
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol. , 11 February 2021
Sec. Molecular and Cellular Pathology
Volume 9 - 2021 | https://doi.org/10.3389/fcell.2021.600344
This article is part of the Research Topic Advances in Genomic and Genetic Tools, and Their Applications for Understanding Embryonic Development and Human Diseases View all 33 articles
 Yang Yang1
Yang Yang1 Xiaohong Wang1
Xiaohong Wang1 Yiqiang Liu2
Yiqiang Liu2 Ying Hu1
Ying Hu1 Zhongwu Li2
Zhongwu Li2 Ziyu Li3
Ziyu Li3 Zhaode Bu3
Zhaode Bu3 Xiaojiang Wu3
Xiaojiang Wu3 Lianhai Zhang1,3*
Lianhai Zhang1,3* Jiafu Ji1,3*
Jiafu Ji1,3*SALL4, a transcriptional factor involved in embryonic stem cell self-renewal and pluripotency, is overexpressed in gastric cancer (GC). However, the association of SALL4 with the survival of GC patients is not well-understood, and the role of SALL4 in cancer progression is still unknown. In the present study, a total of 1,815 GC patients who underwent radical resection at Peking Cancer Hospital were included consecutively from 2015 to 2018, confirming the prognostic value of SALL4 and validating by data from TCGA and GEO. The protein and mRNA expression levels of SALL4 were evaluated by immunohistochemistry and qPCR, respectively. Besides, GSEA and WGCNA were applied to explore the SALL4-related cancer-promoting signaling pathways and gene modules. Our results showed that overexpression of SALL4 was observed in 16.7% of GC patients. SALL4 positivity was associated with male, older age, mixed-type histology, late stages, lymphatic metastasis, vascular invasion, non-cardia location, high AFP level, and no EBV infection background. SALL4 could be served as a marker for prognostic prediction in GC, and SALL4-positive GC was significantly associated with shortened survival. Further, the bioinformatic analysis indicated that the Wnt/β-catenin signaling pathway was activated in SALL4-high cases compared with SALL4-low cases. Expression of SALL4 was also positively correlated with the expression of multiple co-expressed genes, such as TRIB3, which plays an important role in activating the Wnt/β-catenin pathway. Our findings indicate that SALL4 is associated with clinicopathological features related to cancer progression in GC and its function in the Wnt/β-catenin pathway.
Gastric cancer (GC) is the third leading cause of cancer-related mortality worldwide (Bray et al., 2018). The incidence and mortality rates for patients with GC have declined recently. However, despite recent advancements in treatment strategies, the prognosis of patients with GC is still poor, and the 5-year survival rate is <30% (Araki et al., 2018; Cats et al., 2018; Kudou et al., 2018). Therefore, the identification of novel biomarkers related to GC progression is imperative. The ideal markers not only would promote the current understanding of GC pathogenesis but also could reveal new effective strategies for GC treatment to us.
Spalt-like transcription factor 4 (SALL4), located on chromosome 20q13.2, encodes a zinc finger transcription factor that plays a key role in maintaining the pluripotency and self-renewal capacity of embryonic stem cells (Zhang et al., 2014). SALL4 was first described in human acute myeloid leukemia (Ma et al., 2006). Subsequently, overexpression of SALL4 was reported in a variety of cancers, such as breast cancer, lung cancer, liver cancer, endometrial cancer, germ cell cancer, as well as GC (Camparo and Comperat, 2013; Gonzalez-Roibon et al., 2013; Li et al., 2015; Zhang et al., 2015; Tatetsu et al., 2016). Numerous studies have revealed the role of SALL4 in the process of carcinogenesis, including invasion and metastasis, cell proliferation, stemness, and apoptosis (Liu et al., 2015; Kim et al., 2017).
Previous research conducted at the mRNA level has suggested that SALL4 is a factor for poor prognosis in GC (Zhang et al., 2014; Yanagihara et al., 2015). However, the association between SALL4 protein expression and the clinical outcome in patients with GC is not yet defined and the functional role of SALL4 in GC is still unknown. This study aimed to comprehensively analyze SALL4 expression in GC and explore its prognostic value, along with the underlying mechanism. We determined SALL4 protein expression in GC and analyzed its relationship with clinicopathological features. Then, using The Cancer Genome Atlas (TCGA) data, we explored the regulation of SALL4 gene expression by analyzing the correlation between DNA copy number variation (CNV) and aberrant SALL4 mRNA expression in GC. Besides, bioinformatics analysis was used to investigate the relevant pathways and co-expression genes of SALL4 in GC, including gene set enrichment analysis (GSEA), weighted gene co-expression network analysis (WGCNA), and differential expression gene analysis, which provides new insight into the follow-up research.
GC patients, who underwent radical resection at Peking University Cancer Hospital between 2015 and 2018, were consecutively enrolled. Patients without paraffin-embedded clinical tissue specimens or incomplete clinicopathological information were excluded. In total, 1,815 cases were eligible for analyses. TNM stage was determined in accordance with the 7th edition of classification recommended by the American Joint Committee on Cancer (AJCC). All patients underwent follow-up evaluations until October 2020. This investigation was approved by the Ethics Committee of Peking University Cancer Hospital. Informed consent was obtained from each patient at the time of sample collection.
Four-micrometer-thick sections of formalin-fixed paraffin-embedded tissues were mounted on poly-L-lysine-coated slides. Then, the slides were deparaffinized in xylene and rehydrated with a gradient of ethanol and distilled water. Endogenous peroxidase activity was quenched with 3% hydrogen peroxide for 10 min at room temperature. The slides were inactivated by incubation in 10 mmol/L ethylenediaminetetraacetic acid (EDTA; pH 8.0) for 3 min. The sections were incubated overnight at 4°C with mouse anti-SALL4 antibody (1:100) (Zsbio; Beijing, China). The primary antibodies were probed with a two-step Poly-HRP Anti-Mouse/Rabbit IgG Detection System (Zsbio; Beijing, China). Positive and negative controls in immunohistochemistry (IHC) were routinely used. For the negative controls, the primary antibody was replaced with non-immune mouse serum to confirm specificity. We also used an internal positive control (seminoma tissue) for quality assurance.
Reverse transcription-quantitative real-time PCR (RT-qPCR) was performed to determine the relative SALL4 mRNA expression levels in the clinical samples as previously described (Wang et al., 2017). Extracted mRNA was amplified with human SALL4-specific primers by q-PCR. The human GAPDH was included as an internal control. Each sample was assessed in triplicate. Expression of gene was given as the ratio of expression of target gene mRNA to that of GAPDH mRNA.
Evaluation of SALL4 staining was principally based on the scoring criteria described previously (Yong et al., 2013). Briefly, only nuclear reactivity with a diffuse pattern was considered as SALL4 positive. Patchy granular nuclear reactivity was scored as negative (Liu T.-C. et al., 2014). SALL4 expression was classified according to a semi-quantitative score based on the percentage of tumor cells displaying a diffuse nuclear pattern of SALL4 immunoreactivity, as described previously: 0, <5%; 1, 5–30%; 2, 31–50%; 3, 51–80%; 4, >80%. Scores >1 were defined as positive (Cao et al., 2009b; Yong et al., 2013). SALL4 expression in GC tissues was scored independently by two senior pathologists. If any disagreement arose during the evaluation, a third pathologist was consulted.
Gene expression profiles and clinical and survival data of GC patients in the TCGA stomach adenocarcinoma (STAD) dataset (n = 407) were downloaded from the TCGA Data Portal (https://portal.gdc.cancer.gov/). The survival data and microarray datasets of GSE15459, GSE34942, GSE57303, and GSE62254 were obtained from the gene expression omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/), which included a total of 626 Asian GC patients. After array annotations, all samples in four datasets were integrated to reduce deviation and variability by batch normalization. Also, SALL4 mRNA expression and copy number data were obtained from the UCSC Xena browser (http://xena.ucsc.edu/).
