
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol. , 18 January 2021
Sec. Cell Adhesion and Migration
Volume 8 - 2020 | https://doi.org/10.3389/fcell.2020.628222
This article is part of the Research Topic Evolution, Emerging Functions and Structure of Actin-Binding Proteins View all 32 articles
 Sang A. Mun1,2†
Sang A. Mun1,2† Jongseo Park1,2†
Jongseo Park1,2† Kyoung Ryoung Park1,2,3
Kyoung Ryoung Park1,2,3 Youngjin Lee1,2,4
Youngjin Lee1,2,4 Jung Youn Kang1,2
Jung Youn Kang1,2 Taein Park2,5
Taein Park2,5 Minwoo Jin1,2
Minwoo Jin1,2 Jihyeong Yang1,2
Jihyeong Yang1,2 Chang-Duk Jun1
Chang-Duk Jun1 Soo Hyun Eom1,2,5*
Soo Hyun Eom1,2,5*Ca2+ regulates several cellular functions, including signaling events, energy production, and cell survival. These cellular processes are mediated by Ca2+-binding proteins, such as EF-hand superfamily proteins. Among the EF-hand superfamily proteins, allograft inflammatory factor-1 (AIF-1) and swiprosin-1/EF-hand domain-containing protein 2 (EFhd2) are cytosolic actin-binding proteins. AIF-1 modulates the cytoskeleton and increases the migration of immune cells. EFhd2 is also a cytoskeletal protein implicated in immune cell activation and brain cell functions. EFhd1, a mitochondrial fraternal twin of EFhd2, mediates neuronal and pro-/pre-B cell differentiation and mitoflash activation. Although EFhd1 is important for maintaining mitochondrial morphology and energy synthesis, its mechanism of action remains unclear. Here, we report the crystal structure of the EFhd1 core domain comprising a C-terminus of a proline-rich region, two EF-hand domains, and a ligand mimic helix. Structural comparisons of EFhd1, EFhd2, and AIF-1 revealed similarities in their overall structures. In the structure of the EFhd1 core domain, two Zn2+ ions were observed at the interface of the crystal contact, suggesting the possibility of Zn2+-mediated multimerization. In addition, we found that EFhd1 has Ca2+-independent β-actin-binding and Ca2+-dependent β-actin-bundling activities. These findings suggest that EFhd1, an actin-binding and -bundling protein in the mitochondria, may contribute to the Ca2+-dependent regulation of mitochondrial morphology and energy synthesis.
Regulation of the cytoskeleton is essential for cell dynamics, such as the maintenance of cell shape or motility (Egelman, 2004; Wu et al., 2016). Its malfunction promotes muscle weakness, cerebral arteriopathy, cardiomyopathy, and brain abnormalities (Parker et al., 2020). As the major cytoskeletal protein is actin, its regulation is responsible for several cellular functions, including maintenance of cellular morphology and formation of lamellipodia or filopodia (Lee and Dominguez, 2010). In the cytosol, actin monomers form actin filaments, and the actin filament networks are modulated by several actin-binding proteins (ABPs), including profilin and cofilin, which regulate the polymerization of actin and actinin, fascin, allograft inflammatory factor-1 (AIF-1), and EF-hand domain-containing protein 2 (EFhd2), which facilitate actin-bundling or cross-linking (Dubernard et al., 1997; Autieri et al., 2003; Aratyn et al., 2007; Lee and Dominguez, 2010; Kwon et al., 2013; Ali et al., 2016). In the mitochondria, the maintenance of morphology and function requires the mitochondrial actin, β-actin (Xie et al., 2018). β-actin knockout (KO) in mitochondria induces a severe loss of mitochondrial membrane potential, resulting in impaired mitochondrial DNA transcription and large aggregates of nucleoids (Xie et al., 2018). The EF-hand domain-containing protein 1 (EFhd1), a homologous protein of EFhd2, is localized in the mitochondria (Tominaga et al., 2006; Dutting et al., 2011). Since the gene encoding EFhd1 is not present in the mitochondrial DNA, following its translation in cytoplasm, EFhd1 translocates from the cytoplasm to the mitochondria (Anderson et al., 1981).
EFhd2, AIF-1, and EFhd1 have Ca2+-binding EF-hand motifs, which belong to the EF-hand superfamily, but they have distinct subcellular locations (Dutting et al., 2011). For cytosolic EF-hand superfamily, EFhd2 was first identified in lymphocytes, and it regulates cell spreading and the cell migration of immune and epithelial cells by F-actin rearrangement (Vuadens et al., 2004; Aratyn et al., 2007; Ramesh et al., 2009; Kwon et al., 2013). The crystal structure of Ca2+-bound state and EF-hand mutants of EFhd2 have been reported previously (Park et al., 2016). The overall structures of EFhd2 are compact and rigid, comparable to those of Ca2+-calmodulin-peptide complexes; however, EF-hand motifs are flexible in the mutant structures. Since the rigidity of EF-hand motifs in EFhd2 is essential for the F-actin-bundling activity of EFhd2, the mutants cannot bundle F-actin in vitro (Park et al., 2016; Durvanger and Harmat, 2019). AIF-1 is another cytosolic ABP that induces F-actin-bundling to control membrane ruffling in immune cells (Sasaki et al., 2001; Kanazawa et al., 2002; Autieri et al., 2003). The crystal structure of AIF-1 has a similar overall topology to that of EFhd2 (Yamada et al., 2006; Park et al., 2016). Unlike EFhd2 and AIF-1, EFhd1 is localized in the mitochondria and may regulate mitochondrial energy metabolism (Tominaga et al., 2006). EFhd1 modulates the apoptosis and differentiation of neuronal and muscle cells (Tominaga et al., 2006; Dutting et al., 2011). In addition, EFhd1 induces not only mitoflashes but also metabolic changes during the development of pro-/pre-B cells (Hou et al., 2016; Stein et al., 2017). A recent report suggested that EFhd1 affects mitochondrial morphology and energy production in the dorsal root ganglion neurons (Ulisse et al., 2020). However, the mechanism underlying the regulation of how EFhd1 regulates several cellular functions is currently unclear.
Here, we report the crystal structure of the core domain of mouse EFhd1 (CDEFhd1, residues 79–180) in a Ca2+-bound state, comprising a proline-rich (PR) region, two EF-hand motifs, a ligand mimic helix (LM-helix), and a C-terminal linker. The overall structure of CDEFhd1 was similar to that of CDEFhd2 and AIF-1. Intriguingly, we found two Zn2+ ions in the crystal packing interface, suggesting the plausible Zn2+-mediated multimerization. In addition, we identified Ca2+-independent α- and β-actin-binding and Ca2+-dependent β-actin-bundling activities of EFhd1, indicating that EFhd1 might be involved in the Ca2+-dependent regulation of mitochondrial morphology via interactions with β-actin.
The mouse EFhd1 ΔNTD was amplified from full-length EFhd1 using polymerase chain reaction (PCR). The amplified DNA was cloned into a modified pET-28a vector (Novagen) that carried an N-terminal 6 × His (His6)-tobacco etch virus (TEV) protease cleavage site (Glu-Asn-Leu-Tyr-Phe-Gln/Gly). The recombinant plasmid was transformed into Escherichia coli strain BL21 (DE3) cells for protein expression. The cells were cultured at 37°C in Luria-Bertani (LB) broth containing 50 μg/mL kanamycin until the absorbance at 600 nm reached 0.7. Recombinant protein expression was induced with isopropyl β-D-1-thiogalactopyranoside (IPTG) (final concentration of 0.5 mM), and the cells were cultivated for an additional 4 h at 37°C. Cells were harvested by centrifugation at 4,000 × g for 15 min at 4°C. The harvested cell pellet was suspended in a lysis buffer [50 mM HEPES-NaOH pH 7.5, 300 mM NaCl, 20 mM imidazole, 0.4 mM phenylmethylsulfonyl fluoride (PMSF), and 14.3 mM β-mercaptoethanol]. The resuspended cells were disrupted via sonication and centrifuged at 14,000 × g for 50 min at 4°C to discard cell debris. The soluble supernatant was loaded onto a gravity-flow column (Bio-Rad, Hercules, CA, USA) packed with Ni-IDA agarose resin (Elpis), pre-equilibrated, and subsequently washed with the lysis buffer to remove any non-specific proteins. The desired protein was eluted with lysis buffer supplemented with 400 mM imidazole. After concentrating the eluate, the protein solution was incubated with TEV protease overnight at 4°C to cleave the N-terminal His6-TEV tag. To exchange the buffer for crystallization, the final purified protein was passed through a HiLoad 16/60 Superdex 75 gel-filtration column (Pharmacia Biotech) pre-equilibrated with the final buffer (20 mM HEPES-NaOH pH 7.5, 150 mM NaCl, 0.4 mM PMSF, and 14.3 mM β-mercaptoethanol).
