
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Cell Dev. Biol. , 23 November 2020
Sec. Evolutionary Developmental Biology
Volume 8 - 2020 | https://doi.org/10.3389/fcell.2020.586158
MicroRNAs regulate gene expression at post-transcriptional levels. Some of them appear to regulate brain development and are involved in neurodevelopmental disorders. This has led to the suggestion that the role of microRNAs in neuronal development and function may be more central than previously appreciated. Here, we review the data about miR-9 function to depict the subtlety, complexity, flexibility and limited functional conservation of this essential developmental regulatory system. On this basis we propose that species-specific actions of miR-9 could underlie to a large degree species differences in brain size, shape and function.
MicroRNAs (miRNAs, miRs) are short non-coding RNA oligonucleotides (18–25 bases), which regulate gene expression at post-transcriptional levels by binding specific mRNAs and in this way marking them for enzymatic destruction (reviewed in Pasquinelli, 2012). The downregulation of a gene by a miR can result in a reciprocal negative feedback signaling between the specific miR and its target gene(s). It also can form a positive feedback loop when the miR reduces expression of another miR or of an inhibitory factor (Avraham and Yarden, 2012). One miR can theoretically regulate hundreds of target genes because the miR sequence never matches completely the target mRNA sequence. For this reason, target verification for any given miR is a challenge. Several miRs seem to work as fine-tuning regulators of brain development since they reinforce or disrupt developmental or transitional stages (reviewed in Coolen and Bally-Cuif, 2009; Petri et al., 2014; Davis et al., 2015; Rajman and Schratt, 2017). Some of those have been associated with neurodevelopmental disorders such as Autism Spectrum Disorder, Down syndrome, Rett syndrome and schizophrenia (reviewed in Im and Kenny, 2012; Banerjee-Basu et al., 2014; Sun and Shi, 2015). This has led to the suggestion that their role in neuronal development and function may be more central than previously appreciated (Davis et al., 2015). In this mini-review, we will discuss miR-9 actions in the neural tube with a specific focus on the mid-hindbrain-area. We hope to show how complex and flexible the functional conservation of this essential developmental regulatory system is. Our analysis points to a species-specific set of miR-9 interactions, which (1) could crucially hone some aspects of development in some CNS regions; and (2) could therefore achieve species differences in brain size, shape and function.
MiR-9 is conserved from flies to humans (Yuva-Aydemir et al., 2011) and primarily expressed in the central nervous system (CNS) at least in vertebrates (Wienholds et al., 2005; Deo et al., 2006; Kloosterman et al., 2006; Kapsimali et al., 2007; Radhakrishnan and Alwin Prem Anand, 2016). However, the extent of its functional conservation is not known. Studies on the CNS of different species and on neural stem cells (NSCs) have disclosed an important role of miR-9 in balancing proliferation and differentiation of neural progenitor cells (NPCs) and NSCs (Leucht et al., 2008; Packer et al., 2008; Shibata et al., 2008, 2011; Yoo et al., 2009; Zhao et al., 2009; Bonev et al., 2011; Roese-Koerner et al., 2017). MiR-9 knockout (KO) as well as overexpression (OE) experiments result in smaller or larger brains or specific brain regions and confirm the role of miR-9 in neural proliferation-differentiation balance (reviewed in Coolen et al., 2013). MiR-9 is also important to keep the quiescence/activation balance of adult NSCs in zebrafish telencephalon (Katz et al., 2016) and in human and mouse adult NSCs (Zhao et al., 2009; Roese-Koerner et al., 2017). Moreover, miR-9 expression is also upregulated after Zika virus infections in the developing mouse telencephalon (Zhang et al., 2019).
MiR-9 expression is tightly regulated by several genes and by other miRs (Packer et al., 2008; Denli et al., 2009; Bonev et al., 2012; Coolen et al., 2013; Davila et al., 2014). MiR-9 is repressed by the neurogenic repressor REST and its cofactors (SCP1, CoREST) as well as by TLX in mouse cortex (Packer et al., 2008; Zhao et al., 2013). The Notch effector HES1/HAIRY1 also regulates miR-9 in mouse cortex (Bonev et al., 2012; Tan et al., 2012). Ngn1 inhibits astrogliogenesis through induction of miR-9 (Zhao et al., 2015). And ElAV2 counteracts the suppression of miR-9 by binding to U-rich region of Foxg1 mRNA (Shibata et al., 2011). The all trans retinoic acid and retinoic acid have been shown to induce miR-9 (Kutty et al., 2010).
MiR-9, in turn, regulates a variety of genes to balance proliferation and differentiation in telencephalon, hindbrain, spinal cord, and (in vitro) in NSCs (reviewed in Coolen et al., 2013). It induces the switch of BAF5a to BAF53b, an epigenetic regulator (Yoo et al., 2011; Tang et al., 2013). Other reported targets of miR-9 include Foxg1, Foxp2, Gsh2, SIRT1, and REST (Shibata et al., 2008, 2011; Clovis et al., 2012). In the developing mouse cortex, miR-9 targets Foxg1, Nr2e1, Gsh2, and Meis2 (Shibata et al., 2011).
