- 1Centre for Biological Signalling Studies and Centre for Integrative Biological Signalling Studies, University Freiburg, Freiburg, Germany
- 2Department of Immunology, Faculty of Biology, University of Freiburg, Freiburg, Germany
- 3Centre for Chronic Immunodeficiency (CCI), University of Freiburg, Freiburg, Germany
- 4Immunology, Infectious Diseases and Ophthalmology Disease Translational Area, Roche Innovation Center Basel, Basel, Switzerland
- 5State Key Laboratory of Molecular Biology, CAS Center for Excellence in Molecular Cell Science, Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, Shanghai, China
- 6School of Life Science and Technology, ShanghaiTech University, Shanghai, China
Biological membranes consist of hundreds of different lipids that together with the embedded transmembrane (TM) proteins organize themselves into small nanodomains. In addition to this function of lipids, TM regions of proteins bind to lipids in a very specific manner, but the function of these TM region-lipid interactions is mostly unknown. In this review, we focus on the role of plasma membrane cholesterol, which directly binds to the αβ T cell antigen receptor (TCR), and has at least two opposing functions in αβ TCR activation. On the one hand, cholesterol binding to the TM domain of the TCRβ subunit keeps the TCR in an inactive, non-signaling conformation by stabilizing this conformation. This assures that the αβ T cell remains quiescent in the absence of antigenic peptide-MHC (the TCR's ligand) and decreases the sensitivity of the T cell toward stimulation. On the other hand, cholesterol binding to TCRβ leads to an increased formation of TCR nanoclusters, increasing the avidity of the TCRs toward the antigen, thus increasing the sensitivity of the αβ T cell. In mouse models, pharmacological increase of the cholesterol concentration in T cells caused an increase in TCR clustering, and thereby enhanced anti-tumor responses. In contrast, the γδ TCR does not bind to cholesterol and might be regulated in a different manner. The goal of this review is to put these seemingly controversial findings on the impact of cholesterol on the αβ TCR into perspective.
Introduction
A eukaryotic plasma membrane is composed of a variety of lipids and sterols, such as cholesterol. The most common composition of the plasma membrane is 20–50% phosphatidylcholine, 20–25% sphingomyelin, 30–50% cholesterol, 10% phosphatidylserine and 25% phosphatidylethanolamine (van Meer et al., 2008; Marquardt et al., 2015). One important sterol is cholesterol (Figure 1A), that is synthesized by the cells themselves and can be taken up from the environment. It determines membrane fluidity and permeability (Heerklotz and Tsamaloukas, 2006; Subczynski et al., 2017). The tetracyclic structure of cholesterol is planar and rigid. As a result, increase in membrane cholesterol increases lipid packing and stiffness causing decreased fluidity of lipid bilayers.
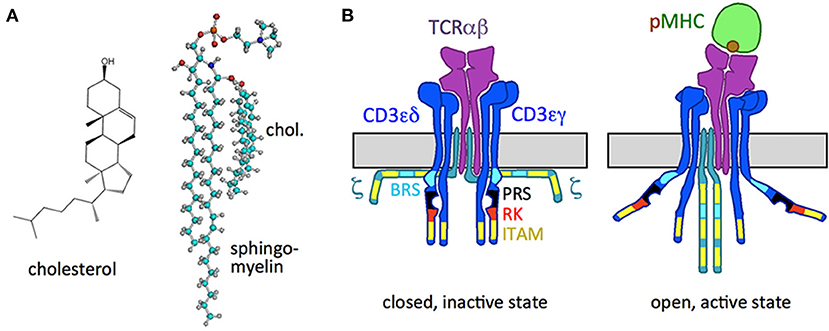
Figure 1. Cholesterol and the TCR. (A) Structure of cholesterol and the cholesterol sphingomyelin pair. (B) Schematic of the resting, inactive TCR, in which the cytoplasmic signaling motifs of the CD3 and ζ subunits are not accessible (right), and of the active TCR with the pMHC ligand bound (left), in which the motifs are exposed. The ITAM, BRS, PRS, and RK motifs are indicated.
Lipids are not randomly distributed within the membrane but are organized. Using model membranes lipid nanodomains called liquid-ordered (Lo) and liquid-disordered (Ld) domains can be distinguished (Veiga et al., 2001; Veatch et al., 2004). It has been argued that these nanodomains are also present in the plasma membrane of living cells, although in a less stable and smaller manner (Eggeling et al., 2009; Levental et al., 2011; Mueller et al., 2011). The Lo domains would correspond to the lipid rafts in cellular membranes and the Ld domains to the non-raft domains (Simons and Ikonen, 1997; Sharma et al., 2004). In cellular membranes the lipid rafts are enriched in sphingolipids and cholesterol and are most likely very small (10–40 nm) and short-lived (microseconds) and hence difficult to characterize. Important for the formation of these domains is the interaction between cholesterol and sphingomyelin that facilitates stable dimers (Figure 1A) (Demel et al., 1977; Veiga et al., 2001; Bjorkbom et al., 2011). In addition to the dimer, free cholesterol and free sphingomyelin also exist (Simons and Ikonen, 1997; Endapally et al., 2019). Rafts concentrate signaling molecules and thus are important for signaling (Simons and Ikonen, 1997). Non-raft domains are rich in unsaturated glycerophospholipids, mostly lack sphingolipids and contain less cholesterol. Lo domains are thicker than Ld domains, due to the loss of kinks in acyl chains (Subczynski et al., 2017). Another factor that contributes to nanodomain formation in cellular membranes is the lipid asymmetry between the outer and the inner leaflet. For example, phosphatidylserine is strongly enriched in the inner leaflet and sphingomyelin is mainly found in the outer leaflet (Fadeel and Xue, 2009). Another well-known asymmetry observed is of that of cholesterol where its affinity toward sphingomyelin leads to its enrichment in the outer layer (Wood et al., 2011), although due to its small hydrophilic group (Figure 1A) it possesses a very high flip-flop rate (Steck et al., 2002).
Transmembrane (TM) proteins are also not randomly distributed on the cell surface, but localize to certain lipid nanodomains. This is most likely dictated by the exact sequence of the TM region that interacts with the lipids, but also by interactions with other proteins. This localization impacts the function of these proteins, as it allows the vicinity to proteins with a similar lipid preference and guarantees a distance to proteins with a different lipid preference. For example, specific interaction of TM proteins with certain lipids has been demonstrated by structural biology for the bacteriorhodopsin-glycolipid S-TGA-1 (Essen et al., 1998), the cytochrome bc1 complex of the mitochondrial respiratory chain (Hunte, 2005), the metarhodopsin-cholesterol (Ruprecht et al., 2004), the β2-adrenergic receptor-cholesterol (Cherezov et al., 2007; Hanson et al., 2008) interactions or by functional assays for the epidermal growth factor receptor (EGFR)-ganglioside GM3 association (Coskun et al., 2011). These interactions might be the underlying reason for their preferential localization to certain lipid domains or not. In addition, these specific TM region-lipid interactions might directly influence the function of the TM protein. One well-studied example is the T cell antigen receptor (TCR)-cholesterol interaction (Schamel et al., 2017, 2019) and this is the focus of this review.
The T Cell Antigen Receptor (TCR)
T cells are important for an adaptive immune response against pathogens and tumors and are involved in autoimmunity. In humans 95% of the T cells express an αβ TCR while 5% express a γδ TCR on their surface. The TCR expression is crucial for their development and activation. The αβ TCR (here denoted as TCR for simplicity) binds to pathogen-, tumor- or host-derived peptides presented on MHC molecules (pMHC) by the host's cells. This binding leads to the activation and proliferation of the T cells and downstream effector functions such as cytokine production, provision of help to B cells, regulation of the T cell response or killing of cells expressing the cognate pMHC.
