- Institute of Molecular Biosciences, J.W. Goethe University, Frankfurt am Main, Germany
Organismic aging is known to be controlled by genetic and environmental traits. Pathways involved in the control of cellular metabolism play a crucial role. Previously, we identified a role of PaCLPP, a mitochondrial matrix protease, in the control of the mitochondrial energy metabolism, aging, and lifespan of the fungal aging model Podospora anserina. Most surprisingly, we made the counterintuitive observation that the ablation of this component of the mitochondrial quality control network leads to lifespan extension. In the current study, we investigated the role of energy metabolism of P. anserina. An age-dependent metabolome analysis of the wild type and a PaClpP deletion strain verified differences and changes of various metabolites in cultures of the PaClpP mutant and the wild type. Based on these data, we generated and analyzed a PaSnf1 deletion mutant and a ΔPaSnf1/ΔPaClpP double mutant. In both mutants PaSNF1, the catalytic α-subunit of AMP-activated protein kinase (AMPK) is ablated. PaSNF1 was found to be required for the development of fruiting bodies and ascospores and the progeny of sexual reproduction of this ascomycete and impact mitochondrial dynamics and autophagy. Most interestingly, while the single PaSnf1 deletion mutant is characterized by a slight lifespan increase, simultaneous deletion of PaSnf1 and PaClpP leads to a pronounced lifespan extension. This synergistic effect is strongly reinforced in the presence of the mating-type “minus”-linked allele of the rmp1 gene. Compared to the wild type, culture temperature of 35°C instead of the standard laboratory temperature of 27°C leads to a short-lived phenotype of the ΔPaSnf1/ΔPaClpP double mutant. Overall, our study provides novel evidence for complex interactions of different molecular pathways involved in mitochondrial quality control, gene expression, and energy metabolism in the control of organismic aging.
Introduction
Aging of biological systems is characterized by a time-dependent decrease of physiological functions and an increase in morbidity and mortality. It has a multifactorial basis, and various molecular pathways are known to control aging, lifespan, and healthspan. Over decades of research, a paramount role of mitochondria, the cellular “power plants,” was repeatedly demonstrated in various systems from fungi to humans (Linnane et al., 1989; Cortopassi and Arnheim, 1990; Wallace, 1992; Melov et al., 1994; Petersen et al., 2003; Osiewacz, 2010; Breitenbach et al., 2012). In general, mitochondrial function declines during aging, although a number of different molecular pathways are active in keeping a “healthy population” of mitochondria over time (Luce et al., 2010; Baker et al., 2011; Fischer et al., 2012).
One of the aging model organisms in which the role of mitochondria was extensively investigated is the filamentous fungus Podospora anserina (Osiewacz, 2002; Scheckhuber and Osiewacz, 2008; Osiewacz et al., 2013). Classical genetic experiments identified an extrachromosomal genetic basis of aging already in the pre-molecular era (Marcou, 1962). Subsequently, this basis was located in mitochondria (Stahl et al., 1978; Cummings et al., 1979). Specifically, it was shown that the mitochondrial DNA (mtDNA) grossly reorganizes during the aging process as a result of the activity of a mobile DNA species that acts as a mutator (Kück et al., 1981, 1985; Osiewacz and Esser, 1984; Sellem et al., 1993; Borghouts et al., 2002). Later on, mitochondrial metabolism via the generation of adenosine triphosphate (ATP) at the inner mitochondrial membrane and the formation of reactive oxygen species (ROS) (Borghouts et al., 1997, 2001; Gredilla et al., 2006) was found to strongly affect the aging process. In more recent years, it was established that various pathways which are involved in the control of functional mitochondria have a strong impact on lifespan. For instance, it was shown that the overexpression of the gene coding for the mitochondrial LON protease leads to a healthspan extension via the increased degradation of damaged (carbonylated) proteins and the conservation of mitochondrial function (Luce and Osiewacz, 2009). While such an effect was expected, results from some other experimental interventions were counterintuitive and unexpected. For instance, the deletion of the gene coding for PaIAP, an AAA protease (ATPases associated with diverse cellular activities) that is located in the inner mitochondrial membrane, was found to lead to a lifespan extension (Weil et al., 2011). The molecular analysis revealed that PaIAP is a protease which, at controlled growth temperature of 27°C, is rather lifespan limiting. In contrast, at increased temperatures, as they are regularly reached during the day under natural conditions, the protease has a protective function. Yet another protease is PaCLPP which, together with the chaperone PaCLPX, is part of a multiprotein PaCLPXP complex located in the mitochondrial matrix. Deletion of the genes coding for both components of this complex resulted in healthspan extension of the corresponding mutants (Fischer et al., 2013; Knuppertz and Osiewacz, 2017). A detailed analysis of PaCLPXP interaction partners identified 47 proteins among which 19 turned out to be high confident substrates (Fischer et al., 2015b). Most of the substrates, including components of the pyruvate dehydrogenase complex, the tricarboxylic acid (TCA) cycle, and the respiratory chain, are involved in mitochondrial energy metabolism. Among these, there are components of the N-module of respiratory complex I which were subsequently also identified in mammals and the plant Arabidopsis thaliana (Szczepanowska et al., 2016; Petereit et al., 2020) suggesting at least a partial conserved role of CLPXP in pathways involved in cellular energy metabolism.
Initial data from studies with P. anserina revealed that ablation of the two components of the PaCLPXP protein complex does not affect cellular ATP levels most probably due to the conservation of cellular energy resulting from the induction of autophagy (Knuppertz and Osiewacz, 2017), a process that is stringently controlled by the nutrient status. In eukaryotes, two protein complexes, “target of rapamycin” (TOR) and “AMP-activated protein kinase” (AMPK), are master regulators of autophagy. The two complexes respond inversely to the cellular energy status. Under ATP-replete conditions, TOR is active and promotes anabolic processes as well as transcription and translation while inhibiting autophagy. Upon ATP depletion and AMP increase, AMPK becomes activated and stimulates various catabolic processes including autophagy (Knuppertz and Osiewacz, 2016; Wen and Klionsky, 2016; Yin et al., 2016).
Here we report an experimental study aimed to shed more light on metabolic aspects linked to the role of PaCLPXP in development and aging. We performed an age-related metabolome analysis of the wild type and the PaClpP deletion strain. Based on the results of this analysis and because of its role in autophagy induction, we investigated the consequences on aging of the ablation of the catalytic AMPK α-subunit (PaSNF1) in the wild-type and ΔPaClpP background. Surprisingly, we found that AMPK activity is not required for the lifespan extension of ΔPaClpP strains. Even more, lifespan extension of the PaClpP deletion mutant appears to be limited by AMPK. Most strikingly, we also observed a strong impact of the mating type, most likely the mating type-linked rmp1 gene of P. anserina, on lifespan in a mutant in which PaSnf1 and PaClpP are both deleted. These observations link protein quality control and the control of cellular energy with this rather poorly characterized RMP1 protein of P. anserina.
Materials and Methods
P. anserina Strains and Cultivation
The P. anserina wild-type (WT) strain “s” (Rizet, 1953), the previously described reporter strain PaSod1::Gfp (Zintel et al., 2010), the PaClpP deletion strain (ΔPaClpP; Luce and Osiewacz, 2009), and the following recently generated strains were used: ΔPaSnf1, ΔPaSnf1/Flag::PaSnf1, ΔPaSnf1/PaSod1::Gfp, ΔPaSnf1/ΔPaClpP, and ΔPaSnf1/ΔPaClpP/PaSod1::Gfp. Double mutants were obtained by crossing single-mutant strains. The triple-mutant ΔPaSnf1/ΔPaClpP/PaSod1::Gfp was obtained by crossing of ΔPaSnf1/ΔPaClpP with PaSod1::Gfp. All mutant strains were constructed in the genetic background of the WT “s.” Unless otherwise stated, strains were grown on complete medium (BMM) containing cornmeal extract (Esser, 1974). To induce the formation of fruiting bodies, cultures with opposite mating types were placed on solid semi-synthetic M2 agar plates (M2) (Osiewacz et al., 2013) and incubated for 10–14 days at 27°C and constant light. For spore germination, the spores were incubated for 2 days at 27°C in the dark on BMM containing 60 mM ammonium acetate. Strains were grown on M2 (0.025% KH2PO4, 0.03% K2HPO4, 0.025% MgSO4, 0.05% urea, 1% dextrin, 2% agar), M2 + glycerol (0.025% KH2PO4, 0.03% K2HPO4, 0.025% MgSO4, 0.05% urea, 1% glycerol, 2% agar), or M2-N (0.025% KH2PO4, 0.03% K2HPO4, 0.025% MgSO4, 1.5% dextrin, 2% agar) or in liquid complete medium [CM: 0.1% KH2PO4, 0.05% KCl, 0.05% MgSO4, 1% glucose, 0.37% NH4Cl, 0.2% tryptone, 0.2% yeast extract, 0.1% stock solution A (0.1% ZnSO4, 0.1% FeCl2, 0.1% MnCl2, pH 6.5 (KOH)), pH 6.5 (KOH)] at 27°C under constant light (Osiewacz et al., 2013).
Metabolic Profiling
To analyze putative changes in the abundance of metabolites in the PaClpP deletion mutant compared to the wild type, a metabolomic profiling was performed with 5- and 20-day-old wild-type and ΔPaClpP strains (n = 4 each). Mycelial extracts were obtained from strains grown on M2 agar plates at 27°C and constant light for a specific time. Subsequently, the mycelium of the aged strains (17-day-old strains) and young strains (2-day-old strains, directly used from the BMM plates with ammonium acetate) were inoculated on M2 agar plates, covered with cellophane, and allowed to overgrow the plates for three additional days at 27°C and constant light. The agar pieces were removed, the mycelium was harvested and portioned in 50-mg aliquots into a 1.5-ml tube. The samples were frozen in liquid nitrogen and stored at −80°C until the final analysis. The analysis was performed by Metabolomic Discoveries (Metabolomic Discoveries GmbH, Potsdam, Germany). The used optimized method allows the unambiguous characterization and relative quantification of the requested metabolites. Metabolite analysis by LC–tandem mass spectrometry was performed using a LCMS-8050 by Shimadzu triple-quadrupole mass spectrometer equipped with an electrospray ionization (ESI) source and operated in the multiple reaction mode (MRM).
Cloning Procedures and Generation of P. anserina Mutants
All strains were constructed in the genetic background of the wild-type strain “s” (Rizet, 1953). To generate the ΔPaSnf1 strain, we proceeded as described by Hamann et al. (2005). First, the flanking regions of PaSnf1 were amplified with the following oligonucleotides: For the 5′-flanking region, Snf1KO1 (CAAGGGCACGGTGCTGTC, Biomers, Ulm, Germany) and Snf1KO2 (TTAAGCTTCATGCTGGCGACGGCTATC, HindIII site is underlined, Biomers, Ulm, Germany) were used. For the 3′-flank, Snf1KO3 (CGCGACTAGTGCGGATGCGGA TTGATTATTG, BcuI site is underlined, Biomers, Ulm, Germany) and Snf1KO4 (AAGCGGCCGCTGTCGATGATA CAGATAG, NotI site is underlined, Biomers, Ulm, Germany) were used. These amplified flanking regions were cloned in the plasmid pKO4 (Hamann et al., 2005; Luce and Osiewacz, 2009), using the restriction enzymes KpnI, BcuI, NotI, and HindIII (Thermo Scientific, Waltham, MA, United States, ER0521, ER1251, ER0591, ER0501; KpnI site is part of the 5′-flank and thus has not to be added through the oligonucleotide). The wild-type PaSnf1 gene in a cosmid selected from a representative cosmid library (Osiewacz, 1994) was replaced in Escherichia coli by a combined blasticidin/phleomycin resistance cassette as described by Hamann et al. (2005). The resulting replacement cosmid was used to transform P. anserina wild-type spheroplasts (Osiewacz et al., 1991). Transformants were selected on a phleomycin- (Genaxxon, Ulm, Germany, M3429) containing medium. For the complementation of the PaSnf1 deletion mutant, and additionally to mark the PaSNF1 protein with a His::FLAG epitope, the sequence of PaSnf1 was fused with the sequence encoding this epitope. To achieve this, the sequence encoding the His::FLAG epitope was introduced in the plasmid pKO7 (Kunstmann and Osiewacz, 2009), which contains a hygromycin B resistance cassette. To include the PaSnf1 gene in this pHisFLAG vector, the gene was amplified in two parts. The first part consists of the promoter region, which was amplified with the oligonucleotides Snf1-4 (AAGCGGCCGCCATGGAAATAAGGACCCAG, NotI site is underlined) and Snf1-11 (TTGGATCCCATGCTGGCGACG GCTAT, BamHI site is underlined). The second part contains the PaSnf1 ORF and 500 bp of the terminator region, which were amplified with the oligonucleotides Snf1-12 (ATGGCCCAGGCCTACGACGACGA, this oligonucleotide contains a phosphate residue at the 5′-ending) and Snf1-9 (ACCTGCAGTGCCAGAATGGCCAGCCTTATG, PstI site is underlined). The PCR product of the PaSnf1 ORF and the pHisFLAG vector was digested with the restriction enzymes SmaI and PstI (Thermo Scientific, Waltham, MA, United States, ER0661, ER0611). The constructed vector pHis-Flag::PaSnf1-1 contains the PaSnf1 ORF fused with a sequence encoding a HIS::FLAG epitope at the 5′-ending. In a second step, the promoter region of PaSnf1 was integrated into the newly constructed plasmid pHis-Flag::PaSnf1-1 using the restriction enzyme sites of NotI and BamHI (Thermo Scientific, Waltham, MA, United States, ER0591, ER0051). The final plasmid pFlag-Snf1 contains the sequence of a HIS::FLAG epitope between the sequence of the promoter region of PaSnf1 and the PaSnf1 ORF with the terminator region and was transformed into P. anserina wild-type spheroplasts as previously described (Osiewacz et al., 1991), and hygromycin B [Merck Millipore (Calbiochem), Burlington, MA, United States, 400051]-resistant transformants were selected. Finally, the newly generated strain Flag::PaSnf1 was crossed with the PaSnf1 deletion mutant to remove the endogenous PaSnf1 sequence. The new ΔPaSnf1/Flag::PaSnf1 strain was selected by the phleomycin and hygromycin B resistance and verified by Southern blot analysis.
