- Laboratory for Bio-Feed and Molecular Nutrition, College of Animal Science and Technology, Southwest University, Chongqing, China
The muscle growth and development of livestock animals is a complex, multistage process, which is regulated by many factors, especially the genes related to muscle development. In recent years, it has been reported frequently that circular RNAs (circRNAs) are involved widely in cell proliferation, cell differentiation, and body development (including muscle development). However, the research on circRNAs in muscle growth and development of livestock animals is still in its infancy. In this paper, we briefly introduce the discovery, classification, biogenesis, biological function, and degradation of circRNAs and focus on the molecular mechanism and mode of action of circRNAs as competitive endogenous RNAs in the muscle development of livestock and poultry. In addition, we also discuss the regulatory mechanism of circRNAs on muscle development in livestock in terms of transcription, translation, and mRNAs. The purpose of this article is to discuss the multiple regulatory roles of circRNAs in the process of muscle development in livestock, to provide new ideas for the development of a new co-expression regulation network, and to lay a foundation for enriching livestock breeding and improving livestock economic traits.
Introduction
Circular RNAs (circRNAs) are widely found in eukaryotic cells; they are a special type of nucleotide sequence containing conserved microRNA (miRNA) binding sites (Westholm et al., 2014). Studies show that most of the circRNAs that have been found so far are non-coding RNAs; this kind of circRNA does not have the coding function of linear RNA, but it plays a regulatory role in various life activities, including the process of muscle development in livestock animals (Das et al., 2020). Throughout the research results of the past decade, we find that circRNAs are gradually becoming an indispensable part of the gene regulatory network.
Muscle is an essential component of human and most meat animal bodies, and it plays an important role in providing exercise, maintaining posture, and generating heat (Silva et al., 2019; Janssen et al., 2000). Livestock muscle development is a crucial link to individual growth and development and an important research direction in modern animal science. The condition of muscle development directly exerts an economic effect on animals. Therefore, an in-depth understanding of muscle development is of great significance to the development of animal husbandry.
With the rapid development of molecular genetics and the development of molecular biology technology, in vitro cell line culture technology, and gene targeting technology, people have become more and more aware of cell growth and development processes at the molecular level (Li D. et al., 2020). For the past few years, scientists have repeatedly pointed out that circRNAs may be an important biomolecule to understand the mechanism of body development. At the same time, circRNAs play a unique role in regulating human and animal muscle development and its related physiological and pathological processes (Das et al., 2020). Liang et al. identify 149 circRNAs that may be related to muscle growth from 3 skeletal muscles of Guizhou miniature pigs (S. scrofa); the gene ontology (GO) and KEGG enrichment analysis of the host gene of the circRNAs indicates that these circRNAs are mainly involved in the growth and development of muscle-related signaling pathways (Liang et al., 2017a). The results show that their host genes are closely related to muscle development, chromatin modification, contraction, ATP hydrolysis-coupled proton transport, and cation homeostasis (Liang et al., 2017a). The above studies show that circRNAs are ubiquitous in muscle and play a critical role in the development process. Therefore, in-depth study of the specific mechanism of circRNAs regulating muscle development has become one of the urgent problems for researchers. Here, we focus on the molecular mechanism and mode of action of circRNA as a competitive endogenous RNA in livestock muscle development. The purpose of this study is to expand researchers’ understanding of the regulation of muscle development by circRNAs, to collect research data for further improvement of the circRNA–miRNA–mRNA (ceRNA) co-expression regulation network, and to provide theoretical support for the improvement of muscle development and economic traits of livestock.
Overview of Circrnas
The Discovery of CircRNAs
CircRNA is a kind of closed circular RNA, which can stably exist in the organism, but it does not have the 5′ terminal hat structure and the 3′ terminal poly (A) tail structure (Westholm et al., 2014). As early as the 1970s and 1980s, researchers proposed in Nature and PANs that circRNA is a kind of covalently closed circular RNA molecule discovered in plant viroids and eukaryotic cells (Sanger et al., 1976; Hsu and Coca-Prados, 1979; Kos et al., 1986). Later, Danan et al. (2012) found that some non-coding RNAs, snoRNAs, and RNase P RNAs could form circRNAs in archaea. Although these early studies clearly document the existence of circRNA molecules, their potential impact was underappreciated. With the advent of some advanced RNA sequencing techniques and methods for calculating non-polyadenylate RNA transcription, more and more circRNAs have been found in paleontological, nematode, zebrafish, mouse, and human cells (Zhang et al., 2014), and regulation of circRNA levels can lead to a variety of molecular and physiological phenotypic changes, which include effects on growth and development (Liu B. et al., 2019), the nervous system (Irie et al., 2019), innate immunity, microRNAs (Yang et al., 2017), and many disease-related pathways (Du et al., 2018). As a result, circRNA became a hot spot.
The Classification of CircRNAs
In order to carry out follow-up research in a more organized way, scholars depend on the genomic loci and the relationship with the connected parental transcript; circRNAs are categorized into five types: exonic (Danan et al., 2012; Salzman et al., 2012; Memczak et al., 2013; Li X. et al., 2019), intronic (Zhang et al., 2013; Das et al., 2019a,b), sense overlapping (Humphreys et al., 2019; Patop et al., 2019), antisense, and intergenic (Qu et al., 2015; Wang et al., 2016). Cao et al. (2018) found 886 circRNAs in the form of introns and exons in the sheep skeletal muscle circRNA library—most of which interact with muscle-specific miRNA involved in muscle growth and development, especially circ776. It is worth noting that there are few reports on other types of circRNAs regulating muscle development. Therefore, this is also a place where we need further study and breakthroughs.
The Biogenesis of CircRNAs
The difference in structure between circRNA and linear RNA means that they are formed in different ways and have different biological functions (Figure 1). Li X. et al. (2019) prove for the first time that the assembly mechanism of the “intro-definition” and “exon-definition” E complex can exist from the point of view of structural biology. On this basis, they point out that some E complexes assembled on the middle exons of yeast EFM5 or HMRA1 can be chased into circRNA, but this requires exons long enough to achieve this, and most eukaryotic circRNAs are catalyzed by classical splice bodies or group I/II ribozymes (Lasda and Parker, 2014; Chen and Yang, 2015). With reference to the existing research, we summarize and sort out the formation of circRNAs: First, the lasso structure drives cyclization, and the lasso structure is a by-product of exon hopping. After the intron in the lasso structure is removed, exons can be connected to form circRNAs. The second is intron pairing driving cyclization, and there is an intron with a reverse complementary sequence at both ends of the ring-shaped exon. The pairing mediation of the reverse complementary sequence of the intron can make the splicing donor and splice recipient of the exon spliced into a ring closer to each other in space, thus forming circRNAs (Li X. et al., 2019). Besides this, the RNA binding protein (RBP) is an important factor in regulating the production of circRNA. RBP can specifically bind to the flanking introns at both ends of RNA, acting as an RBP while narrowing the distance between the splice recipient and the donor, resulting in the formation of circRNAs (Naqvi et al., 2019; Pagliarini et al., 2020; Zhao et al., 2020). It is well-known that CDR1as, as an antisense transcript of cerebellar degeneration associated protein 1, can be used as a specific circRNA of miR-7, so it is also known as CIRS-7. However, it is surprising that ciRS-7/CDR1as biosynthesis in circRNAs is mediated by mammalian scattered repetitive elements mammalian-wide interspersed repeats (MIRs) (Yoshimoto et al., 2020). In addition, a recent study shows that m6A modification can promote the production of circRNAs carrying an open reading frame (ORF) during the development of male germ cells in mice (Tang et al., 2020). Tan et al. (2018) report that the EML4-ALK fusion gene can form F-circrEA. Later, Wu et al. (2019) found that SLC34A2-ROS1 can form F-circrSR1 and FmurcircrSR2 by the fusion gene, and both of them may be used as diagnostic markers of lung cancer. Also, some scholars find that genes that interfere with transcriptional termination contribute to the production of transcriptional read-through products and promote the production of downstream gene-derived circRNAs (Liang et al., 2017; Chen S. et al., 2019). From this point of view, the formation of circRNAs is affected by many biological factors. Therefore, the formation of circRNAs still needs in-depth study by researchers.
The Biological Characteristics of CircRNAs
Many studies show that circRNA is a structure with a missing 5′ end cap and 3′ end poly (A) tail, so it is not easy for circRNAs to be degraded by exonuclease RNAseR (Suzuki et al., 2006; Suzuki and Tsukahara, 2014; Li X. et al., 2019). Further, due to circRNAs being highly conserved in many species, such as humans, nematodes, zebrafish, and mice (Salzman et al., 2012; Li X. et al., 2019), only a small number of them can evolve and change rapidly (Zhang et al., 2013, 2014). Some studies point out that the number of circRNAs found in eukaryotic cells has been as high as more than 20,000 (Cocquerelle et al., 1993; Westholm et al., 2014); a few are formed by direct cyclization of introns, and most of them come from exons (Liu et al., 2020a). In addition, as a kind of non-coding RNA, circRNA can only regulate the formation of proteins at the transcriptional or post-transcriptional level (Westholm et al., 2014), and only a few of them can regulate life activities by encoding proteins (Pamudurti et al., 2017; Yang et al., 2018). Medical researchers believe that circRNAs have certain tissue and disease specificity and have guiding significance for the treatment of many diseases (Cocquerelle et al., 1993).