The purified protein was concentrated using a 10 K centrifugal filter (Millipore) and stored at −80°C. During purification, the presence of EFhd1 protein was confirmed via sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and protein degradation was observed following incubation with TEV protease.
To investigate the actin-binding function, we purified full-length EFhd1 and EFhd2. Full-length EFhd1 was amplified using PCR and cloned into the modified pET-28a vector (Novagen) carrying an N-terminal His6-TEV tag. The overall expression and affinity chromatography procedure of full-length EFhd1 was similar to that of EFhd1 ΔNTD, except for the step involving the incubation of TEV protease. The process for the removal of the N-terminal His6-TEV tag was omitted due to protein degradation. After affinity chromatography, the eluted protein was concentrated. The final purified protein was passed through a HiLoad 16/60 Superdex 75 gel-filtration column pre-equilibrated with the final buffer (20 mM HEPES-NaOH pH 7.5, 150 mM NaCl, 0.8 mM PMSF, and 5 mM DTT). The purified protein was concentrated using 10 K centrifugal filters (Millipore) and stored at −80°C.
Full-length EFhd2 was amplified using PCR and cloned into the modified pET-28a vector carrying an N-terminal His6 tag. The recombinant plasmid was transformed into E. coli strain BL21 (DE3) cells to express the protein. The cells were grown at 37°C in LB broth containing 50 μg/mL kanamycin until the absorbance at 600 nm reached 0.7. The recombinant protein was induced with 0.5 mM IPTG and the cells were cultured for an additional 5 h at 37°C. Cells were harvested via centrifugation at 4,000 × g for 15 min at 4°C, and the harvested cell pellet was suspended in a lysis buffer (50 mM HEPES-NaOH pH 7.5, 300 mM NaCl, 5 mM imidazole, 0.4 mM PMSF, and 14.3 mM β-mercaptoethanol). The resuspended cells were disrupted via sonication and centrifuged at 14,000 × g for 50 min at 4°C to remove cell debris. The soluble supernatant was loaded onto a gravity-flow column packed with Ni-IDA agarose resin previously equilibrated and subsequently washed with the lysis buffer to remove any non-specific proteins. The protein was eluted with lysis buffer supplemented with 400 mM imidazole. After concentrating the eluate, the final purified protein was passed through a HiLoad 16/60 Superdex 75 gel-filtration column pre-equilibrated with the final buffer (20 mM HEPES-NaOH pH 7.5, 150 mM NaCl, 0.8 mM PMSF, and 5 mM DTT). The purified protein was concentrated using 10 K centrifugal filters (Millipore) and stored at −80°C.
Initially, we attempted to crystallize Ca2+-bound EFhd1 ΔNTD (residues 69–240). EFhd1 ΔNTD was incubated for at least 20 min on ice after the addition of 1 mM CaCl2 and then screened using the sitting-drop vapor-diffusion method in a 96-well INTELLI-PLATE (Art Robbins Ins.). We found that EFhd1 ΔNTD was degraded and the core domain (CDEFhd1, residues 79–180) was crystallized. CDEFhd1 formed rod-shaped crystals after 1 week in a reservoir solution containing 80 mM HEPES-NaOH (pH 7.0), 2 mM ZnSO4, and 25% (v/v) Jeffamine ED-2003 (Molecular Dimensions). Additional refinements of crystallization conditions were performed using the sitting-drop vapor-diffusion method, and drops were prepared by mixing 1 μL of protein and 1 μL of reservoir solution. Crystals were obtained using a reservoir solution containing 0.1 M HEPES-NaOH (pH 7.5), 5 mM ZnSO4, and 25% (w/v) Jeffamine ED-2001 (Hampton Research). For data collection, CDEFhd1 crystals were cryoprotected by transferring into a mother liquor containing additional 30% (v/v) glycerol and flash freezing in liquid nitrogen.
X-ray diffraction data of CDEFhd1 were collected at 100 K using synchrotron X-ray sources on beamline 5C at the Pohang Accelerator Laboratory (PAL, South Korea). We collected the best resolution diffraction data for CDEFhd1 at a 2.07 Å resolution. The CDEFhd1 crystal belongs to the space group P212121 with cell dimensions of a = 31.8, b = 47.6, and c = 87.2 Å. The diffraction data were indexed, processed, and scaled using the HKL2000 suite (Otwinowski and Minor, 1997). Template for molecular replacement (MR) of the EFhd1 core domain was generated by the SWISS-MODEL homology-modeling server using the human EFhd2 core domain (PDB ID: 5I2L) as the template (Waterhouse et al., 2018). Using this homology-model, the initial model of EFhd1 was determined via MR using Phaser in CCP4 (McCoy et al., 2007; Winn et al., 2011). Using the initial model, additional model building was performed using the COOT program (Emsley and Cowtan, 2004). Iterative refinement was performed with phenix.refine (Afonine et al., 2012; Liebschner et al., 2019). The details of the data collection and refinement statistics are provided in Table 1.
All structural figures were generated using PyMOL version 1.5.0.4 (Schrödinger LLC). PDBePISA was used to analyze the interface, and the PRODIGY web server was used to predict the binding energies of symmetry-mate molecules (Krissinel and Henrick, 2007; Xue et al., 2016). Multiple sequence alignment was performed using ESPript 3.0 (Robert and Gouet, 2014). The Fobs-Fcalc map was calculated using phenix.maps and converted to the ccp4 format using a phenix.mtz2map (Liebschner et al., 2019).
To measure the precipitation of EFhd1 and EFhd2 in various Zn2+ concentrations, we performed an in vitro precipitation assay. First, 6 μM of His6-TEV tagged full-length EFhd1 and His6 tagged full-length EFhd2 were incubated with 20 μM to 10 mM ZnCl2 in reaction buffer (100 mM KCl, 0.2 mM Tris-HCl, pH 8.0) at 24°C for 30 min. The precipitated proteins were pelleted via centrifugation at 15,000 × g for 10 min at 24°C. Equal volumes of pellet or supernatant solutions were resolved via SDS-PAGE, and the protein bands were visualized via Coomassie Brilliant Blue staining.
To measure the value (concentration of half maximal protein aggregation for Zn2+), we used a spectrophotometric method. First, 6 μM of His6-TEV tagged full-length EFhd1 and His6 tagged full-length EFhd2 were incubated with various concentrations of ZnCl2 (0–1 mM ZnCl2 with EFhd1, 0–20 mM ZnCl2 with EFhd2) in reaction buffer (100 mM KCl, 0.2 mM Tris-HCl, pH 8.0) at 24°C for 30 min. The turbidity of the reacted proteins was monitored by measuring the absorbance at 470 nm using a spectrophotometer (Ultrospec 2000; Pharmacia Biotech). Graphs of absorbance at 470 nm were fitted using OriginPro 9.1 software (OriginLab Corporation, Northampton, MA, USA).