Southern Blot Analysis
Total DNA was isolated as described before (Lecellier and Silar, 1994). Restriction of the DNA, gel electrophoresis, and Southern blotting were performed in accordance with standard protocols (Sambrook et al., 1989). Hybridization and detection were accomplished as described in the manufacturer’s protocol with digoxigenin-labeled hybridization probes (DIG DNA Labeling Mix, Sigma-Aldrich, St. Louis, MO, United States, 11175033910). The PaSnf1-specific probe was generated by PCR with the oligonucleotides Snf1-1 (GCATCGGCGCCTACAACATTGTC, Biomers, Ulm, Germany) and Snf1-2 (GAGACCGAAATCGGCGATCTTG, Biomers, Ulm, Germany). The phleomycin (Ble)-specific probe was generated from the plasmid pKO4 (Luce and Osiewacz, 2009), which contains the phleomycin resistance cassette. The respective fragment was recovered after digestion with BamHI. The resulting 1.3-kb fragment was extracted and purified from the agarose gel. The PaClpP-specific probe was generated according to Fischer et al. (2013).
Quantitative Real-Time PCR (qRT-PCR)
For isolation of total RNA, strains were allowed to overgrow a cellophane foil-covered M2 petri dish for 3 days at 27°C and constant light. The mycelium was subsequently transferred into 2-ml tubes with 0.5-mm glass beads (Bertin Technologies, Montigny-le-Bretonneux, France, KT03961-1-004.2) and disrupted in a Precellys 24 homogenizer (Bertin Technologies; peqlab; Precellys 24 lysis and homogenization, Montigny-le-Bretonneux, France) for 2 × 25 s at 5800 rpm and a 10-s break in between. The isolation was performed using the NucleoSpin RNA Plant kit (Macherey Nagel #740949.250, Düren, Germany) according to the manufacturer’s instruction. Samples of wild type, ΔPaSnf1, ΔPaClpP, and ΔPaSnf1/ΔPaClpP strains were analyzed. 1 μg RNA was used for the reverse transcription with a Revert Aid Reverse transcriptase and Ribolock RNase inhibitor (Thermo Scientific, Waltham, MA, United States, EP0441, EC0381) and oligo(dT)18 (Thermo Scientific, SO132). 2 μl of the resulting cDNA was used for the quantitative real-time PCR (qRT-PCR) reaction (IQ Sybr Green SuperMix, Bio-Rad, Hercules, CA, United States). To determine the expression of PaMdm38, the primers Pa_1_1789-1 (GCCCAGAAGGAAGAGTTTAC) and Pa_1-1780-2 (GTTGGAGACACCATCACTAC) were used. For each gene, the PCR efficiency was determined according to Pfaffl (2001). The relative expression level was normalized to the expression of PaPorin which was determined with the oligonucleotides Porin-RT-for (TCTCCTCCGGCAGCCTTG, Biomers, Ulm, Germany) and Porin-RT-rev (CGGAGGCGGACTTGTGAC, Biomers, Ulm, Germany). The relative expression level was calculated with the following formula:
Relative expression = (E(porin)^CP(porin))/(E(target gene)^CP(target gene));
E = PCR efficiency of the respective primer pair;
CP = crossing point for each transcript.
Growth Rate and Lifespan Analysis
The lifespan and growth rate of monokaryotic isolates at 27 and 35°C were determined using race tubes containing M2 medium as described previously (Osiewacz et al., 2013). Lifespan analyses on M2 with glycerol instead of dextrin, or on the autophagy-inducing M2 medium without the nitrogen source urea but with the 1.5-fold concentration of dextrin (M2-N), were performed in petri dishes. The petri dishes and the race tubes were incubated at 27 or 35°C and constant light. The lifespan is defined as the time period in days (d) of linear growth. The growth rate is defined as the growth in centimeters (cm) per day (d). For measurement, the growth front of the strains was marked every 2–3 days until death.
Microscopy and Spore Analysis
To analyze the content of the fruiting bodies and the spore morphology, confrontational crosses of different strains were performed on M2 medium. The strains were placed 3 cm apart of each other and were incubated for 12 (wild type × wild type; wild type × ΔPaSnf1; ΔPaSnf1/Flag::PaSnf1 × ΔPaSnf1; ΔPaSnf1 × ΔPaSnf1) and 14 days (ΔPaSnf1 × ΔPaSnf1) at 27°C and constant light. The content of a fruiting body was placed in a 20-μl H2O droplet on a slide and covered with a cover slip. The samples were observed with the Leica Biomed Microscope (Leica Biosystems Nussloch GmbH, Nußloch, Germany) and documented with the camera DFC7000 T. To analyze the fruiting body content, the 10×/0.25 objective lens (506088) was used. To visualize spore morphology, the 40×/0.65 objective lens (506099) was used. For quantification of fruiting body content, spores of 10 fruiting bodies of each cross were analyzed (wild type × wild type n = 1691 spores, ΔPaSnf1 × ΔPaSnf1: n = 760 spores; wild type × ΔPaSnf1: n = 1595 spores; ΔPaSnf1/Flag::PaSnf1 × ΔPaSnf1: n = 1423 spores).
Fertility Analysis
Determination of male (♂) or female (♀) fertility was performed by spermatization experiments. Cultures derived from mononucleate ascospores of the wild type (n = 3 of each mating type) and ΔPaSnf1 (n = 3 of mating type “minus” and n = 2 of mating type “plus”) were grown for 7 days on M2 agar plates at 27°C and constant light. To harvest spermatia, the mycelium was overlaid with 5 ml sterile water and incubated for 5 min. Afterward the water containing spermatia was collected from the plate. For spermatization, 300 μl of this suspension was dropped on mycelium of the opposite mating type (n = 5 technical replicates each). After 5 min of incubation, the droplets were removed and the plates were incubated for 5 days at 27°C and constant light. The number of perithecia obtained from wild-type crosses was set to 100%. For each case [wild type (♂) × wild type (♀); wild type (♂) × ΔPaSnf1 (♀), ΔPaSnf1 (♂) × wild type (♀); ΔPaSnf1 (♂) × ΔPaSnf1 (♀)], six different spermatizations were used for quantification.
Fluorescence Microscopy
For this experiment, P. anserina strains were cultivated on glass slides with a central depression containing 160 μl M2 medium for 1 day at 27°C and constant light in a moisture-prone chamber. For microscopic analysis, the Zeiss Cell Observer SD-Spinning Disk Confocal Microscope (Carl Zeiss Microscopy, Jena, Germany) with a 100×/1.46 oil objective lens (Carl Zeiss Microscopy, Jena, Germany, 420792-9800) was used. For visualization of mitochondria, the hyphae were stained with 100 μl of 2 μM MitoTracker Red (Invitrogen, Carlsbad, CA, United States) for 15 min in the dark. Subsequently, the hyphae were washed with 100 μl water. The mitochondria were visualized with the appropriate excitation and emission filters. For image processing, ZEN 2.5 (blue, Carl Zeiss Microscopy, Jena, Germany) was used.
Measurement of Oxygen Consumption Rate
The measurement of oxygen consumption rate was performed at 27°C by high-resolution respirometry (Oxygraph-2k series C and G, OROBOROS Instruments, Innsbruck, Austria). Cultures derived from mononucleate ascospores of wild type, ΔPaSnf1, ΔPaClpP, and ΔPaSnf1/ΔPaClpP were grown on M2 plates covered with a cellophane foil for 2 days, shifted into CM liquid medium, and further incubated for 3 days at 27°C under shaking and constant light. A small piece of mycelium was placed into the Oroboros respirometer chamber into 2 ml fresh CM media. The oxygen consumption rate was determined as previously described in Fischer et al. (2015a). After the measurement, the mycelium was placed into a 2-ml tube and boiled for 10 min at 95°C. Subsequently, the mycelium was air-dried for 2 days, and dry weight was determined. Finally, basal oxygen consumption per mg mycelium was calculated. For the analysis of the data, the DatLab6 software from Oroboros (Innsbruck, Austria) was used.
Western Blot Analysis
For isolation of total protein extracts, the strains were allowed to age on M2 plates at 27°C under constant light, after spore germination on BMM plates with 60 mM ammonium acetate for 2 days at 27°C in the dark. Mycelia from the different P. anserina strains and of different age were allowed to overgrow the cellophane foil-covered solid M2 medium for 3 days at 27°C and finally shifted to CM liquid medium and further incubated for 3 days at 27°C under constant light and shaking. To harvest the mycelium, the cultures were filtered over two layers of gauze. Approximately 250 mg mycelia was transferred in 2-ml tubes with 0.5-mm glass beads (Bertin Technologies, Montigny-le-Bretonneux, France, KT03961-1-004.2), and two volumes of protein extraction buffer (50 mM HEPES, 100 mM NaCl, 5 mM DTT, pH = 7.4) were added. The samples were kept on ice. For homogenization of the mycelia, the Precellys homogenizer (Bertin Technologies; peqlab; Precellys 24 lysis and homogenization, Montigny-le-Bretonneux, France) was used (2 × 25 s at 5800 rpm with a 10-s break in between). Afterward, the samples were centrifuged for 5 min at 9300 × g and 4°C. Subsequently, the supernatant was transferred to a new 1.5-ml reaction tube. The isolation of mitochondrial protein extracts was performed as previously described by Kunstmann and Osiewacz (2008). For western blot analysis, 50–150 μg protein extract was mixed with SDS-loading buffer (6% TRIS/HCl pH 6.8, 48% glycerol, 9% β-mercapto-ethanol, 0.03% bromophenol blue, 6% SDS) and heated up for 10 min at 95°C. The separation of proteins was accomplished by SDS-PAGE by standard protocols (Brust et al., 2010), and the proteins were subsequently transferred to a PVDF membrane (Immobilon-FL PVDF, Millipore, Burlington, MA, United States, IPFL00010). Blocking and antibody treatment of the membrane were performed according to protocols of the Odyssey® Imaging System Western blot analysis (LI-COR Biosciences, Bad Homburg, Germany). The following primary antibodies were used for this study: anti-Phos-αAMPK (Phospho-AMPKα (Thr172) (40H9), rabbit mAb (dilution 1:300, Cell Signaling Technology, Danvers, MA, United States, #2535L), anti-FLAG (mouse, dilution 1:1,000, Sigma, F3165), anti-GFP (mouse, dilution 1:10,000, Sigma-Aldrich, St. Louis, MO, United States, G6795), and anti-PaDNM1 (rabbit, dilution 1:5,000, Sigma (NEP), polyclonal antibody raised against synthetic peptide “Ac-DKGTPEKESIAIRKC-amide,” affinity purification). IRDye® 680RD anti-mouse (goat, dilution 1:15,000, LI-COR Biosciences, Bad Homburg, Germany, 926-68070) and IRDye® 680LT anti-rabbit (goat, dilution 1:20,000, LI-COR Biosciences, Bad Homburg, Germany, 926-68021) were used as secondary antibodies.
Statistical Analysis
The statistical analysis of lifespans was performed with the IBM SPSS statistics 19 software package, and a Kaplan–Meier survival estimate was generated. Significance was evaluated using three independent statistical tests (Breslow, generalized Wilcoxon; log rank, Mantel–Cox; Tarone-Ware). In the figure legends, only the lowest p-values are indicated. A table with the exact values can be found in the Supplementary Tables 2, 3. To determine the significance of the other experiments, two-tailed Student’s t-test was used (∗∗∗: p < 0.001; ∗∗: p < 0.01; ∗: p < 0.05).
Results
Ablation of PaCLPP Affects TCA Cycle, Glucose and Amino Acid Metabolism, and Nucleotide Levels
The previous demonstration of potential substrates of the mitochondrial PaCLPXP complex, the majority of which are associated with metabolic pathways in mitochondria (Fischer et al., 2015b), and the observation that the ablation of the protease and chaperone component of the complex leads to a pronounced lifespan extension (Fischer et al., 2013) directed our interest to more carefully investigate the impact of metabolic processes on aging of P. anserina.
In a first series of experiments, we investigated the metabolome of the ΔPaClpP mutant of 5- and 20-day-old cultures and compared it with that of the wild type of the same chronological age. In addition, the metabolomes of the wild type of 5- and 20-days of age were analyzed. At note, since the two strains are characterized by a different lifespan, their physiological status (i.e., their biological age) at a given chronological age is not identical. For instance, while 20-day-old wild-type strains are almost senescent and functionally impaired, ΔPaClpP mutant strains are rather unaffected.
The metabolome analysis was performed with cell extracts isolated from mycelia of four biological replicates of each strain and age and the relative amount of 50 metabolites was determined by triple-quadrupole mass spectrometry. The complete data set is presented in Supplementary Table 1 and visualized in a heat map (Figure 1). Most striking alterations occur in the abundance of components of the TCA cycle, of nucleotides, glycolysis metabolites, and amino acids between young (5-day-old) mutant and wild-type cultures (Figure 1; left). Interestingly, some of these alterations are also observed in wild-type cultures of different age (Figure 1; WT 20 day vs. 5 day). In particular, the abundance of glycolysis metabolites and of nucleotides changes during aging in the same direction compared to the situation in young mutant vs. young wild-type strains. It appears that these alterations are rather not responsible for the observed lifespan extension of the mutant. In contrast, changes in amino acid and TCA cycle metabolite levels differ in the aging wild type compared to young mutant vs. young wild type. Especially a significant increase in citrate/isocitrate and aconitate levels is observed in the 5-day-old mutant compared to wild type of the same chronological age. No change in abundance of these metabolites is observed in wild-type cultures of 5 and 20 days of age. Moreover, the abundance of several amino acids (arginine, proline, glycine, histidine, and threonine) is significantly lower in 5-day-old mutants compared to wild types of the same chronological age but not in 20-day-old wild types compared to 5-day-old wild-type cultures. In addition, during wild-type aging several metabolite levels (especially glycolysis metabolites) significantly decreased, while during ΔPaClpP aging (20 days vs. 5 days, Figure 1; right) rather an overall increase in metabolite levels is observed. This effect may result from the upregulation of non-selective autophagy in the ΔPaClpP mutant that was observed in a previous study (Knuppertz and Osiewacz, 2017) as a compensatory response to mitochondrial impairments in the mutant. This idea is supported by the observed changes in the abundance of several nucleotides. Most interestingly, AMP is increased while ATP levels decrease in the young mutant compared to young wild type. This increase in the AMP/ATP ratio directed us to a potential induction of AMPK signaling in the ΔPaClpP mutant, which may explain its lifespan-extended phenotype. To address this possibility experimentally, we set out to delete the gene encoding the catalytic α-subunit of the AMPK complex, termed SNF1 in fungi, which becomes activated by an increased AMP/ATP ratio. During the course of our investigation, Li et al. (2020) reported the generation and characterization of such a deletion mutant of P. anserina (in wild-type background “S”) with special emphasis on lignocellulose degradation. In their study, the impact of PaSNF1 on aging and lifespan control was not addressed.
