
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol. , 09 November 2020
Sec. Cell Growth and Division
Volume 8 - 2020 | https://doi.org/10.3389/fcell.2020.605672
This article is part of the Research Topic Editor’s Pick 2021: Highlights in Cell Growth and Division View all 14 articles
Cytokinins (CKs) are a group of adenine-derived, small signaling molecules of crucial importance for growth and multiple developmental processes in plants. Biological roles of classical CKs: isopentenyladenine (iP), trans and cis isomers of zeatin (tZ, cZ), and dihydrozeatin, have been studied extensively and their functions are well defined in many aspects of plant physiology. In parallel, extensive knowledge exists for genes involved in tRNA modifications that lead to the production of tRNA-bound methylthiolated CKs, especially in bacterial and mammalian systems. However, not much is known about the origins, fates, and possible functions of the unbound methylthiolated CKs (2MeS-CKs) in biological systems. 2MeS-CKs are the free base or riboside derivatives of iP or Z-type CKs, modified by the addition of a thiol group (–SH) at position 2 of the adenine ring that is subsequently methylated. Based on the evidence to date, these distinctive CK conjugates are derived exclusively via the tRNA degradation pathway. This review summarizes the knowledge on the probable steps involved in the biosynthesis of unbound 2MeS-CKs across diverse kingdoms of life. Furthermore, it provides examples of CK profiles of organisms from which the presence of 2MeS-CKs have been detected and confirms a close association and balance between the production of classical CKs and 2MeS-CKs. Finally, it discusses available reports regarding the possible physiological functions of 2MeS-CKs in different biological systems.
Cytokinins (CKs) are a group of adenine-derived, small signaling molecules, that comprise a class of phytohormones that are of crucial importance for multiple growth and developmental processes in plants (Spíchal, 2012; Kieber and Schaller, 2018). Plant CKs play a significant role in regulating cell proliferation and differentiation, control of shoot/root balance, transduction of nutritional signals (source/sink distribution), delaying senescence, and increasing crop productivity (Sakakibara, 2006). Cytokinins exist in two main structural forms, depending on the chemistry of the side chain attached at N6 position to the adenine ring – isoprenoid or aromatic. Regarding the structure of their side chain, classical, isoprenoid CKs include isopentenyladenine (iP), zeatin (trans and cis isomers; tZ, cZ), dihydrozeatin (DZ), and their various derivatives and conjugates (Kisiala et al., 2019) that often strongly differ in their biological activity (Spíchal, 2012).
In plants, isoprenoid CKs can be synthesized via two metabolic pathways, the de novo pathway, and the tRNA degradation pathway. In de novo CK biosynthesis, specific isopentenyltransferase (IPT) enzymes (adenylate IPTs) add the isopentenyl sidechains (originating mainly from the methylerythritol phosphate pathway; MEP) to adenosine tri- di- or monophosphates (ATP, ADP, AMP). The de novo pathway, that is often a main source of plant CKs, is localized mainly in cell plastids and results predominantly in the production of iP- and tZ-type CKs that typically demonstrate high biological activity (Sakakibara, 2006).
The other known CK production pathway involves a modification of the adenine base at position 37 of tRNA molecules with the isopentenyl sidechain obtained primarily via the mevalonate pathway (MVA). Production of CKs through tRNA degradation is localized mainly in the cytosol. Generally, tRNA modifications contribute to an increased adaptation to environmental conditions through the control of translational efficiency and fidelity, in addition to reading frame maintenance (Dabravolski, 2020; Liaqat et al., 2020). Following the degradation of the modified tRNA molecules, cZ-type CKs are produced. Although the tRNA degradation pathway is thought to play only a minor role in overall CK production, it contributes significantly to the levels of cZ-CKs in certain plant species (Emery et al., 1998; Frébort et al., 2011; Gajdošová et al., 2011).
The presence of CK metabolites, or the genetic mechanisms required for CK production, have been reported in all kingdoms of life (Björk, 1995; Mayaka et al., 2019). Cytokinin biosynthesis involves conserved mechanisms even among evolutionary distant organisms, and the formation of CKs in bacteria, fungi, plants, or mammals all involve specific gene homologs and their corresponding enzymes, including IPT, adenosine kinase (AK), LONELY GUY (LOG) and a CK-degradation enzyme, cytokinin oxidase/dehydrogenase (CKX; Sakakibara, 2006; Chanclud et al., 2016; Trdá et al., 2017; Daudu et al., 2019). Genes encoding enzymes responsible for subsequent steps of CK metabolism exist in a variety of different organisms, although not all genes have been fully characterized or discovered.
Since the end of the 20th century, significant discoveries in plants have been made; from not even knowing if plants synthesized their own CKs (Holland, 1997), to having nearly comprehensively defined CK pathways, leading to identification of the active CKs and their corresponding conjugates (e.g., cis and trans-zeatin, iP, DZ and nucleotide, riboside, glucoside conjugates) (Kieber and Schaller, 2018). Unknown CK pathways are now rare and include those that may produce aromatic side chain CKs (Frébort et al., 2011) and the seldomly observed conjugates such as lupinic acid, a zeatin metabolite isolated from Lupinus angustifolius seedlings (Guern and Peaud-Lenoel, 2012), mono- and dimethylated isopentenyladenine CKs found in virulent Rhodococcus fascians strains (Jameson, 2019) or discadenine, iP derivative unique for slime mold Dictyostelium discoideum (Aoki et al., 2020). Another curious case is that of the hydrophobic, methylthiolated CKs (2MeS-CKs) (Figure 1). 2MeS-CKs are iP- or Z-type CKs modified by addition of a thiol group (–SH) at the position 2 of the adenine ring and its subsequent methylation (Tarkowski et al., 2010). Based on the evidence to date, these unique CK conjugates are derived exclusively via the tRNA degradation pathway (Koenig et al., 2002; Morrison et al., 2015a, 2017). 2MeS-CKs are commonly observed, yet they are poorly understood in terms of their origins, biological activity and functions. Due to their low quantities in plant tissues, the development of analytical methods with adequate sensitivity to detect these compounds is essential in order to elucidate their biological function (Tarkowski et al., 2010). A survey of CK review literature has yet to offer a place for 2MeS-CK production in the overall CK pathway schemes (Spíchal, 2012; Morrison et al., 2015a, 2017).
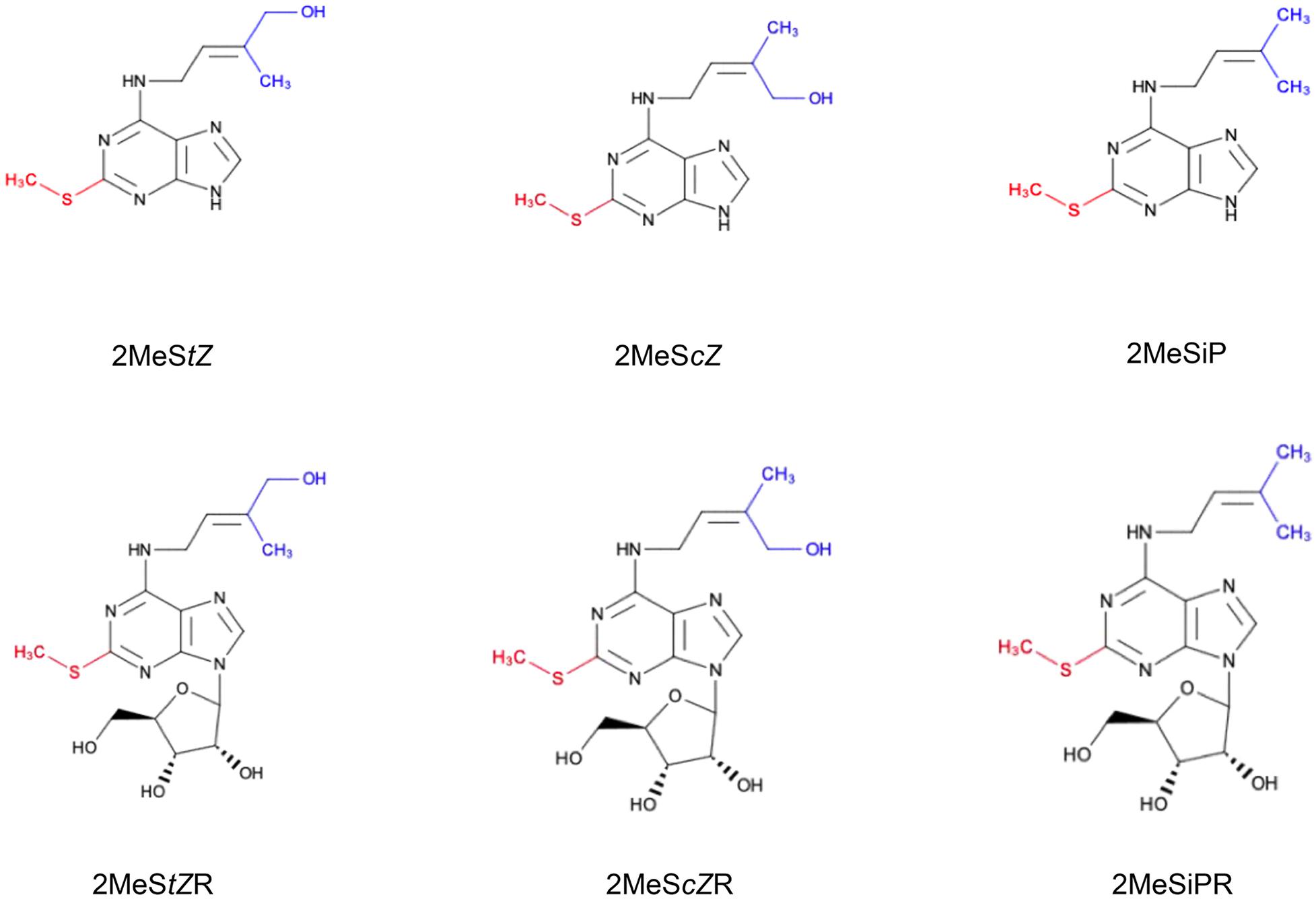
Figure 1. Chemical structures of methylthiolated cytokinins (2MeS-CKs). The attachment of the –S–CH3 group (red) at C2 position of the adenine ring of tRNA-bound CKs is regulated by methylthiotransferase-like enzymes. 2MeS-CK forms differ based on the modifications to their isoprenoid side chain (blue) and include: 2-methylthio-transZeatin (2MeStZ), 2-methylthio-cisZeatin (2MeScZ), and 2-methylthio-isopentenyladenine (2MeSiP), and their riboside derivatives: 2MeStZR, 2MeScZR and 2MeSiPR, respectively.
This review will summarize the knowledge on the probable steps involved in biosynthesis of the unbound 2MeS-CKs. Furthermore, it will provide examples of CK profiles for organisms from which 2MeS-CKs were previously detected and confirm a close association and balance between production of classical CKs and 2MeS-CKs. Finally, it will discuss available reports regarding any possible physiological functions of 2MeS-CKs in different biological systems.