Mir-9 reinforces it’s own expression by targeting REST, TLX, and HES1 in forming auto-regulatory loops (Bonev et al., 2012; Tan et al., 2012; Goodfellow et al., 2014; Roese-Koerner et al., 2017). In spinal cord, FOXP1 (Otaegi et al., 2011) and OC1 (onecut transcription factor) (Luxenhofer et al., 2014) were reported as miR-9 target genes.
In particular, Hes genes are a recurring target of miR-9 in forebrain and in NSCs, and the intensity of their expression oscillates with that of miR-9 to balance neurogenesis and proliferation (Bonev et al., 2012; Tan et al., 2012; Goodfellow et al., 2014; Roese-Koerner et al., 2017). Across vertebrates, Hes1/her6 genes have a conserved 3′UTR binding site for miR-9. MiR-9 targeting of her/hairy/hes is necessary to properly balance progenitor proliferation genes in zebrafish, Xenopus and mouse (Leucht et al., 2008; Bonev et al., 2011, 2012; Coolen et al., 2012). In all three model animals, miR-9 and Hes1 form a regulatory loop. This loop is also active in human neural stem cells (Roese-Koerner et al., 2017) and helps to steady the ultradian Hes oscillation (Kageyama et al., 2008), necessary for controlled neural proliferation.
Particularly interesting is the fact that miR-9 can regulate different target pathways in forebrain and hindbrain, to obtain region-specific results (Bonev et al., 2011).
The function of miR-9 in the development of the CNS has been approached by gain of function (Gof) using oligonucleotide mimics or plasmid vector OE and loss of function (LoF) by KO or anti-miRs experiments in different brain regions and in the spinal cord of animal models from different vertebrate classes. The results show that miR-9 is essential for proper neural differentiation but that its effect is not uniform in all vertebrate models and cannot be easily generalized. As an example, LoF and OE in mouse and Xenopus forebrain suggest that miR-9 is necessary for the production of the early population of neurons (Shibata et al., 2008, 2011; Bonev et al., 2011; Shu et al., 2019a,b). More specifically, in the cortex, miR-9 is essential for the specification of the first-born cortical layers (Shu et al., 2019b). On the contrary, miR-9 is necessary for the differentiation of late born motor neurons of the spinal cord (Otaegi et al., 2011; Luxenhofer et al., 2014).
Since the variety of results on different CNS regions of different models makes it difficult to generalize, here we would like to focus on the midbrain-hindbrain region (MHB). The role of miR-9 in the development of the MHB showcases the major themes of complexity, subtlety and species-specificity.
The Intervening Zone (IZ) is a region rostral to the MHB and separates midbrain from hindbrain (Palmgren, 1921; Vaage, 1969; Bally-Cuif and Wassef, 1994; Wullimann and Knipp, 2000). The IZ expresses Fgf8, undergoes neurogenesis later than neighboring areas and is crucial for MHB maintenance and MH development (Figures 1A,B; Palmgren, 1921; Vaage, 1969; Bally-Cuif and Wassef, 1994; Wullimann and Knipp, 2000). The IZ does not express miR-9 in zebrafish, Xenopus and chick (Leucht et al., 2008; Bonev et al., 2011; Figure 1A). Such a miR-9-free zone has not been explicitly described in the mouse, but it is visible e.g., in the expression patterns published by Shibata et al. (2008). The formation and maintenance of the IZ region in zebrafish are based on active inhibition of neurogenesis and expression of the hairy/E(spl) gene her5 (Geling et al., 2003). Her5 together with the her-like gene “him” suppresses neurogenesis and sustains in this way the growth of the entire mid-hindbrain area (Tallafuss and Bally-Cuif, 2003). MiR-9 OE experiments cause premature neurogenesis in the IZ and rostral hindbrain in zebrafish (Leucht et al., 2008) and chick (Alwin Prem Anand et al., 2018). Interestingly, in chick and Xenopus Hairy1/Hes1 is not expressed in the IZ, although it shows a predicted miR-9 binding site in the 3′UTR (Bonev et al., 2011; Alwin Prem Anand et al., 2018). In chick, FGF8 and EN1 are target genes of miR-9, where the former shows consistent experimental reduction, the latter does not (Alwin Prem Anand et al., 2018).
