- 1School of Medicine and Surgery, University of Milano-Bicocca, Milan, Italy
- 2Centro Ricerca Tettamanti, University of Milano-Bicocca, Milan, Italy
- 3Pediatrics, Fondazione MBBM/Ospedale San Gerardo, University of Milano-Bicocca, Milan, Italy
Several lines of evidence suggest that childhood leukemia, the most common cancer in young age, originates during in utero development. However, our knowledge of the cellular origin of this large and heterogeneous group of malignancies is still incomplete. The identification and characterization of their cell of origin is of crucial importance in order to define the processes that initiate and sustain disease progression, to refine faithful animal models and to identify novel therapeutic approaches. During embryogenesis, hematopoiesis takes place at different anatomical sites in sequential waves, and occurs in both a hematopoietic stem cell (HSC)-dependent and a HSC-independent fashion. Despite the recently described relevance and complexity of HSC-independent hematopoiesis, few studies have so far investigated its potential involvement in leukemogenesis. Here, we review the current knowledge on prenatal origin of leukemias in the context of recent insights in developmental hematopoiesis.
Introduction
Leukemia is the most frequent childhood malignancy and it is characterized by a heterogeneous manifestation (Steliarova-Foucher et al., 2017). Despite improved clinical outcome in recent years, the incidence rate is increasing (Howlader et al., 2020). Therefore, there is an urgent need to develop precision medicine strategies for specific targeting of pediatric leukemias.
Even though the cellular origin of the childhood disease remains unclear, several lines of evidence suggest that its origin is distinct from that of the adult counterpart (Bolouri et al., 2018; Malard and Mohty, 2020). The hypothesis of a prenatal origin of pediatric leukemias was initially proposed in the mid-sixties (MacMahon and Levy, 1964) and various evidences are now supporting this view. Although hematopoietic stem cells (HSCs) sustain the production of most blood cells in adults, the ontogeny of vertebrate hematopoiesis is characterized by the presence of HSC-independent hematopoietic cells that originate sequentially during embryo development and in some cases persist to adulthood (Ghosn et al., 2019). These observations raise the hypothesis that HSC-independent progenitors arising at the embryonic level could be subject to genetic hits leading to childhood leukemia.
Herewith, we will focus on the established evidence of the in utero origin of the disease, and discuss about recent advances in the understanding of embryonic hematopoiesis, which is crucial for the identification of the still elusive origin and features of pre-leukemic clones. We will highlight the ontogeny impact on cell transformation focusing on both HSC-independent and HSC-dependent progenitors and report the recent insights into unique embryonic hematopoietic cell populations potentially providing a permissive environment for cell transformation.
Childhood Acute Leukemias
Acute leukemias are characterized by uncontrolled proliferation of undifferentiated cells, called blasts, which impair normal hematopoiesis, in the bone marrow (BM) and peripheral blood (PB), with secondary infiltration of other tissues. According to morphology and cytochemistry, they are classified into acute myeloid leukemia (AML) and acute lymphoblastic leukemia (ALL). Less frequently, they can show an intermediate phenotype, with features of both diseases, and are defined as mixed lineage leukemias. During childhood, lymphoid phenotypes are predominant over myeloid ones, with a highest age-specific incidence between 2 and 5 years. On the contrary, the incidence of childhood AML is highest in patients younger than 1 year (infants) (Howlader et al., 2020). 5-year survival rate has increased over time and it currently reaches 90% and over 60% for childhood ALL and AML, respectively (Gatta et al., 2014; Malard and Mohty, 2020). Despite presenting with unique clinical and biological features, the outcome of infant AML patients is similar to that of older children (Masetti et al., 2015), whereas infants with ALL tend to manifest a more aggressive course of the disease with an event-free survival lower than 50% (Pieters et al., 2007).
Cytogenetic and molecular lesions diverge in infant, childhood, and adult acute leukemias in terms of type and recurrence. Chromosomal aberrations and especially translocations involving the gene KMT2A (MLL) are the most common genetic lesions in both AML and ALL infant leukemias, but are less frequent in children and adults (Malard and Mohty, 2020; Rice and Roy, 2020). Other frequent translocations in young children (<3 years) with AML involve CBFA2T3 and MNX1, while RUNX1, CBFB, and RARA peak in older children (Bolouri et al., 2018). In infants with ALL, ETV6-RUNX1 (TEL-AML1), and TCF3-PBX1 rearrangements are prevalent, as well as a high-hyperdiploid karyotype (Hein et al., 2020; Malard and Mohty, 2020). Conversely, the BCR-ABL translocation is more frequent in adults with ALL (Malard and Mohty, 2020). Focal mutations in the N/KRAS, KIT, and CBL genes are more frequent in children than adults with AML, whereas IDH1, IDH2, RUNX1, NPM1, DNMT3A, and TP53 mutations are almost exclusively found in the latter category (Papaemmanuil et al., 2016; Bolouri et al., 2018). On a similar line, the genomic landscapes of adult and childhood ALL differ (Liu et al., 2016; Gröbner et al., 2018).
Together, these observations led to speculate that childhood and adult leukemias are biologically distinct and might diverge not only in their molecular landscape but also in their cellular origin. Nevertheless, the underlying mechanisms for these differences and the precise entity of the cell(s) of origin of childhood acute leukemia are still unknown.
Evidences for a Prenatal Origin of Childhood Leukemias
The hypothesis of a prenatal origin of childhood leukemias derived from studies on pairs of monozygotic monochorionic twins with both members affected by the disease, with the first case reported in 1882 (Greaves et al., 2003). A proposed explanation of leukemia concordance (shared disease features) in twins is that preleukemic cells arising in one twin fetus can diffuse via vascular anastomosis of a monochorionic placenta to the other twin (Clarkson and Boyse, 1971). The key piece of evidence which allowed to conclusively demonstrate prenatal initiation of leukemia was provided by the identification of unique clonal markers of leukemic cells, such as chromosome translocations, which can facilitate tracking of preleukemic clones. Interestingly, chromosome breakpoints always occur in a unique intronic region of the genes involved in the rearrangements and differ from patient to patient, although the fusion proteins generated are functionally equivalent. The evaluation of breakpoints in twins with concordant leukemia allowed to demonstrate that these children share the same breakpoints and consequently leukemia originated prenatally (Greaves, 1999). Preleukemic clones generated in utero have been described to evolve to overt leukemia either few days after birth or as long as 14 years later (Ford et al., 1993; Wiemels et al., 1999; Maia et al., 2004).