Actin co-sedimentation assays were performed as previously reported (Kwon et al., 2013). In brief, non-muscle actin (85% β-actin and 15% γ-actin), derived from human platelets, and muscle actin (α-actin), derived from rabbit skeletal muscle (Cytoskeleton Inc.), were mixed in G-buffer (0.2 mM CaCl2, 5 mM Tris-HCl, pH 8.0) to produce actin stock and polymerized in an actin polymerization buffer (100 mM KCl, 2 mM MgCl2, 0.5 mM ATP, 0.2 mM Tris-HCl, pH 8.0) at 24°C for 1 h. Solutions containing polymerized actin (8 μM) were incubated with bovine serum albumin (BSA, 4 μM), EFhd1 (12 μM), or EFhd2 (12 μM) for 30 min at 24°C in the presence of 1 mM ethylene glycol tetraacetic acid (EGTA) or 1 mM CaCl2. Actin filaments with each protein were pelleted via centrifugation at 100,000 × g for 2 h at 24°C (for the actin-binding assay). BSA and EFhd2 were used as negative and positive controls, respectively. Equal amounts of pellet and supernatant were resolved via SDS-PAGE, and the protein bands were visualized by Coomassie Blue staining. The percentage of each protein in the pellet was quantified via densitometry using ImageJ 1.44p, and the percentage of pellet histogram was plotted using OriginPro 9.1 software (OriginLab Corporation, Northampton, MA, USA) (Schneider et al., 2012).
Non-muscle actin (Cytoskeleton Inc.) was polymerized in F-actin buffer containing 100 mM KCl, 2 mM MgCl2, 0.5 mM ATP, and 0.2 mM Tris-HCl at pH 8.0. Mixtures (50 μL) of F-actin (4 μM) and full-length EFhd1 (6 μM) in the presence of 1 mM EGTA or 0.5 mM CaCl2 were allowed to react for 1 h. For grid preparation, 2 μL of reaction mixture was loaded onto the Formvar and metal-coated grids and blotted with filter paper to remove excess samples. The sample-loaded grid was stained using a solution of 1% (w/v) uranyl acetate. The grids were immersed in the stain solution for 20 min, blotted with filter paper to remove excess stain, and air-dried. The samples were analyzed using an FEI Tecnai G2 transmission electron microscope operated at 120 kV.
We determined the crystal structure of the core domain of mouse EFhd1 (CDEFhd1, residues 79–180) at a resolution of 2.07 Å and refined to Rwork = 20.9 (%) and Rfree = 22.9 (%) (Table 1). We initially attempted to crystallize the EFhd1 ΔNTD (residues 69–240) construct, but only the core domain was crystallized due to proteolytic degradation (Figure 1A). The CDEFhd1 structure comprised two EF-hand motifs (residues 91–162), an LM-helix (residues 169–176), a C-terminus of the PR region (residues 79–89) at the N-terminus, and a C-terminal linker (residues 177–180) (Figure 1B). Within the structure, Ca2+ ions were coordinated in each of the two EF-hand motifs of CDEFhd1 (Figures 1C,D). Consensus residues for Ca2+ coordination in the EF-hand consist of 12 amino acids with patterns of 1(X), 3(Y), 5(Z), 7(-Y), 9(-X), and 12(-Z) comprised of the five monodentate ligands and one bidentate ligand for -Z (Lewit-Bentley and Rety, 2000). Consequently, the geometry for Ca2+ coordination of the EF-hand is generally pentagonal bipyramid with a coordination number of seven comprising six oxygen atoms from the side chains and one main-chain carbonyl oxygen of -Y (Lewit-Bentley and Rety, 2000; Grabarek, 2006). In the case of EF-hand 1 of CDEFhd1, two water molecules participated in the Ca2+ coordination instead of the residues in position Y (G106) and -X (D112) (Supplementary Figure 1). Notably, this alternative pattern of Ca2+ coordination formed a distorted pentagonal bipyramid geometry. Unlike EF-hand 1, in the case of EF-hand 2 of CDEFhd1, one water molecule participated in Ca2+ coordination instead of the residues in position -X (S148). The Ca2+ coordination geometry of EF-hand 2 was maintained in the general pentagonal bipyramid. Collectively, EF-hand 1 and EF-hand 2 of CDEFhd1 had a distorted or general geometry for Ca2+ coordination, respectively.
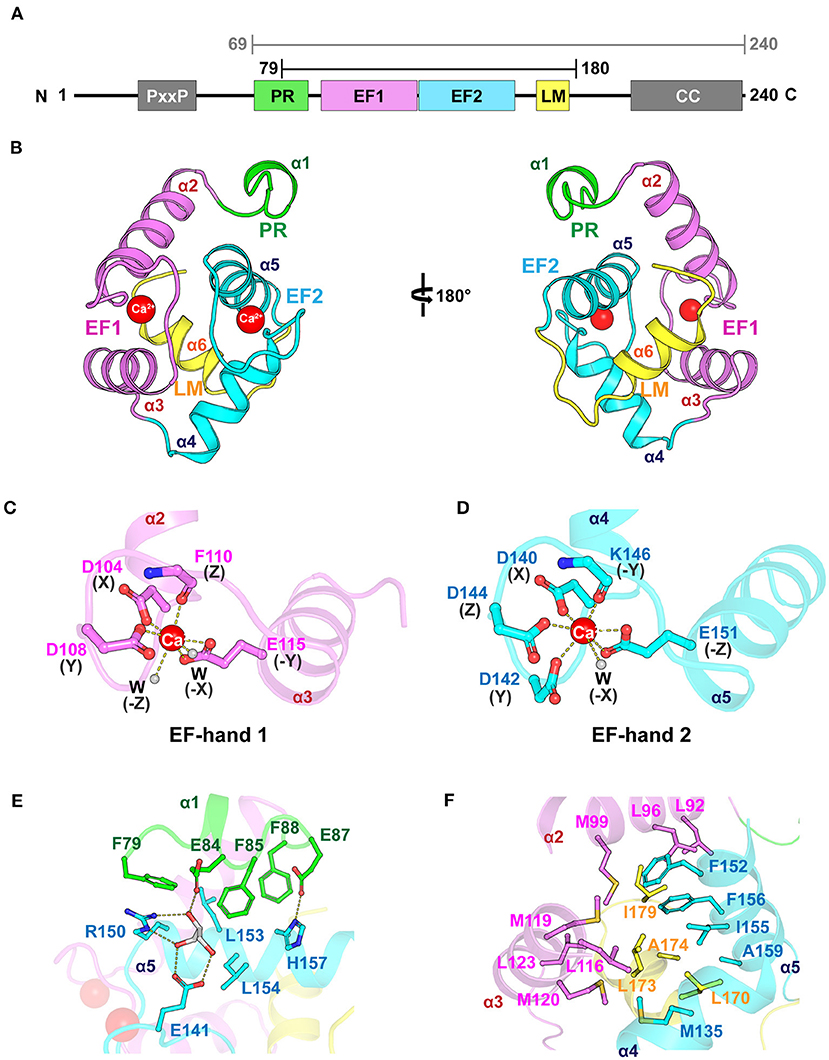
Figure 1. Overall structure of CDEFhd1. (A) Schematic diagram of mouse EFhd1 consisting of a PR (proline-rich) region, EF1 (EF-hand 1), EF2 (EF-hand 2), LM (ligand mimic)-helix, and CC (coiled coil). The upper bars indicate purified regions of EFhd1 (residues 69–240) and crystallized regions of EFhd1 (residues 79–180), respectively. (B) The overall structure of the core domain of EFhd1 (CDEFhd1). The PR region is colored green. EF1 and EF2 are colored violet and cyan, respectively. The LM-helix was colored yellow. (C,D) A cartoon representation of EF1 (C) and EF2 (D) with Ca2+ represented by a red sphere. The residues participating in Ca2+ coordination are represented in the stick form. Ca2+ coordination is marked by dashed lines. (E) Detailed view of the interaction between PR region and EF2. The interacting residues are represented in stick form, and the hydrogen bonds are marked by dashed lines. (F) Detailed view of the interaction between the LM-helix and two EF-hand motifs. The interacting residues are represented in stick form.
