- 1Institute for Research in Molecular Medicine (INFORMM), Universiti Sains Malaysia, Penang, Malaysia
- 2USM-RIKEN International Centre for Aging Science (URICAS), Universiti Sains Malaysia, Penang, Malaysia
Aging is a complex biological process that occurs in all living organisms. Aging is initiated by the gradual accumulation of biomolecular damage in cells leading to the loss of cellular function and ultimately death. Cellular senescence is one such pathway that leads to aging. The accumulation of nucleic acid damage and genetic alterations that activate permanent cell-cycle arrest triggers the process of senescence. Cellular senescence can result from telomere erosion and ribosomal DNA instability. In this review, we summarize the molecular mechanisms of telomere length homeostasis and ribosomal DNA stability, and describe how these mechanisms are linked to cellular senescence and longevity through lessons learned from budding yeast.
Introduction
Mammalian cells possess a finite replicative capacity known as “Hayflick limit” (Hayflick and Moorhead, 1961) which when reached initiates replicative senescence upon irreversible cell cycle arrest (Demidenko and Blagosklonny, 2008). Replicative senescence is a natural process that occurs due to telomere shortening with every cell division (Stewart et al., 2003). However, premature senescence can be induced by cellular exposure to stresses such as oxidative stress or DNA damage (Debacq-Chainiaux et al., 2016). Cellular senescence is thought to be a beneficial protective system against cancer because it limits the proliferation of damaged cells and progression of malignant cells (Muñoz-Espín and Serrano, 2014). Senescent cells remain metabolically active and are viable for a long period of time (Blagosklonny, 2003), and exhibit phenotypes such as enlarged intracellular organelles and increased cell size (Cristofalo et al., 2004; Matsui and Matsuura, 2010)
While aging is caused by structural deteriorations at the organismal level, at the cellular level aging is caused by replicative or premature senescence via genomic instability amongst other factors (Lidzbarsky et al., 2018; Lagunas-Rangel and Bermúdez-Cruz, 2019). In this mini review, we summarize our current understanding of telomere length homeostasis and maintenance of ribosomal DNA (rDNA) stability, the two major contributors to genomic instability.
Maintenance of Telomre Length Homeostasis Prevents Cellular Senescence and Aging
Telomeres at eukaryotic chromosome ends protect the chromosome ends from end-to-end fusion, degradation, and prevent misrecognition of the ends as double-stranded DNA breaks (DSBs) (Tham and Zakian, 2002; Dieckmann et al., 2016). Telomeres have terminal single-stranded (ss) DNA overhangs with 3′ repetitive guanine-rich sequences (termed G tail or G-overhang) (Giraud-Panis et al., 2010; Eugène et al., 2017). Telomeres are marked by tandem repeats such as G3T2A in vertebrates and TG1–3 in budding yeast Saccharomyces cerevisiae (Tran et al., 2011; Wellinger and Zakian, 2012). In budding yeast, telomeres consist of subtelomeric repeats known as X′ element found in all telomeres and Y′ element found in two-thirds of telomeres (Louis and Haber, 1992; Teng and Zakian, 1999; Figure 1A).
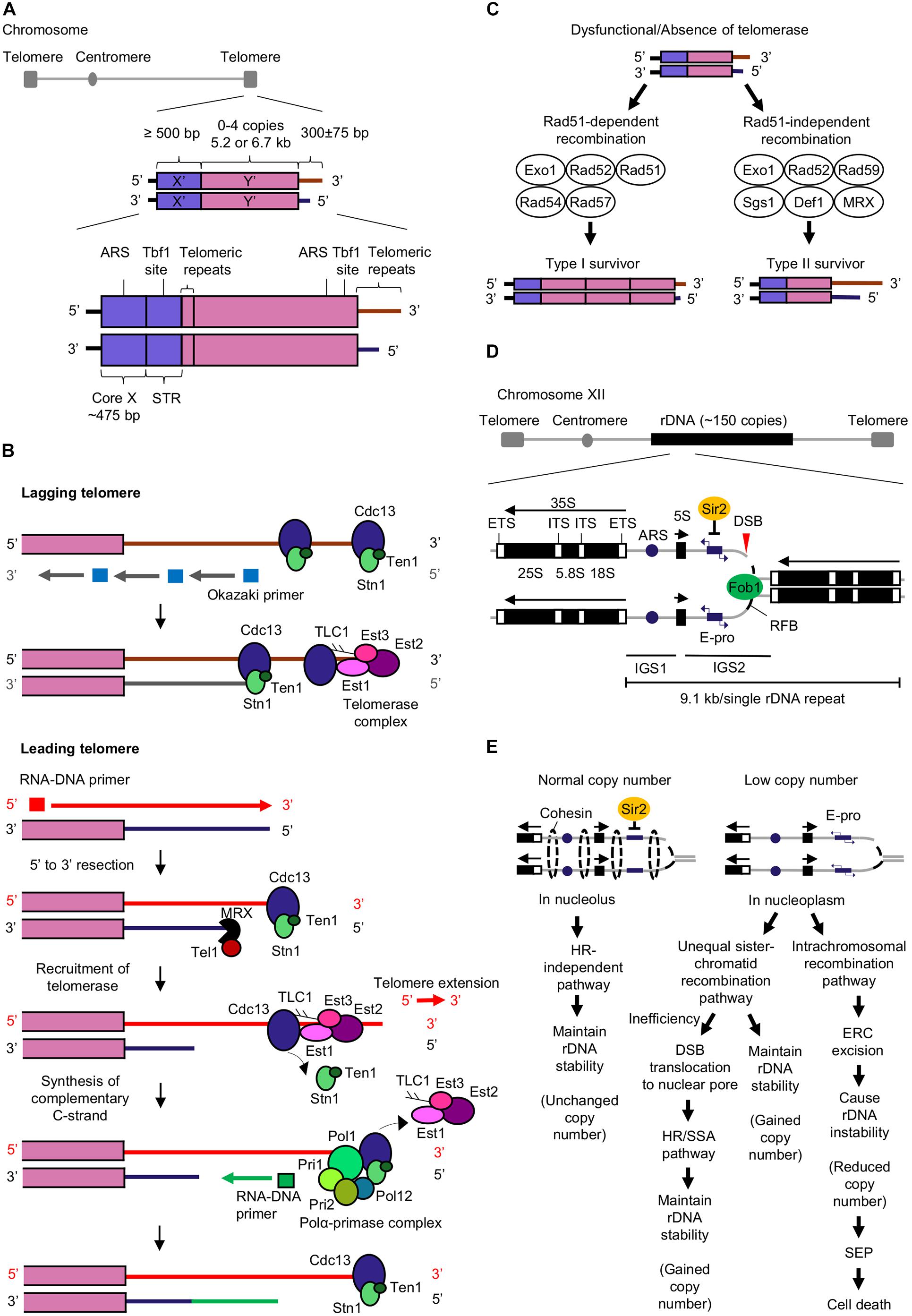
Figure 1. Mechanisms of telomere extension and rDNA copy number maintenance that prevent cellular senescence and aging. (A) The budding yeast telomere structure consists of X′ and Y′ elements, and telomeric repeats. Core X that contains an autonomously replicating sequence (ARS) which is an origin of replication, and subtelomeric repeated elements (STR) that contains a Tbf1 binding site, are found in the X′ element. The ARS and Tbf1 binding site are also found in the Y′ element (Louis et al., 1994; Tham and Zakian, 2002; Wellinger and Zakian, 2012). (B) Telomerase-dependent pathway for telomere extension. Cdc13 and telomerase complex (Est1, Est2, Est3, and Tlc1) bind to both leading and lagging telomeres (Faure et al., 2010). DNA polymerase α (Polα)-primase complex generates RNA-DNA primers that initiate the synthesis of Okazaki fragments by DNA polymerase δ (Pol δ) (McElhinny et al., 2008; Perera et al., 2013) at lagging strand. After removal of the primers, the Okazaki fragments are ligated by DNA ligase I to form complementary lagging strand (Faure et al., 2010; Perera et al., 2013; Liu et al., 2017). Telomere extension mediated by telomerase occurs primarily at the leading telomere (Faure et al., 2010). The RNA-DNA primer is required for initiating synthesis of complementary leading strand by DNA polymerase ε (Pol ε) (Pursell et al., 2007; McElhinny et al., 2008; Perera et al., 2013). CST complex (Cdc13-Stn1-Ten1) bound at telomere end restricts the access of telomerase to telomere end. MRX complex (Mre11-Rad50-Xrs2) induces the binding of Tel1 (Nakada et al., 2003; McGee et al., 2010) to short telomere and executes 5′ to 3′ exonuclease activity to synthesize 3′ overhang (Diede and Gottschling, 2001). Both MRX and Tel1 promote Cdc13-mediated telomerase tethering to telomere (Tseng et al., 2006; Yang et al., 2017). Extension of telomere by telomerase is completed upon the synthesis of the CST complex (Pfeiffer and Lingner, 2013) and telomerase departure from the telomere. Polα-primase complex (Pol1, Pol12, Pri1, and Pri2) (Lue et al., 2014) interacts with CST complex at the telomere (Churikov et al., 2013) and generates an RNA-DNA primer for synthesis of complementary C-strand (Churikov et al., 2013; Pfeiffer and Lingner, 2013). (C) Homologous recombination pathways for telomere extension in the absence of telomerase or when telomerase is dysfunctional. The Rad51-dependent recombination pathway for the generation of type I survivors requires