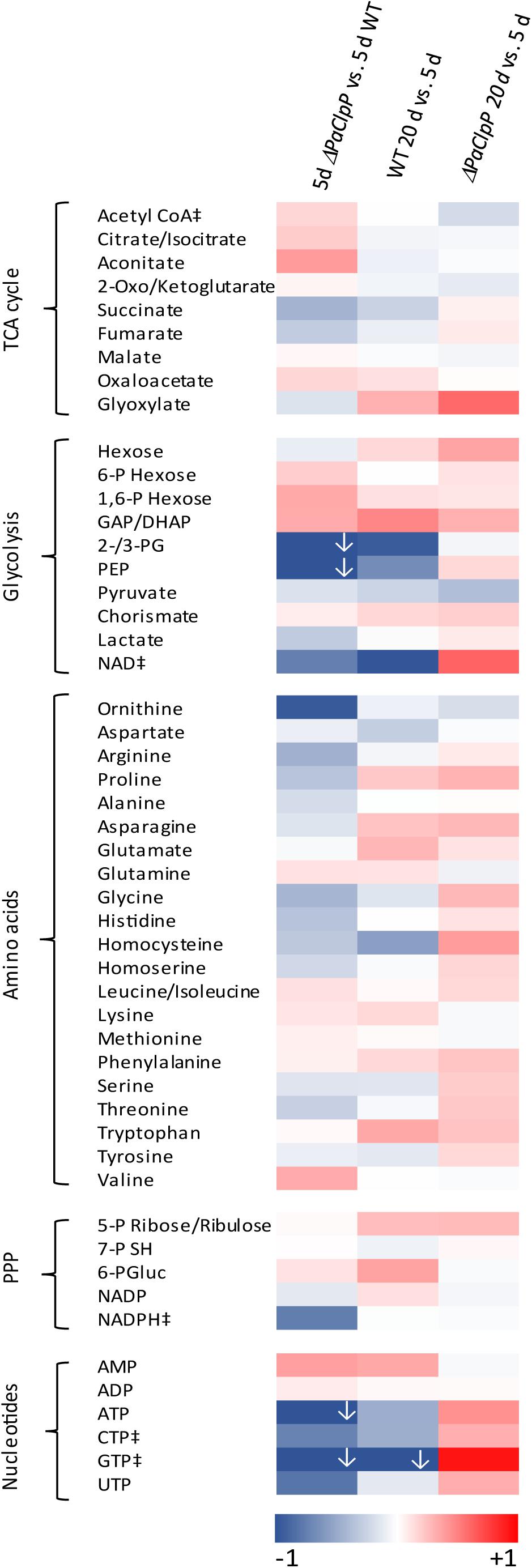
Figure 1. Deletion of PaClpP affects the energy metabolism. Color-coded heat map shows the fold change log(2) of 5-day-old ΔPaClpP vs. wild-type strains, 20-day-old wild-type vs. 5-day-old wild-type strains, and 20-day-old ΔPaClpP vs. 5-day-old ΔPaClpP strains of four biological replicates, each. For better visualization, log2-fold changes lower than 1 (indicated by ↓) are also presented in darkest blue. In Supplementary Table 1, the exact values are provided. “‡”: In at least one biological replicate, this metabolite is at the detection limit. Abbreviations: citrate/isocitrate, citrate or isocitrate; 2-oxo/ketoglutarate, 2-oxoglutarate or 2-ketoglutarate; 6-P hexose, hexose 6-phosphate; 1/6-P hexose, hexose 1,6-bisphosphate, GAP: glyceraldehyde 3-phosphate; DHAP, dihydroxyacetone phosphate; 2-/3-PG, 2- or 3-phosphoglycerate; PEP, phosphoenolpyruvate; 5-P ribose/ribulose, ribose 5-phosphate or ribulose 5-phosphate; 7-P SH, sedoheptulose 7-phosphate; 6-PGluc, 6-phosphogluconate; PPP, pentose phosphate pathway.
Ablation of PaSNF1 Affects Development and Increases Lifespan
Deletion of PaSnf1 was performed following the procedure described in Hamann et al. (2005) in wild-type strain “s” Rizet (1953). The genomic sequence Pa_2_770 annotated to encode PaSNF1 (UniProt B2B4C1) was replaced by a phleomycin resistance cassette. A double mutant, lacking PaSnf1 and PaClpP, was selected from the progeny of crosses of the newly isolated PaSnf1 mutant with the previously isolated ΔPaClpP strain (Fischer et al., 2013). The correct genotype of strains was verified by Southern blot analysis (see Supplementary Figure 1A). In addition, western blot analysis using a commercially available anti-phospho-α-AMPK antibody failed to detect phosphorylated and thus activated AMPK in the PaSnf1 deletion strain (Supplementary Figure 1B). As control, a plasmid encoding FLAG::PaSNF1 was introduced in the ΔPaSnf1 strain. The resulting ΔPaSnf1/Flag::PaSnf1 strain was verified by Southern blot analysis (Supplementary Figure 1C), and production of the FLAG-tagged PaSNF1 protein was demonstrated by western blot analysis (Supplementary Figure 1D).
Except for a significantly reduced growth rate, the PaSnf1 deletion strain does not show any obvious macroscopic differences (i.e., pigmentation, aerial hyphae formation) compared to the wild type grown on M2 standard medium (Figures 2A,B). As described earlier, the ΔPaClpP strain has a wild-type-like growth rate (Fischer et al., 2013). In the double mutant ΔPaSnf1/ΔPaClpP, growth rate is reduced like in ΔPaSnf1 (Figure 2B), suggesting that growth impairment of ΔPaClpP is epistatic. Introduction of the Flag-tagged PaSnf1 copy into the PaSnf1 deletion strain restored the wild-type growth rate (Figures 2A,B). In addition to the reduction in growth rate, the analysis of sexual reproduction revealed severe impairments in the ΔPaSnf1 strain. In our analyses, we investigated the fertility of PaSnf1 deletion strains originating from monokaryotic ascospores. These cultures were either of mating type “plus (+)” or “minus (−)” and form fruiting bodies only after contact with a partner of the opposite mating type. At note, mating of P. anserina is possible by placing mycelial pieces of the two strains of opposite mating type on opposite positions on one agar plate. After the partners contact each other, the male gametes (spermatia) of each of the two strains fertilize the female gametangia (protoperithecia) and fruiting bodies (perithecia) are subsequently formed. This kind of “confrontational cross” allows to investigate the efficiency and morphological differences in the formation of reproductive structures (perithecia, ascospores) but not the discrimination of the functionality of male gametes and female gametangia, respectively. Such discrimination is possible by spermatization of a growing culture of one mating type (the “acceptor” strain) with spermatia of the opposite mating type isolated from the “donor” strain grown on a separate agar plate. In this case, fruiting bodies are only formed when spermatia are poured on the “acceptor” strain and female and male fertility can be clearly attributed and measured.
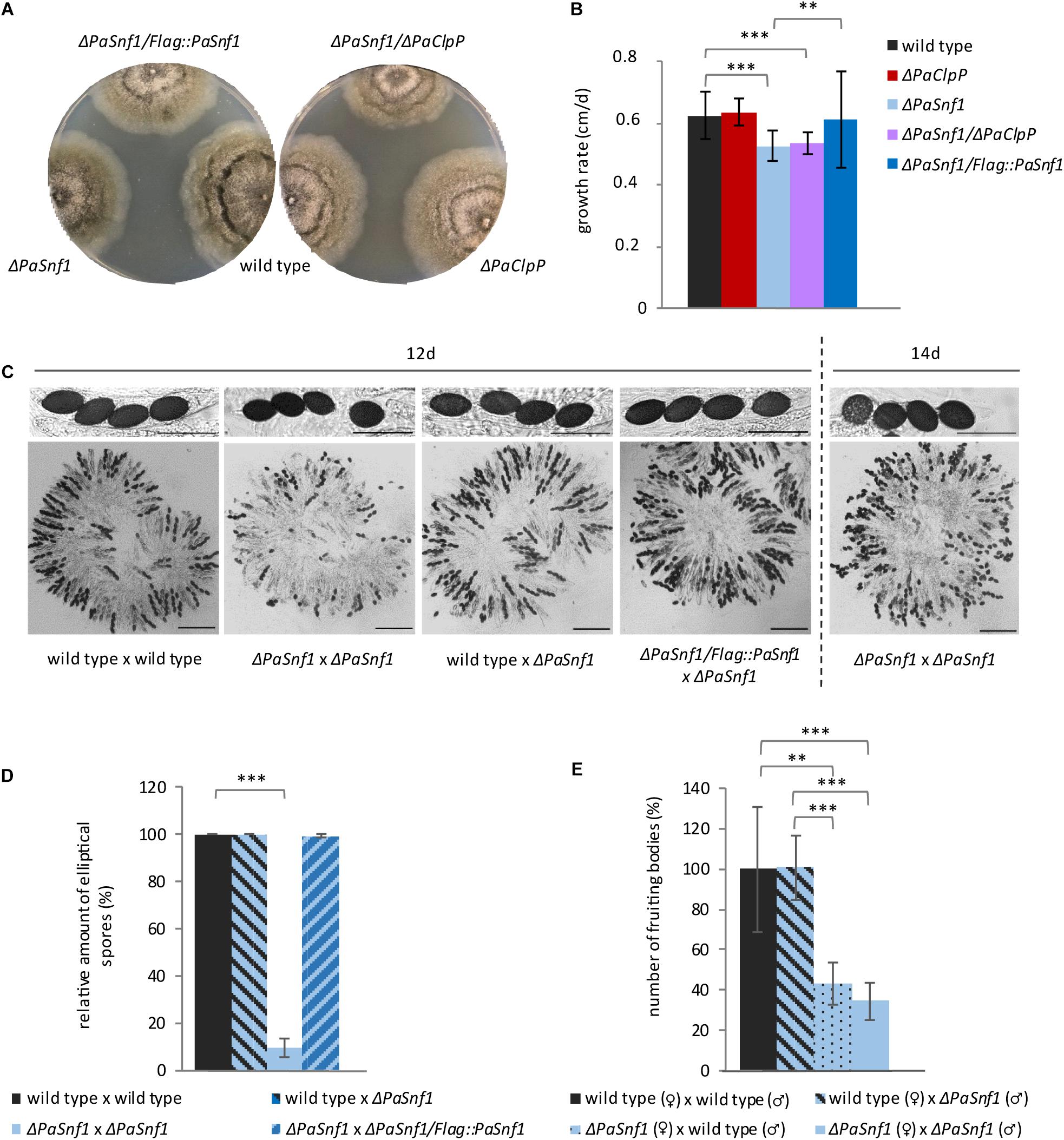
Figure 2. Ablation of PaSNF1 affects growth rate and fertility. (A) Phenotypes of ΔPaSnf1, ΔPaClpP, and ΔPaSnf1/ΔPaClpP; the complementation strain ΔPaSnf1/Flag::PaSnf1; and the wild type at the age of 9 days on M2 medium, cultivated at 27°C and in constant light. (B) Growth rate of ΔPaSnf1 (n = 28), ΔPaClpP (n = 22), ΔPaSnf1/ΔPaClpP (n = 25), ΔPaSnf1/Flag::PaSnf1 (n = 21), and wild type (n = 32) on M2 medium at 27°C and in constant light. (C) Content of fruiting bodies from cultures after a 12-day incubation of pairs of opposite mating types (confrontational crosses) of two wild-type strains, of two ΔPaSnf1 strains, of wild type × ΔPaSnf1, or of ΔPaSnf1/Flag::PaSnf1 × ΔPaSnf1, respectively. (C, right) Content of a fruiting body of the cross of two ΔPaSnf1 after two additional days of incubation (14 day). Scale bar of fruiting body content corresponds to 250 μm and the scale bar of ascus pictures corresponds to 100 μm. (D) Quantification of spore morphology. Only mature black spores (10 fruiting bodies of each cross) were evaluated and the relative amount of elliptical spores is indicated. Evaluated ascospores of specific crosses: wild type × wild type n = 1691 spores, ΔPaSnf1 × ΔPaSnf1: n = 760; wild type × ΔPaSnf1: n = 1595; ΔPaSnf1/Flag::PaSnf1 × ΔPaSnf1: n = 1423. (E) Relative amount of fruiting bodies resulting from the spermatization of the wild type (n = 6) and ΔPaSnf1 (n = 6) with different male or female partners. (B,D,E) The values presented are mean values ± SD (∗∗∗p < 0.001; ∗∗p < 0.01, two-tailed Student’s t-test).
In our study, we first investigated the content of the fruiting bodies derived from confrontational crosses of different strains by light microscopy (Figure 2C). In crosses of two wild-type strains of the opposite mating type, normal mature spores occur after 12 days of incubation at 27°C (constant light). In contrast, after this time, only few mature spores are visible in fruiting bodies of ΔPaSnf1 × ΔPaSnf1 crosses. However, after two additional days of incubation, in the mutant’s fruiting bodies more mature spores occur (Figure 2C, right) demonstrating that ablation of PaSNF1 delays spore development. Even more, the morphology of the ascospores in these crosses is clearly distinct from those in which one parent is wild type. The ascospores derived from a ΔPaSnf1 × ΔPaSnf1 cross are rather round instead of elliptical, suggesting that PaSNF1 is involved in spore morphology regulation. The presence of one PaSnf1 gene within the fruiting bodies is sufficient (e.g., in a wild type × ΔPaSnf1 cross) to induce normal spore morphology (Figures 2C,D). Complementation with an ectopically integrated PaSnf1 gene fused to a Flag tag restores spore morphology (Figures 2C,D).