The TCR is a trans-membrane protein complex composed of non-covalently bound TCRαβ, CD3γε, CD3δε, and ζζ2 dimers (Figure 1B). All subunits are type I membrane proteins that contain either basic amino acid residues (arginine and lysine in TCRα; lysine in TCRβ) or acidic ones in their TM domains (aspartic acid in CD3ε, CD3δ, and ζ; glutamic acid in CD3γ) (Alarcon et al., 2003; Malissen, 2003). It is suggested that the potentially charged amino acids in the TM domains are involved in the interaction between the TCRαβ and CD3 (Call et al., 2002) as are also the ectodomains (Schamel et al., 2019).
With their variable extracellular regions TCRαβ bind to pMHC and the information of ligand binding is transduced through the membrane to the cytosolic tails of CD3 and ζ, that contain intracellular signaling motifs (Figure 1B). These motifs include the receptor-kinase (RK) motif that binds to the TCR's kinase Lck (Hartl et al., 2020), tyrosines in the context of the immunoreceptor tyrosine-based activation motifs (ITAMs) that can be phosphorylated (Reth, 1989; Weiss, 2010) and a proline-rich sequence (PRS) that can associate with the adaptor protein Nck (Gil et al., 2002). Further, basic rich sequences (BRSs) in CD3ε and ζ bind to negatively charged lipids of the inner leaflet of the plasma membrane in the resting TCR (Aivazian and Stern, 2000; Xu et al., 2008; Zhang et al., 2011). pMHC-binding leads to the exposure of these motifs with a consequent phosphorylation of the tyrosines by Lck. These phospho-tyrosines serve as docking sites for signaling proteins with SH2 domains (Acuto et al., 2008; Courtney et al., 2018). The latter then transduce the signal into the cells, causing activation of the T cell and subsequent effector functions.
The αβ TCR Binds To Cholesterol
Compared to techniques to study protein-protein interactions, the ones for identifying specific associations of lipids with the TM regions of proteins are scarce and have limitations. Thus, not much is known about lipid-protein interactions. Useful techniques include the following: (i) In living cells covalent cross-linking of lipid derivatives with a UV light inducible reactive groups to proteins as been successfully used (Thiele et al., 2000; Hulce et al., 2013). However, the lipids are not exactly the natural ones and thus some interactions might not be detected. (ii) Structural studies of membrane proteins, such as NMR or crystallization, might resolve lipids that either were co-purified with the protein or added during the analysis or crystallization (Hunte, 2005). (iii) Although indirect, another approach is to modulate the lipid composition of the membrane, as e.g., is artificial liposomes, and then detect changes on the embedded membrane protein (Coskun et al., 2011). (iv) A complementary method is to use beads coupled to a lipid that are then used for pull-down assays to purify proteins that bind to the lipid (Beck-Garcia et al., 2013). However, this requires solubilisation of the membrane proteins by detergent that might be a source for artifacts. Due to these caveats it is recommended to use at least two complementary techniques. These experiments done with the TCR are described in the next paragraph.
Using a radioactive cross-linkable cholesterol derivative (Thiele et al., 2000), we could show that cholesterol specifically binds to the TCR in living cells, and it did not bind to other receptors tested (Molnar et al., 2012). This binding occurred to the TCRβ chain in the resting, i.e., non-ligand bound TCR (Molnar et al., 2012; Swamy et al., 2016). In a recent first high resolution structure of the complete TCR, bound cholesterol was not seen (Dong et al., 2019), most likely because digitonin was used to solubilize the TCR from the cell membrane, which is known to extract and remove cholesterol from the TCR (Schamel et al., 2005; Alarcon et al., 2006; Molnar et al., 2012). Interestingly, cholesterol sulfate, a naturally occurring derivative of cholesterol, competes with cholesterol in binding to the TCR (Wang et al., 2016).
The cholesterol-TCRβ interaction is dynamic, since only the non-ligand bound TCR associated with cholesterol and the ligand-bound TCR did not (Swamy et al., 2016). These binding characteristics were recapitulated using purified TCRs and cholesterol-coupled beads (Beck-Garcia et al., 2013; Swamy et al., 2016) as only the resting TCR bound to these beads. This demonstrated that the dynamic cholesterol binding is a property of the TCRβ TM region and is not a consequence of altered membrane properties caused by ligand engagement.
In addition to the cholesterol-TM region interaction and as mentioned above, the cytosolic tails of CD3ε and ζ might interact with the head groups of negatively charged lipids, such as phosphatidylserine, in the inner leaflet of the plasma membrane (Aivazian and Stern, 2000; Xu et al., 2008). Since this has already been reviewed by Wu et al. (2016), it will not be discussed.
The γδ TCR Does Not Bind To Cholesterol
At first sight the γδ TCR looks similar to the αβ TCR. It also contains the CD3 and ζ subunits, but instead of TCRαβ it contains the highly related TCRγδ ligand-binding dimer. However, the TM region of TCRγ is partially different to the of TCRβ, and consequently using a radioactive cross-linkable cholesterol derivative (Thiele et al., 2000), we demonstrated that the γδ TCR does not bind to cholesterol (Swamy et al., 2016). Thus, the function of cholesterol on the activity of the TCR that we discuss in this review is limited to the αβ TCR and the γδ TCR must therefore be regulated by different mechanisms. A comparison of both TCRs was published recently (Morath and Schamel, 2020).
Cholesterol Regulates The Allosteric Switch of The αβ TCR
As a prerequisite for allostery, the TCR exists in (at least) two different conformations that differ in their tertiary and/or quaternary structure (Figure 1B). Although most crystal structures of certain isolated domains of TCRαβ and CD3 did not provide information on these changes [Garboczi et al., 1996; Garcia et al., 1996; Rudolph et al., 2006; and the reason for that is discussed in a recent review Schamel et al. (2019)], a number of experiments have detected changes in the TCR structure upon ligand binding. These include NMR (Natarajan et al., 2017; Rangarajan et al., 2018), crystallography (Beddoe et al., 2009), and fluorescence-based or H/D exchange approaches (Beddoe et al., 2009; Hawse et al., 2012; Lee et al., 2015). In addition, biochemistry has provided evidence that the TCR structure changes when pMHC (or stimulating antibodies) are bound. These include limited trypsin digest (Risueno et al., 2008), measuring the distance between two subunits (Lee et al., 2015), accessibility of an antibody epitope (Risueno et al., 2005), cholesterol-binding to TCRβ (Swamy et al., 2016), and exposure of the proline-rich sequence (PRS) (Gil et al., 2002; Minguet et al., 2007), the receptor-kinase (RK) motif (Hartl et al., 2020) and the tyrosines in the cytosolic tails of the CD3 and ζ subunits (Swamy et al., 2016).
The two conformations of the TCR are: (i) the resting, inactive conformation (TCR), in which the CD3ε RK motif cannot bind to the Lck and the cytoplasmic tyrosines are shielded and thus are not phosphorylated; and (ii) the active conformation, which is stabilized after pMHC or antibody binding, in which Lck binds to CD3ε and the exposed cytosolic tyrosines of CD3 and ζ are phosphorylated (Gil et al., 2002, 2005; Minguet et al., 2007; Lee et al., 2015; Swamy et al., 2016; Hartl et al., 2020). The switch to the active conformation is essential for TCR phosphorylation and T cell stimulation. This was confirmed using artificial ligands (Minguet et al., 2007) and TCR mutants that are trapped in the resting conformation (Martinez-Martin et al., 2009; Blanco et al., 2014; Dopfer et al., 2014).
Thus, the αβ TCR is allosterically regulated; binding of pMHC at one site (through the variable regions of TCRαβ) causes structural alterations and dynamic changes at other sites, e.g., in the CD3 subunits. As a side note, the γδ TCR does not show these changes and its activity is regulated in a different manner (Blanco et al., 2014; Dopfer et al., 2014; Juraske et al., 2018; Morath and Schamel, 2020).