Exo1, Rad52 (Chen et al., 2001; Maringele and Lydall, 2004), Rad51, Rad54, and Rad57 (Chen et al., 2001; Claussin and Chang, 2015) while the Rad51-independent recombination pathway for the generation of type II survivors includes Exo1, Rad52 (Chen et al., 2001; Maringele and Lydall, 2004), Rad59, Sgs1, Def1, and MRX complex (Huang et al., 2001; Signon et al., 2001; Chen et al., 2005). (D) The structure of budding yeast rDNA (Poveda et al., 2010; Chand Dakal et al., 2016; Kobayashi and Sasaki, 2017). Abbreviations: ETS, external transcribed spacer; ITS, internal transcribed spacer; IGS, intergenic spacer; ARS, autonomous replication sequence; E-pro, rDNA non-coding promoter; RFB, replication fork barrier site; DSB, double-stranded DNA break. (E) Regulation of rDNA stability by homologous recombination (HR)-independent and -dependent pathways (unequal sister-chromatid or intrachromosomal recombination pathway) (Kobayashi and Sasaki, 2017; Sasaki and Kobayashi, 2017; Horigome et al., 2019; Morlot et al., 2019). Abbreviations: DSB, double-stranded break; ERC, extrachromosomal rDNA circles; SEP, senescence entry point; SSA, single strand annealing.
Telomere Shortening
Telomere shortening (or erosion) causes genomic instability through the breakage-fusion-bridge cycle. The progressive loss of telomere end after cell division (Soudet et al., 2014) initiates DNA break repair (DBR) machinery that repairs shortened telomeres through DNA replication creating a fusion of two sister chromatids. During cell division, the segregation of fused chromosomes will cause a random break, leading to inheritance of deleted or amplified chromosomes by daughter cells. The continuous breakage-fusion-bridge cycle with every cell division leads to genomic instability (McClintock, 1938; Tanaka and Yao, 2009).
Telomere Length Homeostasis
Lagging and leading telomeres are synthesized in the progression of replication fork during telomere replication. Due to the end-replication problem, telomeres are replicated incompletely by DNA polymerases and end with a 3′ overhang. Excessive critically short telomeres will elicit a DNA damage signal causing permanent cell cycle arrest, subsequently cellular senescence and death (Shay and Wright, 2005; Aubert and Lansdorp, 2008).
Additionally, dysfunctional telomeres due to telomere uncapping can cause cellular senescence in an indirect manner. The unprotected telomeres undergo degradation (Vodenicharov and Wellinger, 2006; Ghadaouia et al., 2018) and induce a weak DNA damage response (DDR). Unprotected telomeres are prone to chromosomal end fusion resulting in secondary DNA breaks and genomic instability, eliciting a strong DDR. Consequently, the additional DNA damage causes permanent growth arrest and cellular senescence (Ghadaouia et al., 2018). Therefore, telomere length homeostasis must be maintained by telomere elongation to compensate for the end-replication problem and protect telomere ends. The two pathways involved in telomere elongation are telomerase-dependent pathway and homologous recombination (HR) pathway.
Telomerase-Dependent Pathway
Telomerase is a reverse transcriptase that depends on its internal RNA subunit Tlc1 (Gilson and Géli, 2007) as a template to extend telomeric repeats. Telomerase preferentially extends short telomeres in the late S phase (Wellinger and Zakian, 2012) by applying dNTP synthesized by ribonucleotide reductase (RNR) to add nucleotides at the telomeric 3′ overhang (Maicher et al., 2017) while the complementary strand is synthesized by DNA polymerases (Hug and Lingner, 2006; Figure 1B). A telomerase consists of four subunits Est1, Est2, Est3, and Tlc1 which positively regulate telomerase activity for telomere extension (Lendvay et al., 1996). Est1 enables the access of telomerase to telomere by interacting with Tlc1 and telomeric ssDNA-binding protein Cdc13 (Virta-Pearlman et al., 1996; Evans and Lundblad, 1999; Zhou et al., 2000; Li et al., 2013). Additionally, Est1 stimulates the generation of G-quadruplex at telomeric overhang for telomere extension and protection (Zhang et al., 2010; Tong et al., 2011; Li et al., 2013). Est2 (Counter et al., 1997) and Tlc1 (Singer and Gottschling, 1994; Cohn and Blackburn, 1995) catalyze telomere extension. Est3 which associates with Est1 and Est2, induces Est2’s catalytic activity for telomere extension (Zhang et al., 2010; Mariasina et al., 2018).
Telomerase-Independent Homologous Recombination Pathway
Telomerase deficiency leads to progressive telomere shortening, and consequently cell death (Le et al., 1999). Nevertheless, a subset of cells with telomerase deficiency can still survive and have extended telomeres. These survivors are classified as type I or type II; with their telomeres extended via Rad51-dependent or Rad51-independent homologous recombination (HR) pathways respectively. Both recombination pathways involve Rad52, Exo1, and Pol32 which is a non-essential subunit of DNA polymerase δ (Chen et al., 2001; Maringele and Lydall, 2004; Lydeard et al., 2007; Figure 1C).
The telomeres of type I survivors contain tandemly amplified Y′ elements and short telomeric repeats TG1–3 at their ends while telomeres of type II survivor have amplified telomeric repeats TG1–3 at their ends with heterogenous length (Figure 1C). Although the survivors can depend on HR pathway to maintain telomere length, they possess shorter replicative life spans (RLS). The reactivation of telomerase activity can restore the reduced RLS, revealing the role of telomerase in sustaining cellular RLS possibly by suppressing telomere recombination and maintaining telomere length (Chen et al., 2009). Additionally, other proteins involved in the regulation of telomere length homeostasis in S. cerevisiae are summarized in Table 1.