Next, we performed spermatization experiments and found that female fertility is strongly affected in PaSnf1 deletion strains. While spermatia of the ΔPaSnf1 strain are readily able to induce fruiting body formation on wild-type acceptor strains, the spermatization of ΔPaSnf1 by spermatia of the wild type (reciprocal cross) leads to a reduction in fruiting body formation by 57% (Figure 2E). In crosses of male and female ΔPaSnf1, the number of fruiting bodies is not further reduced, indicating that it is female fertility that is impaired in the mutant.
Finally, we investigated the impact of PaSnf1 deletion on lifespan. We analyzed individual cultures derived from monokaryotic ascospores grown at 27°C on the M2 medium containing glucose as a carbon source (Figure 3A and Supplementary Table 2). As described earlier (Fischer et al., 2013), ΔPaClpP is long-lived. Compared to the wild type, the PaSnf1 deletion strain shows a 22% mean lifespan extension (Figure 3B and Supplementary Table 2, mean lifespan 28 days vs. wild-type mean lifespan of 23 days). Complementation of ΔPaSnf1 with an ectopically integrated Flag-tagged PaSnf1 gene restores wild-type lifespan (Supplementary Figure 1E and Supplementary Table 3). Most surprisingly, in contrast to our initial expectation, the ΔPaClpP/ΔPaSnf1 double mutant did not display a wild-type like lifespan. In fact, the simultaneous deletion of the two genes was found to be synergistic leading to a spectacular extension of the mean lifespan to more than 186 days (Figures 3A,B and Supplementary Table 2). This is an increase of at least 174 and 564%, respectively, in comparison to the PaClpP and PaSnf1 single deletion strains. At note, the exact values could not be calculated yet, since some of the double mutant strains are still alive. It is striking that in the double mutant the mortality rate is strongly reduced in particular after about 50% of the investigated individual cultures died (Figure 3A).
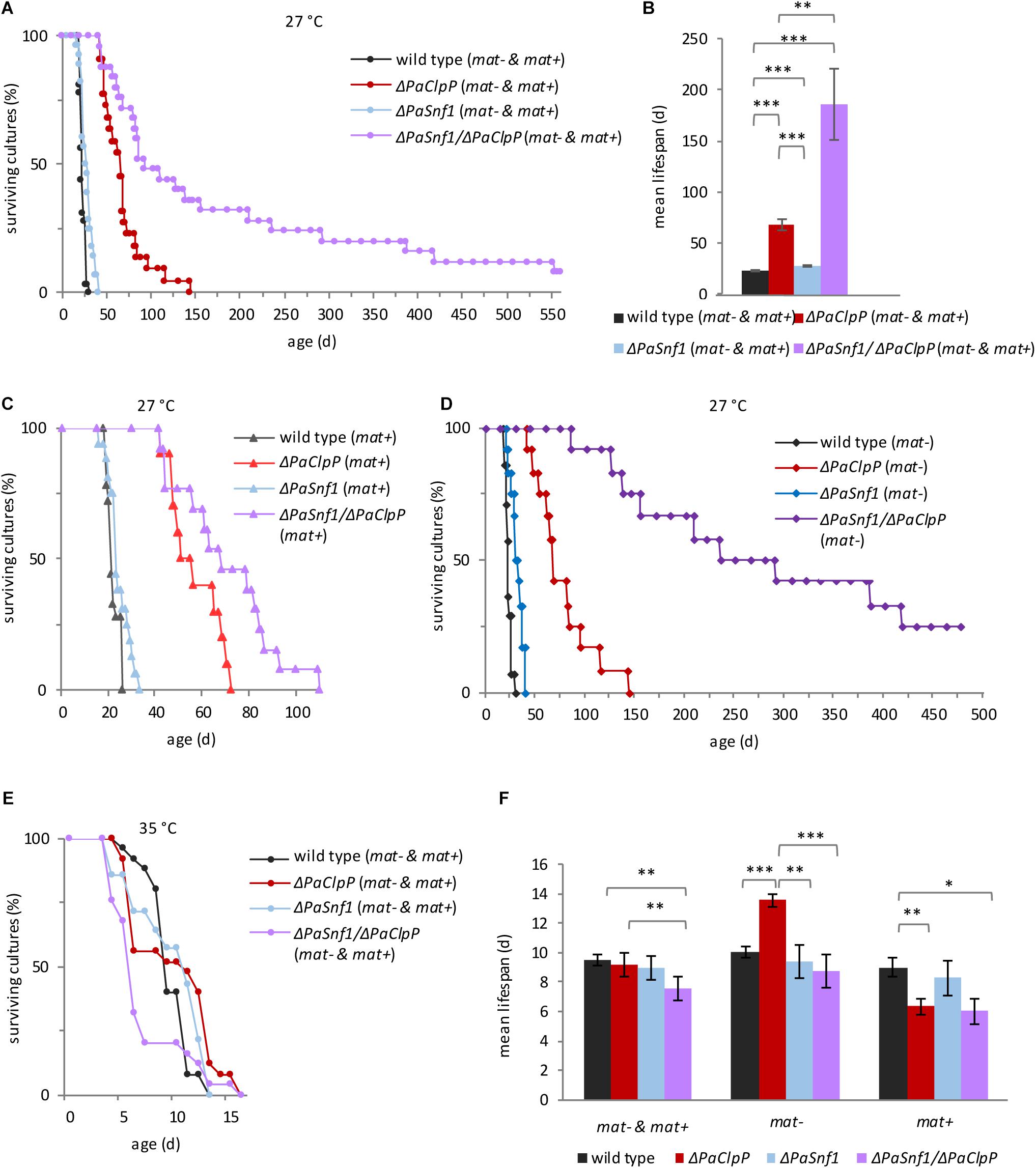
Figure 3. PaSNF1 impacts on lifespan. (A,B) Lifespan analysis at 27°C of ΔPaSnf1 (n = 28, p < 0.001), ΔPaClpP (n = 22, p < 0.001), ΔPaSnf1/ΔPaClpP (n = 25, p < 0.001), and the wild type (n = 32), grown on M2 medium in constant light. (B) Mean lifespan of cultures from panel (A). (C) Lifespan analysis at 27°C of ΔPaSnf1 (mat+) (n = 16, p = 0.0389), ΔPaClpP (mat+) (n = 10, p = 0.0058), ΔPaSnf1/ΔPaClpP (mat+) (n = 13, p < 0.001), and wild type (mat+) (n = 18) on M2 medium in constant light. “(mat+)” represents the mating type “plus” (rmp1-2). (D) Lifespan analysis at 27°C of ΔPaSnf1 (mat–) (n = 12, p < 0.001), ΔPaClpP (mat–) (n = 12, p < 0.001), ΔPaSnf1/ΔPaClpP (mat–) (n = 12, p < 0.001), and wild type (mat–) (n = 14) on M2 medium in constant light. “(mat–)” represents the mating type “minus” (rmp1-1). (E) Lifespan analysis at 35°C of ΔPaSnf1 (n = 17, p = 0.523), ΔPaClpP (n = 28, p = 0.691), ΔPaSnf1/ΔPaClpP (n = 19, p = 0.024), and wild type (n = 25), grown on M2 medium in constant light. (F) Mean lifespan of cultures from panel (E). (A,B,E, and left group in panel F): the data of strains of both mating types are combined (“mat– & mat+”). (A,C–E): p-values of the lifespan curves in comparison to wild type were determined by SPSS with three different statistic tests. A compilation of all p-values is provided in Supplementary Tables 2, 3. (B,F) Shown are mean values ± SEM (∗∗∗p < 0.001; ∗∗p < 0.01; *p < 0.05, two-tailed Student’s t-test).
This biphasic progression of the survival curve is rather unusual, and therefore, we examined the lifespan data in more detail. In P. anserina, it is known that the mating type has an impact on lifespan. In most cases (e.g., wild type), this impact is rather moderate. However, in combination with some genes it can be stronger (Belcour et al., 1991; Contamine et al., 1996, 2004; Sellem et al., 2005; Adam et al., 2012). In fact, this mating type-linked effect is due to two alleles of the rmp1 gene that are located in very close proximity to the mating-type alleles and therefore are not separated by intrachromosomal recombination during sexual reproduction and co-inherited with the mating type of the parental strain (Contamine et al., 2004). We therefore decided to split the combined lifespan curves of Figure 3A in which mating type “plus” and mating type “minus” isolates are both presented into two mating type-specific curves (Figures 3C,D). Significantly, we found a strong effect of the mating type on lifespan of ΔPaClpP/ΔPaSnf1. While the lifespan of mating-type “plus” double mutants are only moderately increased, in mating-type “minus” background lifespan is dramatically increased. Thus, the original biphasic progression of the survival curve is due to the fact that the “plus” strains have a shorter lifespan than the “minus” strains. In contrast, the effect of the mating type on lifespan of the wild type and the PaSnf1 and PaClpP single deletion strains was rather moderate (Figures 3C,D and Supplementary Table 3). Because of this observation, we provide lifespan values for both combined mating types (Supplementary Table 2) and lifespans separated for the two mating types (Supplementary Table 3).
From a previous study, we know that the observed lifespan extension may only be valid under the specific laboratory conditions applied in the corresponding study (Weil et al., 2011). In particular, incubation temperature, which – under natural conditions – strongly varies during the day, may have a clear impact. In order to investigate such an impact, we analyzed cultures derived from monokaryotic spores of all strains incubated at 35°C (Figures 3E,F and Supplementary Tables 2,3). All strains showed a strong reduction in lifespan when grown at this temperature. The mean lifespan of wild type, ΔPaClpP, and ΔPaSnf1 is 10, 9, and 10 days, respectively (Figure 3E and Supplementary Table 2) compared to 23, 68, and 28 days at 27°C (Figure 3B and Supplementary Table 2). In wild type and ΔPaSnf1, the lifespan at 35°C is not affected by the mating type. However, interestingly, strains ablated for PaCLPP with rmp1-2 (mating type “plus”) have a strongly reduced lifespan (Figure 3F). In contrast, the ΔPaClpP strain with rmp1-1 allele (mating type “minus”) has a longer lifespan than wild type “minus” (Figure 3F).
PaSNF1 Is Required for Autophagy
AMP-activated protein kinase is known to activate autophagy through different mechanisms, e.g., directly by phosphorylation of ULK1 and BECLIN1 (the mammalian homologs of ATG1 and ATG6, Kim et al., 2011, 2013) and indirectly by inhibiting TOR (Russell et al., 2014). Therefore, we expected that ablation of PaSNF1 results in impaired autophagy. To test this possibility, we measured autophagy in the wild type, ΔPaSnf1, and ΔPaSnf1/ΔPaClpP using the previously established reporter degradation assay (Knuppertz et al., 2014). In this assay, after autophagic degradation of the cytosolic reporter protein PaSOD1::GFP, the GFP moiety remains stable in the fungal vacuole and can be detected by western blot analyses. The results obtained with this reporter are independent of the mating type, and therefore, we presented the data of “plus” and “minus” isolates together. As expected, in 7-day-old ΔPaSnf1 strains autophagy is significantly impaired and reduced amounts of free GFP are found (Figures 4A,B). At 20 days of age, this difference is reduced and no longer significant, suggesting that aging induces PaSNF1-independent autophagy. Surprisingly, in 7-day-old ΔPaSnf1/ΔPaClpP autophagy is indistinguishable from that of 7-day-old wild type. In contrast, in 20-day-old cultures autophagic flux is significantly lower in the double mutant compared to wild type. Thus, a beneficial induction of autophagy, as it is known for the single PaClpP deletion mutant (Knuppertz and Osiewacz, 2017), is not responsible for the lifespan extension of the double mutant.
