- Department of Pediatric Hemato-Oncology and Cell and Gene Therapy, IRCCS, Bambino Gesù Children’s Hospital, Rome, Italy
Autophagy is an intracellular degradation process involved in the removal of proteins and damaged organelles by the formation of a double-membrane vesicle named autophagosome and degraded through fusion with lysosomes. An intricate relationship between autophagy and the endosomal and exosomal pathways can occur at different stages with important implications for normal physiology and human diseases. Recent researches have revealed that extracellular vesicles (EVs), such as exosomes, could have a cytoprotective role by inducing intracellular autophagy; on the other hand, autophagy plays a crucial role in the biogenesis and degradation of exosomes. Although the importance of these processes in cancer is well established, their interplay in tumor is only beginning to be documented. In some tumor contexts (1) autophagy and exosome-mediated release are coordinately activated, sharing the molecular machinery and regulatory mechanisms; (2) cancer cell-released exosomes impact on autophagy in recipient cells through mechanisms yet to be determined; (3) exosome-autophagy relationship could affect drug resistance and tumor microenvironment (TME). In this review, we survey emerging discoveries relevant to the exosomes and autophagy crosstalk in the context of cancer initiation, progression and recurrence. Consequently, we discuss clinical implications by targeting autophagy-exosomal pathway interaction and how this could lay a basis for the purpose of novel cancer therapeutics.
Autophagy: An Overview
Autophagy is a self-degradative process occurring in all eukaryotic cells for maintaining homeostasis and cell survival. In basal conditions, autophagy degrades cytosolic materials such as long-lived proteins and old organelles for renewal of cellular components. During stressful conditions such as starvation or hypoxia, autophagy is induced to recycle macromolecules, providing energy and nutrients. Autophagy begins with the formation of a unique smooth double-membrane phagophore that traps cytosolic materials; after closure, autophagosome vesicle is formed and upon fusion with lysosomes, the inner membrane and the cargos are degraded and subsequently recycled (Yu et al., 2018). Three morphologically and mechanistically distinct types of autophagy have been described: macroautophagy (here referred to as autophagy), microautophagy and chaperone-mediated autophagy (CMA) (Abdrakhmanov et al., 2020). They differ substantially for cargo selection and delivery mechanism into lysosomes: macroautophagy is characterized by the formation of autophagosome, while during microautophagy the cargos are sequestered by direct invagination of the vacuole membrane. CMA only occurs in mammalian cells and uses chaperones to identify cargo proteins that contain a particular pentapeptide motif that are translocated directly across the lysosomal membrane. Although autophagy was initially thought to be a non-selective degradation mechanism, now it is clear that it allows the removal of specific cellular components such as mitochondria (mitophagy), aggregates (aggrephagy), or invading pathogens (xenophagy) (Gatica et al., 2018).
Autophagosome formation is driven by the autophagy-related (ATG) proteins that are both spatially and temporally controlled and are divided into distinct units: ULK complex, the class III phosphatidylinositol 3-kinase (PI3K) complex, the ATG2-ATG18/WIPI4 complex, ATG9, the ATG12 conjugation system and the ATG8/LC3 conjugation system (Nishimura and Tooze, 2020).
The role of autophagy has been explored in many fields (Yang and Klionsky, 2020). In cancer progression, autophagy has a dual and paradoxical role: while at early stages autophagy acts as a tumor suppressor mechanism, in advanced stages it has a fundamental role in tumor survival being active in response to cellular stress (White, 2015; Keulers et al., 2016; Cotzomi-Ortega et al., 2018; Amaravadi et al., 2019, 2020; Mulcahy Levy and Thorburn, 2020).
Exosomes: Biogenesis, Release and Functions
According to the International Society for Extracellular Vesicles (ISEV) which provides guidelines for the classification of extracellular vesicles (EVs), exosomes are nano-sized (30–120 nm) (Théry et al., 2018). EVs secreted by all cell types that play a role in cell-cell communication through the transfer of active biomolecules such as proteins, lipids, RNAs, DNA and microRNAs (Raposo and Stoorvogel, 2013). Exosome precursors, named intraluminal vesicles (ILVs), derive from the membrane of endocytic cisternae by inward budding of microdomains. Upon ILVs accumulation, the cisternae become multivesicular bodies (MVBs) that undergo exocytic fusion with the plasma membrane followed by release of their ILVs to the extracellular space (Raposo and Stoorvogel, 2013; Cocucci and Meldolesi, 2015). Due to the mechanism of biogenesis, the exosomal membrane has the same orientation as the parental cell plasma membrane, and it is enriched in endosome-related proteins, lipids and tetraspanins. The exact mechanisms involved in exosomes packaging have not been fully elucidated but their secretion requires formation of an endosomal-sorting complex that is required for transport (ESCRT) (Scita and Di Fiore, 2010). ESCRT is comprised of four complexes (ESCRT−0, −I, −II, and −III) and associated proteins (vacuolar protein sorting-associated protein 4 (VPS4), tumor Susceptibility 101 (Tsg101) and ALIX) (Schuh and Audhya, 2014). In addition to ESCRT, which recognizes ubiquitylated proteins, other ESCRT-independent mechanisms operate to generate exosomes (Stuffers et al., 2009). These unconventional ESCRT-independent pathways seem to be driven by the presence of certain lipids, such as ceramides and lysobisphosphatidic acid (Matsuo et al., 2004; Babst, 2011). The release of exosomes into the extracellular environment requires the transport and docking of MVBs as well as their fusion with the plasma membrane (van Niel et al., 2006). These processes need several factors including molecular switches, cytoskeleton, molecular motors and the membrane fusion apparatus. It has been proposed that exosome release is a Ca2+-dependent (Savina et al., 2003) and pH-dependent (Parolini et al., 2009) process. After secretion, exosomes can be transferred to recipient cells via clathrin-mediated endocytosis (Tian et al., 2014), lipid raft-mediated endocytosis (Svensson et al., 2013), heparin sulfate proteoglycans-dependent endocytosis (Christianson et al., 2013), or phagocytosis (Feng et al., 2010). These pathways lead to different sorting and fate of exosomal cargo and the route by which exosomes are internalized appears to be cell and context specific. Tian et al. (2014) have showed that clathrin-mediated endocytosis and macropinocytosis are involved in the up-take of PC12-derived exosomes through a receptor-mediated mechanism. Svensson et al. (2013) have demonstrated that the signaling status of recipient cells is important in determining the pathway by which exosomes are internalized: exosomes derived from glioblastoma (GBM) cells, indeed, trigger lipid raft-mediated endocytosis where ERK activation is required. Furthermore, Christianson and co-workers provide evidences that exosomes produced by GBM cells require heparan sulfate proteoglycans for internalization and this affects the functional effects of exosomes in cancer cells (Christianson et al., 2013). Feng et al. (2010) have highlighted the role of the type of recipient cell in determining how exosomes are internalized: they have showed that phagocytic cells internalize exosomes via phagocytosis while in non-phagocytic cells exosomes attach to the cell membrane. In target cells, molecules carried by exosomes can trigger and influence several processes both in physiological and pathological conditions. In recent years, numerous evidence highlights the involvement of exosomes in angiogenesis promotion (Skog et al., 2008; Hong et al., 2009; Ahmadi and Rezaie, 2020), suppression of immune response (Yu et al., 2007; Clayton et al., 2008), induction of invasive (Luga et al., 2012; Guo et al., 2019; Jabbari et al., 2020a) and metastatic phenotype (Peinado et al., 2012), formation of pre-metastatic niche (Costa-Silva et al., 2015; Colletti et al., 2020). Moreover, tumoral exosomes can induce drug resistance carrying miRNAs that target antiapoptotic and immune-suppressive pathways or ABC transporters, which export chemotherapeutic agents out of recipient cells (Santos and Almeida, 2020). Given their involvement in cancer progression and their presence in different biological fluids, there have been increasing efforts toward their characterization as a source of possible diagnostic and prognostic biomarkers even in pediatric oncology (Colletti et al., 2017, 2019, 2020; Galardi et al., 2019, 2020) and as a delivery tool for biomedical applications (Rezaie et al., 2018; Rahbarghazi et al., 2019; Jabbari et al., 2020b; Wu Z. et al., 2020).
Crosstalk Between Autophagy and Exosome Biogenesis
An intricate relationship among autophagy and the exosome biogenesis (Figure 1A) occurs at different stages (Buratta et al., 2020; Hassanpour et al., 2020; Salimi et al., 2020). If in some cellular contexts autophagy and exosome production act at the same time to counter cellular stress (Kumar et al., 2014), in other circumstances the two processes can compensate each other. In fact, dysfunctional MVBs can be degraded by autophagy and the inhibition of lysosomal function or autophagy restores exosome secretion (Villarroya-Beltri et al., 2016). Moreover, EVs can have a cytoprotective role by inducing intracellular autophagy and, on the other hand, autophagy regulates the biogenesis and degradation of EVs (Xu et al., 2018). Finally, emerging evidence supports a role of both autophagy and exosomes in contributing to the export of cytokines or proteins by an unconventional secretory pathway (Ponpuak et al., 2015; Zhang et al., 2015; Kimura et al., 2017). The main advances about the crosstalk between these pathways are summarized below.
