al. (2004). Salivary transcriptome diagnostics for oral cancer detection. Clin. Cancer Res. 10, 8442–8450. doi: 10.1158/1078-0432.CCR-04-1167
Liu, C., Yu, S., Zinn, K., Wang, J., Zhang, L., Jia, Y., et al. (2006). Murine mammary carcinoma exosomes promote tumor growth by suppression of NK cell function. J. Immunol. 176, 1375–1385. doi: 10.4049/jimmunol.176.3.1375
Liu, Y., Gu, Y., Cao, X., Liu, Y., Gu, Y., and Cao, X. (2015). The exosomes in tumor immunity The exosomes in tumor immunity. Oncoimmunology 4:e1027472. doi: 10.1080/2162402X.2015.1027472
Liu, Y., Xiang, X., Zhuang, X., Zhang, S., Liu, C., Cheng, Z., et al. (2010). Contribution of MyD88 to the tumor exosome-mediated induction of myeloid derived suppressor cells. Am. J. Pathol. 176, 2490–2499. doi: 10.2353/ajpath.2010.090777
Mao, L., Li, X., Gong, S., Yuan, H., Jiang, Y., Huang, W., et al. (2018). Serum exosomes contain ECRG4 mRNA that suppresses tumor growth via inhibition of genes involved in inflammation, cell proliferation, and angiogenesis. Cancer Gene Ther. 25, 248–259. doi: 10.1038/s41417-018-0032-3
Marleau, A. M., Chen, C. S., Joyce, J. A., and Tullis, R. H. (2012). Exosome removal as a therapeutic adjuvant in cancer. J. Transl. Med. 10:134. doi: 10.1186/1479-5876-10-134
Martin, K. C., and Ephrussi, A. (2009). mRNA localization: gene expression in the spatial dimension. Cell 136, 719–730. doi: 10.1016/j.cell.2009.01.044
Masyuk, A. I., Huang, B. Q., Ward, C. J., Gradilone, S. A., Banales, J. M., Masyuk, T. V., et al. (2010). Biliary exosomes influence cholangiocyte regulatory mechanisms and proliferation through interaction with primary cilia. Am. J. Physiol. Gastrointest. Liver Physiol. 299, 990–999. doi: 10.1152/ajpgi.00093.2010
Melo, S. A., Luecke, L. B., Kahlert, C., Fernandez, A. F., Seth, T., Kaye, J., et al. (2015). Glypican1 identifies cancer exosomes and facilitates early detection of cancer. Nature 523, 177–182. doi: 10.1038/nature14581
Mu, W., Rana, S., and Zöller, M. (2013). Host matrix modulation by tumor exosomes promotes motility and invasiveness. Neoplasia 15, 875–887. doi: 10.1593/neo.13786
Muller, L., Mitsuhashi, M., Simms, P., Gooding, W. E., and Whiteside, T. L. (2016). Tumor-derived exosomes regulate expression of immune function-related genes in human T cell subsets. Sci. Rep. 6:20254. doi: 10.1038/srep20254
Murakami, T., Yamamoto, C. M., Akino, T., Tanaka, H., Fukuzawa, N., Suzuki, H., et al. (2018). Bladder cancer detection by urinary extracellular vesicle mRNA analysis. Oncotarget 9, 32810–32821. doi: 10.18632/oncotarget.25998
Nabhan, J. F., Hu, R., Oh, R. S., Cohen, S. N., and Lu, Q. (2012). Formation and release of arrestin domain-containing protein 1-mediated microvesicles (ARMMs) at plasma membrane by recruitment of TSG101 protein. Proc. Natl. Acad. Sci. U.S.A. 109, 4146–4151. doi: 10.1073/pnas.1200448109