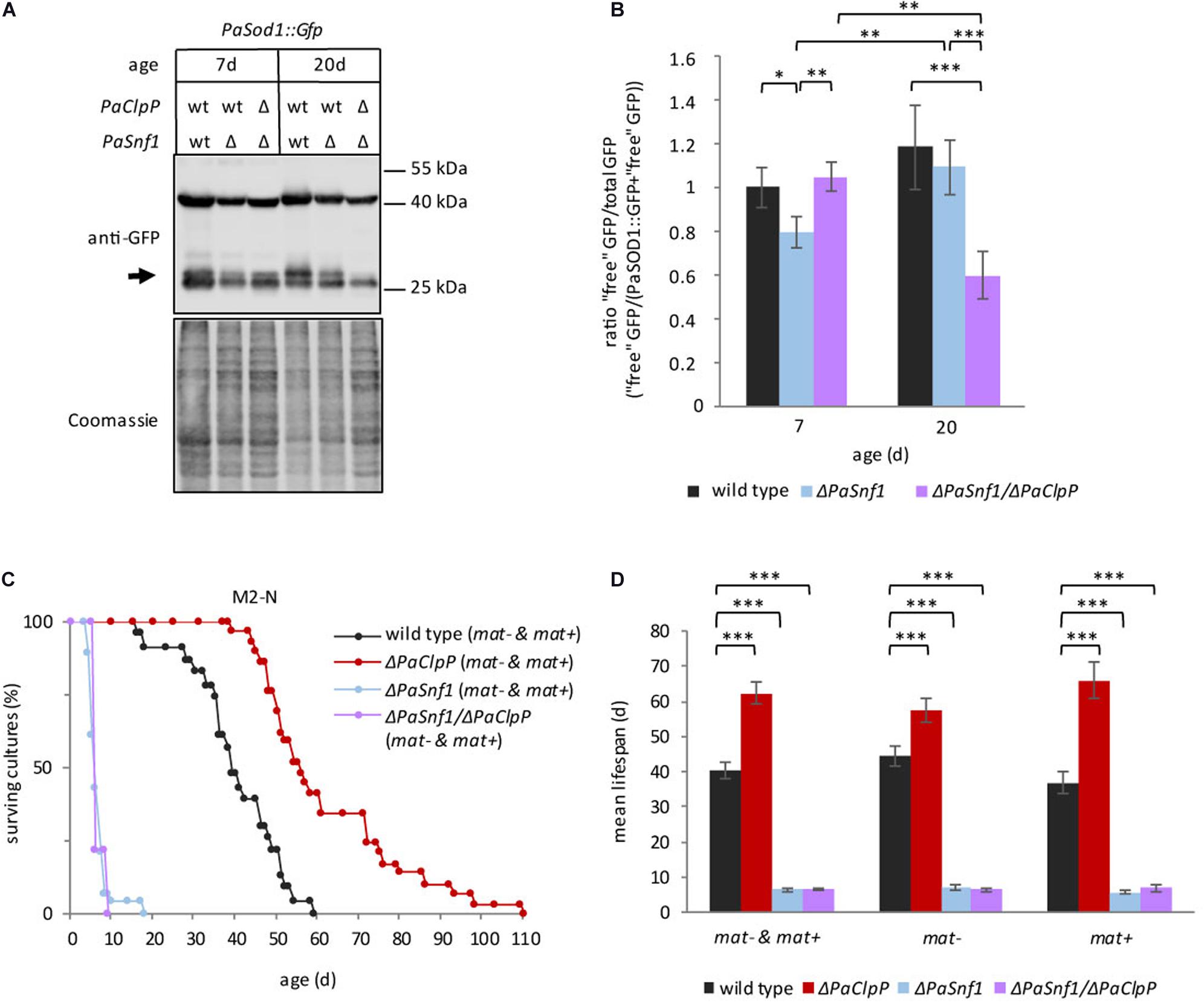
Figure 4. Deletion of PaSnf1 leads to impaired autophagy. (A) Western blot analysis of ΔPaSnf1 and wild type expressing the autophagy marker gene PaSod1::Gfp of 7- (PaSod1::Gfp n = 5; ΔPaSnf1/PaSod1::Gfp n = 4; ΔPaSnf1/ΔPaClpP/PaSod1::Gfp n = 3) and 20-day-old (PaSod1::Gfp n = 5; ΔPaSnf1/PaSod1::Gfp n = 5; ΔPaSnf1/ΔPaClpP/PaSod1::Gfp n = 4) strains. 50 μg of total protein extract was separated in a 12% SDS polyacrylamide gel. After transfer on a PVDF membrane, the gel was stained with Coomassie and serves as loading control. The signal of “free GFP” is marked with an arrow. (B) Quantification of the relative amount of “free GFP”/total GFP. The values shown are mean ± SD. (C) Lifespan analysis of ΔPaSnf1 (n = 28, p < 0.001), ΔPaClpP (n = 29, p < 0.001), ΔPaSnf1/ΔPaClpP (n = 9, p < 0.001), and wild type (n = 23), grown on M2 medium without nitrogen and with 1.5-fold glucose at 27°C (constant light). P-values of the lifespan curves in comparison to wild type were determined by SPSS with three different statistic tests. All p-values are provided in Supplementary Tables 2, 3. (D) Mean lifespan of cultures from (C) ± SEM. “mat– & mat+” represents the mean lifespan taking together both mating types; “mat–” shows the mean lifespan of cultures with mating type “minus” (rmp1-1). “mat+” shows the mean lifespan of cultures with mating type “plus” (rmp1-2). (∗∗∗: p < 0.001; **: p < 0.01; *: p < 0.05, two-tailed Student’s t-test).
Next, we tested whether or not the observed impairment in autophagy of the PaSnf1 deletion strain affects the ability to respond to nitrogen limitation, a well-known condition inducing autophagy in different organisms including P. anserina (Todde et al., 2009; Mukaiyama et al., 2010; Knuppertz et al., 2014; Li et al., 2015). We determined the lifespan of all investigated strains on M2 medium without urea as nitrogen source and supplemented with additional glucose (Figures 4C,D). Nitrogen limitation leads to a pronounced lifespan extension in the wild type (40 days, Figure 4D, vs. 23 days on M2, Figure 3B, respectively, Supplementary Table 2), which is independent of the mating type (Figure 4D and Supplementary Table 3). The mean lifespan of ΔPaClpP is slightly reduced compared to the lifespan on M2 media with nitrogen (62 days, Figure 4D, vs. 68 days on M2, Figure 3B, Supplementary Table 2, respectively) but still longer than that of the wild type. In contrast, under nitrogen-replete conditions both ΔPaSnf1 and ΔPaSnf1/ΔPaClpP are characterized by a strongly reduced lifespan, which is 7 days for both strains (Figure 4D and Supplementary Table 2) compared to 28 days and at least 186 days on M2 (Figure 3B and Supplementary Table 2). These results identified a pivotal role of PaSNF1 in lifespan control under nitrogen-limiting conditions. Moreover, under these conditions, the deletion of PaSnf1 is epistatic over the deletion of PaClpP again indicating a link of PaClpP and PaSnf1 controlled pathways.
Ablation of PaSNF1 Affects Mitochondrial Dynamics and Respiration
Since AMPK is known to be involved in the regulation mitochondrial homeostasis via mitochondrial biogenesis and dynamics (reviewed in Herzig and Shaw, 2018), we next investigated the impact of the deletion of PaSnf1 on mitochondria. To this end, we analyzed the mitochondrial morphology of wild type, the two single deletion mutants, and the ΔPaSnf1/ΔPaClpP double mutant by fluorescence microscopy (Figure 5A). As described earlier, during aging of P. anserina mitochondria become fragmented (Scheckhuber et al., 2007). In 20-day-old wild-type hyphae all mitochondria are fragmented, while in ΔPaClpP some mitochondria remain filamentous (Figure 5A). Interestingly, we found that both ΔPaSnf1 and ΔPaSnf1/ΔPaClpP still contain a substantial fraction of filamentous mitochondria even at 20 days of age – independent of the mating type. Thus, as described in other systems (Toyama et al., 2016; Willis et al., 2018; Chen et al., 2019; Wu et al., 2020), PaSNF1 appears to be crucial for fission of P. anserina mitochondria.
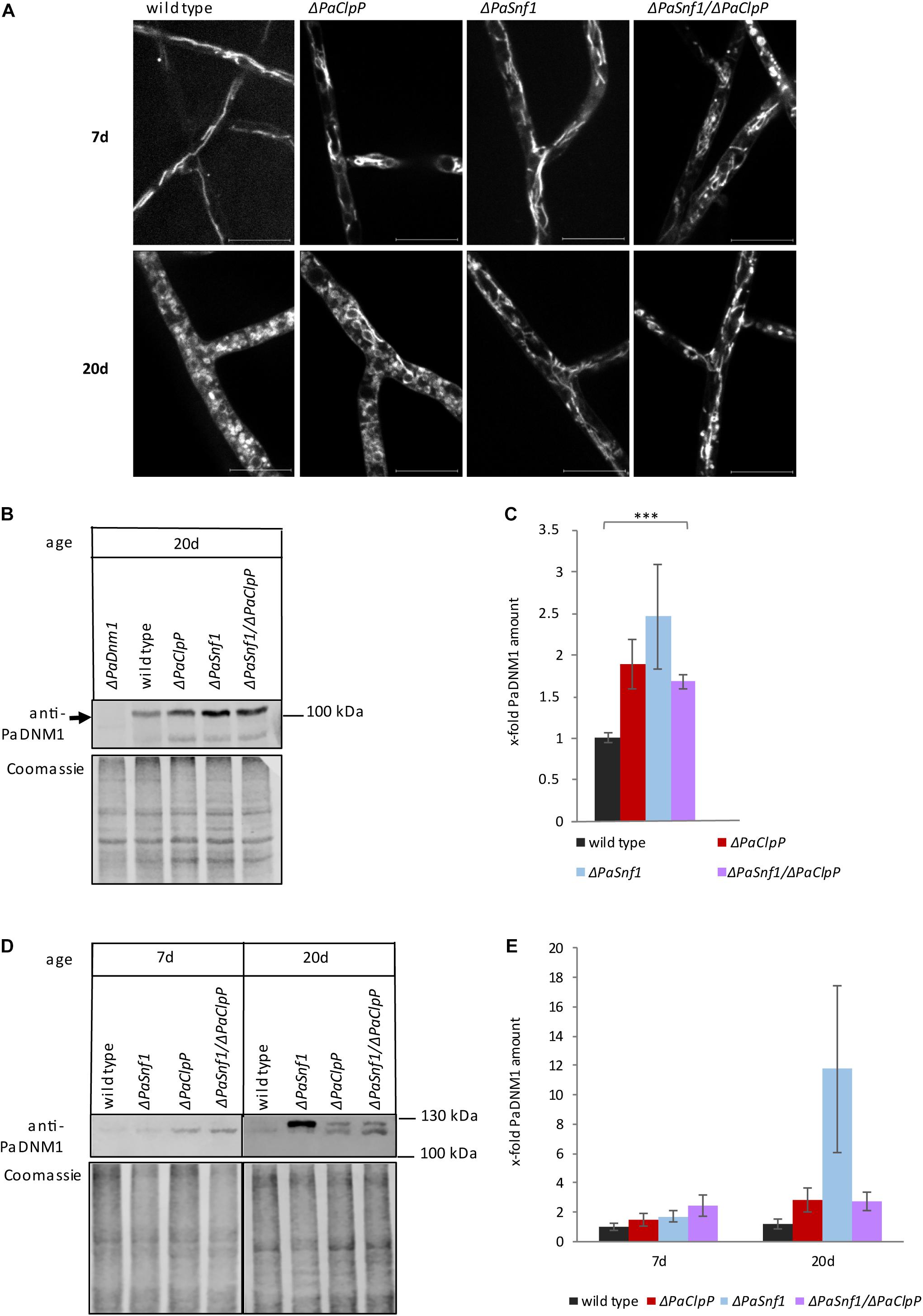
Figure 5. Ablation of PaSNF1 affects mitochondrial morphology. (A) Confocal laser scanning fluorescence microscopy with stained mitochondria of 7- and 20-day-old ΔPaSnf1, ΔPaClpP, ΔPaSnf1/ΔPaClpP, and the wild type. After aging on M2 medium at 27°C (constant light), pieces of mycelium were used to inoculate microscope slides with a central depression filled with M2 media. After a 1-day incubation at 27°C (constant light), the mycelium was incubated with MitoTracker Red (2 μM, 15 min, in the dark) to visualize mitochondria. The scale bar refers to 10 μm. (B) Western blot analysis of 100 μg total protein extracts of 20-day-old ΔPaSnf1 (n = 3), ΔPaClpP (n = 4), ΔPaSnf1/ΔPaClpP (n = 4), and wild-type strains (n = 3) with an anti-PaDNM1 antibody. (C) Quantification of PaDNM1 (B) after normalization to the Coomassie-stained gel. (D) Western blot analysis of 50 μg mitochondrial protein extracts of 7- and 20-day-old ΔPaSnf1 (n = 3), ΔPaClpP (n = 3), ΔPaSnf1/ΔPaClpP (n = 3), and wild type strains (n = 3) with an anti-PaDNM1 antibody. (E) Quantification of PaDNM1 (D) after normalization to the Coomassie-stained gel. (B,D): 8% SDS polyacrylamide gels were used. After transfer, the gels were stained with Coomassie and served as loading control. The values shown are mean ± SEM (∗∗∗p < 0.001, two-tailed Student’s t-test).
Recently, in HeLa cells the translocation of AMPK from the cytosol to mitochondria was demonstrated upon energy stress, thereby regulating mitochondrial division and autophagy (Hu et al., 2020). We wondered whether ablation of PaSNF1 might impair the recruitment of the mitochondrial fission factor PaDNM1 to mitochondria leading to impaired mitochondrial fission. Scheckhuber et al. (2007) showed that the PaDnm1 gene is only poorly expressed in young cultures. Therefore, we first analyzed the amount of PaDNM1 in total protein extracts of 20-day-old cultures (Figures 5B,C). Interestingly, in all mutants, the amount of PaDNM1 in total protein extracts is higher than in the wild type. However, only the increase in the double mutant is significant (Figure 5C). Subsequently, we investigated whether recruitment of PaDNM1 to mitochondria is altered in the mutants in comparison to the wild type. Surprisingly, on first glance, in purified mitochondria of 20-day-old ΔPaSnf1 the amount of PaDNM1 appears to be elevated (Figures 5D,E). However, the different biological replicates show a high variability which is not dependent on the mating type. The differences in the abundance of PaDNM1 are not significant, and the ablation of PaSNF1 thus does not impair PaDNM1 recruitment to the mitochondrial fraction. Nonetheless, PaDNM1 function appears to be affected in the mutant strains. While in all young cultures and in 20-day-old wild types only one PaDNM1 protein of about 120 kDa is visible, in 20-day-old mutant strains an additional band of about 125 kDa can be detected (Figure 5D). This band is very prominent in ΔPaSnf1, suggesting that PaSNF1 ablation may affect post-translational modification of PaDNM1. It thus is possible that this modification may lead to inactivation of PaDNM1 and, as a consequence, to reduced mitochondrial fission and to filamentous mitochondria even in 20-day-old cultures. In the double mutant as well as in ΔPaClpP, both PaDNM1 bands are present in equal amounts. Thus, the observed differences in mitochondrial morphology between these two strains cannot be explained by distinct post-translational modification of PaDNM1.
In search of a possible explanation for the observed differences in mitochondrial morphology, we carefully inspected the transcriptome data of the PaSnf1 deletion mutant which was recently published by Li et al. (2020). Interestingly, one gene, PODANS_1_1780, was found to be strongly upregulated in ΔPaSnf1 compared to the wild type. The corresponding protein, PODANS_1_1780 (hereafter termed PaMDM38), is a homolog of yeast MDM38 and human LETM1 protein (Nowikovsky et al., 2007; Nakamura et al., 2020), proteins which are implicated in the regulation of mitochondrial morphology. Therefore, we investigated PaMdm38 transcription by qRT-PCR and found that the gene is indeed upregulated in ΔPaSnf1 (Supplementary Figure 2), but not in other mutants, suggesting that post-translational modification of PaDNM1 and PaMdm38 expression is linked.
