Introduction
The profilins (PRFs) are low-molecular weight, cytosolic proteins made up of 129–133 amino acids and tightly control cell-cytoskeleton architecture mainly through actin polymerization (Christensen et al., 1996; Pollard and Cooper, 2009). The PRFs are the actin-binding proteins having conserved profilin-actin interacting regions (PAINRs) which are indispensable to the mechanism of actin-polymerization or -depolymerization process (Giehl et al., 1994; Pandey and Chaudhary, 2017). Globally, the size and location of PAINRs vary from 10 to 11 amino acids and between 57 and 128 amino acids in PRFs, respectively. Evidently, a hypervariable region at the C-terminal of PRF is responsible for their complex binding with actin filaments and regulates diverse functions (Sohn and Goldschmidt-Clermont, 1994). Although PRF proteins are present in almost all life forms including lower and higher plants, animals, fungi, protists and viruses, little is explicit about the PRF-mediated molecular interplay and the corresponding functional networks in land plants.
The PRF genes are ancient, evolutionarily conserved and functionally divergent among life kingdoms. Apparently, these genes are responsible for the maintenance of cell-walls by actin sequestration, nucleation and cytokinesis in both prokarya and eukarya (Magdolen et al., 1988). The advent of genomics and biotechnology has emphasized the emerging roles of PRF genes during plant growth and development that reveals their “novel” functional capabilities beyond their traditional contributions to the cytoskeleton maintenance. Their functional attributes in the root elongation, leaf morphology, epidermal expansion, flowering time phenotypes and seed germination have recently been explored in various plant species (Ramachandran et al., 2000; Müssar et al., 2015; Pandey and Chaudhary, 2016). Constitutive overexpression and silencing of PRF genes show strong effects on fiber initiation and elongation in cotton as well as flowering time phenotypes, stress tolerance and regulation of in vitro organogenesis in tobacco. Previously, we have characterized the molecular interactions of PRFs with proline-rich ligands including Arp2/3 complex, ARP4, ARP6 proteins, cell signaling polyphosphoinositides, and other actin-binding proteins during plant growth and development (Supplementary Figure 1) (Pandey and Chaudhary, 2016). These findings decisively endorse the invaluable roles of PRF genes not only in the traditionally established cytoskeleton maintenance but also in the key aspects of plant development which can be further exploited for the crop improvement programs.
Evolution of PRF Gene Family in Land Plants
Comparative PRF phylogenetic analyses of various life forms including lower and higher plants, animals, fungi, protists, and viruses emphasized their ancient origin. Most plant and animal species have several PRF genes on contrary to few such genes in the lower life forms. Interestingly, PRF genes are evolutionarily preserved and emerged prior to the expansion of descendant species. The non-synonymous to synonymous nucleotide substitution ratio (Ka/Ks) reveal that the purifying selection is primarily responsible for the evolutionary stability of PRF genes. In addition, the segmental and tandem duplication events are also prominent for their structural/functional divergences among lineages (Supplementary Figures 2, 3). Such structural variations in PRF genes are probably attributed to the variations in the exon-intron architecture of paralogous and orthologous sequences across species (Bao et al., 2011; Pandey and Chaudhary, 2016). Remarkably, evolutionary succession is largely achieved by the insertion and/or deletion in the exon/intron regions of PRF genes (Rogozin et al., 2003; Babenko et al., 2004). Results show that the evolution of exonic and intronic regions of PRF genes occurred essentially before the divergence of eudicot and monocot species in the Plantae kingdom (Pandey and Chaudhary, 2017) (Supplementary Figure 2). On the contrary to higher plants, lower plant species generally contain one long exon possibly due to intron/exon gain; or exon-fission and it reflects their evolutionary divergence from higher plants. Intron-exon structural organization of orthologs and paralogs provides clues to interpret the functional diversification of PRF gene family with their ancient evolutionary footprints (Pandey and Chaudhary, 2017). This illustrated a fascinating trend of greater sequence diversity among PRF genes with their evolutionary origin in the polyphyletic mode. Furthermore, viral PRFs had probably originated from their ancient relatives through horizontal gene transfer, and it suggests the prevalence of evolutionary conservation of PRF genes among living and non-living. Interestingly, the evolutionary testing of lineage-specific PRFs shows that these proteins are essential in organisms' development and evolved in Paleozoic (545 MYA), Mesozoic (252 MYA) and Cenozoic (66 MYA) life forms. Thus, PRFs are the notable outcome of the continuous evolution under natural selection (Supplementary Figure 3). The PRF genes are prevalent during the evolution of land plants e.g., bryophytes and gymnosperms (~425 and 385 MYA, respectively) (Pandey and Chaudhary, 2017). Bayesian phylogenetic algorithm shows that KYMVIQGE and VIRGKKG amino acid motifs are distinct among PRF-homologs and -orthologs, respectively, and categorized as evolutionary residues. The signatures of residual conservation show distinct and apparent classification patterns comprising three or five amino acids such as LAPTG, PGQCN, MSWQ, GDYL, YVD, AAI, and KKT (Pandey and Chaudhary, 2017).