Nguyen, D. X., Bos, P. D., and Massagué, J. (2009). Metastasis: from dissemination to organ-specific colonization. Nat. Rev. Cancer 9, 274–284. doi: 10.1038/nrc2622
Nishida, N., Yano, H., Nishida, T., Kamura, T., and Kojiro, M. (2006). Angiogenesis in cancer. Vasc. Health Risk Manag. 2, 213–219. doi: 10.2147/vhrm.2006.2.3.213
Nishida-Aoki, N., Tominaga, N., Takeshita, F., Sonoda, H., Yoshioka, Y., and Ochiya, T. (2017). Disruption of circulating extracellular vesicles as a novel therapeutic strategy against cancer metastasis. Mol. Ther. 25, 181–191. doi: 10.1016/j.ymthe.2016.10.009
Noerholm, M., Balaj, L., Limperg, T., Salehi, A., Zhu, L. D., Hochberg, F. H., et al. (2012). RNA expression patterns in serum microvesicles from patients with glioblastoma multiforme and controls. BMC Cancer 12:22. doi: 10.1186/1471-2407-12-22
Nolte’T Hoen, E. N. M., Buermans, H. P. J., Waasdorp, M., Stoorvogel, W., Wauben, M. H. M., and T Hoen, P. A. C. (2012). Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 40, 9272–9285. doi: 10.1093/nar/gks658
Ohno, S. I., Takanashi, M., Sudo, K., Ueda, S., Ishikawa, A., Matsuyama, N., et al. (2013). Systemically injected exosomes targeted to EGFR deliver antitumor microrna to breast cancer cells. Mol. Ther. 21, 185–191. doi: 10.1038/mt.2012.180
O’Neill, C. P., Gilligan, K. E., and Dwyer, R. M. (2019). Role of extracellular vesicles (EVs) in cell stress response and resistance to cancer therapy. Cancers. 11:136. doi: 10.3390/cancers11020136
Ostrowski, M., Carmo, N. B., Krumeich, S., Fanget, I., Goud, B., Benaroch, P., et al. (2010). Rab27a and Rab27b control different steps of the exosome secretion pathway Matias. Nat. Cell Biol. 12, 19–30.
Pan, B. T., and Johnstone, R. M. (1983). Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell. 33, 967–978. doi: 10.1016/0092-8674(83)90040-5
Pant, S., Hilton, H., and Burczynski, M. E. (2012). The multifaceted exosome: biogenesis, role in normal and aberrant cellular function, and frontiers for pharmacological and biomarker opportunities. Biochem. Pharmacol. 83, 1484–1494. doi: 10.1016/j.bcp.2011.12.037
Pastuzyn, E. D., Day, C. E., Kearns, R. B., Kyrke-Smith, M., Taibi, A. V., McCormick, J., et al. (2018). The neuronal gene arc encodes a repurposed retrotransposon gag protein that mediates intercellular RNA transfer. Cell 172, 275–288.e18. doi: 10.1016/j.cell.2017.12.024
Peinado, H. (2012). Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 18, 883–891. doi: 10.1038/nm.2753.Melanoma
Penn, I. (1994). Depressed immunity and the development of cancer. Cancer Detect. Prev. 18, 241–252.