The Monod-Wyman-Changeux model of allostery (Monod et al., 1965) proposes that the αβ TCR can switch spontaneously between the two states in the absence of ligand (Figure 2) (Schamel et al., 2017); experimental evidences support this notion (Mingueneau et al., 2008; de la Cruz et al., 2011; Swamy et al., 2016). The ligand binding can perturb the equilibrium between these two states. In fact, ligand only binds to the active state and thus shifts the equilibrium to the active state (Swamy et al., 2016); consequently the cytoplasmic motifs are exposed and the TCR becomes signaling active (Figure 2). So how does a T cell guarantee that in the absence of ligand not too many TCRs are in the active state? This is achieved by cholesterol, which binds only to the resting TCR, and hence shifts the equilibrium to inactive TCRs (Figure 2) (Swamy et al., 2016). Thus, the TCR has two opposing binding partners, one that leads to the accumulation of inactive TCRs (cholesterol) and the other one that promotes active TCRs (pMHC).
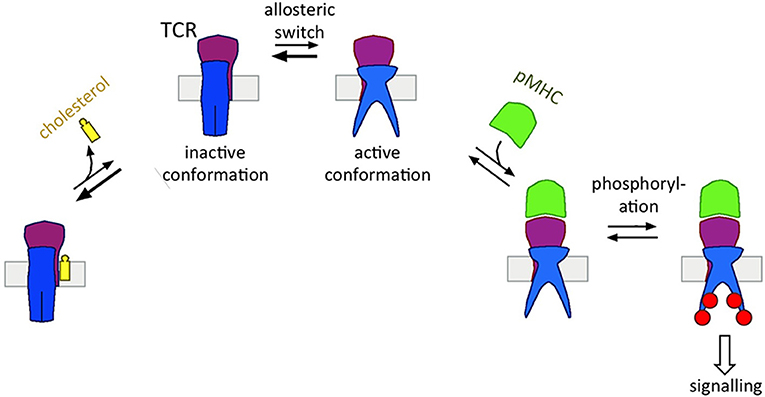
Figure 2. Cholesterol's function of regulating the allosteric switch of the TCR. The TCR can switch spontaneously between the inactive and active state (allosteric switch). Cholesterol binds to the TCRβ subunit only in the inactive TCR, thus shifting the equilibrium to the left side. The pMHC ligand binds to the TCRαβ subunits only in the active TCR, thus shifting the equilibrium to the right side. Only in the active state the TCR can be phosphorylated transmitting the signal of pMHC-binding downstream.
The spontaneous shift of the TCR between its conformations was seen when in the absence of pMHC the cholesterol concentration was lowered (by extraction with methyl-β-cyclodextrin or by oxidation to cholestenone), which caused accumulation of active TCRs and initiated spontaneous TCR signaling (Kabouridis et al., 2000; Rouquette-Jazdanian et al., 2006; Swamy et al., 2016). Although methyl-β-cyclodextrin is commonly used to extract or to deliver cholesterol to membranes, it has several undefined effects on the plasma membrane and cell viability. Apart from increasing membrane permeability, it also depolymerizes the actin cytoskeleton and thereby reduces cell stiffness (Mundhara et al., 2019). Hence, it is important to complement the results obtained by methyl-β-cyclodextrin with other methods. In our previous studies we employed cholesterol oxidase to reduce the amount of available membrane cholesterol and again observed an accumulation of TCRs in the active state (Swamy et al., 2016). Similarly, mutating the TCRβ TM region so that cholesterol can no longer bind led to a shift of the equilibrium toward the active state and low level of T cell stimulation (Petersen et al., 2004; Swamy et al., 2016). These reports show that the TCR TM regions are key regulators of the conformational states of the TCR and that changes at the TM regions are linked to changes at the cytosolic tails.
In conclusion, cholesterol is a natural negative allosteric regulator of the TCR that guarantees that in the absence of ligand most TCRs remain in the resting state.
In another model, the TCR acts as a mechanosensor, in which force that is applied via pMHC to the TCR changes the TCR's structure to a signaling active configuration (Kim et al., 2009; Schamel et al., 2019). Since cholesterol stiffens the membrane, its presence at the TCR might influence these changes.
Cholesterol Regulates αβ TCR Nanoclustering
By using complementary techniques, several studies have suggested that on the surface of a resting T cell, the TCR exists in a monomeric and in a nanoclustered form (Schamel et al., 2005; Lillemeier et al., 2010; Kumar et al., 2011; Sherman et al., 2011; Schamel and Alarcon, 2013; Pageon et al., 2016; Martín-Leal et al., 2020). Other studies only found low amount of TCR nanoclusters and thus concluded that nanoclusters would not exist (James et al., 2011; Rossboth et al., 2018). Thus, the existence of TCR nanoclusters is still controversial, and technical limitations that contribute to this disagreement are discussed in several articles (Schamel and Alarcon, 2013; Platzer et al., 2020). For example, detergents might disrupt nanoclusters when being analyzed biochemically, in microscopy a low labeling efficiency might prevent the detection of nanoclusters or rapid blinking of a fluorophore attached to a TCR might lead to the detection of a nanocluster when in reality there is only one TCR present. Our own studies favor the existence of TCR nanoclusters. In fact, the amount and the size of the nanoclusters depend on the state of the cell (and this might be another confounding factor for detecting or not the nanoclusters). For example, a naïve T cell has less and smaller nanoclusters than an antigen-experienced T cell (Kumar et al., 2011; Schamel and Alarcon, 2013). Likewise, the cholesterol content of these cells increased from naïve to memory cells (Kaech et al., 2002; Kersh et al., 2003; Tani-ichi et al., 2005). These findings suggest that cholesterol is involved in the TCR nanoclustering (Figure 3) and three different approaches have shown that this is the case: (i) solubilisation of the TCR from T cell membranes with detergents that do not extract cholesterol preserved the TCR's nanoclustered form; in contrast, when cholesterol was extracted the nanoclusters disassemble to the monomeric TCRs (Schamel et al., 2005; Alarcon et al., 2006; Molnar et al., 2012). (ii) TCR nanoclusters disassembled when cholesterol was either extracted from the cells or when cholesterol levels were reduced pharmacologically as detected by immuno-gold electron microscopy or super-resolution fluorescence microscopy (Schamel et al., 2005; Alarcon et al., 2006; Molnar et al., 2012; Yang et al., 2016). (iii) Reconstituting the monomeric TCR in liposomes of defined lipid composition only allowed nanoclusters to form when cholesterol and sphingomyelin were present in the otherwise phosphatidylcholine-containing liposomes (Schamel et al., 2005; Alarcon et al., 2006; Molnar et al., 2012). This indicated that membrane proteins other than the TCR and lipids other than the ones mentioned are not required for TCR nanoclustering. Since sphingomyelin is mainly present in the outer leaflet of the plasma membrane, it is possible that cholesterol and sphingomyelin [maybe as a pair (Demel et al., 1977; Veiga et al., 2001; Bjorkbom et al., 2011)] bind to the N-terminal part of the TCRβ TM region. However, this remains to be tested. Concerning the mechanism of how cholesterol and sphingomyelin promote TCR nanoclustering, we suggested that cholesterol and sphingomyelin form a mini-raft-islet at the TCRβ TM region that is not favored to be in contact with the non-raft lipid domains that are around the TCR (Molnar et al., 2012; Beck-Garcia et al., 2015). Thus, these islets from several TCRs would come close to each other to shield each other from the non-raft domains, causing TCR nanoclustering.