Gene set enrichment analysis (GSEA) was used to further elucidate the significant enriched pathway data between high and low SALL4 expression group. Transcription data from four GEO and TCGA datasets were analyzed by GSEA 3.0 software (Broad Institute; Cambridge, Massachusetts, USA), and the number of permutations was set to 1,000. The mean expression value of each gene was used for correlation analysis.
Based on GEO microarray data (GSE15459, GSE34942, GSE57303, and GSE62254), SALL4-related gene modules were identified using WGCNA. Correlation coefficients between the module eigengenes and traits were calculated by using Pearson's approach. High SALL4-related gene modules were defined as those with maximum correlation coefficients. We then constructed a PPI network by the STRING database using Cytoscape (V.3.8.0) for network visualization.
The pathway enrichment analysis was performed on these genes in the most relevant gene modules using Enrichr (https://amp.pharm.mssm.edu/Enrichr/). The DEGs of high SALL4 expression-related gene modules were identified using the R package limma with a threshold of |log2FoldChange| >1 and P < 0.05.
Statistical analysis was performed using SPSS software version 22.0 (IBM, Armonk, NY, USA). The association between SALL4 expression and clinicopathological parameters in GC was assessed by the chi-square test. Univariate analysis of cumulative overall survival was conducted using a Cox regression model. Overall survival and its association with SALL4 protein expression were evaluated by the Kaplan–Meier method followed by the log-rank test. Subsequent multivariate analysis of prognostic factors was conducted using the Cox regression model. Unpaired Student's t-test and a receiver operating characteristic (ROC) curve were performed to assess the differential expression of SALL4 in GC and normal tissues. The correlation between copy number and SALL4 mRNA expression was also analyzed. SALL4 mRNA expression and co-expressed genes were evaluated by linear regression and correlation analyses. The significance threshold is P < 0.05.
A total of 1,815 consecutive patients with GC (1,296 males and 519 females) were included in this study. The mean age at diagnosis was 59 ± 10 years. Most cancers were diagnosed at relatively early stages (27.7% at stage I and 29.6% at stage II), localized to the non-cardia sites of the stomach (75.6%), and classified as Lauren intestinal type (38.3%). Of 1,812 patients for whom lymph node status was available, 1,072 (59.1%) had lymph node metastasis. Vascular invasion was observed in 974 (53.7%) out of 1,811 patients (Supplementary Table 1).
SALL4 staining was localized to the nucleus of cancer cells, and only diffuse nuclear staining was considered positive for SALL4 (Figures 1A–G). Three hundred four (16.7%) tumors were SALL4-positive by IHC. There was rare SALL4 staining in adjacent non-neoplastic tissues (Figures 1A,B). The results of RT-qPCR were concordant with the IHC findings in this sample set (Supplementary Figure 1). Among SALL4-positive and SALL4-negative GCs according to the IHC assessment, the samples were dichotomized into high and low expression subgroups based on RT-qPCR analysis of SALL4 mRNA (R = 0.670, P < 0.001).
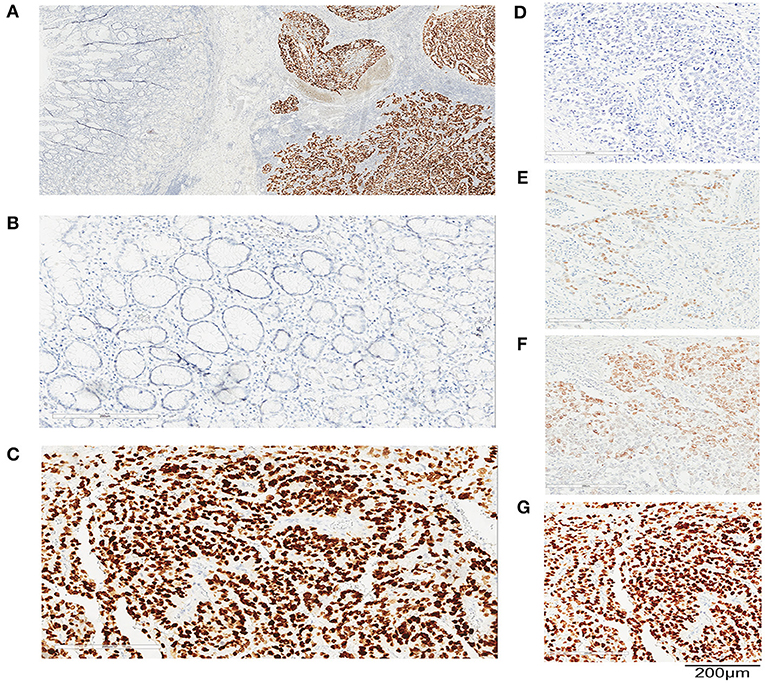
Figure 1. SALL4 expression in human primary gastric cancer (GC) by immunohistochemistry (IHC). Representative images of immunohistochemical staining for SALL4 in GC. Original magnification (×100) (A), adjacent non-tumors gastric mucosa (×200) (B), and cancer (×200) (C). Staining intensity in GC is classified as negative staining (D), weak staining (E), moderate staining (F), and strong staining (G). Original magnification, ×200. Bar = 200 μm.
We observed that the SALL4-positive rate was higher in patients with advanced stage GC (P = 0.006), lymph node metastasis (P = 0.002), non-cardia localization (P = 0.043), and vascular invasion (P = 0.023). Tumor SALL4-positive was also more common in Lauren mixed-type GC (P < 0.001). In addition, SALL4 expression in tumor cells was more frequent in male patients (P = 0.001) and in older patients (P = 0.012).
Among the GC subtypes, SALL4 expression was negatively correlated with Epstein–Barr virus (EBV) infection (P < 0.001), which is also referred to as EBV associated gastric cancer (EBVaGC) and is generally acknowledged to have a favorable prognosis. In addition, another GC subtype, lymphoepithelioma-like carcinoma (LELC) of GC, featured with intense lymphocytic infiltration and partially overlapped with EBVaGC subtype (Yang et al., 2019), was SALL4-negative. SALL4 was frequently expressed in GC patients with high AFP levels (Table 1).
Univariate Cox regression analysis revealed that the TNM stage, Lauren type, degree of differentiation, SALL4 expression, lymph node metastasis, vascular invasion, perineural invasion, location, sex, and distant metastasis were the significant prognostic indicators of GC (P < 0.05, respectively) (Table 2). In the multivariate model, TNM stage, location, vascular invasion, sex, and the degree of differentiation were statistically significant predictors of mortality. In addition, SALL4-positive predicted poorer survival (HR = 2.211, 95% CI, 1.384–3.533, P = 0.001, Table 3).
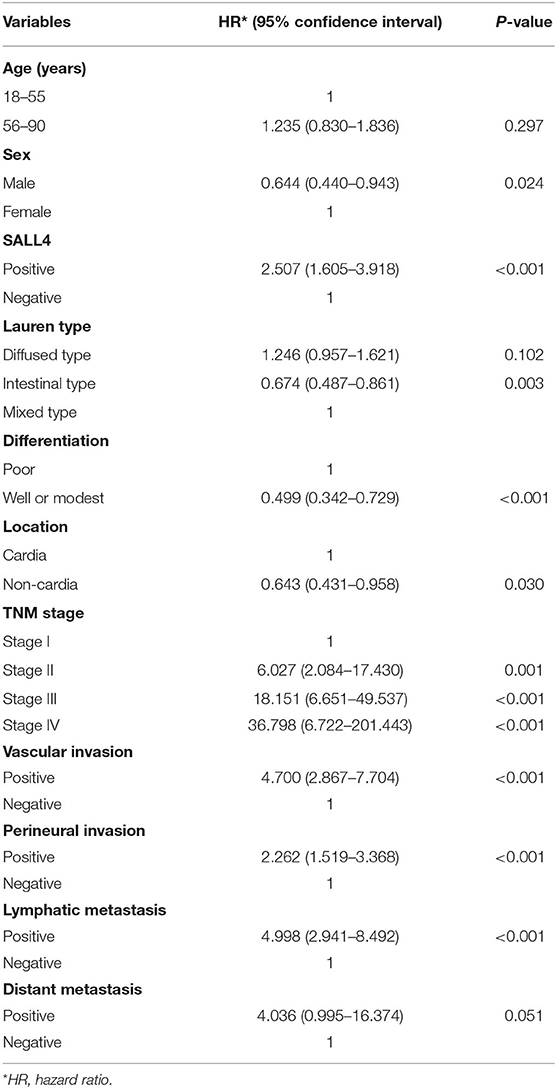
Table 2. Univariate Cox regression analysis of potential poor prognostic factors for gastric cancer patients in our study.