The structure of the motifs in CDEFhd1 was stabilized by hydrophobic intramolecular interactions. In the PR region (helix α1), three Phe residues (F79, F85, and F88) formed hydrophobic interactions with L153 and L154 of helix α5 of EF-hand 2 (Figure 1E). The interaction of the PR region and helix α5 was further stabilized through the hydrogen bond network comprising E84 (PR region), E87 (PR region), E141 (interloop of EF-hand 2), R150 (helix α5), H157 (helix α5), and a glycerol molecule (gray). The LM-helix was stabilized by the intramolecular hydrophobic interaction network comprising the LM-helix (L170, L173, A174), helix α2 (M99), helix α3 (L116, M119, M120, L123), helix α4 (M135), and helix α5 (F152, I155, F156, A159) (Figure 1F). In the case of the C-terminal linker, I179 formed hydrophobic interactions with L92, L96 of helix α2, and F156 of helix α5. Collectively, CDEFhd1 formed a compact and rigid domain structure through these intramolecular interactions.
The genes encoding EFhd1, EFhd2, and AIF-1 evolved from the common ancestral species of Bilateria (Dutting et al., 2011). The sequences of EFhd1 and EFhd2 are highly conserved, with a sequence identity of 65%, but the sequence of AIF-1 is conserved with that of EFhd1 only in the EF-hand motifs with an overall sequence identity of 15% due to the difference in evolutionary branching. Although the sequence conservation was limited to the EF-hand motifs in AIF-1, the overall structures of these proteins for Ca2+-bound states were relatively well-superimposed (RMSD of CDEFhd1 and CDEFhd2 = 0.403 Å for 85 Cα atoms, RMSD of CDEFhd1 and AIF-1 = 2.089 Å for 63 Cα atoms) (Figure 2A). EF-hand motifs of these proteins accommodate a helix (LM-helix in CDEFhd1 and CDEFhd2, and helix E in AIF-1), comparable with the binding mode of the calmodulin-ligand interaction (Durvanger and Harmat, 2019). When we compared the Ca2+-bound CDEFhd1 and CDEFhd2, the LM-helices of both proteins participated in the intramolecular hydrophobic interactions with the hydrophobic groove of the EF-hands, and the hydrophobic interaction networks of CDEFhd1 and CDEFhd2 were structurally conserved (Figures 2B,C) (Park et al., 2016). Unlike the LM-helix of CDEFhd1 and CDEFhd2, the helix E of Ca2+-bound AIF-1 formed intermolecular hydrophobic interactions with the hydrophobic groove of two EF-hands in a symmetry-mate molecule, resulting in the dimer formation of AIF-1 (Yamada et al., 2006). When we compared the intramolecular interactions in CDEFhd1 and the intermolecular interactions in AIF-1, a distinct hydrophobic interaction network between the hydrophobic groove of the EF-hands and the accommodated helix (LM-helix in CDEFhd1 and helix E′ in AIF-1) was found (Figures 2B,D). Collectively, the overall structure of CDEFhd1 was similar to that of CDEFhd2 and AIF-1, but the hydrophobic interaction network between the LM-helix and hydrophobic groove of EF-hands in CDEFhd1 was similar to that of CDEFhd2, but not AIF-1.
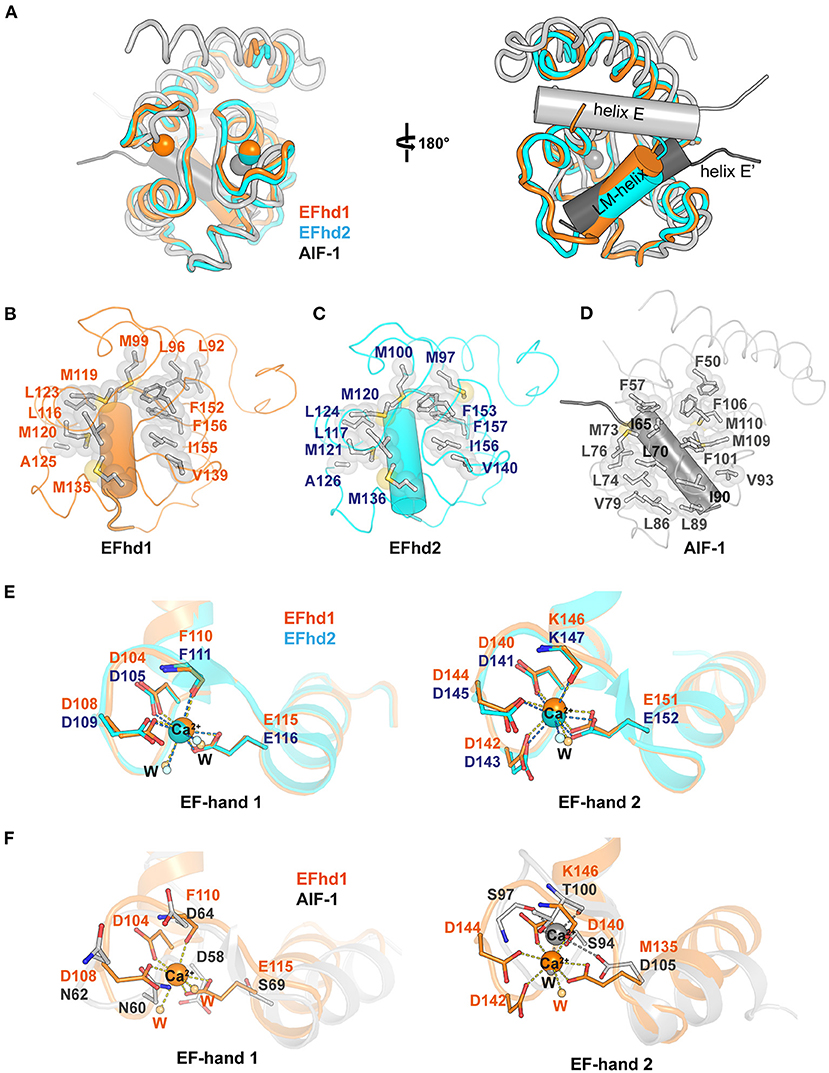
Figure 2. Structural comparison between Ca2+-bound CDEFhd1, CDEFhd2, and AIF-1. (A) Ribbon diagrams of superimposed CDEFhd1 (PDB ID: 7CLT), CDEFhd2 (PDB ID: 5I2L), and AIF-1 (PDB ID: 1WY9). The ribbon diagrams are represented in different colors: orange (CDEFhd1), cyan (CDEFhd2), and gray (AIF-1). The LM-helices of EFhd1 (orange) and EFhd2 (cyan) are presented in a cylindrical diagram. Helix E (gray) and E′ (dark gray) of AIF-1 are presented in the cylindrical diagram. Helix E′ is derived from the symmetry mate molecule of AIF-1. (B–D) Detailed view of the hydrophobic networks of the EFhd1 (B), EFhd2 (C), and AIF-1 (D). The hydrophobic residues are represented in the stick and surface form. The LM-helices of EFhd1 and EFhd2 are colored in orange and cyan, respectively. The helix E′ in AIF-1 is colored in dark gray. The LM-helices and helix E′ are represented in the cylindrical diagram. (E,F) Detailed view of superimposed EF-hand 1 or EF-hand 2 on EFhd1 (orange) and EFhd2 (cyan) (E), or EFhd1 (orange) and AIF-1 (gray) (F). The residues for Ca2+ coordination are represented in the stick form. Ca2+ is marked by orange (EFhd1), cyan (EFhd2), or gray (AIF-1) spheres. Ca2+ coordination is marked by dashed lines.