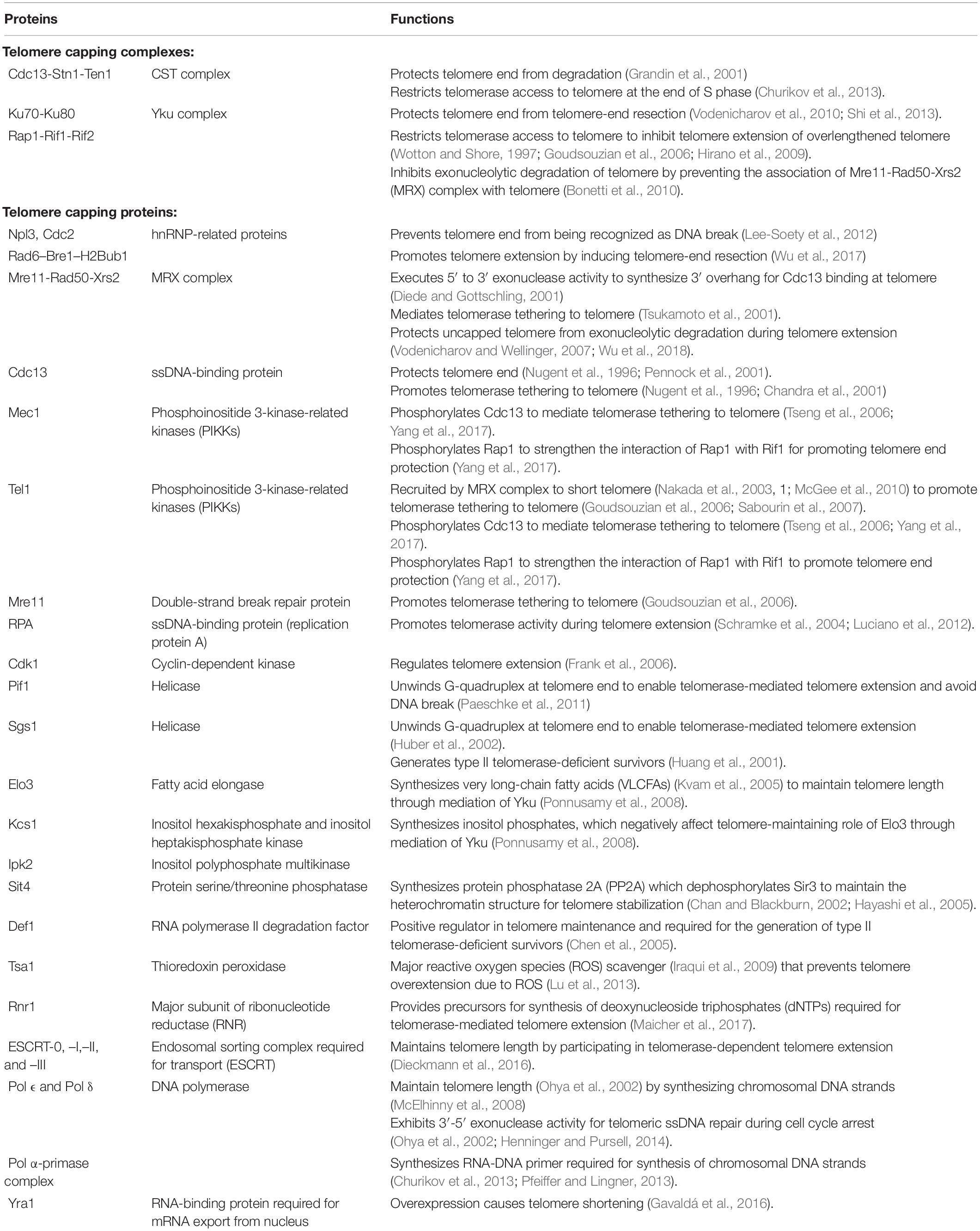
Table 1. Functions of proteins involved in the regulation of telomere length homeostasis in Saccharomyces cerevisiae.
Telomere Shortening and Its Effects on Aging
Telomere shortening decreases life span in mice and humans (Muñoz-Lorente et al., 2019; Whittemore et al., 2019). In mice, reactivation of telomerase activity can rescue premature aging phenotype through restoration of short telomere length and its function (Samper et al., 2001). Telomerase overexpression which promotes telomere extension can prolong life span in mice (Bernardes de Jesus et al., 2012). Furthermore, mice with overlengthened telomeres exhibited less DNA damage, less metabolic aging and an increased life span (Muñoz-Lorente et al., 2019). These findings show that the telomerase-mediated telomere extension can promote life span extension in animal models. Interestingly, telomerase overexpression in mice also reduced cancer incidences (Bernardes de Jesus et al., 2012; Muñoz-Lorente et al., 2019). This highlights the complex link between senescence and cancer suppression because senescence is thought to be a safeguard against cancer.
Overlengthened telomeres has no impact on yeast chronological life span (CLS) (Harari et al., 2017). Conversely, telomere shortening causes RLS extension in budding yeast, possibly due to the relocalization of Sir2/3/4 complex to non-telomeric sites for heterochromatin structure maintenance and increased genomic stability (Austriaco and Guarente, 1997; Liu et al., 2019). Sir2 inhibits extreme CLS extension in yeast (Fabrizio et al., 2005), unlike its importance for RLS maintenance (Kaeberlein et al., 1999). Hence, the opposite roles of Sir2 in CLS and RLS may explain the different effects of telomere length on yeast aging. Furthermore, yeast telomeres do not shorten with age like the mammalian phenotype (D’Mello and Jazwinski, 1991). Therefore, while yeast is useful for telomere maintenance mechanism studies, it may not be the best model to study the telomere-aging link.
Maintenance of rDNA Stability Prevents Cellular Senescence and Replicative Aging
The ribosomal RNA gene (rDNA) is the most abundant RNA gene that encodes for ribosomal RNA. Ribosomal RNA is essential to form ribosomes with ribosomal proteins for protein synthesis (Kobayashi, 2008). S. cerevisiae has ∼150 tandem repeats of a 9.1 kb rDNA unit (Ganley and Kobayashi, 2014), found on chromosome XII locus (Petes, 1979; Figure 1D). These rDNA repeats are the most prone to DNA damage by external (ultraviolet light) and internal factors (replication errors and ROS), causing regional genomic instability. According to rDNA theory of aging, rDNA instability induces an aging signal that triggers DDR and initiate cellular senescence thus limiting cellular life span (Kobayashi, 2008, 2011a, 2014).
Maintenance of rDNA Copy Number for rDNA Stability
In yeast, Fob1 and Sir2 are major regulators of the rDNA maintenance system. Fob1 binds at the replication fork barrier (RFB) site in rDNA repeat to block the progression of the replication fork to the opposite direction of 35S rRNA gene transcription and form a DSB at the blocking site (Kobayashi, 2003). The blockage of the replication fork initiates the DBR either through a HR-independent or -dependent pathway depending on cellular rDNA copy number (Kobayashi and Sasaki, 2017; Sasaki and Kobayashi, 2017; Figure 1E).
The histone deacetylase Sir2 inhibits E-pro transcription when rDNA copy number is maintained at wild-type level. The inhibited E-pro transcription strengthens cohesion association to the broken DNA end and enables DBR which is independent of HR to occur in nucleolus (Saka et al., 2016; Sasaki and Kobayashi, 2017; Horigome et al., 2019). As a result, this repair leads to an unchanged rDNA copy number, maintaining rDNA stability and generating little to no aging signal (Saka et al., 2016).