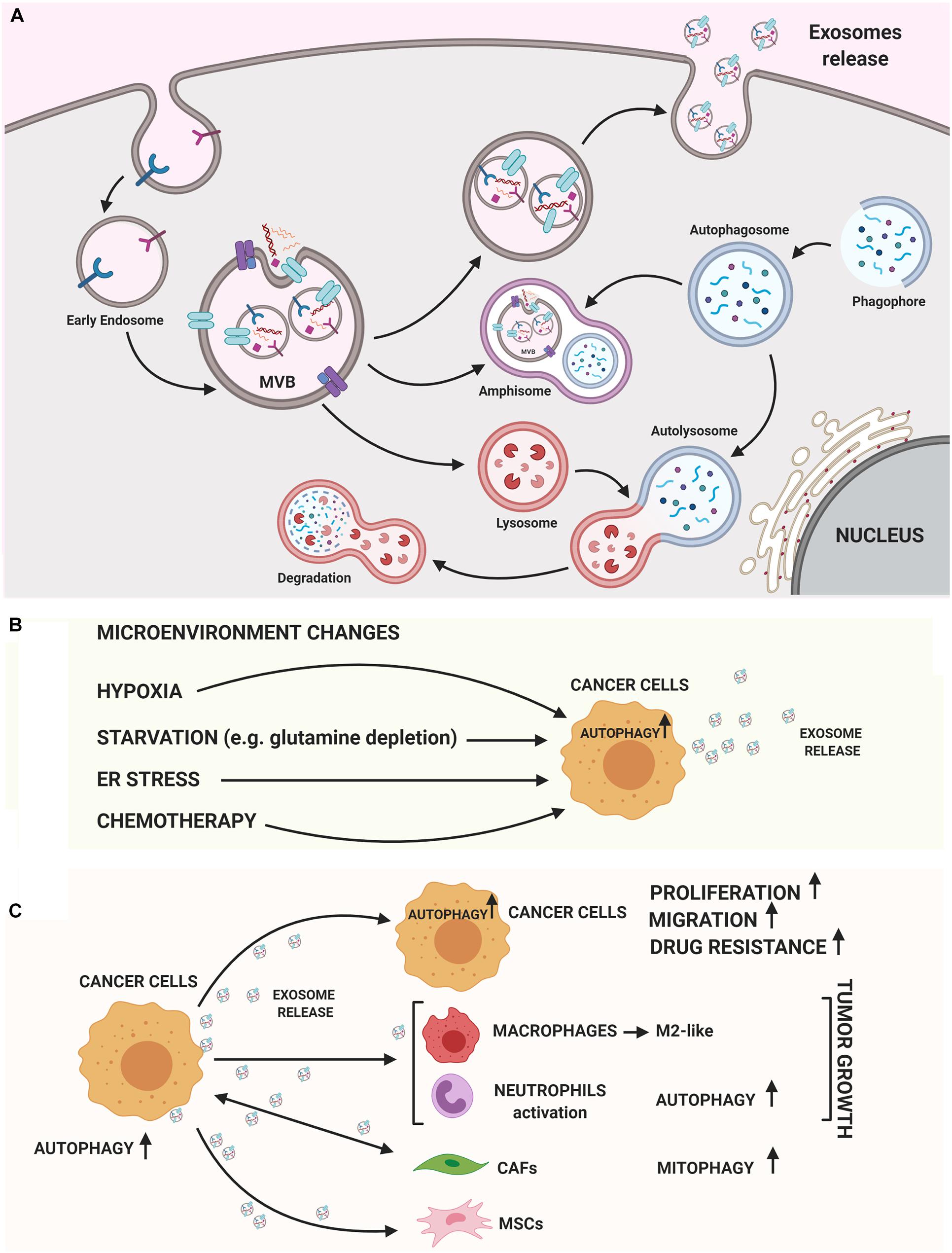
Figure 1. Autophagy and exosomes relationship. (A) A crosstalk between exosome biogenesis and autophagy flux occurs both at molecular level and at membranous vesicles such as amphisomes. In the cytoplasm several Rab-GTPase proteins regulate the movement of vesicles between autophagy and exosomal secretory pathway. On the MVB’s membrane different autophagic proteins such as LC3B, ATG5, and ATG16L1 participate to generate exosomes. Then, exosomes can carry autophagic cargo and secrete this into extracellular milieu. (B) Both autophagy and exosome release are strongly stimulated by TME conditions (hypoxia, starvation, ER stress) or chemotherapy treatments. (C) Exosomes released by cancer cells can induce autophagy in recipient cells, stimulating growth, migration and enhancing drug resistance. On the other hand, cancer cell-released exosomes can activate pro-tumoral stromal or immune cells via autophagy-related mechanisms or MSCs-derived exosomes may induce oncogenic autophagy in recipient cancer cells. The figure was performed with https://biorender.com.
Molecular Interaction Mechanism
Some studies are emerging about how individual ATG proteins can regulate exosome biogenesis and secretion (Salimi et al., 2020). Intriguingly, it has been observed that ATG5, which participates at the stage of autophagosome precursor synthesis (Nishimura and Tooze, 2020), allows the dissociation of vacuolar proton pumps (V1V0-ATPase) from the MVBs, thus inhibiting the acidification of MVB-lumen and contributing to the fusion of MVB with the plasma membrane (Guo et al., 2017) in a canonical autophagy-independent manner. The treatment with V-ATP inhibitors of Atg5 knockout cells demonstrated that luminal pH plays a role in controlling whether MVBs must undergo fusion with lysosomes for degradation or with plasma membrane for exosomes release (Mauthe et al., 2018). Guo et al. (2017) have demonstrated that the down-regulation of both ATG16L1, a core autophagy protein implicated at distinct phases of autophagosome biogenesis (Nishimura and Tooze, 2020), and ATG5 reduces exosome biogenesis in breast cancer cells; this, in turn, decreases tumor metastasis. Moreover, G alpha interacting protein (GAIP) and GAIP interacting protein C-terminus (GIPC), two proteins initially identified for G-protein coupled receptor subunit GI alpha (De Vries et al., 1998), can simultaneously stimulate exosome biogenesis and autophagy flux in pancreatic tumor cells (Bhattacharya et al., 2014).
Murrow et al. (2015) have demonstrated that the inhibition of ATG12–ATG3, a complex essential for a late step of autophagosome formation (Nishimura and Tooze, 2020), changes the form of MVBs, disrupts late endosome trafficking and reduces exosome biogenesis. This occurs through an interaction between ATG12-ATG3 and ALIX, a protein implicated in membrane fission which interacts with ESCRT members involved in exosomes release. ALIX inhibition also reduces basal autophagy flux, indicating a reciprocal regulation between autophagy and exosome biogenesis. Moreover, loss of ALIX or depletion of ATG12-ATG3 complex does not impact on starvation-induced autophagy, specifying the association of different complexes which control basal and stress-induced autophagy (Murrow et al., 2015).
Interestingly, a study perfomed by Bader and collaborators reports that the transmembrane protein ATG9 is implicated in the formation of ILVs in Drosophila melanogaster. In basal conditions, depletion of ATG9 leads to both autophagy inhibition and decrease of the ILVs content of amphisomes and autolysosome (Bader et al., 2015).
One of the key autophagy players is MAP1LC3B, Microtubule Associated Protein 1 Light Chain 3 Beta (or LC3B). LC3B is one of the main autophagy flux markers: in the initiation step, LC3B conjugation complex induces autophagosome biogenesis through ULK activation; during the maturation step, LC3B mediates closure, fusion and transport of the autophagosome (Mizushima et al., 2011). LC3B is incorporated into autophagosome membranes but it is also recruited to single-membrane phagosomes in a process called LC3-associated phagocytosis (LAP), which does not require the formation of autophagosomes (Florey et al., 2011; Martinez et al., 2011). LC3B has been found into exosomes (LC3-I rather than LC3-II) in an ATG7-independent manner, suggesting that the LAP-like lipidation mechanism could share a non-degradative role in exosome secretion. Furthermore, a very recent work has identified a new secretory mechanism where components of LC3 conjugation complex favor the association with RNA binding proteins (RBPs) and small non-coding RNAs into EVs, resulting in their secretion outside of cells (Leidal et al., 2020). This process has been defined as LDELS: LC3-Dependent EV Loading and Secretion. Using a proximity-dependent biotinylation proteomics strategy, the authors found that this mechanism does not require canonical autophagy but only components of the LC3 conjugation machinery, linking exosome secretion pathway, extracellular RNA release and autophagy in a very fascinating way. Finally, although not designated as ATGs, soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins are also an example of the interplay between autophagy and exosome secretion (Zhao and Zhang, 2019). SNARE complexes (such as STX17-SNAP29-VAMP7/VAMP8 or STX7-SNAP29-YKT6) mediate autophagosome-lysosome fusion; however, secretory autophagy requires different SNAREs than degradative autophagy (such as Sec22b), adding further to the complexity of this crosstalk (Zhao and Zhang, 2019).