Pisitkun, T., Shen, R. F., and Knepper, M. A. (2004). Identification and proteomic profiling of exosomes in human urine. Proc. Natl. Acad. Sci. U.S.A. 101, 13368–13373. doi: 10.1073/pnas.0403453101
Raposo, G., Nijman, H. W., Stoorvogel, W., Leijendekker, R., Harding, C. V., Melief, C. J. M., et al. (1996). B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 183, 1161–1172. doi: 10.1084/jem.183.3.1161
Raposo, G., and Stoorvogel, W. (2013). Extracellular vesicles: exosomes, microvesicles, and friends. J. Cell Biol. 200, 373–383. doi: 10.1083/jcb.201211138
Ratajczak, J., Miekus, K., Kucia, M., Zhang, J., Reca, R., Dvorak, P., et al. (2006). Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia 20, 847–856. doi: 10.1038/sj.leu.2404132
Ridder, K., Keller, S., Dams, M., Rupp, A. K., Schlaudraff, J., Del Turco, D., et al. (2014). Extracellular vesicle-mediated transfer of genetic information between the hematopoietic system and the brain in response to inflammation. PLoS Biol. 12:e1001874. doi: 10.1371/journal.pbio.1001874
Salomon, C., Ryan, J., Sobrevia, L., Kobayashi, M., Ashman, K., Mitchell, M., et al. (2013). Exosomal Signaling during hypoxia mediates microvascular endothelial cell migration and Vasculogenesis. PLoS One 8:e0068451. doi: 10.1371/journal.pone.0068451
Sceneay, J., Smyth, M. J., and Möller, A. (2013). The pre-metastatic niche: finding common ground. Cancer Metastasis Rev. 32, 449–464. doi: 10.1007/s10555-013-9420-1
Schorey, J. S., and Bhatnagar, S. (2008). Exosome function: from tumor immunology to pathogen biology. Traffic 9, 871–881. doi: 10.1111/j.1600-0854.2008.00734
Shen, M., and Abate-Shen, C. (2010). Molecular genetics of prostate cancer: new prospects for old challenges. Genes Dev. 24, 1967–2000. doi: 10.1101/gad.1965810
Skog, J., Wurdinger, T., Rijn, S. v., Meijer, D., Gainche, L., Sena-Esteves, M., et al. (2008). Glioblastoma microvesicles transport RNA and protein that promote tumor growth and provide diagnostic biomarkers. Nat. Cell Biol. 176, 1470–1476.
Skokos, D., Botros, H. G., Demeure, C., Morin, J., Peronet, R., Birkenmeier, G., et al. (2003). Mast cell-derived exosomes induce phenotypic and functional maturation of dendritic cells and elicit specific immune responses In Vivo. J. Immunol. 170, 3037–3045. doi: 10.4049/jimmunol.170.6.3037
Skokos, D., Le Panse, S., Villa, I., Rousselle, J.-C., Peronet, R., David, B., et al. (2001). Mast Cell-Dependent B and T Lymphocyte activation is mediated by the secretion of immunologically active exosomes. J. Immunol. 166, 868–876. doi: 10.4049/jimmunol.166.2.868
Sleeman, J. P. (2012). The metastatic niche and stromal progression. Cancer Metastasis Rev. 31, 429–440. doi: 10.1007/s10555-012-9373-9
Tannous, B. A., Kim, D. E., Fernandez, J. L., Weissleder, R., and Breakefield, X. O. (2005). Codon-optimized gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Mol. Ther. 11, 435–443. doi: 10.1016/j.ymthe.2004.10.016
Tauro, B. J., Greening, D. W., Mathias, R. A., Ji, H., Mathivanan, S., Scott, A. M., et al. (2012). Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods 56, 293–304. doi: 10.1016/j.ymeth.2012.01.002
Taylor, D. D., and Gerçel-Taylor, C. (2005). Tumour-derived exosomes and their role in cancer-associated T-cell signalling defects. Br. J. Cancer 92, 305–311. doi: 10.1038/sj.bjc.6602316
Taylor, D. D., Gerçel-Taylor, Ç, Lyons, K. S., Stanson, J., and Whiteside, T. L. (2003). T-Cell apoptosis and suppression of T-Cell Receptor/CD3-ζ by Fas ligand-containing membrane vesicles shed from ovarian Tumors. Clin. Cancer Res. 9, 5113–5119.