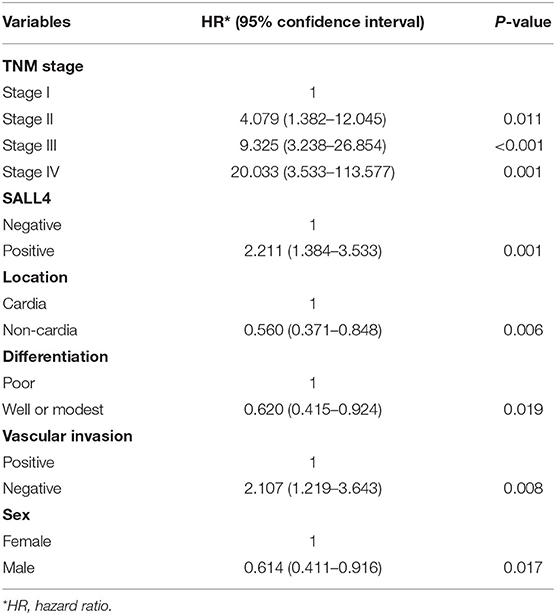
Table 3. Multivariate Cox regression analysis of potential poor prognostic factors for gastric cancer patients in our study.
Patients with GC having SALL4-positive expression showed worse survival in the Kaplan–Meier survival analysis (3-year overall survival, 0.66 vs. 0.93, log-rank test, P < 0.001, Figure 2A). In further analysis, the discrepancy in survival remained significant when patients were stratified by the TNM stage (Figures 2B–D). In stage II GC, patients with SALL4-positive tumors had shorter overall survival than those with SALL4-negative tumors (log-rank test, P < 0.001, Figure 2C).
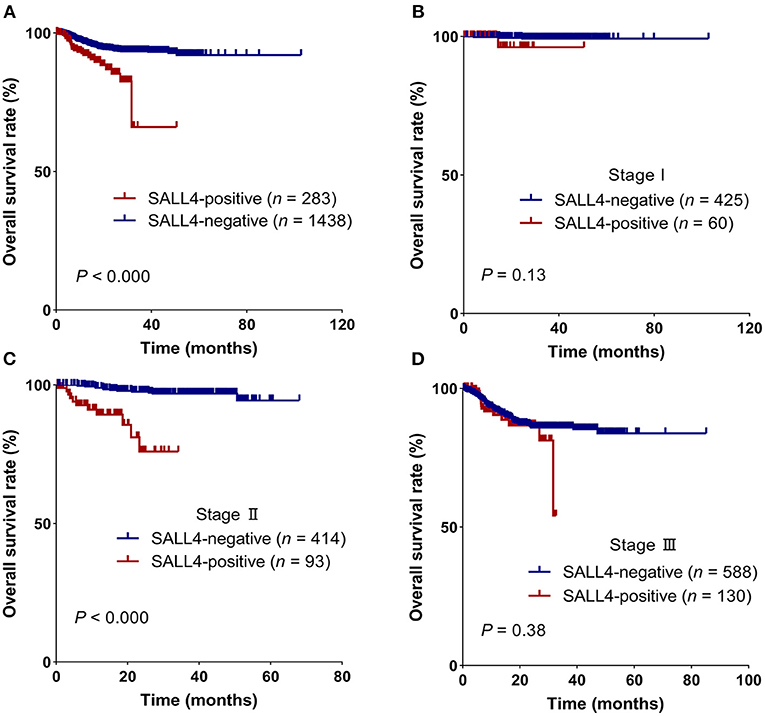
Figure 2. SALL4 is a poor prognostic factor in patients with gastric cancer (GC). Kaplan–Meier survival curves for all patients (A), stage I (B), stage II (C), and stage III (D) in different expression groups, which were stratified by SALL4. SALL4-positive is associated with worse outcome in the subgroup of TNM stage II (C) patients.
To compare with our findings, we analyzed the association between SALL4 mRNA expression and prognosis in the TCGA dataset, which includes 375 GC patients (Supplementary Table 2). SALL4 mRNA expression was significantly increased in primary gastric tumors when compared with adjacent non-tumor tissues (P < 0.001, Figure 4A). We also plotted the area under the ROC curves (AUCs), which illustrated strong separation between the tumor and normal tissues, with an AUC of 0.954 for SALL4 (Figure 4B). We divided GC patients into two groups (high-expression group vs. low-expression group) according to the analysis of X-title. As shown in the Kaplan–Meier survival curve, patients in the high-expression group had shorter survival than those with low or silenced SALL4 expression (3-year overall survival, 0.42 vs. 0.65, log-rank test, P = 0.01, Figure 3A). After being stratified by tumor stage, SALL4 overexpression predicted poor prognosis in stage III GC patients (P = 0.05, Figure 3C), but not in patients with other stages of GC. Note that results at the mRNA level were consistent with our observations at the protein level (Tables 4, 5).
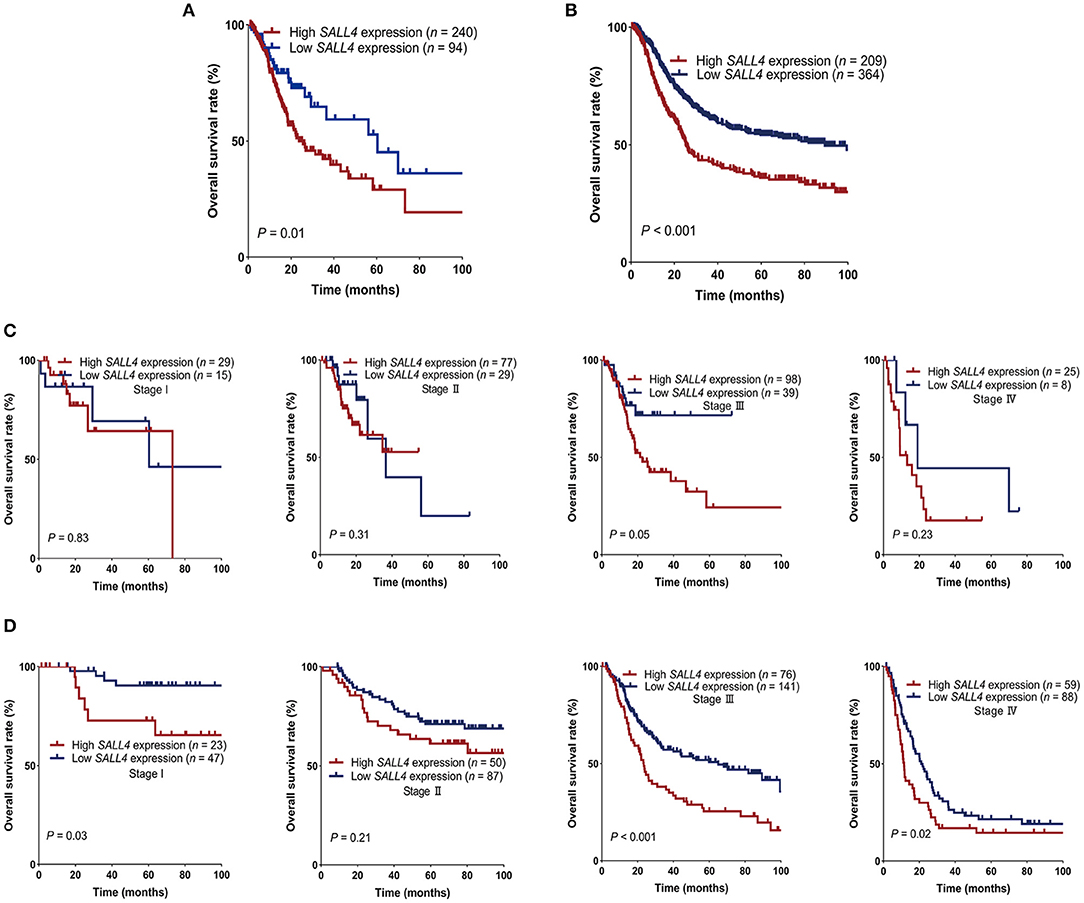
Figure 3. Survival of gastric cancer (GC) patients in The Cancer Genome Atlas (TCGA) and gene expression omnibus (GEO) stratified by SALL4. GC patients with high SALL4 expression had worse prognosis than those with low SALL4 expression in TCGA (A) and GEO (B). Kaplan–Meier survival curves for all stage I, II, III, and IV in different expression groups of TCGA (C) and GEO (D). SALL4 low expression is associated with better outcome in the subgroup of TNM stage III patients.