Our results indicate that ablation of PaCLPP is able to counteract changes in PaDNM1 recruitment and post-translational modification as well as the induction of PaMdm38 expression, suggesting that PaSnf1 affects mitochondrial pathways. We therefore tested mitochondrial function in the different strains by measuring oxygen consumption of fungal mycelium at three different age stages (7, 12, and 20 days) in fresh liquid medium containing 1% glucose (Figure 6A). We found that aging does not significantly affect the oxygen consumption rate (OCR) in the investigated strains. Markedly, the OCR of ΔPaClpP is lower than the OCR of the wild type as described before (Knuppertz and Osiewacz, 2017). In contrast, at these conditions, OCR is not significantly affected in ΔPaSnf1 (Figure 6A). A small but significant increase in OCR is observed in 12-day-old ΔPaSnf1/ΔPaClpP (Figure 6A). This moderate impact on OCR seen upon PaSNF1 ablation is in agreement with the observation that yeast SNF1 only becomes activated upon glucose depletion (Vallejo et al., 2020). If this response is also true in P. anserina, ablation of PaSNF1 should not affect OCR at high glucose concentrations. However, it can be expected that on medium containing non-fermentable glycerol as the sole carbon source, PaSNF1 is important. To test this possibility, we performed a lifespan analysis on M2 medium containing glycerol. On this substrate, all strains show a reduction in mean lifespan. It is 23 days in wild type, 9 days in ΔPaClpP, 10 days in ΔPaSnf1, and 7 days in ΔPaSnf1/ΔPaClpP, compared to 23, 68, 28, and at least 186 days on M2 with 1% glucose (Figures 6B,C vs. Figure 3A,B and Supplementary Table 2). We also tested whether or not rmp1 affects survival on glycerol medium (Supplementary Table 3). Indeed, similar to the situation at 27 and 35°C with 1% glucose, also growth on glycerol is affected by the rmp1 allele (Figure 6C and Supplementary Table 3). Most intriguingly, the lifespan of wild-type strains is clearly higher in the presence of rmp1-1 (mating type “minus”) than rmp1-2 (mating type “plus”). On this medium, rmp1-1 or rmp1-2 has no effect in the mutant strains ΔPaClpP, ΔPaSnf1, and ΔPaSnf1/ΔPaClpP. Thus, rmp1-1 ensures the ability to use the non-fermentable carbon source glycerol in the wild type. This ability is reduced in all deletion mutants, clearly demonstrating that they are characterized by impaired mitochondrial function although mitochondria morphology is filamentous.
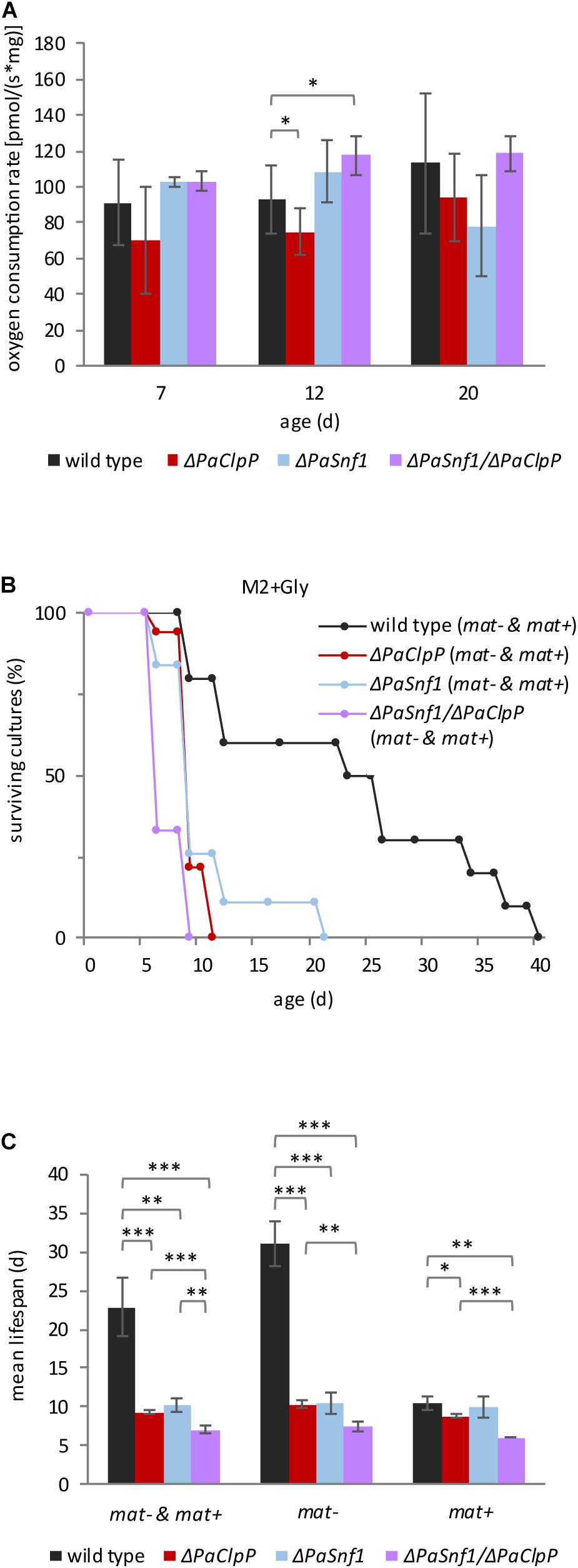
Figure 6. Lack of PaSNF1 does not significantly affect oxygen consumption and impairs growth on glycerol. (A) Oxygen consumption rate (OCR) of mycelium from 7-, 12-, and 20-day-old ΔPaSnf1, ΔPaClpP, ΔPaSnf1/ΔPaClpP, and wild type. [7 days: ΔPaSnf1 (n = 3), ΔPaClpP (n = 6), ΔPaSnf1/ΔPaClpP (n = 4), and wild type (n = 12) // 12 days: ΔPaSnf1 (n = 4), ΔPaClpP (n = 6), ΔPaSnf1/ΔPaClpP (n = 4), and wild type (n = 13) // 20 days: ΔPaSnf1 (n = 3), ΔPaClpP (n = 6), ΔPaSnf1/ΔPaClpP (n = 3), and wild type (n = 10)]. The values presented are mean ± SD. (B) Lifespan analysis of ΔPaSnf1 (n = 19, p < 0.001), ΔPaClpP (n = 18, p < 0.001), ΔPaSnf1/ΔPaClpP (n = 9, p < 0.001), and wild type (n = 10), grown on M2 medium at 27°C with glycerol as carbon source instead of glucose. The p-values of the lifespan curves in comparison to wild type were determined by SPSS with three different statistic tests. A compilation of all p-values can be found in Supplementary Tables 2, 3. (C) Mean lifespan of cultures from panel (B), the values presented are mean ± SEM. “mat– & mat+” represent the combined mean lifespan of both mating types; “mat–” shows the mean lifespan of cultures with mating type “minus” (rmp1-1). “mat+” shows the mean lifespan of cultures with mating type “plus” (rmp1-2) (∗∗∗p < 0.001; ∗∗p < 0.01; ∗p < 0.05, two-tailed Student’s t-test).
From these observations, several conclusions can be drawn: First, the lifespan of P. anserina carrying the rmp1-2 allele is reduced on the medium, which requires functional mitochondria indicating a critical lifespan-limiting role of the rmp1 gene. Second, survival of ΔPaClpP is significantly more strongly compromised on glycerol than survival of the wild type. This demonstrates that mitochondrial function is impaired in this mutant, which is in accordance with the observed reduction in OCR. Thus, ablation of PaCLPP affects mitochondrial function independent of the available carbon source. Third, although ablation of PaSNF1 alone does not reduce OCR at 1% glucose, it strongly impairs survival on glycerol containing M2 (Figures 6B,C). Thus, PaSNF1 seems to control mitochondrial function only in the glycerol medium, which suggests that PaSNF1 is required for the metabolic switch from glycolysis to respiration. Obviously, PaCLPP and PaSNF1 affect mitochondrial function independently. Overall, our results uncover a hitherto unknown mechanism by which PaSNF1, RMP1, and PaCLPP synergistically interact with each other and affect the lifespan of P. anserina.
Discussion
The previously generated P. anserina mutant in which the gene coding for mitochondrial CLPXP complex as part of the mitochondrial protein quality control network is deleted displays an unexpected pronounced lifespan extension (Fischer et al., 2013). In this mutant, clear changes in respiration were observed (Knuppertz and Osiewacz, 2017). Specifically, we found a reduction in complex I- and II-dependent respiration and the induction of an alternative salicyl hydroxamate-sensitive oxidase (AOX), an enzyme that has repeatedly been demonstrated to be induced in a number of different longevity mutants of P. anserina (Scheckhuber and Osiewacz, 2008). Despite that respiration via the alternative pathway is less efficient than that via the standard cytochrome c oxidase (COX)-dependent pathway, we found no significant reduction of the ATP content in the mutant. The observed induction of autophagy in the mutant appears to be responsible for the conservation of cellular ATP.
In the current study, we extended our investigations and first performed a comparative, age-related metabolome analysis of P. anserina wild-type and ΔPaClpP cultures. This analysis revealed changes of various metabolites. Most significantly, compared to young wild types, the metabolites of the TCA cycle, glycolysis, and amino acid level are affected. In addition, the level of AMP is increased, and ATP level is decreased in young ΔPaClpP. On the one hand, this increase in the AMP/ATP ratio suggests that protein kinase SNF1 is activated in the young mutant. However, on the other hand, the amount of 1,6-P hexose is increased in the young mutant compared to wild-type, which might impair SNF1 activation, as it has recently been shown in mammals (Zhang et al., 2017). Thus, based on the metabolome data, the role of SNF1 in ΔPaClpP is unclear. To address this issue, we generated a PaSnf1 deletion mutant in which the catalytic α-subunit of AMPK is ablated and used this mutant to select a ΔPaClpP/ΔPaSnf1 double mutant.
Since SNF1/AMPK is implicated in autophagy induction (Herzig and Shaw, 2018; Tamargo-Gomez and Marino, 2018), we were not surprised to see that ablation of PaSNF1 reduces the autophagic degradation of reporter proteins. Concordantly, the lifespan-extending effect of nitrogen depletion which is visible in wild-type and ΔPaClpP strains is lost upon PaSnf1 deletion. Thus, autophagy induction and the lifespan-promoting role of nitrogen depletion both depend at least partially on PaSNF1.
In our study, we observed a prominent role of PaSNF1 in development. In concordance with a parallel study by Li et al. (2020), we found that the growth rate of cultures is reduced in the PaSnf1 deletion strain and sexual development is affected. However, in contrast to their study, although delayed in comparison to the wild type, we observed the formation of mature ascospores which are functional and able to form colonies. Moreover, we found that spermatization of cultures derived from a mononuclear ascospore of one mating type with spermatia isolated from a culture of a mononuclear ascospore of the opposite mating type leads to the formation of round spores instead of elliptical spores as they are formed in crosses in which one or both parents are wild type. Clearly, PaSNF1 is involved in the control of ascospore formation and morphology. Moreover, we found that it is the formation of female gametangia that is affected in the PaSnf1 deletion strain. The discrepancies in the two studies are probably due to different designs of the experiments. In their study, Li et al. (2020) used P. anserina wild-type strain “S” and we used “s.” Moreover, we used cultures derived from mononucleate small ascospores instead of large binucleate ascospores that, without contact to a second parent, give rise to fruiting body formation. Apart from the investigation of sexual development, Li et al. (2020) investigated the impact of PaSNF1 on sterigmatocystin biosynthesis, lignocellulosic degradation, and stress resistance but did not address the role in aging and lifespan control, the focus of our study.
We found that deletion of PaSnf1 leads to moderate lifespan extension. More surprisingly, the ΔPaClpP/ΔPaSnf1 double mutant was not characterized by a reversion of the long-lived phenotype of the PaClpP deletion mutant as we expected it since the AMP/ATP ratio in this mutant was increased and PaSNF1 is involved in the induction of autophagy and longevity in ΔPaClpP. It thus appears that an AMPK-independent induction of autophagy occurs in the PaClpP deletion mutant and the PaCLPXP- and PaSNF1-controlled pathways act synergistically. Moreover, it is striking that, in comparison to the wild type and the two single deletion mutants, the maximum lifespan of the double mutant is extremely increased (more than 708%, compared to the wild type) due to a strong decrease of the rate of mortality especially in cultures of mating type “minus.” This dependency on the mating type can be attributed to the rmp1 gene (PODANS _1_20180), a gene that is present in two different alleles in P. anserina wild-type strains “s” and “S” and tightly linked to the mating-type locus. In mating type “minus,” the rmp1-1 allele is found, in mating type “plus” rmp1-2. The rmp1-1 allele is assumed to represent the fully functional version of rmp1 (Contamine et al., 2004), which encodes an essential 1000 amino acids protein (UniProt Q70GH5). The protein bears a mitochondrial targeting peptide, and a GFP-tagged version of the protein has been localized to filamentous cellular structures and to the cytosol depending on the cell type and developmental stage (Contamine et al., 2004). Moreover, RMP1 has previously been linked to respiratory complex assembly (Sellem et al., 2005). A BLAST analysis identified sequence homologs of this protein only in multicellular ascomycetes. The protein contains a so-called SLS domain which is part of the Saccharomyces cerevisiae ScSLS1 protein (UniProt P42900), a mitochondrial integral membrane protein whose ablation does not affect growth on glucose but is lethal on a non-fermentable medium (Rouillard et al., 1996). More recently, this protein has been implicated in mitochondrial gene expression (Bryan et al., 2002). ScSLS1 and the mitochondrial matrix protein ScNAM1 together with mtRNA polymerase have been shown to coordinate transcription and translation of mtDNA-encoded gene products ensuring efficient mitochondrial translation (Rodeheffer and Shadel, 2003).
The prominent role of rmp1 in wild-type lifespan control is specifically seen on the medium containing glycerol. In the presence of rmp1-2, the lifespan of wild type is strongly reduced compared to the wild type that contains the rmp1-1 allele, suggesting that mitochondrial function strongly depends on the presence of rmp1-1. Ablation of PaCLPP, PaSNF1, or both, strongly reduces lifespan on glycerol medium independent of rmp1. Thus, ablation of PaCLPP as well as PaSNF1 impairs the ability to use the non-fermentable carbon source glycerol. Interestingly, on first glance, the impaired growth on glycerol does not fit to the morphology of the mitochondria of the PaSnf1 deletion strains which are filamentous and therefore expected to be functional. However, since AMPK/SNF1 is known to be crucial for mitochondrial fission (Toyama et al., 2016; Willis et al., 2018; Chen et al., 2019; Wu et al., 2020), filamentous mitochondria, which are observed even at late age in the PaSnf1 deletion mutant, rather seem to reflect the inability to undergo fission than being indicative for functional mitochondria. Supporting this idea, we observed an altered migration behavior of the fission factor PaDNM1 in the PaSnf1 deletion strains.