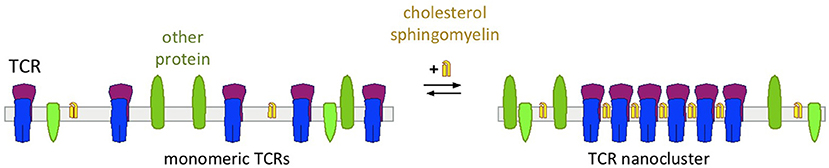
Figure 3. Cholesterol's function of regulating nanoclustering of the TCR. With low levels of cholesterol and sphingomyelin TCRs are expressed as monomers on the cell surface (left)—as it is the case in naïve T cells. With increasing concentrations of cholesterol and sphingomyelin, these lipids bind to the TCR and cause TCR nanoclustering—as it is the case in activated and memory T cells.
By regulating TCR nanoclustering cholesterol defines the sensitivity of the TCR for activation through its ligand; a T cell with more and bigger nanoclusters is easier to activate than a cell with predominantly monomeric TCRs (Kumar et al., 2011). Indeed, it was shown that TCR nanoclusters possess a higher avidity toward multimeric pMHC than monomeric TCRs (Molnar et al., 2012). Further, TCRs within a nanocluster show positive cooperativity, so that if one TCR is stabilized in the active state by ligand-binding also the other TCRs in the nanocluster reside in the active state (Martinez-Martin et al., 2009; Schamel et al., 2017). TCRs in a given nanocluster that are stabilized in inactive state by cholesterol can spontaneously release cholesterol and thereby subsequently switch to the active conformation. Whether it is sufficient that one single TCR within a cluster binds to cholesterol to prevent the switch of all TCRs to the active conformation is not known. Since nanoclusters disassemble when cholesterol is extracted from the cells, the cooperativity of TCR within nanoclusters could be abrogated by cholesterol removal (Martin-Blanco et al., 2018). This result again showed that nanoclusters are required for the TCR cooperativity and that cholesterol is crucial for TCR nanocluster formation.
Studies show that naive T cells contain lower levels of cholesterol than activated T cells (Kersh et al., 2003; Tani-ichi et al., 2005). Indeed, upon activation of T cells cholesterol metabolism is reprogrammed to synthesize more cholesterol by upregulation of the Sterol Regulatory Element-Binding Protein-2 (SREBP-2) pathway and to transport less cholesterol out of the cell by downregulation of Liver X Receptor (LXR) target genes (Bensinger et al., 2008; Wu et al., 2016). Importantly, in antigen-experienced T cells, such as effector or memory T cells, the increased cholesterol levels contribute to enhanced TCR nanoclustering (Kumar et al., 2011). This might be a danger, since the nanoclusters lower the threshold for T cell activation due to increased avidity and cooperativity. Thus, a counter-regulation through cholesterol by keeping the TCRs in the inactive state might prevent spontaneous activation or activation by weak signals, in order to prevent autoimmune diseases. Indeed, elevated cholesterol levels in T cells have been linked to certain autoimmune diseases (see below).
Cholesterol sulfate, which is a low abundant derivative of cholesterol (Bergner and Shapiro, 1981) can bind to the TCR and disrupt the TCR-cholesterol interaction (Wang et al., 2016). This finding suggested that cholesterol sulfate binds to the same region as cholesterol. Interestingly, cholesterol sulfate disrupted TCR nanoclustering in liposomes and in T cells (Wang et al., 2016). The reduced TCR nanoclustering was paralleled by a reduced avidity of the T cells toward multivalent TCR ligands (Wang et al., 2016). Maybe the cholesterol-sphingomyelin pair is required for TCR nanoclustering (see above); and since cholesterol sulfate may not bind to sphingomyelin, it does not promote TCR nanoclustering.
Likewise, the lipid ceramide reduced TCR nanoclustering (Martín-Leal et al., 2020). This was observed in liposomes as well as in T cells that were treated with sphingomyelinase, which hydrolyses sphingomyelin to ceramide. This data suggest that the cholesterol-sphingomyelin pair drives TCR nanoclustering. Interestingly, signaling by the receptor CCR5 reduces ceramide levels in antigen-experienced T cells (Martín-Leal et al., 2020). In these cells, along with reduced ceramide levels, increased membrane cholesterol contributes to enhanced TCR nanocluster formation and increased sensitivity of these cells compared to naive T cells. In conclusion, T cells regulate their membrane lipid composition, in order to tune TCR nanoclustering and thus TCR signaling.
Modulation of Cholesterol Levels To Tune αβ TCR Function In The Treatment of Diseases
As discussed, cholesterol modulates the activity of the TCR. Moreover, since dampening or increasing signaling by the TCR can be used to treat autoimmunity or cancer, respectively, it is not surprising that pharmacologically changing the cholesterol content of T cells has been used to ameliorate certain diseases.
Autoantibodies and the deposition of immune complexes are known to cause the autoimmune disease systemic lupus erythematosus (SLE). Overactive T cells contribute to the pathology by help provided to B cells and by the killing of host cells in a number of organs. Thus, a strong T cell activity contributes to SLE (Moulton and Tsokos, 2015). Importantly, T cells from SLE patients possess increased plasma membrane levels of cholesterol and glycosphingolipids (Jury et al., 2004; McDonald et al., 2014). These could lead to enhanced TCR nanoclustering and formation of signaling-promoting lipid rafts, consequently leading to increased T cell activation and effector functions as observed experimentally (McDonald et al., 2014). Extraction of cholesterol from the membrane of T cells from SLE patients using methyl-β-cyclodextrin indeed reversed the heightened signaling by the TCR (Krishnan et al., 2004). This may be a result of a disintegration of TCR nanoclusters and a partial disruption of lipid rafts. Reduced TCR signaling was also seen when the inhibitor N-butyldeoxynojirimycin was used, which normalized glycosphingolipid levels in T cells of SLE patients (McDonald et al., 2014). Similarly, inhibition of cholesterol biosynthesis in the SLE T cells by Atorvastatin reduced signaling and T cell activation (Jury et al., 2006). Statins are widely prescribed as drugs to reduce cholesterol levels by inhibiting 3-hydroxy-3-methylglutaryl-coenzyme-A (HMG-CoA) reductase, which is a key enzyme in the mevalonate pathway to synthesize cholesterol, but also to generate protein prenylations, such as farnesylation or geranylgeranylation. Thus, statins have multiple effects. Indeed, Simvastatin impairs T cell activation through inhibition of Ras prenylation (Ghittoni et al., 2005) and Lovastatin suppresses T cells proliferation due to reduced farnesol pyrophosphate levels (Chakrabarti and Engleman, 1991; Bietz et al., 2017). These anti-inflammatory effects of statins could be beneficial for autoimmune or inflammatory disorders but would worsen immune responses against cancer. In this review, we focus on the cholesterol-related effects.
In vivo extraction of cholesterol from plasma membrane of T cells using methyl-β-cyclodextrin in a mouse model of SLE delayed disease onset (Deng and Tsokos, 2008). This is line with reducing the T cells' activity by disruption of TCR nanoclusters and of lipid rafts. The latter mechanism was most likely involved, as clustering of lipid rafts in T cells by cholera toxin B promoted disease progression in vivo (Deng and Tsokos, 2008).
To treat cancer by increasing T cell activation has been proven to be a successful strategy (Iwai et al., 2002; Fritz and Lenardo, 2019). The enzyme acyl-CoA cholesterol acyltransferase 1 esterificates cholesterol and thus reduces cholesterol levels in T cells (Chang et al., 2006). Inhibition of this enzyme by Avasimibe led to elevated membrane cholesterol levels in CD8+ T cells and enhanced signaling (Yang et al., 2016) most likely by increased TCR nanoclustering. Importantly, this led to enhanced T cell effector functions resulting in stronger killing of tumor cells in mouse melanoma and lung carcinoma models (Yang et al., 2016). Additionally, combination of Avasimibe and anti PD1 treatments proved to be more potent than either monotherapies against cancer (Yang et al., 2016).