Rearrangements that involve the histone lysine methyltransferase 2A, MLL, at chromosome 11q23, are recurrent events in childhood leukemia, with highest incidence in infants (Wiemels et al., 2002). These rearrangements occur in approximately 50 and 70–80% of infant AML and ALL, respectively, and their frequency decreases with age (Hilden et al., 2006; Pieters et al., 2007; Harrison et al., 2010). Up to 135 MLL partner genes have been identified so far, among which AFF1 (AF4), MLLT3 (AF9), MLLT10 (AF10), MLLT1 (ENL), and ELL are the most prevalent (Meyer et al., 2018). Especially in infant ALL, and in approximately 60% of AML pediatric cases, MLL rearrangements seem sufficient to induce leukemic transformation on their own, since they usually not co-occur with additional mutations (Balgobind et al., 2011; Andersson et al., 2015). Retrospective analysis of blood spots taken at birth (Guthrie cards), which allow for the detection of around 1–20 leukemic cells in a single spot, have shown the acquisition of MLL translocations in utero (Gale et al., 1997), as already suggested by concordance studies on twins (Ford et al., 1993). Furthermore, the in utero appearance of cytogenetic lesions typical of leukemia has been supported by the detection of a MLL-fusion gene in fetal tissue and BM from abortions (Uckun et al., 1998). Another frequent gene fusion detected in pediatric leukemia involves the genes ETV6, on chromosome 12, and RUNX1, on chromosome 21. ETV6-RUNX1 occurs in approximately 25% of B lineage pediatric ALL (Bernard et al., 1996). Monozygotic twins, which both developed ALL before their fifth birthday, have been described to share the same ETV6-RUNX1 sequence (Ford et al., 1998). Additionally, after the evaluation of Guthrie cards of newly diagnosed ALL patients with ETV6-RUNX1 fusion, both twins and singletons of 2–5 years, have been revealed to share unique or clonotypic sequence of the translocation (Wiemels et al., 1999). The AML1-ETO translocation (RUNX1-RUNX1T1) derives from the rearrangement of chromosomes 8 and 21 and has been shown to be the most common rearrangement in both children and adults with AML, suggesting its appearance as a postnatal event. Nevertheless, the prenatal occurrence of the translocation is supported by its detection in blood spots collected at birth, which were still available at the time of AML diagnosis (Wiemels et al., 2002). Altogether, the early onset of the disease, the high concordance rate between twin pairs (5–50% within the range of 0–15 years and less than 1% for adults) (MacMahon and Levy, 1964; Buekley et al., 1996; Greaves et al., 2003; Greaves, 2005), and the presence of the first mutation hit already at birth strongly suggest a prenatal origin of leukemia. More recent studies showed that bone marrow mesenchymal stem cells (MSC) derived from infants with acute leukemia harboring ETV6-RUNX1, E2A-PBX1, and MLL-rearrangements express the fusion genes, suggesting that rearrangements can also occur in early embryonic progenitors before hemogenic specification (Menendez et al., 2009; Shalapour et al., 2010). These and other evidences suggest that the first mutation event can take place at different levels along prenatal hematopoietic development.
Insights Into Embryonic Hematopoiesis
A fine understanding of hematopoietic ontogeny is critical to determine the processes that initiate and sustain the progression of hematopoietic disorders. Increasing knowledge in the field of developmental hematopoiesis unraveled a complex organization of the hematopoietic system during gestation, and showed that embryonic/fetal and adult hematopoiesis differ in many aspects. In this regard, mouse, zebrafish, and chicken models have provided essential information on the dynamics of emergence of hematopoietic stem and progenitor cells in vertebrates (Dzierzak and Bigas, 2018). In adults, HSCs are located in the bone marrow niche and reside at the top of the hematopoietic hierarchy. In embryos, instead, several tissues harbor hematopoietic activity and HSCs appearance is preceded by the emergence of other progenitors endowed with various potency.
In vertebrates, embryonic hematopoietic development occurs in sequential waves (Figure 1). In the mouse, at embryonic day (E) 7.25 the extra-embryonic yolk sac (YS) represents the site of origin of the first hematopoietic wave which gives rise to primitive nucleated erythroid cells, macrophages (which do not transit through a monocyte intermediate), and megakaryocytes (Palis et al., 1999, 2001; Tober et al., 2007). A second wave originates from E8.25 in the YS and possibly other sites. Several progenitors with various potential are generated in this wave, including erythromyeloid progenitors (EMPs), lymphoid primed multipotent progenitors (LMPPs), and progenitors with multi-lineage mesodermal potential (Azzoni et al., 2014; Palis, 2016; Ghosn et al., 2019). All of these cells arise from hemogenic endothelium through a endothelial-to-hematopoietic transition (EHT) (Swiers et al., 2016; Ottersbach, 2019). EMPs differ in their surface marker profile from primitive hematopoietic cells and early definitive progenitors with HSC potential (Table 1). They possess myeloid potential and sustain erythropoiesis, megakaryocytes, and myeloid cell production, including macrophages and neutrophils (McGrath et al., 2015a). As more recently shown, they also have some lymphoid potential as they give rise to cytotoxic natural killer cells (NK) (Dege et al., 2020). Between E10.5 and E11.5, EMPs seed the fetal liver (FL) (Gomez Perdiguero et al., 2015; McGrath et al., 2015a). Similar to HSCs, EMPs are regulated by the c-Kit signaling pathway (Azzoni et al., 2018). Although EMPs were initially considered transient, it is now well accepted that they constitute a source of tissue-resident macrophages which persist and self-renew throughout adulthood, independently of HSCs (Schulz et al., 2012; Epelman et al., 2014; Gomez Perdiguero et al., 2015; Mass et al., 2016). Later than EMPs, but still before HSCs and FL hematopoiesis, LMPPs arise in the YS at around E9.5 (Figure 1 and Table 1). In the developing embryo they contribute to lymphopoiesis and myelopoiesis, and are devoid of the potential to generate erythrocytes, basophils, eosinophils, and tissue resident macrophages (Böiers et al., 2013). The HSC-independent wave is required for embryo survival (Chen M. J. et al., 2011). Given this relevance, it is possible that HSC-independent progenitors are subject to genetic hits leading to childhood leukemia.