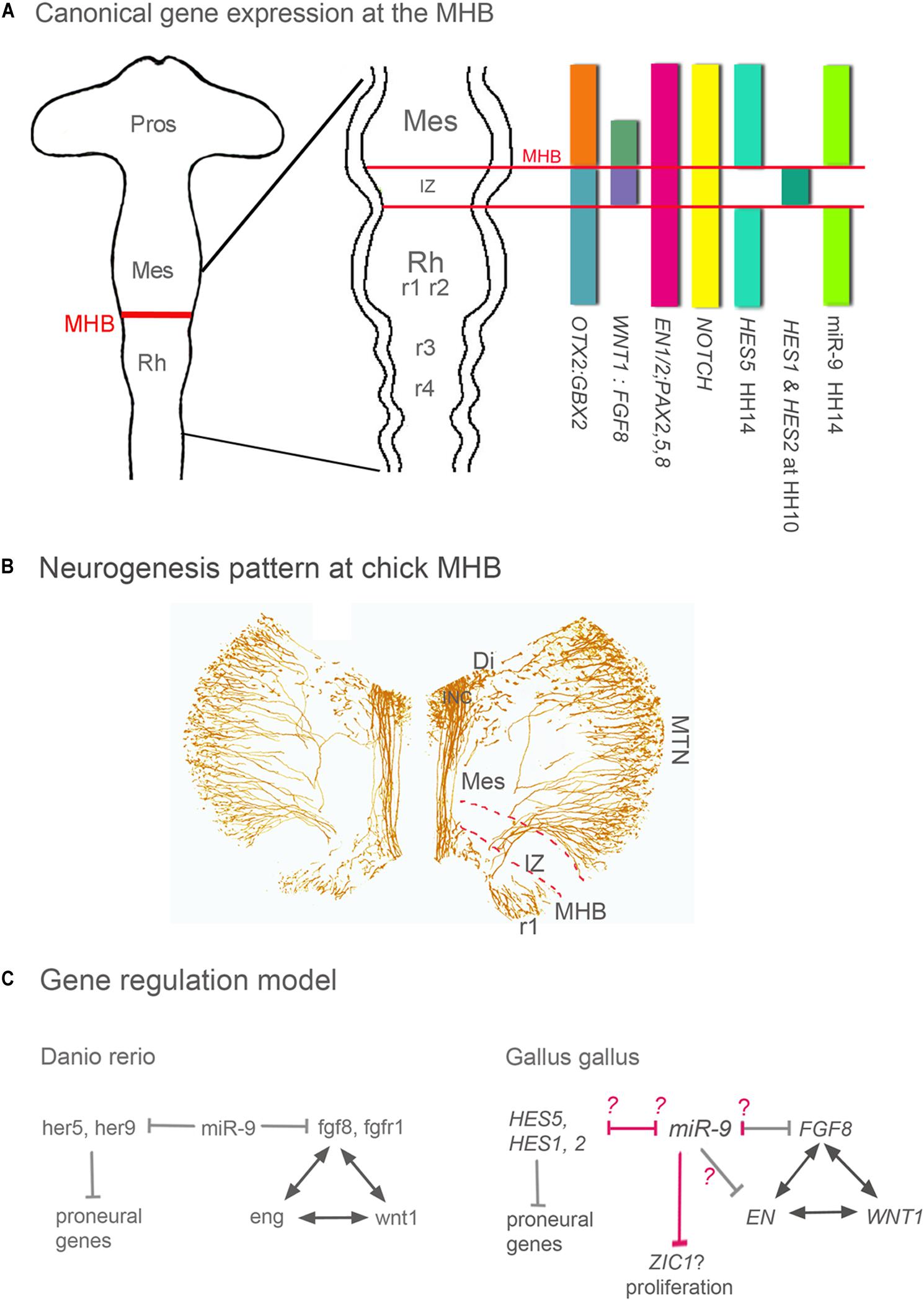
Figure 1. Gene expression, neurogenesis and gene regulation around the MHB. (A) Summary of gene expression patterns around the MHB. Otx2, Gbx2, Wnt1, Fgf8, En1/2, Pax2 -5 -8, and Notch are expressed in a similar pattern in all vertebrates. The expression pattern of Hes genes however differs between vertebrate species. For instance, the pattern of zebrafish her 9 is similar to that of chick HES5. (B) Pattern of neurogenesis around the chick MHB at HH17 (∼E3). Differentiated neurons were labeled with an antibody against medium weight neurofilament (RMO-270). The IZ lacks neurons at this stage, and in midbrain only the dorsally located MTN neurons have developed. ICN neurons are located in ventral diencephalon and form the medial longitudinal tract left and right of the FP. In r1 ventral and dorsal neurons have differentiated. (C) In all vertebrates studied, during the maintenance phase, Wnt1, Fgf8, Pax2/5/8, and En genes regulate each other to maintain the MHB. In zebrafish and chick, miR-9 suppresses Fgf8 expression and thus indirectly the expression of Wnt1, Pax, and En genes. The difference between both species is in the miR-9 targeting. In chick, Fgf8, and En1 are target genes of miR-9; where the former shows experimental reduction and the later is inconsistent (Alwin Prem Anand et al., 2018). In zebrafish, miR-9 promotes neurogenesis by inhibiting different Hes genes around and within the IZ (her9 and her5, respectively). In chick, none of the Hes genes expressed in and around the IZ are miR-9 targets (pink arrows). miR-9 OE in chick resulted in immature neurogenesis only in r1 and IZ but not in midbrain, These results suggests that at least in chick IZ and in hindbrain genes of the Hes pathway could be inhibited by miR-9. In chick midbrain, like in Xenopus telencephalon (Bonev et al., 2011) growth is reduced but without an effect on cell death as in Xenopus, which might be mediate by inhibiting the Wnt activator Zic1. The Models are modified from Rhinn and Brand (2001) and Delaloy and Gao (2008). Di, diencephalon; FP, floor plate; INC, interstitial nucleus of Cajal; IZ, intervening zone; MES, mesencephalon; MHB, mid-hindbrain boundary; MTN, mesencephalic trigeminal nucleus; Pros, prosencephalon; Rh, rhombencephalon; r1 to 4, rhombomeres 1–4.