Tewari, M. (2015). A functional extracellular transcriptome in animals? Implications for biology, disease and medicine. Genome Biol. 16:47. doi: 10.1186/s13059-015-0613-5
Théry, C., Amigorena, S., Raposo, G., and Clayton, A. (2006). Isolation and characterization of exosomes from cell culture supernatants. Curr. Protoc. Cell Biol. Chapter 3, Unit3.22. doi: 10.1002/0471143030.cb0322s30
Théry, C., Boussac, M., Véron, P., Ricciardi-Castagnoli, P., Raposo, G., Garin, J., et al. (2001). Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J. Immunol. 166, 7309–7318. doi: 10.4049/jimmunol.166.12.7309
Théry, C., Regnault, A., Garin, J., Wolfers, J., Zitvogel, L., Ricciardi-Castagnoli, P., et al. (1999). Molecular characterization of dendritic cell-derived exosomes: selective accumulation of the heat shock protein hsc73. J. Cell Biol. 147, 599–610. doi: 10.1083/jcb.147.3.599
Théry, C., Witwer, K. W., Aikawa, E., Alcaraz, M. J., Anderson, J. D., Andriantsitohaina, R., et al. (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 7:1535750. doi: 10.1080/20013078.2018.1535750
Todorova, D., Simoncini, S., Lacroix, R., Sabatier, F., and Dignat-George, F. (2017). Extracellular vesicles in angiogenesis. Circ. Res. 120, 1658–1673. doi: 10.1161/CIRCRESAHA.117.309681
Tominaga, N., Kosaka, N., Ono, M., Katsuda, T., Yoshioka, Y., Tamura, K., et al. (2015). Brain metastatic cancer cells release microRNA-181c-containing extracellular vesicles capable of destructing blood-brain barrier. Nat. Commun. 6:6716. doi: 10.1038/ncomms7716
Trajkovic, K., Hsu, C., Chiantia, S., Rajendran, L., Wenzel, D., Wieland, F., et al. (2008). Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 319, 1244–1247. doi: 10.1126/science.1153124
Valadi, H., Ekström, K., Bossios, A., Sjöstrand, M., Lee, J. J., and Lötvall, J. O. (2007). Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659. doi: 10.1038/ncb1596
Valenti, R., Huber, V., Iero, M., Filipazzi, P., Parmiani, G., and Rivoltini, L. (2007). Tumor-released microvesicles as vehicles of immunosuppression. Cancer Res. 67, 2912–2915. doi: 10.1158/0008-5472.CAN-07-0520
Van Niel, G., D’Angelo, G., and Raposo, G. (2018). Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 19, 213–228. doi: 10.1038/nrm.2017.125
Vergauwen, G., Dhondt, B., Van Deun, J., De Smedt, E., Berx, G., Timmerman, E., et al. (2017). Confounding factors of ultrafiltration and protein analysis in extracellular vesicle research. Sci. Rep. 7:2704. doi: 10.1038/s41598-017-02599-y
Vickers, K., and Remaley, A. T. (2012). Lipid-based carriers of microRNAs and intercellular communication Kasey. Curr. Opin. Lipidol. 23, 91–97.
Villarroya-Beltri, C., Gutiérrez-Vázquez, C., Sánchez-Cabo, F., Pérez-Hernández, D., Vázquez, J., Martin-Cofreces, N., et al. (2013). Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 4:2980. doi: 10.1038/ncomms3980
Vivier, E., Ugolini, S., Blaise, D., Chabannon, C., and Brossay, L. (2017). Targeting natural killer cells and natural killer T cells in cancer. Nat. Rev. Immunol. 12, 239–252.
Webber, J., Steadman, R., Mason, M. D., Tabi, Z., and Clayton, A. (2010). Cancer exosomes trigger fibroblast to myofibroblast differentiation. Cancer Res. 70, 9621–9630. doi: 10.1158/0008-5472.CAN-10-1722
Wei, Z., Batagov, A. O., Schinelli, S., Wang, J., Wang, Y., El Fatimy, R., et al. (2017). Coding and noncoding landscape of extracellular RNA released by human glioma stem cells. Nat. Commun. 8:1145. doi: 10.1038/s41467-017-01196-x
Weigelt, B., Peterse, J. L., and Van’t Veer, L. J. (2005). Breast cancer metastasis: Markers and models. Nat. Rev. Cancer 5, 591–602. doi: 10.1038/nrc1670
Wen, S. W., Sceneay, J., Lima, L. G., Wong, C. S. F., Becker, M., Krumeich, S., et al. (2016). The biodistribution and immune suppressive effects of breast cancer-derived exosomes. Cancer Res. 76, 6816–6827. doi: 10.1158/0008-5472.CAN-16-0868
Whiteside, T. L. (2008). The tumor microenvironment and its role in promoting tumor growth. Oncogene 27, 5904–5912. doi: 10.1038/onc.2008.271
Wieckowski, E., and Whiteside, T. L. (2006). Human tumor-derived vs dendritic cell-derived exosomes have distinct biologic roles and molecular profiles. Immunol. Res. 36, 247–254.