The first step toward 2MeS-CK production involves the initial formation of a CK in the tRNA degradation pathway. It involves the addition of an isopentenyl group to an adenine at position 37 of tRNA molecules, which is known to read codons beginning with uridine (Figure 2). This isopentenyl addition occurs via the tRNA-isopentenyltransferase enzyme (tRNA-IPT) and it creates a tRNA-bound N6-isopentenyladenosine phosphate (iPRP). It also represents the rate limiting step of CK biosynthesis. To date, tRNA-IPT homologs have been identified in bacteria, fungi, plants, insects, and mammals (Pertry et al., 2009; Dabravolski, 2020).
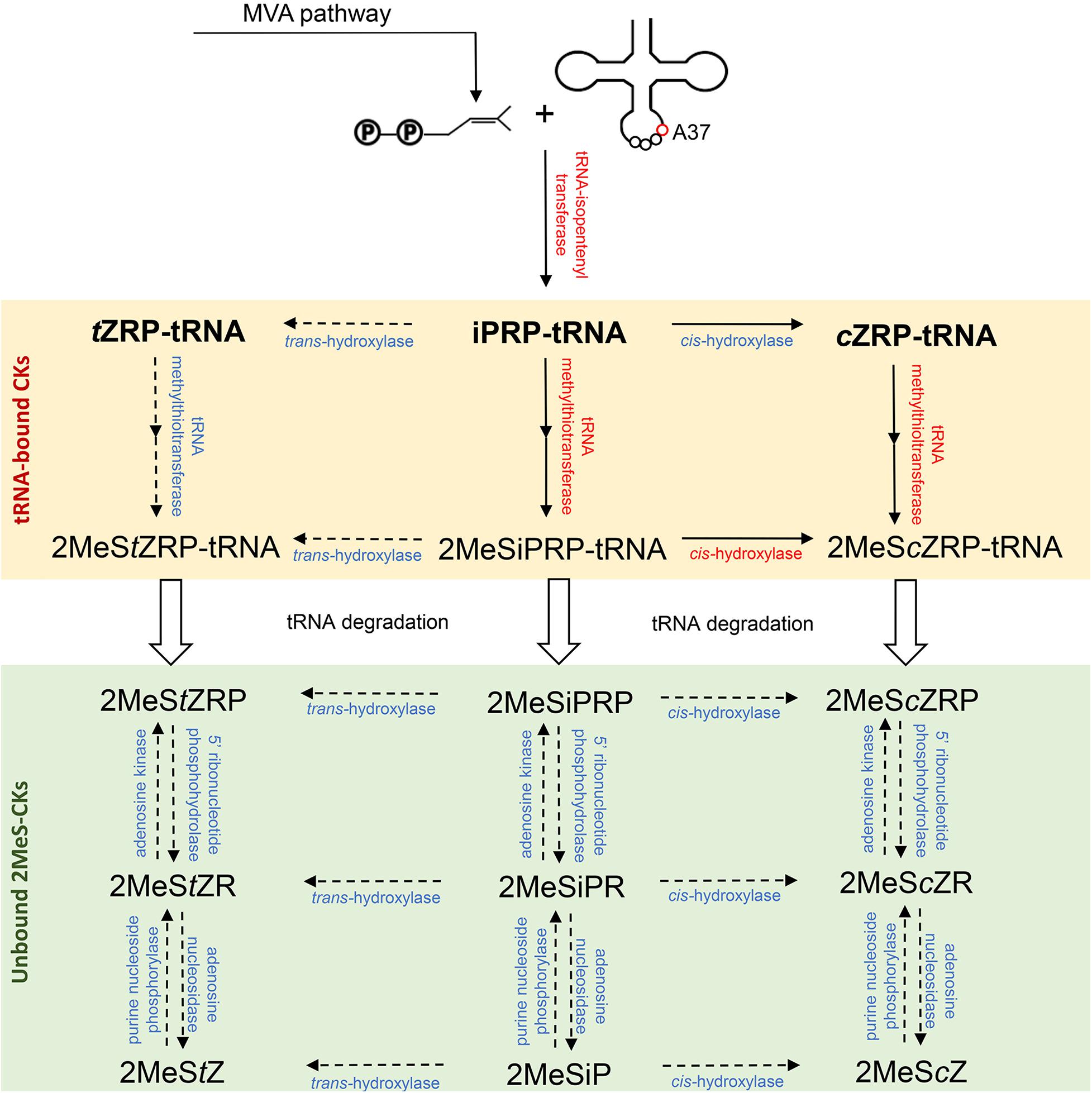
Figure 2. Biosynthesis pathway of the methylthiolated cytokinins (2MeS-CKs) proposed based on the microbial, plant and animal 2MeS-CK metabolite profiles obtained at the Water Quality Centre, Trent University and from other laboratories. Three groups of enzymes (red font) involved in the production of 2MeS-CKs have been previously identified across kingdoms of life including: tRNA-isopentenyltransferases (e.g., miaA, MOD5, IPT2 and 9, TRIT1), tRNA-methylthiotransferases (e.g., miaB + C, CDK5RAP1), and cis-hydroxylases (e.g., miaE) (Dabravolski, 2020). The inferred enzymes involved in further modification of tRNA-bound and free 2MeS-CKs (blue font, dashed arrows) were predicted based on the information available from other CK producing pathways (Sakakibara, 2006; Spíchal, 2012; Morrison et al., 2017; Aoki et al., 2019a). Mevalonate (MVA) pathway is the main source of the isoprenoid substrate for tRNA-isopentenyltranserases; however, a small pool of the prenyl chain molecules can originate from the methylerythritol phosphate (MEP) pathway. If tRNA degradation occurs prior to methylthiolation of the prenylated adenine nucleobase (A37), tRNA-bound iPRP, tZRP, and cZRP (bold font) are released and act as the precursors of classical CKs (iP, tZ and cZ).
In bacteria, the enzyme responsible for tRNA isopentenylation is MiaA (Buck and Ames, 1984; Gray et al., 1996; Koenig et al., 2002). A deletion of the miaA gene in Salmonella typhimurium, a pathogenic, Gram-negative bacterium, caused extensive pleiotropic effects, including a temperature-sensitive growth phenotype (Blum, 1988). miaA mutant strain of Bradorhizobium spp. displayed a significant reduction in CK production but was also characterized by more intense growth in CK-free media (Podlešáková et al., 2013). The isopentenyl side chain of the tRNA-bound iP can be further modified via cis-hydroxylase, forming a tRNA-bound cis isomer of Zeatin (tRNA-cZ) and, upon tRNA degradation, iP- or cZ-type CKs are released (Cherayil and Lipsett, 1977; Morris et al., 1981; McGaw and Burch, 1995). tRNA-derived cZ-CKs are often thought to have significantly reduced biological activity, compared to the de novo synthesized, highly active tZ-type CKs (Sakakibara, 2006; Wang et al., 2020); however, recent studies suggested cZ is strongly involved in plant stress alleviation (Gajdošová et al., 2011; Schäfer et al., 2015; Silva-Navas et al., 2019). trans-zeatin secretion in plant symbiotic Methylobacterium was previously linked to a tRNA source since the presence of Z trans-isomers was detected even in the absence of the genetic machinery for de novo pathway synthesis. The tRNA hydrolysate of the Methylobacterium extorquens miaA mutant also lacked the precursor riboside forms, trans-zeatin riboside (tZR) and isopentenyladenosine (iPR) (Koenig et al., 2002). The presence of significant levels of tZ and its derivatives have since been reported from a wide selection of plant-associated Methylobacterium strains (Jorge et al., 2019), additionally supporting the claim that, unlike plants, some bacteria can derive tZ isomers via a tRNA source. The possibility of obtaining tZ-type CKs via the tRNA-IPT activity was also proposed for a phytopathogenic fungi-plant interaction system (Morrison et al., 2017) while production of tZ-type CK via de novo biosynthesis pathway has been previously identified in a fungal rye pathogen, ergot (Claviceps purpurea) (Hinsch et al., 2015).
The dominance of cZ- and iP-type CKs in the hormone profiles of saprophytic and biotrophic fungal species suggests that the tRNA degradation is the predominant CK pathway in that kingdom (Morrison et al., 2015b; Moffatt et al., 2018). A fungal tRNA-isopentenyltransferase gene homolog (MOD5) was first identified in yeast (Saccharomyces cerevisiae) (Laten et al., 1978; Dihanich et al., 1987). Evidence to support its function in CK production was demonstrated via a Mod5-1 mutation in S. cerevisiae that caused a significant reduction in biosynthesis of iPR from cytoplasmic and mitochondrial tRNA (Laten et al., 1978). Mod5, a yeast tRNA-isopentenyltransferase, is also involved in protein synthesis and sporulation (Laten et al., 1978) and Mod5 mutants displayed reduced resistance to antifungal agents (Suzuki et al., 2012). More recently, a survey confirmed production of zeatin-type CKs in a wide range of yeast species (Streletskii et al., 2019); however, the authors did not resolve their data between the trans- and cis-isomers.
tRNA-IPT genes responsible for CK biosynthesis have been identified in pathogenic fungi and fungal CK production is a crucial factor in strain virulence (Chanclud et al., 2016; Morrison et al., 2017; Trdá et al., 2017). Cytokinin Synthesis 1 (CKS1), a gene encoding a tRNA-IPT was identified in the rice pathogen, Magnaporthe oryzae (Chanclud et al., 2016). cks1 mutants showed normal in vitro growth; however, the deletion resulted in impaired CK production preventing the mutants from maintaining nutrient levels at the site of infection. This in turn, led to the induction of early and strong plant defenses suggesting that fungal CKs contribute to metabolite mobilization and to rice defense inhibition (Chanclud et al., 2016). In Ustilago maydis, a biotrophic fungal pathogen that causes corn smut disease, the tRNA-IPT homolog was designated as UMAG_10043 (Morrison et al., 2017). Deletion of UMAG_10043 resulted in a loss of CK production and significantly reduced virulence of the fungus during infection of maize seedlings and cobs but it also affected fungus filamentous growth and its ability to modify CKs taken up from the media (Morrison et al., 2017). The analysis of LmIPT (XP_003842057) silenced lines of Leptosphaeria maculans, a hemibiotrophic fungal pathogen of oilseed rape, demonstrated a significant reduction in cZ levels (Trdá et al., 2017). However, no other phenotypic effects were observed in the mutants, and total CK levels were not altered, suggesting that another, alternative CK biosynthesis pathway might be active in L. maculans (Trdá et al., 2017).
Another microbial CK producer, slime mold (Dictyostelium discoideum) is a member of the Amoebozoa phylum and possesses three IPT genes, two of which are putative tRNA-IPT genes (iptB and iptC) (Anjard and Loomis, 2008; Nishii et al., 2018). These tRNA-IPT gene candidates are thought to be distantly related to plant IPTs and they were acquired in Dictyostelium by horizontal gene transfer (Eichinger et al., 2005; Nishii et al., 2018). Although the biological role of the two putative tRNA-IPTs in D. discoideum have not been confirmed, the available CK profiles suggest they might be involved in the production of cZ-type and 2Me-S-type CKs by slime mold (Aoki et al., 2019a).