In zebrafish, miR-9 suppresses not only her5 but also genes of the fgf pathway (fgf8, fgfr1, and canopy 1; Figure 1C) at the MHB, thus affecting positioning, establishment, and maintenance of the MHB. Indeed, in zebrafish, miR-9 overexpression can completely abolish fgf8 expression in the MHB and thus the development of the early MHB (Leucht et al., 2008). Chick and zebrafish show miR-9 target binding sites for Fgf8/fgf8 (Leucht et al., 2008; Alwin Prem Anand et al., 2018). The following example is particularly interesting since it reveals a regional specificity in the mechanisms of miR-9 function that depends on regionally expressed downstream genes. In Xenopus, the function of miR-9 promoting neurogenesis and antagonizing proliferation is mediated by decreasing the availability of Hairy1 but while in the forebrain the final effect of this decrease is mediated by Fgf8, in the hindbrain it is mediated by Wnt (Bonev et al., 2011).
In chick, miR-9 OE or LoF resulted in either a smaller or a larger zone of FGF8 expression at the MHB, respectively, but never in a complete loss of the MHB domain and FGF8 expression like in zebrafish (Leucht et al., 2008). Thus, after miR-9 OE (Alwin Prem Anand et al., 2018), the chick MHB continued to express not only FGF8, but also typical regional markers WNT1, EN1, and EN2 (Rhinn and Brand, 2001; Wurst and Bally-Cuif, 2001; Raible and Brand, 2004; Dworkin et al., 2012). Nevertheless, FGF8 expression was affected, as was indirectly WNT expression (Alwin Prem Anand et al., 2018). Thus, one of the tasks of miR-9 in chick seems to restrict FGF-8 expression (Figure 1C) and thus the extent of the IZ. Since the size of the IZ is important for the growth and patterning of the MH area, miR-9 influences the size of that area. Several HES genes are expressed in and around the chick IZ (Figure 1A). So far none comparable to her5 in zebrafish and Her2 in mouse (Leimeister et al., 1999; Tallafuss et al., 2003) has been described. HAIRY1/HES1 and HAIRY2/HES2 are only temporarily expressed in the IZ in chick (Tossell et al., 2011). HES1 seems to be the ortholog of her9 in zebrafish (Leve et al., 2001); however, its expression pattern correlates only transiently with that of her5 in zebrafish. Chick HES5 on the other hand is expressed along the entire MH area except for the IZ and correlates rather with the expression pattern of zebrafish her9 (Figure 1A; Kimura et al., 2004). Alas, so far miR-9 has shown no theoretical target-binding site for chick HES2 or HES5, and in chick we have not observed downregulation of HAIRY1/HES1 expression after miR-9 OE although there is a theoretical target site (Alwin Prem Anand et al., 2018). Nevertheless, miR-9 OE causes premature neurogenesis in posterior MHB, i.e., in the IZ and in anterior hindbrain of the chick (Alwin Prem Anand et al., 2018), as reported in zebrafish MHB (Leucht et al., 2008) and in anterior hindbrain of Xenopus (Bonev et al., 2011). In addition, or instead of HES genes their target NOTCH might be downregulated by miR-9. NOTCH is known to block miR-9 expression in neural stem cells (Roese-Koerner et al., 2016). This suggests that the NOTCH-HES pathway in chick is interrupted and proneural genes are activated.
Although miR-9 has an effect on Fgf8 expression in the MHB both in chick and zebrafish, the consequences of over-expression are never as severe in chick as in zebrafish. This could be an artifact of timing and targeting of the experiments in chick, which were performed unilaterally and only after the MHB was formed (Alvarado-Mallart et al., 1990; Itasaki et al., 1991). In the experiments, miR-9 OE correlated approximately with the beginning of activity of miR-9 around the MHB in chick at Embryonic day (E) 1.5 (or HH 14 (Hamburger and Hamilton, 1951; Alwin Prem Anand et al., 2018; Figure 1A). In contrast, in the zebrafish experiments, miR-9 manipulations were performed in oocytes, long before the MHB is positioned and formed and before miR-9 is expressed in the MH area (Leucht et al., 2008). This early overexpression may explain the more profound effects in zebrafish. These results suggest that an early KO of miR-9 might have additional effects on neural tube development and thus influence MHB development.