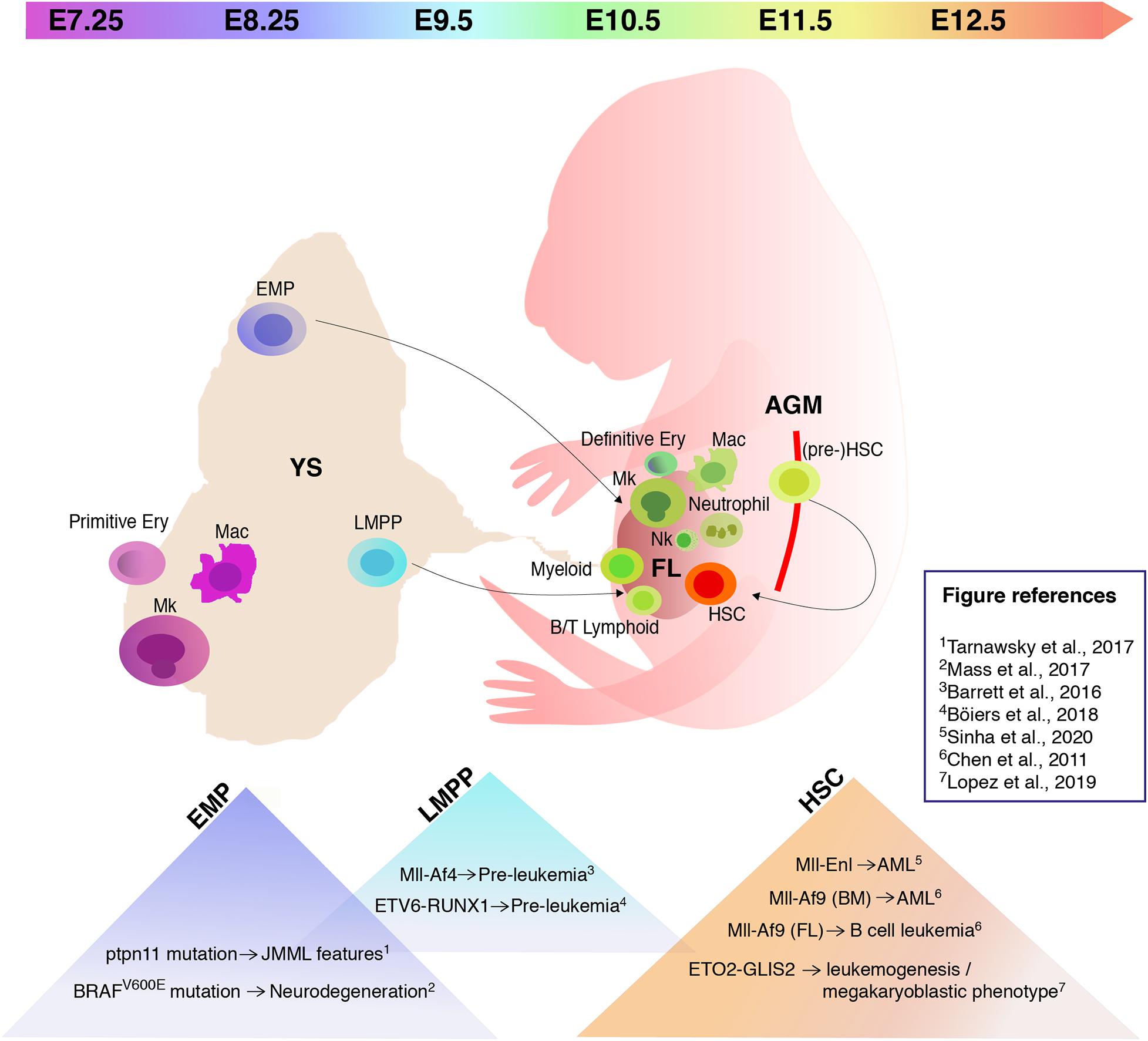
Figure 1. Hematopoiesis in the mouse embryo and embryonic hematopoietic cell populations that provide a permissive environment for cell transformation. Schematic of a mouse embryo showing sites of hematopoiesis (yolk sac/YS), fetal liver/FL, aorta-gonad-mesonephros/AGM) and timeline of appearance of hematopoietic stem/progenitor cells. The triangles in the lower part of the image list some recently studied mutations and translocations, and their effect following expression in specific embryonic hematopoietic cell populations. Mk, megakaryocytes; Mac, macrophages; EMP, erythromyeloid progenitors; LMPP, lymphoid primed multipotent progenitors; Ery, erythrocytes; Nk, natural killer; HSC, hematopoietic stem cells.
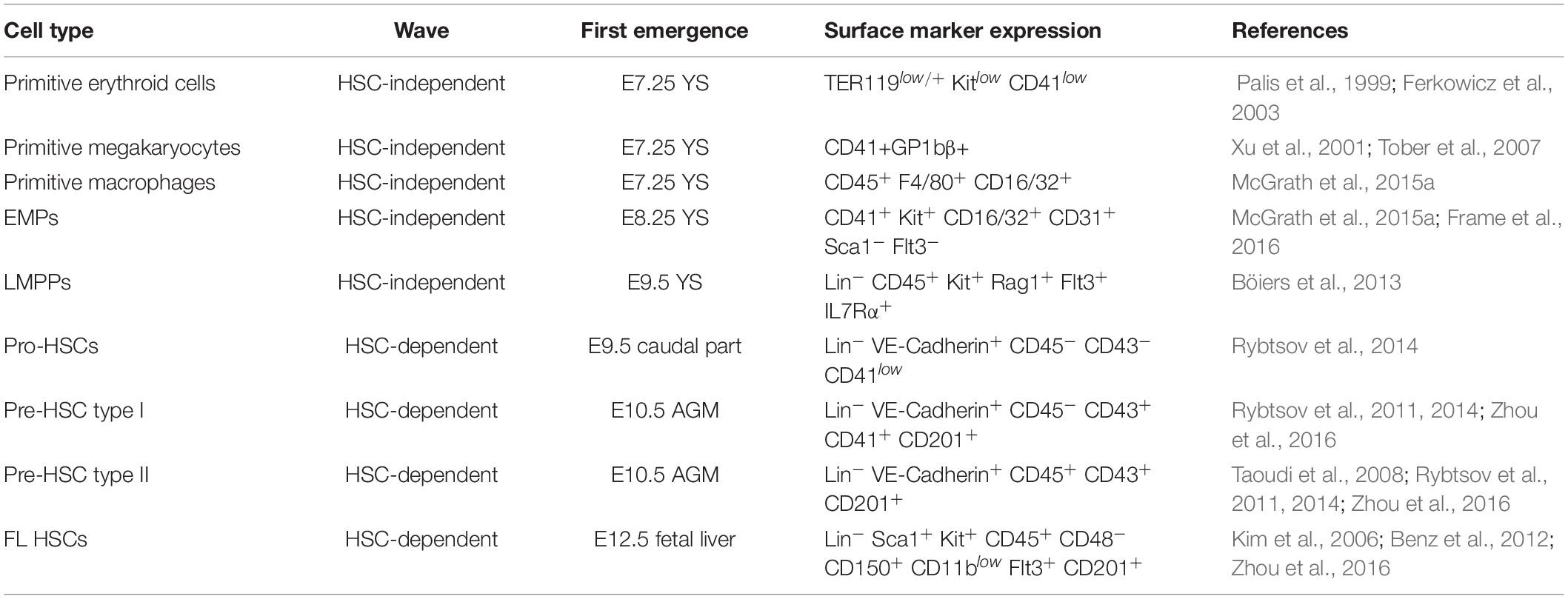
Table 1. List of the main hematopoietic stem and progenitor cell populations in the early mouse embryo, their time and site of first emergence and surface marker expression profile.
HSCs can be detected starting from E10.5 (Figure 1 and Table 1) in the aorta-gonad-mesonephros (AGM) region, budding from the ventral endothelium of the dorsal aorta through EHT (Müller et al., 1994; Medvinsky and Dzierzak, 1996; de Bruijn et al., 2000, 2002; Bertrand et al., 2010; Boisset et al., 2010; Kissa and Herbomel, 2010). Vitelline/umbilical arteries, embryonic head, and placenta represent additional embryonic vascular sites in which HSC activity takes place (de Bruijn et al., 2000; Ottersbach and Dzierzak, 2005; Robin et al., 2010; Ivanovs et al., 2011; Li et al., 2012). Until E12.5, very few transplantable HSCs arise in the embryo, although pro/pre-HSCs are already present from E9.5 (Rybtsov et al., 2016; Table 1). HSC maturation and expansion takes place in the FL from E12; subsequently, HSCs colonize the bone marrow where they reside throughout adult life (Kumaravelu et al., 2002; Rybtsov et al., 2016). Although it is well acknowledged that mammalian hematopoiesis is highly conserved and shares many similarities with the mouse (Enzan, 1964; Tavian et al., 2001; Ivanovs et al., 2011; Julien et al., 2016; Bian et al., 2020), the mechanisms that describe human hematopoiesis are less clear and need to be further investigated (Easterbrook et al., 2019).