In plants, CKs contribute an important role in inducing plant cell division, differentiation and regulation of various aspects of plant development and interaction with the biotic and abiotic environment (Sakakibara, 2006; Kieber and Schaller, 2018). tRNA-IPT homologs were identified in Physcomitrella patens, a moss species that belongs to the early divergent clade of land plants utilizing CKs for growth control (Lindner et al., 2014). In Arabidopsis thaliana, 9 isopentenyltransferase genes have been identified (AtIPT1-9), although only two; AtIPT2 (AT2G27760) and AtIPT9 (AT5G20040) are known to be responsible for modifying tRNA and production of cZ type CKs (Miyawaki et al., 2006). Although cZ activity is greatly limited in comparison with tZ or iP type CKs, the evidence supports the importance of plant tRNA-IPTs. For example, the atipt2 9 double mutants demonstrate reduced cZ CK levels, but their iP and tZ-type CKs are not affected, and yet they often have a chlorotic phenotype (Miyawaki et al., 2006).
The available studies of CKs in the kingdom of Animalia indicate the existence of an active tRNA pathway in insects and mammals. Cytokinin metabolites have been found in insects (e.g., Straka et al., 2010; Andreas et al., 2020); although, it has been postulated that CK presence in insects might be a result of the activity of bacterial symbionts (Giron et al., 2013). In silkworm (Bombyx mori), a candidate tRNA-IPT gene with three alternative splicing isoforms (BmIPT1-BmITP3) was recently identified (Chen et al., 2019). A recombinant vector containing BmIPT1 could restore isopentenylation of tRNA in the IPT-deficient yeast strain MT-8 suggesting BmIPT1 is a functional tRNA-IPT enzyme in B. mori. Although no CK data are available for silkworm to date, the importance of BmIPT1 was demonstrated following the i6A (iPR) modification at position A37 in tRNA, which resulted in severe abnormalities in silk spinning and metamorphosis (Chen et al., 2019).
Cytokinin presence and metabolism has been reported in canine tissues (Seegobin et al., 2018) and human cell cultures (Aoki et al., 2019b), and the CK profiles indicate that tRNA degradation is the only source of unbound mammalian CKs. tRNA-isopentenyltransferase 1 (TRIT1), a homolog of the prokaryotic miaA, facilitates production of tRNA-bound iP-type CKs in human cytosol and mitochondria (Lamichhane et al., 2013; Smaldino et al., 2015). TRIT1 was previously suggested to play a role as a tumor suppressor (Spinola et al., 2005), in selenoprotein regulation (Fradejas et al., 2013) in gene-mediated transcriptional silencing (Smaldino et al., 2015) and amyloid fiber folding (Waller et al., 2017). The product of the TRIT1 gene, iPR, has long been the lone CK type detected as tRNA-bound or as a free mononucleotide in mammals (Persson et al., 1994; Golovko et al., 2000). Since then, more recent studies on CK profiling by High Performance Liquid Chromatography Tandem Mass Spectrometry (HPLC-MS/MS) revealed a presence of seven types of CKs in a range of canine tissues (Seegobin et al., 2018). The presence of unbound CK derivatives in HeLa cell cultures were detected, strengthening the evidence of TRIT1 activity in CK biosynthesis in mammalian cells (Aoki et al., 2019b).
Following isopentenylation, methylthiolation of the adenine ring at the C2 position is the next step of the tRNA degradation pathway that leads to the production of 2MeS-CKs (Figure 2). In bacteria, the gene responsible for methylthiolation is miaB which modifies tRNA-bound i6A (iPR) or io6A (cisZeatin riboside; cZR) to ms2i6A (2-methylthio-isopentenyladenine riboside; 2MeSiPR) and ms2io6A (2-methylthio-cisZeatin riboside; 2MeScZR), respectively (Esberg et al., 1999; more details on hydroxylation of the isoprenoid chain of tRNA-bound CKs will be provided in the section “Hydroxylation of the Isopentenyl Chain of tRNA-Bound 2MeSiP-CKs: tRNA-ms2io6A37-Hydroxylase: MiaE/CYP450”). Methylthiolation requires iron (Fe), cysteine (Cys), S-adenosylmethionine (SAM) and is thought to occur in two steps, initially as thiolation of i6A37 (tRNA-bound iPR) to s2i6A37, and a subsequent methyl transfer gives ms2i6A37 (tRNA-bound 2MeSiPR) (Esberg et al., 1999). It remains unknown whether each reaction is catalyzed by a single enzyme (MiaB + MiaC) or both steps are facilitated by the same enzyme, MiaB (Pierrel et al., 2003). However, Escherichia coli mutant strains that lacked a functional miaB gene were shown to contain only the product of the first step in the tRNA pathway, i6A37; thus it is suggested that the MiaB protein is involved in formation of the C-S bond that can be further methylated via MiaC activity (Esberg et al., 1999). A conserved canonical Cys triad is found both in the N-terminal half of miaB genes and in enzymes such as biotin and lipoate synthases, which are involved in catalyzing C–H to C-S bond conversion reactions. The motif provides Cys ligands for a [4Fe-4S]+2/+1 cluster. The Cys triad and iron cluster are essential for thiolation activity which indicates that MiaB, biotin synthase, and lipoate synthase all utilize similar radical mechanisms to activate sulfur (S) and insert it into the respective substrates (Pierrel et al., 2002). Additionally, MiaB enzymes contain a sequence of 60–80 residues in the C-terminal region that is similar to the ß-barrel RNA-binding domains (Anantharaman et al., 2001). This domain, known as TRAM, was identified as the site likely involved in binding the tRNA substrate (Pierrel et al., 2003). The level of synthesis of the 2-methylthiol group of ms2io6A (2MeScZR) is sensitive to the presence of S or Fe and may function as a signal device for the availability of these elements (Buck and Griffiths, 1982; Buck and Ames, 1984).
The role of the miaC gene has been postulated to exist but never identified (Pierrel et al., 2004). MiaC is thought to function during the methylthiolation reaction as a methyltransferase following the S transfer performed by MiaB. Examples of methyltransferase genes in plant pathogenic bacteria can be found in Streptomyces turgidiscabies and R. fascians which both carry a fas operon (Joshi and Loria, 2007; Pertry et al., 2009). The fas operon contains two open reading frames (ORFs) that code for methyltransferases (mtr-1 and mtr-2) which may be involved in the methylthiolation of bacterial CKs (Frébort et al., 2011).
Unlike bacteria, there are no miaB homologs yet identified from fungi. Correspondingly, 2MeS-CKs have not been found in axenic fungal cultures (Table 1; Hinsch et al., 2015; Chanclud et al., 2016; Morrison et al., 2017; Vedenicheva et al., 2018). Previously, 2MeS-CKs were detected from temperate forest fungi collected in situ (Morrison et al., 2015b); however, these samples were not grown aseptically and would have had an associated microbiome, including bacteria capable of producing 2MeS-CKs.
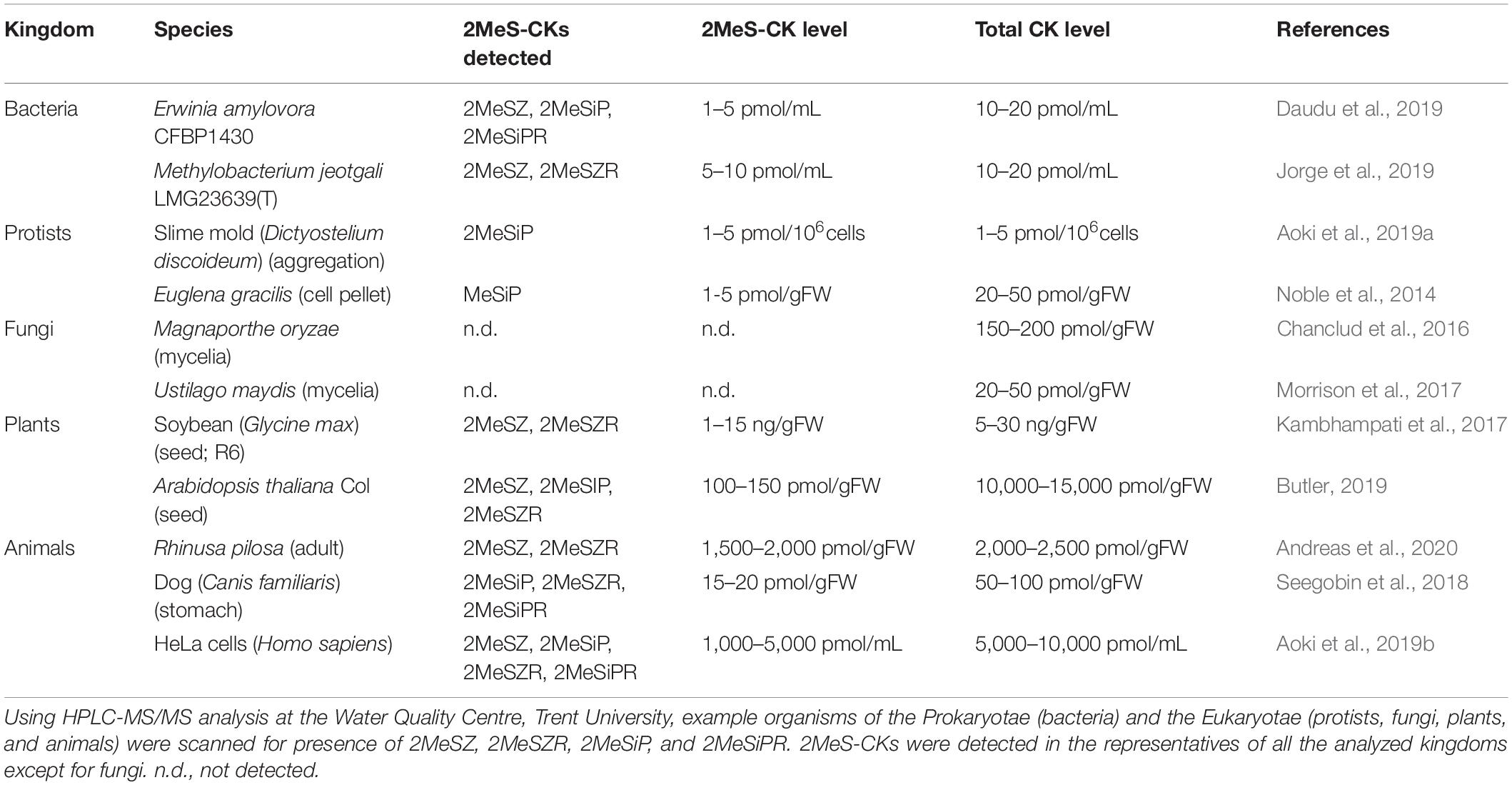
Table 1. Profiles of methylthiolated cytokinins (2MeS-CKs) among representatives of different life kingdoms.