Witwer, K. W., and Théry, C. (2019). Extracellular vesicles or exosomes? On primacy, precision, and popularity influencing a choice of nomenclature. J. Extracell. Vesicles 8:1648167. doi: 10.1080/20013078.2019.1648167
Wysocynski, M., and Ratajczak, M. Z. (2009). Lung cancer secreted microvesicles: underappreciated modulators of microenvironment in expanding tumors. Int J Cancer 125, 1595–1603.
Xu, T., Xiao, D., and Zhang, X. (2013). ECRG4 inhibits growth and invasiveness of Squamous cell carcinoma of the head and neck in vitro and in vivo. Oncol. Lett. 5, 1921–1926. doi: 10.3892/ol.2013.1298
Yáñez-Mó, M., Siljander, P. R. M., Andreu, Z., Zavec, A. B., Borràs, F. E., Buzas, E. I., et al. (2015). Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 4:27066. doi: 10.3402/jev.v4.27066
Yang, C., and Robbins, P. D. (2011). The roles of tumor-derived exosomes in cancer pathogenesis. Clin. Dev. Immunol. 2011:842849. doi: 10.1155/2011/842849
Yang, J., Wei, F., Schafer, C., and Wong, D. T. W. (2014). Detection of tumor cell-specific mRNA and protein in exosome-like microvesicles from blood and saliva. PLoS One 9:e0110641. doi: 10.1371/journal.pone.0110641
Yi, J. S., Cox, M. A., and Zajac, A. J. (2010). T-cell exhaustion: characteristics, causes and conversion. Immunology 129, 474–481. doi: 10.1111/j.1365-2567.2010.03255.x
Yokoi, A., Yoshioka, Y., Yamamoto, Y., Ishikawa, M., Ikeda, S. I., Kato, T., et al. (2017). Malignant extracellular vesicles carrying MMP1 mRNA facilitate peritoneal dissemination in ovarian cancer. Nat. Commun. 8:14470. doi: 10.1038/ncomms14470
Yu, Y., Abudula, M., Li, C., Chen, Z., Zhang, Y., and Chen, Y. (2019). Icotinib-resistant HCC827 cells produce exosomes with mRNA MET oncogenes and mediate the migration and invasion of NSCLC. Respir. Res. 20:217. doi: 10.1186/s12931-019-1202-z
Zaborowski, M. P., Lee, K., Na, Y. J., Sammarco, A., Zhang, X., Iwanicki, M., et al. (2019). Methods for systematic identification of membrane proteins for specific capture of cancer-derived extracellular vesicles. Cell Rep. 27, 255–268. doi: 10.1016/j.celrep.2019.03.003
Keywords: cancer, mRNA, extracellular vesicles, biological function, exosome, EV
Citation: Prieto-Vila M, Yoshioka Y and Ochiya T (2021) Biological Functions Driven by mRNAs Carried by Extracellular Vesicles in Cancer. Front. Cell Dev. Biol. 9:620498. doi: 10.3389/fcell.2021.620498
Received: 23 October 2020; Accepted: 30 July 2021;
Published: 30 August 2021.
Edited by:
Clive R. Bramham, University of Bergen, NorwayReviewed by:
Janusz Rak, McGill University, CanadaCopyright © 2021 Prieto-Vila, Yoshioka and Ochiya. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Takahiro Ochiya, dG9jaGl5YUB0b2t5by1tZWQuYWMuanA=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.