The Biological Function of CircRNAs
In the past decade, research results have shown that circRNAs can directly or indirectly participate in the process of gene expression, such as RNA translation, miRNA bait, RNA translation, protein–protein interaction, and so on (Westholm et al., 2014; Pamudurti et al., 2017; Yang et al., 2018) (Figure 1). Based on the results of previous studies, we summarize the biological functions of circRNAs: (1) CircRNAs can be used as an miRNA sponge to regulate the stability of related mRNAs or protein formation. For example, circHIPK3 acts as a sponge of miR-7 in CRC when the co-expression of miR-7 and circHIPK3 promotes the proliferation of colorectal cancer cells (Zeng et al., 2018). (2) A few circRNAs can encode functional proteins when they contain the m6A motif or IRES. For example, studies show that some circRNAs can carry longer ORFs, and the initiation codon of these ORFs is modified by m6A and binds to ribosomes to form functional proteins (Zhao et al., 2018; Tang et al., 2020). (3) CircRNAs can be spliced with linear RNA and promote the expression of linear mRNA. For example, complementary pairing of CDR1as and CDR1mRNA can enhance the stability of CDR1mRNA (Rong et al., 2017). (4) EIcircRNA or ciRNA interacts with Pol II and U1 snRNP at the promoter of the parent genes, thus promoting the transcription of the parent genes. For example, circEIF3J and circPAIP2 regulate gene expression by forming complexes with U1 snRNP and Pol II, which bind to the promoter region of the host gene (Li D. et al., 2020). (5) Some circRNAs can be used as translation templates for proteins and peptides. For example, some ribo-circRNAs use the start codon of the host mRNA and bind to membrane-related ribosomes to participate in circRNA translation (Granados-Riveron and Aquino-Jarquin, 2016; Pamudurti et al., 2017). (6) CircRNAs in some nuclei are also involved in histone modification (Burd et al., 2010; Kotake et al., 2011) and RNA maturation (Holdt et al., 2016). CircRNAs (mecciRNAs) from humans, and mouse mitochondria can enter mitochondria by interacting with TOM40 and PNPASE (Liu et al., 2020b).
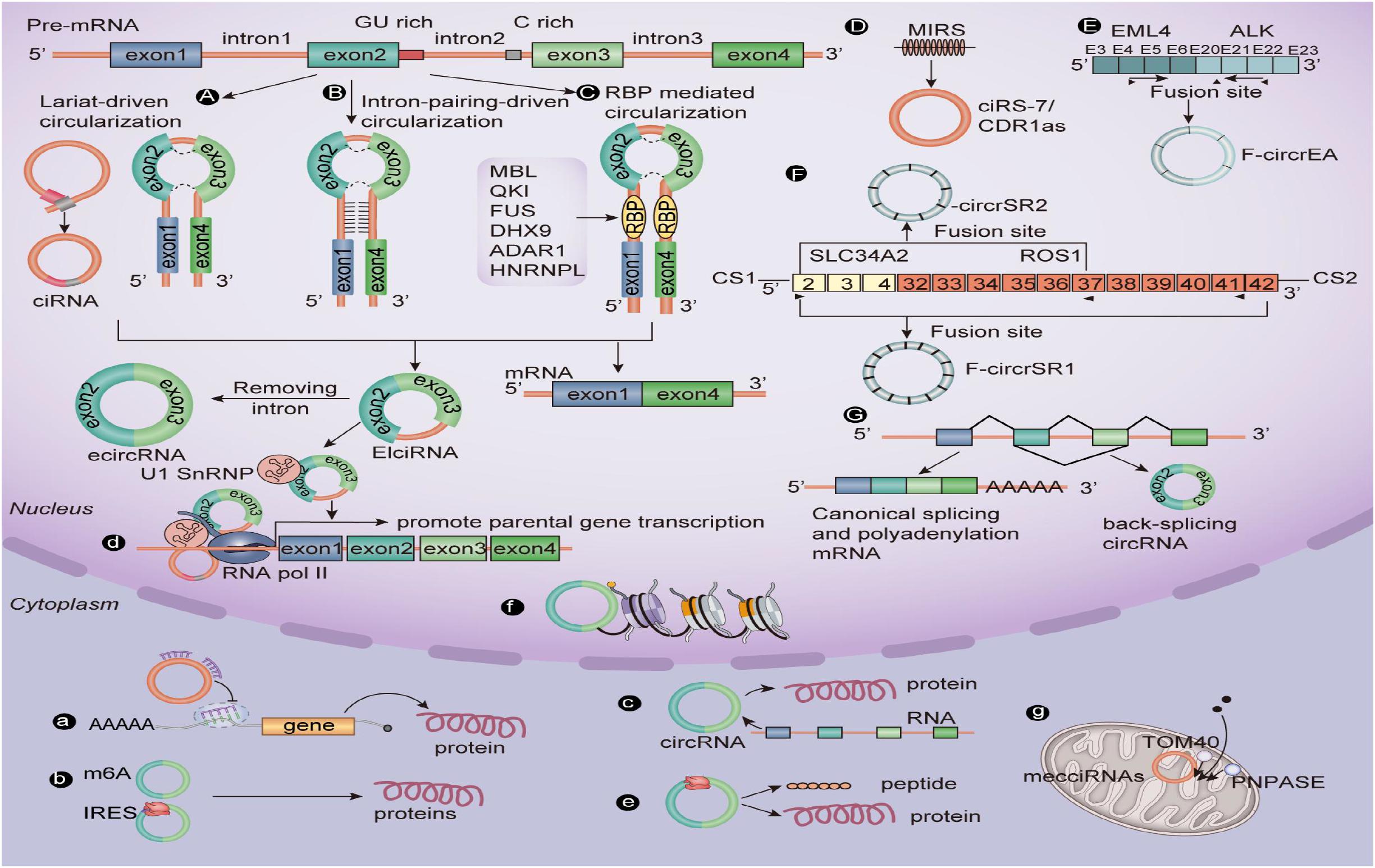
Figure 1. The biogenesis mechanisms and biological roles of circular RNAs. (A) Lariat-driven circularization. (B) Intron pairing–driven circularization. (C) RBP-mediated circularization. (D) Repetitive MIR-mediated circularization. (E) EML4-ALK fusion gene–mediated circularization. (F) SLC34A2-ROS1 fusion gene–mediated circularization. (G) Interfering with gene-mediated circularization that regulates transcription. (a) CircRNAs can sponge miRNAs. (b) A few circRNAs containing the m6A motif or IRES can encode functional proteins. (c) CircRNAs splice with linear RNA to promote the expression of mRNA. (d) EIciRNA can interact with U1 small nuclear ribonucleoproteins and then increase the transcription of their host genes by binding with RNA pol II; ciRNA, and the RNA pol II complex can directly interact and play a role in regulating parental gene transcription. (e) CircRNAs can be translated into peptides or proteins. (f) CircRNA in some nuclei is involved in histone modification. (g) Mitochondrial-derived circRNA can regulate the entry of proteins into mitochondria under the interaction of TOM40 and PNPASE. CiRNA, Circular intronic RNA; EcircRNA, Exonic circRNA; EIciRNA, Exon–intron circRNA; RBP, RNA-binding protein; RNA pol II, RNA polymerase II.
The Degradation of CircRNAs
CircRNAs maintain their cellular homeostasis by highly dynamic and tightly regulated biogenesis and degradation, thereby exerting proper biological functions. Compared with the biogenetic mechanism of circRNA biogenesis, the specific pathway by which cells eventually degrade circRNAs is still in need of continued study. Some studies show that miRNAs may initiate the degradation of circRNAs through Ago2-mediated cleavage. For instance, as the target of CDR1as/ciRS-7, miR-671 can perfectly load Ago2 into CDR1as/ciRS-7, which leads to the cleavage of Ago2 in the nucleus and the subsequent dissolution of RNA in the outer nucleus. However, it is not known whether other miRNAs can regulate the degradation of circRNAs by perfectly matching them with Ago2 (Hansen et al., 2011). A recent study has shown that m6A can mediate circRNA degradation. Park et al. (2019) point out that, when m6A carrying circRNA is used as a marker, m6A read–write protein YTHDF2 and linker protein HRSP12 can be recruited. As a bridge between YTHDF2 and endoribonuclease RNase P/MRP, HRSP12 binds directly to the GGUUC motif on circRNAs, which leads to the initiation of RNase P/MRP and the gradual degradation of circRNAs. In addition, Fischer et al. (2020) find a degradation mechanism of RNA (including circRNAs). He also points out the specific mechanism of selective degradation of high-structure RNA under normal conditions. High overall structure circRNA decay is regulated globally by two RNA binding proteins, UPF1 and G3BP1 (Chen et al., 2020). Because this pathway perceives the whole RNA structure rather than a linear first-order sequence, it is called structure-mediated RNA decay (SRD). Because mammalian RNA decay pathways are widely linked to translation (Tuck et al., 2020), it is also worth exploring whether circRNAs targeted by SRD have potential encoding peptides. Furthermore, Liu C. X. et al. (2019) discovered an endonuclease RNaseL that can degrade circRNAs in a full range, which seems to increase researchers’ understanding of the mechanism of circRNA degradation.
General Situation of Muscle Development of Livestock Animals
Muscle development is a very complex biological process, which mainly depends on the proliferation and hypertrophy of muscle fiber cells (Molkentin and Olson, 1996). Studies show that the number of muscle fibers increases only before birth and does not change much after birth, and the growth of muscle fibers depends on the hypertrophy of muscle fibers (Christ and Brand, 2004). The hypertrophy of muscle fibers includes two aspects: One is the increase of myofibrils, and the other is the increase in the number of nuclei in muscle fibers (Berger et al., 2015). Muscle development is a multistep process regulated by multiple genes, and it is not only regulated by a variety of myogenic regulatory factors, including myogenin (MyoG), myogenic regulatory factor-4 (MRF-4), myosin heavy chain (MyhC), myoblast regulatory factor family myogenic factor-5 (Myf-5), myostatin (MSTN), myocyte enhancer factor2A-D (MEF2A-2D), myogenic differentiation antigen (MyoD), and paired box (Pax) family members Pax 3 and Pax 7 (Chen et al., 2020), but it is also regulated by other related genes, such as insulin-like growth factor-I (IGF-I), insulin-like growth factor-II (IGF-II), forkhead box transcription factor O1 (FoxO1), and mammalian target of rapamycin (mTOR) (Wan et al., 2016). Furthermore, studies confirm that the genes that affect muscle development have two regulatory effects: One is positive promotion; the other is reverse inhibition (McPherron and Lee, 2002; Chen P. R., 2019; Chen et al., 2020). These growth factors play a unique role in regulating muscle development and can regulate cell proliferation, apoptosis, sarcomere activation, and muscle-specific genes at many sites in the muscle (Doynova et al., 2017; Hernández-Hernández et al., 2017) pedigree.