Interestingly, growing evidence indicates that exosomes could contain ATG proteins. For example, Sequestosome 1 (SQSTM1), a classical receptor of autophagy, Neighbor Of BRCA1 Gene 1 Protein (NBR1), a selective autophagy receptor, WD Repeat Domain, Phosphoinositide Interacting 2 (WIPI2), a component of the autophagy machinery, or LC3 were found into exosomal fractions in PC-3 cells; intriguingly, both SQSTM1 and CD63, used as a representative exosomal marker protein, were found in the same MVB-like organelles inside the cells (Hessvik et al., 2016). Minakaki et al. (2018) have discovered that, in neuronal cells, autophagy inhibition increases alpha-synuclein levels in EVs released in human cerebrospinal fluid. These vesicles are biochemically characterized by the presence of both LC3-II and SQSTM1 together with classical MVB-EV markers. This study provides, for the first time, the presence of EVs with a hybrid autophagosome-exosome-like profile.
Vesicular Interaction Mechanism
Antagonist interaction between autophagy and exosomes release has been well-defined in the context of amphisomes biogenesis. Amphisomes are degradative hybrid compartments formed after fusion between autophagosomes and MVBs, which can then fuse with lysosomes (Liou et al., 1997). As an example, autophagy induction supports MVB-autophagosome fusion and leads to a reduction of exosomes release (Fader et al., 2008). On the other hand, autophagy inhibition rescues exosome secretion, suggesting an involvement of autophagy in the lysosome-dependent degradation of MVBs (Villarroya-Beltri et al., 2016).
Recently, using biochemical, electron microscopy and fluorescence microscopy-based approaches, Ariotti et al. (2020) dissect a novel autophagy-based secretion of Caveolin 1 (CAV1) in addition to conventional exosome-based release. In details, in pancreatic cancer cells, they identified a novel class of exosomes, enriched in CAV1 (50–60 copies), and released via a non-canonical secretory autophagy pathway.
Autophagy and Exosomes Relationship in Cancer
Although the importance of autophagy and exosomes in tumor progression is well-documented (Yu et al., 2007; Clayton et al., 2008; Skog et al., 2008; Hong et al., 2009; Svensson et al., 2013; Keulers et al., 2016; Ahmadi and Rezaie, 2020; Amaravadi et al., 2020), in recent years the understanding of their connection and interplay in cancer has aroused a lot of interest (Table 1; Kulshreshtha et al., 2007; Bellot et al., 2009; Chiavarina et al., 2010; Mazure and Pouysségur, 2010; White et al., 2010; Aga et al., 2014; Dutta et al., 2014; Li et al., 2016; Zhang et al., 2018, 2019; Jin et al., 2019; Meng et al., 2019; Shao et al., 2019; Wang et al., 2019, 2020; Yeon et al., 2019; Yuwen et al., 2019; Zeng et al., 2019; Zhou et al., 2019; Dai et al., 2020; Han et al., 2020; Huang et al., 2020; Kulkarni et al., 2020; Kumar and Deep, 2020; Liu et al., 2020; Sung et al., 2020; Wu X. et al., 2020; Yao et al., 2020).
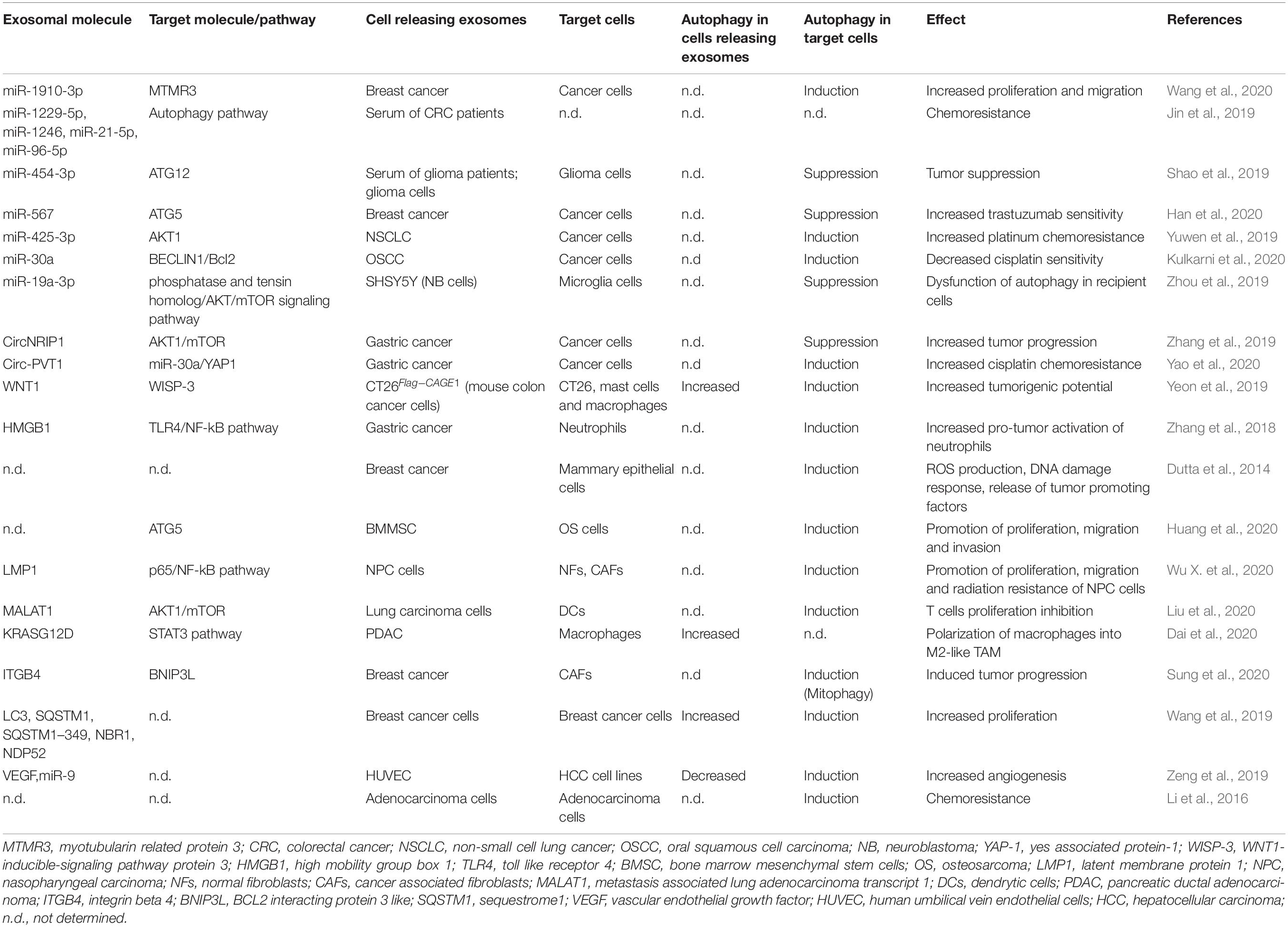
Table 1. Summary of the exosomal molecules that regulate autophagy in target cells or whose release is regulated by autophagy in cancer models.
Are Autophagy and Exosome-Mediated Release Coordinately Induced During Carcinogenesis?
In tumor cells, both autophagy and exosome release are strongly activated, suggesting that both these pathways are a part of cancer cells response (Figure 1B). This coordinated activation may represent an adaptive stress response, although the molecular details are not yet understood. Hypoxic tumor microenvironment (TME) is a key feature in many solid tumors and it is associated with unfavorable prognosis. In tumors, both starvation and hypoxia induce autophagy, which avoids inflammation and cell death (Bellot et al., 2009; Chiavarina et al., 2010; Mazure and Pouysségur, 2010; White et al., 2010). Many studies have shown that cancer cells secrete a higher number of exosomes under hypoxic conditions (Kumar and Deep, 2020) and hypoxia is able to alter the proteomic and nuclear acid profiles of exosomes (Meng et al., 2019). Interestingly, HIF-1α was found in exosomes with transcriptional activity (Aga et al., 2014), representing a potential cancer biomarker. In addition, several miRNAs under the transcriptional control of HIF-1α are enriched in EV derived from hypoxic cells; among these, miR-23a targets BCL2 Interacting Protein 3 Like (BNIP3L), a crucial mitophagy receptor (Kulshreshtha et al., 2007). Moreover, high levels of the HIF1-α transcriptional target BCL2 Interacting Protein 3 (BNIP3) mRNA, another mitophagy receptor, are found in EV produced by hypoxic glioma cells (Kucharzewska et al., 2013).
In cancer cells autophagy and exosome release may be concomitantly up-regulated in response to other cellular stressors such as unfolding protein response (UPR) and endoplasmic reticulum (ER) stress. ER stress is known to increase autophagy in several types of normal and tumor cells (Verfaillie et al., 2010; Corazzari et al., 2017). Kanemoto et al. (2016) found that MVB formation and exosomes release are enhanced by ER stress; furthermore, the down-regulation of both Inositol-Requiring Protein 1 (IRE1α) and PKR-like ER kinase (PERK), two key players of UPR pathway, impacts on exosome production. In addition, the spliced form of X-box binding protein 1 (sXBP1), a key transcription factor that promotes UPR, was found in exosomes, suggesting the transmission outside the cell of UPR mechanism, following exposure to stresses (Hosoi et al., 2018).