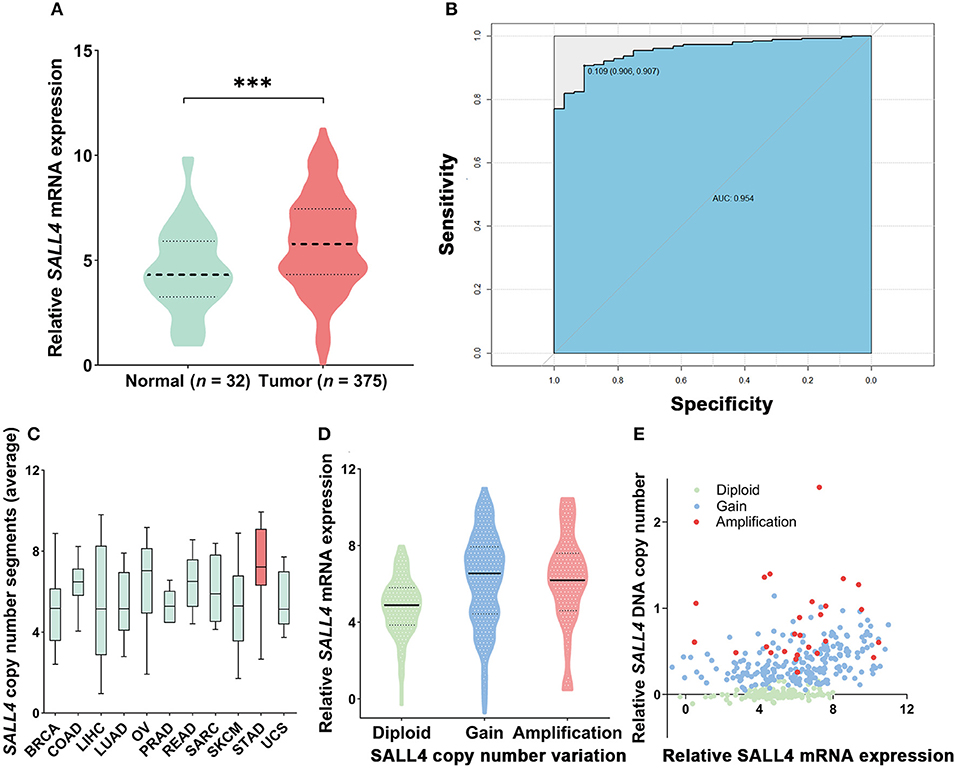
Figure 4. mRNA expression and copy number variant of SALL4 in gastric cancer (GC) in The Cancer Genome Atlas (TCGA). SALL4 overexpressed in tumor compared to adjacent non-tumor gastric mucosa (***P < 0.001) (A). ROC curve for SALL4 expression in normal gastric tissue and GC (B). Copy number variant of SALL4 in gastric cancer (GC). A pan-cancer analysis of SALL4 comparing GC (red) with data from TCGA for other cancer types (green). SALL4 is amplification in various types of cancers, especially GC (C). DNA copy number gain (D) is associated with SALL4 mRNA expression (E) in primary GC (BRCA, breast cancer; COAD, colon adenocarcinoma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; OV, ovarian cancer; PRAD, prostate adenocarcinoma; READ, rectum adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; UCS, uterine carcinosarcoma).
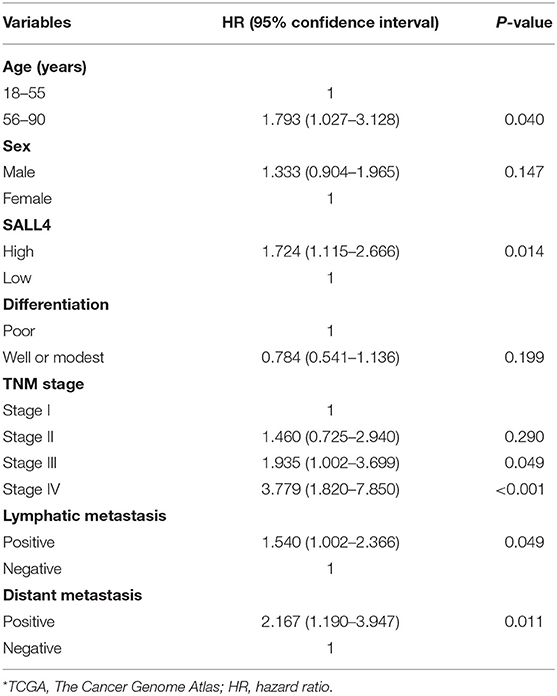
Table 4. Univariate Cox regression analysis of prognostic factors for gastric cancer patients in TCGA*.
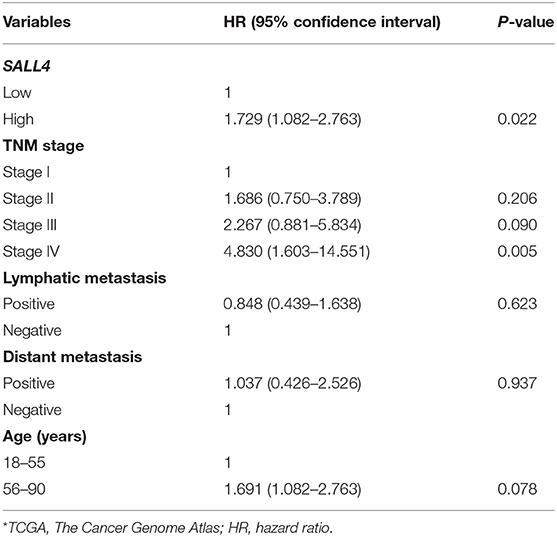
Table 5. Multivariate Cox regression analysis of potential poor prognostic factors for gastric cancer patients in TCGA*.
Since the TCGA dataset includes only a few Asian patients with GC, we downloaded raw data and platform information of four datasets of Asian GC patients (GSE15459, GSE34942, GSE57303, and GSE62254) from the GEO database. To lessen the bias of false-positive findings, four datasets were batch corrected and merged, and raw and normalized data are shown in Supplementary Figure 3. Then, the survival data of 573 GC samples from these four datasets were used to analyze the association between SALL4 mRNA expression and outcomes among Asian GC patients.
Kaplan–Meier survival analysis also indicated SALL4 to be highly correlated with survival (3-year overall survival, 0.43 vs. 0.61, log-rank test, P < 0.001, Figure 3B). SALL4 overexpression predicted poor prognosis in patients with stage I (P = 0.02), stage III (P < 0.001), and stage IV GC (P = 0.02, Figure 3D).