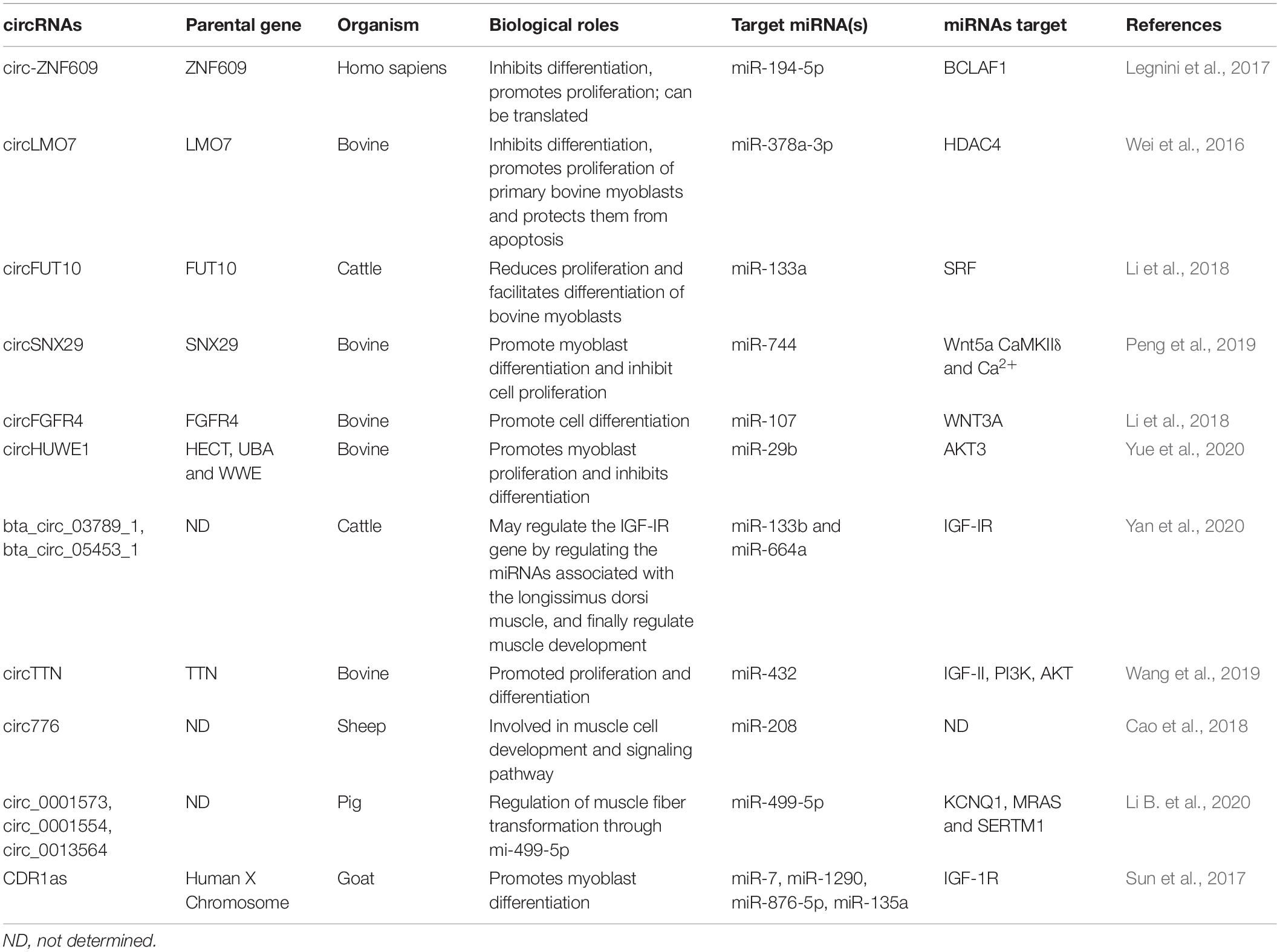
Exploring the regulatory mechanism of muscle cell proliferation and differentiation is one of the research hot spots in developmental biology in recent years. As an important economic trait, the muscle development of livestock has been paid more and more attention by researchers. CircRNAs have been widely studied in the related fields of human medicine and bioinformatics, which provides a new idea for exploring the construction of regulatory networks of circRNAs in animal husbandry. As was mentioned earlier, muscle development is a complex biological process that is affected by many factors. Although researchers have conducted extensive and in-depth studies on circRNAs, some researchers noticed that circRNAs can further guide the process of muscle development by binding to miRNAs or regulating the expression of genes related to muscle development at the transcriptional level (Zhang et al., 2019). Based on previous studies, we summarize the unique role of circRNAs in muscle development (as shown in the Table 1 and Figure 2).
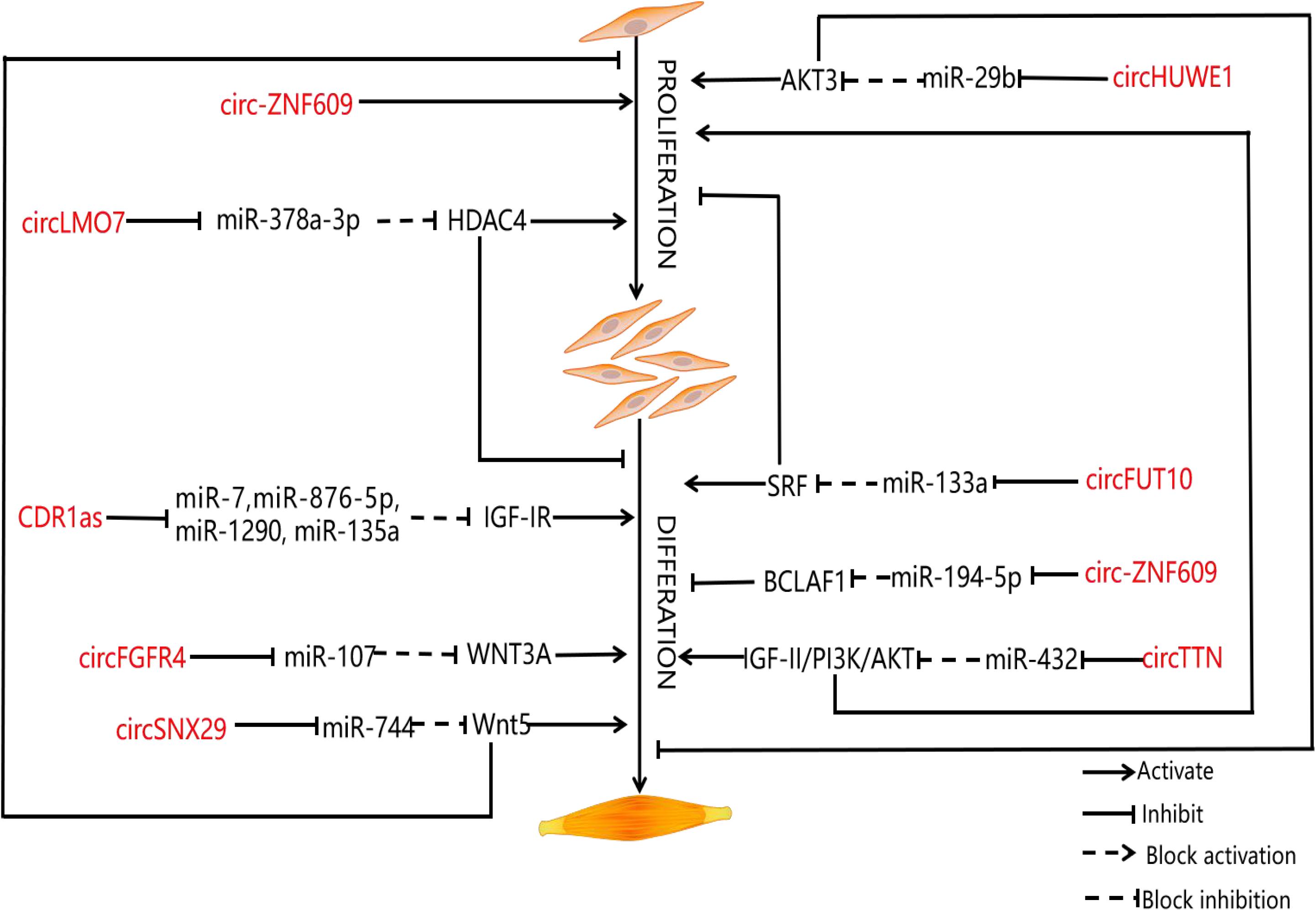
Figure 2. CircRNAs involved in muscle development and their function. Schematic representation of the role of circRNAs in muscle proliferation and differentiation. The diagram shows various circRNAs that regulate the proliferation and differentiation of myoblasts. The circRNAs are represented in red.