There is another interesting difference between species. Although miR-9 overexpression causes premature neurogenesis in the IZ in zebrafish and chick embryo and in anterior hindbrain in zebrafish, chick and Xenopus (Leucht et al., 2008; Bonev et al., 2011; Alwin Prem Anand et al., 2018), neither chick nor Xenopus seem to express Hes1/Hairy1 in the IZ. In the Xenopus hindbrain (Bonev et al., 2011) and possibly also in the chick MHB (Alwin Prem Anand et al., 2018), miR-9 targets zic1, a Wnt activator (Figure 1C), in this way reducing proliferation, a step previous to neurogenesis induction. Is miR-9 then an inductor of neurogenesis? Results obtained in the chick midbrain, immediately rostral to the MHB, where broad ectopic miR-9 OE results in reduced proliferation but no ectopic neurogenesis (Alwin Prem Anand et al., 2018), suggest the opposite. Alternatively, the chick midbrain could have a very strong antagonist to miR-9 to inhibit premature neurogenesis. It will be interesting to see if this is also the case in other species.
Our synopsis of all these results from different species is that miR-9 has essential, complex and time-dependent but only partially conserved functions in vertebrates. These make this intricate system difficult to approach. At the same time, the identification of species-specific regulation of miR-9 expression is yielding new insights on the different mechanisms that regulate the spatiotemporal functions of miR-9. One conclusion that can be drawn is that the miR system has evolved to be flexible, species-specific, subtle and time dependent. On the basis of this mini-review we tentatively propose that (1) maybe there are species-specific sets of miRs governing certain aspects of development; (2) this could be part of the cause of the differences in brain size, shape, and function in different vertebrate classes.
AAPA, GA-B, and AW wrote the manuscript. AAPA and AW designed the figure. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Alvarado-Mallart, R. M., Martinez, S., and Lance-Jones, C. C. (1990). Pluripotentiality of the 2-day-old avian germinative neuroepithelium. Dev. Biol. 139, 75–88. doi: 10.1016/0012-1606(90)90280-v
Alwin Prem Anand, A., Huber, C., Asnet Mary, J., Gallus, N., Leucht, C., Klafke, R., et al. (2018). Expression and function of microRNA-9 in the mid-hindbrain area of embryonic chick. BMC Dev. Biol. 18:3. doi: 10.1186/s12861-017-0159-8
Avraham, R., and Yarden, Y. (2012). Regulation of signalling by microRNAs. Biochem. Soc. Trans. 40, 26–30. doi: 10.1042/bst20110623
Bally-Cuif, L., and Wassef, M. (1994). Ectopic induction and reorganization of Wnt-1 expression in quail/chick chimeras. Development 120, 3379–3394.
Banerjee-Basu, S., Larsen, E., and Muend, S. (2014). Common microRNAs Target Established ASD Genes. Front. Neurol. 5:205. doi: 10.3389/fneur.2014.00205
Bonev, B., Pisco, A., and Papalopulu, N. (2011). MicroRNA-9 reveals regional diversity of neural progenitors along the anterior-posterior axis. Dev. Cell 20, 19–32. doi: 10.1016/j.devcel.2010.11.018
Bonev, B., Stanley, P., and Papalopulu, N. (2012). MicroRNA-9 modulates Hes1 ultradian oscillations by forming a double-negative feedback loop. Cell Rep. 2, 10–18. doi: 10.1016/j.celrep.2012.05.017
Clovis, Y. M., Enard, W., Marinaro, F., Huttner, W. B., and De Pietri Tonelli, D. (2012). Convergent repression of Foxp2 3’UTR by miR-9 and miR-132 in embryonic mouse neocortex: implications for radial migration of neurons. Development 139, 3332–3342. doi: 10.1242/dev.078063
Coolen, M., and Bally-Cuif, L. (2009). MicroRNAs in brain development and physiology. Curr. Opin. Neurobiol. 19, 461–470. doi: 10.1016/j.conb.2009.09.006
Coolen, M., Katz, S., and Bally-Cuif, L. (2013). miR-9: a versatile regulator of neurogenesis. Front. Cell Neurosci. 7:220. doi: 10.3389/fncel.2013.00220