Models for the Study of the Origin of Childhood Leukemias
A comprehensive understanding of the cellular origins of childhood leukemia is fundamental for the establishment of faithful animal models. However, despite intensive investigation of the intrinsic and extrinsic factors regulating HSPC biology and their relationship to leukemogenesis, this has not been achieved so far. Several studies showed that gene fusions recurrent in pediatric leukemias can lead to divergent outcomes in terms of disease aggressiveness, latency, phenotype, and transcriptional features, according to their time of appearance during ontogeny (Chen W. et al., 2011; Horton et al., 2013; Man et al., 2016; Chaudhury et al., 2018; Lopez et al., 2019; Sinha et al., 2020). Differences in lineage specification and disease latency have been clearly shown after the induction of Mll-Af9 in FL and adult BM HSCs. In these two models the translocation gave rise to B cell leukemia with a prolonged latency and AML, respectively (Chen W. et al., 2011). Similarly, transduction of MLL-AF9 and MLL-AF4 in human neonatal cord blood (CB) HSPCs mainly resulted in ALL (Horton et al., 2013; Lin et al., 2016). In contrast, MLL-AF9 transduced human adult BM HSPCs gave rise to non-leukemic myeloid-biased engraftment and MLL-Af4 transduced mouse adult BM cells led to AML (Horton et al., 2013; Lin et al., 2016). Consistent with these findings it has been demonstrated that B-cell committed progenitors harbor transforming potential in ALL, (Castor et al., 2005; Kong et al., 2008), but there are also evidences hinting at a cell of origin at an earlier developmental stage (le Viseur et al., 2008). The inducible expression of the recurrent gene fusion product ETO2-GLIS2, associated with acute megakaryoblastic leukemia, triggered leukemogenesis in both FL HSPCs and adult BM HSPCs, but gave rise to a megakaryoblastic phenotype with a shorter latency and caused more evident transcriptional changes only in the former (Lopez et al., 2019). The induction of the Mll-Enl translocation in FL HSPCs at E12.5 led to an overt, more aggressive AML form than the one triggered in adults, and to a transplantable disease in secondary recipients (Sinha et al., 2020). Overall, these studies suggested that childhood leukemias originating from the FL possess unique features that differentiate them from the ones resulting from the same genetic lesions occurring in the adult. However, they did not precisely address the cellular origin of the disease, as the FL is not a site of de novo hematopoietic generation. Cells seeding the FL during embryogenesis can have multiple origins, most of which are HSC-independent, and could already carry genetic lesions at the time of FL seeding. Besides the cell subtype in which the genetic hit takes place, the developmental stage of the hematopoietic niche also elicits an important contribution in regulating leukemic lineage commitment (Chaudhury et al., 2018; Rowe et al., 2019). Indeed, it has been recently shown how a MLL-rearrangement can differentially give rise to either a mixed lineage or a myeloid leukemia according to the developmental age of the microenvironment in a setting where the cell of origin is the same (Rowe et al., 2019). In addition to the fact that the susceptibility for transformation and the resulting phenotype change during ontogeny, differences in human and mouse embryonic hematopoietic development could also affect the faithful modeling of the human pediatric leukemia.
An HSC-Independent Origin of Pediatric Leukemia?
The potential link between HSC-independent hematopoiesis and leukemogenesis remains so far largely unexplored. Only few studies have investigated which of the unique embryonic hematopoietic cell populations provide a permissive environment for cell transformation. As EMPs sustain fetal myelopoiesis and their progeny persists in the adult (Gomez Perdiguero et al., 2015; McGrath et al., 2015b), it has been recently suggested that EMPs may represent cells of origin of diseases associated with fetal development. Furthermore, in the context of the Mll-Af9 translocation, it has been shown how the proliferation rate is a limiting factor for malignant transformation (Chen et al., 2019). Thus, the rapid proliferation of EMPs (McGrath et al., 2015a) suggests that these cells may be susceptible to oncogenic transformation. A proof of principle of the notion that EMPs could be cells of origin for post-natal diseases derives from a recent study that investigated whether the neurodegeneration observed in patients with histiocytosis could be caused by somatic mutations in the EMP lineage. In particular, the mosaic expression of the BRAFV600E mutation in EMPs at E8.5 was shown to cause expansion of microglia and neurodegeneration in adult mice (Mass et al., 2017). The involvement of HSC-independent hematopoietic progenitors has been assessed in the context of juvenile myelomonocytic leukemia (JMML). EMPs looked like good candidates, given the high relapse rates after HSCs transplantation (Locatelli et al., 2013) and the plausible in utero initiation of the disease (Matsuda et al., 2010). For this purpose, the ptpn11 gain-of-function JMML-initiating mutation had been introduced into EMPs. Although mice demonstrated features of JMML and mutant EMPs engrafted spleens of neonatal recipients, the disease was not transplantable (Tarnawsky et al., 2017).
Even though no studies have so far evaluated the susceptibility of EMPs to acute leukemias-specific genetic hits, some have specifically targeted acute leukemia-related translocations to LMPPs. LMPPs have recently emerged as potential cells of origin of B cell acute lymphoblastic leukemia (B-ALL) (Barrett et al., 2016; Böiers et al., 2018). Conditional activation of the Mll-AF4 translocation in murine embryonic hematopoietic cells before the predominance of HSC-dependent hematopoiesis (E12.5–E14.5) resulted in a pre-leukemic phenotype. Even if mice showed B-cell lymphomas after a long latency, the model was unable to fully recapitulate the disease seen in patients harboring the same genetic alteration (Barrett et al., 2016). A possible counterpart for mouse LMPPs has also been identified in human FL as a IL-7R+ progenitor which maintains both myeloid and lymphoid potential. Evidence of the susceptibility of human fetal B cells progenitors to dysregulation by ETV6-RUNX1 was provided by the introduction of the translocation in human induced pluripotent stem cells (iPSC). This led to a pre-leukemic initiation with expansion of the CD19– IL-7R+ population, suggesting IL-7R progenitors as candidate cells of origin for ETV6-RUNX1 preleukemia (Böiers et al., 2018). Moreover, the introduction of Runx1 and Ezh2 mutations in early thymic progenitors (ETPs), which are closely related to LMPPs, could model ETP leukemia features in mice and lead to acute lympho-myeloid leukemia progression upon introduction of Flt3-ITD (Booth et al., 2018). To summarize, recent work has shown that (i) HSC-independent progenitor cells can be subject to genetic hits occurring prenatally, which can lead at least to a pre-leukemic state; (ii) HSC-independent progenitors are susceptible to pre-leukemic initiation during a limited time frame, and (iii) multiple genetic hits can be required before an overt manifestation of the disease.
Conclusion and Future Perspectives
Prenatal leukemic development in humans is a multifactorial process. Because of the complexity of the events leading to childhood acute leukemia and the difficulty of studying in utero stages, the identification and characterization of the cell(s) of origin is still a challenge. Although recent studies shed light on the potential of HSC-independent hematopoietic progenitor cells to act as the cell of origin for pediatric leukemia, there is urgent need to further investigate this aspect and in particular how leukemia-associated genetic hits may impact early stages of disease development in utero. This knowledge would be critical to better understand the etiology and pathogenesis of the disease, which would enable the refinement of animal models, the identification of new therapeutic approaches and to define preventive measures.
Author Contributions
AC and EA conceived the topic, reviewed the literature, made the figure, and wrote the manuscript. GC, SB, AB, and RM revised and edited the manuscript. All authors read and approved the final version of the manuscript.