We also analyzed the mutation level of SALL4 base on the TCGA dataset. Although SALL4 is rarely mutated (0.1%) in GC, analysis of the TCGA pan-cancer dataset revealed that SALL4 is amplified in GC and other human cancers, including lung squamous cell carcinoma, colon carcinoma, bladder carcinoma, and lung adenocarcinoma (Figure 4C). A positive association between SALL4 copy number gain and SALL4 mRNA expression (R = 0.3615, P < 0.001, Figures 4D,E) was noted in the TCGA dataset. These findings demonstrate that the DNA copy number gain contributes to the up-regulation of SALL4 in GC.
To understand the molecular mechanisms underlying the pro-tumorigenic action of highly expressed SALL4, GESA pathway enrichment analysis was performed based on GEO (GSE15459, GSE34942, GSE57303, and GSE62254) and TCGA datasets. The results showed that SALL4 significantly dysregulated Wnt/β-catenin signaling, KRAS signaling, epithelial–mesenchymal transition and mitotic spindle (Figure 5) in GEO datasets. GSEA results also revealed Wnt signaling, transforming growth factor-beta (TGF-beta) signaling pathways, and gap junction signaling differentially enriched in high SALL4 expression GC in TCGA dataset (Supplementary Figure 4). We herein assumed that up-regulated SALL4 also contributed to the poor prognosis of GC via the Wnt/β-catenin signaling pathway.
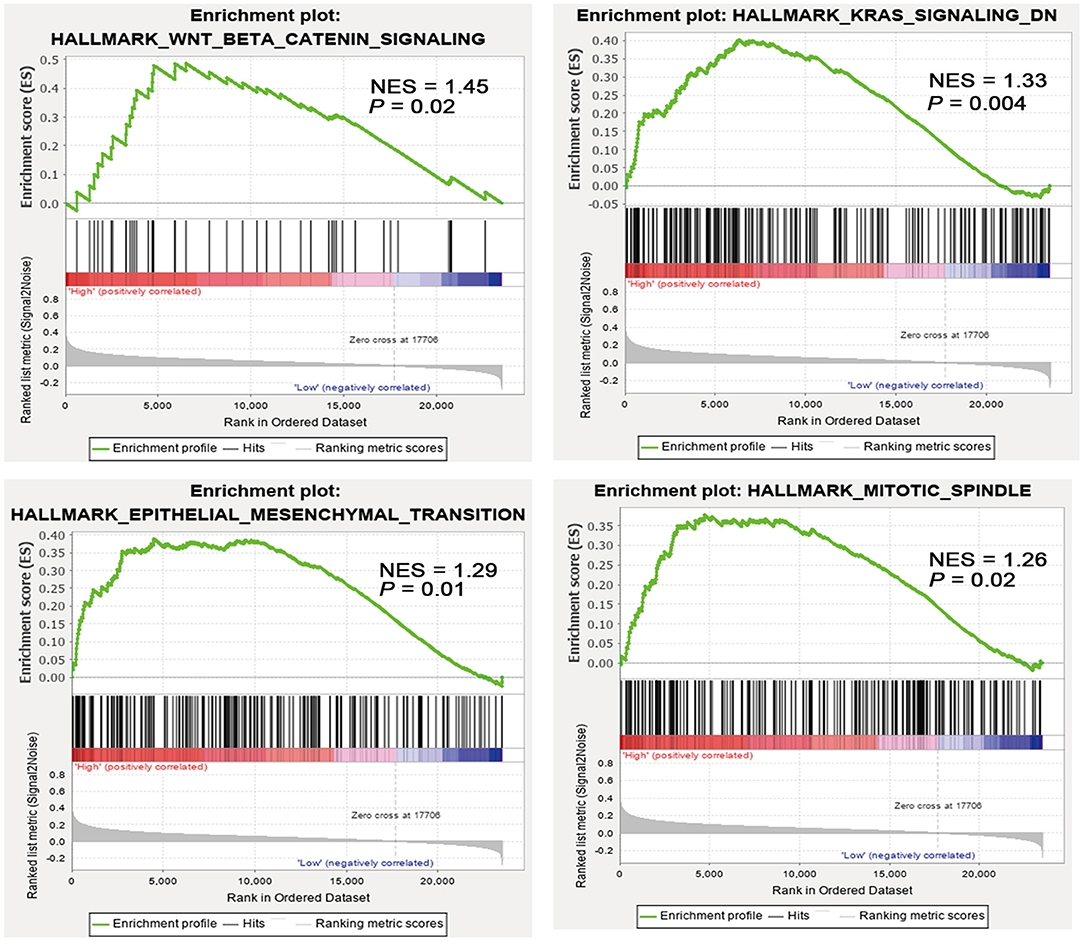
Figure 5. High SALL4 expression promotes tumorigenesis through multiple pathways. Gene set enrichment analysis (GSEA) of gastric cancer (GC) samples from four GEO datasets (GSE15459, GSE34942, GSE57303, and GSE62254) showed that SALL4 expression is positively correlated with Wnt/β-catenin signaling, KRAS signaling, mitotic spindle, and epithelial–mesenchymal transition. Each bar corresponds to one gene.
To further explore the role of SALL4 in Wnt signaling pathway, we conducted an integrated analysis of four GEO datasets to find SALL4-related genes and their function (Figures 6A,B). By using R package WGCNA, we obtain SALL4-related genes that include 16 gene modules. The correlation between the trait and gene modules showed that eight gene modules were related with high SALL4 expression (P < 0.05), especially the red module. The genes in the red module were used to construct the PPI network (Figure 7A). To better understand the biological characteristics of the SALL4-related gene module, we performed pathway analysis using the genes in the red module by Enrichr. The red module genes were enriched in Wnt-mediated activation of DVL (Figure 7B).
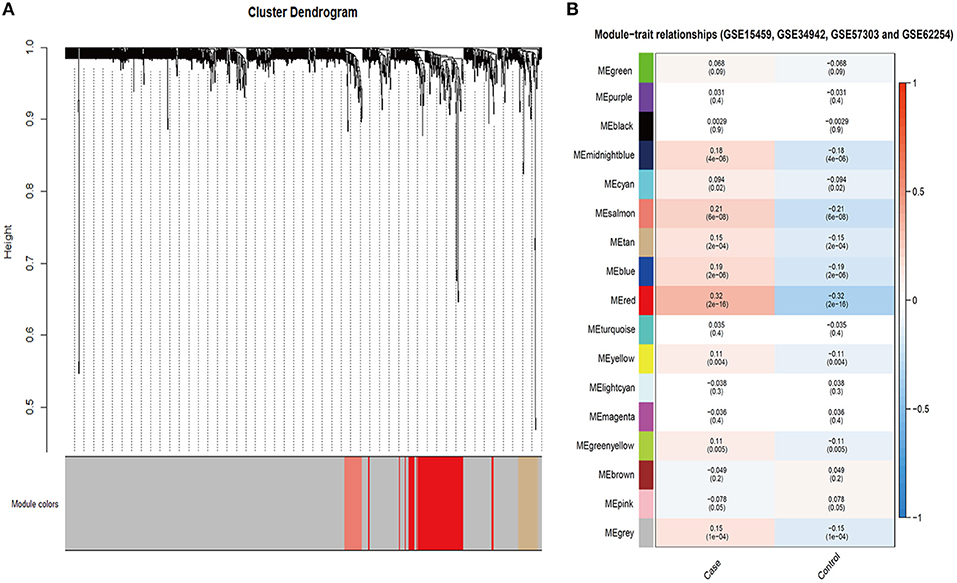
Figure 6. Identification of SALL4-related gene modules. Gene modules identified by WGCNA in GSE15459, GSE34942, GSE57303, and GSE62254 (A). The correlation between gene module and trait in four GEO datasets (B). The case represented high SALL4 expression and control represented low.
