
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Cell Dev. Biol. , 18 January 2021
Sec. Stem Cell Research
Volume 8 - 2020 | https://doi.org/10.3389/fcell.2020.626383
This article is part of the Research Topic Chromatin Regulation in Cell Fate Decisions View all 15 articles
The unique capability of embryonic stem cells (ESCs) to maintain and adjust the equilibrium between self-renewal and multi-lineage cellular differentiation contributes indispensably to the integrity of all developmental processes, leading to the advent of an organism in its adult form. The ESC fate decision to favor self-renewal or differentiation into specific cellular lineages largely depends on transcriptome modulations through gene expression regulations. Chromatin remodeling complexes play instrumental roles to promote chromatin structural changes resulting in gene expression changes that are key to the ESC fate choices governing the equilibrium between pluripotency and differentiation. BAF (Brg/Brahma-associated factors) or mammalian SWI/SNF complexes employ energy generated by ATP hydrolysis to change chromatin states, thereby governing the accessibility of transcriptional regulators that ultimately affect transcriptome and cell fate. Interestingly, the requirement of BAF complex in self-renewal and differentiation of ESCs has been recently shown by genetic studies through gene expression modulations of various BAF components in ESCs, although the precise molecular mechanisms by which BAF complex influences ESC fate choice remain largely underexplored. This review surveys these recent progresses of BAF complex on ESC functions, with a focus on its role of conditioning the pluripotency and differentiation balance of ESCs. A discussion of the mechanistic bases underlying the genetic requirements for BAF in ESC biology as well as the outcomes of its interplays with key transcription factors or other chromatin remodelers in ESCs will be highlighted.
Embryonic stem (ES) cells are pluripotent cells derived from the inner cell mass of blastocyst-stage embryos (Evans and Kaufman, 1981; Martin, 1981; Thomson et al., 1998). Their importance to basic biology and translational medicine derives from two unique characteristics that distinguish them from all other cell types. First, they can be maintained as a self-renewing stem cell population in vitro. Second, they have the capacity to differentiate into every cell type of the body. For decades, the mechanism underlying the self-renewal and pluripotency of ESCs has been the focus of intensive research in the field of stem cell biology.
Mouse ESCs were initially established and maintained by co-culture with mouse embryonic fibroblasts (Evans and Kaufman, 1981; Martin, 1981). Subsequent studies identified leukemia inhibitory factor (LIF) as one of the feeder-cell-derived molecules that support the growth of undifferentiated ESCs through gp130-mediated activation of STAT3 (Smith et al., 1988; Williams et al., 1988; Stewart et al., 1992; Niwa et al., 1998; Matsuda et al., 1999). In contrast to mouse ESCs, LIF and STAT3 appear to be dispensable for the self-renewal of primed human ESCs (Thomson et al., 1998; Reubinoff et al., 2000; Dahéron et al., 2004). Furthermore, serum could be replaced by BMP4, which activates Smad and subsequently induces the expressions of helix–loop–helix ID factors (Ying et al., 2003). ESCs cultivated in a serum-free medium with MAPK/ERK pathway inhibitor PD0325901 and glycogen synthase kinase 3β (GSK3β) pathway inhibitor CHIR99021 (called 2i), and LIF represent naïve state and exhibit greater and homogenous pluripotent gene expression than those cultivated in serum with LIF (Ying et al., 2008). With these developments, it is now possible to grow ESCs with defined factors in the absence of serum or feeder cells.
Numerous studies demonstrate the importance of transcription factors (TFs) on the maintenance of ESCs and their pluripotency, among which OCT4, SOX2, and NANOG form a core transcriptional regulatory circuit (Martello and Smith, 2014). Ablation of their expression disrupts the pluripotency network, leading to the exit from pluripotency and initiation of differentiation of ESCs (Okamoto et al., 1990; Schöler et al., 1990; Nichols et al., 1998; Chambers et al., 2003; Mitsui et al., 2003; Masui et al., 2007; Silva et al., 2009). In addition, downregulation of epiblast-specific TFs such as TBX3, KLF2/4/5, TFCP2L1, and ESRRB disturb the self-renewal of ESCs, demonstrating their supporting roles in the maintenance of ESC identity (Ivanova et al., 2006; Jiang et al., 2008; Festuccia et al., 2012, 2018; Martello et al., 2013; Yeo et al., 2014).
Besides signaling and TFs, chromatin remodeling complexes play instrumental roles on maintaining the identity of ESCs (Papatsenko et al., 2018). At least three epigenetic mechanisms allow regulation of DNA expression and chromatin accessibility, which include DNA methylation (Winata et al., 2018), histone modifications (Lawrence et al., 2016), and ATP-dependent chromatin remodeling (Clapier et al., 2017). This mini-review will focus on the SWI/SNF family of ATP-dependent chromatin remodeling complexes and its role in the maintenance of ESCs and their differentiation.
The ATP-dependent SWI/SNF complexes were first discovered in yeast in genetic screens aimed at uncovering factors responsible for the regulation of mating type switching (Stern et al., 1984) and those being able to allow changing of nutrient sources used for energy supply (Carlson et al., 1981; Neigeborn and Carlson, 1984, 1987), therefore termed SWI/SNF complex (short for SWItch/sucrose non-fermentable) (Alfert et al., 2019). In Drosophila melanogaster, this complex was first discovered in screens to uncover genes that are able to suppress phenotypes caused by mutations in Polycomb genes (PcGs) (Tamkun et al., 1992; Elfring et al., 1994).