In yeast, SNF1 controls glycolytic flux and mitochondrial respiration in a glucose-dependent manner. At 1% glucose, a concentration which we used in M2 medium in our studies with P. anserina, ScSNF1 sustains glycolytic flux and increases respiration (Martinez-Ortiz et al., 2019). Such a function of PaSNF1 in the ΔPaClpP background appears to be not possible because, as revealed by the metabolome analysis, glycolytic flux seems to be impaired in the PaClpP deletion strain leading to the accumulation of C6 sugars and depletion of C3 carbohydrates and because complex I- and complex II-dependent respiration is impaired (Knuppertz and Osiewacz, 2017). We suggest that in this situation functional PaSNF1 is detrimental and deletion of PaSnf1 has a beneficial effect on a medium with 1% glucose. Interestingly, this synergistic effect is strongly reinforced in the presence of the rmp1-1 allele, bringing mitochondrial translation efficiency into play. In mice, CLPP ablation was found to decrease mitochondrial translation (Szczepanowska et al., 2016). In wild-type mice, CLPP regulates the turnover of ERAL1, a putative 12S rRNA chaperone, which, for efficient translation, has to be timely removed from maturating mitoribosomes. Accumulation of ERAL1 due to ablation of CLPP delays maturation of mitoribosomes (Szczepanowska et al., 2016), thereby diminishing protein synthesis. In certain settings, this effect on protein biosynthesis may be beneficial. For instance, in a mouse mutant with dysregulated mitochondrial translation, ClpP deletion has been found to result in a robust lifespan extension (Seiferling et al., 2016). In the corresponding double mutant, more proficient vs. abortive protein synthesis has been observed in mitochondria. It appears that the delay in mitoribosome maturation in ClpP mice allows to overcome erroneous translation and ameliorates the phenotype by increasing the lifespan. It is well documented that not only protein quality control is affected during aging of biological systems but also protein synthesis (Anisimova et al., 2018; Klaips et al., 2018). The latter not only holds true for cytosolic protein synthesis but also is relevant for mitochondrial translation as it has been shown in human skeletal muscle (Rooyackers et al., 1996). In P. anserina, inhibition of mitochondrial translation by streptomycin, tiamulin, and other inhibitors results in a robust lifespan extension (Esser and Tudzynski, 1977; Tudzynski and Esser, 1977, 1979), which is not seen upon application of inhibitors for cytosolic translation (Tudzynski and Esser, 1977). Thus, mitochondrial rather than cytosolic translation seems to critically intervene into aging. Ablation of PaCLPP resulting in a slowdown of mitochondrial translation may mitigate the consequences of age-dependent impairments in mitochondrial translation leading to the observed lifespan extension of the PaClpP deletion mutant. Since ablation of PaSNF1 impairs the switch from glycolysis to respiration, a functional rmp1 allele is not required, explaining the rather minor effect of the mating type on lifespan. However, concomitant ablation of PaCLPP strongly challenges mitochondrial function and, under these conditions, a fully functional rmp1 allele is highly beneficial, leading to an extreme increase in lifespan.
Such a lifespan extension is observed not only under standard growth conditions of 27°C but also under heat stress conditions at 35°C. In mammals, the impact of heat stress on mitochondria has been studied. Wilkening et al. (2018) found mitochondrial translation elongation factor Tu to be highly aggregation-prone even under mild heat stress conditions. Thus, under heat stress conditions, mitochondrial translation becomes reduced. In P. anserina, such a reduction in mitochondrial translation may be beneficial in ΔPaClpP incubated at 35°C leading to lifespan extension (Figure 3F). However, again, lifespan extension strongly depends on the presence of the rmp1-1 allele (mating-type “minus”), indicating that functional RMP1 is required for this effect.
Overall, the identification of interactions between the proteolytic pathway controlled by PaCLPP, RMP1, a putative component of the mitochondrial transcription/translation apparatus, and energy metabolism governed by PaSNF1 as the central cellular energy sensor described in the current study calls for further detailed investigations to unravel the precise underlying molecular mechanisms. It will be interesting to see how far these mechanisms are unique for P. anserina or conserved among species. From the available data, the view is mixed. Common substrates of complex I of the respiratory chain have been identified in P. anserina, mammals, and A. thaliana. Other substrates differ among systems. Moreover, the yeasts S. cerevisiae and Schizosaccharomyces pombe lack CLPP and RMP1, the latter of which appears to be specific for multicellular fungi. Nevertheless, work using different biological systems will be instrumental to unraveling the variety of different biological facets of the molecular pathways and their variations, which have been reported in this study, in aging and lifespan control. Such work will certainly also provide important information concerning the role of CLPXP in human health and disease, which has been found in previous investigation (Gispert et al., 2013; Cole et al., 2015; Seo et al., 2016) and certainly will provide clues for therapeutical interventions.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
HO initiated and supervised this study. DH, AH, and HO designed the experiments and analyzed the data. DH, EK, and AH performed the experiments. AH and HD wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)–Project-ID Os75/15-2 and Project-ID 259130777–(SFB 1177) to HO.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.616520/full#supplementary-material
References
Adam, C., Picard, M., Déquard-Chablat, M., Sellem, C. H., Denmat, S. H., and Contamine, V. (2012). Biological roles of the Podospora anserina mitochondrial Lon protease and the importance of its N-domain. PLoS One 7:e38138. doi: 10.1371/journal.pone.0038138
Anisimova, A. S., Alexandrov, A. I., Makarova, N. E., Gladyshev, V. N., and Dmitriev, S. E. (2018). Protein synthesis and quality control in aging. Aging (Albany NY) 10, 4269–4288. doi: 10.18632/aging.101721
Baker, M. J., Tatsuta, T., and Langer, T. (2011). Quality control of mitochondrial proteostasis. Cold Spring Harb. Perspect. Biol. 3:a007559. doi: 10.1101/cshperspect.a007559
Belcour, L., Begel, O., and Picard-Bennoun, M. (1991). A site-specific deletion in mitochondrial DNA of Podospora is under the control of nuclear genes. Proc. Natl. Acad. Sci. U.S.A. 88, 3579–3583. doi: 10.1073/pnas.88.9.3579
Borghouts, C., Kimpel, E., and Osiewacz, H. D. (1997). Mitochondrial DNA rearrangements of Podospora anserina are under the control of the nuclear gene grisea. Proc. Natl. Acad. Sci. U.S.A. 94, 10768–10773. doi: 10.1073/pnas.94.20.10768
Borghouts, C., Scheckhuber, C. Q., Werner, A., and Osiewacz, H. D. (2002). Respiration, copper availability and SOD activity in P. anserina strains with different lifespan. Biogerontology 3, 143–153. doi: 10.1023/a:1015696404723
Borghouts, C., Werner, A., Elthon, T., and Osiewacz, H. D. (2001). Copper-modulated gene expression and senescence in the filamentous fungus Podospora anserina. Mol. Cell Biol. 21, 390–399. doi: 10.1128/MCB.21.2.390-399.2001
Breitenbach, M., Laun, P., Dickinson, J. R., Klocker, A., Rinnerthaler, M., Dawes, I. W., et al. (2012). “The role of mitochondria in the aging processes of yeast,” in Aging Research in Yeast, eds M. Breitenbach, S. M. Jazwinski, and P. Laun (Dordrecht: Springer Netherlands), 55–78.
Brust, D., Hamann, A., and Osiewacz, H. D. (2010). Deletion of PaAif2 and PaAmid2, two genes encoding mitochondrial AIF-like oxidoreductases of Podospora anserina, leads to increased stress tolerance and lifespan extension. Curr. Genet. 55, 225–235. doi: 10.1007/s00294-010-0295-1
Bryan, A. C., Rodeheffer, M. S., Wearn, C. M., and Shadel, G. S. (2002). Sls1p is a membrane-bound regulator of transcription-coupled processes involved in Saccharomyces cerevisiae mitochondrial gene expression. Genetics 160, 75–82.
Chen, Z., Lei, C., Wang, C., Li, N., Srivastava, M., Tang, M., et al. (2019). Global phosphoproteomic analysis reveals ARMC10 as an AMPK substrate that regulates mitochondrial dynamics. Nat. Commun. 10:104. doi: 10.1038/s41467-018-08004-0
Cole, A., Wang, Z., Coyaud, E., Voisin, V., Gronda, M., Jitkova, Y., et al. (2015). Inhibition of the mitochondrial protease ClpP as a therapeutic strategy for human acute myeloid leukemia. Cancer Cell 27, 864–876. doi: 10.1016/j.ccell.2015.05.004
Contamine, V., Lecellier, G., Belcour, L., and Picard, M. (1996). Premature death in Podospora anserina: sporadic accumulation of the deleted mitochondrial genome, translational parameters and innocuity of the mating types. Genetics 144, 541–555.
Contamine, V., Zickler, D., and Picard, M. (2004). The Podospora rmp1 gene implicated in nucleus-mitochondria cross-talk encodes an essential protein whose subcellular location is developmentally regulated. Genetics 166, 135–150. doi: 10.1534/genetics.166.1.135
Cortopassi, G. A., and Arnheim, N. (1990). Detection of a specific mitochondrial DNA deletion in tissues of older humans. Nucleic Acids Res. 18, 6927–6933. doi: 10.1093/nar/18.23.6927
Cummings, D. J., Belcour, L., and Grandchamp, C. (1979). Mitochondrial DNA from Podospora anserina and the occurrence of multimeric circular DNA in senescent cultures. Mol. Gen. Genet. 171, 239–250. doi: 10.1007/BF00267578
Esser, K. (1974). “Podospora anserina,” in Handbook of Genetics, ed. R. C. King (New York, NY: Plenum Press), 531–551.
Esser, K., and Tudzynski, P. (1977). Prevention of senescence in the ascomycete Podospora anserina by the antibiotic tiamulin. Nature 265, 454–456. doi: 10.1038/265454a0
Fischer, F., Filippis, C., and Osiewacz, H. D. (2015a). RCF1-dependent respiratory supercomplexes are integral for lifespan-maintenance in a fungal ageing model. Sci. Rep. 5:12697. doi: 10.1038/srep12697
Fischer, F., Hamann, A., and Osiewacz, H. D. (2012). Mitochondrial quality control: an integrated network of pathways. Trends Biochem. Sci. 37, 284–292. doi: 10.1016/j.tibs.2012.02.004
Fischer, F., Langer, J. D., and Osiewacz, H. D. (2015b). Identification of potential mitochondrial CLPXP protease interactors and substrates suggests its central role in energy metabolism. Sci. Rep. 5:18375. doi: 10.1038/srep18375
Fischer, F., Weil, A., Hamann, A., and Osiewacz, H. D. (2013). Human CLPP reverts the longevity phenotype of a fungal ClpP deletion strain. Nat. Commun. 4:1397. doi: 10.1038/ncomms2397
Gispert, S., Parganlija, D., Klinkenberg, M., Dröse, S., Wittig, I., Mittelbronn, M., et al. (2013). Loss of mitochondrial peptidase Clpp leads to infertility, hearing loss plus growth retardation via accumulation of CLPX, mtDNA and inflammatory factors. Hum. Mol. Genet. 22, 4871–4887. doi: 10.1093/hmg/ddt338
Gredilla, R., Grief, J., and Osiewacz, H. D. (2006). Mitochondrial free radical generation and lifespan control in the fungal aging model Podospora anserina. Exp. Gerontol. 41, 439–447. doi: 10.1016/j.exger.2006.01.010
Hamann, A., Krause, K., Werner, A., and Osiewacz, H. D. (2005). A two-step protocol for efficient deletion of genes in the filamentous ascomycete Podospora anserina. Curr. Genet. 48, 270–275. doi: 10.1007/s00294-005-0018-1
Herzig, S., and Shaw, R. J. (2018). AMPK: guardian of metabolism and mitochondrial homeostasis. Nat. Rev. Mol. Cell Biol. 19, 121–135. doi: 10.1038/nrm.2017.95
Hu, Y., Chen, H., Zhang, L., Lin, X., Li, X., Zhuang, H., et al. (2020). The AMPK-MFN2 axis regulates MAM dynamics and autophagy induced by energy stresses. Autophagy doi: 10.1080/15548627.2020.1749490 [Epub ahead of print].