Funding
The authors would like to acknowledge Fondazione Cariplo “Biomedical Research conducted by young researchers” grant agreement n. 2018-0102 to EA and Italian Association for Cancer Research (IG2018-21999) to GC.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Andersson, A. K., Ma, J., Wang, J., Chen, X., Gedman, A. L., Dang, J., et al. (2015). The landscape of somatic mutations in infant MLL-rearranged acute lymphoblastic leukemias. Nat. Genet. 47, 330–337. doi: 10.1038/ng.3230
Azzoni, E., Conti, V., Campana, L., Dellavalle, A., Adams, R. H., Cossu, G., et al. (2014). Hemogenic endothelium generates mesoangioblasts that contribute to several mesodermal lineages in vivo. Development 141, 1821–1834. doi: 10.1242/dev.103242
Azzoni, E., Frontera, V., McGrath, K. E., Harman, J., Carrelha, J., Nerlov, C., et al. (2018). Kit ligand has a critical role in mouse yolk sac and aorta–gonad–mesonephros hematopoiesis. EMBO Rep. 19:e45477. doi: 10.15252/embr.201745477
Balgobind, B. V., Zwaan, C. M., Pieters, R., and Van Den Heuvel-Eibrink, M. M. (2011). The heterogeneity of pediatric MLL-rearranged acute myeloid leukemia. Leukemia 25, 1239–1248. doi: 10.1038/leu.2011.90
Barrett, N. A., Malouf, C., Kapeni, C., Bacon, W. A., Giotopoulos, G., Jacobsen, S. E. W., et al. (2016). Mll-AF4 confers enhanced self-renewal and lymphoid potential during a restricted window in development. Cell Rep. 16, 1039–1054. doi: 10.1016/j.celrep.2016.06.046
Benz, C., Copley, M. R., Kent, D. G., Wohrer, S., Cortes, A., Aghaeepour, N., et al. (2012). Hematopoietic stem cell subtypes expand differentially during development and display distinct lymphopoietic programs. Cell Stem Cell 10, 273–283. doi: 10.1016/j.stem.2012.02.007
Bernard, O. A., Romana, S. P., Poirel, H., and Berger, R. (1996). Molecular cytogenetics of t(12;21)(p13;q22). Leuk. Lymphoma 23, 459–465. doi: 10.3109/10428199609054854
Bertrand, J. Y., Chi, N. C., Santoso, B., Teng, S., Stainier, D. Y. R., and Traver, D. (2010). Haematopoietic stem cells derive directly from aortic endothelium during development. Nature 464, 108–111. doi: 10.1038/nature08738
Bian, Z., Gong, Y., Huang, T., Lee, C. Z. W., Bian, L., Bai, Z., et al. (2020). Deciphering human macrophage development at single-cell resolution. Nature 582, 571–576. doi: 10.1038/s41586-020-2316-7
Böiers, C., Carrelha, J., Lutteropp, M., Luc, S., Green, J. C. A., Azzoni, E., et al. (2013). Lymphomyeloid contribution of an immune-restricted progenitor emerging prior to definitive hematopoietic stem cells. Cell Stem Cell 13, 535–548. doi: 10.1016/j.stem.2013.08.012
Böiers, C., Richardson, S. E., Laycock, E., Zriwil, A., Turati, V. A., Brown, J., et al. (2018). A Human IPS model implicates embryonic B-myeloid fate restriction as developmental susceptibility to B acute lymphoblastic leukemia-associated ETV6-RUNX1. Dev. Cell 44, 362. doi: 10.1016/j.devcel.2017.12.005
Boisset, J. C., Van Cappellen, W., Andrieu-Soler, C., Galjart, N., Dzierzak, E., and Robin, C. (2010). In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature 464, 116–120. doi: 10.1038/nature08764
Bolouri, H., Farrar, J. E., Triche, T., Ries, R. E., Lim, E. L., Alonzo, T. A., et al. (2018). The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions. Nat. Med. 24, 103–112. doi: 10.1038/nm.4439
Booth, C. A. G., Barkas, N., Neo, W. H., Boukarabila, H., Soilleux, E. J., Giotopoulos, G., et al. (2018). Ezh2 and Runx1 mutations collaborate to initiate Lympho-myeloid leukemia in early Thymic progenitors. Cancer Cell 33, 274–291.e8. doi: 10.1016/j.ccell.2018.01.006
Buekley, J. D., Buckley, C. M., Breslow, N. E., Draper, G. J., Roberson, P. K., and Mack, T. M. (1996). Concordance for childhood cancer in twins. Med. Pediatr. Oncol. 26, 223–229. doi: 10.1002/(sici)1096-911x(199604)26:4<223::aid-mpo1>3.0.co;2-l
Castor, A., Nilsson, L., Åstrand-Grundström, I., Buitenhuis, M., Ramirez, C., Anderson, K., et al. (2005). Distinct patterns of hematopoietic stem cell involvement in acute lymphoblastic leukemia. Nat. Med. 11, 630–637. doi: 10.1038/nm1253
Chaudhury, S., O’Connor, C., Cañete, A., Bittencourt-Silvestre, J., Sarrou, E., Prendergast, Á., et al. (2018). Age-specific biological and molecular profiling distinguishes paediatric from adult acute myeloid leukaemias. Nat. Commun. 9:5280. doi: 10.1038/s41467-018-07584-1
Chen, M. J., Li, Y., De Obaldia, M. E., Yang, Q., Yzaguirre, A. D., Yamada-Inagawa, T., et al. (2011). Erythroid/myeloid progenitors and hematopoietic stem cells originate from distinct populations of endothelial cells. Cell Stem Cell 9, 541–552. doi: 10.1016/j.stem.2011.10.003
Chen, W., Gerard, O., Sullivan, M., Hudson, W., and Kersey, J. (2011). Modeling human infant MLL leukemia in mice: leukemia from fetal liver differs from that originating in postnatal marrow. Blood 117, 3474–3475. doi: 10.1182/blood-2010-11-317529
Chen, X., Burkhardt, D. B., Hartman, A. A., Hu, X., Eastman, A. E., Sun, C., et al. (2019). MLL-AF9 initiates transformation from fast-proliferating myeloid progenitors. Nat. Commun. 10:5767. doi: 10.1038/s41467-019-13666-5
Clarkson, B. D., and Boyse, E. (1971). Possible explanation of the high concordance for acute leukaemia in monozygotic twins. Lancet 1, 699–701. doi: 10.1016/s0140-6736(71)92705-x
de Bruijn, M., Speck, N. A., Peeters, M. C. E., and Dzierzak, E. (2000). Definitive hematopoietic stem cells first develop within the major arterial regions of the mouse embryo. EMBO J. 19, 2465–2474. doi: 10.1093/emboj/19.11.2465
de Bruijn, M. F. T. R., Ma, X., Robin, C., Ottersbach, K., Sanchez, M. J., and Dzierzak, E. (2002). Hematopoietic stem cells localize to the endothelial cell layer in the midgestation mouse aorta. Immunity 16, 673–683. doi: 10.1016/S1074-7613(02)00313-8
Dege, C., Fegan, K. H., Creamer, J. P., Berrien-Elliott, M. M., Luff, S. A., Kim, D., et al. (2020). Potently cytotoxic natural killer cells initially emerge from Erythro-myeloid progenitors during mammalian development. Dev. Cell 53, 229–239.e7. doi: 10.1016/j.devcel.2020.02.016.