It is reported that the number of circRNAs in animal muscles and muscle cells ranges from 2,000 to 37,000 (Abdelmohsen et al., 2015; Li et al., 2017; Liang et al., 2017b; Wei et al., 2017; Ouyang et al., 2018; Zhang et al., 2018). Li et al. (2017) first used RNA sequencing to detect 6,113 circRNAs from the longissimus dorsi muscle of sheep. The researchers found 12,000 circRNA expressions during muscle aging in monkeys by RNA sequencing (Abdelmohsen et al., 2015). The study found that circRNAs regulated the growth and development of porcine skeletal muscle and the transformation of muscle fiber types at the age of 0–30 days; at the age of 30–240 days, circRNAs regulated the glucose metabolism and calcium signal of porcine skeletal muscle (Liang et al., 2017b). Many reports point out that circRNAs play a unique and irreplaceable role in guiding animal muscle development (Abdelmohsen et al., 2015; Li et al., 2017; Liang et al., 2017b; Wei et al., 2017; Ouyang et al., 2018; Zhang et al., 2018). Liang et al. (2017b) constructed the first miniature pig circRNA database and point out that ssc-ciR-02753, ssc-ciR-03065, ssc-ciR-03066, ssc-ciR-03069, ssc-ciR-04335, ssc-ciR-04348, ssc-ciR-04349, ssc-ciR-04353, and ssc-ciR-04359, can regulate porcine muscle development by affecting cell proliferation and fusion. It is reported that many circRNAs contain binding sites, such as miR-1, miR-133, miR-206, miR-29, miR-378, miR-431 (Ebbesen et al., 2017), miR-7 (Li L. et al., 2019), miR-135a (Greco et al., 2009), miR-1290 (Ng et al., 2015), and miR-876-5p (Cook et al., 2015). Interestingly, these miRNAs are involved in cell proliferation, differentiation, and signal transduction during myogenesis. It lays a theoretical foundation for the construction of a co-expression network of circRNAs regulating the muscle development of livestock. Combined with human medical research, it is not difficult to see that circRNAs and miRNAs can bind and regulate the expression of downstream mRNAs (that is, the ceRNA co-expression regulatory network) in a certain way. A lot of evidence also shows that the muscle development of livestock is indeed regulated by both circRNAs and miRNAs (Abdelmohsen et al., 2015; Li et al., 2017; Liang et al., 2017b; Wei et al., 2017; Ouyang et al., 2018; Zhang et al., 2018). In addition, a very small number of circRNAs can regulate muscle development in livestock at the transcriptional and translational levels (Shen et al., 2019). However, there is almost no relevant research in this field at present. Therefore, it is necessary for us to pay close attention to the related regulatory mechanisms of circRNAs on muscle development in different ways.
Circrnas Regulate Muscle Development of Livestock Animals
CeRNA Co-expression Network Regulates Muscle Development of Livestock Animals
Muscle development of livestock is an important economic character in the development of animal husbandry. Despite the controversy surrounding the ceRNA hypothesis, large amounts of experimental results show that circRNAs can regulate the expression of mRNAs related to the muscle development of livestock through specific miRNAs (Qian et al., 2017). Therefore, researchers have evaluated multiple co-regulatory relationships during muscle development. Among them, Yue et al. (2020) verify the interaction among circHUWE1, miR-29b, and AKT3 with the help of bioinformatics, double luciferase report analysis, and AGO2-RNA immunoprecipitation (RIP) and point out that circHUWE1 can directly interfere with the ability of miR-29b to release AKT3 inhibition and finally activate the AKT signal pathway, thus promoting the proliferation of bovine myoblasts and inhibiting apoptosis and differentiation of bovine myoblasts. For this reason, Yue et al. (2020) build upon previous research results to construct a ceRNA co-expression regulatory network, which not only provides a good idea for the study of muscle development, but also expands our understanding of the function of circRNAs. KEGG pathway analysis shows that hosting genes of circRNAs are related to the muscle development pathway, including the mammalian target of rapamycin signaling pathway, Wnt signaling pathway, MAPK, and transforming growth factor-β signaling pathway (Li et al., 2017). Yan et al. (2020) identify a total of 5,177 circRNAs in the longissimus dorsi samples of Kazakh cattle and Xinjiang brown cattle by establishing a RNA sequence library; of these, 46 are differentially expressed. The identification of differentially expressed genes shows that the process of muscle development is related to differentially expressed circRNAs. In addition, miRanda predictions show that there are 66 interactions between 65 circRNAs and 14 miRNAs. For this reason, Yan et al. (2020) also establish a co-expression network. Further, some miRNAs known to be involved in myoblast regulation, such as miR-133b and miR-664a, are identified. Sun et al. (2017) study the differentially expressed coding genes, lncRNAs, circRNAs, and miRNAs, in muscle tissue of Lantang and Landrace pigs. The results show that there are 1,401 high expressions of circRNAs and 2,959 low expressions of circRNAs in Lantang pig, of which 236 circRNAs are closely related to muscle development, and 40 circRNAs regulate muscle development by participating in a miRNA-mediated ceRNA regulatory network. There are 6,113 differentially expressed circRNAs in the longissimus dorsi of Kazakh sheep at both embryonic and adult stages, and the maternal genes of circRNAs are mainly enriched in the signal pathways related to muscle growth and development. Among them, oar_circ 0000385, oar_circ_0000582, and oar_circ_0001099 have multiple binding sites on miRNAs (miR-143, miR-133, and miR-23) related to muscle development (Li et al., 2017). Wei et al. (2017) detect the circRNA expression profile of bovine skeletal muscle at two developmental periods (embryonic and adult longissimus) for the first time and point out that the overexpression of circLMO7 can competitively absorb miR-378a-3p when the expression of miR-378a-3p is downregulated; the target gene hdac4 is activated, thus promoting muscle cell proliferation and inhibiting muscle cell differentiation. After that, Li et al. (2018) find that circFGFR4 and miR-107 are highly expressed in Qinchuan cattle at the embryonic stage (90 days) and adulthood (24 months old) longissimus dorsi, and circFGFR4 could adsorb miR-107. MiR-107 can weaken the expression of Wnt3a by binding to overexpressed circFGFR4. It can be seen that Wnt3a, as the target of miR-107, plays an important role in inhibiting myotube formation and protecting myoblast apoptosis. To sum up, circFGFR4 can be used as an miR-107 sponge to eliminate the inhibitory effect of miR-107 on the expression of Wnt3a and the differentiation of bovine primary myoblasts. The differential expression of circTitin (circTTN) in bovine skeletal muscle between fetal and adult bovine muscle tissue and the overexpression and inhibition of circTTN induced its promoting effect on the proliferation and differentiation of bovine primary myoblasts because miR-432, the target gene of circTTN, is the regulator of IGF-II. Wang et al. (2019) point out that circTTN can activate the IGF-II/phosphatidylinositol 3-kinase (PI3K)/AKT signal pathway through competitive binding with miR-432, which promotes the proliferation and differentiation of bovine myoblasts. Li et al. (2018) find that circFUT10 in adult bovine muscle can reduce its inhibition on target genes by competitive binding to miR-133a, and the expression of MyHC, MyoD, and MyoG related to muscle development changed synchronously with that of circFUT10 at the mRNA and protein levels. Overexpression of circ-FUT10 can promote MyHC expression, induce myoblast apoptosis, and promote myoblast differentiation. On the contrary, circZfp609 binding to miR-194-5p inhibits the expression of bcl2-related transcription factor 1 (BCLAF1), which also affects the expression of Myf5 and MyoG, and inhibits myoblast differentiation (Li et al., 2018). CircSNX29 exists widely in bovine primary myoblasts, but its expression level in embryonic skeletal muscle is significantly higher than that in adult skeletal muscle. Peng et al. (2019) find that overexpression of circSNX29 promotes myoblast differentiation and inhibits cell proliferation while interfering with circSNX29 inhibits myoblast differentiation and promotes proliferation. Subsequently, using RNAhybrid for bioinformatics prediction, it was found that circSNX29 may adsorb to miR-744 with 9 potential binding sites. Using a double luciferase report assay, the results show that circSNX29 could directly bind to miR-744 competitively and effectively reverse the inhibitory effect of miR-744 on Wnt5a and CaMKIIδ. Importantly, through KEGG pathway enrichment analysis, Western blotting, a calcium fluorescence probe, and CamKII activity detection, it is found that overexpression of Wnt5a and circSNX29 activate the non-classical Wnt/Ca2+ pathway by increasing the activity of CamKII kinase and the phosphorylation level of PKC and then regulate the proliferation and differentiation of bovine myoblasts. These results are helpful to further understand the role of circRNAs and miRNAs in myogenesis. The study further shows that muscle development is more efficient in embryo than in adulthood. Interestingly, Von et al. point out that Wnt5a and CaMKIId are the targets of miR-744, and the expression of miR-744 leads to the activation of the atypical Wnt pathway by inhibiting the expression of Wnt5a and CaMKIId (von Maltzahn et al., 2012). From this, we can see that circSNX29 acts as an miR-744 sponge to upregulate the expression of CaMKIId and Wnt5a through activating the Wnt pathway and promotes myoblast differentiation. In addition, it is reported that circRNA9210-miR-23a-MEF2C and circRNA290-miR27b-Foxj3 networks play a unique role in regulating the conversion of muscle fiber types in porcine skeletal muscle (Shen et al., 2019). Li et al. (2017) find a total of 5,086 differentially expressed circRNAs in the RNA sequences of sheep adult longissimus dorsi (LDM-A) and longissimus dorsi (LDM-E) of which 2,146 are downregulated and 2,940 are upregulated. The results of real-time quantitative PCR show that the expression of circRNA 0000552, circRNA 00002456, circRNA 00004666, circRNA00004676, and circRNA 00004690 in LDM-E is relatively higher than that of LDM-A. The expression of circRNA 0003451, circRNA 0005243, circRNA 0005250, and circRNA 0005256 in LDM-A is relatively higher than that in LDM-E. Thus, the differential expression of circRNAs in sheep muscle is proved. Cao et al. (2018) extracted 75.5 million sequences from the sheep skeletal muscle RNA gene bank. These sequences were mapped to 729 genes in the reference genome of sheep, containing a total of 886 circRNAs. Reverse transcription PCR and DNA sequencing analysis confirm the existence of many kinds of circRNAs and the resistance of sheep circRNAs to RNase R digestion. Finally, Cao et al. (2018) first used RNA-seq to study circRNAs in the longus dorsi muscle of sheep before and after delivery. A total of 6113 circRNAs were detected, of which some circRNAs (circRNA100, circRNA108, circRNA205, circRNA606, circRNA678, circRNA744, and circRNA776) contained at least two conservative targets of miRNAs related to muscle development (miR-29b, miR-133, miR-208, and miR-499, respectively). Thus, it can be seen that most circRNAs interact with muscle-specific miRNAs and then jointly regulate the process of muscle development. The results of GO and KEGG enrichment analysis show that the host gene of circRNAs is involved in muscle cell development and signal transduction (Cao et al., 2018). For this reason, Cao et al. (2018) establish a relatively complete ceRNA network that contains a large number of potential functional circRNA (circRNA 0000385, circRNA 0001099, and circRNA 0000582) and its predicted miRNA targets and downstream regulatory genes. At present, these regulatory networks are an important source of ideas for us to study muscle development. This information may help us to further explore the unique role of circRNAs in muscle development.