These preclinical findings show that the role of cholesterol in promoting TCR signaling (by inducing TCR nanoclustering and formation of lipid rafts) is dominant over its role in suppressing TCR signaling (by stabilizing the inactive TCR state).
Conclusion
Cholesterol specifically binds to the αβ TCR through its TCRβ subunit in the TCR's inactive conformation, thus supressing signaling. Cholesterol also promotes TCR nanoclustering and the formation of lipid rafts, both of which promote signaling. In a translational approach, this knowledge was recently used to pharmacologically enhance cholesterol levels in T cells, which potentiated the anti-tumor function of T cells in mouse models. This suggests that the cholesterol-induced nanoclustering and lipid raft formation are dominant in this setting and hence, cholesterol acted as a positive regulator of TCR signaling. What remains to be understood is, how the balance between positive and negative regulation through cholesterol interaction is regulated, in order to achieve fine-tuning of TCR activation and how this can be translated for the treatment of diseases that depends on the sensitivity of TCR activation.
Most likely, the TCR is an example protein for which its regulation by lipids is beginning to unfold. Most likely the influence of direct lipid-TM region interactions on the functioning of membrane proteins is much more widespread than currently thought.
Author Contributions
SP-C, CD, and WS wrote the first draft of the manuscript and updated the last version. KR, AM, WW, CX, and BO completed and corrected the draft. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the German Research Foundation (DFG) through the Sino-German grant SCHA 976/7-1 and National Natural Science Foundation of China 31861133009. The DFG also funded this work through BIOSS—EXC294 and CIBSS—EXC 2189 to WS, SFB854 (B19 to WS), FOR2799 (SCHA 976/8-1 to WS) and SFB1381 (A9 to WS). SP-C and WS obtained funding through the Roche Access to Distinguished Scientists (ROADS) programme. This work was also supported by Key Research Program of Frontier Sciences, CAS (QYZDB-SSW-SMC043 to BO), and CAS Major Science and Technology Infrastructure Open Research Project to BO and CX.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Acuto, O., Di Bartolo, V., and Michel, F. (2008). Tailoring T-cell receptor signals by proximal negative feedback mechanisms. Nat. Rev. Immunol. 8, 699–712. doi: 10.1038/nri2397
Aivazian, D., and Stern, L. J. (2000). Phosphorylation of T cell receptor ζ is regulated by a lipid dependent folding transition. Nat. Struct. Biol. 7, 1023–1026. doi: 10.1038/80930
Alarcon, B., Gil, D., Delgado, P., and Schamel, W. W. (2003). Initiation of TCR signaling: regulation within CD3 dimers. Immunol. Rev. 191, 38–46. doi: 10.1034/j.1600-065X.2003.00017.x
Alarcon, B., Swamy, M., van Santen, H. M., and Schamel, W. W. A. (2006). T-cell antigen-receptor stoichiometry: pre-clustering for sensitivity. EMBO Rep. 7, 490–495. doi: 10.1038/sj.embor.7400682
Beck-Garcia, E., Beck-Garcia, K., Schlosser, A., and Schamel, W. W. (2013). Analysis of interactions between proteins and fatty acids or cholesterol using a fatty acid/cholesterol pull-down-assay. Anal. Biochem. 436, 75–77. doi: 10.1016/j.ab.2013.01.026
Beck-Garcia, K., Beck-Garcia, E., Bohler, S., Zorzin, C., Sezgin, E., Levental, I., et al. (2015). Nanoclusters of the resting T cell antigen receptor (TCR) localize to non-raft domains. Biochim. Biophys. Acta 1853, 802–809. doi: 10.1016/j.bbamcr.2014.12.017
Beddoe, T., Chen, Z., Clements, C. S., Ely, L. K., Bushell, S. R., Vivian, J. P., et al. (2009). Antigen ligation triggers a conformational change within the constant domain of the alphabeta T cell receptor. Immunity 30, 777–788. doi: 10.1016/j.immuni.2009.03.018
Bensinger, S. J., Bradley, M. N., Joseph, S. B., Zelcer, N., Janssen, E. M., Hausner, M. A., et al. (2008). LXR signaling couples sterol metabolism to proliferation in the acquired immune response. Cell 134, 97–111. doi: 10.1016/j.cell.2008.04.052
Bergner, E. A., and Shapiro, L. J. (1981). Increased cholesterol sulfate in plasma and red blood cell membranes of steroid sulfatase deficient patients. J. Clin. Endocrinol. Metab. 53, 221–223. doi: 10.1210/jcem-53-1-220
Bietz, A., Zhu, H., Xue, M., and Xu, C. (2017). Cholesterol metabolism in T cells. Front. Immunol. 8:1664. doi: 10.3389/fimmu.2017.01664
Bjorkbom, A., Rog, T., Kankaanpaa, P., Lindroos, D., Kaszuba, K., Kurita, M., et al. (2011). N- and O-methylation of sphingomyelin markedly affects its membrane properties and interactions with cholesterol. Biochim. Biophys. Acta 1808, 1179–1186. doi: 10.1016/j.bbamem.2011.01.009
Blanco, R., Borroto, A., Schamel, W. W., Pereira, P., and Alarcon, B. (2014). Conformational changes in the T cell receptor differentially determine T cell subset development in mice. Sci. Signal 7:ra115. doi: 10.1126/scisignal.2005650
Call, M. E., Pyrdol, J., Wiedmann, M., and Wucherpfennig, K. W. (2002). The organizing principle in the formation of the T cell receptor-CD3 complex. Cell 111, 967–979. doi: 10.1016/S0092-8674(02)01194-7
Chakrabarti, R., and Engleman, E. G. (1991). Interrelationships between mevalonate metabolism and the mitogenic signaling pathway in T lymphocyte proliferation. J. Biol. Chem. 266, 12216–12222.