In contrast, Sir2 does not inhibit E-pro transcription when rDNA copy number is less than wild-type level. The active transcription of E-pro dissociates the cohesins from the broken DNA end, stimulating the unequal sister chromatid recombination to occur between the misaligned rDNA repeats for DBR. The unequal sister chromatid recombination is Rad52-dependent (Torres-Rosell et al., 2007) and occurs in nucleoplasm (Horigome et al., 2019). As a result, unequal sister chromatid recombination duplicates and restore rDNA copy number (Kobayashi and Sasaki, 2017).
However, when the unequal sister chromatid recombination mediated DBR is inefficient, the rDNA break ends move to the nuclear pore and interact with nuclear pore complex. These DSBs at the nuclear pore may be repaired either by the HR or single strand annealing pathway. DNA damage checkpoint-associated Mec1/Tel1 kinases, replisome component Tof1 and proteins Tof2, Csm1, and Lrs4 that tether condensins to rDNA assist in the translocation of DSBs to nuclear pore for DBR and maintain rDNA stability (Horigome et al., 2019).
Intrachromosomal recombination can also occur upon activation of E-pro transcription, whereby the broken DNA end recombines with the rDNA copy within the same chromosome. Intrachromosomal recombination is mediated by Rad52 (Park et al., 1999) and results in the formation of extrachromosomal rDNA circles (ERCs) which are segregated from rDNA strand, leading to rDNA copy loss (Kobayashi and Sasaki, 2017). The loss of rDNA copy causes rDNA instability and stimulates cellular senescence (Hein et al., 2012; Saka et al., 2016; Figure 1E).
rDNA Stability and Its Effects on Aging
Extrachromosomal rDNA circles were previously shown to cause aging in yeast and speculated to be the molecular cause of aging in higher species, including mammals (Sinclair and Guarente, 1997). This ERC theory of aging was later disputed by the rDNA theory of aging (Kobayashi, 2008) which affirms that rDNA instability is a major cause of aging independent of ERC accumulation level. Although the accumulation of ERCs and other episomes (plasmids) can shorten yeast RLS, they stimulate rDNA instability, reaffirming rDNA stability as a major life span-determinant (Falcón and Aris, 2003; Ganley et al., 2009; Saka et al., 2013). Nevertheless, ERCs can be markers for rDNA instability.
Asymmetrical segregations of instable rDNAs and ERCs occur more frequently to yeast mother cells, resulting in cellular senescence and aging while stable rDNAs are segregated to daughter cells, allowing daughter cells to undergo rejuvenation (Kobayashi, 2011b; Morlot et al., 2019). However, rejuvenation of daughter cells produced by old mother cells (after their first 40% of RLS) would be affected, thus exhibiting decreased RLS. This is likely due to impaired asymmetrical segregation of aging factors which are constrained as well in daughter cells (Kennedy et al., 1994).
More recently, Morlot et al. (2019) proposed a model that links the accumulation of ERCs to senescence and longevity. ERC-linked senescence is categorized into three stages: ERC excision, ERC self-replication, and post-SEP interval. Extensive ERC excision from rDNA during intrachromosomal recombination and self-replication of ERC leads to the accumulation of ERC. Even though accumulating ERC can upregulate rDNA transcription, ribosome synthesis is not enhanced. Loss of coordination between rDNA transcription and ribosome biogenesis could negatively affect cell growth. When the amount of ERCs reaches a threshold, the cells reach a senescence entry point (SEP). In post-SEP interval (an interval between SEP and cell death), the cells experience a loss of nuclear homeostasis [an increase in nucleus-to-cell area ratio (N/C ratio), increase in histone content and genomic defect] which causes cell death (Morlot et al., 2019; Figure 1E). The relationship between rDNA stability to RLS has been shown with yeast mutants of rDNA stability regulators. Yeast cells lacking SIR2 with more ERCs causing rDNA instability exhibited a ∼50% decrease in RLS (Kaeberlein et al., 1999). Unlike SIR2 mutants, FOB1 mutants has less ERCs, thus enhancing rDNA stability and showed a 70% extension of RLS (Defossez et al., 1999). More recently, a new regulator of rDNA copy number, Eaf3 was discovered (Wakatsuki et al., 2019). Yeast cells lacking Eaf3 exhibited less ERCs which enhances rDNA stability leading to a 30% extension of RLS. Eaf3 likely activates transcription of E-pro to induce unequal sister chromatid recombination and intrachromosomal recombination which results in the formation of ERCs.
The link of rDNA stability to senescence and aging has also been established in mammalian cell studies. For example, genomic imaging revealed rDNA copy loss in senescent human cells and blood from aged individuals, validating the link between human aging and rDNA instability (Ren et al., 2017). The mammalian Sir2 homolog SIRT7 was also found to function like Sir2 to prevent rDNA instability and consequently cellular senescence via chromatin silencing (Paredes et al., 2018).
In summary, DNA replication-based telomere elongation and DBR-based rDNA copy number maintenance are fundamental mechanisms that minimize DNA loss and damage for maintenance of genomic stability, thus inhibiting the onset of cellular senescence and aging. Studies in yeast have identified proteins regulating telomere length homeostasis and the understanding of rDNA copy number maintenance. Current knowledge shows that rDNA instability possibly plays a bigger role than ERCs in aging in yeast and especially higher organisms. Still, yeast will continue to serve as a versatile model for studying rDNA instability and telomere length maintenance mechanisms.
Author Contributions
JL drafted the manuscript. EO revised and edited further. Both authors approved the final version of the manuscript for submission.