Coolen, M., Thieffry, D., Drivenes, O., Becker, T. S., and Bally-Cuif, L. (2012). miR-9 controls the timing of neurogenesis through the direct inhibition of antagonistic factors. Dev. Cell 22, 1052–1064. doi: 10.1016/j.devcel.2012.03.003
Davila, J. L., Goff, L. A., Ricupero, C. L., Camarillo, C., Oni, E. N., Swerdel, M. R., et al. (2014). A positive feedback mechanism that regulates expression of miR-9 during neurogenesis. PLoS One 9:e94348. doi: 10.1371/journal.pone.0094348
Davis, G. M., Haas, M. A., and Pocock, R. (2015). MicroRNAs: not “fine-tuners” but key regulators of neuronal development and function. Front. Neurol. 6:245. doi: 10.3389/fneur.2015.00245
Delaloy, C., and Gao, F. B. (2008). A new role for microRNA-9 in human neural progenitor cells. Cell Cycle 9, 2913–2914. doi: 10.4161/cc.9.15.12699
Denli, A. M., Cao, X., and Gage, F. H. (2009). miR-9 and TLX: chasing tails in neural stem cells. Nat. Struct. Mol. Biol. 16, 346–347. doi: 10.1038/nsmb0409-346
Deo, M., Yu, J. Y., Chung, K. H., Tippens, M., and Turner, D. L. (2006). Detection of mammalian microRNA expression by in situ hybridization with RNA oligonucleotides. Dev. Dyn. 235, 2538–2548. doi: 10.1002/dvdy.20847
Dworkin, S., Darido, C., Georgy, S. R., Wilanowski, T., Srivastava, S., Ellett, F., et al. (2012). Midbrain-hindbrain boundary patterning and morphogenesis are regulated by diverse grainy head-like 2-dependent pathways. Development 139, 525–536. doi: 10.1242/dev.066522
Geling, A., Itoh, M., Tallafuss, A., Chapouton, P., Tannhauser, B., Kuwada, J. Y., et al. (2003). bHLH transcription factor Her5 links patterning to regional inhibition of neurogenesis at the midbrain-hindbrain boundary. Development 130, 1591–1604. doi: 10.1242/dev.00375
Goodfellow, M., Phillips, N. E., Manning, C., Galla, T., and Papalopulu, N. (2014). microRNA input into a neural ultradian oscillator controls emergence and timing of alternative cell states. Nat. Commun. 5:3399. doi: 10.1038/ncomms4399
Hamburger, V., and Hamilton, H. L. (1951). A series of normal stages in the development of the chick embryo. J. Morphol. 88, 49–92. doi: 10.1002/jmor.1050880104
Im, H. I., and Kenny, P. J. (2012). MicroRNAs in neuronal function and dysfunction. Trends Neurosci. 35, 325–334. doi: 10.1016/j.tins.2012.01.004
Itasaki, N., Ichijo, H., Hama, C., Matsuno, T., and Nakamura, H. (1991). Establishment of rostrocaudal polarity in tectal primordium: engrailed expression and subsequent tectal polarity. Development 113, 1133–1144.
Kageyama, R., Ohtsuka, T., and Kobayashi, T. (2008). Roles of Hes genes in neural development. Dev. Growth Differ. 50(Suppl. 1), S97–S103. doi: 10.1111/j.1440-169X.2008.00993.x
Kapsimali, M., Kloosterman, W. P., de Bruijn, E., Rosa, F., Plasterk, R. H., and Wilson, S. W. (2007). MicroRNAs show a wide diversity of expression profiles in the developing and mature central nervous system. Genome Biol. 8:R173. doi: 10.1186/gb-2007-8-8-r173
Katz, S., Cussigh, D., Urban, N., Blomfield, I., Guillemot, F., Bally-Cuif, L., et al. (2016). A nuclear role for miR-9 and argonaute proteins in balancing quiescent and activated neural stem cell states. Cell Rep. 17, 1383–1398. doi: 10.1016/j.celrep.2016.09.088
Kimura, J., Katahira, T., Araki, I., and Nakamura, H. (2004). Possible role of Hes5 for the rostrocaudal polarity formation of the tectum. Dev. Growth Differ. 46, 219–227. doi: 10.1111/j.1440-169X.2004.00739.x
Kloosterman, W. P., Steiner, F. A., Berezikov, E., de Bruijn, E., van de Belt, J., Verheul, M., et al. (2006). Cloning and expression of new microRNAs from zebrafish. Nucleic Acids Res. 34, 2558–2569. doi: 10.1093/nar/gkl278
Kutty, R. K., Samuel, W., Jaworski, C., Duncan, T., Nagineni, C. N., Raghavachari, N., et al. (2010). MicroRNA expression in human retinal pigment epithelial (ARPE-19) cells: increased expression of microRNA-9 by N-(4-hydroxyphenyl)retinamide. Mol. Vis. 16, 1475–1486.