To compare the EF-hand motifs of CDEFhd1, CDEFhd2, and AIF-1, we superimposed the structures based on EF-hand 1 or 2. In CDEFhd1 and CDEFhd2, EF-hand 1 coordinated Ca2+, and the Ca2+ coordinating residues were well-superimposed (RMSD = 0.269 Å for 36 Cα atoms) (Figure 2E). However, the EF-hand 1 of AIF-1 could not coordinate Ca2+ because there was no space for Ca2+ coordination due to the β-turn, which was stabilized by a hydrogen bond network comprised of N60, N62, and D64 (Yamada et al., 2006) (Figure 2F). In addition, the consensus residues for the EF-hand 1 of AIF-1 were not conserved with those of CDEFhd1 and CDEFhd2 (Supplementary Figure 1). Although EF-hand 2 of these proteins could coordinate Ca2+, the geometries for Ca2+ coordination were distinct. The EF-hand 2 of EFhd1 and EFhd2 formed geometries of the pentagonal bipyramid for Ca2+ coordination, but that of AIF-1 formed a trigonal bipyramidal geometry (Figures 2E,F). This originated from the differences in sequences between AIF-1 and EFhd1 or EFhd2.
The canonical EF-hand domain has two helix-loop-helix motifs comprising four helices (helix 1, helix 2, helix 3, and helix 4) and forms two hydrophobic clusters (I and II) (Denessiouk et al., 2014). Helices 1 and 4 form hydrophobic cluster I, comprising three aromatic residues, and helices 2 and 3 usually form hydrophobic cluster II, comprising a combination of aromatic, hydrophobic, and polar amino acids. Based on the conformational changes of the hydrophobic clusters I and II upon Ca2+ binding, EF-hand-containing proteins can be classified into five separate types (Denessiouk et al., 2014). A previous report suggested that CDEFhd2 belongs to type I, which maintains an open conformation, secondary structures, and cluster interactions independent of Ca2+ (Ferrer-Acosta et al., 2013; Park et al., 2016). Although we could not determine the type of CDEFhd1 due to the lack of the structure of the apo-state, we expected that CDEFhd1 also belonged to type I because of its structure and sequence similarity with CDEFhd2 (Figure 2A, Supplementary Figure 2). Indeed, the structures of the EF-hand motifs comprising hydrophobic clusters were highly conserved in CDEFhd1 and CDEFhd2 [RMSDs of EF-hand motifs, cluster I, and cluster II are 0.334 Å (for 66 Cα atoms), 0.266 Å (for 25 Cα atoms), and 0.177 Å (for 22 Cα atoms), respectively]. In addition, the sequences of the EF-hands in both proteins were highly conserved with a sequence identity of 85% (hydrophobic cluster I: F100, F149, and F152 in EFhd1; F101, F150, and F153 in EFhd2; hydrophobic cluster II: L113, L116, and I136 in EFhd1; L114, L117, and I137 in EFhd2). Therefore, CDEFhd1 was classified as type I.
In the electron density map (Fobs-Fcalc) of CDEFhd1, we observed two strong unidentified electron density maps located at the interfaces of the symmetry-mate molecules (Figure 3A). We further analyzed the unidentified electron density based on the metal coordinating geometric analysis performed with the CheckMyMetal and MetalPDB server (Zheng et al., 2017; Putignano et al., 2018). The coordination geometry was predicted to be tetrahedron, which is the major geometry for Zn2+ coordination. In addition, with the addition of 5 mM ZnSO4 to the crystallization condition, we expected that Zn2+ ions (Zn1 and Zn2) would be present in the maps. We identified two Zn2+-mediated interactions between one CDEFhd1 (MolA) and other symmetry-mate molecules (MolB and MolC) (Figures 3B,C). Zn1 was coordinated by H129 (MolB), K133 (MolB), H157 (MolA), and E163 (MolA), and Zn2 was coordinated by D142 (MolA), D144 (MolA), a water molecule, and E166 (MolC). To further analyze the Zn2+-mediated interactions, we compared the interfaces between MolA and MolB (interface 1), and between MolA and MolC (interface 2). The interface areas of interfaces 1 and 2 were 339 and 151 Å2, respectively, and the predicted binding energy of interface 1 (−4.3 kcal mol−1) was lower than that of interface 2 (−3.3 kcal mol−1), suggesting that interface 1 is more energetically stable than interface 2. Therefore, interface 1 might contribute more to the Zn2+-mediated multimerization of the EFhd1 than interface 2.
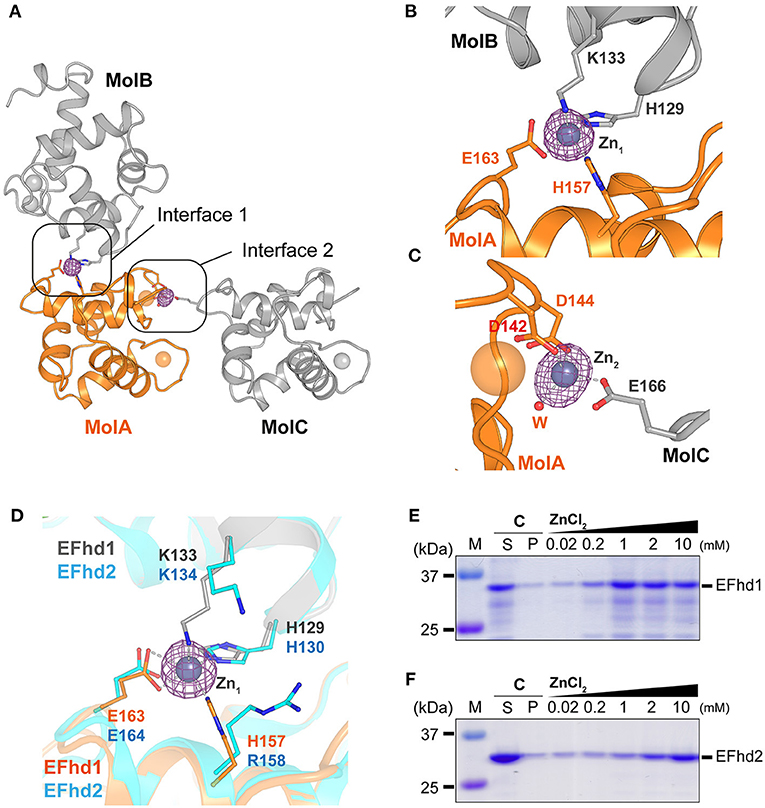
Figure 3. Zn2+-mediated crystal packing interactions of CDEFhd1 and Zn2+-dependent precipitation assay of CDEFhd1 and CDEFhd2. (A) A cartoon representation of the symmetry mate molecules whose interactions are mediated by Zn2+. Interfaces 1 and 2 are formed by MolA and MolB or MolA and MolC, respectively. MolA is colored in orange, and MolB and MolC are colored in gray. The residues for intermolecular interactions are represented in the stick form. The Fobs-Fcalc maps for Zn2+ are marked by the magenta mesh form. (B,C) Detailed view of interface 1 (B) and interface 2 (C). The residues for Zn2+ coordination are represented in stick form. The Zn2+ coordination is marked by dashed lines. (D) Detailed view of superimposed interface 1 of EFhd1 (orange and gray) and EFhd2 (cyan) (PDB ID: 5I2L). The residues for Zn2+ coordination are represented in the stick form. (E,F) SDS-PAGE results of the Zn2+-dependent precipitation assay using EFhd1 (E) or EFhd2 (F). Zn2+ untreated samples are marked by control (C) and were centrifuged to separate the supernatant (S) and precipitant (P) fractions. The samples mixed with various concentrations of ZnCl2 (0.02, 0.2, 1, 2, and 10 mM) were centrifuged to separate supernatant (S) and precipitant (P) fractions, and the P fractions were analyzed via SDS-PAGE.
The Zn1 coordinating residues in EFhd1 (H129, K133, H157, and E163) were highly conserved with those in EFhd2 (H130, K134, R158, and E164), except H157 in EFhd1, which was replaced by R158 in EFhd2 (Figure 3D, Supplementary Figure 1). As histidine is a major ligand for Zn2+, we expected that the Zn2+-mediated multimerization of EFhd1 would be observed to a greater degree than that of EFhd2. To evaluate the difference between the Zn2+-mediated multimerization of EFhd1 and EFhd2, we performed Zn2+-dependent precipitation assays and turbidity measurements. In the precipitation assays, we found that the precipitation ratio of EFhd1 and EFhd2 increased in proportion to [Zn2+] (Figures 3E,F). Consistent with the precipitation assays, the turbidities of both proteins increased with a [Zn2+] (Supplementary Figure 3), and we obtained the (concentration of half maximal protein aggregation for Zn2+) of 0.41 ± 0.02 mM for EFhd1 and 5.9 ± 0.4 mM for EFhd2. Therefore, we concluded that EFhd1 and EFhd2 could be multimerized by Zn2+, and EFhd1 was more sensitive to Zn2+ than EFhd2 for multimerization.