Chang, T. Y., Chang, C. C., Ohgami, N., and Yamauchi, Y. (2006). Cholesterol sensing, trafficking, and esterification. Annu. Rev. Cell Dev. Biol. 22, 129–157. doi: 10.1146/annurev.cellbio.22.010305.104656
Cherezov, V., Rosenbaum, D. M., Hanson, M. A., Rasmussen, S. G., Thian, F. S., Kobilka, T. S., et al. (2007). High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science 318, 1258–1265. doi: 10.1126/science.1150577
Coskun, U., Grzybek, M., Drechsel, D., and Simons, K. (2011). Regulation of human EGF receptor by lipids. Proc. Natl. Acad. Sci. U.S.A. 108, 9044–9048. doi: 10.1073/pnas.1105666108
Courtney, A. H., Lo, W. L., and Weiss, A. (2018). TCR signaling: mechanisms of initiation and propagation. Trends Biochem. Sci. 43, 108–123. doi: 10.1016/j.tibs.2017.11.008
de la Cruz, J., Kruger, T., Parks, C. A., Silge, R. L., van Oers, N. S., Luescher, I. F., et al. (2011). Basal and antigen-induced exposure of the proline-rich sequence in CD3ε. J. Immunol. 186, 2282–2290. doi: 10.4049/jimmunol.1003225
Demel, R. A., Jansen, J. W., van Dijck, P. W., and van Deenen, L. L. (1977). The preferential interaction of cholesterol with different classes of phospholipids. Biochim. Biophys. Acta 465, 1–10. doi: 10.1016/0005-2736(77)90350-9
Deng, G. M., and Tsokos, G. C. (2008). Cholera toxin B accelerates disease progression in lupus-prone mice by promoting lipid raft aggregation. J. Immunol. 181, 4019–4026. doi: 10.4049/jimmunol.181.6.4019
Dong, D., Zheng, L., Lin, J., Zhang, B., Zhu, Y., Li, N., et al. (2019). Structural basis of assembly of the human T cell receptor-CD3 complex. Nature 573, 546–552. doi: 10.1038/s41586-019-1537-0
Dopfer, E. P., Hartl, F. A., Oberg, H. H., Siegers, G. M., Yousefi, O. S., Kock, S., et al. (2014). The CD3 conformational change in the γδ T cell receptor is not triggered by antigens but can be enforced to enhance tumor killing. Cell Rep. 7, 1704–1715. doi: 10.1016/j.celrep.2014.04.049
Eggeling, C., Ringemann, C., Medda, R., Schwarzmann, G., Sandhoff, K., Polyakova, S., et al. (2009). Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature 457, 1159–1162. doi: 10.1038/nature07596
Endapally, S., Frias, D., Grzemska, M., Gay, A., Tomchick, D. R., and Radhakrishnan, A. (2019). Molecular discrimination between two conformations of sphingomyelin in plasma membranes. Cell 176, 1040–1053. doi: 10.1016/j.cell.2018.12.042
Essen, L., Siegert, R., Lehmann, W. D., and Oesterhelt, D. (1998). Lipid patches in membrane protein oligomers: crystal structure of the bacteriorhodopsin-lipid complex. Proc. Natl. Acad. Sci. U.S.A. 95, 11673–11678. doi: 10.1073/pnas.95.20.11673
Fadeel, B., and Xue, D. (2009). The ins and outs of phospholipid asymmetry in the plasma membrane: roles in health and disease. Crit. Rev. Biochem. Mol. Biol. 44, 264–277. doi: 10.1080/10409230903193307
Fritz, J. M., and Lenardo, M. J. (2019). Development of immune checkpoint therapy for cancer. J. Exp. Med. 216, 1244–1254. doi: 10.1084/jem.20182395
Garboczi, D. N., Ghosh, P., Utz, U., Fan, Q. R., Biddison, W. E., and Wiley, D. C. (1996). Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature 384, 134–141. doi: 10.1038/384134a0
Garcia, K. C., Degano, M., Stanfield, R. L., Brunmark, A., Jackson, M. R., Peterson, P. A., et al. (1996). An alphabeta T cell receptor structure at 2.5 A and its orientation in the TCR-MHC complex. Science 274, 209–219. doi: 10.1126/science.274.5285.209
Ghittoni, R., Patrussi, L., Pirozzi, K., Pellegrini, M., Lazzerini, P. E., Capecchi, P. L., et al. (2005). Simvastatin inhibits T-cell activation by selectively impairing the function of Ras superfamily GTPases. FASEB J. 19, 605–607. doi: 10.1096/fj.04-2702fje
Gil, D., Schamel, W. W., Montoya, M., Sanchez-Madrid, F., and Alarcon, B. (2002). Recruitment of Nck by CD3 epsilon reveals a ligand-induced conformational change essential for T cell receptor signaling and synapse formation. Cell 109, 901–912. doi: 10.1016/S0092-8674(02)00799-7
Gil, D., Schrum, A. G., Alarcon, B., and Palmer, E. (2005). T cell receptor engagement by peptide-MHC ligands induces a conformational change in the CD3 complex of thymocytes. J. Exp. Med. 201, 517–522. doi: 10.1084/jem.20042036
Hanson, M. A., Cherezov, V., Griffith, M. T., Roth, C. B., Jaakola, V. P., Chien, E. Y., et al. (2008). A specific cholesterol binding site is established by the 2.8 A structure of the human beta2-adrenergic receptor. Structure 16, 897–905. doi: 10.1016/j.str.2008.05.001
Hartl, F. A., Beck-Garcia, E., Woessner, N. M., Flachsmann, L. J., Cardenas, R. M. V., Brandl, S. M., et al. (2020). Noncanonical binding of Lck to CD3ε promotes TCR signaling and CAR function. Nat. Immunol. 21, 902–913. doi: 10.1038/s41590-020-0732-3
Hawse, W. F., Champion, M. M., Joyce, M. V., Hellman, L. M., Hossain, M., Ryan, V., et al. (2012). Cutting edge: evidence for a dynamically driven T cell signaling mechanism. J. Immunol. 188, 5819–5823. doi: 10.4049/jimmunol.1200952
Heerklotz, H., and Tsamaloukas, A. (2006). Gradual change or phase transition: characterizing fluid lipid-cholesterol membranes on the basis of thermal volume changes. Biophys. J. 91, 600–607. doi: 10.1529/biophysj.106.082669
Hulce, J. J., Cognetta, A. B., Niphakis, M. J., Tully, S. E., and Cravatt, B. F. (2013). Proteome-wide mapping of cholesterol-interacting proteins in mammalian cells. Nat. Methods 10, 259–264. doi: 10.1038/nmeth.2368
Hunte, C. (2005). Specific protein-lipid interactions in membrane proteins. Biochem. Soc. Trans. 33, 938–942. doi: 10.1042/BST0330938
Iwai, Y., Ishida, M., Tanaka, Y., Okazaki, T., Honjo, T., and Minato, N. (2002). Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc. Natl. Acad. Sci. U.S.A. 99, 12293–12297. doi: 10.1073/pnas.192461099
James, J. R., McColl, J., Oliveira, M. I., Dunne, P. D., Huang, E., Jansson, A., et al. (2011). The T cell receptor triggering apparatus is composed of monovalent or monomeric proteins. J. Biol. Chem. 286, 31993–32001. doi: 10.1074/jbc.M111.219212
Juraske, C., Wipa, P., Morath, A., Hidalgo, J. V., Hartl, F. A., Raute, K., et al. (2018). Anti-CD3 fab fragments enhance tumor killing by human γδ T cells independent of nck recruitment to the γδ T cell antigen receptor. Front. Immunol. 9:1579. doi: 10.3389/fimmu.2018.01579
Jury, E. C., Isenberg, D. A., Mauri, C., and Ehrenstein, M. R. (2006). Atorvastatin restores Lck expression and lipid raft-associated signaling in T cells from patients with systemic lupus erythematosus. J. Immunol. 177, 7416–7422. doi: 10.4049/jimmunol.177.10.7416
Jury, E. C., Kabouridis, P. S., Flores-Borja, F., Mageed, R. A., and Isenberg, D. A. (2004). Altered lipid raft-associated signaling and ganglioside expression in T lymphocytes from patients with systemic lupus erythematosus. J. Clin. Invest. 113, 1176–1187. doi: 10.1172/JCI200420345