2-Methylthio-Zeatin riboside (2MeSZR) and 2-methylthio-Zeatin (2MeSZ) accumulated in the infected cobs during the later stages of maize infection by U. maydis but were not present in aseptic fungal cultures or in control plant tissue. This finding suggests there is a fungal-stimulated, plant-origin of 2MeS-CKs that may be involved in promoting tissue proliferation around the site of infection (Morrison et al., 2015a).
Apart from fungi, miaB homolog genes can be found across all kingdoms of life, indicating their evolutionarily conserved character and universal roles in posttranscriptional and posttranslational RNA modifications (Anantharaman et al., 2002). Figure 3 presents a MAFFT alignment (Geneious Prime 2020.2.3) of the characterized and predicted miaB-like proteins in model and non-model species from Archaea, Bacteria, Protista, Planta, and Animalia. The top candidates were selected based on the highest sequence similarity with E. coli miaB protein sequence (accession no. WP_000162747.1) using non-redundant BLASTp searches on 59 selected taxids. Searches were done using the BLOSUM62 matrix and default parameters.1 A signature miaB motif that has three cysteine residues spaced by 3 and 2 amino acids (CxxxCxxC; Figure 3), and that is responsible for iron binding in the process of thiolation (Esberg et al., 1999), was present in all the analyzed sequences. Another highly conserved miaB motif (IVGFPGET; Figure 3; Esberg et al., 1999) was found in 18 species including representatives of Archaea, Bacteria and Protists. The presence of the TRAM domain (PF01938) responsible for nucleic acid binding, has been identified in 54 of the 59 aligned sequences2; however, the remaining five accessions revealed similar sequence structure to those of TRAM domains from the other analyzed taxons (Figure 3).
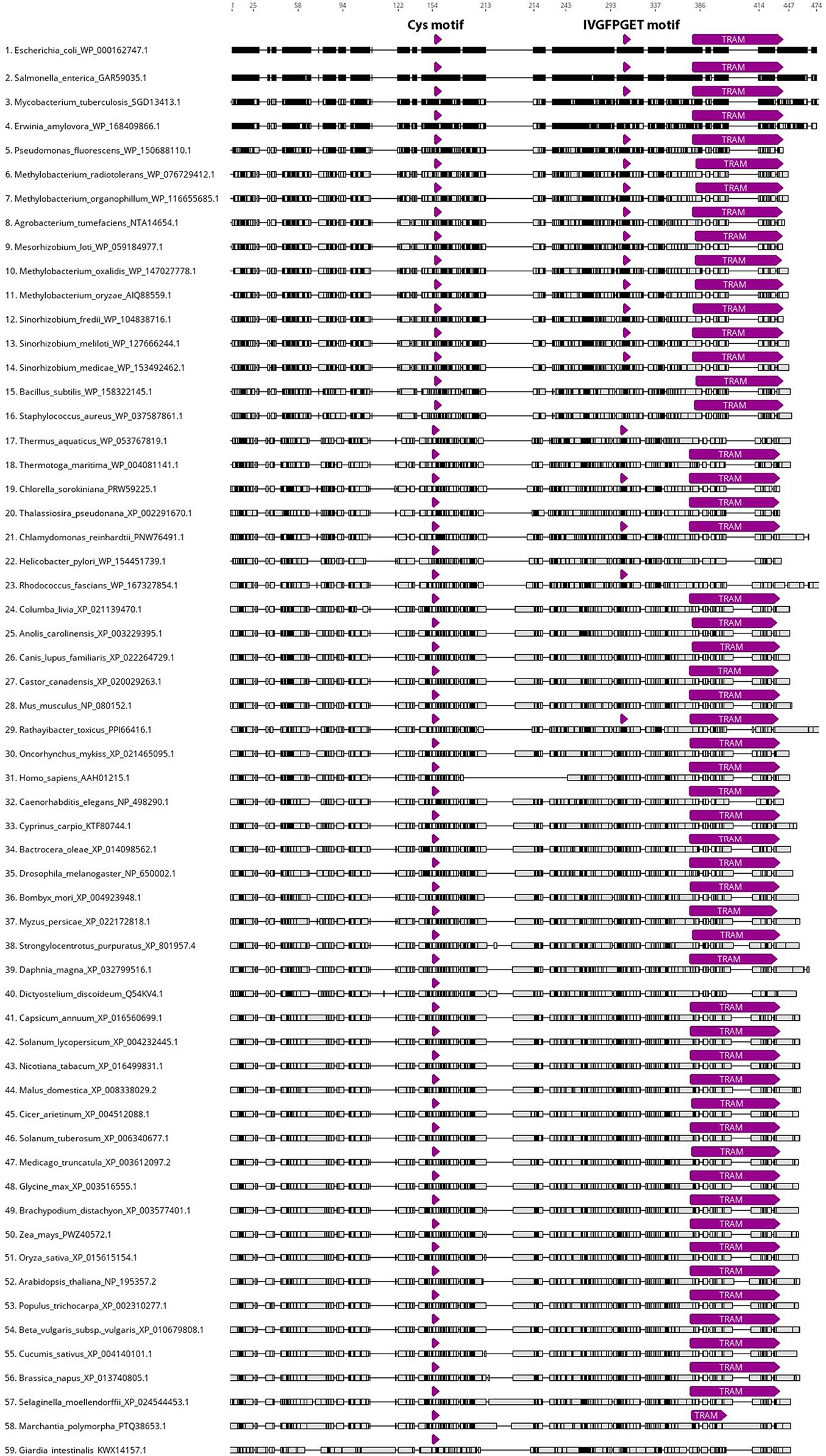
Figure 3. MAFFT alignment of 59 miaB-like protein sequences including representatives of Archaea, Bacteria, Protista, Planta, and Animalia kingdoms. All amino acid sequences were trimmed to the length of E. coli miaB (WP_000162747.1; 1–474 aa). WP_000162747.1 accession was set as reference sequence and all aligned sequences were sorted by the number of differences to reference sequence. Black color indicates regions identical to those of E. coli miaB, gray color indicates differences in the aligned sequences from miaB protein. The locations of three highly conserved miaB regions are highlighted by the purple arrows: Cys motif (CxxxCxxC) – involved in methylthiolation of the prenylated adenine in tRNA 37 position, was identified in all the analyzed sequences; IVGFPGET motif was found in 18 accessions, including representatives of Archaea, Bacteria and Protista; TRAM domain, responsible for nucleic acid binding, was present in 54 out of the 59 aligned sequences. The alignment, motif search and visualization were performed using Align/Assemble function in Geneious Prime 2020.2.3.
The A. thaliana homolog of bacterial miaB gene is known as AT4G36390. Structurally, AT4G36390 resembles miaB as it contains a C terminal RNA-binding TRAM domain and functions similarly to bacterial tRNA methylthiotransferases (Dabravolski, 2020). AT4G36390 is predicted to localize mainly to the mitochondria and carry an [Fe-S]; however, much of its role remains unknown (Dabravolski, 2020).
In mammals, cyclin dependent kinase 5 regulatory subunit-associated protein 1 (CDK5RAP1) is a type of radical SAM enzyme that reductively cleaves S-adenosyl-L-methionine (Atta et al., 2010), with homology to miaB (Kaminska et al., 2008; Reiter et al., 2012). CDK5RAP1 is responsible for methylthiolation of tRNA-bound CKs and has roles in altering stability of tRNA molecules, interactions with ribosomes and translation (Jenner et al., 2010; Horvath and Chinnery, 2015). The presence of unbound 2MeS-CK metabolites was recently reported from the surveyed canine tissues and human cell cultures (Seegobin et al., 2018; Aoki et al., 2019b).
A further step of the 2MeS-CK production via tRNA degradation pathway involves hydroxylation of the isopentenyl side chain which can occur before or after methylthiolation (Dabravolski, 2020) (Figure 2). The biosynthesis of tRNA-bound ms2io6A (2MeScZR) in S. typhimurium involves multiple enzymatic activities and their corresponding genetic loci of which miaE is responsible for the hydroxylation step (Persson and Björk, 1993). The tRNA-(ms2io6A37)-hydroxylase is present in Salmonella under anaerobic conditions; but the hydroxylation reaction does not occur in the absence of oxygen, indicating that it depends on the presence of molecular oxygen (Buck and Ames, 1984).
Bacterial MiaE hydroxylates a terminal methyl group of ms2i6A (2MeSiPR) to form ms2io6A (2MeScZR). Cloning the miaE gene that encodes the tRNA hydroxylase from S. typhimurium and complementing the E. coli strain that is naturally deficient in ms2io6A hydroxylation gene increased the rate of ms2i6A37 (2MeSiPR) hydroxylation by approximately 250-fold (Persson and Björk, 1993). It has been suggested that the miaE gene may encode either the tRNA-(ms2io6A37)-hydroxylase or a cofactor necessary for the hydroxylation reaction. However, it is unlikely that miaE encodes a cofactor needed for only one reaction, and even less likely that E. coli would have kept the hydroxylating enzyme through evolution despite the absence of the necessary cofactor; therefore, it seems more probable that the miaE gene encodes the tRNA-(ms2io6A37)-hydroxylase itself (Persson and Björk, 1993).
Fungi lack the miaE gene as it is not found in eukaryotes (Persson and Björk, 1993; Frébort et al., 2011). In plants, conversion of iPR and 2MeSiPR to cZ and 2MeScZR, respectively, occurs through hydroxylases, which function similarly to bacterial MiaE enzymes (Takei et al., 2004). BLASTp searches could not identify corresponding hydroxylase homologs in mammalian candidates (Seegobin et al., 2018). However, CYP450 enzymes that are involved in hormone metabolism in mammals (Nebert and Russell, 2002; Schmidt et al., 2008) remain to be investigated for their potential role as cis-hydroxylating agents, especially regarding the previously detected 2MeSZR metabolites in both canine tissues and human cells (Seegobin et al., 2018; Aoki et al., 2019b).
Unbound 2MeS-CKs are present in biological systems as a result of degradation of the tRNA molecules modified by the set of genes described in the previous sections. The genes responsible for subsequent steps of 2MeS-CK biosynthesis and the effects of tRNA modifications leading to the formation of bound 2MeS-CKs in microbes and mammals have been known for decades; but the highly sensitive methods of HPLC-MS/MS that enable the precise detection and quantification of unbound, hydrophobic 2MeS-CK metabolites from biological samples have only been available for the last 10 years (Tarkowski et al., 2010; Šimura et al., 2018; Kisiala et al., 2019). Table 1 is a summary of the previously published HPLC-MS/MS results which confirm a recurring presence of 2MeS-CKs among species from various kingdoms of life including microbes, plants, and animals.