Furthermore, PRFs play a vital function in the actin polymerization and depolymerization processes through binding to PAINRs on the protein surface (Giehl et al., 1994). The PAINRs are composed of alanines, isoleucines, glutamine, glutamic acid, lysine, glycine, threonine, proline, and arginine which allow PRFs to interact with actin residues. The Arabidopsis PRF sequence (Accession number-AAG10091) consists of 134 amino acids and 11 amino acids long PAINRs (AIQEKGTPGMR) present at 64, 78–79, 81, 89, 94, 114–116, 120, 124 amino acid positions (Pandey and Chaudhary, 2017). The evolutionary conjunction of PAINRs is identified during the functional evolution of lineage-specific PRFs (Pandey and Chaudhary, 2017). Lineage-specific PRFs consisting of at least 11 residues also show large heterogeneity in the PAINRs of plant, fungal, animal and cyanobacterial origin which confirms their ancient evolution in the primitive life forms (Pandey and Chaudhary, 2017).
Emerging roles of PRF Genes During Plant Development and the Underlying Mechanisms
In response to endogenous or external signals, the cytoskeleton modifications in plants occur in an exceedingly coordinated manner preferably by the PRF-mediated polymerization of sequestered actin monomers and/or depolymerization of actin proteins (Pantaloni and Carlier, 1993). Despite striking discoveries into the genetics of cell-wall organization of plants (Taylor-Teeples et al., 2015), little is explicit about the PRF-mediated molecular interplay and the corresponding gene expression networks in plants. Several noteworthy examples highlighting the novel functions of PRF genes in plants are summarized here:
Cotton Fiber Development
Cotton is one of the most important sources of natural fiber and remarkable result of single epidermal cell extension on the surface of ovules. Modern long and spinnable fibers are the extraordinary product of the evolutionary selection forces such as genomic polyploidy imposed on wild short-fuzz phenotypes during cotton evolution, followed by the millennium of selection under domestication. The comparative transcript profiling of the fiber cells of domesticated diploid and allopolyploid cotton species at three developmental stages with their wild counterparts reveal the expression up-regulation of cell-wall associated PRF genes (Chaudhary et al., 2008, 2009). In cotton, the PRF gene family is structured in its coding and flanking regions as a multigene family consisting of six members. These genes are up-regulated up to ~400 times in the elongating fiber cells of domesticated accessions compared to their wild forms (Bao et al., 2011).
Five PRF genes of the diploid cotton species express at high levels in the elongating fiber cells at 10 days post-anthesis (dpa) compared to the vegetative and floral tissues (Pandey and Chaudhary, 2019). GhPRF1 gene expression at the 10 dpa fiber elongation stage is 9- and 16-fold higher than 5 and 20 dpa fiber tissues, respectively. Higher PRF expression at the fiber elongation stage is primarily important for cell extension and downstream cell signaling which declines very sharply at the maturation stage. Correspondingly, the extent of F-actin is also upregulated with increased filament length in the elongating fibers suggesting that temporal PRF transcript levels and actin filament organization are in equilibrium during fiber elongation (Pandey and Chaudhary, 2019). The temporal expression evolution of PRF paralog/homolog during cotton domestication gives evolutionary impressions of the selection of highly divergent transcription abundance, especially in the fiber development (Pandey and Chaudhary, 2017).