Funding
This work is supported by Universiti Sains Malaysia’s RU Top-Down grant (1001/CIPPM/870038) for the USM-RIKEN International Centre for Aging Science (URICAS) program.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Aubert, G., and Lansdorp, P. M. (2008). Telomeres and aging. Physiol. Rev. 88, 557–579. doi: 10.1152/physrev.00026.2007
Austriaco, N. R., and Guarente, L. P. (1997). Changes of telomere length cause reciprocal changes in the lifespan of mother cells in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 94, 9768–9772. doi: 10.1073/PNAS.94.18.9768
Bernardes de Jesus, B., Vera, E., Schneeberger, K., Tejera, A. M., Ayuso, E., Bosch, F., et al. (2012). Telomerase gene therapy in adult and old mice delays aging and increases longevity without increasing cancer. EMBO Mol. Med. 4, 691–704. doi: 10.1002/emmm.201200245
Blagosklonny, M. V. (2003). Cell senescence and hypermitogenic arrest. EMBO Rep. 4, 358–362. doi: 10.1038/sj.embor.embor806
Bonetti, D., Clerici, M., Anbalagan, S., Martina, M., Lucchini, G., and Longhese, M. P. (2010). Shelterin-like proteins and Yku inhibit nucleolytic processing of Saccharomyces cerevisiae telomeres. PLoS Genet. 6:e1000966. doi: 10.1371/journal.pgen.1000966
Chan, S. W.-L., and Blackburn, E. H. (2002). New ways not to make ends meet: telomerase, DNA damage proteins and heterochromatin. Oncogene 21, 553–563. doi: 10.1038/sj.onc.1205082
Chand Dakal, T., Giudici, P., and Solieri, L. (2016). Contrasting patterns of rDNA homogenization within the Zygosaccharomyces rouxii species complex. PLoS One 11:e0160744. doi: 10.1371/journal.pone.0160744
Chandra, A., Hughes, T. R., Nugent, C. I., and Lundblad, V. (2001). Cdc13 both positively and negatively regulates telomere replication. Genes Dev. 15, 404–414. doi: 10.1101/gad.861001
Chen, Q., Ijpma, A., and Greider, C. W. (2001). Two survivor pathways that allow growth in the absence of telomerase are generated by distinct telomere recombination events. Mol. Cell. Biol. 21, 1819–1827. doi: 10.1128/mcb.21.5.1819-1827.2001
Chen, X.-F., Meng, F.-L., and Zhou, J.-Q. (2009). Telomere recombination accelerates cellular aging in Saccharomyces cerevisiae. PLoS Genet. 5:e1000535. doi: 10.1371/journal.pgen.1000535
Chen, Y. B., Yang, C. P., Li, R. X., Zeng, R., and Zhou, J. Q. (2005). Def1p is involved in telomere maintenance in budding yeast. J. Biol. Chem. 280, 24784–24791. doi: 10.1074/jbc.M413562200
Churikov, D., Corda, Y., Luciano, P., and Geli, V. (2013). Cdc13 at a crossroads of telomerase action. Front. Oncol. 3:39. doi: 10.3389/fonc.2013.00039
Claussin, C., and Chang, M. (2015). The many facets of homologous recombination at telomeres. Microb. Cell 2, 308–321. doi: 10.15698/mic2015.09.224
Cohn, M., and Blackburn, E. H. (1995). Telomerase in yeast. Science 269, 396–400. doi: 10.1126/science.7618104
Counter, C. M., Meyerson, M., Eaton, E. N., and Weinberg, R. A. (1997). The catalytic subunit of yeast telomerase. Proc. Natl. Acad. Sci. U.S.A. 94, 9202–9207. doi: 10.1073/pnas.94.17.9202
Cristofalo, V. J., Lorenzini, A., Allen, R. G., Torres, C., and Tresini, M. (2004). Replicative senescence: a critical review. Mech. Ageing Dev. 125, 827–848. doi: 10.1016/j.mad.2004.07.010
Debacq-Chainiaux, F., Ben Ameur, R., Bauwens, E., Dumortier, E., Toutfaire, M., and Toussaint, O. (2016). “Stress-Induced (Premature) Senescence,” in Cellular Ageing and Replicative Senescence Healthy Ageing and Longevity, eds S. I. S. Rattan and L. Hayflick (Cham: Springer), 243–262. doi: 10.1007/978-3-319-26239-0_13
Defossez, P. A., Prusty, R., Kaeberlein, M., Lin, S. J., Ferrigno, P., Silver, P. A., et al. (1999). Elimination of replication block protein fob1 extends the life span of yeast mother cells. Mol. Cell 3, 447–455. doi: 10.1016/S1097-2765(00)80472-4
Demidenko, Z. N., and Blagosklonny, M. V. (2008). Growth stimulation leads to cellular senescence when the cell cycle is blocked. Cell Cycle 7, 3355–3361. doi: 10.4161/cc.7.21.6919
Dieckmann, A. K., Babin, V., Harari, Y., Eils, R., König, R., Luke, B., et al. (2016). Role of the ESCRT complexes in telomere biology. mBio 7:e01793-16. doi: 10.1128/mBio.01793-16
Diede, S. J., and Gottschling, D. E. (2001). Exonuclease activity is required for sequence addition and Cdc13p loading at a de novo telomere. Curr. Biol. 11, 1336–1340. doi: 10.1016/S0960-9822(01)00400-6
D’Mello, N. P., and Jazwinski, S. M. (1991). Telomere length constancy during aging of Saccharomyces cerevisiae. J. Bacteriol. 173, 6709–6713. doi: 10.1128/jb.173.21.6709-6713.1991
Eugène, S., Bourgeron, T., and Xu, Z. (2017). Effects of initial telomere length distribution on senescence onset and heterogeneity. J. Theor. Biol. 413, 58–65. doi: 10.1016/j.jtbi.2016.11.010
Evans, S. K., and Lundblad, V. (1999). Est1 and Cdc13 as comediators of telomerase access. Science 286, 117–120. doi: 10.1126/science.286.5437.117
Fabrizio, P., Gattazzo, C., Battistella, L., Wei, M., Cheng, C., McGrew, K., et al. (2005). Sir2 blocks extreme life-span extension. Cell 123, 655–667. doi: 10.1016/j.cell.2005.08.042
Falcón, A. A., and Aris, J. P. (2003). Plasmid accumulation reduces life span in Saccharomyces cerevisiae. J. Biol. Chem. 278, 41607–41617. doi: 10.1074/jbc.M307025200
Faure, V., Coulon, S., Hardy, J., and Géli, V. (2010). Cdc13 and telomerase bind through different mechanisms at the lagging- and leading-strand telomeres. Mol. Cell 38, 842–852. doi: 10.1016/j.molcel.2010.05.016
Frank, C. J., Hyde, M., and Greider, C. W. (2006). Regulation of telomere elongation by the cyclin-dependent kinase CDK1. Mol. Cell 24, 423–432. doi: 10.1016/j.molcel.2006.10.020
Ganley, A. R. D., Ide, S., Saka, K., and Kobayashi, T. (2009). The effect of replication initiation on gene amplification in the rDNA and Its relationship to aging. Mol. Cell 35, 683–693. doi: 10.1016/j.molcel.2009.07.012
Ganley, A. R. D., and Kobayashi, T. (2014). Ribosomal DNA and cellular senescence: new evidence supporting the connection between rDNA and aging. FEMS Yeast Res. 14, 49–59. doi: 10.1111/1567-1364.12133
Gavaldá, S., Santos-Pereira, J. M., García-Rubio, M. L., Luna, R., and Aguilera, A. (2016). Excess of Yra1 RNA-binding factor causes transcription-dependent genome instability, replication impairment and telomere shortening. PLoS Genet. 12:e1005966. doi: 10.1371/journal.pgen.1005966
Ghadaouia, S., Olivier, M. A., Martinez, A., Malaquin, N., Cardin, G. B., and Rodier, F. (2018). Telomeric dysfunction triggers an unstable growth arrest leading to irreparable genomic lesions and entry into cellular senescence. bioRxiv [Preprint]. doi: 10.1101/442533
Gilson, E., and Géli, V. (2007). How telomeres are replicated. Nat. Rev. Mol. Cell Biol. 8, 825–838. doi: 10.1038/nrm2259
Giraud-Panis, M. J., Pisano, S., Poulet, A., Du, M. H. L., and Gilson, E. (2010). Structural identity of telomeric complexes. FEBS Lett. 584, 3785–3799. doi: 10.1016/j.febslet.2010.08.004
Goudsouzian, L. K., Tuzon, C. T., and Zakian, V. A. (2006). S. cerevisiae Tel1p and Mre11p are required for normal levels of Est1p and Est2p telomere association. Mol. Cell 24, 603–610. doi: 10.1016/j.molcel.2006.10.005
Grandin, N., Damon, C., and Charbonneau, M. (2001). Ten1 functions in telomere end protection and length regulation in association with Stn1 and Cdc13. EMBO J. 20, 1173–1183. doi: 10.1093/emboj/20.5.1173
Harari, Y., Zadok-Laviel, S., and Kupiec, M. (2017). Long telomeres do not affect cellular fitness in yeast. mBio 8:e01314-17. doi: 10.1128/mBio.01314-17