Very recently, using Drosophila model and human cell lines, Fan et al. (2020) have found that glutamine depletion induces secretion of exosomes carrying exclusive cargos created in Rab11−positive recycling endosomal MVBs. Interestingly, the release of exosomes from glutamine depleted HCT116 cells stimulate angiogenesis and enhances tumor cell proliferation. Glutamine depletion is likely to be an autophagy inducer determining tumor growth (Tan et al., 2017), supporting the concept of a strict connection between autophagy and exosome secretion as a part of neoplastic cells response.
Beside microenvironmental conditions, up-regulation of both autophagy and exosome release has been well-recognized after chemotherapy treatments (Bandari et al., 2018; Yun and Lee, 2018; Ab Razak et al., 2019; Kang et al., 2020) and in mediating chemoresistance (Yun and Lee, 2018; Ender et al., 2019; Nazio et al., 2019; Steinbichler et al., 2019). For example, exosomes can carry functional plasma membrane transporter proteins from resistant cells to their drug-sensitive ones (Lu et al., 2013) or can sequestrate drugs reducing their concentration (Goler-Baron et al., 2012). It is unknown whether up-regulation of both autophagy and exosome secretion is part of the resistance mechanism or a consequence of cellular phenotype changes. Nevertheless, autophagy inhibiiton and modulation of exosome release may serve for therapeutic approaches and needs to be investigated.
How Do Exosomes Released by Tumor Influence Autophagy in Recipient Cells?
Specific cancer exosomal miRNAs and proteins seem to have a crucial role in determining an ATG response (Figure 1C; Jin et al., 2019; Shao et al., 2019; Yuwen et al., 2019; Han et al., 2020; Kulkarni et al., 2020; Wang et al., 2020). Recently, exosomes carrying miR-1910-3p secreted by breast cancer cells have been found to promote tumor development inducing proliferation, migration and autophagy in recipient mammary epithelial cells and breast cancer cells (Table 1; Wang et al., 2020). Several recent studies have shown that specific exosomal miRNAs regulate autophagy-dependent therapy resistance in recipient cells (Kaminskyy et al., 2012; Zhou et al., 2012; Dutta et al., 2014; Zhang et al., 2018; Deng et al., 2019; Huang et al., 2020; Kang et al., 2020). In breast cancer, exosomal miR-567 down-regulates ATG5 and consequently autophagy, reversing trastuzumab resistance (Dutta et al., 2014). In cisplatin-resistant non-small cell lung cancer (NSCLC), exosomal miR-425-3p down-regulates AKT1 inducing autophagy and leading to therapeutic failure both in early and advanced stages (Yuwen et al., 2019). Also, in oral squamous cell carcinoma (OSCC), exosomal miR-30a modulates cisplatin-sensitivity reducing autophagy via Beclin1 and Bcl2 (Kulkarni et al., 2020). These studies are in support for the use of exosome- mediated miRNA delivery as an effective therapeutic approach. In a very recent paper, exosomal circRNA-plasmacytoma variant translocation 1 (circ-PVT1) intensifies cisplatin-resistant gastric cells through modulating autophagy, invasion, and apoptosis; circ-PVT1 negatively controls miR-30a-5p that, in turn, regulates Yes-associated protein 1 (YAP1) levels (Yao et al., 2020). Besides general autophagy, there is some evidence that also specific forms of autophagy could be modulated by exosomes. Liu et al. (2019) showed that hepatitis B virus (HBV)-infected liver cancer cells-derived exosomes promote liver cancer chemoresistance by modulating the CMA pathway. Specifically, an increased expression of lysosome associated-membrane protein type 2A (LAMP2A), a membrane protein that acts as a specific receptor for the CMA, was observed in cells treated with HBV-associated exosomes and this is associated with a down-regulation of cell death after oxaliplatin treatment due to the activation of the CMA pathway. This is the first study investigating the connection between CMA and exosome release in cancer drug resistance; it proposes the targeting of exosomes to increase chemosensitivity in patients with HBV-liver cancer.
In addition to having a role on tumor cells communication, cancer cell-released exosomes are also able to modulate ATG mechanisms in surrounding stromal and immune cells to support tumor progression. In a study performed by Zhang et al. (2018) suggest that gastric cancer cell-derived exosomes induce autophagy and pro-tumor activation of neutrophils, which, in turn, promote gastric cancer cell migration. Other authors have begun to investigate the importance of exosomes-autophagy interplay between normal and neoplastic cells in supporting carcinogenesis. For example, mesenchymal stem cells (MSCs)-derived exosomes have been identified to modulate autophagy in pathological conditions such as during ischemia or spinal cord injury (Baixauli et al., 2014; Tian et al., 2019) and, more recently, in cancer (Huang et al., 2020). Huang et al. (2020) indeed, found that MSCs-derived exosomes promote osteosarcoma development and invasion by inducing autophagy.
Dai et al. (2020) found that extracellular KRASG12D is packaged into exosomes and transferred, through them, from cancer cells to macrophages. In pancreatic ductal adenocarcinoma (PDAC), G12D is the most frequent mutation in KRAS. In this work the authors demonstrate that oxidative-stress induced autophagy regulates KRASG12D protein release from PDAC cells, and this drives macrophages polarization into pro-tumor M2-like tumor-associated macrophages. Given that autophagy can influence exosome release, a novel study discovers a potential strategy to counteract esophangeal squamous cell carcinoma (ESCC) growth by affecting autophagy and exosome-mediated paracrine senescence (Zheng et al., 2020). Sulforaphane, an isothiocyanate derived from cruciferous vegetables, inhibited fusion process between autophagosome and lysosome resulting in significantly higher exosome release; these exosomes evidently trigger senescence of receipt ESCC cells in a ROS-mTOR-dependent manner. This is in line with the idea that defects in autophagy avoid the effective degradation of intracellular aggregates and exosome discharge may be increased to improve the proteotoxic stress. Another study proposes a link between mitochondria-selective autophagy and exosome content in cancer. Sung et al. (2020) reported that triple negative breast cancer-derived exosomal Integrin beta 4 (ITGB4) induces a metabolic reprogramming in cancer-associated fibroblasts (CAFs) that, in turn, supports tumor progression. Exosomal ITGB4 triggers the conversion of pyruvate to lactate in CAFs via BCL2 Interacting Protein 3 Like (BNIP3L)-dependent mitophagy. The produced lactate is released in the extracellular space and then taken-up by breast cells. This study suggests that ITGB4-induced mitophagy could be a novel target for cancer therapy.
Unveiling Exosomal Contents as New Frontier for Autophagy Modulation and Cancer Treatment
In the era of precision medicine, the development of targeted drugs is also addressing several efforts in investigating new pharmaceutical compound that can modulate autophagy, overcoming the stress tolerance of the tumor and undermining the mechanism of survival of tumor cells. In cancer biology, autophagy plays dual role in both tumor promotion and suppression. In this context the choice to induce or inhibit autophagy is related to the role of autophagy in each specific cancer. A large number of clinical trials using autophagy inhibitors (Malhotra et al., 2019) or activators (Geissler et al., 2016; Rodríguez-Perálvarez et al., 2018; Kulka et al., 2020) are ongoing and, when used in association with anti-cancer drugs, can sensitize chemoresistant cells to treatment (Singh et al., 2018). Chloroquine/Hydroxychloroquine is the only autophagy inhibitor that has been approved by the FDA; however, it also has many off-target effects and the majority of clinical trials have been performed in patients with no specific selected criteria beyond the tumor type.
Given the close relationship between autophagy and exosome pathways in cancer, a better understanding of the biological basis of this complex dialog will help to design specific therapeutic strategy (Lin et al., 2019), such as nano-carriers therapy, to modulate autophagy. Although the use of nanotechnology for the delivery of drugs/biological products targeting autophagy is largely unexplored, investigation of exosome cargo contents could offer opportunities for affecting autophagy in a specific tumor context. In particular, for the treatment of personalized cancer, antagonistic oligonucleotides (antagomiRs, anti-miRs) may be designed for the development of autophagy-modulating therapy, increasing cell chemo-sensitivity and overcoming drug resistance. Proof-of-concept studies are required to understand the role of autophagy in each tumoral context and whether triggering or suppressing autophagy (by specific miRNAs/anti-MiRs) could counteract tumor aggressiveness and progression. To this regard, nanoparticles as miRNAs/miRs delivery systems for modulation of autophagy could be a promising therapeutic strategy.
Conclusion
Autophagy and exosome pathways are strictly interconnected at several levels. In cancer, increasing evidence discussed above indicate a crucial interplay between these processes. Although exosomes control of autophagy is context-dependent, targeting the exosomal pathway to modulate autophagy may suggest a basis for aiming novel cancer therapeutics that need to be further studied. Moreover, the biomarker application of the regulatory factors of both autophagy and exosome signaling has been proposed. However, the effects of their interaction are intricate and TME-dependent and therefore need further valuations.
Author Contributions
FN conceptualized the work and together with AD, DC, and MC prepared a draft of the manuscript text. MC contributed to the figures development. FN, AD, and MC critically reviewed and edited the manuscript. All authors discussed the reported information and commented on the manuscript.
Funding
This work was supported by grants from the Ministero della Salute (GR-2016-02364088 to AD) and Ricerca Corrente (to AD and FN). Dr. Marta Colletti (MC) was supported by Fondazione Umberto Veronesi.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Valentina Polcini for proofreading of the manuscript.