Regulation of Flowering Phenotypes
Previously, we show that the ectopic expression of trans-profilin (trans-PRF) gene in tobacco results in the hyperactivation of apical meristem, early flowering and a relative increase in the flower number per plant (Pandey and Chaudhary, 2016). The apical meristem tissues of PRF transgenics show coordinated expression of flowering-associated FT4, SOC1, FLC1, and FT1 genes and a positive flowering regulator AP1 gene. Moreover, protein-protein interactions and expression profiling reveal that Actin-Related Protein 4 (ARP4) and ARP6 genes are upregulated in the vegetative and floral tissues of PRF transgenics. These results establish a novel and systematic functional relationship between trans-PRF gene expression and early flower primordium initiation in PRF trasngenics (Supplementary Figure 1). The PRF-overexpression lines of tobacco also exhibit increased plant height, internode length, leaf size and flower number per inflorescence without yield penalties (Figure 1) (Pandey and Chaudhary, 2016). On the contrary, PRF-downregulation significantly reduces (~40%) the initiation of healthy floral buds, normal stamen and pollen production, and flower growth. Floral organ development is severely defected in PRF silencing lines with shortened staminal tube coverage and filament duration, reduced stamen number, indehiscence of anthers, abrupt and uncoordinated increase in the style duration, and irregular seed shape (Pandey and Chaudhary, 2016). Due to the dominance of aberrant floral phenotypes of PRF-silencing lines, the average number of flowers per branch significantly decreases compared to both PRF-overexpression lines and control tobacco plants (Pandey and Chaudhary, 2016). Besides the traditionally established role of PRFs in the cytoskeleton maintenance, these observations strongly recommend their indispensable roles in novel aspects of plant development. The development of enhanced agronomic traits in the PRF overexpression lines has enhanced our understanding of the novel and vital roles of PRF genes that could be utilized in their future deployment in crop improvement programs (Pandey and Chaudhary, 2016).
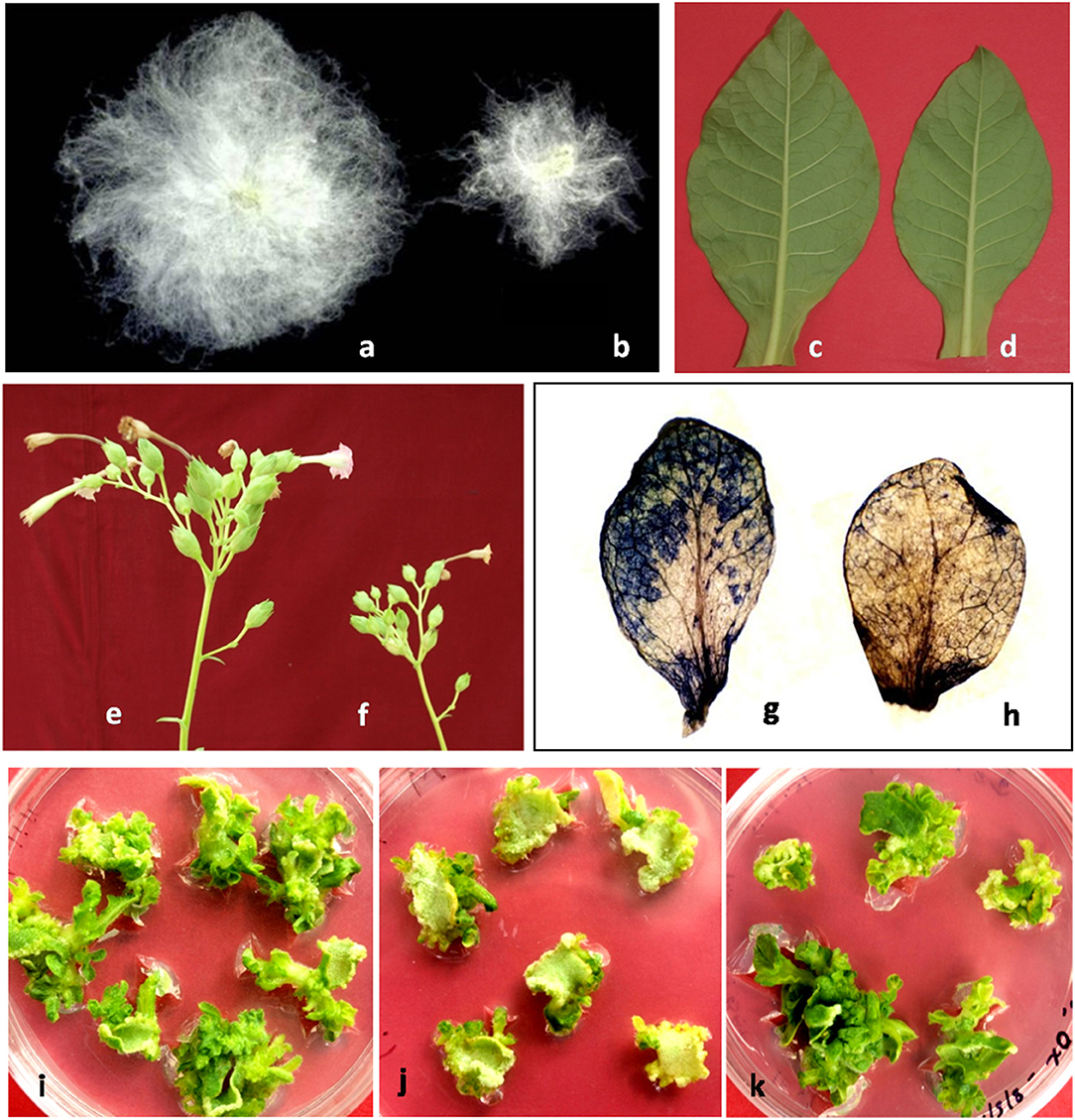
Figure 1. Functional diversity of actin-binding PRFs in plants. (a) domesticated cotton showing increased fibers length having ~400-fold increased PRF gene expression as compared to its wild counterpart (b) wild cotton fibers (c) increased size of leaf lamina in PRF overexpression line of tobacco (d) leaf lamina size of control plant (e) increased number of flowers in PRF overexpression line of tobacco (f) flowers in control plant (g) NBT-stained leaf of untransformed tobacco (grown on 50 mM NaCl concentration) showing higher ROS levels (h) NBT-stained leaf of PRF overexpression line of tobacco (grown on 50 mM NaCl concentration) (i) leaf explants of untransformed tobacco grown on in vitro culture medium (j) leaf explants of gus-transformed tobacco cultured on kanamycin antibiotic selection (k) leaf explants of PRF overexpression line of tobacco cultured on antibiotic selection (images in different panels are not to scale).