As we all know, circRNA CDR1as (CDR1as or CiRS-7) is an antisense transcript of cerebellar degeneration associated protein 1, but in fact, CDR1as is also considered to be related to miRNAs related to muscle development (Geng et al., 2016; Sang et al., 2018). Although CDR1as was identified as the specific circular RNA of miR-7, other miRNAs, such as miR-135a, miR-876-5p, and miR-1290, are also shown to be CDR1 response elements (Geng et al., 2016; Sang et al., 2018). It is worth noting that CDR1as-responsive miRNA and its targeted muscle-derived genes, such as IGF-IR, N-cadherin, ABCG2, WNT5A, EGFR, FAK, and CCNE1, play a key role in normal conditions and muscle diseases, such as DM1, FSHD, and IIM, which makes CDR1 potentially an important regulatory factor in muscle (Geng et al., 2016; Sang et al., 2018; Kyei et al., 2020). First of all, Li L. et al. (2019) discovered that MyoD promotes CDR1as by binding on the CDR1as 5′ flank region; however, the overexpression or knockout of CDR1as can significantly induce or hinder the process of muscle differentiation. Second, CDR1as can reduce the downregulation of IGF-IR induced by miR-7 through competitive binding to miR-7, thus activating muscle differentiation in goat metaphase. The above results further indicate that CDR1as plays an irreplaceable role in the regulation of muscle development. At the same time, these potential CDR1as/miRNAs/mRNA regulatory networks provide a basis for further study of the function of CDR1as in muscle development and other life activities and processes.
In general, these results show that circRNA is a key factor that cannot be ignored in the process of muscle development; it can compete for endogenous miRNAs to form a circRNA–miRNA complex and further relieve the inhibitory effect of miRNAs on mRNA. However, the interaction between circRNAs and endogenous miRNAs needs to be further verified because, in some special cases, the number of miRNA sites that bind to a specific circRNA is limited, and these sites are specific in different species or tissues, so the ceRNA regulatory network is not the whole content of muscle development.
CircRNA Regulates Muscle Development in Livestock Animals at the Transcriptional Level
Because there is no biological original to guide coding protein in the special structure of circRNA, circRNA is considered to be a non-coding RNA in most cases. However, recently, researchers are increasingly finding that a very small number of circRNAs have the ability to encode proteins (Li and Lytton, 1999; Legnini et al., 2017; Yang et al., 2018; Liang et al., 2019). Shen et al. (2019) point out that circRNA41, circRNA69, and circRNA153 differentially expressed in porcine skeletal muscle during oxidation and glycolysis are transcribed from MyH1 (encoding MyHC-2X protein), MyH7 (encoding MyHC-β protein), and MyH2 (encoding MyHC-2A protein) genes, respectively. These results indicate that circRNAs play a unique role in regulating the heterogeneity of muscle fiber types, but at present, the research in this area has not been reported. In addition, previous studies point out that the protein encoded by circRNAs depends on the ORF on the sequence. However, the circRNAs that actually have the function of coding protein should have many necessary conditions at the same time, such as the internal ribosome entry sites (IRES), the biological element activated by translation, and the biological element for detecting protein products (Wang and Wang, 2015; Legnini et al., 2017; Pamudurti et al., 2017). A typical example is circRNA ZNF609, which is one of the earliest endogenous circRNAs that can be translated into a protein driven by RES and regulating myogenesis. Circ-ZNF609 itself does not possess the factors required for cap structure and polyadenosine transcription translation, but it can be involved in muscle cell development by initiating translation under cis-acting elements through a mechanism independent of the cap structure in response to different cellular stresses (Legnini et al., 2017). In addition, a recent study finds that circSamd4 is related to Pura and PURB during muscle development; as myogenic inhibitors, Pura and PURB can inhibit the transcription of the myosin heavy chain (MHC) protein family. Silencing CircSamd4 enhances the binding of PUR protein to the MHC promoter, and overexpression of circSamd4 interfers with the binding of pur protein to the MHC promoter, indicating that circSamd4 could bind to PUR protein and prevent it from interacting with DNA. When using mutant circSamd4 without a PUR binding site, these effects were canceled. In other words, the binding of PUR protein to circSamd4 can promote muscle development by reducing MHC transcription (Pandey et al., 2020). Song et al. (2020) identify 197 differentially expressed circRNAs in the gastrocnemius of Duchenne muscular dystrophy (DMD) mice and predicted their protein coding ability according to the Nmurine 6-methyladenosine motif and ORF of circRNAs. Among them, 189 circRNAs were predicted to have protein coding potential, and 98 circRNAs may be translated into peptides containing 150 or more amino acids, indicating that circRNAs may play a key role in the pathophysiological mechanism of DMD. CircFAM188B contains an ORF that can be translated as circFAM188B-103aa during the skeletal muscle development of broilers to promote the proliferation of chicken SMSC (Yin et al., 2020). To sum up, although researchers have gradually deepened their understanding of circRNAs, there are still few studies on circRNAs regulating muscle development by encoding proteins.
CircRNAs Directly Regulate Muscle Development Through mRNA
As we mentioned earlier, muscle development in livestock is a complex physiological process, and different kinds of circRNAs play different roles in muscle development. Ling et al. (2020) find that cluster16 circRNAs are highly expressed in the early and late stages of muscle development of Anhui white goat (AWG) embryo and are directly involved in the Wnt signal pathway, AMPK signal pathway, and so on. It can be seen that circRNAs can directly regulate the muscle development of livestock. In addition, circQKI (as well as QKI mRNA) depletion is demonstrated to have a negative effect on myoblast differentiation, indicating that both the circRNA and its linear counterpart cooperate in this process. By contrast, although BNC2 mRNA depletion causes an increase in myotube formation, knockdown of its circular counterpart has no effect on differentiation. Interestingly, circBNC2 expression during myoblast differentiation increases at the expense of the corresponding mRNA, suggesting that circBNC2 could contrast the expression of the anti-differentiative BNC2 mRNA (Legnini et al., 2017). Similarly, circEch1 is the most different circRNA in buffalo and beef muscle in vitro. The results of in vitro experiments show that the overexpression of circEch1 inhibits the proliferation of bovine myoblasts but promotes differentiation; in vivo tests show that circEch1 stimulates skeletal muscle regeneration. In general, circEch1 induces myoblast differentiation and skeletal muscle regeneration (Huang et al., 2021). Although there are few studies in this area at present, we believe that with the development of research technology, the relationship between circRNAs and mRNA related to livestock muscle development will become more and more clear.
Concluding Remarks and Perspectives
CircRNAs are a new regulator of muscle development. However, at present, the functional annotation of circRNAs is mainly to predict and analyze source coding genes or possible miRNA binding sites. In this review, we further discuss the relationship between circRNAs and muscle development of livestock by reviewing the discovery, classification, formation, characteristics, biological function, and degradation pathway of circRNAs. It is obvious that most studies focus on circRNAs guiding muscle development through the ceRNA co-expression regulation network. In addition, some studies confirm that circRNAs can regulate muscle development in livestock in transcription and translation, and only a few studies show that circRNAs can directly regulate muscle development in livestock. There is no doubt that our point of view plays a directional role in the study of muscle development in domestic animals; at the same time, we believe that there are other ways for circRNAs to regulate muscle development in livestock.
Up to now, the study of intracellular circRNAs made people further realize the complexity of eukaryotic gene expression regulation. In-depth study of the formation and types of circRNAs and the mechanism of action with target genes and exploring its biological function is of great significance for understanding the growth and development of organisms and disease treatment. Due to the variety of circRNAs, the diversity of action modes, and the constraints of research methods, people still need some time to clarify these genes and their regulatory mechanisms. However, as a kind of non-coding RNA discovered in the post-genome era, it effectively enriches the research model of gene expression regulation. We think that, with the rapid development of modern molecular biology technology, new generation sequencing technology, and bioinformation analysis technology, more and more new circRNAs will be discovered in the future, and people will study its function more and more deeply.