The BAF (BRG1/BRM-associated factor) complex, the mammalian homolog of the SWI/SNF complex, is one of four ATP-dependent chromatin remodeling complex families known in mammals (the other three are INO80/SWR1, ISWI, and CHD complexes) (Clapier and Cairns, 2009). Three mammalian BAF complexes have been identified based on their different subunit compositions. The subunits are encoded by 29 genes (Centore et al., 2020). The PBAF (Polybromo-associated BAF complex) is distinguished from the cBAF (canonical BAF complex) by the incorporation of BAF200 instead of BAF250A/B and of BAF180 (Yan et al., 2005). Furthermore, PBAF lacks SS18 but includes the PBAF-specific subunits BAF45A and BRD7 (Kaeser et al., 2008; Middeljans et al., 2012). Recently, a third class, called ncBAF (for non-canonical BAF complex) or GBAF (after its distinctive subunits GLTSCR1/1L), has been identified, which is characterized by the incorporation of BRD9 and GLTSCR1/1L (Alpsoy and Dykhuizen, 2018), but lacks the cBAF subunits such as BAF47, BAF57, and BAF250 and the PBAF-specific subunits BAF180 and BRD7 (Clapier et al., 2017; Alpsoy and Dykhuizen, 2018; Mashtalir et al., 2018).
BAF complexes are made up of multiple subunits that are assembled in a combinatorial manner to tailor their functions, regulating specific developmental events (Ho and Crabtree, 2010). The BAF complexes in different tissues are distinctive for their specific subunit compositions (Lickert et al., 2004; Lessard et al., 2007; Vogel-Ciernia et al., 2013; Harada et al., 2017; Sokpor et al., 2018; Akerberg and Pu, 2020). Hence, it is not only the BAF complex itself that controls biological processes, but the expressions of distinct BAF complexes with unique subunit compositions are also a major part of the regulatory process.
The assembly of an ESC-specific BAF (esBAF) complex is required for the regulation of the ESC transcriptome, therefore controlling the self-renewal and differentiation of ESCs (Ho et al., 2009a). The esBAF complex depends on BRG1 as the ATPase subunit, as BRM does not express in ESCs (Ho et al., 2009b). Moreover, esBAF can be distinguished by the incorporation of Baf250a not 250b, Baf60a/b not 60c, and a Baf155::155 homodimer instead of a Baf155::170 heterodimer (Kaeser et al., 2008; Ho et al., 2009b). In human ESCs, BAF170, and not BAF155, seems to play an important role in the maintenance of pluripotency (Zhang et al., 2014).
A possible way of elucidating the role and importance of individual subunits of multiprotein complexes is the deletion or downregulation of genes encoding their subunits. Genetic inactivation of specific subunit of BAF complex leads to diverse aberrant phenotypes in ESCs (Table 1).
Both Brg1 and Baf155 knockout mice are lethal at the pre-implantation stage (Bultman et al., 2000; Kim et al., 2001), suggesting that they play a key role in the maintenance of pluripotency. Consistently, depletion of either Brg1 or Baf155 expression in ESCs leads to the downregulation of the key pluripotent TFs Oct4, Sox2, and Nanog, indicating that BAF155 and BRG1 cooperate to maintain ESC identity (Fazzio et al., 2008; Ho et al., 2009b; Kidder et al., 2009). Corresponding to the unique subunit composition of esBAF, neither Brm nor Baf170 overexpression can rescue Brg1 or Baf155 knockout, respectively (Ho et al., 2009b). Different from mouse esBAF complex, human Baf170 deficiency led to the differentiation of human ESCs, demonstrating that the BAF170-containing BAF complex was required for the self-renewal of human ESCs (Zhang et al., 2014). Ho et al. reported that neuro-ectodermal differentiation was impaired and mesodermal differentiation was delayed in Brg1 knockout embryoid bodies (Ho et al., 2009a). In contrast, knockdown of Brg1 in ESCs promoted the expression of differentiation marker genes (Kim et al., 2001). These results might indicate the distinct role of BRG1 in ESCs and differentiating cells. Baf47 knockout mice are also lethal at the pre-implantation stage (Klochendler-Yeivin et al., 2000; Guidi et al., 2001). The negative regulation of Oct4 by Baf47 may control the balance between pluripotency and differentiation of ESCs (You et al., 2013). A recent contradicting report indicates the upregulation of Cdx2 expression in Baf47 KO ESCs (Sakakura et al., 2019). BAF250a and BAF250b are two mutually exclusive esBAF subunits. Inactivation of either of them decreases expression of Oct4 and Sox2 or Nanog, thereby inhibiting the self-renewal of ESCs (Gao et al., 2008; Yan et al., 2008). Knockout of Baf250a upregulates primitive endoderm maker genes, such as Gata4, Gata6, and Sox17 in mouse ESCs but impairs mesodermal lineage differentiation of both mouse and human ESCs (Gao et al., 2008; Lei et al., 2020). In contrast, knockout of Baf250b increased the expression of mesoderm marker genes in mouse ESCs, Gata2 and Esx1 (Yan et al., 2008). This may indicate the balance role of BAF250a- and BAF250b-containing BAF complexes on mesoderm differentiation of ESCs. The deletion of esBAF subunit Baf45d only perturbs the self-renewal of ESCs, whereas its knockout impairs the differentiation of ESCs to all three germ lineages (Zhang et al., 2019).
In addition to the long-known esBAF, the newly discovered ncBAF complex also plays an important role in the regulation of the ESC transcriptome. Inhibition of Brd9, the specific ncBAF subunit, changed the morphology of ESCs to that resembles primed or epiblast ESCs (EpiESCs), reduced colony-forming capability, and downregulated expressions of Nanog and Klf4, indicating that BRD9 has an important role in maintaining the naïve pluripotent state of ESCs (Gatchalian et al., 2018).