Stress Tolerance
The cell-wall is the outermost protective layer of a plant cell involved in the cytoskeleton integrity and provides conditions for the cell growth, development and defense from external stress (Le Gall et al., 2015; Höfte and Voxeur, 2017). Evidently, mass-spectrometry data reveal that up-regulation of PRFs in the root tissues of Cucumis and barley species under the salt treatment conditions (Du et al., 2010; Fatehi et al., 2012). Moreover, PRF gene expression elevation in a succulent halophyte Suaeda aegyptiaca under salinity stress further confirmed the role of PRFs in stress tolerance (Askari et al., 2006). These data provide evidence that PRFs are one of the foremost cytoskeleton-associated proteins that play vital roles during abiotic stress tolerance in plants. We have also investigated the effect of salt stress (up to 50 mM NaCl concentration) on tobacco PRF-overexpression transgenic lines under greenhouse conditions. Remarkably, oxidative stress measurement of these lines shows higher salt tolerance compared with control plants. Apparently, in response to various environmental stress conditions, the increased levels of oxidative stress produce highly reactive chemical molecules known as reactive oxygen species (ROS). Increased accumulation of ROS may cause cellular damages mainly through the degradation of lipids, proteins and nucleic acids (Das and Roychoudhury, 2014). Therefore, the cellular ROS levels of PRF-overexpression lines grown under salt stress conditions are measured by nitro blue tetrazolium (NBT) staining of the target tissues (Grellet Bournonville and Díaz-Ricci, 2011). The leaf tissues of PRF-overexpression tobacco lines grown under salt stress conditions show relatively decreased levels of ROS and establish a novel role of PRF genes in stress management in plants (Figure 1).
Enhanced Magnitude of in vitro Organogenesis
A prerequisite to the successful in vitro micropropagation and genetic modification of a crop species is the availability of an efficient, robust and reproducible regeneration system. In vitro regeneration process largely involves in the transcriptional reprogramming of soma-cells to enable cellular totipotency. We observe that overexpression of trans-PRF gene under a constitutive promoter enhances the number of in vitro shoot formation (up to 14%) in tobacco. The molecular basis of such enhanced emergence of shoot-initial on the edges of explants is directly linked to the relative transcription of CLAVATA1 (CLV1) and WUSCHEL (WUS) genes/trans-factors that alter proportionally with the magnitude of organogenesis in PRF-overexpression lines. The CLV1 and WUS genes are involved in the activation of essential signal transduction pathways required for the activation and formation of shoot primordial during in vitro organogenesis (Supplementary Figure 1) (Pandey and Chaudhary, 2016). Therefore, PRF genes have a direct role in the organogenesis process in vitro which can further be exploited for the improvement of agronomic traits in many crop species.