Hayashi, N., Nomura, T., Sakumoto, N., Mukai, Y., Kaneko, Y., Harashima, S., et al. (2005). The SIT4 gene, which encodes protein phosphatase 2A, is required for telomere function in Saccharomyces cerevisiae. Curr. Genet. 47, 359–367. doi: 10.1007/s00294-005-0577-1
Hayflick, L., and Moorhead, P. S. (1961). The serial cultivation of human diploid cell strains. Exp. Cell Res. 25, 585–621. doi: 10.1016/0014-4827(61)90192-6
Hein, N., Sanij, E., Quin, J., Hannan, K. M., Ganley, A., and Hannan, R. D. (2012). “The nucleolus and ribosomal genes in aging and senescence,” in Senescence, ed. T. Nagata (London: InTech). doi: 10.5772/34581
Henninger, E. E., and Pursell, Z. F. (2014). DNA polymerase ε and its roles in genome stability. IUBMB Life 66, 339–351. doi: 10.1002/iub.1276
Hirano, Y., Fukunaga, K., and Sugimoto, K. (2009). Rif1 and Rif2 inhibit localization of tel1 to DNA ends. Mol. Cell 33, 312–322. doi: 10.1016/j.molcel.2008.12.027
Horigome, C., Unozawa, E., Ooki, T., and Kobayashi, T. (2019). Ribosomal RNA gene repeats associate with the nuclear pore complex for maintenance after DNA damage. PLoS Genet. 15:e1008103. doi: 10.1371/journal.pgen.1008103
Huang, P. H., Pryde, F. E., Lester, D., Maddison, R. L., Borts, R. H., Hickson, I. D., et al. (2001). SGS1 is required for telomere elongation in the absence of telomerase. Curr. Biol. 11, 125–129. doi: 10.1016/S0960-9822(01)00021-5
Huber, M. D., Lee, D. C., and Maizels, N. (2002). G4 DNA unwinding by BLM and Sgs1p: substrate specificity and substrate-specific inhibition. Nucleic Acids Res. 30, 3954–3961. doi: 10.1093/nar/gkf530
Hug, N., and Lingner, J. (2006). Telomere length homeostasis. Chromosoma 115, 413–425. doi: 10.1007/s00412-006-0067-3
Iraqui, I., Kienda, G., Soeur, J., Faye, G., Baldacci, G., Kolodner, R. D., et al. (2009). Peroxiredoxin Tsa1 is the key peroxidase suppressing genome instability and protecting against cell death in Saccharomyces cerevisiae. PLoS Genet. 5:e1000524. doi: 10.1371/journal.pgen.1000524
Kaeberlein, M., McVey, M., and Guarente, L. (1999). The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 13, 2570–2580. doi: 10.1101/gad.13.19.2570
Kennedy, B. K., Austriaco, N. R., and Guarente, L. (1994). Daughter cells of Saccharomyces cerevisiae from old mothers display a reduced life span. J. Cell Biol. 127, 1985–1993. doi: 10.1083/jcb.127.6.1985
Kobayashi, T. (2003). The replication fork barrier site forms a unique structure with Fob1p and inhibits the replication fork. Mol. Cell. Biol. 23, 9178–9188. doi: 10.1128/MCB.23.24.9178-9188.2003
Kobayashi, T. (2008). A new role of the rDNA and nucleolus in the nucleus - rDNA instability maintains genome integrity. Bioessays 30, 267–272. doi: 10.1002/bies.20723
Kobayashi, T. (2011a). How does genome instability affect lifespan? Genes Cells 16, 617–624. doi: 10.1111/j.1365-2443.2011.01519.x
Kobayashi, T. (2011b). Regulation of ribosomal RNA gene copy number and its role in modulating genome integrity and evolutionary adaptability in yeast. Cell. Mol. Life Sci. 68, 1395–1403. doi: 10.1007/s00018-010-0613-2
Kobayashi, T. (2014). Ribosomal RNA gene repeats, their stability and cellular senescence. Proc. Jpn. Acad. Ser. B 90, 119–129. doi: 10.2183/pjab.90.119
Kobayashi, T., and Sasaki, M. (2017). Ribosomal DNA stability is supported by many ‘buffer genes’—introduction to the yeast rDNA stability database. FEMS Yeast Res. 17:fox001. doi: 10.1093/femsyr/fox001
Kvam, E., Gable, K., Dunn, T. M., and Goldfarb, D. S. (2005). Targeting of Tsc13p to nucleus–vacuole junctions: a role for very-long-chain fatty acids in the biogenesis of microautophagic vesicles. Mol. Biol. Cell 16, 3987–3998. doi: 10.1091/mbc.e05-04-0290
Lagunas-Rangel, F. A., and Bermúdez-Cruz, R. M. (2019). “The role of DNA repair in cellular aging process,” in Advances in DNA Repair, ed. M. Mognato (London: IntechOpen). doi: 10.5772/intechopen.84628
Le, S., Moore, J. K., Haber, J. E., and Greider, C. W. (1999). RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics 152, 143–152.
Lee-Soety, J. Y., Jones, J., MacGibeny, M. A., Remaly, E. C., Daniels, L., Ito, A., et al. (2012). Yeast hnRNP-related proteins contribute to the maintenance of telomeres. Biochem. Biophys. Res. Commun. 426, 12–17. doi: 10.1016/j.bbrc.2012.07.144
Lendvay, T. S., Morris, D. K., Sah, J., Balasubramanian, B., and Lundblad, V. (1996). Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics 144, 1399–1412.
Li, Q. J., Tong, X. J., Duan, Y. M., and Zhou, J. Q. (2013). Characterization of the intramolecular G-quadruplex promoting activity of Est1. FEBS Lett. 587, 659–665. doi: 10.1016/j.febslet.2013.01.024
Lidzbarsky, G., Gutman, D., Shekhidem, H. A., Sharvit, L., and Atzmon, G. (2018). Genomic instabilities, cellular senescence, and aging: in vitro, in vivo and aging-like humansyndromes. Front. Med. 5:104. doi: 10.3389/fmed.2018.00104
Liu, B., Hu, J., Wang, J., and Kong, D. (2017). Direct visualization of RNA-DNA primer removal from Okazaki fragments provides support for flap cleavage and exonucleolytic pathways in eukaryotic cells. J. Biol. Chem. 292, 4777–4788. doi: 10.1074/jbc.M116.758599
Liu, J., Wang, L., Wang, Z., and Liu, J. P. (2019). Roles of telomere biology in cell senescence, replicative and chronological ageing. Cells 8:54. doi: 10.3390/cells8010054
Louis, E. J., and Haber, J. E. (1992). The structure and evolution of subtelomeric Y’ repeats in Saccharomyces cerevisiae. Genetics 131, 559–574.
Louis, E. J., Naumova, E. S., Lee, A., Naumov, G., and Haber, J. E. (1994). The chromosome end in yeast: its mosaic nature and influence on recombinational dynamics. Genetics 136, 789–802.
Lu, J., Vallabhaneni, H., Yin, J., and Liu, Y. (2013). Deletion of the major peroxiredoxin Tsa1 alters telomere length homeostasis. Aging Cell 12, 635–644. doi: 10.1111/acel.12085
Luciano, P., Coulon, S., Faure, V., Corda, Y., Bos, J., Brill, S. J., et al. (2012). RPA facilitates telomerase activity at chromosome ends in budding and fission yeasts. EMBO J. 31, 2034–2046. doi: 10.1038/emboj.2012.40
Lue, N. F., Chan, J., Wright, W. E., and Hurwitz, J. (2014). The CDC13-STN1-TEN1 complex stimulates Pol α activity by promoting RNA priming and primase-topolymerase switch. Nat. Commun. 5:5762. doi: 10.1038/ncomms6762
Lydeard, J. R., Jain, S., Yamaguchi, M., and Haber, J. E. (2007). Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature 448, 820–823. doi: 10.1038/nature06047
Maicher, A., Gazy, I., Sharma, S., Marjavaara, L., Grinberg, G., Shemesh, K., et al. (2017). Rnr1, but not Rnr3, facilitates the sustained telomerase-dependent elongation of telomeres. PLoS Genet. 13:e1007082. doi: 10.1371/journal.pgen.1007082
Mariasina, S. S., Efimov, S. V., Petrova, O. A., Rodina, E. V., Malyavko, A. N., Zvereva, M. I., et al. (2018). Chemical shift assignments and the secondary structure of the Est3 telomerase subunit in the yeast Hansenula polymorpha. Biomol. NMR Assign. 12, 57–62. doi: 10.1007/s12104-017-9780-5
Maringele, L., and Lydall, D. (2004). EXO1 plays a role in generating type I and type II survivors in budding yeast. Genetics 166, 1641–1649. doi: 10.1534/genetics.166.4.1641
Matsui, A., and Matsuura, A. (2010). Cell size regulation during telomere-directed senescence in Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 74, 195–198. doi: 10.1271/bbb.90627
McClintock, B. (1938). The Fusion of Broken Ends of Sister Half-Chromatids Following Chromatid Breakage at Meiotic Anaphases. (Columbia, MO: University of Missouri Agricultural Experiment Station Research Bulletin), 290.