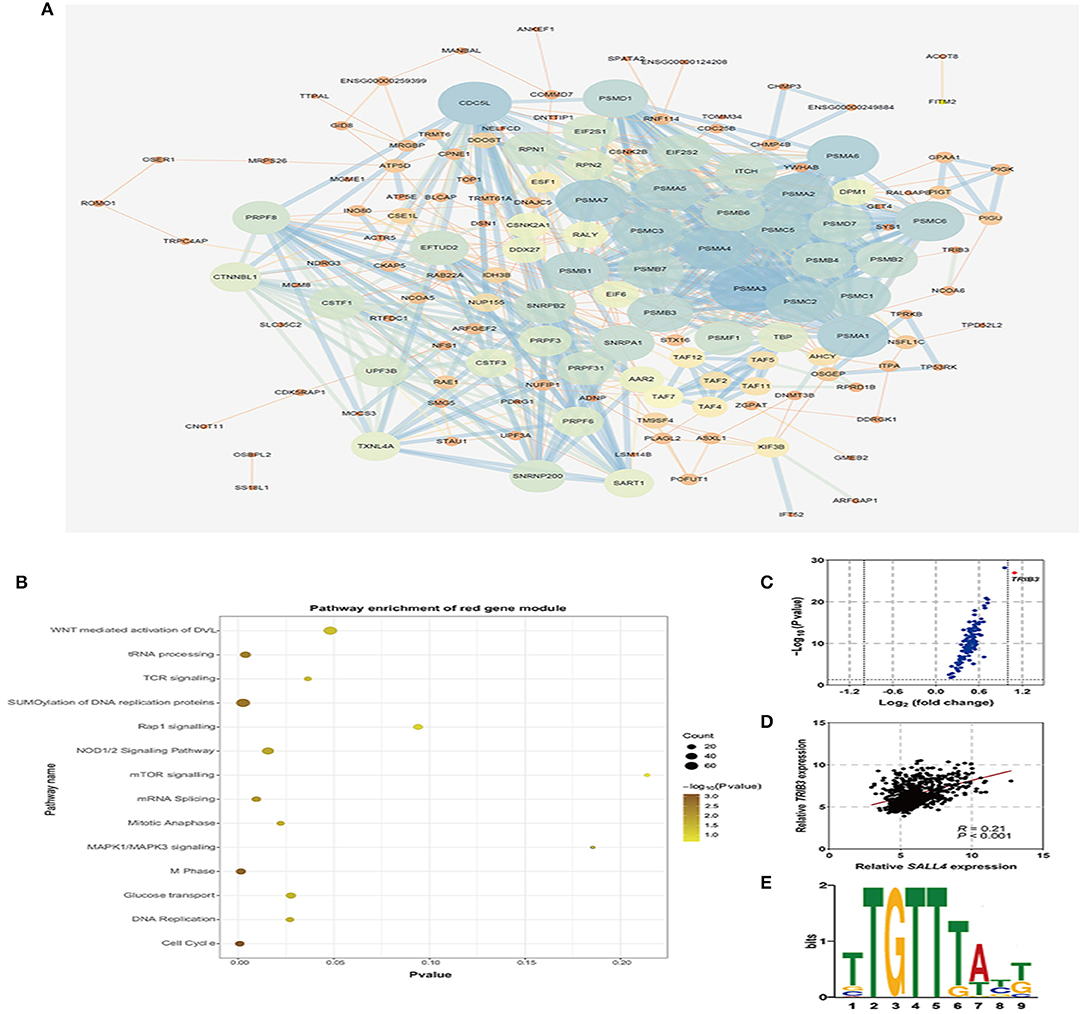
Figure 7. Co-expression genes of SALL4 and its function. Protein–Protein Interaction (PPI) network of the red module, which is the most related gene module of high SALL4 expression (A). Pathway enrichment analysis of genes in the red module in the PPI network (B). The volcano plot of genes in the red module by the R package limma (C). Scatter plot analysis of mRNA expression of SALL4 and its co-expression gene TRIB3 (D). SALL4 binding motif, TTGTTTA(T)T(G)T, was identified by motif analysis tool, MEME-Chip (E).
To find the core gene in the red module, we then analyzed the differential expression of genes (DEGs), and TRIB3 was significantly differentially expressed (Figure 7C). Also, we found that the expression of SALL4 was positively correlated with that of TRIB3 (Figure 7D). TRIB3 interacted with the β-catenin–TCF4 complex, thus activating Wnt/β-catenin signaling (Zhang et al., 2019), pointing that activation of Wnt signaling by SALL4 may need TRIB3. It was evident that SALL4, as a transcriptional regulator, trans-activates CTNNB1 by binding to the promoter of CTNNB1 (Chen et al., 2019), and we wondered whether SALL4 activates Wnt signaling through regulating TRIB3 at the transcriptional level. Using an online motif analysis tool, MEME-Chip, to screen SALL4 binding motifs, one binding motif has been identified: TTGTTTA(T)T(G)T (Figure 7E). Collectively, the present results suggested that the up-regulation of SALL4 may activate the oncogenic Wnt signaling pathway to promote gastric tumorigenesis by influencing the transcriptional mechanisms of TRIB3.
The clinical outcomes of GC differ among patients, and TNM staging is used as a conventional clinical prognostic indicator that only partially reflects biological malignancy. Therefore, prognostic biomarkers of oncogenic potential are required for better risk assessment. This study aimed to investigate whether SALL4, a putative oncofetal gene, influences the prognosis of GC patients, and to investigate the mechanism underlying its effects.
Although SALL4 has been reported as a prognosis-related biomarker for many cancers, such as liver cancer, esophageal cancer, germ cell cancer, and breast cancer (Cao et al., 2009a; Liu T.-C. et al., 2014; Yue et al., 2015; He et al., 2016), the correlation between SALL4 expression and GC prognosis has not been clarified. Previous studies on the prognostic significance of SALL4 in GC are limited by their small sample size (103 cases), and the clinical significance of SALL4 has been evaluated exclusively at the mRNA level, rather than at the protein level (Liu J. et al., 2014). This study, which included a larger series of 1,815 patients, reported that 16.7% of GC tumors were SALL4-positive and that SALL4 positivity was associated with cancer progression-related clinicopathologic parameters, such as advanced stage, lymph node metastasis, and vascular invasion. Our findings are consistent with those of previous reports, which indicated that SALL4 levels were positively associated with lymph node metastasis and that SALL4 is an indicator of metastatic potential in GC (Zhang et al., 2018). A novel finding of our study is that SALL4 is negatively associated with EBV infection in GC. EBVaGC, a molecular subtype classified by TCGA that accounts for approximately 5% of GC cases, has distinct clinicopathological and genetic features, and EBV infection and a lack of SALL4 may mark a group of GC patients with a better prognosis (Yang et al., 2019).
In both univariate and multivariate Cox regression analyses, we noted that SALL4-positive was associated with poor survival in GC. Notably, among patients with stage I and II GC, a SALL4-positive status has a poorer overall survival than a SALL4-negative status. There was no prognostic value among stage III GC patients, implying that SALL4 may predict aggressive types in the early stages of GC. We compared the present results with data from the TCGA STAD dataset and four Asian GC cohorts from GEO by analyzing the association between SALL4 mRNA expression and patient prognosis.
Previous studies have demonstrated the regulation of SALL4 overexpression in cancer. Hypomethylation of the promoter region of SALL4 has been observed in myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML) and is strongly associated with high mRNA levels of SALL4 (Lin et al., 2013; Ma et al., 2013). Also, some studies have demonstrated that miR-16 and lncRNA DANCR mediate the up-regulation of SALL4 (Zhou et al., 2016; Pan et al., 2018). SALL4 is reportedly up-regulated in CDX1-positive intestinal metaplasia of the stomach in both humans and mice (Bard et al., 2009; Fujii et al., 2012). The up-regulated group plays an important role in tumor stemness, drug resistance, apoptosis, cell proliferation, and invasion, highlighting the need to further the current understanding of SALL4 regulation in GC. Several studies on GC have recently suggested gene amplification to be associated with up-regulation (Gorringe et al., 2005; Tsukamoto et al., 2008). In this study, we also found a positive correlation between CN vs. and the mRNA expression of SALL4 in the TCGA dataset, suggesting that the SALL4 copy number gain may contribute to its overexpression in GC.