Author Contributions
DP and BC conceptualized the research and outlines of the article and discussed all results and wrote the manuscript. DP performed all lab experiments cited in the article. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Department of Biotechnology (DBT), Government of India (Grant No. BT/PR8727/AGR/36/777/2013), and Science and Engineering Research Board (SERB), Government of India (Grant No. EMR/2016/005497).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are thankful to the DBT and SERB for providing financial support to carry out this research work.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2020.588689/full#supplementary-material
Supplementary Figure 1. Model of complex interactive network of molecular factors associated to the PRF functional diversity.
Supplementary Figure 2. Evolutionary analyses of full length PRF proteins of various species of plant, animals, fungi, algae, cyanobacteria, and viruses is shown using Bayesian algorithm. Evolutionary distances among PRF sequences are proportional to the branch length on the phylogenetic tree.
Supplementary Figure 3. Existence and natural selection of PRF genes at evolutionary scale.
References
Askari, H., Edqvist, J., Hajheidari, M., Kafi, M., and Salekdeh, G. H. (2006). Effects of salinity levels on proteome of Suaeda aegyptiaca leaves. Proteomics 6, 2542–2554. doi: 10.1002/pmic.200500328
Babenko, V. N., Rogozin, I. B., Mekhedov, S. L., and Koonin, E. V. (2004). Prevalence of intron gain over intron los in the evolution of paralogous gene families. Nucleic Acids Res. 32, 3724–3733. doi: 10.1093/nar/gkh686
Bao, Y., Hu, G., Flagel, L. E., Salmon, A., Bezanilla, M., Paterson, A. H., et al. (2011). Parallel up-regulation of the profilin gene family following independent domestication of diploid and allopolyploid cotton (Gossypium). Proc. Natl. Acad. Sci. U.S.A. 108, 21152–21157. doi: 10.1073/pnas.1115926109
Chaudhary, B., Hovav, R., Flagel, L., Mittler, R., and Wendel, J. F. (2009). Parallel expression evolution of oxidative stress-related genes in fiber from wild and domesticated diploid and polyploid cotton (Gossypium). BMC Genomics 10:378. doi: 10.1186/1471-2164-10-378
Chaudhary, B., Hovav, R., Rapp, R., Verma, N., Udall, J. A., and Wendel, J. F. (2008). Global analysis of gene expression in cotton fibers from wild and domesticated Gossypium barbadense. Evol. Dev. 10, 567–582. doi: 10.1111/j.1525-142X.2008.00272.x
Christensen, H. E. M., Ramachandran, S., Tan, C. T., Surana, U., Dong, C. H., and Chua, N. H. (1996). Arabidopsis profilins are functionally similar to yeast profilins: identification of a vascular bundle-specific profilin and a pollen-specific profilin. Plant J. 10, 269–279. doi: 10.1046/j.1365-313X.1996.10020269.x
Das, K., and Roychoudhury, A. (2014). Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front. Environ. Sci. 2:53. doi: 10.3389/fenvs.2014.00053
Du, C. X., Fan, H. F., Guo, S. R., Tezuka, T., and Li, J. (2010). Proteomic analysis of cucumber seedling roots subjected to salt stress. Phytochemistry 71, 1450–1459. doi: 10.1016/j.phytochem.2010.05.020
Fatehi, F., Hosseinzadeh, A., Alizadeh, H., Brimavandi, T., and Struik, P. C. (2012). The proteome response of salt-resistant and salt-sensitive barley genotypes to long-term salinity stress. Mol. Biol. Rep. 39, 6387–6397. doi: 10.1007/s11033-012-1460-z
Giehl, K., Valenta, R., Rothkegel, M., Ronsiek, M., Mannherz, H. -G., and Jockusch, B. M. (1994). Interaction of plant profilin with mammalian actin. Eur. J. Biochem. 226, 681–689. doi: 10.1111/j.1432-1033.1994.tb20096.x