To date, most research on CK molecules have investigated plant classical CKs (unmodified iP-, tZ-, cZ-, and DZ-types) while not much interest has been put into explaining the presence of unbound 2MeS-CKs in biological systems (Großkinsky et al., 2016). This, in consequence, resulted in a general lack of studies involving mutants impaired in the function of 2MeS-CK biosynthetic genes, as compared to IPT, LOG, or CKX modifications, creating additional challenges for understanding the methylthiolated CK compounds. Nevertheless, the available CK profiling data on miaB knock-out mutant of E. coli (JW0658-1) revealed a lack of 2MeS-CKs while, simultaneously, the levels of iPRP were considerably increased in the culture lysate (Daudu et al., 2019). These findings support other evidence from genetic studies that the pool of available prenyl-tRNA – the precursor molecule of the classical CKs originated via tRNA degradation pathway – is also the source of unbound 2MeS-CKs (Esberg et al., 1999; Morrison et al., 2017; Figure 2). The metabolite profiles reported for microbial, plant and animal-derived 2MeS-CKs often reflect the close association between this group of compounds and select, classical CK types (particularly, iP and its precursor forms), which implies that they originate from a common precursor in the tRNA degradation pathway.
Methylobacterium strains are plant-associated bacteria well known for their plant growth promoting characteristics and unique capabilities to synthesize high levels of biologically active tZ-type CKs. The study of CK profiles of eight Methylobacterium isolates revealed that strains secreting the highest levels of 2MeS-CKs (specifically 2MeSZ) simultaneously had low tZ production (i.e., M. organophillum NBRC103119: tZ – 12.4 pmol/10 mL, 2MeSZ – 119.6 pmol/10 mL, and M. oryzae LMG23582(T): tZ – 212.9 pmol/10 mL, 2MeSZ – 39.9 pmol/10 mL) (Jorge et al., 2019).
In plants, the analysis of CK profiles during the reproductive development of 27 field grown soybean cultivars revealed an association between the levels of 2MeS-CKs and CK nucleotides. Namely, the cultivars characterized by the higher 2MeSZ levels in their pods at R4 growth stage contained lower concentrations of iPRP while high iPRP production was associated with the lower 2MeSZ levels in the developing pods (Pearson’s r = −0.467) (Kambhampati et al., 2017). A clear, offsetting balance between the levels of CK nucleotides and 2MeSCKs occurs during the reproductive growth stages of A. thaliana WT as well as its two tRNA-IPT mutants, ipt2 and ipt9 (Butler, 2019). In detail, in immature siliques, Arabidopsis plants contained prominent levels of iPRP and a relatively low concentrations of a single 2MeS-CK form (2MeSZ). On the other hand, CK profiles of maturing seeds lacked any nucleotide CK precursors while considerable increases in the concentrations of 2MeS-CK derivatives [2MeSZ, 2MeSZR and 2-methylthio-isopentenyladenine (2MeSiP)] were observed (Butler, 2019).
These types of inverse associations were also characteristic for the HPLC-MS/MS profiles of CKs reported from representatives of six orders of Insecta (Andreas et al., 2020). For example, larvae of two stem-boring beetle species, Mecinus janthinus and Mecinus janthiniformis, demonstrated highly elevated concentrations of nucleotide precursors of the classical CKs and only trace levels of 2MeS-CK derivatives. The adult forms of these two weevils had unusually high levels of 2MeS-CKs (2MeSZ and 2MeSiP), and minimal concentrations of the classical CKs (tZ- and iP-type) suggesting there were different activities of the genes in the tRNA modification pathway during subsequent stages of insect development. Similar associations were observed among other insect forms and species (Andreas et al., 2020).
A negative association between the levels of 2MeS-CKs and classical CKs was reported from analyses of mammalian samples (Seegobin et al., 2018). The presence of 2MeS-CKs was identified in 8 out of 21 tested canine organs and tissues, which included: the inner and outer kidney medulla, stomach, appendix, small and large intestine, bile and thyroid. In most of the analyzed organs, iP-type CKs were the predominant CK forms while 2MeS-CKs were generally found at much lower levels. However, in bile, considerably higher concentration of 2MeS-CKs were detected, which was accompanied by lower levels of iP-type CKs (i.e., thyroid: 2MeS-type CKs – 3.82 pmol/gFW, iP-type CKs – 1392.75 pmol/gFW; bile: 2MeS-type CKs – 229.96 pmol/gFW, iP-type CKs – 114.68 pmol/gFW) (Seegobin et al., 2018).
Despite the availability of mass spectrometry methods for the analysis of 2MeS-CKs (Tarkowski et al., 2010; Kisiala et al., 2019) and emerging number of reports of their presence in different biological systems (including mammals, plants, pathogen-host associations, bacteria, and algae), the functional significance of 2MeS-CKs continues to remain poorly understood (Esberg et al., 1999; Morrison et al., 2015a; Kambhampati et al., 2017; Daudu et al., 2019; Koshla et al., 2019). Most knowledge about the role of 2MeS-CKs stems from their occurrence in post translational tRNA modifications which have a role in translational efficiency, fidelity, and specificity of codon/anticodon interactions (Persson and Björk, 1993; Golovko et al., 2000; Koshla et al., 2020). Deletion of key genes in the methylthiolation pathway can result in aberrant phenotypes most often related to impacts on translation (Koshla et al., 2019). However, the impact of the released, methylthiolated CK compounds on the physiology of organisms has yet to be fully understood.
Koshla et al. (2019, 2020) examined the impact of miaA, B and AB null mutants in Streptomyces albus showing that miaA and B deficiency caused significant changes in morphology of Streptomyces and its secondary metabolome profile. Likely caused by less efficient translation, the miaAB mutant was devoid of endogenous antibacterial activity while the parent miaA mutant still displayed some antibacterial activity (Koshla et al., 2019). miaB mutants had delayed biomass production and miaAB mutants produced sparse aerial hyphae without spore chains compared to the parental mutant miaA (Koshla et al., 2020), suggesting that, even in the absence of miaA, miaB functions to maintain cell cycle and morphology. Phenotypic discrimination between the mutated bacterial lines was highly conditional and only possible on specific media during early stages of their lifecycle; indicating that the impact of methylthiolation may be specific to certain stages of the growth cycle under defined nutrient conditions (Koshla et al., 2020). Although the levels of unbound 2MeS-CKs were not analyzed, this study suggests that the removal of genes responsible for thiolation impacts total free 2MeS-CKs and thereby also has an impact on the overall development of the bacteria (Koshla et al., 2020).
Similar to bacteria, knockdown mutations of enzymes responsible for the methylthiolation of tRNA in mammals resulted in changes to cell cycle. Mammalian CDK5RAP1 is related to the regulation and progression of the M phase of the cell cycle (Wang et al., 2015). Deficiency of the CDK5RAP1 enzyme (a homolog of bacterial miaB) in the knockdown mutation of human breast cancer cell line MCF-7 suppressed tumor growth through cell arrest at the G2/M phase and induced cell apoptosis (Wang et al., 2015). Cytokinins have antitumor effects, with exogenous iPR treatment causing autophagic cell death; however, CDK5RAP1 reduced the antitumor effect of endogenous iPR by converting it to 2MeSiPR and protecting glioma initiating cells (GICs) from autophagy triggered by iPR toxicity. Isopentenyladenosine was not converted directly to 2MeSiPR when it was exogenously applied to GICs (Yamamoto et al., 2019).
The hypermodifications of tRNA not only aid in translation and cell cycle functioning, but after degradation of the modified tRNAs, they act as small molecules able to trigger physiological effects and activate CK receptors (Daudu et al., 2017, 2019). The role of histidine kinase (HK) CK receptors from Malus domestica (apple tree) in the perception of classical and methylthiolated CKs were examined utilizing a histidine kinase deletion mutant in yeast (sln1 deletion) (Daudu et al., 2017). Complementation assays and examining yeast cell growth in response to CK addition revealed that MdCHK2 have higher sensitivity to iP and 2MeSiP (1 nM) relative to other CKs tested. Other examined MdCHKs also responded to 2MeS-CKs (5 nM–5 μM depending on receptor) indicating that 2MeS-CKs can be perceived by CK receptors and trigger growth responses (Daudu et al., 2017).
The yeast mutant strain sln1Δ-MdCHK was further utilized as a CK biosensor and was exposed to extracts from a variety of organisms to detect their production of CKs (Daudu et al., 2019). When exposed to supernatant extracts from E. coli miaB deletion mutant, yeast growth was triggered but was considered to be less than the wildtype. Yeast growth was not decreased due to changes in total level of CKs as these were similar between the wildtype (16.6 pmol/mL) and miaB mutant (17.9 pmol/mL). The E. coli miaB deletion strain had higher levels of iPRP and iP relative to the WT strain while it was suggested that the limited yeast growth was due to reduction in 2MeSiP which was not detected in the miaB deletion mutant (Daudu et al., 2019).
The ability of receptors to respond to 2MeS-CKs suggests these unique CKs can trigger downstream impacts such as growth changes. Correlations between 2MeS-CK levels and observed phenotypes can indicate a potential role or influence that 2MeS-CKs may have on a system. In a field trial sampling of twenty-seven soybean cultivars high quantities of 2MeSZR and 2MeSZ were detected at all three stages of seed filling (R4,5,6) of high yielding varieties. There was also a positive correlation of 2MeSZ with all yield components measured suggesting 2MeS-CKs may play a role in seed filling and subsequent yield formation in soybean (Kambhampati et al., 2017).
In plant-microbe interaction systems, 2MeS-type CKs are often suggested to be of microbial origin. Cytokinins, including bacterially produced 2MeS-CKs play a role in the dormancy of annual ryegrass (Lolium rigidum) seeds (Goggin et al., 2015). Removal of the bacterial symbiont resulted in seeds being unable to break dormancy in the dark, suggesting that a complex integration between plant hormones and those produced by bacterial microflora, including 2MeS-CKs, play an important role in controlling seed maturation in L. rigidum (Goggin et al., 2015).
Spikes in 2MeS-CKs have been detected during host-pathogen relations where increased plant growth is a disease symptom. A. thaliana infection by a bacterial phytopathogenic strain of R. fascians had the same spectrum of CKs as the non-pathogenic counterpart; yet the former had higher levels of 2MeScZ, iP and cZ (Pertry et al., 2009). The authors suggested that 2MeS-CKs may be less cytotoxic than classical CKs which could account for their accumulation during infection (Pertry et al., 2009). The low but consistent presence of 2MeS-CKs may be important in eliciting plant responses during disease development (Giron and Glevarec, 2014). The accumulation of cZ and 2MeS-CKs in infected plant tissues and the transient peak in the levels of iP, 2MeSiP, and 2-methylthio-transZeatin (2MeStZ) in the early interaction phase hinted at the biological significance of the identified bacterial 2MeS-CKs (Pertry et al., 2009). These characteristics also suggest that 2MeS-CKs may be more resistant to CK degrading enzymes (CKX) and are capable of escaping attempts by the host to balance CKs (Morrison et al., 2015a). Therefore, 2MeS-CKs are thought to help hijack the plant machinery while avoiding deleterious effects on plant development and activation of CK mediated plant defenses (Giron and Glevarec, 2014). Overall, 2MeS-CKs may be responsible for continuous tissue proliferation and symptom maintenance during plant-pathogen interaction (Pertry et al., 2009). Observations of the Zea mays – U. maydis plant-pathogen system led to the similar conclusion as previously suggested for the 2MeS-CK effect in Arabidopsis – R. fascians interaction. Although the fungus U. maydis does not produce 2MeS-CKs separate from the plant, during the Zea mays – U. maydis relation an accumulation of 2MeS-CKs (specifically 2MeSZR and 2MeSZ) occurred in the infected plant tissue during the later stages of cob infection (days 20-28) while these CKs were not detected in the control cobs (Morrison et al., 2015a, 2017).