Author Contributions
ZY and TH: conceptualization and writing-original draft and supervising, reviewing, and editing. ZY, TH, and QC: editing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National key research and development plan of China (Grant No. 2018YFD0501004), the National Natural Science Foundation of China (Grant No. 31902167), the Chongqing Natural Science Foundation (Basic Research and Frontier Exploration Special Project) General Project (Grant No. cstc2019jcyj-msxmX0524), the National Science Foundation for Post-doctoral Scientists of China (Grant No. 2018M640895), the Special Funding for Postdoctoral Research Projects in Chongqing (Grant No. XMT 2081061), and Excellent Youth Exchange Program of China Association for Science and Technology in 2019 [2019(293)-98].
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are very grateful to Prof. Guozhong Dong of Southwest University and Prof. De Wu of Sichuan Agricultural University for their meticulous guidance in the process of writing the article.
Abbreviations
ceRNA, circRNAs-miRNAs-mRNAs; circRNAs, circular RNAs; FoxO1, Forkhead box transcription Factor O1; IGF-I, Insulin-like growth factor-I; IGF-II, Insulin-like growth factor-II; MEF2A-2D, myocyte enhancer factor2A-D; MIRs, Mammalian-Wide Interspersed Repeats; miRNAs, microRNAs; MRF-4, myogenic regulatory factor-4; MSTN, Myostatin; mTOR, Mammalian target of rapamycinMyf-5: myoblast regulatory factor family myogenic factor-5; Myhc, myosin heavy chain; MyoD, myogenic differentiation antigen; MyoG, myogenin; Pax, paired box; RBP, RNA binding protein; RNA-Seq, RNA sequencing.
References
Abdelmohsen, K., Panda, A. C., De, S., Grammatikakis, I., Kim, J., Ding, J., et al. (2015). Circular RNAs in monkey muscle: age-dependent changes. Aging (Albany N.Y.) 7, 903–910. doi: 10.18632/aging.100834
Berger, J., Hall, T. E., and Currie, P. D. (2015). Novel transgenic lines to label sarcolemma and myofibrils of the musculature. Zebrafish 12, 124–125. doi: 10.1089/zeb.2014.1065
Burd, C. E., Jeck, W. R., Liu, Y., Sanoff, H. K., Wang, Z., and Sharpless, N. E. (2010). Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. Plos Genet. 6:e1001233–e1001247. doi: 10.1371/journal.pgen.1001233
Cao, Y., You, S., Yao, Y., Liu, Z. J., Hazi, W., Li, C. Y., et al. (2018). Expression profiles of circular RNAs in sheep skeletal muscle. Asian Austr. J. Anim. 31, 1550–1557.
Chen, B., You, W., Wang, Y., and Shan, T. (2020). The regulatory role of Myomaker and Myomixer-Myomerger-Minion in muscle development and regeneration. Cell. Mol. Life Sci. 77, 1551–1569. doi: 10.1007/s00018-019-03341-9
Chen, L. L., and Yang, L. (2015). Regulation of circRNA biogenesis. RNA Biol. 12, 381–388. doi: 10.1080/15476286.2015.1020271
Chen, P. R., Suh, Y., Shin, S., Woodfint, R. M., Hwang, S., and Lee, K. (2019). Exogenous expression of an alternative splicing variant of Myostatin prompts leg muscle fiber hyperplasia in Japanese quail. Int. J. Mol. Sci. 20, 4617–4633. doi: 10.3390/ijms20184617
Chen, S., Huang, V., Xu, X., Livingstone, J., Soares, F., Jeon, J., et al. (2019). Widespread and functional RNA circularization in localized prostate cancer. Cell 176, 831–843. doi: 10.1016/j.cell.2019.01.025
Christ, B., and Brand, S. B. (2004). Limb muscle development. Int. J. Dev. Biol. 46, 905–914. doi: 10.1079/IVP2001258
Cocquerelle, C., Mascrez, B., Hétuin, D., and Bailleul, B. (1993). Mis-splicing yields circular RNA molecules. FASEB J. 7, 155–160. doi: 10.1096/fasebj.7.1.7678559
Cook, J. R., MacIntyre, D. A., Samara, E., Kim, S. H., Singh, N., Johnson, M. R., et al. (2015). Exogenous oxytocin modulates human myometrial microRNAs. Am. J. Obstet. Gynecol. 213, 65–65. doi: 10.1016/j.ajog.2015.03.015
Danan, M., Schwartz, S., Edelheit, S., and Sorek, R. (2012). Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res. 40, 3131–3142. doi: 10.1093/nar/gkr1009
Das, A., Das, A., Das, D., Abdelmohsen, K., and Panda, A. C. (2020). Circular RNAs in myogenesis. BBA Gene Regul. Mech. 1863, 194372–194378. doi: 10.1016/j.bbagrm.2019.02.011
Das, A., Rout, P. K., Gorospe, M., and Panda, A. C. (2019a). Past, present, and future of circRNAs. EMBO J. 38, e100836–e100848. doi: 10.3390/ijms20163988
Das, A., Rout, P. K., Gorospe, M., and Panda, A. C. (2019b). Rolling circle cDNA synthesis uncovers circular RNA splice variants. Int. J. Mol. Sci. 20, 3988–3999.
Doynova, M. D., Markworth, J. F., Cameron-Smith, D., Vickers, M. H., and O’Sullivan, J. M. (2017). Linkages between changes in the 3D organization of the genome and transcription during myotube differentiation in vitro. Skelet. Muscle 7, 5–14. doi: 10.1186/s13395-017-0122-1
Du, W. W., Yang, W., Li, X., Awan, F. M., Yang, Z., Fang, L., et al. (2018). A circular RNA circ-DNMT1 enhances breast cancer progression by activating autophagy. Oncogene 37, 5829–5842. doi: 10.1038/s41388-018-0369-y
Ebbesen, K. K., Hansen, T. B., and Kjems, J. (2017). Insights into circular RNA biology. RNA Biol. 14, 1035–1045. doi: 10.1080/15476286.2016.1271524
Fischer, J. W., Busa, V. F., Shao, Y., and Leung, A. (2020). Structure-mediated RNA Decay by UPF1 and G3BP1. Mol. Cell 78, 70–84. doi: 10.1016/j.molcel.2020.01.021
Geng, H. H., Li, R., Su, Y. M., Xiao, J., Pan, M., Cai, X. X., et al. (2016). The circular RNA Cdr1as promotes myocardial infarction by mediating the regulation of miR-7a on its target genes expression. PLoS One 11:e0151753–e0151769. doi: 10.1371/journal.pone.0151753
Granados-Riveron, J. T., and Aquino-Jarquin, G. (2016). The complexity of the translation ability of circRNAs. BBA Gene Regul. Mech. 1859, 1245–1251. doi: 10.1016/j.bbagrm.2016.07.009
Greco, S., De Simone, M., Colussi, C., Zaccagnini, G., Fasanaro, P., Pescatori, M., et al. (2009). Common micro-RNA signature in skeletal muscle damage and regeneration induced by Duchenne muscular dystrophy and acute ischemia. FASEB J. 23, 3335–3346. doi: 10.1096/fj.08-128579
Hansen, T. B., Wiklund, E. D., Bramsen, J. B., Villadsen, S. B., Statham, A. L., Clark, S. J., et al. (2011). miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 30, 4414–4422. doi: 10.1038/emboj.2011.359
Hernández-Hernández, J. M., García-González, E. G., Brun, C. E., and Rudnicki, M. A. (2017). The myogenic regulatory factors, determinants of muscle development, cell identity and regeneration. Semin. Cell Dev. Biol. 72, 10–18. doi: 10.1016/j.semcdb.2017.11.010
Holdt, L. M., Stahringer, A., Sass, K., Pichler, G., Kulak, N. A., Wilfert, W., et al. (2016). Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 7:12429. doi: 10.1038/ncomms12429
Hsu, M. T., and Coca-Prados, M. (1979). Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 280, 339–340. doi: 10.1038/280339a0
Huang, K. W., Chen, M. J., Zhong, D. D., Luo, X., Feng, T., Song, M. M., et al. (2021). Circular RNA profifiling reveals an abundant circEch1 that promotes myogenesis and differentiation of bovine skeletal muscle. J. Agric. Food Chem. 69, 592–601. doi: 10.1021/acs.jafc.0c06400
Humphreys, D. T., Fossat, N., Demuth, M., Tam, P., and Ho, J. (2019). Ularcirc: visualization and enhanced analysis of circular RNAs via back and canonical forward splicing. Nucleic Acids Res. 47, e123–e139. doi: 10.1093/nar/gkz718
Irie, T., Shum, R., Deni, I., Hunkele, A., Le Rouzic, V., Xu, J., et al. (2019). Identification of abundant and evolutionarily conserved opioid receptor circular RNAs in the nervous system modulated by morphine. Mol. Pharmacol. 96, 247–258. doi: 10.1124/mol.118.113977
Janssen, I., Heymsfield, S. B., Wang, Z. M., and Ross, R. (2000). Skeletal muscle mass and distribution in 468 men and women aged 18–88 yr. J. Appl. Physiol. 89, 81–88. doi: 10.1152/jappl
Kos, A., Dijkema, R., Arnberg, A. C., van der Meide, P. H., and Schellekens, H. (1986). The hepatitis delta (δ) virus possesses a circular RNA. Nature 323, 558–560. doi: 10.1038/323558a0
Kotake, Y., Nakagawa, T., Kitagawa, K., Suzuki, S., Liu, N., Kitagawa, M., et al. (2011). Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15 INK4B tumor suppressor gene. Oncogene 30, 1956–1962. doi: 10.1038/onc.2010.568
Kyei, B., Li, L., Yang, L., Zhan, S., and Zhang, H. (2020). CDR1as/miRNAs-related regulatory mechanisms in muscle development and diseases. Gene 730, 144315–144322. doi: 10.1016/j.gene.2019.144315
Lasda, E., and Parker, R. (2014). Circular RNAs: diversity of form and function. RNA 20, 1829–1842. doi: 10.1261/rna.047126.114
Legnini, I., Di Timoteo, G., Rossi, F., Morlando, M., Briganti, F., Sthandier, O., et al. (2017). Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell 66, 22–37. doi: 10.1016/j.molcel.2017.02.017
Li, B., Yin, D., Li, P., Zhang, Z., Zhang, X., Li, H., et al. (2020). Profiling and functional analysis of circular RNAs in porcine fast and slow muscles. Front. Cell Dev. Biol. 8:322–324. doi: 10.3389/fcell.2020.00322
Li, C., Li, X., Yao, Y., Ma, Q., Ni, W., Zhang, X., et al. (2017). Genome-wide analysis of circular RNAs in prenatal and postnatal muscle of sheep. Oncotarget 8, 97165–97177. doi: 10.18632/oncotarget.21835
Li, D., Li, Z., Yang, Y., Zeng, X., Li, Y., Du, X., et al. (2020). Circular RNAs as biomarkers and therapeutic targets in environmental chemical exposure-related diseases. Environ. Res. 180, 108825–108833. doi: 10.1016/j.envres.2019.108825
Li, H., Wei, X., Yang, J., Dong, D., Hao, D., Huang, Y., et al. (2018a). circFGFR4 promotes differentiation of myoblasts via binding miR-107 to relieve its inhibition of Wnt3a. Mol. Ther. Nucleic Acids 11, 272–283. doi: 10.1016/j.omtn.2018.02.012
Li, H., Yang, J., Wei, X., Song, C., Dong, D., Huang, Y., et al. (2018b). CircFUT10 reduces proliferation and facilitates differentiation of myoblasts by sponging miR-133a. J. Cell Physiol. 233, 4643–4651. doi: 10.1002/jcp.26230
Li, L., Chen, Y., Nie, L., Ding, X., Zhang, X., Zhao, W., et al. (2019). MyoD-induced circular RNA CDR1as promotes myogenic differentiation of skeletal muscle satellite cells. BBA Gene Regul. Mech. 1862, 807–821. doi: 10.1016/j.bbagrm.2019.07.001
Li, X. F., and Lytton, J. (1999). A circularized sodium-calcium exchanger exon 2 transcript. J. Biol. Chem. 274, 8153–8160.