EFhd2 and AIF-1, which belong to the EF-hand superfamily proteins, have F-actin-binding and -bundling activities (Sasaki et al., 2001; Schulze et al., 2008; Huh et al., 2013; Kwon et al., 2013). As EFhd1 also belongs to the EF-hand superfamily proteins and has a high sequence similarity with EFhd2, we expected that EFhd1 would also be involved in F-actin-binding and -bundling. To measure the actin-binding activity, we performed in vitro high-speed co-sedimentation assays using β-actin with full-length EFhd1 or EFhd2 (Figure 4A). The co-sedimentation ratios of EFhd1 and EFhd2 in the presence of Ca2+ were 36 ± 2% (all errors of means are at 95% confidence interval) and 24 ± 2%, respectively. In the absence of Ca2+, the co-sedimentation ratios of EFhd1 and EFhd2 were 36 ± 2% and 19 ± 2%, respectively. Both proteins were Ca2+-independently co-sedimented with β-actin, but the sedimentation ratio of EFhd1 was 1.7-fold higher than that of EFhd2. This suggested that EFhd1 has a higher binding affinity for β-actin than EFhd2, independent of Ca2+.
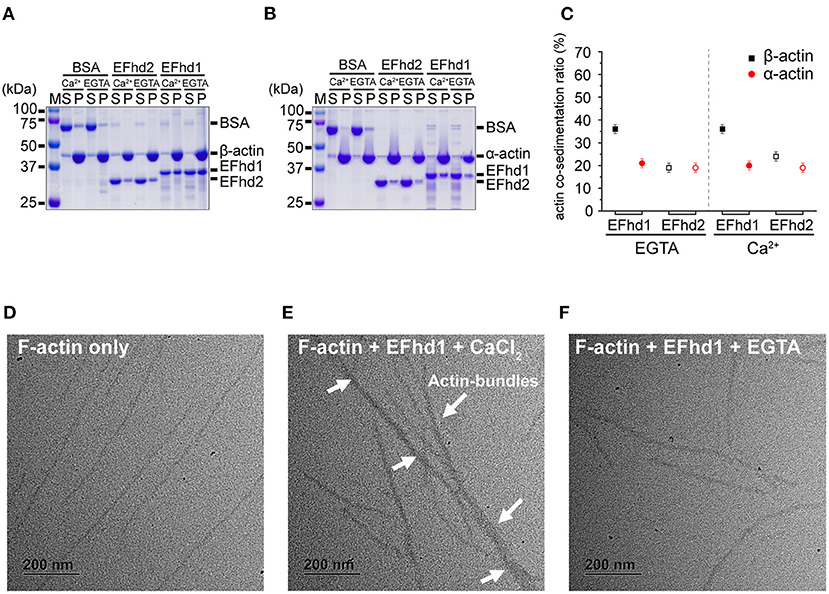
Figure 4. In vitro β/α-actin-binding and -bundling activities of EFhd1. (A,B) SDS-PAGE results for co-sedimentation assays with EFhd1 and filamentous β-actin (A) and α-actin (B). EFhd2 and BSA were used as positive and negative controls, respectively. To control the presence or absence of the Ca2+ environment, 1 mM CaCl2 or 1 mM EGTA was added. (C) Quantification of the results of the co-sedimentation assays. The average values of the co-sedimentation ratios of each experiment are marked by black squares or red spheres. The error bars represent the 95% confidence interval for the mean, which was calculated from 10 independent experiments. Filled and open black squares represent the co-sedimentation ratios of EFhd1 and EFhd2 with β-actin, respectively. Filled and open red spheres represent the co-sedimentation ratios of EFhd1 and EFhd2 with α-actin, respectively. All co-sedimentation ratios were measured in the presence and absence of Ca2+. (D–F) Micrographs of negative staining electron microscopy for F-actin only (D), F-actin with 0.5 mM CaCl2 (E), and F-actin with EFhd1 and 1 mM EGTA (F).
While α-actin is localized in the cytosol, β-actin is localized not only in the cytosol but also in the mitochondrial matrix (Storch et al., 2007; Reyes et al., 2011; Xie et al., 2018). We hypothesized that the binding affinity of EFhd1 for β-actin may differ from that of α-actin because EFhd1 is localized in mitochondria (Tominaga et al., 2006). We performed co-sedimentation assays using α-actin with full-length EFhd1 or EFhd2 (Figure 4B). The co-sedimentation ratios of EFhd1 and EFhd2 in the presence of Ca2+ were 20 ± 2% and 19 ± 2%, respectively. In the absence of Ca2+, the co-sedimentation ratios of EFhd1 and EFhd2 were 21 ± 2% and 19 ± 2%, respectively. Both proteins were similarly co-sedimented with α-actin independent of Ca2+, and the sedimentation ratio of EFhd1 was comparable to that of EFhd2, suggesting similar α-actin-binding affinities of EFhd1 and EFhd2. When we compared the sedimentation ratio of EFhd1 for α- and β-actin, the sedimentation ratio for β-actin was 1.8-fold higher than that for α-actin, suggesting that EFhd1 has higher binding affinities for β-actin than α-actin. In the case of EFhd2, the sedimentation ratio for α- and β-actin was similar, suggesting similar binding affinities of EFhd2 for α- and β-actin. Collectively, EFhd1 had a higher actin-binding affinity for β-actin than that for α-actin regardless of Ca2+, and EFhd2 had a similar actin-binding affinity for α- and β-actin independent of Ca2+ (Figure 4C).
In addition to the co-sedimentation assays, we performed negative staining electron microscopy imaging with EFhd1 and F-actin (β-actin) to identify the actin-bundling activity of EFhd1 (Figures 4D–F). In the electron micrographs, we found the F-actin bundles only in the presence of Ca2+, suggesting the Ca2+-dependent actin-bundling activity of EFhd1. Meanwhile, the F-actin-bundling activity of EFhd1 for β-actin was lower than that of EFhd2 for α-actin (Huh et al., 2013). Collectively, EFhd1 had a Ca2+-dependent β-actin-bundling activity, which is lower than the α-actin-bundling activity of EFhd2.
This study demonstrated the crystal structure of the EFhd1 core domain, whose overall structure was similar to that of EFhd2 and AIF-1. We found two Zn2+ ions in the crystal packing interface, providing new insights into the Zn2+-mediated multimerization of EFhd1. In addition, we first identified the actin-binding and -bundling activities of EFhd1 in vitro. For β-actin, EFhd1 had Ca2+-independent β-actin-binding and Ca2+-dependent β-actin-bundling activities. EFhd1 bound not only to β-actin, but also to α-actin in vitro, which is the primary actin isoform in the cytosol, implying that EFhd1 could bind to α-actin in the cytosol.
We identified the Zn2+-mediated aggregation of EFhd1 with = 0.41 ± 0.02 mM. Mitochondrial [Zn2+] remains controversial, but it is estimated to be in the submicromolar range in the Zn2+ overload state (Sensi et al., 2003; Park et al., 2012; Chabosseau et al., 2014). Although [Zn2+] for half aggregation of EFhd1 in vitro was much higher than that of mitochondrial [Zn2+], and the concentration of EFhd1 may differ between in vitro and physiological conditions, we cannot rule out the possibility of the multimerization of EFhd1 in the Zn2+ overload state because the local spatial and temporal mitochondrial [Zn2+] may be much higher than the reported micromolar range. It will be interesting to study the structural and functional role of Zn2+ ions in actin-binding and -bundling activities. In the cytosol, [Zn2+] is tightly regulated from the picomolar to nanomolar range, suggesting that the Zn2+-mediated multimerization of EFhd1 may not occur in the cytosol (Kambe et al., 2015). Thus, we suggest that EFhd1 binds to actin and is multimerized by Zn2+ in the mitochondria.