McElhinny, S. A. N., Gordenin, D. A., Stith, C. M., Burgers, P. M. J., and Kunkel, T. A. (2008). Division of labor at the eukaryotic replication fork. Mol. Cell 30, 137–144. doi: 10.1016/j.molcel.2008.02.022
McGee, J. S., Phillips, J. A., Chan, A., Sabourin, M., Paeschke, K., and Zakian, V. A. (2010). Reduced Rif2 and lack of Mec1 target short telomeres for elongation rather than double-strand break repair. Nat. Struct. Mol. Biol. 17, 1438–1445. doi: 10.1038/nsmb.1947
Morlot, S., Song, J., Léger-Silvestre, I., Matifas, A., Gadal, O., and Charvin, G. (2019). Excessive rDNA transcription drives the disruption in nuclear homeostasis during entry into senescence in budding yeast. Cell Rep. 28, 408–422. doi: 10.1016/j.celrep.2019.06.032
Muñoz-Espín, D., and Serrano, M. (2014). Cellular senescence: from physiology to pathology. Nat. Rev. Mol. Cell Biol. 15, 482–496. doi: 10.1038/nrm3823
Muñoz-Lorente, M. A., Cano-Martin, A. C., and Blasco, M. A. (2019). Mice with hyper-long telomeres show less metabolic aging and longer lifespans. Nat. Commun. 10:4723. doi: 10.1038/s41467-019-12664-x
Nakada, D., Matsumoto, K., and Sugimoto, K. (2003). ATM-related Tel1 associates with double-strand breaks through an Xrs2-dependent mechanism. Genes Dev. 17, 1957–1962. doi: 10.1101/gad.1099003
Nugent, C. I., Hughes, T. R., Lue, N. F., and Lundblad, V. (1996). Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science 274, 249–252. doi: 10.1126/science.274.5285.249
Ohya, T., Kawasaki, Y., Hiraga, S. I., Kanbara, S., Nakajo, K., Nakashima, N., et al. (2002). The DNA polymerase domain of polϵ is required for rapid, efficient, and highly accurate chromosomal DNA replication, telomere length maintenance, and normal cell senescence in Saccharomyces cerevisiae. J. Biol. Chem. 277, 28099–28108. doi: 10.1074/jbc.M111573200
Paeschke, K., Capra, J. A., and Zakian, V. A. (2011). DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell 145, 678–691. doi: 10.1016/j.cell.2011.04.015
Paredes, S., Angulo-Ibanez, M., Tasselli, L., Carlson, S. M., Zheng, W., Li, T.-M., et al. (2018). The epigenetic regulator SIRT7 guards against mammalian cellular senescence induced by ribosomal DNA instability. J. Biol. Chem. 293, 11242–11250. doi: 10.1074/jbc.AC118.003325
Park, P. U., Defossez, P. A., and Guarente, L. (1999). Effects of mutations in DNA repair genes on formation of ribosomal DNA circles and life span in Saccharomyces cerevisiae. Mol. Cell. Biol. 19, 3848–3856. doi: 10.1128/MCB.19.5.3848
Pennock, E., Buckley, K., and Lundblad, V. (2001). Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 104, 387–396. doi: 10.1016/S0092-8674(01)00226-4
Perera, R. L., Torella, R., Klinge, S., Kilkenny, M. L., Maman, J. D., and Pellegrini, L. (2013). Mechanism for priming DNA synthesis by yeast DNA polymerase α. eLife 2:e00482. doi: 10.7554/eLife.00482
Petes, T. D. (1979). Yeast ribosomal DNA genes are located on chromosome XII. Proc. Natl. Acad. Sci. U.S.A. 76, 410–414. doi: 10.1073/pnas.76.1.410
Pfeiffer, V., and Lingner, J. (2013). Replication of telomeres and the regulation of telomerase. Cold Spring Harb. Perspect. Biol. 5:a010405. doi: 10.1101/cshperspect.a010405
Ponnusamy, S., Alderson, N. L., Hama, H., Bielawski, J., Jiang, J. C., Bhandari, R., et al. (2008). Regulation of telomere length by fatty acid elongase 3 in yeast. J. Biol. Chem. 283, 27514–27524. doi: 10.1074/jbc.M802980200
Poveda, A. M., Clech, M. L., and Pasero, P. (2010). Transcription and replication. Transcription 1, 99–102. doi: 10.4161/trns.1.2.12665
Pursell, Z. F., Isoz, I., Lundström, E. B., Johansson, E., and Kunkel, T. A. (2007). Yeast DNA polymerase ε participates in leading-strand DNA replication. Science 317, 127–130. doi: 10.1126/science.1144067
Ren, R., Deng, L., Xue, Y., Suzuki, K., Zhang, W., Yu, Y., et al. (2017). Visualization of aging-associated chromatin alterations with an engineered TALE system. Cell Res. 27, 483–504. doi: 10.1038/cr.2017.18
Sabourin, M., Tuzon, C. T., and Zakian, V. A. (2007). Telomerase and Tel1p preferentially associate with short telomeres in S. cerevisiae. Mol. Cell 27, 550–561. doi: 10.1016/j.molcel.2007.07.016
Saka, K., Ide, S., Ganley, A. R. D., and Kobayashi, T. (2013). Cellular senescence in yeast is regulated by rDNA noncoding transcription. Curr. Biol. 23, 1794–1798. doi: 10.1016/j.cub.2013.07.048
Saka, K., Takahashi, A., Sasaki, M., and Kobayashi, T. (2016). More than 10% of yeast genes are related to genome stability and influence cellular senescence via rDNA maintenance. Nucleic Acids Res. 44, 4211–4221. doi: 10.1093/nar/gkw110
Samper, E., Flores, J. M., and Blasco, M. A. (2001). Restoration of telomerase activity rescues chromosomal instability and premature aging in Terc–/– mice with short telomeres. EMBO Rep. 2, 800–807. doi: 10.1093/embo-reports/kve174
Sasaki, M., and Kobayashi, T. (2017). Ctf4 prevents genome rearrangements by suppressing DNA double-strand break formation and its end resection at arrested replication forks. Mol. Cell 66, 533–545. doi: 10.1016/j.molcel.2017.04.020
Schramke, V., Luciano, P., Brevet, V., Guillot, S., Corda, Y., Longhese, M. P., et al. (2004). RPA regulates telomerase action by providing Est1p access to chromosome ends. Nat. Genet. 36, 46–54. doi: 10.1038/ng1284
Shay, J. W., and Wright, W. E. (2005). Senescence and immortalization: role of telomeres and telomerase. Carcinogenesis 26, 867–874. doi: 10.1093/carcin/bgh296
Shi, T., Bunker, R. D., Mattarocci, S., Ribeyre, C., Faty, M., Gut, H., et al. (2013). Rif1 and Rif2 shape telomere function and architecture through multivalent Rap1 interactions. Cell 153, 1340–1353. doi: 10.1016/j.cell.2013.05.007