The contribution of CK interplay was examined in the human pathogen, Mycobacterium tuberculosis, the causative agent of tuberculosis. This bacterium secretes several iP- and tZ-type CKs including 2MeSiP, 2MeSiPR and 2MeStZR (Samanovic et al., 2015). A higher activity of M. tuberculosis LOG-like enzyme, Rv105, results in CK accumulation including 2MeSiP. That, in turn, increases sensitivity of bacterial strains to nitric oxide, a defense response from the host cells, detrimental to pathogen survival. These results suggest that among other CKs, 2MeS-CKs play a role in sensitizing the pathogen to the activation of host defense mechanisms (Samanovic et al., 2015).
While most studies focus on the impact of gene deletion and the resulting effects which might then be rescued through gene complementation (Koshla et al., 2020), few have added exogenous 2MeS-CKs to rescue deletion phenotypes or applied exogenous methylthiol CK derivatives to examine their impact on a biological system.
The HPLC-MS/MS analysis of CK profiles in microalgae Chlorella vulgaris revealed that 2MeS-CKs were the prevalent CKs in both culture supernatant and cell pellet, with 2MeSZ being the most abundant in the pellet (4.7 pmol/g) and at high concentrations in the supernatant (220.7 pmol/g) followed by other methylthiol conjugates (Ramphal, 2016). To understand the role of 2MeS-CKs in C. vulgaris cell cycle, the effect of exogenous 2MeSZ treatment on the cell growth and select metabolite production were examined. Culture supplementation with 2MeSZ resulted in growth stimulation and had an impact on the microalgae fatty acid profiles in a manner comparable to those of the other active CKs that were tested (tZ, benzyladenine). In particular, significant increases in linolenic acid (18-carbon chain with three double bonds; 18:3) occurred at 10–7 and 10–6 M 2MeSZ concentrations; 204 and 457% increase relative to the control, respectively (Ramphal, 2016). It indicates that the addition of exogenous methylthiol CKs can impact growth as well as fatty acid metabolism in algae. Modified secondary metabolite production was also a result of the deletion of miaB in the bacterium, S. albus (Koshla et al., 2020). These studies suggest that the methylthiolated CK compounds are not only necessary for efficient translation but likely are also involved in activating downstream metabolic pathways.
Physiological roles of classical CKs (unmodified iP, tZ, cZ, and DZ and their derivatives) have been studied extensively to help uncover the functions of these signaling molecules in different aspects of plant physiology. In parallel, extensive knowledge exists for genes involved in tRNA modifications that lead to the production of tRNA-bound 2MeS-CKs, especially in bacterial and mammalian systems. However, not much is known about the fate and possible function of the unbound 2MeS-CKs, derived via degradation of the modified tRNA molecules in biological systems.
There is a compelling need to further study this most elusive group of CKs as evidenced by their frequent detection, which is often at high concentrations when found in different plant organs and at different developmental stages; moreover, there is an increasing number of reports on 2MeS-CK presence in other, non-plant organisms. Suggestions of the potential metabolic activities of unbound 2MeS-CKs have been made previously. The limited physiological data available indicate that 2MeS-CKs may play a role that is similar to classical CKs; whereby, 2MeS-CKs impact cell division as they are often detected in systems where active proliferation occurs, like plant-pathogen disease development and mammalian cancer cells. In addition, 2MeS-CKs may be able to reduce the pool of more active iP-type CKs resulting in production of less cytotoxic forms of CKs. As discussed above, conversion of iPR in mammals to 2MeSiPR reduced the anticancer effect of iPR on cells thus decreasing the potency of the CK application. Likewise, production of 2MeS-CKs during plant-pathogen disease development may allow for persistent accumulation of forms that are more resistant to degradation and, therefore, can act over a longer period of time. Finally, exogenous application of 2MeS-CKs can impact cell metabolism and growth as seen in C. vulgaris.
The potential impact of 2MeS-CKs should no longer be overlooked. Their functional role needs to be better understood and further investigation into the deletion of key genes in the methylthiolation pathways as well as the exogenous applications of 2MeS-CKs should be continued to determine the physiological roles of these compounds. However, parsing out the translation-efficiency effects of bound 2MeS-CKs and physiological effects of unbound 2MeS-CKs may prove to be quite difficult. With this in mind, the development of CK mutants impaired in functioning of 2MeS-CK biosynthesis genes might create significant challenges and the possible, strong pleiotropic effects of the mutations targeting the genes involved in tRNA modifications have to be considered. tRNA-bound 2MeS-CKs play critical roles in diverse aspects of RNA metabolism and the effect of their disturbance might be difficult to separate from the potential physiological roles of unbound 2MeS-CKs obtained via tRNA degradation. New approaches that separate effects of tRNA-bound and unbound-2MeS-CKs will have to be developed to overcome that challenge, of fully understanding the often-prevalent and enigmatic 2MeS-CKs.
MG and AK wrote the first draft of the manuscript. EM and RE wrote sections of the manuscript. All authors contributed to manuscript revision, read and approved the submitted version.
RE was supported by Discovery Grant from the Natural Sciences and Engineering Research Council of Canada (RGPIN-05436).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Anantharaman, V., Koonin, E. V., and Aravind, L. (2001). TRAM, a predicted RNA-binding domain, common to tRNA uracil methylation and adenine thiolation enzymes. FEMS Microbiol. Lett. 197, 215–221. doi: 10.1111/j.1574-6968.2001.tb10606.x
Anantharaman, V., Koonin, E. V., and Aravind, L. (2002). Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acid Res. 30, 1427–1464. doi: 10.1093/nar/30.7.1427
Andreas, P., Kisiala, A., Emery, R. J. N., Clerck-Floate, D., Tooker, J., and Price, M. III, et al. (2020). Cytokinins are abundant and widespread among insect species. Plants 9:208. doi: 10.3390/plants9020208
Anjard, C., and Loomis, W. F. (2008). Cytokinins induce sporulation in Dictyostelium. Development 135, 819–827. doi: 10.1242/dev.018051
Aoki, M. M., Emery, R. J. N., Anjard, C., Brunetti, C. R., and Huber, R. J. (2020). Cytokinins in Dictyostelia – A unique model for studying the functions of signaling agents from species to kingdoms. Front. Cell Dev. Biol. 8:511. doi: 10.3389/fcell.2020.00511
Aoki, M. M., Kisiala, A. B., Li, S., Stock, N. L., Brunetti, C. R., Huber, R. J., et al. (2019a). Cytokinin detection during the Dictyostelium discoideum life cycle: profiles are dynamic and affect cell growth and spore germination. Biomolecules 9:702. doi: 10.3390/biom9110702
Aoki, M. M., Seegobin, M., Kisiala, A., Noble, A., Brunetti, C., and Emery, R. J. N. (2019b). Phytohormone metabolism in human cells: cytokinins are taken up and interconverted in HeLa cell culture. FASEB BioAdv. 1, 320–331. doi: 10.1096/fba.2018-00032
Atta, M., Mulliez, E., Arragain, S., Forouhar, F., Hunt, J. F., and Fontecave, M. (2010). S-Adenosylmethionine-dependent radical-based modification of biological macromolecules. Curr. Opin. Struc. Biol. 20, 684–692. doi: 10.1016/j.sbi.2010.09.009
Björk, G. R. (1995). Genetic dissection of synthesis and function of modified nucleosides in bacterial transfer RNA. Prog. Nucleic Acid Res. Mol. Biol. 50, 263–338. doi: 10.1016/s0079-6603(08)60817-x
Blum, P. H. (1988). Reduced leu operon expression in a miaA mutant of Salmonella typhimurium. J. Bacteriol. 170, 5125–5133. doi: 10.1128/jb.170.11.5125-5133.1988
Buck, M., and Ames, B. N. (1984). A modified nucleotide in tRNA as a possible regulator of aerobiosis: synthesis of cis-2-methyl-thioribosylzeatin in the tRNA of Salmonella. Cell 36, 523–531. doi: 10.1016/0092-8674(84)90245-9
Buck, M., and Griffiths, E. (1982). Iron mediated methylthiolation of tRNA as a regulator of operon expression in Escherichia coli. Nucleic Acids Res. 10, 2609–2624. doi: 10.1093/nar/10.8.2609
Butler, C. (2019). cis-Cytokinins from the tRNA-Degradation Pathway Impact the Phenotype and Metabolome of Arabidopsis thaliana: Evidence from AtIPT2 and AtIPT9 Null Mutants. master’s thesis, Trent University, Peterborough, ON.