Dzierzak, E., and Bigas, A. (2018). Blood development: hematopoietic stem cell dependence and independence. Cell Stem Cell 22, 639–651. doi: 10.1016/j.stem.2018.04.015
Easterbrook, J., Rybtsov, S., Gordon-Keylock, S., Ivanovs, A., Taoudi, S., Anderson, R. A., et al. (2019). Analysis of the spatiotemporal development of hematopoietic stem and progenitor cells in the early human embryo. Stem Cell Rep. 12, 1056–1068. doi: 10.1016/j.stemcr.2019.03.003
Enzan, H. (1964). Electron microscopic studies of macrophages in early human yolk sacs. Acta Pathol. Jpn. 36, 49–64. doi: 10.1002/bip.1964.360020311
Epelman, S., Lavine, K. J., Beaudin, A. E., Sojka, D. K., Carrero, J. A., Calderon, B., et al. (2014). Embryonic and adult-derived resident cardiac macrophages are maintained through distinct mechanisms at steady state and during inflammation. Immunity 40, 91–104. doi: 10.1016/j.immuni.2013.11.019
Ferkowicz, M. J., Starr, M., Xie, X., Li, W., Johnson, S. A., Shelley, W. C., et al. (2003). CD41 expression defines the onset of primitive and definitive hematopoiesis in the murine embryo. Development 130, 4393–4403. doi: 10.1242/dev.00632
Ford, A. M., Bennett, C. A., Price, C. M., Bruin, M. C. A., Van Wering, E. R., and Greaves, M. (1998). Fetal origins of the TEL-AML1 fusion gene in identical twins with leukemia. Proc. Natl. Acad. Sci. U.S.A. 95, 4584–4588. doi: 10.1073/pnas.95.8.4584
Ford, A. M., Ridge, S. A., Cabrera, M. E., Mahmoud, H., Steel, C. M., Chan, L. C., et al. (1993). In utero rearrangements in the trithorax-related oncogene in infant leukaemias. Nature 363, 358–360. doi: 10.1038/363358a0
Frame, J. M., Fegan, K. H., Conway, S. J., McGrath, K. E., and Palis, J. (2016). Definitive hematopoiesis in the yolk sac emerges from Wnt-responsive hemogenic endothelium independently of circulation and arterial identity. Stem Cells 34, 431–444. doi: 10.1002/stem.2213
Gale, K. B., Ford, A. M., Repp, R., Borkhardt, A., Keller, C., Eden, O. B., et al. (1997). Backtracking leukemia to birth: identification of clonotypic gene fusion sequences in neonatal blood spots. Proc. Natl. Acad. Sci. U.S.A. 94, 13950–13954. doi: 10.1073/pnas.94.25.13950
Gatta, G., Botta, L., Rossi, S., Aareleid, T., Bielska-Lasota, M., Clavel, J., et al. (2014). Childhood cancer survival in Europe 1999-2007: results of EUROCARE-5-a population-based study. Lancet Oncol. 15, 35–47. doi: 10.1016/S1470-2045(13)70548-5
Ghosn, E., Yoshimoto, M., Nakauchi, H., Weissman, I. L., and Herzenberg, L. A. (2019). Hematopoietic stem cell-independent hematopoiesis and the origins of innate-like B lymphocytes. Development 146:dev170571. doi: 10.1242/dev.170571
Gomez Perdiguero, E., Klapproth, K., Schulz, C., Busch, K., Azzoni, E., Crozet, L., et al. (2015). Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature 518, 547–551. doi: 10.1038/nature13989
Greaves, M. (1999). Molecular genetics, natural history and the demise of childhood leukaemia. Eur. J. Cancer 35, 1941–1953. doi: 10.1016/S0959-8049(99)00296-8
Greaves, M. (2005). In utero origins of childhood leukaemia. Early Hum. Dev. 81, 123–129. doi: 10.1016/j.earlhumdev.2004.10.004
Greaves, M. F., Maia, A. T., Wiemels, J. L., and Ford, A. M. (2003). Leukemia in twins : lessons in natural history. Blood 102, 2321–2333. doi: 10.1182/blood-2002-12-3817
Gröbner, S. N., Worst, B. C., Weischenfeldt, J., Buchhalter, I., Kleinheinz, K., Rudneva, V. A., et al. (2018). The landscape of genomic alterations across childhood cancers. Nature 555, 321–327. doi: 10.1038/nature25480
Harrison, C. J., Hills, R. K., Moorman, A. V., Grimwade, D. J., Hann, I., Webb, D. K. H., et al. (2010). Cytogenetics of childhood acute myeloid leukemia: United Kingdom Medical Research Council Treatment Trials AML 10 and 12. J. Clin. Oncol. 28, 2674–2681. doi: 10.1200/JCO.2009.24.8997
Hein, D., Borkhardt, A., and Fischer, U. (2020). Insights into the prenatal origin of childhood acute lymphoblastic leukemia. Cancer Metastasis Rev. 39, 161–171. doi: 10.1007/s10555-019-09841-1
Hilden, J. M., Dinndorf, P. A., Meerbaum, S. O., Sather, H., Villaluna, D., Heerema, N. A., et al. (2006). Analysis of prognostic factors of acute lymphoblastic leukemia in infants: report on CCG 1953 from the Children’s Oncology Group. Blood 108, 441–451. doi: 10.1182/blood-2005-07-3011
Horton, S. J., Jaques, J., Woolthuis, C., Van Dijk, J., Mesuraca, M., Huls, G., et al. (2013). MLL-AF9-mediated immortalization of human hematopoietic cells along different lineages changes during ontogeny. Leukemia 27, 1116–1126. doi: 10.1038/leu.2012.343
Howlader, N., Noone, A., Krapcho, M., Miller, D., Brest, A., Yu, M., et al. (2020). SEER Cancer Statistics Review, 1975-2017. Bethesda, MD: National Cancer Institute.
Ivanovs, A., Rybtsov, S., Welch, L., Anderson, R. A., Turner, M. L., and Medvinsky, A. (2011). Highly potent human hematopoietic stem cells first emerge in the intraembryonic aorta-gonad-mesonephros region. J. Exp. Med. 208, 2417–2427. doi: 10.1084/jem.20111688
Julien, E., El Omar, R., and Tavian, M. (2016). Origin of the hematopoietic system in the human embryo. FEBS Lett. 590, 3987–4001. doi: 10.1002/1873-3468.12389
Kim, I., He, S., Yilmaz, H., Kiel, M. J., and Morrison, S. J. (2006). Enhanced purification of fetal liver hematopoietic stem cells using SLAM family receptors. Blood 108, 737–744. doi: 10.1182/blood-2005-10-4135
Kissa, K., and Herbomel, P. (2010). Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature 464, 112–115. doi: 10.1038/nature08761
Kong, Y., Yoshida, S., Saito, Y., Doi, T., Nagatoshi, Y., Fukata, M., et al. (2008). CD34+CD38+CD19+ as well as CD34+CD38-CD19+ cells are leukemia-initiating cells with self-renewal capacity in human B-precursor ALL. Leukemia 22, 1207–1213. doi: 10.1038/leu.2008.83
Kumaravelu, P., Hook, L., Morrison, A. M., Ure, J., Zhao, S., Zuyev, S., et al. (2002). Quantitative developmental anatomy of definitive haematopoietic stem cells/long-term repopulating units (HSC/RUs): role of the aorta-gonad-mesonephros (AGM) region and the yolk sac in colonisation of the mouse embryonic liver. Development 129, 4891–4899.