Leimeister, C., Externbrink, A., Klamt, B., and Gessler, M. (1999). Hey genes: a novel subfamily of hairy- and Enhancer of split related genes specifically expressed during mouse embryogenesis. Mech. Dev. 85, 173–177. doi: 10.1016/s0925-4773(99)00080-5
Leucht, C., Stigloher, C., Wizenmann, A., Klafke, R., Folchert, A., and Bally-Cuif, L. (2008). MicroRNA-9 directs late organizer activity of the midbrain-hindbrain boundary. Nat. Neurosci. 11, 641–648. doi: 10.1038/nn.2115
Leve, C., Gajewski, M., Rohr, K. B., and Tautz, D. (2001). Homologues of c-hairy1 (her9) and lunatic fringe in zebrafish are expressed in the developing central nervous system, but not in the presomitic mesoderm. Dev. Genes Evol. 211, 493–500. doi: 10.1007/s00427-001-0181-4
Luxenhofer, G., Helmbrecht, M. S., Langhoff, J., Giusti, S. A., Refojo, D., and Huber, A. B. (2014). MicroRNA-9 promotes the switch from early-born to late-born motor neuron populations by regulating Onecut transcription factor expression. Dev. Biol. 386, 358–370. doi: 10.1016/j.ydbio.2013.12.023
Otaegi, G., Pollock, A., Hong, J., and Sun, T. (2011). MicroRNA miR-9 modifies motor neuron columns by a tuning regulation of FoxP1 levels in developing spinal cords. J. Neurosci. 31, 809–818. doi: 10.1523/JNEUROSCI.4330-10.2011
Packer, A. N., Xing, Y., Harper, S. Q., Jones, L., and Davidson, B. L. (2008). The bifunctional microRNA miR-9/miR-9∗ regulates REST and CoREST and is downregulated in Huntington’s disease. J. Neurosci. 28, 14341–14346. doi: 10.1523/JNEUROSCI.2390-08.2008
Palmgren, A. (1921). Embryological and morphological studies on the mid-brain and cerebellum of vertebrates. Acta Zool. 2, 1–94. doi: 10.1111/j.1463-6395.1921.tb00464.x
Pasquinelli, A. E. (2012). MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat. Rev. Genet. 13, 271–282. doi: 10.1038/nrg3162
Petri, R., Malmevik, J., Fasching, L., Akerblom, M., and Jakobsson, J. (2014). miRNAs in brain development. Exp. Cell Res. 321, 84–89. doi: 10.1016/j.yexcr.2013.09.022
Radhakrishnan, B., and Alwin Prem Anand, A. (2016). Role of miRNA-9 in brain development. J. Exp. Neurosci. 10, 101–120. doi: 10.4137/JEN.S32843
Raible, F., and Brand, M. (2004). Divide et Impera–the midbrain-hindbrain boundary and its organizer. Trends Neurosci. 27, 727–734. doi: 10.1016/j.tins.2004.10.003
Rajman, M., and Schratt, G. (2017). MicroRNAs in neural development: from master regulators to fine-tuners. Development 144, 2310–2322. doi: 10.1242/dev.144337
Rhinn, M., and Brand, M. (2001). The midbrain–hindbrain boundary organizer. Curr. Opin. Neurobiol. 11, 34–42. doi: 10.1016/s0959-4388(00)00171-179
Roese-Koerner, B., Stappert, L., Berger, T., Braun Nils, C., Veltel, M., Jungverdorben, J., et al. (2016). Reciprocal Regulation between Bifunctional miR-9/9∗ and its transcriptional modulator notch in human neural stem cell self-renewal and differentiation. Stem Cell Rep. 7, 207–219. doi: 10.1016/j.stemcr.2016.06.008
Roese-Koerner, B., Stappert, L., and Brustle, O. (2017). Notch/Hes signaling and miR-9 engage in complex feedback interactions controlling neural progenitor cell proliferation and differentiation. Neurogenesis 4:e1313647. doi: 10.1080/23262133.2017.1313647
Shibata, M., Kurokawa, D., Nakao, H., Ohmura, T., and Aizawa, S. (2008). MicroRNA-9 modulates Cajal-Retzius cell differentiation by suppressing Foxg1 expression in mouse medial pallium. J. Neurosci. 28, 10415–10421. doi: 10.1523/JNEUROSCI.3219-08.2008
Shibata, M., Nakao, H., Kiyonari, H., Abe, T., and Aizawa, S. (2011). MicroRNA-9 regulates neurogenesis in mouse telencephalon by targeting multiple transcription factors. J. Neurosci. 31, 3407–3422. doi: 10.1523/JNEUROSCI.5085-10.2011
Shu, P., Wu, C., Liu, W., Ruan, X., Liu, C., Hou, L., et al. (2019a). The spatiotemporal expression pattern of microRNAs in the developing mouse nervous system. J. Biol. Chem. 294, 3444–3453. doi: 10.1074/jbc.RA118.004390
Shu, P., Wu, C., Ruan, X., Liu, W., Hou, L., Fu, H., et al. (2019b). Opposing gradients of MicroRNA expression temporally pattern layer formation in the developing neocortex. Dev. Cell 49:e764. doi: 10.1016/j.devcel.2019.04.017