Several lines of evidence suggest that overexpression of SALL4 in human cancers affects multiple cellular processes involved in tumorigenesis, tumor growth, and tumor progression (Ma et al., 2006; Yang et al., 2008; Itou et al., 2013). To explore how SALL4 affects tumor behavior, especially the molecular mechanisms relevant to SALL4 in GC, we identified the relevant pathways of SALL4 and its co-expressed genes by pathway enrichment analysis and WGCNA. By comparing the enrichment of DEGs with SALL4 in GC, we demonstrated that high expression of SALL4 GC was more concentrated in several signaling pathways, such as Wnt/β-catenin, epithelial–mesenchymal transition, KRAS, and TGF-beta signaling pathways. Several studies have shown that the deregulation of CD44 and TGF-beta signaling could be involved in the SALL4-mediated oncogenic mechanisms (Yuan et al., 2016; Zhang et al., 2018). SALL4 expression has also been reported to be regulated by the Wnt signaling pathway in cervical cancer, hepatocellular carcinoma, and esophageal squamous cell carcinoma (Böhm et al., 2006; He et al., 2016). The Wnt signaling pathway is strongly induced in tumorigenesis, activating kinases to transmit extracellular signals that regulate cell growth, differentiation, proliferation, apoptosis, and migration. Meanwhile, the results of correlation analyses indicated that the co-expression genes of SALL4, such as TRIB3, exhibited positive correlations with SALL4 expression. TRIB3 is a core gene in the Wnt signaling pathway by interacting with β-catenin. A previous study demonstrated that SALL4 could bind to the promoter of CTNNB1 (the gene name of β-catenin) and further activate Wnt/β-catenin signaling in cervical cancer cells, and then we explored whether the transcriptional regulator SALL4 could regulate the expression of TRIB3. Using MEME-Chip, we screen the putative SALL4 binding motif: TTGTTTA(T)T(G)T that is found in the promoter region of TRIB3. Together, activation of the Wnt pathway and SALL4-related genes may result in poor survival in GC, giving new ideas for the follow-up study.
In conclusion, an analysis of 1,815 consecutive GC cases revealed that patients with SALL4-positive GC experience significantly worse outcomes than SALL4-negative patients, indicating that SALL4 is a prognostic indicator in GC. The corresponding increase in the regional copy number of SALL4 and SALL4 mRNA overexpression suggests that its overexpression may due to DNA copy number changes. Besides, SALL4, as well as its co-expressed genes, could potentially activate several pathways, especially the Wnt signaling pathway, which is closely associated with a worse prognosis in GC patients.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The studies involving human participants were reviewed and approved by the Ethics Committee of Peking University Cancer Hospital. The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
LZ and JJ were the principal investigator and responsible for the study design. YY, ZiL, ZB, and XWu collected and assembled data. XWa and YY performed data analysis, interpretation, and drafted the manuscript. YH provided gastric tissues from biobank. YL and ZhL contributed to pathologist scoring. All authors have read and approved the final version of the manuscript.
This work was supported by the National High-Tech R&D Program of China (863 Program) and the Grant Number is 2014AA020603.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.600344/full#supplementary-material
Araki, I., Washio, M., Yamashita, K., Hosoda, K., Ema, A., Mieno, H., et al. (2018). Robust vascular invasion concurrent with intense EGFR immunostaining can predict recurrence in patients with stage IB node-negative gastric cancer. Surg. Today 48, 1–8. doi: 10.1007/s00595-017-1611-x
Bard, J. D., Gelebart, P., Amin, H., Young, L., Ma, Y., and Lai, R. (2009). Signal transducer and activator of transcription 3 is a transcriptional factor regulating the gene expression of SALL4. FASEB J. 23, 1405–1414. doi: 10.1096/fj.08-117721
Böhm, J., Sustmann, C., Wilhelm, C., and Kohlhase, J. (2006). SALL4 is directly activated by TCF/LEF in the canonical Wnt signaling pathway. Biochem. Biophys. Res. Commun. 348, 898–907. doi: 10.1016/j.bbrc.2006.07.124
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A., and Jemal, A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424. doi: 10.3322/caac.21492
Camparo, P., and Comperat, E. M. (2013). SALL4 is a useful marker in the diagnostic work-up of germ cell tumors in extra-testicular locations. Virchows Archiv. 462, 337–341. doi: 10.1007/s00428-012-1353-5
Cao, D., Humphrey, P. A., and Allan, R. W. (2009a). SALL4 is a novel sensitive and specific marker for metastatic germ cell tumors, with particular utility in detection of metastatic yolk sac tumors. Cancer 115, 2640–2651. doi: 10.1002/cncr.24308
Cao, D., Li, J., Guo, C. C., Allan, R. W., and Humphrey, P. A. (2009b). SALL4 is a novel diagnostic marker for testicular germ cell tumors. Am. J. Surg. Pathol. 33, 1065–1077. doi: 10.1097/PAS.0b013e3181a13eef
Cats, A., Jansen, E. P., van Grieken, N. C., Sikorska, K., Lind, P., Nordsmark, M., et al. (2018). Chemotherapy vs. chemoradiotherapy after surgery and preoperative chemotherapy for resectable gastric cancer (CRITICS): an international, open-label, randomised phase 3 trial. Lancet Oncol. 19, 616–628. doi: 10.1016/S1470-2045(18)30132-3
Chen, M., Li, L., and Zheng, P. S. (2019). SALL4 promotes the tumorigenicity of cervical cancer cells through activation of the Wnt/β-catenin pathway via CTNNB1. Cancer Sci. 110, 2794–2805. doi: 10.1111/cas.14140
Fujii, Y., Yoshihashi, K., Suzuki, H., Tsutsumi, S., Mutoh, H., Maeda, S., et al. (2012). CDX1 confers intestinal phenotype on gastric epithelial cells via induction of stemness-associated reprogramming factors SALL4 and KLF5. Proc. Natl. Acad. Sci. U.S.A. 109, 20584–20589. doi: 10.1073/pnas.1208651109
Gonzalez-Roibon, N., Katz, B., Chaux, A., Sharma, R., Munari, E., Faraj, S. F., et al. (2013). Immunohistochemical expression of SALL4 in hepatocellular carcinoma, a potential pitfall in the differential diagnosis of yolk sac tumors. Hum. Pathol. 44, 1293–1299. doi: 10.1016/j.humpath.2012.10.017
Gorringe, K. L., Boussioutas, A., and Bowtell, D. D. (2005). Novel regions of chromosomal amplification at 6p21, 5p13, and 12q14 in gastric cancer identified by array comparative genomic hybridization. Genes Chromosomes Cancer 42, 247–259. doi: 10.1002/gcc.20136
He, J., Zhou, M., Chen, X., Yue, D., Yang, L., Qin, G., et al. (2016). Inhibition of SALL4 reduces tumorigenicity involving epithelial-mesenchymal transition via Wnt/β-catenin pathway in esophageal squamous cell carcinoma. J. Exp. Clin. Cancer Res. 35:98. doi: 10.1186/s13046-016-0378-z
Itou, J., Matsumoto, Y., Yoshikawa, K., and Toi, M. (2013). Sal-like 4 (SALL4) suppresses CDH1 expression and maintains cell dispersion in basal-like breast cancer. FEBS Lett. 587, 3115–3121. doi: 10.1016/j.febslet.2013.07.049
Kim, J., Xu, S., Xiong, L., Yu, L., Fu, X., and Xu, Y. (2017). SALL4 promotes glycolysis and chromatin remodeling via modulating HP1α-Glut1 pathway. Oncogene 36:6472. doi: 10.1038/onc.2017.265
Kudou, K., Saeki, H., Nakashima, Y., Sasaki, S., Jogo, T., Hirose, K., et al. (2018). Postoperative development of sarcopenia is a strong predictor of a poor prognosis in patients with adenocarcinoma of the esophagogastric junction and upper gastric cancer. Am J Surg. 217, 757–763. doi: 10.1016/j.amjsurg.2018.07.003
Li, A., Jiao, Y., Yong, K. J., Wang, F., Gao, C., Yan, B., et al. (2015). SALL4 is a new target in endometrial cancer. Oncogene 34:63. doi: 10.1038/onc.2013.529
Lin, J., Qian, J., Yao, D.-,m., Qian, W., Yang, J., Wang, C.-,z., et al. (2013). Aberrant hypomethylation of SALL4 gene in patients with myelodysplastic syndrome. Leukemia Res. 37, 71–75. doi: 10.1016/j.leukres.2012.10.014
Liu, J., Wang, L., Yang, A., Jiang, P., and Wang, M. (2014). Up-regulation of SALL4 associated with poor prognosis in gastric cancer. Hepato Gastroenterol. 61, 1459–1464.