Li, X., Liu, S., Zhang, L., Issaian, A., Hill, R. C., Espinosa, S., et al. (2019). A unified mechanism for intron and exon definition and back-splicing. Nature 573, 375–380. doi: 10.1038/s41586-019-1523-6
Li, X., Xie, S., Qian, L., Cai, C., Bi, H., and Cui, W. (2020). Identification of genes related to skeletal muscle growth and development by integrated analysis of transcriptome and proteome in myostatin-edited Meishan pigs. J. Proteomics 213, 103628–103639. doi: 10.1016/j.jprot.2019.103628
Liang, D., Tatomer, D. C., Luo, Z., Wu, H., Yang, L., Chen, L. L., et al. (2017). The output of protein-coding genes shifts to circular RNAs when the pre-mRNA processing machinery is limiting. Mol. Cell 68, 940–954. doi: 10.1016/j.molcel.2017.10.034
Liang, G., Yang, Y., Niu, G., Tang, Z., and Li, K. (2017a). Circular RNA profiling reveals an abundant circLMO7 that regulates myoblasts differentiation and survival by sponging miR-378a-3p. Cell Death Dis. 8, e3153–e3165. doi: 10.1093/dnares/dsx022
Liang, G., Yang, Y., Niu, G., Tang, Z., and Li, K. (2017b). Genome-wide profiling of Sus scrofa circular RNAs across nine organs and three developmental stages. DNA Res. 24, 523–535.
Liang, W. C., Wong, C. W., Liang, P. P., Shi, M., Cao, Y., Rao, S. T., et al. (2019). Translation of the circular RNA circβ-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol. 20:84. doi: 10.1186/s13059-019-1685-4
Ling, Y., Zheng, Q., Zhu, L., Xu, L., Sui, M., Zhang, Y., et al. (2020). Trend analysis of the role of circular RNA in goat skeletal muscle development. BMC Genomics 21:220. doi: 10.1186/s12864-020-6649-2
Liu, B., Song, F., Yang, Q., Zhou, Y., Shao, C., Shen, Y., et al. (2019). Characterization of tissue-specific biomarkers with the expression of circRNAs in forensically relevant body fluids. Int. J. Legal. Med. 133, 1321–1331. doi: 10.1007/s00414-019-02027-y
Liu, C. X., Li, X., Nan, F., Jiang, S., Gao, X., Guo, S. K., et al. (2019). Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell 177, 865–880. doi: 10.1016/j.cell.2019.03.046
Liu, X., Hu, Z., Zhou, J., Tian, C., Tian, G., He, M., et al. (2020a). Interior circular RNA. RNA Biol. 17, 87–97. doi: 10.1080/15476286.2019.1669391
Liu, X., Wang, X., Li, J., Hu, S., Deng, Y., Yin, H., et al. (2020b). The identification of mecciRNAs and their roles in mitochondrial entry of proteins. Sci. China Life Sci. 63, 1429–1449. doi: 10.1007/s11427-020-1631-9
McPherron, A. C., and Lee, S. J. (2002). Suppression of body fat accumulation in myostatin-deficient mice. J. Clin. Invest. 109, 595–601. doi: 10.1172/JCI13562
Memczak, S., Jens, M., Elefsinioti, A., Torti, F., Krueger, J., Rybak, A., et al. (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338. doi: 10.1038/nature11928
Molkentin, J. D., and Olson, E. N. (1996). Defining the regulatory networks for muscle development. Curr. Opin. Genet. Dev. 6, 445–453. doi: 10.1016/s0959-437x(96)80066-9
Naqvi, A. S., Asnani, M., Black, K. L., Hayer, K. E., Taylor, D., and Thomas-Tikhonenko, A. (2019). The role of SRSF3 splicing factor in generating circular RNAs. bioRxiv [Preprint] 799700. doi: 10.1101/799700
Ng, P. C., Chan, K. Y., Leung, K. T., Tam, Y. H., Ma, T. P., Lam, H. S., et al. (2015). Comparative MiRNA expressional profiles and molecular networks in human small bowel tissues of necrotizing enterocolitis and spontaneous intestinal perforation. PLoS One 10:e0135737–e0135753. doi: 10.1371/journal.pone.0135737
Ouyang, H., Chen, X., Wang, Z., Yu, J., Jia, X., Li, Z., et al. (2018). Circular RNAs are abundant and dynamically expressed during embryonic muscle development in chickens. DNA Res. 25, 71–86. doi: 10.1093/dnares/dsx039
Pagliarini, V., Jolly, A., Bielli, P., Di Rosa, V., De la Grange, P., and Sette, C. (2020). Sam68 binds Alu-rich introns in SMN and promotes pre-mRNA circularization. Nucleic Acids Res. 48, 633–645. doi: 10.1093/nar/gkz1117
Pamudurti, N. R., Bartok, O., Jens, M., Ashwal-Fluss, R., Stottmeister, C., Ruhe, L., et al. (2017). Translation of circRNAs. Mol. Cell 66, 9–21. doi: 10.1016/j.molcel.2017.02.021
Pandey, P. R., Yang, J. H., Tsitsipatis, D., Panda, A. C., and Gorospe, M. (2020). circSamd4 represses myogenic transcriptional activity of PUR proteins. Nucleic Acids Res. 48, 3789–3805. doi: 10.1093/nar/gkaa035
Park, O. H., Ha, H., Lee, Y., Boo, S. H., Kwon, D. H., Song, H. K., et al. (2019). Endoribonucleolytic cleavage of m6A-containing RNAs by RNase P/MRP complex. Mol. Cell 74, 494–507. doi: 10.1016/j.molcel.2019.02.034
Patop, I. L., Wust, S., and Kadener, S. (2019). Past, present, and future of circRNAs. Embo. J. 38:e100836. doi: 10.15252/embj.2018100836
Peng, S., Song, C., Li, H., Cao, X., Ma, Y., Wang, X., et al. (2019). Circular RNA SNX29 sponges miR-744 to regulate proliferation and differentiation of myoblasts by activating the Wnt5a/Ca2+ signaling pathway. Mol. Ther. Nucleic Acids 16, 481–493. doi: 10.1016/j.omtn.2019.03.009
Qian, D. Y., Yan, G. B., Bai, B., Chen, Y., Zhang, S. J., Yao, Y. C., et al. (2017). Differential circRNA expression profiles during the BMP2-induced osteogenic differentiation of MC3T3-E1 cells. Biomed. Pharmacother. 90, 492–499. doi: 10.1016/j.biopha.2017.03.051
Qu, S., Yang, X., Li, X., Wang, J., Gao, Y., and Shang, R. (2015). Circular RNA: a new star of noncoding RNAs. Cancer Lett. 365, 141–148. doi: 10.1016/j.canlet.2015.06.003
Rong, D., Sun, H., Li, Z., Liu, S., Dong, C., Fu, K., et al. (2017). An emerging function of circRNA-miRNAs-mRNA axis in human diseases. Oncotarget 8, 73271–73281. doi: 10.18632/oncotarget.19154
Salzman, J., Gawad, C., Wang, P. L., Lacayo, N., and Brown, P. O. (2012). Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLos One 7:e30733–e30744. doi: 10.1371/journal.pone.0030733
Sang, M., Meng, L., Sang, Y., Liu, S., Ding, P., Ju, Y., et al. (2018). Circular RNA ciRS-7 accelerates ESCC progression through acting as a miR-876-5p sponge to enhance MAGE-A family expression. Cancer Lett. 426, 37–46. doi: 10.1016/j.canlet.2018.03.049
Sanger, H. L., Klotz, G., Riesner, D., Gross, H. J., and Kleinschmidt, A. K. (1976). Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. U.S.A. 73, 3852–3856. doi: 10.1073/pnas.73.11.3852
Shen, L., Gan, M., Tang, Q., Tang, G., Jiang, Y., Li, M., et al. (2019). Comprehensive analysis of lncRNAs and circRNAs reveals the metabolic specialization in oxidative and glycolytic skeletal muscles. Int. J. Mol. Sci. 20, 2855–2872. doi: 10.3390/ijms20122855
Silva, W. J., Graça, F. A., Cruz, A., Silvestre, J. G., Labeit, S., Miyabara, E. H., et al. (2019). miR-29c improves skeletal muscle mass and function throughout myocyte proliferation and differentiation and by repressing atrophy-related genes. Acta Physlol. 226, e13278–e13296. doi: 10.1111/apha.13278
Song, Z. B., Liu, Y. M., Fang, X. B., Xie, M. S., Ma, Z. Y., Zhong, Z. G., et al. (2020). Comprehensive analysis of the expression profile of circRNAs and their predicted protein-coding ability in the muscle of mdx mice. Funct. Integer. Genomic 20, 397–407. doi: 10.1007/s10142-019-00724-w