References
Ab Razak, N. S., Ab Mutalib, N. S., Mohtar, M. A., and Abu, N. (2019). Impact of chemotherapy on extracellular vesicles: understanding the chemo-EVs. Front. Oncol. 9:1113. doi: 10.3389/fonc.2019.01113
Abdrakhmanov, A., Gogvadze, V., and Zhivotovsky, B. (2020). To eat or to die: deciphering selective forms of autophagy. Trends Biochem. Sci. 45, 347–364. doi: 10.1016/j.tibs.2019.11.006
Aga, M., Bentz, G. L., Raffa, S., Torrisi, M. R., Kondo, S., Wakisaka, N., et al. (2014). Exosomal HIF1α supports invasive potential of nasopharyngeal carcinoma-associated LMP1-positive exosomes. Oncogene 33, 4613–4622. doi: 10.1038/onc.2014.66
Ahmadi, M., and Rezaie, J. (2020). Tumor cells derived-exosomes as angiogenenic agents: possible therapeutic implications. J. Transl. Med. 18:249. doi: 10.1186/s12967-020-02426-5
Amaravadi, R. K., Kimmelman, A. C., and Debnath, J. (2019). Targeting autophagy in cancer: recent advances and future directions. Cancer Discov. 9, 1167–1181. doi: 10.1158/2159-8290
Amaravadi, R. K., Kimmelmann, A. C., and White, E. (2020). Recent insights into the function of autophagy in cancer. Genes Dev. 30, 1913–1930. doi: 10.1101/gad.287524.116
Ariotti, N., Wu, Y., Okano, S., Gambin, Y., Follett, J., Rae, J., et al. (2020). An inverted CAV1 (caveolin 1) topology defines novel autophagy-dependent exosome secretion from prostate cancer cells. Autophagy. doi: 10.1080/15548627.2020.1820787 [Epub ahead of print].
Babst, M. (2011). MVB vesicle formation: ESCRT-dependent. ESCRT-independent and everything in between. Curr. Opin. Cell Biol. 23, 452–457. doi: 10.1016/j.ceb.2011.04.008
Bader, C. A., Shandala, T., Ng, Y. S., Johnson, I. R. D., and Brooks, D. A. (2015). Atg9 is required for intraluminal vesicles in amphisomes and autolysosomes. Biol. Open 4, 1345–1355. doi: 10.1242/bio.013979
Baixauli, F., López-Otín, C., and Mittelbrunn, M. (2014). Exosomes and autophagy: coordinated mechanisms for the maintenance of cellular fitness. Front. Immunol. 5:403. doi: 10.3389/fimmu.2014.00403
Bandari, S. K., Purushothaman, A., Ramani, V. C., Brinkley, G. J., Chandrashekar, D. S., Varambally, S., et al. (2018). Chemotherapy induces secretion of exosomes loaded with heparanase that degrades extracellular matrix and impacts tumor and host cell behavior. Matrix Biol. 65, 104–118. doi: 10.1016/j.matbio.2017.09.001
Bellot, G., Garcia-Medina, R., Gounon, P., Chiche, J., Roux, D., Pouysségur, J., et al. (2009). Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol. Cell. Biol. 29, 2570–2581. doi: 10.1128/MCB.00166-09
Bhattacharya, S., Pal, K., Sharma, A. K., Dutta, S. K., Lau, J. S., Yan, I. K., et al. (2014). GAIP interacting protein C-terminus regulates autophagy and exosome biogenesis of pancreatic cancer through metabolic pathways. PLoS One 9:e114409. doi: 10.1371/journal.pone.0114409
Buratta, S., Tancini, B., Sagini, K., Delo, F., Chiaradia, E., Urbanelli, L., et al. (2020). Lysosomal exocytosis, exosome release and secretory autophagy: the autophagic- and endo-lysosomal systems go extracellular. Int. J. Mol. Sci. 21:2576. doi: 10.3390/ijms21072576
Chiavarina, B., Whitaker-Menezes, D., Migneco, G., Martinez-Outschoorn, U. E., Pavlides, S., Howell, A., et al. (2010). HIF1-alpha functions as a tumor promoter in cancer associated fibroblasts, and as a tumor suppressor in breast cancer cells: Autophagy drives compartment-specific oncogenesis. Cell Cycle 9, 3534–3551. doi: 10.4161/cc.9.17.12908
Christianson, H. C., Svensson, K. J., van Kuppevelt, T. H., Li, J. P., and Belting, M. (2013). Cancer cell exosomes depend on cell-surface heparan sulfate proteoglycans for their internalization and functional activity. Proc. Natl. Acad. Sci. U.S.A. 110, 17380–17385. doi: 10.1073/pnas.1304266110
Clayton, A., Mitchell, J. P., Court, J., Linnane, S., Mason, M. D., and Tabi, Z. (2008). Human tumor-derived exosomes down-modulate NKG2D expression. J. Immunol. 180, 7249–7258. doi: 10.4049/jimmunol.180.11.7249
Cocucci, E., and Meldolesi, J. (2015). Ectosomes and exosomes: shedding the confusion between extracellular vesicles. Trends Cell Biol. 25, 364–372. doi: 10.1016/j.tcb.2015.01.004
Colletti, M., Paolini, A., Galardi, A., Di Paolo, V., Pascucci, L., Russo, I., et al. (2019). Expression profiles of exosomal miRNAs isolated from plasma of patients with desmoplastic small round cell tumor. Epigenomics 11, 489–500. doi: 10.2217/epi-2018-0179
Colletti, M., Petretto, A., Galardi, A., Di Paolo, V., Tomao, L., Lavarello, C., et al. (2017). Proteomic analysis of neuroblastoma-derived exosomes: new insights into a metastatic signature. Proteomics 17:1600430. doi: 10.1002/pmic.201600430
Colletti, M., Tomao, L., Galardi, A., Paolini, A., Di Paolo, V., De Stefanis, C., et al. (2020). Neuroblastoma-secreted exosomes carrying miR-375 promote osteogenic differentiation of bone-marrow mesenchymal stromal cells. J. Extracell. Vesicles 9:1774144. doi: 10.1080/20013078.2020.1774144
Corazzari, M., Gagliardi, M., Fimia, G. M., and Piacentini, M. (2017). Endoplasmic reticulum stress, unfolded protein response, and cancer cell fate. Front. Oncol. 7:78. doi: 10.3389/fonc.2017.00078
Costa-Silva, B., Aiello, N. M., Ocean, A. J., Singh, S., Zhang, H., Thakur, B. K., et al. (2015). Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 17, 816–826. doi: 10.1038/ncb3169
Cotzomi-Ortega, I., Aguilar-Alonso, P., Reyes-Leyva, J., and Maycotte, P. (2018). Autophagy and its role in protein secretion: implications for cancer therapy. Mediators Inflamm. 2018:4231591. doi: 10.1155/2018/4231591
Dai, E., Han, L., Liu, J., Xie, Y., Kroemer, G., Klionsky, D. J., et al. (2020). Autophagy-dependent ferroptosis drives tumor-associated macrophage polarization via release and uptake of oncogenic KRAS protein. Autophagy 16, 2069–2083. doi: 10.1080/15548627.2020.1714209
De Vries, L., Lou, X., Zhao, G., Zheng, B., and Farquhar, M. G. (1998). GIPC, a PDZ domain containing protein, interacts specifically with the C terminus of RGS-GAIP. Proc. Natl. Acad. Sci. U.S.A. 95, 12340–12345. doi: 10.1073/pnas.95.21.12340
Deng, S., Shanmugam, M. K., Kumar, A. P., Yap, C. T., Sethi, G., and Bishayee, A. (2019). Targeting autophagy using natural compounds for cancer prevention and therapy. Cancer 125, 1228–1246. doi: 10.1002/cncr.31978
Dutta, S., Warshall, C., Bandyopadhyay, C., Dutta, D., and Chandran, B. (2014). Interactions between exosomes from breast cancer cells and primary mammary epithelial cells leads to generation of reactive oxygen species which induce DNA damage response, stabilization of p53 and autophagy in epithelial cells. PLoS One 9:e97580. doi: 10.1371/journal.pone.0097580
Ender, F., Von Bubnoff, N., and Gieseler, F. (2019). Extracellular vesicles: subcellular organelles with the potential to spread cancer resistance. Anticancer Res. 39, 3395–3404. doi: 10.21873/anticanres.13483
Fader, C. M., Sánchez, D., Furlán, M., and Colombo, M. I. (2008). Induction of autophagy promotes fusion of multivesicular bodies with autophagic vacuoles in k562 cells. Traffic 9, 230–250. doi: 10.1111/j.1600-0854.2007.00677.x
Fan, S. J., Kroeger, B., Marie, P. P., Bridges, E. M., Mason, J. D., McCormick, K., et al. (2020). Glutamine deprivation alters the origin and function of cancer cell exosomes. EMBO J. 39:e103009. doi: 10.15252/embj.2019103009
Feng, D., Zhao, W. L., Ye, Y. Y., Bai, X. C., Liu, R. Q., Chang, L. F., et al. (2010). Cellular internalization of exosomes occurs through phagocytosis. Traffic 11, 675–687. doi: 10.1111/j.1600-0854.2010.01041.x