EFhd1 KO neurons showed alterations in mitochondrial morphology to a shortened shape, and the mitochondrial morphology could be affected by β-actin regulation (Xie et al., 2018; Ulisse et al., 2020). We found that EFhd1 had Ca2+-independent β-actin-binding and Ca2+-dependent β-actin-bundling activities (Figure 4). Therefore, we suggest that EFhd1 binds to β-actin in the resting state and induces β-actin-bundling in the Ca2+ overload state of mitochondria. The regulation of β-actin not only affects mitochondrial morphology but also the energy synthesis of mitochondria. The energy synthesis of mitochondria is reduced when the expression of efhd1 is downregulated (Stein et al., 2017; Ulisse et al., 2020). Therefore, we suggest that EFhd1 induces actin rearrangement in the mitochondria, resulting in changes in energy synthesis.
In this study, we determined the crystal structure of mouse EFhd1 without C-terminal coiled-coil, and proposed Zn2+-mediated EFhd1 multimerization. In addition, we unveiled the actin-binding and -bundling activities of EFhd1. Nevertheless, the C-terminal coiled-coil is important for understanding the actin regulation mechanism of EFhd1. The coiled-coil of EFhd1 is expected to be important for the dimerization of EFhd1 and actin-bundling activity because these regions are highly conserved in EFhd1 and EFhd2 (Kwon et al., 2013). Therefore, to understand the structural and functional role of EFhd1, structural studies on the full-length EFhd1 and EFhd1-actin filament complexes need to be performed.
Atomic coordinates and structure factors of CDEFhd1 have been deposited in the RCSB PDB with accession code 7CLT.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: http://www.wwpdb.org/, 7CLT.
SHE, C-DJ, and SAM planned and organized the experiments. SAM and JYK carried out the gene cloning and expression. SAM performed the purification, crystallization, structure determination and analysis, ensemble refinement, data analysis, in vitro actin co-sedimentation assay, and analysis of electron microscopy. KRP performed in vitro actin co-sedimentation assay and electron microscopy analysis. YL and JP collected the X-ray diffraction data. TP, MJ, and JY performed the structure determination. SHE, SAM, and JP wrote the manuscript with critical editorial input from the C-DJ. All authors contributed to the article and approved the submitted version.
This work was supported by the GIST Research Institute (GRI) IIBR grant funded by the GIST in 2020.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank the staff at beamlines BL-5C, 7A, and 11C of the Pohang Accelerator Laboratory (Pohang, Republic of Korea) for their kind help with data collection. We thank Dr. Yasuhiro Tomooka (Department of Biological Science and Technology, Faculty of Industrial Science and Technology, Tokyo University of Science) for providing the EFhd1 gene.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2020.628222/full#supplementary-material
Afonine, P. V., Grosse-Kunstleve, R. W., Echols, N., Headd, J. J., Moriarty, N. W., Mustyakimov, M., et al. (2012). Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367. doi: 10.1107/S0907444912001308
Ali, M., Heyob, K., Jacob, N. K., and Rogers, L. K. (2016). Alterative expression and localization of profilin 1/VASPpS157 and Cofilin 1/VASPpS239 regulates metastatic growth and is modified by DHA supplementation. Mol. Cancer Ther. 15, 2220–2231. doi: 10.1158/1535-7163.MCT-16-0092
Anderson, S., Bankier, A. T., Barrell, B. G., De Bruijn, M. H., Coulson, A. R., Drouin, J., et al. (1981). Sequence and organization of the human mitochondrial genome. Nature 290, 457–465. doi: 10.1038/290457a0
Aratyn, Y. S., Schaus, T. E., Taylor, E. W., and Borisy, G. G. (2007). Intrinsic dynamic behavior of fascin in filopodia. Mol. Biol. Cell 18, 3928–3940. doi: 10.1091/mbc.e07-04-0346
Autieri, M. V., Kelemen, S. E., and Wendt, K. W. (2003). AIF-1 is an actin-polymerizing and Rac1-activating protein that promotes vascular smooth muscle cell migration. Circ. Res. 92, 1107–1114. doi: 10.1161/01.RES.0000074000.03562.CC
Chabosseau, P., Tuncay, E., Meur, G., Bellomo, E. A., Hessels, A., Hughes, S., et al. (2014). Mitochondrial and ER-targeted eCALWY probes reveal high levels of free Zn2+. ACS Chem. Biol. 9, 2111–2120. doi: 10.1021/cb5004064
Denessiouk, K., Permyakov, S., Denesyuk, A., Permyakov, E., and Johnson, M. S. (2014). Two structural motifs within canonical EF-hand calcium-binding domains identify five different classes of calcium buffers and sensors. PLoS ONE 9:e109287. doi: 10.1371/journal.pone.0109287
Dubernard, V., Arbeille, B. B., Lemesle, M. B., and Legrand, C. (1997). Evidence for an alpha-granular pool of the cytoskeletal protein alpha-actinin in human platelets that redistributes with the adhesive glycoprotein thrombospondin-1 during the exocytotic process. Arterioscler. Thromb. Vasc. Biol. 17, 2293–2305. doi: 10.1161/01.ATV.17.10.2293
Durvanger, Z., and Harmat, V. (2019). Structural diversity in calmodulin–peptide interactions. Curr. Protein Pept. Sci. 20, 1102–1111. doi: 10.2174/1389203720666190925101937
Dutting, S., Brachs, S., and Mielenz, D. (2011). Fraternal twins: Swiprosin-1/EFhd2 and Swiprosin-2/EFhd1, two homologous EF-hand containing calcium binding adaptor proteins with distinct functions. Cell Commun. Signal. 9:2. doi: 10.1186/1478-811X-9-2
Egelman, E. H. (2004). More insights into structural plasticity of actin binding proteins. Structure 12, 909–910. doi: 10.1016/j.str.2004.05.001
Emsley, P., and Cowtan, K. (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132. doi: 10.1107/S0907444904019158
Ferrer-Acosta, Y., Rodriguez Cruz, E. N., Vaquer Adel, C., and Vega, I. E. (2013). Functional and structural analysis of the conserved EFhd2 protein. Protein Pept. Lett. 20, 573–583. doi: 10.2174/0929866511320050011
Grabarek, Z. (2006). Structural basis for diversity of the EF-hand calcium-binding proteins. J. Mol. Biol. 359, 509–525. doi: 10.1016/j.jmb.2006.03.066
Hou, T., Jian, C., Xu, J., Huang, A. Y., Xi, J., Hu, K., et al. (2016). Identification of EFHD1 as a novel Ca2+ sensor for mitoflash activation. Cell Calcium 59, 262–270. doi: 10.1016/j.ceca.2016.03.002
Huh, Y. H., Kim, S. H., Chung, K. H., Oh, S., Kwon, M. S., Choi, H. W., et al. (2013). Swiprosin-1 modulates actin dynamics by regulating the F-actin accessibility to cofilin. Cell Mol. Life Sci. 70, 4841–4854. doi: 10.1007/s00018-013-1447-5
Kambe, T., Tsuji, T., Hashimoto, A., and Itsumura, N. (2015). The physiological, biochemical, and molecular roles of zinc transporters in zinc homeostasis and metabolism. Physiol. Rev. 95, 749–784. doi: 10.1152/physrev.00035.2014
Kanazawa, H., Ohsawa, K., Sasaki, Y., Kohsaka, S., and Imai, Y. (2002). Macrophage/microglia-specific protein Iba1 enhances membrane ruffling and Rac activation via phospholipase C-gamma -dependent pathway. J. Biol. Chem. 277, 20026–20032. doi: 10.1074/jbc.M109218200