Kabouridis, P. S., Janzen, J., Magee, A. L., and Ley, S. C. (2000). Cholesterol depletion disrupts lipid rafts and modulates the activity of multiple signaling pathways in T lymphocytes. Eur. J. Immunol. 30, 954–963. doi: 10.1002/1521-4141(200003)30:3<954::AID-IMMU954>3.0.CO;2-Y
Kaech, S. M., Hemby, S., Kersh, E., and Ahmed, R. (2002). Molecular and functional profiling of memory CD8 T cell differentiation. Cell 111, 837–851. doi: 10.1016/S0092-8674(02)01139-X
Kersh, E. N., Kaech, S. M., Onami, T. M., Moran, M., Wherry, E. J., Miceli, M. C., et al. (2003). TCR signal transduction in antigen-specific memory CD8 T cells. J. Immunol. 170, 5455–5463. doi: 10.4049/jimmunol.170.11.5455
Kim, S. T., Takeuchi, K., Sun, Z. Y., Touma, M., Castro, C. E., Fahmy, A., et al. (2009). The alphabeta T cell receptor is an anisotropic mechanosensor. J. Biol. Chem. 284, 31028–31037. doi: 10.1074/jbc.M109.052712
Krishnan, S., Nambiar, M. P., Warke, V. G., Fisher, C. U., Mitchell, J., Delaney, N., et al. (2004). Alterations in lipid raft composition and dynamics contribute to abnormal T cell responses in systemic lupus erythematosus. J. Immunol. 172, 7821–7831. doi: 10.4049/jimmunol.172.12.7821
Kumar, R., Ferez, M., Swamy, M., Arechaga, I., Rejas, M. T., Valpuesta, J. M., et al. (2011). Increased sensitivity of antigen-experienced T cells through the enrichment of oligomeric T Cell receptor complexes. Immunity 35, 375–387. doi: 10.1016/j.immuni.2011.08.010
Lee, M. S., Glassman, C. R., Deshpande, N. R., Badgandi, H. B., Parrish, H. L., Uttamapinant, C., et al. (2015). A mechanical switch couples t cell receptor triggering to the cytoplasmic juxtamembrane regions of CD3zetazeta. Immunity 43, 227–239. doi: 10.1016/j.immuni.2015.06.018
Levental, I., Grzybek, M., and Simons, K. (2011). Raft domains of variable properties and compositions in plasma membrane vesicles. Proc. Natl. Acad. Sci. U.S.A. 108, 11411–11416. doi: 10.1073/pnas.1105996108
Lillemeier, B. F., Mortelmaier, M. A., Forstner, M. B., Huppa, J. B., Groves, J. T., and Davis, M. M. (2010). TCR and Lat are expressed on separate protein islands on T cell membranes and concatenate during activation. Nat. Immunol. 11, 90–96. doi: 10.1038/ni.1832
Malissen, B. (2003). An evolutionary and structural perspective on T cell antigen receptor function. Immunol. Rev. 191, 7–27. doi: 10.1034/j.1600-065X.2003.00016.x
Marquardt, D., Geier, B., and Pabst, G. (2015). Asymmetric lipid membranes: towards more realistic model systems. Membranes 5, 180–196. doi: 10.3390/membranes5020180
Martin-Blanco, N., Blanco, R., Alda-Catalinas, C., Bovolenta, E. R., Oeste, C. L., Palmer, E., et al. (2018). A window of opportunity for cooperativity in the T cell receptor. Nat. Commun. 9:2618. doi: 10.1038/s41467-018-05050-6
Martinez-Martin, N., Risueno, R. M., Morreale, A., Zaldivar, I., Fernandez-Arenas, E., Herranz, F., et al. (2009). Cooperativity between T cell receptor complexes revealed by conformational mutants of CD3epsilon. Sci. Signal 2:ra43. doi: 10.1126/scisignal.2000402
Martín-Leal, A., Blanco, R., Casas, J., Sáez, M. E., Rodríguez-Bovolenta, E., de Rojas, I., et al. (2020). CCR5 deficiency impairs CD4 + T-cell memory responses and antigenic sensitivity through increased ceramide synthesis. EMBO J. 39:e104749. doi: 10.15252/embj.2020104749
McDonald, G., Deepak, S., Miguel, L., Hall, C. J., Isenberg, D. A., Magee, A. I., et al. (2014). Normalizing glycosphingolipids restores function in CD4+ T cells from lupus patients. J. Clin. Invest. 124, 712–724. doi: 10.1172/JCI69571
Mingueneau, M., Sansoni, A., Gregoire, C., Roncagalli, R., Aguado, E., Weiss, A., et al. (2008). The proline-rich sequence of CD3epsilon controls T cell antigen receptor expression on and signaling potency in preselection CD4+CD8+ thymocytes. Nat. Immunol. 9, 522–532. doi: 10.1038/ni.1608
Minguet, S., Swamy, M., Alarcon, B., Luescher, I. F., and Schamel, W. W. (2007). Full activation of the T cell receptor requires both clustering and conformational changes at CD3. Immunity 26, 43–54. doi: 10.1016/j.immuni.2006.10.019
Molnar, E., Swamy, M., Holzer, M., Beck-Garcia, K., Worch, R., Thiele, C., et al. (2012). Cholesterol and sphingomyelin drive ligand-independent T-cell antigen receptor nanoclustering. J. Biol. Chem. 287, 42664–42674. doi: 10.1074/jbc.M112.386045
Monod, J., Wyman, J., and Changeux, J. P. (1965). On the nature of allosteric transitions: a plausible model. J. Mol. Biol. 12, 88–118. doi: 10.1016/S0022-2836(65)80285-6
Morath, A., and Schamel, W. W. (2020). αβ and γδ T cell receptors: Similar but different. J. Leukoc. Biol. 107, 1045–1055. doi: 10.1002/JLB.2MR1219-233R
Moulton, V. R., and Tsokos, G. C. (2015). T cell signaling abnormalities contribute to aberrant immune cell function and autoimmunity. J. Clin. Invest. 125, 2220–2227. doi: 10.1172/JCI78087
Mueller, V., Ringemann, C., Honigmann, A., Schwarzmann, G., Medda, R., Leutenegger, M., et al. (2011). STED nanoscopy reveals molecular details of cholesterol- and cytoskeleton-modulated lipid interactions in living cells. Biophys. J. 101, 1651–1660. doi: 10.1016/j.bpj.2011.09.006
Mundhara, N., Majumder, A., and Panda, D. (2019). Methyl-β-cyclodextrin, an actin depolymerizer augments the antiproliferative potential of microtubule-targeting agents. Sci. Rep. 21:7638. doi: 10.1038/s41598-019-43947-4
Natarajan, K., McShan, A. C., Jiang, J., Kumirov, V. K., Wang, R., Zhao, H., et al. (2017). An allosteric site in the T-cell receptor Cβ domain plays a critical signalling role. Nat. Commun. 8:15260. doi: 10.1038/ncomms15260
Pageon, S. V., Tabarin, T., Yamamoto, Y., Ma, Y., Bridgeman, J. S., Cohnen, A., et al. (2016). Functional role of T-cell receptor nanoclusters in signal initiation and antigen discrimination. Proc. Natl. Acad. Sci. U.S.A. 113, E5454–E5463. doi: 10.1073/pnas.1607436113
Petersen, T. R., Gulland, S., Bettelli, E., Kuchroo, V., Palmer, E., and Backstrom, B. T. (2004). A chimeric T cell receptor with super-signaling properties. Int. Immunol. 16, 889–894. doi: 10.1093/intimm/dxh098
Platzer, R., Rossboth, B. K., Schneider, M. C., Sevcsik, E., Baumgart, F., Stockinger, H., et al. (2020). Unscrambling fluorophore blinking for comprehensive cluster detection via photoactivated localization microscopy. Nat. Commun. 11:4993. doi: 10.1038/s41467-020-18726-9
Rangarajan, S., He, Y., Chen, Y., Kerzic, M. C., Ma, B., Gowthaman, R., et al. (2018). Peptide-MHC (pMHC) binding to a human antiviral T cell receptor induces long-range allosteric communication between pMHC- and CD3-binding sites. J. Biol. Chem. 293, 15991–16005. doi: 10.1074/jbc.RA118.003832