Florey, O., Kim, S. E., Sandoval, C. P., Haynes, C. M., and Overholtzer, M. (2011). Autophagy machinery mediates macroendocytic processing and entotic cell death by targeting single membranes. Nat. Cell Biol. 13, 1335–1343. doi: 10.1038/ncb2363
Galardi, A., Colletti, M., Di Paolo, V., Vitullo, P., Antonetti, L., Russo, I., et al. (2019). Exosomal MiRNAs in pediatric cancers. Int. J. Mol. Sci. 20:4600. doi: 10.3390/ijms20184600
Galardi, A., Colletti, M., Lavarello, C., Di Paolo, V., Mascio, P., Russo, I., et al. (2020). Proteomic profiling of retinoblastoma-derived exosomes reveals potential biomarkers of vitreous seeding. Cancers 12:1555. doi: 10.3390/cancers12061555
Gatica, D., Lahiri, V., and Klionsky, D. J. (2018). Cargo recognition and degradation by selective autophagy. Nat. Cell Biol. 20, 233–242. doi: 10.1038/s41556-018-0037-z
Geissler, E. K., Schnitzbauer, A. A., Zülke, C., Lamby, P. E., Proneth, A., Duvoux, C., et al. (2016). Sirolimus use in liver transplant recipients with hepatocellular carcinoma: a randomized, multicenter, open-label phase 3 trial. Transplantation 100, 116–125. doi: 10.1097/TP.0000000000000965
Goler-Baron, V., Sladkevich, I., and Assaraf, Y. G. (2012). Inhibition of the PI3K-Akt signaling pathway disrupts ABCG2-rich extracellular vesicles and overcomes multidrug resistance in breast cancer cells. Biochem. Pharmacol. 83, 1340–1348. doi: 10.1016/j.bcp.2012.01.033
Guo, D., Lui, G. Y. L., Lai, S. L., Wilmott, J. S., Tikoo, S., Jackett, L. A., et al. (2019). RAB27A promotes melanoma cell invasion and metastasis via regulation of pro-invasive exosomes. Int. J. Cancer 144, 3070–3085. doi: 10.1002/ijc.32064
Guo, H., Chitiprolu, M., Roncevic, L., Javalet, C., Hemming, F. J., Trung, M. T., et al. (2017). Atg5 disassociates the V1V0-ATPase to promote exosome production and tumor metastasis independent of canonical macroautophagy. Dev. Cell 43, 716–730.e7. doi: 10.1016/j.devcel.2017.11.018
CrossRef Full Text 716-730.e7, | PubMed Abstract | Google Scholar
Han, M., Hu, J., Lu, P., Cao, H., Yu, C., Li, X., et al. (2020). Exosome-transmitted miR-567 reverses trastuzumab resistance by inhibiting ATG5 in breast cancer. Cell Death Dis. 11:43. doi: 10.1038/s41419-020-2250-5
Hassanpour, M., Rezabakhsh, A., Rezaie, J., Nouri, M., and Rahbarghazi, R. (2020). Exosomal cargos modulate autophagy in recipient cells via different signaling pathways. Cell Biosci. 10:92. doi: 10.1186/s13578-020-00455-7
Hessvik, N. P., Øverbye, A., Brech, A., Lyngaas Torgersen, M., Jakobsen, I. S., Alicia, L., et al. (2016). PIKfyve inhibition increases exosome release and induces secretory autophagy. Cell. Mol. Life Sci. 73, 4717–4737. doi: 10.1007/s00018-016-2309-8
Hong, B. S., Cho, J. H., Kim, H., Choi, E. J., Rho, S., Kim, J., et al. (2009). Colorectal cancer cell-derived microvesicles are enriched in cell cycle-related mRNAs that promote proliferation of endothelial cells. BMC Genomics 10:556. doi: 10.1186/1471-2164-10-556
Hosoi, T., Nakashima, M., and Ozawa, K. (2018). Incorporation of the endoplasmic reticulum stress-induced spliced form of XBP1 mRNA in the exosomes. Front. Physiol. 9:1357. doi: 10.3389/fphys.2018.01357
Huang, Y., Liu, W., He, B., Lei, W., Shu, H., Sun, L., et al. (2020). Exosomes derived from bone marrow mesenchymal stem cells promote osteosarcoma development by activating oncogenic autophagy. J. Bone Oncol. 21:100280. doi: 10.1016/j.jbo.2020.100280
Jabbari, N., Akbariazar, E., Feqhhi, M., Rahbarghazi, R., and Rezaie, J. (2020a). Breast cancer-derived exosomes: Tumor progression and therapeutic agents. J. Cell. Physiol. 235, 6345–6356. doi: 10.1002/jcp.29668
Jabbari, N., Karimipour, M., Khaksar, M., Akbariazar, E., Heidarzadeh, M., Mojarad, B., et al. (2020b). Tumor-derived extracellular vesicles: insights into bystander effects of exosomes after irradiation. Lasers Med. Sci. 35, 531–545. doi: 10.1007/s10103-019-02880-8
Jin, G., Liu, Y., Zhang, J., Bian, Z., Yao, S., Fei, B., et al. (2019). A panel of serum exosomal microRNAs as predictive markers for chemoresistance in advanced colorectal cancer. Cancer Chemother. Pharmacol. 84, 315–325. doi: 10.1007/s00280-019-03867-6
Kaminskyy, V. O., Piskunova, T., Zborovskaya, I. B., Tchevkina, E. M., and Zhivotovsky, B. (2012). Suppression of basal autophagy reduces lung cancer cell proliferation and enhances caspase-dependent and -independent apoptosis by stimulating ROS formation. Autophagy 8, 1032–1044. doi: 10.4161/auto.20123
Kanemoto, S., Nitani, R., Murakami, T., Kaneko, M., Asada, R., Matsuhisa, K., et al. (2016). Multivesicular body formation enhancement and exosome release during endoplasmic reticulum stress. Biochem. Biophys. Res. Commun. 480, 166–172. doi: 10.1016/j.bbrc.2016.10.019
Kang, R., Zeh, H., Lotze, M., and Tang, D. (2020). The multifaceted effects of autophagy on the tumor microenvironment. Adv. Exp. Med. Biol. 1225, 99–114. doi: 10.1007/978-3-030-35727-6_7
Keulers, T. G., Schaaf, M. B. E., and Rouschop, K. M. A. (2016). Autophagy-dependent secretion: contribution to tumor progression. Front. Oncol. 25:251. doi: 10.3389/fonc.2016.00251
Kimura, T., Jia, J., Kumar, S., Choi, S. W., Gu, Y., Mudd, M., et al. (2017). Dedicated SNAREs and specialized TRIM cargo receptors mediate secretory autophagy. EMBO J. 36, 42–60. doi: 10.15252/embj.201695081
Kucharzewska, P., Christianson, H. C., Welch, J. E., Svensson, K. J., Fredlund, E., Ringnér, M., et al. (2013). Exosomes reflect the hypoxic status of glioma cells and mediate hypoxia-dependent activation of vascular cells during tumor development. Proc. Natl. Acad. Sci. U.S.A. 110, 7312–7317. doi: 10.1073/pnas.1220998110
Kulka, L. A. M., Fangmann, P. V., Panfilova, D., and Olzscha, H. (2020). Impact of HDAC inhibitors on protein quality control systems: consequences for precision medicine in malignant disease. Front. Cell Dev. Biol. 8:425. doi: 10.3389/fcell.2020.00425
Kulkarni, B., Gondaliya, P., Kirave, P., Rawal, R., Jain, A., Garg, R., et al. (2020). Exosome-mediated delivery of miR-30a sensitize cisplatin-resistant variant of oral squamous carcinoma cells via modulating Beclin1 and Bcl2. Oncotarget 11, 1832–1845. doi: 10.18632/oncotarget.27557
Kulshreshtha, R., Ferracin, M., Wojcik, S. E., Garzon, R., Alder, H., Agosto-Perez, F. J., et al. (2007). A microRNA signature of hypoxia. Mol. Cell. Biol. 27, 1859–1867. doi: 10.1128/MCB.01395-06
Kumar, A., and Deep, G. (2020). Hypoxia in tumor microenvironment regulates exosome biogenesis: molecular mechanisms and translational opportunities. Cancer Lett. 479, 23–30. doi: 10.1016/j.canlet.2020.03.017
Kumar, D., Gupta, D., Shankar, S., and Srivastava, R. K. (2014). Biomolecular characterization of exosomes released from cancer stem cells: possible implications for biomarker and treatment of cancer. Oncotarget 6, 3280–3291. doi: 10.18632/oncotarget.2462
Leidal, A. M., Huang, H. H., Marsh, T., Solvik, T., Zhang, D., Ye, J., et al. (2020). The LC3-conjugation machinery specifies the loading of RNA-binding proteins into extracellular vesicles. Nat. Cell Biol. 22, 187–199. doi: 10.1038/s41556-019-0450-y
Li, X. Q., Liu, J. T., Fan, L. L., Liu, Y., Cheng, L., Wang, F., et al. (2016). Exosomes derived from gefitinib-treated EGFR-mutant lung cancer cells alter cisplatin sensitivity via up-regulating autophagy. Oncotarget 7, 24585–24595. doi: 10.18632/oncotarget.8358
Lin, J., Lu, X., Liao, S., Chen, X., Wang, S., Zhao, C., et al. (2019). Cross-regulation between exosomal and autophagic pathways: promising therapy targets in disease. Discov. Med. 27, 201–210.