Consistent to its functions on the maintenance and differentiation of ESCs, BAF complexes also play important roles in the reprogramming of somatic cells to induced pluripotent stem cells (iPSCs). Depletion of Brg1 was associated with failures in reprogramming (Hansis et al., 2004; Egli and Eggan, 2010). Overexpression of Brg1 and Baf155 achieves euchromatin, enhances binding of OCT4, and increases the reprogramming efficiency of MEFs to iPSCs (Singhal et al., 2010). In contrast, downregulation of Brm and Baf170 improves reprogramming efficiency and promotes complete reprogramming of immature iPSCs (Jiang et al., 2015). Therefore, similar to the distinct roles of different BAF subunits for the maintenance and differentiation of ESCs, different BAF components may play different roles in the reprogramming.
In summary, esBAF complex is crucial for the maintenance of ESCs, with distinct effects from the deletion of different subunits. The knockout of different subunits of esBAF leads to defects of ES differentiations to different lineages, though the precise molecular mechanisms underlying the different phenotypes upon the deletion of different subunits need further investigations.
Inactivation of individual esBAF subunits downregulates expression of pluripotent TFs (Gao et al., 2008; Yan et al., 2008; Ho et al., 2009b; You et al., 2013; Zhang et al., 2019), indicating that esBAF controls the self-renewal of ESCs via regulating pluripotent factors. esBAF subunits BRG1, BAF155, BAF250a, and BAF45d are bound at sites engaged by the core pluripotency TFs OCT4, SOX2, and NANOG (Ho et al., 2009a; Gatchalian et al., 2018; Zhang et al., 2019). The expression of the core TFs Nanog, Oct4, and Sox2 as well as a variety of other factors governs the maintenance of pluripotency in ESCs (Martello and Smith, 2014). Specifically, NANOG, OCT4, and SOX2 have been shown to repress the expression of developmental genes while modulating their own expression levels by binding to each other's promoter regions (Saunders et al., 2013). Both BRG1 and BAF155 are located near the transcriptional starting site (TSS) of core pluripotency factors Oct4, Nanog, and Sox2 (Ho et al., 2009b). The binding of OCT4, SOX2, and NANOG is impaired in Dpf2 KO ESCs (Zhang et al., 2019). Therefore, esBAF complex may collaborate with the core TFs to regulate the expression of pluripotency TFs, thereby controlling the maintenance of ES self-renewal.
LIF/STAT3 pathway is essential for the maintenance of mouse ESCs (Niwa et al., 1998; Matsuda et al., 1999). It also plays a role in naïve or murine-like human ESC pluripotency (Hanna et al., 2010; Buecker et al., 2014). BRG1, DPF2, and STAT3 binding sites display a substantial genome-wide overlap in mouse ESCs. STAT3 binding is considerably impaired in Brg1- or Dpf2 (Baf45d)-depleted ESCs, leading to the downregulation of Stat3 target genes (Ho et al., 2011; Zhang et al., 2019). esBAF stabilizes the binding of STAT3 and thereby helps the maintenance of ES self-renewal (Ho et al., 2011).
esBAF also regulates gene expression in ESCs (Ho et al., 2011; Gatchalian et al., 2018; Zhang et al., 2019). esBAF preferably binds to enhancers and regulates their H3K27ac deposition. Loss of esBAF subunit Dpf2 changes the activity of enhancers and the target gene expression (Ho et al., 2011; Zhang et al., 2019). On the other hand, ncBAF predominantly binds to H3K4-trimethylated promoter regions and is associated with the TFs Klf4 (Kruppel-like factor 4) (Gatchalian et al., 2018), indicating a distinct mechanism to regulate gene expression. One of the most striking differences is, however, that ncBAF binds to TAD (topologically associating domain) boundaries and CTCF sites, potentially contributing to the regulation of genome topology (Gatchalian et al., 2018). Thus, esBAF and ncBAF complexes might regulate ESC identity coordinately via distinct mechanisms that future studies need to elucidate.
The PcG family has first been discovered in Drosophila followed by the observation of male flies with ectopic sex combs (Margueron and Reinberg, 2011). In mammals, the multiprotein-containing Polycomb repressive complex 2 (PRC2) has repressive influence on the genome (Margueron and Reinberg, 2011). PRC2 is dispensable for the maintenance of undifferentiated mouse ESCs, as the deletion of PRC2 components has little effect on their morphology and self-renewal, although a subset of PRC2 target genes are derepressed (Boyer et al., 2006; Pasini et al., 2007; Chamberlain et al., 2008; Shen et al., 2008; Leeb et al., 2010). Similarly, deletion of EZH2, the catalytic subunit of PRC2 complex, in human ESCs also causes misexpression of developmental genes but severely affects the self-renewal of human ESCs (Collinson et al., 2016).
The cooperative function of BAF complex with PRC2 in ESCs has been revealed (Ho et al., 2011; Zhang et al., 2019). Ho et al. report that the core subunit of BAF complex, BRG1, in ESCs potentiates LIF signaling by opposing PRC2 complex (Ho et al., 2011). The opposing regulation of BAF and PRC2 subunits DPF2 and EED on Tbx3 expression is critical to the proper differentiation of ESCs to mesoendoderm. The other PRC2 subunit EZH2 also opposes DPF2-dependent differentiation through a distinct mechanism involving Nanog repression (Zhang et al., 2019). Contrary to the opposing function of BAF and PRC2 complexes, BRG1 facilitates PRC2 to reinforce the repression on its target genes in ESCs. Therefore, esBAF not only simply antagonizes PcG but also acts synergistically with the common goal of supporting pluripotency (Ho et al., 2011).