Sun, J., Xie, M., Huang, Z., Li, H., Chen, T., Sun, R., et al. (2017). Integrated analysis of non-coding RNA and mRNA expression profiles of 2 pig breeds differing in muscle traits. J. Anim. Sci. 95, 1092–1103. doi: 10.2527/jas.2016.0867
Suzuki, H., and Tsukahara, T. (2014). A view of pre-mRNA splicing from RNase R resistant RNAs. Int. J. Mol. Sci. 15, 9331–9342. doi: 10.3390/ijms15069331
Suzuki, H., Zuo, Y., Wang, J., Zhang, M. Q., Malhotra, A., and Mayeda, A. (2006). Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 34, e63–e69. doi: 10.1093/nar/gkl151
Tan, S., Gou, Q., Pu, W., Guo, C., Yang, Y., Wu, K., et al. (2018). Circular RNA F-circEA produced from EML4-ALK fusion gene as a novel liquid biopsy biomarker for non-small cell lung cancer. Cell Res. 28, 693–695. doi: 10.1038/s41422-018-0033-7
Tang, C., Xie, Y., Yu, T., Liu, N., Wang, Z., Woolsey, R. J., et al. (2020). m6A-dependent biogenesis of circular RNAs in male germ cells. Cell Res. 30, 211–228. doi: 10.1038/s41422-020-0279-8
Tuck, A. C., Rankova, A., Arpat, A. B., Liechti, L. A., Hess, D., Iesmantavicius, V., et al. (2020). Mammalian RNA decay pathways are highly specialized and widely linked to translation. Mol. Cell 77, 1222–1236. doi: 10.1016/j.molcel.2020.01.007
von Maltzahn, J., Chang, N. C., Bentzinger, C. F., and Rudnicki, M. A. (2012). Wnt signaling in myogenesis. Trends Cell Biol. 22, 602–609. doi: 10.1016/j.tcb.2012.07.008
Wan, H., Zhu, J., Su, G., Liu, Y., Hua, L., Hu, L., et al. (2016). Dietary supplementation with β-hydroxy-β-methylbutyrate calcium during the early postnatal period accelerates skeletal muscle fibre growth and maturity in intra-uterine growth-retarded and normal-birth-weight piglets. Br. J. Nutr. 115, 1360–1369. doi: 10.1017/S0007114516000465
Wang, F., Nazarali, A. J., and Ji, S. (2016). Circular RNAs as potential biomarkers for cancer diagnosis and therapy. Am. J. Cancer Res. 6, 1167–1176.
Wang, X., Cao, X., Dong, D., Shen, X., Cheng, J., Jiang, R., et al. (2019). Circular RNA TTN acts as a miR-432 sponge to facilitate proliferation and differentiation of myoblasts via the IGF2/PI3K/AKT signaling pathway. Mol. Ther. Nucleic Acids 18, 966–980. doi: 10.1016/j.omtn.2019.10.019
Wang, Y., and Wang, Z. (2015). Efficient backsplicing produces translatable circular mRNAs. RNA Publ RNA Soc. 21, 172–179. doi: 10.1261/rna.048272.114
Westholm, J. O., Miura, P., Olson, S., Shenker, S., Joseph, B., Sanfilippo, P., et al. (2014). Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 9, 966–1980. doi: 10.1016/j.celrep.2014.10.062
Wei, X., Li, H., Yang, J., Hao, D., Dong, D., Huang, Y., et al. (2017). Circular RNA profiling reveals an abundant circLMO7 that regulates myoblasts differentiation and survival by sponging miR-378a-3p. Cell Death Dis. 8, e3153–e3165. doi: 10.1038/cddis.2017.541
Wei, X., Li, H., Zhang, B., Li, C., Dong, D., Lan, X., et al. (2016). miR-378a-3p promotes differentiation and inhibits proliferation of myoblasts by targeting HDAC4 in skeletal muscle development. RNA Biol. 13, 1300–1309. doi: 10.1080/15476286.2016.1239008
Wu, K., Liao, X., Gong, Y., He, J., Zhou, J. K., Tan, S., et al. (2019). Circular RNA F-circSR derived from SLC34A2-ROS1 fusion gene promotes cell migration in non-small cell lung cancer. Mol. Cancer. 18, 98–103. doi: 10.1186/s12943-019-1028-9
Yan, X. M., Zhang, Z., Meng, Y., Li, H. B., Gao, L., Luo, D., et al. (2020). Genome-wide identification and analysis of circular RNAs differentially expressed in the longissimus dorsi between Kazakh cattle and Xinjiang brown cattle. PEER J. 8, 8646–8662. doi: 10.7717/peerj.8646
Yang, Y. B., Gao, X. Y., Zhang, M. L., Yan, S., Sun, C. J., Xiao, F. Z., et al. (2018). Novel role of FBXW7 circular RNA in repressing Glioma Tumorigenesis. J. Natl. Cancer Inst. 110, 304–315. doi: 10.1093/jnci/djx166
Yang, Z. G., Awan, F. M., Du, W. W., Zeng, Y., Lyu, J., Wu, D., et al. (2017). The circular RNA interacts with STAT3, increasing its nuclear translocation and wound repair by modulating Dnmt3a and miR-17 function. Mol. Ther. 25, 2062–2074. doi: 10.1016/j.ymthe.2017.05.022
Yin, H. D., Shen, X. X., Zhao, J., Cao, X. A., He, H. R., Han, S. S., et al. (2020). Circular RNA CircFAM188B encodes a protein that regulates proliferation and differentiation of chicken skeletal muscle satellite cells. Front. Cell Dev. Biol. 8:522588. doi: 10.3389/fcell.2020.522588
Yoshimoto, R., Rahimi, K., Hansen, T. B., Kjems, J., and Mayeda, A. (2020). Biosynthesis of circular RNA ciRS-7/CDR1as is mediated by mammalian-wide interspersed repeats (MIRs). Iscience 23, 101345–101359. doi: 10.1016/j.isci.2020.101345
Yue, B., Wang, J., Ru, W., Wu, J., Cao, X., Yang, H., et al. (2020). The circular RNA circHUWE1 sponges the miR-29b-AKT3 axis to regulate myoblast development. Mol. Ther. Nucleic Acids 19, 1086–1097. doi: 10.1016/j.omtn.2019.12.039
Zeng, K., Chen, X., Xu, M., Liu, X., Hu, X., Xu, T., et al. (2018). CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 9:417. doi: 10.1038/s41419-018-0454-8
Zhang, P., Xu, H., Li, R., Wu, W., Chao, Z., Li, C., et al. (2018). Assessment of myoblast circular RNA dynamics and its correlation with miRNA during myogenic differentiation. Biochem. Cell Biol. 99, 211–218. doi: 10.1016/j.biocel.2018.04.016
Zhang, P. P., Chao, Z., Zhang, R., Ding, R. Q., Wang, Y. L., Wu, W., et al. (2019). Circular RNA Regulation of Myogenesis. Cells 8, 885–897. doi: 10.3390/cells8080885
Zhang, X. O., Wang, H. B., Zhang, Y., Lu, X., Chen, L. L., and Yang, L. (2014). Complementary sequence-mediated exon circularization. Cell 159, 134–147. doi: 10.1016/j.cell.2014.09.001
Zhang, Y., Zhang, X. O., Chen, T., Xiang, J. F., Yin, Q. F., Xing, Y. H., et al. (2013). Circular intronic long noncoding RNAs. Mol. Cell 51, 792–806. doi: 10.1016/j.molcel.2013.08.017
Zhao, J., Wu, J., Xu, T., Yang, Q., He, J., and Song, X. (2018). IRESfinder: identifying RNA internal ribosome entry site in eukaryotic cell using framed k-mer features. J. Genet. Genomics 45, 403–406. doi: 10.1016/j.jgg.2018.07.006
Keywords: circRNAs, livestock animals, muscle development, co-expression regulatory network, transcription and translation
Citation: Yang Z, He T and Chen Q (2021) The Roles of CircRNAs in Regulating Muscle Development of Livestock Animals. Front. Cell Dev. Biol. 9:619329. doi: 10.3389/fcell.2021.619329
Received: 20 October 2020; Accepted: 18 January 2021;
Published: 05 March 2021.
Edited by:
Xiao Han, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Rajnish Kumar, Amity University Uttar Pradesh, Lucknow Campus, IndiaTiande Zou, Jiangxi Agricultural University, China
Copyright © 2021 Yang, He and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhenguo Yang, Z3VvZ3VvMDAwMDJAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Zhenguo Yang
Zhenguo Yang Tianle He
Tianle He Qingyun Chen
Qingyun Chen