Krissinel, E., and Henrick, K. (2007). Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797. doi: 10.1016/j.jmb.2007.05.022
Kwon, M. S., Park, K. R., Kim, Y. D., Na, B. R., Kim, H. R., Choi, H. J., et al. (2013). Swiprosin-1 is a novel actin bundling protein that regulates cell spreading and migration. PLoS ONE 8:e71626. doi: 10.1371/journal.pone.0071626
Lee, S. H., and Dominguez, R. (2010). Regulation of actin cytoskeleton dynamics in cells. Mol Cells 29, 311–325. doi: 10.1007/s10059-010-0053-8
Lewit-Bentley, A., and Rety, S. (2000). EF-hand calcium-binding proteins. Curr. Opin. Struct. Biol. 10, 637–643. doi: 10.1016/S0959-440X(00)00142-1
Liebschner, D., Afonine, P. V., Baker, M. L., Bunkoczi, G., Chen, V. B., Croll, T. I., et al. (2019). Macromolecular structure determination using X-rays, neutrons, and electrons: recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 75, 861–877. doi: 10.1107/S2059798319011471
McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C., and Read, R. J. (2007). Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674. doi: 10.1107/S0021889807021206
Otwinowski, Z., and Minor, W. (1997). Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326. doi: 10.1016/S0076-6879(97)76066-X
Park, J. G., Qin, Y., Galati, D. F., and Palmer, A. E. (2012). New sensors for quantitative measurement of mitochondrial Zn(2+). ACS Chem. Biol. 7, 1636–1640. doi: 10.1021/cb300171p
Park, K. R., Kwon, M. S., An, J. Y., Lee, J. G., Youn, H. S., Lee, Y., et al. (2016). Structural implications of Ca(2+)-dependent actin-bundling function of human EFhd2/Swiprosin-1. Sci. Rep. 6:39095. doi: 10.1038/srep39095
Parker, F., Baboolal, T. G., and Peckham, M. (2020). Actin mutations and their role in disease. Int. J. Mol. Sci. 21:3371. doi: 10.3390/ijms21093371
Putignano, V., Rosato, A., Banci, L., and Andreini, C. (2018). MetalPDB in 2018: a database of metal sites in biological macromolecular structures. Nucleic Acids Res. 46, D459–D464. doi: 10.1093/nar/gkx989
Ramesh, T. P., Kim, Y. D., Kwon, M. S., Jun, C. D., and Kim, S. W. (2009). Swiprosin-1 regulates cytokine expression of human mast cell line HMC-1 through actin remodeling. Immune Netw. 9, 274–284. doi: 10.4110/in.2009.9.6.274
Reyes, A., He, J., Mao, C. C., Bailey, L. J., Di Re, M., Sembongi, H., et al. (2011). Actin and myosin contribute to mammalian mitochondrial DNA maintenance. Nucleic Acids Res. 39, 5098–5108. doi: 10.1093/nar/gkr052
Robert, X., and Gouet, P. (2014). Deciphering key features in protein structures using the new ENDscript server. Nucleic Acids Res. 42, W320–W324. doi: 10.1093/nar/gku316
Sasaki, Y., Ohsawa, K., Kanazawa, H., Kohsaka, S., and Imai, Y. (2001). Iba1 is an actin-crosslinking protein in macrophages/microglia. Biochem. Biophys. Res. Commun. 286, 292–297. doi: 10.1006/bbrc.2001.5388
Schneider, C. A., Rasband, W. S., and Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. doi: 10.1038/nmeth.2089
Schulze, J. O., Quedenau, C., Roske, Y., Adam, T., Schuler, H., Behlke, J., et al. (2008). Structural and functional characterization of human Iba proteins. FEBS J. 275, 4627–4640. doi: 10.1111/j.1742-4658.2008.06605.x
Sensi, S. L., Ton-That, D., Sullivan, P. G., Jonas, E. A., Gee, K. R., and Kaczmarek, L. K.. (2003). Modulation of mitochondrial function by endogenous Zn2+ pools. Proc. Natl. Acad. Sci. U.S.A. 100, 6157–6162. doi: 10.1073/pnas.1031598100
Stein, M., Dutting, S., Mougiakakos, D., Bosl, M., Fritsch, K., Reimer, D., et al. (2017). A defined metabolic state in pre-B cells governs B-cell development and is counterbalanced by Swiprosin-2/EFhd1. Cell Death Differ. 24, 1239–1252. doi: 10.1038/cdd.2017.52
Storch, K. N., Taatjes, D. J., Bouffard, N. A., Locknar, S., Bishop, N. M., and Langevin, H. M. (2007). Alpha smooth muscle actin distribution in the cytoplasm and nuclear invaginations of connective tissue fibroblasts. Histochem. Cell Biol. 127, 523–530. doi: 10.1007/s00418-007-0275-9
Tominaga, M., Kurihara, H., Honda, S., Amakawa, G., Sakai, T., and Tomooka, Y. (2006). Molecular characterization of mitocalcin, a novel mitochondrial Ca2+-binding protein with EF-hand and coiled-coil domains. J. Neurochem. 96, 292–304. doi: 10.1111/j.1471-4159.2005.03554.x
Ulisse, V., Dey, S., Rothbard, D. E., Zeevi, E., Gokhman, I., Dadosh, T., et al. (2020). Regulation of axonal morphogenesis by the mitochondrial protein Efhd1. Life Sci Alliance 3:e202000753. doi: 10.26508/lsa.202000753
Vuadens, F., Rufer, N., Kress, A., Corthesy, P., Schneider, P., and Tissot, J. D. (2004). Identification of swiprosin 1 in human lymphocytes. Proteomics 4, 2216–2220. doi: 10.1002/pmic.200300779
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: homology modeling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303. doi: 10.1093/nar/gky427
Winn, M. D., Ballard, C. C., Cowtan, K. D., Dodson, E. J., Emsley, P., Evans, P. R., et al. (2011). Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242. doi: 10.1107/S0907444910045749
Wu, J., Wang, H., Guo, X., and Chen, J. (2016). Cofilin-mediated actin dynamics promote actin bundle formation during Drosophila bristle development. Mol. Biol. Cell 27, 2554–2564. doi: 10.1091/mbc.e16-02-0084
Xie, X., Venit, T., Drou, N., and Percipalle, P. (2018). In mitochondria? Actin regulates mtDNA transcription and is required for mitochondrial quality control. iScience 3, 226–237. doi: 10.1016/j.isci.2018.04.021
Xue, L. C., Rodrigues, J. P., Kastritis, P. L., Bonvin, A. M., and Vangone, A. (2016). PRODIGY: a web server for predicting the binding affinity of protein-protein complexes. Bioinformatics 32, 3676–3678. doi: 10.1093/bioinformatics/btw514
Yamada, M., Ohsawa, K., Imai, Y., Kohsaka, S., and Kamitori, S. (2006). X-ray structures of the microglia/macrophage-specific protein Iba1 from humans and mice demonstrate novel molecular conformation changes induced by calcium binding. J. Mol. Biol. 364, 449–457. doi: 10.1016/j.jmb.2006.09.027
Keywords: EFhd1, swiprosin-2, crystal structure, β-actin, actin-binding protein, actin-bundling protein
Citation: Mun SA, Park J, Park KR, Lee Y, Kang JY, Park T, Jin M, Yang J, Jun C-D and Eom SH (2021) Structural and Biochemical Characterization of EFhd1/Swiprosin-2, an Actin-Binding Protein in Mitochondria. Front. Cell Dev. Biol. 8:628222. doi: 10.3389/fcell.2020.628222
Received: 12 November 2020; Accepted: 21 December 2020;
Published: 18 January 2021.
Edited by:
Tadahide Furuno, Aichi Gakuin University, JapanReviewed by:
Dirk Mielenz, University of Erlangen Nuremberg, GermanyCopyright © 2021 Mun, Park, Park, Lee, Kang, Park, Jin, Yang, Jun and Eom. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Soo Hyun Eom, ZW9tQGdpc3QuYWMua3I=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.