Inactivation of different subunit of BAF complex differentially affects the expression of pluripotency TFs (Table 1). Furthermore, DPF2 opposingly regulates differentiation of ESCs via controlling different pluripotency TFs with distinct components of PRC2 complex. As a result, distinct BAF subunits may regulate the expression of different pluripotency TFs collaboratively with other TFs and chromatin modifiers and therefore leads to different differentiation defects of ESCs upon the deletion of different BAF components (Figure 1). Interestingly, knockout of Dpf2 only affects about 8% of BRG1 binding sites on the genome (Zhang et al., 2019), indicating that the loss of a specific BAF subunit only affects the binding of a specific portion of BAF complex on genome, which may lead to the specific phenotypes upon the deletion of that subunit. Future studies on the deletion of other specific BAF components on the binding of BRG1 will help to explain the different phenotypes in ESCs that resulted from the deletion of specific subunits of other chromatin complexes.
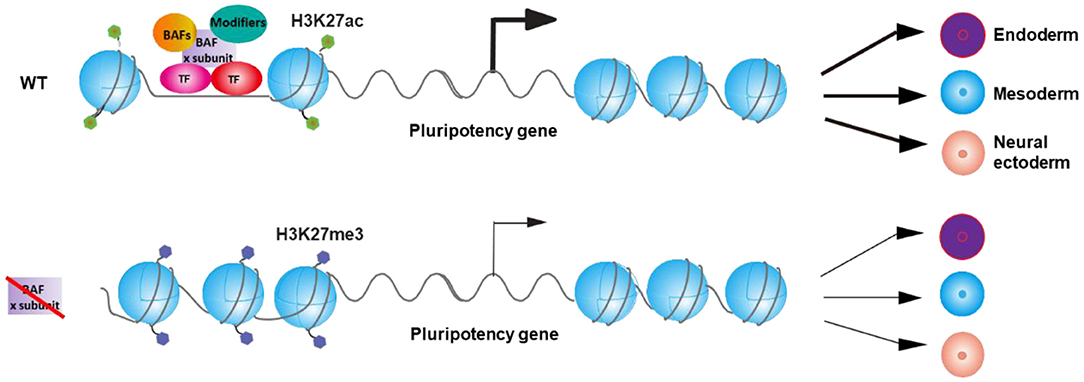
Figure 1. Model for the regulation of the balance between pluripotency and differentiation of embryonic stem cells (ESCs) by BAF complexes via the control of pluripotency gene expression. BAF complex, transcription factors, and other chromatin modifiers regulate the expression of specific pluripotency gene(s) and thereby control the balance between pluripotency and differentiation of ESCs. Inactivation of specific BAF subunit leads to the deregulation of the expression of specific pluripotency gene(s) and therefore results in the differential differentiation defects of ESC.
BAF complex regulates both the maintenance and differentiation of ESCs (Ho and Crabtree, 2010). Knockout of Dpf2 does not change the level of H3K27ac around DPF2-bound lineage markers during differentiation of ESCs. Consistently, overexpression of Dpf2 in ESCs does not lead to the upregulation of endo- and mesodermal markers, supporting an idea that BAF complex regulates ESC differentiation indirectly (Zhang et al., 2019). Tbx3 is a pluripotency TF, and the downregulation of its expression impairs ESC self-renewal (Ivanova et al., 2006). Tbx3 also plays key roles on ESC differentiation. Deregulation of its expression impairs the differentiation of ESCs (Lu et al., 2011; Weidgang et al., 2014; Waghray et al., 2015; Zhang et al., 2019). Dpf2 participates in the self-renewal and differentiation of ESCs via precisely regulating Tbx3 expression in both ESCs and differentiating cells (Zhang et al., 2019). As a core pluripotency TF, Nanog represses expression of differentiation marker genes and maintains the self-renewal of ESCs (Niwa, 2007). Dpf2 regulates the expression of Nanog with PRC2 subunit Ezh2, thereby controlling the proper differentiation of ESCs (Zhang et al., 2019).
BAF47 controls the differentiation of ESCs via regulating Oct4 expression, which provides another example to demonstrate how BAF complex controls the balance between pluripotency and differentiation (You et al., 2013). The controversial result from a recent work upon the deletion of Baf47 in ESCs indicates that more studies are required to clarify the discrepancy (Sakakura et al., 2019). Changed expression of pluripotency genes in ESCs upon the deletion of other BAF subunits has been reported (Gao et al., 2008; Ho et al., 2009b; Ho and Crabtree, 2010). It will be of interest to carry out systematic studies to determine whether and how other subunits of BAF complex regulate the expression of specific pluripotency genes and thereby control the balance between pluripotency and differentiation.
BAF complex is functionally important for the self-renewal and differentiation of ESCs. Knockout of different subunits of BAF complex changes the expression of different pluripotency TFs and impairs the differentiation of ESCs differently. Thus, it is of particular importance to explore how BAF complex regulates the balance between the maintenance of identity of ESCs and their differentiation to three germ layers. We have outlined studies that described functions of specific subunits of various BAF complexes in ESCs. Moreover, our recent study demonstrates an attractive mechanism that distinct BAF subunit controls the integrity of only a part of BAF complex on the genome, and therefore, its deletion only affects the binding of a part of BAF complex, which directly changes the expression of distinct pluripotency TFs in both ESCs and differentiating cells with other TFs and chromatin modifiers (Zhang et al., 2019). Consistently, another recent report demonstrates that the loss of a single subunit of the BAF complex in cancer cells did not destroy the entire complex but will change the composition of the BAF complex (Schick et al., 2019). BAF complex regulates ESC differentiation via controlling the expressions of pluripotency TFs, with different subunits affecting ESC differentiation via regulating different TFs. Further systematic studies of other subunits of BAF complex are needed to warrant the mechanism, which may also explain the distinct phenotypes that resulted from the deletion of various subunits of chromatin remodeling complex.
YY, XC, and WZ conceived the study and wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was funded by a grant from the National Natural Science Foundation of China (31970812).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are indebted to all the cited authors for their work. The authors gratefully acknowledge the comments on the manuscript by Dr. Li Li.