le Viseur, C., Hotfilder, M., Bomken, S., Wilson, K., Röttgers, S., Schrauder, A., et al. (2008). In childhood acute lymphoblastic leukemia, blasts at different stages of immunophenotypic maturation have stem cell properties. Cancer Cell 14, 47–58. doi: 10.1016/j.ccr.2008.05.015.In
Li, Z., Lan, Y., He, W., Chen, D., Wang, J., Zhou, F., et al. (2012). Mouse embryonic head as a site for hematopoietic stem cell development. Cell Stem Cell 11, 663–675. doi: 10.1016/j.stem.2012.07.004
Lin, S., Luo, R. T., Ptasinska, A., Kerry, J., Assi, S. A., Wunderlich, M., et al. (2016). Instructive role of MLL-fusion proteins revealed by a model of t(4;11) Pro-B Acute Lymphoblastic Leukemia. Cancer Cell 30, 737–749. doi: 10.1016/j.ccell.2016.10.008
Liu, Y. F., Wang, B. Y., Zhang, W. N., Huang, J. Y., Li, B. S., Zhang, M., et al. (2016). Genomic profiling of adult and pediatric B-cell acute lymphoblastic leukemia. EBioMedicine 8, 173–183. doi: 10.1016/j.ebiom.2016.04.038
Locatelli, F., Crotta, A., Ruggeri, A., Eapen, M., Wagner, J. E., MacMillan, M. L., et al. (2013). Analysis of risk factors influencing outcomes after cord blood transplantation in children with juvenile myelomonocytic leukemia: a EUROCORD, EBMT, EWOG-MDS,CIBMTR study. Blood 122, 2135–2141. doi: 10.1182/blood-2013-03-491589
Lopez, C. K., Noguera, E., Stavropoulou, V., Robert, E., Aid, Z., Ballerini, P., et al. (2019). Ontogenic changes in hematopoietic hierarchy determine pediatric specificity and disease phenotype in fusion oncogene– driven myeloid leukemia. Cancer Discov. 9, 1736–1753. doi: 10.1158/2159-8290.CD-18-1463
MacMahon, B., and Levy, M. A. (1964). Prenatal origin of childhood leukemia. N. Engl. J. Med. 270, 1082–1085. doi: 10.1056/NEJM196405212702102
Maia, A. T., Koechling, J., Corbett, R., Metzler, M., Wiemels, J. L., and Greaves, M. (2004). Protracted postnatal natural histories in childhood leukemia. Genes Chromosom. Cancer 39, 335–340. doi: 10.1002/gcc.20003
Malard, F., and Mohty, M. (2020). Acute lymphoblastic leukaemia. Lancet 395, 1146–1162. doi: 10.1016/S0140-6736(19)33018-1
Man, N., Sun, X. J., Tan, Y., García-Cao, M., Liu, F., Cheng, G., et al. (2016). Differential role of Id1 in MLL-AF9-driven leukemia based on cell of origin. Blood 127, 2322–2326. doi: 10.1182/blood-2015-11-677708
Masetti, R., Vendemini, F., Zama, D., Biagi, C., Pession, A., and Locatelli, F. (2015). Acute myeloid leukemia in infants: biology and treatment. Front. Pediatr. 3:37. doi: 10.3389/fped.2015.00037
Mass, E., Ballesteros, I., Farlik, M., Halbritter, F., Günther, P., Crozet, L., et al. (2016). Specification of tissue-resident macrophages during organogenesis. Science 353:aaf4238. doi: 10.1126/science.aaf4238
Mass, E., Jacome-Galarza, C. E., Blank, T., Lazarov, T., Durham, B. H., Ozkaya, N., et al. (2017). A somatic mutation in erythro-myeloid progenitors causes neurodegenerative disease. Nature 549, 389–393. doi: 10.1038/nature23672
Matsuda, K., Sakashita, K., Taira, C., Tanaka-Yanagisawa, M., Yanagisawa, R., Shiohara, M., et al. (2010). Quantitative assessment of PTPN11 or RAS mutations at the neonatal period and during the clinical course in patients with juvenile myelomonocytic leukaemia. Br. J. Haematol. 148, 593–599. doi: 10.1111/j.1365-2141.2009.07968.x
McGrath, K. E., Frame, J. M., Fegan, K. H., Bowen, J. R., Conway, S. J., Catherman, S. C., et al. (2015a). Distinct sources of hematopoietic progenitors emerge before HSCs and provide functional blood cells in the mammalian embryo. Cell Rep. 11, 1892–1904. doi: 10.1016/j.celrep.2015.05.036
McGrath, K. E., Frame, J. M., and Palis, J. (2015b). Early hematopoiesis and macrophage development. Semin. Immunol. 27, 379–387. doi: 10.1016/j.smim.2016.03.013
Medvinsky, A., and Dzierzak, E. (1996). Definitive hematopoiesis is autonomously initiated by the AGM region. Cell 86, 897–906. doi: 10.1016/S0092-8674(00)80165-8
Menendez, P., Catalina, P., Rodríguez, R., Melen, G. J., Bueno, C., Arriero, M., et al. (2009). Bone marrow mesenchymal stem cells from infants with MLL-AF4+ acute leukemia harbor and express the MLL-AF4 fusion gene. J. Exp. Med. 206, 3131–3141. doi: 10.1084/jem.20091050
Meyer, C., Burmeister, T., Gröger, D., Tsaur, G., Fechina, L., Renneville, A., et al. (2018). The MLL recombinome of acute leukemias in 2017. Leukemia 32, 273–284. doi: 10.1038/leu.2017.213
Müller, A. M., Medvinsky, A., Strouboulis, J., Grosveld, F., and Dzierzakt, E. (1994). Development of hematopoietic stem cell activity in the mouse embryo. Immunity 1, 291–301. doi: 10.1016/1074-7613(94)90081-7
Ottersbach, K. (2019). Endothelial-to-haematopoietic transition: an update on the process of making blood. Biochem. Soc. Trans. 47, 591–601. doi: 10.1042/BST20180320
Ottersbach, K., and Dzierzak, E. (2005). The murine placenta contains hematopoietic stem cells within the vascular labyrinth region. Dev. Cell 8, 377–387. doi: 10.1016/j.devcel.2005.02.001
Palis, J. (2016). Hematopoietic stem cell-independent hematopoiesis: emergence of erythroid, megakaryocyte, and myeloid potential in the mammalian embryo. FEBS Lett. 590, 3965–3974. doi: 10.1002/1873-3468.12459
Palis, J., Chan, R. J., Koniski, A., Patel, R., Starr, M., and Yoder, M. C. (2001). Spatial and temporal emergence of high proliferative potential hematopoietic precursors during murine embryogenesis. Proc. Natl. Acad. Sci. U.S.A. 98, 4528–4533. doi: 10.1073/pnas.071002398
Palis, J., Robertson, S., Kennedy, M., Wall, C., and Keller, G. (1999). Development of erythroid and myeloid progenitors in the yolk sac and embryo proper of the mouse. Development 126, 5073–5084.