Kim, J., Kim, Y. C., Fang, C., Russell, R. C., Kim, J. H., Fan, W., et al. (2013). Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell 152, 290–303. doi: 10.1016/j.cell.2012.12.016
Kim, J., Kundu, M., Viollet, B., and Guan, K.-L. (2011). AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 13, 132–141. doi: 10.1038/ncb2152
Klaips, C. L., Jayaraj, G. G., and Hartl, F. U. (2018). Pathways of cellular proteostasis in aging and disease. J. Cell Biol. 217, 51–63. doi: 10.1083/jcb.201709072
Knuppertz, L., Hamann, A., Pampaloni, F., Stelzer, E., and Osiewacz, H. D. (2014). Identification of autophagy as a longevity-assurance mechanism in the aging model Podospora anserina. Autophagy 10, 822–834. doi: 10.4161/auto.28148
Knuppertz, L., and Osiewacz, H. D. (2016). Orchestrating the network of molecular pathways affecting aging: role of nonselective autophagy and mitophagy. Mech. Ageing Dev. 153, 30–40. doi: 10.1016/j.mad.2016.01.003
Knuppertz, L., and Osiewacz, H. D. (2017). Autophagy compensates impaired energy metabolism in CLPXP-deficient Podospora anserina strains and extends healthspan. Aging Cell 16, 704–715. doi: 10.1111/acel.12600
Kück, U., Osiewacz, H. D., Schmidt, U., Kappelhoff, B., Schulte, E., Stahl, U., et al. (1985). The onset of senescence is affected by DNA rearrangements of a discontinuous mitochondrial gene in Podospora anserina. Curr. Genet. 9, 373–382. doi: 10.1007/BF00421608
Kück, U., Stahl, U., and Esser, K. (1981). Plasmid-like DNA is part of mitochondrial DNA in Podospora anserina. Curr. Genet. 3, 151–156. doi: 10.1007/BF00365719
Kunstmann, B., and Osiewacz, H. D. (2008). Over-expression of an S-adenosylmethionine-dependent methyltransferase leads to an extended lifespan of Podospora anserina without impairments in vital functions. Aging Cell 7, 651–662. doi: 10.1111/j.1474-9726.2008.00412.x
Kunstmann, B., and Osiewacz, H. D. (2009). The S-adenosylmethionine dependent O-methyltransferase PaMTH1: a longevity assurance factor protecting Podospora anserina against oxidative stress. Aging (Albany NY) 1, 328–334. doi: 10.18632/aging.100029
Lecellier, G., and Silar, P. (1994). Rapid methods for nucleic acids extraction from Petri dish-grown mycelia. Curr. Genet. 25, 122–123. doi: 10.1007/BF00309536
Li, D., Song, J. Z., Li, H., Shan, M. H., Liang, Y., Zhu, J., et al. (2015). Storage lipid synthesis is necessary for autophagy induced by nitrogen starvation. FEBS Lett. 589, 269–276. doi: 10.1016/j.febslet.2014.11.050
Li, Y., Yan, P., Lu, X., Qiu, Y., Liang, S., Liu, G., et al. (2020). Involvement of PaSNF1 in fungal development, tterigmatocystin biosynthesis, and lignocellulosic degradation in the filamentous fungus Podospora anserina. Front. Microbiol. 11:1038. doi: 10.3389/fmicb.2020.01038
Linnane, A. W., Marzuki, S., Ozawa, T., and Tanaka, M. (1989). Mitochondrial DNA mutations as an important contributor to ageing and degenerative diseases. Lancet 1, 642–645. doi: 10.1016/s0140-6736(89)92145-4
Luce, K., and Osiewacz, H. D. (2009). Increasing organismal healthspan by enhancing mitochondrial protein quality control. Nat. Cell Biol. 11, 852–858. doi: 10.1038/ncb1893
Luce, K., Weil, A. C., and Osiewacz, H. D. (2010). Mitochondrial protein quality control systems in aging and disease. Adv. Exp. Med. Biol. 694, 108–125. doi: 10.1007/978-1-4419-7002-2_9
Marcou, D. (1962). Notion de longevite et nature cytoplasmatique du determinant de senescence chez quelques champignons. Ann. Sci. Nat. Biol. Bot. Veg. 2, 653–764.
Martinez-Ortiz, C., Carrillo-Garmendia, A., Correa-Romero, B. F., Canizal-Garcia, M., Gonzalez-Hernandez, J. C., Regalado-Gonzalez, C., et al. (2019). SNF1 controls the glycolytic flux and mitochondrial respiration. Yeast 36, 487–494. doi: 10.1002/yea.3399
Melov, S., Hertz, G. Z., Stormo, G. D., and Johnson, T. E. (1994). Detection of deletions in the mitochondrial genome of Caenorhabditis elegans. Nucleic Acids Res. 22, 1075–1078. doi: 10.1093/nar/22.6.1075
Mukaiyama, H., Nakase, M., Nakamura, T., Kakinuma, Y., and Takegawa, K. (2010). Autophagy in the fission yeast Schizosaccharomyces pombe. FEBS Lett. 584, 1327–1334. doi: 10.1016/j.febslet.2009.12.037
Nakamura, S., Matsui, A., Akabane, S., Tamura, Y., Hatano, A., Miyano, Y., et al. (2020). The mitochondrial inner membrane protein LETM1 modulates cristae organization through its LETM domain. Commun. Biol. 3, 99. doi: 10.1038/s42003-020-0832-5
Nowikovsky, K., Reipert, S., Devenish, R. J., and Schweyen, R. J. (2007). Mdm38 protein depletion causes loss of mitochondrial K+/H+ exchange activity, osmotic swelling and mitophagy. Cell Death Differ. 14, 1647–1656. doi: 10.1038/sj.cdd.4402167
Osiewacz, H. D. (1994). A versatile shuttle cosmid vector for the efficient construction of genomic libraries and for the cloning of fungal genes. Curr. Genet. 26, 87–90. doi: 10.1007/BF00326309
Osiewacz, H. D. (2002). Aging in fungi: role of mitochondria in Podospora anserina. Mech. Ageing Dev. 123, 755–764. doi: 10.1016/s0047-6374(01)00421-3
Osiewacz, H. D. (2010). Role of mitochondria in aging and age-related disease. Exp. Gerontol. 45:465. doi: 10.1016/j.exger.2010.05.001
Osiewacz, H. D., and Esser, K. (1984). The mitochondrial plasmid of Podospora anserina: a mobile intron of a mitochondrial gene. Curr. Genet. 8, 299–305. doi: 10.1007/BF00419728
Osiewacz, H. D., Hamann, A., and Zintel, S. (2013). Assessing organismal aging in the filamentous fungus Podospora anserina. Methods Mol. Biol. 965, 439–462. doi: 10.1007/978-1-62703-239-1_29
Osiewacz, H. D., Skaletz, A., and Esser, K. (1991). Integrative transformation of the ascomycete Podospora anserina: identification of the mating-type locus on chromosome VII of electrophoretically separated chromosomes. Appl. Microbiol. Biotechnol. 35, 38–45. doi: 10.1007/BF00180633
Petereit, J., Duncan, O., Murcha, M. W., Fenske, R., Cincu, E., Cahn, J., et al. (2020). Mitochondrial CLPP2 assists coordination and homeostasis of respiratory complexes. Plant Physiol. 184, 148–164. doi: 10.1104/pp.20.00136
Petersen, K. F., Befroy, D., Dufour, S., Dziura, J., Ariyan, C., Rothman, D. L., et al. (2003). Mitochondrial dysfunction in the elderly: possible role in insulin resistance. Science 300, 1140–1142. doi: 10.1126/science.1082889
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45
Rizet, G. (1953). Impossibility of obtaining uninterrupted and unlimited multiplication of the ascomycete Podospora anserina. C. R. Hebd. Seances Acad. Sci. 237, 838–855.
Rodeheffer, M. S., and Shadel, G. S. (2003). Multiple interactions involving the amino-terminal domain of yeast mtRNA polymerase determine the efficiency of mitochondrial protein synthesis. J. Biol. Chem. 278, 18695–18701. doi: 10.1074/jbc.M301399200
Rooyackers, O. E., Adey, D. B., Ades, P. A., and Nair, K. S. (1996). Effect of age on in vivo rates of mitochondrial protein synthesis in human skeletal muscle. Proc. Natl. Acad. Sci. U.S.A. 93, 15364–15369. doi: 10.1073/pnas.93.26.15364
Rouillard, J. M., Dufour, M. E., Dujardin, G., Lacroute, F., Theunissen, B., and Mandart, E. (1996). SLS1, a new Saccharomyces cerevisiae gene involved in mitochondrial metabolism, isolated as a synthetic lethal in association with an SSM4 deletion. Mol. Gen. Genet. 252, 700–708. doi: 10.1007/BF02173976
Russell, R. C., Yuan, H.-X., and Guan, K.-L. (2014). Autophagy regulation by nutrient signaling. Cell Res. 24, 42–57. doi: 10.1038/cr.2013.166
Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. Cold Spring Harbour: Cold Spring Harbour Press.
Scheckhuber, C. Q., Erjavec, N., Tinazli, A., Hamann, A., Nyström, T., and Osiewacz, H. D. (2007). Reducing mitochondrial fission results in increased life span and fitness of two fungal ageing models. Nat. Cell Biol. 9, 99–105. doi: 10.1038/ncb1524
Scheckhuber, C. Q., and Osiewacz, H. D. (2008). Podospora anserina: a model organism to study mechanisms of healthy ageing. Mol. Genet. Genomics 280, 365–374. doi: 10.1007/s00438-008-0378-6
Seiferling, D., Szczepanowska, K., Becker, C., Senft, K., Hermans, S., Maiti, P., et al. (2016). Loss of CLPP alleviates mitochondrial cardiomyopathy without affecting the mammalian UPRmt. EMBO Rep. 17, 953–964. doi: 10.15252/embr.201642077
Sellem, C. H., Lecellier, G., and Belcour, L. (1993). Transposition of a group II intron. Nature 366, 176–178. doi: 10.1038/366176a0
Sellem, C. H., Lemaire, C., Lorin, S., Dujardin, G., and Sainsard-Chanet, A. (2005). Interaction between the oxa1 and rmp1 genes modulates respiratory complex assembly and life span in Podospora anserina. Genetics 169, 1379–1389. doi: 10.1534/genetics.104.033837
Seo, J. H., Rivadeneira, D. B., Caino, M. C., Chae, Y. C., Speicher, D. W., Tang, H. Y., et al. (2016). The mitochondrial unfoldase-peptidase complex ClpXP controls bioenergetics stress and metastasis. PLoS Biol. 14:e1002507. doi: 10.1371/journal.pbio.1002507
Stahl, U., Lemke, P. A., Tudzynski, P., Kuck, U., and Esser, K. (1978). Evidence for plasmid like DNA in a filamentous fungus, the ascomycete Podospora anserina. Mol. Gen. Genet. 162, 341–343. doi: 10.1007/BF00268860
Szczepanowska, K., Maiti, P., Kukat, A., Hofsetz, E., Nolte, H., Senft, K., et al. (2016). CLPP coordinates mitoribosomal assembly through the regulation of ERAL1 levels. EMBO J. 35, 2566–2583. doi: 10.15252/embj.201694253
Tamargo-Gomez, I., and Marino, G. (2018). AMPK: regulation of metabolic dynamics in the context of autophagy. Int. J. Mol. Sci. 19:3812. doi: 10.3390/ijms19123812
Todde, V., Veenhuis, M., and van der Klei, I. J. (2009). Autophagy: principles and significance in health and disease. Biochim. Biophys. Acta 1792, 3–13. doi: 10.1016/j.bbadis.2008.10.016
Toyama, E. Q., Herzig, S., Courchet, J., Lewis, T. L., Losón, O. C., Hellberg, K., et al. (2016). AMP-activated protein kinase mediates mitochondrial fission in response to energy stress. Science 351, 275–281. doi: 10.1126/science.aab4138
Tudzynski, P., and Esser, K. (1977). Inhibitors of mitochondrial function prevent senescence in the ascomycete Podospora anserina. Mol. Gen. Genet. 153, 111–113. doi: 10.1007/BF01036003
Tudzynski, P., and Esser, K. (1979). Chromosomal and extrachromosomal control of senescence in the ascomycete Podospora anserina. Mol. Gen. Genet. 173, 71–84. doi: 10.1007/BF00267692
Vallejo, B., Peltier, E., Garrigós, V., Matallana, E., Marullo, P., and Aranda, A. (2020). Role of Saccharomyces cerevisiae nutrient signaling pathways during winemaking: a phenomics approach. Front. Bioeng. Biotechnol. 8:853. doi: 10.3389/fbioe.2020.00853
Wallace, D. C. (1992). Mitochondrial genetics: a paradigm for aging and degenerative diseases? Science 256, 628–632. doi: 10.1126/science.1533953
Weil, A., Luce, K., Dröse, S., Wittig, I., Brandt, U., and Osiewacz, H. D. (2011). Unmasking a temperature-dependent effect of the P. anserina i -AAA protease on aging and development. Cell Cycle 10, 4280–4290. doi: 10.4161/cc.10.24.18560
Wen, X., and Klionsky, D. J. (2016). An overview of macroautophagy in yeast. J. Mol. Biol. 428, 1681–1699. doi: 10.1016/j.jmb.2016.02.021
Wilkening, A., Rüb, C., Sylvester, M., and Voos, W. (2018). Analysis of heat-induced protein aggregation in human mitochondria. J. Biol. Chem. 293, 11537–11552. doi: 10.1074/jbc.RA118.002122
Willis, S. D., Stieg, D. C., Ong, K. L., Shah, R., Strich, A. K., Grose, J. H., et al. (2018). Snf1 cooperates with the CWI MAPK pathway to mediate the degradation of Med13 following oxidative stress. Microb. Cell 5, 357–370. doi: 10.15698/mic2018.08.641
Wu, X., Luo, J., Liu, H., Cui, W., Guo, K., Zhao, L., et al. (2020). Recombinant adiponectin peptide ameliorates brain injury following intracerebral hemorrhage by suppressing astrocyte-derived inflammation via the inhibition of Drp1-mediated mitochondrial fission. Transl. Stroke Res. 11, 924–939. doi: 10.1007/s12975-019-00768-x
Yin, Z., Pascual, C., and Klionsky, D. J. (2016). Autophagy: machinery and regulation. Microb. Cell 3, 588–596. doi: 10.15698/mic2016.12.546
Zhang, C.-S., Hawley, S. A., Zong, Y., Li, M., Wang, Z., Gray, A., et al. (2017). Fructose-1,6-bisphosphate and aldolase mediate glucose sensing by AMPK. Nature 548, 112–116. doi: 10.1038/nature23275
Keywords: Podospora anserina, CLP protease, SNF1, AMPK, RMP1, aging, development
Citation: Heinz D, Krotova E, Hamann A and Osiewacz HD (2021) Simultaneous Ablation of the Catalytic AMPK α-Subunit SNF1 and Mitochondrial Matrix Protease CLPP Results in Pronounced Lifespan Extension. Front. Cell Dev. Biol. 9:616520. doi: 10.3389/fcell.2021.616520
Received: 12 October 2020; Accepted: 27 January 2021;
Published: 04 March 2021.
Edited by:
Dmitry A. Knorre, Lomonosov Moscow State University, RussiaReviewed by:
Lianjun Zhang, Center of Systems Medicine, Chinese Academy of Medical Sciences, Suzhou Institute of Systems Medicine (ISM), ChinaYong Liu, Xuzhou Medical University, China
Copyright © 2021 Heinz, Krotova, Hamann and Osiewacz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Heinz D. Osiewacz, b3NpZXdhY3pAYmlvLnVuaS1mcmFua2Z1cnQuZGU=
 Daniela Heinz
Daniela Heinz Andrea Hamann
Andrea Hamann Heinz D. Osiewacz
Heinz D. Osiewacz