Liou, W., Geuze, H. J., Geelen, M. J. H., and Slot, J. W. (1997). The autophagic and endocytic pathways converge at the nascent autophagic vacuoles. J. Cell Biol. 136, 61–70. doi: 10.1083/jcb.136.1.61
Liu, D. X., Li, P. P., Guo, J. P., Li, L. L., Guo, B., Jiao, H. B., et al. (2019). Exosomes derived from HBV-associated liver cancer promote chemoresistance by upregulating chaperone-mediated autophagy. Oncol. Lett. 17, 323–331. doi: 10.3892/ol.2018.9584
Liu, Y., Yin, Z., Lu, P., Ma, Y., Luo, B., Xiang, L., et al. (2020). Lung carcinoma cells secrete exosomal MALAT1 to inhibit dendritic cell phagocytosis, inflammatory response, costimulatory molecule expression and promote dendritic cell autophagy via AKT/mTOR pathway. Onco Targets Ther. 13, 10693–10705. doi: 10.2147/OTT.S256669
Lu, J. F., Luk, F., Gong, J., Jaiswal, R., Grau, G. E., and Bebawy, M. (2013). Microparticles mediate MRP1 intercellular transfer and the re-templating of intrinsic resistance pathways. Pharmacol. Res. 76, 77–83. doi: 10.1016/j.phrs.2013.07.009
Luga, V., Zhang, L., Viloria-Petit, A. M., Ogunjimi, A. A., Inanlou, M. R., Chiu, E., et al. (2012). Exosomes mediate stromal mobilization of autocrine Wnt-PCP signaling in breast cancer cell migration. Cell 151, 1542–1556. doi: 10.1016/j.cell.2012.11.024
Malhotra, J., Jabbour, S., Orlick, M., Riedlinger, G., Guo, Y., White, E., et al. (2019). Phase Ib/II study of hydroxychloroquine in combination with chemotherapy in patients with metastatic non-small cell lung cancer (NSCLC). Cancer Treat. Res. Commun. 21:100158. doi: 10.1016/j.ctarc.2019.100158
Martinez, J., Almendinger, J., Oberst, A., Ness, R., Dillon, C. P., Fitzgerald, P., et al. (2011). Microtubule-associated protein 1 light chain 3 alpha (LC3)-associated phagocytosis is required for the efficient clearance of dead cells. Proc. Natl. Acad. Sci. U.S.A. 108, 17396–17401. doi: 10.1073/pnas.1113421108
Matsuo, H., Chevallier, J., Mayran, N., Le Blanc, I., Ferguson, C., Fauré, J., et al. (2004). Role of LBPA and Alix in multivesicular liposome formation and endosome organization. Science 303, 531–534. doi: 10.1126/science.1092425
Mauthe, M., Orhon, I., Rocchi, C., Zhou, X., Luhr, M., Hijlkema, K. J., et al. (2018). Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy 14, 1435–1455. doi: 10.1080/15548627.2018.1474314
Mazure, N. M., and Pouysségur, J. (2010). Hypoxia-induced autophagy: cell death or cell survival? Curr. Opin. Cell Biol. 22, 177–180. doi: 10.1016/j.ceb.2009.11.015
Meng, W., Hao, Y., He, C., Li, L., and Zhu, G. (2019). Exosome-orchestrated hypoxic tumor microenvironment. Mol. Cancer 18:57. doi: 10.1186/s12943-019-0982-6
Minakaki, G., Menges, S., Kittel, A., Emmanouilidou, E., Schaeffner, I., Barkovits, K., et al. (2018). Autophagy inhibition promotes SNCA/alpha-synuclein release and transfer via extracellular vesicles with a hybrid autophagosome-exosome-like phenotype. Autophagy 14, 98–119. doi: 10.1080/15548627.2017.1395992
Mizushima, N., Yoshimori, T., and Ohsumi, Y. (2011). The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27, 107–132. doi: 10.1146/annurev-cellbio-092910-154005
Mulcahy Levy, J. M., and Thorburn, A. (2020). Autophagy in cancer: moving from understanding mechanism to improving therapy responses in patients. Cell Death Differ. 27, 843–857. doi: 10.1038/s41418-019-0474-7
Murrow, L., Malhotra, R., and Debnath, J. (2015). ATG12–ATG3 interacts with Alix to promote basal autophagic flux and late endosome function. Nat. Cell Biol. 17, 300–310. doi: 10.1038/ncb3112
Nazio, F., Bordi, M., Cianfanelli, V., Locatelli, F., and Cecconi, F. (2019). Autophagy and cancer stem cells: molecular mechanisms and therapeutic applications. Cell Death Differ. 26, 690–702. doi: 10.1038/s41418-019-0292-y
Nishimura, T., and Tooze, S. A. (2020). Emerging roles of ATG proteins and membrane lipids in autophagosome formation. Cell Discov. 6:32. doi: 10.1038/s41421-020-0161-3
Parolini, I., Federici, C., Raggi, C., Lugini, L., Palleschi, S., De Milito, A., et al. (2009). Microenvironmental pH is a key factor for exosome traffic in tumor cells. J. Biol. Chem. 284, 34211–34222. doi: 10.1074/jbc.M109.041152
Peinado, H., Aleèkoviæ, M., Lavotshkin, S., Matei, I., Costa-Silva, B., Moreno-Bueno, G., et al. (2012). Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 18, 883–891. doi: 10.1038/nm.2753
Ponpuak, M., Mandell, M. A., Kimura, T., Chauhan, S., Cleyrat, C., and Deretic, V. (2015). Secretory autophagy. Curr. Opin. Cell Biol. 35, 106–116. doi: 10.1016/j.ceb.2015.04.016
Rahbarghazi, R., Jabbari, N., Sani, N. A., Asghari, R., Salimi, L., Kalashani, S. A., et al. (2019). Tumor-derived extracellular vesicles: reliable tools for Cancer diagnosis and clinical applications. Cell Commun. Signal. 17:73. doi: 10.1186/s12964-019-0390-y
Raposo, G., and Stoorvogel, W. (2013). Extracellular vesicles: exosomes, microvesicles, and friends. J. Cell Biol. 200, 373–383. doi: 10.1083/jcb.201211138
Rezaie, J., Ajezi, S., Avci, C. B., Karimipour, M., Geranmayeh, M. H., Nourazarian, A., et al. (2018). Exosomes and their application in biomedical field: difficulties and advantages. Mol. Neurobiol. 55, 3372–3393. doi: 10.1007/s12035-017-0582-7
Rodríguez-Perálvarez, M., Guerrero, M., Barrera, L., Ferrín, G., Álamo, J. M., Ayllón, M. D., et al. (2018). Impact of early initiated everolimus on the recurrence of hepatocellular carcinoma after liver transplantation. Transplantation 102, 2056–2064. doi: 10.1097/TP.0000000000002270
Salimi, L., Akbari, A., Jabbari, N., Mojarad, B., Vahhabi, A., Szafert, S., et al. (2020). Synergies in exosomes and autophagy pathways for cellular homeostasis and metastasis of tumor cells. Cell Biosci. 10:64. doi: 10.1186/s13578-020-00426-y
Santos, P., and Almeida, F. (2020). Role of exosomal miRNAs and the tumor microenvironment in drug resistance. Cells 9:1450. doi: 10.3390/cells9061450
Savina, A., Furlán, M., Vidal, M., and Colombo, M. I. (2003). Exosome release is regulated by a calcium-dependent mechanism in K562 cells. J. Biol. Chem. 278, 20083–20090. doi: 10.1074/jbc.M301642200
Schuh, A. L., and Audhya, A. (2014). The ESCRT machinery: from the plasma membrane to endosomes and back again. Crit. Rev. Biochem. Mol. Biol. 49, 242–261. doi: 10.3109/10409238.2014.881777
Scita, G., and Di Fiore, P. P. (2010). The endocytic matrix. Nature 463, 464–473. doi: 10.1038/nature08910
Shao, N., Xue, L., Wang, R., Luo, K., Zhi, F., and Lan, Q. (2019). miR-454-3p is an exosomal biomarker and functions as a tumor suppressor in glioma. Mol. Cancer Ther. 18, 459–469. doi: 10.1158/1535-7163.mct-18-0725
Singh, S. S., Vats, S., Chia, A. Y. Q., Tan, T. Z., Han, M. S., Ravi, M., et al. (2018). Dual role of autophagy in hallmarks of cancer. Oncogene 37, 1142–1158. doi: 10.1038/s41388-017-0046-6
Skog, J., Würdinger, T., van Rijn, S., Meijer, D. H., Gainche, L., Sena-Esteves, M., et al. (2008). Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 10, 1470–1476. doi: 10.1038/ncb1800
Steinbichler, T. B., Dudás, J., Skvortsov, S., Ganswindt, U., Riechelmann, H., and Skvortsova, I. I. (2019). Therapy resistance mediated by exosomes. Mol. Cancer 18:58. doi: 10.1186/s12943-019-0970-x
Stuffers, S., Sem Wegner, C., Stenmark, H., and Brech, A. (2009). Multivesicular endosome biogenesis in the absence of ESCRTs. Traffic 10, 925–937. doi: 10.1111/j.1600-0854.2009.00920.x