Akerberg, B. N., and Pu, W. T. (2020). Genetic and epigenetic control of heart development. Cold Spring Harb Perspect Biol. 12:a036756. doi: 10.1101/cshperspect.a036756
Alfert, A., Moreno, N., and Kerl, K. (2019). The BAcomplex, F. in development and disease. Epigenetics Chromatin. 12:19. doi: 10.1186/s13072-019-0264-y
Alpsoy, A., and Dykhuizen, E. C. (2018). Glioma tumor suppressor candidate region gene 1 (GLTSCR1) and its paralog GLTSCR1-like form SWI/SNchromatin, F. remodeling subcomplexes. Biol. J. Chem. 293, 3892–3903. doi: 10.1074/jbc.RA117.001065
Boyer, L. A., Plath, K., Zeitlinger, J., Brambrink, T., Medeiros, L. A., Lee, T. I., et al. (2006). Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441, 349–353. doi: 10.1038/nature04733
Buecker, C., Srinivasan, R., Wu, Z., Calo, E., Acampora, D., Faial, T., et al. (2014). Reorganization of enhancer patterns in transition from naive to primed pluripotency. Cell Stem Cell 14, 838–853. doi: 10.1016/j.stem.2014.04.003
Bultman, S., Gebuhr, T., Yee, D., La Mantia, C., Nicholson, J., Gilliam, A., et al. (2000). A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol. Cell 6, 1287–1295. doi: 10.1016/S1097-2765(00)00127-1
Carlson, M., Osmond, B. C., and Botstein, D. (1981). Mutants of yeast defective in sucrose utilization. Genetics 98, 25–40.
Centore, R. C., Sandoval, G. J., Soares, L. M. M., Kadoch, C., and Chan, H. M. (2020). Mammalian SWI/SNF chromatin remodeling complexes: emerging mechanisms and therapeutic strategies. Trends Genet. 36, 936–950. doi: 10.1016/j.tig.2020.07.011
Chamberlain, S. J., Yee, D., and Magnuson, T. (2008). Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells 26, 1496–1505. doi: 10.1634/stemcells.2008-0102
Chambers, I., Colby, D., Robertson, M., Nichols, J., Lee, S., Tweedie, S., et al. (2003). Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113, 643–655. doi: 10.1016/S0092-8674(03)00392-1
Clapier, C. R., and Cairns, B. R. (2009). The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78, 273–304. doi: 10.1146/annurev.biochem.77.062706.153223
Clapier, C. R., Iwasa, J., Cairns, B. R., and Peterson, C. L. (2017). Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell Biol. 18, 407–422. doi: 10.1038/nrm.2017.26
Collinson, A., Collier, A. J., Morgan, N. P., Sienerth, A. R., Chandra, T., Andrews, S., et al. (2016). Deletion of the polycomb-group protein EZH2 leads to compromised self-renewal and differentiation defects in human embryonic stem cells. Cell Rep. 17, 2700–2714. doi: 10.1016/j.celrep.2016.11.032
Dahéron, L., Opitz, S. L., Zaehres, H., Lensch, M. W., Andrews, P. W., Itskovitz-Eldor, J., et al. (2004). LIF/STAT3 signaling fails to maintain self-renewal of human embryonic stem cells. Stem Cells 22, 770–778. doi: 10.1634/stemcells.22-5-770
Egli, D., and Eggan, K. (2010). Recipient cell nuclear factors are required for reprogramming by nuclear transfer. Development 137, 1953–1963. doi: 10.1242/dev.046151
Elfring, L. K., Deuring, R., McCallum, C. M., Peterson, C. L., and Tamkun, J. W. (1994). Identification and characterization of Drosophila relatives of the yeast transcriptional activator SNF2/SWI2. Mol. Cell Biol. 14, 2225–2234. doi: 10.1128/MCB.14.4.2225
Evans, M. J., and Kaufman, M. H. (1981). Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154–156. doi: 10.1038/292154a0
Fazzio, T. G., Huff, J. T., and Panning, B. (2008). An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell 134, 162–174. doi: 10.1016/j.cell.2008.05.031
Festuccia, N., Halbritter, F., Corsinotti, A., Gagliardi, A., Colby, D., Tomlinson, S. R., et al. (2018). Esrrb extinction triggers dismantling of naïve pluripotency and marks commitment to differentiation. EMBO J. 37:e95476. doi: 10.15252/embj.201695476
Festuccia, N., Osorno, R., Halbritter, F., Karwacki-Neisius, V., Navarro, P., Colby, D., et al. (2012). Esrrb is a direct Nanog target gene that can substitute for Nanog function in pluripotent cells. Cell Stem Cell 11, 477–490. doi: 10.1016/j.stem.2012.08.002
Gao, X., Tate, P., Hu, P., Tjian, R., Skarnes, W. C., and Wang, Z. (2008). Ecell, S. pluripotency and germ-layer formation require the SWI/SNF chromatin remodeling component BAF250a. Proc. Natl. Acad. Sci. U. S. A. 105, 6656–60. doi: 10.1073/pnas.0801802105
Gatchalian, J., Malik, S., Ho, J., Lee, D. S., Kelso, T. W. R., Shokhirev, M. N., et al. (2018). A non-canonical BRD9-containing BAF chromatin remodeling complex regulates naive pluripotency in mouse embryonic stem cells. Nat. Commun. 9:5139. doi: 10.1038/s41467-018-07528-9
Guidi, C. J., Sands, A. T., Zambrowicz, B. P., Turner, T. K., Demers, D. A., Webster, W., et al. (2001). Disruption of Ini1 leads to peri-implantation lethality and tumorigenesis in mice. Mol. Cell Biol. 21, 3598–3603. doi: 10.1128/MCB.21.10.3598-3603.2001
Hanna, J., Cheng, A. W., Saha, K., Kim, J., Lengner, C. J., Soldner, F., et al. (2010). Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc. Natl. Acad. Sci. U. S. A. 107, 9222–9227. doi: 10.1073/pnas.1004584107
Hansis, C., Barreto, G., Maltry, N., and Niehrs, C. (2004). Nuclear reprogramming of human somatic cells by xenopus egg extract requires BRG1. Curr. Biol. 14, 1475–1480. doi: 10.1016/j.cub.2004.08.031
Harada, A., Ohkawa, Y., and Imbalzano, A. N. (2017). Temporal regulation of chromatin during myoblast differentiation. Semin. Cell Dev. Biol. 72, 77–86. doi: 10.1016/j.semcdb.2017.10.022