Chanclud, E., Kisiala, A., Emery, R. J., Chalvon, V., Ducasse, A., Romiti-Michel, C., et al. (2016). Cytokinin production by the rice blast fungus is a pivotal requirement for full virulence. PLoS Pathog. 12:e1005457. doi: 10.1371/journal.ppat.1005457
Chen, Y., Bai, B., Yan, H., Wen, F., Qin, D., Jander, G., et al. (2019). Systemic disruption of the homeostasis of transfer RNA isopentenyltransferase causes growth and development abnormalities in Bombyx mori. Insect Mol. Biol. 28, 380–391. doi: 10.1111/imb.12561
Cherayil, J. D., and Lipsett, M. N. (1977). Zeatin ribonucleosides in the transfer ribonucleic acid of Rhizobium leguminosarum, Agrobacterium tumefaciens, Corynebacterium fascians, and Erwinia amylovora. J. Bacteriol. 131, 741–744. doi: 10.1128/jb.131.3.741-744.1977
Dabravolski, S. (2020). Multi-faceted nature of the tRNA isopentenyltransferase. Funct. Plant Biol. 47, 475–485. doi: 10.1071/FP19255
Daudu, D., Allion, E., Liesecke, F., Papon, N., Courdavault, V., Dugé de Bernonville, T., et al. (2017). CHASE-containing histidine kinase receptors in apple tree: from a common receptor structure to divergent cytokinin binding properties and specific functions. Front. Plant Sci. 8:1614. doi: 10.3389/fpls.2017.01614
Daudu, D., Kisiala, A., Ribeiro, C. W., Mélin, C., Perrot, L., Clastre, M., et al. (2019). Setting-up a fast and reliable cytokinin biosensor based on a plant histidine kinase receptor expressed in Saccharomyces cerevisiae. J. Biotechnol. 289, 103–111. doi: 10.1016/j.jbiotec.2018.11.013
Dihanich, M. E., Najarian, D., Clark, R., Gillman, E. C., Martin, N. C., and Hopper, A. K. (1987). Isolation and characterization of MOD5, a gene required for isopentenylation of cytoplasmic and mitochondrial tRNAs of Saccharomyces cerevisiae. Mol. Cell. Biol. 7, 177–184. doi: 10.1128/mcb.7.1.177
Eichinger, L., Pachebat, J. A., Glöckner, G., Rajandream, M. A., Sucgang, R., Berriman, M., et al. (2005). The genome of the social amoeba Dictyostelium discoideum. Nature 435, 43–57. doi: 10.1038/nature03481
Emery, R. J. N., Leport, L., Barton, J. E., Turner, N. C., and Atkins, C. A. (1998). Cis -isomers of cytokinins predominate in chickpea seeds throughout their development. Plant Physiol. 117, 1515–1523. doi: 10.1104/pp.117.4.1515
Esberg, B., Leung, H.-C. E., Tsui, H.-C. T., Björk, G. R., and Winkler, M. E. (1999). Identification of the miaB gene, involved in methylthiolation of isopentenylated A37 derivatives in the tRNA of Salmonella typhimurium and Escherichia coli. J. Bacteriol. 181, 7256–7265. doi: 10.1128/jb.181.23.7256-7265.1999
Fradejas, N., Carlson, B. A., Rijntjes, E., Becker, N.-P., Tobe, R., and Schweizer, U. (2013). Mammalian Trit1 is a tRNA([Ser]Sec)-isopentenyl transferase required for full selenoprotein expression. Biochem. J. 450, 427–432. doi: 10.1042/BJ20121713
Frébort, I., Kowalska, M., Hluska, T., Frébortová, J., and Galuszka, P. (2011). Evolution of cytokinin biosynthesis and degradation. J. Exp. Bot. 62, 2431–2452. doi: 10.1093/jxb/err004
Gajdošová, S., Spíchal, L., Kamínek, M., Hoyerová, K., Novák, O., Dobrev, P. I., et al. (2011). Distribution, biological activities, metabolism, and the conceivable function of cis-zeatin-type cytokinins in plants. J. Exp. Bot. 62, 2827–2840. doi: 10.1093/jxb/erq457
Giron, D., Frago, E., Glevarec, G., Pieterse, C. M. J., and Dicke, M. (2013). Cytokinins as key regulators in plant–microbe–insect interactions: connecting plant growth and defence. Funct. Ecol. 27, 599–609. doi: 10.1111/1365-2435.12042
Giron, D., and Glevarec, G. (2014). Cytokinin-induced phenotypes in plant-insect interactions: learning from the bacterial world. J. Chem. Ecol. 40, 826–835. doi: 10.1007/s10886-014-0466-5
Goggin, D. E., Emery, R. N., Kurepin, L. V., and Powles, S. B. (2015). A potential role for endogenous microflora in dormancy release, cytokinin metabolism and the response to fluridone in Lolium rigidum seeds. Ann. Bot. 115, 293–301. doi: 10.1093/aob/mcu231
Golovko, A., Hjälm, G., Sitbon, F., and Nicander, B. (2000). Cloning of a human tRNA isopentenyl transferase. Gene 258, 85–93. doi: 10.1016/s0378-1119(00)00421-2
Gray, J., Gelvin, S. B., Meilan, R., and Morris, R. O. (1996). Transfer RNA is the source of extracellular isopentenyladenine in a Ti-plasmidless strain of Agrobacterium tumefaciens. Plant Physiol. 110, 431–438. doi: 10.1104/pp.110.2.431
Großkinsky, D. K., Tafner, R., Moreno, M. V., Stenglein, S. A., García de Salamone, I. E., Nelson, L. M., et al. (2016). Cytokinin production by Pseudomonas fluorescens G20-18 determines biocontrol activity against Pseudomonas syringae in Arabidopsis. Sci. Rep. 6, 23310. doi: 10.1038/srep23310
Guern, J., and Peaud-Lenoel, C. (2012). Metabolism and Molecular Activities of Cytokinins: Proceedings of the International Colloquium of the Centre National de la Recherche Scientifique held at Gif-sur-Yvette (France) 2–6 September 1980. Cham: Springer Science and Business Media.
Hinsch, J., Vrabka, J., Oeser, B., Novák, O., Galuszka, P., and Tudzynski, P. (2015). De novo biosynthesis of cytokinins in the biotrophic fungus Claviceps purpurea. Environ. Microbiol. 17, 2935–2951. doi: 10.1111/1462-2920.12838
Holland, M. A. (1997). Occam’s razor applied to hormonology (Are cytokinins produced by plants?). Plant Physiol. 115, 865–868. doi: 10.1104/pp.115.3.865
Horvath, R., and Chinnery, P. F. (2015). Modifying mitochondrial tRNAs: delivering what the cell needs. Cell Metab. 21, 351–352. doi: 10.1016/j.cmet.2015.02.012
Jameson, P. E. (2019). Virulent Rhodococcus fascians produce unique methylated cytokinins. Plants 8:582. doi: 10.3390/plants8120582
Jenner, L. B., Demeshkina, N., Yusupova, G., and Yusupov, M. (2010). Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat. Struct. Mol. Biol. 17, 555–560. doi: 10.1038/nsmb.1790
Jorge, G. L., Kisiala, A., Morrison, E., Aoki, M., Nogueira, A. P. O., and Emery, R. J. N. (2019). Endosymbiotic Methylobacterium oryzae mitigates the impact of limited water availability in lentil (Lens culinaris Medik.) by increasing plant cytokinin levels. Environ Exp. Bot. 162, 525–540. doi: 10.1016/j.envexpbot.2019.03.028
Joshi, M. V., and Loria, R. (2007). Streptomyces turgidiscabies possesses a functional cytokinin biosynthetic pathway and produces leafy galls. MPMI 20, 751–758. doi: 10.1094/MPMI-20-7-0751
Kambhampati, S., Kurepin, L., Kisiala, A., Bruce, K., Cober, E., Morrison, M., et al. (2017). Yield associated traits correlate with cytokinin profiles in developing pods and seeds of field-grown soybean cultivars. Field Crops Res. 214, 175–184. doi: 10.1016/j.fcr.2017.09.009
Kaminska, K. H., Baraniak, U., Boniecki, M., Nowaczyk, K., Czerwoniec, A., and Bujnicki, J. M. (2008). Structural bioinformatics analysis of enzymes involved in the biosynthesis pathway of the hypermodified nucleoside ms(2)io(6)A37 in tRNA. Proteins 70, 1–18. doi: 10.1002/prot.21640
Kieber, J. J., and Schaller, G. E. (2018). Cytokinin signaling in plant development. Development 145:dev149344. doi: 10.1242/dev.149344
Kisiala, A., Kambhampati, S., Stock, N. L., Aoki, M., and Emery, R. J. N. (2019). Quantification of cytokinins using high-resolution accurate-mass orbitrap mass spectrometry and parallel reaction monitoring (PRM). Anal. Chem. 91, 15049–15056. doi: 10.1021/acs.analchem.9b03728
Koenig, R. L., Morris, R. O., and Polacco, J. C. (2002). tRNA is the source of low-level trans-Zeatin production in Methylobacterium spp. J. Bacteriol. 184, 1832–1842. doi: 10.1128/JB.184.7.1832-1842.2002
Koshla, O., Kravets, V., Dacyuk, Y., Ostash, I., Süssmuth, R., and Ostash, B. (2020). Genetic analysis of Streptomyces albus J1074 mia mutants suggests complex relationships between post-transcriptional tRNAXXA modifications and physiological traits. [published online ahead of print, 2020 Jul 17]. Folia Microbiol. [Epub ahead of print]. doi: 10.1007/s12223-020-00811-7
Koshla, O., Yushchuk, O., Ostash, I., Dacyuk, Y., Myronovskyi, M., Jäger, G., et al. (2019). Gene miaA for post-transcriptional modification of tRNAXXA is important for morphological and metabolic differentiation in Streptomyces. Mol. Microbiol. 112, 249–265. doi: 10.1111/mmi.14266
Lamichhane, T. N., Mattijssen, S., and Maraia, R. J. (2013). Human cells have a limited set of tRNA anticodon loop substrates of the tRNA isopentenyltransferase TRIT1 tumor suppressor. Mol. Cell Biol. 33, 4900–4908. doi: 10.1128/MCB.01041-13
Laten, H., Gorman, J., and Bock, R. M. (1978). Isopentenyladenosine deficient tRNA from an antisuppressor mutant of Saccharomyces cerevisiae. Nucleic Acids Res. 5, 4329–4342. doi: 10.1093/nar/5.11.4329
Liaqat, A., Stiller, C., Michel, M., Sednev, M. V., and Höbartner, C. (2020). N6-isopentenyladenosine in RNA determines the cleavage site of endonuclease deoxyribozymes. Angew. Chem. 132, 18786–18790. doi: 10.1002/ange.202006218
Lindner, A.-C., Lang, D., Seifert, M., Podlešáková, K., Novák, O., Strnad, M., et al. (2014). Isopentenyltransferase-1 (IPT1) knockout in Physcomitrella together with phylogenetic analyses of IPTs provide insights into evolution of plant cytokinin biosynthesis. J. Exp. Bot. 65, 2533–2543. doi: 10.1093/jxb/eru142
Mayaka, J. B., Huang, Q., Xiao, Y., Zhong, Q., Ni, J., and Shen, Y. (2019). The Lonely Guy (LOG) homologue SiRe_0427 from the thermophilic Archaeon Sulfolobus islandicus REY15A is a phosphoribohydrolase representing a novel group. Appl. Environ. Microbiol. 85:e01739-19. doi: 10.1128/AEM.01739-19
McGaw, B. A., and Burch, L. R. (1995). “Cytokinin biosynthesis and metabolism,” in Plant Hormones: Physiology, Biochemistry, and Molecular Biology, 2nd Edn, ed. P. J. Davies (Dodrecht: Kluwer Academic Publishers), 98–117. doi: 10.1007/978-94-011-0473-9_5