Papaemmanuil, E., Gerstung, M., Bullinger, L., Gaidzik, V. I., Paschka, P., Roberts, N. D., et al. (2016). Genomic classification and prognosis in acute myeloid leukemia. N. Engl. J. Med. 374, 2209–2221. doi: 10.1056/NEJMoa1516192
Pieters, R., Schrappe, M., De Lorenzo, P., Hann, I., De Rossi, G., Felice, M., et al. (2007). A treatment protocol for infants younger than 1 year with acute lymphoblastic leukaemia (Interfant-99): an observational study and a multicentre randomised trial. Lancet 370, 240–250. doi: 10.1016/S0140-6736(07)61126-X
Rice, S., and Roy, A. (2020). MLL-rearranged infant leukaemia : a ‘ thorn in the side ’ of a remarkable success story. Biochim. Biophys. Acta Gene Regul. Mech. 1863:194564. doi: 10.1016/j.bbagrm.2020.194564
Robin, C., Bollerot, K., Mendes, S., Haak, E., Crisan, M., Cerisoli, F., et al. (2010). Human placenta is a potent hematopoietic niche containing hematopoietic stem and progenitor cells throughout development. Cell Stem Cell 5, 385–395. doi: 10.1016/j.stem.2009.08.020
Rowe, R. G., Da Rocha, E. L., Sousa, P., Missios, P., Morse, M., Marion, W., et al. (2019). The developmental stage of the hematopoietic niche regulates lineage in MLL-rearranged leukemia. J. Exp. Med. 216, 527–538. doi: 10.1084/jem.20181765
Rybtsov, S., Batsivari, A., Bilotkach, K., Paruzina, D., Senserrich, J., Nerushev, O., et al. (2014). Tracing the origin of the HSC hierarchy reveals an SCF-dependent, IL-3-independent CD43- embryonic precursor. Stem Cell Rep. 3, 489–501. doi: 10.1016/j.stemcr.2014.07.009
Rybtsov, S., Ivanovs, A., Zhao, S., and Medvinsky, A. (2016). Concealed expansion of immature precursors underpins acute burst of adult HSC activity in foetal liver. Development 143, 1284–1289. doi: 10.1242/dev.131193
Rybtsov, S., Sobiesiak, M., Taoudi, S., Souilhol, C., Senserrich, J., Liakhovitskaia, A., et al. (2011). Hierarchical organization and early hematopoietic specification of the developing HSC lineage in the AGM region. J. Exp. Med. 208, 1305–1315. doi: 10.1084/jem.20102419
Schulz, C., Perdiguero, E. G., Chorro, L., Szabo-Rogers, H., Cagnard, N., Kierdorf, K., et al. (2012). A lineage of myeloid cells independent of myb and hematopoietic stem cells. Science 335, 86–90. doi: 10.1126/science.1219179
Shalapour, S., Eckert, C., Seeger, K., Pfau, M., Prada, J., Henze, G., et al. (2010). Leukemia-associated genetic aberrations in mesenchymal stem cells of children with acute lymphoblastic leukemia. J. Mol. Med. 88, 249–265. doi: 10.1007/s00109-009-0583-8
Sinha, R., Porcheri, C., d’Altri, T., González, J., Ruiz-Herguido, C., Rabbitts, T., et al. (2020). Development of embryonic and adult leukemia mouse models driven by MLL-ENL translocation. Exp. Hematol. 85, 13–19. doi: 10.1016/j.exphem.2020.04.008
Steliarova-Foucher, E., Colombet, M., Ries, L. A. G., Moreno, F., Dolya, A., Bray, F., et al. (2017). International incidence of childhood cancer, 2001–10: a population-based registry study. Lancet Oncol. 18, 719–731. doi: 10.1016/S1470-2045(17)30186-9
Swiers, G., Rode, C., Azzoni, E., and de Bruijn, M. F. T. R. (2016). A short history of hemogenic endothelium. Blood Cells Mol. Dis. 51, 206–212. doi: 10.1016/j.bcmd.2013.09.005
Taoudi, S., Gonneau, C., Moore, K., Sheridan, J. M., Blackburn, C. C., Taylor, E., et al. (2008). Extensive hematopoietic stem cell generation in the AGM region via maturation of VE-Cadherin+CD45+ pre-definitive HSCs. Cell Stem Cell 3, 99–108. doi: 10.1016/j.stem.2008.06.004
Tarnawsky, S. P., Yoshimoto, M., Deng, L., Chan, R. J., and Yoder, M. C. (2017). Yolk sac erythromyeloid progenitors expressing gain of function PTPN11 have functional features of JMML but are not sufficient to cause disease in mice. Dev. Dyn. 246, 1001–1014. doi: 10.1002/dvdy.24598
Tavian, M., Robin, C., Coulombel, L., and Péault, B. (2001). The human embryo, but not its yolk sac, generates lympho-myeloid stem cells: mapping multipotent hematopoietic cell fate in intraembryonic mesoderm. Immunity 15, 487–495. doi: 10.1016/S1074-7613(01)00193-5
Tober, J., Koniski, A., McGrath, K. E., Vemishetti, R., Emerson, R., De Mesy-Bentley, K. K. L., et al. (2007). The megakaryocyte lineage originates from hemangioblast precursors and is an integral component both of primitive and of definitive hematopoiesis. Blood 109, 1433–1441. doi: 10.1182/blood-2006-06-031898
Uckun, F., Herman-Hatten, K., Crotty, M., Sensel, M., Sather, H., Tuel-Ahlgren, L., et al. (1998). Clinical significance of MLL-AF4 fusion transcript expression in the absence of a cytogenetically detectable t(4;11)(q21;q23) chromosomal translocation. Blood 810–821.
Wiemels, J. L., Cazzaniga, G., Daniotti, M., Eden, O. B., Addison, G. M., Masera, G., et al. (1999). Prenatal origin of acute lymphoblastic leukaemia in children. Lancet 354, 1499–1503. doi: 10.1016/S0140-6736(99)094039
Wiemels, J. L., Xiao, Z., Buffler, P. A., Maia, A. T., Ma, X., Dicks, B. M., et al. (2002). In utero origin of t(8;21) AML1-ETO translocations in childhood acute myeloid leukemia. Blood 99, 3801–3805. doi: 10.1182/blood.V99.10.3801
Xu, M. J., Matsuoka, S., Yang, F. C., Ebihara, Y., Manabe, A., Tanaka, R., et al. (2001). Evidence for the presence of murine primitive megakarycytopoiesis in the early yolk sac. Blood 97, 2016–2022. doi: 10.1182/blood.V97.7.2016
Keywords: pediatric leukemia, cell of origin, hematopoiesis, hematopoietic stem cells, eryhtro-myeloid progenitors
Citation: Cazzola A, Cazzaniga G, Biondi A, Meneveri R, Brunelli S and Azzoni E (2021) Prenatal Origin of Pediatric Leukemia: Lessons From Hematopoietic Development. Front. Cell Dev. Biol. 8:618164. doi: 10.3389/fcell.2020.618164
Received: 16 October 2020; Accepted: 15 December 2020;
Published: 12 January 2021.
Edited by:
Simone Pacini, University of Pisa, ItalyReviewed by:
Arndt Borkhardt, Heinrich Heine University of Düsseldorf, GermanyVeronica Ramos-Mejia, Junta de Andalucía de Genómica e Investigación Oncológica (GENYO), Spain
Copyright © 2021 Cazzola, Cazzaniga, Biondi, Meneveri, Brunelli and Azzoni. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Emanuele Azzoni, ZW1hbnVlbGUuYXp6b25pQHVuaW1pYi5pdA==
 Anna Cazzola
Anna Cazzola Giovanni Cazzaniga
Giovanni Cazzaniga Andrea Biondi
Andrea Biondi Raffaella Meneveri
Raffaella Meneveri Silvia Brunelli
Silvia Brunelli Emanuele Azzoni
Emanuele Azzoni