Liu, L., Zhang, J., Yang, X., Fang, C., Xu, H., and Xi, X. (2015). SALL4 as an epithelial-mesenchymal transition and drug resistance inducer through the regulation of c-Myc in endometrial cancer. PLoS ONE 10:e0138515. doi: 10.1145/2818302
Liu, T.-C., Vachharajani, N., Chapman, W. C., and Brunt, E. M. (2014). SALL4 immunoreactivity predicts prognosis in western hepatocellular carcinoma patients but is a rare event-a study of 236 cases. Am. J. Surg. Pathol. 38:966. doi: 10.1097/PAS.0000000000000218
Ma, J.-,c., Qian, J., Lin, J., Qian, W., Yang, J., Wang, C.-,z., et al. (2013). Aberrant hypomethylation of SALL4 gene is associated with intermediate and poor karyotypes in acute myeloid leukemia. Clin. Biochem. 46, 304–307. doi: 10.1016/j.clinbiochem.2012.11.018
Ma, Y., Cui, W., Yang, J., Qu, J., Di, C., Amin, H. M., et al. (2006). SALL4, a novel oncogene, is constitutively expressed in human acute myeloid leukemia (AML) and induces AML in transgenic mice. Blood 108, 2726–2735. doi: 10.1182/blood-2006-02-001594
Pan, L., Liang, W., Gu, J., Zang, X., Huang, Z., Shi, H., et al. (2018). Long noncoding RNA DANCR is activated by SALL4 and promotes the proliferation and invasion of gastric cancer cells. Oncotarget 9, 1915–1930. doi: 10.18632/oncotarget.23019
Tatetsu, H., Kong, N. R., Chong, G., Amabile, G., Tenen, D. G., and Chai, L. (2016). SALL4, the missing link between stem cells, development and cancer. Gene 584, 111–119. doi: 10.1016/j.gene.2016.02.019
Tsukamoto, Y., Uchida, T., Karnan, S., Noguchi, T., Nguyen, L., Tanigawa, M., et al. (2008). Genome-wide analysis of DNA copy number alterations and gene expression in gastric cancer. J. Pathol. J. Pathol. Soc. Great Britain Ireland 216, 471–482. doi: 10.1002/path.2424
Wang, X., Fu, R., Hu, Y., Du, H., Li, S., Li, Z., et al. (2017). EGFR gene status predicts response and survival benefit in a preclinical gastric cancer trial treating patient-derived xenografts with cetuximab. Oncol. Rep. 38, 2387–2393. doi: 10.3892/or.2017.5907
Yanagihara, N., Kobayashi, D., Kuribayashi, K., Tanaka, M., Hasegawa, T., and Watanabe, N. (2015). Significance of SALL4 as a drug-resistant factor in lung cancer. Int. J. Oncol. 46, 1527–1534. doi: 10.3892/ijo.2015.2866
Yang, J., Chai, L., Gao, C., Fowles, T. C., Alipio, Z., Dang, H., et al. (2008). SALL4 is a key regulator of survival and apoptosis in human leukemic cells. Blood 112, 805–813. doi: 10.1182/blood-2007-11-126326
Yang, Y., Liu, Y. Q., Wang, X. H., Ji, K., Li, Z. W., Bai, J., et al. (2019). Clinicopathological and molecular characteristics of Epstein-Barr virus associated gastric cancer: a single center large sample case investigation. Beijing Da Xue Xue Bao Yi Xue Ban 51, 451–458. doi: 10.19723/j.issn.1671-167X.2019.03.012
Yong, K. J., Gao, C., Lim, J. S., Yan, B., Yang, H., Dimitrov, T., et al. (2013). Oncofetal gene SALL4 in aggressive hepatocellular carcinoma. N. Engl. J. Med. 368, 2266–2276. doi: 10.1056/NEJMoa1300297
Yuan, X., Zhang, X., Zhang, W., Liang, W., Zhang, P., Shi, H., et al. (2016). SALL4 promotes gastric cancer progression through activating CD44 expression. Oncogenesis 5:e268. doi: 10.1038/oncsis.2016.69
Yue, X., Xiao, L., Yang, Y., Liu, W., Zhang, K., Shi, G., et al. (2015). High cytoplasmic expression of SALL4 predicts a malignant phenotype and poor prognosis of breast invasive ductal carcinoma. Neoplasma 62, 980–988. doi: 10.4149/neo_2015_119
Zhang, L., Xu, Z., Xu, X., Zhang, B., Wu, H., Wang, M., et al. (2014). SALL4, a novel marker for human gastric carcinogenesis and metastasis. Oncogene 33:5491. doi: 10.1038/onc.2013.495
Zhang, X., Yuan, X., Zhu, W., Qian, H., and Xu, W. (2015). SALL4: an emerging cancer biomarker and target. Cancer Lett. 357, 55–62. doi: 10.1016/j.canlet.2014.11.037
Zhang, X., Zhang, P., Shao, M., Zang, X., Zhang, J., Mao, F., et al. (2018). SALL4 activates TGF-β/SMAD signaling pathway to induce EMT and promote gastric cancer metastasis. Cancer Manag. Res. 10, 4459–4470. doi: 10.2147/CMAR.S177373
Zhang, X., Zhong, N., Li, X., and Chen, M. B. (2019). TRIB3 promotes lung cancer progression by activating β-catenin signaling. Eur. J. Pharmacol. 863:172697. doi: 10.1016/j.ejphar.2019.172697
Keywords: gastric cancer, SALL4, immunohistochemistry, prognosis, weighted gene co-expression network analysis, enrich pathway analysis, Wnt signaling pathway
Citation: Yang Y, Wang X, Liu Y, Hu Y, Li Z, Li Z, Bu Z, Wu X, Zhang L and Ji J (2021) Up-Regulation of SALL4 Is Associated With Survival and Progression via Putative WNT Pathway in Gastric Cancer. Front. Cell Dev. Biol. 9:600344. doi: 10.3389/fcell.2021.600344
Received: 29 August 2020; Accepted: 11 January 2021;
Published: 11 February 2021.
Edited by:
Mourad Assidi, King Abdulaziz University, Saudi ArabiaReviewed by:
Maximilian Schuff, NEXTCLINIC IVF Zentren Prof. Zech, AustriaCopyright © 2021 Yang, Wang, Liu, Hu, Li, Li, Bu, Wu, Zhang and Ji. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lianhai Zhang, emxoemxoQGhvdG1haWwuY29t; Jiafu Ji, amlqaWFmdUBoc2MucGt1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.