Grellet Bournonville, C. F., and Díaz-Ricci, J. C. (2011). Quantitative determination of superoxide in plant leaves using a modified NBT staining method. Phytochem. Anal. 22, 268–271. doi: 10.1002/pca.1275
Höfte, H., and Voxeur, A. (2017). Plant cell walls. Curr. Biol. 27, R865–R870. doi: 10.1016/j.cub.2017.05.025
Le Gall, H., Philippe, F., Domon, J. M., Gillet, F., Pelloux, J., and Rayon, C. (2015). Cell wall metabolism in response to abiotic stress. Plants 4, 112–166. doi: 10.3390/plants4010112
Magdolen, V., Oechsner, U., Müller, G., and Bandlow, W. (1988). The intron-containing gene for yeast profilin (PFY) encodes a vital function. Mol. Cell. Biol. 8, 5108–5115. doi: 10.1128/MCB.8.12.5108
Müssar, K. J., Kandasamy, M. K., McKinney, E. C., and Meagher, R. B. (2015). Arabidopsis plants deficient in constitutive class profilins reveal independent and quantitative genetic effects. BMC Plant Biol. 15:177. doi: 10.1186/s12870-015-0551-0
Pandey, D. K., and Chaudhary, B. (2016). Domestication-driven Gossypium profilin 1 (GhPRF1) gene transduces early flowering phenotype in tobacco by spatial alteration of apical/floral-meristem related gene expression. BMC Plant Biol. 16:201310. doi: 10.1186/s12870-016-0798-0
Pandey, D. K., and Chaudhary, B. (2017). Evolutionary expansion and structural functionalism of the ancient family of profilin proteins. Gene 626, 70–86. doi: 10.1016/j.gene.2017.05.024
Pandey, D. K., and Chaudhary, B. (2019). Synchronous transcription of cytoskeleton-associated genes is critical to cotton fiber elongation. J. Plant Growth Regul. 38, 1037–1061. doi: 10.1007/s00344-019-09913-0
Pantaloni, D., and Carlier, M. F. (1993). How profilin promotes actin filament assembly in the presence of thymosin β4. Cell 75, 1007–1014. doi: 10.1016/0092-8674(93)90544-Z
Pollard, T. D., and Cooper, J. A. (2009). Actin, a central player in cell shape and movement. Science 326, 1208–1212. doi: 10.1126/science.1175862
Ramachandran, S., Christensen, H. E. M., Ishimaru, Y., Dong, C. H., Chao-Ming, W., Cleary, A. L., et al. (2000). Profilin plays a role in cell elongation, cell shape maintenance, and flowering in Arabidopsis. Plant Physiol. 124, 1637–1647. doi: 10.1104/pp.124.4.1637
Rogozin, I. B., Wolf, Y. I., Sorokin, A. V., Mirkin, B. G., and Koonin, E. V. (2003). Remarkable interkingdom conservation of intron positions and massive, lineage-specific intron loss and gain in eukaryotic evolution. Curr. Biol. 13, 1512–1517. doi: 10.1016/S0960-9822(03)00558-X
Sohn, R. H., and Goldschmidt-Clermont, P. J. (1994). Profilin: at the crossroads of signal transduction and the actin cytoskeleton. BioEssays 16, 465–472. doi: 10.1002/bies.950160705
Keywords: profilin, actin, evolution, expression, genomics, plants
Citation: Pandey DK and Chaudhary B (2020) Evolution of Functional Diversity Among Actin-Binding Profilin Genes in Land Plants. Front. Cell Dev. Biol. 8:588689. doi: 10.3389/fcell.2020.588689
Received: 29 July 2020; Accepted: 19 November 2020;
Published: 16 December 2020.
Edited by:
Chang-Duk Jun, Gwangju Institute of Science and Technology, South KoreaReviewed by:
Thomas Fath, Macquarie University, AustraliaCopyright © 2020 Pandey and Chaudhary. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bhupendra Chaudhary, Ymh1cGVuZHJhQGdidS5hYy5pbg== orcid.org/0000-0001-9593-1016
†Present address: Dhananjay K. Pandey, Department of FF21, University of California, San Diego, CA, United States
 Dhananjay K. Pandey
Dhananjay K. Pandey Bhupendra Chaudhary
Bhupendra Chaudhary