Ho, L., and Crabtree, G. R. (2010). Chromatin remodelling during development. Nature 463, 474–484. doi: 10.1038/nature08911
Ho, L., Jothi, R., Ronan, J. L., Cui, K., Zhao, K., and Crabtree, G. R. (2009a). An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc. Natl. Acad. Sci. U. S. A. 106, 5187–5191. doi: 10.1073/pnas.0812888106
Ho, L., Miller, E. L., Ronan, J. L., Ho, W. Q., Jothi, R., and Crabtree, G. R. (2011). esBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signalling and by regulating polycomb function. Nat. Cell Biol. 13, 903–913. doi: 10.1038/ncb2285
Ho, L., Ronan, J. L., Wu, J., Staahl, B. T., Chen, L., Kuo, A., et al. (2009b). An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc. Natl. Acad. Sci. U. S. A. 106, 5181–5186. doi: 10.1073/pnas.0812889106
Ivanova, N., Dobrin, R., Lu, R., Kotenko, I., Levorse, J., DeCoste, C., et al. (2006). Dissecting self-renewal in stem cells with RNA interference. Nature 442, 533–538. doi: 10.1038/nature04915
Jiang, J., Chan, Y. S., Loh, Y. H., Cai, J., Tong, G. Q., Lim, C. A., et al. (2008). A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat. Cell Biol. 10, 353–360. doi: 10.1038/ncb1698
Jiang, Z., Tang, Y., Zhao, X., Zhang, M., Donovan, D. M., and Tian, X. C. (2015). Knockdown of Brm and Baf170, components of chromatin remodeling complex, facilitates reprogramming of somatic cells. Stem Cells Dev. 24, 2328–2336. doi: 10.1089/scd.2015.0069
Kaeser, M. D., Aslanian, A., Dong, M. Q., Yates, J. R. 3rd, and Emerson, B. M. (2008). BRD7, a novel PBAF-specific SWI/SNF subunit is required for target gene activation and repression in embryonic stem cells. Biol. J. Chem. 283, 32254–32263. doi: 10.1074/jbc.M806061200
Kidder, B. L., Palmer, S., and Knott, J. G. (2009). SWI/SNF-Brg1 regulates self-renewal and occupies core pluripotency-related genes in embryonic stem cells. Stem Cells 27, 317–328. doi: 10.1634/stemcells.2008-0710
Kim, J. K., Huh, S. O., Choi, H., Lee, K. S., Shin, D., Lee, C., et al. (2001). Srg3, a mouse homolog of yeast SWI3, is essential for early embryogenesis and involved in brain development. Mol. Cell Biol. 21, 7787–7795. doi: 10.1128/MCB.21.22.7787-7795.2001
Klochendler-Yeivin, A., Fiette, L., Barra, J., Muchardt, C., Babinet, C., and Yaniv, M. (2000). The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression. EMBO Rep. 1, 500–506. doi: 10.1093/embo-reports/kvd129
Lawrence, M., Daujat, S., and Schneider, R. (2016). Lateral thinking: how histone modifications regulate gene expression trends. Genet. 32, 42–56. doi: 10.1016/j.tig.2015.10.007
Leeb, M., Pasini, D., Novatchkova, M., Jaritz, M., Helin, K., and Wutz, A. (2010). Polycomb complexes act redundantly to repress genomic repeats and genes. Genes Dev. 24, 265–276. doi: 10.1101/gad.544410
Lei, I., Tian, S., Chen, V., Zhao, Y., and Wang, Z. (2020). SWI/SNF component BAF250a coordinates OCT4 and WNT signaling pathway to control cardiac lineage differentiation. Front. Cell Dev. Biol. 7:358. doi: 10.3389/fcell.2019.00358
Lessard, J., Wu, J. I., Ranish, J. A., Wan, M., Winslow, M. M., Staahl, B. T., et al. (2007). An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron 55, 201–215. doi: 10.1016/j.neuron.2007.06.019
Lickert, H., Takeuchi, J. K., Von Both, I., Walls, J. R., McAuliffe, F., Adamson, S. L., et al. (2004). Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature 432, 107–112. doi: 10.1038/nature03071
Lu, R., Yang, A., and Jin, Y. (2011). Dual functions of T-box 3 (Tbx3) in the control of self-renewal and extraembryonic endoderm differentiation in mouse embryonic stem cells. Biol. J. Chem. 286, 8425–36. doi: 10.1074/jbc.M110.202150
Margueron, R., and Reinberg, D. (2011). The Polycomb complex PRC2 and its mark in life. Nature 469, 343–349. doi: 10.1038/nature09784
Martello, G., Bertone, P., and Smith, A. (2013). Identification of the missing pluripotency mediator downstream of leukaemia inhibitory factor. EMBO J. 32, 2561–2574. doi: 10.1038/emboj.2013.177
Martello, G., and Smith, A. (2014). The nature of embryonic stem cells. Annu. Rev. Cell Dev. Biol. 30, 647–675. doi: 10.1146/annurev-cellbio-100913-013116
Martin, G. R. (1981). Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. U. S. A. 78, 7634–7638. doi: 10.1073/pnas.78.12.7634
Mashtalir, N., D'Avino, A. R., Michel, B. C., Luo, J., Pan, J., Otto, J. E., et al. (2018). Modular organization and assembly of SWI/SNF family chromatin remodeling complexes. Cell 175, 1272–1288.e20. doi: 10.1016/j.cell.2018.09.032
Masui, S., Nakatake, Y., Toyooka, Y., Shimosato, D., Yagi, R., Takahashi, K., et al. (2007). Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat. Cell Biol. 9, 625–635. doi: 10.1038/ncb1589
Matsuda, T., Nakamura, T., Nakao, K., Arai, T., Katsuki, M., Heike, T., et al. (1999). STAT3 activation is sufficient to maintain an undifferentiated state of mouse embryonic stem cells. EMBO J. 18, 4261–4269. doi: 10.1093/emboj/18.15.4261
Middeljans, E., Wan, X., Jansen, P. W., Sharma, V., Stunnenberg, H. G., and Logie, C. (2012). SS18 together with animal-specific factors defines human BAF-type SWI/SNF complexes. PLoS ONE 7:e33834. doi: 10.1371/journal.pone.0033834
Mitsui, K., Tokuzawa, Y., Itoh, H., Segawa, H., Murakami, M., Takahashi, K., et al. (2003). The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113, 631–642. doi: 10.1016/S0092-8674(03)00393-3
Neigeborn, L., and Carlson, M. (1984). Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics 108, 845–858.