Miyawaki, K., Tarkowski, P., Matsumoto-Kitano, M., Kato, T., Sato, S., Tarkowska, D., et al. (2006). Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. PNAS 103, 16598–16603. doi: 10.1073/pnas.0603522103
Moffatt, K., Monroy Flores, C., Andreas, P., Kisiala, A., and Emery, R. J. N. (2018). Phytohormone profiling reveals fungal signatures and strong manipulation of infection cycle in the Gymnosporangium juniper-virginianae dual-host plant system. Botany 96, 57–65. doi: 10.1139/cjb-2017-0178
Morris, R. O., Regier, D. A., Olson, R. M., Struxness, L. A., and Armstrong, D. J. (1981). Distribution of cytokinin-active nucleosides in isoaccepting transfer ribonucleic acids from Agrobacterium tumefaciens. Biochemistry 20, 6012–6017. doi: 10.1021/bi00524a014
Morrison, E. N., Emery, R. J., and Saville, B. (2015a). Phytohormone involvement in the Ustilago maydis – Zea mays pathosystem: relationships between abscisic acid and cytokinin levels and strain virulence in infected cob tissue. PLoS One 10:e0130945. doi: 10.1371/journal.pone.0130945
Morrison, E. N., Knowles, S., Hayward, A., Thorn, R. G., Saville, B. J., and Emery, R. J. N. (2015b). Detection of phytohormones in temperate forest fungi predicts consistent abscisic acid production and a common pathway for cytokinin biosynthesis. Mycologia 107, 245–257. doi: 10.3852/14-157
Morrison, E. N., Emery, R. J. N., and Saville, B. J. (2017). Fungal derived cytokinins are necessary for normal Ustilago maydis infection of maize. Plant Pathol. 66, 726–742. doi: 10.1111/ppa.12629
Nebert, D. W., and Russell, D. W. (2002). Clinical importance of the cytochromes P450. Lancet 360, 1155–1162. doi: 10.1016/S0140-6736(02)11203-7
Nishii, K., Wright, F., Chen, Y.-Y., and Möller, M. (2018). Tangled history of a multigene family: the evolution of ISOPENTENYLTRANSFERASE genes. PLoS One 13:e0201198. doi: 10.1371/journal.pone.0201198
Noble, A., Kisiala, A., Galer, A., Clysdale, D., and Emery, R. J. N. (2014). Euglena gracilis (Euglenophyceae) produces abscisic acid and cytokinins and responds to their exogenous application singly and in combination with other growth regulators. Eur. J. Phycol. 49, 244–254. doi: 10.1080/09670262.2014.911353
Persson, B. C., and Björk, G. R. (1993). Isolation of the gene (miaE) encoding the hydroxylase involved in the synthesis of 2-methylthio-cis-ribozeatin in tRNA of Salmonella typhimurium and characterization of mutants. J. Bacteriol. 175, 7776–7785. doi: 10.1128/jb.175.24.7776-7785.1993
Persson, B. C., Esberg, B., Olafsson, O., and Björk, G. R. (1994). Synthesis and function of isopentenyl adenosine derivatives in tRNA. Biochimie 76, 1152–1160. doi: 10.1016/0300-9084(94)90044-2
Pertry, I., Václavíková, K., Depuydt, S., Galuszka, P., Spíchal, L., Temmerman, W., et al. (2009). Identification of Rhodococcus fascians cytokinins and their modus operandi to reshape the plant. PNAS 106, 929–934. doi: 10.1073/pnas.0811683106
Pierrel, F., Björk, G. R., Fontecave, M., and Atta, M. (2002). Enzymatic modification of tRNAs: MiaB is an iron-sulfur protein. J. Biol. Chem. 277, 13367–13370. doi: 10.1074/jbc.C100609200
Pierrel, F., Douki, T., Fontecave, M., and Atta, M. (2004). MiaB protein is a bifunctional radical-S-adenosylmethionine enzyme involved in thiolation and methylation of tRNA. J. Biol. Chem. 279, 47555–47563. doi: 10.1074/jbc.M408562200
Pierrel, F., Hernandez, H. L., Johnson, M. K., Fontecave, M., and Atta, M. (2003). MiaB protein from Thermotoga maritima. Characterization of an extremely thermophilic tRNA-methylthiotransferase. J. Biol. Chem. 278, 29515–29524. doi: 10.1074/jbc.M301518200
Podlešáková, K., Fardoux, J., Patrel, D., Bonaldi, K., Novák, O., Strnad, M., et al. (2013). Rhizobial synthesized cytokinins contribute to but are not essential for the symbiotic interaction between photosynthetic Bradyrhizobia and Aeschynomene legumes. MPMI 26, 1232–1238. doi: 10.1094/MPMI-03-13-0076-R
Ramphal, K. (2016). Hormonal algae: A Source of Functional Fatty Acids. master’s thesis, Trent University, Peterborough, ON.
Reiter, V., Matschkal, D. M., Wagner, M., Globisch, D., Kneuttinger, A. C., Müller, M., et al. (2012). The CDK5 repressor CDK5RAP1 is a methylthiotransferase acting on nuclear and mitochondrial RNA. Nucleic Acids Res. 40, 6235–6240. doi: 10.1093/nar/gks240
Sakakibara, H. (2006). Cytokinins: activity, biosynthesis, and translocation. Ann. Rev. Plant Biol. 57, 431–449. doi: 10.1146/annurev.arplant.57.032905.105231
Samanovic, M. I., Tu, S., Novák, O., Iyer, L. M., McAllister, F. E., Aravind, L., et al. (2015). Proteasomal control of cytokinin synthesis protects Mycobacterium tuberculosis against nitric oxide. Mol. Cell. 57, 984–994. doi: 10.1016/j.molcel.2015.01.024
Schäfer, M., Brütting, C., Canales, I. M., Großkinsky, D. K., Vankova, R., Baldwin, I. T., et al. (2015). The role of cis-zeatin-type cytokinins in plant growth regulation and mediating responses to environmental interactions. J. Exp. Bot. 66, 4873–4884. doi: 10.1093/jxb/erv214
Schmidt, U., Ahmed, J., Michalsky, E., Hoepfner, M., and Preissner, R. (2008). Comparative Vegf receptor tyrosine kinase modeling for the development of highly specific inhibitors of tumor angiogenesis. Genome Inform. 20, 243–251. doi: 10.11234/gi1990.20.243
Seegobin, M., Kisiala, A., Noble, A., Kaplan, D., Brunetti, C., and Emery, R. J. N. (2018). Canis familiaris tissues are characterized by different profiles of cytokinins typical of the tRNA degradation pathway. FASEB J. [Epub ahead of print]. doi: 10.1096/fj.201800347
Silva-Navas, J., Conesa, C. M., Saez, A., Navarro-Neila, S., Garcia-Mina, J. M., Zamarreño, A. M., et al. (2019). Role of cis-zeatin in root responses to phosphate starvation. New Phytol. 224, 242–257. doi: 10.1111/nph.16020
Šimura, J., Antoniadi, I., Široká, J., Tarkowská, D., Strnad, M., Ljung, K., et al. (2018). Plant hormonomics: multiple phytohormone profiling by targeted metabolomics. Plant Physiol. 177, 476–489. doi: 10.1104/pp.18.00293
Smaldino, P. J., Read, D. F., Pratt-Hyatt, M., Hopper, A. K., and Engelke, D. R. (2015). The cytoplasmic and nuclear populations of the eukaryote tRNA-isopentenyl transferase have distinct functions with implications in human cancer. Gene 556, 13–18. doi: 10.1016/j.gene.2014.09.049
Spíchal, L. (2012). Cytokinins – recent news and views of evolutionally old molecules. Funct. Plant Biol. 39, 267–284. doi: 10.1071/fp11276
Spinola, M., Galvan, A., Pignatiello, C., Conti, B., Pastorino, U., Nicander, B., et al. (2005). Identification and functional characterization of the candidate tumor suppressor gene TRIT1 in human lung cancer. Oncogene 24, 5502–5509. doi: 10.1038/sj.onc.1208687
Straka, J., Hayward, A., and Emery, R. J. (2010). Gall-inducing Pachypsylla celtidis (Psyllidae) infiltrate hackberry trees with high concentrations of phytohormones. J. Plant Interact. 5, 197–203. doi: 10.1080/17429145.2010.484552
Streletskii, R. A., Kachalkin, A. V., Glushakova, A. M., Yurkov, A. M., and Demin, V. V. (2019). Yeasts producing zeatin. PeerJ 7:e6474. doi: 10.7717/peerj.6474
Suzuki, G., Shimazu, N., and Tanaka, M. (2012). A yeast prion, Mod5, promotes acquired drug resistance and cell survival under environmental stress. Science 336, 355–359. doi: 10.1126/science.1219491
Takei, K., Yamaya, T., and Sakakibara, H. (2004). Arabidopsis CYP735A1 and CYP735A2 encode cytokinin hydroxylases that catalyze the biosynthesis of trans-Zeatin. J. Biol. Chem. 279, 41866–41872. doi: 10.1074/jbc.M406337200
Tarkowski, P., Václavíková, K., Novák, O., Pertry, I., Hanuš, J., Whenham, R., et al. (2010). Analysis of 2-methylthio-derivatives of isoprenoid cytokinins by liquid chromatography-tandem mass spectrometry. Anal. Chim. Acta 680, 86–91. doi: 10.1016/j.aca.2010.09.020
Trdá, L., Barešová, M., Šašek, V., Nováková, M., Zahajská, L., Dobrev, P. I., et al. (2017). Cytokinin metabolism of pathogenic fungus Leptosphaeria maculans involves isopentenyltransferase, adenosine kinase and cytokinin oxidase/dehydrogenase. Front. Microbiol. 8:1374. doi: 10.3389/fmicb.2017.01374
Vedenicheva, N. P., Al-Maali, G. A., Bisko, N. A., Shcherbatiuk, M. M., Lomberg, M. L., Mytropolska, N. Y., et al. (2018). Comparative analysis of cytokinins in mycelial biomass of medicinal mushrooms. Int. J. Med. Mushrooms 20, 837–847. doi: 10.1615/IntJMedMushrooms.2018027797
Waller, T. J., Read, D. F., Engelke, D. R., and Smaldino, P. J. (2017). The human tRNA-modifying protein, TRIT1, forms amyloid fibers in vitro. Gene 612, 19–24. doi: 10.1016/j.gene.2016.10.041
Wang, H., Wei, L., Li, C., Zhou, J., and Li, Z. (2015). CDK5RAP1 deficiency induces cell cycle arrest and apoptosis in human breast cancer cell line by the ROS/JNK signaling pathway. Oncol. Rep. 33, 1089–1096. doi: 10.3892/or.2015.3736
Wang, X., Lin, S., Liu, D., Gan, L., McAvoy, R., Ding, J., et al. (2020). Evolution and roles of cytokinin genes in angiosperms 1: do ancient IPTs play housekeeping while non-ancient IPTs play regulatory roles? Hortic. Res. 7:28. doi: 10.1038/s41438-019-0211-x
Keywords: 2MeSZ, 2MeSiP, methylthiolated cytokinins, methylthiotransferase, tRNA degradation pathway
Citation: Gibb M, Kisiala AB, Morrison EN and Emery RJN (2020) The Origins and Roles of Methylthiolated Cytokinins: Evidence From Among Life Kingdoms. Front. Cell Dev. Biol. 8:605672. doi: 10.3389/fcell.2020.605672
Received: 12 September 2020; Accepted: 19 October 2020;
Published: 09 November 2020.
Edited by:
Andrew Burgess, Anzac Research Institute, AustraliaReviewed by:
Wendy A. Stirk, University of KwaZulu-Natal, South AfricaCopyright © 2020 Gibb, Kisiala, Morrison and Emery. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anna B. Kisiala, YW5uYWtpc2lhbGFAdHJlbnR1LmNh
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.