Signon, L., Malkova, A., Naylor, M. L., Klein, H., and Haber, J. E. (2001). Genetic requirements for RAD51- and RAD54-independent break-induced replication repair of a chromosomal double-strand break. Mol. Cell. Biol. 21, 2048–2056. doi: 10.1128/MCB.21.6.2048-2056.2001
Sinclair, D. A., and Guarente, L. (1997). Extrachromosomal rDNA circles - a cause of aging in yeast. Cell 91, 1033–1042. doi: 10.1016/S0092-8674(00)80493-6
Singer, M. S., and Gottschling, D. E. (1994). TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science 266, 404–409. doi: 10.1126/science.7545955
Soudet, J., Jolivet, P., and Teixeira, M. T. (2014). Elucidation of the DNA end-replication problem in Saccharomyces cerevisiae. Mol. Cell 53, 954–964. doi: 10.1016/j.molcel.2014.02.030
Stewart, S. A., Ben-Porath, I., Carey, V. J., O’Connor, B. F., Hahn, W. C., and Weinberg, R. A. (2003). Erosion of the telomeric single-strand overhang at replicative senescence. Nat. Genet. 33, 492–496. doi: 10.1038/ng1127
Tanaka, H., and Yao, M. C. (2009). Palindromic gene amplification - an evolutionarily conserved role for DNA inverted repeats in the genome. Nat. Rev. Cancer 9, 215–224. doi: 10.1038/nrc2591
Teng, S. C., and Zakian, V. A. (1999). Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 19, 8083–8093. doi: 10.1128/mcb.19.12.8083
Tham, W. H., and Zakian, V. A. (2002). Transcriptional silencing at Saccharomyces telomeres: implications for other organisms. Oncogene 21, 512–521. doi: 10.1038/sj.onc.1205078
Tong, X. J., Li, Q. J., Duan, Y. M., Liu, N. N., Zhang, M. L., and Zhou, J. Q. (2011). Est1 protects telomeres and inhibits subtelomeric Y′-element recombination. Mol. Cell. Biol. 31, 1263–1274. doi: 10.1128/MCB.00831-10
Torres-Rosell, J., Sunjevaric, I., De Piccoli, G., Sacher, M., Eckert-Boulet, N., Reid, R., et al. (2007). The Smc5–Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat. Cell Biol. 9, 923–931. doi: 10.1038/ncb1619
Tran, P. L. T., Mergny, J. L., and Alberti, P. (2011). Stability of telomeric G-quadruplexes. Nucleic Acids Res. 39, 3282–3294. doi: 10.1093/nar/gkq1292
Tseng, S. F., Lin, J. J., and Teng, S. C. (2006). The telomerase-recruitment domain of the telomere binding protein Cdc13 is regulated by Mec1p/Tel1p-dependent phosphorylation. Nucleic Acids Res. 34, 6327–6336. doi: 10.1093/nar/gkl786
Tsukamoto, Y., Taggart, A. K. P., and Zakian, V. A. (2001). The role of the Mre11-Rad50-Xrs2 complex in telomerase- mediated lengthening of Saccharomyces cerevisiae telomeres. Curr. Biol. 11, 1328–1335. doi: 10.1016/S0960-9822(01)00372-4
Virta-Pearlman, V., Morris, D. K., and Lundblad, V. (1996). Est1 has the properties of a single-stranded telomere end-binding protein. Genes Dev. 10, 3094–3104. doi: 10.1101/gad.10.24.3094
Vodenicharov, M. D., Laterreur, N., and Wellinger, R. J. (2010). Telomere capping in non-dividing yeast cells requires Yku and Rap1. EMBO J. 29, 3007–3019. doi: 10.1038/emboj.2010.155
Vodenicharov, M. D., and Wellinger, R. J. (2006). DNA degradation at unprotected telomeres in yeast is regulated by the CDK1 (Cdc28/Clb) cell-cycle kinase. Mol. Cell 24, 127–137. doi: 10.1016/j.molcel.2006.07.035
Vodenicharov, M. D., and Wellinger, R. J. (2007). The cell division cycle puts up with unprotected telomeres: cell cycle regulated telomere uncapping as a means to achieve telomere homeostasis. Cell Cycle 6, 1161–1167. doi: 10.4161/cc.6.10.4224
Wakatsuki, T., Sasaki, M., and Kobayashi, T. (2019). Defects in the NuA4 acetyltransferase complex increase stability of the ribosomal RNA gene and extend replicative lifespan. Genes Genet. Syst. 94, 197–206. doi: 10.1266/ggs.19-00022
Wellinger, R. J., and Zakian, V. A. (2012). Everything you ever wanted to know about saccharomyces cerevisiae telomeres: beginning to end. Genetics 191, 1073–1105. doi: 10.1534/genetics.111.137851
Whittemore, K., Vera, E., Martínez-Nevado, E., Sanpera, C., and Blasco, M. A. (2019). Telomere shortening rate predicts species life span. Proc. Natl. Acad. Sci. U.S.A. 116, 15122–15127. doi: 10.1073/pnas.1902452116
Wotton, D., and Shore, D. (1997). A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev. 11, 748–760. doi: 10.1101/gad.11.6.748
Wu, Z., He, M. H., Zhang, L. L., Liu, J., Zhang, Q. D., and Zhou, J. Q. (2018). Rad6-Bre1 mediated histone H2Bub1 protects uncapped telomeres from exonuclease Exo1 in Saccharomyces cerevisiae. DNA Repair 72, 64–76. doi: 10.1016/j.dnarep.2018.09.007
Wu, Z., Liu, J., Zhang, Q. D., Lv, D. K., Wu, N. F., and Zhou, J. Q. (2017). Rad6–Bre1-mediated H2B ubiquitination regulates telomere replication by promoting telomere-end resection. Nucleic Acids Res. 45, 3308–3322. doi: 10.1093/nar/gkx101
Yang, C. W., Tseng, S. F., Yu, C. J., Chung, C. Y., Chang, C. Y., Pobiega, S., et al. (2017). Telomere shortening triggers a feedback loop to enhance end protection. Nucleic Acids Res. 45, 8314–8328. doi: 10.1093/nar/gkx503
Zhang, M. L., Tong, X. J., Fu, X. H., Zhou, B. O., Wang, J., Liao, X. H., et al. (2010). Yeast telomerase subunit Est1p has guanine quadruplex–promoting activity that is required for telomere elongation. Nat. Struct. Mol. Biol. 17, 202–209. doi: 10.1038/nsmb.1760
Keywords: aging, longevity, rDNA stability, Saccharomyces cerevisiae, senescence, telomere length homeostasis
Citation: Lee JW and Ong EBB (2021) Genomic Instability and Cellular Senescence: Lessons From the Budding Yeast. Front. Cell Dev. Biol. 8:619126. doi: 10.3389/fcell.2020.619126
Received: 19 October 2020; Accepted: 15 December 2020;
Published: 12 January 2021.
Edited by:
Elizabeth Lara Ostler, University of Brighton, United KingdomReviewed by:
Alan Morgan, University of Liverpool, United KingdomOliver Bischof, Institut Pasteur, France
Copyright © 2021 Lee and Ong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Eugene Boon Beng Ong, ZXVnZW5lQHVzbS5teQ==
 Jee Whu Lee
Jee Whu Lee Eugene Boon Beng Ong
Eugene Boon Beng Ong