Sun, E., and Shi, Y. (2015). MicroRNAs: small molecules with big roles in neurodevelopment and diseases. Exp. Neurol. 268, 46–53. doi: 10.1016/j.expneurol.2014.08.005
Tallafuss, A., Adolf, B., and Bally-Cuif, L. (2003). Selective control of neuronal cluster size at the forebrain/midbrain boundary by signaling from the prechordal plate. Dev. Dyn. 227, 524–535. doi: 10.1002/dvdy.10329
Tallafuss, A., and Bally-Cuif, L. (2003). Tracing of her5 progeny in zebrafish transgenics reveals the dynamics of midbrain-hindbrain neurogenesis and maintenance. Development 130, 4307–4323. doi: 10.1242/dev.00662
Tan, S. L., Ohtsuka, T., Gonzalez, A., and Kageyama, R. (2012). MicroRNA9 regulates neural stem cell differentiation by controlling Hes1 expression dynamics in the developing brain. Genes Cells 17, 952–961. doi: 10.1111/gtc.12009
Tang, J., Yoo, A. S., and Crabtree, G. R. (2013). Reprogramming human fibroblasts to neurons by recapitulating an essential microRNA-chromatin switch. Curr. Opin. Genet. Dev. 23, 591–598. doi: 10.1016/j.gde.2013.07.001
Tossell, K., Kiecker, C., Wizenmann, A., Lang, E., and Irving, C. (2011). Notch signalling stabilises boundary formation at the midbrain-hindbrain organiser. Development 138, 3745–3757. doi: 10.1242/dev.070318
Vaage, S. (1969). The segmentation of the primitive neural tube in chick embryos (Gallus domesticus). A morphological, histochemical and autoradiographical investigation. Ergeb. Anat. Entwicklungsgesch. 41, 3–87.
Wienholds, E., Kloosterman, W. P., Miska, E., Alvarez-Saavedra, E., Berezikov, E., de Bruijn, E., et al. (2005). MicroRNA expression in zebrafish embryonic development. Science 309, 310–311. doi: 10.1126/science.1114519
Wullimann, M. F., and Knipp, S. (2000). Proliferation pattern changes in the zebrafish brain from embryonic through early postembryonic stages. Anat. Embryol. 202, 385–400. doi: 10.1007/s004290000115
Wurst, W., and Bally-Cuif, L. (2001). Neural plate patterning: upstream and downstream of the isthmic organizer. Nat. Rev. Neurosci. 2, 99–108. doi: 10.1038/35053516
Yoo, A. S., Staahl, B. T., Chen, L., and Crabtree, G. R. (2009). MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature 460, 642–646. doi: 10.1038/nature08139
Yoo, A. S., Sun, A. X., Li, L., Shcheglovitov, A., Portmann, T., Li, Y., et al. (2011). MicroRNA-mediated conversion of human fibroblasts to neurons. Nature 476, 228–231. doi: 10.1038/nature10323
Yuva-Aydemir, Y., Simkin, A., Gascon, E., and Gao, F. B. (2011). MicroRNA-9: functional evolution of a conserved small regulatory RNA. RNA Biol. 8, 557–564. doi: 10.4161/rna.8.4.16019
Zhang, H., Chang, Y., Zhang, L., Kim, S. N., Otaegi, G., Zhang, Z., et al. (2019). Upregulation of MicroRNA miR-9 Is associated with microcephaly and zika virus infection in mice. Mol. Neurobiol. 56, 4072–4085. doi: 10.1007/s12035-018-1358-1354
Zhao, C., Sun, G., Li, S., and Shi, Y. (2009). A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat. Struct. Mol. Biol. 16, 365–371. doi: 10.1038/nsmb.1576
Zhao, C., Sun, G., Ye, P., Li, S., and Shi, Y. (2013). MicroRNA let-7d regulates the TLX/microRNA-9 cascade to control neural cell fate and neurogenesis. Sci. Rep. 3:1329. doi: 10.1038/srep01329
Keywords: neural development, mid-hindbrain boundary, danio, gallus, xenopus, Fgf8, miR-9, hairy-1
Citation: Alwin Prem Anand A, Alvarez-Bolado G and Wizenmann A (2020) MiR-9 and the Midbrain-Hindbrain Boundary: A Showcase for the Limited Functional Conservation and Regulatory Complexity of MicroRNAs. Front. Cell Dev. Biol. 8:586158. doi: 10.3389/fcell.2020.586158
Received: 22 July 2020; Accepted: 23 October 2020;
Published: 23 November 2020.
Edited by:
David Ellard Keith Ferrier, University of St Andrews, United KingdomReviewed by:
Sebastian Kittelmann, Oxford Brookes University, United KingdomCopyright © 2020 Alwin Prem Anand, Alvarez-Bolado and Wizenmann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andrea Wizenmann, YW5kcmVhLndpemVubWFubkB1bmktdHVlYmluZ2VuLmRl
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.