Sung, J. S., Kang, C. W., Kang, S., Jang, Y., Chae, Y. C., Kim, B. G., et al. (2020). ITGB4-mediated metabolic reprogramming of cancer-associated fibroblasts. Oncogene 39, 664–676. doi: 10.1038/s41388-019-1014-0
Svensson, K. J., Christianson, H. C., Wittrup, A., Bourseau-Guilmain, E., Lindqvist, E., Svensson, L. M., et al. (2013). Exosome uptake depends on ERK1/2-heat shock protein 27 signaling and lipid Raft-mediated endocytosis negatively regulated by caveolin-1. J. Biol. Chem. 288, 17713–17724. doi: 10.1074/jbc.M112.445403
Tan, H. W. S., Sim, A. Y. L., and Long, Y. C. (2017). Glutamine metabolism regulates autophagy-dependent mTORC1 reactivation during amino acid starvation. Nat. Commun. 8:338. doi: 10.1038/s41467-017-00369-y
Théry, C., Witwer, K. W., Aikawa, E., Alcaraz, M. J., Anderson, J. D., Andriantsitohaina, R., et al. (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 7:1535750. doi: 10.1080/20013078.2018.1535750
Tian, J., Popal, M. S., Zhao, Y., Liu, Y., Chen, K., and Liu, Y. (2019). Interplay between exosomes and autophagy in cardiovascular diseases: novel promising target for diagnostic and therapeutic application. Aging Dis. 10, 1302–1310. doi: 10.14336/AD.2018.1020
Tian, T., Zhu, Y. L., Zhou, Y. Y., Liang, G. F., Wang, Y. Y., Hu, F. H., et al. (2014). Exosome uptake through clathrin-mediated endocytosis and macropinocytosis and mediating miR-21 delivery. J. Biol. Chem. 289, 22258–22267. doi: 10.1074/jbc.M114.588046
van Niel, G., Porto-Carreiro, I., Simoes, S., and Raposo, G. (2006). Exosomes: a common pathway for a specialized function. J Biochem. 140, 13–21. doi: 10.1093/jb/mvj128
Verfaillie, T., Salazar, M., Velasco, G., and Agostinis, P. (2010). Linking ER stress to autophagy: potential implications for cancer therapy. Int J. Cell Biol. 2010:930509. doi: 10.1155/2010/930509
Villarroya-Beltri, C., Baixauli, F., Mittelbrunn, M., Fernaìndez-Delgado, I., Torralba, D., Moreno-Gonzalo, O., et al. (2016). ISGylation controls exosome secretion by promoting lysosomal degradation of MVB proteins. Nat. Commun. 7:13588. doi: 10.1038/ncomms13588
Wang, B., Mao, J. H., Wang, B. Y., Wang, L. X., Wen, H. Y., Xu, L. J., et al. (2020). Exosomal miR-1910-3p promotes proliferation, metastasis, and autophagy of breast cancer cells by targeting MTMR3 and activating the NF-κB signaling pathway. Cancer Lett. 489, 87–99. doi: 10.1016/j.canlet.2020.05.038
Wang, X., Yin, X., and Yang, Y. (2019). Rasal2 suppresses breast cancer cell proliferation modulated by secretory autophagy. Mol. Cell. Biochem. 462, 115–122. doi: 10.1007/s11010-019-03615-7
White, E. (2015). The role for autophagy in cancer. J. Clin. Invest. 125, 42–46. doi: 10.1172/JCI73941
White, E., Karp, C., Strohecker, A. M., Guo, Y., and Mathew, R. (2010). Role of autophagy in suppression of inflammation and cancer. Curr. Opin. Cell Biol. 212–217. doi: 10.1016/j.ceb.2009.12.008
Wu, Z., He, D., and Li, H. (2020). Bioglass enhances the production of exosomes and improves their capability of promoting vascularization. Bioact. Mater. 6, 823–835. doi: 10.1016/j.bioactmat.2020.09.011
Wu, X., Zhou, Z., Xu, S., Liao, C., Chen, X., Li, B., et al. (2020). Extracellular vesicle packaged LMP1-activated fibroblasts promote tumor progression via autophagy and stroma-tumor metabolism coupling. Cancer Lett. 478, 93–106. doi: 10.1016/j.canlet.2020.03.004
Xu, J., Camfield, R., and Gorski, S. M. (2018). The interplay between exosomes and autophagy – partners in crime. J. Cell Sci. 131:jcs215210. doi: 10.1242/jcs.215210
Yang, Y., and Klionsky, D. J. (2020). Autophagy and disease: unanswered questions. Cell Death Differ. 27, 858–871. doi: 10.1038/s41418-019-0480-9
Yao, W., Guo, P., Mu, Q., and Wang, Y. (2020). Exosome-derived Circ-PVT1 contributes to cisplatin resistance by regulating autophagy, invasion, and apoptosis Via miR-30a-5p/YAP1 axis in gastric cancer Cells. Cancer Biother. Radiopharm. doi: 10.1089/cbr.2020.3578 [Epub ahead of print].
Yeon, M., Lee, S., Lee, J. E., Jung, H. S., Kim, Y., and Jeoung, D. (2019). CAGE-miR-140-5p-Wnt1 axis regulates autophagic flux, tumorigenic potential of mouse colon cancer cells and cellular interactions mediated by exosomes. Front. Oncol. 9:1240. doi: 10.3389/fonc.2019.01240
Yu, L., Chen, Y., and Tooze, S. A. (2018). Autophagy pathway: Cellular and molecular mechanisms. Autophagy 14, 207–215. doi: 10.1080/15548627.2017.1378838
Yu, S., Liu, C., Su, K., Wang, J., Liu, Y., Zhang, L., et al. (2007). Tumor exosomes inhibit differentiation of bone marrow dendritic cells. J. Immunol. 178, 6867–6875. doi: 10.4049/jimmunol.178.11.6867
Yun, C. W., and Lee, S. H. (2018). The roles of autophagy in cancer. Int. J. Mol. Sci. 19:3466. doi: 10.3390/ijms19113466
Yuwen, D., Ma, Y., Wang, D., Gao, J., Li, X., Xue, W., et al. (2019). Prognostic role of circulating exosomal miR-425-3p for the response of NSCLC to platinum-based chemotherapy. Cancer Epidemiol. Biomarkers Prev. 28, 163–173. doi: 10.1158/1055-9965.epi-18-0569
Zeng, Y., Yao, X., Liu, X., He, X., Li, L., Liu, X., et al. (2019). Anti-angiogenesis triggers exosomes release from endothelial cells to promote tumor vasculogenesis. J. Extracell. Vesicles 8:1629865. doi: 10.1080/20013078.2019.1629865
Zhang, M., Kenny, S. J., Xu, K., and Schekman, R. (2015). Translocation of interleukin-1β into a vesicle intermediate in autophagy-mediated secretion. eLife 4:e11205. doi: 10.7554/eLife.11205
Zhang, X., Shi, H., Yuan, X., Jiang, P., Qian, H., and Xu, W. (2018). Tumor-derived exosomes induce N2 polarization of neutrophils to promote gastric cancer cell migration. Mol. Cancer 17:146. doi: 10.1186/s12943-018-0898-6
Zhang, X., Wang, S., Wang, H., Cao, J., Huang, X., Chen, Z., et al. (2019). Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol. Cancer 18:20. doi: 10.1186/s12943-018-0935-5
Zhao, Y. G., and Zhang, H. (2019). Autophagosome maturation: an epic journey from the ER to lysosomes. J. Cell Biol. 218, 757–770. doi: 10.1083/jcb.201810099
Zheng, K., Ma, J., Wang, Y., He, Z., and Deng, K. (2020). Sulforaphane inhibits autophagy and induces exosome-mediated paracrine senescence via regulating mTOR/TFE3. Mol. Nutr. Food Res. 64:e1901231. doi: 10.1002/mnfr.201901231
Zhou, J., Hu, S. E., Tan, S. H., Cao, R., Chen, Y., Xia, D., et al. (2012). Andrographolide sensitizes cisplatin-induced apoptosis via suppression of autophagosome-lysosome fusion in human cancer cells. Autophagy 8, 338–349. doi: 10.4161/auto.18721
Keywords: autophagy, exosome, microenvironment, cancer, target therapy
Citation: Colletti M, Ceglie D, Di Giannatale A and Nazio F (2021) Autophagy and Exosomes Relationship in Cancer: Friends or Foes? Front. Cell Dev. Biol. 8:614178. doi: 10.3389/fcell.2020.614178
Received: 05 October 2020; Accepted: 16 December 2020;
Published: 12 January 2021.
Edited by:
Daniel Hector Grasso, Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), ArgentinaReviewed by:
Ravi Manjithaya, Jawaharlal Nehru Centre for Advanced Scientific Research, IndiaJafar Rezaie, Urmia University of Medical Sciences, Iran
Copyright © 2021 Colletti, Ceglie, Di Giannatale and Nazio. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Angela Di Giannatale, YW5nZWxhLmRpZ2lhbm5hdGFsZUBvcGJnLm5ldA==; Francesca Nazio, ZnJhbmNlc2NhLm5hemlvQG9wYmcubmV0
†These authors have contributed equally to this work
 Marta Colletti†
Marta Colletti† Donatella Ceglie
Donatella Ceglie