Risueno, R. M., Gil, D., Fernandez, E., Sanchez-Madrid, F., and Alarcon, B. (2005). Ligand-induced conformational change in the T-cell receptor associated with productive immune synapses. Blood 106, 601–608. doi: 10.1182/blood-2004-12-4763
Risueno, R. M., Schamel, W. W., and Alarcon, B. (2008). T cell receptor engagement triggers its CD3epsilon and CD3zeta subunits to adopt a compact, locked conformation. PLoS ONE 3:e1747. doi: 10.1371/journal.pone.0001747
Rossboth, B., Arnold, A. M., Ta, H., Platzer, R., Kellner, F., Huppa, J. B., et al. (2018). TCRs are randomly distributed on the plasma membrane of resting antigen-experienced T cells. Nat. Immunol. 19, 821–827. doi: 10.1038/s41590-018-0162-7
Rouquette-Jazdanian, A. K., Pelassy, C., Breittmayer, J. P., and Aussel, C. (2006). Revaluation of the role of cholesterol in stabilizing rafts implicated in T cell receptor signaling. Cell. Signal 18, 105–122. doi: 10.1016/j.cellsig.2005.03.024
Rudolph, M. G., Stanfield, R. L., and Wilson, I. A. (2006). How TCRs bind MHCs, peptides, and coreceptors. Annu. Rev. Immunol. 24, 419–466. doi: 10.1146/annurev.immunol.23.021704.115658
Ruprecht, J. J., Mielke, T., Vogel, R., Villa, C., and Schertler, G. F. (2004). Electron crystallography reveals the structure of metarhodopsin I. EMBO J. 23, 3609–3620. doi: 10.1038/sj.emboj.7600374
Schamel, W. W., and Alarcon, B. (2013). Organization of the resting TCR in nanoscale oligomers. Immunol. Rev. 251, 13–20. doi: 10.1111/imr.12019
Schamel, W. W., Alarcon, B., Hofer, T., and Minguet, S. (2017). The allostery model of TCR regulation. J. Immunol. 198, 47–52. doi: 10.4049/jimmunol.1601661
Schamel, W. W., Alarcon, B., and Minguet, S. (2019). The TCR is an allosterically regulated macromolecular machinery changing its conformation while working. Immunol. Rev. 291, 8–25. doi: 10.1111/imr.12788
Schamel, W. W., Arechaga, I., Risueno, R. M., van Santen, H. M., Cabezas, P., Risco, C., et al. (2005). Coexistence of multivalent and monovalent TCRs explains high sensitivity and wide range of response. J. Exp. Med. 202, 493–503. doi: 10.1084/jem.20042155
Sharma, P., Varma, R., Sarasij, R. C., Ira, G., ousset, K., Krishnamoorthy, G., Rao, M., et al. (2004). Nanoscale organization of multiple GPI-anchored proteins in living cell membranes. Cell 116, 577–589. doi: 10.1016/S0092-8674(04)00167-9
Sherman, E., Barr, V., Manley, S., Patterson, G., Balagopalan, L., Akpan, I., et al. (2011). Functional nanoscale organization of signaling molecules downstream of the T cell antigen receptor. Immunity 35, 705–720. doi: 10.1016/j.immuni.2011.10.004
Simons, K., and Ikonen, E. (1997). Functional rafts in cell membranes. Nature 387, 569–572. doi: 10.1038/42408
Steck, T. L., Ye, J., and Lange, Y. (2002). Probing red cell membrane cholesterol movement with cyclodextrin. Biophys. J. 83, 2118–2125. doi: 10.1016/S0006-3495(02)73972-6
Subczynski, W. K., Pasenkiewicz-Gierula, M., Widomska, J., Mainali, L., and Raguz, M. (2017). High cholesterol/low cholesterol: effects in biological membranes: a review. Cell Biochem. Biophys. 75, 369–385. doi: 10.1007/s12013-017-0792-7
Swamy, M., Beck-Garcia, K., Beck-Garcia, E., Hartl, F. A., Morath, A., Yousefi, O. S., et al. (2016). A cholesterol-based allostery model of T cell receptor phosphorylation. Immunity 44, 1091–1101. doi: 10.1016/j.immuni.2016.04.011
Tani-ichi, S., Maruyama, K., Kondo, N., Nagafuku, M., Kabayama, K., Inokuchi, J., et al. (2005). Structure and function of lipid rafts in human activated T cells. Int. Immunol. 17, 749–758. doi: 10.1093/intimm/dxh257
Thiele, C., Hannah, M. J., Fahrenholz, F., and Huttner, W. B. (2000). Cholesterol binds to synaptophysin and is required for biogenesis of synaptic vesicles. Nat. Cell Biol. 2, 42–49. doi: 10.1038/71366
van Meer, G., Voelker, D. R., and Feigenson, G. W. (2008). Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9, 112–124. doi: 10.1038/nrm2330
Veatch, S. L., Polozov, I. V., Gawrisch, K., and Keller, S. L. (2004). Liquid domains in vesicles investigated by NMR and fluorescence microscopy. Biophys. J. 86, 2910–2922. doi: 10.1016/S0006-3495(04)74342-8
Veiga, M. P., Arrondo, J. L., Goni, F. M., Alonso, A., and Marsh, D. (2001). Interaction of cholesterol with sphingomyelin in mixed membranes containing phosphatidylcholine, studied by spin-label ESR and IR spectroscopies. A possible stabilization of gel-phase sphingolipid domains by cholesterol. Biochemistry 40, 2614–2622. doi: 10.1021/bi0019803
Wang, F., Beck-Garcia, K., Zorzin, C., Schamel, W. W., and Davis, M. M. (2016). Inhibition of T cell receptor signaling by cholesterol sulfate, a naturally occurring derivative of membrane cholesterol. Nat. Immunol. 17, 844–850. doi: 10.1038/ni.3462
Weiss, A. (2010). The right team at the right time to go for a home run: tyrosine kinase activation by the TCR. Nat. Immunol. 11, 101–104. doi: 10.1038/ni0210-101
Wood, W. G., Igbavboa, U., Müller, W. E., and Eckert, G. P. (2011). Cholesterol asymmetry in synaptic plasma membranes. J. Neurochem. 116, 684–689. doi: 10.1111/j.1471-4159.2010.07017.x
Wu, W., Shi, X., and Xu, C. (2016). Regulation of T cell signalling by membrane lipids. Nat. Rev. Immunol. 16, 690–701. doi: 10.1038/nri.2016.103
Xu, C., Gagnon, E., Call, M. E., Schnell, J. R., Schwieters, C. D., Carman, C. V., et al. (2008). Regulation of T cell receptor activation by dynamic membrane binding of the CD3ε cytoplasmic tyrosine-based motif. Cell 135, 702–713. doi: 10.1016/j.cell.2008.09.044
Yang, W., Bai, Y., Xiong, Y., Zhang, J., Chen, S., Zheng, X., et al. (2016). Potentiating the antitumour response of CD8+ T cells by modulating cholesterol metabolism. Nature 531, 651–655. doi: 10.1038/nature17412
Keywords: cholesterol, lipid, TCR, signaling, T cell, nanocluster, allostery
Citation: Pathan-Chhatbar S, Drechsler C, Richter K, Morath A, Wu W, OuYang B, Xu C and Schamel WW (2021) Direct Regulation of the T Cell Antigen Receptor's Activity by Cholesterol. Front. Cell Dev. Biol. 8:615996. doi: 10.3389/fcell.2020.615996
Received: 10 October 2020; Accepted: 09 December 2020;
Published: 08 January 2021.
Edited by:
Enrique Aguado, University of Cádiz, SpainReviewed by:
Elizabeth C. Jury, University College London, United KingdomHannes Stockinger, Medical University of Vienna, Austria
Copyright © 2021 Pathan-Chhatbar, Drechsler, Richter, Morath, Wu, OuYang, Xu and Schamel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wolfgang W. Schamel, d29sZmdhbmcuc2NoYW1lbEBiaW9sb2dpZS51bmktZnJlaWJ1cmcuZGU=
 Salma Pathan-Chhatbar
Salma Pathan-Chhatbar Carina Drechsler1,2,3
Carina Drechsler1,2,3 Kirsten Richter
Kirsten Richter Wei Wu
Wei Wu Bo OuYang
Bo OuYang Chenqi Xu
Chenqi Xu Wolfgang W. Schamel
Wolfgang W. Schamel