Neigeborn, L., and Carlson, M. (1987). Mutations causing constitutive invertase synthesis in yeast: genetic interactions with snf mutations. Genetics 115, 247–253.
Nichols, J., Zevnik, B., Anastassiadis, K., Niwa, H., Klewe-Nebenius, D., Chambers, I., et al. (1998). Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor. Cell 95, 379–91. doi: 10.1016/S0092-8674(00)81769-9
Niwa, H. (2007). How is pluripotency determined and maintained? Development 134, 635–646. doi: 10.1242/dev.02787
Niwa, H., Burdon, T., Chambers, I., and Smith, A. (1998). Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 12, 2048–2060. doi: 10.1101/gad.12.13.2048
Okamoto, K., Okazawa, H., Okuda, A., Sakai, M., Muramatsu, M., and Hamada, H. (1990). A novel octamer binding transcription factor is differentially expressed in mouse embryonic cells. Cell 60, 461–472. doi: 10.1016/0092-8674(90)90597-8
Papatsenko, D., Waghray, A., and Lemischka, I. R. (2018). Feedback control of pluripotency in embryonic stem cells: signaling, transcription and epigenetics. Stem Cell Res. 29, 180–188. doi: 10.1016/j.scr.2018.02.012
Pasini, D., Bracken, A. P., Hansen, J. B., Capillo, M., and Helin, K. (2007). The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol. Cell Biol. 27, 3769–3779. doi: 10.1128/MCB.01432-06
Reubinoff, B. E., Pera, M. F., Fong, C. Y., Trounson, A., and Bongso, A. (2000). Embryonic stem cell lines from human blastocysts: somatic differentiation in vitro. Nat. Biotechnol. 18, 399-404. doi: 10.1038/74447
Sakakura, M., Ohta, S., Yagi, M., Tanaka, A., Norihide, J., Woltjen, K., et al. (2019). Smarcb1 maintains the cellular identity and the chromatin landscapes of mouse embryonic stem cells. Biochem. Biophys. Res. Commun. 519, 705–713. doi: 10.1016/j.bbrc.2019.09.054
Saunders, A., Faiola, F., and Wang, J. (2013). Concise review: pursuing self-renewal and pluripotency with the stem cell factor. Stem Cells 31, 1227–1236. doi: 10.1002/stem.1384
Schaniel, C., Ang, Y. S., Ratnakumar, K., Cormier, C., James, T., Bernstein, E., et al. (2009). Smarcc1/Baf155 couples self-renewal gene repression with changes in chromatin structure in mouse embryonic stem cells. Stem Cells 27, 2979–2991. doi: 10.1002/stem.223
Schick, S., Rendeiro, A. F., Runggatscher, K., Ringler, A., Boidol, B., Hinkel, M., et al. (2019). Systematic characterization of BAF mutations provides insights into intracomplex synthetic lethalities in human cancers. Nat. Genet. 51, 1399–1410. doi: 10.1038/s41588-019-0477-9
Schöler, H. R., Dressler, G. R., Balling, R., Rohdewohld, H., and Gruss, P. (1990). Oct-4: a germline-specific transcription factor mapping to the mouse t-complex. EMBO J. 9, 2185–2195. doi: 10.1002/j.1460-2075.1990.tb07388.x
Shen, X., Liu, Y., Hsu, Y. J., Fujiwara, Y., Kim, J., Mao, X., et al. (2008). EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol. Cell. 32, 491–502. doi: 10.1016/j.molcel.2008.10.016
Silva, J., Nichols, J., Theunissen, T. W., Guo, G., van Oosten, A. L., Barrandon, O., et al. (2009). Nanog is the gateway to the pluripotent ground state. Cell 138, 722–737. doi: 10.1016/j.cell.2009.07.039
Singhal, N., Graumann, J., Wu, G., Araúzo-Bravo, M. J., Han, D. W., Greber, B., et al. (2010). Chromatin-remodeling components of the BAF complex facilitate reprogramming. Cell 141, 943–955. doi: 10.1016/j.cell.2010.04.037
Smith, A. G., Heath, J. K., Donaldson, D. D., Wong, G. G., Moreau, J., Stahl, M., et al. (1988). Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature 336, 688–690. doi: 10.1038/336688a0
Sokpor, G., Castro-Hernandez, R., Rosenbusch, J., Staiger, J. F., and Tuoc, T. (2018). ATP-dependent chromatin remodeling during cortical neurogenesis. Front. Neurosci. 12:226. doi: 10.3389/fnins.2018.00226
Stern, M., Jensen, R., and Herskowitz, I. (1984). Five SWI genes are required for expression of the Hgene, O. in yeast. Mol. J. Biol. 178, 853–868. doi: 10.1016/0022-2836(84)90315-2
Stewart, C. L, Kaspar, P., Brunet, L. J., Bhatt, H., Gadi, I., Köntgen, F., et al. (1992). Blastocyst implantation depends on maternal expression of leukaemia inhibitory factor. Nature 359, 76–79. doi: 10.1038/359076a0
Tamkun, J. W., Deuring, R., Scott, M. P., Kissinger, M., Pattatucci, A. M., Kaufman, T. C., et al. (1992). brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell 68, 561–572. doi: 10.1016/0092-8674(92)90191-E
Thomson, J. A., Itskovitz-Eldor, J., Shapiro, S. S., Waknitz, M. A., Swiergiel, J. J., Marshall, V. S., et al. (1998). Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147. doi: 10.1126/science.282.5391.1145
Vogel-Ciernia, A., Matheos, D. P., Barrett, R. M., Kramár, E. A., Azzawi, S., Chen, Y., et al. (2013). The neuron-specific chromatin regulatory subunit BAF53b is necessary for synaptic plasticity and memory. Nat. Neurosci. 16, 552–561. doi: 10.1038/nn.3359
Waghray, A., Saiz, N., Jayaprakash, A. D., Freire, A. G., Papatsenko, D., Pereira, C. F., et al. (2015). Tbx3 controls Dppa3 levels and exit from pluripotency toward mesoderm. Stem Cell Rep. 5, 97–110. doi: 10.1016/j.stemcr.2015.05.009
Weidgang, C. E., Russell, R., Tata, P. R., Kühl, S. J., Illing, A., Müller, M., et al. (2014). TBX3 directs cell-fate decision toward mesendoderm. Stem Cell Rep. 2:747. doi: 10.1016/j.stemcr.2014.04.011
Williams, R. L., Hilton, D. J., Pease, S., Willson, T. A., Stewart, C. L., Gearing, D. P., et al. (1988). Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature 336, 684–687. doi: 10.1038/336684a0
Winata, P., William, M., Keena, V., Takahashi, K., and Cheng, Y. Y. (2018). “DNA methylation in mammalian cells,” in Gene Expression and Regulation in Mammalian Cells - Transcription Toward the Establishment of Novel Therapeutics, ed F. Uchiumi (IntechOpen). doi: 10.5772/intechopen.72991
Yan, Z., Cui, K., Murray, D. M., Ling, C., Xue, Y., Gerstein, A., et al. (2005). PBAF chromatin-remodeling complex requires a novel specificity subunit, BAF200, to regulate expression of selective interferon-responsive genes. Genes Dev. 19, 1662–1667. doi: 10.1101/gad.1323805
Yan, Z., Wang, Z., Sharova, L., Sharov, A. A., Ling, C., Piao, Y., et al. (2008). BAF250B-associated SWI/SNF chromatin-remodeling complex is required to maintain undifferentiated mouse embryonic stem cells. Stem Cells 26, 1155–1165. doi: 10.1634/stemcells.2007-0846
Yeo, J. C., Jiang, J., Tan, Z. Y., Yim, G. R., Ng, J. H., Göke, J., et al. (2014). Klf2 is an essential factor that sustains ground state pluripotency. Cell Stem Cell 14, 864–872. doi: 10.1016/j.stem.2014.04.015
Ying, Q. L., Nichols, J., Chambers, I., and Smith, A. (2003). BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell 115, 281–92. doi: 10.1016/S0092-8674(03)00847-X
Ying, Q. L., Wray, J., Nichols, J., Batlle-Morera, L., Doble, B., Woodgett, J., et al. (2008). The ground state of embryonic stem cell self-renewal. Nature 453, 519–523. doi: 10.1038/nature06968
You, J. S., De Carvalho, D. D., Dai, C., Liu, M., Pandiyan, K., Zhou, X. J., et al. (2013). SNF5 is an essential executor of epigenetic regulation during differentiation. PLoS Genet. 9:e1003459. doi: 10.1371/journal.pgen.1003459
Zhang, W., Chronis, C., Chen, X., Zhang, H., Spalinskas, R., Pardo, M., et al. (2019). The BAF and PRC2 complex subunits Dpf2 and Eed antagonistically converge on Tbx3 to control ESC differentiation. Cell Stem Cell 24, 138–152.e8. doi: 10.1016/j.stem.2018.12.001
Zhang, X., Li, B., Li, W., Ma, L., Zheng, D., Li, L., et al. (2014). Transcriptional repression by the BRG1-SWI/SNF complex affects the pluripotency of human embryonic stem cells. Stem Cell Rep. 3, 460–74. doi: 10.1016/j.stemcr.2014.07.004
Keywords: SWI/SNF (BAF) complex, embryonic stem cells, pluripotency, differentiation, chromatin remodeling complex
Citation: Ye Y, Chen X and Zhang W (2021) Mammalian SWI/SNF Chromatin Remodeling Complexes in Embryonic Stem Cells: Regulating the Balance Between Pluripotency and Differentiation. Front. Cell Dev. Biol. 8:626383. doi: 10.3389/fcell.2020.626383
Received: 05 November 2020; Accepted: 30 November 2020;
Published: 18 January 2021.
Edited by:
Bruno Di Stefano, Baylor College of Medicine, United StatesReviewed by:
Davide Seruggia, Boston Children's Hospital, United StatesCopyright © 2021 Ye, Chen and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xi Chen, Y2hlbng5QHN1c3RlY2guZWR1LmNu; Wensheng Zhang, emhhbmd3ZW5zaGVuZ0BzdWRhLmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.